Short abstract
Esophageal adenocarcinoma displays a poor prognosis and current treatments are often not curative. Pathological TNM-stage is a prognostic parameter, but a better understanding of the pathophysiology of esophageal adenocarcinoma is needed to better predict survival. Recent work in other malignancies indicated an important role for the regulator microRNA-126 (miR-126) in tumors. The aim of this study was to investigate the function of miR-126 in esophageal adenocarcinoma and to correlate expression of miR-126 with tumor cell behavior and patient survival. Functional assays were performed in esophageal adenocarcinoma cell lines (OE33) in vitro by overexpressing or antagonizing miR-126 and assessing cellular processes linked to the hallmarks of cancer. In vivo pre-treatment biopsies of 58 patients with esophageal adenocarcinoma who underwent neoadjuvant chemoradiotherapy and surgery were analyzed for miR-126 expression in tumor cells by qRT-PCR and patient survival was analyzed by Kaplan–Meier and Cox regression. In OE33 cancer cells, stable overexpression of miR-126 modest though significantly altered expression of genes related to cell death (MEK1) and DNA repair (POLB and TERF1) was observed. Also the secretion of the angiogenic and pro-inflammatory factors, VEGF, IL-1β, and IL-6 were regulated by miR-126 (P < 0.029). Importantly, miR-126 was found to be a regulator of cell viability in OE33 cells. Overexpressing (P = 0.043) and antagonizing (P = 0.035) miR-126 showed reciprocal effects on tumor cell viability and significantly regulated expression of pro- and anti-apoptotic genes, TP53, and GATA6 (P < 0.031). In patients, high levels of miR-126 expression in pre-treatment tumors were significantly associated with poor survival (P = 0.031). In multivariable analysis, high miR-126 (P = 0.038) together with ypN-stage (P = 0.048) were shown to be independent risk factors for poor survival. In conclusion, high expression of miR-126 in esophageal adenocarcinoma prevents tumor-cell death and is associated with poor patient survival. This study warrants further analysis of miR-126 as biomarker or potential therapeutic target for OAC.
Impact statement
Esophageal adenocarcinoma is a common form of cancer of the esophagus. It has an increasing health impact as it is associated with very poor patient survival. A better understanding of the pathophysiology of this cancer is needed to identify better treatment strategies and to provide a better prognosis for these patients. MicroRNAs have emerged as important molecular regulators of cancer cell viability and proliferation. The aim of our study was to investigate the role of one very well established microRNA, miR-126, in esophageal adenocarcinoma. Our research shows clear experimental evidence that miR-126 controls cell viability of esophageal adenocarcinoma cells. High (over)expression of miR-126 increased the viability of these cells. Our preclinical data were shown to be clinically relevant for this field of oncology. In an independent validation study of esophageal adenocarcinoma biopsies, we confirmed that high miR-126 expression in tumor cells was an independent risk factor for poor patient survival.
Keywords: Esophageal cancer, multimodality treatment, miR-126
Introduction
Cancers of the esophagus are often diagnosed at an advanced stage, which results in less than half of the patients being eligible for potentially curative treatment at diagnosis.1 Incidences of esophageal cancer, and especially of esophageal adenocarcinoma (OAC) are rising in Western Europe.2 Neoadjuvant chemoradiotherapy (nCRT) followed by surgery is considered standard treatment for locoregional disease (cT1N1 and cT2-4a, cN0-N3, M0). Multimodality treatment is able to downstage the tumor, it facilitates a resection with tumor negative margins and may cure (locoregional) micrometastases at an early stage. Recently, the CROSS trial showed that these expectations are achieved and that patients who underwent nCRT followed by surgery had a better survival and had reduced locoregional recurrences as compared to surgery alone.3–6 There is an ongoing need to identify objective biomarkers to predict response to nCRT or to predict survival. Currently, a promising approach for molecular characterization of tumors is the understanding of the role that microRNAs (miRNAs) play in cancer.
MiRNAs are small non-coding RNAs who are able to regulate gene expression. Approximately one-third of all human genes is directly regulated by miRNA.7,8 The miRNA, by interaction with a complementary sequence in mRNA, causes inhibition of post-transcriptional translation or induces targeted mRNA degradation.9,10 MiRNAs regulate many important cellular processes such as cell proliferation, migration, and apoptosis. Dysregulation of miRNAs occurs in a number of pathological conditions most prominent in cancer.7,8,11
Several cancer-associated miRNAs have been now identified, of which miRNA-126 (miR-126) is one of the most established and broad acting ones. MiR-126 is implicated to play an important role in cancer biology of different cancer types including breast-, gastric-, and pancreatic neoplasms.12 Two pathways that have been linked with miR-126 are angiogenesis and cell death.13,14 More recently, a role in mitochondrial function and metabolism has been demonstrated.15,16 In non-small cell lung cancer patients, low miR-126 expression in either biopsies or resection specimens was correlated with a poorer survival compared to tumors with high expression.17 In these tumors, high miR-126 expression, combined with VEGF co-expression, is prognostically unfavorable for patients and points more towards an oncogenic role.18 Furthermore, in patients with colon cancer, high expression of miR-126 in the resection specimen is related to a better overall survival.19
The role of miR-126 in esophageal cancer is less well established. Three known target genes of miR-126, namely VEGFA, TP53 and GATA6, are often amplified in the genome of OAC tumor cells.20,21 In squamous cell carcinoma, miR-126 expression was decreased when compared to normal squamous epithelium in the esophagus.22 In OAC resection specimens, miR-126 expression is associated with poor prognostics factors including tumor cell dedifferentiation and lymphatic dissemination.23 The exact impact of miR-126 expression on the prognosis of survival in patients with OAC remains yet to be established. Moreover, it is still unknown which cancer signaling pathways are involved. The aim of this study is to investigate the functional role of miR-126 in OAC cells and profile if the level of miR-126 has associations with patient survival.
Materials and methods
Patients, disease staging, and treatment
Some 58 patients with histologically proven adenocarcinoma of the intrathoracic esophagus or gastro-esophageal junction who underwent nCRT followed by surgery were identified from a prospectively collected institutional database. All patients were treated at the Erasmus MC University Medical Centre Rotterdam, which is a tertiary referral center for patients with esophageal carcinoma in the Netherlands. The use of tissue biopsies for this research has been approved by the local medical ethical committee at the Erasmus MC Rotterdam and was judged not WMO obligatory. Tumors were staged according to the 7th UICC-AJCC TNM staging manual.24 An upper gastro-intestinal endoscopy with biopsies, endoscopic ultrasonography (EUS) with fine needle aspiration (FNA) when indicated, external ultrasonography of the neck with FNA and computed tomography (CT) of the neck, chest, and abdomen was performed in every patient. Bronchoscopy was only indicated for tumors of the intrathoracic esophagus when there was suspicion of infiltration of the tracheobronchial tree. Patients received chemoradiotherapy prior to surgery according to CROSS.5 In short, carboplatin and paclitaxel were administered intravenously on a weekly basis consisting of five courses with concurrent irradiation with 23 fractions of 1.8 Gy each. Surgical resection of the esophagus was performed within four to six weeks of completing the nCRT regimen. Patient’s survival was calculated from date of surgery till date of death or date of follow-up, updated in March 2015, irrespective of cause of death.
