Abstract
We describe several notable aspects of our structure predictions using Rosetta in CASP12 in the free modeling (FM) and refinement (TR) categories. First, we had previously generated (and published) models for most large protein families lacking experimentally determined structures using Rosetta guided by co-evolution based contact predictions, and for several targets these models proved better starting points for comparative modeling than any known crystal structure—our model database thus starts to fulfill one of the goals of the original protein structure initiative. Second, while our “human” group simply submitted ROBETTA models for most targets, for six targets expert intervention improved predictions considerably; the largest improvement was for T0886 where we correctly parsed two discontinuous domains guided by predicted contact maps to accurately identify a structural homolog of the same fold. Third, Rosetta all atom refinement followed by MD simulations led to consistent but small improvements when starting models were close to the native structure, and larger but less consistent improvements when starting models were further away.
Keywords: ab initio prediction, co-evolution, protein structure prediction, refinement, Rosetta
1 |. INTRODUCTION
The Rosetta hybrid protocol, originally developed for homology modeling1 to combine multiple input templates and alignments, can optimize discontinuous chain segments in both Cartesian and internal coordinate space through Rosetta’s core kinematic system. In fully internal coordinate representations, small perturbations can result in large atomic movements due to lever-arm effects; the hybrid protocol gets around this problem by sampling individual chain segment geometries in internal coordinate space, and interactions between the segments, in Cartesian space. Multiple alternative conformations of individual chain segments are maintained and can be recombined in Cartesian space; low energy models are fed back into the protocol for further recombination and iterative refinement. The protocol has been successfully applied to a broad range of refinement problems starting from de novo or homology models in combination with experimental2–4 and co-evolution data.5
In the previous CASP11 experiment, we used the hybrid protocol to refine de novo models generated using NOE and/or co-evolution data. Post-analysis revealed that the greatest source of error came from incorrect local fragments, resulting from incorrectly predicted secondary structure, and also from incorrect domain parsing. For the CASP12 experiment we sought to remedy these problems by using coevolution-derived contact predictions directly, if available, for both picking discontinuous fragments and domain parsing. Also, given that structural matches can occur in the absence of detectable sequence similarity, we sought to identify distant structural homologs based on the match between predicted contact maps and known structures.
For the refinement category predictions, we used an adaptation of the hybrid protocol alone and in combination with explicit water MD simulations.6,7 As the Rosetta energy function is an implicit solvent model, we sought to test whether inclusion of explicit waters could improve model accuracy when the starting model was close to the native structure.
2 |. MATERIALS AND METHODS
2.1 |. Hybridization protocol
The protocol is described in detail in ref. 1, and here we give a general overview. The protocol works with single or multiple templates; here we describe the more general multiple template scenario. The protocol starts by randomly selecting a single template (biased by input weight), the remaining templates are then superpositioned with the selected template, bringing all input templates to a common global frame. Rosetta’s symmetric sampling protocol is used if the selected template has multi-chain symmetry. During sampling, the secondary structure segments (“chunks”) are recombined between templates. Additional fragment replacement within chunks is performed, allowing local sampling without lever-arm effects. The sampling (chunk recombination or fragment replacement) can be limited to specific regions, or biased to a certain direction using residue-pair restraints.
We also developed an iterative version of the hybridization protocol for problems requiring more aggressive sampling or further refinement. The overall iterative process is guided by an evolutionary algorithm applying hybridization as mutation or crossover operations at each iteration and controlling diversity within the structural pool to prevent rapid convergence. The objective function within the iterative process is the Rosetta all-atom energy,8,9 with additional restraints if any information is available (for example, co-evolution data). Iterative hybridization is briefly introduced in ref. 8, and will be reported in more detail elsewhere (manuscript in preparation).
2.2 |. Contact prediction
For contact prediction we use GREMLIN10 with a multiple sequence alignment (MSA) generated using HHblits (from HHsuite version 2.0.15; -id 90 -cov 75 -n 8 -maxfilt ∞ -neffmax 20 -nodiff -realign_max ∞ -e [e-value]).11 The MSA is generated by using an iterative procedure with a stopping criteria to avoid attracting distant homologs at early stages or when they are not necessary (if enough sequences can be recruited with a lower e-value). The protocol starts with an e-value 1E-80 increasing by a factor of 1E10 until either the number of effective sequences reaches 128 Nf or the e-value reaches 1E-10. Nf is a metric for the number of effective sequences used for the MSA, and considered as sufficient for accurate model building when it is >64.8 After 1E-10, the e-value is increased by a factor of 1E2 until the Nf reaches 64 or e-value of 1E-4 is reached. If <64 Nf sequences are reached we try to enrich the alignment using additional sequences from metagenomes.8 hmmbuild (from HMMER version 3.1b1)12 is used to construct a hidden Markov model (HMM) of the MSA and hmmsearch (with bit-score of 27) is used to search against the database. The enriched alignment is filtered to reduce the redundancy to 90% and to remove sequences that do not cover at least 75% of the query (HHfilter -id 90 -cov 75). Positions that have >50% gaps are removed before the alignment is fed into GREMLIN. The protocol for restraint generation was the same one used in ref 8.
