Key Points
Question
Can a genome-wide association study of neurophysiological and neurocognitive endophenotypes identify genes related to the underlying molecular dysfunctions in schizophrenia?
Findings
A total of 7 regions exceeding a conventional genome-wide significance threshold (P < 5 × 10−8) were associated with various endophenotype deficits, and the genes within these regions largely implicate glutamate and axonal dysfunctions as well as other neurobiologically salient targets. Many regions of interest were identified, including 39 previously associated with a schizophrenia diagnosis.
Meaning
These results suggest that the use of quantitative endophenotypes in genomic studies may provide an important, strong inference-based strategy for understanding the molecular basis of schizophrenia and may suggest novel treatment targets.
This genome-wide association study reports 11 schizophrenia-related endophenotypes in an independent cohort of patients with schizophrenia and healthy comparison participants within the Consortium on the Genetics of Schizophrenia study population.
Abstract
Importance
The Consortium on the Genetics of Schizophrenia (COGS) uses quantitative neurophysiological and neurocognitive endophenotypes with demonstrated deficits in schizophrenia as a platform from which to explore the underlying neural circuitry and genetic architecture. Many of these endophenotypes are associated with poor functional outcome in schizophrenia. Some are also endorsed as potential treatment targets by the US Food and Drug Administration.
Objective
To build on prior assessments of heritability, association, and linkage in the COGS phase 1 (COGS-1) families by reporting a genome-wide association study (GWAS) of 11 schizophrenia-related endophenotypes in the independent phase 2 (COGS-2) cohort of patients with schizophrenia and healthy comparison participants (HCPs).
Design, Setting, and Participants
A total of 1789 patients with schizophrenia and HCPs of self-reported European or Latino ancestry were recruited through a collaborative effort across the COGS sites and genotyped using the PsychChip. Standard quality control filters were applied, and more than 6.2 million variants with a genotyping call rate of greater than 0.99 were available after imputation. Association was performed for data sets stratified by diagnosis and ancestry using linear regression and adjusting for age, sex, and 5 principal components, with results combined through weighted meta-analysis. Data for COGS-1 were collected from January 6, 2003, to August 6, 2008; data for COGS-2, from June 30, 2010, to February 14, 2014. Data were analyzed from October 28, 2016, to May 4, 2018.
Main Outcomes and Measures
A genome-wide association study was performed to evaluate association for 11 neurophysiological and neurocognitive endophenotypes targeting key domains of schizophrenia related to inhibition, attention, vigilance, learning, working memory, executive function, episodic memory, and social cognition.
Results
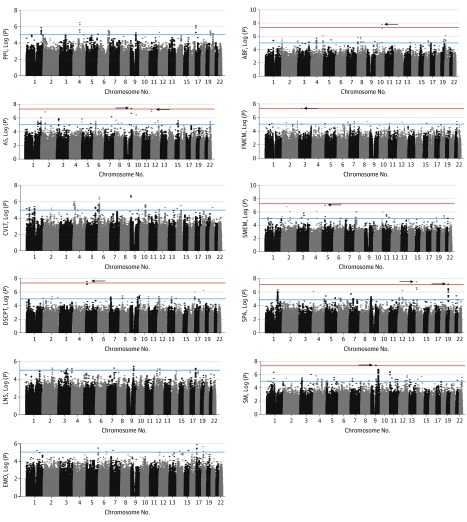
The final sample of 1533 participants included 861 male participants (56.2%), and the mean (SD) age was 41.8 (13.6) years. In total, 7 genome-wide significant regions (P < 5 × 10−8) and 2 nearly significant regions (P < 9 × 10−8) containing several genes of interest, including NRG3 and HCN1, were identified for 7 endophenotypes. For each of the 11 endophenotypes, enrichment analyses performed at the level of P < 10−4 compared favorably with previous association results in the COGS-1 families and showed extensive overlap with regions identified for schizophrenia diagnosis.
Conclusions and Relevance
These analyses identified several genomic regions of interest that require further exploration and validation. These data seem to demonstrate the utility of endophenotypes for resolving the genetic architecture of schizophrenia and characterizing the underlying biological dysfunctions. Understanding the molecular basis of these endophenotypes may help to identify novel treatment targets and pave the way for precision-based medicine in schizophrenia and related psychotic disorders.
Introduction
Schizophrenia is a severe and persistent psychotic disorder with a prevalence of approximately 1% and a heritability of as much as 80%.1 Large-scale genome-wide association studies (GWAS) using a case-control design have detected many common variants that collectively account for as much as 7% of risk for schizophrenia.2 Several of the implicated genes are consistent with leading hypotheses regarding pathophysiology, such as DRD2 (OMIM 126450), the target of most effective antipsychotics, and genes involved in glutamate signaling (eg, GRM3 [OMIM, 601115], GRIN2A [OMIM, 138253], and GRIA1 [OMIM, 138248]). The identification of the complement component 4 gene (C4 [OMIM, 120810]) within the major histocompatibility complex region and its relation to synaptic pruning is particularly intriguing.3 However, most of the genetic risk for schizophrenia remains unexplained, and the pathways by which the implicated variants contribute to the clinically observable signs and symptoms are largely unknown to date.
To achieve large samples for GWAS, clinically diverse patients are typically grouped together by diagnosis, ignoring the broad range of deficits associated with schizophrenia and possibly obscuring salient genetic signals. As stable biomarkers of the underlying brain dysfunctions, endophenotypes hold promise for parsing the clinical heterogeneity of schizophrenia and refining the genetic signal.4 Because endophenotypes provide laboratory-based measures of specific neurobiological functions with characteristic impairment in schizophrenia (eg, inhibition, memory, and executive function), they are less subjective than the clinical symptoms used for diagnosis. As quantitative measures of genetic liability to schizophrenia, endophenotypes provide increased efficiency and as much as a 10-fold increase in power compared with comparably sized case-control studies.5,6 Endophenotypes are also amenable to the use of animal models and neuroimaging in humans to facilitate direct investigations of underlying neurobiological processes.7,8 Finally, many neurocognitive endophenotypes are associated with poor real-world functioning and have been endorsed by the Measurement and Treatment Research to Improve Cognition in Schizophrenia (MATRICS) initiative and the US Food and Drug Administration as targets for the development of new treatments in schizophrenia.9 For example, targeted cognitive training has been shown to improve verbal memory in patients with schizophrenia, with a resultant increase in real-world functioning.10 The endophenotype strategy thus complements large-scale genomic investigations of diagnosis and provides important neurobiological context.
The Consortium on the Genetics of Schizophrenia (COGS)11 has previously reported the significant heritability of 11 neurophysiological and neurocognitive endophenotypes related to dysfunction across key domains, including aspects of inhibition, attention, learning, episodic and working memory, executive function, complex cognition, and social cognition. Such endophenotypes are intrinsic to the clinical presentation of schizophrenia, with clear deficits observed in patients with schizophrenia compared with healthy comparison participants (HCPs) and intermediate values observed for unaffected first-degree relatives.11,12,13,14,15,16 Candidate gene association and linkage analyses of the COGS phase 1 (COGS-1) families implicated a network of genes related to neuregulin and glutamate signaling pathways and synaptic plasticity as underlying endophenotype deficits in schizophrenia,17,18,19 consistent with the findings of other studies evaluating common, rare, and de novo variation in relation to a schizophrenia diagnosis.2,20,21 Herein we report the initial findings from the GWAS of these endophenotypes in the independent phase 2 (COGS-2) cohort of patients with schizophrenia and HCPs, which confirm and extend previous results and show consistency with studies of schizophrenia diagnosis in large samples.
