Abstract
AIMS:
The aim of the study was to evaluate the immune response triggered by the first contact of human monocytes with two T. cruzi strains from distinct discrete typing units (DTUs) IV and V, and whether co-infection with these strains leads to changes in monocyte immune profiles, which could in turn influence the subsequent infection outcome.
METHODS AND RESULTS:
We evaluated the influence of in vitro single- and co-infection with AM64 and 3253 strains on immunological characteristics of human monocytes. Single-infection of monocytes with AM64 or 3253 induced opposing anti-inflammatory and inflammatory responses, respectively. Co-infection was observed in over 50% of monocytes after 15 hours of culture, but this percentage dropped ten-fold after 72 hours. Co-infection led to high monocyte activation, and an increased percentage of both IL-10 and TNF. The decreased percentage of co-infected cells observed after 72 hours was associated with a decreased frequency of TNF-expressing cells.
CONCLUSION:
Our results show that the exacerbated response observed in co-infection with immune-polarizing strains is associated with a decreased frequency of co-infected cells, suggesting that the activated response favors parasite control. These findings may have implications for designing new Chagas disease preventive strategies.
Keywords: Trypanosoma cruzi, strains, discrete typing units, co-infection, human, monocytes, parasitemia
INTRODUCTION
Chagas disease, caused by infection with the protozoan Trypanosoma cruzi, is a major health problem, with over 7 million infected and millions more at risk. During the chronic phase, 60–70% of patients remain asymptomatic (indeterminate form), while about 30% develop severe cardiac and/or digestive manifestations. Parasite and host-related factors play key roles in defining clinical outcome (1, 2).
Inflammatory cytokines such as IFN-gamma and TNF-alpha, play a role in controlling parasite infection, while IL-10 is crucial for preventing the exacerbation of the immune response during the chronic indeterminate clinical form. Non-excluding hypotheses argue that T-cell exhaustion is the cause of the severe clinical form of Chagas disease (3, 4), and that the balance between the inflammatory (TNF) and anti-inflammatory (IL-10) cytokines is crucial for the clinical outcome (5–13). Although T cells from Chagas patients can produce TNF and IL-10 (11, 14, 15), monocytes are the main source of these cytokines in Chagas disease (16).
The diverse T. cruzi population is currently classified into six Discrete Typing Units (DTUs) (17–19), which display distinct geographic and biological properties (19–22). The heterogeneity of T. cruzi strains has been considered a critical factor for the distinct clinical evolution of Chagas disease (23–26). Although the majority of studies performed in vitro and in experimental models employ infections with single strains of T. cruzi, different strains coexist dynamically in nature. Infection with more than one strain has been described in vectors (27, 28) and patients (25, 29, 30).
Initial contact of immune cells with the parasite is critical for shaping the subsequent adaptive response during infection, and we have previously shown that the initial contact with different T. cruzi strains can interfere with monocyte and neutrophil immune profiles (31, 32). We hypothesize that two strains from different DTUs, associated with mild or severe clinical manifestations of Chagas disease, induce different immune responses in human monocytes and may influence one another during co-infection. Thus, we used AM64 (DTU TcIV, associated with mild disease) (33, 34) and 3253 (DTU TcV, associated with severe disease) (19, 35), strains to infect human monocytes and evaluate their immunological profile in single and co-infected cultures.
We observed that single infection with AM64 or 3253 strains induced opposing anti-inflammatory and inflammatory cytokine profiles in human monocytes, respectively. Co-infection led to high activation of monocytes, as shown by increased expression of TLR-2 and CD80. Interestingly, the frequency of both TNF and IL-10 were increased in co-infected cells, as compared to single-infected ones. Although over 50% of monocytes were co-infected after 15 hours, this percentage decreased ten-fold after 72 hours of infection. This decrease was directly associated with a decrease in the percentage of TNF+ cells. Thus, the phenotypic and functional activated profile generated by co-infection may favor parasite control. These findings may aid in the design of new Chagas disease preventive strategies.
HUMAN SAMPLES, MATERIALS AND METHODS
Human samples
The donors included in our studies were non-Chagas healthy individuals (n=8), as determined by negative specific serological test for Chagas disease. Individuals were from the city of Belo Horizonte, state of Minas Gerais, Brazil, with ages ranging between 18 and 49 years (average ± SD: 31.6±10.8). We excluded from our study individuals with any chronic inflammatory disease, diabetes, heart and circulatory illnesses (including hypertension) or bacterial infections. All individuals included in this work were volunteers and provided written informed consent. This work was approved by the Ethical Committee of the Universidade Federal de Minas Gerais, under the protocol# ETIC077/06. Peripheral blood was collected from the donors by venipuncture.
Parasites
Parasites used in the assays are isolates of AM64 and 3253 strains (36, 37). Tissue culture-derived trypomastigotes (TCT) of AM64 (TcIV) and 3253 (TcV) strains were isolated from infected monolayers of Lewis-lung cells (LLC). LLCs were infected using five TCT/host cells and kept in DMEM enriched with 1% with inactivated fetal calf serum (FCS), supplemented with antibiotics (penicillin at 500μ/mL and streptomycin at 0.5 mg/mL). After approximately six days, the TCTs were collected from the supernatant, washed once by centrifugation with phosphate-buffered saline (PBS) pH 7.2 at 1000 × g for 10 min at 4°C and resuspended in RPMI to 6 ×107 TCT/mL.
Trypomastigotes were labeled with Alexa Fluor 647 or CFSE. For the staining with Alexa Fluor 647 (Thermo Fisher Scientific), 108 TCTs were incubated with 3.2μg of Alexa Fluor 647 for 30 min at 37°C under 5% CO2. Labeled parasites were washed two times with PBS by centrifugation at 1000g for 10 min at 4°C. Trypomastigotes were labeled with CFSE (carboxyfluorescein diacetate succinimidyl ester – Molecular Probes C1157) using a protocol previously reported by us (31). Briefly, 6,0 × 107 parasites were incubated with 5μM CFSE for 15 min at 37°C under 5% CO2. Labeled parasites were washed three times with cold PBS + 10% of inactivated fetal bovine serum by centrifugation at 1000 × g for 10 min at 4°C.
Adherent cell preparation, infection and confocal analysis
Adherent cells were used solely to confirm the infectivity of monocytes by the different strains using confocal microscopy. Peripheral blood mononuclear cells (PBMC) were purified as previously done by us (9). Briefly, heparinized blood was diluted 1:1 with PBS and applied over a Ficoll gradient. The mixture was centrifuged for 40 min at 600 × g. PBMCs were collected at the interface between the plasma and the Ficoll. Cells were washed three times by centrifugation (600 × g, 10min) with PBS and resuspended in RPMI supplemented 5% of human sera, antibiotics (penicillin at 500U/ml and streptomycin at 05 mg/ml) and 1mΜ of L-glutamine. To obtain adherent cells, 2 ×106 PBMC/well were plated on 13-mm round coverslips in RPMI enriched with 5% of inactivated human sera, antibiotics (penicillin at 500U/ml and streptomycin at 05 mg/mL) and 1mΜ of L-glutamine and incubated for 3 hours at 37°C, 5% CO2. After incubation, non-adherent cells were removed by carefully washing the wells with warm PBS. As previously determined by us, adherent cells obtained using this protocol are approximately 85% CD11b+ or CD14+(9).
