Abstract
Introduction:
High density lipoprotein (HDL) particles are heterogeneous and their proteome is complex and distinct from HDL cholesterol However, it is largely unknown whether HDL proteins are associated with cardiovascular protection.
Areas covered:
HDL isolation techniques and proteomic analyses are reviewed. A list of HDL proteins reported in 37 different studies was compiled and the effects of different isolation techniques on proteins attributed to HDL is discussed. Mass spectrometric techniques used for HDL analysis and the need for precise and robust methods for quantification of HDL proteins is discussed.
Expert opinion:
Proteins associated with HDL have the potential to be used as biomarkers and/or help to understand HDL functionality. To achieve this, large cohorts must be studied using precise quantification methods. Key factors in HDL proteome quantification are the isolation methodology and the mass spectrometry technique employed. Isolation methodology affects what proteins are identified in HDL and the specificity of association with HDL particles needs to be addressed. Shotgun proteomics yields imprecise quantification, but the majority of HDL studies relied in this approach. Few recent studies used targeted tandem mass spectrometry to quantify HDL proteins, and it is imperative that future studies focus on application of these precise techniques.
Keywords: HDL, proteomics, shotgun, targeted LC-MS/MS methods, parallel reaction monitoring, Selected reaction monitoring, data independent analysis
1. Introduction
Cardiovascular disease (CVD) is the number one cause of morbidity and mortality worldwide [1]. However, reliable diagnostic tests of CVD risk are lacking and in nearly 1/3 of patients, the first indication of CVD is an acute cardiovascular event (i.e. myocardial infarction). Epidemiological studies demonstrated inverse association of CVD risk with plasma concentration of HDL-C [2], but recent failures of clinical trials that pharmacologically elevate HDL-C [3,4], association of high HDL-C with cardiovascular and all-cause mortality [5], together with Mendelian Randomization genetic studies[6] provided strong evidence against a causal relationship between HDL-C levels and cardiovascular risk. However, numerous studies have shown that HDL-C does not capture HDL diverse functions [7] and therefore other HDL metrics need to be explored. Whether HDL proteins [8–10] are associated with the cardiovascular protection and whether they play a role in determining HDL antiatherogenic properties remains largely unknown [11].
HDL-C levels associate negatively with CVD risk, and HDL proteome is less complex than plasma to be analyzed (e.g., ~100 vs. thousands of proteins and 4 vs. >10 order of magnitude concentration range) [12,13], making HDL an attractive target to look for surrogate markers of atheroprotective function. A large number of studies have analyzed HDL protein cargo and its changes in a number of disease conditions. However, there is little congruency in proteins found in HDL. The major challenges stem from HDL complexity, definition, different approaches for isolation, and the heterogeneity of its particles. Moreover, technical challenges for the analysis of HDL proteome come from the high phospholipids content of HDL (30% of HDL by weight and ~100x molar excess over average protein in HDL)[12], as phospholipids are recognized electrospray ionization (ESI) suppressants.
In this review, we have attempted to reconcile proteomic studies presented up to May 2019, and to address challenging questions in HDL field, concerning the identity of HDL proteins, how different isolation techniques affect proteins attributed to HDL, and how to handle the issue of contaminant proteins in HDL. Lastly, we review mass spectrometric (MS) techniques applied to HDL analyses and suggest a guidance for future translational studies, which could provide definitive answers about utility of HDL proteome as a marker or mediator of CVD. Because several excellent reviews [10,14] provide thorough overview of HDL measurement techniques [15,16], isolation methodologies [9,17], as well as association of the HDL proteome with various disease conditions[18], these aspects will not be discussed here in detail.
