Abstract
Purpose:
Poly (ADP-ribose) polymerase (PARP) inhibitors are approved for the treatment of high-grade serous ovarian cancers (HGSOCs). Therapeutic resistance, resulting from restoration of homologous recombination (HR) repair or replication fork stabilization, is a pressing clinical problem. We assessed the activity of prexasertib, a checkpoint kinase 1 (CHK1) inhibitor known to cause replication catastrophe, as monotherapy and in combination with the PARP inhibitor olaparib in preclinical models of HGSOC, including those with acquired PARP inhibitor resistance.
Experimental Design:
Prexasertib was tested as a single agent or in combination with olaparib in 14 clinically annotated and molecularly characterized luciferized HGSOC patient-derived xenograft (PDX) models and in a panel of ovarian cancer cell lines. The ability of prexasertib to impair HR repair and replication fork stability was also assessed.
Results:
Prexasertib monotherapy demonstrated anti-tumor activity across the 14 PDX models. Thirteen models were resistant to olaparib monotherapy, including 4 carrying BRCA1 mutation. The combination of olaparib with prexasertib was synergistic and produced significant tumor growth inhibition in an olaparib-resistant model and further augmented the degree and durability of response in the olaparib-sensitive model. HGSOC cell lines, including those with acquired PARP inhibitor resistance, were also sensitive to prexasertib, associated with induction of DNA damage and replication stress. Prexasertib also sensitized these cell lines to PARP inhibition and compromised both HR repair and replication fork stability.
Conclusions:
Prexasertib, exhibits monotherapy activity in PARP inhibitor-resistant HGSOC PDX and cell line models, reverses restored HR and replication fork stability and synergizes with PARP inhibition.
Keywords: CHK1 inhibition, PARP1 inhibition, ovarian cancer, homologous recombination repair, replication fork stability, patient-derived xenograft models
INTRODUCTION
High grade serous ovarian cancer (HGSOC) is the most frequent and aggressive subtype of epithelial ovarian cancer (EOC), with high recurrence rates despite initial responses to platinum-based chemotherapy (1, 2). Approximately 50% of the EOCs exhibit genetic and epigenetic alterations in gene members of the homologous recombination (HR) DNA repair pathway including BRCA1/2 and other Fanconi Anemia/BRCA pathway genes (3, 4). Based on the synthetic lethal interaction of defective HR repair and poly (ADP-ribose) polymerase (PARP) inhibition, HR-deficient EOCs are highly sensitive to PARP inhibitors (5). Currently, olaparib, rucaparib and niraparib are all FDA-approved for HGSOC (6); in the recurrent disease setting, these agents have been primarily used in patients with tumors harboring BRCA alterations with substantial response rates. Additionally, these agents are now commonly considered in the maintenance setting, after a response to a platinum-based chemotherapy, where PARP inhibition has significantly increased progression-free survival and the chemotherapy-free interval for many patients (7, 8).
With more common PARP inhibitor use, acquired resistance has emerged as an unmet medical need. The major mechanisms of PARP inhibitor resistance include restoration of HR repair or stabilization and protection of replication forks (9-12). There is a critical need for combinations utilizing agents that can inhibit HR and/or reverse replication fork stability and thereby sensitize HGSOC tumors to PARP inhibition. Such approaches may also extend the use of PARP inhibitors to include HR-proficient, BRCA wild-type tumors.
Due to the endogenous DNA damage caused by HR deficiency, HGSOC tumors require an intact ataxia telangiectasia and Rad3-related (ATR)–checkpoint kinase 1 (CHK1) pathway to allow time for alternative DNA repair pathways to function and maintain genome integrity. Additionally, HGSOCs often demonstrate genomic alterations suggestive of replicative stress. For example, ∼20% of HGSOCs may express high levels of cyclin E (4, 13), often as a result of gene amplification. Tumors with replication stress are also dependent on the ATR-CHK1 pathway, in this case for maintaining replication fork stability (14). Based on its role in DNA-damage induced checkpoint control (15, 16) and control of replication origin firing (17-19), CHK1 is an attractive therapeutic target for HGSOC. CHK1 deficiency can exacerbate both DNA damage and replication stress, leading to cell death (16, 20-23) . Additionally, the roles of CHK1 in both replication fork stability (18, 24) and HR repair (23, 25) suggest that CHK1 deficiency may also reverse PARP inhibitor resistance.
Prexasertib (LY2606368) is a potent inhibitor of CHK1, and to a lesser extent, CHK2, and is currently being tested in phase 1 and 2 clinical trials (26, 27). Among patients with HGSOC, preliminary activity has been demonstrated in BRCA wild-type, platinum-resistant and refractory disease (27). As a single agent, prexasertib causes replication catastrophe, DNA double-strand breaks, HR defects, and apoptosis (28, 29). We therefore investigated the efficacy of prexasertib as a monotherapy and in combination with PARP inhibition in fourteen HGSOC patient-derived xenograft (PDX) models, and in a panel of ovarian cancer cell lines. Prexasertib showed broad preclinical monotherapy activity and was synergistic with PARP inhibition in several models, including those representing BRCA1-mutant, PARP inhibitor-resistant disease, where there was reversal of both restored HR repair and of replication fork stability.
MATERIALS AND METHODS
Establishment and treatment of patient-derived tumor xenografts (PDXs)
Tumor ascites was collected from patients with suspected or established ovarian cancer at the Brigham and Women’s Hospital or the Dana-Farber Cancer Institute (DFCI) under IRB-approved protocols conducted in accordance with the Declaration of Helsinki and the Belmont Report. Written informed consent was obtained from patients when required. Fourteen ascites-derived ovarian PDX models were established, luciferized and propagated at the DFCI and have been described previously (30). All animal studies were performed in accordance with DFCI Institutional Animal Care and Use Committee guidelines per DFCI-approved animal protocols. Treatment studies in mice bearing PDX tumors were performed as described previously (30, 31). Approximately 2–10 × 106 luciferized PDX cells derived from mouse ascites were injected intraperitoneally into 8-week old female NSG mice. Tumors were typically established 1–2 weeks after implantation. Tumor burden was measured by bioluminescence imaging (BLI) using a Xenogen IVIS-200 system (Xenogen). Mice were randomized into treatment groups and disseminated disease progression in the peritoneal cavity was measured serially once per week by BLI.
Prexasertib (mesylate monohydrate salt of LY2606368) (Eli Lilly) was formulated in 20% Captisol® (CyDex Inc, Lenexa, KS) pH 4.0 and administered subcutaneously at 8 mg/kg twice-daily for 3 days, followed by four days of rest and repeated for two additional cycles. Olaparib (ChemExpress) was formulated in PBS containing 10% DMSO and 10% (wt/vol) 2-hydroxy-propyl-β-cyclodextrin (Sigma-Aldrich) and administered orally at 100 mg/kg daily for 3 weeks. Tumor-bearing mice were monitored for survival and tumor burden for approximately 100–150 days. For pharmacodynamic (PD) studies, mice were euthanized at specific time points and tumor cells were harvested from ascites.
Cell lines
OVCAR3, UWB1.289, UWB1.289+ BRCA1, ES2, OV90, SKOV3, PA-1, SW-626, SKOV3 and CAOV3 cell lines were purchased from American Type Culture Collection (ATCC). KURAMOCHI, OVSAHO and OVTOKO cell lines were purchased from JCRB. TOV21G, TOV21G+FANCF, PEO1, PEO4, COV362, A2780, JHOS2, JHOS4 and CAOV4 cell lines were obtained from other laboratories. Cell line identities were confirmed by STIR profiling. The generation of UWB1.289-SYR12 and UWB1.289-SYR14 olaparib-resistant cell lines has been described previously (32). UWB1.289 cells were grown in RPMI (ATCC-R-30–2001™) and MEGM bullet kit (Lonza CC-3150) at a 1:1 ratio, supplemented with 3% fetal bovine serum and 1% penicillin/streptomycin (Lonza 17–602E). All other cell lines were cultured in RPMI (GIBCO 11875) supplemented with 10% fetal bovine serum (GIBCO 7471) and 1% penicillin/streptomycin (Lonza 17–602E).
siRNAs
The siRNA sequence targeting 53BP1 was sense 5´-AGAACGAGGAGACGGUAAUAGUGGG-3´, antisense 5´- CCCACUAUUACCGUCUCCUCGUUCU-3´(33). The control non-targeting siRNA was purchased from Dharmacon.
