Abstract
Receptor activator of nuclear factor κB ligand (RANKL) plays a crucial role in bone metabolism. RANKL gene misregulation has been implicated in several bone and cancer diseases. Here, we aimed to identify novel transcription regulators of RANKL expression. We discovered that transcription factors, sex-determining region Y (SRY) and c-Myb, regulate RANKL expression. We demonstrated that c-Myb increases and male-specific SRY decreases RANKL expression through direct binding to its 5’-proximal promoter. These results are corroborated by the gene expression in human bone samples. In osteoporotic men, expression of RANKL is 17-fold higher, which correlates with the drastically reduced expression (200-fold) of Sry, suggesting that in osteoporotic men, the upregulation of RANKL is caused by a decrease of Sry. In healthy men, the expression of RANKL is 20% higher than that in healthy women. Our data suggest that gender differences in RANKL expression and bone quality could be due to the sex-specific transcription factor SRY.
Subject terms: Osteoporosis, Genetics, Metabolic bone disease
Bone health: Insight into gender differences in osteoporosis
A male-specific gene offers clues to diagnosis and treatment of age-related osteoporosis. Osteoporosis was known to be linked to higher expression levels of RANKL, a gene that induces bone resorption, but the details were poorly understood. Nika Lovsin at the University of Ljubljana in Slovenia and co-workers searched for the genetic switches that control RANKL levels. They found that SRY, a gene on the male-specific Y chromosome, was a strong repressor of RANKL. In bone samples from osteoporotic men, expression levels of SRY levels were low and those of RANKL were high, suggesting that in men, when SRY fails to keep the bone-resorbing RANKL in check, osteoporosis results. SRY shows promise as an osteoporosis marker in men, or for development of treatment for both genders. Future research could address what triggers decreased SRY expression in men.
Introduction
Receptor activator of nuclear factor κB ligand (RANKL) is part of the RANKL/RANK/OPG system and is one of the most important molecules in bone remodeling. RANKL was identified as a key signaling molecule for the differentiation of bone macrophages into mature osteoclasts and for the activation of these cells, which leads to bone resorption. RANKL functions by binding to the RANK receptor on osteoclasts. RANKL belongs to the tumor necrosis factor (TNF) family of cytokines and was also named TNFSF111. RANKL is found as membrane-bound and soluble forms, the latter of which is produced by alternative splicing or by proteolytic cleavage of membrane-bound RANKL2–4. Osteoprotegerin (OPG) is a RANKL decoy receptor that can bind RANKL and prevent its function5,6. In addition to cells involved in bone formation (e.g., osteoblasts, their precursors, and osteocytes), there are other cells that express RANKL, such as T and B lymphocytes, mammary epithelial cells, vascular endothelial cells, synovial fibroblasts, chondrocytes, keratinocytes, malignant cells, cells in periodontal tissue, and other cell types7. Studies have shown that RANKL is involved in numerous physiological processes, as well as bone metabolism, such as in T cell functions and lymph node and mammary gland development5,8–10. In addition, RANKL is involved in pathological processes, such as the migration of tumor cells and the development of bone metastases in diseases, such as breast, lung and prostate cancer11–15.
While the biological functions of RANKL have been extensively studied, much less is known about the regulation of the RANKL gene at the transcription level. RANKL is regulated by transcription factors that bind to distal and proximal regulatory regions. The major expression regulator of RANKL is parathyroid hormone (PTH), which acts on the 76,000-bp-distant cAMP-response elements (CREs) through the protein kinase A–cAMP pathway, which requires CREB7,16–21. CREB is also required for 1,25(OH)2D3 and oncostatin M (OSM)-stimulated RANKL expression19. In addition to the binding of CREB, the same DNA region also contains a RUNX2 binding site. These binding sites and their importance were confirmed with electrophoretic mobility shift assays (EMSAs) and ChIP and transgenic mouse models16,19,22. The same distant enhancer also contains 1,25(OH)2D3 response elements–the VDREs19,22,23. VDREs were also found at −96, −87, −75, −25, and −20 kb upstream of the transcription start site23–25. Other known RANKL transcription regulators acting through distal regulatory regions are interleukin (IL)-6-type cytokines26,27, molecules of the Wnt/β-catenin signaling pathway28–35 and c-Fos in activated T cells36. Previous studies have indicated that Vitamin D, PTH, and other transcriptional regulators could also regulate RANKL gene expression through its proximal promoter37,38. A recent study revealed the involvement of C/EBPβ in the transcriptional regulation of RANKL through binding to a CCAAT-box in the region −56/−51 upstream of the transcription start site of the RANKL gene39.
Overall, very little is known about the factors that regulate RANKL expression at the 5’-proximal region of the gene. The aim of the present study was therefore to identify other transcription factors that bind to this 5’-proximal region and regulate the expression of RANKL and to evaluate their in vivo significance. Two novel transcription factors were identified here: SRY and c-Myb. SRY is encoded by the sex determining region Y (Sry) gene that initiates testis development. SRY is implicated in sex determination and belongs to the SRY-box (Sox) gene family. SRY is not expressed in females, and this protein is responsible for the development of the male phenotype in therian mammals40. c-Myb is a transcription factor that has previously been associated with bone formation and chondrogenesis41–43. We hypothesized that SRY-mediated RANKL gene expression might be the molecular mechanism underlying the sex differences in bone metabolism and bone diseases, such as osteoporosis.
Materials and methods
Patients
Bone tissue samples from 112 Slovenian patients were included in this study44. Fifty-eight patients (46 female, 12 male) were undergoing hip arthroplasty because of low energy femoral neck fracture (osteoporosis; OP), and 42 patients (29 female, 13 male) had primary hip osteoarthritis (osteoarthritis; OA). Twelve bone tissue samples were collected from male autopsy subjects (control; CTL). All subjects included in the study signed written informed consent prior to inclusion. The study was approved by the Republic of Slovenia National Medical Ethics Committee (consent numbers: KME 91-93/08/10, KME 106/03/06, KME 137/03/06, KME 45/10/16, KME 91/08/10, KME 45/10/16) and is summarized in Supplementary Table 1. Detailed descriptions of the patient samples have been reported previously44. Briefly, the exclusion criteria for all of these patients included a history of systemic or metabolic diseases that impact bone or mineral metabolism or taking any drugs known to impact bone or mineral metabolism in the 12 months prior to the surgery. Patients with any diseases or drug use that might influence bone metabolism were excluded from the study.
Cell experiments
Human bone osteosarcoma (HOS) cells (ATCC CRL-1543) were grown in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, Thermo Fisher Scientific, Massachusetts, USA) supplemented with 10% fetal bovine serum (FBS) (Gibco, Thermo Fisher Scientific, Massachusetts, USA), 1% L-glutamine (Gibco, Thermo Fisher Scientific, Massachusetts, USA) and 1% antibiotic/ antimycotic (Gibco, Thermo Fisher Scientific, Massachusetts, USA) at 37 °C under 5% CO2. Primary human osteoblasts (Promocell, Heidelberg, Germany) were grown in Osteoblast Growth Medium (Ready-to-use; Promocell, Heidelberg, Germany) at 37 °C under 5% CO2 and were subcultured according to the manufacturer’s instructions. Briefly, after thawing, the cells were grown until they reached 70 to 90% confluency, and then the cells were subcultured using DetachKit (Promocell, Heidelberg, Germany). The osteoblast growth medium was changed every 2 days to 3 days. All experiments were performed between the third and the fifth passages. The presence of osteoblast markers was confirmed by quantitative real-time polymerase chain reaction (q-PCR) (Supplementary Fig. 1).
Plasmid construction and site-specific mutagenesis
Ensembl ENST00000398795 (Human GRCh38.p2) was used as the reference sequence for the RANKL gene. Five different fragments of the human RANKL proximal promoter region were PCR amplified from the genomic DNA with a 5’-primer harboring a BglII site and a reverse primer complementary to the sequences near the transcription start site of the RANKL gene harboring a HindIII site. The primers used for PCR amplifications of the fragments are listed in Supplementary Table 1. The resulting PCR fragments were digested with BglII and HindIII and cloned into the pGL3 basic vector (Promega, Madison, USA). With PCR amplification, five fragments of the RANKL proximal promoter were obtained, with sizes of 898 bp (pGL3-F4), 762 bp (pGl3-F3), 538 bp (pGl3-F2), 346 bp (pGl3-F1), and 1769 bp (pGl3-F5). Mutants in the putative Sex-determining region Y (Sry) binding site (−732/−726 bp upstream of the RANKL transcription start, −731 AA > CC, pGl3-F4-SRY) and putative Myb proto-oncogene C (c-Myb) binding site (691/−674 bp upstream of the RANKL transcription start, −680 TT > GG, pGl3-F4-c-Myb) were prepared using a Quick-Change II Site-Directed Mutagenesis kit (Stratagene, San Diego, California, USA) and the primers listed in Supplementary Table 2.
The pCIneo-hc-Myb-HA expression vector was kindly donated by Prof. Odd Stokke Gabrielsen (Department of Biosciences, University of Oslo, Oslo, Norway). The pcDNA3-hSRY expression vector was constructed from the pcDNA3-FLAG-hSRY vector, which was kindly donated by Dr. Vincent Harley (Hudson Institute of Medical Research, Melbourne, Victoria, Australia), through the removal of the FLAG tag by PCR using the primers listed in Supplementary Table 2. The pEGFP-SRY plasmid was constructed by amplifying the Sry open reading frame from plasmid pcDNA3-FLAG-hSRY, kindly donated by Dr. Vincent Harley (Prince Henry’s Institute, Australia), using the primers listed in Supplementary Table 1 to add the restriction sites for ligation into the pEGFP-N1 plasmid (Clontech, CA, USA). All constructs were verified by DNA sequencing (GATC, Konstanz, Germany).
Western blotting
Human bone osteosarcoma cells were seeded onto 10-cm2 dishes and transfected with empty pcDNA3 expression vector, Sry-FLAG expression vector, or c-Myb-HA expression vector. The cells were lysed after 24 h and resolved by SDS-PAGE. The proteins were transferred to nitrocellulose membranes, subsequently blocked in 5% milk/TTBS, and incubated overnight with the primary antibodies against FLAG (clone M2; Sigma-Aldrich, St. Louis, Missouri, USA) or HA (H3663; Sigma-Aldrich, St. Louis, Missouri, USA). After incubation with the primary antibodies, the membranes were washed with TTBS and incubated with secondary antibodies bound to HRP for 1 h. Substrate was subsequently added, and the proteins were visualized using UVITEC Alliance. Antibodies against beta-actin (A2228, Sigma-Aldrich, St. Louis, Missouri, USA) were used for loading controls. Whole WB membranes are shown in Supplementary Fig. 2.
