Abstract
Akt signaling is an important regulator of neural development, but the distinctive function of Akt isoforms in brain development presents a challenge. Here we show Siah1 as an ubiquitin ligase that preferentially interacts with Akt3 and facilitates ubiquitination and degradation of Akt3. Akt3 is enriched in the axonal shaft and branches but not growth cone tips, where Siah1 is prominently present. Depletion of Siah1 enhanced Akt3 levels in the soma and axonal tips, eliciting multiple branching. Brain-specific somatic mutation in Akt3-E17K escapes from Siah1-mediated degradation and causes improper neural development with dysmorphic neurons. Remarkably, coexpression of Siah1 with Akt3-WT restricted disorganization of neural development is caused by Akt3 overexpression, whereas forced expression of Siah1 with the Akt3-E17K mutant fails to cope with malformation of neural development. These findings demonstrate that Siah1 limits Akt3 turnover during brain development and that this event is essential for normal organization of the neural network.
Keywords: E3 ubiquitin ligase, ubiquitylation (ubiquitination), protein degradation, Akt PKB, signal transduction
Introduction
Akt (protein kinase B) signaling contributes an important node to multiple neural signaling, including neural survival, axon/dendrite specification and growth, and synapse formation (1). Although three Akt isoforms display some functional redundancy, they also perform distinctive functions (2, 3). Akt3 expression is mainly restricted to the brain and testes, whereas Akt1 is widely expressed, and Akt2 is predominantly expressed in the liver and adipose tissue. Especially in the human fetal brain, Akt3 expression is higher than Akt3 expression in any other tissues, whereas Akt1 and Akt2 show comparable or lower levels of expression in other tissues (4). Akt1 promotes cell growth and survival (5). Akt2-null mice show defective glucose transport in response to insulin (6), and Akt2 up-regulates cell survival and cell cycle progression through specific regulation of nucleophosmin/B23 stability (7, 8). Akt3-null mice exhibit impaired brain development with an around 25% smaller brain size and corpus callosum disorganization, and Akt3 has an important role in oligodendrocyte genesis in glial cells (9–11). Duplication of Akt3 causes macrocephaly and focal cortical dysplasia (12), and germline and somatic mutation of Akt3 is associated with megalencephaly, focal malformation of cortical development, epilepsy, and hypoglycemia (13–16), suggesting that its primary role in brain development and appropriate expression of Akt3 is essential for normal brain development. Although high functional redundancy of Akt isoforms is suggested in homeostasis and development, the specific functions of each Akt isoform in neural development in the brain have not been fully elucidated.
Although the research on Akt has been focused on the role of Akt phosphorylation and how this phosphorylation is regulated, recent studies have suggested that, in addition to phosphorylation, ubiquitination is an important posttranslational mechanism of Akt regulation. For example, upon growth factor stimulation, Akt1 and Akt2 (but not Akt3) undergo ubiquitination by tumor necrosis factor receptor–associated factor 6 (TRAF 6), S phase kinase–associated protein 2 (Skp2), and NEDD4-1, contributing to recruitment and activation of Akt but do not direct Akt for degradation (17–19). Other E3 ligases, which contain a RING finger domain, tetratricopeptide repeat domain 3 (TTC3), and mitochondrial ubiquitin ligase activator of NF-κB (MULAN), bind to Akt and facilitate its ubiquitination and degradation (20, 21). During neural development, selective degradation of Akt by the ubiquitin–proteasome system (UPS)3 induces asymmetric Akt activation and axon localization (22). However, the full scope of ubiquitination in Akt regulation awaits further exploration.
The seven in absentia homolog (Siah) family of E3 ubiquitin ligases are RING finger ubiquitin ligases composed of a catalytic RING domain, two zinc finger domains, and a substrate binding domain. A growing number of Siah1-binding proteins have been identified, and they are mostly associated with fundamental cellular processes, including hypoxia (PHDs, FIH, HIPK2, and AKAP1), the DNA damage response (HIPK2, TRF2, and TIN2), cancer (PHDs/HIF, C/EBPd, HBK, and Sprouty2), and neural functions (α-synuclein, synphilin-1, and PARD3) (23). In addition, we have reported previously that Siah1 interacts with nucleophosmin, impeding its function in neuronal death, thereby enhancing neuronal survival (7). In this study, we demonstrated that Siah1 acts as an E3 ligase for the Akt3 isoform. Siah1 binds to the kinase domain and leads to proteasomal degradation of Akt3 but fails to bind to the Akt3-E17K mutant, which has frequently observed somatic mutations in Akt3 in various brain diseases. During neural development, Siah1 contributes to proper axon growth and branching, restricting Akt3 distribution in the growing neuron. Our study proposes a contribution of possible specific E3 ligases to the individual Akt isoform and specific degradation of the Akt3 isoform for proper neural development.
Results
Siah1 is a binding partner for Akt
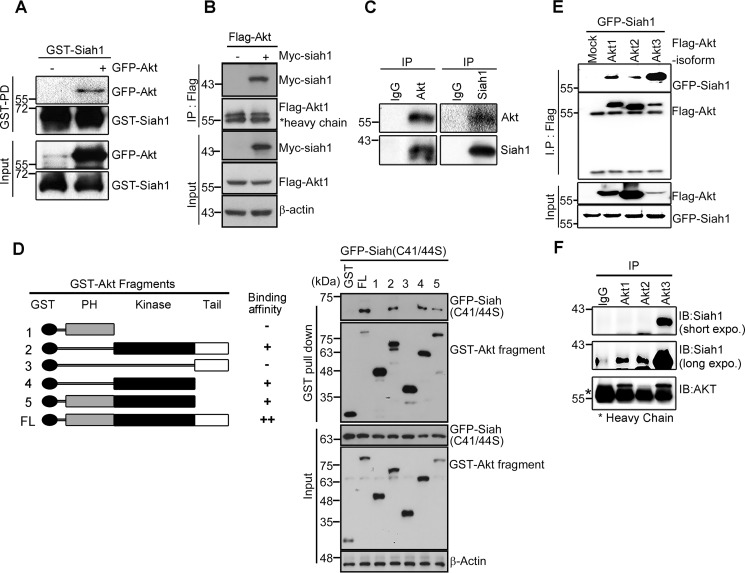
As Akt shares several binding partners with Siah1, including B23, GAPDH, MAPK8, and α-synuclein (7, 24–27), we wondered whether Akt interacts directly with Siah1. To determine the interaction between Siah1 and Akt, we incubated full-length GST-Siah1 fusion protein with soluble extracts from the rat pheochromocytoma PC12 cell line following transfection with GFP-Akt. PC12 cells are comparable with adrenal chromaffin cells of neural crest origin (28). A GST pulldown assay showed that Siah1 physically associated with GFP-Akt (Fig. 1A). Reciprocal immunoprecipitation analysis with lysate from mammalian FLAG-Akt–expressing cells with or without lysate from Myc-Siah1–expressing cells showed interaction between Akt and Siah1 (Fig. 1B). Moreover, we confirmed that endogenous Akt interacts with Siah1 in PC12 cells (Fig. 1C).
Figure 1.
