Summary
Microbial communities from harsh environments hold great promise as sources of biotechnologically relevant strains and compounds. In the present work, we have characterized the microorganisms from the supralittoral and splash zone in three different rocky locations of the Western Mediterranean coast, a tough environment characterized by high levels of irradiation and large temperature and salinity fluctuations. We have retrieved a complete view of the ecology and functional aspects of these communities and assessed the biotechnological potential of the cultivable microorganisms. All three locations displayed very similar taxonomic profiles, with the genus Rubrobacter and the families Xenococcaceae, Flammeovirgaceae, Phyllobacteriaceae, Rhodobacteraceae and Trueperaceae being the most abundant taxa; and Ascomycota and halotolerant archaea as members of the eukaryotic and archaeal community respectively. In parallel, the culture‐dependent approach yielded a 100‐isolates collection, out of which 12 displayed high antioxidant activities, as evidenced by two in vitro (hydrogen peroxide and DPPH) and confirmed in vivo with Caenorhabditis elegans assays, in which two isolates, CR22 and CR24, resulted in extended survival rates of the nematodes. This work is the first complete characterization of the Mediterranean splash‐zone coastal microbiome, and our results indicate that this microbial niche is home of an extremophilic community that holds biotechnological potential.
Introduction
The interphase between marine and land environments is an ecologically complex habitat in which selection pressures from both environments can co‐occur. Some of those pressures are high salinity, dehydration, wind and sun exposition, extreme temperature oscillations and mechanical stress associated with seawater splash, often with sand or pebbles, with strong abrasive effects. The aquatic to land transition has been reported to be linked to a narrow gradient in species distribution in function of the distance to the water line, as for example in cyanobacteria in an English lake (Pentecost, 2014). Regarding marine environments, the microbial ecology of rocky shores has previously been analysed (Chan et al., 2003; Langenheder and Ragnarsson, 2007; Pinedo et al., 2007; Brandes et al. 2015), including its links with oil spills and biodegradation (Alonso‐Gutiérrez et al., 2009). However, and in contrast with the well‐studied microbial ecology of the intertidal zone (for a review, see Mitra et al., 2014), a holistic study on the microbial ecology of the marine supralittoral Mediterranean rocky shore has not been addressed previously.
Harsh, extremophilic environments can be sources of biotechnologically relevant bacteria and therefore hold great promise for the biotechnological industry (Raddadi et al., 2015). For example, extremophilic microorganisms can yield enzymes such as lipases and esterases that can be used under a wide range of conditions and may have relevant applications in the food, detergent and biofuel industries (Fuciños et al., 2012). There are many other examples of biotechnologically relevant microorganisms from extreme environments, including the well‐known case of Thermus aquaticus, which produces the widely used Taq polymerase; or the hyperthermophilic biofuel‐producing archaea that live in deep‐sea hydrothermal vents (Chien et al., 1976; Nishimura and Sako, 2009).
The present study focuses on the microorganisms that inhabit the rocky areas of the supralittoral zone (the area just above the tide line that is subjected regularly to splash but is not permanently underwater) of the Mediterranean coast. Surface‐associated microbial communities that are sun‐exposed are often rich in microorganisms that produce pigments, including carotenoids (Dorado‐Morales et al., 2015; Kumar et al., 2015; : Shindo and Misawa, 2014; Tanner et al., 2017). These pigments play a key role in radiation tolerance (Tian and Hua, 2010; Klindworth et al., 2013; Sandmann, 2015; Tanner et al., 2018), and they are valuable for the food, pharmacological and cosmetic industries as colourants, antioxidants and protectors against solar radiation respectively (Sandmann, 2015). Therefore, we hypothesized that rough conditions of the supratidal zone may be associated with the presence of biotechnologically relevant microbial taxa. From this hypothesis, we have, in the present work, compared three different supralittoral coastal locations of the Mediterranean West coast and combined culturing techniques and high throughput sequencing data (16S rRNA amplicon and metagenomic sequencing) in order to shed light on the taxonomic composition of these communities, and to explore the biotechnological potential of the culturable strains.
Results
High‐throughput 16S rRNA analysis
High‐throughput 16S rRNA sequencing of the samples revealed that, based on the comparison of the richness value (number of different species; Fig. 1A) and the diversity (Shannon index; Fig. 1B), the alpha diversity was not significantly different among the locations. Moreover, the shape of the rarefaction curve at OTU level (Operational Taxonomic Unit) showed that the sequences covered the majority of taxa present in the samples (Fig. S1).
Figure 1.
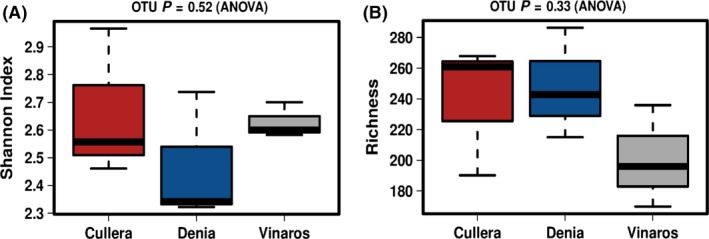
Box plots showing the values of alpha diversity indexes in the sampled locations on the Mediterranean rocky‐shore. (A) Observed richness at OTU level (number of OTUs). (B) Shannon index of diversity.
However, the composition of the bacterial communities varied depending on the location, as represented in the Principal Coordinates Analysis (PCoA; Fig. 2A). Samples from Dénia showed the highest intragroup homogeneity, whereas samples from Vinaròs and Cullera displayed higher differences between replicates. Nevertheless, samples from all three locations could be distinguished in the plot. The variability explained by both axes is high enough to conclude that the microbial communities among the three locations are different. Moreover, the representation of the relative abundances (TSS) of the top 30 most abundant genera showed that the microbial composition was generally similar along the locations (Fig. 2B), although some taxa such as the genus Rubrobacter in Vinaròs or the genus Rubricoccus in Dénia allowed the differentiation of specific regions (Table 1). Eleven out of the 30 most abundant genus were significantly different at least in one location. A list of the 30 more significantly different genus is shown in (Table S1). The original data have been deposited with the NCBI SRA accession number PRJNA556782.
Figure 2.
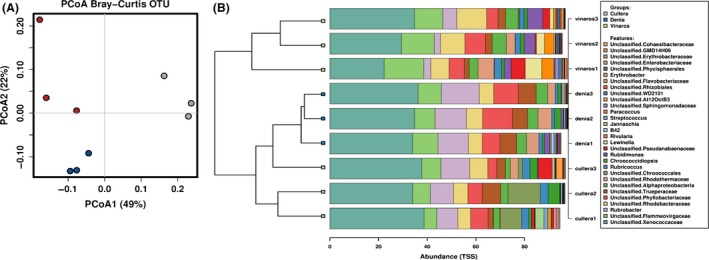
(A) Principal coordinates analysis (PCoA) based on Bray–Curtis distances between OTUs in bacterial communities of three different locations. (B) Clustered‐Barchart showing the top 30 most abundant genera in terms of relative abundance.
Table 1.
