Abstract
Mammalian olfactory sensory neurons (OSNs) form the primary elements of the olfactory system. Inserted in the olfactory mucosa lining of the nasal cavity, they are exposed to the environment and their lifespan is brief. Several reports say that OSNs are regularly regenerated during the entire life and that odorant environment affects the olfactory epithelium. However, little is known about the impact of the odorant environment on OSNs at the cellular level and more precisely in the context of early postnatal olfactory exposure. Here we exposed MOR23-green fluorescent protein (GFP) and M71-GFP mice to lyral or acetophenone, ligands for MOR23 or M71, respectively. Daily postnatal exposure to lyral induces plasticity in the population of OSNs expressing MOR23. Their density decreases after odorant exposure, whereas the amount of MOR23 mRNA and protein remain stable in the whole epithelium. Meanwhile, quantitative PCR indicates that each MOR23 neuron has higher levels of olfactory receptor transcripts and also expresses more CNGA2 and phosphodiesterase 1C, fundamental olfactory transduction pathway proteins. Transcript levels return to baseline after 4 weeks recovery. Patch-clamp recordings reveal that exposed MOR23 neurons respond to lyral with higher sensitivity and broader dynamic range while the responses' kinetics were faster. These effects are specific to the odorant–receptor pair lyral–MOR23: there was no effect of acetophenone on MOR23 neurons and no effect of acetophenone and lyral on the M71 population. Together, our results clearly demonstrate that OSNs undergo specific anatomical, molecular, and functional adaptation when chronically exposed to odorants in the early stage of life.
Keywords: development, electrophysiology, mice, molecular biology, olfaction, plasticity
Introduction
Olfactory sensory neurons (OSNs) constitute the primary elements of the olfactory system. They convert the chemical information into an electrical signal, which is then processed by the brain. Each OSN expresses one receptor selected from a large multigene family (Buck and Axel, 1991). These receptors are G-protein-coupled receptors and the vast majority of OSNs rely on the canonical cAMP transduction pathway. This pathway involves activation of some key elements, including olfactory-specific G-protein, adenylyl cyclase type III (ACIII), and cyclic nucleotide-gated (CNG) channel (Kleene, 2008).
Embedded in the epithelium lining of the nasal cavity, OSNs are continuously exposed to environmental factors, such as odorant molecules and pollutants. So far, the effects of the olfactory environment on the olfactory system have been primarily analyzed in terms of olfactory behaviors and at the olfactory bulb (OB) level. Odorant enrichment improves the general olfactory discrimination abilities without specificity to the enriched odor (Mandairon et al., 2006a,b). This enhancement of olfactory abilities is correlated with an increase in survival and activity of OB interneurons and a modulation of mitral cell responses. Olfactory enrichment promotes the survival of interneurons both in the adult OB (Rochefort et al., 2002) and during postnatal development (Woo et al., 2006). The survival of interneurons is associated with an increase of their activity (Mandairon et al., 2008). As a result, mitral cell activity is reduced (Buonviso et al., 1998).
By contrast, fewer reports have discussed the effects of odor enrichment on OSNs. Enrichment improves the refinement of the glomerular map (Kerr and Belluscio, 2006) and targeting of OSN projections (Col et al., 2007). Repeated stimulations increase olfactory sensitivity at the periphery (Wang et al., 1993). This increase suggests survival of stimulated OSNs and the induction of the expression of odorant-specific receptors (Wang et al., 1993). Another report shows that exposure promotes survival of stimulated OSNs (Zhao and Reed, 2001). Regarding the effect on a specific OSN population, studies show opposite results: odorant exposure either promotes survival of OSNs (Watt et al., 2004; Santoro and Dulac, 2012; François et al., 2013; Dias and Ressler, 2014) or decreases the number of OSNs (Cavallin et al., 2010). The consequences of the odorant may also depend on the context of the exposure: associated with a stress situation, olfactory exposure leads to an increase of a specific OSN population that is transmitted to subsequent generations (Dias and Ressler, 2014). Last, there is no data on the functional and molecular consequences of odor exposure on specific populations of OSNs at the cellular level.
Here we report that daily postnatal exposure to a specific odorant, lyral, induces plasticity at the cellular and molecular level in OSNs. Such exposure leads to a strong decrease of the MOR23-expressing OSN population. Surprisingly, this decrease is not correlated with the amount of MOR23 mRNA and protein, which remains stable at the entire epithelium level. Similarly, EOG recordings in response to lyral and other odorants were not altered by lyral exposure, indicating that the olfactory epithelium is not globally affected by the lyral exposition. Meanwhile, using quantitative PCR (qPCR) at the cellular level, we show that each MOR23 neuron expresses more MOR23 olfactory receptors, CNGA2, and phosphodiesterase 1C (PDE1C), which are all key elements of the transduction pathway. Using patch-clamp recordings, we show that exposed MOR23 neurons respond to lyral with higher sensitivity, broader dynamic range, faster rise time, and shorter responses, while the overall amplitude did not change. MOR23 neurons exposed to nonligand odorant acetophenone do not exhibit any change. Interestingly M71 neurons exposed either to acetophenone, its ligand, or to lyral do not exhibit any change. Our results clearly demonstrate that postnatal odorant exposure induces molecular and cellular remodeling in olfactory sensory neurons at the cellular level. Through this plasticity, OSNs can adapt to their environment.
Materials and Methods
Animals
Gene-targeted MOR23-IRES-tauGFP mice were used to monitor the MOR23 neurons. These neurons express the green fluorescent protein (GFP) under the control of the promoter of the MOR23 odorant receptor (Vassalli et al., 2002). Similarly, gene-targeted M71-IRES-tauGFP mice were used to monitor the M71 neurons (Bozza et al., 2002). Animals were allowed access to food and water ad libitum and were kept on a 12 h light/dark cycle, with a constant temperature. All procedures were approved by the Université de Bourgogne ethics committee.
Odorant exposure
Odors were presented using a tea ball hanging from the cover of the cage (dimensions of the cage, 265 × 160 × 142 mm, with a filter top on the cage), 3 cm above the bedding and containing 20 μl of the pure odorant on a cotton ball. After exposure, the cages were kept open in the room near a fume hood to let the odorant in the cage dissipate. Animals were exposed to the odorant 1 h each day for 3 weeks. We used the term “control animals” for nonexposed mice since we observed no effect of either an empty tea ball or an empty tea ball containing cotton or containing cotton impregnated with mineral oil hanging in the cage (see Results).
For recovery, animals were kept for another 4 weeks without further odorant exposure; control animals were nonexposed from postnatal day (P) 0 to P21 and kept for another 4 weeks. These two “recovery” groups were therefore 7 weeks old at the time of analysis.
Unless specified, all chemicals were from Sigma-Aldrich. Lyral was provided as a generous gift from International Fragrances and Flavors.
Histology
Olfactory epithelium.
To check whether GFP-containing cells visible in the flat-mount preparation were in fact OSNs, we performed cryosections of P21 MOR23 GFP mice. Briefly mice were decapitated following anesthesia and the head was immersed in PBS containing 4% paraformaldehyde and 0.2% glutaraldehyde overnight at 4°C, rinsed with PBS, immersed in decalcification solution (0.5 m EDTA) for 2 d, and cryoprotected with 30% sucrose in PBS. Ten-micrometer-thick cryosections were mounted between slide and coverslip using Vectashield containing DAPI (Vector Labs). Olfactory epithelium slices were visualized using a sp2 confocal microscope and a 63× oil-immersion objective. GFP and DAPI were excited at 488 and 405 nm, respectively.
Flat-mount preparation.
Septal olfactory epithelia were removed and washed in cold Ringer's solution, which contained the following (in mm): 124 NaCl, 3 KCl, 1.3 MgSO4, 2 CaCl2, 26 NaHCO3, 1.25 NaH2PO4, 15 glucose, pH 7.6 and 305 mOsm. The epithelia were further immersed in PBS containing 4% paraformaldehyde and 0.2% glutaraldehyde for 1 h at room temperature. They were subsequently washed in PBS and mounted using DAKO mounting medium between a slide and a coverslip. The septal epithelia were visualized using a Olympus BX51WI microscope (10× objective; Olympus UPLFLN10x; numerical aperture, 0.30) coupled to a Olympus DP72 camera with a 4140 × 3096 resolution. GFP-containing neurons were observed using a GFP filter after excitation through a BP460-490 filter with light from a mercury HBO 100W Olympus lamp. Images covering the entire epithelium were taken and the whole tissue was reconstituted using the MosaicJ plugins of ImageJ software (National Institutes of Health) and the neuronal density was calculated as the ratio of the number of GFP-containing neurons against the total olfactory sensory epithelium surface. We used SigmaPlot (Systat) to produce a graph representing neuronal densities in both control and exposed conditions and to implement the statistics (two-sample, two-tailed homoscedastic t tests). Data were expressed as neurons/mm2 of olfactory sensory epithelium ± SE. Note that the picture for Figure 1C was taken from a flat-mounted septal olfactory epithelium using a sp2 confocal microscope (Leica) and a 63× oil-immersion objective. Figure 1D,E was taken using the same microscope but with a 20× objective. In both cases GFP was excited at 488 nm.
Figure 1.
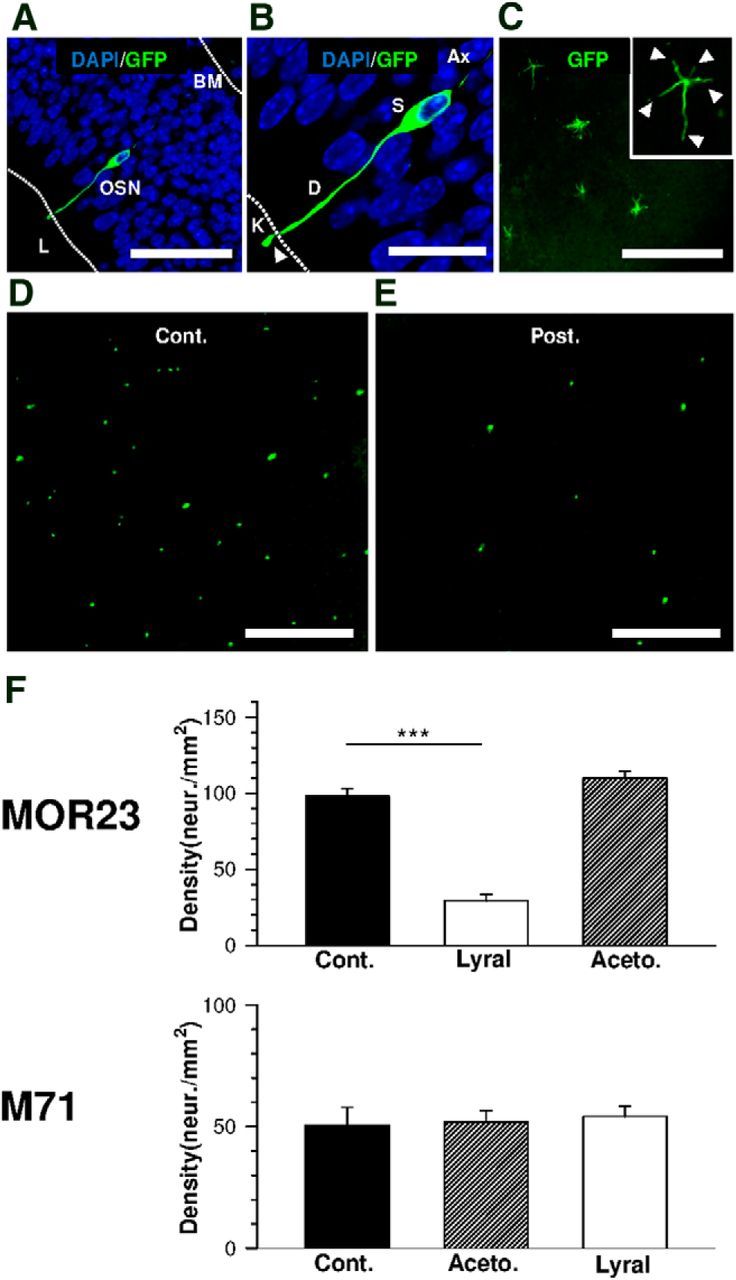
Postnatal exposure to lyral significantly reduces the density of MOR23-GFP olfactory sensory neurons. A, Confocal image showing an individual MOR23-GFP OSN within the septal olfactory epithelium. BM, Basal membrane; L, lumen. Scale bar, 50 μm. B, Magnification of the OSN presented in A. The somas (S) of OSNs are located within the epithelium in nuclei-rich area. Individual neurons project to the surface through a dendritic process (D). Knob (K) carries cilia (white arrow). Scale bar, 20 μm. C, Confocal image showing individual MOR23-GFP OSN knobs in a septal olfactory epithelium flat-mount preparation. A close-up look at an individual knob can be seen in the right upper corner of the picture (white arrows indicate the cilia). Scale bar, 50 μm. D, E, Confocal images showing a decrease in MOR23-GFP neuron density when mice are exposed to lyral between P0 and P21. D, Control (Cont.). E, Postnatal exposure (Post.). Scale bar, 200 μm. F, Bar graphs summarizing the effects of odorant exposure on MOR23 and M71 population. The neuronal density was calculated as the number of neurons per epithelium/total surface of the epithelium (see Materials and Methods). Top, Summary of the density of the MOR23 population after postnatal exposure to lyral (n = 20) or acetophenone (Aceto.; n = 12); n = 23 for control (Cont.). Bottom, Summary of the density of the M71 population after postnatal exposure to lyral (n = 5) or acetophenone (n = 7); n = 10 for control. ***p < 0.001.