2. How is HDL defined?
Lipoproteins are non-covalent complexes of proteins (generally called apolipoproteins) and several classes of lipids, in which the neutral lipid core (composed of cholesteryl esters and triglycerides) is surrounded by a monolayer of polar lipids, primarily phospholipids and unesterified cholesterol, together with protein(s). Lipoproteins were described as a distinct complex of proteins and lipids for the first time in 1929 [19]. HDL constitutes the most heterogeneous and protein enriched class of lipoproteins. The definition of HDL as a unique class of lipoprotein particle comes from studies using density ultracentrifugation (UC). Given its high proportion of protein to lipids, HDL floats at a density range of 1.063 to 1.21 g/mL, denser than larger and more buoyant classes of lipoproteins [20–22]. Since 1950’s, definition of HDL particles by their density range is the gold standard in the field. Lipoproteins also have characteristic electrophoretic mobility, with HDL migrating predominantly with alpha electrophoretic mobility [21]. Soon after isolation and characterization of HDL, it became evident that HDL particles carry proteins (namely apolipoprotein A-I (APOA1) and apolipoprotein A-II (APOA2)) distinct from those carried by larger lipoproteins, low-density lipoprotein (LDL) and very-low density lipoprotein (VLDL) [23]. This discovery lead Alaupovic to propose definition of lipoproteins based on their apolipoprotein composition rather than their density range [23–26]. Alaupovic defined three major classes of HDL particles – those containing only APOA1 (LpA-I), those containing APOA1 and APOA2 (LpA-I/LpA-II) and those particles containing only APOA2 (LpA-II)[24,27]. From these studies, it became evident that HDL is composed by a heterogeneous mixture of particles, and that some particles containing APOA1 are not present in the density range of 1.063–1.21 g/mL, and thus do not fit HDL’s density definition.
Based on density, HDL can be separated into two distinct classes – a more dense HDL3 (1.125–1.21 g/ml), and a lighter HDL2 (1.063–1.125 g/ml)[28,29] Density isolated HDL can be further fractionated by size on non-denaturing gradient gel electrophoresis. This technique distinguishes 5 individual particle species, 2 of them comprised in the HDL2 range (designated HDL2a,b), and 3 species in HDL3 range (designated HDL3a,b,c)[30.]. These 5 classes of HDL can also be isolated by isopycnic UC [31]. Notably, these classes of HDL defined by density do not directly correspond to HDL classes defined by apolipoprotein composition, with both LpA-I and the LpA-I/A-II particles spanning the entire size range of HDL on gradient gel electrophoresis [32]. The distinction between definition of HDL by density or by apolipoprotein composition was corroborated by immunoaffinity purification of different density ranges of HDL [33], and further confirmed using native two-dimensional (2D) gel electrophoresis [34,35]. In native 2D gel electrophoresis, lipoprotein particles are first separated based on their charge (pI) in agarose gel and in the second dimension based on their size using gradient gel electrophoresis. Native 2D gel electrophoresis applied to plasma provides separation of a number of unique APOA1 containing species that include, but are not limited to small, very dense pre-beta-1 particles, large particles with pre-beta electrophoretic mobility, as well as four species with alpha-electrophoretic mobility, including spherical HDL particles [35,36].
3. Approaches to HDL isolation
To analyze HDL and categorize its subspecies, i.e. by using 1D or 2D electrophoresis or size distribution, it is not necessary to isolate HDL particles from plasma. In this case, lipid or protein specific (e.g. anti-APOA1 antibody) staining can detect HDL subspecies. In contrast, to perform proteomic analysis, HDL has to be first isolated from plasma. A number of different techniques have been used to accomplish this task.
The density ultracentrifugation (UC) has been a “golden standard” method since it was first used to define lipoprotein classes [22]. Many variations of UC methods have been developed over the years. Thus, sequential density UC uses a sequence of distinct steps to separate HDL from plasma proteins and from apolipoprotein B (apoB)-containing lipoproteins [37]. In contrast, gradient density UC, also called isopycnic ultracentrifugation, separates HDL in a single spin. This technique builds a density gradient by carefully overlaying layers of different densities with a single spin run until isopycnic equilibrium is reached [29]. Moreover, the specific density can be adjusted using different chemicals. Most frequently, aqueous KBr has been used, but achieving the density for HDL isolation using this salt results in high ionic strength of the solution that may exert possible adverse effects on stability of HDL. Besides, the sheer force particles experience in the ultracentrifuge may cause stripping of proteins from HDL particles. Indeed, several studies suggested that UC might partially disrupt HDL particles primarily by stripping a fraction of APOA1 and possibly other exchangeable apolipoproteins from the particles [38–40]. It should be noted that most of these studies used extremely long centrifugation times (24 – 96 h). Moreover, different extent of HDL sheering was observed in sequential versus gradient density UC [41] and different subpopulations of HDL particles may be affected to a different extent. For example, APOA1 only containing particles (LpA-I) appear to be more unstable than LpA-I/A-II [42]. Attempts have been made to alleviate these problems by using agents that minimize or eliminate increased ionic strength. Thus, D2O in combination with KBr, sucrose, or heavy metal salts (e.g. CsCl) have been used to attenuate the ionic strength of the solution [38,43]. Another alternative is the use of iodixanol, that produces iso-osmotic solution at all densities[44]. However, data demonstrating how well each one of these techniques can separate HDL from plasma proteins are scarce. In some studies, a band at 66–70 kDa in gel electrophoresis strongly suggested the presence of albumin after HDL isolation [39], while in others the separation appears free of plasma contamination [43]. Consequently, the wide range of UC approaches used for HDL isolation may result in substantial variations in HDL proteome, either by stripping proteins from HDL particles or by co-isolating a variable number of plasma proteins as well as more buoyant lipoproteins (LDL). Moreover, dense fractions of larger lipoproteins may overlap in density with HDL, e.g. lipoprotein(a) (Lp(a)).