Tissue handling and laser capture microdissection
Pre-treatment formalin-fixed paraffin-embedded (FFPE) biopsies were requested from the tissue-bank in the Erasmus Medical Centre Rotterdam. A rotation microtome was used to cut the biopsies to 4 μm thickness. The first two slides and the last one were collected on normal StarFrost glass-slides to identify the tumor locations before laser capture micro dissection. The in between slides were collected and placed on to special membrane slides (Zeiss Membraneslide 1.0 PEN, Zeiss, Breda, the Netherlands). The next day these slides were deparaffinized and shortly (1–2 s) stained with haematoxylin (Mayer's hemalum solution for microscopy), followed by a short rehydration through graded concentrations of ethanol in water. After drying of the slides, the tumor areas in the biopsies were selected by a pathologist before the laser capture micro dissection.
Laser capture microdissection was performed with the PALM laser micro dissector (Carl Zeiss Micro Imaging, Breda, the Netherlands). The micro dissected tumor pieces were collected in the cap of a 500 μL autoclaved tube within a drop (40 μL) of digestion buffer (RecoverAll Total Nucleic Acid Isolation Kit for FFPE, Applied Biosystems, Fisher Scientific, Landsmeer, the Netherlands). More digestion buffer (160 μL) was added, together with 4 μL protease. The tubes were placed in a micro centrifuge and spinned shortly to collect all the liquid with tissue to the bottom of the tube. The samples were then incubated in a heat block for 15 min at 50°C, followed by 15 min at 80°C and stored at −20°C.
RNA-isolation and quantitative reverse transcript PCR
RNA isolation was performed according to the manufacturer’s instructions (RecoverAll Total Nucleic Acid Isolation Kit for FFPE from Applied Biosystems, Fisher Scientific, Landsmeer, the Netherlands). MicroRNA-specific cDNA was synthesized using a Taqman MicroRNA Reverse Transcription Kit (Applied Biosystems) containing for every reaction 0.4 µL 100 mM dNTP’s, 1.35 µL multiscribe RT, 2.0 µL 10× RT Buffer, 0.25 µL RNase inhibitor and 1.0 µL of the primer for miR-126 (UCGUACCGUGAGUAAUAAUGCG) and RNU43 as endogenous control for data normalization. Real-time qRT-PCR was carried out in duplicate and each reaction consisted of 10 µL TaqMan Universal PCR Master Mix (TaqMan® Universal PCR Master Mix No AmpErase® UNG), 0.5 µL miR-specific PCR primer (Applied Biosystems), and 5.0 µL of diluted cDNA. The reaction ran in a 40 cycle-schedule: 10 min at 95°C, followed by 40 times 15 s 95°C for 1 min 60°C (iQ5 multicolor real-time PCR detection system Bio-Rad Laboratories, Veenendaal, the Netherlands).
MiR-126 expression was quantified based on the 2−ΔCt method where relative levels were normalized to levels of reference non-coding RNA, RNU43. Groups were created based on expression of miR-126, where a cut-off was made at the 75th percentile to discriminate high (top25%) versus low expression.
Transfection of cell lines
To establish a stable cell line overexpressing miR-126, OE33 cells were transfected via Lipofectamine (Applied Biosystems) with 10 ng of the plasmid carrying the miR-126 sequence 5′-CGUACCGUGAGUAAUAAUGCG-or an empty plasmid as control (vector control) (OriGene, Herford, Germany). Continuous selection was carried out with 0.5 mg/mL G418 (Sigma-Aldrich, Zwijndrecht, the Netherlands) added to the cell culture media after 24 h after transfection. G418-resistant clones were maintained in RPMI (+10% FCS) with 0.5 mg/mL G418. MiR-126 expression levels were measured via qRT-PCR analysis normalized to RNU43 as reference gene, to evaluate the degree of overexpression per experiment. A transient overexpression of miR-126 in OE33 cells was established within 48 h with similar transduction except without continuous selection. To antagonize miR-126, OE33 cells were transiently transfected after overnight adherence with 5 µM of antimiR-126 or a scrambled control (both fluorescein-labeled, Exiqon, QIAGEN Benelux B.V. Venlo The Netherlands) using Dharmafect I (Dharmacon, Cambrigde, United Kingdom). Transfection efficiency was measured after 24 h with flow cytometry.
Gene expression analysis in OE33 (cancer pathways)
To screen which cancer pathways miR-126 was altering, a cancer pathway finder pathway gene array analysis was performed which included 84 genes, representing 9 cancer pathways (angiogenesis, apoptosis, cell cycle, cellular senescence, DNA damage and repair, epithelial to mesenchymal transition, hypoxia signaling, metabolism, and telomeres and telomerase) (RT2 Profiler PCR Array, Qiagen, Manchester, UK). Three independent batches of paired stably overexpressing miR-126 and accompanying empty vector controls were cultured and RNA was isolated from the cells. cDNA was made according to the manufacturer’s protocol (Bioscript, Berlin, Germany) from 1 µg input RNA. The qRT-PCR analysis was performed with a customized SYBR green master mix according to the manufacturer’s protocol and ran for 40 cycles (Applied Biosystems 7900 HT Fast Real-Time PCR system) to obtain relative expression levels.
Based on the results of the cancer pathway analysis, validation of the expression levels of the following genes was conducted by q-RT-PCR: CDH1, CTNNBL1, SNAI1, SNAI2, TWIST1 and ZEB2 for epithelial to mesenchymal transition (EMT), SMUG1, PARP1, MLH1 and MMS19 for DNA damage and repair; 18S was used as a reference gene. Furthermore, pro-apoptotic gene TP53 and anti-apoptotic gene GATA6 were measured in each of the samples by qRT-PCR.21,25,26
ELISA detection of inflammation and angiogenesis factors
In order to assess if miR-126 affected the secretion of inflammatory and angiogenic proteins, MSD Multispot ELISA was performed (Meso Scale Diagnostics, Rockville, Maryland, USA). The inflammatory panel included IL-1β, IL-2, IL-10, IL-6, MCP-1, MIP-3a, Gro-a, MMP2, MMP9, and TNF-α. The angiogenic panel included VEGF, Ang-1, Ang-2, bFGF, PAI-1, sVCAM, and sICAM. Conditioned culture medium of untransfected and transfected OE33 cells was analyzed using this multiplex panel according to the manufacturer’s instructions.
Flowcytometry
After 24 h of transient antagonizing, transfection cells were harvested and 7AAD (BD Biosciences, Vianen, the Netherlands) was added prior to flowcytometry. Controls were taken into account for compensations (OE33 cells unstained, OE33 cells + 10% DMSO + 7 AAD for single staining in Per-Cyp-5 channel and OE33 cells + antimiR-126 for single staining in FITC channel). By adding 7AAD, apoptosis was measured for transfected cells in the FITC channel. Similar to this method, transient overexpressing transfected cells were analyzed for cell death.