Methods
Participants
Recruitment for the COGS study was conducted in 2 phases and reflects a carefully coordinated effort across multiple academic sites with standardized methods for diagnostic evaluation and endophenotype testing.22 The COGS-2 sample of 726 unrelated patients with schizophrenia and 667 HCPs of self-reported European or admixed European ancestry was collected through recruitment at the University of California, San Diego; UCLA (University of California, Los Angeles); Icahn School of Medicine at Mount Sinai, New York, New York; University of Pennsylvania, Philadelphia; and University of Washington, Seattle. We also included a sample of 396 HCPs of self-reported European ancestry collected as an adjunct to the COGS-1 family sample, which additionally involved recruitment at the University of Colorado Health Sciences Center, Denver, and Harvard University, Boston, Massachusetts. All participants underwent standardized psychiatric interviews using the Structured Clinical Interview for DSM-IV to obtain consensus diagnoses.23 All patients with schizophrenia met criteria for schizophrenia, and HCPs were excluded for psychosis, bipolar disorder, current depression, alcohol and/or substance abuse, or a first-degree relative with a history of psychosis. Participants ranged from 18 to 65 years of age and received urine toxicology screens before assessment. The HCPs were matched as a group to the patients with schizophrenia on age, sex, ancestry, and parental educational attainment. Written informed consent was obtained for each participant after a detailed description of study participation. This study was approved by the local institutional review board at each COGS site.
Data for COGS-1 were collected from January 6, 2003, to August 6, 2008; data for COGS-2, from June 30, 2010, to February 14, 2014. Detailed descriptions of the rationale and assessment procedures for the 11 endophenotypes have been previously published.24,25 Prepulse inhibition of the startle response at 60 milliseconds was used to evaluate inhibition. The proportion correct on the antisaccade task was used to evaluate oculomotor inhibition and directed attention. The degraded stimulus version of the Continuous Performance Test signal/noise discrimination was used to measure perceptual-load vigilance. Total correct responses on the California Verbal Learning Test, trials 1 to 5, and correct reordering on the Letter-Number Span were used as measures of learning and working memory, respectively. The University of Pennsylvania Computerized Neurocognitive Battery included tests of abstraction and mental flexibility, face memory, spatial memory, spatial processing, sensorimotor dexterity, and emotion recognition to measure functioning across additional cognitive domains, including executive function, episodic memory, complex cognition, and social cognition. All endophenotype data underwent a rigorous quality assurance process before analysis. The correlations between the endophenotypes are presented in the eTable in Supplement 1.
Genotyping
The 1789 participants were genotyped using the PsychChip, which was developed by the Psychiatric Genomics Consortium and contains a 240 000 genome-wide marker grid, 240 000 exome markers, and 50 000 custom content markers for psychiatric illness. Genotype calls from AutoCall, Birdseed, and zCall algorithms were merged to form a single consensus file. Using the Psychiatric Genomics Consortium RICOPILI (Rapid Imputation and Computational Pipeline; https://github.com/Ripkelab/ricopili), standard quality control filters were applied to remove 41 participants with poor genotype call rates (<0.98). An additional 27 participants were removed for sex discrepancies, sampling error, or relatedness. Single-nucleotide polymorphisms (SNPs) were removed for low minor allele frequency (<0.01), poor genotype call rates (<0.98), and deviation from Hardy-Weinberg equilibrium among HCPs (P < 10−6). Ancestry was confirmed via comparison with the 1000 Genomes Project Consortium phase 3 and Human Genome Diversity Project reference populations.26,27 After the removal of 188 population outliers, the largest ancestral groups included participants of European (523 patients with schizophrenia and 506 HCPs from COGS-2 and 321 HCPs from COGS-1; >95% European) and Latino (100 patients with schizophrenia and 83 HCPs from COGS-2; approximately 60% European and 30% Native American and >90% European and Native American combined) ancestry. Genotype data were imputed to the 1000 Genomes Project Consortium phase 3 reference panel,26 and a second round of quality control filters was applied as above to generate a final data set consisting of 1533 participants (623 patients with schizophrenia and 910 HCPs) and more than 6.2 million variants with a genotyping call rate of greater than 0.99.
Statistical Analysis
Data were analyzed from October 28, 2016, to May 4, 2018. The final data set was stratified by study (COGS-1 or COGS-2), case status (SZ case or HCP), and ancestry (European or Latino) for analysis. Principal components were calculated for each data set separately to correct for residual population stratification and genotype pool discrepancies. Association was performed using linear regression in PLINK, adjusting for age, sex, and the first 5 principal components, with a λ of less than 1.1 observed for each data set.28 Results were combined across data sets using weighted meta-analysis to adjust for sample size.29 Quantile-quantile plots for each analysis are provided in the eFigure in Supplement 1.
The combined sample has 80% power to detect a single variant accounting for 2.5% of the variance in a given endophenotype at the standard genome-wide significance threshold of P < 5 × 10−8. To the extent that the endophenotypes follow a polygenic model influenced by common variation, we expect that the same alleles contributing to endophenotype deficits in schizophrenia when aggregated in high density also influence endophenotype variation in the general population. As such, the inclusion of HCPs provides greater phenotypic range and increased power to detect associated variants, whereas the inclusion of patients with schizophrenia provides the appropriate context for the analyses. GWAS are intended to be hypothesis generating, yet a stringent statistical threshold designed to eliminate all false-positive results at a certain probability necessarily comes at the expense of an increased false-negative rate, obscuring any true associations that might fail to reach genome-wide significance. Indeed, several studies have demonstrated that subthreshold GWAS findings (P < 10−4) are enriched for SNPs that regulate gene expression.30,31 We have thus used an additional threshold of P < 10−4 to generate a broader set of SNPs potentially enriched for true associations, allowing for the exploration of patterns across endophenotypes and facilitating cross-study comparisons. A complete listing of SNPs exceeding the P < 10−4 threshold for each endophenotype is provided in Supplement 2.
Results
We included a total of 1533 participants (672 women [43.8%] and 861 men [56.2%]; mean [SD] age, 41.8 [13.6] years). Seven regions exceeding a conventional genome-wide significance threshold of P < 5 × 10−8 were identified, along with 2 regions nearly reaching genome-wide significance, as shown in Figure 1 and detailed in Table 1. The antisaccade task produced a significant peak (P = 3.5 × 10−8) on chromosome 9q31 within an olfactory gene family region and a nearly significant peak (P = 5.3 × 10−8) on chromosome 12p13 that spanned the cation-dependent PHC1 (OMIM 602978) and M6PR (OMIM 154540) genes. The Continuous Performance Test produced a significant peak on chromosome 5p15 near the BASP1 gene(OMIM 605940). Multiple associations were also observed for the Computerized Neurocognitive Battery measures. Abstraction and mental flexibility produced a significant (P = 1.5 × 10−8) on chromosome 10q23 near the NRG3 gene (OMIM 605533). Face memory produced a significant peak (P = 4.2 × 10−8) on chromosome 3p21 in the KIF15 gene (OMIM 617569). Spatial processing produced significant peaks on chromosome 14q11 in the AJUBA gene (OMIM 609066; P = 1.7 × 10−8) and chromosome 19q12 near the CCNE1 gene (OMIM 123837; P = 3.8 × 10−8). Spatial memory produced a nearly significant peak (P = 8.3 × 10−8) on chromosome 5p12 near the HCN1 gene (OMIM 602780). Sensorimotor dexterity produced a significant association (P = 4.0 × 10−8) with the C9orf135 gene (HGNC 31422) on chromosome 9q21.
