Abstract
Prior studies have demonstrated that fibroblast receptor 3 (FGFR3)-mutant urothelial cancers (UCs) are associated with decreased T-cell infiltration. As FGFR3 mutations are enriched in luminal-like UC and luminal-like UC has been shown to be relatively less responsive to PD-1/PD-L1 inhibition (checkpoint inhibition [CPI]), these data have led to the speculation that FGFR3 mutations may be causally related to poor T-cell infiltration and that UC patients harboring FGFR3 mutations may be suboptimal candidates for CPI. Using data derived from two clinical trials exploring CPI in metastatic UC, we demonstrate no statistically significant difference in response rates in patients with FGFR3-mutant versus wild-type UC. We present hypothesis-generating data, suggesting that similar response rates may be explained by a “balancing out” of previously identified independent positive and negative predictors of CPI sensitivity; that is, compared with FGFR3 wild-type UC, FGFR3-mutant UC is associated with a similar tumor mutational burden, lower T-cell infiltration, but also lower stromal/transforming growth factor beta (TGF-β) signals. Based on our findings, FGFR3 mutation status is not a biomarker of resistance to CPI. Indeed, the single-agent activity of both FGFR3 inhibitors and CPI in FGFR3-mutant UC, and potential non–cross resistance provide a strong pragmatic rationale for combination approaches.
Keywords: Bladder cancer, Immune checkpoint blockade, FGFR3, PD-1 blockade, PD-L1 blockade, Urothelial cancer
Patient summary
In this report, we examined the impact of a mutated gene found in a subset of urothelial cancers on response to treatment with immunotherapy. We found that patients with tumors harboring mutations in the gene FGFR3 respond to immunotherapy similarly to patients without such mutations.
Immune checkpoint inhibition (CPI) has changed the landscape of treatment for metastatic urothelial cancer (mUC). However, a minority of patients respond to treatment, prompting the pursuit of biomarkers and mechanisms underlying resistance to guide combination approaches. Prior studies have shown that luminal I, or luminal papillary, urothelial cancer (UC) harbors lower T-cell infiltration compared with other subtypes, and is also associated with lower response rates with CPI [1,2]. Further, an in silico analysis previously reported a correlation between fibroblast receptor 3 (FGFR3) alterations and decreased T-cell infiltration [3]. As FGFR3 mutations (mFGFR3) are enriched in luminal UC, these data have led to the speculation that mFGFR3 may causally be related to poor T-cell infiltration. These observations have further fueled the conjecture that UC patients harboring mFGFR3 may be suboptimal candidates for CPI and have stimulated interest in combining CPI with FGFR3 inhibition as a means of overcoming CPI resistance.
