Abstract
The response of cancer cells to therapeutic drugs that cause DNA damage depends on genes playing a role in DNA repair. RecQ-like helicase 1 (RECQ1), a DNA repair helicase, is critical for genome stability, and loss-of-function mutations in the RECQ1 gene are associated with increased susceptibility to breast cancer. In this study, using a CRISPR/Cas9-edited cell-based model, we show that the genetic or functional loss of RECQ1 sensitizes MDA-MB-231 breast cancer cells to gemcitabine, a nucleoside analog used in chemotherapy for triple-negative breast cancer. RECQ1 loss led to defective ATR Ser/Thr kinase (ATR)/checkpoint kinase 1 (ChK1) activation and greater DNA damage accumulation in response to gemcitabine treatment. Dual deficiency of MUS81 structure-specific endonuclease subunit (MUS81) and RECQ1 increased gemcitabine-induced, replication-associated DNA double-stranded breaks. Consistent with defective checkpoint activation, a ChK1 inhibitor further sensitized RECQ1-deficient cells to gemcitabine and increased cell death. Our results reveal an important role for RECQ1 in controlling cell cycle checkpoint activation in response to gemcitabine-induced replication stress.
Keywords: DNA helicase, DNA damage response, anticancer drug, breast cancer, checkpoint control, DNA repair, gemcitabine, genome stability, missense mutation, RecQ, replication stress
Introduction
RecQ helicases are a highly conserved class of proteins with important roles in genome maintenance and DNA repair (1). The loss of DNA repair functions and SNP in the RECQ1 gene is linked to cancer predisposition and increased resistance to chemotherapeutic drugs (2–4). Recent whole-genome sequencing efforts revealed that rare, recurrent RECQ1 (also known as RECQL or RECQL1) mutations increased the risk of breast cancer in French Canadian and Polish populations (5). The association of RECQ1 mutations with breast cancer was also confirmed in a Chinese population, suggesting that RECQ1 mutations are not limited to specific populations (6). In a subsequent report, further studies were recommended to establish a better association of increased breast cancer risk in individuals carrying RECQ1 loss-of-function variants (7).
RECQ1 is the most abundant member of the five human RecQ helicases (1). It consists of a helicase and RecQ C-terminal (RQC)2 domain similar to that of Escherichia coli RecQ (8). RECQ1 is a DNA-stimulated ATPase and a helicase capable of binding and unwinding structural intermediates of DNA replication and repair (9). RECQ1 unwinds duplex DNA and catalyzes ATP-dependent branch migration on Holliday junctions and mobile D-loop substrates (9, 10). In addition to unwinding DNA, RECQ1 promotes annealing of complementary single-stranded DNA in an ATP-independent manner (9). Consistent with these biochemical activities, RECQ1 interacts with proteins known to function in DNA replication and repair, such as FEN1 (11), RPA (12, 13), Ku70/80 (14), and PARP1 (4, 13, 15), as well as with mismatch repair proteins (MLH1 and MSH2/6) that regulate genetic recombination (16). RECQ1−/− null mice obtained by targeting the helicase domain IV and part of domain V in the RECQ1 gene displayed spontaneously increased chromosomal instability in primary embryonic fibroblasts (17). RECQ1 deletion in human cells has not yet been reported, and the cellular functions of RECQ1 have been investigated by using siRNAs or stable short hairpin RNA–mediated knockdown (4, 15, 18–21). The depletion of RECQ1 causes decreased cell proliferation, increased sensitivity to replication blocking agents, and increased DNA damage accumulation (18, 19). The increase in chromosomal rearrangements in RECQ1-depleted cells upon replication stress suggests that RECQ1 is involved in the resolution of stalled replication forks (17, 18). RECQ1 governs RPA availability during replication stress (20), and the catalytic activity of RECQ1 is required for the restoration of stalled forks induced by camptothecin (4), clearing the way for replication to resume after the block is removed. RECQ1 catalyzes strand exchange on stalled replication structures in vitro, and the ATPase function of RECQ1 is critical in facilitating branch migration of the intermediates formed upon fork stalling (19, 22). Collectively, these studies suggest that RECQ1 functions to restore productive DNA replication following stress and prevents subsequent genomic instability.
Cells respond to blocked DNA synthesis by activating the DNA damage checkpoint (23, 24). This response is necessary for replication fork stabilization and DNA repair, which is necessary for the subsequent restart of replication (25). When a replication fork stalls, the uncoupling of replicative helicase from the polymerase complexes leads to the generation of single-stranded DNA (ssDNA) that is coated by RPA (replication protein A) (26, 27). The RPA-coated ssDNA acts as a scaffold and recruits the key kinases ATR (ataxia-telangiectasia and RAD3-related) that activate the downstream effectors ChK1 (Checkpoint kinase 1) to facilitate DNA repair and cell survival (28). Failure in this protective cascade results in the irreversible collapse of the replication fork, which can result in a DNA double-stranded break, which is the most potent form of DNA lesions (25). Furthermore, these protective systems are severely challenged by exogenous sources of DNA damage, such as cancer therapeutics that block replication (29).
In this study, we investigated how the genetic or functional loss of RECQ1 in breast cancer MDA-MB-231 cells impacts the cellular response to chemotherapeutic drugs, such as camptothecin and gemcitabine, which are known to interfere with DNA replication. Gemcitabine is a nucleoside analog that disrupts replication by incorporating into DNA and inhibiting ribonucleotide reductase, resulting in the depletion of the dNTP pool (30). SNP in RECQ1 has been associated with the overall survival of patients who received gemcitabine-based therapy (2). To understand the molecular functions of RECQ1 in DNA repair and how the mutations in this gene promote tumorigenesis, we utilized CRISPR-Cas9 gene editing to generate an isogenic pair of RECQ1 WT and RECQ1 knockout (KO) MDA-MB-231 cell lines. Furthermore, we investigated the replication stress response of MDA-MB-231 cells that expressed RECQ1 variants with individual missense mutations (A195S, R215Q, R455C, M458K, and T562I) that were associated with breast cancer susceptibility (5, 6) and demonstrated to be deficient in helicase activity (6).
Results
Establishing breast cancer cell-line models of the genetic and functional loss of RECQ1
We used the CRISPR-Cas9 technique to generate an isogenic pair of MDA-MB-231 breast cancer cell lines that either expressed (RECQ1-WT) or lacked (RECQ1-KO) RECQ1 expression. We designed single guide RNA (gRNA) targeting the RECQ1 gene on exon 3, cloned the gRNA into the pENTR221 gRNA cloning vector, and confirmed the correct insertion of the target sites by DNA sequencing. The efficiency of the gRNA to generate indels was validated by SURVEYOR assay. After co-transfection of the MDA-MB-231 cells by RECQ1-gRNA and the corresponding vectors, followed by puromycin selection, the isolated clones were screened for RECQ1 expression at the protein level by Western blotting analysis, using a specific antibody against RECQ1. The results confirmed the complete loss of RECQ1 protein in the RECQ1-KO cell line (Fig. 1A). The results of the Sanger sequencing revealed that in the WT clones, the RECQ1 genomic sequence at the target site was intact and the same as that of the unedited parental MDA-MB-231 control. In the KO clones, RECQ1 genomic sequences at the target site showed insertions and deletions compared with the unedited control (Fig. S1A). In the KO clones obtained, each clone had distinct genome sequences at the target site. All the genomic sequences in the KO clones contained indels that resulted in the nonfunctional gene products, which eventually led to the loss of the protein.
Figure 1.
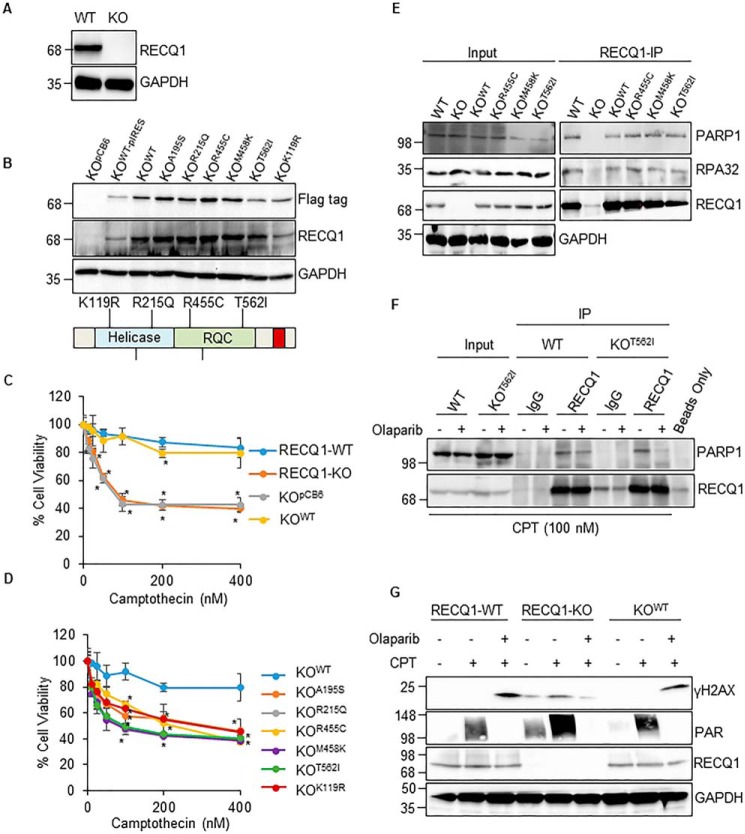
Functional or genetic loss of RECQ1 sensitizes MDA-MB-231 cells to camptothecin. A, immunoblot detection of RECQ1 in cell lysates from MDA-MB-231 CRISPR-Cas9–derived RECQ1-WT and RECQ1-KO clones. GAPDH is used as a loading control. B, stable expression of RECQ1 WT and missense mutants in the RECQ1-KO cell line was validated by Western blotting analysis using specific antibodies against FLAG tag and RECQ1. GAPDH is used as a loading control. The schematic diagram represents the location of missense mutations in helicase and RQC domains of RECQ1. C and D, drug sensitivity to camptothecin in RECQ1-WT, RECQ1-KO, and complemented cell lines. Cell viability data are presented as means (± S.D.) from three independent experiments. An asterisk denotes the statistical significance of cell viability changes in RECQ1-WT versus other groups (p < 0.05). Differences are not statistically significant unless denoted by an asterisk. E, co-immunoprecipitation analysis of RECQ1 interaction with PARP1 using whole cell extracts from RECQ1-WT, RECQ1-KO, KOWT, KOR455C, KOM458K, and KOT562I cells. IP with antibodies specific for RECQ1 and IgG are indicated. Eluted proteins in immunoprecipitate were analyzed by Western blotting using antibodies against RECQ1, PARP1, and RPA32. Input represents 10% of cell lysates used for co-immunoprecipitation assays. GAPDH is used as a loading control. F, RECQ1-IP from RECQ1-WT and KOT562I cells following camptothecin (CPT) (100 nm for 2 h) treatment and ± PARP inhibitor (10 μm olaparib for 2 h). G, Western blotting analysis of camptothecin (100 nm for 2 h) induced γH2AX ± PARP inhibitor (olaparib, 10 μm for 2 h) in RECQ1-WT, RECQ1-KO, and KOWT cells. PAR levels were tested to confirm PARP inhibition by olaparib. GAPDH is used as a loading control. Molecular mass (in kDa) is shown to the left of the Western blots. IP, immunoprecipitation.
To compare the functions of the WT RECQ1 protein with those of the RECQ1 variant in an isogenic background, we reconstituted the RECQ1-KO cell line using engineered pCB6 vectors that stably expressed either the empty vector or the full-length WT RECQ1 or RECQ1 variants with individual missense mutations (A195S, R215Q, R455C, M458K, and T562I), which were reported to increase breast cancer susceptibility. Based on the crystal structure and conserved functions of RecQ helicases (1), Ala-195 is involved in dimer interaction (31); Arg-215 is located near the ADP-binding pocket and is expected to weaken ATP hydrolysis (32, 33); the conserved residues Arg-455 and Met-458 are located in the zinc-binding subdomain, which is important in maintaining protein stability (34); and Thr-562 is located in a β-hairpin, which is required for DNA unwinding (31, 35). Compared with the ability of WT RECQ1 helicase to unwind forked DNA substrates, the RECQ1 variants R215Q, R455C, M458K, and T562I show complete loss of helicase activity, and A195S variant shows very weak helicase activity in vitro (6). Therefore, we also included a K119R variant that has not yet been associated with cancer risk but has been biochemically characterized as helicase-dead (9, 32).