2.3 |. Fold recognition and fragment picking using contact maps
The query contact map generated by GREMLIN is filtered to only include the top 1.5 × sequence-length predictions (sorted by coupling strength) and contacts between residue pairs with sequence separation ≥3. To standardize the values, the coupling strength are converted to probabilities based on the number of effective sequences and length.5,13 This predicted contact map is aligned to the contact maps of known structures in the PDB for fold recognition. For the contact map search database, a non-redundant list of PDBs from HHsearch (pdb70; May 2016) was used.14 A distance cutoff of 5 Å between any two heavy atoms (with sequence seperation ≥3) was used to define a contact. A contact map alignment tool called map_align8 was used to search against this data. Before the search, the query contact map is manually analyzed for any clear domain boundaries, and the contact map is trimmed into domains if detected. Global and local matches are measured by Rc score for each hit; Rc score is the (# of contacts made)/(# of expected contacts), where the expected number of contacts is estimated by taking the sum of the computed probabilities. For the local Rc score, the expected number of contacts are only computed over the aligned regions (global Rc is calculated using the full query contact map). The results are ranked by global Rc, and the top 20 hits are extracted as templates for modeling. In addition to the 20 hits, we also extract the top 10 hits with (local Rc ≥0.8 and length ≥100) or (local Rc ≥0.9 and length ≥50), sorted by local Rc score. In the hybridization protocol, each of the templates is weighted by the global Rc score, and if they do not align to the global frame (due to a mismatched or partial starting template), the fragments from these templates are still used for internal coordinate sampling.
2.4 |. Robetta: fully automated structure prediction server
Since 18 of the 39 free modeling submissions were directly copied from the Robetta server without replacement, here we briefly describe the four major improvements made to the server pipeline since the CASP11 experiment.
First, the iterative hybridization protocol was automated and used to refine challenging targets (estimated GDT-TS < 0.61), when the length of the protein was <200 with no co-evolution data or <300 with sufficient co-evolution data (Nf > 32). This automation was able to reproduce our highlights from the CASP11 experiment made by the human group (T0806,T0824).5 Second, the Rosetta all-atom energy function was updated to the recent significantly improved REF2015.9 Third, a set of models for 614 protein families (Pfams) from our recent large-scale modeling efforts using co-evolution data8,13 was collected into a model database (MDB), and served as additional templates for modeling. These Pfams did not have any homologous structures in the PDB. The models were only added to our HHsearch database. Finally, the model quality assessment program ProQ2 was used for ranking the final five models.
2.5 |. Human modeling
Our main focus for the human submission efforts was to correct the inputs to our modeling framework. For 21 free-modeling domains, 6 domains were reparsed and modeled using co-evolution data, 3 were remodeled using known functional data (beside co-evolution data), and 12 included additional sampling using the hybridization protocol.
2.6 |. Overview of our approach on the refinement category
In the CASP12 refinement category, we designed and tested an integrated approach that runs the Rosetta hybridization protocol with a provided starting model, followed by extra MD-based refinement.6,7 Rosetta hybridization was applied in two separate ways as in CASP11,15 namely high- and low-resolution protocols (described below), depending on how close the starting model is to the native structure (as provided by organizers in GDT-HA). In the high-resolution protocol, Rosetta hybridization was applied to rebuild local regions estimated to contain errors, while in the low-resolution protocol, the iterative version was applied to rebuild the whole structure while more intensively focusing on less reliable regions.16 The criteria for running the high-resolution protocol is (1) if the starting model’s GDT-HA ≥ 55 or (2) the protein size is >400 amino acids (even if starting GDTHA< 55). Otherwise the low-resolution protocol was chosen. The basis for splitting targets into two categories stems from quite distinct characteristics of errors in the starting models;15 while errors in high-resolution starting models occur at local backbone regions that can be refined through a conservative approach, errors in low-resolution starting models occur at multiple regions that can be only fixed by allowing more aggressive backbone sampling throughout the entire model. In the following sections we describe more details on each component applied.
2.7 |. High-resolution refinement protocol
Our high-resolution protocol was designed based on the hypothesis15 that fixing local errors and core-refinement should play a complementary role for refining moderately accurate starting models. Local regions to rebuild are identified following Park et al.16 which uses a well-known correlation between residue-level fluctuation and structural error. The maximum number of regions to rebuild is set to 3 for a protein smaller than 100 amino acids and otherwise set to 5. Rosetta hybridization is repeated 2,000 times independently to intensively sample around the identified local regions. The representatives of five lowest energy clusters from the generated models are selected and subject to further MD-refinement for core-refinement.
2.8 |. Low-resolution refinement protocol
Our low-resolution protocol is designed for large-scale energy-guided refinement of an inaccurate model using the iterative version of hybridization. Because the main driving force for structural change is the all-atom energy function,9,15 and only weak restraints to the starting model are used, the success strongly depends on the accuracy of our energy function with sufficient sampling. More details on the low-resolution refinement protocol based on hybridization will be reported elsewhere. Representatives of the five lowest energy clusters after the iterative process are selected for further MD-refinement.