Statistical analysis
The probability of survival over time was estimated with the Kaplan–Meier method and the log-rank test was used to determine statistical differences between groups. To determine which variables affect survival, all variables with a significance P < 0.100 together with clinically relevant variables were included in a multivariable logistic regression model. Statistical significance was set at the 5% level. Statistical analysis was performed with the use of SPSS software, version 22.0 (SPSS, IBM, New York, USA).
Results
MiR-126 overexpression provides subtle changes in MAP kinase gene expression and cytokines and vascular growth factor production in OAC cells
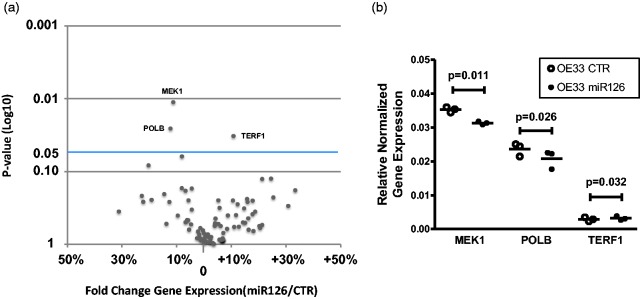
Using OE33 esophageal cancer cells stably overexpressing miR-126, a cancer pathway analysis was performed which included 84 specific genes representing nine different cancer pathways including programmed cell death, cell cycle, DNA damage and repair, angiogenesis, and epithelial to mesenchymal transition (EMT). As shown in Figure 1(a), overall effects of miR-126 overexpression were relatively modest and significance in gene expression was detected for three genes. As shown in Figure 1(b), MAP kinase MEK1 was significantly downregulated in samples overexpressing miR-126 compared to its empty vector control (P = 0.011). The MEK1 MAP kinase pathway is a known regulator of cell death in various tumor cell types. This effect was not validated by conventional RT-PCR as the exact sequence of these PCR primers is not disclosed. Additional findings from cancer pathway analysis were two genes related to DNA damage responses, POLB (DNA polymerase beta) and TERF1 (telomeric repeat binding factor 1) (Figure 1(b)). Based on these results, further validation of selected genes related to epithelial to mesenchymal transition (EMT) and DNA damage responses was performed by targeted qRT-PCR. In this validation none of the EMT or DNA damage response genes were significantly regulated by miR-126 (data not shown). Further, the effect of miR-126 on the production and secretion of pro-inflammatory and angiogenic factors by EO33 cells was evaluated using a multispot ELISA essay. Medium of control OE33 cells and miR-126 overexpressing cells were tested for a panel of 10 pro-inflammatory factors and 7 angiogenic factors. Of all angiogenic factors, only proangiogenic factor VEGF was significantly lower in OE33 cells stably transfected with miR-126 (25% reduction, P = 0.009 paired t test, data not shown). Furthermore, levels of the two pro-inflammatory cytokines IL-1β and IL-6 were significantly higher in miR-126 overexpressing OE33 cells (97% and 112% increase respectively, P < 0.029 paired t test, data not shown).
Figure 1.
MiR-126 regulated MAP kinase and DNA damage-related genes. Cancer pathway analysis was performed which includes 84 genes, representing nine major cancer pathways. Three independent experiments of paired OE33 cells either stably overexpressing miR-126 and an empty vector controls (CTR). (a) Shown is fold change in normalized gene expression and the P-value of statistical analyses (paired T-test). (b) MAP kinase MEK1 and DNA polymerase beta (POLB) were downregulated and telomeric repeat binding factor 1 (TERF1) significantly upregulated in samples overexpressing miR-126 compared to its empty vector control.
MiR-126 affects cell death and regulates cell-death-related gene expression in OAC cells
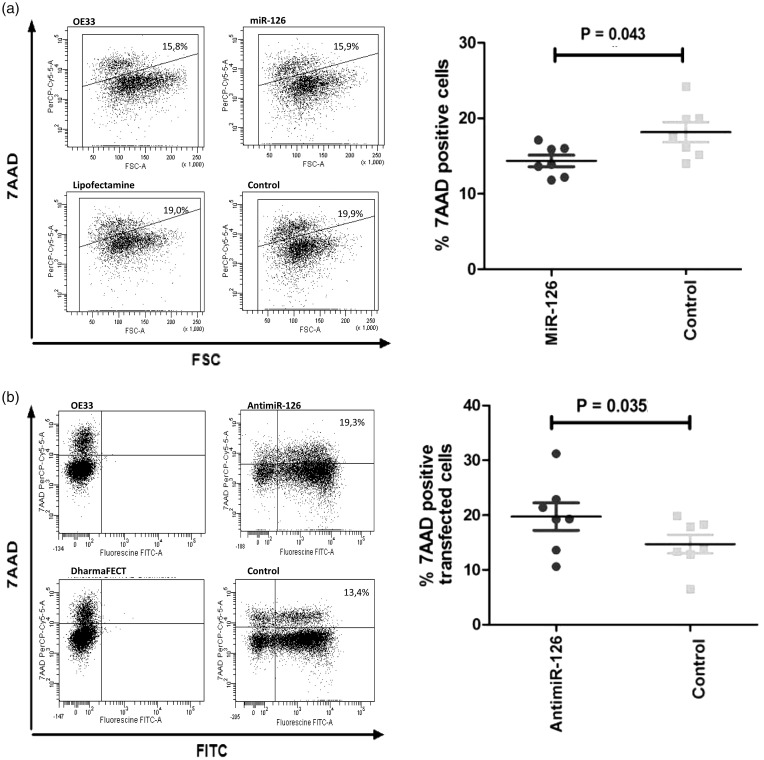
To further investigate the role of miR-126 in regulating programmed cell death, quantification of cell vitality and cell death was performed using flow cytometry. To this end, OE33 cells were transiently transfected with a plasmid containing miR-126 or an empty vector control. As shown in Figure 2(a), there was less death of OE33 cells overexpressing miR-126. The reverse effects were seen when OE33 cells were transfected with an antimiR-126 resulting in a significantly increased death of OE33 cells as compared to cells transfected with a non-specific antimiR control (Figure 2(b)). This effect was only observed in cells with detectable antimiR levels as shown by fluorescein labeling.
Figure 2.
