Abstract
The relationships between inflammation and cancer are varied and complex. An important connection linking inflammation to cancer development is DNA damage. During inflammation reactive oxygen and nitrogen species (RONS) are created to combat pathogens and to stimulate tissue repair and regeneration, but these chemicals can also damage DNA, which in turn can promote mutations that initiate and promote cancer. DNA repair pathways are essential for preventing DNA damage from causing mutations and cytotoxicity, but RONS can interfere with repair mechanisms, reducing their efficacy. Further, cellular responses to DNA damage, such as damage signaling and cytotoxicity, can promote inflammation, creating a positive feedback loop. Despite coordination of DNA repair and oxidative stress responses, there are nevertheless examples whereby inflammation has been shown to promote mutagenesis, tissue damage, and ultimately carcinogenesis. Here, we discuss the DNA damage-mediated associations between inflammation, mutagenesis and cancer.
1. Introduction
As early as 1863, Rudolf Virchow recognized the inextricable connections between the immune system and cancer development. Virchow’s prediction that cancer arises at sites of “lymphoreticular infiltrate” has been confirmed many times over, as pancreatitis, hepatitis, colitis, and other chronic inflammatory diseases are now known to be major risk factors for cancer in those tissues (1-6). In fact, chronic inflammation is often necessary for tumor development (1, 7-9). The inflammatory environment promotes cellular proliferation (10-12) and survival (13), degradation and remodeling of the extracellular matrix (13-15), and weakening of vascular barriers to facilitate immune cell migration (16), all of which enable cancer progression (17, 18). In addition, collateral damage from inflammation can result in apoptosis (19), necrosis (20, 21), and mutations (12, 22-24), driving the tissue further away from homeostasis and accelerating transformation. Inflammation is so pro-tumorigenic that tumors can even generate their own inflammatory microenvironment to facilitate growth (7, 25, 26). Accordingly, mitigating inflammation is often an effective strategy for slowing or even preventing neoplasia (3, 27, 28). There are many excellent reviews on a wide range of connections between inflammation and cancer (7, 17,18, 26, 29). Here, we will focus on how inflammation and DNA damage contribute to each other, as well as to the development of cancer.
The many departures from normal behavior that cancer cells exhibit, such as unchecked proliferation and aberrant migration, can be traced to alterations in DNA that accumulate over time. Mutations generally arise from damaged DNA, and inflammation can cause high levels of mutagenic DNA damage. A key feature of inflammation is the production of reactive chemicals designed to destroy pathogens, and while these chemicals are essential for protecting the body from infection, they can damage host biomolecules as well, including DNA. Efficient repair of DNA damage is crucial to prevent mutations in the genetic code. As such, several DNA repair mechanisms have evolved to address the many types of DNA damage. However, DNA repair can be fallible or unable to handle excessive damage, allowing mutations to occur. Since cancer develops from accumulated mutations, it logically follows that unrepaired DNA damage from inflammation contributes to cancer development by increasing mutagenesis.
It is broadly appreciated that genomic instability is a hallmark of cancer, and that inflammation contributes to genomic instability (30-33). The human haploid genome contains 3.2×109 base pairs, and the spontaneous rate of mutation in normal human cells is estimated to be on the order of 1×10−10 nucleotides/cell/division (34, 35). Several driver mutations are required within the same cell lineage for it to become malignant, and although the number of necessary mutations has not been defined and likely depends on the type of cancer, the minimum number is around three (36, 37). There are approximately 3×1013 cells in the human body, and the average human lifespan comprises 1016 cell divisions (BNIDs 100379, 108562). According to Loeb, the probability of any one cell acquiring three mutations is approximated by (1×10−10 spontaneous mutations/cell/division)3(1016 cell divisions/lifetime) = 10−14 potential cancer cells/lifetime (35). In other words, the prevalence of cancer in the population should be orders of magnitude less frequent than the observed probability of developing cancer, which is greater than 30% (38). In order to accumulate multiple mutations within a cell lineage, additional factors such as genetics, environmental exposures, and physiology (including inflammatory conditions) must increase mutation frequency beyond the normal rate, a quality known as a mutator phenotype (35). Thus, while only a few mutations are needed to promote cancer, the increased mutation rate not only leads to the required cancer driving mutations, but also dozens if not hundreds of associated mutations. A continually growing body of evidence indicates that inflammation is one such factor that contributes to accelerated mutagenesis and genomic instability.
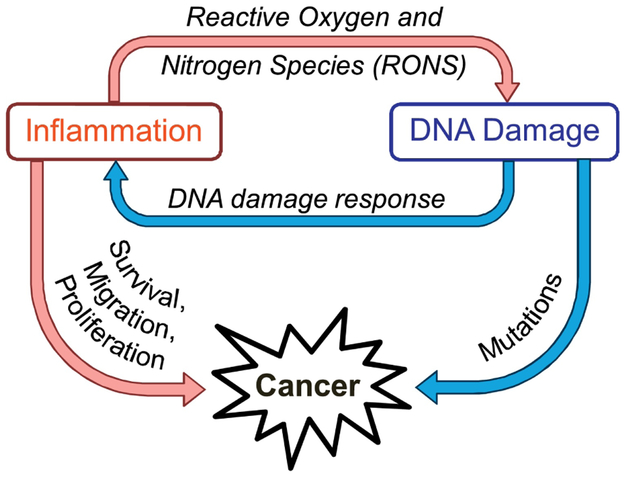
Inflammation and genomic instability have a complex relationship (a simplified model is shown in Figure 1). Inflammation contributes to mutagenesis through production of RONS that can damage DNA, and DNA damage can also exacerbate inflammation. This positive feedback loop is carefully regulated by a network of DNA repair pathways, transcription factors, and cellular signals. Due to the intricate relationships linking inflammation, DNA damage, and DNA repair, these processes can easily become dysregulated, leading to cancer.
Figure 1.
Diagram describing the relationship between inflammation and DNA damage and how they contribute to cancer.
2. Inflammation Leads to DNA Damage and Mutations
Most inflammation-induced DNA damage is caused by RONS, which are evolved by immune cells to destroy pathogens, but which can also damage nearby human cells. Importantly, the damage that RONS inflict upon DNA can be potently mutagenic. One pleiotropic RONS chemical is nitric oxide (NO), which is an essential signaling molecule (39, 40) at concentrations below 400 nM (41). During inflammation, however, innate immune cells produce NO at high levels (approaching μM levels (42, 43)). In addition, neutrophils and macrophages produce superoxide (O•2−) and numerous enzymes contribute to a cascade of chemical reactions that produce a range of RONS (Figure 2), including radicals (e.g., superoxide, hydroxyl radical •OH, and nitrogen dioxide NO•2), anions (e.g., peroxynitrite ONOO−, and nitrosoperoxycarbonate ONOOCO2−), anhydrides (e.g., nitrous anhydride N2O3), hypohalous acids (e.g., hypochlorous acid HOCl and hypobromous acid HOBr) and hydrogen peroxide (H2O2) (44-46). In addition to the RONS produced by immune cells, pro-inflammatory cytokines can stimulate intracellular RONS production (47-49). For excellent reviews on RONS and their chemistry, please see (50, 51).
Figure 2.
Many reactive oxygen and nitrogen species are produced or derived from innate immune cells.
To understand the associations between inflammation and mutagenesis, one must first recognize the chemical modifications to DNA and the mechanisms for repair of those lesions. We begin with a discussion of the DNA damage produced from RONS, including nucleobase oxidation, deamination, halogenation, and alkylation, as well as strand breaks of the phosphodiester backbone. Subsequent sections will review the major repair pathways for these types of damage, the mechanisms that balance DNA damage, DNA repair, and inflammation, and the implications for carcinogenesis.
2.1. Oxidation
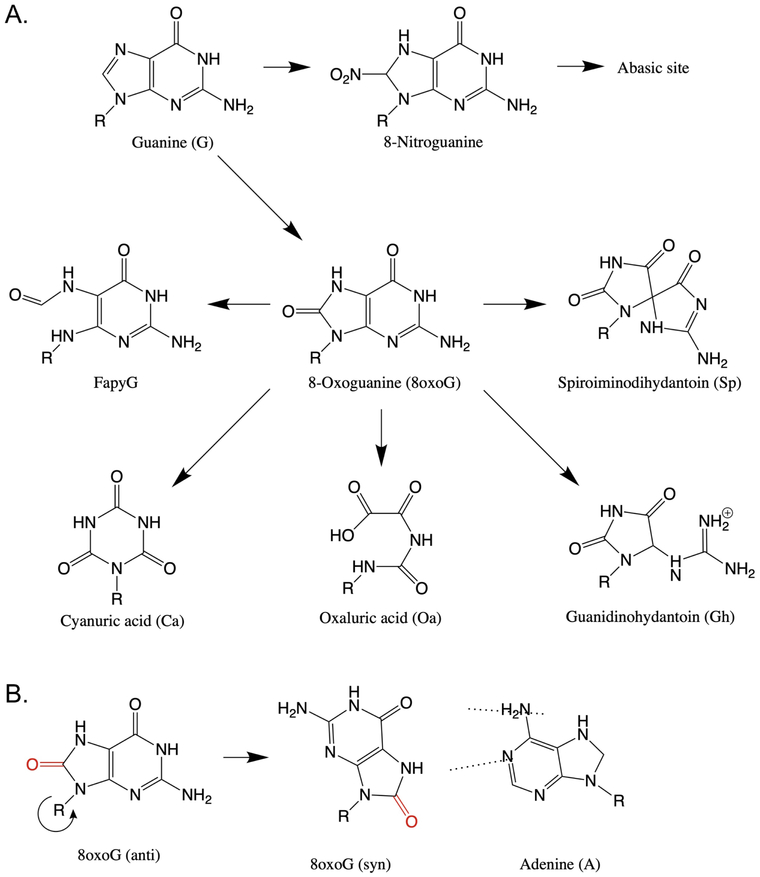
Many RONS are potent oxidizing agents and can produce a variety of DNA lesions. While there are many potential products of DNA oxidation (52), guanine is the most easily oxidized DNA base (51, 53) and is therefore the primary target for reaction with nucleophilic RONS and the focus of this section. Primary oxidation of guanine by RONS produces 8-oxo-guanine (8oxoG, which is mutagenic) and 8-nitro-guanine (which is unstable and quickly becomes an abasic site) (Figure 3A). 8oxoG in the normal anti conformation pairs with cytosine, but rotation of the glycosidic bond to the syn position allows human polymerases to pair it with A (Figure 3B) thus, 8oxoG can lead to G→T transversions (54-57). The potential for 8oxoG to cause this mutation depends on the polymerase that encounters it, as will be described in later sections (56-58). 8oxoG is ~1000 times more prone to oxidation than its parent guanine, leading to production of several more stable and mutagenic secondary products (Figure 3A) (59). These include spiroiminodihydantoin (Sp), guanidinohydantoin (Gh), oxazolone (Oz), oxaluric acid (Oa), and cyanuric acid (Ca) (see Figure 3 for examples). Oz, Oa and Ca have been shown to produce G→T transversions with much higher potency than the parent 8oxoG in E. coli (60). Gh primarily leads to G→C mutations, and Sp causes both →T and G→C mutations, both with frequencies at least an order of magnitude higher than that of 8oxoG in E. coli (61). While Gh is relatively easily bypassed (albeit mutagenically), Sp blocks replication (61) and leads to strand slippage, producing broken replication forks and single base pair deletions in addition to transversions in vitro (61-65).
Figure 3.
Products of guanine oxidation. A. Primary nitrosation of guanine leads to an abasic site. Primary oxidation of guanine produces 8oxoG, and oxidation of 8oxoG leads to a variety of secondary oxidation products. B. Rotation of the glycosidic bond allows 8oxoG to mispair with A.
In addition to secondary oxidation, 8oxoG can be reduced, opening the imidazole ring to form a 2,6-diamino-4-hydroxy-5-formamidopyrimidine (FapyG). FapyG produces G→T transversions in mammalian cells (66), and some studies suggest it may be more mutagenic than the parent 8oxoG (67).
2.2. Deamination
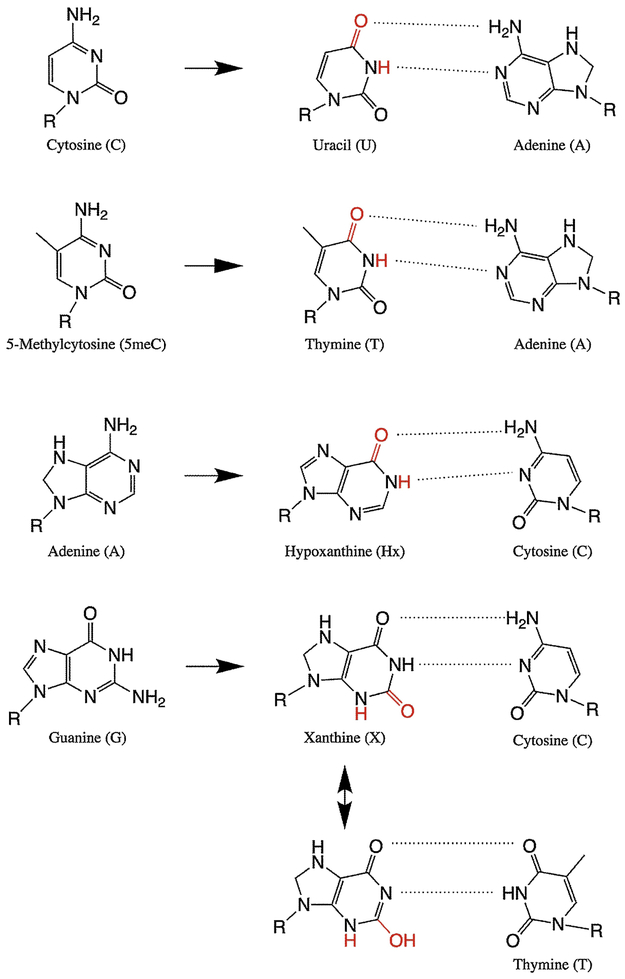
In addition to oxidation, nitrosative RONS can deaminate DNA bases. Deamination products are particularly mutagenic because the chemistry occurs on the functional groups that determine hydrogen bonding, altering the pattern of H-bond donors and acceptors and leading to base mispairing. The chemical primarily responsible for base deamination is thought to be nitrous anhydride (50), generating the products from canonical bases shown in Figure 4 (note that hypoxanthine (Hx) is called deoxyinosine (dI) in DNA). Many in vitro studies have shown that NO chemistry causes high levels of U, Hx and X in DNA (68, 69), and Hx and X are also elevated in inflamed tissues (70).
Figure 4.
Products of DNA deamination and subsequent base mispairing.
Deamination of cytosine or its methylated form 5meC changes the base into uracil or thymine, respectively, causing C→T transitions. The mutational signature corresponding to spontaneous deamination of 5meC has been found in all cancers, and the abundance of these mutations correlates with age of cancer diagnosis, supporting the hypothesis that cancer-associated inflammation causes deamination and accumulation of mutations (71). Recently it has been shown in human cells that deamination of adenine to produce Hx can cause either A→G transitions or deletions, depending on the cell type and whether the lesion is on the leading or lagging strand (72). Finally, guanine deamination to X is generally not mutagenic, as X preferentially pairs with C, but polymerases sometimes incorrectly pair X with T to cause G→A transitions in vitro (73, 74), possibly by tautomerization of X to an enol form (see Figure 4). Nitrosative deamination of G or A at the N7 position can also cause depurination to form an abasic site (75).
2.3. Halogenation
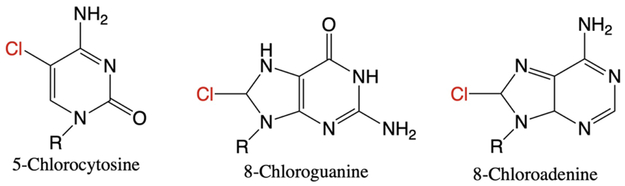
In addition to reactive oxygen and nitrogen species, inflammatory cells can also generate hypohalous acids. Neutrophils secrete the enzyme myeloperoxidase to produce hypochlorous acid (HOCl) (76-78), and eosinophils secrete eosinophil peroxidase to produce hypobromous acid (HOBr) (79, 80) (Figure 2). These hypohalous acids readily react with DNA during inflammation to form the adducts shown in Figure 5 (81). Interestingly, the most abundant halogenated nucleobase, 5-chlorocytosine (5ClC) (81, 82), accumulates to a greater degree than oxidative, deamination or lipid peroxidation (LPO, described below) DNA lesions in mouse models (70, 83). Due to its significant and persistent accumulation, perhaps a result of inefficient repair, 5ClC has been designated a biomarker for chronic inflammation (70, 83, 84).
Figure 5.
Products of DNA halogenation
The mutagenicity of halogenated DNA has only recently been demonstrated. Halogenation had previously been shown to impact epigenetic modifications (85), but now 5ClC has been shown to cause C→T transitions when encountered by any class of DNA polymerase (84). These mutations are frequently observed in tissues under inflammatory stress as well as in inflammation-associated cancers (83, 84, 86), further supporting the designation of 5ClC as a pertinent biomarker for inflammation.
2.4. Lipid Peroxidation-Derived Adducts
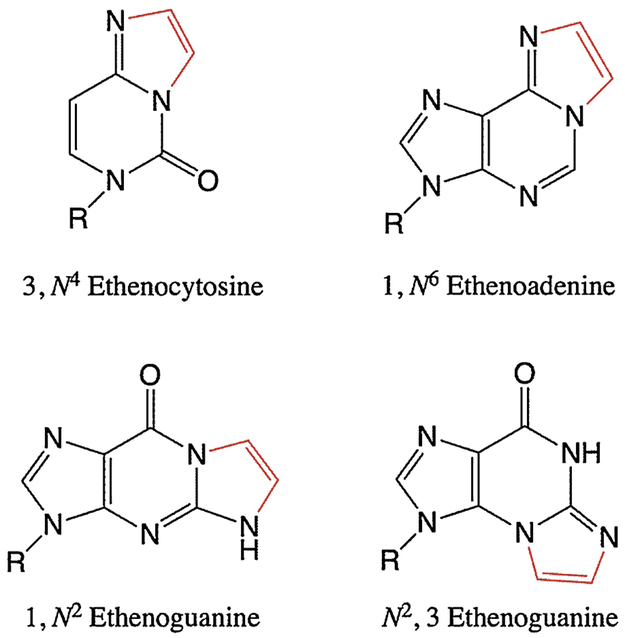
Oxidative chemicals can inflict damage on DNA bases themselves, as described above, but they can also cause indirect damage by creating reactive species from other biomolecules. Specifically, when RONS encounter polyunsaturated fatty acids, they cause lipid peroxidation to generate electrophilic, DNA-reactive aldehyde species. The best-studied of these aldehyde species are 4-hydroxy-2-nonenal (HNE), malondialdehyde (MDA), acrolein, and crotonaldehyde (87), of which HNE and MDA have been directly implicated in carcinogenesis (88, 89). The resulting LPO-induced DNA adducts are exocyclic additions of two (ε, etheno-) or three (P, propano-) carbons onto a base (87, 89, 90). Examples of etheno adducts are shown in Figure 6. Many studies have demonstrated significant accumulation of etheno adducts as a result of inflammation in mammals, and this is often associated with increased incidence of cancers (23, 70, 89, 91, 92).
Figure 6.
Products of DNA alkylation following electrophilic attack by lipid peroxidation products
The most abundant etheno adduct found in DNA is N2,3-εG (92-94). N2,3-εG is a potent inducer of G→A transitions, and its isomer 1,N2-εG causes G→T and G→C transversions with E. coli and mammalian polymerases (93, 95-97). In mammalian cells, 1,N6-εA causes primarily A→G and A→T mutations (98, 99), and 3,N4-εC potently induces C→A and C→T mutations (90, 100). All etheno lesions can block replication to some extent (93, 96, 100), which can lead to larger-scale mutations (discussed below).
2.5. Single Strand Breaks (SSBs)
While lesions on nucleobases are an important source of mutations, breakage of the DNA backbone is a far greater threat to genomic integrity. Strand breaks potentiate large scale sequence rearrangement mutations, such as deletions, insertions, and translocations, and they can also cause stress signaling, cell cycle arrest, and cytotoxicity if not efficiently repaired (101-105). During inflammation, SSBs can arise from direct reaction with RONS: for example, radicals can hydrolyze the phosphodiester backbone, and peroxynitrite produces single strand breaks and abasic sites (106). SSBs also occur naturally as intermediates of some DNA repair pathways, including pathways involved with repair of inflammation-associated damage (described below).
2.6. Double Strand Breaks (DSBs)
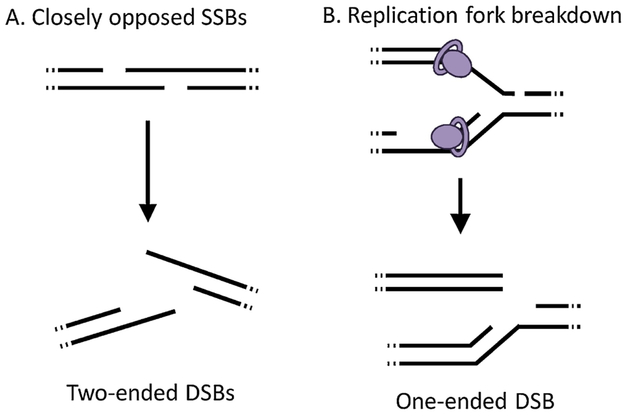
Double strand breaks (DSBs) also arise from a variety of sources (see (107) for a review). Some enzymes, such as endonucleases and topoisomerases, cut the backbone to produce DSBs (108, 109). SSBs can also potentiate DSBs (110, 111): for example, two opposed SSBs less than 7 base pairs apart reduce the structural integrity of the DNA duplex, causing breakage into two DSB ends (112) (Figure 7A). Additionally, if a polymerase encounters a SSB during replication, the replication machinery cannot synthesize past the gap, and the replication fork collapses, leaving a one-ended DSB (Figure 7B) (113). Similarly, replication-blocking lesions (such as εA and εC) can cause DSBs (114-116), possibly because the stress of a replication fork encountering a lesion that cannot be bypassed causes breakage of the backbone, though the exact mechanism(s) for this breakage remains unclear (117, 118). Both DSBs and SSBs have been observed in cells co-cultured with activated macrophages, confirming that phagocyte-associated RONS can cause these lesions (119). While both DSBs and SSBs can be mutagenic, DSBs are one of the most deleterious types of DNA damage (102, 120). Strand breaks potentiate large-scale mutations, including insertions, deletions, translocations, and sequence rearrangements (102, 116, 121-125).
Figure 7.
Modes by which single strand breaks may lead to double strand breaks.
3. Tolerance and Repair of Inflammation-Induced DNA Lesions
In order to minimize mutations, multiple mechanisms have evolved to address different types of DNA damage. The simplest way to prevent mutations is through accurate replication of DNA. Polymerases read the template strand and select the correct nucleotide, based on the template base’s shape and hydrogen bonding pattern, to extend the strand being synthesized. When the hydrogen bonding of a base is altered (e.g., by deamination) or blocked (e.g., by etheno adducts), the cell first attempts to synthesize past the lesion (translesion synthesis, or TLS), often using a low-fidelity polymerase, which is only sometimes accurate (67, 116, 126). Since persistent signals of DNA damage can lead to cytotoxicity, mutagenic TLS can be favorable to enable cell survival. However, some lesions cannot be efficiently bypassed by TLS, causing the replication fork to stall (127). Several mechanisms are capable of protecting (128) and restoring stalled replication forks (please see (129-131) for reviews), some of which leave the lesion in place (117), but inefficient fork restoration can cause strand breakage as described above. For reviews of TLS in eukaryotes, please see (132) and (133).