Overexpression of Sry-FLAG and c-Myb-HA in human primary osteoblasts
Male and female human primary osteoblasts were plated in 24-well tissue-culture plates (5 × 104 cells per well), incubated overnight, and transfected with 500 ng pcDNA3, Sry-FLAG, or c-Myb-HA using Lipofectamine 2000 (Thermo Fisher Scientific, Massachusetts, USA), according to the manufacturer’s instructions. RANKL expression was analyzed by q-PCR after 48 h, as described below. At least three biological replicates were analyzed for each gender.
Electroporation of human primary osteoblasts
Human primary osteoblasts were trypsinized with 0.05% Trypsin/EDTA, resuspended in fresh osteoblast growth medium (Promocell) and counted. Then, 1 × 106 cells per nucleofection were transferred to fresh tubes and centrifuged at 100 × g for 5 min. After removing the supernatant, the cell pellets were resuspended in 100 μL Mouse Primary Neurons Nucleofector Solution (Amaxa Biosystems) per transfection, with 2 μg pEGFP-SRY added to the cell suspensions, which were transferred to Amaxa electrode cuvettes and electroporated in Amaxa Nucleofector Device II using program T-030. Immediately thereafter, the cells were diluted in 500 μL Osteoblast Growth Medium supplemented with 20% FBS and seeded onto glass coverslips in 12-well plates. The medium was replaced 2 h after nucleofection with 750 μL osteoblast growth medium supplemented with 10% FBS.
RNA interference knock-down of gene expression
siRNAs for silencing of Sry (sc-38443), c-Myb (sc-29855) and a negative control, nc (sc-37007), were from Santa Cruz Biotechnology. Here, 30-pmol siRNA (final concentration, 300 nM) was nucleofected into 1 × 106 human primary osteoblasts, as described above. After 24 h and 48 h of nucleofection, the growth medium was changed. Then, at 72 h after nucleofection, the RNA was isolated using PeqGold RNA extraction kits (VWR), reverse transcribed, and subjected to quantitative PCR for the genes of interest. Three biological replicates were analyzed.
Luciferase reporter assay
Luciferase activity was measured with the dual-luciferase reporter assay system (Promega, Madison, WI, USA) according to the manufacturer’s protocol. Briefly, 3.5 × 104 cells were plated per well in 24-well tissue culture plates. After 24 h, cells were transfected with X-tremeGENE HP DNA Transfection Reagent (Roche Diagnostics, Mannheim, Germany), with 500 ng DNA per well. In the case of human primary osteoblasts, 2 µg of pDNA was nucleofected using the Amaxa System as described above for silencing experiments. A plasmid containing the Renilla luciferase gene was used as an internal control. Luciferase activity was measured at 24 h after transfection (BIO-TEK Synergy HT, Fisher Scientific, Pittsburgh, PA, USA). All experiments were carried out at least three times independently.
Electrophoretic mobility shift assays
Electrophoretic mobility shift assays were performed using LightShift Chemiluminescent EMSA kits (Thermo Fisher Scientific, Waltham, Massachusetts, USA) according to the manufacturer’s instructions. HOS and HeLa nuclear extracts were prepared using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific, Waltham, Massachusetts, USA) according to the manufacturer’s instructions. Double-stranded biotinylated oligonucleotides were prepared by hybridization in a C1000 thermal cycler by stepwise 0.5 °C decreases in the temperature, ranging from 95 °C to 35 °C, over 120 cycles, each 30 s long. The binding reactions between oligonucleotides and SRY/c-Myb were performed in 20 µL reactions containing 2 µL 10× binding buffer, 1 µL 100 mM MgCl2, 2 µL Poly(dI-dC), 2 µL 50% glycerol, 3 µL nuclear extract, and 20 fmol hybridized oligonucleotides (c-Myb) or 4 µmol hybridized oligonucleotides (SRY). Control reactions were performed without nuclear extracts. The specificity of the DNA-protein complexes was verified by dilution of the nuclear extracts and by competitive binding with nonbiotinylated DNA or mutated nonbiotinylated DNA. A primary rabbit polyclonal antibody against c-Myb (117635, Abcam, Cambridge, UK) was used to confirm the identity of the protein. For SRY, the mouse monoclonal anti-FLAG clone M2 was used (Sigma-Aldrich, St. Louis, Missouri, USA). The biotinylated oligonucleotides listed in Supplementary Table 1 were used in the EMSA experiments.
After the binding reactions, the samples were loaded onto electrophoretic gels and run for 1 h (100 V), then transferred to nylon membranes (30 min, 100 V), and cross-linked on a transilluminator. Biotin-labeled nuclear probes were detected with Chemiluminescent Nucleic Acid Detection Module kits (Pierce, Thermo Scientific, Wilmington, DE, USA) according to the manufacturer’s instructions.
Immunofluorescence
Cells were fixed with 3% paraformaldehyde (Sigma Aldrich, MO, USA) in phosphate-buffered saline (PBS) (Sigma Aldrich, MO, USA) at 37 °C with 0.2 μg/mL Hoechst 33342 (Sigma Aldrich, MO, USA) for 20 min. The cells were permeabilized with 0.1% Triton X-100 (Sigma Aldrich, MO, USA) in PBS for 5 min, stained with the primary antibodies anti-cMyb (ab117635, Abcam, Cambridge, UK) for 30 min, washed 3 times with PBS, stained with the secondary antibodies anti-rabbit CF568 (20102, Biotium, Fremont, CA, USA) for 30 min, washed 3 times with PBS, and mounted on glass slides with prolong anti-fade with DAPI (Thermo Fischer Scientific, MA, USA). Images were taken with a laser-scanning confocal microscope (710 META; Carl Zeiss Ltd, Germany) at 40× magnification using ZEN software (Carl Zeiss Ltd).
Human bone tissue immunohistochemistry and immunofluorescence
Bone samples were collected as described previously44. Briefly, trabecular bone from the intertrochanteric region of patients undergoing hemi- or total hip arthroplasty due to low-energy osteoporotic fracture or primary osteoarthritis or post-mortem donors with no musculoskeletal disorders were fixed in neutral buffered formalin for up to 24 h and decalcified with 4% EDTA for 2 to 6 weeks. Wax sections were prepared following a routine procedure at the Institute of Pathology, University of Ljubljana (Slovenia). Sections were dewaxed and rehydrated, and antigen retrieval was performed using 0.01 M sodium citrate buffer (pH 6) for 45 min in a microwave. Immunohistochemistry was performed using mouse-specific and rabbit-specific HRP/DAB detection IHC kits (ab64264; Abcam, Cambridge, UK), according to the manufacturer’s procedure, and TBS with Triton X-100 (Sigma-Aldrich, Steinheim, Germany) for washing. The samples were incubated overnight at 4 °C in a humidified chamber with primary rabbit polyclonal antibodies against c-Myb and SRY (ab117635 and ab135239, respectively; Abcam, Cambridge, UK) diluted 1:100 in TBS buffer. Control slides were prepared for each sample using TBS buffer only. The tissue sections were counterstained with hematoxylin solution (Thermo Shandon, Pittsburgh, USA) and examined under a microscope (BX50; Olympus, Hamburg, Germany). The intensity of the staining in osteoblasts, lining cells and osteocytes was compared between all of the samples by two independent blinded evaluators. Images were acquired using a digital camera (Olympus XC50) and the CellSens Dimension program 1.6.0 (Olympus, Hamburg, Germany).
Immunofluorescence for colocalization studies was performed using the following primary antibodies: SRY ab135239 (1:100) and c-Myb ab226470 (1:500) (both Abcam, Cambridge, UK); CD3 D7A6E (1:50) (Cell Signaling Technology, Danvers, Massachusetts, USA); and CD19 sc-19650 (1:50) and CD3 sc-20047 (1:50) (both Santa Cruz Biotechnology, Dallas, Texas, USA). Primary antibodies were applied overnight. The secondary antibodies of goat anti-rabbit Alexa Fluor 633 and goat anti-mouse Alexa Fluor 488 (1:500) (both Thermo Fisher Scientific Waltham, Massachusetts, USA) were applied for 1 h. The slides were mounted with ProLong Gold Antifade Mountant with DAPI (Thermo Fisher Scientific Waltham, Massachusetts, USA), and images were taken using a laser-scanning confocal microscope (710 META; Carl Zeiss Ltd, Germany) at 40× magnification using ZEN software (Carl Zeiss Ltd).
Quantitative PCR
The expression of RANKL was analyzed using qPCR with RNA samples obtained in gain-of-function and loss-of-function experiments. The RNA was extracted from cells, and the complementary DNAs (cDNAs) were synthesized, with gene expression analyses performed as described below. The expression of RANKL, Sry, and c-Myb was also analyzed using qPCR assays with the bone samples of Slovenian patients. RNA was extracted from the bone samples and the complementary DNA (cDNA) synthesized according to our previously described procedure44,45. Pairs of oligonucleotides for RANKL, RPLP0, GAPDH, and Sry were designed using the Primer Designing Tool (NCBI) (Supplementary Table 1), while for c-Myb, previously optimized oligonucleotides were used46. For qPCR, 5× Hot FirePol EvaGreen qPCR Mix Plus (Solis, BioDyne, Tartu, Estonia) was used, following the manufacturer recommendations, on a LightCycler 480 (Roche Diagnostics, Rotkreuz, Switzerland). All samples were quantified in triplicate. Dilution series of cDNA were prepared to create a relative standard curve, and absolute quantification of the data was performed using the second derivative maximum method (LightCycler 480, software version 1.5; Roche Diagnostics, Rotkreuz, Switzerland). All data were normalized to the internal housekeeping genes of ribosomal protein, large, P0 (RPLP0) or glyceraldehyde 3-phosphate dehydrogenase (GAPDH).
Statistical analysis and bioinformatics
The TF Search program was used to predict transcription factor binding to specific DNA regions. Matrices for vertebrates and threshold scores of more than 85 were used. The results for the luciferase activities were first transformed by calculating their z-scores and then normalized from 0 to 1 using unity scaling. The normalized results were then compared using ANOVA. Gene expression results were logarithmically transformed where needed to obtain normal distributions. The results were compared using Student’s t-tests or ANOVA.
