Abstract
Background:
Genetic risk scores (GRSs) have been associated with CHD events and coronary artery calcium (CAC). We sought to evaluate the ability of a GRS to improve CAC as a screening test.
Methods:
Using the results of the most recent genome-wide association studies, we calculated a GRS in 6660 individuals from the Multi-Ethnic Study of Atherosclerosis and used it to determine the optimal age for an individual to undergo CAC screening.
Results:
This 157-SNP GRS was predictive of non-zero CAC in individuals aged 44-54 and improved the positive yield of CAC as a screening test in this age group. The GRS was predictive of CAC in the entire multiethnic cohort and in each self-identified ethnic group (European American, Chinese American, African American, and Hispanic American) assessed individually. Given a specified target yield rate of non-zero CAC, an equation was derived to calculate an individual’s optimal age to undergo CAC screening. In addition, a “direct-to-consumer” GRS consisting of only risk SNPs or their proxies that are directly genotyped on the 23andMe v5 chip (102-SNP GRS) was assessed in the European American population and was predictive of non-zero CAC in younger individuals.
Conclusion:
A GRS is associated with non-zero CAC in a multi-ethnic cohort and can be used to calculate the age of a person’s first calcium scan, given a target threshold for CAC discovery. Furthermore, an inexpensive and widely available “direct-to-consumer” GRS was found to be a viable option to calculate the optimal age for CAC screening.
Keywords: coronary heart disease, coronary artery calcium, screening, genetics, genetic risk score
1. Introduction
Coronary heart disease (CHD), primarily caused by atherosclerosis, affects 15.5 million Americans, and caused approximately 1 out of every 7 deaths in the United States in 2013.1Because CHD is a common, multigenic disorder, genome-wide association studies (GWAS) have been conducted to identify single nucleotide polymorphisms (SNPs) associated with disease.2–10 These SNPs individually confer small additional risk of disease but can be combined into a single genetic risk score (GRS), summarizing a person’s overall genetic predisposition for CHD.11–13 Over the past ten years, several GRSs derived from GWAS for CHD have been associated with the incidence of CHD events.11–17 Mega et al found that in primary prevention trials, the number needed to treat with statins to prevent one CHD event was more than double among those of low genetic risk compared to high risk individuals.16
GRS for CHD and its clinical risk factors have also been associated with coronary artery calcium (CAC) in asymptomatic individuals.14,17–19 However, these findings have been limited to cohorts of European ancestry. Additionally, recent analyses using the UK Biobank and CARDIoGRAMplusC4D cohorts identified 64 novel genomic loci associated with CHD, bringing the total number of known CHD loci to 161.10
CAC is a phenotypic risk factor that can be measured non-invasively and is correlated with total atherosclerotic burden.20–22 Computed tomography (CT)-based CAC scoring is an established method for assessing subclinical atherosclerosis:23 increased CAC is associated with a higher incidence of CHD events,24–27 whereas a CAC score of 0 is linked with very low risk.28,29 CAC has also been shown to improve risk classification when added to traditional risk factors.30
Current guidelines from the American College of Cardiology and the American Heart Association provide CAC with a Class IIa recommendation in individuals deemed of intermediate (7.5-20%) 10-year risk according to traditional measures such as the Framingham Risk Score (FRS) or Atherosclerotic Cardiovascular Disease (ASCVD) risk score31. Yeboah et al. found that, when combined with FRS, CAC was the best predictor of CHD events in individuals of intermediate (5-20%) risk, compared to four other non-traditional risk markers as well as family history of CHD.32 Although CAC increases with age,33 CAC remains highly predictive of CHD events even after stratification by 10-year age group, “suggesting that once CAC is known chronologic age has less importance”.34 Moreover, absolute CAC scores predict CHD events better than scores adjusted for age and sex,35 and CAC presence in younger individuals has been associated with a 5-fold increase in CHD event risk.36
Additionally, establishing when a person should begin preventative statin therapy remains a significant clinical question. In a recent study using coronary CT angiography (CCTA), one-half of patients with non-obstructive CAD and one-third with obstructive CAD did not qualify for statin therapy according to the 2013 ACC/AHA guidelines.37 These guidelines state that additional risk factors, including CAC, can be considered when a traditional “risk-based treatment decision is uncertain”.38 When CAC is considered within statin eligibility group, CHD event prediction improves.39
These results indicate that CAC is a valuable screening test for determining CHD risk and provides improved insight into an individual’s need for preventive statin therapy; however, because coronary artery calcification progresses with age, scanning all asymptomatic young individuals will result in very low positive detection, and scanning older individuals will result in CAC detection greater than 45% after age 55. To be economically valuable, a screening test must have a reasonably high prevalence of positive detection coupled with an appropriate response,40 in this case, onset of statin therapy. This led us to ask: is it possible to use a GRS derived from SNPs associated with CHD to increase the positive yield of CAC as a screening test?
The goal of this study was to determine the optimal age for an individual to undergo CAC screening based on a GRS derived from previously identified CHD risk loci. The evaluation was in a multi-ethnic cohort, and further analyses were done within each ethnic group.
2. Methods
2.1. Multi-Ethnic Study of Atherosclerosis
The Multi-Ethnic Study of Atherosclerosis (MESA) was designed to study the characteristics of subclinical atherosclerosis in a diverse cohort of asymptomatic individuals. Its details have been previously published.27,41,42 In short, participants were enrolled and initially examined at one of six participating clinics throughout the United States between July 2000 and July 2002. Participants were between the ages of 44 and 84 and free from clinical cardiovascular disease at the first exam. Data from two ancillary studies, the MESA Family Study, which enrolled subjects between May 2004 and May 2007, and MESA Air were also included in the analysis. The study was approved by the institutional review committee at each participating institution, and all subjects gave informed consent.