To facilitate their representation in the figures and in the text, the RECQ1-KO cells reconstituted by these plasmids are labeled KOpCB6, KOWT, KOA195S, KOR215Q, KOR455C, KOM458K, KOT562I, and KOK119R (Fig. 1B). The results of a Western blotting analysis using protein lysates prepared from individual cell lines indicated that each mutant RECQ1 protein was expressed at a level comparable with its WT counterpart; however, the K119R mutant is expressed at a lower level (Fig. 1B).
In the subsequent experiments in this study, we utilized this panel of MDA-MB-231 cell lines to assess the roles of WT RECQ1 and breast cancer risk–associated RECQ1 variants in DNA damage response to replication blocking agents.
Loss of RECQ1 protein expression or catalytic function sensitizes MDA-MB-231 cells to camptothecin
Camptothecin is a DNA topoisomerase inhibitor that blocks DNA replication, leading to replication stress in cancer cells (36). Prior studies established that the knockdown of RECQ1 expression by siRNA or short hairpin RNA increases cellular sensitivity to camptothecin (4, 18–20), but the consequences of the complete loss of RECQ1 protein in human cells are unknown. The CCK-8 assay to measure the cell viability demonstrated the increased sensitivity of RECQ1-KO cells to camptothecin (0–400 nm; 24 h). Camptothecin treatment induced a dose-dependent decrease in cell viability up to 100 nm; however, further increase in camptothecin dose to 200 and 400 nm did not result in corresponding decrease in cell viability. When they were exposed to 100 nm camptothecin, ∼91% viable cells were observed in RECQ1-WT compared with only 41% viable cells in RECQ1-KO, indicating that their ability to respond to replication stress was compromised (Fig. 1C). Compared with the RECQ1-WT group, statistically significant sensitivity to camptothecin was observed in the RECQ1-KO and KOpCB6 groups (p < 0.05). We next tested whether camptothecin sensitivity of RECQ1-KO could be corrected by the re-expression of WT and/or mutant RECQ1 protein. Genetic complementation with WT RECQ1 (KOWT cells) rescued the camptothecin sensitivity of RECQ1-KO to a level comparable with RECQ1-WT cells, whereas the expression of an empty vector (pCB6) did not rescue the viability of cells lacking RECQ1 (KOpCB6) (Fig. 1C). In contrast to the RECQ1-KO cells reconstituted with WT RECQ1 (KOWT), increased camptothecin sensitivity was displayed by the RECQ1-KO cells that had been complemented with cancer risk-associated RECQ1 variants (KOA195S, KOR215Q, KOR455C, KOM458K, and KOT562I), as well as helicase-dead KOK119R (Fig. 1D), which was statistically significant (p < 0.05). Among the different mutants tested for camptothecin sensitivity, compared with the KOWT cells, the KOT562I and KOK119R cells displayed greater sensitivity at 100–400 nm camptothecin (p < 0.001). Overall, our results showed that the loss of RECQ1 protein expression or catalytic function sensitized cancer cells to camptothecin and suggested that the helicase function of RECQ1 is important for cell survival following camptothecin-induced DNA damage.
Breast cancer risk–associated RECQ1 variants retain interaction with PARP1
The physical and functional interaction of RECQ1 protein with poly(ADP)ribosyl polymerase 1 (PARP1) is important for the restoration of stalled replication forks caused by camptothecin treatment and general genome maintenance (4, 13, 15). The direct interaction between RECQ1 and PARP1 is mediated by the zinc-binding and winged helix region, which constitutes the RQC domain in RECQ1 (13). Because three of the RECQ1 mutations (R455C, M458K, and T562I) map to the RQC region, we next tested whether the RECQ1 variants interacted with PARP1 in co-immunoprecipitation experiments (Fig. 1E). The RECQ1 antibody specifically co-precipitated PARP1 from whole cell lysates prepared from RECQ1-WT cells, as previously reported (4, 13). RECQ1 also immunoprecipitated RPA, which is known to interact with RECQ1 (12, 13) and was used in these experiments as positive control. PARP1 and RPA were also pulled down in RECQ1 immunoprecipitates from the lysates obtained from KOR455C, KOM458K, and KOT562I cells expressing RECQ1 variants, which suggests that the mutations identified in breast cancer patients do not disrupt the RECQ1-PARP1 interaction (Fig. 1E). Similar immunoprecipitation using cellular extracts prepared from RECQ1-KO cells failed to pull down PARP1, indicating the specificity of RECQ1 antibody.
We next examined whether mutant RECQ1 protein interacted with PARP1 upon camptothecin treatment and whether this interaction required PARP1 activity (Fig. 1F). We compared the RECQ1-PARP1 pulldowns from lysates prepared from RECQ1-WT and KOT562I cells treated with camptothecin in the absence or presence of the PARP inhibitor olaparib (10 μm) (4). The results of the co-immunoprecipitation experiment demonstrated that RECQ1 and PARP1 remain associated following camptothecin treatment in the RECQ1-WT and KOT562I cells (Fig. 1F). In the camptothecin-treated cells, the inhibition of PARP1 activity by olaparib reduced the interaction of RECQ1-PARP1 by ∼50% in the RECQ1-WT cells and by almost 80% in the KOT562I cells compared with their interaction in the absence of PARP inhibition (Fig. 1F). These results suggest that T562I mutation in the RQC domain of RECQ1 does not interfere with PARP1 interaction, but their interaction depends on PARP1 activity in cells treated with camptothecin.
PARP1 activity is required for the slowing of replication forks, and combined with RECQ1, it influences the formation of double-stranded breaks upon camptothecin treatment (4). To determine whether RECQ1 loss and PARP1 activity affected the accumulation of double-stranded breaks in response to camptothecin treatment, we examined γH2AX induction by Western blotting analysis (Fig. 1G). The camptothecin treatment in the RECQ1-WT and KOWT cells did not yield a detectable signal of γH2AX. The further inhibition of PARP1 upon DNA damage induced by camptothecin led to an increase in γH2AX by 5.8- and 5.2-fold in the RECQ1-WT and KOWT cells, respectively. These results are consistent with previous studies (4) demonstrating that PARP inhibition caused the accumulation of reversed forks, leading to the increased accumulation of double-stranded breaks upon camptothecin treatment (4, 37, 38). However, in the RECQ1-KO cells, the camptothecin treatment induced γH2AX by 2.8-fold, and inhibition of PARP1 decreased γH2AX to a level comparable with that of untreated conditions (Fig. 1G). This result indicated that RECQ1 loss prevents double-stranded break formation following PARP inhibition. The DNA damage induced by camptothecin activated PARP1, which was evidenced by the increased signal of poly(ADP)ribosyl polymer (PAR). In addition, the inhibition of PARP1 by olaparib abolished the PAR signal, indicating that the concentration of olaparib used in the experiment indeed inhibited PARP1 activity (Fig. 1G).
Overall, these results showed that RECQ1-KO cells exhibit similar response to camptothecin-induced replication stress as reported previously in RECQ1 knockdown cells (4, 18, 19). Therefore, these cells could be used to investigate cellular functions of RECQ1.
RECQ1 modulates cell survival following gemcitabine treatment
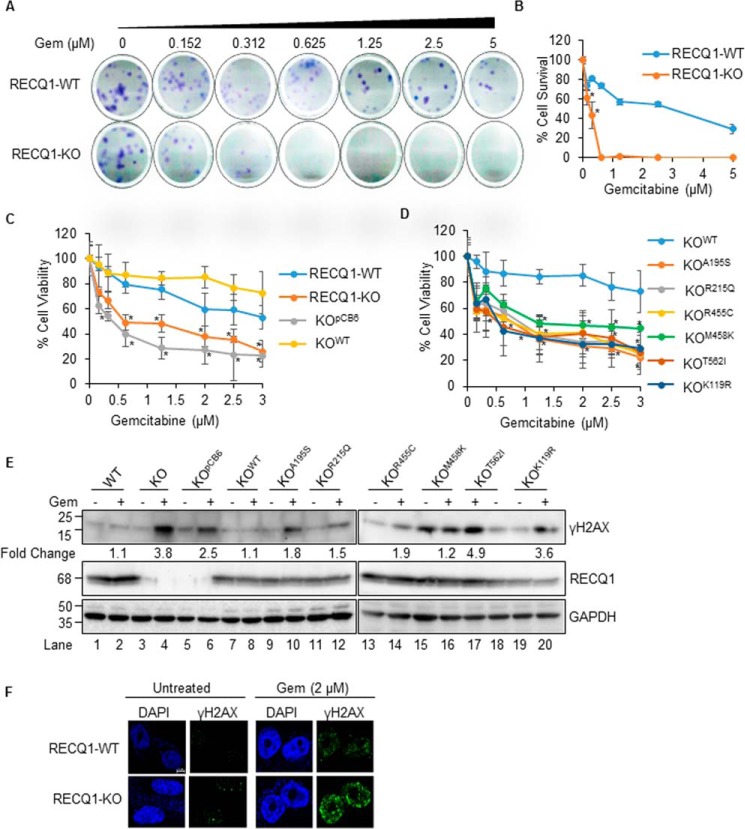
Because of the association of RECQ1 expression with chemotherapeutic response (3, 4, 19) and its relevance to breast cancer (21), we decided to test whether RECQ1 modulated the cellular response to gemcitabine, a nucleoside analog that induces replication-associated DNA damage and is used in the treatment of triple negative breast cancer (30, 39). Clonogenic assays with increasing concentrations of gemcitabine (0–5 μm for 2 h) showed a significantly reduced number of colonies in the RECQ1-KO cells compared with the RECQ1-WT cells (Fig. 2, A and B), indicating decreased survival in the RECQ1-KO cells. Compared with the RECQ1-WT group, the RECQ1-KO cells exhibited greater sensitivity to gemcitabine (p < 0.05) (Fig. 2B). The results of the CCK-8 assay to determine cell viability demonstrated that the RECQ1-KO cells were sensitive to gemcitabine and that the expression of WT RECQ1 in RECQ1-KO cells (KOWT) could reduce gemcitabine sensitivity and increase cell viability, whereas the expression of an empty vector (pCB6) did not rescue the viability of cells lacking RECQ1 (KOpCB6) (Fig. 2C). Furthermore, the cells expressing cancer risk-associated RECQ1 variants (KOA195S, KOR215Q, KOR455C, KOM458K, and KOT562I) or the helicase-dead KOK119R mutant were more sensitive to gemcitabine than those expressing WT RECQ1 (KOWT) (Fig. 2D), indicating that the catalytic activity of RECQ1 may be essential in resolving the DNA damage induced by gemcitabine and improving cell survival. Statistical significance measured using RECQ1-WT cells as a control indicated that the different cell lines (except the KOWT) were sensitive to gemcitabine with p value <0.05. Moreover, RECQ1-KO, KOpCB6, KOT562I, and KOK119R displayed greater differences in sensitivity at 2 μm and 2.5 μm gemcitabine with a p value <0.001 (Fig. 2D).
Figure 2.
Catalytic functions of RECQ1 are important in resolving DNA damage induced by nucleoside analog gemcitabine in MDA-MB-231 cells. A, representative images of colony formation assay of RECQ1-WT and RECQ1-KO cells treated with gemcitabine (Gem). B, quantification of clonogenic survival assay after gemcitabine treatment in RECQ1-WT and RECQ1-KO cells. The graph represents the means (± S.D.) from three independent experiments, and the statistical significance (p < 0.05) between the two cell types is indicated by an asterisk. C and D, drug sensitivity to gemcitabine in RECQ1-WT, RECQ1-KO, and complemented lines. Cell viability data are presented as means (± S.D.) from three independent experiments. The statistical significance of cell viability changes among RECQ1-WT versus other groups is indicated as an asterisk (p < 0.05). Differences are not statistically significant unless denoted by an asterisk. E, Western blotting analysis of γH2AX in the indicated cells treated with gemcitabine (0.1 μm for 24 h). GAPDH is used as a loading control. The load order for untreated and gemcitabine-treated samples from KOT562I cells is reversed. Fold change in gemcitabine-induced γH2AX compared with the respective untreated condition was determined by quantification of signal intensities using ImageJ. F, representative immunostaining images of γH2AX in RECQ1-WT and RECQ1-KO cells untreated or treated with gemcitabine (2 μm for 2 h). DAPI is used as a nuclear stain. The scale bar is 5 μm and represents all images in Fig. 2F. Molecular mass (in kDa) is shown to the left of the Western blots.
