Key Points
Question
Can tape strips serve as a minimally invasive approach to assess biomarkers for early-onset pediatric atopic dermatitis?
Findings
In this cross-sectional study of 51 children younger than 5 years with and without atopic dermatitis, the use of tape strips, a minimally invasive approach for skin sampling, detected the cutaneous immune and barrier abnormalities of early-onset atopic dermatitis in infants and young children and defined biomarkers that are associated with disease severity, pruritus, and transepidermal water loss.
Meaning
Minimally invasive tape strips can be used to broadly characterize immune and epidermal barrier biomarkers of the lesional and nonlesional skin of children with early-onset pediatric atopic dermatitis, providing a useful, noninvasive approach for pediatric clinical trials and longitudinal studies.
Abstract
Importance
Molecular profiling of skin biopsies is the criterion standard for evaluating the cutaneous atopic dermatitis (AD) phenotype. However, skin biopsies are not always feasible in children. A reproducible minimally invasive approach that can track cutaneous disease in pediatric longitudinal studies or clinical trials is lacking.
Objective
To assess a minimally invasive approach using tape strips to identify skin biomarkers that may serve as a surrogate to biomarkers identified using whole-tissue biopsies.
Design, Setting, and Participants
This cross-sectional study of 51 children younger than 5 years recruited children with moderate to severe AD and children without AD from the dermatology outpatient clinics at a children’s hospital. Sixteen tape strips were serially collected from the nonlesional and lesional skin of 21 children who had AD and were less than 6 months from disease initiation and from the normal skin of 30 children who did not have AD between January 22, 2016, and April 20, 2018.
Main Outcomes and Measures
Gene and protein expression were evaluated using quantitative real-time polymerase chain reaction and immunohistochemistry.
Results
A total of 51 children younger than 5 years were included in the study; 21 children had moderate to severe AD with less than 6 months of disease duration, and 30 children did not have AD. Of the 21 children with AD, the mean (SD) age was 1.7 (1.7) years, and most were male (15 [71.4%] and white (15 [71.4%]). Of the 30 children without AD, the mean (SD) age was 1.8 (2.0) years, and most were female (20 [66.7%]) and white (22 [73.3%]). Seventy-seven of 79 evaluated immune and barrier gene products were detected (gene detection rate, 97%) in 70 of 71 tape strips (sample detection rate, 99%), with 53 of 79 markers differentiating between children with lesional and/or nonlesional AD from children without AD. Many cellular markers of T cells (CD3), AD-related dendritic cells (Fc ε RI and OX40 ligand receptors), and key inflammatory (matrix metallopeptidase 12), innate (interleukin 8 [IL-8] and IL-6), helper T cell 2 (TH2; IL-4, IL-13, and chemokines CCL17 and CCL26), and TH17/TH22 (IL-19, IL-36G, and S100A proteins) genes were significantly increased in lesional and nonlesional AD compared with tape strips from normal skin. For example, IL-4 mean (SE) for lesional was −15.2 (0.91) and normal was −19.5 (0.48); P < .001. Parallel decreases occurred in epidermal barrier gene products (FLG, CLDN23, and FA2H) and negative immune regulators (IL-34 and IL-37). For example, the decrease for FLG lesional was mean (SE) −2.9 (0.42) and for normal was 2.2 (0.45); P < .001. Associations were found between disease severity or transepidermal water loss and TH2 (IL-33 and IL-4R) and TH17/TH22 (IL-36G and S100As) products in lesional and nonlesional AD skin (evaluated using the SCORing Atopic Dermatitis, Eczema Area and Severity Index, and Pruritus Atopic Dermatitis Quickscore tools).
Conclusions and Relevance
In this study, tape strips provide a minimally invasive alternative for serially evaluating AD-associated cutaneous biomarkers and may prove useful for tracking pediatric AD therapeutic response and predicting future course and comorbidities.
This cross-sectional study examines whether tape strips might be used to detect immune and barrier abnormalities and to define biomarkers associated with atopic dermatitis in children younger than 5 years with early-onset atopic dermatitis.
Introduction
Atopic dermatitis (AD) has a growing therapeutic pipeline,1,2,3,4,5,6,7,8,9 largely owing to increased understanding of AD mechanisms in adults.2,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20 Previous studies identified treatment-response biomarkers, which are important for understanding molecular tissue responses and their association with clinical severity.6,15,16,18,20,21,22,23 However, AD onset usually occurs in children who are younger than 5 years.24,25,26 Recently, AD-associated biomarkers were identified by whole-skin profiling from children younger than 5 years 27,28 and adolescents.29 However, biopsies are not always practical in children, and much less invasive blood phenotyping cannot capture the complex AD skin phenotype.30,31,32,33,34,35 Minimally invasive approaches that accurately capture key immune and barrier biomarkers in the skin of patients with early-onset pediatric AD are needed.
Research has been directed toward identifying AD biomarkers using tape strips, a minimally invasive method that captures the stratum corneum.36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69 However, most tape-stripping studies have focused on adults with chronic AD,45,47,52,53,54,56,57,58,63,66,70,71 studying a limited panel of protein analytes, including antimicrobials (human β-defensin 4 [hBD4] and antimicrobial peptide cathelicidin LL-37), serine proteases (kallikrein-related peptidase 5 [KLK5] and KLK7),45,52,57,72 lipids,46 and inflammatory proteins (interleukin 4 [IL-4] and chemokine 17 [CCL17]).53,70
Two recent pediatric studies60,61 used proteomic immune assays to evaluate stratum corneum biomarkers in tape strips from the nonlesional skin of infants with moderate to severe AD compared with infants without AD61 and from the skin of children aged 0 to 12 years with mild to moderate AD compared with those without AD,60 detecting 19 of 2761 and 13 of 2860 of all evaluated inflammatory mediators, respectively. Although a few AD biomarkers, such as helper T cell 2 (TH2) CCL17 and CCL22, were upregulated in tape strips from AD skin compared with tape strips from normal skin, key AD biomarkers, including those recently reported to be significantly upregulated in pediatric AD skin biopsies compared with normal skin biopsies (IL-4, IL-13, and IL-5),27,34,35 were either not detected or showed lower protein expression in AD skin.61
Two studies performed transcriptomic RNA analyses using Ion AmpliSeq sequencing (ThermoFisher Scientific), primarily focusing on nonlesional skin.58,73 One study that involved adults with mild to severe AD and adults without AD identified 29 differentially expressed genes.58 Another study involved children and adolescents aged 8 to 16 years with mild to severe AD, some of whom had food allergies (FAs) and nonatopic dermatitis, reporting that those with AD and FAs had a greater number of barrier- and TH2-related abnormalities.73 However, these transcriptomic studies had detection rates of only 18% to 52% for normal skin, 26% to 60% for nonlesional skin, and 57% to 89% for lesional skin (total success rate, approximately 45%), rendering this approach questionable for therapeutic or longitudinal studies.58,73 A reproducible minimally invasive messenger RNA (mRNA)–based tape-strip profiling approach that defines early-onset pediatric AD biomarkers that can track cutaneous disease in pediatric longitudinal studies or clinical trials is lacking.
To evaluate whether tape strips accurately reflected AD activity in skin, we performed mRNA profiling on tape strips from the lesional and nonlesional skin of 21 children who had moderate to severe AD and were less than 6 months from disease initiation and 30 children without AD, all younger than 5 years. We used an expanded panel of 79 genes, including those previously associated with immune and barrier AD abnormalities8,9,27,33,34,35,74,75 and those detected in previous AD tape-strip studies.42,45,47,51,52,53,54,56,57,58,60,63,66,70,71
Methods
A total of 51 children younger than 5 years were enrolled in the study. Of those, 21 children had new-onset (disease duration <6 months) moderate to severe AD and 30 children did not have AD or a history of personal or family atopy (Table 1). Patients were recruited from the dermatology outpatient clinics at the Ann & Robert H. Lurie Children’s Hospital of Chicago between January 22, 2016, and April 20, 2018. Patients with active skin infections or who used systemic immunosuppressants within 4 weeks, topical steroids or immunomodulators within 1 week, or moisturizers within 12 hours before evaluation were excluded. Transepidermal water loss (TEWL) was measured, and AD severity was assessed using the SCORing Atopic Dermatitis (SCORAD), Eczema Area and Severity Index (EASI), and Pruritus Atopic Dermatitis Quickscore (ADQ) tools (Table 1). Filaggrin gene (FLG) loss-of-function mutations were not evaluated. This study was approved by the institutional review board of the Feinberg School of Medicine, Northwestern University (Chicago, Illinois), and all parents signed written consent forms that were approved by the institutional review board.
