Abstract
Aim:
We investigated associations of prenatal socioeconomic status (SES) with DNA methylation at birth, and to explore persistence of associations into early (∼3 years) and mid-childhood (∼7 years) among 609 mother–child pairs in a Boston-area prebirth cohort.
Materials & methods:
First, we created a prenatal SES index comprising individual- and neighborhood-level metrics and examined associations of low (lowest 10%) versus high (upper 90%) SES with genome-wide DNA methylation in cord blood via the Infinium HumanMethylation450 BeadChip. Next, we evaluated persistence of associations detected in cord blood with DNA methylation of the same CpG sites measured in peripheral leukocytes in early- and mid-childhood.
Results & conclusion:
Low prenatal SES was associated with methylation at CpG sites near ACSF3, TNRC6C-AS1, MTMR4 and LRRN4. The relationship with LRRN4 persisted into early childhood.
Keywords: : developmental origins of health and disease, DNA methylation, epigenome-wide association study, pediatric cohort, socioeconomic status
In hierarchical societies of both humans and animals, social status is a pervasive determinant of health and fitness [1]. A longstanding and extensively-reviewed social sciences literature [2–5], in conjunction with mechanistic studies in animal models [6,7] link lower social status – for example, lower socioeconomic position in humans or lower social rank in baboons – to a range of adverse physiological and psychological phenotypes.
Growing awareness of the Developmental Origins of Health and Disease (DOHaD) paradigm over the last two decades has forged interest in: the relevance of timing of exposures – namely, the importance of exposures during sensitive periods of development (typically, early in the life course), and biological mechanisms linking social exposures to disease risks [8]. Both lines of inquiry have placed a spotlight on epigenetic processes, with DNA methylation being the most widely studied, as a mechanism underlying DOHaD phenomena [9]. The relevance of epigenetics in DOHaD stems from the fact that sensitive windows of development coincide with higher lability of the epigenome. For instance, shortly after fertilization, a genome-wide demethylation event occurs, followed by systematic re-establishment of DNA methylation marks [10]. The process of remethylation is sensitive to environmental disturbances, thereby earmarking the in utero period as a sensitive developmental time frame during which environmental factors – including the mother’s social environment – can impact the fetus’ long-term health. Evidence for this notion has been documented in nonhuman primates; for example, where researchers have detected differential DNA methylation at >25,000 genomic locations in placenta of low versus high ranking female baboons [11]. In humans, maternal education level (a proxy for socioeconomic position) has been associated with methylation of specific genes involved in growth and stress physiology in placentae [12], cord blood [13] and infant blood [14], as well as with global DNA methylation in cord blood [15].
However, the existing evidence base is not without shortcomings. First, the majority of published human studies have focused on individual socioeconomic metrics (i.e., household income, maternal education level), which may not reflect the real-life experience of social status. Second, many analyses are limited by relatively small sample size and restricted markers of DNA methylation (e.g., candidate gene approaches and proxies of global DNA methylation). Third, while a few small-scale (i.e., N <100 [16–18]) and one larger study in the Multi Ethnic Study of Atherosclerosis cohort [19] compared the effect of childhood versus adulthood socioeconomic status (SES) on DNA methylation profiles during adulthood, little is known regarding whether the effects of social exposures during the prenatal period ‘persist’ into later life, which is central to evaluating the extent to which this developmental stage is critical to future health outcomes. To our knowledge, we are aware of only one study that has attempted to do this. In a recent publication, Alfano et al. investigated persistent effects of early life SES on genome-wide DNA methylation from birth through adolescence among 860 mother–child pairs in the Avon Longitudinal Study of Parents and Children (ALSPAC) [20]. The investigators found that while maternal education was associated with differential methylation of four CpG sites in cord blood and 20 CpG sites in adolescent blood, there was no overlap in the CpG sites across life stages. However, a shortfall of ALSPAC is the predominantly White ethnic composition (4.1% non-White participants in the overall cohort), which limits generalizability of findings.
Here, we sought to address key gaps in literature using data from >600 participants in Project Viva, a large multi-ethnic prebirth cohort in the greater Boston area. First, we created an SES status index based on individual- and neighborhood-level characteristics during the prenatal period. Next, we carried out an epigenome-wide association study (EWAS) to identify differentially methylated CpG sites in cord blood associated with prenatal SES. Then, we investigated whether associations of prenatal SES with the CpG sites of interest remained apparent during early and mid-childhood. At last, we examined associations of prenatal SES with global DNA methylation, hydroxymethylation and the ratio of the two during all three life stages, to complement EWAS findings.
Materials & methods
Study population
This study includes participants of Project Viva, an ongoing prebirth cohort recruited from a multispecialty group practice in eastern Massachusetts (Atrius Harvard Vanguard Medical Associates). Details on study design and recruitment are reported elsewhere [21]. Of 2128 enrolled Project Viva mother–child pairs, this study included 609 pairs for whom we had data on DNA methylation at birth (n = 422), during early childhood (age: 3–6 years; n = 108), or in mid-childhood (age: 6–10 years; n = 389); 53 participants have data during all three life stages. The Institutional Review Board of Harvard Pilgrim Health Care approved all study protocols. All mothers provided written informed consent.
Exposure: SES during the prenatal period
The exposure of interest in this study was SES during the prenatal period. We created an SES index based on individual- and neighborhood-level metrics that are either associated with health outcomes of the child in Project Viva (e.g., maternal education, marital status, annual household income), or have been implicated as determinants of health in the literature (e.g., neighborhood-level income [22], use of welfare programs [23]). Individual-level SES metrics included maternal education level, marital status, self-reported income at enrollment and receipt of public assistance. These data were collected through interviewer-administered questionnaires during the first trimester of pregnancy. We extracted neighborhood-level metrics, including the median neighborhood income and percentage of neighborhood below the poverty line, based on the census tract of the residential address at enrollment.
To create the SES index, we first weighted participant responses for each metric on a scale of 0 (lowest SES) to 3 (highest SES) as follows: mother’s education level (no college = 0; some college = 1; college degree = 2; graduate degree = 3), mother’s marital status (not married/cohabiting = 0; married/cohabiting = 3), mother’s self-reported household income at the initial enrollment visit during the first trimester (<$20,000 = 0; >$20,000–40,000 = 1; >$40,000–70,000 = 2; >$70,000 = 3), receipt of public assistance (Yes = 0; No = 3), quartiles of median neighborhood income based on the census tract (Q1 = 0; Q2 = 1; Q3 = 2; Q4 = 3), and quartiles of percentage of neighborhood below the poverty line according to the census tract (Q4 = 0; Q3 = 1; Q2 = 2; Q1 = 3). Next, we entered the six variables into an unsupervised principal components analysis and retained the first factor as the SES index score which accounted for 68.6% of variability in the original components of the score. This factor represents a continuous, normally distributed weighted score of the six SES components based on their natural intercorrelations. A higher score for this index is indicative of higher SES during the prenatal period and vice versa. For the analysis, we dichotomized prenatal SES as the lowest 10% versus the upper 90% (referent) of the index based on initial evidence of a threshold effect with respect to the epigenome-wide association analysis and the global measures of DNA methylation at the lower end of the distribution. This is likely related to the fact that Project Viva participants are relatively well-educated and a mid- to upper-class population, making it so that an effect of SES is detectable only at extreme ends of the distribution. This phenomenon has been observed in other studies exploring associations of SES with DNA methylation in socioeconomically homogenous populations [24].
When exploring associations of individual components of the prenatal SES index, we used the same categorization scheme for each variable as they were used in the index with two exceptions: we collapsed ‘no college’ with ‘some college’ for maternal education level, and ‘<$20,000’ with ‘$20,000–40,000’ due to small cell sizes.
