Abstract
Destruction of the cartilage matrix in joints is an important feature of arthritis. Proteolytic degradation of cartilage glycoproteins can contribute to the loss of matrix integrity. Human inter-α-inhibitor (IαI), which stabilizes the extracellular matrix, is composed of the light-chain serine proteinase inhibitor bikunin and two homologous heavy chains (HC1 and HC2) covalently linked through chondroitin 4-sulfate. Inflammation promotes the transfer of HCs from chondroitin 4-sulfate to hyaluronan by tumor necrosis factor–stimulated gene-6 protein (TSG-6). This reaction generates a covalent complex between the heavy chains and hyaluronan that can promote leukocyte invasion. This study demonstrates that both IαI and the HC–hyaluronan complex are substrates for the extracellular matrix proteases ADAMTS-5 and matrix metalloprotease (MMP) -3, -7, and -13. The major cleavage sites for all four proteases are found in the C terminus of HC2. ADAMTS-5 and MMP-7 displayed the highest activity toward HC2. ADAMTS-5 degradation products were identified in mass spectrometric analysis of 29 of 33 arthropathic patients, indicating that ADAMTS-5 cleavage occurs in synovial fluid in arthritis. After cleavage, free HC2, together with TSG-6, is able to catalyze the transfer of heavy chains to hyaluronan. The release of extracellular matrix bound HC2 is likely to increase the mobility of the HC2/TSG-6 catalytic unit and consequently increase the rate of the HC transfer reaction. Ultimately, ADAMTS-5 cleavage of HC2 could alter the physiological and mechanical properties of the extracellular matrix and contribute to the progression of arthritis.
Keywords: arthritis, hyaluronan, ADAMTS, extracellular matrix protein, metalloprotease, bikunin, inter-α-inhibitor
Introduction
Human inter-α-inhibitor (IαI)3 is a heterotrimer consisting of the serine protease inhibitor bikunin (UniProt P02760) and two homologous heavy chains referred to as heavy chain 1 (HC1) (UniProt P19827) and heavy chain 2 (HC2) (UniProt P19823) (1). Bikunin is a proteoglycan with an undersulfated chondroitin 4-sulfate (CS) chain attached to Ser-10 by a typical tetrasaccharide linkage (Fig. 1) (2, 3). The HCs contain both N- and O-linked glycans (4) and are covalently linked to the CS chain by a unique ester bond between the α-carboxyl group of the C-terminal Asp residues and the C6 atoms of internal N-acetylgalactosamines in the CS chain. This interaction is referred to as a protein–glycosaminoglycan–protein (PGP) cross-link (2, 3, 5). HC2 is positioned closer to bikunin than HC1 on the CS chain, and the two HCs are attached one to two disaccharides apart (6, 7). The unique covalent linkages between the HCs and the CS are assembled intracellularly (6) and are referred to as PGP cross-links (2). Bikunin CS can also link to a third heavy chain (HC3) (UniProt Q06033) to form the heterodimeric pre-α-inhibitor (PαI) (1, 2). In addition, indirect evidence based on sequence identity suggests that the related HC5 (UniProt C9J2H1) (8) and possibly HC6 (UniProt Q6UXX5), but not HC4 (UniProt Q14624), can also become attached to the bikunin CS chain.
Figure 1.
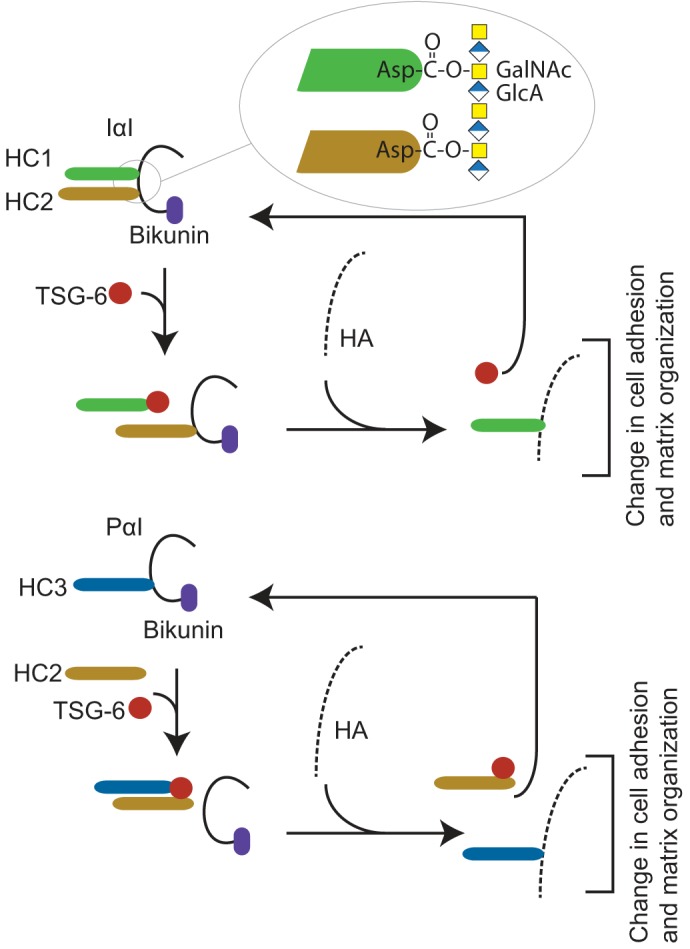
Overview of the human IαI- and TSG-6–mediated HC transfer. IαI is composed of bikunin and two homologous heavy chains (HC1 and HC2) (top). The HCs are covalently linked to the CS chain by a unique ester bond between the C terminus and N-acetylgalactosamines in the CS chain (inset) referred to as a PGP cross-link (2, 3, 5). In the ECM, IαI stabilizes HA-rich matrixes in a process requiring TSG-6 expressed during inflammatory disease states (13–15). TSG-6 catalyzes the transfer of HCs from IαI to HA, generating covalent HC–HA complexes (16). PαI is composed of bikunin linked to HC3 (bottom) (1, 2). The presence of HC2 is essential for the TSG-6–mediated transfer of HC3 to HA (12). The HC transfer reaction takes place in the presence of divalent cations and when the TSG-6 and bikunin proteins colocalize, e.g. during inflammation (17). The transfer of HC to HA changes the characteristics of the HA matrix and affects cell adhesive properties and matrix organization (18–20).
IαI is produced primarily in the liver and is found in plasma at concentrations up to ∼0.5 mg/ml (9). This protein is involved in diverse biological processes such as ovulation, cell migration, and inflammation (10) in addition to playing roles in the stabilization and remodeling of extracellular matrix (ECM) (11). In the ECM, IαI stabilizes hyaluronan (HA)-rich matrixes in a process requiring HC2 (12) and the tumor necrosis factor–stimulated gene-6 protein (TSG-6) (UniProt P98066) (13–15). HC2/TSG-6 catalyzes two sequential transesterifications, thereby transferring HCs from the CS of IαI or PαI to HA, generating a covalent HC–HA complex (Fig. 1) (16). The presence of HC2/TSG-6 and divalent cations is essential for this catalytic activity (12). The HC transfer reaction takes place during inflammation and inflammation-like processes in which TSG-6 expression is induced and colocalizes with IαI (17). The functional characteristics of HA are modified within HC–HA complexes, which can consequently change cell adhesive properties as well as matrix organization (18–20). A documented role of the HC–HA complex is in the formation and stabilization of the ECM in the expanding cumulus oocyte complex. The cumulus matrix contains HA, which becomes modified by the covalent attachment of HCs. Bikunin- or TSG-6–deficient mice are unable to form a stable ECM around their oocytes, resulting in naked oocytes and female infertility (21–24). The inflammatory process associated with rheumatoid arthritis generates abundant HC–HA complexes in synovial fluid (11, 25), which can increase the infiltration of leukocytes in the inflamed joints (20).
The proteolytic destruction of articular cartilage, including the two major components, collagen type II and aggrecan, is a key characteristic of arthritis. This tissue destruction follows aggrecan degradation initiated by members of the ADAMTS family and collagen degradation by matrix metalloproteinases (MMPs), leading to the loss of cartilage matrix (26). The expression, activation, inhibition, and clearance of each protease involved is strictly controlled (27, 28). However, during rheumatoid arthritis (RA) and osteoarthritis (OA), the level of active proteases increases (27). The main aggrecan-degrading enzymes that play a role during the development of RA and OA appear to be ADAMTS-4 and ADAMTS-5 (27). Only a few human proteins are able to cleave fibrillar collagen type II, including MMP-1 and MMP-13. In view of the preferred specificity for type II collagen and its inducible expression by chondrocytes, MMP-13 is presumed to be a key factor in the progression of arthritis (26).