Table 1. Baseline Demographics and Clinical Characteristics.
| Characteristic | Children With AD (n = 21)a | Children Without AD (n = 30) | P Value |
|---|---|---|---|
| Age, mean (SD), y | 1.7 (1.7) | 1.8 (2.0) | .85 |
| Sex, No. (%) | |||
| Female | 6 (28.6) | 20 (66.7) | .02 |
| Male | 15 (71.4) | 10 (33.3) | |
| Race/ethnicity, No. (%) | |||
| Asian/Pacific Islander | 2 (9.5) | 6 (20.0) | .30 |
| African American | 4 (19.0) | 2 (6.7) | |
| White | 15 (71.4) | 22 (73.3) | |
| Clinical severity disease scores, mean (SD) | |||
| SCORAD | 54.4 (21.2) | NA | NA |
| EASI | 20.9 (12.6) | NA | NA |
| TEWL, lesional, g/h/m2 | 65.4 (49.2) | NA | NA |
| TEWL, nonlesional, g/h/m2 | 28.1 (18.1) | NA | NA |
| Pruritus ADQ | 16.6 (8.1) | NA | NA |
| Patient history | |||
| Age at onset of AD, mean (SD), mo | 2.3 (1.3) | NA | NA |
| History of atopy, No. (%) | 6 (28.6) | NA | NA |
| Family history of AD, No. (%) | 18 (87.7) | NA | NA |
Abbreviations: AD, atopic dermatitis; ADQ, Atopic Dermatitis Quickscore; EASI, Eczema Area and Severity Index; NA, not applicable; SCORAD, SCORing Atopic Dermatitis; TEWL, transepidermal water loss.
Normal value. Mean (SD) TEWL at age 2 months in children without AD has been reported as 10.97 (7.98) g/h/m2 and at age 6 months has been reported as 10.71 (7.1) g/h/m2.76
Among children with AD, 16 consecutive, large D-Squame tape strips (CuDerm Corp) were collected from the lesional skin of the antecubital fossa (ie, the triangular region in the forearm on the anterior surface of the elbow) when possible. Nonlesional skin was sampled from nearby skin on the same arm. Skin from children without AD was sampled from the same areas at the same times. Tape strips were consecutively labeled, and the first 2 tape strips were discarded (eMethods 1 in the Supplement).52,58,60,63,72,77
We extracted RNA for quantitative real-time polymerase chain reaction (qRT-PCR) analysis through the miRNeasy Mini Kit (Qiagen), and we used 500 pg total RNA (eMethods 2 in the Supplement). Preamplification was performed on all samples; 1 nonlesional sample was undetectable. TaqMan Low Density Array cards (ThermoFisher Scientific) were used for qRT-PCR, as reported.78,79 Cycle threshold (Ct) values were normalized to the housekeeping gene RPLP0 by negatively transforming the Ct values to −dCt. Undetected expression values for each gene were estimated as 20% of the minimum expression across all samples. Primers are listed in eTable 1 in the Supplement.
Immunohistochemistry was performed on frozen tissue sections from children with early pediatric AD (lesional and nonlesional) and children without AD (n = 3 for each group) using purified mouse and rabbit antihuman monoclonal antibodies (eTable 2 in the Supplement).
Statistical Analysis
Statistical analyses were performed using R software, version 3.6.1 (R Foundation for Statistical Analysis), and a linear mixed-effects model was used to analyze log2 qRT-PCR data. Mean expressions of all markers were summarized in a heat map, in which unsupervised clustering was performed using euclidean distance and average agglomeration criteria. Spearman correlation coefficients were used to evaluate the association between inflammatory markers and disease severity, with P < .05 considered significant. To evaluate the performance of an AD classifier, we used the receiver operating characteristic area under the curve (AUC); details are provided in eMethods 3 in the Supplement.
Results
Study Participants
Tape strips were collected from a total of 51 children younger than 5 years; 21 children had moderate to severe AD (SCORAD mean, 54.4) and a disease duration of less than 6 months, and 30 children did not have AD. Of the 21 children with AD, the mean (SD) age was 1.7 (1.7) years, and most were male (15 [71.4%]) and white (15 [71.4%]). Of the 30 children without AD, the mean (SD) age was 1.8 (2.0) years, and most were female (20 [66.7%]) and white (22 [73.3%]). Tape stripping was well tolerated without clinical sequelae. An analysis of a large panel of 79 immune and barrier mediators, including key AD biomarkers2,21,80,81 in lesional and nonlesional AD skin and normal skin, had a 99% success rate for samples (mRNA was undetectable in only 1 of 71 samples).
Cellular AD Biomarkers
To understand whether tape strips could assess the cellular profile of early AD skin, we analyzed a panel of 15 cellular markers.58,62,65 We detected markers of monocytes and macrophages,10,82,83 T cells, activated TH2 cells, dendritic cells and dendritic-cell subsets,84,85 and Langerhans cells (langerin protein; Figure 1 and eFigure 1). Most cellular markers, with the exception of CD83 and CD11c, showed significant differences between lesional AD skin and normal skin. Some markers, such as the OX40 ligand (OX40L) receptor (associated with atopic dendritic cells),86,87 the inducible T-cell costimulatory activation marker (ICOS), CD209, CD123, and langerin protein, were also significantly increased in nonlesional AD skin (Figure 1; eFigure 1 and eTable 4 in the Supplement).58,60,61 Only colony-stimulating factor 1 (CSF1) and CSF2 showed significant differences between lesional and nonlesional AD skin.
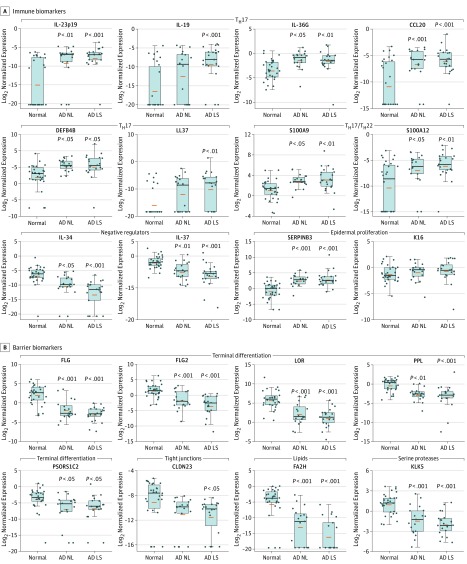
Figure 1. AD Biomarkers Detected in Tape Strips.
A, Cellular biomarkers. B, Immune biomarkers. A set of cellular biomarkers and immune biomarkers were validated and detected using tape strips in normal skin, atopic dermatitis (AD) nonlesional (NL) skin, and AD lesional skin (LS). CSF indicates colony-stimulating factor; ICOS, inducible T-cell costimulator. Boxes depict interquartile range. Whiskers depict the minimum and maximum intervals; the blue dots represent individual samples; orange bars represent means; horizontal black lines represent medians. P values denote significance of AD nonlesional or AD lesional versus normal skin.
aP < .05; significance between AD lesional versus nonlesional skin.
Immune Activation
We next evaluated the mRNA expression of 64 immune and barrier markers previously associated with AD in skin biopsies.10,11,12 Many of these markers were below the detection level of microarrays or whole transcriptome shotgun sequencing (RNA-Seq),10,11,58,61 and many were not detected in previous tape-strip studies (eTable 3 in the Supplement).53,58,60,61
The evaluation of immune biomarkers included general inflammation (matrix metallopeptidase 12 [MMP12]), epidermal proliferation, innate immunity, TH2-associated, TH1 natural killer T-cell (NKT) activation cytokine, TH1-associated, TH17-induced, TH17/TH22-associated, regulatory T (Treg) cell, and negative regulator markers (Figure 1 and Figure 2; eFigure 1, eTable 2, and eTable 4 in the Supplement). Overall key innate immune (IL-8 and IL-6), TH2-related (IL-13, IL-4, CCL17, and CCL26), and TH17/TH22 (IL-23p19, IL-19, IL-36G, CCL20, β-defensin 4 [DEFB4], cathelicidin LL-37, and S100A proteins [S100As]) measures were significantly upregulated, whereas negative regulators were significantly downregulated across AD vs normal skin. Similar to our recent reports regarding early-onset pediatric AD whole-skin biopsies, and unlike the skin from adults with chronic AD,27,35 TH1-associated mRNAs (with the exception of CCL2 and the STAT 1 signaling pathway) were not significantly increased in tape strips from children with early-onset AD (Figure 1; eFigure 1 and eFigure 2 in the Supplement).
Figure 2. Key Immune and Barrier AD Biomarkers Detected in Tape Strips.
A, Immune biomarkers. Epidermis-derived barrier biomarkers show differential expression across atopic dermatitis (AD) skin vs normal skin. B, Barrier biomarkers. Immune biomarkers show greater differential expression across AD skin vs normal skin in tape strips in normal skin, AD nonlesional (NL) skin, and AD lesional skin (LS). Boxes depict interquartile range. Whiskers depict the minimum and maximum intervals; the blue dots represent individual samples; orange bars represent means; horizontal black lines represent medians. P values denote significance of AD nonlesional or AD lesional versus normal skin.
Barrier Biomarkers
We also evaluated markers of epidermal differentiation, tight junctions, lipids, and serine proteases. Several epidermal differentiation markers showed significantly reduced expression in both lesional and nonlesional AD tape strips vs normal tape strips. These included FLG, FLG2, loricrin (LOR), periplakin (PPL), and psoriasis susceptibility 1 candidate 2 (PSORS1C2; Figure 2 and eTable 4 in the Supplement).