The monocytes (adherent cells) were infected with TCTs from both strains previously stained with Alexa Fluor 647 and CFSE (AM64 and 3253 respectively) at a ratio of 5:5:1 (AM64/3253/monocytes) in triplicates and incubated for additional 3 hours. After the incubation period, the monolayers were washed with PBS to remove extracellular TCTs and re-incubated for 69 hours in RPMI enriched with 5% of inactivated human sera, antibiotics (penicillin at 500U/mL and streptomycin at 05 mg/mL) and 1mΜ of L-glutamine, completing a total of 72 hours of culture. At the end of the culture time, cells were fixed by incubating the slides with 300 μL of paraformaldehyde for 15 min at room temperature, washed three times with PBS, stained with 4’6’-diamino-2-phenylindole (DAPI) diluted 1:300 in PBS for 15 min at room temperature, and mounted using Vectashield (Vector laboratories). Confocal analyses were performed using a confocal Zeiss 5-live running ZEN 2009 coupled to an Axio observer Z1 using an oil immersion objective (63X, 1.2 numerical aperture, Oberkochen, Germany).
Infection of peripheral blood cells
Infection of peripheral blood (2mL) was used for all experiments of surface molecule and cytokine analysis. The single infection was performed using 10 TCT/cells. TCTs from AM64 and 3253 were previously stained with CFSE as described above, and used separately to infect blood cells by incubation at 37°C in 5% CO2 for 3 hours with RPMI enriched with 5% of inactivated human serum, antibiotics (penicillin at 500U/mL and streptomycin at 05 mg/mL) and 1mΜ of L-glutamine. After this period, samples were washed and re-incubated for a total of 15 hours. These infections were used to evaluate the immunological characteristics of single-infected cultures (Figures 1 and 2).
Figure 1: Monocyte single infection with CFSE-labeled trypomastigotes from AM64 (Tc IV) and 3253 (Tc V) strains.
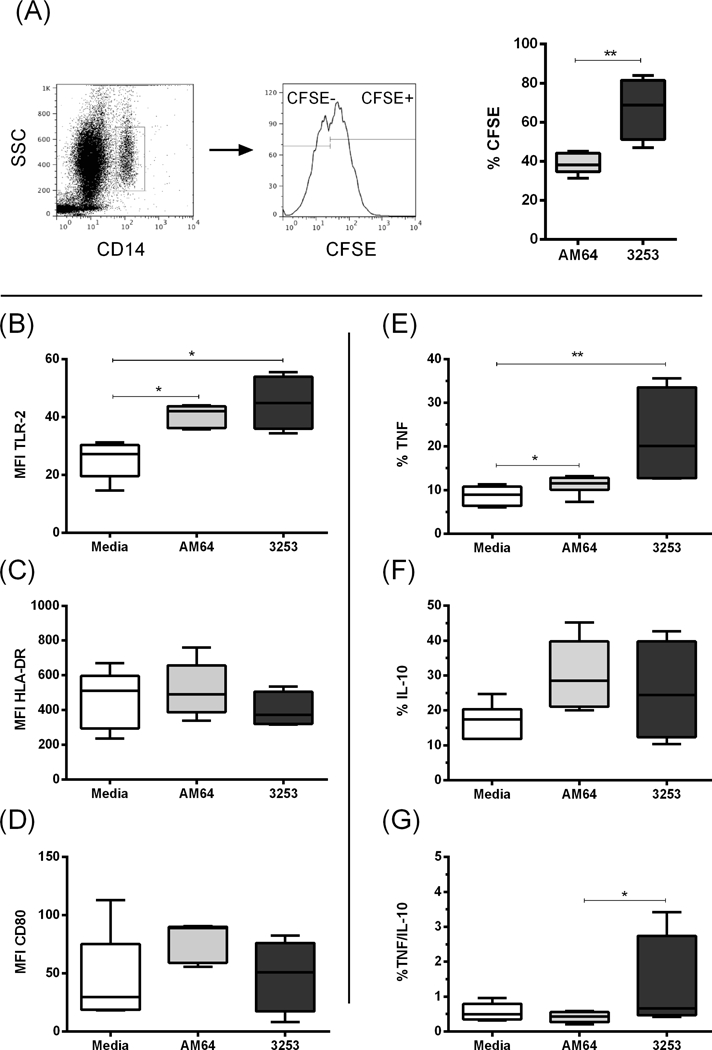
Total human blood samples from non-Chagas donors were cultured for 15 hours with either AM64 or 3254 trypomastigotes previously labeled with CFSE, and monocyte population was analyzed by flow cytometry as described in the materials and methods section. (A) First panel shows representative dot plot of CD14 expression versus granularity (SSC), indicating the selection of monocytes (CD14high). Second plot shows representative histogram for selection of CFSE+ monocytes. Graphs are gated in infected CFSE+ monocytes, and the % of CFSE+ cells exposed to the different strains are shown in the third graph. (B) Mean fluorescence intensity (MFI) of TLR-2 expression in CFSE+ monocytes; (C) MFI of HLA-DR expression in CFSE+ monocytes; (D) MFI of CD80 expression in CFSE+ monocytes; (E) Percentage of TNF+ in CFSE+ monocytes; (F) Percentage of IL-10 in CFSE+ monocytes and (G) Percentage of TNF/IL-10 ratio in CFSE+ monocytes. White boxes represent total blood incubated with media; grey and black boxes represent blood incubated in the presence of CFSE-labeled AM64 or 3253 trypomastigotes, respectively. Results are expressed as box and whisker plots showing minimum and maximum values. Comparison between groups were performed with using Kruskal-Wallis test as described in material and methods section. *p<0.05, **p<0.01
Figure 2: Percentage of human monocytes single- and co-infected with trypomastigotes from AM64 and 3253 strains.
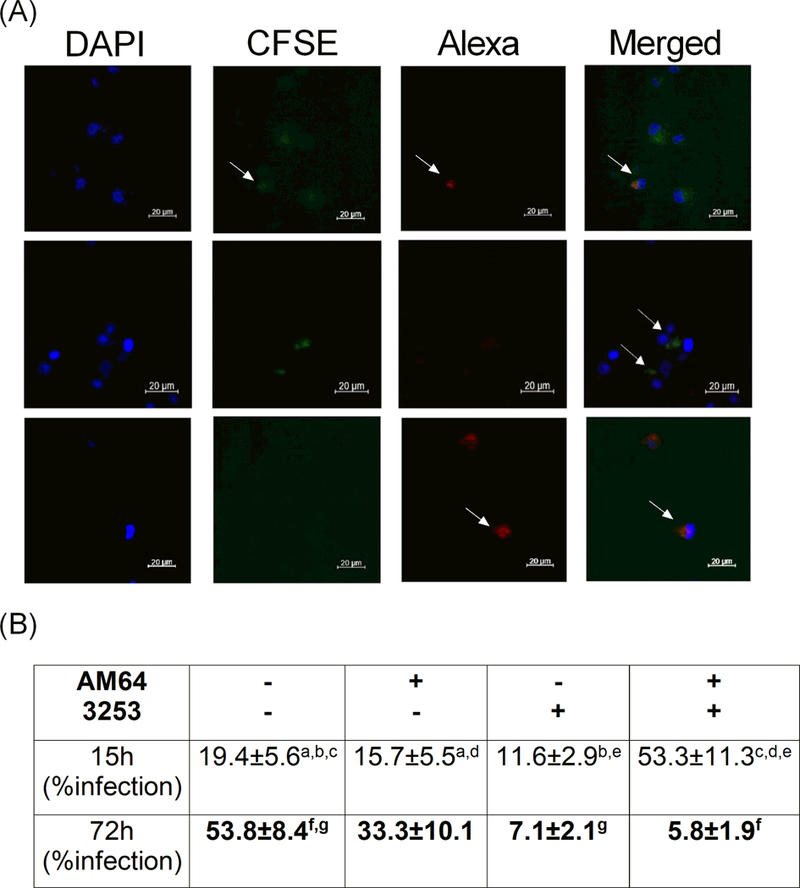
Adherent cells obtained by plating PBMC for 1h (Panel A) or total human blood (Panel B) from non-Chagas donors were co-cultured with AM64 or 3254 trypomastigotes previously labeled with Alexa-Fluo or CFSE, respectively, as described in material and methods. (A) shows representative confocal microscopy analysis, showing double infected monocytes (first row, last figure - merged), CFSE-labeled 3253 single-infected (second row) and Alexa-labeled AM64 single-infected (third row) monocytes after 72 hours of infection. Arrows indicate monocytes co-infected with AM64 and 3253 (first row), single infected with 3253 (second row) and single infected with AM64 (third row); (B) Percentage of infection after 15 or 72 hours of culture obtained in whole blood incubated with both strains and acquired by flow cytometry. Results are expressed as average ± standard deviation. Identical letters indicate statistical significance, a: p= 0.04, b: p= 0.043, c: p= 0.007, d: p= 0.003, e: p= 0.005, g: p= 0.036 and f: p= 0.008.