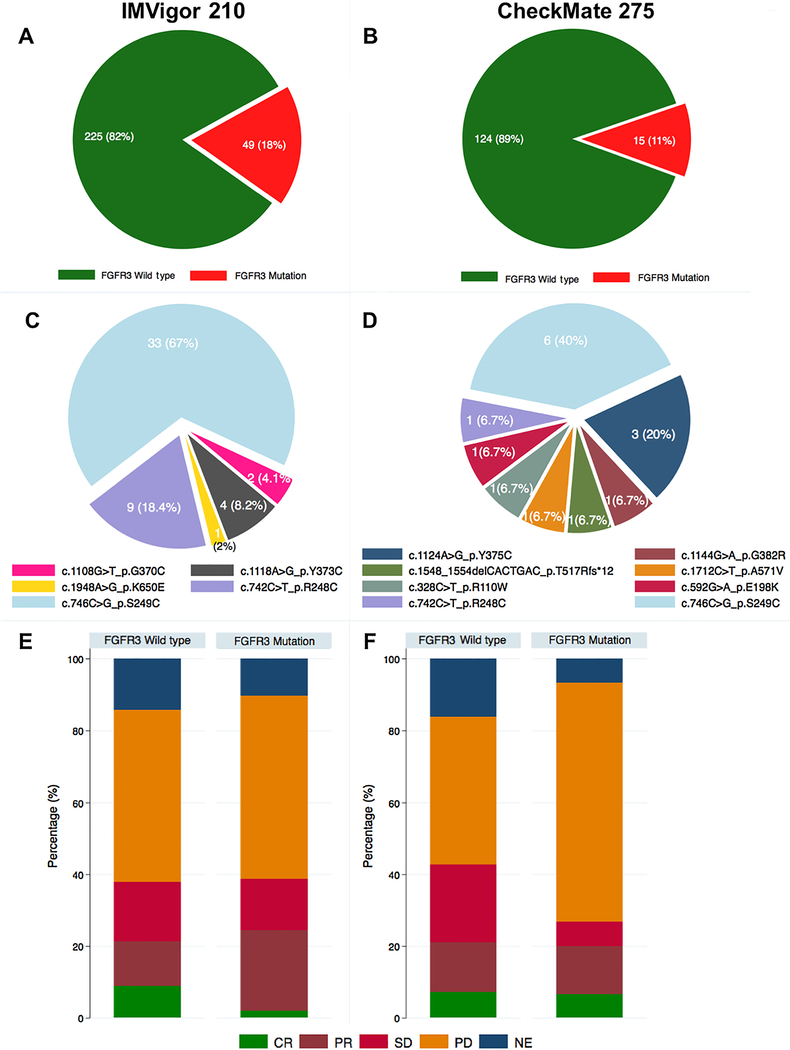
We used the IMVigor 210 cohorts 1 and 2, a phase 2 trial exploring the PD-L1 inhibitor atezolizumab in patients with mUC, to assess the association between mFGFR3 and response to CPI [4]. The characteristics of IMVigor 210 have been described [4] and cohort characteristics are summarized in Supplementary Table 1. Hybrid capture-based next-generation sequencing data were available for 274 patients, among whom 49 had tumors harboring mFGFR3. There was no statistically significant difference in objective response rate with CPI or overall survival (OS) in patients with and without mFGFR3 in an analysis combining IMVigor 210 cohorts 1 and 2 (Fig. 1, Supplementary Table 2, and Supplementary Fig. 1 and 2), or when cohorts 1 and 2 were analyzed separately (Supplementary Fig. 3). We confirmed these findings in a second cohort utilizing whole exome sequencing data from CheckMate 275 [2], a single-arm phase 2 trial exploring the PD-1 inhibitor nivolumab in patients with mUC (Fig. 1, Supplementary material, Supplementary Table 1, and Supplementary Fig. 1 and 2).
Fig. 1 –
Association between FGFR3 mutations and response to treatment with atezolizumab in IMVigor 210 or nivolumab in CheckMate 275 cohorts. (A) Targeted exome sequencing data were available for 274 patients (Foundation One; Foundation Medicine) from the IMVigor 210 cohort, among whom 49 had tumors harboring FGFR3 mutations. (B) Whole exome sequencing data were available for 139 patients from the CheckMate 275 cohort, among whom 15 had tumors harboring FGFR3 mutations. Distribution of specific FGFR3 mutations among (C) 49 tumors in IMVigor 210 and (D) 15 tumors in CheckMate 275. (E) Objective response rate in patients with FGFR3-mutant versus wild-type tumors with atezolizumab in the IMVigor 210 cohort. The response rates (complete and partial responses) were 24% (95% CI: 14%, 39%) and 21% (95% CI: 16%, 27%) in FGFR3-mutant group and wild-type group, respectively (p = 0.8). (F) Objective response rate to nivolumab in the CheckMate 275 cohort. When considering only known FGFR3 hotspot mutations in the CheckMate 275 cohort, 12/139 (8.6%) harbored mutations and the objective response rate (complete and partial responses) in patients with FGFR3-mutant tumors was 20% (95% CI: 6%, 51%) versus 21% (95% CI: 15%, 29%) in patients with FGFR3 wild-type tumors (p = 0.2). All p values are based on the chi-square test. CI = confidence interval; CR = complete response; NE = not evaluable; PD = progression of disease; PR = partial response; SD = stable disease.
