Abstract
Background:
Attempts to identify opioid users at increased risk of escalating to opioid use disorder have had limited success. Data from a variety of sources suggest that genetic variation may mediate the subjective response to opioid drugs, and therefore contribute to their abuse potential. The goal of the current study was to observe the relationship between select genetic polymorphisms and the subjective effects of oxycodone under controlled clinical laboratory conditions.
Methods:
Non-dependent, volunteers with some history of prescription opioid exposure (N=36) provided a blood sample for analyses of variations in the genes that encode for the μ-, κ- and δ-opioid receptors, and the dopamine metabolizing enzyme, catechol-O-methyltransferase (COMT). Participants then completed a single laboratory test session to evaluate the subjective and analgesic effects of oral oxycodone (0, 10, and 20 mg, cumulative dose = 30 mg).
Results:
Oxycodone produced typical μ-opioid receptor agonist effects, such as miosis, and decreased pain perception. Oxycodone also produced dose-dependent increases in positive subjective responses such as: drug “Liking” and “Good Effect.” Genetic variants in the μ- (rs6848893) and δ-opioid receptor (rs581111) influenced the responses to oxycodone administration. Additionally, self-reported “Stimulated” effects of oxycodone varied significantly as a function of COMT rs4680 genotype.
Discussion:
The current study shows that the euphoric and stimulating effects of oxycodone can vary as a function of genetic variation. Though the relationship between the stimulating effects of opioids and their abuse liability is not well established, we know that the ability of opioids to provide intense feelings of pleasure is a significant motivator for continued use. If replicated, specific genetic variants may be useful in predicting who is at increased risk of developing maladaptive patterns of use following medical exposure to opioid analgesics.
Introduction
The clinical utility of opioid analgesics is offset by their potential for misuse. It is now recognized that the diversion and misuse of opioid analgesics is a significant pathway to heroin use and a substantial contributor to the current opioid public health crisis, which is estimated to have taken over 48,000 lives in the United States during the past year (Centers for Disease Control (CDC), 2018; Cicero et al., 2014; Daily et al., 2012; Muhuri et al., 2013). While many factors influence the initiation of opioid use, the opioids’ ability to provide intense feelings of pleasure is a significant motivator for non-medical use of these drugs (Adinoff et al., 2004; de Witt and Phillips, 2012; Ridenour et al., 2003; Peltz and Südhof, 2018).
The rewarding and positive subjective effects of opioids have typically been attributed to their activation of the μ-opioid receptor (MOR), which also triggers dopamine (DA) release in the brain’s reward center (Fields and Margolis, 2015; Pasternak, 2011; Wise, 1998; 2002). By contrast, the κ-opioid receptor (KOR) is thought to mediate dysphoria. The role of the δ-opioid receptor (DOR) remains unclear, but research suggests that DOR modulation has a minimal role in reward processes (Le Merrer et al., 2011; 2012). Some data show that opioid effects on the DOR may be negatively reinforcing by producing anxiolytic or anti-depressant effects (Margois et al., 2008; Pradhan et al., 2011).
It has been estimated that genetic factors contribute up to 80% of the vulnerability to Opioid Use Disorder (OUD), so they are obvious targets in the search for risk and protective factors (Goldman et al., 2005; Li and Burmeister, 2009). Variation in the genes that encode the three opioid receptors has been associated with maladaptive opioid use (Levran et al., 2008; Li et al., 2006; Li and Zhang, 2013; Zhang et al., 2008). Candidate-gene investigations have found data to suggest that select genetic variant may moderate various pharmacodynamic effects of opioids. For example, variation in the genes encoding the μ-opioid receptor (OPRM1) is associated with heroin-induced subjective responses (Zhang et al., 2007). Meanwhile, variation in the genes that encode the δ- and κ -opioid receptors (OPRD1, OPRK1) have been associated with the efficacy of OUD pharmacotherapy (Crist et al., 2013; 2018 B) and severity of opioid withdrawal (Jones et al., 2016 A). Furthermore, genetic variation in dopamine receptor polymorphisms has been linked to cue-elicited opioid craving (Shao et al., 2006).
Although opioid analgesic prescriptions have decreased in recent years, it is estimated that over 200 million opioid analgesic prescriptions are written in the United States each year (CDC, 2017; IQVIA Institute, 2018). Despite promising findings regarding the genetic etiology of OUD and the pharmacogenetics of opioid treatment, no methods currently exist for identifying individuals who are at increased risk of developing OUD. An ability to identify individuals who are at risk of maladaptive patterns of opioid use would potentially be of great benefit in the prevention of OUD.
The current study examines the relationship between opioidergic genetic variation and the subjective and analgesic response to oxycodone among human research volunteers. Because of the noted role of dopaminergic pathways in the neurobiology of drug reinforcement, dopaminergic genetic polymorphisms were also assessed. It is anticipated that genetic variants associated with the effects of oxycodone can provide insight into the addictive properties of this widely prescribed and misused opioid analgesic.
Methods
The investigators assessed the potential influence of a select number of genetic variants on the pharmacodynamic effects of oral oxycodone (OXY), using a human clinical laboratory model. To limit the risk of Type-1 error, we used a candidate-gene approach, in which we tested the effects of 11 genetic markers. Although more exploratory genetic approaches can be used to evaluate large numbers of polymorphisms, this method requires hundreds of participants to correct for multiple testing (Balding, 2006).
Participant Screening and Selection
Participants were recruited from the New York City metropolitan area through various print media advertisements. Respondents who met preliminary study inclusion/exclusion criteria, based upon an initial telephone interview, were scheduled for additional screening procedures at the New York State Psychiatric Institute. Screening consisted of both self-report and clinical interviews administered by a team of research assistants, psychologists, nurses, and physicians. The assessments examined drug use, general health, psychiatric functioning, and medical history. Clinical laboratory testing (hematology, blood chemistry panel, liver and thyroid functioning, and urinalysis) was also performed.
Participants were required to be physically and mentally healthy volunteers between the ages of 21 and 55 years. Potential participants were excluded from the study if they met DSM-IV criteria for drug abuse or dependence or were seeking treatment for any substance use disorder (except nicotine). To test for physiological opioid dependence, naloxone (0.2–0.8 mg) was administered intramuscularly during screening and a trained research nurse assessed the presence of withdrawal signs (Wang, 1974). Rapid urine toxicology screens were used to assess recent drug use at each screening visit. Participants were also excluded if they reported chronic pain or had a severe Axis I psychiatric diagnosis that could make participation hazardous. Participants were compensated $25 for each screening visit and $125 to complete the laboratory testing session. The Institutional Review Board of the New York State Psychiatric Institute approved the study procedures (Study ID: 6400) and this work was carried out in accordance with the code of ethics of the World Medical Association (Declaration of Helsinki).
