Summary
Pichia pastoris KM71H (MutS) is an efficient producer of hard‐to‐express proteins such as the membrane protein P‐glycoprotein (Pgp), an ATP‐powered efflux pump which is expressed properly, but at very low concentration, using the conventional induction strategy. Evaluation of different induction strategies indicated that it was possible to increase Pgp expression by inducing the culture with 20% media containing 2.5% methanol. By quantifying methanol, formaldehyde, hydrogen peroxide and formate, and by measuring alcohol oxidase, catalase, formaldehyde dehydrogenase, formate dehydrogenase, malate dehydrogenase, isocitrate dehydrogenase and α‐ketoglutarate dehydrogenases, it was possible to correlate Pgp expression to the induction strategy. Inducing the culture by adding methanol with fresh media was associated with decreases in formaldehyde and hydrogen peroxide, and increases in formaldehyde dehydrogenase, formate dehydrogenase, isocitrate dehydrogenase and α‐ketoglutarate dehydrogenases. At these conditions, Pgp expression was 1400‐fold higher, an indication that Pgp expression is affected by increases in formaldehyde and hydrogen peroxide. It is possible that Pgp is responsible for this behaviour, since the increased metabolite concentrations and decreased enzymatic activities were not observed when parental Pichia was subjected to the same growth conditions. This report adds information on methanol metabolism during expression of Pgp from P. pastoris MutS strain and suggests an expression procedure for hard‐to‐express proteins from P. pastoris.
Introduction
Pichia pastoris is an efficient producer of recombinant membrane proteins for structural and functional studies (Bill et al., 2011; Lee et al., 2016; Habeck et al., 2017). Unlike prokaryotes, this eukaryotic organism can produce correctly folded proteins with all the required disulfide bond and post‐translational modifications (Wang et al., 2017), and the transgene is stably integrated in the host genome ensuring production stability (Athmaram et al., 2013). Pichia pastoris contains an efficient methanol‐inducible promoter of the alcohol oxidase gene (AOX) that supports high expression of recombinant proteins (Liu et al., 2019). In addition, this yeast can grow to high biomass levels on both complex and minimal media (Liu et al., 2016; Matthews et al., 2018), and does not contain potentially oncogenic or viral nucleic acids which are present in mammalian cells, or cell wall pyrogens present in E. coli (Ciofalo et al., 2006; Noseda et al., 2013). Over one thousand proteins have been successfully expressed in P. pastoris with yields as high as 22 g l−1 for intracellular production of recombinant hydroxynitrile lyase and as high as 14.8 g l−1 for extracellular production of recombinant gelatines (Werten et al., 1999; Ren and Yuan, 2005). Three phenotypes of P. pastoris are being used: Mut+, where both AOX1 (encodes alcohol oxidase I) and AOX2 (encodes alcohol oxidase II) genes are intact; MutS, a mutant with an inactivated AOX1 gene; and Mut−, where both AOX1 and AOX2 genes are disrupted (Juturu and Wu, 2018). The Mut+ and MutS strain, able to metabolize methanol as the only carbon source, are the most commonly used strains (Looser et al., 2015).
Unlike other recombinant proteins highly expressed in P. pastoris by the conventional strategy (Kastilan et al., 2017; Bußwinkel et al., 2018; Moser et al., 2018), the membrane protein mouse P‐glycoprotein (Pgp) was not expressed well in bioreactors, unless the growth media was replaced prior to the induction phase (replacement strategy). Pgp is a 170 kDa ABC transporter and functions as an ATP hydrolysis‐driven efflux pump to rid the cell of structurally unrelated diverse hydrophobic amphipathic compounds, which could lead to multidrug resistance (MDR) and failure of chemotherapy (Esser et al., 2017; De Vera et al., 2019). The replacement strategy is clearly not a practical approach to obtain proteins in bioreactors, and an improved production strategy needs to be developed. In the current study, three methanol feeding strategies were investigated: a conventional strategy based on direct methanol addition to the growing culture (Sun et al., 2018); a replacement strategy, where media was replaced before the induction phase; and a supplemented strategy, where methanol supplemented with fresh media was added at induction time. The three induction approaches were followed by measuring methanol and the associated metabolites – formaldehyde, hydrogen peroxide and formate – and by measuring the activities of alcohol oxidase (AOX), catalase (CAT), NAD+‐dependent formaldehyde dehydrogenase (FLD) and NAD+‐dependent formate dehydrogenase (FDH). NAD+‐dependent enzymes involved in TCA cycle, malate dehydrogenase (MDH), isocitrate dehydrogenase (IDH) and α‐ketoglutarate dehydrogenases (α‐KGDHs) were also analysed. The study provides insight not only on Pgp expression in P. pastoris, but also on production related to cell growth, methanol consumption, metabolite concentrations and enzymatic activity related to methanol metabolism and energy regeneration.
Results
The common approach for expressing recombinant proteins from P. pastoris is by implementing a conventional two‐phase process. In the first phase, biomass was accumulated by growing the cells on glycerol, and in the second phase, the expression of the desired protein was induced by adding methanol (Viña?Gonzalez, et al., 2018). However, at these conditions Pgp was minimally expressed. Significantly higher expression was achieved by implementing a replacement strategy, when methanol was added to a culture that was first grown on glycerol and then centrifuged and resuspended in fresh media (Esser et al., 2017). Although this media replacement approach is a workable procedure, it is not a practical production strategy for a culture grown in a bioreactor, and therefore, an understanding of what prevents Pgp expression was needed. In this work, three production strategies were compared. In the conventional strategy, Pgp was traditionally expressed by adding methanol to the growing cells. In the replacement strategy, methanol was added to culture after media replacement. Finally, in the supplemented strategy, methanol was added together with fresh media to the culture that was grown initially on glycerol.
Biomass accumulation after induction
Methanol induction was initiated at a biomass concentration of 4 g l−1 dry cell weight (20 OD600), Fig. 1A. The supplemented strategy supported the cells to grow to an OD600 of 28 for 36 h of induction and kept constant at an average OD600 of 28 ± 0.35, which is 29% higher than the culture induced with the conventional strategy, and 11% higher than the level achieved using the replacement strategy (P < 0.001, n = 3). The culture induced using the conventional strategy showed a peak biomass of 24 OD600 after 24 h, and the culture induced using the replacement strategy showed a biomass concentration of 27 OD600 after 36 h. At the same time, a significant decrease in biomass was observed during the last 36 h of the conventional and replacement strategies (P < 0.001, n = 3).
