Abstract
The number of patients with Alzheimer’s disease (AD) is rapidly increasing in Asia. Mutations in the amyloid protein precursor (APP), presenilin-1 (PSEN1), and presenilin-2 (PSEN2) genes can cause autosomal dominant forms of early-onset AD (EOAD). Although these genes have been extensively studied, variant classification remains a challenge, highlighting the need to colligate mutations across populations. In this study, we performed a genetic screening for mutations in the APP, PSEN1, and PSEN2 genes in 200 clinically diagnosed EOAD patients across four Asian countries, including Thailand, Malaysia, the Philippines, and Korea, between 2009 and 2018. Thirty-two (16%) patients presented pathogenic APP, PSEN1, or PSEN2 variants; eight (25%), 19 (59%), and five (16%) of the 32 patients presented APP, PSEN1, and PSEN2 variants, respectively. Among the 21 novel and known non-synonymous variants, five APP variants were found in Korean patients and one APP variant was identified in a Thai patient with EOAD. Nine, two, and one PSEN1 mutation was found in a Korean patient, Malaysian siblings, and a Thai patient, respectively. Unlike PSEN1 mutations, PSEN2 mutations were rare in patients with EOAD; only three variants were found in Korean patients with EOAD. Comparison of AD-causative point mutations in Asian countries; our findings explained only a small fraction of patients, leaving approximately 84% (p = 0.01) of autosomal dominant pedigrees genetically unexplained. We suggest that the use of high-throughput sequencing technologies for EOAD patients can potentially improve our understanding of the molecular mechanisms of AD.
Keywords: Alzheimer’s disease, Asian, genetics, mutation, EOAD
1. Introduction
Alzheimer’s disease (AD) is one of the most common neurodegenerative disorders, which accounts for up to 75% of all dementia cases [1,2]. AD can be categorized into two major types: Early-onset AD (EOAD) and late-onset AD (LOAD). EOAD is usually inherited autosomal dominantly, and occurs before the age of 60–65 years. Presenilin-1 (PSEN1; MIM #104311) [3], presenilin-2 (PSEN2; MIM #600759) [4], and amyloid protein precursor (APP; MIM #104760) gene mutations [5,6] and duplications [7] can cause autosomal-dominant EOAD. Mutations in these genes have been relatively rarely observed [8,9,10,11,12], since the prevalence is estimated to be 5.3 per 100,000 individuals [13]. The significance of APP, PSEN1, and PSEN2 in AD were confirmed by different genetic studies, and majority of these mutations share a common feature of exhibiting increased production of the Aβ1-42 peptide, associated with altered gamma secretase activity [5,14]. Among these three genes, PSEN1 mutations were more frequently observed in AD, since approximately 252 different mutations were reported (http://www.alzforum.org/mutations, accessed in June 2019). Mutations in APP and PSEN2 were less frequently observed, since only 35 pathogenic APP mutations and 20 pathogenic PSEN2 mutations have been reported (http://www.alzforum.org/mutations, accessed in June 2019).
Despite the fact that genomic sequencing and bioinformatics have dramatically improved the identification of other genetic risk factors over the last few years, the interpretation of rare variants remains a challenge [1,2,8,10,15,16]. Remarkably, the age of onset and disease progression is not only influenced by genetics, but also by both lifestyle and environmental factors [17,18,19,20,21]. These factors may cause altered gene expression by epigenetic modifications, thereby affecting AD pathology [1,17,19,22]. Although majority of these mutations of these three genes are associated with familial EOAD, follow the Mendelian rules, several de novo cases of AD have been reported in patients without any family history of dementia [10,23].
The fastest increase in the number of elderly individuals has been observed in the East Asian countries. Approximately 60% of all patients diagnosed with dementia inhabit the Asian countries [24]. However, the genetics of EOAD are not well characterized, since only a few reports are available regarding mutations in EOAD causative genes (Figure 1) [25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44]. Therefore, the aim of the present study was to report mutations in additional cases, including sporadic ones, since our last update from 2009 for Asian patients with EOAD. We performed a genetic screening for mutations in the PSEN1, PSEN2, and APP genes in 200 patients with EOAD.
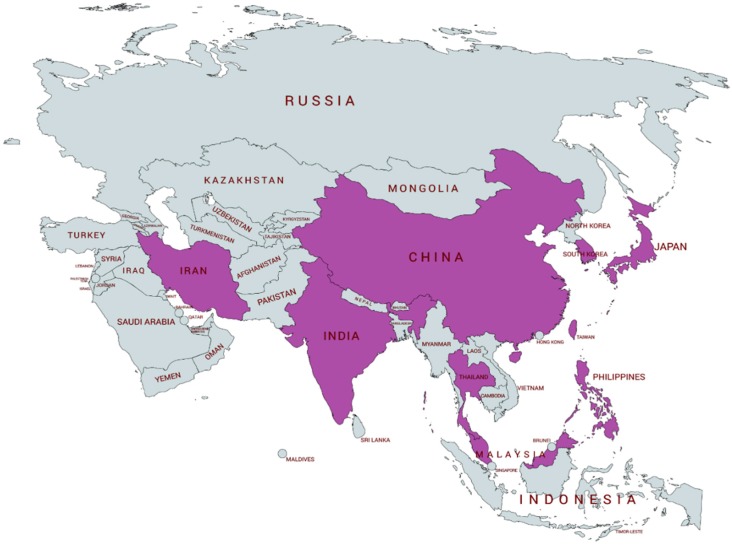
Figure 1.
Distribution of APP, PSEN1, and PSEN2 mutations in Asian countries. The fastest increase in the number of elderly individuals has been observed in Asian countries with approximately 60% of all patients diagnosed with dementia. The countries from which the gene mutations are reported are shown in purple.
2. Results
2.1. Identified Gene Mutations of APP
Considering that the genetic background of EOAD in the Asian population is not well characterized [10,15], we reported non-synonymous mutations in 200 clinically diagnosed patients with EOAD across four Asian countries, including Thailand, Malaysia, the Philippines, and Korea between 2009 and 2018. A total of 32 mutation carriers, including affected relatives in EOAD families and sporadic cases, were found among the 200 patients. From the 21 novel and known non-synonymous variants, five APP variants were found in Korean patients, and one APP variant was identified in a Thai patient. Nine, two, and one PSEN1 variant was identified in Korean patients, Malaysian siblings, and a Thai patient, respectively. Unlike PSEN1 mutations, PSEN2 mutations were a rare in EOAD, with only three variants identified in Korean patients (Table 1). Moreover, the mutation spectrum associated with AD for all Asian countries is shown in Table 2 [29,31,32,45,46,47,48,49,50,51,52,53].
Table 1.
APP, PSEN1, and PSEN2 mutations discovered in Asian early-onset Alzheimer’s disease (EOAD) patients between 2009 and 2018.
| Gene | Protein Change | Nucleotide Change | Exon | APOE | AOO (Years) | Gender | Family History | Pathogenicity Prediction | Clinical Significance | Population | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PolyPhen | SIFT | ||||||||||
| APP | p.Glu145Lys | c.433G > A | 4 | ε3/ε3 | 55 | F | Y | D: 0.932 | T:0.496 | Located outside of the amyloid progressing region | Korean |
| p.Val225Ala | c.674T > C | 7 | ε3/ε3 | 65 | F | Y | D: 932 | T: 0.496 | |||
| p.Thr297Met | c.890C > T | 7 | ε3/ε3 | 60 | F | Y | D: 0.98 | D: 0.0 | |||
| p.Pro484Ser | c.1450C > T | 11 | ε4/ε4 | 61 | F | Y | P: 0.765 | T: 0.063 | |||
| p.Val604Met | c.1810C > T | 14 | ε3/ε3 | 55 | M | Y | B: 0.450 | T: 0.095 | Thai | ||
| p.Val669Leu | c.2005G > C | 17 | ε3/ε3 | 55 | F | Y | B: 0.017 | T: 0.16 | Novel mutation, may cause EOAD | Korean | |
| PSEN1 | p.Val96Phe | c.286G > T | 4 | ε3/ε4 | 40 | M | Y | D: 1.0 | T: 0.002 | Known pathogenic mutation (EOAD) | Malaysian |
| ε3/ε4 | 40 | F | N | ||||||||
| p.Thr116Ile | c.335C > T | 5 | ε3/ε3 | 38 | F | Y | D: 1.00 | D: 0 | Known pathogenic mutation (EOAD) | Korean | |
| ε3/ε3 | 41 | F | Y | ||||||||
| p.Thr119Ile | c.356C > T | ε3/ε3 | 64 | F | Y | D: 1.00 | D: 0 | Novel mutation, may be involved in EOAD | |||
| p.His163Pro | c.488A > C | 4 | ε3/ε3 | 37 | F | Y | D: 1.00 | D: 0 | Novel mutation, may be involved in EOAD | ||
| p.Trp165Cys | c.695G > T | 6 | ε3/ε3 | 53 | M | Y | D: 1.00 | D: 0.001 | Known pathogenic mutation (EOAD) | ||
| p.Glu184Gly | c.551A > G | 7 | ε3/ε3 | 37 | F | Y | D: 0.878 | D: 0.005 | Known pathogenic mutation (EOAD) | ||
| p.Gly209Ala | c.626G > C | 7 | ε3/ε3 | 54 | F | Y | D: 1.00 | D: 0 | Novel mutation, may be involved in EOAD | ||
| p.Leu226Phe | CTC > TTC | 7 | ε3/ε3 | 37 | F | Y | D: 1.00 | D: 0 | Known pathogenic mutation (EOAD) | ||
| p.Leu232Pro | c.695T > C | 7 | ε3/ε3 | 37 | M | Y | D: 1.00 | D: 0 | Novel mutation, may be involved in EOAD | ||
| p.Glu280Lys | c.826G > A | 8 | ε3/ε3 | 48 | M | Y | D: 1.00 | D: 0 | Novel mutation, may be involved in EOAD | Malaysian | |
| ε3/ε3 | 55 | F | Y | ||||||||
| ε3/ε3 | 57 | M | Y | ||||||||
| p.Ala285Val | c.854C > T | 8 | ε3/ε3 | 46 | F | N | D: 1.0 | D: 0.015 | Known pathogenic mutation (EOAD) | Korean | |
| p.Gly417Ala | c.1250G > C | 12 | ε3/ε3 | 37 | M | N | D: 1.00 | D: 0 | Novel mutation, may be involved in EOAD | ||
| PSEN2 | p.Arg62Cys | c.184C > T | 5 | ε3/ε3 | 49 | M | N | D: 0.877 | D: 0.05 | Known mutation, may be involved AD | Korean |
| p.His169Asn | c.505C > A | 6 | ε3/ε3 | 56 | F | Y | D: 1.00 | D: 0 | Known mutation, May be involved AD | ||
| p.Val214Leu | c.640G > A | 7 | ε3/ε3 | 56 | M | Y | D: 0.836 | D: 0.09 | May be involved AD | ||
| ε3/ε4 | 70 | F | Y | D: 0.836 | D: 0.09 | May be involved AD | |||||
Abbreviations: MC, number of mutations carriers in the family; AOO, age of onset ranges in the family; DD, disease duration (at death or last examination); APOE, apolipoprotein E genotype; F, familial; S, sporadic; Y, yes, U, unknown; D, damaging; AD, Alzheimer’s disease; EOAD: early-onset Alzheimer’s disease.
