Abstract
Proteins are relatively easy to synthesize, compared to nucleic acids and it is likely that there existed a stage prior to the RNA world which can be called the protein world. Some of the three-dimensional (3D) peptide structures in these proteins have, we argue, been conserved since then and may constitute the oldest biological relics in existence. We focus on 3D peptide motifs consisting of up to eight or so amino acid residues. The best known of these is the ‘nest’, a three- to seven-residue protein motif, which has the function of binding anionic atoms or groups of atoms. Ten per cent of amino acids in typical proteins belong to a nest, so it is a common motif. A five-residue nest is found as part of the well-known P-loop that is a recurring feature of many ATP or GTP-binding proteins and it has the function of binding the phosphate part of these ligands. A synthetic hexapeptide, ser–gly–ala–gly–lys–thr, designed to resemble the P-loop, has been shown to bind inorganic phosphate. Another type of nest binds iron–sulfur centres. A range of other simple motifs occur with various intriguing 3D structures; others bind cations or form channels that transport potassium ions; other peptides form catalytically active haem-like or sheet structures with certain transition metals. Amyloid peptides are also discussed. It now seems that the earliest polypeptides were far from being functionless stretches, and had many of the properties, both binding and catalytic, that might be expected to encourage and stabilize simple life forms in the hydrothermal vents of ocean depths.
Keywords: origin of life, proteins, peptides, hydrogen bonds, nest, amyloid
1. Introduction
Many protein three-dimensional (3D) structures are very old [1–3], and may be the oldest biological relics that exist. It is true that the sequences of most proteins change fairly steadily over evolutionary time, but the main chain 3D structures are often highly conserved and once formed hardly change; they can also be re-used for various functional purposes. This applies to domain structures and to the small structural motifs that are the subject of this review.
Amino acids, and even peptides, turn up fairly regularly in the various experimental broths that have been allowed to be cooked from simple chemicals, whereas nucleotides do not. This is because nucleotides are far more difficult to synthesize chemically than amino acids. It is our premise that, at the very earliest stage of the origin of life on our planet, there must have been a ‘protein world’ [4] or at least a ‘polypeptide world’ [5]. This is by no means to contradict the existence of the ‘RNA world’ [6], followed later by the ‘DNA world’, but these would have taken a substantial amount of time to appear, such that a protein world of some sort at the very beginning seems inevitable, as in figure 1. This is argued to have taken place at the ‘lost city’ type of hydrothermal vents at mid-ocean ridges where warm alkaline reduced fluid issues into an acidulous sea [7,8].
Figure 1.
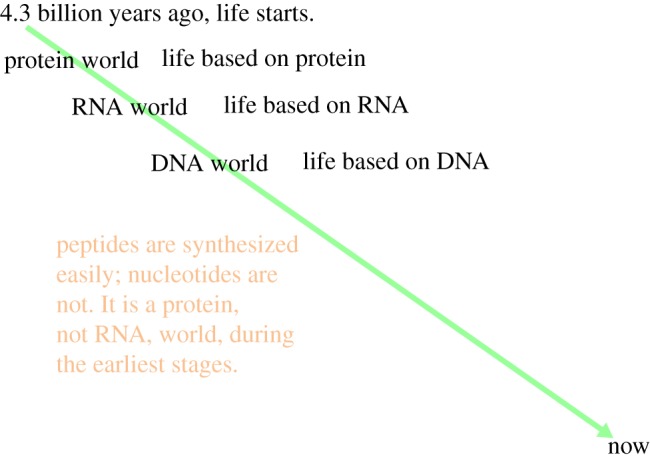
Evolutionary time diagram. (Online version in colour.)
We have been interested in detecting and analysing small motifs [9–14], commonly occurring in crystal structures of proteins and polypeptides, consisting of only a few (less than six or so) amino acid residues and held together via hydrogen bonds involving main chain rather than side chain atoms. These motifs can be examined interactively on two websites that are commonly available [12]. Four categories of such motif will be described that are especially likely to have been prevalent at the origin of life: (i) nests, (ii) potassium channels, (iii) amyloid/α-sheet, and (iv) metal–peptides.
2. Nests
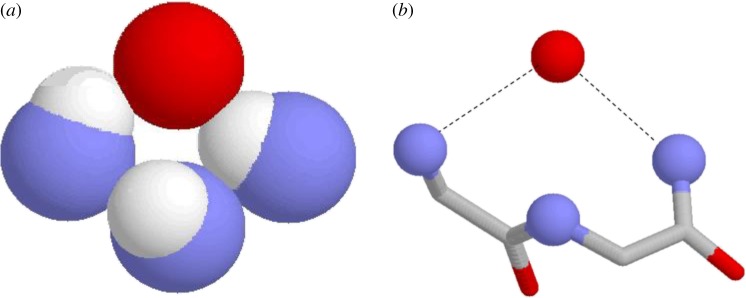
Simple nests [9] are formed by tripeptides in which the main chain NH groups form a concavity into which an anionic or partially anionic atom, sometimes called the ‘egg’ [15], may fit. This atom is hydrogen bonded to, and thus bridges, the first and third main chain NH groups of the tripeptide. The NH of the second nest residue is sometimes hydrogen bonded too, but usually it points slightly away from the anionic atom. This is seen in figure 2. The side chain atoms if any are usually not involved in the nest. Considering databases of soluble folded protein structures, it turns out [9] that 8% of amino acids belong to a nest, so these features are very common indeed; in a high proportion of nests in proteins, but by no means all, the egg atom is a carbonyl oxygen.
Figure 2.
A simple RL tripeptide nest. Nitrogen atoms are blue, oxygens red, hydrogens white, carbons grey; hydrogen bonds are shown as dashed lines. (a) Only the NH groups. (b) The main chain atoms of the first two residues but only the NH atoms of the third residue. The red spherical atom is the ‘egg’. The ϕ and ψ angles of the first two residues are given by the orange circles in figure 3. (Online version in colour.)
