Short abstract
It is important to find specific and easily detectable diagnostic markers in acute stage of spinal cord injury for guiding treatment and estimating prognosis. Although, microRNAs are attractive biomarkers, there is still no uniform standard for clinical evaluation of spinal cord injury based on “free circulation” miRNA spectrum. The reason may be that miRNA analysis from biological fluids is influenced by many pre-analysis variables. Exosome miRNAs are widely distributed in body fluids and have many advantages comparing with free miRNAs. The specific miRNAs in the central nervous system can be transported to the peripheral circulation and concentrated in exosomes. Therefore, we hypothesized that there might be some physiological changes associated with spinal cord injury in serum exosomal miRNAs. Using next-generation sequencing, miRNA profiles in serum exosomes of sham and acute spinal cord injury rats were analyzed, and integrative bioinformatics were used to analyze the function and regulation of putative target genes. The results showed that acute spinal cord injury can lead to changes in miRNA expression in the circulating exosomes. The changed miRNAs and their associated pathways may explain the pathology of acute spinal cord injury. More importantly, we determined serum exosomal miR-125b-5p, miR-152-3p, and miR-130a-3p are specific and easily detectable diagnostic markers in acute spinal cord injury. More interestingly, we also found some valuable known and novel miRNAs. Further bioinformatics analysis and functional research will be of great help to make clear their role in the pathological process of spinal cord injury and judging whether they can be used as diagnostic markers.
Impact statement
This research hypothesized that there might be some physiological changes associated with SCI in serum exosomal miRNAs. Using next-generation sequencing, miRNA profiles in serum exosomes of sham and acute SCI rats were analyzed, and integrative bioinformatics were used to analyze the function and regulation of putative target genes. The results showed that acute SCI can lead to changes in miRNA expression in the circulating exosomes. The changed miRNAs and their associated pathways may explain the pathology of acute SCI. More importantly, we determined serum exosomal miR-125b-5p, miR-152-3p, and miR-130a-3p are specific and easily detectable diagnostic markers in acute SCI.
Keywords: Spinal cord injury, serum, exosomes, microRNAs
Introduction
Spinal cord injury (SCI) is defined as a variety of injuries to the spinal cord. With the rapid development of modern transportation, construction, and industry, the incidence and disability rate of SCI are increasing year by year.1 Accurately and timely judging SCI in the early stage has great significance for guiding treatment and estimating prognosis.2,3 At present, the most significant clinical indicators reflecting the prognosis of SCI include neurological function assessment, spinal cord imaging, biochemical indicators, etc.4–7 However, these methods have their own drawbacks: SCI scores are often quite inconsistent with the final results of SCI patients;8 spinal cord imaging examinations, such as CT scans, lack of sensitivity to spinal cord edema and cervical SCI without fracture and dislocation, and MRI cannot accurately predict the outcome of the disease;9 biochemical indicators, such as lactate dehydrogenase and protein kinase C, lack specificity, and non-neurological damage can also cause changes in the measured values, which reduces their clinical application value.10,11 Although, the biochemical markers specific to the central nervous system (CNS), such as GFAP, NFL and S100B, were found to be elevated in cerebrospinal fluid of patients with SCI, but the increase of these markers is too late to be suitable for early monitoring and treatment.12 Therefore, it is important to find specific and easily detectable diagnostic markers in acute stage of SCI.
MicroRNAs (miRNAs) are a kind of endogenous non-coding RNAs with a length of 20–24 nucleotides, which could post-transcriptionally regulate gene expression. MiRNAs have become an attractive candidate for biomarkers due to its tissue specificity in body fluid.13,14 Hachisuka et al.15 found that there are several miRNAs increased in spinal cord injured mouse serum in a severity-dependent manner. In a pig model, Tigchelaar et al.16 showed that some serum miRNAs can reflect injury severity. In human, miRNAs in cerebrospinal fluid and serum could also reflect injury severity in acute traumatic SCI.17 However, up to now, there is no uniform standard for clinical evaluation of SCI based on “free circulation” miRNA spectrum. The reason may be that miRNA analysis from biological fluids is influenced by many pre-analysis variables, such as sample collection and processing methods, stability, and differences in serum and plasma coagulation processes.18
Extracellular vesicles (EVs) are all kinds of vesicles with membrane structure released by cells. The diameter of these vesicles can range from 30 nm to several micrometers.19,20 There are different subgroups of EVs, and the exosome is under intense investigation at present.21 Exosome is one of the smaller EVs with a diameter of 30–150 nm.22,23 Although exosomes were first discovered in 1983, they have long been regarded as a waste of cells. However, in recent years, it has been found that these tiny vesicles contain cell-specific proteins, nucleic acids and lipids, which can be transmitted to other cells as signaling molecules to change their functions.24 Exosomes can play important roles in many physiological and pathological aspects, such as antigen presentation, growth and migration of tumors, repair of tissue damage, and so on.25 In addition, exosomes secreted by different cells have unused components and functions, which can be used as biomarkers for disease diagnosis.26–28
Exosome miRNAs are widely distributed in blood, saliva, milk, urine, and other body fluids. As an important source of biomarkers, compared with free miRNAs, exosome miRNAs have the following advantages:29–31 (1) Exosomes can be used as reliable carriers for the study of miRNAs because they contain almost all kinds of miRNAs; (2) The bilayer membrane structure of exosome can enhance the stability of miRNAs, enhance the sensitivity of miRNAs amplification, and reduce the probability of negative results; (3) Exosomes can penetrate the blood–brain barrier or the blood–spinal cord barrier. The specific miRNAs in the CNS can be transported to the peripheral circulation system and concentrated in exosomes. Therefore, we hypothesized that there might be some physiological changes associated with SCI in serum exosomal miRNAs. Using next-generation sequencing, miRNA profiles in serum exosomes of sham and spinal cord injured rats were analyzed, and integrative bioinformatics were used to analyze the function and regulation of putative target genes. The key miRNAs and their targeted signal pathways were identified to provide new indicators for reflecting the diagnosis and prognosis of SCI.
Materials and methods
Animals
A total of 15 healthy adult SD female rats (220–250 g) were used to model SCI. The study was carried out in accordance with the recommendations of the Guide for the Care and Use of Laboratory Animals, the Association for Assessment and Accreditation of Laboratory Animal Care international. The protocol was approved by the Institutional Committee on Animal Care, Use and Research of the Bengbu Medical College.
Contusive SCI
A T9 moderate contusive SCI model was performed by using an Infinite Horizon impactor (Precision Systems & Instrumentation, Lexington, KY) according to our previous method.32 In SCI group, the exposed spinal cord was impacted with a force of 120 kdynes and a dwell time of zero seconds. In sham group, the rats only received a laminectomy without contusion.
Serum collection and preparation
Blood was collected by using BD Vacutainer™ Tube (Catalog # 367955, BD, USA) at 6 h post-injury under deep anesthesia (80 mg/kg pentobarbital, intraperitoneal). A total 8 mL blood sample was obtained from each rat by using puncture of main abdominal vein with a 23-gauge butterfly needle. Serum-gel tubes were centrifuged at 3000g, 4°C for 10 min (Thermo, centrifugal gradient set to 7). The resulting serum was transferred into 15 mL tubes and centrifuged at 1800g for 10 min (Thermo, centrifugal gradient set to 5). The serum sample was aliquoted into 1.5 mL microcentrifuge tubes (509-GRD-Q, AXYGEN) and stored at −80°C.
Hematoxylin and eosin staining
After serum collection at the indicated time post-injury, the spinal cord segments containing injured center were removed, postfixed, frozen, sectioned and stained with hematoxylin and eosin (HE) staining according to our previous method.33
Isolation and identification of exosomes
A Total Exosome Isolation Reagent (from serum) Kit (Invitrogen™ 4478360) was used to isolate exosomes from serum according to the manufacturer's instructions. The morphology of exosomes was characterized with transmission electron microscopy (TEM) (HITACHI, HT7700, Hitachi, Ltd, Japan), NanoSight analysis, and Western blot. The size distribution of the exosomes was analyzed using NanoSight NS300 (Malvern Instruments Ltd, UK). The exosome markers, CD9, CD63, CD81, and TSG101, were identified using Western Blot. The primary antibodies were Rabbit anti-CD9 [EPR2949] (ab92726, Abcam), Rabbit anti-CD63 (25682–1-AP, Proteintech), Rabbit anti-CD81 [EPR4244] (ab109201, Abcam), and Rabbit anti-TSG101 (14497–1-AP, Proteintech), respectively. HRP-conjugated goat anti-rabbit (ab205718) secondary antibody was used to detect the specific binding primary antibodies. Finally, the bands were observed by enhanced chemiluminescence (ECL).
Exosomal RNA isolation, quantification, and qualification
Total exosomal RNA was extracted using Trizol-LS kit (Invitrogen, CA) from exosome fractions. After treating with DNase I to remove genomic DNA, RNA purity, concentration, and integrity were assessed according to our previous methods.34
Small RNA library preparation and sequencing
Small RNA library preparation, sequencing, and bioinformatic analyses were performed at BGI (China). The libraries were sequenced using BGISEQ-500 technology.35
Prediction and expression of miRNA
We used miRDeep2 to predict novel miRNA by exploring the characteristic hairpin structure of miRNA precursor.36 The small RNA expression level is calculated by transcripts per kilobase million (TPM).37
Screening differentially expressed miRNAs
The DEmiRNAs of two groups were analyzed using the DESeq software (http://www.bioconductor.org/). The P-values calculated for each gene were adjusted to Q-values for multiple testing corrections by two alternative strategies.38,39 To improve accuracy of DEmiRNAs result, we defined a gene as a DEG when reads number foldchange ≥2 and Q-value ≤0.001.