Co-infection was performed using 5 TCTs of each strain/cell. TCTs from AM64 and 3253 were previously stained with Alexa Fluor 647 or CFSE, respectively, as described above. Cells and TCTs were incubated at 37°C in 5% CO2 for 3 hours with RPMI enriched with 5% of inactivated human serum, antibiotics (penicillin at 500U/mL and streptomycin at 05 mg/mL) and 1mΜ of L-glutamine. After the incubation period, cells were washed by centrifugation with PBS at 600 × g for 10 min at 4°C to remove extracellular TCTs. For the incubation of “15 hours” and “72 hours”, we re-incubated the cultures for additional 12 and 69 hours, respectively, after washing off the free TCTs. Brefeldin A (1μg/mL) was added for the last four hours of infection in both groups (15 and 72 hours) to prevent protein secretion. These infected cultures were used to analyze immunological characteristics of single and co-infected cells present in the same sample, using the strategy of analysis shown in Figure 3A and B.
Figure 3: Intensity of expression of TLR-2, HLA-DR and CD80 by human monocytes single- and co-infected with trypomastigotes from AM64 and 3253 strains after 15 or 72 hours of infection.
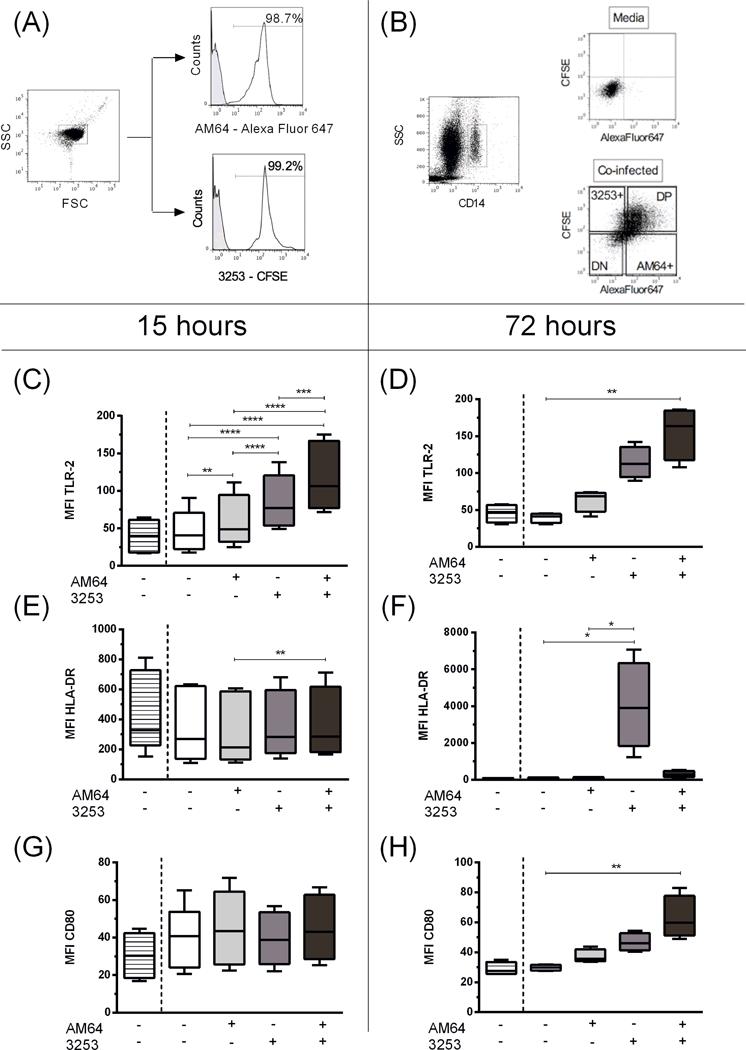
Trypomastigotes from AM64 and 3253 were stained with Alexa Fluor 647 and CFSE, respectively, and co-cultured for 15 or 72 hours with whole blood from healthy donors as described in material and methods. (A) shows representative plots of AlexaFluor 647- or CFSE-labeled trypomastigotes, indicating the selection of trypomastigotes in a forward (FSC) and side scatter (SSC) graph. In the histograms, the grey-shaded lines represent media control with no staining and white lines show representative staining for AM64 labeled with Alexa fluor 647 (top histogram) and 3253 labeled with CFSE (botton histogram); (B) Representative figure of gating strategy for the experiments with mixed infection. First plot shows the selection of monocytes that was performed in a similar way to the single-infection analysis (SSC versus CD14, selecting CD14high cells). After selection of monocytes, we divided the subpopulations of non-infected double negative monocytes (DN – AF647-CFSE-), AM64 single infection (AM64+: AF647+CFSE-), 3253 single infection (CFSE+: AF647-CFSE+), and double infected, double positive (DP - AF647+CFSE+) monocytes using the plot AF647 versus CFSE. Plots show representative selection of the four subpopulations in blood incubated with media and with both strains (top and bottom plot respectively). In graphs C-H striped box show media incubation, white, light grey, dark grey and black plot show DN, AM64 single infected, 3253 single infected and double-infected monocytes from incubation with AM64 and 3253 in the same tube. (C) Mean fluorescent intensity (MFI) of TLR-2 expression in monocytes after 15 hours of infection; (D) MFI of TLR-2 expression in monocytes after 72 hours of infection; (E) MFI of HLA-DR expression in monocytes after 15 hours of infection. (F) MFI of HLA-DR expression in monocytes after 72 hours of infection. (G) MFI of CD80 expression in monocytes after 15 hours of infection. (H) MFI of CD80 expression in monocytes after 72 hours of infection. Results are expressed in box and whisker plots showing minimum and maximum values. Comparisons between groups were performed with using Kruskal-Wallis test as described in material and methods section. *p<0.05, **p<0.01, ***p<0.001 and ****p<0.0001.
