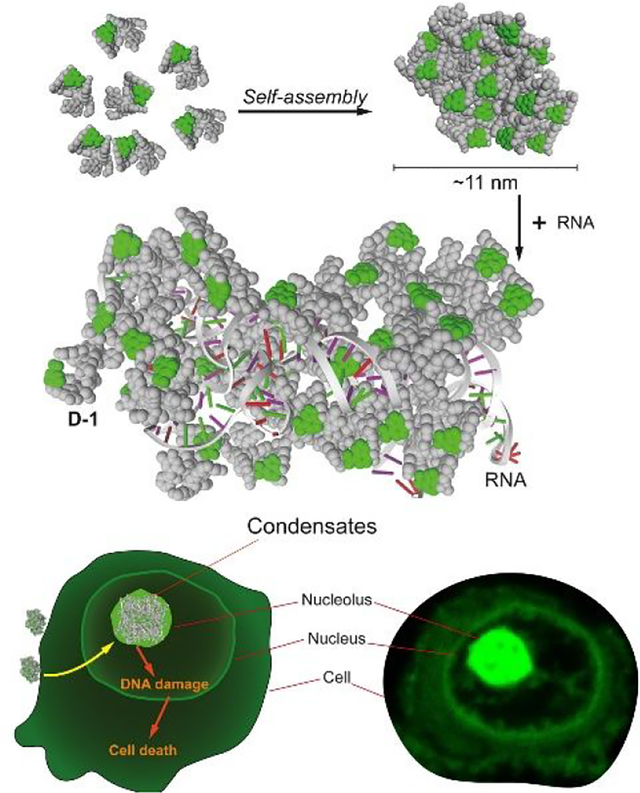
Abstract
Selectively targeting cell nucleolus remains a challenge. Here we report the first case that D-peptides form membraneless molecular condensates with RNA for targeting cell nucleolus. A D-peptide derivative, enriched with lysine and hydrophobic residues, self-assembles to form nanoparticles, which enter cells through clathrin dependent endocytosis and mainly accumulate at cell nucleolus. Structural analogue of the D-peptide reveals that particle morphology of the assemblies, which depends on the side chain modification, favors the cellular uptake. Contrasting to those of the D-peptide, the assemblies of the corresponding L-enantiomer largely localize in cell lysosomes. Preliminary mechanism study suggests that the D-peptide nanoparticles interact with RNA to form membraneless condensates in the nucleolus, which further induces DNA damage and results in cell death. This work illustrates a new strategy for rationally designing supramolecular assemblies of D-peptides for targeting subcellular organelles.
Graphical Abstract
Nucleolus is the most prominent structure within the nucleus of eukaryotic cells.1 Consisting of proteins, DNA, and RNA,2 nucleolus acts as the site of ribosome biogenesis, participates in the formation of signal recognition particles, and responds to stress.3 Nucleolus has been considered as a target for cancer chemotherapy.4–5 Although delivery of cargo molecules into nucleus is feasible by utilizing nuclear location sequences (NLS),6–9 it is, however, difficult to target nucleolus. A major challenge has been proteolysis of the peptides made of L-amino acids. For example, a prominent recent result is the use of poly(dipeptide) (20 repeats) encoded by C9orf72 to bind nucleolus for killing cancer cells.10 Despite it demonstrates the feasibility for targeting nucleolus, the poly(dipeptide), however, undergoes rapid proteolysis in cell culture so that the poly(dipeptide) has to be replenished every 2 h.10 The authors also found that replacing glycine to proline improves the stability of the poly(dipeptide). Another facile approach for minimizing proteolysis is to use D-peptides.11–12 While there is a report of D-peptides entering nucleus,13 using D-peptide for targeting nucleolus has not yet to be explored.
We have been investigating the self-assembly of D-peptides for in vivo applications,14 especially the assemblies of D-peptides in cellular environment.15 Comparing with monomeric D-peptides, the assemblies of D-peptides, as demonstrated by our studies, are advantageous in several aspects, including cellular uptake, reaction-diffusion controlled bioactivity, aggregation enhanced retention, and specific organelle targeting.15–17 Moreover, we and several labs have found that the morphologies of the peptide assemblies dictate cellular uptake efficiency and cellular location of the peptides.18–21 Despite these progresses, there are few reports on D-peptide to target nucleolus preferentially. To address the aforementioned challenge, we decided to explore the use of the assemblies of D-peptides for targeting nucleolus via forming membraneless condensates with RNA.