Verification of miRNAs by RT-qPCR
RT-qPCR was used to determine the changes of randomly selected DEmiRNAs. Stem-loop method (Sangon Biotech Company, Shanghai, China) was used to synthesize cDNA from miRNAs. The reaction system of qPCR was performed by using miRNAs qPCR Kit (SYBR Green Method) (Sangon Biotech Company, Shanghai, China). The reverse transcription and PCR primer sequences are listed in Table 1; miR-191-5p was used to be housekeeping miRNA.40 According to the results of qPCR, the relative quantitative of miRNAs was calculated by using ΔΔCt.41
Table 1.
Primers used in the study.
| miRNA id | Sequence (mature) | RT primer | Forward primer | Common reverse primer |
|---|---|---|---|---|
| rno-miR-500-3p | AATGCACCTGGGCAAGGGTTCA | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTGAACC | CGAATGCACCTGGGCAAG | AGTGCAGGGTCCGAGGTATT |
| rno-miR-3589 | GAGGAAACCAGCAAGTGTTGAC | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGTCAAC | GCGGAGGAAACCAGCAAGT | |
| rno-miR-200b-5p | CATCTTACTGGGCAGCATTGGA | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCCAAT | GCGCATCTTACTGGGCAGC | |
| rno-miR-1839-5p | AAGGTAGATAGAACAGGTCTTG | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCAAGAC | CGCGCGAAGGTAGATAGAACAG | |
| rno-miR-152-3p | TCAGTGCATGACAGAACTTGG | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCCAAGT | CGCGTCAGTGCATGACAGA | |
| novel_mir2064 | cagtagcttagagttgggc | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGCCCAA | CGCGCGCAGTAGCTTAGAG | |
| novel_mir2027 | tgtctctggatcctgggcctttc | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGAAAGG | CGTGTCTCTGGATCCTGGG | |
| novel_mir51 | ttgtctgtgtgtatgtccatg | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCATGGA | GCGCGTTGTCTGTGTGTATG | |
| rno-miR-191-5p | CAACGGAATCCCAAAAGCAGCTG | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCAGCTG | CGCAACGGAATCCCAAAAG |
Target gene prediction
Two types of software, miRanda42 and RNAhybrid,43 were used to find more accurate targets. The default parameters were as follows: miRanda: -en -20 –strict; RNAhybrid: -b 100 -c -f 2,8 -m 100,000 -v 3 -u 3 -e -20 -p 1 -s 3utr_human.
Gene ontology and Kyoto encyclopedia of genes and genomes database pathway enrichment analysis
Gene ontology (GO) enrichment analysis was used to reveal the cellular component, biological process, and molecular function of the target mRNAs (http://www.geneontology.org/). DAVID Bioinformatics Resources 6.7 (https://david-d.ncifcrf.gov/) was used to perform KEGG pathway enrichment analysis.44–46 The P-value was corrected by using the Bonferroni method,47 a corrected P-value ≤ 0.05 was taken as a threshold.
Results
Histopathological characteristics of injured spinal cords
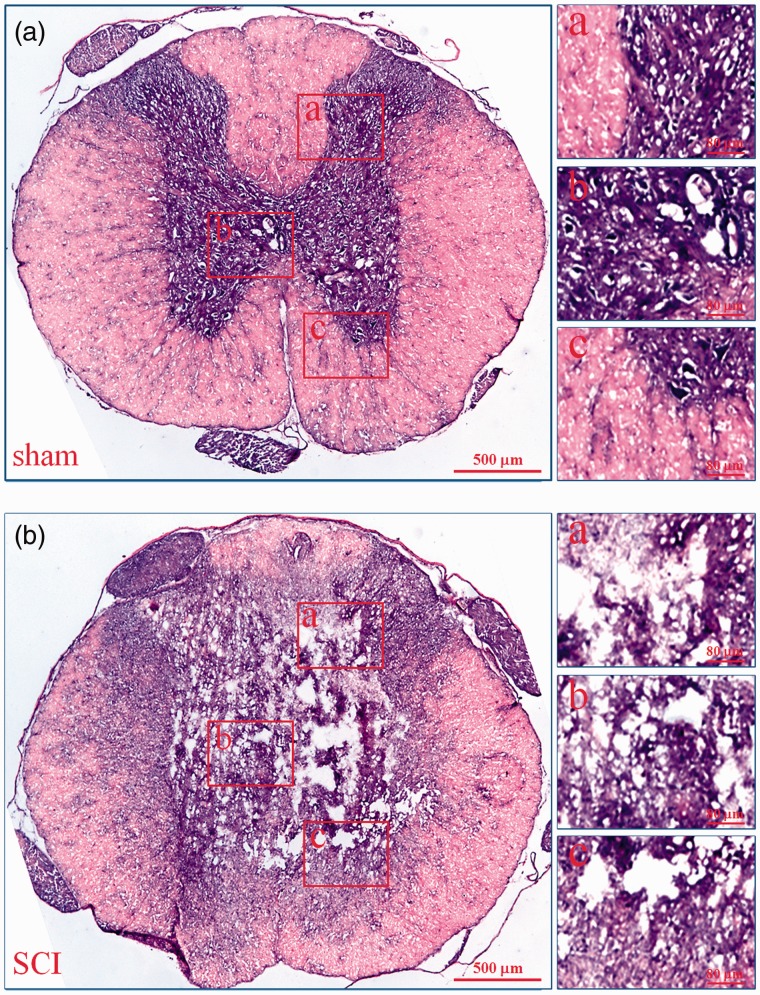
To prove the success of SCI model, we used H&E staining to observe the histopathological characteristics of injured spinal cord. As shown in Figure 1, in the sham group (A), the gray matter and white matter were complete. However, in the SCI group (B), massive hemorrhage occurred around capillaries, arterioles, and venules at the site of injury, the gray matter and white matter were destroyed. In injury epicenter, the neurons and glias were lost. In the periphery, a rim of spared white matter could be found. These are the typical histopathological changes in acute phase of SCI in rats.
Figure 1.
Histopathological changes of injured spinal cords. The histopathological changes of injured spinal cords at 6 h after SCI were evaluated by HE staining. In every group, the low microscopic images of T9 spinal cord whole coronal sections were given and three high magnification photographs from dorsal to ventral (a–c) were also displayed on the right. a: sham; b: SCI. (A color version of this figure is available in the online journal.)
Identification of exosomes from serum
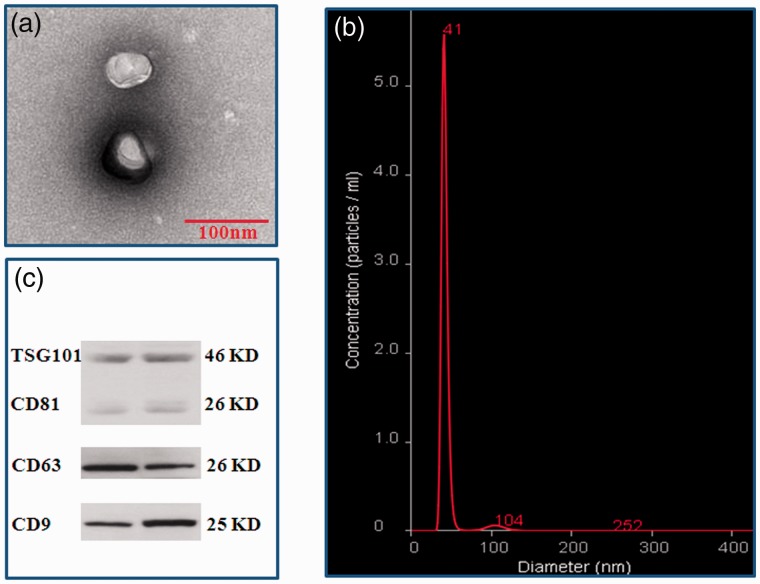
To identify the serum exosomes, the morphology, size, and biomarkers were detected by TEM, NanoSight, and Western Blot, respectively. TEM showed that the vesicles were round (“cup”) or oval (“dish”) with a diameter of 30 to 120 nm (Figure 2(a)). NanoSight revealed that the main peak of isolated particle size was 41 nm, the average diameter was 48.0 nm, and more than 99% of the particles were 30 to 150 nm in diameter (Figure 2(b)). Western Blot demonstrated that the expressions of 4 common exosome markers, CD9, CD63, CD81 and TSG101 could be found in all vesicles (Figure 2(c)). These are the typical features of exosomes.
Figure 2.
Identification of exosomes from serum. Vesicles isolated from serum were identified by TEM, NanoSight, and Western Blot. (a) Vesicles with round-(“cup”) or oval-shaped (“dish”) shapes ranged from 30 to 120 nm were observed by TEM; (b) NanoSight profile of seurm exosomes; (c) Representative results of exosome markers (CD9, CD63, CD81, and TSG101) in seurm exosomes indentified by Western blot. (A color version of this figure is available in the online journal.)