2.2. Genotyping
Genotype information for 8296 participants was obtained from the NHLBI MESA SNP Health Association Resource (SHARe) on dbGaP (Study Accession: phs000420.v6.p3). Participants were genotyped using the Affymetrix Genome-Wide Human SNP 6.0. Samples with <95% call rate or observed heterozygosity greater than 3 standard deviations from the mean were removed, and the data was filtered to remove related individuals based on self-identified familial relationships, leaving 6660 participants with genotype data and a CAC score. At the variant level, SNPs with minor allele frequency <0.01 or call rate <95% were removed. To obtain most of the reported coronary artery disease (CAD)10 risk loci, the genotype data was further imputed using the SNPs that passed quality control. The imputation was conducted via the Michigan Imputation Server v1.0.2, phased with ShapeIT, and imputed with IMPUTE2, using the 1000 genomes phase 3 reference panel and genome build hg19.43
2.3. Calculation of the GRS
A GRS was calculated for each individual using the SNPs associated with CHD in the recent meta-analysis.10 4 SNPs were excluded because they were not in the imputed dataset (rs7797644) or had low imputation quality (R2<0.6) (rs116843064, rs6511720, rs7412), leaving 157 SNPs. The GRS for each individual was calculated by summing the number of risk alleles (Xi) multiplied by the previously reported odds ratio for CHD for each of the 157 SNPs (equation 1).
| (1) |
2.4. CAC Assessment
CAC assessment in MESA was performed as per Carr et al.44 Briefly, CAC was measured using electron-beam CT at three sites and a four-detector row helical CT at the other three sites. Prospective electrocardiographic triggering was used at all sites. Each individual received two consecutive scans, and the results were averaged. All scans were assessed at a central CT reading center, and CAC was quantified using the Agatston method.23
2.5. Statistical analysis
6660 individuals with genetic information and a CAC score were included in the analysis. All analyses were done using R v3.4.0 (R Foundation for Statistical Computing, Vienna, Austria). As per Mega et al,16 participants were divided into low (GRS quintile 1), intermediate (GRS quintiles 2-4), and high (GRS quintile 5) genetic risk categories. Traditional risk factors were assessed for individuals aged 44-54 and compared between the high and low risk groups using the student t-test for continuous variables and the X2 test for categorical variables. The entire cohort was then separated into 10-year age groups (44-54, 55-64, 65-74, 75-84), and the rate of calcium presence, defined as a non-zero CAC score, was determined for each genetic risk category. Because current guidelines suggest CAC measurement in individuals of intermediate (7.5-20%) risk and initiation of stain therapy in individuals of high (>20%) risk, the utility of the GRS was also assessed specifically in individuals of all ages who were identified as low (<7.5%) risk according to the FRS.
In the youngest age group (44-54), the odds ratio for non-zero CAC was calculated within each quintile of genetic risk in a univariate model and in a multivariate model with age and sex included as covariates. A normalized GRS was then calculated by taking an individual’s raw GRS, subtracting the total population mean, and normalizing by the standard deviation of the total population (equation 2):
| (2) |
where GRS is an individual’s raw GRS calculated from equation 1, μ is the mean GRS of the total population, and σ is the total population standard deviation. Normalized GRS was assessed as a continuous variable in the entire cohort for its association with non-zero CAC in a multivariate model with age and sex included as covariates. Using the model and a target discovery rate of non-zero CAC in the population, a simple equation was derived to calculate the appropriate age for a person to receive their first calcium scan, given the individual’s age-independent risk factors (sex and GRS).
After analysis of overall genetic predisposition, we also sought to determine if any specific SNPs were driving the predictive power of the GRS. Therefore, we assessed the predictive ability of each GRS SNP individually using GCTA software.45,46 Analysis was done within each ethnic group separately and using a mixed linear model with the genetic relatedness matrix to account for any remaining population substructure.
The advent of direct-to-consumer personal genotyping services, such as 23andMe, has caused widespread interest in genetic tests that use chips which include many SNPs in the GRS developed here. However, imputing the remaining SNPs not genotyped directly requires substantial time and data storage space, limiting usefulness for consumers. 23andMe represents one of the largest profiled populations for personal genotyping; therefore, we assessed the predictive ability of two GRSs derived solely from SNPs genotyped directly on the chip currently used by 23andMe (23andMe v5, Illumina Infinium Global Screening Array). The first of these GRSs consisted of CHD risk SNPs genotyped directly on 23andMe v5 (n=37) in addition to CHD risk SNP proxies (n=65) genotyped directly on 23andMe v5, resulting in a 102-SNP “direct-to-consumer” GRS. For each CHD risk SNP not genotyped on 23andMe v5, the LDproxy module in LDlink47 was used to search for proxy SNPs in the 5 European populations represented in the 1000 genomes phase 3 v5 reference set. If a queried risk SNP had a proxy in high linkage disequilibrium (R2>0.8) and that proxy was genotyped directly on 23andMe v5, the proxy was included in the GRS. Because European populations were referenced to search for proxies, GRS effectiveness was analyzed in the European American population. The second of these GRSs included only the 37 SNPs genotyped directly on 23andMe v5 and was analyzed in all ethnic populations.
3. Results
Genotype information was retrieved for 6660 individuals that also had reported CAC scores. A GRS was calculated for each individual by weighting the genotype at each SNP by the associated odds ratio determined via previous CHD GWAS. All SNPs included in the GRS were in the original dataset (n=30) or imputed (n=127) with an R2>0.6 (Supplementary Table 1). The mean GRS for the population was 3.735±0.187.
Baseline characteristics of traditional risk factors were assessed for the high GRS group (quintile 5), intermediate risk group (quintiles 2-4), and low risk group (quintile 1) (Table 1) in the youngest age group (44-54 years). All traditional risk factors except for BMI, systolic blood pressure, total cholesterol, and LDL cholesterol did not differ significantly by genetic risk group. The high GRS group had a lower mean BMI (27.3 vs 29.9 kg/m^2, p=2.9×10−10) and systolic blood pressure (114.7 vs 118.2 mmHg, p=0.007) and higher total cholesterol (197.5 vs 190.5 mg/dl, p=0.010) and LDL cholesterol (121.2 vs 115.6 mg/dl, p=0.018) than the low GRS group.
Table 1.