Study Design
Participants who passed the physiological and psychiatric screening completed a test session during which the subjective and physiological effects of oral oxycodone (0 mg, 10, mg, 20 mg = cumulative dose of 30 mg) were examined under double-blind conditions. A cumulative dosing procedure was used, as it has been shown to be an efficient and reliable strategy for assessing dose-response within a single session (Walker et al., 2001; Walker and Zacny, 1999).
Participants were required to have not used alcohol recently (confirmed by breathalyzer) and provide a negative drug urine drug screen (except for marijuana because it can be detected in the urine for up to a month) on the day of the laboratory test session. Marijuana use could not have been recent (within 2 days of the laboratory session) based on self-report, and the participant could not appear to be acutely intoxicated. No test sessions were rescheduled due to evidence of drug use.
Laboratory sessions began at 1000 hrs and took approximately 5–6 hours to complete (Table 1). After the test session, if participants successfully completed a field sobriety test, they were transported home via taxi or a car service. A follow-up safety visit was conducted 30 days after completion of the test session. During the follow-up visit, participants met with a research psychologist to discuss any subsequent changes in their drug use patterns, and a nurse assessed the presence of study-related adverse events.
Table 1:
Laboratory Session Schedule
| Time | Event |
|---|---|
| −90 | Urine drug toxicology, breathalyzer, pregnancy test |
| −30 | Physiological monitoring begins, Subjective effects, pupils, CPT* |
| 0 | Dose administration #1 (Placebo) |
| 15 | Subjective effects, pupils |
| 30 | Subjective effects, pupils, CPT |
| 45 | Dose administration #2 (10 mg oxycodone) |
| 60 | Subjective effects, pupils |
| 75 | Subjective effects, pupils, CPT |
| 90 | Dose administration #3 (20 mg oxycodone) |
| 105 | Subjective effects, pupils |
| 120 | Subjective effects, pupils, CPT |
| 135 | Subjective effects, pupils |
| 150 | Subjective effects, pupils |
| 165 | Subjective effects, pupils, CPT |
| 180 | Subjective effects, pupils, CPT |
| 195 | Subjective effects, pupils |
| 210 | Subjective effects, pupils, CPT |
| 255 | Subjective effects, pupils, CPT |
| 315 | Subjective effects, pupils, CPT |
Cold Pressor Test (CPT) of pain threshold and pain tolerance.
Laboratory Session Measures
Subjective Effects:
Nineteen Visual Analog Scale (VAS) items were used to assess various subjective drug effects (e.g., “Good Effect,” “High,” “Liking,” “Stimulated”; see Table 3 for the comprehensive list of items). Participants rated each item on the scale from “Not at all” (0 mm) to “Extremely” (100 mm), with the exception of the “Would Pay” item, which ranged from “$0” (0 mm) to “$25” (100 mm). A comprehensive VAS assessment of subjective drug effects is an FDA-recommended metric when assessing the abuse potential of drugs within human subjects (FDA, 2017). Our division has regularly used this tool in clinical abuse liability assessments for over 20 years (Fischman and Foltin, 1996).
Table 3:
Subjective and Physiological Effects of Oxycodone
| Baseline (−30 mins) | Peak | |
|---|---|---|
| Visual Analog Scale: Mean 0–100 (SD) | ||
| Alert | 57.9 (27.8) | 62.1 (25.9) |
| Anxious | 27.1 (27.4) | 29.9 (28.3) |
| Bad Effect | 0.62 (1.24) | 26.8 (33.3)*** |
| Depressed | 7.30 (15.1) | 10.6 (15.7) |
| Energetic | 34.1 (25.5) | 46.9 (27.9) |
| Good Effect | 1.33 (2.7) | 43.8 (34.2)*** |
| High | 3.50 (10.3) | 44.6 (34.9)*** |
| Irritable | 7.80 (12.9) | 28.5 (29.4)** |
| Liking | 12.5 (26.6) | 37.8 (33.7)** |
| Mellow | 47.0 (31.8) | 62.3 (27.4) |
| Nauseated | 3.80 (11.6) | 33.2 (34.2)** |
| Potent | 11.4 (25.4) | 40.1 (32.1)** |
| Quality | 15.0 (27.1) | 37.2 (32.2)** |
| Restless | 19.8 (22.8) | 40.4 (31.5)** |
| Sedated | 10.9 (19.3) | 36.5 (32.6)** |
| Stimulated | 11.0 (16.3) | 32.9 (27.3)** |
| Sleepy | 20.8 (30.3) | 66.9 (31.6)** |
| Talkative | 29.2 (29.5) | 36.8 (26.2) |
| Would Pay | 1.50 (4.70) | 4.40 (6.10) |
| Pupil Diameter: Mean (SD) | ||
| Trough (mm) | 3.20 (0.98) | 3.00 (0.78) |
| Cold Pressor Test (SD) | ||
| Latency to Withdraw (sec) | 38.6 (42.9) | 54.8 (62.4)** |
| Latency to Feel Pain (sec) | 26.8 (51.3) | 41.0 (65.0)** |
p <0.01,
p < 0.001
Analgesic Effects:
The analgesic effects of OXY were evaluated with experimentally induced pain using the cold pressor test (CPT). Crushed ice was added to a cold tank and warm water was placed in a warm tank. The temperature was maintained at 4°C in the cold tank (additional ice was added, if necessary) and 37°C in the warm tank. Each participant was asked first to immerse a hand in the warm tank for 2 min (to equalize baseline skin temperature across participants). Next, the participant was asked to immerse the same hand in the cold tank for up to 2 min. Standard instructions were read to each participant before administration of the CPT. Objective dependent measures included: pain latency (time to the first report of pain) and pain tolerance (time until removal of the hand from water). The CPT is a commonly used and validated model to assess the analgesic effects of opioids in the clinical laboratory (Conley et al., 1997; Zacny et al., 1996).