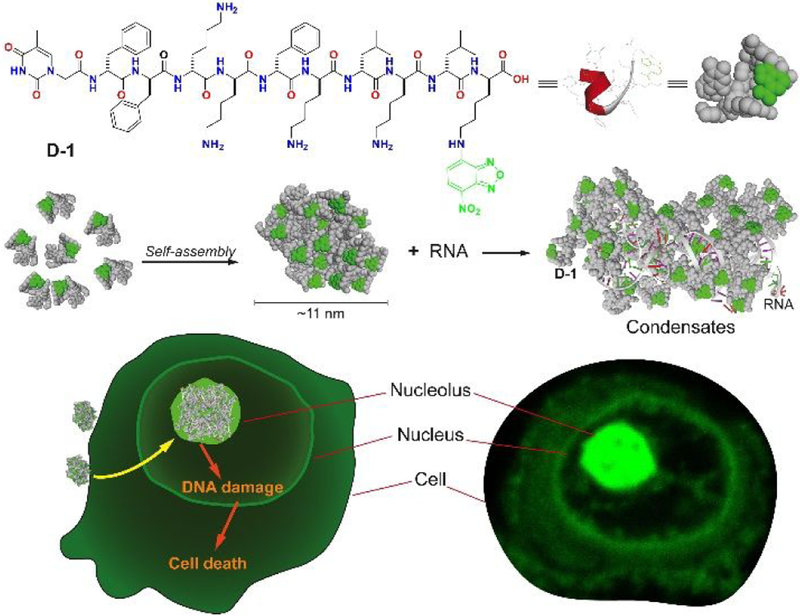
Here, we report the first case of nucleolus targeting by the assemblies of D-peptide, which consists of hydrophobic residues, positive charged lysine residues, and 4-nitro-2,1,3-benzoxadiazole (NBD) attached to the side chain of the C-terminal lysine (Scheme 1). Adopting α-helix dominant conformation and self-assembles to form nanoparticles, the D-peptides interact with RNA to form molecular condensatesin nucleolus preferentially, confirmed by a nucleolus marker.22 Treating the cells with the structural analogs of the D-peptides reveals that the α-helix dominant conformation of the D-peptides and the particle morphology of the assemblies of the D-peptides are critical for entering the cells to target nucleolus. Providing a new D-peptide sequence to target nucleolus, this work illustrates a new strategy to explore supramolecular assemblies for mimicking biomolecular condensates23 and for targeting specific organelles of live cells.
Scheme 1.
Molecular structures of D-1 and the illustration of membraneless condensates formation of D-1 and RNA.
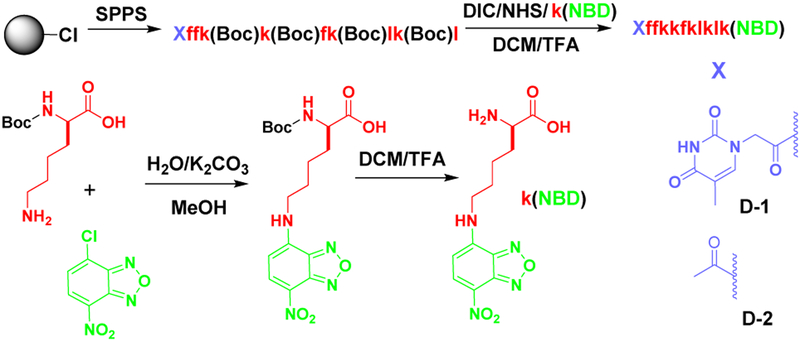
Molecule D-1 (or D-2) consists of three main components: 1) Diphenylalanine, the well-known self-assembly motif;24–25 2) an environment-sensitive fluorophore NBD for visualizing the peptide assemblies in live cells;26 3) a heptapeptide, kkfklkl (D-lysine: k; D-phenylalanine: f; D-leucine: l), which exhibits affinity to adenosine triphosphate,27 for interacting with RNA. We chose D-peptides because of their proteolytic resistance and low immunogenicity.28 Scheme 2 shows the synthetic route of D-1 (or D-2), D-1 is a molecule (NP1-NBD) reported in our previous work.27 We chose the thymine as capping group because it could interact with the DNA and RNA in the nucleolus. We first used solid-phase peptide synthesis (SPPS) to synthesize all the side chain protected D-peptides. After connecting NBD at the ɛ-amine of D-lysine to yield k(NBD), we used N-hydroxysuccinimide (NHS) to activate the C-terminal carboxylic group of the D-peptide to react with k(NBD). After the deprotection, purification by using reversed-phase high performance liquid chromatography (HPLC) gives the final product D-1 (or D-2).
Scheme 2.
Synthetic procedure of D-1 or D-2.
Red color and lower case letter in molecular structure represents the D-amino acid residue.