Table 2.
The spectrum of APP, PSEN1, and PSEN2 mutations found in Asian countries.
| Gene | Exon | Codon, Mutation | Location in the Protein | Age of Onset, Clinical Characteristics | Pathogenic Nature | Country | References |
|---|---|---|---|---|---|---|---|
| APP | 3 | p.Glu145Lys | N-terminal | 50s/Familial, EOAD | Located outside of the amyloid progressing region | Korea | This study |
| 4 | p.Val225Ala | N-terminal | 65/Familial, EOAD | This study | |||
| 7 | pThr297Met | N-terminal | 60s/Familial, EOAD | This study | |||
| 8 | p. Pro484Ser | N-terminal | 60s/Familial, EOAD | This study | |||
| 14 | p.Val604Met | N-terminal | 55/Familial, EOAD | Pathogenic | Thailand | This study | |
| 16 | p.Val669Leu | N-terminal | 56 years; AD with a positive family history | Located nearby the β-secretase cleavage site of APP, right next to the Swedish APP (Lys, Met670/671Asn, Leu) mutation | Korea | This study | |
| p.Asp678Asn | N-terminal | 59–65 years/familial, EOAD | Probably pathogenic, may enhance the toxic amyloid oligomer formation | Japan | Wakutani et al., 2004 [25] | ||
| 17 | p.Glu693del | N-terminal | 44 years/familial, EOAD/MCI | Enhances the toxic amyloid oligomer formation | Japan | Tomiyama et al., 2008 [26] | |
| p. Val710Gly | TM-I | 65–82 years/Familial, AD, Parkinsonism | Probably pathogenic | China, Taiwan | Thajeb et al. 2009 [27] | ||
| p. Thr714Ala | TM-I | 47–55 years/Familial, EOAD, epilepsy | Probably pathogenic | Iran | Pasalar et al. 2002 [28] | ||
| p.Val715Met | TM-I | 41 years/ Familial EOAD |
Expressed in HEK293 cells, revealed 2* decrease in Abeta 40 levels. Might destroy the cleavage of gamma secretase at site at Abeta40 | Korea | Park et al., 2008 [29] | ||
| p.Val717Ile | TM-I | 53 years/Familial, EOAD | Increased Abeta42/Abeta40 ratio in CHO and HEK293 cells | Japan | Yoshioka et al., 1991 [30] | ||
| 54 years/unknown, EOAD | Thailand | Jiao et al., 2014 [31] | |||||
| p. Ile718Leu | TM-I | 65–82 years/Familial, AD, Parkinsonism | Probably pathogenic | China, Taiwan | Thajeb et al., 2009 [27] | ||
| p.Leu720Ser | TM-I | 65–82 years/Familial, AD, Parkinsonism | Probably pathogenic | China, Taiwan | Thajeb et al. 2009 [27] | ||
| 4 | p.Leu85Pro | TM-I | 26 years, Juvenile EOAD | Abeta42/Abeta40 ratio increased in HEK293 | Japan | Ataka et al. 2004 [54] | |
| p. Val96Phe | TM-I | EOAD, 49–60 years | 2.1 * increased Abeta 42/40 ratio in COS-1 cells | Japan | Kamino et al. 1996 [32] | ||
| p.Val97Leu | TM-I | EOAD | Higher beta secretase activity in human neuroblastoma cells | China | Fang et al. 2006 [33] | ||
| p. Phe105Cys | HL-I | 59 years/Familial, EOAD | Survival of mutant neuroblastoma cells dropped | China | Jiao et al., 2014 [31] | ||
| 5 | p. Gly111Val | HL-I | EOAD; 59 years/Familial | Increased ratios of secreted Aβ42/Aβ40 in vitro study | China | Qiu et al., 2019 [53] | |
| p. Thr116Ile | HL-I | Late 30s–early-40s years; EOAD with a probable familial | Possible pathogenic mechanisms of mutation | Korea | This study | ||
| p. Thr119Ile | HL-I | 49–64 years; EOAD with a probable familial | |||||
| p.Glu120Lys | HL-I | 40–65 years/Familial, EOAD | Probably pathogenic | Iran | Akbari et al., 2013 [34] | ||
| p.Glu123Lys | HL-I | 26–45 years, EOAD, myoclonus, epilepsy | Abeta42/total Abeta increased in COS-1 cells (2.7 *) and in HEK293 (4 *) cells | Japan | Yasuda et al. 1999 [35] | ||
| p.Ala136Gly | TM-II | Unknown, EOAD | Survival of mutant neuroblastoma cells dropped, deleterious effects | China | Fang et al., 2007 [36] | ||
| p.Met139Ile | TM-II | 38 years/Familial, EOAD | Ratio of Abeta42/total Abeta increased in COS-1 cell lines. | Korea | Kim et al., 2010 [37] | ||
| p. Ile143Thr | TM-II | 26–45 years, EOAD, myoclonus, epilepsy | Abeta42/total Abeta increased in COS-1 cells (2.7 *) and in HEK293 (4 *) cells | Japan | Arai et al., 2008 [38] | ||
| p.Tyr154Asn | TM-II | 40–60 years, EOAD, spastic paraparesis | Pathogenic nature might be associated with the missing aromatic ring. | Japan | Hattori et al., 2004 [47] | ||
| 6 | p.His163Arg | HL-II | 43–50 years/5 Japanese families, both familial and de novo cases | Abeta42/Abeta40 ratio increased 2 * in COS1 cell lines | Japan | Kamino et al., 1996 [32] | |
| p.His163Arg | HL-II | 43–50 years/5 Japanese families, both familial and de novo cases | Abeta42/Abeta40 ratio increased 2 * in COS1 cell lines | Korea | Hong et al., 1997 [48] | ||
| p.His163Pro | HL-II | 35 years/de novo EOAD, parkinsonism | The rigid proline might result abnormalities in the border of HL-II and TM-III | Korea | This study | ||
| p.Trp165Gly | TM-III | 34–38 years; EOAD with strong familiar | The small glycine is a rare amino acid in the helix | Japan | Higuchi et al., 2000 [55] | ||
| p.Trp165Cys | TM-III | 55 years; memory decline, followed by difficulty in finding ways and had a strong family history of dementia | Increased Aβ42 and decreased Aβ40 production in vitro; elevated Aβ42/Aβ40 ratio | Korea | This study | ||
| 45 years; EOAD, a severe form of the illness, with cerebral and cerebellar atrophies and rapid deterioration | India | Syama et al., 2018 [49] | |||||
| p.Ile167del | TM-III | 38 years/familial; EOAD, spastic paraparesis | Deletion might result abnormal conformation in TM-III | China | Jiao et al., 2014 [31] | ||
| p.Ser169del | TM-III | EOAD, 42–50 years/familial | Missing –OH group might result a missing H-bound in the TM-III | China | Guo et al., 2010 [43] | ||
| p.Leu173Phe | TM-III | 47–50/familial; EOAD with parkinsonism | Elevated Abeta42 levels and Abeta42/Abeta40 ration in neuroblastoma cells | Japan | Kasuga et al. 2009 [50] | ||
| 7 | p.Glu184Asp | HL-III | 40s years; EOAD, DLB-like phenotype | The smaller asparatic acid might change the loop conformation | Japan | Yasuda et al. 1997 [35] | |
| p.Glu184Gly | HL-III | 40s years; probable autosomal dominant EOAD, frontal variant form | Resulting potential functional alterations; may also disturb the splicing near exon 7 | Thailand | This study | ||
| p.Gly206Ser | TM-IV | 30–35 years/familial, EOAD | Probably pathogenic | Korea | Park et al., 2008 [29] | ||
| p.Gly209Arg | TM-IV | 46–53 years, EOAD | Arginine might result extra stress inside the helix and form abnormal hydrogen bonds | Japan | Sugiyama et al., 1999 [44] | ||
| p.Gly209Ala | TM-IV | 54 years; MCI with depression, followed by progressive deterioration in verbal and visual memory | The extra –CH3 group in alanine might result extra stress inside the TM-IV region | Korea | This study | ||
| p.Ile213Thr | TM-IV | 42–47 years, EOAD | Increased (1.7 * Abeta) | Japan | Kamino et al., 1996 [32] | ||
| p.Gly217Asp | HL-IV | 42–47 years/familial, EOAD | Increased (1.7 * Abeta) | Japan | Takao et al., 2002 [52] | ||
| p.Leu226Phe | TM-V | 37 years; de novo, Aβ plaques observed | Results elevated Abeta42/Abeta40 ratio in HEK293 cells | Korea | This study | ||
| p.Leu226Arg | TM-V | 60 years/familial, EOAD | Probably pathogenic | China | Ma et al., 2019 [41] | ||
| p.Glu311Arg | TM-V | > 65 years, familial, LOAD | Overproducing toxic Aβ species and enhancing tau phosphorylation | China | Dong et al., 2017 [56] | ||
| p.Leu232Pro | TM-V | 37 years/familial; EOAD | The rigid proline might result serious torsion in the TM-V since proline is helix breaker | Korea | This study | ||
