Abstract
Background & Aims:
As an NAD+-dependent deacetylase and a key epigenetic regulator, sirtuin 6 (SIRT6) has been implicated in the regulation of metabolism, DNA repair, and inflammation. However, the role of SIRT6 in alcoholic liver disease (ALD) remains unclear. The aim of this study was to investigate the function and mechanism of SIRT6 in ALD pathogenesis.
Methods:
We developed and characterized Sirt6 knockout (KO) and transgenic (Tg) mouse models that were treated with either control or ethanol diet. Hepatic steatosis, inflammation, and oxidative stress were analyzed using biochemical and histological methods. Gene regulation was analyzed by luciferase reporter and chromatin immunoprecipitation assays.
Results:
The Sirt6 KO mice developed severe liver injury manifested by a remarkable increase of oxidative stress and inflammation whereas the Sirt6 Tg mice were protected from ALD via normalization of hepatic lipids, inflammatory response, and oxidative stress. Our molecular analysis has identified a number of novel Sirt6-regulated genes that are involved in anti-oxidative stress, including metallothionein 1 and 2 (Mt1 and Mt2). Mt1/2 genes were down-regulated in the livers of Sirt6 KO mice and alcoholic hepatitis patients. Overexpression of Mt1 in the liver of Sirt6 KO mice improved ALD by reducing hepatic oxidative stress and inflammation. We also identified a critical link between SIRT6 and metal regulatory transcription factor 1 (Mtf1) via a physical interaction and functional coactivation. Mt1/2 promoter reporter assays showed a strong synergistic effect of SIRT6 on the Mtf1 transcriptional activity.
Conclusions:
Our data suggest that SIRT6 plays a critical protective role against ALD and it may serve as a potential therapeutic target for ALD.
Lay summary:
Liver, the primary organ for ethanol metabolism, can be damaged by the byproducts of ethanol metabolism including reactive oxygen species. In this study, we have identified a key epigenetic regulator SIRT6 that plays a critical role in protecting liver from the oxidative stress-induced liver injury. Thus, our data suggest that SIRT6 may be a potential therapeutic target for alcohol-related liver disease.
Keywords: SIRT6, Sirtuin, alcoholic liver disease, liver injury, oxidative stress, epigenetic regulation
Graphical Abstract
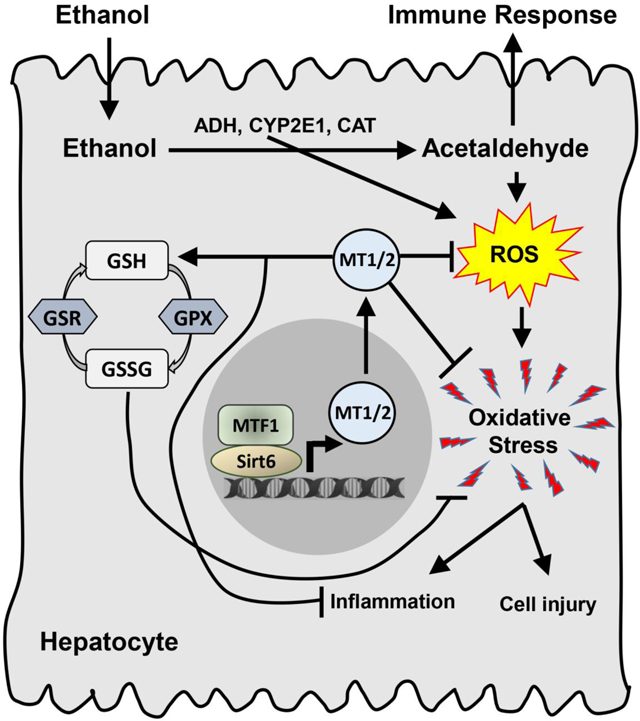
Introduction
Chronic and excessive alcohol consumption causes nearly half of liver cirrhosis-associated mortality in the United States, but there remains no effective treatment for the underlying liver disorder [1]. An early stage of alcohol-related liver disease (ALD), which is featured as simple hepatic steatosis, is reversible; however, chronic and excessive alcohol consumption can lead to progressive steatohepatitis (ASH) and fibrosis, and in some cases, the disease further progresses to cirrhosis and even hepatocellular carcinoma [2, 3]. Alcohol-induced hepatic steatosis initially manifests as lipid droplet accumulation in the liver, but as the liver tissue gets injured by the lipid overload, circulated and resident immune cells including Kupffer cells and infiltrated macrophages and neutrophils respond to the liver injury by producing inflammatory cytokines, such as TNF-α and IL-1β. Chronic alcohol drinking and drinking patterns such as binges can lead to repeated liver injury, inflammation, and oxidative stress [2, 4]. Therefore, there is a clinical need for the better understanding of ALD pathogenesis and identification of therapeutic targets.
As a critical epigenetic regulator of nicotinamide adenine dinucleotide (NAD+)-dependent histone deacetylase, sirtuin 6 (SIRT6) has been implicated in the regulation of longevity, genome stability, metabolism and inflammation [5–24]. Regarding metabolic functions of SIRT6, it decreases hepatic triglycerides (TG) and total cholesterol (TC) by suppressing their biosynthesis-related genes [9, 17, 18, 25–27]. Sirt6 systemic knockout (KO) leads to severe hypoglycemia and premature death [8, 23]. Hepatocyte-specific Sirt6 knockout mice manifest hepatic steatosis even on a regular chow diet [9]. In addition, SIRT6 also plays a critical role as an anti-inflammatory regulator by suppressing pro-inflammatory cytokines such as interleukin (IL)-1β, IL-6, and tumor necrosis factor (TNF) α [7, 22, 28, 29]. Furthermore, SIRT6 has been shown to reduce oxidative stress in the ischemic brain, nonalcoholic fatty liver, and mesenchymal stem cells via regulation of nuclear factor erythroid 2-related factor 2 (NRF2) [11, 30, 31]. However, the role of SIRT6 in ALD remains unclear. In this study, we aimed to examine the hepatic function of SIRT6 using preclinical ALD mouse models and identify the underlying molecular mechanism.
Materials and Methods
Human hepatic tissue samples
Human liver samples were obtained from control subjects and patients with alcoholic cirrhosis (AC) or alcoholic hepatitis (AH) under the institutional review board protocols approved by the Indiana University School of Medicine and Johns Hopkins University School of Medicine, respectively (Supplementary Tables S1 and S2).
Animals
All animal care and experimental procedures were approved by the Institutional Animal Care and Use Committee of Indiana University School of Medicine and Xinxiang Medical University in accordance with National Institutes of Health guidelines for the care and use of laboratory animals. As the phenotypes were similar in males and females, the data presented here were primarily from male mice.
Statistical analysis
Data were presented as mean ± standard error (SEM). Comparisons between two groups were performed using nonparametric Mann-Whitney U tests and comparisons between multiple groups were analyzed using nonparametric Kruskal-Wallis tests (GraphPad, La Jolla, CA). Differences with a p value < 0.05 were considered statistically significant. Error bars represent ± SEM.
For further details regarding the materials and methods used in this study, please refer to the CTAT table and supplementary data.
