Abstract
Background:
Crimean-Congo hemorrhagic fever (CCHF) is an acute viral zoonotic disease, with a mortality rate of 30-50%. There is no approved vaccine or any specific antiviral treatment for CCHF; therefore, the rapid diagnosis seems to be crucial for both efficient supportive therapy and control of infection spread. In this study, the potency of recombinant nucleoprotein of virus expressed in prokaryotic system was investigated for diagnosis of the infection.
Methods:
The DNA sequence of complete nucleoprotein ORF was codon optimized based on E. coli codon usage and synthesized commercially. The gene was subcloned in pCA4 vector and expressed in E. coli BL21 (DE3). Refolding and simultaneous purification of nucleoprotein were performed using protein folding liquid chromatography method. The recombinant nucleoprotein was analyzed by Western blotting, ELISA, immunofluorescence assay, and circular dichroism. Forty eight human samples, in three IgM positive and three negative control groups, were evaluated using recombinant nucleoprotein in a capture ELISA setting. Serum from healthy individuals, those suspected to viral hemorrhagic fevers, and positive samples of Chikungunya and Dengue were considered as negative controls.
Results:
The existence and structure of recombinant nucleoprotein were verified and confirmed. Capture IgM ELISA detected all positive samples (sensitivity of 100%), but none of the 25 negative samples was detected as positive (specificity of 100%). The test also detected all the included genotypes of virus.
Conclusion:
Our recombinant nucleoprotein can be used in IgM capture ELISA for easy and efficient detection of CCHF in any lab in endemic regions.
Key Words: Crimean congo hemorrhagic fever, Enzyme-linked immunosorbent assay, Nucleoprotein
INTRODUCTION
Crimean-Congo hemorrhagic fever (CCHF) is an acute, viral, zoonotic disease, with a mortality rate of 30-50%, considered as one of the infectious pathogens with a high risk of epidemic in near future[1,2]. The causative agent of the disease is one of the most important viruses classified under Nairoviridae family and Orthonairovirus genus carried and spread by tick[3,4]. Despite its high death rate and transmissibility among human beings, currently there are no approved vaccines or specific therapeutics against this virus. Therefore, rapid diagnosis of the disease seems to be crucial for both effective treatment of patients and control of infection transmission. It worth to mention that CCHF symptoms are not specific; therefore, the only way of precise diagnosis is laboratory tests.
The routine diagnostic tests include virus’s genome detection by RT-PCR or evaluation of serum IgM by ELISA[5]. Although highly sensitive and specific, the genome-based detection method is usually difficult since the collection of samples in a short viremia period (one week) is problematic. This is mainly due to the occurrence of most infections in rural, far away regions. IgM, which is detectable in blood for 4-6 months, can play an important role in the diagnosis process[6]. Several ELISA methods have been developed to detect anti-CCHF virus IgM; many of these methods utilize inactivated whole virus particle as an antigen. BSL-4 laboratory is necessary for virus culture, which is not available in most endemic regions. The recombinant expression of necessary antigens is a possible solution for this shortcoming[7]. CCHF virus nucleoprotein is the most important structural protein against which the highest rates of antibodies are raised. Therefore, it can be considered as a reliable candidate for designing serological assays such as ELISA[8].
Although there are some available commercial ELISA kits to detect the specific IgM against CCHF in human sera, the highest sensitivity reported is still 87%, indicating that they can be improved[9]. The aim of this study was to produce a recombinant viral nucleoprotein antigen in a prokaryotic expression system, to develop a safe, low-cost and more sensitive diagnostic platform.
MATERIALS AND METHODS
Plasmid and reagents
pAC4 vector (Cat No: PAC4) and AbCA-AbC mab antibody conjugated to sepharose resin (Cat No: AbCA) were purchased from Avidity, LLC. (Aurora, Colorado, USA). VectoCrimean-CHF-Ag (Cat No: D-5056) and VectoCrimean-CHF-IgM (Cat No: D-5054) were obtained from Vector-Best (Vector-Novosibirsk, Russia). NcoI (Cat No: ER0571) and BamHI (Cat No: ER0051) were procured from Thermo Fisher Scientific Inc., USA. PVDF membrane (Cat No: 03010040001) was from Roche Diagnostics (GmbH, Germany). Extra Thick Blot Paper (Cat No: 1703966) were purchased from Bio-Rad (Bio-Rad Laboratories Inc., USA). Immune ascitic fluid against CCHF (MAF4, Institute Pasteur, Dakar, Senegal), goat anti-mouse IgG, rabbit anti-human IgM (Anti-µ-Chain; ICN, Ohio, USA [Cat Nos: 55558 and 55017]), and other chemicals were acquired from Merck and/or Sigma, USA.