Traditional risk factors for individuals ages 44-54 in the high, intermediate, and low GRS groups. Mean and standard deviation are presented for continuous variables. Categorical variables are presented as percentages.
| High Genetic Risk (Q5) | Int Genetic Risk (Q2-Q4) | Low Genetic Risk (Q1) | p (High vs Low) | |
|---|---|---|---|---|
| Age (years) | 49.9 ± 3.0 | 49.8 ± 2.9 | 49.9 ± 2.7 | 0.86 |
| Male (%) | 46.8 | 47.1 | 47.2 | 0.97 |
| BMI (kg/m2) | 27.3 ± 5.6 | 28.9 ± 6.0 | 29.9 ± 5.8 | 2.9×10−10 |
| Smoking (%) | 16.4 | 19.5 | 19.9 | 0.26 |
| Diabetes mellitus (%) | 7.2 | 7.9 | 10.7 | 0.13 |
| Systolic blood pressure (mmHg) | 114.7 ± 17.7 | 117.0 ± 17.0 | 118.2 ± 17.7 | 0.007 |
| Antihypertensive therapy (%) | 17.9 | 20.9 | 23.1 | 0.093 |
| Total cholesterol (mg/dl) | 197.5 ± 36.8 | 194.6 ± 35.5 | 190.5 ± 36.8 | 0.010 |
| LDL cholesterol (mg/dl) | 121.2 ± 30.6 | 119.2 ± 30.2 | 115.6 ± 33.1 | 0.018 |
| HDL cholesterol (mg/dl) | 49.1 ± 13.8 | 49.9 ± 14.3 | 49.1 ± 13.5 | 0.996 |
| Triglycerides (mg/dl) | 136.7 ± 89.2 | 129.2 ± 95.9 | 131.3 ± 100.9 | 0.43 |
| Statin (%) | 7.0 | 6.8 | 4.8 | 0.27 |
Q5, quintile 5; BMI, body mass index, LDL, low density lipoprotein, HDL, high density lipoprotein.
For the entire cohort (all ages), CAC presence increased between low (40.8%), intermediate (51.1%), and high (58.4%) GRS score (Table 2). This trend was also observed among each 10-year age group (44-54, 55-64, 65-74, 75-84) with a lower overall prevalence but stronger genetic effect in the younger age groups. In the youngest age group (44-54), CAC presence in the high GRS group was approximately double that of the low GRS group (31.6% vs 15.8%) while the absolute percentage increase in ages 75-84 was 13.8% (72.4% in low risk subjects vs 86.2% in high risk subjects).
Table 2.
Prevalence of non-zero CAC within each genetic risk group, stratified by 10-year age category
| Age | All | 44-54 | 55-64 | 65-74 | 75-84 |
|---|---|---|---|---|---|
| Total people | 6660 | 1827 | 1908 | 1975 | 935 |
| Total CAC>0 | 3365 | 446 | 867 | 1284 | 762 |
| Low GRS: CAC>0 % | 544/1332 (40.8%) | 59/377 (15.8%) | 131/385 (34.0%) | 219/383 (57.2%) | 134/185 (72.4%) |
| Int GRS: CAC>0% | 2043/3996 (51.1%) | 269/1076 (25.0%) | 517/1134 (45.6%) | 788/1215 (64.9%) | 465/561 (82.9%) |
| High GRS: CAC>0% | 778/1332 (58.4%) | 118/374 (31.6%) | 219/389 (56.3%) | 277/377 (73.5%) | 163/189 (86.2%) |
| Total CAC>0 rate | 50.5% | 24.4% | 45.4% | 65.0% | 81.5% |
| High/Low | 1.43 | 2.02 | 1.65 | 1.28 | 1.19 |
CAC, coronary artery calcium; GRS, genetic risk score.
6166 individuals of all ages had complete data for standard clinical risk assessment by the FRS. Of these individuals, 1985 were deemed to be of low (<7.5% 10-year CHD event) risk by the FRS. Among the low risk individuals, CAC presence in the high GRS group was nearly double that of the low GRS group (32.8% vs 16.5%) (Table 3).
Table 3.
Prevalence of non-zero CAC among individuals of all ages classified as low (<7.5% 10-year) risk according to the Framingham Risk Score
| FRS <7.5% | |
|---|---|
| Total people | 1985 |
| Total CAC>0 | 501 |
| Low GRS: CAC>0 % | 65/393 (16.5%) |
| Int GRS: CAC>0 % | 301/1192 (25.6%) |
| High GRS: CAC>0 % | 131/400 (32.8%) |
| Total CAC>0 rate | 25.2% |
| High/Low | 1.98 |
CAC, coronary artery cacium; GRS, genetic risk score.
Among individuals aged 44-54, the odds ratio for CAC generally increased with increasing GRS quintile in a univariate model and in a multivariate model adjusted for age and sex (Figure 1) and was statistically significant in both models for quintiles 2-5 (compared to quintile 1). To ensure these findings were not being driven by any single ethnic group, the GRS was assessed in each ethnic group separately. The GRS was predictive of CAC in each ethnic group (Appendix 2).
Figure 1:
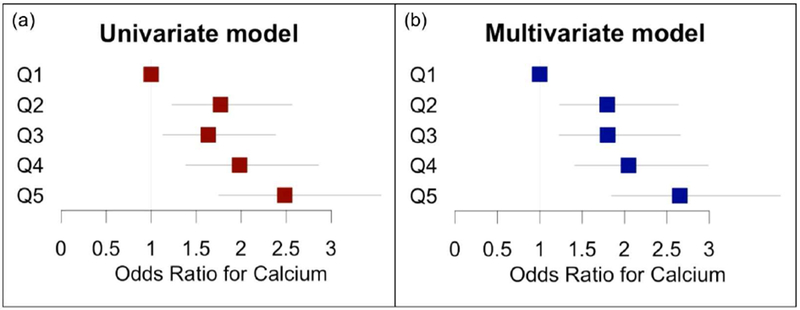
Odds ratio for risk of non-zero CAC by GRS quintile in individuals ages 44-54. (a) Univariate model. (b) Multivariate model adjusted for age and sex.