Outcome: DNA methylation
Blood collection
At delivery, obstetricians and midwives collected blood from the umbilical cord vein from approximately half of births. Collection of umbilical cord blood was based on participant consent and occurred at one of two delivery hospitals that accounted for approximately three-fourths of deliveries in Project Viva. At the early childhood and mid-childhood visits, trained research assistants collected a blood sample from the antecubital vein. All samples were refrigerated immediately, processed within 24 h (including extraction of DNA using Qiagen Puregene Kit, CA, USA), and stored at -80°C until the time of analysis.
Epigenome-wide DNA methylation analysis
Methods for genome-wide DNA methylation analysis have been published [25]. In brief, we bisulfite-converted buffy coat DNA (EZ DNA Methylation-Gold Kit, Zymo Research, CA, USA) in cord blood, and in peripheral leukocytes from early- and mid-childhood. We shipped samples to Illumina Inc., where they are analyzed using the Infinium HumanMethylation450 BeadChip (Illumina, CA, USA). This array is widely-used in epidemiologic studies and has been previously validated [26]. Upon receiving the data, we removed technical replicates, samples with low quality, genotype mismatch and sex mismatch. We also excluded low-quality probes with detection p-values greater than 0.05, those on sex chromosomes, non-CpG probes and nonspecific and previously-identified cross-reactive probes [27]. We further removed probes within ten base pairs of a known SNP (UCSC Human Feb. 2009 [GRCh37/hg19] Assembly), and those with a minor allele frequency ≥1%. After the quality control, a total of 394,460 probes were included for subsequent analysis. We used the normal-exponential out-of-band method for background correction and dye bias adjustment [28] and normalized our sample with Beta Mixture Quantile Dilation [29]. To control for technical variability across sample plates we applied the R package ComBat [30]. For the analysis, we examined the percent of methylation for each CpG site, calculated as the signal intensity of methylated cytosines over the signal intensity of methylated plus unmethylated cytosines at the 5C position.
Global DNA methylation & hydroxymethylation
Using DNA extracted from cord blood, and from blood collected at the early and mid-childhood visits, we carried out the following procedure. First, we isolated 1 μg of DNA from the buffy coat. Next, we spiked extracted DNA with an internal standard mixture, dried it and then hydrolyzed it at 37°C for at least 8 h with 10 μl of digest mixture containing phosphodiesterase I, ALP and Benzonase® Nuclease to Tris-HCl buffer (Sigma-Aldrich, Diegem, Belgium). We then prepared stock solution of 5mC, 5hmC and cytosine (C) by dissolving Sigma-Aldrich commercial standards in 22 liquid chromatography mass spectrometry-grade water to prepare the calibration standards. At last, we measured global genomic DNA methylation and hydroxymethylation of cytosine nucleotides simultaneously for each sample using ultra-performance liquid chromatography tandem mass spectrometry, as previously described [31]. We have previously published more details on analytical conditions, calibration curves and validation parameters [32].
We calculated percent global DNA methylation (%5mC) as 5mC over the sum of 5mC, 5hmC and C (%5mC = 5mC/[5hmC+5mC+C]). Likewise, percent global DNA hydroxymethylation (%5hmC) was derived as 5hmC/(5hmC+5mC+C). We also calculated the ratio of global DNA methylation to hydroxymethylation (%5mC:%5hmC) and log2-transformed %5mC:%5hmC for the analysis due to non-normal distributions.
Covariates
At enrollment, we collected information on the mother’s race/ethnicity and age. At enrollment, during mid-pregnancy, and at delivery, we asked the women about pregnancy smoking habits via an interviewer-administered questionnaire. We extracted child sex and date of delivery from the medical record. We used date of delivery to calculate gestational age at delivery by subtracting from this value the date of the last menstrual period as reported by the woman, or the estimated last menstrual period based on second trimester ultrasound if gestational age between the two sources differed by more than 10 days. Based on these calculations, we included continuous gestational age at delivery as a covariate in the EWAS analysis, and also evaluated this variable dichotomously as preterm versus term (<37 vs ≥37 gestational weeks). Prenatal medical records provided information on perinatal characteristics including mother’s prenatal glucose tolerance status, including diagnoses of gestational diabetes mellitus. To identify women with hypertensive disorders of pregnancy, we reviewed outpatient charts for blood pressure and urine protein results, and created a four-level variable with the categories normotensive, chronic hypertension, gestational hypertension and preeclampsia [33]. The child’s age at the early and mid-childhood visits were calculated as the difference between the date of the visit and date of birth. Cell type composition at each research visit was estimated from genome-wide DNA methylation arrays (HumanMethylation450 BeadChip; Illumina) using the minfi package in R (version 3.3.0, R Core Team) [34]. We used an adult reference DNA methylation panel to estimate leukocyte composition in early and mid-childhood and a cord blood DNA methylation reference panel, which includes nucleated red blood cells, to estimate nucleated cell types at birth [35].
Data analysis
Prior to formal analysis, we assessed univariate distributions of continuous variables and frequency distributions of categorical or ordinal variables to assess for deviations from normality and missing data, and transformed variables as needed. At this stage, we also assessed background and SES-related characteristics of the three subsamples of Project Viva participants with 450 K methylation data at birth (n = 422), during early childhood (n = 108) and during mid-childhood (n = 389) to qualitatively assess for notable discrepancies that would compromise comparability across the three overlapping subsamples (n = 53 children have DNA methylation data available at all three time points). Then, we carried out the main analysis in three steps, conforming to the study objectives.
Step 1: Identify differentially-methylated CpGs in cord blood associated with prenatal SES
We dichotomized the prenatal SES index as the lowest 10% versus upper 90% (referent) based on exploratory analyses indicating a threshold effect of prenatal SES on DNA methylation outcomes at the lowest end of the distribution. For the EWAS, we used robust linear regression to examine associations and assess statistical significance of the relationship between prenatal SES and DNA methylation of individual CpG sites in cord blood on the M-value scale. However, to facilitate interpretability, we re-ran the top hits on the β-scale and report estimates of association in terms of β-values (% difference in DNA methylation) and standard errors (SEs) in the results and tables.
In the models, we also accounted for maternal age at enrollment, race/ethnicity and smoking habits during pregnancy; gestational age at delivery; infant sex; and cell type composition. Of note, while smoking during pregnancy could be part of the low SES effect, we were interested in the effect of SES independent of maternal smoking. Thus, we included this variable in the EWAS to maintain consistency with previous studies in this cohort [25,32], and based on expert consensus [36]. We considered a CpG site to be of interest in our analysis of persistence of associations (Step 2) if its effect estimate reached statistical significance (p < 0.05) after false discover rate (FDR)-correction based on Benjamini and Hochberg’s method [37]. However, in our discussion of findings regarding associations in cord blood, we focus on CpG sites that were statistically significant (p < 0.05) after the more conservative Bonferroni correction. In our results, we logit-transformed the β-values on the M-value scale such that results are interpreted as a percentage difference in DNA methylation.
Step 2: Examine persistence of associations during early childhood & mid-childhood
In a second step, we examined associations of prenatal SES with CpG sites that were statistically significant after FDR correction in Step 1, measured in leukocytes during early and mid-childhood using ComBat to account for technical variability. We accounted for the same set of covariates as in Step 1, except rather than adjusting for gestational age at birth, we adjusted for current child age and estimated leukocytes composition from the same time point. We considered an association to be persistent if the p-value was significant after a Bonferroni correction for the number of CpG sites tested at 3 and 7 years.