Results
The region −662/−798 bp in the RANKL proximal promoter appears to contain binding sites for the transcription factors
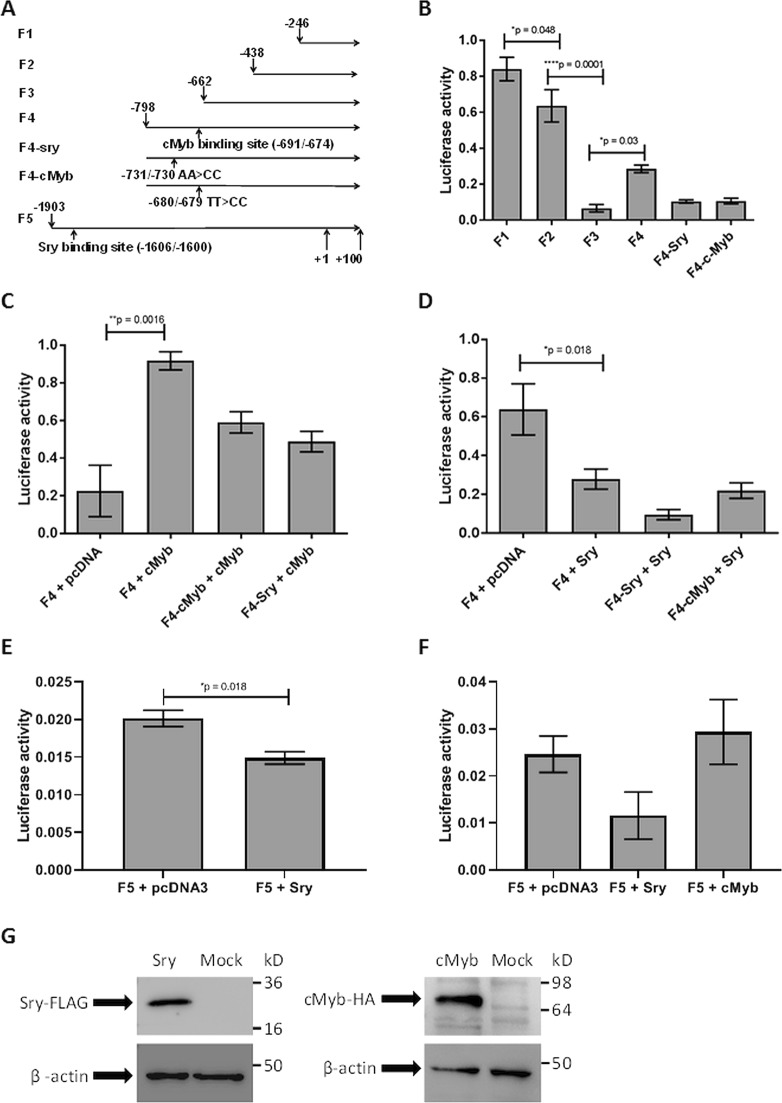
In this study, we aimed to characterize the proximal promoter region of the RANKL gene. First, to analyze the contributions of particular regions of the 5’-proximal promoter of the RANKL gene to its activation, various lengths of the proximal promoter region were cloned in front of the luciferase reporter gene (pGl3-Basic), and luciferase activity was measured in the HOS cells (Fig. 1a). These data showed that the F2 region (−438 bp to +100 bp) has 20% lower luciferase activity (p = 0.048) compared to that of the F1 region (−246 bp to +100 bp), and the F3 region (−662 bp to +100 bp) has 57% lower luciferase activity (p = 0.0001) compared to that of the F2 region. This result suggests that the region from −662 to −246 contains binding sites for transcriptional repressors. Surprisingly, the luciferase activity of the F4 region (−798 bp to +100 bp) was increased by 22% compared to that in the F3 region (p = 0.03), which suggested that the region from −798 to −662 of the RANKL gene contains binding sites for transcriptional activators.
Fig. 1. c-Myb increases and SRY decreases the luciferase activity of the RANKL promoter region.
a Different lengths of the RANKL proximal promoter region (F1, F2, F3, F4, F5) were cloned into the luciferase reporter vector pGL3-basic. Mutations were induced in the predicted SRY binding site (−731 AA > CC) and the predicted c-Myb binding site (−680 TT > GG) in the F4 vector using site-directed mutagenesis. b HOS (human bone osteosarcoma) cells were transfected with pGL3-F1/-F2/-F3/-F4/-F4-SRY/-F4-c-Myb and pRL normalization vector. Luciferase activities were measured 24 h after transfection. c Luciferase assays of HOS cells cotransfected with the pGl3-F4 RANKL promoter region and pcDNA3 (empty) or c-Myb-HA and cotransfected with the mutated pGl3-F4 RANKL promoter regions and c-Myb-HA. d Luciferase assays of HOS cells cotransfected with the pGl3-F4 RANKL promoter region and pcDNA3 (empty) or Sry-FLAG and cotransfected with the mutated pGl3-F4 RANKL promoter regions and Sry-FLAG. e Luciferase assays of HOS cells cotransfected with the pGl3-F5 RANKL promoter region and pcDNA3 (empty) or Sry-FLAG. f Luciferase assays of human primary osteoblasts nucleofected with pGL3-F5 RANKL promoter region and pcDNA3 (empty) or Sry-FLAG or c-Myb-HA. g Western blotting of human bone osteosarcoma cells after transfection with Sry-FLAG or c-Myb-HA expression vector and of empty human bone osteosarcoma cells. All of the data are presented as the mean luciferase activities after normalization ± SEM. Asterisks indicate significant differences between luciferase activities
Next, to identify potential transcription factors that bind to the RANKL proximal promoter, the F4 region of the RANKL promoter (Fig. 1a) was analyzed using the TF SEARCH program. SRY (possibly binding to position −732/−726 bp upstream of the RANKL transcription start; sequence AAACTAA; score 94.5) and c-Myb (possibly binding to position −691/−674 bp upstream of the RANKL transcription start; sequence TTTCCTGACTGTTGGGTG; score 85.2) were identified as probable candidates. To examine whether SRY or c-Myb putative binding sites contribute to RANKL promoter activity, the putative binding sites in the F4 region were mutated. At the putative SRY binding site, mutation −731/−730 AA > CC was created, and at the putative c-Myb binding site, mutation −680/−679 TT > CC was created (35–37). The results of the luciferase assays showed that the mutations introduced at the putative c-Myb (−691/−674) or SRY (−732/−726) binding sites abolished the effects of region −798/−622, as the promoter activity decreased to the activity of the F3 region (Fig. 1b), indicating that both SRY and c-Myb binding sites are important regulatory regions in the RANKL proximal promoter.
SRY represses the activity of the RANKL proximal promoter
Next, we examined whether SRY affects the activity of the RANKL proximal promoter by measuring the luciferase activity of the F4 region of the RANKL promoter in the presence of the overexpressed SRY protein (pcDNA3-hSry) in HOS cells. In the presence of SRY, the activity of the F4 region of the RANKL proximal promoter decreased compared to that of the empty vector. However, mutation of the putative SRY binding site (−731/−730) did not alleviate the inhibitory action of SRY (Fig. 1d). In contrast, the luciferase activity of the mutated promoter region was lower than in the presence of the F4 RANKL promoter region, which suggested that another site is important for SRY regulation of RANKL promoter activity. We identified another possible SRY binding site, 5-[AT]AACAA[AT]-3, at position −1606/−1600 in the proximal RANKL promoter region.
Next, we examined whether SRY affects the activity of a longer RANKL proximal promoter region by measuring the luciferase activity of the longer proximal RANKL promoter region (F5 region, from −1669 to +100) in the presence of the overexpressed SRY protein (pcDNA3-hSry) in HOS cells. In the presence of SRY, the activity of the F5 region of the RANKL proximal promoter decreased by 30%, which suggested that this site is important for RANKL promoter regulation (Fig. 1e). To verify that SRY affects the RANKL promoter in human primary cells, human POB were also employed in the luciferase assay, resulting in the same effect (Fig. 1f), which suggests that SRY decreases RANKL promoter activity, regardless of the host cells, implying the role of SRY in the regulation of RANKL expression. Indeed, the importance of the −1606/−1600 SRY binding site was further confirmed by EMSA (Fig. 2) and silencing experiments (Fig. 3). Western blotting was used to confirm the presence of overexpressed SRY in HOS cells (Fig. 1g).
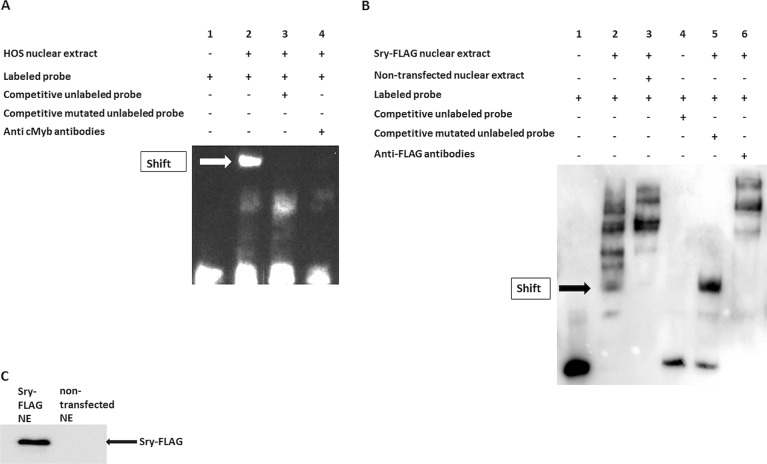
Fig. 2. c-Myb binds to the site −691/−674 bp upstream of the RANKL transcription start, and SRY binds to the site −1606/−1600 bp upstream of the RANKL transcription start.