As in our early-onset pediatric AD biopsies,27,28 tape-strip mRNA expression of some lipid (fatty acid 2-hydroxylase [FA2H] and fatty acyl-CoA reductase 2 [FAR2]) and tight junction (CLDN8 and CLDN23) products was downregulated in both lesional and nonlesional AD skin compared with normal skin (Figure 2 and eFigure 2 in the Supplement). Other lipid markers showed no differential expression in tape strips (eFigure 2). The serine proteases KLK5 and KLK7 were significantly downregulated in AD vs normal skin (Figure 2; eTable 4 and eFigure 2 in the Supplement). Only 2 markers (CCL11 and ELOVL fatty acid elongase 3 [ELOVL3]) were undetectable.
A summary heat map of all 79 immune and barrier measures depicts mRNA expression differences (as fold changes) among lesional and nonlesional AD tape-stripped skin samples compared with normal tape-stripped skin samples (Figure 3 and eTable 4 in the Supplement). Significantly downregulated products in AD skin include barrier measures (ie, LOR, PPL, FLG, CLDN23, and FA2H) and negative regulators (IL-34 and IL-37). Markers significantly upregulated in AD skin include multiple immune genes representing dendritic cells (CD11b [ITGAM] and OX40L), T-cell activation (ICOS), TH2 (CCL17, IL-4, and IL-13), and TH17/TH22 (IL-19, IL-23p19, IL36G, S100As, and CCL20) (Figure 3). Means, P values, SEs, and CIs are summarized in eTable 4 in the Supplement.
Figure 3. Heat Map of Immune and Barrier Atopic Dermatitis (AD) Biomarkers Detected in Tape Strips .
Mean expression levels of all 79 barrier and inflammatory mediators detected using tape strips (measured by real-time polymerase chain reaction). The table shows biomarkers with signed fold changes in nonlesional (NL) AD skin vs normal (N) skin, lesional (LS) AD skin vs normal skin, and LS AD skin vs NL AD skin. The yellow box indicates the significant downregulation of biomarkers in LS and NL AD skin compared with normal skin; the green box indicates the significant upregulation of biomarkers in LS and NL AD skin compared with normal skin.
aP <.05.
bP < .01.
cP < .001.
Because older children and adolescents with AD and FAs were reported to have differences in tape strips compared with those with AD and no FAs, we conducted a sensitivity subanalysis of 6 children with FAs compared with 15 children without FAs to evaluate whether the coexistence of atopic comorbidities was associated with changes in the skin phenotype. Data were similar between children with AD with and without other atopic manifestations. A sensitivity subanalysis evaluating sex as a confounder found no sex-related differences.
Protein Validation of Selected Mediators
We further validated the protein expression of dendritic cell (CD11b) and epithelial cytokine (IL-33 and IL-17C) markers in tissue sections from children aged 0 to 5 years with moderate to severe pediatric AD and those without AD using immunohistochemistry.27,28 Although most CD11b+ dendritic cells are located in the dermis, we detected foci of cellular infiltrates in the outer epidermis, primarily in AD lesions (eFigure 3 in the Supplement). Immunostaining for epidermal cytokines (IL-33 and IL-17C)28,88,89 showed diffuse, more intense epidermal staining in both lesional and nonlesional AD skin, with fainter staining that was more localized to the lower epidermis in normal skin (n = 3 for all tissues; eFigure 3 in the Supplement).
Tape-Strip Biomarkers and Clinical Disease
To determine how molecular and cellular tape-strip biomarkers from early-onset pediatric AD lesional and nonlesional skin correlate with clinical severity (based on SCORAD, EASI, and Pruritus ADQ assessments) and epidermal barrier function (based on TEWL assessment), we performed a Spearman correlation coefficient analysis (Table 2 and eFigure 4 in the Supplement). The SCORAD assessment showed the greatest number of significant correlations with lesional biomarkers. Significant correlations with SCORAD and EASI assessments were noted with the expression of lesional TH2 (IL-33) and TH17 (IL-23p19) cytokines (Table 2 and eTable 4 in the Supplement). Key TH17/TH22 (IL-19, S100As, PI3, IL-36G, β-defensin 4B [DEFB4B], STAT 3, and cathelicidin LL-37; eFigure 4 in the Supplement), innate (IL-17C), hyperplasia (epidermal proliferation marker K16), TH2 (IL-4R), and cellular (CD11b and CD11c) biomarkers in lesional skin were also significantly associated with disease severity (Table 2 and eTable 4 in the Supplement). Significant correlations were found between pruritus and lesional AD markers (cellular markers: CD209, CD11b, CSF2, and CD3; TH2–associated markers: CCR4, CCL18, IL-10, and IL-13; and TH1-associated markers: immune interferon [IFNγ] and chemokines CXCL9 and CXCL11). Transepidermal water loss was positively associated with several immune markers (TH1, IFNγ, TH17/TH22, IL-19, S100A7, TH2, and CCL26) and negatively associated with LOR (Table 2).
Table 2. Spearman Correlations of Atopic Dermatitis (AD) Biomarkers.
| Correlation Rank | Assessment Tool | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SCORAD | EASI | Pruritus ADQ | TEWL | |||||||||
| Marker | ρ | P Value | Marker | ρ | P Value | Marker | ρ | P Value | Marker | ρ | P Value | |
| Correlations With Clinical Indices of AD in Pediatric Lesional Skin Using Tape Strips | ||||||||||||
| 1 | EASI | 0.784 | <.001 | SCORAD | 0.784 | <.001 | CD209 | 0.814 | <.001 | IFNγ | 0.597 | .02 |
| 2 | SERPINB3 | 0.676 | .003 | IL-33 | 0.498 | .04 | CCR4 | 0.670 | .01 | IL-19 | 0.570 | .03 |
| 3 | IL-33 | 0.651 | .005 | IL-23p19 | 0.494 | .04 | LL37 | 0.635 | .02 | S100A7 | 0.565 | .04 |
| 4 | IL-36G | 0.608 | .01 | Pruritus ADQ | 0.482 | .08 | CD11b | 0.579 | .03 | Pruritus ADQ | 0.564 | .10 |
| 5 | CD11b | 0.594 | .01 | CCR4 | 0.475 | .047 | CCL18 | 0.572 | .03 | ELOVL3 | 0.563 | .04 |
| 6 | IL-4R | 0.594 | .01 | K16 | 0.455 | .06 | IL-10 | 0.570 | .03 | IL-33 | 0.469 | .09 |
| 7 | S100A9 | 0.591 | .01 | NA | NA | NA | IFNγ | 0.564 | .04 | CCL26 | 0.453 | .10 |
| 8 | S100A8 | 0.578 | .02 | DEFB4B | 0.552 | .04 | FLG | −0.495 | .08 | |||
| 9 | DEFB4B | 0.571 | .02 | CSF2 | 0.542 | .045 | LOR | −0.543 | .048 | |||
| 10 | PI3 | 0.564 | .02 | CXCL9 | 0.531 | .05 | NA | NA | NA | |||
| 11 | K16 | 0.564 | .02 | CXCL11 | 0.530 | .05 | ||||||
| 12 | IL-23p19 | 0.548 | .02 | CD3 | 0.526 | .05 | ||||||
| 13 | STAT3 | 0.531 | .03 | IL-13 | 0.524 | .05 | ||||||
| 14 | ELOVL5 | 0.531 | .03 | |||||||||
| 15 | S100A12 | 0.527 | .03 | |||||||||
| 16 | IL-17C | 0.521 | .03 | |||||||||
| 17 | KLK7 | 0.513 | .04 | |||||||||
| 18 | CD11c | 0.484 | .049 | |||||||||
| 19 | LL37 | 0.478 | .05 | |||||||||
| 20 | IL-19 | 0.473 | .06 | |||||||||
| Correlations With Clinical Indices of AD in Pediatric Nonlesional Skin Using Tape Strips | ||||||||||||
| 1 | EASI | 0.774 | .001 | SCORAD | 0.774 | .001 | CXCL9 | 0.797 | .002 | DGAT2 | 0.629 | .03 |
| 2 | IL-23p19 | 0.643 | .01 | IL-17C | 0.642 | .01 | IL-19 | 0.709 | .01 | S100A7 | 0.622 | .04 |
| 3 | FAR2 | 0.633 | .01 | PI3 | 0.632 | .01 | CXCL10 | 0.616 | .03 | SERPINB3 | 0.552 | .07 |
| 4 | S100A12 | 0.626 | .01 | DEFB4B | 0.629 | .01 | IL-12/IL-23p40 | 0.586 | .045 | IL-4R | 0.545 | .07 |
| 5 | IL-17C | 0.597 | .02 | S100A9 | 0.609 | .01 | EASI | 0.567 | .05 | S100A8 | 0.524 | .08 |
| 6 | IL-36G | 0.584 | .02 | IL-36G | 0.606 | .02 | IL-2 | 0.555 | .06 | K16 | 0.524 | .08 |
| 7 | PI3 | 0.579 | .02 | SERPINB3 | 0.594 | .02 | PI3 | 0.550 | .06 | IL-1RA | 0.524 | .08 |
| 8 | DEFB4B | 0.552 | .03 | Pruritus ADQ | 0.567 | .06 | LL37 | 0.519 | .08 | CD11c | 0.517 | .09 |
| 9 | IL-19 | 0.471 | .08 | K16 | 0.556 | .03 | S100A9 | 0.511 | .09 | FcεRI | 0.503 | .10 |
| 10 | SERPINB3 | 0.427 | .11 | FAR2 | 0.550 | .03 | IL-37 | −0.494 | .10 | SCD | −0.609 | .04 |
| 11 | NA | NA | NA | S100A12 | 0.550 | .03 | FLG2 | −0.529 | .08 | NA | NA | NA |
| 12 | S100A8 | 0.526 | .04 | CCL22 | −0.539 | .07 | ||||||
| 13 | IL-19 | 0.513 | .04 | FLG | −0.620 | .03 | ||||||
| 14 | S100A7 | 0.500 | .05 | LOR | −0.683 | .01 | ||||||
| 15 | IL-23p19 | 0.487 | .06 | NA | NA | NA | ||||||
Abbreviations: ADQ, Atopic Dermatitis Quickscore; EASI, Eczema Area and Severity Index; SCORAD, SCORing Atopic Dermatitis; TEWL, transepidermal water loss; NA, not applicable.