Synthetic gene and subcloning
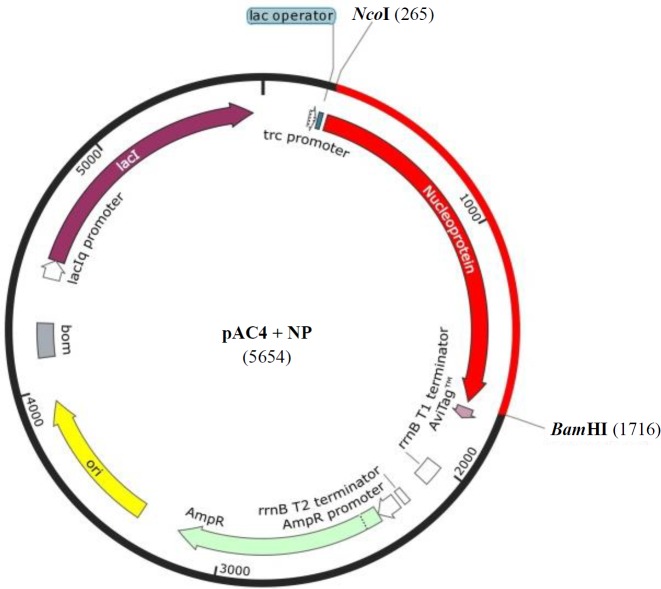
The DNA sequence of complete open reading frame of nucleocapsid protein (1454 bp) from Matin strain (accession number: AF527810.1) was codon optimized for Escherichia coli (BL21 [DE3]) and synthesized in pBSK(+) simple Amp by Biomatik (Cambridge, Ontario, Canada). NcoI and BamHI restriction enzymes were included for 5’ and 3’ ends, respectively. The embedded ATG in NcoI restriction site (C↓CATGG) simplified the subsequent step of subcloning and protein expression in the right frame. The gene was subcloned in pAC4 expression vector, which adds AviTag to the C-terminus of nucleoprotein as a fusion (Fig. 1).
Fig. 1.
Schematic view of the cloning strategy showing the restriction sites, NcoI, and BamHI used for cloning
Expression, purification, and refolding of recombinant nucleoprotein
For optimal growth and recombinant nucleoprotein production, 5 ml of fresh cultured bacteria was inoculated into 500 ml of Luria-Bertani containing 100 µg/ml of ampicillin and was incubated at 37 °C in a shaking incubator at 250 rpm until OD600 of medium reached 0.4. IPTG with the final concentration of 1 mM was added to the culture and incubated at 37 °C for 4 hours. Medium was centrifuged at 4 °C, 5,000 g for 5 min, and the pellet was washed twice with PBS and lysed with a sonication using a lysis buffer (140 mM of NaCl, 2.7 mM of KCl, 10 mM of Na2HPO4, and 1.8 mM of KH2PO4, pH 7.3). The cell lysate was analyzed on SDS-PAGE. Inclusion bodies (IBs) were rinsed twice with the lysis buffer, followed by 2× washing with 50 mM of Tris-HCl, 2.5 mM of EDTA, 0.1% Tween-20, and then 2× washing with 50 mM of Tris-HCl, 2.5 mM of EDTA, and 1 M of NaCl. The pallet was subsequently washed 2× with 50 mM of Tris-HCl, 2.5 mM of EDTA, and 2 M of urea, and the final washing was carried out with 1× 50 mM of Tris-HCl and 2.5 mM of EDTA, followed by washing with double distilled water. To resuspend the pellet after each wash, a brief sonication was applied. The washed pellet was finally solubilized in a solubilization buffer (50 mM of Tris-HCl, 5 mM of EDTA, and 6 M of urea) by incubating at 4 °C overnight[10-12]. Refolding and simultaneous purification of nucleoprotein were performed with 4 ml of AbCA-AbC resin using protein folding liquid chromatography method. The column was equilibrated with 40 ml (10 CV) of PBS containing additional 50 mM of NaCl, at 4 °C. The solubilized protein was loaded on column and incubated with gentle flip flop shaking at room temperature for 2 hours. The column was washed with gradient buffers at 4 °C as follows: 15 ml of buffer 1 (50 mM of Tris-HCl, 5 mM of EDTA, 50 mM of NaCl, and 4 M of urea), 15 ml of buffer 2 (50 mM of Tris-HCl, 5 mM of EDTA, 50 mM of NaCl, and 2 M of urea), and 10 ml of PBS. The resin was incubated with 5 ml of glycine (100 mM) as the elution buffer at 4 °C for one hour, and the protein was eluted with another 5 ml of glycine. For desalting and concentrating the recombinant protein, Amicon centrifugal filter device, with a cut-off size of 30 MWCO, was used. According to device user's guide, the eluted protein was added to the device and then centrifuged at 4 °C at 4000 g for 10 min.
SDS-PAGE analysis and Western blotting of the recombinant protein
Recombinant protein was analyzed on 12% SDS-PAGE with Coomassie blue staining. For Western blotting, gel was soaked in the protein transfer buffer (25 mM of Tris-base, 198 mM of glycine, and 200 ml/L of methanol) for 15 min. PVDF membrane was activated with methanol and equilibrated in the protein transfer buffer for 15 min. The transfer sandwich was assembled according to the manufacturer’s instructions (Mini Trans-Blot SD semi-dry transfer cell, Bio-Rad, CA, USA). The band-transfer was carried out at 12 V for 25 min. Following transfer, non-specific binding sites on the membrane were blocked using the blocking buffer (5% skim milk, 25 mM of Tris, 0.15 M of NaCl, 0.1% Tween-20, pH 7.4) at room temperature for 1 hour. The blot was incubated with mouse hyperimmune ascitic fluid against CCHF virus (1: 3000 dilution, MAF4, Institute Pasteur, Dakar) at 4 °C overnight. The blot was subsequently washed five times with blotting buffer (25 mM of Tris, 0.15 M of NaCl, 0.1% Tween-20, pH 7.4). The blot was incubated with horse radish proxidase-conjugated goat anti-mouse IgG (ICN ,Cat No: 55558), for 1 hour, then was washed with the blotting buffer. Next, the enzyme substrate 3-3’diaminobenzidine (DAB) solution (0.007 g of DAB, 10 ml of PBS, 8 µl of H2O2) was added to the blot and incubated in darkness until the desired band intensity was appeared[13].