Top 30 most abundant genera and P‐values for the One‐Way ANOVA statistical analysis of their distributions among the three sampled locations
| Taxa | P labelA | P (Tukeys) Dénia‐Cullera | P (Tukeys) Vinaròs‐Cullera | P (Tukeys) Vinaròs‐Dénia |
|---|---|---|---|---|
| Rubrobacter | 0.0011* | 0.083 | 0.0096* | 0.00091* |
| Rubricoccus | 0.0018* | 0.0014* | 0.057 | 0.026* |
| Unclassified Flammeovirgaceae | 0.006* | 0.22 | 0.0051* | 0.04* |
| Rubidimonas | 0.0075* | 0.58 | 0.0078* | 0.024* |
| Unclassified Erythrobacteraceae | 0.0085* | 0.011* | 0.018* | 0.9 |
| Unclassified Alphaproteobacteria | 0.019* | 0.052 | 0.02* | 0.71 |
| Unclassified Cohaesibacteraceae | 0.021* | 0.021* | 0.062 | 0.66 |
| Rivularia | 0.026* | 1 | 0.04* | 0.038* |
| Unclassified Rhodobacteraceae | 0.037* | 1 | 0.058 | 0.052 |
| Unclassified WD2101 | 0.039* | 1 | 0.06 | 0.054 |
| Unclassified Chroococcales | 0.045* | 0.063 | 0.068 | 1 |
| Lewinella | 0.052 | 1 | 0.073 | 0.075 |
| Unclassified Trueperaceae | 0.066 | 0.29 | 0.45 | 0.056 |
| Unclassified Phyllobacteriaceae | 0.091 | 0.094 | 0.87 | 0.18 |
| Unclassified Xenococcaceae | 0.1 | 0.85 | 0.1 | 0.21 |
| Unclassified GMD14H09 | 0.12 | 1 | 0.17 | 0.16 |
| Unclassified Sphingomonadaceae | 0.13 | 0.88 | 0.24 | 0.13 |
| Unclassified Flavobacteriaceae | 0.13 | 1 | 0.16 | 0.17 |
| B42 | 0.18 | 0.19 | 0.95 | 0.28 |
| Unclassified Rhodothermaceae | 0.24 | 0.23 | 0.47 | 0.82 |
| Unclassified Rhizobiales | 0.25 | 0.72 | 0.55 | 0.22 |
| Chroococcidiopsis | 0.28 | 0.4 | 0.3 | 0.97 |
| Erythrobacter | 0.28 | 0.29 | 0.42 | 0.95 |
| Unclassified Phycisphaerales | 0.32 | 0.29 | 0.66 | 0.73 |
| Unclassified Pseudanabaenaceae | 0.34 | 0.51 | 0.93 | 0.33 |
| Jannaschia | 0.39 | 0.44 | 0.47 | 1 |
| Paracoccus | 0.44 | 0.78 | 0.41 | 0.78 |
| Unclassified At12OctB3 | 0.46 | 0.49 | 0.55 | 0.99 |
| Streptococcus | 0.54 | 0.91 | 0.52 | 0.76 |
| Unclassified Enterobacteriaceae | 0.77 | 0.91 | 0.75 | 0.95 |
Global P‐values and P‐values for the comparison by pairs is shown. Significant results are marked by an asterisk.
Shotgun metagenomic analysis
The three locations exhibited similar taxonomic profiles according to the metagenomics analysis. The most abundant bacterial phyla were the same ones observed with high‐throughput 16S rRNA sequencing, with Cyanobacteria being the most abundant in all three locations. Moreover, other taxa, such as the families Rhodobacteraceae, Flammeovirgaceae, Trueperaceae and the genus Rubrobacter, belonging to the phyla Proteobacteria, Bacteroidetes, Deinococcus‐Thermus and Actinobacteria respectively, were also detected (Figs S2A, S3A and S4A). Metagenomic sequencing allowed the identification of abundant taxa in the Cyanobacteria phylum, including the genera Staniera, Pleurocapsa, Myxosarcina and Xenococcus, in contrast to the high‐throughput 16S rRNA, which mainly showed unclassified Xennococcaceae taxa.
Archaeal and eukaryotic communities proved very diverse, with a high number of salt‐adapted microorganisms in the former and a large fraction of Ascomycota in the latter (Figs S2, S3, S4B and C). Salt‐adapted archaea included members of Halococcus, Halobacteriaceae (Haladaptatus and Halalkalicoccus), Haloarculaceae, Haloferaceae, Halorubraceae and Natrialbaceae families, as well as methanogenic archaea (members of the Methanosarcinaceae family; Figs S2B, S3B and S4B). Among the diversity of Ascomycota, the most abundant taxa were Glonium stellatum, Cenococcum geophilum, Coniosporium apollinis and Lepidopterella palustris (Figs S2C, S3C and S4C).
The functional analysis of the samples revealed a high representation of enzymes related to oxidative stress, being peroxiredoxin (EC 1.11.1.15) and peroxidase (EC 1.11.1.7) the most abundant activities, and displaying the highest values in Cullera and Vinaròs respectively. Thioredoxin‐related enzymatic activities (EC 1.8.4.8; EC 1.8.1.9; EC 1.8.4.10) were homogeneously represented in all three samples, as well as superoxide dismutase (EC 1.15.1.1). Other enzymes such as glutathione transferase (EC 2.5.1.18) or glutathione peroxidase (EC 1.11.1.9) varied among locations, with the former being more represented in Vinaròs and Cullera than in Dénia, and the latter being more abundant in Vinaròs. Among the genes related to carotenoid biosynthetic routes, the abscisic acid 8′‐hydroxylase (EC 1.14.14.137) was particularly represented in Cullera, whereas sphingolipid‐related genes such as glucosylceramidase proved to be frequent in Vinaròs (EC 3.2.1.45; Fig. 3). The original data have been deposited with the NCBI SRA accession number PRJNA556786.
Figure 3.
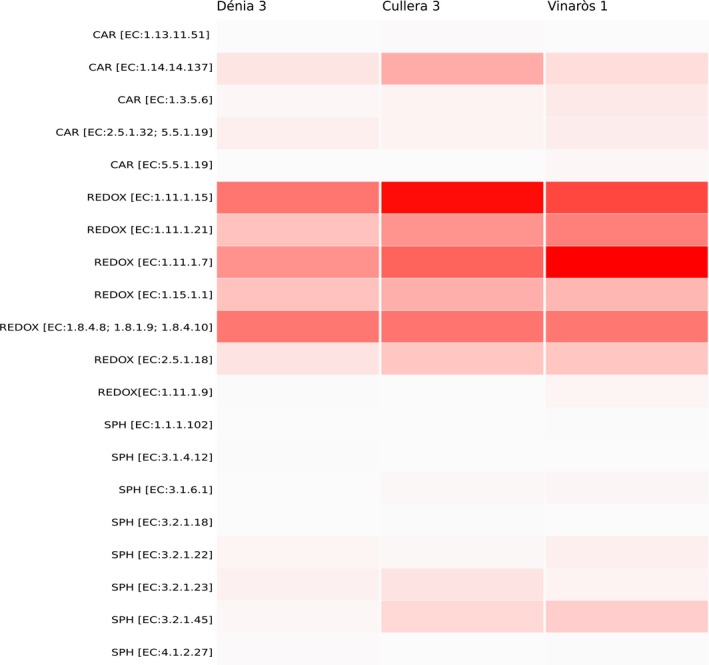
Heatmap representing the functional analysis carried out through metagenomics sequencing. Enzymes related to carotenoid biosynthesis (CAR), oxidative stress (REDOX) and sphingolipid biosynthesis (SPH) are shown in the Y‐axis.