2.9 |. Succeeding MD-based refinement and model ranking
MD-based refinement6,7 is applied to the output models from both the high- or low-resolution Rosetta protocol with the same set of parameters. The AMBER12SB force field5,17 is used for the simulation. The protein is solvated in a periodic boundary box filled with the TIP3P explicit water model.18 During the simulation, harmonic restraints are applied to the Ca atoms at their starting coordinates of MD. For each of five selected Rosetta-refined models, 10 ns of 5 independent Langevin dynamics simulations are run, and structures on the trajectories are averaged (without filtering) to pick the representative model. This process is repeated separately for each of five Rosetta models to produce five structure-averaged models, which are then regularized by Rosetta FastRelax19 with hard positional restraints to backbone atoms.
Five MD-refined models are ranked by the ensemble properties observed from their corresponding MD trajectories. The feature primarily used for ranking is the ensemble-average Rosetta energy (average Rosetta energy across the MD trajectory). The only exception is when the structural convergence within the ensemble of the second-ranked model (in the energy-based metric) is better than that of the first-ranked model by >10%; structural convergence within the ensemble structures is measured by the percent of residues with root-mean-squared fluctuation (RMSF) lower than 1.0 Ang in the corresponding MD trajectory. The concept of this model ranking can be understood as cross-validation between two orthogonal components—the first energy-based metric evaluates the quality of ensemble structures sampled by the molecular mechanics force field using the Rosetta energy, and the second convergence-based metric validates the quality of a Rosetta model by running further MD simulations.
3 |. RESULTS
3.1 |. Free modeling (FM) and free modeling/template-based modeling (FM/TBM) results
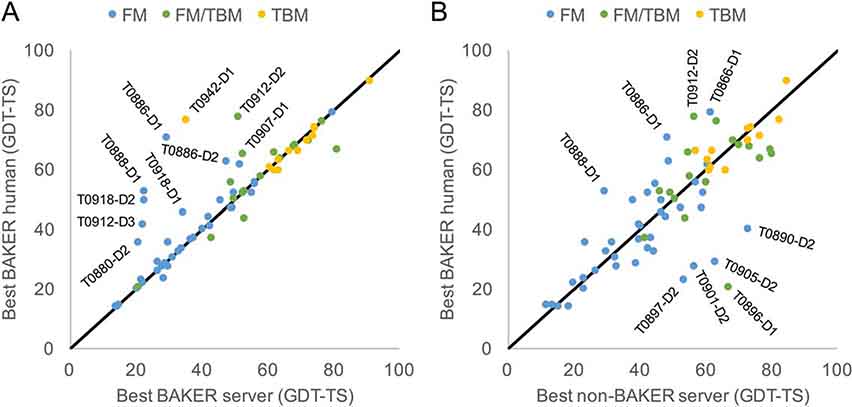
In Figure 1A, the model quality of all-group targets is shown for our server models and human-guided models. For 37 out 69 domains of the “all group” target, the human group simply submitted the Robetta models. In the remaining cases, human efforts generally added extra value: six had significant improvements (ΔGDT-TS > 20) and additional six had modest improvements (ΔGDT-TS > 10). Most of the significantly improved cases involved more sophisticated use of co-evolution data with human intervention. Comparison of our human submission to all other non-BAKER server submission (See Figure 1B) highlights failures that were not fixed with our human efforts. All five domains having significant failures (ΔGDT-TS < −20) were due to domain parsing issues. In one case T0942-D1, the human group was able to correct the domain parse, bringing the GDT-TS from 35 to 77. Below we summarize a few highlights where there was a significant improvement over other server submissions.
FIGURE 1.