Dissociation and cell collection for molecular biology
The olfactory epithelium was incubated in an enzymatic dissociation solution (papain from Carica papaya, 0.2 mg/tube) at 37°C for 10 min while gently shaking. The epithelium was then mechanically dissociated and the nondissociated fragments and debris were removed by passing the sample through a cell strainer (40 μm). The cells were incubated at room temperature for 20 min on polyethylenimine-coated glass and washed before collection. Borosilicate glass capillaries were pulled using a Sutter P97 pipette puller (Sutter Instruments). The diameter of the pipette tip was 5.2 μm. Single GFP cells were collected under a microscope (Olympus BX51WI) equipped with a micromanipulator and fluorescence microscopy capabilities. After collecting a single GFP cell, the pipette was removed from the holder, and the tip was broken into a standard PCR tube containing 2 μl of single-cell lysis buffer solution (SideStep Lysis and Stabilization Buffer, Stratagene). PCR tubes containing single cells were stored on ice during collection and stored at −80°C until reverse transcription. As a control for contamination and inhibition of downstream reactions, 1 μl of medium surrounding the cells was collected and analyzed along with single-cell samples.
Preparation of cDNA
For cDNA from seven cells, all processes were performed in one tube to minimize sample loss. Following single-cell lysis, we added 3.4 μl of shrimp DNase solution (0.2U DNase shrimp in 40 mm Tris-HCl, pH 8, 14 mm MgCl2; Affymetrix) and mixed. The sample solution was heated to 37°C for 10 min to enable digestion of genomic DNA. Reverse transcription was performed using 115U of Superscript III reverse transcriptase (Invitrogen) and random hexamers according to the manufacturer's instructions.
For cDNA from the entire olfactory epithelium, total RNA was extracted using the Quiagen RNeasy Plus Mini Kit. One hundred nanograms of total RNA in the final volume of 10 μl were subjected to reverse transcription as described for single-cell experiments.
RNA analysis (seven cell samples and whole olfactory epithelium)
qPCR amplifications were performed on StepOnePlus Fast Real-Time PCR system (Applied Biosystems) in a final volume of 10 μl with 5 μl of SsoFast EvaGreen Supermix (Bio-Rad Laboratories), 2 μl of cDNA (from collected cells), and 0.2 μm primers according to the manufacturer's specification.
For the whole olfactory epithelium, qPCRs were performed in a final volume of 20 μl with 10 μl of Sso Fast Eva Green Supermix (Bio-Rad Laboratories), 0.01 μg of cDNA, and 0.2 μm primers according to the manufacturer's specification The following cycling parameters were applied: 95°C for 20 s and 40 cycles of amplification (95°C for 3 s; 60°C for 30 s) directly followed by melt curve acquisition from 65 to 95°C. Gene-specific primers (Table 1) were designed according to standard recommendations with primer design software (Primer Express 3.0). Primer sequence specificities were analyzed using Primer-Blast. The uniqueness of PCR products obtained was controlled at the end of amplification using melting curve analysis. All amplicons were also sequenced. The relative expression levels of genes of interest were analyzed using the 2−ΔΔCt method by normalization with succinyl-CoA ligase (Susclg) gene expression and presented as relative quantity (RQ) compared with the control group. Statistical analysis (two-sample, two-tailed homoscedastic t tests) was performed using the built-in macro in Excel.
Table 1.
PCR conditions, slope, correlation coefficient (R2) and efficiency values (E) for each primer
| Gene (Consensus Coding Sequence identification number) | Sequence | Product length (bp) | Annealing (°C) | Slope | R2 | E |
|---|---|---|---|---|---|---|
| Olf16 (15521.1) | F: 5′-CTTGTTGGTTTGTGGGTCTTTAGG-3′; R: 5′-CAAAAGGGCAAATGGAACATG-3′ | 80 | 60 | −3.33 | 0.99 | 1.99 |
| Ac3 (49017.1) | F: 5′-AGGCGACTGTTACTACTGCATCTG-3′; R: 5′-ACGTACGAGATGGCCTCTACCA-3′ | 109 | 65 | −3.4 | 0.99 | 1.96 |
| Cnga2 (30184.1) | F: 5′-GGCACCAAAAAGAAATTTGAACTG-3′; R: 5′-CAGGCTCTGGCTACCAACAGA-3′ | 101 | 62 | −3.28 | 0.99 | 2.01 |
| Pde1c (51784.1) | F: 5′-GTCCCAGCGTCGTGATTAGC-3′; R: 5′-TCATGACATCTCGAGCAAGTCTTT-3′ | 66 | 60 | −3.26 | 0.99 | 1.99 |
| Gapdh (51913.1) | F: 5′-CCTGGAGAAACCTGCCAAGTAT-3′; R: 5′-CTGTTGAAGTCGCAGGAGACAA-3′ | 77 | 62 | |||
| Suclg (27271.1) | F: 5′-GCGTCATCAACCCTGGAGAA-3′; R: 5′-CTGCCCCAATCCGACTTG-3′ | 139 | 60 | −3.2 | 0.99 | 2.05 |
Analysis of 42 genes
The expression of 42 genes (plus two endogenous control genes) was analyzed using qPCR with the custom TaqMan Array Fast Plate (StepOnePlus Fast Real-Time PCR system, Applied Biosystems). The RQ gene expression data were analyzed using Data Assist Software (Applied Biosystems). A two-sample, two-tailed Student's t test comparing the ΔCt values of the two groups was performed and a p value was calculated. The p value was adjusted with the Benjamini–Hochberg method (Benjamini and Hochberg, 1995), and pairs with p < 0.05 were considered as the statistically significant pairs. β-Actin and hypoxanthine ribosyltransferase (HPRT) were used to normalize the data. Data from nonexposed groups were considered as the reference. A gene was considered in Data Assist Software significantly upregulated or downregulated if both following conditions were present: (1) the value for RQ was >1.5 (upregulation) or <0.5 (downregulation) and (2) the p value was <0.05.
Cellular protein extracts and immunoblotting analysis
Cellular protein extracts were prepared using a cell lysis buffer containing 50 mm tris-HCl, 1% SDS, 1 mm EDTA, 1 mm EGTA, 1 mm NaF, and 1 mm Na3VO4, and supplemented with a protease inhibitor mixture (Calbiochem). Protein samples (25 μg) were next subjected to PAGE in a 12% gel and electrophoretically transferred to PVDF membrane (Trans-blot transfer pack, Bio-Rad Laboratories). After blocking with Tris-buffer saline containing 0.5% casein and 0.1% Tween 20 for 1 h at room temperature, membranes were incubated overnight at 4°C with primary Ab (monoclonal rabbit anti-GFP, 1:350; reference number ab6556, Abcam). After incubation with HRP-conjugated secondary Ab (polyclonal goat anti-rabbit, 1:200; Dako) for 1 h at room temperature, immunolabeled proteins were visualized using an HRP chemiluminescent substrate. Images were acquired with a CCD-based system (chemiDoc TM XRS+, Bio-Rad Laboratories) and bands quantified with ImageLab software (Bio-Rad Laboratories). The signal intensities of the GFP band of exposed mice (n = 3) was compared with that of control mice (n = 3). Relative quantification of GFP protein level was performed after normalization to β-actin used as a loading control (see Fig. 6A1). Data were normalized to control mice GFP band [set to 1 arbitrary unit (a.u.)]. Statistical analysis (two-sample, two-tailed homoscedastic t tests) was performed using the built-in macro in Excel.
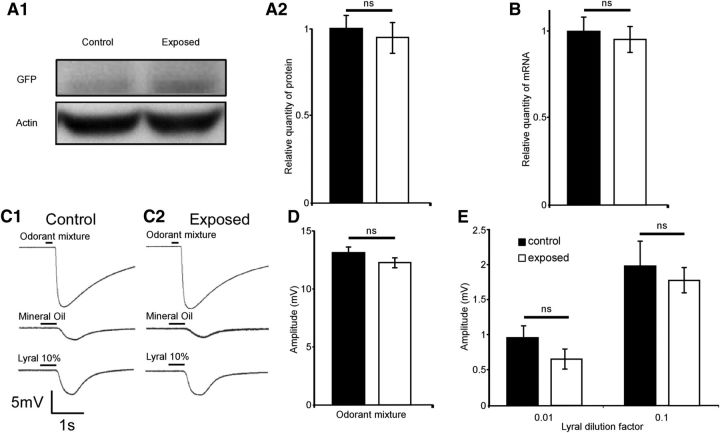
Figure 6.
Postnatal exposure to lyral does not affect the expression of MOR23 at the olfactory epithelium level and does not modify the response of the epithelium to lyral. A1, Western blot of exposed and control epitheliums for GFP and the control protein actin. Band densitometry was performed using Image Lab software (Bio-Rad Laboratories) gels for quantification of signals from three control and three exposed animals. A2, Results are presented in GFP band volume normalized to actin, a loading control. B, Quantification by reverse-transcription qPCR of the level of MOR23 mRNA in olfactory epithelium from exposed or nonexposed mice. Results were normalized to Suclg for each sample. The results are presented in arbitrary units and the values obtained for nonexposed mice were set at 1 (n = 10 for both groups). C–E, Consequences of postnatal exposure to lyral on EOG recordings in response to an odor mixture, mineral oil, and lyral. C, Representative recordings in response to 200 ms stimulus of 1:1000 dilution of the odor mixture (top), 500 ms stimulus with mineral oil (middle), and 500 ms stimulus of lyral 10% dilution (bottom) in a control mouse (C1) and an exposed mouse (C2). D, Quantification histogram of the maximum amplitude of the response to the 1:1000 dilution of the mixture in control mice (black bar, n = 17) and exposed mice (white bar, n = 36). E, Quantification histogram of the maximum amplitude of the response to 1% (left; n = 17 for control mice; n = 44 for exposed mice) and 10% (right; n = 17 for control mice; n = 37 for exposed mice) dilution of lyral. ns, Not significant; error bars indicate SEM.
