SUMMARY
Mitochondrial apoptosis can be effectively targeted in lymphoid malignancies with the FDA-approved B-cell lymphoma 2 (BCL-2) inhibitor venetoclax, but resistance to this agent is emerging. We show that venetoclax resistance in chronic lymphocytic leukemia is associated with complex clonal shifts. To identify determinants of resistance, we conducted parallel genome-scale screens of the BCL-2-driven OCI-Ly1 lymphoma cell line after venetoclax exposure along with integrated expression profiling and functional characterization of drug-resistant and engineered cell lines. We identified regulators of lymphoid transcription and cellular energy metabolism as drivers of venetoclax resistance in addition to the known involvement by BCL-2 family members, which were confirmed in patient samples. Our data support the implementation of combinatorial therapy with metabolic modulators to address venetoclax resistance.
Graphical Abstract

INTRODUCTION
Mitochondrial apoptosis is governed by the B-cell lymphoma 2 (BCL-2) family including both pro- and anti-apoptotic proteins. Across cancers, apoptosis dysregulation can result from overexpression of the anti-apoptotic BCL-2 protein that can sequester certain pro-apoptotic BH3-only proteins (BIM, BID) to avoid oligomerization of pore-forming proteins (BAX, BAK) and subsequent mitochondrial outer membrane permeabilization. Within B-cell tumors, BCL-2 dysregulation commonly arises from genetic abnormalities. These include the translocation t(14;18)(q32;q21), which places BCL2 under the control of IGH promoter, in follicular lymphoma (Tsujimoto et al., 1985; Kridel et al., 2012) or focal deletion of chromosome 13 (del[13q14]), which leads to loss of miR-15a/16-1, a negative regulatory microRNA of BCL-2, in chronic lymphocytic leukemia (CLL) (Cimmino et al., 2005). Thus, BCL-2 has been a rational therapeutic target in lymphoid cancers.
Venetoclax is an FDA-approved BCL-2 inhibitor for the treatment of CLL (Roberts et al., 2016) and for acute myeloid leukemia (AML) (DiNardo et al., 2018). It displaces pro-apoptotic BH3-only proteins from BCL-2, allowing them to activate the pro-apoptotic BAX or BAK proteins (Del Gaizo Moore et al., 2008). Despite its potent clinical activity in even those CLL cases failing control with chemotherapy regimens such as those carrying disruption of TP53 (Roberts et al., 2016), disease progression on venetoclax is an emerging therapeutic challenge (Anderson et al., 2017; Mato et al., 2018). Here, we aimed to uncover the determinants of venetoclax resistance in lymphoid malignancies.
RESULTS
Resistance to venetoclax in patients is associated with complex patterns of clonal evolution
Gene mutations are well-known mechanisms of resistance to targeted therapy in CLL (Woyach et al., 2014). To determine whether consistent genetic alterations could be observed in CLL patients demonstrating venetoclax resistance, we performed matched whole-exome sequencing (WES) of DNA from leukemia samples collected from 6 patients before venetoclax treatment and at the time of relapse compared to germline DNA (Figure 1A, Tables S1, S2 and S3). All patients had been previously treated with chemoimmunotherapy. After starting venetoclax, all experienced partial response and progressed after a median of 12.3 months (range, 5.1-22.8 months) on therapy. Progression was assessed in blood for 3 patients, in marrow for 2 patients and in lymph node for 1 patient.
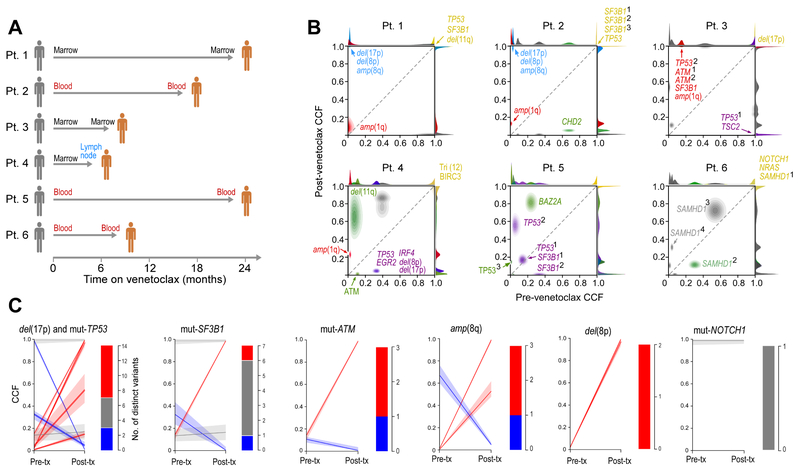
Figure 1. CLL cells from patients developing resistance to venetoclax undergo clonal evolution and exhibit complex trajectories.
(A) Schema of the 6 studied patients (Pt) with timing and sites of pre- and post-treatment sample collections indicated.
(B) Comparison of the shifts in cancer cell fraction (CCF) in pre-treatment and relapse samples, demonstrating clonal evolution and diverse changes in subclonal composition across the 6 patients. Driver mutations associated with each clone are indicated. Superscripted numbers indicate distinct mutations of the same gene per patient. Del, deletion; amp, amplification.
(C) Comparison (modal CCF with 95% confidence interval) between pre-treatment and relapse samples for select drivers previously reported as recurrently observed in CLL. Mut, mutations.
Analysis of the WES data revealed no differences in the median mutation rate between baseline and relapse samples across patients (Figure S1A). We did not identify any somatic single-nucleotide variations (sSNVs) in BCL2 itself at baseline or at progression, nor in any coding region of BCL-2 family members (Table S4). Inference of subclonal architecture revealed marked clonal shifts in all patients (Figure 1B, Figure S1B-S1C, Table S4). Numerous instances of shifting cancer cell fractions of subclones with CLL drivers such as ATM, TP53 or SF3B1 mutation were observed, even of distinct subclones with differing mutated alleles of the same gene within an individual (Figure 1C), but no sSNVs of known CLL driver mutations were consistently selected with resistance.
We noted that Patients 1 and 2 share a common evolutionary trajectory where resistance was associated with a large expansion of a subclone carrying del(17p) [TP53], amp(8q) and del(8p) [encompasses the TRAIL receptor genes] together with truncal SF3B1 and TP53 mutations (Figure 1B-1C, Figure S1C). We previously described enrichment of del(8p) in the setting of resistance to the BTK inhibitor ibrutinib (Burger et al., 2016), and BCL-2 inhibition has been shown to sensitize human cancer cells to TRAIL-induced apoptosis by inducing the expression of its receptor (Song et al., 2008). Our finding of amp(8q) (MYC locus) together with the recently reported del(9p) (CDKN2A/B locus) in leukemia samples from venetoclax resistant CLL patients (Herling et al., 2018) supports the idea that changes in cell cycle regulation contribute to resistance. Altogether, these results suggested that venetoclax resistance is not solely driven by any particular sSNV or somatic copy number variation (sCNV) but rather involves multiple complex changes. Such observations prompted us to undertake an unbiased genome-wide screening approach to uncover drivers of venetoclax resistance.
BCL-2 family members, lymphoid transcription regulators and components of AMP-dependent pathways are candidate drivers of venetoclax resistance
We performed parallel genome-scale loss-of-function (LOF) and gain-of-function (GOF) genetic modifier screens, using the BCL-2-driven OCI-Ly1 lymphoma cell line (Figure 2A). This cell line was selected for its sensitivity to venetoclax and ability to be transduced, amongst the 6 lymphoid lines evaluated (Figure S2A). For the LOF screen, OCI-Ly1 cells modified to stably express Cas9 were infected with the Brunello lentiviral library of 76,441 sgRNAs targeting 19,114 genes and 1,000 control sgRNAs (Doench et al., 2016), and treated with venetoclax (or DMSO, as control) for 14 days (Figure S2B). We then evaluated changes in sgRNA representation following drug treatment by targeted sequencing of genomic DNA isolated from viable cells on day 14. Although average sgRNA representation was overall decreased in the transduced cells treated with venetoclax compared to duplicate sets of untreated and DMSO-treated transduced cells, a clear subset of drug-treated transduced cells was enriched (Figure 2B). By analysis with the STARS software (v1.3, Broad Institute) (Doench et al., 2016), sgRNAs corresponding to 11 genes were consistently enriched over 2 replicates after 14 days of drug treatment compared to DMSO (STARS score > 4; Figure 2C, Table S5), with high consistency in the changes in representation amongst the 4 sgRNAs per gene (Figure S2C). Not surprisingly, four of 11 knocked-out genes encoded pro-apoptotic BCL-2-family proteins (PMAIP1, BAX, BAK1, BCL2L11). Moreover, the level of representation by sgRNAs corresponding to the other BCL-2 members matched their known functions as either pro- or anti-apoptotic family members (Figure S2D). Notably, the other 7 candidate hit genes were not BCL-2-family genes but were involved in pathways highly relevant to lymphoid biology. These included NFKBIA, encoding an inhibitor of the NF-κB pathway, and genes encoding lymphoid transcription factors and modulators (IKZF5, ID3, EP300, NFIA) or components of the processes of ubiquitination (OTUD5, UBR5). Several of the discovered genes are recurrently mutated in B-cell malignancies: EP300 in 10-20% of follicular lymphomas (Morin et al., 2011; Pastore et al., 2015), ID3 in 68% of Burkitt lymphomas (Love et al., 2012), NFKBIA in 20% of Hodgkin lymphomas (Weniger and Küppers, 2016), and UBR5 in 18% of mantle cell lymphomas (Meissner et al., 2013).
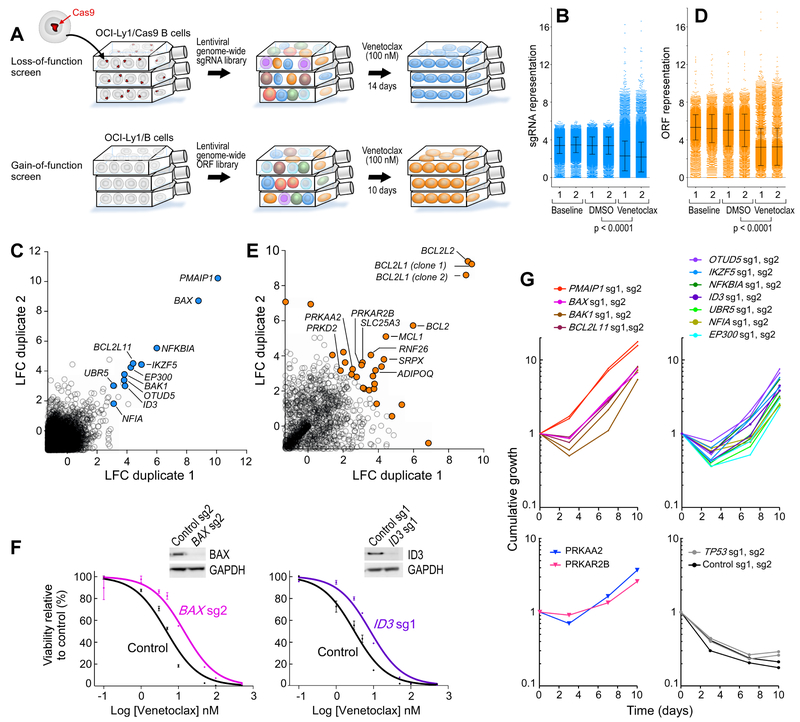
Figure 2. BCL-2 family members, lymphoid transcription regulators and components of AMP-dependent pathways are candidate drivers of venetoclax resistance.
(A) Experimental schema of the parallel knockout and overexpression screens using the BCL-2 driven OCI-Ly1 cell line (two biologically independent experiments per screen).
(B) sgRNAs frequencies at different timepoints during the screens (two independent experiments shown), black bars are mean +/− s.d., p value is from two-sided t-test.
(C) Scatter plots showing the average log2fold-change (LFC) for each gene in both duplicates of the loss-of-function screens (only genes with LFC > −1 are shown). Genes with a significant increase of sgRNAs representation (using the gene-ranking algorithm STARS, Broad Institute) are highlighted.
(D) ORF frequencies at different timepoints during the screens (two independent experiments shown), black bars are mean +/− s.d., p value is from two-sided t-test.
(E) Scatter plots showing the average log2fold-change (LFC) for each gene in both duplicates of the GOF screens (only genes with LFC > −1 are shown). Genes within the top-30 ORFs are highlighted.
(F) Dose-response curves to venetoclax of 2 representative single-knockout OCI-Ly1 cells with related western-blots for quantification of the target protein. Data are mean +/− s.e.m.
(G) Cumulative growth over time of each of the genetically perturbed OCI-Ly1 cells.
In an analogous fashion, we performed a GOF screen by using a genome-scale library including 17,255 barcoded ORFs encoding 12,952 unique proteins with at least 99% nucleotide and protein match to comprehensively identify genes that confer resistance to venetoclax when overexpressed in OCI-Ly1 cells (Figure 2A, Figure S2B). A clear shift of ORF representation was observed (Figure 2D,) and a total of 71 ORFs (arising from 70 genes) had a log2fold change (LFC) greater than 2 (Figure 2E, Table S6). The top four genes that generated resistance when overexpressed were those encoding known anti-apoptotic proteins (BCL2L1, BCL2L2, BCL2, MCL1). Included among the top 50 genes identified in the screen were those encoding components of the energy-stress sensor PKA/AMPK signaling pathway (ADIPOQ, PRKAR2B, PRKAA2), of mitochondrial energy metabolism (SLC25A3) or vesicle transport/autophagy (RNF26, DNM2, PRKD2, ATG5), ribosomal proteins (RPL17, RPS4Y1, RPS15A), or components of ubiquitination (OTUD6A, FBXO9, USP54) (Figure 2E).
To confirm the LOF screen results, we generated single-gene knockout OCI-Ly1 cell lines for each of the 11 hits (2 cell lines per gene, generated from the 2 most efficient sgRNAs per gene). We also generated control lines corresponding to 2 non-targeting sgRNAs and for 2 sgRNAs targeting TP53 (Figure S2E). From the GOF screen, we detected genes encoding two protein kinases components from related signaling pathways (PRKAR2B, PRKAA2). We hence prioritized the generation of 2 overexpression lines from OCI-Ly1 cells, one with the regulatory subunit of cAMP-dependent protein kinase (protein kinase A, PKA) encoded by PRKAR2B and the other the catalytic subunit of the AMP-activated protein kinase (AMPK) encoded by PRKAA2 (Figure S2F). Both are key regulators of cellular energy metabolism (Lark et al., 2015; Lin and Hardie, 2018).
Across the individual knockout cell lines, we observed diminished sensitivity to venetoclax, with median increase in IC50 of 2.1-fold (range, 1.3- to 13.8-fold; p < 0.05, extra-sum-of-squares F test) compared to the two cell lines transduced with non-targeting control sgRNAs (Figure 2F, Figure S2G). Moreover, all of the knockout and overexpression lines showed increased cumulative growth over 10 days of in vitro venetoclax exposure compared to control lines (Figure 2G), which was not observed in the absence of venetoclax (Figure S2H). Exposure of the knockout cell lines to venetoclax increased the degree of depletion of the targeted protein and enriched the frequency of frameshift indels generated by the introduction of sgRNAs into the cell lines (Figure S2E and S2I). Altogether, these results confirmed the on-target effects of the sgRNAs and the ORFs identified through the parallel genome-wide screens.
Venetoclax resistance involves MCL-1 overexpression and changes in cellular energy metabolism
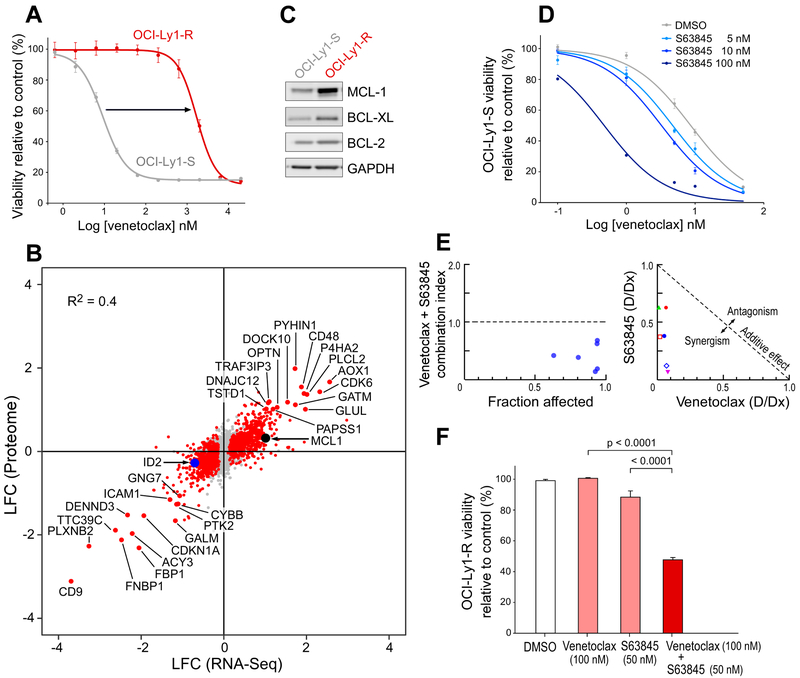
In complementary experiments, we characterized the transcript and protein expression profiles of a venetoclax-resistant cell line (OCI-Ly1-R, IC50 1 μM), generated through chronic in vitro drug exposure of the parental (OCI-Ly1-S, IC50 4 nM) cells (Figure 3A). By RNA-sequencing (RNA-Seq) of the resistant and sensitive cell lines, we identified 19 upregulated and 28 downregulated genes (adjusted p value < 0.05; ∣LFC∣ > 2) (Figure S3A). Comparison of the transcriptomes and the proteomes (identified via mass spectrometry-based proteomics) of the OCI-Ly1-S and OCI-Ly1-R cells revealed close concordance across these two platforms (R2 = 0.4, all genes; R2 = 0.8, restricted to differentially expressed genes; Figure 3B), with 14 upregulated and 13 downregulated proteins (adjusted p value < 0.05, ∣LFC∣ > 2). MCL-1 emerged as the only significantly and coordinately deregulated transcript and protein that also overlapped with the gene hits from the genome-scale screens (Figure 3B). MCL-1 overexpression has been previously reported in the characterizations of cancer cell lines rendered resistant to BCL-2 inhibition and has been described to sequester the pro-apoptotic BIM protein (Deng et al., 2007; Yecies et al., 2010). We confirmed the relative increase in protein expression of MCL-1 in OCI-Ly1-R cells compared to OCI-Ly1-S cells (Figure 3C), and observed in vitro synergy between venetoclax and the MCL-1 inhibitor S63845 (Kotschy et al., 2016) on OCI-Ly1-S cells (combination index <1, Figure 3D-3E). MCL-1 inhibition furthermore restored venetoclax sensitivity to the OCI-Ly1-R cells (Figure 3F). These results confirm a key role of MCL-1 overexpression in mediating venetoclax resistance.