Western blotting and Antibodies
Cells were lysed in a buffer containing 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% Triton, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM sodium vanadate and 1 μg/ml leupeptin (Cell Signaling 9803S) with protease inhibitor PMSF (Cell Signaling 8553S). Western blots were performed with following antibodies: CHK1 (G-4; Santa Cruz Biotechnology SC-8408), Phospho-CHK1 [Ser345] (Cell Signaling Technology 2348S), phospho-histone H2A.X [S139] (EMD 05–636), phospho-RPA32 [S4/S8] (Bethyl A300–245A), phospho-KAP1 [S824] (Abcam ab84077), vinculin (Santa Cruz Biotechnology SC-25336), RAD51 (EMD PC-130), RAD51C (Novus) and Cyclin E (Santa Cruz Biotechnology SC-247).
Drug Sensitivity assays
Stock solutions of olaparib (Selleck S1060; 100 mM) and prexasertib (40 mM) were prepared in dimethyl sulfoxide (DMSO) and stored in aliquots at −800 C. Appropriate dilutions were prepared in culture medium. For viability assays, cells were seeded in 96-well plates (2,000–4,000 cells/well) and exposed to DMSO or graded concentrations of prexasertib, olaparib or a combination of both drugs for 3–6 days. Cell survival was determined by CellTiter-Glo (Promega G7571). Cell viability was calculated relative to DMSO-treated samples and dose response curves or IC50 plots were generated using GraphPad Prism. Synergy between prexasertib and olaparib was calculated using Combenefit software as previously described (34). For colony formation assays, 500–2000 cells/well were plated in 6-well plates along with graded concentrations of drugs and cultured for 10–14 days for colony growth. Colonies were then fixed with methanol for 10 min at −20°C and stained with crystal violet. Plates were imaged with ImageQuant Las4000 (GE Healthcare) and quantification of the cell growth area was performed with ImageJ.
Immunohistochemistry
Ascites-derived tumor cells were harvested from euthanized mice, washed with saline and transferred to DMEM-F12 supplemented with 10% fetal bovine serum. Samples were divided into two and one of the aliquots was subjected to 10 Gy γ-irradiation (IR). Both unirradiated and irradiated samples were incubated for 4 hours at 37°C and fixed in 4% neutral buffered formalin for 10 min. Post-fixation, cells were washed with PBS and encapsulated in Histogel (Richard Alen Scientific) embedded in paraffin and sectioned for immunohistochemistry with antibodies to RAD51 (35), 53BP1 (Millipore MAB3802) or phospho-histone H2AX (MiliporeSigma 05–636).
Genomic analysis
Genomic DNA was isolated from ascites-derived tumor cells from PDX models and the Molecular Inversion Probe (MIP) assay (OncoSan3 platform) was performed. Resulting data were processed using Chromosome Analysis Suite (ChAS). Allelic copy number analysis was performed using Nexus Express (Affymetrix). Whole-exome sequencing of genomic DNA from ascites-derived tumor cells was performed using the Illumina HiSeq 4000 and analyzed by members of the DFCI Center for Cancer Computational Biology.
For bi-sulfite sequencing, genomic DNA was treated with bi-sulfite using EZ DNA methylation-direct kit (ZYMO Research) and then the promoter region of RAD51C was amplified using specific primers (Forward primer: ATGGTGTATAAGTGTGAAAATTTATAAGA; Reverse primer: CCTCTAAAAATTCCTCAACAATCTAAA). The PCR product was sequenced by Sanger sequencing.
To confirm the 14 bp deletion at the exon-intron boundary and the mutation identified at the TP53BP1 locus in DF59 PDX model, PCR was performed on genomic DNA using primers flanking the deletion site (Primer sequence 53BP1 exon12-intron12 boundary: GCCCATGCCTCACAAAGCTT, AAGCATGCAGTTCTCGCCAA). To verify the presence of the mutation, mutation specific primers were used, including primer sequence 53BP1 exon 26 T>C (p.L1806P) specific forward primer: AACACAGCTTACCAGTGTCC; and consensus reverse primer: ACGGTAGTTCTGGAGCTGGT. Real-time PCR analysis was performed using total RNA isolated from DF83, DF20 and DF181 PDX models. Primer sequences for RAD51 expression were: TGCGCCCACAACCCATTTCAC, TCAATGTACAACCCATTTCCTTCAC. Primer sequences for RAD51C expression were: AAGACGTTCCGCTTTGAA, GCATTGCCAGAGGAAAAC. CCNE1 expression in PDX tumor cells was determined by quantitative real-time PCR (qPCR) using a TaqMan kit (Applies Biosystems 4331182).
Immunofluorescence for analysis of RAD51 foci
Cells were plated on autoclaved coverslips and allowed to grow for 24 hrs before exposure to drugs or IR, followed by fixation in 4% paraformaldehyde for 15 min at room temperature. The cells were then washed with PBS and permeabilized using cold methanol for 2 min. After blocking with 10% normal goat serum containing 0.3% Triton X-100 for 1 hr, cells were stained with primary anti-RAD51 antibody (EMD, PC130, diluted 1:1000 in PBS containing 1% BSA and 0.3% triton X-100) followed by staining with goat anti-rabbit IgG (H+L) Alexa Fluor 488 labelled secondary antibody (ThermoFisher Scientific A11034, diluted 1:1500 in PBS containing 1% BSA and 0.3% Triton X-100). Coverslips were then mounted with a solution containing DAPI (ThermoFisher Scientific p36935) and cells were visualized under the fluorescence microscope.
DNA fiber assay
Replication fork stability of UWB1.289 cells was determined by the DNA fiber assay as previously described (36) using FiberComb machine (Genomic Vision). 200,000 cells/well were plated in 6-well plates and cultured for 48 hrs before treatment with 5-Iodo-2’-deoxyuridine (IdU) (Sigma I7125) for 45 min, 5-Chloro-2’-deoxyuridine (CldU) (Sigma C6891) for 45 min, followed by hydroxyurea (HU) with or without prexasertb for 4.5 hrs, with 3 PBS washes between each treatment. Cells were then embedded in low melting point agarose plugs, followed by proteinase K treatment and agarose digestion, after which DNA samples were combed onto silanized coverslips. The labeling of CldU and IdU was detected by staining with rat anti-BrdU antibody (clone BU1/75-ICR specific to CldU (Abcam ab6326) and mouse anti-BrdU Antibody specific to IdU, (BD Biosciences 347580), followed by staining with chicken anti-mouse IgG Alexa Fluor 594 (Red, Life TechnologyA-11031) and goat anti-rat IgG Alexa Fluor 488 (Green, Life Technology A-11006) secondary antibodies. DNA fibers were visualized by fluorescence microscopy and images were taken of at least 100 fibers per condition, quantified with ImageJ and graphed.
Statistical Analysis
For monotherapy efficacy studies in vivo, a group size of n=5 was utilized in order to provide >95% power to detect a difference of at least 50% in tumor growth inhibition between vehicle-treated mice and drug-treated mice using the Mann-Whitney-Wilcoxon test. For combinatorial efficacy studies, a group size of n=9 or 10 was utilized in order to provide > 95% power to detect a difference of at least 30% in tumor growth inhibition between combination and monotherapy-treated mice or drug- and vehicle-treated mice using the Mann-Whitney-Wilcoxon test. Survival estimates were computed using the Kaplan-Meier method and survival curves were compared between monotherapy and combination treated groups. GraphPad Prism was used to graph the data and carry out statistical analyses. A P value < 0.05 was considered statistically significant.
RESULTS
PARP inhibitor-resistant BRCA1-mutant PDX models of HGSOCs exhibit restoration of homologous recombination (HR) repair
In order to determine the preclinical activity of prexasertib in ovarian cancer, we used PDX models of HGSOC. Fourteen ascites-derived PDX models from patients who had received platinum chemotherapy have been clinically-annotated, luciferized, characterized by immunohistochemistry (IHC) and analyzed for BRCA status and other genomic changes (30) (Fig. 1A and Supplementary Table S1). These models retain genetic alterations observed in the corresponding original tumors. One BRCA wild-type model was sensitive to the PARP inhibitor olaparib. However, the remaining 13 models, including four derived from BRCA1 germline carriers (DF59, DF68, DF86 and DF101), were resistant to olaparib (Fig. 1A and Supplementary Fig. S1A).