a Electrophoretic mobility shift assays using human bone osteosarcoma cell nuclear extracts and biotinylated oligonucleotide probes containing site −691/−674 bp upstream of the RANKL transcription start. Lane 1, negative control without nuclear extract; lane 2, nuclear extract added; lane 3, competitive unlabeled probes added; lane 4, anti-c-Myb antibodies added; lane 5, mutated competitive unlabeled probes added. b Electrophoretic mobility shift assays using human bone osteosarcoma cell nuclear extracts and biotinylated oligonucleotide probes containing site −1606/−1600 bp upstream of the RANKL transcription start. Lane 1, negative control without nuclear extract; lane 2, Sry-FLAG transfected nuclear extract added; lane 3, nontransfected nuclear extract added; lane 4, competitive unlabeled probes added; lane 5, mutated competitive unlabeled probes added; lane 6, anti-FLAG antibodies added. c Western blotting confirmation of Sry-FLAG transfection in the nuclear extract (NE) used for the electrophoretic mobility shift assays in b
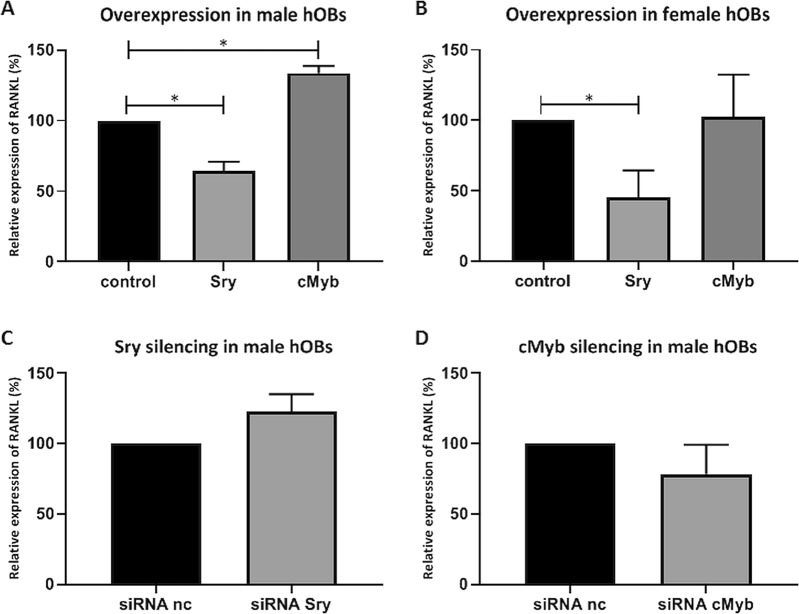
Fig. 3. c-Myb and SRY affect the expression of the RANKL gene in human primary osteoblasts.
a, b Female (a) and male (b) human primary osteoblasts were transfected with empty expression vector (ctl) and Sry-FLAG or c-Myb-HA. RANKL gene expression was measured at 48 h after treatment using q-PCR. c, d Male human primary osteoblasts were nucleofected with negative control siRNA and siRNAs against c-Myb (c) or Sry (d). RANKL gene expression was measured at 72 h after treatment using q-PCR. All of the data are presented as the mean relative RANKL expression after normalization ± SEM. Asterisks indicate significant differences between two samples. Data are representative of at least three independent experiments
c-Myb increases the activity of the RANKL proximal promoter
After identification of the putative c-Myb binding site in the RANKL proximal promoter region, we investigated whether c-Myb impacts the activity of the RANKL promoter. We cotransfected HOS cells with the c-Myb expressing vector (pcDNA3-c-Myb) and the pGl3-F4 reporter vector. In the presence of c-Myb, the activity of the F4 RANKL promoter increased 4-fold compared to that of the empty vector. Mutation in the putative c-Myb binding site (−680/−679 TT > CC) partially decreased the RANKL promoter activity, which indicated that c-Myb activates the RANKL promoter (Fig. 1c, f). Western blotting was used to confirm the presence of overexpressed c-Myb in HOS cells (Fig. 1g).
c-Myb and SRY bind to the RANKL proximal promoter region
Electrophoretic mobility shift assays were used to examine whether the transcription factor c-Myb binds to the site −691/−674 bp upstream of the RANKL transcription start. Figure 2a shows the appearance of the shifted biotinylated oligonucleotides in the presence of nuclear extracts from HOS cells compared to the control without nuclear extract (Fig. 2a, lanes 2, 1), which indicated that a protein from the nuclear extract binds to the oligonucleotides. The specificity of the binding was confirmed using competitive oligonucleotides without biotin in 200-fold molar excess (Fig. 2a, lane 3). The identification of c-Myb as the binding protein was confirmed using an antibody against c-Myb that prevented the interaction between protein and DNA (Fig. 2a, lane 4). The effects of binding site mutation were tested with the mutated competitive oligonucleotides in 200-fold excess. The shifted biotinylated oligonucleotides indicated that the mutated unlabeled competitive oligonucleotides did not bind the c-Myb protein, which confirmed that the site −691/−674 bp upstream of the RANKL transcription start is responsible for c-Myb binding (Fig. 2a, lane 5).
Electrophoretic mobility shift assays were also used to determine whether the transcription factor SRY binds to the site −1606/−1600 bp upstream of the RANKL transcription start site. Figure 2b shows the appearance of the shifted biotinylated oligonucleotides in the presence of the nuclear extracts from Sry-FLAG-transfected HOS cells compared to the control without the nuclear extract (Fig. 2b, lanes 2, 1), which indicated that a protein from the transfected nuclear extract binds to the oligonucleotides. The specificity of the binding was confirmed using nontransfected nuclear extracts (Fig. 2b, lane 3) and using competitive oligonucleotides without biotin in 200-fold molar excess (Fig. 2b, lane 4). The effects of binding site mutations were tested with mutated competitive oligonucleotides in 200-fold excess. The shifted biotinylated oligonucleotides indicated that the mutated unlabeled competitive oligonucleotides did not bind the Sry-FLAG protein, which confirmed that the site −1606/−1600 bp upstream of the RANKL transcription start is responsible for the SRY binding (Fig. 2b, lane 5). The identification of Sry-FLAG as the binding protein was confirmed using an antibody against FLAG, which caused a supershift (Fig. 2b, lane 6). Western blotting was used to confirm the presence of Sry-FLAG in the nuclear extract used for EMSA (Fig. 2c).
Our results demonstrate that both c-Myb and SRY directly bind to the RANKL proximal promoter region and could consequently directly impact RANKL gene expression.
Overexpression of Sry in human female osteoblasts lowers the expression of RANKL
As c-Myb increased and SRY decreased the activity of the RANKL promoter in our luciferase assays (Fig. 1) and as both of these transcription factors bind to the RANKL proximal promoter (EMSA; Fig. 2), we hypothesized that these transcription factors affect the expression of the RANKL gene in primary human osteoblasts. To test this hypothesis, Sry-FLAG and c-Myb-HA were transfected into male and female human primary osteoblasts, and at 48 h after transfection, RANKL expression was measured. As shown in Fig. 3a, overexpression of Sry decreased and overexpression of c-Myb increased expression of the RANKL gene in male human osteoblasts. In female human osteoblasts (Fig. 3b), overexpression of Sry decreased expression of the RANKL gene. These results indicated that SRY and c-Myb can regulate RANKL expression in human primary osteoblasts. To further confirm this hypothesis, RNA interference assays were performed using siRNAs against c-Myb or SRY in human primary osteoblasts. Figure 3c, d show the nonsignificant differences in expression of the RANKL gene upon silencing of c-Myb (Fig. 3c) and Sry (Fig. 3d). However, there was a trend of decreased expression of the RANKL gene upon silencing of c-Myb and increased expression of the RANKL gene upon silencing of Sry, which might suggest that SRY downregulates and c-Myb upregulates the expression of RANKL in human primary osteoblasts.
SRY and c-Myb localize in human primary osteoblasts
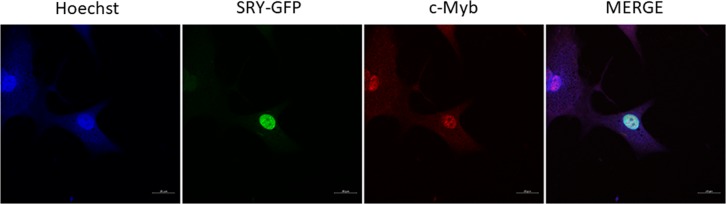
To examine whether both transcription factors localize to the nucleus of human primary osteoblasts, which is a prerequisite for their activity on the RANKL promoter, we examined the subcellular localization of SRY and cMyb in primary osteoblasts of male origin. We designed a fusion between SRY and green fluorescent protein (GFP) and overexpressed the fusion protein in male primary osteoblasts. The SRY-GFP localized to the nucleus and did not show any other localization phenotype (Fig. 4, Supplementary Fig. 3). The localization of endogenous cMyb was also examined by staining with an antibody. Endogenous cMyb localized to the nucleus; however, the intensity was low. c-Myb is an oncogene, and therefore high levels would not be expected in primary cells. Our data suggest that both the transcription factors SRY and c-Myb localize to the nucleus in human primary osteoblasts, which are prerequisites the activation/inhibition of the RANKL promoter.
Fig. 4. Localization of SRY and cMyb in primary osteoblast cells of male origin.
