Abstract
The effect of co-culturing white-rot fungus Phlebia brevispora with growth-promoting bacterial strains Enterobacter sp. TN3W-14 and Pseudomonas sp. TN3W-8 on the degradation of polycyclic aromatic hydrocarbons (PAHs) was evaluated in liquid culture. In axenic cultures, P. brevispora strains TN3F and TMIC33929 showed high degradation of phenanthrene (> 98%) within 15 days, and degraded 65% and 63% of pyrene, and 12% and 8% of benzo(a)pyrene, respectively. This low level of degradation ability toward benzo(a)pyrene was improved significantly by co-culturing the fungi with a mixture of bacterial strains TN3W-8 and TN3W-14 (mixed bacterial co-culture; MBC). Within 15 days, P. brevispora TN3F with MBC achieved about 86% pyrene and 53% benzo(a)pyrene degradation; P. brevispora TMIC33929 with MBC showed 92% pyrene and 72% benzo(a)pyrene degradation. The MBC alone degraded little PAH, as its growth was inhibited by PAH; however, its co-culture with P. brevispora improved mycelial growth of the fungus, which led to improved degradation of the PAHs. A possible dihydrodiol metabolite of pyrene from fungal cultures suggests that hydroxylation was the first step in the degradation of pyrene by P. brevispora.
Keywords: Fungal growth management, Microbial degradation, Phlebia brevispora, Polycyclic aromatic hydrocarbon
Introduction
Polycyclic aromatic hydrocarbons (PAHs), also called polynuclear aromatic hydrocarbons, are a group of more than 100 different organic priority pollutants consisting of two or more fused benzene rings, which can occur naturally, or be manufactured (Haritash and Kaushik 2009). Released mainly by incomplete combustion of fossil fuels, PAHs transform to a wide range of toxic, carcinogenic and recalcitrant products (Kästner et al. 1994) in air, soil, and water (Johnsen et al. 2005). Their adverse effects on human health and ecosystems have led to research and guidelines aimed at regulating their release, as well as removing them from the environment (Johnsen et al. 2005).
A dominant form of inexpensive but effective PAH transformation in the environment is biological degradation by microbes (Gibson et al. 1975; Kotterman et al. 1998), but difficulty in removing high-molecular-weight (HMW) PAHs remains a major obstacle to bioremediation. The highest molecular weight PAHs mineralized by bacteria contain four benzene rings. Bacterial strains such as Mycobacterium sp. (Bouchez et al. 1995; Heitkamp et al. 1988; Kästner et al. 1994), Rhodococcus sp. (Walter et al. 1991) and Burkholderia cepacia (Juhasz et al. 1996, 1997) are only able to partially degrade five-benzene-ring PAHs to oxidized products. Fungal metabolism has thus been widely examined for potential detoxification of HMW PAHs. White-rot fungi, naturally growing on wood, are able to decompose lignin, organic (Buswell 1991) and even recalcitrant substances by means of their ligninolytic enzymes which have broad substrate specificity (Buswell 1991; Tortella et al. 2005), unlike bacterial intracellular enzymes. However, no particular microorganism is able to completely mineralize a particular organic pollutant like PAHs; in nature, bioremediation depends on cooperative metabolic activities of mixed microbial populations (Wang et al. 2011). For this, research has focused on developing microbial consortia consisting of various bacteria and fungi, with biodegradation performances superior to those of single microbial strains (Boonchan et al. 2000; Meulenberg et al. 1997).
Bioremediation often involves the manipulation of environmental and physical parameters to allow microbial growth and degradation to proceed at a faster rate. In most cases, as in bioaugmentation, naturally or genetically engineered bacterial strains with unique metabolic profiles are introduced to degrade the target compound(s). However, contrary to this common approach, our approach is to achieve higher degradation by applying bacterial strains whose interactions improve mycelial growth of the white-rot fungus Phlebia brevispora but which are not themselves extensively involved in the degradation. P. brevispora is one of the most studied species that can degrade organohalogen compounds such as polychlorinated biphenyls, polychlorinated dioxins, and pesticides (Kamei et al. 2005, 2006; Xiao et al. 2011). Our preliminary study (Harry-asobara and Kamei 2018) focused on isolation and characterization of bacteria co-isolated with P. brevispora strain TN3F, with an emphasis on selecting strains capable of enhancing the growth and morphology of mycelia of the fungus, along with mycelia of P. brevispora strain TMIC33929 from a dissimilar environment. We noted that bacterial strains designated TN3W-8 and TN3W-14, which showed high identity with the genera Pseudomonas and Enterobacter, respectively, were able to improve the mycelial growth of P. brevispora in liquid medium. Recently, some bacteria that were isolated with white-rot fungi from rotting wood were proven to possess enhancement activity for mycelial growth and wood degradation (Kamei 2017; Kamei et al. 2012). With the hypothesis that co-cultivation with each of the growth-promoting bacteria will result in improved degradation traits of the white-rot fungus, the present study evaluated the biodegradation efficiency and population dynamics of various combinations of P. brevispora strains TN3F and TMIC33929 with the growth-promoting bacteria TN3W-8 and TN3W-14, in the degradation of pyrene, phenanthrene, and benzo(a)pyrene.
