Abstract
Genetic variants of chemokine and regulatory cytokines play functional roles in chronic HBV infection. The objective of the study, was to evaluate the association between the CCR5D32, CCR5-2459A/G, MCP-1-2518A/G, VDR-APa1A/C, VDR-Taq1T/C SNPs and HBV susceptibility, in samples of Iranian populations. The CCR5D32, CCR5-2459A/G, MCP1-2518A/G, VDR-APa1A/C, VDR-Taq1T/C polymorphisms were analyzed by polymerase chain reaction and PCR-RFLP using 100 chronic HBV infected (HBV) patients, 40 spontaneously recovered HBV (SR) subjects and 100 healthy controls (C). Also, serum levels of protein were monitored. The study showed that the existence of CCR5-2459A, MCP1-2518G and VDR-CC alleles significantly increased risk of chronic HBV infection. In addition, WtAGCC haplotype had a higher frequency in HBV patients than C and SR groups and might relate to the natural history of the infection. Statistical analysis indicated positive correlations between CCR5-2459A/G, MCP1-2518A/G, VDR-APa1A/C, VDR-Taq1T/C genotypes and serum levels of the CCR5, MCP-1 and VDR in HBV patients. According to the statistical analysis, significant associations with susceptibility to chronic HBV infection was observed with CCR5-2459A/G, MCP1-2518A/G, VDR-APa1A/C, VDR-Taq1T/C polymorphisms. In addition, no association of the CCR5D32 SNP with the disease was found.
Keywords: Chemokine, HBV, Single nucleotide polymorphism
Introduction
Hepatitis B virus (HBV) infection is a serious public health problem worldwide and especially in Asia and Africa. HBV is a hepato-tropic virus, exists in more than 350 million people. Disease can develop into chronic hepatitis, cirrhosis, hepatocellular cancer and finally leads to death. Iran is located in the low endemicity of HBV infection and just like some other country, the chronic HBV is prevalent [1].
Different functions of the host immune system cause various clinical presentations of HBV infection. However, the actual mechanism for the chronic infection not fully understood, but it might be depend on host immune response and genetic factors. Macrophages, T cytotoxic lymphocytes and natural killer (NK) cells are potential elements which can produce the host immune responses against HBV antigens [2].
Chemokines, as defensive compounds have receptors on the surface of the immune cells. Cooperation between chemokine and its receptor, induces the migration of the immune cells to the site of the infection and regulates innate and adaptive immune responses [2].
Genetic variations such as single nucleotide polymorphisms (SNPs) of genes encoding the cytokines and chemokines can affect the clinical presentation of the hepatitis [3] and other inflammations [4]. In addition, results evidenced that polymorphisms of the IL-10 [5], IL-28B [6, 7], macrophage migration factor [8], interferon gamma (IFN-γ) [4], tumour necrosis factor-alpha [9] might change the susceptibility to the chronic inflammations.
CC-chemokine receptor 5 (CCR5) is a regulator of T cell through activation of T helper and cytotoxic T cells. CCR5 can bind to regulated on activation normal T cell expressed and secreted (RANTES). Vitamin D regulates the production of RANTES. The combination of CCR5 with some ligands (CCL3, CCL4, and CCL5) lead to immune response to viral infections. This chemokine induces immune cells to migrate to the inflamed liver [10]. It means that down-regulation of CCR5 might cause insufficient immune responses against infection.
The gene encoded CCR5 is located on chromosome 3p21.31. CCR5D32 deletion in exon 1 of the gene blocks the protein synthesis and associated with the increased susceptibility to the HCV. Results showed that CCR5D32-MtMt genotype might associated to the increased risk of HCV. This SNP is prevalent in different ethnic and geographical populations. How CCR5 change the immune response is unknown, but the risk of chronic HBV infection might be depend on the CCR5 polymorphisms [11]. Monocyte chemoattractant protein 1 (MCP-1) is a multifunctional cytokine, which is expressed on leukocytes. In vitro studies showed that MCP-1 had chemotactic activity. MCP-1 that is released from hepatic stellate cells can regulate leukocyte trafficking, which means that MCP-1 has direct profibrogenic action [12]. MCP-1 gene is located on chromosome 17q12. In regard to the MCP-1 polymorphisms in the promoter region, results indicated that − 2518G allele was associated with higher production of MCP-1 [13]. Moreover, in HCV patients, − 2518G allele was reported as a risk factor to hepatic inflammation and fibrosis [14]. Park et al. [15] reported that MCP-1 − 2518A/G polymorphism was contributed to HBV clearance and the frequency of − 2518A allele was higher in HBV patients compared to the spontaneously recovered people. On the other hand, Cheong et al. [16] study did not reveal any relationship between MCP-1 − 2518A/G SNP and HBV infection.
The active form of vitamin D has immunomodulatory action and regulates calcium metabolism, cellular growth and differentiation. This hormone inhibits lymphocyte proliferation, activates monocytes, blocks cytokine synthesis and affects programmed cell death through vitamin D receptor (VDR) [17]. Vitamin D as an immunoregulatory hormone, bind to VDR and mediate immuno-regulatory functions. This receptor is expressed on macrophages, active T lymphocytes and monocytes. Complex of the vitamin D and VDR induces the production of some cytokines (TNF-a, IL-1) and inhibits the production of IFN-ϒ and IL-2. VDR gene is located on chromosome 12q13 and has four polymorphisms with Pathological functions related to the immune mediated diseases such as HBV [18].
In the present study, it was hypothesized that genetic variants of CCR5, MCP-1 and VDR play functional roles in chronic HBV infection. Therefore, the objective of our research was to study the association between the CCR5D32, CCR5-2459A/G (rs1799987), MCP1-2518A/G (rs1024611), VDR-APa1 A/C (rs7975232), VDR-Taq1 T/C (rs731236) SNPs and HBV susceptibility, in a sample of Iranians.
Materials and Methods
Subjects
One hundred chronic HBV patients showing positive HBsAg and antibodies against anti-HBc and impaired liver function test (≥ 2 times the upper limit of normal) for at least six months were enrolled into the study (age: 29.03 ± 5.710, age range of 18–43). Serological tests (presence of HBsAg by enzyme-linked immunosorbent assay (ELISA) and HBV-DNA by reverse transcription polymerase chain reaction (RT-PCR) and clinical findings were compatible with chronic liver disease. 40 subjects with negative HBsAg and anti-HCV and positive antibodies against anti-HBs and anti-HBc were also included as spontaneously recovered (SR) group (age: 29.72 ± 5.517, age range of 18–42). The control group comprised 100 HBsAg, anti-HBs and anti-HBc negative, healthy volunteers with normal values for alanine transaminase (ALT), without any history of hepatitis B infection (age: 30.44 ± 4.539, age range of 22–45). All subjects were in the same geographical area. All participants were unrelated Iranians. There was no difference between groups in terms of age, gender and ethnicity. Subjects with HCV, HEV, HAV, HIV, alcohol consumption, drug abuse, liver diseases were excluded. Between July and December 2015, the study population was recruited, and they were enrolled from the outpatient clinic of the Blood Transfusion Organization clinics in Zahedan, Iran. The study was approved by the Institutional Ethics Committee of the Zahedan University of Medical Sciences (IR.ZAUMS.REC.1395.159, grant number: 7868) and carried out in Infectious Diseases and Tropical Medicine Research Center, Zahedan, Iran. Written informed consent was obtained from each participant.
