Abstract
Purpose:
The detection rate of breast ductal carcinoma in situ (DCIS) has increased significantly, raising the concern that DCIS is over-diagnosed and over-treated. Therefore, there is an unmet clinical need to better predict the risk of progression among DCIS patients. Our hypothesis is that by combining molecular signatures with clinicopathologic features, we can elucidate the biology of breast cancer progression, and risk-stratify patients with DCIS.
Methods:
Targeted exon-sequencing with a custom panel of 223 genes/regions was performed for 125 DCIS cases. Among them, 60 were from cases having concurrent or subsequent invasive breast cancer (IBC) (DCIS+IBC group), and 65 from cases with no IBC development over a median follow-up of 13 years (DCIS-only group). Copy number alterations in chromosome 1q32, 8q24, 11q13 were analyzed using fluorescence in situ hybridization (FISH). Multivariable logistic regression models were fit to the outcome of DCIS progression to IBC as a function of demographic and clinical features.
Results:
We observed recurrent variants of known IBC-related mutations, and the most commonly mutated genes in DCIS were PIK3CA (34.4%) and TP53 (18.4%). There was an inverse association between PIK3CA kinase domain mutations and progression (Odds Ratio [OR]=10.2, p<0.05). Copy number variations in 1q32 and 8q24 were associated with progression (OR=9.3 and 46, respectively; both p<0.05).
Conclusions:
PIK3CA kinase domain mutations and the absence of copy number gains in DCIS are protective against progression to IBC. These results may guide efforts to distinguish low-risk versus high-risk DCIS.
Keywords: Ductal carcinoma in situ, PIK3CA, copy number variant, breast cancer
INTRODUCTION
Ductal carcinoma in situ (DCIS) of the breast is a risk factor as well as a precursor lesion for invasive breast cancer (IBC). The current standard of treatment for DCIS involves surgery in combination with radiation therapy and/or endocrine therapy [1–4]. However, this treatment paradigm was developed based on the natural history of DCIS and IBC in the pre-mammography screening era. In recent decades, DCIS detection rates have increased significantly due to contemporary, advanced screening imaging modalities [5]. Concurrently, multiple studies have demonstrated that only 13-52% of patients with DCIS eventually develop subsequent IBC [3,6], suggesting that many patients with DCIS may not require extensive treatment, and raising the concern that some DCIS patients are being over-treated.
Therefore, a precision medicine approach to stratify risk of developing IBC among DCIS patients is critically needed. The discovery of genomic features that correlate with high-risk and low-risk DCIS would be important for tailored IBC screening and prevention. A number of studies, including ours, have examined the changes in the genome during the progression of breast neoplasia to IBC. The findings suggest that single nucleotide variations (SNVs) and copy number alterations (CNAs) both are acquired over a series of genomic events that occurs over the course of development of the IBC [7–11].
In our prior studies, we have examined genomic changes in hyperplasia, DCIS and IBC by targeted sequencing [8], whole genome sequencing[7], and fluorescence in situ hybridization (FISH) [12] to identify genomic changes. These studies have found recurrent genomic changes in pre-invasive neoplasia, both CNAs and SNVs, which have also been identified in IBC. Some events, like PIK3CA mutations, can occur quite early in the neoplastic timeline [13] while others, like ERBB2 amplification, occur later [14]. However, our hypothesis is that there is no single genomic feature that correlates with the transition from DCIS to IBC; instead, it is likely that a constellation of features or higher order features, such as genome complexity or gene pathway alterations, are driving progression.
In this study, we investigated the mutational profiles of DCIS cases that do not develop IBC over a long follow up interval (median 13 years), and DCIS cases that are initially associated with IBC or later develop IBC. We hypothesize that by combining molecular signatures with clinicopathologic features, we can elucidate the biology of breast cancer progression, and risk-stratify patients with DCIS.
MATERIALS AND METHODS
Patient population
Available cases were identified in the Department of Pathology at Stanford University Hospital (SUH) from 2000 to June 2011. DCIS cases with adequate tissue for research sampling and confirmed follow-up were categorized as follows: 1) DCIS-only group: DCIS and no development of IBC over a median follow-up of 13 years (average 11 years) or 2) DCIS+IBC group: DCIS with concurrent or subsequent IBC present. There is no restrain for the size of associated invasive cancer or the detection methods (screen-detect v.s. palpable mass, etc). Surgical samples with sufficient tissue were collected with Health Insurance Portability and Accountability Act (HIPAA)-compliant Stanford University Institutional Review Board (IRB) approval (Protocol number 32496). Clinical data were obtained from the Oncoshare breast cancer research database, which has been described previously [15,16].
Generation of Targeted-capture libraries
Hematoxylin and eosin stained slides were reviewed to confirm the diagnosis of DCIS. The areas with aboundant amout of DCIS were chosen for examination. For cases with DCIS and invasive carcinoma present in the same specimen, we carefully selected the area with abundant DCIS away from the invasive component. The DCIS samples were acquired by taking 3-10 2mm cores from the corresponding areas of paraffin blocks (from cases on tissue microarrays TA 239, 419, 420, and 445). The thickness of the tissue in each core is approximately 2-3 mm, so we do not anticipate a major contamination from invasive carcinoma in deeper content of the core. Only tumor samples were analyzed, and no paired normal samples were included. The DNA was extracted using RecoverAll Total Nucleic Acid Isolation kit (Ambion). Targeted libraries of genomic DNAs were generated using the Agilent SureSelect XT kit and Agilent Automation Systems NGS system. Additional information is provided in the supplementary materials and methods.