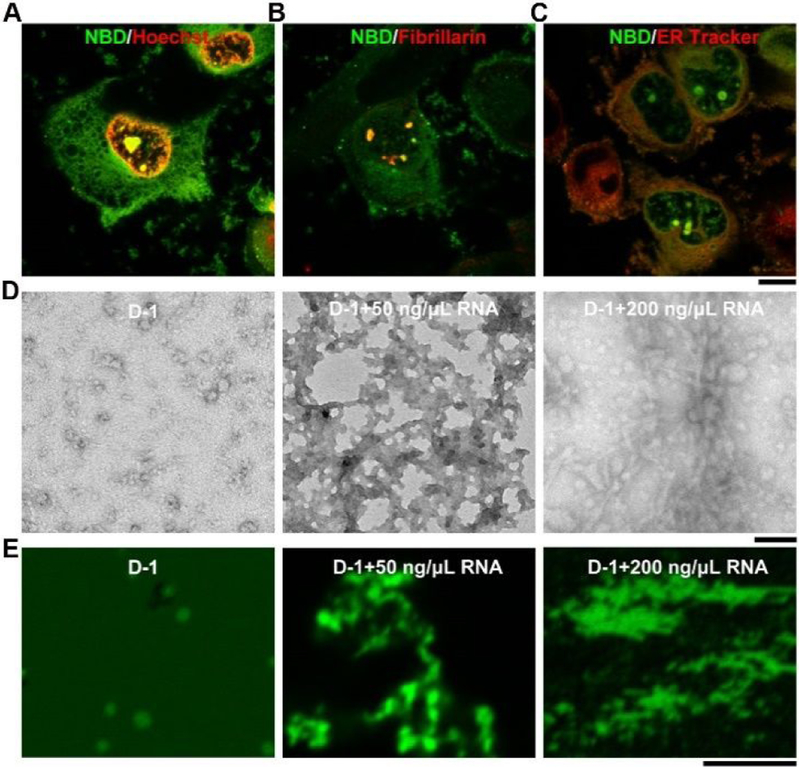
To investigate the cellular distribution of D-1, we incubated T98G cells with D-1 at different concentrations. While D-1 hardly enters the cells at the concentration of 100 μM (Figure S1), increasing the concentration of D-1 to 200 μM results in bright fluorescent puncta in cell nucleus, in addition to diffusive fluorescence in cytoplasm (Figures 1A and S1). The fluorescent puncta of D-1 in the nucleus largely co-localizes with red fluorescence of fibrillarin,10 a biomarker of nucleolus (Figure 1B) that interacts with RNA.22 This result indicates that D-1 accumulates in cell nucleolus and interact with RNA. To confirm the interaction between D-1 and RNA, we extracted total RNA from T98G cells and incubated it with D-1 in PBS buffer. The results (Figure S2) show that D-1 interacts with RNA to form precipitates that resist ribonuclease, suggesting the strong interaction between D-1 and RNA. Although we cannot obtain the accurate binding constant between D-1 and RNA, the precipitate and CLSM image of D-1 and RNA indeed indicate the strong interactions between these two. Fluorescent images show that minor amount of D-1 also presents in endoplasmic reticulum (ER) (Figure 1C), likely due the positive charges of the peptides. To better understand the interactions between assemblies of D-1 and RNA, we investigated the morphologies of the assemblies of D-1 without and with the addition of RNA. As revealed by transmission electron microscopy (TEM), D-1 forms nanoparticles with diameters about 11±2 nm (Figure 1D). TEM and CLSM images indicate that RNA interact with D-1 to form more droplet-like molecular condensates with increasing the concentration of RNA from 50 ng/μL to 200 ng/μL (Figure 1D and E), which is one of feature of molecular condensates.29 These results indicate that after D-1 enters cell nucleolus, it is likely to interact with negatively charged RNA to form assemblies, in which the assemblies D-1 likely acts as scaffolds and RNA as the clients.29
Figure 1.
Fluorescent images of T98G cells treated with D-1 for 2 h and then following by (A) Hoechst 33342 staining; (B) immunofluorescence of fibrillarin (abcam, ab5821); (C) ER Tracker staining. Scale bar is 10 μm. (D) TEM and (E) the corresponding confocal laser scanning microscopy (CLSM) images of D-1, and D-1 plus the RNA extracted from T98G cells. All the compounds are dissolved in PBS buffer (final pH=7.4) at concentration of 200 μM. Scale bar in TEM is 100 nm, and 5 μm in CLSM. For better comparing, we changed the color of the Hoechst 33342 to red.