Figure 1.
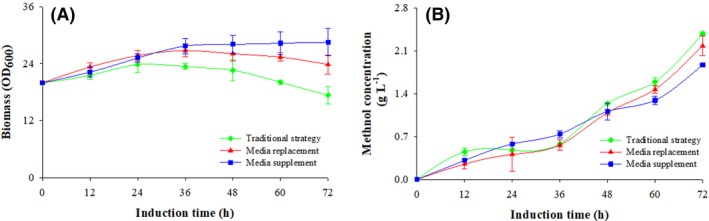
Biomass and methanol concentration in response to different induction strategies. Conventional strategy (filled green diamond), replacement strategy (filled red triangle) and supplemented strategy (filled blue circle). Time zero indicates the initiation of the methanol induction phase.
(A) Optical cell density (OD 600); (B) methanol concentration.
Methanol utilization
Figure 1B shows the methanol concentrations in the different cultures. Approximately 12 g methanol was added throughout the process, and no significant differences in the methanol concentration were observed at the three different strategies (P > 0.1, n = 3). When the supplemented strategy was used, the methanol concentration increased gradually, and after 72 h, it was 1.87 g l−1, which is 0.51 and 0.31 g l−1 lower than the concentration with the conventional and the replacement strategy respectively (P < 0.05, n = 3). There were no differences in methanol consumption between the different processes (P > 0.1, n = 3), and approximately 67%, 70% and 74% of the methanol added to the cultures was consumed with the conventional, replacement and the supplemented strategies respectively.
Pgp expression
Volumetric production of Pgp at the three induction strategies is shown in Fig. 2A. The production was negligible when the conventional strategy was implemented (P < 0.001, n = 3), and only 0.15 mg l−1 of Pgp was accumulated throughout the 72 h of induction. In comparison, 47.4 mg l−1 of Pgp was produced after 24 h of induction using the replacement strategy; however, the concentration declined to 32.5 mg l−1 after 72 h. When the induction was conducted by the supplemented strategy, 49.2 mg l−1 of the enzymatically active Pgp was produced after 36 h, Fig. 2B. ATPase activity was negligible when the culture was induced using the conventional strategy (P < 0.001, n = 3), and 0.021 × 102 U l−1 was observed throughout the 72 h. When the replacement strategy was used, the enzymatic activity reached 8.1 × 102 U l−1 after 24 h but declined to 5.5 × 102 U l−1 after 72 h of induction. However, when the supplemented strategy was implemented the ATPase activity reached 8.4 × 102 U l−1 after 36 h of induction, and the enzymatic activity remained constant throughout the 72 h of induction process.
Figure 2.
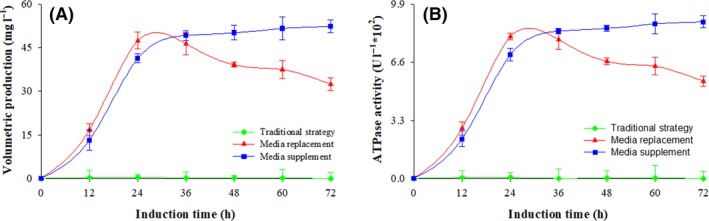
Volumetric Pgp production and ATPase activity at three fermentation processes. Conventional strategy (filled green diamond), replacement strategy (filled red triangle) and supplemented strategy (filled blue circle). Time zero indicates the initiation of the methanol induction phase.
(A) Volumetric Pgp production; (B) ATPase activity.
Intracellular metabolite concentration and enzymatic activity
To understand the reasons for the different Pgp expression levels with the three induction processes, methanol metabolites and activities of enzymes associated with methanol metabolism and energy regeneration were analysed. The results are shown in Figs 3, 4, 5 and Table 1. Compared with the replacement and the supplemented strategies, the conventional strategy was associated with higher intracellular concentrations of formaldehyde and hydrogen peroxide, lower activity of FLD, FDH, IDH and α‐KGDHs, and higher CAT activity. No difference in the AOX or MDH activities was observed (P > 0.1, n = 3).
Figure 3.
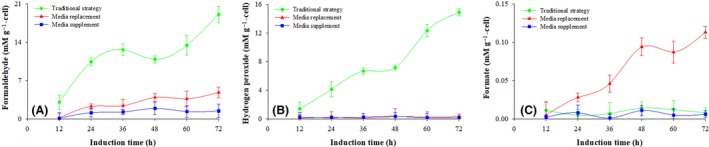
Concentrations of methanol metabolites in response to various induction strategies. Conventional strategy (filled green diamond), replacement strategy (filled red triangle) and supplemented strategy (filled blue circle). Time zero indicates the initiation of the methanol induction phase.
(A) Formaldehyde concentration; (B) hydrogen peroxide concentration; (C) formate concentration.
Figure 4.
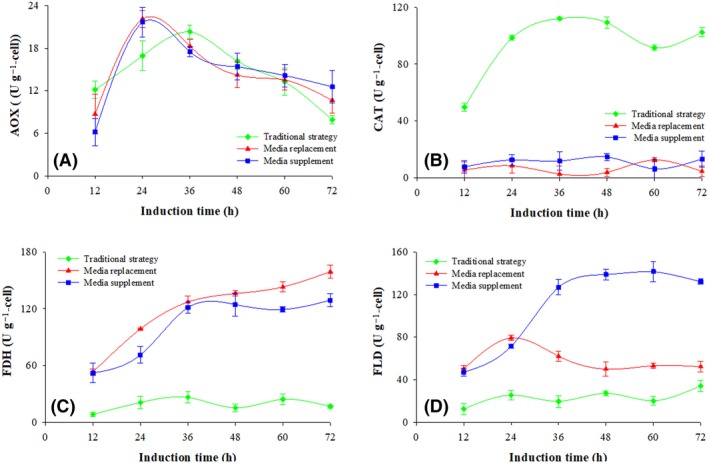
Specific activities of enzymes related to methanol metabolism at three fermentation processes. Conventional strategy (filled green diamond), replacement strategy (filled red triangle) and supplemented strategy (filled blue circle). Time zero indicates the initiation of the methanol induction phase.
(A) Alcohol oxidase (AOX); (B) catalase (CAT); (C) formaldehyde dehydrogenase (FLD); (D) formate dehydrogenase (FDH) .