Localization of SRY-GFP fusion protein in male primary osteoblasts. Color legend for merged image: blue, Hoechst 33342 nuclear stain; green, SRY-GFP fusion protein; red, antibody targeting endogenous cMyb. POB primary osteoblasts; GFP green fluorescent protein. Scale bar: 10 μm
SRY and c-Myb are present in diverse human bone cells
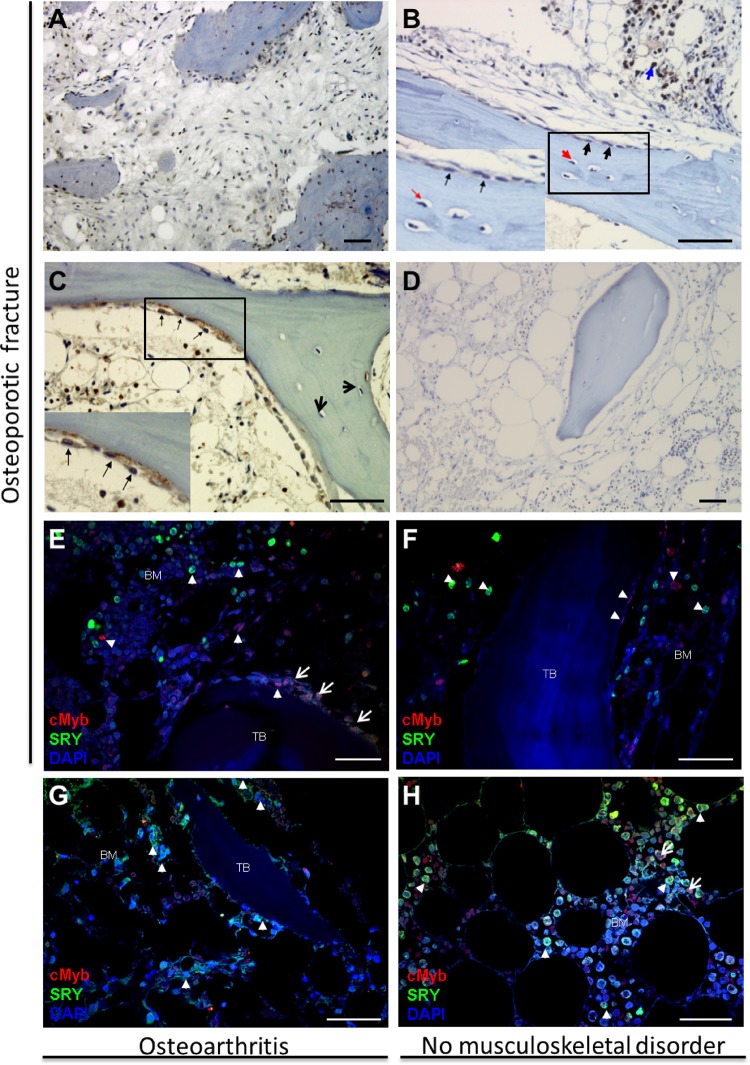
Since SRY and c-Myb impacted RANKL promoter activity and gene expression in human primary osteoblasts and HOS cells, we investigated whether these proteins are present in physiologically relevant human bone tissues where they could actually influence the bone phenotype. To this end, we studied the localization of c-Myb and SRY in human bone tissues after an osteoporotic fracture using immunohistochemistry (Fig. 5). Intensive c-Myb staining was observed in the nuclei of lining cells and in individual bone marrow cells, but very little staining was observed in osteocytes (Fig. 5a). In dormant bone, intensive SRY staining was observed in various cells of the bone marrow (Fig. 5b, blue arrow) and with less staining of the lining cells (Fig. 5b, black arrows), while osteocytes were mostly negative for SRY (Fig. 5b, red arrow), similar to the results observed for c-Myb. During the process of active bone remodeling, SRY was observed in bone marrow cells, in active osteoblasts (Fig. 5c, black arrows), and in the osteocytes located just beneath the trabecular bone surface (Fig. 5c, red arrows), which suggested a role for SRY in bone remodeling involving RANKL. No positive SRY staining was observed in female patients (Fig. 5d), which confirmed the specificity of the SRY antibody. Double staining of SRY and c-Myb using human tissue samples from male patients with osteoporotic fractures (Fig. 5e, f) and osteoarthritis (Fig. 5g) or no such disorder (i.e., controls, Fig. 5h) showed double-positive and single-positive cells. The SRY and c-Myb double-positive cells were observed in osteoblast-like cells (Fig. 5e, arrows) on the surface of trabecular bone, while single-positive SRY or c-Myb cells (Fig. 5e, f, arrowheads) were present in bone marrow, and c-Myb single-positive cells were also present on the surface of trabecular bone (Fig. 5e).
Fig. 5. Immunolocalization of cMyb and SRY in human bone tissue.
a–c Bone tissue of a male patient with osteoporotic fracture. a cMyb staining is observed in the nuclei of lining cells, individual bone marrow cells, and osteocytes. b In inactive bone, SRY staining is observed in various cells of the bone marrow (blue arrow) and in the lining cells (black arrows), while osteocytes are negative for SRY (red arrow), similar to what was observed with cMyb. c In bone with active processes of bone remodeling, in addition to the cells in bone marrow, SRY was observed in active osteoblasts (closed arrows) and osteocytes (open arrows) just beneath the trabecular bone surface, which suggests its role in bone remodeling via RANKL. d No positive SRY staining was observed in female patients with osteoporotic fracture. e, f Bone tissue of a male patient with osteoporotic fractures showing costaining of cMyb and SRY in osteoblast-like cells (arrows) on the surface of trabecular bone (e); single-positive SRY or cMyb cells (arrowheads, e, f) are present in bone marrow (BM); and cMyb single-positive cells are also present on the surface of trabecular bone (TB) (e, f). g, h Costaining for SRY and cMyb in bone tissue of a male patient with osteoarthritis (g) and in a male donor with no musculoskeletal disease (h) show similar patterns of localization; i.e., SRY single-positive cells (arrowheads) and SRY and cMyb double-positive cells (arrows) in bone marrow (BM). Immunohistochemistry (a–d) and double immunofluorescence (e–h) of human bone tissue. N = 3 donors per group (fracture, osteoarthritis and control group). Scale bars: 50 µm
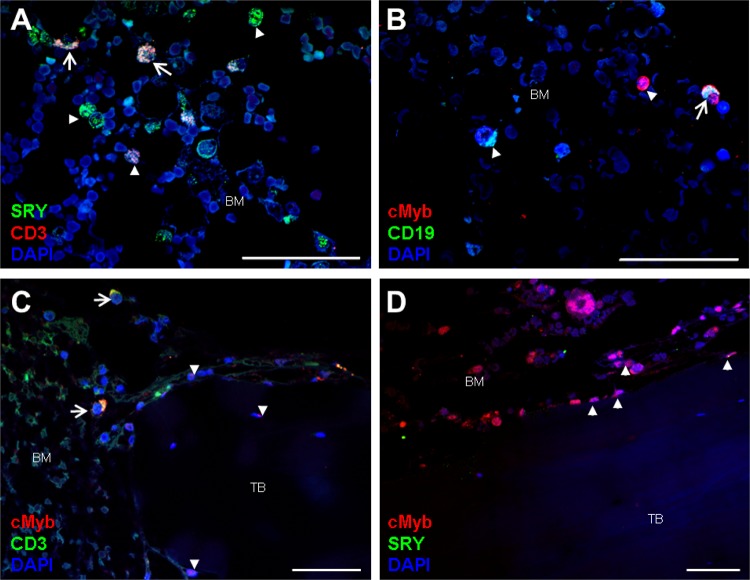
Immunolocalization studies showed both SRY and c-Myb single-positive cells and the colocalization of these proteins with the T-lymphocyte marker CD3 in bone marrow cells (Fig. 6a, c, Supplementary Fig. 4A–D). Moreover, c-Myb colocalization with the B-lymphocyte marker CD19 was shown (Fig. 6b, Supplementary Fig. 4E–H). The results of SRY and CD19 colocalization, which required different CD19 antibodies, showed nonspecific staining of CD19 (data not shown). Bone tissue from a female patient with osteoporotic fracture showed negative staining for SRY and similar patterns of c-Myb staining as for the male osteoporotic fracture tissue (Fig. 6d). The gene expression of these markers in human bone tissue is shown in Fig. 7.
Fig. 6. Colocalization of SRY and cMyb with T-lymphocyte and B-lymphocyte markers in human bone tissue.
a Bone tissue of a male patient with osteoporotic fracture showing costaining of SRY with the T-lymphocyte marker CD3 (arrow) in bone marrow (BM) cells. Double-positive cells (arrows) and single-positive cells for SRY or CD3 (arrowhead) are observed within bone marrow cells. b Bone tissue of a male patient with osteoporotic fracture showing costaining of cMyb with the B-lymphocyte marker CD19. Double-positive cells (arrow) and single-positive cells for cMyb or CD19 (arrowhead) are observed within bone marrow (BM) cells. c Bone tissue of a male patient with osteoporotic fracture showing costaining of cMyb with the T-lymphocyte marker CD3 (arrow) in bone marrow (BM) cells. Double-positive cells (arrows) are observed within bone marrow cells. Single-positive cells for cMyb (arrowhead) are observed within bone marrow cells, on the trabecular bone (TB) surface, and in a few osteocytes. d Bone tissue of female patients with osteoporotic fracture showing negative staining for SRY and similar patterns of cMyb staining as in the male osteoporotic fracture tissue. Scale bars: 50 µm
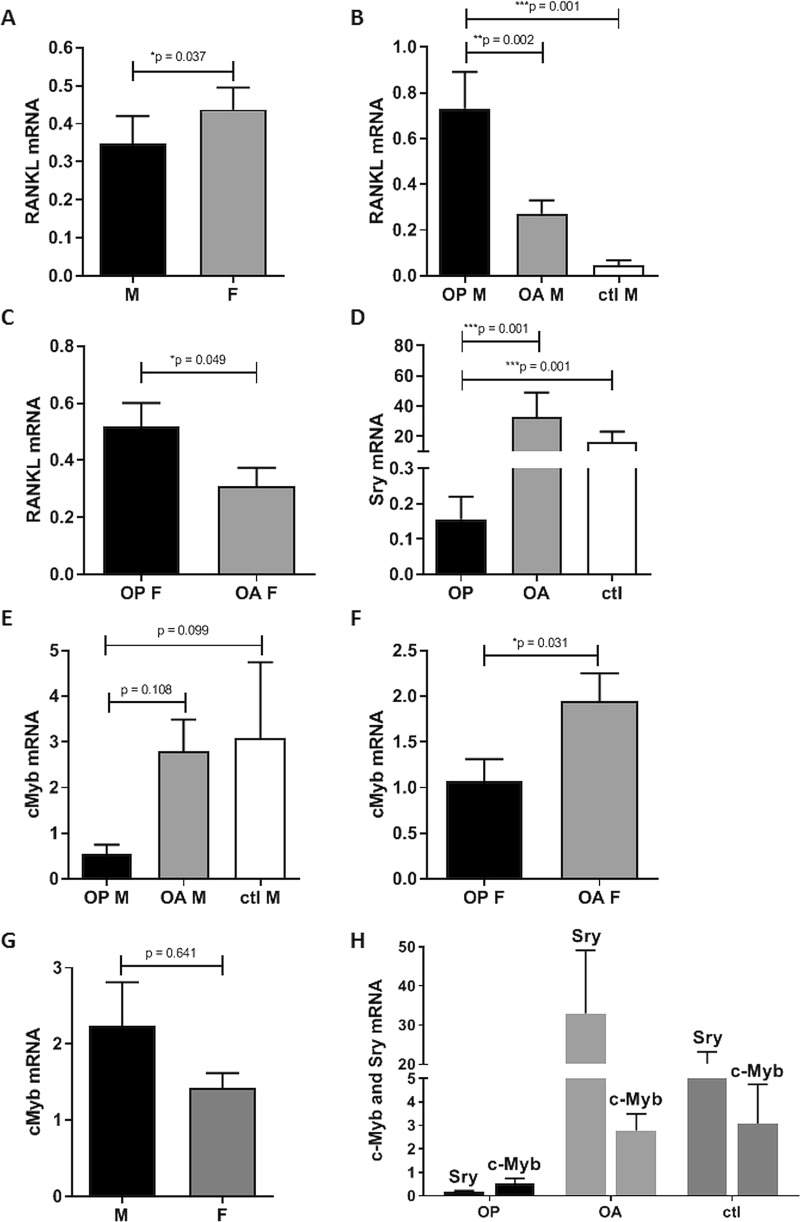
Fig. 7. Expression of RANKL, c-Myb, and Sry in human bone tissues is dependent on gender and health state.
