Abstract
Calligrapha is a New World leaf beetle genus that includes several unisexual species in northeastern North America. Each unisexual species had an independent hybrid origin involving different combinations of bisexual species. However, surprisingly, they all cluster in a single mtDNA clade and with some individuals of their parental species, which are in turn deeply polyphyletic for mtDNA. This pattern is suggestive of a selective sweep which, together with mtDNA taxonomic incongruence and occurrence of unisexuality in Calligrapha, led to hypothesize that Wolbachia might be responsible. I tested this hypothesis studying the correlation between diversity of Wolbachia and well‐established mtDNA lineages in >500 specimens of two bisexual species of Calligrapha and their derived unisexual species. Wolbachia appears highly prevalent (83.4%), and fifteen new supergroup‐A strains of the bacteria are characterized, belonging to three main classes: wCallA, occupying the whole species ranges, and wCallB and wCallC, narrowly parapatric, infecting beetles with highly divergent mtDNAs where they coexist. Most beetles (71.6%) carried double infections of wCallA with another sequence class. Bayesian inference of ancestral character states and association tests between bacterial diversity and the mtDNA genealogy show that each mtDNA lineage of Calligrapha has specific types of infection. Moreover, shifts can be explained by horizontal or vertical transfer from local populations to an expanding lineage and cytoplasmic incompatibility between wCallB and wCallC types, suggesting that the symbionts hitchhike with the host and are not responsible for selective mtDNA sweeps. Lack of evidence for sweeps and the fact that individuals in the unisexual clade are uninfected or infected by the widespread wCallA type indicate that Wolbachia does not induce unisexuality in Calligrapha, although they may manipulate host reproduction through cytoplasmic incompatibility.
Keywords: cryptic species, cytoplasmic incompatibility, mtDNA, parthenogenesis, selective sweep, superinfection
Calligrapha has several unisexual species which arose independently through interspecific hybridization of different parentals, yet they are all monophyletic for mtDNA. In this article, I test the hypothesis that this may be the result of a selective sweep induced by Wolbachia and that these endosymbionts may be responsible for the transition to unisexuality in these beetles. The answer to this question is that the bacteria are associated neither with the unisexual mtDNA type nor with the reproductive transition to unisexuality in Calligrapha, although they seem responsible for geographic patterns of mtDNA through cytoplasmic incompatibility.

1. INTRODUCTION
Alphaproteobacteria of the genus Wolbachia (Rickettsiales: Anaplasmataceae) have been shown to live as endosymbionts of a large proportion of arthropods and also nematodes (Hilgenboecker, Hammerstein, Schlattmann, Telschow, & Werren, 2008; Weinert, Araujo, Ahmed, & Welch, 2015; Zug, Koehncke, & Hammerstein, 2012). In insects, these bacteria have been typically found or predominantly studied in germline cells (Dobson et al., 1999; Toomey, Panaram, Fast, Beatty, & Frydman, 2013; Werren, 1997), although they occur in somatic tissues of infected individuals too (e.g., Pietri, DeBruhl, & Sullivan, 2016). Their highly specialized natural history, adapted to the intracellular environment, makes their evolutionary survival often conditional on vertical inheritance from one host generation to the next, although horizontal transfer, particularly for species with intimate ecological contact and among related species, is a prevalent phenomenon as well (e.g., Heath, Butcher, Whitfield, & Hubbard, 1999; Huigens, Almeida, Boons, Luck, & Stouthamer, 2004; Russell et al., 2009). In the case of vertical transmission, it occurs via host maternal transmission through a complex interaction of the bacteria with the microtubule network of the oocytes (Ferree et al., 2005), since they are effectively removed from male gametes in late stages of spermatogenesis (Clark, Veneti, Bourtzis, & Karr, 2002). The way in which the association between the prokaryote and the eukaryotic host was established, whereby only female hosts transmit the infection, has led to one of the most Machiavellian behaviors and types of interaction of the natural world. Wolbachia can provide with physiological benefits for the host, such as nutritional mutualism, protection against viral infections, or even correct development of oogenesis (Dedeine et al., 2001; Hosokawa, Koga, Kikuchi, Meng, & Fukatsu, 2010; Martinez et al., 2014). Yet, they can also manipulate host reproduction and exert an influence on female reproductive fitness and sex ratio demography. As a result, they can effectively disconnect the history of mitochondrial DNA (mtDNA), also with maternal inheritance, from the history of the host. These so‐called selective sweeps have serious implications for phylogeographic and taxonomic enquiry of the host based precisely on mtDNA sequences. Moreover, the intimate association of host and endosymbiont can promote some degree of coevolution or parallel evolution, so that the confinement of the bacteria in a particular evolutionary line of the host may result in nonrandom associations of genetic diversity of both host and endosymbiont (Jiggins, Bentley, Majerus, & Hurst, 2002; Russell et al., 2009).
Wolbachia has evolved four strategies to attain more efficient invasive rates in their host populations (Charlat, Hurst, & Merçot, 2003; Werren, Baldo, & Clark, 2008): (a) induction of parthenogenetic reproduction (Ma & Schwander, 2017); (b) feminization of male embryos (Kageyama, Nishimura, Hoshizaki, & Ishikawa, 2002; Rousset, Bouchon, Pintureau, Juchault, & Solignac, 1992); (c) killing of male embryos (Fialho & Stevens, 2000; Hurst et al., 1999); and/or (d) sterilization of uninfected females by infected males (cytoplasmic incompatibility, also at play when different strains of the bacteria infect each parent; Engelstädter & Telschow, 2009). Because of disparate effects depending on host sexes, the potential impact of the evolutionary selfishness of these intracellular bacteria on the demography of host populations ranges from distorted sex ratios to male eradication (Werren & Beukeboom, 1998). These effects can be transitory, and “curing” the infection with antibiotics can revert them (Li, Floate, Fields, & Pang, 2014). However, they can have long‐lasting consequences, and these master manipulators have been associated, for example, with the evolutionary transition from bisexual to unisexual reproductive modes (Ma & Schwander, 2017). When the endosymbiont successfully manipulates host reproduction, a resulting side effect is that mtDNA hitchhikes with the symbiont, thus establishing a linkage disequilibrium relationship (Turelli, Hoffmann, & McKechnie, 1992). The fact that spreading of mtDNA in natural populations may be conditioned by the bacteria dynamics has dramatic consequences on the reliability of mtDNA for population, phylogeographic, and phylogenetic studies (Hurst & Jiggins, 2005). For example, mtDNA selective sweeps driven by intracellular bacteria can reduce genetic diversity, similarly to population bottleneck effects (Gompert, Forister, Fordyce, & Nice, 2008), and symbiont‐induced mtDNA introgression following interspecific hybridization may confound mtDNA‐based inference of species relationships (Galtier, Nabholz, Glémin, & Hurst, 2009; Hurst & Jiggins, 2005; Smith et al., 2012), occasionally to an exacerbated degree (Schmidt & Sperling, 2008). Similar disequilibrium effects also occur between different strains of Wolbachia, so that host mtDNA diversity may mirror bacterial genotypic diversity rather than true host population structure (Charlat et al., 2009; Ilinsky, 2013).
Leaf beetles in the genus Calligrapha are an interesting group of organisms to investigate for the presence and potential effects of reproductive manipulation by Wolbachia. In the first place, it is exceptional among beetles for including several examples of unisexual species, each derived from interspecific hybridization events involving different bisexual species in North America (Gómez‐Zurita & Cardoso, 2019; Gómez‐Zurita, Funk, & Vogler, 2006; Gómez‐Zurita, Vogler, & Funk, 2004; Montelongo & Gómez‐Zurita, 2015; Robertson, 1966). Moreover, it displays abnormally high levels of mtDNA paraphyly for most North American species, particularly those identified as involved in the origin of unisexual lineages (Gómez‐Zurita & Cardoso, 2019; Gómez‐Zurita et al., 2006; Montelongo & Gómez‐Zurita, 2015). Thus, bisexual species of Calligrapha are deeply polyphyletic for mtDNA, with individuals in at least two highly divergent clades, one with populations with normal sex ratios and named B‐clade and one exclusively comprised of females, also including representatives of the unisexual species evolutionarily derived from them, or U‐clade (Gómez‐Zurita & Cardoso, 2019; Montelongo & Gómez‐Zurita, 2015). This pattern is interpreted as each of the unisexual species of Calligrapha having had a history with a minimum of two waves of interspecific hybridization, with available evidence suggesting that sex was not lost immediately (Montelongo & Gómez‐Zurita, 2015). The last hybridization events leading to unisexual species occurred recently, plausibly after the Last Glacial Maximum (LGM), and they involved female‐only lineages of the otherwise bisexual parental species, suggesting that unisexuality may have actually predated the origin of unisexual species (Gómez‐Zurita & Cardoso, 2019). In this model, although hybridization appears as a necessary and important condition, this mechanism alone is not sufficient to explain the origin of unisexuality.
Given that a particular type of ancestral mtDNA is associated with unisexuality in Calligrapha, this is suggestive that the same process that introgressed this specific mtDNA could have simultaneously transferred a factor that would eventually lead to the evolution of unisexuality. There is no satisfactory explanation for this idea. However, among the possible hypotheses worth testing, the most prominent, linking both the origins of unisexuality and also selective sweeps of mtDNA, advocates for the potential role of intracellular symbionts in the process (Montelongo & Gómez‐Zurita, 2015). As highlighted above, the involvement of these endosymbionts in the evolution of Calligrapha and important life‐history traits, such as reproductive mode, could additionally explain other observations in this system, such as the highly distorted sex ratios in some populations (Brown, 1945; Robertson, 1966) or evolutionary lineages (Gómez‐Zurita & Cardoso, 2019), and extensive species mtDNA polyphyly (Gómez‐Zurita et al., 2006; Montelongo & Gómez‐Zurita, 2015), among others.
In order to test the hypothesis that Wolbachia may be responsible for these observations, this work shows a replicated study in several species of Calligrapha for which there is a detailed understanding of their genetic diversity and phylogeographic processes (Gómez‐Zurita & Cardoso, 2019). The study will try to establish the association between their mtDNA diversity, their geographic distribution, and the apparent conflict with their taxonomic boundaries with the presence of Wolbachia. This will be accomplished after screening first the same samples studied by Gómez‐Zurita and Cardoso (2019) for the presence of Wolbachia, which has not been attempted before in the genus, and characterizing their strains. Based on this information, the main objective of the study will be testing for links between Wolbachia and unisexuality in Calligrapha, and more generally investigating if and how Wolbachia may have affected mtDNA diversity in this host.