When the GRS was treated as a continuous variable, the odds ratio for CAC was 1.37 (1.29-1.45) per standard deviation from the population mean in the multivariate model adjusted for age and sex. Figure 2 illustrates the change in probability of CAC as a function of age at increasing GRS values.
Figure 2:
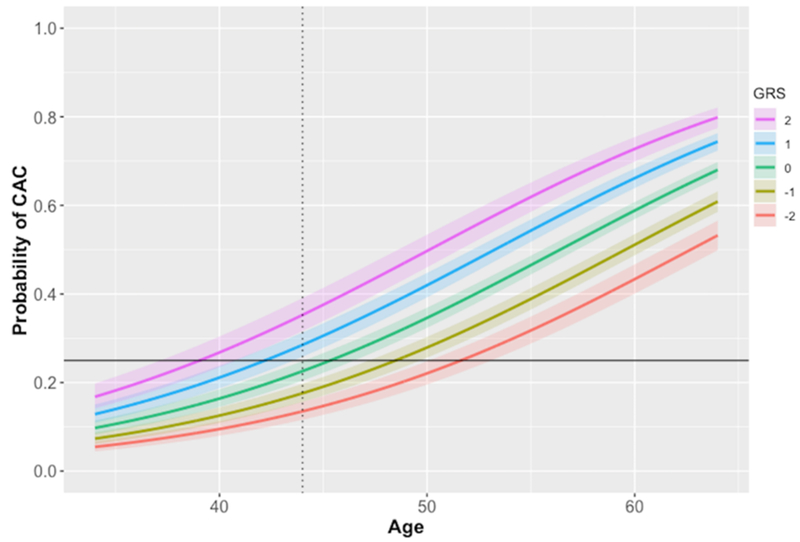
Probability of coronary artery calcium (CAC) presence as a function of age, presented for increasing normalized genetic risk score (GRS) values. Shaded area indicates 95% confidence interval. Probability curves left of the dotted line represent predictions outside the age range from which the regression was performed. Solid line represents a 25% rate of positive CAC.
Using the multivariate model, the age of a first scan can be calculated, given a particular yield rate of CAC, according to the following equation:
| (3) |
where r is the target rate of non-zero CAC detected in the population, GRS is the individual’s normalized genetic risk score, and s is the patient’s sex (0 for females, 1 for males). If we screen individuals with an anticipated CAC rate of 25% for males, this equation reduces to:
| (4) |
and therefore, for a male with a GRS 2 standard deviations above the mean, the model suggests an age of 39.0 for a first scan, whereas for a male with a GRS 2 standard deviations below the mean, the model suggests an age of 51.6. However, it should be noted that the model was created with data from individuals ages 44-84. At a non-zero CAC rate of 25% for females, the equation reduces to:
| (5) |
and the model predicts an age of first scan at 49.5 years for a female 2 standard deviations above the population mean and 62.1 years for a female 2 standard deviations below.
To assess the predictive ability of each GRS SNP individually, we analyzed each ethnic group separately and used a mixed linear model to account for remaining population substructure. In the European American, Chinese American, and Hispanic American populations, no SNPs were significant predictors (p<0.05) of non-zero CAC after false discover rate (FDR) correction.48 In the African American population, one SNP, rs1887318 located on chromosome 10, was a significant predictor of non-zero CAC after FDR correction. Q-q plots show observed p-values that follow a uniform distribution (Figure 3).
Figure 3:
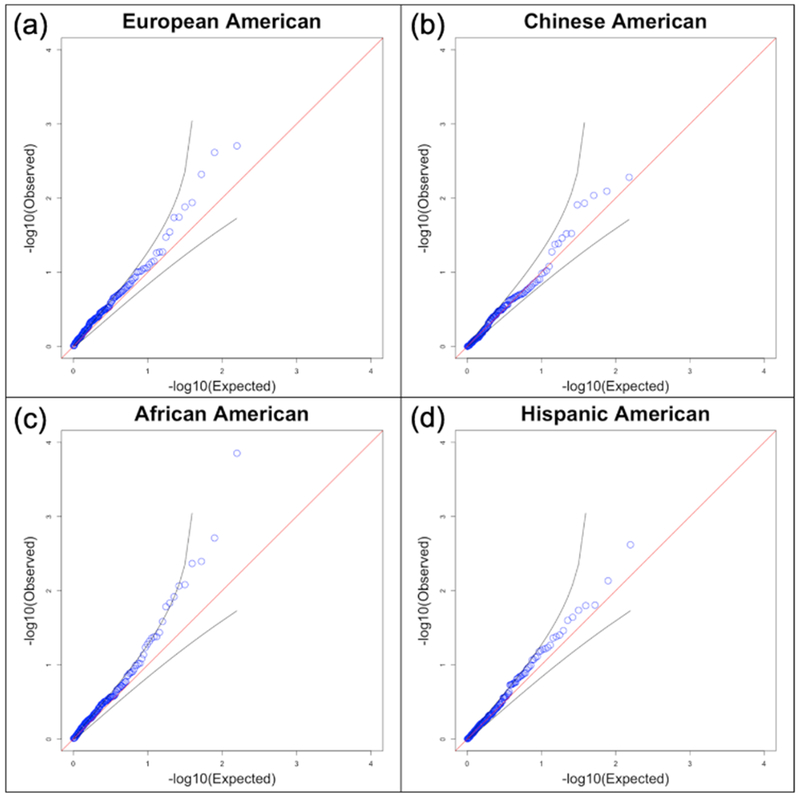
Q-q plots comparing the observed p-value for each SNP with expected p-values.
Given the rising popularity of direct-to-consumer genotyping services, we sought to evaluate the utility of a commercially available test without imputing additional SNPs. Because 23andMe is one of the largest companies to offer this service, we evaluated the effectiveness of a GRS derived solely from SNPs genotyped directly on the 23andMe v5 chip. This 102-SNP GRS included 37 SNPs from our original GRS and 65 proxy SNPs that were derived as per the Methods section (Appendix 1). This 102-SNP GRS had a mean of 2.91±0.157 in the European population and can be calculated using a simple python tool (https://github.com/LaurenSeverance/GRS). In the youngest age group, individuals in quintiles 2-5 had significantly increased risk (p<0.05) when compared to quintile 1, and in all ages, individuals in quintiles 3-5 had significantly increased risk (Figure 4).