qPCR was used to measure gene expression in human bone samples from male osteoporotic (OP, n = 12), osteoarthritic (OA, n = 13) and healthy (ctl. n = 12) patients and from female osteoporotic (OP, n = 46) and osteoarthritic (OA, n = 29) patients. Mean mRNA levels were normalized to RPLP0. Asterisk, significant differences between groups. a Comparison of RANKL gene expression in bone samples from male and female patients. b Comparison of RANKL gene expression in bone samples from OP, OA, and ctl male patients. c Comparison of RANKL gene expression in bone samples from OP and OA female patients. d Comparison of Sry gene expression in bone samples from OP, OA, and ctl male patients. e Comparison of c-Myb gene expression in bone samples from OP, OA, and ctl male patients. f Comparison of c-Myb gene expression in bone samples from OP and OA female patients. g Comparison of c-Myb gene expression in bone samples from male and female patients. h Comparison of Sry: c-Myb expression ratios in bone samples from OP, OA, and ctl male patients
Osteoporotic males show significantly decreased expression of Sry and highly increased expression of RANKL
As male-specific transcription factor SRY decreased the activity of the RANKL promoter in vitro, we hypothesized that SRY downregulates RANKL expression in vivo and that lower RANKL expression will be observed in males. To quantify the expression of RANKL, Sry, and c-Myb in human bone samples, real-time PCR was used (Fig. 7). Indeed, the expression of RANKL was 20% lower in males than in females, which supported our hypothesis (p = 0.037; Fig. 7a). When comparing RANKL expression between osteoporotic, osteoarthritic, and healthy individuals, RANKL was significantly increased in both male and female osteoporotic patients. Indeed, in male osteoporotic patients, RANKL expression was 17-fold higher than that in healthy controls (p = 0.001; Fig. 7b) and 3-fold higher than that in osteoarthritic patients (p = 0.002; Fig. 7b). In female osteoporotic patients, RANKL expression was almost twice that in osteoarthritic patients (p = 0.049; Fig. 7c). Interestingly, the expression of RANKL in male osteoporotic patients was 40% higher than that in female osteoporotic patients. These data indicate that the expression of RANKL is upregulated in male osteoporotic patients and that the expression of RANKL increases more in male than female osteoporotic patients, which associates the expression of RANKL with osteoporosis.
As our in vitro data suggested that male-specific transcription factor SRY represses the activity of the RANKL promoter, the expression of Sry was examined in osteoporotic patients and compared with healthy and osteoarthritic individuals. Sry expression was 210-fold lower in osteoporotic men compared to osteoarthritic men (p = 0.001) and 105-fold lower compared to healthy men (p = 0.001; Fig. 7d). These data demonstrate that Sry expression is significantly downregulated in osteoporotic patients. Together with the in vitro data, our results suggest that downregulation of Sry causes upregulation of RANKL and may consequently cause the development of osteoporosis in male patients. Moreover, our data suggest that SRY is one of the regulators of RANKL expression in males and might protect men from osteoporosis.
Given that our in vitro data indicate that RANKL promoter activity is controlled by the transcription factor c-Myb, we next examined the expression of c-Myb in osteoporotic and osteoarthritic patients and healthy individuals. The expression of c-Myb appeared to be lower in male osteoporotic patients compared to that in male osteoarthritic patients and healthy males; however, these data were not significant (Fig. 7e). The expression of c-Myb was half in osteoporotic than osteoarthritic females (p = 0.031; Fig. 7f). However, no significant differences in c-Myb expression were observed between male and female bone samples (p = 0.641; Fig. 7g). These data demonstrate that c-Myb is slightly downregulated in osteoporotic patients and that both female and male patients express similar levels of c-Myb, which indicates that the expression of c-Myb is not gender dependent.
Figure 7h shows the comparison of expression of Sry and c-Myb in male patients. Again, a drastic downregulation of Sry was observed in osteoporotic males compared to osteoarthritic and healthy individuals. Intriguingly, in osteoporotic males, the expression of c-Myb also appeared lower than that in osteoarthritic patients and healthy individuals; however, these data were not significant. In healthy and osteoarthritic male subjects, the expression of Sry was 5-fold and 12-fold higher, respectively, than the expression of c-Myb. On the other hand, in osteoporotic patients, the expression of c-Myb was 3-fold higher than the expression of Sry. Altogether, these data reveal that in osteoporotic male patients, the expression of Sry was downregulated.
Discussion
RANKL is a protein that is involved in bone metabolism and modulates the activation and differentiation of osteoclasts. Its activation leads to increased bone resorption. In bone diseases, such as osteoporosis, RANKL expression is highly increased; therefore, RANKL represents a great target for osteoporosis treatment. Indeed, a monoclonal antibody against RANKL (denosumab) is currently used for osteoporosis treatment and prevention of skeletal-related events in patients with bone metastases. Playing a central role in bone remodeling, RANKL represents a good drug target for the development of novel cheaper approaches for the prevention of fragility fractures and osteoporosis. Thus, deciphering the regulation of RANKL gene expression is necessary. The expression of RANKL is regulated by transcription factors that bind to its distal and proximal promoter regions. Here, we aimed to identify transcription factors that bind to the RANKL proximal promoter region and can regulate its expression. We confirmed that the −662/−798 bp region in the RANKL proximal promoter contains binding sites for transcription factors, c-Myb and SRY. c-Myb is one of the oncogenes previously shown to be involved in bone formation and chondrogenesis41–43. SRY is a sex-specific transcription factor, expressed only in men and responsible for sex determination. We demonstrated that c-Myb increased and SRY decreased the activity of the RANKL promoter (Fig. 1). Using EMSA, we confirmed that c-Myb directly binds to the −691/−674 bp site (Fig. 2a) and showed that SRY binds to the −1606/−1600 bp region (Fig. 2b) of the RANKL promoter. When SRY was overexpressed in female human primary osteoblasts, the expression of RANKL was significantly reduced, which indicates that SRY decreases RANKL expression in human primary osteoblasts (Fig. 3a). Gene expression in the human bone samples revealed that in women bone tissues RANKL expression is 20% higher compared to that in men bone tissues, which suggested that higher expression of RANKL accounts for lower bone mineral density in women. This finding is in agreement with observations that men have higher bone mineral density47–49 and suffer from osteoporosis less frequently than women50,51. Importantly, in our osteoporotic patients, the expression of RANKL was highly upregulated and dependent on gender. In osteoporotic men, the expression of RANKL was significantly increased (17-fold). We demonstrated that the expression levels of RANKL were highly inversely correlated with the expression levels of Sry, which implies that the gender differences in the RANKL expression levels might be due to the sex-specific transcription factor SRY. Moreover, the drastic downregulation of Sry in male osteoporotic patients together with the upregulation of RANKL suggests that lower Sry expression might cause upregulation of RANKL and cause the development of osteoporosis in males. The mechanism behind the downregulation of Sry in men, leading to the upregulation of RANKL and osteoporosis, remains unknown. This study is the first to identify the gender-specific transcription factor SRY as a regulator of RANKL expression by directly binding to the RANKL promoter region. Our results indicate that SRY could be one of the reasons for gender differences in bone mineral density and for gender differences in the pathogenesis of osteoporosis. Previous studies have shown differences in the microarchitecture between osteoporotic and osteoarthritic bone52 could now be explained by SRY. Strong correlations between the reduced levels of Sry and incidence of osteoporosis in male osteoporotic patients suggest that SRY can serve as a biomarker for male osteoporosis. Moreover, our results provide a novel target for the development of new therapeutic approaches for the prevention of bone loss in men and women. Namely, by the stimulation of SRY-mediated downregulation of RANKL expression or by application of SRY analogs, the expression of RANKL could be lowered, and consequently, bone resorption prevented.
One of the major gender differences that contributes to bone phenotype and is dependent on sex is hormones. The role of hormones in bone remodeling has been well established53. Indeed, the decline of estrogens in postmenopausal women is associated with a decrease in bone mineral density, which indicates that lower estrogen levels influence bone mineral density53. Estrogen protection of bone mass is mediated via estrogen receptor α53. However, this mechanism is not limited to females. Estrogen metabolism has also been demonstrated to influence bone density in males, which indicates that estrogen is a regulator of bone remodeling both in males and females (reviewed in54). On the other hand, androgen hormones are required for the maintenance of trabecular bone in males, and their role has also been implicated in female bone remodeling53. Notably, RANKL-mediated increases in bone resorption have already been associated with sex hormones55. Although bone mass, size and quality differ between genders, no gender-distinctive transcriptional regulation mechanism that affects bone remodeling has been described to date. Given the important contribution of hormones to bone remodeling in our male patients, changes in estrogen or androgen levels might have contributed to the onset of osteoporosis. However, this explanation is less probable as patients with abnormal levels of hormones were excluded from the study. Here, we unveil the first gender-related transcriptional mechanism that distinctly regulates bone remodeling in males.
The role of SRY in bone biology is plausible because the Sry gene is part of the SOX gene family with an HMG box DNA binding domain56,57 involved in bone development and turnover58–60. Mutations of Sry cause XY gonadal dysgenesis (Swyer syndrome), in which a genetically male fetus fails to develop testes, which consequently leads to the development of female genitalia with a defective reproductive system. Hormonal replacement therapy is necessary to assure a normal menstrual cycle and secondary sex characteristics and to reduce the risks of osteopenia, which suggests the importance of SRY in normal bone development and hormonal status61–63. The tissue widespread expression of Sry in adult men has also been observed, which indicates functions of SRY other than the development of testes64. Importantly, RANKL upregulation and c-Myb downregulation were also observed in osteoporotic women (Fig. 7c, f), which suggested that in females, another transcription factor might have a similar role as SRY in males. Further studies are required to determine the transcription factors that regulate RANKL expression in women. We speculate that other members of the SOX family might be involved in c-Myb–dependent regulation of RANKL in women. Altogether, this is the first study that demonstrates the importance of SRY in the regulation of RANKL expression in males.
Here, we show that c-Myb activates the RANKL promoter by direct binding and causes upregulation of RANKL expression in primary osteoblasts. Moreover, we demonstrate that c-Myb is expressed in human bone tissues by immunohistochemistry and Q-PCR, suggesting the importance of c-Myb in bone biology. Indeed, it has already been shown that c-Myb is involved in bone biology and, more specifically, in novel bone formation41–43 and chondrogenesis65. In addition to the upregulation of RANKL in osteoporosis, dysregulation of RANKL has also been associated with tumorigenesis66 and metastases67 in several cancers. The importance of RANKL in tumorigenesis has been underpinned by clinical trials where denosumab decreased the risk of metastasis development68–70. c-Myb has recently been associated with the Wnt/β-catenin pathway as a mediator of metastasis in breast cancer71, and has been shown to promote migration and invasion of mammary tumor cells through regulation of cathepsin D and matrix metalloproteinase 972. Our data demonstrate that c-Myb overexpression activates the RANKL promoter in vitro and might be the reason for the elevated levels of RANKL in bone cancer diseases.