Materials and methods
Chemicals
Pyrene, phenanthrene, benzo(a)pyrene, and acetonitrile were purchased from Wako Pure Chemical Industries, Tokyo, Japan. PAHs were all of environmental analysis grade. The solvent used was HPLC grade, and all other chemicals were of the highest commercially available grade.
Microorganisms and culture conditions
White-rot fungus P. brevispora strain TN3F and its growth-promoting bacterial strains TN3W-8 and TN3W-14 were isolated from a basidiomycetous fruit body and associated white-rot-decayed cedar wood, respectively in the Tano Forest Science Station at the University of Miyazaki in our previous study (Harry-asobara and Kamei 2018). Amplification of rRNA genes was as described by Hiraishi (1992). PCR and sequencing were performed according to our previous paper (Kamei et al. 2012). The gene sequences were compared with those in the GenBank database using the BLAST search engine of the DNA Data Bank of Japan (DDBJ, Tokyo, Japan). P. brevispora strain TMIC33929 was obtained from Tottori Mycological Institute (TMI), Japan. Microbes were maintained on 9-cm diameter potato dextrose agar (PDA) plates at 25 °C.
PAH degradation, extraction and analytical procedures
Potato extract (PE) medium containing 0.5% (w/v) glucose was prepared according to our previous study (Harry-asobara and Kamei 2018). For axenic fungal cultures, 100-ml Erlenmeyer flasks containing 10 ml PE medium were autoclaved, then inoculated with 2-cm diameter mycelial disks from PDA. For fungal-bacterial co-cultures, mycelial disks were inoculated into 8 ml of PE medium and then 2 ml of either bacterial culture preincubated in PE medium was inoculated with approximately 106 cells ml−1 of initial bacterial cell population. To determine the impact of mixing of the two bacteria together on the growth and degradation efficiency of the white-rot fungi, cultures were formulated by adding 1 ml each of the bacterial pre-incubated cultures to 8 ml of growth medium with or without fungal mycelial disks. Inoculated PE medium was incubated for 5 days on a 120-rpm rotary shaker at 25 °C. Then, 200 μl of 5 mM substrate solution (pyrene, phenanthrene or benzo(a)pyrene; final concentration 0.025 mM) in N,N-dimethylformamide was added. The headspace of each flask was flushed with oxygen, and flasks were sealed with a glass stopper and sealing tape and incubated in the dark on a rotary shaker at 120 rpm and 25 °C for further 5, 10 and 15 days. Abiotic cultures with each substrate served as controls.
After 5, 10 and 15 days of substrate addition, culture was homogenized with 10 ml of acetonitrile, and the biomass was removed by centrifugation (Centrifuge 5430 R, Germany) at 15,294×g for 10 min after addition of internal standard (6 and 4 μl of 5 mM phenanthrene for samples of pyrene and benzo(a)pyrene, respectively; 20 μl of 5 mM pyrene for phenanthrene samples). To determine the amount of substrate recovery, the resulting supernatants were analyzed by high-performance liquid chromatography (HPLC) performed on a Shimadzu LC-10AD fitted with an STR ODS-II 250 × 4.6 mm column and with an SPD-20A UV/VIS detector. The solvent consisted of water/acetonitrile (0.6:1) at a flow-rate of 0.8 ml min−1. Compounds in the eluate were detected at 254 nm. PAH disappearance was interpreted as biodegradation since the amount of PAH recovered in abiotic controls always exceeded recovery from biotic samples.
Biomass determination
To determine the relationships between microbial growth and PAH degradation, fungal and bacterial growth was investigated from inoculation throughout the 15-day experimental period. For fungal growth determination, at each 5-day sampling interval, cultures were filtered through a 1G2 glass filter (pore size 40–100 μm) and washed with 100 ml of deionized water and then dried to constant weight in an oven at 105 °C. For the determination of bacterial growth, absorbance of the culture samples at 600 nm (OD600) was measured every 2 days using a V-630 bio spectrophotometer (JASCO Corporation, Japan).
Metabolite detection
After analysis by HPLC, the homogenates of relevant cultures, including 10 ml acetonitrile, were acidified to pH 2.5 with HCl, then transferred to separating funnels and extracted twice with an equal volume of ethyl acetate. The organic extracts were dried over anhydrous sodium sulfate, evaporated to near dryness with a rotary evaporator, and then dried completely using nitrogen gas. Dried samples were dissolved in 1 ml ethyl acetate for gas chromatography-mass spectrometry (GC–MS) analysis. GC–MS was performed on an Agilent Technologies 5975C (mass system) linked with an Agilent Technologies 7890A (gas system) equipped with an Agilent Technologies HP-5MS column (30 m × 0.250 mm). The oven temperature was programmed to increase from 80 to 300 °C at 20 °C min−1. Triplicate samples were used for the extraction and analysis of PAH degradation products.