Figure 5.
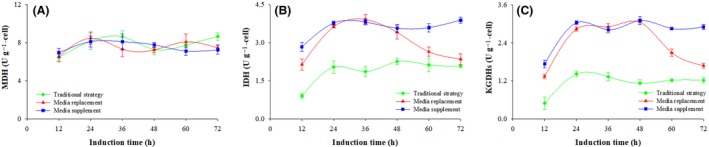
Specific activities of enzymes related to energy regeneration at three fermentation processes. Conventional strategy (filled green diamond), replacement strategy (filled red triangle) and supplemented strategy (filled blue circle). Time zero indicates the initiation of the methanol induction phase.
(A) Malate dehydrogenase (MDH); (B) isocitrate dehydrogenase (IDH); (C) α‐Ketoglutarate dehydrogenases (α‐KGDHs).
Table 1.
The peak value of methanol metabolites (mM g−1 dry cell) and enzymatic activities (U g−1 dry cell) during three processes. (h‐sampling time)
| Fermentation strategy | Conventional strategy | Replacement strategy | Supplemented strategy |
|---|---|---|---|
| Metabolites | |||
| Formaldehyde (CH2O) | 19.1 (72 h) | 4.8 (72 h) | 1.9 (48 h) |
| Hydrogen peroxide (H2O2) | 14.9 (72 h) | 0.3 (48 h) | 0.4 (48 h) |
| Formate (CH2O2) | 0.014 (48 h) | 0.11 (72 h) | 0.011 (48 h) |
| Enzymes | |||
| Alcohol oxidase (AOX) | 20.3 (36 h) | 22.1 (24 h) | 21.6 (24 h) |
| Catalase (CAT) | 112.1 (36 h) | 12.6 (60 h) | 14.7 (48 h) |
| Formaldehyde dehydrogenase (FLD) | 20.2 (60 h) | 53.2 (60 h) | 126.9 (36 h) |
| Formate dehydrogenase (FDH) | 26.5 (36 h) | 127.2 (36 h) | 124.4 (48 h) |
| Malate dehydrogenase (MDH) | 8.6 (72 h) | 8.5 (24 h) | 8.1 (24 h) |
| Isocitrate dehydrogenase (IDH) | 2.3 (48 h) | 3.9 (36 h) | 3.9 (72 h) |
| α‐Ketoglutarate dehydrogenases (α‐KGDHs) | 1.4 (24 h) | 3.1 (48 h) | 3.1 (48 h) |
The intracellular concentrations of methanol metabolites are shown in Fig. 3A–C. When the supplemented strategy was used, the formaldehyde, hydrogen peroxide and formate concentrations were 1.2 ± 0.6, 0.2 ± 0.07 and 0.006 ± 0.004 mM g−1 (DCW) respectively. When the replacement strategy was implemented, hydrogen peroxide and formaldehyde concentrations were similar (P > 0.1, n = 3), but formate concentration was 11‐fold higher and reached 0.1 mM g−1 after 72 h of induction. However, when the conventional strategy was implemented, formaldehyde concentration was ninefold higher (19.1 ± 1.5 mM g−1) and hydrogen peroxide was 33‐fold higher (14.9 ± 0.5 mM g−1) than the concentrations obtained when the supplemented strategy was used.
Measuring the activities of key enzymes associated with methanol utilization showed no difference in AOX activity among the three expression strategies. It increased in the first 24 h and then decreased slowly during the rest of the induction, with an average activity of 14.6 U g−1 (DCW) (P > 0.1, n = 3) (Fig. 4A). Catalase activity was higher with the conventional strategy; it reached 93.4 ± 22.9 U g−1, which is 90% higher than that observed with both the replacement and the supplemented strategies (Fig. 4B). The pattern of FDH activity was different; it was fivefold lower (102.9 ± 32.6 U g−1) with the conventional strategy than with both the replacement and the supplemented strategies (Fig. 4C). However, FLD activity was higher only with the supplemented strategy process and increased from 47.2 (at 12 h) to 126.9 U g−1 (at 36 h). At the same time, the average activities of the FLD at the last 24 h with the conventional and the replacement strategies were 80% and 62% respectively lower than with the supplemented strategy (Fig. 4D).
Measuring the activities of the key enzymes associated with the TCA cycle showed no difference related to MDH (P > 0.1, n = 3). The average activity was 7.5 ± 0.5 U g−1 for the three production strategies (Fig. 5A). On the other hand, the activities of IDH and α‐KGDHs with the conventional strategy were lower than those observed with the replacement and the supplemented strategies. The activities of IDH and α‐KGDHs were 1.9 ± 0.2 and 1.1 ± 0.1 U g−1, which are 48% and 58% lower than the activities observed with the supplemented strategy. Also, there was a significant decrease in both IDH and α‐KGDHs activities in the last 24 h of induction using the replacement strategy compared with the supplemented strategy (P < 0.001, n = 3). The activities of IDH and α‐KGDH decreased to 2.4 and 1.7 U g−1 at an induction time of 72 h, which are 39% and 42% respectively lower than those observed with supplemented strategy (Fig. 5B and C).
Performance of the control strains
To evaluate whether the observed methanol metabolism, energy regeneration and metabolic activities of the organism are related to the gene expressed, three control strains were utilized. One strain was without recombinant protein (KM71H); one strain expressed Pgp without ATPase (GS115‐ZA‐Pgp m); and one strain expressed the glycoside hydrolase LXYL‐P1‐2. The results are seen in Figures 6, 7, 8, S1 and S2.
Figure 6.
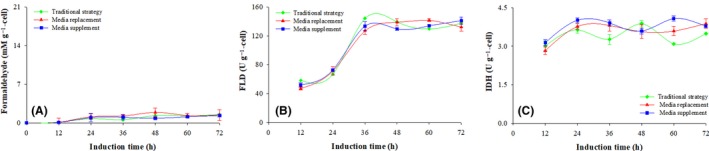
Formaldehyde concentration, FLD and IDH enzymatic activities of the parental strain KM71H. Conventional strategy (filled green diamond), replacement strategy (filled red triangle) and supplemented strategy (filled blue circle). Time zero indicates the initiation of the methanol induction phase.
(A) Formaldehyde concentration; (B) FLD activity; (C) IDH activity.
Figure 7.
