Summary
Aminoglycoside antibiotics require proton motive force (PMF) for bacterial internalization. In non-respiring populations, PMF drops below the level required for drug influx, limiting the utility of aminoglycosides against strict and facultative anaerobes. We recently demonstrated that rhamnolipids (RLs), biosurfactant molecules produced by Pseudomonas aeruginosa, potentiate aminoglycoside activity against Staphylococcus aureus. Here, we demonstrate that RLs induce PMF-independent aminoglycoside uptake to restore sensitivity to otherwise tolerant persister, biofilm, small colony variant, and anaerobic populations of S. aureus. Furthermore, we show that this approach prevents the rise of resistance, restores sensitivity to highly resistant clinical isolates, and is effective against other gram-positive pathogens. Finally, while other membrane-acting agents can synergize with aminoglycosides, induction of PMF-independent uptake is uncommon, and distinct to RLs among several compounds tested. In all, small molecule induction of PMF-independent aminoglycoside uptake circumvents phenotypic tolerance, overcomes genotypic resistance, and expands the utility of aminoglycosides against intrinsically recalcitrant bacterial populations.
Graphical Absatract

eTOC Blurb
Aminoglycoside efficacy is limited against non-respiring bacterial populations due to deficiencies in uptake. Here we show that chemical induction of PMF-independent aminoglycoside uptake with rhamnolipids sensitizes non-respiring populations of S. aureus to aminoglycoside killing, expanding the utility of this important class of antibiotics.
Introduction
Environmental heterogeneity within a host elicits a spectrum of activities and metabolic states that render a pathogen recalcitrant to antibiotic killing (Radlinski and Conlon, 2018). For the facultative anaerobe, Staphylococcus aureus, antibiotic treatment failure occurs in approximately 1 in 5 patients, leading to an estimated 20,000 deaths annually in the United States alone (Kourtis et al., 2019). Paradoxically, clinical isolates from these infections often exhibit sensitivity to administered antibiotics, as measured in vitro using minimum inhibitory concentration (MIC) assays (Sakoulas et al., 2004; Walraven et al., 2011). This suggests that resistance alone cannot fully account for treatment failure, but rather that environmental factors present in vivo may influence the pathogen’s susceptibility to antibiotic killing. S. aureus frequently colonizes microaerophillic or anaerobic niches during infection of the bone, within an abscess, or in the late-stage cystic fibrosis (CF) lung (Tong et al., 2015). The absence of a terminal electron acceptor, such as oxygen or nitrate, pushes S. aureus into a fermentative lifestyle associated with diminished PMF and intracellular ATP (Balasubramanian et al., 2017).
Aminoglycosides are broad-spectrum antibiotics that exemplify the disconnect between in vitro and in vivo antibiotic efficacy. Aminoglycoside internalization is a multi-step process that begins with ionic interaction with the plasma membrane. This association displaces magnesium cations that stabilize the phospholipid bilayer, leading to increased permeability to ionic molecules (Hancock et al., 1981). This promotes proton motive force (PMF)-mediated aminoglycoside diffusion into the cell, followed by an exponential increase of drug influx as aminoglycosides disrupt translation, and misfolded proteins insert into and further destabilize membrane integrity (Andry 1974; Davis 1987; Taber et al. 1987a). Thus, slow growing or non-respiring bacterial populations can withstand high levels of aminoglycosides as the initial stage of drug uptake is hindered by diminished PMF (Bryan et al., 1979). In vitro, aminoglycosides are potent killers of growing, respiring bacteria, yet these antibiotics are often dismissed as viable therapeutics for chronic infections because efficacy is severely limited against anaerobic, small colony variant (SCV), biofilm-associated, and persister populations (Garcia et al., 2013; Lechner et al., 2012; Waters et al., 2016).
The observation that aminoglycoside efficacy is impeded by defects in drug uptake has prompted several groups to pursue novel means for overcoming this barrier. Clinically, aminoglycosides are often prescribed in combination with cell wall-acting antibiotics such as β-lactams (Eliopoulos, 1989; Rhodes et al., 2017; Scaglione et al., 1995). β-lactam treatment interferes with peptidoglycan crosslinking at the gram-positive cell surface(Wise et al., 1965). This action has been demonstrated to reduce S. aureus cell surface charge, and may promote interaction with positively charged aminoglycosides (Mehta et al., 2012; Taber et al., 1987). Though this synergy has been reported in vitro, clinical reports documenting this combinational therapy are conflicting (Tamma et al., 2012). Similarly, groups have investigated other means of stimulating aminoglycoside uptake, including pH-mediated stimulation of ∆ψ, metabolite-mediated stimulation of PMF in persisters, or the use of other antimicrobial or anti-biofilm peptides and compounds (Allison et al., 2011; Farha et al., 2013; Lebeaux et al., 2014; Pletzer et al., 2018). Apparent synergy has also been reported between aminoglycosides and compounds that destabilize the bacterial membrane, such as antimicrobial lipids and retinoids, however the mechanisms of potentiation are unclear (Hess et al., 2014; Kim et al., 2018).
We recently demonstrated that P. aeruginosa-produced rhamnolipids (RLs) synergize with tobramycin to improve efficacy against S. aureus (Radlinski et al., 2017). RLs are a class of amphiphilic biosurfactants composed of two β-hydroxy fatty acid tails of varying lengths connected to one or two rhamnose sugar molecules (Figure 1A)(Haba et al., 2014, 2003). Previous studies have shown that at high concentrations (>300µg/mL)(Aleksic et al., 2017), RLs exert bactericidal activity against certain gram-positive species by intercalating into the cytoplasmic membrane and forming pores (Sotirova et al., 2008). Despite demonstrating broad-spectrum antimicrobial activity against gram-positive pathogens, RLs are often dismissed as a potential therapeutic because the concentrations necessary to exert bactericidal activity are cytotoxic to host cells. However, here we show that sub-cytotoxic concentrations of RLs significantly stimulate tobramycin uptake in S. aureus, and precipitate rapid and complete sterilization of S. aureus cultures. We further demonstrate that small molecule targeting of membrane permeability represents a novel and viable strategy for overcoming aminoglycoside tolerance and resistance in S. aureus and other clinically relevant pathogens.
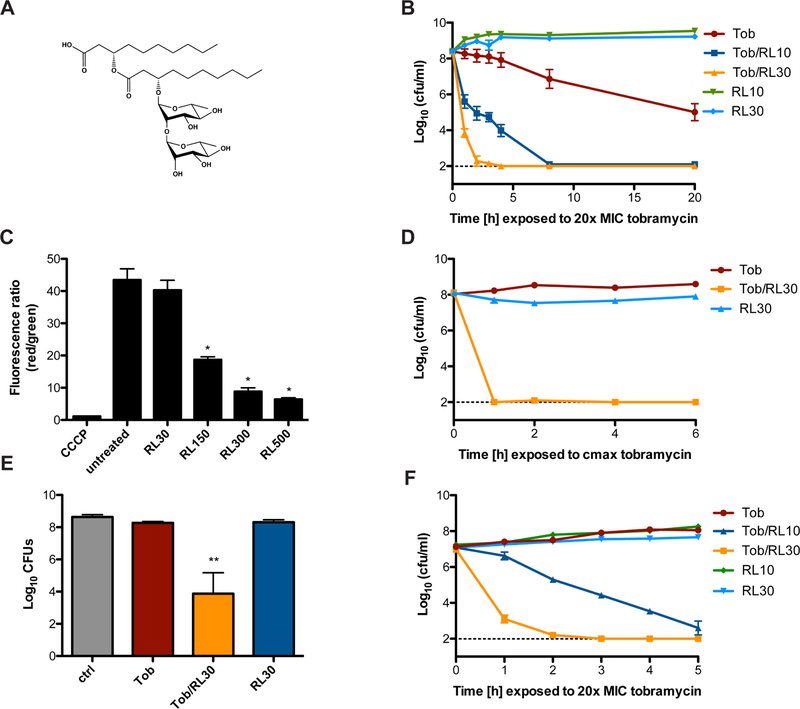
Figure 1. Rhamnolipids synergize with aminoglycosides against tolerant S. aureus populations.
A. Chemical structure of a representative di-rhamnolipid congener, di-rhamnolipid C10C10. B. S. aureus strain HG003 was grown to mid-exponential phase and challenged with 20x the MIC (15.6µg/mL) of tobramycin +/− 10 or 30µg/ml RLs. Survivors were enumerated at the indicated time points. C. S. aureus HG003 membrane potential was measured by calculating the red/green ratio using the population mean fluorescence intensities (MFI) for HG003 incubated with 30µM DiOC2(3) for 30 minutes. Prior to staining, cells were grown to mid-exponential phase, then treated for 30 minutes with the indicated concentration of RLs or for 5 minutes with 5µM CCCP as a control. *p<0.05 (One-way ANOVA with Tukey’s multiple comparison post test). D. Mid-exponential phase HG003 was challenged with the Cmax concentration of tobramycin (58µg/mL) +/− 30µg/ml RLs under anaerobic conditions. Survivors were enumerated at the indicated time points. E. Biofilm-associated S. aureus HG003 was challenged with 58µg/mL tobramycin +/− 30µg/ml RLs for 24 hours prior to CFU enumeration. F. S. aureus SCV strain HG003 menD::tn was grown to mid-exponential phase and challenged with 15.6µg/mL tobramycin +/− 10 or 30µg/ml RLs. CFU enumeration occurred at the indicated time points. All experiments were performed in biological triplicate. **p<0.005 (Student’s t-test). Error bars represent mean +/− SD. Limit of detection is indicated by the horizontal dashed line. See also Figure S1 and Figure S2.