Analysis of the Serum Parameters
Five ml of blood was taken in tubes containing EDTA for determining biochemical parameters. The measurement of the serum level of CCR5, MCP1, VDR was done used the sensitive sandwich ELISA technique (EASTBIOPHARM, China), according to the manufacturer’s instructions.
Peripheral DNA Isolation
Genomic DNA was extracted from the 500 μl of peripheral blood leukocytes using the salting-out method. These samples were stored at − 80 °C until they were used for the study.
Polymorphism Genotyping
The CCR5D32 polymorphism was detected by polymerase chain reaction (PCR) amplification and CCR5-2459A/G, MCP-1-2518A/G, VDR-APa1A/C, VDR-Taq1T/C SNPs were detected by polymerase chain reaction-restriction fragment-length polymorphism (PCR-RFLP).
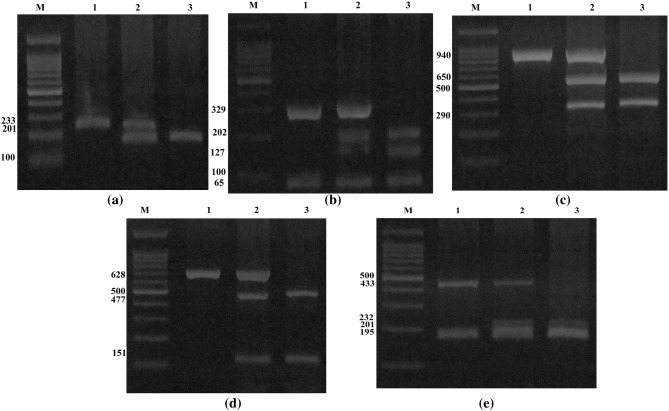
The sequence of the primers, restriction enzymes and the fragment length after digestion are shown in Table 1. Every reaction contained 1 μl of each primer, 100 ng of template DNA and 10 μl of 2× Prime Taq Premix (Genet Bio, Korea) and 7 μl ddH2O in a 20 μl of total reaction volume. The PCR conditions were as follows: initial denaturation at 95 °C for 5 min followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at 56 °C for CCR5D32, 58 °C for CCR5-2459A/G, 60 °C for MCP-1-2518A/G and 61.5 °C for VDR-APa1A/C and VDR-Taq1T/C polymorphisms for 30 s and extension at 72 °C for 30 s, followed by a final extension step at 72 °C for 5 min. Finally, the PCR products digested by the restriction enzymes (Fermentas, Vilnius, Lithuania) and digested products were resolved by electrophoresis in 2–4% agarose gel and stained with ethidium bromide (Fig. 1a–e).
Table 1.
PCR-RFLP-based assay of CCR5D32, CCR5-2459A/G, MCP1-2518A/G, VDR-APa1A/C, VDR-Taq1T/C SNPs
| Polymorphism | Primers | Annealing temperature (°C) | Restriction enzyme | Allele phenotype (bp) |
|---|---|---|---|---|
| CCR5D32 | F: TGTTTGCGTCTCTCCCAG | 56 | – | Wt: 233 |
| R: CACAGCCCTGTGCCTCTT | Mt: 201 | |||
| CCR5-2459A/G | F: AAAATCCCCACTAAGATCCTG | 58 | Bsp1286 I | A: 329/65 |
| R: ATTCATCTAGTCAAAAGCCCAC | G: 202/127/65 | |||
| MCP1-2518A/G | F: CTTTCCCTTGTGTGTCCCC | 60 | PvuII | C: 940 |
| R: TTACTCCTTTTCTCCCCAACC | G: 650/290 | |||
| VDR-APa1A/C | F: CTAGGTCTGG ATCCTAAATGCA | 61.5 | Apa1 | A: 628 |
| R: TTAGGTTGGACAGGAGAGAGAA | C: 477/151 | |||
| VDR-Taq1T/C | F: CTAGGTCTGG ATCCTAAATGCA | 61.5 | Taq 1 | T: 433/195 |
| R: TTAGGTTGGACAGGAGAGAGAA | C: 232/201/195 |
Fig. 1.
The electrophoresis pattern of CCR5D32, CCR5-2459A/G, MCP-1-2518A/G, VDR-APa1A/C, VDR-Taq1T/C polymorphisms. a The electrophoresis pattern of CCR5D32 SNP on 3% agarose gel; M marker 100 bp, 1 genotype WtWt, 2 genotype WtMt, 3 genotype MtMt. b The digestion pattern of Bsp1286 I restriction enzyme on 4% agarose gel at CCR5-2459A/G SNP; M marker 100 bp, 1 genotype AA, 2 genotype AG, 3 genotype GG.c The digestion pattern of PvuII restriction enzyme on 2% agarose gel at MCP-1-2518A/G SNP, M marker 100 bp, 1 genotype AA, 2 genotype AG, 3 genotype GG. d The digestion pattern of Apa1 restriction enzyme on 2% agarose gel at VDR-APa1A/C SNP; M marker 100 bp, 1 genotype AA, 2 genotype AC, 3 genotype CC. e The digestion pattern of Taq 1 restriction enzyme on 4% agarose gel at VDR-Taq1T/C SNP; M marker 100 bp, 1 genotype TT, 2 genotype TC, 3 genotype CC
Statistical Analysis
Statistical analysis was performed by SPSS 20.0. Computing the odds ratio (OR) and 95% confidence intervals (95% CI) from logistic regression analyses were used for assessing the relationship between genotypes and HBV. The distributions of genotypes were tested with the Chi-square analyses. Data were expressed as mean ± SD. A P value of < 0.05 indicated the statistical significance.
Results
Clinical characteristics of study populations are listed in Table 2. Study subjects were divided into three groups: patients with chronic HBV infection (female/male. 43.0/57.0), patients that spontaneously recovered from infection (female/male. 37.5/62.5) and healthy people (female/male. 49.0/51.0). The gender ratio (female/male), ages and ethnicities were not significantly different between the 3 groups (P = 0.427, P = 0.125, P = 0.292).
Table 2.