2. MATERIAL AND METHODS
2.1. Sampling and major phylogeographic lineages of Calligrapha
This study used the specimens, DNA, and cox1 and Wg sequence data previously analyzed by Gómez‐Zurita and Cardoso (2019). The sample included representatives of two bisexual species of Calligrapha with transcontinental ranges in North America, C. philadelphica, and two subspecies of C. multipunctata, as well as two unisexual species, C. vicina and C. suturella, evolutionarily derived from the unisexual populations of each of the previous two, respectively (Montelongo & Gómez‐Zurita, 2015). A total of 506 specimens were studied, including 322 individuals of C. multipunctata bigsbyana, 22 of C. multipunctata s. str., 148 of C. philadelphica, and seven each of C. suturella and C. vicina (Table S1). MtDNA and nuclear Wg data were consistent in identifying seven major and 13 geographically concordant evolutionary lineages in the sample (Gómez‐Zurita & Cardoso, 2019). In bisexual C. multipunctata, four major lineages were distinguished (Figure 1a): (a) evolutionary branch associated with the Mississippi basin (m ML) with one lineage expanding northwest, to establish populations in the Rocky Mountains (m ML1), and one expanding north to the Northern Great Plains in the Upper Mississippi and to the Alleghany region along the Ohio River Basin (m ML2); (b) branch established and expanding in the Northern Great Plains (m CL); (c) northern branch (m NL) with three main lineages, one reaching the Pacific Northwest (m NL1), one established in the Northern Great Plains (m NL2), and one in New England and the northern Atlantic regions (m NL3), the latter hybridizing with C. philadelphica in its easternmost front of expansion in the Holocene; and (d) unisexual lineage of C. multipunctata (m UL), expanding from the Great Lakes region, giving rise to C. suturella upon hybridization with C. ignota after the LGM (Montelongo & Gómez‐Zurita, 2015). In C. philadelphica, three major lineages were present (Figure 1b): (a) western lineage, west of the Saint Lawrence River and reaching southeastern plains of Alberta (p WL); (b) eastern lineage, east of the Saint Lawrence River (p EL), with one lineage expanding northeast (p EL1) and one expanding east and southwest (p EL2) along the western Appalachian slopes; and (c) unisexual lineage of C. philadelphica (p UL), with independent branches expanding south to Georgia along central and southern Appalachians (p UL1), west to Minnesota (p UL2), and east to New England (p UL3), the latter giving rise to C. vicina via hybridization with ancestral populations of C. rowena after the LGM (Montelongo & Gómez‐Zurita, 2015).
Figure 1.

Schematic summary of phylogeographic lineages of Calligrapha considered in this work and based on the results of Gómez‐Zurita and Cardoso (2019), including (a) four main lineages (m ML, m CL, m NL, and m UL) in C. multipunctata and (b) three main lineages (p WL, p EL, and p UL) in C. philadelphica. The m UL and p UL lineages also include the unisexual species C. suturella and C. vicina, respectively. The asterisk marks the hypothetical area where m NL3 and p EL1 lineages met and hybridized, establishing a population of C. philadelphica with typical C. multipunctata mtDNA
2.2. MLST genotyping of Wolbachia
Each individual DNA extraction of Calligrapha was used as template to investigate the presence and strain of Wolbachia infecting every beetle and using the multilocus sequence typing system (MLST) designed for Wolbachia by Baldo et al. (2006). We used the same loci (gatB, coxA, hcpA, fbpA, and ftsZ) and PCR amplification protocols of Baldo et al. (2006), which worked well in our hands, except for the ftsZ locus, which was amplified in most cases using combinations of nondegenerate custom primers that amplified fragments between 522 and 624 bp and encompassing the whole region of interest: ftsZf2 (5′‐GTCTTGGTGCTGGTGCTTTG) or ftsZf3 (5′‐GGTGCTTTGCCTGATGTTGG) with ftsZr2 (5′‐TGGATATTGCAGCTTCCGCA) or ftsZr3 (5′‐TGGCTGCAGCATCAACTTCA). PCR products were purified with ammonium acetate 5M and isopropanol and sequenced in both directions using the same PCR primers and the Sanger method with BigDye Terminator 3.1 technology and capillary sequencing on a 3730xl DNA Analyzer (Applied Biosystems). Sequences were trimmed to their respective MLST fragment size (Baldo et al., 2006), aligned using the L‐INS‐i algorithm of MAFFT 7 (Katoh & Standley, 2013), and filtered to allele sequence data using analytical tools in Geneious R9 (Biomatters Ltd.). Each allele was identified with reference to the curated Wolbachia MLST Database (update: 17 April 2017) using the associated PubMLST online public resource (https://pubmlst.org/wolbachia/; Jolley & Maiden, 2010), but since the curation of Wolbachia data was very unfortunately discontinued (J. Werren, personal communication), the identity of the obtained alleles was additionally checked against the latest release of GenBank too (31 July 2018). A representative allele of each of these sequences was deposited in GenBank, with sequence accession numbers LR135794–LR135810.
2.3. Association test between mtDNA and Wolbachia diversity: contingency tests
Gómez‐Zurita and Cardoso (2019) identified a number of mtDNA lineages in the species of Calligrapha under study, their distribution, and the history of their range changes based on the genealogy of cox1 and a major association of these lineages with different allelic and genotypic characteristics of Wg (Figure 1). These lineages were established in part based on an unrooted cox1 genealogy obtained under statistical parsimony and a nested‐clade design resulting from the implementation of Templeton, Boerwinkle, and Sing (1987) algorithm. For visualization purposes in the current study, the same nested clades of Gómez‐Zurita and Cardoso (2019) were mapped on the phylogeny of cox1 haplotypes obtained under maximum likelihood with RAxML 7.2.8 (Stamatakis, 2006), specifying a GTR+G+I evolutionary model and 10 replicates of random addition of taxa, and assessing node support based on 100 bootstrap pseudoreplicates. The association between the occurrence and type of Wolbachia infections and mtDNA evolutionary diversity in Calligrapha, as well as a possible scenario for the history of this association by reference to the phylogeographic inferences of Gómez‐Zurita and Cardoso (2019), was explored using permutation contingency tests of discrete variables. To carry out this analysis, the nested clades of the cox1 haplotype statistical parsimony genealogy of Gómez‐Zurita and Cardoso (2019), retaining here the same numbering as in that work for easier cross‐reference, were transformed into a categorical variable to test the association of these groups of related haplotypes with different Wolbachia genotypes. In turn, Wolbachia infections were transformed into three types of categorical variables: infection status and type of infection (uninfected vs. single infection vs. double infection); genotype group (one of three main groups); or specific genotype. Permutation chi‐square exact contingency tests of these variables on the structure defined by the cox1 genealogy, best known as nested‐clade analysis (Templeton & Sing, 1993), were carried out for all contingency matrices where both variables showed observations. The tests and their significance were carried out using 9,999 data permutations for each contingency matrix using the function perm.ind.test of the R package wPerm 1.0.1 (Weiss, 2015). Permutation of the contingency matrices simulated the null hypothesis of no association between mtDNA genetic differences and the type and characteristics of the infection with Wolbachia, and a significant result was interpreted as the existence of statistical differences between the infections of tested mtDNA groups.
When a significant association was obtained in the previous tests, the diversity of Wolbachia was tested also for an association with the Wg genotypes of the same individuals involved in the original test to find whether the association was specific for mtDNA or if there was signal of association with the nuclear marker as well.
2.4. Association test between mtDNA and Wolbachia diversity: ancestral states
Bayesian inference of ancestral states (Ronquist, 2004) was used as a complementary tool to investigate the association of specific mtDNA evolutionary lineages in Calligrapha and the type of Wolbachia they carried. Bayesian trait analysis and ancestral character inference were carried out with BEAST 1.8.4 (Drummond, Suchard, Xie, & Rambaut, 2012) on a reduced matrix where each different combination of taxon, cox1 haplotype, and associated Wolbachia sequence type(s) was considered. This matrix was analyzed under an HKY+I+G model and a relaxed lognormal clock. An additional partition was created to represent Wolbachia sequence type traits associated with each species/haplotype and allowing for ambiguous codes to account for double infections. Two coding schemes were used: one considering each ST or ST combination as well as the uninfected state (31 states), and one grouping sequence types in three major groups as recognized in this study (four states). This trait was allowed to change under a symmetric substitution model and a random local clock, independent from the nucleotide sequence evolution clock. Both nucleotide sequence and infection partitions were linked to a single tree estimated under a constant‐size coalescent model. Infection states for all ancestors, including the root of the tree, were deduced and annotated on the obtained tree topologies. Tree searches were based on two independent MCMC chains of 70 million generations each, sampling one every 7,000 trees, and the results of the search were analyzed with Tracer 1.6.0 (Rambaut, Suchard, Xie, & Drummond, 2014) and TreeAnnotator 1.8.4 (Drummond et al., 2012), excluding 10% of the initial, stabilizing phase, and visualized with FigTree 1.4.3 (available from http://tree.bio.ed.ac.uk/software/figtree/).
3. RESULTS
3.1. Incidence of infection and strains of Wolbachia
Most samples of Calligrapha (83.4%) were confirmed for infection with Wolbachia by consistent amplification of MLST markers, and 71.6% of them showed evidence of double infections, presenting double sequence peaks for all markers. The availability of clean allele sequences for each gene in numerous specimens and the expected double‐peak patterns of their combinations allowed to separate easily the alleles of coinfecting strains in all but one instance of gatB, found in one individual of C. multipunctata bigsbyana from Quyon (Quebec). A number of samples (9.1%), upon several PCR trials, failed to amplify any of the five MLST markers of Wolbachia, indicating the absence of infection, an idea reinforced by their trend to affect coherent sets of samples (e.g., sharing a certain cox1 haplotype, coming from the same locality or belonging to the same evolutionary lineage). Some samples produced faint bands for one (7.3%) or two (0.2%) of the loci tested, but inconsistently and only in some of the repeated trials. Nonetheless, we also tried to sequence these PCR products, always producing low‐quality sequences and at most for one of the strands. These results could represent idiosyncrasies of PCR or template DNA quality issues, but their inconsistency and unrepeatability were suggestive of PCR artifacts or contamination surfacing when forcing PCR conditions.
In total, the sample of Wolbachia yielded four gatB, two coxA, three hcpA, three ftsZ, and five fbpA alleles, of which only one allele each of coxA, hcpA, and fbpA and two of ftsZ had been described previously (Table S2). Previously uncharacterized alleles were evolutionarily close to already known variants (Figure 2; Table S2). These 18 alleles produced 15 genotypes or sequence types (ST) belonging to three main groups, called here wCallA to wCallC, mostly defined by the gatB allele, with all genotypes from one group separated by mutations in two or more MLST loci from genotypes in other such group (Table 1; Figure 2). Half of these genotypes were recognized directly from singly infected individuals. In turn, deduced genotypes differed from one of the former in just one of the five MLST loci (Table 1), making their separation straightforward by discounting the state of the corresponding observed genotype from the polymorphism established empirically (Table 2). All the alleles and their combinations could be referred to the supergroup A of Wolbachia.
Figure 2.