Results
SIRT6 is decreased in the livers of alcoholic cirrhosis patients and ALD mice
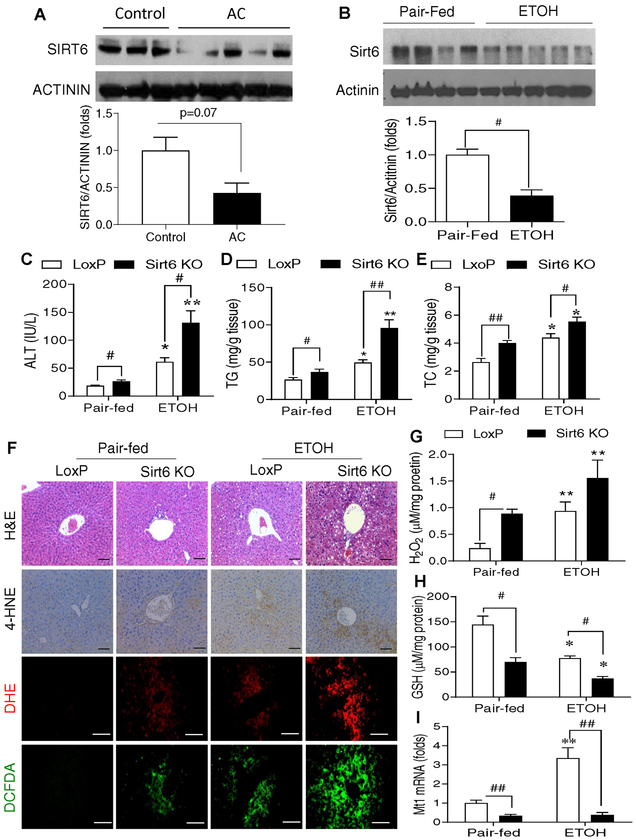
To assess the relevance of hepatic SIRT6 to ALD, we analyzed SIRT6 protein in hepatic tissues of control subjects and alcoholic cirrhosis (AC) patients (Supplementary Table S1) as well as mouse models of ALD. Our data showed that hepatic SIRT6 protein levels were significantly decreased in the livers of AC patients compared to those in controls (Fig. 1A). For the ALD mouse models, we tested two ethanol feeding protocols modified from a previous report by Bin Gao’s laboratory [32]. In the first protocol, wild-type (WT) mice were fed a Lieber-DeCarli liquid diet containing 5% (vol/vol) ethanol for 4 weeks. In the second protocol, WT mice were fed a 6% (vol/vol) ethanol diet for 15 days and administered a single binge of ethanol (6 g/kg) on day 16. In both ALD models, hepatic Sirt6 protein levels were decreased 50–60% in the ethanol-treated mice compared to the pair-fed mice (Fig. 1B and Fig. S1A).
Fig. 1. Hepatic SIRT6 protein levels are decreased in the patients with alcoholic cirrhosis and hepatic Sirt6 deletion aggravates oxidative stress in an ALD mouse model.
(A) Western blot and quantification analysis of SIRT6 protein in the liver of normal controls and alcoholic cirrhosis (AC) patients (n=3–5/group). (B) Western blot and quantification analysis of Sirt6 in hepatic tissues from WT male mice that were fed with a control diet (Pair-fed) or 5% ethanol (ETOH, vol/vol) diet (ETOH) for 4 weeks. (C-I) Control LoxP and Sirt6 KO male mice were pair-fed or ethanol-fed (6% vol/vol) for 15 days plus a single binge (6 g/kg) on day 16. (C) Serum ALT measurements. (D, E) Hepatic triglycerides (TG) and total cholesterol (TC) measurements. (F) Mouse hepatic staining by H&E, IHC detection of 4-HNE, and fluorescence analysis of ROS by DHE and DCFDA dyes. (G) Hepatic H2O2 measurements. (H) Hepatic GSH measurements. (I) Hepatic Mt1 mRNA analysis by qPCR. Data are presented as mean ± S.E.M. Nonparametric Mann-Whitney U tests were used for statistical analysis. In panel B, #p < 0.05 (n = 4–5/group). In panels C-I, #p < 0.05 and ##p < 0.01 for LoxP vs. Sirt6 KO; *p < 0.05 and **p < 0.01 for Pair-fed vs. ETOH for the same genotype (n=4–6/group). Images were captured by light microscopy for H&E and IHC staining (200× magnification), and fluorescence images were obtained using a fluorescence microscope (200× magnification). Scale bars: 50 μm. ALT, alanine transaminase; DCFDA, dichlorofluorescin diacetate; DHE, dihydroethidium; GSH, glutathione; 4-HNE, 4-hydroxynonenal; IHC, immunohistochemistry; qPCR, quantitative PCR; ROS, reactive oxygen species.
Deletion of hepatic Sirt6 exacerbates liver injury and ALD.
To further investigate the role of Sirt6 in the ALD pathogenesis, we used the loss-of-function approach by generating an inducible Sirt6 knockout (Sirt6 KO) mouse model using a floxed Sirt6 mouse strain and an Mx1-Cre line. Upon exposure to synthetic poly(I:C), the Cre transgene is activated via an interferon-inducible mechanism. Numerous reports have shown that the Mx1-Cre is very efficient for hepatic gene manipulation due to an exposure to a high-concentration of poly(I:C) in the liver [33–37]. To confirm that hepatic Sirt6 was deleted in the Sirt6 KO mice, we performed immunoblot analysis of Sirt6 in multiple tissues including liver, heart, skeletal muscle, spleen, white adipose tissue, brain, and lung. Our data showed that the Sirt6 gene was efficiently deleted in the liver but not in other tissues analyzed (Fig. S1C–E). As a functional validation, a Sirt6 substrate — acetylated histone H3 lysine 9 (H3K9Ac) — was elevated in the liver due to the Sirt6 deficiency (Fig. S1C,D). To investigate the role of Sirt6 in ALD, control floxed Sirt6 (LoxP) and Sirt6 KO mice were subjected to either a control (pair-fed) or a Lieber-DeCarli diet containing 6% (vol/vol) ethanol for 15 days and administered an oral gavage of 6 g/kg ethanol on day 16. Diet consumption was comparable in both dietary groups (data not shown). Body weights for both genotypes on the same diet were not significantly different although the LoxP animals on the ethanol diet were lighter than those on the control diet at multiple time points during the experiment (Fig. S2A). Liver weights or liver to body weight ratios were not significantly different between LoxP and Sirt6 KO mice (Fig. S2B,C). Serum ALT levels were remarkably increased in the Sirt6 KO mice on either diet compared to the LoxP mice (Fig. 1C). Hepatic triglycerides and cholesterol were also elevated in the Sirt6 KO mice on both diets compared to the control mice (Fig. 1D,E). This is consistent with elevated expression of lipogenic genes including Srebp1c and Fasn (Fig. S3A). Histological analysis revealed that Sirt6 KO mice had higher levels of oxidative stress in the liver, as indicated by an elevation of a lipid peroxidation marker 4-HNE and reactive oxygen species (ROS) shown by DHE and DCFDA staining (Fig. 1F). Biochemical analysis of hepatic H2O2 and glutathione (GSH) also showed increased oxidative stress and decreased antioxidant capacity (Fig. 1G,H). Interestingly, real-time qPCR analysis showed that induction of metallothionein 1 (Mt1), an anti-oxidative stress gene, by ethanol was significantly impaired in the liver of Sirt6 KO mice (Fig. 1I). Additionally, mRNA levels of a number of anti-oxidant genes showed a trend of downregulation in the liver of Sirt6 KO mice, including glutathione-disulfide reductase (Gsr), catalase (Cat), and superoxide dismutase 1 (Sod1) (Fig. S3B).