Strain collection and identification
Culturing the samples on LB and Marine Agar yielded a large diversity of colonies in terms of colour, shape and morphology. A total of 100 strains were isolated and named with a code, after the location (C: Cullera, D: Dénia, V: Vinaròs) and the origin (M: Marine water, R: Rock surface). In our conditions, there was no significant fungal growth in any of the samples. The colonies observed on Marine Agar displayed the widest range of colours (wine‐red, red, pink and orange, among others) in comparison with the ones observed on LB media, which were mostly yellowish and cream‐coloured. Due to the known relation between the presence of pigments and antioxidant power, the main criterion for colony selection was the colour (Pawar et al., 2015).
A collection of the 100 selected isolates in pure culture was established. A total of 34 isolates were initially identified through colony PCR and 16S rRNA Sanger sequencing. Although an initial step of incubation at 100°C was added to the PCR protocol of the isolates whose amplification had failed, some remained non‐identified and therefore their total DNA was extracted to repeat the PCR. Finally, 56 of the isolates remained unidentified. Among the identified isolates, there were many Bacillus spp. (B. oleronius, B. licheniformis, B. marisflavi, B. salsus and B. altitudinis) and Halobacillus spp. (H. trueperi and H. faecis) as well as other species such as Micrococcus antarcticus, Micrococcus luteus, Staphylococcus pasteuri, Vibrio tubiashii and Virgibacillus halodenitrificans (Table S2).
Antioxidant activity
In order to select and establish a collection of isolates with antioxidant properties, a high‐throughput screening of the 100 isolates was performed by growing them on solid media containing H2O2. Planomicrobium glaciei and E. coli JM109 were used as positive and negative controls respectively. Strain JM109, with no known reports of antioxidant effect, exhibited a weak growth in the first (OD600 1) and, sometimes, second dilution (OD600 10−1). This led us to the criterion to consider positive antioxidant producers those strains able to grow on H2O2‐containing plates at least up to threefold dilutions (OD600 10−2). A total of 12 isolates were thus selected (Table 2) based on their ability to grow on 1 mM H2O2 plates as described above.
Table 2.
List of selected isolates, percentage of identity with the closest type strain, sequence similarity and results obtained in the H2O2 assay
| Sample | Closest type strain | % | H2O2 Assay (dilution at which the isolate remains viable) |
|---|---|---|---|
| CR10 | Micrococcus luteus (CP001628) | 99.77 | 3 |
| CR17 | Virgibacillus halodenitrificans (AY543169) | 99.58 | 7 |
| CR21 | Non‐identified | – | 4 |
| CR22 | Virgibacillus halodenitrificans (AY543169) | 99.37 | 4 |
| CR24 | Halobacillus trueperi (AJ310349) | 98.31 | 6 |
| CR28 | Virgibacillus halodenitrificans (AY543169) | 100 | 6 |
| CR37 | Bacillus marisflavi (LGUE01000011) | 100 | 4 |
| CR44 | Non‐identified | – | 3 |
| CR67 | Bacillus oleronius (X82492) | 97.32 | 4 |
| DM10 | Non‐identified | – | 3 |
| DR12 | Non‐identified | – | 3 |
| VR1 | Bacillus altitudinis (ASJC011000029) | 100 | 6 |
| VR2 | Micrococcus luteus (CP001628) | 99.35 | 3 |
| Positive control | P. glaciei | 8 | |
| Negative control | E. coli (JM109) | 1 |
DPPH‐based assays are widely used to detect and quantify the antioxidant power of plants or bacterial extracts. These assays are based on the decrease of DPPH absorbance at 517 nm in presence of antioxidant factors. The oxidative stress‐resistant isolates selected from the H2O2 assay (shown in Table 2) were further tested using this method. CR17, CR21 and CR57 could not be tested due to poor growth in liquid culture, which made it impossible to obtain a concentrated extract, prepared as described in Experimental Procedures. 16S rRNA sequences were compared using NCBI BLAST tool. Isolates CR10‐VR2 and CR22‐CR28 were 100% identical in their 16S rRNA sequence, and therefore only one of them was selected for further assays (CR10 and CR22 respectively).
The test resulted in a general decrease in absorbance in all the samples, suggesting that the extracts were able to scavenge the DPPH. The isolates that proved more effective as antioxidants were CR22, CR24 and CR28, with values of scavenged DPPH over 30% (Fig. 4A). DR12 displayed low DPPH scavenging values maybe due to failure of the pigment extraction. Surprisingly, the control samples P. glaciei and JM109 did not display the expected effect. A set of three strains that had previously shown a protective effect against oxidative stress in a Caenorhabditis elegans model and a set of three E. coli strains (JM109, HB101 and DH5α) were also tested (Fig. 4D).
Figure 4.
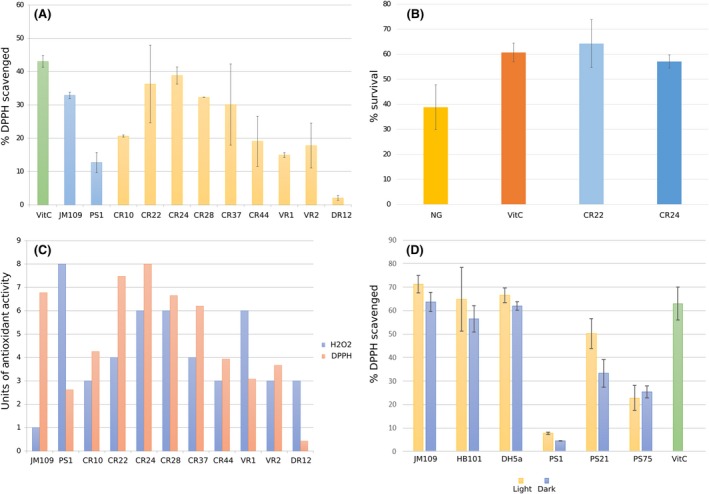
(A) Antioxidant activity as measured through DPPH assay as described in EP. Absorbance was measured at 517 nm after 30 min of incubation with DPPH 50 μM. DPPH scavenged (%) is represented in Y‐axis. VitC, vitamin C (0.5 μg ml−1 solution). (B) Antioxidant activity in vivo (using the model organism C. elegans). Y‐axis indicates percentage of surviving worms after 5 h of incubation under oxidative stress (H2O2). Worms were treated with either a control diet (NG), a diet supplemented with the known antioxidant vitamin C as a positive control (VitC), or a diet supplemented with the selected strains CR22 and CR24. (C) Comparative analysis of the results obtained with H2O2 and DPPH assays. Values in Y‐axis are normalized with respect to the highest value obtained in both assays. (D) DPPH assay with positive and negative controls. Absorbance was measured at 517 nm after 30 min of incubation with DPPH 50 μM. DPPH scavenged percentage is represented in Y‐axis. VitC, Vitamin C, 0.5 μg ml−1 solution. Light and dark conditions are represented.