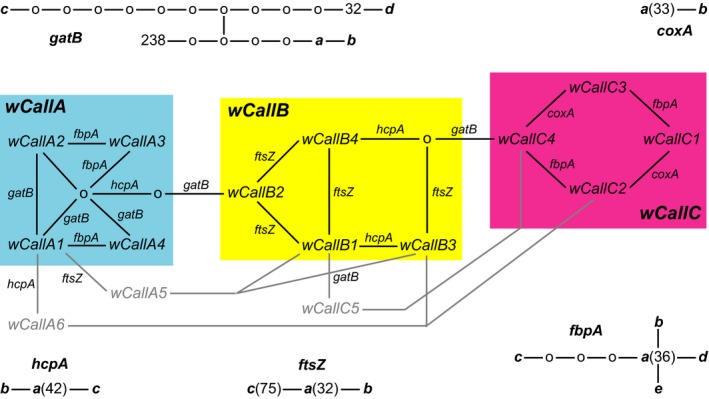
Intralocus and MLST evolutionary relationships of Wolbachia characterized in the sample of four species of Calligrapha. Relationships among alleles of individual MLST loci (identified with the codes given in Table S2) and among wCall sequence types are represented using statistical parsimony networks where edges identify individual mutations in the former and allele changes in the latter. Three main groups of wCall MLSTs are identified based on changes occurring in two or more individual loci
Table 1.
Wolbachia sequence types (ST) observed in the sample of Calligrapha in the case of single infections or deduced from individuals with double infections
| Wolbachia ST | Allelesa | N | Calligrapha cox1 haplotypes or coinfecting ST |
|---|---|---|---|
| Uninfected | – | 83 | 11B1, 1B6, 2B14, 1B15, 2B18, 1B27, 4B28, 1B29, 2B31, 1B32, 2B33, 1B35, 5B46, 3B47, 1B48, 3B57, 2B58, 3B59, 1B69, 1B73, 9U1, 1U2, 2U3, 4U3, 1U4, 6U5, 4U5, 1U7, 6U8, 1U13 |
| Observed | |||
| wCallA1 | a,33,42,32,36 | 61 | 13B1, 1B12, 2B18, 1B38, 1B45, 1B46, 16B49, 1B54, 3B63, 6U1, 2U3, 1U5, 4U9, 2U10, 4U11, 2U12, 1U13 |
| wCallA2 | b,33,42,32,36 | 27 | 3B1, 1B7, 23B14 |
| wCallA3 | b,33,42,32,d | 3 | 1B39, 1B40, 1B41 |
| wCallA4 | a,33,42,32,e | 1 | 1B1 |
| wCallB1 | c,33,c,b,b | 16 | 2B14, 8B33, 2B34, 2B35, 2B36 |
| wCallB2 | c,33,c,32,b | 6 | 1B33, 2B34, 2B35, 1B56 |
| wCallC2 | d,33,b,75,c | 6 | 3B59, 3B68 |
| Deduced | |||
| wCallA5 | a,33,42,b,36 | – | wCallB4, wCallC1 |
| wCallA6 | a,33,b,32,36 | – | wCallA1, wCallC2 |
| wCallB3 | c,33,b,b,b | – | wCallA1 |
| wCallB4 | c,33,c,75,b | – | wCallA1, wCallA5 |
| wCallC1 | d,b,b,75,c | – | wCallA1, wCallA4, wCallA5 |
| wCallC3 | d,b,b,75,b | – | wCallA1 |
| wCallC4 | d,33,b,75,b | – | wCallA1 |
| wCallC5 | d,33,c,b,b | – | wCallA1 |
In the case of the latter, coinfecting STs are listed. In the case of the former and uninfected individuals, the cox1 haplotypes and the frequency of beetle individuals where the Wolbachia ST is found are given, with an indication of species identity (round font: C. multipunctata bigsbyana; italics: C. multipunctata s. str.; bold: C. philadelphica; bold italics: C. suturella and C. vicina).
Characteristics of alleles and their repositories described in Table S2.
Table 2.
Double Wolbachia infections in the sample of Calligrapha, identifying coinfecting MLST types, their frequency, and the cox1 haplotypes of affected individuals and their proportion in the sample (their taxonomic identity is coded as in Table 1)
| ST1 | ST2 | Freq. | Calligrapha cox1 haplotypes |
|---|---|---|---|
| wCallA1 | wCallA6 | 1 | 1B63 |
| wCallA1 | wCallB1 | 99 | 1B1, 1B7, 1B8, 4B9, 1B10, 4B11, 1B13, 2B16, 10B24, 1B25, 1B26, 13B29, 1B37, 8B42, 1B44, 25B49, 2B50, 1B51, 1B52, 10B53, 1B55, 1B63, 1B64, 6U1, 1U3 |
| wCallA1 | wCallB3 | 1 | 1B49 |
| wCallA1 | wCallB4 | 1 | 1B29 |
| wCallA1 | wCallC1 | 116 | 68B1, 1B2, 1B3, 1B5, 40B18, 1B19, 1B20, 1B21, 1U5, 1U6 |
| wCallA1 | wCallC2 | 27 | 15B59, 1B60, 1B61, 3B62, 3B65, 1B66, 1B71, 1B72, 1U1 |
| wCallA1 | wCallC3 | 3 | 2B1, 1B18 |
| wCallA1 | wCallC4 | 1 | 1B65 |
| wCallA1 | wCallC5 | 1 | 1B4 |
| wCallA2 | wCallB1 | 36 | 4B1, 1B7, 6B14, 5B17, 1B22, 1B23, 2B26, 7B27, 7B28, 2B30 |
| wCallA2 | wCallC2 | 4 | 4B71 |
| wCallA4 | wCallC1 | 1 | 1B1 |
| wCallA5 | wCallB4 | 3 | 1B29, 2U1 |
| wCallA5 | wCallC1 | 1 | 1B1 |
| wCallA6 | wCallC2 | 4 | 3B67, 1B70 |
| wCallA1 | wCallB1r | 3 | 2B42, 1B43 |
| wCallB1 a | wCallC1 a | 1 | 1B18 |
wCall STs could not be determined because of uncertainty in establishing the gatB alleles. This is the only individual in the sample where strains wCallB and wCallC coexist, apparently. wCallB1r shows an abnormal gatB sequence which could be interpreted as a mosaic between c and d alleles.
There were few cases of intralocus and interlocus recombinants in the genetic sample of Wolbachia. The former were represented by three specimens of Calligrapha multipunctata bigsbyana from the Grass Island County (New York), where the gatB locus was seemingly a chimera between “c” and “d” alleles, whereby “c” is also present in the population (Table 2). Moreover, up to three rare STs (wCallA5, wCallA6, and wCallC5), observed in ten individuals, could be proposed of recombinant origin between loci, as they were responsible for loops in an ST evolutionary network connecting at least two divergent STs (Figure 2). Most double infections detected in Calligrapha always involved one of the wCallA STs with either a wCallB or a wCallC strain of Wolbachia. The only exceptions were a single case of double infection inferred between two strains of group wCallA in one individual of C. philadelphica from Pinchot Lake in York County (New York), and an uncertain ST because of the ambiguous resolution of gatB polymorphisms mentioned above and seemingly involving wCallB and wCallC variants, both present in the same population (Table 2).
3.2. Geographic ranges and prevalence of Wolbachia strains
The most prevalent Wolbachia ST in Calligrapha was wCallA1, infecting alone or in combination with other strains up to 74.2% of individuals, including representatives of all the species analyzed, except C. multipunctata s. str. Correspondingly, this type of Wolbachia appeared widely distributed throughout most of the geographic range considered here, in the eastern half of North America, from Saskatchewan to Nova Scotia and south to NE Texas, Georgia, and Alabama (Figure S1a). The next most abundant wCallA type in the sample was wCallA2, infecting 15.8% of specimens, including C. philadelphica and a genetically coherent group of C. m. bigsbyana, and it was distributed in the northern periphery of the studied range, in isolated populations along the Pacific coast, southern Alberta, and Maine and New Brunswick (Figure S1b). The second most abundant ST, either in single or in double infections of all species but C. suturella, was wCallB1, and it was present in 35.7% of infected individuals, also occupying most of the studied range across the continent, including the interior mountain beetle populations in Oregon and Utah, those in the upper Mississippi, the James and Missouri rivers, and as far south as the localities studied in western North Carolina (Figure S1c). All other wCallB types were much less prevalent and occupying narrower taxonomic and geographic areas within this same range, always found in localities where wCallB1 was present (Figure S1c). wCallC1 was very abundant, present in 27.9% of individuals analyzed, but exclusively found doubly infecting C. m. bigsbyana and C. suturella from eastern Ontario to Nova Scotia and south to Massachusetts, with one isolated occurrence in the northern Michigan peninsula (Figure S1d). Finally, wCallC2 was the fifth in prevalence in the sample (9.7% of infected Calligrapha), and it was only found in C. philadelphica from nearly the same area as wCallC1, from the Lake Ontario (southernmost locality) to New Brunswick and the Gaspe Peninsula (Figure S1e). In fact, the wCallC type was very coherent geographically, occupying an area from the Great Lakes area to Nova Scotia (Figure S2). Unisexual species of Calligrapha were either uninfected (57.1%) or infected with the wCallA1 type, solely (21.4%) or simultaneously with wCallC1 (14.3%, only C. suturella) or wCallB1 (7.1%, only C. vicina).
Individuals with single and double infections, as well as lacking any sign of infection, could be found nearly throughout the whole studied range (Figure S3). However, doubly infected individuals were absent in samples captured in the Rockies and in southern populations of the studied range, with the southernmost double infections detected in western North Carolina (Figure S3). The individuals in these southern or mountainous areas were either uninfected or singly infected by wCallA or wCallB Wolbachia. Other uninfected individuals were found occasionally within infected populations (Figure S3a).
3.3. Association of genetic diversity of Calligrapha and Wolbachia
The maximum‐likelihood tree of cox1 haplotype data was consistent with the multifurcate unrooted topology of Gómez‐Zurita and Cardoso (2019), with very few supported clades beyond the clear separation of bisexual and unisexual groups (B‐ and U‐clades, respectively; Figure 3). Figure 3 also shows the frequency, type, and infecting strains of Wolbachia characterized for individuals with each haplotype, as well as the arrangement of these haplotypes in relatedness groups mapped on the maximum‐likelihood tree. A number of nonrandom patterns emerged from the visual inspection of this tree: (a) Infection with Wolbachia was widespread in Calligrapha and typically as double infections, with wCallA being dominant in this pattern; (b) some groups of related haplotypes, including m ML1 (B2‐4, B2‐5, B2‐6), part of m ML2 (B2‐9, B2‐11, B2‐12), and to a good extent the unisexual lineages m UL and p UL (U2‐1), showed an alternative pattern, with dominance of uninfected individuals or infected by a single type of Wolbachia; and (c) wCallC‐type infections appeared restricted to haplotypes in the m NL3 (B2‐1) and p EL (B2‐7, B2‐8) lineages, and marginally in the unisexual U2‐1 group (e.g., m UL), usually contributing the majority of double infections with wCallA bacteria.
Figure 3.