Demographic data of chronic hepatitis B (HBV) patients, spontaneously recovered (SR) subjects and control group (C)
| Parameters | C, N (%) | SR, N (%) | HBV, N (%) | P |
|---|---|---|---|---|
| Age (years) | 30.44 ± 4.539 | 29.72 ± 5.517 | 29.03 ± 5.710 | 0.125 |
| Sex | ||||
| Male | 51 (51.0) | 25 (62.5) | 57 (57.0) | 0.427 |
| Female | 49 (49.0) | 15 (37.5) | 43 (43.0) | |
| Ethnicities | ||||
| Sistani | 46 (46.0) | 18 (45.0) | 41 (41.0) | 0.292 |
| Baluch | 18 (18.0) | 13 (32.5) | 22 (22.0) | |
| Others | 36 (36.0) | 9 (22.5) | 37 (37.0) | |
Genotype Analysis
The genotype frequencies for each polymorphism are shown in Tables 3 and 4. Genotype distributions at all SNPs was in Hardy–Weinberg equilibrium (P > 0.05). Genotype distribution of CCR5-2459A/G, MCP-1-2518A/G, VDR-APa1A/C, VDR-Taq1T/C SNPs among the chronic HBV patients was found to be significantly different compared to healthy controls (C + SR) (P = 0.001, P = 0.009, P = 0.003, respectively). Likewise, compared to healthy controls (C + SR), allele distributions in chronic HBV patients were significantly different (P = 0.001, P = 0.002, P = 0.001, respectively). There were a higher rate of CCR5-2459AA, MCP-1-2518GG, VDR-APa1CC and VDR-Taq1CC genotypes (42.0, 19.0, 22.0%) in the chronic HBV patients compared to the healthy controls (20.0, 8.6, 8.6%). Carriage of the A, G, C, C alleles at position CCR5-2459A/G, MCP-1-2518A/G, VDR-APa1A/C, VDR-Taq1T/C were associated with chronic HBV infection. No significant difference was found between the 3 groups (P > 0.05) in the genotype distribution of CCR5D32 polymorphism.
Table 3.
The frequency of genotypes and alleles of the CCR5D32, CCR5-2459A/G, MCP-1-2518A/G, VDR-APa1A/C, VDR-Taq1T/C polymorphisms between chronic hepatitis B (HBV) patients, spontaneously recovered (SR) subjects and control groups (C)
| Chemokine polymorphisms | C (%) | SR (%) | P value | Odds ratio | HBV (%) | P value | Odds ratio |
|---|---|---|---|---|---|---|---|
| CCR5D32 | |||||||
| WtWt | 96 (96.0) | 38 (95.0) | Ref = 1 | – | 97 (97.0) | Ref = 1 | – |
| WtMt | 3 (3.0) | 1 (2.5) | 0.883 | 0.842 (0.085–8.350) | 2 (2.0) | 0.653 | 0.660 (0.108–4.037) |
| MtMt | 1 (1.0) | 1 (2.5) | 0.516 | 2.526 (0.154–41.426) | 1 (1.0) | 0.994 | 0.990 (0.061–16.051) |
| WtMt + MtMt | 4 (4.0) | 2 (5.0) | 0.792 | 1.263 (0.222–7.185) | 3 (3.0) | 0.701 | 0.742 (0.162–3.405) |
| Wt | 195 (97.5) | 77 (96.2) | Ref = 1 | – | 196 (98.0) | Ref = 1 | – |
| Mt | 5 (2.5) | 3 (3.8) | 0.573 | 1.519 (0.354–6.513) | 4 (2.0) | 0.737 | 0.796 (0.211–3.008) |
| CCR5-2459A/G | |||||||
| AA | 21 (21.0) | 7 (17.5) | 0.825 | 0.889 (0.314–2.518) | 42 (42.0) | 0.001 | 3.636 (1.738–7.608) |
| AG | 39 (39.0) | 18 (45.0) | 0.617 | 1.231 (0.545–2.780) | 36 (36.0) | 0.141 | 1.678 (0.842–3.346) |
| GG | 40 (40.0) | 15 (37.5) | Ref = 1 | – | 22 (22.0) | Ref = 1 | – |
| AA + AG | 60 (60.0) | 25 (62.5) | 0.784 | 1.111 (0.522–2.364) | 78 (78.0) | 0.007 | 2.364 (1.272–4.392) |
| A | 81 (40.5) | 32 (40.0) | 0.939 | 0.979 (0.577–1.662) | 120 (60.0) | 0.001 | 2.204 (1.478–3.286) |
| G | 119 (59.5) | 48 (60.0) | Ref = 1 | – | 80 (40.0) | Ref = 1 | – |
| MCP-1-2518A/G | |||||||
| AA | 53 (53.0) | 20 (50.0) | Ref = 1 | – | 35 (35.0) | Ref = 1 | – |
| AG | 40 (40.0) | 15 (37.5) | 0.988 | 0.994 (0.453–2.179) | 46 (46.0) | 0.071 | 1.741 (0.954–3.178) |
| GG | 7 (7.0) | 5 (12.5) | 0.320 | 1.893 (0.538–6.657) | 19 (19.0) | 0.004 | 4.110 (1.564–10.799) |
| GG + AG | 47 (47.0) | 20 (50.0) | 0.748 | 1.128 (0.541–2.349) | 65 (65.0) | 0.011 | 2.094 (1.186–3.697) |
| A | 146 (73.0) | 55 (68.8) | Ref = 1 | – | 116 (58.0) | Ref = 1 | – |
| G | 54 (27.0) | 25 (31.2) | 0.476 | 1.229 (0.697–2.166) | 84 (42.0) | 0.002 | 1.958 (1.287–2.979) |
| VDR-APa1A/C | |||||||
| AA | 51 (51.0) | 19 (47.5) | Ref = 1 | – | 33 (33.0) | Ref = 1 | – |
| AC | 42 (42.0) | 16 (40.0) | 0.955 | 1.023 (0.469–2.232) | 45 (45.0) | 0.103 | 1.656 (0.902–3.039) |
| CC | 7 (7.0) | 5 (12.5) | 0.312 | 1.917 (0.542–6.778) | 22 (22.0) | 0.001 | 4.857 (1.866–12.643) |
| AC + CC | 49 (49.0) | 21 (52.5) | 0.708 | 1.150 (0.552–2.397) | 67 (67.0) | 0.010 | 2.113 (1.192–3.745) |
| A | 144 (72.0) | 54 (67.5) | Ref = 1 | – | 111 (55.5) | Ref = 1 | – |
| C | 56 (28.0) | 26 (32.5) | 0.455 | 1.238 (0.707–2.169) | 89 (44.5) | 0.001 | 2.062 (1.360–3.125) |
| VDR-Taq1T/C | |||||||
| TT | 51 (51.0) | 19 (47.5) | Ref = 1 | – | 33 (33.0) | Ref = 1 | – |
| TC | 42 (42.0) | 16 (40.0) | 0.955 | 1.023 (0.469–2.232) | 45 (45.0) | 0.103 | 1.656 (0.902–3.039) |
| CC | 7 (7.0) | 5 (12.5) | 0.312 | 1.917 (0.542–6.778) | 22 (22.0) | 0.001 | 4.857 (1.866–12.643) |
| TC + CC | 49 (49.0) | 21 (52.5) | 0.708 | 1.150 (0.552–2.397) | 67 (67.0) | 0.010 | 2.113 (1.192–3.745) |
| T | 144 (72.0) | 54 (67.5) | Ref = 1 | – | 111 (55.5) | Ref = 1 | – |
| C | 56 (28.0) | 26 (32.5) | 0.455 | 1.238 (0.707–2.169) | 89 (44.5) | 0.001 | 2.062 (1.360–3.125) |
Table 4.