The two strains with the best results in the in vitro assays (CR22 and CR24) were selected for further in vivo antioxidant assays in the model organism C. elegans, where both proved able to display an important antioxidant activity (Fig. 4B). Nematodes subjected to oxidative stress after being treated with isolates CR22 and CR24 displayed survival rates higher than the untreated worms and similar to those observed in the worms treated with vitamin C (survival rates of around 55%–65%).
Discussion
We report here, for the first time, and by using culture‐dependent and independent (NGS) techniques, the microbiomes of the rocky‐coastal surface of the supralittoral zone in three regions on the Mediterranean western coast. The three sampled sites, covering a coast line of about 260 km, displayed remarkably similar taxonomic profiles in terms of richness and microbial diversity, but still could perfectly be differentiated thanks to the significant difference in abundances of specific taxa, which suggest that the microbial composition of the Mediterranean supratidal zone, at least in eastern Spain, is stable but not identical within rocky locations. The studied communities were particularly dominated by bacterial strains previously described as thermophilic, halotolerant or radioresistant, such as the species within the genus Rubrobacter (Jurado et al., 2012), and pigmented isolates, as is the case of species within the Flameovirgaceae family, like Tunicatimonas pelagia and Porifericola rhodea (Yoon et al., 2011, 2012).
Truepera radiovictrix, characterized by an optimum growth temperature of 50°C and an extreme resistance to ionizing radiation, was first isolated from a hot spring in a geothermal area close to the Azores (Albuquerque et al., 2005; Ivanova et al., 2011). Moreover, the Truepera genus has been previously found in Lake Lucero Playa (New Mexico, USA), a particularly hostile environment as the lake dries periodically (Sirisena et al., 2018). This is, to the best of our knowledge, the first report of sea‐inhabiting Truepera in a non‐thermal environment, and it is tempting to hypothesize that the genus Truepera might have a similar ecological niche (radiation‐ and desiccation‐resistance) than Deinococcus, but in saline environments, as a consequence of both its radiation resistance and halotolerance (Albuquerque et al., 2005).
Shotgun metagenomic analysis confirmed the similarity between the communities of the three sampled locations, as discussed above from the high‐throughput 16S rRNA results, particularly at higher taxonomic (i.e. family) levels. Nevertheless, the results at lower taxonomic levels varied considerably among sequencing techniques. One of the largest differences at the species level was observed within the Cyanobacterial group. In particular, high‐throughput 16S rRNA revealed a large abundance of Xennococcaceae, whereas shotgun metagenomic sequencing revealed a more diverse population including members of Pleurocapsa, Myxosarcina, Stanieria and Xenococcus, as previously reported for marine environments (Burns et al., 2004; Alex et al., 2012; Yu et al., 2015; Brito et al., 2017).
The eukaryotic fraction of the samples was mainly composed of Ascomycota, such as Glonium stellatum. The genus Glonium includes saprophytic Dothideomycetes that produce darkly pigmented apothecia, which could contribute to the dark colour of the sampled rocks (Spatafora et al., 2012). Other species detected in the samples included as follows: Cenococcum geophilum, an ectomycorrhizal fungus previously described in coastal forest soils (Matsuda et al., 2015) and previously demonstrated to grow at up to 100 mM of NaCl (Obase et al., 2010); Coniosporium apollinis, a rock‐inhabiting fungi previously isolated from the Mediterranean basin (Sterflinger et al., 1997); and Lepidopterella palustris, typically a freshwater fungus (Shearer et al., 2009), with this being, to the best of our knowledge, the first description of this species in a salt water habitat.
Taken together, the results obtained from both high‐throughput 16S rRNA and metagenomic sequencing suggest that the sampled communities are composed of a diverse array of fungi (mainly belonging to the phylum Ascomycota), cyanobacteria (mainly S. cyanosphaera and Pleurocapsa spp., but also Myxosarcina spp. and Xenococcus spp.) and salt‐adapted archaea, which remain rather stable among the three different sampled locations.
From the functional point of view, metagenomics sequencing showed abundance of enzymes involved in oxidative stress, mainly peroxidase, peroxiredoxin and thioredoxin, but also catalase and glutathione transferase. In contrast with this, enzymes involved in carotenoid or sphingolipid biosynthesis, which also play a role in the protection against oxidative stress, were less abundant and varied among locations, being abscisic acid 8′‐hydroxylase (EC 1.14.14.137) in Cullera and glucosylceramidase (EC 3.2.1.45) in Vinaròs the ones with the highest values.
From the collection of cultured microorganisms, a total of 12 isolates were selected for their high antioxidant activity as measured by the oxidative stress assay performed with H2O2. Of those, M. luteus has been reported to encode genes related to resistance and tolerance to oxidative stress (superoxide dismutase and NADP reductase; Lafi et al., 2017). The DPPH assay was performed to dismiss false positives through the H2O2 assay. In general, the results correlated well with the ones previously observed in the H2O2 assay. It is important to note that, although DR12 displayed low scavenging in the DPPH assay, the extraction of pigments from this isolate was sub‐optimal, since the pellet remained pink‐coloured after the extraction process. Surprisingly, the control samples P. glaciei and JM109 did not display the expected effect in terms of antioxidant activity. On one hand, P. glaciei was expected to be one of the most antioxidant isolates, as its antioxidant activity was demonstrated in previous in vivo assays in C. elegans (Tanner et al., 2019) and in the H2O2 assay. Nevertheless, it was the worst strain in terms of DPPH scavenging. On the other hand, E. coli JM109, with no previous reports on antioxidant activity, resulted in high DPPH scavenging. This raises concerns on the suitability of DPPH‐methods in bioprospecting for the determination of antioxidant activity and highlights the importance of using several alternative methods as the best option to have a proxy of the in vivo antioxidant effects. Nevertheless, the in vivo antioxidant assay performed in C. elegans allowed to confirm the antioxidant activity detected in the DPPH and H2O2 tests. Specifically, CR22 and CR24 displayed an antioxidant activity similar to the one observed in Vitamin C (Fig. 4B).
In general, though, the correlation between both methods was good, as the isolates with higher survival in the presence of H2O2 also displayed higher DPPH‐scavenging ability (Fig. 4C). Nevertheless, there were some isolates that displayed different results depending on the method, in particular VR1 and CR37. Differences in VR1 could be the result of catalase activity, which may have enhanced its growth on the H2O2‐supplemented plates. On the contrary, differences between both methods for CR37 could be caused by a deficient growth in solid medium. Once again, these results highlight the limitation of using a single screening technique for the selection of microbial strains with antioxidant activities.