Analysis of small non-coding RNA
In order to evaluate the quality of sequencing data, we established six small non-coding RNA (sncRNA) libraries, which included sham (A1, A2, and A3) and SCI (B1, B2, and B3). The sequence data have been deposited into Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra/PRJNA545354). Before data analysis was carried out, low-quality tags were removed. Table 2 summarized the sequencing data for each sample. The results showed that there were 29,268,292 to 53,333,333 raw tags for each library. After filtering out the low-quality reads, the clean tags were from 23,888,725 to 24,933,748, with the percentages of 45.95% to 82.65%. These clean tags were mapped to sRNA database such as miRBase, Rfam, siRNA, piRNA, snoRNA, etc. [http://www.mirbase.org/; http://rfam.xfam.org/]. The separate mapping rate for each sample is listed in Table 3. The miRNAs were used for further analyses.
Table 2.
Summary of sequencing data for each sample.
| Sample name | Sequence type | Raw tag count | Low quality tag count | Invalid adapter tag count | PolyA tag count | Short valid length tag | Clean tag count | Q20 of clean tag (%) | Percentage of clean tag (%) |
|---|---|---|---|---|---|---|---|---|---|
| A1 | SE50 | 33,802,816 | 798,386 | 1,928,938 | 968 | 6,926,317 | 2,414,8207 | 99.2 | 71.44 |
| A2 | SE50 | 53,333,333 | 1,461,394 | 8,263,994 | 2188 | 19,097,315 | 24,508,442 | 99.3 | 45.95 |
| A3 | SE50 | 33,333,333 | 759,589 | 3,743,393 | 882 | 4,940,744 | 23,888,725 | 99.4 | 71.67 |
| B1 | SE50 | 44,444,444 | 1,158,469 | 5,440,175 | 938 | 13,658,455 | 24,186,407 | 99.4 | 54.42 |
| B2 | SE50 | 30,168,691 | 698,531 | 2,603,094 | 2321 | 1,930,997 | 24,933,748 | 99.1 | 82.65 |
| B3 | SE50 | 29,268,292 | 726,845 | 1,906,737 | 405 | 2,532,111 | 24,102,194 | 99.5 | 82.35 |
Note: Percentage (%) = clean tag count/raw tag count.
Table 3.
Alignment statistics of tags align to reference genome.
| Sample name | Total tag | Mapped tag | Percentage (%) |
|---|---|---|---|
| A1 | 24,148,207 | 22,613,121 | 93.64 |
| A2 | 24,508,442 | 22,888,377 | 93.39 |
| A3 | 23,888,725 | 20,320,753 | 85.06 |
| B1 | 24,186,407 | 21,240,123 | 87.82 |
| B2 | 24,933,748 | 23,155,923 | 92.87 |
| B3 | 24,102,194 | 20,207,475 | 83.84 |
Screening DEmiRNAs between sham and SCI groups
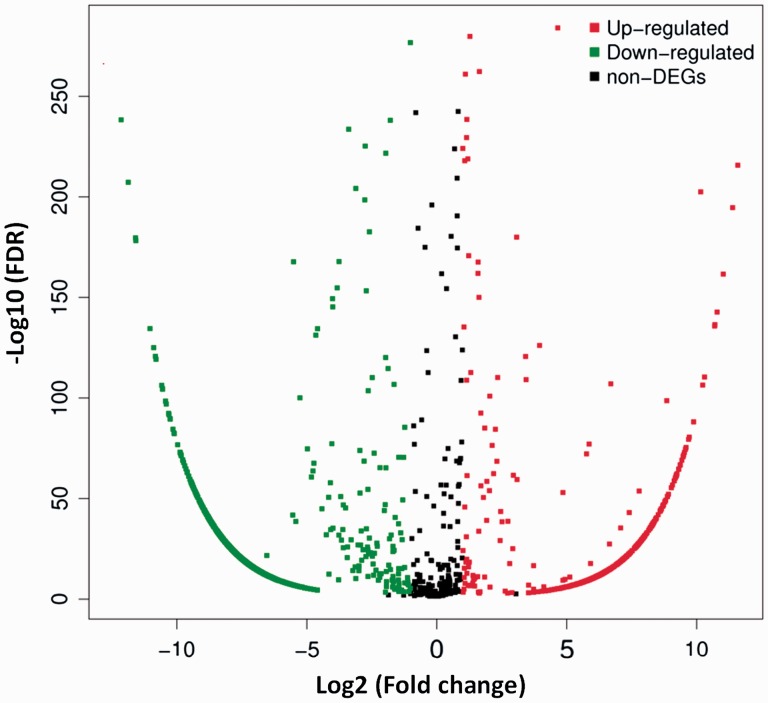
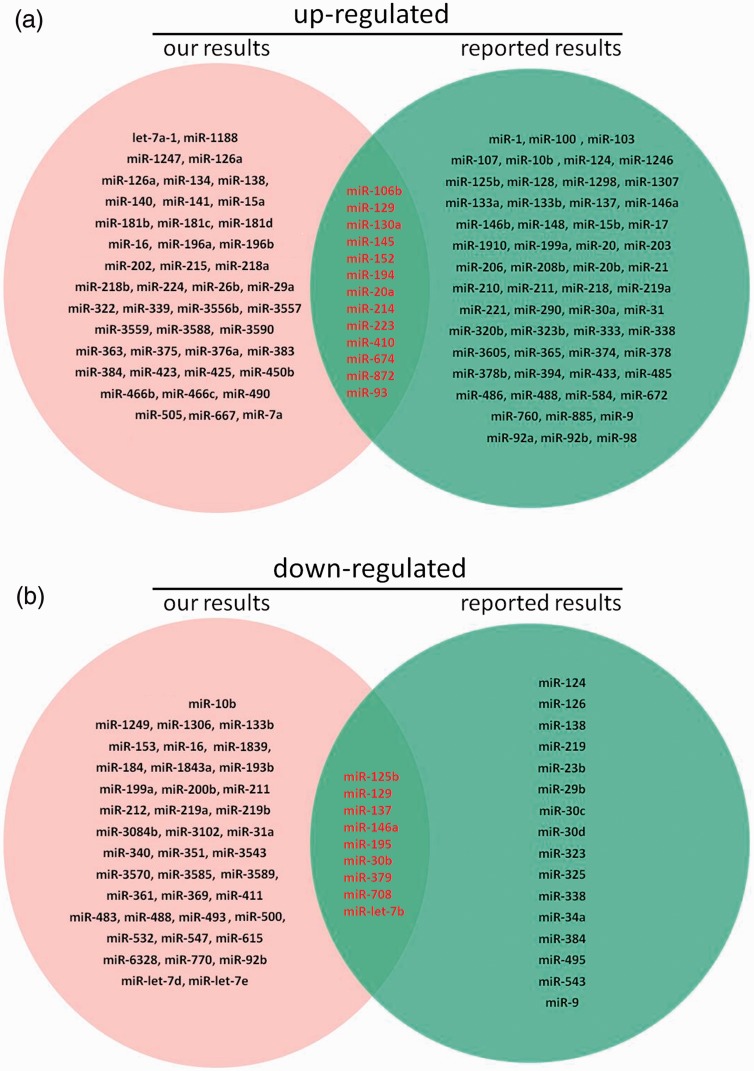
To find DEmiRNAs between samples and do the further analysis, DEGseq was used. In SCI group, there were 1807 DEmiRNAs (899 up-regulated and 908 down-regulated) comparing with sham group (Figure 3 and Table S1). In order to further find the valuable DEmiRNAs, miRNAs with less than 10 Read counts per sample were filtered out (Table S2). There were 217 miRNAs leaved (78 up-regulated and 139 down-regulated). In the up-regulated miRNAs, there were 18 novel miRNAs and 60 known miRNAs. Based on log2Ratio (SCI_6h/sham_6h), the top 10 most up-regulated miRNAs were novel_mir39, novel_mir2064, rno-miR-126a-5p, novel_mir603, rno-miR-152-3p, novel_mir51, rno-miR-130a-3p, rno-miR-466b-5p, rno-miR-1188-5p, and novel_mir1387 (Table 4). In the down-regulated miRNAs, there were 85 novel miRNAs and 54 known miRNAs. Based on log2Ratio (SCI_6h/sham_6h), the top 10 most down-regulated miRNAs were novel_mir803, novel_mir1945, novel_mir1541, novel_mir513, novel_mir1985, novel_mir4, rno-miR-125b-5p, novel_mir564, novel_mir429, and novel_mir2364 (Table 4). In addition, the filtered known miRNAs were compared with recently reported miRNAs which can be used as biomarkers after SCI.16,17,48 As shown in Figure 4, our novel results intersected with the reported results in 13 of the up-regulated (A) and 9 of the down-regulated miRNAs (B). Further comparing these miRNAs with the miRNAs in Table 3, we found that rno-miR-152-3p, rno-miR-130a-3p, and rno-miR-125b-5p belong to the top 10 up-regulated or down-regulated miRNAs. Next, eight DEmiRNAs were selected (three up-regulated and five down-regulated) for the next identification.
Figure 3.
Distribution of DEmiRNAs. The X axis is the log2 (Fold change) of DEmiRNAs, the Y axis is the −log10 (FDR) of DEmiRNAs. The red points indicate 899 up-regulated DEmiRNAs [log2 (Fold change) ≥1 and FDR ≤0.001]. The green points indicate 908 down-regulated DEmiRNAs [log2 (Fold change) ≤ −1 and FDR ≤ 0.001]. (A color version of this figure is available in the online journal.)
Table 4.