Because our data suggested a RECQ1-specific role, we next questioned whether the observed gemcitabine sensitivity is due to increased DNA damage accumulation. To test this, we treated different cell lines used in the study with gemcitabine (0.1 μm for 24 h), prepared total cell lysates, and performed Western blotting analysis for γH2AX as a marker of DNA damage (Fig. 2E). Under untreated conditions, RECQ1-WT and RECQ1-KO cells exhibit comparable basal DNA damage, as indicated by the low γH2AX signal (Fig. 2E, lanes 1 and lane 3). The intensity of γH2AX signal in gemcitabine-treated RECQ1-WT cells was similar to the untreated condition, indicating that these cells efficiently resolve the damage (Fig. 2E, lane 1 versus lane 2). In contrast, gemcitabine treatment induced a 3.8- and 2.5-fold increase in γH2AX in RECQ1-KO and KOpCB6 cells compared with untreated (Fig. 2E, lanes 3 and 5 versus lanes 4 and 6). Reintroduction of WT RECQ1 in RECQ1-KO cells (KOWT) suppressed DNA damage by gemcitabine treatment and displayed γH2AX levels similar to that of RECQ1-WT cells (Fig. 2E, lanes 7 and 8 compared with lanes 1 and 2). Immunostaining of γH2AX in untreated and gemcitabine-treated (2 μm for 2 h) RECQ1-WT and RECQ1-KO cells further demonstrated that the RECQ1-KO cells accumulate gemcitabine-induced DNA damage (Fig. 2F).
Remarkably, cells expressing various RECQ1 variants responded distinctly with respect to the level of gemcitabine-induced DNA damage. RECQ1 variants KOA195S, KOR215Q, and KOR455C displayed up to 2-fold more γH2AX following gemcitabine treatment as compared with RECQ1-WT or KOWT (Fig. 2E, lanes 10, 12, 14, and 16 versus lanes 2 and 8). Although KOM458K cells were sensitive to gemcitabine (Fig. 2D) and exhibit constitutively elevated γH2AX signal, the γH2AX signal intensity in response to gemcitabine was similar to that of RECQ1-WT (Fig. 2E, lane 16 versus lane 2). Consistent with increased sensitivity, the greatest accumulation of gemcitabine-induced γH2AX was seen in KOT562I and KOK119R cells (Fig. 2E, lanes 17 and 20) expressing a RECQ1 mutation in the tyrosine residue in β-hairpin and ATPase-deficient mutation known to be critical for DNA substrate specificity, respectively (9, 31, 35) (Fig. 2D). Collectively, our results suggest that RECQ1 deficiency sensitizes MDA-MB-231 cells to gemcitabine, and the helicase function of RECQ1 may contribute to resolving gemcitabine-induced replication stress and preventing DNA damage.
RECQ1 contributes to effective ATR–ChK1 activation upon gemcitabine treatment
We next sought to examine gemcitabine-induced DNA damage response in RECQ1-WT and RECQ1-KO cells. Previous studies have demonstrated that stable knockdown of RECQ1 increases phosphorylation of RPA32 subunit of RPA and ChK1 in response to camptothecin treatment (19, 20). ATR is activated by phosphorylation of Ser-428, and upon activation, it mediates ChK1 activation by phosphorylating at Ser-317 and Ser-345 residues (28). The inability of RECQ1-KO cells to recover from replication stress indicated by reduced cell survival to gemcitabine raised the possibility that RECQ1 may play role in DNA repair of stalled forks and activation of the ATR–ChK1 pathway. Therefore, we looked at phosphorylation of RPA32, ATR, and ChK1 proteins (Fig. 3).
Figure 3.
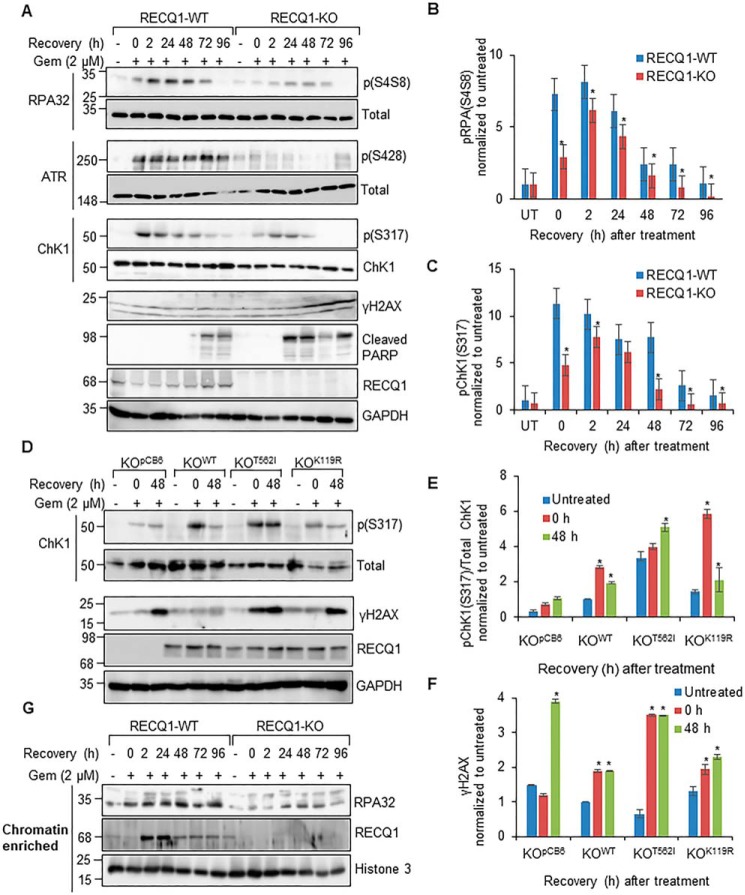
RECQ1 directs the replication stress response induced by gemcitabine in MDA-MB-231 cells via the ATR–ChK1 pathway. A, Western blotting analysis of pRPA32 (Ser-4/Ser-8), RPA32, pATR (Ser-428), ATR, pChK1 (Ser-317), ChK1, γH2AX, and RECQ1 and cleaved PARP in RECQ1-WT and RECQ1-KO cells treated with gemcitabine (Gem, 2 μm for 2 h) followed by recovery in drug-free medium as indicated. GAPDH is used as a loading control. B and C, quantitation of Western blotting signal intensities of pRPA32 (Ser-4/Ser-8) and pChK1 (Ser-317) as represented in Fig. 3A and presented as means (± S.D.) from three independent experiments. UT, untreated. D, Western blotting analysis of pChK1 (Ser-317), ChK1, γH2AX, and RECQ1 in KOpCB6, KOWT, KOT562I, and KOK119R cells treated with gemcitabine (2 μm for 2 h) followed by recovery in drug-free medium as indicated. GAPDH is used as a loading control. E and F, quantitation of Western blotting signal intensities of pChK1 (Ser-317)/total ChK1 and γH2AX as represented in Fig. 3D and presented as means (± S.D.) from three independent experiments. G, Western blotting analysis of RPA32 and RECQ1 in chromatin-enriched fractions of RECQ1-WT and RECQ1-KO cells treated with gemcitabine (2 μm for 2 h) followed by recovery in drug-free medium as indicated. Histone 3 is used as loading control. Molecular mass (in kDa) is shown to the left of the Western blots.
The cells were treated with gemcitabine (2 μm for 2 h) and allowed to recover in drug-free medium for the indicated time points (0, 2, 24, 48, 72, and 96 h) (Fig. 3A). This treatment is known to cause replication fork arrest in U2OS cells (40). The gemcitabine treatment (i.e. 0 h recovery) induced the phosphorylation of RPA32 (at Ser-4/Ser-8, pRPA32), ATR (at Ser-428, pATR), and ChK1 (at Ser-317, pChK1) in the RECQ1-WT cells. During the recovery of the RECQ1-WT cells in the drug-free medium, pRPA32 increased at 2 h, persisted until 48 h, and gradually decreased at 72 and 96 h. The level of pATR did not change appreciably during the recovery, whereas the pChK1 signal decreased gradually during the recovery. In contrast, the gemcitabine treatment induced significantly less (2-fold less than the RECQ1-WT) pRPA32 in the RECQ1-KO cells and failed to activate ATR, which was evidenced by a weak signal of pATR compared with the RECQ1-WT cells (Fig. 3A). Consistent with impaired ATR activation, the RECQ1-KO cells also failed to activate ChK1 robustly, which was seen in their WT counterparts. The level and kinetics of the phosphorylation of RPA32 and ChK1 in response to the gemcitabine treatment in the RECQ1-KO cells were distinct from the RECQ1-WT cells (Fig. 3, B and C). The levels of the total proteins (ATR, ChK1, and RPA32) were comparable in the RECQ1-WT and RECQ1-KO cells (Fig. 3A).
To eliminate the possibility of clonal variation, we utilized another set of RECQ1-WT and RECQ1-KO clones that were obtained by CRISPR-Cas9 for treatment with gemcitabine and followed the recovery to 96 h (Fig. S1, B–D). The RECQ1-KO4 cells displayed a decreased signal of pRPA32 and pChK1, indicating decreased checkpoint activation and further accumulated DNA damage. Because the reduced ChK1 phosphorylation in response to gemcitabine in the RECQ1-KO cells was demonstrated in two different clones, it is unlikely that the genetic background of cells influenced this specific phenotype. Although the ATR–ChK1 pathway plays a primary role in protection against gemcitabine-induced replication stress, gemcitabine treatment is also known to activate the ATM-ChK2 signaling pathways (41). Although the difference between RECQ1-WT and RECQ1-KO cells was not as striking, reduced phosphorylation of ATM (at Ser-1981, pATM) and ChK2 (at Thr-68, pChK2) was observed in RECQ1-KO cells upon gemcitabine treatment and recovery (Fig. S1E).
Consistent with defective ATR–ChK1 activation, the RECQ1-KO cells accumulated progressively increased γH2AX, indicating that the repair of gemcitabine-induced DNA damage was compromised during the recovery period and that they underwent increased apoptosis, which was evidenced by cleaved PARP, a widely used apoptotic marker (42) (Fig. 3A). The RECQ1-WT cells did not exhibit a significant increase in DNA damage, which was indicated by the γH2AX signal during recovery. Moreover, in the RECQ1-WT cells, the cleaved PARP was observed only at 72 and 96 h after the gemcitabine treatment (Fig. 3A).
We next examined ATR–ChK1 activation in cells expressing RECQ1 variant that was defective in helicase activity using T562I (cancer risk-associated) and K119R (biochemically characterized) mutants as representatives (Fig. 3D). RECQ1-KO cells that stably expressed empty vector pCB6 (KOpCB6), WT RECQ1 (KOWT), T562I (KOT562I), or K119R (KOK119R) were subjected to gemcitabine treatment (2 μm for 2 h) and allowed to recover for 48 h in a drug-free medium. The total ChK1 protein level in the cells expressing mutant RECQ1 (i.e. KOT562I and KOK119R) was lower than those expressing WT RECQ1 (i.e. KOWT) or lacking RECQ1 (i.e. KOpCB6) (Fig. 3D). Although the ChK1 activation in the KOT562I cells was comparable with KOWT, which was shown by the pChK1 signal (Fig. 3E), they accumulated DNA damage similar to the KOpCB6 cells, which was evidenced by a γH2AX signal following the gemcitabine treatment and recovery (Fig. 3F). This result suggests that the presence of RECQ1 protein (WT or mutant) is enough to activate the ATR–ChK1 pathway in response to replication stress; however, RECQ1 helicase activity is indispensable in resolving gemcitabine-induced DNA damage in MDA-MB-231 cells.
RPA binding to ssDNA stretches triggers the activation of the ATR–ChK1 axis (24, 27); therefore, we examined RPA levels in the chromatin fractions from RECQ1-WT and RECQ1-KO cells treated with gemcitabine, followed by recovery at indicated time points (Fig. 3G). In the RECQ1-WT cells, the treatment with gemcitabine resulted in increased RPA on chromatin (Fig. 3G). In contrast, ∼50% reduced RPA was detected in the chromatin-enriched fractions of the RECQ1-KO cells compared with the RECQ1-WT cells (Fig. 3G). Greater RPA levels were also seen in the chromatin fractions of RECQ1-KO cells expressing WT RECQ1 (KOWT) as compared with those expressing empty vector (KOpCB6) during gemcitabine-induced replication stress (Fig. S2). This suggests that the impaired RPA recruitment on chromatin contributed to defective checkpoint activation in RECQ1-KO cells. RECQ1 mRNA levels are induced 2–3-fold in response to gemcitabine as we have previously reported (43). Gemcitabine treatment also enriched RECQ1 on chromatin during recovery from the gemcitabine treatment (Fig. 3G).