Figure 3. Expression changes related to acquisition of venetoclax resistance involves MCL-1 and cellular energy metabolism.
(A) Dose-response curve of the drug-resistant OCI-Ly1-R and the drug-sensitive OCI-Ly1-S cell lines. Data are mean +/− s.e.m.
(B) Scatter plot reporting log2fold-change (LFC) of expression levels of transcripts (X-axis) and proteins (Y-axis) levels between OCI-Ly1-S and OCI-Ly1-R cells. Red dots – events with adjusted p value < 0.05 at the protein level.
(C) Western-blot showing MCL-1, BCL-XL and BCL-2 protein levels in OCI-Ly1-S and OCI-Ly1-R cells.
(D) Dose-response curves of OCI-Ly1-S to venetoclax and varying doses of the MCL-1 inhibitor S63845. Data are mean +/− s.e.m.
(E) Combination index according to the fraction affected (left) and normalized isobologram (right), Chou-Talalay method.
(F) Viability of the OCI-Ly1-R line 24 hours after exposure to venetoclax 100 nM, S63845 50 nM and both drugs (and DMSO as control), data are mean +/− s.e.m. from three biologically independent experiments; p value is from ANOVA test with adjustment for multiple comparisons.
See also Figure S3.
Geneset enrichment analysis (GSEA) based on RNA-Seq data revealed 35 significantly enriched pathways (nominal p value < 0.05, FDR < 0.25). Consistent with the results from our survival screens, top positively regulated pathways included lymphoid differentiation and chromatin maintenance, while top negatively regulated pathways related to metabolism and the endoplasmic reticulum (nominal p value < 0.002, FDR < 0.9) (Figure 4A, Table S7). In addition, the most coordinately upregulated transcripts and proteins originated from genes critical to cellular metabolism (AOX1, GLUL, PAPSS1, GATM, TSTD1, GALM, FBP1) (Figure 3B). Glutamine synthetase (encoded by GLUL) plays a key role in cell survival (Bott et al., 2015), while repression of fructose-bisphosphatase 1 (encoded by FBP1) was previously shown to efficiently promote glycolysis (Guo et al., 2018). Other mechanisms of potential interest, highlighted by other upregulated transcripts/proteins, included cell cycle regulation (CDK6, CDKN1A [encoding p21], TT39C), B-cell biology (DOCK10) as well as autophagy or mitophagy, a process for removing damaged mitochondria through autophagy (DENND3, OPTN) and reactive oxygen species generation (CYBB).
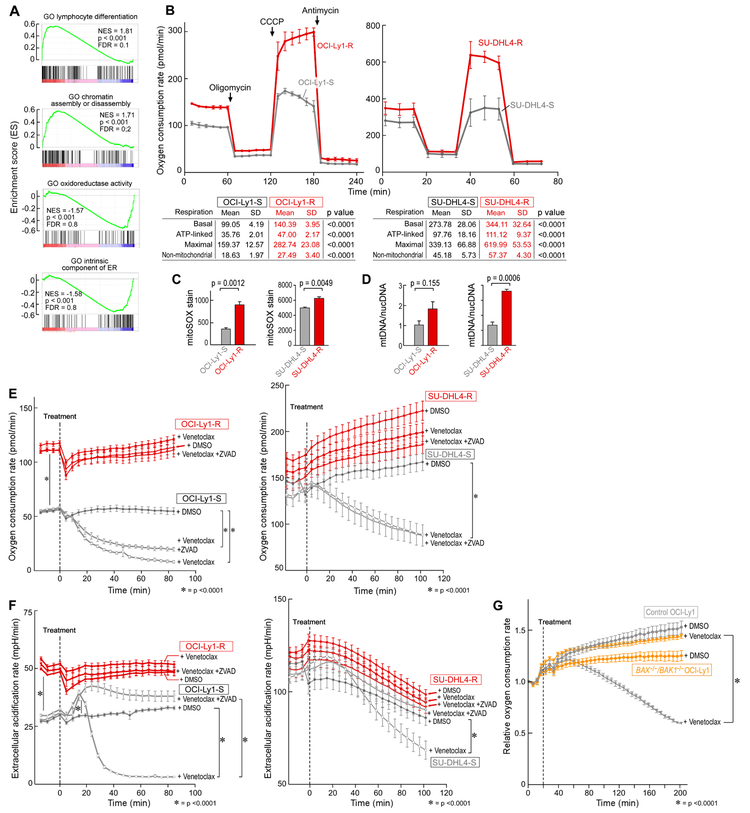
Figure 4. Increased oxidative phosphorylation drives resistance to BCL-2 inhibition.
(A) Selected gene set enrichment analysis plots based on differential RNA expression between OCI-Ly1-S and OCI-Ly1-R.
(B) Oxygen consumption rate (OCR) as a function of time in the OCI-Ly1 and SU-DHL4 lines, with exposure to inhibitors of the electron transport chain and oxidative phosphorylation (OXPHOS) to derive bioenergetics parameters of mitochondrial respiration.
(C) Quantification of the reactive oxygen species superoxide by flow cytometry in resistant vs parental B-cell lines.
(D) Ratio of mitochondrial DNA (mtDNA) over nuclear DNA (nucDNA) in resistant vs parental B-cell lines.
(E and F) Oxygen consumption rate (OCR) (E) and extracellular acidification rate (ECAR) (F) over time in the OCI-Ly1 and SU-DHL4 lines upon treatment by venetoclax, with or without prior zVAD treatment, or DMSO as control.
(G) Relative oxygen consumption rate (OCR) and over time in the control OCI-Ly1 and BAX/BAK1 double knockout OCI-Ly1 cell lines upon the treatment by venetoclax or DMSO as control.
Data are mean +/− s.e.m. from three biologically independent experiments (panel C) and one representative experiment of three biological replicates (panel B, D, E, F and G), * denotes p < 0.0001. p value is from two-sided t-test.
Metabolic reprogramming plays a critical role in the resistance to BCL-2 inhibition
Given the dysregulation of proteins critical to AMPK signaling and energy metabolism in both the GOF screen and in OCI-Ly1-R cells, we hypothesized that metabolic reprogramming contributes to resistance of malignant B cells to venetoclax. A recent genome-wide CRISPR screen identified AMPK subunits as positive regulators of oxidative phosphorylation (OXPHOS) (Arroyo et al., 2016). We therefore evaluated mitochondrial respiration of the OCI-Ly1-R cells compared to OCI-Ly1-S cells, as well as of venetoclax-resistant SU-DHL4 lymphoma cells (IC50 2.2 μM), which we generated through chronic in vitro drug exposure of the parental cell line (IC50 301 nM) (Figure S3B). For both sets of cell lines, we measured the oxygen consumption rate at baseline and over time in response to modulators of the mitochondrial electron transport chain (ETC) and OXPHOS. Both resistant cell lines demonstrated markedly higher rates of basal and maximal oxygen consumption, nearly all of which was coupled to ATP production by OXPHOS. Uncoupled rates of respiration were also markedly higher, suggesting an overall increased capacity for respiration and OXPHOS (Figure 4B, p < 0.0001). Resistant lines also demonstrated higher steady state levels of reactive oxygen species and higher mitochondrial membrane potential than parental lines (Figure 4C, Figure S3C). This was a result in part of an increased mass of mitochondria per cell in the resistant cells, since the quantity of mitochondrial DNA (mtDNA) was greater in the drug-resistant than –sensitive cells (Figure 4D).
To investigate whether venetoclax directly affects cellular energy metabolism, we measured oxygen consumption of both sets of OCI-Ly1 and SU-DHL4 resistant and parental cell lines following acute venetoclax exposure. Consistent with impairment of OXPHOS by venetoclax, we observed an immediate decrease in oxygen consumption (by 2-fold within 20 minutes after drug exposure for OCI-Ly1) (Figure 4E, p < 0.0001). OCI-Ly1-R exhibited a higher basal level of glycolysis, as assessed by extracellular acidification rate (ECAR) (Figure 4F, p < 0.0001), and we observed an immediate, yet transitory, increase in ECAR following venetoclax in the sensitive but not in the resistant cells (Figure 4F). In these experiments, the impact of venetoclax on OXPHOS was independent of caspase activation-related apoptotic cell death, as it was not fully prevented with the caspase inhibitor zVAD. The impact of venetoclax was however dependent on mitochondrial outer membrane permeabilization since exposure of the BAX/BAK1 double knockout OCI-Ly1 cell line to venetolcax did not result in demonstrable decrease in OXPHOS (Figure 4G, Figure S3D).
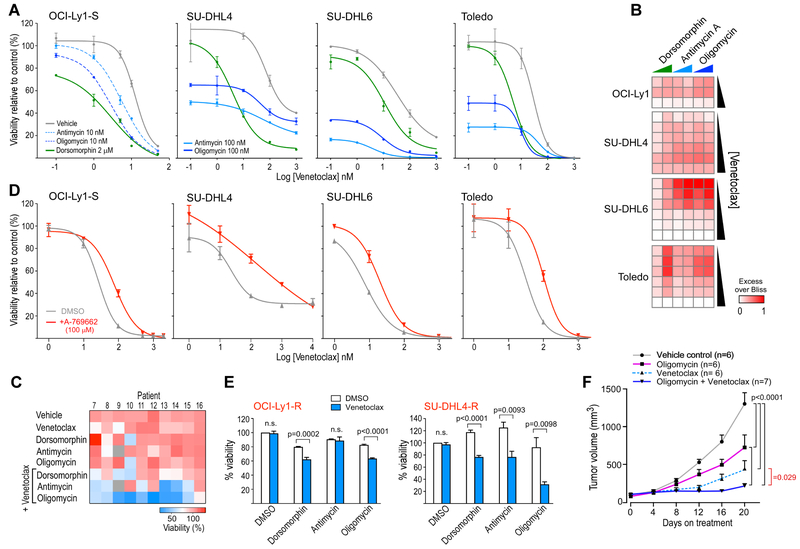
To investigate whether targeting AMPK and OXPHOS can impact venetoclax resistance, we treated multiple cell lines with venetoclax along with inhibitors and activators targeting the implicated pathways. The AMPK inhibitor dorsomorphin (compound C), the inhibitors of the ETC complex III (antimycin A) and complex V (oligomycin) increased venetoclax sensitivity when applied to the OCI-Ly1, SU-DHL4, SU-DHL6 lymphoma cell lines and the Toledo AML cell line (Figure 5A, Figure 5B, Figure S4A). These 3 compounds also increased the sensitivity to venetoclax in 10 of 10 primary CLL samples tested (Figure 5C, Figure S4B, Table S1). Conversely, treatment of cell lines with the AMPK activator A-769662 lowered sensitivity to venetoclax (Figure 5D). Furthermore, dorsomorphin, oligomycin and antimycin A could restore sensitivity to venetoclax in both the OCI-Ly1-R and SU-DHL4-R cells (Figure 5E). In vivo, we noted a higher efficacy of the oligomycin A and venetoclax combination compared to either drug alone when administered to OCI-Ly1 xenografts in NSG mice (Figure 5F). Altogether, these results implicate AMPK activation and mitochondrial metabolic reprogramming in resistance to venetoclax both in vitro and in vivo.
Figure 5. Targeting AMPK or the mitochondrial electron transport chain modulates sensitivity to venetoclax.
(A) Dose-response curves of OCI-Ly1, SU-DHL4, SU-DHL6 and Toledo cell lines to increasing doses of venetoclax alone or in combination with the AMPK inhibitor dorsomorphin, the inhibitor of electron transport chain complex 3 antimycin and the F1Fo-ATPase inhibitor oligomycin.
(B) Heatmap showing the Excess Over Bliss value related to the indicated combinations - venetoclax 5, 10 and 50 nM for OCI-Ly1 and 10, 50, 100, 5000, 1000, 5000 nM for SU-DHL4, SU-DHL6 and Toledo, dorsomorphin 1 and 2 μM, oligomycin 10 and 100 nM, antimycin 10 and 100 nM.
(C) Heatmap of the mean viability from duplicate experiments of primary tumor cells collected from 10 CLL patients after exposure to various drugs combinations, as indicated (venetoclax 1 nM, dorsomorphin 2 μM, oligomycin 10 nM, antimycin 10 nM).
(D) Dose-response curves of indicated cell lines to increasing doses of venetoclax with and without exposure to the AMPK-activator A-769662.
(E) Viability of the resistant OCI-Ly1-R and SU-DHL4-R cell lines after exposure to venetoclax 500 nM with and without dorsomorphin 1 μM, antimycin 10 nM or oligomycin 10 nM (DMSO as control).
For (A, C-E) viabilities were assessed at 24h of drugs exposure. For (A, D and E) the data shown represent mean +/− s.e.m. p value is calculated using a two-sided t-test.
(F) Tumor volume measurements of NSG OCI-Ly1 xenografts treated for 20 days, with vehicle control (black line), Oligomycin (200 mg/kg, i.p., pink line), Venetoclax (25 mg/kg, p.o., dashed light blue line), or their combination (dark blue line). Data are mean +/− s.d. p values result from Repeated-Measures ANOVA test with Tukey’s correction.
Venetoclax resistance involves a resistance circuit of ID3, AMPK, and PKA
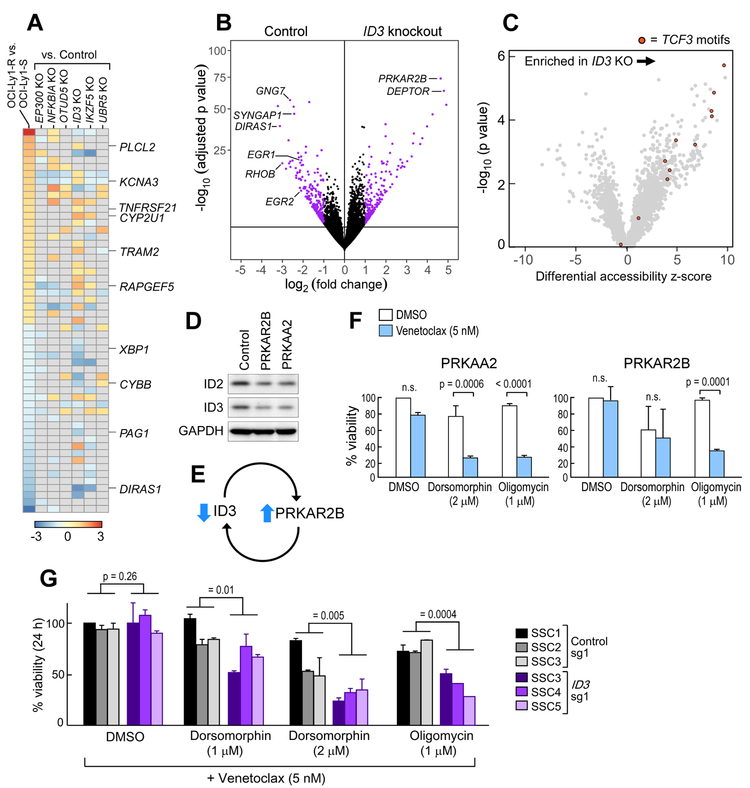
To define possible mechanisms by which the candidate hit genes identified from the survival screens were related to venetoclax resistance, we first evaluated MCL-1 expression in the respective knockout and overexpression cell lines. None of 14 tested cell lines demonstrated elevated MCL-1 levels before or after 14 days of treatment with venetoclax (Figure S5A-S5B) nor clear sensitivity to MCL-1 inhibition (Figure S5C), suggesting their engagement in alternative resistance mechanisms. Transcriptome profiling of each of the knockout cell lines corresponding to the 6 non-BCL-2 family member gene candidates revealed the greatest similarity between the knockout line of the lymphoid transcription regulator ID3 and the OCI-Ly1-R line (Figure 6A). Shared were transcripts involved in lymphoid signaling (PLCL2, KCNA3, PAG1) or transcription (XBP1), metabolism (CYP2U1, CYBB), apoptosis (TNFRSF21) and Ras signaling (DIRAS1, GNG7, RAPGEF5). Of note, ID2 (an ID3 paralog) was amongst the coordinately deregulated transcripts and proteins in the OCI-Ly1-R cell line (indicated in Figure 3B). As for the OCI-Ly1 cells, we confirmed that knockout of ID3 led to a survival advantage in SU-DHL4 cells exposed to venetoclax as well (Figure S5D-S5E-S5F).