In addition to polymerase selectivity and proofreading, several DNA repair pathways have evolved to correct lesions before and during replication. The rate of incorrect base incorporation (point mutation) for replicative polymerases is on the order of 10−6 to 10−8 nucleotides (134, 135), and with 3.2×109 bases in the genome, at least ten point mutations would occur with every cell division. DNA repair processes reduce point mutation frequency to 10−10 (34, 35) and also protect against large-scale mutations such as insertions, deletions and rearrangements (116, 123-125). DNA repair is of particular importance during inflammation because of (i) the large amount of reactive chemicals produced (91) and (ii) the stimulation of cellular proliferation to regenerate damaged tissue (12).
Most DNA damage from inflammation is addressed by the Direct Reversal (DR), Base Excision Repair (BER), and Homologous Recombination (HR) pathways. Here we will briefly describe each pathway and discuss their interactions for robust repair of inflammation-derived damage.
3.1. Direct Reversal (DR)
Some DNA alkyl adducts can be directly removed from the base in a process called Direct Reversal, leaving the original base intact. For example, the O6-methylguanine DNA methyltransferase (MGMT) protein repairs O6-methylguanine by transferring the methyl group to a cysteine residue in its active site (136, 137). The other mammalian DR enzymes belong to the ALKBH family, which utilize oxygen, Fe2+ and α-ketoglutarate as co-factors for oxidative dealkylation, releasing the alkyl lesion as an aldehyde and restoring the original base (138, 139). Importantly, ALKBH enzymes can repair etheno adducts, and therefore supplement BER during inflammation (91, 140, 141).
3.2. Base Excision Repair (BER)
In general, Base Excision Repair deals with single-base lesions that do not significantly distort the DNA helix; since this encompasses most inflammation-derived lesions, BER is the primary pathway responsible for DNA repair during inflammation. Indeed, BER activity has been shown to be key in removing DNA lesions and protecting against mutations in animal models of inflammation (23, 91).
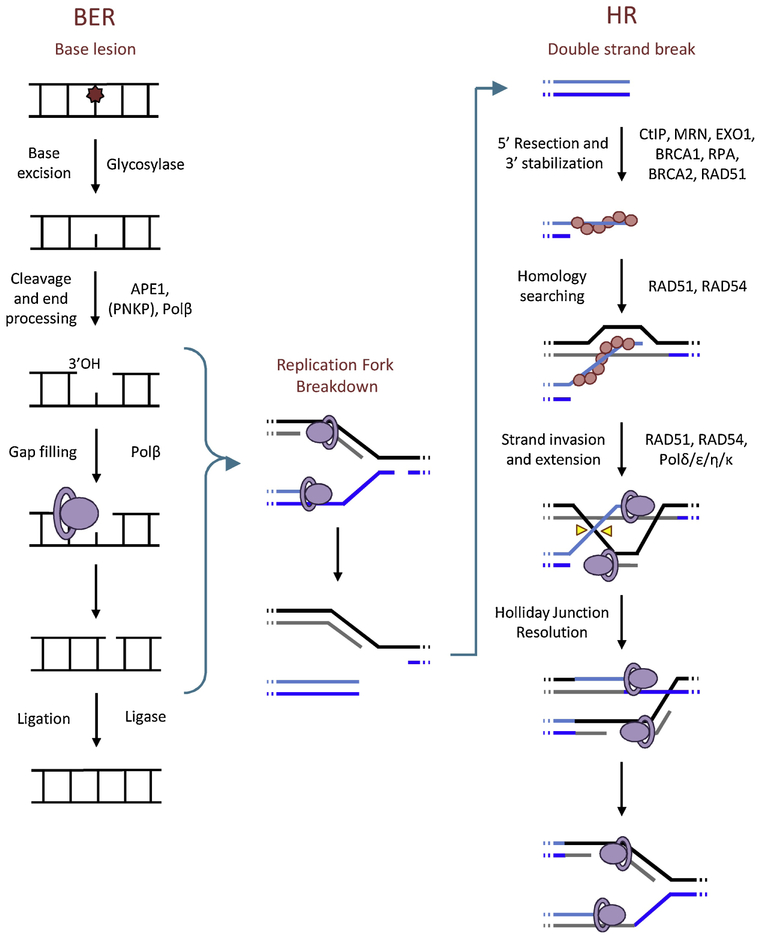
An overview of the BER pathway is shown on the left side of Figure 8 and reviewed in (142) and (143). BER enzymes scan DNA for base lesions through processive searching (144). Upon recognition of a substrate, the first step of BER is removal of the damaged base by one of several DNA glycosylases. Monofunctional glycosylases remove only the DNA base, leaving an abasic site (as shown in Figure 8), whereas bifunctional glycosylases also nick the DNA backbone. In the case of monofunctional glycosylases, the second step is to nick the backbone by AP endonuclease-1 (APE1). The two ends of the single strand break must then be processed to produce a 3’OH (with APE1, Polβ, or PNKP) capable of extension and a 5’PO4 (with Polβ or PNKP) capable of ligation. The gap is filled in with a polymerase (Polβ for single nucleotides, and Polδ or Polε for larger gaps (145)), and ligase seals the nick to complete the process. The elements of BER downstream of the glycosylase can also contribute to repair of SSBs.
Figure 8.
Base excision repair pathway, homologous recombination pathway, and how BER intermediates may lead to HR
Each glycosylase has its own repertoire of substrates, and there is some redundancy in substrate recognition to ensure a robust response. Some of the most important DNA glycosylases for repairing damage from inflammation are 8-oxo-guanine glycosylase (OGG1), which removes 8oxoG and FapyG (146), and alkyladenine glycosylase (AAG, also known as MPG or ANPG), which removes εA, 1,N2 -εG, Hx, and 8oxoG, among others (91, 147, 148). AAG also recognizes and binds to εC, but it cannot excise the base, instead blocking replication and contributing to its increased genotoxicity (100). Damaged pyrimidines may be excised by pyrimidine-specific glycosylases such as MBD4, UNG, TDG, and SMUG1 (147). The enzyme MYH also uniquely protects against 8oxoG mutagenesis, because it specifically recognizes and excises A misincorporated across from 8oxoG during replication (149-151). Other BER glycosylases, such as NTHL1, NEIL1, and NEIL2, can recognize a broad range of substrates, including FapyG, hydantoin lesions, 8oxoG, and oxidized pyrimidines (63, 147, 152-154), providing robust repair of single-base lesions.
Importantly, every intermediate of the BER pathway contains a potentially toxic lesion: either an abasic site or some kind of strand break. Normally, the cell is able to complete BER without toxicity, but this is not always the case. If there is a large amount of damage, the cell may be unable to efficiently or adequately repair the DNA, leading to an accumulation of cytotoxic lesions and subsequent apoptosis (111, 155). Alternatively, if a replication fork encounters a BER intermediate, the fork may break down, creating a DSB (110, 156). Thus, while BER is essential for repairing DNA damage during inflammation, accelerated BER can sometimes have negative effects on mutagenesis and survival due to the production of toxic intermediates (157-159).
3.3. Double Strand Break Repair
Although DSBs generally arise as secondary lesions in inflammation (e.g., from replication fork breakdown or nearby opposing SSBs) these lesions are one of the most toxic types of DNA damage (102, 120-122, 160). Here, we will briefly describe the two major pathways responsible for DSB repair: Non-Homologous End Joining (NHEJ) and Homologous Recombination (HR). A review on the significance of DSBs and DSB repair in mutagenesis and carcinogenesis can be found here (120).
NHEJ, the dominant repair mechanism in G1 phase (161), functions by joining two DSB ends with the Ku70/80 and DNA-PK complexes, and ligating the strands together (162). This process occurs quickly but is very prone to error. For example, if the two DSB ends are from different chromosomes, translocations occur. However, since DSBs are generally rare in G1, NHEJ sufficiently preserves genomic integrity during this cell cycle phase.
Homologous Recombination repairs DSBs much more accurately than NHEJ, but it acts more slowly and functions mainly during the S and G2 phases of the cell cycle (163, 164). Since the risk for mutations is highest during replication, and since HR can rescue replication stress, this pathway is of greater importance for inflammation-induced DNA damage. Induction of HR during inflammation has been shown in mice (12).
HR comprises multiple subpathways with distinct mechanisms, so here we will provide a basic overview of the major steps (Figure 8, right side). HR begins by ATM-assisted recognition of the DSB by CtIP and the MRN complex (consisting of MRE11, NBS1 and RAD50). The 5’ end of the DNA is resected by EXO1 to leave a 3’ overhang of single stranded DNA, which is stabilized by the protein RPA. BRCA2 displaces the RPA, replacing it with RAD51, and this 3’ nucleoprotein filament searches nearby DNA for a homologous sequence. Usually, the cell identifies the correct homologous sequence on the nearby sister chromatid, which it invades and utilizes as a template to resume extension. There are several sub-pathways of HR, but the most common is synthesis-dependent strand annealing (SDSA) (for more detailed descriptions and digital animations of HR mechanisms, please see (165)). In SDSA, once enough DNA has been synthesized from the template, the overhang will re-hybridize with its original strand, any remaining gaps will be filled, and DNA ligation completes repair of the DSB.
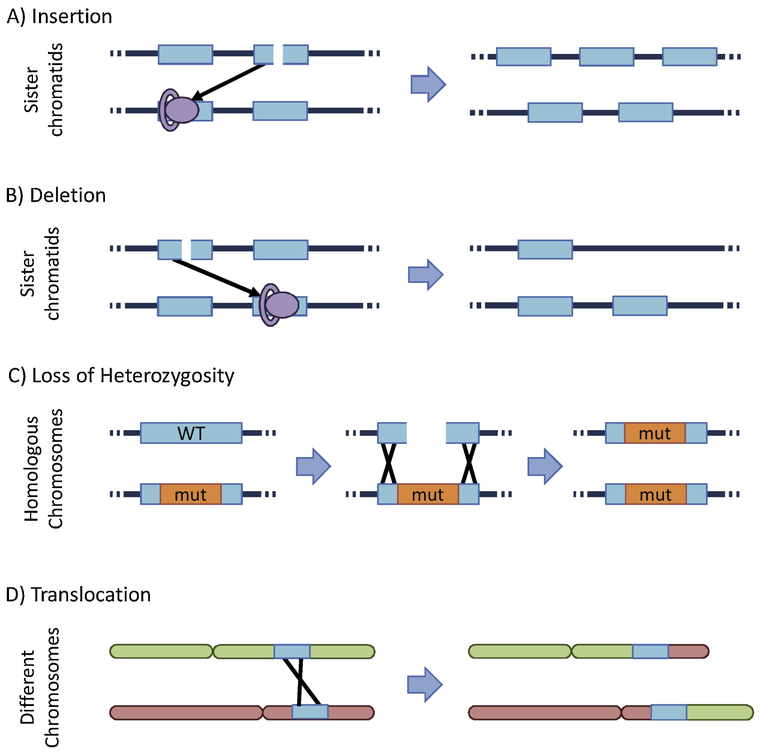
Since HR utilizes a homologous region of DNA as a template, this process is mostly error-free as long as the cell identifies the correct sequence in the sister chromatid. However, identification of homology in the homologous chromosome rather than the sister chromatid can lead to loss of heterozygosity, a significant source of tumor suppressor inactivation (125, 166, 167). Furthermore, a significant portion of the genome has been identified as repetitive or repeat-derived: nearly 10% of the genome consists of Alu repeats (168, 169), and a recent analysis estimates 2/3 of the genome consists of repetitive elements (170). Thus, there are many opportunities for HR to identify a homologous sequence in the wrong location. Aberrant HR can lead to sequence rearrangement mutations, such as translocations, deletions, insertions (171-176). Some of the mechanisms for HR-derived mutations are illustrated in Figure 9. In addition to these large-scale mutations, HR can also produce point mutations, because the polymerases that participate in HR are often error-prone (127, 177-179).
Figure 9.
Several mechanisms for HR-derived mutations.
As mentioned earlier, an important source of DSBs during inflammation is broken replication forks, which generate only one DSB end (see Figure 7), and thus cannot be accurately repaired by NHEJ. Therefore, lesions that cause replication fork breakdown (including replication blocking lesions and SSBs) necessitate HR to restore the replication fork (114, 115, 180). Notably, many of the intermediates of the BER pathway are SSBs, so if replication forks encounter these breaks before BER is completed, HR may be initiated as well (Figure 8) (111, 181).
For more information on HR mechanisms and the role of HR in cancer, please see (182, 183), and for a review of HR-related signaling, repair, and cell fate, refer to (184).
4. Coordination of DNA Repair During Inflammation
During inflammation, DNA repair must be coordinated and balanced to ensure that mutagenic lesions are corrected efficiently and accurately. In this section, we will illustrate the relationships and balance between different DNA repair pathways and discuss mechanisms by which inflammatory processes help coordinate DNA repair processes.
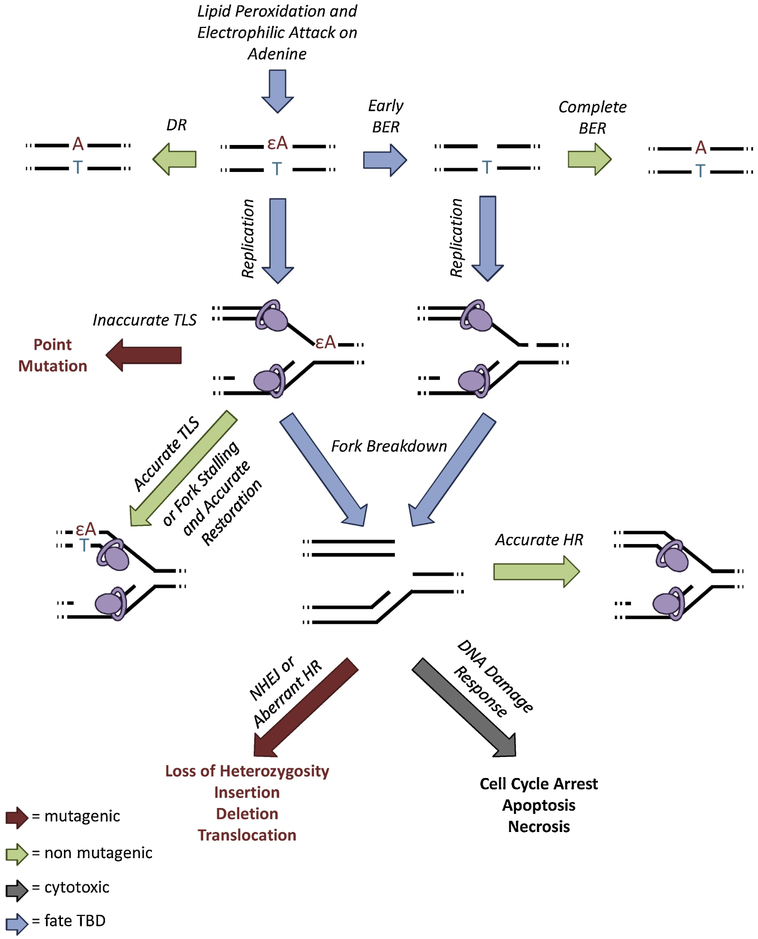
4.1. Fate of the Lesion
To illustrate the relationships among the DNA repair mechanisms described here, we consider the mammalian response to εA (Figure 10), an LPO lesion that accumulates during inflammation. Efficient repair of εA can be accomplished with either DR, which removes the etheno lesion and leaves the undamaged base intact, or BER, which excises the entire base and re-synthesizes the DNA properly (91, 141, 148, 185). However, if εA is encountered during replication, mutagenic consequences can arise. Translesion synthesis by an error-prone polymerase allows the putative development of a point mutation (116); if the εA lesion is repaired following inaccurate TLS, the mutation will become fixed. εA may also block some polymerases (100, 116), causing the replication fork to stall. Stalled replication forks can be restarted by several mechanisms (117), some of which leave the lesion intact (131), but they may also break down to produce a one-ended DSB. Accurate restoration of the replication fork by HR prevents mutagenesis, but aberrant HR or NHEJ causes large-scale mutations, such as translocations, insertions and deletions (some examples are shown in Figure 9) (121, 173, 174, 180). DSBs are also highly cytotoxic, so persistence or abundance of DSBs in the cell can signal for it to undergo apoptosis (101, 120, 186, 187). Thus, a single εA lesion creates the opportunity for many types of mutations through multiple repair pathways.
Figure 10.
Pathways leading from a single εA lesion to multiple types of mutations.
4.2. Intra-Pathway Balance
The importance of balance within a repair pathway has been illustrated with studies of modulated expression of BER proteins. Mice that contain a transgene for increased expression of Aag are more sensitive to alkylation damage, whereas Aag−/− mice are more resistant (157). This varied sensitivity stems from the fact that BER intermediates (abasic sites and SSBs) are often more toxic than the original lesion itself (188). If the downstream repair proteins do not keep up with glycosylase activity, intermediates accumulate and increase mutagenesis (189, 190) or signal cell death (111). Accordingly, elevated AAG expression in humans has been associated with lung cancer risk (191, 192) and poor glioma prognosis (193).
4.3. Inter-Pathway Redundancy and Crosstalk
To account for potential imbalances or perturbations within repair pathways, there must also be inter-pathway crosstalk, regulation and redundancy. The coordination of multiple DNA repair pathways allows the network to address lesions quickly and compensate with other repair mechanisms if the pathway is not resolved efficiently.
There is often redundancy within and between pathways so that multiple mechanisms can correct the same lesion with equivalent efficacy. For example, supplementary to BER, direct reversal enzymes ALKBH2 and ALKBH3 can repair etheno lesions (91, 194), and nucleotide excision repair (NER) can contribute to repair of the hydantoin lesions Sp and Gh (195, 196). Redundancy between pathways is so important that mice lacking AAG, ALKBH2 and ALKBH3 are unable to survive even one episode of colonic inflammation (91).
In other cases, potentially redundant repair pathways are coordinated by cell cycle and crosstalk between repair machinery. For example, NHEJ is the dominant mechanism to repair DSBs during G0 and G1 phases of the cell cycle, when non-specific ligation is sufficiently accurate and there is no sister chromatid present for homology-directed repair (161). During replication, however, there are likely to be more DSBs and therefore a much greater risk that non-specific ligation will produce a mutation, and so the slower but more accurate mechanism HR is preferred (163, 164). Thus, NHEJ and HR activities are regulated in part by cell cycle, and many of the proteins that recognize and bind DSBs contribute to the choice of which pathway will complete repair (197-201).
Finally, repair processes may compensate for each other if the pathway that initiated repair is not resolved efficiently. A particularly relevant example is that BER intermediates can cause replication forks to break down, inducing HR for repair (Figure 8) (111, 181, 202). Kiraly et al. showed that wild type levels of the BER protein AAG caused a greater accumulation of HR-driven mutations compared to Aag−/− mice (181), indicating that even normal BER glycosylase activity may result in aberrant DSB repair. Further, HR-driven mutations increased synergistically when alkylation damage occurred during proliferation (12, 181). When Aag activity exceeds the capacity of downstream enzymes, as described above, accumulated BER intermediates can cause stress signaling and apoptosis (157), or they can elicit responses from other pathways, providing the opportunity for different types of mutations than the initial lesion might have created (189, 190, 203). Of course, there are many ways in which intermediates of some pathways may be substrates for other pathways, but for the scope of this review we confine this example to BER and HR (203).
4.4. ROS promote DNA repair through ATM activation
A key mechanism for stimulating DNA repair during inflammation is through oxidation of ATM. When the inactive ATM dimer is activated at DNA DSBs, ATM is monomerized (204) and promotes DNA repair via HR (205), p53 (206), and checkpoint activation (207-209). However, oxidation of the ATM dimer (e.g., by ROS) allows it to promote DNA repair and/or apoptosis via p53 and CHK2 pathways, independent of the presence DNA DSBs (210, 211). Oxidized ATM can also be exported from the nucleus to activate NFκB signaling (212), which is a major transcriptional regulator of inflammation, and which enhances some DNA repair pathways (described below). The dual roles of ATM in DNA damage and redox sensing are highlighted by the fact that different amino acid residues are involved in activation by DSB detection vs oxidation (210). Thus, stimulation of cells with oxidative chemicals such as ROS during inflammation elicits a modified DNA damage response by activating ATM, regardless of DNA DSB induction (213).
4.5. Transcriptional regulation of DNA repair during inflammation
To cope with the barrage of DNA damage, inflammatory signaling includes upregulation of DNA repair. Indeed, increasing severity of inflammation and precancerous lesions correlates positively with DNA damage response indicators (203, 214, 215). Elements of BER and HR are promoted in inflammatory environments (214-219), and they have been shown to protect against inflammation-driven mutations (23, 220-222). Here we will describe a few major examples of transcription factors that moderate inflammation and their roles in promoting DNA repair.
One of the most important responses to oxidative stress is mediated by NRF2, a transcription factor that regulates the expression of antioxidants and other cytoprotective elements. Briefly, NRF2 is sequestered by KEAP1 in the cytoplasm until stimulation by oxidative stress (223, 224), whereupon NRF2 translocates to the nucleus (225) to bind antioxidant response elements (AREs) in target gene promoters (226-228). Target genes of NRF2 include glutathione S-transferase (GST) (229, 230), NADPH quinone oxidoreductase 1 (NQO1) (231, 232) and superoxide dismutase (SOD) (233, 234), known collectively as the adaptive response to oxidative and electrophilic damage (227, 235). Neutralizing RONS to less reactive species protects DNA from much of the deleterious reactions described earlier. Interestingly, increases in the pro-inflammatory cytokine IL6 has been shown to reduce DNA damage after radiation, in part through upregulation of the NRF2 pathway to reduce oxidative stress (236), and possibly through promoting DNA DSB repair (237, 238).
In addition to antioxidant and metabolic enzymes, AREs have also recently been identified in promoters of several DNA repair genes, including components of HR (216), NHEJ (239), and BER (240). Indeed, disruption of NRF2 activity has been shown to result in decreased DNA repair activity (216) and increased levels of DNA adducts (241-243). Thus, when present, NRF2 helps to manage the self-damage from inflammation by increasing expression of genes that neutralize oxidative stressors and promote DNA repair.
Several other transcriptional regulators of inflammation may also play roles in promoting DNA repair. NFκB is a key transcription factor with roles in induction, propagation, and eventual downregulation of inflammation, and DNA damage is one of many signals that can activate it (244-246). There is also evidence that NFκB enhances HR by stabilizing the CtIP-BRCA1 complex (218). Similarly, several members of the interferon regulatory factor (IRF) transcription factor family promote DNA repair (247, 248). For example, recognition of DSBs can induce IRF1 signaling (247), and some of its target genes include elements of HR and BER (217).
Overall, because inflammation produces a great deal of DNA damage, and the pathogenic insults that cause inflammation also can cause DNA damage, transcriptional regulation of inflammation includes increased DNA repair. Indeed, the only way an organism could evolve to have a self-damaging physiological response like inflammation is to also be capable of efficiently repairing the damage it causes.