Results
P. aeruginosa rhamnolipids potentiate aminoglycoside killing of S. aureus.
Using a mix of mono- and di-rhamnolipid congeners (RL90; AGAE), we first identified a RL concentration that potentiated tobramycin killing without itself exerting bactericidal activity against S. aureus (Figure S1A), and demonstrated that RL/tobramycin combinational therapy mediates the rapid and total sterilization of S. aureus HG003 cultures in a dose-dependent manner (Figure 1B). Interestingly, RL treatment did not lead to substantial dissipation of cellular PMF until the applied concentration approached concentrations necessary to inhibit S. aureus growth (Figure 1C, Figure S1A). Other commercially available RL molecules including two purified monorhamnose RL molecules, Rha-C10C10 and Rha-C12C12 (Glycosurf), as well as a purified mix of 90% di-rhamnose RL molecules of various carbon tail lengths (RL95D90; AGAE) similarly potentiated at least a 4-fold increase in tobramycin killing of S. aureus (Figure S1E). RL concentrations that potentiated aminoglycosides fell well below what were cytotoxic to host cells (Figure S2A). Similar potentiation was observed against the MRSA strain JE2 (Figure S2B), as well as with the other aminoglycosides including gentamicin, amikacin, neomycin, and kanamycin, demonstrating that RL/aminoglycoside synergy is not strain-dependent, or restricted to tobramycin (Figure S2C–F). RLs did not potentiate killing by ciprofloxacin, rifampicin or oxacillin, and 30µg/mL RLs did not change the MIC of ciprofloxacin (0.3125µg/mL), rifampicin (0.008µg/mL), oxacillin (0.5µg/mL), or the bacteriostatic antibiotic tetracycline (0.25µg/mL), suggesting that the synergy observed is specific to aminoglycosides. (Figure S2G–I).
Aminoglycosides lack activity against non-respiring bacterial populations, as these cells maintain a low PMF (Bryan et al., 1979). S. aureus frequently experiences oxygen limitation during infection, particularly in biofilms, and biofilm associated S. aureus is highly recalcitrant to antibiotics (Waters et al., 2016). We hypothesized that RLs could restore aminoglycoside sensitivity to non-respiring populations of S. aureus by facilitating PMF-independent, passive transport. S. aureus was grown to mid-exponential phase under anaerobiosis or overnight in conditions that promote biofilm formation, then challenged with the maximum attainable serum concentration (Cmax, 58µg/mL) of tobramycin (Burkhardt, 2006). As expected, tobramycin alone was ineffective against anaerobic or biofilm-associated S. aureus (Figure 1D, E). However, RL/tobramycin combinational therapy led to the complete sterilization of S. aureus HG003 anaerobic cultures within one hour (Figure 1D). Similarly, combinational therapy led to approximately a 4-log reduction in biofilm-associated CFU (Figure 1E).
S. aureus SCVs are notoriously difficult to treat and are isolated from at least 25% of patients with CF (Proctor et al., 1998). SCVs are often auxotrophic for the biosynthesis of thiamine, menadione or hemin, and consequently are defective in respiration and resistant to aminoglycosides (Lannergard et al., 2008; Proctor et al., 2006; Schaaff et al., 2003). To test whether RLs could be used to re-sensitize S. aureus SCVs to tobramycin killing, we generated a menadione (menD) auxotroph in S. aureus strain HG003. The menD mutant demonstrated high-level resistance (MIC = 50µg/ml) to tobramycin, however RLs reduced this MIC to 0.0975µg/ml. Similarly, though HG003 menD grew unabated in the presence of tobramycin (Figure 1F), RL treatment facilitated tobramycin-mediated sterilization of the cultures in a dose-dependent manner (Figure 1F). Taken together, these data demonstrate that by bypassing the requirement of PMF for aminoglycoside uptake, RLs re-sensitize S. aureus anaerobic, biofilm, and SCV populations to aminoglycoside killing.
Rhamnolipid/tobramycin combinational therapy eradicates S. aureus persisters
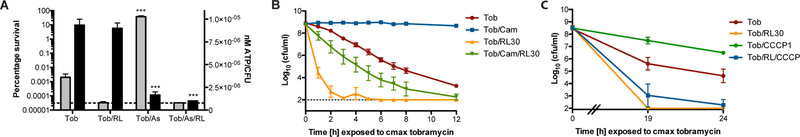
Bactericidal antibiotics corrupt active cellular processes such as cell wall or protein synthesis to facilitate killing (Kohanski et al., 2010). Persisters are subpopulations of bacteria that stochastically enter a phenotypically dormant state and can thus tolerate bactericidal concentrations of antibiotics (Lewis, 2010). S. aureus persister cell formation is associated with a decrease in intracellular ATP levels (Conlon et al., 2016). We previously showed that a population-wide state of antibiotic tolerance can be induced in S. aureus by treating cells with potassium arsenate (AsKO2) to reduce ATP levels (Conlon et al., 2016). As expected, exposing a susceptible population of S. aureus to AsKO2 reduced the intracellular ATP concentration of the population, and induced tobramycin tolerance (Figure 2A). Conversely, this protective effect was abrogated in the presence of RLs, suggesting that RLs allow tobramycin to overcome the level of protection conferred by ATP depletion, and strengthened our conclusion that increasing tobramycin uptake allows us to target low-energy S. aureus persister populations.
Figure 2. Rhamnolipid/aminoglycoside combinational therapy targets S. aureus persisters.
A. Mid-exponential phase S. aureus strain HG003 was treated with 5mM AsKO2 for 30 min prior to addition of 58µg/mL tobramycin. Intracellular ATP was measured using a BacTiter-Glo cell viability assay immediately prior to antibiotic challenge (black bars). An aliquot of each culture was removed after 24hr, washed, and plated to enumerate survivors (grey bars). ***p<0.005 (Student’s t-test, calculated relative to cultures treated with tobramycin alone). B, C. Exponential phase populations of HG003 were exposed to B. 30µg/mL chloramphenicol or C. 1µM CCCP for 30 min prior to challenge with 58µg/mL tobramycin +/− 30µg/ml RLs. At the indicated time points an aliquot was removed, washed and plated to enumerate survivors. All experiments were performed in biological triplicate. Error bars represent mean +/− SD. Limit of detection is indicated by the horizontal dashed line. See also Figure S3.
Aminoglycoside bactericidal activity requires protein synthesis. Consequently, populations demonstrating a decreased rate of translation should be slower to succumb to killing. Chloramphenicol is bacteriostatic antibiotic that inhibits, rather than corrupts, protein synthesis and thus at certain concentrations will cause cessation of growth (Kohanski et al., 2010). We aimed to slow the rate of translation by exposing cells to chloramphenicol and assess the impact on tobramycin/RL killing. We measured translation using a xylose inducible GFP reporter plasmid, pCLON46, to confirm that addition of chloramphenicol to growing S. aureus cultures slowed the rate of GFP synthesis (Figure S3A). Remarkably, while chloramphenicol treatment protected S. aureus from tobramycin killing, tobramycin/RL combinational therapy facilitated eradication of chloramphenicol-treated S. aureus cultures, albeit at a diminished rate (Figure 2B). Similar results were observed following linezolid pre-treatment (Figure S3B). RLs also potentiated tobramycin killing in the presence of the protonophore and uncoupling agent, carbonyl cyanide 3-chlorophenylhydrazone (CCCP), which causes collapse of PMF and prevents tobramycin uptake. This confirmed that RLs facilitate PMF-independent tobramycin uptake and resensitize PMF-depleted populations to tobramycin killing (Figure 2C). Together, these results demonstrate that RL/tobramycin combinational therapy targets persisters by overcoming the requirement of PMF for drug influx and lowering the threshold energy and translation levels required for killing. As low PMF, ATP, and antibiotic target activity represent the primary barriers proposed to reduce to antibiotic efficacy in persisters, RL induction of aminoglycoside uptake may significantly improve treatment of otherwise tolerant persister cell populations.
Rhamnolipids repress the rise of tobramycin resistance, and re-sensitize resistant isolates to killing
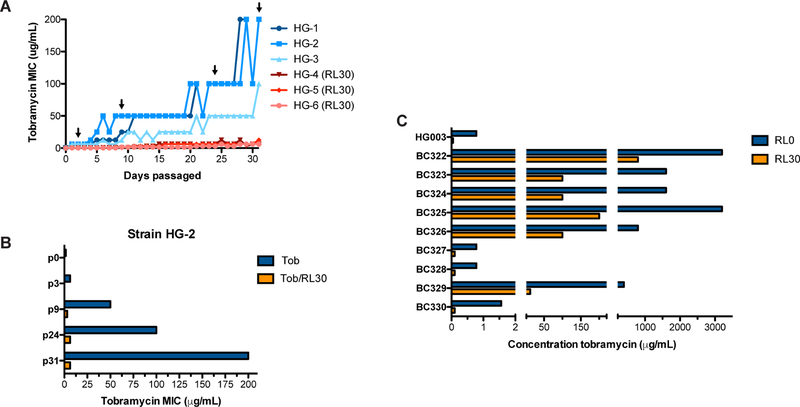
Subinhibitory RL concentrations significantly decreased the MIC of tobramycin for S. aureus HG003 WT from 0.78 to 0.0975µg/ml. For S. aureus, the predominant mechanisms of aminoglycoside resistance include decreased uptake through the adoption of a non-respiring SCV phenotype, or horizontal transfer of an aminoglycoside-modifying enzyme (Vakulenko and Mobashery, 2003). Because RLs allow PMF-independent drug uptake, we hypothesized that tobramycin/RL combinational therapy could prevent the rise of tobramycin resistance due to poor drug penetration during long-term tobramycin exposure. To test this, 6 independent S. aureus HG003 strains were passaged daily in sub-MIC concentrations of tobramycin with or without 30µg/mL of RL. Serial passage with tobramycin alone led to a 256–500-fold increase in tobramycin MIC, yielding a final MIC of 200µg/ml for HG-1 and HG-2, and 100µg/ml for HG-3 (Figure 3A). Conversely, serial passage in tobramycin + RLs generated a maximum MIC increase to 6.25 or 12.5µg/ml (Figure 3A). Two of these strains (HG-4, HG-6) remained below the clinical breakpoint for intravenous tobramycin (8µg/ml). Furthermore, subsequent treatment with RLs restored tobramycin susceptibility to resistant mutant strains that arose during passage in tobramycin alone (Figure 3B). Together, these findings indicate that when used in combination with aminoglycosides, RLs slow the rise of aminoglycoside resistance and re-sensitize aminoglycoside resistant isolates to killing.