RECQ1 and MUS81 resolve gemcitabine-induced DNA damage in MDA-MB-231 breast cancer cells
RECQ1 helicase functions in the restarting of stalled replication forks, defective processing of which may lead to double-stranded breaks (1, 4). Therefore, we next carried out a DNA comet assay under neutral conditions to measure gemcitabine-induced double-stranded breaks in RECQ1-WT and RECQ1-KO cells and their repair during recovery (Fig. 4, A and B). We observed a negligible difference in the comet tail moments in the RECQ1-WT and RECQ1-KO cells under untreated conditions. The results of the comet assay showed that double-stranded breaks occurred within 2 h following the treatment with gemcitabine, and the pattern of distribution of the comet tail length differed in both cell lines. Significantly greater DNA damage was seen in RECQ1-KO cells upon the gemcitabine treatment and up to 96 h of recovery in a drug-free medium compared with the RECQ1-WT cells (p < 0.05) (Fig. 4A). The analysis of the percentage of DNA in the tail under each condition revealed that up to 60% of the double-stranded breaks were repaired in the RECQ1-WT cells by 96 h compared with only 10% in the RECQ1-KO cells (Fig. 4B).
Figure 4.
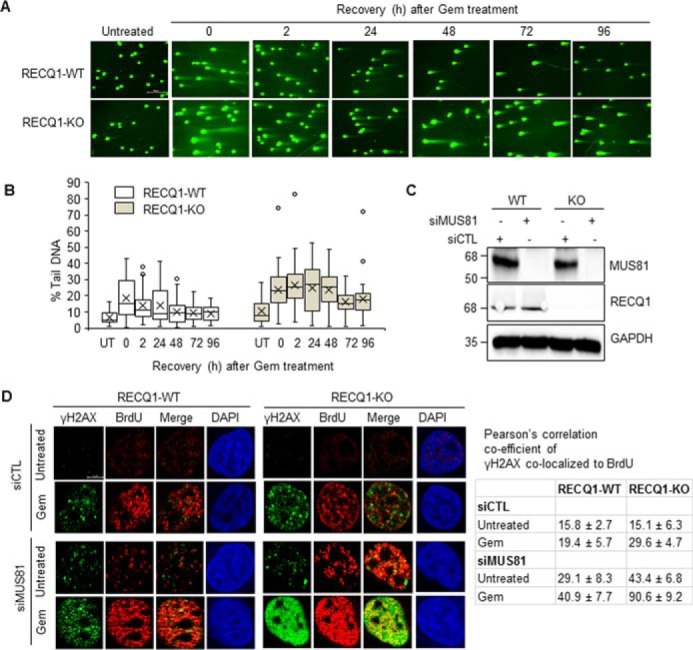
Loss of RECQ1 and depletion of MUS81 accumulates gemcitabine-induced DNA damage in MDA-MB-231 cells. A, RECQ1-WT and RECQ1-KO cells were treated with gemcitabine (Gem, 2 μm for 2 h) followed by recovery in drug-free medium as indicated. Double-stranded breaks, and repair were analyzed using a neutral comet assay. The scale bar is 200 μm and represents all images in Fig. 4A. B, DNA damage in RECQ1-WT and RECQ1-KO cells was quantified as % tail DNA using the Open Comet plugin in ImageJ. In the box-whisker plot, the median divides the box and error bars are shown as whiskers. At least 50 comets were scored in each condition. C, MUS81 knockdown in control (siCTL) or MUS81 (siMUS81) siRNA transfected RECQ1-WT and RECQ1-KO cells was verified by Western blotting analysis. GAPDH is used as a loading control. D, representative images of γH2AX and BrdU staining in siCTL or siMUS81 transfected RECQ1-WT and RECQ1-KO cells upon ± gemcitabine treatment (2 μm for 2 h). DAPI is used as a nuclear stain. Shown are Pearson's correlation co-efficients concerning the co-localization of γH2AX and BrdU expressed as percentages under the indicated test conditions in RECQ1-WT and RECQ1-KO cells. The scale bar is 5 μm and represents all images in Fig. 4D. Molecular mass (in kDa) is shown to the left of the Western blots.
MUS81 is a structure-specific endonuclease that plays a critical role in replication fork rescue by converting stalled replication forks into double-stranded breaks that can be processed by homologous recombination repair (44, 45). Therefore, we next tested whether MUS81 mediates the formation of replication associated double-stranded breaks in RECQ1-KO cells (Fig. 4, C and D). Under our experimental conditions, MUS81 siRNA reduced MUS81 protein expression by >90% in the RECQ1-WT and RECQ1-KO cells (Fig. 4C). 48 h after control or MUS81 siRNA transfection, replication fork arrest was induced by gemcitabine (2 μm for 2 h), and the replication-associated double-stranded breaks were measured by co-localization of γH2AX and BrdU foci using immunostaining (Fig. 4D). In both cell lines, the treatment with gemcitabine induced BrdU foci that represented replicating cells (Fig. 4D). MUS81 knockdown led to a higher frequency of γH2AX-BrdU co-localizing foci in both RECQ1-WT and RECQ1-KO cells. MUS81 knockdown and gemcitabine treatment in RECQ1-WT cells resulted in ∼2-fold increase in γH2AX-BrdU co-localizing foci when compared with the gemcitabine-treated RECQ1-WT cells transfected with control siRNA. As compared with gemcitabine-treated RECQ1-KO cells transfected with control siRNA, ∼3-fold increase in γH2AX-BrdU co-localizing foci were observed in gemcitabine-treated RECQ1-KO cells transfected with MUS81 siRNA, indicating greater accumulation of replication-associated double-stranded breaks (Fig. 4D). This suggests that RECQ1 helicase and MUS81 nuclease resolve arrested replication forks and promote replication restart, thus preventing the generation of double-stranded breaks from aberrant processing of stalled forks. In the dual absence of RECQ1 and MUS81, other structure specific endonucleases like MRE11, CtIP, EXO1, and DNA2 may promote processing of regressed replication forks leading to the formation of double-stranded breaks (37, 46, 47).
ChK1 inhibition synergizes with gemcitabine and leads to death in RECQ1 knockout cells
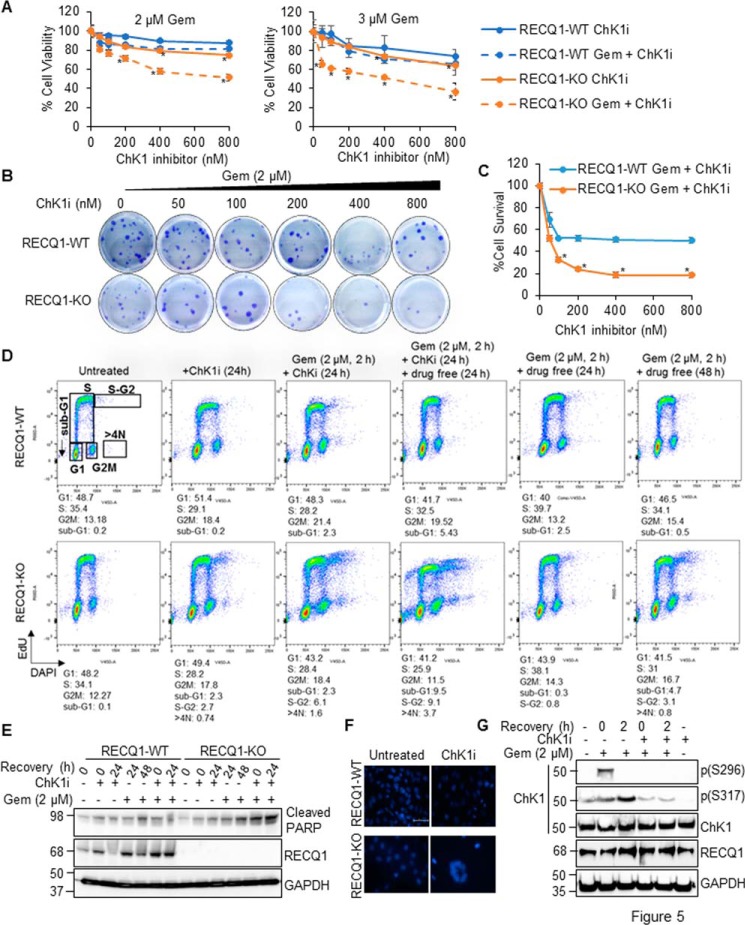
Targeting ChK1 can augment the effect of gemcitabine-based chemotherapy on breast cancer cells (48). We treated RECQ1-WT, RECQ1-KO, and complemented cell lines with gemcitabine (2 μm or 3 μm for 2 h), followed by the addition of increasing concentrations of ChK1 inhibitor (LY2603618) or ChK1 inhibitor alone. We then measured cell viability after 24 h by CCK-8 assay. When cultured for 24 h in the presence of the ChK1 inhibitor, the viability of the RECQ1-KO cells was comparable with the RECQ1-WT cells at lower doses (p > 0.05). A statistically significant difference in viability of RECQ1-WT and RECQ1-KO cells was observed at higher doses of the ChK1 inhibitor (400–800 nm) (p < 0.05) (Fig. 5A). In the combined treatments using 2 μm gemcitabine, RECQ1-KO displayed significant sensitivity (p < 0.05) at all concentrations of the ChK1 inhibitor compared with the similarly treated RECQ1-WT cells. Further reduction in proliferation of RECQ1-KO cells were observed when a higher concentration of gemcitabine (3 μm) was used in combination with ChK1 inhibitor, indicating potential synergism between gemcitabine and ChK1 inhibitor (Fig. 5A). The KOpCB6 cells displayed increased sensitivity to combination treatment (ChK1 inhibitor plus gemcitabine) when compared with ChK1 inhibitor alone, and this was partially rescued by WT RECQ1 (KOWT), indicating a RECQ1 specific function in resolving stress induced by gemcitabine via the ATR–ChK1 axis. Cells expressing RECQ1 variants did not display sensitivity to ChK1 inhibitor alone or combination treatment to gemcitabine and ChK1 inhibitor (Fig. S3). We next tested the effect of combined treatment of gemcitabine (2 μm) and ChK1 inhibitor on cell survival by colony formation assay (Fig. 5, B and C). As compared with the RECQ1-WT cells, the RECQ1-KO cells displayed significantly reduced survival over the range of ChK1 inhibitor tested in combination with gemcitabine (p < 0.05) (Fig. 5, B and C).
Figure 5.
Loss of RECQ1 combined with ChK1 inhibitor markedly potentiates cytotoxicity of gemcitabine in MDA-MB-231 cells. A, sensitivity of RECQ1-WT and RECQ1-KO cells treated with increasing concentrations of ChK1 inhibitor (ChK1i) alone or a combination of gemcitabine (Gem, 2 μm or 3 μm for 2 h) and ChK1i was measured by CCK-8 reagent. The data represent means (± S.D.) from three independent experiments. The statistical significance of cell viability changes between RECQ1-WT versus RECQ1-KO was determined for each treatment and is indicated as an asterisk (p < 0.05). B, representative images of colony formation assay in RECQ1-WT and RECQ1-KO cells treated with gemcitabine (2 μm for 2 h) followed by increasing concentrations of ChK1i (24 h). C, quantification of clonogenic survival after gemcitabine plus ChK1i treatment in RECQ1-WT and RECQ1-KO cells. Statistical significance is indicated as an asterisk (p < 0.05). D, flow cytometry analysis of the cell cycle profiles of RECQ1-WT and RECQ1-KO cells treated with ChK1i alone, gemcitabine + ChK1i, or gemcitabine alone followed by recovery as indicated. The cells were pulse-labeled with EdU for 90 min before harvesting, and the cell cycle was tested with a Click-iT EdU kit by FACS. Scatter plots represent EdU labeling (y axis) and DAPI (x axis). The percentage of cells in each cell cycle phase was gated based on the intensity of EdU staining and DNA content, as represented here for the untreated RECQ1-WT cells and is indicated below each histogram. E, Western blotting analysis of cleaved PARP in RECQ1-WT and RECQ1-KO cells treated as in D to indicate apoptosis. F, representative images of DAPI-stained nuclei of RECQ1-WT and RECQ1-KO cells treated with ChK1i (100 nm for 24 h). The scale bar is 200 μm and represents all images in F. G, Western blotting analysis of phosphorylated ChK1 (pSer-296 and pSer-317) in RECQ1-WT cells treated with gemcitabine (2 μm for 2 h) ± ChK1i (100 nm for 2 h) confirm ChK1 inhibition by LYS2603618 under stated experimental conditions. GAPDH is used as a loading control. Molecular mass (in kDa) is shown to the left of the Western blots.
