Summary
Seaweeds have been used as a source of natural fertilizer and biostimulant in agriculture for centuries. However, their effects on soil and crop root microbiota remain unclear. Here, we used a commercially available Ascophyllum nodosum extract (ANE) to test its effect on bacterial and fungal communities of rhizospheric soils and roots of pepper and tomato plants in greenhouse trials. Two independent trials were conducted in a split‐block design. We used amplicon sequencing targeting fungal ITS and bacterial 16S rRNA gene to determine microbial community structure changes. We find that productivity parameters of root, shoot and fruit biomass were positively and significantly influenced by the ANE amendment. In addition, a‐diversity differed significantly between amended and control plants, but only in some of the experimental conditions. Species composition among sites (b‐diversity) differed according to the amendment treatment in all four communities (fungal‐root, fungal‐soil, bacterial‐root and bacterial‐soil). Finally, we identified a number of candidate taxa most strongly correlated with crop yield increases. Further studies on isolation and characterization of these microbial taxa linked to the application of liquid seaweed extract may help to enhance crop yield in sustainable agro‐ecosystems.
Introduction
Seaweeds (also known as marine macroalgae) have been used as a source of organic matter and mineral nutrients for centuries, especially in coastal areas (Khan et al., 2009; Craigie, 2011). Liquid seaweed extracts, developed in the 1950s in order to concentrate plant growth‐stimulating compounds, facilitate their usage (Milton, 1952). Today, most commercially available extracts are made from the brown algae Ascophyllum nodosum, Ecklonia maxima or Laminaria spp. Unlike modern chemical fertilizers, seaweed extracts are biodegradable, non‐toxic and come from a renewable resource (Dhargalkar and Pereira, 2005). Therefore, they represent an attractive tool of sustainable crop management programmes (Craigie, 2011; du Jardin, 2015).
Several comprehensive reviews have described the effects of seaweed extracts on agricultural plant productivity (Khan et al., 2009; Craigie, 2010, 2011; Battacharyya et al., 2015). The science points to wide‐ranging effects from biotic to abiotic resistance, effects on growth and development, and ultimately, to their impact on plant establishment, crop yield and/or quality. At the physiological level, these extracts have been found to influence hormones levels that, in turn, influence physiological processes even at very low concentrations (Wally et al., 2013). They impact plant‐signalling mechanisms through a multitude of plant processes and cellular modifications including osmotic/oxidative stresses such as salinity, freezing and drought stress (Jithesh et al., 2012). Contrary to the effects of ANE on plant development, the effect of seaweed extracts on the biology of the rhizosphere is still largely unknown. Yet, previous work has showed that the application of biofertilizer (containing fermented Bacillus and pig manure) can reshape the rhizosphere community and may help to control diseases (Shen et al., 2015, 2019). The rhizosphere harbours a large microbial biodiversity where numerous microbial taxa form biofilm that contributes to the aggregation of particles, enhances nutrient cycling and delivery to plants, degrades toxic substances, allows better soil water retention and plays a role in plant disease management (reviewed by Pandin et al., 2017). For example, ANE applications increased strawberry root and shoot growth, berry yield and rhizosphere microbial diversity and physiological activity (Alam et al., 2013). Similar results were found in carrots (Alam et al., 2014) and showed a strong relationship between plant growth and microbial activity. As such, in‐depth examination of sustainable products that influence microbial interactions between plant roots and soil biota will in turn help to further understand plant–microbe dynamics.
The recent development of culture‐independent molecular techniques and high‐throughput sequencing should permit to circumvent the inherent biases of culture‐based approaches by targeting the ubiquitous component of life, DNA. In turn, this will lead to a better understanding of the microbial response to seaweed extract. DNA barcoding targeting specific regions of the genome (e.g. ITS for Fungi and 16S ribosomal RNA for Bacteria) is now regarded as a prerequisite procedure to comprehensively document the diversity and ecology of microorganisms (Toju et al., 2012; Klindworth et al., 2013).
Here, the objective was to quantify the impact of a commercial seaweed extract on plant growth and test how the fungal and bacterial communities responded to the addition of these extracts. We also aimed to identify specific taxa positively correlated with increases in plant productivity following ANE amendments. We hypothesized that the addition of liquid seaweed extracts would improve productivity and alter significantly the fungal and bacterial communities. We used a commercially available ANE, Stella Maris®, developed by Acadian Seaplants Ltd (NS, Canada) and derived from the marine algae A. nodosum, harvested in Eastern Canada. We tested the effect of ANE amendment on two agricultural plants commonly grown in greenhouse conditions (tomato and pepper). Several traits related to plant productivity were measured, and soil and root bacterial and fungal diversity were quantified using high‐throughput sequencing.
Results
Experimental design
Greenhouse trials were set up in large trays using tomato (Solanum lycopersicum L.) and pepper (Capsicum annuum L.) crops. For each species, a randomized split‐block design (Table S1) was used with four trays set up per block and eight blocks for each trial. Half of the trays were amended with ANE, and half of the trays were also planted (planting effect) with four plants per tray, while the other trays were not planted. This allowed a direct comparison of fungal and bacteria‐soil communities with respect to the amendment and planting effects (see Experimental procedure for more details).
Effects of the amendment treatment on productivity
The effects of the amendment treatment on tomato (hen manure + ANE) and pepper (ANE) were determined by measuring six agronomic parameters (fruit number, average fruit weight, shoots’ fresh weight, shoots’ dry weight, roots’ fresh weight and roots’ dry weight). We observed a significant increase in almost all agronomic parameters (LMM, P‐value < 0.005, Fig. 1) for amended plants except for the average fruit fresh weight for tomato that did not differ between amended and control plants (LMM, F (1,23) = 1.81, P‐value = 0.19, Fig. 1 and Fig. S1). The amendment effect was stronger in the tomato plants (fold changes between amended and control plants shown in Fig. 1), likely due to the fact that these plants were fertilized with both hen manure and ANE.
Figure 1.