Both SCORAD and EASI measurements were correlated with biomarkers in nonlesional AD skin, including innate (IL-17C), TH17/TH22 (IL-23p19, IL-36G, DEFB4B, phosphatidylinositol 3 [PI3], and S100A12; eFigure 4 in the Supplement), and lipid (FAR2) measures (Table 2 and eTable 4 in the Supplement). Additional TH17/TH22 (IL-19, S100A8, and S100A9) and epidermal hyperplasia/proliferation (K16 and serine protease inhibitor B3 [SERPINB3]) biomarkers were positively correlated with EASI measurements (eTable 4 in the Supplement). Pruritus ADQ showed significant positive correlations with TH1 NKT–associated (CXCL9 and CXCL10), and TH17-related (IL-12, IL-23p40, and IL-19) mediators, and significant negative correlations with differentiation markers (FLG and LOR). Transepidermal water loss was significantly positively associated with TH17/TH22, S100A7, and diglyceride acyltransferase 2 (DGAT2; Table 2).
Finally, we evaluated whether any profiled biomarker in tape strips could accurately discriminate between pediatric AD skin and normal skin. The epidermal negative regulator cytokine IL-34 was the best single-gene classifier, discriminating AD from normal skin with almost 100% accuracy (AUC, 0.94). Epidermal barrier markers (FLG, FLG2, LOR, and FA2H) were also effective discriminators (AUC, 0.93-0.90; eFigure 5 in the Supplement). Integration of the top differentially expressed immune and barrier markers (defined as markers with AUC values greater than 0.65) resulted in a combined score that discriminated AD skin from normal skin with nearly 100% accuracy (AUC, 1). The predictor included key AD biomarkers that contributed positively (CSF1, MMP12, CCL26, CCL20, IL-13, S100A7, and S100A9) as well as negatively (IL-34 and barrier genes, FLG, FLG2, LOR, PSORS1C2, FA2H, and FAR2; eFigure 5 in the Supplement) to the score. Only lesional biomarkers were able to discriminate between AD and normal skin.
Discussion
To our knowledge, this is the first study to provide a broad characterization of immune and barrier abnormalities in the skin of children with early-onset (aged <5 years) AD skin using a minimally invasive tape-strip approach. Because tissue biopsies are considered the criterion standard for evaluating dysregulation in AD lesional and nonlesional skin,2,6,7,8,9,10,12,16,17,18,20,27,28,29,78,86,90,91,92 it is crucial to understand whether tape-strip profiling can accurately yield key AD-related biomarkers. While this minimally invasive approach can be useful across AD endotypes,8,9,14,21,27,28,29,58,78,90 it is particularly important for studying pediatric AD skin owing to the impracticality of performing biopsies in children. By defining a minimally invasive method for biomarker detection in early-onset pediatric AD skin, we can potentially help evaluate disease progression in longitudinal studies and monitor early disease reversal in clinical trials.6,18,19
Most previous tape-strip studies primarily focused on protein profiling in the skin of adults with chronic AD (eTable 3 in the Supplement)42,53,54,57,58,69 and evaluated a limited panel of inflammatory and/or barrier-related proteins. Two studies analyzed protein expression in tape strips from children with AD.60,61 While these studies used nearly identical methods to measure protein expression, their results and conclusions were largely dissimilar. Hulshof et al75 detected 13 of the 28 assessed immune markers in tape strips from the lesional AD, nonlesional AD, and normal skin of children aged 0 to 12 years, with no significant increases in nonlesional AD skin compared with normal skin and with significant downregulation in 3 markers (IL-1α, CCL4, and CXCL10). Seven cytokines (eg, IL-1β, CCL17, CCL18, and IL-8) were significantly increased in lesional vs nonlesional and normal skin.
In contrast, McAleer et al61 studied only the nonlesional skin of infants with early-onset AD vs infants without AD, detecting 19 of the 27 analyzed inflammatory products, with significant increases in 11 cytokines in nonlesional AD vs normal tape-stripped skin. The study also detected significantly decreased expression of key TH2 and TH17 cytokines (IL-13, IL-5, IL-12, IL-23p40, and CSF2).45 Some key TH2 (IL-4, CCL13, and CCL26), TH1 (IFNγ), and other cytokines could not be detected by these studies. Significant correlations between AD severity (based on SCORAD assessment) with CCL17 and IL-8 expression in nonlesional and/or lesional skin were reported (r = 0.4; P < .05 for both comparisons).60,61 Two recent studies evaluated RNA profiling using Ion AmpliSeq sequencing (ThermoFisher Scientific) to analyze tapes 11 to 20.58,73 One study focused on gene expression in adults with nonlesional mild to severe AD vs adults without AD58 and detected 29 differentially expressed genes (false discovery rate, <0.05),58 including several immune genes (HLA-DOB, S100A7A, MMP9, MMP10, and CXCL6). A subset of 9 patients with higher inflammation and disease severity was identified, with upregulation of TH2 products (IL-13, IL-4R, CCL22, and CCR4). Detection rates for nonlesional AD and normal skin were 60% and 52%, respectively. Another recent study73 evaluated primarily nonlesional skin from children and adolescents aged 8 to 16 years with mild to severe AD, focusing on those with and without concurrent FAs (n = 21 and n = 19, respectively) vs children and adolescents without AD (n = 22). The overall success rate for transcriptome sequencing was 45% of all samples (18% for normal skin, <40% for nonlesional skin, and 57%-89% for lesional skin detection). The study reported higher expression of TH2 products (IL-4R and thymic stromal lymphopoietin [TSLP] receptor) and lower expression of FLG breakdown products and lipids in the nonlesional skin of children and adolescents with AD and FAs compared with those with AD and no FAs and those without AD.73
This study expands on previous profiling studies,42,45,53,54,57,58,66,68,69,93 including 2 RNA-Seq studies that reported a 45% overall detection rate.58,73 This study also identifies the largest set of differentially expressed products between AD (both lesional and nonlesional) and normal tape-stripped skin, with a 99% sample detection rate (only 1 of 71 samples was undetectable), which is critical for paired sample analysis. Given our high sample detection rate, it is probable that 2 sequential samples in clinical trials or longitudinal studies will both generate measures.10,11,12,27,28,51,53,54,57,58,60,61,69,72,78,84,94,95 Seventy-seven of 79 evaluated immune and barrier gene products were detected (gene detection rate, 97%) from 70 of 71 tape-strip samples (sample detection rate, 99%), with 53 of 79 markers significantly different between children with lesional and/or nonlesional AD and children without AD. Among these mediators were key AD biomarkers, many of which were not detected or evaluated in previous AD tape-strip studies.53,54,58,60,61 These biomarkers included measures of cellular infiltrates, including those associated with T cells and T-cell activation (CD3 and ICOS), atopic dendritic cells (Fc ε RI and OX40L),84,86 and key inflammatory markers of general inflammation (MMP12), innate immunity (IL-8 and IL-1RA), TH2 (IL-4, IL-13, CCL17, and CCL26), TH17/TH22 (IL-19, IL-23p19, IL-36G, CCL20, S100As, and DEFB4), Treg cells (FOXP3 and IL-10), negative regulators (IL-34 and IL-37), and epidermal proliferation (SERPINB3). The novel epidermal cytokines IL-33 and IL-17C, which are currently targeted in clinical trials of patients with AD, were also highlighted as novel tape-strip biomarkers88,96 and demonstrated significant correlations with AD severity. Interestingly, CCL20 has also been reported to be associated with commensal microbes in early life that induce Treg cells,97 which had increased markers in our study.
