Abstract
A constructed, variable-flow treatment wetland was evaluated for its ability to reduce microbial loads from the Banklick Creek, an impacted recreational waterway in Northern Kentucky. For this study, levels of traditional (Escherichia coli and enterococci measured by culture and molecular techniques) and alternative fecal indicators (infectious somatic and F+ coliphage, Clostridium spp. and Clostridium perfringens by culture), potential pathogens (molecular signal of Campylobacter spp.) as well as various microbial source tracking (MST) markers (human fecal marker HF183 and avian fecal marker GFD) were monitored during the summer and early fall through five treatment stages within the Banklick Creek Wetland. No difference in concentrations of traditional or alternative fecal indicators were observed in any of the sites monitored. Microbial source tracking markers were employed to identify sources of fecal contamination within the wetland. Human marker HF183 concentrations at beginning stages of treatment were found to be significantly higher (P value range: 0.0016–0.0003) than levels at later stages. Conversely, at later stages of treatment where frequent bird activity was observed, Campylobacter and avian marker (GFD) signals were detected at significantly higher frequencies (P value range: 0.024 to <0.0001), and both signals were strongly correlated (P = 0.0001). Our study suggests constructed wetlands are an effective means for removal of microbial contamination in ambient waters, but reliance on general fecal indicators is not ideal for determining system efficacy or assessing appropriate remediation efforts.
Keywords: Microbial source tracking, Wetland, Fecal indicator, Pathogens
Introduction
An estimated 3.5 million illnesses each year are attributed to contact with sewagecontaminated recreational water in the United States (Dorfman. 2004). Anthropogenic effects such as wastewater treatment plant discharge and failing or inadequate sewage and septage systems can be a source of continued water quality issues to receiving waterways. Due to increased enforcement of treatment standards for municipal wastewater treatment facilities, the effect of these point sources of pollution have been greatly reduced (US Environmental Protection Agency. 2018b). Point and non-point sources of contamination such as combined sewer overflows (CSO), sanitary sewer overflows (SSO), storm water runoff and failing septic systemsare a continuing concern for contamination present in receiving waterbodies. To minimize the impacts of pollution events, watershed based approaches are increasingly being considered, fostering the development of innovative solutions for water quality management.
Constructed wetlands have demonstrated an ability to offer a low cost, natural means of alleviating microbial contamination of wastewater prior to discharge into environmental receiving waters (Hench et al., 2003, Tanner et al., 1995, Toet et al., 2005). Wetlands act as natural biofilters and have demonstrated effectiveness in removal of physical, chemical and biological contaminants (Cheng et al., 2002, Hench et al., 2003, Stottmeister et al., 2003). Although exact processes are not fully understood, factors such as sedimentation, UV degradation, and predation are thought to influence decay and removal of contaminants within these systems. Wetland treatment technologies offer a low cost means of reducing pollutant levels in wastewaters, offering a more cost-effective means of treatment compared to traditional gray infrastructure (Kurzbaum et al., 2012).
In the United States, over 772 cities and 40 million U.S. citizens rely on (SSO/CSO) collection systems as their primary means of sewage collection (US Environmental Protection Agency, 2017). These systems are designed to transport both residential sewage and storm water to wastewater treatment facilities for decontamination. During heavy rain events the holding capacity of these systems can be exceeded and overflows of raw untreated wastewater are diverted and released into neighboring waterbodies. Sewer overflows are a common issue to receiving waterbodies in Northern Kentucky, contaminating surface waters used for drinking water, while creating a public health risk to individuals using these waterbodies for recreation. Recognizing this, Sanitation District No. 1 (SD1) sought to investigate the potential benefits of using a treatment-based constructed wetland for improving water quality along a segment of the Banklick Creek, one of 16 watersheds within SD1′s service area in Kentucky. Banklick Creek watershed within the Licking River Basin (Fig. 1a) has been identified as a 303 (d) impaired waterbody due to the numerous CSOs, SSOs as well as nonpoint sources thought to influence the observed water quality (elevated nutrients and indicator bacteria) within the watershed (US Environmental Protection Agency, 2018a). In 2011, SD1 completed construction of a 12-acre pilot wetland adjacent to Banklick Creek in Northern Kentucky, diverting a portion of the creek into the wetland in an attempt to improve downstream water quality (Scott et al., 2013). The use of constructed wetlands to improve recreational water quality is a relatively new application for this treatment technology. Diverting a portion of an impaired waterway through a treatment wetland could allow for the reduction of biological and chemical contaminants, improving downstream water quality upon discharge from the wetland. Because the application of treatment wetlands for this purpose has not been thoroughly investigated to date, an in-depth analysis and characterization of such systems is greatly needed.
Figure 1a.
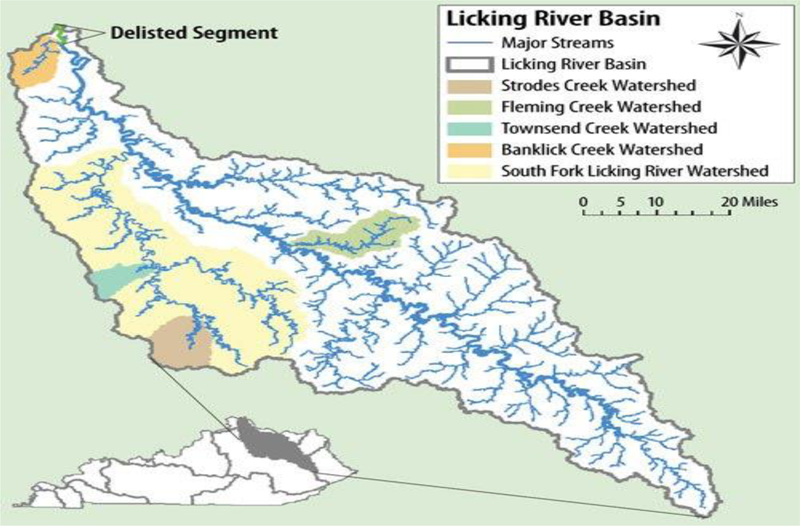
Licking River Basin overview. Banklick Creek Watershed highlighted in orange.