Immunoaffinity chromatography (immunosorption) with antibodies specific for APOA1 is an alternative method for HDL isolation reflecting Alaupovic’s apolipoprotein based definition of lipoprotein classes, here defined by the presence of APOA1 (LpA-I particles). After the first reports of the existence of particle defined by APOA1 (Lp-A-I) [45,46], a number of studies in 70’s and early 80’s employed immunosorption to characterize the distribution of different apolipoproteins and to define lipoprotein particles based on apolipoproteins. However, use of immunoaffinity chromatography on preparative scale was first reported by Albers[47] and Kane[48] laboratories in 1984. In their approach, purified antibodies against APOA1 were immobilized on Sepharose beads and captured APOA1-containing particles were eluted using gentle acidic conditions, after extensive washing of unbound apoB-containing lipoproteins and plasma proteins. Nearly quantitative recovery of plasma APOA1 was achieved, and characterization of the LpA-I particles revealed they were relatively enriched in total protein content, at the expense of decrease in phospholipid and cholesterol content, when compared to particles isolated by UC [48]. Cheung further showed LpA-I particles can be subfractionated to APOA1 only (LpA-I) and APOA1/ APOA2 (LpA-I/LpA-II) containing particles. Each of these particle populations is unique with distinct size distribution and composition from each other as well as from HDL2 and HDL3 isolated based on density [47]. In contrast to UC, the immunoaffinity isolated LpA-I particles include pre-beta HDL, a unique particle that contains over 90% of protein in the form of APOA1[49]. Furtado et al extended subfractionation of LpA-I particle beyond APOA2 and used anti-APOA1 immunosorption combined with immunoaffinity columns against 16 HDL proteins to define 16 unique HDL particles [50]. Collectively, the immunoaffinity isolation is a method of isolation of LpA-I particles that reflect the native particles as present in serum better than HDL isolated by density UC and include particles with density higher than 1.21 g/mL (sometimes termed very-high density lipoprotein, VHDL). However, it should be noted that APOA1 is found also on APOB containing lipoproteins and the immunoaffinity approach will co-purify these large APOA1 containing particles [51,52]. Furthermore, an analysis of subjects with APOA1 deficiency demonstrated that population of particles that do not contain APOA1 exists in the HDL size and density range [53] and these particles are not captured by immunoaffinity aimed at APOA1. Furthermore, establishing specific association with immunoaffinity beads is not trivial and requires careful control of washing as well as controls for non-specific binding to the carrier beads as well as to the non-specific binding to the antibody itself [54].
In addition to isolation based on density or APOA1 content, it has been suggested that HDL can be isolated based on its size using size exclusion chromatography (SEC) with fast protein liquid chromatography (FPLC) systems [55,56]. Fractions eluting from SEC are collected and HDL range is selected based on phospholipid and cholesterol in the HDL size range. Although SEC separates proteins in their native forms, many plasma proteins have molecular weights in the same range as HDL, e.g. albumin dimer (~135 kDa), IgG (150–180 kDa), complement C3 (180 kDa), and many exist in protein complexes that would fall into the same range. Thus, the SEC merely enriches plasma for HDL. To circumvent this problem, Davidson lab extended this approach by applying the isolated HDL enriched fractions to lipid binding resin, a lipid removal agent (LRA) which efficiently binds lipids as well as lipid binding proteins [57]. Unfortunately, the proteins bound cannot be easily dissociated from the resin, and the method is only suitable for proteome analysis after digestion directly on the resin [57,58]. Moreover, a number of other proteins in plasma bind lipids although they are not considered to be classically defined as associated with HDL.