The top 10 most up-regulated and down-regulated miRNAs.
| miRNA id | Read count(sham) | Read count(SCI) | Expression(sham) | Expression(SCI) | log2Ratio(SCI/sham) | Up or down | P value | Q value |
|---|---|---|---|---|---|---|---|---|
| novel_mir39 | 45 | 1372 | 0.654 | 20.857 | 4.657 | UP | 9.01E-285 | 9.79E-285 |
| novel_mir2064 | 27 | 537 | 0.381 | 7.543 | 4.587 | UP | 5.70E-135 | 3.45E-135 |
| rno-miR-126a-5p | 35 | 657 | 0.507 | 10.000 | 3.957 | UP | 1.2096E-126 | 7.1451E-127 |
| novel_mir603 | 48 | 628 | 0.697 | 9.554 | 3.437 | UP | 1.29E-109 | 6.86E-110 |
| rno-miR-152-3p | 36 | 370 | 0.524 | 5.483 | 3.088 | UP | 1.02E-59 | 3.23E-60 |
| novel_mir51 | 113 | 1149 | 1.641 | 17.290 | 3.073 | UP | 1.42E-180 | 1.06E-180 |
| rno-miR-130a-3p | 13 | 110 | 0.197 | 1.597 | 2.808 | UP | 2.22E-17 | 1.98E-18 |
| rno-miR-466b-5p | 42 | 290 | 0.611 | 4.144 | 2.515 | UP | 1.67E-38 | 3.35E-39 |
| rno-miR-1188-5p | 51 | 339 | 0.741 | 4.917 | 2.460 | UP | 1.19E-43 | 2.71E-44 |
| novel_mir1387 | 34 | 223 | 0.494 | 3.347 | 2.440 | UP | 4.76E-29 | 6.80E-30 |
| novel_mir803 | 1579 | 94 | 21.411 | 1.397 | −4.343 | DOWN | 0.00E+00 | 0.00E+00 |
| novel_mir1945 | 177 | 10 | 2.694 | 0.141 | −4.419 | DOWN | 5.10E-45 | 1.22E-45 |
| novel_mir1541 | 521 | 25 | 7.064 | 0.404 | −4.654 | DOWN | 9.23E-132 | 5.54E-132 |
| novel_mir513 | 263 | 12 | 3.567 | 0.171 | −4.727 | DOWN | 7.65851E-68 | 2.71864E-68 |
| novel_mir1985 | 247 | 11 | 3.351 | 0.154 | −4.762 | DOWN | 5.45E-64 | 1.85E-64 |
| novel_mir4 | 234 | 10 | 3.550 | 0.141 | −4.822 | DOWN | 5.75E-61 | 1.85E-61 |
| rno-miR-125b-5p | 46817 | 1897 | 706.663 | 29.787 | −4.898 | DOWN | 0.00E+00 | 0.00E+00 |
| novel_mir564 | 287 | 11 | 3.944 | 0.154 | −4.979 | DOWN | 4.62E-75 | 1.88E-75 |
| novel_mir429 | 382 | 12 | 5.181 | 0.171 | −5.266 | DOWN | 1.62E-100 | 7.99E-101 |
Figure 4.
The Venn pictures of our novel results intersected with the reported results.16,17,48 (a): up-regulated; (b): down-regulated. (A color version of this figure is available in the online journal.)
Identification of DEmiRNAs by RT-qPCR
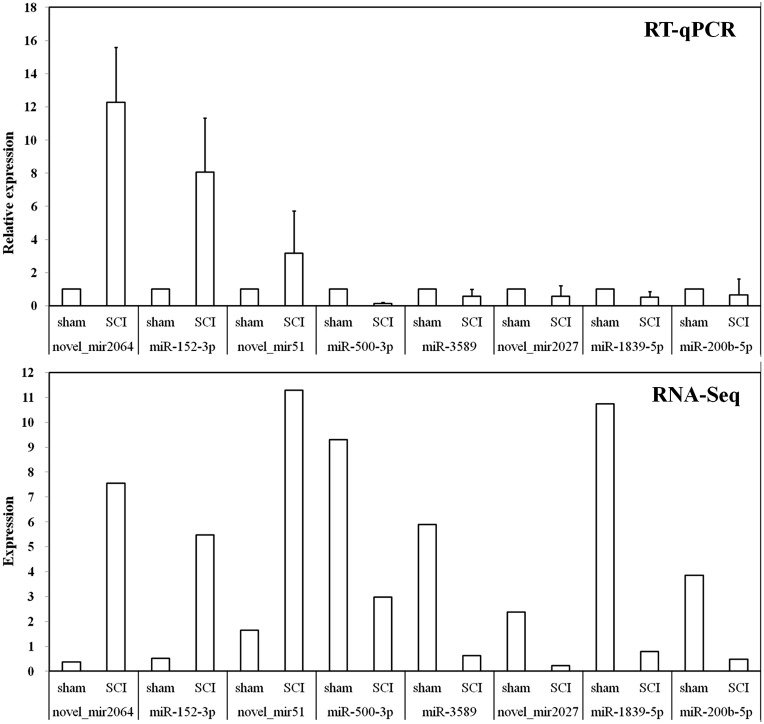
In order to verify the small RNA-sequencing (sRNA-Seq) results, we selected eight miRNAs (three up-regulated and five down-regulated) as mentioned above for RT-qPCR analysis (Table 1). The results indicated that these DEmiRNAs’ expression trends were similar in sRNA-Seq and RT-qPCR (Figure 5).
Figure 5.
RT-qPCR validations of DEmiRNAs characterized by sRNA-Seq. The ΔΔCt method was used to calculate the relative expression levels of DEmiRNAs in SCI group versus sham group (designated as 1). All data were calculated with mean ± standard deviation (n = 3). The longitudinal coordinates in RNA-Seq were the expression levels of DEmiRNAs calculated by using TPM, which can be used directly to compare gene expression differences between the samples.
DEmiRNAs target prediction
We used multiple types of software to find the target gene of DEmiRNAs. The results of top 10 up-regulated and down-regulated are listed in Table S3 and S4, and used for further analysis.
GO analysis of DEmiRNA targets
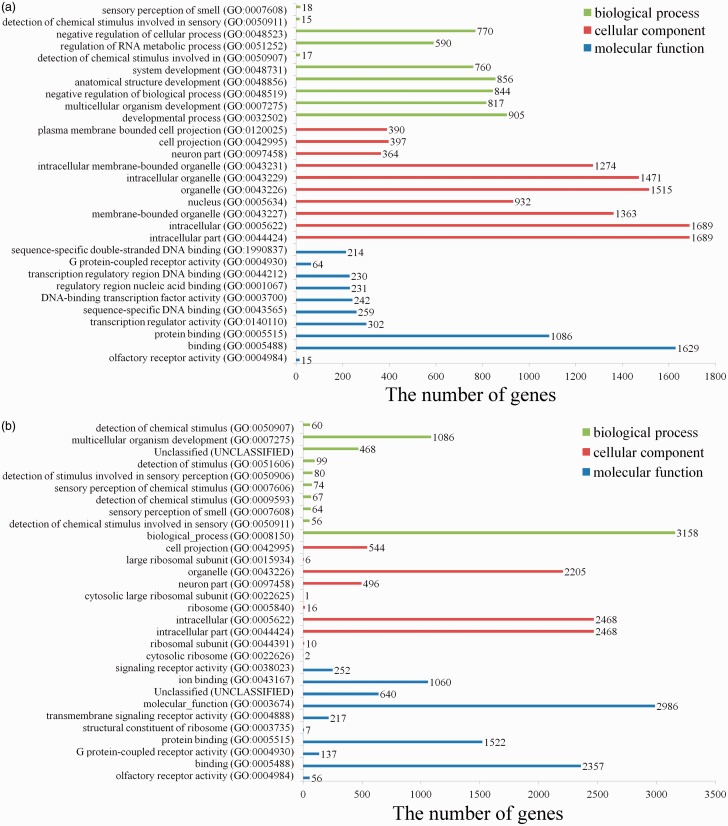
Comparing with sham group, we found 825 GO terms in up-regulated miRNAs (Table S5) and 524 GO terms in down-regulated miRNAs (Table S6) in SCI (PBS) group. The top 10 terms in domains of GO analysis of up-regulated DEmiRNAs indicated the following (Figure 6(a)): (1) In terms of molecular functions, the meaningful terms were protein binding, transcription regulator activity, sequence-specific DNA binding, DNA-binding transcription factor activity, regulatory region nucleic acid binding, G protein-coupled receptor activity, and sequence-specific double-stranded DNA binding. (2) In terms of cellular components, the meaningful terms were intracellular part, membrane-bounded organelle, intracellular membrane-bounded organelle, neuron part and plasma membrane bounded cell projection. (3) In terms of biological processes, the meaningful terms were developmental process, negative regulation of biological process, regulation of RNA metabolic process, and negative regulation of cellular process. The top 10 terms in domains of GO analysis of down-regulated DEmiRNAs indicated the following (Figure 6(b)): (1) In terms of molecular functions, the meaningful terms were olfactory receptor activity, G protein-coupled receptor activity, protein binding, transmembrane signaling receptor activity, ion binding, and signaling receptor activity. (2) In terms of cellular components, the meaningful terms were cytosolic ribosome, intracellular part, neuron part, organelle, and cell projection. (3) In terms of biological processes, the meaningful terms were detection of chemical stimulus, sensory perception of chemical stimulus, and multicellular organism development.
Figure 6.
GO analysis of the targets of up-regulated or down-regulated DEmiRNAs. The 10 most enriched GO terms in the categories of biological process, cellular component, and molecular function are shown. (A color version of this figure is available in the online journal.)