Figure 6. A circuit of ID3 repression and PKA-AMPK deregulation is implicated in venetoclax resistance.
(A) Heatmap of differentially expressed transcripts between the OCI-Ly1-S and OCI-Ly1-R cells, and of knockout (KO) cell lines from the screen hits vs cell lines with KO using non-targeting sgRNAs. Relevant genes affected in common with the OCI-Ly1-S and –R cells and in the ID3 KO line are indicated.
(B) Volcano plot of transcripts changes in ID3 KO OCI-Ly1 cells compared to non-targeting sgRNA transduced OCI-Ly1 cells.
(C) Volcano plot of enriched accessible transcription factor motifs comparing ID3 KO vs control.
(D) Western-blot quantification of ID2 and ID3 proteins in PRKAR2B and PRKAA2 overexpressing OCI-Ly1 cell lines.
(E) Schema of the ID3 and PRKAR2B resistance circuit.
(F) Sensitivity of PRKAA2 and PRKAR2B overexpressing OCI-Ly1 cells to venetoclax when used in combination with dorsomorphin (2 μM) and oligomycin (1 μM), compared to DMSO control. Data are mean +/− s.e.m. from three biologically independent experiments, p value is from two-sided t-test.
(G) Viability at 24 hours of single-cell clones from ID3 knockout OCI-Ly1 cells compared to non-targeting sgRNAs transduced OCI-Ly1 cells after exposure to dorsomorphin and oligomycin in addition to venetoclax. Data are mean +/− s.d. from three biologically independent experiments and p values are from ANOVA test.
See also Figure S5.
The most significantly upregulated gene in the ID3 knockout cells in OCI-Ly1 cells was PRKAR2B, a PKA subunit which we previously uncovered in the GOF screen; we further confirmed its overexpression in ID3 KO SU-DHL4 cells (Figure 6B, Figure S5G). To clarify how the dominant-negative transcription factor ID3 regulates PRKAR2B expression, we performed ATAC-seq of the ID3 knockout (vs control) OCI-Ly1 lines in order to determine differential signatures of chromatin accessibility and transcription factor engagement. Notably, we identified increased accessibility associated with TCF3 motifs genome-wide in the ID3 knockout cells. TCF3 is a lymphoid transcription regulator previously reported to interact with ID3 (Figure 6C) (Wang and Baker, 2015), as well as to be involved in the transcription of ADIPOQ, which was detected in the GOF screen (Doran et al., 2008). A similar increase in TCF3 motif accessibility was also observed when comparing OCI-Ly1-R vs OCI-Ly1-S cells (Figure S5H). TCF3 binding sites were confirmed to be present within putative enhancer regions of PRKAR2B in a B cell context, based on the analysis of chromatin immunoprecipitation data available for the GM12878 line (ENCODE Project Consortium, 2012) and regions of open chromatin in our manipulated OCI-Ly1 lines (Figure S5I). Conversely, overexpression of PRKAR2B or PRKAA2, which we observed to provide survival advantage to OCI-Ly1, SU-DHL4 and SU-DHL6 lines following venetoclax (Figure 2G, Figure S5J-S5K), led to reduced ID2 and ID3 proteins (Figure 6D-6E. Confirming the involvement of cellular energy metabolism in this circuit, venetoclax sensitivity of the PRKAA2 and PRKAR2B overexpressing cells was restored by either dorsomorphin or oligomycin (Figure 6F, Figure S5L). Finally, single cell clones derived from the ID3 knockout cell line (Figure S5M) exhibited greater sensitivity to the combination of venetoclax and either dorsomorphin or oligomycin than control cells (Figure 6G). These observations link the results of each screen and identify a venetoclax resistance circuit involving AMPK dependency and mitochondrial respiratory-chain adaptations related to ID3 repression and PRKAR2B overexpression.
MCL-1 and AMPK deregulation drive venetoclax resistance in patients
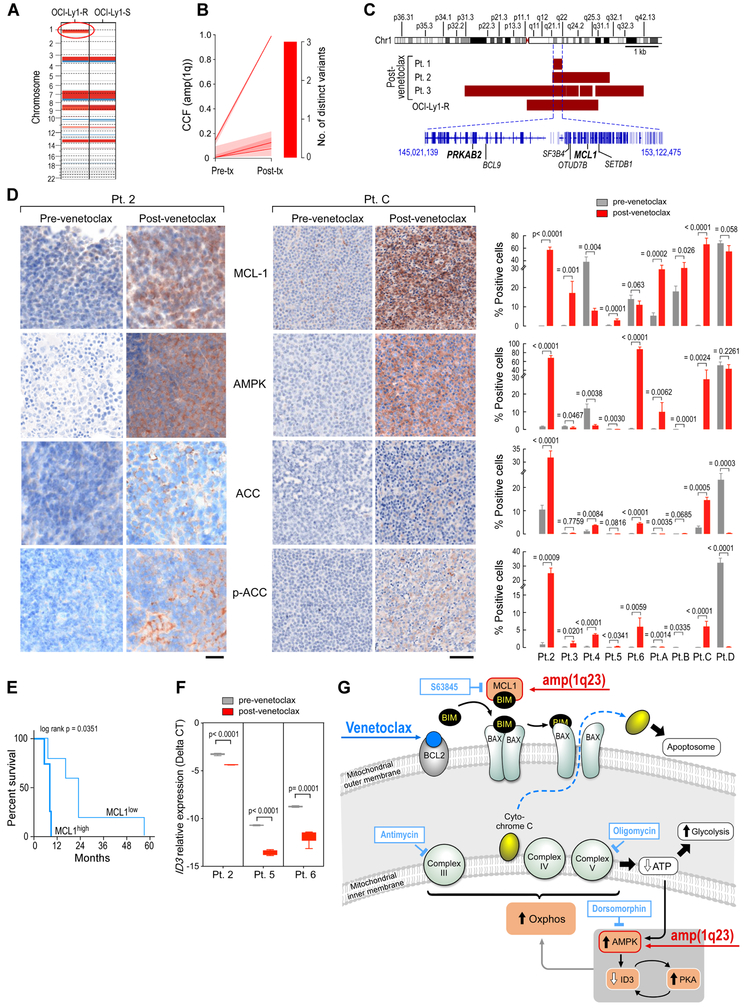
To determine if there was a genetic basis for the observed drug resistance in the OCI-Ly1-R cells, we compared the results of WES of DNA isolated from the OCI-Ly1-R and OCI-Ly1-S cell lines. No non-silent sSNVs overlapping with our screen data were identified except in BCL2, a target of aberrant somatic hypermutation frequently affected in B lymphomas and most B-cell lines (Table S8) (Chapuy et al., 2018). However, none involved the BH3 domain residues previously linked to venetoclax resistance in cell lines or in patients (Blombery et al., 2018; Fresquet et al., 2014). Regarding sCNVs, a clear region was amplified on chromosome 1q23 (amp[1q]) in the OCI-Ly1-R but not in the OCI-Ly1-S cells. No other differential sCNVs were identified (Figure 7A, Figure S6A).
Figure 7. Deregulated MCL-1 and AMPK signaling detected in CLL samples from patients developing resistance to venetoclax.
(A) Comparison of somatic copy number variations in the OCI-Ly1-S and OCI-Ly1-R cells. The red oval indicates amplification 1q as the main difference between the two lines. Red-gain; blue - loss.
(B) Comparison (modal CCF with 95%CI) between pre-treatment and relapse leukemia samples from patients 1, 2 and 3 for amp(1q).
(C) Representation of the minimal gained region in the 1q locus across the OCI-Ly1-R cell line and relapsed samples from Patients 1, 2 and 3.
(D) Immunohistochemical stains of patient samples before and after progression on venetoclax for MCL-1, AMPK, ACC, and p-ACC, with representative images from Patient C and 2 (left, the scale bar represents 20 μm in Patient 2 and 50 μm in Patient C), and quantification of % positively staining cells before (grey) and after (red) venetoclax treatment for Patients 2-6 (right). Data are mean +/− s.e.m. from replicates; p value is from Welch t-test.
(E) Progression-free survival according to MCL-1 expression (low, <10%; high, > or = 10%). p value is from log rank test.
(F) Relative expression of ID3 by qPCR from patient sample RNA before (grey) and after (red) venetoclax treatment from Patients 2, 5, and 6; p value is from Welch t-test. The boxes extend from the 25th to 75th percentiles and the whiskers from min to max.
(G) Proposed model for venetoclax resistance in lymphoid malignancies.
In line with the cell line data, we observed growing subclones with amp(1q) in 4 of 6 aforementioned cases with venetoclax-resistant CLL (Patients 1, 2, 3 and 4; Figures 1B, 7B, Tables S1-S4). We confirmed a common region of gain of 8 Mb amongst the resistant cell line and Patients 1, 2, and 3. Contained within this amplified region were several genes of interest including MCL1 and PRKAB2, encoding a regulatory subunit of AMPK (Figure 7C). To confirm the involvement of MCL-1 overexpression and AMPK signaling in patients, we investigated changes in protein expression by immunohistochemistry staining on biopsies obtained from 5 of 6 patients (Patients 2-6) for which pre- and post-venetoclax tissues were available. We extended this cohort to include tissues from 4 additional CLL patients with relapsed disease following venetoclax (Patients A-D) (Tables S1 and S2, Figure S6B). As expected, MCL-1 and AMPK were expressed at higher levels on formalin-fixed OCI-Ly1-R cells than on OCI-Ly1-S cells, and furthermore, we observed elevated expression of the downstream target of AMPK, acetyl-CoA carboxylase (ACC) and its phosphorylated form (p-ACC) on the resistant cell line (all p < 0.03, Figure S6C). In patient tissue samples, we detected increased MCL-1 expression in post-venetoclax tissue samples from 6 of 9 patients (Pt 2, 3, 5, A, B and C) (all p = or < 0.026), but also that pre-treatment tissues from Patients 4, 6 and D already exhibited elevated MCL-1 expression (Figure 7D). In our series, high pretreatment MCL-1 expression (>10% of positive cells) was associated with shorter progression-free survival (p= 0.017, Figure 7E); these findings support further evaluation of this association in larger cohorts. Elevations in AMPK expression with relapse were clearly evident for Patients 2, 6, A and C (all p = or < 0.0062) and AMPK signaling activation was supported by the observations of increased ACC and p-ACC in tissue from Patients 2, 4, 6 and C (Figure 7D). In line with our cell line data implicating a resistance circuit involving overexpression of AMPK and reduction of ID3, quantitative PCR of RNA extracted from formalin-fixed tissue from 3 of 3 paired patient samples (Pts 2, 5 and 6) revealed decreased relative expression of ID3 in association with relapse (p< or =0.0001, Figure 7F). These results support the involvement of both MCL-1 and AMPK in venetoclax resistance in CLL patient samples with acquired venetoclax resistance and reveal a genetic basis for MCL-1 and AMPK overexpression.
DISCUSSION
Diversity within a cancer cell population has been shown to fuel clonal evolution and subsequent therapeutic resistance. In line with that model, our study implicates major clonal shifts and heterogeneous evolutionary trajectories in patients with CLL relapsing on venetoclax, thereby involving multiple potential means of escape.
Discrete mutations in the gene targets of therapeutic inhibitors have been long-identified as a common mechanism of resistance to targeted therapies (Ellis and Hicklin, 2009; Gorre et al., 2001; Woyach et al., 2014). Recently, a BCL2 mutation impairing the venetoclax binding site of BCL-2 was described in a small cohort of CLL patients relapsing on venetoclax (Blombery et al., 2018). However, this mutation was commonly detected at only subclonal levels in this series, and was found in only a subset of patients, suggesting the presence of alternative non-mutational mechanisms of resistance to venetoclax. Indeed, neither we nor a previous study (Herling et al., 2018) detected this mutation.
Our systematic characterization of venetoclax resistance revealed dramatic alterations in the expression profiles of cells and complex genomic changes. On the one hand, our aggregate results underscore that alterations leading to overexpression of MCL-1, and thus the ability to sequester BIM and therefore to compensate BCL-2 inhibition, is a consistent adaptive mechanism of resistance for this class of BCL-2 inhibitor (Konopleva et al., 2006; Lin et al., 2016; Yecies et al., 2010). We detected MCL-1 overexpression and amplification in patient samples, not previously reported. On the other hand, our genome-wide screens together with functional characterization of resistant cell lines have uncovered deregulated energy metabolism via altered expression of components of the AMPK signaling pathway in this process. In support of the notion of a dual impact of venetoclax on the mitochondria, we notably identified a recurrence of amp(1q23), encompassing both MCL1 and the AMPK pathway component PRKAB2 in both representative cell lines and patient samples. Our work thus characterizes venetoclax resistance as involving both reprogramming of the biology of the mitochondria outer membrane, leading to expression changes in BCL-2 family members, as well as increased OXPHOS activity which is resident in the organelle’s inner membrane (Figure 7G). We note however, that other important mechanisms could be also involved, and could include other energy pathways, lymphoid differentiation, endoplasmic reticulum, mitochondrial biogenesis or mitophagy.
Our findings thus demonstrate that venetoclax imposes a broader scope of ‘mitochondrial stress’ than previously suggested. In AML, leukemia stem cells have dependency on oxidative phosphorylation which can be efficiently targeted by BCL-2 inhibition (Jones et al., 2018; Lagadinou et al., 2013; Pollyea et al., 2018). We confirm that venetoclax also targets OXPHOS in lymphoid malignancies. Furthermore, our study reveals that increased OXPHOS is a resistance mechanism to BCL-2 inhibition, a finding of potential relevance to other malignancies as well. Our work adds to the growing evidence of crosstalk between the BCL-2 family and the ETC. Early studies demonstrated that even prior to downstream activation of caspases, the apoptosis program results in a relatively rapid defect in the ETC (Mootha et al., 2001). Maintenance of ETC integrity was recently shown to allow cells to proliferate after apoptosis induction (Jiang et al., 2016). Conversely, the BCL-2 family proteins can participate in non-apoptotic functions such as regulation of mitochondrial morphology, autophagy or metabolism (Chen and Pervaiz, 2007; Lagadinou et al., 2013; Giménez-Cassina and Danial, 2015; Gross and Katz, 2017). Our data support the notion that venetoclax perturbs the cellular respiration process through the process of mitochondrial outer membrane permeabilization, leading to the release of cytochrome c, a crucial component of the inner membrane (Figure 7G).
We further find that the regulation of such optimized metabolism depends on repression of the ID family of lymphoid transcriptional regulators. These results mesh well with recent work demonstrating that lymphoid transcription factors function as metabolic gatekeepers by limiting the amount of cellular ATP to levels that are insufficient for malignant transformation (Chan et al., 2017). Hence, although our studies focused on the impact of ID3, a broader involvement of altered lymphoid differentiation state on venetoclax resistance is anticipated since reduction of additional lymphoid transcription factors (IKZF5, EP300) were identified in our knockout screen.
These insights have the potential to provide therapeutic opportunities. We point out that mutations affecting these factors have been detected across B-cell malignancies, and thus we expect them to confer venetoclax resistance. Our findings may therefore guide the further development of BCL-2 inhibitors in diseases lacking these mutations. Our data support the idea that targeting MCL-1 with emerging inhibitors would be a rationale and effective way to counteract venetoclax resistance. Given the association with higher OXPHOS status with resistance, this adaptive activity can be exploited to overcome venetoclax resistance. Our study further casts light on the mechanisms underlying diverse strategies previously evaluated to overcome resistance to BCL-2 inhibition, which have included hypoxia, caloric restriction (Meynet et al., 2013) and the use of the kinase inhibitors (including AMPK) (Kerkela et al., 2009; Oppermann et al., 2016) or mitochondrial ETC modulators (Chukkapalli et al., 2018).
STAR METHODS TEXT
Lead Contact and Materials Availability
Further information and requests for resources and reagents should be directed to and fulfilled by the Lead Contact, Catherine J Wu (cwu@partners.org). This study did not generate new unique reagents.
Experimental Model and Subject Details
Animals
Eight-week old (at time of transplant) female NSG (strain name: NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ) were used for in vivo studies. Randomization prior to treatment was performed according to tumor measurements by caliper. Weight of animals was monitored thrice weekly throughout the treatment. All animals were housed at Dana-Farber Cancer Institute (DFCI). All animal procedures were completed in accordance with the Guidelines for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committees at DFCI.