The TSG-6–catalyzed formation of HC–HA complexes during inflammation means that IαI is located in an environment where proteolytic activity apparently overwhelms natural protease regulation. We hypothesized that ADAMTS or MMPs involved in cartilage degradation cleave IαI as part of the progression of arthritis. This hypothesis is supported by the observation of truncated HCs in both arthritic cartilage and synovial fluid (29). In addition, ADAMTS-5 and IαI colocalize in equine degenerative suspensory ligament desmitis, and truncated versions of the HCs are observed (30). In the present study, we show that IαI is a substrate for ADAMTS-5 and MMP-3, -7 and -13. The main cleavage site is found in the C terminus of HC2, and the corresponding degradation product is present in arthritic synovial fluid. Furthermore, we present data showing that the ADAMTS-5–released HC2 retains the capacity to form an active HC2/TSG-6 complex and is therefore able to influence arthritis by increasing the catalytic transfer of heavy chains to hyaluronan.
Results
IαI is a substrate for metalloproteinases
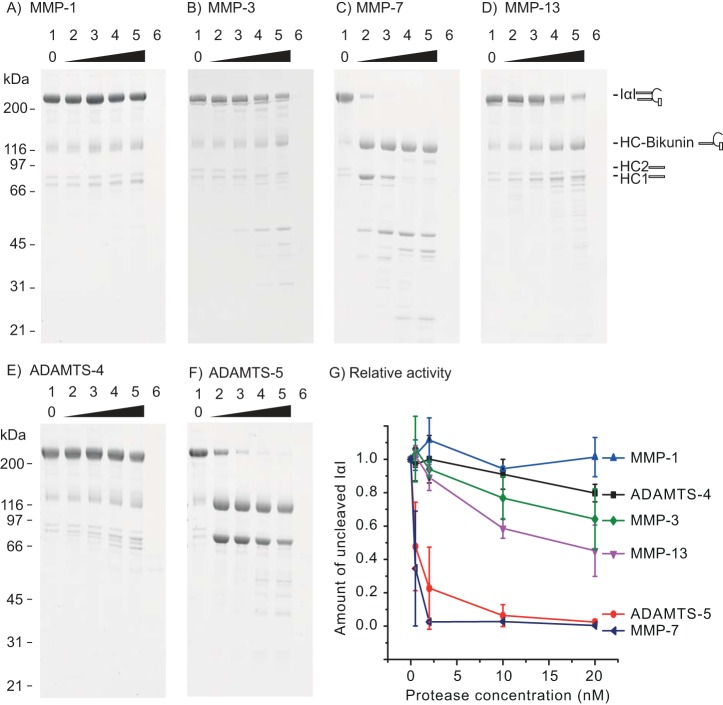
We initially tested whether IαI was proteolytically processed by MMP-1, MMP-3, MMP-7, MMP-13, ADAMTS-4, or ADAMTS-5. The ability of the proteases to carry out cleavage was determined by titrating a fixed amount of IαI with the different proteases at 0–20 nm (Fig. 2). IαI was cleaved by MMP-7 or ADAMTS-5 at 0.5 nm (Fig. 2, C and F, lane 2). MMP-3 and MMP-13 cleaved IαI to a lesser extent, whereas MMP-1 and ADAMTS-4 showed no or a very low level of activity toward IαI.
Figure 2.
IαI is a substrate for MMP-3, MMP-7, MMP-13, and ADAMTS-5. A fixed amount of IαI was incubated with increasing amounts of the indicated proteases for 16 h at 37 °C before being analyzed by SDS-PAGE and stained with Coomassie Blue (A–F). The concentration of IαI was 0.9 μm, and the concentration of the protease was 0, 0.5, 2, 10, and 20 nm (lanes 1–5). The protease alone at 20 nm was loaded as a control (lane 6). Molecular mass size markers are indicated on the left side (kDa). The schematics on the right side of the gels show the migration of intact IαI, HC–bikunin, HC2, and HC1 alone. In lane 1, IαI appears together with HC–bikunin and free HCs as a result of nonenzymatic autohydrolysis of the ester in the PGP cross-link. The relative rate of IαI proteolysis by the tested proteases was evaluated by densitometry (G). The amount of noncleaved IαI at different protease concentrations was normalized to the IαI control (lane 1). The error bars represent the standard deviation of at least three experiments. In conclusion, these data show that MMP-3, MMP-7, MMP-13, and ADAMTS-5 are able to cleave IαI, which is demonstrated by the progressive reduction of the IαI level, accompanied by the occurrence of proteolytic degradation products.
To evaluate the relative rates of proteolysis, the amount of noncleaved IαI was measured by densitometry at different protease concentrations and normalized to nontreated IαI (Fig. 2G). From these data, it was inferred that ADAMTS-5 and MMP-7 are the most efficient proteases, whereas MMP-3 and MMP-13 display intermediate rates, and MMP-1 and ADAMTS-4 exhibit little or no activity toward IαI.
The proteases that cleaved IαI produced two initial reaction products of ∼80 and 116 kDa, which were further degraded at higher protease concentrations. The 80- and 116-kDa reaction products displayed migration similar to that of free HCs (80 kDa) and also migrated as a single HC linked to bikunin (120 kDa), indicating that the initial cut was near the C terminus of one of the two IαI HCs.
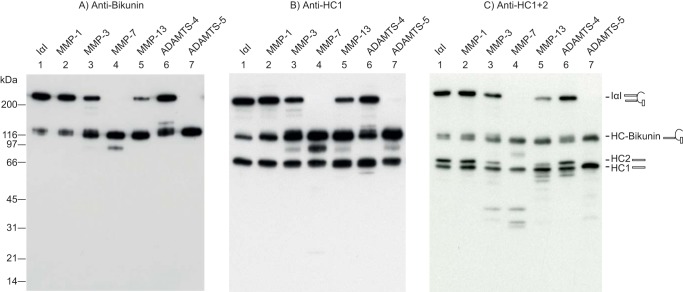
To further characterize the reaction products, IαI treated with MMP-1, MMP-3, MMP-7, MMP-13, ADAMTS-4, or ADAMTS-5 was analyzed by immunoblotting using a bikunin antiserum (Fig. 3A), an HC1 antiserum (Fig. 3B), or an antiserum recognizing both HC1 and HC2 (HC1+2) to visualize HC2 by subtraction (Fig. 3C). As nonenzymatic autohydrolysis of the ester in the PGP cross-link releases free full-length HCs, the protease digest was compared with IαI incubated for a similar time at a similar temperature to the protease-treated samples (lane 1). When the samples were probed with the bikunin antibody, one major product and full-length IαI were observed (Fig. 3A). The migration of the major degradation product corresponded to bikunin linked to a single HC. Additional bikunin reactive products were observed after MMP-7 treatment (Fig. 3A, lane 4), indicating additional cleavage sites in the HC–bikunin product. Bikunin remained associated with an HC (the 120-kDa band), suggesting that only one of the HCs contained a C-terminal cleavage site through which the 80-kDa band was released. The HC1-specific antibody revealed two major bands at 80 and 116 kDa (Fig. 3B). These bands were also observed after autohydrolysis of the PGP cross-link and represent free HC1 (80 kDa) and HC1–bikunin (Fig. 3B, lane 1). However, the intensity of the HC1–bikunin band was significantly increased after MMP-3, MMP-7, MMP-13, or ADAMTS-5 treatment (Fig. 3B, lanes 3, 4, 5, and 7), indicating that the HC–bikunin product contains HC1–bikunin (Fig. 3B, lane 6). The digest analyzed with the HC1+2 antibody exhibited the HC–bikunin species, the free HCs, and some minor degradation products after MMP-3, MMP-7, or MMP-13 treatment (Fig. 3C). These additional degradation products all present lower molecular weights than the full-length HCs and are not observed when an anti-HC1 antibody is used, indicating that these products are HC2-related. Moreover, the free HC2 band observed after autohydrolysis (Fig. 3C, lane 1) was reduced or disappeared after treatment with MMP-3, MMP-7, MMP-13, or ADAMTS-5 (Fig. 3C, lanes 3, 4, 5, and 7). These data show that the initial and major cleavage site in IαI is within the C terminus of HC2. After cleavage, the released HC2 most likely comigrates with HC1.
Figure 3.