The intra–S-phase checkpoint is regulated by the checkpoint kinases ATR and ChK1 (49). Our results showed that the loss of RECQ1 resulted in decreased ATR–ChK1 activation in response to replication stress (Fig. 3A). Moreover, the combination of gemcitabine and the ChK1 inhibitor increased the sensitivity of the RECQ1-KO cells (Fig. 5, A–C), which suggest the combined role of RECQ1 and ChK1 in mediating the response to gemcitabine-induced DNA damage. To further investigate the mechanism by which the ChK1 inhibitor increased the effect of gemcitabine on cell viability, we analyzed cell cycle distribution and apoptosis induction in response to drug treatments. RECQ1-WT and RECQ1-KO cells were treated with gemcitabine alone, the ChK1 inhibitor alone, or a combination of gemcitabine and the ChK1 inhibitor, followed by recovery in a drug-free medium as indicated (Fig. 5D). The resumption of DNA replication in the drug-treated cells was determined by monitoring the cell cycle progression of the EdU-labeled cells, which is a widely used marker of cell division (50).
The unperturbed RECQ1-WT and RECQ1-KO cells exhibited comparable cell cycle distribution, indicating that RECQ1 loss does not alter the cell cycle profile of MDA-MB-231 cells in the absence of DNA damage (Fig. 5D). The single treatment with the ChK1 inhibitor (100 nm for 24 h) caused the transition of S-phase cells into G2M phases in both the RECQ1-WT and RECQ1-KO cells compared with their respective untreated controls. This result was evidenced by a 1.75-fold increase in the G2M population and a reduction in the S-phase population of ∼10% in both cell lines. Compared with the RECQ1-WT cells, the ChK1 inhibitor treatment resulted in a distinct increase in the S-G2 population (2.7%) and in the >4N population (0.74%) in the RECQ1-KO cells (Fig. 5D).
The gemcitabine treatment (2 μm for 2 h) followed by the ChK1 inhibitor resulted in an increase in the G2M population from 13.18% (untreated) to 21.4% in the RECQ1-WT cells. This result is consistent with earlier reports that in the context of DNA damage by replication inhibitors, ATR is involved in the S-phase checkpoint, and ChK1 inhibition results in the abrogation of the S-phase checkpoint, which forces the cells to enter the G2M phase (49, 51). In the RECQ1-WT cells, recovery in the drug-free medium for an additional 24 h caused an increase in the sub-G1 population (5%); however, the surviving cells were able to resume the cell cycle, and their profiles resembled a normal untreated condition, which indicated an efficient response to DNA damage in the event of gemcitabine treatment and ChK1 inhibition (Fig. 5D). In contrast, when the RECQ1-KO cells were treated with gemcitabine followed by continuous incubation with the ChK1 inhibitor, these cells incorporated less EdU into their DNA (28.4% compared with 34.1% in the untreated cells). However, they showed the increased accumulation of cells in S-G2 (6.1%), >4N phases (1.6%), and sub-G1 (2.3%) populations. This increase suggests that RECQ1-KO cells fail to resume normal DNA replication and are forced to enter mitosis with under replicated DNA (premature mitotic entry). Further recovery in the drug-free medium clearly increased the S-G2 (9.5%), >4N (3.7%), and sub-G1 (9.1%) populations. The increase in the sub-G1 suggests that the population of RECQ1-KO cells that underwent premature mitotic entry failed to complete cell division and succumbed to cell death.
Our results suggest that in the RECQ1-KO cells, the combined gemcitabine and ChK1 inhibitor caused the abrogation of the S-phase checkpoint and induced the premature entry of the S-phase cells to mitosis, which may lead to the phenomenon of the mitotic catastrophe, which is considered a lethal event that progresses to apoptosis (52, 53). Consistent with the results of the experiments on cell survival, the loss of RECQ1 notably increased the percentage of the sub-G1 cells produced by the combined gemcitabine and ChK1 inhibitor treatment compared with gemcitabine alone. However, during the single treatment with gemcitabine, the RECQ1-WT cells enriched in the S phase at 24 h recovery and resumed regular cell cycle distribution at 48 h recovery following treatment. In contrast, the RECQ1-KO cells failed to return to their untreated cell cycle distribution at 48 h of recovery from gemcitabine treatment. We did not observe a >4N population (premature mitosis) in the RECQ1-WT cells treated with gemcitabine alone, the ChK1 inhibitor alone, or the combination of gemcitabine and the ChK1 inhibitor. Additionally, the combination of gemcitabine and ChK1 inhibition in RECQ1-KO cells increased apoptosis, which was evidenced by the increased signal of cleaved PARP (Fig. 5E). These findings suggest that in the absence of RECQ1, cells accumulate DNA damage and undergo cell death upon ChK1 inhibition subsequent to gemcitabine treatment. We then assessed the percentage of apoptotic cells after ChK1 inhibition by conducting quantitative microscopy of the DAPI-stained cells. We observed an increased number of RECQ1-KO cells with multinucleus upon ChK1 inhibition, which suggested failed mitosis or cell death by mitotic catastrophe (52) (Fig. 5F). To confirm the inactivation of ChK1, we treated RECQ1-WT cells with gemcitabine followed by the ChK1 inhibitor, and the cell lysates were analyzed by Western blotting (Fig. 5G). The gemcitabine treatment induced phosphorylation of ChK1 (at Ser-296, pSer-296) and (at Ser-317, pSer-317) levels in MDA-MB-231 cells, and the addition of the ChK1 inhibitor abolished the pChK1(S296) levels in response to gemcitabine-induced DNA damage, thus confirming the activity of the ChK1 inhibitor under our experimental conditions (Fig. 5G).
Overall, our results suggest that RECQ1-KO MDA-MB-231 cells have defective ATR–ChK1 activation in response to gemcitabine-induced replication stress and that the combined treatment with the ChK1 inhibitor and gemcitabine induces significant apoptosis and polyploidy in RECQ1-KO cells. Based on these results, we speculate that RECQ1's cellular functions are critical for the optimal resolution of stalled forks and the activation of kinases that respond to damage, thus maintaining genome integrity.
Discussion
The discovery that germ-line mutations compromise RECQ1 helicase activity, thus increasing breast cancer susceptibility, suggests that the catalytic activities of RECQ1 are important in DNA repair and genome maintenance (5, 6). In this study, we hypothesized that the expression and molecular functions of RECQ1 are important for the repair of gemcitabine-induced DNA damage. To test this hypothesis, we established a novel panel of MDA-MB-231 cell lines that either express or lack RECQ1 expression or express RECQ1 variants that are known to increase breast cancer risk. The results of using this model system suggest that RECQ1 is required for efficient checkpoint activation in response to gemcitabine treatment. We showed that RECQ1 loss sensitized MDA-MB-231 cells to gemcitabine treatment and that subsequent treatment with the ChK1 inhibitor forced these cells to undergo cell death. RECQ1 appears to modulate ATR activation by facilitating the phosphorylation of ATR on Ser-428 and the subsequent phosphorylation of its downstream target ChK1 on Ser-317. Furthermore, RECQ1-deficient cells exhibit more γH2AX and increased signals for cleaved PARP, suggesting that defective ATR activation upon RECQ1 loss leads to increased genomic instability and apoptosis.
Overall, our results suggest the significant role of RECQ1 in signaling the response to DNA damage in cancer cells (Fig. 6). ATR activation is a multistep process in which the RPA-coated ssDNA recruits ATR–ATRIP (ATR-interacting protein) and localizes ATR to the sites of DNA damage (54). The ATR–ATRIP complex then interacts with the Rad9–Rad51–Hus1 (9-1-1) complex, and a series of phosphorylation events in the involved proteins eventually leads to ATR activation (24, 26). Activated ATR (i.e. ATR phosphorylated at Ser-428) activates ChK1 by phosphorylation on serine residues 317 and 345 in response to replication arrest and mediates the down-regulation of replication origin firing, cell cycle arrest, and DNA repair (28). The defective activation of ATR in RECQ1-KO cells raises the possibility that RECQ1 may cooperate with ATR, which is the main responder in the DNA damage response during replication stress (29), to elicit a rapid cellular response to resolve stalled replication forks. The observation that the loss of RECQ1 decreased RPA phosphorylation suggests that RECQ1 either functions upstream of RPA or in the same step as RPA activation or recruitment to chromatin. In contrast to earlier reports that the stable knockdown of RECQ1 results in the hyperactivation of ChK1 in response to replication stress (19, 20), we found that the complete loss of RECQ1 expression in MDA-MB-231 cells resulted in the reduced activation of ChK1, which was indicated by the phosphorylation of Ser-317.
Figure 6.
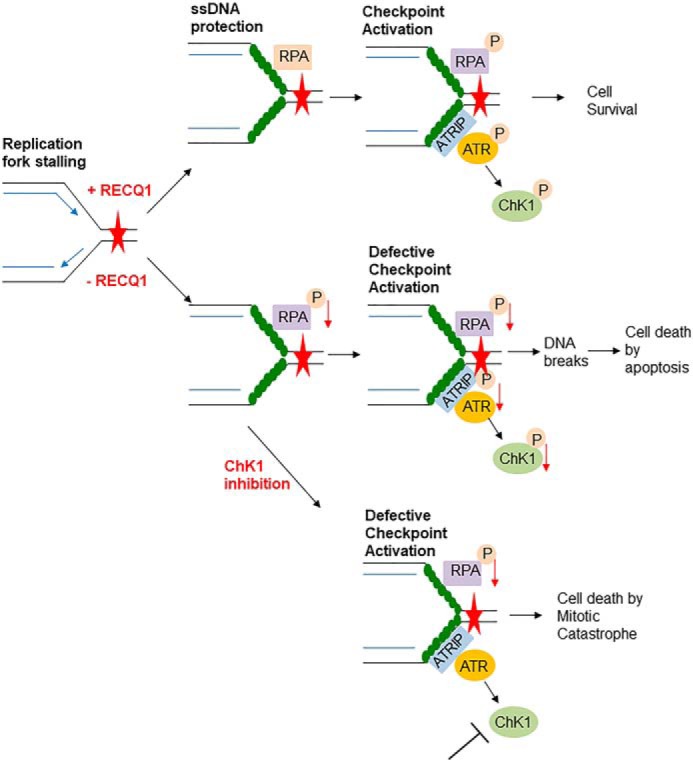
Model/summary of RECQ1 as a mediator of replication stress response induced by gemcitabine in MDA-MB-231 cells. RECQ1 is involved in RPA recruitment to DNA damaged sites and facilitates activation of ATR–ChK1 pathway to resolve the DNA damage induced by gemcitabine. In the absence of RECQ1, RPA recruitment to damage sites is decreased, leading to defective checkpoint activation accompanied by DNA damage accumulation that eventually leads to cell death by apoptosis. RECQ1 loss in combination with gemcitabine and ChK1 inhibitor facilitates cell death by mitotic catastrophe.
Multiple mechanisms have been proposed to show how DNA checkpoint inhibitors function (55, 56). Emphasizing the importance of how the time and dosage of gemcitabine treatment and ChK1 inhibition modulates the mechanism by which a cancer cell undergoes apoptosis, the Eastman lab recently reported that gemcitabine treatment followed by the late administration of ChK1 inhibition resulted in cell death by replication catastrophe caused by the exhaustion of RPA instead of premature mitotic entry in MDA-MB-231 cells (51). Because our results suggest that RECQ1 loss decreases RPA recruitment at damage sites that are expected to expose ssDNA, we cannot eliminate the possibility that replication catastrophe is a mechanism of gemcitabine-induced cytotoxicity in RECQ1-KO cells. The Brosh lab reported the role of RECQ1 in maintaining free RPA availability to coat nascent ssDNA during replication stress and therefore in the repair of replication-associated double-stranded breaks (20). Because RECQ1 efficiently binds and unwinds synthetic fork duplexes in vitro (9, 19) and a physical and functional interaction exists between RECQ1 and RPA (10, 12, 13), it is plausible that RECQ1 stabilizes stalled forks and promotes RPA accumulation at exposed ssDNA, thereby activating the ATR–ChK1 pathway to promote DNA repair. Moreover, our results indicate that RECQ1 loss induces cell death by premature mitotic entry when MDA-MB-231 cells are treated with a combination of gemcitabine and the ChK1 inhibitor, which further confirms the potential benefits of strategies that induce forced mitotic entry (57).