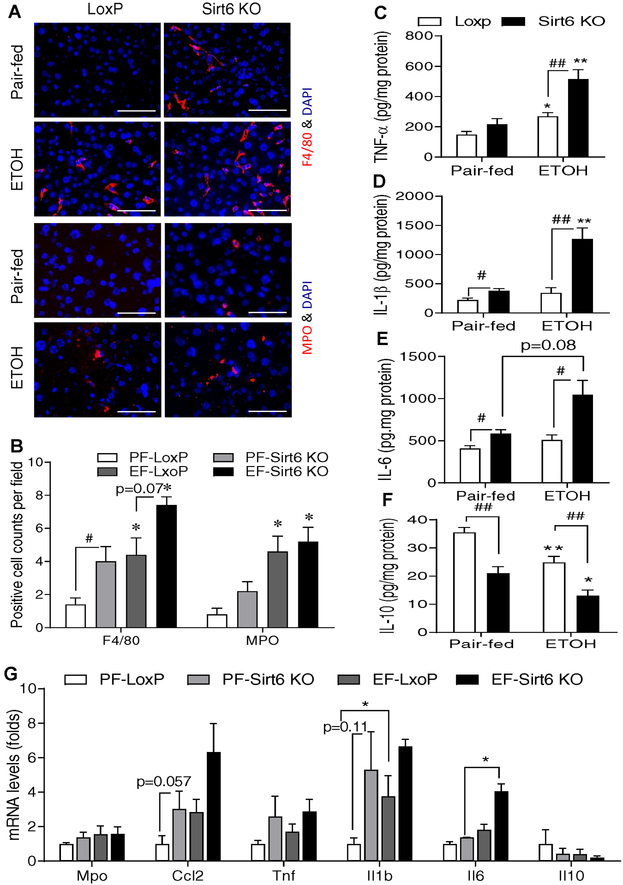
To assess the role of hepatic Sirt6 deficiency in hepatic inflammation, we first performed immunofluorescence (IF) analysis of F4/80 (a marker for macrophage/Kupffer cell) and MPO (a marker for neutrophil) in the liver sections of LoxP and Sirt6 KO mice. Sirt6 deficiency increased the number of F4/80-positive cells even on the control diet and the ethanol diet further increased that number whereas the ethanol diet increased the number of neutrophils in both LoxP and Sirt6 KO mice to the similar levels. (Fig. 2A,B and Fig. S4A–C). Additionally, pro-inflammatory cytokines including TNF-α, IL-1β, and IL-6 in the liver were significantly increased in the ethanol diet-fed Sirt6 KO mice while anti-inflammatory IL-10 was decreased in the Sirt6 KO livers compared to the LoxP livers (Fig. 2C–F). Hepatic mRNA levels of Ccl2, Il1b, and Il6 genes were also highly induced in the ethanol-treated Sirt6 KO livers (Fig. 2G).
Fig. 2. Ethanol-induced inflammation is worsened in hepatic Sirt6 deficient mice.
Control LoxP and Sirt6 KO male mice were pair-fed or ethanol-fed (6% vol/vol) for 15 days plus a single binge (6 g/kg) on day 16. (A) Representative immunofluorescent staining of F4/80 and MPO in mouse liver sections. (B) Quantification of positive staining cells in Panel A. (C-F) Hepatic cytokine measurements: TNF-α (C), IL-1β (D), IL-6 (E), and IL-10 (F). (G) mRNA analysis of inflammation related genes in the liver of control LoxP and Sirt6 KO mice by qPCR. Data are presented as means ± S.E.M. #p < 0.05 and ##p < 0.01 for LoxP vs. Sirt6 KO; *p < 0.05 and **p < 0.01 for Pair-fed vs. ETOH for the same genotype (n=4–6/group). IF images were obtained using a fluorescence microscope (400× magnification). Scale bars: 50 μm. MPO, myeloperoxidase; qPCR, quantitative PCR.
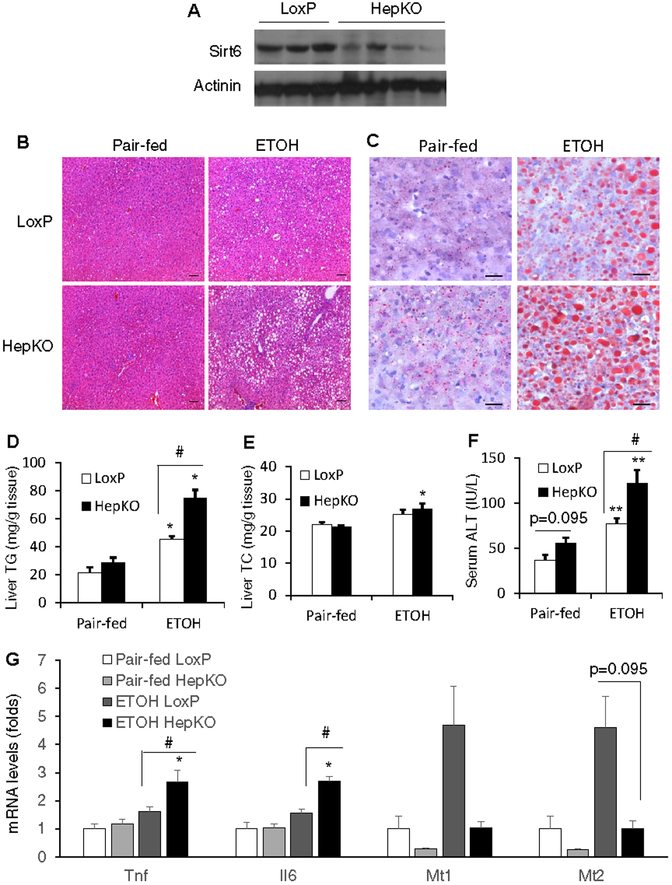
To examine the hepatocyte-specific function of Sirt6, we also generated another Sirt6 knockout mouse model using Alb-Cre (Sirt6-HepKO). The Sirt6-HepKO mice also developed fatty liver disease and liver injury on a 5% (vol/vol) ethanol Lieber-DeCarli diet, confirming the role of hepatic Sirt6 in the protection against the alcohol-induced hepatic steatosis and liver injury (Fig. 3A–F). Real-time qPCR analysis of Tnf, Il6, Mt1 and Mt2 genes showed that the expression of the Tnf and Il6 genes was elevated in the ethanol-treated Sirt6-HepKO livers and the induction of the Mt1 and Mt2 genes was impaired in the Sirt6-HepKO livers (Fig. 3G). RNA-seq analysis revealed that 116 and 136 genes were significantly upregulated in the Sirt6-HepKO livers compared to WT livers under the control or ethanol diet condition, respectively, and 68 of them were common for both dietary conditions (Fig. S5A,B). In addition, 42 and 90 genes were significantly down-regulated in the Sirt6-HepKO livers under the control or ethanol diet condition, respectively, and 10 of them were shared by both dietary conditions (Fig. S5A,B). Pathway analysis further revealed that pathways like steroid hormone biosynthesis, retinol metabolism, and ascorbate and aldarate metabolism were up-regulated and pathways involved in innate immune response and defense response were down-regulated (Fig. S6).
Fig. 3. Hepatocyte-specific Sirt6 knockout (HepKO) mice develop alcoholic liver disease.
Control LoxP and Sirt6 HepKO female mice were pair-fed or ethanol-fed (5% vol/vol) for 10 days plus a single binge (5 g/kg) on day 11. (A) Western blot analysis of Sirt6 in the liver of control LoxP and HepKO mice. (B) Representative H&E stained liver sections (100× magnification). (C) Representative liver sections stained by oil Red O (200× magnification). (D, E) Hepatic TG and TC measurements. (F) Serum ALT measurements. (G) mRNA analysis of Tnf, Il6, Mt1, and Mt2 genes in the liver of control LoxP and HepKO mice. Data are presented as means ± S.E.M. #p < 0.05 for LoxP vs. Sirt6 HepKO; *p < 0.05, **p < 0.01 for Pair-fed vs. ETOH for the same genotype (n=4–5/group). Scale bars: 50 μm. TC, total cholesterol; TG, triglyceride.
SIRT6 knockdown in a cell model of ALD leads to dysregulation of lipid homeostasis and oxidative stress.
