Abstract
Microsatellite repeat expansion disease loci can exhibit pleiotropic clinical and biological effects depending on repeat length. Large expansions in C9orf72 (100s-1000s of units) are the most common genetic cause of amyotrophic lateral sclerosis (ALS) and frontotemporal degeneration (FTD). However, whether intermediate expansions also contribute to neurodegenerative disease is not well understood. Several studies have identified intermediate repeats in Parkinson’s disease patients, but the association was not found in autopsy confirmed cases. We hypothesized that intermediate C9orf72 repeats are a genetic risk factor for corticobasal degeneration (CBD), a neurodegenerative disease that can be clinically similar to Parkinson’s but has distinct tau protein pathology. Indeed, intermediate C9orf72 repeats were significantly enriched in autopsy-proven CBD (n=354 cases, odds ratio=3.59, p-value=0.00024). While large C9orf72 repeat expansions are known to decrease C9orf72 expression, intermediate C9orf72 repeats result in increased C9orf72 expression in human brain tissue and CRISPR/cas9 knockin iPSC derived neural progenitor cells. In contrast to cases of FTD/ALS with large C9orf72 expansions, CBD with intermediate C9orf72 repeats was not associated with pathologic RNA foci or dipeptide repeat protein aggregates. Knock-in cells with intermediate repeats exhibit numerous changes in gene expression pathways relating to vesicle trafficking and autophagy. Additionally, overexpression of C9orf72 without the repeat expansion leads to defects in autophagy under nutrient starvation conditions. These results raise the possibility that therapeutic strategies to reduce C9orf72 expression may be beneficial for the treatment of CBD.
Keywords: Neurodegeneration, Corticobasal degeneration, C9orf72 repeat expansion, Parkinsonism, Autophagy
Introduction
Large hexanucleotide repeat expansions (G4C2) in C9orf72 are the most common genetic cause of the neurodegenerative diseases amyotrophic lateral sclerosis (ALS) and frontotemporal degeneration (FTD)[19, 49, 60]. ALS/FTD is characterized neuropathologically by the presence of TDP-43 inclusions which are tightly linked to neurodegeneration[35]. While the pathophysiologic mechanisms that lead to TDP-43 aggregation and neurodegeneration in the setting of large C9orf72 repeat expansions are still unclear, both a loss of normal C9orf72 function and a toxic gain of function attributed to repetitive RNA and protein aggregates have been proposed[19, 36, 60, 63, 67]. At the protein level, C9orf72 regulates several vesicle trafficking pathways [4, 26, 79] and is necessary for proper myeloid cell function in mice, but knockout of the protein does not cause neurodegeneration[12, 58]. On the other hand, the presence of the repeat expansion is sufficient to induce neurodegeneration in several mouse models[15, 42]. There is also uncertainty in the repeat size threshold necessary to cause disease. Repeat expansions that cause ALS/FTD are typically hundreds to thousands of units long and can differ substantially between tissues[10, 70], making it difficult to accurately determine the pathogenic repeat size threshold. There have been reports of pathogenic repeats as small as 55–100 units in patient blood[23, 28]. In contrast, non-diseased individuals typically harbor repeats of 2–8 units[20]. Several studies have explored whether intermediate C9orf72 expansions (e.g. those larger than controls but not large enough to cause ALS/FTD) are associated with risk for other neurodegenerative diseases, and these have often yielded conflicting results[1, 20, 30, 32, 37, 56, 80, 84, 86]. For instance, a significant association between intermediate expansion carriers and Parkinson’s Disease (PD) was reported in clinically diagnosed PD[56] and confirmed in a large meta-analysis[73], but these associations were not found in an autopsy confirmed PD cohort[57]. Other studies with very small numbers of cases have also suggested that intermediate C9orf72 repeats are associated with atypical Parkinsonian syndromes[13, 64]. Thus, there is some evidence that intermediate expansions in C9orf72 may confer risk for neurodegenerative diseases other than ALS/FTD, but more exploration is needed to better understand the molecular pathways and exact neurodegenerative diseases that are affected.
Here, we sought to determine whether intermediate expansions in C9orf72 are a risk factor for corticobasal degeneration (CBD), a rare neurodegenerative disease that shares some similar clinical features with PD such as limb rigidity, bradykinesia, tremor and apraxia[5]. Patients that present with these neurologic features are often diagnosed with corticobasal syndrome (CBS) because these symptoms can also be caused by several other neurodegenerative diseases; definitive diagnosis of CBD requires neuropathology autopsy examination[33]. CBD is characterized by hyper-phosphorylated tau aggregates in neurons and astrocytes that are morphologically distinct from those found in Alzheimer’s disease and other tauopathies[21]. Only ~25–50% of patients diagnosed with CBS are confirmed as CBD at autopsy[11, 39, 40], making clinically defined cohorts difficult to use for genetic studies of CBD due to underlying neuropathological heterogeneity. In this study, we determine whether C9orf72 intermediate expansions may be a risk factor for CBD by measuring repeat size in the largest cohort of autopsy confirmed cases ever tested. In addition, we use post mortem CBD brain tissue to confirm the genetic findings, use CRISPR/cas9 to modify the repeat number in human iPS cells at the endogenous locus, and determine the effects of C9orf72 expression on autophagy. In doing so, we identify a novel mechanism for how intermediate repeat expansions may affect CBD pathogenesis that is distinct from large expansions that cause ALS/FTD.
Materials and Methods
Case Samples
Genomic DNA for repeat expansion size screening was obtained from 354 autopsy confirmed CBD cases from 17 institutions. Institutions contributing the vast majority of cases included Mayo Clinc, University College London and the University of Pennsylvania. CBD patient cerebellum tissue for RNA expression studies was obtained from the Center for Neurodegenerative Disease Research brain bank at the University of Pennsylvania and from the Mayo Clinic brain bank. Brain autopsy material is not considered human subjects research. However, legal consent for autopsy was obtained in all instances. As a control cohort, we utilized a previously published global cohort of healthy controls for which C9orf72 repeat sizing was available[73].
Assessment of Repeat Expansion Size and SNP Genotyping
The CBD cohort was screened for intermediate C9orf72 repeat expansions using fluorescent labeled touchdown PCR followed by fragment-length analysis on an ABI 3130xl Genetic Analyzer (Applied Biosystems, Foster City, CA). PCR used 50ng genomic DNA in a final volume of 20ul containing 0.5U of Amplitaq Gold (Applied Biosystems) and a final concentration of 1x Amplitaq Buffer I, 5uM reverse primer, 5uM 6FAM-fluorescent labeled forward primer, 1M Betaine, 5% DMSO, 0.25mM 7-deaza-dGTP, and 0.25mM of each dNTP. For detection of full expansions using repeat-primed PCR, the Roche FastStart (Roche, Basel, Switzerland) polymerase system was used to amplify 1ug of genomic DNA in a reaction with 0.9 mM MgCl2, 200nM dNTP mix, 180nM 7-deaza-dGTP, 1.4uM flanking primers, 0.175uM repeat primer, 7% DMSO, .93M Betaine and 0.125 U polymerase. For both intermediate and full expansion detection, a touchdown PCR cycling program was used where the annealing temperature was gradually lowered from 70°C to 56°C in 2°C increments with a 3-minute elongation time at 72°C for each cycle. SNP genotyping was carried out using Taqman probes as previously described[19]. All primer sequences are listed in Supplemental Table 1.
Detection of RNA foci
RNA foci detection was performed on paraffin embedded cerebellum sections as follows: Slides were deparaffinized with Xylene, followed by Xylene/Ethanol (1:1) and then rehydrated with an ethanol series (100%, 90%, 80%,70% for 3 mins each). Slides were incubated for 5 minutes in water, then digested with proteinase K (20ug/mL) for 10 minutes at 37 degrees in a buffer containing 10mM Tris-HCL pH 8 and 0.5% SDS in DEPC-treated water. Slides were then washed twice with water then dehydrated with increasing ethanol series and allowed to air dry. Slides were then prehybridized for 1 hour at 66 degrees in hybridization buffer containing 50% Foramide, 2X SSC, 50mM NaHPO4 , and 10% Dextran sulfate in DEPC-treated water. Slides were hybridized with 40nM probe (LNA 5TYE563C4G2; Exiqon (Denmark) # 300500, batch # 621440) at 66 degrees overnight. Slides were then washed once with 2X SSC + 0.1% Tween for 5 mins at room temperature, then 3 times with 0.1X SSC for 10 minutes at 65 degrees. Slides were then rinsed with water, stained with DAPI and mounted with coverslips using Prolong glass antifade mountant (Invitrogen, Carlsbad, CA). Cases for RNA foci detection included 12 non-expanded CBD, 9 intermediate CBD, 4 non-expanded control cases (negative control) and 5 full expansion FTD/ALS cases (positive control).
Detection of Dipeptide Repeat proteins and Measurement of DNA methylation
Paraffin embedded cerebellum sections were deparaffinized and stained with antibodies against p62, GA (1:7500), GP (1:7500) or GR (1:100) as previously described[48]. 116 non-expanded CBD, 9 intermediate CBD, 4 non-expanded control cases (negative control) and 5 full expansion FTD/ALS cases (positive control) were stained. For measurement of DNA methylation in CBD patient genomic DNA, 100ng DNA was digested using the restriction enzymes HhaI and HaeIII (New England Biolabs, Ipswich, MA) followed by qPCR as previously described[41].
RNA Expression Analysis from Postmortem Brain and repeat edited NPCs
RNA was extracted from approximately 100mg of cerebellum tissue using Trizol reagent (Life Technologies, Carlsbad, CA). RNA integrity was tested on an Agilent 2100 bioanalyzer using the RNA Nanochip 6000 kit (Agilent, Santa Clara, CA). Only RNA samples with RIN values > 6 were included in this study. For repeat edited NPCs, cells were pelleted at day 12 post induction and RNA was extracted using the RNeasy Kit (Qiagen). cDNA was prepared using random hexamers and Superscript III (Invitrogen) according to manufacturer’s protocol. RT-qPCR was performed using SYBR Green reagents (Roche) on a StepOne Plus Real-Time PCR Machine (Life Technologies). RNA levels were normalized to the geometric mean of two housekeeping genes (ACTB, GPS1) using the ΔΔCt method. All primer sequences are listed in Supplemental Table 1.
CRISPR Editing of Repeat Size
CRISPR gRNAs were designed using the MIT CRISPR design tool (http://crispr.mit.edu/) and cloned into the Cas9-GFP PX458 vector (Addgene, Cambridge, MA) as previously described[59]. Repair templates were generated via PCR cloning using genomic DNA from intermediate repeat carriers. PCR was carried out using the FastStart Polymerase (Roche) with additives for GC rich templates (360nM 7-deaza-dGTP, 7% DMSO, 930mM Betaine). PCR products containing 2 or 28 repeats were cloned into PGEM Easy-T vector (Promega, Madison, WI) and verified by sequencing. A 2 bp substitution was made at the PAM site in the repair template using QuickChange site directed mutagenesis (Aligent) in order to prevent Cas9 cutting of the repair template and to incorporate a restriction enzyme site (See Supplemental Figure 3). PX458 and repair template vectors were co-transfected into WT iPSCs[46] using Viafect reagent (Promega). On day 2 post transfection, GFP+ cells were sorted via FACS (BD FACS Aria II) to 96 well plates for clonal isolation. DNA was extracted from 24 well plates of confluent clones using 0.2% SDS lysis buffer followed by phenol-chloroform extraction. DNA from clones was PCR amplified and screened for homology directed repair by restriction enzyme digest. Repeat size was measured as outlined above on positive clones, and all CRISPR edited cell lines were Sanger sequenced via PCR cloning (10 clones/cell line).