Maximum‐likelihood tree (likelihood score = −1,869.796071) of cox1 bisexual (B) and unisexual (U) haplotypes of Calligrapha multipunctata s.l. (thinner branches) and C. philadelphica (thicker branches). Bootstrap support > 70% is shown next to the corresponding node. Haplotype groups consistent with phylogeographic lineages of Figure 1 and with the 2‐ and 3‐level nested clades of Gómez‐Zurita and Cardoso (2019), retained here for easier cross‐referencing, are shown on the right panel. Bubble graphs show the proportion of individuals with a given haplotype which are uninfected, infected by a single strain of Wolbachia, with double infections, and infected by Wolbachia of sequence types wCallA, wCallB, and wCallC
The existence of those and other more subtle nonrandom patterns was also confirmed statistically on the evolutionary lineages of Calligrapha defined by the nested design of Gómez‐Zurita and Cardoso (2019). Permutation contingency tests of haplotype groups and different ways to code the genetic diversity of infecting Wolbachia showed statistically significant associations at all higher hierarchical levels (whole dataset, 5‐ and 4‐level clades) in the mtDNA genealogy, and progressively fewer significant associations at lower levels: 83.0% of 3‐level, 31.0% of 2‐level, and 18.5% of 1‐level clades. These associations were generally found for all coding strategies of infection, including its presence/absence, whether it was unique or by two strains of Wolbachia, but also considering the main groups of Wolbachia (wCallA–wCallC), and the specific sequence types associated with each cox1 haplotype group or Wg genotypes (Table 3). Of particular interest was the statistically significant difference found between bisexual and unisexual mtDNA lineages, the latter characterized by the lack of infection or by single infections by wCallA Wolbachia, omnipresent in the system. This result alone, where the shift to unisexuality appears associated with the loss, not the gain, of an infection, makes it very unlikely that the endosymbiont was responsible for the shift in reproductive mode in Calligrapha.
Table 3.
Results of permutation contingency tests between categorical variables defined by the nested groups within each higher‐level nesting category as defined in Gómez‐Zurita and Cardoso (2019) and shown in Figure 3, and different partitioning of the characteristics of Wolbachia infections
| Clade | N | Infection level | Infection level and group | Group | Genotypea | Genotype | Infection level and genotypea | Infection level and genotype | N | Infection level and Wg genotype | N | Wolbachia and Wg genotypes |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B1‐1 | 114 | 10.5062ns | 26.3025ns | 33.9207ns | 152.1504* | 135.5373* | 149.6708ns | 131.9611ns | 99 | 72.3118ns | 92 | 417.9370* |
| B1‐3 | 41 | 30.9013*** | 30.9013** | 21.9500* | 49.5733*** | 30.1257*** | 69.0863*** | 51.9296*** | 38 | 26.4478ns | 37 | 49.7538ns |
| B1‐12 | 18 | 0.1324ns | 0.1324ns | 0.1328ns | 17.8763* | 17.3022* | 18.0000* | 17.0000* | 18 | 2.1176ns | 17 | 19.0269ns |
| B1‐20 | 9 | 3.6000ns | 12.6000* | 8.0357ns | 8.7500ns | NA | 12.6000* | NA | 9 | 3.7500ns | 9 | 15.2500ns |
| B1‐23 | 13 | 23.0000** | 23.0000** | 24.7500* | 35.7500* | 13.1250ns | 39.0000*** | 24.0000** | 13 | 18.2963ns | 12 | 23.6667ns |
| B2‐1 | 220 | 61.4040*** | 188.9107*** | 125.3078*** | 292.1368*** | 285.1533*** | 345.3811*** | 325.4300*** | 196 | 164.0585ns | 229 | 940.5087* |
| B2‐3 | 37 | 2.9321ns | 2.9321ns | 3.1912ns | 28.6671*** | 25.6945*** | 26.6591*** | 23.9338*** | 36 | 8.0000ns | 30 | 32.3180ns |
| B2‐7 | 43 | 4.1658ns | 30.9205** | 14.5367* | 23.7644** | 21.3997** | 37.4397*** | 34.0385*** | 37 | 21.3215ns | 34 | 63.8681ns |
| B3‐1 | 232 | 2.4728ns | 40.2968*** | 32.8445*** | 36.7553** | 34.1012** | 79.9876*** | 73.1354*** | 207 | 170.0170ns | 240 | 961.3259* |
| B3‐3 | 31 | 16.0423*** | 16.0423*** | 16.0423*** | 16.8095** | 2.4561ns | 16.8095** | 2.4561ns | 28 | 19.8333* | 20 | 12.6191ns |
| B3‐4 | 58 | 0.7942ns | 2.7529ns | 1.3826ns | 27.4388*** | 27.4371*** | 30.3779*** | 30.8729*** | 52 | 20.0018ns | 47 | 120.6781ns |
| B3‐5 | 28 | 43.5436*** | 43.5436*** | 39.6610*** | 55.7508*** | 16.5128** | 52.4103*** | 24.1641*** | 26 | 22.2897ns | 19 | 23.4069ns |
| B4‐1 | 269 | 10.4523** | 73.6027*** | 60.9202*** | 76.1527*** | 76.4096*** | 82.1639*** | 85.1756*** | 243 | 190.9150ns | 270 | 1,187.9560* |
| B4‐2 | 89 | 36.0953*** | 73.8563*** | 98.5158*** | 98.8854*** | 99.1529*** | 73.8563*** | 73.0000*** | 80 | 89.1380** | 67 | 281.3043ns |
| B4‐3 | 86 | 20.8517*** | 20.8517*** | 23.2303*** | 32.0679*** | 9.9204* | 29.7042*** | 10.4934* | 84 | 55.4334ns | 77 | 87.0427ns |
| B5‐1 | 444 | 29.7130*** | 233.0242*** | 85.6091*** | 443.6518*** | 451.1974*** | 515.8262*** | 529.3976*** | 407 | 280.4885** | 415 | 2,768.9090* |
| U2‐1 | 69 | 37.5108*** | 42.9322*** | 33.2761*** | 39.6440* | 26.53214* | 46.5465*** | 33.6566** | 62 | 48.0701** | 31 | 73.3656* |
| Total | 506 | 82.6303*** | 96.0976*** | 118.8766*** | 136.1692*** | 46.55018** | 129.9034*** | 97.6395*** | 469 | 370.2899*** | 446 | 2,834.5360* |
“Infection level” considers single, double, and lack of infection; “group,” any of wCallA to wCallC types; and “genotype,” individual MLST types. Only nesting categories with any of the tests producing significant results are shown. The last four columns show the results of permutation contingency tests of Wolbachia infection type and genotypes relative to the Wg genotypes of the individuals involved as characterized by Gómez‐Zurita and Cardoso (2019). In every case, N represents the number of individual observations considered in the test.
The test includes a category for uninfected individuals.
.05 ≥ p > .01
.01 ≥ p > .001
p ≤ .001 (Significance).
3.4. Temporal buildup of the association of Calligrapha with Wolbachia
The close association between mtDNA genealogy (and Wg diversity to some extent) and Wolbachia infections showed that it was possible to estimate a dominant ancestral infection state for major groups of haplotypes, strengthening the perception of a tightly linked association between the evolution of mtDNA in Calligrapha and their endosymbionts (Figure 4). For most major lineages, their ancestor was inferred as carrying symbionts compatible with their extant infection status, or in other words, closely related haplotypes typically showed similar infection status. For example, the most recent common ancestor of haplotypes B59‐B73 of C. philadelphica (p EL lineage; clade B3‐4 of Gómez‐Zurita & Cardoso, 2019) was inferred with 51.6% probability of carrying a double Wolbachia infection of the wCallA and wCallC types, as found in 63.8% of the individuals examined (the next possible state inferred was 14.7% probability of lack of infection). Also, the ancestor of the large clade including the m CL and m NL lineages (clade B3‐1) was inferred a 56.7% combined probability of carrying a wCallA‐type Wolbachia alone or, most likely, doubly infecting with wCallB‐type Wolbachia. The most recent common ancestor of the U‐clade showed a highest probability of being either uninfected (37.7%) or infected by a wCallA‐type Wolbachia (32.5%), reinforcing the idea of the lack of association of unisexuality in Calligrapha with any particular type of Wolbachia infection.
Figure 4.
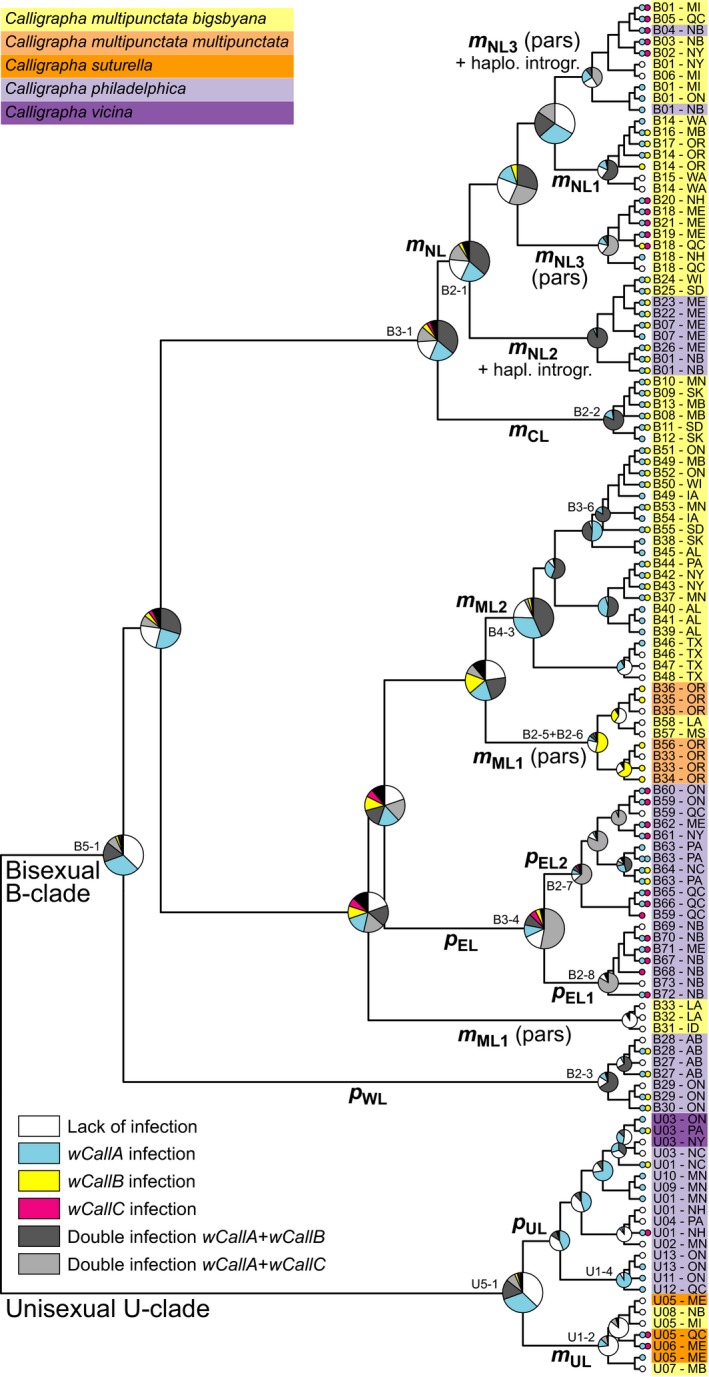
Evolution of the level and type of Wolbachia infection as deduced from a Bayesian trait analysis of Wolbachia associations on the cox1 genealogy of Calligrapha. The specific infection status of each terminal in the genealogy is shown with colored symbols using the same code as in the legend, and pie charts at each node show the relative posterior probabilities for each type of infection inferred for a particular ancestor. The major phylogeographic lineages from Figure 1 that can be identified in the genealogy are labeled accordingly, as well as their correspondence with the nested clades of Gómez‐Zurita and Cardoso (2019)
4. DISCUSSION
4.1. Calligrapha beetles host a diverse and partially incompatible array of Wolbachia
The first relevant, novel result from this study is confirming that Wolbachia occurs with high prevalence in natural populations of Calligrapha beetles. Fifteen closely related supergroup‐A MLST variants of Wolbachia (Lo, Casiraghi, Salati, Bazzocchi, & Bandi, 2002) were characterized in a continental sample of four species of Calligrapha. None of these variants had been previously recorded in other insect hosts, although similar STs, sharing allele combinations for up to four MLST markers, had been described already for other beetles. These include variants Cobs_A in the weevil Ceutorhynchus obstrictus (Floate, Coghlin, & Dosdall, 2011) and Ocac_A in the leaf beetle Oreina cacaliae (Montagna et al., 2014). The diversity of Wolbachia associated with Calligrapha is relatively low and exclusive, suggesting limited divergence within the host. However, the relative conservation of MLST loci implies a certain risk to misjudge horizontal transfer between distant Calligrapha species or from other hosts (e.g., Kraaijeveld, Franco, Knijff, Stouthamer, & Alphen, 2011). The successful recognition of meaningful evolutionary and geographic patterns in the data is interpreted here as limited or absent random horizontal transfer events, which would obscure these patterns.