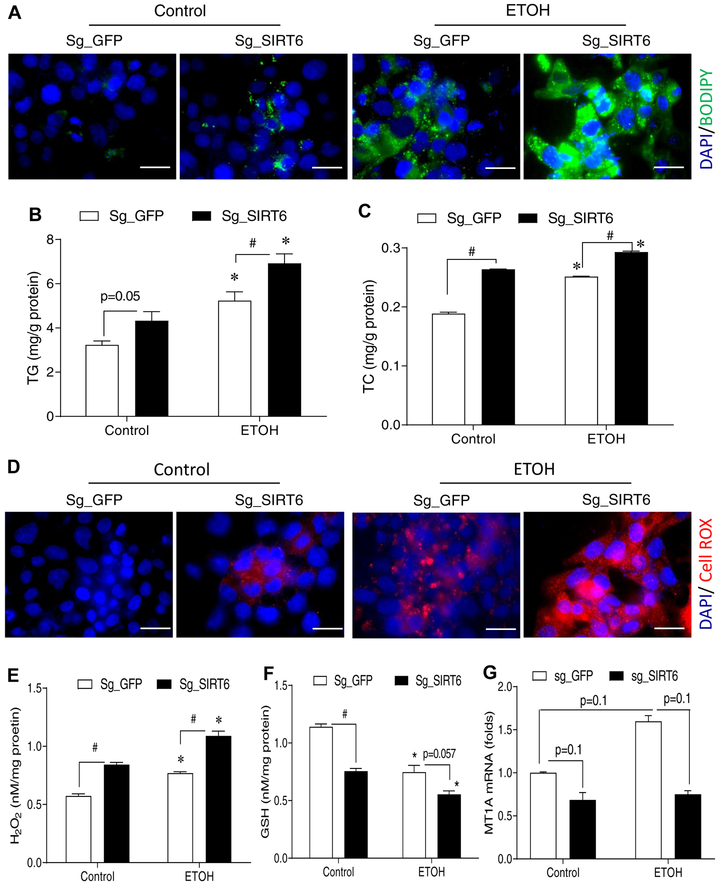
To verify the animal data in a human liver cell model, we created a stable SIRT6 knockdown cell line of VL-17A, which is derived from human HepG2 cell line but stably expresses human alcohol dehydrogenase (ADH) and cytochrome P450 family 2 subfamily E member 1 (CYP2E1), using the CRISPR-Cas9 approach. Both IF imaging and Western blot analysis confirmed that SIRT6 was efficiently knocked down (Fig. S7A–C), and ethanol (50 mM) treatment for 48 hrs further decreased SIRT6 protein and enzymatic activity (Fig. S7B–D). SIRT6 deficiency increased neutral lipid accumulation (BODIPY staining) and intracellular TG and TC under the control and ethanol treatment conditions (Fig. 4A–C). We also measured ROS in live cells using CellROX fluorescent probe (red fluorescence), and our data showed that SIRT6 deficiency dramatically increased ROS levels after the ethanol treatment (Fig. 4D). Cellular H2O2 levels were significantly increased in the SIRT6 knockdown cells under both control and ethanol treatments (Fig. 4E); whereas cellular GSH levels were decreased in the SIRT6 knockdown cells under both control and ethanol conditions (Fig. 4F). Real-time qPCR analysis also showed a trend of downregulation of MT1A gene in the SIRT6-deficient cells under either control or ethanol condition (Fig. 4G). Overall, our data from the VL-17A cell model are consistent with the findings in the Sirt6 KO mice.
Fig. 4. Knockdown of SIRT6 in an ALD cell model leads to lipid dysregulation and oxidative stress.
(A) BODIPY staining of neutral lipids in VL-17A cells that were transduced with control sg_GFP or sg_SIRT6 lentiviruses. (B, C) Cellular TG and TC measurements. (D) ROS detection by CellROX fluorescent probe in live VL-17A cells. (E-G) VL-17A cells were either not treated or treated with 50 mM ethanol for 48 hrs before they were harvested for measurements of intracellular H2O2 (E), GSH (F), or MT1A mRNA (G). Data were presented as mean ± S.E.M (n = 3–4/group). #p < 0.05 for sg_GFP vs. sg_Sirt6; *p < 0.05 for Control vs. ETOH for the same sgRNA. Fluorescence images were obtained using a fluorescence microscope (630× magnification). Scale bars: 10 μm. GSH, glutathione; ROS, reactive oxygen species; TC, total cholesterol; TG, triglyceride.
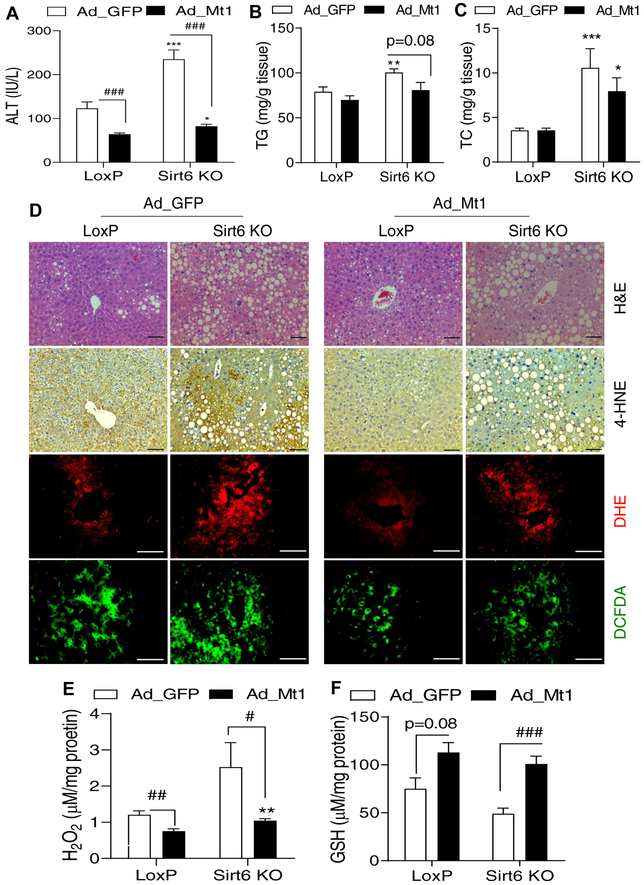
Hepatic Mt1 overexpression ameliorates ethanol-induced liver injury and ALD in Sirt6-deficient mice.
As Mt genes play a critical role in anti-oxidative stress, we hypothesized that overexpression of Mt1 may improve ALD in the Sirt6 KO mice. To test this hypothesis, we generated the Sirt6 knockout mice by injection of poly(I:C) and initiated the ethanol feeding protocol (Fig. S8A). On day 12 after the ethanol feeding, control LoxP and Sirt6 KO mice were injected with either adenoviral GFP (control) or Mt1 (1 × 109 pfu/mouse). Three days later, the animals were given a single ethanol (6 g/kg) gavage. Mt1 mRNA levels were increased 150-fold in LoxP and 78-fold in Sirt6 KO mouse livers, respectively (Fig. S8B). Ethanol-induced liver injury was largely normalized in both LoxP and Sirt6 KO mice by Mt1 overexpression as indicated by serum ALT levels (Fig. 5A). Hepatic TG and TC in the Sirt6 KO mice tended to decrease after the Mt1 overexpression (Fig. 5B,C). Histological analysis also showed significant improvement of lipid peroxidation (4-HNE staining) and ROS levels (DHE and DCFDA staining) in both control and Sirt6 KO mice after the Mt1 overexpression (Fig. 5D). Biochemical analysis also confirmed that Mt1 overexpression decreased hepatic H2O2 and increased GSH levels (Fig. 5E,F). Real-time qPCR analysis showed that Mt1 overexpression also increased expression of antioxidant genes such as Gsr and Cat but had no effect on lipid metabolism genes (Fig. S8D,E).
Fig. 5. Hepatic Mt1 overexpression ameliorates ethanol-induced oxidative stress in the liver.