Figure 3. Rhamnolipids repress the rise of tobramycin resistance and restore sensitivity to resistant isolates.
A. Six independent lineages of HG003 were passaged daily in subinhibitory concentrations of tobramycin +/− 30µg/mL RLs and monitored for the spontaneous occurrence of tobramycin resistant mutants through changes in MIC. B, C. Minimum tobramycin concentration necessary to inhibit the growth of B. resistant isolates from passaged strain HG-2 (Figure 3A, black arrows) or C. CF clinical isolates, +/− 30µg/mL RLs.
Patients with CF are routinely administered high doses of inhaled tobramycin therapy (300mg of aerosolized tobramycin twice daily for 28 days, reaching sputum concentrations of 737µg/ml) to reduce P. aeruginosa burden during periods of clinical exacerbation (Chmiel et al., 2014). Co-infecting S. aureus isolates exposed to inhaled tobramycin therapy can become highly tobramycin resistant. We measured the MIC of tobramycin against a panel of S. aureus CF isolates, and found that 6 of 8 isolates grew in concentrations of tobramycin ranging from 800–1600µg/ml (Figure 3C). Aminoglycoside-modifying enzymes allow non-SCV S. aureus to inactivate intracellular drug and grow in high concentrations of aminoglycosides. All of our resistant isolates were non-SCV, and thus it is likely that these isolates had either acquired an aminoglycoside modifying enzyme or had mutated ribosomal binding sites with lower binding affinity for tobramycin. Regardless of mechanism, we hypothesized that significantly increasing intracellular concentrations of tobramycin with RLs could overwhelm both mechanisms of resistance to restore some level of susceptibility to these highly resistant isolates. Indeed, we found that RLs synergized with tobramycin to reduce the MIC 8 to 32-fold among our isolates (Figure 3C). Further, RLs reduced the concentration of tobramycin necessary to inhibit S. aureus growth in these isolates to below the clinical achievable concentration following aerosolized delivery (~673µg/ml per dose)(Chmiel et al., 2014) (Figure 3C). In all, these findings demonstrate that combinational therapy can slow the rise of resistance in a closed system, and re-sensitize even the most highly resistant clinical isolates.
Rhamnolipids sensitize other gram-positive pathogens to aminoglycoside killing
We next asked whether targeting membrane permeability to induce aminoglycoside uptake is a valid therapeutic approach against other bacterial pathogens. Consistent with previous findings, we observed that RLs demonstrated no bactericidal activity, and thus no aminoglycoside-potentiating effects against gram-negative Escherichia coli, presumably due to the presence of an additional outer membrane (Table 1)(Sotirova et al., 2008). For gram-positive species, we found that RLs lowered the MIC of tobramycin for Enterococcus faecalis, Bacillus subtilis, Listeria monocytogenes and Clostridioides difficile (Table 1). Of note, RLs reduced the MIC of C. difficile from 400µg/ml to under 0.39µg/ml, sensitizing what is otherwise a highly resistant, strictly anaerobic species. RLs were ineffective against Streptococcus pneumoniae.
Table 1.
Tobramycin/rhamnolipid MIC values for other gram positive and negative bacterial species (µg/mL)
| Tobramycin | Tobramycin + RLs | Fold change | RL MIC | |
|---|---|---|---|---|
| Gram negative | ||||
| Escherichia coli | 0.78 | 0.78 | 0 | >2400 |
| Gram positive | ||||
| Bacillus subtilis | 0.39 | 0.0061 | 64 | 62.5 |
| Streptococcus pneumoniae | 12.5 | 12.5 | 0 | >2400 |
| Enterococcus faecalis | 100 | 0.78 | 128 | 125 |
| Listeria monocytogenes | 1.56 | 0.049 | 32 | 125 |
| Clostridioides difficile | 400 | 0.39 | 1025 | 31.25 |
Rhamnolipids induce distinct modifications to the S. aureus membrane to promote tobramycin uptake
To better understand the molecular mechanism(s) for how RLs and other cell envelope-acting agents (CEAAs) potentiate aminoglycosides, we selected 3 other putative aminoglycoside adjuvants and compared their impact on S. aureus membrane physiology. Antimicrobial monoglycerides such as glycerol monolaurate (GML) target the plasma membrane and were recently demonstrated to synergize with aminoglycosides against S. aureus biofilm (Hess et al., 2014; Yoon et al., 2018). Similarly, the retinoid adarotene targets bacterial membranes and synergizes with aminoglycosides against S. aureus persisters (Kim et al., 2018). Finally, β-lactam/aminoglycoside synergy has long been theorized to occur in vivo, with limited in vitro support and conflicting clinical reports (Figure S4A)(Tamma et al., 2012).
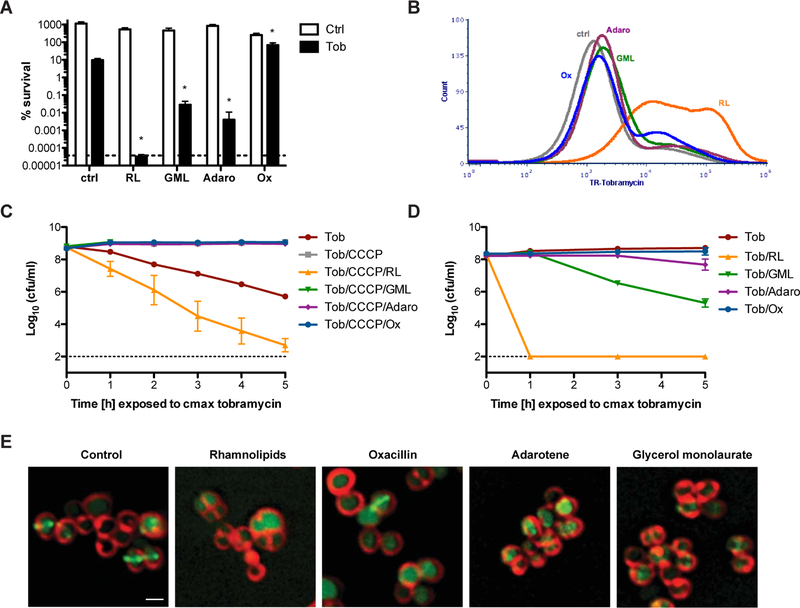
For each compound we attempted to identify a concentration that potentiated tobramycin killing without exerting bactericidal activity alone (Figure S1B–D). Treatment with 3.2µg/mL of adarotene and 40µg/mL of GML facilitated 100 to 1000-fold increase in tobramycin killing (Figure 4A). Surprisingly we observed that sub-bactericidal concentrations of oxacillin conferred a moderate but significant protective effect against tobramycin (Figure S1D, Figure 4A). RLs were the most powerful potentiator of tobramycin killing, sterilizing cultures to the limit of detection (Figures S1A, 4A). These findings were supported by traditional checkerboard synergy assays where synergy (FICI ≤ 0.5) was observed between tobramycin and RLs, GML, and adarotene; but not between tobramycin and oxacillin (Figure S5, Table S1). We then used flow cytometry to measure changes in Texas Red-conjugated tobramycin uptake following exposure to each CEAA. Strikingly, we found that only RL treatment lead to significant changes in tobramycin uptake post antibiotic exposure despite the fact that RLs, GML and adarotene all potentiated tobramycin killing at these concentrations (Figures 4B, S4B, C).
Figure 4. Rhamnolipids specifically induce PMF-independent aminoglycoside uptake to resensitize tolerant S. aureus.
A. S. aureus was grown to mid-exponential phase then challenged with 58µg/mL tobramycin alone, or in combination with 30µg/mL RLs, 40µg/mL GML, 3.2µg/mL adarotene, or 0.5µg/mL oxacillin (black bars). An aliquot of each culture was removed after 24hr, washed, and plated to enumerate survivors. White bars represent S. aureus survivors following 24hr treatment with each cell envelope-acting agent without tobramycin. B. Texas Red-conjugated tobramycin was added to S. aureus cultures with our without each compound. Following 1hr, Texas Red-tobramycin uptake was measured by flow cytometry. C, D. S. aureus HG003 was C. treated with 1µM CCCP or D. grown anaerobically prior to treatment with 58µg/mL tobramycin +/− each cell envelope acting compound individually. Survivors were enumerated at the indicated time points. E. S. aureus strain SH1000 harboring an inducible PzapA::gfp cell division reporter was grown to OD600=0.5, then treated with 30 RLs, 40µg/mL GML 3.2µg/mL adarotene, or 0.5µg/mL oxacillin for 1 hr. Cells were washed, then treated with the membrane dye, FM4–64. Changes to membrane morphology and ZapA localization relative to control cultures were visualized using a GE Applied Precision DeltaVision Elite de-convolution fluorescence microscope equipped with a Photometrics CoolSnap HQ2 camera. Scale bar: 1µm. *p<0.05 (Student’s t-test). All experiments were performed in biological triplicate. Error bars represent mean +/− SD. See also Figure S4.