Comparison of human group submissions to server submissions. Shown are 68 all-group targets. A, Comparing the BAKER server submission to the BAKER human submission. Highlighted are the domains where large improvements were observed. The three colors are used to distinguish the Free-modeling (FM), FM/Template-based-modeling (FM/TBM) and Template-based-modeling (TBM) targets. B, Comparing against non-BAKER server submission. Highlighted are domains where the human submission did significantly worse than the best non-baker server. Targets T0896-D1, T0897-D2, T0890-D2, and T0905-D2 all suffered due to domain parsing issues. For T0942-D1, the domain parsing was corrected for the human submission
3.2 |. Domain parsing and structural homology search using co-evolution contacts
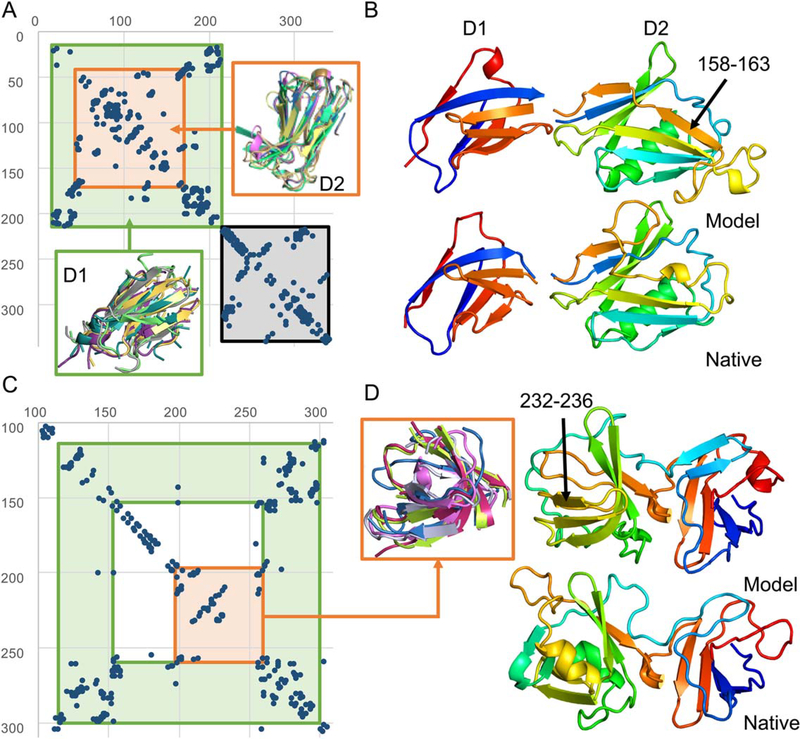
For T0886, human intervention corrected domain parsing and improved coevolution-guided homolog search (Figure 2A). Inspection of the predicted contact map suggested it has at least three domains (Figure 2A). For domain 1 and 2, map_align was run to see if there were any structural homologs. For these we found strong hits to flagellar proteins, with little sequence homology (6% identity between 4ut1_A and D1, 20% between 4nx9_A and D2) and different chain connectivity. While domain 1 is discontinuous in the query, it is continuous in the structural homolog, which made it difficult for the server to take full advantage of co-evolution analysis. Domain 3 was sampled using the Rosetta Abinitio protocol since it was all alpha-helical and no hits were found using map_align, but unfortunately, most of domain 3 does not appear in the crystal structure and thus was not used for evaluation. For T0886-D1 and D2, the GDT-TS is 71 and (next best server 29, our 23) 63 (next best server 49, our 48), respectively (side by side structural comparison in Figure 2B).
FIGURE 2.
Contact-guided domain parsing and fold-recognition allows accurate modeling. A, Based on the contact map, T0886 was parsed into three domains (green, orange and grey; green is a discontinuous domain). Here, we focus on the first two N-terminal domains (green and orange). Most of the third C-terminal domain (grey) was not present in the crystal structure, hence is not evaluated here. The partial threads from the top 5 map_align hits are shown for the two domains. B, Comparing the model (top) to native (bottom) structure. Each domain is superpositioned into the corresponding native domains for illustration. C, Based on the contact map (trimmed to 100–310), the initial parsing of two domains (green and orange; also green is a discontinuous domain) are shown. Each of these domains were modeled separately and then refined as a whole. D, Comparing the model (top) to native (bottom). One of the sheets (indicated with black arrow) was incorrectly predicted. The partial threads from the top 5 map-align hits are shown in the orange box
For T0912, expert intervention also improved domain parsing and modeling guided by co-evolution data. Using the contact map, we parsed T0912 into 2 initial domains: (114–153, 267–300) and (197–260) (Figure 2C). Similar to T0886, discontinuity in the domain1 sequence caused difficulty in using co-evolution data at the server stage. The two domains were modeled separately, docked guided by the extensive interdomain contacts, and then refined. Due to time limitation, little effort was made to model the remaining parts of the protein. In this case, the domains were over-parsed, resulting in errors for the remainder of the defined domains. Nonetheless, both had significant improvement over our or other server models, GDT-TS being 78 (next best server 56, our 51) and 42 (next best server 40, our 22) for D2 and D3, respectively. For the two assembled domains as defined by the assessors, the GDT-TS is 49 (next best submission 29; see Figure 2D for side by side comparison; the GDT-TS of the two assembled domains using our domain definitions is 56 compared to 36 for the next best submission).
T0886 and T0912 illustrate the importance of discontinuous domain modeling and domain parsing using contact maps. Without parsing these domains, map_align would not have been able to find the best templates, due to strong gap penalties, but in the case of T0912, over-parsing may have limited proper sampling of sheets in the remainder of T0912-D3. The map_align hit, formed a sheet pairing at 232–236, preventing further sampling in that region. In the case of T0886, “human intuition” to select models that have most sheets hurt our prediction for one of the loops (158–163) in T0886-D2. For both targets, there were no contacts predicted indicating that these sheets should be paired, for future experiments it would be worth deleting regions not restrained by contacts and resampling these.