Figure 1. Results of the Genome-wide Association Analyses of 11 Endophenotypes for Schizophrenia.
Significance is indicated on the y-axis as the –log(P). A total of 7 regions exceeded the conventional genome-wide significance threshold of P < 5 × 10−8 (red line) with 2 additional regions nearly reaching significance (P < 9 × 10−8). The blue line indicates P = 1 × 10−5. The University of Pennsylvania Computerized Neurocognitive Battery consisted of the following tests: abstraction and mental flexibility (ABF), emotional recognition (EMO), face memory (FMEM), sensorimotor dexterity (SM), spatial memory (SMEM), and spatial processing (SPA). AS indicates antisaccade task; CVLT, California Verbal Learning Test; DSCPT, degraded stimulus Continuous Performance Test; LNS, Letter-Number Span; and PPI, prepulse inhibition of the startle response.
Table 1. Summary of Significantly Associated Regions.
| Endophenotype | Chromosome | Positiona | Lead SNP in Region | Effect Size (95% CI)b | P Value for Meta-analysisc | P Value for EAc | Nearest Gene | Genes Within 100 kb |
|---|---|---|---|---|---|---|---|---|
| Face memory | 3p21 | 44813350 to 44874489 | rs73076581 | −0.72 (−1.00 to −0.45) |
4.2 × 10−8 | 4.2 × 10−8 | KIF15 | ZNF502, ZNF501, TMEM42, TGM4 |
| Continuous Performance Test | 5p15 | 17439886 to 17443654 | rs7706512 | 0.21 (0.14 to 0.28) |
2.5 × 10−8 | 1.2 × 10−6 | BASP1 | None |
| Spatial memory | 5p12 | 44791335 to 45215941 | rs187763903 | −0.72 (−0.98 to −0.47) |
8.3 × 10−8 | 6.8 × 10−6 | HCN1 | None |
| Sensorimotor dexterity | 9q21 | 72488418 | rs140830767 | −1.11 (−1.58 to −0.64) |
4.0 × 10−8 | 3.3 × 10−6 | C9orf135 | None |
| Antisaccade task | 9q31 | 107334694 to 107499546 | rs77038697 | −0.24 (−0.31 to −0.16) |
3.5 × 10−8 | 3.5 × 10−8 | OR13C9 | OR13C4, OR13C3, OR13C8, OR13C5, OR13C2, OR13D1, NIPSP3A |
| Abstraction and mental flexibility | 10q23 | 85685084 to 85719133 | rs11200733 | −0.56 (−0.20 to −0.07) |
1.5 × 10−8 | 6.1 × 10−8 | NRG3 | None |
| Antisaccade task | 12p13 | 9082391 to 9135281 | rs117302528 | −0.20 (−0.28 to −0.13) |
5.4 × 10−8 | 3.7 × 10−7 | PHC1 | A2ML1, M6PR, KLRG1 |
| Spatial processing | 14q11 | 23364106 to 23473706 | rs45543740 | −0.62 (−0.82 to −0.41) |
1.7 × 10−8 | 5.8 × 10−8 | AJUBA | REM2, RBM23, PRMT5, HAUS4, PSMB5, PSMB11, ACIN1 |
| Spatial processing | 19q12 | 44813350 to 44874489 | rs4805491 | −0.46 (−0.61 to −0.31) |
3.8 × 10−8 | 6.0 × 10−7 | CCNE1 | URI1 |
Abbreviations: EA, European ancestry; SNP, single-nucleotide polymorphism.
Indicates hg19 region spanned by SNPs tagged by the lead SNP. The boundaries of each gene are expanded by 20 kb on each side to capture putative regulatory elements.
Calculated from the lead SNP from the meta-analysis and the corresponding lower and upper bounds, respectively, of the 95% CI.
Indicates P value of lead SNP.
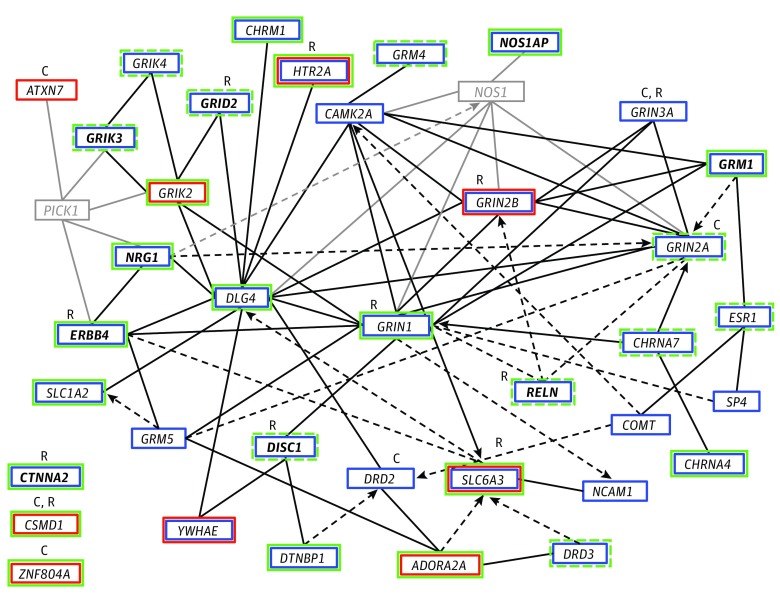
In total, 1070 independent regions of association were identified at an enrichment threshold of P < 10−4, including 128 regions of shared association across 2 or more endophenotypes. These regions are summarized in Supplement 2 and include the major histocompatibility complex region on chromosome 6, as well as several other candidate genes and regions previously identified for schizophrenia diagnosis. Of the 108 regions previously identified for schizophrenia diagnosis by the Psychiatric Genomics Consortium Schizophrenia Working Group, 39 were associated with 1 or more COGS endophenotypes, as shown in Table 2.2 In addition, as shown in Figure 2, most genes in the previously reported gene network defined through linkage and association studies of the COGS-1 families were at least nominally significant (P < .01) in the independent COGS-2 GWAS of patients with schizophrenia and HCPs, including all 8 genes that were found to be pleiotropic across multiple endophenotypes.17
Table 2. Regions of Association Overlap Between the COGS Endophenotypes and Schizophrenia Diagnosisa.