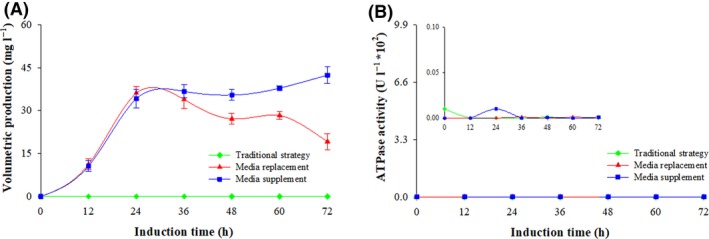
ATPase activity and volumetric production of Pgpm at three fermentation processes. Conventional strategy (filled green diamond), replacement strategy (filled red triangle) and supplemented strategy (filled blue circle). Time zero indicates the initiation of the methanol induction phase.
(A) ATPase activity; (B) volumetric Pgpm production.
Figure 8.
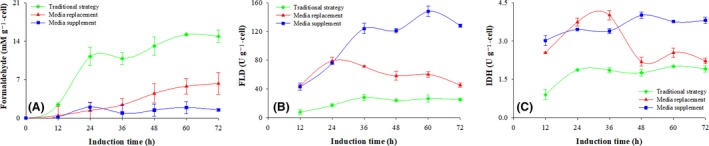
Formaldehyde concentration, FLD and IDH enzymatic activities of GS115‐ZA‐Pgp m at various processes. Conventional strategy (filled green diamond), replacement strategy (filled red triangle) and supplemented strategy (filled blue circle). Time zero indicates the initiation of the methanol induction phase.
(A) Formaldehyde concentration; (B) FLD activity; (C) IDH activity.
The metabolic activities of the parental strain (KM71H) without the recombinant gene under the three induction strategies are seen in Fig. 6. The intracellular formaldehyde concentration reached 0.9 ± 0.4 mM g−1 , and no significant differences were observed between the three induction strategies (P > 0.1, n = 3) (Fig. 6A). Key enzymes associated with methanol dissimilation at the different strategies also did not show major differences; FLD increased to a value of 138.9 ± 7.5 U g−1 in the first 36 h and stayed constant at an average activity of 136.0 ± 2.2 U g−1 (P > 0.1, n = 3) (Fig. 6B.) Isocitrate dehydrogenase activity was 3.4 ± 0.3 U g−1 with the conventional strategy and increased to 3.8 ± 0.3 U g−1 (Fig. 6C) when the supplemented strategy was implemented.
The protein expression and the metabolic activity of the Pichia strain expressing Pgp without ATPase activity (GS115‐ZA‐Pgp m) was like the behaviour of the strain expressing active Pgp (GS115‐ZA‐Pgp). As shown in Fig. 7A, the production of Pgpm was negligible with the conventional strategy, but over 42.3 mg ml−1 of Pgpm was produced after 72 h of induction using the supplemented strategy (Fig. 7B) and 17.1 mg ml−1 was produced when the induction was done using the replacement strategy. Regarding the methanol metabolites, when the conventional strategy was implemented the formaldehyde concentration reached 14.9 mM g−1; it was 3.5 ± 2.4 mM g−1 when the replacement strategy was implemented and 1.3 ± 0.7 mM g−1 when the supplemented strategy was used (Fig. 8A). The activities of the enzymes associated with methanol metabolism and energy regeneration showed a similar pattern to those obtained from the strain expressing active Pgp. As shown in Fig. 8B–C, the enzymatic activities of FLD and IDH in the ATPase free strain, GS115‐ZA‐Pgpm, were lower using the conventional strategy and higher when the supplemented strategy was implemented.
When the behaviour of the engineered P. pastoris strain expressing glycoside hydrolase LXYL‐P1‐2 was tested under the three induction strategies, no difference was observed regarding the expression of LXYL‐P1‐2. The production of recombinant LXYL‐P1‐2 and β‐glucosidase activity gradually increased to the maximum with the three different methanol induction strategies, and the formaldehyde concentrations and enzymatic activities of FLD and IDH were like those obtained from the parental strain KM71H (Fig. 6, S1 and S2).
Discussion
The quantities and qualities of recombinant proteins expressed in P. pastoris are affected by the methanol feeding strategy, and therefore, various strategies have been implemented for efficient expression processes (Ahmad et al., 2014; Looser et al., 2015; Mears et al., 2017). Since it was impossible to achieve efficient expression of Pgp in P. pastoris by using the traditional strategy where methanol is added directly into the culture, an alternative approach was required. The expression was significantly improved when the induction was done by methanol supplemented with fresh media (supplemented strategy) (Fig. 1). Compared with the traditional procedure (conventional strategy), twofold higher biomass was obtained, and 52.3 mg l−1 Pgp with 8.9 × 102 U l−1 was produced after 72 h of induction (Fig. 2).
It is known that methanol metabolites and energy generation affect P. pastoris growth and protein expression (Jahic et al., 2002; Plantz et al., 2006; Gao et al., 2015). In this work, careful evaluation of metabolites and enzyme activities showed that low Pgp expression was associated with higher concentrations of formaldehyde and hydrogen peroxide, together with lower enzymatic activities of formaldehyde dehydrogenase (FLD), formate dehydrogenase (FDH), isocitrate dehydrogenase (IDH) and α‐ketoglutarate dehydrogenases (α‐KGDHs) (Figs 3, 4, 5). At the same time, no significant difference in alcohol oxidase (AOX) activity was observed, as shown by the methanol concentration in the culture (Fig. 1B). It was therefore reasonable to assume that the lower expression of Pgp is not related to methanol oxidation. Instead, the increased concentration of formaldehyde and hydrogen peroxide, which are highly toxic (Theron et al., 2018), could be responsible for the limited cell growth and lower Pgp expression. It is likely that formaldehyde did not metabolize to nicotinamide adenine dinucleotide (NADH) and adenosine triphosphate because of a weakened or blocked metabolic flux caused by the over accumulation of formaldehyde.