Siah1 is a binding partner for Akt. A, purified GST-Siah1 protein was obtained using GST resin dialyzed against PBS. GST-Siah1 protein (500 ng) was prebound to GST resin and reacted with lysate from PC12 cells transiently transfected with GFP-Akt. B, PC12 cells were transfected with Myc-Siah1 and GST-Akt. After 24-h transfection, cell lysates were subjected to a GST pulldown assay. IP, immunoprecipitation. C, the cell lysates were immunoprecipitated with indicated antibodies and analyzed by immunoblotting. D, schematic of Akt full-length and fragments (left panel). PC12 cells were cotransfected with Myc-tagged Siah1 C41S/C44S and GST-Akt full-length (FL) or fragment proteins. Cell lysates were used for a GST pulldown assay and immunoblotted with the indicated antibodies (right panel). The total amount of protein was checked with anti-actin antibody. PH, pleckstrin homology. E, cells were transfected with GFP-Siah1 and FLAG-Akt isoforms. After 24-h transfection, the cell lysates were subjected to immunoprecipitation assay. The total amount of protein was checked with anti-actin antibody. F, mouse brain lysates were subjected to immunoprecipitation with anti-Akt1, Akt2, and Akt3 antibodies. IB, immunoblot.
To verify the specificity of this interaction, we performed a mapping analysis with a series of Akt deletion mutants and Myc-tagged Siah1 C41S/C44S. The latter is known to have lost E3 ligase activity but still bind to the target (29) because overexpression of WT Siah1 leads to undetectable expression of kinase domain–containing fragments of Akt (Fig. S1A), reflecting that Siah1 probably targets the kinase domain of Akt for degradation. Our in vitro binding assay demonstrated that the interaction between Siah1 and Akt was primarily mediated through the kinase of Akt, whereas the C-terminal tail domain or N-terminal pleckstrin homology domain is dispensable for the interaction with Siah1 (Fig. 1D).
Recently, we and others have shown that individual isoforms of Akt (Akt1, Akt2, and Akt3) possess specific binding partners and/or play a differential function in a wide range of cellular events. Therefore, we attempted to test whether Siah1 interacts with specific isoforms of Akt. Using an Akt isoform–specific immunoprecipitation assay, we found that the strongest interaction occurs between GFP-Siah1 and the FLAG-Akt3 isoform, whereas a basal level of interaction was observed with Akt1 and Akt2 in PC12 cells (Fig. 1E). A similar result was obtained when using FLAG-Akt1/2/3 with Myc-Siah1-C41S/C44S, which is no longer degraded by the proteasome and, thus, stably expressed, showing strong interaction between Siah1 and Akt3 (Fig. S1B). Notably, our immunoprecipitation assay with mouse brain lysate demonstrated that Akt3 interacts with Siah1 in vivo, although long-time exposure showed basal levels of interaction with Akt1 or Akt2 (Fig. 1F). These data demonstrated that the interaction between Siah1 and Akt isoforms was most prominent with Akt3, at least in brain tissues. As both Akt3 and Siah1 are largely confined to the central nervous system, we focused our investigation of biological significance of this interaction by using Akt3 in neuronal cells unless otherwise specified.
Siah1 is a novel E3 ligase for Akt3
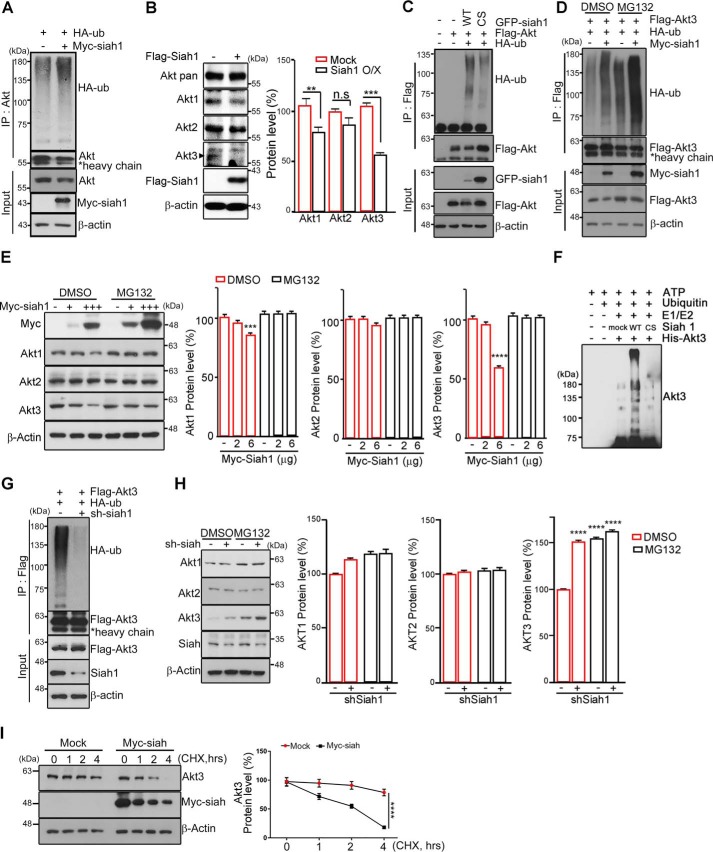
As the Siah1 gene encodes a canonical E3 RING motif at the C terminus end, and we observed that overexpression of Siah1 reduced the protein levels of Akt (Fig. S1A), we determined whether Siah1 is able act on Akt3 as an E3 ubiquitin ligase by conducting a ubiquitination assay in PC12 cells. Ectopic expression of Siah1 promoted endogenous Akt ubiquitination (Fig. 2A). As Siah1 elicits marked polyubiquitination of endogenous Akt, we wondered whether Siah1 targets any specific isoforms of Akt. Using a set of antibody preparations that detect each of the specific Akt isoforms, we observed that overexpression of FLAG-Siah1 resulted in a notable reduction of endogenous Akt3 proteins, although it also slightly affected the protein levels of Akt1 and merely reduced Akt2, reflecting the strong interaction between Akt3 and Siah1 (Fig. 2B). Accordingly, ubiquitination of Akt3 by Siah1 was confirmed by the presence of evident anti-HA immunoreactivity, displayed as a smear on the gel, which is characteristic of polyubiquitinated proteins, whereas Akt3 ubiquitination did not occur by the ligase-deficient Siah1-C41S/C44S mutant (Fig. 2C).
Figure 2.