Physiological Measures:
Miosis was assessed as a physiological indicator of μ agonist effects using a NeurOptics™ Pupillometer (Neuroptics INC. Irvine, CA) under ambient lighting conditions. For safety, a pulse oximeter was used to continuously monitor oxygen saturation (%SpO2) during the test session, while respiration (breaths per minute), heart rate, and blood pressure (systolic and diastolic) were measured and recorded every 5 min.
Drugs
Oxycodone HCL tablets (5 mg) were purchased from TYCO Healthcare (Princeton, NJ). For blinding, the New York State Psychiatric Institute Pharmacy over-encapsulated the tablets prior to dispensing them to study staff. At each dosing time point, participants were given 4 capsules consisting of active drug and/or lactose-filled placebo.
Naloxone HCl (Narcan) for IM injection, used to evaluate the presence of physiological dependence, was obtained from the International Medication System Limited Amphastar (South Elmonte, CA).
Genotyping
Thirty milliliters of venous blood was collected in 8.5 ml ACD vacutainer tubes for genotyping. Within 48 hr of their collection, blood samples were transferred to Columbia’s Human Genetics Research Core where DNA was isolated and stored at −20°C. To ensure the quality of the samples, once extracted, DNA was checked for purity at an OD280 ratio of 1.8 – 2.0. Extracted DNA was genotyped for the variants of interest by LGC Genomics using PCR-based methods.
Genetic Polymorphisms of Interest
We examined variation in the genes encoding the μ-opioid receptor (OPRM1: rs1799971, rs6848893), δ-opioid receptor (OPRD1: rs10753331, rs2234918, rs581111, rs678849), κ-opioid receptor (OPRK1: rs6473797, rs963549) and the major DA-metabolizing enzyme, catechol-O-methyltransferase (COMT: rs165599, rs4680, rs737865) as independent variables. These polymorphisms were selected based upon functional relevance, a minor allele frequency of >10%, and/or prior evidence that they have affect the variables of interest (Bond et al., 1998; Crist et al., 2013, 2018a,b; Crettol et al., 2008; Drakenberg et al., 2006; Jones et al., 2015,2016A; Mayer et al., 1997; Oosterhuis et al., 2008; Vandenbergh et al., 1997).
Statistical Analyses
Continuous and categorical demographic variables were summarized descriptively in terms of means, standard deviations, and proportions. The distributions of all continuous variables were checked for normality before parametric comparison techniques were employed. When assessing general effects of OXY (0, 10, 20 mg), analysis of variance (ANOVA) was used to compare the time course of drug effects over the various time points throughout the session. If ANOVA revealed a significant main effect of “Time” T-tests were used to compare the baseline (pre-drug) measurement to the maximal post-OXY measurement (peak= highest or trough= lowest) to determine where significant differences existed. Peak/trough analysis is the recommended method of analyzing time-course subjective effects data in human abuse potential studies (Comer et al., 2012). All tests were performed at a 2-tail probability. For statistical tests without genetic independent variables, a conservative p-value of <0.01 was considered statistically significant.
To assess the influence of the genetic variants of interest, a factorial ANOVA was used to compare peak or trough OXY effect between each of the genotypes of interest (alone and in combination). The target genetic polymorphisms were coded as binary variables, homozygous major allele carriers vs minor allele carriers. Generally, the p-value threshold that is considered statistically significant in biomedical research is 0.05. However, this p-value is not appropriate when testing many independent variables in a pharmacogenetic analysis as the frequency of Type-I error is greatly increased (Peters et al., 2010). Therefore, a Bonferroni correction was applied (Shaffer et al., 1995) dividing the significance cut-off by the number of independent variables (i.e., p= 0.05/11 genetic variants = adjusted p<.004). In cases where the overall ANOVA was significant for a variant of interest. All data analyses were performed using SPSS version 25 (IBM Corp., 2017). Genotype frequencies for all single nucleotide polymorphisms (SNPs) were tested for consistency with Hardy–Weinberg equilibrium expectations.
Results
Participants
Between 2012 and 2015, 36 participants completed the test session. The average age of the participants was 36.5 years (± 9.7) and included 33 men and 3 women. The racial breakdown was as follows: 12 African-American /Black, 18 Caucasian/White, 3 Multiracial, 1 Native-American or Alaskan, 2 chose not to report. Ethnically, 22% of the sample identified as Hispanic or Latino.
All participants had previous exposure to opioid analgesics, with 35% (n=13) reporting exclusively medical use. Among participants who currently used opioids recreationally (65%, n=23), 70% reported doing so on a less than monthly basis, 12% monthly, and 18% weekly. All of the recreational opioid users reported using opioids orally, with the exception of 2 who used via both oral and intranasal routes. In addition to their opioid use, 65% of the total sample (n=23) reported alcohol use (10 weekly and 13 monthly or less), 51% were marijuana users (6 daily, 5 weekly, 7 monthly or less), 53% were tobacco smokers (8 daily, 11 weekly), and 30% used cocaine (all monthly or less). The subjects reported negligible use of heroin, sedatives (benzodiazepines or barbiturates), club drugs (ecstasy, GHB, ketamine) or hallucinogens (LSD or PCP). A comparison of participants with and without a history of non-medical opioid use found no significant differences in oxycodone effects with the exception of greater oxycodone-induced nausea among those with no nonmedical use.
Allele Frequencies
All genotype frequencies tested were in conformity with Hardy–Weinberg equilibrium, p > 0.10 (Table 2: Rodriguez et al., 2009). There were no significant differences in allele frequencies between the two major racial groups (Caucasians & African-Americans; Supplementary Table).
Table 2:
Observed Allele Frequencies
| Gene | SNP ID | Minor-Allele Homozygote | Heterozygous | Major-Allele Homozygote |
|---|---|---|---|---|
| OPRM1 | ||||
| rs1799971 | --- | 14.3% (AG) | 85.7% (AA) | |
| rs6848893 | --- | 19.0% (CT) | 81.0% (TT) | |
| OPRD1 | ||||
| rs10753331 | 19.0% (AA) | 33.3% (AG) | 47.7% (GG) | |
| rs2234918 | 28.6% (TT) | 33.3% (CT) | 38.1% (CC) | |
| rs581111* | 19.0% (TT) | 28.6% (CT) | 47.6% (CC) | |
| rs678849 | 33.3% (TT) | 23.8% (CT) | 42.9% (CC) | |
| OPRK1 | ||||
| rs6473797 | 23.8% (TT) | 38.1% (CT) | 38.1% (CC) | |
| rs963549 | 19.0% (TT) | 28.6% (CT) | 52.4% (CC) | |
| COMT | ||||
| rs165599 | 23.8% (AA) | 38.1% (AG) | 38.1% (GG) | |
| rs4680 | 14.3% (AA) | 61.9% (AG) | 23.8% (GG) | |
| rs737865 | 4.80% (CC) | 23.8% (CT) | 71.4% (TT) | |
Genotype missing for one participant.