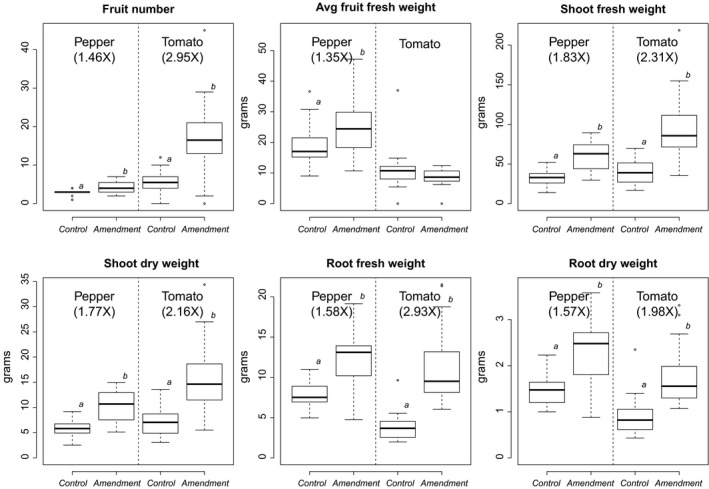
Measures of plant productivity. a and b subscripts above box plots denote significant differences (P‐value < 0.005) according to the amendment effect (tomato: hen manure + ANE; pepper: ANE). Fold changes between the mean of the control and amended plants were also noted for significant differences (for pepper and tomato separately).
Amplicon sequencing
A total of 2.7 million paired‐end raw reads were obtained for all samples combined (976 000 for fungi‐soil, 920 000 for fungi‐root, 309 000 for bacteria‐soil and 535 000 for bacteria‐root, Table S4). On average, 47 664 paired‐end reads were obtained per sample. After quality filters were applied, including removing chimeras, and paired‐end reads were merged, an average of 19 690 sequences remained per sample. From 192 soil samples for fungi and bacteria, and 96 root samples for fungi and bacteria, three fungi‐soil samples, 15 fungi‐root samples and one bacteria‐root sample were removed because they had few reads based on our strict quality thresholds.
The dada2 pipeline inferred 6112 fungal‐soil, 845 fungal‐root, 9352 bacterial‐soil and 2023 bacterial‐root ASV (Table S4). In bacteria‐soil, we further removed a total of 79 ASV whose taxonomy corresponded to mitochondria or chloroplast and represented 0.1% of all sequencing reads. In bacteria‐root samples, we removed a total of 284 ASV that corresponded to mitochondria or chloroplast and represented 89% of all sequencing reads. After filtering out rare ASV, we retained 413, 106, 807 and 262 ASV respectively for fungal‐soil, fungal‐root, bacterial‐soil and bacterial‐root. These retained ASV comprised 94%, 95%, 89% and 11% of all filtered‐merged sequences assigned to ASV by the dada2 pipeline in the fungal‐soil, fungal‐root, bacterial‐soil and bacterial‐root samples respectively.
Fungal and bacterial diversity in root and soil biotopes
The microbial community structures of soil and root samples were analysed, and the relative abundance of their taxa was determined at the family level (Figs 2 and 3). Nectriaceae dominated the fungal communities, in both the root and soil samples, while the bacterial family Bacillaceae dominated to a lesser extent the soil samples. Bacterial‐root communities harboured a number of different families: Streptomycetaceae, Sphingomonadaceae, Rhizobiaceae and Pseudomonadaceae, among others.
Figure 2.
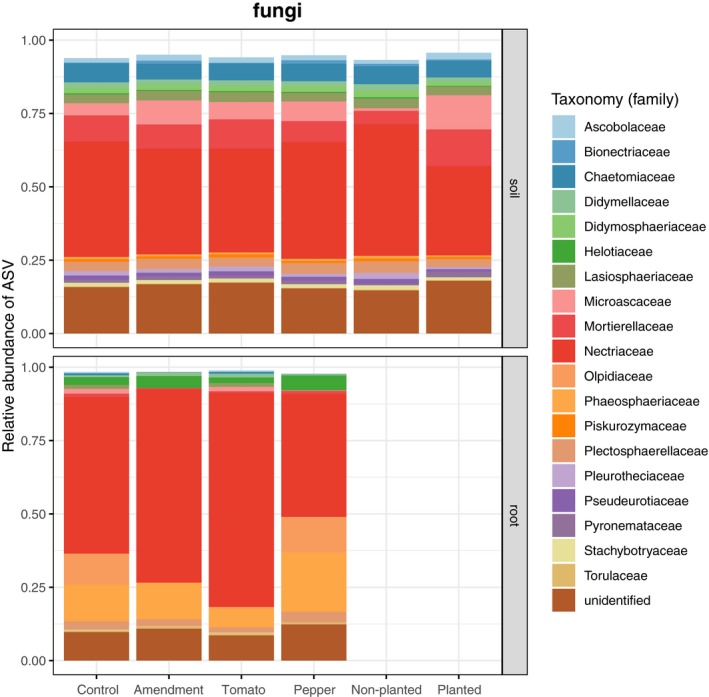
Barplots of the relative abundance of fungal ASV for fungi.
Figure 3.
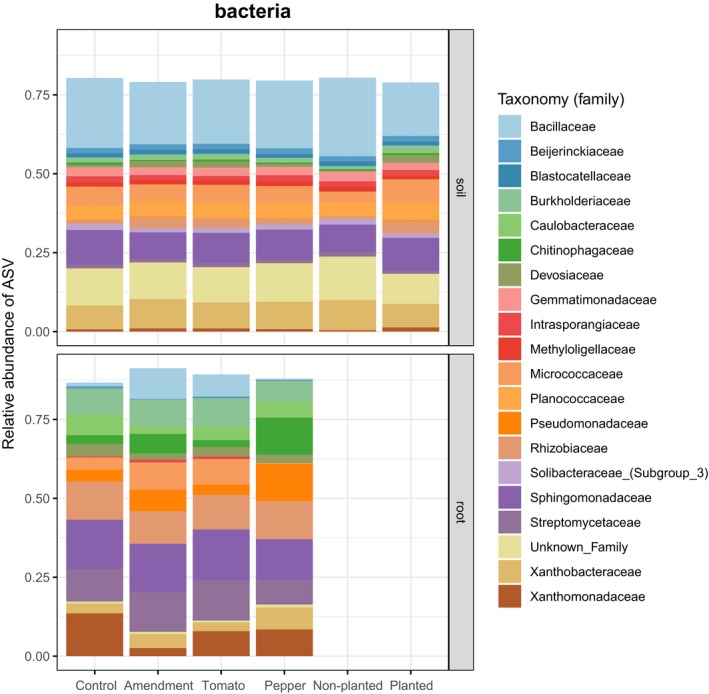
Barplots of the relative abundance of bacterial ASV for bacteria.
Local (a‐diversity)
The a‐diversity was calculated separately for each sample, under each experimental condition (fungi‐soil, fungi‐root, bacteria‐soil and bacteria‐root for both tomato and pepper, Fig. 4). Linear mixed‐effects models showed that the a‐diversity (inverse Simpson index) was significantly higher in the soil biotope than in the roots for both fungi (mean a‐diversity fungi‐soil = 2.88 vs. mean a‐diversity fungi‐root = 27.3, F (1,239) = 899.5, P‐value < 0.0001) and bacteria (mean a‐diversity bacteria‐soil = 4.7 vs. mean a‐diversity bacteria‐root = 69.2, F (1,223) = 1198.1, P‐value < 0.0001).