Results
PAH degradation by P. brevispora and growth-promoting bacterial isolates
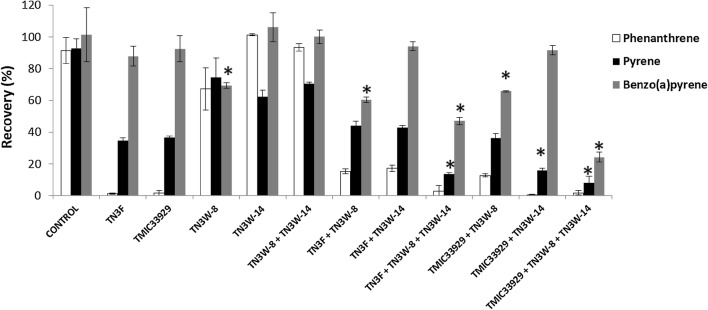
Figure 1 shows the recovery of substrates pyrene, phenanthrene, and benzo(a)pyrene from cultures with or without fungal mycelia or bacteria after 15 days of incubation. Note that substrate was added on day 5 of the experiments. Over 90% of each substrate was recovered from the abiotic controls. Axenic cultures of P. brevispora strains TN3F and TMIC33929 showed similar degradation traits to each other. Both fungi were able to degrade phenanthrene (the recovery of phenanthrene from each culture was < 5%, meaning > 95% of the phenanthrene was degraded). A four-benzene-ring PAH, pyrene, was also degraded significantly by both fungi (35% and 37% recovery from cultures of TN3F and TMIC33939, respectively). However, the degradation of benzo(a)pyrene by the fungal axenic cultures was very low; as much as 88% and 92% of this compound was recovered from cultures of strains TN3F and TMIC33929 respectively after 15 days.
Fig. 1.
Phenanthrene, pyrene and benzo(a)pyrene recovery from cultures of Phlebia brevispora strains TN3F and TMIC33929, bacterial strains Pseudomonas sp. TN3W-8 and Enterobacter sp. TN3W-14, and fungal-bacterial co-cultures over a 15-day incubation period. Asterisks represent sample values significantly different from the corresponding fungal axenic culture TN3F or TMIC33929 (p < 0.01). Values are mean ± SD of triplicate samples
In axenic culture, Pseudomonas sp. strain TN3W-8 showed slight degradation of pyrene (25%), phenanthrene (33%), and benzo(a)pyrene (31%). Although Enterobacter sp. TN3W-14 also showed slight degradation of pyrene (38%), no degradation of phenanthrene or benzo(a)pyrene was observed over 15 days. When the two bacterial strains were mixed and used for degradation experiments, the degradation traits for each PAH were similar to those of axenic culture of TN3W-14.
PAH degradation by co-cultures of P. brevispora and growth-promoting bacteria
We evaluated the effects of co-culturing growth-promoting bacteria with strains of the white-rot fungus P. brevispora on the degradation of PAHs. The experimental conditions for the co-culture of a single fungus and a single bacterium are designated TN3F + TN3W-8, TN3F + TN3W-14, TMIC33929 + TN3W-8, and TMIC33929 + TN3W-14, respectively. Co-cultures of a single fungus and mixed-bacteria (i.e., a mixture of TN3W-8 and TN3W-14) are designated TN3F + TN3W-8 + TN3W-14 and TMIC33929 + TN3W-8 + TN3W-14.
TN3F + TN3W-14 and TN3F + TN3W-8 co-cultures had no significant effect on pyrene degradation compared with axenic TN3F culture (Fig. 1). TMIC33929 + TN3W-14 co-culture degraded pyrene significantly better (p <0.01) than axenic TMIC33929 culture (final recovery 16%) (Fig. 1). However, recovery of pyrene from culture of TMIC33929 + TN3W-14 was higher than that from TMIC33929 + TN3W-8 + TN3W-14 and TN3F + TN3W-8 + TN3W-14, meaning pyrene degradation was enhanced by co-culturing the fungus with both bacteria.
Axenic cultures of P. brevispora strains TN3F and TMIC33929 showed little degradation of benzo(a)pyrene. Co-cultivation of bacterial strain TN3W-8 with each fungal strain resulted in significant degradation of benzo(a)pyrene (Fig. 1; TN3F + TN3W-8 and TMIC33939 + TN3W-8 showed significantly lower recovery than TN3F). However, co-cultivation of bacterial strain TN3W-14 with each fungal strain had no significant effect on the degradation of benzo(a)pyrene (Fig. 1, TN3F + TN3W-14 and TMIC33939 + TN3W-14). Co-cultivation of both bacteria (i.e. mixed culture) with each fungus resulted in significant (p <0.01) degradation of benzo(a)pyrene compared with degradation by each of the fungal strains. The culture TN3F + TN3W-8 + TN3W-14 showed higher degradation of benzo(a)pyrene than TN3F + TN3W-8; TMIC33929 + TN3W-8 + TN3W-14 showed higher degradation than TMIC33929 + TN3W-8. Overall, TMIC33929 + TN3W-8 + TN3W-14 co-culture achieved the highest degradation of both pyrene and benzo(a)pyrene over a 15-day period, followed by TN3F + TN3W-8 + TN3W-14.