5. DNA Damage Promotes Inflammation in a Positive Feedback Loop
Many pathogens can cause DNA damage (219, 249-253), which may be why DNA damage can promote inflammation. Regardless of the reason for this evolved mechanism, the proteins that detect and respond to DNA damage can trigger cell cycle arrest, apoptosis, senescence, and necrosis, of which the latter two can promote inflammatory signaling (254-261). Further, proteins involved in DNA repair can serve to promote NFκB transcription of pro-inflammatory genes (246, 262-264). Unfortunately, DNA damage and inflammation can therefore create a positive feedback loop, which can be difficult to regulate. Additionally, response to DNA damage in one cell can induce damage in nearby cells through extracellular signals and epigenetic modifications (265-268). The propagation of genomic instability to such “bystander” cells, along with systemic inflammatory signals, may contribute to genotoxicity in off-target tissues during inflammation (269-272).
5.1. DNA Damage Signals for Inflammation
There are many elements of the DNA damage response that promote inflammation. For example, PARP1 (poly(ADP-ribose) polymerase 1) detects and binds SSBs to recruit BER machinery (273-277), and it has been closely linked to inflammation promotion. PARP1 builds branched polymers of ADP-ribose moieties out of NAD+ (called PARylation) at strand breaks to help recruit BER proteins and initiate repair. PARP1 is also capable of post-translational PARylation of proteins to modify their activity, notably including the key inflammatory regulator NFκB (244-246, 278). Indeed, several components of the NFκB complex display increased activity following PARylation (244, 246).
Inhibition of PARP1 has repeatedly been demonstrated to decrease the severity of inflammation in the intestines (279), pancreas (280), heart (281), brain (282), liver (283), and many other tissues and model systems (284-288). In particular, studies have shown that inhibition of PARP1 results in decreased inflammatory cytokine expression (244, 280, 289, 290) decreased adhesion molecule production (280, 285, 289), decreased inflammation-associated enzyme activity, including iNOS, COX2 and NADPH oxidase (285, 291-294), and decreased immune cell infiltration (280, 290, 295). These molecular and cellular alterations all support a model wherein PARP1 activity contributes to increased inflammatory signaling, and its inhibition protects the organism from inflammation-associated damage.
PARP inhibitors have shown significant clinical success as adjuvants in cancer therapy. PARP inhibition is particularly effective in HR-deficient cancers due to synthetic lethality (296), and many other DNA damage-related chemotherapeutics show increased efficacy in combination with PARP inhibitors (297-299). PARP inhibitors can also enhance anticancer activity of PD-1/PD-L1 or CTLA-4 inhibition (300, 301), demonstrating that in addition to DNA damage and repair impacts, the immunogenic effects of PARP can also be exploited for cancer therapy (302, 303). For reviews on PARP inhibitor cancer therapies, please refer to (304, 305).
The BER glycosylase OGG1 can also augment inflammation through NFκB. OGG1 bound to 8oxoG facilitates NFκB binding (306, 307) and increases expression of its pro-inflammatory target genes (262). Similar to inhibition of PARP1, downregulation, knockout, or inhibition of OGG1 can also reduce expression of pro-inflammatory genes and inflammation severity (263, 308, 309) through prevention of NFκB transcription (263, 264, 306). Interestingly, the free OGG1-8oxoG complex also acts as a guanine exchange factor and activates Ras family GTPases, further promoting inflammation through the MEK/ERK pathway (310). Together, the profound effects of BER proteins OGG1 and PARP1 in the transcription of pro-inflammatory genes demonstrate the importance of BER in regulation of inflammatory responses.
Proteins involved in double strand break repair have also been implicated in pro-inflammatory signaling. DSBs can be recognized and bound by ATM, and RPA-coated single stranded DNA (an early intermediate of HR) is recognized by ATR. Multiple studies have shown that ATM (218, 255, 256, 311) and ATR (311) can both promote NFκB signaling independent of downstream DNA damage responses. Accordingly, ATM/ATR activity results in increased cytokine production (255-257, 312), and knockdown of ATM or ATR inhibits the production of the immune cell-activating ligand NKG2D (311).
Inflammation can also be promoted by generalized DNA damage, independent of BER or HR initiation. For instance, the growth arrest and DNA damage-inducible protein 34 (GADD34) is upregulated in response to multiple types of cellular stress, including DNA damage and ER-stress (259). In two models of DNA damage-induced cancer, mice knocked out for GADD34 display significantly decreased levels of pro-inflammatory cytokines, immune cell infiltration and malignant lesions (313) (314). Genomic instability often involves the generation of micronuclei, composed of tiny fragments of DNA encased by nuclear envelopes separate from the nucleus, which can promote inflammation as well. Breakdown of micronuclear envelopes release DNA into the cytosol (315), which can trigger activation of the pro-inflammatory cytokines IL-1β (316) and IFNγ (317-319). Cytosolic DNA also activates the cGAS-STING pathway, activating of IRF3 and inducing IFNβ expression (320, 321).
5.2. DNA Damage Promotes Inflammation by Cell Death and Senescence
In addition to the direct upregulation of inflammation by the DNA damage response, DNA damage also indirectly promotes inflammation through cytotoxicity (322). Too much DNA damage, especially during a proliferative phase, cannot be processed sufficiently by DNA repair pathways, so the cell is eventually forced to undergo apoptosis, necroptosis, necrosis, or senescence.
DNA damage-induced apoptosis is generally not considered pro-inflammatory, and indeed most forms of apoptotic cell death are not inflammatory. However, there are exceptions. For example, the Fas ligand acts as a DNA damage sensor, activating the Fas-mediated apoptosis pathway (323), and studies have shown that FasL-mediated apoptosis promotes inflammation (324, 325).
Necroptosis is another form of programmed cell death that closely resembles necrosis and is far more pro-inflammatory than apoptosis. One type of necroptosis, known as parthanatos, is triggered by excessive PARP1 activation at DNA single strand breaks. It has widely been hypothesized that persistent or extensive PARP1 activation can cause cellular NAD+ and ATP depletion and bioenergetic collapse, leading to necrosis (20, 326, 327). However, more recent studies show that PARP1 is a mediator of parthanatos, in which excessive PARP1 activation leads to PAR accumulation in the cytosol, translocation of AIF from mitochondria to the nucleus (328) and activation of RIP1/RIP3-mediated necroptosis (329-332). Necroptosis resembles necrosis in that both involve swelling and plasma membrane rupture, releasing intracellular contents and promoting inflammation. Molecules that would not normally be found outside the cell are known as damage-associated molecular patterns (DAMPs). Extracellular DAMPs, such as IL1-family cytokines, uric acid, ATP, or HMGB1, trigger activation of immune cells, including macrophages, mast cells, neutrophils, and B-cells (261). Thus, PARP-mediated necroptosis is a significant source of pro-inflammatory signals.
In addition to cell death, DNA damage can lead to senescence (reviewed here (333) and here (334)), wherein the cell permanently arrests growth and secretes inflammatory cytokines (255). DNA double strand breaks activate p53, persistent activation of which can lead to either apoptosis or senescence in vivo (335). Senescent cells develop a senescence-associated secretory phenotype (SASP), which includes release of many inflammatory cytokines (255, 336). Over time, senescence is maintained by SASP signaling as well as persistent DNA damage-related foci, such as activated ATM, γH2AX, 53BP1, and CHK2 (336). In fact, ATM has been found to be required for maintenance of senescent-associated production of cytokine IL-6 (255). Depending on biological context, SASP components reinforce growth arrest (337) or promote cancer cell growth and invasion (338-340). For a more detailed discussion of the complex and varied consequences of senescence in inflammation and tumor development, see (341).
The positive feedback relationship between DNA repair and inflammation is somewhat unexpected, because repair of inflammation-derived lesions can promote inflammation further. This mechanism may have evolved to ensure that inflammation persists long enough to thoroughly remove the perceived insult before down-regulation to baseline levels. However, the processes by which this positive feedback loop is suppressed have yet to be revealed.
6. Inflammation Impairs Some DNA Repair Processes
In addition to direct damage by RONS, pro-inflammatory cytokines have also been shown to contribute to DNA damage. This signal-induced damage is due in part to the cytokine-stimulated increase of intracellular RONS (47-49) and in part to impairment of some DNA repair components (342-350). Since RONS readily react with cysteine residues on proteins, many cellular functions are susceptible to disruption from RONS (351). Indeed, Jaiswal et al. demonstrated that inflammatory cytokines were capable of both inducing DNA damage (as measured by the comet assay) and impairing DNA repair activity (as measured by radiolabel incorporation) via a NO-dependent mechanism (342).
More targeted studies have defined particular components of DNA repair that are disrupted by inflammation. Reaction of NO with glutathione (GSH) produces S-nitrosoglutathione (GSNO), which acts as an NO reservoir as well as a vehicle for cysteine nitrosylation on other proteins. S-nitrosylation can have a variety of consequences for protein function, such as altering activity or localization (352), and notable targets include DNA repair enzymes. One of the most thoroughly studied targets of S-nitrosylation is the Direct Reversal protein MGMT, which removes methyl lesions from nucleotides by directly transferring the alkyl group onto a cysteine residue (136, 353). Nitrosylation of MGMT’s active cysteine disables the enzyme (354), causing an accumulation of the genotoxic lesion O6-methylguanine (347). While DNA methylation is not a major type of inflammation-derived damage, all DNA repair pathways are essential for genomic maintenance, so inhibition of MGMT can increase stress on other repair pathways to compensate. It is noteworthy that inflammation can increase sensitivity to types of DNA damage that are not produced by inflammation.
Ironically, while BER is responsible for repairing many of the inflammation-induced DNA lesions, it is also especially susceptible to disruption by inflammation. For instance, S-nitrosylation of OGG1 decreases its activity (348, 349, 355), and S-nitrosylation of APE1 causes it to be exported from the nucleus (344, 349, 356). Thus, two key steps in repairing oxidative damage can be impeded by excess NO. Additionally, a common genetic variant of OGG1 in humans (Ser326Cys), associated with increased lung cancer risk (357), is susceptible to inactivation by intracellular RONS following pro-inflammatory stimulation (343). The relationship between OGG1 and inflammation is thus quite complex: OGG1 repairs the most common type of oxidative DNA damage, and it promotes transcription of pro-inflammatory genes, but it is also prone to inactivation by inflammation. These seemingly contradictory functions may help serve to regulate the positive feedback loop between inflammation and DNA damage.
Interestingly, and underscoring the complexity of the equilibrium between inflammation and DNA repair, S-nitrosylation of AAG slightly increases its activity (349, 356). However, rather than accelerating BER, increased AAG activity can produce more BER intermediates than the downstream enzymes can efficiently process, leading to increased tissue damage and mutations (181, 188). Indeed, excess AAG has been shown to contribute to microsatellite instability in the inflamed colon (203), highlighting the importance of balance among DNA repair components in maintaining genomic integrity.
Finally, rejoining broken DNA strands via ligase is the final step of nearly all DNA repair pathways, and evidence suggests that S-nitrosylation reduces ligase activity (350, 355), causing accumulation of unresolved strand breaks that can be cytotoxic or mutagenic.
7. Inflammation ←→ DNA Damage → Mutations → Cancer
The studies described here all support to the paradigm that inflammation and DNA damage contribute to each other and to the development of mutations and cancer, but few have explicitly demonstrated their direct connections. First, we integrate the collective importance inflammation and DNA damage to mutagenesis. This is perhaps exemplified by the fact that DNA damage during periods of increased cell division, such as inflammation-associated proliferation, has been shown to greatly increase mutation frequency (12). Frequency of cellular transformation in livers treated with the DNA alkylating agent N-methyl-N-nitrosourea (MNU) was shown to correlate directly with the number of proliferating cells in the tissue at the time of dosing (358). Further, two closely related studies showed that DNA damage occurring during a period of increased proliferation synergistically increases the frequency of mutant cell populations within the pancreas (12, 181). The first of these studies showed that MNU-induced DNA damage and hormone-induced proliferation both increase the frequency of mutant populations in the tissue, but when MNU was given during proliferation, the frequency of mutant clusters increased synergistically (181). A follow-up study then demonstrated that overlapping bouts of inflammation, wherein the DNA damage of one bout of inflammation coincides with the regenerative proliferation of an earlier bout, also synergistically increased the frequency of mutant populations compared to non-overlapping bouts of inflammation (12). These studies show that proliferation coinciding with DNA damage, whether induced exogenously or from inflammation, vastly increases the frequency of mutant cells in a tissue.
Second, studies from the Samson laboratory have demonstrated that DNA damage from inflammation drives mutations as well as cancer development (23, 359). In one such study, mice lacking the BER glycosylase Aag were treated with a common colitis model of DSS in drinking water. The Aag−/− animals developed far more severe tissue damage and neoplasia than wild type, which correlated with a dramatic increase in DNA damage. Oxidation, deamination, and LPO-derived base lesions accumulated, particularly εA, and some genetic deletions were observed, suggesting aberrant repair of broken replication forks. Sequencing tumors revealed point mutations in oncogenes that correspond with the mutational signatures associated with inflammation (23). Similar results were obtained in stomach tissue when Aag−/− and wild type animals were treated with H. pylori, a prevalent source of gastric inflammation and cancer (23, 360). Another study demonstrated the importance of BER on deamination products during inflammation, observing more severe pathology in DSS-treated Mbd4−/− mice compared to wild type (359). Further research revealed that DNA repair is essential to tolerating inflammation, as mice lacking multiple repair enzymes could not tolerate even a single bout of DSS-induced colitis (91). With these targeted studies, the Samson lab has uniquely contributed to this field by directly demonstrating the importance of DNA damage and its repair for tolerating inflammation Further such research revealing the presence of DNA strand breaks and oxidation, deamination, lipid peroxidation and halogenation base lesions, combined with analyses of DNA repair pathway activity and mutagenic consequences, will provide a more thorough understanding of the specific DNA transactions that occur during inflammation.
8. Conclusions
The relationships between inflammation and DNA damage are mediated by many factors, including a complex network of chemical reactions, DNA repair and tolerance pathways, cell cycle arrest mechanisms, and intra- and extracellular signaling pathways (Figure 11). Inflammation causes DNA damage primarily via RONS, which can produce DNA base and backbone lesions both by direct reaction or via reactive LPO intermediates. The detection and response to DNA damage by BER (e.g., PARP, OGG1), HR (e.g., ATM/ATR), or generalized damage recognition can signal for increased inflammation, creating a positive feedback loop. Inflammatory transcription factors such as NRF2 can mitigate the damage to DNA by neutralizing reactive chemicals with antioxidants and upregulating DNA repair pathways. However, nitric oxide can impair some repair enzymes, complicating the network and increasing the potential for dysregulation. Failure to repair DNA damage can lead to the mutations that initiate cancer, and additional physiological processes involved in inflammation (e.g., proliferation, migration) also promote cancer development. Given the number of mechanisms by which DNA damage and inflammation stimulate each other, it is no wonder that autoimmune and inflammatory diseases are so prevalent, difficult to control, and carcinogenic.
Figure 11.
Expanded paradigm describing relationships between inflammation, DNA damage and cancer
Acknowledgements
This work was supported in part by Superfund Research Program grant P42ES027707, NIEHS Training Grant T32-ES007020, and NCI Program Project Grant P01-CA026731.
Footnotes
The authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Guerra C, Schuhmacher AJ, Canamero M, Grippo PJ, Verdaguer L, Perez-Gallego L, Dubus P, Sandgren EP, Barbacid M. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer cell. 2007;11(3):291–302. doi: 10.1016/j.ccr.2007.01.012. PubMed PMID: 17349585. [DOI] [PubMed] [Google Scholar]
- 2.Bansal P, Sonnenberg A. Pancreatitis is a risk factor for pancreatic cancer. Gastroenterology. 1995;109(1):247–51. PubMed PMID: 7797022. [DOI] [PubMed] [Google Scholar]
- 3.Guerra C, Collado M, Navas C, Schuhmacher AJ, Hernandez-Porras I, Canamero M, Rodriguez-Justo M, Serrano M, Barbacid M. Pancreatitis-induced inflammation contributes to pancreatic cancer by inhibiting oncogene-induced senescence. Cancer cell. 2011;19(6):728–39. doi: 10.1016/j.ccr.2011.05.011. PubMed PMID: 21665147; PMCID: PMC4890723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rutter M, Saunders B, Wilkinson K, Rumbles S, Schofield G, Kamm M, Williams C, Price A, Talbot I, Forbes A. Severity of inflammation is a risk factor for colorectal neoplasia in ulcerative colitis. Gastroenterology. 2004;126(2):451–9. PubMed PMID: 14762782. [DOI] [PubMed] [Google Scholar]
- 5.Ohata K, Hamasaki K, Toriyama K, Matsumoto K, Saeki A, Yanagi K, Abiru S, Nakagawa Y, Shigeno M, Miyazoe S, Ichikawa T, Ishikawa H, Nakao K, Eguchi K. Hepatic steatosis is a risk factor for hepatocellular carcinoma in patients with chronic hepatitis C virus infection. Cancer. 2003;97(12):3036–43. doi: 10.1002/cncr.11427. PubMed PMID: 12784339. [DOI] [PubMed] [Google Scholar]
- 6.Beasley RP. Hepatitis B virus. The major etiology of hepatocellular carcinoma. Cancer. 1988;61(10):1942–56. PubMed PMID: 2834034. [DOI] [PubMed] [Google Scholar]
- 7.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420(6917):860–7. Epub 2002/12/20. doi: 10.1038/nature01322. PubMed PMID: 12490959; PMCID: 2803035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erdman SE, Rao VP, Poutahidis T, Rogers AB, Taylor CL, Jackson EA, Ge Z, Lee CW, Schauer DB, Wogan GN, Tannenbaum SR, Fox JG. Nitric oxide and TNF-alpha trigger colonic inflammation and carcinogenesis in Helicobacter hepaticus-infected, Rag2-deficient mice. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(4):1027–32. Epub 2009/01/24. doi: 10.1073/pnas.0812347106. PubMed PMID: 19164562; PMCID: 2633549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anuja K, Roy S, Ghosh C, Gupta P, Bhattacharjee S, Banerjee B. Prolonged inflammatory microenvironment is crucial for pro-neoplastic growth and genome instability: a detailed review. Inflammation research : official journal of the European Histamine Research Society [et al. ]. 2017;66(2):119–28. doi: 10.1007/s00011-016-0985-3. PubMed PMID: 27653961. [DOI] [PubMed] [Google Scholar]
- 10.Pikarsky E, Porat RM, Stein I, Abramovitch R, Amit S, Kasem S, Gutkovich-Pyest E, Urieli-Shoval S, Galun E, Ben-Neriah Y. NF-kappaB functions as a tumour promoter in inflammation-associated cancer. Nature. 2004;431(7007):461–6. Epub 2004/08/27. doi: 10.1038/nature02924. PubMed PMID: 15329734. [DOI] [PubMed] [Google Scholar]
- 11.Maeda S, Kamata H, Luo JL, Leffert H, Karin M. IKKbeta couples hepatocyte death to cytokine-driven compensatory proliferation that promotes chemical hepatocarcinogenesis. Cell. 2005;121(7):977–90. doi: 10.1016/j.cell.2005.04.014. PubMed PMID: 15989949. [DOI] [PubMed] [Google Scholar]