Analysis of expression of surface molecules and cytokines by peripheral blood cells using flow cytometry
After 15 and 72 hours of incubation with parasite strains, the erythrocytes were lysed using RBC “Lysing buffer” (Bio Legend) at 20mL/1mL of peripheral blood. The tubes were incubated for 15 min at 20°C in the dark. After the incubation, cells were washed three times with PBS by centrifugation at 600 × g for 10 min at 4°C and resuspended in PBS to 107cells/ml. Cells were then immunostained and analyzed using multiparametric flow cytometry. 250.000 cells were incubated for 15 min at 4°C with different antibody combinations. Samples were washed three times in PBS-1% bovine serum albumin (BSA) and fixed by 20-min incubation with 2% formaldehyde solution. After removal of the fixing solution by centrifugation and washing once with PBS, we permeabilized the cells by incubation for 10 min with 0.5% saponin solution, centrifuged, and incubated with antibodies to intracellular molecules for 30 min at 20°C.
The antibodies to surface molecules used were: anti-TLR-2 clone TLR2.1 – labeled with PE; anti-CD14 clone M5E2 – labeled with BV421; anti-HLA-DR clone L243 – labeled with BV510 and anti-CD80 clone 2D10 – labeled with PE-Cy7. For intracellular staining we used the following antibodies: anti-TNF-alpha clone Mab11 – labeled with PE and anti-IL-10 clone JES3–9D7 – labeled with PECY7. All antibodies were purchased from BioLegend (San Diego, CA, USA). After intracellular staining, cells were washed and resuspended in PBS and acquired using a FACSCanto II (Becton & Dickinson, San Jose, CA, USA). A total of 200,000 events were acquired and the parameters were analyzed in the monocyte population using the FloJo software. Digital compensation matrices were performed for all analysis, providing adequate compensation to eliminate any potential spill over. In order to select the monocytes population, we gated on CD14high cells in plot of SSC versus CD14. For the analysis of single infection we selected the monocytes population and further gated on CFSE+ and CFSE- subpopulation (Figure 1A). For the analysis of mixed infection we selected the monocytes population and further gated on Alexa Fluor 647 versus CFSE, selecting non-infected monocytes (Alexa Fluor647-CFSE-) AM64 single-infected monocytes (Alexa Fluor647+CFSE-), 3253 single-infected monocytes (Alexa Fluor647-CFSE+) and double-infected monocytes (Alexa Fluor647+CFSE+) (Figure 3B). Appropriate isotype controls, uninfected cells stained with the same antibody combinations as used in infected cells, as well as unlabeled parasites were used in all experiments as controls of unspecific binding.
Statistical analysis
We compared our results using One-Way Anova or Kruskal-Wallis test according to Kolmogorov-Smirnov normality test. All analyses were performed using Graph Pad Prism Software (La Jolla, CA, USA). Differences that returned p values equal or less than 0.05 were considered statistically significant from one another. For the principal component analysis (PCA), we used Clustvis software (38).
RESULTS
Single infections with AM64 (TcIV) and 3253 (TcV) induce polar responses by human monocytes
To evaluate whether infection with strains belonging to DTUs TcIV and TcV induce different immune response in human monocytes, we infected human peripheral blood with CFSE-labeled AM64 (TcIV) and 3253 (TcV) strains individually. Figure 1A shows that 3253 (TcV) infected a higher frequency of monocytes in comparison with AM64 (TcIV). Based on the expression of surface molecules associated with monocyte activation in CFSE+ cells, no differences were observed in the expression of HLA-DR or CD80 by infected monocytes (Figure 1 C and D) regardless of the strain. Infection with either strain led to high monocyte activation, as measured by the high intensity of expression of TLR-2 (Figure 1 B).
Although both strains led to a similar level of monocyte activation, we observed that their induced cytokine profiles were distinct. AM64 induced high TNF expression as compared to media. Infection with 3253 induced a higher TNF percentage in comparison with both media and AM64 infection (Figure 1E). There was no statistical difference in the percentage of IL-10 after single infection with either strain (Figure 1F). The ratio between the percentages of the inflammatory cytokine, TNF, and the anti-inflammatory cytokine, IL-10, showed that infection with 3253 induced a more inflammatory response in infected monocytes, with a greater TNF/IL-10 ratio (Figure 1G).
Mixed infection with AM64 and 3253 predominates in relation to single infection of human monocytes
During natural transmission of T. cruzi, the same vector could be infected with more than one strain, therefore, the patient would have a mixed infection. It is possible that this mixed infection would induce a different immune response compared to a single infection. Thus, we sought to investigate how mixed infection with two strains (AM64 and 3253) that induce polar cytokine profiles would affect the immune response of human monocytes upon initial contact.
In order to perform the co-infection we stained TCT from the AM64 and 3253 strains with Alexa Fluor 647 and CFSE, respectively, and incubated total human blood from non-Chagas donors with trypomastigotes from both strains at the same time. It is important to note that all cells were in contact with both 3253 and AM64 but, as expected, after the incubation time not all monocytes were infected. Therefore, in the final culture it was possible to identify monocytes that were non-infected (CFSE- and AF647-), infected with only AM64 (CFSE- and AF647+), infected with only 3253 (CFSE+ and AF647-) and infected with both AM64 and 3253 (CFSE+ and AF647+).
Figure 2A shows representative images, obtained by confocal microscopy, of monocytes that were single infected with either AM64 or 3253, and monocytes that were double-infected.
Figure 2B shows the percentage, obtained by flow cytometry, of single and mixed infections. After 15 hours of infection, the majority of monocytes were co-infected with both strains (Figure 2B). Moreover, similar rates of single infected monocytes with AM64 or 3253 were observed, while only 19% of the monocytes were not infected with either strain (Figure 2B). After 72 hours of infection, we noted a shift in the monocytes infection profile, with 53% uninfected monocytes, followed by 33% AM64 single-infected monocytes (Figure 2B). The percentage of monocytes infected with 3253 and double-infected monocytes decreased drastically (Figure 2B).
Double-infected monocytes display higher activation, as compared to single-infected monocytes
Next we investigated if there was a difference in monocyte activation after mixed infection with AM64 and 3253. To achieve this goal, we assessed the expression of TLR-2, HLA-DR and CD80 in double-infected cells. Figure 3A and B shows the gating strategy employed in these analyses. Our results show that after 15 hours, double-infected monocytes showed a higher intensity of TLR-2 expression compared with single-infected and non-infected monocytes (Figure 3C). Furthermore, single-infected monocytes with both strains also displayed a higher intensity of TLR-2 expression compared to non-infected cells. In addition, 3253 single-infected monocytes expressed higher intensity of TLR-2 compared to monocytes single-infected with AM64 strain (Figure 3C). Regarding the intensity of HLA-DR expression after 15 hours of infection, it was observed that double-infected monocytes express higher intensity of this molecule when compared to AM64 single-infected monocytes (Figure 3E). After 15 hours of infection, there were no differences in the intensity of CD80 expression between single or co-infected cells (Figure 3G).
Analysis of the expression of activation molecules after 72 hours of infection showed a greater intensity of TLR-2 expression in double infected compared to non-infected monocytes (Figure 3D). 3253 single-infected monocytes induced a greater intensity of HLA-DR expression compared to non-infected and AM64 single-infected monocytes (Figure 3F). Finally, double-infected monocytes showed increased intensity of CD80 expression compared to non-infected monocytes (Figure 3H).
Double-infected monocytes express a higher frequency of TNF and IL-10 producing cells
We then questioned whether monocyte activation triggered by infection had an influence on the percentage of TNF and IL-10 producing cells, as examples of inflammatory and anti-inflammatory cytokines, respectively.