Sequencing data analysis
The sequencing data were analyzed using a custom pipeline. In brief, reads were aligned to the hg19 human reference genome assembly using BWA [17]. Duplications were marked using Picard Tools V1.118 (http://broadinstitute.github.io/picard). Cases with less than 92M aligned reads were discarded. Insertion-deletion realignment and base recalibration were achieved using GATK v.3.3-0 [18]. The somatic variant calls were carried out using an ensemble approach with four variant callers: MuTect[19], VarScan2 [19], VarDict [20], and Freebayes [21] Calls present in at least two out four callers were accepted. The variant annotation was done using ANNOVAR [22] and custom scripts. The full list of nonsynonemous variants was provided in Supplmentary Table S6. Bedtools coverage was used to create a histogram of coverage for each feature in the BED file and a summary histogram of all of features in the BED file. These histograms were then plotted using R to show the percentage of capture regions covered at any given depth for every individual sample. The sequencing data was uploaded to the NCBI Sequence Read Archive.
Fluorescence in situ hybridization (FISH)
FISH was performed as previously described [12], and additional details is provided in the Supplmentary Material and Methods. Total test probe green counts (1q32.1, 8q24.21, 11q13.11) were compared with red (2q37.3) control counts, which are frequently unaltered in breast cancer [24]. The signals were evaluated according to two parameters: signals per cell and ratio of test probe to control probes. Cases were scored as gain at the locus if the target to control probe ratio was greater than 1.5 or the number of test signals was greater than three per cell.
Statistical Analysis
Descriptive statistics were presented of baseline characteristics among the two DCIS groups (DCIS-only group and DCIS+IBC group). The student t-test was used to assess whether differences in mutation burden existed between the DCIS alone and DCIS with IBC cohorts.
Two main multivariable logistic regression models were fit to characterize the association between DCIS progression to IBC and clinical and demographic features. The first model fit DCIS progression to IBC as a function of categorical copy number status, age at diagnosis, race/ethnicity, DCIS nuclear grade, tumor size, margins, surgery type, and an indicator for lack of PIK3CA mutation in the kinase region (PIK3CA-KD). The second model additionally adjusted for ER+ status and HER2+ status. Odds ratios and 95% confidence intervals were reported. Further details on additional models fit are available in the supplementary materials and methods. All statistical analyses were performed in R (Version 3.2.2, Vienna, Austria) [26].
RESULTS
Patient population and demographic data
Cases of DCIS and documented follow-up in two cohorts were identified for genomic studies: 1) DCIS and no development of IBC (DCIS-only group) or 2) DCIS with concurrent/subsequent IBC events (DCIS+IBC group). A total of 125 DCIS cases were successfully analyzed, including 65 cases (52%) in the DCIS-only group and 60 cases (48%) in the DCIS+IBC group. In the DCIS+IBC group, 5 DCIS cases had IBC detected at a later time point. All patients were female, with median age at DCIS diagnosis of 51 years (range 29-89 years, Table 1). DCIS was characterized histologically and immunophenotypically; more than half (77 cases, 62%) demonstrated high-grade nuclei. The median tumor size was 2.4 cm (range 0.4-13.0 cm). The majority (72%) of cases had tumor margins of 0.2 cm and above. Sixty-one percent of surgeries were mastectomies. Eighty-one cases (65%) were positive for estrogen receptor (ER). Seventy-two cases (58%) were negative for HER2 (0, 1+, or 2+ by immunohistochemistry), and 27 cases (21.6%) were positive for HER2 (3+ by immunohistochemistry). The univariable analysis of these parameters for association with IBC was presented in supplementary Table S2, and the detailed clinicopathologic data was provided in supplementary Table S7.
Table 1.
Baseline features of the DCIS cases
| Characteristic | DCIS-only (n=65) | DCIS+IBC (n=60) | Total (n=125) |
|---|---|---|---|
| Age at diagnosis (years) | |||
| Less than 40 | 10 (15%) | 11 (18%) | 21 (17%) |
| 40-49 | 19 (29%) | 15 (25%) | 34 (27%) |
| 50-64 | 23 (35%) | 23 (38%) | 46 (37%) |
| 65 and older | 13 (20%) | 11 (18%) | 24 (19%) |
| Race / ethnicity | |||
| NH Asian/Pacific Islander | 11 (17%) | 10 (17%) | 21 (17%) |
| NH white | 47 (72%) | 43 (72%) | 90 (72%) |
| Other | 6 (9%) | 4 (7%) | 10 (8%) |
| Surgery type | |||
| Lumpectomy | 27 (42%) | 21 (35%) | 48 (38%) |
| Mastectomy | 37 (57%) | 39 (65%) | 76 (61%) |
| No surgery | 1 (2%) | 0 | 1 (1%) |
| ER status† | |||
| Positive | 46 (71%) | 35 (58%) | 81 (65%) |
| Negative | 10 (15%) | 19 (32%) | 29 (23%) |
| Missing | 9 (14%) | 6 (10%) | 15 (12%) |
| HER2 status† | |||
| Positive | 15 (23%) | 12 (20%) | 27 (22%) |
| Negative | 37 (57%) | 35 (58%) | 72 (58%) |
| Missing | 13 (20%) | 13 (22%) | 26 (21%) |
| Tumor size (cm) | |||
| Median (range) | 2.4 (0.4-10.9) | 2.4 (0.7-13.0) | 2.4 (0.4-13.0) |
| Tumor margins | |||
| < 0.2 cm | 19 (29%) | 14 (23%) | 33 (26%) |
| 0.2 cm + | 46 (71%) | 44 (73%) | 90 (72%) |
| DCIS nuclear grade | |||
| Low | 5 (8%) | 2 (3%) | 7 (6%) |
| Intermediate | 22 (34%) | 16 (27%) | 38 (30%) |
| High | 38 (58%) | 39 (65%) | 77 (62%) |
| Copy number variations | |||
| None | 20 (31%) | 5 (8%) | 25 (20%) |
| Gene 1q only | 10 (15%) | 13 (22%) | 23 (18%) |
| Gene 8q24 only | 1 (2%) | 6 (10%) | 7 (6%) |
| Gene 11q13 only | 2 (3%) | 1 (2%) | 3 (2%) |
| Two of three gains | 10 (15%) | 20 (33%) | 30 (24%) |
| All three gains | 10 (15%) | 12 (20%) | 22 (18%) |
Note: DCIS = ductal carcinoma in situ; IBC = invasive breast cancer; NH = non-Hispanic.