Step 3: Examine associations of prenatal SES with global DNA methylation, hydroxymethylation & global:hydroxymethylation ratio
At last, we examined associations of prenatal SES with global DNA methylation (%5mC), hydroxymethylation (%5hmC) and log2-transformed %5mC:%5hmC during all three life stages. In the multivariable linear regression models, we accounted for the same set of covariates as in Steps 1 and 2. In addition to assessing the prenatal SES index, we also examined individual components of the index in relation to the global measures of DNA methylation. In sensitivity analyses, we assessed the impact of further adjustment for preterm delivery status (<37 vs ≥37 gestational weeks at birth), maternal gestational diabetes and hypertensive disorders of pregnancy.
All analyses were performed using SAS 9.4 (SAS Institute Inc., NC, USA), except for analyses in Steps 1 and 2, which were carried out in R.
Results
At enrollment, median age of the mothers was 32.9 years (range: 17.8–44.9 years). The majority (74.2%) of women were White, with 10.7% Black and 6.9% Hispanic participants. Approximately half (49.4%) of the children were female. Mean ± standard deviation (SD) gestational age at delivery was 39.7 ± 1.6 weeks. Mean ± SD age of the children at the early and mid-childhood research visits was 3.4 ± 0.5 and 7.9 ± 0.8 years, respectively. Mean ± SD of the SES index was 0.00 ± 0.94 (median: 0.11; range: -1.65–1.53). Table 1 shows additional background characteristics of the study sample, as well as bivariate associations of the characteristics with the prenatal SES index. Supplementary Table 1 shows background characteristics of participants based on availability of DNA methylation data at birth (n = 422), during early childhood (n = 108) and during mid-childhood (n = 389), and the exposure of interest (prenatal SES index). The relative distribution of characteristics are similar across the three subsamples, with the exception that there is a higher proportion of women who reported an annual household income ≤$20,000 in the mid-childhood sample (5.1%) as compared with the birth (1.9%) and early childhood samples (0%).
Table 1. . Sociodemographic characteristics of 609 Project Viva mother–child pairs.
| % (N) | Mean ± SD prenatal SES index score | |
|---|---|---|
| Maternal and perinatal characteristics | ||
| Mother’s age at enrollment: | ||
| – <25 years | 8.1% (49) | -0.78 ± 0.74 |
| – 25–<35 years | 59.4% (362) | 0.03 ± 0.92 |
| – ≥35 years | 32.5% (198) | 0.14 ± 0.92 |
| – p-trend | <0.0001 | |
| Mother’s prepregnancy weight status: | ||
| – Underweight (<18.5 kg/m2) | 2.8% (17) | -0.82 ± 0.72 |
| – Normal weight (18.5–24.9 kg/m2) | 61.7% (375) | 0.10 ± 0.91 |
| – Overweight (25.0–29.9 kg/m2) | 21.9% (133) | -0.08 ± 0.94 |
| – Obese (≥30 kg/m2) | 13.7% (83) | -0.19 ± 0.98 |
| – p-trend | 0.11 | |
| Pregnancy smoking status: | ||
| – Never | 68.3% (416) | -0.05 ± 0.94 |
| – Former | 21.0% (128) | 0.28 ± 0.87 |
| – Smoked during pregnancy | 10.7% (65) | -0.24 ± 0.91 |
| – p-trend | 0.0002 | |
| Race/ethnicity: | ||
| – Black | 10.7% (65) | -0.77 ± 0.87 |
| – Hispanic | 6.9% (42) | -0.46 ± 0.99 |
| – White | 74.2% (452) | 4.19 ± 0.86 |
| – Other | 8.2% (50) | -0.37 ± 0.94 |
| – p-difference | <0.0001 | |
| Child sex: | ||
| – Female | 49.4% (301) | 0.03 ± 0.92 |
| – Male | 50.6% (308) | 0.03 ± 0.96 |
| – p-difference | 0.48 | |
| Socioeconomic characteristics during the prenatal period | ||
| Mother was married or cohabitating: | ||
| – No | 7.1% (43) | -0.47 ± 0.76 |
| – Yes | 92.9% (566) | 0.04 ± 0.94 |
| – p-difference | 0.0006 | |
| Mother’s education level: | ||
| – No college | 7.4% (45) | -0.29 ± 0.91 |
| – Some college | 21.5% (131) | -0.25 ± 0.94 |
| – College degree | 37.9% (231) | 0.25 ± 0.91 |
| – Graduate degree | 33.2% (202) | -0.06 ± 0.91 |
| – p-trend | 0.03 | |
| Mother-reported annual household income: | ||
| – ≤$20,000 | 3.8% (23) | -0.53 ± 0.89 |
| – >$20,000–40,000 | 11.7% (71) | -0.67 ± 0.84 |
| – >$40,000–70,000 | 22.0% (134) | -0.16 ± 0.89 |
| – >$70,000 | 62.6% (381) | 0.21 ± 0.89 |
| – p-trend | <0.0001 | |
| Receipt of public assistance: | ||
| – Yes | 11.3% (69) | -0.12 ± 0.96 |
| – No | 88.7% (540) | 0.02 ± 0.93 |
| – p-trend | 0.25 | |
| Median neighborhood income (census tract): | ||
| – Q1 ($15,374–41,716) | 20.7% (126) | -1.32 ± 0.24 |
| – Q2 ($41,771–53,855) | 24.6% (150) | -0.48 ± 0.32 |
| – Q3 ($54,013–68,438) | 26.1% (159) | 0.37 ± 0.43 |
| – Q4 ($68,564–121,899) | 28.6% (174) | 1.04 ± 0.31 |
| – p-trend | <0.0001 | |
| % of neighborhood below poverty line (census tract): | ||
| – Q1 (highest poverty, >14–48%) | 19.5% (119) | -1.33 ± 0.25 |
| – Q2 (>7–14%) | 26.4% (161) | -0.53 ± 0.33 |
| – Q3 (>3.6–7%) | 24.8% (151) | 0.35 ± 0.33 |
| – Q4 (lowest poverty, ≤3.6%) | 29.2% (178) | 1.08 ± 0.23 |
| – p-trend | <0.0001 | |
| Prenatal SES index†: | ||
| – Lowest 10% | 9.5% (58) | -1.53 ± 0.08 |
| – Upper 90% | 90.5% (551) | 0.16 ± 0.83 |
Based on a weighted index comprising maternal self-reported household income, receipt of public assistance, education level, marital status, median household income (census tract), % of neighborhood below poverty line (census tract).
SD: Standard deviation; SES: Socioeconomic status.
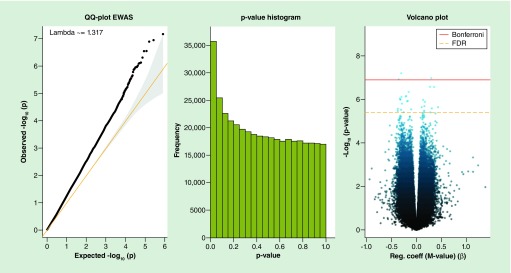
In EWAS of low versus high prenatal SES and CpG-by-CpG DNA methylation (see Figure 1 for Q-Q plot and volcano plot; Lamda = 1.317), we identified 29 differentially methylated CpGs (FDR < 0.05), three of which were also significant after Bonferroni correction: ACSF3 (β ± SE = −0.40 ± 0.07 %5mC), TNRC6C-AS1 (β ± SE = 0.98 ± 0.18 %5mC), and MTMR4 (β ± SE = −0.27 ± 0.06 %5mC; Table 2). At this step, we noted that many of the CpG sites had percentage methylation values close to 0 or 100, thus reflecting an overall pattern of low variability in DNA methylation (Table 2).