3253 single-infected monocytes showed an increased percentage of TNF compared to non-infected and AM64 single-infected monocytes. However, an even greater percentage of TNF positive cells was observed in double-infected monocytes (Figure 4A), as compared to all other conditions, after 15 hours of infection. A significant decrease in TNF producing cells was observed after 72 hours of infection as compared to 15 hours, and no statistical difference was observed comparing the different groups after 72 hours of culture (Figure 4B).
Figure 4: Percentage of TNF and IL-10 expression by human monocytes single- and co-infected with trypomastigotes from AM64 and 3253 strains after 15 or 72 hours of infection.
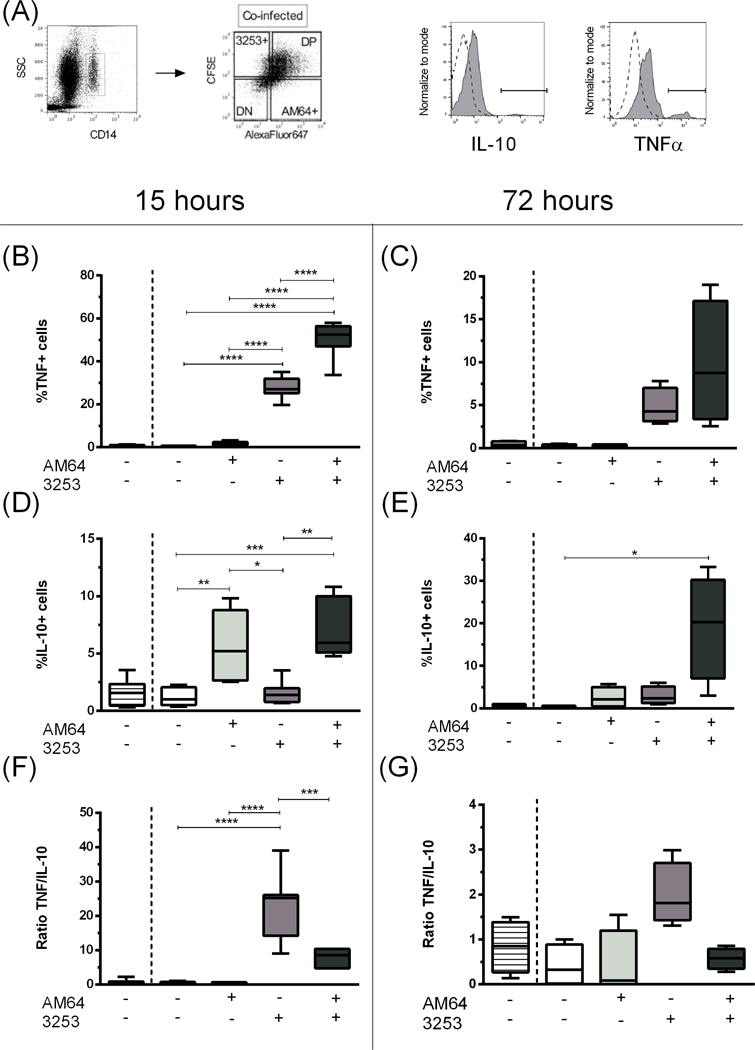
Trypomastigotes from AM64 and 3253 were stained with Alexa Fluor 647 and CFSE, respectively, and co-cultured for 15 or 72 hours with whole blood from healthy donors as described in material and methods. The percentage of cytokine expression in each subpopulation was evaluated. (A) shows the gating strategy employed, where monocytes were selected using the plot SSC x CD14 (first plot) followed by the selection of the subpopulations of double-negative (DN), AM64 single-infection (AM64+), 3253 single-infection (3253+) and double-positive (DP) through the plot Alexa-Fluor 647 versus CFSE (second plot); histograms show the isotype control for intracellular staining (dashed lines), as well as a representative curve for IL-10 and TNF expression (solid grey curves). Determination of percentage of (B) TNF+ in monocytes after 15 hours of infection (C) TNF+ in monocytes after 72 hours of infection; (D) IL-10+ in monocytes after 15 hours of infection; (E) IL-10+ in monocytes after 72 hours of infection; (F) TNF/IL-10 ratio in monocytes after 15 hours of infection; (G) TNF/IL-10 ratio in monocytes after 72 hours of infection. Results are expressed in box and whisker plots showing minimum and maximum values. Comparisons between groups were performed with using Kruskal-Wallis test as described in material and methods section. *p<0.05, **p<0.01, ***p<0.001 and ****p<0.0001.
Interestingly, an opposite profile was observed for the percentage of IL-10 producing cells, regarding single-infected cells. AM64 single-infected monocytes exhibited a greater percentage of IL-10 producing cells in comparison to non-infected and 3253 single-infected monocytes (Figure 4C). A similar result was observed in double-infected monocytes. After 72 hours of infection, double-infected monocytes showed a much greater percentage of IL-10 producing cells compared to all other conditions (Figure 4D). No differences were observed in the MIF of TNF or IL-10.
When we analyzed the ratio between TNF/IL-10 percentage, we observed that 3253 single infected monocytes present an increased inflammatory profile compared to all other conditions, with a much higher TNF/IL-10 ratio (Figure 4 E and F).
Principal Component Analysis (PCA) confirms distinct, non-overlapping, profiles between monocytes infected with two polar T. cruzi strains
In order to evaluate the similarity between non-infected, single-infected and double-infected monocytes we performed principal component analysis (PCA) using either surface molecule expression or cytokine percentage. Figure 5A shows the PCA segregation for principal component 1 (PC1) vs. principal component 2 (PC2) for activation molecules (TLR-2, HLA-DR, CD80), while figure 5B shows the PCA segregation for cytokines (TNF and IL-10). Overlapping populations are seen using analysis of surface molecules with greater similarity between non-infected and AM64 infected monocytes, as compared to the double infected and 3253 single-infected monocytes (Figure 5A). Strikingly, when performing PCA and plotting the populations using cytokine production, a clear segregation (non-overlapping) is seen between the 3253 and AM64 infected monocytes (Figure 5B). Moreover, co-infected monocytes segregate from AM64 and non-infected monocytes, and are more similar -yet separate from- 3253 infected monocytes. Figure 5 C and D show plots obtained from non-instructed analysis (tSNE - t-distributed Stochastic Neighbor Embedding), which is an unsupervised computer learning analysis that uses all characteristics analyzed to cluster the different populations (single and co-infected, as well as non-infected) based on similarities. Figure 5C shows the distribution of the 4 conditions, showing the double-infected cells in between the single infected populations. Figure 5D clearly shows the bias of TNF and IL-10 into 3253 and AM64 populations, respectively.
Figure 5: Principal component and t-distributed stochastic neighbor embedding (t-SNE) analysis of non-infected, AM64 single-infected, 3253 single-infected and double-infected monocytes.