Patch-clamp recordings
These recordings were performed as described earlier (Grosmaitre et al., 2006). Male or female gene-targeted 3-week-old mice were anesthetized by injection of ketamine HCl and xylazine (150 and 10 mg/kg body weight, respectively), and then decapitated. The head was immediately put into ice-cold Ringer's solution, which contained the following (in mm): 124 NaCl, 3 KCl, 1.3 MgSO4, 2 CaCl2, 26 NaHCO3, 1.25 NaH2PO4, glucose 15, pH 7.6 and 305 mOsm. The pH was kept at 7.4 by bubbling with 95% O2 and 5% CO2. The nose was dissected out en bloc. The olfactory epithelium attached to the nasal septum and the dorsal recess was removed and kept in oxygenated Ringer's solution. Right before starting the recording session, the entire epithelium was peeled away from the underlying bone and transferred to a recording chamber with the mucus layer facing up. Oxygenated Ringer's solution was continuously perfused at room temperature.
The dendritic knobs of OSNs were visualized through an upright microscope (Olympus BX51WI) equipped with an Olympus DP72 camera and a 40× water-immersion objective (numerical aperture, 0.8). An extra 2× magnification was achieved by a magnifying lens in the light path. The GFP-labeled cells were visualized under fluorescent illumination. Superimposition of the fluorescent and bright-field images allowed identification of the fluorescent cells under bright field, which directed the recording pipettes. Electrophysiological recordings were controlled by an EPC-10 USB amplifier combined with Patchmaster software (HEKA Electronic). Perforated patch clamp was performed on the dendritic knobs by including 260 μm nystatin in the recording pipette, which was filled with the following solution (in mm): 70 KCl, 53 KOH, 30 methanesulfonic acid, 5 EGTA, 10 HEPES, 70 sucrose, pH 7.2 (KOH) and 310 mOsm. The junction potential was ∼9 mV and was corrected in all experiments off-line. For odorant-induced transduction currents, signals were sampled at 20 kHz. Under voltage-clamp mode, the signals were initially filtered at 10 kHz and then at 2.9 kHz.
A seven-barrel pipette was used to deliver stimuli by pressure ejection through a picospritzer (Pressure System IIe, Toohey). The stimulus electrode was placed ∼20 μm downstream from the recording site. Distance and pressure were adjusted to minimize mechanical responses (Grosmaitre et al., 2007). All stimuli were delivered at a 138 kPa (∼20 psi) pressure indicated on the picospritzer with 300 ms pulse length. Odorant stimuli were prepared in 0.5 m solution in dimethylsulfoxide and kept at −20°C. Final solutions were prepared before each experiment by adding Ringer's solution.
Data were analyzed using Fitmaster (HEKA). Maximum amplitude of the response and kinetics characteristics were measured. Dose–response curves were drafted and fitted using Origin software (OriginLab). Statistical analysis (t tests and Kolmogorov–Smirnov tests followed by Mann–Whitney U tests for two independent samples) were performed using Origin software (OriginLab).
EOG recordings
EOG recordings were made from the olfactory mucosa in an opened nasal cavity configuration on mouse hemiheads as described earlier (Negroni et al., 2012). The hemihead was placed in a recording chamber under an upright Olympus SZ51 stereo microscope equipped with a low-magnification objective (0.8 to 4×) and two MX-160 micromanipulators (Siskiyou). The odor stimulation device was modified from Scott and Brierley (1999). The hemihead was kept under a constant flow of humidified filtered air (∼1000 ml/min) delivered close to the septum through a 9 mm glass tube. This tube was positioned ∼2 cm from the epithelial surface. Odor stimulations were performed by blowing air puffs (200 ms, 200 ml/min) through an exchangeable Pasteur pipette enclosed in the glass tube containing a filter paper impregnated with 20 μl of the odorant. Odorants were diluted in mineral oil (Sigma-Aldrich). To prevent variable accumulations of the odorant in the pipette, an air flush was applied to the Pasteur pipette before it was placed in the glass tube. The odorants used were either a mixture of 12 odorants (equimolar mixture of anisole, citral, heptanal, isoamyl acetate, lyral, lilial, octanol, 1-4-cineol, isomenthone, limonene, l-carvone, and pyridine) diluted to a final concentration of 10−3 m; lyral from 1:100 to 1:10. As lyral vapor pressure point is very low compared with other odorants (from 103 to 104 times lower than classical odorants used in the odorant mixture), we had to perform longer stimulation (500 ms, 200 ml/min) and used longer filter paper impregnated with 100 μl of lyral to get signal significantly higher than signal with only mineral oil.
EOG voltage signals were recorded using an XtraCell two-channel amplifier (Dipsi) used in a DC current-clamp configuration (I = 0), low-pass Bessel filtered at 1 kHz, and digitized at a rate of 2 kHz using a Digidata 1322a analog-to-digital converter (Molecular Devices) interfaced to a Pentium PC and Pclamp 9.2 software (Molecular Devices). A reference Ag/AgCl electrode was placed on the frontal bone overlaying the OB. Recordings were made with glass micropipettes of 4–5 MΩ filled with a saline solution. EOGs were recorded from two separated positions localized centrally on the septum. These positions gave robust, reproducible, and long-lasting EOG recordings ranging from 10 to 17 mV when stimulated with the mixture of odorants. Odorant-free air stimulation (with mineral oil) always produced signals of ∼1 mV amplitude. Analyses were performed using Clampfit 9.2 (Molecular Devices). We measured the peak amplitude, onset slope (from 10 to 90%), fast-decay slopes (from 100 to 50%), and slow-decay slopes(from 40 to 10%). Data shown in Results represent the amplitude of response to lyral minus the amplitude with mineral oil alone. Statistical analysis (two-sample, two-tailed heteroscedastic t tests) was performed using the built-in macro in Excel.
Results
Postnatal exposure to lyral reduces the density of MOR23 neurons on the septum epithelium
First, we checked whether GFP labeling was indeed seen in OSNs. We performed cryosections on P21 MOR23 mouse olfactory epithelium. As depicted in Figure 1A,B, GFP-containing cells corresponding to MOR23 neurons presented all the characteristics of mature olfactory sensory neurons, i.e., a dendritic knob, a dendrite, a soma, and an axon projecting to the OB (data not shown). Olfactory cilia were only fully visible when the septal olfactory epithelium was flat mounted as depicted in Figure 1C (see Materials and Methods). To analyze the consequences of the odor exposure on the MOR23 population, we used the very same flat-mount preparation of the dorsal septal epithelium. In this preparation, the total number of GFP-labeled OSNs located on the dorsoseptal epithelium can be monitored (Fig. 1D). Using the total surface of the epithelium, we could then calculate the density of GFP-labeled OSNs in number of neurons per square millimeter. In control MOR23 mice, the density of MOR23-GFP neurons was 98.1 ± 5.1 neurons/mm2 (n = 23), with no difference between the left and the right side. In lyral-exposed mice, the MOR23-GFP neuronal density dropped to 29.3 ± 4.1 neurons/mm2 (n = 10), a 70% reduction, as depicted in Figure 1E,F (top). Postnatal exposure to lyral induced a significant decrease (p < 0.001) in the MOR23-GFP population in the dorsal septal epithelium. We also exposed MOR23 mice to acetophenone, a nonligand odorant for MOR23. No difference was ever seen between the control animals (MOR23 neuronal density, 98.1 ± 5.1 neurons/mm2; n = 23) and the acetophenone-treated animals (MOR23 neuronal density, 109.9 ± 4.5 neurons/mm2; n = 12; p = 0.14; Fig. 1F, top). Note that no significant differences were found between the control values and when mice were in presence of the tea ball infuser containing a cotton pad (density, 90.3 ± 8.8 neurons/mm2; n = 8) or a cotton pad impregnated with mineral oil (density, 93.9 ± 4.3 neurons/mm2; n = 14). The decrease in MOR23 neuronal density was partially reversible, returning to 73.0% of the control 4 weeks after postnatal exposure.
We further exposed M71-GFP transgenic mice to lyral or acetophenone following the same protocol from P0 to P21. No difference was ever seen between the control animals (M71 neuronal density, 50.6 ± 7.2 neurons/mm2; n = 10) and the lyral-treated animals (M71 neuronal density, 54.2 ± 4.3 neurons/mm2; n = 5; p = 0.75). Interestingly, no effect was seen also after acetophenone exposure with no difference between the control and the acetophenone-treated animals (M71 neuronal density, 51.7 ± 4.8 neurons/mm2; n = 7; p = 0.91). Together, our results indicate that postnatal exposure to lyral induced an odorant-specific MOR23 neuronal loss during early postnatal development. Since the exposure to the M71 ligand did not induce any change in the M71 population, this effect is not similar among all olfactory receptors.
Postnatal exposure to lyral triggers an increase in mRNA-level expression of MOR23, CNGA2, and PDE1C at cellular level in MOR23-expressing neurons
To analyze mRNA at the neuronal level, we developed a method of reverse-transcription qPCR analysis on dissociated cells (see Materials and Methods). We performed these experiments on small samples of seven neurons to minimize mRNA expression variations between neurons: seven neurons were collected and pooled per animal. Our cell dissociation method allowed us to observe and harvest specifically the GFP-labeled OSNs (Fig. 2A). Transcript levels for olfactory receptor MOR23, cyclic nucleotide gated channel (CNGA2), ACIII, and PDE1C were analyzed. mRNA levels for target genes were normalized to that of Suclg used as a reference gene. Samples containing isolated cells show qPCR amplification signal for all the genes of interest compared with medium used as a negative control. For the determination of a stable endogenous gene, three genes commonly used for normalization were tested: Suclg (ligase Krebs cycle), HPRT (involved in metabolic recycling of nucleotides), and β-actin (cytoskeletal component). The SD calculated by BestKeeper program for all the reference genes tested in this study revealed low value for Suclg (SD, 0.76; n = 16; Fig. 2B,C, table), indicating that this gene is suited and stable as a reference gene (Pfaffl et al., 2004). We can observe that the median cycle threshold values for Suclg between control and exposed cells (34.54 vs 34.52, n = 5 and n = 12, p = 0.4) are more stable than those of β-actin (35.14 vs 34.1, n = 7 and n = 5, p = 0.1, Fig. 2D).
Figure 2.