The frequency of genotypes and alleles of the CCR5D32, CCR5-2459A/G, MCP-1-2518A/G, VDR-APa1A/C, VDR-Taq1T/C polymorphisms between chronic hepatitis B (HBV) patients and spontaneously recovered (SR) subjects/control group (C) (C + SR)
| Chemokine polymorphisms | HBV (%) | Healthy (C + SR) (%) | P value | Odds ratio |
|---|---|---|---|---|
| CCR5D32 | ||||
| WtWt | 97 (97.0) | 134 (95.7) | Ref = 1 | – |
| WtMt | 2 (2.0) | 4 (2.9) | 0.673 | 0.691 (0.124–3.847) |
| MtMt | 1 (1.0) | 2 (1.4) | 0.764 | 0.691 (0.062–7.726) |
| WtMt + MtMt | 3 (3.0) | 6 (4.3) | 0.607 | 0.691 (0.169–2.830) |
| Wt | 196 (98.0) | 272 (97.1) | Ref = 1 | – |
| Mt | 4 (2.0) | 8 (2.9) | 0.555 | 0.694 (0.206–2.337) |
| CCR5-2459A/G | ||||
| AA | 42 (42.0) | 28 (20.0) | 0.000 | 3.750 (1.885–7.460) |
| AG | 36 (36.0) | 57 (40.7) | 0.166 | 1.579 (0.827–3.015) |
| GG | 22 (22.0) | 55 (39.3) | Ref = 1 | – |
| AA + AG | 78 (78.0) | 85 (60.7) | 0.005 | 2.294 (1.282–4.106) |
| A | 120 (60.0) | 113 (40.4) | 0.000 | 2.217 (1.531–3.210) |
| G | 80 (40.0) | 167 (59.6) | Ref = 1 | – |
| MCP-1-2518A/G | ||||
| AA | 35 (35.0) | 73 (52.1) | Ref = 1 | – |
| AG | 46 (46.0) | 55 (39.3) | 0.052 | 1.744 (0.995–3.060) |
| GG | 19 (19.0) | 12 (8.6) | 0.005 | 3.302 (1.444–7.554) |
| GG + AG | 65 (65.0) | 67 (47.9) | 0.009 | 2.023 (1.193–3.431) |
| A | 116 (58.0) | 201 (71.8) | Ref = 1 | – |
| G | 84 (42.0) | 79 (28.2) | 0.002 | 1.842 (1.256–2.702) |
| VDR-APa1A/C | ||||
| AA | 33 (33.0) | 70 (50.0) | Ref = 1 | – |
| AC | 45 (45.0) | 58 (41.4) | 0.086 | 1.646 (0.932–2.905) |
| CC | 22 (22.0) | 12 (8.6) | 0.001 | 3.889 (1.719–8.795) |
| AC + CC | 67 (67.0) | 70 (50.0) | 0.009 | 2.030 (1.192–3.458) |
| A | 111 (55.5) | 198 (70.7) | Ref = 1 | – |
| C | 89 (44.5) | 82 (29.3) | 0.001 | 1.936 (1.325–2.830) |
| VDR-Taq1T/C | ||||
| TT | 33 (33.0) | 70 (50.0) | Ref = 1 | – |
| TC | 45 (45.0) | 58 (41.4) | 0.086 | 1.646 (0.932–2.905) |
| CC | 22 (22.0) | 12 (8.6) | 0.001 | 3.889 (1.719–8.795) |
| TC + CC | 67 (67.0) | 70 (50.0) | 0.009 | 2.030 (1.192–3.458) |
| T | 111 (55.5) | 198 (70.7) | Ref = 1 | – |
| C | 89 (44.5) | 82 (29.3) | 0.001 | 1.936 (1.325–2.830) |
Analysis of linkage disequilibrium indicated that there was a significant association between CCR5-2459A/G, MCP-1-2518A/G polymorphisms (D′ = 0.556, R2 = 0.310). Interestingly, the results suggested complete linkage disequilibrium between VDR-APa1A/C, VDR-Taq1T/C SNPs (D′ = 1, R2 = 1). Linkage disequilibrium did not observe between CCR5D32 and CCR5-2459A/G polymorphisms (D′ = 0.134, R2 = 0.018).
We also calculated the CCR5D32, CCR5-2459A/G, MCP-1-2518A/G, VDR-APa1A/C, and VDR-Taq1T/C haplotype association analysis, and found 5 genotypes. As shown in Tables 5 and 6, the distribution frequency of haplotypes showed significant association between chronic HBV infection compared to the C and SR groups (P < 0.05). In our study, WtAGCC haplotype had a significantly higher frequency in HBV patients than C and SR groups. Also, results indicated enrichment of WtAGCC haplotype among patients than healthy controls (C + SR) (4.0 vs 14.3%, P = 0.000). It means that this haplotype might relate to the natural history of the infection.
Table 5.
Haplotype frequencies (D32/2459/MCP-1/Apa1/Taq1) in chronic hepatitis B (HBV) patients, spontaneously recovered (SR) subjects and control group (C)
| Haplotypes | C group n (%) |
SR group n (%) |
P value | Odds ratio | HBV group n (%) |
P value | Odds ratio |
|---|---|---|---|---|---|---|---|
| WtAGCC | 12 (12.0) | 8 (20.0) | 0.349 | 1.692 (0.563–5.089) | 40 (40.0) | 0.000 | 8.462 (3.406–21.021) |
| WtGAAT | 33 (33.0) | 13 (32.5) | Ref = 1 | – | 13 (13.0) | Ref = 1 | – |
| MtAGCC | 5 (5.0) | 3 (7.5) | 0.599 | 1.523 (0.317–7.311) | 4 (4.0) | 0.343 | 2.031 (0.470–8.771) |
| WtAAAC | 31 (31.0) | 7 (17.5) | 0.295 | 0.573 (0.202–1.624) | 26 (26.0) | 0.073 | 2.129 (0.931–4.867) |
| WtGGCT | 19 (19.0) | 9 (22.5) | 0.723 | 1.202 (0.433–3.335) | 17 (17.0) | 0.079 | 2.271 (0.908–5.680) |
| TOTAL | 100 (100.0) | 40 (100.0) | 100 (100.0) |
Table 6.