Figure 1. . QQ plot, p-value histogram and volcano plot for associations of prenatal socioeconomic status (lowest 10% vs upper 90% of the socioeconomic status index) with cord blood EWAS.
The yellow dashed line and the red line in the volcano plot represent FDR and Bonferroni correction cutoffs, respectively.
EWAS: Epigenome-wide association study; FDR: False discover rate.
Table 2. . Associations of prenatal socioeconomic status (lowest 10% vs upper 90% of socioeconomic status index) with cord blood DNA methylation (n = 422).
| CpG | CHR | MAPINFO | Gene | Genomic location | Gene region | CpG position (from TSS) | β† | SE† | p-value‡ | % methylated | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | ||||||||||
| cg16746620 | 16 | 89178589 | ACSF3§ | Open Sea | Inside | 18372 | -0.40% | 0.07 | 6.39E–08 | 0.98 | 0.01 |
| cg02283662 | 17 | 76106981 | TNRC6C-AS1¶ | Open Sea | Inside | 899 | 0.98% | 0.18 | 1.07E–07 | 0.94 | 0.02 |
| cg02961795 | 17 | 56588344 | MTMR4§ | N. Shelf | Downstream | 173781 | -0.27% | 0.06 | 1.22E–07 | 0.99 | 0.01 |
| cg12453539 | 20 | 6033866 | LRRN4§ | S. Shore | Downstream | 45968 | 2.34% | 0.49 | 2.69E–07 | 0.14 | 0.03 |
| cg20057398 | 11 | 114493710 | NXPE2¶ | Open Sea | Upstream | 55490 | 0.81% | 0.16 | 2.72E–07 | 0.96 | 0.01 |
| cg01346596 | 10 | 11868146 | C10orf47§ | S. Shelf | Inside | 2749 | -0.47% | 0.10 | 4.65E–07 | 0.96 | 0.01 |
| cg04312358 | 19 | 41197166 | NUMBL§ | S. Shore | Downstream | 25624 | -0.26% | 0.05 | 7.12E–07 | 0.02 | 0.01 |
| cg10430625 | 1 | 59320924 | JUN¶ | Open Sea | Upstream | 71139 | -1.12% | 0.23 | 7.46E–07 | 0.92 | 0.02 |
| cg27467787 | 20 | 36160512 | NNAT¶ | S. Shelf | Downstream | 10905 | -0.35% | 0.08 | 7.70E–07 | 0.98 | 0.01 |
| cg08492625 | 22 | 29978158 | NIPSNAP1§ | S. Shore | Promoter | 832 | -1.09% | 0.20 | 9.12E–07 | 0.96 | 0.02 |
| cg12503835 | 8 | 143324660 | TSNARE§ | Open Sea | Downstream | 44943 | -0.47% | 0.12 | 1.03E–06 | 0.98 | 0.01 |
| cg23495031 | 7 | 151372668 | PRKAG2§ | Open Sea | Inside | 201648 | -0.66% | 0.15 | 1.03E–06 | 0.97 | 0.01 |
| cg23928910 | 5 | 89705806 | CETN3§ | Island | Upstream | 64875 | 0.25% | 0.05 | 1.07E–06 | 0.03 | 0.00 |
| cg19113686 | 3 | 101406347 | RPL24§ | S. Shore | Downstream | 11073 | -0.29% | 0.06 | 1.19E–06 | 0.98 | 0.01 |
| cg24313932 | 15 | 35591670 | DPH6¶ | Open Sea | Downstream | 246734 | -0.71% | 0.14 | 1.24E–06 | 0.94 | 0.01 |
| cg04127903 | 3 | 186335125 | AHSG§ | Open Sea | Upstream | 46793 | 0.09% | 0.02 | 1.31E–06 | 0.01 | 0.00 |
| cg00747160 | 16 | 57286437 | ARL2BP§ | Open Sea | Inside | 7399 | -0.36% | 0.09 | 1.59E–06 | 0.98 | 0.01 |
| cg22281207 | 2 | 85765696 | MAT2A§ | Island | Inside | 313 | 0.29% | 0.06 | 2.08E–06 | 0.03 | 0.00 |
| cg14543995 | 17 | 30036582 | MIR365B¶ | Open Sea | Downstream | 134152 | 0.33% | 0.06 | 2.31E–06 | 0.99 | 0.01 |
| cg21502786 | 17 | 56234476 | MSX2P1§ | Island | Inside | 156 | 0.12% | 0.03 | 2.33E–06 | 0.02 | 0.00 |
| cg18437144 | 1 | 10788928 | CASZ1§ | Open Sea | Inside | 67805 | 1.05% | 0.23 | 2.62E–06 | 0.94 | 0.02 |
| cg27523842 | 17 | 3911067 | ZZEF1§ | S. Shelf | Inside | 135186 | -0.26% | 0.07 | 2.76E–06 | 0.99 | 0.00 |
| cg19674232 | 2 | 242662301 | ING5§ | N. Shore | Upstream | 35918 | -0.18% | 0.04 | 2.93E–06 | 0.98 | 0.00 |
| cg26524547 | 1 | 201820022 | IPO9§ | Open Sea | Inside | 21734 | -0.39% | 0.09 | 3.06E–06 | 0.98 | 0.01 |
| cg21444639 | 1 | 171711847 | VAMP4§ | S. Shore | Promoter | 468 | 0.16% | 0.04 | 3.37E–06 | 0.02 | 0.00 |
| cg23371877 | 7 | 152067904 | MLL3§ | S. Shelf | Inside | 65186 | -0.40% | 0.10 | 3.45E–06 | 0.99 | 0.01 |
| cg16532399 | 1 | 38031726 | DNALI1§ | Open Sea | Inside | 9206 | -0.36% | 0.09 | 3.97E–06 | 0.98 | 0.01 |
| cg06506007 | 13 | 36699873 | DCLK1§ | Open Sea | Inside | 5641 | 4.30% | 0.93 | 4.00E–06 | 0.63 | 0.06 |
| cg01479122 | 15 | 32163262 | OTUD7A¶ | Open Sea | Promoter | 270 | 0.25% | 0.06 | 4.07E–06 | 0.02 | 0.00 |
Estimates are on the β-value scale and are adjusted for maternal race/ethnicity, age at enrollment and smoking habits during pregnancy; infant sex and gestational age at birth; and cell type distribution.
The p-values are from EWAS run on the M-value scale. All CpG sites in this table are FDR significant; those in bolded font are statistically significant after Bonferroni correction.
Gene names referenced from Illumina annotation
Gene names referenced from University of California Santa Cruz Genome Browser
CHR: Chromosome; CpG: Cytosine–phosphate–guanine site; EWAS: Epigenome-wide association study; FDR: False discover rate; SD: Standard deviation; SE: Standard error.
When we examined associations of prenatal SES with DNA methylation of the FDR significant CpG sites in peripheral leukocyte DNA collected during early childhood, only one association (cg12453539) remained apparent: β ± SE for low versus high prenatal SES in relation to LRRN4 methylation at birth was 2.34 ± 0.49 %5mC, and during early childhood was 5.3 ± 1.1 %5mC. None of the associations between prenatal SES and the 29 CpG sites were detected during mid-childhood.
We did not find any associations of prenatal SES with %5mC, %5hmC or log2-transformed %5mC:%5hmC at any life stage (Table 3).