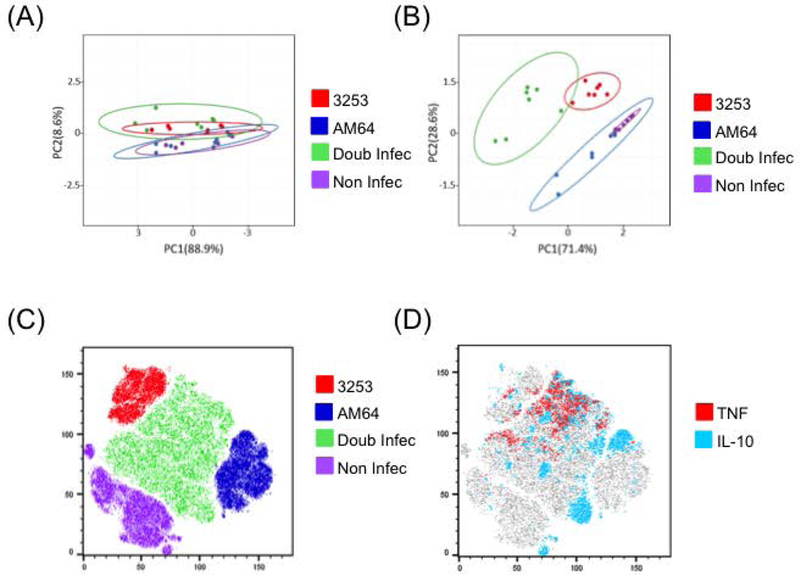
Principal component analysis (PCA) was performed with data collected after 15 hours of incubation with AM64 and 3253 trypomastigotes. (A) shows cluster associations between intensity of expression of activation molecules (TLR-2, HLA-DR and CD80), and (B) shows cluster associations between percentage of cytokines (TNF and IL-10) for non-infected (purple), AM64 (blue) and 3253 (red) single-infected or double-infected cells (green). (C) shows tSNE plot identifying the distribution of the single and double infected, as well as non-infected samples and (D) shows the bias of TNF and IL-10 toward 3253 and AM64 strains, respectively.
DISCUSSION
The T. cruzi population is currently classified into six DTUs (named TcI-VI). In general, different DTUs exhibit distinct biological properties, yet with some overlap between strains. In this study, we evaluated the influence of single and co-infection with isolates belonging to DTU TcIV and TcV on the immunological characteristics of human monocytes. TcIV occurs in North and South America and is associated with human Chagas disease in Venezuela and in the Brazilian Amazon (15, 21, 31, 34, 39–41). It is speculated that TcIV parasites are responsible for the low parasitemia and morbidity seen in chronic Chagas disease in Amazonas State in Brazil, despite a high seroprevalence (34). It is also known that Amazonian TcIV isolates induce low parasitemia with less virulence and pathogenicity in mice as compared to TcII strains(33, 36). On the other hand, TcV is strongly associated with human Chagas disease in the southern cone (19, 35, 42). TcV is also associated with megacolon (35) and cardiac manifestations (19). Given that TcIV strains are associated with mild disease and TcV are associated with severe manifestations, we investigated if isolates representative of these DTUs altered differently the immune profile of human monocytes, whether alone or in a co-infection in vitro system.
We demonstrated that AM64 and 3253 have different infection rates in monocytes. Also, single infection with AM64 (TcIV) and 3253 (TcV) induced an opposite TNF/IL-10 balance. Infection with AM64 induced a more regulatory environment with low TNF/IL-10 ratios, while infection with 3253 induced high TNF/IL-10 ratios. Previous work from our group and others have demonstrated that infection with strains from DTUs TcI and TcII also induced distinct cytokine profiles in human monocytes (25, 31). We cannot rule out that the difference in the cytokine profile induced after infection with either strain could be due to the different infection rate observed. However, the inflammatory and anti-inflammatory profiles induced by 3253 and AM64, respectively, may be related to the clinical outcomes reported to TcV and TcIV strains, strengthening the hypothesis that the early interactions of distinct parasites with the host may influence the clinical outcome of Chagas disease. Also, these results indicate the possible use of immunological characteristics, in association with the other biological characteristics already in use, as additional criteria for T. cruzi classification.
Evidence suggests that the majority of T. cruzi populations isolated from hosts and vectors are multiclonal (39, 43). The possibility of infections involving more than one DTU in the same host is one of the most interesting and challenging aspects considering the clinical outcome of Chagas disease. To assess whether the polar profiles of cytokine observed when monocytes were single-infected with AM64 or 3253 would remain during a mixed infection, we labeled AM64 with Alexa Fluor 647 and 3253 with CFSE. This strategy allowed us to separate single-infected monocytes with AM64 or 3253 (AlexaFluor647+CFSE- and AlexaFluor647-CFSE+) from double-infected monocytes and non-infected monocytes (AlexaFluor647+CFSE+ and AlexaFluor647-CFSE-). These dyes stably label the parasites with distinct fluorescent signals allowing for later identification on the flow cytometer. Because they complex with cell components, once the staining is performed, it is stable.
Our results also showed that after 15 hours of infection the majority of monocytes were infected with the two strains (Figure 2B). However, the percentage of double-infected monocytes drastically decreased after 72 hours. The frequency of 3253 single-infected monocytes also decreased, although not as much as double-infected cells. On the other hand, the percentage of AM64 single-infected monocytes increased after 72 hours of infection compared to 15 hours. This difference in the percentage of monocytes infected with the two strains may represent compensation, but may also reflect a selection of the AM64 strain during infection. This idea is in accordance with the paper by Pena et al., who demonstrated a selection of TcII strains during mixed infection with TcI and TcII using both human and murine macrophages (44).
Interestingly, AM64 and 3253 single-infected monocytes, as well as double infected monocytes showed greater expression of the activation molecule TLR-2. This is consistent with the findings that TLR-2 is related to the internalization of trypomastigotes in murine macrophages via activation of Rab-5 (40).
Surprisingly, the cytokine profile observed when the monocytes were infected only with AM64 or 3253 was significantly amplified in co-infected cells. The 3253 single-infected monocytes exhibited high percentage of TNF and low percentage of IL-10, while the AM64 single-infected monocytes had the opposite profile. Double-infected monocytes did not present an intermediate response. Instead, they presented an even higher percentage of percentage of both TNF and IL-10. The difference between the expression of activation molecules and percentage of cytokines from infected monocytes exposed to only one strain (Figure 1), and single infected monocytes that were exposed to both strains (Figure 3), could be due to an amplification effect triggered by the presence of both strains at the same time (Figure 3).
The fact that we observed a decrease in double-infected, as well as 3253 single infected monocytes after 72 hours of culture suggests that this reduction is due to the high activation profile, consistent with the high intensity of expression of the activation molecules TLR-2, HLA-DR and a higher percentage of the pro-inflammatory cytokine TNF observed in those cells. In fact, the decrease in infected cells was also accompanied by a decrease in TNF+ cells, suggesting that these cells may undergo apoptosis, as previous studies have shown that TNF can induce apoptosis (41, 45). We have also shown that T. cruzi infection induces apoptosis in neutrophils, which is associated with TNF production (32). In addition to this hypothesis, it is also possible that heavily infected cells are dying due to parasite-induced rupture. If this is the case, it is interesting that it preferentially occurred with 3253 infected cells.
To further analyze characteristics of the single and double infection monocytes, we performed a principle component analysis (PCA) with activation molecules and cytokines. PCA showed that 3253 single-infected and double-infected monocytes grouped together, while AM64 single-infected and non-infected monocytes were more related to each other. This was seen most dramatically when analyzing cytokine production by the subpopulations, where the mixed infection population most closely resembles the 3253 infection. Moreover, this analysis also confirms that infection by 3253 and AM64 induce polar responses that are clearly segregated based on the PCA analysis.