| PGC Region | Peak PGC P Value | Gene | Endophenotype Association |
|---|---|---|---|
| Chromosome 1: 2372401 to 2402501 |
8.7 × 10−10 | PLCH2b | PPI (rs114330159, P = 2.5 × 10−5)c; LNS (rs12744097, P = 3.0 × 10−5) |
| Chromosome 1: 97792625 to 98559084 |
3.4 × 10−19 | DPYD,b MIR137 | FMEM (rs1520668, P = 7.4 × 10−6)d; EMO (rs78891607, P = 5.2 × 10−5) |
| Chromosome 1: 243503719 to 244002945 |
3.7 × 10−9 | AKT3,e SDCCAG8 | AS (rs55777167, P = 3.2 × 10−6)d |
| Chromosome 2: 185601420 to 185785420 |
1.5 × 10−12 | ZNF804Ab | AS (rs151076669, P = 5.2 × 10−5) |
| Chromosome 2: 225334096 to 225467796 |
1.1 × 10−8 | CUL3b | SM (rs140109470, P = 5.5 × 10−7) |
| Chromosome 3: 2532786 to 2561686 |
2.7 × 10−11 | CNTN4b | SM (rs11129200, P = 2.0 × 10−6); PPI (rs146541530, P = 9.1 × 10−5)b |
| Chromosome 3: 52541105 to 52903405 |
4.3 × 10−11 | NISCH, STAB1, NT5DC2, SMIM4, PBRM1,e GNL3,e GLT8D1,e SPCS,e NEK4,b ITIH1,e ITIH3,e ITIH4,e MUSTN1,e TMEM110e | EMO (rs34184137, P = 4.0 × 10−5) |
| Chromosome 4: 103146888 to 103198090 |
8.0 × 10−15 | SLC39A8b | CVLT (rs192068, P = 9.8 × 10−5) |
| Chromosome 4: 176851001 to 176875801 |
9.5 × 10−9 | GPM6Ab | SM (rs114201782, P = 6.9 × 10−7)b |
| Chromosome 5: 45291475 to 45393775 |
5.1 × 10−9 | HCN1e | SMEM (rs187763903, P = 8.3 × 10−8); SM (rs73917002, P = 4.0 × 10−5) |
| Chromosome 5: 88581331 to 88854331 |
4.6 × 10−9 | MEF2Cc,e | SPA (rs114097435, P = 4.7 × 10−5)b |
| Chromosome 5: 137598121 to 137948092 |
4.7 × 10−9 | GFRA3, CDC25C, FAM53C, KDM3B,e REEP2,e EGR1,e ETF1,b HSPA9,e CTNNA1 | CVLT (rs154067, P = 4.3 × 10−6) |
| Chromosome 5: 140023664 to 140222664 |
4.9 × 10−8 | CD14,b TMCO6,e NDUFA2,e IK,e WDR55,e DND1,e HARS,e HARS2,e ZMAT2,e PCDHA1-10 | SPA (rs12659563, P = 7.8 × 10−5) |
| Chromosome 5: 151941104 to 152797656 |
1.1 × 10−10 | GRIA1c,e | FMEM (rs177521, 4.5 × 10−5) |
| Chromosome 6: 73132701 to 73171901 |
2.7 × 10−8 | RIMS1b | AS (rs10665509, P = 8.0 × 10−5) |
| Chromosome 6: 84279922 to 84407274 |
8.2 × 10−10 | SNAP91b | FMEM (rs1171121, P = 9.4 × 10−6); SMEM (rs583637, P = 2.9 × 10−5) |
| Chromosome 6: 96459651 to 96459651 |
1.6 × 10−9 | FUT9b | SM (rs4240598, P = 9.6 × 10−6); EMO (rs9399876, P = 5.0 × 10−5) |
| Chromosome 7: 1896096 to 2190096 |
8.2 × 10−15 | MAD1L1b | AS (rs6969832, P = 1.5 × 10−5) |
| Chromosome 7: 24619494 to 24832094 |
2.9 × 10−8 | MPP6, DFNA5, OSBPL3b | SMEM (rs2391082, P = 1.9 × 10−5) |
| Chromosome 7: 110034393 to 111205915 |
3.0 × 10−13 | IMMP2Lb,f | FMEM (rs35648000, P = 7.4 × 10−5) |
| Chromosome 8: 4177794 to 4192544 |
1.1 × 10−8 | CSMD1b | CVLT (rs2688282, P = 8.2 × 10−5)d; ABF (rs17066292, P = 3.6 × 10−5); SMEM (rs72507671, P = 6.1 × 10−6); EMO (rs10081576, P = 7.9 × 10−5) |
| Chromosome 8: 89340626 to 89753626 |
1.2 × 10−8 | MMP16e | LNS (rs78075875, P = 8.1 × 10−5) |
| Chromosome 9: 84630941 to 84813641 |
3.6 × 10−9 | TLE1b | SM (rs149799159, P = 5.3 × 10−5) |
| Chromosome 10: 18681005 to 18770105 |
2.0 × 10−12 | CACNB2e | AS (rs11012693, P = 5.0 × 10−6) |
| Chromosome 11: 46342943 to 46751213 |
1.3 × 10−11 | CREB3L1,e DGKZ,e MDK,e CHRM4,b AMBRA1,e HARBI1, ATG13, ARHGAP1 ZNF408, F2, CKAP5 | SMEM (rs61882689, P = 8.3 × 10−5) |
| Chromosome 11: 133808069 to 133852969 |
3.9 × 10−11 | IGSF9Be | FMEM (rs117439990, P = 6.9 × 10−5) |
| Chromosome 12: 2321860 to 2523731 |
3.2 × 10−18 | CACNA1Cb | SM (rs7956107, P = 1.9 × 10−6) |
| Chromosome 12: 92243186 to 92258286 |
4.6 × 10−8 | C12orf79b,f | SM (rs78503948, P = 9.3 × 10−5)d |
| Chromosome 12: 103559855 to 103616655 |
4.8 × 10−8 | C12orf42e | LNS (rs11838301, P = 9.3 × 10−5) |
| Chromosome 14: 30189985 to 30190316 |
1.4 × 10−8 | PRKD1b | CVLT (rs45525431, 6.3 × 10−6); SPA (rs138940224, P = 9.1 × 10−6) |
| Chromosome 14: 72417326 to 72450526 |
4.9 × 10−9 | RGS6b | ABF (rs2108709, P = 1.8 × 10−5) |
| Chromosome 15: 70573672 to 70628872 |
1.8 × 10−8 | TLE3e | AS (rs35122286, P = 5.8 × 10−5) |
| Chromosome 15: 78803032 to 78926732 |
2.4 × 10−13 | IREB2,b AGPHD1,e PSMA4,e CHRNA5,e CHRNA3, CHRNB4 | EMO (rs117028191, P = 5.6 × 10−6) |
| Chromosome 16: 9875519 to 9970219 |
1.3 × 10−18 | GRIN2Ab | AS (rs4782090, P = 5.3 × 10−5) |
| Chromosome 16: 29924377 to 30144877 |
4.6 × 10−11 | SEZ6L2,e ASPHD1,e KCTD13,e TMEM219,e TAOK2,b HIRIP3,e INO80E,e DOC2A,e C16orf92,e FAM57B,e ALDOA,e PPP4C,e TBX6,e YPEL3, GDPD3, MAPK3 | SM (rs10445105, P = 9.1 × 10−5) |
| Chromosome 19: 19374022 to 19658022 |
3.6 × 10−10 | NCAN,e HAPLN4,e TM6SF2,e SUGP1,b MAU2,e GATAD2A,e TSSK6, NDUFA13, CILP2, PBX4 | SPA (rs78844461, P = 9.9 × 10−5) |
| Chromosome 19: 50067499 to 50135399 |
4.7 × 10−8 | RCN3,b NOSIP,e PRRG2,e PRR12,e RRAS, SCAF1 | ABF (rs148832708, P = 1.0 × 10−5)d |
| Chromosome 20: 37361494 to 37485994 |
1.5 × 10−11 | SLC32A1,e ACTR5,b PPP1R16Be | PPI (rs111764110, P = 8.5 × 10−5); SPA (chromosome 20: 37254033, P = 5.3 × 10−5)g |
| Chromosome 22: 42315744 to 42689414 |
1.7 × 10−9 | SREBF2, TNFRSF13C, SHISA8, CENPM, SEPT3,e WBP2NL,e NAGA,e FAM109B,e SMDT1,e NDUFA6,e CYP2D6,e TCF20b | AS (rs11703217, P = 3.0 × 10−6) |
Abbreviations: ABF, abstraction and mental flexibility; AS, antisacccade task; COGS, Consortium on the Genetics of Schizophrenia; CVLT, California Verbal Learning Test; EMO, emotion recognition; FMEM, face memory; LNS, Letter-Number Span; PGC, Psychiatric Genomics Consortium; PPI, prepulse inhibition of the startle response; SM, sensorimotor dexterity; SMEM, spatial memory; SNP, single-nucleotide polymorphism; SPA, spatial processing.