Table 3. . Associations of prenatal socioceonomic status with global DNA methylation (%5mC), hydroxymethylation (%5hmC) and the ratio of global DNA methlyation to hydroxymethylation (%5mC:%5hmC) among Project Viva participants at birth, during early childhood (median age 3 years) and mid-childhood (median age 7 years).
| Prenatal SES§ | N | β (95% CI)† in cord blood DNA methylation (birth) | ||
|---|---|---|---|---|
| % 5mC | % 5hmC | % 5mC: % 5hmC ratio‡ | ||
| Mean (SD) = 5.67 (2.07) | Mean (SD) = 0.25 (0.15) | Mean (SD) = 4.67 (0.70) | ||
| Lowest 10% | 45 | -0.44 (−1.08, 0.20) | 0.00 (-0.05, 0.04) | −0.05 (-0.26, 0.17) |
| Upper 90% | 368 | 0.00 (Reference) | 0.00 (Reference) | 0.00 (Reference) |
| p-value | 0.18 | 0.86 | 0.66 | |
| N | β (95% CI)† in peripheral leukocyte DNA methylation during early childhood (∼3 years) | |||
| %5mC | % 5hmC | %5mC:%5hmC ratio‡ | ||
| Mean (SD) = 5.11 (1.77) | Mean (SD) = 0.17 (0.11) | Mean (SD) = 5.10 (0.75) | ||
| Lowest 10% | 9 | -0.28 (-1.46, 0.91) | 0.02 (-0.06, 0.10) | -0.32 (-0.86, 0.22) |
| Upper 90% | 94 | 0.00 (Reference) | 0.00 (Reference) | 0.00 (Reference) |
| p-value | 0.64 | 0.57 | 0.25 | |
| N | β (95% CI)† in peripheral leukocyte DNA methylation during mid-childhood (∼7 years) | |||
| %5mC | % 5hmC | %5mC:%5hmC ratio‡ | ||
| Mean (SD) = 5.19 (1.94) | Mean (SD) = 0.19 (0.11) | Mean (SD) = 4.95 (0.64) | ||
| Lowest 10% | 35 | 0.21 (-0.50, 0.91) | -0.01 (-0.05, 0.03) | 0.10 (-0.13, 0.33) |
| Upper 90% | 332 | 0.00 (Reference) | 0.00 (Reference) | 0.00 (Reference) |
| P-value | 0.56 | 0.75 | 0.39 | |
Models include SES indicator of interest, maternal age at enrollment, race/ethnicity and smoking habits during pregnancy; child’s sex and age at the time of DNA methylation assessment (at birth, this variable was gestational age at delivery); and cell type distribution.
%5mC:%5hmC is Log2 transformed to meet normality assumptions.
Based on a weighted index comprising maternal self-reported household income, receipt of public assistance, education level, marital status, median household income (census tract), % of neighborhood below poverty line (census tract).
SD: Standard deviation; SES: Socioeconomic status.
Supplementary Tables 2–4 show relations of the prenatal SES index components with %5mC, %5hmC and log2-transformed %5mC:%5hmC. At birth, women who were married/cohabiting had infants with lower cord blood %5mC and %5hmC. We also observed an inverse association between maternal education and %5mC in early and mid-childhood, but we did not note any other consistent associations of the individual SES indicators with the DNA methylation outcomes across the three life stages.
In all conventional regression models (i.e., Table 3, Supplementary Tables 2–4) further adjustment for maternal pre- and perinatal conditions (preterm delivery, gestational glucose tolerance during pregnancy, hypertensive disorders) did not change the direction, magnitude or precision of the associations (data not shown; available upon request).
Discussion
In this study of 609 mother–child pairs in Project Viva, we examined associations of maternal SES during pregnancy (‘prenatal SES’, dichotomized as lowest 10% vs upper 90% of an SES index) with epigenome-wide DNA methylation, as well as liquid chromatography-based assessments of global DNA methylation, global hydroxymethylation and the ratio of the two at birth (in cord blood), and during early and mid-childhood (in peripheral leukocytes collected at median age 3 and 7 years, respectively). We identified associations of prenatal SES with DNA methylation at 29 CpG sites after FDR correction. However, only three of these sites, which are associated with the genes ACSF3, TNRC6C-AS1 and MTMR4, were significant after Bonferroni correction. When we explored relations of prenatal SES with the 29 CpG sites that were FDR-significant during early (∼age 3 years) and mid-childhood (∼age 7 years), only one estimate – higher LRRN4 methylation for low versus high prenatal SES – persisted into early childhood. No associations were detected during mid-childhood.
Associations of prenatal SES with CpG sites in cord blood (EWAS)
After FDR adjustment, prenatal SES was associated with methylation at 29 sites in cord blood. Of them, only three were statistically significant after Bonferroni correction. We describe the direction of associations below, as well as potential ways in which dysregulation of genes at these sites may link SES to development of noncommunicable diseases.
ACSF3
As compared with participants with high prenatal SES, those categorized as having low SES during the prenatal period had lower DNA methylation of a CpG on chromosome 16 located in the body of the ACSF3 gene [38–40]. ACSF3 encodes a synthetase protein that is essential for fatty acid production in mammalian mitochondria [41–43]. Deficiencies in ACSF3 enzymes are associated with combined malonic and methylmalonic aciduria, a pathology related to delayed growth and dysmetabolism in children and psychiatric disease in adults [44]. If SES-related disparities in DNA methylation are associated with transcription and translation of ACSF3, it might be possible that this epigenetic modification may provide a molecular biomarker between the documented associations of SES and development of metabolic and psychiatric disease [4,5,45,46].
TNRC6C-AS1
Low prenatal SES was associated with higher methylation of a CpG on chromosome 17 in the body of TNRC6C-AS1 gene [38–40]. The TNRC6C-AS1 gene is expressed as an antisense noncoding RNA predominately in the spleen and lymph nodes [47,48], and regulates expression of TNRC6C protein which is itself an important regulator of proliferation, apoptosis and migration of certain cancer cells [49]. Dysregulation of TNRC6C-AS1 and TNRC6C have been implicated in some cancers [50,51].
MTMR4
We also found that low prenatal SES was related to lower methylation on chromosome 17 in the body of the MTMR4 [52]. MTMR4 is ubiquitously expressed in many tissues and modulates cellular signalling through dephosphorylation of proteins involved in cell differentiation, growth and death [52–55]. Inadequate MTMR4 expression can disrupt metabolic homeostasis and autophagy in cells [56], which are basic physiological precursors of noncommunicable pathologies, like cancer, metabolic syndrome and neurodegenerative disease [57–59] that disproportionately afflict persons of low socioeconomic position [60,61]. Thus, the effects of prenatal SES on early life epigenetic modifications that are related to basic cellular processes, like autophagy, may contribute to the development later life disease and mortality.
Step 2: Persistence of associations into early & mid-childhood
The three genes described above did not remain differentially methylated during early or mid-childhood. However, of the 29 CpG sites that passed FDR correction, the association of low prenatal SES with higher methylation of a CpG site on chromosome 20 in the 5′ untranslated region of LRRN4 [38–40] remained apparent in early childhood. An early mouse study that characterized LRRN4 found that expression of this gene is critical for long-term memory formation in the hippocampus [62]. LRRN4, which also is highly expressed in the striatum of the brain beginning around late childhood, shows a pattern of hypomethylation in cell lines derived from patients with schizophrenia [63]. Furthermore, researchers have also linked expression of LRRN4 in cardiac tissue with heart disease [64]. The relationship between prenatal SES, DNA methylation and expression of the LRRN4 gene, and the development of cardiac pathology and schizophrenia is intriguing as these diseases unduly afflict socially-disadvantaged persons [5,45,65–67]. So, while the positive association between prenatal SES and LRRN4 DNA methylation was significant only after FDR correction (but not Bonferroni correction), the potential biological relevance of this CpG site is interesting given the persistence of the SES ‘effect’ from birth through early childhood, as well as the functional role LRRN4 in SES-related pathologies.