Figure 4.
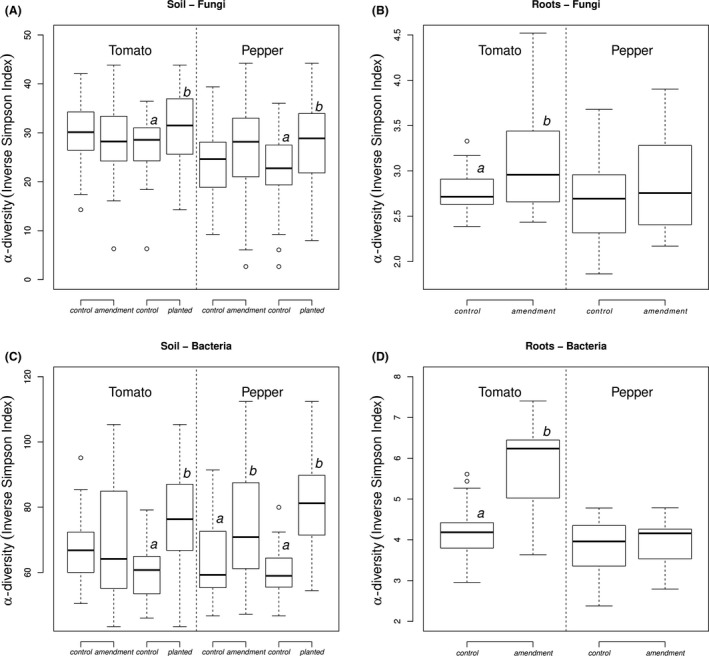
Box plot of a‐diversity according to the amendment and planting effect for fungal‐root, fungal‐soil, bacteria‐soil and bacteria‐root for tomato and pepper. a and b subscripts above box plots denote significant differences (P‐value < 0.05).
In soil samples, fungal a‐diversity was not significantly different in amended versus control plants for neither tomato (F (1,66) = 1.6, P‐value = 0.21) nor pepper (F (1,69) = 1.2, P‐value = 0.05). In root samples, fungal a‐diversity was significantly different in amended versus control plants for tomato (F (1,21) = 10.2, P‐value = 0.004), but not pepper (F (1,56) = 3.1, P‐value = 0.10).
In soil samples, bacterial a‐diversity was significantly different in amended versus control plants for pepper (F (1,69) = 31.5, P‐value < 0.0001), but not tomato (F (1,69) = 1.9, P‐value = 0.17). In root samples, bacterial a‐diversity was significantly different in amended versus control plants for tomato (F (1,22) = 39.7, P‐value < 0.0001), but not pepper (F (1,4) = 0.17, P‐value = 0.70).
Differences in species composition among sites
Using a PERMANOVA, we identified that the ANE amendment treatment had a highly significant effect on both fungal and bacterial community structures (Table 1). This effect was stronger in the root (9–30% of variance explained in the models) than in the soil (3–6% of variance explained in the models). Planting also had a significant effect on fungal and bacterial community structures (12–24% of variance explained in the models).
Table 1.
Variance explained by the terms in the PERMANOVA models.
| Amendment | Planting | Amendment:planting | |
|---|---|---|---|
| Fungi‐soil (tomato) | 0.05*** | 0.24*** | 0.02** |
| Fungi‐root (tomato) | 0.29*** | NA | NA |
| Bacteria‐soil (tomato) | 0.06*** | 0.17*** | 0.04** |
| Bacteria‐root (tomato) | 0.33*** | NA | NA |
| Fungi‐soil (pepper) | 0.03** | 0.2*** | 0.02* |
| Fungi‐root (pepper) | 0.1*** | NA | NA |
| Bacteria‐soil (pepper) | 0.06*** | 0.12*** | 0.02* |
| Bacteria‐root (pepper) | 0.19 | NA | NA |
NA, not applicable.
r 2 (fraction of variance explained by the term in the model).
*P‐value < 0.05, **< 0.005, ***< 0.0005.
Redundancy analyses (RDAs, Fig. 5 for fungi and Fig. 6 for bacteria) illustrated that roots’ fresh weight, shoots’ fresh weight and fruit number responded similarly, while average fruit weight behaved differentially as noted previously (in fact nearly orthogonally to the other three parameters in most ordinations). Note that we excluded the shoots’ and roots’ dry weights as constraints to simplify the model. In addition, these were highly collinear with the fresh weight already included as constraints (r 2 = 0.98 and 0.76 for shoot dry/fresh weights and root dry/fresh weights, respectively). In addition, RDAs showed that fertilized samples clustered together and were positively correlated with increases in productivity. All RDA models tested were significant (F (4,10) > 1.4, P‐value < 0.03 for all models).
Figure 5.
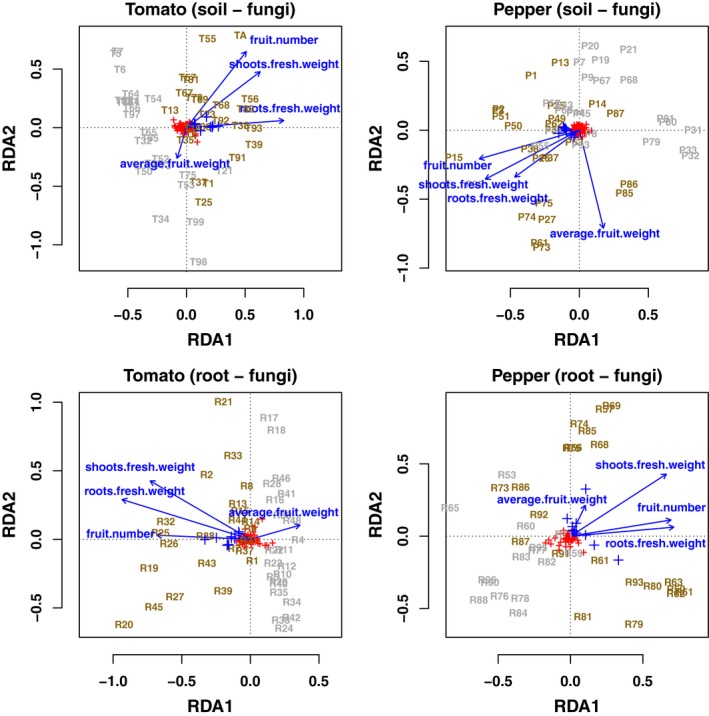
Redundancy analyses (RDAs) for fungal ASV (species scaling). Labelled samples were coloured in grey (unfertilized) or dark yellow (fertilized). Red + signs represent individual ASV, while blue + signs are the ten ASV most closely associated with the three productivity measures of root fresh weight, shoot fresh weight and fruit number. Blue arrows are the four productivity measures used as constraints in the ordinations.