- 12.Kiraly O, Gong G, Olipitz W, Muthupalani S, Engelward BP. Inflammation-induced cell proliferation potentiates DNA damage-induced mutations in vivo. PLoS genetics. 2015;11(2):e1004901. doi: 10.1371/journal.pgen.1004901. PubMed PMID: 25647331; PMCID: PMC4372043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burns JM, Summers BC, Wang Y, Melikian A, Berahovich R, Miao Z, Penfold ME, Sunshine MJ, Littman DR, Kuo CJ, Wei K, McMaster BE, Wright K, Howard MC, Schall TJ. A novel chemokine receptor for SDF-1 and I-TAC involved in cell survival, cell adhesion, and tumor development. The Journal of experimental medicine. 2006;203(9):2201–13. Epub 2006/08/31. doi: 10.1084/jem.20052144. PubMed PMID: 16940167; PMCID: PMC2118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chung AS, Kao WJ. Fibroblasts regulate monocyte response to ECM-derived matrix: the effects on monocyte adhesion and the production of inflammatory, matrix remodeling, and growth factor proteins. J Biomed Mater Res A. 2009;89(4):841–53. Epub 2009/05/14. PubMed PMID: 19437738; PMCID: PMC3795518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J, Chen L, Xiao M, Wang C, Qin Z. FSP1+ fibroblasts promote skin carcinogenesis by maintaining MCP-1-mediated macrophage infiltration and chronic inflammation. The American journal of pathology. 2011;178(1):382–90. Epub 2011/01/13. doi: 10.1016/j.ajpath.2010.11.017. PubMed PMID: 21224075; PMCID: PMC3070559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wedmore CV, Williams TJ. Control of vascular permeability by polymorphonuclear leukocytes in inflammation. Nature. 1981;289(5799):646–50. Epub 1981/02/19. PubMed PMID: 7464931. [DOI] [PubMed] [Google Scholar]
- 17.Allavena P, Garlanda C, Borrello MG, Sica A, Mantovani A. Pathways connecting inflammation and cancer. Current opinion in genetics & development. 2008;18(1):3–10. Epub 2008/03/08. doi: 10.1016/j.gde.2008.01.003. PubMed PMID: 18325755. [DOI] [PubMed] [Google Scholar]
- 18.Shacter E, Weitzman SA. Chronic inflammation and cancer. Oncology (Williston Park). 2002;16(2):217–26, 29; discussion 30–2. PubMed PMID: 11866137. [PubMed] [Google Scholar]
- 19.Savill JS, Wyllie AH, Henson JE, Walport MJ, Henson PM, Haslett C. Macrophage phagocytosis of aging neutrophils in inflammation. Programmed cell death in the neutrophil leads to its recognition by macrophages. The Journal of clinical investigation. 1989;83(3):865–75. Epub 1989/03/01. doi: 10.1172/JCI113970. PubMed PMID: 2921324; PMCID: PMC303760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sosna J, Voigt S, Mathieu S, Lange A, Thon L, Davarnia P, Herdegen T, Linkermann A, Rittger A, Chan FK, Kabelitz D, Schutze S, Adam D. TNF-induced necroptosis and PARP-1-mediated necrosis represent distinct routes to programmed necrotic cell death. Cell Mol Life Sci. 2014;71(2):331–48. doi: 10.1007/s00018-013-1381-6. PubMed PMID: 23760205; PMCID: PMC3889832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Westbrook AM, Wei B, Hacke K, Xia M, Braun J, Schiestl RH. The role of tumour necrosis factor-alpha and tumour necrosis factor receptor signalling in inflammation-associated systemic genotoxicity. Mutagenesis. 2012;27(1):77–86. Epub 2011/10/08. doi: 10.1093/mutage/ger063. PubMed PMID: 21980144; PMCID: 3241942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujita M, Matsubara N, Matsuda I, Maejima K, Oosawa A, Yamano T, Fujimoto A, Furuta M, Nakano K, Oku-Sasaki A, Tanaka H, Shiraishi Y, Mateos RN, Nakai K, Miyano S, Tomita N, Hirota S, Ikeuchi H, Nakagawa H. Genomic landscape of colitis-associated cancer indicates the impact of chronic inflammation and its stratification by mutations in the Wnt signaling. Oncotarget. 2018;9(1):969–81. doi: 10.18632/oncotarget.22867. PubMed PMID: 29416670; PMCID: PMC5787528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meira LB, Bugni JM, Green SL, Lee CW, Pang B, Borenshtein D, Rickman BH, Rogers AB, Moroski-Erkul CA, McFaline JL, Schauer DB, Dedon PC, Fox JG, Samson LD. DNA damage induced by chronic inflammation contributes to colon carcinogenesis in mice. The Journal of clinical investigation. 2008;118(7):2516–25. Epub 2008/06/04. doi: 10.1172/JCI35073. PubMed PMID: 18521188; PMCID: 2423313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neeley WL, Essigmann JM. Mechanisms of formation, genotoxicity, and mutation of guanine oxidation products. Chem Res Toxicol. 2006;19(4):491–505. doi: 10.1021/tx0600043. PubMed PMID: 16608160. [DOI] [PubMed] [Google Scholar]
- 25.Coussens LM, Raymond WW, Bergers G, Laig-Webster M, Behrendtsen O, Werb Z, Caughey GH, Hanahan D. Inflammatory mast cells up-regulate angiogenesis during squamous epithelial carcinogenesis. Genes & development. 1999; 13(11):1382–97. Epub 1999/06/11. doi: 10.1101/gad.13.11.1382. PubMed PMID: 10364156; PMCID: PMC316772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Balkwill F, Mantovani A. Inflammation and cancer: back to Virchow? Lancet. 2001;357(9255):539–45. Epub 2001/03/07. doi: 10.1016/S0140-6736(00)04046-0. PubMed PMID: 11229684. [DOI] [PubMed] [Google Scholar]
- 27.Erdman SE, Poutahidis T, Tomczak M, Rogers AB, Cormier K, Plank B, Horwitz BH, Fox JG. CD4+ CD25+ regulatory T lymphocytes inhibit microbially induced colon cancer in Rag2-deficient mice. The American journal of pathology. 2003;162(2):691–702. Epub 2003/01/28. doi: 10.1016/S0002-9440(10)63863-1. PubMed PMID: 12547727; PMCID: 1851156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rayburn ER, Ezell SJ, Zhang R. Anti-Inflammatory Agents for Cancer Therapy. Mol Cell Pharmacol. 2009;1(1):29–43. doi: 10.4255/mcpharmacol.09.05. PubMed PMID: 20333321; PMCID: PMC2843097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454(7203):436–44. Epub 2008/07/25. doi: 10.1038/nature07205. PubMed PMID: 18650914. [DOI] [PubMed] [Google Scholar]
- 30.Colotta F, Allavena P, Sica A, Garlanda C, Mantovani A. Cancer-related inflammation, the seventh hallmark of cancer: links to genetic instability. Carcinogenesis. 2009;30(7):1073–81. doi: 10.1093/carcin/bgp127. PubMed PMID: 19468060. [DOI] [PubMed] [Google Scholar]
- 31.Pikor L, Thu K, Vucic E, Lam W. The detection and implication of genome instability in cancer. Cancer Metastasis Rev. 2013;32(3-4):341–52. doi: 10.1007/s10555-013-9429-5. PubMed PMID: 23633034; PMCID: PMC3843371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability--an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11(3):220–8. doi: 10.1038/nrm2858. PubMed PMID: 20177397. [DOI] [PubMed] [Google Scholar]
- 33.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. doi: 10.1016/j.cell.2011.02.013. PubMed PMID: 21376230. [DOI] [PubMed] [Google Scholar]
- 34.DeMars R, Held KR. The spontaneous azaguanine-resistant mutants of diploid human fibroblasts. Humangenetik. 1972;16(1):87–110. PubMed PMID: 4647448. [DOI] [PubMed] [Google Scholar]
- 35.Loeb LA. A mutator phenotype in cancer. Cancer research. 2001;61(8):3230–9. PubMed PMID: 11309271. [PubMed] [Google Scholar]
- 36.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458(7239):719–24. doi: 10.1038/nature07943. PubMed PMID: 19360079; PMCID: PMC2821689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr., Kinzler KW. Cancer genome landscapes. Science. 2013;339(6127):1546–58. doi: 10.1126/science.1235122. PubMed PMID: 23539594; PMCID: PMC3749880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ahmad AS, Ormiston-Smith N, Sasieni PD. Trends in the lifetime risk of developing cancer in Great Britain: comparison of risk for those born from 1930 to 1960. British journal of cancer. 2015;112(5):943–7. Epub 2015/02/04. doi: 10.1038/bjc.2014.606. PubMed PMID: 25647015; PMCID: PMC4453943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bredt DS, Snyder SH. Nitric oxide: a physiologic messenger molecule. Annu Rev Biochem. 1994;63:175–95. doi: 10.1146/annurev.bi.63.070194.001135. PubMed PMID: 7526779. [DOI] [PubMed] [Google Scholar]
- 40.Moncada S, Palmer RM, Higgs EA. Nitric oxide: physiology, pathophysiology, and pharmacology. Pharmacol Rev. 1991;43(2):109–42. PubMed PMID: 1852778. [PubMed] [Google Scholar]
- 41.Thomas DD, Ridnour LA, Isenberg JS, Flores-Santana W, Switzer CH, Donzelli S, Hussain P, Vecoli C, Paolocci N, Ambs S, Colton CA, Harris CC, Roberts DD, Wink DA. The chemical biology of nitric oxide: implications in cellular signaling. Free radical biology & medicine. 2008;45(1):18–31. doi: 10.1016/j.freeradbiomed.2008.03.020. PubMed PMID: 18439435; PMCID: PMC2572721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lewis RS, Tamir S, Tannenbaum SR, Deen WM. Kinetic analysis of the fate of nitric oxide synthesized by macrophages in vitro. The Journal of biological chemistry. 1995;270(49):29350–5. PubMed PMTD: 7493969. [DOI] [PubMed] [Google Scholar]
- 43.Stuehr DJ, Marletta MA. Synthesis of nitrite and nitrate in murine macrophage cell lines. Cancer research. 1987;47(21):5590–4. PubMed PMTD: 3117354. [PubMed] [Google Scholar]
- 44.Takahashi R, Edashige K, Sato EF, Inoue M, Matsuno T, Utsumi K. Luminol chemiluminescence and active oxygen generation by activated neutrophils. Archives of biochemistry and biophysics. 1991;285(2):325–30. Epub 1991/03/01. PubMed PMID: 1654772. [DOI] [PubMed] [Google Scholar]
- 45.Badwey JA, Karnovsky ML. Active oxygen species and the functions of phagocytic leukocytes. Annu Rev Biochem. 1980;49:695–726. Epub 1980/01/01. doi: 10.1146/annurev.bi.49.070180.003403. PubMed PMID: 6250449. [DOI] [PubMed] [Google Scholar]
- 46.Van den Worm E, Beukelman CJ, Van den Berg AJ, Kroes BH, Labadie RP, Van Dijk H. Effects of methoxylation of apocynin and analogs on the inhibition of reactive oxygen species production by stimulated human neutrophils. Eur J Pharmacol. 2001;433(2-3):225–30. Epub 2002/01/05. PubMed PMID: 11755156. [DOI] [PubMed] [Google Scholar]
- 47.Colin TM, Poncin S, Leveque P, Gallez B, Gerard AC. Differential regulation of the production of reactive oxygen species in Th1 cytokine-treated thyroid cells. Thyroid. 2014;24(3):441–52. doi: 10.1089/thy.2013.0142. PubMed PMID: 24073824. [DOI] [PubMed] [Google Scholar]
- 48.Sundaresan M, Yu ZX, Ferrans VJ, Sulciner DJ, Gutkind JS, Irani K, Goldschmidt-Clermont PJ, Finkel T. Regulation of reactive-oxygen-species generation in fibroblasts by Rac1. The Biochemical journal. 1996;318 ( Pt 2):379–82. PubMed PMID: 8809022; PMCID: PMC1217632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang D, Elner SG, Bian ZM, Till GO, Petty HR, Elner VM. Pro-inflammatory cytokines increase reactive oxygen species through mitochondria and NADPH oxidase in cultured RPE cells. Exp Eye Res. 2007;85(4):462–72. doi: 10.1016/j.exer.2007.06.013. PubMed PMID: 17765224; PMCID: PMC2094037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dedon PC, Tannenbaum SR. Reactive nitrogen species in the chemical biology of inflammation. Archives of biochemistry and biophysics. 2004;423(1):12–22. PubMed PMID: 14989259. [DOI] [PubMed] [Google Scholar]
- 51.Lonkar P, Dedon PC. Reactive species and DNA damage in chronic inflammation: reconciling chemical mechanisms and biological fates. International journal of cancer Journal international du cancer. 2011;128(9):1999–2009. Epub 2011/03/10. doi: 10.1002/ijc.25815. PubMed PMID: 21387284; PMCID: 3334345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dizdaroglu M Oxidatively induced DNA damage: mechanisms, repair and disease. Cancer letters. 2012;327(1-2):26–47. doi: 10.1016/j.canlet.2012.01.016. PubMed PMID: 22293091. [DOI] [PubMed] [Google Scholar]
- 53.Steenken S How Easily Oxidizable Is DNA? One-Electron Reduction Potentials of Adenosine and Guanosine Radicals in Aqueous Solution. Journal of the American Chemical Society. 1997;119(3):617–8. [Google Scholar]
- 54.Shibutani S, Takeshita M, Grollman AP. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349(6308):431–4. doi: 10.1038/349431a0. PubMed PMID: 1992344. [DOI] [PubMed] [Google Scholar]
- 55.Cheng KC, Cahill DS, Kasai H, Nishimura S, Loeb LA. 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G----T and A----C substitutions. The Journal of biological chemistry. 1992;267(1):166–72. PubMed PMID: 1730583. [PubMed] [Google Scholar]
- 56.Duarte V, Muller JG, Burrows CJ. Insertion of dGMP and dAMP during in vitro DNA synthesis opposite an oxidized form of 7,8-dihydro-8-oxoguanine. Nucleic acids research. 1999;27(2):496–502. PubMed PMID: 9862971; PMCID: PMC148206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hsu GW, Ober M, Carell T, Beese LS. Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase. Nature. 2004;431(7005):217–21. doi: 10.1038/nature02908. PubMed PMID: 15322558. [DOI] [PubMed] [Google Scholar]
- 58.Markkanen E, Castrec B, Villani G, Hubscher U. A switch between DNA polymerases delta and lambda promotes error-free bypass of 8-oxo-G lesions. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(50):20401–6. doi: 10.1073/pnas.1211532109. PubMed PMID: 23175785; PMCID: PMC3528542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Uppu RM, Cueto R, Squadrito GL, Salgo MG, Pryor WA. Competitive reactions of peroxynitrite with 2'-deoxyguanosine and 7,8-dihydro-8-oxo-2'-deoxyguanosine (8-oxodG): relevance to the formation of 8-oxodG in DNA exposed to peroxynitrite. Free radical biology & medicine. 1996;21(3):407–11. PubMed PMID: 8855454. [DOI] [PubMed] [Google Scholar]
- 60.Henderson PT, Delaney JC, Gu F, Tannenbaum SR, Essigmann JM. Oxidation of 7,8-dihydro-8-oxoguanine affords lesions that are potent sources of replication errors in vivo. Biochemistry. 2002;41(3):914–21. PubMed PMID: 11790114. [DOI] [PubMed] [Google Scholar]
- 61.Henderson PT, Delaney JC, Muller JG, Neeley WL, Tannenbaum SR, Burrows CJ, Essigmann JM. The hydantoin lesions formed from oxidation of 7,8-dihydro-8-oxoguanine are potent sources of replication errors in vivo. Biochemistry. 2003;42(31):9257–62. doi: 10.1021/bi0347252. PubMed PMID: 12899611. [DOI] [PubMed] [Google Scholar]
- 62.Fenn D, Chi LM, Lam SL. Effect of hyperoxidized guanine on DNA primer-template structures: spiroiminodihydantoin leads to strand slippage. FEBS letters. 2008;582(30):4169–75. doi: 10.1016/j.febslet.2008.11.021. PubMed PMID: 19041867. [DOI] [PubMed] [Google Scholar]
- 63.Zhao X, Krishnamurthy N, Burrows CJ, David SS. Mutation versus repair: NEIL1 removal of hydantoin lesions in single-stranded, bulge, bubble, and duplex DNA contexts. Biochemistry. 2010;49(8):1658–66. doi: 10.1021/bi901852q. PubMed PMID: 20099873; PMCID: PMC2872175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Neeley WL, Delaney S, Alekseyev YO, Jarosz DF, Delaney JC, Walker GC, Essigmann JM. DNA polymerase V allows bypass of toxic guanine oxidation products in vivo. The Journal of biological chemistry. 2007;282(17):12741–8. doi: 10.1074/jbc.M700575200. PubMed PMID: 17322566. [DOI] [PubMed] [Google Scholar]
- 65.Delaney S, Neeley WL, Delaney JC, Essigmann JM. The substrate specificity of MutY for hyperoxidized guanine lesions in vivo. Biochemistry. 2007;46(5):1448–55. doi: 10.1021/bi061174h. PubMed PMID: 17260974. [DOI] [PubMed] [Google Scholar]
- 66.Kalam MA, Haraguchi K, Chandani S, Loechler EL, Moriya M, Greenberg MM, Basu AK. Genetic effects of oxidative DNA damages: comparative mutagenesis of the imidazole ring-opened formamidopyrimidines (Fapy lesions) and 8-oxo-purines in simian kidney cells. Nucleic acids research. 2006;34(8):2305–15. doi: 10.1093/nar/gkl099. PubMed PMID: 16679449; PMCID: PMC1458282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Asagoshi K, Terato H, Ohyama Y, Ide H. Effects of a guanine-derived formamidopyrimidine lesion on DNA replication: translesion DNA synthesis, nucleotide insertion, and extension kinetics. The Journal of biological chemistry. 2002;277(17):14589–97. doi: 10.1074/jbc.M200316200. PubMed PMID: 11839760. [DOI] [PubMed] [Google Scholar]
- 68.Dong M, Wang C, Deen WM, Dedon PC. Absence of 2'-deoxyoxanosine and presence of abasic sites in DNA exposed to nitric oxide at controlled physiological concentrations. Chem Res Toxicol. 2003;16(9):1044–55. doi: 10.1021/tx034046s. PubMed PMID: 12971791. [DOI] [PubMed] [Google Scholar]
- 69.Taghizadeh K, McFaline JL, Pang B, Sullivan M, Dong M, Plummer E, Dedon PC. Quantification of DNA damage products resulting from deamination, oxidation and reaction with products of lipid peroxidation by liquid chromatography isotope dilution tandem mass spectrometry. Nature protocols. 2008;3(8):1287–98. Epub 2008/08/21. doi: 10.1038/nprot.2008.119. PubMed PMID: 18714297; PMCID: PMC2832793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mangerich A, Knutson CG, Parry NM, Muthupalani S, Ye W, Prestwich E, Cui L, McFaline JL, Mobley M, Ge Z, Taghizadeh K, Wishnok JS, Wogan GN, Fox JG, Tannenbaum SR, Dedon PC. Infection-induced colitis in mice causes dynamic and tissue-specific changes in stress response and DNA damage leading to colon cancer. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(27):E1820–9. Epub 2012/06/13. doi: 10.1073/pnas.1207829109. PubMed PMID: 22689960; PMCID: 3390855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.COSMIC [Internet] [cited November 28, 2017]. Available from: http://cancer.sanger.ac.uk/cosmic/signatures.
- 72.DeVito S, Woodrick J, Song L, Roy R. Mutagenic potential of hypoxanthine in live human cells. Mutation research. 2017;803–805:9-16. doi: 10.1016/j.mrfmmm.2017.06.005. PubMed PMID: 28704682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Caulfield JL, Wishnok JS, Tannenbaum SR. Nitric oxide-induced deamination of cytosine and guanine in deoxynucleosides and oligonucleotides. The Journal of biological chemistry. 1998;273(21):12689–95. PubMed PMID: 9582291. [DOI] [PubMed] [Google Scholar]
- 74.Wuenschell GE, O'Connor TR, Termini J. Stability, miscoding potential, and repair of 2'-deoxyxanthosine in DNA: implications for nitric oxide-induced mutagenesis. Biochemistry. 2003;42(12):3608–16. doi: 10.1021/bi0205597. PubMed PMID: 12653565. [DOI] [PubMed] [Google Scholar]
- 75.Dong M, Dedon PC. Relatively small increases in the steady-state levels of nucleobase deamination products in DNA from human TK6 cells exposed to toxic levels of nitric oxide. Chem Res Toxicol. 2006;19(1):50–7. doi: 10.1021/tx050252j. PubMed PMID: 16411656; PMCID: PMC2515361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.van der Veen BS, de Winther MP, Heeringa P. Myeloperoxidase: molecular mechanisms of action and their relevance to human health and disease. Antioxidants & redox signaling. 2009;11(11):2899–937. doi: 10.1089/ARS.2009.2538. PubMed PMID: 19622015. [DOI] [PubMed] [Google Scholar]
- 77.Winterbourn CC, Hampton MB, Livesey JH, Kettle AJ. Modeling the reactions of superoxide and myeloperoxidase in the neutrophil phagosome: implications for microbial killing. The Journal of biological chemistry. 2006;281(52):39860–9. doi: 10.1074/jbc.M605898200. PubMed PMID: 17074761. [DOI] [PubMed] [Google Scholar]
- 78.Henderson JP, Byun J, Heinecke JW. Molecular chlorine generated by the myeloperoxidase-hydrogen peroxide-chloride system of phagocytes produces 5-chlorocytosine in bacterial RNA. The Journal of biological chemistry. 1999;274(47):33440–8. PubMed PMID: 10559226. [DOI] [PubMed] [Google Scholar]
- 79.Henderson JP, Byun J, Williams MV, McCormick ML, Parks WC, Ridnour LA, Heinecke JW. Bromination of deoxycytidine by eosinophil peroxidase: a mechanism for mutagenesis by oxidative damage of nucleotide precursors. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(4):1631–6. doi: 10.1073/pnas.041146998. PubMed PMID: 11172002; PMCID: PMC29308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wu W, Samoszuk MK, Comhair SA, Thomassen MJ, Farver CF, Dweik RA, Kavuru MS, Erzurum SC, Hazen SL. Eosinophils generate brominating oxidants in allergen-induced asthma. The Journal of clinical investigation. 2000;105(10):1455–63. doi: 10.1172/JCI9702. PubMed PMID: 10811853; PMCID: PMC315470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kawai Y, Morinaga H, Kondo H, Miyoshi N, Nakamura Y, Uchida K, Osawa T. Endogenous formation of novel halogenated 2'-deoxycytidine. Hypohalous acid-mediated DNA modification at the site of inflammation. The Journal of biological chemistry. 2004;279(49):51241–9. doi: 10.1074/jbc.M408210200. PubMed PMID: 15364942. [DOI] [PubMed] [Google Scholar]
- 82.Kang JI Jr., Sowers LC. Examination of hypochlorous acid-induced damage to cytosine residues in a CpG dinucleotide in DNA. Chem Res Toxicol. 2008;21(6):1211–8. doi: 10.1021/tx800037h. PubMed PMID: 18826175. [DOI] [PubMed] [Google Scholar]
- 83.Knutson CG, Mangerich A, Zeng Y, Raczynski AR, Liberman RG, Kang P, Ye W, Prestwich EG, Lu K, Wishnok JS, Korzenik JR, Wogan GN, Fox JG, Dedon PC, Tannenbaum SR. Chemical and cytokine features of innate immunity characterize serum and tissue profiles in inflammatory bowel disease. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(26):E2332–41. Epub 2013/06/12. doi: 10.1073/pnas.1222669110. PubMed PMID: 23754421; PMCID: 3696795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fedeles BI, Freudenthal BD, Yau E, Singh V, Chang SC, Li D, Delaney JC, Wilson SH, Essigmann JM. Intrinsic mutagenic properties of 5-chlorocytosine: A mechanistic connection between chronic inflammation and cancer. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(33):E4571–80. doi: 10.1073/pnas.1507709112. PubMed PMID: 26243878; PMCID: PMC4547254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lao VV, Herring JL, Kim CH, Darwanto A, Soto U, Sowers LC. Incorporation of 5-chlorocytosine into mammalian DNA results in heritable gene silencing and altered cytosine methylation patterns. Carcinogenesis. 2009;30(5):886–93. doi: 10.1093/carcin/bgp060. PubMed PMID: 19279184; PMCID: PMC2675655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sato Y, Takahashi S, Kinouchi Y, Shiraki M, Endo K, Matsumura Y, Kakuta Y, Tosa M, Motida A, Abe H, Imai G, Yokoyama H, Nomura E, Negoro K, Takagi S, Aihara H, Masumura K, Nohmi T, Shimosegawa T. IL-10 deficiency leads to somatic mutations in a model of IBD. Carcinogenesis. 2006;27(5):1068–73. Epub 2006/01/13. doi: 10.1093/carcin/bgi327. PubMed PMID: 16407368. [DOI] [PubMed] [Google Scholar]