For a subset of the 108 regions associated with schizophrenia diagnosis in the the most current data set from the Psychiatric Genomics Consortium Schizophrenia Work Group, the peak P value and regional coordinates are indicated as originally reported, along with all genes within 100 kb, arranged according to genomic position.2 For each COGS endophenotype meeting the enrichment threshold of P < 1 × 10−4, the lead SNP and corresponding P value are listed.
Indicates gene within 20 kb of a COGS lead SNP.
The association is within 100 kb of the gene.
Association of the indicated endophenotype to this region derives from a single SNP with P < 1 × 10−4.
Indicates gene within 100 kb of a COGS lead SNP.
PGC-SCZ2–associated region lists the nearest gene within 500 kb although the endophenotype association is more proximate.
The association is within 100 kb of the ACTR5 gene.
Figure 2. Interaction Network of Genes Identified for the 11 Endophenotypes Through Association and Linkage Analysis of the Consortium on the Genetics of Schizophrenia (COGS) Samples.
Genes are represented as nodes with solid lines representing direct protein-protein interactions and dashed arrows representing more indirect effects on expression, activation, or inhibition. Genes and interactions in black revealed association (blue box) or linkage (red box) with 1 or more endophenotypes, whereas those in gray represent other interacting genes. Those with pleiotropic associations to at least 4 endophenotypes in the COGS families are shown in bold. Genes with additional evidence of association in the genome-wide association study (GWAS) of the independent cases with schizophrenia and healthy comparison participants are indicated in green, with a solid box representing genes meeting the enrichment threshold of P < 1 × 10−4 and a dashed box representing nominal significance (P < .01). C indicates genes identified as associated with a schizophrenia diagnosis through GWAS of common variants; R, those identified by studies of rare and de novo variation. CTNNA2, CSMD1, and ZNF804A were prominent genes in these analyses with external support from studies of schizophrenia diagnosis but do not directly connect into the defined network.
Discussion
These initial GWAS of neurophysiological and neurocognitive deficits in schizophrenia have identified promising associations for multiple genomic regions and genes involved in critical neurodevelopmental processes. At an enrichment threshold, we found support for the gene network identified through candidate gene association and linkage studies of the COGS-1 families.17,18,19 In addition to implicating glutamate dysfunction in schizophrenia, many of the genes in this network and those newly identified through GWAS of these endophenotypes form part of the DISC1 (OMIM 605210) regulome for which rare, disruptive variants have been associated with schizophrenia and low cognitive ability in childhood.32 Such convergence at the pathway level may be more likely to be observed and more meaningful than concordance at the level of an individual gene or SNP in illuminating the underlying neurobiological dysfunction in schizophrenia. Finally, 39 of the genomic regions associated in this study of schizophrenia-related endophenotypes overlap with the 108 regions previously identified by the Psychiatric Genomics Consortium study of schizophrenia diagnosis and may be helpful in refining the association signal.2
By and large, the regions meeting genome-wide significance harbor genes that appear to play a role in key cellular and neurodevelopmental processes and have previously shown association with schizophrenia and related disorders, such as bipolar disorder, autism spectrum disorder, and intellectual dysfunction. For example, the chromosome 9q31 region found to be significantly associated with oculomotor antisaccade dysfunction has previously been implicated in a linkage analysis of psychotic bipolar disorder (log of odds, 3.55),33 and de novo chromosomal abnormalities in this region have been observed in patients with schizophrenia.34,35 The associated SNPs are located within a region containing several genes from the olfactory receptor family 13, subfamily C. Olfactory dysfunction has been observed in schizophrenia and may even serve as an early warning or prodromal sign of schizophrenia onset.36 The antisaccade task also produced a nearly significant association to chromosome 12p13 in the PHC1 gene, which functions in early neurodevelopment and is associated with microcephaly.37 M6PR, located 10 kb downstream of the lead SNP in this region, plays a critical role in lysosome function and is associated with neuronal ceroid lipofuscinosis, for which vision loss is an early sign.38,39 The Continuous Performance Test, a measure of vigilance, produced a significant peak on chromosome 5p15 near BASP1, which is involved in neuronal axon growth and guidance and has previously been identified in a GWAS of mood-incongruent psychotic symptoms in bipolar disorder.40 A long history of findings implicate neurodevelopmental axon formation dysfunction in schizophrenia,41,42,43 and bipolar disorder shows substantial clinical and genetic overlap with schizophrenia as part of the psychosis dimension.44,45 Face memory produced a significant peak on chromosome 3p21 in KIF15, which colocalizes in dendrites and growth cones of axons with microtubules and is differentially expressed in fetal vs mature astrocytes.46 KIF15 is also a target of microRNA miR-192-5p, which has been shown to be downregulated in schizophrenia.47
Spatial processing produced a significant association to 14q11 within the AJUBA gene, which encodes a scaffold protein that is expressed in the cerebellum, cortex, hippocampus, and retina. AJUBA interacts with SLC1A2 (OMIM 600300), a candidate gene for schizophrenia, and plays a role in the regulation of microRNA-mediated gene silencing.48 Spatial processing also produced a significant association to CCNE1 on 19q12, which is part of the DISC1 regulome and has been shown to be upregulated in schizophrenia.32,49 Sensorimotor dexterity produced a significant peak on chromosome 9q21 in C9orf135, which is associated with cognitive function.50 However, this association derived from a single SNP and requires further validation.