3.3 |. Contact-guided model database (MDB) for additional templates
Prior to CASP12, we had carried out and published a comprehensive set of structure models for large protein families without solved crystal structures. The accuracy of this large scale modeling effort has been highlighted on numerous occasions since the article was published as six newly solved crystal structures published more recently are all very close to our models. For CASP12, we tested the use of this set of models (collected in a database we call MDB) as a starting point for comparative modeling. This resembles the goal of the original protein structure initiative (PSI)–to produce a set of models that provided starting points for reliable comparative modeling of any protein sequence.
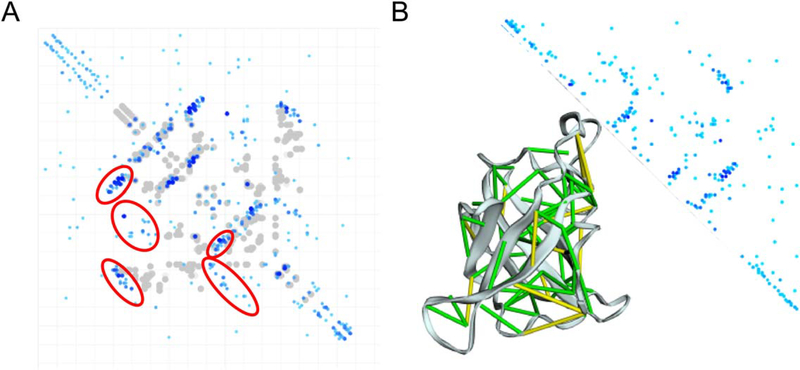
Three targets had a significant hit to one of the MDB models: T0866, T0907 and T0918. T0907 is a homolog of T0866 and contains 3 copies of this domain. Our model for T0866-D1 had a GDT-TS of 80, which far exceeds the best GDT-TS of 62 available by typical sequence-based homolog search. The top HHsearch hit (pdb:1lpl_A) for T0866-D1 with probability of 17.8% made very few of the predicted contacts due to a flipped beta hairpin (Figure 3A). In contrast, we found a strong hit (HHsearch probability of 100%) from the MDB making most of the predicted contacts (Figure 3B). For T0918-D2 and D3, our MDB hit also provided much better starting templates, resulting in final model GDT-TS of 50 and 56, compared to the next best server of 38 and 45, for D2 and D3, respectively.
FIGURE 3.
Comparing the Model Data Bank (MDB) and Protein Data Bank (PDB) hits for T0866. A, The overlay of the predicted contacts over the top HHsearch PDB hit 1lpl_A (Prob: 17.8%). In blue are the top 1.5 × length predicted contacts. The regions with most violations are circled in red. B, The overlay of the predicted contact over top MDB HHsearch hit (Prob: 100%)
3.4 |. Incorporating known functional information for fold recognition
We reasoned that T0880 and T0888 could have folds similar to previously solved adenovirus head structures, and used HHsearch to generate threadings of their sequence onto these structures. The alignments were poor, but were improved by hybridization at the modeling stage; during chunk recombination, we allowed stochastic sequence registry shifts up to 4 amino acids in either direction. For T0880-D1, the GDT-TS is 62 (next best server 60, our 51). For T0880-D2 the GDT-TS is 36 (next best server 32, our 21). For T0888-D1 the GDT-TS is 53 (next best server 29, our 23).
3.5 |. Refinement of close to native targets
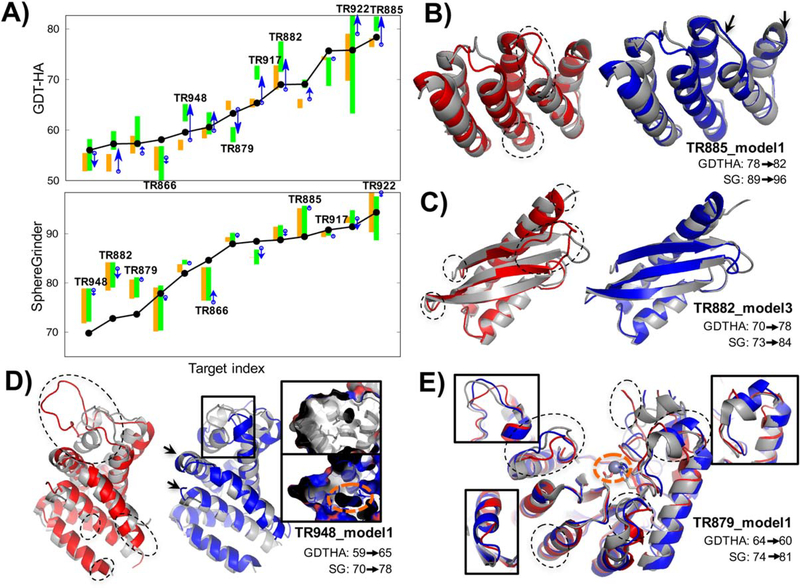
Our high-resolution protocol combining reconstruction by Rosetta hybridization protocol with subsequent MD simulation was run for 18 of 42 targets. In Figure 4, the models generated by Rosetta local reconstruction and MD are individually compared to the starting model using GDT-HA and SphereGrinder (SG).18,20 With the combined protocol, Model 1 improved over the starting model in half the cases as assessed by both GDT-HA and SG.