Changes in the growth of P. brevispora and the growth-promoting bacteria over the degradation period
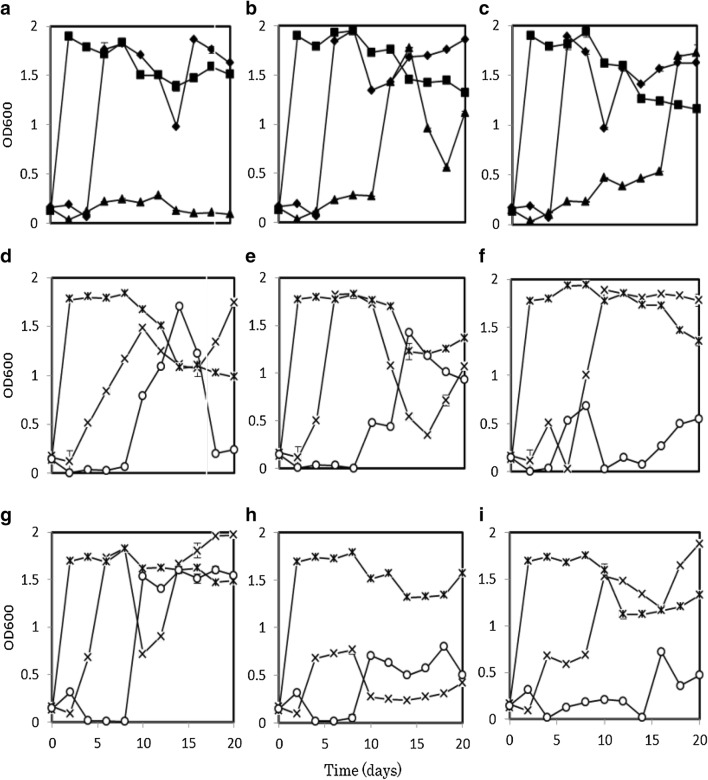
To investigate changes in the population structures of the microbes during PAH degradation, time courses of the fungal biomass and bacterial densities were estimated in axenic cultures, fungal-single bacterial co-cultures, and fungal-mixed bacterial co-cultures. Figure 2 shows bacterial growth in PAH-treated cultures (as estimated by OD600). In axenic culture, the growth of TN3W-8 peaked after 2 days of incubation, then the cell density decreased slightly after 8 days of incubation. A similar trend was observed for axenic TN3W-8 cultures with each substrate (Fig. 2a–c). The growth of TN3W-14 was slow, but picked just immediately after substrate addition. The growth was, however, not sustained, and was unstable compared with that of TN3W-8 (Fig. 2a–c). However, the cell growth of mixed bacteria TN3W-8 and TN3W-14 was inhibited in cultures containing PAH substrates compared with those of axenic TN3W-8 and TN3W-14. This phenomenon might be why the mixed bacterial culture showed lower degradation of phenanthrene and benzo(a)pyrene than the axenic bacterial cultures (Fig. 1).
Fig. 2.
Growth curves of bacterial strains TN3W-14 and TN3W-8 on phenanthrene (a, d and g), pyrene (b, e and h), and benzo(a)pyrene (c, f and i). a–c show absorbance values at OD600 for axenic bacterial cultures and mixed bacterial culture. d–f and g–i show data for co-cultures with P. brevispora strains TN3F and TMIC33929 respectively. Key: TN3W-14 (diamonds); TN3W-8 (squares); TN3W-14 + TN3W-8 (triangles); TN3F + TN3W-14 or TMIC33929 + TN3W-14 (time symbols); TN3F + TN3W-8 or TMIC33929 + TN3W-8 (asterisks); TN3F + TN3W-8 + TN3W-14 or TMIC33929 + TN3W-8 + TN3W-14 (circles). Values are mean ± SD of triplicate samples
The growth of bacterium TN3W-8 was not affected by co-culturing with the fungus TN3F, showing the same trend as the bacterial monoculture with all substrates. However, the growth of TN3W-14 was inhibited by co-cultivation with fungal strains TN3F and TMIC33929 (Fig. 2d–i). In phenanthrene-added culture with fungus TN3F or TMIC33939, the growth of the mixed bacteria was higher than the growth of the mixed bacteria in cultures without fungus (Fig. 2a–d). Growth of mixed bacteria in pyrene- and benzo(a)pyrene-added cultures with the fungi TN3F or TMIC33929 was observed in the later stages of the incubation; however, the maximum OD600 in the mixed bacterial cultures with fungus did not reach the maximum values of the mixed cultures without fungus.
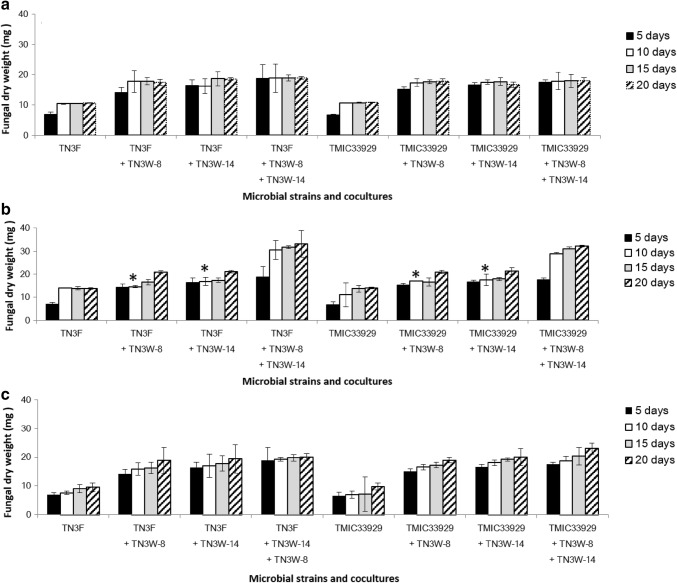
The growth of P. brevispora strains TN3F and TMIC33929 on the PAHs varied in axenic and fungal-bacterial co-cultures (Fig. 3a–c). Although there was little mycelial growth in liquid medium in fungal axenic cultures (about 7 mg at 5 days, i.e., immediately before substrate addition), fungal dry weights eventually increased to 13.7 and 14.0 mg for TN3F and TMIC33929, respectively on pyrene (Fig. 3a), 10.6 and 10.8 mg for TN3F and TMIC33929 respectively on phenanthrene (Fig. 3b), and 9.6 and 9.8 mg for TN3F and TMIC33929, respectively on benzo(a)pyrene (Fig. 3c) after 20 days of total incubation. Growth of strains TN3F and TMIC33929 was observed 5 days after substrate addition in axenic cultures with phenanthrene and pyrene, but growth in axenic cultures with benzo(a)pyrene was inhibited.