OCI-Ly1 xenografts were generated in 30 NSG mice that were injected subcutaneously into the left flank with 5×106 OCI-Ly1 cells per animal, resuspended 1:1 in PBS:Matrigel (Sigma-Aldrich). Tumors were measured by caliper twice weekly, starting one week after transplant, and volumes calculated according to the formula: LxW2/2. Animals were randomized for treatment on day 10 post-transplant (median 93-122 mm3 in each group; animals with tumors <30 mm3 were not included in the treatment study), and assigned to the four treatment cohorts (6-7 mice/group) receiving either vehicle control; 200 μg/kg oligomycin-A (Sigma-Aldrich) in PBS-10% ethanol; 25 mg/kg venetoclax (LC Laboratories) in 60% phosal 50 propylene glycol, 30% propylene glycol 400 and 10% ethanol (Souers et al., 2013) or their combination, for 20 days. Tumors were measures twice weekly throughout treatment.
Human Samples
Specimens were collected from CLL patients enrolled on clinical trials of venetoclax treatment (Patients 1-5, A and C from ; Patients 6 and B from ) or from general practice (Patients D and 7-16), conducted in accordance with the principles of the Declaration of Helsinki and with the approval of the Institutional Review Boards (IRB) of the University of Texas M.D. Anderson Cancer Center (MDACC; Patients 1, 3, 4) or of Dana-Farber Cancer Institute (DFCI; Patient 2, 5-16, A-D). Informed consent was obtained from all subjects. Their clinical characteristics are reported in Table S1. For patient 1-6, blood and/or tissue tumor samples were collected at baseline, before initiation of venetoclax therapy, and at relapse or progression on venetoclax (Table S2). For both PBMC (unsorted) or tissue, we first confirmed that the tumor purity was >80% by flow cytometry or immunohistochemistry. Genomic DNA (patient 1-6) was isolated with the DNAeasy Blood and Tissue Kit (Qiagen). Total RNA was purified from formalin-fixed, paraffin-embedded tissue sections with the RNeasy FFPE Kit (Qiagen). For patient 7-16, fresh CLL cells were isolated from peripheral blood using the Ficoll method.
Microbe Strains
NEB-5alpha Competent E. coli were transformed based on the high efficiency transformation protocol and grown on plates of LB agar with ampicillin 100 microg/mL for plasmid selection at 37°C.
Cell Lines
OCI-Ly1 cells (male) were cultured in Iscove's Modified Dulbecco's Media (Gibco) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin/glutamine. Cell lines resistant to venetoclax were generated over 10 weeks by exposing the cells to increasing doses of venetoclax starting at 10 nM, and then doubling this dose when the cells were able to grow at a rate equivalent to the parental lines until the cells were able to tolerate 1 μM of venetoclax. SU-DHL4 (male), SU-DHL6 (male) and Toledo (female) cells were cultured in Roswell Park Memorial Institute media (Gibco) supplemented with 10% FBS and 1% penicillin/streptomycin. All cell lines were incubated at 37°C.
Primary Cell Culture
Isolated CLL cells from patients described in the human samples section (patient 7-16, Table S1) were cultured in Roswell Park Memorial Institute media (Gibco) supplemented with 10% FBS and 1% penicillin/streptomycin for in vitro drug testing. Samples were collected in accordance with the principles of the Declaration of Helsinki and with the approval of the Institutional Review Boards (IRB) of Dana-Farber Cancer Institute. Informed consent was obtained from all subjects.
Method Details
Cell viability assay
The Cell Titer-Glo Luminescent Cell Viability Assay (Promega) was used to determine the relative number of viable cells after drug treatment. 0.2 × 106 cells/mL were seeded in a 24 well-plate and treated with drugs for 24 or 48 hours. The viability assay was conducted using the manufacturer’s protocol after treatment. Values were normalized to DMSO-treated cells. Plates were read on a SpectraMax M3 reader (Molecular Devices).
Western blotting
Total protein from cells was isolated using RIPA Buffer (20 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% NP-40, 1% sodium deoxycholate, 2.5 mM sodium pyrophosphate, 1 mM ß-glycerophosphate, 1 mM Na3VO4, 1 μg/mL leupeptin), supplemented with protease inhibitors (Thermo Fisher Scientific) and phosphatase inhibitors (Thermo Fisher Scientific). Protein concentration was determined with the BCA Protein Assay (Pierce). Protein samples (25 μg) were separated on either 4-12% Bis-Tris gels (proteins <250 kDa) or Tris-acetate gels (proteins >250 kDa). Protein was transferred to a nitrocellulose or PVDF membrane (Life Technologies) using the iBlot2 system (Life Technologies). Membranes were incubated overnight with primary antibodies. After incubation in the appropriate secondary antibodies, protein was detected using chemiluminescence (Western Lightning Plus-ECL, Perkin Elmer).
Genome-scale screens
Conduct of the genome-wide CRISPR-screen
The strategy used was similar in approach as previously reported (Shalem et al., 2014). 300 × 106 Cas9-OCI-Ly1 cells were suspended in media supplemented with 8 μg/mL polybrene and seeded into 9 12-well plates (1 mL per well). Titration of the dose of puromycin and of polybrene on OCI-Ly1 cells was undertaken to achieve 100% and minimal death of non-infected cells, respectively. The BRUNELLO sgRNA viral library in lentiGuide-puro (Genetic Perturbation Platform, Broad Institute) was added to each well (200 μL/mL), titrated to achieve an infection rate of 30% without excessive cell death and to minimize multiplicity of infection. The plates were spun at 2000 rpm for 2 hr at 37°C and incubated at 37°C for 24 hr. Polybrene was diluted by adding 2 mL of standard media to each well. Puromycin selection (1 μg/mL) was initiated 48 hours post-transduction and sustained for 5 days. Two days after puromycin selection, transduced OCI-Ly1 cells were treated with venetoclax (100 nM -- a dose identified to be growth suppressive at day 14) or DMSO as control for 14 days in T225 flasks. Cells were counted and re-split every three days, maintaining a concentration of 200,000 cells/mL Approximately 40 million cells were frozen before and after venetoclax or DMSO selection for sequencing. This experiment was performed in duplicates.
Genomic DNA was isolated (Maxiprep kits, Qiagen), and PCR and barcoded sgRNA or ORF-sequencing were performed, as previously described.(Doench et al., 2016) Samples were sequenced on a HiSeq2000 (Illumina). For analysis, the read counts were normalized to reads per million and then log2 transformed. The log2 fold-change of each sgRNAs was determined relative to the initial time point for each. Significance of the sgRNAs’ enrichment was assessed using the STARS software (v1.3, Broad Institute).
Conduct of the genome-wide ORF-screen
The approach used was based on previous reports (Johannessen et al., 2013; Rotem et al., 2015). Optimal transduction conditions were determined in order to achieve 30-50% infection efficiency, corresponding to a multiplicity of infection (MOI) of ~0.5 - 1. Briefly, 1.5 × 106 of OCI-Ly1 cells were spun-infected in 6 wells of a 12-well plates with different virus volumes. The day after, OCI-Ly1 cells were seeded in 2 wells of a 6-well plate, each with complete medium, one supplemented with 1 μg/mL of puromycin. Cells were counted 4 days postselection to determine the infection efficiency, comparing survival with and without puromycin selection. Volumes of virus that yielded ~30 - 50% infection efficiency were used for screening.
We used an ORFeome barcoded library that contains 17,255 barcoded ORFs overexpressing 12,952 unique genes (Broad Institute). Large-scale infections were performed in 12-well format as the viral titration described above using the optimized volume of virus, and pooled 24 hours post-centrifugation. Infections were performed with an adequate number of cells to achieve a representation of at least 1000 cells per ORF following puromycin selection (~2 × 107 surviving cells containing 17,255 ORFs). ~24 hours after infection, all wells within a replicate were pooled and were split into T225 flasks. 48 hours after infection, cells were selected with puromycin for 72 hours to remove uninfected cells. Seventy-two hours after selection, OCI-Ly1 cells were treated with either DMSO or 100 nM venetoclax and passaged in fresh media containing either DMSO or drug every 3-4 days. Cells were harvested 10 days after initiation of treatment. Isolation of genomic DNA, sequencing and analyses were performed as for the CRISPR-Cas9 screen. The log2 fold-change of each ORF was determined relative to the initial time point for each. This experiment was performed in duplicates.
Generation of CRISPR-Cas9 engineered isogenic cell lines and overexpression cell lines
Cloning of sgRNA vectors
Two of 4 sgRNAs per target were selected from the BRUNELLO genome-scale library (based on highest levels of representation from the genome-wide screen) and related DNA oligonucleotides were synthesized (Gene Link; Table S5), along with oligonucleotides corresponding to 2 control non-targeting sgRNAs per gene. Oligonucleotides were phosphorylated and annealed using T4 PNK (New England Biolabs). The backbone vector (pLKO5.sgRNA.EFS.GFP) (Addgene Plasmid #57822) (Heckl et al., 2014) was digested with FastDigest BsmBI (Thermo Scientific), and the vector and oligonucleotides were ligated with T7 DNA ligase (New England Biolabs). The ligation reaction was treated with Plasmid-Safe exonuclease (Epicentre) to prevent unwanted recombination products. The final product (1 μL) was transformed into 25 μL of DH5α competent cells (New England Biolabs). Colonies were selected and sequenced before undergoing plasmid DNA extraction (Endotoxin-Free Plasmid Maxiprep, Qiagen).
Cloning of ORFs vectors
The ORFs for PRKAR2B and PRKAA2 (clone ID: TRCN0000480583 and TRCN0000492160, respectively; Broad Institute Genetic Perturbation Platform ORFeome library (https://portals.broadinstitute.org/gpp/public/)) were cloned into the pLX_TRC317. This is a lentiviral expression vector that encodes a puromycin resistance cassette and an ORF expression cassette under control of the EF1-alpha promoter.
Generation of a Cas9 overexpressing cell line
Stable Cas9-expressing OCI-Ly1 and SU-DHL4 cells were generated by transducing parental cells with lentivirus prepared with lentiCas9-Blast pXR101 (Addgene plasmid # 52962) (Sanjana et al., 2014) encoding Cas9 and blasticidin resistance. Selection with blasticidin (10 μg/mL) was initiated 48 hr after transduction and sustained throughout culture of Cas9-expressing cell lines. Cas9 activity was checked as previously reported by using the pXPR-011 vector (Addgene plasmid # 59702) (Doench et al., 2014).
Lentivirus production and purification
To produce lentivirus, ~800,000 HEK293T cells were seeded per well in a 6-well plate in 2.7 mL of antibiotic-free DMEM supplemented with 10% FBS. For each well, 150 μL of OptiMEM (Life Technologies) was mixed with 5 μg of pLKO5_sgRNA plasmid, 0.4 μg of pVSV.G (Addgene #12259), and 1.5 μg of psPAX2 (Addgene #12260). Separately, 9 μl of Lipofectamine 2000 (Life Technologies) was diluted in 150 μl OptiMEM. After a 15 min incubation at room temperature, the DNA and Lipofectamine mixes were combined and incubated together at room temperature for 30 min before being added to the cells. After 12 hr, the media was changed to DMEM supplemented with 20% FBS. 48 hr post-transfection, 3 mL of media was removed and filtered through a 0.45 μm low protein binding membrane (Millipore Steriflip HV/PVDF) and added to 1 mL of LentiX Concentrator (Clontech). This mixture was then incubated at 4°C for 2 h, and centrifuged at 1500×g for 45 min at 4°C. The pellet was resuspended in 100 μL of PBS and stored in aliquots at −80°C.
Generation of engineered cell lines
0.5 × 106 target cells (OCI-Ly1, SU-DHL4, SU-DHL6) were suspended in media supplemented with 8 μg/mL polybrene and seeded into 6-well plates (1 mL per well), to which lentivirus was added (50 μL/mL to each well). The plates were spun at 2000 rpm for 2 hr at 37°C and incubated at 37°C for 24 hr. The polybrene-containing media was then replaced by 2 mL of fresh media per well. After 3 days, transduced cells were selected (i.e. by puromycin (1 μg/mL for OCI-Ly1, 0.5 μg/mL for SU-DHL4 and SU-DHL6) for 1 week for the ORF-overexpressing cells) or sorted (i.e. based GFP or mCherry expression for the CRISPR gene-edited lines) and cryopreserved for further experiments. As confirmation that the engineered cell lines expressed the expected alterations, we: (i) evaluated the expression of the targeted protein by western-blotting (Figure S2E-F, Figure S5D-J), and (ii) for the CRISPR-Cas9 engineered cell lines, performed targeted DNA sequencing for the CRISPR target sites before and after 2 weeks of exposure to venetoclax (100 nM), and assessed the proportion of frameshift indels (Figure S2I). In brief, we used a two-step touchdown PCR protocol. Genomic DNA from the pre- and post-treated samples was PCR-amplified using KAPA HiFi DNA polymerase and primers specific for the target sequence of the sgRNAs. Products from the first reaction were barcoded with Illumina sequencing adaptor sequences and indexes during a second round of PCR. Following PCR, samples were purified with Agencourt AMPure XP beads (Beckman Coulter) and quantified on a Bioanalyzer (Agilent) with High Sensitivity DNA chips. Sample libraries were diluted to 4 nM, pooled, and ran on the Illumina MiSeq platform using single-end sequencing with the following parameters: read 1: 296nt, index 1: 6nt.
Single-cell cloning of the ID3 knockout OCI-Ly1 cells was performed using dose-limiting dilution strategy. Cells from the bulk ID3 OCI-Ly1 were seeded at a concentration of 0.5 cells/well in a 96-well plate (5 plates per cell line). To investigate the presence of mutations, 6 clones per sgRNA were analyzed by PCR and Sanger sequencing using primers flanking the target sites for the sgRNAs. The absence of ID3 protein were confirmed in clones harboring loss-of-function mutations by western blot (Figure S5M). Three different knockout single-cell clones for each sgRNA were used for further functional studies.
Generation of OCI-Ly1 Double-knock out for BAX and BAK1
To achieve stable down-regulation of both BAX and BAK proteins in the OCI-Ly1 cell line, a two-step transduction and selection process was performed. First, OCI-Ly1 cells were transduced with lentivirus expressing sgRNA targeting BAX (5’-TCGGAAAAAGACCTCTCGGG-3’), together with a GFP marker, selected by cell sorting after 5 days, and cultured in vitro for at least a week before western blot assessment of successful BAX targeting; then, the cell line was further transduced with lentivirus expressing sgRNA targeting BAK1 (5’-GCTCACCTGCTAGGTTGCAG-3’), together with an mCherry marker. Five days after transduction, double-positive cells were sorted, and cultured for 8 days before assessment of protein expression by Western Blot. A similar two-step transduction was obtained in the OCI-Ly control line, transduced with non-targeting sgRNA-expressing lentivirus (one carrying GFP, one carrying mCherry).
Whole-exome sequencing and data analyses
Library construction from CLL and matched germline DNA of Patients 1–6 was performed as previously described (Fisher et al., 2011), with the following modifications: (i) initial genomic DNA input into shearing was reduced from 3 μg to 10–100 ng in 50 μl of solution; (ii) for adapter ligation, Illumina paired-end adapters were replaced with palindromic forked adapters (from Integrated DNA Technologies), with unique dual-indexed molecular barcode sequences included in the adapter sequence to facilitate downstream pooling. With the exception of the palindromic forked adapters, the reagents used for end repair, A-base addition, adapter ligation and library enrichment PCR were purchased from KAPA Biosciences in 96-reaction kits, (iii) during the post-enrichment solid-phase reversible immobilization (SPRI) cleanup, elution volumes were reduced to 30 μL to maximize library concentration, and a vortexing step was added to maximize the amount of template eluted. Any libraries with concentrations below 40 ng/ml (per PicoGreen assay, automated on an Agilent Bravo) were considered failures and reworked from the start of the protocol. Following library construction, hybridization and capture were performed using the relevant components of Illumina’s Nextera Rapid Capture Exome Kit and following the manufacturer’s suggested protocol. with the following exceptions: first, all libraries within a library construction plate were pooled prior to hybridization. Second, the Midi plate from Illumina’s Nextera Rapid Capture Exome Kit was replaced with a skirted PCR plate to facilitate automation. All hybridization and capture steps were automated on the Agilent Bravo liquid handling system. After post-capture enrichment, library pools were quantified using qPCR (automated assay on the Agilent Bravo), using a kit purchased from KAPA Biosystems with probes specific to the ends of the adapters. On the basis of qPCR quantification, libraries were normalized to 2 nM, and then denatured using 0.1N NaOH on the Hamilton Starlet. After denaturation, libraries were diluted to 20pM using hybridization buffer purchased from Illumina.
Cluster amplification of denatured templates was performed according to the manufacturer’s protocol (Illumina) using HiSeq 4000 cluster chemistry and HiSeq 4000 flowcells. The flowcells are then analyzed using RTA v.1.18.64 or later. Each pool of whole exome libraries was run on paired 76bp runs, reading the dual-indexed sequences to identify molecular indices and sequenced across the number of lanes needed to meet coverage for all libraries in the pool.
Alignments to hg19 using bwa version 0.5.9-r16 (Li and Durbin, 2010) and quality control were performed using the Picard (http://picard.sourceforge.net/) and Firehose (pipelines at the Broad Institute). Firehose is a framework combining workflows for the analysis of cancer-sequencing data. The workflows perform quality control, local re-alignment, mutation calling, small insertion and deletion identification, rearrangement detection and coverage calculations, among other analyses. Average coverage and quality control metrics are listed in Table S3.