Another relevant finding recognizes that most Calligrapha specimens studied here support double infections of Wolbachia and that coexisting strains are not random, but consist of simultaneous infections of wCallA with either wCallB or wCallC bacteria. Very early, since the pioneering screenings of invertebrates for the presence of Wolbachia (Perrot‐Minnot, Guo, & Werren, 1996), it became apparent that the host could support two—and in some cases up to three, as in the adzuki bean beetle (Kondo, Ijichi, Shimada, & Fukatsu, 2002)—strains of the endosymbiont in their tissues. Double infections, maintained in certain circumstances by offering a selective advantage to both symbionts and the host (Engelstädter, Hammerstein, & Hurst, 2007; Vautrin, Charles, Genieys, & Vavre, 2007), were soon interpreted under the light of the effects on host phenotype as well as the compatibility dynamics between bacterial strains affecting the outcome of host reproduction. These effects of superinfections range from feminization of males (Hiroki, Tagami, Miura, & Kato, 2004) to reproductive incompatibility of male individuals with double infections with uninfected females or females infected by a single strain, with each strain individually incompatible with the other, a phenomenon dubbed as cytoplasmic incompatibility (Merçot, Llorente, Jacques, Atlan, & Montchamps‐Moreau, 1995; Perrot‐Minnot et al., 1996).
4.2. Elements supporting cytoplasmic incompatibility in the system
Cytoplasmic incompatibility is one of the biological processes that have been studied more intensively for Wolbachia (Bourtzis, Braig, & Karr, 2003; Charlat, Bourtzis, & Merçot, 2001; Engelstädter & Telschow, 2009). At present, we know that several strains of Wolbachia infect the studied species of Calligrapha, and a wealth of indirect evidence points at the existence of incompatibilities between these strains of Wolbachia. Unfortunately, proving the existence of cytoplasmic incompatibility in the system would require breeding experiments, which are currently unattainable. One line of reasoning considers the prevalence of double infections in Calligrapha. This condition is only stable when the bacteria induce cytoplasmic incompatibility or increase the fitness of the host, with selection favoring in both cases individuals carrying high symbiotic diversity (Vautrin et al., 2007). Additional evidence for cytoplasmic incompatibility clearly emerges when the distribution of each type of Wolbachia is examined within each pool of individuals where the conflict should chiefly operate, that is, each single host species. In C. multipunctata bigsbyana, the wCallA type is widespread and always syntopic (generally as double infections) with the types wCallB and wCallC (Figure S2a). However, the latter two are allopatric, with wCallB distributed from the Pacific coast to the easternmost limit of Lake Ontario, and wCallC distributed between this region and the Atlantic shores (Figure S2b). This clear‐cut geographic pattern is suggestive of a tension zone limiting each strain to spread over the range of the other, likely because of cytoplasmic incompatibility as shown for other systems, including other leaf beetles in North America (Roehrdanz & Levine, 2007). Alternatively, this pattern could reflect some kind of environmental filtering for Wolbachia (Keller, Windsor, Saucedo, & Werren, 2004), but this explanation is at present more speculative than cytoplasmic incompatibility, and it clashes with the following observations. In particular, the analogous pattern for C. philadelphica offers additional indirect evidence for the incompatibility between strains wCallB and wCallC. As before, wCallA occupies the whole range of this species, nearly always coexisting with wCallB and wCallC, in turn broadly parapatric (Figure S2c). However, the latter coexist in New England and eastern Canada (Figure S2d), but they do in the area of sympatry of two deeply divergent mtDNA lineages of C. philadelphica: one group of haplotypes shared (B1) or closely allied (B4, B7) to these in C. m. bigsbyana (in the m NL lineage), and carrying the wCallB type; and one clearly separated from C. m. bigsbyana (haplotypes B67–B73 in p EL1) and ancestrally carrying the wCallC type. In this case, incompatibility as a postmating isolation mechanism may be also in the interest of the host retaining the identity of each divergent lineage, and a possible driver for speciation (e.g., Brucker & Bordenstein, 2012; Rokas, 2000; Telschow, Hilgenboecker, Hammerstein, & Werren, 2014; Wade, 2001).
4.3. History of Calligrapha mtDNA expansion and its association with Wolbachia
Gómez‐Zurita and Cardoso (2019) demonstrated that species paraphyly for mtDNA in Calligrapha is not at odds with the existence of clear phylogeographic patterns and also coherence with the nuclear genomic background (as represented by the single‐copy gene marker wingless, Wg) for individual mtDNA lineages. Several major independent lineages modified and expanded their ranges in different areas of North America in a coherent manner, influenced by glaciation cycles contracting them to a number of refugia from where secondary expansions occurred after the LGM (Gómez‐Zurita & Cardoso, 2019). Interestingly, these lineages have statistically demonstrated specific associations with Wolbachia too. Figure 5 condenses the phylogeographic history of these Calligrapha lineages, their association with Wolbachia, and the inferred changes in this association throughout the history of this group of beetles. The mere existence of the association suggests that Wolbachia could have influenced mtDNA evolution in the analyzed species of Calligrapha, as proposed in several studies of similar geographic (Baudry, Bartos, Emerson, Whitworth, & Werren, 2003; Raychoudhury et al., 2010) and/or taxonomic scopes (Jäckel, Mora, & Dobler, 2013; Roehrdanz & Levine, 2007). In fact, it was suggested that the occurrence of more or less dramatic symbiont‐driven effects on mtDNA diversity patterns would be the norm when this association exists (Hurst & Jiggins, 2005). However, it is also possible that this association reveals a superimposed combination of ancestral founder events of the endosymbiont, hitchhiking and expanding its own range thanks to the expansion of the beetle populations. Three lines of evidence support that the association may have weak or only local effects on the mtDNA evolution of Calligrapha: (a) mtDNA diversity is high and without the signature for selective sweeps or bottlenecks at least for major groups and based on the implementation of Tajima's test of neutrality (Gómez‐Zurita & Cardoso, 2019); (b) mtDNA lineages within each species are also distinctive for a nuclear marker, wingless, which reinforces the idea of historical separation of these groups (Gómez‐Zurita & Cardoso, 2019) and not specific selection of mtDNA by endosymbionts; and (c) mtDNA lineages associated with specific sequence types of Wolbachia, when expanding their range in an area with a different strain of Wolbachia, shift their association, usually incorporating the local strain, independently of their mtDNA type. This could easily occur via paternal transmission from local residents to colonists uninfected or carrying the wCallA type only, to account for symbiont incompatibility dynamics, as explained above. The examples below also consider that these shifts could occur via horizontal transmission from the environment or other species infected by the local Wolbachia strains without altering the idea of the endosymbiont not affecting mtDNA evolution.
Figure 5.

Phylogeographic history of the B‐ and U‐clades of Calligrapha multipunctata, C. philadelphica, and their derived unisexual species, C. suturella and C. vicina, as inferred by Gómez‐Zurita and Cardoso (2019), showing the current association of each lineage with the dominant type of Wolbachia infection and the inferred changes in this association through time and space
In the case of the B‐clade, C. philadelphica p WL and p EL lineages originated in the Northern Appalachians during the Middle Pleistocene (Gómez‐Zurita & Cardoso, 2019). Lineage p WL spread west, reaching southern Alberta, and it is statistically different from other lineages regarding the type of Wolbachia infection. p WL individuals are typically characterized by double infections with wCallA and wCallB Wolbachia, with an ancestor that likely showed this condition already. An evolutionary branch (p EL1) of linage p EL colonized and spread in the northern Appalachians and northeastern Atlantic region, also typically doubly infected with Wolbachia, but in this case with wCallA and wCallC bacteria. The other evolutionary branch (p EL2) expanded southwest along the Appalachians, reaching the Blue Ridge Mts., accompanied by a derived shift from a wCallA/wCallC to wCallA/wCallB infection, consistent with the geographic background of the bacteria. This clearly exemplifies the uncoupling of the characteristics of the Wolbachia association from the evolution of mtDNA in Calligrapha, which shows the history of the host, not a symbiont‐driven mtDNA sweep.
The remaining B‐clade lineages include most of the samples of C. multipunctata, with a southern group associated with the Mississippi basin (m ML), and a northern group distributed from Eastern Canada and New England to the Pacific Northwest (m NL and m NC). The m ML1 branch of the southern Mississippian lineage is exclusively formed by uninfected individuals or individuals only infected by wCallB‐type Wolbachia. It is different from its sister lineage, m ML2, which spread north in the Late Pleistocene and where individuals display either simple infections with wCallA‐type Wolbachia or double wCallA/wCallB infections. It is uncertain whether the ancestor of the m ML2 lineage carried the double infection of the wCallA and wCallB types or if it was only host to the former, but the first option received higher probability (56.2%) than the second (32.8%), suggesting a possible early acquisition of the wCallA infection during its range expansion north. Derived populations and haplotypes of this mtDNA lineage are mainly distributed around the Great Lakes area and show a generalized double wCallA/wCallB infection, statistically different relative to ancestral sources and populations in southernmost locations, with a relative dominance of lack of infection or single wCallA infections. The changes in the association with Wolbachia in this case would be also congruent with the history and range expansion of the beetles, without noticeable effects of the symbiont on mtDNA diversity or hierarchy.