MiR-126 regulates cell death of OAC cells. (a) Flowcytometric analysis of cell death in untreated OE33 cells, transfected OE33 cells overexpressing miR-126, OE33 cells treated with the transfection reagent Lipofectamin alone and transfected OE33 cells overexpressing an empty vector. Shown are representative plots with on the y-axis 7-AAD and on the x-axis the FSC (forward side scatter). The gated 7-AAD positive cells represent the fraction of dead cells. Results of seven independent experiments show significantly less dead cells when overexpressing miR-126 compared to its empty vector. (b) Shown are representative plots of untreated OE33 cells, OE33 cells transfected with antagonized miR-126 (antimiR-126), OE33 cells treated with the transfection reagent Darmafect alone, and OE33 cells transfected with irrelevant antimiR control. On the y-axis 7-AAD and on the x-axis, the FITC-channel is shown. Dead cells are represented in the double positive fraction (upper right corner). Results from seven independent experiments show significantly more dead cells when antagonizing miR-126 compared to the control antimiR treated cells.
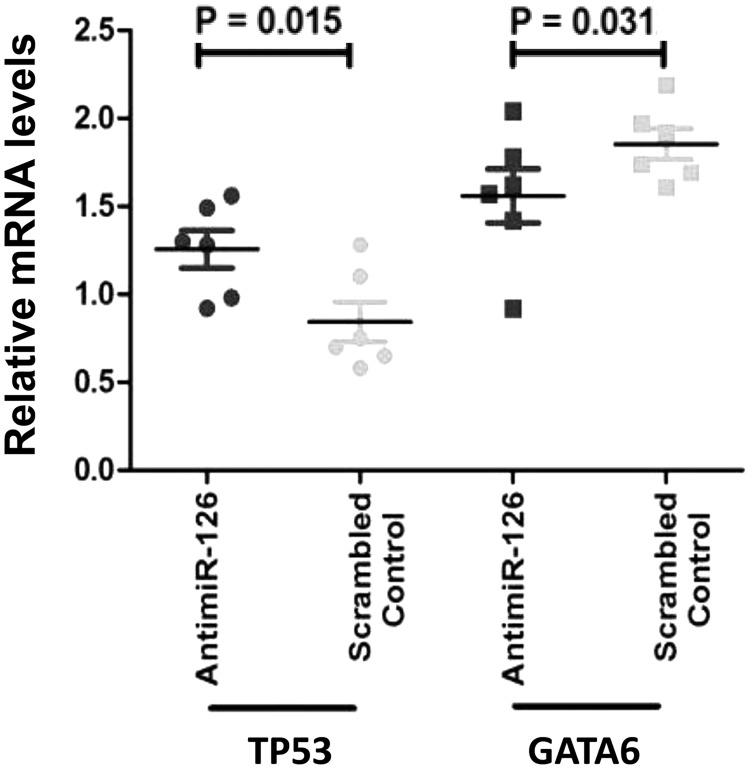
Subsequently, gene expression analysis was performed for the pro- and anti-apoptotic genes TP53 and GATA6. As shown in Figure 3, OE33 cells transfected with an antimiR-126 showed increased mRNA levels of the pro-apoptotic tumor suppressor gene TP53 (P = 0.015). This indicates that miR-126 downregulates expression of TP53 in OAC. OE33 cells transfected with an antimiR-126 showed a significant decreased mRNA level of the anti-apoptotic gene GATA6, as compared to control-treated cells (P = 0.031). This indicates that miR-126 indirectly upregulates expression of GATA6 in this OAC cell line. Together, these data indicate that miR-126 may be an important regulator of cell death in this OAC cell line model.
Figure 3.
MiR-126 regulates genes relevant for apoptosis. OE33 cells transfected with antagonized miR-126 (antimiR-126) were compared to transfected OE33 cells with irrelevant antimiR (scrambled) control. TP53 and GATA6 are plotted as fold changes. Results of six experiments are plotted showing antimiR-126 treatment significantly upregulates expression of TP53. A significantly lower expression of anti-apoptotic transcription factor GATA6 was seen in cells treated with antimiR-126.
High miR-126 expression in tumor is associated with poor patient survival
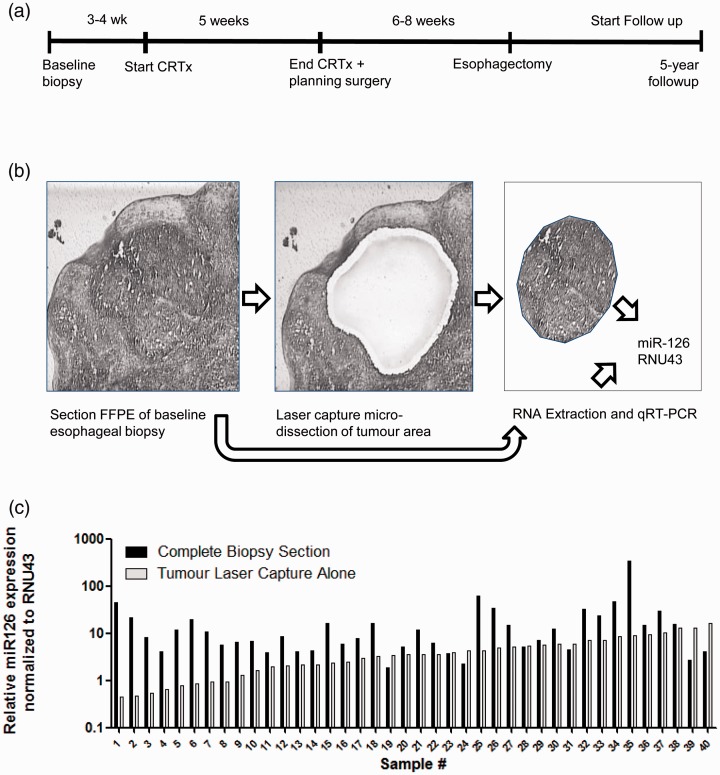
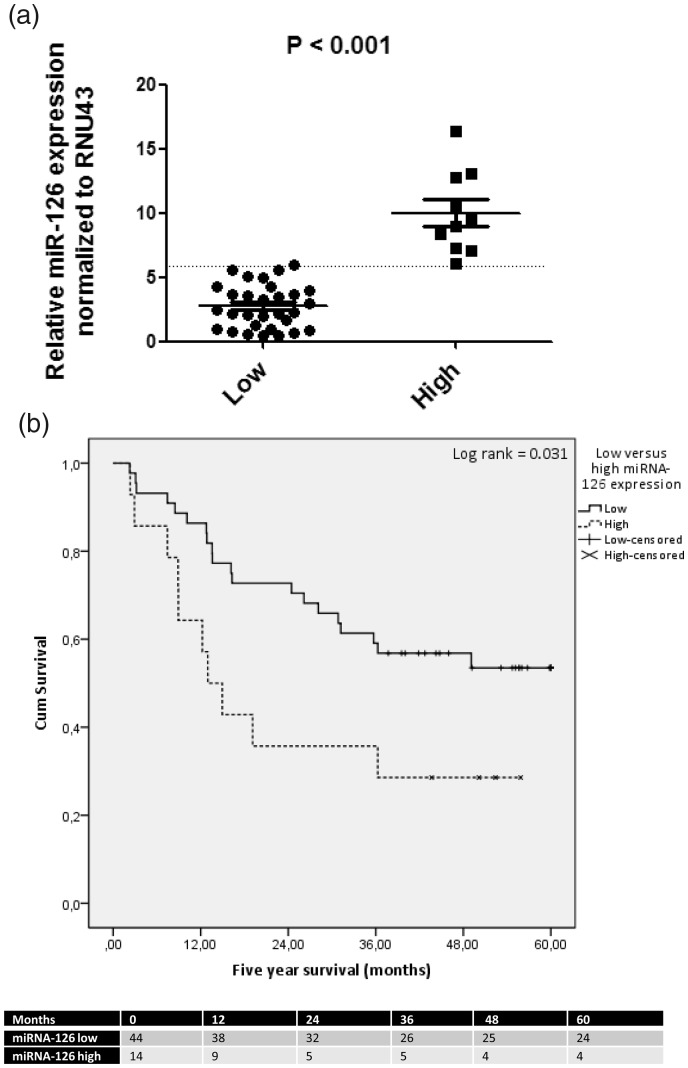
Next we evaluated the role of miR-126 in OAC patient samples, all treated with nCRT followed by surgery (Figure 4(a)). These samples were analyzed for miR-126 expression levels in their baseline pre-treatment biopsies. Laser capture microdissection was performed in order to obtain tumor-specific RNA (Figure 4(b)). Pre- and post-treatment patient and tumor characteristics are shown in Table 1. The majority of the patients were down staged by the nCRT, where 34% had a pathologically complete response. As shown in Figure 4(c), the relative expression of miR-126 in the laser capture tumor tissue greatly varied per patient. In tumor tissue with the lowest miR-126 level (lowest 25%), the relative expression in the complete biopsy section, which included RNA from both tumor and non-tumorous squamous epithelial cells, was a mean 16.3-fold higher than tumor alone (P = 0.002). In contrast, in tumors with the highest relative miR-126 levels (top 25%), there was no significant difference compared to the complete samples containing both tumor and squamous epithelial cells (P = 0.08). This suggests that the average level of miR-126 in the majority of OAC tumor cells is reduced compared to non-tumorous esophagus squamous epithelial cells.