A collection of both positive and negative controls (in terms of theoretical antioxidant activity) were tested using both assays (H2O2 and DPPH). PS1, PS21 and PS75 (P. glaciei 423, 97.38% ID; Rhodobacter maris JA276, 98.89% ID; and Bacillus megaterium NBRC 15308, 100% ID respectively) were the three control strains selected, all of them recovered from solar panels and previously tested in C. elegans for in vivo protection against oxidative stress (Tanner et al., 2019). Three different strains of E. coli were chosen as negative controls (JM109, BH101, DH5α). For the DPPH assay, the isolates were grown under both light and dark conditions, in order to determine whether the light had a negative impact on the production of pigments or other antioxidant factors, as it is known that many pigments, particularly carotenoids, are prone to photodegradation (Boon et al., 2010). For the E. coli strains, no significant differences were observed between growth in dark and light conditions, whereas PS21 proved very sensitive to light (Fig. 4D). Moreover, the scavenging effect of the JM109 strain was also observed in the other two E. coli strains, confirming that the extracts obtained from E. coli contain compounds that are indeed able to react with DPPH. Even though R. maris and B. megaterium displayed better antioxidant properties than P. glaciei, which was again comparable to the negative control of methanol, they yielded lower DPPH‐based activity than E. coli strains.
The biotechnological potential of extremophiles is well known, and saline environments are no exception to this rule (de Lourdes Moreno et al., 2013). However, and in contrast with the well‐studied intertidal zone (Mitra et al., 2014), the supralittoral zone has been poorly studied to date. Interestingly, this zone experiences much higher selection pressures than the intertidal zone since while the intertidal zone is basically a marine environment which is only transiently and partially exposed to land conditions, the supralittoral zone forces organisms to adapt to a sea/land intermediate habitat where both marine and land stresses are present.
This work is the first holistic (using culture‐dependent, culture‐independent and biological activity assays) approach studying the microbial ecology and biotechnological potential, in terms of antioxidant properties, of the supralittoral zone of the Mediterranean rocky shore. Our results suggest that the western coastline of the Mediterranean Sea harbours a stable microbial community that is conserved among different locations, with cyanobacteria as the majoritarian bacterial taxon, followed by members of the Flameovirgaceae family and members of the Rubrobacter genus, as well as eukaryotic and archaeal members, such Ascomycota and halotolerant archaea. Furthermore, in vitro and in vivo assays demonstrate that this environment is a potential source of microorganisms with antioxidant activities that could hold potential for a wide range of applications in the food, cosmetic or pharmacological industries.
Experimental procedures
Sampling
Samples were collected from three different locations on the Mediterranean Western coast, in Eastern Spain: Vinaròs (Castelló), Cullera (València) and Dénia (Alacant). Three samples of dark‐stained rock, at least two metres apart from each other and thus considered as biological replicates, were collected from the supralittoral (splash) zone of each location by scraping the surface with a sterile blade. Samples of the adjacent marine water were also taken, and both types of samples (scrapped rock and sea water samples) were separately stored in Falcon tubes in 15% glycerol, transported to the laboratory on ice and then stored at −20°C until required.
High‐throughput rRNA and metagenomic sequencing
Total DNA was isolated from the samples with the PowerSoil DNA Isolation kit (MO BIO laboratories, Carlsbad, CA, USA) following the manufacturer's instructions. The quantity and quality of the isolated DNA was assessed using a Nanodrop‐100 Spectrophotometer (Thermo Scientific, Wilmington, DE, USA) and purified DNA samples were sequenced by Life Sequencing SL (València, Spain). On one hand, the hypervariable V3‐V4 regions of the 16S rRNA gene was amplified as described by Klindworth et al. (2013) and sequenced on the high‐throughput NextSeq 500 (Illumina) platform. Greengenes database was used for the taxonomic analysis. The statistical analysis was carried out with Calypso web tool (version 8.84; http://cgenome.net). The statistical comparison of the relative abundances between locations at the genus level was calculated through One‐Way Anova test (Tables 1 and S1). Richness and Shannon index box plots, PCoA, relative abundances clustering and rarefaction curve were also constructed with Calypso.
On the other hand, shotgun metagenomic sequencing was performed on the NextSeq500 Illumina platform, with paired‐end sequences and reads of 150 base pairs. The obtained sequences were filtered by using ‘BBtools’ version 37.28 (https://jgi.doe.gov/data-and-tools/bbtools/) in order to avoid ends holding quality values under the Q20 standards. Lectures coming from human contamination were also dismissed by mapping them against the reference human genome (GRCh37d5) version 0.7.15. Assembly was carried out with ‘SPAdes’ (Bankevich et al., 2012) version 3.9. ORFs prediction was carried out by ‘MegaGeneMArk’ (Zhu et al., 2010) version 3.38 and rRNA prediction, by ‘RNAmmer’ (Lagesen et al., 2007) version 1.2. Functional annotation of the predicted CDS was carried out with BLAST2go (Conesa et al., 2005) version 4.1.9.
The Clustergrammer on‐line software (Fernández et al., 2017) was used for the functional analysis heatmap construction, by using a correlation type distance and average linkage.
Isolation and identification of bacterial strains
Three different growth media were used for this study: Lysogenic Broth (LB, composition in g l−1: 10 tryptone, 10 NaCl, 5.0 yeast extract, 15 agar); Reasoner's 2A agar (R2A, composition in g l−1: peptone 0.5, casaminoacids 0.5, yeast extract 0.5, dextrose 0.5, soluble starch 0.5, K2HPO4 0.3, MgSO4 0.05, sodium pyruvate 0.3, 15 agar); and Marine Agar (composition in g l−1: peptone 5.0, yeast extract 1.0, ferric citrate 0.1, NaCl 19.45, MgCl2 5.9, Na2SO4 3.24, CaCl2 1.8, KCl 0.55, NaHCO3 0.16, KBr 0.08, SrCl2 0.034, H3BO3 0.022, Na4O4Si 0.004, NaF 0.024, NH4NO3 0.0016, Na2HPO4 0.008, 15 agar). The scraped rock samples were homogenized in the Falcon tube by vigorously mixing with a vortex, and serial dilutions were cultured on the different media and incubated at room temperature for 7 days. Marine water samples were also cultured in the same conditions. After 1 week of incubation, individual colonies were selected based on colony pigmentation and isolated by independent re‐streaking on fresh medium. Pure cultures were then cryo‐preserved at −80°C in 20% glycerol (vol:vol) until required.
Colony PCR and, were needed, DNA extracts of each of the isolated strains, were used for taxonomic identification through 16S rRNA gene sequencing using universal primers 28F (5′‐GAG TTT GAT CNT GGC TCA G‐3′) and 519R (5′‐GTN TTA CNG CGG CKG CTG‐3′). Colony PCR was performed with an initial step of incubation at 95°C for 5 min to lyse cells followed by PCR amplification (30 cycles of 30 s at 95°C, 30 s at 54°C, 30 s at 72°C, followed by 10 min at 72°C). The DNA extraction was done following the Latorre et al. (1986) protocol. Amplifications were verified by electrophoresis in a 0.8% agarose gel and then amplicons were precipitated overnight in isopropanol 1:1 (vol:vol) and potassium acetate 1:10 (vol:vol; 3 M, pH 5). DNA pellets were washed with 70% ethanol and resuspended in 30 μl Milli‐Q water. BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Carlsbad, CA, USA) was used to tag amplicons, which were sequenced with the Sanger method by the Sequencing Service (SCSIE) of the University of Valencia (Spain). All sequences were manually edited with Pregap4 (Staden Package, 2002) to eliminate low‐quality base calls, and final sequences were compared by EzBioCloud 16S tool (https://sourceforge.net/projects/staden/).