KEGG analysis of DEmiRNAs target
Table S7 and S8 showed the all up-regulated and down-regulated pathways, respectively. Comparing with sham group, in the up-regulated DEmiRNAs, the enriched signaling pathways were axon guidance, melanogenesis, Wnt signaling pathway, and long-term potentiation (Table 5). In the down-regulated DEmiRNAs, the enriched signaling pathways were ECM-receptor interaction and focal adhesion (Table 5).
Table 5.
KEGG pathway analysis of DESs targets.
| Pathway | Count | % | P-value | Bonferroni | FDR |
|---|---|---|---|---|---|
| Axon guidance | 33 | 0.2 | 6.4E-7 | 1.1E-4 | 7.8E-4 |
| Melanogenesis | 27 | 0.1 | 6.9E-7 | 1.2E-4 | 8.4E-4 |
| Wnt signaling pathway | 33 | 0.2 | 1.4E-5 | 2.3E-3 | 1.6E-2 |
| Long-term potentiation | 20 | 0.1 | 2.1E-5 | 3.6E-3 | 2.6E-2 |
| ECM-receptor interaction | 33 | 0.1 | 1.5E-7 | 2.8E-5 | 1.9E-4 |
| Focal adhesion | 59 | 0.2 | 3.1E-7 | 5.7E-5 | 3.8E-4 |
Note: Red: up-regulated. Blue: down-regulated.
The P-value is corrected by using the Bonferroni method, a corrected P-value (FDR) ≤ 0.05 is taken as a threshold. (A color version of this table is available in the online journal.)
Discussion
In this study, we identified the serum exosomal miRNAs in rats with acute SCI by using sRNA-Seq technology. Our aims were to investigate the changes of miRNAs in serum exosomes, to find specific and easily detectable diagnostic markers in acute stage of SCI, and to explore their target molecules and pathways.
In the present study, 217 valuable DEmiRNAs were found in the serum exosomal miRNAs between sham and acute spinal cord injured rats, including 78 up-regulated and 139 down-regulated. Among them, 18 novel miRNAs and 60 known miRNAs were up-regulated and 85 novel miRNAs and 54 known miRNAs were down-regulated. In order to verify the small sRNA-Seq results, eight DEmiRNAs were selected for RT-qPCR analysis. We demonstrated that the expression patterns of these DEmiRNAs were similar between sRNA-Seq and RT-qPCR. These support the reliability of our sRNA-seq analysis. In addition, we also compared the filtered known miRNAs with recently reported miRNAs which can be used as biomarkers after SCI.16,17,48 We found that our results intersected with the reported results in 22 miRNAs (13 up-regulated and 9 down-regulated). Further comparing these intersected miRNAs with the top 20 most variable miRNAs (10 up-regulated and 10 down-regulated), we were surprised to find that miR-130a-3p, miR-152-3p, and miR-125b-5p belong to these most variable miRNAs. Although, our results are in good agreement with those reported in the literature, these reported altered miRNAs are distributed in different parts of different species of animals, such as spinal cord, cerebrospinal fluid, or serum. However, as far as we know, this is the first time to detect the changes of miRNAs in serum exosomes after SCI. These will provide strong evidence for further investigation.
First of all, we believe that the top 10 most variable miRNAs, especially the three reported miRNAs (miR-130a-3p, miR-152-3p, and miR-125b-5p), can be used as specific markers of acute SCI in serum exosomes.
MicroRNA-152 belongs to the miR-148/152 family, which is a negative regulator of innate immune response and antigen presentation.49 A large number of literatures have reported that serum miR-152-3p is an important marker in the diagnosis, treatment, and prognosis of tumors.50–53 Its plasma levels are also been reported being associated with dengue infection,54 Type 2 diabetic nephropathy,55 and Alzheimer disease.56 A recent report shows that the expression of miR-152 is increased in the injured area after SCI,57 which supports our experimental results. This indicates that serum miR-152-3p, especially serum exosomal miR-152-3p, is a promising marker in the diagnosis and prognosis of acute SCI when excluding tumors, infection, diabetic nephropathy, Alzheimer disease, etc.
MicroRNA-130a was first found to be highly expressed in primary glioblastomas.58 It was also been proved to be involved in the substance P synthesis and release of mesenchymal stem cells (MSC)-derived neuronal cells.59 Chen and Gorski60 confirmed that miR-130a can inhibit GAX and HOXA5 expressions in human umbilical vein endothelial cells. Most of the studies on miR-130a-3p focus on its functions. For example, it can regulate the insulin sensitivity and hepatic steatosis,61 regulate cell migration and invasion in gemcitabine-resistant hepatoma cells,62 suppress cell migration and invasion in human gastric carcinoma,63 suppress cell viability, proliferation, and invasion in nasopharyngeal carcinoma,64 increase cisplatin resistance in non-small-cell lung cancer,65 regulate gemcitabine resistance in cholangiocarcinoma,66 attenuate activation, and induces apoptosis of hepatic stellate cells.67 The bioinformatics analyses using mice SCI model also found that miR-130a was significantly increased in the injured spinal cords.68 This indicates that serum miR-130a-3p, especially serum exosomal miR-130a-3p, is also a promising marker in the diagnosis and prognosis of acute SCI.
MicroRNA-125b was also first found to be significantly upregulated in primary glioblastomas.58 It was also found to be highly expressed in mouse models of cardiac hypertrophy and idiopathic end-stage failing human hearts.69 MicroRNA-125b was also found to be decreased in primary neuroblastoma tumors,70 psoriasis, and atopic eczema.71 In the 3-prime UTRs of zebrafish and human p53, there are highly conserved miR-125b response elements. During development and stress response, miR-125b plays an important negative regulator of p53 and p53-induced apoptosis.72 In fact, apoptosis is also important in the pathological process of SCI. After SCI, apoptosis can occur in astrocytes, oligodendrocytes, neurons, and microglia.73 Here, we confirmed that miR-125b was significantly decreased in the acute phase of SCI, which indicated that its anti-apoptotic role is limited in the injured area. Therefore, the low expression of miR-125b in serum exosomes not only can explain the pathological mechanism of SCI, but also indicate that it is also a promising marker in SCI diagnosis and prognosis.
In addition to the three miRNAs mentioned above, we found that there are other significantly up-regulated known miRNAs. Although we do not know whether these miRNAs are involved in the pathological process of SCI, they have been the research hotspots in other diseases, especially tumors. The next step is to determine the relationship between these altered miRNAs as potential biomarkers and the severity of SCI. The more detailed research on this subject will be performed in combination with animal models and clinical practice. More interestingly, we also found some valuable new miRNAs, such as the up-regulated novel_mir39, novel_mir2064, novel_mir603, novel_mir51, novel_mir1387, and the down-regulated novel_mir803, novel_mir1945, novel_mir1541, novel_mir513, novel_mir1985, novel_mir4, novel_mir564, and novel_mir429. Further bioinformatics analysis and functional research will be of great help to make clear their role in the pathological process of SCI and in judging whether they can be used as diagnostic markers.
In order to further analyze these DEmiRNAs, we used multiple types of software to find the target gene of DEmiRNAs, and for further GO and KEGG analysis. The results showed that the targets of up-regulated DEmiRNAs in SCI versus sham were most enriched in protein or DNA binding, transcription regulator activity, negative regulation of biological process, etc. Interestingly, the targets of down-regulated DEmiRNAs in SCI versus sham were most enriched in receptor activity, protein binding, cytosolic ribosome, intracellular part, neuron part, organelle and cell projection, chemical stimulus and multicellular organism development, etc. These were consistent with many well-known reports that demonstrate the diverse roles of miRNAs in many physiological and pathological processes.74–79 These suggest that the localized changes of miRNAs after acute SCI are definitely related to the pathological process of SCI, and whether peripheral circulating miRNAs are involved in this process is uncertain, but they can indeed be used as markers of acute SCI.
The KEGG analysis found that the most enriched pathways of the targets of up-regulated DEmiRNAs were axon guidance, melanogenesis, Wnt signaling pathway, and long-term potentiation. The most enriched pathways of the targets of down-regulated DEmiRNAs were ECM-receptor interaction and focal adhesion. Among these inhibited signal pathways, some have been reported to be related to SCI, such as axon guidance,34,80–82 Wnt signaling pathway,81–84 long-term potentiation,85,86 ECM-receptor interaction,87 and focal adhesion.88–90 Interestingly, we can detect the changes of peripheral circulating miRNAs in the acute phase of SCI, which not only indicates that these miRNAs can be used as markers for the rapid diagnosis of SCI, but also suggests that they may participate in the pathological process of acute SCI by regulating the SCI-related signaling pathways.
Conclusions
Acute SCI can lead to changes of serum exosomal miRNAs. These miRNAs and their target-regulated pathways are related to the pathological mechanism of acute SCI. More importantly, serum exosomal miR-130a-3p, miR-152-3p, and miR-125b-5p are specific and easily detectable diagnostic markers in acute SCI. More interestingly, we also found some valuable known and novel miRNAs. Further bioinformatics analysis and functional research will be of great help to make clear their function in the pathological process of SCI and in judging whether they can be used as diagnostic markers.