Control LoxP and Sirt6 KO male mice were pair-fed or ethanol-fed (6% vol/vol) for 15 days plus a single binge (6 g/kg) on day 16. Adenoviral vectors were injected on day 12 of ethanol feeding. (A) Serum ALT levels. (B, C) Hepatic TG and TC measurements. (D) H&E staining, IHC analysis of 4-HNE, and fluorescence analysis of ROS using DHE and DCFDA dyes in mouse liver sections. (E, F) Hepatic H2O2 and GSH measurements. Data were presented as mean ± S.E.M (n = 8/group). #p < 0.05, ##p < 0.01, and ###p < 0.001 for Ad_ GFP vs. Ad_Mt1; *p < 0.05, **p < 0.01, and ***p < 0.001 for LoxP vs. Sirt6 KO for the same adenoviral vector. Images were captured by light microscopy for H&E and IHC staining (200× magnification), and fluorescence images were obtained using a fluorescence microscope (200× magnification). Scale bars: 50 μm. ALT, alanine transaminase; DCFDA, dichlorofluorescin diacetate; DHE, dihydroethidium; GSH, glutathione; 4-HNE, 4-hydroxynonenal; IHC, immunohistochemistry; qPCR, quantitative PCR; ROS, reactive oxygen species; TC, total cholesterol; TG, triglyceride.
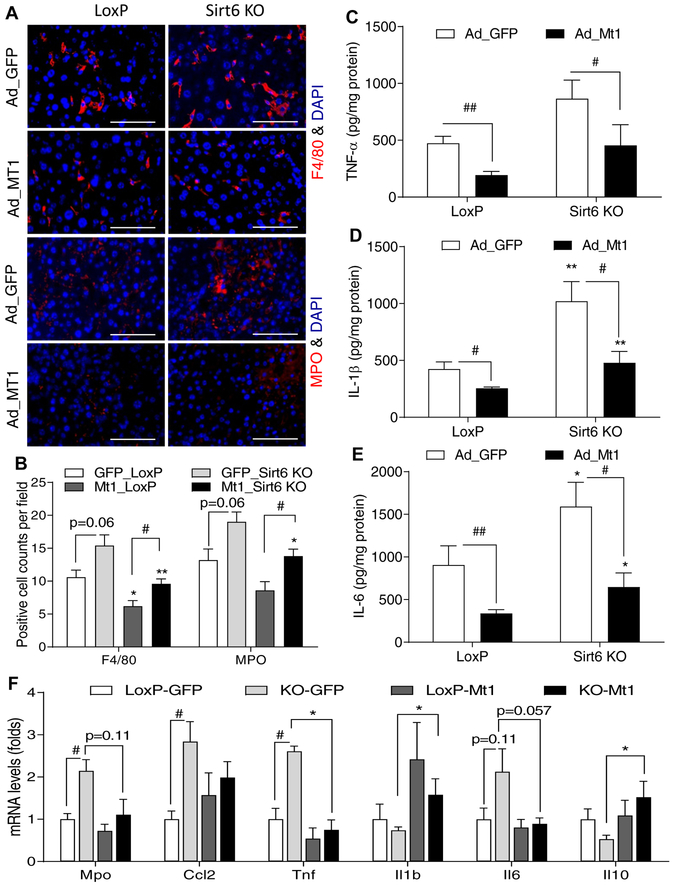
In addition, we also analyzed the effect of Mt1 overexpression on hepatic inflammation. F4/80 and MPO imaging analysis showed that Mt1 overexpression reduced the number of macrophage and neutrophils in the liver tissue (Fig. 6A,B). Mt1 overexpression also significantly lowered TNF-α, IL-1β, and IL-6 in control and Sirt6 KO livers (Fig. 6C–E). Real-time qPCR analysis showed a significant down-regulation of proinflammatory cytokine Tnf and an upregulation of anti-inflammatory cytokine Il10 (Fig. 6F).
Fig. 6. Hepatic Mt1 overexpression dampens ethanol-induced inflammation.
Control LoxP and Sirt6 KO male mice were pair-fed or ethanol-fed (6% vol/vol) for 15 days plus a single binge (6 g/kg) on day 16. Adenoviral vectors were injected on day 12 of ethanol feeding. (A) Representative images of F4/80 and MPO staining in the liver sections. (B) Quantification of positively stained cells in Panel A (n=5). (C-E) Measurements of proinflammatory cytokines including TNF-α, IL-1β, and IL-6 in the mouse liver tissues (n=8). (F) mRNA analysis of inflammation related genes in the liver of LoxP and Sirt6 KO mice (n=4). Data are presented as means ± S.E.M. #p < 0.05 and ##p < 0.01 for LoxP vs. Sirt6 KO; *p < 0.05 and **p < 0.01 for Ad_ GFP vs. Ad_Mt1 for the same genotype. Fluorescence images were obtained using a fluorescence microscope (400× magnification). Scale bars: 50 μm. MPO, myeloperoxidase.
Hepatic Sirt6 overexpression in mice protects against ALD.
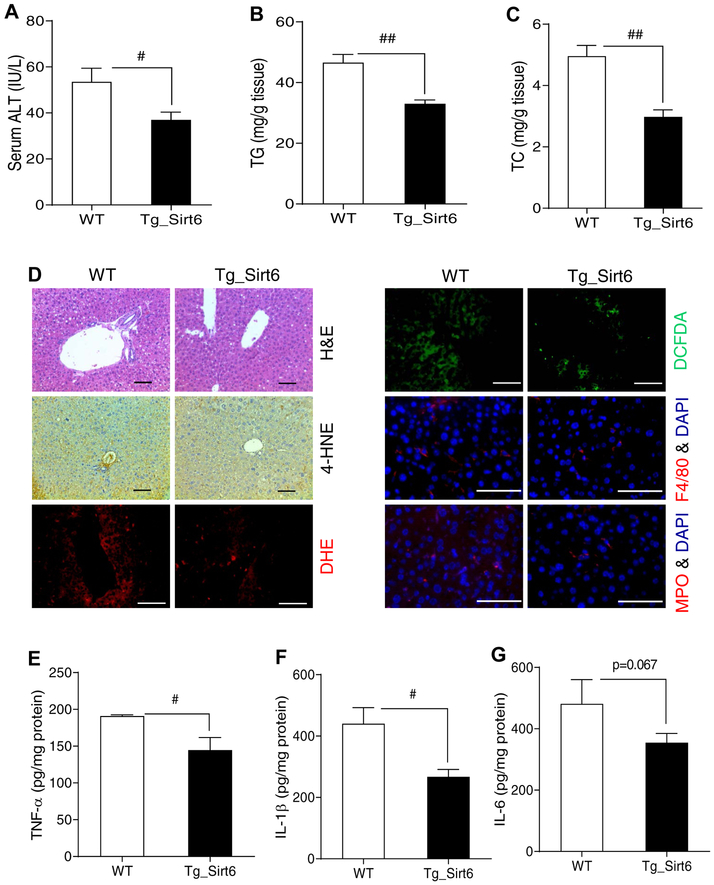
To confirm that Sirt6 indeed has a protective role in ALD, we also generated a hepatocyte-specific Sirt6 transgenic mice using a floxed STOP cassette in front of the Sirt6 transgene and Alb-Cre. Sirt6 protein was increased by 4-fold in the transgenic mouse liver (Fig. S9A,B). After challenged with an ethanol diet (6% vol/vol) and a single ethanol binge (6 g/kg), Sirt6 transgenic mice had lower serum ALT and hepatic TG and TC (Fig. 7A–C). Histological analysis also showed that Sirt6 transgenic mice had lower lipid peroxidation and ROS levels and less inflammation (Fig. 7D). Mt1 gene expression tended to increase in the liver of Sirt6 transgenic mice (Fig. S9C). Hepatic H2O2 levels were decreased and GSH levels were increased in the Sirt6 transgenic liver (Fig. S9D,E). The inflammatory gene Ccl2 was downregulated in the Sirt6 transgenic liver (Fig. S9F). Anti-oxidative stress genes including Gpx3, Gsr, Sod1, and Sod2 were upregulated and lipogenic genes like Srebp1c and Fasn were downregulated in the liver of Sirt6 transgenic mice (Fig. S9H). Hepatic cytokine analysis also showed a significant decrease in the levels of TNF-α and IL-1β and a trend of decrease in IL-6 in the Sirt6 transgenic liver (Fig. 7E–G).