Oxycodone-Induced Effects
Positive Subjective Effects:
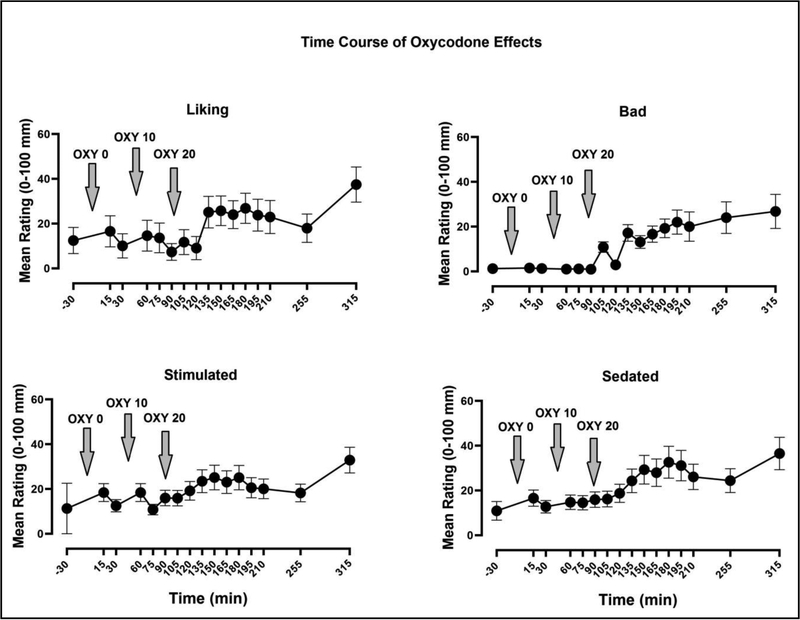
VAS assessments of positive subjective effects including: “High” (p< 0.001, η2 = 0.48), “Good Effect” (p< 0.001, η2 = 0.47), “Liking” (p< 0.01, η2 = 0.39: Figure 1), and “High Quality” (p< 0.01, η2 = 0.42) increased significantly as a function of Time. This main effect of “Time” indicates an increasing OXY dose-response function.
Figure 1:
Mean (± SEM) Visual Analog Scale ratings of oxycodone “Liking,” “Bad Effect,” “Stimulated,” and “Sedated” shown throughout the test session. Downward arrows indicate the time at which each oral oxycodone dose was administered.
Aversive Subjective Effects:
VAS ratings of “Bad Effect,” “Nauseated” and “Irritable” significantly increased later in the session (Time, p< 0.01, η2 = 0.36). There was no difference from baseline in self-reports of “Anxious,” though baseline levels were modest. Ratings of “Depression” were minimal throughout the session and did not vary as a function of Time.
Sedating Effects:
Ratings of “Sedated” (p< 0.01, η2 = 0.38)” and “Sleepy” (p<0.01, η2 = 0.39)” significantly increased after administration of oxycodone, but ratings of “Mellow” did not significantly differ from baseline.
Stimulating Effects:
Subjective reports of “Potent,” “Restless,” and “Stimulated” increased as a function of Time, though changes on these measures did not reach statistical significance. No significant changes in ratings of “Alert” and “Energetic” were found. Mean baseline and peak post-drug ratings for all measures are shown in Table 3.
Experimental pain/CPT:
Analyses revealed that as the laboratory session progressed (and the OXY dose increased) there was a significant increase in the length of time that participants were able to keep their hand immersed in 4° water. In comparison to baseline and placebo (earlier time points), active doses of oxycodone significantly increased participants’ latency to withdraw their hand from cold water and latency to report feeling pain (p’s < 0.01, η2 = 0.40).
Physiological Effects:
Pupillary diameter, heart rate, and respiratory rate decreased from baseline, but not significantly. No significant effects on blood pressure were observed.
Genetic Predictors of Oxycodone Effects
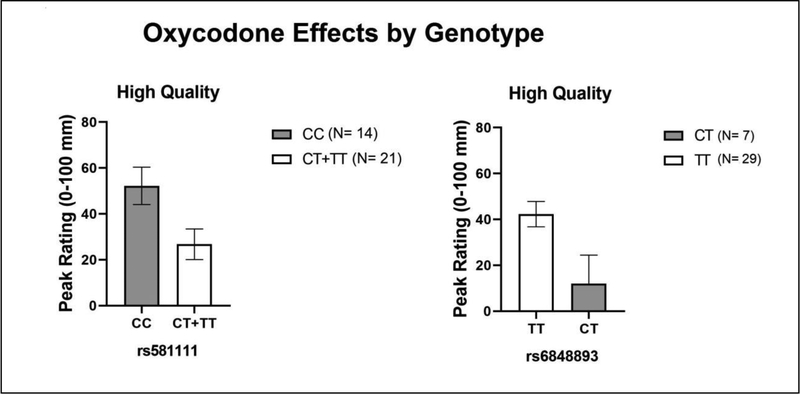
Concerning the positive subjective effects of oxycodone, an interaction was observed between the OPRD1 SNP rs581111 and the OPRM1 SNP rs6848893 on ratings of “High Quality,” (p<.004, η2 = 0.41). Rs581111 minor allele carriers (CT+TT) and rs6848893 minor allele carriers (CT*) rated OXY as less “High Quality,” [*there were no minor allele homozygotes (CC) for this genotype, Figure 2]. The only other significant effect was observed on subjective reports of “Stimulated.” Minor allele carriers of COMT SNP rs4680 (AA + AG) reported a greater response (p<.004, η2 = 0.42).
Figure 2:
Mean Peak (± SEM) Visual Analog Scale ratings of oxycodone-induced “High as a function of major and minor allele carriers of OPRD1 (rs581111) and OPRM1 (rs6848893) SNPs. * indicates p<0.05 and ** p <0.01.