We hypothesized that the unique ability of RLs to promote rapid aminoglycoside influx might stem from how RLs interact with the bacterial membrane. Aminoglycoside internalization begins with ionic interaction with the cell surface, followed by PMF-mediated diffusion into the cell (Taber et al., 1987). We reasoned that RLs might promote aminoglycoside uptake by altering membrane charge and/or permeability. Similar to oxacillin, RL treatment decreased S. aureus surface charge, and both RLs and adarotene stimulated a lasting increase in membrane fluidity (Figure S4D, E). However, only RLs significantly stimulated leakage of intracellular ATP into the medium, suggesting that at these concentrations only RLs induce small molecule permeability of the membrane (Figure S4F). Overall, RLs simultaneously altered surface charge, membrane fluidity, and small molecule permeability, which may explain their unique capacity to stimulate PMF-independent aminoglycoside uptake.
As GML and adarotene did not promote tobramycin influx during the initial phase of tobramycin uptake, we hypothesized that these CEAAs may instead potentiate aminoglycoside killing downstream of initial antibiotic influx. If true, then RLs alone should restore tobramycin susceptibility to CCCP treated cells, as PMF-depleted cultures exclude aminoglycosides altogether. As expected, CCCP-treatment induced tolerance to tobramycin that was overcome with RLs (Figure 4C). However, combinational therapy with GML, adarotene, or oxacillin were unable to overcome CCCP-mediated PMF depletion, confirming that these compounds do not bypass the initial phase of tobramycin uptake and instead likely synergize with tobramycin after antibiotic has penetrated the cell, possibly by rendering S. aureus more sensitive to downstream membrane damage resulting from protein synthesis corruption (Figure 4C). Physiologically this distinction is critical, because synergy with GML or adarotene will likely be limited to respiring, high PMF S. aureus populations in vivo. Indeed, when grown anaerobically, adarotene no longer potentiated aminoglycoside killing, and the synergy observed for GML was minor compared to the rapid eradication observed following RL treatment (Figure 4D).
We used high-resolution fluorescent microscopy to visualize the effects of each compound on the S. aureus plasma membrane. We observed that treatment with RLs at concentrations that potentiate aminoglycoside killing, resulted in a population of viable cells that retained overall shape and membrane morphology, however localization of membrane-associated cell division machinery, measured with the FtsZ proxy, ZapA-GFP, was perturbed, suggesting that RLs interfere with membrane physiology without completely destabilizing it (Figure 4E, Figure S4G). Adarotene and GML treatment resulted in membrane clumping indicative of more pronounced membrane destabilization at concentrations that induced tobramycin synergy (Figure 4E). All compounds stimulated mislocalization of ZapA-GFP from the membrane (Figure S4G). Taken together, these data suggest that RLs distinctly modify the plasma membrane to induce tobramycin uptake, while the potentiation effects observed from other CEAAs may occur through general destabilization of the membrane during aminoglycoside-induced membrane stress.
Discussion
Aminoglycoside efficacy is limited by the environmental context of infection (Radlinski and Conlon, 2018). Minor fluctuations in nutrient and oxygen availability have dramatic implications for drug penetration, rendering aminoglycosides useless against certain types of infections (Bryan et al., 1979; Taber et al., 1987). Here, we demonstrated that inducing PMF-independent aminoglycoside uptake with RLs facilitates the eradication of anaerobic, biofilm, SCV and persister populations, represses the rise of aminoglycoside resistance, and restores susceptibility to highly resistant isolates. Furthermore, we showed that while other cell membrane-targeting agents can synergize with aminoglycosides, stimulating the initial phase of aminoglycoside influx is rare, and unique to RLs under these conditions. Consequently, RLs demonstrated the greatest efficacy as a combinational therapy against aminoglycoside-tolerant S. aureus.
The specific mechanisms that induce antibiotic tolerance in the host are poorly understood. In vitro, bacterial tolerance is associated with a stochastic or deterministic drop in ATP-dependent antibiotic target activity below the threshold required to facilitate death (Conlon et al., 2016; Shan et al., 2017). Aminoglycoside tolerance, however, is further complicated by the fact that PMF-dependent drug influx is also contingent on the metabolic state of the cell. Consequently, it is difficult to delineate between deficiencies in drug penetration or low intracellular ATP when examining an aminoglycoside tolerant population. RLs remove the prerequisite of PMF for drug influx, and allow us to specifically study the relationship between ATP depletion and aminoglycoside tolerance. Strikingly, we found that RLs allowed for tobramycin-mediated eradication of S. aureus populations exhibiting minimal intracellular ATP and translation activity, suggesting that the primary obstacle to aminoglycoside efficacy lies in drug influx. This is in contrast to other antibiotics that diffuse freely across the membrane or act on the cell surface, where ATP depletion is sufficient to induce tolerance (Conlon et al., 2016). With these antibiotics, we suggest that the primary barrier to efficacy likely lies in the reduction of active cellular targets such as DNA replication (fluoroquinolones), transcription (rifamycins) or cell wall synthesis (β-lactams) among tolerant populations. Recent work suggests that “dormant” persister populations maintain low level metabolic activity (Lechner et al., 2014; Prax and Bertram, 2014; Pu et al., 2016). Although protein synthesis is likely reduced in persisters, our results suggest that the number of active ribosomal targets in these populations is sufficient for aminoglycosides to precipitate cell death if they are accessible to the antibiotic. This is supported by the recent finding that growth-arrested bacterial populations actively synthesize protein for several days after entering stationary phase (Gefen et al., 2014). By stimulating aminoglycoside influx, RLs allow us to target and eradicate persister cells with extremely low-level rates of translation.
To date, the molecular details of how RLs interact with the bacterial membrane are poorly understood. Data from biophysical studies suggest that the inverted cone-like structure that arises from a large polar head group and smaller hydrophilic tail causes RLs to induce a positive curvature in the membrane that leads to the formation of pores (Ortiz et al., 2006). Indeed, at high bactericidal concentrations, RL treatment causes catastrophic pore formation in B. subtilis (Sotirova et al., 2008). We suspect that the subinhibitory RL concentrations used here elicit sufficient membrane destabilization to permit diffusion of aminoglycosides into the cell without inducing bactericidal pore formation and membrane dissolution. This conclusion is supported by the fact that we observed increased small molecule permeability and aminoglycoside uptake, but not total dissipation of PMF when cells were treated with subinhibitory concentrations of RLs. Importantly, RLs synergized with aminoglycosides against a panel of gram positive pathogens, all of which produce a range of different phospholipid molecules and cell membrane components (Sohlenkamp and Geiger, 2016), indicating that this potentiating effect is not unique to the S. aureus membrane composition.
Destabilizing the bacterial plasma membrane during aminoglycoside therapy represents a promising approach that is complicated by off-target effects to host cell membranes. While RLs facilitated tobramycin-mediated eradication of S. aureus at concentrations far below what was cytotoxic to host cells, cytotoxicity at higher concentrations may limit the therapeutic potential of these molecules. Furthermore, it remains to be seen whether this therapeutic approach increases the toxicity of aminoglycosides against eukaryotic host cells. An ideal aminoglycoside adjuvant would exhibit broad affinity for the bacterial membrane and limited affinity for eukaryotic membranes. Here we used a commercially available mix of mono- and di-rhamnolipids, however a recent study revealed that chemical derivatization of purified RL congeners significantly alters the antibacterial and cytotoxic properties of these molecules (Aleksic et al., 2017). In particular, semi-synthetic amide RL derivatives demonstrated greatly increased antibacterial activity against S. aureus. Furthermore, certain antimicrobial peptides are postulated to disrupt cell membranes via a mechanism that is similar to RLs (Hallock et al., 2003; Ortiz et al., 2006). In their paper, Lin et al. demonstrate that cationic antimicrobial peptides synergize with the macrolide antibiotic azithromycin to potentiate activity against gram negative pathogens (Lin et al., 2015). Similarly, aminoglycoside/antimicrobial peptide synergism was demonstrated against Enterococcus faecium in a murine cutaneous abscess model (Pletzer et al., 2018). In both of these cases the proposed mechanism of potentiation is increased antibiotic uptake. Further investigation to uncover a specific RL derivative or a novel small molecule inducer of permeability that demonstrates minimal cytotoxicity may yield the ideal combinational therapy for targeting recalcitrant bacterial populations within a host. Importantly, however, our findings demonstrate that potential adjuvants must operate at the initial phase of aminoglycoside uptake to induce PMF-independent influx. Otherwise, compounds that require PMF-mediated aminoglycoside uptake for synergy will fail to clear physiologically relevant PMF-depleted, aminoglycoside tolerant populations.
STAR Methods
Lead contact and materials availability
Further information and requests for resources and reagents should be direct to and will be fulfilled by the Lead Contact, Brian Conlon (bconlon@med.unc.edu).