In balanced growth, P. pastoris produces formaldehyde from methanol oxidation. As seen in Fig. 9, part of the formaldehyde is metabolized through a dissimilation pathway (pathway A), generating ATP that is required for cell growth and protein biosynthesis (Vanz et al., 2012; Yamada et al., 2019). Another part of the formaldehyde is metabolized through the assimilation pathway (pathway B), and the resulting ATP from the TCA cycle is mainly used to support biosynthetic purposes (Maghsoudi et al., 2012; Fam et al., 2017). In cases where the formaldehyde dissimilation pathway is weakened or blocked, the energy regeneration and cell functions are supported by the assimilation pathway (Hu et al., 2009; Gao et al., 2015). As summarized in Fig. 9, when the expression was induced by methanol supplemented with fresh media (supplemented strategy), higher activities of FLD, FDH (involved in pathway A) and IDH, and α‐KGDHs (involved in pathway B/TCA cycle) were observed (141.6, 128.9 and 3.9, 3.1 U g−1 respectively), together with lower concentrations of formaldehyde and hydrogen peroxide (1.2 ± 0.6 and 0.2 ± 0.07 mM g−1).
Figure 9.
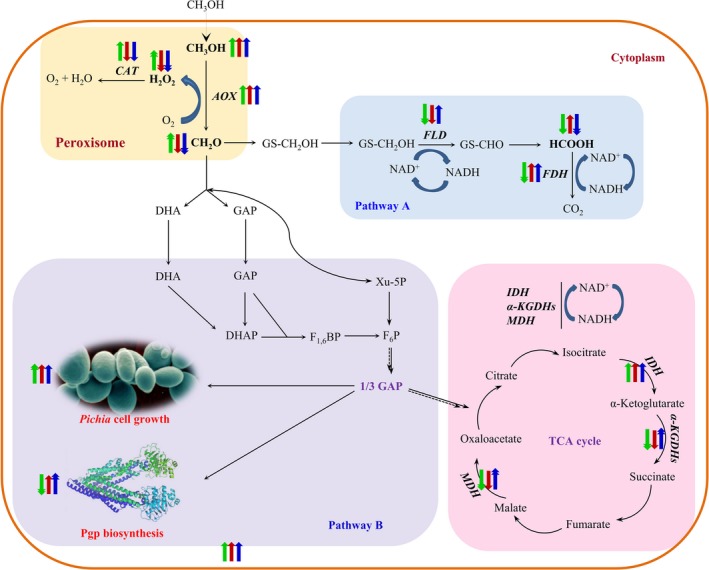
Simplified methanol metabolic networks and the performance of three fermentation processes. Pathway A is formaldehyde dissimilation pathway, and pathway B is formaldehyde assimilation pathway. The green arrow indicates conventional strategy, red arrow indicates replacement strategy, and blue arrow indicates supplemented strategy. The upward arrow indicates upregulated, and the downward arrow indicates downregulated. One arrow indicates small changes, and two arrows indicate strong changes. Abbreviations (enzymes): AOX, alcohol oxidase; CTA, catalase; FLD, NAD +‐dependent formaldehyde dehydrogenase; FDH, NAD +‐dependent formate dehydrogenase; IDH, NAD +‐dependent isocitrate dehydrogenase; MDH, NAD +‐dependent malate dehydrogenase; α‐KGDHs, NAD +‐dependent α‐ketoglutarate dehydrogenases.
To investigate the effect of the expressed proteins on the yeast metabolism, the parental strain KM71H, the strain expressing Pgp without ATPase activity (Pgp m) and the strain expressing LXYL‐P1‐2 were tested with the three growth strategies. When the conventional strategy was used, both KM71H and GS115‐3.5K‐P1‐2 generated low formaldehyde concentration and high enzymatic activity of FLD and IDH (Fig. 6, S2). At this condition, LXYL‐P1‐2 expression was high (Fig. S1); however, very little recombinant Pgpm was produced which was similar to the behaviour of KM71H‐ZA‐Pgp cultured at the same conditions (Fig. 7). It is reasonable to conclude that the methanol metabolism and energy regeneration were influenced by this specific membrane protein, whether the protein was ATPase active or not. But when the protein was expressed using the supplemented strategy, the formaldehyde concentration was low and the enzymatic activity of FLD and IDH was high, indicating that the media supplement strategy is necessary for efficient expression of Pgp. Therefore, this strategy could be used to express other, hard‐to‐express, proteins from this yeast.
Compared with soluble proteins, expression of membrane proteins is a complex process because the proteins need to go through an assembly process. Once synthesis of a membrane protein begins, a secretory machinery is engaged, and the protein must be targeted and inserted into the membrane (Loll, 2003). It is likely that Pgp accumulation affects the host cell metabolism and energy regeneration, and the modified induction strategy somehow prevents this accumulation and allows efficient expression.
Experimental procedures
Strains
The KM71H‐ZA‐Pgp (MutS) strain was prepared by transforming P. Pastoris KM71H with the pPICZA‐Pgp plasmid harbouring mouse Pgp (Esser et al., 2017). The GS115‐ZA‐Pgp m strain (Mut+) was carrying mutated Pgp gene at the nucleotide‐binding domains (NBD) (Esser et al., 2017). The GS115‐3.5K‐P1‐2 strain contains the sequence encoding a glycoside hydrolase LXYL‐P1‐2 (Cheng, et al., 2013, Liu et al., 2016).
Fermentation process
The strain harbouring Pgp or Pgp m gene was streaked on a YPDS‐Zeocin agar plate containing 1% yeast extract, 2% peptone, 2% glucose, 2% agar (m/v), 1 M sorbitol and 100 μg ml−1 ZeocinR and incubated at 30°C for 3 days. Single colonies were transferred to 500 ml baffled shake flasks containing 50 ml BMGYH (100 mM potassium phosphate buffer, pH 6, 13.4 g yeast nitrogen base with ammonium sulfate and without amino acids, 400 μg biotin, 40 mg l−1 L‐histidine and 100 ml of 10% glycerol per litter). The flasks were incubated at 28°C at 220 rpm. Methanol induction was carried using three different strategies in bench‐top bioreactors (Sartorius Stedim Biotech, Germany, interfaced to Sartorius MFCS/win 3.0). (i) Conventional strategy: overnight culture from shake flasks was transferred to the bioreactor containing BMGYH media, when the culture in the bioreactor reached an OD600 of 20. A solution containing 2.5% (v/v) methanol and 4.35 ml l−1 PTM in 100 mM potassium phosphate buffer (pH6) was added at a rate of 8.5 ml l−1 h−1 for 72 h [peristaltic pump (101 U/R; Watson‐Marlow Limited, Falmouth, UK)]. (ii) Replacement strategy: after reaching an OD600 of 20, the culture was centrifuged at 5000 rpm, 4°C for 15 min and the cells were transferred back to the bioreactor containing fresh 1 l BMMYH; at that point, the same solution used at the conventional strategy was pumped into bioreactor at a rate of 8.5 ml−1 l−1h−1 for 72 h. (iii) Supplemented strategy: after reaching an OD600 of 20, fresh BMMYH media containing 2.5% (v/v) methanol and 4.35 ml l−1 PTM was added into the culture at a rate of 8.5 ml l−1 h−1 for 72 h. The growth conditions for the three strategies were as follows: 29°C, pH kept at 5.0 with 7‐7.5% ammonium hydroxide, and DO was controlled at 30% saturation by adjusting the RPM and airflow (100–700 rpm; 0.8–1.5 vvm) with 0.03% (v/v) P2000 antifoam. PTM solution was the same as described previously (Liu and Zhu, 2015). All experiments and analysis were done in triplicates. The results shown are the mean ± standard deviation (SD) from three independent experiments.