Haplotype frequencies (D32/2459/MCP-1/Apa1/Taq1) in chronic hepatitis B (HBV) patients and spontaneously recovered (SR) subjects/control group (C) (C + SR)
| Haplotypes | Healthy)C + SR) n (%) | HBV group n (%) |
P value | Odds ratio |
|---|---|---|---|---|
| WtAGCC | 20 (14.3) | 40 (40.0) | 0.000 | 7.077 (3.127–16.016) |
| WtGAAT | 46 (32.9) | 13 (13.0) | Ref = 1 | – |
| MtAGCC | 8 (5.7) | 4 (4.0) | 0.407 | 1.769 (0.459–6.817) |
| WtAAAC | 38 (27.1) | 26 (26.0) | 0.290 | 2.421 (0.096–5.347) |
| WtGGCT | 28 (20.0) | 17 (17.0) | 0.082 | 2.148 (0.908–5.084) |
| Total | 140 (100.0) | 100 (100.0) |
Analysis of Serum Parameters
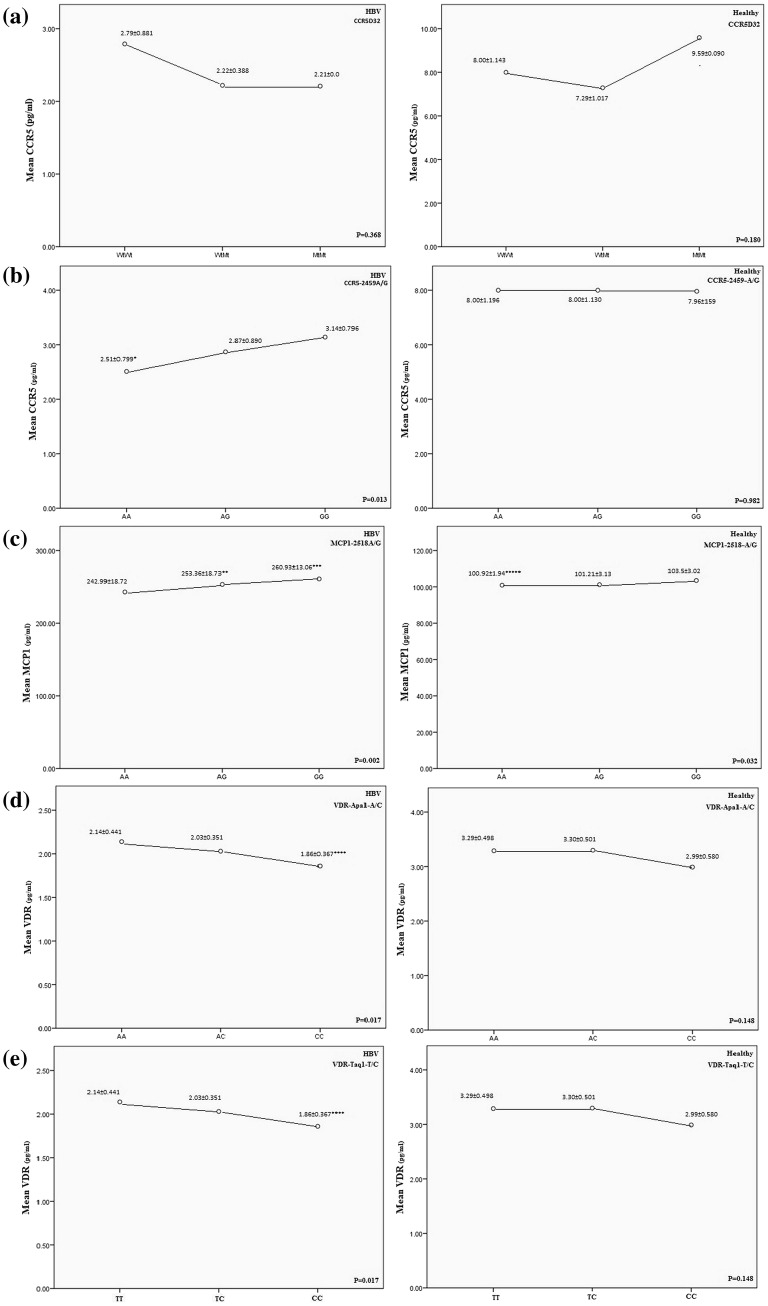
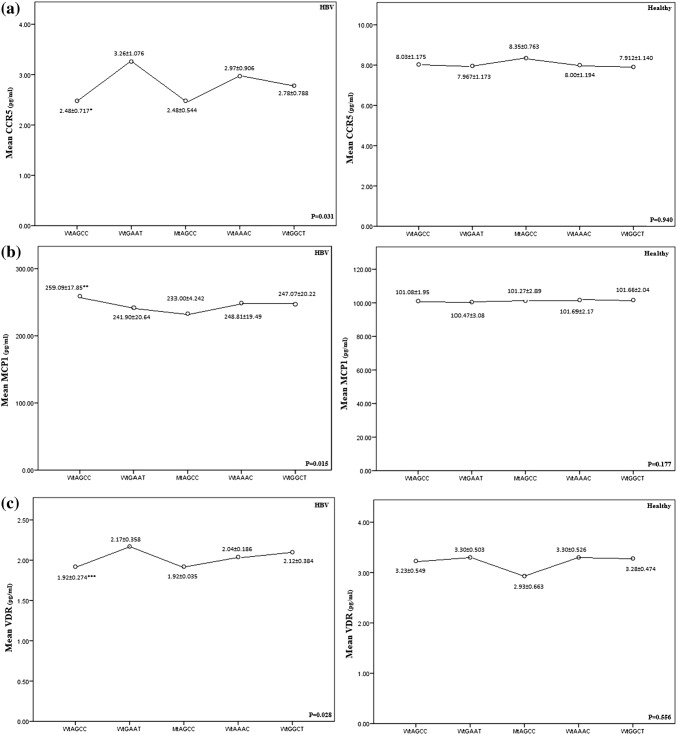
Serum levels of chemokine were different between 3 groups (Fig. 2). Serum levels of the MCP-1 were significantly increased and CCR5 and VDR reduced in the chronic HBV patients compared to the healthy groups (C + SR) (P < 0.05) (Fig. 3). Statistical analysis indicated positive correlations between CCR5, MCP-1 and VDR genotypes and serum levels of the CCR5, MCP-1 and VDR in HBV patients (Fig. 3). Chronic HBV patients with MCP-1-2518AG and MCP-1-2518GG genotypes had higher levels of MCP-1 compared to MCP-1-2518AA genotype (P = 0.029, P = 0.002, respectively). In this regard, healthy groups (C + SR) with MCP-1-251AA genotype had reduced levels of MCP-1 compared to MCP-1-2518GG genotype (P = 0.024). Also, Chronic HBV patients with CCR5-2459AA genotype had reduced levels of CCR5 compared to CCR5-2459GG (P = 0.013). Lower levels of VDR had seen in HBV patients with VDR-aa/tt genotypes compare to the VDR-AA/TT (P = 0.012). The results showed that the risk of HBV infection was significantly increased in subjects with WtAGCC genotype that had higher levels of MCP-1 and reduced levels of CCR5 and VDR, compared to the WtGAAT genotype (P = 0.015, P = 0.034, P = 0.047, respectively) (Fig. 4).