In addition to providing the expected benefits of water treatment mentioned above, wetlands provide many ancillary benefits to both humans and wildlife such as aesthetic improvements, habitat restoration, and the addition of public use areas. Because constructed wetlands closely mimic natural wetlands, it is no surprise that they attract a wide variety of wildlife. Depending on the amount and species of wildlife present, wetland treatment efficacy could be negatively affected due to wildlife activities (sediment disruption, direct defecation, etc.). SD1 has observed elevated fecal indicator microbial densities in one (east cell) of the two wetland treatment cells of the Banklick Creek Wetland (bird deterrents have been installed in west cell) leading scientist to speculate that elevated bird activity could be impacting the effectiveness of wetland treatment processes (Scott et al., 2013). SD1 has monitored water quality within the Banklick Wetland using standard tests for E. coli (Colilert®). These tests do not reveal the source of the microbial contamination, nor do they provide direct measurements of potential microbial pathogens. The goal of our research was to: 1) monitor levels of both traditional (E. coli and enterococci) and alternative (somatic and F+ coliphage, Clostridium spp. and Clostridium perfringens) culturable fecal indicators and genetic markers for Campylobacter, E. coli, and enterococci through select sites within the wetland treatment system and; 2) determine the sources of pollution present at the Banklick Creek Wetland by applying microbial source tracking (MST) assays for genetic markers for both human (HF183) and avian inputs (GFD).
Materials and Methods
Study Site
Banklick Creek Wetland encompasses a 12-acre land area adjacent to the Banklick Creek located in Fort Wright, Kentucky (39°01′14.2″N 84°31′42.1″W). Water from the Banklick Creek is pumped into the wetland using a series of variable-frequency pumps that are programmed by a logic controller whose operation is dictated by water levels of the Banklick Creek. To ensure flow is not disrupted in the Banklick Creek, the wetland’s pumping system is designed to withdraw only when creek flow is between 1.5 ft3/s and 31 ft3/s. Pumping rate is a graduated function of creek flow with minimum pumping rates of 0.5 ft3/s to maximum rates of 7 ft3/s. A series of three pumps direct creek water into an equalization basin (0.7 acre), where water is stored to allow for removal of suspended solids while also equalizing the flow of water prior to entering the wetland treatment channels. From the equalization basin, water is separated and gravity fed through a series of perforated pipes into one of two wetland treatment channels of approximately 3 acres each. During the 2017 operational season, the west treatment channel was decommissioned to perform routine maintenance, so sampling occurred in the remaining (east) treatment cell. Naturally occurring vegetative cover, along the banks of the east treatment channel consisted of various native and invasive species including; cattails (Typha latifolia), arrow arum (Peltandra virginica) and water primrose (Ludwigia grandiflora). From the treatment channel, water is gravity fed into an outlet basin (0.25 acre) before being discharged back into the Banklick Creek downstream of the pump inlet (Fig. 1b).
Figure 1b.
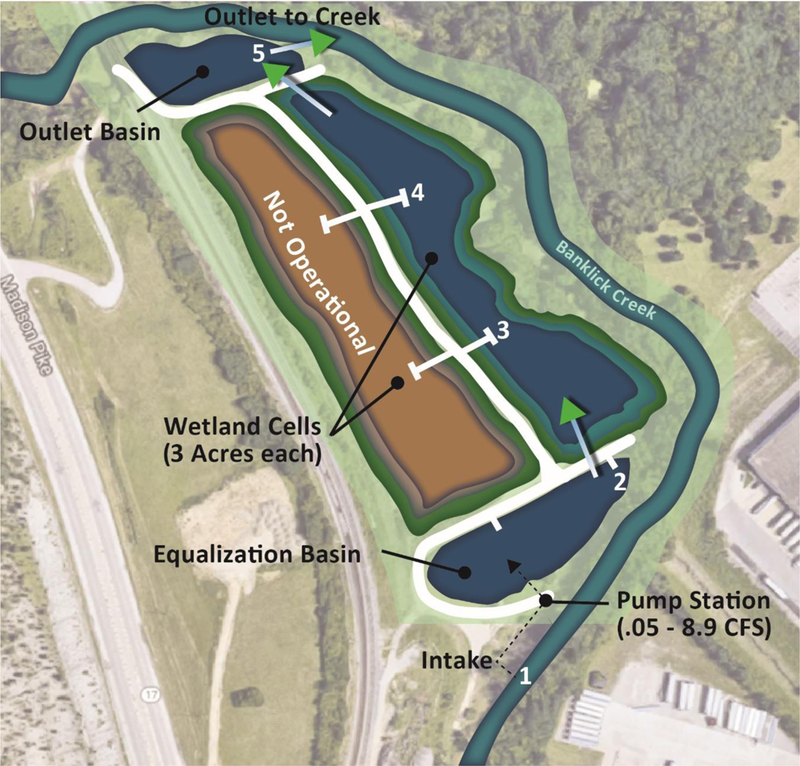
Banklick Creek Wetland site overview with sampling locations (1–5). Access roads and wetland cell sampling platforms are highlighted in white.
Sampling
The wetland was sampled on a weekly basis at five sampling locations through the Banklick Creek Wetland system (Fig. 1b), from June through September of 2017. Sampling locations were chosen to best assess effects of treatment within the wetland, beginning with the Banklick Creek source water (site #1) and ending with the outlet basin discharge (site #5). At each sampling point prior to sample collection, physical and chemical parameters of the water column (total suspended solids, pH, temperature, dissolved oxygen and conductivity) were measured using a YSI ProDSS multiparameter water quality meter (YSI Inc., Yellow Springs, OH), while total nitrogen and phosphorus levels were measured using a Hach hand-held colorimeter II (Hach, Loveland, CO) following manufacturer’s instructions. While taking these measurements, numbers of birds were recorded at each of the five wetland sample sites. Each week, sample volumes of 10 L were collected in sterilized carboys from each of the five sampling locations, over a 1-hour long sampling event, and immediately transported to the laboratory for processing.
Bacterial Indicators (Culture Based Analyses)
Water samples were passed through a 47 mm cellulose acetate filter with a nominal pore size of 0.45 μm. Samples for E. coli were processed according to EPA Method 1603 (US Environmental Protection Agency, 2009) using a modified mTEC agar (Difco, Sparks, MD). Plates were incubated at 35 °C for two hours, followed by overnight incubation at 44.5 °C. Red or magenta-colored colonies signifying E. colipresence were enumerated and expressed as CFU/100 mL. Samples for enterococciwere processed according to EPA Method 1600 (US Environmental Protection Agency, 1997), using mEI agar (Difco, Sparks, MD), incubated overnight at 41 °C, and all resulting colonies with blue halos indicative of enterococci growth were recorded and data were expressed as CFU/100 mL. Samples for Clostridium spp. and Clostridium perfringens were first pasteurized at 60 °C for 15 min to remove vegetative cells, and then were isolated by membrane filtration and cultured on mCP agar (Oxoid, Nepean, ON). Plates were inverted and incubated under anaerobic conditions for 18–24 h at 45 °C. All colonies having yellow/straw-colored appearance were enumerated as total Clostridium. Colonies were then exposed to ammonium hydroxide fumes, and all colonies turning pink/magenta were enumerated as Clostridium perfringens. All resulting Clostridium spp. positives were reported as CFU/100 mL.