Terminal differentiation mRNAs (FLG, FLG2, LOR, and PPL) had decreased expression in lesional and nonlesional early-onset AD tape-stripped vs normal tape-stripped skin, similar to whole biopsies from chronic adult AD skin10,11,27,28,84 and in contrast with the normal expression of terminal differentiation products in our recent biopsy studies of early-onset pediatric AD.27,28 Lipid (FA2H) and tight junction (CLDN8 and CLDN23) mRNAs showed similar decreases in early-onset AD tape strips and biopsies.27,28 Terminal differentiation measures were generally not reduced in adult AD tape-strip genomic and proteomic studies.54,58 Few lipid and FLG degradation products showed lower expressions in children and adolescents with AD and FAs compared with those with AD and no FAs and those without AD. The effect of the epidermal depth on expression patterns in serial tape strips deserves further study.40,52,53,57,61
Our tape-strip data support the view that the nonlesional skin of patients with early-onset AD is widely abnormal. The significant correlations between the expression of AD-associated biomarkers (TH2, IL-33, IL-4R, CCR4, and CCL18) in nonlesional skin and severity indices (based on SCORAD, EASI, Pruritus ADQ, and TEWL assessments) further suggest that nonlesional abnormalities may be directed by the extent of active disease. This tape-strip method may be useful, together with TEWL assessment, for early studies of children who are at high risk of developing AD to dissect the sequence of epidermal vs immune changes leading to AD initiation.
Tape strips provide a perfect classifier (defined as an AUC score of 1, with 0 indicating a classifier with no power) to discriminate between early-onset AD skin and normal pediatric skin, including biomarkers pathogenically linked to AD (ie, IL-13, CCL26, S100As, and FLG) and an almost perfect molecular classifier using the negative regulator cytokine IL-34.10,98 This classifier may be useful in future longitudinal studies that evaluate the resolution of AD. Interleukin 34, a primarily epidermal cytokine and negative regulator of inflammation,98 has been recently suggested as a treatment-response biomarker in the skin of adults with AD.6
Limitations
The study had several limitations, primarily related to tape stripping. First, it is unclear whether tape strips in AD vs normal skin attain similar epidermal depth, especially given the greater mRNA yield in AD skin. Thus, differential expression of some genes may have merely reflected a greater depth of tape stripping within the stratum corneum.57,58,73 Second, mRNA extraction from tape strips is much more labor intensive compared with biopsies, potentially limiting widespread use. Third, some AD biomarkers, such as the hyperplasia-related K16, which is characteristically upregulated in AD skin,2,12,20,27,28 were not upregulated in early-onset AD tape strips, although K16 tape-strip mRNA remained correlated with AD severity. Tape strips allow detection of molecular abnormalities in superficial keratinocytes, whereas most K16 induction takes place in basal and suprabasal keratinocytes. Fourth, some AD biomarkers, such as IL-22, are restricted to the dermis9,10,84,95 and cannot be detected using tape strips. Fifth, our study did not have a direct comparison with whole skin biopsy biomarkers from the same patients. We plan to determine the correlation between skin biopsy and tape-stripping findings in future studies. Sixth, mRNA contained in corneocytes may be less stable than the protein products it encodes; thus, protein staining may show more precise differences, as reported in this article.
Conclusions
Through the use of a minimally invasive tape-strip method, we reported extensive gene-profiling characterization of early-onset moderate to severe pediatric AD skin. This characterization identified the molecular measures of disease activity that precede the onset of clinically visible lesions and may be useful for identifying biomarkers associated with AD. Because tape stripping is painless, nonscarring, and allows repeated sampling, it may be associated with benefits for longitudinal pediatric studies and clinical trials, in which serial measures are needed to identify predictors of response, course, and comorbidities.
eMethods 1. Tape-Strip Collection
eMethods 2. RNA Extraction, Quality Control, and Quantitative Real-Time PCR
eMethods 3. Statistical Analysis
eTable 1. qRT-PCR Primers for 79 Immune and Barrier Mediators
eTable 2. Immunohistochemistry Antibodies
eTable 3. Summary of Past Tape-Strip Studies in Patients With Atopic Dermatitis
eTable 4. Means, Standard Errors, and Confidence Intervals for All Immune and Barrier Biomarkers
eFigure 1. Gene Expression of Inflammatory Biomarkers in Tape Strips
eFigure 2. Immune and Barrier AD Biomarkers Detected in Tape Strips
eFigure 3. Histology
eFigure 4. Expression of Biomarkers in Tape Strips Correlates With Clinical Severity
eFigure 5. Receiver Operating Characteristic Area Under the Curve (ROC AUC) in Tape Strips
eReferences
References
- 1.Simpson EL, Bieber T, Guttman-Yassky E, et al. ; SOLO 1 and SOLO 2 Investigators . Two phase 3 trials of dupilumab versus placebo in atopic dermatitis. N Engl J Med. 2016;375(24):2335-2348. doi: 10.1056/NEJMoa1610020 [DOI] [PubMed] [Google Scholar]
- 2.Guttman-Yassky E, Bissonnette R, Ungar B, et al. . Dupilumab progressively improves systemic and cutaneous abnormalities in patients with atopic dermatitis. J Allergy Clin Immunol. 2019;143(1):155-172. doi: 10.1016/j.jaci.2018.08.022 [DOI] [PubMed] [Google Scholar]
- 3.Guttman-Yassky E, Silverberg JI, Nemoto O, et al. . Baricitinib in adult patients with moderate-to-severe atopic dermatitis: a phase 2 parallel, double-blinded, randomized placebo-controlled multiple-dose study. J Am Acad Dermatol. 2019;80(4):913-921.e9. doi: 10.1016/j.jaad.2018.01.018 [DOI] [PubMed] [Google Scholar]
- 4.Guttman-Yassky E, Brunner PM, Neumann AU, et al. . Efficacy and safety of fezakinumab (an IL-22 monoclonal antibody) in adults with moderate-to-severe atopic dermatitis inadequately controlled by conventional treatments: a randomized, double-blind, phase 2a trial. J Am Acad Dermatol. 2018;78(5):872-881.e6. doi: 10.1016/j.jaad.2018.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wollenberg A, Howell MD, Guttman-Yassky E, et al. . Treatment of atopic dermatitis with tralokinumab, an anti–IL-13 mAb. J Allergy Clin Immunol. 2019;143(1):135-141. doi: 10.1016/j.jaci.2018.05.029 [DOI] [PubMed] [Google Scholar]
- 6.Brunner PM, Pavel AB, Khattri S, et al. . Baseline IL-22 expression in patients with atopic dermatitis stratifies tissue responses to fezakinumab. J Allergy Clin Immunol. 2019;143(1):142-154. doi: 10.1016/j.jaci.2018.07.028 [DOI] [PubMed] [Google Scholar]
- 7.Czarnowicki T, Krueger JG, Guttman-Yassky E. Novel concepts of prevention and treatment of atopic dermatitis through barrier and immune manipulations with implications for the atopic march. J Allergy Clin Immunol. 2017;139(6):1723-1734. doi: 10.1016/j.jaci.2017.04.004 [DOI] [PubMed] [Google Scholar]
- 8.Brunner PM, Guttman-Yassky E, Leung DY. The immunology of atopic dermatitis and its reversibility with broad-spectrum and targeted therapies. J Allergy Clin Immunol. 2017;139(4S):S65-S76. doi: 10.1016/j.jaci.2017.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brunner PM, Leung DYM, Guttman-Yassky E. Immunologic, microbial, and epithelial interactions in atopic dermatitis. Ann Allergy Asthma Immunol. 2018;120(1):34-41. doi: 10.1016/j.anai.2017.09.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Esaki H, Ewald DA, Ungar B, et al. . Identification of novel immune and barrier genes in atopic dermatitis by means of laser capture microdissection. J Allergy Clin Immunol. 2015;135(1):153-163. doi: 10.1016/j.jaci.2014.10.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ewald DA, Malajian D, Krueger JG, et al. . Meta-analysis derived atopic dermatitis (MADAD) transcriptome defines a robust AD signature highlighting the involvement of atherosclerosis and lipid metabolism pathways. BMC Med Genomics. 2015;8:60. doi: 10.1186/s12920-015-0133-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suárez-Fariñas M, Ungar B, Correa da Rosa J, et al. . RNA sequencing atopic dermatitis transcriptome profiling provides insights into novel disease mechanisms with potential therapeutic implications. J Allergy Clin Immunol. 2015;135(5):1218-1227. doi: 10.1016/j.jaci.2015.03.003 [DOI] [PubMed] [Google Scholar]