Differentiation of iPSCs to Neural Progenitor Cells
IPSCs were differentiated via dual SMAD inhibition as previously described[66]. Single cells were plated at high density (2×105/cm2) in mTeSR1 (StemCell Technologies, Vancouver, Canada) + 10uM ROCK inhibitor Y27632 (ATCC) on Matrigel (Corning, Corning, NY) coated 6 well plates. The following day, media was changed to neural induction media (see Shi et al.[66] for complete formulation) containing 10uM SB431542 (Tocris, Bristol, United Kingdom) and 1uM dorsomorphin (Sigma, St. Louis, MO). Neural induction media was changed each day. At day 6, cells were split 1:2 into neural induction media + ROCK inhibitor and media changes were continued until day 12. Day 12 NPCs were characterized for expression of NPC markers by immunofluorescence and RT-qPCR. For western blot analysis, NPCs were passaged again at day 12 and collected at day 16 post induction.
Western Blots and Antibodies
For detection of C9orf72, day 16 NPCs were pelleted, lysed (50mM Tris-HCl, 750mM NaCl, 5mM EDTA, 2% SDS) and sonicated using a probe sonicator (Sonics Vibra-cell), then spun at 21,000 g for 30 mins. BCA analysis (Pierce, Thermo Fisher) was performed on cleared lysate to quantify protein expression. 50ug of protein was run on an 8% Tris-Glycine gel, transferred using the Transblot Turbo system (Biorad, Hercules, CA) to a nitrocellulose membrane and blocked for 1 hour at room temperature with Odyssey Licor Blocking buffer (Licor, Lincoln, NE). Membranes were blotted with the following antibodies overnight at 4 degrees: anti-C9orf72 1:1000 (Millipore, Burlington, MA, ABN1645), anti-GAPDH 1:1000 (Cell signaling Technology, Danvers, MA 2118S). IR dye secondary antibodies (Licor) were used to detect protein on a Licor Odyssey. Protein was quantified using Image Studio software (Licor).
For C9orf72 expression and autophagy experiments, cell lysates were prepared in ice-cold radio immunoprecipitation assay (RIPA) buffer containing 150 mM NaCl, 1% TritonX-100, 0.5% sodium deoxycholate, 0.1% SDS and 50 mM Tris (pH8.0) supplemented with protease and phosphatase inhibitors (PhosphoSTOP, Roche). Cells were lysed for 20 min and centrifuged for 10 min at 13,500 rpm. Protein concentration of the soluble fractions were determined using BCA reagent (Thermo Fisher Scientific). 20–70ug of total protein was resolved using 10% or 12% denaturing polyacrylamide gel electrophoresis and transferred to PVDF membrane (Immobilon-P, PVDF). Proteins on PVDF membranes were blocked using 5% low-fat milk in TBST containing 0.1% Tween-20 and incubated with primary antibody in SuperBlock (TBS, Thermo Fisher Scientific) overnight at 4°C. The primary antibodies used were: LC3 (ProteinTech, 14600–1-AP, 1:1000), and GAPDH (ProteinTech, 60004–1-Ig, 1:5000). The membranes were washed in TBST. Anti-rabbit or anti-mouse horse-radish peroxidase (HRP) conjugated secondary antibodies (1:3000, Thermo Fisher Scientific) in 5% milk in TBST was incubated with the membranes for 1 hour at room temperature. The membranes were incubated with WestPico or WestFemto chemiluminescent substrate (Thermo Fisher Scientific) and exposed to Amersham Hyperfilm ECL films (GE Healthcare Life Sciences). The films were developed using Konica Minolta film developer. Protein band intensity was quantified using LI-COR Image Studio Lite.
RNA sequencing in knockin intermediate repeat cells
50 base pair paired end RNA sequencing was carried out on day 12 post induction isogenic NPC lines (See Supplemental Table 2) with 3 biological replicates per group and 2 technical replicates per cell line. Libraries were prepared using the TrueSeq Stranded mRNA and Ribo-zero rRNA removal kits (illumina, San Diego, CA). Sequencing was carried out on a Hiseq 2500 (illumina) and at least 100 million reads were generated per biological replicate. Adapter sequences were removed using Cutadapt[50]. Reads were aligned to the genome (GRCh38) using STAR (v2.6) [22] with the default mapping parameters and the read counts per gene were counted using the quantMode function and genecode v29 annotation. Differentially expressed genes between the short repeat group and intermediate repeat group were called using the DEseq2 program (v1.22)[45] with default parameters. Over enrichment analysis of biological pathways was carried out using WebGestalt (2019) on genes that were either upregulated or downregulated (adjusted p-value < 0.05) in the intermediate repeat group. For comparison of genes dysregulated in full expansion carriers, differentially expressed gene lists (adjusted p > 0.05 and >2 fold change) from two studies[24, 63] that compared gene expression in iPSC derived neurons with full expansions vs. low repeat controls were compared against the differentially expressed genes in this study (intermediate repeat vs low repeat).
Generation of stable C9orf72 expressing cell lines, transfection and drug treatments
The generation of an inducible single-copy C9orf72 transgene using Flp-In TRex-HeLa cells was as described previously[31]. All cell lines used in this study were maintained in DMEM (GE Healthcare HyClone, SH30243.01) with 10% FBS (GE Healthcare HyClone, SV30160.03) and 1% Pen-Strep (Thermo Fisher Scientific, 15070063) at 37°C and 5% CO2. Tetracycline (Sigma-Aldrich, T7660) was added (100 ug/ml) for 18 h to induce the expression of C9orf72 transgene. Parental HeLa cells treated with tetracycline were used as controls in the experiments shown in Figure 6b and 7b. All plasmid transfections in cell lines were performed using Lipofectamine 2000 (Thermo Fisher Scientific, 11668019) or TurboFect (Thermo Fisher Scientific, R0531) as per the manufacturer’s instruction. For autophagy experiments, Chloroquine (CQ, Sigma-Aldrich) was dissolved in water (20 mM) and used at 40μM in the media. Autophagy induction using Earle’s Balanced Salt Solution (EBSS, Gibco) was incubated on cells following two successive PBS rinses. Incubation time for each condition is stated in the respective figure legends.
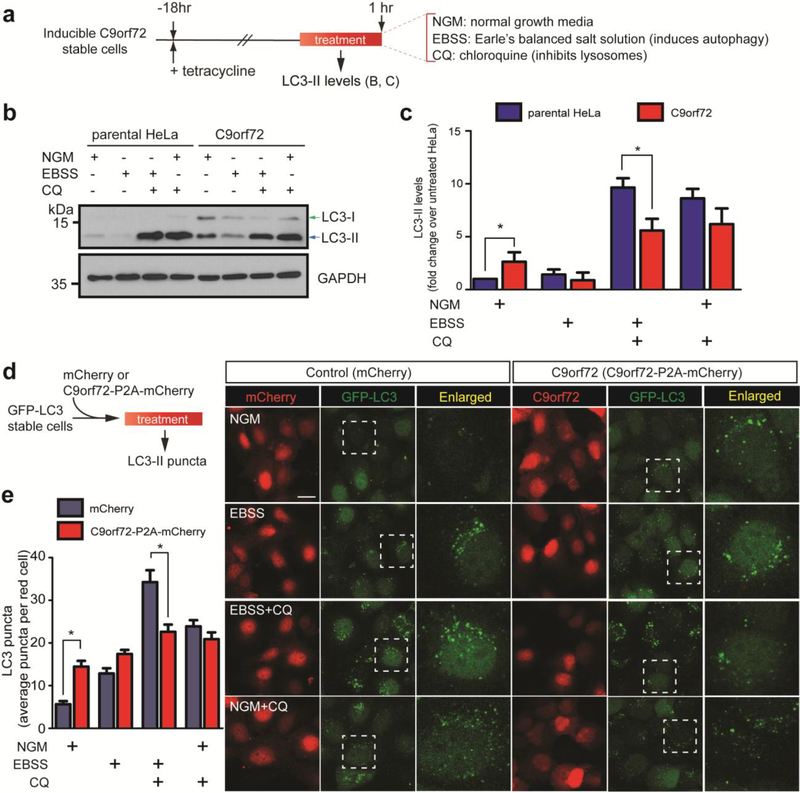
Figure 6: Autophagy defects in inducible C9orf72 over-expressing cells.
a) Schematic of experimental procedure for LC3-II protein detection in C9orf72 HeLa stable cells. b) Representative western blot showing LC3-II expression levels of parental HeLa cells and C9orf72 cells first treated with treated with tetracycline to induce C9orf72 transgene expression then with EBSS and/or CQ for 1 hour. c) Bar chart corresponding to (a) showing pooled data of LC3-II protein levels expressed as fold change over parental HeLa cells in normal growth media without CQ treatment. Data represents three independent experiments with * p<0.05, Student’s t-test. d) Schematic of experimental procedure for the detection of LC3-II puncta using fluorescence microscopy in parental HeLa cells overexpressing GFP-LC3 and mCherry (control) or C9orf72-P2A-mCherry plasmids. The cells were treated with NGM, EBSS, EBSS+CQ or NGM+CQ for 2 hours and green puncta associated with GFP-LC3 in mCherry-overexpressing cells were analyzed using ImageJ particle count. At least 25 cells were analyzed per treatment group. Scale bar = 20 μm. e) Bar chart corresponds to the average LC3 puncta count in (d) with * p < 0.05, Student’s t-test. Error bars represent standard error of the mean (S.E.M).
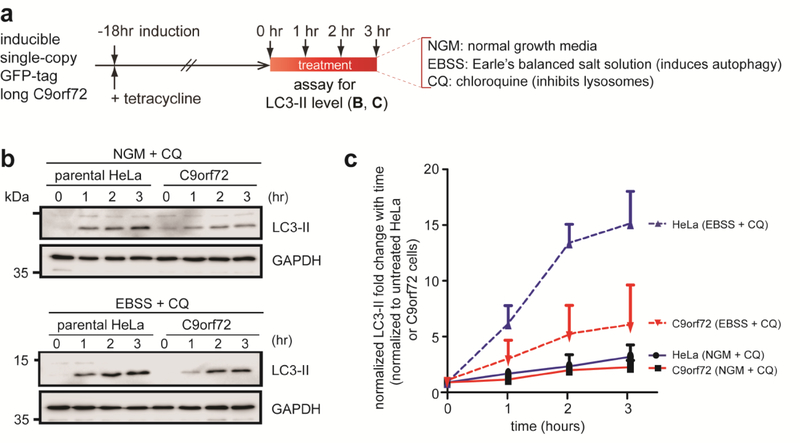
Figure 7. Kinetics of starvation induced autophagy in C9orf72 over-expressing cells.
a) Schematic of time-course experiments on autophagy in C9orf72 HeLa stable cells. b) Representative western blot showing LC3-II expression levels of parental HeLa cells and C9orf72 cells first treated with tetracycline to induce C9orf72 transgene expression then with either normal growth media (NGM)+CQ or EBSS+CQ for 0, 1, 2, and 3 hours. c) Time course chart corresponding to (b) showing pooled data of LC3-II protein levels expressed as fold change over respective controls, i.e. parental HeLa cells or C9orf72 cells grown in normal growth media without CQ treatment. Data represents three independent experiments with error bars representing standard error of the mean (S.E.M).