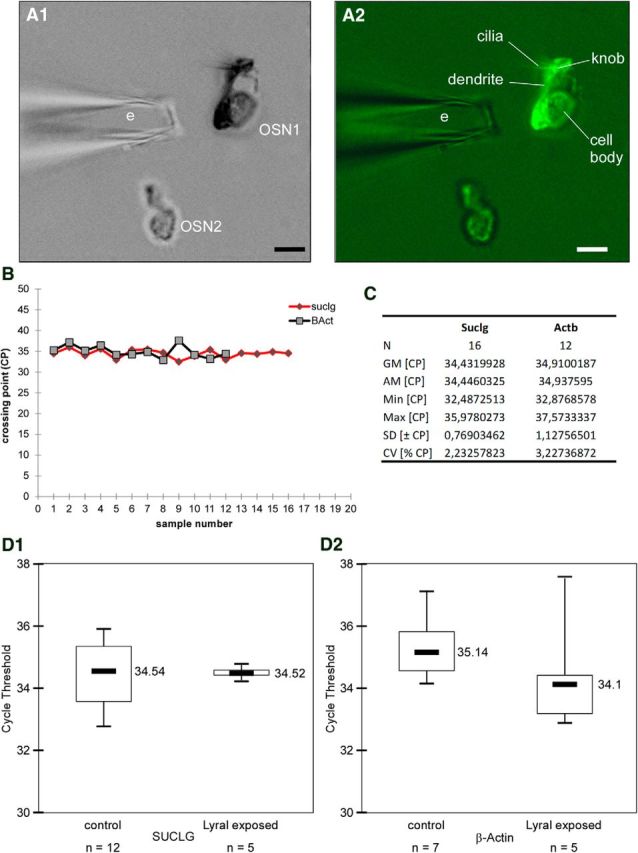
GFP-labeled cells harvesting and design of valid reference genes for reverse-transcription qPCR on small samples. A, Isolated cells can be selected for their fluorescence: photographs of dissociated neurons are seen in bright field (A1) and under fluorescence (A2). Two OSNs are visible: OSN1 expresses GFP and MOR23 and is fluorescent while OSN2 is not fluorescent and expresses another type of receptor. The pipette used to harvest fluorescent OSNs is visible (e) while the different anatomical features of the OSN can be observed (cilia, knob, dendrite, and cell body). Scale bar, 5 μm. B, Graph representing the crossing point values of the reference genes in harvested MOR23 neurons. Sample number equals single MOR23 neuron. C, Table summarizing the data of candidate housekeeping genes. N, Number of samples; GM [CP], the geometric mean of crossing points (CP); AM [CP], the arithmetic mean of CP; Min [CP] and Max [CP], the extreme values of CP; SD [±CP], the SD of the CP; CV [%CP], the coefficient of variance expressed as a percentage of the CP level. D, Boxplot representation of reference gene expression variability for each individual group in the analysis for the two genes tested, Suclg (D1) and β-actin (D2). Statistically significant differences were not found among the level expression in both groups. On the y-axis cycle threshold values are represented. Median values are also shown. Statistical analysis was performed using the Student's t test, considering p < 0.05 statistically significant.
Postnatal exposure to lyral increased the MOR23 receptor mRNA level by a factor of 2.98 ± 0.8 (n = 6, p < 0.05; Fig. 3A1). Postnatal exposure to lyral also induces an increase in mRNA levels for CNGA2 and PDE1 respectively by 6.88 ± 2.5 (n = 4; p < 0.05) and 15.79 ± 7 (n = 4, p < 0.05; Fig. 3A2). The level of ACIII transcripts did not change significantly although it increased by a factor 1.51 ± 0.53 (n = 5, p = 0.13). The ΔCt distribution values of each gene between both groups were significantly different. This means that there are limited biological variations of the genes of interest between cells from a single individual.
Figure 3.
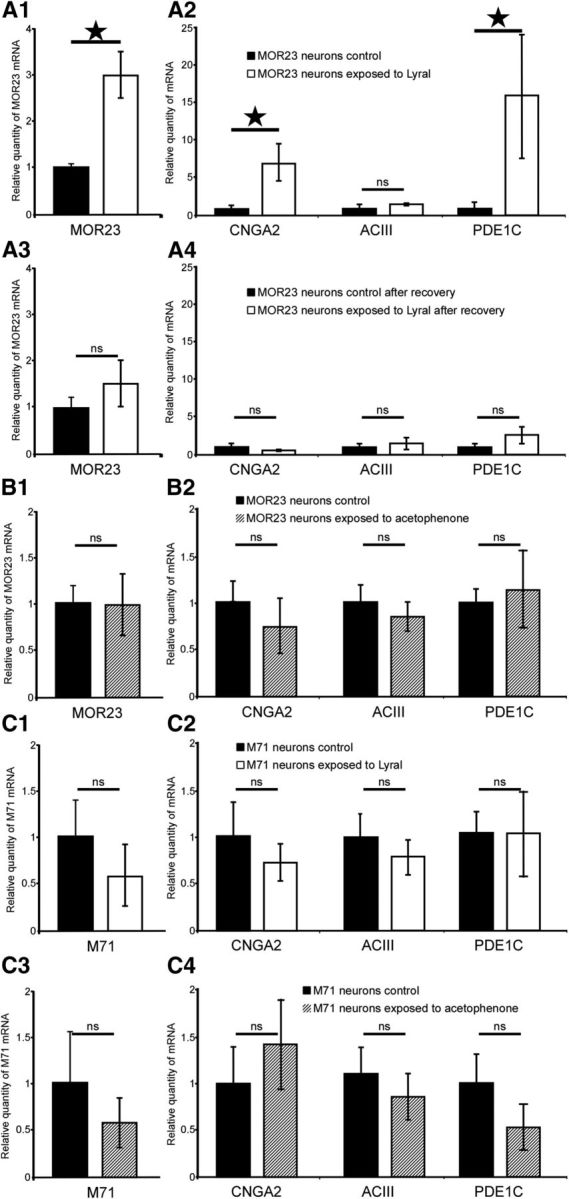
Postnatal exposure to lyral triggers an increase in mRNA level for MOR23, CNGA2, and PDE1C at the cellular level only in MOR23 neurons. A1, A2, Histogram representations of reverse-transcription qPCR measurements of mRNA levels for MOR23 (A1) and for CNGA2, ACIII, and PDE1C (A2) in MOR23 OSNs from lyral-exposed or nonexposed MOR23 mice. Results were normalized to Suclg for each sample. Results are presented in arbitrary units and the values obtained for nonexposed mice were set at 1 (n = 6 for MOR23 gene analysis, 5 for ACIII, 4 for ACIII and PDE1C). A3, A4, Histogram representations of reverse-transcription qPCR measurements of mRNA levels for MOR23 (A3) and for CNGA2, ACIII, and PDE1C (A4) in MOR23 OSNs from lyral-exposed and nonexposed mice after recovery. Results were normalized to Suclg for each sample. Results are presented in arbitrary units and the values obtained for nonexposed mice were set at 1 (n = 7 for control and n = 8 for recovery for all genes). B1, B2, Histogram representations of reverse-transcription qPCR measurements of mRNA levels for MOR23 (B1) and for CNGA2, ACIII, and PDE1C (B2) in MOR23 OSNs from acetophenone-exposed or nonexposed MOR23 mice. Results were normalized to Suclg for each sample. Results are presented in arbitrary units and the values obtained for nonexposed mice were set at 1 (n = 9 for exposed neurons, 11 for control neurons for all genes). C1, C2, Histogram representations of reverse-transcription qPCR measurements of mRNA levels for M71 (C1) and for CNGA2, ACIII, and PDE1C (C2) in M71 OSNs from lyral-exposed or nonexposed M71 mice. Results were normalized to Suclg for each sample. Results are presented in arbitrary units and the values obtained for nonexposed mice were set at 1 (n = 6 for exposed neurons, 6 for control neurons for all genes). C3, C4, Histogram representations of reverse-transcription qPCR measurements of mRNA levels for M71 (C3) and for CNGA2, ACIII, and PDE1C (C4) in M71 OSNs from acetophenone-exposed or nonexposed M71 mice. Results were normalized to Suclg for each sample. Results are presented in arbitrary units and the values obtained for nonexposed mice were set at 1 (n = 10 for exposed neurons, 7 for control neurons for all genes). *p < 0.05; ns, not significant; error bars indicate SEM.
The increase of mRNA transcript induced by postnatal lyral exposure is transient and reversible: after 4 weeks of recovery, the MOR23 transcript level in exposed MOR23 neurons (1.48 ± 0.2, n = 8) did not differ from that of control neurons (1.02 ± 0.53, n = 7, p = 0.604; Fig. 3A3). Similarly, after recovery, the level of CNGA2, PDE1C, and ACIII in exposed mice (0.5 ± 0.12, 2.56 ± 1.1, 1.43 ± 0.4, respectively; n = 8) did not differ from that of control neurons (1 ± 0.45, 1 ± 0.4, 1 ± 0.82, respectively; n = 7; Fig. 3A4).
Is the effect on MOR23 neurons specific to an exposure to lyral? To address this question, we exposed MOR23 mice to acetophenone. Postnatal exposure to acetophenone did not induce any difference in the level of mRNA transcripts analyzed in MOR23 neurons. The MOR23 transcript level in acetophenone-exposed MOR23 neurons (1.00 ± 0.3, n = 9) did not differ from that of control neurons (1.01 ± 0.2, n = 11, p = 0.982; Fig. 3B1). Similarly, after acetophenone exposure, the level of CNGA2, PDE1C, and ACIII in acetophenone-exposed MOR23 neurons (0.74 ± 0.3, 1.14 ± 0.4, 0.85 ± 0.2, respectively; n = 9) did not differ from that of control MOR23 neurons (1 ± 0.23, p = 0.65; 1 ± 0.15, p = 0.84; 1 ± 0.18, p = 0.36, respectively; n = 11; Fig. 3B2).
Postnatal exposure to odorants does not modulate mRNA-level expressions of M71, CNGA2, ACIII, and PDE1C at cellular level in M71-expressing neurons
To test the hypothesis that the effect of lyral could be generalized to other olfactory receptors, we then analyzed the consequences of lyral on M71. The M71 transcript level in lyral-exposed M71 neurons (0.58 ± 0.34, n = 6) did not differ from that of control neurons (1.00 ± 0.39, n = 6, p = 0.49; Fig. 3C1). Similarly, after lyral exposure, the level of CNGA2, PDE1C, and ACIII in lyral-exposed mice (0.72 ± 0.2, 1.03 ± 0.5, 0.78 ± 0.2, respectively; n = 6) did not differ from that of control neurons (1 ± 0.4, p = 0.53; 1 ± 0.27, p = 0.96; 1 ± 0.25, p = 0.49, respectively; n = 6; Fig. 3C2). In short, postnatal exposure to lyral did not induce any difference in the level of mRNA transcripts analyzed in M71 neurons.
Is the effect of odorant exposure similar for all ligand–olfactory receptor pairs? To address this question, we analyzed the consequences of acetophenone on M71 OSN molecular properties. The M71 transcript level in acetophenone-exposed M71 neurons (1.00 ± 0.55, n = 10) did not differ from that of control neurons (1.00 ± 0.55, n = 7, p = 0.45; Fig. 3C3). Similarly, after acetophenone exposure, the level of CNGA2, PDE1C, and ACIII in exposed M71 neurons (1.41 ± 0.5, 0.53 ± 0.2, 0.86 ± 0.25, respectively; n = 10) did not differ from that of control M71 neurons (1 ± 0.48, p = 0.7; 1 ± 0.31, p = 0.2; 1 ± 0.29, p = 0.55, respectively; n = 7; Fig. 3C4). In short, postnatal exposure to acetophenone did not induce any difference in the level of mRNA transcripts analyzed in M71 neurons.
Postnatal exposure to lyral changes the sensitivity and kinetics of responses to lyral in MOR23 cells
To monitor the effect of postnatal odorant exposure on the odorant-elicited responses in MOR23 neurons, we performed patch-clamp recordings. These recordings were performed on the dendritic knob of MOR23 neurons in an intact preparation as described previously (Grosmaitre et al., 2006). We measured the following characteristics of the responses: maximum amplitude, rise time (duration in milliseconds from 0 to 90% of the maximum amplitude), time at 50% (duration in milliseconds of the response's width at 50% of the maximum amplitude), total current elicited (area under the curve in picoamperes.seconds). In voltage-clamp mode, MOR23 neurons respond to increasing lyral concentration stimuli with inward currents showing increasing maximum amplitude in acetophenone-exposed (Fig. 4A1), control (Fig. 4A2), and lyral-exposed cells (Fig. 4A3). The peak transduction currents versus the concentration are plotted and fitted with the Hill equation: I = Imax/[1 + (K1/2/C)n], where I represents the peak current, Imax the maximum response at saturating concentrations, K1/2 the concentration at which half of the maximum response was reached, C the concentration of odorant, and n the Hill coefficient (Fig. 4A4). We recorded the dose–response curves from 12 MOR23 OSNs in control mice, 10 MOR23 OSNs in lyral-exposed mice, and 7 MOR23 OSNs in acetophenone-exposed mice. The average maximum amplitude elicited by saturating concentrations in control or in lyral-exposed mice did not change significantly [142 ± 18 pA vs 112.9 ± 18 pA, respectively; p = 0.16, Kolmogorov–Smirnov (KS) test]. The K1/2 did not vary between the control and lyral-exposed cells (3.2 ± 1 μm vs 2.4 ± 1 μm, p = 0.67, KS test). However, we observed that the dynamic range of lyral-exposed neurons was much broader compared with dynamic range of control neurons: the Hill coefficient was significantly smaller in lyral-exposed cells (0.64 ± 0.06) than in control cells (1.01 ± 0.12, p = 0.03, KS test; p = 0.027, Mann–Whitney U test). Also some lyral-exposed neurons (5 of 10 neurons) responded at lower concentrations (10−8 m) and all responded at 10−7 m while control neurons started to respond at 10−7 m.