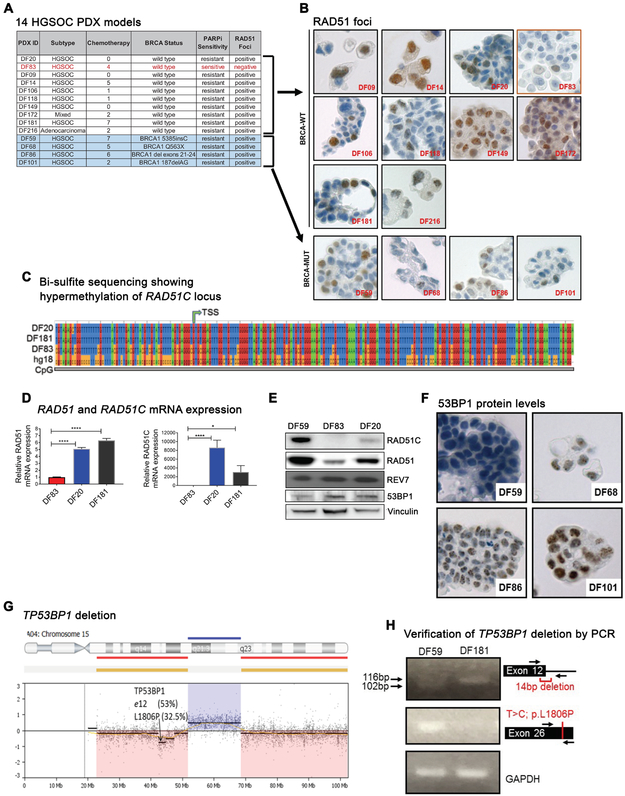
Figure 1. PARP inhibitor resistant BRCA1-deficient ovarian cancer PDX models exhibit restoration of homologous recombination.
(A) BRCA status and PARP inhibitor sensitivity of 14 HGSOC PDX models. Four of the 14 models are BRCA1-mutated and 13 of the models are olaparib resistant. (B) RAD51 immunohistochemical (IHC) staining in irradiated tumor cells from PDX models showing presence of RAD51 foci in 4 BRCA1-mutated PDX models and absence of RAD51 foci in the BRCA-WT DF83 model. FFPE tissue sections of the fourteen PDX models were stained using an anti-RAD51 antibody and representative images (60X) are shown. (C) Bisulfite sequencing of tumor cells from PDX models showing hypermethylation of the RAD51C locus in the DF83 model. TSS indicates the transcription start site. Human genome build (hg18) was used as a control. (D) qRT-PCR analysis showing absence of RAD51C mRNA expression (right panel) and reduced RAD51 mRNA expression (left panel) in the DF83 PDX model. **** p value <0.0001, *p value =0.0210 using the one-way Anova (n=4) (E) Western blots of tumor lysates from the DF59, DF83 and DF20 PDX models. (F) 53BP1 immunohistochemical (IHC) staining of irradiated tumor cells from BRCA1-mutant PDX models showing absence of 53BP1 protein levels in the DF59 PDX model. FFPE tissue sections of the four BRCA1-mutant models were stained using an anti-53BP1 antibody. Representative images (60X) are shown. Sub-nuclear 53BP1 foci are present in all models except DF59. (G) MIP-SNP array (OncoScan3 platform) analysis showing allelic copy number loss of the 15q segment harboring the TP53BP1 locus in ascites-derived tumor cells from the DF59 PDX model. Exome sequencing identified two mutations in the ascites-derived DF59 cells; 53% of the reads showed a 14bp deletion at the end of Exon12 and 32.5% of the reads carried a missense mutation L1806P. A schematic representation of these events is shown. Results were verified using genomic PCR (H) The 14bp deletion in ~50% of the genomic DNA was observed as a lower migrating band in the PCR (top panel). A mutation-specific primer was used to verify the presence of the T>C mutation in the DF59 model (middle panel). DF181 was used as the control in the PCR analyses and GAPDH was used as the loading control.
To assess the HR-mediated DNA repair status in these models, we developed a RAD51-based immunohistochemical (IHC) assay. Freshly derived tumor cells from PDX-bearing mice were exposed to radiation and stained with an anti-RAD51 antibody. Consistent with the olaparib response, the 13 olaparib-resistant models exhibited DNA-damage induced RAD51 foci (Fig. 1B). The presence of RAD51 foci in the BRCA1-mutant models suggested that restoration of HR had occurred. In contrast, the olaparib-sensitive PDX model DF83 showed absence of RAD51 foci (Fig. 1B). Bisulfite sequencing demonstrated hypermethylation of the RAD51C locus (Fig. 1C) along with low expression of both RAD51C and RAD51 mRNAs (Fig. 1D) and their encoded proteins (Fig. 1E) indicating that RAD51C, a known FA gene (FANCO) and regulator of HR repair (37, 38), is silenced in this model, accounting for HR deficiency and PARP inhibitor sensitivity.
Because loss of 53BP1 is known to restore HR (9, 39) and confer PARP inhibitor resistance in BRCA1-mutant cancer cells (9), we assessed the levels of 53BP1 in the four BRCA1-mutant, olaparib-resistant models. DF59 showed low expression of 53BP1 staining by IHC (Fig. 1F). CGH and MIP-SNP array analysis showed allelic deletion in the TP53BP1 locus (30). Whole- exome sequencing of this tumor identified two mutations in the TP53BP1 locus. A 14-base pair deletion at the end of exon 12 was present in 53% and the missense mutation L1806P was present in 32.5% of the reads, respectively (Fig. 1G), confirmed by PCR (Fig. 1H). These results confirm that BRCA1-mutant HGSOCs can restore HR and become PARP inhibitor- resistant by mutation of TP53BP1.
Prexasertib exhibits monotherapy activity in PARP inhibitor-resistant HGSOC PDX models
We next sought to determine whether prexasertib has activity in the PARP inhibitor-resistant PDX models. Prexasertib monotherapy demonstrated significant antitumor activity across all of the models (Supplementary Fig. S1A), including the four that are BRCA1-mutated, PARP inhibitor-resistant, as well as the remaining BRCA wild-type models, several of which demonstrate moderately elevated CCNE1 mRNA expression and one with extremely high expression (DF172) (Supplementary Table S1 and Supplementary Fig. S1B). Of note, prexasertib monotherapy activity was not restricted to the models with the highest levels of CCNE1 expression.
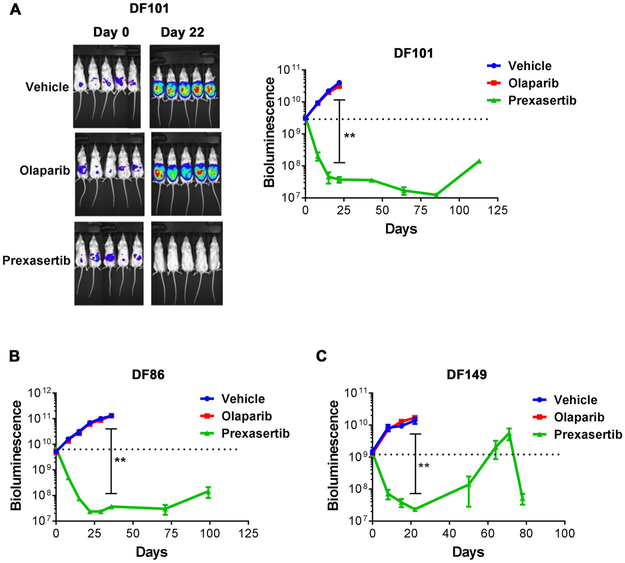
For 3 models, the activity was confirmed in larger cohorts of mice, including two BRCA1-mutated models (DF-101 and DF-86) and one BRCA wild-type model with high MYC expression (DF-149) (Fig. 2 and Supplementary Table S1). These 3 models were resistant to olaparib as expected, but sensitive to prexasertib, with profound and durable tumor regression. Prexasertib was overall well tolerated with 5–15% loss of body weight during the treatment period (Supplementary Fig. S2A). Collectively, these results demonstrate that prexasertib has monotherapy activity in PARP inhibitor-resistant HGSOC PDX models, including BRCA-mutant tumors and tumors with genomic features indicative of replication stress.
Figure 2. Prexasertib exhibits monotherapy activity in PARP inhibitor-resistant HGSOC PDX models.