Results
Screening of CBD patient DNA for C9orf72 expansions
To test the hypothesis that intermediate size expansions in C9orf72 are a risk factor for CBD, genomic DNA from 354 autopsy confirmed CBD cases was screened using a PCR-based sizing assay (Figure 1a). We compared CBD patients to controls from a previously published global meta-analysis of C9orf72 repeat size [73]. We found that 1.84% of alleles from CBD cases (corresponding to 3.7%, or 13 cases) harbored intermediate length repeat expansions compared to 0.52% of controls (Figure 1b–c, OR = 3.59, p = 0.00024). This analysis was based on a minimum size cutoff of 17 which was determined a priori and based on prior independent studies[7, 73]. The increase in C9orf72 intermediate repeat allele frequency was not due to an increase in the frequency of the C9orf72 repeat expansion haplotype between CBD patients verses non-diseased controls or general global populations (Supplemental Table 3), and we did not detect any expansions larger than 29 repeats (Supplemental Table 4). We conducted a post-hoc analysis on our CBD cohort for each repeat length cut off below 18 repeats which revealed that repeats as low as 10 units was associated with CBD (p = 0.026, OR = 1.36), where 61 out of 354 CBD cases had C9orf72 repeat alleles that were at least 10 units. This included 5 cases which had two alleles at 10 or greater. Although this post-hoc analysis is considered exploratory[9], there appeared to be a somewhat graded increase in CBD risk with increasing repeat size (Figure 1d, Supplemental Table 5). Overall these results indicate that intermediate expansions in C9orf72 may be a genetic risk factor for CBD, and the risk may be additive with higher repeat sizes conferring increased CBD risk.
Figure 1: C9orf72 repeat sizing of CBD.
a) Example capillary electrophoresis traces of CBD cases measured from genomic DNA using a fluorescent PCR assay with genotypes of 2,2 (top), 2,18 (middle) or 9,20 (bottom). b) Summary statistics for repeat size screening of CBD cases (n = 708 alleles) and previously published controls (n = 11,774 alleles)[73]. Fisher’s exact test: OR = 3.59, 95% CI = 1.80 – 6.65, p = 0.000238. c) Allele frequencies of repeat sizes in CBD cases and controls. The number of alleles in the CBD cohort for each size is depicted above the bars. d) Results of Fisher’s exact test between CBD cases and controls at each allele size cutoff. Dotted gray line represents p < 0.05 significance level.
C9orf72 expression from intermediate expansion carriers
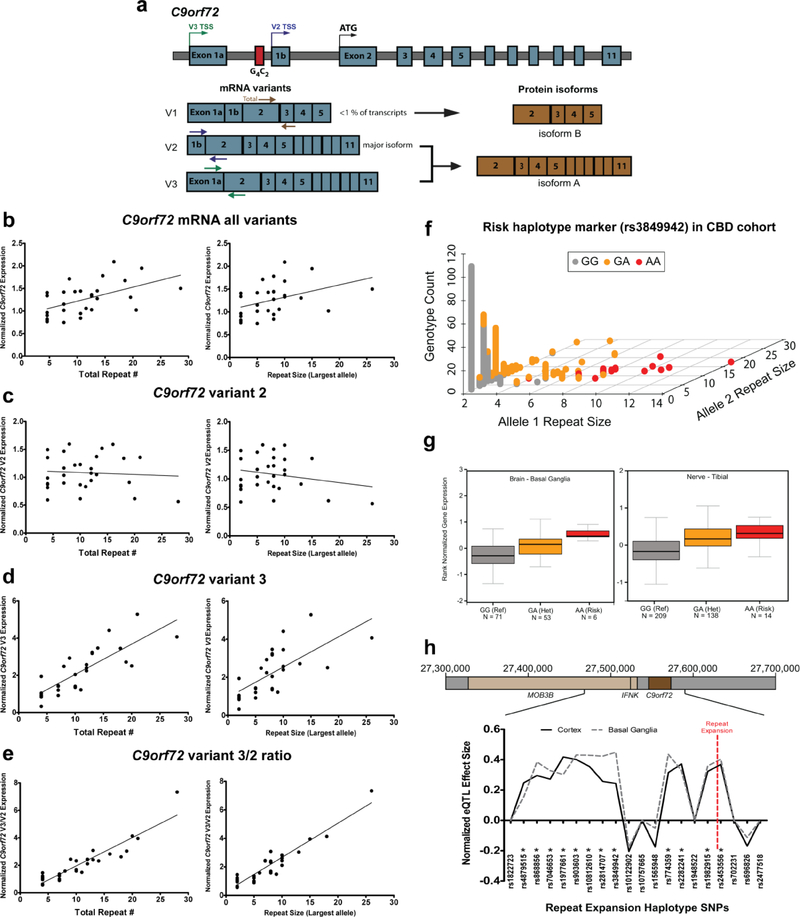
Next, we determined how intermediate repeat expansions affect expression of the C9orf72 gene. Previous studies have shown that large expansions in C9orf72, which cause ALS/FTD, reduces mRNA from the expanded allele[8, 19, 77]. This reduction in expression is often associated with hypermethylation of the C9orf72 promoter region[41, 52, 61, 81]. To test whether intermediate expansions alter C9orf72 DNA methylation and mRNA levels, we analyzed post mortem cerebellum samples from a subset of 47 CBD cases for which frozen tissue was readily available. We measured repeat expansion size, DNA methylation and C9orf72 mRNA levels of the two major isoforms[76] (Figure 2a, V2 and V3 isoforms). In this subgroup analysis, we again observed high frequencies of intermediate repeat expansions between 17 and 29 units (Supplemental Figure 1). Intermediate size expansions did not correlate with hypermethylation of the promoter region, as is observed in full expansion cases (Supplemental Figure 2). Surprisingly, total C9orf72 mRNA levels were positively correlated with total C9orf72 repeat size (Figure 2b left, R2 = 0.276, p = 0.0034).This positive correlation was not observed for the major isoform, V2, which has a transcription start site downstream from the repeat expansion (Figure 2c left, R2 = 0.00496, p = 0.717). In contrast, V3 has a transcription start site upstream from the repeat expansion and shows a robust positive correlation between repeat expansion size and mRNA levels (Figure 2d left, R2 = 0.667, p < 0.0001). Cases with >17 total repeats exhibited ~2.5 to 5 fold increase in variant 3 mRNA compared to cases with shorter repeats. Finally, analysis of the variant 3 to variant 2 ratio also reveals a striking positive correlation with larger repeat sizes (Figure 2e left, R2 = 0.849, p < 0.0001). Similar results were obtained when using the largest repeat allele as opposed to the total C9orf72 repeat length (Figure 2b–d, right). However, total repeat number produced a stronger positive correlation to C9orf72 mRNA levels, supporting an additive effect of repeat size on expression.
Figure 2: Effect of intermediate repeats on C9orf72 expression.
a) Diagram of the C9orf72 locus with mRNA and protein isoforms. Transcription start sites (TSS) are labeled for each mRNA variant on the genomic DNA. Colored arrows on the mRNA depict primers used for RT-qPCR. b) Correlation between total C9orf72 repeat size (left) or largest allele (right) and C9orf72 mRNA levels from all variants measured by RT-qPCR using RNA from CBD patient cerebellum (Total repeats: n = 29, R2 = 0.276, p = 0.0034; Largest allele: n = 29, R2 = 0.1613, p = 0.0308). Data is normalized to samples with 4 total repeats. c) Correlation between total C9orf72 repeat size (left) or largest allele (right) and variant 2 mRNA levels (Total repeats: n = 29, R2 = 0.00496, p = 0.717; Largest allele: n = 29, R2 = 0.04432, p = 0.2730). d) Correlation between total C9orf72 repeat size (left) or largest allele (right) and variant 3 mRNA levels (Total repeats: n = 29, R2 = 0.667, p < 0.0001; Largest allele: n = 29, R2 = 0.4874, p < 0.0001). e) Correlation between total C9orf72 repeat size (left) or largest allele (right) and the variant 3 mRNA to variant 2 mRNA ratio (Total repeats: n = 29, R2 = 0.849, p < 0.0001; Largest allele: n = 29, R2 = 0.8869, p < 0.0001). f) Genotyping results for rs3849942 in the CBD patient genomic DNA cohort (n = 708 alleles). The A allele is associated with intermediate and full length repeat expansions. g) SNP eQTL data from the Gtex project showing the effect of rs3849942 on C9orf72 expression in basal ganglia (left, p = 9.00E-07) and tibial nerve (right, p = 2.80E-16). For box plots, center line indicates median, boxes are upper and lower quartiles, error bars are standard error of the mean. Numbers of samples are indicated below each group. h) Normalized GTEx eQTL effect size from cortex and basal ganglia on C9orf72 expression for all 20 SNPs in the consensus repeat expansion risk haplotype (from Mok et al. 2012[53]). * indicates SNPs with a significant effect on C9orf72 expression. Red dotted line indicates the location of the C9orf72 repeat expansion.
To confirm the positive relationship between C9orf72 repeat size and mRNA expression, we analyzed publicly available SNP eQTL data from the Genotype-Tissue Expression (GTEx) Project[44]. Large C9orf72 repeat expansion mutations invariably occur within a 140kb haplotype block with a defined series of SNPs[49, 53]. The SNP rs3849942 is a commonly used marker for this haplotype block in which the G allele is associated with small non-expanded C9orf72 alleles, and the A allele is associated with both larger intermediate to full repeat expansions[19]. We observe the same trend in our CBD genomic DNA cohort where the rs3849942 A risk allele was associated with larger C9orf72 repeat sizes (Figure 2f). Using rs3849942 as a marker for longer/intermediate repeats (as full expansions are rare in unselected populations), we used GTEx data to determine if this SNP is associated with altered C9orf72 expression. We found that the risk allele is significantly associated with increased C9orf72 expression across numerous tissue types, with the largest effect size occurring in neural tissues (Figure 2g). Expanding the analysis to 20 haplotype defining SNPs[53], we found that 12 of the SNPs on the risk allele are associated with increased C9orf72 expression in both the cortex and basal ganglia (Figure 2h ). In contrast, there were no significant eQTL SNPs that led to a decrease in expression. These results provide additional evidence that intermediate expansions lead to increased C9orf72 expression.
CRISPR Editing of C9orf72 expansion size in iPSCs
In order to confirm the previous findings experimentally and to determine whether the risk haplotype versus the intermediate C9orf72 repeats themselves are driving increased C9orf72 gene expression, CRISPR/cas9 was used to modify the repeat size in previously characterized induced pluripotent stem cells (iPSCs) from a non-diseased individual[46]. A repair template containing either 2 or 28 repeats was used to alter the size of the repeat expansion in iPSCs that normally have alleles of either 2 or 6 repeats (Figure 3a–b; Supplemental Figure 3). Clonal lines were derived by FACS and were screened for changes in repeat size and for the presence of the knocked-in allele by PCR followed by restriction enzyme digest (Figure 3b). Genotype, including repeat size, was confirmed by repeat primed PCR (Figure 3c) and Sanger sequencing (Supplemental Figure 4). Despite using a repair template with 28 repeats, several different iPSC clones with repeat sizes ranging from 23 to 28 were obtained. Additional isogenic iPSC clones were also obtained with two or six C9orf72 repeats.
Figure 3: CRISPR/cas9 knock-in of intermediate repeats into iPSCs.
a) Diagram of C9orf72 genomic locus and double stranded repair templates used for knocking in intermediate repeats. Black triangle represents the CRISPR/cas9 cut site. Black arrows indicate the PCR primers used in (b). Red boxes indicate the G4C2 repeat. Blue boxes indicate the 2 bp substitution included in the repair templates to mutate the Cas9 PAM site and generate a PvuII restriction enzyme site. b) PCR (top) and PvuII digest (bottom) of CRISPR edited iPS cell lines using PCR primers spanning the CRISPR cas9 cut site and repeat expansion. Digestion of the PCR product indicates that HDR occurred. c) Capillary electrophoresis traces of repeat primed PCR from CRISPR edited iPSCs. d-g) RT-qPCR from the indicated variants of C9orf72 from neural progenitor cells differentiated from CRISPR edited iPSCs. Light bars correspond to low repeat lines while black bars correspond to intermediate repeat lines. N = 3 biological replicates per cell line; ANOVA (All variants: F = 18.3, p = 0.0001; Variant 2: F= 2.9, p = 0.08; Variant 3: F = 216.0, p < 0.0001; V2/V3: F = 460.0, p < 0.0001) followed by Bonferroni post hoc test between low and intermediate repeat cell lines. **p < 0.01, ***p < 0.001, ****p < 0.0001. Error bars are standard error of the mean.