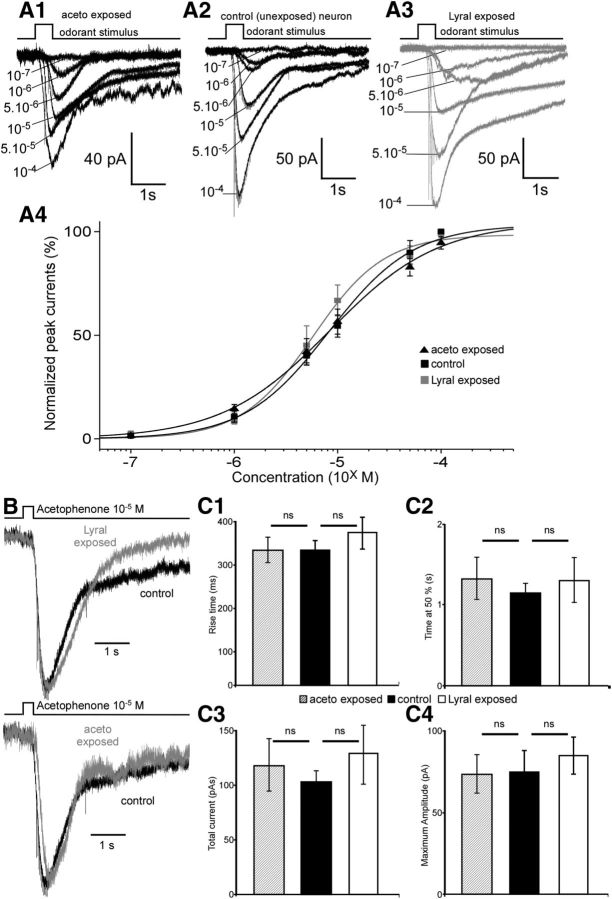
Figure 4.
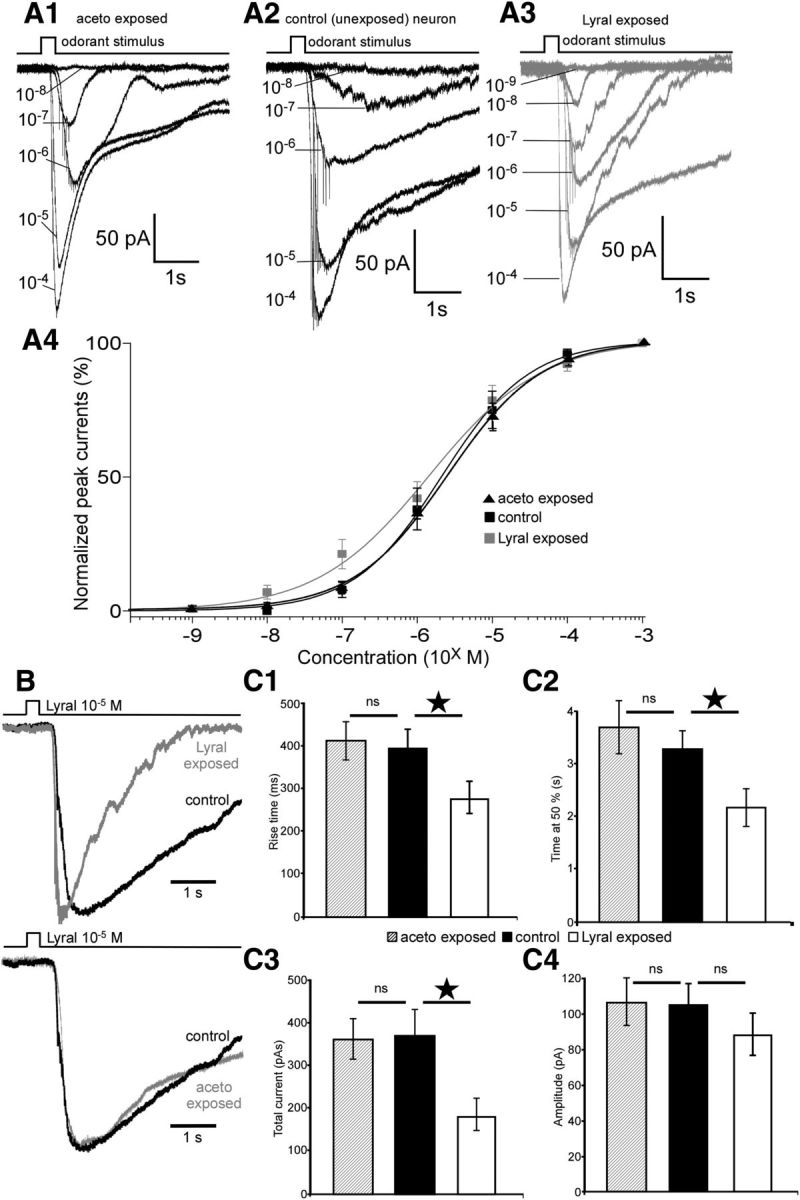
Postnatal exposure to lyral changes the sensitivity and kinetics of responses to lyral in MOR23 cells. A1–A3, Dose–response relationships of MOR23 cells in response to lyral: examples of an acetophenone-exposed cell (A1) and a control cell (A2) start to respond at 10−7 m while a lyral-exposed cell (A3) starts to respond at 10−8 m. A4, Average dose–response curves of the amplitude currents normalized to the maximum amplitude fitted with Hill equation in lyral-exposed (n = 10), acetophenone-exposed (n = 7), and control cells (n = 12). B, Top, Representative responses of a lyral-exposed (gray) and a control (black) MOR23 neuron to 10−5 m lyral normalized to the same peak amplitude. Bottom, Representative responses of an acetophenone-exposed (gray) and a control (black) MOR23 neuron to 10−5 m lyral normalized to the same peak amplitude. C, Quantification of the kinetics characteristics of control (n = 18), lyral-exposed (n = 19), and acetophenone-exposed (n = 12) MOR23 neurons in response to 10−5 m: analysis of the rise time (C1), the time at 50% (C2), the total current elicited (C3), and the maximum amplitude (C4). All recordings performed in perforated patch and at a membrane potential of −65 mV. *p < 0.05; ns, no significant; error bars indicate SEM.
After exposure to acetophenone, MOR23 neurons responded to lyral with dose–response characteristics similar to those of control MOR23 neurons: the average maximum amplitude elicited by saturating concentration, Imax, was not different between acetophenone-exposed and control MOR23 neurons (175 ± 24 pA vs 142 ± 18 pA, respectively; p = 0.75, KS test); the K1/2 did not vary between the control and acetophenone-exposed cells (3.2 ± 1 μm vs 4.48 ± 1 μm, p = 0.33, KS test); the dynamic range did not change since the Hill coefficient did not vary between the control and acetophenone-exposed cells (1.01 ± 0.12 vs 0.95 ± 0.1, p = 0.7, KS test).
We also noticed that the shape of the responses differed only in lyral-exposed cells compared with control cells (Fig. 4B). In fact, inward currents in lyral-exposed neurons were characterized by a faster return to the baseline compared with control neurons. We therefore analyzed the kinetics of the responses for a specific concentration, 10−5 m, close to the K1/2. All MOR23 neurons tested responded at this concentration. The kinetics parameters changed dramatically between lyral-exposed and control mice. First, the responses were faster in lyral-exposed mice: the rise-time dropped to 278 ms (±38 ms, n = 19) from 396 ms (±40 ms, n = 18, p = 0.022, KS test; p = 0.02, Mann–Whitney U test; Fig. 4C1) in control animals. Second, the responses were shorter since the width at 50% decreased from 3.3 s (±0.3 s, n = 18) in control mice to 2.2 s (±0.4, n = 19, p = 0.045, KS test; p = 0.011, Mann–Whitney U test; Fig. 4C2). Third, the total current elicited was also smaller in lyral-exposed mice (183 ± 35 pA.s, n = 19) compared with the total current in control mice (366 ± 92 pA.s, n = 18, p = 0.04, KS test; p = 0.039, Mann–Whitney U test; Fig. 4C3), further indicating that the responses were shorter. Meanwhile, the maximum amplitude did not change significantly between cells from control mice (105 ± 12 pA, n = 18) and cells from lyral-exposed mice (76 ± 12 pA, n = 19, p = 0.28, KS test; Fig. 4C4).
After acetophenone exposure, the kinetics of the responses of MOR23 neurons to 10−5 m lyral did not change significantly (Fig. 4B,C): the rise time did not vary between control and acetophenone-exposed neurons (396 ± 40 ms, n = 18, vs 409 ± 44 ms, n = 12, p = 0.45, KS test; Fig. 4C1); the width at 50% did not vary between control and acetophenone-exposed neurons (3.3 ± 0.3 s, n = 18, vs 3.8 ± 0.5 s, n = 12; p = 0.78, KS test; Fig. 4C2); the total current did not vary between control and acetophenone-exposed neurons (366 ± 92 pA.s, n = 18, vs 362 ± 47 pA.s, n = 12; p = 0.70, KS test; Fig. 4C3); finally, the maximum amplitude did not change significantly between cells from control mice (105 ± 12 pA, n = 18) and cells from acetophenone-exposed mice (107 ± 13 pA, n = 12, p = 0.61; Fig. 4C4).
In summary, postnatal exposure to lyral induces a higher sensitivity at lower concentration and a broader dynamic range in MOR23 neurons. Also after postnatal exposure, the responses to medium-range concentrations become both faster and shorter without change in the maximum amplitude. This effect is specific to the lyral exposure since the exposition to acetophenone, a nonligand odorant for MOR23, did not change significantly the response of MOR23 neurons.
Postnatal exposure to acetophenone or lyral did not change the characteristics of the responses to acetophenone by M71 cells
To evaluate whether the effect observed in MOR23 OSNs after lyral exposure could be nonspecific, we performed perforated patch clamp on the dendritic knob of GFP-labeled M71 neurons as described above for MOR23 neurons. In voltage-clamp mode, all M71 neurons respond to increasing acetophenone concentration stimuli with inward currents showing increasing maximum amplitude. This was observed in acetophenone-exposed (Fig. 5A1), control (Fig. 5A2), and lyral-exposed M71 cells (Fig. 5A3). The peak transduction currents versus the concentration are plotted and fitted with the Hill equation (Fig. 5A4). We recorded the dose–response curves from four M71 OSNs in control mice, six M71 OSNs in lyral-exposed mice, and eight M71 OSNs in acetophenone-exposed mice. After exposure to lyral or acetophenone, M71 neurons responded to acetophenone with dose–response characteristics similar to those of control M71 neurons. The average maximum amplitude elicited by saturating concentration, Imax, was not different between acetophenone-exposed (101 ± 18 pA, p = 0.51) or lyral-exposed M71 neurons (127 ± 20 pA) and control M71 neurons (119 ± 22 pA, p = 0.51 and p = 0.92, respectively; KS test). The K1/2 did not vary between acetophenone-exposed (10.0 ± 3 μm) or lyral-exposed M71 neurons (10.1 ± 4 μm) and control M71 neurons (9.4 ± 4 μm; p = 0.99 and p = 0.92, respectively, KS test). Finally, the dynamic range did not change since the Hill coefficient did not vary between acetophenone-exposed cells (1.1 ± 0.1) or lyral-exposed M71 neurons (1.3 ± 0.2) and control M71 cells (0.88 ± 0.17; p = 0.22 and p = 0.29, respectively, KS test).