- 13.Brunner PM, Emerson RO, Tipton C, et al. . Nonlesional atopic dermatitis skin shares similar T-cell clones with lesional tissues. Allergy. 2017;72(12):2017-2025. doi: 10.1111/all.13223 [DOI] [PubMed] [Google Scholar]
- 14.Brunner PM, Guttman-Yassky E. Racial differences in atopic dermatitis. Ann Allergy Asthma Immunol. 2019;122(5):449-455. doi: 10.1016/j.anai.2018.11.015 [DOI] [PubMed] [Google Scholar]
- 15.Guttman-Yassky E, Krueger JG, Lebwohl MG. Systemic immune mechanisms in atopic dermatitis and psoriasis with implications for treatment. Exp Dermatol. 2018;27(4):409-417. doi: 10.1111/exd.13336 [DOI] [PubMed] [Google Scholar]
- 16.Tintle S, Shemer A, Suarez-Farinas M, et al. . Reversal of atopic dermatitis with narrow-band UVB phototherapy and biomarkers for therapeutic response. J Allergy Clin Immunol. 2011;128(3):583-593.e1-4. doi: 10.1016/j.jaci.2011.05.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rozenblit M, Suarez-Farinas M, Shemer A, et al. . Residual genomic profile after cyclosporine treatment may offer insights into atopic dermatitis reoccurrence. J Allergy Clin Immunol. 2014;134(4):955-957. doi: 10.1016/j.jaci.2014.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khattri S, Brunner PM, Garcet S, et al. . Efficacy and safety of ustekinumab treatment in adults with moderate-to-severe atopic dermatitis. Exp Dermatol. 2017;26(1):28-35. doi: 10.1111/exd.13112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Renert-Yuval Y, Guttman-Yassky E. Systemic therapies in atopic dermatitis: the pipeline. Clin Dermatol. 2017;35(4):387-397. doi: 10.1016/j.clindermatol.2017.03.012 [DOI] [PubMed] [Google Scholar]
- 20.Brunner PM, Khattri S, Garcet S, et al. . A mild topical steroid leads to progressive anti-inflammatory effects in the skin of patients with moderate-to-severe atopic dermatitis. J Allergy Clin Immunol. 2016;138(1):169-178. doi: 10.1016/j.jaci.2015.12.1323 [DOI] [PubMed] [Google Scholar]
- 21.Thijs J, Krastev T, Weidinger S, et al. . Biomarkers for atopic dermatitis: a systematic review and meta-analysis. Curr Opin Allergy Clin Immunol. 2015;15(5):453-460. doi: 10.1097/ACI.0000000000000198 [DOI] [PubMed] [Google Scholar]
- 22.Cabanillas B, Brehler AC, Novak N. Atopic dermatitis phenotypes and the need for personalized medicine. Curr Opin Allergy Clin Immunol. 2017;17(4):309-315. doi: 10.1097/ACI.0000000000000376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thijs JL, Drylewicz J, Fiechter R, et al. . EASI p-EASI: utilizing a combination of serum biomarkers offers an objective measurement tool for disease severity in atopic dermatitis patients. J Allergy Clin Immunol. 2017;140(6):1703-1705. doi: 10.1016/j.jaci.2017.06.046 [DOI] [PubMed] [Google Scholar]
- 24.Weidinger S, Beck LA, Bieber T, Kabashima K, Irvine AD. Atopic dermatitis. Nat Rev Dis Primers. 2018;4(1):1. doi: 10.1038/s41572-018-0001-z [DOI] [PubMed] [Google Scholar]
- 25.Mortz CG, Andersen KE, Dellgren C, Barington T, Bindslev-Jensen C. Atopic dermatitis from adolescence to adulthood in the TOACS cohort: prevalence, persistence and comorbidities. Allergy. 2015;70(7):836-845. doi: 10.1111/all.12619 [DOI] [PubMed] [Google Scholar]
- 26.Kelleher M, Dunn-Galvin A, Hourihane JO, et al. . Skin barrier dysfunction measured by transepidermal water loss at 2 days and 2 months predates and predicts atopic dermatitis at 1 year. J Allergy Clin Immunol. 2015;135(4):930-935.e1. doi: 10.1016/j.jaci.2014.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Brunner PM, Israel A, Zhang N, et al. . Early-onset pediatric atopic dermatitis is characterized by TH2/TH17/TH22-centered inflammation and lipid alterations. J Allergy Clin Immunol. 2018;141(6):2094-2106. doi: 10.1016/j.jaci.2018.02.040 [DOI] [PubMed] [Google Scholar]
- 28.Esaki H, Brunner PM, Renert-Yuval Y, et al. . Early-onset pediatric atopic dermatitis is TH2 but also TH17 polarized in skin. J Allergy Clin Immunol. 2016;138(6):1639-1651. doi: 10.1016/j.jaci.2016.07.013 [DOI] [PubMed] [Google Scholar]
- 29.Cole C, Kroboth K, Schurch NJ, et al. . Filaggrin-stratified transcriptomic analysis of pediatric skin identifies mechanistic pathways in patients with atopic dermatitis. J Allergy Clin Immunol. 2014;134(1):82-91. doi: 10.1016/j.jaci.2014.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Czarnowicki T, Krueger JG, Guttman-Yassky E. Skin barrier and immune dysregulation in atopic dermatitis: an evolving story with important clinical implications. J Allergy Clin Immunol Pract. 2014;2(4):371-379. doi: 10.1016/j.jaip.2014.03.006 [DOI] [PubMed] [Google Scholar]
- 31.Czarnowicki T, Gonzalez J, Shemer A, et al. . Severe atopic dermatitis is characterized by selective expansion of circulating TH2/TC2 and TH22/TC22, but not TH17/TC17, cells within the skin-homing T-cell population. J Allergy Clin Immunol. 2015;136(1):104-115.e7. doi: 10.1016/j.jaci.2015.01.020 [DOI] [PubMed] [Google Scholar]
- 32.Czarnowicki T, Esaki H, Gonzalez J, et al. . Early pediatric atopic dermatitis shows only a cutaneous lymphocyte antigen (CLA)(+) TH2/TH1 cell imbalance, whereas adults acquire CLA(+) TH22/TC22 cell subsets. J Allergy Clin Immunol. 2015;136(4):941-951.e3. doi: 10.1016/j.jaci.2015.05.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Esaki H, Czarnowicki T, Gonzalez J, et al. . Accelerated T-cell activation and differentiation of polar subsets characterizes early atopic dermatitis development. J Allergy Clin Immunol. 2016;138(5):1473-1477.e5. doi: 10.1016/j.jaci.2016.04.052 [DOI] [PubMed] [Google Scholar]
- 34.Czarnowicki T, Esaki H, Gonzalez J, et al. . Alterations in B-cell subsets in pediatric patients with early atopic dermatitis. J Allergy Clin Immunol. 2017;140(1):134-144.e9. doi: 10.1016/j.jaci.2016.09.060 [DOI] [PubMed] [Google Scholar]
- 35.Czarnowicki T, Gonzalez J, Bonifacio KM, et al. . Diverse activation and differentiation of multiple B-cell subsets in patients with atopic dermatitis but not in patients with psoriasis. J Allergy Clin Immunol. 2016;137(1):118-129.e5. doi: 10.1016/j.jaci.2015.08.027 [DOI] [PubMed] [Google Scholar]
- 36.Wood LC, Feingold KR, Sequeira-Martin SM, Elias PM, Grunfeld C. Barrier function coordinately regulates epidermal IL-1 and IL-1 receptor antagonist mRNA levels. Exp Dermatol. 1994;3(2):56-60. doi: 10.1111/j.1600-0625.1994.tb00047.x [DOI] [PubMed] [Google Scholar]
- 37.Morhenn VB, Chang EY, Rheins LA. A noninvasive method for quantifying and distinguishing inflammatory skin reactions. J Am Acad Dermatol. 1999;41(5 Pt 1):687-692. doi: 10.1016/S0190-9622(99)70002-2 [DOI] [PubMed] [Google Scholar]
- 38.Wong R, Tran V, Morhenn V, et al. . Use of RT-PCR and DNA microarrays to characterize RNA recovered by non-invasive tape harvesting of normal and inflamed skin. J Invest Dermatol. 2004;123(1):159-167. doi: 10.1111/j.0022-202X.2004.22729.x [DOI] [PubMed] [Google Scholar]
- 39.Benson NR, Papenfuss J, Wong R, et al. . An analysis of select pathogenic messages in lesional and non-lesional psoriatic skin using non-invasive tape harvesting. J Invest Dermatol. 2006;126(10):2234-2241. doi: 10.1038/sj.jid.5700412 [DOI] [PubMed] [Google Scholar]
- 40.Wong R, Tran V, Talwalker S, Benson NR. Analysis of RNA recovery and gene expression in the epidermis using non-invasive tape stripping. J Dermatol Sci. 2006;44(2):81-92. doi: 10.1016/j.jdermsci.2006.08.007 [DOI] [PubMed] [Google Scholar]
- 41.Escobar-Chávez JJ, Merino-Sanjuán V, López-Cervantes M, et al. . The tape-stripping technique as a method for drug quantification in skin. J Pharm Sci. 2008;11(1):104-130. doi: 10.18433/J3201Z [DOI] [PubMed] [Google Scholar]