Coliphage Enumeration
Samples from each of the wetland sampling sites were filtered in the laboratory using a previously described dead-end hollowfiber ultrafiltration and single agar layer (D-HFUF-SAL) method (McMinn et al., 2017). Briefly, 4L of water was passed through a single-use 15S Asahi Kasei Rexeed ultrafilter (Dial Medical Supply, Chester Springs, PA) using a peristaltic pump. Each filter was eluted using 200 mL of an elution solution (0.01% Tween 80, 0.01% sodium hexametaphosphate, 0.001% Antifoam Y-30) (Sigma-Aldrich, St. Louis, MO). The elution solution was circulated through the filter in a clockwise (1 min.), counter clockwise (1 min.) and finally a clockwise (1 min.) pattern. The resulting eluate (∼200 mL) was divided evenly (100 mL for somatic assay and 100 mL for F+ coliphage assay) and processed using the SAL procedure, as previously described (McMinn et al., 2017). Briefly, the filter eluate (100 mL) was mixed with equal parts of a 2X concentration of molten tryptic soy agar containing appropriate concentrations of log-phase host bacteria, antibiotics (nalidixic acid for host ΦX174 for or streptomycin/ampicillin for host MS2) and MgCl and spread over five 150 mm plates. Agar plates were incubated at 37 °C for 16–18 h, resulting coliphage plaques were enumerated the following day, and data were adjusted and expressed as PFU/100 mL.
Sample Processing Molecular Analytes
Duplicate 200 mL volumes of water from five sites at the Banklick Creek Wetland were vacuum-filtered through 0.45 μm Polyvinylidene difluoride (PVDF) membranes (Millipore, Burlington, MA) within six hours of collection. Each membrane was transferred to a Lysing Matrix A bead tube (MP Biomedicals, Santa Ana, CA), and the resulting filter tubes were stored at −80 °C until extraction. Additional 50 or 100 mL volumes of water were filtered through 0.4 μm polycarbonate membranes, which were subsequently transferred to corresponding glass bead tubes (GeneRite, North Brunswick, NJ), according to EPA Method 1609.1 (US Environmental Protection Agency, 2015) and stored at −80 °C until extraction.
Nucleic Acid Extraction
Nucleic acids were extracted from the PVDF filters using the AllPrep DNA/RNA Mini Kit (Qiagen, Germantown, MD), with an initial bead beating step. Briefly, after 600 μL RLT Plus buffer was added to each filter tube, the tube underwent bead beating (5000 reciprocations/minute) for 30 s, cooling on ice for 5 min, exposure to another round of bead beating, and centrifugation for 5 min at 12,000 RPM. The supernatant was then processed though the AllPrep Kit following manufacturer’s instructions. Crude DNA extracts for enterococci, were obtained from the polycarbonate filter tubes, as described in EPA Method 1609.1 (US Environmental Protection Agency, 2015). In brief, polycarbonate filters were bead beaten for 30 s, then centrifuged at 12,000 × g for 1 min. Four hundred microliters of supernatant were transferred to a 1.5 mL tube and centrifuged for an additional 5 min as 12,000×g. The clarified supernatant of approximately 350 μL was stored at 4 °C for under 24 h prior to analysis.
qPCR Analysis
Campylobacter genus (Lund et al., 2004), human-associated Bacteroides species (HF183) (Green et al., 2014), avian-associated Helicobacter species (GFD) (Green et al., 2012) and Escherichia coli (Chern et al., 2011) gene copy levels in the AllPrep DNA extracts were determined by qPCR, along with enterococci gene copy levels (US Environmental Protection Agency, 2015) in the crude DNA extracts. The 20 μL qPCR reactions were prepared with Power SYBR-Green Master Mix (Bio-Rad, Hercules, CA) for the GFD assay and with TaqMan Environmental Master Mix 2.0 (Life Technologies, Grand Island, NY) for the Campylobacter, HF183, and E. coli assays, and were supplemented with 0.2 mg/mL of BSA. The 25 μL qPCR reactions for the enterococci assay also utilized the TaqMan Master Mix with 0.2 mg/mL of BSA. Individual primer and probe sequences for each assay are listed in Appendix Table 1. The thermocycler program for the HF183, Campylobacter, enterococci, and Sketa22 salmon assays was as follows: 2 min at 50 °C, and 10 min at 95 °C; 40 cycles of 15 s at 95 °C, and 1 min at 60 °C. The thermocycler program for the E. coli assay was as follows: 2 min at 50 °C, and 10 min at 95 °C; 40 cycles of 15 s at 95 °C, and 1 min at 56 °C. The thermocycler program used to perform the GFD assay was: 2 min at 50 °C, and 10 min at 95 °C; 40 cycles of 15 s at 95 °C, 30 s at 57 °C, and 30 s at 72 °C.
Quality Control qPCR
As described in EPA Method 1609.1 (US Environmental Protection Agency, 2015), two quality control assays were incorporated into the enterococci assay. Briefly, salmon DNA added to the crude extract was used as a sample processing control (SPC) to assess sample matrix interference. The SPC was amplified with the Sketa22 primers and probe under the same qPCR reaction conditions as the enterococci assay. In addition, a second probe UC1P1, or internal amplification control (IAC), was included in each enterococci assay reaction to determine the presence of inhibition during qPCR amplification. SPC and IAC offset Ct values for each sample were used to classify the enterococci results as acceptable, or unacceptable due to low target sequence recoveries and/or significant qPCR inhibition. Offset Ct values were calculated by subtracting the mean Ct of the calibrator samples from the mean Ct estimate of each sample. Sample results were considered unacceptable for an SPC offset Ct of >3.0 and/or an IAC offset Ct of >1.5. IAC assay results were also required to meet the established acceptability criterion relative to the filter blank samples. No samples were determined to have SPC/IAC results outside of acceptable criteria during the study (data not shown).
Data analysis
Significant changes in concentrations of microbial targets (traditional fecal indicators both culture and molecular, alternative fecal indicators, pathogens and MST markers) between wetland treatment stages were determined using a one-way ANOVA (GraphPad StatMate version 2.0 Windows, San Diego, CA). Correlations were identified between microbial target concentrations, treatment stage and physical chemical measurements using the Pearson correlation coefficient in GraphPad StatMate.
Results
Monthly Precipitation and Pumping Frequency
Monthly rainfall amounts during the 2017 operational season were above average for June and July + 1.22″ and + 2.05″, respectively, while in August and September, lower than average precipitation was recorded, −1.08″ and −0.22″, respectively (NOAA, 2018). During the 2017 season, the wetland pumping system was in operation a total of 20 days or 67% of the total days in June, 20 days (65%) in July, 24 days (77%) in August and 26 days (87%) in September. Through the 2017 operational season, a total of 116 million gallons of water of Banklick Creek water was pumped into and treated though the wetland.