Fig. 7. Hepatic Sirt6 overexpression protects against ALD in mice.
WT and Sirt6 transgenic (Tg_Sirt6) male mice were fed with a 6% ethanol (vol/vol) diet for 15 days plus a single binge (6 g/kg) on day 16. (A) Serum ALT measurements. (B, C) Hepatic TG and TC measurements. (D) H&E staining, IHC analysis of 4-HNE, ROS analysis by DHE and DCFDA dyes, and F4/80 and MPO staining. (E-G) Measurements of proinflammatory cytokines including TNF-α, IL-1β, and IL-6 in the liver tissues of WT and Sirt6 transgenic mice. Data are presented as means ± S.E.M (n = 4–6/group). #p < 0.05 and ##p < 0.01 for WT vs. Tg_Sirt6. H&E and IHC images were captured using a light microscope (200× magnification), and fluorescence images were obtained using a fluorescence microscope (200× magnification). Scale bars: 50 μm. ALT, alanine transaminase; DCFDA, dichlorofluorescin diacetate; DHE, dihydroethidium; 4-HNE, 4-hydroxynonenal; IHC, immunohistochemistry; ROS, reactive oxygen species; TC, total cholesterol; TG, triglyceride.
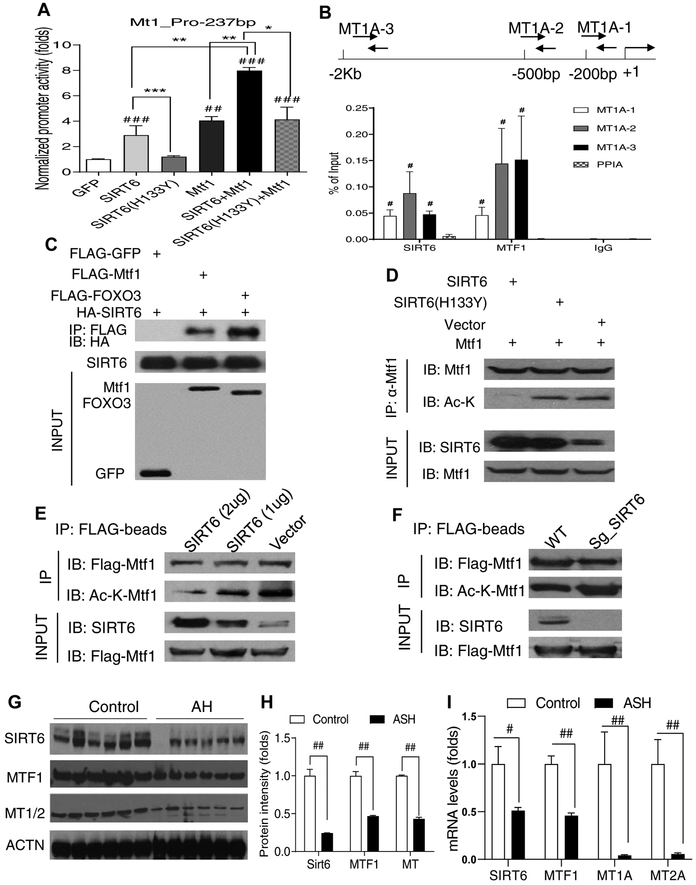
Hepatic Mt1 gene is directly regulated by SIRT6 through activation of Mtf1.
To investigate whether SIRT6 directly controls Mt gene expression, we performed luciferase reporter assays for the proximal promoters of mouse Mt1 and Mt2 genes. Indeed, both Mt1 and Mt2 promoters were activated by SIRT6. Moreover, SIRT6 and metal regulatory transcription factor 1 (Mtf1) had a synergistic effect on those Mt gene promoters (Fig. 8A and Fig. S10A,B). The activation by SIRT6 required its enzymatic activity as the catalytically inactive SIRT6 mutant (H133Y) could not activate the Mt1 gene promoter (Fig. 8A). Chromatin immunoprecipitation (ChIP) qPCR analysis further confirmed that SIRT6 was associated with the chromatin in the promoters of human MT1A and MT2A genes in VL-17A cells (Fig. 8B and Fig. S10C). To assess whether SIRT6 and Mtf1 physically interact, we performed coimmunoprecipitation (Co-IP) analysis by co-transfection of the tagged constructs in HEK 293T cells. Indeed, SIRT6 interacted with Mtf1 as well as FOXO3 (a positive control) (Fig. 8C). Additionally, wild-type but not catalytically inactive mutant SIRT6 (H133Y) deacetylated Mtf1 (Fig. 8D). Furthermore, SIRT6 overexpression decreased and SIRT6 knockdown increased the Mtf1 acetylation levels, suggesting that Mtf1 acetylation is modulated by SIRT6 (Fig. 8E,F).
Fig. 8. SIRT6 induces hepatic Mt1 gene expression by interaction and activation of Mtf1.
(A) Mt1 gene promoter activity analysis by transfection of luciferase reporters and GFP, SIRT6, SIRT6 (H133Y), or Mtf1 plasmids in HEK 293T cells (n =6–7/group). (B) ChIP qPCR analysis of the association of SIRT6 and MTF1 with the proximal promoter of the human MT1A gene under the ethanol treatment condition in VL-17A (n =4/group). (C) Co-IP analysis of an interaction between SIRT6 and Mtf1 in HEK 293T cells. (D) Mtf1 deacetylation analysis by cotransfection of Mtf1 and SIRT6 or SIRT6 (H133Y) in VL-17A cells. (E) Dose-dependent Mtf1 deacetylation analysis in VL-17A cells. (F) Mtf1 acetylation analysis in SIRT6-deficient VL-17A cells. (G, H) Western blot and quantification analysis of SIRT6, MTF1, and MT1/2 proteins in the liver of normal controls and AH patients (n=6). (I) qPCR analysis of SIRT6, MTF1, MT1A, and MT2A mRNAs in the liver of normal controls and AH patients (n=6). Data are presented as mean ± S.E.M. In panels A and B: #p < 0.05, ##p < 0.01, and ###p < 0.001 vs. GFP or PPIA; *p < 0.05, **p<0.01, and ***p<0.001 for the indicated promoter reporter assays. In panels H and I: #p < 0.05 and ##p < 0.01 for AH vs. Control. ACTN, actinin; AH, alcoholic hepatitis; ChIP, chromatin immunoprecipitation; Co-IP, co-immunoprecipitation; qPCR, quantitative PCR.
To examine the relevance of the newly identified SIRT6-MTF1-MTs pathway in human ALD, we obtained 12 human liver samples including 6 normal controls and 6 alcoholic hepatitis (AH) explants (Supplementary Table S2). Western blot analysis showed that SIRT6, MTF1, and MT1/2 protein levels were decreased more than 50% compared to the normal controls (Fig. 8G,H). Real-time qPCR analysis also revealed that mRNA levels of SIRT6, MTF1, MT1A, MT2A genes were also decreased ranging from 50% (SIRT6 and MTF1) to over 90% (MT1A and MT2A) (Fig. 8I). These data suggest that SIRT6, MTF1, and MTs may be involved in human ALD.