Abstraction and mental flexibility produced a significant peak on chromosome 10q23 near NRG3, which influences neuroblast proliferation, migration, and differentiation by signaling through ERBB4 (OMIM 600543). Many studies have implicated NRG3 as a susceptibility locus for schizophrenia.51,52,53,54 Specifically, genetic variants of NRG3 are associated with cognitive and psychotic symptom severity in schizophrenia and may result from differential prefrontal cortical expression of the gene.51,55,56 Finally, NRG3 knockout mice exhibit behaviors that are consistent with psychotic disorders in humans.57 These data are consistent with prior reports of association for NRG1 and ERBB4, suggesting that pathogenic variation in the ErbB-neuregulin pathway may underlie cognitive functioning and risk for psychosis.17,19,54,58,59,60
Spatial memory produced a nearly significant peak on 5p12 near HCN1, which is highly expressed in the frontal cortex and regulates neuronal excitability, rhythmic activity, and synaptic plasticity. Several lines of evidence in human and animal studies have suggested that hyperpolarization-activated cyclic nucleotide–gated (HCN) channels may play a role in psychiatric and neurological disorders. Common variation in HCN1 is associated with schizophrenia and educational attainment, and rare inherited and de novo variants are associated with a broad phenotypic spectrum that includes neonatal epileptic encephalopathy, generalized seizures, intellectual disability, and autistic spectrum disorders.2,61,62,63 Interestingly, spatial memory also produced an association with SHANK3 (OMIM 606230), an interacting partner of HCN1, mutations of which have similarly been shown to predispose to autism.64 Sensorimotor dexterity revealed an association with another HCN channel, HCN2 (OMIM 602781). Studies suggest that a reduction of HCN function through knockout of HCN1 or HCN2 in the dorsal hippocampus induces antidepressant behaviors in rodents, suggesting a potential treatment target for depression.65,66 Of particular relevance to the observed association in this study, studies of HCN1 knockouts have revealed a role for this gene in spatial learning and memory.67,68 An imbalance of HCN1 and HCN2 expression in the hippocampal CA1 area is also associated with impairments in spatial learning and memory associated with drug abuse.69
Strengths and Limitations
Although shared genetic substrates appear likely, this is not a study of schizophrenia but rather a study of neurophysiological and neurocognitive deficits that occur in the general population but are more pronounced in the context of schizophrenia and have implications for treatment.70 In addition, although our sample may appear modest by current case-control GWAS standards, it is suitable for the identification of genomic substrates underlying deficits in quantitative schizophrenia-related endophenotypes, which are labor intensive to acquire but provide increased efficiency compared with case-control designs.5,6 Future analyses of the complete COGS sample and other ongoing collaborations designed to increase power will provide opportunities to gain additional mechanistic insight into schizophrenia. GWAS of the COGS-1 families and the COGS-2 patients with schizophrenia and HCPs of African ancestry may allow us to replicate and extend these initial results as well as increase power for enrichment analyses. Sequencing of the COGS-1 families is pending completion to allow for an assessment of common vs rare variation in relation to family history, endophenotype deficit load, and clinical severity.71
The question of when and how to adjust for multiple tests remains challenging. In recent years, GWAS have converged toward use of P < 5 × 10−8 as a default threshold for genome-wide significance to facilitate comparisons across studies and avoid the alternative in which each study selects its own correction from a possible multitude of complex algorithms that increase opportunities for “p-hacking,” that is, choosing which analytical strategy to report based on the significance of the resulting P values. We have similarly used this threshold to account for the multiple testing of SNPs, although no correction was made for the multiple testing of endophenotypes, because each represents a distinct subject of scientific inquiry in numerous independent studies and publications. Although we believe the GWAS of each endophenotype has intrinsic interest of its own and could be published separately, we have opted to combine these results into a single report to allow for patterns of association to emerge. Also, as we previously demonstrated, P values are so highly variable that even under ideal circumstances, an initial P < 5 × 10−8 can vary from 2 × 10−16 to .007 in an identical replication study, even without considering multiple testing issues.72,73 This observation holds true for other thresholds, suggesting that any significance threshold is somewhat arbitrary and cannot eliminate the possibility of false-positive and/or false-negative findings.
Conclusions
Although some important considerations exist for interpretation of these findings, the present study has identified neurobiologically relevant genomic substrates for key neurophysiological and neurocognitive deficits in schizophrenia, reinforcing the goals of endophenotype-based projects.74,75 As many of the 11 endophenotypes have been endorsed as targets for the development of novel treatments for schizophrenia, a better understanding of the corresponding cellular and molecular processes may pave the way for precision-based medicine in schizophrenia and perhaps other psychiatric illnesses with a shared genetic liability.
eTable. Correlations Among All 11 Endophenotypes
eFigure. Quantile-Quantile Plots for Each of the 11 Endophenotypes
Summary of All Regions Identified Through Lead SNPs With P < 1 × 10−4
References
- 1.Sullivan PF. The genetics of schizophrenia. PLoS Med. 2005;2(7):e212. doi: 10.1371/journal.pmed.0020212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schizophrenia Working Group of the Psychiatric Genomics Consortium Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511(7510):421-427. doi: 10.1038/nature13595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sekar A, Bialas AR, de Rivera H, et al. ; Schizophrenia Working Group of the Psychiatric Genomics Consortium . Schizophrenia risk from complex variation of complement component 4. Nature. 2016;530(7589):177-183. doi: 10.1038/nature16549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braff DL. The importance of endophenotypes in schizophrenia research. Schizophr Res. 2015;163(1-3):1-8. doi: 10.1016/j.schres.2015.02.007 [DOI] [PubMed] [Google Scholar]
- 5.Blangero J, Williams JT, Almasy L. Novel family-based approaches to genetic risk in thrombosis. J Thromb Haemost. 2003;1(7):1391-1397. doi: 10.1046/j.1538-7836.2003.00310.x [DOI] [PubMed] [Google Scholar]
- 6.Lee SH, Wray NR. Novel genetic analysis for case-control genome-wide association studies: quantification of power and genomic prediction accuracy. PLoS One. 2013;8(8):e71494. doi: 10.1371/journal.pone.0071494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Swerdlow NR, Braff DL, Geyer MA. Sensorimotor gating of the startle reflex: what we said 25 years ago, what has happened since then, and what comes next. J Psychopharmacol. 2016;30(11):1072-1081. doi: 10.1177/0269881116661075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roalf DR, Ruparel K, Gur RE, et al. Neuroimaging predictors of cognitive performance across a standardized neurocognitive battery. Neuropsychology. 2014;28(2):161-176. doi: 10.1037/neu0000011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nuechterlein KH, Green MF, Kern RS, et al. The MATRICS Consensus Cognitive Battery, part 1: test selection, reliability, and validity. Am J Psychiatry. 2008;165(2):203-213. doi: 10.1176/appi.ajp.2007.07010042 [DOI] [PubMed] [Google Scholar]