Siah1 is a novel E3 ligase for Akt. A, PC12 cells were transfected with HA-ubiquitin (HA-ub) and Myc-Siah1. The cell lysates were immunoprecipitated (IP) with anti-Akt antibody. The ubiquitinated Akt was detected by immunoblotting using an anti-HA antibody. The total amount of protein was checked with anti-actin antibody. B, PC12 cells were transfected with FLAG control and FLAG-Siah1. The endogenous levels of the Akt isoform proteins were detected with specific antibodies as indicated. The total amount of protein was checked with anti-actin antibody. The bar graphs show quantified protein levels of Akt1, Akt2, and Akt3 (right panel). Data are shown as mean ± S.E. of three independent experiments; n.s., not significant; **, p = 0.0064; ***, p = 0.0005. O/X, overexpression. C, PC12 cells were transfected with the indicated constructs. The cell lysates were subjected to immunoprecipitation with the anti-FLAG antibody. Ubiquitinated Akt was detected using anti-HA antibody. C/S, C41S/C44S. D, PC12 cells were transfected with the indicated constructs. After transfection for 24 h, cells were treated with 10 μm MG132 for 8 h. The cell lysates were immunoprecipitated with anti-FLAG antibody. The ubiquitination was checked with anti-HA antibody. E, PC12 cells were transfected with the indicated amounts of Myc-Siah1 and treated with 10 μm MG132 for 8 h. The levels of Akt isoforms were detected by immunoblot. The bar graph shows quantification of Akt isoform protein levels (right panel). Data are shown as mean ± S.E. of three independent experiments. ***, p < 0.0005; ****, p < 0.0001. F, in vitro ubiquitination of Akt3 by Siah-1 was performed with ubiquitin, E1, E2-UbcH5a, His-Akt3, and GST-Siah1 (WT or C41S/C44S). After addition of ATP, the samples were incubated at 30 °C for 2 h. The reactions were terminated with SDS loading buffer and boiling for 10 min. The Coomassie-stained SDS-PAGE gel is shown in Fig. S1D. G, PC12 cells were cotransfected with shSiah1 and FLAG-Akt. The cell lysates were subjected to an immunoprecipitation assay. The total amount of protein was checked with anti-actin antibody. H, PC12 cells were transfected with shSiah1and treated with 10 μm MG132 for 8 h. The protein levels of Akt isoforms were detected by immunoblot. The bar graphs represent quantification of the protein levels of the Akt isoforms (right panel). ****, p < 0.0001. I, PC12 cells were transfected with Myc and Myc-Siah1 and treated with 10 μm cycloheximide (CHX) for the indicated times (left panel). The bar graphs show quantified protein levels of Akt3 (right panel). Data are shown as mean ± S.E. of three independent experiments; one way analysis of variance; ****, p < 0.0001.
If Siah1-mediated ubiquitination marks Akt3 for proteasomal degradation, then inhibition of the UPS would be expected to stabilize ubiquitin–Akt3 conjugates. Indeed, treatment of PC12 cells with the proteasomal inhibitor MG132 stabilized the ubiquitinated forms of Akt3 (Fig. 2D). Moreover, Siah1-mediated degradation of Akt depended on the concentration of Siah1 in PC12 cells, especially with virtual abolition of the Akt3 isoform (Fig. 2E). To clarify whether Siah1 mediates Akt3 ubiquitination directly, we performed in vitro ubiquitination assays using recombinant E1 and E2 enzymes with purified His-Akt3. Siah1 efficiently facilitated Akt3 ubiquitination in this system, in contrast to the Siah1-C41S/C44S mutant, which failed to catalyze Akt3 ubiquitination (Fig. 2F and Fig. S1C). GST-Siah1 WT and the GST-Siah C41S/C44S mutant were purified by affinity chromatography (Fig. S1D). Our data indicate that Siah1-mediated polyubiquitination of Akt3 directs it to the proteasomal degradation pathway.
To further verify whether Siah1 physiologically regulates Akt3 ubiquitination, we eliminated Siah1 using shRNA. In PC12 cells, depletion of Siah1 largely decreased Akt3 ubiquitination (Fig. 2G). In addition, the reduction of Siah1 protein expression remarkably augmented Akt3 protein levels, whereas the protein levels of Akt1 and Akt2 changed insignificantly (Fig. 2H), implying that, presumably, Siah1 preferentially targets Akt3 for degradation.
The half-life of Akt3 was decreased in Myc-Siah1–expressing cells, whereas Akt3 stability did not change much in control vector–expressing cells after cycloheximide treatment in PC12 cells (Fig. 2I). Taken together, our data indicate that Siah1 functions as a specific E3 ligase for polyubiquitination and subsequent proteasomal degradation of Akt3.
Akt3 contributes to proper axon growth and branches during neural development
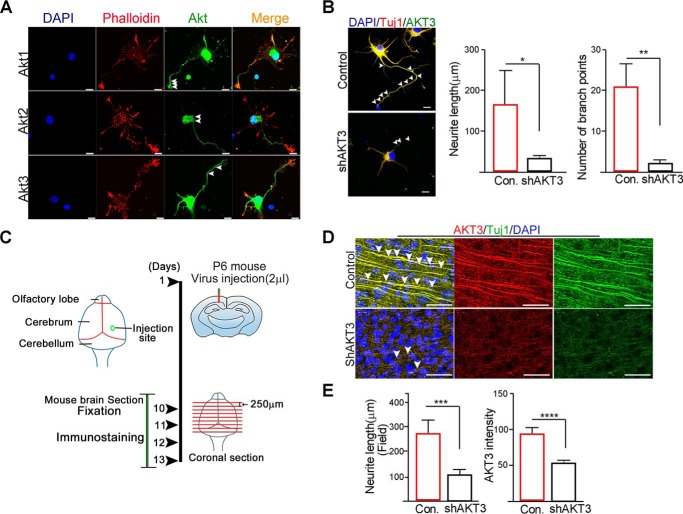
Although the distinctive functions and differential expression of Akt isoforms have been known, and the importance of Akt3 expression in axonal length has been suggested (30), in developing neurons, distinctive expression profiles of Akt isoforms and functions are not well-defined, and the specific roles in axonal growth and the regulatory effects of the UPS are not well-understood. Thus, we wondered whether Akt3 expression in the developing neuron is regionally restricted because of the consequence of Akt3 degradation by Siah1. We first examined the spatial distribution of each Akt isoform in cultured embryonic hippocampal neurons using isoform-specific antibodies. In accordance with our previous finding and the importance of Akt1 in axon growth and growth cone formation (31), Akt1 was found in proximal axons, and expression tapered off along the distal axon; however, the signals were strikingly intense at the distal part of the growth cone, revealing notable codistribution with phalloidin-labeled F-actin in growing axons. However, Akt2 was highly accumulated in the soma and almost invisible at the distal part of axon or dendrites. Expression of Akt3 was observed in the distal part of growing axon and branches but less in tips of growth cones, implying that Akt3 might contribute to sprouting of new branches of growing axons (Fig. 3A). Indeed, depletion of Akt3 using Akt3-specific siRNA in the developing hippocampal neuron largely impaired not only elongation but also branching of growing axons, along with a severe reduction of branching numbers (Fig. 3B). Axons were identified based on their morphology and expression of the neuronal marker Tuj1.
Figure 3.
Akt3 contributes to proper axon growth and branches during neural development. A, a neuron was immunostained with Phalloidin (F-actin marker, red) and Akt isoforms (green) at DIV 3. Arrowheads indicate Akt1, Akt2, and Akt3 signals. Scale bars = 10 μm. DAPI, 4′,6-diamidino-2-phenylindole. B, hippocampal neurons were transfected with control (Con) or shAkt3 at DIV 4 and fixed at DIV 6. A neuron was immunostained with Akt3 (green) and Tuj1(red). Arrowheads indicate branch points. Scale bars = 10 μm. The bar graphs represent quantification of neurite length and number of branch points (right panel). Data are shown as mean ± S.E. of three independent experiments. *, p < 0.05; **, p < 0.005. C, experimental schematic. D, brain sections were stained with anti-Akt3 (red) or Tuj1 (green) antibody. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI, blue). Arrowheads indicate Tuj1-positive neurites. Scale bars = 50 μm. E, bar graphs represent quantification of neurite length and Akt3 intensity. Data are shown as mean ± S.E. of three independent experiments. ***, p = 0.0002; ****, p < 0.0001.