| p.Met233Thr | TM-V | 34 years/de novo, EOAD, rapid progressive memory impairment | Elevated (3.2 *) Abeta42/Abeta40 levels in CHO cells | Korea | Park HK et al., 2008 [29] | ||
| p.Phe237Ile | TM-V | 35 years/de novo, EOAD, spastic paraparesis | Probably pathogenic | Japan | Sodeyama et al. 2001 [57] | ||
| p.Leu248Pro | TM-VI | 42 years/familial, EOAD | Proline is a helix breaker, resulting in torsion in TM-IV | China | Jiao et al., 2014 [31] | ||
| p.Leu250Val | TM-VI | 40–51 years/Familial, EOAD, myoclonus, seizures | Probably pathogenic | Japan | Furuya t al., 2003 [58] | ||
| 8 | p.Ala260Val | TM-VI | 27–46 years/Familial, EOAD, Pick inclusions | 1.5 * Increased Abeta42/total Abeta in COS1 cells | Japan | Ikeda et al., 1996 [59] | |
| p.Gly266Ser | HL-VI(a) | 35–44 years, EOAD, spastic paraparesis, aphasia | Probably pathogenic | Japan | Matsubara-Tsutsui et al., 2002 [60] | ||
| p.Arg 269His | HL-VI(a) | 46–67 years/Familial, EOAD, myoclonus | Unknown | Japan | Kamimura el al., 1998 [61] | ||
| p.Glu273Ala | HL-VI(a) | 46–67 years/Familial, EOAD, myoclonus | Unknown | Japan | Kamimura el al., 1998 [61] | ||
| p.Glu280Ala | HL-VI (MA) | 48–57 years/Familial, EOAD, parkinsonism | Probably pathogenic | Japan | Tanahashi et al., 1996 [62] | ||
| p.Glu280Lys | HL-VI (MA) | 48–57; EOAD | Probably pathogenic | Malaysia | This study | ||
| p.Leu282Phe | HL-VI (MA) | 51 years, familial, EOAD | Probably pathogenic | Japan | Hamaguchi et al., 2009 [63] | ||
| p.Pro284Leu | HL-VI (MA) | 32 years, cotton-wool plaques and neurofibrillary tangles or amyloid angiopathy in brain | Probably pathogenic | Japan | Tabira et al., 2002 [64] | ||
| p.Ala285Val | HL-VI (MA) | 46 year/de novo, EOAD | The Abeta42/total Abeta ratio increased; Abeta40/total Abeta and Abeta38/total Abeta ratios decreased | Korea | This study | ||
| 50.5 years, two families | Japan | Ikeuchi et al., 2008 [65] | |||||
| p.Leu286Val | HL-VI (MA) | 47 years | Increases in the Abeta42/total Abeta ratio (1.5 *) and Abeta42/Abeta40 ratio (2.1 *) | Japan | Ikeuchi et al., 2008 [65] | ||
| Intron 8 | Exon9 del | - | 47.5 years, in EOAD with spastic paraparesis | elevated Abeta42 levels and Abeta42/40 ratio were observed | Japan | Tabira et al., 2002 [64] | |
| 10 | p.Arg352Cys | HL-VI (b) | 56–62 years, EOAD, psychiatric, behavioral symptoms | Cysteine could result intramolecular disulfide bound | China | Jiang et al., 2015 [66] | |
| 11 | p.Gly378Glu | TM-VII | 37 years, EOAD, familiar positive | Abeta42/Abeta40 ratio increased (3.2 *) | Japan | Ikeda et al., 1996 [59] | |
| p.Leu381Val | TM-VII | 30s years, AD and spastic paraparesis | Abeta42/Abeta40 ratio increased (1.9 *) | Japan | Ikeuchi et al., 2008 [65] | ||
| p.Gly384Ala | TM-VII | 31–37 years, EOAD, senile plaques and tangles inside proband’s brain | Beta40 and the Abeta42/Abeta40 ratio decreased and increased significantly. Abeta42/total Abeta ratio increased (3.8 *) | Japan | Kamimura et al. 1998 [61] | ||
| p.Leu392Val | TM-VII | 42 years, EOAD | Abeta42/Abeta40 ratio (2.4*). An increase in the Abeta42/Abeta40 ratio (2.9 *) | Japan | Ikeuchi et al. 2008 [65] | ||
| p.Asn405Ser | HL-VII | EOAD, the patient has several senile plaques and tangles in the brain | It caused disturbances in the motor neuronal systems, leading to spastic paraparesis | Japan | Yasuda et al., 2000 [46] | ||
| p.Gly417Ala | HL-VIII | 37 years; EOAD, parkinsonism, positive familiar | Pathogenic mechanism | Korea | This study | ||
| 12 | p.Ala431Val | HL-VIII | 16 months, t-tau and phospho-Tau levels increased in the CSF, and metabolic deficits were detected in several parts of the brain | Possibly pathogenic | Japan | Matsushita et al., 2002 [45] | |
| p.Ala434Thr | HL-VIII | 38 years, EOAD, Hallucinations, delusions |
Threonine might result extramolecular or intramolecular hydrogen bound | China | Jiao et al., 2014 [31] | ||
| p.Thr440del | HL-VIII | 52 years, strong familiar history, EOAD and parkinsonism | Probably pathogenic, may alter the normal amyloid production | Japan | Ishikawa et al., 2005 [42] | ||
| PSEN2 | 4 | p.Arg62Cys | N-term | 49 years, EOAD | Possibly pathogenic, may alter the normal amyloid production. | Korea | This study |
| 40–65 years, EOAD | Iran | Akbari et al., 2013 [34] | |||||
| 5 | p.Asn141Tyr | TM-II | 43–49 years, EOAD | No functional data | China | Niu et al., 2014 [39] | |
| 6 | p.His169Asn | TM-III | 50 years; de novo | It may result in major helix torsion due to histidine to asparagine substitution | Korea | This study | |
| 62 years; AD, de novo | China | Shi Z et al., 2015 [40] | |||||
| 68 years; FTD, progressive nonfluent aphasia, Familial | |||||||
| 63 years/Familial, LOAD | China | Ma et al., 2018 [41] | |||||
| 7 | p.Val214Leu | TM-IV | 56–70 years; AD | The extra CH3 group in leucine could result extra stress in the TM-IV region | Korea | This study |
Abbreviation: APP, amyloid precursor protein; PSEN1, presenilin-1; PSEN2, presenilin-2; AD, Alzheimer’s disease; EOAD, early-onset Alzheimer’s disease; LOAD, late-onset Alzheimer’s disease; MCI, mild cognitive impairment; DLB, dementia with Lewy bodies; FTD, frontotemporal dementia; HEK293, human embryonic kidney 293; CHO, Chinese hamster ovary; COS-1, cercopithecus aethiops kidney; TM, transmembrane domain; * multiplication sign.
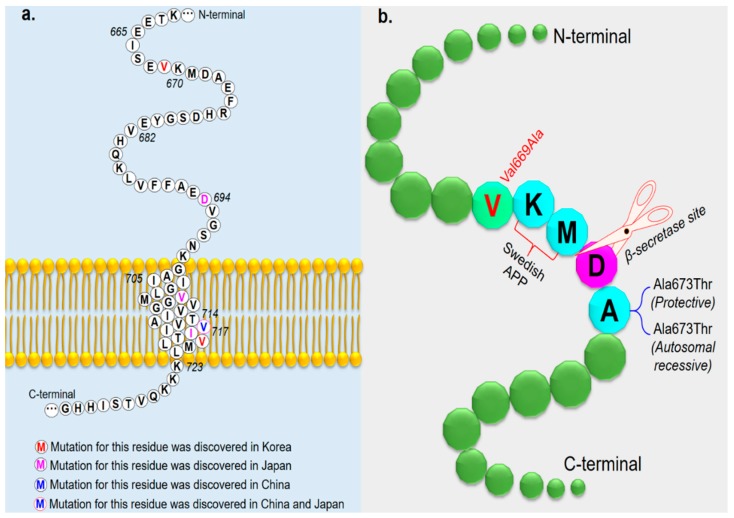
Six novel APP mutations were found in six out of 200 EOAD patients (Table 1). A novel mutation, c.2005G > C, p.(Val669Leu) substitution, present in a 56-year-old Korean female and two of her daughters [67]. The clinical features were typical of AD with aggravated diffuse brain atrophy and a small vessel ischemic lesion. The c.1810C > T, p.(Val604Met) mutation was found in a Thai patient with EOAD [12]. The patient was diagnosed in 2013 with AD presenting logopenic aphasia, and this variant appeared to be associated with the phenotype [12]. Three APP variants—c.674T > C (p.Val225Ala); c.1450C > T, (p.Pro484Ser); and c.890C > T, (p.Thr297Met)—were found in Korean patients with EOAD at an onset age between 60 and 65 years. Although these three variants have not been previously reported in the literature, their allele frequencies in the ExAC database are 0.00002471, 0.00003304, and 0.0002062, respectively. Only one novel APP mutation—c.1810C > T p.(Val604Met)—was identified during this screen in a Thai patient with EOAD; this mutation is presumed to be associated with altered APP function due to increased hydrophobicity of methionine in the helix [12]. Over 35 APP variants have been discovered in exons 16 and 17; of them, 10 have been reported in Asia (Table 2, Figure 2a). Remarkably, a novel mutation in the APP gene, Val669Leu, was discovered in a Korean female patient with AD [67]. She developed cognitive decline at the age of 56 years, and MRI scans showed mild global atrophy with medial temporal lobe predominance and hippocampal atrophy. The patient may have a positive history of the disease, since her mother was also diagnosed. APP V669L was predicted as the non-damaging variant by the PolyPhen2 and Sorting Intolerant From Tolerant (SIFT) tools. APP mutations are rare in Korean populations because of the presence of only one mutation in APP, V715M (V715M). APP V669L is located near the β-secretase cleavage site, adjacent to the Swedish APP (KM670/671NL) mutation (Figure 2b) [68]. This mutation may disrupt amyloid-beta metabolism.
Figure 2.