Figure 6.
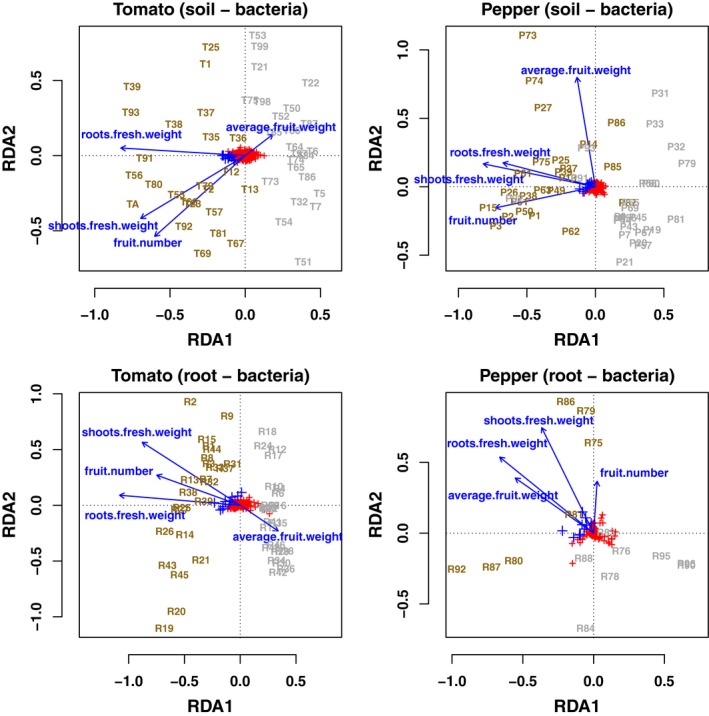
Redundancy analyses (RDAs) for bacterial ASV (species scaling). Labelled samples were coloured in grey (unfertilized) or dark yellow (fertilized). Red + signs represent individual ASV, while blue + signs represent the ten ASV most closely associated with the three productivity measures of root fresh weight, shoot fresh weight and fruit number. Blue arrows are the four productivity measures used as constraints in the ordinations.
Next, we identified, for each RDA, the ten ASV most closely related to the three constraints of the model (roots’ fresh weight, shoots’ fresh weight and fruit number). These ASV were considered as putative candidate taxa most positively impacted by increases in productivity due to the ANE amendment treatment. We further analysed the corresponding sequences for these 80 candidate ASV (10 candidates × eight ordinations) in two separate alignments (one for fungi and one for bacterial ASV) and their accompanying phylogenetic trees.
In fungi, we identified a number of ASV positively associated with productivity (Fig. S2). Notably, five different ASV taxonomically assigned to the family Microascaceae (phylum Ascomycota) in all conditions except the pepper‐root were positively associated with productivity. In addition, two ASV assigned to Mortierella spp (soil saprotrophs in the phylum Mucoromycota) and a cluster of six closely related fungal ASV in tomato‐soil (ASV67 & ASV132), tomato‐root (ASV10, ASV1017, ASV1018, ASV1019) and pepper‐soil (ASV67) were positively associated with productivity in both tomato and pepper roots. Given that no taxonomy was assigned to these sequences through the dada2 RDP bootstrap approach, we used a blastn (Altschul et al., 1997) approach to identify the most closely related sequences against NCBI nr. The most closely related fungal reference sequences were from an uncultured fungus clone (blastn, 86% identity, e‐value = 9e‐58, sequence ID: EU517002.1). Similarly, two unknown ASV (ASV61 & ASV81) also matched an uncultured fungus clone (BLASTn, 94% identity, e‐value = 4e‐165, sequence ID: DQ900965.1). Finally, another cluster of ASV in the pepper‐root was assigned to Olpidium brassicae, a putative fungal parasite belonging to flagellate fungi (Lay et al., 2018).
In bacteria‐root, we identified a large diversity of ASV positively correlated (increased abundance of these ASV) with productivity (Fig. S3), Among others, we identified Rhizobium, Sphingomonas, Sphingobium and Bradyrhizobium in both the soil and root biotopes and tomato and pepper species.
Discussion
In the current study, we investigated the effects of A. nodosum extracts on root, shoot and fruit biomass in addition to bacterial and fungal communities. Overall, parameters related to plant growth significantly increased in both tomato and pepper in response to amendment treatment. These results corroborate previous studies documenting the impact of ANE on productivity in strawberries (Alam et al., 2013) and carrots (Alam et al., 2014).
In the tomato experimental set‐up, the effect of the amendment treatment was especially high, likely due to the fact that plants were also fertilized with hen manure (Fig. 1). In fact, in tomatoes the amounts of N, P and K supplied via the application of ANE were 200–1000 times less than from the hen manure itself. As such, these nutrients were given at very low concentrations relative to the crop requirements and are not expected to significantly impact growth relative to a regular agricultural fertility programme (Bruulsema et al., 2012; Alam et al., 2013). Instead, organic molecules such as betaines, polyamines, cytokinins, auxins, oligosaccharides, amino acids and vitamins present in ANE have been found to have overall beneficial productivity effects on plant growth (Khan et al., 2009; Craigie, 2010, 2011; Battacharyya et al., 2015).
Most ASV identified were rare and unique to one or a few sample. In fact, approximately 90% of all ASV were discarded given that they were found in singletons or present in very few samples and were thus not representative of a particular experimental treatment. These ‘rare’ ASV comprised a small minority of all sequencing reads (approximately 5% of all sequences), a pattern reminiscent of the early species abundance models showing that in most ecological communities, few species are exceptionally abundant whereas most are rare (Fisher et al., 1943). In addition, a large fraction of the sequencing reads in the root bacterial communities likely originated from the plants themselves (identified as chloroplast or mitochondria). This may be partly explained by the fact that most of root biomass collected was from large roots (Fig. S1B), rather than fine root hair where most microbial biological activity likely takes place (Pregitzer et al., 2002).