- 87.Nair U, Bartsch H, Nair J. Lipid peroxidation-induced DNA damage in cancer-prone inflammatory diseases: a review of published adduct types and levels in humans. Free radical biology & medicine. 2007;43(8):1109–20. Epub 2007/09/15. doi: 10.1016/j.freeradbiomed.2007.07.012. PubMed PMID: 17854706. [DOI] [PubMed] [Google Scholar]
- 88.Zarkovic N 4-hydroxynonenal as a bioactive marker of pathophysiological processes. Mol Aspects Med. 2003;24(4-5):281–91. PubMed PMID: 12893006. [DOI] [PubMed] [Google Scholar]
- 89.Nair J, Gansauge F, Beger H, Dolara P, Winde G, Bartsch H. Increased etheno-DNA adducts in affected tissues of patients suffering from Crohn's disease, ulcerative colitis, and chronic pancreatitis. Antioxidants & redox signaling. 2006;8(5-6):1003–10. doi: 10.1089/ars.2006.8.1003. PubMed PMID: 16771690. [DOI] [PubMed] [Google Scholar]
- 90.Moriya M, Zhang W, Johnson F, Grollman AP. Mutagenic potency of exocyclic DNA adducts: marked differences between Escherichia coli and simian kidney cells. Proceedings of the National Academy of Sciences of the United States of America. 1994;91(25):11899–903. PubMed PMID: 7991554; PMCID: PMC45343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Calvo JA, Meira LB, Lee CY, Moroski-Erkul CA, Abolhassani N, Taghizadeh K, Eichinger LW, Muthupalani S, Nordstrand LM, Klungland A, Samson LD. DNA repair is indispensable for survival after acute inflammation. The Journal of clinical investigation. 2012;122(7):2680–9. doi: 10.1172/JCI63338. PubMed PMID: 22684101; PMCID: PMC3386829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pang B, Zhou X, Yu H, Dong M, Taghizadeh K, Wishnok JS, Tannenbaum SR, Dedon PC. Lipid peroxidation dominates the chemistry of DNA adduct formation in a mouse model of inflammation. Carcinogenesis. 2007;28(8):1807–13. doi: 10.1093/carcin/bgm037. PubMed PMID: 17347141. [DOI] [PubMed] [Google Scholar]
- 93.Chang SC, Fedeles BI, Wu J, Delaney JC, Li D, Zhao L, Christov PP, Yau E, Singh V, Jost M, Drennan CL, Marnett LJ, Rizzo CJ, Levine SS, Guengerich FP, Essigmann JM. Next generation sequencing reveals the biological significance of the N(2),3-ethenoguanine lesion in vivo. Nucleic acids research. 2015;43(11):5489–500. doi: 10.1093/nar/gkv243. PubMed PMID: 25837992; PMCID: PMC4477646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Swenberg JA, Lu K, Moeller BC, Gao L, Upton PB, Nakamura J, Starr TB. Endogenous versus exogenous DNA adducts: their role in carcinogenesis, epidemiology, and risk assessment. Toxicol Sci. 2011; 120 Suppl 1:S130–45. doi: 10.1093/toxsci/kfq371. PubMed PMID: 21163908; PMCID: PMC3043087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shrivastav N, Li D, Essigmann JM. Chemical biology of mutagenesis and DNA repair: cellular responses to DNA alkylation. Carcinogenesis. 2010;31(1):59–70. doi: 10.1093/carcin/bgp262. PubMed PMID: 19875697; PMCID: PMC2802671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Langouet S, Muller M, Guengerich FP. Misincorporation of dNTPs opposite 1,N2-ethenoguanine and 5,6,7,9-tetrahydro-7-hydroxy-9-oxoimidazo[1,2-a]purine in oligonucleotides by Escherichia coli polymerases I exo- and II exo-, T7 polymerase exo-, human immunodeficiency virus-1 reverse transcriptase, and rat polymerase beta. Biochemistry. 1997;36(20):6069–79. doi: 10.1021/bi962526v. PubMed PMID: 9166777. [DOI] [PubMed] [Google Scholar]
- 97.Zhao L, Pence MG, Christov PP, Wawrzak Z, Choi JY, Rizzo CJ, Egli M, Guengerich FP. Basis of miscoding of the DNA adduct N2,3-ethenoguanine by human Y-family DNA polymerases. The Journal of biological chemistry. 2012;287(42):35516–26. Epub 2012/08/23. doi: 10.1074/jbc.M112.403253. PubMed PMID: 22910910; PMCID: PMC3471744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pandya GA, Moriya M. 1,N6-ethenodeoxyadenosine, a DNA adduct highly mutagenic in mammalian cells. Biochemistry. 1996;35(35):11487–92. doi: 10.1021/bi960170h. PubMed PMID: 8784204. [DOI] [PubMed] [Google Scholar]
- 99.Levine RL, Yang IY, Hossain M, Pandya GA, Grollman AP, Moriya M. Mutagenesis induced by a single 1,N6-ethenodeoxyadenosine adduct in human cells. Cancer research. 2000;60(15):4098–104. PubMed PMID: 10945616. [PubMed] [Google Scholar]
- 100.Gros L, Maksimenko AV, Privezentzev CV, Laval J, Saparbaev MK. Hijacking of the human alkyl-N-purine-DNA glycosylase by 3,N4-ethenocytosine, a lipid peroxidation-induced DNA adduct. The Journal of biological chemistry. 2004;279(17):17723–30. doi: 10.1074/jbc.M314010200. PubMed PMID: 14761949. [DOI] [PubMed] [Google Scholar]
- 101.Ryan AJ, Squires S, Strutt HL, Johnson RT. Camptothecin cytotoxicity in mammalian cells is associated with the induction of persistent double strand breaks in replicating DNA. Nucleic acids research. 1991;19(12):3295–300. PubMed PMID: 1905803; PMCID: PMC328325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pommier Y, Zwelling LA, Kao-Shan CS, Whang-Peng J, Bradley MO. Correlations between intercalator-induced DNA strand breaks and sister chromatid exchanges, mutations, and cytotoxicity in Chinese hamster cells. Cancer research. 1985;45(7):3143–9. PubMed PMID: 2988762. [PubMed] [Google Scholar]
- 103.Nelson WG, Kastan MB. DNA strand breaks: the DNA template alterations that trigger p53-dependent DNA damage response pathways. Molecular and cellular biology. 1994;14(3):1815–23. PubMed PMID: 8114714; PMCID: PMC358539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Szabo C, Zingarelli B, O'Connor M, Salzman AL. DNA strand breakage, activation of poly (ADP-ribose) synthetase, and cellular energy depletion are involved in the cytotoxicity of macrophages and smooth muscle cells exposed to peroxynitrite. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(5):1753–8. PubMed PMID: 8700830; PMCID: PMC39853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dasika GK, Lin SC, Zhao S, Sung P, Tomkinson A, Lee EY. DNA damage-induced cell cycle checkpoints and DNA strand break repair in development and tumorigenesis. Oncogene. 1999;18(55):7883–99. doi: 10.1038/sj.onc.1203283. PubMed PMID: 10630641. [DOI] [PubMed] [Google Scholar]
- 106.Kiziltepe T, Yan A, Dong M, Jonnalagadda VS, Dedon PC, Engelward BP. Delineation of the chemical pathways underlying nitric oxide-induced homologous recombination in mammalian cells. Chemistry & biology. 2005;12(3):357–69. Epub 2005/03/31. doi: 10.1016/j.chembiol.2004.12.011. PubMed PMID: 15797220. [DOI] [PubMed] [Google Scholar]
- 107.Cannan WJ, Pederson DS. Mechanisms and Consequences of Double-Strand DNA Break Formation in Chromatin. J Cell Physiol. 2016;231(1):3–14. Epub 2015/06/05. doi: 10.1002/jcp.25048. PubMed PMID: 26040249; PMCID: PMC4994891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ashour ME, Atteya R, El-Khamisy SF. Topoisomerase-mediated chromosomal break repair: an emerging player in many games. Nature reviews Cancer. 2015;15(3):137–51. doi: 10.1038/nrc3892. PubMed PMID: 25693836. [DOI] [PubMed] [Google Scholar]
- 109.Bryant PE, Johnston PJ. Restriction-endonuclease-induced DNA double-strand breaks and chromosomal aberrations in mammalian cells. Mutation research. 1993;299(3-4):289–96. PubMed PMID: 7683096. [DOI] [PubMed] [Google Scholar]
- 110.Yang N, Galick H, Wallace SS. Attempted base excision repair of ionizing radiation damage in human lymphoblastoid cells produces lethal and mutagenic double strand breaks. DNA repair. 2004;3(10):1323–34. Epub 2004/09/01. doi: 10.1016/j.dnarep.2004.04.014. PubMed PMID: 15336627. [DOI] [PubMed] [Google Scholar]
- 111.Sobol RW, Kartalou M, Almeida KH, Joyce DF, Engelward BP, Horton JK, Prasad R, Samson LD, Wilson SH. Base excision repair intermediates induce p53-independent cytotoxic and genotoxic responses. The Journal of biological chemistry. 2003;278(41):39951–9. doi: 10.1074/jbc.M306592200. PubMed PMID: 12882965. [DOI] [PubMed] [Google Scholar]
- 112.D'Souza DI, Harrison L. Repair of clustered uracil DNA damages in Escherichia coli. Nucleic acids research. 2003;31(15):4573–81. PubMed PMID: 12888518; PMCID: PMC169883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hashimoto Y, Puddu F, Costanzo V. RAD51- and MRE11-dependent reassembly of uncoupled CMG helicase complex at collapsed replication forks. Nat Struct Mol Biol. 2011;19(1):17–24. Epub 2011/12/06. doi: 10.1038/nsmb.2177. PubMed PMID: 22139015; PMCID: PMC4306020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Muris DF, Vreeken K, Carr AM, Murray JM, Smit C, Lohman PH, Pastink A. Isolation of the Schizosaccharomyces pombe RAD54 homologue, rhp54+, a gene involved in the repair of radiation damage and replication fidelity. J Cell Sci. 1996;109 ( Pt 1):73–81. Epub 1996/01/01. PubMed PMID: 8834792. [DOI] [PubMed] [Google Scholar]
- 115.Lambert S, Watson A, Sheedy DM, Martin B, Carr AM. Gross chromosomal rearrangements and elevated recombination at an inducible site-specific replication fork barrier. Cell. 2005;121(5):689–702. Epub 2005/06/07. doi: 10.1016/j.cell.2005.03.022. PubMed PMID: 15935756. [DOI] [PubMed] [Google Scholar]
- 116.Tolentino JH, Burke TJ, Mukhopadhyay S, McGregor WG, Basu AK. Inhibition of DNA replication fork progression and mutagenic potential of 1, N6-ethenoadenine and 8-oxoguanine in human cell extracts. Nucleic acids research. 2008;36(4):1300–8. doi: 10.1093/nar/gkm1157. PubMed PMID: 18184697; PMCID: PMC2275085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yeeles JT, Poli J, Marians KJ, Pasero P. Rescuing stalled or damaged replication forks. Cold Spring Harb Perspect Biol. 2013;5(5):a012815. doi: 10.1101/cshperspect.a012815. PubMed PMID: 23637285; PMCID: PMC3632063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nature cell biology. 2014;16(1):2–9. doi: 10.1038/ncb2897. PubMed PMID: 24366029; PMCID: PMC4354890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ponath V, Kaina B. Death of Monocytes through Oxidative Burst of Macrophages and Neutrophils: Killing in Trans. PloS one. 2017;12(1):e0170347. Epub 2017/01/19. doi: 10.1371/journal.pone.0170347. PubMed PMID: 28099491; PMCID: PMC5242493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.van Gent DC, Hoeijmakers JH, Kanaar R. Chromosomal stability and the DNA double-stranded break connection. Nature reviews Genetics. 2001;2(3):196–206. doi: 10.1038/35056049. PubMed PMID: 11256071. [DOI] [PubMed] [Google Scholar]
- 121.Bishop AJ, Schiestl RH. Homologous recombination as a mechanism for genome rearrangements: environmental and genetic effects. Human molecular genetics. 2000;9(16):2427–334. PubMed PMID: 11005798. [DOI] [PubMed] [Google Scholar]
- 122.Costantino L, Sotiriou SK, Rantala JK, Magin S, Mladenov E, Helleday T, Haber JE, Iliakis G, Kallioniemi OP, Halazonetis TD. Break-induced replication repair of damaged forks induces genomic duplications in human cells. Science. 2014;343(6166):88–91. doi: 10.1126/science.1243211. PubMed PMID: 24310611; PMCID: PMC4047655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Shibata A, Kamada N, Masumura K, Nohmi T, Kobayashi S, Teraoka H, Nakagama H, Sugimura T, Suzuki H, Masutani M. Parp-1 deficiency causes an increase of deletion mutations and insertions/rearrangements in vivo after treatment with an alkylating agent. Oncogene. 2005;24(8):1328–37. Epub 2004/12/21. doi: 10.1038/sj.onc.1208289. PubMed PMID: 15608683. [DOI] [PubMed] [Google Scholar]
- 124.Yang Y, Sterling J, Storici F, Resnick MA, Gordenin DA. Hypermutability of damaged single-strand DNA formed at double-strand breaks and uncapped telomeres in yeast Saccharomyces cerevisiae. PLoS genetics. 2008;4(11):e1000264. Epub 2008/11/22. doi: 10.1371/journal.pgen.1000264. PubMed PMID: 19023402; PMCID: PMC2577886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ogiwara H, Kohno T, Nakanishi H, Nagayama K, Sato M, Yokota J. Unbalanced translocation, a major chromosome alteration causing loss of heterozygosity in human lung cancer. Oncogene. 2008;27(35):4788–97. Epub 2008/04/15. doi: 10.1038/onc.2008.113. PubMed PMID: 18408757. [DOI] [PubMed] [Google Scholar]
- 126.Buisson R, Niraj J, Pauty J, Maity R, Zhao W, Coulombe Y, Sung P, Masson JY. Breast cancer proteins PALB2 and BRCA2 stimulate polymerase eta in recombination-associated DNA synthesis at blocked replication forks. Cell Rep. 2014;6(3):553–64. doi: 10.1016/j.celrep.2014.01.009. PubMed PMID: 24485656; PMCID: PMC4162405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.McIlwraith MJ, Vaisman A, Liu Y, Fanning E, Woodgate R, West SC. Human DNA polymerase eta promotes DNA synthesis from strand invasion intermediates of homologous recombination. Molecular cell. 2005;20(5):783–92. doi: 10.1016/j.molcel.2005.10.001. PubMed PMID: 16337601. [DOI] [PubMed] [Google Scholar]
- 128.Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145(4):529–42. Epub 2011/05/14. doi: 10.1016/j.cell.2011.03.041. PubMed PMID: 21565612; PMCID: PMC3261725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Liao H, Ji F, Helleday T, Ying S. Mechanisms for stalled replication fork stabilization: new targets for synthetic lethality strategies in cancer treatments. EMBO reports. 2018;19(9). Epub 2018/08/16. doi: 10.15252/embr.201846263. PubMed PMID: 30108055; PMCID: PMC6123652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Rickman K, Smogorzewska A. Advances in understanding DNA processing and protection at stalled replication forks. The Journal of cell biology. 2019;218(4):1096–107. Epub 2019/01/24. doi: 10.1083/jcb.201809012. PubMed PMID: 30670471; PMCID: PMC6446843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Marians KJ. Lesion Bypass and the Reactivation of Stalled Replication Forks. Annu Rev Biochem. 2018;87:217–38. Epub 2018/01/04. doi: 10.1146/annurev-biochem-062917-011921. PubMed PMID: 29298091; PMCID: PMC6419508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hedglin M, Benkovic SJ. Eukaryotic Translesion DNA Synthesis on the Leading and Lagging Strands: Unique Detours around the Same Obstacle. Chem Rev. 2017;117(12):7857–77. Epub 2017/05/13. doi: 10.1021/acs.chemrev.7b00046. PubMed PMID: 28497687; PMCID: PMC5662946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jain R, Aggarwal AK, Rechkoblit O. Eukaryotic DNA polymerases. Curr Opin Struct Biol. 2018;53:77–87. Epub 2018/07/14. doi: 10.1016/j.sbi.2018.06.003. PubMed PMID: 30005324. [DOI] [PubMed] [Google Scholar]
- 134.Schaaper RM. Base selection, proofreading, and mismatch repair during DNA replication in Escherichia coli. The Journal of biological chemistry. 1993;268(32):23762–5. Epub 1993/11/15. PubMed PMID: 8226906. [PubMed] [Google Scholar]
- 135.Kunkel TA. DNA replication fidelity. The Journal of biological chemistry. 2004;279(17):16895–8. doi: 10.1074/jbc.R400006200. PubMed PMID: 14988392. [DOI] [PubMed] [Google Scholar]
- 136.Olsson M, Lindahl T. Repair of alkylated DNA in Escherichia coli. Methyl group transfer from O6-methylguanine to a protein cysteine residue. The Journal of biological chemistry. 1980;255(22):10569–71. Epub 1980/11/25. PubMed PMID: 7000780. [PubMed] [Google Scholar]
- 137.Lindahl T, Demple B, Robins P. Suicide inactivation of the E. coli O6-methylguanine-DNA methyltransferase. The EMBO journal. 1982;1(11):1359–63. Epub 1982/01/01. PubMed PMID: 6765195; PMCID: PMC553217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Duncan T, Trewick SC, Koivisto P, Bates PA, Lindahl T, Sedgwick B. Reversal of DNA alkylation damage by two human dioxygenases. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(26):16660–5. doi: 10.1073/pnas.262589799. PubMed PMID: 12486230; PMCID: PMC139200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Begley TJ, Samson LD. AlkB mystery solved: oxidative demethylation of N1-methyladenine and N3-methylcytosine adducts by a direct reversal mechanism. Trends Biochem Sci. 2003;28(1):2–5. PubMed PMID: 12517444. [DOI] [PubMed] [Google Scholar]
- 140.Fu D, Calvo JA, Samson LD. Balancing repair and tolerance of DNA damage caused by alkylating agents. Nature reviews Cancer. 2012;12(2):104–20. doi: 10.1038/nrc3185. PubMed PMID: 22237395; PMCID: PMC3586545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ringvoll J, Moen MN, Nordstrand LM, Meira LB, Pang B, Bekkelund A, Dedon PC, Bjelland S, Samson LD, Falnes PO, Klungland A. AlkB homologue 2-mediated repair of ethenoadenine lesions in mammalian DNA. Cancer research. 2008;68(11):4142–9. Epub 2008/06/04. doi: 10.1158/0008-5472.CAN-08-0796. PubMed PMID: 18519673; PMCID: PMC4725713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kim YJ, Wilson DM 3rd. Overview of base excision repair biochemistry. Curr Mol Pharmacol. 2012;5(1):3–13. Epub 2011/11/30. PubMed PMID: 22122461; PMCID: PMC3459583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Krokan HE, Bjoras M. Base excision repair. Cold Spring Harb Perspect Biol. 2013;5(4):a012583. Epub 2013/04/03. doi: 10.1101/cshperspect.a012583. PubMed PMID: 23545420; PMCID: PMC3683898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Howard MJ, Wilson SH. DNA scanning by base excision repair enzymes and implications for pathway coordination. DNA repair. 2018;71:101–7. Epub 2018/09/06. doi: 10.1016/j.dnarep.2018.08.013. PubMed PMID: 30181039; PMCID: PMC6340770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Fortini P, Pascucci B, Parlanti E, Sobol RW, Wilson SH, Dogliotti E. Different DNA polymerases are involved in the short- and long-patch base excision repair in mammalian cells. Biochemistry. 1998;37(11):3575–80. doi: 10.1021/bi972999h. PubMed PMID: 9530283. [DOI] [PubMed] [Google Scholar]
- 146.Krishnamurthy N, Haraguchi K, Greenberg MM, David SS. Efficient removal of formamidopyrimidines by 8-oxoguanine glycosylases. Biochemistry. 2008;47(3):1043–50. doi: 10.1021/bi701619u. PubMed PMID: 18154319; PMCID: PMC2424258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Jacobs AL, Schar P. DNA glycosylases: in DNA repair and beyond. Chromosoma. 2012;121(1):1–20. doi: 10.1007/s00412-011-0347-4. PubMed PMID: 22048164; PMCID: PMC3260424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Engelward BP, Weeda G, Wyatt MD, Broekhof JL, de Wit J, Donker I, Allan JM, Gold B, Hoeijmakers JH, Samson LD. Base excision repair deficient mice lacking the Aag alkyladenine DNA glycosylase. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(24):13087–92. PubMed PMID: 9371804; PMCID: PMC24267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Michaels ML, Cruz C, Grollman AP, Miller JH. Evidence that MutY and MutM combine to prevent mutations by an oxidatively damaged form of guanine in DNA. Proceedings of the National Academy of Sciences of the United States of America. 1992;89(15):7022–5. Epub 1992/08/01. doi: 10.1073/pnas.89.15.7022. PubMed PMID: 1495996; PMCID: PMC49637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Al-Tassan N, Chmiel NH, Maynard J, Fleming N, Livingston AL, Williams GT, Hodges AK, Davies DR, David SS, Sampson JR, Cheadle JP. Inherited variants of MYH associated with somatic G:C-->T:A mutations in colorectal tumors. Nature genetics. 2002;30(2):227–32. doi: 10.1038/ng828. PubMed PMID: 11818965. [DOI] [PubMed] [Google Scholar]
- 151.Jones S, Emmerson P, Maynard J, Best JM, Jordan S, Williams GT, Sampson JR, Cheadle JP. Biallelic germline mutations in MYH predispose to multiple colorectal adenoma and somatic G:C-->T:A mutations. Human molecular genetics. 2002;11(23):2961–7. PubMed PMID: 12393807. [DOI] [PubMed] [Google Scholar]
- 152.Hailer MK, Slade PG, Martin BD, Rosenquist TA, Sugden KD. Recognition of the oxidized lesions spiroiminodihydantoin and guanidinohydantoin in DNA by the mammalian base excision repair glycosylases NEIL1 and NEIL2. DNA repair. 2005;4(1):41–50. doi: 10.1016/j.dnarep.2004.07.006. PubMed PMID: 15533836. [DOI] [PubMed] [Google Scholar]
- 153.Chakraborty A, Wakamiya M, Venkova-Canova T, Pandita RK, Aguilera-Aguirre L, Sarker AH, Singh DK, Hosoki K, Wood TG, Sharma G, Cardenas V, Sarkar PS, Sur S, Pandita TK, Boldogh I, Hazra TK. Neil2-null Mice Accumulate Oxidized DNA Bases in the Transcriptionally Active Sequences of the Genome and Are Susceptible to Innate Inflammation. The Journal of biological chemistry. 2015;290(41):24636–48. doi: 10.1074/jbc.M115.658146. PubMed PMID: 26245904; PMCID: PMC4598976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Krishnamurthy N, Zhao X, Burrows CJ, David SS. Superior removal of hydantoin lesions relative to other oxidized bases by the human DNA glycosylase hNEIL1. Biochemistry. 2008;47(27):7137–46. doi: 10.1021/bi800160s. PubMed PMID: 18543945; PMCID: PMC2574819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Trivedi RN, Almeida KH, Fornsaglio JL, Schamus S, Sobol RW. The role of base excision repair in the sensitivity and resistance to temozolomide-mediated cell death. Cancer research. 2005;65(14):6394–400. doi: 10.1158/0008-5472.CAN-05-0715. PubMed PMID: 16024643. [DOI] [PubMed] [Google Scholar]