Circular dichroism (CD) analysis
Secondary structure of the recombinant nucleoprotein was analyzed with CD system. Nucleoprotein (0.3 mg/mL) in 50 mM of Na3PO4, pH 7.5, was tested in the CD system. Spectra were recorded in 180-260 nm on a circular dichroism spectrometer Model 215, Aviv, USA at a scan speed of 1 nm/s. The test was performed in triplicate, and the results were analyzed by CDNN software (Circular Dichroism analysis using Neural Networks).
ELISA detection of nucleoprotein
In order to detect the recombinant nucleoprotein, VectoCrimean-CHF-Ag ELISA kit (Vector-Best, Russia) in which the antibodies against CCHF virus are coated on the plate, was used. Recombinant nucleoprotein was added instead of sera sample. Three different dilutions of nucleoprotein (7, 0.7, and 0.07 mg/ml in PBS) were added to the wells, and the rest of the reaction was performed according to kit’s manual. Briefly, 100 µl from each dilution of nucleoprotein was added to each well, along with positive and negative controls, and the ELISA plate was incubated at 37 °C for 60 min. After washing, 100 µl of anti-CCHF antibodies labeled with HRP was added and incubated at 37 °C for another 60 min. 3,3’,5,5’-tetramethyl-benzidine substrate was added to the wells and incubated for 15 min. The reaction was stopped by the addition of 100 μl of 4N H2SO4, and the OD450 value was recorded.
Immunofluorescence assay
The recombinant nucleoprotein was coated on wells at 37 °C for 1 hour. Unspecific binding sites were blocked using 1% BSA at 37 °C for two hours. Mouse hyperimmune ascitic fluid against CCHF virus solution in phosphate buffered saline (1:600) was added and incubated at 37 °C for 45 min. Wells were washed five times with PBST. Anti-mouse IgG conjugated with fluorescein-5-isothiocyanate solution in PBST (1:1000) was added and incubated at 37 °C for 30 min. Wells were washed five times and observed under an Axiovert 200M inverted microscope, Zeiss, Germany.
Diagnostic value of nucleoprotein for detecting IgM antibody
The possibility of using the recombinant nucleoprotein to detect CCHF-specific IgM in ELISA tests was studied. A panel of three IgM-positive groups and three negative control groups were selected. Based on the result of IgM ELISA obtained by VectoCrimean -CHF-IgM kit, three positive groups were sorted as weak, medium, and high immune response. Negative control groups included healthy group, suspected to viral hemorrhagic fever, and the group of Chikungunya- and Dengue-positive samples (other frequent arboviruses in Iran and neighboring countries). All the samples in the panel were tested in parallel with molecular and serological methods to confirm positive and negative results. The CCHF virus strain in all the positive samples were sequenced and determined. The CCHF virus contains seven district clades with a relatively high diversity at the nucleotide and amino acid levels. Iran is a miniature model of the world for genotypes of CCHF virus[14]. The serum samples were chosen in a way to cover all the circulating CCHF genotypes in Iran: AsiaI, AsiaII, Kerman-22, EuropeI, and EuropeII[1,12,15]. In IgM capture ELISA test, the wells were coated overnight with diluted anti-µ chain in PBS (1:900). For each sample, two wells were assigned, test well and control well. Wells were washed with PBST (Tween-20, 0.3%) three times and blocked with PBSTM (PBST containing 3% skim milk) for 1 hour. Diluted serum in PBSTM (1:100) was incubated at 37 °C for 45 min and after washing with PBST recombinant nucleoprotein (40 µg/mL) or clear cell lysate from uninduced culture, it was incubated at 37 °C for 100 min in test or control wells, respectively. Mouse hyperimmune ascitic fluid against CCHF virus solution in PBSTM (1:600) was added to the wells and incubated at 37 °C for 45 min. Mouse anti-IgG HRP conjugate in PBSTM (1:350) was added and incubated at 37 °C for 45 min. After 10 minutes of incubation with 3,3’,5,5’-tetramethyl-benzidine substrate the reaction was terminated with 4 N H2SO4. Results were read in 450 nm with a reference of 620 nm. The wells were then washed three times with 300 µl of PBST and dried after each step. The cut-off value of the test was calculated based on Lardeux et al.[16]:
Cut-off = a. x̅ + f. SD
Where x̅ is the mean, and SD is the standard deviation of negative controls, a and f are two multipliers, set as a = 2 and f = 0.
Sensitivity and specificity of IgM capture ELISA were calculated as follows:
RESULTS
Synthetic gene and sub-cloning
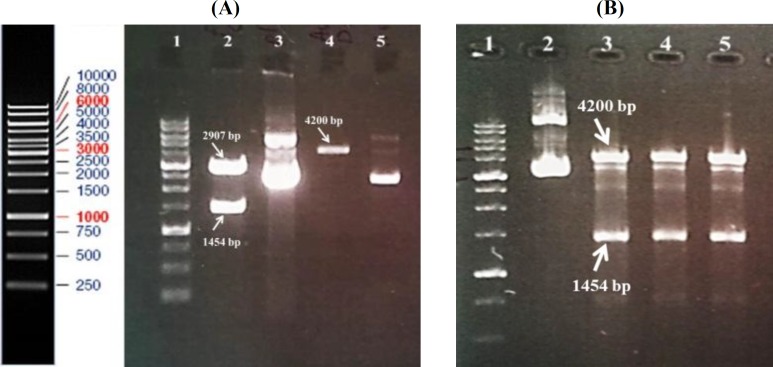
The codon-optimized synthetic gene (1454-bp) was delivered in pBSK (+) simple-Amp vector. The gene was sub-cloned in NcoI/BamHI site of pAC4 expression vector (Fig. 2). The correct cloning procedure was confirmed by restriction mapping and DNA sequencing of the final vector (Fig. 2B and data not shown).