Supplemental Material
Supplemental material, Supplemental Material1 for Identification of serum exosomal microRNAs in acute spinal cord injured rats by Shu-Qin Ding, Jing Chen, Sai-Nan Wang, Fei-Xiang Duan, Yu-Qing Chen, Yu-Jiao Shi, Jian-Guo Hu and He-Zuo Lü in Experimental Biology and Medicine
Supplemental Material
Supplemental material, Supplemental Material2 for Identification of serum exosomal microRNAs in acute spinal cord injured rats by Shu-Qin Ding, Jing Chen, Sai-Nan Wang, Fei-Xiang Duan, Yu-Qing Chen, Yu-Jiao Shi, Jian-Guo Hu and He-Zuo Lü in Experimental Biology and Medicine
Supplemental Material
Supplemental material, Supplemental Material3 for Identification of serum exosomal microRNAs in acute spinal cord injured rats by Shu-Qin Ding, Jing Chen, Sai-Nan Wang, Fei-Xiang Duan, Yu-Qing Chen, Yu-Jiao Shi, Jian-Guo Hu and He-Zuo Lü in Experimental Biology and Medicine
Supplemental Material
Supplemental material, Supplemental Material4 for Identification of serum exosomal microRNAs in acute spinal cord injured rats by Shu-Qin Ding, Jing Chen, Sai-Nan Wang, Fei-Xiang Duan, Yu-Qing Chen, Yu-Jiao Shi, Jian-Guo Hu and He-Zuo Lü in Experimental Biology and Medicine
Supplemental Material
Supplemental material, Supplemental Material5 for Identification of serum exosomal microRNAs in acute spinal cord injured rats by Shu-Qin Ding, Jing Chen, Sai-Nan Wang, Fei-Xiang Duan, Yu-Qing Chen, Yu-Jiao Shi, Jian-Guo Hu and He-Zuo Lü in Experimental Biology and Medicine
Supplemental Material
Supplemental material, Supplemental Material6 for Identification of serum exosomal microRNAs in acute spinal cord injured rats by Shu-Qin Ding, Jing Chen, Sai-Nan Wang, Fei-Xiang Duan, Yu-Qing Chen, Yu-Jiao Shi, Jian-Guo Hu and He-Zuo Lü in Experimental Biology and Medicine
Supplemental Material
Supplemental material, Supplemental Material7 for Identification of serum exosomal microRNAs in acute spinal cord injured rats by Shu-Qin Ding, Jing Chen, Sai-Nan Wang, Fei-Xiang Duan, Yu-Qing Chen, Yu-Jiao Shi, Jian-Guo Hu and He-Zuo Lü in Experimental Biology and Medicine
Supplemental Material
Supplemental material, Supplemental Material8 for Identification of serum exosomal microRNAs in acute spinal cord injured rats by Shu-Qin Ding, Jing Chen, Sai-Nan Wang, Fei-Xiang Duan, Yu-Qing Chen, Yu-Jiao Shi, Jian-Guo Hu and He-Zuo Lü in Experimental Biology and Medicine
Authors’ contributions
HZL and JGH participated in the design of the study. SQD, JC, SNW and YQC performed experimental procedures. FXD and YJS conducted data analysis. All authors read and approved the final manuscript.
Availability of supporting data
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declaration of conflicting interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by grants from the National Natural Science Foundation of China (Nos. 81571194, 81772321).
Supplemental material
Supplemental material for this article is available online.
References
- 1.Friedli L, Rosenzweig ES, Barraud Q, Schubert M, Dominici N, Awai L, Nielson JL, Musienko P, Nout-Lomas Y, Zhong H, Zdunowski S, Roy RR, Strand SC, van den Brand R, Havton LA, Beattie MS, Bresnahan JC, Bezard E, Bloch J, Edgerton VR, Ferguson AR, Curt A, Tuszynski MH, Courtine G. Pronounced species divergence in corticospinal tract reorganization and functional recovery after lateralized spinal cord injury favors primates. Sci Transl Med 2015; 7:302ra134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Furlan JC, Fehlings MG. Role of screening tests for deep venous thrombosis in asymptomatic adults with acute spinal cord injury: an evidence-based analysis. Spine 2007; 32:1908–16 [DOI] [PubMed] [Google Scholar]
- 3.Fehlings MG, Tator CH. An evidence-based review of decompressive surgery in acute spinal cord injury: rationale, indications, and timing based on experimental and clinical studies. J Neurosurg 1999; 91:1–11 [DOI] [PubMed] [Google Scholar]
- 4.Battistuzzo CR, Smith K, Skeers P, Armstrong A, Clark J, Agostinello J, Cox S, Bernard S, Freeman BJ, Dunlop SA, Batchelor PE. Early rapid neurological assessment for acute spinal cord injury trials. J Neurotrauma 2016; 33:1936–45 [DOI] [PubMed] [Google Scholar]
- 5.Kwon BK, Streijger F, Fallah N, Noonan VK, Belanger LM, Ritchie L, Paquette SJ, Ailon T, Boyd MC, Street J, Fisher CG, Dvorak MF. Cerebrospinal fluid biomarkers to stratify injury severity and predict outcome in human traumatic spinal cord injury. J Neurotrauma 2017; 34:567–80 [DOI] [PubMed] [Google Scholar]
- 6.Badhiwala JH, Ahuja CS, Fehlings MG. Time is spine: a review of translational advances in spinal cord injury. J Neurosurg Spine 2018; 30:1–18 [DOI] [PubMed] [Google Scholar]
- 7.Talbott JF, Whetstone WD, Readdy WJ, Ferguson AR, Bresnahan JC, Saigal R, Hawryluk GW, Beattie MS, Mabray MC, Pan JZ, Manley GT, Dhall SS. The brain and spinal injury center score: a novel, simple, and reproducible method for assessing the severity of acute cervical spinal cord injury with axial T2-weighted MRI findings. J Neurosurg Spine 2015; 23:495–504 [DOI] [PubMed] [Google Scholar]
- 8.Wilson JR, Tetreault LA, Kwon BK, Arnold PM, Mroz TE, Shaffrey C, Harrop JS, Chapman JR, Casha S, Skelly AC, Holmer HK, Brodt ED, Fehlings MG. Timing of decompression in patients with acute spinal cord injury: a systematic review. Global Spine J 2017; 7:95S–115S [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fagerlund MK. Acute neuroradiology: methods, indications and timing. Ann Med 1995; 27:657–62 [DOI] [PubMed] [Google Scholar]
- 10.Martinez-Useros J, Garcia-Foncillas J. Obesity and colorectal cancer: molecular features of adipose tissue. J Transl Med 2016; 14:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adegunsoye A, Balachandran J. Inflammatory response mechanisms exacerbating hypoxemia in coexistent pulmonary fibrosis and sleep apnea. Mediators Inflamm 2015; 2015:510105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Winnerkvist A, Anderson RE, Hansson LO, Rosengren L, Estrera AE, Huynh TT, Porat EE, Safi HJ. Multilevel somatosensory evoked potentials and cerebrospinal proteins: indicators of spinal cord injury in thoracoabdominal aortic aneurysm surgery. Eur J Cardiothorac Surg 2007; 31:637–42 [DOI] [PubMed] [Google Scholar]
- 13.Hombach S, Kretz M. Non-coding RNAs: classification, biology and functioning. Adv Exp Med Biol 2016; 937:3–17 [DOI] [PubMed] [Google Scholar]
- 14.Ma L, Bajic VB, Zhang Z. On the classification of long non-coding RNAs. RNA Biol 2013; 10:925–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hachisuka S, Kamei N, Ujigo S, Miyaki S, Yasunaga Y, Ochi M. Circulating microRNAs as biomarkers for evaluating the severity of acute spinal cord injury. Spinal Cord 2014; 52:596–600 [DOI] [PubMed] [Google Scholar]
- 16.Tigchelaar S, Streijger F, Sinha S, Flibotte S, Manouchehri N, So K, Shortt K, Okon E, Rizzuto MA, Malenica I, Courtright-Lim A, Eisen A, Keuren-Jensen KV, Nislow C, Kwon BK. Serum MicroRNAs reflect injury severity in a large animal model of thoracic spinal cord injury. Sci Rep 2017; 7:1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tigchelaar S, Gupta R, Shannon CP, Streijger F, Sinha S, Flibotte S, Rizzuto MA, Street J, Paquette S, Ailon T, Charest-Morin R, Dea N, Fisher C, Dvorak MF, Dhall S, Mac-Thiong JM, Parent S, Bailey C, Christie S, Van Keuren-Jensen K, Nislow C, Kwon BK. MicroRNA biomarkers in cerebrospinal fluid and serum reflect injury severity in human acute traumatic spinal cord injury. J Neurotrauma 2019; 36:2358–71 [DOI] [PubMed] [Google Scholar]
- 18.Ebrahimkhani S, Vafaee F, Young PE, Hur SSJ, Hawke S, Devenney E, Beadnall H, Barnett MH, Suter CM, Buckland ME. Exosomal microRNA signatures in multiple sclerosis reflect disease status. Sci Rep 2017; 7:14293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chuo ST, Chien JC, Lai CP. Imaging extracellular vesicles: current and emerging methods. J Biomed Sci 2018; 25:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sork H, Corso G, Krjutskov K, Johansson HJ, Nordin JZ, Wiklander OPB, Lee YXF, Westholm JO, Lehtio J, Wood MJA, Mager I, El Andaloussi S. Heterogeneity and interplay of the extracellular vesicle small RNA transcriptome and proteome. Sci Rep 2018; 8:10813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vagner T, Chin A, Mariscal J, Bannykh S, Engman DM, Di Vizio D. Protein composition reflects extracellular vesicle heterogeneity. Proteomics 2019; 19:e1800167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gheinani AH, Vogeli M, Baumgartner U, Vassella E, Draeger A, Burkhard FC, Monastyrskaya K. Improved isolation strategies to increase the yield and purity of human urinary exosomes for biomarker discovery. Sci Rep 2018; 8:3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zeringer E, Barta T, Li M, Vlassov AV. Strategies for isolation of exosomes. Cold Spring Harb Protoc 2015; 2015:319–23 [DOI] [PubMed] [Google Scholar]