In the northern lineage of C. multipunctata, one group of descendants (m NL2) remained in the upper courses of the Mississippi and the Missouri Rivers, and they uniformly display a wCallA/wCallB association with Wolbachia, as other mtDNA lineages in the area. Another group (m NL1) crossed the Rocky Mountains and arrived to the Pacific Northwest, evolving a different association with Wolbachia by losing the wCallB type. Contemporarily, another lineage (m NL3) spread east reaching to Nova Scotia. This evolutionary branch is polymorphic for the type of Wolbachia infections, with individuals showing single wCallA or double wCallA/wCallB infections, as supposedly carried by the ancestor of the lineage. However, most of them show a double wCallA/wCallC infection, consistent with the colonization over the area that seems the natural range of the wCallC strain of Wolbachia. At the edge of this expansion, in Maine and New Brunswick, this C. multipunctata bigsbyana lineage hybridized with C. philadelphica (Gómez‐Zurita & Cardoso, 2019). Interestingly, the specimens of C. philadelphica embedded in this group also inherited with very high probability a double wCallA/wCallB infection, the ancestral condition for this whole C. multipunctata lineage, and this trait, together with their divergent mtDNA, differentiates them from sympatric C. philadelphica, typically with a wCallA/wCallC infection. The m CL evolutionary branch of the northern C. multipunctata bigsbyana lineage colonized the Northern Great Plains and survived the LGM locally. This lineage was ancestrally associated with wCallA‐ and wCallB‐type Wolbachia, coexisting and sharing endosymbiotic makeup with descendants of two divergent mtDNA lineages, as discussed previously, reinforcing the idea of Calligrapha changing or adopting the diversity of Wolbachia present in the ranges they expand to, without obvious selective sweeps for mtDNA.
As mentioned above, high genetic diversity of the host organelle genome is usually interpreted as Wolbachia not having an influence on extant mtDNA diversity. However, it has been shown also analytically that the mere presence of Wolbachia in a system already has an impact on mtDNA polymorphism and nonsynonymous substitution rates, among others (Cariou, Duret, & Charlat, 2017). Moreover, there are potential alternative explanations for this pattern without challenging the role of Wolbachia in manipulating mtDNA diversity, for example, considering multiple sweeps associated with multiple infections by divergent strains (Frost, Hernández‐Marín, Smith, & Hughes, 2010; Symula et al., 2013). Nevertheless, in the case of Calligrapha, the observed pattern is consistent with a dynamics of multiple infection, extinction, or turnover involving few strains of Wolbachia, where the strain of bacteria can change, but without evidence for loss or replacement of mtDNA diversity in the host, at least beyond the establishment of the main lineages. The results obtained for Calligrapha, where geography or even demography plays a major role in the structure and diversification of mtDNA despite of Wolbachia, parallel those obtained for spider mites across China (Chen, Zhang, Du, Jin, & Hong, 2016), or to some extent those for the oak gallwasp in Europe (Rokas, Atkinson, Brown, West, & Stone, 2001), increasing the body of Wolbachia literature reporting lack of evidence for selective sweeps, or at least showing that these are not a dominant feature for the mtDNA evolution of the host (Bereczki, Rácz, Varga, & Tóth, 2015; Keller et al., 2004).
4.4. Wolbachia is not responsible for current unisexuality in Calligrapha
The main motivation of the current study was examining whether reproductive manipulation by Wolbachia could explain the deep mtDNA split and the apparent association of one type of mtDNA with unisexuality in Calligrapha (Montelongo & Gómez‐Zurita, 2015). The lack of strong evidence for selective mtDNA sweeps in Calligrapha already suggests that Wolbachia is not manipulating reproduction in this system in ways that condition mtDNA evolution, at least in recent times. These sweeps are only expected to occur when Wolbachia alters the outcome of host reproduction inducing unisexuality (Hurst & Jiggins, 2005; Johnstone & Hurst, 1996). Indeed, the lines of Calligrapha with the most dramatic alterations in reproduction, that is, those including unisexual taxa in the U‐clade, are either devoid of Wolbachia or are infected by the most abundant and widespread strain of Wolbachia, which is also present in populations that do not show any reproductive bias.
The history of the U‐clade and the evolution of its association (or lack thereof) with Wolbachia reinforce the idea of this association being disconnected from the origin of hybrid unisexual species. In the case of unisexual Calligrapha species, Wolbachia may at most benefit from this reproductive mode, as has been suggested, for instance, for unisexual Eusomus weevils in Europe (Mazur et al., 2016). Taxonomically recognizable unisexual species of Calligrapha derive from a female‐only lineage of bisexual Calligrapha (Gómez‐Zurita & Cardoso, 2019). The common ancestor of this lineage was inferred in the Great Lakes area during the Late Pleistocene (Gómez‐Zurita & Cardoso, 2019), and with high probability, it was not associated with Wolbachia, or only with the wCallA strain. This group includes two sublineages, one with C. multipunctata bigsbyana and C. suturella (m UL) and one with C. philadelphica and C. vicina (p UL), and they also show statistically significant differences in the type of association with Wolbachia. The first is typically free from Wolbachia, and the second includes a lineage that expanded south, also without Wolbachia, and one that spread east and west in the Late Pleistocene and typically with wCallA Wolbachia. Despite relatively small samples sizes, both unisexual species embedded in this unisexual group have individuals uninfected, infected by Wolbachia of the wCallA type, and doubly infected. The latter are with the wCallB type in the case of C. vicina and the wCallC type in the case of C. suturella, consistent with the segregated ranges of these Wolbachia types, and ruling out any cause/effect association between extant Wolbachia and reproductive strategy. Both species originated in the Holocene, and the short evolutionary time since the establishment of these unisexual lineages suggests that their polymorphic association with Wolbachia could be the result of independent founder events. This scenario was plausibly considered by Gómez‐Zurita and Cardoso (2019) and would imply multiple origins from maternal parentals with different associations with the bacteria. The evidence amounts to a change in reproductive mode in the analyzed unisexual lineages irrespective of the hypothetical effects of Wolbachia, which would not be in principle responsible for this evolutionary transition in Calligrapha, as also recognized for other systems (Ma & Schwander, 2017).
4.5. Was evidence for the role of Wolbachia in the reproductive transition lost?
So far, all available information was interpreted to argue against Wolbachia being responsible for the transition from bi‐ to unisexuality in Calligrapha. This interpretation considers the relatively strong evidence against mtDNA selective sweeps in this system and the current and inferred ancestral lack of Wolbachia or special strain of Wolbachia in the unisexual lineage and unisexual species of these beetles. However, there is a particular scenario that would still require the participation of the symbiont for the transition in reproductive mode, yet erasing the required proof for this involvement. This scenario considers that the observed reduced prevalence of Wolbachia or trend to single wCallA infections of the unisexual mtDNA branch of the Calligrapha tree is not the ancestral, but a derived state for this lineage. If unisexual reproduction is a stable condition in this evolutionary lineage of Calligrapha, their reproductive independence from males may have relaxed the selective pressures imposed by cytoplasmic incompatibility, which gives advantage to infected females and the spread of superinfections in the populations (Turelli, 1994; Vautrin et al., 2007). This relaxation, together with evolutionary independence from bisexual populations of conspecific Calligrapha (in turn potentially reinforced by cytoplasmic incompatibility between superinfected B‐type individuals and uninfected U‐type individuals), could explain the gradual loss of Wolbachia in the lineage, as predicted by the theory (Hurst, Jiggins, & Pomiankowski, 2002; Zug et al., 2012). In support to these forces being at play, it is worth noting that there are 14 localities where individuals with B‐ and U‐clade haplotypes are sympatric, often collected on the same plant, and in most of these localities (78.6%), each mtDNA lineage shows a different infection status, whereby the B‐type individuals are usually (79.3%) doubly infected and the U‐type individuals are either uninfected (52.6%) or exclusively infected with wCallA Wolbachia (28.9%). These data suggest that horizontal or sexual transmission of Wolbachia between these divergent lines is unlikely. In these circumstances, considering the relatively old age of the split between the U and the B mtDNA types, dated at some 2.20–9.46 Ma (Gómez‐Zurita & Cardoso, 2019) and in the same range of spread/extinction dynamics of Wolbachia proposed by Bailly‐Bechet et al. (2017), it is possible that the record of the ancestral infection status of the U‐lineage has been lost. With this loss, we may have also lost evidence for the actual liability of Wolbachia and of a particular strain not characterized in this study to explain an ancestral sweep (but allowing for subsequent accumulation of mtDNA polymorphism) and transition to unisexuality, responsible for the divergent evolutionary pathways in the first place (see, e.g., Dyer, Burke, & Jaenike, 2011). In this case, the putatively extinct symbiont, which should have been able to induce unisexuality, would have spread in a number of lineages of Calligrapha in a wave of multiple hybridization events that also introgressed the taxonomically unascribable U mtDNA type (Montelongo & Gómez‐Zurita, 2015). However, while this possibility is theoretically possible, in the absence of additional evidence, it is safer to dissociate Wolbachia from the origin of unisexuality in Calligrapha.
CONFLICT OF INTEREST
The author declares no conflict of interest in relation to the work described in this article.
AUTHOR CONTRIBUTIONS
JG‐Z designed the study, prepared and analyzed the data, processed and interpreted the results, and wrote the manuscript.
Supporting information
ACKNOWLEDGMENTS
This work has clearly benefited from a number of anonymous reviewers who have helped me through the process of increasing focus, rigor, and relevance of this text, and their help is much appreciated. Anabela Cardoso and Tinguaro Montelongo (IBE, Barcelona), Daniel J. Funk (Vanderbilt Univ., Tennessee), and Daniel P. Duran (Drexler University, Pennsylvania) helped me over the years to achieve the sampling that allowed this study. The help of Josep A. Roca (IBE, Barcelona) was crucial to obtain most of the sequence data from Wolbachia presented in this work, and I am profoundly indebted to him. Anabela Cardoso (IBE, Barcelona) also contributed sequences for this work and discussion of the results. This study was possible specifically thanks to funds made available by the Spanish Ministry of Economy (MINECO) through Grant No. CGL2014‐52937‐P, but it also benefited from samples acquired thanks to previous projects funded by the same Organism, including Grant Nos. CGL2011‐23820 and CGL2008‐00007, the latter with contribution from the European Union Regional Development Fund (ERDF).
Gómez‐Zurita J. Assessment of the role of Wolbachia in mtDNA paraphyly and the evolution of unisexuality in Calligrapha (Coleoptera: Chrysomelidae). Ecol Evol. 2019;9:11198–11214. 10.1002/ece3.5621
DATA AVAILABILITY STATEMENT
Vouchers of the specimens used in the study are part of the research collection of the author at the Institute of Evolutionary Biology (CSIC‐Universitat Pompeu Fabra) in Barcelona (Spain). Sequences have been deposited in the public nucleotide sequence repository GenBank under accession numbers LR135794–LR135810.