NSG mice bearing luciferized PDXs were treated with vehicle, olaparib (100 mg/kg, daily) or prexasertib (8 mg/kg, BIDx3, rest 4 days) (n=5 mice per group for each model) for 3 weeks. Tumor growth was monitored by weekly bioluminescence imaging of the mice for approximately 125 days. Tumor measurements are represented as mean ± SE. P values were determined using the Mann-Whitney test. (A) Left panel. Representative images of mice bearing DF101 tumors. Right panel. Tumor growth in mice bearing DF-101 xenografts. ** P = 0.0079, for vehicle vs. prexasertib at day 22. (B) Tumor growth in mice bearing DF86 xenografts. ** P = 0.0079, for vehicle vs. prexasertib at day 36. (C) Tumor growth in mice bearing DF149 xenografts. ** P = 0.0079, for vehicle vs. prexasertib at day 22.
Prexasertib induces DNA damage and replication stress in HGSOC PDX models
To determine whether prexasertib causes DNA damage and replication stress in HGSOC PDX models, we assessed pharmacodynamic markers of prexasertib activity, using DF-59, the PARP inhibitor-resistant model with absent TP53BP1 expression (Fig. 3A). Mice bearing DF59 xenografts were exposed to vehicle, prexasertib, olaparib or the combination. Lysates from tumors harvested at variable timepoints were subjected to Western blotting for p-CHK1[S345], a pharmacodynamic marker of CHK1 inhibition, markers of DNA damage, including p-KAP1 and γ-H2AX, as well as a marker of replication stress, p-RPA32. Prexasertib-mediated CHK1 inhibition is expected to result in increased phosphorylation at the S345 ATR phosphorylation site (28, 29). As expected, olaparib alone did not cause significant DNA damage in this model, although small increases in RAD51 and γ-H2AX foci were observed by immunohistochemistry (Supplementary Fig. S3). In contrast, prexasertib, either alone or in combination with olaparib, caused DNA damage as early as 6 hrs, with continued DNA damage and replication stress evident after 52 hrs of exposure (Fig. 3A, Supplementary Fig. S3).
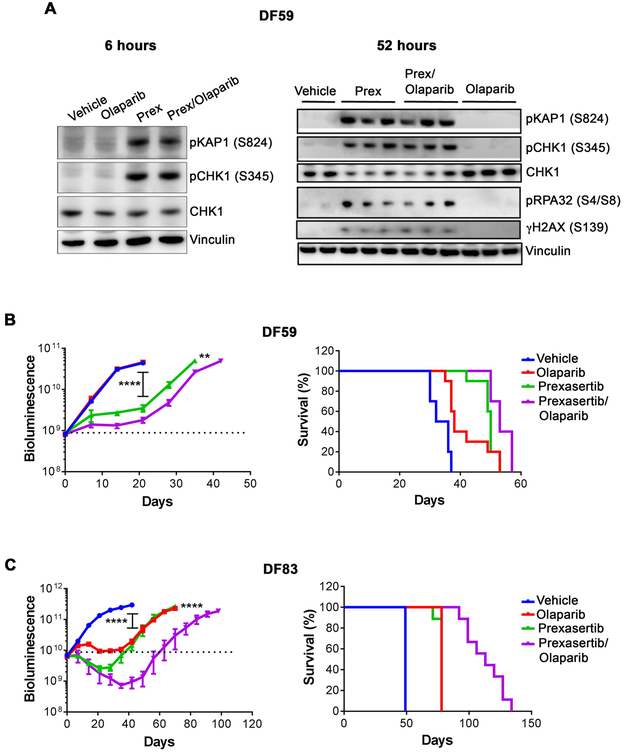
Figure 3. Prexasertib causes DNA damage and synergizes with PARP inhibition in ovarian cancer HGSOC PDX models.
(A) Prexasertib causes DNA damage in PDX tumors. NSG mice bearing luciferized PDX model DF59 were treated with vehicle (n=2 mice), olaparib (100 mg/kg) (n=3 mice), prexasertib (8 mg/kg, BID) (n=3 mice) or olaparib (100 mg/kg) + prexasertib (8 mg/kg, BID) (n=3 mice). Lysates from individual tumors harvested at 6 hrs (Left panel) and 52 hrs (Right panel) after dosing were subjected to Western blotting with the indicated antibodies. In left panel, data are from a single mouse in each group. (B, C) Tumor growth (left panels) and survival (right panels) in mice bearing DF59 or DF83 PDX tumors. NSG mice bearing luciferized PDXs, namely DF59 (n=10 mice/group) or DF83 (n=9 mice/ group) were treated with vehicle, olaparib (100 mg/kg, daily), prexasertib (8 mg/kg, BIDx3, rest 4 days) or olaparib (100 mg/kg, daily) + prexasertib (8 mg/kg, BIDx3, rest 4 days) for 3 weeks. Tumor growth by weekly bioluminescence imaging and survival of tumor-bearing mice were monitored. Tumor measurements are represented as mean ± SE. In panels B and C, P values for tumor growth curves were determined using the Mann-Whitney test. In the DF59 model (B), ** P = 0.002 for prexasertib vs. prexasertib+olaparib at day 35; **** P <0.0001 for prexasertib vs. vehicle at day 21. In the DF83 model (C), **** P <0.0001 for prexasertib vs. prexasertib+olaparib at day 70 and for vehicle vs. olaparib, as well as vehicle vs. prexasertib group at day 42. In the right-hand grahps, log-rank survival plots were generated using prizm graphpad. In the DF59 model (B), P = 0.0005 for vehicle vs. olaparib; P < 0.0001 for vehicle vs prexasertib; P = 0.0045 for prexasertib vs. prexasertib+olaparib; and P = 0.0010 for olaparib vs. prexasertib+olaparib. In the DF83 model (C), P <0.0001 between all groups except olaparib vs. prexasertib.
Combined prexasertib and olaparib is synergistic in a subset of HGSOC PDX models
We next tested the efficacy of the combination of prexasertib and olaparib in the DF-59 model (Fig. 3B). Olaparib alone had no activity; however, the combination of prexasertib and olaparib demonstrated substantial tumor growth inhibition, greater than that observed with prexasertib alone, along with prolonged survival, consistent with CHK1 inhibitor-mediated sensitization to PARP inhibition. Combination treatment was well tolerated with approximately 10% loss of body weight during the time of drug exposure (Supplementary Fig. S2B). We also assessed the combination in the HR-deficient olaparib-sensitive PDX model, DF-83 (Fig. 3C and Supplementary Fig. S2B). Here, the combination augmented the degree and duration of response compared to either of the monotherapies, again translating to improved survival. Collectively, these results suggest that the combination of prexasertib and olaparib may be relevant in both PARP inhibitor-resistant and sensitive backgrounds.
Prexasertib reduces viability across a panel of HGSOC cell lines
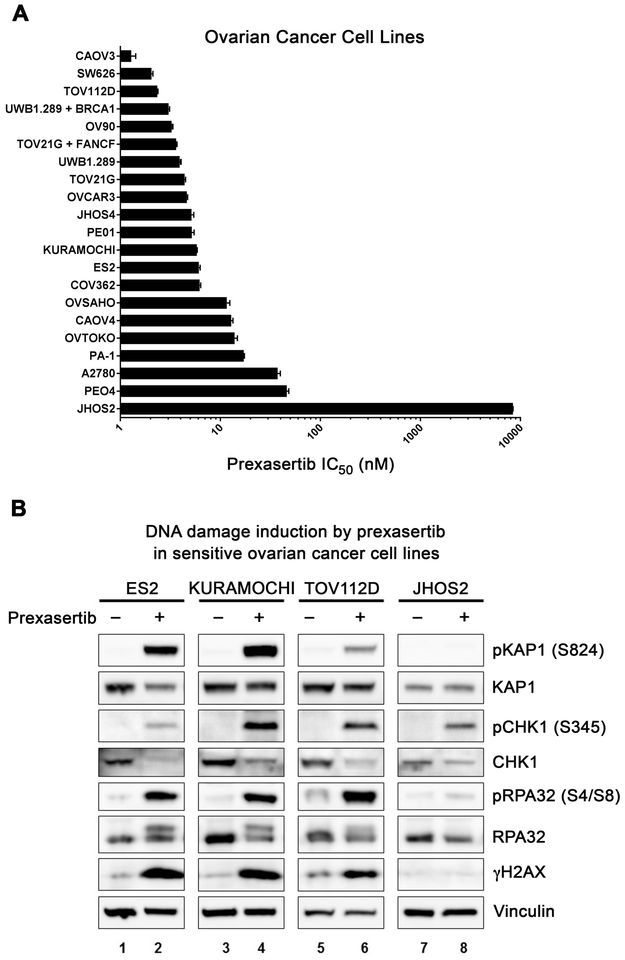
We next evaluated prexasertib activity in a panel of ovarian cancer cell lines including BRCA wild-type lines, BRCA-mutant lines, and cell lines with high cyclin E expression suggestive of replication stress. Prexasertib was cytotoxic in most of the ovarian cancer cell lines with IC50 values of 1–10 nM in a Cell Titer-Glo viability assay (Fig. 4A), regardless of BRCA status or cyclin E expression (Supplementary Fig. S4). The activity of prexasertib was lowest in the BRCA1-mutant, TP53-mutant JHOS2 cell line, which demonstrated an IC50 of 8.4 μM (Fig. 4A). Prexasertib exposure resulted in increased levels of pKAP1, pRPA32 and γ-H2AX in sensitive cell lines, whereas these events were absent in resistant JHOS2 cells (Fig. 4B), suggesting that monotherapy activity may be mediated by the induction of DNA damage and replication stress.