DNA repair pathways that may alter the cytotoxicity of gemcitabine have not been well-elucidated. Brief gemcitabine treatment inhibits ribonucleotide reductase, thereby depleting the dNTP pool (30), which may lead to nucleotide misincorporation, potentially causing errors in DNA replication, such as single-base substitution, insertion, or deletion (30). Previous studies demonstrated that at earlier time points, the repair of gemcitabine-induced DNA damage depends on the mismatch repair pathway, whereas recovery in a drug-free medium at later time points requires a homologous recombination (58). RECQ1 is known to interact with the mismatch repair proteins MSH1 and MSH6 (16). Therefore, we cannot rule out the potentially altered mismatch repair of RECQ1-KO cells treated with gemcitabine. The catalytic functions of RECQ1 are implicated in the homologous recombination pathway, which contributes to the repair of damaged replication forks by resolving aberrant DNA structure intermediates (19, 22). RECQ1 has a striking preference for promoting fork restoration (reversal), which may serve to prevent chromosome breakage upon exogenous replication stress and DNA damage (4). Our results suggest that RECQ1 and MUS81 function in alternate pathways to resolve gemcitabine-induced DNA damage. Nucleases that mediate gemcitabine-induced double-stranded breaks in RECQ1-KO cells remain to be examined.
A large gap in the understanding of RECQ1-associated breast cancer susceptibility is the extent to which disease pathogenesis is governed by a defective DNA metabolism in the functional absence of RECQ1 helicase versus other poorly characterized noncatalytic roles of RECQ1 protein. In this context, by employing the RECQ1 missense mutants (i.e. identified patient mutations), our results suggest that the catalytic activities of RECQ1 may not be involved in ATR–ChK1 activation. Although the breast cancer risk–associated RECQ1 T562I variant was able to induce robust ChK1 phosphorylation, cells expressing this variant failed to resolve the accumulated DNA damage as evidenced by increased γH2AX accumulation, indicating that the RECQ1 T562I mutation uncouples RECQ1's role in ChK1 activation from DNA damage in response to gemcitabine treatment under our experimental conditions. Our results demonstrated that missense mutations in the catalytic domain of RECQ1 sensitize cancer cells to camptothecin and gemcitabine and compromise DNA repair. However, our results showing that RECQ1 variants associate with PARP1 and RPA, two well-characterized partners of RECQ1 (4, 12, 13, 20), demand a more comprehensive investigation of the effects of RECQ1 variants on the cross-talk between the proteins involved in the response to DNA damage. The poly(ADP)ribosylation activity of PARP1 inhibits fork reversal by RECQ1 in vivo to prevent the premature restarting of regressed forks, and the RECQ1-PARP1 complex stabilizes regressed forks until the repair is complete (4). Although the tested mutations in RECQ1 did not disrupt its association with PARP1 in unperturbed cells, the association of the RECQ1-T562I variant with PARP1 under replication stress was found to be dependent on PARP1 activity.
Overall, our results indicate that RECQ1 functions to facilitate cell survival upon gemcitabine-induced replication stress. When replication forks stall under gemcitabine treatment, RECQ1 helicase aids in RPA accumulation by revealing/generating tracts of ssDNA at stalled forks, resulting in checkpoint activation, DNA damage repair, and cell survival. In the absence of RECQ1, RPA recruitment at ssDNA and checkpoint activation is reduced, leading to an increased number of DNA breaks and cell death (Fig. 6). Although ATR and ChK1 inhibitors are included in clinical trials (59), there is a need for biomarkers that can be used to identify patients who would most likely benefit from anti-ATR/anti-ChK1 therapy. The results of our study suggest that RECQ1 expression may have the potential to stratify patients and to be used as a biomarker in breast cancer to predict their survival outcomes based on ChK1 inhibition or gemcitabine treatment. Our results also suggest that targeting RECQ1 with a small molecule inhibitor in combination with ChK1 inhibition could be beneficial. The effects of many chemotherapeutics are modulated by DNA damage responses; therefore, it may be relevant to target RECQ1 to improve therapeutic strategies to treat cancer and transfer this knowledge to clinical practice.
Experimental procedures
Generation of RECQ1-KO MDA-MB-231 breast cancer cell-line by CRISPR-Cas9 technique
Human breast cancer MDA-MB-231 cells (ER−, PR−, and HER2−) in a 6-well plate at 80–90% confluency were transfected with vectors encoding RECQ1 gRNA corresponding to the target site in Exon 3 (2 μg), Cas9 nuclease (WT, 2 μg), transposon (puromycin selection marker, 500 ng), and transposase (500 ng) at a ratio of 4:4:1:1 using Lipofectamine 2000 as described by the manufacturer. 48 h post-transfection, the transformants were selected by puromycin (2 μg/ml) for an additional 48 h. Following puromycin selection, the cells were harvested using trypsin and then diluted using the serial dilution method. The diluted cells were plated for 3 weeks in a 96-well plate at a cell density of 1 cell/well in puromycin (2 μg/ml) containing medium. Single clones were marked and then expanded in 24 wells for further screening. Protein extracts from the isolated clones were prepared by resuspending the cell pellets in radioimmune precipitation assay buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 2 mm EDTA, 1% Nonidet P-40, 0.5% sodium deoxycholate) supplemented by protease inhibitors and incubated on ice for 30 min. After incubation, the cells were centrifuged at 15,000 rpm for 20 min at 4 °C, and the supernatant was collected as the protein lysate. Equal amounts of protein from each sample were separated by 10% SDS-PAGE (135V for 2 h), transferred to polyvinylidene difluoride membrane (135 V for 2 h at 4 °C), blocked with 5% milk (room temperature, 1 h), and probed by Western blotting with antibodies specific to RECQ1 (1:1000, Santa Cruz Biotechnology) and GAPDH (1:1000, Cell Signaling Technologies). The genomic DNA was isolated from different clones, and the indels in the knockout clones were identified by comparing them with the parental WT sequence analyzed by Sanger sequencing.
Cloning and site-directed mutagenesis
cDNA encoding full-length human RECQ1 was cloned into pCB6 plasmid to be expressed as the HA-FLAG dual-tagged fusion protein using BamH1 and EcoRV as restriction enzymes. Individual point mutations (A195S, R215Q, R455C, M458K, and T562I) were introduced by site-directed mutagenesis using a QuikChange II XL site-directed mutagenesis kit (Agilent Technologies) according to the manufacturer's instructions. The mutagenic oligonucleotide primers for each desired mutation were designed using the primer design tool produced by Agilent Technologies, as indicated in Table 1. Using pCB6-RECQ1-WT as a template, the PCR was set up as follows: 5 μl of 10× reaction buffer, 1 μl (10 ng) of pCB6 RECQ1-WT template, 1.25 μl (125 ng) of oligonucleotide primer 1, 1.25 μl (125 ng) of oligonucleotide primer 2, 1 μl of dNTP mix, 3 μl of QuikSolution, 1 μl of Pfu Ultra HF DNA polymerase (2.5 units/μl), and double-distilled H2O to a final volume of 50 μl. The PCR cycling parameters were as follows: step 1, 95 °C for 1 min; step 2, 95 °C for 50 s, 60 °C for 50 s, 68 °C for 5 min; step 2 repeated for 18 cycles; and step 3, 68 °C for 7 min. After the PCR was complete, 1 μl of DpnI was added to the PCR product and incubated at 37 °C for 1 h. After digestion with DpnI, the transformation was set up using XL10-Gold Ultracompetent cells. To increase the transfection efficiency, 2 μl of β-mercaptoethanol was added to 40 μl of XL10-Gold Ultracompetent cells and incubated on ice for 10 min. Subsequently, 2 μl of DpnI-digested mix was added to the competent cells and incubated on ice for 30 min. The tubes were heat-pulsed at 42 °C for 30 s followed by incubation on ice for 2 min. Then 0.5 ml of SOC medium was added to the transformation mix, and the tubes were incubated at 37 °C for 1 h on a rotor. The transformation mix was plated on LB ampicillin agar plates followed by incubation at 37 °C for 16 h. Individual colonies were propagated in LB broth containing ampicillin. Plasmid DNA was isolated using a Qiagen mini prep kit, and the isolated plasmid DNA was sequenced by Sanger sequencing using the appropriate primers.
Table 1.
Primers used to generate site-directed mutants by site-directed mutagenesis
| Desired mutation | Sequence 5′ → 3′ | Melting temperature (Tm) |
|---|---|---|
| A195S | ||
| RECQ1_583_FW | ACTCCAGAGAAAATTTCAAAAAGCAAAATGT | 62.7 |
| RECQ1_583_RV | ACATTTTGCTTTTTGAAATTTTCTCTGGAGT | 62.7 |
| R215Q | ||
| RECQ1_644_FW | CAAGGAGATTTACTCAAATTGCTGTGGATGA | 65.1 |
| RECQ1_644_RV | TCATCCACAGCAATTTGAGTAAATCTCCTTG | 65.1 |
| R455C | ||
| RECQ1_1363_FW | ATAAGCAAATGTCGTTGTGTGTTGATGGCTC | 67.4 |
| RECQ1_1363_RV | GAGCCATCAACACACAACGACATTTGCTTAT | 67.4 |
| M458K | ||
| RECQ1_1373_FW | GTCGTCGTGTGTTGAAGGCTCAACATTTTGA | 69.1 |
| RECQ1_1373_RV | TCAAAATGTTGAGCCTTCAACACACGACGAC | 69.1 |
| T562I | ||
| RECQ1_1685_FW | AAGACTACAGTTTTATAGCTTATGCTACCAT | 60.8 |
| RECQ1_1685_RV | ATGGTAGCATAAGCTATAAAACTGTAGTCTT | 60.8 |
Drug sensitivity assays
RECQ-WT cells, RECQ1-KO cells, and stable lines expressing either an empty vector or WT RECQ1 or RECQ1 variants were plated in triplicate in 96-well plates (3,000 cells/well). 24 h after plating, the cells were treated with increasing concentrations of camptothecin (Calbiochem, 0–400 nm for 24 h) or gemcitabine (Selleck Chemicals, 0–3 μm for 48 h). For the treatment of ChK1 inhibitor (Selleck Chemicals, LY2603618) alone, the cells were treated with increasing concentrations of with ChK1 inhibitor (0–800 nm for 24 h). In the combination treatments, the cells were treated first with gemcitabine (2 or 3 μm for 2 h), followed by the addition of increasing concentrations of ChK1 inhibitor for subsequent 24 h. After the indicated treatment, the cell viability was assayed by adding 10 μl of CCK-8 reagent (Dojindo Technologies) to each well containing 100 μl of growth medium. The plates were incubated at 37 °C, and absorbance was measured at 450 nm every hour for 4 h. The percentage of cell viability was calculated by normalizing the absorbance values to untreated condition in each cell type.
Clonogenic assays
RECQ1-WT and RECQ1-KO cells were plated in a 12-well plate at 70% confluency. 24 h after plating, the cells were treated with increasing concentrations of gemcitabine (0–5 μm for 2 h). Following the treatment, the cells were harvested using trypsin, and 100 cells for each condition were plated in 12-well plates in triplicate and allowed to grow in drug-free medium for 10 days. On the day of the analysis, the growth medium was aspirated, and the wells were washed with 1× PBS. The cells were fixed in 3.75% paraformaldehyde for 30 min at room temperature, washed in 1× PBS, and stained in 0.5% methylene blue for 60 min, followed by destaining using distilled water. The plates were allowed to dry, and the colonies were counted. For the ChK1 inhibitor and gemcitabine combined treatments, the RECQ1-WT and RECQ1-KO cells were treated with gemcitabine (2 μm for 2 h) followed by the indicated doses of the ChK1 inhibitor (0–800 nm for 24 h). Following the treatments, the cells were allowed to recover in a drug-free medium for 10 days, and the protocol for staining colonies was followed as previously described. The percentage of cell survival was calculated by counting the number of colonies and normalizing to untreated in each cell type.