- 24.Nagarajah S. Exosome secretion – more than simple waste disposal? Implications for physiology, diagnostics and therapeutics. J Circ Biomark 2016; 5:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Isola AL, Chen S. Exosomes: the messengers of health and disease. Curr Neuropharmacol 2017; 15:157–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jalalian SH, Ramezani M, Jalalian SA, Abnous K, Taghdisi SM. Exosomes, new biomarkers in early cancer detection. Anal Biochem 2019; 571:1–13 [DOI] [PubMed] [Google Scholar]
- 27.Chan BD, Wong WY, Lee MM, Cho WC, Yee BK, Kwan YW, Tai WC. Exosomes in inflammation and inflammatory disease. Proteomics 2019; 19:e1800149. [DOI] [PubMed] [Google Scholar]
- 28.Bei Y, Yu P, Cretoiu D, Cretoiu SM, Xiao J. Exosomes-based biomarkers for the prognosis of cardiovascular diseases. Adv Exp Med Biol 2017; 998:71–88 [DOI] [PubMed] [Google Scholar]
- 29.Rezaie J, Ajezi S, Avci CB, Karimipour M, Geranmayeh MH, Nourazarian A, Sokullu E, Rezabakhsh A, Rahbarghazi R. Exosomes and their application in biomedical field: difficulties and advantages. Mol Neurobiol 2018; 55:3372–93 [DOI] [PubMed] [Google Scholar]
- 30.Revenfeld AL, Baek R, Nielsen MH, Stensballe A, Varming K, Jorgensen M. Diagnostic and prognostic potential of extracellular vesicles in peripheral blood. Clin Ther 2014; 36:830–46 [DOI] [PubMed] [Google Scholar]
- 31.Martirosyan NL, Carotenuto A, Patel AA, Kalani MY, Yagmurlu K, Lemole GM, Jr., Preul MC, Theodore N. The role of microRNA markers in the diagnosis, treatment, and outcome prediction of spinal cord injury. Front Surg 2016; 3:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen J, Wu Y, Duan FX, Wang SN, Guo XY, Ding SQ, Zhou JH, Hu JG, Lu HZ. Effect of M2 macrophage adoptive transfer on transcriptome profile of injured spinal cords in rats. Exp Biol Med 2019; 244:880–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu HZ, Xu L, Zou J, Wang YX, Ma ZW, Xu XM, Lu PH. Effects of autoimmunity on recovery of function in adult rats following spinal cord injury. Brain Behav Immun 2008; 22:1217–30 [DOI] [PubMed] [Google Scholar]
- 34.Shi LL, Zhang N, Xie XM, Chen YJ, Wang R, Shen L, Zhou JS, Hu JG, Lu HZ. Transcriptome profile of rat genes in injured spinal cord at different stages by RNA-sequencing. BMC Genomics 2017; 18:173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 2009; 10:57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Friedlander MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, Rajewsky N. Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol 2008; 26:407–15 [DOI] [PubMed] [Google Scholar]
- 37.T Hoen PA, Ariyurek Y, Thygesen HH, Vreugdenhil E, Vossen RH, de Menezes RX, Boer JM, van Ommen GJ, den Dunnen JT. Deep sequencing-based expression analysis shows major advances in robustness, resolution and inter-lab portability over five microarray platforms. Nucleic Acids Res 2008; 36:e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grant GR, Liu J, Stoeckert CJ., Jr. A practical false discovery rate approach to identifying patterns of differential expression in microarray data. Bioinformatics 2005; 21:2684–90 [DOI] [PubMed] [Google Scholar]
- 39.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A 2003; 100:9440–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bell E, Watson HL, Bailey S, Murray MJ, Coleman N. A robust protocol to quantify circulating cancer biomarker MicroRNAs. Methods Mol Biol 2017; 1580:265–79 [DOI] [PubMed] [Google Scholar]
- 41.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001; 25:402–8 [DOI] [PubMed] [Google Scholar]
- 42.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol 2004; 2:e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kruger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res 2006; 34:W451–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y. KEGG for linking genomes to life and the environment. Nucl Acids Res 2008; 36:D480–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4:44–57 [DOI] [PubMed] [Google Scholar]
- 46.Dennis G, Jr., Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol 2003; 4:P3. [PubMed] [Google Scholar]
- 47.Lesack K, Naugler C. An open-source software program for performing Bonferroni and related corrections for multiple comparisons. J Pathol Inform 2011; 2:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rodrigues LF, Moura-Neto V, Tcls ES. Biomarkers in spinal cord injury: from prognosis to treatment. Mol Neurobiol 2018; 55:6436–48 [DOI] [PubMed] [Google Scholar]
- 49.Liu X, Zhan Z, Xu L, Ma F, Li D, Guo Z, Li N, Cao X. MicroRNA-148/152 impair innate response and antigen presentation of TLR-triggered dendritic cells by targeting CaMKIIalpha. J Immunol 2010; 185:7244–51 [DOI] [PubMed] [Google Scholar]
- 50.Stokowy T, Gawel D, Wojtas B. Differences in miRNA and mRNA profile of papillary thyroid cancer variants. Int J Endocrinol 2016; 2016:1427042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Han Y, Liu M, Wang Z, Huang M, Xu N, Wu L. Serum MicroRNAs related with chemoradiotherapy resistance in Advanced-Stage cervical squamous cell carcinoma. Transl Oncol 2017; 10:378–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moya L, Meijer J, Schubert S, Matin F, Batra J. Assessment of miR-98-5p, miR-152-3p, miR-326 and miR-4289 expression as biomarker for prostate cancer diagnosis. Int J Mol Sci 2019; 20:1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.You W, Zhang X, Ji M, Yu Y, Chen C, Xiong Y, Liu Y, Sun Y, Tan C, Zhang H, Li J, Chen W, Li R. MiR-152-5p as a microRNA passenger strand special functions in human gastric cancer cells. Int J Biol Sci 2018; 14:644–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ouyang X, Jiang X, Gu D, Zhang Y, Kong SK, Jiang C, Xie W. Dysregulated serum MiRNA profile and promising biomarkers in dengue-infected patients. Int J Med Sci 2016; 13:195–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roux M, Perret C, Feigerlova E, Mohand Oumoussa B, Saulnier PJ, Proust C, Tregouet DA, Hadjadj S. Plasma levels of hsa-miR-152-3p are associated with diabetic nephropathy in patients with type 2 diabetes. Nephrol Dial Transplant 2018; 33:2201–7 [DOI] [PubMed] [Google Scholar]
- 56.Lugli G, Cohen AM, Bennett DA, Shah RC, Fields CJ, Hernandez AG, Smalheiser NR. Plasma exosomal miRNAs in persons with and without alzheimer disease: altered expression and prospects for biomarkers. PLoS One 2015; 10:e0139233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Y, Jiao J, Zhang S, Zheng C, Wu M. RIP3 inhibition protects locomotion function through ameliorating mitochondrial antioxidative capacity after spinal cord injury. Biomed Pharmacother 2019; 116:109019. [DOI] [PubMed] [Google Scholar]
- 58.Ciafre SA, Galardi S, Mangiola A, Ferracin M, Liu CG, Sabatino G, Negrini M, Maira G, Croce CM, Farace MG. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun 2005; 334:1351–8 [DOI] [PubMed] [Google Scholar]
- 59.Greco SJ, Rameshwar P. MicroRNAs regulate synthesis of the neurotransmitter substance P in human mesenchymal stem cell-derived neuronal cells. Proc Natl Acad Sci U S A 2007; 104:15484–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen Y, Gorski DH. Regulation of angiogenesis through a microRNA (miR-130a) that down-regulates antiangiogenic homeobox genes GAX and HOXA5. Blood 2008; 111:1217–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xiao F, Yu J, Liu B, Guo Y, Li K, Deng J, Zhang J, Wang C, Chen S, Du Y, Lu Y, Xiao Y, Zhang Z, Guo F. A novel function of microRNA 130a-3p in hepatic insulin sensitivity and liver steatosis. Diabetes 2014; 63:2631–42 [DOI] [PubMed] [Google Scholar]
- 62.Liu Y, Li Y, Wang R, Qin S, Liu J, Su F, Yang Y, Zhao F, Wang Z, Wu Q. MiR-130a-3p regulates cell migration and invasion via inhibition of Smad4 in gemcitabine resistant hepatoma cells. J Exp Clin Cancer Res 2016; 35:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang S, Han H, Hu Y, Yang W, Lv Y, Wang L, Zhang L, Ji J. MicroRNA-130a-3p suppresses cell migration and invasion by inhibition of TBL1XR1-mediated EMT in human gastric carcinoma. Mol Carcinog 2018; 57:383–92 [DOI] [PubMed] [Google Scholar]