(a) Mutated APP residues identified in Asian countries are shown in different colors. (b) Location of Val669Leu in APP and mutations located near the β-secretase cleavage site. The nearest mutation is the “Swedish APP” mutation. Additional mutations located near the β-secretase cleavage site are the protective Ala673Thr and the pathogenic Ala673Val.
2.2. Identified Gene Mutations of PSEN1
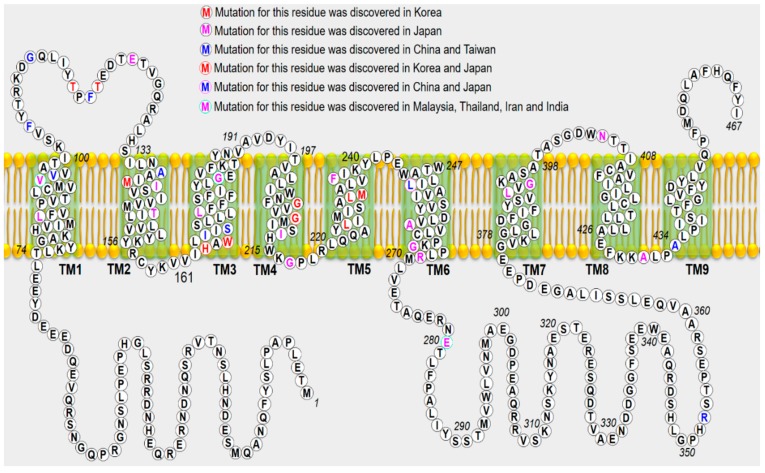
Twelve PSEN1 mutations were identified in 20 patients with EOAD (Table 1). Remarkably, a previously reported PSEN1 mutation—Val96Phe—was identified in two siblings from Malaysia. This mutation was reported previously in a Japanese family with disease onset in the late 40s or 50s [32]. Similar to Japanese patients, the disease onset in these siblings was in the 40s, and they presented symptomatic changes in behavior and personality, such as apathy and withdrawal. In addition, seven additional novel or known PSEN1 mutations, including Thr116Ile, Thr119Ile His163Pro, Leu226Phe, Gly209Ala, Leu232Pro [69], and Gly417Ala [11], have been identified in Korean patients with AD. Importantly, even though PSEN1 is the most commonly involved gene, with > 231 mutations reported as pathogenic in the Alzforum database (www.alzforum.org/mutations), this study did not find any PSEN1 mutation in the Thai and Philippine cohorts. Moreover, only three Malaysian patients with AD have been identified to carry a novel mutation, Glu280Lys [70]. As Korea is one of the fastest “aging countries” in the world, the number of AD, including EOAD, patients will rapidly increase [24]. The carriers of PSEN1 mutation presented with isolated and progressive cognitive decline. Another patient carrying the PSEN1 p.Gly417Ala substitution also exhibited an atypical presentation: Cerebellar ataxia and extra pyramidal with pessimism syndrome. According to the Alzheimer’s Research Forum database, more than 230 PSEN1 variants have been identified worldwide (www.alzforum.org/mutations). Among them, > 55 variants have been identified in Japan, Korea, the People’s Republic of China, Malaysia, and Thailand (Figure 3).
Figure 3.
Mutated PSEN1 residues identified in the Asian countries are shown in different colors. The predicted membrane topology of PSEN1 with the nine transmembrane domains (green shaded boxes) and boundaries between coding exons is shown. TM, transmembrane domain.
2.3. Identified Gene Mutations of PSEN2
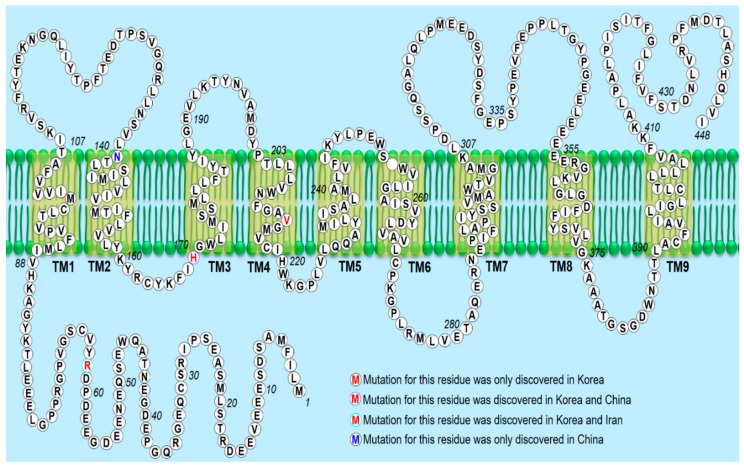
We also discovered the following three PSEN2 mutations in Korean patients for the first time: Arg62Cys, His169Asn [9], and Val214Leu. Arg62Cys (CGC TGC) was discovered in the Asian population for the first time by our research group. The mutation was identified in a patient with dementia. Memory impairment, personality change, and disorientation appeared at the age of 49 years. Val214Leu was one of the first PSEN2 mutations identified in an Asian population. In addition, it is the first mutation identified in the TM-IV region of PSEN2. Val214Leu mutation was identified in the following two unrelated patients: A 70-year-old patient with AD-type dementia and a 56-year-old patient with memory impairment. The exact family history is unknown for both patients. A pathogenic mutation p.His169Asn in the PSEN2 gene in a Korean patient with EOAD has also been identified [9]. PolyPhen-2 and SIFT software analyses predicted this mutation to be a probable damaging variant. The mutation was identified in a 58-year-old woman who was presented with progressive memory decline in her 50s. The patient had an apolipoprotein E genotype (APOE) ε 3/3 polymorphism. The family history of the proband generations was negative for any neurological disease, indicative of a de novo case of AD. All living family members declined genetic testing. Interestingly, PSEN2 p.His169Asn mutation was previously identified in one patient with familial LOAD and one patient with sporadic frontotemporal dementia (FTD) from People’s Republic of China [40]; however, the pathogenic nature has not been clarified yet. Compared with the two Chinese patients, the Korean patient showed similar clinical manifestation with the proband with frontal variant AD. Although no additional mutation was reported at residue 169 of the PSEN2 protein, the p.His169Asn mutation was found in the conserved TM-III region of PSEN2, containing the pathogenic variants (p.M174V and p.S175C), based on the algorithms to predict the pathogenicity of the mutations described by Guerreiro et al. [71] More than 40 missense and frameshift mutations in the PSEN2 gene have been reported so far; however, until 2019, no pathogenic mutation has been found in PSEN2 in any Asian country. The findings of this study as well as those of recent studies revealed novel and known PSEN2 variants in Korean and Chinese patients (Figure 4).
Figure 4.
Mutated PSEN2 residues identified in the Asian countries are shown in different colors. The predicted membrane topology of PSEN2 with the nine transmembrane domains (yellow shaded boxes) and boundaries between coding exons are shown. TM, transmembrane domain.
3. Discussion
In this study, we performed genetic screening for mutations in the APP, PSEN1, and PSEN2 genes in 200 clinically diagnosed EOAD patients across four Asian countries, including Thailand, Malaysia, the Philippines, and Korea from 2009 to 2018, and identified 21 novel and known missense mutations. According to Guerreiro’s algorithm [71], pathogenicity was considered as “definite” for two APP, “probable” for nine PSEN1, and “possible” for three PSEN2 mutations. The pathological effect of the known mutations deserves discussion because of incomplete penetrance, nonpathogenicity, or a wide range of age onset [41,47,52,56,57,58,59,60,61,63,64,65].
This study detected six mutations in APP, among patients under 65 years of age. Among them, only one APP, the Val669Leu mutation, was located in the amyloid processing area. The patient with APP Val669Leu presented had a progressive short-term memory impairment, as observed in typical AD. However, atypical symptoms of AD, including focal signs and symptoms, were also observed. Frontal lobe impairment (depression, apathy, and disinhibition), epileptic seizures, and myoclonus were also observed. As APP is responsible for the disease it is located relatively near to the beta secretase cleavage site; therefore, Val669Leu may interfere with the normal proteolytic processing of APP. This mechanism is thought to involve alternative proteolytic processing pathways [5,6,7,47,72].
PSEN1 c.286G > T, p.(Val96Phe) substitution was identified in two siblings from Malaysia. This was the second report of the PSEN1 Val96Phe mutation among EOAD patients in Asia. Patients presented similar phenotypes like the previously described Japanese patients: The disease onset was in their 40s, and they presented a symptomatic change in behavior and personality, such as apathy and withdrawal. PSEN1 Val96Phe mutation is considered pathogenic and can lead to an increase in Aβ42 level and Aβ42/Aβ40 ratio in cell cultures [32]. These findings suggest that mutations in TM-I may be responsible for pathogenic mutations in EOAD. Cellular studies with different mutations (including Val96Phe) suggest that TM-I plays a significant role in APP trafficking and amyloid peptide cleavage. Therefore, we speculate about an underestimation of its frequency. In addition, two PSEN1 mutations—Trp165Cys [73] and p.Ala285Val—were identified in a 53-year-old male who presented memory decline, followed by disorientation, and in a 46-year-old woman who presented with progressive memory dysfunction, respectively. Both patients had probable EOAD, and the family history was positive in them. Both mutations were previously shown to have increased Aβ42 and decreased Aβ40 levels. Moreover, both mutations could elevate the Aβ42/Aβ40 ratio by impairing the gamma secretase functions [49,59,72,74,75,76].
Similar to APP and PSEN1 mutations, PSEN2 mutations can also enhance Aβ production and contribute to AD development. An extensive literature search for PSEN2 mutations was conducted. Thirty-eight PSEN2 mutations have been reported yet, and most of these mutations were identified in European and African populations. Until now, only five missense mutations have been reported in Asian populations. Asn141Tyr was identified in a Chinese Han patient with EOAD [77], Gly34Ser was found in a Japanese patient [4], and three possibly pathogenic mutations—Arg62Cys, Val214Leu, and His169Asn—were reported in this study. PSEN2 mutations are associated with variable penetrance and a wide age range of disease onset, from 45 to 88 years [78,79]. PSEN2 is a transmembrane protein and a component of γ-secretase intramembrane protease, and is involved in various signaling pathways in AD development [80,81].