Equivocal is included in the negative category.
The genomic profile of DCIS is similar to IBC
Targeted sequencing for common mutations in IBC was performed on 125 cases of DCIS. The targeted regions include SNPs and coding exons of known breast-cancer/pan-cancer related genes, including APC, AR, ATM, BAP1, BRAF, CCND1, CHD1, CDKN1A, CTNNB1, DICER1, DNMT3A, EGFR, ERBB2, FOXA1, GATA3, IDH1/2, KRAS, MED12, MYB, NF1, NOTCH1, PIK3CA, PTEN, RB1, VHL, WT1 (full list in Supplementary material and methods). The average read count obtained per case was 5,604,491. Additional quality control data are presented in Supplementary Table S5 and Supplementary Figure 1. We excluded calls with coverage less than 30 reads. To focus on mutations having tumor suppressive or oncogenic effects, we limited the analysis to recurrent position mutations identified in the TCGA dataset that are frequently mutated in IBC. The most commonly mutated genes were PIK3CA (34.4%) and TP53 (18.4%) (Figure 1A). We observed no significant difference of mutational burdens in known breast cancer associated genes between the two groups of DCIS cases (p>0.05 for all the genes) (Figure 1B). We further analyzed variants based on their locations within the functional domains in each gene [27–29]. We identified “hotspots” of somatic mutations in the helical domain and kinase domain of PIK3CA for both DCIS-only and DCIS+IBC groups (Figure 2). There was a significant enrichment of PIK3CA kinase domain mutations (PIK3CA-KD mutations) in the DCIS-alone group (p=0.029). Analysis of other domains in PIK3CA genes found no statistically significant differences in recurrent mutations outside the kinase domain. Similar domain analysis was also performed for other frequently mutated genes, TP53 and GATA3, and no further predictive mutational profiles were identified.
Figure 1:

Genomic landscape of DCIS. (A) Distributions of known recurrent IBC-associated variants are (A) displayed in a bar graph with the number of cases denoted above the bars; (B) displayed in a heat map. In the group of DCIS+IBC, cases with subsequent IBC events are highlighted in the boxes.
Figure 2:
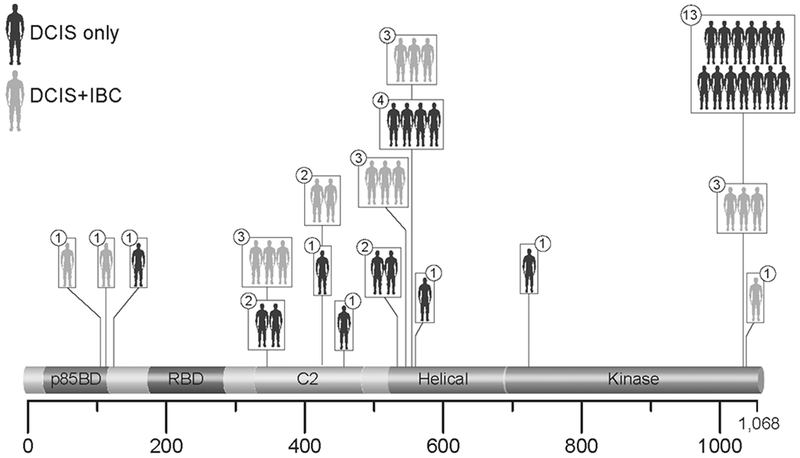
Distribution of PIK3CA mutations in two groups of DCIS cases. The number in the circles depicts the number of DCIS cases harboring the mutation in that particular position. To note, one of the DCIS-only cases exhibited two PIK3CA-KD variants.
In addition to SNV and small insertion-deletion mutations identified by targeted exon sequencing, we also interrogated larger-scale copy number variations in selected “hotspot” genomic areas in the two DCIS groups (Table 1 and Supplementary Table S1). Three chromosomal loci were measured by FISH, 1q32, 8q24, and 11q13, based on prior genomic data on invasive breast cancer [30] and DCIS [12]. The current study cohort consists of 73 cases (58.4%) from the previously published cohort [12], and 52 new cases that have not been analyzed before. Consistent with our prior data, frequency of 1q32 gain is the highest in the cohort (59.0%), followed by 8q24 (48.2%) and 11q13 (32.5%).
Multivariable analysis demonstrate strong correlation between PIK3CA-KD mutation and risk of progression
We performed multivariable analysis, examining the association of IBC dependent on variables including PIK3CA-KD mutational status, copy number gains, age, race, nuclear grade, tumor size, margins, and surgery type (Table 2). After removing cases with missing data, we had 97 complete DCIS cases. We identified a statistically significant association between lack of PIK3CA–KD mutation and increased risk of IBC (p<0.05). Patients without PIK3CA-KD mutations were 4.52 times (confidence interval: 1.05-25.27) as likely to have IBC compared to subjects with the mutation. This association was also statistically significant in the univariable analysis (Supplementary Table S2). When subdividing the DCIS+IBC group into the cases with synchronous or recurrent IBC (Supplementary Table S8), similar trend was noticed. The number of cases in the group of DCIS+ subsequent IBC is not powerful enough to draw a conclusion.