Discussion
The current study sought to determine whether candidate genetic polymorphisms were associated with subjective responses to oral OXY among non-dependent healthy volunteers. OXY produced typical μ-opioid receptor agonist effects, including a slight decrease in pupillary diameter, and decreased pain perception. OXY also produced dose-dependent increases in self-reported positive effects and drug-induced nausea. These responses to opioids are similar to those previously reported among comparable samples (Jones et al., 2016B; Zacny et al., 2003; 2009). These findings confirm the abuse potential common to opioid analgesics that often results in diversion to non-medical use, which can lead to the development of OUD. Research has shown that there is significant variability in subjective responses to opioids, with greater euphoric effects during initial experiences with opioid analgesics being associated with the development of maladaptive patterns of use (Bieber et al., 2008). If we are able to predict those individuals who are most vulnerable to aberrant opioid use behaviors, it could greatly improve our ability to circumvent the development of OUD. For example, the CDC has issued recommendations for treating pain without opioids in order to reduce the number of people who misuse these drugs (CDC, 2016). Within this context, candidate-gene testing could be incorporated into risk assessment and mitigation strategies when prescribers are making decisions on how to treat pain.
Genetic variation has been shown to moderate both subjective and physiological responses to opioids, so they could be useful in identifying individuals at increased risk for OUD (Agarwal et al., 2017; Berrettini et al., 2017). In the current study, we found that SNPs in the gene encoding the μ-opioid (rs6848893) and δ-opioid receptors (rs581111) influenced OXY-induced positive subjective effects. Although the functional significance of the intron three variant in the μ receptor is unknown, in previous case-control studies it has been associated with opioid use disorder (Zhang et al., 2006; 2007) and the severity of opioid withdrawal (Jones et al., 2016A).
OPRM1 is the gene that has been studied most extensively as a risk factor for opioid misuse, given its central role in the neurobiology of opioid reward. Therefore, it is not surprising that variation in the gene was associated with the positive subjective effects of OXY in the current study. Notably, we found no effect of the OPRM1 variant, rs1799971. This SNP alters the μ-opioid receptor’s amino acid sequence by substituting aspartic acid for asparagine and has previously been associated with both the sensitivity to opioid analgesics, risk of OUD, and the response to medication-assisted treatment of OUD (Crettol et al., 2008; Ren et al., 2015; Schwantes et al., 2016; Szeto et al., 2001). In the current study, the most robust predictor of opioid reward was a SNP in the δ-opioid receptor (rs581111). Though its specific contribution to opioid reward is unknown, rs581111 has been associated with heroin use disorder and was predictive of buprenorphine treatment outcome among female OUD patients (Clarke et al., 2014: Nelson et al., 2014).
The COMT variant, rs4680, found to influence “Stimulated” response to OXY. The minor A-allele of this polymorphism decreases catabolic enzyme activity by 25%, resulting in more DA in the prefrontal cortex (Chen et al., 2004). The contribution of the stimulating effects of opioids to their abuse potential is poorly understood. However, some researchers have proposed dopamine-mediated reward deficiency as a motivator for substance abuse (Blum et al., 2000; 2012). Although opioids are generally considered to be central nervous system depressants, a subset of individuals report that they experience a stimulating effect upon initial use of opioids (Cicero and Ellis, 2017). This activating effect could constitute an endophenotype (Gottesman and Gould, 2003) that identifies a motivation for non-medical opioid use. Although previous studies have shown an association of rs4680 with variation in opioid use in post-operative and chronic pain patients (Hu et al., 2018), we found no association of the SNP with the analgesic effects of OXY.
The current study has several limitations that should be noted. Foremost, because a racially diverse sample was tested, we are unable to account for population admixture, which has been shown to have a confounding effect in association genetic studies (Liu et al., 2013). Though we cannot rule out the potentially confounding effects of race, we found no significant differences in allele frequency among the two racial groups that constitute 83% of the study sample. Another concern is that examination of the time course of OXY effects seems to indicate that ratings on several measures may have continued to increase past the last point of data collection. Therefore, the complete time course and true maximal drug effect may not have been captured. Additionally, combining data from participants with and without current non-medical opioid use could have diminished the significance of association with measures of the abuse potential of OXY. Additionally, because of the poor representation of women, sex could not be examined as a potential moderator. Finally, as the study recruited a population in their 30’s, with exposure to opioids but who did not develop OUD (or any drug addiction), we may have self-selected a sample that is resilient to the development of substance use disorders. A younger, more drug-naïve sample may have been more generalizable to the population to which these data may be relevant (i.e., those at risk of non-medical opioid use).
In conclusion, prescription opioid misuse is a critical health problem in the United States. While many factors, both individual and environmental, influence the initiation of opioid use, opioids’ ability to produce intense feelings of pleasure is a significant motivator for their use. Data from this study suggest that genetic variation may moderate the subjective responses to oxycodone. Thus, if replicated, specific genetic variants may be useful in predicting who is at increased risk of developing maladaptive patterns of use following medical exposure to opioid analgesics.
Supplementary Material
Highlights.
Genetic influence on the pharmacodynamics of oxycodone is tested.
Oxycodone produced typical μ-opioid receptor agonist effects.
Oxycodone produced dose-dependent increases in positive subjective responses.
Euphoric and stimulating effects of oxycodone varied as a function of genotype.
Acknowledgements
The medical assistance of Janet Murray and Claudia Tindall, along with the technical assistance of Gabriella Madera, Brian Wade, Rachel Luba, and Andrew Segoshi, is gratefully acknowledged.
Funding Source: Financial support for this study was provided by the National Institute on Drug Abuse grant K01DA030446 to JDJ.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
Conflicts of Interest: Only the authors listed are responsible for the content and preparation of this manuscript. Drs. Comer, Mogali, and Manubay have received compensation (in the form of partial salary support) from studies supported by Alkermes, Braeburn Pharmaceuticals, Cerecor Inc., Corbus, Endo Pharmaceuticals, Go Medical, Indivior PLC/Reckitt-Benckiser Pharmaceuticals, Intra-cellular Therapies, Johnson & Johnson Pharmaceutical Research & Development, Lyndra, MediciNova, Omeros, and Schering-Plough Corporation. In addition, Dr. Comer has consulted for: Alkermes, Charleston Labs, Clinilabs, Collegium, Daiichi Sankyo, Depomed, Egalet, Endo, Epiodyne, Inspirion Delivery Sciences, Janssen, KemPharm, Mallinckrodt, Neurolixis, Newron, Opiant, Otsuka, Pfizer, and Sun Pharma. She also has received honoraria from the World Health Organization (WHO).