- 156.Ensminger M, Iloff L, Ebel C, Nikolova T, Kaina B, Lbrich M. DNA breaks and chromosomal aberrations arise when replication meets base excision repair. The Journal of cell biology. 2014;206(1):29–43. doi: 10.1083/jcb.201312078. PubMed PMID: 24982429; PMCID: PMC4085701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Calvo JA, Moroski-Erkul CA, Lake A, Eichinger LW, Shah D, Jhun I, Limsirichai P, Bronson RT, Christiani DC, Meira LB, Samson LD. Aag DNA glycosylase promotes alkylation-induced tissue damage mediated by Parp1. PLoS genetics. 2013;9(4):e1003413. Epub 2013/04/18. doi: 10.1371/journal.pgen.1003413. PubMed PMID: 23593019; PMCID: 3617098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Calvo JA, Allocca M, Fake KR, Muthupalani S, Corrigan JJ, Bronson RT, Samson LD. Parp1 protects against Aag-dependent alkylation-induced nephrotoxicity in a sex-dependent manner. Oncotarget. 2016;7(29):44950–65. doi: 10.18632/oncotarget.10440. PubMed PMID: 27391435; PMCID: PMC5216697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Allocca M, Corrigan JJ, Fake KR, Calvo JA, Samson LD. PARP inhibitors protect against sex- and AAG-dependent alkylation-induced neural degeneration. Oncotarget. 2017;8(40):68707–20. doi: 10.18632/oncotarget.19844. PubMed PMID: 28978150; PMCID: PMC5620290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Krejci L, Altmannova V, Spirek M, Zhao X. Homologous recombination and its regulation. Nucleic acids research. 2012;40(13):5795–818. doi: 10.1093/nar/gks270. PubMed PMID: 22467216; PMCID: PMC3401455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kim JS, Krasieva TB, Kurumizaka H, Chen DJ, Taylor AM, Yokomori K. Independent and sequential recruitment of NHEJ and HR factors to DNA damage sites in mammalian cells. The Journal of cell biology. 2005;170(3):341–7. doi: 10.1083/jcb.200411083. PubMed PMID: 16061690; PMCID: PMC2171485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Chang HHY, Pannunzio NR, Adachi N, Lieber MR. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat Rev Mol Cell Biol. 2017;18(8):495–506. Epub 2017/05/18. doi: 10.1038/nrm.2017.48. PubMed PMID: 28512351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Sonoda E, Hochegger H, Saberi A, Taniguchi Y, Takeda S. Differential usage of non-homologous end-joining and homologous recombination in double strand break repair. DNA repair. 2006;5(9-10):1021–9. doi: 10.1016/j.dnarep.2006.05.022. PubMed PMID: 16807135. [DOI] [PubMed] [Google Scholar]
- 164.Takata M, Sasaki MS, Sonoda E, Morrison C, Hashimoto M, Utsumi H, Yamaguchi-Iwai Y, Shinohara A, Takeda S. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. The EMBO journal. 1998;17(18):5497–508. Epub 1998/09/16. doi: 10.1093/emboj/17.18.5497. PubMed PMID: 9736627; PMCID: PMC1170875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Helleday T, Lo J, van Gent DC, Engelward BP. DNA double-strand break repair: from mechanistic understanding to cancer treatment. DNA repair. 2007;6(7):923–35. doi: 10.1016/j.dnarep.2007.02.006. PubMed PMID: 17363343. [DOI] [PubMed] [Google Scholar]
- 166.Gupta PK, Sahota A, Boyadjiev SA, Bye S, Shao C, O'Neill JP, Hunter TC, Albertini RJ, Stambrook PJ, Tischfield JA. High frequency in vivo loss of heterozygosity is primarily a consequence of mitotic recombination. Cancer research. 1997;57(6):1188–93. Epub 1997/03/15. PubMed PMID: 9067291. [PubMed] [Google Scholar]
- 167.Bishop AJ, Schiestl RH. Homologous recombination as a mechanism of carcinogenesis. Biochimica et biophysica acta. 2001;1471(3):M109–21. Epub 2001/03/16. PubMed PMID: 11250067. [DOI] [PubMed] [Google Scholar]
- 168.Kolomietz E, Meyn MS, Pandita A, Squire JA. The role of Alu repeat clusters as mediators of recurrent chromosomal aberrations in tumors. Genes Chromosomes Cancer. 2002;35(2):97–112. doi: 10.1002/gcc.10111. PubMed PMID: 12203773. [DOI] [PubMed] [Google Scholar]
- 169.Ohshima K, Hattori M, Yada T, Gojobori T, Sakaki Y, Okada N. Whole-genome screening indicates a possible burst of formation of processed pseudogenes and Alu repeats by particular L1 subfamilies in ancestral primates. Genome Biol. 2003;4(11):R74. doi: 10.1186/gb-2003-4-11-r74. PubMed PMID: 14611660; PMCID: PMC329124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.de Koning AP, Gu W, Castoe TA, Batzer MA, Pollock DD. Repetitive elements may comprise over two-thirds of the human genome. PLoS genetics. 2011;7(12):e1002384. doi: 10.1371/journal.pgen.1002384. PubMed PMID: 22144907; PMCID: PMC3228813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Furmaga WB, Ryan JL, Coleman WB, Cole SR, Tsongalis GJ. Alu profiling of primary and metastatic nonsmall cell lung cancer. Exp Mol Pathol. 2003;74(3):224–9. PubMed PMID: 12782008. [DOI] [PubMed] [Google Scholar]
- 172.Gupta PK, Shao C, Zhu Y, Sahota A, Tischfield JA. Loss of heterozygosity analysis in a human fibrosarcoma cell line. Cytogenet Cell Genet. 1997;76(3-4):214–8. PubMed PMID: 9186527. [DOI] [PubMed] [Google Scholar]
- 173.Pal J, Bertheau R, Buon L, Qazi A, Batchu RB, Bandyopadhyay S, Ali-Fehmi R, Beer DG, Weaver DW, Shmookler Reis RJ, Goyal RK, Huang Q, Munshi NC, Shammas MA. Genomic evolution in Barrett's adenocarcinoma cells: critical roles of elevated hsRAD51, homologous recombination and Alu sequences in the genome. Oncogene. 2011;30(33):3585–98. Epub 2011/03/23. doi: 10.1038/onc.2011.83. PubMed PMID: 21423218; PMCID: 3406293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Strout MP, Marcucci G, Bloomfield CD, Caligiuri MA. The partial tandem duplication of ALL1 (MLL) is consistently generated by Alu-mediated homologous recombination in acute myeloid leukemia. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(5):2390–5. PubMed PMID: 9482895; PMCID: PMC19353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Roy R, Chun J, Powell SN. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nature reviews Cancer. 2012;12(1):68–78. doi: 10.1038/nrc3181. PubMed PMID: 22193408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Gu W, Zhang F, Lupski JR. Mechanisms for human genomic rearrangements. Pathogenetics. 2008;1(1):4. doi: 10.1186/1755-8417-1-4. PubMed PMID: 19014668; PMCID: PMC2583991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Sebesta M, Burkovics P, Juhasz S, Zhang S, Szabo JE, Lee MY, Haracska L, Krejci L. Role of PCNA and TLS polymerases in D-loop extension during homologous recombination in humans. DNA repair. 2013;12(9):691–8. doi: 10.1016/j.dnarep.2013.05.001. PubMed PMID: 23731732; PMCID: PMC3744802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Sharma S, Hicks JK, Chute CL, Brennan JR, Ahn JY, Glover TW, Canman CE. REV1 and polymerase zeta facilitate homologous recombination repair. Nucleic acids research. 2012;40(2):682–91. doi: 10.1093/nar/gkr769. PubMed PMID: 21926160; PMCID: PMC3258153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.McVey M, Khodaverdian VY, Meyer D, Cerqueira PG, Heyer WD. Eukaryotic DNA Polymerases in Homologous Recombination. Annu Rev Genet. 2016;50:393–421. doi: 10.1146/annurev-genet-120215-035243. PubMed PMID: 27893960; PMCID: PMC5295669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Arnaudeau C, Lundin C, Helleday T. DNA double-strand breaks associated with replication forks are predominantly repaired by homologous recombination involving an exchange mechanism in mammalian cells. Journal of molecular biology. 2001;307(5):1235–45. Epub 2001/04/09. doi: 10.1006/jmbi.2001.4564. PubMed PMID: 11292338. [DOI] [PubMed] [Google Scholar]
- 181.Kiraly O, Gong G, Roytman MD, Yamada Y, Samson LD, Engelward BP. DNA glycosylase activity and cell proliferation are key factors in modulating homologous recombination in vivo. Carcinogenesis. 2014;35(11):2495–502. doi: 10.1093/carcin/bgu177. PubMed PMID: 25155011; PMCID: PMC4216056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Chen CC, Feng W, Lim PX, Kass EM, Jasin M. Homology-Directed Repair and the Role of BRCA1, BRCA2, and Related Proteins in Genome Integrity and Cancer. Annu Rev Cancer Biol. 2018;2:313–36. Epub 2018/10/23. doi: 10.1146/annurev-cancerbio-030617-050502. PubMed PMID: 30345412; PMCID: PMC6193498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol. 2010;11(3):196–207. Epub 2010/02/24. doi: 10.1038/nrm2851. PubMed PMID: 20177395; PMCID: PMC3261768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Nickoloff JA. Paths from DNA damage and signaling to genome rearrangements via homologous recombination. Mutation research. 2017;806:64–74. doi: 10.1016/j.mrfmmm.2017.07.008. PubMed PMID: 28779875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Fu D, Samson LD. Direct repair of 3,N(4)-ethenocytosine by the human ALKBH2 dioxygenase is blocked by the AAG/MPG glycosylase. DNA repair. 2012;11(1):46–52. Epub 2011/11/15. doi: 10.1016/j.dnarep.2011.10.004. PubMed PMID: 22079122; PMCID: PMC3253959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Lips J, Kaina B. DNA double-strand breaks trigger apoptosis in p53-deficient fibroblasts. Carcinogenesis. 2001;22(4):579–85. Epub 2001/04/04. doi: 10.1093/carcin/22.4.579. PubMed PMID: 11285192. [DOI] [PubMed] [Google Scholar]
- 187.Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nature genetics. 2001;27(3):247–54. doi: 10.1038/85798. PubMed PMID: 11242102. [DOI] [PubMed] [Google Scholar]
- 188.Ebrahimkhani MR, Daneshmand A, Mazumder A, Allocca M, Calvo JA, Abolhassani N, Jhun I, Muthupalani S, Ayata C, Samson LD. Aag-initiated base excision repair promotes ischemia reperfusion injury in liver, brain, and kidney. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(45):E4878–86. doi: 10.1073/pnas.1413582111. PubMed PMID: 25349415; PMCID: PMC4234618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Coquerelle T, Dosch J, Kaina B. Overexpression of N-methylpurine-DNA glycosylase in Chinese hamster ovary cells renders them more sensitive to the production of chromosomal aberrations by methylating agents--a case of imbalanced DNA repair. Mutation research. 1995;336(1):9–17. PubMed PMID: 7528899. [DOI] [PubMed] [Google Scholar]
- 190.Klapacz J, Lingaraju GM, Guo HH, Shah D, Moar-Shoshani A, Loeb LA, Samson LD. Frameshift mutagenesis and microsatellite instability induced by human alkyladenine DNA glycosylase. Molecular cell. 2010;37(6):843–53. doi: 10.1016/j.molcel.2010.01.038. PubMed PMID: 20347426; PMCID: PMC2894629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Crosbie PA, Watson AJ, Agius R, Barber PV, Margison GP, Povey AC. Elevated N3-methylpurine-DNA glycosylase DNA repair activity is associated with lung cancer. Mutation research. 2012;732(1-2):43–6. doi: 10.1016/j.mrfmmm.2012.01.001. PubMed PMID: 22266085. [DOI] [PubMed] [Google Scholar]
- 192.Leitner-Dagan Y, Sevilya Z, Pinchev M, Kramer R, Elinger D, Roisman LC, Rennert HS, Schechtman E, Freedman L, Rennert G, Livneh Z, Paz-Elizur T. N-methylpurine DNA glycosylase and OGG1 DNA repair activities: opposite associations with lung cancer risk. J Natl Cancer Inst. 2012;104(22):1765–9. doi: 10.1093/jnci/djs445. PubMed PMID: 23104324; PMCID: PMC3502197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Liu C, Tu Y, Yuan J, Mao X, He S, Wang L, Fu G, Zong J, Zhang Y. Aberrant expression of N-methylpurine-DNA glycosylase influences patient survival in malignant gliomas. J Biomed Biotechnol. 2012;2012:760679. doi: 10.1155/2012/760679. PubMed PMID: 22496614; PMCID: PMC3303893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Zdzalik D, Domanska A, Prorok P, Kosicki K, van den Born E, Falnes PO, Rizzo CJ, Guengerich FP, Tudek B. Differential repair of etheno-DNA adducts by bacterial and human AlkB proteins. DNA repair. 2015;30:1–10. doi: 10.1016/j.dnarep.2015.02.021. PubMed PMID: 25797601; PMCID: PMC4451939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Shafirovich V, Kropachev K, Anderson T, Liu Z, Kolbanovskiy M, Martin BD, Sugden K, Shim Y, Chen X, Min JH, Geacintov NE. Base and Nucleotide Excision Repair of Oxidatively Generated Guanine Lesions in DNA. The Journal of biological chemistry. 2016;291(10):5309–19. doi: 10.1074/jbc.M115.693218. PubMed PMID: 26733197; PMCID: PMC4777862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.McKibbin PL, Fleming AM, Towheed MA, Van Houten B, Burrows CJ, David SS. Repair of hydantoin lesions and their amine adducts in DNA by base and nucleotide excision repair. Journal of the American Chemical Society. 2013;135(37):13851–61. doi: 10.1021/ja4059469. PubMed PMID: 23930966; PMCID: PMC3906845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Escribano-Diaz C, Orthwein A, Fradet-Turcotte A, Xing M, Young JT, Tkac J, Cook MA, Rosebrock AP, Munro M, Canny MD, Xu D, Durocher D. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Molecular cell. 2013;49(5):872–83. doi: 10.1016/j.molcel.2013.01.001. PubMed PMID: 23333306. [DOI] [PubMed] [Google Scholar]
- 198.Chapman JR, Taylor MR, Boulton SJ. Playing the end game: DNA double-strand break repair pathway choice. Molecular cell. 2012;47(4):497–510. doi: 10.1016/j.molcel.2012.07.029. PubMed PMID: 22920291. [DOI] [PubMed] [Google Scholar]
- 199.Shibata A, Moiani D, Arvai AS, Perry J, Harding SM, Genois MM, Maity R, van Rossum-Fikkert S, Kertokalio A, Romoli F, Ismail A, Ismalaj E, Petricci E, Neale MJ, Bristow RG, Masson JY, Wyman C, Jeggo PA, Tainer JA. DNA double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Molecular cell. 2014;53(1):7–18. doi: 10.1016/j.molcel.2013.11.003. PubMed PMID: 24316220; PMCID: PMC3909494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Jachimowicz RD, Goergens J, Reinhardt HC. DNA double-strand break repair pathway choice - from basic biology to clinical exploitation. Cell Cycle. 2019:1–12. Epub 2019/05/23. doi: 10.1080/15384101.2019.1618542. PubMed PMID: 31116084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18(1):134–47. Epub 2007/12/25. doi: 10.1038/cr.2007.111. PubMed PMID: 18157161. [DOI] [PubMed] [Google Scholar]
- 202.Kondo N, Takahashi A, Ono K, Ohnishi T. DNA damage induced by alkylating agents and repair pathways. J Nucleic Acids. 2010;2010:543531. doi: 10.4061/2010/543531. PubMed PMID: 21113301; PMCID: PMC2989456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Hofseth LJ, Khan MA, Ambrose M, Nikolayeva O, Xu-Welliver M, Kartalou M, Hussain SP, Roth RB, Zhou X, Mechanic LE, Zurer I, Rotter V, Samson LD, Harris CC. The adaptive imbalance in base excision-repair enzymes generates microsatellite instability in chronic inflammation. The Journal of clinical investigation. 2003;112(12):1887–94. Epub 2003/12/18. doi: 10.1172/JCI19757. PubMed PMID: 14679184; PMCID: 296999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421(6922):499–506. Epub 2003/01/31. doi: 10.1038/nature01368. PubMed PMID: 12556884. [DOI] [PubMed] [Google Scholar]
- 205.Bryant HE, Helleday T. Inhibition of poly (ADP-ribose) polymerase activates ATM which is required for subsequent homologous recombination repair. Nucleic acids research. 2006;34(6):1685–91. Epub 2006/03/25. doi: 10.1093/nar/gkl108. PubMed PMID: 16556909; PMCID: PMC1410911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Banin S, Moyal L, Shieh S, Taya Y, Anderson CW, Chessa L, Smorodinsky NI, Prives C, Reiss Y, Shiloh Y, Ziv Y. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281(5383):1674–7. Epub 1998/09/11. PubMed PMID: 9733514. [DOI] [PubMed] [Google Scholar]
- 207.Barlow C, Brown KD, Deng CX, Tagle DA, Wynshaw-Boris A. Atm selectively regulates distinct p53-dependent cell-cycle checkpoint and apoptotic pathways. Nature genetics. 1997;17(4):453–6. Epub 1997/12/17. doi: 10.1038/ng1297-453. PubMed PMID: 9398849. [DOI] [PubMed] [Google Scholar]
- 208.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316(5828):1160–6. Epub 2007/05/26. doi: 10.1126/science.1140321. PubMed PMID: 17525332. [DOI] [PubMed] [Google Scholar]
- 209.Lavin MF. Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signalling and cancer. Nat Rev Mol Cell Biol. 2008;9(10):759–69. Epub 2008/09/25. doi: 10.1038/nrm2514. PubMed PMID: 18813293. [DOI] [PubMed] [Google Scholar]
- 210.Guo Z, Kozlov S, Lavin MF, Person MD, Paull TT. ATM activation by oxidative stress. Science. 2010;330(6003):517–21. Epub 2010/10/23. doi: 10.1126/science.1192912. PubMed PMID: 20966255. [DOI] [PubMed] [Google Scholar]
- 211.Guo Z, Deshpande R, Paull TT. ATM activation in the presence of oxidative stress. Cell Cycle. 2010;9(24):4805–11. Epub 2010/12/15. doi: 10.4161/cc.9.24.14323. PubMed PMID: 21150274; PMCID: PMC3047807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Fang L, Choudhary S, Zhao Y, Edeh CB, Yang C, Boldogh I, Brasier AR. ATM regulates NF-kappaB-dependent immediate-early genes via RelA Ser 276 phosphorylation coupled to CDK9 promoter recruitment. Nucleic acids research. 2014;42(13):8416–32. Epub 2014/06/25. doi: 10.1093/nar/gku529. PubMed PMID: 24957606; PMCID: PMC4117761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Ditch S, Paull TT. The ATM protein kinase and cellular redox signaling: beyond the DNA damage response. Trends Biochem Sci. 2012;37(1):15–22. Epub 2011/11/15. doi: 10.1016/j.tibs.2011.10.002. PubMed PMID: 22079189; PMCID: PMC3259275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Lin R, Xiao D, Guo Y, Tian D, Yun H, Chen D, Su M. Chronic inflammation-related DNA damage response: a driving force of gastric cardia carcinogenesis. Oncotarget. 2015;6(5):2856–64. doi: 10.18632/oncotarget.3091. PubMed PMID: 25650663; PMCID: PMC4413622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.He H, Tian D, Guo J, Liu M, Chen Z, Hamdy FC, Helleday T, Su M, Ying S. DNA damage response in peritumoral regions of oesophageal cancer microenvironment. Carcinogenesis. 2013;34(1):139–45. doi: 10.1093/carcin/bgs301. PubMed PMID: 23027622. [DOI] [PubMed] [Google Scholar]
- 216.Jayakumar S, Pal D, Sandur SK. Nrf2 facilitates repair of radiation induced DNA damage through homologous recombination repair pathway in a ROS independent manner in cancer cells. Mutation research. 2015;779:33–45. doi: 10.1016/j.mrfmmm.2015.06.007. PubMed PMID: 26133502. [DOI] [PubMed] [Google Scholar]
- 217.Frontini M, Vijayakumar M, Garvin A, Clarke N. A ChIP-chip approach reveals a novel role for transcription factor IRF1 in the DNA damage response. Nucleic acids research. 2009;37(4):1073–85. doi: 10.1093/nar/gkn1051. PubMed PMID: 19129219; PMCID: PMC2651779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Volcic M, Karl S, Baumann B, Salles D, Daniel P, Fulda S, Wiesmuller L. NF-kappaB regulates DNA double-strand break repair in conjunction with BRCA1-CtIP complexes. Nucleic acids research. 2012;40(1):181–95. doi: 10.1093/nar/gkr687. PubMed PMID: 21908405; PMCID: PMC3245919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Li N, Parrish M, Chan TK, Yin L, Rai P, Yoshiyuki Y, Abolhassani N, Tan KB, Kiraly O, Chow VT, Engelward BP. Influenza infection induces host DNA damage and dynamic DNA damage responses during tissue regeneration. Cell Mol Life Sci. 2015;72(15):2973–88. doi: 10.1007/s00018-015-1879-1. PubMed PMID: 25809161; PMCID: PMC4802977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Mani RS, Amin MA, Li X, Kalyana-Sundaram S, Veeneman BA, Wang L, Ghosh A, Aslam A, Ramanand SG, Rabquer BJ, Kimura W, Tran M, Cao X, Roychowdhury S, Dhanasekaran SM, Palanisamy N, Sadek HA, Kapur P, Koch AE, Chinnaiyan AM. Inflammation-Induced Oxidative Stress Mediates Gene Fusion Formation in Prostate Cancer. Cell Rep. 2016;17(10):2620–31. doi: 10.1016/j.celrep.2016.11.019. PubMed PMID: 27926866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Chan MK, Ocampo-Hafalla MT, Vartanian V, Jaruga P, Kirkali G, Koenig KL, Brown S, Lloyd RS, Dizdaroglu M, Teebor GW. Targeted deletion of the genes encoding NTH1 and NEIL1 DNA N-glycosylases reveals the existence of novel carcinogenic oxidative damage to DNA. DNA repair. 2009;8(7):786–94. doi: 10.1016/j.dnarep.2009.03.001. PubMed PMID: 19346169; PMCID: PMC4894318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Klungland A, Rosewell I, Hollenbach S, Larsen E, Daly G, Epe B, Seeberg E, Lindahl T, Barnes DE. Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(23):13300–5. PubMed PMID: 10557315; PMCID: PMC23942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Zhang DD, Hannink M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Molecular and cellular biology. 2003;23(22):8137–51. Epub 2003/10/31. doi: 10.1128/mcb.23.22.8137-8151.2003. PubMed PMID: 14585973; PMCID: PMC262403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Wakabayashi N, Dinkova-Kostova AT, Holtzclaw WD, Kang MI, Kobayashi A, Yamamoto M, Kensler TW, Talalay P. Protection against electrophile and oxidant stress by induction of the phase 2 response: fate of cysteines of the Keap1 sensor modified by inducers. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(7):2040–5. Epub 2004/02/07. doi: 10.1073/pnas.0307301101. PubMed PMID: 14764894; PMCID: PMC357048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Velichkova M, Hasson T. Keap1 regulates the oxidation-sensitive shuttling of Nrf2 into and out of the nucleus via a Crm1-dependent nuclear export mechanism. Molecular and cellular biology. 2005;25(11):4501–13. Epub 2005/05/19. doi: 10.1128/MCB.25.11.4501-4513.2005. PubMed PMID: 15899855; PMCID: PMC1140621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Rushmore TH, Morton MR, Pickett CB. The antioxidant responsive element. Activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. The Journal of biological chemistry. 1991. ;266(18):11632–9. PubMed PMID: 1646813. [PubMed] [Google Scholar]