- 42.Broccardo CJ, Mahaffey SB, Strand M, Reisdorph NA, Leung DY. Peeling off the layers: skin taping and a novel proteomics approach to study atopic dermatitis. J Allergy Clin Immunol. 2009;124(5):1113-1115.e1-11. doi: 10.1016/j.jaci.2009.07.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kezic S, Kammeyer A, Calkoen F, Fluhr JW, Bos JD. Natural moisturizing factor components in the stratum corneum as biomarkers of filaggrin genotype: evaluation of minimally invasive methods. Br J Dermatol. 2009;161(5):1098-1104. doi: 10.1111/j.1365-2133.2009.09342.x [DOI] [PubMed] [Google Scholar]
- 44.Onoue A, Kabashima K, Kobayashi M, Mori T, Tokura Y. Induction of eosinophil- and Th2-attracting epidermal chemokines and cutaneous late-phase reaction in tape-stripped skin. Exp Dermatol. 2009;18(12):1036-1043. doi: 10.1111/j.1600-0625.2009.00899.x [DOI] [PubMed] [Google Scholar]
- 45.Voegeli R, Rawlings AV, Breternitz M, Doppler S, Schreier T, Fluhr JW. Increased stratum corneum serine protease activity in acute eczematous atopic skin. Br J Dermatol. 2009;161(1):70-77. doi: 10.1111/j.1365-2133.2009.09142.x [DOI] [PubMed] [Google Scholar]
- 46.Jungersted JM, Høgh JK, Hellgren LI, Jemec GB, Agner T. Skin barrier response to occlusion of healthy and irritated skin: differences in trans-epidermal water loss, erythema and stratum corneum lipids. Contact Dermatitis. 2010;63(6):313-319. doi: 10.1111/j.1600-0536.2010.01773.x [DOI] [PubMed] [Google Scholar]
- 47.Kezic S, O’Regan GM, Yau N, et al. . Levels of filaggrin degradation products are influenced by both filaggrin genotype and atopic dermatitis severity. Allergy. 2011;66(7):934-940. doi: 10.1111/j.1398-9995.2010.02540.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wachsman W, Morhenn V, Palmer T, et al. . Noninvasive genomic detection of melanoma. Br J Dermatol. 2011;164(4):797-806. doi: 10.1111/j.1365-2133.2011.10239.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Emson CL, Fitzmaurice S, Lindwall G, et al. . A pilot study demonstrating a non-invasive method for the measurement of protein turnover in skin disorders: application to psoriasis. Clin Transl Med. 2013;2:12. doi: 10.1186/2001-1326-2-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Westman M, Al-Bader T, Merinville E, et al. . In vivo cosmetic product efficacy testing by analyzing epidermal proteins extracted from tape strips. Cosmetics. 2014;1(1):29-36. doi: 10.3390/cosmetics1010029 [DOI] [Google Scholar]
- 51.Riethmuller C, McAleer MA, Koppes SA, et al. . Filaggrin breakdown products determine corneocyte conformation in patients with atopic dermatitis. J Allergy Clin Immunol. 2015;136(6):1573-1580.e2. doi: 10.1016/j.jaci.2015.04.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Clausen ML, Slotved HC, Krogfelt KA, Agner T. Tape stripping technique for stratum corneum protein analysis. Sci Rep. 2016;6:19918. doi: 10.1038/srep19918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koppes SA, Brans R, Ljubojevic Hadzavdic S, Frings-Dresen MH, Rustemeyer T, Kezic S. Stratum corneum tape stripping: monitoring of inflammatory mediators in atopic dermatitis patients using topical therapy. Int Arch Allergy Immunol. 2016;170(3):187-193. doi: 10.1159/000448400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Winget JM, Finlay D, Mills KJ, et al. . Quantitative proteomic analysis of stratum corneum dysfunction in adult chronic atopic dermatitis. J Invest Dermatol. 2016;136(8):1732-1735. doi: 10.1016/j.jid.2016.03.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Azimi A, Ali M, Kaufman KL, Mann GJ, Fernandez-Penas P. Tape stripped stratum corneum samples prove to be suitable for comprehensive proteomic investigation of actinic keratosis. Proteomics Clin Appl. 2019;13(3):e1800084. doi: 10.1002/prca.201800084 [DOI] [PubMed] [Google Scholar]
- 56.Berdyshev E, Goleva E, Bronova I, et al. . Lipid abnormalities in atopic skin are driven by type 2 cytokines. JCI Insight. 2018;3(4):98006. doi: 10.1172/jci.insight.98006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Clausen ML, Slotved HC, Krogfelt KA, Agner T. Measurements of AMPs in stratum corneum of atopic dermatitis and healthy skin-tape stripping technique. Sci Rep. 2018;8(1):1666. doi: 10.1038/s41598-018-20204-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dyjack N, Goleva E, Rios C, et al. . Minimally invasive skin tape strip RNA sequencing identifies novel characteristics of the type 2-high atopic dermatitis disease endotype. J Allergy Clin Immunol. 2018;141(4):1298-1309. doi: 10.1016/j.jaci.2017.10.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hochart G, Farcette F, Bonnel D, Stauber J, Stamatas GN. 688 Human keratinocyte biomarker mapping on skin tape strips using mass spectrometry imaging. J Investigative Derm. 2018;138(5):S117. doi: 10.1016/j.jid.2018.03.697 [DOI] [Google Scholar]
- 60.Hulshof L, Hack DP, Hasnoe QCJ, et al. . A minimally invasive tool to study immune response and skin barrier in children with atopic dermatitis. Br J Dermatol. 2019;180(3):621-630. doi: 10.1111/bjd.16994 [DOI] [PubMed] [Google Scholar]
- 61.McAleer MA, Jakasa I, Hurault G, et al. . Systemic and stratum corneum biomarkers of severity in infant atopic dermatitis include markers of innate and T helper cell-related immunity and angiogenesis. Br J Dermatol. 2019;180(3):586-596. doi: 10.1111/bjd.17088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pfannes EKB, Weiss L, Hadam S, et al. . Physiological and molecular effects of in vivo and ex vivo mild skin barrier disruption. Skin Pharmacol Physiol. 2018;31(3):115-124. doi: 10.1159/000484443 [DOI] [PubMed] [Google Scholar]
- 63.Tham EH, Dyjack N, Kim BE, et al. . Expression and function of the ectopic olfactory receptor OR10G7 in patients with atopic dermatitis. J Allergy Clin Immunol. 2019;143(5):1838-1848.e4. doi: 10.1016/j.jaci.2018.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dickel H, Kreft B, Kuss O, et al. . Increased sensitivity of patch testing by standardized tape stripping beforehand: a multicentre diagnostic accuracy study. Contact Dermatitis. 2010;62(5):294-302. doi: 10.1111/j.1600-0536.2010.01710.x [DOI] [PubMed] [Google Scholar]
- 65.Dickel H, Gambichler T, Kamphowe J, Altmeyer P, Skrygan M. Standardized tape stripping prior to patch testing induces upregulation of Hsp90, Hsp70, IL-33, TNF-α and IL-8/CXCL8 mRNA: new insights into the involvement of ‘alarmins’. Contact Dermatitis. 2010;63(4):215-222. doi: 10.1111/j.1600-0536.2010.01769.x [DOI] [PubMed] [Google Scholar]
- 66.Leitch CS, Natafji E, Yu C, et al. . Filaggrin-null mutations are associated with increased maturation markers on Langerhans cells. J Allergy Clin Immunol. 2016;138(2):482-490.e7. doi: 10.1016/j.jaci.2015.11.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.de Jongh CM, Verberk MM, Spiekstra SW, Gibbs S, Kezic S. Cytokines at different stratum corneum levels in normal and sodium lauryl sulphate-irritated skin. Skin Res Technol. 2007;13(4):390-398. doi: 10.1111/j.1600-0846.2007.00242.x [DOI] [PubMed] [Google Scholar]
- 68.Koppes SA, Kemperman P, Van Tilburg I, et al. . Determination of natural moisturizing factors in the skin: Raman microspectroscopy versus HPLC. Biomarkers. 2017;22(6):502-507. doi: 10.1080/1354750X.2016.1256428 [DOI] [PubMed] [Google Scholar]
- 69.Amarbayasgalan T, Takahashi H, Dekio I, Morita E. Interleukin-8 content in the stratum corneum as an indicator of the severity of inflammation in the lesions of atopic dermatitis. Int Arch Allergy Immunol. 2013;160(1):63-74. doi: 10.1159/000339666 [DOI] [PubMed] [Google Scholar]
- 70.Morita E, Takahashi H, Niihara H, et al. . Stratum corneum TARC level is a new indicator of lesional skin inflammation in atopic dermatitis. Allergy. 2010;65(9):1166-1172. doi: 10.1111/j.1398-9995.2010.02361.x [DOI] [PubMed] [Google Scholar]