Fig. 2.
Enzyme digestion of plasmids. (A) Lane 1, 1 kb DNA ladder; lane 2, construct [pBSK (+) simple-Amp-nucleoprotein gene] digested with restriction enzymes of NcoI and BamHI; lane 3, undigested pBSK (+) simple-Amp; lane 4, linearized pCA4 vector; lane 5, undigested pCA4. (B) Lane 1, 1 kb DNA ladder; lane 2, plasmids extracted from clones after ligation reaction; Lanes 3-5, ligate digested with NcoI and BamHI
Expression, purification, and refolding of recombinant nucleoprotein
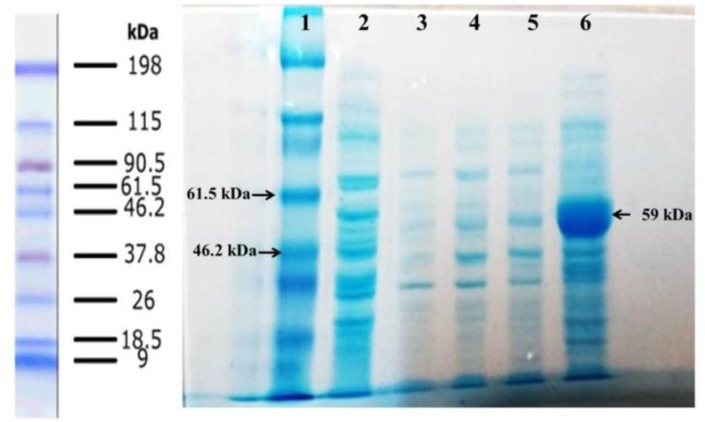
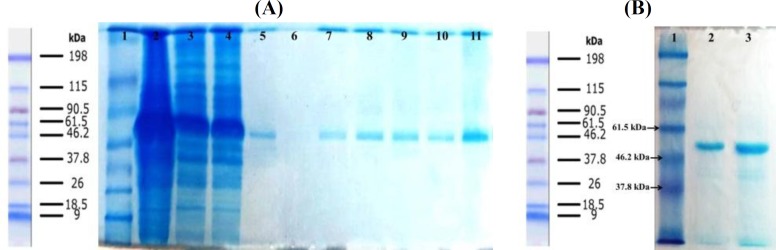
The nucleoprotein was expressed under the control of the trc promoter and induced by IPTG. SDS-PAGE analysis indicated that the recombinant nucleoprotein was appeared as IB. The band on gel was matched with the predicted 59 kDa molecular weight. Figure 3 shows the steps of washing IBs, and Figure 4A indicates protein purification processes on AbCA-AbC column. The purified protein was concentrated with Amicon centrifugal filter (Fig. 4B). Refolded protein structure was examined by Western blot, Circular dichroism, ELISA, and immunofluorescence test.
Fig. 3.
Washing steps of inclusion bodies. Lane 1, protein marker; lane 2, supernatant of second wash with lysis buffer; lane 3, supernatant of second wash with Tris EDTA Tween-20; lane 4, supernatant of second wash with Tris EDTA NaCl; lane 5, supernatant of second wash with Tris EDTA urea; lane 6, solubilized protein in solubilization buffer
Fig. 4.
Protein purification on AbCA-AbC column. (A) lane1, protein marker; lane 2, extracted protein from cultured media before washing steps; lanes 3 and 4, inclusion bodies after washing steps; lanes 5 and 6, washing the column with refolding buffers; lanes 7-11, protein eluted from column with the elution buffer. (B) Lane 1, protein marker; lane 2 purified protein; lane 3 concentrated protein using Amicon column
Western blotting
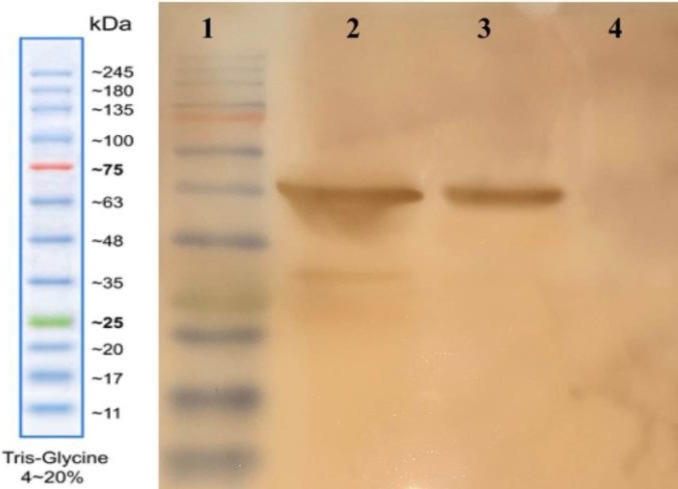
The expression of nucleoprotein was confirmed with the interaction of specific antibody against CCHF virus produced in mouse using Western blot test. Negative control, proteins extracted from E. coli (BL21 [DE3]) containing expression vector cultured without induction, did not show any specific band. Two samples of recombinant nucleoprotein showed visible specific bands in expected positions (Fig. 5). Positive controls were inactivated virus extracted from mouse brain in two different batches and had a clear band in accordance with the recombinant nucleoprotein. A slight difference in the size of positive controls and recombinant nucleoprotein was due to 21 amino acids added as spacer and AviTag to recombinant nucleoprotein.
Fig. 5.