Figure 4.
Evaluation of miR-126 levels in esophageal biopsies of patients with OAC. (a) The time-line of treatment of patients with OAC and their follow-up are shown. In total, 58 patients were treated with combined neoadjuvant chemoradiotherapy and surgical resection and patient overall survival was monitored for five years post treatment. (b) Esophageal biopsies taken at baseline containing both tumor and non-tumorous tissue including the squamous epithelium. Shown is a representative tissue section with HE-staining within the middle a tumor-island (Left panel). Tumor-specific RNA was obtained by laser capture microdissection microscopy (Middle panel). Following microdissection, the tumor-piece was captured in a small tube ready for RNA isolation. Also, RNA was extracted from the complete tissue section (Right panel). (c) Shown is the relative level of miR-126 normalized to RNU43 of 40 patients. Levels in tumor showed a wide variation (ranging from 0.4 to 16.4 normalized miR-126 values).
Table 1.
Patient and tumor characteristics at baseline and post-surgery of 58 patients receiving neoadjuvant chemoradiotherapy followed by surgery.
| Characteristic | Pre-treatment | Post-surgery |
|---|---|---|
| Gender (male) | 50 (86.2%) | 50 (86.2%) |
| Age (mean (range)) | 60 (36–78) | 60 (36–78) |
| Histology (AC) | 58 (100%) | 38 (65.5%) |
| Location | ||
| Mid | 5 (8.6%) | – |
| Distal | 34 (58.6%) | – |
| Gastro-esophageal junction | 19 (32.8%) | – |
| (yp)T–stage | ||
| 0 | 0 | 20 (34.5%) |
| 1 | 0 | 9 (15.5%) |
| 2 | 20 (34.5%) | 6 (10.3%) |
| 3 | 37 (63.8%) | 20 (34.5%) |
| 4 | 1 (1.7%) | 0 |
| Missing | 0 | 3 (5.2%) |
| (yp)N–stage | ||
| 0 | 17 (29.3%) | 40 (69%) |
| 1 | 25 (43.1%) | 12 (21%) |
| 2 | 15 (25.9%) | 6 (10%) |
| 3 | 1 (1.7%) | |
| (yp)M–stage | ||
| 0 | 58 (100%) | 57 (98.3%) |
| 1 | 0 | 1 (1.7%) |
| Grading | ||
| Good | 2 (3.4%) | 1 (1.7%) |
| Moderate | 33 (56.9%) | 22 (37.9%) |
| Poor | 21 (36.2%) | 10 (17.2%) |
| Missing | 2 (3.4%) | 25 (43.1%) |
| Radicality | ||
| R0 | – | 53 (91%) |
| R1 | – | 5 (9%) |
| Tumor regression grade | ||
| TRG 1 | – | 20 (34.5%) |
| TRG 2 | – | 12 ( 20.7%) |
| TRG 3 | – | 15 (25.9%) |
| TRG 4 | – | 11 (19.0%) |
| Recurrence | ||
| No | – | 34 (58.6%) |
| Yes | – | 24 (41.4%) |
AC: adenocarcinoma; yp: after neoadjuvant treatment; after resection; p: pathology; R0, R1: radicality of the resection; TRG: tumor regression grade.
In 14 patients with high relative expression of miR-126 (top 25%), the average expression level was 2.3-fold higher compared to 44 patients with a low relative expression (Figure 5(a)). Tumor or whole biopsy miR-126 expression was not predictive of response to neoadjuvant therapy with 95% CI including one for all four tumor regression grades (data not shown). The median five-year survival after surgery of all 58 patients was 36 months (IQR). No statistically significant association was observed between miR-126 levels in total biopsy sections and patient survival (data not shown). However, for tumor-specific miR-126 levels, patients with high miR-126 expression had a median survival of 14 months (IQR 37 months) compared to 41 months (IQR 39 months) for patients with low relative expression of miR-126 in tumors (Log rank P = 0.031, Figure 5(b)). Univariable and multivariable analysis of pre- and on-treatment characteristics which affect survival are shown in Table 2. The pre-treatment tumor miR-126 levels and the T stage (ypT) and N stage (ypN) (both derived from the resection specimen) were all associated with five-year survival. Also, post-surgery recurrence of disease significantly affected survival (HR 12.9, P < 0.0001, not shown). The relatively low number of patients limited the number of variables to be included in multivariable analysis. Taken the three most significant univariables, only pre-treatment miR-126 (HR 2.5, P = 0.038) and N-stage (ypN2, HR 4.6, P = 0.048) were independent risk factors for poor survival in multivariable analysis (Table 2). Taken together, these data indicate that high miR-126 levels in tumor cells are associated with a more progressive cancer resulting in reduced patient survival despite nCRT and esophagectomy treatments.
Figure 5.
High tumor miR-126 levels are associated with poor survival in patients with OAC.