- 71.Reisdorph N, Armstrong M, Powell R, et al. . Quantitation of peptides from non-invasive skin tapings using isotope dilution and tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2018;1084:132-140. doi: 10.1016/j.jchromb.2018.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Clausen ML, Jungersted JM, Andersen PS, Slotved HC, Krogfelt KA, Agner T. Human β-defensin-2 as a marker for disease severity and skin barrier properties in atopic dermatitis. Br J Dermatol. 2013;169(3):587-593. doi: 10.1111/bjd.12419 [DOI] [PubMed] [Google Scholar]
- 73.Leung DYM, Calatroni A, Zaramela LS, et al. . The nonlesional skin surface distinguishes atopic dermatitis with food allergy as a unique endotype. Sci Transl Med. 2019;11(480):1-13. doi: 10.1126/scitranslmed.aav2685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gittler JK, Krueger JG, Guttman-Yassky E. Atopic dermatitis results in intrinsic barrier and immune abnormalities: implications for contact dermatitis. J Allergy Clin Immunol. 2013;131(2):300-313. doi: 10.1016/j.jaci.2012.06.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sullivan M, Silverberg NB. Current and emerging concepts in atopic dermatitis pathogenesis. Clin Dermatol. 2017;35(4):349-353. doi: 10.1016/j.clindermatol.2017.03.006 [DOI] [PubMed] [Google Scholar]
- 76.Kelleher MM, Dunn-Galvin A, Gray C, et al. . Skin barrier impairment at birth predicts food allergy at 2 years of age. J Allergy Clin Immunol. 2016;137(4):1111-1116.e8. doi: 10.1016/j.jaci.2015.12.1312 [DOI] [PubMed] [Google Scholar]
- 77.Dapic I, Kobetic R, Brkljacic L, Kezic S, Jakasa I. Quantification of free fatty acids in human stratum corneum using tandem mass spectrometry and surrogate analyte approach. Biomed Chromatogr. 2018;32(2):e4056. doi: 10.1002/bmc.4056 [DOI] [PubMed] [Google Scholar]
- 78.Noda S, Suarez-Farinas M, Ungar B, et al. . The Asian atopic dermatitis phenotype combines features of atopic dermatitis and psoriasis with increased TH17 polarization. J Allergy Clin Immunol. 2015;136(5):1254-1264. doi: 10.1016/j.jaci.2015.08.015 [DOI] [PubMed] [Google Scholar]
- 79.Sanyal RD, Pavel AB, Glickman J, et al. . Atopic dermatitis in African American patients is TH2/TH22-skewed with TH1/TH17 attenuation. Ann Allergy Asthma Immunol. 2019;122(1):99-110.e6. doi: 10.1016/j.anai.2018.08.024 [DOI] [PubMed] [Google Scholar]
- 80.Zhou L, Leonard A, Pavel AB, et al. . Age-specific changes in the molecular phenotype of patients with moderate-to-severe atopic dermatitis. J Allergy Clin Immunol. 2019;144(1):144-156. doi: 10.1016/j.jaci.2019.01.015 [DOI] [PubMed] [Google Scholar]
- 81.Brunner PM, Israel A, Leonard A, et al. . Distinct transcriptomic profiles of early-onset atopic dermatitis in blood and skin of pediatric patients. Ann Allergy Asthma Immunol. 2019;122(3):318-330.e3. doi: 10.1016/j.anai.2018.11.025 [DOI] [PubMed] [Google Scholar]
- 82.Shiomi A, Usui T. Pivotal roles of GM-CSF in autoimmunity and inflammation. Mediators Inflamm. 2015;2015:568543. doi: 10.1155/2015/568543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Szegedi K, Lutter R, Res PC, et al. . Cytokine profiles in interstitial fluid from chronic atopic dermatitis skin. J Eur Acad Dermatol Venereol. 2015;29(11):2136-2144. doi: 10.1111/jdv.13160 [DOI] [PubMed] [Google Scholar]
- 84.Fujita H, Shemer A, Suarez-Farinas M, et al. . Lesional dendritic cells in patients with chronic atopic dermatitis and psoriasis exhibit parallel ability to activate T-cell subsets. J Allergy Clin Immunol. 2011;128(3):574-582.e1-12. doi: 10.1016/j.jaci.2011.05.016 [DOI] [PubMed] [Google Scholar]
- 85.Dos Santos VG, Orfali RL, de Oliveira Titz T, da Silva Duarte AJ, Sato MN, Aoki V. Evidence of regulatory myeloid dendritic cells and circulating inflammatory epidermal dendritic cells-like modulated by Toll-like receptors 2 and 7/8 in adults with atopic dermatitis. Int J Dermatol. 2017;56(6):630-635. doi: 10.1111/ijd.13537 [DOI] [PubMed] [Google Scholar]
- 86.Suárez-Farinas M, Dhingra N, Gittler J, et al. . Intrinsic atopic dermatitis shows similar TH2 and higher TH17 immune activation compared with extrinsic atopic dermatitis. J Allergy Clin Immunol. 2013;132(2):361-370. doi: 10.1016/j.jaci.2013.04.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zheng R, Chen FH, Gao WX, et al. . The TH2-polarizing function of atopic interleukin 17 receptor B-positive dendritic cells up-regulated by lipopolysaccharide. Ann Allergy Asthma Immunol. 2017;118(4):474-482.e1. doi: 10.1016/j.anai.2016.12.011 [DOI] [PubMed] [Google Scholar]
- 88.Guttman-Yassky E, Krueger JG. IL-17C: a unique epithelial cytokine with potential for targeting across the spectrum of atopic dermatitis and psoriasis. J Invest Dermatol. 2018;138(7):1467-1469. doi: 10.1016/j.jid.2018.02.037 [DOI] [PubMed] [Google Scholar]
- 89.Dajnoki Z, Beke G, Mocsai G, et al. . Immune-mediated skin inflammation is similar in severe atopic dermatitis patients with or without filaggrin mutation. Acta Derm Venereol. 2016;96(5):645-650. doi: 10.2340/00015555-2272 [DOI] [PubMed] [Google Scholar]
- 90.Chan TC, Sanyal RD, Pavel AB, et al. . Atopic dermatitis in Chinese patients shows TH2/TH17 skewing with psoriasiform features. J Allergy Clin Immunol. 2018;142(3):1013-1017. doi: 10.1016/j.jaci.2018.06.016 [DOI] [PubMed] [Google Scholar]
- 91.Czarnowicki T, Malajian D, Khattri S, et al. . Petrolatum: barrier repair and antimicrobial responses underlying this “inert” moisturizer. J Allergy Clin Immunol. 2016;137(4):1091-1102.e7. doi: 10.1016/j.jaci.2015.08.013 [DOI] [PubMed] [Google Scholar]
- 92.Simpson EL, Imafuku S, Poulin Y, et al. . A phase 2 randomized trial of apremilast in patients with atopic dermatitis. J Invest Dermatol. 2019;139(5):1063-1072. doi: 10.1016/j.jid.2018.10.043 [DOI] [PubMed] [Google Scholar]
- 93.Angelova-Fischer I, Mannheimer AC, Hinder A, et al. . Distinct barrier integrity phenotypes in filaggrin-related atopic eczema following sequential tape stripping and lipid profiling. Exp Dermatol. 2011;20(4):351-356. doi: 10.1111/j.1600-0625.2011.01259.x [DOI] [PubMed] [Google Scholar]
- 94.Guttman-Yassky E, Suárez-Fariñas M, Chiricozzi A, et al. . Broad defects in epidermal cornification in atopic dermatitis identified through genomic analysis. J Allergy Clin Immunol. 2009;124(6):1235-1244.e58. doi: 10.1016/j.jaci.2009.09.031 [DOI] [PubMed] [Google Scholar]
- 95.Guttman-Yassky E, Lowes MA, Fuentes-Duculan J, et al. . Major differences in inflammatory dendritic cells and their products distinguish atopic dermatitis from psoriasis. J Allergy Clin Immunol. 2007;119(5):1210-1217. doi: 10.1016/j.jaci.2007.03.006 [DOI] [PubMed] [Google Scholar]
- 96.Ogg G. Proof-of-concept phase-2a clinical trial of ANB020 (anti–IL-33 antibody) in the treatment of moderate-to-severe adult atopic dermatitis. Paper presented at: American Academy of Dermatology 77th Annual Meeting; February 16-20, 2018; San Diego, CA. https://www2.anaptysbio.com/wp-content/uploads/AAD-Presentation-Ogg-021718.pdf Accessed December 26, 2018.
- 97.Ali N, Zirak B, Rodriguez RS, et al. . Regulatory T cells in skin facilitate epithelial stem cell differentiation. Cell. 2017;169(6):1119-1129.e11. doi: 10.1016/j.cell.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Boulakirba S, Pfeifer A, Mhaidly R, et al. . IL-34 and CSF-1 display an equivalent macrophage differentiation ability but a different polarization potential. Sci Rep. 2018;8(1):256. doi: 10.1038/s41598-017-18433-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eMethods 1. Tape-Strip Collection
eMethods 2. RNA Extraction, Quality Control, and Quantitative Real-Time PCR
eMethods 3. Statistical Analysis
eTable 1. qRT-PCR Primers for 79 Immune and Barrier Mediators
eTable 2. Immunohistochemistry Antibodies
eTable 3. Summary of Past Tape-Strip Studies in Patients With Atopic Dermatitis
eTable 4. Means, Standard Errors, and Confidence Intervals for All Immune and Barrier Biomarkers
eFigure 1. Gene Expression of Inflammatory Biomarkers in Tape Strips
eFigure 2. Immune and Barrier AD Biomarkers Detected in Tape Strips
eFigure 3. Histology
eFigure 4. Expression of Biomarkers in Tape Strips Correlates With Clinical Severity
eFigure 5. Receiver Operating Characteristic Area Under the Curve (ROC AUC) in Tape Strips
eReferences