To further define the specific role of Akt3 in vivo, we injected shAkt3 or a control vector expressing lentiviruses into mouse brains on postnatal day 6, allowed the mice to survive 10 more days (P16), and then performed morphometric and histological analyses (Fig. 3C). Compared with control vectors infected with nontargeted shRNA, Akt3 shRNA significantly reduced Akt3 expression in mouse brain slices. Tuj1 immunohistochemistry revealed aberrant gross layering and severely distorted alignment of neurons with short processes of axons and branching in the absence of Akt3 compared with the control (Fig. 3D). Accordingly, morphometric analysis showed that the total length of neurites (Tuj1-positive neurite length) was notably abrogated in Akt3 shRNA lentivirus–infected brain sections (Fig. 3E). Taken together, our data demonstrate that Akt3 is essential for proper axon elongation and branching during neural development.
Siah1-mediated Akt3 degradation controls axonal branches and growth
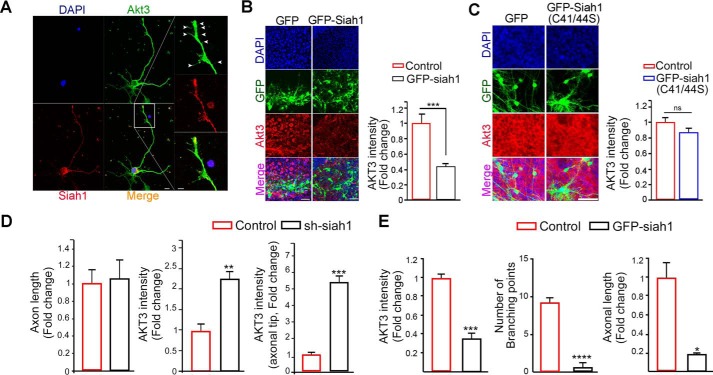
Accounting for the prominent expression of Akt3 in branches of axons and its contribution to normal neural development, we found that expression of Siah1 was visualized not only in the soma but also in the tips of axons and/or neurites in the early stages of differentiation, revealing codistribution with phalloidin-labeled F-actin in the growing axon but relatively much less so in branches. Expression of the neuronal β-tubulin III marker Tuj1 was used for verification of axons (most long growing neurites were considered axons) (Fig. S2, A and B). To evaluate whether the distribution and function of Akt3 in the developing neuron are mediated by Siah1, we conducted dual immunostaining of Akt3 and Siah1 in developing hippocampal neurons. Consistently, the intensity of Akt3 was much weaker in axonal tips of growth cones, whereas Siah1 was profoundly expressed, and Akt3 expression was relatively enriched in axonal branches, where Siah1 expression was less visible (Fig. 4A), implying that the distinctive localization of Akt3 could be controlled by Siah1 during neural development. To ensure Akt3 is a physiological substrate of Siah1 in the brain, we employed organotypic slice culture of embryonic brain tissue, which allows study of cortical development (i.e. neurogenesis) as well as neuronal differentiation in situ (32–34). Cultured brain slices were infected with GFP-Siah1 or a GFP control vector expressing adenoviruses. GFP-Siah1–expressing neurons were identified with neuronal marker NeuN staining (Fig. S3A). In contrast to GFP-control–expressing adenovirus-infected brain slices, Akt3 expression was drastically impaired in GFP-Siah1–expressing neurons (Fig. 4B). However, comparable with control GFP-adenovirus expression, the ligase-deficient mutant of Siah1-C41S/C44S expression merely affected Akt3 expression in brain slices (Fig. 4C). These results support the notion that Siah1 restricts Akt3 distribution in neurons through local degradation for normal neural development.
Figure 4.
Siah1-mediated Akt3 degradation controls axonal branches and growth. A, a neuron was immunostained with Siah1 (red) and Akt3 (green) at DIV 3. Scale bar = 10 μm. An enlargement of the boxed area is shown on the right. Scale bar = 5 μm. The images shown here are representative of at least three independent experiments. Arrowheads indicate Akt3 in the branch. B and C, embryo brain slices were infected with ad-GFP, ad-GFP-Siah1(WT), or GFP-Siah1 (C41S/C44S) at DIV 5 and fixed at DIV 10. The embryo brain slices were stained with Akt3 (red). Scale bars = 50 μm. The bar graphs show Akt3 signal intensity. Data are shown as mean ± S.E. of three independent experiments. ***, p = 0.0001; ns, not significant. The images shown here are representative of at least three independent experiments. D, hippocampal neurons were infected with adeno-GFP or adeno-shSiah1 virus at DIV 2 and fixed at DIV 4. The bar graphs show length and Akt3 signal intensity. Data are shown as mean ± S.E. of three independent experiments. **, p = 0.0026; ***, p = 0.0003. E, hippocampal neurons were transfected with GFP or GFP-Siah1 at DIV 2 and fixed at DIV 4. The bar graphs show Akt3 signal intensity, axonal length, and a number of branch points. Data are shown as mean ± S.E. of three independent experiments. ***, p = 0.0006; ****, p < 0.0001.
To clarify the physiological role of Siah1 in regulation of Akt3 in developing neurons, we infected hippocampal neurons on day 1 in culture with an shSiah1-expressing adenovirus and analyzed the cells by immunocytochemistry using an isoform-specific Akt3 antibody. Depletion of Siah1 did not statistically influence axon growth but slightly enhanced axon length. However, it conspicuously increased the expression of Akt3 not only at the tips of axons but also in the shaft and cell body compared with neurons expressing the control vector (Fig. 4D and Fig. S3B). Especially at the axonal tip, an obvious increase in Akt3 (5-fold) was observed (Fig. 4D, right panel). In contrast, overexpression of Siah1 in cultured hippocampal neurons notably decreased Akt3 intensity, revealing shortened axons and reduced numbers of branching points (Fig. 4E and Fig. S3C). These results lend support to a unique role of Akt3 in branching and axon elongation and the hypothesis that its local distribution in growing neurites is regulated by Siah1 through UPS-mediated degradation in axon development.
Brain-specific somatic mutation in Akt3 debilitates Siah1-mediated Akt3 degradation
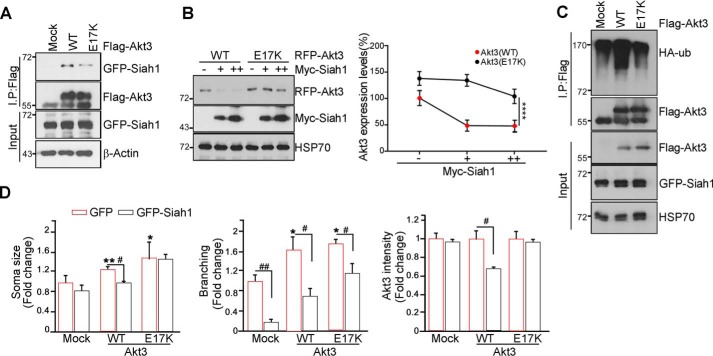
Somatic mutation in Akt3 (c.49G>A;p.E17K-, hereafter called Akt3-E17K) was identified in focal malformations of cortical development, which are characterized by enlarged, malformed cerebral hemisphere, and this gain-of-function mutation is brain-specific (35). As we determined that Akt3 is responsible for proper axon growth and branching, we wondered whether the Akt3-E17K mutation is not be properly degraded during neural development, causing cytomegalic neurons and the presence of dysmorphia. To examine whether the Akt3-E17K mutant has a differential binding affinity with Siah1, we transfected FLAG-tagged Akt3-WT or the Akt3-E17K mutant. Compared with Akt3-WT, the Akt3-E17K mutant weakened the interaction with Siah1 (Fig. 5A). Increased expression of Siah1 decreased the protein level of Akt3-WT. In contrast, the Akt3-E17K mutant was less susceptible to Siah1-mediated degradation (Fig. 5B). Indeed, compared with Akt3-WT, the Akt3-E17K mutant was less ubiquitinated (Fig. 5C). Hence, these data suggest that Akt3 E17K mutation decreases the interaction with Siah1, protecting itself from proteasomal degradation. Perhaps the unexpected expression of Akt3 E17K mutant elicits improper neuron morphology and disorganization of neural network.