Fig. 3.
Mycelial growth of P. brevispora strains TN3F and TMIC33929 on phenanthrene (a), pyrene (b) and benzo(a)pyrene (c). Data for “5 days” represents growth on liquid medium immediately before substrate addition. Asterisks represent values not significantly different from the control TN3F or TMIC33929 at p < 0.01 (n = 4)
Mycelial growth in fungal + single bacterial co-cultures with PAHs was higher than that in fungal axenic cultures (Fig. 3a–c). In fungal + single bacterial co-cultures, mycelia of P. brevispora strain TN3F increased on the PAHs, achieving 20.7 and 21.1 mg fungal dry weight on pyrene in co-cultures TN3F + TN3W-8 and TN3F + TN3W-14 respectively (Fig. 3a), 17.5 and 18.6 mg on phenanthrene respectively (Fig. 3b), and 18.8 and 19.4 mg on benzo(a)pyrene respectively (Fig. 3c) over the period of cultivation. Quite similar rates of mycelial growth on the PAHs were achieved by co-cultures TMIC33929 + TN3W-14 and TMIC33929 + TN3W-8 (Fig. 3a–c). However, 5 days after substrate addition (i.e. on day 10), their mycelial growth on pyrene was not significantly different from those of the fungal axenic cultures (Fig. 2a). Fungal + mixed bacterial co-cultures gave higher increases in fungal dry weight over the incubation period than fungal + single bacterial co-cultures. With growth on pyrene, TN3F + TN3W-8 + TN3W-14 yielded 33.0 mg fungal dry weight (Fig. 3a), 18.9 mg fungal dry weight on phenanthrene (Fig. 3b), and 20.1 mg fungal dry weight on benzo(a)pyrene (Fig. 3c), while TMIC33929 + TN3W-8 + TN3W-14 gave 32.2 mg fungal dry weight on pyrene (Fig. 3a), 18.0 mg on phenanthrene (Fig. 3b), and 23.0 mg on benzo(a)pyrene (Fig. 3c), suggesting overall higher mycelial growth on pyrene, followed by benzo(a)pyrene, then phenanthrene. As observed in our previous study (Harry-asobara and Kamei 2018), adsorption of the bacterial cells to glass filter, and to fungal mycelium, was minimal.
Mycelial growth of P. brevispora in relation to sequential degradation of each of pyrene and benzo(a)pyrene
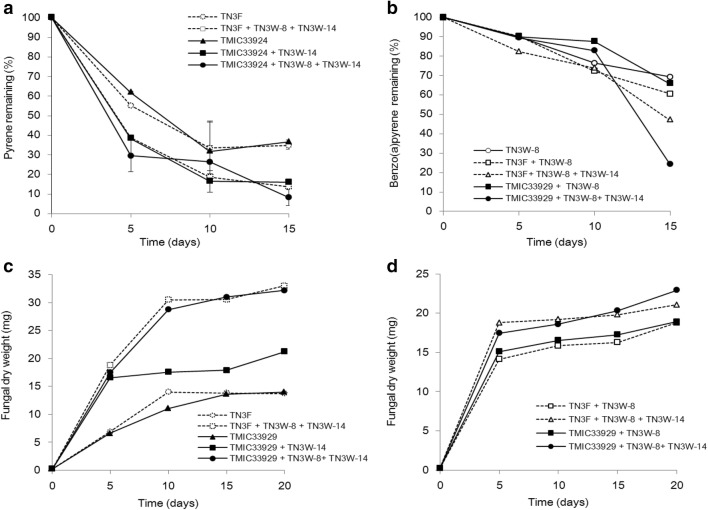
Due to observed increases in fungal growth, especially in fungal-bacterial cultures, on the high molecular weight PAHs over the incubation period, time course for the degradation of each of pyrene and benzo(a)pyrene was studied along with mycelial increases. Much of the pyrene degradation (around 70%) occurred within 5 days of substrate addition (Fig. 4a), the same time period (day 10, i.e. 5 days after substrate addition) in which we observed significant growth (p <.01) of the fungal mycelia in TMIC33929 + TN3W-8 + TN3W-14 co-culture (Fig. 4c). More so, higher pyrene degradation by TMIC33929 + TN3W-14 co-culture compared with axenic TMIC33929 fungal culture corresponded to higher mycelial growth in the co-culture (Fig. 4c), suggesting that pyrene degradation was contemporaneous with mycelial growth. Similarly, early mycelial growth on pyrene (Fig. 4c) in TN3F + TN3W-8 + TN3W-14 co-culture following substrate addition translated into about 60% pyrene degradation at 5 days after substrate addition (Fig. 4a).