Western blotting analysis
The cells were plated in a 6-well plate at 70% confluency. 24 h after plating, the cells were treated with gemcitabine (2 μm for 2 h) and allowed to recover in drug-free medium for indicated time points. At the end of each recovery period, the cells were harvested by trypsinization, and whole-cell protein lysates were made using radioimmune precipitation assay buffer. The protein concentration was estimated using a Bio-Rad DC protein assay reagent: 50 μg of protein was loaded on 4–20% SDS-PAGE and subjected to Western blotting detection using specific antibodies against pRPA32 (Ser-4/Ser-8) (1:1000, Bethyl Laboratories), RPA32 (1:1000, Bethyl Laboratories), RECQ1 (1:1000, Santa Cruz Biotechnology), cleaved PARP (1:1000, BD Pharmingen), pNBS1 (Ser-95) (1:1000, Novus Biologicals), pATR (Ser-428) (1:1000), pChK1 (Ser-317) (1:1000), ChK1 (1:1000), ATR (1:1000), γH2AX (1:1000), pATM (Ser-1981) (1:1000), pChK2 (Thr-68, 1:1000), and GAPDH (1:1000) (all from Cell Signaling Technologies). The band intensities were quantified using ImageJ software.
Chromatin fractionation
To isolate the chromatin fractions, RECQ1-WT and RECQ1-KO cells were plated in a 6-well plate at 70% confluency and treated with gemcitabine (2 μm for 2 h), followed by recovery in drug-free medium at the indicated time points. At each time point, the cells were harvested by scraping in ice-cold 1× PBS and then were centrifuged at 1,200 rpm for 10 min. The cell pellets were resuspended in buffer A (20 mm Tris-HCl, pH 7.4, 2.5 mm MgCl2, 0.5% Nonidet P-40, 1 mm phenylmethylsulfonyl fluoride, and 1 mm DTT and protease inhibitors) and then incubated on ice for 30 min. The samples were centrifuged at 10,000 rpm for 5 min at 4 °C, and the supernatant was collected as the soluble fraction. The pellets were washed twice in 1× PBS, resuspended in 2× Laemmli sample loading buffer with β-mercaptoethanol, sonicated, and centrifuged at 15,000 rpm for 15 min at room temperature, and the supernatant was collected as the chromatin-enriched fraction.
siRNA depletion and immunostaining
RECQ1-WT and RECQ1-KO cells were transfected with a pool of siCTL and siMUS81 siRNA (20 nm) (Dharmacon) using Lipofectamine RNAimax according to the manufacturer's instructions. 48 h after transfection, the cells were treated with gemcitabine (2 μm for 2 h) followed by the addition of 10 μm BrdU for 90 min prior to harvesting. Following treatment, the cells were fixed in 3.75% paraformaldehyde for 15 min at room temperature. The cells were permeabilized with 0.5% Triton X-100 in 1× PBS for 5 min at room temperature, followed by blocking in 1% BSA for 1 h at room temperature. The coverslips were incubated overnight with BrdU (1:500, Life Technologies) and γH2AX (1:500, Cell Signaling Technologies) antibodies at 4 °C. The coverslips were washed three times and then incubated with the secondary antibodies Alexa 594 anti-mouse (1:300, Invitrogen) and Alexa 488 anti-rabbit (1:300, Invitrogen) for 45 min at room temperature. The coverslips were then mounted using DAPI prolong gold anti-fade reagent, and the images were captured using a Zeiss LSM 880 NLO Airyscan confocal microscope. Fifty cells were counted for each condition, and cells with more than 10 foci for either BrdU or γH2AX were scored positive. The Pearson's correlation co-efficient of γH2AX co-localized to BrdU was calculated using the Zen Blue software.
Neutral comet assay
The cells were treated with gemcitabine (2 μm for 2 h) 24 h after plating at 70% confluency and allowed to recover in drug-free medium for the indicated time periods. Following treatment, the cells were harvested by trypsinization, and comet assay was performed following the manufacturer's (Trevigen) instructions. The cell pellets were resuspended carefully in 500 μl of low-melting agarose, and 50 μl of this mixture was spread over a comet slide and placed at 4 °C for 30 min to solidify. The slides were incubated in lysis buffer for 1 h at 4 °C. Subsequently, the slides were submerged in neutral electrophoresis buffer for 30 min at 4 °C, and the samples were electrophoresed in the same buffer for 45 min at 21 V. After electrophoresis, the slides were incubated in a DNA precipitation solution for 30 min, followed by consecutive incubation in 70% ethanol for 30 min at room temperature and allowed to air-dry. The comets were stained with SYBRGold for 30 min, and the slides were rinsed twice with water and left to air-dry at room temperature. The images were captured using Nikon Eclipse Ti fluorescent microscope, and at least 50 comets were quantitated using the Open Comet plugin in ImageJ software. The percentage of DNA in the tail was the parameter selected to describe each comet.
EdU labeling and FACS analysis
The cells were treated with the ChK1 inhibitor (100 nm for 24 h), gemcitabine (2 μm for 2 h), or a combination of gemcitabine and the ChK1 inhibitor, followed by recovery in a drug-free medium or in a medium containing the ChK1 inhibitor as indicated. The cells were pulse-labeled with 10 μm EdU for 90 min before harvesting. EdU staining using the Click-IT EdU kit (Invitrogen) was performed according to the manufacturer's protocol. Briefly, the cells were harvested by trypsinization, washed with 1× PBS, and fixed in Click-iT fixative buffer for 15 min. The cells were subsequently permeabilized in a saponin-based permeabilization buffer and stained by the Click-iT staining solution for 30 min. The DAPI-stained DNA content was measured using flow cytometry. At least 20,000 events in each sample were recorded using the BD FACSCanto II machine and analyzed using FlowJo software.
DAPI staining
RECQ1-WT and RECQ1-KO cells grown on coverslips were treated with ChK1 inhibitor (100 nm for 24 h). Following the treatment, the cells were fixed in 3.75% paraformaldehyde for 15 min at room temperature and permeabilized with 0.5% Triton X-100 in 1× PBS for 5 min at room temperature, followed by blocking in 1% BSA for 1 h at room temperature. The coverslips were then mounted using DAPI prolong gold anti-fade reagent, and the images were captured using a Nikon Eclipse Ti fluorescent microscope.
Statistical analysis
At least three independent experiments and three replicates per experiment were performed. For the cell viability and cell survival experiments, all statistical tests were conducted using SPSS Biostatistics software (SPSS Inc., Chicago, IL). The results are presented as the means ± S.D. of three independent experiments. Statistical significance between groups was assessed by one-way analysis of variance followed by Bonferroni post hoc analysis, and p < 0.05 was considered statistically significant.
Author contributions
S. P. validation; S. P. and S. S. investigation; S. P. writing-original draft; S. P. and S. S. writing-review and editing; S. S. conceptualization; S. S. resources; S. S. formal analysis; S. S. supervision; S. S. methodology; S. S. project administration.
Supplementary Material
Acknowledgments
We thank Drs. Ashish lal, Xiao Ling Li, and Ferenc Livak and Langston Lim of NCI, National Institutes of Health for help and resources.
This work was supported by NIGMS, National Institutes of Health Grant SC1GM093999 and National Science Foundation Grant NSF1832163. This work was also supported by NIA, National Institutes of Health Grant 1R25 AG047843. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S3.
- RQC
- RecQ C-terminal
- ssDNA
- single-stranded DNA
- ChK1i
- checkpoint kinase 1 inhibitor
- KO
- knockout
- gRNA
- guide RNA
- PARP1
- poly(ADP)ribosyl polymerase 1
- PAR
- poly(ADP)ribosyl polymer
- BrdU
- bromodeoxyuridine
- DAPI
- 4′,6′-diamino-2-phenylindole
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase.
References
- 1. Croteau D. L., Popuri V., Opresko P. L., and Bohr V. A. (2014) Human RecQ helicases in DNA repair, recombination, and replication. Annu. Rev. Biochem. 83, 519–552 10.1146/annurev-biochem-060713-035428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Li D., Frazier M., Evans D. B., Hess K. R., Crane C. H., Jiao L., and Abbruzzese J. L. (2006) Single nucleotide polymorphisms of RecQ1, RAD54L, and ATM genes are associated with reduced survival of pancreatic cancer. J. Clin. Oncol. 24, 1720–1728 10.1200/JCO.2005.04.4206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Viziteu E., Klein B., Basbous J., Lin Y. L., Hirtz C., Gourzones C., Tiers L., Bruyer A., Vincent L., Grandmougin C., Seckinger A., Goldschmidt H., Constantinou A., Pasero P., Hose D., and Moreaux J. (2017) RECQ1 helicase is involved in replication stress survival and drug resistance in multiple myeloma. Leukemia 31, 2104–2113 10.1038/leu.2017.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Berti M., Ray Chaudhuri A., Thangavel S., Gomathinayagam S., Kenig S., Vujanovic M., Odreman F., Glatter T., Graziano S., Mendoza-Maldonado R., Marino F., Lucic B., Biasin V., Gstaiger M., Aebersold R., et al. (2013) Human RECQ1 promotes restart of replication forks reversed by DNA topoisomerase I inhibition. Nat. Struct. Mol. Biol. 20, 347–354 10.1038/nsmb.2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cybulski C., Carrot-Zhang J., Kluzniak W., Rivera B., Kashyap A., Wokolorczyk D., Giroux S., Nadaf J., Hamel N., Zhang S., Huzarski T., Gronwald J., Byrski T., Szwiec M., Jakubowska A., et al. (2015) Germline RECQL mutations are associated with breast cancer susceptibility. Nat. Genet. 47, 643–646 10.1038/ng.3284 [DOI] [PubMed] [Google Scholar]
- 6. Sun J., Wang Y., Xia Y., Xu Y., Ouyang T., Li J., Wang T., Fan Z., Fan T., Lin B., Lou H., and Xie Y. (2015) Mutations in RECQL gene are associated with predisposition to breast cancer. PLoS Genet. 11, e1005228 10.1371/journal.pgen.1005228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bowden A. R., and Tischkowitz M. (2019) Clinical implications of germline mutations in breast cancer genes: RECQL. Breast Cancer Res. Treat. 174, 553–560 10.1007/s10549-018-05096-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Puranam K. L., and Blackshear P. J. (1994) Cloning and characterization of RECQL, a potential human homologue of the Escherichia coli DNA helicase RecQ. J. Biol. Chem. 269, 29838–29845 [PubMed] [Google Scholar]
- 9. Sharma S., Sommers J. A., Choudhary S., Faulkner J. K., Cui S., Andreoli L., Muzzolini L., Vindigni A., and Brosh R. M. Jr. (2005) Biochemical analysis of the DNA unwinding and strand annealing activities catalyzed by human RECQ1. J. Biol. Chem. 280, 28072–28084 10.1074/jbc.M500264200 [DOI] [PubMed] [Google Scholar]
- 10. Cui S., Klima R., Ochem A., Arosio D., Falaschi A., and Vindigni A. (2003) Characterization of the DNA-unwinding activity of human RECQ1, a helicase specifically stimulated by human replication protein A. J. Biol. Chem. 278, 1424–1432 10.1074/jbc.M209407200 [DOI] [PubMed] [Google Scholar]
- 11. Sami F., Lu X., Parvathaneni S., Roy R., Gary R. K., and Sharma S. (2015) RECQ1 interacts with FEN-1 and promotes binding of FEN-1 to telomeric chromatin. Biochem. J. 468, 227–244 10.1042/BJ20141021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cui S., Arosio D., Doherty K. M., Brosh R. M. Jr., Falaschi A., and Vindigni A. (2004) Analysis of the unwinding activity of the dimeric RECQ1 helicase in the presence of human replication protein A. Nucleic Acids Res. 32, 2158–2170 10.1093/nar/gkh540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sharma S., Phatak P., Stortchevoi A., Jasin M., and Larocque J. R. (2012) RECQ1 plays a distinct role in cellular response to oxidative DNA damage. DNA Repair (Amst.) 11, 537–549 10.1016/j.dnarep.2012.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Parvathaneni S., Stortchevoi A., Sommers J. A., Brosh R. M. Jr., and Sharma S. (2013) Human RECQ1 interacts with Ku70/80 and modulates DNA end-joining of double-strand breaks. PLoS One 8, e62481 10.1371/journal.pone.0062481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Woodrick J., Gupta S., Camacho S., Parvathaneni S., Choudhury S., Cheema A., Bai Y., Khatkar P., Erkizan H. V., Sami F., Su Y., Schärer O. D., Sharma S., and Roy R. (2017) A new sub-pathway of long-patch base excision repair involving 5′ gap formation. EMBO J. 36, 1605–1622 10.15252/embj.201694920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Doherty K. M., Sharma S., Uzdilla L. A., Wilson T. M., Cui S., Vindigni A., and Brosh R. M. Jr. (2005) RECQ1 helicase interacts with human mismatch repair factors that regulate genetic recombination. J. Biol. Chem. 280, 28085–28094 10.1074/jbc.M500265200 [DOI] [PubMed] [Google Scholar]