REFERENCES
- Bailly‐Bechet, M. , Martins‐Simões, P. , Szöllõsi, G. J. , Mialdea, G. , Sagot, M.‐F. , & Charlat, S. (2017). How long does Wolbachia remain on board? Molecular Biology and Evolution, 34, 1183–1193. 10.1093/molbev/msx073 [DOI] [PubMed] [Google Scholar]
- Baldo, L. , Dunning Hotopp, J. C. , Jolley, K. A. , Bordenstein, S. R. , Biber, S. A. , Choudhury, R. R. , … Werren, J. H. (2006). Multilocus sequence typing system for the endosymbiont Wolbachia pipientis . Applied Environmental Microbiology, 72, 7098–7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudry, E. , Bartos, J. , Emerson, K. , Whitworth, T. , & Werren, J. H. (2003). Wolbachia and genetic variability in the birdnest blowfly Protocalliphora sialia . Molecular Ecology, 12, 1843–1854. [DOI] [PubMed] [Google Scholar]
- Bereczki, J. , Rácz, R. , Varga, Z. , & Tóth, J. P. (2015). Controversial patterns of Wolbachia infestation in the social parasitic Maculinea butterflies (Lepidoptera: Lycaenidae). Organisms Diversity & Evolution, 15, 591–607. [Google Scholar]
- Bourtzis, K. , Braig, H. R. , & Karr, T. L. (2003). Cytoplasmic incompatibility In Bourtzis K., & Miller T. A. (Eds.), Insect symbiosis (pp. 217–246). Boca Raton, FL: CRC Press. [Google Scholar]
- Brown, W. J. (1945). Food‐plants and distribution of the species of Calligrapha in Canada, with descriptions of new species (Coleoptera, Chrysomelidae). The Canadian Entomologist, 77, 117–133. [Google Scholar]
- Brucker, R. M. , & Bordenstein, S. R. (2012). Speciation by symbiosis. Trends in Ecology & Evolution, 27, 443–451. [DOI] [PubMed] [Google Scholar]
- Cariou, M. , Duret, L. , & Charlat, S. (2017). The global impact of Wolbachia on mitochondrial diversity and evolution. Journal of Evolutionary Biology, 30, 2204–2210. [DOI] [PubMed] [Google Scholar]
- Charlat, S. , Bourtzis, K. , & Merçot, H. (2001). Wolbachia induced cytoplasmic incompatibility In Seckbach J. (Ed.), Symbiosis (pp. 621–644). Dordrecht, The Netherlands: Kluwer Academic Publishers. [Google Scholar]
- Charlat, S. , Duplouy, A. , Hornett, E. A. , Dyson, E. A. , Davies, N. , Roderick, G. K. , … Hurst, G. D. D. (2009). The joint evolutionary histories of Wolbachia and mitochondria in Hypolimnas bolina . BMC Evolutionary Biology, 9, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlat, S. , Hurst, G. D. D. , & Merçot, H. (2003). Evolutionary consequences of Wolbachia infections. Trends in Genetics, 19, 217–223. [DOI] [PubMed] [Google Scholar]
- Chen, Y.‐T. , Zhang, Y.‐K. , Du, W.‐X. , Jin, P.‐Y. , & Hong, X.‐Y. (2016). Geography has a greater effect than Wolbachia infection on population genetic structure in the spider mite, Tetranychus pueraricola . Bulletin of Entomological Research, 106, 685–694. [DOI] [PubMed] [Google Scholar]
- Clark, M. E. , Veneti, Z. , Bourtzis, K. , & Karr, T. L. (2002). The distribution and proliferation of the intracellular bacteria Wolbachia during spermatogenesis in Drosophila . Mechanisms of Development, 111, 3–15. [DOI] [PubMed] [Google Scholar]
- Dedeine, F. , Vavre, F. , Fleury, F. , Loppin, B. , Hochberg, M. E. , & Boulétreau, M. (2001). Removing symbiotic Wolbachia bacteria specifically inhibits oogenesis in a parasitic wasp. Proceedings of the National Academy of Sciences of the United States of America, 98, 6247–6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson, S. L. , Bourtzis, K. , Braig, H. R. , Jones, B. F. , Wg, Z. , Rousset, F. , & O'Neill, S. L. (1999). Wolbachia infections are distributed throughout insect somatic and germ line tissues. Insect Biochemistry and Molecular Biology, 29, 153–160. [DOI] [PubMed] [Google Scholar]
- Drummond, A. J. , Suchard, M. A. , Xie, D. , & Rambaut, A. (2012). Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology and Evolution, 29, 1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer, K. A. , Burke, C. , & Jaenike, J. (2011). Wolbachia‐mediated persistence of mtDNA from a potentially extinct species. Molecular Ecology, 20, 2805–2817. [DOI] [PubMed] [Google Scholar]
- Engelstädter, J. , Hammerstein, P. , & Hurst, G. D. D. (2007). The evolution of endosymbiont density in doubly infected host species. Journal of Evolutionary Biology, 20, 685–695. [DOI] [PubMed] [Google Scholar]
- Engelstädter, J. , & Telschow, A. (2009). Cytoplasmic incompatibility and host population structure. Heredity, 103, 196–207. [DOI] [PubMed] [Google Scholar]
- Ferree, P. M. , Frydman, H. M. , Li, J. M. , Cao, J. , Wieschaus, E. , & Sullivan, W. (2005). Wolbachia utilizes host microtubules and dynein for anterior localization in the Drosophila oocyte. PLoS Pathogens, 1, e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fialho, R. F. , & Stevens, L. (2000). Male‐killing Wolbachia in a flour beetle. Proceedings of the Royal Society of London, Series B: Biological Sciences, 267, 1469–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floate, K. D. , Coghlin, P. C. , & Dosdall, L. (2011). A test using Wolbachia bacteria to identify Eurasian source populations of cabbage seedpod weevil, Ceutorhynchus obstrictus (Marsham), in North America. Environmental Entomology, 40, 818–823. [DOI] [PubMed] [Google Scholar]
- Frost, C. L. , Hernández‐Marín, H. , Smith, J. E. , & Hughes, W. O. H. (2010). Multiple gains and losses of Wolbachia symbionts across a tribe of fungus‐growing ants. Molecular Ecology, 19, 4077–4085. [DOI] [PubMed] [Google Scholar]
- Galtier, N. , Nabholz, B. , Glémin, S. , & Hurst, G. D. D. (2009). Mitochondrial DNA as a marker of molecular diversity: A reappraisal. Molecular Ecology, 18, 4541–4550. [DOI] [PubMed] [Google Scholar]
- Gómez‐Zurita, J. , & Cardoso, A. (2019). Phylogeographic assessment of mtDNA paraphyly and the evolution of unisexuality in Calligrapha (Coleoptera: Chrysomelidae). Journal of Zoological Systematics and Evolutionary Research, 57, 561–579. [Google Scholar]
- Gómez‐Zurita, J. , Funk, D. J. , & Vogler, A. P. (2006). The evolution of unisexuality in Calligrapha leaf beetles: Molecular and ecological insights on multiple origins via interspecific hybridization. Evolution, 60, 328–347. [PubMed] [Google Scholar]
- Gómez‐Zurita, J. , Vogler, A. P. , & Funk, D. J. (2004). Diagnosing an overlooked North American taxon: Biological observations and mitochondrial insights on Calligrapha suturella Schaeffer, 1933 new status (Coleoptera, Chrysomelidae). Annals of the Entomological Society of America, 97, 28–36. [Google Scholar]
- Gompert, Z. , Forister, M. L. , Fordyce, J. A. , & Nice, C. C. (2008). Widespread mito‐nuclear discordance with evidence for introgressive hybridization and selective sweeps in Lycaeides . Molecular Ecology, 17, 5231–5244. [DOI] [PubMed] [Google Scholar]
- Heath, B. D. , Butcher, R. D. J. , Whitfield, W. G. F. , & Hubbard, S. F. (1999). Horizontal transfer of Wolbachia between phylogenetically distant insect species by a naturally occurring mechanism. Current Biology, 9, 313–316. [DOI] [PubMed] [Google Scholar]
- Hilgenboecker, K. , Hammerstein, P. , Schlattmann, P. , Telschow, A. , & Werren, J. H. (2008). How many species are infected with Wolbachia? – A statistical analysis of current data. FEMS Microbiology Letters, 281, 215–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiroki, M. , Tagami, Y. , Miura, K. , & Kato, Y. (2004). Multiple infection with Wolbachia inducing different reproductive manipulations in the butterfly Eurema hecabe . Proceedings of the Royal Society of London, Series B: Biological Sciences, 271, 1751–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa, T. , Koga, R. , Kikuchi, Y. , Meng, X. Y. , & Fukatsu, T. (2010). Wolbachia as a bacteriocyte‐associated nutritional mutualist. Proceedings of the National Academy of Sciences of the United States of America, 107, 769–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huigens, M. E. , De Almeida, R. P. , Boons, P. A. H. , Luck, R. F. , & Stouthamer, R. (2004). Natural interspecific and intraspecific horizontal transfer of parthenogenesis‐inducing Wolbachia in Trichogramma wasps. Proceedings of the Royal Society of London, Series B: Biological Sciences, 271, 509–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst, G. D. D. , & Jiggins, F. M. (2005). Problems with mitochondrial DNA as a marker in population, phylogeographic and phylogenetic studies: The effects of inherited symbionts. Proceedings of the Royal Society of London, Series B: Biological Sciences, 272, 1525–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst, G. D. D. , Jiggins, F. M. , Graf von der Schulenburg, J. H. , Bertrand, D. , West, S. A. , Goriacheva, I. I. , … Majerus, M. E. N. (1999). Male‐killing Wolbachia in two species of insect. Proceedings of the Royal Society of London, Series B: Biological Sciences, 266, 735–740. [Google Scholar]
- Hurst, G. D. D. , Jiggins, F. M. , & Pomiankowski, A. (2002). Which way to manipulate host reproduction? Wolbachia that cause cytoplasmic incompatibility are easily invaded by sex ratio‐distorting mutants. The American Naturalist, 160, 360–373. [DOI] [PubMed] [Google Scholar]
- Ilinsky, Y. (2013). Coevolution of Drosophila melanogaster mtDNA and Wolbachia genotypes. PLoS ONE, 8, e54373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäckel, R. , Mora, D. , & Dobler, S. (2013). Evidence for selective sweeps by Wolbachia infections: Phylogeny of Altica leaf beetles and their reproductive parasites. Molecular Ecology, 22, 4241–4255. [DOI] [PubMed] [Google Scholar]
- Jiggins, F. M. , Bentley, J. K. , Majerus, M. E. N. , & Hurst, G. D. D. (2002). Recent changes in phenotype and patterns of host specialization in Wolbachia bacteria. Molecular Ecology, 11, 1275–1283. [DOI] [PubMed] [Google Scholar]
- Johnstone, R. A. , & Hurst, G. D. D. (1996). Maternally inherited male‐killing microorganisms may confound interpretation of mtDNA variation in insects. Biological Journal of the Linnean Society, 53, 453–470. [Google Scholar]