FIGURE 4.
Automated high-resolution refinement combining local rebuilding by Rosetta and MD refinement. A, Decomposition of contribution to refinement by Rosetta and MD in A, GDT-HA (top) and SphereGrinder (bottom). Thirteen targets with native structures available are shown here. For each target, starting model quality is shown in black dots, range of five models by Rosetta modeling and succeeding MD refinement in orange and green bars, respectively, and change in quality of model1 from Rosetta stage to MD refinement by blue arrow. B-E, Structures for the targets with successful refinement or targets showing lessons for future direction. Native, starting model, and refined model structures are shown in gray, red, blue cartoons, respectively. Regions automatically detected and reconstructed are shown in dotted circles. Improvements in secondary structure orientations are highlighted by black arrows. B-C, High-end refinement targets, TR885 and TR882, were successful with our protocol. D, TR948, reconstruction on long region (large circle on top, residue 53–78) put helix at roughly correct position but had poorer hydrophobic packing in our model (below) compared to the native structure (top); void shown as orange circle on the inset panel. E, TR879 was the only target significantly worsen by MD refinement; decrease in GDT-HA solely comes from MD refinement stage, presumably due to ignoring metal binding (black arrow). Reconstructed at five regions, and three of these reproduced correct loop conformations (inset) which led to improvement to SG
The most successful cases (TR885, TR882, TR922, and TR917) were those for which the input models were already quite accurate (GDT-HA > 65). In control experiments with TR882 and TR885 using the MD simulation protocol alone, poorer results were obtained (Combined protocol GDT-HA/SG +5/+8 and +4/+9; MDonly,+3/0 and 0/0; for TR882 and TR885, respectively); the enhanced sampling brought about by the Rosetta refinement evidently improves performance.
Reconstruction of large unreliable regions was attempted for multiple targets (TR948, TR947, TR909, and TR912) but the models were either partially refined or remained unrefined because of insufficient sampling. TR948 (Figure 4D) is a good example; a 25-residue-loop forming loop-helix-loop motif was roughly refined by intensive reconstruction but the model still contained an obvious error having a void inside the hydrophobic core (orange dotted circle in the figure). Other targets similarly had issues with hydrophobic packing. To enhance the sampling of the hydrophobic core, it should be possible to introduce metrics for hydrophobic packing to the convergence check, or use more sophisticated structural operators and restraints.21
Another source of error in high-resolution refinement was neglect of interactions with other subunits in homo-oligomers, ordered water molecules, and small-molecules/metals (an example is TR879 shown in Figure 4E). Consideration of the full biological unit using information available from templates with more aggressive local error corrections should improve high-resolution refinement.
Model ranking using MD trajectory was generally helpful but can be improved in the future. The best-of-five model was correctly selected as model1 for a number of targets: TR885, TR922, TR948 among close-to-native targets. Despite the small sample size, analysis on these targets suggests that successful sampling improves model selection; selection failures may be associated with poor sampling. An exception was TR882 (Figure 4C), which was sampled successfully but ranked incorrectly due to inaccuracy in the Rosetta energy function, suggesting future directions in model ranking.
3.6 |. Refinement of distant from native targets
For the more challenging 24 of the 42 targets, we ran aggressive energy guided model rebuilding by Rosetta followed by MD refinement. In Figure 5A, the overall results are again compared to their starting models in GDT-HA and SphereGrinder (SG). Substantial improvement in SG were found for multiple targets (>10% for 7 targets), but improvements in GDT-HA was observed only in a handful cases (TR894, TR594, and TR942). The overall fraction of targets for which model1 improved over starting model is 50% and 58% (75% and 71% with best of five models) in GDT-HA and SphereGrinder, respectively.
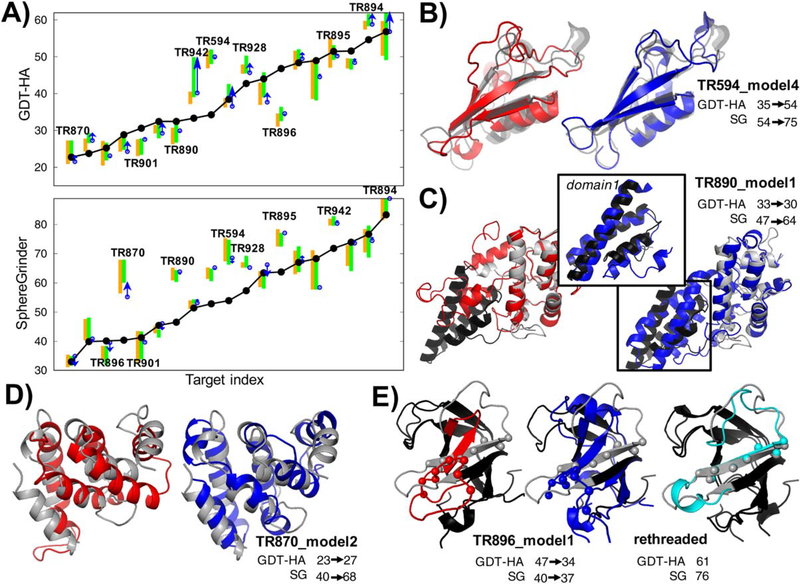
FIGURE 5.