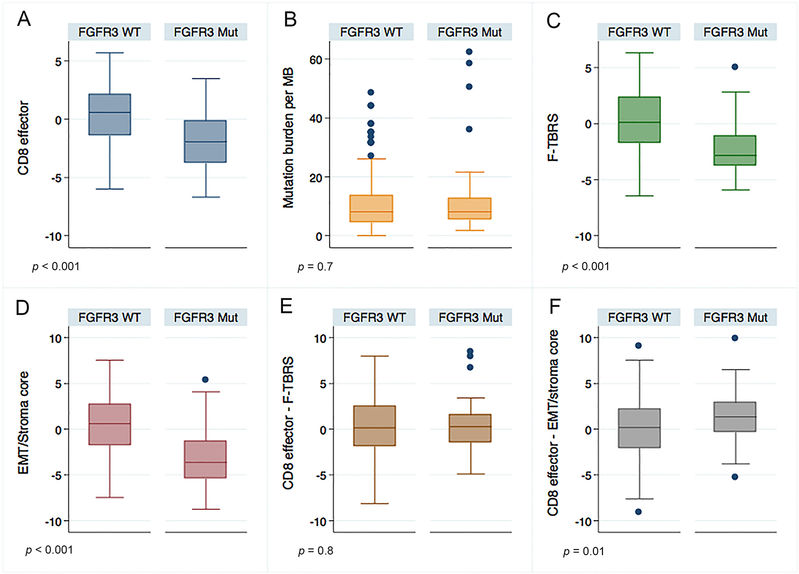
We sought to reconcile our observations from these two clinical trial cohorts with prior observations correlating FGFR3 alterations with decreased T-cell infiltration. We first focused on FGFR3 gene expression because (1) prior studies correlated increased FGFR3 gene expression, along with mutations, with decreased T-cell infiltration [3] and (2) FGFR3 gene expression may encompass other mechanisms of increased FGFR3 signaling (eg, amplifications, gene fusions) [5]. We confirmed that mFGFR3 was associated with increased FGFR3 gene expression compared with wild-type (WT) UC in the IMVigor 210 cohort (Supplementary Fig. 4). Consistent with prior studies, FGFR3 expression, or mFGFR3, was also negatively correlated with a T-cell gene signature (Supplementary Fig. 5 and Fig. 2). However, there was no statistically significant difference in response rates or OS with CPI among groups separated based on FGFR3 gene expression (Supplementary Fig. 6 and 7).
Fig. 2 –
Relationship between FGFR3 mutations and previously identified independent predictors of response to PD-1/PD-L1 blockade. Compared with wild-type tumors, neoplasms with FGFR3 mutations in the IMVigor 210 cohort were associated with (A) lower CD8 T-cell gene signature expression (estimated difference of CD8 effector expression level −2.34 [95% CI: −3.14, −1.54; p < 0.001]), (B) no statistically significant difference in tumor mutational burden (estimated difference of mutation burden: 1.88 [95% CI: −2.09, 5.86; p = 0.7]), and (C) lower fibroblast TGF-β response signature (F-TBRS) (estimated difference of F-TBRS: −2.43 [95% CI: −3.21, −1.65; p < 0.001]) and (D) EMT/Stroma_core signature expression (estimated difference of EMT/Stroma_core: −3.63 [95% CI: −4.62, −2.64; p < 0.001]). The balance of CD8 T-cell gene signature expression and expression of the stromal signatures was either (E) not significantly different in the FGFR3-mutant versus wild-type tumors (F-TBRS; estimated difference 0.09 [95% CI: −0.79, 0.97, p = 0.8]) or (F) higher in FGFR3-mutant versus wild-type tumors (EMT/Stroma- core signature; estimated difference: 1.28 [95% CI: 0.42, 2.14; p = 0.01]). Gene expression is log2 transformed. All p values are based on the Wilcoxon test. CI = confidence interval; Mut = mutant; TGF-β = transforming growth factor beta; WT = wild type.