Table 2.
Multivariable associations with DCIS progression to IBC (n=97)
| Characteristic | Odds Ratio (95% CI) |
|---|---|
| Copy number variations | |
| None | reference |
| Gene 1q only | 5.44** (1.16, 30.46) |
| Gene 8q24 only | 21.92** (2.00, 603.68) |
| Gene 11q13 only | 1.71 (0.06, 29.98) |
| Two of three gains | 7.61*** (1.90, 36.86) |
| All three gains | 2.73 (0.58, 14.82) |
| Age at diagnosis | |
| Less than 40 | 0.45 (0.09, 2.17) |
| 40-49 | 0.86 (0.25, 2.88) |
| 50-64 | reference |
| 65 and older | 0.88 (0.21, 3.76) |
| Race / ethnicity | |
| NH Asian/Pacific Islander | 1.67 (0.19, 16.46) |
| NH white | 1.55 (0.26, 10.58) |
| Other | reference |
| DCIS nuclear grade | |
| Low | reference |
| Intermediate | 0.54 (0.05, 6.33) |
| High | 0.78 (0.07, 9.41) |
| Tumor size | 1.08 (0.85, 1.40) |
| Tumor margins < 0.2 cm | 1.61 (0.53, 5.10) |
| Lumpectomy | 0.77 (0.26, 2.21) |
| No PIK3CA-KD mutation | 4.52** (1.05, 25.27) |
Note:
p < 0.10
p < 0.05
p < 0.01
When additionally controlling for ER and HER2 status (complete case number=81, Table 3), the association between lack of PIK3CA-KD mutation and IBC risk remained statistically significant: DCIS patients without the PIK3CA-KD mutations were 10.22 times as likely to be progress to IBC as compared to DCIS with PIK3CA-KD mutations (p<0.05, Table 3). Similar results were observed when DCIS nuclear grades were grouped into a two-tiered system (low grade vs not low grade DCIS; high grade vs not high grade DCIS) (data not shown). In addition, the inverse association between any PIK3CA mutation and IBC risk became statistically significant (OR=4.66, p<0.05, data not shown).
Table 3.
Multivariable associations with DCIS progression to IBC include ER and HER2 status (n=81)
| Characteristic | Odds Ratio (95% CI) |
|---|---|
| Copy number variations | |
| None | reference |
| Gene 1q only | 9.31** (1.57, 76.66) |
| Gene 8q24 only | 45.96** (2.58, 1,917.18) |
| Gene 11q13 only | 2.89 (0.07, 83.36) |
| Two of three gains | 15.68*** (2.75, 129.49) |
| All three gains | 10.85** (1.36, 120.55) |
| Age at diagnosis | |
| Less than 40 | 0.32 (0.05, 2.06) |
| 40-49 | 0.78 (0.17, 3.47) |
| 50-64 | reference |
| 65 and older | 1.77 (0.32, 10.77) |
| Race / ethnicity | |
| NH Asian/Pacific Islander | 0.91 (0.07, 12.41) |
| NH white | 2.74 (0.34, 28.03) |
| Other | reference |
| DCIS nuclear grade | |
| Low | reference |
| Intermediate | 0.40 (0.02, 6.56) |
| High | 0.68 (0.03, 13.12) |
| No PIK3CA-KD mutation | 10.22** (1.61, 101.71) |
| ER positive | 0.57 (0.11, 2.75) |
| HER2 positive | 0.30 (0.07, 1.23) |
| Tumor size | 0.84 (0.59, 1.16) |
| Tumor margins < 0.2 cm | 2.04 (0.54, 8.93) |
| Lumpectomy | 0.83 (0.20, 3.24) |
Note:
p < 0.10
p < 0.05
p < 0.01
The presence of genomic copy number gain (1q32 only, 8q24 only, or two or three of three gains) was also associated with increased risk of progression to IBC (Table 3). In addition, there were a trend, but not statistically significant, that overexpression of HER2 is inversely associated with the risk of IBC (OR= 0.28, p<0.1). This trend became statistically significant (OR= 0.24, p<0.05) when modeling any PIK3CA mutations instead of PIK3CA-KD mutations in the multivariable analysis (data not shown).
ER status and DCIS nuclear grade are important pathological features that are routinely examined in the clinical setting. Therefore, we investigated the interaction between ER/nuclear grade and the PIK3CA mutation status. The association between lack of PIK3CA-KD mutations and progression to IBC was not modified by ER status (Supplementary Table S3). The association between lack of any PIK3CA mutations (including PIK3CA-KD mutations and other PIK3CA mutations) and progression to IBC was modified by ER status (p<0.05, Supplementary Table S4). There was a statistically significant interaction between DCIS nuclear grade and the presence of PIK3CA-KD mutation for the association with IBC (data not shown). The effect of PIK3CA-KD mutation appeared to be dependent on the nuclear grade: the association with no progression to IBC was stronger in the group of high-grade DCIS patients.