Figure 5.
Brain-specific somatic mutation in Akt3 debilitates Siah1-mediated Akt3 degradation. A, cells were transfected with GFP-Siah1 and FLAG-Akt-WT or E17K. After 24-h transfection, the cell lysates were subjected to an immunoprecipitation (IP) assay. The total amount of protein was checked with anti-actin antibody. B, PC12 cells were transfected with the indicated amounts of Myc-Siah1. The level of Akt3-WT or E17K was detected by immunoblot. The graph shows the percentage of Akt3-WT and E17K protein levels (right panel). Data are shown as mean ± S.E. of three independent experiments; one way analysis of variance. ****, p < 0.0001. C, PC12 cells were transfected with the indicated constructs. After transfection for 24 h, cells were treated with 10 μm MG132 for 8 h. The cell lysates were immunoprecipitated with the anti-FLAG antibody. The ubiquitination was checked with the anti-HA antibody. D, the hippocampal neurons were transfected with the indicated constructs at DIV 3 and fixed at DIV 5. Data are shown as mean ± S.E. of three independent experiments. *, p < 0.05 versus control; **, p < 0.005 versus control; #, p < 0.05. The images shown here are representative of at least three independent experiments.
To investigate whether introduction of the Akt3-E17K mutation is sufficient to cause a defect in neural development and whether expression of Siah1 can rescue improper differentiation of neurons, we introduced the red fluorescent protein-Akt3-WT or RFP-Akt3-E17K construct with or without GFP-Siah1 into primary hippocampal cultures. In the absence of Siah1, overexpression of Akt3 enlarged the soma size with multiple branching. In contrast, coexpression of Akt3 with Siah1 decreased neuronal crowding and revealed a relatively typical morphology of neurons compared with overexpression of Akt3 only, reflecting a lesser intensity of the RFP-Akt3 signal. Akt3-E17K mutant–electroporated primary hippocampal neurons showed more dysmorphic neurons with numerous branching and much enlarged somata compared with Akt3-WT. Unlike Akt3-WT, in the presence of GFP-Siah1, the intensity of the Akt3-E17K mutant was not altered, and morphological defects and crowding of neurons were not rescued by Siah1 (Fig. 5D and Fig. S4). Taken together, these results suggest that the dysmorphic defects of neurons caused by Akt3-E17K are due to the higher protein stability of this mutant form escaping from UPS-dependent degradation by Siah1.
Discussion
Given the important role of Akt signaling in many biological events, including neuronal survival and neural development, deregulation of the Akt pathway is associated with various human pathological conditions. Thus, a comprehensive understanding of how Akt is regulated is of importance and of relevance regarding the specific roles and regulation of each Akt isoform. In this study, we show for the first time that Siah1 acts as an E3 ligase that ubiquitinates and degrades Akt3, leading to proper distribution of Akt3 in neural development. A somatic mutation of Akt3-E17K known to be enriched in focal malformations of cortical development disrupted the interaction with Siah1 and enhanced its protein stability, leading to abnormal neural development with excess numbers of neurons and enlarged somata with multiple neurites in the developing brain.
Although overexpression of Siah1 in hippocampal neurons specifically degraded Akt3 in growing neurites and branching abrogated axon growth (Fig. 4D), knockdown of Siah1 did not largely alter axon growth, although depletion of Siah1 increased Akt3 expression (Fig. 4C). This might be because many downstream targets of Siah1 are degraded by overexpression of Siah1, eliciting defects in neural development, including axon growth. On the other hand, axon growth is regulated cross-talk of multiple signaling not only Siah1/Akt3 signaling but also many of intracellular signaling is involved, reflecting that depletion of Siah1 itself could not alter axon growth. For each emerging target of study, it is important to identify the specific ubiquitin ligases that govern its turnover. Neuronal polarity and plasticity appear to develop through dynamic changes in molecular signaling within synapses, regulated through protein degradation within the ubiquitin–proteasome system (36, 37). Moreover, the tissue distribution of Siah1 is located predominantly in neuronal cells and has shown functional relevance in the regulation of synaptic plasticity by selective degradation of group 1 metabotropic glutamate receptors (38, 39), and Siah1 is known to be implicated in neurodegenerative disorders targeting synphilin 1and α-synuclein in neurons (24, 40). Although we cannot rule out the possibility of other targets of Siah1 in the neuron that are not yet identified could contribute together in addition to SAkt3, for neural development, at least partially if not all, Siah1 functions an E3 ligase for Akt3 isoform controlling axonal branching and growth by regulating turnover of Akt3 for proper neural development.
The three isoforms of Akt display considerable redundancy in their physiological functions, as revealing specific depletion of the isoforms individually has no detectable influence on neuronal polarity. However, the restriction of Akt3 expression to brain is consistent with specific roles of this isoform; reduced axonal growth in hippocampal neurons and Akt3 is known to regulate brain size (30). Recent studies also reported that mutations of Akt3 are associated with developmental disorders, including megalencephaly, dysplasia, and epilepsy (35, 41). Interestingly, many Akt3 mutations have been detected in malformations of the brain, with large, constitutively activate Akt3 (41). Akt3 was observed in axonal shafts and branches but not in axonal tips, where Siah1 expression was prominently detected (Fig. 4A and Fig. S2), although all Akt isoforms localized with Siah1 in the axons of neuron. It has been reported that Akt is preferentially localized at the axon after establishment of polarity (22). More importantly, depletion of Siah1 enhanced the intensity of Akt3 not only in the soma but also in the distal axon, along with multiple axonal branching points (Fig. 4 and Fig. S3). Presumably, knockdown of Siah1 in developing neurons allows evasion of Akt3 from degradation and permits Akt3 to induce more abnormal branching and overgrowth of axons. This could be probable paralogous cause like germline mutation of Akt3 to bring brain malformation out such as hemimegalencephaly.
A frequently observed somatic activating mutation in Akt3 is the E17K variation, which causes a broader spectrum of developmental brain disorders, including hemimegalencephaly. We found that the Akt3-E17K mutant markedly alleviated binding to Siah1 and elevated the protein stability of Akt3 (Fig. 5, A and B). Electroporating a somatic gain-of-function mutation of the Akt3-E17K mutant form into primary hippocampal neurons, we demonstrated that forced expression of Siah1 is restricting malformations of neural morphology showing reductions in Akt3-WT expression. In contrast, Siah1 was not sufficient to recapitulate the altered differentiation and excessive neuron size observed in Akt3-E17K mutant–expressing slices. Thus, our data encountered one possible explanation for how the Akt3-E17K mutation might acquire enhanced life expectancy of proteins and mislocalization in overgrowing neurites in brain pathology. Further studies should facilitate possible targeted design for future human clinical trials.
