Abstract
Until recently, small cell lung cancer (SCLC) was described as SCLC and SCLC variant, based upon cellular morphology and loss of neuroendocrine markers in the SCLC variant. However, based on recent research advances, driven in part by the increase in comprehensive genomic data, it has become clear that there are multiple SCLC subtypes including an ASCL1 and NEUROD1 low, YAP1 high (SCLC-Y) subtype enriched for WT RB1. Comparing morphological and other features of this SCLC subtype to neuroendocrine negative RB1, KEAP1, STK11 WT LCNEC raises a number of important questions with diagnostic and therapeutic implications.
Keywords: : CDK4/6 inhibitor, LCNEC, neuroendocrine negative, RB1 WT, SCLC
Practice points.
Similarity between some subtypes of small cell lung cancer (SCLC) and large cell neuroendocrine carcinoma (LCNEC) can be even higher than between SCLC and LCNEC in general.
Neuroendocrine markers negative/low RB1 WT SCLC-Y subtype and neuroendocrine markers negative/low WT RB1, WT KEAP1, WT STK11 LCNEC subtype are highly similar and might be in fact the same entity.
Unanimous agreement between expert pathologists is around 50% in differentiating between SCLC and LCNEC, indicating high level of diagnostic variability.
Patients with neuroendocrine markers negative/low WT RB1, WT KEAP1, WT STK11 LCNEC subtype might be candidates for clinical trial-based treatment with CDK4/6 inhibitors.
Lung anatomic locations could be important information to record along with genomic information and neuroendocrine markers staining.
About 10% of tumors diagnosed as SCLC are RB1 WT, such tumors may have sensitivity to treatments which are not applicable to majority of SCLC with inactivated RB1, therefore, confirming RB1 status should be an important consideration.
Small cell lung cancer (SCLC) is an aggressive form of lung cancer with limited therapeutic options, a very high mortality rate and is characterized, in most cases, by neuroendocrine features. SCLC accounts for approximately 15% of lung cancers. The majority of SCLC are genetically characterized by bi-allelic inactivation of RB1 (∼90%) and TP53 (∼98%) tumor suppressor genes [1–6]. The prevailing hypothesis is that inactivation of RB1 in SCLC leads to increase in cellular proliferation due to loss of cell cycle control and inactivation of TP53 prevents oncogene-induced senescence. SCLC diagnosis is commonly based on morphological features of biopsy or cytology samples. A panel of neuroendocrine markers (CHGA, NCAM1, SYP) may also be utilized [7,8]. In a noticeable fraction of cases, SCLC is present along with other lung cancer subtype(s) such as: large cell neuroendocrine carcinoma (LCNEC), large cell carcinoma (LCC), adenocarcinoma and/or squamous cell carcinoma [9].
Currently, SCLC is divided into four subtypes: ASCL1 high (SCLC-A), NEUROD1 high (SCLC-N), POU2F3 high (SCLC-P) and YAP1 high (SCLC-Y) subtype enriched for WT RB1 [10–13]. SCLC-Y subtype has low or absent expression of ASCL1, NEUROD1 and other neuroendocrine markers and accounts for approximately 5–10% SCLC tumors [11,12]. The SCLC-Y subtype is enriched for CCND1 amplification and CDKN2A inactivation, these alterations may play similar role to RB1 inactivation resulting in cell cycle control defects [11].
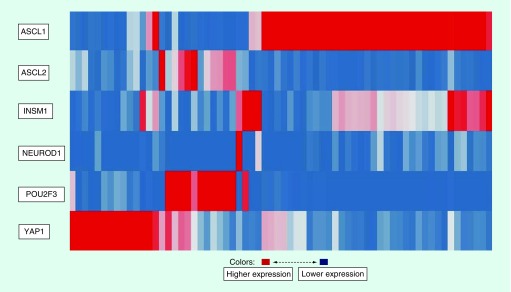
LCNEC is a relatively rare lung cancer, accounting for approximately 3% of lung cancer cases. In general, LCNEC is characterized by neuroendocrine morphology and markers; similarly to SCLC, patients with LCNEC have a poor prognosis [14]. About 90% of LCNEC have bi-allelic TP53 inactivation, ∼40% have bi-allelic RB1 inactivation and ∼40% have KEAP1 and/or bi-allelic STK11 inactivation; bi-allelic RB1 inactivation is generally mutually exclusive with KEAP1/STK11 bi-allelic inactivation [15,16]. From genetic alterations, prospective LCNEC with bi-allelic RB1 and TP53 inactivation are considered ‘SCLC like’ and LCNEC with KEAP1/STK11 bi-allelic inactivation are considered ‘NSCLC like’. About 4–8% of LCNEC are ASCL1 and NEUROD1 low with WT RB1, WT KEAP1, WT STK11 and bi-allelic TP53 inactivation. It is interesting to note that clustering gene expression LCNEC data from George et al. using key ‘SCLC’ transcription factors results in the gene expression pattern shown in Figure 1 which is very similar to one observed in SCLC [10,16].