Analytical methods
Preparation of supernatant and cell‐free extract
After centrifugation (12 000 g for 3 min), supernatant samples were collected and stored at −80°C. Cell‐free extract was prepared as previously described (Suye et al., 1990; Jin et al., 2010).
Quantifications of methanol, formaldehyde, hydrogen peroxide and formate
Methanol was measured using Analox GM8 Micro‐Stat Analyzer (Analox Instruments, Hammersmith, UK). Formaldehyde, hydrogen peroxide and formate were measured using assay kits (Sigma‐Aldrich, St. Louis, MO, USA, MAK131; Sigma‐Aldrich, MAK165; R‐Biopharm AG, Darmstadt, Germany, 10979732035).
Activity assays of enzymes related to methanol metabolism and TCA cycle
Alcohol oxidase (AOX, EC 1.1.3.13) activity was spectrophotometrically measured in the present of 2.2‐azino‐bis(3‐ethylbenzothiazoline‐6‐sulfonic acid) and horseradish peroxidase (Arrocha et al., 2014). Formaldehyde dehydrogenase (FLD, EC 1.2.1.46) and formate dehydrogenase (FDH, EC 1.2.1.2) activities were determined according to the previously described method (Allais et al., 1983; Kato, 1990; Tran et al., 2015). α‐Ketoglutarate dehydrogenases (α‐KGDHs) was assayed by colorimetrical methods (Jin et al., 2010). The enzymatic activities of catalase (CAT, EC 1.11.1.6), malate dehydrogenase (MDH, EC 1.1.1.37) and isocitrate dehydrogenase (IDH, EC 1.1.1.42) were determined using enzyme kits (E‐100, E‐124 and E‐138) from Biomedical Research Service Center, University of Buffalo, State University of New York, USA. All other reagents and chemicals were purchased from Sigma‐Aldrich or Thermo Fisher Scientific, USA.
Biomass measurement
Optical density at 600 nm (OD600) of the appropriately diluted culture was determined using a spectrophotometer. Dry cell weight (DCW, mg ml−1) was measured by a moisture determination balance (MB 200; Ohaus Corporation, Florham Park, NJ, USA) after centrifugation and washing at 95°C (Liu and Zhu, 2018).
Pgp concentration and ATPase activity
Pgp was analysed by electrophoresis on NuPAGE 12% (w/v) Bis‐Tris gel (Invitrogen Co, San Diego, CA, USA) and Western blotting on Immun‐Blot PVDF membranes sandwiches (Bio‐Rad Laboratories, Hercules, CA, USA). Primary antibody was Pgp‐specific monoclonal antibody C219 that was visualized by chemiluminescent HRP antibody detection reagent (Denville Scientific Inc, Metuchen, NJ, USA) (Esser et al., 2017). The volumetric production of Pgp was determined by scanning the area on the Western blotting membranes with ImageQuant TL software (GE Healthcare, Piscataway, NJ, USA) using purified Pgp as a reference. ATPase activity was determined using a high‐throughput colorimetric ATPase assay kit (Innova Biosciences, Cambridge, UK, 6010120). The ATPase activity of the parental P. pastoris strain was deducted before calculating the ATPase activity of the recombinant Pgp. One ATPase unit was defined as the amount of enzyme that catalyses 1 μmol of ATP per minute. For all data, replicates from three parallel measurements or independent assays were measured and the mean ± standard error (SD) was calculated. Student's t‐test in spss 17.0 (SPSS Inc., Chicago, IL, USA) was used for two‐group comparisons, and P < 0.05 was considered statistically significant.
Conflict of interest
None declared.
Supporting information
Fig. S1. Volumetric production and enzymatic activity of LXYL‐P1‐2 at three fermentation processes. Conventional strategy (filled green diamond), replacement strategy (filled red triangle) and supplemented strategy (filled blue circle). Time zero indicates the initiation of the methanol induction phase. A. Volumetric production; B. enzymatic activity
Fig. S2. Formaldehyde concentration, FLD and IDH enzymatic activities of GS115‐3.5K‐P1‐2 at various processes. Conventional strategy (filled green diamond), replacement strategy (filled red triangle) and supplemented strategy (filled blue circle). Time zero indicates the initiation of the methanol induction phase. A. Formaldehyde concentration; B. FLD activity; C. IDH activity.
Acknowledgements
The research was supported by the intramural research programme of NIDDK and NCI, National Institutes of Health. The authors would like to thank the NIH Library Writing Center for manuscript editing assistance.
Microb Biotechnol (2019) 12(6), 1226–1236
Funding Information
The research was supported by the intramural research programme of NIDDK and NCI, National Institutes of Health.