Experimental procedures
Primary neuron and cell culture
The brains of embryonic day 18 rat embryos were dissected, and hippocampi were removed and placed in a 15-ml tube with 14 ml of Hanks' balanced salt solution on ice. The medium was carefully aspirated, leaving 2 ml of medium in the tube with the hippocampi. Trypsin–EDTA was added to digest the tissue. Digestion was stopped by washing the hippocampi twice with 4 ml of complete (10% FBS) medium. Then, 3 ml of Neurobasal medium (Invitrogen, 21103-049)/B27 (Invitrogen, 17504-044) was added, and the tissue was dissociated by gently triturating the hippocampi through a fire-polished Pasteur pipette. The cell mixture was diluted to 10 ml with Neurobasal medium/B27 and then filtered through a 40-μm strainer. HEK293T cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% FBS and 100 units of penicillin/streptomycin. PC12 cells were maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum, 5% horse serum, and 100 units of penicillin/streptomycin at 37 °C in a 5% CO2 atmosphere.
Embryo brain slice culture
Embryo brain slice cultures were prepared from embryonic day 15 mouse brains. The 280-μm-thick brain slices were obtained by vibratome sectioning (VT1200, Leica Biosystems) in chilled MEMp (50% (v/v) MEM, 25 mm HEPES, and 2 mm glutamine without antibiotics, adjusted to pH 7.2–7.3 with 1 m NaOH). The slices were transferred onto semiporous membrane inserts (Millipore, 0.4-μm pore diameter, Schwalbach, Germany). Intact slices were cultured at 37 °C and 5% CO2 in maintenance medium (Neurobasal medium, 2% B27, 2 mm glutamine, 1% penicillin/streptomycin solution, 0.5% glucose, and gentamycin). The medium was changed every other day. The slices were infected after DIV 5 and cultured for an additional 5 days. The slices were fixed with 4% paraformaldehyde at DIV 10.
Antibodies, shRNA, and chemicals
Anti-phospho-Ser473 Akt, anti-pan Akt, anti-Akt1, anti-Akt2, and anti-Akt3 antibodies were purchased from Cell Signaling Technology (Danvers, MA). Anti-Siah1, anti-GFP, anti-GST, anti-MYC, anti-actin, and anti-ubiquitin were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The anti-Siah1 antibody was purchased from Novus Biologicals and Abcam (Cambridge, MA). The anti-FLAG M2 antibody was purchased from Sigma. Alexa Fluor–tagged secondary antibodies were purchased from Invitrogen/Molecular Probes (Eugene, OR). All other chemicals were purchased from Sigma-Aldrich (St. Louis, MO). Oligomers for shSiah1 (forward, GATCCGCAATTTAGGCATCAATGTAAGCTTACATTGATGCCTAAATTGCTTTTTT; reverse, CTAGAAAAAAGCAATTTAGGCATCAATGTAAGCTTACATTGATGCCTAAATTGCG) were cloned into a pGE1 vector.
Coimmunoprecipitation assay and in vitro binding assay
For coimmunoprecipitation, cells were rinsed with PBS and lysed in buffer (50 mm Tris-Cl (pH 7.4), 150 mm NaCl, 1 mm EDTA, 0.5% Triton X-100, 1.5 mm Na3VO4, 50 mm sodium fluoride, 10 mm sodium pyrophosphate, 10 mm β-glycerolphosphate, 1 mm PMSF, and protease mixture (Calbiochem, San Diego, CA). Cell lysates (0.5–1 mg of protein) were mixed with a primary antibody with protein G/A beads and incubated for 3 h at 4 °C with gentle agitation. The beads were then washed in lysis buffer, mixed with 2× SDS sample buffer, boiled, and analyzed by immunoblotting. For the in vitro binding assay, proteins were bacterially expressed, purified with GST resin, and dialyzed against PBS. Samples of intact proteins (500 ng) were reacted at 4 °C for 1 h with gentle agitation and immunoprecipitated with the indicated antibodies.
GST pulldown assay
Cells were rinsed with PBS and lysed in buffer as described above. Cell lysates (0.5–1 mg of protein) were mixed with GSH-Sepharose beads and incubated for 3 h at 4 °C with gentle agitation. The beads were then washed in lysis buffer, mixed with 2× SDS sample buffer, boiled, and analyzed by immunoblotting.
Immunofluorescence
Cells grown on coverslips in 24-well plates were fixed in 4% paraformaldehyde for 15 min, permeabilized in PBS containing 0.25% Triton X-100 for 10 min, and blocked in 2% BSA for 30 min. Cells were immunostained using primary antibodies and the appropriate Alexa Fluor 488– or Alexa Fluor 594–conjugated secondary antibodies. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole. Immunostained images were acquired using a laser-scanning confocal microscope (LSM 710, Carl Zeiss). The confocal microscope was controlled using ZEN software.
Bacterial protein purification
Siah-1 (WT and C41S/C44S) was cloned in a pGEX 4T-1 vector. The proteins were overexpressed in the Escherichia coli BL21(DE3) strain. Cells were grown at 37 °C until they reached A600 of 0.6–0.8 and were then induced with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside for 4 h at 25 °C.
In vitro ubiquitination assay
The in vitro ubiquitination experiment was carried out at a final volume of 30 μl, including E1 (5 μm, BostonBiochem), E2-UbcH5a (25 μm, BostonBiochem), FLAG-ubiquitin (5 μg, BostonBiochem), His-Akt3 (1 μg, Bioscience), and purified GST–Siah-1 (WT or C41S/C44S). The assay was performed in ubiquitination buffer containing 100 mm NaCl, 1 mm DTT, 5 mm MgCl2, 25 mm Tris-Cl (pH 7.5), and activated 10 mm ATP and incubated at 30 °C for 2 h. To stop the ubiquitination reaction, the samples were incubated for 10 min at 95 °C after addition of SDS loading buffer. Ubiquitination reactions were analyzed by Western blotting using AKT3 primary antibody (Cell Signaling Technology) and Siah1 (Abcam).
Ethics
All animal experiments were reviewed and approved by the Institutional Animal Care and Use Committee of Sungkyunkwan University School of Medicine (SKKUIACUC2018-09-13-1). All experimental procedures were carried out in accordance with the guidelines of the Institutional Animal Care and Use Committee of Sungkyunkwan University.
Statistical analyses
Graphs and associated statistical analyses were generated using GraphPad Prism (GraphPad, La Jolla, CA). The data were generated by performing the experiments at least three times. All data are presented as mean ± S.E. Statistical significance of two groups was assessed by unpaired t test; p < 0.05 was considered statistically significant.
Author contributions
H. R. K., S. B. L., S.-W. C., K. W. P., and J.-Y. A. data curation; H. R. K., S.-W. C., and K. W. P. formal analysis; H. R. K., E.-J. J., S. B. L., C. K. K., T. Y., and J.-Y. A. investigation; H. R. K. project administration; S. B. L., S.-W. C., and J.-Y. A. conceptualization; J.-Y. A. funding acquisition; J.-Y. A. methodology; J.-Y. A. writing-original draft; J.-Y. A. writing-review and editing.