Figure 1. . Gene expression clustering of large cell neuroendocrine carcinoma by key ‘small cell lung cancer’ transcription factors.
Thus, there are noticeable genetics, genomics and phenotypical similarities between the RB1 WT SCLC-Y subtype and the WT RB1, WT KEAP1, WT STK11 LCNEC subtype, where each subtype displays low or absent expression of ASCL1 and NEUROD1. In order to provide a more detailed background for comparison, some of the key features of neuroendocrine negative (NE-) SCLC are listed in Table 1. For comparison purposes, features of classical neuroendocrine positive (NE+) SCLC are also listed. Properties of LCNEC listed in Table 1 are generic and not necessarily specific to neuroendocrine negative (NE-) LCNEC subtype with WT RB1, WT KEAP1, WT STK11; it is possible that such LCNEC subtype has morphological properties even more similar to neuroendocrine negative (NE-) SCLC.
Table 1. . Morphological properties of large cell neuroendocrine carcinoma, neuroendocrine negative small cell lung cancer and classical neuroendocrine positive small cell lung cancer.
| Feature | LCNEC | SCLC NE- | SCLC NE+ |
|---|---|---|---|
| Mostly smokers | Yes | Yes | Yes |
| Median age | ∼67 years | ∼65 years | ∼65 years |
| Cell size | Typical of NSCLC | Larger than classical SCLC | Small |
| Cytoplasm | Abundant | More plentiful than classical SCLC | Scarce |
| Nucleoli | Prominent | Prominent | Not prominent |
| Mitotic rate | High | High | High |
| Nuclear chromatin | Less uniform | More uniform | More uniform |
| Cell borders | More distinct | Less distinct | Less distinct |
LCNEC: Large cell neuroendocrine carcinoma; NE: Neuroendocrine; NSCLC: Non-small-cell lung carcinoma; SCLC: Small cell lung cancer.
In general, SCLC are mostly located in central lung location and LCNEC in peripheral or midzone lung location [17–19]. A recent study from Zhou et al. suggests the possibility of differences between LCNEC in peripheral versus central locations [20]. Unfortunately, data regarding tumor location in the lung is limited for the neuroendocrine-negative SCLC subtype and for neuroendocrine-negative LCNEC with WT RB1, KEAP1, STK11.
There is currently no definitive marker(s) for LCNEC and diagnosis is based on exclusion. In part due to this and significant variabilities in LCNEC and SCLC phenotypes, there is limited agreement among pathologists on the LCNEC diagnosis. Ha et al. indicated that, among five expert thoracic pathologists, unanimous agreement in the diagnosis of LCNEC vs SCLC was achieved in only 40% of cases, other studies indicate similar levels of unanimous agreement between expert pathologists in SCLC and LCNEC diagnosis [21–23]. As can be seen from Table 1, neuroendocrine negative SCLC and LCNEC have a number of similarities and potential differences that are rather subtle, which makes it even more difficult to achieve agreement between pathologists.
In the Nicholson et al. study, out of 100 surgical biopsies or resections with a diagnosis of SCLC, 28 showed evidence of an NSCLC component in addition to SCLC, with LCC being the largest mixture component present in 16 cases of combined SCLC. Out of these 16 SCLC/LCC combined cases, 6 had LCNEC subtype of LCC [24]. In most instances of NSCLC combined with SCLC there is strong evidence of clonality between SCLC and NSCLC components, likely indicating a common precursor cell in such cases [25–29]. Pulmonary neuroendocrine cells (PNECs) and perhaps common pulmonary stem cells are potential cells of origin for SCLC and LCNEC [30–32].
Keeping in mind a likely common cell of origin and very similar genetics, genomics and morphology of the RB1 WT SCLC-Y subtype and the WT RB1, WT KEAP1, WT STK11 LCNEC subtype with low or absent expression of ASCL1 and NEUROD1, it is tempting to suggest that perhaps these two lung cancer subtypes are in-fact two faces of the same entity. If this is indeed the case, there are important diagnostic and therapeutic implications. From a diagnostic prospective, it should be important to note that such tumors share SCLC and LCNEC properties. Such acknowledgment has a potential therapeutic implication in relation to CDK4/6 inhibitors. Due to their lack of functional RB1, the clear majority of SCLC models are insensitive to the CDK4/6 inhibitors [33–36]. In contrast, some SCLC models with functional RB1 are sensitive to the CDK4/6 inhibitors [11,12,36,37]. Following these preclinical studies, a clinical trial of a CDK4/6 inhibitor in RB1 WT SCLC is being developed. Based on the discussion in this perspective, patients diagnosed with LCNEC subtype with WT RB1, WT KEAP1, WT STK11 and loss of neuroendocrine markers may benefit from being considered eligible for such clinical trial.