HC2 contains the initial cleavage site. IαI was treated with the indicated proteases. The products were analyzed by SDS-PAGE and visualized by immunoblotting using an antibody recognizing bikunin (A), HC1 (B), or HC1+2 (C). Due to the hydrolysis of the PGP cross-link, IαI alone (lane 1) contained some HC–bikunin and free HCs when analyzed by SDS-PAGE. The schematics on the right side of the gels show the migration of intact IαI, HC–bikunin, HC2, and HC1 alone. In the anti-bikunin blot (A), MMP-3, MMP-7, MMP-13, and ADAMTS-5 treatment (lanes 3–5 and 7) reduced the level of IαI and increased the amount of a product migrating similarly to HC–bikunin. Using anti-HC1 (B), we similarly observed an increase in HC–bikunin, whereas the intensity of free HC1 appeared to be stable (lanes 3–5 and 7). The anti-HC1+2 blot (C) revealed a reduction in the intensity or a complete absence of the HC2 band (lanes 3–5 and 7). The variation in the intensity of the HC1-containing bands between B and C is a result of antibody efficiency and exposure time and does not represent variations in the amount of HC1. The proteolytic digestion of IαI was repeated more than 10 times, whereas immunoblotting was repeated two times for A and B and more than six times for C. Taken together, the observations indicate that the major MMP-3, MMP-7, MMP-13, and ADAMTS-5 cleavage sites are located in the C terminus of HC2.
The C terminus of HC2 is susceptible to proteolysis
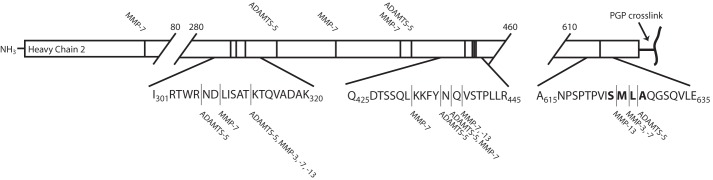
An N-terminomics approach was applied to identify the neo-N termini after protease treatment. Purified IαI was titrated with MMP-3, MMP-7, MMP-13, or ADAMTS-5, and all N termini were acetylated to label the neo-N termini. The labeled samples were digested with either trypsin or a combination of trypsin and the endoprotease Glu-C and subsequently analyzed by LC-MS/MS. N termini originating from MMP or ADAMTS cleavage events were identified as acetylated N termini, which were not observed in a control sample of nontreated IαI (Table 1). To verify the N-terminomics results, we determined the N-terminal sequence by Edman degradation of the major HC–bikunin product after transfer to polyvinylidene difluoride membranes (Fig. 2 and Table 1). In line with the previous results, all proteases preferred a substrate site in the C terminus of HC2. Significantly, residues Ser-625 to Ala-628 were particularly susceptible to proteolysis. At this site, MMP-13 uniquely cleaved Ser-625↓Met-626, MMP-3 and MMP-7 cleaved Met-626↓Leu-627, and ADAMTS-5 cleaved the Leu-627↓Ala-628 peptide bond (Fig. 4). In addition to the main cleavage sites, there are two other regions in HC2 (308–331 and 436–438) that are susceptible to cleavage to a minor degree. The 420–438 region has previously been shown to be exposed and available for transglutaminase cross-linking (31).
Table 1.
Identification of metalloproteinase cleavage sites in IαI
| Concentration |
Edman sequencinga | |||
|---|---|---|---|---|
| 2 nm | 10 nm | 20 nm | ||
| MMP-3 | HC2: 627 | HC2: 313, 627 | L_QGSQVLE__PP (HC2) | |
| MMP-7 | HC2: 308,b 313, 397, 627 | HC2: 308, 313, 397, 432, 436, 511, 571, 574, 627 | HC2: 65, 308, 313, 362, 397, 432, 437, 438, 511, 574, 627 | LAQG_QVLES_PP (HC2) |
| HC1: 461 | HC1: 399, 461, 572 | HC1: 22, 367, 399, 572 | ||
| MMP-13 | HC2: 438 | HC2: 308, 313, 438, 626 | ML_QGSQVLE__P (HC2) | |
| HC1: 461, 572 | ||||
| ADAMTS-5 | HC2: 313 437 628 | HC2: 313, 330, 436, 437, 628 | HC2: 305, 313, 314, 330, 403, 436, 437, 628 | AQGSQVLES_PPP (HC2) |
a Thr-637 is glycosylated and is not identified by Edman degradation. Unidentified residues are indicated wtih “__”.
b Bold residues are major cleavage sites based on the intensity of the MS signal.
Figure 4.
Overview of the neo-N termini in heavy chain 2 after proteolytic treatment. A summary of the N-terminal sequencing results and LC-MS/MS data are given in Table 1. The schematics represent HC2 with the location of the neo-N termini indicated with vertical lines, and the relevant protease is indicated. The major and initial cleavage sites are located in the C terminus close to the PGP cross-link. Cleavage in the C terminus of HC2 releases HC2 from the IαI complex and is likely to facilitate HC2/TSG-6 activity and increase the rate of the HC transfer reaction.
HC2 is a substrate for MMP-3, MMP-7, MMP-13, and ADAMTS-5 when covalently linked to HA
It has previously been shown that MMP-3, MMP-13, ADAMTS-5, and IαI are present in the cartilage and synovial fluid of arthropathic patients (27, 29, 32, 33). Although these proteases are able to act directly on IαI, the major IαI-related substrate is most likely the HC–HA complex, which shows increased abundance in arthritic patients. Consequently, we analyzed whether the proteases were able to cleave the HCs when linked to HA (Fig. 5). Preincubation with IαI (Fig. 5, lane 1), TSG-6, and a short HA oligomer was performed for 4 h (Fig. 5, lane 3) before proteases were added to the sample (Fig. 5, lanes 4–7). Subsequently, all samples were incubated for 16 h (lanes 3–7) and analyzed by SDS-PAGE and immunoblotting using antibodies recognizing both HC1 and HC2. IαI, TSG-6, and HA oligomers generated two bands corresponding to a single HA oligomer linked to either HC1 or HC2 (Fig. 5, lane 3). The four proteases that cleaved IαI (MMP-3, MMP-7, MMP-13, and ADAMTS-5) also specifically cleaved HC2 when transferred to HA as demonstrated by a significant reduction in the HC2–HA band (Fig. 5, lanes 4–7).
Figure 5.
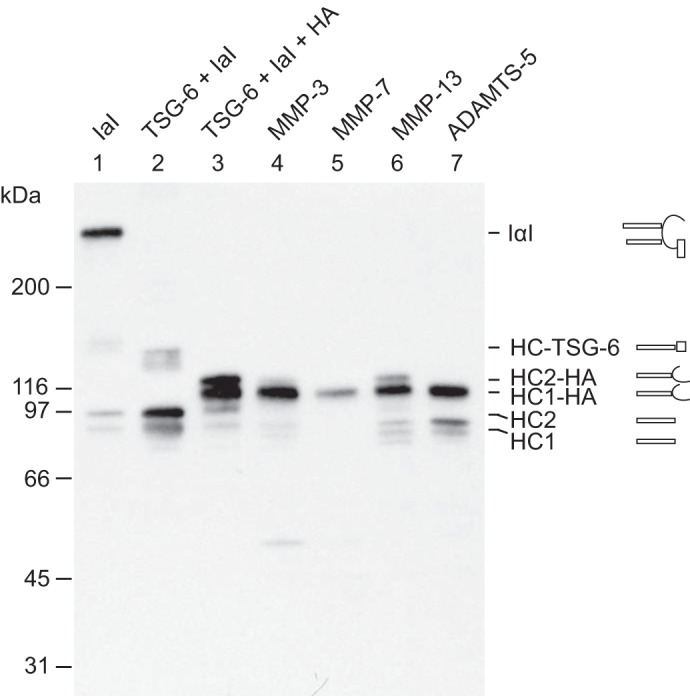
ADAMTS-5, MMP-3, MMP-7, and MMP-13 are able to cleave HC2 when covalently linked to HA. IαI (lane 1) was incubated with TSG-6 alone (lane 2) or with both TSG-6 and HA oligomers (lane 3). After 4 h at 37 °C, the protease was added to the HA-containing sample (lanes 4–7), which was then incubated for 16 h in parallel with the control sample (lane 3). All samples were analyzed by SDS-PAGE and immunoblotting using an anti-HC antibody. Schematics of the subunit compositions of the bands are shown on the right side of the gel. As expected, the migration of the HCs transferred to HA by HC2/TSG-6 (lane 3) was slower than that of the free HCs. The migration of the free HCs is shown in lane 1 as a result of nonenzymatic autohydrolysis of the ester in the PGP cross-link. Following proteolysis (lanes 4–7), the bands corresponding to HC2–HA are reduced or absent. The degradation products can be observed following MMP-13 or ADAMTS-5 treatment (lanes 6 and 7). MMP-3 and -7 (lanes 4 and 5) apparently cleave HC2 into fragments that are not recognized by the antibody. The data represent four experimental repeats. These data show that HC2 cross-linked to HA still acts as a substrate for MMP-3, MMP-7, MMP-13, and ADAMTS-5.