References
- Adinoff B. Neurobiologic processes in drug reward and addiction. Harvard Rev. Psychiatry 2004; 12: 305–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal D, Udoji MA, and Trescot A. Genetic Testing for Opioid Pain Management: A Primer Pain Ther. 2017; 6(1): 93–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balding DJ. A tutorial on statistical methods for population association studies. Nat Rev Genet. 2006; 7: 781–791. [DOI] [PubMed] [Google Scholar]
- Barratt DT, Coller JK, Somogyi AA. Association between the DRD2 A1 allele and response to methadone and buprenorphine maintenance treatments. Am. J. Med. Genet. B. Neuropsychiatr. Genet 2006; 141B: 323–331. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Statistical Society. 1995; 57: 289–300. [Google Scholar]
- Berrettini W. A brief review of the genetics and pharmacogenetics of opioid use disorders Dialogues Clin Neurosci. 2017; 19(3): 229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieber CM, Fernandez K, Borsook D, Brennan MJ, Butler SF, Jamison RN, Osgood E, Sharpe-Potter J, Thomson HN, Weiss RD, Katz NP. Retrospective accounts of initial subjective effects of opioids in patients treated for pain who do or do not develop opioid addiction: a pilot case-control study. Exp Clin Psychopharmacol. 2008; 16(5): 429–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum K, Braverman ER, Holder JM, et al. Reward deficiency syndrome: a biogenetic model for the diagnosis and treatment of impulsive, addictive, and compulsive behaviors. J Psychoactive Drugs. 2000; 32(Suppl i–iv): 1–112. [DOI] [PubMed] [Google Scholar]
- Blum K, Gardner E, Oscar-Berman M, et al. “Liking” and “wanting” linked to reward deficiency syndrome (RDS): hypothesizing differential responsivity in brain reward circuitry. Curr Pharm Des. 2012; 18: 113–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention Wide-ranging online data for epidemiologic research (WONDER). Atlanta, GA: CDC, National Center for Health Statistics; 2018. Available at http://wonder.cdc.gov. [Google Scholar]
- Centers for Disease Control and Prevention. Vital Signs: Changes in Opioid Prescribing in the United States, 2006–2015. MMWR 2017; 66(26):697–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. 2016. Treating Chronic Pain without Opioids Applying CDC’s Guideline for Prescribing Opioids. https://www.cdc.gov/drugoverdose/training/nonopioid/508c/index.html
- Chen J, Lipska BK, Halim N, Ma QD, Matsumoto M, Melhem S. Functional analysis of genetic variation in catechol-O-methyltransferase (COMT): Effects on mRNA, protein, and enzyme activity in postmortem human brain. Am J Hum Genet. 2004; 75: 807–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicero TJ, Ellis MS. Understanding the demand side of the prescription opioid epidemic: Does the initial source of opioids matter? Drug Alcohol Depend. 2017; 173 Suppl 1: S4–S10. [DOI] [PubMed] [Google Scholar]
- Cicero TJ, Ellis MS, Surratt HL, Kurtz SP The changing face of heroin use in the United States: a retrospective analysis of the past 50 years. JAMA Psychiatry. 2014; 71(7): 821–826. [DOI] [PubMed] [Google Scholar]
- Clarke TK, Crist RC, Ang A, Ambrose-Lanci LM, Lohoff FW, Saxon AJ, Ling W, Hillhouse MP, Bruce RD, Woody G, Berrettini WH, Genetic variationin OPRD1 and the response to treatment for opioid dependence with buprenor-phine in European-American females. Pharmacogenomics J. 2014a; 14: 303–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comer SD, et al. , (2012) Core outcome measures for opioid abuse liability laboratory assessment studies in humans: IMMPACT recommendations. Pain. 2012; 153(12): 2315–2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crist RC, Clarke TK., Ang A, Ambrose-Lanci LM, Lohoff, FW, Saxon AJ, Ling W, Hillhouse MP, Bruce RD, Woody G, Berrettini WH. An intronic variant in OPRD1 predicts treatment outcome for opioid dependence in African-Americans. Neuropsychopharmacolgy. 2013a; 38: 2003–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crist RC, Li J, Doyle GA, Gilbert A, Dechairo BM, Berrettini WH. Pharmacogenetic analysis of opioid dependence treatment dose and dropout rate. Am J Drug Alcohol Abuse. 2018a; 44(4): 431–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crist RC, Phillips KA, Furnari MA, Moran LM, Doyle GA, McNicholas LF, Cornish JW, Kampman KM, Preston KL, Berrettini WH. Replication of the pharmacogenetic effect of rs678849 on buprenorphine efficacy in African-Americans with opioid use disorder. Pharmacogenomics J. 2018b, doi: 10.1038/s41397-018-0065-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crettol S, Besson J, Croquette-Krokar M, Hämmig R, Gothuey I, Monnat M, Déglon JJ, Preisig M, Eap CB. Association of dopamine and opioid receptor genetic polymorphisms with response to methadone maintenance treatment. Prog Neuropsychopharmacol. Biol Psychiatry. 2008; 32: 1722–1727. [DOI] [PubMed] [Google Scholar]
- Conley KM, Toledano AY, Apfelbaum JL, Zacny JP Modulating effects of a cold water stimulus on opioid effects in volunteers. Psychopharmacol. 1997; 131: 313–320. [DOI] [PubMed] [Google Scholar]
- Crettol S, Besson J, Croquette-Krokar M, Hämmig R, Gothuey I, Monnat M, Déglon JJ, Preisig M, Eap CB. Association of dopamine and opioidreceptor genetic polymorphisms with response to methadone maintenance treatment. Prog. Neuropsychopharmacol. Biol Psychiatry. 2008; 32, 1722–1727. [DOI] [PubMed] [Google Scholar]
- Daly EM. With rise in young painkiller abusers, officials see more heroin overdoses. California Watch. August 15, 2012. https://www.scpr.org/programs/madeleine-brand/20 [Google Scholar]