DISCUSSION
While the genomic landscape of IBC has been extensively studied [31–35], the molecular profiling of DCIS is still under investigation. Limited data have been published for DCIS genomic profiling, using either next-generation sequencing or array CGH [9,36–41]. To our knowledge, this is the largest cohort of genomic profiling with longitudinal clinical follow-up of DCIS cases that did not progress. PIK3CA, TP53, and GATA3 are among the most commonly mutated genes in DCIS, and chromosome 1q and 8q copy number gains are frequently identified in DCIS, as seen in prior studies [36–38,41]. Of note, in these previous reports, there was no recurrent mutation in a single gene that could stratify the risk of progression to IBC. However, further analysis based on protein domains was not performed in these previous publications.
We demonstrated a novel finding that the somatic mutations in the PIK3CA kinase domain provide predictive value of DCIS progression to IBC. The presence of mutations in this particular domain is associated with lower risk of concurrent or subsequent IBC. Previous studies have investigated the role of PIK3CA mutations in in situ breast cancers. In a small cohort of lobular carcinoma in situ (LCIS) without invasive lobular carcinoma (ILC) versus LCIS with associated ILC, the presence of PIK3CA mutations was not correlated with progression [42]. One of the studies showed that in ER-positive/HER2-negative DCIS, PIK3CA “hotspot mutations” were more prevalent in DCIS associated with IBC, compared with DCIS alone [43]. However, these “hotspot mutations” queried in this study includes mutations in C2, helical, and kinase domains. This difference could account for the different conclusion drawn in our study, as we demonstrated that specifically PIK3CA kinase domain mutations are associated with lack of progress.
The finding that activating mutations in PIK3CA and overexpression of HER2 oncogenes are correlated with a tendency not to progress to IBC is unexpected. Variants of these two oncogenes are quite prevalent in IBC. In cultures and animal model systems, they demonstrate biologic influences that promote neoplastic growth, invasion, or metastasis [44–46]. Given that conventional models of cancer associate progression with oncogenic mutations, it is surprising that two prominent oncogenes would be inversely correlated with the progression of DCIS to IBC.
One possible explanation for our findings is that alterations in specific pathways (such as HER2, PIK3CA) allow the cells to overcome immediate biological constraints in the process of tumorigenicity, rather than specifically promoting the DCIS to IBC transition. After the initial biologic challenges have been overcome, their influence on progression may be diminished. That they remain at high incidence in the invasive carcinoma may be in part due to the genomic difficulty or biological ambivalence in removing these somatic changes. In fact, PIK3CA mutations are extremely common in hyperplastic lesions of the breast[47]. HER2 is often amplified in DCIS and other non-invasive breast lesions, with a higher rate of amplifications in preinvasive lesions than in IBC [48–51]. A large cohort study with long-term follow-up data from Sweden showed that HER2-positive DCIS has lower risk of progression to IBC compared to HER2-negative DCIS [52]. Similar results have been reported independently [53]. For PIK3CA, previous researchers have found that in a subset of paired DCIS alone and DCIS with IBC samples, the PIK3CA mutations were present in DCIS alone but not DCIS with IBC, or with lower alternative allele frequency in the IBC component [39,43,54,55]. PIK3CA KD mutations (exon 20 mutations) have also been observed in pre-neoplastic lesions (usual ductal hyperplasia, columnar cell change, or atypical ductal hyperplasia) while paired IBC lesions lack such mutations [47]. These results and our current findings suggest that selection for HER2 amplification or PIK3CA mutations may address neoplastic challenges that occur well before the transition from in situ to invasive cancer.
Notably, the genomic features we found to correlate with risk of progression consisted of aneuploides or large amplicons. While there are a number of known and suspected oncogenes present in the chromosomal regions with recurrent copy number alterations, it is not clear whether a single driving event is responsible for these somatic changes. We speculate that higher-order function attributable to the gross change in genomic composition (e.g., copy number gains that likely have widespread effects on cellular function and genomic instability) may influence progression rather than a more precise influence on a specific gene function or related pathway. This observation has also been made by other studies in the literature [9,56]. In our prior whole genome sequencing study, clonally-related progression was marked by recurrent events of aneuploidy at the earliest stages and successive DNA copy number events throughout progression to invasion [7].
There are some limitations of our study. First, while some associations in the multivariable analyses were significant, those with large confidence intervals should be carefully interpreted. An independent cohort is required to validate these findings. Also, more than half (57%) of the DCIS-only patient received mastectomy specimen. The choice of mastectomy over lumpectomy potentially could drastically decrease the rate subsequent IBC event. In deed, in our cohort, only 5 patients subsequently developed invasive disease. In addition, without paired normal controls, we could not reliably distinguish between germline and somatic events. However, we focused on ‘hotspot’ mutations with known impacts on protein functions, such as PIK3CA kinase domain mutations, that are easily recognized. Moreover, there are several other genes that are mutated at frequencies that are likely to be significant for a clinical classifier, such as TP53, GATA3 and MAPK3. But beyond these, most genes are mutated at less than 1% and are thus unlikely to be useful as clinical biomarkers of progression.
The strength of our cohort is the long-term comprehensive follow-up data, to ensure DCIS-only cases were indeed without invasive or metastatic events. These cases are considered “low-risk” clinically. It is important to identify this type of DCIS patients, who carry low-risk of progression and could consider forgoing extensive treatment. In this study, we hypothesized that the molecular signatures of the “low-risk” DCIS are distinct from the “high-risk” DCIS. DCIS patients with either subsequent or synchronous IBC are considered “high-risk.” We acknowledge that the risk of progress in the “high-risk” DCIS group may be heterogeneous, that DCIS with synchronous IBC have even higher risk and may carry a different pathological mechanism and molecular profiles compared the DCIS with subsequent IBC. This is an interesting, important, yet separate hypothesis that is beyond the scope of this current study.