Cleavage of HC2 by ADAMTS-5 is identified in synovial fluid from arthropathic patients as the major degradation product
Our in vitro analysis revealed that IαI is a substrate for MMP-3, MMP-7, MMP-13, and ADAMTS-5 as they all cleave HC2 within three residues, with P1 located at Ser-625 (MMP-13), Met-626 (MMP-3 and -7), or Leu-627 (ADAMTS-5). These proteases are known to be involved in tissue degradation during pathological conditions such as RA and OA. In addition, inflammation increases the number of HCs linked to HA. To determine whether HC2 is cleaved by MMPs or ADAMTS during inflammatory conditions in vivo, an MS-based assay was employed. The cleaved C-terminal HC2 peptide was isolated after hydrolysis of the PGP cross-link in synovial fluid from 10 OA patients, nine RA patients, and 14 spondyloarthropathy (SpA) patients and identified by MS (Table 2). The ADAMTS-5 cleavage site was detected in 29 of the 33 patient samples (Table 2) (OA, 9 of 10; RA, 8 of 9; and SpA, 12 of 14). The ADAMTS-5 degradation product was found in all types of arthritic synovial fluid examined. In three of the 33 samples, we observed the MMP-3/-7 degradation product (two RA and one SpA), whereas we did not identify the MMP-13 degradation product in any of the synovial fluid samples. The in vivo identification of the ADAMTS-5 reaction product in synovial fluid from arthropathic patients supports the relevance of the observation to the pathogenesis of arthropathy.
Table 2.
Identification of HC2 cleavage mediated by ADAMTS-5 or MMPs in patients with arthritis
| Arthropathy | No. of patients | No. of patients with ADAMTS-5 product | No. of patients with MMP-3/-7 product | Male/female | Age (average) | BMIa (average) |
|---|---|---|---|---|---|---|
| Osteoarthritis | 10 | 9 | 0 | 5/5 | 25–79 (55.8) | 15.8–33.6 (26.8) |
| Rheumatoid arthritis | 9 | 8 | 2 | 2/7 | 31–79 (53.6) | 18.4–33.8 (23.6) |
| Spondyloarthropathy | 14 | 12 | 1 | 7/7 | 21–77 (46.8) | 21.6–38.1 (26.9) |
| Total | 33 | 29 | 3 | 14/19 | 21–79 (51.1) | 18.4–38.1 (25.9) |
a Information not available for all patients (OA, 8 of 10; RA, 8 of 9; SpA, 10 of 14).
ADAMTS-5 releases functional HC2 from the HA-binding articular cartilage scaffold
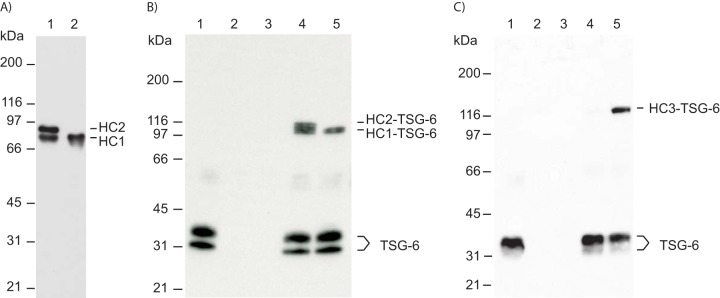
We have previously shown that complex formation between TSG-6 and HC3 depends on HC2 (12). Although the PGP cross-link is necessary for transesterification, HC2 in its free form can induce complex formation between glycosaminoglycan (HA or CS)-bound HCs and TSG-6 (12). To determine whether HC2 released by ADAMTS-5 proteolysis was able to induce complex formation between HCs and TSG-6, we incubated TSG-6 with IαI cleaved by ADAMTS-5 (Fig. 6). Following incubation with ADAMTS-5, all the HC2 was cleaved (Fig. 6A, lane 2). After incubation with TSG-6, the samples were separated by SDS-PAGE and visualized by immunoblotting using a TSG-6 antibody (Fig. 6B). TSG-6 incubated with IαI usually forms complexes with both HCs (Fig. 6B, lane 4). When IαI was preincubated with ADAMTS-5, only a single complex corresponding to HC1–TSG-6 was apparent. We cannot exclude the possibility that HC1–bikunin is able to form a complex with TSG-6 independent of HC2. Consequently, ADAMTS-5–cleaved HC2 was isolated and incubated with PαI and TSG-6 (Fig. 6C). A previous study showed that TSG-6 and PαI alone are unable to form the HC3–TSG-6 complex (Fig. 6C, lane 4). However, when purified and cleaved HC2 was included, the HC3–TSG-6 complex was observed (Fig. 6C, lane 5). Because HC2 is required for complex formation, we conclude that ADAMTS-5–released HC2 retains the ability to induce the HC3–TSG-6 complex.
Figure 6.
C-terminally truncated HC2 maintains the ability to induce HC–TSG-6 complex formation. A, IαI was pretreated with ADAMTS-5 to release all HC2. IαI (lane 1) and IαI treated with ADAMTS-5 (lane 2) were incubated at high pH to hydrolyze the ester in the PGP cross-link. The samples were analyzed by SDS-PAGE and visualized by immunoblotting using an anti-HC1+2 antibody. After ADAMTS-5 treatment, no full-length HC2 was present (lane 2). B, TSG-6 (seen alone in lane 1) was incubated with either IαI (lane 4) or IαI pretreated with ADAMTS-5 (lane 5). The samples were analyzed by SDS-PAGE and immunoblotting using an anti-TSG-6 antibody. IαI alone and IαI pretreated with ADAMTS-5 are shown in lanes 2 and 3, respectively. Prior to ADAMTS-5 treatment, both HC1 and HC2 form a complex with TSG-6 (lane 4). Significantly, following ADAMTS-5 treatment, cleaved HC2 remains able to catalyze the formation of the HC1–TSG-6 complex (lane 5). C, to verify that cleaved HC2 is able to mediate HC–TSG-6 complex formation, cleaved HC2 was isolated after ADAMTS-5 treatment with IαI. Purified HC2 was used to evaluate complex formation between TSG-6 and HC3. TSG-6 (seen alone in lane 1) was incubated with PαI (lane 4) or PαI and purified cleaved HC2 (lane 5). The samples were analyzed by SDS-PAGE and immunoblotting using an anti-TSG-6 antibody. It is only in the presence of HC2 that TSG-6 and HC3 from PαI form the HC3–TSG-6 complex. PαI and purified HC2 are shown in lanes 2 and 3, respectively. Because complex formation between TSG-6 and HC3 is HC2-dependent, these data show that the ADAMTS-5–released HC2 is still functional.
Discussion
In this study, we demonstrated that IαI is a substrate of ADAMTS-5 in vivo. HC2 was cleaved in vitro at an enzyme concentration (2 nm) equivalent to the physiological ADAMTS-5 concentration (34). In addition, MMP-3, -7, and -13 cleaved IαI in vitro. The major cleavage sites of all of these proteases are located within three residues (VIS625↓MMP-13M↓MMP-3/7L↓ADAMTS-5AQGSQ). After the initial cleavage, additional N- and C-terminal cleavage may occur. To identify degradation in vivo, we used a novel isolation technique exploiting the ability to selectively release HCs from HA by hydrolyzing the PGP cross-link using 100 mm NaOH (2). In synovial fluid from SpA, RA, and OA patients, we were able to identify the ADAMTS-5–cleaved C terminus of HC2 in 33 of 37 patient samples.
In general, MMPs prefer to cleave peptide bonds preceding residues containing hydrophobic side chains (P1′: Leu, Ile, Met, Phe, or Tyr) and those with a proline as the third residue prior to the cleavage site (P3) (36). For both MMP-3 and MMP-7, Leu is favored in the P1′ position. Based on the substrate cleavage sites reported in MEROPS (37), the frequencies of Leu at P1′ for MMP-3 and MMP-7 are 0.28 and 0.55, respectively, which is in agreement with the cleavage site in HC2. MMP-13 also shows specificity toward Leu (frequency 0.34), whereas the frequency of Met at P1′, as observed in HC2, is only 0.08. The best-described ADAMTS-5 substrate is aggrecan, containing five sites, all of which harbor a Glu residue at P1 and a Gly, Ala, or Leu residue at P1′. Among the 38 ADAMTS-5 substrate sites reported in MEROPS (37), the frequency of Glu at P1 is 0.21, whereas Leu is only observed at 5% of the substrate sites in HC2. However, the frequency of Ala at P1′ is 0.18 and is only superseded by that of Gly, with a frequency of 0.21. The amino acid sequence of the HC2 region susceptible to proteolysis is in agreement with the specificity of MMP-3 and MMP-7, whereas the preferred substrate recognition residues of MMP-13 and ADAMTS-5 are absent. In addition, the location of the cleavage sites indicates that CS could be involved in facilitating the proteolytic event. A hypothesis supported by a previous study showing that the activity of ADAMTS-4 and -5 depends on the chondroitin sulfate chains in aggrecan (38).