Experimental model and subject details
Bacterial strains and growth conditions
S. aureus strains HG003 or JE2 were cultured aerobically in Mueller-Hinton (MHB) or tryptic soy (TSB) broth at 37°C with shaking at 225 rpm. For anaerobic growth, overnight cultures were washed twice with PBS and diluted into 5 ml of pre-warmed (37°C) TSB to an OD 600 of 0.05. Cultures were prepared in triplicate in 16×150mm glass tubes containing 1mm stir bars. Following dilution, cultures were immediately transferred into a Coy anaerobic chamber and grown at 37°C with stirring. CF isolates were collected from patients at the UNC medical center. Isolates were cultured from sputum or bronchoalveolar lavage (BAL) from patients with CF after obtaining informed consent. HG003 menD::erm was generated through transduction using 80α as described previously (Grosser and Richardson, 2014), using a published COL menD::erm mutant strain (Joshi et al., 2011) as the donor and HG003 WT was the recipient. A modified version of the E. coli/S. aureus cloning vector pEPSA5(Forsyth et al., 2002) carrying an erythromycin selection cassette and a xylose-inducible GFP was constructed as follows: pEPSA5 vector was linearized through PCR amplification using primers SR43 and SR44 at the 5’ and 3’ sites immediately flanking the chloramphenicol cassette, which was then replaced with an erythromycin resistance cassette amplified using primers specific to the bursa aurealis erythromycin resistant transposon from the NARSA library (SR41, SR42) (Bae et al., 2004). The gfpuvr (Clontech)(Kahl et al., 2000) was amplified by primers which added a 5’ EcoRI site, 3’ KpnI site, and a sarA ribosome binding site and spacer. This amplicon was digested with KpnI and EcoRI (NEB) and ligated into similarly digested pEPSA5-erm to yield plasmid pCLON46. S. aureus SH1000 harboring a pRB42 plasmid-based cadmium-inducible copy of zapA-gfp (Eswara et al., 2018), was grown in tryptic soy broth (TSB) with 5 µg/ml erythromycin (erm) for plasmid maintenance. Streptococcus pneumoniae wild type 262 [CIP 104340] was cultured in MHB plus 5% sheep’s blood (Hemostat Laboratories) at 37°C plus 5% CO2. Enterococcus faecalis wild type OG1 and Listeria monocytogenes wild type 10403s were cultured in BHI at 37°C. Escherichia coli wild-type MG1655 and Bacillus subtilis wild-type BS49 were cultured in MHB at 37°C. Clostridioides difficile wild-type R20291 was cultured anaerobically in BHI supplemented with 2% yeast at 37°C.
Cell lines
J774A.1 (ATCC TIB-67) macrophage-like cells were maintained in Dulbecco’s modified Eagle medium (DMEM) supplemented with 4.5 g/liter glucose, 10% fetal bovine serum, 2mM L-glutamine, and 1mM sodium pyruvate.
Method Details
Antibiotic survival assays
HG003 was grown to ~3×108cfu/ml in 3ml MHB or in 5ml TSB under aerobic and anaerobic conditions, respectively. An aliquot was plated to enumerate cfu before antibiotic challenge. Tobramycin was added at either the concentration similar to the Cmax in humans at recommended dosing (58µg/ml)(Burkhardt, 2006) or at 20x the MIC (15.6 µg/ml). Non-aminoglycoside antibiotics were added at 20x MIC concentrations as stated. At indicated times, culture aliquots were removed, washed with 1% NaCl, then serially diluted and plated to enumerate survivors. Where indicated, RLs (RL90, AGAE), GML (Sigma), adarotene (Sigma) or oxacillin (Fisher) was added with antibiotics. For plate-based killing assays, HG003 was grown to ~3×108 CFU/mL and 100µl of each culture was seeded into wells of a 96-well plate pre-loaded with 100µL MHB containing 2x the desired concentration of each CEAA +/− 15.6µg/mL tobramycin (final concentration=7.8µg/mL). Plates were incubated with shaking for 24hr at 37°C. Cells were then pelleted, washed, and plated to enumerate survivors.
Biofilm susceptibility assays
Overnight cultures (~18hr) of HG003 were subcultured 1:200 in BHI, seeded in a 96-well microtiter plate and incubated statically at 37°C overnight. After 24hrs, wells were washed 2x with 1% NaCl and overlaid with 200µl of BHI with or without tobramycin at 58µg/ml +/− RLs and incubated overnight at 37°C. 24 hr later, wells were washed 2x, overlaid with 100µl of 1% NaCl, and sonicated for 10 minutes to disrupt biofilm. Sonicated samples were then serially diluted and plated for CFU enumeration.
ATP assays
HG003 was grown to ~3×108cfu/ml in 3ml MHB and treated with 5mM sodium arsenate dibasic heptahydrate (Sigma) for 30min +/− 30µg/mL RLs. ATP levels were measured using a Promega BacTiter Glo kit according to the manufacturer’s instructions. For small molecule permeability assays, exponential phase cultures of HG003 were treated with CEAAs for 1hr, then 1mL aliquots of each culture was pelleted and the supernatant used to measure ATP concentration.
Minimum inhibitory concentration (MIC) assays
MICs were determined using the microdilution method. Briefly, ~5×105cfu were incubated with varying concentrations of tobramycin in a total volume of 200µl MHB in a 96-well plate. MICs were determined following incubation at 37°C for 24hr. MICs were performed in BHI for E. faecalis and L. monocytogenes, and in BHI supplemented with 2% yeast extract for C. difficile to support growth. Where indicated, MICs were performed in the presence of 30µg/ml RLs. To monitor the rise in spontaneous tobramycin resistant mutants over time, six independent lineages of HG003 were grown in varying concentrations of tobramycin in a 96-well plate. After 24hrs of static incubation at 37°C, wells with the highest concentr ation of tobramycin that permitted significant bacterial growth (OD600≥ 0.1) were used to inoculate fresh MHB for the next passage at a bacterial density of approximately 5×105 CFU/mL. Three independent lines (HG1–3) were serially passaged with tobramycin alone, and three lineages (HG4–6) were passaged in tobramycin + RL at 30µg/ml for a total of 31 days. At every third passage, strains were collected and stored at −80°C in a 20% glycerol stock. As a control, the MIC of wild-type HG003 was determined concurrently with each passage of strains HG1–6.
Checkerboard synergy assays
Dual antibiotic synergy was assayed using plate-based checkerboard assay as described previously(Orhan et al., 2005), where 2-fold serial dilutions of each compound added to 2-fold dilutions of tobramycin to create an 8×8 matrix in a 96-well plate. The Fractional Inhibitory Concentration Index (FICI) was calculated as: FICI= (MIC of compound A in combination/MIC compound A alone) + (MIC compound B in combination/MIC compound B alone). FICI ≤ 0.5 = synergy, 0.5 < FICI ≤4 = no interaction, 4<FICI = antagonism.
Texas Red-tobramycin uptake
Texas Red-succinimidyl ester (Invitrogen) was dissolved in high-quality anhydrous N,N-dimethylformamide at a final concentration of 20mg/ml. Tobramycin was resuspended in 100mM K2CO3, pH 8.5 at a final concentration of 10mg/ml. On ice, 10µl of Texas Red was slowly added to 350µl tobramycin solution at 30 molar excess to allow the conjugation reaction to occur, and to maximize the formation of single-label tobramycin as described (Sandoval et al., 1998). HG003 was grown to mid-exponential phase and then treated with the indicated compound + Texas Red-tobramycin at a final concentration of 15.6µg/ml. After 1hr, an aliquot of cells was removed, washed twice in 1% NaCl and plated to enumerate survivors. The remaining aliquot was analyzed for Texas Red uptake on a Thermo Fisher Attune NxT flow cytometer. 30,000 events were recorded. Figures were generated using FSC Express 6 Flow.
Membrane potential measurements
Bacterial membrane potential was measured with the BacLight Bacterial Membrane Potential Kit as per the manufacturers instructions. Briefly, overnight cultures of HG003 were subcultured 1:100 in MHB and grown for 3.5 hrs before a 30 min exposure to the indicated concentration of RLs. Cultures were then diluted 1:100 in 1mL PBS to approximately 1×106 CFU/mL and treated with 30µM DiOC2 for approximately 30 minutes before analysis on a Thermo Fisher Attune NxT flow cytometer. 30,000 events were recorded and relative membrane potential was calculated by taking the ratio of population red and green linear mean fluorescence intensity (MFI) values.
Cytochrome C binding assays
S. aureus surface charge was measured using a modified protocol described previously (Peschel et al., 1999). HG003 was subcultured 1:100 in TSB and grown 2.5hr at 37°C with shaking prior to a 1hr incubation with indicated compounds. Cells were harvested from 1mL of each cultures normalized to OD600=0.3, then resuspended in 150µL 20mM MOPS pH 7.0. Samples were treated with 200µg/mL of purified cytochrome C (MP Bio) for 10 minutes. Cells were then pelleted and 100µL of supernatant was transferred to a 96-well plate where absorbance was measured at 530nM.
Laurdan GP Membrane Fluidity
Overnight cultures of HG003 were diluted 1:100 in MHB and grown for 4hrs at 37°C with shaking. Laurdan staining was performed as described(Scheinpflug et al., 2017), and carried out in a 37°C climate-controlled room to en sure stable temperatures. Laurdan was added to each culture at a final concentration of 10µM for 10 min, while shaking in the dark. Cells were harvested by centrifugation in pre-warmed microtubes and washed 4x with pre-warmed wash buffer (137mM NaCl, 2.7mM KCl, 10mM Na2HPO4, 0.2% glucose, 1% DMF). Following wash steps, cells were resuspended in wash buffer to an OD600=0.8. 100µL of stained cells were added to 100µL of wash buffer containing 2x the desired concentration of each compound in a pre-warmed black wall, clear bottom plate alongside a baseline control. Fluorescence was measured immediately in 2min intervals over 30mins (excitation: 350nM, emission: 460nM and 500nM. Laurdan GP = (I460−I500)/(I460+I500).