Nest structure is best appreciated by considering the main chain dihedral angles, ϕ and ψ, which are measures of the rotation about the N–Cα and the Cα–C bonds, respectively. The orange circles in the graph in figure 3 give the ϕ and ψ angles of the first two nest residues. One circle is labelled R and one L. The reader will gather that two kinds of nest are possible, one with R followed by L, called RL, and the other with L followed by R, called LR.
Figure 3.
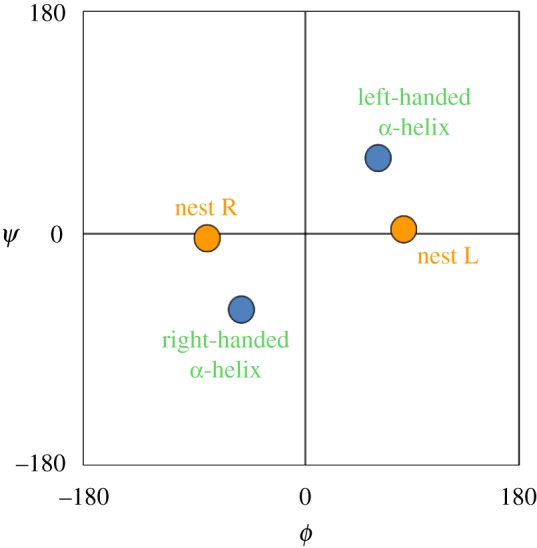
Ramachandran plot, ϕ versus ψ. (Online version in colour.)
It is worth adding that tripeptide motifs called crowns that bind anions or δ-atoms in the same way as nests, but with all ϕ and ψ angles negative for the first two residues, have been described [16]. They are far less abundant than nests but should be distinguished from them.
In proteins, about 80% of nests are RL, while 20% are LR [9]. Considering only the main chain atoms, these two types of nest are chiral or enantiomeric (mirror images to non-scientists). This sort of chirality is conformational in nature rather than configurational. d and l amino acids exhibit configurational chirality around the Cα atom, which involves the side chains; this remains whatever the conformation because it is constrained by the covalent bonds. It is useful to distinguish these two types of chirality. Configurational chirality often impinges on conformational chirality, but the two manifestations of chirality are distinct.
Graphs like those in figure 3 are called Ramachandran plots. Points that lie equidistant from, and on either side of, the central ϕ = 0, ψ = 0 point exhibit conformational chirality (with regard to the main chain atoms of a peptide). In figure 3, the two orange circles are enantiomeric conformers of this sort, and so are the two blue circles.
Consideration of figure 3 suggests the occurrence of longer nests, with alternating RLR, RLRL, LRL, etc., residues. In proteins, about 25% of simple nests are part of a longer, compound, nest. In these features, the NH groups all face inwards, towards a single point, resulting in a wider nest that can bind anionic groups rather than single atoms. For example, an RLRL nest consists of five residues and could be regarded as three overlapping simple nests. Geometrically, these motifs can be regarded as incomplete rings.
A functional example of a simple nest is given by the well-known ‘oxyanion hole’ [17] found in serine proteases, enzymes that cleave certain peptide bonds. This cleavage is enhanced by the attraction of the carbonyl oxygen of the substrate's peptide bond to an LR nest in trypsin. In figure 4, which is taken from a crystal structure of trypsin bound to a peptide bond substrate (below), the blue atoms of the nitrogens of the nest (above), also known as the oxyanion hole, are ready to accept the carbonyl oxygen of the peptide bond via N-H … O = C hydrogen bonding.
Figure 4.
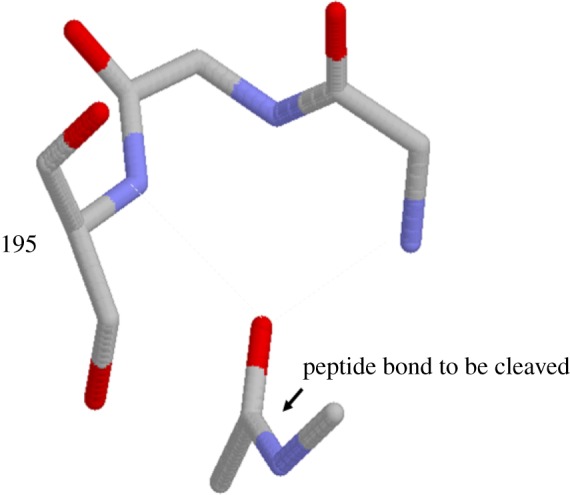
Trypsin active site nest peptide bound to substrate. Coordinates from the trypsin (above) - pancreatic trypsin inhibitor (below) complex crystal structure PDB code: 2ptc. (Online version in colour.)
The P-loop [18] is a characteristic feature of what is by far the most common group of enzymes that bind ATP or GTP. These enzymes all cleave the terminal (γ) phosphate; some transfer it to another molecule (kinases) and some transfer it to a water molecule (ATPases, GTPases). From substrate-bound crystal structures, the P-loop is seen to be a short polypeptide that loops around, and forms multiple hydrogen bonds to, the middle (β) phosphate by means of an LRLR nest, as seen in figure 5.
Figure 5.