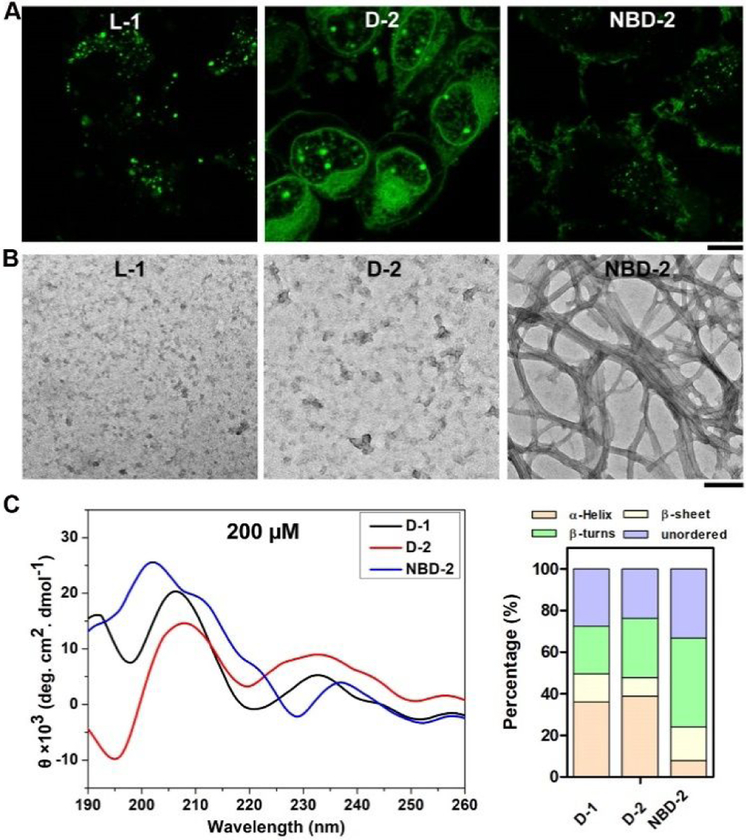
To examine how the structures of the peptides affect the ability of the assemblies for targeting nucleolus, we tested several analogs of D-1. We used L-1, the enantiomer of D-1, to co-incubate with T98G cells for testing the contribution of stereochemistry of peptide assemblies. While D-1 only slightly localized in the lysosome (Figure S3), most of L-1 forms fluorescent dots entrapped in lysosome (Figures 2A, S4 and S5). This result confirms that the biostability of the peptide assemblies in cells is indispensable for targeting the nucleolus. Although it is plausible that cell uptake pathways of L- and D-enantiomer of assemblies of peptides are different, our results provide a facile approach using D-enantiomer of peptide for targeting cell nucleolus. Acetyl group replacing thymine at the N-terminal of D-1 results in D-2. Being incubated with T98G cells, D-2 shows the similar cellular distribution as that of D-1 (Figure 2A), suggesting that the N-terminal thymine capping group is replaceable by acetyl group. Interestingly, NBD-2, an analog of D-1 formed by capping with NBD at the N-terminal of D-peptide, exhibits a completely different cellular distribution from that of D-1: Fluorescent fibrous structures formed by NBD-2 cover the surface of T98G cells (Figures 2A and S6) with little fluorescence in the cytoplasm of the cells. This result suggests that the peptide sequence unlikely is the sole factor for determining the cellular distribution of the assemblies of the D-peptides.
Figure 2.
Fluorescent images T98G cells treated with (A) L-1, D-2, or NBD-2 for 2 h. Scale bar is 10 μm. (B) TEM images of the assemblies of L-1, D-2, or NBD-2. The concentration of all the molecules is 200 μM. Scale bar is 100 nm. (C) CD spectra of D-1, D-2, and NBD-2 at the concentration of 200 μM, and the percentage of secondary structures at the concentration of 200 μM.
To further understand the factors that result in different cellular distributions of these peptide assemblies, we investigated the morphologies and secondary structures of the assemblies. As revealed by TEM, L-1 and D-2 both are able to forms nanoparticles with diameters about 11±2 nm (Figure 2B). But NBD-2 forms uniform nanofibers with several hundred nanometers in length and 4±2 nm in width, which further entangle with each other to form fibrous bundles (Figure 2B). Since L-1 undergoes proteolysis, we only examined the secondary structures of these assemblies of D-1, D-2, and NBD-2 by circular dichroism (CD) spectra and used DichroWeb30 to infer the secondary structure of the D-peptides. The results indicate that D-1 presents predominantly in α-helical conformation (36%), plus 13% of β-sheet, 23% of β-turns, and 28% of unordered structures (Figure 2C). D-2 exhibits the similar trend as that of D-1, 39% of α-helix plus 9% of β-sheet, 28% β-turns, and 24% of unordered structures. In contrast, assemblies of NBD-2 exhibits dominantly 43% of β-turns with 16% of β-sheet and 33% of unordered structures, but only 8% of α-helical conformation. These results together suggest that β-turn and β-sheet conformations favor the formation of nanofibers, which disfavors the cellular uptake of the peptides. This observation agrees with the recent reports19,21,31 that nanoparticles promote endocytosis.