Antioxidant activity
Hydrogen peroxide assay
The collection of isolates was initially screened for antioxidant activity by applying oxidative stress to the isolated colonies through the addition of hydrogen peroxide (H2O2) to the growth medium. In order to do so, isolates were grown on solid media for 4 days or until reaching enough biomass. Then, the optical density at 600 nm (OD600) was measured, adjusted to a value of 1, and serial dilutions prepared up to seven times fold. Two microlitres of each dilution were placed on a LB or Marine Agar place, to which 1 mM H2O2 had been previously added. The plates were incubated at room temperature and in the dark to avoid degradation of the H2O2, and results were recorded after two, four and six days. Two strains were used as controls for the assay: PS1 (Planomicrobium glaciei 423, 97.38% ID) and Escherichia coli JM109 as a positive and negative control for antioxidant activity respectively. Planomicrobium glaciei is a pigmented microorganism whose antioxidant activity has previously been reported in vivo using a Caenorhabditis elegans model (Tanner et al., 2019).
DPPH assay
Since the H2O2 assay can result in false‐positive results due to catalase activity, a second assay using 2,2‐diphenyl‐1‐picrylhydrazyl (DPPH) was performed to dismiss false positives in the H2O2 assay and to confirm the antioxidant activity of the selected strains (the ones with the best antioxidant activity according to the previous assay). Pigments were extracted from the isolates based on the protocols described by Brand‐Williams et al. (1995), von Gadow et al. (1997) and Su et al. (2015), with the modifications suggested by Sharma and Bhat (2009). Briefly, the isolates were grown overnight in liquid LB medium and OD600 was measured and normalized at a value of 1.2. Cells were then harvested by centrifugation at 11,300 g for 3.5 min, and the pellets resuspended in 500 μL of methanol, vigorously vortexed and sonicated for 5 min (Ultrasonic bath XUBA1, Grant Instruments, Royston, UK). The supernatant was collected after centrifugation at 11,300 g for 3 min and kept in the dark until the assay was performed. The extraction was repeated as described until a colourless pellet was obtained.
For the DPPH assay, 600 μl of the extract in methanol were mixed with 400 μl of DPPH solution (50 μM in methanol) and incubated for 30 min in the dark. The negative control sample consisted of DPPH mixed with methanol. Absorbance was measured at 517 nm (Ultrospec 200 UV/V Visible Spectrophotometer, Pharmacia Biotech, Piscataway Township, NJ, USA).
A standard curve with a control antioxidant, ascorbic acid (vitamin C) was performed at 10, 5, 1, 0.5, 0.1, 0.05 and 0.01 μg ml−1 concentrations in methanol. The detection threshold was established at 0.5 μg ml−1 of vitamin C, as lower concentrations of vitamin C did not change DPPH absorbance (data not shown).
DPPH scavenging ability was quantified by measuring the decrease in the absorbance of this compound at 517 nm, and the percentage of scavenged DPPH was calculated using the following formula:
In vivo oxidative stress assays with C. elegans
Wild‐type C. elegans strain N2 (Bristol, UK) was routinely propagated at 20°C on Nematode Growth Medium (NGM) plates supplemented with E. coli strain OP50 as the regular food source.
Nematodes were synchronized by isolating eggs from gravid adults at 20°C. Synchronization was performed on NGM plates with different treatments: E. coli OP50 was supplied as a negative control; E. coli OP50 plus vitamin C (vitC) at 10 μg ml−1 as a positive control; and, finally, E. coli OP50 plus one of the selected isolates was used in order to test the effect of administrating the selected strains. Duplicates were performed for every condition. Bacterial strains were grown overnight in liquid LB medium at 28°C and 11,300 g. Then, OD600 was adjusted to 30 and 50 μl of the bacterial suspension were added to the plates.
The synchronized worms were incubated for 3 days on the previously described plates, until reaching young adult stage. Then, young adult worms were selected for each treatment (n = 50) and incubated at 20°C on the corresponding treatment, until reaching 5‐day adult stage. The selected worms were then transferred to plates containing basal medium supplemented with 2 mM H2O2 and incubated for 5 h at 20°C. After incubation, survival rates for each condition (negative control, positive control and bacteria‐fed worms) were recorded by manually counting the number of living versus dead worms.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
MP conceived the work. MP, KT and EMM collected the samples. EMM, KT and ÀVV performed the culture‐based characterization, and KT carried out the bioinformatic analysis. All authors (MP, KT, ÀVV, EMM and JP) analysed the results, wrote and approved the manuscript.
Supporting information
Fig. S1. Rarefaction curve at OTU level.
Fig. S2. Main bacterial (A), archaeal (B) and eukaryotic (C) groups identified in the sample obtained from Vinaròs and analysed through metagenomics sequencing.
Fig. S3. Main bacterial (A), archaeal (B) and eukaryotic (C) groups identified in the sample obtained from Cullera and analysed through metagenomics sequencing.
Fig. S4. Main bacterial (A), archaeal (B) and eukaryotic (C) groups identified in the sample obtained from Dénia and analysed through metagenomics sequencing.
Table S1. Top 30 most significant genera and P‐values for the One‐Way ANOVA statistical analysis of their distributions among the three sampled locations. Global P‐values and P‐values for the comparison by pairs is shown. Significant results are marked by an asterisk.
Table S2. List of the strains identified in the collection, with the closest type strain, accession number, ID percentage and the GenBank accession number for the 16S rRNA sequences. The identification code of the strains corresponds to the location from which it was isolated (V: Vinaròs, C: Cullera, D: Dènia), the sample type (R: rock, M: marine water) and a number.
Acknowledgements
We thank Adriel Latorre and Darwin Bioprospecting Excellence S.L. (Valencia, Spain) for their assistance with the bioinformatic analysis. We thank ADM‐Biopolis S.L. for granting us access to their laboratory and materials for the C. elegans assays.
Microbial Biotechnology (2019) 12(6), 1359–1370
Funding information
Financial support from the Spanish Government (Grant Helios, Reference: BIO2015‐66960‐C3‐1‐R co‐financed by FEDER funds and Ministerio de Ciencia, Innovación y Universidades) and from the European CSA on biological standardization BIOROBOOST (EU grant number 820699) is acknowledged. EMM is funded with a Formación de Profesorado Universitario (FPU) grant from the Spanish Government (Ministerio de Ciencia, Innovación y Universidades), with reference FPU17/04184. KT is a recipient of a Doctorado Industrial fellowship from the Ministerio de Ciencia, Innovación y Universidades (Spain), with reference DI‐16‐08976. ÀVV is funded with a Formación de Profesorado Universitario (FPU) grant from the Spanish Government (Ministerio de Ciencia, Innovación y Universidades), with reference FPU18/02578.