In an EOAD patient cohort, the estimated mutation frequencies for the three genes were < 1% for APP, 6% for PSEN1, and 1% for PSEN2 [82]. Together, they explain only 5% to 10% of the mutational profile in patients with EOAD [82,83]; however, approximately 90% of the mutations remain genetically unexplained [1,84]. With the exception of Korea, the People’s Republic of China, Taiwan, and Japan, limited reports are available on EOAD-associated mutations in Asian countries (Table 2). Two APP mutations have been identified in patients from Thailand [12] and Iran [85]. Recently, a novel PSEN1 mutation was reported in a Malaysian family [70]. Our primary goal was to provide clinicians a list of variants that can be accurately used in genetic counseling. Considering our whole cases, this goal is achieved for 9% mutations reported in the Asian population. Limited reports are available regarding EOAD-associated mutations in other East Asian countries. Hence, our investigators have begun efforts for screening AD-related mutations across Asian countries through collaborations. Compared with Caucasian patients, over 30 novel EOAD-associated mutations have been found in the APP, PSEN1, and PSEN2 genes in Asian patients (http://www.alzforum.org/mutations). Since the overall population and aging population in most Asian countries is increasing, genetic testing of patients with AD and other types of dementia is important for the diagnosis of dementia.
A limitation of this study is the absence of functional assessment of the possible and probable pathogenic variants, which could simplify their classification [8]. Moreover, only three genes were analyzed. It is possible that de novo mutations in other genes are also involved in the genetic determination of sporadic forms [16,18,86,87,88,89,90]. The limited number of resolved pedigrees and large number of genetically unexplained EOAD patients indicate that additional causal genes remain to be discovered. The next step involves performing whole exome/genome sequencing on negatively screened families and sporadic cases.
In conclusion, among the distinct mutations in the Asian patients and isolated cases in the Asian population, definite pathogenicity accounted for less than 16%, leaving a large group of autosomal dominant pedigrees genetically unexplained. In addition, our findings suggest that continuing the investigation of families harboring known mutations and the elucidation of the missing genetic etiology in unexplained EOAD patients has a vast potential to improve our understanding about the complexity of AD [1,10,15,90,91]. We also suggest that the use of high-throughput sequencing technologies for patients with EOAD and data integration from other -omics analyses (epigenomics, proteomics, transcriptomics, and metabolomics) might help in better understanding the underlying molecular mechanisms of AD.
4. Materials and Methods
Two-hundred patients with EOAD from the University Hospitals of Korea, Malaysia, Thailand, and the Philippines were recruited between 2009 and 2018. All patients underwent a comprehensive clinical examination, including personal medical and family history assessment and neuropsychological assessment. For each patient, AD diagnosis was established using the National Institute of Aging–Alzheimer’s Association (NIA–AA) criteria [92]. The project received ethics approval from the Seoul National University College of Medicine of Seoul National Bundang Hospital (SNUH), and written informed consent was obtained from all participants according to the requirements of the Seoul National Bundang Human Research Committee (B-1302/192-006, approval date: 15/03/2013). All procedures involving human participants were conducted in accordance with the ethical standards of the institutional and/or national research committee and 1964 Helsinki Declaration and its later amendments.
4.1. Genetic Analyses
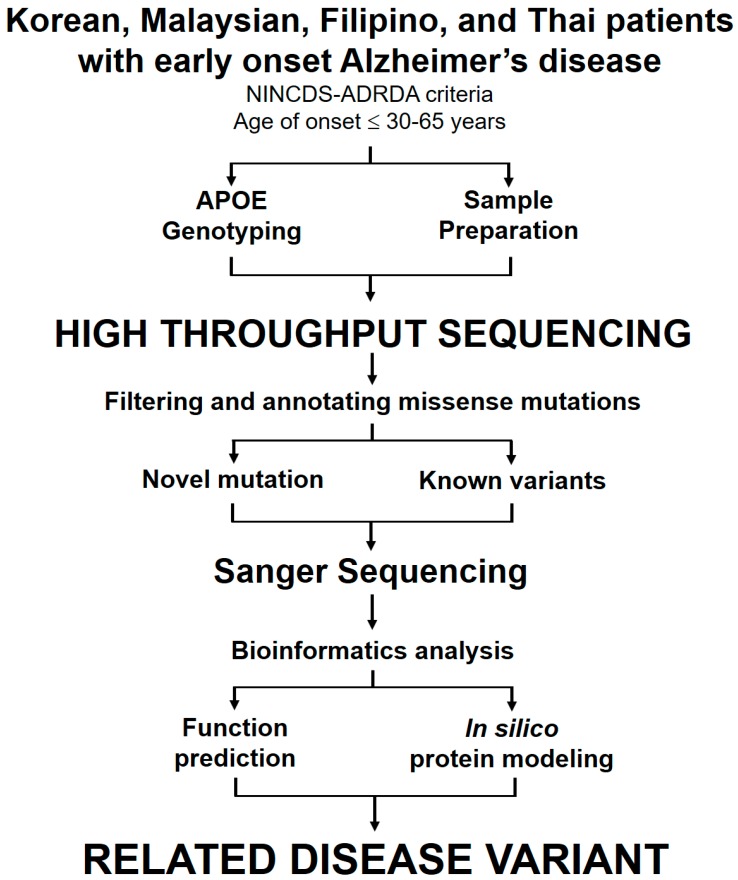
Genetic analyses were performed on DNA extracted from whole blood. Sanger sequencing, next-generation sequencing (NGS) and whole exon sequencing (WES) were employed to search for mutations in the APP, PSEN1, and PSEN2 genes in patients with both sporadic and family history of AD. APOE genotypes comprising the APOE ε2, ε3, and ε4 alleles were assayed [15]. To confirm the presence of the identified mutations, standard sequencing was performed in both directions using the previously used primer set [1,2]. Prior to sequencing, PCR products were purified using the GeneAll PCR protocol kit (Seoul, Korea), following the manufacturer’s protocol. Big Dye Terminator Cyclic sequencing was performed on an ABI 3730XL DNA Analyzer (http://eng.bioneer.com/home.aspx, Bioneer Inc., Dajeon, Korea). The sequenced product was aligned using the NCBI Blast tool (http://blast.ncbi.nlm.nih.gov/Blast.cgi), and chromatograms were screened using the DNA BASER (http://www.dnabaser.com). Mutations and sequence variants were identified from the NCBI Gene (http://www.ncbi.nlm.nih.gov/gene) and UniProt (http://www.uniprot.org) databases. Briefly, patients with EOAD were analyzed by high-throughput sequencing, following the schematic diagram shown in Figure 5.
Figure 5.
High-throughput sequencing strategy for identifying gene variants in AD patients. The flow chart illustrates the major steps of the working procedure from patient sample analysis to the identification of mutation.
4.2. Bioinformatics
To determine whether APP, PSEN1, and PSEN2 variants presented rare or common polymorphisms, the variants were checked in the Korean Genome Reference Database (http://152.99.75.168/KRGDB/menuPages/firstInfo.jsp) for their novelty. The full genome sequences of 622 asymptomatic individuals were obtained by whole genome sequencing. In addition, variants were also checked in other large-scale genome reference databases, including the 1000 Genomes (http://www.internationalgenome.org/) and Exome Aggregation Consortium (ExAC; http://exac.broadinstitute.org) databases. Polymorphism phenotype v2 (PolyPhen-2) and Sorting Intolerant From Tolerant (SIFT) were used to predict whether the amino acid change would be disruptive to the encoded protein.
Acknowledgments
The authors would like to thank Gaik-Siew Ch’ng (Department of Genetics, Kuala Lumpur Hospital, Kuala Lumpur, Malaysia), Vorapun Senanarong (Faculty of Medicine Siriraj Hospital, Mahidol University, Bangkok, Thailand), and Antonio D. Ligsay (Institute for Neurosciences, St. Luke’s Medical Center, Philippine) for their continued collaboration for research on neurodegeneration.
Author Contributions
Conceptualization: V.V.G., E.B., S.S.A.A., Y.C.Y., and S.Y.K.; Investigation: V.V.G., S.S.A.A., Y.C.Y., and S.Y.K.; Formal Analysis: V.V.G, and E.B.; Writing-Original Draft Preparation and Writing-Review & Editing: V.V.G; Supervision and project administration: S.S.A.A., Y.C.Y. and S.Y.K.