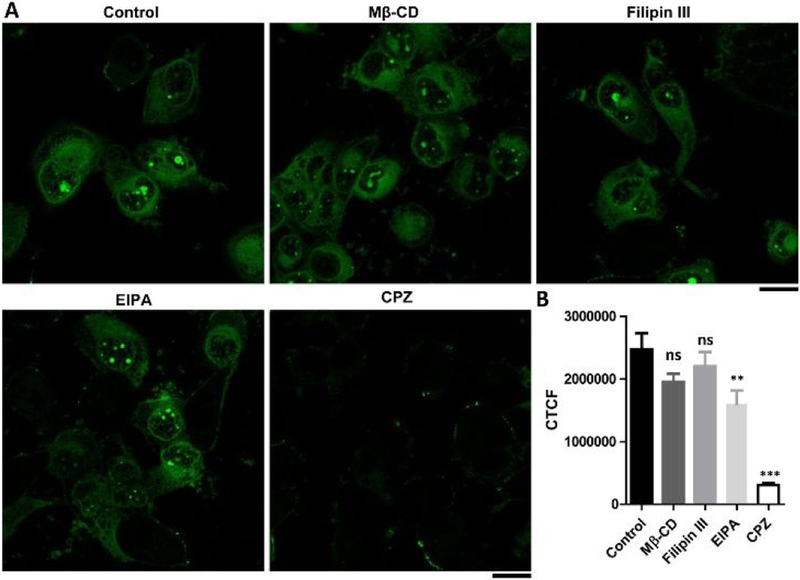
We next examined the modes of endocytosis that involved in the cellular uptake of D-1. After incubating D-1 with T98G cells at 4 °C for 1 h, the cells hardly exhibit any fluorescence in or outside cells (Figure S7), indicating that the internalization of D-1 is an energy-dependent process. We further used several established endocytosis inhibitors to infer the modes of the endocytosis of D-1 into the T98G cells. As shown in Figure 3, M-β-cyclodextrin (Mβ-CD) and Filipin III,32–33 inhibitors of lipid raft/caveolae dependent endocytosis, slightly reduce the cellular uptake of D-1. Being an inhibitor of Na+/H+ pump, ethyl-isopropyl-amiloride (EIPA),34 the cells reduce the uptake of D-1 for about 35.2%, indicating the cellular uptake of D-1 partially involves macropinocytosis and phagocytosis. The addition of chlorpromazine (CPZ),35 inhibitor of clathrin dependent endocytosis, significantly reduces the cellular uptake of D-1 for about 88%, suggesting the major mode of the internalization of D-1 could be clathrin mediated endocytosis. Though it is not possible to rule out that the use of one endocytotic inhibitor could force cells to use other endocytotic pathways, our work, nevertheless, provides a promising strategy for targeting cell nucleolus. Interestingly, a negative charged peptide (Nap-FFDALDLTD), derived from the other surface of the crystal structure of ASCPYD,36 blocks the cellular uptake of D-1 (Figure S8), suggesting that the charge interactions between D-1 and cell surface molecules likely contribute to the endocytosis of D-1.37
Figure 3.
(A) Fluorescent images and (B) the corrected total cell fluorescence (CTCF, quantified from the gray scale of CLSM images of 20 cells) of the T98G cells treated with D-1 (200 μM) for 1 h in the absence (control) or presence of the inhibitors M-βCD (5 mM), Filipin III (5 μg/mL), EIPA (100 μM) and CPZ (30 μM). Scale bar is 20 μm. Differences between the group of control and other groups are determined using one-way ANOVA analysis. **: p < 0.005, ***: p < 0.001.