Automated low-resolution refinement by large-scale energy guided sampling. A, Decomposition of contribution to refinement by Rosetta and MD in GDT-HA (top) and SphereGrinder (bottom). Bars, dots, arrows are drawn in the same way as in Figure 4A. Nineteen targets with native structures available are shown. B-E, Structures for the targets with successful refinement or targets showing lessons for future direction. Native, starting model, and refined model structures are shown in gray, red, blue cartoons, respectively. B, Successful refinement on TR594. C-E, Challenging refinement targets in CASP12. C, TR890, separate colors are used for the two domains in the native structure; black for domain1 and white for domain2. Domain1, which was poor in the input model, was correctly refined (inset; superimposed onto domain1), but its relative orientation to domain2 was wrong and did not lead to an increase in GDT-HA. D, TR870; secondary structures and their rough orientations were fixed, but precise positioning was incorrect. E, Incorrect register shift by six residues at one of the strands (highlighted by spheres) was not fixed by refinement, and this error propagated to mis-prediction at the other parts of the submitted model. When started from the correct threading to the starting model, the output of refinement converged close to native (cyan, left panel)
The largest improvement was for TR594 (Figure 5B). Refinement of this target was attempted within the context of the whole complex built from the starting models generated for TR594, TR894, and TR895. Iterative Rosetta reconstruction applied to the complex recovered all the secondary structure segments and orientations of TR594, and also improved the other two targets in the complex. Refinement was also successful for a couple other large proteins (TR928, 381 aas; TR942, 387 aas), which are larger than the proteins in the benchmark set used to previously test the method (200 aas).
Analysis of results on a number of very challenging targets revealed limitations in our large-scale sampling approach not observed in our previous benchmarks. 6 targets in CASP12 were especially challenging; TR869, TR870, and TR898 had starting SG< 25, GDT-HA< 30, protein size> 100 aas, and TR890, TR901 and TR905 had starting SG< 40, GDT-HA < 35, protein size > 180 aas. Refined models for TR870 and TR890 (Figure 5C,D) were partially successful but also highlighted missing aspects in our sampling protocol. For TR890, the intra-domain conformation at domain1 was corrected (inset of Figure 5C) but the orientation with respect to domain2 was made worse, which led to an increase in SG but not in GDT-HA. Improvement for TR870 was also only in SG because while interactions between pairs of contacting helices were improved, their overall global positioning was not. For the other four targets, none of our models improved even partially over the starting model. All these pathologies are likely related to insufficient sampling—native structures have clearly better energy than what we sampled—and provide useful challenges for future methods development.
Our method also performed poorly in correcting sequence alignment errors. Starting models for two targets (TR896 and TR921) had incorrect beta-strand registers. As a post analysis, we carried out a control experiment on TR896 after the release of the native structure to see how correcting this error could make difference. The same low-resolution protocol was repeated but with a starting model rethreaded following secondary structure prediction.6 The control refinement yielded a significantly improved structure (Figure 5E) compared to our CASP submission, as not only the Rosetta energy significantly dropped but also clear convergence was found in final models. In spite of this encouraging result, correcting sequence alignments through refinement will require some effort given uncertainty in secondary structure prediction and identification of a small number of alternative alignments.
4 |. DISCUSSION
4.1 |. Overcoming mispredicted secondary structure, domain parsing, and sampling complex topologies using contact information
Due to the vast search space required for free modeling targets, Rosetta modeling methods such as hybridization or AbInitio in the past were limited to small and simple folds. Challenges to adequate sampling are at both local and global levels: in regions where the local structure is strained by surrounding residues or otherwise poorly modeled by an ensemble of short fragments with similar local sequences, and complex topologies with many non-local interactions. Our results here and in ref. 8 show that if the protein belongs to a sufficiently large sequence family, the second problem – modeling proteins with complex toplogies – can be largely overcome by using contact information and a population based evolutionary algorithm. Given the large number of independent MC trajectories, it is likely that at least some correct non-local interactions are sampled in each trajectory. By iteratively recombining these models guided by contact information, we expect to sample new structures that contain all the non-local interactions.
The first problem – correcting local structures – is not easily solved. Local-window-based secondary structure prediction approaches such as PSIPRED6 are sometimes not able to capture secondary structure preferences that are depend on long-range interaction (sequence separation > 15). Errors are often seen in terminal and buried beta-sheets that do not follow a typical alternating hydrophobic/hydrophilic pattern. In the CASP12 experiment, we explored the possibility of correcting the second problem by using contact information to search for discontinuous fragments that make a significant fraction of the contacts, independent of sequence or predicted secondary structure. After developing the method, we found that in some cases it was able to recover entire folds in the PDB that make a majority of the predicted contacts, allowing us to bypass the AbInitio stage altogether. This was facilitated by manual domain parsing of the contact maps into regions strongly connected by the predicted contacts. Future development will focus on automating contact map based domain parsing and on combining partial discontinuous hits during sampling.