Fig. 2.
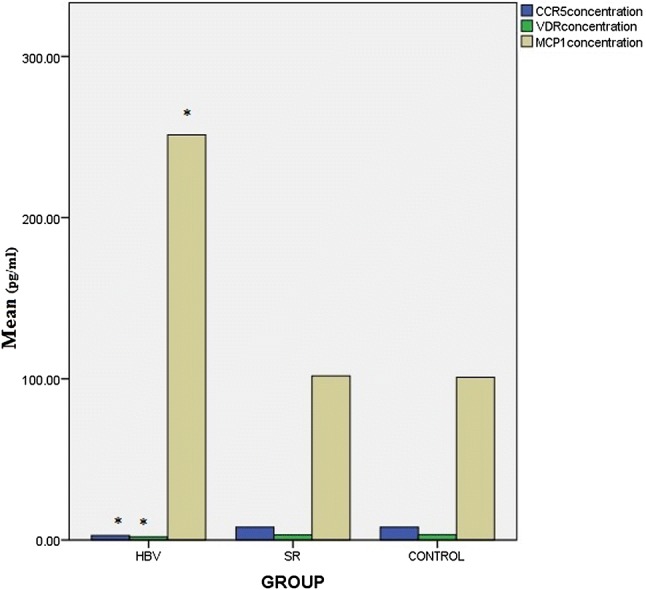
Comparison of CCR5, MCP-1, VDR serum levels between chronic hepatitis B (HBV) patients, spontaneously recovered (SR) subjects and control group (C). P < 0.05 considered significant
Fig. 3.
Serum levels of chemokines of chronic hepatitis B (HBV) patients and spontaneously recovered (SR) subjects/control group (C) (C + SR), stratified for the a CCR5D32, b CCR5-2459A/G, c MCP-1-2518A/G, d VDR-APa1A/C, e VDR-Taq1T/C polymorphism. The dots show the mean value with standard deviation. A line was inserted to illustrate the direction of the association. *P = 0.013, compared with genotype GG. **P = 0.029, compared with genotype AA; ***P = 0.002, compared with genotype AA; *****P = 0.024, compared with genotype GG. ****P = 0.012, compared with genotype AA. ****P = 0.012, compared with genotype TT
Fig. 4.
Association of haplotypes of D32/2459/MCP-1/Apa1/Taq1 polymorphisms with serum a CCR5, b MCP-1, c VDR level between chronic hepatitis B (HBV) patients and spontaneously recovered (SR) subjects/control group (C) (C + SR). The dots show the mean value with standard deviation. A line was inserted to illustrate the direction of the association. *P = 0.034, Compared with genotype WtGAAT. **P = 0.015, Compared with genotype WtGAAT. ***P = 0.047, Compared with genotype WtGAAT
Discussion
This is a case–control study, analyzing the five SNPs in chemokines and regulatory proteins in a sample of Iranian. SNPs were monitored in 100 Iranian chronic HBV infected patients and 140 healthy controls comprised from spontaneously recovered from HBV and healthy subjects. The genotype and allele frequencies of CCR5-2459A/G, MCP-1-2518A/G, VDR-APa1A/C and VDR-Taq1T/C polymorphisms in HBV patients were significantly different from the two other groups (SR and C). No significant differences in the distribution of CCR5D32 variations were found between HBV patients and healthy subjects in this study. The current study emphasized the critical role of chemokines in chronic HBV infection and the results showed that CCR5-2459AA, MCP-1-2518GG, VDR-APa1CC and VDR-Taq1CC genotypes were completely associated with the existence of disease.
Recently we have reported that some cytokine SNPs affected the susceptibility to HBV infection [6, 8, 19, 20]. It is well known that chemokines as regulators of host immune system mediate inflammation and immune responses against pathogens [21].
Although the positive relationship between CCR5 polymorphisms and HCV infection had been shown [11], but the main function of CCR5 in chronic HBV infection is not clear.
Chang et al. [22], Ahn et al. [23], indicated that CCR5-2459A/G polymorphism might be a risk factor for HBV infected patients. In accordance with our finding, they reported that CCR5-2459A allele was common in chronic HBV patients compared to the healthy controls (SR + C) and was associated with an increased risk of chronic infection. Also, they did not find significant differences between groups in regard to the CCR5D32 because CCR5-32 bp deletion mutation did not exist in participants. Among the participants in our study, only 9 people (4C, 2SR, 3hbv) had CCR5-32 bp deletion, but the frequency of genotypes and alleles was not statistically significant. In this study, the frequency of CCR5-2489G allele was similar between the SR group and HBV group, which means that CCR5-2489G allele might not have a protective role against HBV infection.
Also, we found lower levels of CCR5 in serum of the HBV patients carrying the CCR5-2459A allele. In regard to the HBV, lower levels of CCR5 might lead to the lack of suitable ligand binding to the CCR5, which caused a deficiency in immune responses against HBV infection [24]. Therefore, CCR5, as a mediator of immune signaling cascades, should be considered in drug design. In addition, CCR5-2459A/G polymorphism located at the downstream of promoter region which is responsible for the transcriptional activity of the CCR5 gene [25]. In accordance with our results, Paxton et al. [26] demonstrated the association of the level of CCR5 expression and mRNA production with the risk of HBV infection.
Another explanation for our findings is that, the risk of HBV is associated with Th1 and Th2 responses against the HBV antigens partly through the CCR5. CCR5 is expressed in lymphocytes in the liver. In other words, incomplete activity of Th1 and Th2 cells may be responsible for the HBV survival [24]. Also, genetic variations such as SNPs in promoter, which has transcription activity, might cause changes in Th1 and Th2 biological activity and reduce or enhance immune response in hepatic tissue. Our findings indicated that infection was enhanced with the CCR5-2459AA homozygous patients, which reduce the expression of CCR5. This phenomenon leads to the exacerbation of the hepatic inflammation. However, the results of Cheong et al. [16] did not show statistically significant association between CCR5 promoter SNPs and susceptibility to persistent HBV infection. Conflicting Results may be due to differences in the study design.