In inflammatory conditions such as arthritis, increased TSG-6 expression facilitates the generation of HC–HA complexes (11). In addition, MMP activity is increased, and the HC–HA complexes therefore become likely to undergo metalloprotease degradation. Indeed, the in vitro results showed that the HC–HA complexes were substrates for MMP-3, MMP-7, MMP-13, and ADAMTS-5. Furthermore, it is known that MMP-13 and ADAMTS-5 are involved in tissue degradation during osteoarthritis (27, 36), indicating that ADAMTS-5 might be responsible for the observed in vivo processing of HC2 in the joints of OA, RA, and SpA patients. In a previous study, truncated versions of the HCs were detected in synovial fluid and osteoarthritic cartilage. It was suggested that they might have been synthesized by the chondrocytes (29). Alternatively, these truncated HC species, which migrated with apparent molecular masses between 55 and 75 kDa, might result from ADAMTS-5 or MMP proteolysis as described in the present study.
The ADAMTS-5–mediated release of HC2 from either HA or IαI is likely to have a number of functional implications (Fig. 7). This HC–HA processing may change the stability as well as the cell-binding properties of HA with multiple downstream consequences (20, 23). In addition, the bioavailability of HC2 and TSG-6 will be affected as HC2 is released from the HA-coated ECM surface (Fig. 7). This is likely to affect the kinetics of the HC transfer reactions and, thus, the physical properties of HA. In synovial fluids from RA patients, up to five HCs are found attached to a single HA chain of 2 × 106 Da. This covalent HC modification influences the propensity of HA to aggregate compared with unmodified HA and may, thus, alter HC–HA matrix stabilization (25). In addition, the transfer of HCs to HA in arthritis (11) increases the binding of HA to its major cell surface receptor, CD44, on leukocytes (20), although it is not clear how this might influence the pathology of arthritis (15, 39).
Figure 7.
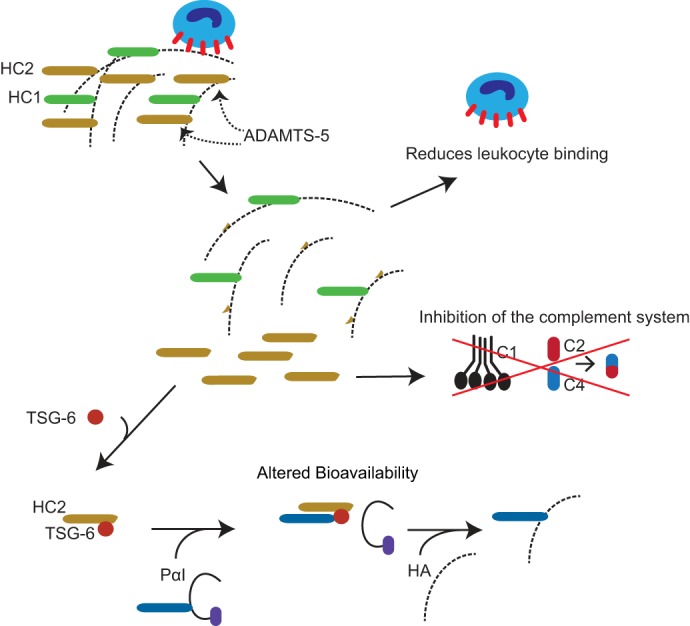
Potential consequences of ADAMTS-5–mediated HC2 release from HA. ADAMTS-5–mediated release of HC2 may change the stability and cell-binding properties of HA. In addition, it has previously been shown that the transfer of HCs to HA increases the binding of HA to its primary cell surface receptor, CD44, on leukocytes (20). Consequently, the release of HC2 may inhibit leukocyte adhesion to the HA-rich matrix. HC2 also affects complement activation (42), and the ADAMTS-5–mediated release of HC2 may therefore provide a mechanism for local regulation of the immune response. Furthermore, the bioavailability and mobility of the HC2–TSG-6 complex will be affected when HC2 is released from HA in the ECM.
One of the most significant physiological functions of the HCs is stabilization of the ECM in the expanding cumulus oocyte complex. During the maturation of the oocyte, the granulosa cells in the follicle expand the ECM and form an HC-dependent HA-rich matrix that is essential for fertilization (21–23). In a study analyzing the expression of ADAMTS-5 in the cumulus oocyte complex, ADAMTS-5 was expressed by granulosa cells in most follicles (40). However, after ovulation, ADAMTS-5 expression was absent in the released oocytes, whereas it was still expressed during follicular atresia. The ovulation-dependent regulation of ADAMTS-5 may therefore govern the formation of the HC–HA-rich matrix by cleaving and, thus, releasing HC2 from the ECM. Matured follicles that are not ovulated undergo atresia (apoptosis), and ADAMTS-5 may participate in this process by dissolving the hyaluronan-rich matrix, offering an explanation and role for the continued expression of ADAMTS-5.
It has been shown previously that the HC–HA complex mediates cellular adhesion to the HA-rich matrix (18–20). This interaction is regulated by thrombin-induced cleavage of HC1 and reduces leukocyte binding to an HA-rich matrix during inflammation (41). The ADAMTS-5–mediated release of HC2 could have a similar effect and, hence, regulate leukocyte adhesion and migration by dissolving the HC–HA matrix. An additional study has shown that the HCs of IαI inhibit the classical pathway of the complement system, with HC2 acting as the most potent inhibitor (42). We speculate that the release of HC2 acts as a mechanism for local control of the activation of the complement system. In conclusion, the release of HC2 by ADAMTS-5 could influence inflammation by regulating both leukocyte infiltration and complement activation.
In conclusion, the data presented in this study show that HC2 from IαI is a substrate for matrix-degrading proteases, with ADAMTS-5 being the most efficient. Our in vitro data are supported by the identification of the ADAMTS-5 degradation product in synovial fluids from arthritic patients. The release of HC2 can influence both the mechanical and physiological properties of the ECM.
Experimental procedures
Materials
IαI and PαI were purified from human plasma obtained from Aarhus University Hospital, Skejby, Denmark (1). Human TSG-6 was expressed in insect cells and purified as described previously (43). MMP-1, -3, -7, -13 (44–46), ADAMTS-4 lacking the C-terminal spacer domain, and ADAMTS-5 lacking the C-terminal thrombospondin domain (38) were expressed, activated, and purified as described previously. HA oligomers were produced and purified as described previously (12). Briefly, a 13-disaccharide-long HA oligosaccharide was generated and purified following limited chondroitinase ABC digestion of HA. Subsequently, HA was fractionated by anion exchange and desalted by size-exclusion chromatography. All reagents and chemicals were from Sigma-Aldrich/Merck unless otherwise stated.
SDS-PAGE and immunoblotting
Samples were boiled in SDS sample buffer in the presence of 50 mm dithiothreitol (DTT). SDS-PAGE was performed in 5–15% gradient gels (10 × 10 × 0.15 cm) using the glycine/2-amino-2-methyl-1,3-propanediol/HCl system (47). The gels were either stained for protein using Coomassie Blue or electroblotted to an Immobilon-P membrane (48). Immunoblotting was carried out using standard protocols and a rabbit anti-bikunin (3), rabbit anti-TSG-6 (49), or rabbit anti-HC antibody (3) as the primary antibody. The secondary antibody was goat anti-rabbit IgG-peroxidase. The membranes were developed using ECL reagents (GE Healthcare). Densitometry measurements of the Coomassie Blue–stained gels were performed using NIH ImageJ 1.52 (50).
Proteolytic digestion of IαI
Fixed concentrations of IαI (1.1 μm) were incubated with either fixed (20 nm) or increasing amounts of activated MMP-1, MMP-3, MMP-7, MMP-13, ADAMTS-4, or ADAMTS-5 (0, 0.5, 2, 10, and 20 nm). All samples were dissolved in 50 mm Tris-HCl, 150 mm NaCl, 10 mm CaCl2, pH 7.6, supplemented with 0.05% Brij-35. The samples were incubated for 16 h at 37 °C and analyzed by either LC-MS/MS or SDS-PAGE.