Given that both neurons and glial cells are affected in CBD[5], we used the dual SMAD inhibition method[14, 66] to differentiate edited iPSCs with intermediate C9orf72 expansions and isogenic controls with small repeats into neural progenitor cells (NPCs) as determined by uniform expression of the NPC marker Pax6 (Supplemental Figure 5). RT-qPCR of these edited NPC lines showed a remarkably consistent pattern of higher C9orf72 mRNA expression in clones with intermediate C9orf72 repeats compared to isogenic controls with small repeats (Figure 3d). Consistent with the data from CBD post mortem brain tissue, we again saw no change in the amount of variant 2 mRNA (Figure 3e) but a robust increase in variant 3 mRNA levels in all intermediate C9orf72 repeat cell lines (Figure 3f–g). This effect was not an artifact of neuronal differentiation, as undifferentiated iPSCs also exhibited increased C9orf72 mRNA levels (Supplemental Figure 6). Thus, these experiments demonstrate a causal relationship between intermediate C9orf72 expansions and increased C9orf72 mRNA levels.
Intermediate repeat carriers do not have RNA foci or DPR pathology
Full expansion carriers have characteristic neuropathology in post mortem brain and spinal cord tissue that includes nuclear RNA foci of G4C2 transcripts[19] and aggregates of dipeptide repeat proteins (DPRs) that co-stain with p62[6]. Staining for DPRs using p62 antibody or DPR specific antibodies (GA, GP and GR) was carried out on 125 CBD cases in hippocampus and/or cerebellum, including 9 cases with intermediate repeats (repeat sizes 17–29, Figure 4a). Interestingly, all of these cases were negative for the aggregates typically seen in full expansion cases (Figure 4a). Additionally, RNA foci were not detected in CBD cerebellum tissues with intermediate repeats, nor were they detected in cases with low repeat numbers (Figure 4b). In comparison, full expansion FTD/ALS cases had abundant DPR aggregates together with RNA foci in ~18% of nuclei. Thus, the major pathologic hallmarks of full expansion carriers are not present in CBD cases, leading us to hypothesize that the underlying mechanisms of intermediate expansions and CBD risk may be distinct from the large repeat expansions that cause ALS/FTD.
Figure 4: Absence of repeat pathology in intermediate expansion cases.
a) Representative Immunohistochemistry images of non-expanded control (n=4 cases) or non-expanded CBD (n = 116 cases), intermediate expansion CBD (n = 9 cases), and full expansion FTD/ALS (n = 5 cases) post mortem cerebellar samples using anti-GA or p62 antibodies. Intermediate expansion carriers were also negative for GP/GR aggregates. Scale bar 10μm. b) Representative fluorescence in situ hybridization images of non-expanded control (n=4 cases) or non-expanded CBD (n = 12 cases), intermediate expansion CBD (n = 5 cases), and full expansion FTD/ALS (n = 5 cases) post mortem cerebellar samples using a probe complimentary to the G4C2 repeat. Scale bar 5μm.
Elevated C9orf72 protein and global gene expression changes in intermediate repeat cell lines
To test whether C9orf72 protein expression is higher in cells with intermediate repeats, protein expression was measured in isogenic repeat edited NPCs. NPC lines with intermediate repeats exhibited a consistent ~40% increase in C9orf72 protein levels when compared to isogenic low repeat number controls (Figure 5a–b). Thus, intermediate size repeat expansions increase both C9orf72 mRNA and protein levels.
Figure 5: Elevated C9orf72 protein levels and gene expression changes in intermediate repeat NPCs.
a) Western blot of protein expression from CRISPR edited NPCs. N = 2 low repeat and 3 intermediate repeat number cell lines. C9orf72 antibody was validated in HEK293T cells transfected with C9orf72 overexpression plasmid (Lanes 1–2). Representative blot of 2 technical replicates is shown. b) Quantification of C9orf72 protein levels from repeat edited NPCs. Protein expression was normalized to GAPDH as a loading control and the average of 2 technical replicates is reported. N = 2 low repeat and 3 intermediate repeat number cell lines. Two tailed t-test, p = 0.0145. Error bars are standard error of the mean. c) RNA sequencing analysis of 3 low repeat NPC lines (max repeat size 2 or 6) and 3 intermediate repeat (max repeat size 23, 27 or 28) NPC lines. Differentially expressed genes with an adjusted p-value < 0.05 and greater than 2 fold change are depicted as red dots. d-e) Overrepresentation analysis of biological processes on differentially downregulated genes (d) or upregulated genes (e) in NPCs with intermediate repeats. Labels highlight the top 10 most significant biological processes. f) Changes in gene expression of genes involved in vesicular trafficking. All genes shown are significantly differentially expressed (adjusted p < 0.05) between low repeat and intermediate repeat NPCs. g) Changes in gene expression of genes involved in autophagy. All genes shown are significantly differentially expressed (adjusted p < 0.05) between low repeat and intermediate repeat NPCs. h) Comparison of overlap in differentially expressed genes between low repeat NPCs versus intermediate repeat NPCs (this study) or low repeat vs full expansion neurons from patient derived iPSCs from two previous studies [24, 63]. Only genes with adjusted p < 0.05 and fold change > 2 were used in this comparison.
To determine whether overexpression of C9orf72 in the context of intermediate repeats alters expression of other genes, RNA sequencing was performed on 3 edited intermediate NPC lines and 3 low repeat isogenic control lines (Supplemental table 2, Supplmental file 1). We found 1,307 genes significantly downregulated (adjusted p-value < 0.05 and fold change > 2) in intermediate repeat cell lines and 959 genes upregulated (Figure 5c). Next, overrepresentation analysis was performed on gene ontology pathways in the differentially expressed genes. Genes down regulated in intermediate repeat cell lines (Figure 5d) are highly enriched for metabolic processes, especially those involving nutrient catabolism. Genes upregulated in intermediate repeat cell lines (Figure 5e) are enriched in vesicle trafficking and protein degradation pathways including golgi vesicle transport, response to ER stress and autophagy. We then looked in more detail at specific genes within the most highly dysregulated biological processes. When categorizing vesicle trafficking genes according to their function[69], there is modest increase in expression of numerous endosomal/lysomal genes, including C9orf72 (Figure 5f). Interestingly, there is a consistent and sizable decrease in expression of genes involved in secretory pathways in intermediate repeat cell lines, indicating a possible defect in vesicle secretion (Figure 5f). Additionally, key genes involved in autophagy are modestly increased in intermediates (Figure 5g). However, several genes that have been shown to regulate autophagy in response to nutrient starvation[3, 71, 85] and ER stress[78] such as TMEM74 and TMEM74B are markedly downregulated in intermediate repeat cells (Figure 5g), suggesting a defect in autophagy under stress conditions. Finally, we compared the differentially expressed genes in our dataset to previously published[24, 63] gene expression studies between control (low repeat) and full expansion cells derived from ALS/FTD patients. Interestingly, we see a distinct group of dysregulated genes between control and intermediate repeat cells than between control and full expansion cells (Figure 5h), indicating that different processes are affected by intermediate expansions versus full expansions.
Over-expression of C9orf72 impairs starvation-induced autophagy
Next, a cell culture model was used to further validate the observed changes in biological pathways in the RNA sequencing data and determine if these were in fact caused by C9orf72 over expression. Since C9orf72 is known to regulate autophagy[16, 18, 65, 74] and this pathway was altered in the RNA sequencing data, we generated an inducible isogenic HeLa cell line expressing a single-copy of human C9orf72 long isoform without the repeat expansion tagged with GFP and 6xHis-FLAG tag. The transgene expression level is comparable, i.e., 1:1, to that of endogenous C9orf72 and is under the tight control of tetracycline (Supplmental Figure 7 and [31]). Importantly, the transgene also associates with the autophagy initiation complex, thereby confirming that the GFP-tagged C9orf72 recapitulates the known endogenous gene function[31]. As a readout of autophagy, we utilized an LC3 (microtubule-associated protein light chain 3)-based assay (Figure 6a). During autophagosome assembly, LC3-I undergoes lipidation via conjugation with a phosphatidylethanolamine group to become LC3-II [54, 82]. LC3-II is normally rapidly degraded during autophagy via fusion with the lysosome, but blocking lysosome fusion allows for accumulation of LC3-II to serve as a marker of autophagic activity[72]. Immunoblotting in C9orf72 overexpressing cells revealed that basal LC3-II levels are ~20% higher in C9orf72 overexpressing cells compared to parental HeLa cells (lane NGM, normal growth medium, Figure 6b–c), indicating that either autophagosome production is increased or lysomal degradation of LC3-II is reduced under normal conditions. When autophagosome-lysosome fusion was blocked with chloroquine (CQ)[51], LC3-II accumulates at a higher level in CQ-treated condition when compared to their untreated counterparts, and at a similar level between the parental HeLa and C9orf72 overexpressing cells (Lane NGM+CQ, Figure 6b–c). Interestingly, when autophagy was induced using nutrient-deficient medium, EBSS (Earle’s balance salt solution), the accumulation of LC3-II was reduced by ~40% in the C9orf72 overexpressing cells when compared with parental HeLa cells (Lane EBSS+CQ, Figure 6b–c), suggesting that C9orf72 overexpression blocks starvation-induced autophagy. To further corroborate the biochemical findings, a GFP-LC3 stable HeLa cell line[87] was transfected with C9orf72 construct linked via P2A peptide to mCherry to better visualize the transfected cells. Consistent with the immunoblotting results above, ~50% more LC3-II puncta were observed in the C9orf72 overexpressing conditions in normal growth media. (NGM panel, Figure 6d–e). However, there was ~30% reduction of LC3-II puncta in C9orf72 overexpressing cells under starvation (EBSS+CQ panel, Figure 6d–e), again indicating that there is a failure of autophagy initiation in starvation conditions.
To more accurately measure the actual autophagic flux or rate, a time course without and with chloroquine treatment was performed (Figure 7a). With the lysosomes blocked, LC3-II accumulated at a similar rate in C9orf72 overexpressing cells under normal conditions as the parental HeLA cells (Figure 7b–c, top blot). Under starvation conditions, however, the rate of accumulation was markedly reduced (Figure 7b–c, bottom blot) compared to the parental HeLa cells. Taken together, these results suggest that C9orf72 overexpression promotes autophagy under nutrient rich conditions but limits the cell’s ability to induce autophagy during times of nutrient stress.
Discussion
In this study, we demonstrate that intermediate repeat expansions in C9orf72 may be a genetic risk factor for CBD, thus broadening the spectrum of neurodegenerative diseases implicated by this gene. This study highlights the importance of using autopsy confirmed cases to better understand the genetics of neurodegenerative diseases that have similar or overlapping clinical manifestations. Although intermediate repeats are found in only a small percentage of CBD patients, the magnitude of risk conferred by intermediate repeats is comparable or greater than other known genetic risk factors for CBD[34]. This study assembled the most CBD cases yet tested, and due to the rarity of CBD, a replication cohort is not yet available for follow-up analysis. While we established an a priori hypothesis based on the previously established but arbitrary cutoff of 17 repeat units or larger to define intermediate expansion carriers, our findings suggest that both CBD risk and C9orf72 expression increases in an additive manner with increasing repeat length where perhaps C9orf72 intermediate expansions could be defined as at least 10 units. Interestingly, initial reports of C9orf72 repeat mutation cohorts, defined at that time as at least 30 repeats, included one case of autopsy-confirmed CBD[68]. We did not detect any expansions larger than 29 repeats in our CBD cohort, and intermediate expansion carriers did not have the characteristic RNA foci and dipeptide repeat pathology seen in full expansions. Future studies will need to further define the size threshold that determines whether carriers are more likely to develop risk for CBD versus ALS/FTD.