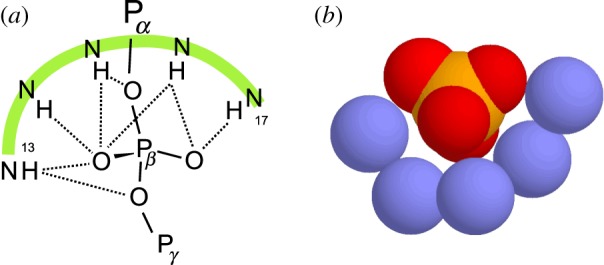
The main chain NH atoms of the P-loop nest peptide bound to the β-phosphate of GDP. Coordinates from GDP-bound P21ras crystal structure PDB code :5p21. (a) Diagram with hydrogen bonds as dashed lines. (b) Nitrogens blue, phosphorus orange, oxygens red, hydrogens omitted. (Online version in colour.)
In the GTPase G-protein [19] named P21ras, the LRLR residues correspond to particular amino acids in the sequence, as in figure 6. This sequence is familiar to many biochemists as in almost the entire family, it is conserved as GxxxxGKS/T (capital letters denote individual amino acids; small x means a variable amino acid). Of course the conserved lysine side chain amino group binds the phosphate as well as the nest, but here the focus is on the main chain feature that overlaps it.
Figure 6.
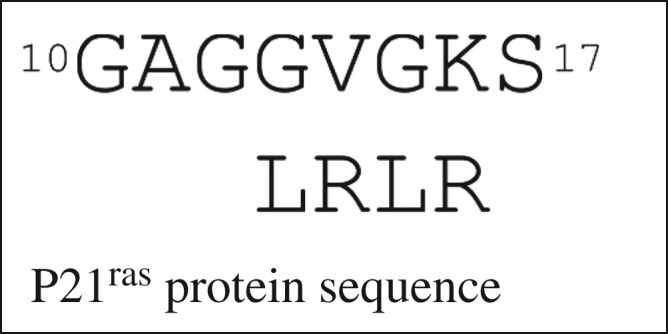
How the LRLR nest residues of P21ras fit with the sequence.
A hexapeptide was synthesized [20] with the sequence SGAGKS (the serine at the beginning was to increase the peptide's solubility). It was found to bind phosphate strongly at neutral pH values, K = 63 000 M–1. Since in all proteins, the P-loop occurs at the N-terminus of an α-helix, it had previously been assumed that an important aspect of phosphate binding was its situation. However, the hexapeptide is too short to form an α-helix and so it has to be the LRLR nest rather than the helix that gives rise to the strong phosphate binding.
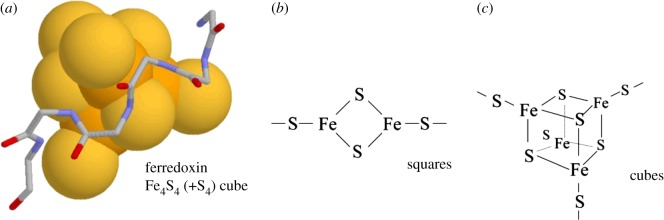
Iron–sulfur centres are diverse, in comparison to phosphate ions. They occur in proteins as three types: single Fe, Fe2S2 (square), four Fe2S4 (cuboid). In each type, the iron or iron–sulfur centre is bonded covalently to four cysteine sulfur atoms. About 50% of these centres are also bound to a nest [9], often a compound nest. An example from the protein ferredoxin is illustrated in figure 7a. The main chain NH groups are hydrogen bonded to sulfur rather than oxygen atoms. The figure also reveals the characteristic sinuous or s-shape exhibited by the main chain atoms of nests.
Figure 7.
Iron–sulfur centres in proteins. (a) The main chain atoms (no hydrogens) are shown as sticks for the nest surrounding the iron–sulfur centre, shown as spheres (iron atoms rust, sulfurs yellow); coordinates from ferredoxin crystal structure PDB code: 2fxn; (b,c) types of iron–sulfur centre. (Online version in colour.)
Both the phosphate binding and the iron–sulfur-centre binding nests are expected to have been functionally useful at the origin of life [11,21,22], as illustrated in figure 8.
Figure 8.
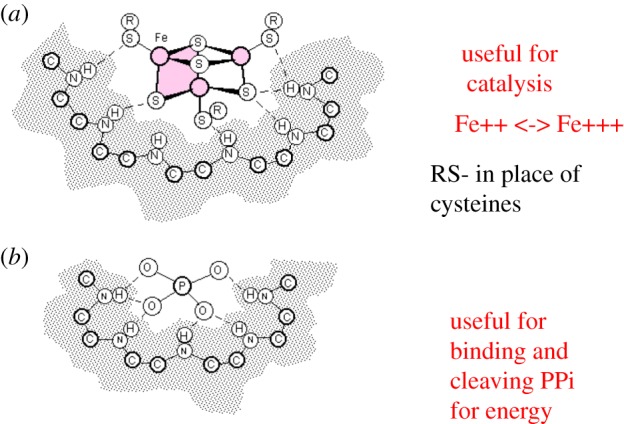
Functional peptide nests. (a) Fe3S4 in peptide nest and (b) phosphate in peptide nest. (Online version in colour.)
Iron–sulfur centres at the earliest stage of the origin of life are not expected to bind to cysteines as in present-day proteins. However, the hydrothermal fluid issuing from the hydrothermal vents where life is thought to have evolved is relatively rich in organic sulfides such as methanethiol and these would be expected to complete the iron–sulfur centre complexes. The nest function is twofold; on the one hand, it binds and stabilizes the iron–sulfur centres; on the other hand, it modulates the oxidation–reduction potential of the potentially catalytically active Fe++/Fe+++ atoms, altering their ability to receive or donate single electrons.
Phosphate-binding nests could have been useful in the provision of energy for metabolism. We have previously argued that polyphosphate hydrolysis (providing energy) coupled to polypeptide synthesis is a candidate for the earliest form of metabolism [23,57]. Since P-loop enzymes catalyse hydrolysis of polyphosphate as mentioned earlier, this functionality may have been retained since the earliest stages of life.