In 14 patients with high relative miR-126 levels in tumor (top25%), as compared to 44 patients with a median or low relative level of tumor miR-126, the average difference was 2.3-fold (P<0.001). (b) Shown is five-year overall survival of the 58 patients treated with neoadjuvant chemoradiotherapy followed by surgery. High expression level of miR-126 in tumor tissue was significantly associated with poorer survival in patients with OAC. Cox regression analysis was used (Log rank P=0.031). The number of patients in each group at 12, 24, 36, 48, and 60 months post-treatments is shown at the bottom.
Table 2.
Univariable (UV) and multivariable (MV) analysis of prognostic (pre- and on-treatment) characteristics associated with poor five-year survival of 58 patients after combined neoadjuvant chemoradiotherapy and surgery.
| Parameters | HR (95% CI), P value, UV | HR (95% CI), P value, MV |
|---|---|---|
| Gender | ||
| Male | 1 (Ref) | x |
| Female | 0.664 (0.201–2.191), p = 0.502 | x |
| Age | ||
| <65 year | 1 (Ref) | x |
| ≥65 year | 1.698 (0.817–3.531), p = 0.156 | x |
| Location | ||
| Mid | 1 (Ref) | x |
| Distal | 0.406 (0.116–1.471), p = 0.157 | x |
| Junction | 0.809 (0.230–2.845), p = 0.741 | x |
| ypT-stagea | ||
| ypT0 | 1 (Ref) | 1 (Ref) |
| ypT1 | 1.607 (0.470–5.495), p = 0.449 | 0.880 (0.950–8.154), p = 0.910 |
| ypT2 | 1.010 (0.210–4.871), p = 0.990 | 0.617 (0.056–6.808), p = 0.693 |
| ypT3 | 3.965 (1.611–9.755), p = 0.003 | 2.019 (0.435–9.365), p = 0.369 |
| ypN-stagea | ||
| ypN0 | 1 (Ref) | 1 (Ref) |
| ypN1 | 1.245 (0.494–3.139), p = 0.642 | 1.299 (0.385–4.389), p = 0.673 |
| ypN2 | 6.336 (2.015–19.923), p = 0.002 | 4.606 (1.013–20.936), p = 0.048 |
| ypN3 | 4.917 (1.085–22.273), p = 0.039 | 1.995 (0.334–11.898), p = 0.449 |
| Radicality | ||
| R0 | 1 (Ref) | x |
| R1 | 2.506 (0.870–7.218), p = 0.089 | x |
| Tumor regression grade | ||
| TRG 1 | 1 (Ref) | x |
| TRG 2 | 2.453 (0.859–7.008), p = 0.094 | x |
| TRG 3 | 2.234 (0.829–6.020), p = 0.112 | x |
| TRG 4 | 2.637 (0.921–7.551), p = 0.071 | X |
| MiR-126 expressiona | ||
| Low | 1 (Ref) | 1 (ref) |
| High | 2.259 (1.053–4.846), p = 0.036 | 2.558 (1.052–6.220), p = 0.038 |
yp: after neoadjuvant treatment; after resection; p: pathology; R0, R1: radicality of the resection; TRG: tumor regression grade.
Taken into account for multivariable analysis.
Discussion
In this study, the function and expression of miR-126 in OAC were assessed in vitro and in vivo. The results show a role for miR-126 in regulating tumor cell death and that high miR-126 expression is associated with poor survival. The first step in this process was to create an OE33 cell line overexpressing miR-126 by stable transfection. VEGF production and secretion were significantly downregulated by miR-126. This is in line with previous studies, where overexpression of miR-126 mimics causes a downregulation of VEGF in ovarian, hepatocellular, and lung cancer.27–30 With the cancer pathway finder, testing 9 different cancer pathways included total 84 specific genes, we found that only MAP kinase (MEK1) and DNA damage (POLB and TERF1) genes were slightly though significantly affected by miR-126 overexpression in univariable analysis. Interestingly, a recent study showed that miR-126 (mimic) increases telomere length, consistent with the observed increase of TERF1 gene expression in EO33 cells (Figure 1).31 In statistics, the problem of multiple comparisons, like in the case for the cancer pathway assay, the Bonferroni correction or related methods have been proposed. However, tests like the Bonferroni correction reduce the statistical power and come at the cost of increasing the probability of producing false negatives. There is not a definitive consensus on how to correct for multiplicity and is therefore not used in our analyses.
The exact mechanism by which miR-126 regulated expression of these genes is not clear and may involve either direct or indirect effects. Direct regulation involves the miRNA binding to the 3′-UTR of the mRNA molecule suppressing the translation or promoting degradation. Indirect regulation would involve interaction of miR-126 with an mRNA of regulatory proteins, like transcription factors or signaling molecules, indirectly suppressing down-stream genes. Indeed, some of the gene regulation is likely to be indirectly, as the mRNA levels of TERF1 (Figure 1) and GATA6 (Figure 3) are increased, rather than suppressed, this would suggest indirect regulation by miR-126. This can be explained by miR-126 suppressing the translation of regulatory proteins that normally suppress the expression of TERF1 or GATA6. Regarding the targeting of the 3′-UTR of genes by miRNAs, bioinformatics databases are available. According to the Targetscan database (http://www.targetscan.org/),32 TERF1, VEGFA, and IL-6 are confirmed targets of miR-126. Related to the increased production of IL-1β by miR-126 overexpression in EO33 cells, the type I interleukin 1 receptor (IL1R1) and the interleukin 1α (IL1A) genes are predicted targets of miR-126 and maybe involved in indirect regulation IL-1β.
In the cancer pathway analysis, particular MEK1 showed a statistically significant lower expression in cells where miR-126 was overexpressed compared to the vector control. According to bioinformatics data, MEK1 is not a direct target of regulation by miR-126. However, miR-126 is a predicted target of the SMEK1 and SMEK2 genes which may indirectly effect MEK1.32 MEK1, also known as MAP2K1, was earlier identified as a factor in the development of cancer in general, but also in OAC.33,34 MEK1 is an essential component of the MAP kinase signal transduction pathway, regulating among others cellular homeostasis and apoptosis.35 When miR-126 was overexpressed in hepatocellular carcinoma cell lines, expression of EGFL7, ERK, Bcl-2, and P-ERK was suppressed, and expression levels of apoptotic-associated proteins Fas/FasL and Caspase-3 were increased. Furthermore, it induced apoptosis and inhibited cell proliferation.36 It is known that cancer cells are resistant to programmed cell death and can exhibit cellular senescence.37
Regarding the effects of miR-126 on OAC cell death, in the experiments where miR-126 was transiently overexpressed or antagonized, miR-126 showed reciprocal effects on tumor cell death. Indeed, in earlier studies, miR-126 is linked to apoptosis in the setting of rheumatoid arthritis by inhibiting the PI3K/AKT signal pathway and thereby inhibiting apoptosis.30,31,38,39 Furthermore, miR-126 significantly affected cell death regulating genes, TP53 and GATA6.21,25,31 Regarding the role of TP53, a recent study showed in kidney mesangial cells showed that miR-126 overexpression reduced TP53 protein expression.31 In accordance with this observation, in OE33 cells, TP53 gene expression was increased by antagonizing miR-126 by antimiR-126 (Figure 3). Though there is no evidence that miR-126 directly targets the 3′-UTR of TP53 mRNA, several TP53-related genes are predicted targets. These include TP53 apoptosis effector (PERP), TP53 inducible ribonucleotide reductase (RRM2B), TP53 inducible nuclear protein (TP53INP1), and TP53 regulating kinase (P53RK).32 Regarding the role of GATA6, an earlier study showed that silencing of GATA6 induces apoptosis in OAC cells but not in esophageal squamous cells. Moreover, OAC patients whose tumors carry a GATA6 gene amplification showed poorer survival.21 Finally, there is new compelling evidence for a role of IL-6 signaling in OAC cell death.40 Using a xenografting model of OE33 cells in mice, it was showed that anti-IL-6Rα antibody, Tocilizumab, suppressed tumor growth in vivo. This tumor suppression was associated with increased cell death of OE33 cells in vivo as shown by increased cleaved-caspase 3 staining. These findings are consistent with our in vitro results, showing that overexpression of miR-126 in OE33 cells increased IL-6 production and reduced cell death. Further experiments would need to show the role of IL-6 signaling in relation to cell death in miR-126 overexpressing OE33 cells.