Identification of somatic mutations
Sequencing output was processed with the Picard and GATK toolkits developed at the Broad Institute, a process that involves marking duplicate reads, recalibrating base qualities and realigning around somatic small insertions and deletions (sINDELs). All BAM files were generated by aligning with bwa version 0.5.9 to the NCBI Human Reference Genome Build hg19. Prior to variant calling, the impact of oxidative damage (oxoG) and FFPE damage to DNA during sequencing was quantified according to Costello et al. (Costello et al., 2013). The cross-sample contamination was measured with ContEst based on the allele fraction of homozygous SNPs (Cibulskis et al., 2011), and this measurement was used in MuTect. From the aligned BAM files, somatic alterations were identified using a set of tools developed at the Broad Institute (www.broadinstitute.org/cancer/cga). The details of our sequencing data processing have been described elsewhere (Berger et al., 2011; Chapman et al., 2011).
Following our standard procedure, sSNVs were detected using MuTect (version 1.1.6) (Cibulskis et al., 2013); sINDELs were detected using Strelka (Saunders et al., 2012). We then applied a stringent set of filters to improve the specificity of our sSNV and sINDEL calls and remove likely FFPE artifacts. We applied an allele fraction specific panel-of-normals filter, which compares the detected variants to a large panel of normal exomes and removes variants that were observed in the panel-of-normals. We then applied a realignment based filter, which removes variants that can be attributed entirely to ambiguously mapped reads. All filtered events in candidate CLL genes were also manually reviewed using the Integrated Genomics Viewer (IGV) (Robinson et al., 2011). In the matched sample sets from 6 individuals, we utilized “forced calling” to quantify the number of reads supporting the alternate and reference alleles at sites which were detected in any sample from that individual. Estimation of and correction for tumour contamination in normal was performed using the deTiN algorithm (Taylor-Weiner et al., 2018) to recover somatic mutations that would have otherwise been filtered out due to evidence of the mutation in the normal. To address the lack of a matched normal sample (in Patient 3) we used a stringent panel-of-normals and population allele frequency criteria, and excluded non-coding variants from analysis. To address the lack of a matched normal sample (in Patient 6), we used a stringent panel-of-normals and population allele frequency criteria, and excluded non-coding variants from analysis. Furthermore, parental OCI-Ly1-S cells were used as a source control DNA in order to highlight sSNVs that were acquired in the resistant OCI-Ly1-R cells. Reference lists for sSNVs and sINDELs in known putative CLL driver genes as well as for recurrent CNAs were concatenated based on previous sequencing studies of large CLL cohorts (Wang et al., 2011; Quesada et al., 2012; Landau et al., 2013; Puente et al., 2015; Landau et al., 2015). Total copy number was measured using ReCapSeg (Ramos et al., 2015), then segmented into allelic copy number with AllelicCapSeg (Stachler et al., 2015) based on heterozygous germline sites detected with HaplotypeCaller according to the protocol described previously (http://archive.broadinstitute.org/cancer/cga/acsbeta).
Estimation of mutation cancer cell fraction using ABSOLUTE and clonal evolution mapping
The cancer cell fraction (represented as a probability density distribution ∈ [0, 1]) of individual somatic alterations were estimated using the ABSOLUTE algorithm (v1.5) which calculates the sample purity, ploidy, and local absolute DNA copy-number of each mutation, as previously described (Carter et al., 2012; Landau et al., 2013). CCFs were clustered as previously described (Landau et al., 2013) to delineate distinct subclonal populations. Phylogenetic relationships between these populations were inferred using patterns of shared mutations and CCF using the PhylogicNDT analysis (Leshchiner et al., 2018).
RNA sequencing
cDNA Library Construction
Total RNA was quantified using the Quant-iT™ RiboGreen® RNA Assay Kit and normalized to 5 ng/μl Following plating, 2 μL of ERCC controls (using a 1:1000 dilution) were spiked into each sample. An aliquot of 200ng for each sample was transferred into library preparation which uses an automated variant of the Illumina TruSeq™ Stranded mRNA Sample Preparation Kit. This method preserves strand orientation of the RNA transcript. It uses oligo dT beads to select mRNA from the total RNA sample, followed by heat fragmentation and cDNA synthesis from the RNA template. The resultant 400bp cDNA then goes through dual-indexed library preparation: ‘A’ base addition, adapter ligation using P7 adapters, and PCR enrichment using P5 adapters. After enrichment, the libraries were quantified using Quant-iT PicoGreen (1:200 dilution). After normalizing samples to 5 ng/μL, the set was pooled and quantified using the KAPA Library Quantification Kit for Illumina Sequencing Platforms.
The entire process was in a 96-well format and all pipetting is done by either Agilent Bravo or Hamilton Starlet.
Illumina Sequencing
Pooled libraries were normalized to 2 nM and denatured using 0.1 N NaOH prior to sequencing. Flowcell cluster amplification and sequencing were performed according to the manufacturer’s protocols using either the HiSeq 2000 or HiSeq 2500 instrument. Each run generated a 101bp paired-end with an eight-base index barcode read. Data was analyzed using the Broad Picard Pipeline, which includes de-multiplexing and data aggregation.
Data analyses
RNA-seq data were aligned to GRCh38.p5 with STAR-2.5.1b (Dobin et al., 2013). Gene expression was quantified with RSEM-1.2.31 (Li and Dewey, 2011). DESeq2 (Love et al., 2014) was applied to call differentially expressed genes between each cell line and control group. Pathway enrichment analysis was performed with GSEA (Subramanian et al., 2005) in GenePattern (Reich et al., 2006).
Mass spectrometry-based proteome investigations
In Solution Digestion
OCI-Ly1 cell pellets were lysed at 4 °C in 8 M urea, 50 mM Tris-HCl pH 8.0, 150 mM NaCl, 1 mM EDTA, 2 μg/μl aprotinin (Sigma-Aldrich), 10 μg/μl leupeptin (Roche), and 1 mM phenylmethylsulfonyl fluoride (PMSF) (Sigma). Protein concentration was determined using a bicinchoninic acid (BCA) protein assay (Pierce). Proteins were reduced with 5 mM (DTT) for 45 min at room temperature (RT), followed by alkylation with 10 mM iodoacetamide for 30 min at room temperature in the dark. Urea concentration was reduced to 2 M with 50 mM Tris-HCl, pH 8. Samples were pre-digested for 2 hr at 30 °C with endoproteinase Lys-C (Wako Laboratories) at an enzyme-to-substrate ratio of 1:50. Samples were digested overnight at 37°C with sequencing grade trypsin (Promega) at an enzyme-to-substrate ratio of 1:50. Following overnight digest, samples were acidified with neat formic acid to a final concentration of 1%. Acidified samples were subsequently desalted on a 100 mg tC18 Sep-Pak SPE cartridge (Waters). Briefly, cartridges were conditioned with 1 mL of 100% MeCN, 1 mL of 50% MeCN/0.1% FA, and 4x with 1 mL of 0.1% TFA. The sample was loaded, and washed 3x with 1 mL of 0.1% TFA, 1x with 1 mL of 1% FA, and eluted 2x with 600 μl of 50% MeCN/0.1% FA. Following desalting, 100 μg of the sample was dried to completion and stored at −80 °C.
TMT labeling of peptides
Desalted peptides were labeled with TMT 10-plex isobaric mass tagging reagents (Thermo Fisher Scientific) as previously described (Mertins et al., 2018). Each TMT reagent was resuspended in 41 μL of MeCN. Peptides were resuspended in 100 μL of 50 mM HEPES and combined with TMT reagent. Samples were incubated at RT for 1 hr while shaking. The TMT reaction was quenched with 8 μL of 5% hydroxylamine at RT for 15 min with shaking. TMT labeled samples were combined, dried to completion, reconstituted in 100 μL of 0.1% FA, and desalted on StageTips or 100 mg SepPak columns as described above.
Basic Reverse Phase (bRP) Fractionation
The TMT labeled samples were fractionated using offline high pH reversed-phase chromatography (bRP) as previously described (Mertins et al., 2018). Samples were fractionated using Zorbax 300 Extend C18 column (4.6 × 250 mm, 300 Å, 5 μm, Agilent) on an Agilent 1100 series high-pressure liquid chromatography (HPLC) system. Samples were reconstituted in 900 μL of 5 mM ammonium formate (pH 10.0)/2% (vol/vol) acetonitrile (MeCN) (bRP solvent A). Samples were injected with Solvent A at a flow rate of 1 mL/min and separated using a 96 min gradient. The gradient consisted of an initial increase to 16% solvent B (90% MeCN, 5 mM ammonium formate, pH 10), followed by 60 min linear gradient from 16% solvent B to 40% B and successive ramps to 44% and 60% at a flow rate of 1 mL/min. Fractions were collected in a 96-deep well plate (GE Healthcare) and pooled in a non-contiguous manner into final 24 proteome fractions. Pooled fractions were dried to completeness using a SpeedVac concentrator.
Liquid chromatography and mass spectrometry
Desalted peptides were resuspended in 9 μL of 3% MeCN/0.1% FA and analyzed by online nanoflow liquid chromatography tandem mass spectrometry (LC-MS/MS) using Q-Exactive + mass spectrometer (Thermo Fisher Scientific) coupled on-line to a Proxeon Easy-nLC 1200 (Thermo Fisher Scientific) as previously described (Mertins et al., 2018). Briefly, 4 μL of each sample was loaded onto a microcapillary column (360 μm outer diameter × 75 μm inner diameter) containing an integrated electrospray emitter tip (10 μm), packed to approximately 22 cm with ReproSil-Pur C18-AQ 1.9 μm beads (Dr. Maisch GmbH) and heated to 50 °C. Samples were analyzed with 110 min method. The 110 min method contained a mobile phase with a flow rate of 200 nL/min, comprised of 3% acetonitrile/0.1% formic acid (Solvent A) and 90% acetonitrile /0.1% formic acid (Solvent B), with the following gradient profile: (min:%B) 0:2; 1:6; 85:30; 94:60; 95:90; 100:90; 101:50; 110:50 (the last two steps at 500 nL/min flow rate). The Q-Exactive+ MS was operated in the data-dependent mode acquiring HCD MS/MS scans (r = 35,000) after each MS1 scan (r = 70,000) on the 12 most abundant precursor ions using an MS1 target of 3 × 106 and an MS2 target of 5 × 104. The maximum ion time utilized for MS/MS scans was 120ms; the HCD-normalized collision energy was set to 30; the dynamic exclusion time was set to 20 s, isotope exclusion function was enabled, and peptide match function was set to preferred. Charge exclusion was enabled for charge states that were unassigned, 1 and >6.
Data Analysis
All data were analyzed using Spectrum Mill software package v 6.1 pre-release (Agilent Technologies). Similar MS/MS spectra acquired on the same precursor m/z within +/− 60 s were merged. MS/MS spectra were excluded from searching if they were not within the precursor MH+ range of 750-4000 Da or if they failed the quality filter by not having a sequence tag length >0. MS/MS spectra were searched against UniProt human database. All spectra were allowed +/− 20 ppm mass tolerance for precursor and product ions, 30% minimum matched peak intensity, and “trypsin allow P” enzyme specificity with up to 2 missed cleavages. The fixed modifications were carbamidomethylation at cysteine, and TMT at N-termini and internal lysine residues. Variable modifications included oxidized methionine and N-terminal protein acetylation. Individual spectra were automatically designated as confidently assigned using the Spectrum Mill autovalidation module. Specifically, a target-decoy based FDR scoring threshold criteria via a two-step auto threshold strategy at the spectral and protein levels was used. First, peptide mode was set to allow automatic variable range precursor mass filtering with score thresholds optimized to yield a spectral level FDR of <1.2%. A protein polishing autovalidation was applied to further filter the peptide spectrum matches using a target protein-level FDR threshold of 0. Following autovalidation, a protein-protein comparison table was generated, which contained experimental ratios. For all experiments, non-human contaminants and reversed hits were removed. Furthermore, data were filtered to only consider proteins with 2 or more unique peptides and was median normalized.
For statistical analysis, a moderated t-test was implemented in R-Shiny using the limma R library. Correction for multiple testing was performed using the Benjamini-Hochberg false discovery rate method.
Metabolic assays
Measurement of oxygen consumption and extracellular acidification rate
OCI-Ly and SU-DHL4 cells were counted and plated on a Seahorse 96-well plate at a density of 1.25×105 or 5×105 cells, respectively in Seahorse XF DMEM media (Agilent). OCR and ECAR were simultaneously recorded using a Seahorse XF96 Analyzer (Agilent) for 12-38 consecutive measurements. OCR is measured before and after the addition of inhibitors to assess mitochondrial function by deriving several parameters of mitochondrial respiration: (i) basal respiration, (ii) ATP-linked respiration and proton leak respiration (after 3 μM oligomycin [Sigma], a complex V inhibitor) and (iii) maximal respiration (after 1 μM carbonyl cyanide m-chlorophenyl hydrazine (CCCP) [Sigma], a protonophore). Mitochondrial respiration is finally inhibited by 1 μM antimycin A (Sigma), a complex III inhibitor.
To measure the effect of venetoclax on OCR/ECAR, cells were pre-treated for 1h with 10μM of zVAD-FMK (Abcam) or left untreated and OCR/ECAR were recorded for 4 measurements before injection of venetoclax at a final concentration of 100 nM. DMSO injection was used as control.
Mitochondrial DNA copy number
Mitochondrial DNA copy number was determined using a multiplexed qPCR assay previously reported (Bao et al., 2016). QPCR quantification of relative abundance of mitochondrial (mtDNA) and nuclear (nucDNA) DNA was performed with the following primers and probes: ND2 forward (5’-tgttggttatacccttcccgtacta-3’), ND2 reverse (5’-cctgcaaagatggtagagtagatga-3’), and probe (5’-ccctggcccaaccc-3’, 6FAM on the 5’ end and MGBNFQ on the 3’ end) for detection of mtDNA; AluYb8 forward (5’-cttgcagtgagccgagatt-3’), AluYb8 reverse (5’-gagacggagtctcgctctgtc-3’) and probe (5’-actgcagtccgcagtccggcct-3’, VIC on 5’, MGBNFQ on 3’) for detection of nucDNA. Cycling conditions for qPCR were the following: 95C for 10 minutes followed by 40 cycles of 95C 15sec, 55C 15sec, 60C 1 min). Ten nanograms of genomic DNA extracted from OCI-LY1 S, OCI-LY1 R, SU-DHL4 S, SU-DHL4 R were used for this assay, in technical triplicates.
Measurement of mitochondrial superoxide levels
To measure relative levels of mitochondrial superoxide, cells were stained with 5 μM MitoSOX Red (Invitrogen, cat# M36008) for 10 min at 37°C. Cells were then washed three times with the media. Using a flow cytometer (FACSymphony, BD), MitoSOX Red was excited at 488 nm and fluorescence emission at 610/20nm (BB630) was measured. Relative fluorescence intensity from biological triplicate of 10,000 cells were used as an indicator of mitochondrial superoxide levels.
Mitochondrial membrane potential
Cells were stained with 2.5 μM JC-1 (ThermoFisher Scientific, cat# T3168) for 30 min at 37 °C. Cells were then washed three times with the media and subjected to the flow cytometry (FACS-Canto) following manufacturer instruction. Briefly, JC-1 was excited at 488 nm and its emission at both 525 nm (FITC-A) and 585 nm (PE-A) were measured. By comparing the ratio of emission at 585nm/525nm, relative levels of mitochondrial membrane potential were determined from 10,000 cells in biological triplicate.
ATAC sequencing
ATAC-seq was performed on 50,000 cells as described (Buenrostro et al., 2013). Transposition reactions were performed for 1 hr at 37° C, followed by purification and sample barcoding by PCR. Samples were sequenced on an Illumina NextSeq 500, 35 base pair cycles in paired-end mode. ATAC-seq reads in fastq format were trimmed to remove adapter sequences using skewer v0.2.2.
Trimmed reads were aligned to hg19 using bowtie2 v2.3.3.1 (Langmead and Salzberg, 2012) with the following parameters:
'-D 20 -R 3 -N 1 -L 20 --local -i S,1,0.50 --rdg 5,1 --rfg 5,1 -X 2000'. Alignments were filtered using samtools v1.9 (Li and Durbin, 2009) to remove duplicate reads and retain only proper pairs with mapping quality greater than 30. Reads mapping to blacklisted regions were removed using samtools v1.9. Reads were converted to bed format and reads on the forward and reverse strands were shifted +4 and −5 bp respectively. MACS2 v2.1.1 (Zhang et al., 2008) was then used to call peaks and summits using the following parameters: '--nomodel --extsize 200 --shift −100 --nolambda --keep-dup all −q 0.1'. IDR v2.0.4.2 was used to filter enriched regions at an IDR of less than 0.05 to produce high-confidence peak and summit sets. High-confidence summits were expanded to 500bp and a count matrix was generated with Rsubread v1.30.9 (Chen et al., 2016). This matrix was imported to R and chromVAR (Schep et al., 2017) was used in conjunction with motifs in the JASPAR2018 (10.18129/B9.bioc.JASPAR2018) (Khan et al., 2018) and chromVARmotifs (https://github.com/GreenleafLab/chromVARmotifs) R packages to generate motif-level measures of differential accessibility. Signal tracks were generated from bigwig files created from aligned bams using Deeptools v3.1.1 bamCoverage command (Ramírez et al., 2016) with the following parameters: "--Offset 1 -bs 50 --smoothLength 150 --maxFragmentLength 2000 --scaleFactor SCALEFACTOR". Scaling factors for each sample were generated by quantifying total cutsites within each sample's IDR-filtered peaks and dividing by 1e6. Signal tracks for combined replicates were generated by first concatenating replicate fastqs, then applying the same processing steps described above.