Fig. 4.
Sequential degradation of each of pyrene and benzo(a)pyrene in relation to mycelial growth of P. brevispora strains TN3F and TMIC33929, each in co-culture with growth-promoting bacteria TN3W-14 or TN3W-8 or both, on pyrene (a, c) and benzo(a)pyrene (b, d) over 15 days of incubation
The co-cultures generally degraded benzo(a)pyrene slowly (Fig. 4b), consistent with the low fungal biomass yield after the initial mycelial growth, i.e. 5 days before substrate addition (Fig. 4d). Only about 20% degradation was achieved at 5 days, and about 25% at 10 days. Compared with mycelia from axenic TMIC33929 culture, higher mycelial growth in TMIC33929 + TN3W-8 + TN3W-14 co-culture (Fig. 4d) in the later period of the incubation saw a sharp increase in benzo(a)pyrene degradation, to about 52% by the 15th day after substrate addition (Fig. 4b). For TMIC33929 + TN3W-8 and TN3F + TN3W-8 co-cultures, higher increases in mycelial mass over the incubation period (Fig. 4d) compared with mycelia from corresponding axenic fungus resulted to increases in benzo(a)pyrene degradation (Fig. 4b). TMIC33929 + TN3W-8 + TN3W-14 and TN3F + TN3W-8 + TN3W-14 co-cultures, which had the highest mycelial growth, achieved the most benzo(a)pyrene degradation over time (Fig. 4b, d).
Metabolites of pyrene and benzo(a)pyrene
To ascertain the products of degradation of pyrene and benzo(a)pyrene by fungal and bacterial cultures, extracts from axenic cultures, fungal + single bacterial co-cultures, and fungal + mixed-bacterial co-cultures were analyzed by GC–MS.
The total ion chromatograms of co-cultures of TN3F + TN3W-8 + TN3W-14, TMIC33929 + TN3W-8 + TN3W-14, and fungal axenic cultures at 5 days after substrate addition, showed a common pyrene metabolic peak I, at 12.265 min (not shown). This compound was neither detected in control cultures nor cultures of bacterial strains lacking fungus. The mass spectrum of this compound showed a molecular ion (M+) peak at m/z 234 (molecular mass of pyrene [202] + 32 mass, most intense peak at m/z 202, and other significant fragment ions at m/z 43, m/z 69, m/z 81 and m/z 95). The m/z difference of 32 is indicative of oxidation (O2), and the appearance of a molecular ion at m/z 234 (the molecular weight of pyrene dihydrodiol) suggests compound I is a dihydrodiol metabolite.
Following slow degradation of benzo(a)pyrene, extracts from 15-day (after substrate addition) cultures were analyzed by GC–MS. From the total ion chromatograms of TN3F + TN3W-8 co-culture (not shown), a metabolite peak II was detected at 8.684 min. This compound was not detected in axenic TN3 W-8 culture, but its detection was enhanced in fungal + mixed bacterial cultures. The mass spectrum of this compound showed an M+ peak at m/z 282, most intense peak at m/z 254, and other significant fragment ions at m/z 126 and m/z 113.
Discussion
Recent research focused on bioremediation has involved the isolation and integration of different microorganisms (Pino and Peñuela 2011; Kim et al. 2009) as well as manipulation of environmental parameters (Sutherland 1992), to enable microbial growth and degradation to proceed at a faster rate. To achieve high degradation of contaminants, microorganisms are usually isolated from PAH-contaminated sites (Pino and Peñuela 2011; Boonchan et al. 2000), sludges, slurries (Kim et al. 2009) etc., and screened on the basis of their ability to degrade a target compound(s), followed by integration of different microorganisms to harness the unique potentials of each. In the present study, however, higher PAH degradability by white-rot fungus P. brevispora strains TN3F and TMIC33929 was instead achieved through mycelial growth management by co-culture with bacterial strains TN3W-8 and TN3W-14. The inhibited bacterial population in the co-cultures suggested that the increased mycelial growth might be the result of interactions among the bacterial and fungal cells; bacterial growth may not be necessary.
In the present study, axenic cultures of white-rot fungi TN3F and TMIC33929 degraded phenanthrene completely, even with low mycelial growth on the PAH, but did not extensively degrade benzo(a)pyrene. Phenanthrene is listed among the PAHs susceptible to fungal biodegradation (Cerniglia 1997). The long lag period of pure bacterial strains in the presence of phenanthrene suggests inability of the strains to use phenanthrene for growth. Also, the low growth of mixed-bacterial culture on phenanthrene suggests little synergy between TN3W-14 and TN3W-8 in phenanthrene degradation. In the present study, the bacterial population in mixed-bacterial culture could only grow on phenanthrene when co-cultured with either of the fungal strains, although growth did not exceed the maximal growth of the pure bacterial strains. Sutherland (1992) reported that a bacterial consortium from one bioremediation site grew on a PAH only alongside a fungal strain.