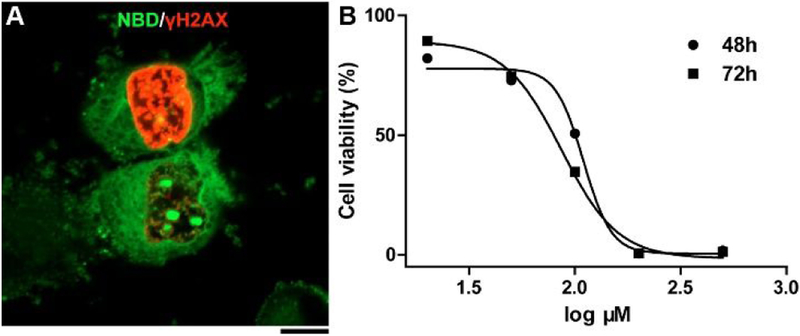
To investigate the generality of the assemblies of D-1 for targeting cell nucleolus, we incubated D-1 with four other cell lines including three cancer cell lines (HeLa, MES-SA/Dx5, and Saos-2). The results (Figure S9) indicate that D-1 is able to target nucleoli in different mammalian cells. Moreover, the nucleus staining by Hoechst (Figure 1A) shows partly disintegrated cell nucleus, indicating DNA damage and disruption of cell cycle. Immunostaining by phospho-Ser139-H2AX (γH2AX)38 (an early indicator in cellular response to DNA double strand break) on the T98G cells incubated with D-1 for 2 h reveals that D-1 leads to γH2AX foci formation in some of the T98G cells (Figure 4A), while the γH2AX foci are undetectable without D-1 treatment (Figure S10). This result confirms that the assemblies of D-1 result in DNA damage. Cell viability studies show that D-1 and D-2 exhibit similar cytotoxicity on cancer cells (T98G, HeLa) and normal bone stromal cell (HS-5), while NBD-2 is less toxic to these cells (Figures S11 and S12). The IC50 of D-1 (Figure 4B, 72h) is 86.2 μM, which appears to be higher than the reported IC50 of poly(GR), 8.4 μM (against U2OS cells and requiring fresh peptide every 2h10). But the need of 12 times replenishment of poly(GR) in 24 h, in fact, makes D-1 be more potent than the poly(GR). Although change (GR)20 to PR20 improves the stability of the polypeptide, the molecular weight also increases considerably. Because D-1 is readily accessible by solid-phase synthesis, the strategy reported in this work warrants further exploration, such as integrating enzyme-instructed self-assembly for minimizing acquired drug resistance.39–40
Figure 4.
(A) Fluorescent image of T98G cells treated with D-1 (200 μM) for 2 h and then analyzed by immunofluorescence of γH2AX. Scale bar is 10 μm. (B) Cytotoxicity of D-1 against T98G cell lines for 48 and 72 h.
In summary, this work illustrated a new strategy for targeting cell nucleolus by the assemblies of D-peptides, which interact with RNA to form membraneless condensates in the nucleolus. In our previous work,27 we found that D-1 could co-stain with ATP in cell level at low concentration (50 uM), which is innocuous to the cells. However, when co-incubating D-1 with cells at a higher concentration (200 uM), the assemblies in cell medium could change the permeability of cell membrane, and result in more uptake of D-1 in cytoplasm. Although other cell penetrating peptides can also interact with RNA, however, they lack the ability for targeting cell nucleolus. Being an alternative of the recent works of poly-dipeptides that impede RNA biogenesis,10 this work provides a new approach for designing functional assemblies through noncovalent synthesis.41–43 Moreover, this work not only illustrates D-peptide assemblies in complex conditions (e.g., live cells) for controlling cell behaviors, but also provides an alternative tool for modulating and understanding biological condensates in specific organelle.23
Supplementary Material
ACKNOWLEDGMENT
This work was partially supported by NIH (CA142746) and NSF (DMR-1420382). ZF thanks the Dean’s fellowship and NIH (F99CA234746).
Footnotes
Supporting Information
Materials and detailed experimental procedures, and additional figures.
The authors declare no competing financial interest.