3. Potassium channels
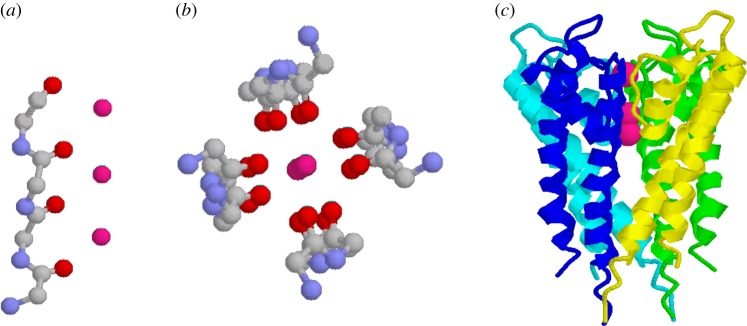
Referring back to figure 3, in nests the main chain atoms of successive amino acids are enantiomeric as shown by the orange circles. If an apparently small change is made such that main chain atoms of successive amino acids are enantiomeric in the way shown by the blue circles, the conformation alters such that the nest concavity is lost and the peptide becomes linear, as shown in figure 9a. This is the conformation found [10,24] in the selectivity filter of the potassium channel protein. Instead of NH groups binding an anion as in concave nests, here the CO groups bind a cation. Four peptides form a ring, as in figure 9b,c, pointing their CO groups inwards to create a channel.
Figure 9.
Potassium channel and selectivity filter. Coordinates are from protein crystal structure PDB code: 1bl8; three potassium atoms in the channel are shown as magenta spheres. Hydrogens and side chains are omitted. (a) One linear peptide from selectivity filter, (b) four peptides of selectivity filter and (c) potassium channel transmembrane domain. (Four colours indicate the four identical subunits. The K+ ions indicate the selectivity filter position.) (Online version in colour.)
4. Amyloid/α-sheet
Almost all proteins [25], under certain conditions, readily clump into characteristic stable insoluble amyloid fibres, made of β-sheet blocks. Amyloid fibres occur in, and are thought to cause: Alzheimer's, Parkinson's, BSE, type II diabetes and other diseases. Amyloid is sticky. Small polypeptides form amyloid just as well as proteins. So, early polypeptides are likely to have gummed up the FeS membranes of the very earliest ‘cells’, making them fairly impermeable. It is thought that eventually multi-sheet amyloid would have become the main component of the membranes [26,27] during the earliest stages of evolution, given that phospholipid membranes evolved at a relatively later stage in evolution.
Intermediates in amyloid formation, rather than amyloid itself, are what are toxic to cells. Evidence from various [28–30] different types of studies on proteins show β-sheet readily interconverts with α-sheet. This occurs by the process of peptide-plane flipping [31] whereby the CO–NH peptide plane rotates by 180° without too much alteration to the neighbouring atoms. It is proposed that α-sheet, shown in figure 10, is the main component of these intermediates. An α-sheet strand has the linear nest-like main chain conformation of the K+ channel.
Figure 10.
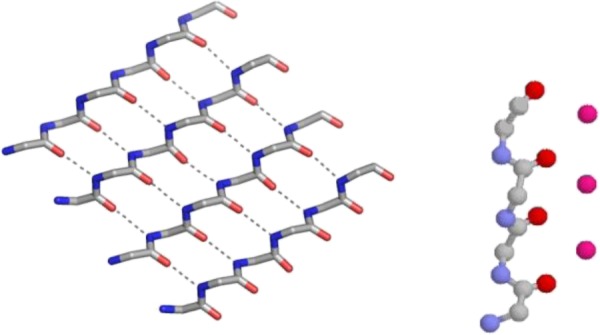
Comparison of α-sheet with potassium channel selectivity filter peptide. Side chains are omitted. The α-sheet is modelled. (Online version in colour.)
The toxicity of α-sheet for phospholipid cell membranes provides an explanation [29,32] why the conformation is rarely found in present-day folded proteins. Natural selection eliminates it.
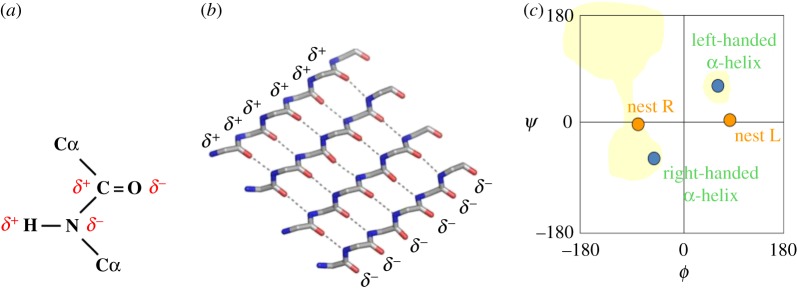
The partial charges on the main chain atoms of polypeptides mean that α-sheet differs from β-sheet in being more polar. One edge has a net positive charge and the other edge is net negative, as seen in figure 11. This may be the reason for its toxicity for present-day cells. It is possible that the positive edge may be attracted to the negative charges of phospholipids and that this causes membrane damage.
Figure 11.
α-Sheet. (a,b) α-Sheet partial (δ+, δ−) charge distribution. (c) Ramachandran plot, as in figure 3, except that the yellow shading shows areas favoured for l-amino acids. (Online version in colour.)
Another aspect of membranes made of α-sheet is that potassium channels are to be expected at the junctions between contiguous α-sheet blocks, where two or more of the net negative edges happen to be contiguous. The selectivity filter of the potassium channel may be regarded as a relic of this, except that the rest of the protein consists of α-helix rather than any sort of sheet. Diagrams showing the arrangement of blocks are in fig. 3 of [11].