Future perspective
We hope that analysis presented in this article would help to increase awareness about highly similar subtypes of SCLC and LCNEC and potential clinical implications which might became even more relevant in future. We also hope our work would help to rejuvenate efforts on collecting information on lung anatomic locations of SCLC, LCNEC (and perhaps other lung tumors) in addition to genomic information and neuroendocrine markers staining. It is possible that paying attention to lung location such as: peripheral, midzone or central along with genomic information and neuroendocrine status may help to better characterize lung malignancies.
Acknowledgments
The views expressed in this article are the personal opinions of the authors and do not necessarily reflect policy of the US National Cancer Institute.
Footnotes
Financial & competing interests disclosure
This article is a US Government work and is in the public domain in the USA. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Open Access
This work is licensed under the Attribution-NonCommercial-NoDerivatives 4.0 Unported License
References
Papers of special note have been highlighted as: • of interest
- 1.George J, Lim JS, Jang SJ. et al. Comprehensive genomic profiles of small cell lung cancer. Nature 524(7563), 47–53 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Provides comprehensive genomic profiles of small cell lung cancer (SCLC), including exome and RNA sequencing.
- 2.van Meerbeeck JP, Fennell DA, De Ruysscher DKM. Small-cell lung cancer. Lancet 378(9804), 1741–1755 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Weinberg RA. The retinoblastoma protein and cell cycle control. Cell 81(3), 323–330 (1995). [DOI] [PubMed] [Google Scholar]
- 4.Knudson AG. Mutation and cancer: statistical study of retinoblastoma. Proc. Natl Acad. Sci. USA 68(4), 820–823 (1971). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goodrich DW. The retinoblastoma tumor-suppressor gene, the exception that proves the rule. Oncogene 25(38), 5233–5243 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nat. Rev. Cancer 9(10), 749–758 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rekhtman N. Neuroendocrine tumors of the lung: an update. Arch. Pathol. Lab. Med. 134(11), 1628–1638 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Travis WD. Update on small cell carcinoma and its differentiation from squamous cell carcinoma and other non-small cell carcinomas. Mod. Pathol. 25, S18–S30 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Nicholson SA, Beasley MB, Brambilla E. et al. Small cell lung carcinoma (SCLC): a clinicopathologic study of 100 cases with surgical specimens. Am. J. Surg. Pathol. 26(9), 1184–1197 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Rudin CM, Poirier JT, Byers LA. et al. Molecular subtypes of small cell lung cancer: a synthesis of human and mouse model data. Nat. Rev. Cancer 19, 289–297 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Reviews current knowledge on SCLC and proposes genomics-based classification.
- 11.Sonkin D, Vural S, Thomas A, Teicher BA. Neuroendocrine negative SCLC is mostly RB1 WT and may be sensitive to CDK4/6 inhibition. bioRxiv. (2019) (Epub ahead of print). [Google Scholar]; • Provides detailed genomic analysis of SCLC subtype with high YAP1 expression, loss of neuroendocrine markers and WT RB1.
- 12.McColl K, Wildey G, Sakre N. et al. Reciprocal expression of INSM1 and YAP1 defines subgroups in small cell lung cancer. Oncotarget 8(43), 73745–73756 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Describes SCLC subtype with high YAP1 expression, loss of neuroendocrine markers and enrichment for WT RB1.
- 13.Huang Y-H, Klingbeil O, He X-Y. et al. POU2F3 is a master regulator of a tuft cell-like variant of small cell lung cancer. Genes Dev. 32(13–14), 915–928 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fasano M, Della Corte CM, Papaccio F, Ciardiello F, Morgillo F. Pulmonary large-cell neuroendocrine carcinoma: from epidemiology to therapy. J. Thorac. Oncol. 10(8), 1133–1141 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rekhtman N, Pietanza MC, Hellmann MD. et al. Next-generation sequencing of pulmonary large cell neuroendocrine carcinoma reveals small cell carcinoma-like and non-small cell carcinoma-like subsets. Clin. Cancer Res. 22(14), 3618–3629 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.George J, Walter V, Peifer M. et al. Integrative genomic profiling of large-cell neuroendocrine carcinomas reveals distinct subtypes of high-grade neuroendocrine lung tumors. Nat. Commun. 9(1), 1048 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Provides comprehensive genomic profiles of large cell neuroendocrine carcinoma (LCNEC) and outlines existence of distinct subtypes.