- de Wit H, Phillips TJ. Do initial responses to drugs predict future use or abuse? Neurosci Biobehav Rev. 2012; 36:1565–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drakenberg K, Nikoshkov A, Horvath MC, Fagergren P, Gharibyan A, Saarelainen K, Rahman S, Nylander I, Bakalkin G, Rajs J, Keller E, Hurd YL. Mu opioid receptor A118G polymorphism in association with striatal opioid neuropeptide gene expression in heroin abusers. PNAS. 2006; 103(20): 7883–7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields HL, Margolis EB. Understanding opioid reward. Trends Neurosci. 2015; 38(4): 217–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltin RW, Fischman MW. Effects of methadone or buprenorphine maintenance on the subjective and reinforcing effects of intravenous cocaine in humans. J Pharmacol Exp Ther. 1996; 278(3): 1153–1164. [PubMed] [Google Scholar]
- Food and Drug Administration (2017) https://www.fda.gov/downloads/drugs/guidancecomplianceregulatoryinformation/guidances/ucm198650.pdf Accessed 6 AUG, 2019
- Goldman D, Oroszi G, Ducci F. The genetics of addictions: uncovering the genes. Nat Rev Genet. 2005; 6: 521–532. [DOI] [PubMed] [Google Scholar]
- Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry. 2003; 60(4): 636–645. [DOI] [PubMed] [Google Scholar]
- IBM Corp. Released 2017. IBM SPSS Statistics for Windows, Version 25.0. Armonk, NY: IBM Corp. [Google Scholar]
- IQVIA Institute (2018) Medicine Use and Spending in the U.S.: A Review of 2017 and Outlook to 2022. https://www.iqvia.com/institute/reports/medicine-use-and-spending-in-the-us-review-of-2017-outlook-to-2022
- Hu B, Zhang X, Xu G, Zhang Q, Qian P, Liu S, Zhu J, Shen R. Association between COMT Polymorphism Val158Met and Opioid Consumption in Patients with Postoperative Pain: A Meta-Analysis Neurosignals. 2018; 26:11–21. [DOI] [PubMed] [Google Scholar]
- Jones JD, Luba RL, Vogelman J, Comer SD. Evidence of Genetic Modulation of Opioid Withdrawal Severity. Am J Addictions. 2016a; 25(1):41–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Sullivan MA, Manubay J, Metz V, Mogali S, Comer SD. The Effects of Pioglitazone, a PPARγ Receptor Agonist, on the Abuse Liability of Oxycodone. Physiology & Behavior. 2016b; 159:33–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Comer SD. A Review of Pharmacogenetic Studies of Drug Use Disorders. Drug and Alcohol Dependence. 2015. 152:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Merrer J, Plaza-Zabala A, Del Boca C, Matifas A, Maldonado R, Kieffer BL: Deletion of the delta opioid receptor gene impairs place conditioning but preserves morphine reinforcement. Biol Psychiatry. 2011; 69: 700–703. [DOI] [PubMed] [Google Scholar]
- Le Merrer J, Faget L, Matifas A, Kieffer BL: Cues predicting drug or food reward restore morphine-induced place conditioning in mice lacking delta opioid receptors. Psychopharmacol (Berl) 2012; 223: 99–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levran O, Londono D, O’Hara K, Nielsen DA, Peles E, Rotrosen J. Genetic susceptibility to heroin addiction: a candidate gene association study. Genes Brain Behav. 2008; 7: 720–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MD, Burmeister M. New insights into the genetics of addiction. Nat Rev Genet. 2009; 10: 225–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Shao C, Zhang D, Zhao M, Lin L, Yan P, Xie Y, Jiang K, Jin L. The effect of dopamine D2, D5 receptor and transporter (SLC6A3) polymorphisms on the cue-elicited heroin craving in Chinese. Am J Med Genet B Neuropsychiatr Genet. 2006; 141: 269–273. [DOI] [PubMed] [Google Scholar]
- Li Z, Zhang H. Analyzing Interaction of μ-, δ- and κ-opioid Receptor Gene Variants on Alcohol or Drug Dependence Using a Pattern Discovery-based Method. J Addict Res Ther. 2013; Suppl 7: 007. [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lewinger JP, Gilliland FD, Gauderman WJ, Conti DV. Confounding and heterogeneity in genetic association studies with admixed populations. Am J Epidemiol. 2013; 177(4): 351–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lötsch J, Geisslinger G. (2006) Relevance of frequent mu-opioid receptor polymorphisms for opioid activity in healthy volunteers. Pharmacogenomics J. 6(3): 200–210. [DOI] [PubMed] [Google Scholar]
- Margolis EB, Fields HL, Hjelmstad GO, Mitchell JM: Delta-opioid receptor expression in the ventral tegmental area protects against elevated alcohol consumption. J Neurosci. 2008; 28: 12672–12681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer P, Rochlitz H, Rauch E, Rommelspacher H, Hasse HE, Schmidt S. Höllt V. Association between a delta opioid receptor gene polymorphism and heroin dependence in man. Neuroreport. 1997; 8: 2547–2550. [DOI] [PubMed] [Google Scholar]
- Muhuri PK, Gfroerer JC, Davies MC. Associations of Nonmedical Pain Reliever Use and Initiation of Heroin Use in the United States. https://www.samhsa.gov/data/sites/default/files/DR006/DR006/nonmedical-pain-reliever-use-2013.htm [Google Scholar]
- Nelson EC, Lynskey MT, Heath AC, Wray N, Agrawal A, Shand FL, Henders AK, Wallace L, Todorov AA, Schrage AJ, Madden PA, Degenhardt L, Martin NG, Montgomery GW. Association of OPRD1 polymorphisms with heroin dependence in a large case-control series. Addict Biol. 2014; 19(1): 111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoshkov A, Drakenberg K, Wang X, et al. Opioid neuropeptide genotypes in relation to heroin abuse: Dopamine tone contributes to reversed mesolimbic proenkephalin expression. Proc Natl Acad Sci USA 2008; 105: 786–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosterhuis BE, LaForge KS, Proudnikov D, Ho A, Nielsen DA, Gianotti R, Barral S, Gordon D, Leal SM, Ott J, Kreek MJ. Catechol-O-methyltransferase (COMT) gene variants: possible association of the Val158Met variant with opiate addiction in Hispanic women. Am J Med Genet B Neuropsychiatr Genet. 2008; 147B(6): 793–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak GW, editor. The Opiate Receptors. Humana Press Totowa, NJ: Humana; 2011. [Google Scholar]