Figure 5.
Postnatal exposure to acetophenone or lyral does not change the sensitivity or kinetics of responses to acetophenone in M71 cells. A1–A3, Dose–response relationships of M71 cells in response to acetophenone: examples of an acetophenone-exposed cell (A1), a control cell (A2), and a lyral-exposed cell (A3) start to respond at 10−6 m. A4, Average dose–response curves of the amplitude currents normalized to the maximum amplitude fitted with Hill equation in lyral-exposed (n = 6), acetophenone-exposed (n = 8), and control cells (n = 4). B, Top, Representative responses of a lyral-exposed (gray) and a control (black) M71 neuron to 10−5 m acetophenone normalized to the same peak amplitude. Bottom, Representative responses of an acetophenone-exposed (gray) and a control (black) M71 neuron to 10−5 m acetophenone normalized to the same peak amplitude. C, Quantification of the kinetics characteristics of control (n = 6), lyral-exposed (n = 10), and acetophenone-exposed (n = 10) M71 neurons in response to 10−5 m: analysis of the rise time (C1), the time at 50% (C2), the total current elicited (C3), and the maximum amplitude (C4). All recordings performed in perforated patch and at a membrane potential of −65 mV. ns, Not significant; error bars indicate SEM.
We then analyzed the kinetics of the responses for a specific concentration of acetophenone, 10−5 m, close to the K1/2. All M71 neurons tested responded at this concentration. After lyral or acetophenone exposure, the kinetics of the responses of M71 neurons to 10−5 m acetophenone did not change significantly (Fig. 5B,C). The rise time did not vary between acetophenone-exposed (328 ± 30 ms, n = 10) or lyral-exposed M71 neurons (370 ± 39 ms, n = 10) and control M71 neurons (330 ± 21 ms, n = 6; p = 0.96 and p = 0.89 respectively, KS test; Fig. 5C1). The width at 50% did not vary between acetophenone-exposed (1.326 ± 0.26 s, n = 10) or lyral-exposed M71 neurons (1.306 ± 0.28 s, n = 10) and control M71 neurons (1.145 ± 0.12 s, n = 6; p = 0.99 and p = 0.87 respectively, KS test; Fig. 5C2). The total current did not vary between acetophenone-exposed (117 ± 24 pA.s, n = 10) or lyral-exposed M71 neurons (129 ± 29 pA.s, n = 10) and control M71 neurons (102 ± 10 pA.s, n = 6; p = 0.35 and p = 0.43, respectively, KS test; Fig. 5C3). Finally the maximum amplitude did not vary between acetophenone-exposed (73 ± 12 pA, n = 10) or lyral-exposed M71 neurons (84 ± 11 pA, n = 10) and control M71 neurons (74 ± 13 pA, n = 6; p = 0.89 and p = 0.19, respectively, KS test; Fig. 5C4).
In summary, postnatal exposure to odorants did not modify the dose–response curves and kinetics of currents in response to acetophenone in M71 neurons. This result was observed after exposure to acetophenone, a ligand for M71, or to lyral, a molecule nonligand for M71. The functional plasticity observed in MOR23 neurons after lyral exposure seems specific for the odorant–receptor pair lyral–MOR23.
Postnatal exposure to lyral does not induce molecular or functional changes at the level of the entire epithelium
After demonstrating that postnatal exposure to lyral induces changes in the anatomy, the molecular properties, and the responses of MOR23 neurons, we then analyzed the properties of the entire olfactory epithelium after lyral exposure. Does lyral exposure affect the level of MOR23 receptor at the entire epithelium level? Does lyral exposure modify the electrical properties of the entire olfactory epithelium or only at the level of individual MOR23 neurons?
There are several ways to compare the expression level of the gene MOR23. Western blot evaluates protein expression and reverse-transcription qPCR monitors changes in amounts of mRNA for genes of interest. Antibodies specific for olfactory receptors are scarce and not extremely reliable. We therefore performed a relative semiquantification of MOR23 expression using GFP as a reporter of MOR23 expression level. On immunoblots, we compared the signal intensities of the GFP band of exposed mice (n = 3) with those of control mice (n = 3). Relative quantification of GFP protein level was performed after normalization to β-actin used as a loading control (Fig. 6A1). Data were normalized to control mice GFP band (set to 1 a.u.). We observed no difference in protein expression level (1 ± 0.07 a.u. for control vs 0.95 ± 0.08 a.u. for exposed mice; p = 0.64; Fig. 6A2). We then performed a comparative analysis of mRNA levels for the MOR23 gene by reverse-transcription qPCR. To avoid genomic DNA amplification, we designed primers specific for exon–exon junctions (Table 1). We compared relative MOR23 mRNA levels in exposed (n = 10) and control mice (n = 10). Postnatal lyral exposure does not affect the overall amounts of MOR23 mRNA in the entire olfactory epithelium (1 ± 0.14 a.u. for control mice vs 1.02 ± 0.14 a.u. for exposed mice; p = 0.4; Fig. 6B).
To assess the physiological consequences of postnatal exposure to lyral on the entire tissue, we then performed EOGs on the dorsal septal epithelium. We measured and compared the maximum amplitude reached by the signal in response to different stimuli: an odorant mixture (composed of 12 odorants each diluted in mineral oil at 10−3 m), 1 and 10% lyral in mineral oil, and mineral oil alone. Representative responses to the odorant mixture, mineral oil, and 10% lyral are shown on Figure 6C in a control mouse (C1) and in an exposed mouse (C2). The measurements of the amplitude revealed that postnatal lyral exposure does not affect the amplitude in response to the odorant mixture and to the two lyral concentrations tested (Fig. 6D,E). For the responses to the mixture, we observe no difference in the maximum amplitude elicited between the control (13.03 ± 0.45 mV, n = 16) and exposed mice (12.16 ± 0.39 mV, n = 36, p = 0.156; Fig. 6D). For the response to 1% lyral, we observed no difference in the maximum amplitude elicited between the control (0.95 ± 0.17 mV, n = 16) and exposed mice (0.65 ± 0.15 mV, n = 36, p = 0.51; Fig. 6E, left). For the response to 10% lyral, we observed no difference in the maximum amplitude elicited between the control (2 ± 0.33 mV, n = 16) and exposed mice (1.8 ± 0.4 mV, n = 36, p = 0.19; Fig. 6E, right). In our experimental conditions, postnatal odorant exposure did not affect the general physiological properties of the olfactory epithelium.
To further assess on a larger scale the transcriptomic status of the olfactory epithelium of lyral-exposed MOR23 mice, we performed a comparative analysis of mRNA level for genes of interest using TaqMan Gene Expression Assays. Our analysis included six groups of genes: genes of the cytoskeleton; growth factor genes; detoxification genes; odorant-binding protein (OBP) and lipocalins genes; calcium binding, cAMP pathway genes; and IP3 pathway genes (Table 2). Four mice from both control and exposed groups were used for this study. Data obtained are presented as a table (Table 2). The table shows gene identification, RQs between exposed and control groups, and the p value of the statistical test indicating the significance of the difference observed. A gene was considered significantly upregulated or downregulated if both of the following conditions were present: (1) the RQ value of >1.5 (upregulation) or <0.5 (downregulation), and (2) a p value of <0.05. Postnatal exposure to lyral does not affect the transcript levels for genes involved in olfactory neurogenesis, such as IGF1, LIF (leukemia inhibitory factor), and others (TGFα, HSF-1, and NGF). This is also the case for intermediate filaments (cytokeratin, gelsolin, vimentin) that maintain cytoskeleton structure of olfactory sensory neurons and of the olfactory epithelium. Gelsolin is also required in many other cellular processes, such as apoptosis. Vimentin plays an important role in the regeneration of neurons. No difference was observed for detoxification genes, such as the cytochrome P450 family (in particular CYP1A2 and CYP2A5), and for enzymes of the second phase of detoxification (Gst m, α, and π). No difference was observed for odorant-binding proteins (OBP1A and MUP-5). Finally, we showed that the level of transcripts coding for cAMP and IP3 transduction pathways did not change after postnatal exposure to lyral. In brief, 41 of the 42 analyzed genes were not affected by postnatal lyral exposure. For more detailed indication of fold change for each gene, see Table 2. Only calbindin 2 was downregulated (RQ, 0.44; p < 0.05).
Table 2.