C9orf72 joins a list of other repeat expansion containing genes in which the length of the repeat confers a pleiotropic effect on the disease phenotype. For instance, intermediate length CGG repeats in the gene FMR1 result in the neurodegenerative disease Fragile X-associated tremor/ataxia syndrome (FXTAS) linked to toxicity of repeat RNA[29], whereas full expansions lead to near complete silencing of FMR1 and the neurodevelopmental disorder Fragile X Syndrome[75]. Similarly, intermediate CAG expansions in Ataxin-2 are a risk factor for ALS[25], whereas full expansions cause spinocerebellar ataxia type 2[62]. For C9orf72, large expansions cause ALS/FTD[19, 60], whereas we demonstrate that intermediate expansions confer ~3.5 fold higher risk for CBD. Full expansions mutations in C9orf72 have been associated with reduction of C9orf72 gene expression and a gain of repeat associated pathology including nuclear repeat RNA accumulation and dipeptide repeat protein production. [19, 27, 67, 76] In contrast, we show that intermediate expansions cause an increase in C9orf72 mRNA levels in both CBD post mortem cerebellum and knock-in isogenic neural cell lines. This mechanism is also supported by publicly available SNP eQTL data, which shows that numerous SNPs on the expansion haplotype are associated with increased C9orf72 expression. Thus, it appears that maintaining proper levels of C9orf72 may be necessary for proper neuronal function, with increased expression observed in CBD patients and decreased expression observed in ALS/FTD patients.
C9orf72 is thought to function as Rab guanine exchange factor[38], which participates in a host of vesicle trafficking pathways[55]. C9orf72 has been shown to regulate endocytosis[26], vesicle secretion[4], intracellular trafficking[4, 26, 58, 67] and autophagy[16–18, 74, 79, 83]. Indeed, we observe numerous gene expression changes in all of these vesicle trafficking pathways in knock-in NPCs with intermediate expansions. The large number of significant gene expression changes detected in this study is likely due to the depth of sequencing (~100 million reads/sample) and the use of isogenic CRISPR edited controls, which removes variability due to genetic background. Particularly striking changes were observed in genes involved in vesicle secretion and autophagy. The involvement of autophagy pathways in numerous neurodegenerative diseases has been well documented [55], and increased levels of C9orf72 may enhance CBD risk by disrupting autophagy pathways necessary for responding to stress. C9orf72 has been shown to regulate autophagy initiation through the ULK1 complex[31, 65, 79], and we observe increased expression of autophagy initiation genes in the intermediate repeat knockin NPCs. Interestingly, over expression of C9orf72 seems to increase autophagy initiation under normal conditions, but inhibits autophagy induction under nutrient stress. Several other studies have implicated C9orf72 as regulator of the cellular stress response[2, 16, 43, 47], and we observe dysregulation of genes involved in the ER stress response and nutrient biosynthesis pathways in intermediate repeat knockin NPCs. In particular, the gene TMEM74 and its paralog TMEM74B were high downregulated in C9orf72 NPCs with intermediate repeats. TMEM74 has been shown to be a potent activator of autophagy induced via nutrient starvation[85] and is thought to function via a unique pathway that does not require initiation via the canonical ULK1 autophagy initiation complex[71]. This alternative initiation pathway may be affected by increased C9orf72 levels and could explain why we observe an autophagy defect in C9orf72 over expressing cells under nutrient starvation. Future studies will need to further define the molecular mechanisms by which C9orf72 overexpression alters specific autophagy pathways.
Overall, the findings presented here illustrate the complexity of C9orf72 repeat expansion mutations, with the length of expansion conferring risk for different neurodegenerative diseases via opposing mechanisms. Notably, the proportion of CBD cases with intermediate C9orf72 repeats is still relatively low (~4%) indicating that the overall genetic risk associated with intermediate repeats is modest. Likewise, given that CBD is a rare disease, an otherwise normal individual harboring an intermediate repeat is unlikely to develop CBD. Finally, these results indicate that autophagy pathways are dysregulated by elevated C9orf72 levels and suggest a link between C9orf72 expression and neurodegenerative disease.
Supplementary Material
Acknowledgements
We acknowledge the contributions of the Los Angeles VA Hospital (University of California Los Angeles) and the Harvard Brain Tissue Resource Center (McLean Hospital) for contributing cases to this study. This study was supported by grants from the NIH (R01 NS095793, E.B.L.; R25 GM071745, M.P.; P01 AG017586, E.B.L., V.M.V.D, V.M.-Y.L, J.Q.T., G.S.; P30 AG010124, E.B.L., V.M.V.D, V.M.-Y.L, J.Q.T.; P30 AG10133, B.G.; UG3 NS104095, D.W.D.; U54 NS100693, D.W.D., G.S.; P30 AG012300 C.L.W.; P50 AG023501 and P01 AG019724 W.W.S., B.L.M.; P50 NS038377 and P50 AG05146 J.C.T.; P50 AG025688 M.G.), CurePSP (D.W.D.), the Tau Consortium (D.W.D. and W.W.S.), CBD Solutions (H.L. and K.Y.M.), National Medical Research Council, Singapore (NMRC/OFIRG/0001/2016 and NMRC/OFIRG/0042/2017 to S.C.L.), Ministry of Education, Singapore (MOE2016-T2–1-024 to S.C.L), the Reta Lila Weston Trust (K.Y.M.), the Bluefield Project to Cure FTD (W.W.S.), the UK Medical Research Council (G0400074 to C.M.M.), NIHR Newcastle Biomedical Research Center (C.M.M.), the Alzheimer’s Society and Alzheimer’s Research UK as part of the Brains for Dementia Research project (C.M.M.). The London Neurodegenerative Diseases Brain Bank receives funding from the UK Medical Research Council (MR/L016397/1) and as part of the Brains for Dementia Research programme, jointly funded by Alzheimer’s Research UK and the Alzheimer’s Society. We would also like to acknowledge Beth Dombroski, and EunRan Suh for their assistance, and the patients and families without which this research would not be possible.
Footnotes
Declaration of interests: The authors declare no competing interests.
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
References
- 1.Akimoto C, Forsgren L, Linder J, Birve A, Backlund I, Andersson J, Nilsson AC, Alstermark H, Andersen PM (2013) No GGGGCC-hexanucleotide repeat expansion in C9ORF72 in parkinsonism patients in Sweden. Amyotroph Lateral Scler Front Degener 14:26–29. doi: 10.3109/17482968.2012.725415 [DOI] [PubMed] [Google Scholar]
- 2.Amick J, Tharkeshwar AK, Amaya C, Ferguson SM (2018) WDR41 supports lysosomal response to changes in amino acid availability. Mol Biol Cell 29:2213–2227. doi: 10.1091/mbc.E17-12-0703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antonelli M, Barilà D, Manic G, Brandi R, Sambucci M, Arisi I, Stagni V, D’Onofrio M, Strappazzon F, Vitale I (2017) ATM kinase sustains breast cancer stem-like cells by promoting ATG4C expression and autophagy. Oncotarget 8:21692–21709. doi: 10.18632/oncotarget.15537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aoki Y, Manzano R, Lee Y, Dafinca R, Aoki M, Douglas AGL, Varela MA, Sathyaprakash C, Scaber J, Barbagallo P, Vader P, Mäger I, Ezzat K, Turner MR, Ito N,Gasco S, Ohbayashi N, El Andaloussi S, Takeda S, Fukuda M, Talbot K, Wood MJA (2017) C9orf72 and RAB7L1 regulate vesicle trafficking in amyotrophic lateral sclerosis and frontotemporal dementia. Brain 140:887–897. doi: 10.1093/brain/awx024 [DOI] [PubMed] [Google Scholar]
- 5.Armstrong MJ, Litvan I, Lang AE, Bak TH, Bhatia KP, Borroni B, Boxer AL, Dickson DW, Grossman M, Hallett M, Josephs KA, Kertesz A, Lee SE, Miller BL, Reich SG, Riley DE, Tolosa E, Tröster AI, Vidailhet M, Weiner WJ (2013) Criteria for the diagnosis of corticobasal degeneration. Neurology 80:496–503. doi: 10.1212/WNL.0b013e31827f0fd1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ash PEA, Bieniek KF, Gendron TF, Caulfield T, Lin WL, DeJesus-Hernandez M, Van Blitterswijk MM, Jansen-West K, Paul JW, Rademakers R, Boylan KB, Dickson DW, Petrucelli L (2013) Unconventional Translation of C9ORF72 GGGGCC Expansion Generates Insoluble Polypeptides Specific to c9FTD/ALS. Neuron 77:639–646. doi: 10.1016/j.neuron.2013.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beck J, Poulter M, Hensman D, Rohrer JD, Mahoney CJ, Adamson G, Campbell T, Uphill J, Borg A, Fratta P, Orrell RW, Malaspina A, Rowe J, Brown J, Hodges J, Sidle K, Polke JM, Houlden H, Schott JM, Fox NC, Rossor MN, Tabrizi SJ, Isaacs AM, Hardy J, Warren JD, Collinge J, Mead S (2013) Large C9orf72 hexanucleotide repeat expansions are seen in multiple neurodegenerative syndromes and are more frequent than expected in the UK population. Am J Hum Genet 92:345–353. doi: 10.1016/j.ajhg.2013.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belzil VV, Bauer PO, Prudencio M, Gendron TF, Stetler CT, Yan IK, Pregent L, Daughrity L, Baker MC, Rademakers R, Boylan K, Patel TC, Dickson DW, Petrucelli L (2013) Reduced C9orf72 gene expression in c9FTD/ALS is caused by histone trimethylation, an epigenetic event detectable in blood. Acta Neuropathol 126:895–905. doi: 10.1007/s00401-013-1199-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bender R, Lange S (2001) Adjusting for multiple testing - When and how? J Clin Epidemiol 54:343–349. doi: 10.1016/S0895-4356(00)00314-0 [DOI] [PubMed] [Google Scholar]