In the present study, higher bacterial growth in mixed-bacterial cultures on pyrene than on benzo(a)pyrene indicated that the mixed culture could use pyrene better as a secondary carbon source than benzo(a)pyrene. Microbial consortia were shown to grow differently on different PAHs (Cerniglia 1997) and fungal-bacterial co-cultures that used pyrene as a sole carbon and energy source could not grow on benzo(a)pyrene (Boonchan et al. 2000). Furthermore, increases in cell growth synonymous with substrate removal or degradation have been established for bacterial strains with good metabolic profiles for PAH degradation (Boonchan et al. 2000; Bouchez et al. 1995; Kim et al. 2009). However, in the present study, bacterial growth in co-cultures with fungal strains was inhibited, and the rate of bacterial growth did not correspond with the trend in substrate degradation. This might either suggest a pattern of growth-preceding degradation (Kim et al. 2009), or limited involvement of the bacterial strains in achieving the observed degradation in fungal-mixed bacterial co-cultures. The situation is evidenced in the detection of pyrene metabolic compound I in the chromatogram of fungal axenic cultures as well as from that of fungal-bacterial co-cultures at 5 days after substrate addition. Similarly, detection of metabolic compound II from benzo(a)pyrene samples of fungal-mixed bacterial cultures (15 days after substrate addition) was contemporaneous with sharp increases in fungal mycelial growth. Although the degradation of a portion of benzo(a)pyrene by Pseudomonas sp. strain TN3W-8 in axenic culture in the present study indicated that it could possess a benzo(a)pyrene catabolic pathway, the inhibited growth of the bacterium in co-culture with each of the white-rot fungi indicates little involvement of this bacterium in the rapid degradation of benzo(a)pyrene in the later days of the incubation period.
Detected metabolic product of pyrene provided evidence of the microbial degradation pathway of the aromatic compound, suggesting that hydroxylation was the first step in the degradation of pyrene by the fungi. Our data show that growth of the white-rot fungus P. brevispora can be managed for higher degradation of aromatic compounds. The strategy was to isolate bacterial strains that could singly, or by interactions, induce white-rot fungal mycelial growth and morphological enhancement in liquid medium, as previously reported (Harry-asobara and Kamei 2018). The enhanced mycelia in turn enhanced remediation of the aromatic compounds. The results obtained in the present study are significant since the two fungal strains were not isolated from the same environment. This suggests that the bacterial strains TN3W-14 and TN3W-8 may also be able to improve the degradation ability of other strains of Phlebia through mycelial growth enhancement, but this requires verification.
Acknowledgements
This work was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (Grant Nos. 18H02257 and 17K19296). We thank Edanz Group (www.edanzediting.com/ac) for editing a draft of this manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
References
- Boonchan S, Britz ML, Stanley GA. Degradation and mineralization of high-molecular-weight polycyclic aromatic hydrocarbons by defined fungal-bacterial cocultures. Appl Environ Microbiol. 2000;66:1007–1019. doi: 10.1128/AEM.66.3.1007-1019.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchez M, Blanchet D, Vandecasteele JP. Degradation of polycyclic aromatic hydrocarbons by pure strains and by defined strain associations: inhibition phenomena and cometabolism. Appl Microbiol Biotechnol. 1995;43:156–164. doi: 10.1007/BF00170638. [DOI] [PubMed] [Google Scholar]
- Buswell JA. Fungal degradation of lignin. In: Arora K, Mukerij KG, Knudsen G, editors. Handbook of applied mycology. New York: Marcel Dekker; 1991. pp. 425–480. [Google Scholar]
- Cerniglia CE. Fungal metabolism of polycyclic aromatic hydrocarbons: past, present and future applications in bioremediation. J Ind Microbiol Biotechnol. 1997;19(5–6):324–333. doi: 10.1038/sj.jim.2900459. [DOI] [PubMed] [Google Scholar]
- Gibson DT, Mahadevan V, Jerina DM, Yagi H, Yeh HJC. Oxidation of the carcinogens benzo[a]pyrene and benzo[a]anthracene to dihydrodiols by a bacterium. Science. 1975;189(4199):295–297. doi: 10.1126/science.1145203. [DOI] [PubMed] [Google Scholar]
- Haritash AK, Kaushik CP. Biodegradation aspects of polycyclic aromatic hydrocarbons (PAHs): a review. J Hazard Mater. 2009;169(1–3):1–15. doi: 10.1016/j.jhazmat.2009.03.137. [DOI] [PubMed] [Google Scholar]