Eukaryotic cytotoxicity assays
Rhamnolipid cytotoxicity activity was measured for J774A.1 macrophage-like cells using the CellTiter-Blue Cell Viability assay according to the manufacturer’s instructions. Briefly, J774A.1 cells were seeded in a black wall, clear bottom 96-well plate at 25,000 cells/well in high-glucose DMEM (Gibco) supplemented with 10% FBS and 2mM L-glutamine, and allowed to incubate overnight at 37°C and 5% CO2. After 24hr, the media was removed from wells and replaced with media + the indicated concentration of RLs and cells were returned to incubate overnight. After 24hr, 100µL of CellTiter-Blue reagent was added to each well and allowed to incubate at 37°C for 2 hours before fluorescence was read at 560/590 nm (excite/emit).
Fluorescent Microscopy
Overnight cultures of S. aureus SH1000 harboring a pRB42 plasmid-based cadmium-inducible copy of zapA-gfp (Eswara et al., 2018), were grown at 22°C in tryptic soy broth (TSB) with 5 µg/ml erythromycin (erm) for plasmid maintenance, and then subsequently diluted 1:10 into fresh TSB+erm. Cultures were then grown at 37°C and growth was monitored by the measurement of absorbance. At mid-log phase, when optical density at 600 nm reached 0.5, 1.25 mM cadmium chloride was added to the cultures to induce the expression of zapA-gfp. Cells were then treated with either RLs (30µg/ml), GML (40µg/ml), adarotene (3.2µg/ml), or oxacillin (0.5µg/ml) for 1 h. Untreated cells and cells treated with DMSO were used as controls. Following the incubation period, 1 ml cells were washed three times in PBS, and then resuspended in 100 ml of PBS containing 1 mg/ml membrane stain (FM6–64). Aliquots of 5 ml culture were pipetted onto a glass bottom dish (MatTek), and sample was covered with a 1% agarose pad. Microscopy was performed at room temperature using a GE Applied Precision DeltaVision Elite deconvolution fluorescence microscope equipped with a Photometrics CoolSnap HQ2 camera.
Quantification and Statistical Analysis
Experiments were performed with three biological replicates from at least two independent experiments when possible. Statistical significance is reported in Figure Legends, and data are presented as mean +/− SD as indicated. All statistical analysis was performed with Graphpad Prism software.
Supplementary Material
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial Strains and Plasmids | ||
| Staphylococcus aureus HG003 wild-type | (Herbert et al., 2010) | N/A |
| Staphylococcus aureus HG003 harboring plasmid pCLON46 | This study | N/A |
| Staphylococcus aureus HG003 menD::erm | This study | N/A |
| Staphylococcus aureus tobramycin-resistant isolates HG1-HG6 | Derived from HG003 wild-type, this study | N/A |
| Staphylococcus aureus COL menD::erm | (Joshi et al., 2011) | N/A |
| Staphylococcus aureus SH1000 harboring plasmid pRB42 | (Eswara, 2018) | N/A |
| Staphylococcus aureus CF isolates BC322-BC330 | Isolated from patients, this study | N/A |
| Staphylococcus aureus JE2 wild-type | (Bae et al., 2004) | N/A |
| Escherichia coli wild-type MG1655 | ATCC | ATCC 47076 |
| Bacillus subtilis wild-type BS49 | (Christie et al., 1987) | N/A |
| Streptococcus pneumoniae wild-type 262 [CIP 104340] | ATCC | ATCC 49619 |
| Enterococcus faecalis wild-type OG1 | ATCC | ATCC 47077 |
| Listeria monocytogenes wild-type 10403s | (Portnoy et al., 1988) | N/A |
| Clostridioides difficile wild-type R20291 | (Stabler et al., 2009) | N/A |
| pEPSA5 E. coli/S. aureus cloning vector | (Forsyth et al., 2002) | N/A |
| pCLON46, pEPSA5 derivative | This study | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Mueller-Hinton broth | Thermo Scientific Oxoid | Cat# OXCM0405B |
| Tryptic Soy broth | BD | Cat# DF0370–17-3 |
| Brain heart infusion broth | Oxoid | Cat# CM1135 |
| Gentamicin | Fisher BioReagents | Cat# BP918–1 |
| Rifampicin | Fisher BioReagents | Cat# BP2679250 |
| Ciprofloxacin | Acros Organics | Cat# 449620050 |
| Oxacillin | Acros Organics | Cat# 455440050 |
| Tobramycin | Sigma-Aldrich | Cat# T4014 |
| Tetracycline | Thermo Fisher | Cat#ICN10301125 |
| Erythromycin | Thermo Fisher | Cat# AC227330050 |
| Rhamnolipids, 90% | AGAE | Cat# RL90 |
| Rhamnolipids, 95% (90% di-rhamnolipid | AGAE | Cat# R95D90 |
| Rhamnolipid, 98% mono-rhamnolipid C10C10 | Glycosurf | Cat# Rha-C10C10 |
| Rhamnolipid, 98% mono-rhamnolipid C12C12 | Glycosurf | Cat# Rha-C12C12 |
| Glycerol monolaurate | Sigma-Aldrich | Cat# M1765 |
| Adarotene | Sigma-Aldrich | Cat# SML2061 |
| Texas Red- succinimidyl ester | Thermo Fisher | Cat# T6134 |
| Sodium arsenate dibasic hepahydrate | Sigma-Aldrich | Cat# A6756 |
| N,N-dimethylformamide | Sigma-Aldrich | Cat# 227056 |
| Cytochrome C (horse heart) | MP Bio | Cat# ICN10146701 |
| Laurdan | Cayman Chemicals | Cat# 19706 |
| Dulbecco’s Modified Essential Media, high glucose | Gibco | Cat# 11965092 |
| Fetal bovine serum | EMD Milipore | Cat# TMS-013-B |
| L-glutamine | Gibco | Cat# 25030081 |
| Critical Commercial Assays | ||
| BacTiter-Glo Microbial Cell Viability Kit | Promega | Cat# G8232 |
| BacLight Bacterial Membrane Potential Kit | Thermo Fisher | Cat# B34950 |
| CellTiter Blue Cell Viability Assay | Promega | Cat# G8080 |
| Oligonucleotides | ||
| Primer SR41- Forward AAGAAAGCAGACAAGTAAGATTCCAAATGCGTAATG | This Study | N/A |
| Primer SR42- Reverse TCATATTATAAAAGCCATTATTTCCTCCCGTTAAATAATAG | This Study | N/A |
| Primer SR43- Forward TGGCTTTTATAATATGAGATAATGCCG | This Study | N/A |
| Primer SR44- Reverse TCTTACTTGTCTGCTTTCTTCATTAG | This Study | N/A |
| Software and Algorithms | ||
| Graphpad Prism 5 | N/A | N/A |
| FCS Express Flow Cytometry Data Analysis | N/A | N/A |
| ChemDraw Professional 16.0 | N/A | N/A |
Highlights.
Tolerance to aminoglycosides is primarily mediated by inhibition of uptake
Rhamnolipids (RL) induce PMF-independent aminoglycoside uptake in S. aureus
RL/aminoglycoside therapy targets tolerant and resistant bacterial populations
Significance.
The widespread onset of multidrug resistant pathogenic strains, coupled with an evaporating pipeline of new antibiotics reaching market emphasizes the importance of maximizing the efficacy of current antibiotics. As such, novel means for overcoming antibiotic tolerance and re-sensitizing resistant strains are desperately needed to combat infection. Aminoglycosides demonstrate broad-spectrum bactericidal activity against actively growing bacterial populations, but their activity is limited against non-respiring pathogen populations commonly found within the host. This limits the usefulness of this class of antibiotics against difficult-to-treat infections. Our study demonstrates that chemically inducing aminoglycoside uptake by promoting small molecule permeability in the membrane with RLs overcomes the barriers imposed during PMF depletion to extend the reach of aminoglycosides against recalcitrant pathogen populations. This approach is broadly applicable to gram-positive pathogens, and sensitizes biofilm, SCV, and anaerobic populations of S. aureus to aminoglycoside killing. Furthermore, this combinational therapy demonstrates remarkably potent bactericidal activity against persisters, a bacterial population that is notorious tolerant to killing, regardless of antibiotic class. Chemically inducing PMF-independent aminoglycoside uptake represents a promising new approach for resolving chronic or relapsing infection, improving patient health, and slowing the spread of resistance.
Acknowledgements
This work was funded by the US NIH (F31AI140520 (LCR), R01AI137273 (BPC), and R01GM128037 (PJE)) as well as by a start-up grant from the University of South Florida (PJE). We thank Nikki Wagner, Jenna Beam, Natalia Maldonado-Vazquez, and Ashelyn Sidders for advice with this project.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
LCR, SER and BPC are inventors on a patent describing rhamnolipid potentiation of aminoglycoside killing (Radlinski LC, Conlon SR, Conlon BP, Potentiation of antibiotic effect, US 62/534450 Filed July 19 2017). The other authors declare no competing interests.
Data and code availability
This study did not generate or analyze data sets or code.