The amendment effect on bacterial community composition (b‐diversity) was relatively small (3–33% of variance explained in the models, Table 1) but significant, implying that the addition of ANE (pepper) or ANE and hen manure (tomato) is, at least partly, responsible for shaping microbial communities. We also tested the effect of plant species identity on community structure on a combined data set comprised of both the tomato and pepper plants. In the root biotope, we find that this effect (Table S5) is in line with numerous studies reporting how plants select their microbial communities (Chaparro et al., 2014; Reinhold‐Hurek et al., 2015). Nevertheless, we recognize that the current experimental set‐up precludes any strong conclusion regarding the plant species’ effect of community structure, as it does not explicitly disentangle the species effect from the ‘addition of hen manure’ effect.
Nectriaceae, a family of fungi in the order Hypocreales and often encountered as saprotrophs on decaying organic matter, comprised most of the diversity in both the soil and plant roots (Fig. 2). With respect to bacterial communities of the soil, these comprised many different families (Fig. 3). We found one cluster of ASV taxonomically assigned to Mortierella (soil saprotrophs) positively correlated to productivity in both tomato and pepper roots. Interestingly, Li et al. (2018) found that a closely related species (M. elongata) can improve soil health and stimulate production of plant growth hormones. In their study, Chung et al. (2007) showed how increases in productivity led to greater microbial biomass and greater number of saprophytic and arbuscular mycorrhizal fungi. Perhaps, this is explained by the fact that an increase in plant productivity can lead to greater substrate availability, potentially increasing the activity of saprophytic fungi feeding on this organic matter substrate.
Surprisingly, a putative plant pathogenic fungus (Olpidium brassicaceae, Fig. S2) was positively associated with increased productivity. However, O. brassicae only leads to decreased plant growth when present in large amount (Lay et al., 2018). In addition, O. brassicae is likely a species complex that constitutes a large proportion of the roots or rhizosphere fungal community in many different systems, particularly in Brassicaceae crops (Lay et al., 2018).
In bacteria‐root samples, a diverse number of ASV were positively impacted by the amendment treatment (Fig. S3) and many of those are known to be present in the root endosphere (Tkacz and Poole, 2015). For example, Rhizobium and Bradyrhizobium spp. can promote plant growth, P solubilization, N fixation and overall plant productivity (Antoun et al., 1998; Avis et al., 2008).
It is now well established that biofertilizers can have an impact of the rhizospheric community and agricultural plant productivity (Trivedi et al., 2017; Shen et al., 2019; Thomashow et al., 2019). In fact, plants and microbes should likely be redefined as holobionts, an assemblage of different species that forms an ecological unit (Margulis and Fester, 1991). In this study, we showed that the addition of ANE increased plant productivity. It also increased, by a small, but significant margin, the fungal and bacterial (only in the rhizosphere) biodiversity and changed the microbial community structure in the rhizosphere of both tomato and pepper plants. Finally, we identified bacterial and fungal taxa, especially saprotroph positivity associated with plant productivity. Further studies, for example using inoculum of a consortium of the candidate bacterial and fungal species linked to increases in productivity (Baez‐Rogelio et al., 2017) that we identified, may help to identify a causative link between liquid seaweed extracts, microbes and productivity.
Experimental procedure
Experimental design
Greenhouse trials were set up in large trays (60 × 30 × 18 cm L × W × H) using two different crops: tomato (Solanum lycopersicum L.) and pepper (Capsicum annuum L.). Tomato cultivar Totem Hybrid#A371 was planted on 16 November 2015 and pepper cultivar Ace Hybrid#318 was planted on 9 December 2015. Tomato and pepper seeds were purchased from William Dam Seeds Ltd (ON, Canada). These cultivars were selected for greenhouse production. Soil was collected from an agricultural field under organic regime at the IRDA research station in St. Bruno (QC, Canada, 45°32′59.6″N, 73°21′08.0″W) on 7 October 2015. The soil was a loamy sand and was collected from the 15 cm top layer. Natural soil was homogenized and put into trays, and filled to 15 cm in height. Soil analysis was done using a commercial service provided by EnvironeX (formerly Agridirect, Longueuil, QC), and soil characteristics are shown in Table S2. Eight seeds per tray were planted, and after germination, only four seedlings per tray were kept.
Two different amendment regimes were used according to the plant species. For tomatoes, plants were amended using multipurpose organic fertilizer (pure hen manure, 18 g per tray repeated every 4 weeks, 5‐3‐2) from Acti‐Sol (Notre‐Dame‐du‐Bon‐Conseil, QC) in addition to Stella Maris® (3.5 ml per 1 l, each tray received 250 ml, repeated every 2 weeks) for the duration of the experiment. The other half were not treated, but watered with 250 ml per tray instead. The physico‐chemical composition of Stella Maris® is shown in Table S3. For the pepper experiment, the amendment treatment consisted solely of Stella Maris® (3.5 ml per 1 l, each tray received 250 ml, repeated every 2 weeks) for the duration of the experiment. The other half was not amended, but watered with 250 ml per tray instead. Both experiments were managed under organic farming practices. Thrips were controlled using Neoseiulus cucumeris (syn. Amblyseius cucumeris; 1 bag per plant), and Fungus gnats were also controlled using predatory mite Gaeolaelaps gillespiei (1L; Natural Insect Control, ON). Plants were treated once a week with MilStop, a potassium bicarbonate‐based foliar fungicide to control the powdery mildew on both crops.
Plant productivity
Tomato and pepper experiments were harvested on 29 March 2016. The following traits assessed plant productivity: fruit number, fruit weight, shoots’ fresh weight and roots’ fresh weight. Traits were measured on three plants chosen randomly per tray for each amended/control plant, crop (tomato/pepper) and block (eight blocks) for a total of 96 samples. In addition, both shoot and root samples were dried in a 70 degrees drying oven, and dry weights were quantified after 48 h.
Sample preparation, DNA extraction and high‐throughput sequencing
Soil and root samples were taken for both experiments. Soil DNA was extracted using NucleoSpin® Soil DNA extraction kit (Macherey‐Nagel, BioLinx, ON, Canada) on 250 mg of soil, following the manufacturer's protocol. Roots were first washed with tap water and rinsed with sterile water. Chopped root subsamples (100 mg) were subjected to DNA extraction using DNeasy Plant Mini kit (Qiagen Inc, Toronto, ON, Canada), following the manufacturer's recommendations. Amplicon sequencing targeting bacterial 16S rRNA gene and fungal ITS was performed on both root and soil samples.
For fungal ITS, we used the following primers with the universal CS1 and CS2 adapters: CS1_ITS3_KYO2 (5′‐ACA CTG ACG ACA TGG TTC TAC AGA TGA AGA ACG YAG YRA A‐3′) and CS2_ITS4_KYO3 (5′‐TAC GGT AGC AGA GAC TTG GTC TCT BTT VCC KCT TCA CTC G‐3′) to produce a final amplicon size of approximately 430 bp including adapters (Toju et al., 2012).