In conclusion, we identified two novel transcription factors, SRY and c-Myb, that regulate the expression of RANKL in human primary osteoblasts and that might influence the expression of RANKL in vivo. Importantly, we unveiled a novel gender-specific transcriptional mechanism that contributes to the regulation of RANKL in a sex-specific manner and that might contribute to gender differences in bone phenotypes.
Supplementary information
Acknowledgements
We would like to thank the Slovenian Research Agency (ARRS) for funding our work through grant J3-7245 and P-0298 and through the postgraduate research funding of young researchers. M.N.L. was funded also through Interreg ITA-SLO project ARTE. We thank Klemen Mezgec and Ajda Ogrin for assistance with the experiments. We thank anonymous reviewers for their fruitful comments that helped to significantly improve our paper. We would like to thank Chris Berrie for scientific English editing of the paper. We would also like to thank Dr. Odd Stokke Gabrielsen and Dr. Vincent Harley for plasmids.
Author contributions
Cloning site, specific mutagenesis, plasmid preparation, sequencing, and luciferase assays: V.M., K.K., M.N.L.; immunohistochemistry and immunolocalization: J.Z., V.M., T.K.; EMSA: V.M., K.K., M.N.L.; sample collection and RNA extraction: J.Z., R.K., J.M.; real-time PCR: J.Z., K.K., V.M.; statistical analysis: J.Z., V.M., K.K., N.M.L.; study design and conceiving the research: V.M., M.N.L., J.M.; paper preparation: V.M., K.K., M.N.L., J.Z., J.M., R.K.; critical review: V.M., K.K., M.N.L., J.Z., J.M., R.K.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Vid Mlakar, Janja Marc, Nika Lovšin
Supplementary information
Supplementary information accompanies this paper at 10.1038/s12276-019-0294-3.
References
- 1.NCBI. TNFSF11 tumor necrosis factor superfamily member 11 [Homo sapiens (human)]; Gene ID: 8600 (2016).
- 2.Hikita A, et al. Negative regulation of osteoclastogenesis by ectodomain shedding of receptor activator of NF-kappaB ligand. J. Biol. Chem. 2006;281:36846–36855. doi: 10.1074/jbc.M606656200. [DOI] [PubMed] [Google Scholar]
- 3.Ikeda T, Kasai M, Utsuyama M, Hirokawa K. Determination of three isoforms of the receptor activator of nuclear factor-kappaB ligand and their differential expression in bone and thymus. Endocrinology. 2001;142:1419–1426. doi: 10.1210/endo.142.4.8070. [DOI] [PubMed] [Google Scholar]
- 4.Nagai M, Kyakumoto S, Sato N. Cancer cells responsible for humoral hypercalcemia express mRNA encoding a secreted form of ODF/TRANCE that induces osteoclast formation. Biochem. Biophys. Res Commun. 2000;269:532–536. doi: 10.1006/bbrc.2000.2314. [DOI] [PubMed] [Google Scholar]
- 5.Kong Y-Y, et al. OPGL is a key regulator of osteoclastogenesis, lymphocyte development and lymph-node organogenesis. Nature. 1999;397:315–323. doi: 10.1038/16852. [DOI] [PubMed] [Google Scholar]
- 6.Lacey DL, et al. Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell. 1998;93:165–176. doi: 10.1016/S0092-8674(00)81569-X. [DOI] [PubMed] [Google Scholar]
- 7.O’Brien CA. Control of RANKL gene expression. Bone. 2010;46:911–919. doi: 10.1016/j.bone.2009.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong BR, et al. TRANCE is a novel ligand of the tumor necrosis factor receptor family that activates c-Jun N-terminal kinase in T cells. J. Biol. Chem. 1997;272:25190–25194. doi: 10.1074/jbc.272.40.25190. [DOI] [PubMed] [Google Scholar]
- 9.Dougall WC, et al. RANK is essential for osteoclast and lymph node development. Genes Dev. 1999;13:2412–2424. doi: 10.1101/gad.13.18.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fata JE, et al. The osteoclast differentiation factor osteoprotegerin-ligand is essential for mammary gland development. Cell. 2000;103:41–50. doi: 10.1016/S0092-8674(00)00103-3. [DOI] [PubMed] [Google Scholar]
- 11.Zhang J, et al. Osteoprotegerin inhibits prostate cancer–induced osteoclastogenesis and prevents prostate tumor growth in the bone. J. Clin. Invest. 2001;107:1235–1244. doi: 10.1172/JCI11685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown JM, et al. Osteoprotegerin and rank ligand expression in prostate cancer. Urology. 2001;57:611–616. doi: 10.1016/S0090-4295(00)01122-5. [DOI] [PubMed] [Google Scholar]
- 13.Huang L, Cheng YY, Chow LT, Zheng MH, Kumta SM. Tumour cells produce receptor activator of NF-kappaB ligand (RANKL) in skeletal metastases. J. Clin. Pathol. 2002;55:877–878. doi: 10.1136/jcp.55.11.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farrugia AN, et al. Receptor activator of nuclear factor-kappaB ligand expression by human myeloma cells mediates osteoclast formation in vitro and correlates with bone destruction in vivo. Cancer Res. 2003;63:5438–5445. [PubMed] [Google Scholar]
- 15.Jones DH, et al. Regulation of cancer cell migration and bone metastasis by RANKL. Nature. 2006;440:692–696. doi: 10.1038/nature04524. [DOI] [PubMed] [Google Scholar]
- 16.Ducy P, Karsenty G. Two distinct osteoblast-specific cis-acting elements control expression of a mouse osteocalcin gene. Mol. Cell Biol. 1995;15:1858–1869. doi: 10.1128/MCB.15.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fan X, et al. Regulation of RANKL promoter activity is associated with histone remodeling in murine bone stromal cells. J. Cell Biochem. 2004;93:807–818. doi: 10.1002/jcb.20217. [DOI] [PubMed] [Google Scholar]
- 18.Kabe Y, et al. NF-Y is essential for the recruitment of RNA polymerase II and inducible transcription of several CCAAT box-containing genes. Mol. Cell Biol. 2005;25:512–522. doi: 10.1128/MCB.25.1.512-522.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu Q, Manolagas SC, O’Brien CA. Parathyroid hormone controls receptor activator of NF-kappaB ligand gene expression via a distant transcriptional enhancer. Mol. Cell Biol. 2006;26:6453–6468. doi: 10.1128/MCB.00356-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kondo H, Guo J, Bringhurst FR. Cyclic adenosine monophosphate/protein kinase A mediates parathyroid hormone/parathyroid hormone-related protein receptor regulation of osteoclastogenesis and expression of RANKL and osteoprotegerin mRNAs by marrow stromal cells. J. Bone Min. Res. 2002;17:1667–1679. doi: 10.1359/jbmr.2002.17.9.1667. [DOI] [PubMed] [Google Scholar]
- 21.Onal M, St John HC, Danielson AL, Pike JW. Deletion of the Distal Tnfsf11 RL-D2 Enhancer That Contributes to PTH-Mediated RANKL Expression in Osteoblast Lineage Cells Results in a High Bone Mass Phenotype in Mice. J. Bone Min. Res. 2016;31:416–429. doi: 10.1002/jbmr.2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galli C, et al. Targeted deletion of a distant transcriptional enhancer of the receptor activator of nuclear factor-kappaB ligand gene reduces bone remodeling and increases bone mass. Endocrinology. 2008;149:146–153. doi: 10.1210/en.2007-0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim S, Yamazaki M, Zella LA, Shevde NK, Pike JW. Activation of Receptor Activator of NF-κB Ligand Gene Expression by 1,25-Dihydroxyvitamin D(3) Is Mediated through Multiple Long-Range Enhancers. Mol. Cell Biol. 2006;26:6469–6486. doi: 10.1128/MCB.00353-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nerenz RD, Martowicz ML, Pike JW. An enhancer 20 kilobases upstream of the human receptor activator of nuclear factor-kappaB ligand gene mediates dominant activation by 1,25-dihydroxyvitamin D3. Mol. Endocrinol. 2008;22:1044–1056. doi: 10.1210/me.2007-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martowicz ML, Meyer MB, Pike JW. The mouse RANKL gene locus is defined by a broad pattern of histone H4 acetylation and regulated through distinct distal enhancers. J. Cell Biochem. 2011;112:2030–2045. doi: 10.1002/jcb.23123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Brien CA, Gubrij I, Lin SC, Saylors RL, Manolagas SC. STAT3 activation in stromal/osteoblastic cells is required for induction of the receptor activator of NF-kappaB ligand and stimulation of osteoclastogenesis by gp130-utilizing cytokines or interleukin-1 but not 1,25-dihydroxyvitamin D3 or parathyroid hormone. J. Biol. Chem. 1999;274:19301–19308. doi: 10.1074/jbc.274.27.19301. [DOI] [PubMed] [Google Scholar]
- 27.Jilka RL, et al. Increased osteoclast development after estrogen loss: mediation by interleukin-6. Science. 1992;257:88–91. doi: 10.1126/science.1621100. [DOI] [PubMed] [Google Scholar]
- 28.Holmen SL, et al. Essential role of beta-catenin in postnatal bone acquisition. J. Biol. Chem. 2005;280:21162–21168. doi: 10.1074/jbc.M501900200. [DOI] [PubMed] [Google Scholar]
- 29.Spencer GJ, Utting JC, Etheridge SL, Arnett TR, Genever PG. Wnt signalling in osteoblasts regulates expression of the receptor activator of NFkappaB ligand and inhibits osteoclastogenesis in vitro. J. Cell Sci. 2006;119:1283–1296. doi: 10.1242/jcs.02883. [DOI] [PubMed] [Google Scholar]
- 30.Fujita K, Janz S. Attenuation of WNT signaling by DKK-1 and -2 regulates BMP2-induced osteoblast differentiation and expression of OPG, RANKL and M-CSF. Mol. Cancer. 2007;6:71. doi: 10.1186/1476-4598-6-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang FS, et al. Knocking down dickkopf-1 alleviates estrogen deficiency induction of bone loss. A histomorphological study in ovariectomized rats. Bone. 2007;40:485–492. doi: 10.1016/j.bone.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 32.Aicher A, et al. The Wnt antagonist Dickkopf-1 mobilizes vasculogenic progenitor cells via activation of the bone marrow endosteal stem cell niche. Circ. Res. 2008;103:796–803. doi: 10.1161/CIRCRESAHA.107.172718. [DOI] [PubMed] [Google Scholar]