Figure 4:
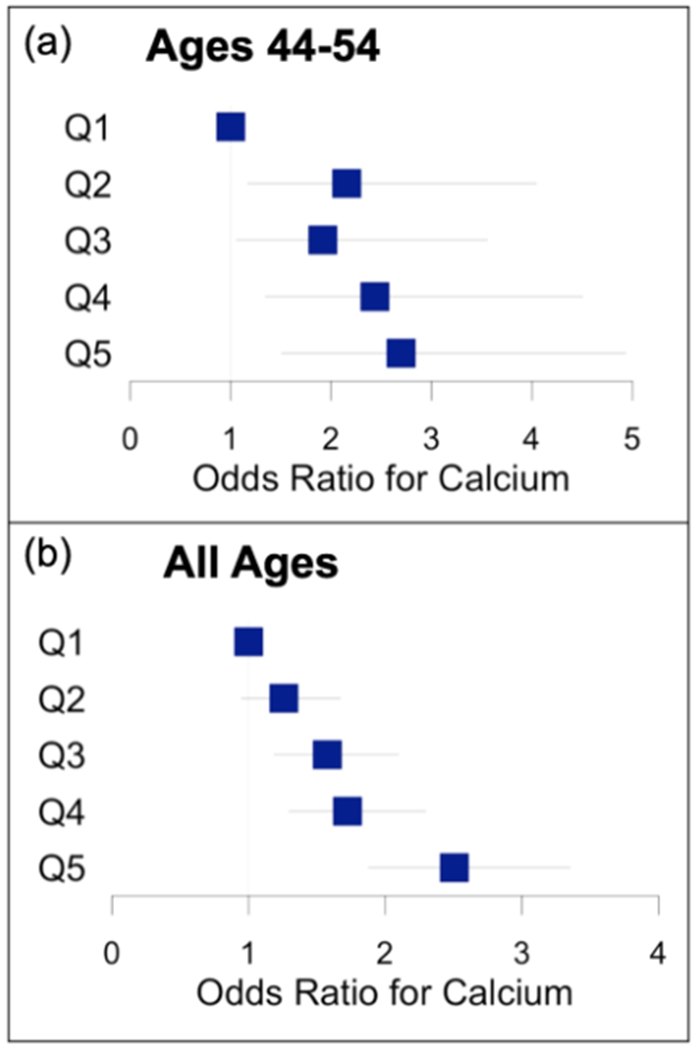
Odds ratio for non-zero coronary artery calcium by quintile of “direct-to-consumer” genetic risk score (n=102 SNPs) in European American group. (a) Ages 44-54. (b) All ages.
With the 102-SNP GRS treated as a continuous variable, an age of scan equation was derived for the European population. At a 25% non-zero CAC rate for males, the equation reduces to:
| (6) |
and for females it reduces to:
| (7) |
where GRSn is the individual’s 102-SNP GRS normalized to the European population mean and standard deviation. When proxy SNPs were excluded and a 37-SNP GRS based only on CHD risk SNPs measured directly on the v5 chip was assessed among ages 44-54 in each ethnic population individually, it was not predictive of CAC in any population.
4. Discussion
Current guidelines provide CAC with a class IIa recommendation for cardiovascular risk assessment in asymptomatic individuals of intermediate risk, and it is clear that CAC provides valuable insight of CHD risk. However, screening all younger individuals will result in a low CAC discovery rate and consequently low cost effectiveness. Therefore, we sought to evaluate a GRS derived from SNPs that predict CHD outcomes for its ability to improve the positive yield of CAC as a screening test in a group of higher risk individuals. In this multi-ethnic cohort of clinically asymptomatic individuals, subjects with a higher GRS had an increased rate of non-zero CAC. The GRS was predictive of calcium despite the fact that SNPs were initially derived from GWAS for CHD events. These findings are in agreement with previous analyses14,17–19 performed in cohorts of strictly European ancestry and which did not include the novel loci discovered during the most recent meta-analysis of SNPs associated with CHD.
Our findings are consistent among all four 10-year age groups with the strongest effect in the youngest group (44-54). In this group, we found a significant increase in risk for non-zero CAC in GRS quintiles 2-5 when compared to quintile 1. We also demonstrate that the GRS is effective in stratifying individuals of all ages who are classified as low risk according to the FRS. According to current guidelines, CAC scanning is not recommended in these individuals, yet the rate of non-zero CAC is nearly double in the high GRS group compared to the low GRS group. While this finding highlights the utility of the GRS beyond traditional risk scoring, the optimal integration of GRS with traditional risk factors, other than age, that change over time remains an open clinical question.
When GRS was analyzed as a continuous variable, we found that a 45 year old male with a GRS 2 standard deviations above the population mean has the same probability of CAC as a 57 year old male with a GRS 2 standard deviations below the mean. This suggests that consideration of a GRS could increase the positive yield of CAC screening by identifying individuals of high genetic risk – in fact, the GRS could be used to define the age at which the probability of non-zero CAC crosses a predetermined threshold.
Thus, we used the model to derive an equation for an individual to receive a first CAC scan, given a target non-zero CAC discovery rate and the individual’s age-independent risk factors of GRS and sex. Given this equation, an individual with a GRS 2 standard deviations below the population mean has a recommended age of first scan approximately 12 and a half years later than an individual with a GRS 2 standard deviations above (51.6 years vs 39.0 years for males and 62.1 years vs 49.5 years for females). The GRS could also be used clinically to encourage early statin therapy in younger individuals with high genetic predisposition for disease and a non-zero CAC score.
Although the GRS was derived from SNPs discovered primarily in populations of European ancestry and is most effective in stratifying the European American population, it was predictive of CAC in this multi-ethnic cohort. It remained predictive even when assessed in each ethnic group separately. However, further studies are needed to validate these findings in younger individuals, and GWAS among populations of more diverse ancestry may improve GRS utility.