Discussion
In this work, we have demonstrated a critical role of SIRT6 in anti-oxidative stress against the ethanol-induced liver injury and ALD using both cell and animal models. Our investigation has begun from an interesting observation that ethanol significantly down-regulates hepatic SIRT6 expression in both human alcoholic cirrhosis and mouse ALD livers. By using two Sirt6 knockout mouse models, we have confirmed that animals with hepatic Sirt6 deficiency are predisposed to alcoholic liver disease. Mx1-Cre mediated Sirt6 gene knockout is generally believed to target poly(I:C) and interferon α/β-responsive cells including hematopoietic cell lineages and liver cells [33–40]. Previous studies have shown that within a few days of induction of the Mx1-Cre, the floxed sequences can be almost completely deleted in the liver and lymphocytes whereas deletions are partial in other tissues [33]. Since both Mx1-Cre- and Alb-Cre-mediated Sirt6 knockout mice share the general phenotype of ALD, we believe that hepatic Sirt6 deficiency is largely responsible for the ethanol-induced hepatic steatosis, oxidative stress, and inflammation. However, we cannot rule out that Sirt6-deficient Kupffer cells and other immune cells might also play a significant role in the pathogenesis of ALD in the Sirt6 KO mouse model. To test that hypothesis, macrophage/Kupffer cell-specific Sirt6 knockout mouse model will be needed for future investigation.
Once ingested, ethanol is rapidly absorbed by stomach and intestine and delivered to liver for metabolism. The first biochemical reaction is conversion of ethanol to acetaldehyde mainly by alcohol dehydrogenases and to a lesser extent by CYP2E1 and catalase [41]. Acetaldehyde is very toxic and can cause cellular damage and trigger immune reaction. Additionally, under conditions of excessive or chronic intake of ethanol, CYP2E1 not only generates ROS including superoxide anion radical and H2O2, but also produces highly reactive conjugated adducts [41]. Thus, excessive ethanol intake is believed to exhaust hepatic capacity of endogenous antioxidants including the GSH-GSSG system [2]. As an adaptive response to the oxidative stress environment, MT genes are strongly induced by ethanol intake. MTF1 has been previously shown to be the key transcription factor for the induction of MT genes [42, 43]. In this work, we have identified a novel coactivator of MTF1 — SIRT6. Our data suggest that as an NAD+-dependent deacetylase SIRT6 can activate MTF1 by deacetylation whereas the catalytic inactive mutant of SIRT6 cannot activate MTF1. The induction of MT genes by the synergistic action of SIRT6 and MTF1 can boost the hepatic defense mechanism against the ethanol-induced oxidative stress, tissue damage, and inflammation (Fig. S11). Our MT1 overexpression data support a critical role of MT1 in the protection against ALD. Previous studies using MT1 transgenic mice have found similar benefits in the ALD animal model [44].
As SIRT6 is decreased in the liver of ALD mice and AC/AH human patients, it suggests that SIRT6 can be a potential therapeutic target for ALD. Our Sirt6 transgenic mouse data strongly support the salutary effects of Sirt6 against ALD, including a decrease in hepatic triglycerides and cholesterol, reduction of ROS and inflammatory cytokines, and an increase in hepatic antioxidant capacity (elevated GSH and Gsr, Gpx3, Sod1, and Sod2 gene expression). Since SIRT6 is an NAD+-dependent enzyme, an increase in cellular NAD+ level is expected to boost the SIRT6 enzymatic activity. Therefore, modulation of SIRT6 protein levels or its enzymatic activity can be a useful strategy for improving ALD. Moreover, additional study is needed to elucidate the detailed mechanisms that orchestrate the regulation from ethanol intake to epigenetic regulation of MT1/2 gene transcription and anti-oxidative stress by SIRT6.
In summary, here we have reported the novel findings on hepatic Sirt6 function in the ALD animal models and AC/AH human livers. Hepatic Sirt6 is down-regulated in ALD mice and AC/AH patients, and hepatic Sirt6 deficiency exacerbates dyslipidemia, inflammation, oxidative stress, and tissue injury in the liver. Hepatic Sirt6 overexpression reverses the ethanol-induced damages in mice. As one of the potential mechanisms by which hepatic Sirt6 protects against ALD, Sirt6 enhances the transcriptional induction of Mt1 and Mt2 genes by coactivating Mtf1, and Mt1/2 boost the liver defense system against alcoholic toxicity by ameliorating ROS, inflammation, and tissue injury. Therefore, SIRT6 may serve as a promising therapeutic target for ALD.
Supplementary Material
Highlights.
SIRT6 deficiency is predisposed to the development of alcoholic liver disease.
SIRT6 overexpression ameliorates the alcoholic liver disease.
SIRT6 induces metallothionein genes to protect against oxidative stress.
SIRT6 coactivates metal regulatory transcription factor 1.
Acknowledgements
We wish to thank Dr. Rongya Tao, Dr. Xiaolin Zhong, and Zhigang Fang for the assistance with some experiments in this work.
Financial support: This study was supported in part by the following funding sources: NIH R21AA024550 (X. Charlie Dong), R01DK107682 (Suthat Liangpunsakul and X. Charlie Dong), R01DK091592 (X. Charlie Dong), R56DK091592 (X. Charlie Dong), National Natural Science Foundation of China project 81670526 (Xiwen Xiong), Showalter Scholar Award (X. Charlie Dong), R24AA025017 (Zhaoli Sun), Indiana Diabetes Research Center grant NIH P30DK097512, and Indiana Clinical and Translational Sciences Institute funded from the NIH NCATS CTSA UL1TR002529.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest: All authors report no conflict of interest.
References
- [1].Singal AK, Bataller R, Ahn J, Kamath PS, Shah VH. ACG Clinical Guideline: Alcoholic Liver Disease. Am J Gastroenterol 2018;113:175–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Seitz HK, Bataller R, Cortez-Pinto H, Gao B, Gual A, Lackner C, et al. Alcoholic liver disease. Nat Rev Dis Primers 2018;4:16. [DOI] [PubMed] [Google Scholar]
- [3].Louvet A, Mathurin P. Alcoholic liver disease: mechanisms of injury and targeted treatment. Nat Rev Gastroenterol Hepatol 2015;12:231–242. [DOI] [PubMed] [Google Scholar]
- [4].Gao B, Bataller R. Alcoholic liver disease: pathogenesis and new therapeutic targets. Gastroenterology 2011;141:1572–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Qin K, Zhang N, Zhang Z, Nipper M, Zhu Z, Leighton J, et al. SIRT6-mediated transcriptional suppression of Txnip is critical for pancreatic beta cell function and survival in mice. Diabetologia 2018;61:906–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Zhang P, Tu B, Wang H, Cao Z, Tang M, Zhang C, et al. Tumor suppressor p53 cooperates with SIRT6 to regulate gluconeogenesis by promoting FoxO1 nuclear exclusion. Proc Natl Acad Sci U S A 2014;111:10684–10689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Xiao C, Wang RH, Lahusen TJ, Park O, Bertola A, Maruyama T, et al. Progression of Chronic Liver Inflammation and Fibrosis Driven by Activation of c-JUN Signaling in Sirt6 Mutant Mice. J Biol Chem 2012;287:41903–41913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Xiao C, Kim HS, Lahusen T, Wang RH, Xu X, Gavrilova O, et al. SIRT6 deficiency results in severe hypoglycemia by enhancing both basal and insulin-stimulated glucose uptake in mice. J Biol Chem 2010;285:36776–36784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kim HS, Xiao C, Wang RH, Lahusen T, Xu X, Vassilopoulos A, et al. Hepatic-specific disruption of SIRT6 in mice results in fatty liver formation due to enhanced glycolysis and triglyceride synthesis. Cell Metab 2010;12:224–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zhang R, Li H, Guo Q, Zhang L, Zhu J, Ji J. Sirtuin6 inhibits c-triggered inflammation through TLR4 abrogation regulated by ROS and TRPV1/CGRP. J Cell Biochem 2018. [DOI] [PubMed] [Google Scholar]