References
- Aleksic I, Petkovic M, Jovanovic M, Milivojevic D, Vasiljevic B, Nikodinovic-Runic J, Senerovic L, 2017. Anti-biofilm Properties of Bacterial Di-Rhamnolipids and Their Semi-Synthetic Amide Derivatives. Front. Microbiol 8, 2454 10.3389/fmicb.2017.02454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison KR, Brynildsen MP, Collins JJ, 2011. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature 473, 216–220. 10.1038/nature10069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andry K, Bockrath RC, 1974. Dihydrostreptomycin accumulation in E. coli. Nature 251, 534–536. 10.1038/251534a0 [DOI] [PubMed] [Google Scholar]
- Bae T, Banger AK, Wallace A, Glass EM, Aslund F, Schneewind O, Missiakas DM, 2004. Staphylococcus aureus virulence genes identified by bursa aurealis mutagenesis and nematode killing. Proc. Natl. Acad. Sci 101, 12312–12317. 10.1073/pnas.0404728101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian D, Harper L, Shopsin B, Torres VJ, 2017. Staphylococcus aureus pathogenesis in diverse host environments. Pathog. Dis 75 10.1093/femspd/ftx005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan LE, Kowand SK, Van Den Elzen HM, 1979. Mechanism of aminoglycoside antibiotic resistance in anaerobic bacteria: Clostridium perfringens and Bacteroides fragilis. Antimicrob. Agents Chemother 15, 7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhardt O, 2006. Once-daily tobramycin in cystic fibrosis: better for clinical outcome than thrice-daily tobramycin but more resistance development? J. Antimicrob. Chemother 58, 822–829. 10.1093/jac/dkl328 [DOI] [PubMed] [Google Scholar]
- Chmiel JF, Aksamit TR, Chotirmall SH, Dasenbrook EC, Elborn JS, LiPuma JJ, Ranganathan SC, Waters VJ, Ratjen FA, 2014. Antibiotic management of lung infections in cystic fibrosis. I. The microbiome, methicillin-resistant Staphylococcus aureus, gram-negative bacteria, and multiple infections. Ann. Am. Thorac. Soc 11, 1120–9. 10.1513/AnnalsATS.201402-050AS [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon BP, Rowe SE, Gandt AB, Nuxoll AS, Donegan NP, Zalis EA, Clair G, Adkins JN, Cheung AL, Lewis K, 2016. Persister formation in Staphylococcus aureus is associated with ATP depletion. Nat. Microbiol 1, 16051 10.1038/nmicrobiol.2016.51 [DOI] [PubMed] [Google Scholar]
- Davis BD, 1987. Mechanism of bactericidal action of aminoglycosides. Microbiol. Rev 51, 341–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliopoulos GM, 1989. Synergism and antagonism. Infect. Dis. Clin. North Am 3, 399–406. [PubMed] [Google Scholar]
- Eswara PJ, Brzozowski RS, Viola MG, Graham G, Spanoudis C, Trebino C, Jha J, Aubee JI, Thompson KM, Camberg JL, Ramamurthi KS, 2018. An essential Staphylococcus aureus cell division protein directly regulates FtsZ dynamics. Elife 7 10.7554/eLife.38856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farha MA, Verschoor CP, Bowdish D, Brown ED, 2013. Collapsing the Proton Motive Force to Identify Synergistic Combinations against Staphylococcus aureus. Chem. Biol 20, 1168–1178. 10.1016/j.chembiol.2013.07.006 [DOI] [PubMed] [Google Scholar]
- Forsyth RA, Haselbeck RJ, Ohlsen KL, Yamamoto RT, Xu H, Trawick JD, Wall D, Wang L, Brown-Driver V, Froelich JM, C., K.G., King P, McCarthy M, Malone C, Misiner B, Robbins D, Tan Z, Zhu Z, Carr G, Mosca DA, Zamudio C, Foulkes JG, Zyskind JW, 2002. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol. Microbiol 43, 1387–1400. 10.1046/j.1365-2958.2002.02832.x [DOI] [PubMed] [Google Scholar]
- Garcia LG, Lemaire S, Kahl BC, Becker K, Proctor RA, Denis O, Tulkens PM, Van Bambeke F, 2013. Antibiotic activity against small-colony variants of Staphylococcus aureus: review of in vitro, animal and clinical data. J. Antimicrob. Chemother 68, 1455–1464. 10.1093/jac/dkt072 [DOI] [PubMed] [Google Scholar]
- Garneau-Tsodikova S, Labby KJ, 2016. Mechanisms of Resistance to Aminoglycoside Antibiotics: Overview and Perspectives. Medchemcomm 7, 11–27. 10.1039/C5MD00344J [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gefen O, Fridman O, Ronin I, Balaban NQ, 2014. Direct observation of single stationary-phase bacteria reveals a surprisingly long period of constant protein production activity. PNAS 111 10.1073/pnas.1314114111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosser MR, Richardson AR, 2014. Method for Preparation and Electroporation of S. aureus and S. epidermidis Humana Press, New York, NY, pp. 51–57. 10.1007/7651_2014_183 [DOI] [PubMed] [Google Scholar]
- Haba E, Pinazo A, Jauregui O, Espuny MJ, Infante MR, Manresa A, 2003. Physicochemical characterization and antimicrobial properties of rhamnolipids produced byPseudomonas aeruginosa 47T2 NCBIM 40044. Biotechnol. Bioeng 81, 316–322. 10.1002/bit.10474 [DOI] [PubMed] [Google Scholar]
- Haba E, Pinazo A, Pons R, Pérez L, Manresa A, 2014. Complex rhamnolipid mixture characterization and its influence on DPPC bilayer organization. Biochim. Biophys. Acta - Biomembr 1838, 776–783. 10.1016/J.BBAMEM.2013.11.004 [DOI] [PubMed] [Google Scholar]
- Hallock KJ, Lee D-K, Ramamoorthy A, 2003. MSI-78, an Analogue of the Magainin Antimicrobial Peptides, Disrupts Lipid Bilayer Structure via Positive Curvature Strain. Biophys. J 84, 3052–3060. 10.1016/S0006-3495(03)70031-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock RE, Raffle VJ, Nicas TI, 1981. Involvement of the outer membrane in gentamicin and streptomycin uptake and killing in Pseudomonas aeruginosa. Antimicrob. Agents Chemother 19, 777–85. 10.1128/aac.19.5.777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess DJ, Henry-Stanley MJ, Wells CL, 2014. Antibacterial synergy of glycerol monolaurate and aminoglycosides in Staphylococcus aureus biofilms. Antimicrob. Agents Chemother 58, 6970–3. 10.1128/AAC.03672-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi GS, Spontak JS, Klapper DG, Richardson AR, 2011. ACME encoded speG abrogates the unique hypersensitivity of Staphylococcus aureus to exogenous polyamines. Mol. Microbiol 82, 9 10.1111/J.1365-2958.2011.07809.X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahl BC, Goulian M, van Wamel W, Herrmann M, Simon SM, Kaplan G, Peters G, Cheung AL, 2000. Staphylococcus aureus RN6390 replicates and induces apoptosis in a pulmonary epithelial cell line. Infect. Immun 68, 5385–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Zhu W, Hendricks GL, Van Tyne D, Steele AD, Keohane CE, Fricke N, Conery AL, Shen S, Pan W, Lee K, Rajamuthiah R, Fuchs BB, Vlahovska PM, Wuest WM, Gilmore MS, Gao H, Ausubel FM, Mylonakis E, 2018. A new class of synthetic retinoid antibiotics effective against bacterial persisters. Nature 556, 103–107. 10.1038/nature26157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohanski MA, Dwyer DJ, Collins JJ, 2010. How antibiotics kill bacteria: from targets to networks. Nat. Rev. Microbiol 8, 423–435. 10.1038/nrmicro2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourtis AP, Hatfield K, Baggs J, Mu Y, See I, Epson E, Nadle J, Kainer MA, Dumyati G, Petit S, Ray SM, Ham D, Capers C, Ewing H, Coffin N, McDonald LC, Jernigan J, Cardo D, 2019. Vital Signs: Epidemiology and Recent Trends in Methicillin-Resistant and in Methicillin-Susceptible Staphylococcus aureus Bloodstream Infections — United States. MMWR. Morb. Mortal. Wkly. Rep 68, 214–219. 10.15585/mmwr.mm6809e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lannergard J, von Eiff C, Sander G, Cordes T, Seggewiss J, Peters G, Proctor RA, Becker K, Hughes D, 2008. Identification of the Genetic Basis for Clinical Menadione-Auxotrophic Small-Colony Variant Isolates of Staphylococcus aureus. Antimicrob. Agents Chemother 52, 4017–4022. 10.1128/AAC.00668-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebeaux D, Chauhan A, Létoffé S, Fischer F, de Reuse H, Beloin C, Ghigo J-M, 2014. pH-Mediated Potentiation of Aminoglycosides Kills Bacterial Persisters and Eradicates In Vivo Biofilms. J. Infect. Dis 210, 1357–1366. 10.1093/infdis/jiu286 [DOI] [PubMed] [Google Scholar]
- Lechner S, Lewis K, Bertram R, 2012. Staphylococcus aureus persisters tolerant to bactericidal antibiotics. J. Mol. Microbiol. Biotechnol 22, 235–44. 10.1159/000342449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner S, Prax M, Lange B, Huber C, Eisenreich W, Herbig A, Nieselt K, Bertram R, 2014. Metabolic and transcriptional activities of Staphylococcus aureus challenged with high-doses of daptomycin. Int. J. Med. Microbiol 304, 931–940. 10.1016/j.ijmm.2014.05.008 [DOI] [PubMed] [Google Scholar]