References
- Ahmad, M. , Hirz, M. , Pichler, H. , and Schwab, H. (2014) Protein expression in Pichia pastoris: recent achievements and perspectives for heterologous protein production. Appl Microbiol Biotechnol 98: 5301–5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allais, J. , Louktibi, A. , and Baratti, J. (1983) Oxidation of methanol by the yeast, Pichia pastoris, purification and properties of the formaldehyde dehydrogenase. Agric Biol Chem 47: 1509–1516. [Google Scholar]
- Arrocha, A.A. , Cano‐Castillo, U. , Aguila, S.A. , and Vazquez‐Duhalt, R. (2014) Enzyme orientation for direct electron transfer in an enzymatic fuel cell with alcohol oxidase and laccase electrodes. Biosens Bioelectron 61: 569–574. [DOI] [PubMed] [Google Scholar]
- Athmaram, T. , Singh, A.K. , Saraswat, S. , Srivastava, S. , Misra, P. , Rao, M.K. , et al (2013) A simple Pichia pastoris fermentation and downstream processing strategy for making recombinant pandemic swine origin influenza a virus hemagglutinin protein. J Ind Microbiol Biotechnol 40: 245–255. [DOI] [PubMed] [Google Scholar]
- Bill, R.M. , Henderson, P.J. , Iwata, S. , Kunji, E.R. , Michel, H. , Neutze, R. , et al (2011) Overcoming barriers to membrane protein structure determination. Nat Biotechnol 29: 335–340. [DOI] [PubMed] [Google Scholar]
- Bußwinkel, F. , Goñi, O. , Cord‐Landwehr, S. , O'Connell, S. , and Moerschbacher, B.M. (2018) Endochitinase 1 (Tv‐ECH1) from Trichoderma virens has high subsite specificities for acetylated units when acting on chitosans. Int J Biol Macromol 114: 453–461. [DOI] [PubMed] [Google Scholar]
- Cheng, H.L. , Zhao, R.Y. , Chen, T.J. , Yu, W.B. , Wang, F. , Cheng, K.D. , and Zhu, P. (2013) Cloning and characterization of the glycoside hydrolases that remove xylosyl groups from 7‐β‐xylosyl‐10‐deacetyltaxol and its analogues. Mol Cell Proteomics 12: 2236–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciofalo, V. , Barton, N. , Kreps, J. , Coats, I. , and Shanahan, D. (2006) Safety evaluation of a lipase enzyme preparation, expressed in Pichia pastoris, intended for use in the degumming of edible vegetable oil. Regul Toxicol Pharmacol 45: 1–8. [DOI] [PubMed] [Google Scholar]
- De Vera, A.A. , Gupta, P. , Lei, Z. , Liao, D. , Narayanan, S. , Teng, Q. , et al (2019) Immuno‐oncology agent IPI‐549 is a modulator of P‐glycoprotein (P‐gp, MDR1, ABCB1)‐mediated multidrug resistance (MDR) in cancer: In vitro and in vivo. Cancer Lett 442: 91–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esser, L. , Zhou, F. , Pluchino, K.M. , Shiloach, J. , Ma, J. , Tang, W.K. , et al (2017) Structures of the multidrug transporter P‐glycoprotein reveal asymmetric ATP binding and the mechanism of polyspecificity. J Biol Chem 292: 446–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fam, J.P. , Sabri, S. , Baharum, S.N. , Salleh, A.B. , and Oslan, S.N. (2017) Metabolomics study in methylotrophic yeast: a minireview. J Life Sci 11: 11–18. [Google Scholar]
- Gao, M.J. , Zhan, X.B. , Gao, P. , Zhang, X. , Dong, S.J. , Li, Z. , et al (2015) Improving performance and operational stability of porcine interferon‐α production by Pichia pastoris with combinational induction strategy of low temperature and methanol/sorbitol co‐feeding. Appl Biochem Biotechnol 176: 493–504. [DOI] [PubMed] [Google Scholar]
- Habeck, M. , Kapri‐Pardes, E. , Sharon, M. , and Karlish, S.J. (2017) Specific phospholipid binding to Na, K‐ATPase at two distinct sites. Proc Natl Acad Sci USA 114: 2904–2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, H. , Qian, J. , Chu, J. , Wang, Y. , Zhuang, Y. , and Zhang, S. (2009) Optimization of L‐methionine feeding strategy for improving S‐adenosyl‐L‐methionine production by methionine adenosyltransferase overexpressed Pichia pastoris . Appl Microbiol Biotechnol 83: 1105. [DOI] [PubMed] [Google Scholar]
- Jahic, M. , Rotticci‐Mulder, J. , Martinelle, M. , Hult, K. , and Enfors, S.O. (2002) Modeling of growth and energy metabolism of Pichia pastoris producing a fusion protein. Bioprocess Biosyst Eng 24: 385–393. [Google Scholar]
- Jin, H. , Liu, G. , Ye, X. , Duan, Z. , Li, Z. , and Shi, Z. (2010) Enhanced porcine interferon‐α production by recombinant Pichia pastoris with a combinational control strategy of low induction temperature and high dissolved oxygen concentration. Biochem Eng J 52: 91–98. [Google Scholar]
- Juturu, V. , and Wu, J.C. (2018) Heterologous protein expression in Pichia pastoris: latest research progress and applications. ChemBioChem 19: 7–21. [DOI] [PubMed] [Google Scholar]
- Kastilan, R. , Boes, A. , Spiegel, H. , Voepel, N. , Chudobová, I. , Hellwig, S. , et al (2017) Improvement of a fermentation process for the production of two Pf AMA1‐DiCo‐based malaria vaccine candidates in Pichia pastoris . Sci Rep 7: 11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, N. (1990) Formaldehyde dehydrogenase from methylotrophic yeasts. Methods Enzymol 188: 455–459. [DOI] [PubMed] [Google Scholar]
- Lee, J.Y. , Kinch, L.N. , Borek, D.M. , Wang, J. , Wang, J. , Urbatsch, I.L. , et al (2016) Crystal structure of the human sterol transporter ABCG5/ABCG8. Nature 533: 561–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, W. and Zhu, P. (2015) Pilot studies on scale‐up biocatalysis of 7‐β‐xylosyl‐10‐deacetyltaxol and its analogues by an engineered yeast. J Ind Microbiol Biotechnol 42: 867–876. [DOI] [PubMed] [Google Scholar]
- Liu, W. and Zhu, P. (2018) Demonstration‐scale high‐cell‐density fermentation of Pichia pastoris In Recombinant Glycoprotein Production. Berlin, Germany: Springer, pp. 109–116. [DOI] [PubMed] [Google Scholar]