Western blot analysis. Lane 1, protein marker; lane 2, positive control (inactivated virus); lane 3, recombinant nucleoprotein; lane 4, negative control (un-induced cell lysate)
Circular dichroism test
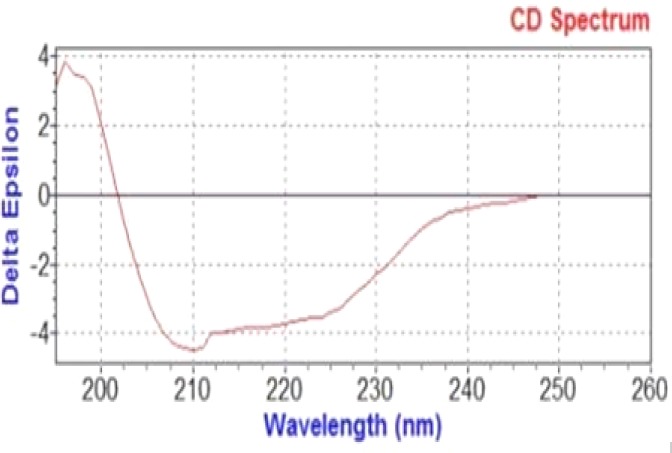
CD test was performed to find the evidence of nucleoprotein secondary structure. Analyzed data with CDNN demonstrated α-helix, random coil, and β-turn percentages with the frequencies of 59.2%, 20%, and 11%, respectively (Fig. 6).
Fig. 6.
Results of CD analysis by CDNN software (Circular Dichroism analysis using Neural Networks). Nucleoprotein (0.3 mg/mL) in 50 mM of Na3PO4, pH 7.5 was tested in the CD system. Spectra were recorded in 180-260 nm on a Circular Dichroism Spectrometer Model-215, Aviv, USA at a scan speed of 1 nm/sec
ELISA detection of nucleoprotein
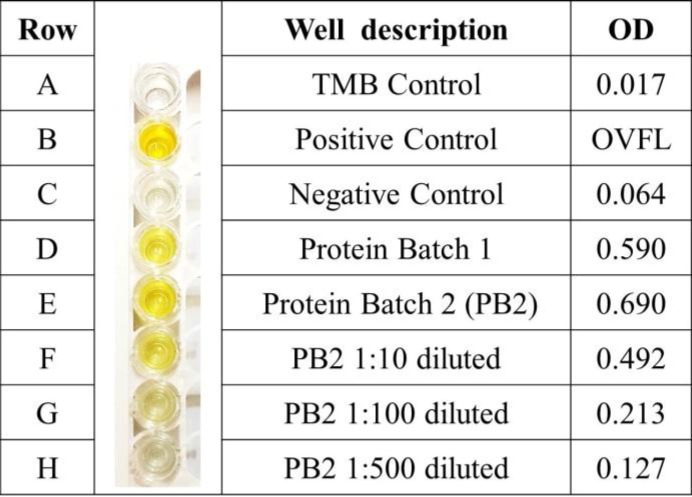
Refolded nucleoprotein in the dilutions of 1:10, 1:100, and 1:500 in PBS along with positive and negative controls were tested by using VectoCrimean-CHF-Ag. The results are exhibited in Figure 7.
Fig. 7.
Validation of recombinant nucleoprotein by using standard antigen detection kit of CCHF
IgM capture ELISA
Serum samples (n = 48) were tested using IgM capture ELISA. The test was blind for operator, i.e. positive and negative samples were arranged randomly. The cut-off for test was calculated to be 0.242, and capture IgM ELISA specifically identified all the positive samples. Of 23 positive samples, all were detected as positive with our ELISA test, which demonstrated 100% sensitivity, while none of the 25 negative samples were found as positive. All genotypes of the virus were detected by the test. Results are summarized in Table 1.
Table 1.
Results of IgM capture ELISA in panel groups
| Panel group | Total samples |
Genotype of
CCHF virus |
Mean OD with IgM capture ELISA | Detected by IgM capture ELISA | ||
|---|---|---|---|---|---|---|
| +ive | -ive | |||||
| Positive-weak immune response | 7 | AsiaI-Kerman22-EuropeI-EuropeII | 0.359 | 7 | 0 | |
| Positive-medium immune response | 8 | AsiaI-EuropeI-EuropeII | 0.730 | 8 | 0 | |
| Positive-high immune response | 8 | AsiaI-AsiaII-Kerman22-EuropeI-EuropeII | 0.980 | 8 | 0 | |
| Negative-healthy | 7 | NA | 0.150 | 0 | 7 | |
| Negative-suspected to other VHFs | 10 | NA | 0.113 | 0 | 10 | |
| Negative-other arboviruses (Den, Chik) IgM positives | 8 | NA | 0.107 | 0 | 8 | |
NA, not assigned
Immunofluorescence assay
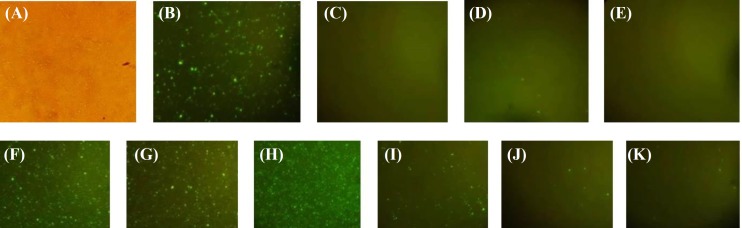
Immunofluorescence assay test revealed a significant decrease in florescent intensity in accordance with coated nucleoprotein concentration. Negative controls showed no florescent dot. Results are shown in Figure 8.
Fig. 8.