Immunohistochemistry and image acquisition
Immunohistochemical staining (IHC) for MCL1 (Clone 22, 1:100, Santa Cruz), Acetyl-CoA Carboxylase (Clone C83B10, 1:50, Cell Signaling), Phospho-Acetyl-CoA Carboxylase (Clone D7D11, 1:50, Cell Signaling) or AMPKα (clone D5A2, 1:50, Cell Signaling) was performed using an automated staining system (BondRX, Leica Biosystems, Buffalo Grove, IL) according to the manufacturer’s protocol and as previously described (Roemer et al., 2016). Briefly, 4-μm thick formalin-fixed paraffin-embedded (FFPE) tissue sections were baked for 3h at 60 °C before loading onto the BOND RX. Slides were deparaffinized (BOND DeWax Solution, Leica Biosystems), rehydrated and antigen retrieval was performed onto the autostainer. Antigen retrieval was performed in BOND Epitope Retrieval Solution 1 (ER1, Leica Biosystems) at pH 6 for 30 min at 98 °C or Epitope Retrieval Solution 2 (ER2, Leica Biosystems) at pH 9 for 20 min at 98 °C as indicated above for each primary antibody. Next, slides were incubated with primary antibodies for 40 min. Subsequently, a secondary antibody of anti-rabbit Polymeric Horseradish Peroxidase (PolyHRP, BOND Polymer Refine Detection Kit, Leica Biosystems) was applied for 10 min. As a last step, slides were counterstained by hematoxylin for 5 minutes.
Image acquisition was performed using the Mantra imaging platform (Akoya PerkinElmer), as previously described (Carey et al., 2017). At least 5 or more representative regions of interest per section at 20x magnification were chosen by the pathologist within the tumor area. Areas without tumor or normal tissue were excluded from the analysis. Image Analysis was performed using the InForm 2.4 Image Analysis Software (PerkinElmer). All nucleated tumor cells within a field of view were identified via nuclear hematoxylin staining. Image analysis algorithms were trained by a pathologist to identify tumor cells that were positive for each of the targets above. A percentage of the total tumor cells identified that are positive for the stain of interest is given.
Quantitative PCR
The ID3 and PRKAR2B quantitative PCR assay was designed using a nested design, with outer primers for preamplification and inner primers for qPCR detection. ACTB and B2M were used as housekeeping genes. For cDNA reaction, 200 ng FFPE RNA or 100 ng fresh frozen RNA were dispensed to a PCR tube. Six qPCR replicates were performed per sample.
Quantification and Statistical Analysis
Data analyses were carried out using GraphPad Prism version 7 and R software. The data are summarized as mean ± s.e.m. or s.d. as indicated, and presented as individual values, scatter plots, heatmap or with column bar graphs. Data were analyzed using Student’s t-tests. One-way ANOVA was used to compare three or more groups in time point analyses. Differences were considered significant when p < 0.05.
Synergy analysis for venetoclax drug combinations were performed using the Chou-Talalay method based on the median-effect principle (Chou, 2010). Combination index values and isobolograms were generated using CompuSyn. Excess over Bliss scores were calculated based on the Bliss Independence model of drug synergy (Bliss, 1939). Trajectory analyses for effect of venetoclax on metabolism were performed by looking at the least squares regression of OCR/ECAR vs time for each cell line/treatment. Survival time data were calculated using the Kaplan-Meier method for and were compared by log-rank testing.
Data and Code Availability
The RNA seq and ATAC seq data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE 128563 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE128563).
The original mass spectra and sequence database used for searches may be downloaded from MassIVE (http://massive.ucsd.edu) using the identifier: MSV000083512. The dataset is directly accessible via ftp://massive.ucsd.edu/MSV000083512.
dbGaP accession number for the depositing of WES data for this study is phs001875.v1.p1.
Supplementary Material
Table S5 CRISPR-Cas9 screen results, Related to Figure 2.
Table S6. ORF screen results, Related to Figure 2.
Table S8. OCI-Ly1 sSNVs report, Related to Figure 7.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Bcl2 | abcam | Cat# ab59348, RRID:AB_2064155 |
| Mcl-1 | Santa Cruz Biotechnology | Cat# sc-12756, RRID:AB_627915 |
| Bcl-xL | Santa Cruz Biotechnology | Cat# sc-8392, RRID:AB_626739 |
| Bim | Cell Signaling Technology | Cat# 2819, RRID:AB_10692515 |
| Bak | Cell Signaling Technology | Cat# 3814, RRID:AB_2290287 |
| Bax | Cell Signaling Technology | Cat# 2772, RRID:AB_10695870 |
| Pegasus | Santa Cruz Biotechnology | Cat# sc-517137 |
| OTUD5 | Cell Signaling Technology | Cat# 20087, RRID:AB_2798833 |
| NOXA | Santa Cruz Biotechnology | Cat# sc-56169, RRID:AB_784877 |
| Id3 | Cell Signaling Technology | Cat# 9837, RRID:AB_2732885 |
| Id2 | Cell Signaling Technology | Cat# 3431, RRID:AB_2122877 |
| p300 | Santa Cruz Biotechnology | Cat# sc-48343, RRID:AB_628075 |
| UBR5 | Cell Signaling Technology | Cat# 65344, RRID:AB_2799679 |
| IkappaB-alpha | Cell Signaling Technology | Cat# 9242, RRID:AB_331623 |
| NF-1 | Santa Cruz Biotechnology | Cat# sc-74444, RRID:AB_2153048 |
| AMPK-alpha | Cell Signaling Technology | Cat# 2532, RRID:AB_330331 |
| AMPK-alpha | Cell Signaling Technology | Cat #5831 |
| PKA beta | Abcam | Cat# ab94612, RRID:AB_10675826 |
| GAPDH | Cell Signaling Technology | Cat# 2118, RRID:AB_561053 |
| Acetyl-CoA Carboxylase | Cell Signaling Technology | Cat# 3676 |
| Phospho-Acetyl-CoA Carboxylase (Ser79) | Cell Signaling Technology | Cat# 11818 |
| Bacterial and Virus Strains | ||
| NEB® 5-alpha Competent E. coli | New England BioLabs | C2987I |
| Biological Samples | ||
| Human CLL Primary Peripheral Blood Mononuclear Cells or Tissue | DFCI, MDACC | N/A |
| Chemicals, Peptides and Recombinant Proteins | ||
| Venetoclax ABT-199 | Selleck Chemicals | S8048; CAS: 1257044-40-8 |
| Venetoclax ABT-199 | LC-Laboratories | CAS#1257044-40-8 |
| Dorsomorphin | Sigma | P5499; CAS: 866405-64-3 |
| Oligomycin A | Sigma | 75351; CAS: 579-13-5 |
| Antimycin A | Sigma | A8674; CAS: 1397-94-0 |
| MCL-1 inhibitor S63845 | Chemietek | S-63845; CAS: CT-S63845 |
| AMPK activator A-769662 | Santa Cruz Biotechnology | A-769662; CAS: 844499-71-4 |
| Cristical Commercial Assays | ||
| DNAeasy Blood and Tissue Kit | Qiagen | #69504 |
| RNeasy FFPE Kit | Qiagen | #73504 |
| Cell Titer-Glo Luminescent Cell Viability Assay | Promega | G7571 |
| Pierce™ BCA Protein Assay Kit | ThemoFisher | #23225 |
| QIAGEN Plasmid Maxi Kit | Qiagen | #12162 |
| KAPA Biosciences 96-reaction kits | Illumina | KK8234 |
| Quant-iT™ PicoGreen™ dsDNA Assay Kit | ThemoFisher | P11496 |
| Nextera Rapid Capture Exome Kit | Illumina | #20020616 |
| Quant-iT™ RiboGreen™ RNA Assay Kit | ThemoFisher | R11490 |
| BOND Polymer Refine Detection Ki | Leica Biosystems | DS9800 |
| Deposited Data | ||
| RNA seq and ATAC seq data | This paper | NCBI's Gene Expression Omnibus GSE 128563 |
| Proteomics data | This paper | MassIVE database MSV000083512 |
| WES data | This paper | dbGaP phs001875.v1.p1 |
| Experimental Models: Cell Lines | ||
| OCI-Ly1 | DSMZ | ACC 722 |
| SU-DHL-4 | Letai lab, DFCI, Boston MA | ATCC® CRL-2957™ |
| SU-DHL-6 | Letai lab, DFCI, Boston MA | ATCC® CRL-2959™ |
| Toledo | ATCC | ATCC® CRL-2631™ |
| Mouse: NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ | The Jackson Laboratory | JAX : 005557 |
| Oligonucleotides | ||
| Primer: ID3 Single-cell Screening Forward: 5’-TGACAAGTTCCGGAGTGAGC-3’ | This paper | N/A |
| Primer: ID3 Single-cell Screening Reverse: 5’-CGGTATCAGCGCTTCCTCAT-3’ | This paper | N/A |
| ND2 forward: 5’-tgttggttatacccttcccgtacta-3’ | Bao et al., 2016 | N/A |
| ND2 reverse: 5’-cctgcaaagatggtagagtagatga-3’ | Bao et al., 2016 | N/A |
| mtDNA probe: 5’-ccctggcccaaccc-3’ | Bao et al., 2016 | N/A |
| AluYb8 forward: 5’-cttgcagtgagccgagatt-3’ | Bao et al., 2016 | N/A |
| AluYb8 reverse: 5’-gagacggagtctcgctctgtc-3’ | Bao et al., 2016 | N/A |
| nucDNA probe: 5’-actgcagtccgcagtccggcct-3’ | Bao et al., 2016 | N/A |
| ID3 quantitative PCR Outer Forward: 5'-cgaCCACCTTCCCATCCAgaCA-3' | This paper | N/A |
| ID3 quantitative PCR Outer Reverse: 5'-tcgTCCTTTTGTCGTTGGAGATGACAA-3' | This paper | N/A |
| ID3 quantitative PCR inner Forward: 5'-acgCCAgaCAGCCGAGCTCA-3' | This paper | N/A |
| ID3 quantitative PCR inner Reverse: 5'-cgaGTTGGAGATGACAAGTTCCGGA-3' | This paper | N/A |
| PRKAR2B quantitative PCR Outer Forward: 5'-cgaGAGAGCTTGCCCTGGTAACTAA-3' | This paper | N/A |
| PRKAR2B quantitative PCR Outer Reverse: 5'-cgaCAAATGCTTGCACATCCATTgcTAA-3' | This paper | N/A |
| PRKAR2B quantitative PCR inner Forward: 5'-cgtCTAACAAACCTCGAGCAGCTTCT-3' | This paper | N/A |
| PRKAR2B quantitative PCR inner Reverse: 5'-cgaCATTgcTAAACATTTGACAGTCCCAA-3' | This paper | N/A |
| BAK1 gRNA1: 5'-AAGACCCTTACCAGAAGCAG-3' | This paper | N/A |
| BAK1 gRNA2: 5'-GGTAGACGTGTAGGGCCAGA-3' | This paper | N/A |
| BAX gRNA1: 5'-TCGGAAAAAGACCTCTCGGG-3' | This paper | N/A |
| BAX gRNA2: 5'-AGTAGAAAAGGGCGACAACC-3' | This paper | N/A |
| NFKBIA gRNA1: 5'-GGTTGGTGATCACAGCCAAG-3' | This paper | N/A |
| NFKBIA gRNA1: 5'-CTGGACGACCGCCACGACAG-3' | This paper | N/A |
| PMAIP1 gRNA1: 5'-TCGAGTGTGCTACTCAACTC-3' | This paper | N/A |
| PMAIP1 gRNA2: 5'-TTCTTGCGCGCCTTCTTCCC-3' | This paper | N/A |
| IKZF5 gRNA1: 5'-AGTTACTTCGATCACTGCAG-3' | This paper | N/A |
| IKZF5 gRNA2: 5'-ATCAGCTCTCGACTCTAGCA-3' | This paper | N/A |
| ID3 gRNA1: 5'-CTGGTACCCGGAGTCCCGAG-3' | This paper | N/A |
| ID3 gRNA2: 5'-TGGCCAGACTGCGTTCCGAC-3' | This paper | N/A |
| EP300 gRNA1: 5'-GTGGCACGAAGATATTACTC-3' | This paper | N/A |
| EP300 gRNA2: 5'-ATGGTGAACCATAAGGATTG-3' | This paper | N/A |
| NFIA gRNA1: 5'-TCCAGACTTTATCTGCCTGG-3' | This paper | N/A |
| NFIA gRNA2: 5'-CCCTGGATTAGAGCATTGTG-3' | This paper | N/A |
| OTUD5 gRNA1: 5'-CGGCGCAGGCTACAACAGTG-3' | This paper | N/A |
| OTUD5 gRNA2: 5'-GGTTGTGCGAAAGCATTGCA-3' | This paper | N/A |
| BCL2L11 gRNA1: 5'-TATGGATCGCCCAAGAGTTG-3' | This paper | N/A |
| BCL2L11 gRNA2: 5'-GTTCTGATGCAGCTTCCATG-3' | This paper | N/A |
| UBR5 gRNA1: 5'-TTGTTGTCTAAGAACGACGA-3' | This paper | N/A |
| UBR5 gRNA2: 5'-GCAACCAAGATAATGCTAGT-3' | This paper | N/A |
| TP53 gRNA1: 5'-GGATGATTTGATGCTGTCCC-3' | This paper | N/A |
| TP53 gRNA4: 5'-AGACGGAAACCGTAGCTGCC-3' | This paper | N/A |
| BAK1 gRNA1 target site F1 : TTGTGGAAAGGACGAAACACCGCCCATCTCTGCTTTTTCTCG | This paper | N/A |
| BAK1 gRNA1 target site R1 : TCTACTATTCTTTCCCCTGCACTGTTGATTGGCCAGCTTACTTCC | This paper | N/A |
| BAK1 gRNA2 target site F1 : TTGTGGAAAGGACGAAACACCGGGGCTCTCCACCAATGCTA | This paper | N/A |
| BAK1 gRNA2 target site R1 : TCTACTATTCTTTCCCCTGCACTGTGTACCACAAACTGGCCCAAC | This paper | N/A |
| BAX gRNA1 target site F1 : TTGTGGAAAGGACGAAACACCGCTTGGGGCTCAGTCTCCTTA | This paper | N/A |
| BAX gRNA1 target site R1 : TCTACTATTCTTTCCCCTGCACTGTCCTGAGAGCAGGGATGTAGC | This paper | N/A |
| BAX gRNA2 target site F1 : TTGTGGAAAGGACGAAACACCGGCACTGGTTCTCCTCTCTCCT | This paper | N/A |
| BAX gRNA2 target site R1 : TCTACTATTCTTTCCCCTGCACTGTCACTTTACTTCACCCCTGCAC | This paper | N/A |
| NFKBIA gRNA1 target site F1 : TTGTGGAAAGGACGAAACACCGCCTGTCTAGGAGGAGCAGCA | This paper | N/A |
| NFKBIA gRNA1 target site R1 : TCTACTATTCTTTCCCCTGCACTGTGCATCCAATAGGCACTTTGC | This paper | N/A |
| NFKBIA gRNA2 target site F1 : TTGTGGAAAGGACGAAACACCGCCAGCCAGCGTTTATAGGG | This paper | N/A |
| NFKBIA gRNA2 target site R1 : TCTACTATTCTTTCCCCTGCACTGTCTTATGCAACCGGGGACTT | This paper | N/A |
| PMAIP1 gRNA1 target site site F1 : TTGTGGAAAGGACGAAACACCGCAGGTGCACATAAAGCCAAA | This paper | N/A |