- Lewis K, 2010. Persister Cells. Annu. Rev. Microbiol 64, 357–372. 10.1146/annurev.micro.112408.134306 [DOI] [PubMed] [Google Scholar]
- Lin L, Nonejuie P, Munguia J, Hollands A, Olson J, Dam Q, Kumaraswamy M, Rivera H Jr., Corriden R, Rohde M, Hensler ME, Burkart MD, Pogliano J, Sakoulas G, Nizet V, 2015. Azithromycin Synergizes with Cationic Antimicrobial Peptides to Exert Bactericidal and Therapeutic Activity Against Highly Multidrug-Resistant Gram-Negative Bacterial Pathogens. EBioMedicine 2, 690 10.1016/J.EBIOM.2015.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta S, Singh C, Plata KB, Chanda PK, Paul A, Riosa S, Rosato RR, Rosato AE, 2012. β-Lactams increase the antibacterial activity of daptomycin against clinical methicillin-resistant Staphylococcus aureus strains and prevent selection of daptomycin-resistant derivatives. Antimicrob. Agents Chemother 56, 6192–200. 10.1128/AAC.01525-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orhan G, Bayram A, Zer Y, Balci I, 2005. Synergy tests by E test and checkerboard methods of antimicrobial combinations against Brucella melitensis. J. Clin. Microbiol 43, 140–3. 10.1128/JCM.43.1.140-143.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz A, Teruel JA, Espuny MJ, Marqués A, Manresa Á, Aranda FJ, 2006. Effects of dirhamnolipid on the structural properties of phosphatidylcholine membranes. Int. J. Pharm 325, 99–107. 10.1016/j.ijpharm.2006.06.028 [DOI] [PubMed] [Google Scholar]
- Peschel A, Otto M, Jack RW, Kalbacher H, Jung G, Götz F, 1999. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem 274, 8405–10. 10.1074/JBC.274.13.8405 [DOI] [PubMed] [Google Scholar]
- Pletzer D, Mansour SC, Hancock REW, 2018. Synergy between conventional antibiotics and anti-biofilm peptides in a murine, sub-cutaneous abscess model caused by recalcitrant ESKAPE pathogens. PLOS Pathog 14, e1007084 10.1371/journal.ppat.1007084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prax M, Bertram R, 2014. Metabolic aspects of bacterial persisters. Front. Cell. Infect. Microbiol 4 10.3389/FCIMB.2014.00148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor RA, Kahl B, von Eiff C, Vaudaux PE, Lew DP, Peters G, 1998. Staphylococcal small colony variants have novel mechanisms for antibiotic resistance. Clin. Infect. Dis 27 Suppl 1, S68–74. [DOI] [PubMed] [Google Scholar]
- Proctor RA, von Eiff C, Kahl BC, Becker K, McNamara P, Herrmann M, Peters G, 2006. Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat. Rev. Microbiol 4, 295–305. 10.1038/nrmicro1384 [DOI] [PubMed] [Google Scholar]
- Pu Y, Zhao Z, Li Y, Zou J, Ma Q, Zhao Y, Ke Y, Zhu Y, Chen H, Baker MAB, Ge H, Sun Y, Xie XS, Bai F, 2016. Enhanced Efflux Activity Facilitates Drug Tolerance in Dormant Bacterial Cells. Mol. Cell 62, 284–294. 10.1016/j.molcel.2016.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radlinski L, Conlon BP, 2018. Antibiotic efficacy in the complex infection environment. Curr. Opin. Microbiol 42, 19–24. 10.1016/J.MIB.2017.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radlinski L, Rowe SE, Kartchner LB, Maile R, Cairns BA, Vitko NP, Gode CJ, Lachiewicz AM, Wolfgang MC, Conlon BP, 2017. Pseudomonas aeruginosa exoproducts determine antibiotic efficacy against Staphylococcus aureus. PLOS Biol 15, e2003981 10.1371/journal.pbio.2003981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes A, Evans LE, Alhazzani W, Levy MM, Antonelli M, Ferrer R, Kumar A, Sevransky JE, Sprung CL, Nunnally ME, Rochwerg B, Rubenfeld GD, Angus DC, Annane D, Beale RJ, Bellinghan GJ, Bernard GR, Chiche J-D, Coopersmith C, De Backer DP, French CJ, Fujishima S, Gerlach H, Hidalgo JL, Hollenberg SM, Jones AE, Karnad DR, Kleinpell RM, Koh Y, Lisboa TC, Machado FR, Marini JJ, Marshall JC, Mazuski JE, McIntyre LA, McLean AS, Mehta S, Moreno RP, Myburgh J, Navalesi P, Nishida, , Osborn TM, Perner A, Plunkett CM, Ranieri M, Schorr CA, Seckel MA, Seymour CW, Shieh L, Shukri KA, Simpson SQ, Singer M, Thompson BT, Townsend SR, Van der Poll T, Vincent J-L, Wiersinga WJ, Zimmerman JL, Dellinger RP, 2017. Surviving Sepsis Campaign: International Guidelines for Management of Sepsis and Septic Shock: 2016. Intensive Care Med 43, 304–377. 10.1007/s00134-017-4683-6 [DOI] [PubMed] [Google Scholar]
- Sakoulas G, Moise-Broder PA, Schentag J, Forrest A, Moellering RC, Eliopoulos GM, 2004. Relationship of MIC and bactericidal activity to efficacy of vancomycin for treatment of methicillin-resistant Staphylococcus aureus bacteremia. J. Clin. Microbiol 42, 2398–402. 10.1128/JCM.42.6.2398-2402.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval R, Leiser J, Molitoris BA, 1998. Aminoglycoside antibiotics traffic to the Golgi complex in LLC-PK1 cells. J. Am. Soc. Nephrol 9. [DOI] [PubMed] [Google Scholar]
- Scaglione F, Dugnani S, Demartini G, Arcidiacono MM, Cocuzza CE, Fraschini F, 1995. Bactericidal Kinetics of an in vitro Infection Model of Once-Daily Ceftriaxone plus Amikacin against Gram-Positive and Gram-Negative Bacteria. Chemotherapy 41, 239–246. 10.1159/000239351 [DOI] [PubMed] [Google Scholar]
- Schaaff F, Bierbaum G, Baumert N, Bartmann P, Sahl H-G, 2003. Mutations are involved in emergence of aminoglycoside-induced small colony variants of Staphylococcus aureus. Int. J. Med. Microbiol 293, 427–435. 10.1078/1438-4221-00282 [DOI] [PubMed] [Google Scholar]
- Scheinpflug K, Krylova O, Strahl H, 2017. Measurement of Cell Membrane Fluidity by Laurdan GP: Fluorescence Spectroscopy and Microscopy, in: Methods in Molecular Biology (Clifton, N.J.). pp. 159–174. 10.1007/978-1-4939-6634-9_10 [DOI] [PubMed] [Google Scholar]
- Shan Y, Brown Gandt A, Rowe SE, Deisinger JP, Conlon BP, Lewis K, 2017. ATP-Dependent Persister Formation in Escherichia coli. MBio 8, e02267–16. 10.1128/mBio.02267-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohlenkamp C, Geiger O, 2016. Bacterial membrane lipids: diversity in structures and pathways. FEMS Microbiol. Rev 40, 133–159. 10.1093/femsre/fuv008 [DOI] [PubMed] [Google Scholar]
- Sotirova AV, Spasova DI, Galabova DN, Karpenko E, Shulga A, 2008. Rhamnolipid–Biosurfactant Permeabilizing Effects on Gram-Positive and Gram-Negative Bacterial Strains. Curr. Microbiol 56, 639–644. 10.1007/s00284-008-9139-3 [DOI] [PubMed] [Google Scholar]
- Taber HW, Mueller JP, Miller PF, Arrow AS, 1987. Bacterial uptake of aminoglycoside antibiotics. Microbiol. Rev 51, 439–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamma PD, Cosgrove SE, Maragakis LL, 2012. Combination Therapy for Treatment of Infections with Gram-Negative Bacteria. Clin. Microbiol. Rev 25, 450–470. 10.1128/CMR.05041-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong SYC, Davis JS, Eichenberger E, Holland TL, Fowler VG Jr, 2015. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev 28, 603–61. 10.1128/CMR.00134-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakulenko SB, Mobashery S, 2003. Versatility of aminoglycosides and prospects for their future. Clin. Microbiol. Rev 16, 430–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walraven CJ, North MS, Marr-Lyon L, Deming P, Sakoulas G, Mercier R-C, 2011. Site of infection rather than vancomycin MIC predicts vancomycin treatment failure in methicillin-resistant Staphylococcus aureus bacteraemia. J. Antimicrob. Chemother 66, 2386–2392. 10.1093/jac/dkr301 [DOI] [PubMed] [Google Scholar]
- Waters EM, Rowe SE, O’Gara JP, Conlon BP, 2016. Convergence of Staphylococcus aureus Persister and Biofilm Research: Can Biofilms Be Defined as Communities of Adherent Persister Cells? PLOS Pathog 12, e1006012 10.1371/journal.ppat.1006012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise EM, Park JT, Park JT, 1965. Penicillin: its basic site of action as an inhibitor of a peptide cross-linking reaction in cell wall mucopeptide synthesis. Proc. Natl. Acad. Sci. U. S. A 54, 75–81. 10.1073/pnas.54.1.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon B, Jackman J, Valle-González E, Cho N-J, 2018. Antibacterial Free Fatty Acids and Monoglycerides: Biological Activities, Experimental Testing, and Therapeutic Applications. Int. J. Mol. Sci 19, 1114 10.3390/ijms19041114 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.