Future studies are necessary to provide deeper understanding of this novel finding of the PIK3CA-KD predictive value in DCIS progression. Previous literature has suggested that different PIK3CA variants could cause different downstream signaling pathway alteration and biological functions in in vitro systems [57,58]. One key goal is to map the occurrence and evolution of these genomic alterations in early neoplastic and precursor lesions, as well as in paired invasive and metastatic samples. This would help us to understand the roles of PIK3CA-KD mutations and copy number gains in breast cancer progression. It is also imperative to combine transcriptional and proteomics analyses, in order to interrogate the downstream effects related to the PIK3CA-KD mutations.
Our novel findings that PIK3CA-KD mutations are associated with relative lack of DCIS progression to IBC, coupled with other traditional and novel risk factors, contributes to knowledge of the sequence and mechanisms of breast cancer progression. These data also begin to demonstrate the possibility of an integrated clinico-pathologic-molecular risk classifier of DCIS. For example, women with low nuclear grade, ER+, PIK3CA-KD mutant DCIS may have particularly low risk of progression. Larger studies with more complete histologic, immunohistochemical, proteomic, molecular, treatment and long term follow-up data are necessary to build precision risk assessment models to counsel patients, tailor therapy, and reduce overtreatment of the more indolent forms of DCIS.
Supplementary Material
Supplmentary Figure 1. Coverage of sequencing data
Table S1. Performance of cytogenetic combinations as predictors of invasive breast cancer
Table S2. Univariable associations with DCIS progression to IBC (n=104)
Table S3. Multivariable associations with DCIS progression to IBC using PIK3CA-KD mutations vs no PIK3CA-KD mutations as one of the variables (n=86)
Table S4. Multivariable analysis of associations with DCIS progression to IBC using PIK3CA mutations vs no PIK3CA mutations as one of the variables (n=86)
Table S5. QC data
Table S6. Nonsynonemous variants with annotation
Table S7. Clinicopathogic data
Table S8. Multivariable associations with DCIS progression to IBC (n=97), comparing between DCIS alone vs IBC recurrence as well as DCIS alone vs synchronous. Data comparing DCIS alone with DCIS+IBC (both recurrence and synchronous) from Table 2 are also included for comparison.
ACKNOWLEDGEMENT
Short read sequencing assays were performed by the OHSU Massively Parallel Sequencing Shared Resource. We thank Norman Cyr for artistic work on Figure 2.
Funding
This work is supported by NIH R01CA193694, the Susan and Richard Levy Gift Fund; the Suzanne Pride Bryan Fund for Breast Cancer Research; the Breast Cancer Research Foundation; the Jan Weimer Junior Faculty Chair in Breast Oncology; the BRCA Foundation and the National Cancer Institute’s Surveillance, Epidemiology and End Results Program under contract HHSN261201000140C awarded to the Cancer Prevention Institute of California. The project was supported by an NIH CTSA award number UL1 RR025744. The collection of cancer incidence data used in this study was supported by the California Department of Health Services as part of the statewide cancer reporting program mandated by California Health and Safety Code Section 103885; the National Cancer Institute’s Surveillance, Epidemiology, and End Results Program under contract HHSN261201000140C awarded to the Cancer Prevention Institute of California, contract HHSN261201000035C awarded to the University of Southern California, and contract HHSN261201000034C awarded to the Public Health Institute; and the Centers for Disease Control and Prevention’s National Program of Cancer Registries, under agreement #1U58 DP000807-01 awarded to the Public Health Institute. The ideas and opinions expressed herein are those of the authors, and endorsement by the University or State of California, the California Department of Health Services, the National Cancer Institute, or the Centers for Disease Control and Prevention or their contractors and subcontractors is not intended nor should be inferred.
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
SUPPLIMENTARY MATERIALS
Supplimentary materials and methods, supplementary Table S1–S8, and supplementary Figure 1 are available online.
Additional information is avaialbe online:
Conflict of Interest
The authors declare that they have no competing interests.
COMPLIANCE WITH ETHICAL STANDARDS
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. As stated in the materials and methods, the study was performed with Health Insurance Portability and Accountability Act (HIPAA)-compliant Stanford University Institutional Review Board (IRB) approval (Protocol number 32496). Because archival tissue was used, a waiver of consent was obtained.