- 10.Thomas ML, Bismark AW, Joshi YB, et al. Targeted cognitive training improves auditory and verbal outcomes among treatment refractory schizophrenia patients mandated to residential care. Schizophr Res. 2018;202:378-384. doi: 10.1016/j.schres.2018.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greenwood TA, Braff DL, Light GA, et al. Initial heritability analyses of endophenotypic measures for schizophrenia: the Consortium on the Genetics of Schizophrenia. Arch Gen Psychiatry. 2007;64(11):1242-1250. doi: 10.1001/archpsyc.64.11.1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gur RC, Braff DL, Calkins ME, et al. Neurocognitive performance in family-based and case-control studies of schizophrenia. Schizophr Res. 2015;163(1-3):17-23. doi: 10.1016/j.schres.2014.10.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee J, Green MF, Calkins ME, et al. Verbal working memory in schizophrenia from the Consortium on the Genetics of Schizophrenia (COGS) study: the moderating role of smoking status and antipsychotic medications. Schizophr Res. 2015;163(1-3):24-31. doi: 10.1016/j.schres.2014.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nuechterlein KH, Green MF, Calkins ME, et al. Attention/vigilance in schizophrenia: performance results from a large multi-site study of the Consortium on the Genetics of Schizophrenia (COGS). Schizophr Res. 2015;163(1-3):38-46. doi: 10.1016/j.schres.2015.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Radant AD, Millard SP, Braff DL, et al. Robust differences in antisaccade performance exist between COGS schizophrenia cases and controls regardless of recruitment strategies. Schizophr Res. 2015;163(1-3):47-52. doi: 10.1016/j.schres.2014.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stone WS, Mesholam-Gately RI, Braff DL, et al. California Verbal Learning Test-II performance in schizophrenia as a function of ascertainment strategy: comparing the first and second phases of the Consortium on the Genetics of Schizophrenia (COGS). Schizophr Res. 2015;163(1-3):32-37. doi: 10.1016/j.schres.2014.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greenwood TA, Lazzeroni LC, Murray SS, et al. Analysis of 94 candidate genes and 12 endophenotypes for schizophrenia from the Consortium on the Genetics of Schizophrenia. Am J Psychiatry. 2011;168(9):930-946. doi: 10.1176/appi.ajp.2011.10050723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greenwood TA, Swerdlow NR, Gur RE, et al. Genome-wide linkage analyses of 12 endophenotypes for schizophrenia from the Consortium on the Genetics of Schizophrenia. Am J Psychiatry. 2013;170(5):521-532. doi: 10.1176/appi.ajp.2012.12020186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greenwood TA, Lazzeroni LC, Calkins ME, et al. Genetic assessment of additional endophenotypes from the Consortium on the Genetics of Schizophrenia Family Study. Schizophr Res. 2016;170(1):30-40. doi: 10.1016/j.schres.2015.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walsh T, McClellan JM, McCarthy SE, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320(5875):539-543. doi: 10.1126/science.1155174 [DOI] [PubMed] [Google Scholar]
- 21.Kirov G, Pocklington AJ, Holmans P, et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol Psychiatry. 2012;17(2):142-153. doi: 10.1038/mp.2011.154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Calkins ME, Dobie DJ, Cadenhead KS, et al. The Consortium on the Genetics of Endophenotypes in Schizophrenia: model recruitment, assessment, and endophenotyping methods for a multisite collaboration. Schizophr Bull. 2007;33(1):33-48. doi: 10.1093/schbul/sbl044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.First MB, Spitzer RL, Gibbon M, Williams JB. Structured Clinical Interview for DSM-IV Axis I Disorders–Patient Edition (SCID-I/P, Version 2.0). New York: New York State Psychiatric Institute; 1995. [Google Scholar]
- 24.Turetsky BI, Calkins ME, Light GA, Olincy A, Radant AD, Swerdlow NR. Neurophysiological endophenotypes of schizophrenia: the viability of selected candidate measures. Schizophr Bull. 2007;33(1):69-94. doi: 10.1093/schbul/sbl060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gur RE, Calkins ME, Gur RC, et al. The Consortium on the Genetics of Schizophrenia: neurocognitive endophenotypes. Schizophr Bull. 2007;33(1):49-68. doi: 10.1093/schbul/sbl055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abecasis GR, Auton A, Brooks LD, et al. ; 1000 Genomes Project Consortium . An integrated map of genetic variation from 1092 human genomes. Nature. 2012;491(7422):56-65. doi: 10.1038/nature11632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jakobsson M, Scholz SW, Scheet P, et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451(7181):998-1003. doi: 10.1038/nature06742 [DOI] [PubMed] [Google Scholar]
- 28.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559-575. doi: 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190-2191. doi: 10.1093/bioinformatics/btq340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morrow JD, Cho MH, Platig J, et al. Ensemble genomic analysis in human lung tissue identifies novel genes for chronic obstructive pulmonary disease. Hum Genomics. 2018;12(1):1. doi: 10.1186/s40246-018-0132-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X, Tucker NR, Rizki G, et al. Discovery and validation of sub-threshold genome-wide association study loci using epigenomic signatures. Elife. 2016;5:5. doi: 10.7554/eLife.10557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teng S, Thomson PA, McCarthy S, et al. Rare disruptive variants in the DISC1 Interactome and Regulome: association with cognitive ability and schizophrenia. Mol Psychiatry. 2018;23(5):1270-1277. doi: 10.1038/mp.2017.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park N, Juo SH, Cheng R, et al. Linkage analysis of psychosis in bipolar pedigrees suggests novel putative loci for bipolar disorder and shared susceptibility with schizophrenia. Mol Psychiatry. 2004;9(12):1091-1099. doi: 10.1038/sj.mp.4001541 [DOI] [PubMed] [Google Scholar]
- 34.Inayama Y, Yoneda H, Fukushima K, Sakai J, Asaba H, Sakai T. Paracentric inversion of chromosome 9 with schizoaffective disorder. Clin Genet. 1997;51(1):69-70. doi: 10.1111/j.1399-0004.1997.tb02419.x [DOI] [PubMed] [Google Scholar]
- 35.Park JP, Moeschler JB, Berg SZ, Wurster-Hill DH. Schizophrenia and mental retardation in an adult male with a de novo interstitial deletion 9(q32q34.1). J Med Genet. 1991;28(4):282-283. doi: 10.1136/jmg.28.4.282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turetsky BI, Hahn CG, Borgmann-Winter K, Moberg PJ. Scents and nonsense: olfactory dysfunction in schizophrenia. Schizophr Bull. 2009;35(6):1117-1131. doi: 10.1093/schbul/sbp111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Awad S, Al-Dosari MS, Al-Yacoub N, et al. Mutation in PHC1 implicates chromatin remodeling in primary microcephaly pathogenesis. Hum Mol Genet. 2013;22(11):2200-2213. doi: 10.1093/hmg/ddt072 [DOI] [PubMed] [Google Scholar]
- 38.Hille-Rehfeld A. Mannose 6-phosphate receptors in sorting and transport of lysosomal enzymes. Biochim Biophys Acta. 1995;1241(2):177-194. doi: 10.1016/0304-4157(95)00004-B [DOI] [PubMed] [Google Scholar]
- 39.Ouseph MM, Kleinman ME, Wang QJ. Vision loss in juvenile neuronal ceroid lipofuscinosis (CLN3 disease). Ann N Y Acad Sci. 2016;1371(1):55-67. doi: 10.1111/nyas.12990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goes FS, Hamshere ML, Seifuddin F, et al. ; Bipolar Genome Study (BiGS) . Genome-wide association of mood-incongruent psychotic bipolar disorder. Transl Psychiatry. 2012;2:e180. doi: 10.1038/tp.2012.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumra S, Ashtari M, Cervellione KL, et al. White matter abnormalities in early-onset schizophrenia: a voxel-based diffusion tensor imaging study. J Am Acad Child Adolesc Psychiatry. 2005;44(9):934-941. doi: 10.1097/01.chi.0000170553.15798.94 [DOI] [PubMed] [Google Scholar]
- 42.Szeszko PR, Christian C, Macmaster F, et al. Gray matter structural alterations in psychotropic drug-naive pediatric obsessive-compulsive disorder: an optimized voxel-based morphometry study. Am J Psychiatry. 2008;165(10):1299-1307. doi: 10.1176/appi.ajp.2008.08010033 [DOI] [PubMed] [Google Scholar]
- 43.Yang JM, Shen CJ, Chen XJ, et al. erbb4 Deficits in chandelier cells of the medial prefrontal cortex confer cognitive dysfunctions: implications for schizophrenia. Cereb Cortex. 2018. doi: 10.1093/cercor/bhy316 [DOI] [PubMed] [Google Scholar]