References
- Albuquerque, L. , Simoes, C. , Nobre, M.F. , Pino, N.M. , Battista, J.R. , Silva, M.T. , et al (2005) Truepera radiovictrix gen. nov., sp. nov., a new radiation resistant species and the proposal of Trueperaceae fam. nov. FEMS Microbiol Lett 247: 161–169. [DOI] [PubMed] [Google Scholar]
- Alex, A. , Vasconcelos, V. , Tamagnini, P. , Santos, A. , and Antunes, A. (2012) Unusual symbiotic cyanobacteria association in the genetically diverse intertidal marine sponge Hymeniacidon perlevis (demospongiae, halichondrida). PLoS ONE 7: e51834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso‐Gutiérrez, J. , Figueras, A. , Albaigés, J. , Jiménez, N. , Viñas, M. , Solanas, A.M. , and Novoa, B. (2009) Bacterial communities from shoreline environments (Costa da Morte, northwestern Spain) affected by the prestige oil spill. Appl Environ Microbiol 75: 3407–3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich, A. , Nurk, S. , Antipov, D. , Gurevich, A.A. , Dvorkin, M. , Kulikov, A.S. , et al (2012) SPAdes: a new genome assembly algorithm and its applications to single‐cell sequencing. J Comput Biol 19: 455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon, C.S. , McClements, D.J. , Weiss, J. , and Decker, E.A. (2010) Factors influencing the chemical stability of carotenoids in foods. Crit Rev Food Sci Nutr 50: 515–532. [DOI] [PubMed] [Google Scholar]
- Brandes, M. , Albach, D.C. , Vogt, J.C. , Mayland-Quellhorst, E. , Mendieta-Leiva, G. , Golubic, S. , et al (2015) Supratidal extremophiles-Cyanobacterial diversity in the rock pools of the Croatian Adria. Microb Ecol 70: 876–888. [DOI] [PubMed] [Google Scholar]
- Brand‐Williams, W. , Cuvelier, M. , and Berset, C. (1995) Use of a free radical method to evaluate antioxidant activity. Lebensmittel‐Wissenschaft Technol 28: 25–30. [Google Scholar]
- Brito, A. , Ramos, V. , Mota, R. , Lima, S. , Santos, A. , Vieira, J. , et al (2017) Description of new genera and species of marine Cyanobacteria from the Portuguese Atlantic coast. Mol Phylogenet Evol 111: 18–34. [DOI] [PubMed] [Google Scholar]
- Burns, B.P. , Goh, F. , Allen, M. , and Neilan, B.A. (2004) Microbial diversity of extant stromatolites in the hypersaline marine environment of Shark Bay, Australia. Environ Microbiol 6: 1096–1101. [DOI] [PubMed] [Google Scholar]
- Chan, B.K. , Chan, W.K. , and Walker, G. (2003) Patterns of biofilm succession on a sheltered rocky shore in Hong Kong. Biofouling. 19: 371–380. [DOI] [PubMed] [Google Scholar]
- Chien, A. , Edgar, D.B. , and Trela, J.M. (1976) Deoxyribonucleic acid polymerase from the extreme thermophile Thermus aquaticus . J Bacteriol 127: 1550–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa, A. , Götz, S. , García‐Gómez, J.M. , Terol, J. , Talón, M. , and Robles, M. (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676. [DOI] [PubMed] [Google Scholar]
- Dorado‐Morales, P. , Vilanova, C. , Peretó, J. , Codoñer, F.M. , Ramón, D. , and Porcar, M. (2015) A highly diverse, desert‐like microbial biocenosis on solar panels in a Mediterranean city. Sci Rep‐UK 6: 29235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández, N.F. , Gundersen, G.W. , Rahman, A. , Grimes, M.L. , Rikova, K. , Hornbeck, P. , and Ma'ayan, A. (2017) Clustergrammer, a web‐based heatmap visualization and analysis tool for high‐dimensional biological data. Scientific Data 4: 170151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuciños, P. , González, R. , Atanes, E. , Sestelo, A.B. , Pérez‐Guerra, N. , Pastrana, L. , et al (2012) Lipases and esterases from extremophiles: overview and case example of the production and purification of an esterase from Thermus thermophilus HB27. Methods Mol Biol 861: 239–266. [DOI] [PubMed] [Google Scholar]
- von Gadow, A. , Elizabeth, J. , and Hansmann, C. (1997) Comparison of the antioxidant activity of aspalathin with that of other plant phenols of rooibos tea (Aspalathus linearis), r‐tocopherol, BHT, and BHA. J Agric Food Chem 45: 632–638. [Google Scholar]
- Ivanova, N. , Rohde, C. , Munk, C. , Nolan, M. , Lucas, S. , Rio, T.G. , et al (2011) Complete genome sequence of Truepera radiovictrix . Stan Genomic Sci 4: 91–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurado, V. , Miller, A. , Alias‐Villegas, C. , Laiz, L. , and Saiz‐Jimenez, C. (2012) Rubrobacter bracarensis sp. nov., a novel member of the genus Rubrobacter isolated from a biodeteriorated monument. Syst Appl Microbiol 35: 306–309. [DOI] [PubMed] [Google Scholar]
- Klindworth, A. , Pruesse, E. , Schweer, T. , Peplies, J. , Quast, C. , Horn, M. , et al (2013) Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next generation sequencing‐based diversity studies. Nucleic Acids Res 41: e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, V.B.N. , Kampe, B. , Rösch, P. , and Popp, J. (2015) Characterization of carotenoids in soil bacteria and investigation of their photo degradation by UVA radiation via resonance Raman spectroscopy. Analyst 140: 4584–4593. [DOI] [PubMed] [Google Scholar]
- Lafi, F.F. , Ramírez‐Prado, J.S. , Alam, I. , Bajic, V.B. , Hirt, H. , and Saad, M.M. (2017) Draft genome sequence of plant growth‐promoting Micrococcus luteus strain K39 isolated from Cyperus conglomeratus in Saudi Arabia. Genome Announc 5: pii: e01520‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagesen, K. , Hallin, P. F. , Rødland, E. , Stærfeld, H.H. , Rognes, T. , and Ussery, D.W. (2007) RNAmmer: consistent annotation of rRNA genes in genomic sequences. Nucleic Acids Res 35: 3100–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenheder, S. , and Ragnarsson, H. (2007) The role of environmental and spatial factors for the composition of aquatic bacterial communities. Ecology 88: 2154–2161. [DOI] [PubMed] [Google Scholar]
- Latorre, A. , Moya, A. , and Ayala, F. (1986) Evolution of mitochondrial DNA in Drosophila subobscura . P Natl Acad Sci 83: 8649–8653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lourdes Moreno, M. , Pérez, D. , García, M.T. , and Mellado, E. (2013) Halophilic bacteria as a source of novel hydrolytic enzymes. Life (Basel) 3: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda, Y. , Takeuchi, K. , Obase, K. , and Ito, S. (2015) Spatial distribution and genetic structure of Cenococcum geophilum in coastal pine forests in Japan. FEMS Microbiol Ecol 91:fiv108. [DOI] [PubMed] [Google Scholar]