REFERENCES
- 1.Meijnen P, Oldenburg HS, Peterse JL, et al. Clinical outcome after selective treatment of patients diagnosed with ductal carcinoma in situ of the breast. Ann Surg Oncol 2008; 15: 235–243. [DOI] [PubMed] [Google Scholar]
- 2.Cutuli B, Cohen-Solal-le Nir C, de Lafontan B, et al. Breast-conserving therapy for ductal carcinoma in situ of the breast: the French Cancer Centers’ experience. Int J Radiat Oncol Biol Phys 2002; 53: 868–879. [DOI] [PubMed] [Google Scholar]
- 3.Wapnir IL, Dignam JJ, Fisher B, et al. Long-term outcomes of invasive ipsilateral breast tumor recurrences after lumpectomy in NSABP B-17 and B-24 randomized clinical trials for DCIS. J Natl Cancer Inst 2011; 103: 478–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cuzick J, Sestak I, Pinder SE, et al. Effect of tamoxifen and radiotherapy in women with locally excised ductal carcinoma in situ: long-term results from the UK/ANZ DCIS trial. Lancet Oncol 2011; 12: 21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ernster VL, Barclay J, Kerlikowske K, et al. Incidence of and treatment for ductal carcinoma in situ of the breast. JAMA 1996; 275: 913–918. [PubMed] [Google Scholar]
- 6.Fisher ER, Dignam J, Tan-Chiu E , et al. Pathologic findings from the National Surgical Adjuvant Breast Project (NSABP) eight-year update of Protocol B-17: intraductal carcinoma. Cancer 1999; 86: 429–438. [DOI] [PubMed] [Google Scholar]
- 7.Newburger DE, Kashef-Haghighi D, Weng Z, et al. Genome evolution during progression to breast cancer. Genome Res 2013; 23: 1097–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weng Z, Spies N, Zhu SX, et al. Cell-lineage heterogeneity and driver mutation recurrence in pre-invasive breast neoplasia. Genome Med 2015; 7: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson CE, Gorringe KL, Thompson ER, et al. Identification of copy number alterations associated with the progression of DCIS to invasive ductal carcinoma. Breast Cancer Res Treat 2012; 133: 889–898. [DOI] [PubMed] [Google Scholar]
- 10.Rane SU, Mirza H, Grigoriadis A, et al. Selection and evolution in the genomic landscape of copy number alterations in ductal carcinoma in situ (DCIS) and its progression to invasive carcinoma of ductal/no special type: a meta-analysis. Breast Cancer Res Treat 2015; 153: 101–121. [DOI] [PubMed] [Google Scholar]
- 11.Kroigard AB, Larsen MJ, Laenkholm AV, et al. Clonal expansion and linear genome evolution through breast cancer progression from pre-invasive stages to asynchronous metastasis. Oncotarget 2015; 6: 5634–5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Afghahi A, Forgo E, Mitani AA, et al. Chromosomal copy number alterations for associations of ductal carcinoma in situ with invasive breast cancer. Breast Cancer Res 2015; 17: 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Troxell ML, Brunner AL, Neff T, et al. Phosphatidylinositol-3-kinase pathway mutations are common in breast columnar cell lesions. Mod Pathol 2012; 25: 930–937. [DOI] [PubMed] [Google Scholar]
- 14.Xu R, Perle MA, Inghirami G, et al. Amplification of Her-2/neu gene in Her-2/neu-overexpressing and -nonexpressing breast carcinomas and their synchronous benign, premalignant, and metastatic lesions detected by FISH in archival material. Mod Pathol 2002; 15: 116–124. [DOI] [PubMed] [Google Scholar]
- 15.Weber SC, Seto T, Olson C, et al. Oncoshare: lessons learned from building an integrated multi-institutional database for comparative effectiveness research. AMIA Annu Symp Proc 2012; 2012: 970–978. [PMC free article] [PubMed] [Google Scholar]
- 16.Kurian AW, Mitani A, Desai M, et al. Breast cancer treatment across health care systems: linking electronic medical records and state registry data to enable outcomes research. Cancer 2014; 120: 103–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009; 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010; 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 2013; 31: 213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lai Z, Markovets A, Ahdesmaki M, et al. VarDict: a novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic Acids Res 2016; 44: e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. arXiv:12073907 2012. [Google Scholar]
- 22.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010; 38: e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chabon JJ, Simmons AD, Lovejoy AF, et al. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat Commun 2016; 7: 11815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao Y, Niu Y, Wang X, et al. Genetic changes at specific stages of breast cancer progression detected by comparative genomic hybridization. J Mol Med (Berl) 2009; 87: 145–152. [DOI] [PubMed] [Google Scholar]
- 25.Wolff AC, Hammond ME, Hicks DG, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol 2013; 31: 3997–4013. [DOI] [PubMed] [Google Scholar]
- 26.Team RDC. R: A Language and Environment for Statistical Computing. Vienna, Austria: : the R Foundation for Statistical Computing; In., 2015. [Google Scholar]
- 27.Porta-Pardo E, Godzik A. e-Driver: a novel method to identify protein regions driving cancer. Bioinformatics 2014; 30: 3109–3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Porta-Pardo E, Garcia-Alonso L, Hrabe T, et al. A Pan-Cancer Catalogue of Cancer Driver Protein Interaction Interfaces. PLoS Comput Biol 2015; 11: e1004518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang F, Petsalaki E, Rolland T, et al. Protein domain-level landscape of cancer-type-specific somatic mutations. PLoS Comput Biol 2015; 11: e1004147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Curtis C, Shah SP, Chin SF, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012; 486: 346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cancer Genome Atlas N Comprehensive molecular portraits of human breast tumours. Nature 2012; 490: 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horlings HM, Weigelt B, Anderson EM, et al. Genomic profiling of histological special types of breast cancer. Breast Cancer Res Treat 2013; 142: 257–269. [DOI] [PubMed] [Google Scholar]
- 33.Ciriello G, Gatza ML, Beck AH, et al. Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer. Cell 2015; 163: 506–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Desmedt C, Zoppoli G, Gundem G, et al. Genomic Characterization of Primary Invasive Lobular Breast Cancer. J Clin Oncol 2016; 34: 1872–1881. [DOI] [PubMed] [Google Scholar]