MCP-1 induces the inflammatory monocytes to produce the immune responces. The MCP-1-2518 G allele upregulates serum MCP-1 levels, so HBV infected patients have an increased frequency of MCP-1-2518 GG genotype [13]. In current study, HBV infected patients, SR and C groups had different frequency of MCP-12518 variants, which means this SNP has significant role in susceptibility to HBV infection. It has been made clear that MCP-1-2518 SNP was associated with various diseases such as HBV clearance [15], the distribution genotypes were different between studies. In line with our study, Park et al. [15] analyzed MCP-1-2518 polymorphism and found a significant relationship between MCP-1-2518A allel and chronic hepatitis B virus. They implied A allele as a risk factor for HBV. In contrast, Cheong et al. [16] reported that MCP-1-2518 polymorphism did not associate with the outcome of hepatitis B virus infection. On the other hand, MCP-1 gene SNP was associated with severity of HCV related liver disease [14]. Such differences may be due to variations in the immunopathogenesis of HBV and HCV infection.
Some SNPs in the VDR gene with biological effects were identified. Also, the positive association of VDR variations with chronic diseases such as autoimmune hepatitis [27], Crohn’s disease [28] and hepatitis B [29] were investigated. Interestingly, we found that VDR Apa1 and VDR Taq1 polymorphisms are in complete linkage disequilibrium in which that all subjects with VDR Apa1 CC genotype also has VDR Taq1 CC variants. The co-existence of these two alleles associated with an increased susceptibility to HBV infection and also reduced serum levels of VDR in patients. In the current study, the combination of the polymorphisms revealed a correlation between allelic combination and disease severity. The frequency of WtAGCC (CCR5D32, CCR5-2459, VDR Apa1 and Taq1, respectively) haplotype was significantly higher in HBV patients. In 1994, Morrison et al. [30] suggested that VDR Apa1 and Taq1 genotypes and alleles were completely in linkage disequilibrium. They also indicated that VDR polymorphisms affect mRNA production and VDR mRNA stability. Our study reveals that CC genotype is associated with HBV infection which might be caused by reduced levels of VDR and varied mRNA stability.
Some SNPs studied have important effects on the susceptibility and outcomes of HBV infection. Also, we found that chemokine levels probably be depend on the severity of infection. Determination of the real effects of chemokine polymorphisms needs more detailed studies. The restrictions that are leading to contradictory results, likely include: sample size, patients’ selection, different cultural backgrounds, epidemiological and geographical factors, study conditions (such as; number and characteristics of the subjects and HBV genotype variations) and different gene–gene interactions. In conclusion, the results have shown the role of genetic polymorphisms of CCR5D32, CCR5-2459A/G, MCP-1-2518A/G, VDR-APa1A/C, VDR-Taq1T/C in the HBV infection, and elucidated an association between HBV infection and chemokines gene polymorphisms.
Acknowledgements
The authors would like to thank all participants who willingly participated in this study. We appreciate all who helped us in this work, especially in the Blood Transfusion Research Center of Zahedan, Iran.
Authors’ Contribution
B. Moudi, Z. Heidari, H. Mahmoudzadeh-Sagheb conceived and co-designed the study, supervised all the experimental design, analyzed the results, and drafted the manuscript. All authors read, modified and approved the final version of the manuscript. These authors equally contributed to this work.
Financial Support
This project was supported by the vice chancellor of Research and Technology of Zahedan University of Medical Sciences (ZUMS) and Infectious Diseases and Tropical Medicine Research Center, Zahedan University of Medical Sciences.
Compliance with Ethical Standards
Conflict of interest
The authors report no declarations of interest.
Compliance with Ethical Standards
This study was approved by the Institutional Ethics Committee of the Zahedan University of Medical Sciences (IR.ZAUMS.REC.1395.159, Grant Number 7868).
Ethical Approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed Consent
Informed consent was written and signed by all individual participants included in the study.
Contributor Information
Bita Moudi, Email: bita.moudi@yahoo.com.
Zahra Heidari, Phone: 98-5433295794, Email: histology_iri@yahoo.com.
Hamidreza Mahmoudzadeh-Sagheb, Email: histology@ymail.com.
References
- 1.Moudi B, Heidari Z, Mahmoudzadeh-Sagheb H, Alavian SM, Lankarani KB, Farrokh P, et al. Concomitant use of heat-shock protein 70, glutamine synthetase and glypican-3 is useful in diagnosis of HBV-related hepatocellular carcinoma with higher specificity and sensitivity. Eur J Histochem: EJH. 2018;62(1):2859. doi: 10.4081/ejh.2018.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miyara M, Wing K, Sakaguchi S. Therapeutic approaches to allergy and autoimmunity based on FoxP3 + regulatory T-cell activation and expansion. J Allergy Clin Immunol. 2009;123(4):749–755. doi: 10.1016/j.jaci.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 3.Moudi B, Heidari Z, Mahmoudzadeh-Sagheb H. Impact of host gene polymorphisms on susceptibility to chronic hepatitis B virus infection. Infect Genet Evol. 2016;44:94–105. doi: 10.1016/j.meegid.2016.06.043. [DOI] [PubMed] [Google Scholar]
- 4.Heidari Z, Mahmoudzadeh-Sagheb H, Hashemi M, Ansarimoghaddam S, Moudi B, Sheibak N. Association between IFN-gamma +874A/T and IFN-gammaR1 (−611A/G, +189T/G, and +95C/T) gene polymorphisms and chronic periodontitis in a sample of Iranian population. Int J Dent. 2015;2015:375359. doi: 10.1155/2015/375359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moudi B, Heidari Z, Mahmoudzadeh-Sagheb H, Moudi M. Analysis of interleukin-10 gene polymorphisms in patients with chronic periodontitis and healthy controls. Dent Res J. 2018;15(1):71–79. doi: 10.4103/1735-3327.223614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heidari Z, Moudi B, Mahmoudzadeh-Sagheb H, Hashemi M. The correlation between interferon lambda 3 gene polymorphisms and susceptibility to hepatitis B virus infection. Hepat Mon. 2016;16(3):e34266. doi: 10.5812/hepatmon.34266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heidari Z, Moudi B, Mahmoudzadeh-Sagheb H, Moudi M. The association between interleukin-28B gene polymorphisms as a potential biomarker and the risk of chronic Periodontitis in an Iranian population. Head Face Med. 2017;13(1):16. doi: 10.1186/s13005-017-0148-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moudi B, Heidari Z, Mahmoudzadeh-Sagheb H, Hashemi M. Gene polymorphisms of macrophage migration inhibitory factor affect susceptibility to chronic hepatitis B virus infection in an Iranian cohort. Microbiol Immunol. 2016;60(6):390–396. doi: 10.1111/1348-0421.12382. [DOI] [PubMed] [Google Scholar]