The idea that α-sheet is sufficiently stable to act as the precursor has not yet been accepted by all workers in the field, but several papers [32–41] now provide evidence in its favour. The pale yellow shaded area in figure 11c shows areas that are favoured by l-amino acids in proteins [29]. Note that the region labelled left-handed α-helix is tolerated. The misconception that this area is only tolerated by, or preferentially favours, glycines is a reason why α-sheet is claimed by some to be an unlikely conformation for amyloid precursors of present-day proteins and polypeptides.
5. Metal–peptides
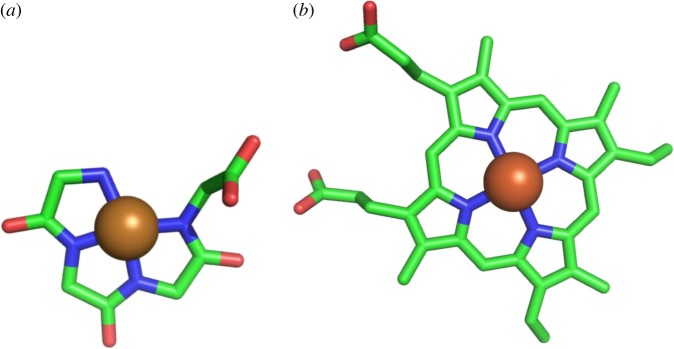
Metal–peptides, coloured complexes that were much studied in the 1960s [42,43], can be readily made by mixing Ni, Cu, Fe or Co with peptides in alkali. Present-day biological examples [22,44] occur in the prion protein and acetyl CoA synthase. With ϕ, ψ angles of 180°, 0° the main chain atoms lie flat in one plane and have no conformational chirality, as in figure 12a. Metallopeptides such as these would be expected to have performed the catalytic functions of haems. Some cobalt tripeptides [15] (Co-gly-gly-his) have been found that act as electrocatalysts for hydrogen evolution from water at neutral pH values with high efficiency.
Figure 12.
Haem-like metal peptide. (a) Ni-tetraglycine. (b) Haem. (Online version in colour.)
In a different vein, peptides of around eight residues can be synthesized [46] that self-assemble into amyloid-like β-sheets, and are catalytically active, much like enzymes, in the presence of Zn++. Other not dissimilar self-assembling histidine-containing heptapeptides, in the presence of Cu++, have been shown [47] to catalyse oxidation reactions involving molecular oxygen.
6. Peptides in early evolution: are there relics?
When the first protein structures were determined by X-ray crystallography and NMR, the question arose as to whether short peptides of less than 10 or so amino acid residues derived from them would also exhibit the same 3D structures. In general, the answer was that they did not. Typically, they seemed to be exploring a large number of different conformations and were moving so quickly between these different states that no fixed conformation was detectable. Even when the peptides were crystallized, they did not always exhibit the same conformation from one crystal form to the next. This conformational variability provided some explanation why many proteins are so big: only in the fully folded state is there a single fixed conformation. The comparison between small peptides and proteins may have led to a certain lack of interest in small peptide motifs as some common features [9–13,20,48,49] have only been reported recently.
Many current proteins are composed of internal structural repeats, giving rise to the idea that they developed over evolution by duplication and repetition of simpler peptide structures that might be referred to as minimal domains. Much interesting work has provided evidence for these repetitions [41,50] and defined the nature of the smallest domains, which can be regarded as relics [1,51–54]. Here, we take a different approach by focusing on the most common small hydrogen-bonded 3D motifs in proteins, consisting of just a few amino acid residues, especially those with functionalities involving just main chain atoms, to consider whether some might be vestiges of primordial peptides.
In pre-RNA evolution, peptides are thought to have had a high proportion of glycines because they are the easiest to synthesize from simple molecules like ammonia and CO2, and that amino acids with side chains were correspondingly less frequent [55]. The chiral amino acids that did exist were a mixture of l and d forms. Peptides with alternating l/d amino acids have been shown [14,23], as might be expected, to favour enantiomeric main chain conformations: repeating dipeptides in which each amino acid residue has a main chain conformation that is the mirror image of that of the adjacent residue. This applies to three types of structure that have been discussed in this article: nests, potassium channels and α-sheet. We have pointed out that such peptides would have been a dominant aspect of 3D structure in the short peptides occurring at the very earliest period in evolution, before nucleic acids and the genetic code were invented. This is in contrast to present-day proteins and peptides whose structure is dominated by regularly repeating 3D structures such as α-helix and β-sheet in which each amino acid residue has the same main chain conformation as its neighbour. However, relics of the earlier types of polypeptide remain to this day within existing proteins. They are still central to function, and are the focus of this review.
Nests are the most convincing relic candidate structures. They occur within P-loop protein enzymes that perform the ATPase (more generally, the phosphotransferase) reaction, which is regarded as the most basic energy-garnering reaction in the whole of biology. The component five-residue nests have been conserved because they selectively bind, and help transfer the terminal phosphate of, ATP. At this early point in evolution, these enzymes would have acted upon polyphosphates rather than nucleotide triphosphates as nucleotides had not evolved, but the selective pressure to synthesize peptides with this capability would ensure their conservation and propagation. It is striking that the phosphotransferases with P-loops are often those that are central to all metabolisms, sometimes called housekeeping enzymes, so probably evolved in early evolution. On the other hand, ATP-selective phosphotransferases that are known to have evolved much later on as eukaryote inventions, such as tyrosine kinases and the various enzymes involved with inositol phosphates, do not have P-loops. It appears that, once the genetic code for amino acids was established, it became easier to employ positively charged side chains for phosphate-binding sites rather than the main chain atoms of nests.