For bacterial 16S, we used the following primers with CS1 and CS2 universal adapters: 341F (5′‐CCT ACG GGN GGC WGC AG‐3′) and 805R (5′‐GAC TAC CAG GGT ATC TAA TC‐3′) to produce a final amplicon size of approximately 460 bp and targeting specifically the bacterial V3–V4 region of the 16S ribosomal gene (Klindworth et al., 2013).
DNA samples were then barcoded, pooled and sequenced (2 × 300 bp, paired‐end) using an Illumina (San Diego, CA, USA) MiSeq sequencer through a commercial service provided by the Genome Quebec Innovation Centre (Montreal, QC, Canada). Sequences were demultiplexed by the sequencing facility and further processed as described below.
Bioinformatics
All bioinformatics, statistical and graphical analyses further described were performed in r 3.5.1 (R Core Team, 2018), and detailed scripts are available here (https://github.com/seb951/Acadian_Seaplants).
We used the r package dada2 (Callahan et al., 2016) to infer amplicon sequence variants (ASV). dada2 offers accurate sample inference from amplicon data with single‐nucleotide resolution in an open‐source environment. Unlike the operational taxonomic unit (OTU) approach (e.g. Schloss et al., 2009; Caporaso et al., 2010), ASV are not treated as cluster of sequences defined with an ad hoc sequence similarity threshold. Instead, after sequences are quality‐trimmed and error‐corrected, dada2 reveals the unique members of the sequenced community, thus allowing sequences and abundance counts to be comparable among studies (Callahan et al., 2016).
First, sequences were trimmed following strict quality thresholds (removing primers and low‐quality nucleotides, see parameter details in the accompanying r scripts). Following this, we applied the error model algorithm of dada2, which incorporates quality information after filtering, unlike other OTU‐based methods. Then, dereplication, sample inference, merging of paired‐end reads and removal of chimera were performed in order to obtain a sequence (ASV) table of abundance per sample. Taxonomy was assigned through the dada2 pipeline using the Ribosomal Database Project (RDP) Naive Bayesian Classifier algorithm from Wang et al. (2007). Depending on support (minimum bootstrap support of 80), we assigned taxonomy from kingdom to species. We used the Silva database formatted for dada2 to infer bacterial taxa (Callahan, 2018). We used the UNITE (UNITE Community, 2018) fasta release (including singletons) to infer fungal taxa after formatting it to the dada2 format using a custom r script. The pipeline was run on a multithreaded (48 CPUs) computer infrastructure provided by WestGrid (https://www.westgrid.ca/support/systems/cedar) and Compute Canada (www.computecanada.ca). Note that the pipeline was run separately for fungal‐root, fungal‐soil, bacteria‐soil and bacteria‐root samples given that these were sequenced separately and therefore a specific error model for each data set was calculated.
Statistical analyses – plant productivity
Each plant species (tomato and pepper) were analysed separately. We tested for the amendment effect (tomato: hen manure + ANE; pepper: ANE) on six plant productivity measures (fruit number, average fruit weight, shoots’ fresh weight, roots’ fresh weight, shoots’ dry weight and roots’ dry weight). We used linear mixed‐effects models (LMMs) in the R package nlme (Pinheiro et al., 2017), which are more appropriate than an analysis of variance (ANOVA) given the current block design (blocks and replicates were treated as random variables). All six plant productivity measures were either square root‐ or log‐transformed in order to help satisfy the assumption of normality and homogeneity of the variance of the residuals in the LMM statistical framework. For the variables, fruit number and average fruit weight, we also verified statistical significance using a permutation‐based 2‐way ANOVA (Anderson and Legendre, 1999) given that the residuals of the LMM were not normally distributed. Results were similar according to the 2‐way ANOVA.
Statistical analyses – microbial and fungal diversity
For each fungal‐root, fungal‐soil, bacterial‐root and bacterial‐soil data sets, we removed samples that showed poor sequencing output and contained few ASV. In addition, for bacterial‐root and soil data sets, we removed ASV that were taxonomically assigned to mitochondria or chloroplast given that these were likely sequences from the plants themselves. To remove low‐quality samples, we first summed the abundance of all ASV for each sample ( ASV) and eliminated samples that had fewer that a summed abundance of 1000. In addition, we removed ASV from our data set that were present in fewer than 5% of the samples (< 10 individuals in the soil samples or less than five in the root samples). This was done to remove very rare ASV unique to a block or replicate, but not found in the majority of samples.
We then conducted community‐based analyses looking at the amendment effect on ASV abundance in the tomato and pepper experiments separately. To visualize communities and reduce the complexity of the data sets, relative abundance of all taxa was calculated per family using the r package dplyr (Wickham et al., 2015) and barplots were drawn using ggplot2 (Wickham, 2016). ASV alpha (a)‐diversity was calculated based on all ASV (excluding rare ASV, see paragraph above) for each sample using the inverse Simpson diversity index in vegan (Oksanen et al., 2013). The effect of the amendment and planting for soil communities was assessed using a linear mixed‐effects model (LMM) in the R package nlme (Pinheiro et al., 2017), given the unbalanced, replicated block design. Alpha diversity was log‐transformed in order to help satisfy the assumption of normality of the residuals in the LMM statistical framework.
Using the community matrix data of ASV abundance, we performed PERmutational Multivariate ANalysis Of VAriance tests (PERMANOVA; Anderson, 2001) to identify relationships between the communities according to the experimental design. Data were analysed separately for fungal‐root, fungal‐soil, bacterial‐root and bacterial‐soil in tomatoes and peppers. The ASV abundance matrix was Hellinger‐transformed, and significance was assessed using 10 000 permutations in vegan (Oksanen et al., 2013). Blocks and replicates were factored as strata in the model. We also performed redundancy analyses (RDAs) using the Hellinger‐transformed ASV abundance matrix in vegan (Oksanen et al., 2013) to visually assess the grouping of samples, ASV and their association with productivity variables (species scaling based on ASV matrix). Data were analysed separately for fungal‐root, fungal‐soil, bacterial‐root and bacterial‐soil in tomatoes and peppers, giving a total of eight RDAs. Statistical significance of the RDAs was tested using an ANOVA‐like permutation test (10 000 permutations) in vegan. Data were constrained based on four productivity measures (fruit number, average fruit weight, shoots’ fresh weight and roots’ fresh weight).