When we assessed the predictive ability of each GRS SNP individually, we found that in all populations except the African American population, no individual SNPs were significant predictors of non-zero CAC. However, when the effects of these SNPs were combined into a single GRS, the result was predictive. This finding is consistent with current hypotheses that individual variants confer very low risk, but their effects are additive and can be combined into a measure of cumulative genetic risk. The only SNP to show significant association with non-zero CAC was rs1887318 near the KIA1462 locus on chromosome 10 in the African American population. To our knowledge, this SNP has not previously been associated with CHD or CAC in African Americans, including in a meta-analysis of African American cohorts by Wojczynski et al which evaluated risk of CAC among SNPs previously associated with CHD in European American cohorts.49
With the rising popularity of personal genotyping services, many individuals can now access the raw data needed to calculate their own GRS but are limited by the imputation. Therefore, we asked, can a GRS derived only from SNPs directly genotyped on the 23andMe v5 chip be used to determine an individual’s risk of non-zero CAC? This 102-SNP “direct-to-consumer” GRS was predictive of non-zero CAC in the European American population and can be calculated directly from 23andMe v5 raw data with a simple formula. However, when the GRS was reduced to 37 SNPs directly genotyped on the chip and no proxies, it was not predictive of CAC in younger individuals of any ethnic group.
This analysis had several limitations. First, the youngest age group was limited to ages 44-54, and approximately one quarter of the group already had calcium. Additional studies are necessary to validate this work in even younger individuals and to further understand the age at which calcium growth begins, particularly in high-risk individuals. Second, although CAC is a well-established, non-invasive potential screening tool for subclinical atherosclerosis, it does not measure soft plaques, which may be more prevalent in this younger population. Finally, analyses were stratified by self-identified ethnicity.
5. Conclusion
In summary, we have shown that a GRS derived from SNPs discovered via GWAS to be associated with CHD events identifies younger individuals at an increased risk of non-zero CAC. The GRS is also effective in stratifying those who are categorized as low risk according to the FRS and who would not be recommended for a CAC scan under current guidelines. From this group, the GRS identifies individuals who are at an increased risk of non-zero CAC. For a CAC screening program with a target positive scan discovery rate, the GRS can be used to calculate an appropriate age for an individual to receive their first scan. The GRS varies by ethnic group due to the difference in risk SNP prevalence within each ethnic group. While the GRS works well for the European American population, small sample size and lower prevalence of risk SNPs in the other populations indicate that further studies relating genetic variants to both CHD events and CAC are needed within these groups. While individual SNPs are not useful in stratifying the cohort, their cumulative effect, summarized in the GRS, is predictive of CAC, and direct-to-consumer genotyping results can be used to calculate a GRS with significant predictive utility.
Supplementary Material
Acknowledgements
MESA and the MESA SHARe project are conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with MESA investigators. Support for MESA is provided by contracts N01-HC-95159, N01-HC95160, N01-HC95162, N01-HC95163, N01-HC95164, N01-HC95165, N01-HC95166, N01-HC95167, N01-HC95168, N01-HC95169 and CTSA UL1-RR-024156.
MESA Family is conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with MESA investigators. Support is provided by grants and contracts R01HL071051, R01HL071205, R01HL071250, R01HL071251, R01HL071258, R01HL071259, UL1-RR0025005, by the National Center for Research Resources, Grant UL1RR033176, and the National Center for Advancing Translational Sciences, Grant UL1TR000124.
The Multi-Ethnic Study of Atherosclerosis and Air Pollution (MESA Air) is supported by the U.S. Environmental Protection Agency (EPA) under Science to Achieve Results (STAR) Program Grant # RD831697. Although the research described in this presentation has been funded wholly or in part by the United States Environmental Protection Agency through RD831697 to the University of Washington, it has not been subjected to the Agency’s required peer and policy review and therefore does not necessarily reflect the views of the Agency and no official endorsement should be inferred.
Funding for SHARe genotyping was provided by NHLBI Contract N02-HL-64278. Genotyping was performed at Affymetrix (Santa Clara, California, USA) and the Broad Institute of Harvard and MIT (Boston, Massachusetts, USA) using the Affymetrix Genome-Wide Human SNP Array 6.0.
This manuscript was not prepared in collaboration with MESA investigators and does not necessarily reflect the opinions or views of MESA, or the NHLBI.
Funding Sources
This work was supported by grants from the National Institutes of Health, USA (NIH grant T32 HL 105373, K01 HL 143113).