- 10.van Blitterswijk M, DeJesus-Hernandez M, Niemantsverdriet E, Murray ME, Heckman MG, Diehl NN, Brown PH, Baker MC, Finch NCA, Bauer PO, Serrano G, Beach TG, Josephs KA, Knopman DS, Petersen RC, Boeve BF, Graff-Radford NR, Boylan KB, Petrucelli L, Dickson DW, Rademakers R (2013) Association between repeat sizes and clinical and pathological characteristics in carriers of C9ORF72 repeat expansions (Xpansize-72): A cross-sectional cohort study. Lancet Neurol 12:978–988. doi: 10.1016/S1474-4422(13)70210-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boeve BF, Maraganore DM, Parisi JE, Ahlskog JE, Graff-Radford N, Caselli RJ, Dickson DW, Kokmen E, Petersen RC (1999) Pathologic heterogeneity in clinically diagnosed corticobasal degeneration. Neurology 53:795–800. doi: 10.1212/WNL.53.4.795 [DOI] [PubMed] [Google Scholar]
- 12.Burberry A, Suzuki N, Wang JY, Moccia R, Mordes DA, Stewart MH, Suzuki-Uematsu S, Ghosh S, Singh A, Merkle FT, Koszka K, Li QZ, Zon L, Rossi DJ, Trowbridge JJ, Notarangelo LD, Eggan K (2016) Loss-of-function mutations in the C9ORF72 mouse ortholog cause fatal autoimmune disease. Sci Transl Med 8:93. doi: 10.1126/scitranslmed.aaf6038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cannas A, Solla P, Borghero G, Floris GL, Chio A, Mascia MM, Modugno N, Muroni A, Orofino G, Di Stefano F, Calvo A, Moglia C, Restagno G, Meloni M, Farris R, Ciaccio D, Puddu R, Vacca MI, Melis R, Murru MR, Tranquilli S, Corongiu D, Rolesu M, Cuccu S, Marrosu MG, Marrosu F (2015) C9ORF72 intermediate repeat expansion in patients affected by atypical Parkinsonian syndromes or parkinson’s disease complicated by psychosis or dementia in a Sardinian population. J Neurol 262:2498–2503. doi: 10.1007/s00415-015-7873-6 [DOI] [PubMed] [Google Scholar]
- 14.Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L (2009) Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol 27:275–280. doi: 10.1038/nbt.1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chew J, Gendron TF, Prudencio M, Sasaguri H, Zhang YJ, Castanedes-Casey M, Lee CW, Jansen-West K, Kurti A, Murray ME, Bieniek KF, Bauer PO, Whitelaw EC, Rousseau L, Stankowski JN, Stetler C, Daughrity LM, Perkerson EA, Desaro P, Johnston A, Overstreet K, PL (2015) C9ORF72 repeat expansions in mice cause TDP-43 pathology, neuronal loss, and behavioral deficits. Science (80- ) 348:1151–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chitiprolu M, Jagow C, Tremblay V, Bondy-Chorney E, Paris G, Savard A, Palidwor G, Barry FA, Zinman L, Keith J, Rogaeva E, Robertson J, Lavallée-Adam M, Woulfe J, Couture J-F, Côté J, Gibbings D (2018) A complex of C9ORF72 and p62 uses arginine methylation to eliminate stress granules by autophagy. Nat Commun 9:2794. doi: 10.1038/s41467-018-05273-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ciura S, Sellier C, Campanari ML, Charlet-Berguerand N, Kabashi E (2016) The most prevalent genetic cause of ALS-FTD, C9orf72 synergizes the toxicity of ATXN2 intermediate polyglutamine repeats through the autophagy pathway. Autophagy 12:1406–1408. doi: 10.1080/15548627.2016.1189070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corbier C, Sellier C (2016) C9ORF72 is a GDP/GTP exchange factor for Rab8 and Rab39 and regulates autophagy. Small GTPases 00–00. doi: 10.1080/21541248.2016.1212688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dejesus-hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Gilmer F, Adamson J, Kouri N, Wojtas A, Sengdy P, Hsiung GR, Karydas A, Seeley WW, Josephs KA, Geschwind DH, Wszolek ZK, Feldman H, Petersen R, Miller BL, Dickson D, Boylan K (2011) Expanded GGGGCC hexanucleotide repeat in non-coding region of C9ORF72 causes chromosome 9p-linked frontotemporal dementia and amyotrophic lateral sclerosis. Neuron 72:245–256. doi: 10.1016/j.neuron.2011.09.011.Expanded [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeJesus-Hernandez M, Rayaprolu S, Soto-Ortolaza AI, Rutherford NJ, Heckman MG, Traynor S, Strongosky A, Graff-Radford N, Van Gerpen J, Uitti RJ, Shih JJ, Lin SC, Wszolek ZK, Rademakers R, Ross OA (2013) Analysis of the C9orf72 repeat in Parkinson’s disease, essential tremor and restless legs syndrome. Park Relat Disord 19:198–201. doi: 10.1016/j.parkreldis.2012.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dickson DW, Bergeron C, Chin SS, Duyckaerts C, Horoupian D, Ikeda K, Jellinger K, Lantos PL, Lippa CF, Mirra SS, Tabaton M, Vonsattel JP, Wakabayashi K, Litvan I (2002) Office of rare diseases neuropathologic criteria for corticobasal degeneration. J Neuropathol Exp Neurol 61:935–946. doi: 10.1093/jnen/61.11.935 [DOI] [PubMed] [Google Scholar]
- 22.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. doi: 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dobson-Stone C, Hallupp M, Loy CT, Thompson EM, Haan E, Sue CM, Panegyres PK, Razquin C, Seijo-Martínez M, Rene R, Gascon J, Campdelacreu J, Schmoll B, Volk AE, Brooks WS, Schofield PR, Pastor P, Kwok JBJ (2013) C9ORF72 Repeat Expansion in Australian and Spanish Frontotemporal Dementia Patients. PLoS One 8. doi: 10.1371/journal.pone.0056899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Donnelly CJ, Zhang P, Pham JT, Heusler AR, Mistry NA, Vidensky S, Daley EL, Poth EM, Fines DM, Maragakis N, Tienari PJ, Petrucelli L, Traynor BJ, Wang J, Rigo F, Bennett CF, Blackshaw S (2013) RNA Toxicity from the ALS/FTD C9ORF72 Expansion Is Mitigated by Antisense Intervention. Neuron 80:415–428. doi: 10.1016/j.neuron.2013.10.015.RNA [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elden AC, Kim H, Hart MP, Chen-plotkin AS, Johnson S, Fang X, Armakola M, Geser F, Greene R, Min M, Padmanabhan A, Clay D, Mccluskey L, Elman L, Juhr D, Gruber J, Rüb U, Auburger G, Trojanowski JQ, Lee VM, Van Deerlin M, Bonini NM, Gitler AD (2011) Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature 466:1069–1075. doi: 10.1038/nature09320.Ataxin-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farg MA, Sundaramoorthy V, Sultana JM, Yang S, Atkinson RAK, Levina V, Halloran MA, Gleeson PA, Blair IP, Soo KY, King AE, Atkin JD (2014) C9ORF72, implicated in amytrophic lateral sclerosis and frontotemporal dementia, regulates endosomal trafficking. Hum Mol Genet 23:3579–3595. doi: 10.1093/hmg/ddu068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frick P, Sellier C, Mackenzie IRA, Cheng C, Tahraoui-bories J, Martinat C, Pasterkamp RJ, Prudlo J, Edbauer D, Oulad-abdelghani M, Feederle R (2018) Novel antibodies reveal presynaptic localization of C9orf72 protein and reduced protein levels in C9orf72 mutation carriers. Acta Neuropathol Commun 6:1–17. doi: 10.1186/s40478-018-0579-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gijselinck I, Van Mossevelde S, van der Zee J, Sieben A, Engelborghs S, De Bleecker J, Ivanoiu A, Deryck O, Edbauer D, Zhang M, Heeman B, Bäumer V, Van den Broeck M, Mattheijssens M, Peeters K, Rogaeva E, De Jonghe P, Cras P, Martin J-J, de Deyn PP, Cruts M, Van Broeckhoven C (2015) The C9orf72 repeat size correlates with onset age of disease, DNA methylation and transcriptional downregulation of the promoter. Mol Psychiatry 21:1–13. doi: 10.1038/mp.2015.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hagerman RJ, Leehey M, Heinrichs W, Tassone F, Wilson R, Hills J, Grigsby J, Gage B, Hagerman PJ (2001) Intention tremor, parkinsonism, and generalized brain atrophy in male carriers of fragile X. Neurology 57:127–130. doi: 10.1212/WNL.57.1.127 [DOI] [PubMed] [Google Scholar]
- 30.Harms MB, Neumann D, Benitez BA, Cooper B, Carrell D, Racette BA, Perlmutter JS, Goate A, Cruchaga C (2013) Parkinson disease is not associated with C9ORF72 repeat expansions. Neurobiol Aging 34:1519. doi: 10.1016/j.neurobiolaging.2012.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ho WY, Tai YK, Chang J-C, Liang J, Tyan S-H, Chen S, Guan J-L, Zhou H, Shen H-M, Koo E, Ling S-C (2019) The ALS-FTD-linked gene product, C9orf72, regulates neuronal morphogenesis via autophagy. Autophagy 1–16. doi: 10.1080/15548627.2019.1569441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiao B, Guo J, Wang Y, Yan X, Zhou L, Liu X, Zhang F, Zhou Y, Xia K, Tang B, Shen L (2013) C9orf72 mutation is rare in Alzheimer’s disease, Parkinson’s disease, and essential tremor in China. Front Cell Neurosci 7:164. doi: 10.3389/fncel.2013.00164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kouri N, Murray ME, Hassan A, Rademakers R, Uitti RJ, Boeve BF, Graff-Radford NR, Wszolek ZK, Litvan I, Josephs KA, Dickson DW (2011) Neuropathological features of corticobasal degeneration presenting as corticobasal syndrome or Richardson syndrome. Brain 134:3264–3275. doi: 10.1093/brain/awr234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kouri N, Ross OA, Dombroski B, Younkin CS, Serie DJ, Soto-Ortolaza A, Baker M, Finch NCA, Yoon H, Kim J, Fujioka S, McLean CA, Ghetti B, Spina S, Cantwell LB, Farlow MR, Grafman J, Huey ED, Ryung Han M, Beecher S, Geller ET, Kretzschmar HA, Roeber S, Gearing M, Juncos JL, Vonsattel JPG, Van Deerlin VM, Grossman M, Hurtig HI, Gross RG, Arnold SE, Trojanowski JQ, Lee VM, Wenning GK, White CL, Höglinger GU, Müller U, Devlin B, Golbe LI, Crook J, Parisi JE, Boeve BF, Josephs KA, Wszolek ZK, Uitti RJ, Graff-Radford NR, Litvan I, Younkin SG, Wang L-S, Ertekin-Taner N, Rademakers R, Hakonarsen H, Schellenberg GD, Dickson DW (2015) Genomewide association study of corticobasal degeneration identifies risk variants shared with progressive supranuclear palsy. Nat Commun 6:7247. doi: 10.1038/ncomms8247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee EB, Lee VMY, Trojanowski JQ (2012) Gains or losses: Molecular mechanisms of TDP43-mediated neurodegeneration. Nat Rev Neurosci 13:38–50. doi: 10.1038/nrn3121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee YB, Chen HJ, Peres JN, Gomez-Deza J, Attig J, Štalekar M, Troakes C, Nishimura AL, Scotter EL, Vance C, Adachi Y, Sardone V, Miller JW, Smith BN, Gallo JM, Ule J, Hirth F, Rogelj B, Houart C, Shaw CE (2013) Hexanucleotide repeats in ALS/FTD form length-dependent RNA Foci, sequester RNA binding proteins, and are neurotoxic. Cell Rep 5:1178–1186. doi: 10.1016/j.celrep.2013.10.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lesage S, Le Ber I, Condroyer C, Broussolle E, Gabelle A, Thobois S, Pasquier F, Mondon K, Dion PA, Rochefort D, Rouleau GA, Dürr A, Brice A (2013) C9orf72 repeat expansions are a rare genetic cause of parkinsonism. Brain 136:385–391. doi: 10.1093/brain/aws357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levine TP, Daniels RD, Gatta AT, Wong LH, Hayes MJ (2013) The product of C9orf72, a gene strongly implicated in neurodegeneration, is structurally related to DENN Rab-GEFs. Bioinformatics 29:499–503. doi: 10.1093/bioinformatics/bts725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ling H, O’Sullivan SS, Holton JL, Revesz T, Massey LA, Williams DR, Paviour DC, Lees AJ (2010) Does corticobasal degeneration exist? A clinicopathological re-evaluation. Brain 133:2045–2057. doi: 10.1093/brain/awq123 [DOI] [PubMed] [Google Scholar]