- [11].Ka SO, Bang IH, Bae EJ, Park BH. Hepatocyte-specific sirtuin 6 deletion predisposes to nonalcoholic steatohepatitis by up-regulation of Bach1, an Nrf2 repressor. FASEB J 2017;31:3999–4010. [DOI] [PubMed] [Google Scholar]
- [12].Cui X, Yao L, Yang X, Gao Y, Fang F, Zhang J, et al. SIRT6 regulates metabolic homeostasis in skeletal muscle through activation of AMPK. Am J Physiol Endocrinol Metab 2017;313:E493–E505. [DOI] [PubMed] [Google Scholar]
- [13].Zhang N, Li Z, Mu W, Li L, Liang Y, Lu M, et al. Calorie restriction-induced SIRT6 activation delays aging by suppressing NF-kappaB signaling. Cell Cycle 2016;15:1009–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Xiong X, Wang G, Tao R, Wu P, Kono T, Li K, et al. Sirtuin 6 regulates glucose-stimulated insulin secretion in mouse pancreatic beta cells. Diabetologia 2016;59:151–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Xiong X, Sun X, Wang Q, Qian X, Zhang Y, Pan X, et al. SIRT6 protects against palmitate-induced pancreatic beta-cell dysfunction and apoptosis. J Endocrinol 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Song MY, Wang J, Ka SO, Bae EJ, Park BH. Insulin secretion impairment in Sirt6 knockout pancreatic beta cells is mediated by suppression of the FoxO1-Pdx1-Glut2 pathway. Scientific reports 2016;6:30321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Tao R, Xiong X, DePinho RA, Deng CX, Dong XC. FoxO3 transcription factor and Sirt6 deacetylase regulate low density lipoprotein (LDL)-cholesterol homeostasis via control of the proprotein convertase subtilisin/kexin type 9 (Pcsk9) gene expression. J Biol Chem 2013;288:29252–29259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Tao R, Xiong X, DePinho RA, Deng CX, Dong XC. Hepatic SREBP-2 and cholesterol biosynthesis are regulated by FoxO3 and Sirt6. J Lipid Res 2013;54:2745–2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kanfi Y, Naiman S, Amir G, Peshti V, Zinman G, Nahum L, et al. The sirtuin SIRT6 regulates lifespan in male mice. Nature 2012;483:218–221. [DOI] [PubMed] [Google Scholar]
- [20].Dominy JE Jr., Lee Y, Jedrychowski MP, Chim H, Jurczak MJ, Camporez JP, et al. The deacetylase Sirt6 activates the acetyltransferase GCN5 and suppresses hepatic gluconeogenesis. Mol Cell 2012;48:900–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Zhong L, D’Urso A, Toiber D, Sebastian C, Henry RE, Vadysirisack DD, et al. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell 2010;140:280–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kawahara TL, Michishita E, Adler AS, Damian M, Berber E, Lin M, et al. SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell 2009;136:62–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell 2006;124:315–329. [DOI] [PubMed] [Google Scholar]
- [24].Xiong X, Zhang C, Zhang Y, Fan R, Qian X, Dong XC. Fabp4-Cre-mediated Sirt6 deletion impairs adipose tissue function and metabolic homeostasis in mice. J Endocrinol 2017;233:307–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Elhanati S, Ben-Hamo R, Kanfi Y, Varvak A, Glazz R, Lerrer B, et al. Reciprocal Regulation between SIRT6 and miR-122 Controls Liver Metabolism and Predicts Hepatocarcinoma Prognosis. Cell reports 2016;14:234–242. [DOI] [PubMed] [Google Scholar]
- [26].Masri S, Rigor P, Cervantes M, Ceglia N, Sebastian C, Xiao C, et al. Partitioning circadian transcription by SIRT6 leads to segregated control of cellular metabolism. Cell 2014;158:659–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Elhanati S, Kanfi Y, Varvak A, Roichman A, Carmel-Gross I, Barth S, et al. Multiple regulatory layers of SREBP1/2 by SIRT6. Cell reports 2013;4:905–912. [DOI] [PubMed] [Google Scholar]
- [28].Luo P, Qin C, Zhu L, Fang C, Zhang Y, Zhang H, et al. Ubiquitin-specific Peptidase 10 (USP10) Inhibits Hepatic Steatosis, Insulin Resistance and Inflammation via Sirt6. Hepatology 2018. [DOI] [PubMed] [Google Scholar]
- [29].Wu Y, Chen L, Wang Y, Li W, Lin Y, Yu D, et al. Overexpression of Sirtuin 6 suppresses cellular senescence and NF-kappaB mediated inflammatory responses in osteoarthritis development. Scientific reports 2015;5:17602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Cardinale A, de Stefano MC, Mollinari C, Racaniello M, Garaci E, Merlo D. Biochemical Characterization of Sirtuin 6 in the Brain and Its Involvement in Oxidative Stress Response. Neurochem Res 2014. [DOI] [PubMed] [Google Scholar]
- [31].Zhang W, Wei R, Zhang L, Tan Y, Qian C. Sirtuin 6 protects the brain from cerebral ischemia/reperfusion injury through NRF2 activation. Neuroscience 2017;366:95–104. [DOI] [PubMed] [Google Scholar]
- [32].Bertola A, Mathews S, Ki SH, Wang H, Gao B. Mouse model of chronic and binge ethanol feeding (the NIAAA model). Nature protocols 2013;8:627–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science 1995;269:1427–1429. [DOI] [PubMed] [Google Scholar]
- [34].Lee AH, Scapa EF, Cohen DE, Glimcher LH. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science 2008;320:1492–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kent LN, Bae S, Tsai SY, Tang X, Srivastava A, Koivisto C, et al. Dosage-dependent copy number gains in E2f1 and E2f3 drive hepatocellular carcinoma. J Clin Invest 2017;127:830–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Nagahama Y, Sone M, Chen X, Okada Y, Yamamoto M, Xin B, et al. Contributions of hepatocytes and bile ductular cells in ductular reactions and remodeling of the biliary system after chronic liver injury. Am J Pathol 2014;184:3001–3012. [DOI] [PubMed] [Google Scholar]
- [37].Onoyama I, Suzuki A, Matsumoto A, Tomita K, Katagiri H, Oike Y, et al. Fbxw7 regulates lipid metabolism and cell fate decisions in the mouse liver. J Clin Invest 2011;121:342–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Wei J, Yuan Y, Chen L, Xu Y, Zhang Y, Wang Y, et al. ER-associated ubiquitin ligase HRD1 programs liver metabolism by targeting multiple metabolic enzymes. Nat Commun 2018;9:3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Levantini E, Lee S, Radomska HS, Hetherington CJ, Alberich-Jorda M, Amabile G, et al. RUNX1 regulates the CD34 gene in haematopoietic stem cells by mediating interactions with a distal regulatory element. EMBO J 2011;30:4059–4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Yu WM, Liu X, Shen J, Jovanovic O, Pohl EE, Gerson SL, et al. Metabolic regulation by the mitochondrial phosphatase PTPMT1 is required for hematopoietic stem cell differentiation. Cell Stem Cell 2013;12:62–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Sugimoto K, Takei Y. Pathogenesis of alcoholic liver disease. Hepatol Res 2017;47:70–79. [DOI] [PubMed] [Google Scholar]
- [42].Ghoshal K, Jacob ST. Regulation of metallothionein gene expression. Progress in nucleic acid research and molecular biology 2001;66:357–384. [DOI] [PubMed] [Google Scholar]
- [43].Laity JH, Andrews GK. Understanding the mechanisms of zinc-sensing by metal-response element binding transcription factor-1 (MTF-1). Arch Biochem Biophys 2007;463:201–210. [DOI] [PubMed] [Google Scholar]
- [44].Zhou Z, Sun X, James Kang Y. Metallothionein protection against alcoholic liver injury through inhibition of oxidative stress. Exp Biol Med (Maywood) 2002;227:214–222. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.