Figure 4. Ovarian cancer cells are highly sensitive to prexasertib.
(A) IC50 of prexasertib in ovarian cancer cell lines. Cells were grown for 24 hrs before exposure to graded concentrations of prexasertib. Viability was assessed after 3 days of treatment using Cell Titer- Glo. IC50 values for growth inhibition were calculated by Prizm graphpad and mean ± SE (n=4) are shown. The experiments were done at least two times for each cell line, and the data from a representative experiment are shown. (B) Western blots of lysates from prexasertib-sensitive ovarian cancer cells (ES2, KURAMOCHI and TOV112D) and prexasertib-resistant ovarian cancer cells (JHOS2) treated with DMSO or prexasertib (40 nM) for 18 hrs.
Prexasertib and olaparib are synergistic in PARP inhibitor-resistant HGSOC cell lines
Several of the cell lines were relatively PARP inhibitor-resistant, with olaparib IC50 values in the 4–10 μM range, including BRCA wild-type TOV112D and ES2 (Fig. 5A and B). The combination of prexasertib and olaparib resulted in synergistic cytotoxicity in these and other cell lines (Fig. 5A and B and Supplementary Fig. S5).
Figure 5. Prexasertib increases sensitivity of ovarian cancer cells to olaparib and disrupts homologous recombination.
(A, B) Synergy between prexasertib and PARP inhibition in TOV112D (A) and ES2 (B) ovarian cancer cells. Cells were grown for 24 hrs in triplicate before exposure to graded concentrations of prexasertib and olaparib. Viability was assessed at 6 days using CellTiter- Glo. Synergy/antagonism between the drugs was determined using Combenefit software. (Left panels) Survival plots. (Middle panels) Bliss synergy/antagonism levels in a matrix format. (Right panels) Bliss synergy/antagonism levels on the experimental combination dose response surface. Bliss scores greater than zero (green/blue shading) indicate synergy between prexasertib and olaparib. (C) Prexasertib inhibits homologous recombination activity in a DR-GFP reporter assay in U2OS cells. 24 hrs after transfection with a plasmid encoding SceI endonuclease, U2OS-DR-GFP cells were exposed to graded concentrations of prexasertib for 48 hrs, and GFP expression analyzed by flow cytometry. The relative GFP-positive cells normalized to vehicle-treated cells are shown. **** P <0.0001 using the one-way Anova (n=3). (D) Prexasertib inhibits radiation-induced RAD51 foci in U2OS cells. Cells were pre-treated with prexasertib for 16 hrs before exposing them to radiation. Six hours after radiation, cells were analyzed for RAD51 foci by immunofluorescence. Representative images at 63X (Left panel) and quantitation (Right panel) of RAD51 foci are shown. At least 100 cells with foci were scored. **** P < 0.0001 using the one-way Anova. (E) Prexasertib inhibits radiation-induced RAD51 foci in ES2 ovarian cancer cells. Cells were pre-treated with prexasertib for 1 hr before exposing them to radiation. Six hours after radiation, cells were analyzed for RAD51 foci by immunofluorescence as in panel D. **** P <0.0001 using the one-way Anova (n=3). (F) (Left panel) Western blots of the lysates from BRCA1-mutated COV362 cells after siRNA-mediated knockdown of 53BP1. (Middle and Right panels) Survival of COV362 cells with siRNA-mediated knockdown of 53BP1 after olaparib or prexasertib treatment, showing resistance to olaparib. Forty-eight hrs after siRNA transfection, cells were exposed to graded concentrations of olaparib or prexasertib in 96-well plates. Viability was assessed at 6 days by CellTiter- Glo and IC50 values were calculated by Prizm graphpad. IC50 values for olaparib in siControl and si53BP1 cells were 2.7 μM and 14.3 μM, respectively. IC50 values for prexasertib in siControl and si53BP1 cells were 3.2 nM and 5.1 nM, respectively (n=4). (G) Synergy between prexasertib and PARP inhibition in COV362 ovarian cancer cells after siRNA-mediated knockdown of 53BP1. Forty-eight hrs after siRNA transfection, cells were exposed to graded concentrations of prexasertib and olaparib in triplicates in 96-well plates. Viability was assessed at 6 days using CellTiter- Glo. Survival and synergy were determined as described in panels A and B. (H) Olaparib-induced RAD51 foci in COV362 cells are increased after depletion of 53BP1. Forty-eight hrs after siRNA transfection, cells were pre-treated with vehicle or olaparib for 24 hrs before exposing them to vehicle or prexasertib. Six hours after the second exposure, cells were analyzed for RAD51 foci by immunofluorescence. Quantitation of RAD51 foci is shown. At least 100 cells with foci were scored. **** P <0.0001 using the one-way Anova (n=3). The experiments in all panels except panel D were done two or three times, and the data from a representative experiment are shown.
To demonstrate that prexasertib directly compromises HR repair as a mechanism of PARP inhibitor sensitization, we utilized U2OS osteosarcoma cells engineered to express GFP during HR repair in response to a restriction enzyme-induced double-strand break. Disruption of HR activity by prexasertib was demonstrated in this assay (Fig. 5C). Consistent with this finding, prexasertib exposure reduced γ-irradiation-induced RAD51 foci in U2OS (Fig. 5D) and ES2 ovarian cancer (Fig. 5E) cells in a dose-dependent manner. Similarly, we studied BRCA1-mutant COV362 cells rendered olaparib-resistant by siRNA-mediated depletion of TP53BP1. The PARP inhibitor-resistant derivative retained sensitivity to prexasertib (Fig. 5F). Prexasertib was synergistic with olaparib in these cells and compromised induction of RAD51 foci after olaparib exposure (Fig. 5G and H). Taken together, these results suggest that prexasertib compromises HR repair, sensitizing resistant HGSOC cells to PARP inhibition. Of note, BRCA1−/− retinal pigmented epithelial (RPE) cells (36) rendered resistant to PARP inhibition via HR restoration by TP53PB1 depletion also retained sensitivity to prexasertib (Supplementary Fig. S6A-D).
Because prexasertib disrupts HR, we asked whether brief exposure of cells to prexasertib prior to olaparib would further augment cytotoxicity. As shown in Supplementary Fig. S6E, there was no improvement over the degree of synergism achieved with concomitant exposure to both drugs at the outset of the experiment.