List of the 46 genes whose expression was analyzed in the entire olfactory epithelium
| Assay | Type | Official full name | Relative quantity | p value | Reference sequence |
|---|---|---|---|---|---|
| 18S-Hs99999901_s1 | Control | 18S ribosomal RNA | 1.0862 | 0.8443 | X03205.1 |
| Actb-Mm00607939_s1 | Control | Actin, β | 1.0482 | 0.6167 | NM_007393.3 |
| Adcy3-Mm00460371_m1 | Target | Adenylate cyclase 3 | 0.8747 | 0.2633 | NM_001159536.1 |
| Ano1-Mm00724407_m1 | Target | Anoctamin 1, calcium-activated chloride channel | 1.0192 | 0.9142 | NM_178642.4 |
| Ano2-Mm00463894_m1 | Target | Anoctamin 2 | 0.7848 | 0.0667 | NM_153589.2 |
| Anxa5-Mm01293059_m1 | Target | Annexin A5 | 0.8888 | 0.1664 | NM_009673.2 |
| Calb2-Mm00801461_m1 | Target | Calbindin 2 | 0.4367 | 0.0228 | NM_007586.1 |
| Calm1-Mm01336281_g1 | Target | Calmodulin 1 | 0.7655 | 0.3614 | NM_009790.4 |
| Camk2a-Mm00437967_m1 | Target | Calcium/calmodulin-dependent protein kinase II α | 1.052 | 0.8047 | NM_009792.3 |
| Camk2b-Mm00432284_m1 | Target | Calcium/calmodulin-dependent protein kinase II β | 0.7051 | 0.0077 | NM_001174053.1 |
| Camk2d-Mm00499266_m1 | Target | Calcium/calmodulin-dependent protein kinase II Δ | 0.7895 | 0.1653 | NM_001025438.1 |
| Camk2g-Mm00618054_m1 | Target | Calcium/calmodulin-dependent protein kinase II γ | 0.8476 | 0.0733 | NM_001039138.1 |
| Clca5-Mm00724513_m1 | Target | Chloride channel calcium activated 5 | 0.9895 | 0.9683 | NM_178697.4 |
| Cnga2-Mm01281967_m1 | Target | Cyclic nucleotide gated channel α 2 | 0.7496 | 0.0807 | NM_007724.2 |
| Cyp1a2-Mm00487224_m1 | Target | Cytochrome P450, family 1, subfamily A, polypeptide 2 | 0.9007 | 0.6488 | NM_009993.3 |
| Cyp2a4;Cyp2a5-Mm00487248_g1 | Target | Cytochrome P450, family 2, subfamily a, polypeptide 4/polypeptide 5 | 0.8635 | 0.4644 | NM_007812.4 |
| Gapdh-Mm99999915_g1 | Control | Glyceraldehyde-3-phosphate dehydrogenase | 0.7807 | 0.7887 | NM_008084.2 |
| Gnal-Mm01258217_m1 | Target | Guanine nucleotide binding protein (G protein), α activating activity polypeptide, olfactory type | 1.1539 | 0.2368 | NM_010307.2 |
| Gsn-Mm00456679_m1 | Target | Gelsolin | 0.7076 | 0.0935 | NM_146120.3 |
| Gsta2;Gm3776;Gsta1;Gm10639-Mm04207463_mH | Target | Glutathione S-transferase α2/α1 | 1.043 | 0.8034 | M_008182.3 |
| Gstm1-Mm00833915_g1 | Target | Glutathione S-transferase μ 1 | 0.6953 | 0.0158 | NM_010358.5 |
| Gstp1-Mm04213618_gH | Target | Glutathione S-transferase π 1 | 0.764 | 0.1837 | NM_013541.1 |
| Guca1b-Mm00524884_m1 | Target | Guanylate cyclase activator 1B | 0.9697 | 0.9192 | NM_146079.1 |
| Hprt-Mm01545399_m1 | Control | Hypoxanthine guanine phosphoribosyl transferase | 0.9671 | 0.7042 | NM_013556.2 |
| Hsf1-Mm01201402_m1 | Target | Heat shock transcription factor 1 | 0.6147 | 0.0677 | NM_008296.2 |
| Hsp90b1-Mm00441926_m1 | Target | Heat shock protein 90, β (Grp94), member 1 | 0.7454 | 0.0012 | NM_011631.1 |
| Igf1-Mm00439560_m1 | Target | Insulin-like growth factor 1 | 1.536 | 0.2825 | NM_001111274.1 |
| Itpr3-Mm01306070_m1 | Target | Inositol 1,4,5-trisphosphate receptor, type 3 | 1.1158 | 0.3789 | NM_080553.3 |
| Krt18-Mm01601704_g1 | Target | Keratin 18 | 0.6707 | 0.0647 | NM_010664.2 |
| Lif-Mm00434762_g1 | Target | Leukemia inhibitory factor | 0.7579 | 0.4066 | NM_001039537.1 |
| Mup5-Mm01702733_g1 | Target | Major urinary protein 5 | 0.9304 | 0.8599 | NM_008649.2 |
| Ngf-Mm00443039_m1 | Target | Nerve growth factor | 1.1036 | 0.8304 | NM_001112698.1 |
| Obp1a-Mm00500903_m1 | Target | Odorant binding protein IA | 1.6581 | 0.3328 | NM_008754.2 |
| Olfr124-Mm00529996_s1 | Target | Olfactory receptor 124 | 0.6214 | 0.0665 | NM_147062.2 |
| Olfr16-Mm00493258_s1 | Target | Olfactory receptor 16 MOR23 | 1.0909 | 0.6025 | NM_008763.2 |
| Omp-Mm00448081_s1 | Target | Olfactory marker protein | 0.683 | 0.3664 | NM_011010.2 |
| Pdc-Mm00445900_m1 | Target | Phosducin | 0.3253 | 0.2505 | NM_001159730.1 |
| Pde1c-Mm00478051_m1 | Target | Phosphodiesterase 1C, calmodulin-dependent 70kDa | 0.8198 | 0.0878 | NM_001025568.2 |
| Pde4a-Mm01147149_m1 | Target | Phosphodiesterase 4A, cAMP specific | 0.859 | 0.5779 | NM_019798.5 |
| Plcb2-Mm01338057_m1 | Target | Phospholipase C, β 2 | 1,0971 | 0,5261 | NM_177568.2 |
| Prkaca-Mm00660092_m1 | Target | Protein kinase, cAMP dependent, catalytic, alpha | 0.739 | 0.012 | NM_008854.4 |
| Prkg1-Mm00440954_m1 | Target | Protein kinase, cGMP dependent, type I | 1.691 | 0.1051 | NM_001013833.2 |
| Sucla2-Mm01310541_m1 | Target | Succinate-CoA ligase, ADP-forming, β subunit | 0.9291 | 0.3629 | NM_011506.3 |
| Tgfa-Mm00446232_m1 | Target | Transforming growth factor, α | 0.5134 | 0.0607 | NM_031199.3 |
| Trpc6-Mm01176083_m1 | Target | Transient receptor potential cation channel, subfamily C, member 6 | 0.9146 | 0.8477 | NM_013838.2 |
| Vim-Mm01333430_m1 | Target | Vimentin | 0.7773 | 0.0323 | NM_011701.4 |
Discussion
Here we report that plasticity induced by the environment occurs at the level of olfactory sensory neurons. Postnatal exposure to lyral induces a drop in the density of MOR23-expressing OSNs, while a similar exposure to acetophenone did not. After a recovery period, the density returns partially to its control level, indicating that the process is reversible.
Lyral exposure effect is not based on toxicity
Some toxic chemicals present in the environment do affect the olfactory epithelium. Notably, inhalation of such toxic compounds as H2S induces the deaths of olfactory neuron cells in rats (Brenneman et al., 2000) and acetone selectively damages cells of the olfactory epithelium in mice (Buron et al., 2009). Here we exposed mice to lyral, a commonly used fragrance of household products known to cause allergies, especially through skin contact (Militello and James, 2005). Could the effects observed here be due to a degree of toxicity of lyral on the olfactory epithelium? To address this question, we measured the effect of postnatal exposure on the transcriptomic level of 42 genes in the epithelium, including transcripts for genes involved in detoxification from the P450 cytochrome family, such as CYP1A2 and CYP2A5 (Ling et al., 2004). We saw no large-scale modification. Similarly, the exposure to lyral does not affect another population of OSNs expressing M71. Finally, lyral does not affect the odorant-evoked responses of the epithelium as shown by EOG recordings (Fig. 6). Overall, these results suggest that the postnatal exposure paradigm we used did not damage the general properties of the epithelium. The decrease in MOR23 OSN density might therefore be a specific response for this ligand–receptor pair.
Diversity of OSN population modulation induced by odorant exposure
It is now clearly established that olfactory stimulation can increase the survival of OSNs (Zhao and Reed, 2001; Watt et al., 2004; Jones et al., 2008; Santoro and Dulac, 2012; François et al., 2013). It also induces higher sensitivity of the epithelium to the odorant used for the exposure, suggesting an increase in the specific olfactory receptor expression (Wang et al., 1993). However, some authors reports no effect of odorization (Kerr and Belluscio, 2006) if not a decrease as observed with the population of M72-expressing neurons after exposure to acetophenone, a ligand of M72 (Cavallin et al., 2010). All those reports, along with our data, suggest that the consequences of an odorant exposure on OSN populations vary depending on the ligand–receptor pair, the concentration, and the exposure paradigm used. Indeed, here we showed a decrease in survival of MOR23-expressing neurons exposed to a pure concentration of lyral 1 h per day. This seems to contradict other research showing an increase in survival for MOR23-expressing neurons after continuous exposure to diluted lyral (Santoro and Dulac, 2012). Such discrepancy is correlated with a recent study showing that lower concentration of odorant is more efficient to promote the activity-driven survival of OSNs (François et al., 2013). Finally, the context of odorant exposure has now clearly been demonstrated to have a major impact on OSN population that may furthermore last through generations (Dias and Ressler, 2014).
The mechanism involved in the decrease of density in M72 (Cavallin et al., 2010) or MOR23 neurons in the present study after a passive postnatal exposure to their ligand remains unclear. Repeated stimulations by odorants leads to excitotoxicity, as observed in OSNs (French and Heberlein, 2009) and in central neurons (Bano and Nicotera, 2007). One hypothesis emerges: that we could evoke such excitotoxicity using pure odorants.
Plasticity of OSNs at the cellular level after odorant exposure
While the density of MOR23 neurons decreases, the protein and mRNA transcript levels for the MOR23 odorant receptor is not affected by postnatal exposure at the level of the whole olfactory epithelium. To maintain the same level of MOR23 mRNA with fewer MOR23 OSNs, the expression of MOR23 in each neuron has to increase after lyral exposure. To test this hypothesis, we used reverse-transcription qPCR on small samples of collected MOR23 OSNs. Sensitive, specific, and reproducible reverse-transcription qPCR can detect small changes in gene expression (Bustin, 2000). Such reverse-transcription qPCR was until recently performed on large samples. A recent report shows that high-throughput expression profiling from single neuronal cells can be performed (Citri et al., 2011). However, we present here, to our knowledge, the first report of mRNA-level quantification in a small sample of specific OSNs. Unlike OSNs automatically selected through other techniques, single harvested OSNs exhibit all the features of healthy neurons (Fig. 2A2). Consequently, we can exclude variations in mRNA levels due to a change in cell integrity. We observe that postnatal exposure to lyral induces an increase of MOR23 transcript levels in MOR23 neurons. Similarly transcripts for CNGA2 and PDE1C also increased while ACIII transcript levels remain unchanged. Does such change in mRNA level related to the olfactory transduction pathway alter the physiology of individual MOR23 neurons? Using patch-clamp recordings, we show that MOR23 neurons become more sensitive to lyral and that their responses exhibit a faster onset and a shorter duration. We can link the level of transcripts to the physiology of the neuron. Indeed, the faster onset of the response is consistent with a higher level of MOR23 and CNGA2 as the receptor and the CNGA2 are among the proteins responsible for the beginning of the response, initiating the depolarization of the cell. Similarly, as PDE1C is involved in the extinction of the signal by degradation of cAMP in AMP, the higher level of PDE1C transcripts present in MOR23 OSNs after lyral exposure is consistent with their shorter response to lyral.
Is this difference based on change of OSN properties or on survival of a subpopulation of MOR23 neurons? Indeed, while they express the same receptor, MOR23 neurons are heterogeneous in their response to lyral (Grosmaitre et al., 2006). This heterogeneity could be partly explained by the maturation status of the neurons: mature neurons exhibit faster onset and shorter overall response compared with immature or olfactory marker protein (OMP) knock-out OSNs (Lee et al., 2011), OMP being present only in mature OSNs. This suggests that most of the OSNs we recorded after odorant exposure display characteristics of mature neurons. Further studies are required to decipher whether pure lyral exposure leads to selection of fewer but more effective MOR23 neurons.
Plasticity of OSN at the cellular level is olfactory receptor–ligand specific.
It has been shown previously that the lack of activity by naris closure or genetic approaches induces changes in OSN populations. These changes will be different depending on the olfactory receptor expressed (Zhao et al., 2013). Here we observe a modulation of the molecular properties in MOR23-expressing OSNs by lyral, the ligand of the MOR23 receptor (Figs. 3, 4). In another population of OSNs, those expressing M71, exposure to the ligand of the receptor does not induce any changes in their molecular (Fig. 3) or in their physiological (Fig. 5) properties. We propose that odorant-induced activity will have different consequences depending on the olfactory receptor–ligand pair. Further studies on other odorant–receptor pairs will be useful to confirm the specificity of the effect. For instance, there are some broadly tuned receptors reported. What would be the effect of odorant exposure on these broadly tuned receptors?
While we present here the first data on how odorant exposure modulates OSN properties, others have shown that naris occlusion modifies the response to lyral of MOR23 neurons and increases their sensitivity (He et al., 2012). In the open side, MOR23 neurons exhibit a decrease of sensitivity (He et al., 2012) induced either by olfactory overstimulation or by an increase in damage induced by increased airflow (Coppola, 2012). Here we report an increase in sensitivity toward lower concentrations while the dynamic range is also increased: there is no shift of the entire dose–response curve to the left but it is rather flattened by the odorant exposure. Compared with the open side in sensory-deprivation experiments, we precisely target the MOR23 neurons by lyral exposure. Our activation of MOR23 neurons might have specific consequences on these neurons that a general overstimulation might not elicit.