Measured Water Parameters
Monthly temperature averages remained relatively consistent throughout each site within the wetland measuring averaging 23 ± 0.84 °C. Average dissolved oxygenconcentrations for all sites was 71 ± 13% with the highest levels recorded at site #1 (83 ± 4.80%) and at site #2 (85 ± 15%). Conductivity readings averaged 533.4 ± 18 μs/cm for all sites during the operational season, with peak average levels recorded at site #5 (557 ± 109 μs/cm). Average pH for the wetland was 7.83 ± 0.17, with the highest recorded averages observed in site #1 (8.04 ± 0.2) with general trends of pH decreases through the remaining wetland sites. Average turbidity readings recorded for the wetland was 15 ± 4.3 FNU, with the lowest levels of turbidity averaging 8.0 ± 5.6 FNU recorded at site #4. Total nitrogen averages recorded at the wetland site were 0.13 ± 0.03 ppm, with levels gradually increasing in concentration through later stages of the wetland treatment system. Total phosphorus averages recorded for the site were 0.27 ± 0.07 ppm, and unlike total nitrogen, phosphorus levels fluctuated in concentration through the wetland treatment system. Canada goose (Branta canadensis) and Mottled duck (Anas fulvigula) were the most commonly occurring species of waterfowl. The lowest bird activity totals for the season were recorded at site #1 (1 bird) and site #5 (0 birds), while the highest numbers were recorded at site #3 (33 birds) and site #4 (16 birds).
Traditional Fecal Indicators
Concentrations of traditional indicators (enterococci and E. coli) measured by both culture and molecular techniques followed similar concentration pattern shifts through the wetland treatment system (Fig. 2). Indicator levels recorded at site #1 (Banklick Creek at intake) were used as baseline concentrations and to evaluate later stages of wetland treatment (Fig. 1b). Both E. coli culture and genetic markerconcentrations did not significantly change through the different stages of the wetland treatment system with averages ranging from 2.37 ± 0.61 to 2.91 ± 0.63 log10CFU (100% sample positives) and from 3.07 ± 0.55 to 3.53 ± 0.47 log10 gene copies/100 mL (92% of samples with measurable signal), respectively through the operational season. Average concentrations of enterococci through the wetland treatment for both culture and molecular signal ranged from 1.91 ± 0.56 to 2.82 ± 0.55 log10 CFU/100 mL, with 100% sample positives for culture and average ranges of 3.40 ± 0.41–3.96 ± 0.41 log10 gene copies/100 mL, with 100% of samples with measurable molecular signal. Average enterococci culture and molecular levels were found to significantly decrease (−0.611 log10 CFU per 100 mL, P = 0.0320, and −0.560 log10 gene copies per 100 mL, P = 0.0284, respectively) in concentrations measured at sample site #1 (creek) compared to concentrations observed at site #2, while a significant increase of culture based concentrations were observed from site #2 to site #5 (+0.909 log10 CFU per 100 mL, P = 0.0003).
Figure 2.
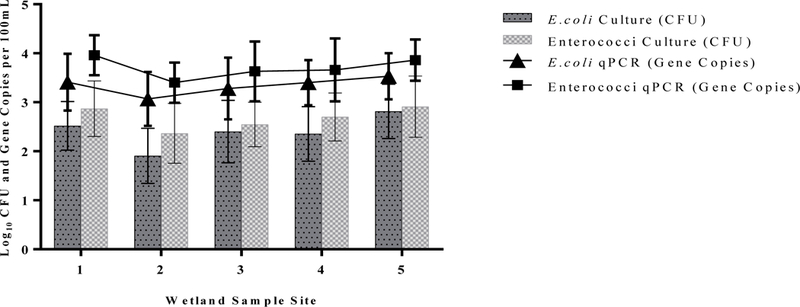
Average Log10 concentrations, error bars depict standard deviation of culturable fecal indicators (bars) and qPCR fecal indicator signals (lines) in Banklick Creek Wetland during the 2017 operational season for each of the 5 sample sites.
Alternative Fecal Indicators
Levels of infectious somatic and F+ coliphage did not significantly change through the wetland treatment system (Fig. 3). Average concentrations of somatic coliphage ranged from 1.04 ± 0.52 log10 to 1.65 ± 1.04 log10 PFU/100 mL and F+ coliphage concentrations ranged from 0.063 ± 1.12 log10 to 0.14 ± 0.15 log10 PFU/100 mL during the operating season. Like coliphage, levels of total Clostridium and Clostridium perfringens, were not found to significantly fluctuate between sites within the wetland, with average concentrations ranging from; 1.47 ± 0.27 log10 to 1.57 ± 0.30 log10 CFU/100 mL and 0.65 ± 0.50 log10 and 0.83 ± 0.48 log10 CFU/100 mL, respectively.
Figure 3.
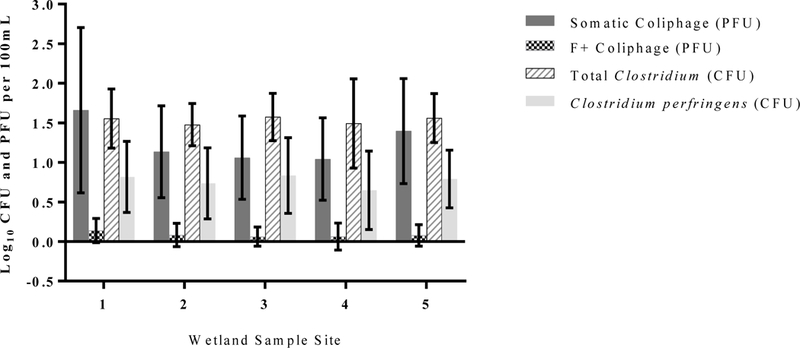
Bars represent average log10 concentrations, and error bars depict standard deviation of culturable alternative fecal indicators in Banklick Creek Wetland during the 2017 operational season for each of the 5 sample sites.
Microbial Source Tracking Markers
Human-associated MST marker HF183 (Table 1) and avian-associated marker GFD (Table 2) were used to compare levels of source signal from the intake at site #1 through the wetland treatment system. For the HF183 marker, quantifiable levels in the creek at site #1 averaged; 2.60 ± 0.73 log10 gene copies/100 mL, with 8 of 15 samples (53%) collected having quantifiable levels of human marker present. At site #2, only 3 of 15 (20%) samples taken during the study had quantifiable levels of human marker detected averaging; 2.66 ± 0.40 log10 gene copies/100 mL. For samples taken downstream of site #2, only a single sample collected on 8/9/2017 (site #3), contained a quantifiable human signal (2.31 ± 0.28 average gene copies/100 mL). Overall, frequency of detection of the HF183 marker was found to be significantly higher at site #1, compared to levels determined at sites #3, #4 and #5 (P value range: 0.0016–0.0003).
Table 1.