Funding
This research was supported by a National Research Foundation of Korea (NRF) grant awarded by the Korean government (NRF-2017R1A2B4012636 & NRF-2019R1G1A109740011).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Giau V.V., Bagyinszky E., An S.S.A., Kim S. Clinical genetic strategies for early onset neurodegenerative diseases. Mol. Cell. Toxicol. 2018;14:123–142. [Google Scholar]
- 2.Van Giau V., An S.S.A., Bagyinszky E., Kim S. Gene panels and primers for next generation sequencing studies on neurodegenerative disorders. Mol. Cell. Toxicol. 2015;11:89–143. [Google Scholar]
- 3.Sun L., Zhou R., Yang G., Shi Y. Analysis of 138 pathogenic mutations in presenilin-1 on the in vitro production of Abeta42 and Abeta40 peptides by gamma-secretase. Proc. Nat. Acad. Sci. USA. 2017;114:E476–E485. doi: 10.1073/pnas.1618657114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai Y., An S.S.A., Kim S. Mutations in presenilin 2 and its implications in Alzheimer’s disease and other dementia-associated disorders. Clin. Interv. Aging. 2015;10:1163–1172. doi: 10.2147/CIA.S85808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goate A., Chartier-Harlin M.C., Mullan M., Brown J., Crawford F., Fidani L., Giuffra L., Haynes A., Irving N., James L., et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer’s disease. Nature. 1991;349:704–706. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- 6.Chartier-Harlin M.C., Crawford F., Houlden H., Warren A., Hughes D., Fidani L., Goate A., Rossor M., Roques P., Hardy J., et al. Early-onset Alzheimer’s disease caused by mutations at codon 717 of the beta-amyloid precursor protein gene. Nature. 1991;353:844–846. doi: 10.1038/353844a0. [DOI] [PubMed] [Google Scholar]
- 7.Rovelet-Lecrux A., Hannequin D., Raux G., le Meur N., Laquerriere A., Vital A., Dumanchin C., Feuillette S., Brice A., Vercelletto M., et al. APP locus duplication causes autosomal dominant early-onset Alzheimer disease with cerebral amyloid angiopathy. Nat. Genet. 2006;38:24–26. doi: 10.1038/ng1718. [DOI] [PubMed] [Google Scholar]
- 8.Giau V.V., Lee H., Shim K.H., Bagyinszky E., An S.S.A. Genome-editing applications of CRISPR-Cas9 to promote in vitro studies of Alzheimer’s disease. Clin. Interv. Aging. 2018;13:221–233. doi: 10.2147/CIA.S155145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giau V.V., Pyun J.-M., Bagyinszky E., An S.S.A., Kim S. A pathogenic PSEN2 p.His169Asn mutation associated with early-onset Alzheimer’s disease. Clin. Interv. Aging. 2018;13:1321–1329. doi: 10.2147/CIA.S170374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giau V.V., Senanarong V., Bagyinszky E., An S.S.A., Kim S. Analysis of 50 Neurodegenerative Genes in Clinically Diagnosed Early-Onset Alzheimer’s Disease. Int. J. Mol. Sci. 2019;20:1514. doi: 10.3390/ijms20061514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giau V.V., Wang M.J., Bagyinszky E., Youn Y.C., An S.S.A., Kim S. Novel PSEN1 p.Gly417Ala mutation in a Korean patient with early-onset Alzheimer’s disease with parkinsonism. Neurobiol. Aging. 2018;72:188.e13–188.e17. doi: 10.1016/j.neurobiolaging.2018.08.003. [DOI] [PubMed] [Google Scholar]
- 12.Van Giau V., Senanarong V., Bagyinszky E., Limwongse C., An S.S.A., Kim S. Identification of a novel mutation in APP gene in a Thai subject with early-onset Alzheimer’s disease. Neuropsychiatr. Dis. Treat. 2018;14:3015–3023. doi: 10.2147/NDT.S180174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campion D., Dumanchin C., Hannequin D., Dubois B., Belliard S., Puel M., Thomas-Anterion C., Michon A., Martin C., Charbonnier F., et al. Early-onset autosomal dominant Alzheimer disease: Prevalence, genetic heterogeneity, and mutation spectrum. Am. J. Hum. Genet. 1999;65:664–670. doi: 10.1086/302553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Selkoe D.J. Alzheimer’s disease: Genotypes, phenotypes, and treatments. Science. 1997;275:630–631. doi: 10.1126/science.275.5300.630. [DOI] [PubMed] [Google Scholar]
- 15.Giau V.V., Bagyinszky E., Yang Y.S., Youn Y.C., An S.S.A., Kim S.Y. Genetic analyses of early-onset Alzheimer’s disease using next generation sequencing. Sci. Rep. 2019;9:8368. doi: 10.1038/s41598-019-44848-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen L., An S.S.A., Bagyinszky E., van Giau V., Choi S.H., Kim S.Y. Novel GRN mutations in Koreans with Alzheimer’s disease. Mol. Cell. Toxicol. 2019;15:345–352. [Google Scholar]
- 17.Bagyinszky E., Giau V.V., Shim K., Suk K., An S.S.A., Kim S. Role of inflammatory molecules in the Alzheimer’s disease progression and diagnosis. J. Neurol. Sci. 2017;376:242–254. doi: 10.1016/j.jns.2017.03.031. [DOI] [PubMed] [Google Scholar]
- 18.Bagyinszky E., Kang M.J., Pyun J., Giau V.V., An S.S.A., Kim S. Early-onset Alzheimer’s disease patient with prion (PRNP) p.Val180Ile mutation. Neuropsychiatr. Dis. Treat. 2019;15:2003–2013. doi: 10.2147/NDT.S215277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giau V.V., Bagyinszky E., An S.S.A. Potential Fluid Biomarkers for the Diagnosis of Mild Cognitive Impairment. Int. J. Mol. Sci. 2019;20:4149. doi: 10.3390/ijms20174149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giau V.V., Wu S.Y., Jamerlan A., An S.S.A., Kim S.Y., Hulme J. Gut Microbiota and Their Neuroinflammatory Implications in Alzheimer’s Disease. Nutrients. 2018;10:1765. doi: 10.3390/nu10111765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang M.J., Yi S., Han J.Y., Park S.Y., Jang J.W., Chun I.K., Giau V.V., Bagyinszky E., Lim K.T., Kang S.M., et al. Analysis of Cerebrospinal Fluid and [11C]PIB PET Biomarkers for Alzheimer’s Disease with Updated Protocols. J. Alzheimer’s Dis. JAD. 2016;52:1403–1413. doi: 10.3233/JAD-160143. [DOI] [PubMed] [Google Scholar]
- 22.Van Giau V., An S.S.A., Hulme P.J. Mitochondrial therapeutic interventions in Alzheimer’s disease. J. Neurol. Sci. 2018;395:62–70. doi: 10.1016/j.jns.2018.09.033. [DOI] [PubMed] [Google Scholar]
- 23.Lanoiselee H.M., Nicolas G., Wallon D., Rovelet-Lecrux A., Lacour M., Rousseau S., Richard A.C., Pasquier F., Rollin-Sillaire A., Martinaud O., et al. APP, PSEN1, and PSEN2 mutations in early-onset Alzheimer disease: A genetic screening study of familial and sporadic cases. PLoS Med. 2017;14:e1002270. doi: 10.1371/journal.pmed.1002270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bagyinszky E., Youn Y.C., An S.S.A., Kim S. Mutations, associated with early-onset Alzheimer’s disease, discovered in Asian countries. Clin. Interv. Aging. 2016;11:1467–1488. doi: 10.2147/CIA.S116218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wakutani Y., Watanabe K., Adachi Y., Wada-Isoe K., Urakami K., Ninomiya H., Saido T.C., Hashimoto T., Iwatsubo T., Nakashima K. Novel amyloid precursor protein gene missense mutation (D678N) in probable familial Alzheimer’s disease. J. Neurol. Neurosurg. Psychiatry. 2004;75:1039–1042. doi: 10.1136/jnnp.2003.010611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tomiyama T., Nagata T., Shimada H., Teraoka R., Fukushima A., Kanemitsu H., Takuma H., Kuwano R., Imagawa M., Ataka S., et al. A new amyloid beta variant favoring oligomerization in Alzheimer’s-type dementia. Ann. Neurol. 2008;63:377–387. doi: 10.1002/ana.21321. [DOI] [PubMed] [Google Scholar]
- 27.Thajeb P., Wang P., Chien C.L., Harrigan R. Novel polymorphisms of the amyloid precursor protein (APP) gene in Chinese/Taiwanese patients with Alzheimer’s disease. J. Clin. Neurosci. 2009;16:259–263. doi: 10.1016/j.jocn.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 28.Pasalar P., Najmabadi H., Noorian A.R., Moghimi B., Jannati A., Soltanzadeh A., Krefft T., Crook R., Hardy J. An Iranian family with Alzheimer’s disease caused by a novel APP mutation (Thr714Ala) Neurology. 2002;58:1574–1575. doi: 10.1212/wnl.58.10.1574. [DOI] [PubMed] [Google Scholar]
- 29.Park H.K., Na D.L., Lee J.H., Kim J.W., Ki C.S. Identification of PSEN1 and APP gene mutations in Korean patients with early-onset Alzheimer’s disease. J. Korean Med. Sci. 2008;23:213–217. doi: 10.3346/jkms.2008.23.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshioka K., Miki T., Katsuya T., Ogihara T., Sakaki Y. The 717Val—Ile substitution in amyloid precursor protein is associated with familial Alzheimer’s disease regardless of ethnic groups. Biochem. Biophys. Res. Commun. 1991;178:1141–1146. doi: 10.1016/0006-291x(91)91011-z. [DOI] [PubMed] [Google Scholar]
- 31.Jiao B., Tang B., Liu X., Xu J., Wang Y., Zhou L., Zhang F., Yan X., Zhou Y., Shen L. Mutational analysis in early-onset familial Alzheimer’s disease in Mainland China. Neurobiol. Aging. 2014;35:1957.e1–1957.e6. doi: 10.1016/j.neurobiolaging.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 32.Kamino K., Sato S., Sakaki Y., Yoshiiwa A., Nishiwaki Y., Takeda M., Tanabe H., Nishimura T., Ii K., George-Hyslop P.H.S., et al. Three different mutations of presenilin 1 gene in early-onset Alzheimer’s disease families. Neurosci. Lett. 1996;208:195–198. doi: 10.1016/0304-3940(96)12587-8. [DOI] [PubMed] [Google Scholar]