Prexasertib reverses stabilization of replication forks in PARP inhibitor-resistant HGSOC cells
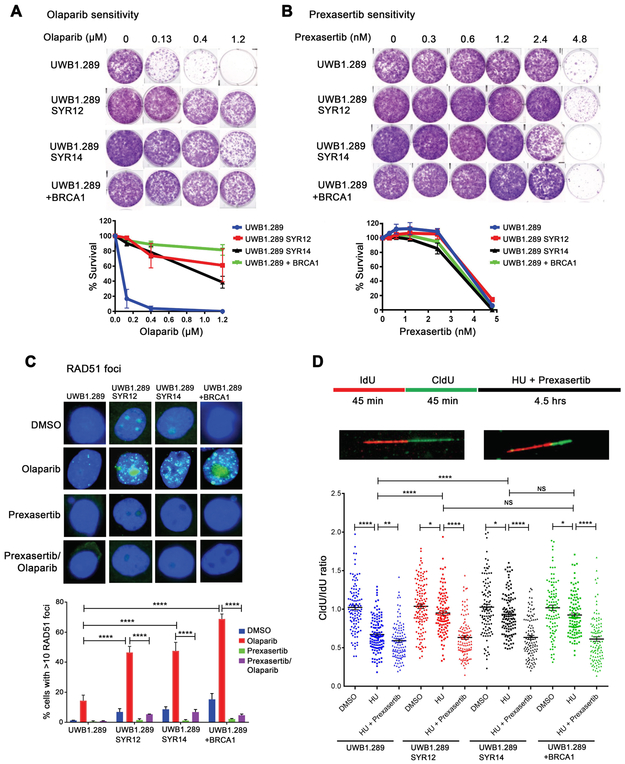
UWB1.289 BRCA1-mutated HGSOC ovarian cancer cells rendered PARP inhibitor-resistant after prolonged exposure to drug have been demonstrated to simultaneously exhibit multiple mechanisms of resistance, including restoration of HR, as well as replication fork stabilization (32) . Similar to the engineered COV362 cells (Fig. 5F), the UWB1.289 olaparib-resistant derivatives SYR12 and SYR14 (32) retained sensitivity to prexasertib, comparable to that of parental cells (Fig. 6A and B, Supplementary Fig. S7A). These subclones demonstrated partial restoration of HR, assessed by RAD51 focus assembly in response to olaparib compared to parental cells complemented with wild-type BRCA1 cDNA. As expected, prexasertib compromised the induction of RAD51 in response to olaparib (Fig 6C).
Figure 6. Prexasertib has activity in BRCA1-mutated ovarian cancer cells that have restored fork stability and acquired PARP inhibitor resistance.
(A, B) UWB1.289 SYR12 and UWB1.289 SYR14 ovarian cancer cells with acquired PARP inhibitor resistance are sensitive to prexasertib. BRCA1-mutated parental UWB1.289 cells, resistant derivatives SYR12 and SYR14 and BRCA1 add-back cells were exposed to olaparib or prexasertib in 6-well plates for 12 days and stained with crystal violet. Representative colony formation assays (n=3) after olaparib or prexasertib exposure are shown in panels A and B, respectively. (C) Prexasertib blocks restored homologous recombination in PARP inhibitor-resistant UWB1.289 SYR12 and UWB1.289 SYR14 ovarian cancer cells. The indicated cell lines were pre-treated with olaparib (5 μM) for 24 hrs before exposing to prexasertib (60 nM) for 6 hrs and analyzed for RAD51 foci by immunofluorescence. Representative images (Upper panel) and quantitation (Lower panel) of RAD51 foci are shown. At least 100 cells were scored. **** P <0.0001 using the one-way Anova, (n=3) (D) Prexasertib reactivates degradation of stalled replication forks in PARP-inhibitor resistant cells as determined by DNA fiber assays. Schematic of IdU, CldU, hydroxyurea (HU) and prexasertib treatment (Upper panel), representative images of normal and stalled fibers (Middle panel) and graphical quantitation of DNA fiber analysis of stalled forks (degraded forks) with each fiber (Lower panel) are shown. Fiber assays were repeated twice and data from a representative experiment are shown. For fiber assays, newly synthesized DNA was sequentially labeled with IdU for 45 min and CldU for 45 min. Cells were then treated with DMSO, replication stalling agent HU (2 mM), or HU (2 mM) + prexasertib (100 nM) for 4.5 hrs. At least 100 DNA fibers were analyzed per condition. IdU labelling was detected in red and CldU labelling was detected in green and fork degradation was assessed by measuring the ratio of CldU to IdU labelling, with a ration < 1 indicating fork degradation. **** P <0.0001, ** P = 0.0023, * P < 0.05 using the Mann-Whitney test. The experiments in panels A-D were done at least two times, and the data from a representative experiment are shown.
As evidenced by DNA fiber assays, the SYR12 and SYR14 derivatives also demonstrated stabilization of replication forks after hydroxyurea-induced stalling. Prexasertib also reversed fork stabilization in these cells (Fig. 6D). Consequently, prexasertib re-sensitized these resistant derivatives to olaparib (Supplementary Fig. S7B). Taken together, these results indicate that prexasertib can induce cytotoxicity in BRCA1-mutated PARP inhibitor-resistant HGSOC cells and reverse the two major mechanisms of acquired PARP inhibitor resistance.
DISCUSSION
In the current report, we demonstrate that the CHK1 inhibitor prexasertib has activity as monotherapy and in combination with PARP inhibition in preclinical EOC in vivo models. Prexasertib has activity in both HR repair-deficient and -proficient models and is effective in killing tumor cells with de novo or acquired PARP inhibitor resistance.
As a monotherapy, prexasertib-mediated CHK1 inhibition compromises checkpoint control and repair in response to endogenous DNA damage, as well as increases replication stress in tumor cells. Cells with TP53 deficiency, which occurs nearly universally in HGSOC, coupled with HR deficiency (3, 4), may be highly dependent on the ATR-CHK1 pathway for maintenance of genomic stability, as evidenced by the response to prexasertib of the RAD51C-deficient PDX model DF83. Additionally, cells with unstable replication forks, as can occur with BRCA-deficiency, may require the ATR-CHK1 pathway to prevent replication fork collapse. In our previous work in HGSOC organoid cultures, those with baseline unstable replication forks were highly sensitive to CHK1 inhibition (40). Alternatively, in cells with a high baseline level of replication stress, including those driven by CCNE1 or MYC copy number gain, amplification or overexpression of the corresponding proteins, further exacerbation of replication stress mediated by CHK1 inhibition may be lethal, accounting for the sensitivity of such cells both in preclinical models tested here and in clinical trials (26, 27).
Importantly, prexasertib also demonstrates activity in PARP inhibitor-resistant BRCA1-mutant cells both as monotherapy and in combination with olaparib. The four BRCA1-mutant PDX models have clearly demonstrated restored HR, as evidenced by formation of DNA damage-induced RAD51 foci; in the case of one model, restored HR is the result of TP53BP1 mutation and reduced expression, which is known to facilitate BRCA1-independent DNA end resection (9). A variety of other mechanisms may restore HR in PARP inhibitor-resistant cells (41), including BRCA reversion mutation (42), stabilization of mutant BRCA proteins (43), or depletion of proteins such REV7 (10) and other components of the Shieldin Complex (44) and DYNLL1 (45). Irrespective of the mechanism of HR restoration, in HGSOC cell lines, prexasertib compromises the formation of RAD51 foci in response to DNA damage. It has been previously shown that CHK1 phosphorylates RAD51 (23), an event required for filament formation that is blocked by CHK1 inhibition.
As shown by UWB1.289 BRCA1-mutant HGSOC cancer cells rendered PARP inhibitor-resistant, multiple mechanisms of resistance may occur simultaneously, so that restored HR may be accompanied by replication fork stabilization (32). The latter event typically results from the exclusion of nucleases, as occurs with reduced expression of PTIP, compromising the recruitment of the MRE11 nuclease (11). Similar to previously reported results with ATR inhibition (32), CHK1 inhibition compromises replication fork stability in PARP inhibitor-resistant cells with stabilized replication forks, likely by preventing RAD51 accumulation at sites of replication fork stalling. In this regard, prexasertib reverses a second major mechanism of PARP inhibitor resistance and sensitizes cells with acquired resistance to PARP inhibition.
Recently, the preliminary activity of prexasertib monotherapy in BRCA wild-type HGSOC was reported (27). Most patients had platinum-resistant or refractory disease; among 24 evaluable patients, eight achieved RECIST partial response. Four of the eight patients with partial tumor responses had both CCNE1 amplification or copy number gain and CCNE1 mRNA upregulation, indicating that prexasertib may have potential activity in this population. While PDX models with high CCNE1 expression and cell lines with high cyclin E levels used in our study were sensitive to prexasertib, activity was not restricted to this subset of models.
In the ovarian cancer study, prexasetib carried substantial hematologic toxicity resulting in transient grade 4 neutropenia and grade 3/4 thrombocytopenia in 79% and 25% of patients, respectively (27). Nonetheless, based on this study, a larger study of prexasertib in platinum-resistant and refractory disease is underway (). One cohort in this study will also evaluate patients with BRCA-mutant tumors with platinum-resistant disease, who have already received a PARP inhibitor. Results in the preclinical BRCA1-mutant models evaluated here suggest promise for this approach.