To identify the clinical relevance of the possible role of miR-126 in tumor cell viability, patients diagnosed with OAC, treated with curative intent with nCRT followed by surgery were selected. In their pre-treatment biopsies, tumor-RNA was analyzed for miR-126 expression. Patients with a relatively higher expression of miR-126 had significantly worse prognosis than patients with lower expression of miR-126. These results were in line with the in vitro experiments with OE33 cells, showing that high (over)expression of miR-126 was associated with less cancer cell death in a cell viability assay (Figure 2). Both TNM-stage and miR-126 expression were significantly associated with patient survival (Table 2).
When comparing our findings to studies performed in other cancer types, somewhat contradictory results are observed. For instance, in non-small cell lung cancer patients, low miR-126 expression, in either biopsies or resection specimens, was correlated with a poorer survival compared to tumors with high expression.17 Similarly, in patients with colon cancer, high expression of miR-126 in the resection specimen was correlated to a better overall survival.19 These differences may be explained by a different biological role of miR-126 in these cancers and a difference in methodology used for miRNA quantification. On the other hand, our results are supported by an earlier study looking at the relationship between miRNA expression and survival in the patients with OAC. Using in situ hybridization with digoxigenin-labeled miRNA probes on tumor microarrays from nearly 100 patients, high expression of miR-126 was significantly associated with tumor cell dedifferentiation and lymph node metastasis, and with a non-significant trend towards poorer overall patient survival.23 Further, prospective studies are required to validate these observations and show the exact role miR-126 on disease progression in patients with OAC.
Although this study supports a role of miR-126 in regulating cell viability in OAC, there are several limitations to address. An adequate multivariable analysis was limited due to the sample size; however, it revealed miR-126 as an independent factor influencing survival. It was unfortunately not feasible to enlarge the group or to validate the results in an independent cohort, due to the limited availability of tissue for tumor-RNA isolation (laser capture microdissection). In vitro data could not be easily compared to other studies where miR-126 was overexpressed or silenced in different cancer types. This study focused on different pathways to elucidate the in vivo findings and makes it therefore a valuable result to share. In conclusion, this is the first study demonstrating a link between miR-126 and cell viability in OAC and this warrants further study of miR-126 as biomarker or potential therapeutic target for OAC.
ACKNOWLEDGEMENTS
The authors would like to thank Kelly Meijsen for help with experiments.
Authors’ contributions
All authors participated in the design of the study and editing of the manuscript; ELAT, NLL, KB, GD and PER conducted the experiments, analyzed and interpreted data, KB, JJBL, JVR and BPLW provided clinical input, ELAT JOS and LJWL wrote the manuscript, JOS and LJWL supervised the project.
DECLARATION OF CONFLICTING INTERESTS
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
FUNDING
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study has been financially supported by the foundations “Stichting Prof. Michaël-van Vloten Fonds” and “Stichting De Drie Lichten,” the Netherlands.
References
- 1.Pennathur A, Gibson MK, Jobe BA, Luketich JD. Oesophageal carcinoma. Lancet 2013; 381:400–12 [DOI] [PubMed] [Google Scholar]
- 2.Dikken JL, Lemmens VE, Wouters MW, Wijnhoven BP, Siersema PD, Nieuwenhuijzen GA. Increased incidence and survival for oesophageal cancer but not for gastric cardia cancer in the Netherlands. Eur J Cancer 2012; 48:1624–32 [DOI] [PubMed] [Google Scholar]
- 3.Oppedijk V, van der Gaast A, van Lanschot JJ, van Hagen P, van Os R, van Rij CM. Patterns of recurrence after surgery alone versus preoperative chemoradiotherapy and surgery in the CROSS trials. J Clin Oncol 2014; 32:385–91 [DOI] [PubMed] [Google Scholar]
- 4.Sjoquist KM, Burmeister BH, Smithers BM, Zalcberg JR, Simes RJ, Barbour A. Survival after neoadjuvant chemotherapy or chemoradiotherapy for resectable oesophageal carcinoma: an updated meta-analysis. Lancet Oncol 2011; 12:681–92 [DOI] [PubMed] [Google Scholar]
- 5.van Hagen P, Hulshof M, van Lanschot JJB, Steyerberg EW, Henegouwen MI, van B, Wijnhoven B. Preoperative chemoradiotherapy for esophageal or junctional cancer. N Engl J Med 2012; 366:2074–84 [DOI] [PubMed] [Google Scholar]
- 6.Shapiro J, van Lanschot JJB, Hulshof Mccm van Hagen P, van Berge Henegouwen MI, Wijnhoven B. Neoadjuvant chemoradiotherapy plus surgery versus surgery alone for oesophageal or junctional cancer (CROSS): long-term results of a randomised controlled trial. Lancet Oncol 2015; 16:1090–8 [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116:281–97 [DOI] [PubMed] [Google Scholar]
- 8.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer 2006; 6:857–66 [DOI] [PubMed] [Google Scholar]
- 9.Gu S, Kay MA. How do miRNAs mediate translational repression? Silence 2010; 1:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 2010; 11:597–610 [DOI] [PubMed] [Google Scholar]
- 11.Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science 2005; 310:1817–21 [DOI] [PubMed] [Google Scholar]
- 12.Meister J, Schmidt MH. miR-126 and miR-126*: new players in cancer. Sci World J 2010; 10:2090–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saito Y, Friedman JM, Chihara Y, Egger G, Chuang JC, Liang G. Epigenetic therapy upregulates the tumor suppressor microRNA-126 and its host gene EGFL7 in human cancer cells. Biochem Biophys Res Commun 2009; 379:726–31 [DOI] [PubMed] [Google Scholar]