Degradation of HA-associated HCs
IαI (2.5 μg), TSG-6 (0.8 μg), and 25 μg of HA oligomers (HA14) were incubated for 4 h at 37 °C in 50 mm Tris-HCl, 100 mm NaCl, 1 mm MgCl2, pH 7.4. Subsequently, MMP-3, MMP-7, MMP-13, or ADAMTS-5 was added to a final concentration of 20 nm. Additionally, the samples were supplemented with 0.05% Brij-35 and 10 mm CaCl2. The samples were incubated for 16 h at 37 °C before they were analyzed by SDS-PAGE and immunoblotting using an anti-HC1+2 antibody.
Isolation of ADAMTS-5–cleaved HC2
Limited proteolysis of IαI using 160 nm ADAMTS-5 completely released HC2 from IαI as confirmed by Western blotting. The released HC2 was separated from HC1–bikunin by anion-exchange chromatography using a Mono Q 4.6/100 PE column (GE Healthcare) equilibrated in 20 mm Tris-HCl, pH 7.4. Proteins were eluted using a linear gradient from 0 to 0.6 m NaCl over 60 min with a flow rate of 1 ml/min. Fractions containing HC2 were visualized by SDS-PAGE and identified by MS. Fractions containing cleaved HC2 were pooled and concentrated by ultrafiltration (30 kDa; Amicon).
HC–TSG-6 complex formation
IαI (0.6 μg), IαI (0.6 μg) pretreated with ADAMTS-5 (160 nm), or PαI (0.6 μg) was incubated with 0.2 μg of TSG-6 for 2 h at 37 °C. The samples were dissolved in 50 mm Tris-HCl, 100 mm NaCl, pH 7.4. To verify the activity of cleaved HC2, purified HC2 was incubated with TSG-6 and PαI under the same conditions. The samples were analyzed by SDS-PAGE and immunoblotting using an anti-TSG-6 antibody.
N-terminal sequencing
Automated Edman degradation was performed in a PPSQ-31B protein sequencer (Shimadzu Biotech) with in-line phenylthiohydantoin analysis in an LC-20AT HPLC system equipped with a CTO-20A column heater and an SPD20A UV detector (Shimadzu Biotech). SDS-PAGE–separated samples were transferred to polyvinylidene difluoride membranes (48) and applied to TFA-treated glass fiber membranes that had been precycled with Polybrene (Shimadzu Biotech). Data were recorded using Shimadzu PPSQ-31B software, and the sequence was determined by visual inspection of the UV 269 nm chromatograms.
Identification of cleavage sites by N-terminomics
IαI was titrated with increasing amounts of MMP-3, MMP-7, MMP-13, or ADAMTS-5 (0, 2, 10, and 20 nm) for 4 h at 37 °C in 50 mm phosphate, 100 mm NaCl, 1 mm MgCl2, 0.05% Brij-35, 10 mm CaCl2, pH 7.4. The reactions were quenched by adding EDTA to a final concentration of 10 mm. Subsequently, all N termini and lysine residues were acetylated using 10 mm sulfo-NHS-acetate. The samples were incubated for 60 min at 22 °C before the reaction was quenched with 150 mm Tris-HCl, pH 7.4. The sample was denatured and reduced in 6 m urea containing 5 mm DTT for 1 h and subsequently alkylated with 15 mm iodoacetamide for 1 h. The urea concentration was reduced by diluting the sample 8× with 50 mm NH4HCO3. The samples were treated either with trypsin alone or with endoproteinase Glu-C followed by trypsin. The proteases were added at a ratio of 1:20 (w/w), and the samples were incubated for 16 h at 37 °C. The digested samples were analyzed by LC-MS/MS.
Qualitative identification of degradation products in arthropathic patient samples
Synovial fluid from 10 OA patients, nine RA patients, and 14 SpA patients was collected at Aarhus University Hospital and the Musculoskeletal Tissue Bank of the University of Liverpool, UK. The collection of patient samples was approved by the Danish Data Protection Agency (2011-41-6863), the Regional Ethics Committee, Denmark (1-10-72-291-12), and the National Research Ethics Service, UK (15/NW/0661). The subjects' written consent was obtained according to the Declaration of Helsinki.
Proteins and hyaluronic acid were precipitated by adding 9 volumes of ice-cold 90% ethanol followed by incubation at −18 °C for 16 h. The pellet was isolated by centrifugation at 17,000 × g for 30 min. The C-terminal product resulting from the in vivo cleavage of HC2 was released from HA and/or CS by hydrolyzing the ester bond with 100 mm NaOH for 30 min on ice. Subsequently, undigested protein and HA were removed by precipitation with TCA and centrifugation at 17,000 × g for 30 min. The supernatant containing the potential C terminus of HC2 was analyzed by LC-MS/MS.
LC-MS/MS analysis
Mass spectrometry
All samples were desalted by micropurification using EmporeTM solid-phase extraction Disks of C18 octadecyl packed in 10-μl pipette tips (51). LC-MS/MS was performed using either an EASY-nLC 1000 system (Thermo Scientific) connected to a QExactive+ Hybrid Quadrupole-Orbitrap mass spectrometer (Thermo Scientific) through a Nanospray FlexTM ion source (Thermo Scientific) or an Eksigent nanoLC 415 system (Sciex) connected to a TripleTOF 6600 mass spectrometer (Sciex) equipped with a NanoSpray III source (Sciex). Peptides were dissolved in 0.1% formic acid, injected, trapped, and desalted in a ReproSil-Pur C18-AQ trap column (2 cm × 100-μm inner diameter) packed in-house with 3-μm resin (Dr. Maisch GmbH, Ammerbuch-Entringen, Germany). The peptides were eluted from the trap column and separated in a 15-cm analytical column (75-μm inner diameter) packed in-house in a pulled emitter with ReproSil-Pur C18-AQ 3-μm resin (Dr. Maisch GmbH). Peptides were eluted using a flow rate of 250 nl/min and a 50-min gradient from 5 to 35% phase B (0.1% formic acid, 90% acetonitrile or 0.1% formic acid, 90% acetonitrile, 5% DMSO). The collected MS files were converted to generic Mascot format (MGF) using the Sciex MS Data Converter beta 1.1 or RawConverter (35).
Analysis of MS data
The data were searched against IαI sequences using a local Mascot search engine and the following settings: MS error tolerance of 10 ppm, MS/MS error tolerance of 0.1 Da, semitryptic, and iodoacetamide as fixed modification. Oxidized Met and glycosylation (hexose, N-acetylhexoseamine, and N-acetylneuraminic acid) of Ser and Thr were selected as variable modifications. Additional settings for the identification of N termini using sulfo-NHS-acetate were fixed acetylation of Lys and variable acetylation of the peptide N terminus.
Author contributions
C. S., M. R., S. F., G. B.-G., K. Y., B. D., and J. J. E. conceptualization; C. S., E. C. P., I. B. T., and K. Y. formal analysis; C. S. and J. J. E. supervision; C. S., E. C. P., I. B. T., and J. J. E. investigation; C. S., I. B. T., and J. J. E. methodology; C. S. writing-original draft; E. C. P., M. R., S. F., G. B.-G., K. Y., and B. D. data curation; M. R., S. F., G. B.-G., K. Y., B. D., and J. J. E. resources; J. J. E. funding acquisition; J. J. E. project administration.
Acknowledgment
We are grateful to the Liverpool Musculoskeletal Bank for the synovium fluid of osteoarthritis patients.
This work was supported by the Independent Research Fund of Denmark and the Velux Foundations. The authors declare that they have no conflicts of interest with the contents of this article.
- IαI
- inter-α-inhibitor
- HC1
- heavy chain 1
- HC2
- heavy chain 2
- CS
- chondroitin 4-sulfate
- ECM
- extracellular matrix
- TSG-6
- tumor necrosis factor–stimulated gene-6 protein
- ADAMTS
- a disintegrin and metalloproteinase with thrombospondin type 1 motifs
- MMP
- matrix metalloproteinase
- HA
- hyaluronan
- RA
- rheumatoid arthritis
- OA
- osteoarthritis
- SpA
- spondyloarthropathy
- PGP
- protein–glycosaminoglycan–protein
- PαI
- pre-α-inhibitor.