- 17. Sharma S., Stumpo D. J., Balajee A. S., Bock C. B., Lansdorp P. M., Brosh R. M. Jr., and Blackshear P. J. (2007) RECQL, a member of the RecQ family of DNA helicases, suppresses chromosomal instability. Mol. Cell Biol. 27, 1784–1794 10.1128/MCB.01620-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sharma S., and Brosh R. M. Jr. (2007) Human RECQ1 is a DNA damage responsive protein required for genotoxic stress resistance and suppression of sister chromatid exchanges. PLoS One 2, e1297 10.1371/journal.pone.0001297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Popuri V., Croteau D. L., Brosh R. M. Jr., and Bohr V. A. (2012) RECQ1 is required for cellular resistance to replication stress and catalyzes strand exchange on stalled replication fork structures. Cell Cycle 11, 4252–4265 10.4161/cc.22581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Banerjee T., Sommers J. A., Huang J., Seidman M. M., and Brosh R. M. Jr. (2015) Catalytic strand separation by RECQ1 is required for RPA-mediated response to replication stress. Curr. Biol. 25, 2830–2838 10.1016/j.cub.2015.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Arora A., Parvathaneni S., Aleskandarany M. A., Agarwal D., Ali R., Abdel-Fatah T., Green A. R., Ball G. R., Rakha E. A., Ellis I. O., Sharma S., and Madhusudan S. (2017) Clinicopathological and functional significance of RECQL1 helicase in sporadic breast cancers. Mol. Cancer Ther. 16, 239–250 10.1158/1535-7163.MCT-16-0290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bugreev D. V., Brosh R. M. Jr., Mazin A. V. (2008) RECQ1 possesses DNA branch migration activity. J. Biol. Chem. 283, 20231–20242 10.1074/jbc.M801582200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhou B. B., and Elledge S. J. (2000) The DNA damage response: putting checkpoints in perspective. Nature 408, 433–439 10.1038/35044005 [DOI] [PubMed] [Google Scholar]
- 24. Awasthi P., Foiani M., and Kumar A. (2016) ATM and ATR signaling at a glance. J. Cell Sci. 129, 1285 10.1242/jcs.188631 [DOI] [PubMed] [Google Scholar]
- 25. Lambert S., Froget B., and Carr A. M. (2007) Arrested replication fork processing: interplay between checkpoints and recombination. DNA Repair (Amst.) 6, 1042–1061 10.1016/j.dnarep.2007.02.024 [DOI] [PubMed] [Google Scholar]
- 26. Cortez D. (2019) Replication-coupled DNA repair. Mol. Cell 74, 866–876 10.1016/j.molcel.2019.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cortez D. (2005) Unwind and slow down: checkpoint activation by helicase and polymerase uncoupling. Genes Dev. 19, 1007–1012 10.1101/gad.1316905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhao H., and Piwnica-Worms H. (2001) ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol. Cell Biol. 21, 4129–4139 10.1128/MCB.21.13.4129-4139.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hengel S. R., Spies M. A., and Spies M. (2017) Small-molecule inhibitors targeting DNA repair and DNA repair deficiency in research and cancer therapy. Cell Chem. Biol. 24, 1101–1119 10.1016/j.chembiol.2017.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Plunkett W., Huang P., Xu Y. Z., Heinemann V., Grunewald R., and Gandhi V. (1995) Gemcitabine: metabolism, mechanisms of action, and self-potentiation. Semin. Oncol. 22, 3–10 [PubMed] [Google Scholar]
- 31. Lucic B., Zhang Y., King O., Mendoza-Maldonado R., Berti M., Niesen F. H., Burgess-Brown N. A., Pike A. C., Cooper C. D., Gileadi O., and Vindigni A. (2011) A prominent β-hairpin structure in the winged-helix domain of RECQ1 is required for DNA unwinding and oligomer formation. Nucleic Acids Res. 39, 1703–1717 10.1093/nar/gkq1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sami F., Gary R. K., Fang Y., and Sharma S. (2016) Site-directed mutants of human RECQ1 reveal functional importance of the zinc binding domain. Mutat. Res. 790, 8–18 10.1016/j.mrfmmm.2016.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mirzaei H., and Schmidt K. H. (2012) Non-Bloom syndrome-associated partial and total loss-of-function variants of BLM helicase. Proc. Natl. Acad. Sci. U.S.A. 109, 19357–19362 10.1073/pnas.1210304109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu J. L., Rigolet P., Dou S. X., Wang P. Y., and Xi X. G. (2004) The zinc finger motif of Escherichia coli RecQ is implicated in both DNA binding and protein folding. J. Biol. Chem. 279, 42794–42802 10.1074/jbc.M405008200 [DOI] [PubMed] [Google Scholar]
- 35. Pike A. C., Shrestha B., Popuri V., Burgess-Brown N., Muzzolini L., Costantini S., Vindigni A., and Gileadi O. (2009) Structure of the human RECQ1 helicase reveals a putative strand-separation pin. Proc. Natl. Acad. Sci. U.S.A. 106, 1039–1044 10.1073/pnas.0806908106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pommier Y., Leo E., Zhang H., and Marchand C. (2010) DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem. Biol. 17, 421–433 10.1016/j.chembiol.2010.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Thangavel S., Berti M., Levikova M., Pinto C., Gomathinayagam S., Vujanovic M., Zellweger R., Moore H., Lee E. H., Hendrickson E. A., Cejka P., Stewart S., Lopes M., and Vindigni A. (2015) DNA2 drives processing and restart of reversed replication forks in human cells. J. Cell Biol. 208, 545–562 10.1083/jcb.201406100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zellweger R., Dalcher D., Mutreja K., Berti M., Schmid J. A., Herrador R., Vindigni A., and Lopes M. (2015) Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells. J. Cell Biol. 208, 563–579 10.1083/jcb.201406099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang J., Wang Z., Hu X., Wang B., Wang L., Yang W., Liu Y., Liu G., Di G., Hu Z., Wu J., and Shao Z. (2015) Cisplatin and gemcitabine as the first line therapy in metastatic triple negative breast cancer. Int. J. Cancer 136, 204–211 10.1002/ijc.28966 [DOI] [PubMed] [Google Scholar]
- 40. Jones R. M., Kotsantis P., Stewart G. S., Groth P., and Petermann E. (2014) BRCA2 and RAD51 promote double-strand break formation and cell death in response to gemcitabine. Mol. Cancer Ther. 13, 2412–2421 10.1158/1535-7163.MCT-13-0862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Karnitz L. M., Flatten K. S., Wagner J. M., Loegering D., Hackbarth J. S., Arlander S. J., Vroman B. T., Thomas M. B., Baek Y. U., Hopkins K. M., Lieberman H. B., Chen J., Cliby W. A., and Kaufmann S. H. (2005) Gemcitabine-induced activation of checkpoint signaling pathways that affect tumor cell survival. Mol. Pharmacol. 68, 1636–1644 10.1124/mol.105.012716 [DOI] [PubMed] [Google Scholar]
- 42. Kaufmann S. H., Desnoyers S., Ottaviano Y., Davidson N. E., and Poirier G. G. (1993) Specific proteolytic cleavage of poly(ADP-ribose) polymerase: an early marker of chemotherapy-induced apoptosis. Cancer Res. 53, 3976–3985 [PubMed] [Google Scholar]
- 43. Parvathaneni S., Lu X., Chaudhary R., Lal A., Madhusudan S., and Sharma S. (2017) RECQ1 expression is upregulated in response to DNA damage and in a p53-dependent manner. Oncotarget. 8, 75924–75942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Regairaz M., Zhang Y. W., Fu H., Agama K. K., Tata N., Agrawal S., Aladjem M. I., and Pommier Y. (2011) Mus81-mediated DNA cleavage resolves replication forks stalled by topoisomerase I-DNA complexes. J. Cell Biol. 195, 739–749 10.1083/jcb.201104003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hanada K., Budzowska M., Davies S. L., van Drunen E., Onizawa H., Beverloo H. B., Maas A., Essers J., Hickson I. D., and Kanaar R. (2007) The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nat. Struct. Mol. Biol. 14, 1096–1104 10.1038/nsmb1313 [DOI] [PubMed] [Google Scholar]
- 46. Symington L. S. (2016) Mechanism and regulation of DNA end resection in eukaryotes. Crit. Rev. Biochem. Mol. Biol. 51, 195–212 10.3109/10409238.2016.1172552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lemaçon D., Jackson J., Quinet A., Brickner J. R., Li S., Yazinski S., You Z., Ira G., Zou L., Mosammaparast N., and Vindigni A. (2017) MRE11 and EXO1 nucleases degrade reversed forks and elicit MUS81-dependent fork rescue in BRCA2-deficient cells. Nat. Commun. 8, 860 10.1038/s41467-017-01180-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Montano R., Thompson R., Chung I., Hou H., Khan N., and Eastman A. (2013) Sensitization of human cancer cells to gemcitabine by the Chk1 inhibitor MK-8776: cell cycle perturbation and impact of administration schedule in vitro and in vivo. BMC Cancer 13, 604 10.1186/1471-2407-13-604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cliby W. A., Lewis K. A., Lilly K. K., and Kaufmann S. H. (2002) S phase and G2 arrests induced by topoisomerase I poisons are dependent on ATR kinase function. J. Biol. Chem. 277, 1599–1606 10.1074/jbc.M106287200 [DOI] [PubMed] [Google Scholar]
- 50. Salic A., and Mitchison T. J. (2008) A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc. Natl. Acad. Sci. U.S.A. 105, 2415–2420 10.1073/pnas.0712168105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Warren N. J. H., and Eastman A. (2019) Inhibition of checkpoint kinase 1 following gemcitabine-mediated S phase arrest results in CDC7- and CDK2-dependent replication catastrophe. J. Biol. Chem. 294, 1763–1778 10.1074/jbc.RA118.005231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Imreh G., Norberg H. V., Imreh S., and Zhivotovsky B. (2011) Chromosomal breaks during mitotic catastrophe trigger γH2AX-ATM-p53-mediated apoptosis. J. Cell Sci. 124, 2951–2963 10.1242/jcs.081612 [DOI] [PubMed] [Google Scholar]
- 53. Mansilla S., Priebe W., and Portugal J. (2006) Mitotic catastrophe results in cell death by caspase-dependent and caspase-independent mechanisms. Cell Cycle 5, 53–60 10.4161/cc.5.1.2267 [DOI] [PubMed] [Google Scholar]
- 54. Cortez D., Guntuku S., Qin J., and Elledge S. J. (2001) ATR and ATRIP: partners in checkpoint signaling. Science 294, 1713–1716 10.1126/science.1065521 [DOI] [PubMed] [Google Scholar]
- 55. Sakurikar N., Thompson R., Montano R., and Eastman A. (2016) A subset of cancer cell lines is acutely sensitive to the Chk1 inhibitor MK-8776 as monotherapy due to CDK2 activation in S phase. Oncotarget 7, 1380–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shaltiel I. A., Krenning L., Bruinsma W., and Medema R. H. (2015) The same, only different: DNA damage checkpoints and their reversal throughout the cell cycle. J. Cell Sci. 128, 607–620 10.1242/jcs.163766 [DOI] [PubMed] [Google Scholar]
- 57. Parsels L. A., Tanska D. M., Parsels J. D., Zabludoff S. D., Cuneo K. C., Lawrence T. S., Maybaum J., and Morgan M. A. (2016) Dissociation of gemcitabine chemosensitization by CHK1 inhibition from cell cycle checkpoint abrogation and aberrant mitotic entry. Cell Cycle. 15, 730–739 10.1080/15384101.2016.1148841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Im M. M., Flanagan S. A., Ackroyd J. J., Knapp B., Kramer A., and Shewach D. S. (2016) Late DNA damage mediated by homologous recombination repair results in radiosensitization with gemcitabine. Radiat. Res. 186, 466–477 10.1667/RR14443.1 [DOI] [PubMed] [Google Scholar]
- 59. Qiu Z., Oleinick N. L., and Zhang J. (2018) ATR/CHK1 inhibitors and cancer therapy. Radiother. Oncol. 126, 450–464 10.1016/j.radonc.2017.09.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.