Our findings do not stand alone. A recent EWAS found that components of SES – specifically, family income and parental education – during early childhood (∼5 years of age) was associated with differential methylation of genes involved in immune function and growth at age 9 years [68]. Additionally, three other EWAS analyses revealed that childhood SES, more than adult SES, is associated with DNA methylation of genes involved in inflammation and cell signaling during adulthood [16,17,19]. At last, a large longitudinal cohort study in the UK (the ALSPAC Study) recently reported that prenatal maternal education corresponded with differential methylation in cord blood and adolescence, albeit the authors found no evidence of persistence effects from birth through adolescence [20]. These studies, in conjunction with our results, suggest an influence of early social factors on DNA methylation of CpG sites involved in inflammation, cell signaling and immune function.
While the magnitude of effects that we detected in the EWAS are small, they are comparable to the effect of prenatal smoking [69], which is recognized as an important determinant of cord blood and offspring DNA methylation. Additionally, a recent study of studies conducted within Children’s Environmental Health and Disease Prevention Research Centers across the USA support the relevance of small effect sizes in environmental epigenetic studies given that they are replicable over time and across populations [70]. Thus, while the functionality of small differences in DNA methylation remain yet to be elucidated, such findings should not be disregarded, but rather, explored in other settings.
Step 3: Associations of prenatal SES with global DNA methylation & hydroxymethylation
The prenatal SES index was not associated with global measures of DNA methylation at any time points. However, we noted that specific individual components of the SES index were both positively and inversely associated with global measures of DNA methylation, but these associations were not consistent across the three life stages with the exception of the inverse relation of maternal education and %5mC during early and mid-childhood. This finding was unexpected as we anticipated that children born to more highly educated mothers would have higher global DNA methylation. While a number of explanations could explain the relationships between components of the SES index and global DNA methylation (i.e., residual confounding by life style characteristics such as micronutrient intake [24] and/or exposure to environmental toxicants [71]), we are cautious to not over interpret the biological importance of any specific association within this realm given the overall null results of the prenatal SES index with the global measures of DNA methylation, the number of tests carried out and the lack of consistency of associations across the individual SES components. Moreover, the literature surrounding associations of SES and global DNA methylation is mixed: some studies reported null associations [72], although others have reported positive [24] and inverse associations [15]. These discrepancies may be attributable to the timing and type of SES assessment [73], the type of metric used to evaluate global DNA methylation (i.e., liquid chromatography-based techniques vs repetitive elements like LINE-1 and Alu) [74], as well as the complexity that is embedded both in social environments and the epigenome.
Strengths & limitations
A key strength of our study is our ability to prospectively investigate associations of SES with genome-wide DNA methylation via the Infinium HumanMethylation450 BeadChip in 609 individuals at birth, and to explore persistence of epigenetic marks into early and mid-childhood, thereby covering three sensitive periods of development for chronic diseases. We were also able to examine associations of early life SES with liquid chromatography-based measures of global DNA methylation and hydroxymethylation, which may capture minute alterations to DNA methylation processes that are impactful at the genome-wide level. In addition, we were able to control for a number of important perinatal, sociodemographic and lifestyle covariates based on the detailed cohort follow-up information.
However, this study is not without its limitations. First, with respect to the EWAS, the majority of our associations had low variabililty in DNA methylation and within CpG sites/regions were close to 0 or 100% DNA methylation. This may lead to higher risk of false positive findings that are found statistically significant, but with no true biological effects. Second, Project Viva participants are relatively well-educated and all had health insurance. Thus, our findings may not be generalizable to more disadvantaged segments of the population that are most vulnerable to SES-related chronic conditions. Third, related to the previous point, we dichotomized the SES index as the lowest 10% versus upper 90% to achieve maximum discriminatory capacity between low versus high SES while also considering statistical power. As such, our findings may not be relevant to other populations with a different distribution of socioeconomic characteristics. Future studies with a broader range of SES are warranted. Fourth, we did not adjust for paternal education due to a large number of missing values for this variable. While paternal characteristics such as education level may be key contributors to prenatal SES, we surmise that their effect is captured, to some extent, by mother-reported annual household income and the census-tract variables of neighborhood income and poverty. Fifth, while it would be ideal to explore our research questions in a subsample of Project Viva with information on prenatal SES and DNA methylation across all three life stages, this only applies to 53 participants (of whom n = 3 had low prenatal SES), which are too few to conduct longitudinal analyses of DNA methylation. Thus, we acknowledge that the associations explored are not in the same sample of participants, which may limit generalizability of EWAS findings to the later life stages. However, an assessment of background and sociodemographic characteristics of the three subsamples did not indicate any marked discrepancies across the subsamples. At last, our findings have not been validated in an external population, which is an important next step for any high-dimensional ‘omics anlayses.
Conclusion
In this contemporary pre-birth cohort, prenatal SES was associated with cord blood DNA methylation of genes involved in basic physiological processes, like mitochondrial fatty acid oxidation (ACSF3), cell differentiation and apoptosis (TNRC6C-AS1) and cellular signaling (MTMR4). We also noted a positive association between prenatal SES and DNA methylation of the LRRN4 gene from birth through early childhood. This finding is noteworthy given that LRRN4 has been implicated in risk of chronic diseases that disparately afflict low-SES populations, including cardiovascular disease and psychiatric conditions. We did not find any associations of prenatal SES with global DNA methylation or hydroxymethylation.
Future perspective
Additional research is warranted to explore long-term health outcomes of altered DNA methylation of the above-mentioned genes; to explore the opposing nature of the associations of individual prenatal SES metrics with the global measures of DNA methylation at two different life stages; and to evaluate the extent to which the effect sizes detected in this study contribute to SES-patterned health disparities.
Summary points.
The early life social environment can impart lasting effects on phenotype and health, and DNA methylation may serve as a mechanistic link.
Few studies have examined associations of the prenatal social environment with DNA methylation patterns during multiple life stages.
First, we examined associations of maternal socioeconomic status during the prenatal period (‘prenatal socioeconomic status’) with DNA methylation in cord blood via an epigenome-wide association study among participants of Project Viva, a Boston-area prebirth cohort.
Next, we evaluated whether significant hits in cord blood persisted into early childhood (age 3–6 years) and mid-childhood (age 6–10 years).
We identified four CpG sites/regions of interest: ACSF3, TNRC6C-AS1, MTMR4 and LRRN4.
Of these genes, only the association of lower socioeconomic status with higher LRRN4 methylation persisted into early childhood.
The role of these genes in regulation of basic cellular processes implicated in chronic diseases sheds light on potential mechanistic processes that underlie development of chronic diseases that disproportionately afflict low-socioeconomic status populations.
Future studies are necessary to validate our findings and ascertain associations of these CpG regions with health outcomes.
Supplementary Material
Footnotes
Author contributions
W Perng and ZM Laubach conceived research idea and led manuscript writing. W Perng, ZM Laubach, S Rifas-Shiman and A Cardenas carried out data analysis. E Oken, D DeMeo, AA Litonjua, RC Duca, L Godderis and A Baccarelli contributed funds to the laboratory assays. E Oken, D DeMeo, AA Litonjua, RC Duca, L Godderis, A Baccarelli and MF Hivert provided critical feedback on the manuscript.