Conflicts of Interest:
Dr. Fan is under employment with Healthlytix, which is a for-profit corporation that holds patent of genetic prediction algorithms. Dr. Dale is a founder of and holds equity in CorTech Labs, Inc. and serves on its Scientific Advisory Board. He is a member of the Scientific Advisory Board of Human Longevity, Inc., and receives funding through research grants with General Electric Healthcare. The terms of these arrangements have been reviewed by and approved by the University of California, San Diego in accordance with its conflict of interest policies. Dr. McVeigh is a founder and shareholder of MR Interventions Inc, and receives funding through research grants with General Electric Healthcare, Pacesetter Inc., Tendyne Holdings Inc.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mozaffarian D, Benjamin EJ, Go AS, et al. Heart Disease and Stroke Statistics-2016 Update a Report from the American Heart Association. Vol 133; 2016. doi: 10.1161/CIR.0000000000000350. [DOI] [PubMed] [Google Scholar]
- 2.Samani NJ, Erdmann J, Hall AS, et al. Genomewide Association Analysis of Coronary Artery Disease. N Engl J Med. 2007;357(5):443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Helgadottir A, Thorleifsson G, Manolescu A, et al. A Common Variant on Chromosome 9p21 Affects the Risk of Myocardial Infarction. Science (80-). 2007;316(5830): 1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 4.McPherson R, Pertsemlidis A, Kavaslar N, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316(5830):1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schunkert H, König IR, Kathiresan S, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011. ;43(4):333–340. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nikpay M, Goel A, Won H-H, et al. A comprehensive 1000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015;47(10):1121–1130. doi: 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Verweij N, Eppinga RN, Hagemeijer Y, Van Der Harst P. Identification of 15 novel risk loci for coronary artery disease and genetic risk of recurrent events, atrial fibrillation and heart failure. Sci Rep. 2017;7(1):1–9. doi: 10.1038/s41598-017-03062-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Howson JMM, Zhao W, Barnes DR, et al. Fifteen new risk loci for coronary artery disease highlight arterial-wall-specific mechanisms. Nat Genet. 2017;49(7):1113–1119. doi: 10.1038/ng.3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nelson CP, Goel A, Butterworth AS, et al. Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat Genet. 2017;49(9):1385–1391. doi: 10.1038/ng.3913. [DOI] [PubMed] [Google Scholar]
- 10.van der Harst P, Verweij N. The Identification of 64 Novel Genetic Loci Provides an Expanded View on the Genetic Architecture of Coronary Artery Disease. Circ Res. 2017;(November 2017):CIRCRESAHA117p. 312086. doi: 10.1161/CIRCRESAHA.117.312086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kathiresan S, Voight BF, Purcell S, et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet. 2009;41(3):334–341. doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ripatti S, Tikkanen E, Orho-Melander M, et al. A multilocus genetic risk score for coronary heart disease: case-control and prospective cohort analyses. Lancet. 2010;376(9750): 1393–1400. doi: 10.1016/S0140-6736(10)61267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davies RW, Dandona S, Stewart AFR, et al. Improved Prediction of Cardiovascular Disease Based on a Panel of Single Nucleotide Polymorphisms Identified Through Genome-Wide Association Studies. Circ Cardiovasc Genet. 2010;3(5):468–474. doi: 10.1161/CIRCGENETICS.110.946269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thanassoulis G, Peloso GM, Pencina MJ, et al. A Genetic Risk Score Is Associated With Incident Cardiovascular Disease and Coronary Artery Calcium: The Framingham Heart Study. Circ Cardiovasc Genet. 2012;5(1):113–121. doi: 10.1161/CIRCGENETICS.111.961342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vaarhorst AAM, Lu Y, Heijmans BT, et al. Literature-Based Genetic Risk Scores for Coronary Heart Disease: The Cardiovascular Registry Maastricht (CAREMA) Prospective Cohort Study. Circ Cardiovasc Genet. 2012;5(2):202–209. doi: 10.1161/CIRCGENETICS.111.960708. [DOI] [PubMed] [Google Scholar]
- 16.Mega JL, Stitziel NO, Smith JG, et al. Genetic risk, coronary heart disease events, and the clinical benefit of statin therapy: an analysis of primary and secondary prevention trials. Lancet. 2015;385(9984):2264–2271. doi: 10.1016/S0140-6736(14)61730-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Natarajan P, Young R, Stitziel NO, et al. Polygenic Risk Score Identifies Subgroup With Higher Burden of Atherosclerosis and Greater Relative Benefit From Statin Therapy in the Primary Prevention Setting. Circulation. 2017;135(22):2091–2101. doi: 10.1161/CIRCULATIONAHA.116.024436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salfati E, Nandkeolyar S, Fortmann SP, et al. Susceptibility Loci for Clinical Coronary Artery Disease and Subclinical Coronary Atherosclerosis Throughout the Life-Course. Circ Cardiovasc Genet. 2015;8(6):803–811. doi: 10.1161/CIRCGENETICS.114.001071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khera AV, Emdin CA, Drake I, et al. Genetic Risk, Adherence to a Healthy Lifestyle, and Coronary Disease. N Engl J Med. 2016;375(24):2349–2358. doi: 10.1056/NEJMoa1605086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rumberger JA, Simons DB, Fitzpatrick LA, Sheedy PF, Schwartz RS. Coronary Artery Calcium Area by Electron-Beam Computed Tomography and Coronary Atherosclerotic Plaque Area: A Histopathologic Correlative Study. Circulation. 1995;92(8):2157–2162. doi: 10.1161/01.CIR.92.8.2157. [DOI] [PubMed] [Google Scholar]
- 21.Sangiorgi G, Rumberger JA, Severson A, et al. Arterial Calcification and Not Lumen Stenosis Is Highly Correlated With Atherosclerotic Plaque Burden in Humans: A Histologic Study of 723 Coronary Artery Segments Using Nondecalcifying Methodology. J Am Coll Cardiol. 1998;31 (1):126–133. doi: 10.1016/S0735-1097(97)00443-9. [DOI] [PubMed] [Google Scholar]
- 22.Rosen BD, Fernandes V, McClelland RL, et al. Relationship Between Baseline Coronary Calcium Score and Demonstration of Coronary Artery Stenoses During Follow-Up. JACC Cardiovasc Imaging. 2009;2(10):1175–1183. doi: 10.1016/j.jcmg.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Agatston AS, Janowitz WR, Hildner FJ, Zusmer NR, Viamonte M, Detrano R. Quantification of coronary artery calcium using ultrafast computed tomography. J Am Coll Cardiol. 1990;15(4):827–832. doi: 10.1016/0735-1097(90)90282-T. [DOI] [PubMed] [Google Scholar]