References
- 1. Enghild J. J., Thøgersen I. B., Pizzo S. V., and Salvesen G. (1989) Analysis of inter-α-trypsin inhibitor and a novel trypsin inhibitor, pre-α-trypsin inhibitor, from human plasma. Polypeptide chain stoichiometry and assembly by glycan. J. Biol. Chem. 264, 15975–15981 [PubMed] [Google Scholar]
- 2. Enghild J. J., Salvesen G., Hefta S. A., Thøgersen I. B., Rutherfurd S., and Pizzo S. V. (1991) Chondroitin 4-sulfate covalently cross-links the chains of the human blood protein pre-α-inhibitor. J. Biol. Chem. 266, 747–751 [PubMed] [Google Scholar]
- 3. Enghild J. J., Salvesen G., Thøgersen I. B., Valnickova Z., Pizzo S. V., and Hefta S. A. (1993) Presence of the protein-glycosaminoglycan-protein covalent cross-link in the inter-α-inhibitor-related proteinase inhibitor heavy chain 2/bikunin. J. Biol. Chem. 268, 8711–8716 [PubMed] [Google Scholar]
- 4. Olsen E. H., Rahbek-Nielsen H., Thogersen I. B., Roepstorff P., and Enghild J. J. (1998) Posttranslational modifications of human inter-α-inhibitor: identification of glycans and disulfide bridges in heavy chains 1 and 2. Biochemistry 37, 408–416 10.1021/bi971137d [DOI] [PubMed] [Google Scholar]
- 5. Morelle W., Capon C., Balduyck M., Sautiere P., Kouach M., Michalski C., Fournet B., and Mizon J. (1994) Chondroitin sulphate covalently cross-links the three polypeptide chains of inter-α-trypsin inhibitor. Eur. J. Biochem. 221, 881–888 10.1111/j.1432-1033.1994.tb18803.x [DOI] [PubMed] [Google Scholar]
- 6. Thøgersen I. B., and Enghild J. J. (1995) Biosynthesis of bikunin proteins in the human carcinoma cell line HepG2 and in primary human hepatocytes. Polypeptide assembly by glycosaminoglycan. J. Biol. Chem. 270, 18700–18709 10.1074/jbc.270.31.18700 [DOI] [PubMed] [Google Scholar]
- 7. Ly M., Leach F. E. 3rd, Laremore T. N., Toida T., Amster I. J., and Linhardt R. J. (2011) The proteoglycan bikunin has a defined sequence. Nat. Chem. Biol. 7, 827–833 10.1038/nchembio.673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Martin J., Midgley A., Meran S., Woods E., Bowen T., Phillips A. O., and Steadman R. (2016) Tumor necrosis factor-stimulated gene 6 (TSG-6)-mediated interactions with the inter-α-inhibitor heavy chain 5 facilitate tumor growth factor β1 (TGFβ1)-dependent fibroblast to myofibroblast differentiation. J. Biol. Chem. 291, 13789–13801 10.1074/jbc.M115.670521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Steinbuch M. (1976) The inter-α-trypsin inhibitor. Methods Enzymol. 45, 760–772 10.1016/S0076-6879(76)45069-3 [DOI] [PubMed] [Google Scholar]
- 10. Zhuo L., and Kimata K. (2008) Structure and function of inter-α-trypsin inhibitor heavy chains. Connect Tissue Res. 49, 311–320 10.1080/03008200802325458 [DOI] [PubMed] [Google Scholar]
- 11. Zhao M., Yoneda M., Ohashi Y., Kurono S., Iwata H., Ohnuki Y., and Kimata K. (1995) Evidence for the covalent binding of SHAP, heavy chains of inter-α-trypsin inhibitor, to hyaluronan. J. Biol. Chem. 270, 26657–26663 10.1074/jbc.270.44.26657 [DOI] [PubMed] [Google Scholar]
- 12. Sanggaard K. W., Sonne-Schmidt C. S., Krogager T. P., Lorentzen K. A., Wisniewski H. G., Thøgersen I. B., and Enghild J. J. (2008) The transfer of heavy chains from bikunin proteins to hyaluronan requires both TSG-6 and HC2. J. Biol. Chem. 283, 18530–18537 10.1074/jbc.M800874200 [DOI] [PubMed] [Google Scholar]
- 13. Rugg M. S., Willis A. C., Mukhopadhyay D., Hascall V. C., Fries E., Fülöp C., Milner C. M., and Day A. J. (2005) Characterization of complexes formed between TSG-6 and inter-α-inhibitor that act as intermediates in the covalent transfer of heavy chains onto hyaluronan. J. Biol. Chem. 280, 25674–25686 10.1074/jbc.M501332200 [DOI] [PubMed] [Google Scholar]
- 14. Sanggaard K. W., Karring H., Valnickova Z., Thøgersen I. B., and Enghild J. J. (2005) The TSG-6 and IαI interaction promotes a transesterification cleaving the protein-glycosaminoglycan-protein (PGP) cross-link. J. Biol. Chem. 280, 11936–11942 10.1074/jbc.M409016200 [DOI] [PubMed] [Google Scholar]
- 15. Day A. J., and Milner C. M. (2018) TSG-6: a multifunctional protein with anti-inflammatory and tissue-protective properties. Matrix Biol. 78–79, 60–83 10.1016/j.matbio.2018.01.011 [DOI] [PubMed] [Google Scholar]
- 16. Sanggaard K. W., Sonne-Schmidt C. S., Krogager T. P., Kristensen T., Wisniewski H. G., Thøgersen I. B., and Enghild J. J. (2008) TSG-6 transfers proteins between glycosaminoglycans via a Ser28-mediated covalent catalytic mechanism. J. Biol. Chem. 283, 33919–33926 10.1074/jbc.M804240200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Milner C. M., Higman V. A., and Day A. J. (2006) TSG-6: a pluripotent inflammatory mediator? Biochem Soc. Trans. 34, 446–450 10.1042/BST0340446 [DOI] [PubMed] [Google Scholar]
- 18. de la Motte C. A., Hascall V. C., Drazba J., Bandyopadhyay S. K., and Strong S. A. (2003) Mononuclear leukocytes bind to specific hyaluronan structures on colon mucosal smooth muscle cells treated with polyinosinic acid:polycytidylic acid: inter-α-trypsin inhibitor is crucial to structure and function. Am. J. Pathol. 163, 121–133 10.1016/S0002-9440(10)63636-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Selbi W., de la Motte C. A., Hascall V. C., Day A. J., Bowen T., and Phillips A. O. (2006) Characterization of hyaluronan cable structure and function in renal proximal tubular epithelial cells. Kidney Int. 70, 1287–1295 10.1038/sj.ki.5001760 [DOI] [PubMed] [Google Scholar]
- 20. Zhuo L., Kanamori A., Kannagi R., Itano N., Wu J., Hamaguchi M., Ishiguro N., and Kimata K. (2006) SHAP potentiates the CD44-mediated leukocyte adhesion to the hyaluronan substratum. J. Biol. Chem. 281, 20303–20314 10.1074/jbc.M506703200 [DOI] [PubMed] [Google Scholar]
- 21. Fülöp C., Szántó S., Mukhopadhyay D., Bárdos T., Kamath R. V., Rugg M. S., Day A. J., Salustri A., Hascall V. C., Glant T. T., and Mikecz K. (2003) Impaired cumulus mucification and female sterility in tumor necrosis factor-induced protein-6 deficient mice. Development 130, 2253–2261 10.1242/dev.00422 [DOI] [PubMed] [Google Scholar]
- 22. Sato H., Kajikawa S., Kuroda S., Horisawa Y., Nakamura N., Kaga N., Kakinuma C., Kato K., Morishita H., Niwa H., and Miyazaki J. (2001) Impaired fertility in female mice lacking urinary trypsin inhibitor. Biochem. Biophys. Res. Commun. 281, 1154–1160 10.1006/bbrc.2001.4475 [DOI] [PubMed] [Google Scholar]
- 23. Zhuo L., Yoneda M., Zhao M., Yingsung W., Yoshida N., Kitagawa Y., Kawamura K., Suzuki T., and Kimata K. (2001) Defect in SHAP-hyaluronan complex causes severe female infertility. A study by inactivation of the bikunin gene in mice. J. Biol. Chem. 276, 7693–7696 10.1074/jbc.C000899200 [DOI] [PubMed] [Google Scholar]
- 24. Mukhopadhyay D., Hascall V. C., Day A. J., Salustri A., and Fülöp C. (2001) Two distinct populations of tumor necrosis factor-stimulated gene-6 protein in the extracellular matrix of expanded mouse cumulus cell-oocyte complexes. Arch. Biochem. Biophys. 394, 173–181 10.1006/abbi.2001.2552 [DOI] [PubMed] [Google Scholar]
- 25. Yingsung W., Zhuo L., Morgelin M., Yoneda M., Kida D., Watanabe H., Ishiguro N., Iwata H., and Kimata K. (2003) Molecular heterogeneity of the SHAP-hyaluronan complex. Isolation and characterization of the complex in synovial fluid from patients with rheumatoid arthritis. J. Biol. Chem. 278, 32710–32718 10.1074/jbc.M303658200 [DOI] [PubMed] [Google Scholar]