Likewise, other nests occur in many proteins as features that help bind iron–sulfur centres, rather as they can for phosphates. The relevance of this observation is that iron sulfides are expected to have been a prominent aspect of the hydrothermal vents where the earliest evolution is considered to have occurred. Peptides with some capability, in conjunction with methyl sulfides, to bind such centres were likely to have been under selection pressure. Another putative relic, which could be regarded as a flattened form of the nest, is the selectivity filter of the potassium channel. However, being a membrane protein, it can only have operated in conjunction with the membranes (perhaps amyloid, see below) that existed in early evolution.
When considering amyloid proteins in early evolution, talking of relics seems inappropriate because amyloid proteins and peptides in general are damaging to present-day cells and so are expected to have been subject to negative rather than positive, selection. On the other hand, it seems inescapable that the polypeptides that existed at the earliest stage often adopted amyloid structures, consisting of multi-sheet β-sheet or α-sheet or a mixture of the two. In spite of the current uncertainties about this, many of the earliest proteins would surely have been amyloid structures. Not only could they have acted as membranes incorporating potassium channels (phospholipids being a much later invention), but recent work shows that, in conjunction with transition metal ions, certain peptides with this structure exhibit various [47] catalytic activities. In addition, haem-like non-amyloid Cu++ tripeptides have been made [45] that catalyse hydrogen evolution from water.
The main aim of this article is to point out that short peptides can and do have more functionality, both in terms of binding and catalysis, than is generally appreciated, and also that relics of these peptides can be discerned in present-day proteins. Just as some of the metal features within present-day proteins are thought to have their roots in geology [22,56], some of these small-scale features of proteins have been retained from the earliest period in the evolution of life before the genetic takeover by RNA.
Data accessibility
This article does not contain any additional data.
Competing interests
I declare I have no competing interests.
Funding
I received no funding for this study.
References
- 1.Ponting CP, Russell RR. 2002. Natural history of protein domains. Annu. Rev. Biophys. Biomol. Struct. 31, 45–71. ( 10.1146/annurev.biophys.31.082901.134314) [DOI] [PubMed] [Google Scholar]
- 2.Orengo CA, Thornton JM. 2005. Protein families and their evolution—a structural perspective. Annu. Rev. Biochem. 74, 867–900. ( 10.1146/annurev.biochem.74.082803.133029) [DOI] [PubMed] [Google Scholar]
- 3.Alva V, Soding J, Lupas AN. 2015. A vocabulary of ancient peptides at the origin of folded proteins. eLife 4, e09410 ( 10.7554/eLife.09410) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caetano-Anolles G, Wang M, Caetano-Anolles D, Mittenthal JE. 2009. The origin, evolution and structure of the protein world. Biochem. J. 417, 621–637. ( 10.1042/BJ20082063) [DOI] [PubMed] [Google Scholar]
- 5.Plankensteiner K, Reiner H, Rode BM. 2005. Prebiotic chemistry: the amino acid and peptide world. Curr. Org. Chem. 9, 1107–1114. ( 10.2174/1385272054553640) [DOI] [Google Scholar]
- 6.Gilbert W. 1986. Origin of life: the RNA world. Nature 319, 618 ( 10.1038/319618a0) [DOI] [Google Scholar]
- 7.Russell MJ, Hall AJ. 1997. The emergence of life from iron monosulphide bubbles at a submarine hydrothermal redox and pH front. J. Geol. Soc. Lond. 154, 377–402. ( 10.1144/gsjgs.154.3.0377) [DOI] [PubMed] [Google Scholar]
- 8.Martin W, Russell MJ. 2007. On the origin of biochemistry at an alkaline hydrothermal vent. Phil. Trans. R. Soc. B 362, 1887–1925. ( 10.1098/rstb.2006.1881) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Watson JD, Milner-White EJ. 2002. A novel anion binding site in proteins: the nest. A particular combination of ϕ, ψ values in successive residues gives rise to anion binding sites that occur commonly and are often found at functionally important regions. J. Mol. Biol. 315, 187–198. ( 10.1006/jmbi.2001.5227) [DOI] [PubMed] [Google Scholar]
- 10.Watson JD, Milner-White EJ. 2002. The conformations of polypeptide chains where the main chain parts of alternating residues are enantiomeric. Their occurrence in cation and anion binding regions of proteins. J. Mol. Biol. 315, 199–207. ( 10.1006/jmbi.2001.5228) [DOI] [PubMed] [Google Scholar]
- 11.Milner-White EJ, Russell MJ. 2008. Predicting the conformations of peptides and proteins in early evolution. Biol. Direct 3, 3 ( 10.1186/1745-6150-3-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leader DP, Milner-White EJ. 2009. Motivated proteins: a web application for studying small three-dimensional protein motifs. BMC Bioinf. 10, 60 ( 10.1186/1471-2105-10-60) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Afzal AM, Al-Shubailly F, Leader DP, Milner-White EJ. 2014. Bridging of anions by hydrogen bonds in nest motifs and its significance for Schellman loops and other larger motifs within proteins. Proteins 82, 3023–3031. ( 10.1002/prot.24663) [DOI] [PubMed] [Google Scholar]