- 44.Craddock N, Owen MJ. The beginning of the end for the Kraepelinian dichotomy. Br J Psychiatry. 2005;186:364-366. doi: 10.1192/bjp.186.5.364 [DOI] [PubMed] [Google Scholar]
- 45.Ivleva E, Thaker G, Tamminga CA. Comparing genes and phenomenology in the major psychoses: schizophrenia and bipolar 1 disorder. Schizophr Bull. 2008;34(4):734-742. doi: 10.1093/schbul/sbn051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y, Sloan SA, Clarke LE, et al. Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron. 2016;89(1):37-53. doi: 10.1016/j.neuron.2015.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vachev TI, Popov NT, Marchev D, Ivanov H, Stoyanova VK. Characterization of micro RNA signature in peripheral blood of schizophrenia patients using μParafloTM miRNA microarray assay. Int J Curr Microbiol Appl Sci. 2016;5(6):503-512. doi: 10.20546/ijcmas.2016.507.055 [DOI] [Google Scholar]
- 48.Marie H, Billups D, Bedford FK, et al. The amino terminus of the glial glutamate transporter GLT-1 interacts with the LIM protein Ajuba. Mol Cell Neurosci. 2002;19(2):152-164. doi: 10.1006/mcne.2001.1066 [DOI] [PubMed] [Google Scholar]
- 49.Sun L, Cheng Z, Zhang F, Xu Y. Gene expression profiling in peripheral blood mononuclear cells of early-onset schizophrenia. Genom Data. 2015;5:169-170. doi: 10.1016/j.gdata.2015.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seshadri S, DeStefano AL, Au R, et al. Genetic correlates of brain aging on MRI and cognitive test measures: a genome-wide association and linkage analysis in the Framingham study. BMC Med Genet. 2007;8(suppl 1):S15. doi: 10.1186/1471-2350-8-S1-S15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kao WT, Wang Y, Kleinman JE, et al. Common genetic variation in neuregulin 3 (NRG3) influences risk for schizophrenia and impacts NRG3 expression in human brain. Proc Natl Acad Sci U S A. 2010;107(35):15619-15624. doi: 10.1073/pnas.1005410107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen PL, Avramopoulos D, Lasseter VK, et al. Fine mapping on chromosome 10q22-q23 implicates neuregulin 3 in schizophrenia. Am J Hum Genet. 2009;84(1):21-34. doi: 10.1016/j.ajhg.2008.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morar B, Dragović M, Waters FA, Chandler D, Kalaydjieva L, Jablensky A. Neuregulin 3 (NRG3) as a susceptibility gene in a schizophrenia subtype with florid delusions and relatively spared cognition. Mol Psychiatry. 2011;16(8):860-866. doi: 10.1038/mp.2010.70 [DOI] [PubMed] [Google Scholar]
- 54.Benzel I, Bansal A, Browning BL, et al. Interactions among genes in the ErbB-neuregulin signalling network are associated with increased susceptibility to schizophrenia. Behav Brain Funct. 2007;3:31. doi: 10.1186/1744-9081-3-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tost H, Callicott JH, Rasetti R, et al. Effects of neuregulin 3 genotype on human prefrontal cortex physiology. J Neurosci. 2014;34(3):1051-1056. doi: 10.1523/JNEUROSCI.3496-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Paterson C, Law AJ. Transient overexposure of neuregulin 3 during early postnatal development impacts selective behaviors in adulthood. PLoS One. 2014;9(8):e104172. doi: 10.1371/journal.pone.0104172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hayes LN, Shevelkin A, Zeledon M, et al. Neuregulin 3 knockout mice exhibit behaviors consistent with psychotic disorders. Mol Neuropsychiatry. 2016;2(2):79-87. doi: 10.1159/000445836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stefansson H, Sigurdsson E, Steinthorsdottir V, et al. Neuregulin 1 and susceptibility to schizophrenia. Am J Hum Genet. 2002;71(4):877-892. doi: 10.1086/342734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hatzimanolis A, McGrath JA, Wang R, et al. Multiple variants aggregate in the neuregulin signaling pathway in a subset of schizophrenia patients. Transl Psychiatry. 2013;3:e264. doi: 10.1038/tp.2013.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Law AJ, Kleinman JE, Weinberger DR, Weickert CS. Disease-associated intronic variants in the ErbB4 gene are related to altered ErbB4 splice-variant expression in the brain in schizophrenia. Hum Mol Genet. 2007;16(2):129-141. doi: 10.1093/hmg/ddl449 [DOI] [PubMed] [Google Scholar]
- 61.Nava C, Dalle C, Rastetter A, et al. ; EuroEPINOMICS RES Consortium . De novo mutations in HCN1 cause early infantile epileptic encephalopathy. Nat Genet. 2014;46(6):640-645. doi: 10.1038/ng.2952 [DOI] [PubMed] [Google Scholar]
- 62.Okbay A, Beauchamp JP, Fontana MA, et al. ; LifeLines Cohort Study . Genome-wide association study identifies 74 loci associated with educational attainment. Nature. 2016;533(7604):539-542. doi: 10.1038/nature17671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marini C, Porro A, Rastetter A, et al. HCN1 mutation spectrum: from neonatal epileptic encephalopathy to benign generalized epilepsy and beyond. Brain. 2018;141(11):3160-3178. doi: 10.1093/brain/awy263 [DOI] [PubMed] [Google Scholar]
- 64.Yi F, Danko T, Botelho SC, et al. Autism-associated SHANK3 haploinsufficiency causes Ih channelopathy in human neurons. Science. 2016;352(6286):aaf2669. doi: 10.1126/science.aaf2669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim CS, Chang PY, Johnston D. Enhancement of dorsal hippocampal activity by knockdown of HCN1 channels leads to anxiolytic- and antidepressant-like behaviors. Neuron. 2012;75(3):503-516. doi: 10.1016/j.neuron.2012.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lewis AS, Vaidya SP, Blaiss CA, et al. Deletion of the hyperpolarization-activated cyclic nucleotide-gated channel auxiliary subunit TRIP8b impairs hippocampal Ih localization and function and promotes antidepressant behavior in mice. J Neurosci. 2011;31(20):7424-7440. doi: 10.1523/JNEUROSCI.0936-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nolan MF, Malleret G, Dudman JT, et al. A behavioral role for dendritic integration: HCN1 channels constrain spatial memory and plasticity at inputs to distal dendrites of CA1 pyramidal neurons. Cell. 2004;119(5):719-732. [DOI] [PubMed] [Google Scholar]
- 68.Thuault SJ, Malleret G, Constantinople CM, et al. Prefrontal cortex HCN1 channels enable intrinsic persistent neural firing and executive memory function. J Neurosci. 2013;33(34):13583-13599. doi: 10.1523/JNEUROSCI.2427-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou M, Luo P, Lu Y, et al. Imbalance of HCN1 and HCN2 expression in hippocampal CA1 area impairs spatial learning and memory in rats with chronic morphine exposure. Prog Neuropsychopharmacol Biol Psychiatry. 2015;56:207-214. doi: 10.1016/j.pnpbp.2014.09.010 [DOI] [PubMed] [Google Scholar]
- 70.Lencz T, Knowles E, Davies G, et al. Molecular genetic evidence for overlap between general cognitive ability and risk for schizophrenia: a report from the Cognitive Genomics Consortium (COGENT). Mol Psychiatry. 2014;19(2):168-174. doi: 10.1038/mp.2013.166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gulsuner S, Walsh T, Watts AC, et al. ; Consortium on the Genetics of Schizophrenia (COGS); PAARTNERS Study Group . Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell. 2013;154(3):518-529. doi: 10.1016/j.cell.2013.06.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lazzeroni LC, Lu Y, Belitskaya-Lévy I. P-values in genomics: apparent precision masks high uncertainty. Mol Psychiatry. 2014;19(12):1336-1340. doi: 10.1038/mp.2013.184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lazzeroni LC, Lu Y, Belitskaya-Lévy I. Solutions for quantifying P-value uncertainty and replication power. Nat Methods. 2016;13(2):107-108. doi: 10.1038/nmeth.3741 [DOI] [PubMed] [Google Scholar]
- 74.Calkins ME, Merikangas KR, Moore TM, et al. The Philadelphia Neurodevelopmental Cohort: constructing a deep phenotyping collaborative. J Child Psychol Psychiatry. 2015;56(12):1356-1369. doi: 10.1111/jcpp.12416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tamminga CA, Ivleva EI, Keshavan MS, et al. Clinical phenotypes of psychosis in the Bipolar-Schizophrenia Network on Intermediate Phenotypes (B-SNIP). Am J Psychiatry. 2013;170(11):1263-1274. doi: 10.1176/appi.ajp.2013.12101339 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eTable. Correlations Among All 11 Endophenotypes
eFigure. Quantile-Quantile Plots for Each of the 11 Endophenotypes
Summary of All Regions Identified Through Lead SNPs With P < 1 × 10−4