- 26. Troeberg L., and Nagase H. (2012) Proteases involved in cartilage matrix degradation in osteoarthritis. Biochim. Biophys. Acta 1824, 133–145 10.1016/j.bbapap.2011.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Murphy G., and Nagase H. (2008) Reappraising metalloproteinases in rheumatoid arthritis and osteoarthritis: destruction or repair? Nat. Clin. Pract. Rheumatol. 4, 128–135 10.1038/ncprheum0727 [DOI] [PubMed] [Google Scholar]
- 28. Yamamoto K., Murphy G., and Troeberg L. (2015) Extracellular regulation of metalloproteinases. Matrix Biol. 44–46, 255–263 10.1016/j.matbio.2015.02.007 [DOI] [PubMed] [Google Scholar]
- 29. Yoshihara Y., Plaas A., Osborn B., Margulis A., Nelson F., Stewart M., Rugg M. S., Milner C. M., Day A. J., Nemoto K., and Sandy J. D. (2008) Superficial zone chondrocytes in normal and osteoarthritic human articular cartilages synthesize novel truncated forms of inter-α-trypsin inhibitor heavy chains which are attached to a chondroitin sulfate proteoglycan other than bikunin. Osteoarthr. Cartil. 16, 1343–1355 10.1016/j.joca.2008.04.004 [DOI] [PubMed] [Google Scholar]
- 30. Plaas A., Sandy J. D., Liu H., Diaz M. A., Schenkman D., Magnus R. P., Bolam-Bretl C., Kopesky P. W., Wang V. M., and Galante J. O. (2011) Biochemical identification and immunolocalization of aggrecan, ADAMTS5 and inter-α-trypsin-inhibitor in equine degenerative suspensory ligament desmitis. J. Orthop. Res. 29, 900–906 10.1002/jor.21332 [DOI] [PubMed] [Google Scholar]
- 31. Scavenius C., Sanggaard K. W., Nikolajsen C. L., Bak S., Valnickova Z., Thøgersen I. B., Jensen O. N., Højrup P., and Enghild J. J. (2011) Human inter-α-inhibitor is a substrate for factor XIIIa and tissue transglutaminase. Biochim. Biophys. Acta 1814, 1624–1630 10.1016/j.bbapap.2011.08.017 [DOI] [PubMed] [Google Scholar]
- 32. Konttinen Y. T., Ainola M., Valleala H., Ma J., Ida H., Mandelin J., Kinne R. W., Santavirta S., Sorsa T., López-Otín C., and Takagi M. (1999) Analysis of 16 different matrix metalloproteinases (MMP-1 to MMP-20) in the synovial membrane: different profiles in trauma and rheumatoid arthritis. Ann. Rheum. Dis. 58, 691–697 10.1136/ard.58.11.691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kida D., Yoneda M., Miyaura S., Ishimaru T., Yoshida Y., Ito T., Ishiguro N., Iwata H., and Kimata K. (1999) The SHAP-HA complex in sera from patients with rheumatoid arthritis and osteoarthritis. J. Rheumatol. 26, 1230–1238 [PubMed] [Google Scholar]
- 34. Karakose M., Demircan K., Tutal E., Demirci T., Arslan M. S., Sahin M., Celik H. T., Kazanci F., Karakaya J., Cakal E., and Delibasi T. (2016) Clinical significance of ADAMTS1, ADAMTS5, ADAMTS9 aggrecanases and IL-17A, IL-23, IL-33 cytokines in polycystic ovary syndrome. J. Endocrinol. Invest. 39, 1269–1275 10.1007/s40618-016-0472-2 [DOI] [PubMed] [Google Scholar]
- 35. He L., Diedrich J., Chu Y. Y., and Yates J. R. 3rd. (2015) Extracting accurate precursor information for tandem mass spectra by RawConverter. Anal. Chem. 87, 11361–11367 10.1021/acs.analchem.5b02721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Murphy G., and Nagase H. (2008) Progress in matrix metalloproteinase research. Mol. Aspects Med. 29, 290–308 10.1016/j.mam.2008.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rawlings N. D., Waller M., Barrett A. J., and Bateman A. (2014) MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 42, D503–D509 10.1093/nar/gkt953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gendron C., Kashiwagi M., Lim N. H., Enghild J. J., Thøgersen I. B., Hughes C., Caterson B., and Nagase H. (2007) Proteolytic activities of human ADAMTS-5: comparative studies with ADAMTS-4. J. Biol. Chem. 282, 18294–18306 10.1074/jbc.M701523200 [DOI] [PubMed] [Google Scholar]
- 39. Baranova N. S., Inforzato A., Briggs D. C., Tilakaratna V., Enghild J. J., Thakar D., Milner C. M., Day A. J., and Richter R. P. (2014) Incorporation of pentraxin 3 into hyaluronan matrices is tightly regulated and promotes matrix cross-linking. J. Biol. Chem. 289, 30481–30498 10.1074/jbc.M114.568154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Richards J. S., Hernandez-Gonzalez I., Gonzalez-Robayna I., Teuling E., Lo Y., Boerboom D., Falender A. E., Doyle K. H., LeBaron R. G., Thompson V., and Sandy J. D. (2005) Regulated expression of ADAMTS family members in follicles and cumulus oocyte complexes: evidence for specific and redundant patterns during ovulation. Biol. Reprod. 72, 1241–1255 10.1095/biolreprod.104.038083 [DOI] [PubMed] [Google Scholar]
- 41. Petrey A. C., and de la Motte C. A. (2016) Thrombin cleavage of inter-α-inhibitor heavy chain 1 regulates leukocyte binding to an inflammatory hyaluronan matrix. J. Biol. Chem. 291, 24324–24334 10.1074/jbc.M116.755660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Okroj M., Holmquist E., Sjölander J., Corrales L., Saxne T., Wisniewski H. G., and Blom A. M. (2012) Heavy chains of inter α inhibitor (IαI) inhibit the human complement system at early stages of the cascade. J. Biol. Chem. 287, 20100–20110 10.1074/jbc.M111.324913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wisniewski H. G., Burgess W. H., Oppenheim J. D., and Vilcek J. (1994) TSG-6, an arthritis-associated hyaluronan binding protein, forms a stable complex with the serum protein inter-α-inhibitor. Biochemistry 33, 7423–7429 10.1021/bi00189a049 [DOI] [PubMed] [Google Scholar]
- 44. Chung L., Dinakarpandian D., Yoshida N., Lauer-Fields J. L., Fields G. B., Visse R., and Nagase H. (2004) Collagenase unwinds triple-helical collagen prior to peptide bond hydrolysis. EMBO J. 23, 3020–3030 10.1038/sj.emboj.7600318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yamamoto K., Higashi S., Kioi M., Tsunezumi J., Honke K., and Miyazaki K. (2006) Binding of active matrilysin to cell surface cholesterol sulfate is essential for its membrane-associated proteolytic action and induction of homotypic cell adhesion. J. Biol. Chem. 281, 9170–9180 10.1074/jbc.M510377200 [DOI] [PubMed] [Google Scholar]
- 46. Yu Z., Visse R., Inouye M., Nagase H., and Brodsky B. (2012) Defining requirements for collagenase cleavage in collagen type III using a bacterial collagen system. J. Biol. Chem. 287, 22988–22997 10.1074/jbc.M112.348979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bury A. F. (1981) Analysis of protein and peptide mixtures: evaluation of three sodium dodecyl sulphate-polyacrylamide gel electrophoresis buffer systems. J. Chromatogr. A 213, 491–500 10.1016/S0021-9673(00)80500-2 [DOI] [Google Scholar]
- 48. Matsudaira P. (1987) Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J. Biol. Chem. 262, 10035–10038 [PubMed] [Google Scholar]
- 49. Lee T. H., Wisniewski H. G., and Vilcek J. (1992) A novel secretory tumor necrosis factor-inducible protein (TSG-6) is a member of the family of hyaluronate binding proteins, closely related to the adhesion receptor CD44. J. Cell Biol. 116, 545–557 10.1083/jcb.116.2.545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rueden C. T., Schindelin J., Hiner M. C., DeZonia B. E., Walter A. E., Arena E. T., and Eliceiri K. W. (2017) ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 18, 529 10.1186/s12859-017-1934-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rappsilber J., Mann M., and Ishihama Y. (2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 10.1038/nprot.2007.261 [DOI] [PubMed] [Google Scholar]