The substantial activity of prexasertib monotherapy in the PDX models made it difficult to demonstrate synergism with olaparib in all cases. Nonetheless, in the TP53BP1-deficient, HR-proficient, BRCA1-mutated model, combined prexasertib and olaparib was superior to monotherapy treatment. Additionally, several cell lines with either de novo or acquired PARP inhibitor resistance demonstrated synergism between prexasertib and olaparib, similar to previous reports (29, 46). This occurred in both BRCA wild-type cells, but also in BRCA1-mutated cells with acquired PARP inhibitor resistance. For this reason, a phase 1 trial combining prexasertib and olaparib has been initiated, focusing on BRCA-mutant, PARP inhibitor-resistant HGSOC (). Although overlapping hematological toxicity has not permitted administration of full doses of both agents, tolerable attenuated doses of the drugs used in combination concurrently have been achieved, with responses observed in this population (47). Nonetheless, ultimately, in the PARP inhibitor-resistant setting, the efficacy of prexasertib monotherapy and combined prexasertib/olaparib may need to be directly compared. Mechanistically, it is possible that combined prexasertib/olaparib may compromise stabilized replication forks to a greater degree than prexasertib alone. We have previously observed this phenomenon when prexasertib combined with a replication stress-inducing agent such as gemcitabine produced greater fork instability than prexasertib alone in HGSOC organoids with stabilized replication forks (40).
In addition to work in the PARP inhibitor-resistant setting, combined prexasertib and olaparib was superior to either monotherapy in the RAD51C-mutant, PARP inhibitor-sensitive model. These results suggest that comparison of the combination to olaparib monotherapy in this setting may also be warranted, and that prexasertib may play a role in delaying the development of resistance.
Furthermore, several cell lines examined in this work, including TOV112D and ES2, may not represent classic HGSOC (48), but rather may demonstrate characteristics of clear cell or endometrioid ovarian cancer (49, 50). These cancers do not typically respond well to standard treatments, suggesting that prexasertib as monotherapy or in combination with olaparib could be relevant therapeutic options in these subtypes as well.
In summary, prexasertib, either as monotherapy or in combination with olaparib, has extensive activity in in vivo and cell line models of HGSOC, in BRCA wild-type and mutant backgrounds and in PARP inhibitor-sensitive and resistant settings. These results are translating to ongoing clinical trials designed to fully assess the potential role of prexasertib in the HGSOC armamentarium.
Supplementary Material
Translational Relevance.
The CHK1 inhibitor prexasertib is currently in clinical development as a single agent and in combination with both cytotoxic and targeted agents. In high-grade serous ovarian cancer (HGSOC), prexasertib monotherapy has demonstrated activity in patients with BRCA wild-type, platinum-resistant disease. Here, we tested the activity of prexasertib in both BRCA wild-type and BRCA1-mutated, poly (ADP-ribose) polymerase (PARP) inhibitor-resistant HGSOC patient-derived xenograft (PDX) models and cell lines. These preclinical models were routinely sensitive to prexasertib, likely related to induction of DNA damage and replication stress. Additionally, prexasertib impaired both homologous recombination repair and replication fork stability, addressing two major mechanisms of resistance to PARP inhibition. Consequently, prexasertib was synergistic with olaparib in multiple models. These data suggest that prexasertib, either as monotherapy or in combination with olaparib, has broad activity against HGSOCs, including those harboring BRCA1 mutation with acquired PARP inhibitor resistance, informing additional ovarian cancer populations for clinical trials.
ACKNOWLEDGMENTS
This work was supported by Lilly-Dana-Farber Cancer Institute Program Grant A09640 (A.D.D., G.I.S.), and Stand Up To Cancer-Ovarian Cancer Research Fund Alliance-National Ovarian Cancer Coalition Dream Team Translational Cancer Research Grant (SU2C-AACR-DT16–15). Stand Up To Cancer is a division of the Entertainment Industry Foundation. Research grants are administered by the American Association for Cancer Research, the Scientific Partner of SU2C. This work was also supported by U.S. National Institutes of Health grants R37 HL052725 (A.D.D), P01 HL048546 (A.D.D.), P50 CA168504 (A.D.D, G.I.S.) and R01 CA090687 (G.I.S.), U.S. Department of Defense grants BM110181 (A.D.D.) and BC151331P1 (A.D.D.), the Breast Cancer Research Foundation (A.D.D.) and the Fanconi Anemia Research Fund (A.D.D.).
Footnotes
Conflict of Interest Disclosure: A.D.D. receives research funding from Eli Lilly, Merck KGaA-EMD Serono, and Sierra Oncology, has served on advisory boards for Eli Lilly, Merck KGaA-EMD Serono, Sierra Oncology, Intellia, Formation Biologics and holds equity in Ideaya Inc, Cyteir Therapeutics, and Cedilla Therapeutics, Inc. G.I.S. receives research funding from Eli Lilly, Merck KGaA-EMD Serono, Sierra Oncology, Merck, Pfizer and Array Biopharma and has served on advisory boards for Eli Lilly, Merck KGaA-EMD Serono, Sierra Oncology, Pfizer, Astex, Almac, Roche, Bicycle Therapeutics, Fusion Pharmaceuticals, G1Therapeutics, Ipsen, Cybrex Therapeutics, Daiichi-Sankyo and Angiex.
REFERENCES
- 1.Ramalingam P. Morphologic, Immunophenotypic, and Molecular Features of Epithelial Ovarian Cancer. Oncology (Williston Park) 2016;30:166–76. [PubMed] [Google Scholar]
- 2.Bowtell DD, Bohm S, Ahmed AA, Aspuria PJ, Bast RC Jr., , Beral V, et al. Rethinking ovarian cancer II: reducing mortality from high-grade serous ovarian cancer. Nat Rev Cancer 2015;15:668–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature 2011;474:609–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Konstantinopoulos PA, Ceccaldi R, Shapiro GI, D’Andrea AD. Homologous Recombination Deficiency: Exploiting the Fundamental Vulnerability of Ovarian Cancer. Cancer Discov 2015;5:1137–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lord CJ, Ashworth A. PARP inhibitors: Synthetic lethality in the clinic. Science 2017;355:1152–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matulonis UA. Management of newly diagnosed or recurrent ovarian cancer. Clin Adv Hematol Oncol 2018;16:426–37. [PubMed] [Google Scholar]
- 7.Moore K, Colombo N, Scambia G, Kim BG, Oaknin A, Friedlander M, et al. Maintenance Olaparib in Patients with Newly Diagnosed Advanced Ovarian Cancer. N Engl J Med 2018;379:2495–505. [DOI] [PubMed] [Google Scholar]
- 8.Mirza MR, Pignata S, Ledermann JA. Latest clinical evidence and further development of PARP inhibitors in ovarian cancer. Annals Oncol 2018;29:1366–76. [DOI] [PubMed] [Google Scholar]
- 9.Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 2010;141:243–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu G, Chapman JR, Brandsma I, Yuan J, Mistrik M, Bouwman P, et al. REV7 counteracts DNA double-strand break resection and affects PARP inhibition. Nature 2015;521:541–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ray Chaudhuri A, Callen E, Ding X, Gogola E, Duarte AA, Lee JE, et al. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature 2016;535:382–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rondinelli B, Gogola E, Yucel H, Duarte AA, van de Ven M, van der Sluijs R, et al. EZH2 promotes degradation of stalled replication forks by recruiting MUS81 through histone H3 trimethylation. Nature Cell Biol 2017;19:1371–8. [DOI] [PubMed] [Google Scholar]