- 40.Litvan I, Agid Y, Goetz C, Jankovic J, Wenning GK, Brandel JP, Lai EC, Verny M, Ray-Chaudhuri K, McKee a, Jellinger K, Pearce RK, Bartko JJ (1997) Accuracy of the clinical diagnosis of corticobasal degeneration: a clinicopathologic study. Neurology 48:119–25. doi: 10.1212/WNL.48.1.119 [DOI] [PubMed] [Google Scholar]
- 41.Liu EY, Russ J, Wu K, Neal D, Suh E, McNally AG, Irwin DJ, Van Deerlin VM, Lee EB (2014) C9orf72 hypermethylation protects against repeat expansion-associated pathology in ALS/FTD. Acta Neuropathol 128:525–541. doi: 10.1007/s00401-014-1286-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y, Pattamatta A, Zu T, Reid T, Bardhi O, Borchelt DR, Yachnis AT, Ranum LPW (2016) C9orf72 BAC Mouse Model with Motor Deficits and Neurodegenerative Features of ALS/FTD. Neuron 90:521–534. doi: 10.1016/j.neuron.2016.04.005 [DOI] [PubMed] [Google Scholar]
- 43.Liu Y, Wang T, Ji YJ, Johnson K, Liu H, Johnson K, Bailey S, Suk Y, Lu Y-N, Liu M, Wang J (2018) A C9orf72-CARM1 axis regulates lipid metabolism under glucose starvation-induced nutrient stress. Genes Dev 32:1380–1397. doi: 10.1101/gad.315564.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, Shad S, Hasz R, Walters G, Garcia F, Young N, Foster B, Moser M, Karasik E, Gillard B, Ramsey K, Sullivan S, Bridge J, Magazine H, Syron J, Fleming J, Siminoff L, Traino H, Mosavel M, Barker L, Jewell S, Rohrer D, Maxim D, Filkins D, Harbach P, Cortadillo E, Berghuis B, Turner L, Hudson E, Feenstra K, Sobin L, Robb J, Branton P, Korzeniewski G, Shive C, Tabor D, Qi L, Groch K, Nampally S, Buia S, Zimmerman A, Smith A, Burges R, Robinson K, Valentino K, Bradbury D, Cosentino M, Diaz-Mayoral N, Kennedy M, Engel T, Williams P, Erickson K, Ardlie K, Winckler W, Getz G, DeLuca D, MacArthur Daniel, Kellis M, Thomson A, Young T, Gelfand E, Donovan M, Meng Y, Grant G, Mash D, Marcus Y, Basile M, Liu J, Zhu J, Tu Z, Cox NJ, Nicolae DL, Gamazon ER, Im HK, Konkashbaev A, Pritchard J, Stevens M, Flutre T, Wen X, Dermitzakis ET, Lappalainen T, Guigo R, Monlong J, Sammeth M, Koller D, Battle A, Mostafavi S, McCarthy M, Rivas M, Maller J, Rusyn I, Nobel A, Wright F, Shabalin A, Feolo M, Sharopova N, Sturcke A, Paschal J, Anderson JM, Wilder EL, Derr LK, Green ED, Struewing JP, Temple G, Volpi S, Boyer JT, Thomson EJ, Guyer MS, Ng C, Abdallah A, Colantuoni D, Insel TR, Koester SE, Roger Little A, Bender PK, Lehner T, Yao Y, Compton CC, Vaught JB, Sawyer S, Lockhart NC, Demchok J, Moore HF (2013) The Genotype-Tissue Expression (GTEx) project. Nat Genet 45:580–585. doi: 10.1038/ng.2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maguire JA, Gagne AL, Jobaliya CD, Gandre-Babbe S, Gadue P, French DL (2016) Generation of human control iPS cell line CHOPWT10 from healthy adult peripheral blood mononuclear cells. Stem Cell Res 16:338–341. doi: 10.1016/j.scr.2016.02.017 [DOI] [PubMed] [Google Scholar]
- 47.Maharjan N, Künzli C, Buthey K, Saxena S (2017) C9ORF72 Regulates Stress Granule Formation and Its Deficiency Impairs Stress Granule Assembly, Hypersensitizing Cells to Stress. Mol Neurobiol 54:3062–3077. doi: 10.1007/s12035-016-9850-1 [DOI] [PubMed] [Google Scholar]
- 48.Mahoney CJ, Beck J, Rohrer JD, Lashley T, Mok K, Shakespeare T, Yeatman T, Warrington EK, Schott JM, Fox NC, Rossor MN, Hardy J, Collinge J, Revesz T, Mead S, Warren JD (2012) Frontotemporal dementia with the C9ORF72 hexanucleotide repeat expansion: Clinical, neuroanatomical and neuropathological features. Brain 135:736–750. doi: 10.1093/brain/awr361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Majounie E, Renton AE, Mok K, Dopper EGP, Waite A, Rollinson S, Chiò A, Restagno G, Nicolaou N, Simon-Sanchez J, van Swieten JC, Abramzon Y, Johnson JO, Sendtner M, Pamphlett R, Orrell RW, Mead S, Sidle KC, Houlden H, Rohrer JD, Morrison KE, Pall H, Talbot K, Ansorge O, Hernandez DG, Arepalli S, Sabatelli M, Mora G, Corbo M, Giannini F, Calvo A, Englund E, Borghero G, Floris GL, Remes AM, Laaksovirta H, McCluskey L, Trojanowski JQ, Van Deerlin VM, Schellenberg GD, Nalls MA, Drory VE, Lu CS, Yeh TH, Ishiura H, Takahashi Y, Tsuji S, Le Ber I, Brice A, Drepper C, Williams N, Kirby J, Shaw P, Hardy J, Tienari PJ, Heutink P, Morris HR, Pickering-Brown S, Traynor BJ (2012) Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: A cross-sectional study. Lancet Neurol 11:323–330. doi: 10.1016/S1474-4422(12)70043-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17:10–12 [Google Scholar]
- 51.Mauthe M, Orhon I, Rocchi C, Zhou X, Luhr M, Hijlkema K-J, Coppes RP, Engedal N, Mari M, Reggiori F (2018) Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy 14:1435–1455. doi: 10.1080/15548627.2018.1474314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McMillan CT, Russ J, Wood EM, Irwin DJ, Grossman M, Mccluskey L, Elman L, Van Deerlin V, Lee EB (2015) C9orf72 promoter hypermethylation is neuroprotective: Neuroimaging and neuropathologic evidence. Neurology 84:1622–1630. doi: 10.1212/WNL.0000000000001495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mok K, Traynor BJ, Schymick J, Tienari PJ, Laaksovirta H, Peuralinna T, Myllykangas L, Chiò A, Shatunov A, Boeve BF, Boxer AL, DeJesus-Hernandez M, Mackenzie IR, Waite A, Williams N, Morris HR, Simón-Sánchez J, van Swieten JC, Heutink P, Restagno G, Mora G, Morrison KE, Shaw PJ, Rollinson PS, Al-Chalabi A, Rademakers R, Pickering-Brown S, Orrell RW, Nalls MA, Hardy J (2012) The chromosome 9 ALS and FTD locus is probably derived from a single founder. Neurobiol Aging 33:209.e3–209.e8. doi: 10.1016/j.neurobiolaging.2011.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nakatogawa H, Ichimura Y, Ohsumi Y (2007) Atg8, a Ubiquitin-like Protein Required for Autophagosome Formation, Mediates Membrane Tethering and Hemifusion. Cell 130:165–178. doi: 10.1016/j.cell.2007.05.021 [DOI] [PubMed] [Google Scholar]
- 55.Nassif M, Woehlbier U, Manque PA (2017) The enigmatic role of C9ORF72 in autophagy. Front Neurosci 11:1–10. doi: 10.3389/fnins.2017.00442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nuytemans K, Bademci G, Kohli MM, Beecham GW, Wang L, Young JI, Nahab F, Martin ER, Gilbert JR, Benatar M, Haines JL, Scott WK, Züchner S, Pericak-Vance MA, Vance JM (2013) C9orf72 intermediate repeat copies are a significant risk factor for parkinson disease. Ann Hum Genet 77:351–363. doi: 10.1111/ahg.12033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nuytemans K, Inchausti V, Beecham GW, Wang L, Dickson DW, Trojanowski JQ, Lee VMY, Mash DC, Frosch MP, Foroud TM, Honig LS, Montine TJ, Dawson TM, Martin ER, Scott WK, Vance JM (2014) Absence of C9ORF72 expanded or intermediate repeats in autopsy-confirmed Parkinson’s disease. Mov Disord 29:827–830. doi: 10.1002/mds.25838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.O’Rourke JG, Bogdanik L, Yáñez A, Lall D, Wolf AJ, Muhammad AKMG, Ho R, Carmona S, Vit JP, Zarrow J, Kim KJ, Bell S, Harms MB, Miller TM, Dangler CA, Underhill DM, Goodridge HS, Lutz CM, Baloh RH (2016) C9orf72 is required for proper macrophage and microglial function in mice. Science (80- ) 351:1324–1329. doi: 10.1126/science.aaf1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F (2013) Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8:2281–2308. doi: 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Renton A, Majounie E, Waite A, Simón-Sánchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, Kalimo H, Paetau A, Abramzon Y, Remes AM, Kaganovich A, Scholz SW, Duckworth J, Ding J, Harmer DW, Hernandez DG, Johnson JO, Mok K, Ryten M, Trabzuni D, Guerreiro RJ, Orrell RW, Neal J, Murray A, Pearson J, Jansen IE, Sondervan D, Seelaar H, Blake D, Young K, Halliwell N,Callister JB, Toulson G, Richardson A, Gerhard A, Snowden J, Mann D, Neary D, Nalls MA, Peuralinna T, Jansson L, Isoviita VM, Kaivorinne AL, Hölttä-Vuori M, Ikonen E, Sulkava R, Benatar M, Wuu J, Chiò A, Restagno G, Borghero G, Sabatelli M, Heckerman D, Rogaeva E, Zinman L, Rothstein JD, Sendtner M, Drepper C, Eichler EE, Alkan C, Abdullaev Z, Pack SD, Dutra A, Pak E, Hardy J, Singleton A, Williams NM, Heutink P, Pickering-Brown S, Morris HR, Tienari PJ, Traynor BJ (2011) A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72:257–268. doi: 10.1016/j.neuron.2011.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Russ J, Liu EY, Wu K, Neal D, Suh ER, Irwin DJ, McMillan CT, Harms MB, Cairns NJ, Wood EM, Xie SX, Elman L, McCluskey L, Grossman M, Van Deerlin VM, Lee EB (2015) Hypermethylation of repeat expanded C9orf72 is a clinical and molecular disease modifier. Acta Neuropathol 129:39–52. doi: 10.1007/s00401-014-1365-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sanpei K, Takano H, Igarashi S, Sato T, Oyake M, Sasaki H, Wakisaka A, Tashiro K, Ishida Y, Ikeuchi T, Koide R, Saito M, Sato A, Tanaka T, Hanyu S, Takiyama Y, Nishizawa M, Shimizu N, Nomura Y, Segawa M, Iwabuchi K, Eguchi I, Tanaka H, Takahashi H, Tsuji S (1996) Identification of the spinocerebellar ataxia type 2 gene using a direct identification of repeat expansion and cloning technique, DIRECT. Nat Genet 14:277–284. doi: 10.1038/ng1196-277 [DOI] [PubMed] [Google Scholar]