- 14.Milner-White EJ, Nissink WM, Allen FH, Duddy WJ. 2004. Recurring main-chain anion-binding motifs in short polypeptides: nests. Acta Cryst. D60, 1935–1942. ( 10.1107/s0907444904021390) [DOI] [PubMed] [Google Scholar]
- 15.Pal D, Sühnel J, Weiss MS. 2002. New principles of protein structure: nests, eggs and then what? Angew. Chem. 41, 4663–4665. ( 10.1002/anie.200290009) [DOI] [PubMed] [Google Scholar]
- 16.Leader DP, Milner-White EJ. 2015. Bridging of partially negative atoms by hydrogen bonds from main-chain NH groups in proteins: the crown motif. Proteins 83, 2067–2076. ( 10.1002/prot.24923) [DOI] [PubMed] [Google Scholar]
- 17.Ménard R, Storer AC. 1992. Oxyanion hole interactions in serine and cysteine proteases . Biol. Chem. Hoppe-Seyler 373, 393–400. ( 10.1515/bchm3.1992.373.2.393) [DOI] [PubMed] [Google Scholar]
- 18.Saraste M, Sibbald PR, Wittinghofer A. 1990. The P-loop—a common motif in ATP- and GTP-binding proteins. Trends Biochem. Sci. 15, 430–434. ( 10.1016/0968-0004(90)90281-F) [DOI] [PubMed] [Google Scholar]
- 19.Pai EF, Krengel U, Petsko GA, Goody RS, Kabsch W, Wittinghofer A. 1990. Refined crystal structure of the triphosphate conformation of H-ras p21 at 1.3 A. EMBO J. 9, 2351–2359. ( 10.1002/j.1460-2075.1990.tb07409.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bianchi A, Giorgi C, Ruzza P, Toniolo C, Milner-White EJ. 2012. A synthetic hexapeptide designed to resemble a proteinaceous P-loop nest is shown to bind inorganic phosphate. Proteins 80, 1418–1424. ( 10.1002/prot.24038) [DOI] [PubMed] [Google Scholar]
- 21.Milner-White EJ, Russell MJ. 2005. Sites for phosphates and iron–sufur thiolates in the first membranes: 3–6 residue anion-binding motifs (nests). Origins Life Evol. Biospheres 35, 19–27. ( 10.1007/s11084-005-4582-7) [DOI] [PubMed] [Google Scholar]
- 22.Nitschke W, McGlynn SE, Milner-White EJ, Russell MJ. 2013. On the antiquity of metalloenzymes and their substrates in bioenergetics. Biochim. Biophys. Acta 1827, 871–881. ( 10.1016/j.bbabio.2013.02.008) [DOI] [PubMed] [Google Scholar]
- 23.Milner-White EJ, Russell MJ. 2011. Functional capabilities of the earliest peptides and the emergence of life. Genes 2, 671–688. ( 10.3390/genes2040671) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doyle DA, Cabral JM, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. 1998. The structure of the potassium channel: molecular basis of K+-conduction and selectivity. Science 280, 69–77. ( 10.1126/science.280.5360.69) [DOI] [PubMed] [Google Scholar]
- 25.Fandrich M, Fletcher MA, Dobson CM. 2001. Amyloid fibrils from muscle myoglobin—even an ordinary globular protein can assume a rogue guise if conditions are right. Nature 410, 165–166. ( 10.1038/35065514) [DOI] [PubMed] [Google Scholar]
- 26.Zhang S, Holmes T, Lockshin C, Rich A. 1993. Spontaneous assembly of a self-complimentary oligopeptide to form a stable macroscopic membrane. Proc. Natl Acad. Sci. USA 90, 3334–3338. ( 10.1073/pnas.90.8.3334) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Childers WS, Ni R, Mehta AK, Lynn DG. 2009. Peptide membranes in chemical evolution. Curr. Opin. Chem. Biol. 13, 652–659. ( 10.1016/j.cbpa.2009.09.027) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Armen RS, DeMarco ML, Alonso DOV, Daggett V. 2004. Pauling and Corey's α-pleated sheet structure may define the prefibrillar amyloidogenic intermediate in amyloid disease. Proc. Natl Acad. Sci. USA 101, 11 622–11 627. ( 10.1073/pnas.0401781101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramachandran GN, Sasiekharan V. 1968. Conformation of polypeptides and proteins. Adv. Prot. Chem. 23, 283–438. ( 10.1016/S0065-3233(08)60402-7) [DOI] [PubMed] [Google Scholar]
- 30.Hayward S, Milner-White EJ. 2011. Simulation of the β- to α-sheet transition results in a twisted sheet for antiparallel and an a nanotube for parallel strands: implications for amyloid formation. Proteins 79, 3193–3207. ( 10.1002/prot.23154) [DOI] [PubMed] [Google Scholar]
- 31.Hayward S. 2001. Peptide plane flipping in proteins. Protein Sci. 10, 2219–2227. ( 10.1110/ps.23101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daggett V. 2006. Alpha-sheet: the toxic conformer in amyloid diseases? Acc. Chem. Res. 39, 594–602. ( 10.1021/ar0500719) [DOI] [PubMed] [Google Scholar]
- 33.Kellock J, Hopping G, Caughey G, Daggett V. 2016. Peptides composed of alternating l- and d-amino acids inhibit amyloidogenesis in three distinct amyloid systems independent of sequence. J. Mol. Biol. 428, 2317–2328. ( 10.1016/j.jmb.2016.03.013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paranjapye N, Daggett V. 2018. De novo designed α-sheet peptides inhibit functional amyloid formation of Streptococcus mutans biofilms. J. Mol. Biol. 430, 3764–3773. ( 10.1016/j.jmb.2018.07.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bleem A, Francisco R, Bryers JD, Daggett V. 2017. Designed α-sheet peptides suppress amyloid formation in Staphylococcus aureus biofilms. npj Biofilm Microbiom. 