- 9.Heidari Z, Moudi B, Mahmoudzadeh Sagheb H, Moudi M. Association of TNF-alpha gene polymorphisms with production of protein and susceptibility to chronic hepatitis B infection in the South East Iranian population. Hepat Mon. 2016;16(11):e41984. doi: 10.5812/hepatmon.41984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ajuebor MN, Carey JA, Swain MG. CCR5 in T cell-mediated liver diseases: what’s going on? J Immunol. 2006;177(4):2039–2045. doi: 10.4049/jimmunol.177.4.2039. [DOI] [PubMed] [Google Scholar]
- 11.Woitas RP, Ahlenstiel G, Iwan A, Rockstroh JK, Brackmann HH, Kupfer B, et al. Frequency of the HIV-protective CC chemokine receptor 5-Delta32/Delta32 genotype is increased in hepatitis C. Gastroenterology. 2002;122(7):1721–1728. doi: 10.1053/gast.2002.33660. [DOI] [PubMed] [Google Scholar]
- 12.Marra F, Romanelli RG, Giannini C, Failli P, Pastacaldi S, Arrighi MC, et al. Monocyte chemotactic protein-1 as a chemoattractant for human hepatic stellate cells. Hepatology. 1999;29(1):140–148. doi: 10.1002/hep.510290107. [DOI] [PubMed] [Google Scholar]
- 13.Rovin BH, Lu L, Saxena R. A novel polymorphism in the MCP-1 gene regulatory region that influences MCP-1 expression. Biochem Biophys Res Commun. 1999;259(2):344–348. doi: 10.1006/bbrc.1999.0796. [DOI] [PubMed] [Google Scholar]
- 14.Muhlbauer M, Bosserhoff AK, Hartmann A, Thasler WE, Weiss TS, Herfarth H, et al. A novel MCP-1 gene polymorphism is associated with hepatic MCP-1 expression and severity of HCV-related liver disease. Gastroenterology. 2003;125(4):1085–1093. doi: 10.1016/S0016-5085(03)01213-7. [DOI] [PubMed] [Google Scholar]
- 15.Park BL, Kim YJ, Cheong HS, Kim LH, Choi YH, Lee HS, et al. Association of common promoter polymorphisms of MCP1 with hepatitis B virus clearance. Exp Mol Med. 2006;38(6):694–702. doi: 10.1038/emm.2006.82. [DOI] [PubMed] [Google Scholar]
- 16.Cheong JY, Cho SW, Choi JY, Lee JA, Kim MH, Lee JE, et al. RANTES, MCP-1, CCR2, CCR5, CXCR1 and CXCR4 gene polymorphisms are not associated with the outcome of hepatitis B virus infection: results from a large scale single ethnic population. J Korean Med Sci. 2007;22(3):529–535. doi: 10.3346/jkms.2007.22.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aranda A, Pascual A. Nuclear hormone receptors and gene expression. Physiol Rev. 2001;81(3):1269–1304. doi: 10.1152/physrev.2001.81.3.1269. [DOI] [PubMed] [Google Scholar]
- 18.Suzuki T, Sano Y, Sotozono C, Kinoshita S. Regulatory effects of 1alpha,25-dihydroxyvitamin D(3) on cytokine production by human corneal epithelial cells. Curr Eye Res. 2000;20(2):127–130. doi: 10.1076/0271-3683(200002)2021-DFT127. [DOI] [PubMed] [Google Scholar]
- 19.Moudi B, Heidari Z, Mahmoudzadeh-Sagheb H, Hashemi M. Association between IL-10 gene promoter polymorphisms (−592 A/C, −819 T/C, −1082 A/G) and susceptibility to HBV infection in an Iranian population. Hepat Mon. 2016 doi: 10.5812/hepatmon.32427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heidari Z, Mahmoudzadeh-Sagheb H, Hashemi M, Ansarimoghaddam S, Moudi B, Sheibak N. Association between IFN-γ +874 A/T and IFN-γR1 (−611A/G, +189T/G and +95C/T) gene polymorphisms and chronic periodontitis in a sample of Iranian population. Int J Dent. 2015 doi: 10.1155/2015/375359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rossi D, Zlotnik A. The biology of chemokines and their receptors. Annu Rev Immunol. 2000;18:217–242. doi: 10.1146/annurev.immunol.18.1.217. [DOI] [PubMed] [Google Scholar]
- 22.Chang HY, Ahn SH, Kim DY, Shin JS, Kim YS, Hong SP, et al. Association between CCR5 promoter polymorphisms and hepatitis B virus infection. Korean J Hepatol. 2005;11(2):116–124. [PubMed] [Google Scholar]
- 23.Ahn SH, Kim DY, Chang HY, Hong SP, Shin JS, Kim YS, et al. Association of genetic variations in CCR5 and its ligand, RANTES with clearance of hepatitis B virus in Korea. J Med Virol. 2006;78(12):1564–1571. doi: 10.1002/jmv.20739. [DOI] [PubMed] [Google Scholar]
- 24.Rehermann B. Immune responses in hepatitis B virus infection. Semin Liver Dis. 2003;23(1):21–38. doi: 10.1055/s-2003-37586. [DOI] [PubMed] [Google Scholar]
- 25.Liu R, Zhao X, Gurney TA, Landau NR. Functional analysis of the proximal CCR5 promoter. AIDS Res Hum Retrovir. 1998;14(17):1509–1519. doi: 10.1089/aid.1998.14.1509. [DOI] [PubMed] [Google Scholar]
- 26.Paxton WA, Liu R, Kang S, Wu L, Gingeras TR, Landau NR, et al. Reduced HIV-1 infectability of CD4 + lymphocytes from exposed-uninfected individuals: association with low expression of CCR5 and high production of beta-chemokines. Virology. 1998;244(1):66–73. doi: 10.1006/viro.1998.9082. [DOI] [PubMed] [Google Scholar]
- 27.Vogel A, Strassburg CP, Manns MP. Genetic association of vitamin D receptor polymorphisms with primary biliary cirrhosis and autoimmune hepatitis. Hepatology. 2002;35(1):126–131. doi: 10.1053/jhep.2002.30084. [DOI] [PubMed] [Google Scholar]
- 28.Simmons JD, Mullighan C, Welsh KI, Jewell DP. Vitamin D receptor gene polymorphism: association with Crohn’s disease susceptibility. Gut. 2000;47(2):211–214. doi: 10.1136/gut.47.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bellamy R, Ruwende C, Corrah T, McAdam KP, Thursz M, Whittle HC, et al. Tuberculosis and chronic hepatitis B virus infection in Africans and variation in the vitamin D receptor gene. J Infect Dis. 1999;179(3):721–724. doi: 10.1086/314614. [DOI] [PubMed] [Google Scholar]
- 30.Morrison NA, Qi JC, Tokita A, Kelly PJ, Crofts L, Nguyen TV, et al. Prediction of bone density from vitamin D receptor alleles. Nature. 1994;367(6460):284–287. doi: 10.1038/367284a0. [DOI] [PubMed] [Google Scholar]