REFERENCES
- (1).Shaw P; Jordan E The nucleolus Annu. Rev. Cell Dev. Biol 1995, 11, 93. [DOI] [PubMed] [Google Scholar]
- (2).O’Sullivan JM; Pai DA; Cridge AG; Engelke DR; Ganley AR The nucleolus: a raft adrift in the nuclear sea or the keystone in nuclear structure? Biomol. Concepts 2013, 4, 277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Raška I; Shaw PJ; Cmarko D Structure and function of the nucleolus in the spotlight Curr. Opin. Cell Biol 2006, 18, 325. [DOI] [PubMed] [Google Scholar]
- (4).Quin JE; Devlin JR; Cameron D; Hannan KM; Pearson RB; Hannan RD Targeting the nucleolus for cancer intervention Biochim. Biophys. Acta, Mol. Basis Dis 2014, 1842, 802. [DOI] [PubMed] [Google Scholar]
- (5).Hein N; Hannan KM; George AJ; Sanij E; Hannan RD The nucleolus: an emerging target for cancer therapy Trends Mol. Med 2013, 19, 643. [DOI] [PubMed] [Google Scholar]
- (6).Arisaka A; Mogaki R; Okuro K; Aida T Caged Molecular Glues as Photoactivatable Tags for Nuclear Translocation of Guests in Living Cells J. Am. Chem. Soc 2018, 140, 2687. [DOI] [PubMed] [Google Scholar]
- (7).Tang R; Wang M; Ray M; Jiang Y; Jiang Z; Xu Q; Rotello VM Active Targeting of the Nucleus Using Nonpeptidic Boronate Tags J. Am. Chem. Soc 2017, 139, 8547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Cheng Y; Sun C; Liu R; Yang J; Dai J; Zhai T; Lou X; Xia F A multifunctional peptide conjugated AIEgen for efficient and sequential targeted gene delivery into the nucleus Angew. Chem., Int. Ed 2019, 5049. [DOI] [PubMed] [Google Scholar]
- (9).Kong J; Wang YF; Zhang JX; Qi W; Su RX; He ZM Rationally Designed Peptidyl Virus-Like Particles Enable Targeted Delivery of Genetic Cargo Angew. Chem., Int. Ed 2018, 57, 14032. [DOI] [PubMed] [Google Scholar]
- (10).Kwon I; Xiang S; Kato M; Wu L; Theodoropoulos P; Wang T; Kim J; Yun J; Xie Y; McKnight SL Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells Science 2014, 345, 1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Wei X; Zhan C; Shen Q; Fu W; Xie C; Gao J; Peng C; Zheng P; Lu W A D‐peptide ligand of nicotine acetylcholine receptors for brain‐targeted drug delivery Angew. Chem., Int. Ed 2015, 54, 3023. [DOI] [PubMed] [Google Scholar]
- (12).Wang H; Feng Z; Xu B D-amino acid-containing supramolecular nanofibers for potential cancer therapeutics Adv. Drug Delivery Rev 2017, 110, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Medina SH; Schneider JP Cancer cell surface induced peptide folding allows intracellular translocation of drug J. Controlled Release 2015, 209, 317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Liang G; Yang Z; Zhang R; Li L; Fan Y; Kuang Y; Gao Y; Wang T; Lu WW; Xu B Supramolecular hydrogel of a D-amino acid dipeptide for controlled drug release in vivo Langmuir 2009, 25, 8419. [DOI] [PubMed] [Google Scholar]
- (15).Wang H; Feng Z; Xu B Bioinspired assembly of small molecules in cell milieu Chem. Soc. Rev 2017, 46, 2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Feng Z; Wang H; Wang S; Zhang Q; Zhang X; Rodal AA; Xu B Enzymatic Assemblies Disrupt the Membrane and Target Endoplasmic Reticulum for Selective Cancer Cell Death J. Am. Chem. Soc 2018, 140, 9566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Feng Z; Zhang T; Wang H; Xu B Supramolecular catalysis and dynamic assemblies for medicine Chem. Soc. Rev 2017, 46, 6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Feng Z; Wang H; Chen X; Xu B Self-assembling ability determines the activity of enzyme-instructed self-assembly for inhibiting cancer cells J. Am. Chem. Soc 2017, 139, 15377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Lock LL; Reyes CD; Zhang P; Cui H Tuning cellular uptake of molecular probes by rational design of their assembly into supramolecular nanoprobes J. Am. Chem. Soc 2016, 138, 3533. [DOI] [PubMed] [Google Scholar]
- (20).Kalafatovic D; Nobis M; Son J; Anderson KI; Ulijn RV MMP-9 triggered self-assembly of doxorubicin nanofiber depots halts tumor growth Biomaterials 2016, 98, 192. [DOI] [PubMed] [Google Scholar]
- (21).Zhan J; Cai Y; Ji S; He S; Cao Y; Ding D; Wang L; Yang Z Spatiotemporal control of supramolecular self-assembly and function ACS Appl. Mater. Interfaces 2017, 9, 10012. [DOI] [PubMed] [Google Scholar]
- (22).Reichow SL; Hamma T; Ferré-D’Amaré AR; Varani G The structure and function of small nucleolar ribonucleoproteins Nucleic Acids Res. 2007, 35, 1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Kato M; McKnight SL In Annual Review of Biochemistry, Vol 87; Kornberg RD, Ed. 2018; Vol. 87, p 351. [DOI] [PubMed] [Google Scholar]