The results presented here showed that AM64, representative of the TcIV DTU, which is related to mild disease, induces a regulated profile in human monocytes, while 3253 (TcV) strain, associated with severe disease, induces an inflammatory profile. This emphasizes the importance of the parasite strain in influencing the immune response and, thus, disease outcome. In addition, strains that induce less activation and production of anti-inflammatory cytokines could be positively selected at the beginning of the infection, leading to parasite persistence and milder disease, as suggested by the persistence of AM64 infected cells seen here. We also found that mixed infection led to higher activation and percentage of inflammatory cytokines, which may be inducing cell death and, as a consequence, may help control early infection. Thus, it is possible that a mixed infection with polarizing strains induces an activated response, which will lead to control of parasitemia, while selecting a less pathogenic population, which in turn could be beneficial for the host. These findings provide information about the role of T. cruzi strains in the immune response that may influence pathology, and shed light on how co-infection with different strains from T. cruzi may change the immune response during acute infection.
ACKNOWLEDGEMENTS:
EC, LMCG, RCG, KJG, WOD are CNPq fellows; LMDM LSAP and CCK are CAPES fellows. This project was funded by FAPEMIG (Universal 2014), INCT-DT/CNPq, NIH R01AI138230.
Footnotes
DISCLOSURES: none
DATA SHARING: The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES:
- 1.Pérez-Molina JA, Molina I. Chagas disease. Lancet. 2018;391(10115):82–94. [DOI] [PubMed] [Google Scholar]
- 2.Dutra WO, Menezes CA, Magalhães LM, Gollob KJ. Immunoregulatory networks in human Chagas disease. Parasite Immunol. 2014;36(8):377–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Natale MA, Cesar G, Alvarez MG, Castro Eiro MD, Lococo B, Bertocchi G, et al. Trypanosoma cruzi-specific IFN-γ-producing cells in chronic Chagas disease associate with a functional IL-7/IL-7R axis. PLoS Negl Trop Dis. 2018;12(12):e0006998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laucella SA, Postan M, Martin D, Hubby Fralish B, Albareda MC, Alvarez MG, et al. Frequency of interferon- gamma -producing T cells specific for Trypanosoma cruzi inversely correlates with disease severity in chronic human Chagas disease. J Infect Dis. 2004;189(5):909–18. [DOI] [PubMed] [Google Scholar]
- 5.Reis DD, Jones EM, Tostes S, Lopes ER, Gazzinelli G, Colley DG, et al. Characterization of inflammatory infiltrates in chronic chagasic myocardial lesions: presence of tumor necrosis factor-alpha+ cells and dominance of granzyme A+, CD8+ lymphocytes. Am J Trop Med Hyg. 1993;48(5):637–44. [DOI] [PubMed] [Google Scholar]
- 6.Dutra WO, Gollob KJ, Pinto-Dias JC, Gazzinelli G, Correa-Oliveira R, Coffman RL, et al. Cytokine mRNA profile of peripheral blood mononuclear cells isolated from individuals with Trypanosoma cruzi chronic infection. Scand J Immunol. 1997;45(1):74–80. [DOI] [PubMed] [Google Scholar]
- 7.Higuchi MD, Ries MM, Aiello VD, Benvenuti LA, Gutierrez PS, Bellotti G, et al. Association of an increase in CD8+ T cells with the presence of Trypanosoma cruzi antigens in chronic, human, chagasic myocarditis. Am J Trop Med Hyg. 1997;56(5):485–9. [DOI] [PubMed] [Google Scholar]
- 8.Abel LC, Rizzo LV, Ianni B, Albuquerque F, Bacal F, Carrara D, et al. Chronic Chagas’ disease cardiomyopathy patients display an increased IFN-gamma response to Trypanosoma cruzi infection. J Autoimmun. 2001;17(1):99–107. [DOI] [PubMed] [Google Scholar]
- 9.Souza PE, Rocha MO, Rocha-Vieira E, Menezes CA, Chaves AC, Gollob KJ, et al. Monocytes from patients with indeterminate and cardiac forms of Chagas’ disease display distinct phenotypic and functional characteristics associated with morbidity. Infect Immun. 2004;72(9):5283–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Talvani A, Rocha MO, Barcelos LS, Gomes YM, Ribeiro AL, Teixeira MM. Elevated concentrations of CCL2 and tumor necrosis factor-alpha in chagasic cardiomyopathy. Clin Infect Dis. 2004;38(7):943–50. [DOI] [PubMed] [Google Scholar]
- 11.Villani FN, Rocha MO, Nunes MoC, Antonelli LR, Magalhães LM, dos Santos JS, et al. Trypanosoma cruzi-induced activation of functionally distinct αβ and γδ CD4- CD8- T cells in individuals with polar forms of Chagas’ disease. Infect Immun. 2010;78(10):4421–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costa I, Linhares AC, Cunha MH, Tuboi S, Argüello DF, Justino MC, et al. Sustained Decrease in Gastroenteritis-related Deaths and Hospitalizations in Children Less Than 5 Years of Age After the Introduction of Rotavirus Vaccination: A Time-Trend Analysis in Brazil (2001–2010). Pediatr Infect Dis J. 2016;35(6):e180–90. [DOI] [PubMed] [Google Scholar]
- 13.Araujo FF, Gomes JA, Rocha MO, Williams-Blangero S, Pinheiro VM, Morato MJ, et al. Potential role of CD4+CD25HIGH regulatory T cells in morbidity in Chagas disease. Front Biosci. 2007;12:2797–806. [DOI] [PubMed] [Google Scholar]
- 14.de Araújo FF, Corrêa-Oliveira R, Rocha MO, Chaves AT, Fiuza JA, Fares RC, et al. Foxp3+CD25(high) CD4+ regulatory T cells from indeterminate patients with Chagas disease can suppress the effector cells and cytokines and reveal altered correlations with disease severity. Immunobiology. 2012;217(8):768–77. [DOI] [PubMed] [Google Scholar]
- 15.Menezes CA, Sullivan AK, Falta MT, Mack DG, Freed BM, Rocha MO, et al. Highly conserved CDR3 region in circulating CD4(+)Vβ5(+) T cells may be associated with cytotoxic activity in Chagas disease. Clin Exp Immunol. 2012;169(2):109–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Souza PE, Rocha MO, Menezes CA, Coelho JS, Chaves AC, Gollob KJ, et al. Trypanosoma cruzi infection induces differential modulation of costimulatory molecules and cytokines by monocytes and T cells from patients with indeterminate and cardiac Chagas’ disease. Infect Immun. 2007;75(4):1886–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brisse S, Barnabé C, Tibayrenc M. Identification of six Trypanosoma cruzi phylogenetic lineages by random amplified polymorphic DNA and multilocus enzyme electrophoresis. Int J Parasitol. 2000;30(1):35–44. [DOI] [PubMed] [Google Scholar]
- 18.Tibayrenc M Genetic epidemiology of parasitic protozoa and other infectious agents: the need for an integrated approach. Int J Parasitol. 1998;28(1):85–104. [DOI] [PubMed] [Google Scholar]
- 19.Zingales B, Miles MA, Campbell DA, Tibayrenc M, Macedo AM, Teixeira MM, et al. The revised Trypanosoma cruzi subspecific nomenclature: rationale, epidemiological relevance and research applications. Infect Genet Evol. 2012;12(2):240–53. [DOI] [PubMed] [Google Scholar]