HF183 qPCR (log10 gene copies/100mL) Concentrations Detected at 5 Sample Locations
| Updated on 4/8/18 |
|||||
|---|---|---|---|---|---|
| Date | Site 1 | Site 2 | Site 3 | Site 4 | Site 5 |
| 6/14/2017 | |||||
| 6/21/2017 | |||||
| 6/28/2017 | 2.97±0.08 | ||||
| 7/12/2017 | 2.06±0.02 | ||||
| 7/17/2017 | |||||
| 7/26/2017 | |||||
| 8/2/2017 | 2.31±0.06 | 2.20±0.14 | |||
| 8/9/2017 | 3.28±0.10 | 2.95±0.10 | 2.32±0.28 | ||
| 8/16/2017 | 2.10±0.11 | ||||
| 8/23/2017 | 4.04±0.01 | 2.82±0.09 | |||
| 8/30/2017 | 2.06±0.07 | ||||
| 9/5/2017 | |||||
| 9/13/2017 | 2.28±0.15 | ||||
| 9/20/2017 | |||||
Table 2.
GFD qPCR (average log10 gene copies ± SD/100mL) Concentrations Detected at 5 Wetland Sample Sites. Shaded areas represent samples with non-detectable target.
| Date | Site 1 | Site 2 | Site 3 | Site 4 | Site 5 |
|---|---|---|---|---|---|
| 6/14/2017 | |||||
| 6/21/2017 | 2.82±0.26 | ||||
| 6/28/2017 | 2.71±0.01 | ||||
| 7/13/2017 | 2.42±0.07 | 3.85±0.00 | |||
| 7/17/2017 | 2.71±1.45 | 2.59±0.07 | 2.59±0.07 | 2.73±0.18 | |
| 7/26/2017 | 3.04±0.00 | 2.32±0.02 | |||
| 8/2/2017 | |||||
| 8/9/2017 | 2.76±0.04 | 2.65±0.14 | |||
| 8/16/2017 | 3.09±0.08 | 2.88±0.07 | |||
| 8/23/2017 | 2.28±0.25 | 3.09±0.04 | |||
| 8/30/2017 | 2.39±0.24 | 2.75±0.07 | 2.69±0.03 | ||
| 9/5/2017 | 2.35±0.03 | 2.98±0.10 | 2.23±0.43 | ||
| 9/13/2017 | 2.19±0.58 | 2.73±0.02 | 4.08±0.05 | 4.08±0.05 | |
| 9/20/2017 | 2.29±0.15 | 2.71±0.02 | 2.43±0.21 | 3.09±0.01 | |
| 9/27/2017 | 2.43±0.26 | 2.97±0.14 | 3.93±0.11 | ||
A total of 4 of 15 samples (27%) collected had measurable concentrations of the GFD marker at site #1, averaging; 2.49 ± 0.41 log10 gene copies/100 mL. At site #2, the GFD signal was detected in only 2 of 15 samples (13%), averaging 2.50 ± 0.30 log10 gene copies/100 mL of the avian marker. At site #3, 7 of 17 samples (47%) had measurable quantities of GFD signal averaging 2.57 ± 0.17 log10 gene copies/100 mL. Quantifiable levels of the GFD marker were more frequently detected in downstream wetland sites #4 and #5, with 8 of 15 samples (53%) averaging 2.96 ± 0.50 log10 gene copies/100 mL, and 12 of 15 samples (80%) averaging 3.02 ± 0.62 log10 gene copies/100 mL positive for avian signal, respectively. Levels of GFD signal were found to be significantly lower at sites #1 and #2 than those recorded at site #5, P = 0.0033 (mean difference of −1.75 log10 gene copies/100 mL) and 0.0003 (mean difference of −2.08 log10 gene copies/100 mL), respectively.
Potential Pathogen Concentrations
Levels of Campylobacter averaged 2.78 ± 0.37 log10 gene copies/100 mL at site #1 and were found to increase through the treatment system, with peak concentrations occurring at site #5, averaging 3.41 ± 0.58 log10 gene copies/100 mL (Table 3). Overall, measurable Campylobacter signal was present in 57% of samples collected from the wetland. Levels of Campylobacter signal were found to be significantly lower at site #1 (27% of samples positive) than those of both #4 (73% of samples positive, +1.51 log10gene copies per 100 mL) and #5 (93% of samples positive, +2.44 log10 genomic copies per 100 mL), were P = 0.024 and P = <0.0001, respectively. Significant increases in Campylobacter signal were also recorded from levels at site #2 to those determined in both #4 (P = 0.02, +1.57 log10 gene copies per 100 mL) and at site #5 (P = <0.0001, +2.50 log10 gene copies per 100 mL).
Table 3.
Campylobacter qPCR (average log10 gene copies ± SD/100mL) Concentrations Detected at 5 Wetland Sample Sites. Shaded areas represent samples with non-detectable target.
| Date | Site 1 | Site 2 | Site 3 | Site 4 | Site 5 |
|---|---|---|---|---|---|
| 6/14/2017 | 2.74±0.06 | 3.28±0.01 | 2.82±0.08 | ||
| 6/21/2017 | 3.65±0.07 | 2.8±0.10 | |||
| 6/28/2017 | 3.22±0.36 | 3.57±0.07 | 3.48±0.10 | ||
| 7/13/2017 | 2.38±0.20 | 2.79±0.00 | 2.82±0.05 | 4.55±0.05 | |
| 7/17/2017 | 2.23±0.06 | 4.34±0.11 | 3.40±0.03 | 3.47±0.01 | |
| 7/26/2017 | 3.26±0.07 | ||||
| 8/2/2017 | 2.43±0.09 | 2.69±0.13 | 2.63±0.07 | ||
| 8/9/2017 | 2.34±0.07 | 3.48±0.13 | 2.84±0.07 | ||
| 8/16/2017 | 2.70±0.04 | 3.14±0.08 | |||
| 8/23/2017 | 3.20±0.10 | 3.75±0.11 | |||
| 8/30/2017 | 2.58±0.02 | 2.59±0.01 | 3.05±0.07 | ||
| 9/5/2017 | 2.59±0.12 | 2.94±0.21 | 2.71±0.01 | ||
| 9/13/2017 | 2.98±0.17 | 3.67±0.03 | 2.70±0.30 | 4.15±0.03 | |
| 9/20/2017 | 2.75±0.17 | 3.09±0.00 | 2.91±0.10 | 3.44±0.07 | |
| 9/27/2017 | 3.21±0.14 | 4.21±0.04 | |||
Predictive Relationship between Microorganisms
Microbial and physical/chemical measurements pooled for all sites were subjected to Pearson product momentum correlation analyses to identify existing positive or negative correlations within the data set. The strength of correlations was divided into 3 categories based on the strength of the resulting statistical association; Strong 0.7–1, Moderate 0.4–0.7 and Weak <0.4. All associations between physical/chemical measurements and microbial data resulted in either a statistically weak or nonexistent correlation. F+ coliphage, Clostridium spp., Clostridium perfringens, and human marker HF183 had no statistically significant correlations with other microbial or water quality data parameter. Both E. coli and enterococci culture had moderate to strong positive associations with their respective qPCR data measurements (P = <0.0001, R2 = 0.502 and P = <0.0001, R2 = 0.712, respectively). Levels of traditional fecal indicators, when compared to one another, displayed strong positive correlations for both culturable (P = <0.0001, R2 = 0.758) and molecular (P = <0.0001, R2 = 0.713) measurements. For alternative fecal indicators, somatic coliphage was found to have a moderately strong positive correlation to levels of E. coli determined by culture (P = <0.0001, R2 = 0.486). Lastly, there was a moderately strong, positive correlation between the presence of the Campylobacter qPCR signal and that of the GFD avian marker, P = <0.0001, R2 = 0.592.