Immunofluorescence assay test results. Different concentrations of recombinant nucleoprotein is used, and description of controls are summarized as follows: (A) positive control with nucleoprotein without any dilution obtained under microscope with halogen filter; (B) experiment A repeated using the UV filter; (C) negative control coated with BSA instead of nucleoprotein; (D) negative control with ascitic fluid from non-infected mice; (E) negative control with nucleoprotein and FITC-labeled anti-mouse IgG; recombinant nucleoprotein used without any dilution (F), with 1:2 dilution in PBS (G), with 10-1 dilution (H), with 10-5 dilution (I), with 10-7 dilution (J), with 10-8 dilution (K)
DISCUSSION
According to WHO blueprint, CCHF is among the public health threatening diseases. It bears the risk of an outbreak in near future, which can be attributed to the characteristics and epidemiology of the virus, its high genetic diversity, animal to human and human to human transmission, high mortality rates, and weak health infrastructures in endemic regions. In addition, the lack of proper measures to deal with virus or inadequacy of existing measures has introduced this disease as a major threat to humanity. With these facts in mind, one of the significant factors in counter-measures remains with the diagnostic methods[17].
In this study, a recombinant nucleoprotein was expressed in E. coli, and its potency for detecting specific IgM by ELISA was evaluated using positive and negative samples. Viral nucleocapsid was selected because it is the major viral protein and is detected in large amounts in the serum of infected patients. This protein also stimulates severe immune responses as an immunogen, and the majority of antibodies in serum of infected patients and livestock are raised against this protein. It is worth mentioning that viral isolates from all over the world demonstrate antigenic resemblance in their nucleocapsids, and recombinant nucleocapsid produced from S segment of any viral isolate can be used to detect all the isolates[2,6,8]. Problems encountered with antigen production in eukaryotic systems such as low yields and high costs make these tests infeasible in every laboratory. Therefore, the necessity of a convenient low-cost method for scale-up production is obvious.
pAC4 vector was used for recombinant nucleoprotein expression in bacterial expression system. This vector adds the AviTag peptide (GLNDIFEAQKIEWHE) to protein C-terminal so that it can bind to biotin in this site by BirA enzyme and also interact with AbC antibody on affinity chromatography resin. In this way, nucleoprotein can be immobilized in a manageable way to carry on different studies based on biotin-streptavidin interactions. The expression vector, pAC4, contains trc promoter and the protein expression can be induced by adding IPTG. This vector is designed for high rates of expression in E. coli[18].
Recombinant nucleoprotein expressed in E. coli expression system was in the form of IBs. Washing IBs in successive steps led to high purity, which had a significant positive effect on the quality and yield of final purification and refolding by affinity chromatography. Further purification and simultaneous refolding of the protein was performed using protein folding liquid chromatography strategy by affinity column of AbCA-AbC mab antibody conjugated to sepharose resin made by Avidity Company. In this column, monoclonal antibody against AviTag is fixed on agarose resins and is able to detect and bind to both biotinylated and non-biotinylated proteins. Binding AviTagged nucleoprotein to the column minimizes the aggregation of denatured proteins and enhances refolding efficiency. When denatured proteins bind to the column by their AviTag end, due to sufficient space available to each molecule to refold, hydrophobic interactions between molecules reduce, leading to the prevention of their aggregation. On the other hand, the denaturing agent, urea, is washed away gradually and passes through the column along with other proteins and impurities[19,20].
Western blot and ELISA Ag detection verified the structure of recombinant nucleoprotein after refolding. Results of nucleoprotein secondary structure analysis by CD are in good accordance with the results of Carter et al.[21]. As standard nucleoprotein was not available, conditions and buffers were set based on Carter et al.'s[21] study, and the results were then compared. According to their reports, 66% of structure is in form of α-helix and 29% in form of random coil. Our CD results showed α-helix, random coil, and β-turn percentages of 59.2%, 20%, and 11%, respectively[21].
Recombinant viral nucleoprotein has previously been used for diagnosis of CCHF antibodies. In 1994, scientists used a bacterial expression system without codon optimization with a fusion protein to form three regions inside nucleoprotein ORF[22]. Emmerich et al.[23] produced recombinant nucleoprotein conjugated with HRP, which was then used in IgM/IgG ELISA to evaluate 15 kosovar. In the present study, codon optimization was employed for more effective expression in prokaryotic system, and the whole ORF was synthesized. Using a codon optimized recombinant nucleoprotein expressed in bacteria, Samudzi et al.[24] detected specific IgG antibody against 12 CCHF patients in South Africa. Sas et al.[25] utilized double antigen ELISA to detect specific antibodies against CCHF virus with multispecies approach and tested three different genotypes of the virus. The CCHF virus has seven district clades with a relatively high diversity at the nucleotide and amino acid levels. It is probable that specific antibodies against one strain would not be able to detect strains from different clades. In this study, five different genotypes circulating in Iran were analyzed, and the IgM capture ELISA could detect all the positive samples in different clades corresponding to 100% specificity.
In our research, the capture ELISA was used to increase the sensitivity of test. Meanwhile, cross-reactivity appears with some viruses, i.e. arboviruses like Dengue, Chikungunya, Zika, and West-Nile. CCHF virus has limited cross-reactivity with other nonpathogen viruses, like Hazara, in human. The non-pathogenonic character of this virus in human decreases the chance of cross-reactivity. In conclusion, we believe that our recombinant nucleoprotein can be used in any ELISA-based technique to detect human IgM antibody in any lab in endemic regions.