- 13.Etemadmoghadam D, Weir BA, Au-Yeung G, Alsop K, Mitchell G, George J, et al. Synthetic lethality between CCNE1 amplification and loss of BRCA1. Proc Natl Acad Sci USA 2013;110:19489–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nature Cell Biol 2014;16:2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bucher N, Britten CD. G2 checkpoint abrogation and checkpoint kinase-1 targeting in the treatment of cancer. Br J Cancer 2008;98:523–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson R, Eastman A. The cancer therapeutic potential of Chk1 inhibitors: how mechanistic studies impact on clinical trial design. Br J Clin Pharmacol 2013;76:358–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feijoo C, Hall-Jackson C, Wu R, Jenkins D, Leitch J, Gilbert DM, et al. Activation of mammalian Chk1 during DNA replication arrest: a role for Chk1 in the intra-S phase checkpoint monitoring replication origin firing. J Cell Biol 2001;154:913–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scorah J, McGowan CH. Claspin and Chk1 regulate replication fork stability by different mechanisms. Cell Cycle 2009;8:1036–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Molecular Cell 2010;37:492–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho SH, Toouli CD, Fujii GH, Crain C, Parry D. Chk1 is essential for tumor cell viability following activation of the replication checkpoint. Cell Cycle 2005;4:131–9. [DOI] [PubMed] [Google Scholar]
- 21.Syljuasen RG, Sorensen CS, Hansen LT, Fugger K, Lundin C, Johansson F, et al. Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Mol Cell Biol 2005;25:3553–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Toledo LI, Altmeyer M, Rask MB, Lukas C, Larsen DH, Povlsen LK, et al. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell 2013;155:1088–103. [DOI] [PubMed] [Google Scholar]
- 23.Sorensen CS, Hansen LT, Dziegielewski J, Syljuasen RG, Lundin C, Bartek J, et al. The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nature Cell Biol 2005;7:195–201. [DOI] [PubMed] [Google Scholar]
- 24.Forment JV, Blasius M, Guerini I, Jackson SP. Structure-specific DNA endonuclease Mus81/Eme1 generates DNA damage caused by Chk1 inactivation. PloS One 2011;6:e23517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bahassi EM, Ovesen JL, Riesenberg AL, Bernstein WZ, Hasty PE, Stambrook PJ. The checkpoint kinases Chk1 and Chk2 regulate the functional associations between hBRCA2 and Rad51 in response to DNA damage. Oncogene 2008;27:3977–85. [DOI] [PubMed] [Google Scholar]
- 26.Hong DS, Moore KN, Patel MR, Grant SC, Burris HA, William WN, et al. Evaluation of Prexasertib, a Checkpoint Kinase 1 Inhibitor, in a Phase Ib Study of Patients with Squamous Cell Carcinoma. Clin Cancer Res 2018; 24:3263–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee JM, Nair J, Zimmer A, Lipkowitz S, Annunziata CM, Merino MJ, et al. Prexasertib, a cell cycle checkpoint kinase 1 and 2 inhibitor, in BRCA wild-type recurrent high-grade serous ovarian cancer: a first-in-class proof-of-concept phase 2 study. Lancet Oncol 2018;19:207–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.King C, Diaz HB, McNeely S, Barnard D, Dempsey J, Blosser W, et al. LY2606368 Causes Replication Catastrophe and Antitumor Effects through CHK1-Dependent Mechanisms. Mol Cancer Ther 2015;14:2004–13. [DOI] [PubMed] [Google Scholar]
- 29.Brill E, Yokoyama T, Nair J, Yu M, Ahn YR, Lee JM. Prexasertib, a cell cycle checkpoint kinases 1 and 2 inhibitor, increases in vitro toxicity of PARP inhibition by preventing Rad51 foci formation in BRCA wild type high-grade serous ovarian cancer. Oncotarget 2017;8:111026–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu JF, Palakurthi S, Zeng Q, Zhou S, Ivanova E, Huang W, et al. Establishment of patient-derived tumor xenograft models of epithelial ovarian cancer for pre-clinical evaluation of novel therapeutics. Clinical Cancer Res 2017;23:1263–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matulonis UA, Wulf GM, Barry WT, Birrer M, Westin SN, Farooq S, et al. Phase I dose escalation study of the PI3kinase pathway inhibitor BKM120 and the oral poly (ADP ribose) polymerase (PARP) inhibitor olaparib for the treatment of high-grade serous ovarian and breast cancer. Annals Oncol 2017;28:512–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yazinski SA, Comaills V, Buisson R, Genois MM, Nguyen HD, Ho CK, et al. ATR inhibition disrupts rewired homologous recombination and fork protection pathways in PARP inhibitor-resistant BRCA-deficient cancer cells. Genes Dev 2017;31:318–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee DH, Acharya SS, Kwon M, Drane P, Guan Y, Adelmant G, et al. Dephosphorylation enables the recruitment of 53BP1 to double-strand DNA breaks. Molecular Cell 2014;54:512–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Di Veroli GY, Fornari C, Wang D, Mollard S, Bramhall JL, Richards FM, et al. Combenefit: an interactive platform for the analysis and visualization of drug combinations. Bioinformatics 2016;32:2866–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kochupurakkal B, Parmar K, Lazaro JB, Unitt C, Zeng Q, Reavis H, Ganesa C, Zhou S, Liu J, Palakurthi S, Strickland K, Howitt B, Konstantinopoulos P, Kirschmeier P, Geradts J, Drapkin R, Matulonis U, D’Andrea A, Shapiro G. Development of a RAD51-based assay for determining homologous recombination proficiency and PARP inhibitor sensitivity. Cancer Res 2017;77(13 supplement):2796. [Google Scholar]
- 36.Lim KS, Li H, Roberts EA, Gaudiano EF, Clairmont C, Sambel LA, et al. USP1 Is Required for Replication Fork Protection in BRCA1-Deficient Tumors. Molecular Cell 2018;72:925–41.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levy-Lahad E Fanconi anemia and breast cancer susceptibility meet again. Nat Genet 2010;42:368–9. [DOI] [PubMed] [Google Scholar]
- 38.Meindl A, Hellebrand H, Wiek C, Erven V, Wappenschmidt B, Niederacher D, et al. Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat Genet 2010;42:410–4. [DOI] [PubMed] [Google Scholar]
- 39.Bouwman P, Aly A, Escandell JM, Pieterse M, Bartkova J, van der Gulden H, et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nature Struct Mol Biol 2010;17:688–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hill SJ, Decker B, Roberts EA, Horowitz NS, Muto MG, Worley MJ Jr., , et al. Prediction of DNA Repair Inhibitor Response in Short-Term Patient-Derived Ovarian Cancer Organoids. Cancer Discov 2018;8:1404–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Francica P, Rottenberg S. Mechanisms of PARP inhibitor resistance in cancer and insights into the DNA damage response. Genome Med 2018;10:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin KK, Harrell MI, Oza AM, Oaknin A, Ray-Coquard I, Tinker AV, et al. BRCA Reversion Mutations in Circulating Tumor DNA Predict Primary and Acquired Resistance to the PARP Inhibitor Rucaparib in High-Grade Ovarian Carcinoma. Cancer Discov 2019; 9:210–19. [DOI] [PubMed] [Google Scholar]
- 43.Johnson N, Johnson SF, Yao W, Li YC, Choi YE, Bernhardy AJ, et al. Stabilization of mutant BRCA1 protein confers PARP inhibitor and platinum resistance. Proc Natl Acad Sci USA 2013;110:17041–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gupta R, Somyajit K, Narita T, Maskey E, Stanlie A, Kremer M, et al. DNA Repair Network Analysis Reveals Shieldin as a Key Regulator of NHEJ and PARP Inhibitor Sensitivity. Cell 2018;173:972–88 e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He YJ, Meghani K, Caron MC, Yang C, Ronato DA, Bian J, et al. DYNLL1 binds to MRE11 to limit DNA end resection in BRCA1-deficient cells. Nature 2018;563:522–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sen T, Tong P, Stewart CA, Cristea S, Valliani A, Shames DS, et al. CHK1 Inhibition in Small-Cell Lung Cancer Produces Single-Agent Activity in Biomarker-Defined Disease Subsets and Combination Activity with Cisplatin or Olaparib. Cancer Res 2017;77:3870–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Do KHS, Kochupurakkal B, Supko JG, Gannon C, Anderson A, Muzikansky A, Wolanski A, Hedglin J, Parmar K, Lazaro JB, Liu J, Campos S, Matulonis U, D’Andrea AD, Shapiro GI. Phase I combination study of the CHK1 inhibitor prexasertib (LY2606368) and olaparib in patients with high-grade serous ovarian cancer and other advanced solid tumors. Proc Am Assoc Cancer Res 2019;abstract CT232. [Google Scholar]
- 48.Domcke S, Sinha R, Levine DA, Sander C, Schult N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nat Commun 2013;4:2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ince TA, Sousa AD, Jones MA, Harrell JC, Agoston ES, Krohn M, et al. Characterization of twenty-five ovarian tumour cell lines that phenocopy primary tumours. Nature Commun 2015;6:7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anglesio MS, Wiegand KC, Melnyk N, Chow C, Salamanca C, Prentice LM, et al. Type-specific cell line models for type-specific ovarian cancer research. PloS One 2013;8:e72162. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.