- Pan ZZ. mu-Opposing actions of the kappa-opioid receptor. Trends Pharmacol. Sci 1998; 19 (3): 94–98. [DOI] [PubMed] [Google Scholar]
- Peltz G, Südhof TC. The Neurobiology of Opioid Addiction and the Potential for Prevention Strategies. JAMA. 2018; 319(20): 2071–2072. [DOI] [PubMed] [Google Scholar]
- Peters BJ, Rodin AS, de Boer A, Maitland-van der Zee AH. Methodological and statistical issues in pharmacogenomics. J Pharm Pharmacol. 2010; 62(2):161–166. [DOI] [PubMed] [Google Scholar]
- Pradhan AA, Befort K, Nozaki C, Gaveriaux-Ruff C, Kieffer BL: The delta opioid receptor: an evolving target for the treatment of brain disorders. Trends Pharmacol Sci 2011; 32: 581–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren ZY, Xu XQ, Bao YP, He J, Shi L, Deng JH, Gao XJ, Tang HL, Wang YM, Lu L. The impact of genetic variation on sensitivity to opioid analgesics in patients with postoperative pain: a systematic review and meta-analysis. Pain Physician. 2015; 18(2): 131–152. [PubMed] [Google Scholar]
- Ridenour TA, Cottler LB, Compton WM, Spitznagel EL, Cunningham-Williams RM. Is there a progression from abuse disorders to dependence disorders? Addiction. 2003; 98: 635–644. [DOI] [PubMed] [Google Scholar]
- Ridenour TA, Maldonado-Molina M, Compton WM, Spitznagel EL, Cottler LB. Factors associated with the transition from abuse to dependence among substance abusers: implications for a measure of addictive liability. Drug and Alcohol Dependence. 2005; 80: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez S, Gaunt TR, and Day INM. Hardy-Weinberg Equilibrium Testing of Biological Ascertainment for Mendelian Randomization Studies. American Journal of Epidemiology. 2009; 169(4): 505–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JR, Rutter D,Welsh K, Joel SP, Goller K, Wells AU, Du Bois R, Riley J. Clinical response to morphine in cancer patients and genetic variation in candidate genes. Pharmacogenomics J. 2005; 5: 324–336. [DOI] [PubMed] [Google Scholar]
- Genetics Silicon (2003) Multiple Testing Corrections. http://nebc.nerc.ac.uk/courses/GeneSpring/GS_Mar2006/Multiple%20testing%20corrections.pdf [Google Scholar]
- Schwantes et al. ,(2016) Association of the OPRM1 Variant rs1799971 (A118G) with Non-Specific Liability to Substance Dependence in a Collaborative de novo Meta-Analysis of European-Ancestry Cohorts. Behav Genet. 2016; 46(2): 151–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer JP. Multiple hypothesis testing. Ann Rev Psychol 1995; 46: 561–584 [Google Scholar]
- Shao C, Li Y, Jiang K, Zhang D, Xu Y, Lin L, Wang Q, Zhao M, Jin L. Dopamine D4 receptor polymorphism modulates cue-elicited heroin craving in Chinese. Psychopharmacol (Berl). 2006; 186: 185–190. [DOI] [PubMed] [Google Scholar]
- Szeto CY, Tang NL, Lee DT, Stadlin A. Association between mu opioid receptor gene polymorphisms and Chinese heroin addicts. Neuroreport. 2001; 12(6): 1103–1106. [DOI] [PubMed] [Google Scholar]
- Vandenbergh DJ, Rodriguez LA, Miller IT, Uhl GR, Lachman HM. High-activity catechol-O-methyltransferase allele is more prevalent in polysubstance abusers. Am J Med Genet. 1997; 74(4): 439–442. [PubMed] [Google Scholar]
- Walker DJ, Zacny JP. Subjective, psychomotor, and physiological effects of cumulative doses of opioid mu agonists in healthy volunteers. J Pharmacol Exp Ther 1999; 289(3): 1454–1464. [PubMed] [Google Scholar]
- Walker DJ, Zacny JP, Galva KE, Lichtor JL. Subjective, psychomotor, and physiological effects of cumulative doses of mixed-action opioids in healthy volunteers. Psychopharmacology (Berl) 2001; 155(4): 362–371. [DOI] [PubMed] [Google Scholar]
- Wang JB, Johnson PS, Persico AM, Hawkins AL, Griffin CA, Uhl GR. Human mu opiate receptor. cDNA and genomic clones, pharmacologic characterization and chromosomal assignment. FEBS Lett. 1994; 338. [DOI] [PubMed] [Google Scholar]
- Wang RA. Rating the presence and severity of opiate dependence. Clin Pharmacol Therapeu. 1974; 16(4): 653–658. [DOI] [PubMed] [Google Scholar]
- Wise RA. Drug-activation of brain reward pathways. Drug Alcohol Depend. 1998; 51(1–2): 13–22. [DOI] [PubMed] [Google Scholar]
- Wise RA. Brain reward circuitry: insights from unsensed incentives. Neuron. 2002; 36(2): 229–240. [DOI] [PubMed] [Google Scholar]
- Zacny JP, McKay MA, Toledano AY, Marks S, Young CJ, Klock PA. The effects of a cold-water immersion stressor on the reinforcing and subjective effects of fentanyl in healthy volunteers. Drug Alcohol Depend. 1996; 42: 133–142. [DOI] [PubMed] [Google Scholar]
- Zacny JP, Gutierrez S. Characterizing the subjective, psychomotor, and physiological effects of oral oxycodone in non- drug-abusing volunteers. Psychopharmacol 2003; 170: 242–254. [DOI] [PubMed] [Google Scholar]
- Zacny JP, Gutierrez S. Within-subject comparison of the psychopharmacological profiles of oral hydrocodone and oxycodone combination products in non-drug-abusing volunteers. Drug Alc Depend 2009; 101: 107–14. [DOI] [PubMed] [Google Scholar]
- Zhang H, Kranzler HR, Yang BZ, Luo X, Gelernter J. The OPRD1 and OPRK1 loci in alcohol or drug dependence: OPRD1 variation modulates substance dependence risk. Mol. Psychiatry 2008; 13: 531–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Luo X, Kranzler HR, Lappalainen J, Yang BZ, Krupitsky E, Zvartau E, Gelernter J. Association between two mu opioid receptor gene (OPRM1) haplotype blocks and drug or alcohol dependence. Hum Mol Genet. 2006; 15: 807–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Shao C, Shao M, Yan P, Wang Y, Liu Y, Liu W, Lin T, Xie Y, Zhao Y, Lu D, Li Y, Jin L. Effect of mu-opioid receptor gene polymorphisms on heroin-induced subjective responses in a Chinese population. Biol Psychiatry 2007; 61: 1244–1251. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.