- 63.Sareen D, O’Rourke JG, Meera P, Muhammad AKMG, Grant S, Simpkinson M, Bell S, Carmona S, Ornelas L, Sahabian A, Gendron T, Petrucelli L, Baughn M, Ravits J, Harms MB, Rigo F, Bennett CF, Otis TS, Svendsen CN, Baloh RH (2013) Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Sci Transl Med 5:208ra149. doi: 10.1126/scitranslmed.3007529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schottlaender LV, Polke JM, Ling H, MacDoanld ND, Tucci A, Nanji T, Pittman A, de Silva R, Holton JL, Revesz T, Sweeney MG, Singleton AB, Lees AJ, Bhatia KP, Houlden H (2015) The analysis of C9orf72 repeat expansions in a large series of clinically and pathologically diagnosed cases with atypical parkinsonism. Neurobiol Aging 36:1221.e1–1221.e6. doi: 10.1016/j.neurobiolaging.2014.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sellier C, Campanari M-L, Julie Corbier C, Gaucherot A, Kolb-Cheynel I, Oulad-Abdelghani M, Ruffenach F, Page A, Ciura S, Kabashi E, Charlet-Berguerand N (2016) Loss of C9ORF72 impairs autophagy and synergizes with polyQ Ataxin-2 to induce motor neuron dysfunction and cell death. EMBO J 35:1–22. doi: 10.15252/embj.201593350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shi Y, Kirwan P, Livesey FJ (2012) Directed differentiation of human pluripotent stem cells to cerebral cortex neurons and neural networks. Nat Protoc 7:1836–1846. doi: 10.1038/nprot.2012.116 [DOI] [PubMed] [Google Scholar]
- 67.Shi Y, Lin S, Staats KA, Li Y, Chang W-H, Hung S-T, Hendricks E, Linares GR, Wang Y, Son EY, Wen X, Kisler K, Wilkinson B, Menendez L, Sugawara T, Woolwine P, Huang M, Cowan MJ, Ge B, Koutsodendris N, Sandor KP, Komberg J, Vangoor VR, Senthilkumar K, Hennes V, Seah C, Nelson AR, Cheng T-Y, Lee S-JJ, August PR, Chen JA, Wisniewski N, Hanson-Smith V, Belgard TG, Zhang A, Coba M, Grunseich C, Ward ME, van den Berg LH, Pasterkamp RJ, Trotti D, Zlokovic BV, Ichida JK (2018) Haploinsufficiency leads to neurodegeneration in C9ORF72 ALS/FTD human induced motor neurons. Nat Med. doi: 10.1038/nm.4490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Snowden JS, Rollinson S, Thompson JC, Harris JM, Stopford CL, Richardson AMT, Jones M, Gerhard A, Davidson YS, Robinson A, Gibbons L, Hu Q, DuPlessis D, Neary D, Mann DMA, Pickering-Brown SM (2012) Distinct clinical and pathological characteristics of frontotemporal dementia associated with C9ORF72 mutations. Brain 135:693–708. doi: 10.1093/brain/awr355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stenmark H (2009) Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol 10:513–525. doi: 10.1038/nrm2728 [DOI] [PubMed] [Google Scholar]
- 70.Suh ER, Lee EB, Neal D, Wood EM, Toledo JB, Rennert L, Irwin DJ, McMillan CT, Krock B, Elman LB, McCluskey LF, Grossman M, Xie SX, Trojanowski JQ, Van Deerlin VM (2015) Semi-automated quantification of C9orf72 expansion size reveals inverse correlation between hexanucleotide repeat number and disease duration in frontotemporal degeneration. Acta Neuropathol 130:363–372. doi: 10.1007/s00401-015-1445-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sun Y, Chen Y, Zhang J, Cao L, He M, Liu X, Zhao N, Yin A, Huang H, Wang L (2017) TMEM74 promotes tumor cell survival by inducing autophagy via interactions with ATG16L1 and ATG9A. Cell Death Dis 8:e3031. doi: 10.1038/cddis.2017.370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tanida I, Minematsu-Ikeguchi N, Ueno T, Kominami E (2005) Lysosomal Turnover, but Not a Cellular Level, of Endogenous LC3 is a Marker for Autophagy. Autophagy 1:84–91. doi: 10.4161/auto.1.2.1697 [DOI] [PubMed] [Google Scholar]
- 73.Theuns J, Verstraeten A, Sleegers K, Wauters E, Gijselinck I, Smolders S, Crosiers D, Corsmit E, Elinck E, Sharma M, Kruger R, Lesage S, Brice A, Chung SJ, Kim M-J, Kim YJ, Ross OA, Wszolek ZK, Rogaeva E, Xi Z, Lang AE, Klein C, Weissbach A, Mellick GD, Silburn PA, Hadjigeorgiou GM, Dardiotis E, Hattori N, Ogaki K, Tan E-K, Zhao Y, Aasly J, Valente EM, Petrucci S, Annesi G, Quattrone A, Ferrarese C, Brighina L, Deutschlander A, Puschmann A, Nilsson C, Garraux G, LeDoux MS, Pfeiffer RF, Boczarska-Jedynak M, Opala G, Maraganore DM, Engelborghs S, De Deyn PP, Cras P, Cruts M, Van Broeckhoven C (2014) Global investigation and meta-analysis of the C9orf72 (G4C2)n repeat in Parkinson disease. Neurology 83:1906–1913. doi: 10.1212/WNL.0000000000001012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ugolino J, Ji YJ, Conchina K, Chu J, Nirujogi RS, Pandey A, Brady NR, Hamacher-Brady A, Wang J (2016) Loss of C9orf72 Enhances Autophagic Activity via Deregulated mTOR and TFEB Signaling. PLoS Genet 12. doi: 10.1371/journal.pgen.1006443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Verkerk AJMH, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DPA, Pizzuti A, Reiner O, Richards S, Victoria MF, Zhang F, Eussen BE, van Ommen GJB, Blonden LAJ, Riggins GJ, Chastain JL, Kunst CB, Galjaard H, Thomas Caskey C, Nelson DL, Oostra BA, Warran ST (1991) Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell 65:905–914. doi: 10.1016/0092-8674(91)90397-H [DOI] [PubMed] [Google Scholar]
- 76.Viodé A, Fournier C, Camuzat A, Fenaille F, Latouche M, Elahi F, Ber I Le, Junot C, Lamari F, Anquetil V, Becher F (2018) New Antibody-Free Mass Spectrometry-Based Quantification Reveals That C9ORF72 Long Protein Isoform Is Reduced in the Frontal Cortex of Hexanucleotide-Repeat Expansion Carriers. Front Neurosci 12:1–11. doi: 10.3389/fnins.2018.00589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Waite AJ, Bäumer D, East S, Neal J, Morris HR, Ansorge O, Blake DJ (2014) Reduced C9orf72 protein levels in frontal cortex of amyotrophic lateral sclerosis and frontotemporal degeneration brain with the C9ORF72 hexanucleotide repeat expansion. Neurobiol Aging 35:1779.e5–1779.e13. doi: 10.1016/j.neurobiolaging.2014.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang N, Tan HY, Li S, Feng Y (2017) Atg9b deficiency suppresses autophagy and potentiates endoplasmic reticulum stress-associated hepatocyte apoptosis in hepatocarcinogenesis. Theranostics 7:2325–2338. doi: 10.7150/thno.18225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Webster CP, Smith EF, Bauer CS, Moller A, Hautbergue GM, Ferraiuolo L, Myszczynska MA, Higginbottom A, Walsh MJ, Whitworth AJ, Kaspar BK, Meyer K, Shaw PJ, Grierson AJ, De Vos KJ (2016) The C9orf72 protein interacts with Rab1a and the ULK1 complex to regulate initiation of autophagy. EMBO J 35:1656–1676. doi: 10.15252/embj.201694401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xi Z, Zinman L, Grinberg Y, Moreno D, Sato C, Bilbao JM, Ghani M, Hernández I, Ruiz A, Boada M, Morón FJ, Lang AE, Marras C, Bruni A, Colao R, Maletta RG, Puccio G, Rainero I, Pinessi L, Galimberti D, Morrison KE, Moorby C, Stockton JD, Masellis M, Black SE, Hazrati L-N, Liang Y, van Haersma de With J, Fornazzari, Villagra R, Rojas-Garcia R, Clarimón J, Mayeux R, Robertson J, St George-Hyslop P, Rogaeva E (2012) Investigation of C9orf72 in 4 Neurodegenerative Disorders. Arch Neurol 69:1583. doi: 10.1001/archneurol.2012.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xi Z, Zinman L, Moreno D, Schymick J, Liang Y, Sato C, Zheng Y, Ghani M, Dib S, Keith J, Robertson J, Rogaeva E (2013) Hypermethylation of the CpG island near the G4C2 repeat in ALS with a C9orf72 expansion. Am J Hum Genet 92:981–989. doi: 10.1016/j.ajhg.2013.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xie Z, Nair U, Klionsky DJ (2008) Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell 19:3290–8. doi: 10.1091/mbc.e07-12-1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang M, Liang C, Swaminathan K, Herrlinger S, Lai F, Shiekhattar R, Chen J-F (2016) A C9ORF72/SMCR8-containing complex regulates ULK1 and plays a dual role in autophagy. Sci Adv 1–17. doi: 10.1126/sciadv.1601167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yeh TH, Lai SC, Weng YH, Kuo HC, Wu-Chou YH, Huang CL, Chen RS, Chang HC, Traynor B, Lu CS (2013) Screening for C9orf72 repeat expansions in parkinsonian syndromes. Neurobiol Aging 34:1311.e3–1311.e4. doi: 10.1016/j.neurobiolaging.2012.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yu C, Wang L, Lv B, Lu Y, Zeng L, Chen Y, Ma D, Shi T, Wang L (2008) TMEM74, a lysosome and autophagosome protein, regulates autophagy. Biochem Biophys Res Commun 369:622–629. doi: 10.1016/j.bbrc.2008.02.055 [DOI] [PubMed] [Google Scholar]
- 86.van der Zee J, Gijselinck I, Dillen L, Van Langenhove T, Theuns J, Engelborghs S, Philtjens S, Vandenbulcke M, Sleegers K, Sieben A, Bäumer V, Maes G, Corsmit E, Borroni B, Padovani A, Archetti S, Perneczky R, Diehl-Schmid J, de Mendonça A, Miltenberger-Miltenyi G, Pereira S, Pimentel J, Nacmias B, Bagnoli S, Sorbi S, Graff C, Chiang HH, Westerlund M, Sanchez-Valle R, Llado A, Gelpi E, Santana I, Almeida MR, Santiago B, Frisoni G, Zanetti O, Bonvicini C, Synofzik M, Maetzler W, vom Hagen JM, Schöls L, Heneka MT, Jessen F, Matej R, Parobkova E, Kovacs GG, Ströbel T, Sarafov S, Tournev I, Jordanova A, Danek A, Arzberger T, Fabrizi GM, Testi S, Salmon E, Santens P, Martin JJ, Cras P, Vandenberghe R, De Deyn PP, Cruts M, Van Broeckhoven C, Ramirez A, Kurzwelly D, Sachtleben C, Mairer W, Firmo C, Antonell A, Molinuevo J, Forsell C, Lillius L, Kinhult Ståhlbom A, Thonberg H, Nennesmo I, Börjesson-Hanson A, Bessi V, Piaceri I, Helena Ribeiro M, Oliveira C, Massano J, Garret C, Pires P, Danel A, Ferrari S, Cavallaro T (2013) A Pan-European Study of the C9orf72 Repeat Associated with FTLD: Geographic Prevalence, Genomic Instability, and Intermediate Repeats. Hum Mutat 34:363–373. doi: 10.1002/humu.22244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhou J, Tan SH, Nicolas V, Bauvy C, Di Yang N, Zhang J, Xue Y, Codogno P, Shen HM (2013) Activation of lysosomal function in the course of autophagy via mTORC1 suppression and autophagosome-lysosome fusion. Cell Res 23:508–523. doi: 10.1038/cr.2013.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.