Discussion
Wetland treatment systems have shown effectiveness in reducing levels of microbial and chemical contamination, in storm water runoff and wastewater prior to environmental discharge (Kaseva, 2004, Keffala and Ghrabi, 2005, Knight et al., 2000, Matamoros and Rodriguez, 2017, Walker and Hurl, 2002). The use of a treatment wetland for reducing microbial contamination levels in an impacted waterway is still a new and relatively unexplored area for this application (Jia et al., 2016, Lin et al., 2015). During this study, we monitored wetland treatment by measuring reductions in average concentrations of both traditional and alternative fecal indicators, which are bacteria or viruses found in fecal material of most warm-blooded animals, and are typically used for evaluating the sanitary condition of recreational waters.
For traditional and alternative fecal indicators, some treatment effect was observed between inlet (site #1) and the equalization basin (site #2). This observation is in contrast with the remaining, later stages of wetland treatment where concentrations either remained consistent or increased. Although it is possible that increases in fecal indicator concentrations in the wetland could be attributed to regrowth within the wetland system (McLain and Williams, 2008, VanKempen-Fryling et al., 2015), elevated bird activity recorded corresponded to wetland treatment stages where increases in fecal indicator concentrations were observed. Because constructed wetlands are designed to mimic natural wetlands, providing rich biodiversity and ideal habitats (vegetative cover, plentiful food sources, undisturbed nestingenvironments), it is not surprising that they attract all types of native wildlife including waterfowl (Beutel et al., 2013, Rea et al., 2015,; Shellenbarger et al., 2008). High bird activity can be a source of fecal contamination within these man-made systems, impairing the ability of a treatment wetland to effectively reduce microbial contamination levels (Andersen et al., 2003, Orosz-Coghlan et al., 2006, Shellenbarger et al., 2008). Unsuccessful reduction in fecal indicator levels could be attributed to wildlife inputs into the wetland either by direct defecation (Grant et al., 2001), resuspension of bacterial laden sediment (Karim et al., 2004, Rea et al., 2015) or indirectly through runoff (Evanson and Ambrose, 2006). When using general fecal indicator concentrations to measure wetland treatment efficacy, fecal microbes actively deposited by birds within the system can lead to false assumptions as to the overall wetland system effectiveness at reducing microbial contamination. This reasoning is supported by the idea that the traditional and alternative fecal indicators analyzed in this study are not source specific (found in all warm-blooded animals) and waterfowl are known to carry levels of these indicators (Middleton and Ambrose, 2005, Rea et al., 2015).
Monitoring levels of bird-associated microorganisms such as Campylobacter can give an indication as to the source of contamination especially in waterbodies having high bird activity, but more precise analyses, such as MST, can identify a panoply of different fecal sources. MST describes a science based approach capable of detecting gene sequences of certain fecal microorganisms that are specific to a host organism (Harwood et al., 2014). MST markers have historically been used for identifying sources of fecal contamination in a variety of matrices such as wastewater (Mayer et al., 2016, Mayer et al., 2015), groundwater (Diston et al., 2015, Tran et al., 2015) and soil and sediments (Madeja, 2015, Marti et al., 2017, Reischer et al., 2008), but most commonly for cases of recreational and beach water quality impairment (Brownell et al., 2007, Harwood et al., 2014). For this study, MST markers (specific to human and avian fecal contamination) were employed to identify possible sources of contamination in the inflow to the Banklick Creek Wetland as well as within the system itself. Although this is not the first application of MST for the identification of possible fecal sources affecting a constructed wetland (Orosz-Coghlan et al., 2006), it is the first to use source specific markers HF183 (human) and GFD (avian) for fecal input source identification and confirmation.
Levels of HF-183 marker were most consistently detected in creek inflow samples entering the wetland and in early stages of wetland treatment. SD1 has identified numerous SSO discharges and septic systems upstream of the wetland inlet along Banklick Creek that are likely a source of the human signal detected during this study (Scott et al., 2013). Notably, comparing the frequency of detection of the HF-183 signal from inlet samples, it was observed to decrease by 33% in the equalization basin, and a further 46% at the upper wetland cell, while disappearing entirely from all later stages of wetland treatment. The successive loss of the HF-183 signal through the wetland signifies that treatment is occurring within the wetland resulting in the removal of human fecal signal.
Avian marker GFD and Campylobacter-specific qPCR assays were employed to assess the presence and level of fecal contamination due to waterfowl activity. Levels of both GFD and Campylobacter signal were detected at moderate frequency in inlet samples, 27% and 33%, respectively. The frequency of detection of the GFD marker and Campylobacter signal in these samples could be attributed to the wetlands being downstream from a community park (adjacent to the Banklick Creek), which commonly attracts local waterfowl with its abundant vegetative cover. Detection frequencies for both GFD and Campylobacter signal within the wetland increased in the upper wetland channel (site #3) and through the remaining downstream treatment stages, with peak detection frequencies observed at the wetland outlet (site #5). Increases in avian-associated signal (GFD) and avian-related bacterial species (Campylobacter) signal from levels observed in the intake suggest active avian fecal input from within the wetland system. Locations of bird activity observed at the wetland during weekly sampling events corresponded to the areas of the wetland having increased bird signal (upper and lower treatment channel) providing additional evidence of waterfowl contaminating the wetlands directly.
SD1 has historically monitored the levels of E. coli at the wetland following its installation in 2011, observing that levels of these bacteria remained relatively consistent within the east wetland treatment cell. Although SD1 researchers had observed high bird activity at this site, they were unable to definitively point to the actual source of contamination impairing perceived wetland treatment effect (Scott et al., 2013). Adapting emerging technologies such as MST enable the identification of likely contamination sources within open water treatment systems such as wetlands, aiding in effective management and remediation efforts.
Conclusions
A performance assessment was conducted during the summer of 2017 on a constructed variable-flow wetland to evaluate removal of microbiological signal (culturable and molecular) through the different stages of treatment. Key findings include:
Both culturable and molecular based FIB levels in the wetland remained relatively consistent during the experiment regardless of treatment stage.