- 14.Zhao C, Li Y, Zhang M, Yang Y, Chang L. miR-126 inhibits cell proliferation and induces cell apoptosis of hepatocellular carcinoma cells partially by targeting Sox2. Hum Cell 2015; 28:91–9 [DOI] [PubMed] [Google Scholar]
- 15.Tomasetti M, Neuzil J, Dong L. MicroRNAs as regulators of mitochondrial function: role in cancer suppression. Biochim Biophys Acta 2014; 1840:1441–53 [DOI] [PubMed] [Google Scholar]
- 16.Tomasetti M, Nocchi L, Staffolani S, Manzella N, Amati M, Goodwin J. MicroRNA-126 suppresses mesothelioma malignancy by targeting irs1 and interfering with the mitochondrial function. Antioxid Redox Signal 2014; 21:2109–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang J, Lan H, Huang X, Liu B, Tong Y. MicroRNA-126 inhibits tumor cell growth and its expression level correlates with poor survival in non-small cell lung cancer patients. PLoS One 2012; 7:e42978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donnem T, Lonvik K, Eklo K, Berg T, Sorbye SW, Al-Shibli K. Independent and tissue-specific prognostic impact of miR-126 in nonsmall cell lung cancer: coexpression with vascular endothelial growth factor-A predicts poor survival. Cancer 2011; 117:3193–200 [DOI] [PubMed] [Google Scholar]
- 19.Hansen T, Kjaer-Frifeldt S, Morgenthaler S, Blondal T, Lindebjerg J, Jakobsen A. The prognostic value of microRNA-126 and microvessel density in patients with stage II colon cancer: results from a population cohort. J Transl Med 2014; 12:254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cancer Genome Atlas Research Network J, Analysis Working Group: Asan University R, BC Cancer Agency AJ, Brigham and Women’s Hospital AG, Broad Institute RD, Brown University AD. Integrated genomic characterization of oesophageal carcinoma. Nature 2017; 541:169–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin L, Bass AJ, Lockwood WW, Wang Z, Silvers AL, Thomas DG. Activation of GATA binding protein 6 (GATA6) sustains oncogenic lineage-survival in esophageal adenocarcinoma. Proc Natl Acad Sci U S A 2012; 109:4251–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu SG, Qin XG, Zhao BS, Qi B, Yao WJ, Wang TY. Differential expression of miRNAs in esophageal cancer tissue. Oncol Lett 2013; 5:1639–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu Y, Correa AM, Hoque A, Guan B, Ye F, Huang J. Prognostic significance of differentially expressed miRNAs in esophageal cancer. Int J Cancer 2011; 128:132–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rice TW, Blackstone EH, Rusch VW. 7th edition of the AJCC Cancer Staging Manual: esophagus and esophagogastric junction. Ann Surg Oncol 2010; 17:1721–4 [DOI] [PubMed] [Google Scholar]
- 25.Sharpless NE, DePinho RA. p53: good cop/bad cop. Cell 2002; 110:9–12 [DOI] [PubMed] [Google Scholar]
- 26.Wang S, Aurora AB, Johnson BA, Qi X, McAnally J, Hill JA. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell 2008; 15:261–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo J, Zhu C, Wang H, Yu L, Zhou J. MicroRNA-126 affects ovarian cancer cell differentiation and invasion by modulating expression of vascular endothelial growth factor. Oncol Lett 2018; 15:5803–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu M-H, Ma C-Y, Wang X-M, Ye C-D, Zhang G-X, Chen L. MicroRNA-126 inhibits tumor proliferation and angiogenesis of hepatocellular carcinoma by down-regulating EGFL7 expression. Oncotarget 2016; 7:66922–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sasahira T, Kurihara M, Bhawal UK, Ueda N, Shimomoto T, Yamamoto K. Downregulation of miR-126 induces angiogenesis and lymphangiogenesis by activation of VEGF-A in oral cancer. Br J Cancer 2012; 107:700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuan Y, Shen C, Zhao S-L, Hu Y-J, Song Y, Zhong Q-J. MicroRNA-126 affects cell apoptosis, proliferation, cell cycle and modulates VEGF/TGF-β levels in pulmonary artery endothelial cells. Eur Rev Med Pharmacol Sci 2019; 23:3058–69 [DOI] [PubMed] [Google Scholar]
- 31.Cao D-W, Jiang C-M, Wan C, Zhang M, Zhang Q-Y, Zhao M, et al. Upregulation of MiR-126 delays the senescence of human glomerular mesangial cells induced by high glucose via telomere-p53-p21-Rb signaling pathway. Curr Med Sci 2018; 38:758–64 [DOI] [PubMed] [Google Scholar]
- 32.TargetScanHuman 7.2, www.targetscan.org/vert_72/ (accessed 30 May 2019)
- 33.Choi YL, Soda M, Ueno T, Hamada T, Haruta H, Yamato A. Oncogenic MAP2K1 mutations in human epithelial tumors. Carcinogenesis 2012; 33:956–61 [DOI] [PubMed] [Google Scholar]
- 34.Keld RR, Ang YS. Targeting key signalling pathways in oesophageal adenocarcinoma: a reality for personalised medicine? World J Gastroenterol 2011; 17:2781–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wada T, Penninger JM. Mitogen-activated protein kinases in apoptosis regulation. Oncogene 2004; 23:2838–49 [DOI] [PubMed] [Google Scholar]
- 36.Gong C, Fang J, Li G, Liu H-H, Liu Z-S. Effects of microRNA-126 on cell proliferation, apoptosis and tumor angiogenesis via the down-regulating ERK signaling pathway by targeting EGFL7 in hepatocellular carcinoma. Oncotarget 2017; 8:52527–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okada H, Mak TW. Pathways of apoptotic and non-apoptotic death in tumour cells. Nat Rev Cancer 2004; 4:592–603 [DOI] [PubMed] [Google Scholar]
- 38.Gao J, Zhou X-L, Kong R-N, Ji L-M, He L-L, Zhao D-B. microRNA-126 targeting PIK3R2 promotes rheumatoid arthritis synovial fibro-blasts proliferation and resistance to apoptosis by regulating PI3K/AKT pathway. Exp Mol Pathol 2016; 100:192–8 [DOI] [PubMed] [Google Scholar]
- 39.Chang L, Liang J, Xia X, Chen X. miRNA-126 enhances viability, colony formation, and migration of keratinocytes HaCaT cells by regulating PI3 K/AKT signaling pathway. Cell Biol Int 2019; 43:182–91 [DOI] [PubMed] [Google Scholar]
- 40.Karakasheva TA, Lin EW, Tang Q, Qiao E, Waldron TJ, Soni M, et al. IL-6 Mediates Cross-Talk between Tumor Cells and Activated Fibroblasts in the Tumor Microenvironment. Cancer Res 2018; 78:4957–70 [DOI] [PMC free article] [PubMed] [Google Scholar]