Supplementary Material
This research was supported by National Research Foundation of Korea (NRF), grants funded by the Korean government, Ministry of Science, ICT and Future Planning (MSIP) 2016R1A5A2945889 and NRF-2017R1A2B4001846 (to J.-Y. A.) and a grant from the Korea Health Technology R&D Project through Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health and Welfare, Republic of Korea, Grant HI17C0227 (to J.-Y. A.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S4.
- UPS
- ubiquitin–proteasome system
- DIV
- day(s) in vitro.
References
- 1. Manning B. D., and Toker A. (2017) AKT/PKB signaling: navigating the network. Cell 169, 381–405 10.1016/j.cell.2017.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhang L., Li Bs., Ma W., Barker J. L., Chang Y. H., Zhao W., and Rubinow D. R. (2002) Dehydroepiandrosterone (DHEA) and its sulfated derivative (DHEAS) regulate apoptosis during neurogenesis by triggering the Akt signaling pathway in opposing ways. Brain Res. Mol. Brain Res. 98, 58–66 10.1016/S0169-328X(01)00315-1 [DOI] [PubMed] [Google Scholar]
- 3. Manning B. D., and Cantley L. C. (2007) AKT/PKB signaling: navigating downstream. Cell 129, 1261–1274 10.1016/j.cell.2007.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wu C., Orozco C., Boyer J., Leglise M., Goodale J., Batalov S., Hodge C. L., Haase J., Janes J., Huss J. W. 3rd, Su A. I. (2009) BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 10, R130 10.1186/gb-2009-10-11-r130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen W. S., Xu P. Z., Gottlob K., Chen M. L., Sokol K., Shiyanova T., Roninson I., Weng W., Suzuki R., Tobe K., Kadowaki T., and Hay N. (2001) Growth retardation and increased apoptosis in mice with homozygous disruption of the Akt1 gene. Genes Dev. 15, 2203–2208 10.1101/gad.913901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bae S. S., Cho H., Mu J., and Birnbaum M. J. (2003) Isoform-specific regulation of insulin-dependent glucose uptake by Akt/protein kinase B. J. Biol. Chem. 278, 49530–49536 10.1074/jbc.M306782200 [DOI] [PubMed] [Google Scholar]
- 7. Lee S. B., Kim C. K., Lee K. H., and Ahn J. Y. (2012) S-nitrosylation of B23/nucleophosmin by GAPDH protects cells from the SIAH1-GAPDH death cascade. J. Cell Biol. 199, 65–76 10.1083/jcb.201205015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kim C. K., Nguyen T. L., Lee S. B., Park S. B., Lee K. H., Cho S. W., and Ahn J. Y. (2011) Akt2 and nucleophosmin/B23 function as an oncogenic unit in human lung cancer cells. Exp. Cell Res. 317, 966–975 10.1016/j.yexcr.2010.12.013 [DOI] [PubMed] [Google Scholar]
- 9. Tschopp O., Yang Z. Z., Brodbeck D., Dummler B. A., Hemmings-Mieszczak M., Watanabe T., Michaelis T., Frahm J., and Hemmings B. A. (2005) Essential role of protein kinase Bγ (PKBγ/Akt3) in postnatal brain development but not in glucose homeostasis. Development 132, 2943–2954 10.1242/dev.01864 [DOI] [PubMed] [Google Scholar]
- 10. Easton R. M., Cho H., Roovers K., Shineman D. W., Mizrahi M., Forman M. S., Lee V. M., Szabolcs M., de Jong R., Oltersdorf T., Ludwig T., Efstratiadis A., and Birnbaum M. J. (2005) Role for Akt3/protein kinase Bγ in attainment of normal brain size. Mol. Cell. Biol. 25, 1869–1878 10.1128/MCB.25.5.1869-1878.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Boland E., Clayton-Smith J., Woo V. G., McKee S., Manson F. D., Medne L., Zackai E., Swanson E. A., Fitzpatrick D., Millen K. J., Sherr E. H., Dobyns W. B., and Black G. C. (2007) Mapping of deletion and translocation breakpoints in 1q44 implicates the serine/threonine kinase AKT3 in postnatal microcephaly and agenesis of the corpus callosum. Am. J. Hum. Genet. 81, 292–303 10.1086/519999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang D., Zeesman S., Tarnopolsky M. A., and Nowaczyk M. J. (2013) Duplication of AKT3 as a cause of macrocephaly in duplication 1q43q44. Am. J. Med. Genet. A 161A, 2016–2019 10.1002/ajmg.a.35999 [DOI] [PubMed] [Google Scholar]
- 13. Riviere J. B., Mirzaa G. M., O'Roak B. J., Beddaoui M., Alcantara D., Conway R. L., St-Onge J., Schwartzentruber J. A., Gripp K. W., Nikkel S. M., Worthylake T., Sullivan C. T., Ward T. R., Butler H. E., Kramer N. A., et al. (2012) De novo germline and postzygotic mutations in AKT3, PIK3R2 and PIK3CA cause a spectrum of related megalencephaly syndromes. Nat. Genet. 44, 934–940 10.1038/ng.2331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee J. H., Huynh M., Silhavy J. L., Kim S., Dixon-Salazar T., Heiberg A., Scott E., Bafna V., Hill K. J., Collazo A., Funari V., Russ C., Gabriel S. B., Mathern G. W., and Gleeson J. G. (2012) De novo somatic mutations in components of the PI3K-AKT3-mTOR pathway cause hemimegalencephaly. Nat. Genet. 44, 941–945 10.1038/ng.2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nellist M., Schot R., Hoogeveen-Westerveld M., Neuteboom R. F., van der Louw E. J., Lequin M. H., Bindels-de Heus K., Sibbles B. J., de Coo R., Brooks A., and Mancini G. M. (2015) Germline activating AKT3 mutation associated with megalencephaly, polymicrogyria, epilepsy and hypoglycemia. Mol. Genet Metab. 114, 467–473 10.1016/j.ymgme.2014.11.018 [DOI] [PubMed] [Google Scholar]
- 16. Baek S. T., Copeland B., Yun E. J., Kwon S. K., Guemez-Gamboa A., Schaffer A. E., Kim S., Kang H. C., Song S., Mathern G. W., and Gleeson J. G. (2015) An AKT3-FOXG1-reelin network underlies defective migration in human focal malformations of cortical development. Nat. Med. 21, 1445–1454 10.1038/nm.3982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yang W. L., Wang J., Chan C. H., Lee S. W., Campos A. D., Lamothe B., Hur L., Grabiner B. C., Lin X., Darnay B. G., and Lin H. K. (2009) The E3 ligase TRAF6 regulates Akt ubiquitination and activation. Science 325, 1134–1138 10.1126/science.1175065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chan C. H., Li C. F., Yang W. L., Gao Y., Lee S. W., Feng Z., Huang H. Y., Tsai K. K., Flores L. G., Shao Y., Hazle J. D., Yu D., Wei W., Sarbassov D., Hung M. C., et al. (2012) The Skp2-SCF E3 ligase regulates Akt ubiquitination, glycolysis, herceptin sensitivity, and tumorigenesis. Cell 149, 1098–1111 10.1016/j.cell.2012.02.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fan C. D., Lum M. A., Xu C., Black J. D., and Wang X. (2013) Ubiquitin-dependent regulation of phospho-AKT dynamics by the ubiquitin E3 ligase, NEDD4-1, in the insulin-like growth factor-1 response. J. Biol. Chem. 288, 1674–1684 10.1074/jbc.M112.416339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Suizu F., Hiramuki Y., Okumura F., Matsuda M., Okumura A. J., Hirata N., Narita M., Kohno T., Yokota J., Bohgaki M., Obuse C., Hatakeyama S., Obata T., and Noguchi M. (2009) The E3 ligase TTC3 facilitates ubiquitination and degradation of phosphorylated Akt. Dev. Cell 17, 800–810 10.1016/j.devcel.2009.09.007 [DOI] [PubMed] [Google Scholar]