In the beginning stages of treatment (intake from Banklick Creek and equalization basin), human marker HF-183 was most commonly detected, but was absent from all later stages of wetland treatment.
Loss of HF-183 signal within the Banklick Wetland suggest treatment is occurring at the site and that human input is likely not the source of elevated FIB levels observed in the wetland.
The presence of avian molecular marker GFD and avian associated Campylobacterspp. significantly correlated, with both signals increasingly detected at later treatment stages corresponding to areas of elevated bird activity, likely the source of elevated FIB levels observed.
The application of MST to distinguish avian and human contamination within a wetland is the first of its kind and was an effective means to establish sources of contamination while also assisting with future remediation efforts at the site.
Supplementary Material
Footnotes
Disclaimer
The United States Environmental Protection Agency through its Office of Research and Development funded and managed the research described here. It has been subjected to Agency’s administrative review and approved for publication. The views expressed in this article are those of the author(s) and do not necessarily represent the views or policies of the U.S. Environmental Protection Agency. Mention of trade names or commercial products does not constitute endorsement or recommendation for use.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Andersen DC, Sartoris JJ, Thullen JS, Reusch PG, 2003. The effects of bird use on nutrient removal in a constructed wastewater-treatment wetland. Wetlands 23 (2), 423–435. [Google Scholar]
- Beutel MW, Whritenour V, Brouillard E, 2013. Fecal coliform removal in a lightly loaded surface-flow constructed treatment wetland polishing agricultural runoff. Water Sci. Technol. 68 (4), 909–915. [DOI] [PubMed] [Google Scholar]
- Brownell MJ, Harwood VJ, Kurz RC, McQuaig SM, Lukasik J, Scott TM, 2007. Confirmation of putative stormwater impact on water quality at a Florida beach by microbial source tracking methods and structure of indicator organism populations. Water Res. 41 (16), 3747–3757. [DOI] [PubMed] [Google Scholar]
- Cheng SP, Grosse W, Karrenbrock F, Thoennessen M, 2002. Efficiency of constructed wetlands in decontamination of water polluted by heavy metals. Ecol. Eng. 18 (3), 317–325. [Google Scholar]
- Chern EC, Siefring S, Paar J, Doolittle M, Haugland RA, 2011. Comparison of quantitative PCR assays for Escherichia coli targeting ribosomal RNA and single copy genes. Lett. Appl. Microbiol. 52 (3), 298–306. [DOI] [PubMed] [Google Scholar]
- Diston D, Sinreich M, Zimmermann S, Baumgartner A, Felleisen R, 2015. Evaluation of molecular- and culture-dependent MST markers to detect fecal contamination and indicate viral presence in good quality groundwater. Environ. Sci. Technol. 49 (12), 7142–7151. [DOI] [PubMed] [Google Scholar]
- Dorfman M, 2004. Swimiming in Sewage, The Growing Problem of Sewage Pollution and How the Bush Administration is Putting Our Health and Environment. at Risk. [Google Scholar]
- Evanson M, Ambrose RF, 2006. Sources and growth dynamics of fecal indicator bacteria in a coastal wetland system and potential impacts to adjacent waters. Water Res. 40 (3), 475–486. [DOI] [PubMed] [Google Scholar]
- Grant SB, Sanders BF, Boehm AB, Redman JA, Kim JH, Mrse RD, Chu AK, Gouldin M, McGee CD, Gardiner NA, Jones BH, Svejkovsky J, Leipzig GV, Brown A, 2001. Generation of enterococci bacteria in a coastal saltwater marsh and its impact on surf zone water quality. Environ. Sci. Technol. 35 (12), 2407–2416. [DOI] [PubMed] [Google Scholar]
- Green HC, Dick LK, Gilpin B, Samadpour M, Field KG, 2012. Genetic markers for rapid PCR-based identification of gull, Canada goose, duck, and chicken fecal contamination in water. Appl. Environ. Microbiol. 78 (2), 503–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green HC, Haugland RA, Varma M, Millen HT, Borchardt MA, Field KG, Walters WA, Knight R, Sivaganesan M, Kelty CA, Shanks OC, 2014. Improved HF183 quantitative real-time PCR assay for characterization of human fecal pollution in ambient surface water samples. Appl. Environ. Microbiol. 80 (10), 3086–3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood VJ, Staley C, Badgley BD, Borges K, Korajkic A, 2014. Microbial source tracking markers for detection of fecal contamination in environmental waters: relationships between pathogens and human health outcomes. FEMS Microbiol. Rev. 38 (1), 1–40. [DOI] [PubMed] [Google Scholar]
- Hench KR, Bissonnette GK, Sexstone AJ, Coleman JG, Garbutt K, Skousen JG, 2003. Fate of physical, chemical, and microbial contaminants in domestic wastewater following treatment by small constructed wetlands. Water Res. 37 (4), 921–927. [DOI] [PubMed] [Google Scholar]
- Jia Z, Tang S, Luo W, Hai Y, 2016. Water quality improvement through five constructed serial wetland cells and its implications on nonpoint-source pollution control. Hydrol. Sci. J. J. Des. Sci. Hydrol. 61 (16), 2946–2956. [Google Scholar]
- Karim MR, Manshadi FD, Karpiscak MM, Gerba CP, 2004. The persistence and removal of enteric pathogens in constructed wetlands. Water Res. 38 (7), 1831–1837. [DOI] [PubMed] [Google Scholar]
- Kaseva ME, 2004. Performance of a sub-surface flow constructed wetland in polishing pre-treated wastewater-a tropical case study. Water Res. 38 (3), 681–687. [DOI] [PubMed] [Google Scholar]
- Keffala C, Ghrabi A, 2005. Nitrogen and bacterial removal in constructed wetlands treating domestic waste water. Desalination 185 (1–3), 383–389. [Google Scholar]
- Knight RL, Payne VWE, Borer RE, Clarke RA, Pries JH, 2000. Constructed wetlands for livestock wastewater management. Ecol. Eng. 15 (1–2), 41–55. [Google Scholar]
- Kurzbaum E, Kirzhner F, Armon R, 2012. Improvement of water quality using constructed wetland systems. Rev. Environ. Health 27 (1), 59–64. [DOI] [PubMed] [Google Scholar]
- Lin JL, Tu YT, Chiang PC, Chen SH, Kao CM, 2015. Using aerated gravel-packed contact bed and constructed wetland system for polluted river water purification: a case study in Taiwan. J. Hydrol. 525, 400–408. [Google Scholar]