- Jolley, K. A. , & Maiden, M. C. J. (2010). BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics, 11, 595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama, D. , Nishimura, G. , Hoshizaki, S. , & Ishikawa, Y. (2002). Feminizing Wolbachia in an insect Ostrinia furnacalis (Lepidoptera: Crambidae). Heredity, 88, 444–449. [DOI] [PubMed] [Google Scholar]
- Katoh, K. , & Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Molecular Biology and Evolution, 30, 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller, G. P. , Windsor, D. M. , Saucedo, J. M. , & Werren, J. H. (2004). Reproductive effects and geographical distribution of two Wolbachia strains infecting the Neotropical beetle, Chelymorpha alternans Boh. (Chrysomelidae, Cassidinae). Molecular Ecology, 13, 2405–2420. [DOI] [PubMed] [Google Scholar]
- Kondo, N. , Ijichi, N. , Shimada, M. , & Fukatsu, T. (2002). Prevailing triple infection with Wolbachia in Callosobruchus chinensis (Coleoptera: Bruchidae). Molecular Ecology, 11, 167–180. [DOI] [PubMed] [Google Scholar]
- Kraaijeveld, K. , Franco, P. , de Knijff, P. , Stouthamer, R. , & van Alphen, J. J. M. (2011). Clonal genetic variation in a Wolbachia‐infected asexual wasp: Horizontal transmission or historical sex? Molecular Ecology, 20, 3644–3652. 10.1111/j.1365-294X.2011.05150.x [DOI] [PubMed] [Google Scholar]
- Li, Y.‐Y. , Floate, K. D. , Fields, P. G. , & Pang, B.‐P. (2014). Review of treatment methods to remove Wolbachia bacteria from arthropods. Symbiosis, 62, 1–15. [Google Scholar]
- Lo, N. , Casiraghi, M. , Salati, E. , Bazzocchi, C. , & Bandi, C. (2002). How many Wolbachia supergroups exist? Molecular Biology and Evolution, 19, 341–346. 10.1093/oxfordjournals.molbev.a004087 [DOI] [PubMed] [Google Scholar]
- Ma, W.‐J. , & Schwander, T. (2017). Patterns and mechanisms in instances of endosymbiont‐induced parthenogenesis. Journal of Evolutionary Biology, 30, 868–888. [DOI] [PubMed] [Google Scholar]
- Martinez, J. , Longdon, B. , Bauer, S. , Chan, Y. S. , Miller, W. J. , Bourtzis, K. , … Jiggins, F. M. (2014). Symbionts commonly provide broad spectrum resistance to viruses in insects: A comparative analysis of Wolbachia strains. PLoS Pathogens, 10, e1004369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazur, M. A. , Holecová, M. , Lachowska‐Cierlik, D. , Lis, A. , Kubisz, D. , & Kajtoch, L. (2016). Selective sweep of Wolbachia and parthenogenetic host genomes – the example of the weevil Eusomus ovulum . Insect Molecular Biology, 25, 701–711. [DOI] [PubMed] [Google Scholar]
- Merçot, H. , Llorente, B. , Jacques, M. , Atlan, A. , & Montchamps‐Moreau, C. (1995). Variability within the Seychelles cytoplasmic incompatibility system in Drosophila simulans . Genetics, 141, 1015–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagna, M. , Chouaia, B. , Sacchi, L. , Porretta, D. , Martin, E. , Giorgi, A. , … Epis, S. (2014). A new strain of Wolbachia in an alpine population of the viviparous Oreina cacaliae (Coleoptera: Chrysomelidae). Environmental Entomology, 43, 913–922. [DOI] [PubMed] [Google Scholar]
- Montelongo, T. , & Gómez‐Zurita, J. (2015). Nonrandom patterns of genetic admixture expose the complex historical hybrid origin of unisexual leaf beetle species in the genus Calligrapha . The American Naturalist, 185, 113–134. [DOI] [PubMed] [Google Scholar]
- Perrot‐Minnot, M.‐J. , Guo, L. R. , & Werren, J. H. (1996). Single and double infections with Wolbachia in the parasitic wasp Nasonia vitripennis: Effects on compatibility. Genetics, 143, 961–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietri, J. E. , DeBruhl, H. , & Sullivan, W. (2016). The rich somatic life of Wolbachia . MicrobiologyOpen, 5, 923–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut, A. , Suchard, M. A. , Xie, D. , & Drummond, A. J. (2014). Tracer v1.6. Retrieved from http://beast.bio.ed.ac.uk/Tracer [Google Scholar]
- Raychoudhury, R. , Grillenberger, B. K. , Gadau, J. , Bijlsma, R. , van de Zande, L. , Werren, J. H. , & Beukeboom, L. W. (2010). Phylogeography of Nasonia vitripennis (Hymenoptera) indicates mitochondrial‐Wolbachia sweep in North America. Heredity, 104, 318–326. [DOI] [PubMed] [Google Scholar]
- Robertson, J. G. (1966). The chromosomes of bisexual and parthenogenetic species of Calligrapha (Coleoptera: Chrysomelidae) with notes on sex ratio, abundance and egg number. Canadian Journal of Genetics and Cytology, 8, 695–732. [Google Scholar]
- Roehrdanz, R. L. , & Levine, E. (2007). Wolbachia bacterial infections linked to mitochondrial DNA reproductive isolation among populations of Northern Corn Rootworm (Coleoptera: Chrysomelidae). Annals of the Entomological Society of America, 100, 522–531. [Google Scholar]
- Rokas, A. (2000). Wolbachia as a speciation agent. Trends in Ecology & Evolution, 15, 44–45. [DOI] [PubMed] [Google Scholar]
- Rokas, A. , Atkinson, R. J. , Brown, G. S. , West, S. A. , & Stone, G. N. (2001). Understanding patterns of genetic diversity in the oak gallwasp Biorhiza pallida: Demographic history or a Wolbachia selective sweep? Heredity, 87, 294–304. 10.1046/j.1365-2540.2001.00872.x [DOI] [PubMed] [Google Scholar]
- Ronquist, F. (2004). Bayesian inference of character evolution. Trends in Ecology & Evolution, 19, 475–481. [DOI] [PubMed] [Google Scholar]
- Rousset, F. , Bouchon, D. , Pintureau, B. , Juchault, P. , & Solignac, M. (1992). Wolbachia endosymbionts responsible for various alterations of sexuality in arthropods. Proceedings of the Royal Society of London, Series B: Biological Sciences, 250, 91–98. [DOI] [PubMed] [Google Scholar]
- Russell, J. A. , Goldman‐Huertas, B. , Moreau, C. S. , Baldo, L. , Stahlhut, J. K. , Werren, J. H. , & Pierce, N. E. (2009). Specialization and geographic isolation among Wolbachia symbionts from ants and lycaenid butterflies. Evolution, 63, 624–640. [DOI] [PubMed] [Google Scholar]
- Schmidt, B. C. , & Sperling, F. A. H. (2008). Widespread decoupling of mtDNA variation and species integrity in Grammia tiger moths (Lepidoptera: Noctuidae). Systematic Entomology, 33, 613–634. [Google Scholar]
- Smith, M. A. , Bertrand, C. , Crosby, K. , Eveleigh, E. S. , Fernández‐Triana, J. , Fisher, B. L. , … Zhou, X. (2012). Wolbachia and DNA barcoding insects: Patterns, potential, and problems. PLoS ONE, 7, e36514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis, A. (2006). RAxML‐VI‐HPC: Maximum likelihood‐based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics, 22, 2688–2690. [DOI] [PubMed] [Google Scholar]
- Symula, R. E. , Alam, U. , Brelsfoard, C. , Wu, Y. , Echodu, R. , Okedi, L. M. , … Caccone, A. (2013). Wolbachia association with the tsetse fly, Glossina fuscipes fuscipes, reveals high levels of genetic diversity and complex evolutionary dynamics. BMC Evolutionary Biology, 13, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telschow, A. , Hilgenboecker, K. , Hammerstein, P. , & Werren, J. H. (2014). Dobzhansky‐Muller and Wolbachia‐induced incompatibilities in a diploid genetic system. PLoS ONE, 9, e95488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton, A. R. , Boerwinkle, E. , & Sing, C. F. (1987). A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping. I. Basic theory and an analysis of alcohol dehydrogenase activity in Drosophila . Genetics, 117, 343–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton, A. R. , & Sing, C. F. (1993). A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping. IV. Nested analyses with cladogram uncertainty and recombination. Genetics, 134, 659–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toomey, M. E. , Panaram, K. , Fast, E. M. , Beatty, C. , & Frydman, H. M. (2013). Evolutionarily conserved Wolbachia‐encoded factors control pattern of stem‐cell niche tropism in Drosophila ovaries and favor infection. Proceedings of the National Academy of Sciences of the United States of America, 110, 10788–10793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli, M. (1994). Evolution of incompatibility‐inducing microbes and their hosts. Evolution, 48, 1500–1513. [DOI] [PubMed] [Google Scholar]
- Turelli, M. , Hoffmann, A. A. , & McKechnie, S. W. (1992). Dynamics of cytoplasmic incompatibility and mtDNA variation in natural Drosophila simulans populations. Genetics, 132, 713–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vautrin, E. , Charles, S. , Genieys, S. , & Vavre, F. (2007). Evolution and invasion dynamics of multiple infections with Wolbachia investigated using matrix based models. Journal of Theoretical Biology, 245, 197–209. [DOI] [PubMed] [Google Scholar]
- Wade, M. J. (2001). Infectious speciation. Nature, 409, 675–677. [DOI] [PubMed] [Google Scholar]
- Weinert, L. A. , Araujo, E. V. , Ahmed, M. Z. , & Welch, J. J. (2015). The incidence of bacterial endosymbionts in terrestrial arthropods. Proceedings of the Royal Society of London, Series B: Biological Sciences, 282, 20150249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss, N. A. (2015). wPerm: Permutation tests. R package version 1.0.1. Retrieved from https://CRAN.R-project.org/package=wPerm [Google Scholar]
- Werren, J. H. (1997). Biology of Wolbachia . Annual Review of Entomology, 42, 587–609. [DOI] [PubMed] [Google Scholar]
- Werren, J. H. , Baldo, L. , & Clark, M. E. (2008). Wolbachia: Master manipulators of invertebrate biology. Nature Reviews. Microbiology, 6, 741–751. [DOI] [PubMed] [Google Scholar]
- Werren, J. H. , & Beukeboom, L. W. (1998). Sex determination, sex ratios, and genetic conflict. Annual Review of Ecology and Systematics, 29, 233–261. [Google Scholar]
- Zug, R. , Koehncke, A. , & Hammerstein, P. (2012). Epidemiology in evolutionary time: The case of Wolbachia horizontal transmission between arthropod species. Journal of Evolutionary Biology, 25, 2149–2160. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Vouchers of the specimens used in the study are part of the research collection of the author at the Institute of Evolutionary Biology (CSIC‐Universitat Pompeu Fabra) in Barcelona (Spain). Sequences have been deposited in the public nucleotide sequence repository GenBank under accession numbers LR135794–LR135810.