- 33.Fang B., Jia L., Jia J. Chinese Presenilin-1 V97L mutation enhanced Abeta42 levels in SH-SY5Y neuroblastoma cells. Neurosci. Lett. 2006;406:33–37. doi: 10.1016/j.neulet.2006.06.072. [DOI] [PubMed] [Google Scholar]
- 34.Akbari L., Noroozian M., Azadfar P., Shaibaninia S., Assarzadegan F., Houshmand M. Investigation of PSEN1, 2 Hot Spots in Iranian Early-Onset Alzheimer’s Disease Patients. Zahedan J. Res. Med. Sci. 2015;17:57–59. [Google Scholar]
- 35.Yasuda M., Maeda K., Hashimoto M., Yamashita H., Ikejiri Y., Bird T.D., Tanaka C., Schellenberg G.D. A pedigree with a novel presenilin 1 mutation at a residue that is not conserved in presenilin 2. Arch. Neurol. 1999;56:65–69. doi: 10.1001/archneur.56.1.65. [DOI] [PubMed] [Google Scholar]
- 36.Fang B.Y., Jia J.P. The effect of two newly Chinese presenilin-1 mutations on the sensitivity to trophic factor withdrawal in human neuroblastoma cells. Zhonghua Yi Xue Za Zhi. 2007;87:336–340. [PubMed] [Google Scholar]
- 37.Kim H.J., Kim H.Y., Ki C.S., Kim S.H. Presenilin 1 gene mutation (M139I) in a patient with an early-onset Alzheimer’s disease: Clinical characteristics and genetic identification. Neurol. Sci. 2010;31:781–783. doi: 10.1007/s10072-010-0233-5. [DOI] [PubMed] [Google Scholar]
- 38.Arai N., Kishino A., Takahashi Y., Morita D., Nakamura K., Yokoyama T., Watanabe T., Ida M., Goto J., Tsuji S. Familial cases presenting very early onset autosomal dominant Alzheimer’s disease with I143T in presenilin-1 gene: Implication for genotype-phenotype correlation. Neurogenetics. 2008;9:65–67. doi: 10.1007/s10048-007-0104-2. [DOI] [PubMed] [Google Scholar]
- 39.Niu F., Yu S., Zhang Z., Yi X., Ye L., Tang W., Qiu C., Wen H., Sun Y., Gao J., et al. Novel mutation in the PSEN2 gene (N141Y) associated with early-onset autosomal dominant Alzheimer’s disease in a Chinese Han family. Neurobiol. Aging. 2014;35:2420.e1–2420.e5. doi: 10.1016/j.neurobiolaging.2014.04.011. [DOI] [PubMed] [Google Scholar]
- 40.Shi Z., Wang Y., Liu S., Liu M., Liu S., Zhou Y., Wang J., Cai L., Huo Y.R., Gao S., et al. Clinical and neuroimaging characterization of Chinese dementia patients with PSEN1 and PSEN2 mutations. Dement. Geriatr. Cogn. Disord. 2015;39:32–40. doi: 10.1159/000366272. [DOI] [PubMed] [Google Scholar]
- 41.Ma L., Zhang J., Shi Y., Wang W., Ren Z., Xia M., Zhang Y., Yang M. Gene mutations in a Han Chinese Alzheimer’s disease cohort. Brain Behav. 2018;9:e01180. doi: 10.1002/brb3.1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ishikawa A., Piao Y.S., Miyashita A., Kuwano R., Onodera O., Ohtake H., Suzuki M., Nishizawa M., Takahashi H. A mutant PSEN1 causes dementia with Lewy bodies and variant Alzheimer’s disease. Ann. Neurol. 2005;57:429–434. doi: 10.1002/ana.20393. [DOI] [PubMed] [Google Scholar]
- 43.Guo J., Wei J., Liao S., Wang L., Jiang H., Tang B. A novel presenilin 1 mutation (Ser169del) in a Chinese family with early-onset Alzheimer’s disease. Neurosci. Lett. 2010;468:34–37. doi: 10.1016/j.neulet.2009.10.055. [DOI] [PubMed] [Google Scholar]
- 44.Sugiyama N., Suzuki K., Matsumura T., Kawanishi C., Onishi H., Yamada Y., Iseki E., Kosaka K. A novel missense mutation (G209R) in exon 8 of the presenilin 1 gene in a Japanese family with presenile familial Alzheimer’s disease. Hum. Mutat. 1999;14:90. doi: 10.1002/(SICI)1098-1004(1999)14:1<90::AID-HUMU19>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 45.Matsushita S., Arai H., Okamura N., Ohmori T., Takasugi K., Matsui T., Maruyama M., Iwatsubo T., Higuchi S. Clinical and biomarker investigation of a patient with a novel presenilin-1 mutation (A431V) in the mild cognitive impairment stage of Alzheimer’s disease. Biol. Psychiatry. 2002;52:907–910. doi: 10.1016/s0006-3223(02)01386-0. [DOI] [PubMed] [Google Scholar]
- 46.Yasuda M., Maeda S., Kawamata T., Tamaoka A., Yamamoto Y., Kuroda S., Maeda K., Tanaka C. Novel presenilin-1 mutation with widespread cortical amyloid deposition but limited cerebral amyloid angiopathy. J. Neurol. Neurosurg. Psychiatry. 2000;68:220–223. doi: 10.1136/jnnp.68.2.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hattori S., Sakuma K., Wakutani Y., Wada K., Shimoda M., Urakami K., Kowa H., Nakashima K. A novel presenilin 1 mutation (Y154N) in a patient with early onset Alzheimer’s disease with spastic paraparesis. Neurosci. Lett. 2004;368:319–322. doi: 10.1016/j.neulet.2004.07.057. [DOI] [PubMed] [Google Scholar]
- 48.Hong K.S., Kim S.P., Na D.L., Kim J.G., Suh Y.L., Kim S.E., Kim J.W. Clinical and genetic analysis of a pedigree of a thirty-six-year-old familial Alzheimer’s disease patient. Biol. Psychiatry. 1997;42:1172–1176. doi: 10.1016/s0006-3223(97)00347-8. [DOI] [PubMed] [Google Scholar]
- 49.Syama A., Sen S., Kota L.N., Viswanath B., Purushottam M., Varghese M., Jain S., Panicker M.M., Mukherjee O. Mutation burden profile in familial Alzheimer’s disease cases from India. Neurobiol. Aging. 2018;64:158.e7–158.e13. doi: 10.1016/j.neurobiolaging.2017.12.002. [DOI] [PubMed] [Google Scholar]
- 50.Kasuga K., Ohno T., Ishihara T., Miyashita A., Kuwano R., Onodera O., Nishizawa M., Ikeuchi T. Depression and psychiatric symptoms preceding onset of dementia in a family with early-onset Alzheimer disease with a novel PSEN1 mutation. J. Neurol. 2009;256:1351–1353. doi: 10.1007/s00415-009-5096-4. [DOI] [PubMed] [Google Scholar]
- 51.Yasuda M., Maeda K., Ikejiri Y., Kawamata T., Kuroda S., Tanaka C. A novel missense mutation in the presenilin-1 gene in a familial Alzheimer’s disease pedigree with abundant amyloid angiopathy. Neurosci. Lett. 1997;232:29–32. doi: 10.1016/s0304-3940(97)00569-7. [DOI] [PubMed] [Google Scholar]
- 52.Takao M., Ghetti B., Hayakawa I., Ikeda E., Fukuuchi Y., Miravalle L., Piccardo P., Murrell J.R., Glazier B.S., Koto A. A novel mutation (G217D) in the Presenilin 1 gene (PSEN1) in a Japanese family: Presenile dementia and parkinsonism are associated with cotton wool plaques in the cortex and striatum. Acta Neuropathol. 2002;104:155–170. doi: 10.1007/s00401-002-0536-6. [DOI] [PubMed] [Google Scholar]
- 53.Qiu Q., Jia L., Wang Q., Zhao L., Jin H., Li T., Quan M., Xu L., Li B., Li Y., et al. Identification of a novel PSEN1 Gly111Val missense mutation in a Chinese pedigree with early-onset Alzheimer’s disease. Neurobiol. Aging. 2019 doi: 10.1016/j.neurobiolaging.2019.05.018. in press. [DOI] [PubMed] [Google Scholar]
- 54.Ataka S., Tomiyama T., Takuma H., Yamashita T., Shimada H., Tsutada T., Kawabata K., Mori H., Miki T. A novel presenilin-1 mutation (Leu85Pro) in early-onset Alzheimer disease with spastic paraparesis. Arch. Neurol. 2004;61:1773–1776. doi: 10.1001/archneur.61.11.1773. [DOI] [PubMed] [Google Scholar]
- 55.Higuchi S., Yoshino A., Matsui T., Matsushita S., Satoh A., Limura T., Ishikawa M., Arai H., Shirakura K. A novel PS1 mutation (W165G) in a Japanese family with early-onset Alzheimer’s disease. Alzheimer’s Rep. 2000;3:227–231. [Google Scholar]
- 56.Dong J., Qin W., Wei C., Tang Y., Wang Q., Jia J. A Novel PSEN1 K311R Mutation Discovered in Chinese Families with Late-Onset Alzheimer’s Disease Affects Amyloid-beta Production and Tau Phosphorylation. J. Alzheimer’s Dis. JAD. 2017;57:613–623. doi: 10.3233/JAD-161188. [DOI] [PubMed] [Google Scholar]
- 57.Sodeyama N., Iwata T., Ishikawa K., Mizusawa H., Yamada M., Itoh Y., Otomo E., Matsushita M., Komatsuzaki Y. Very early onset Alzheimer’s disease with spastic paraparesis associated with a novel presenilin 1 mutation (Phe237Ile) J. Neurol. Neurosurg. Psychiatry. 2001;71:556–557. doi: 10.1136/jnnp.71.4.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Furuya H., Yasuda M., Terasawa K.J., Tanaka K., Murai H., Kira J., Ohyagi Y. A novel mutation (L250V) in the presenilin 1 gene in a Japanese familial Alzheimer’s disease with myoclonus and generalized convulsion. J. Neurol. Sci. 2003;209:75–77. doi: 10.1016/S0022-510X(02)00466-5. [DOI] [PubMed] [Google Scholar]
- 59.Ikeda M., Sharma V., Sumi S.M., Rogaeva E.A., Poorkaj P., Sherrington R., Nee L., Tsuda T., Oda N., Watanabe M., et al. The clinical phenotype of two missense mutations in the presenilin I gene in Japanese patients. Ann. Neurol. 1996;40:912–917. doi: 10.1002/ana.410400614. [DOI] [PubMed] [Google Scholar]
- 60.Matsubara-Tsutsui M., Yasuda M., Yamagata H., Nomura T., Taguchi K., Kohara K., Miyoshi K., Miki T. Molecular evidence of presenilin 1 mutation in familial early onset dementia. Am. J. Med. Genet. 2002;114:292–298. doi: 10.1002/ajmg.10250. [DOI] [PubMed] [Google Scholar]
- 61.Kamimura K., Tanahashi H., Yamanaka H., Takahashi K., Asada T., Tabira T. Familial Alzheimer’s disease genes in Japanese. J. Neurol. Sci. 1998;160:76–81. doi: 10.1016/S0022-510X(98)00219-6. [DOI] [PubMed] [Google Scholar]