3, 16 ( 10.1038/s41522-017-0025-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hilaire MR, Ding B, Mukherjee D, Chen J, Gai F. 2018. Possible existence of α-sheets in the amyloid fibrils formed by a TTR105−115 mutant. J. Am. Chem. Soc. 40, 629−635. ( 10.1021/jacs.7b09262) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Balapuri A, Choi K-E, Kang NS. 2019. Computational insights into the role of α-strand/sheet in aggregation of α-synuclein. Sci. Rep. 9, 59 ( 10.1038/s41598-018-37276-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maris NL, Shea D, Bleem A, Bryers JD, Daggett V. 2018. Chemical and physical variability in structural isomers of an l/d α-sheet peptide designed to inhibit amyloidogenesis. Biochemistry 57, 507–510. ( 10.1021/acs.biochem.7b00345) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grillo-Bosch D, Carulla N, Cruz M, Sanchez L, Pujol-Pina R, Madurga S, Rabanal F, Giralt E. 2009. Retro-Enantio N-methylated peptides as β-amyloid aggregation inhibitors. ChemMedChem 4, 1488–1494. ( 10.1002/cmdc.200900191) [DOI] [PubMed] [Google Scholar]
- 40.Shea D, et al. 2019. α-Sheet secondary structure in amyloid β-peptide drives aggregation and toxicity in Alzheimer's disease. Proc. Natl Acad. Sci. USA 116, 8895–8900. ( 10.1073/pnas.1820585116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andrade MA, Perez-Iratxeta C, Ponting CP. 2001. Protein repeats: structures, functions and evolution. J. Struct. Biol. 134, 117–131. ( 10.1006/jsbi.2001.4392) [DOI] [PubMed] [Google Scholar]
- 42.Harford C, Sarkar B. 1999. Amino terminal Cu(II) and Ni(II) binding ATCUN motif of proteins and peptides. Acc. Chem. Res. 30, 123–130. ( 10.1021/ar9501535) [DOI] [Google Scholar]
- 43.Freeman HC. 1967. Crystal structures of metal-peptide complexes. Adv. Prot. Chem. 22, 257–424. ( 10.1016/S0065-3233(08)60043-1) [DOI] [PubMed] [Google Scholar]
- 44.Darnault C, Volbeda A, Kim EJ, Legrand P, Vernède X, Lindahl PA, Fontecilla-Camps JC.. 2003. Ni–Zn-[Fe4–S4] and Ni–Ni–[Fe4–S4] clusters in closed and open α subunits of acetyl-CoA synthase/carbon monoxide dehydrogenase. Nat. Struct. Biol. 10, 271–279. ( 10.1038/nsb912) [DOI] [PubMed] [Google Scholar]
- 45.Kandemir B, Kubie L, Guo Y, Sheldon B, Bren KL. 2019. Hydrogen evolution from water under aerobic conditions catalyzed by a cobalt ATCUN metallopeptide. Inorg. Chem. 55, 1355–1357. ( 10.1021/acs.inorgchem.5b02157) [DOI] [PubMed] [Google Scholar]
- 46.Al-Garawi ZS, McIntosh BA, Neill-Hall D, Hatimy AA, Sweet SM, Bagley MC, Serpell LC. 2017. The amyloid architecture provides a scaffold for enzyme-like catalysis. Nanoscale 9, 10 773–10 783. ( 10.1039/C7NR02675G) [DOI] [PubMed] [Google Scholar]
- 47.Makhlynets OV, Gosavski PM, Korendovych IV. 2016. Short self-assembling peptides are able to bind to copper and activate oxygen. Angew. Chem. 55, 9017–9020. ( 10.1002/anie.201602480) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Milner-White EJ, Watson JD, Qi G, Hayward S. 2006. Amyloid formation may involve α- to β-sheet interconversion via peptide plane flipping. Structure 14, 1369–1377. ( 10.1016/j.str.2006.06.016) [DOI] [PubMed] [Google Scholar]
- 49.Tompa P, Davey NE, Gibson TJ, Babu MM. 2014. A million peptide motifs for the molecular biologist. Mol. Cell 55, 161–169. ( 10.1016/j.molcel.2014.05.032) [DOI] [PubMed] [Google Scholar]
- 50.Bliven SE, Lafita A, Rose PW, Capitani G, Prlic A, Bourne PE. 2019. Analyzing the symmetrical arrangement of structural repeats in proteins with CE-symm. PLoS Comp. Biol. 15, e1006842 ( 10.1371/journal.pcbi.1006842) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lupas AN, Ponting CP, Russell RB. 2001. On the evolution of protein folds: are similar motifs in different protein folds the result of convergence, insertion or relics of an ancient world? J. Struct. Biol. 134, 191–203. ( 10.1006/jsbi.2001.4393) [DOI] [PubMed] [Google Scholar]
- 52.Koonin EV. 2003. Comparative genomics, minimal gene-sets and the last universal common ancestor. Nat. Rev. Microbiol. 1, 127–136. ( 10.1038/nrmicro751) [DOI] [PubMed] [Google Scholar]
- 53.Ranea JAG, Sillero A, Thornton JM, Orengo CA. 2006. Protein superfamily evolution and the last universal common ancestor (LUCA). J. Mol. Evol. 63, 513–525. ( 10.1007/s00239-005-0289-7) [DOI] [PubMed] [Google Scholar]
- 54.Ma BG, et al. 2008. Characters of very ancient proteins. Biochem. Biophys. Res. Commun. 366, 607–611. ( 10.1016/j.bbrc.2007.12.014) [DOI] [PubMed] [Google Scholar]
- 55.Solis AD. 2019. Reduced alphabet of prebiotic amino acids optimally encodes the conformational space of diverse extant protein folds. BMC Evol. Biol. 19, Article number 158 ( 10.1186/s12862-019-1464-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martin W, Russell MJ. 2003. On the origin of cells: an hypothesis for the evolutionary transitions from abiotic geochemistry to chemoautorophic prokaryotes, and from prokaryotes to nucleated cells. Phil. Trans. R. Soc. Lond B 358, 27–85. ( 10.1098/rstb.2002.1183) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Milner-White EJ, Russell MJ. 2010. Polyphosphate–peptide synergy and the organic takeover at the emergence of life. J. Cosmol. 10, 3217–3229. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article does not contain any additional data.