- (24).Zhang Y; Kuang Y; Gao Y; Xu B Versatile small-molecule motifs for self-assembly in water and the formation of biofunctional supramolecular hydrogels Langmuir 2010, 27, 529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Reches M; Gazit E Casting metal nanowires within discrete self-assembled peptide nanotubes Science 2003, 300, 625. [DOI] [PubMed] [Google Scholar]
- (26).Gao Y; Shi J; Yuan D; Xu B Imaging enzyme-triggered self-assembly of small molecules inside live cells Nat. Commun 2012, 3, 1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Wang H; Feng Z; Qin Y; Wang J; Xu B Nucleopeptide Assemblies Selectively Sequester ATP in Cancer Cells to Increase the Efficacy of Doxorubicin Angew. Chem., Int. Ed 2018, 57, 4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Chong P; Sia C; Tripet B; James O; Klein M Comparative immunological properties of enantiomeric peptides. Lett Pept. Sci 1996, 3, 99. [Google Scholar]
- (29).Alberti S; Gladfelter A; Mittag T Considerations and Challenges in Studying Liquid-Liquid Phase Separation and Biomolecular Condensates Cell 2019, 176, 419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Whitmore L; Wallace B DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data Nucleic Acids Res. 2004, 32, W668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Shimada T; Lee S; Bates FS; Hotta A; Tirrell M Wormlike micelle formation in peptide-lipid conjugates driven by secondary structure transformation of the headgroups J. Phys. Chem. B 2009, 113, 13711. [DOI] [PubMed] [Google Scholar]
- (32).Ros-Baro A; Lopez-Iglesias C; Peiro S; Bellido D; Palacin M; Zorzano A; Camps M Lipid rafts are required for GLUT4 internalization in adipose cells Proceedings of the National Academy of Sciences of the United States of America 2001, 98, 12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Monis GF; Schultz C; Ren RY; Eberhard J; Costello C; Connors L; Skinner M; Trinkaus-Randall V Role of endocytic inhibitory drugs on internalization of amyloidogenic light chains by cardiac fibroblasts American Journal of Pathology 2006, 169, 1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Nakase I; Niwa M; Takeuchi T; Sonomura K; Kawabata N; Koike Y; Takehashi M; Tanaka S; Ueda K; Simpson JC; Jones AT; Sugiura Y; Futaki S Cellular uptake of arginine-rich peptides: Roles for macropinocytosis and actin rearrangement Molecular Therapy 2004, 10, 1011. [DOI] [PubMed] [Google Scholar]
- (35).Inal J; Miot S; Schifferli JA The complement inhibitor, CRIT, undergoes clathrin-dependent endocytosis Experimental Cell Research 2005, 310, 54. [DOI] [PubMed] [Google Scholar]
- (36).Lu A; Magupalli VG; Ruan J; Yin Q; Atianand MK; Vos MR; Schröder GF; Fitzgerald KA; Wu H; Egelman EH Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes Cell 2014, 156, 1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Versluis F; van Elsland DM; Mytnyk S; Perrier DL; Trausel F; Poolman JM; Maity C; le Sage VA; van Kasteren SI; van Esch JH Negatively charged lipid membranes catalyze supramolecular hydrogel formation J. Am. Chem. Soc 2016, 138, 8670. [DOI] [PubMed] [Google Scholar]
- (38).Rogakou EP; Pilch DR; Orr AH; Ivanova VS; Bonner WM DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139 J. Biol. Chem 1998, 273, 5858. [DOI] [PubMed] [Google Scholar]
- (39).Zhou J; Xu B Enzyme-instructed self-assembly: a multistep process for potential cancer therapy Bioconjugate Chem. 2015, 26, 987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Wang H; Feng Z; Yang C; Liu J; Medina JE; Aghvami SA; Dinulescu DM; Liu J; Fraden S; Xu B Unraveling the Cellular Mechanism of Assembling Cholesterols for Selective Cancer Cell Death Mol. Cancer Res 2018, molcanres. 0931.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Medina SH; Miller SE; Keim AI; Gorka AP; Schnermann MJ; Schneider JP An Intrinsically Disordered Peptide Facilitates Non‐Endosomal Cell Entry Angew. Chem., Int. Ed 2016, 55, 3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Wang Y; de Kruijff RM; Lovrak M; Guo X; Eelkema R; van Esch JH Access to metastable gel states using seeded self assembly of low molecular weight gelators Angew. Chem., Int. Ed 2018, 58, 3800. [DOI] [PubMed] [Google Scholar]
- (43).Zhang C; Shafi R; Lampel A; MacPherson D; Pappas CG; Narang V; Wang T; Maldarelli C; Ulijn RV Switchable Hydrolase Based on Reversible Formation of Supramolecular Catalytic Site Using a Self‐Assembling Peptide Angew. Chem., Int. Ed 2017, 56, 14511. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.