- 21. Bae S., Kim S. Y., Jung J. H., Yoon Y., Cha H. J., Lee H., Kim K., Kim J., An I. S., Kim J., Um H. D., Park I. C., Lee S. J., Nam S. Y., Jin Y. W., et al. (2012) Akt is negatively regulated by the MULAN E3 ligase. Cell Res. 22, 873–885 10.1038/cr.2012.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yan D., Guo L., and Wang Y. (2006) Requirement of dendritic Akt degradation by the ubiquitin-proteasome system for neuronal polarity. J. Cell Biol. 174, 415–424 10.1083/jcb.200511028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Qi J., Kim H., Scortegagna M., and Ronai Z. A. (2013) Regulators and effectors of Siah ubiquitin ligases. Cell Biochem. Biophys. 67, 15–24 10.1007/s12013-013-9636-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee S. B., Xuan Nguyen T. L., Choi J. W., Lee K. H., Cho S. W., Liu Z., Ye K., Bae S. S., and Ahn J. Y. (2008) Nuclear Akt interacts with B23/NPM and protects it from proteolytic cleavage, enhancing cell survival. Proc. Natl. Acad. Sci. U.S.A. 105, 16584–16589 10.1073/pnas.0807668105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Baba T., Kobayashi H., Kawasaki H., Mineki R., Naito H., and Ohmori D. (2010) Glyceraldehyde-3-phosphate dehydrogenase interacts with phosphorylated Akt resulting from increased blood glucose in rat cardiac muscle. FEBS Lett. 584, 2796–2800 10.1016/j.febslet.2010.05.015 [DOI] [PubMed] [Google Scholar]
- 26. Chung J. Y., Lee S. J., Lee S. H., Jung Y. S., Ha N. C., Seol W., and Park B. J. (2011) Direct interaction of α-synuclein and AKT regulates IGF-1 signaling: implication of Parkinson disease. Neurosignals 19, 86–96 10.1159/000325028 [DOI] [PubMed] [Google Scholar]
- 27. Ong T., and Solecki D. J. (2017) Seven in absentia E3 ubiquitin ligases: central regulators of neural cell fate and neuronal polarity. Front. Cell Neurosci. 11, 322 10.3389/fnhum.2017.00322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Greene L. A., and Tischler A. S. (1976) Establishment of a noradrenergic clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Proc. Natl. Acad. Sci. U.S.A. 73, 2424–2428 10.1073/pnas.73.7.2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Winter M., Sombroek D., Dauth I., Moehlenbrink J., Scheuermann K., Crone J., and Hofmann T. G. (2008) Control of HIPK2 stability by ubiquitin ligase Siah-1 and checkpoint kinases ATM and ATR. Nat. Cell Biol. 10, 812–824 10.1038/ncb1743 [DOI] [PubMed] [Google Scholar]
- 30. Diez H., Garrido J. J., and Wandosell F. (2012) Specific roles of Akt iso forms in apoptosis and axon growth regulation in neurons. PLoS ONE 7, e32715 10.1371/journal.pone.0032715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ko H. R., Kwon I. S., Hwang I., Jin E. J., Shin J. H., Brennan-Minnella A. M., Swanson R., Cho S. W., Lee K. H., and Ahn J. Y. (2016) Akt1-Inhibitor of DNA binding2 is essential for growth cone formation and axon growth and promotes central nervous system axon regeneration. Elife 5, e20799 10.7554/eLife.20799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Daza R. A., Englund C., and Hevner R. F. (2007) Organotypic slice culture of embryonic brain tissue. CSH Protoc. 2007, 10.1101/pdb.prot4914 [DOI] [PubMed] [Google Scholar]
- 33. de Anda F. C., Meletis K., Ge X., Rei D., and Tsai L. H. (2010) Centrosome motility is essential for initial axon formation in the neocortex. J. Neurosci. 30, 10391–10406 10.1523/JNEUROSCI.0381-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ge X., Frank C. L., Calderon de Anda F., and Tsai L. H. (2010) Hook3 interacts with PCM1 to regulate pericentriolar material assembly and the timing of neurogenesis. Neuron 65, 191–203 10.1016/j.neuron.2010.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Poduri A., Evrony G. D., Cai X., Elhosary P. C., Beroukhim R., Lehtinen M. K., Hills L. B., Heinzen E. L., Hill A., Hill R. S., Barry B. J., Bourgeois B. F., Riviello J. J., Barkovich A. J., Black P. M., et al. (2012) Somatic activation of AKT3 causes hemispheric developmental brain malformations. Neuron 74, 41–48 10.1016/j.neuron.2012.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Malinow R., and Malenka R. C. (2002) AMPA receptor trafficking and synaptic plasticity. Annu. Rev. Neurosci. 25, 103–126 10.1146/annurev.neuro.25.112701.142758 [DOI] [PubMed] [Google Scholar]
- 37. Hegde A. N. (2004) Ubiquitin-proteasome-mediated local protein degradation and synaptic plasticity. Prog. Neurobiol. 73, 311–357 10.1016/j.pneurobio.2004.05.005 [DOI] [PubMed] [Google Scholar]
- 38. Della N. G., Senior P. V., and Bowtell D. D. (1993) Isolation and characterisation of murine homologues of the Drosophila seven in absentia gene (sina). Development 117, 1333–1343 [DOI] [PubMed] [Google Scholar]
- 39. Moriyoshi K., Iijima K., Fujii H., Ito H., Cho Y., and Nakanishi S. (2004) Seven in absentia homolog 1A mediates ubiquitination and degradation of group 1 metabotropic glutamate receptors. Proc. Natl. Acad. Sci. U.S.A. 101, 8614–8619 10.1073/pnas.0403042101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nagano Y., Yamashita H., Takahashi T., Kishida S., Nakamura T., Iseki E., Hattori N., Mizuno Y., Kikuchi A., and Matsumoto M. (2003) Siah-1 facilitates ubiquitination and degradation of synphilin-1. J. Biol. Chem. 278, 51504–51514 10.1074/jbc.M306347200 [DOI] [PubMed] [Google Scholar]
- 41. Alcantara D., Timms A. E., Gripp K., Baker L., Park K., Collins S., Cheng C., Stewart F., Mehta S. G., Saggar A., Sztriha L., Zombor M., Caluseriu O., Mesterman R., Van Allen M. I., et al. (2017) Mutations of AKT3 are associated with a wide spectrum of developmental disorders including extreme megalencephaly. Brain 140, 2610–2622 10.1093/brain/awx203 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.