ACKNOWLEDGMENTS
We gratitude our cordial thanks to Mr. Alborz Naseri for his collaboration and to all the members of Arboviruses and Viral Hemorrhagic Fevers Department (National Ref. Lab), Pasteur Institute of Iran, Tehran for their technical assistance. The authors are also thankful for Deputy of Research, Shahed University, Tehran, Iran for financial support of this project.
CONFLICT OF INTEREST
None declared.
References
- 1.Salehi-Vaziri M, Baniasadi V, Jalali T, Mirghiasi SM, Azad-Manjiri S, Zarandi R, Mohammadi T, Khakifirouz S, Fazlalipour M. The first fatal case of Crimean-Congo hemorrhagic fever caused by the AP92-like strain of the Crimean-Congo hemorrhagic fever virus. Japanese journal of infectious diseases. 2016;69(4):344–346. doi: 10.7883/yoken.JJID.2015.533. [DOI] [PubMed] [Google Scholar]
- 2.Zivcec M, Safronetz D, Scott DP, Robertson S, Feldmann H. Nucleocapsid protein-based vaccine provides protection in mice against lethal Crimean-Congo hemorrhagic fever virus challenge. PLoS neglected tropical diseases. 2018;12(7):e0006628. doi: 10.1371/journal.pntd.0006628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maes P, Alkhovsky SV, Bào Y, Beer M, Birkhead M, Briese T, Buchmeier MJ, Calisher CH, Charrel RN, Choi IR, Clegg CS, de la Torre JC, Delwart E, DeRisi JL, Di Bello PL, Di Serio F, Digiaro M, Dolja VV, Drosten C, Druciarek TZ, Du J, Ebihara H, Elbeaino T, Gergerich RC, Gillis AN, Gonzalez JJ, Haenni AL, Hepojoki J, Hetzel U, Hồ T, Hóng N, Jain RK, Jansen van Vuren P, Jin Q, Jonson MG, Junglen S, Keller KE, Kemp A, Kipar A, Kondov NO, Koonin EV, Kormelink R, Korzyukov Y, Krupovic M, Lambert AJ, Laney AG, LeBreton M, Lukashevich IS, Marklewitz M, Markotter W, Martelli GP, Martin RR, Mielke-Ehret N, Mühlbach HP, Navarro B, Ng TFF, Nunes MRT, Palacios G, Pawęska JT, Peters CJ, Plyusnin A, Radoshitzky SR, Romanowski V, Salmenperä P, Salvato MS, Sanfaçon H, Sasaya T, Schmaljohn C, Schneider BS, Shirako Y, Siddell S, Sironen TA, Stenglein MD, Storm N, Sudini H, Tesh RB, Tzanetakis IE, Uppala M, Vapalahti O, Vasilakis N, Walker PJ, Wáng G, Wáng L, Wáng Y, Wèi T, Wiley MR, Wolf YI, Wolfe ND, Wú Z, Xú W, Yang L, Yāng Z, Yeh SD, Zhāng YZ, Zhèng Y, Zhou X, Zhū C, Zirkel F, Kuhn JH. Taxonomy of the family Arenaviridae and the order Bunyavirales: update 2018. Archives of virology. 2018;163(8):2295–2310. doi: 10.1007/s00705-018-3843-5. [DOI] [PubMed] [Google Scholar]
- 4.Spengler JR, Estrada-Peña A. Host preferences support the prominent role of Hyalomma ticks in the ecology of Crimean-Congo hemorrhagic fever. PLoS neglected tropical diseases. 2018;12(2):e0006248. doi: 10.1371/journal.pntd.0006248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fazlalipour M, Baniasadi V, Mirghiasi SM, Jalali T, Khakifirouz S, Azad-Manjiri S, Mahmoodi V, Naderi HR, Zarandi R, Salehi-Vaziri M. Crimean-Congo Hemorrhagic Fever Due to Consumption of Raw Meat: Case Reports From East-North of Iran. Japanese journal of infectious diseases. 2016;69(3):270–271. doi: 10.7883/yoken.JJID.2015.498. [DOI] [PubMed] [Google Scholar]
- 6.Bente DA, Forrester NL, Watts DM, McAuley AJ, Whitehouse CA, Bray M. Crimean-Congo hemorrhagic fever: history, epidemiology, pathogenesis, clinical syndrome and genetic diversity. Antiviral research. 2013;100(1):159–189. doi: 10.1016/j.antiviral.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 7.Al-Abri SS, Al Abaidani I, Fazlalipour M, Mostafavi E, Leblebicioglu H, Pshenichnaya N, Memish ZA, Hewson R, Petersen E, Mala P, Nhu Nguyen TM, Rahman Malik M, Formenty P, Jeffries R. Current status of Crimean-Congo haemorrhagic fever in the World Health Organization Eastern Mediterranean Region: issues, challenges, and future directions. International journal of infectious diseases. 2017;58:82–89. doi: 10.1016/j.ijid.2017.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dowall S, Buttigieg K, Findlay-Wilson S, Rayner E, Pearson G, Miloszewska A, Graham VA, Carroll MW, Hewson R. A Crimean-Congo hemorrhagic fever (CCHF) viral vaccine expressing nucleoprotein is immunogenic but fails to confer protection against lethal disease. Human vaccines and immunotherapeutics. 2016;12(2):519–527. doi: 10.1080/21645515.2015.1078045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vanhomwegen J, Alves MJ, Županc TA, Bino S, Chinikar S, Karlberg H, Korukluoğlu G, Korva M, Mardani M, Mirazimi A, Mousavi M, Papa A, Saksida A, Sharifi-Mood B, Sidira P, Tsergouli K, Wölfel R, Zeller H, Dubois P. Diagnostic assays for Crimean-Congo hemorrhagic fever. Emerging infectious diseases. 2012;18(12):1958–1965. doi: 10.3201/eid1812.120710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vemula S, Vemula S, Dedaniya A, Ronda SR. In vitro refolding with simultaneous purification of recombinant human parathyroid hormone (rhPTH 1–34) from Escherichia coli directed by protein folding size exclusion chromatography (PF-SEC): implication of solution additives and their role on aggregates and renaturation. Analytical bioanalytical chemistry. 2016;408(1):217–229. doi: 10.1007/s00216-015-9097-0. [DOI] [PubMed] [Google Scholar]