- 35.Yates LR, Gerstung M, Knappskog S, et al. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat Med 2015; 21: 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abba MC, Gong T, Lu Y, et al. A Molecular Portrait of High-Grade Ductal Carcinoma In Situ. Cancer Res 2015; 75: 3980–3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim SY, Jung SH, Kim MS, et al. Genomic differences between pure ductal carcinoma in situ and synchronous ductal carcinoma in situ with invasive breast cancer. Oncotarget 2015; 6: 7597–7607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pang JB, Savas P, Fellowes AP, et al. Breast ductal carcinoma in situ carry mutational driver events representative of invasive breast cancer. Mod Pathol 2017; 30: 952–963. [DOI] [PubMed] [Google Scholar]
- 39.Hernandez L, Wilkerson PM, Lambros MB, et al. Genomic and mutational profiling of ductal carcinomas in situ and matched adjacent invasive breast cancers reveals intra-tumour genetic heterogeneity and clonal selection. J Pathol 2012; 227: 42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liao S, Desouki MM, Gaile DP, et al. Differential copy number aberrations in novel candidate genes associated with progression from in situ to invasive ductal carcinoma of the breast. Genes Chromosomes Cancer 2012; 51: 1067–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hwang ES, Lal A, Chen YY, et al. Genomic alterations and phenotype of large compared to small high-grade ductal carcinoma in situ. Hum Pathol 2011; 42: 1467–1475. [DOI] [PubMed] [Google Scholar]
- 42.Shah V, Nowinski S, Levi D, et al. PIK3CA mutations are common in lobular carcinoma in situ, but are not a biomarker of progression. Breast Cancer Res 2017; 19: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sakr RA, Weigelt B, Chandarlapaty S, et al. PI3K pathway activation in high-grade ductal carcinoma in situ--implications for progression to invasive breast carcinoma. Clin Cancer Res 2014; 20: 2326–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao JJ, Liu Z, Wang L, et al. The oncogenic properties of mutant p110alpha and p110beta phosphatidylinositol 3-kinases in human mammary epithelial cells. Proc Natl Acad Sci U S A 2005; 102: 18443–18448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zardavas D, Phillips WA, Loi S. PIK3CA mutations in breast cancer: reconciling findings from preclinical and clinical data. Breast Cancer Res 2014; 16: 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fry EA, Taneja P, Inoue K. Oncogenic and tumor-suppressive mouse models for breast cancer engaging HER2/neu. Int J Cancer 2017; 140: 495–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ang DC, Warrick AL, Shilling A , et al. Frequent phosphatidylinositol-3-kinase mutations in proliferative breast lesions. Mod Pathol 2014; 27: 740–750. [DOI] [PubMed] [Google Scholar]
- 48.Allred DC, Clark GM, Molina R, et al. Overexpression of HER-2/neu and its relationship with other prognostic factors change during the progression of in situ to invasive breast cancer. Hum Pathol 1992; 23: 974–979. [DOI] [PubMed] [Google Scholar]
- 49.Latta EK, Tjan S, Parkes RK, et al. The role of HER2/neu overexpression/amplification in the progression of ductal carcinoma in situ to invasive carcinoma of the breast. Mod Pathol 2002; 15: 1318–1325. [DOI] [PubMed] [Google Scholar]
- 50.Jang M, Kim E, Choi Y, et al. FGFR1 is amplified during the progression of in situ to invasive breast carcinoma. Breast Cancer Res 2012; 14: R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park K, Han S, Kim HJ, et al. HER2 status in pure ductal carcinoma in situ and in the intraductal and invasive components of invasive ductal carcinoma determined by fluorescence in situ hybridization and immunohistochemistry. Histopathology 2006; 48: 702–707. [DOI] [PubMed] [Google Scholar]
- 52.Borgquist S, Zhou W, Jirstrom K, et al. The prognostic role of HER2 expression in ductal breast carcinoma in situ (DCIS); a population-based cohort study. BMC Cancer 2015; 15: 468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hoque A, Sneige N, Sahin AA, et al. Her-2/neu gene amplification in ductal carcinoma in situ of the breast. Cancer Epidemiol Biomarkers Prev 2002; 11: 587–590. [PubMed] [Google Scholar]
- 54.Miron A, Varadi M, Carrasco D, et al. PIK3CA mutations in in situ and invasive breast carcinomas. Cancer Res 2010; 70: 5674–5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dunlap J, Le C, Shukla A, et al. Phosphatidylinositol-3-kinase and AKT1 mutations occur early in breast carcinoma. Breast Cancer Res Treat 2010; 120: 409–418. [DOI] [PubMed] [Google Scholar]
- 56.Gorringe KL, Hunter SM, Pang JM, et al. Copy number analysis of ductal carcinoma in situ with and without recurrence. Mod Pathol 2015; 28: 1174–1184. [DOI] [PubMed] [Google Scholar]
- 57.Bhat-Nakshatri P, Goswami CP, Badve S, et al. Molecular Insights of Pathways Resulting from Two Common PIK3CA Mutations in Breast Cancer. Cancer Res 2016; 76: 3989–4001. [DOI] [PubMed] [Google Scholar]
- 58.Dogruluk T, Tsang YH, Espitia M, et al. Identification of Variant-Specific Functions of PIK3CA by Rapid Phenotyping of Rare Mutations. Cancer Res 2015; 75: 5341–5354. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplmentary Figure 1. Coverage of sequencing data
Table S1. Performance of cytogenetic combinations as predictors of invasive breast cancer
Table S2. Univariable associations with DCIS progression to IBC (n=104)
Table S3. Multivariable associations with DCIS progression to IBC using PIK3CA-KD mutations vs no PIK3CA-KD mutations as one of the variables (n=86)
Table S4. Multivariable analysis of associations with DCIS progression to IBC using PIK3CA mutations vs no PIK3CA mutations as one of the variables (n=86)
Table S5. QC data
Table S6. Nonsynonemous variants with annotation
Table S7. Clinicopathogic data
Table S8. Multivariable associations with DCIS progression to IBC (n=97), comparing between DCIS alone vs IBC recurrence as well as DCIS alone vs synchronous. Data comparing DCIS alone with DCIS+IBC (both recurrence and synchronous) from Table 2 are also included for comparison.