- 64.Qu R, Sun Y, Li Y, Hu C, Shi G, Tang Y, Guo D. MicroRNA-130a-3p suppresses cell viability, proliferation and invasion in nasopharyngeal carcinoma by inhibiting CXCL12. Am J Transl Res 2017; 9:3586–98 [PMC free article] [PubMed] [Google Scholar]
- 65.Hu B, Zhang H, Wang Z, Zhang F, Wei H, Li L. LncRNA CCAT1/miR-130a-3p axis increases cisplatin resistance in non-small-cell lung cancer cell line by targeting SOX4. Cancer Biol Ther 2017; 18:974–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Asukai K, Kawamoto K, Eguchi H, Konno M, Asai A, Iwagami Y, Yamada D, Asaoka T, Noda T, Wada H, Gotoh K, Nishida N, Satoh T, Doki Y, Mori M, Ishii H. Micro-RNA-130a-3p regulates gemcitabine resistance via PPARG in cholangiocarcinoma. Ann Surg Oncol 2017; 24:2344–52 [DOI] [PubMed] [Google Scholar]
- 67.Wang Y, Du J, Niu X, Fu N, Wang R, Zhang Y, Zhao S, Sun D, Nan Y. MiR-130a-3p attenuates activation and induces apoptosis of hepatic stellate cells in nonalcoholic fibrosing steatohepatitis by directly targeting TGFBR1 and TGFBR2. Cell Death Dis 2017; 8:e2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhu Z, Wang D, Jiao W, Chen G, Cao Y, Zhang Q, Wang J. Bioinformatics analyses of pathways and gene predictions in IL-1alpha and IL-1beta knockout mice with spinal cord injury. Acta Histochem 2017; 119:663–70 [DOI] [PubMed] [Google Scholar]
- 69.van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, Richardson JA, Olson EN. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci U S A 2006; 103:18255–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Laneve P, Di Marcotullio L, Gioia U, Fiori ME, Ferretti E, Gulino A, Bozzoni I, Caffarelli E. The interplay between microRNAs and the neurotrophin receptor tropomyosin-related kinase C controls proliferation of human neuroblastoma cells. Proc Natl Acad Sci U S A 2007; 104:7957–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sonkoly E, Wei T, Janson PC, Saaf A, Lundeberg L, Tengvall-Linder M, Norstedt G, Alenius H, Homey B, Scheynius A, Stahle M, Pivarcsi A. MicroRNAs: novel regulators involved in the pathogenesis of psoriasis?. PLoS One 2007; 2:e610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Le MT, Teh C, Shyh-Chang N, Xie H, Zhou B, Korzh V, Lodish HF, Lim B. MicroRNA-125b is a novel negative regulator of p53. Genes Dev 2009; 23:862–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Beattie MS, Farooqui AA, Bresnahan JC. Review of current evidence for apoptosis after spinal cord injury. J Neurotrauma 2000; 17:915–25 [DOI] [PubMed] [Google Scholar]
- 74.Pinchi E, Frati A, Cantatore S, D'Errico S, Russa R, Maiese A, Palmieri M, Pesce A, Viola RV, Frati P, Fineschi V. Acute spinal cord injury: a systematic review investigating miRNA families involved. Int J Mol Sci 2019; 20:1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang Y, Yuan Y, Gao Y, Li X, Tian F, Liu F, Du R, Li P, Wang F, Xu S, Wu X, Wang C. MicroRNA-31 regulating apoptosis by mediating the phosphatidylinositol-3 kinase/protein kinase B signaling pathway in treatment of spinal cord injury. Brain Dev 2019; 41:649–61 [DOI] [PubMed] [Google Scholar]
- 76.Li F, Zhou MW. MicroRNAs in contusion spinal cord injury: pathophysiology and clinical utility. Acta Neurol Belg 2019; 119:21–7 [DOI] [PubMed] [Google Scholar]
- 77.Wang E. MicroRNA, the putative molecular control for mid-life decline. Ageing Res Rev 2007; 6:1–11 [DOI] [PubMed] [Google Scholar]
- 78.Barnes MR, Deharo S, Grocock RJ, Brown JR, Sanseau P. The micro RNA target paradigm: a fundamental and polymorphic control layer of cellular expression. Expert Opin Biol Ther 2007; 7:1387–99 [DOI] [PubMed] [Google Scholar]
- 79.Ichimura A, Ruike Y, Terasawa K, Tsujimoto G. miRNAs and regulation of cell signaling. Febs J 2011; 278:1610–8 [DOI] [PubMed] [Google Scholar]
- 80.Hollis ER., 2nd. Axon guidance molecules and neural circuit remodeling after spinal cord injury. Neurotherapeutics 2016; 13:360–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Onishi K, Hollis E, Zou Y. Axon guidance and injury-lessons from Wnts and Wnt signaling. Curr Opin Neurobiol 2014; 27:232–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Clark CE, Liu Y, Cooper HM. The yin and yang of wnt/ryk axon guidance in development and regeneration. Sci China Life Sci 2014; 57:366–71 [DOI] [PubMed] [Google Scholar]
- 83.Garcia AL, Udeh A, Kalahasty K, Hackam AS. A growing field: the regulation of axonal regeneration by Wnt signaling. Neural Regen Res 2018; 13:43–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lambert C, Cisternas P, Inestrosa NC. Role of Wnt signaling in Central nervous system injury. Mol Neurobiol 2016; 53:2297–311 [DOI] [PubMed] [Google Scholar]
- 85.Farooqui AA, Horrocks LA. Involvement of glutamate receptors, lipases, and phospholipases in long-term potentiation and neurodegeneration. J Neurosci Res 1994; 38:6–11 [DOI] [PubMed] [Google Scholar]
- 86.Chen R, Cohen LG, Hallett M. Nervous system reorganization following injury. Neuroscience 2002; 111:761–73 [DOI] [PubMed] [Google Scholar]
- 87.Zhou J, Xiong Q, Chen H, Yang C, Fan Y. Identification of the spinal expression profile of non-coding RNAs involved in neuropathic pain following spared nerve injury by sequence analysis. Front Mol Neurosci 2017; 10:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Graham ZA, Qin W, Harlow LC, Ross NH, Bauman WA, Gallagher PM, Cardozo CP. Focal adhesion kinase signaling is decreased 56 days following spinal cord injury in rat gastrocnemius. Spinal Cord 2016; 54:502–9 [DOI] [PubMed] [Google Scholar]
- 89.Hao M, Ji XR, Chen H, Zhang W, Zhang LC, Zhang LH, Tang PF, Lu N. Cell cycle and complement inhibitors may be specific for treatment of spinal cord injury in aged and young mice: transcriptomic analyses. Neural Regen Res 2018; 13:518–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chuang YC, Lee CH, Sun WH, Chen CC. Involvement of advillin in somatosensory neuron subtype-specific axon regeneration and neuropathic pain. Proc Natl Acad Sci U S A 2018; 115:E8557–E66 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Supplemental Material1 for Identification of serum exosomal microRNAs in acute spinal cord injured rats by Shu-Qin Ding, Jing Chen, Sai-Nan Wang, Fei-Xiang Duan, Yu-Qing Chen, Yu-Jiao Shi, Jian-Guo Hu and He-Zuo Lü in Experimental Biology and Medicine
Supplemental material, Supplemental Material2 for Identification of serum exosomal microRNAs in acute spinal cord injured rats by Shu-Qin Ding, Jing Chen, Sai-Nan Wang, Fei-Xiang Duan, Yu-Qing Chen, Yu-Jiao Shi, Jian-Guo Hu and He-Zuo Lü in Experimental Biology and Medicine
Supplemental material, Supplemental Material3 for Identification of serum exosomal microRNAs in acute spinal cord injured rats by Shu-Qin Ding, Jing Chen, Sai-Nan Wang, Fei-Xiang Duan, Yu-Qing Chen, Yu-Jiao Shi, Jian-Guo Hu and He-Zuo Lü in Experimental Biology and Medicine
Supplemental material, Supplemental Material4 for Identification of serum exosomal microRNAs in acute spinal cord injured rats by Shu-Qin Ding, Jing Chen, Sai-Nan Wang, Fei-Xiang Duan, Yu-Qing Chen, Yu-Jiao Shi, Jian-Guo Hu and He-Zuo Lü in Experimental Biology and Medicine
Supplemental material, Supplemental Material5 for Identification of serum exosomal microRNAs in acute spinal cord injured rats by Shu-Qin Ding, Jing Chen, Sai-Nan Wang, Fei-Xiang Duan, Yu-Qing Chen, Yu-Jiao Shi, Jian-Guo Hu and He-Zuo Lü in Experimental Biology and Medicine
Supplemental material, Supplemental Material6 for Identification of serum exosomal microRNAs in acute spinal cord injured rats by Shu-Qin Ding, Jing Chen, Sai-Nan Wang, Fei-Xiang Duan, Yu-Qing Chen, Yu-Jiao Shi, Jian-Guo Hu and He-Zuo Lü in Experimental Biology and Medicine
Supplemental material, Supplemental Material7 for Identification of serum exosomal microRNAs in acute spinal cord injured rats by Shu-Qin Ding, Jing Chen, Sai-Nan Wang, Fei-Xiang Duan, Yu-Qing Chen, Yu-Jiao Shi, Jian-Guo Hu and He-Zuo Lü in Experimental Biology and Medicine
Supplemental material, Supplemental Material8 for Identification of serum exosomal microRNAs in acute spinal cord injured rats by Shu-Qin Ding, Jing Chen, Sai-Nan Wang, Fei-Xiang Duan, Yu-Qing Chen, Yu-Jiao Shi, Jian-Guo Hu and He-Zuo Lü in Experimental Biology and Medicine
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.