- Harry-asobara JL, Kamei I. Bacterial strains isolated from Cedar wood improve the mycelial growth and morphology of white rot fungus Phlebia brevispora on agar and liquid medium. J Wood Sci. 2018;64(4):444–450. doi: 10.1007/s10086-018-1723-y. [DOI] [Google Scholar]
- Heitkamp MA, Freeman JP, Miller DW, Cerniglia CE. Pyrene-degradation by a Mycobacterium sp.: identification of ring oxidation and ring fission products. Appl Environ Microbiol. 1988;54(10):2556–2565. doi: 10.1128/aem.54.10.2556-2565.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraishi A. Direct automated sequencing of 16S rDNA amplified by polymerase chain reaction from bacterial cultures without DNA purification. Lett Appl Microbiol. 1992;15(5):210–213. doi: 10.1111/j.1472-765X.1992.tb00765.x. [DOI] [PubMed] [Google Scholar]
- Johnsen AR, Wick LY, Harms H. Principles of microbial PAH-degradation in soil. Environ Pollut. 2005;133(1):71–84. doi: 10.1016/j.envpol.2004.04.015. [DOI] [PubMed] [Google Scholar]
- Juhasz AL, Britz ML, Stanley GA. Degradation of high molecular weight polycyclic aromatic hydrocarbons by Pseudomonas cepacia. Biotechnol Lett. 1996;18(5):577–582. doi: 10.1007/BF00140206. [DOI] [Google Scholar]
- Juhasz AL, Britz ML, Stanley GA. Degradation of fluoranthene, pyrene, benzo[a]anthracene and dibenz[a, h]anthracene by Burkholderia cepacia. J Appl Microbiol. 1997;83(2):189–198. doi: 10.1046/j.1365-2672.1997.00220.x. [DOI] [Google Scholar]
- Kamei I. Co-culturing effects of coexisting bacteria on wood degradation by Trametes versicolor. Curr Microbiol. 2017;74(1):125–131. doi: 10.1007/s00284-016-1162-1. [DOI] [PubMed] [Google Scholar]
- Kamei I, Suhara H, Kondo R. Phylogenetical approach to isolation of white-rot fungus capable of degrading polychlorinated dibenzo-p-dioxin. Appl Microbiol Biotechnol. 2005;69(3):358–366. doi: 10.1007/s00253-005-0052-4. [DOI] [PubMed] [Google Scholar]
- Kamei I, Sonoki S, Haraguchi K, Kondo R. Fungal bioconversion of toxic polychlorinated biphenyls by white-rot fungus P. brevispora. Appl Microbiol Biotechnol. 2006;73(4):932–940. doi: 10.1007/s00253-006-0529-9. [DOI] [PubMed] [Google Scholar]
- Kamei I, Yoshida T, Enami D, Meguro S. Coexisting Curtobacterium bacterium promotes growth of white-rot fungus Stereum sp. Curr Microbiol. 2012;64(2):173–178. doi: 10.1007/s00284-011-0050-y. [DOI] [PubMed] [Google Scholar]
- Kästner M, Breuer-Jammali M, Mahro B. Enumeration and characterization of the soil microflora from hydrocarbon- contaminated soil sites able to mineralize polycyclic aromatic hydrocarbons (PAH) Appl Microbiol Biotechnol. 1994;41:267–273. doi: 10.1007/BF00186971. [DOI] [Google Scholar]
- Kim YM, Ahn CK, Woo SH, Jung GY, Park JM. Synergic degradation of phenanthrene by consortia of newly isolated bacterial strains. J Biotechnol. 2009;144(4):293–298. doi: 10.1016/j.jbiotec.2009.09.021. [DOI] [PubMed] [Google Scholar]
- Kotterman MJJ, Vis EH, Field JA. Successive mineralization and detoxification of benzo[a]pyrene by the white rot fungus Bjerkandera sp. strain BOS55 and indigenous microflora. Appl Environ Microbiol. 1998;64(8):2853–2858. doi: 10.1128/aem.64.8.2853-2858.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulenberg R, Rijnaarts HHM, Doddema HJ, Field JA. Partially oxidized polycyclic aromatic hydrocarbons show an increased bioavailability and biodegradability. FEMS Microbiol Lett. 1997;15(1):245–249. doi: 10.1111/j.1574-6968.1997.tb10407.x. [DOI] [PubMed] [Google Scholar]
- Pino N, Peñuela G. Simultaneous degradation of the pesticides methyl parathion and chlorpyrifos by an isolated bacterial consortium from a contaminated site. Int Biodeterior Biodegrad. 2011;65(6):827–831. doi: 10.1016/j.ibiod.2011.06.001. [DOI] [Google Scholar]
- Sutherland JB. Detoxification of polycyclic aromatic hydrocarbons by fungi. J Ind Microbiol. 1992;9(1):53–61. doi: 10.1007/BF01576368. [DOI] [PubMed] [Google Scholar]
- Tortella GR, Diez MC, Duran N. Fungal diversity and use in decomposition of environmental pollutants. Crit Rev Microbiol. 2005;31(4):197–212. doi: 10.1080/10408410500304066. [DOI] [PubMed] [Google Scholar]
- Walter U, Beyer M, Klein J, Rehm HJ. Degradation of pyrene by Rhodococcus sp. UW1. Appl Microbiol Biotechnol. 1991;34(5):671–676. doi: 10.1007/BF00167921. [DOI] [Google Scholar]
- Wang W, Yan L, Cui Z, Gao Y, Wang Y, Jing R. Characterization of a microbial consortium capable of degrading lignocellulose. Bioresour Technol. 2011;102(19):9321–9324. doi: 10.1016/j.biortech.2011.07.065. [DOI] [PubMed] [Google Scholar]
- Xiao P, Mori T, Kamei I, Kondo R. Metabolism of organochlorine pesticide heptachlor and its metabolite heptachlor epoxide by white rot fungi belonging to genus Phlebia. FEMS Microbiol Lett. 2011;314(2):140–146. doi: 10.1111/j.1574-6968.2010.02152.x. [DOI] [PubMed] [Google Scholar]