- 33.Kubota T, et al. Lrp6 hypomorphic mutation affects bone mass through bone resorption in mice and impairs interaction with Mesd. J. Bone Min. Res. 2008;23:1661–1671. doi: 10.1359/jbmr.080512. [DOI] [PubMed] [Google Scholar]
- 34.Qiang YW, et al. Myeloma-derived Dickkopf-1 disrupts Wnt-regulated osteoprotegerin and RANKL production by osteoblasts: a potential mechanism underlying osteolytic bone lesions in multiple myeloma. Blood. 2008;112:196–207. doi: 10.1182/blood-2008-01-132134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki A, et al. PTH/cAMP/PKA signaling facilitates canonical Wnt signaling via inactivation of glycogen synthase kinase-3beta in osteoblastic Saos-2 cells. J. Cell Biochem. 2008;104:304–317. doi: 10.1002/jcb.21626. [DOI] [PubMed] [Google Scholar]
- 36.Bishop KA, Coy HM, Nerenz RD, Meyer MB, Pike JW. Mouse Rankl expression is regulated in T cells by c-Fos through a cluster of distal regulatory enhancers designated the T cell control region. J. Biol. Chem. 2011;286:20880–20891. doi: 10.1074/jbc.M111.231548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park HJ, Baek K, Baek JH, Kim HR. The cooperation of CREB and NFAT is required for PTHrP-induced RANKL expression in mouse osteoblastic cells. J. Cell Physiol. 2015;230:667–679. doi: 10.1002/jcp.24790. [DOI] [PubMed] [Google Scholar]
- 38.Kitazawa S, Kajimoto K, Kondo T, Kitazawa R. Vitamin D3 supports osteoclastogenesis via functional vitamin D response element of human RANKL gene promoter. J. Cell Biochem. 2003;89:771–777. doi: 10.1002/jcb.10567. [DOI] [PubMed] [Google Scholar]
- 39.Ng PK, et al. CCAAT/enhancer binding protein beta is up-regulated in giant cell tumor of bone and regulates RANKL expression. J. Cell Biochem. 2010;110:438–446. doi: 10.1002/jcb.22556. [DOI] [PubMed] [Google Scholar]
- 40.Capel B. Vertebrate sex determination: evolutionary plasticity of a fundamental switch. Nat. Rev. Genet. 2017;18:675–689. doi: 10.1038/nrg.2017.60. [DOI] [PubMed] [Google Scholar]
- 41.Bhattarai G, Lee YH, Lee MH, Yi HK. Gene delivery of c-myb increases bone formation surrounding oral implants. J. Dent. Res. 2013;92:840–845. doi: 10.1177/0022034513497753. [DOI] [PubMed] [Google Scholar]
- 42.Ess KC, Witte DP, Bascomb CP, Aronow BJ. Diverse developing mouse lineages exhibit high-level c-Myb expression in immature cells and loss of expression upon differentiation. Oncogene. 1999;18:1103–1111. doi: 10.1038/sj.onc.1202387. [DOI] [PubMed] [Google Scholar]
- 43.Matalova E, et al. Expression and characterization of c-Myb in prenatal odontogenesis. Dev. Growth Differ. 2011;53:793–803. doi: 10.1111/j.1440-169X.2011.01287.x. [DOI] [PubMed] [Google Scholar]
- 44.Dragojevic J, et al. Triglyceride metabolism in bone tissue is associated with osteoblast and osteoclast differentiation: a gene expression study. J. Bone Min. Metab. 2013;31:512–519. doi: 10.1007/s00774-013-0445-x. [DOI] [PubMed] [Google Scholar]
- 45.Logar DB, et al. Expression of bone resorption genes in osteoarthritis and in osteoporosis. J. Bone Min. Metab. 2007;25:219–225. doi: 10.1007/s00774-007-0753-0. [DOI] [PubMed] [Google Scholar]
- 46.Nakata Y, et al. c-Myb contributes to G2/M cell cycle transition in human hematopoietic cells by direct regulation of cyclin B1 expression. Mol. Cell Biol. 2007;27:2048–2058. doi: 10.1128/MCB.01100-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lim S, et al. Body composition changes with age have gender-specific impacts on bone mineral density. Bone. 2004;35:792–798. doi: 10.1016/j.bone.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 48.Group, T. E. P. O. S. & O'Neill, T. The relationship between bone density and incident vertebral fracture in men and women. J. Bone Miner. Res.17, 2214–2221 (2002). [DOI] [PubMed]
- 49.Araneta MR, von Muhlen D, Barrett-Connor E. Sex differences in the association between adiponectin and BMD, bone loss, and fractures: the Rancho Bernardo study. J. Bone Min. Res. 2009;24:2016–2022. doi: 10.1359/jbmr.090519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnell O, Kanis JA. An estimate of the worldwide prevalence and disability associated with osteoporotic fractures. Osteoporos. Int. 2006;17:1726–1733. doi: 10.1007/s00198-006-0172-4. [DOI] [PubMed] [Google Scholar]
- 51.Wade SW, Strader C, Fitzpatrick LA, Anthony MS, O’Malley CD. Estimating prevalence of osteoporosis: examples from industrialized countries. Arch. Osteoporos. 2014;9:182. doi: 10.1007/s11657-014-0182-3. [DOI] [PubMed] [Google Scholar]
- 52.Zupan J, et al. Osteoarthritic versus osteoporotic bone and intra-skeletal variations in normal bone: evaluation with microCT and bone histomorphometry. J. Orthop. Res. 2013;31:1059–1066. doi: 10.1002/jor.22318. [DOI] [PubMed] [Google Scholar]
- 53.Manolagas SC, O’Brien CA, Almeida M. The role of estrogen and androgen receptors in bone health and disease. Nat. Rev. Endocrinol. 2013;9:699–712. doi: 10.1038/nrendo.2013.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karasik D, Ferrari SL. Contribution of gender-specific genetic factors to osteoporosis risk. Ann. Hum. Genet. 2008;72:696–714. doi: 10.1111/j.1469-1809.2008.00447.x. [DOI] [PubMed] [Google Scholar]
- 55.Kearns AE, Khosla S, Kostenuik PJ. Receptor activator of nuclear factor kappaB ligand and osteoprotegerin regulation of bone remodeling in health and disease. Endocr. Rev. 2008;29:155–192. doi: 10.1210/er.2007-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wallis MC, Waters PD, Graves JA. Sex determination in mammals–before and after the evolution of SRY. Cell Mol. Life Sci. 2008;65:3182–3195. doi: 10.1007/s00018-008-8109-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bowles J, Schepers G, Koopman P. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev. Biol. 2000;227:239–255. doi: 10.1006/dbio.2000.9883. [DOI] [PubMed] [Google Scholar]
- 58.Rivadeneira F, et al. Twenty bone-mineral-density loci identified by large-scale meta-analysis of genome-wide association studies. Nat. Genet. 2009;41:1199–1206. doi: 10.1038/ng.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Estrada K, et al. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat. Genet. 2012;44:491–501. doi: 10.1038/ng.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang X, Karsenty G. Transcription factors in bone: developmental and pathological aspects. Trends Mol. Med. 2002;8:340–345. doi: 10.1016/S1471-4914(02)02340-7. [DOI] [PubMed] [Google Scholar]
- 61.Veitia R, et al. Mutations and sequence variants in the testis-determining region of the Y chromosome in individuals with a 46,XY female phenotype. Hum. Genet. 1997;99:648–652. doi: 10.1007/s004390050422. [DOI] [PubMed] [Google Scholar]
- 62.Massanyi EZ, Dicarlo HN, Migeon CJ, Gearhart JP. Review and management of 46,XY disorders of sex development. J. Pedia. Urol. 2013;9:368–379. doi: 10.1016/j.jpurol.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 63.National Institutes of Health, National Library of Medicine, Genetics Home Reference. Swyer syndrome. https://ghr.nlm.nih.gov/condition/swyersyndrome (2015).
- 64.Clepet C, et al. The human SRY transcript. Hum. Mol. Genet. 1993;2:2007–2012. doi: 10.1093/hmg/2.12.2007. [DOI] [PubMed] [Google Scholar]
- 65.Oralova V, et al. Role of c-Myb in chondrogenesis. Bone. 2015;76:97–106. doi: 10.1016/j.bone.2015.02.031. [DOI] [PubMed] [Google Scholar]
- 66.Nolan E, et al. RANK ligand as a potential target for breast cancer prevention in BRCA1-mutation carriers. Nat. Med. 2016;22:933–939. doi: 10.1038/nm.4118. [DOI] [PubMed] [Google Scholar]
- 67.Dougall WC, Holen I, Gonzalez Suarez E. Targeting RANKL in metastasis. Bone. Rep. 2014;3:519. doi: 10.1038/bonekey.2014.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stopeck AT, et al. Denosumab compared with zoledronic acid for the treatment of bone metastases in patients with advanced breast cancer: a randomized, double-blind study. J. Clin. Oncol. 2010;28:5132–5139. doi: 10.1200/JCO.2010.29.7101. [DOI] [PubMed] [Google Scholar]
- 69.Fizazi K, et al. Denosumab versus zoledronic acid for treatment of bone metastases in men with castration-resistant prostate cancer: a randomised, double-blind study. Lancet. 2011;377:813–822. doi: 10.1016/S0140-6736(10)62344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Henry DH, et al. Randomized, double-blind study of denosumab versus zoledronic acid in the treatment of bone metastases in patients with advanced cancer (excluding breast and prostate cancer) or multiple myeloma. J. Clin. Oncol. 2011;29:1125–1132. doi: 10.1200/JCO.2010.31.3304. [DOI] [PubMed] [Google Scholar]
- 71.Li Y, et al. c-Myb Enhances Breast Cancer Invasion and Metastasis through the Wnt/beta-Catenin/Axin2 Pathway. Cancer Res. 2016;76:3364–3375. doi: 10.1158/0008-5472.CAN-15-2302. [DOI] [PubMed] [Google Scholar]
- 72.Knopfova L, et al. c-Myb regulates matrix metalloproteinases 1/9, and cathepsin D: implications for matrix-dependent breast cancer cell invasion and metastasis. Mol. Cancer. 2012;11:15. doi: 10.1186/1476-4598-11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.