- 11.Mansour AA, Mousavi SL, Rasooli I, Nazarian S, Amani J, Farhadi N. Cloning, high level expression and immunogenicity of 1163-1256 residues of C-terminal heavy chain of C. botulinum neurotoxin type E. Biologicals. 2010;38(2):260–264. doi: 10.1016/j.biologicals.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 12.Guo JQ, Li QM, Zhou JY, Zhang GP, Yang YY, Xing GX, Zhao D, You SY, Zhang CY. Efficient recovery of the functional IP10-scFv fusion protein from inclusion bodies with an on-column refolding system. Protein expression purification. 2006;45(1):168–174. doi: 10.1016/j.pep.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 13.Mahmood T, Yang PC. Western blot: technique, theory, and trouble shooting. North American journal of medical sciences. 2012;4(9):429–434. doi: 10.4103/1947-2714.100998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chinikar S, Bouzari S, Shokrgozar MA, Mostafavi E, Jalali T, Khakifirouz S, Nowotny N, Fooks AR, Shah-Hosseini N. Genetic diversity of Crimean Congo hemorrhagic fever virus strains from Iran. Journal of arthropod-borne diseases. 2016;10(2):127–140. [PMC free article] [PubMed] [Google Scholar]
- 15.Fazlalipour M, Mohsenpour B, Baniasadi V, Jalali T, Mohammadi T, Azad-Manjiri S, Jamshisi Y, Azizizadeh S, Hosseini M, Khakifirouz S, Salehi-Vaziri M. A Case report of Crimean-Congo hemorrhagic fever virus Europe-1 genotype in the West of Iran. Iranian Red Crescent medical journal. 2017 In press. [Google Scholar]
- 16.Lardeux F, Torrico G, Aliaga C. Calculation of the ELISA’s cut-off based on the change-point analysis method for detection of Trypanosoma cruzi infection in Bolivian dogs in the absence of controls. Memórias do Instituto Oswaldo Cruz. 2016;111(8):501–504. doi: 10.1590/0074-02760160119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.WHO Research and Development Blueprint 2018. Annual review of diseases prioritized under the Research and Development Blueprint. Retreieved from: https://www.who.int/emergencies/diseases/2018prioritization-report.pdf?ua=1.
- 18.Amann E, Ochs B, Abel KJ. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene. 1988;69(2):301–315. doi: 10.1016/0378-1119(88)90440-4. [DOI] [PubMed] [Google Scholar]
- 19.Bai Q, Geng X. Protein Folding Liquid Chromatography. In: Evans, Jr, Xu MQ., editors. Heterologous Gene Expression in E.coli. Methods in Molecular Biology (Methods and Protocols), vol 705. Berlin: Springer; 2011. [DOI] [PubMed] [Google Scholar]
- 20.Geng X, Wang C. Protein folding liquid chromato-graphy and its recent developments. Journal of chromatography B. 2007;849(1-2):69–80. doi: 10.1016/j.jchromb.2006.10.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carter S, Barr J, Edwards T. Expression, purification and crystallization of the Crimean-Congo haemorrhagic fever virus nucleocapsid protein. Acta crystallographica section F: structural biology and crystallization communications. 2012;68(Pt 5):569–573. doi: 10.1107/S1744309112009736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marriott AC, Polyzoni T, Antoniadis A, Nuttall PA. Detection of human antibodies to Crimean-Congo haemorrhagic fever virus using expressed viral nucleocapsid protein. Journal of general virology. 1994;75(Pt 9):2157–2161. doi: 10.1099/0022-1317-75-9-2157. [DOI] [PubMed] [Google Scholar]
- 23.Emmerich P, Emmerich P, von Possel R, Rackow A, Liu Y, Schmitz H, Günther S, Sherifi K, Halili B, Jakupi X, Berisha L. Sensitive and specific detection of Crimean-Congo Hemorrhagic Fever Virus (CCHFV)—Specific IgM and IgG antibodies in human sera using recombinant CCHFV nucleoprotein as antigen in μ-capture and IgG immune complex (IC) ELISA tests. PLoS neglected tropical diseases. 2018. Reterieved from: https://journals.plos.org/plosntds/article?id=10.1371/journal.pntd.0006366. [DOI] [PMC free article] [PubMed]
- 24.Samudzi RR, Leman PA, Paweska JT, Swanepoel R, Burt FJ. Bacterial expression of Crimean-Congo hemorrhagic fever virus nucleoprotein and its evaluation as a diagnostic reagent in an indirect ELISA. Journal of virological methods. 2012;179(1):70–76. doi: 10.1016/j.jviromet.2011.09.023. [DOI] [PubMed] [Google Scholar]
- 25.Sas MA, Comtet L, Donnet F, Mertens M, Vatansever Z, Tordo N, Pourquier P, Groschup MH. A novel double-antigen sandwich ELISA for the species-independent detection of Crimean-Congo hemorrhagic fever virus-specific antibodies. Antiviral research. 2018;151:24–26. doi: 10.1016/j.antiviral.2018.01.006. [DOI] [PubMed] [Google Scholar]