High tumor mutational burden (TMB) and T-cell gene signatures have been shown to be independently associated with response to CPI [4]. Further, our group and others have demonstrated that gene signatures derived from stromal elements are independently associated with CPI resistance [4,6]. We hypothesized that the lack of association between mFGFR3 and response to CPI, despite a negative correlation with T-cell infiltration, might be due to an imbalance in these other parameters. In the IMVigor 210 cohort, we found no statistically significant difference in TMB between mFGFR3 versus WT tumors; however, mFGFR3 tumors demonstrated significantly lower expression of a fibroblast TGF-β response signature or our epithelial to mesenchymal transition (EMT)/stromal signature versus WT tumors (Fig. 2). These findings support the concept that the lower T-cell infiltration in mFGFR3 tumors may be counterbalanced by a lower level of stromal-mediated immune suppression (Fig. 2) culminating in similar sensitivity to CPI in mFGFR3 and WT UC.
Given that FGFR3 inhibition is being pursued in combination with CPI in clinical trials, we sought to explore the spectrum of gene expression perturbed by mFGFR3 and FGFR3 inhibition in vitro. We obtained gene expression data for a panel of FGFR3 WT and mFGFR3 cell lines (Supplementary material) and also performed RNA sequencing of mFGFR3 cell lines (RT4, SW780, and MGHU3) after knocking down FGFR3 with two independent siRNAs (Supplementary Fig. 8) [7]. Compared with FGFR3 WT cells, mFGFR3 cells were associated with decreased expression of several immune-related genes particularly related to interferon response; conversely, expression of these genes increased with FGFR3 siRNA (Supplementary Fig. 9). FGFR3-mutant cells also demonstrated decreased expression of EMT- and TGF-β–related genes compared with WT cells; again, these gene signatures were increased in cells treated with FGFR3 siRNA (Supplementary Fig. 9). Similar to our data derived from human samples, these findings highlight the possibility of a complex relationship between the downstream impacts of mFGFR3 and antitumor immunity, and underscore the importance of considering the totality of the immunomodulatory effects of FGFR3 inhibition.
At least two retrospective analyses of patients with mFGFR3 UC enrolled in phase I/II trials of FGFR3 inhibitors have indicated infrequent responses to prior CPI in such patients [8,9]; however, given that the majority of patients do not respond to CPI, analyses centered on patients seeking enrollment on trials are at a risk of selection bias for CPI-progressing patients. An analysis of the IMVigor 211 phase 3 study previously revealed a relatively low response rate with atezolizumab in patients with mFGFR3 tumors, although with overlapping 95% confidence intervals for response rate compared with FGFR3 WT tumors [10].
There are potential limitations to our study. Despite confirming that FGFR3 alterations were associated with decreased inferred T-cell infiltration in the IMVigor dataset and decreased interferon response gene signatures in cell culture, these findings do not confirm a causal relationship. We lacked information on FGFR3 gene fusions, but probed the relationship between FGFR3 expression and response to CPI to try and encompass mechanisms of increased FGFR3 signaling beyond mutations. Our cell culture data are hypothesis generating, and the lack of a tumor microenvironment in such systems precludes a comprehensive understanding of the immunomodulatory effects of FGFR3 inhibition and contextualization of changes in gene signatures largely ascribed to stromal cells in studies utilizing bulk human tumor transcriptome data (ie, EMT, TGF-β). However, we hypothesize that cross-talk likely exists between the epithelial and stromal compartments with regard to such signatures. The source of specimens from which sequencing data (eg, primary tumors vs metastases, bladder vs upper urinary tract, etc.) and intratumoral heterogeneity were derived could potentially impact our results. Further, we observed a different prevalence of mFGFR3 in the two clinical trials, which may be at least in part related to technical differences in the next-generation sequencing approaches employed.