- 62.Tanahashi H., Mitsunaga Y., Takahashi K., Tasaki H., Watanabe S., Tabira T. Missense mutation of S182 gene in Japanese familial Alzheimer’s disease. Lancet. 1995;346:440. doi: 10.1016/s0140-6736(95)92810-3. [DOI] [PubMed] [Google Scholar]
- 63.Hamaguchi T., Morinaga A., Tsukie T., Kuwano R., Yamada M. A novel presenilin 1 mutation (L282F) in familial Alzheimer’s disease. J. Neurol. 2009;256:1575–1577. doi: 10.1007/s00415-009-5154-y. [DOI] [PubMed] [Google Scholar]
- 64.Tabira T., Chui D.H., Nakayama H., Kuroda S., Shibuya M. Alzheimer’s disease with spastic paresis and cotton wool type plaques. J. Neurosci. Res. 2002;70:367–372. doi: 10.1002/jnr.10392. [DOI] [PubMed] [Google Scholar]
- 65.Ikeuchi T., Kaneko H., Miyashita A., Nozaki H., Kasuga K., Tsukie T., Tsuchiya M., Imamura T., Ishizu H., Aoki K., et al. Mutational analysis in early-onset familial dementia in the Japanese population. The role of PSEN1 and MAPT R406W mutations. Dement. Geriatr. Cogn. Disord. 2008;26:43–49. doi: 10.1159/000141483. [DOI] [PubMed] [Google Scholar]
- 66.Jiang H.Y., Li G.D., Dai S.X., Bi R., Zhang D.F., Li Z.F., Xu X.F., Zhou T.C., Yu L., Yao Y.G. Identification of PSEN1 mutations p.M233L and p.R352C in Han Chinese families with early-onset familial Alzheimer’s disease. Neurobiol. Aging. 2015;36:1602.e3–1602.e6. doi: 10.1016/j.neurobiolaging.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 67.Bagyinszky E., Kang M.J., van Giau V., Shim K., Pyun J.-M., Suh J., An S.S.A., Kim S. Novel Amyloid Precursor Protein mutation, Val669Leu (“Seoul APP”), in a Korean Early onset Alzheimer’s disease patient. Neurobiol. Aging. 2019 doi: 10.1016/j.neurobiolaging.2019.08.026. in press. [DOI] [PubMed] [Google Scholar]
- 68.Johnston J.A., Cowburn R.F., Norgren S., Wiehager B., Venizelos N., Winblad B., Vigo-Pelfrey C., Schenk D., Lannfelt L., O’Neill C. Increased beta-amyloid release and levels of amyloid precursor protein (APP) in fibroblast cell lines from family members with the Swedish Alzheimer’s disease APP670/671 mutation. Febs Lett. 1994;354:274–278. doi: 10.1016/0014-5793(94)01137-0. [DOI] [PubMed] [Google Scholar]
- 69.Park J., An S.S.A., Giau V.V., Shim K., Youn Y.C., Bagyinszky E., Kim S. Identification of a novel PSEN1 mutation (Leu232Pro) in a Korean patient with early-onset Alzheimer’s disease and a family history of dementia. Neurobiol. Aging. 2017;56:212.e11–212.e17. doi: 10.1016/j.neurobiolaging.2017.04.012. [DOI] [PubMed] [Google Scholar]
- 70.Ch’ng G.-S., An S.S.A., Bae S.O., Bagyinszky E., Kim S. Identification of two novel mutations, PSEN1 E280K and PRNP G127S, in a Malaysian family. Neuropsychiatr. Dis. Treat. 2015;11:2315–2322. doi: 10.2147/NDT.S86334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guerreiro R.J., Baquero M., Blesa R., Boada M., Bras J.M., Bullido M.J., Calado A., Crook R., Ferreira C., Frank A., et al. Genetic screening of Alzheimer’s disease genes in Iberian and African samples yields novel mutations in presenilins and APP. Neurobiol. Aging. 2010;31:725–731. doi: 10.1016/j.neurobiolaging.2008.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hardy J., Guerreiro R. A new way APP mismetabolism can lead to Alzheimer’s disease. EMBO Mol. Med. 2011;3:247–248. doi: 10.1002/emmm.201100139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.van Giau V., Pyun J.-M., Suh J., Bagyinszky E., An S.S.A., Kim S.Y. A pathogenic PSEN1 Trp165Cys mutation associated with early-onset Alzheimer’s disease. BMC Neurol. 2019;19:1–10. doi: 10.1186/s12883-019-1419-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wallon D., Rousseau S., Rovelet-Lecrux A., Quillard-Muraine M., Guyant-Marechal L., Martinaud O., Pariente J., Puel M., Rollin-Sillaire A., Pasquier F., et al. The French series of autosomal dominant early onset Alzheimer’s disease cases: Mutation spectrum and cerebrospinal fluid biomarkers. J. Alzheimer’s Dis. JAD. 2012;30:847–856. doi: 10.3233/JAD-2012-120172. [DOI] [PubMed] [Google Scholar]
- 75.Tanahashi H., Kawakatsu S., Kaneko M., Yamanaka H., Takahashi K., Tabira T. Sequence analysis of presenilin-1 gene mutation in Japanese Alzheimer’s disease patients. Neurosci. Lett. 1996;218:139–141. doi: 10.1016/s0304-3940(96)13138-4. [DOI] [PubMed] [Google Scholar]
- 76.Yang Y., Giau V.V., An S.S.A., Kim S. Plasma Oligomeric Beta Amyloid in Alzheimer’s Disease with History of Agent Orange Exposure. Dement. Neurocogn. Disord. 2018;17:41–49. doi: 10.12779/dnd.2018.17.2.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zatti G., Ghidoni R., Barbiero L., Binetti G., Pozzan T., Fasolato C., Pizzo P. The presenilin 2 M239I mutation associated with familial Alzheimer’s disease reduces Ca2+ release from intracellular stores. Neurobiol. Dis. 2004;15:269–278. doi: 10.1016/j.nbd.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 78.Bird T.D., Levy-Lahad E., Poorkaj P., Sharma V., Nemens E., Lahad A., Lampe T.H., Schellenberg G.D. Wide range in age of onset for chromosome 1-related familial Alzheimer’s disease. Ann. Neurol. 1996;40:932–936. doi: 10.1002/ana.410400619. [DOI] [PubMed] [Google Scholar]
- 79.Sherrington R., Froelich S., Sorbi S., Campion D., Chi H., Rogaeva E.A., Levesque G., Rogaev E.I., Lin C., Liang Y., et al. Alzheimer’s disease associated with mutations in presenilin 2 is rare and variably penetrant. Hum. Mol. Genet. 1996;5:985–988. doi: 10.1093/hmg/5.7.985. [DOI] [PubMed] [Google Scholar]
- 80.Hsu S., Gordon B.A., Hornbeck R., Norton J.B., Levitch D., Louden A., Ziegemeier E., Laforce R., Chhatwal J., Day G.S., et al. Discovery and validation of autosomal dominant Alzheimer’s disease mutations. Alzheimer’s Res. Ther. 2018;10:67. doi: 10.1186/s13195-018-0392-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chávez-Gutiérrez L., Bammens L., Benilova I., Vandersteen A., Benurwar M., Borgers M., Lismont S., Zhou L., Van Cleynenbreugel S., Esselmann H., et al. The mechanism of gamma-Secretase dysfunction in familial Alzheimer disease. EMBO J. 2012;31:2261–2274. doi: 10.1038/emboj.2012.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Brouwers N., Sleegers K., van Broeckhoven C. Molecular genetics of Alzheimer’s disease: An update. Ann. Med. 2008;40:562–583. doi: 10.1080/07853890802186905. [DOI] [PubMed] [Google Scholar]
- 83.Wingo T.S., Lah J.J., Levey A.I., Cutler D.J. Autosomal recessive causes likely in early-onset Alzheimer disease. Arch. Neurol. 2012;69:59–64. doi: 10.1001/archneurol.2011.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cacace R., Sleegers K., van Broeckhoven C. Molecular genetics of early-onset Alzheimer’s disease revisited. Alzheimer’s Dement. 2016;12:733–748. doi: 10.1016/j.jalz.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 85.Finckh U., Kuschel C., Anagnostouli M., Patsouris E., Pantes G.V., Gatzonis S., Kapaki E., Davaki P., Lamszus K., Stavrou D., et al. Novel mutations and repeated findings of mutations in familial Alzheimer disease. Neurogenetics. 2005;6:85–89. doi: 10.1007/s10048-005-0211-x. [DOI] [PubMed] [Google Scholar]
- 86.Nguyen T.T., Giau V.V., Vo T.K. Current advances in transdermal delivery of drugs for Alzheimer’s disease. Indian J. Pharmacol. 2017;49:145–154. doi: 10.4103/0253-7613.208143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Van Giau V., An S.S.A. Optimization of specific multiplex DNA primers to detect variable CLU genomic lesions in patients with Alzheimer’s disease. BioChip J. 2015;9:278–284. [Google Scholar]
- 88.Youn Y.C., Lim Y.K., Han S.H., Giau V.V., Lee M.K., Park K.Y., Kim S., Bagyinszky E., An S.S.A., Kim H.R. Apolipoprotein epsilon7 allele in memory complaints: Insights through protein structure prediction. Clin. Interv. Aging. 2017;12:1095–1102. doi: 10.2147/CIA.S131172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bagyinszky E., Giau V.V., Youn Y.C., An S.S.A., Kim S. Characterization of mutations in PRNP (prion) gene and their possible roles in neurodegenerative diseases. Neuropsychiatr. Dis. Treat. 2018;14:2067–2085. doi: 10.2147/NDT.S165445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Giau V.V., Bagyinszky E., Youn Y.C., An S.S.A., Kim S.Y. Genetic Factors of Cerebral Small Vessel Disease and Their Potential Clinical Outcome. Int. J. Mol. Sci. 2019;20:4298. doi: 10.3390/ijms20174298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bagyinszky E., Yang Y., Giau V.V., Youn Y.C., An S.S.A., Kim S. Novel prion mutation (p.Tyr225Cys) in a Korean patient with atypical Creutzfeldt-Jakob disease. Clin. Interv. Aging. 2019;14:1387–1397. doi: 10.2147/CIA.S210909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.McKhann G.M., Knopman D.S., Chertkow H., Hyman B.T., Jack C.R., Jr., Kawas C.H., Klunk W.E., Koroshetz W.J., Manly J.J., Mayeux R., et al. The diagnosis of dementia due to Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimer’s Dement. J. Alzheimer’s Assoc. 2011;7:263–269. doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]