- 20.Laurent JP, Barnabe C, Quesney V, Noel S, Tibayrenc M. Impact of clonal evolution on the biological diversity of Trypanosoma cruzi. Parasitology. 1997;114 ( Pt 3):213–8. [DOI] [PubMed] [Google Scholar]
- 21.Revollo S, Oury B, Laurent JP, Barnabé C, Quesney V, Carrière V, et al. Trypanosoma cruzi: impact of clonal evolution of the parasite on its biological and medical properties. Exp Parasitol. 1998;89(1):30–9. [DOI] [PubMed] [Google Scholar]
- 22.de Lana M, da Silveira Pinto A, Barnabé C, Quesney V, Noël S, Tibayrenc M. Trypanosoma cruzi: compared vectorial transmissibility of three major clonal genotypes by Triatoma infestans. Exp Parasitol. 1998;90(1):20–5. [DOI] [PubMed] [Google Scholar]
- 23.Vago AR, Andrade LO, Leite AA, d’Avila Reis D, Macedo AM, Adad SJ, et al. Genetic characterization of Trypanosoma cruzi directly from tissues of patients with chronic Chagas disease: differential distribution of genetic types into diverse organs. Am J Pathol. 2000;156(5):1805–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lages-Silva E, Ramírez LE, Pedrosa AL, Crema E, da Cunha Galvão LM, Pena SD, et al. Variability of kinetoplast DNA gene signatures of Trypanosoma cruzi II strains from patients with different clinical forms of Chagas’ disease in Brazil. J Clin Microbiol. 2006;44(6):2167–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poveda C, Fresno M, Gironès N, Martins-Filho OA, Ramírez JD, Santi-Rocca J, et al. Cytokine profiling in Chagas disease: towards understanding the association with infecting Trypanosoma cruzi discrete typing units (a BENEFIT TRIAL sub-study). PLoS One. 2014;9(3):e91154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oliveira MT, Branquinho RT, Alessio GD, Mello CGC, Nogueira-de-Paiva NC, Carneiro CM, et al. TcI, TcII and TcVI Trypanosoma cruzi samples from Chagas disease patients with distinct clinical forms and critical analysis of in vitro and in vivo behavior, response to treatment and infection evolution in murine model. Acta Trop. 2017;167:108–20. [DOI] [PubMed] [Google Scholar]
- 27.Noireau F, Diosque P, Jansen AM. Trypanosoma cruzi: adaptation to its vectors and its hosts. Vet Res. 2009;40(2):26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pena DA, Eger I, Nogueira L, Heck N, Menin Á, Báfica A, et al. Selection of TcII Trypanosoma cruzi population following macrophage infection. J Infect Dis. 2011;204(3):478–86. [DOI] [PubMed] [Google Scholar]
- 29.Monje-Rumi MM, Brandán CP, Ragone PG, Tomasini N, Lauthier JJ, Alberti D’Amato AM, et al. Trypanosoma cruzi diversity in the Gran Chaco: mixed infections and differential host distribution of TcV and TcVI. Infect Genet Evol. 2015;29:53–9. [DOI] [PubMed] [Google Scholar]
- 30.Breniere SF, Waleckx E, Barnabe C. Over Six Thousand Trypanosoma cruzi Strains Classified into Discrete Typing Units (DTUs): Attempt at an Inventory. PLoS Negl Trop Dis. 2016;10(8):e0004792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Magalhães LM, Viana A, Chiari E, Galvão LM, Gollob KJ, Dutra WO. Differential Activation of Human Monocytes and Lymphocytes by Distinct Strains of Trypanosoma cruzi. PLoS Negl Trop Dis. 2015;9(7):e0003816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Magalhães LMD, Viana A, de Jesus AC, Chiari E, Galvão L, Gomes JA, et al. Distinct Trypanosoma cruzi isolates induce activation and apoptosis of human neutrophils. PLoS One. 2017;12(11):e0188083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Monteiro WM, Magalhães LK, de Sá AR, Gomes ML, Toledo MJ, Borges L, et al. Trypanosoma cruzi IV causing outbreaks of acute Chagas disease and infections by different haplotypes in the Western Brazilian Amazonia. PLoS One. 2012;7(7):e41284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Monteiro WM, Magalhães LK, Oliveira JC, Guerra JA, Silveira H, Ferreira LC, et al. Biological behavior of Trypanosoma cruzi stocks obtained from the State of Amazonas, Western Brazilian Amazon, in mice. Rev Soc Bras Med Trop. 2012;45(2):209–14. [DOI] [PubMed] [Google Scholar]
- 35.Virreira M, Serrano G, Maldonado L, Svoboda M. Trypanosoma cruzi: typing of genotype (sub)lineages in megacolon samples from bolivian patients. Acta Trop. 2006;100(3):252–5. [DOI] [PubMed] [Google Scholar]
- 36.Dos Reis D, Monteiro WM, Bossolani GD, Teston AP, Gomes ML, de Araújo SM, et al. Biological behaviour in mice of Trypanosoma cruzi isolates from Amazonas and Paraná, Brazil. Exp Parasitol. 2012;130(4):321–9. [DOI] [PubMed] [Google Scholar]
- 37.Martins K, Andrade CeM, Barbosa-Silva AN, do Nascimento GB, Chiari E, Galvão LM, et al. Trypanosoma cruzi III causing the indeterminate form of Chagas disease in a semi-arid region of Brazil. Int J Infect Dis. 2015;39:68–75. [DOI] [PubMed] [Google Scholar]
- 38.Metsalu T, Vilo J. ClustVis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015;43(W1):W566–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sales-Campos H, Kappel HB, Andrade CP, Lima TP, de Castilho A, Giraldo LE, et al. Trypanosoma cruzi DTU TcII presents higher blood parasitism than DTU TcI in an experimental model of mixed infection. Acta Parasitol. 2015;60(3):435–41. [DOI] [PubMed] [Google Scholar]
- 40.Aoki MP, Carrera-Silva EA, Cuervo H, Fresno M, Gironès N, Gea S. Nonimmune Cells Contribute to Crosstalk between Immune Cells and Inflammatory Mediators in the Innate Response to Trypanosoma cruzi Infection. J Parasitol Res. 2012;2012:737324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brenner D, Blaser H, Mak TW. Regulation of tumour necrosis factor signalling: live or let die. Nat Rev Immunol. 2015;15(6):362–74. [DOI] [PubMed] [Google Scholar]
- 42.Cardinal MV, Lauricella MA, Ceballos LA, Lanati L, Marcet PL, Levin MJ, et al. Molecular epidemiology of domestic and sylvatic Trypanosoma cruzi infection in rural northwestern Argentina. Int J Parasitol. 2008;38(13):1533–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Macedo AM, Pena SD. Genetic Variability of Trypanosoma cruzi:Implications for the Pathogenesis of Chagas Disease. Parasitol Today. 1998;14(3):119–24. [DOI] [PubMed] [Google Scholar]
- 44.Pena DA, Eger I, Nogueira L, Heck N, Menin A, Bafica A, et al. Selection of TcII Trypanosoma cruzi population following macrophage infection. J Infect Dis. 2011;204(3):478–86. [DOI] [PubMed] [Google Scholar]
- 45.Chaves AT, de Assis Silva Gomes Estanislau J, Fiuza JA, Carvalho AT, Ferreira KS, Fares RC, et al. Immunoregulatory mechanisms in Chagas disease: modulation of apoptosis in T-cell mediated immune responses. BMC Infect Dis. 2016;16:191. [DOI] [PMC free article] [PubMed] [Google Scholar]