There are several implications of our findings: (1) Patients with mUC harboring mFGFR3 should not be denied treatment with CPI. Despite prior analyses demonstrating an association between mFGFR3 and decreased T-cell infiltration, and hypothesis-generating data presented in our analysis highlighting at least one potential causative mechanism underlying such observations, there is currently no direct experimental or clinical evidence for FGFR3 signaling driving CPI resistance. (2) Clinical trials evaluating combinations with CPI should probe the balance of immunomodulatory effects of FGFR3 inhibition.
Several trials combining CPI with FGFR3 inhibitors have been initiated. Despite a potentially complex relationship between mFGFR3 and antitumor immunity, given that non–cross resistance of drugs with single-agent activity may underlie the benefit of most combination regimens [11], the finding that patients with mFGFR3 UC are responsive to CPI similarly to those with WT UC may provide an even greater pragmatic rationale for moving such combinations forward.
Supplementary Material
Acknowledgments
Financial disclosures: Matthew D. Galsky certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: Dr. Andrea Necchi reports grants and personal fees from Merck and Incyte; and personal fees from Bayer, BMS, Rainier Therapeutics, Clovis, Astra Zeneca, and Janssen, outside the submitted work. Matthew D. Galsky has served on an advisory board for Janssen, Dendreon, Merck, GlaxoSmithKline, Lilly, Astellas, Genentech, Bristol-Myers Squibb, Novartis, Pfizer, EMD Serono, AstraZeneca, Seattle Genetics, Incyte, Aileron Therapeutics, Dracen, and Inovio Pharmaceuticals; has received research funding from Janssen, Dendreon, Novartis, Bristol-Myers Squibb, Merck, AstraZeneca, and Genentech/Roche; and is a cofounder of RAPPTA Therapeutics.
Funding/Support and role of the sponsor: None.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Rosenberg JE, Hoffman-Censits J, Powles T, et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet (London, England) 2016;387:1909–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Sharma P, Retz M, Siefker-Radtke A, et al. Nivolumab in metastatic urothelial carcinoma after platinum therapy (CheckMate 275): a multicentre, single-arm, phase 2 trial. Lancet Oncol 2017;18:312–22. [DOI] [PubMed] [Google Scholar]
- [3].Sweis RF, Spranger S, Bao R, et al. Molecular drivers of the non-T-cell-inflamed tumor microenvironment in urothelial bladder cancer. Cancer Immunol Res 2016;4:563–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Mariathasan S, Turley SJ, Nickles D, et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018;554:544–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Mahe M, Dufour F, Neyret‐Kahn H, et al. An FGFR3/MYC positive feedback loop provides new opportunities for targeted therapies in bladder cancers. EMBO Mol Med 2018;10:e8163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Wang L, Saci A, Szabo PM, et al. EMT- and stroma-related gene expression and resistance to PD-1 blockade in urothelial cancer. Nat Commun 2018;9:3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Barretina J, Caponigro G, Stransky N, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012;483:603–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Siefker-Radtke AO, Necchi A, Rosenbaum E, et al. Efficacy of programmed death 1 (PD-1) and programmed death 1 ligand (PD-L1) inhibitors in patients with FGFR mutations and gene fusions: results from a data analysis of an ongoing phase 2 study of erdafitinib (JNJ-42756493) in patients (pts) with advanced urothelial cancer (UC). J Clin Oncol 2018;36(6_suppl):450. [Google Scholar]
- [9].Joerger M, Cassier P, Penel N, et al. Rogaratinib treatment of patients with advanced urothelial carcinomas prescreened for tumor FGFR mRNA expression. J Clin Oncol 2018;36(6_suppl) :494. [Google Scholar]
- [10].Galsky MD, Banchereau R, Kadel EE, et al. 902PBiological features and clinical outcomes in atezolizumab (atezo)-treated patients (pts) with metastatic urothelial cancer (mUC) of the upper vs lower urinary tract (UTUC vs LTUC). Ann Oncol 2018;29(suppl_8):mdy283.111. [Google Scholar]
- [11].Palmer AC, Sorger PK. Combination cancer therapy can confer benefit via patient-to-patient variability without drug additivity or synergy. Cell 2017;171:1678–91.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.