- Liu, W. , Gong, T. , Wang, Q.H. , Liang, X. , Chen, J.J. and Zhu, P. (2016) Scaling‐up fermentation of Pichia pastoris to demonstration‐scale using new methanol‐feeding strategy and increased air pressure instead of pure oxygen supplement. Sci Rep 6: 18439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, W. , Inwood, S. , Gong, T. , Sharma, A. , Yu, L. and Zhu, P. (2019) Fed‐batch high‐cell‐density fermentation strategies for Pichia pastoris growth and production. Crit Rev Biotechnol 39: 258–271. [DOI] [PubMed] [Google Scholar]
- Loll, P.J. (2003) Membrane protein structural biology: the high throughput challenge. J Struct Biol 142: 144–153. [DOI] [PubMed] [Google Scholar]
- Looser, V. , Bruhlmann, B. , Bumbak, F. , Stenger, C. , Costa, M. , Camattari, A. , et al (2015) Cultivation strategies to enhance productivity of Pichia pastoris: a review. Biotechnol Adv 33: 1177–1193. [DOI] [PubMed] [Google Scholar]
- Maghsoudi, A. , Hosseini, S. , Shojaosadati, S.A. , Vasheghani‐Farahani, E. , Nosrati, M. , and Bahrami, A. (2012) A new methanol‐feeding strategy for the improved production of β‐galactosidase in high cell‐density fed‐batch cultures of Pichia pastoris Mut+ strains. Biotechnol Bioprocess Eng 17: 76–83. [Google Scholar]
- Matthews, C.B. , Kuo, A. , Love, K.R. , and Love, J.C. (2018) Development of a general defined medium for Pichia pastoris . Biotechnol Bioeng 115: 103–113. [DOI] [PubMed] [Google Scholar]
- Mears, L. , Stocks, S.M. , Sin, G. , and Gernaey, K.V. (2017) A review of control strategies for manipulating the feed rate in fed‐batch fermentation processes. J Biotechnol 245: 34–46. [DOI] [PubMed] [Google Scholar]
- Moser, S. , Strohmeier, G.A. , Leitner, E. , Plocek, T.J. , Vanhessche, K. , and Pichler, H. (2018) Whole‐cell(+)‐ambrein production in the yeast Pichia pastoris . Metab Eng Com 7: e00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noseda, D.G. , Recúpero, M.N. , Blasco, M. , Ortiz, G.E. , and Galvagno, M.A. (2013) Cloning, expression and optimized production in a bioreactor of bovine chymosin B in Pichia (Komagataella) pastoris under AOX1 promoter. Protein Expr Purif 92: 235–244. [DOI] [PubMed] [Google Scholar]
- Plantz, B.A. , Sinha, J. , Villarete, L. , Nickerson, K.W. , and Schlegel, V.L. (2006) Pichia pastoris fermentation optimization: energy state and testing a growth‐associated model. Appl Microbiol Biotechnol 72: 297–305. [DOI] [PubMed] [Google Scholar]
- Ren, H. , and Yuan, J. (2005) Model‐based specific growth rate control for Pichia pastoris to improve recombinant protein production. J Chem Technol Biotechnol 80: 1268–1272. [Google Scholar]
- Sun, Q. , Chen, F. , Geng, F. , Luo, Y. , Gong, S. , and Jiang, Z. (2018) A novel aspartic protease from Rhizomucor miehei expressed in Pichia pastoris and its application on meat tenderization and preparation of turtle peptides. Food Chem 245: 570–577. [DOI] [PubMed] [Google Scholar]
- Suye, S.I. , Ogawa, A. , Yokoyama, S. , and Obayashi, A. (1990) Screening and identification of Candida methanosorbosa as alcohol oxidase‐producing methanol using yeast. Agric Biol Chem 54: 1297–1298. [Google Scholar]
- Theron, C.W. , Berrios, J. , Delvigne, F. , and Fickers, P. (2018) Integrating metabolic modeling and population heterogeneity analysis into optimizing recombinant protein production by Komagataella (Pichia) pastoris . Appl Microbiol Biotechnol 102: 63–80. [DOI] [PubMed] [Google Scholar]
- Tran, S. , Nowicki, M. , Chatterjee, D. , and Gerlai, R. (2015) Acute and chronic ethanol exposure differentially alters alcohol dehydrogenase and aldehyde dehydrogenase activity in the zebrafish liver. Prog Neuropsychopharmacol Biol Psychiatry 56: 221–226. [DOI] [PubMed] [Google Scholar]
- Vanz, A. , Lünsdorf, H. , Adnan, A. , Nimtz, M. , Gurramkonda, C. , Khanna, N. , and Rinas, U. (2012) Physiological response of Pichia pastoris GS115 to methanol‐induced high level production of the Hepatitis B surface antigen: catabolic adaptation, stress responses, and autophagic processes. Microb Cell Fact 11: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viña‐Gonzalez, J. , Elbl, K. , Ponte, X. , Valero, F. , and Alcalde, M. (2018) Functional expression of aryl‐alcohol oxidase in Saccharomyces cerevisiae and Pichia pastoris by directed evolution. Biotechnol Bioeng 115: 1666–1674. [DOI] [PubMed] [Google Scholar]
- Wang, Q.H. , Liang, L. , Liu, W. , Gong, T. , Chen, J.J. , Hou, Q. , et al (2017) Enhancement of recombinant BmK AngM1 production in Pichia pastoris by regulating gene dosage, co‐expressing with chaperones and fermenting in fed‐batch mode. J Asian Nat Prod Res 19: 1–14. [DOI] [PubMed] [Google Scholar]
- Werten, M.W. , van den Bosch, T.J. , Wind, R.D. , Mooibroek, H. , and de Wolf, F.A. (1999) High‐yield secretion of recombinant gelatins by Pichia pastoris . Yeast 15: 1087–1096. [DOI] [PubMed] [Google Scholar]
- Yamada, R. , Ogura, K. , Kimoto, Y. , and Ogino, H. (2019) Toward the construction of a technology platform for chemicals production from methanol: D‐lactic acid production from methanol by an engineered yeast Pichia pastoris . World J Microbiol Biotechnol 35: 37. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Volumetric production and enzymatic activity of LXYL‐P1‐2 at three fermentation processes. Conventional strategy (filled green diamond), replacement strategy (filled red triangle) and supplemented strategy (filled blue circle). Time zero indicates the initiation of the methanol induction phase. A. Volumetric production; B. enzymatic activity
Fig. S2. Formaldehyde concentration, FLD and IDH enzymatic activities of GS115‐3.5K‐P1‐2 at various processes. Conventional strategy (filled green diamond), replacement strategy (filled red triangle) and supplemented strategy (filled blue circle). Time zero indicates the initiation of the methanol induction phase. A. Formaldehyde concentration; B. FLD activity; C. IDH activity.