| PMAIP1 gRNA1 target site site R1 : TCTACTATTCTTTCCCCTGCACTGTCAAGGGTCTTCCATTCTTGC | This paper | N/A |
| PMAIP1 gRNA2 target site F1 : TTGTGGAAAGGACGAAACACCGCAGGACTGTTCGTGTTCAGC | This paper | N/A |
| PMAIP1 gRNA2 target site R1 : TCTACTATTCTTTCCCCTGCACTGTGCGAGGAGGAAAGAGAGGAG | This paper | N/A |
| IKZF5 gRNA1 target site site F1 : TTGTGGAAAGGACGAAACACCGTGCTTATGAGCGTCATCTGG | This paper | N/A |
| IKZF5 gRNA1 target site site R1 : TCTACTATTCTTTCCCCTGCACTGTTCAGGGCAGGGAACTACATC | This paper | N/A |
| IKZF5 gRNA2 target site F1 : TTGTGGAAAGGACGAAACACCGCCACGAAATCCCAAATATCC | This paper | N/A |
| IKZF5 gRNA2 target site R1 : TCTACTATTCTTTCCCCTGCACTGTCTTGGCTGGCTGTTTCCTAT | This paper | N/A |
| ID3 gRNA1 target site site F1 : TTGTGGAAAGGACGAAACACCGAGCGGGACTTCTTTTGGTTT | This paper | N/A |
| ID3 gRNA1 target site site R1 : TCTACTATTCTTTCCCCTGCACTGTGCCGTTTAAACCTCCCTCTC | This paper | N/A |
| ID3 gRNA2 target site F1 : TTGTGGAAAGGACGAAACACCGAGCGGGACTTCTTTTGGTTT | This paper | N/A |
| ID3 gRNA2 target site R1 : TCTACTATTCTTTCCCCTGCACTGTCTGGGTGTTCAGCCCTGT | This paper | N/A |
| EP300 gRNA1 target site site F1 : TTGTGGAAAGGACGAAACACCGTTGGTGACCCCTTTTTGAAG | This paper | N/A |
| EP300 gRNA1 target site site R1 : TCTACTATTCTTTCCCCTGCACTGTTGAGCTGAGGCCTAGTTTTTC | This paper | N/A |
| EP300 gRNA2 target site F1 : TTGTGGAAAGGACGAAACACCGCTGTCTTTGTGAACTTGGAAGTGA | This paper | N/A |
| EP300 gRNA2 target site R1 : TCTACTATTCTTTCCCCTGCACTGTCTCCTGTTTGGATCAACATGC | This paper | N/A |
| NFIA gRNA1 target site site F1 : TTGTGGAAAGGACGAAACACCGCGACCCGAATATCGAGAGG | This paper | N/A |
| NFIA gRNA1 target site site R1 : TCTACTATTCTTTCCCCTGCACTGTCTCAGGAAGAATCCGGCATA | This paper | N/A |
| NFIA gRNA2 target site F1 : TTGTGGAAAGGACGAAACACCGAGTCTGGAGGTTGGACCTTG | This paper | N/A |
| NFIA gRNA2 target site R1 : TCTACTATTCTTTCCCCTGCACTGTAGGGTTTTCACGAGGCTTCT | This paper | N/A |
| OTUD5 gRNA1 target site site F1 : TTGTGGAAAGGACGAAACACCGGTGTGGGTGGTGCCGTAG | This paper | N/A |
| OTUD5 gRNA1 target site site R1 : TCTACTATTCTTTCCCCTGCACTGTCAGGGACTGGAATGACGACT | This paper | N/A |
| OTUD5 gRNA2 target site F1 : TTGTGGAAAGGACGAAACACCGGGCATCTCTGACCCTAGCTG | This paper | N/A |
| OTUD5 gRNA2 target site R1 : TCTACTATTCTTTCCCCTGCACTGTTCCCATGGAATGTGTTGATG | This paper | N/A |
| BCL2L11 gRNA1 target site site F1 : TTGTGGAAAGGACGAAACACCGTGGACCACAATGTGATTTTTG | This paper | N/A |
| BCL2L11 gRNA1 target site site R1 : TCTACTATTCTTTCCCCTGCACTGTTGCACAAGGGGAGTACAGAA | This paper | N/A |
| BCL2L11 gRNA2 target site F1 : TTGTGGAAAGGACGAAACACCGGAGTGTGTGAGATGGGCTTG | This paper | N/A |
| BCL2L11 gRNA2 target site R1 : TCTACTATTCTTTCCCCTGCACTGTGTCTGTGCTCATAAAATTCCTTTGC | This paper | N/A |
| UBR5 gRNA1 target site site F1 : TTGTGGAAAGGACGAAACACCGCCAATTCATAAAGCAGTAGCTTAGG | This paper | N/A |
| UBR5 gRNA1 target site site R1 : TCTACTATTCTTTCCCCTGCACTGTCATCACAAATGCTCAAAGAGAG | This paper | N/A |
| UBR5 gRNA2 target site F1 : TTGTGGAAAGGACGAAACACCGTCCTACCCCCTATGCCTTCT | This paper | N/A |
| UBR5 gRNA2 target site R1 : TCTACTATTCTTTCCCCTGCACTGTCTCTGACTGCACCACTGGAA | This paper | N/A |
| TP53 gRNA1 target site site F1 : TTGTGGAAAGGACGAAACACCGCTGGTAAGGACAAGGGTTGG | This paper | N/A |
| TP53 gRNA1 target site site R1 : TCTACTATTCTTTCCCCTGCACTGTGCCAAAGGGTGAAGAGGAAT | This paper | N/A |
| TP53 gRNA2 target site F1 : TTGTGGAAAGGACGAAACACCGGAAGACCCAGGTCCAGATGA | This paper | N/A |
| TP53 gRNA2 target site R1 : TCTACTATTCTTTCCCCTGCACTGTTCAAAAGCCAAGGAATACACG | This paper | N/A |
| CRISPR target site, second PCR forward primer : AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTTTGTGGAAAGGACGAAACACCG | This paper | N/A |
| CRISPR target site, second PCR reverse primer (pre-venetoclax): CAAGCAGAAGACGGCATACGAGATATTGGCGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTTCTACTATTCTTTCCCCTGCACTGT | This paper | N/A |
| CRISPR target site, second PCR reverse primer (post-venetoclax): CAAGCAGAAGACGGCATACGAGATTACAAGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTTCTACTATTCTTTCCCCTGCACTGT | This paper | N/A |
| Recombinant DNA | ||
| pLKO5.sgRNA.EFS.GFP | Heckl et al., 2014 | Addgene Plasmid #57822 |
| pLX_TRC317 | Broad Institute | https://portals.broadinstitute.org/gpp/public/vector/details?vector=pLX_TRC317 |
| lentiCas9-Blast pXR101 | Sanjana et al., 2014 | Addgene Plasmid #52962 |
| pXPR_011 | Doench et al., 2014 | Addgene Plasmid #59702 |
| pMD2.G | Trono's Lab | Addgene Plasmid #12259 |
| psPAX2 | Trono's Lab | Addgene Plasmid #12260 |
| ORF PRKAR2B | Broad Institute | Clone ID TRCN0000480583 |
| ORF PRKAA2 | Broad Institute | Clone ID TRCN0000492160 |
| Human CRISPR Knockout Pooled Library (Brunello) | Doench et al., 2016 | Addgene Pooled Library #73179, #73179-LV, #73178, #73178-LV, 73178-LVC |
| ORFeome library | Broad Institute | https://portals.broadinstitute.org/gpp/public/dir?dirpath=orf_annot |
| Software and Algorithms | ||
| RSEM-1.2.31 | Li and Dewey et al., 2011 | https://deweylab.github.io/RSEM/ |
| DESeq2 | Love et al., 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| GSEA | Subramanian et al., 2005 | http://software.broadinstitute.org/gsea/index.jsp |
| GenePattern | Reich et al., 2006 | http://software.broadinstitute.org/cancer/software/genepattern/ |
| Spectrum Mill software package v 6.1 pre-release | Agilent Technologies | http://proteomics.broadinstitute.org |
| bowtie2 v2.3.3.1 | Langmead B et al., 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Samtools v1.9 | Li et al., 2009 | http://samtools.sourceforge.net/ |
| MACS2 v2.1.1 | Zhang et al., 2008 | https://github.com/taoliu/MACS/ |
| Rsubread v1.30.9 | Chen et al., 2016 | https://bioconductor.org/packages/release/bioc/html/Rsubread.html |
| chromVAR | Schep et al., 2017 | https://github.com/GreenleafLab/chromVARmotifs |
| JASPAR2018 | Khan et al., 2018 | http://jaspar.genereg.net/ |
| Deeptools v3.1.1 | Ramirez et al., 2016 | https://deeptools.readthedocs.io/en/develop/ |
| InForm 2.4 Image Analysis Software | PerkinElmer | https://www.perkinelmer.com |
| GraphPad Prism version 7 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| CompuSyn | Chou et al., 2010 | http://www.combosyn.com/ |
| STARS software (v1.3, Broad Institute) | Broad Institute | https://portals.broadinstitute.org/gpp/public/software/stars |
| Picard tookit | Broad Institute | https://broadinstitute.github.io/picard/ |
| GATK toolkit | Broad Institute | https://software.broadinstitute.org/gatk/ |
| bwa version 0.5.9 | Li and Durbin, 2010 | http://bio-bwa.sourceforge.net/ |
| ContEst | Cibulskis et al., 2011 | www.broadinstitute.org/cancer/cga |
| MuTect (version 1.1.6) | Cibulskis et al., 2013 | www.broadinstitute.org/cancer/cga |
| Strelka | Saunders et al., 2012 | www.broadinstitute.org/cancer/cga |
| Integrated Genomics Viewer (IGV) | Robinson et al., 2011 | https://software.broadinstitute.org/software/igv/ |
| deTiN algorithm | Taylor-Weiner et al., 2018 | www.broadinstitute.org/cancer/cga |
| ReCapSeg | Ramos et al., 2015 | www.broadinstitute.org/cancer/cga |
| AllelicCapSeg | Stachler et al., 2015 | www.broadinstitute.org/cancer/cga |
| ABSOLUTE algorithm (v1.5) | Carter et al., 2012 | www.broadinstitute.org/cancer/cga |
| PhylogicNDT | Leshchiner et al., 2018) | N/A |
| Other |
SIGNIFICANCE.
BCL-2 inhibition by venetoclax is approved for the treatment of myeloid and lymphoid leukemias. Here, we identify a mechanism by which lymphoid cancer cells can escape this compound by reprogramming cellular energy metabolism and overexpressing MCL-1. Our results provide important mechanistic insights as well as critical preclinical evidence supporting the use of metabolic modulators in combination with BCL-2 inhibitors in patients with lymphoid malignancies.
HIGHLIGHTS.
B-lymphoid cancer cells can escape to venetoclax by overexpressing MCL-1
Modulation of AMPK/PKA axis and lymphoid transcription drive venetoclax resistance
Venetoclax resistance involves changes in cellular energy metabolism such as OXPHOS
Metabolic modulators can cooperate with venetoclax to overcome resistance
Guièze et al. show that resistance to the BCL-2 inhibitor venetoclax in chronic lymphocytic leukemia is associated with complex clonal shifts and identify, in addition to the known involvement by BCL-2 family members, regulators of lymphoid transcription and cellular energy metabolism as resistance drivers.
ACKNOWLEDGMENTS
We are grateful to Drs. Ta-Ming Liu, Jing Deng and Levi Garraway for constructive and valuable discussion. We are appreciative of Dr. Guo Wei (Broad Institute) for providing the MCL-1 inhibitor S63845. We acknowledge Carrie Cibulskis, Adam Tracy, Lan Nguyen, Candace Patterson and Alicia Wong for expert project management. We are grateful to Amy Goodale for her skilled execution of the ORF screen, and to Ben Ferland for the immunohistochemical staining studies.
The authors would like to acknowledge support from NCI-PO1CA206978, and particularly, generous support of the Gerstner Family Foundation for the genetic screens.
R.G. was generously supported by the Fishman Family Fund. V.M.L. received support from the American Society of Hematology HONORS Award, the Harvard Medical School Scholars in Medicine Office, and the HST Research Assistant Award. A.A.J. is supported by a fellowship from the Swiss National Science Foundation. M.H-S. is supported by a grant from FEHH/Janssen (‘Sociedad Española de Hematología y Hemoterapia’). E.T.H is a Special Fellow of the Leukemia and Lymphoma Society (grant #3380-1 8), and a Scholar of the American Society of Hematology. J-M.H. is supported by a Sara Elizabeth O’Brien Trust Postdoctoral Fellowship. J.B.I. acknowledges support by the NIH (5T32-HL007627). D.B.K. is supported by NIH/NCIR21 CA216772-01A1 (to D.B.K.). C.J.O acknowledges support from NCI (R00CA190861) and a Scholar Award from the American Society of Hematology. J.R.B. acknowledges support from NCI (R01CA213442; 2P01CA081534) and from Susan and Gary Rosenbach Fund for Lymphoma Research. J.W.H. was supported by R37 NS083524 and R01 GM095567. S.A.C. acknowledges support by grants from the National Cancer Institute (NCI) Clinical Proteomic Tumor Analysis Consortium grants NIH/NCI U24-CA210986 and NIH/NCI U01 CA214125. V.K.M. is supported by a grant from NIGMS (R35GM122455) and is an investigator of the Howard Hughes Medical Institute. C.J.W. is a Scholar of the Leukemia and Lymphoma Society (LLS) and acknowledges support from NCI (1RO1CA155010-01A1; 1U10CA180861-01).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
C.J.W. is a co-founder of Neon Therapeutics, Inc and is a member of its scientific advisory board, and receives research funding from Pharmacyclics. R.G., Abbvie (honoraria, travel funds), Janssen (honoraria, travel funds), Gilead (honoraria, travel fund) and Roche (travel funds). J.R.B. serves as a consultant for Abbvie, Genentech, Astra-Zaneca, Janssen, Pharmacyclics, Gilead, Verastem, TG Therapeutics, Sunesis and Loxo and receives research funding from Gilead, Verastem and Sun. J.W.H. is co-founder of Rheostat Therapeutics and is member of its scientific advisory board and is also a member the scientific advisory board of X-Chem, Inc (honoraria). A.G.L. discloses consulting and laboratory research support from AbbVie, Novartis, and Astra-Zeneca; he is an equity-holding co-founder of Flash Therapeutics and Vivid Biosciences. V.K.M. is on the Scientific Advisory Board of Janssen Pharmaceuticals, is a venture partner consultant to 5AM Ventures, and a founder and equity holder in Raze Therapeutics. R.G. and C.J.W. disclose a patent related to this work ((U.S. Provisional Application No. 62/744,081). All other authors declare no competing interest.