Finally, we identified the ten ASV most positively associated with the measures of fruit number, shoots’ fresh weight and roots’ fresh weight from each RDA for a total of 40 fungal and 40 bacterial candidate ASV. We aligned candidate sequences from these candidates ASV using the Bioconductor r package decipher (Wright, 2016) and build pairwise distances matrices using a JC69 substitution models of DNA sequence evolution (equal base frequencies, Jukes and Cantor, 1969) in phangorn (Schliep, 2010). Phylogenetic trees (neighbour‐joining) for bacteria and fungi were plotted using ape (Paradis et al., 2004). This permitted to identify if similar candidate ASV were found under different experimental conditions (soil/root, pepper/tomato), thus reinforcing their role in productivity increase and increasing the probability that they are true positives.
Conflict of interest
None declared.
Supporting information
Table S1. Randomized split block design for the tomato and pepper experiments.
Table S2. Soil characteristics (in ppm unless specified otherwise).
Table S3. Stella Maris® characteristics.
Table S4. Summary of sequencing statistics and bioinformatics identification of ASV.
Table S5. Variance explained by the terms in the PERMANOVA models.
Fig. S1. Plant productivity.
Fig. S2. Neighbor‐Joining trees of candidates ASV (fungi) most positively associated with productivity measures as identified in Fig. 5.
Fig. S3. Neighbor‐Joining trees of candidates ASV (bacteria) most positively associated with productivity measures as identified in Fig. 6.
Acknowledgements
We thank Mengxuan Kong for technical assistance in setting up the greenhouse experiment and measuring productivity; Mulan Dai for performing preliminary microbiome analysis; and Simon Morvan for discussion about bioinformatics analyses and seaweed extracts. Research funding was provided by the Quebec Centre for Biodiversity Science [Fonds de recherche du Québec – Nature et Technologies (FRQNT)] to SR, FRQNT to JM and the Natural Sciences and Engineering Research Council of Canada (NSERC) to MH. The authors declare that they have received in‐kind contribution from Acadian Seaplants Ltd in the form of seaweed extract and technical support along with cash contribution from NSERC. The authors SB, JA and MH are not employed nor consultants for Acadian Seaplants Ltd, and they are not patenting any of the results presented in this study. The authors confirm that Acadian Seaplants Ltd did not influence the conclusions of the study.
Microbial Biotechnology (2019) 12(6), 1346–1358
Funding information
Fonds Québécois de la Recherche sur la Nature et les Technologies; Natural Sciences and Engineering Research Council of Canada.
References
- Alam, M.Z. , Braun, G. , Norrie, J. , and Hodges, D.M. (2013) Effect of ascophyllum extract application on plant growth, fruit yield and soil microbial communities of strawberry. Can J Plant Sci 93: 23–36. [Google Scholar]
- Alam, M.Z. , Braun, G. , Norrie, J. , and Hodges, D.M. (2014) Ascophyllum extract application can promote plant growth and root yield in carrot associated with increased root‐zone soil microbial activity. Can J Plant Sci 94: 337–348. [Google Scholar]
- Altschul, S.F. , Madden, T.L. , Schäffer, A.A. , Zhang, J. , Zhang, Z. , Miller, W. , and Lipman, D.J. (1997) Gapped blast and psi‐blast: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, M.J. (2001) A new method for non‐parametric multivariate analysis of variance. Austral Ecol 26: 32–46. [Google Scholar]
- Anderson, M.J. , and Legendre, P. (1999) An empirical comparison of permutation methods for tests of partial regression coefficients in a linear model. J Stat Comput Simul 62: 271–303. [Google Scholar]
- Antoun, H. , Beauchamp, C.J. , Goussard, N. , Chabot, R. , and Lalande, R. (1998) Potential of rhizobium and bradyrhizobium species as plant growth promoting rhizobacteria on non‐legumes: effect on radishes (Raphanus sativus L.) In Molecular Microbial Ecology of the Soil. Dordrecht: Springer; pp. 57–67. [Google Scholar]
- Avis, T.J. , Gravel, V. , Antoun, H. , and Tweddell, R.J. (2008) Multifaceted beneficial effects of rhizosphere microorganisms on plant health and productivity. Soil Biol Biochem 40: 1733–1740. [Google Scholar]
- Baez‐Rogelio, A. , Morales‐García, Y.E. , Quintero‐Hernández, V. , and Muñoz‐Rojas, J. (2017) Next generation of microbial inoculants for agriculture and bioremediation. Microb Biotechnol 10: 19–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battacharyya, D. , Babgohari, M.Z. , Rathor, P. , and Prithiviraj, B. (2015) Seaweed extracts as biostimulants in horticulture. Sci Hortic 196: 39–48. [Google Scholar]
- Bruulsema, T.W. , Heffer, P. , Welch, R. , Cakmak, I. , and Moran, K. (2012) Fertilizing crops to improve human health: a scientific review. Better Crops 2: 96. [Google Scholar]
- Callahan, B. (2018) Silva for dada2: Silva taxonomic training data formatted for dada2 (silva version 132). Zenodo.