Financial & competing interests disclosure
This study was funded by the NIH R01 HD034568, R01 HL111108, R01 NR013945 and UH3 OD023286. The funders played no role in the analysis or interpretation of data in this study. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Sapolsky RM. Social status and health in humans and other animals. Ann. Rev. Anthropol. 33(1), 393–418 (2004). [Google Scholar]
- 2.Mclaren L. Socioeconomic status and obesity. Epidemiol. Rev. 29, 29–48 (2007). [DOI] [PubMed] [Google Scholar]
- 3.De Mestral C, Stringhini S. Socioeconomic status and cardiovascular disease: an update. Curr. Cardiol. Rep. 19(11), 115 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Allen J, Balfour R, Bell R, Marmot M. Social determinants of mental health. Int. Rev. Psychiatry 26(4), 392–407 (2014). [DOI] [PubMed] [Google Scholar]; • Discusses the role of the social environment on mental health.
- 5.Muntaner C, Eaton WW, Miech R, O'Campo P. Socioeconomic position and major mental disorders. Epidemiol. Rev. 26, 53–62 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Tung J, Barreiro LB, Johnson ZP. et al. Social environment is associated with gene regulatory variation in the rhesus macaque immune system. Proc. Natl Acad. Sci USA. 109(17), 6490 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sapolsky RM. Adrenocortical function, social rank, and personality among wild baboons. Biol. Psychiatry 28(10), 862–878 (1990). [DOI] [PubMed] [Google Scholar]
- 8.Laubach ZM, Perng W, Dolinoy DC, Faulk CD, Holekamp KE, Getty T. Epigenetics and the maintenance of developmental plasticity: extending the signalling theory framework. Biol. Rev. Camb. Philos. Soc. 93(3), 1323–1338 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Waterland RA, Michels KB. Epigenetic epidemiology of the developmental origins hypothesis. Annu. Rev. Nutr. 27, 363–388 (2007). [DOI] [PubMed] [Google Scholar]; • Reviews the role of epigenetics in the study of developmental origins of health and disease (DOHaD).
- 10.Faulk C, Dolinoy DC. Timing is everything: the when and how of environmentally induced changes in the epigenome of animals. Epigenetics 6(7), 791–797 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Massart R, Suderman MJ, Nemoda Z. et al. The signature of maternal social rank in placenta deoxyribonucleic acid methylation profiles in rhesus monkeys. Child Dev. 88(3), 900–918 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Appleton AA, Armstrong DA, Lesseur C. et al. Patterning in placental 11-B hydroxysteroid dehydrogenase methylation according to prenatal socioeconomic adversity. PLoS ONE 8(9), e74691 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.King K, Murphy S, Hoyo C. Epigenetic regulation of Newborns' imprinted genes related to gestational growth: patterning by parental race/ethnicity and maternal socioeconomic status. J. Epidemiol. Community Health 69(7), 639–647 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Obermann-Borst SA, Heijmans BT, Eilers PHC. et al. Periconception maternal smoking and low education are associated with methylation of INSIGF in children at the age of 17 months. J. Dev. Orig. Health Dis. 3(5), 315–320 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Coker ES, Gunier R, Huen K, Holland N, Eskenazi B. DNA methylation and socioeconomic status in a Mexican–American birth cohort. Clin. Epigenetics 10, 61 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Study of socioeconomic status (SES) and cord blood repetitive element (LINE-1 and Alu) DNA methylation in a Hispanic population.
- 16.Borghol N, Suderman M, Mcardle W. et al. Associations with early-life socio-economic position in adult DNA methylation. Int. J. Epidemiol. 41(1), 62–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]; • A genome-wide DNA methylation study that observes stronger associations of childhood socioeconomic position than adult socioeconomic position in relation to adult DNA methylation profiles.
- 17.Lam LL, Emberly E, Fraser HB. et al. Factors underlying variable DNA methylation in a human community cohort. Proc. Natl Acad. Sci. USA 109(Suppl. 2), 17253–17260 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tehranifar P, Wu HC, Fan X. et al. Early life socioeconomic factors and genomic DNA methylation in mid-life. Epigenetics 8(1), 23–27 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Needham BL, Smith JA, Zhao W. et al. Life course socioeconomic status and DNA methylation in genes related to stress reactivity and inflammation: the multi-ethnic study of atherosclerosis. Epigenetics 10(10), 958–969 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alfano R, Guida F, Galobardes B. et al. Socioeconomic position during pregnancy and DNA methylation signatures at three stages across early life: epigenome-wide association studies in the ALSPAC birth cohort. Int. J. Epidemiol. 48(1), 30–44 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oken E, Baccarelli AA, Gold DR. et al. Cohort profile: project viva. Int. J. Epidemiol. 44(1), 37–48 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang C, Guttmann A, To T, Dick PT. Neighborhood income and health outcomes in infants: how do those with complex chronic conditions fare? Arch. Pediatr. Adolesc. Med. 163(7), 608–615 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Slack KS, Holl JL, Yoo J, Amsden LB, Collins E, Bolger K. Welfare, work, and health care access predictors of low-income children's physical health outcomes. Child. Youth Serv. Rev. 29(6), 782–801 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perng W, Rozek LS, Mora-Plazas M. et al. Micronutrient status and global DNA methylation in school-age children. Epigenetics 7(10), 1133–1141 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Finds strong associations of family-level SES during childhood with global DNA methylation according to LINE-1 and Alu methylation. This study additionally finds associations of SES and DNA methylation only at the extreme upper end of the distribution (complementary to the present study which detects associations only at the extreme lower end of the SES distribution).