REFERENCES
- Anderson MA, Tam C, Lew TE, Juneja S, Juneja M, Westerman D, Wall M, Lade S, Gorelik A, Huang DCS, et al. (2017). Clinicopathological features and outcomes of progression of CLL on the BCL2 inhibitor venetoclax. Blood 129, 3362–3370. [DOI] [PubMed] [Google Scholar]
- Arroyo JD, Jourdain AA, Calvo SE, Ballarano CA, Doench JG, Root DE, and Mootha VK (2016). A Genome-wide CRISPR Death Screen Identifies Genes Essential for Oxidative Phosphorylation. Cell Metab. 24, 875–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao XR, Ong S-E, Goldberger O, Peng J, Sharma R, Thompson DA, Vafai SB, Cox AG, Marutani E, Ichinose F, et al. (2016). Mitochondrial dysfunction remodels one-carbon metabolism in human cells. ELife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, Sboner A, Esgueva R, Pflueger D, Sougnez C, et al. (2011). The genomic complexity of primary human prostate cancer. Nature 470, 214–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliss CI (1939). THE TOXICITY OF POISONS APPLIED JOINTLY1. Ann. Appl. Biol 26, 585–615. [Google Scholar]
- Blombery P, Anderson MA, Gong J-N, Thijssen R, Birkinshaw RW, Thompson ER, Teh CE, Nguyen T, Xu Z, Flensburg C, et al. (2018). Acquisition of the recurrent Gly101Val mutation in BCL2 confers resistance to venetoclax in patients with progressive chronic lymphocytic leukemia. Cancer Discov. [DOI] [PubMed] [Google Scholar]
- Bott AJ, Peng I-C, Fan Y, Faubert B, Zhao L, Li J, Neidler S, Sun Y, Jaber N, Krokowski D, et al. (2015). Oncogenic Myc Induces Expression of Glutamine Synthetase through Promoter Demethylation. Cell Metab. 22, 1068–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, and Greenleaf WJ (2013). Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger JA, Landau DA, Taylor-Weiner A, Bozic I, Zhang H, Sarosiek K, Wang L, Stewart C, Fan J, Hoellenriegel J, et al. (2016). Clonal evolution in patients with chronic lymphocytic leukaemia developing resistance to BTK inhibition. Nat. Commun 7, 11589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey CD, Gusenleitner D, Lipschitz M, Roemer MGM, Stack EC, Gjini E, Hu X, Redd R, Freeman GJ, Neuberg D, et al. (2017). Topological analysis reveals a PD-L1-associated microenvironmental niche for Reed-Sternberg cells in Hodgkin lymphoma. Blood 130, 2420–2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, Laird PW, Onofrio RC, Winckler W, Weir BA, et al. (2012). Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol 30, 413–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan LN, Chen Z, Braas D, Lee J-W, Xiao G, Geng H, Cosgun KN, Hurtz C, Shojaee S, Cazzaniga V, et al. (2017). Metabolic gatekeeper function of B-lymphoid transcription factors. Nature 542, 479–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, Harview CL, Brunet J-P, Ahmann GJ, Adli M, et al. (2011). Initial genome sequencing and analysis of multiple myeloma. Nature 471, 467–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapuy B, Stewart C, Dunford AJ, Kim J, Kamburov A, Redd RA, Lawrence MS, Roemer MGM, Li AJ, Ziepert M, et al. (2018). Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat. Med 24, 679–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZX, and Pervaiz S (2007). Bcl-2 induces pro-oxidant state by engaging mitochondrial respiration in tumor cells. Cell Death Differ. 14, 1617–1627. [DOI] [PubMed] [Google Scholar]
- Chen Y, Lun ATL, and Smyth GK (2016). From reads to genes to pathways: differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Research 5, 1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou T-C (2010). Drug Combination Studies and Their Synergy Quantification Using the Chou-Talalay Method. Cancer Res. 70, 440–446. [DOI] [PubMed] [Google Scholar]
- Chukkapalli V, Gordon LI, Venugopal P, Borgia JA, and Karmali R (2018). Metabolic changes associated with metformin potentiates Bcl-2 inhibitor, Venetoclax, and CDK9 inhibitor, BAY1143572 and reduces viability of lymphoma cells. Oncotarget 9, 21166–21181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibulskis K, McKenna A, Fennell T, Banks E, DePristo M, and Getz G (2011). ContEst: estimating cross-contamination of human samples in next-generation sequencing data. Bioinforma. Oxf. Engl 27, 2601–2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, and Getz G (2013). Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol 31, 213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et al. (2005). miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. U. S. A 102, 13944–13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello M, Pugh TJ, Fennell TJ, Stewart C, Lichtenstein L, Meldrim JC, Fostel JL, Friedrich DC, Perrin D, Dionne D, et al. (2013). Discovery and characterization of artifactual mutations in deep coverage targeted capture sequencing data due to oxidative DNA damage during sample preparation. Nucleic Acids Res. 41, e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Gaizo Moore V, Schlis KD, Sallan SE, Armstrong SA, and Letai A (2008). BCL-2 dependence and ABT-737 sensitivity in acute lymphoblastic leukemia. Blood 111, 2300–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J, Carlson N, Takeyama K, Dal Cin P, Shipp M, and Letai A (2007). BH3 profiling identifies three distinct classes of apoptotic blocks to predict response to ABT-737 and conventional chemotherapeutic agents. Cancer Cell 12, 171–185. [DOI] [PubMed] [Google Scholar]
- DiNardo CD, Pratz KW, Letai A, Jonas BA, Wei AH, Thirman M, Arellano M, Frattini MG, Kantarjian H, Popovic R, et al. (2018). Safety and preliminary efficacy of venetoclax with decitabine or azacitidine in elderly patients with previously untreated acute myeloid leukaemia: a non-randomised, open-label, phase 1b study. Lancet Oncol. 19, 216–228. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinforma. Oxf. Engl 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Hartenian E, Graham DB, Tothova Z, Hegde M, Smith I, Sullender M, Ebert BL, Xavier RJ, and Root DE (2014). Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat. Biotechnol 32, 1262–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, et al. (2016). Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol 34, 184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doran AC, Meller N, Cutchins A, Deliri H, Slayton RP, Oldham SN, Kim JB, Keller SR, and McNamara CA (2008). The helix-loop-helix factors Id3 and E47 are novel regulators of adiponectin. Circ. Res 103, 624–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, and Lash AE (2002). Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis LM, and Hicklin DJ (2009). Resistance to Targeted Therapies: Refining Anticancer Therapy in the Era of Molecular Oncology. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res 15, 7471–7478. [DOI] [PubMed] [Google Scholar]
- ENCODE Project Consortium (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher S, Barry A, Abreu J, Minie B, Nolan J, Delorey TM, Young G, Fennell TJ, Allen A, Ambrogio L, et al. (2011). A scalable, fully automated process for construction of sequence-ready human exome targeted capture libraries. Genome Biol. 12, R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fresquet V, Rieger M, Carolis C, García-Barchino MJ, and Martinez-Climent JA (2014). Acquired mutations in BCL2 family proteins conferring resistance to the BH3 mimetic ABT-199 in lymphoma. Blood 123, 4111–4119. [DOI] [PubMed] [Google Scholar]
- Giménez-Cassina A, and Danial NN (2015). Regulation of mitochondrial nutrient and energy metabolism by BCL-2 family proteins. Trends Endocrinol. Metab. TEM 26, 165–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorre ME, Mohammed M, Ellwood K, Hsu N, Paquette R, Rao PN, and Sawyers CL (2001). Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science 293, 876–880. [DOI] [PubMed] [Google Scholar]
- Gross A, and Katz SG (2017). Non-apoptotic functions of BCL-2 family proteins. Cell Death Differ. 24, 1348–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B, Huang X, Lee MR, Lee SA, and Broxmeyer HE (2018). Antagonism of PPAR-γ signaling expands human hematopoietic stem and progenitor cells by enhancing glycolysis. Nat. Med [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckl D, Kowalczyk MS, Yudovich D, Belizaire R, Puram RV, McConkey ME, Thielke A, Aster JC, Regev A, and Ebert BL (2014). Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nat. Biotechnol 32, 941–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herling CD, Abedpour N, Weiss J, Schmitt A, Jachimowicz RD, Merkel O, Cartolano M, Oberbeck S, Mayer P, Berg V, et al. (2018). Clonal dynamics towards the development of venetoclax resistance in chronic lymphocytic leukemia. Nat. Commun 9, 727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Li L, Ying Z, Pan C, Huang S, Li L, Dai M, Yan B, Li M, Jiang H, et al. (2016). A Small Molecule That Protects the Integrity of the Electron Transfer Chain Blocks the Mitochondrial Apoptotic Pathway. Mol. Cell 63, 229–239. [DOI] [PubMed] [Google Scholar]
- Johannessen CM, Johnson LA, Piccioni F, Townes A, Frederick DT, Donahue MK, Narayan R, Flaherty KT, Wargo JA, Root DE, et al. (2013). A melanocyte lineage program confers resistance to MAP kinase pathway inhibition. Nature 504, 138–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CL, Stevens BM, D’Alessandro A, Reisz JA, Culp-Hill R, Nemkov T, Pei S, Khan N, Adane B, Ye H, et al. (2018). Inhibition of Amino Acid Metabolism Selectively Targets Human Leukemia Stem Cells. Cancer Cell 34, 724–740.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkela R, Woulfe KC, Durand J-B, Vagnozzi R, Kramer D, Chu TF, Beahm C, Chen MH, and Force T (2009). Sunitinib-induced cardiotoxicity is mediated by off-target inhibition of AMP-activated protein kinase. Clin. Transl. Sci 2, 15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A, Fornes O, Stigliani A, Gheorghe M, Castro-Mondragon JA, van der Lee R, Bessy A, Chèneby J, Kulkarni SR, Tan G, et al. (2018). JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 46, D1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopleva M, Contractor R, Tsao T, Samudio I, Ruvolo PP, Kitada S, Deng X, Zhai D, Shi Y-X, Sneed T, et al. (2006). Mechanisms of apoptosis sensitivity and resistance to the BH3 mimetic ABT-737 in acute myeloid leukemia. Cancer Cell 10, 375–388. [DOI] [PubMed] [Google Scholar]
- Kotschy A, Szlavik Z, Murray J, Davidson J, Maragno AL, Le Toumelin-Braizat G, Chanrion M, Kelly GL, Gong J-N, Moujalled DM, et al. (2016). The MCL1 inhibitor S63845 is tolerable and effective in diverse cancer models. Nature 538, 477–482. [DOI] [PubMed] [Google Scholar]
- Kridel R, Sehn LH, and Gascoyne RD (2012). Pathogenesis of follicular lymphoma. J. Clin. Invest 122, 3424–3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagadinou ED, Sach A, Callahan K, Rossi RM, Neering SJ, Minhajuddin M, Ashton JM, Pei S, Grose V, O’Dwyer KM, et al. (2013). BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell Stem Cell 12, 329–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, Lawrence MS, Sougnez C, Stewart C, Sivachenko A, Wang L, et al. (2013). Evolution and Impact of Subclonal Mutations in Chronic Lymphocytic Leukemia. Cell 152, 714–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau DA, Tausch E, Taylor-Weiner AN, Stewart C, Reiter JG, Bahlo J, Kluth S, Bozic I, Lawrence M, Böttcher S, et al. (2015). Mutations driving CLL and their evolution in progression and relapse. Nature 526, 525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lark DS, Reese LR, Ryan TE, Torres MJ, Smith CD, Lin C-T, and Neufer PD (2015). Protein Kinase A Governs Oxidative Phosphorylation Kinetics and Oxidant Emitting Potential at Complex I. Front. Physiol 6, 332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leshchiner I, Livitz D, Gainor JF, Rosebrock D, Spiro O, Martinez A, Mroz E, Lin JJ, Stewart C, Kim J, et al. (2018). Comprehensive analysis of tumour initiation, spatial and temporal progression under multiple lines of treatment. BioRxiv 508127. [Google Scholar]
- Li B, and Dewey CN (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, and Durbin R (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinforma. Oxf. Engl 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, and Durbin R (2010). Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinforma. Oxf. Engl 26, 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S-C, and Hardie DG (2018). AMPK: Sensing Glucose as well as Cellular Energy Status. Cell Metab. 27, 299–313. [DOI] [PubMed] [Google Scholar]
- Lin KH, Winter PS, Xie A, Roth C, Martz CA, Stein EM, Anderson GR, Tingley JP, and Wood KC (2016). Targeting MCL-1/BCL-XL Forestalls the Acquisition of Resistance to ABT-199 in Acute Myeloid Leukemia. Sci. Rep 6, 27696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love C, Sun Z, Jima D, Li G, Zhang J, Miles R, Richards KL, Dunphy CH, Choi WWL, Srivastava G, et al. (2012). The genetic landscape of mutations in Burkitt lymphoma. Nat. Genet 44, 1321–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mato AR, Thompson M, Allan JN, Brander DM, Pagel JM, Ujjani CS, Hill BT, Lamanna N, Lansigan F, Jacobs R, et al. (2018). Real world outcomes and management strategies for venetoclax-treated chronic lymphocytic leukemia patients in the United States. Haematologica. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner B, Kridel R, Lim RS, Rogic S, Tse K, Scott DW, Moore R, Mungall AJ, Marra MA, Connors JM, et al. (2013). The E3 ubiquitin ligase UBR5 is recurrently mutated in mantle cell lymphoma. Blood 121, 3161–3164. [DOI] [PubMed] [Google Scholar]
- Mertins P, Tang LC, Krug K, Clark DJ, Gritsenko MA, Chen L, Clauser KR, Clauss TR, Shah P, Gillette MA, et al. (2018). Reproducible workflow for multiplexed deep-scale proteome and phosphoproteome analysis of tumor tissues by liquid chromatography-mass spectrometry. Nat. Protoc [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meynet O, Zunino B, Happo L, Pradelli LA, Chiche J, Jacquin MA, Mondragón L, Tanti J-F, Taillan B, Garnier G, et al. (2013). Caloric restriction modulates Mcl-1 expression and sensitizes lymphomas to BH3 mimetic in mice. Blood 122, 2402–2411. [DOI] [PubMed] [Google Scholar]
- Mootha VK, Wei MC, Buttle KF, Scorrano L, Panoutsakopoulou V, Mannella CA, and Korsmeyer SJ (2001). A reversible component of mitochondrial respiratory dysfunction in apoptosis can be rescued by exogenous cytochrome c. EMBO J. 20, 661–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD, Johnson NA, Severson TM, Chiu R, Field M, et al. (2011). Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature 476, 298–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppermann S, Ylanko J, Shi Y, Hariharan S, Oakes CC, Brauer PM, Zúñiga-Pflücker JC, Leber B, Spaner DE, and Andrews DW (2016). High-content screening identifies kinase inhibitors that overcome venetoclax resistance in activated CLL cells. Blood 128, 934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastore A, Jurinovic V, Kridel R, Hoster E, Staiger AM, Szczepanowski M, Pott C, Kopp N, Murakami M, Horn H, et al. (2015). Integration of gene mutations in risk prognostication for patients receiving first-line immunochemotherapy for follicular lymphoma: a retrospective analysis of a prospective clinical trial and validation in a population-based registry. Lancet Oncol. 16, 1111–1122. [DOI] [PubMed] [Google Scholar]
- Pollyea DA, Stevens BM, Jones CL, Winters A, Pei S, Minhajuddin M, D’Alessandro A, Culp-Hill R, Riemondy KA, Gillen AE, et al. (2018). Venetoclax with azacitidine disrupts energy metabolism and targets leukemia stem cells in patients with acute myeloid leukemia. Nat. Med 24, 1859–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puente XS, Beà S, Valdés-Mas R, Villamor N, Gutiérrez-Abril J, Martín-Subero JI, Munar M, Rubio-Pérez C, Jares P, Aymerich M, et al. (2015). Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature 526, 519–524. [DOI] [PubMed] [Google Scholar]
- Quesada V, Conde L, Villamor N, Ordóñez GR, Jares P, Bassaganyas L, Ramsay AJ, Beà S, Pinyol M, Martínez-Trillos A, et al. (2012). Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat. Genet 44, 47–52. [DOI] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, and Manke T (2016). deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos AH, Lichtenstein L, Gupta M, Lawrence MS, Pugh TJ, Saksena G, Meyerson M, and Getz G (2015). Oncotator: cancer variant annotation tool. Hum. Mutat 36, E2423–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, and Mesirov JP (2006). GenePattern 2.0. Nat. Genet 38, 500–501. [DOI] [PubMed] [Google Scholar]
- Roberts AW, Davids MS, Pagel JM, Kahl BS, Puvvada SD, Gerecitano JF, Kipps TJ, Anderson MA, Brown JR, Gressick L, et al. (2016). Targeting BCL2 with Venetoclax in Relapsed Chronic Lymphocytic Leukemia. N. Engl. J. Med 374, 311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, and Mesirov JP (2011). Integrative genomics viewer. Nat. Biotechnol 29, 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roemer MGM, Advani RH, Redd RA, Pinkus GS, Natkunam Y, Ligon AH, Connelly CF, Pak CJ, Carey CD, Daadi SE, et al. (2016). Classical Hodgkin Lymphoma with Reduced β2M/MHC Class I Expression Is Associated with Inferior Outcome Independent of 9p24.1 Status. Cancer Immunol. Res 4, 910–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotem A, Janzer A, Izar B, Ji Z, Doench JG, Garraway LA, and Struhl K (2015). Alternative to the soft-agar assay that permits high-throughput drug and genetic screens for cellular transformation. Proc. Natl. Acad. Sci. U. S. A 112, 5708–5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, and Zhang F (2014). Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders CT, Wong WSW, Swamy S, Becq J, Murray LJ, and Cheetham RK (2012). Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinforma. Oxf. Engl 28, 1811–1817. [DOI] [PubMed] [Google Scholar]
- Schep AN, Wu B, Buenrostro JD, and Greenleaf WJ (2017). chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat. Methods 14, 975–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, et al. (2014). Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JH, Kandasamy K, and Kraft AS (2008). ABT-737 induces expression of the death receptor 5 and sensitizes human cancer cells to TRAIL-induced apoptosis. J. Biol. Chem 283, 25003–25013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souers AJ, Leverson JD, Boghaert ER, Ackler SL, Catron ND, Chen J, Dayton BD, Ding H, Enschede SH, Fairbrother WJ, et al. (2013). ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat. Med 19, 202–208. [DOI] [PubMed] [Google Scholar]
- Stachler MD, Taylor-Weiner A, Peng S, McKenna A, Agoston AT, Odze RD, Davison JM, Nason KS, Loda M, Leshchiner I, et al. (2015). Paired exome analysis of Barrett’s esophagus and adenocarcinoma. Nat. Genet 47, 1047–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor-Weiner A, Stewart C, Giordano T, Miller M, Rosenberg M, Macbeth A, Lennon N, Rheinbay E, Landau D-A, Wu CJ, et al. (2018). DeTiN: overcoming tumor-in-normal contamination. Nat. Methods 15, 531–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujimoto Y, Cossman J, Jaffe E, and Croce CM (1985). Involvement of the bcl-2 gene in human follicular lymphoma. Science 228, 1440–1443. [DOI] [PubMed] [Google Scholar]
- Wang L-H, and Baker NE (2015). E Proteins and ID Proteins: Helix-Loop-Helix Partners in Development and Disease. Dev. Cell 35, 269–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Lawrence MS, Wan Y, Stojanov P, Sougnez C, Stevenson K, Werner L, Sivachenko A, DeLuca DS, Zhang L, et al. (2011). SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N. Engl. J. Med 365, 2497–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weniger MA, and Küppers R (2016). NF-κB deregulation in Hodgkin lymphoma. Semin. Cancer Biol 39, 32–39. [DOI] [PubMed] [Google Scholar]
- Woyach JA, Furman RR, Liu T-M, Ozer HG, Zapatka M, Ruppert AS, Xue L, Li DH-H, Steggerda SM, Versele M, et al. (2014). Resistance mechanisms for the Bruton’s tyrosine kinase inhibitor ibrutinib. N. Engl. J. Med 370, 2286–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yecies D, Carlson NE, Deng J, and Letai A (2010). Acquired resistance to ABT-737 in lymphoma cells that up-regulate MCL-1 and BFL-1. Blood 115, 3304–3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S5 CRISPR-Cas9 screen results, Related to Figure 2.
Table S6. ORF screen results, Related to Figure 2.
Table S8. OCI-Ly1 sSNVs report, Related to Figure 7.
Data Availability Statement
The RNA seq and ATAC seq data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE 128563 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE128563).
The original mass spectra and sequence database used for searches may be downloaded from MassIVE (http://massive.ucsd.edu) using the identifier: MSV000083512. The dataset is directly accessible via ftp://massive.ucsd.edu/MSV000083512.
dbGaP accession number for the depositing of WES data for this study is phs001875.v1.p1.