- 17.Oshiro Y, Kusumoto M, Matsuno Y. et al. CT findings of surgically resected large cell neuroendocrine carcinoma of the lung in 38 patients. AJR Am. J. Roentgenol. 182(1), 87–91 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Jung KJ, Lee KS, Han J. et al. Large cell neuroendocrine carcinoma of the lung: clinical, CT, and pathologic findings in 11 patients. J. Thorac. Imaging 16(3), 156–162 (2001). [DOI] [PubMed] [Google Scholar]
- 19.Rosado-de-Christenson ML, Templeton PA, Moran CA. Bronchogenic carcinoma: radiologic–pathologic correlation. RadioGraphics 14(2), 429–446 (1994). [DOI] [PubMed] [Google Scholar]
- 20.Zhou F, Hou L, Ding T. et al. Distinct clinicopathologic features, genomic characteristics and survival of central and peripheral pulmonary large cell neuroendocrine carcinoma: From different origin cells? Lung Cancer Amst. Neth. 116, 30–37 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Travis WD, Gal AA, Colby TV, Klimstra DS, Falk R, Koss MN. Reproducibility of neuroendocrine lung tumor classification. Hum. Pathol. 29(3), 272–279 (1998). [DOI] [PubMed] [Google Scholar]
- 22.den Bakker MA, Willemsen S, Grünberg K. et al. Small cell carcinoma of the lung and large cell neuroendocrine carcinoma interobserver variability. Histopathology 56(3), 356–363 (2010). [DOI] [PubMed] [Google Scholar]; • Describes challenges in differentiating between SCLC and LCNEC and corresponding high diagnostic variability.
- 23.Ha SY, Han J, Kim W-S, Suh BS, Roh MS. Interobserver variability in diagnosing high-grade neuroendocrine carcinoma of the lung and comparing it with the morphometric analysis. Korean J. Pathol. 46(1), 42–47 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicholson SA, Beasley MB, Brambilla E. et al. Small cell lung carcinoma (SCLC): a clinicopathologic study of 100 cases with surgical specimens. Am. J. Surg. Pathol. 26(9), 1184–1197 (2002). [DOI] [PubMed] [Google Scholar]
- 25.D'Adda T, Pelosi G, Lagrasta C. et al. Genetic alterations in combined neuroendocrine neoplasms of the lung. Mod. Pathol. 21(4), 414–422 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Fellegara G, D'Adda T, Pilato FP. et al. Genetics of a combined lung small cell carcinoma and large cell neuroendocrine carcinoma with adenocarcinoma. Virchows Arch. 453(1), 107–115 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Huang J, Behrens C, Wistuba II, Gazdar AF, Jagirdar J. Clonality of combined tumors. Arch. Pathol. Lab. Med. 126(4), 437–441 (2002). [DOI] [PubMed] [Google Scholar]
- 28.Murase T, Takino H, Shimizu S. et al. Clonality analysis of different histological components in combined small cell and non-small cell carcinoma of the lung. Hum. Pathol. 34(11), 1178–1184 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Wagner PL, Kitabayashi N, Chen Y-T, Saqi A. Combined small cell lung carcinomas: genotypic and immunophenotypic analysis of the separate morphologic components. Am. J. Clin. Pathol. 131(3), 376–382 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Pathology and genetics of tumours of the lung, pleura, thymus and heart. World Health Organization classification of tumors. Travis WD, Brambilla E, Müller-Hermelink HK, Harris CC (). IARC Press, Lyon, France: (2004). [Google Scholar]
- 31.Swarts DRA, Ramaekers FCS, Speel E-JM. Molecular and cellular biology of neuroendocrine lung tumors: evidence for separate biological entities. Biochim. Biophys. Acta. 1826(2), 255–271 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Sutherland KD, Berns A. Cell of origin of lung cancer. Mol. Oncol. 4(5), 397–403 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Leary B, Finn RS, Turner NC. Treating cancer with selective CDK4/6 inhibitors. Nat. Rev. Clin. Oncol. 13(7), 417–430 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Baughn LB, Di Liberto M, Wu K. et al. A novel orally active small molecule potently induces G1 arrest in primary myeloma cells and prevents tumor growth by specific inhibition of cyclin-dependent kinase 4/6. Cancer Res. 66(15), 7661–7667 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Fry DW, Harvey PJ, Keller PR. et al. Specific inhibition of cyclin-dependent kinase 4/6 by PD 0332991 and associated antitumor activity in human tumor xenografts. Mol. Cancer Ther. 3(11), 1427–1438 (2004). [PubMed] [Google Scholar]
- 36.Polley E, Kunkel M, Evans D. et al. Small cell lung cancer screen of oncology drugs, investigational agents, and gene and microRNA expression. J. Natl Cancer Inst. 108(10) (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barretina J, Caponigro G, Stransky N. et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483(7391), 603–607 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]