4.2 |. Future directions in refinement method development
Unifying our low- and high-resolution refinement protocol into a single general refinement framework will be an important research direction. A unified approach would use hybridization to refine unreliable regions and core parts simultaneously, while adjusting the magnitude of perturbations at the core parts depending on the reliability of the input model or the convergence of sampling. Many of the targets that underwent the high-resolution protocol had relatively poor starting model quality (55< GDT-HA < 65) and still contained considerable structural deviations from their natives. For these, iterative refinement on the entire structure should, in principle, be more appropriate than our current conservative approach.
ACKNOWLEDGMENTS
The authors thank Darwin Alonso for developing the computational and network infrastructure and Rosetta@home participants for providing the computing resources necessary for this work. They also thank the CASP12 organizers, the structural biologists who generously provided targets, and the authors of ProQ2. The work was supported by the NIH through grant number R01 GM092802.
Footnotes
AVAILABILITY
Robetta and GREMLIN are available for non-commercial use at http://robetta.bakerlab.org and http://gremlin.bakerlab.org, respectively. The Rosetta software suite can be downloaded from http://www.rosettacommons.org.
REFERENCES
- [1].Song Y, DiMaio F, Wang RY-R, et al. High-resolution comparative modeling with RosettaCM. Structure. 2013;21(10):1735–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].DiMaio F, Echols N, Headd JJ, Terwilliger TC, Adams PD, Baker D. Improved low-resolution crystallographic refinement with Phenix and Rosetta. Nat Methods. 2013;10(11):1102–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].DiMaio F, Song Y, Li X, et al. Atomic-accuracy models from 4.5-Å cryo-electron microscopy data with density-guided iterative local refinement. Nat Methods. 2015;12(4):361–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ovchinnikov S, Park H, Kim DE, Liu Y, Wang RY-R, Baker D. Structure prediction using sparse simulated NOE restraints with Rosetta in CASP11. Proteins. 2016;84(Suppl 1):181–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ovchinnikov S, Kim DE, Wang RY-R, Liu Y, DiMaio F, Baker D. Improved de novo structure prediction in CASP11 by incorporating coevolution information into Rosetta. Proteins. 2016;84(Suppl 1): 67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16(4):404–405. [DOI] [PubMed] [Google Scholar]
- [7].Mirjalili V, Feig M. Protein structure refinement through structure selection and averaging from molecular dynamics ensembles. J Chem Theory Comput. 2013;9(2):1294–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ovchinnikov S, Park H, Varghese N, et al. Protein structure determination using metagenome sequence data. Science. 2017;355(6322): 294–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Park H, Bradley P, Greisen P Jr, et al. Simultaneous optimization of biomolecular energy functions on features from small molecules and macromolecules. J Chem Theory Comput. 2016;12(12):6201–6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kamisetty H, Ovchinnikov S, Baker D. Assessing the utility of coevolution-based residue–residue contact predictions in a sequence-and structure-rich era. Proc Natl Acad Soc. 2013;110(39): 15674–15679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Remmert M, Biegert A, Hauser A, Söding J. HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat Methods. 2011;9(2):173–175. [DOI] [PubMed] [Google Scholar]
- [12].Eddy SR. Accelerated Profile HMM Searches. PLoS Comput Biol. 2011;7(10):e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ovchinnikov S, Kinch L, Park H, et al. Large-scale determination of previously unsolved protein structures using evolutionary information. eLife. 2015;4:e09248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Söding J Protein homology detection by HMM–HMM comparison. Bioinformatics. 2005;21(7):951–960. [DOI] [PubMed] [Google Scholar]
- [15].Park H, DiMaio F, Baker D. CASP11 refinement experiments with ROSETTA. Proteins. 2016;84(Suppl 1):314–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Park H, Seok C. Refinement of unreliable local regions in template-based protein models. Proteins. 2012;80(8):1974–1986. [DOI] [PubMed] [Google Scholar]
- [17].Case DA, Darden TA, Cheatham TE III, et al. AMBER 12. University of California, San Francisco, 2012. [Google Scholar]
- [18].Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79(2):926–935. [Google Scholar]
- [19].Conway P, Tyka MD, DiMaio F, Konerding DE, Baker D. Relaxation of backbone bond geometry improves protein energy landscape modeling. Protein Sci. 2014;23(1):47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Antczak PLM, Ratajczak T, Blazewicz J, Lukasiak P, Blazewicz J. SphereGrinder - reference structure-based tool for quality assessment of protein structural models. In: 2015 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) 2015. p. 665–668. [Google Scholar]
- [21].Perez A, MacCallum JL, Dill KA. Accelerating molecular simulations of proteins using Bayesian inference on weak information. Proc Natl Acad Sci U S A. 2015;112(38):11846–11851. [DOI] [PMC free article] [PubMed] [Google Scholar]