- Lund M, Nordentoft S, Pedersen K, Madsen M, 2004. Detection of Campylobacter spp. in chicken fecal samples by real-time PCR. J. Clin. Microbiol. 42 (11), 5125–5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeja J, 2015. A new tool to trace past human presence from lake sediments: the human-specific molecular marker Bacteroides strain HF 183. J. Quat. Sci. 30 (4), 349–354. [Google Scholar]
- Marti R, Ribun S, Aubin JB, Colinon C, Petit S, Marjolet L, Gourmelon M, Schmitt L, Breil P, Cottet M, Cournoyer B, 2017. Human-driven microbiological contamination of benthic and hyporheic sediments of an intermittent peri-urban river assessed from MST and 16S rRNA genetic structure analyses. Front. Microbiol. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matamoros V, Rodriguez Y, 2017. Influence of seasonality and vegetation on the attenuation of emerging contaminants in wastewater effluent-dominated streams A preliminary study. Chemosphere 186, 269–277. [DOI] [PubMed] [Google Scholar]
- Mayer RE, Bofill-Mas S, Egle L, Reischer GH, Schade M, Fernandez-Cassi X, Fuchs W, Mach RL, Lindner G, Kirschner A, Gaisbauer M, Piringer H, Blaschke AP, Girones R, Zessner M, Sommer R, Farnleitner AH, 2016. Occurrence of human-associated Bacteroidetes genetic source tracking markers in raw and treated wastewater of municipal and domestic origin and comparison to standard and alternative indicators of faecal pollution. Water Res. 90, 265–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer RE, Vierheilig J, Egle L, Reischer GH, Saracevic E, Mach RL, Kirschner AK, Zessner M, Sommer R, Farnleitner AH, 2015. Automated sampling procedures supported by high persistence of bacterial fecal indicators and Bacteroidetes genetic microbial source tracking markers in municipal wastewater during short-term storage at 5 Degrees C. Appl. Environ. Microbiol. 81 (15), 5134–5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLain JE, Williams CF, 2008. Seasonal variation in accurate identification of Escherichia coli within a constructed wetland receiving tertiary-treated municipal effluent. Water Res. 42 (15), 4041–4048. [DOI] [PubMed] [Google Scholar]
- McMinn BR, Huff EM, Rhodes ER, Korajkic A, 2017. Concentration and quantification of somatic and F+ coliphages from recreational waters. J. Virol. Methods 249, 58–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton JH, Ambrose A, 2005. Enumeration and antibiotic resistance patterns of fecal indicator organisms isolated from migratory Canada geese (Branta canadensis). J. Wildl Dis. 41 (2), 334–341. [DOI] [PubMed] [Google Scholar]
- NOAA, 2018, Climate Data Online Search. [Google Scholar]
- Orosz-Coghlan PA, Rusin PA, Karpiscak MM, Gerba CP, 2006. Microbial source tracking of Escherichia coli in a constructed wetland. Water Environ. Res. 78 (3), 227–232. [DOI] [PubMed] [Google Scholar]
- Rea CL, Bisesi MS, Mitsch W, Andridge R, Lee J, 2015. Human health-related ecosystem services of avian-dense coastal wetlands adjacent to a Western Lake Erie swimming beach. EcoHealth 12 (1), 77–87. [DOI] [PubMed] [Google Scholar]
- Reischer GH, Haider JM, Sommer R, Stadler H, Keiblinger KM, Hornek R, Zerobin W, Mach RL, Farnleitner AH, 2008. Quantitative microbial faecal source tracking with sampling guided by hydrological catchment dynamics. Environ. Microbiol. 10 (10), 2598–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott BB, Gibson JP, Frye C, 2013, Moving the Water Quality Needle: Effectiveness of the Banklick Creek Regional Wetland for Water Quality Improvement as Part of an Integrated Watershed Approach to Water Resource Management, pp. 3133–3145, Water Environment Federation. [Google Scholar]
- Shellenbarger GG, Athearn ND, Takekawa JY, Boehm AB, 2008. Fecal indicator bacteria and Salmonella in ponds managed as bird habitat, San Francisco Bay, California, USA. Water Res. 42 (12), 2921–2930. [DOI] [PubMed] [Google Scholar]
- Stottmeister U, Wiessner A, Kuschk P, Kappelmeyer U, Kastner M, Bederski O, Muller RA, Moormann H, 2003. Effects of plants and microorganisms in constructed wetlands for wastewater treatment. Biotechnol Adv 22 (1–2), 93–117. [DOI] [PubMed] [Google Scholar]
- Tanner CC, Clayton JS, Upsdell MP, 1995. Effect of loading rate and planting on treatment of dairy farm wastewaters in constructed wetlands. 1. Removal of oxygen demand, suspended-solids and fecal-coliforms. Water Res. 29 (1), 17–26. [Google Scholar]
- Toet S, Bouwman M, Cevaal A, Verhoeven JT, 2005. Nutrient removal through autumn harvest of Phragmites australis and Thypha latifolia shoots in relation to nutrient loading in a wetland system used for polishing sewage treatment plant effluent. J. Environ. Sci. Health A Tox Hazard. Subst Environ. Eng. 40 (6–7), 1133–1156. [DOI] [PubMed] [Google Scholar]
- Tran NH, Gin KY, Ngo HH, 2015. Fecal pollution source tracking toolbox for identification, evaluation and characterization of fecal contamination in receiving urban surface waters and groundwater. Sci. Total Environ. 538, 38–57. [DOI] [PubMed] [Google Scholar]
- US Environmental Protection Agency (1997) Method 1600: Membrane Filter Test Method for Enterococci in Water, Washington, D.C. [Google Scholar]
- US Environmental Protection Agency, 2009. Method 1603: Escherichia coli (E. coli) in Water by Membrane Filtration Using Modified membrane-Thermotolorant Escherichia coli Agar (Modified mTEC), Washington, D.C. [Google Scholar]
- US Environmental Protection Agency, 2015. Method 1609.1: Enterococci in Water by TaqMan Quantitative Polymerase Chain Reaction (qPCR) with Internal Amplification Control (IAC) Assay. Water O.o. (ed), Washington, D.C. [Google Scholar]
- US Environmental Protection Agency. What are Combined Sewer Overflows (CSOs)? 2017 Boston MA. [Google Scholar]
- US Environmental Protection Agency, 2018a, 303 (d) Impaired Waterbody History Report, Washington, D.C. [Google Scholar]
- US Environmental Protection Agency, 2018b, National Pollutant Discharge Elimination System (NPDES), Washington, D.C. [Google Scholar]
- VanKempen-Fryling RJ, Stein OR, Camper AK, 2015. Presence and persistence of wastewater pathogen Escherichia coli O157:H7 in hydroponic reactors of treatment wetland species. Water Sci. Technol. 72 (1), 135–140. [DOI] [PubMed] [Google Scholar]
- Walker DJ, Hurl S, 2002. The reduction of heavy metals in a stormwater wetland. Ecol. Eng. 18 (4), 407–414. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