- 25.Peng C, Cardenas A, Rifas-Shiman SL. et al. Epigenome-wide association study of total serum immunoglobulin E in children: a life course approach. Clin. Epigenetics 10, 55 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sandoval J, Heyn H, Moran S. et al. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics 6(6), 692–702 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Chen YA, Lemire M, Choufani S. et al. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics 8(2), 203–209 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Triche TJ, Jr., Weisenberger DJ, Van Den Berg D, Laird PW, Siegmund KD. Low-level processing of Illumina Infinium DNA Methylation BeadArrays. Nucleic Acids Res. 41(7), e90 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teschendorff AE, Marabita F, Lechner M. et al. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics 29(2), 189–196 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8(1), 118–127 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Godderis L, Schouteden C, Tabish A. et al. Global methylation and hydroxymethylation in DNA from blood and saliva in healthy volunteers. Biomed Res. Int. 2015, 845041 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cardenas A, Rifas-Shiman SL, Godderis L. et al. Prenatal exposure to mercury: associations with global DNA methylation and hydroxymethylation in cord blood and in childhood. Environ. Health Perspect. 125(8), 087022 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perng W, Stuart J, Rifas-Shiman SL, Rich-Edwards JW, Stuebe A, Oken E. Preterm birth and long-term maternal cardiovascular health. Ann. Epidemiol. 25(1), 40–45 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aryee MJ, Jaffe AE, Corrada-Bravo H. et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 30(10), 1363–1369 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bakulski KM, Feinberg JI, Andrews SV. et al. DNA methylation of cord blood cell types: Applications for mixed cell birth studies. Epigenetics 11(5), 354–362 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Breton CV, Marsit CJ, Faustman E. et al. Small-magnitude effect sizes in epigenetic end points are important in children's environmental health studies: the children's environmental health and disease prevention research center's epigenetics working group. Environ. Health Perspect. 125(4), 511–526 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Methodol. 57(1), 289–300 (1995). [Google Scholar]
- 38.Casper J, Zweig AS, Villarreal C. et al. The UCSC Genome Browser database: 2018 update. Nucleic Acids Res. 46(D1), D762–D769 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kent WJ, Sugnet CW, Furey TS. et al. The human genome browser at UCSC. Genome Res. 12(6), 996–1006 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lander ES, Linton LM, Birren B. et al. Initial sequencing and analysis of the human genome. Nature 409(6822), 860–921 (2001). [DOI] [PubMed] [Google Scholar]
- 41.Watkins PA, Maiguel D, Jia Z, Pevsner J. Evidence for 26 distinct acyl-coenzyme A synthetase genes in the human genome. J. Lipid Res. 48(12), 2736–2750 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Witkowski A, Thweatt J, Smith S. Mammalian ACSF3 protein is a malonyl-CoA synthetase that supplies the chain extender units for mitochondrial fatty acid synthesis. J. Biol. Chem. 286(39), 33729–33736 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.National Library of Medicine (Us). Genetics Home Reference: ACSF3 gene (2018). www.ncbi.nlm.nih.gov/gene/197322
- 44.Sloan JL, Johnston JJ, Manoli I. et al. Exome sequencing identifies ACSF3 as a cause of combined malonic and methylmalonic aciduria. Nat. Genet. 43(9), 883–886 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clark AM, Desmeules M, Luo W, Duncan AS, Wielgosz A. Socioeconomic status and cardiovascular disease: risks and implications for care. Nat. Rev. Cardiol. 6(11), 712–722 (2009). [DOI] [PubMed] [Google Scholar]
- 46.Everson SA, Maty SC, Lynch JW, Kaplan GA. Epidemiologic evidence for the relation between socioeconomic status and depression, obesity, and diabetes. J. Psychosom. Res. 53(4), 891–895 (2002). [DOI] [PubMed] [Google Scholar]
- 47.Fagerberg L, Hallstrom BM, Oksvold P. et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteomics 13(2), 397–406 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.National Library of Medicine (Us). Genetics Home Reference. TNRC6C-AS1 gene (2018). www.ncbi.nlm.nih.gov/gene/100131096
- 49.Zipprich JT, Bhattacharyya S, Mathys H, Filipowicz W. Importance of the C-terminal domain of the human GW182 protein TNRC6C for translational repression. RNA 15(5), 781–793 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hou S, Lin Q, Guan F, Lin C. LncRNA TNRC6C-AS1 regulates UNC5B in thyroid cancer to influence cell proliferation, migration, and invasion as a competing endogenous RNA of miR-129-5p. J. Cell. Biochem. 119(10), 8304–8316 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Muhanhali D, Zhai T, Jiang J, Ai Z, Zhu W, Ling Y. Long non-coding antisense RNA TNRC6C-AS1 is activated in papillary thyroid cancer and promotes cancer progression by suppressing TNRC6C Expression. Front. Endocrinol. 9, 360 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.National Library of Medicine (US). Genetics Home Reference. MTMR4 gene (2018). www.ncbi.nlm.nih.gov/gene/9110
- 53.Yu J, He X, Chen YG. et al. Myotubularin-related protein 4 (MTMR4) attenuates BMP/Dpp signaling by dephosphorylation of Smad proteins. J. Biol. Chem. 288(1), 79–88 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu J, Pan L, Qin X. et al. MTMR4 attenuates transforming growth factor beta (TGFbeta) signaling by dephosphorylating R-Smads in endosomes. J. Biol. Chem. 285(11), 8454–8462 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao R, Qi Y, Chen J, Zhao ZJ. FYVE-DS P2, a FYVE domain-containing dual specificity protein phosphatase that dephosphorylates phosphotidylinositol 3-phosphate. Exp. Cell Res. 265(2), 329–338 (2001). [DOI] [PubMed] [Google Scholar]
- 56.Pham HQ, Yoshioka K, Mohri H. et al. MTMR4, a phosphoinositide-specific 3′-phosphatase, regulates TFEB activity and the endocytic and autophagic pathways. Genes Cells (2018) (Epub ahead of print). [DOI] [PubMed] [Google Scholar]
- 57.Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N. Engl. J. Med. 368(7), 651–662 (2013). [DOI] [PubMed] [Google Scholar]
- 58.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell 132(1), 27–42 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature 451(7182), 1069–1075 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mackenbach JP, Stirbu I, Roskam AJ. et al. Socioeconomic inequalities in health in 22 European countries. N. Engl. J. Med. 358(23), 2468–2481 (2008). [DOI] [PubMed] [Google Scholar]
- 61.Marmot M. Social determinants of health inequalities. Lancet 365(9464), 1099–1104 (2005). [DOI] [PubMed] [Google Scholar]
- 62.Bando T, Sekine K, Kobayashi S. et al. Neuronal leucine-rich repeat protein 4 functions in hippocampus-dependent long-lasting memory. Mol. Cell. Biol. 25(10), 4166–4175 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vitale AM, Matigian NA, Cristino AS. et al. DNA methylation in schizophrenia in different patient-derived cell types. NPJ Schizophr. 3, 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li R FJ, Huo B, Su Y. et al. Leucine-rich repeat neuronal protein 4 (LRRN4) potentially functions in dilated cardiomyopathy. Int. J. Clin. Exp. Pathol. 10(9), 9925–9933 (2017). [PMC free article] [PubMed] [Google Scholar]
- 65.Min YI, Anugu P, Butler KR. et al. Cardiovascular disease burden and socioeconomic correlates: findings from the Jackson Heart Study. J. Am. Heart Assoc. 6(8), pii: e004416 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Werner S, Malaspina D, Rabinowitz J. Socioeconomic status at birth is associated with risk of schizophrenia: population-based multilevel study. Schizophr. Bull. 33(6), 1373–1378 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wicks S, Hjern A, Gunnell D, Lewis G, Dalman C. Social adversity in childhood and the risk of developing psychosis: a national cohort study. Am. J. Psychiatry 162(9), 1652–1657 (2005). [DOI] [PubMed] [Google Scholar]
- 68.Bush NR, Edgar RD, Park M. et al. The biological embedding of early-life socioeconomic status and family adversity in children's genome-wide DNA methylation. Epigenomics 10(11), 1445–1461 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Joubert BR, Haberg SE, Nilsen RM. et al. 450K epigenome-wide scan identifies differential DNA methylation in newborns related to maternal smoking during pregnancy. Environ. Health Perspect. 120(10), 1425–1431 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]; •• A highly-cited study as a reference for the effect of maternal behavior on offspring cord blood DNA methylation.
- 70.Breton CV, Marsit CJ, Faustman E. et al. Small-magnitude effect sizes in epigenetic end points are important in children's environmental health studies: the children's environmental health and disease prevention research center's epigenetics working group. Environ. Health Perspect. 125(4), 511–526 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Emphasizes the relevance of small effect sizes in environmnetal studies of pediatric cohorts, and the importance of adjusting for maternal smoking in studies of perinatal exposures on infant and child health.
- 71.Martin EM, Fry RC. Environmental influences on the epigenome: exposure- associated DNA methylation in human populations. Annu. Rev. Public Health 39, 309–333 (2018). [DOI] [PubMed] [Google Scholar]
- 72.Subramanyam MA, Diez-Roux AV, Pilsner JR. et al. Social factors and leukocyte DNA methylation of repetitive sequences: the multi-ethnic study of atherosclerosis. PLoS ONE 8(1), e54018 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Braveman PA, Cubbin C, Egerter S. et al. Socioeconomic status in health research: one size does not fit all. JAMA 294(22), 2879–2888 (2005). [DOI] [PubMed] [Google Scholar]
- 74.Vryer R, Saffery R. What's in a name? Context-dependent significance of ‘global’ methylation measures in human health and disease. Clin. Epigenetics 9, 2 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.