- 24.Greenland P, LaBree L, Azen SP, Doherty TM, Detrano RC. Coronary artery calcium score combined with Framingham score for risk prediction in asymptomatic individuals. JAMA. 2004;291(2):210–215. doi: 10.1001/jama.291.2.210. [DOI] [PubMed] [Google Scholar]
- 25.Detrano R, Guerci AD, Carr JJ, et al. Coronary Calcium as a Predictor of Coronary Events in Four Racial or Ethnic Groups. N Engl J Med. 2008;358(13):1336–1345. doi: 10.1056/NEJMoa072100. [DOI] [PubMed] [Google Scholar]
- 26.Silverman MG, Blaha MJ, Krumholz HM, et al. Impact of coronary artery calcium on coronary heart disease events in individuals at the extremes of traditional risk factor burden: the Multi-Ethnic Study of Atherosclerosis. Eur Heart J. 2014;35(33):2232–2241. doi: 10.1093/eurheartj/eht508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olson JL, Bild DE, Kronmal RA, Burke GL. Legacy of MESA. Glob Heart. 2016;11(3):269–274. doi: 10.1016/j.gheart.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sarwar A, Shaw LJ, Shapiro MD, et al. Diagnostic and Prognostic Value of Absence of Coronary Artery Calcification. JACC Cardiovasc Imaging. 2009;2(6):675–688. doi: 10.1016/j.jcmg.2008.12.031. [DOI] [PubMed] [Google Scholar]
- 29.Blaha MJ, Cainzos-Achirica M, Greenland P, et al. Role of Coronary Artery Calcium Score of Zero and Other Negative Risk Markers for Cardiovascular Disease. Circulation. 2016;133(9):849–858. doi: 10.1161/CIRCULATIONAHA.115.018524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Polonsky TS, McClelland RL, Jorgensen NW, et al. Coronary Artery Calcium Score and Risk Classification for Coronary Heart Disease Prediction. JAMA. 2010;303(16):1610–1616. doi: 10.1001/jama.2010.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grundy SM, Stone NJ, Chair V, et al. Cholesterol Clinical Practice GuidelinesAHA/ACC/AACVPR/AAPA/ABC/ACPM/ADA/AGS/APhA/ASPC/NLA/P CNA guideline on the management of blood cholesterol: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Gui. Circulation. 2018;(November). doi: 10.1161/CIR.0000000000000625. [DOI] [Google Scholar]
- 32.Yeboah J, Mcclelland RL, Polonsky TS, et al. Comparison of Novel Risk Markers for Improvement in Cardiovascular Risk Assessment in Intermediate-Risk Individuals. Jama. 2012;308(8):788–795. doi: 10.1001/jama.2012.9624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McClelland RL, Chung H, Detrano R, Post W, Kronmal RA. Distribution of coronary artery calcium by race, gender, and age: Results from the Multi-Ethnic Study of Atherosclerosis (MESA). Circulation. 2006;113(1):30–37. doi: 10.1161/CIRCULATIONAHA.105.580696. [DOI] [PubMed] [Google Scholar]
- 34.Tota-Maharaj R, Blaha MJ, Blankstein R, et al. Association of Coronary Artery Calcium and Coronary Heart Disease Events in Young and Elderly Participants in the Multi-Ethnic Study of Atherosclerosis. Mayo Clin Proc. 2014;89(10):1350–1359. doi: 10.1016/j.mayocp.2014.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Budoff MJ, Nasir K, McClelland RL, et al. Coronary Calcium Predicts Events Better With Absolute Calcium Scores Than Age-Sex-Race/Ethnicity Percentiles. J Am Coll Cardiol. 2009;53(4):345–352. doi: 10.1016/j.jacc.2008.07.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carr JJ, Jacobs DR, Terry JG, et al. Association of Coronary Artery Calcium in Adults Aged 32 to 46 Years With Incident Coronary Heart Disease and Death. JAMA Cardiol. 2017;2(4):391–399. doi: 10.1001/jamacardio.2016.5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pursnani A, Mayrhofer T, Ferencik M, Hoffmann U. The 2013 ACC/AHA cardiovascular prevention guidelines improve alignment of statin therapy with coronary atherosclerosis as detected by coronary computed tomography angiography. Atherosclerosis. 2014;237(1):314–318. doi: 10.1016/j.atherosclerosis.2014.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stone NJ, Robinson JG, Lichtenstein AH, et al. 2013 ACC/AHA Guideline on the Treatment of Blood Cholesterol to Reduce Atherosclerotic Cardiovascular Risk in Adults. Circulation. 2014;129(25 suppl 2):S1–S45. doi: 10.1161/01.cir.0000437738.63853.7a. [DOI] [PubMed] [Google Scholar]
- 39.Nasir K, Bittencourt MS, Blaha MJ, et al. Implications of Coronary Artery Calcium Testing among Statin Candidates According to American College of Cardiology/American Heart Association Cholesterol Management Guidelines MESA (Multi-Ethnic Study of Atherosclerosis). J Am Coll Cardiol. 2015;66(15):1657–1668. doi: 10.1016/j.jacc.2015.07.066. [DOI] [PubMed] [Google Scholar]
- 40.Herman CR, Gill HK, Eng J, Fajardo LL. Screening for Preclinical Disease: Test and Disease Characteristics. Am J Roentgenol. 2002;179(4):825–831. doi: 10.2214/ajr.179.4.1790825. [DOI] [PubMed] [Google Scholar]
- 41.Bild DE, Bluemke DA, Burke GL, et al. Multi-Ethnic Study of Atherosclerosis: objectives and design. Am J Epidemiol. 2002;156(9):871–881. doi: 10.1093/aje/kwf113. [DOI] [PubMed] [Google Scholar]
- 42.Burke G, Lima J, Wong ND, Narula J. The Multiethnic Study of Atherosclerosis. Glob Heart. 2016;11(3):267–268. doi: 10.1016/j.gheart.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 43.Das S, Forer L, Schonherr S, et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48(10):1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carr JJ, Nelson JC, Wong ND, et al. Calcified Coronary Artery Plaque Measurement with Cardiac CT in Population-based Studies: Standardized Protocol of Multi-Ethnic Study of Atherosclerosis (MESA) and Coronary Artery Risk Development in Young Adults (CARDIA) Study. Radiology. 2005;234(1):35–43. doi: 10.1148/radiol.2341040439. [DOI] [PubMed] [Google Scholar]
- 45.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 46.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: A tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88(1):76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Machiela MJ, Chanock SJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015;31(21):3555–3557. doi: 10.1093/bioinformatics/btv402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J R Stat Soc Ser B. 2009;57(1):289–300. [Google Scholar]
- 49.Wojczynski MK, Li M, Bielak LF, et al. Genetics of coronary artery calcification among African Americans, a meta-analysis. BMC Med Genet. 2013;14(1):75.doi: 10.1186/1471-2350-14-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