We report an adaptation of the MOR23 population by a loss of many MOR23 neurons when exposed to lyral while the EOG signal remained unchanged. The olfactory system is based on combinatorial coding (Malnic et al., 1999). Therefore, there are probably other receptors, in addition to MOR23, able to detect lyral. The olfactory system might be sufficiently redundant to adapt in order to compensate for the loss of MOR23 neurons. Two possibilities can be proposed: (1) an increase of another population of neurons sensitive to that odorant, or (2) an increase in the activity of other neurons sensitive to lyral. We also describe a lower detection threshold in MOR23 neurons. This indicates that, at low concentrations of odorants, MOR23 neurons themselves could take part in the compensation of their lower density by sending more information to the OB. The plasticity described here could have a role in maintaining an efficient olfactory system in different environmental conditions.
In summary, MOR23 OSNs in an odorant-enriched situation present an adaptive capacity to their environment: the olfactory epithelium may reduce the energy expenditure necessary to generate many OSNs sensitive to that odorant while still detecting it. Meanwhile, in a normal environment where an odorant is scarce or novel, every single neuron able to identify it will be significant for its detection. This phenomenon might be similar to the “novelty detection” present at the OB's mitral cell level (Kato et al., 2012). In total, these results indicate that postnatal exposure to odorants can induce plasticity in OSN populations at the cellular level and that this plasticity is reversible and olfactory receptor–ligand specific.
Footnotes
This work was supported by Centre National de la Recherche Scientifique (CNRS) through an Action Thématique et Incitative sur Programme grant (to X.G.), by the Conseil Régional de Bourgogne [FABER (Favoriser l'accueil en Bourgogne d'équipes de recherche) and PARI (Le plan d'actions régional pour l'innovation) grants], by Université de Bourgogne [BQR (Le Bonus Qualité Reserche) program], by a postdoctoral fellowship from FEDER (European Funding for Regional Economical Development) (for A.M.); I.A. received a doctoral fellowship from the E2S graduate program from the Université de Bourgogne. We thank Dr. Peter Mombaerts for the gift of MOR23-IRES-tauGFP mice and M71-IRES-tauGFP mice; Anne Lefranc and the Centre des Sciences du Goût et de l'Alimentation animal facility for excellent animal care; Christine Arnould at the Dijon Institut National de la Recherche Agronomique imaging core facility for the confocal imaging technical assistance; undergraduate students Juliette Portier, Audrey Loisy, Olympe Arevalo, and Emma Granon for temporary technical help; and Dr. Ingrid Jakob and two anonymous reviewers for insightful comments on this manuscript.
The authors declare no competing financial interests.
References
- Bano D, Nicotera P. Ca2+ signals and neuronal death in brain ischemia. Stroke. 2007;38:674–676. doi: 10.1161/01.STR.0000256294.46009.29. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300. [Google Scholar]
- Bozza T, Feinstein P, Zheng C, Mombaerts P. Odorant receptor expression defines functional units in the mouse olfactory system. J Neurosci. 2002;22:3033–3043. doi: 10.1523/JNEUROSCI.22-08-03033.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenneman KA, James RA, Gross EA, Dorman DC. Olfactory neuron loss in adult male CD rats following subchronic inhalation exposure to hydrogen sulfide. Toxicol Pathol. 2000;28:326–333. doi: 10.1177/019262330002800213. [DOI] [PubMed] [Google Scholar]
- Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-X. [DOI] [PubMed] [Google Scholar]
- Buonviso N, Gervais R, Chalansonnet M, Chaput M. Short-lasting exposure to one odour decreases general reactivity in the olfactory bulb of adult rats. Eur J Neurosci. 1998;10:2472–2475. doi: 10.1046/j.1460-9568.1998.00266.x. [DOI] [PubMed] [Google Scholar]
- Buron G, Hacquemand R, Pourié G, Brand G. Inhalation exposure to acetone induces selective damage on olfactory neuroepithelium in mice. Neurotoxicology. 2009;30:114–120. doi: 10.1016/j.neuro.2008.11.005. [DOI] [PubMed] [Google Scholar]
- Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol. 2000;25:169–193. doi: 10.1677/jme.0.0250169. [DOI] [PubMed] [Google Scholar]
- Cavallin MA, Powell K, Biju KC, Fadool DA. State-dependent sculpting of olfactory sensory neurons is attributed to sensory enrichment, odor deprivation, and aging. Neurosci Lett. 2010;483:90–95. doi: 10.1016/j.neulet.2010.07.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citri A, Pang ZP, Südhof TC, Wernig M, Malenka RC. Comprehensive qPCR profiling of gene expression in single neuronal cells. Nat Protoc. 2011;7:118–127. doi: 10.1038/nprot.2011.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Col JA, Matsuo T, Storm DR, Rodriguez I. Adenylyl cyclase-dependent axonal targeting in the olfactory system. Development. 2007;134:2481–2489. doi: 10.1242/dev.006346. [DOI] [PubMed] [Google Scholar]
- Coppola DM. Studies of olfactory system neural plasticity: the contribution of the unilateral naris occlusion technique. Neural Plast. 2012;2012:351752. doi: 10.1155/2012/351752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias BG, Ressler KJ. Parental olfactory experience influences behavior and neural structure in subsequent generations. Nat Neurosci. 2014;17:89–96. doi: 10.1038/nn.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- François A, Laziz I, Rimbaud S, Grebert D, Durieux D, Pajot-Augy E, Meunier N. Early survival factor deprivation in the olfactory epithelium enhances activity-driven survival. Front Cell Neurosci. 2013;7:271. doi: 10.3389/fncel.2013.00271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French RL, Heberlein U. Glycogen synthase kinase-3/Shaggy mediates ethanol-induced excitotoxic cell death of Drosophila olfactory neurons. Proc Natl Acad Sci U S A. 2009;106:20924–20929. doi: 10.1073/pnas.0910813106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosmaitre X, Vassalli A, Mombaerts P, Shepherd GM, Ma M. Odorant responses of olfactory sensory neurons expressing the odorant receptor MOR23: a patch clamp analysis in gene-targeted mice. Proc Natl Acad Sci U S A. 2006;103:1970–1975. doi: 10.1073/pnas.0508491103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosmaitre X, Santarelli LC, Tan J, Luo M, Ma M. Dual functions of mammalian olfactory sensory neurons as odor detectors and mechanical sensors. Nat Neurosci. 2007;10:348–354. doi: 10.1038/nn1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Tian H, Lee AC, Ma M. Postnatal experience modulates functional properties of mouse olfactory sensory neurons. Eur J Neurosci. 2012;36:2452–2460. doi: 10.1111/j.1460-9568.2012.08170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SV, Choi DC, Davis M, Ressler KJ. Learning-dependent structural plasticity in the adult olfactory pathway. J Neurosci. 2008;28:13106–13111. doi: 10.1523/JNEUROSCI.4465-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato HK, Chu MW, Isaacson JS, Komiyama T. Dynamic sensory representations in the olfactory bulb: modulation by wakefulness and experience. Neuron. 2012;76:962–975. doi: 10.1016/j.neuron.2012.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr MA, Belluscio L. Olfactory experience accelerates glomerular refinement in the mammalian olfactory bulb. Nat Neurosci. 2006;9:484–486. doi: 10.1038/nn1673. [DOI] [PubMed] [Google Scholar]
- Kleene SJ. The electrochemical basis of odor transduction in vertebrate olfactory cilia. Chem Senses. 2008;33:839–859. doi: 10.1093/chemse/bjn048. [DOI] [PubMed] [Google Scholar]
- Lee AC, He J, Ma M. Olfactory marker protein is critical for functional maturation of olfactory sensory neurons and development of mother preference. J Neurosci. 2011;31:2974–2982. doi: 10.1523/JNEUROSCI.5067-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling G, Gu J, Genter MB, Zhuo X, Ding X. Regulation of cytochrome P450 gene expression in the olfactory mucosa. Chem Biol Interact. 2004;147:247–258. doi: 10.1016/j.cbi.2004.02.003. [DOI] [PubMed] [Google Scholar]
- Malnic B, Hirono J, Sato T, Buck LB. Combinatorial receptor codes for odors. Cell. 1999;96:713–723. doi: 10.1016/S0092-8674(00)80581-4. [DOI] [PubMed] [Google Scholar]
- Mandairon N, Stack C, Kiselycznyk C, Linster C. Enrichment to odors improves olfactory discrimination in adult rats. Behav Neurosci. 2006a;120:173–179. doi: 10.1037/0735-7044.120.1.173. [DOI] [PubMed] [Google Scholar]
- Mandairon N, Stack C, Linster C. Olfactory enrichment improves the recognition of individual components in mixtures. Physiol Behav. 2006b;89:379–384. doi: 10.1016/j.physbeh.2006.07.013. [DOI] [PubMed] [Google Scholar]
- Mandairon N, Didier A, Linster C. Odor enrichment increases interneurons responsiveness in spatially defined regions of the olfactory bulb correlated with perception. Neurobiol Learn Mem. 2008;90:178–184. doi: 10.1016/j.nlm.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Militello G, James W. Lyral: a fragrance allergen. Dermatitis. 2005;16:41–44. doi: 10.2310/6620.2005.04033. [DOI] [PubMed] [Google Scholar]
- Negroni J, Meunier N, Monnerie R, Salesse R, Baly C, Caillol M, Congar P. Neuropeptide y enhances olfactory mucosa responses to odorant in hungry rats. PloS One. 2012;7:e45266. doi: 10.1371/journal.pone.0045266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26:509–515. doi: 10.1023/B:BILE.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- Rochefort C, Gheusi G, Vincent JD, Lledo PM. Enriched odor exposure increases the number of newborn neurons in the adult olfactory bulb and improves odor memory. J Neurosci. 2002;22:2679–2689. doi: 10.1523/JNEUROSCI.22-07-02679.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro SW, Dulac C. The activity-dependent histone variant H2BE modulates the life span of olfactory neurons. eLife. 2012;1:e00070. doi: 10.7554/eLife.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JW, Brierley T. A functional map in rat olfactory epithelium. Chemical senses. 1999;24:679–690. doi: 10.1093/chemse/24.6.679. [DOI] [PubMed] [Google Scholar]
- Vassalli A, Rothman A, Feinstein P, Zapotocky M, Mombaerts P. Minigenes impart odorant receptor-specific axon guidance in the olfactory bulb. Neuron. 2002;35:681–696. doi: 10.1016/S0896-6273(02)00793-6. [DOI] [PubMed] [Google Scholar]
- Wang HW, Wysocki CJ, Gold GH. Induction of olfactory receptor sensitivity in mice. Science. 1993;260:998–1000. doi: 10.1126/science.8493539. [DOI] [PubMed] [Google Scholar]
- Watt WC, Sakano H, Lee ZY, Reusch JE, Trinh K, Storm DR. Odorant stimulation enhances survival of olfactory sensory neurons via MAPK and CREB. Neuron. 2004;41:955–967. doi: 10.1016/S0896-6273(04)00075-3. [DOI] [PubMed] [Google Scholar]
- Woo CC, Hingco EE, Taylor GE, Leon M. Exposure to a broad range of odorants decreases cell mortality in the olfactory bulb. Neuroreport. 2006;17:817–821. doi: 10.1097/01.wnr.0000215780.84226.2d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Reed RR. X inactivation of the OCNC1 channel gene reveals a role for activity-dependent competition in the olfactory system. Cell. 2001;104:651–660. doi: 10.1016/S0092-8674(01)00262-8. [DOI] [PubMed] [Google Scholar]
- Zhao S, Tian H, Ma L, Yuan Y, Yu CR, Ma M. Activity-dependent modulation of odorant receptor gene expression in the mouse olfactory epithelium. PloS One. 2013;8:e69862. doi: 10.1371/journal.pone.0069862. [DOI] [PMC free article] [PubMed] [Google Scholar]