- 227.Ma Q Role of nrf2 in oxidative stress and toxicity. Annu Rev Pharmacol Toxicol. 2013;53:401–26. doi: 10.1146/annurev-pharmtox-011112-140320. PubMed PMID: 23294312; PMCID: PMC4680839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Kobayashi M, Yamamoto M. Nrf2-Keap1 regulation of cellular defense mechanisms against electrophiles and reactive oxygen species. Adv Enzyme Regul. 2006;46:113–40. doi: 10.1016/j.advenzreg.2006.01.007. PubMed PMID: 16887173. [DOI] [PubMed] [Google Scholar]
- 229.Rushmore TH, King RG, Paulson KE, Pickett CB. Regulation of glutathione S-transferase Ya subunit gene expression: identification of a unique xenobiotic-responsive element controlling inducible expression by planar aromatic compounds. Proceedings of the National Academy of Sciences of the United States of America. 1990;87(10):3826–30. Epub 1990/05/01. doi: 10.1073/pnas.87.10.3826. PubMed PMID: 2160079; PMCID: PMC53996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Friling RS, Bensimon A, Tichauer Y, Daniel V. Xenobiotic-inducible expression of murine glutathione S-transferase Ya subunit gene is controlled by an electrophile-responsive element. Proceedings of the National Academy of Sciences of the United States of America. 1990;87(16):6258–62. Epub 1990/08/01. doi: 10.1073/pnas.87.16.6258. PubMed PMID: 2166952; PMCID: PMC54512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Favreau LV, Pickett CB. Transcriptional regulation of the rat NAD(P)H:quinone reductase gene. Identification of regulatory elements controlling basal level expression and inducible expression by planar aromatic compounds and phenolic antioxidants. The Journal of biological chemistry. 1991;266(7):4556–61. Epub 1991/03/05. PubMed PMID: 1900296. [PubMed] [Google Scholar]
- 232.Li Y, Jaiswal AK. Regulation of human NAD(P)H:quinone oxidoreductase gene. Role of AP1 binding site contained within human antioxidant response element. The Journal of biological chemistry. 1992;267(21):15097–104. Epub 1992/07/25. PubMed PMID: 1340765. [PubMed] [Google Scholar]
- 233.Dreger H, Westphal K, Weller A, Baumann G, Stangl V, Meiners S, Stangl K. Nrf2-dependent upregulation of antioxidative enzymes: a novel pathway for proteasome inhibitor-mediated cardioprotection. Cardiovasc Res. 2009;83(2):354–61. Epub 2009/04/09. doi: 10.1093/cvr/cvp107. PubMed PMID: 19351736. [DOI] [PubMed] [Google Scholar]
- 234.Kwak MK, Itoh K, Yamamoto M, Sutter TR, Kensler TW. Role of transcription factor Nrf2 in the induction of hepatic phase 2 and antioxidative enzymes in vivo by the cancer chemoprotective agent, 3H-1, 2-dimethiole-3-thione. Molecular medicine. 2001;7(2):135–45. Epub 2001/07/27. PubMed PMID: 11471548; PMCID: PMC1950021. [PMC free article] [PubMed] [Google Scholar]
- 235.Nguyen T, Nioi P, Pickett CB. The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. The Journal of biological chemistry. 2009;284(20):13291–5. doi: 10.1074/jbc.R900010200. PubMed PMID: 19182219; PMCID: PMC2679427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Matsuoka Y, Nakayama H, Yoshida R, Hirosue A, Nagata M, Tanaka T, Kawahara K, Sakata J, Arita H, Nakashima H, Shinriki S, Fukuma D, Ogi H, Hiraki A, Shinohara M, Toya R, Murakami R. IL-6 controls resistance to radiation by suppressing oxidative stress via the Nrf2-antioxidant pathway in oral squamous cell carcinoma. British journal of cancer. 2016;115(10):1234–44. doi: 10.1038/bjc.2016.327. PubMed PMID: 27736845; PMCID: PMC5104896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Chen Y, Zhang F, Tsai Y, Yang X, Yang L, Duan S, Wang X, Keng P, Lee SO. IL-6 signaling promotes DNA repair and prevents apoptosis in CD133+ stem-like cells of lung cancer after radiation. Radiat Oncol. 2015;10:227. doi: 10.1186/s13014-015-0534-1. PubMed PMID: 26572130; PMCID: PMC4647293. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 238.Duan S, Tsai Y, Keng P, Chen Y, Lee SO, Chen Y. IL-6 signaling contributes to cisplatin resistance in non-small cell lung cancer via the up-regulation of anti-apoptotic and DNA repair associated molecules. Oncotarget. 2015;6(29):27651–60. Epub 2015/08/28. doi: 10.18632/oncotarget.4753. PubMed PMID: 26313152; PMCID: PMC4695015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Kim SB, Pandita RK, Eskiocak U, Ly P, Kaisani A, Kumar R, Cornelius C, Wright WE, Pandita TK, Shay JW. Targeting of Nrf2 induces DNA damage signaling and protects colonic epithelial cells from ionizing radiation. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(43):E2949–55. doi: 10.1073/pnas.1207718109. PubMed PMID: 23045680; PMCID: PMC3491493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Singh B, Chatterjee A, Ronghe AM, Bhat NK, Bhat HK. Antioxidant-mediated up-regulation of OGG1 via NRF2 induction is associated with inhibition of oxidative DNA damage in estrogen-induced breast cancer. BMC cancer. 2013;13:253. doi: 10.1186/1471-2407-13-253. PubMed PMID: 23697596; PMCID: PMC3665669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Aoki Y, Hashimoto AH, Amanuma K, Matsumoto M, Hiyoshi K, Takano H, Masumura K, Itoh K, Nohmi T, Yamamoto M. Enhanced spontaneous and benzo(a)pyrene-induced mutations in the lung of Nrf2-deficient gpt delta mice. Cancer research. 2007;67(12):5643–8. doi: 10.1158/0008-5472.CAN-06-3355. PubMed PMID: 17575130. [DOI] [PubMed] [Google Scholar]
- 242.Kwak MK, Egner PA, Dolan PM, Ramos-Gomez M, Groopman JD, Itoh K, Yamamoto M, Kensler TW. Role of phase 2 enzyme induction in chemoprotection by dithiolethiones. Mutation research. 2001;480-481:305–15. PubMed PMID: 11506823. [DOI] [PubMed] [Google Scholar]
- 243.Ramos-Gomez M, Dolan PM, Itoh K, Yamamoto M, Kensler TW. Interactive effects of nrf2 genotype and oltipraz on benzo[a]pyrene-DNA adducts and tumor yield in mice. Carcinogenesis. 2003;24(3):461–7. PubMed PMID: 12663505. [DOI] [PubMed] [Google Scholar]
- 244.Ba X, Gupta S, Davidson M, Garg NJ. Trypanosoma cruzi induces the reactive oxygen species-PARP-1-RelA pathway for up-regulation of cytokine expression in cardiomyocytes. The Journal of biological chemistry. 2010;285(15):11596–606. doi: 10.1074/jbc.M109.076984. PubMed PMID: 20145242; PMCID: PMC2857037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Stilmann M, Hinz M, Arslan SC, Zimmer A, Schreiber V, Scheidereit C. A nuclear poly(ADP-ribose)-dependent signalosome confers DNA damage-induced IkappaB kinase activation. Molecular cell. 2009;36(3):365–78. doi: 10.1016/j.molcel.2009.09.032. PubMed PMID: 19917246. [DOI] [PubMed] [Google Scholar]
- 246.Kameoka M, Ota K, Tetsuka T, Tanaka Y, Itaya A, Okamoto T, Yoshihara K. Evidence for regulation of NF-kappaB by poly(ADP-ribose) polymerase. The Biochemical journal. 2000;346 Pt 3:641–9. PubMed PMID: 10698690; PMCID: PMC1220896. [PMC free article] [PubMed] [Google Scholar]
- 247.Pamment J, Ramsay E, Kelleher M, Dornan D, Ball KL. Regulation of the IRF-1 tumour modifier during the response to genotoxic stress involves an ATM-dependent signalling pathway. Oncogene. 2002;21(51):7776–85. doi: 10.1038/sj.onc.1205981. PubMed PMID: 12420214. [DOI] [PubMed] [Google Scholar]
- 248.Kim TK, Kim T, Kim TY, Lee WG, Yim J. Chemotherapeutic DNA-damaging drugs activate interferon regulatory factor-7 by the mitogen-activated protein kinase kinase-4-cJun NH2-terminal kinase pathway. Cancer research. 2000;60(5):1153–6. PubMed PMID: 10728664. [PubMed] [Google Scholar]
- 249.Lee RB, Hassane DC, Cottle DL, Pickett CL. Interactions of Campylobacter jejuni cytolethal distending toxin subunits CdtA and CdtC with HeLa cells. Infection and immunity. 2003;71(9):4883–90. PubMed PMID: 12933829; PMCID: 187314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Cuevas-Ramos G, Petit CR, Marcq I, Boury M, Oswald E, Nougayrede JP. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(25):11537–42. doi: 10.1073/pnas.1001261107. PubMed PMID: 20534522; PMCID: PMC2895108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Ge Z, Schauer DB, Fox JG. In vivo virulence properties of bacterial cytolethal-distending toxin. Cellular microbiology. 2008;10(8):1599–607. Epub 2008/05/21. doi: 10.1111/j.1462-5822.2008.01173.x. PubMed PMID: 18489725. [DOI] [PubMed] [Google Scholar]
- 252.Chan TK, Loh XY, Peh HY, Tan WN, Tan WS, Li N, Tay IJ, Wong WS, Engelward BP. House dust mite-induced asthma causes oxidative damage and DNA double-strand breaks in the lungs. J Allergy Clin Immunol. 2016;138(1):84–96 e1. doi: 10.1016/j.jaci.2016.02.017. PubMed PMID: 27157131. [DOI] [PubMed] [Google Scholar]
- 253.Rai P, He F, Kwang J, Engelward BP, Chow VT. Pneumococcal Pneumolysin Induces DNA Damage and Cell Cycle Arrest. Sci Rep. 2016;6:22972. doi: 10.1038/srep22972. PubMed PMID: 27026501; PMCID: PMC4812240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, Cordon-Cardo C, Lowe SW. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445(7128):656–60. doi: 10.1038/nature05529. PubMed PMID: 17251933; PMCID: PMC4601097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Rodier F, Coppe JP, Patil CK, Hoeijmakers WA, Munoz DP, Raza SR, Freund A, Campeau E, Davalos AR, Campisi J. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nature cell biology. 2009;11(8):973–9. Epub 2009/07/15. doi: 10.1038/ncb1909. PubMed PMID: 19597488; PMCID: 2743561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Biton S, Ashkenazi A. NEMO and RIP1 control cell fate in response to extensive DNA damage via TNF-alpha feedforward signaling. Cell. 2011;145(1):92–103. doi: 10.1016/j.cell.2011.02.023. PubMed PMID: 21458669. [DOI] [PubMed] [Google Scholar]
- 257.Brzostek-Racine S, Gordon C, Van Scoy S, Reich NC. The DNA damage response induces IFN. J Immunol. 2011;187(10):5336–45. doi: 10.4049/jimmunol.1100040. PubMed PMID: 22013119; PMCID: PMC3246365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Yang X, Wang H, Zhang M, Liu J, Lv B, Chen F. HMGB1: a novel protein that induced platelets active and aggregation via Toll-like receptor-4, NF-kappaB and cGMP dependent mechanisms. Diagnostic pathology. 2015;10:134. Epub 2015/08/08. doi: 10.1186/s13000-015-0348-3. PubMed PMID: 26245198; PMCID: 4527107. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 259.Aggen JB, Nairn AC, Chamberlin R. Regulation of protein phosphatase-1. Chemistry & biology. 2000;7(1):R13–23. PubMed PMID: 10662690. [DOI] [PubMed] [Google Scholar]
- 260.Kim SB, Bozeman RG, Kaisani A, Kim W, Zhang L, Richardson JA, Wright WE, Shay JW. Radiation promotes colorectal cancer initiation and progression by inducing senescence-associated inflammatory responses. Oncogene. 2016;35(26):3365–75. doi: 10.1038/onc.2015.395. PubMed PMID: 26477319; PMCID: PMC4837107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Davidovich P, Kearney CJ, Martin SJ. Inflammatory outcomes of apoptosis, necrosis and necroptosis. Biol Chem. 2014;395(10):1163–71. doi: 10.1515/hsz-2014-0164. PubMed PMID: 25153241. [DOI] [PubMed] [Google Scholar]
- 262.Ba X, Bacsi A, Luo J, Aguilera-Aguirre L, Zeng X, Radak Z, Brasier AR, Boldogh I. 8-oxoguanine DNA glycosylase-1 augments proinflammatory gene expression by facilitating the recruitment of site-specific transcription factors. J Immunol. 2014;192(5):2384–94. doi: 10.4049/jimmunol.1302472. PubMed PMID: 24489103; PMCID: PMC3943862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Aguilera-Aguirre L, Bacsi A, Radak Z, Hazra TK, Mitra S, Sur S, Brasier AR, Ba X, Boldogh I. Innate inflammation induced by the 8-oxoguanine DNA glycosylase-1-KRAS-NF-kappaB pathway. J Immunol. 2014;193(9):4643–53. doi: 10.4049/jimmunol.1401625. PubMed PMID: 25267977; PMCID: PMC4201976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Visnes T, Cazares-Korner A, Hao W, Wallner O, Masuyer G, Loseva O, Mortusewicz O, Wiita E, Sarno A, Manoilov A, Astorga-Wells J, Jemth AS, Pan L, Sanjiv K, Karsten S, Gokturk C, Grube M, Homan EJ, Hanna BMF, Paulin CBJ, Pham T, Rasti A, Berglund UW, von Nicolai C, Benitez-Buelga C, Koolmeister T, Ivanic D, Iliev P, Scobie M, Krokan HE, Baranczewski P, Artursson P, Altun M, Jensen AJ, Kalderen C, Ba X, Zubarev RA, Stenmark P, Boldogh I, Helleday T. Small-molecule inhibitor of OGG1 suppresses proinflammatory gene expression and inflammation. Science. 2018;362(6416):834–9. doi: 10.1126/science.aar8048. PubMed PMID: 30442810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Chaudhry MA. Bystander effect: biological endpoints and microarray analysis. Mutation research. 2006;597(1-2):98–112. doi: 10.1016/j.mrfmmm.2005.04.023. PubMed PMID: 16414093. [DOI] [PubMed] [Google Scholar]
- 266.Nagasawa H, Little JB. Unexpected sensitivity to the induction of mutations by very low doses of alpha-particle radiation: evidence for a bystander effect. Radiation research. 1999;152(5):552–7. PubMed PMID: 10521933. [PubMed] [Google Scholar]
- 267.Rugo RE, Mutamba JT, Mohan KN, Yee T, Chaillet JR, Greenberger JS, Engelward BP. Methyltransferases mediate cell memory of a genotoxic insult. Oncogene. 2011;30(6):751–6. doi: 10.1038/onc.2010.480. PubMed PMID: 21057543; PMCID: 3044496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Rugo RE, Almeida KH, Hendricks CA, Jonnalagadda VS, Engelward BP. A single acute exposure to a chemotherapeutic agent induces hyper-recombination in distantly descendant cells and in their neighbors. Oncogene. 2005;24(32):5016–25. doi: 10.1038/sj.onc.1208690. PubMed PMID: 15856014. [DOI] [PubMed] [Google Scholar]
- 269.Rao VP, Poutahidis T, Ge Z, Nambiar PR, Boussahmain C, Wang YY, Horwitz BH, Fox JG, Erdman SE. Innate immune inflammatory response against enteric bacteria Helicobacter hepaticus induces mammary adenocarcinoma in mice. Cancer research. 2006;66(15):7395–400. Epub 2006/08/04. doi: 10.1158/0008-5472.CAN-06-0558. PubMed PMID: 16885333. [DOI] [PubMed] [Google Scholar]
- 270.Westbrook AM, Wei B, Braun J, Schiestl RH. Intestinal mucosal inflammation leads to systemic genotoxicity in mice. Cancer research. 2009;69(11):4827–34. doi: 10.1158/0008-5472.CAN-08-4416. PubMed PMID: 19487293; PMCID: 2709766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Westbrook AM, Wei B, Braun J, Schiestl RH. Intestinal inflammation induces genotoxicity to extraintestinal tissues and cell types in mice. International journal of cancer Journal international du cancer. 2011;129(8):1815–25. Epub 2011/04/27. doi: 10.1002/ijc.26146. PubMed PMID: 21520038; PMCID: 3197753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Poutahidis T, Cappelle K, Levkovich T, Lee CW, Doulberis M, Ge Z, Fox JG, Horwitz BH, Erdman SE. Pathogenic intestinal bacteria enhance prostate cancer development via systemic activation of immune cells in mice. PloS one. 2013;8(8):e73933. doi: 10.1371/journal.pone.0073933. PubMed PMID: 23991210; PMCID: PMC3753256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Satoh MS, Poirier GG, Lindahl T. Dual function for poly(ADP-ribose) synthesis in response to DNA strand breakage. Biochemistry. 1994;33(23):7099–106. PubMed PMID: 8003475. [DOI] [PubMed] [Google Scholar]
- 274.Langelier MF, Planck JL, Roy S, Pascal JM. Structural basis for DNA damage-dependent poly(ADP-ribosyl)ation by human PARP-1. Science. 2012;336(6082):728–32. doi: 10.1126/science.1216338. PubMed PMID: 22582261; PMCID: PMC3532513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Bryant HE, Petermann E, Schultz N, Jemth AS, Loseva O, Issaeva N, Johansson F, Fernandez S, McGlynn P, Helleday T. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. The EMBO journal. 2009;28(17):2601–15. doi: 10.1038/emboj.2009.206. PubMed PMID: 19629035; PMCID: PMC2738702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Hegde ML, Hazra TK, Mitra S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res. 2008;18(1):27–47. doi: 10.1038/cr.2008.8. PubMed PMID: 18166975; PMCID: PMC2692221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Haince JF, Kozlov S, Dawson VL, Dawson TM, Hendzel MJ, Lavin MF, Poirier GG. Ataxia telangiectasia mutated (ATM) signaling network is modulated by a novel poly(ADP-ribose)-dependent pathway in the early response to DNA-damaging agents. The Journal of biological chemistry. 2007;282(22):16441–53. doi: 10.1074/jbc.M608406200. PubMed PMID: 17428792. [DOI] [PubMed] [Google Scholar]
- 278.Oliver FJ, Menissier-de Murcia J, Nacci C, Decker P, Andriantsitohaina R, Muller S, de la Rubia G, Stoclet JC, de Murcia G. Resistance to endotoxic shock as a consequence of defective NF-kappaB activation in poly (ADP-ribose) polymerase-1 deficient mice. The EMBO journal. 1999;18(16):4446–54. doi: 10.1093/emboj/18.16.4446. PubMed PMID: 10449410; PMCID: PMC1171519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Altmeyer M, Barthel M, Eberhard M, Rehrauer H, Hardt WD, Hottiger MO. Absence of poly(ADP-ribose) polymerase 1 delays the onset of Salmonella enterica serovar Typhimurium-induced gut inflammation. Infection and immunity. 2010;78(8):3420–31. doi: 10.1128/IAI.00211-10. PubMed PMID: 20515923; PMCID: PMC2916260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Mazzon E, Genovese T, Di Paola R, Muia C, Crisafulli C, Malleo G, Esposito E, Meli R, Sessa E, Cuzzocrea S. Effects of 3-aminobenzamide, an inhibitor of poly (ADP-ribose) polymerase, in a mouse model of acute pancreatitis induced by cerulein. Eur J Pharmacol. 2006;549(1-3):149–56. doi: 10.1016/j.ejphar.2006.08.008. PubMed PMID: 16979620. [DOI] [PubMed] [Google Scholar]
- 281.Chopra M, Sharma AC. Distinct cardiodynamic and molecular characteristics during early and late stages of sepsis-induced myocardial dysfunction. Life sciences. 2007;81(4):306–16. doi: 10.1016/j.lfs.2007.05.021. PubMed PMID: 17612571; PMCID: PMC1986677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Czapski GA, Cakala M, Gajkowska B, Strosznajder JB. Poly(ADP-ribose) polymerase-1 inhibition protects the brain against systemic inflammation. Neurochem Int. 2006;49(8):751–5. doi: 10.1016/j.neuint.2006.06.006. PubMed PMID: 16904242. [DOI] [PubMed] [Google Scholar]
- 283.Mota RA, Hernandez-Espinosa D, Galbis-Martinez L, Ordonez A, Minano A, Parrilla P, Vicente V, Corral J, Yelamos J. Poly(ADP-ribose) polymerase-1 inhibition increases expression of heat shock proteins and attenuates heat stroke-induced liver injury. Crit Care Med. 2008;36(2):526–34. doi: 10.1097/01.CCM.0000299735.43699.E9. PubMed PMID: 18091544. [DOI] [PubMed] [Google Scholar]
- 284.Garcia S, Bodano A, Gonzalez A, Forteza J, Gomez-Reino JJ, Conde C. Partial protection against collagen antibody-induced arthritis in PARP-1 deficient mice. Arthritis Res Ther. 2006;8(1):R14. doi: 10.1186/ar1865. PubMed PMID: 16356201; PMCID: PMC1526570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.von Lukowicz T, Hassa PO, Lohmann C, Boren J, Braunersreuther V, Mach F, Odermatt B, Gersbach M, Camici GG, Stahli BE, Tanner FC, Hottiger MO, Luscher TF, Matter CM. PARP1 is required for adhesion molecule expression in atherogenesis. Cardiovasc Res. 2008;78(1):158–66. doi: 10.1093/cvr/cvm110. PubMed PMID: 18093987. [DOI] [PubMed] [Google Scholar]
- 286.Boulares AH, Zoltoski AJ, Sherif ZA, Jolly P, Massaro D, Smulson ME. Gene knockout or pharmacological inhibition of poly(ADP-ribose) polymerase-1 prevents lung inflammation in a murine model of asthma. American journal of respiratory cell and molecular biology. 2003;28(3):322–9. doi: 10.1165/rcmb.2001-0015OC. PubMed PMID: 12594058. [DOI] [PubMed] [Google Scholar]
- 287.Veres B, Gallyas F Jr., Varbiro G, Berente Z, Osz E, Szekeres G, Szabo C, Sumegi B. Decrease of the inflammatory response and induction of the Akt/protein kinase B pathway by poly-(ADP-ribose) polymerase 1 inhibitor in endotoxin-induced septic shock. Biochem Pharmacol. 2003;65(8):1373–82. PubMed PMID: 12694878. [DOI] [PubMed] [Google Scholar]
- 288.Mabley JG, Jagtap P, Perretti M, Getting SJ, Salzman AL, Virag L, Szabo E, Soriano FG, Liaudet L, Abdelkarim GE, Hasko G, Marton A, Southan GJ, Szabo C. Anti-inflammatory effects of a novel, potent inhibitor of poly (ADP-ribose) polymerase. Inflammation research : official journal of the European Histamine Research Society [et al. ]. 2001;50(11):561–9. doi: 10.1007/PL00000234. PubMed PMID: 11766996. [DOI] [PubMed] [Google Scholar]
- 289.Haddad M, Rhinn H, Bloquel C, Coqueran B, Szabo C, Plotkine M, Scherman D, Margaill I. Anti-inflammatory effects of PJ34, a poly(ADP-ribose) polymerase inhibitor, in transient focal cerebral ischemia in mice. Br J Pharmacol. 2006;149(1):23–30. Epub 2006/07/26. doi: 10.1038/sj.bjp.0706837. PubMed PMID: 16865091; PMCID: PMC1629400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Hasko G, Mabley JG, Nemeth ZH, Pacher P, Deitch EA, Szabo C. Poly(ADP-ribose) polymerase is a regulator of chemokine production: relevance for the pathogenesis of shock and inflammation. Molecular medicine. 2002;8(5):283–9. PubMed PMID: 12359959; PMCID: PMC2039990. [PMC free article] [PubMed] [Google Scholar]