- Mitra, S. , Sana, B. , and Mukherjee, J. (2014) Ecological roles and biotechnological applications of marine and intertidal microbial biofilms. Adv Biochem Eng Biotechnol 146: 163–205. [DOI] [PubMed] [Google Scholar]
- Nishimura, H. , and Sako, Y. (2009) Purification and characterization of the oxygen‐thermostable hydrogenase from the aerobic hyperthermophilic archaeon Aeropyrum camini . J Biosci Bioeng 108: 299–303. [DOI] [PubMed] [Google Scholar]
- Obase, K. , Lee, J.K. , Lee, S.K. , Lee, S.Y. , and Chun, K.W. (2010) Variation in sodium chloride resistance of Cenococcum geophilum and Suillus granulatus isolates in liquid culture. Mycobiology 38: 225–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawar, R. , Mohandass, C. , Sivaperumal, E. , Sabu, E. , Rajasabapathy, R. , and Jagtap, T. (2015) Epiphytic marine pigmented bacteria: a prospective source of natural antioxidants. Braz J Microbiol 46: 29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pentecost, A. (2014) Distribution and ecology of cyanobacteria in the rocky littoral of an English lake district water body, devoke water. Life (Basel) 4: 1026–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinedo, S. , García, M. , Satta, M. P. , and de Torres,, M. , Ballesteros, E. (2007) Rocky-shore communities as indicators of water quality: a case study in the Northwestern Mediterranean. Mar Pollut Bull 55: 126–135. [DOI] [PubMed] [Google Scholar]
- Raddadi, N. , Cherif, A. , Daffonchio, D. , Neifar, M. , and Fava, F. (2015) Biotechnological applications of extremophiles, extremozymes and extremolytes. Appl Microbiol Biotechnol 99: 7907. [DOI] [PubMed] [Google Scholar]
- Sandmann, G. (2015) Carotenoids of biotechnological importance. Adv Biochem Eng Biot 148: 449–467. [DOI] [PubMed] [Google Scholar]
- Sharma, O.P. , and Bhat, T.K. (2009) DPPH antioxidant assay revisited. Food Chem 113: 1202–1205. [Google Scholar]
- Shearer, C.A. , Raja, H.A. , Miller, A.N. , Nelson, P. , Tanaka, K. , Hirayama, K. , et al (2009) The molecular phylogeny of freshwater Dothideomycetes . Stud Mycol 64: 145–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo, K. , and Misawa, N. (2014) New and rare carotenoids isolated from marine bacteria and their antioxidant activities. Mar Drugs 12: 1690–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirisena, K.A. , Ramírez, S. , Steele, A. , and Glamodija, M. (2018) Microbial diversity of hypersaline sediments from lake Lucero playa in white sands national monument, New Mexico, USA. Microb Ecol 76: 404–418. [DOI] [PubMed] [Google Scholar]
- Spatafora, J.W. , Owensby, C.A. , Douhan, G.W. , Boehm, E.W. , and Schoch, C.L. (2012) Phylogenetic placement of the ectomycorrhizal genus Cenococcum in Gloniaceae (Dothideomycetes). Mycologia 104: 758–765. [DOI] [PubMed] [Google Scholar]
- Sterflinger, K. , De Baere, R. , de Hoog, G.S. , De Wachter, R. , Krumbein, W.E. , and Haase, G. (1997) Coniosporium perforans and C. apollinis, two new rock‐inhabiting fungi isolated from marble in the Sanctuary of Delos (Cyclades, Greece). Antonie Van Leeuwenhoek 72: 349–363. [DOI] [PubMed] [Google Scholar]
- Su, J. , Wang, T. , Li, Y.‐Y. , Li, J. , Zhang, Y. , Wang, Y. , et al (2015) Antioxidant properties of wine lactic acid bacteria: Oenococcus oeni . Appl Microbiol Biotechnol 99: 5189–5202. [DOI] [PubMed] [Google Scholar]
- Tanner, K. , Vilanova, C. , and Porcar, M. (2017) Bioprospecting challenges in unusual environments. Microb Biotechnol 10: 671–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner, K. , Martí, J.M. , Belliure, J. , Fernández‐Méndez, M. , Molina‐Menor, E. , Peretó, J. , and Porcar, M. (2018) Polar solar panels: arctic and Antarctic microbiomes display similar taxonomic profiles. Env Microbiol Rep 10: 75–79. [DOI] [PubMed] [Google Scholar]
- Tanner, K. , Martorell, P. , Genovés, S. , Ramón, D. , Zacarías, L. , Rodrigo, M.J. , et al (2019) Bioprospecting the solar panel microbiome: high‐throughput screening for antioxidant bacteria in a Caenorhabditis elegans model. Front Microbiol 10: 986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, B. , and Hua, Y. (2010) Carotenoid biosynthesis in extremophilic Deinoccocus‐Thermus bacteria. Trends Microbiol 18: 512–520. [DOI] [PubMed] [Google Scholar]
- Yoon, J. , Oku, N. , Park, S. , Kasai, H. , and Yokota, A. (2011) Porifericola rhodea gen. nov., sp. nov., a new member of the phylum Bacteroidetes isolated by the bait‐streaked agar technique. Antonie Van Leeuwenhoek 100: 145–153. [DOI] [PubMed] [Google Scholar]
- Yoon, J. , Oku, N. , Park, S. , Katsuta, A. , and Kasai, H. (2012) Tunicatimonas pelagia gen. nov., sp. nov., a novel representative of the family Flammeovirgaceae isolated from a sea anemone by the differential growth screening method. Antonie Van Leeuwenhoek 101: 133–140. [DOI] [PubMed] [Google Scholar]
- Yu, C.H. , Lu, C. K. , Su, H.M. , Chiang, T.Y. , Hwang, C.C. , Liu, T. , and Chen, Y.M. (2015) Draft genome of Myxosarcina sp. strain GI1, a baeocytous cyanobacterium associated with the marine sponge Terpios hoshinota . Stand Genomic Sci 10: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, W. , Lomsadze, A. , and Borodovsky, M. (2010) Ab initio gene identification in metagenomic sequences. Nucleic Acids Res 38: e132. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Rarefaction curve at OTU level.
Fig. S2. Main bacterial (A), archaeal (B) and eukaryotic (C) groups identified in the sample obtained from Vinaròs and analysed through metagenomics sequencing.
Fig. S3. Main bacterial (A), archaeal (B) and eukaryotic (C) groups identified in the sample obtained from Cullera and analysed through metagenomics sequencing.
Fig. S4. Main bacterial (A), archaeal (B) and eukaryotic (C) groups identified in the sample obtained from Dénia and analysed through metagenomics sequencing.
Table S1. Top 30 most significant genera and P‐values for the One‐Way ANOVA statistical analysis of their distributions among the three sampled locations. Global P‐values and P‐values for the comparison by pairs is shown. Significant results are marked by an asterisk.
Table S2. List of the strains identified in the collection, with the closest type strain, accession number, ID percentage and the GenBank accession number for the 16S rRNA sequences. The identification code of the strains corresponds to the location from which it was isolated (V: Vinaròs, C: Cullera, D: Dènia), the sample type (R: rock, M: marine water) and a number.