- Callahan, B.J. , McMurdie, P.J. , Rosen, M.J. , Han, A.W. , Johnson, A.J.A. , and Holmes, S.P. (2016) DADA2: high‐resolution sample inference from illumina amplicon data. Nat Methods 13: 581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso, J.G. , Kuczynski, J. , Stombaugh, J. , Bittinger, K. , Bushman, F.D. , Costello, E.K. , et al (2010) QIIME allows analysis of high‐throughput community sequencing data. Nat Methods 7: 335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaparro, J.M. , Badri, D.V. , and Vivanco, J.M. (2014) Rhizosphere microbiome assemblage is affected by plant development. ISME J 8: 790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, H. , Zak, D.R. , Reich, P.B. , and Ellsworth, D.S. (2007) Plant species richness, elevated co2, and atmospheric nitrogen deposition alter soil microbial community composition and function. Glob Change Biol 13: 980–989. [Google Scholar]
- Craigie, J.S. (2010) Seaweed extract stimuli in plant science and agriculture. J Appl Phycol 23: 371–393. [Google Scholar]
- Craigie, J.S. (2011) Seaweed extract stimuli in plant science and agriculture. J Appl Phycol 23: 371–393. [Google Scholar]
- Dhargalkar, V. , and Pereira, N. (2005) Seaweed: promising plant of the millennium. Sci Cult 71: 60–66. [Google Scholar]
- Fisher, R.A. , Corbet, A.S. , and Williams, C.B. (1943) The relation between the number of species and the number of individuals in a random sample of an animal population. J Anim Ecol 12: 42–58. [Google Scholar]
- du Jardin, P. (2015) Plant biostimulants: definition, concept, main categories and regulation. Sci Hortic 196: 3–14. [Google Scholar]
- Jithesh, M.N. , Wally, O.S. , Manfield, I. , Critchley, A.T. , Hiltz, D. , and Prithiviraj, B. (2012) Analysis of seaweed extract‐induced transcriptome leads to identification of a negative regulator of salt tolerance in arabidopsis. HortScience 47: 704–709. [Google Scholar]
- Jukes, T. , and Cantor, C. (1969) Evolution of protein molecules In Mammalian Protein Metabolism. Munro H.N. (ed.). New York, NY: Academic Press, pp. 21–132. [Google Scholar]
- Khan, W. , Rayirath, U.P. , Subramanian, S. , Jithesh, M.N. , Rayorath, P. , Hodges, D.M. , et al (2009) Seaweed extracts as biostimulants of plant growth and development. J Plant Growth Regul 28: 386–399. [Google Scholar]
- Klindworth, A. , Pruesse, E. , Schweer, T. , Peplies, J. , Quast, C. , Horn, M. , and Glöckner, F.O. (2013) Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next‐generation sequencing‐based diversity studies. Nucleic Acids Res 41: e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lay, C.‐Y. , Hamel, C. , and St‐Arnaud, M. (2018) Taxonomy and pathogenicity of olpidium brassicae and its allied species. Fung Biol 122: 837–846. [DOI] [PubMed] [Google Scholar]
- Li, F. , Chen, L. , Redmile‐Gordon, M. , Zhang, J. , Zhang, C. , Ning, Q. , and Li, W. (2018) Mortierella Elongata's roles in organic agriculture and crop growth promotion in a mineral soil. Land Degrad Dev 29: 1642–1651. [Google Scholar]
- Margulis, L. , and Fester, R. (1991) Symbiosis as a Source of Evolutionary Innovation: Speciation and Morphogenesis. Cambridge, MA: MIT Press. [PubMed] [Google Scholar]
- Milton, R. (1952) Improvements in or relating to horticultural and agricultural fertilizers. British Patent 664989:
- Oksanen, J. , Blanchet, F.G. , Kindt, R. , Legendre, P. , Minchin, P.R. , O'Hara, R. , et al (2013) Vegan: Community ecology package. R package version 1.17.2. R software.
- Pandin, C. , Le Coq, D. , Canette, A. , Aymerich, S. , and Briandet, R. (2017) Should the biofilm mode of life be taken into consideration for microbial biocontrol agents? Microb Biotechnol 10: 719–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis, E. , Claude, J. , and Strimmer, K. (2004) APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20: 289–290. [DOI] [PubMed] [Google Scholar]
- Pinheiro, J. , Bates, D. , DebRoy, S. , Sarkar, D. , and R Core Team (2017) Nlme: Linear and nonlinear mixedeffects models. R package version 3.1‐128. R software.
- Pregitzer, K.S. , DeForest, J.L. , Burton, A.J. , Allen, M.F. , Ruess, R.W. , and Hendrick, R.L. (2002) Fine root architecture of nine north American trees. Ecol Monogr 72: 293–309. [Google Scholar]
- R Core Team (2018) R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Reinhold‐Hurek, B. , Bünger, W. , Burbano, C.S. , Sabale, M. , and Hurek, T. (2015) Roots shaping their microbiome: global hotspots for microbial activity. Annu Rev Phytopathol 53: 403–424. [DOI] [PubMed] [Google Scholar]
- Schliep, K.P. (2010) Phangorn: phylogenetic analysis in R. Bioinformatics 27: 592–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss, P.D. , Westcott, S.L. , Ryabin, T. , Hall, J.R. , Hartmann, M. , Hollister, E.B. , et al (2009) Introducing mothur: open‐source, platform‐independent, community‐supported software for describing and comparing microbial communities. Appl Environ Microbiol 75: 7537–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, Z. , Ruan, Y. , Chao, X. , Zhang, J. , Li, R. , and Shen, Q. (2015) Rhizosphere microbial community manipulated by 2 years of consecutive biofertilizer application associated with banana fusarium wilt disease suppression. Biol Fertil Soils 51: 553–562. [Google Scholar]
- Shen, Z. , Wang, B. , Zhu, J. , Hu, H. , Tao, C. , Ou, Y. , et al (2019) Lime and ammonium carbonate fumigation coupled with bio‐organic fertilizer application steered banana rhizosphere to assemble a unique microbiome against Panama disease. Microb Biotechnol 12: 515–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow, L.S. , LeTourneau, M.K. , Kwak, Y.‐S. , and Weller, D.M. (2019) The soil‐borne legacy in the age of the holobiont. Microb Biotechnol 12: 51–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkacz, A. , and Poole, P. (2015) Role of root microbiota in plant productivity. J Exp Bot 66: 2167–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toju, H. , Tanabe, A.S. , Yamamoto, S. , and Sato, H. (2012) High‐coverage its primers for the dna‐based identification of ascomycetes and basidiomycetes in environmental samples. PLoS ONE 7: e40863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi, P. , Schenk, P.M. , Wallenstein, M.D. , and Singh, B.K. (2017) Tiny microbes, big yields: enhancing food crop production with biological solutions. Microb Biotechnol 10: 999–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UNITE Community (2018) UNITE general fasta release. Version 01.12.2017.
- Wally, O.S. , Critchley, A.T. , Hiltz, D. , Craigie, J.S. , Han, X. , Zaharia, L.I. , et al (2013) Regulation of phytohormone biosynthesis and accumulation in arabidopsis following treatment with commercial extract from the marine macroalga Ascophyllum nodosum . J Plant Growth Regul 32: 324–339. [Google Scholar]
- Wang, Q. , Garrity, G.M. , Tiedje, J.M. , and Cole, J.R. (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73: 5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham, H. (2016) Ggplot2: Elegant Graphics for Data Analysis. Houston: Springer. [Google Scholar]
- Wickham, H. , Francois, R. , Henry, L. and Müller, K. (2015) Dplyr: A grammar of data manipulation. R package version 0.4 3:
- Wright, E.S. (2016) Using decipher v2.0 to analyze big biological sequence data in R. R Journal 8: 352–359. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Randomized split block design for the tomato and pepper experiments.
Table S2. Soil characteristics (in ppm unless specified otherwise).
Table S3. Stella Maris® characteristics.
Table S4. Summary of sequencing statistics and bioinformatics identification of ASV.
Table S5. Variance explained by the terms in the PERMANOVA models.
Fig. S1. Plant productivity.
Fig. S2. Neighbor‐Joining trees of candidates ASV (fungi) most positively associated with productivity measures as identified in Fig. 5.
Fig. S3. Neighbor‐Joining trees of candidates ASV (bacteria) most positively associated with productivity measures as identified in Fig. 6.