- 291.Ba X, Garg NJ. Signaling mechanism of poly(ADP-ribose) polymerase-1 (PARP-1) in inflammatory diseases. The American journal of pathology. 2011;178(3):946–55. doi: 10.1016/j.ajpath.2010.12.004. PubMed PMID: 21356345; PMCID: PMC3069822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Thiemermann C, Bowes J, Myint FP, Vane JR. Inhibition of the activity of poly(ADP ribose) synthetase reduces ischemia-reperfusion injury in the heart and skeletal muscle. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(2):679–83. Epub 1997/01/21. doi: 10.1073/pnas.94.2.679. PubMed PMID: 9012844; PMCID: PMC19573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Koh SH, Park Y, Song CW, Kim JG, Kim K, Kim J, Kim MH, Lee SR, Kim DW, Yu HJ, Chang DI, Hwang SJ, Kim SH. The effect of PARP inhibitor on ischaemic cell death, its related inflammation and survival signals. Eur J Neurosci. 2004;20(6):1461–72. Epub 2004/09/10. doi: 10.1111/j.1460-9568.2004.03632.x. PubMed PMID: 15355313. [DOI] [PubMed] [Google Scholar]
- 294.Kim YH, Koh JY. The role of NADPH oxidase and neuronal nitric oxide synthase in zinc-induced poly(ADP-ribose) polymerase activation and cell death in cortical culture. Exp Neurol. 2002;177(2):407–18. Epub 2002/11/14. PubMed PMID: 12429187. [DOI] [PubMed] [Google Scholar]
- 295.Szabo C, Lim LH, Cuzzocrea S, Getting SJ, Zingarelli B, Flower RJ, Salzman AL, Perretti M. Inhibition of poly (ADP-ribose) synthetase attenuates neutrophil recruitment and exerts antiinflammatory effects. The Journal of experimental medicine. 1997;186(7):1041–9. PubMed PMID: 9314553; PMCID: PMC2199068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Davar D, Beumer JH, Hamieh L, Tawbi H. Role of PARP inhibitors in cancer biology and therapy. Curr Med Chem. 2012;19(23):3907–21. PubMed PMID: 22788767; PMCID: PMC3421454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Jiang Y, Dai H, Li Y, Yin J, Guo S, Lin SY, McGrail DJ. PARP inhibitors synergize with gemcitabine by potentiating DNA damage in non-small-cell lung cancer. International journal of cancer Journal international du cancer. 2019;144(5):1092–103. Epub 2018/08/29. doi: 10.1002/ijc.31770. PubMed PMID: 30152517; PMCID: PMC6320711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Muvarak NE, Chowdhury K, Xia L, Robert C, Choi EY, Cai Y, Bellani M, Zou Y, Singh ZN, Duong VH, Rutherford T, Nagaria P, Bentzen SM, Seidman MM, Baer MR, Lapidus RG, Baylin SB, Rassool FV. Enhancing the Cytotoxic Effects of PARP Inhibitors with DNA Demethylating Agents - A Potential Therapy for Cancer. Cancer cell. 2016;30(4):637–50. Epub 2016/10/12. doi: 10.1016/j.ccell.2016.09.002. PubMed PMID: 27728808; PMCID: PMC5201166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Curtin NJ, Wang LZ, Yiakouvaki A, Kyle S, Arris CA, Canan-Koch S, Webber SE, Durkacz BW, Calvert HA, Hostomsky Z, Newell DR. Novel poly(ADP-ribose) polymerase-1 inhibitor, AG14361, restores sensitivity to temozolomide in mismatch repair-deficient cells. Clinical cancer research : an official journal of the American Association for Cancer Research. 2004;10(3):881–9. Epub 2004/02/12. PubMed PMID: 14871963. [DOI] [PubMed] [Google Scholar]
- 300.Jiao S, Xia W, Yamaguchi H, Wei Y, Chen MK, Hsu JM, Hsu JL, Yu WH, Du Y, Lee HH, Li CW, Chou CK, Lim SO, Chang SS, Litton J, Arun B, Hortobagyi GN, Hung MC. PARP Inhibitor Upregulates PD-L1 Expression and Enhances Cancer-Associated Immunosuppression. Clinical cancer research : an official journal of the American Association for Cancer Research. 2017;23(14):3711–20. Epub 2017/02/09. doi: 10.1158/1078-0432.CCR-16-3215. PubMed PMID: 28167507; PMCID: PMC5511572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Higuchi T, Flies DB, Marjon NA, Mantia-Smaldone G, Ronner L, Gimotty PA, Adams SF. CTLA-4 Blockade Synergizes Therapeutically with PARP Inhibition in BRCA1-Deficient Ovarian Cancer. Cancer Immunol Res. 2015;3(11):1257–68. Epub 2015/07/04. doi: 10.1158/2326-6066.CIR-15-0044. PubMed PMID: 26138335; PMCID: PMC4984269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Brown JS, Sundar R, Lopez J. Combining DNA damaging therapeutics with immunotherapy: more haste, less speed. British journal of cancer. 2018;118(3):312–24. Epub 2017/11/11. doi: 10.1038/bjc.2017.376. PubMed PMID: 29123260; PMCID: PMC5808021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Ding L, Chen X, Xu X, Qian Y, Liang G, Yao F, Yao Z, Wu H, Zhang J, He Q, Yang B. PARP1 Suppresses the Transcription of PD-L1 by Poly(ADP-Ribosyl)ating STAT3. Cancer Immunol Res. 2019;7(1):136–49. Epub 2018/11/08. doi: 10.1158/2326-6066.CIR-18-0071. PubMed PMID: 30401677. [DOI] [PubMed] [Google Scholar]
- 304.Mirza MR, Pignata S, Ledermann JA. Latest clinical evidence and further development of PARP inhibitors in ovarian cancer. Ann Oncol. 2018;29(6):1366–76. Epub 2018/05/12. doi: 10.1093/annonc/mdy174. PubMed PMID: 29750420. [DOI] [PubMed] [Google Scholar]
- 305.Sonnenblick A, de Azambuja E, Azim HA Jr., Piccart M. An update on PARP inhibitors--moving to the adjuvant setting. Nature reviews Clinical oncology. 2015;12(1):27–41. Epub 2014/10/08. doi: 10.1038/nrclinonc.2014.163. PubMed PMID: 25286972. [DOI] [PubMed] [Google Scholar]
- 306.Pan L, Hao W, Zheng X, Zeng X, Ahmed Abbasi A, Boldogh I, Ba X. OGG1-DNA interactions facilitate NF-kappaB binding to DNA targets. Sci Rep. 2017;7:43297. doi: 10.1038/srep43297. PubMed PMID: 28266569; PMCID: PMC5339705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Pan L, Zhu B, Hao W, Zeng X, Vlahopoulos SA, Hazra TK, Hegde ML, Radak Z, Bacsi A, Brasier AR, Ba X, Boldogh I. Oxidized Guanine Base Lesions Function in 8-Oxoguanine DNA Glycosylase-1-mediated Epigenetic Regulation of Nuclear Factor kappaB-driven Gene Expression. The Journal of biological chemistry. 2016;291(49):25553–66. doi: 10.1074/jbc.M116.751453. PubMed PMID: 27756845; PMCID: PMC5207254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Bacsi A, Aguilera-Aguirre L, Szczesny B, Radak Z, Hazra TK, Sur S, Ba X, Boldogh I. Down-regulation of 8-oxoguanine DNA glycosylase 1 expression in the airway epithelium ameliorates allergic lung inflammation. DNA repair. 2013;12(1):18–26. doi: 10.1016/j.dnarep.2012.10.002. PubMed PMID: 23127499; PMCID: PMC3678389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Mabley JG, Pacher P, Deb A, Wallace R, Elder RH, Szabo C. Potential role for 8-oxoguanine DNA glycosylase in regulating inflammation. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2005;19(2):290–2. doi: 10.1096/fj.04-2278fje. PubMed PMID: 15677345. [DOI] [PubMed] [Google Scholar]
- 310.Boldogh I, Hajas G, Aguilera-Aguirre L, Hegde ML, Radak Z, Bacsi A, Sur S, Hazra TK, Mitra S. Activation of ras signaling pathway by 8-oxoguanine DNA glycosylase bound to its excision product, 8-oxoguanine. The Journal of biological chemistry. 2012;287(25):20769–73. doi: 10.1074/jbc.C112.364620. PubMed PMID: 22568941; PMCID: PMC3375501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Gasser S, Orsulic S, Brown EJ, Raulet DH. The DNA damage pathway regulates innate immune system ligands of the NKG2D receptor. Nature. 2005;436(7054):1186–90. doi: 10.1038/nature03884. PubMed PMID: 15995699; PMCID: PMC1352168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Dunphy G, Flannery SM, Almine JF, Connolly DJ, Paulus C, Jonsson KL, Jakobsen MR, Nevels MM, Bowie AG, Unterholzner L. Non-canonical Activation of the DNA Sensing Adaptor STING by ATM and IFI16 Mediates NF-kappaB Signaling after Nuclear DNA Damage. Molecular cell. 2018;71(5):745–60 e5. doi: 10.1016/j.molcel.2018.07.034. PubMed PMID: 30193098; PMCID: PMC6127031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Tanaka Y, Ito S, Oshino R, Chen N, Nishio N, Isobe K. Effects of growth arrest and DNA damage-inducible protein 34 (GADD34) on inflammation-induced colon cancer in mice. British journal of cancer. 2015;113(4):669–79. Epub 2015/07/22. doi: 10.1038/bjc.2015.263. PubMed PMID: 26196182; PMCID: 4647681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Chen N, Nishio N, Ito S, Tanaka Y, Sun Y, Isobe K. Growth arrest and DNA damage-inducible protein (GADD34) enhanced liver inflammation and tumorigenesis in a diethylnitrosamine (DEN)-treated murine model. Cancer immunology, immunotherapy : CII. 2015;64(6):777–89. Epub 2015/04/03. doi: 10.1007/s00262-015-1690-8. PubMed PMID: 25832002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Mackenzie KJ, Carroll P, Martin CA, Murina O, Fluteau A, Simpson DJ, Olova N, Sutcliffe H, Rainger JK, Leitch A, Osborn RT, Wheeler AP, Nowotny M, Gilbert N, Chandra T, Reijns MAM, Jackson AP. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature. 2017;548(7668):461–5. doi: 10.1038/nature23449. PubMed PMID: 28738408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Dombrowski Y, Peric M, Koglin S, Kammerbauer C, Goss C, Anz D, Simanski M, Glaser R, Harder J, Hornung V, Gallo RL, Ruzicka T, Besch R, Schauber J. Cytosolic DNA triggers inflammasome activation in keratinocytes in psoriatic lesions. Sci Transl Med. 2011;3(82):82ra38. doi: 10.1126/scitranslmed.3002001. PubMed PMID: 21562230; PMCID: PMC3235683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Ferguson BJ, Mansur DS, Peters NE, Ren H, Smith GL. DNA-PK is a DNA sensor for IRF-3-dependent innate immunity. Elife. 2012;1:e00047. doi: 10.7554/eLife.00047. PubMed PMID: 23251783; PMCID: PMC3524801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Kondo T, Kobayashi J, Saitoh T, Maruyama K, Ishii KJ, Barber GN, Komatsu K, Akira S, Kawai T. DNA damage sensor MRE11 recognizes cytosolic double-stranded DNA and induces type I interferon by regulating STING trafficking. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(8):2969–74. doi: 10.1073/pnas.1222694110. PubMed PMID: 23388631; PMCID: PMC3581880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Shen YJ, Le Bert N, Chitre AA, Koo CX, Nga XH, Ho SS, Khatoo M, Tan NY, Ishii KJ, Gasser S. Genome-derived cytosolic DNA mediates type I interferon-dependent rejection of B cell lymphoma cells. Cell Rep. 2015;11(3):460–73. doi: 10.1016/j.celrep.2015.03.041. PubMed PMID: 25865892. [DOI] [PubMed] [Google Scholar]
- 320.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339(6121):786–91. doi: 10.1126/science.1232458. PubMed PMID: 23258413; PMCID: PMC3863629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Cai X, Chiu YH, Chen ZJ. The cGAS-cGAMP-STING pathway of cytosolic DNA sensing and signaling. Molecular cell. 2014;54(2):289–96. doi: 10.1016/j.molcel.2014.03.040. PubMed PMID: 24766893. [DOI] [PubMed] [Google Scholar]
- 322.Roos WP, Kaina B. DNA damage-induced cell death by apoptosis. Trends Mol Med. 2006;12(9):440–50. doi: 10.1016/j.molmed.2006.07.007. PubMed PMID: 16899408. [DOI] [PubMed] [Google Scholar]
- 323.Kasibhatla S, Brunner T, Genestier L, Echeverri F, Mahboubi A, Green DR. DNA damaging agents induce expression of Fas ligand and subsequent apoptosis in T lymphocytes via the activation of NF-kappa B and AP-1. Molecular cell. 1998;1(4):543–51. PubMed PMID: 9660938. [DOI] [PubMed] [Google Scholar]
- 324.Umemura M, Kawabe T, Shudo K, Kidoya H, Fukui M, Asano M, Iwakura Y, Matsuzaki G, Imamura R, Suda T. Involvement of IL-17 in Fas ligand-induced inflammation. Int Immunol. 2004;16(8):1099–108. doi: 10.1093/intimm/dxh111. PubMed PMID: 15237105. [DOI] [PubMed] [Google Scholar]
- 325.Miwa K, Asano M, Horai R, Iwakura Y, Nagata S, Suda T. Caspase 1-independent IL-1beta release and inflammation induced by the apoptosis inducer Fas ligand. Nat Med. 1998;4(11):1287–92. doi: 10.1038/3276. PubMed PMID: 9809553. [DOI] [PubMed] [Google Scholar]
- 326.Zong WX, Ditsworth D, Bauer DE, Wang ZQ, Thompson CB. Alkylating DNA damage stimulates a regulated form of necrotic cell death. Genes & development. 2004;18(11):1272–82. doi: 10.1101/gad.1199904. PubMed PMID: 15145826; PMCID: PMC420353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Ha HC, Snyder SH. Poly(ADP-ribose) polymerase is a mediator of necrotic cell death by ATP depletion. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(24):13978–82. PubMed PMID: 10570184; PMCID: PMC24176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Yu SW, Wang H, Poitras MF, Coombs C, Bowers WJ, Federoff HJ, Poirier GG, Dawson TM, Dawson VL. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science. 2002;297(5579):259–63. Epub 2002/07/13. doi: 10.1126/science.1072221. PubMed PMID: 12114629. [DOI] [PubMed] [Google Scholar]
- 329.Zhao L, Lin H, Chen S, Chen S, Cui M, Shi D, Wang B, Ma K, Shao Z. Hydrogen peroxide induces programmed necrosis in rat nucleus pulposus cells through the RIP1/RIP3-PARP-AIF pathway. J Orthop Res. 2018;36(4):1269–82. Epub 2017/09/30. doi: 10.1002/jor.23751. PubMed PMID: 28960436. [DOI] [PubMed] [Google Scholar]
- 330.Xu Y, Huang S, Liu ZG, Han J. Poly(ADP-ribose) polymerase-1 signaling to mitochondria in necrotic cell death requires RIP1/TRAF2-mediated JNK1 activation. The Journal of biological chemistry. 2006;281(13):8788–95. Epub 2006/02/01. doi: 10.1074/jbc.M508135200. PubMed PMID: 16446354. [DOI] [PubMed] [Google Scholar]
- 331.Fatokun AA, Dawson VL, Dawson TM. Parthanatos: mitochondrial-linked mechanisms and therapeutic opportunities. Br J Pharmacol. 2014;171(8):2000–16. doi: 10.1111/bph.12416. PubMed PMID: 24684389; PMCID: PMC3976618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Andrabi SA, Dawson TM, Dawson VL. Mitochondrial and nuclear cross talk in cell death: parthanatos. Annals of the New York Academy of Sciences. 2008;1147:233–41. doi: 10.1196/annals.1427.014. PubMed PMID: 19076445; PMCID: PMC4454457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.d'Adda di Fagagna F Living on a break: cellular senescence as a DNA-damage response. Nature reviews Cancer. 2008;8(7):512–22. Epub 2008/06/25. doi: 10.1038/nrc2440. PubMed PMID: 18574463. [DOI] [PubMed] [Google Scholar]
- 334.Campisi J, Aging, cellular senescence, and cancer. Annu Rev Physiol. 2013;75:685–705. Epub 2012/11/13. doi: 10.1146/annurev-physiol-030212-183653. PubMed PMID: 23140366; PMCID: PMC4166529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.te Poele RH, Okorokov AL, Jardine L, Cummings J, Joel SP. DNA damage is able to induce senescence in tumor cells in vitro and in vivo. Cancer research. 2002;62(6):1876–83. PubMed PMID: 11912168. [PubMed] [Google Scholar]
- 336.Rodier F, Munoz DP, Teachenor R, Chu V, Le O, Bhaumik D, Coppe JP, Campeau E, Beausejour CM, Kim SH, Davalos AR, Campisi J. DNA-SCARS: distinct nuclear structures that sustain damage-induced senescence growth arrest and inflammatory cytokine secretion. J Cell Sci. 2011;124(Pt 1):68–81. doi: 10.1242/jcs.071340. PubMed PMID: 21118958; PMCID: PMC3001408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Kuilman T, Michaloglou C, Vredeveld LC, Douma S, van Doorn R, Desmet CJ, Aarden LA, Mooi WJ, Peeper DS. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell. 2008;133(6):1019–31. Epub 2008/06/17. doi: 10.1016/j.cell.2008.03.039. PubMed PMID: 18555778. [DOI] [PubMed] [Google Scholar]
- 338.Bavik C, Coleman I, Dean JP, Knudsen B, Plymate S, Nelson PS. The gene expression program of prostate fibroblast senescence modulates neoplastic epithelial cell proliferation through paracrine mechanisms. Cancer research. 2006;66(2):794–802. Epub 2006/01/21. doi: 10.1158/0008-5472.CAN-05-1716. PubMed PMID: 16424011. [DOI] [PubMed] [Google Scholar]
- 339.Krtolica A, Parrinello S, Lockett S, Desprez PY, Campisi J. Senescent fibroblasts promote epithelial cell growth and tumorigenesis: a link between cancer and aging. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(21):12072–7. Epub 2001/10/11. doi: 10.1073/pnas.211053698. PubMed PMID: 11593017; PMCID: PMC59769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Liu D, Hornsby PJ. Senescent human fibroblasts increase the early growth of xenograft tumors via matrix metalloproteinase secretion. Cancer research. 2007;67(7):3117–26. Epub 2007/04/06. doi: 10.1158/0008-5472.CAN-06-3452. PubMed PMID: 17409418. [DOI] [PubMed] [Google Scholar]
- 341.Ruhland MK, Coussens LM, Stewart SA. Senescence and cancer: An evolving inflammatory paradox. Biochimica et biophysica acta. 2016;1865(1):14–22. doi: 10.1016/j.bbcan.2015.10.001. PubMed PMID: 26453912; PMCID: PMC4733607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Jaiswal M, LaRusso NF, Burgart LJ, Gores GJ. Inflammatory cytokines induce DNA damage and inhibit DNA repair in cholangiocarcinoma cells by a nitric oxide-dependent mechanism. Cancer research. 2000;60(1):184–90. PubMed PMID: 10646872. [PubMed] [Google Scholar]
- 343.Morreall J, Limpose K, Sheppard C, Kow YW, Werner E, Doetsch PW. Inactivation of a common OGG1 variant by TNF-alpha in mammalian cells. DNA repair. 2015;26:15–22. doi: 10.1016/j.dnarep.2014.11.007. PubMed PMID: 25534136; PMCID: PMC4308426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Qu J, Liu GH, Huang B, Chen C. Nitric oxide controls nuclear export of APE1/Ref-1 through S-nitrosation of cysteines 93 and 310. Nucleic acids research. 2007;35(8):2522–32. doi: 10.1093/nar/gkl1163. PubMed PMID: 17403694; PMCID: PMC1885639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Laval F, Wink DA. Inhibition by nitric oxide of the repair protein, O6-methylguanine-DNA-methyltransferase. Carcinogenesis. 1994;15(3):443–7. PubMed PMID: 8118926. [DOI] [PubMed] [Google Scholar]
- 346.Wink DA, Laval J. The Fpg protein, a DNA repair enzyme, is inhibited by the biomediator nitric oxide in vitro and in vivo. Carcinogenesis. 1994;15(10):2125–9. PubMed PMID: 7955043. [DOI] [PubMed] [Google Scholar]
- 347.Wei W, Li B, Hanes MA, Kakar S, Chen X, Liu L. S-nitrosylation from GSNOR deficiency impairs DNA repair and promotes hepatocarcinogenesis. Sci Transl Med. 2010;2(19):19ra3. doi: 10.1126/scitranslmed.3000328. PubMed PMID: 20371487; PMCID: PMC3085984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Jaiswal M, LaRusso NF, Nishioka N, Nakabeppu Y, Gores GJ. Human Ogg1, a protein involved in the repair of 8-oxoguanine, is inhibited by nitric oxide. Cancer research. 2001;61(17):6388–93. PubMed PMID: 11522631. [PubMed] [Google Scholar]
- 349.Jones LE Jr., Ying L, Hofseth AB, Jelezcova E, Sobol RW, Ambs S, Harris CC, Espey MG, Hofseth LJ, Wyatt MD. Differential effects of reactive nitrogen species on DNA base excision repair initiated by the alkyladenine DNA glycosylase. Carcinogenesis. 2009;30(12):2123–9. doi: 10.1093/carcin/bgp256. PubMed PMID: 19864471; PMCID: PMC2792317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Graziewicz M, Wink DA, Laval F. Nitric oxide inhibits DNA ligase activity: potential mechanisms for NO-mediated DNA damage. Carcinogenesis. 1996;17(11):2501–5. PubMed PMID: 8968069. [DOI] [PubMed] [Google Scholar]
- 351.Wink DA, Nims RW, Darbyshire JF, Christodoulou D, Hanbauer I, Cox GW, Laval F, Laval J, Cook JA, Krishna MC, et al. Reaction kinetics for nitrosation of cysteine and glutathione in aerobic nitric oxide solutions at neutral pH. Insights into the fate and physiological effects of intermediates generated in the NO/O2 reaction. Chem Res Toxicol. 1994;7(4):519–25. PubMed PMID: 7981416. [DOI] [PubMed] [Google Scholar]
- 352.Foster MW, Hess DT, Stamler JS. Protein S-nitrosylation in health and disease: a current perspective. Trends Mol Med. 2009;15(9):391–404. doi: 10.1016/j.molmed.2009.06.007. PubMed PMID: 19726230; PMCID: PMC3106339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Pegg AE, Wiest L, Foote RS, Mitra S, Perry W. Purification and properties of O6-methylguanine-DNA transmethylase from rat liver. The Journal of biological chemistry. 1983;258(4):2327–33. Epub 1983/02/25. PubMed PMID: 6822564. [PubMed] [Google Scholar]
- 354.Liu L, Xu-Welliver M, Kanugula S, Pegg AE. Inactivation and degradation of O(6)-alkylguanine-DNA alkyltransferase after reaction with nitric oxide. Cancer research. 2002;62(11):3037–43. PubMed PMID: 12036910. [PubMed] [Google Scholar]
- 355.Tang CH, Wei W, Liu L. Regulation of DNA repair by S-nitrosylation. Biochimica et biophysica acta. 2012;1820(6):730–5. doi: 10.1016/j.bbagen.2011.04.014. PubMed PMID: 21571039; PMCID: PMC3170507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Parrish MC, Chaim IA, Nagel ZD, Tannenbaum SR, Samson LD, Engelward BP. Nitric oxide induced S-nitrosation causes base excision repair imbalance. DNA repair. 2018;68:25–33. Epub 2018/06/22. doi: 10.1016/j.dnarep.2018.04.008. PubMed PMID: 29929044; PMCID: PMC6436541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Sugimura H, Kohno T, Wakai K, Nagura K, Genka K, Igarashi H, Morris BJ, Baba S, Ohno Y, Gao C, Li Z, Wang J, Takezaki T, Tajima K, Varga T, Sawaguchi T, Lum JK, Martinson JJ, Tsugane S, Iwamasa T, Shinmura K, Yokota J. hOGG1 Ser326Cys polymorphism and lung cancer susceptibility. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 1999;8(8):669–74. PubMed PMID: 10744126. [PubMed] [Google Scholar]
- 358.Maguire S, Rabes HM. Transformation sensitivity in early S-phase and clonogenic potential are target-cell characteristics in liver carcinogenesis by N-methyl-N-nitrosourea. International journal of cancer Journal international du cancer. 1987;39(3):385–9. PubMed PMID: 2950063. [DOI] [PubMed] [Google Scholar]
- 359.Yu AM, Calvo JA, Muthupalani S, Samson LD. The Mbd4 DNA glycosylase protects mice from inflammation-driven colon cancer and tissue injury. Oncotarget. 2016;7(19):28624–36. doi: 10.18632/oncotarget.8721. PubMed PMID: 27086921; PMCID: PMC5053750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Parkin DM. The global health burden of infection-associated cancers in the year 2002. International journal of cancer Journal international du cancer. 2006;118(12):3030–44. doi: 10.1002/ijc.21731. PubMed PMID: 16404738. [DOI] [PubMed] [Google Scholar]