Ionic properties of HDL (as an alpha migrating lipoproteins) have been explored in an attempt to isolate HDL using anion-exchange chromatography [59–61]. Although this method separates HDL from other lipoproteins, it does not isolate it from other plasma proteins with similar ionic properties. Analogous to SEC Davidson’s group applied the LRA lipid capture approach to the anion exchange separated phospholipid containing fractions [62].
4. Mass spectrometry techniques applied to HDL proteomics
A typical workflow for HDL proteomic analysis starts with an untargeted approach to determine HDL protein composition in a given studied condition. The most common approach is referred to as bottom-up or shotgun proteomics[63] and uses unbiased data-dependent acquisition (DDA). HDL proteins are enzymatically digested into peptides that are then analyzed by liquid chromatography tandem mass spectrometry (LCMS). In this nominally unbiased discovery strategy, a full MS1 spectrum of peptides at a given time in the gradient elution is acquired, followed by tandem (MS2) mass spectra of most abundant precursor ions detected in the MS1 spectrum. The number of MS2 scans will depend on the instrument used (typically ranging from 5 – 20 scans). During chromatographic separation, the mass spectrometer cycles between the acquisition of MS1 scans and MS2 dependent scans, stochastically acquiring MS2 scans on as many precursor ions as possible, to achieve identification of large number of peptides and thereby proteins. Excellent reviews are available regarding shotgun methodology [64,65].
Identification of proteins is then accomplished by matching the experimental MS2 spectra to theoretical MS2 spectra derived from in silico digestion of proteins in protein databases. Common, vendor-free computational proteomic platforms include MaxQuant, that uses Andromeda as the search algorithm [66], Mascot database search [67], Trans-Proteomic Pipeline using Sequest [68], Comet [69], or X!Tandem [70] search engines, openMS [71], with further validation of peptide-spectrum-matches (PSM) and protein identifications with tools like those included in Trans-Proteomic Pipeline [72–75]. Vendor specific search engines are also available commercially. Further in depth discussion of peptide and protein identification can be found elsewhere [76]. The DDA approach has been so far the most common approach employed for HDL proteome studies.
5. HDL proteome
Although approximately 90% of HDL protein mass is derived from two proteins, APOA1 and APOA2, the list of proteins reported to reside on HDL has been growing steadily. In 2015, the Davidson/Shah Laboratory compiled a list that contained 95 HDL proteins detected by 17 different studies (http://homepages.uc.edu/~davidswm/HDLproteome.html). In this review, we aimed to update the list of proteins associated with HDL and ask questions about the relationship of the HDL proteome and its methods of isolation and analysis. We reviewed studies reporting HDL proteome up to April 2019 and compiled data from 37 different studies on HDL (43 lists of proteins, as some studies reported different isolation methodologies to isolate HDL). Studies that had made HDL protein list available on-line, or that provided the list upon our request are included (Supplemental Table 1) (Supplemental data is also available at HDL Proteome page, http://faculty.washington.edu/tvaisar/). The compiled list includes a wide range of techniques used for isolation of HDL from plasma. Twenty-six studies used UC as the isolation methodology either for direct analysis [77–89] or with subsequent fractionation by 1D [90–92], 2D [93,94], or native gel electrophoresis [84], OFF-gel fractionation [95,96], anion exchange chromatography [97] or a combination of fractionation techniques [84,98–100]. While most studies used the 1.21 g/mL density cutoff, one study used ultracentrifugation with a density cutoff of 1.24 g/mL in an attempt to capture VHDL [101]. Immunoaffinity isolation with antibodies against APOA1 was used in 5 studies [50,83,102–104]. Size exclusion chromatography (SEC) isolating fractions corresponding to the size range of HDL (based on cholesterol and phospholipid content) was used in 9 studies [55–57,62,105–109]. From those, 4 studies performed LCMS analysis directly on the collected fractions [55,56,108,109], while five used further enrichment of lipid binding proteins by capture on calcium silicate hydrate resin. A single study used an approach where a histidine-tagged APOA1 was used as a bait for enrichment of the HDL from plasma using a metal chelate chromatography (AALP method) [82].
The consolidated list of proteins identified in each study is available in Supplemental Table 2. We classified the methods of isolation as ultracentrifugation (UC), immunoaffinity for APOA1 (AFF), size exclusion chromatography (SEC), size exclusion chromatography in combination with lipid binding resin (SEC_LRA) and the use of histidine-tagged APOA1 (AALP). If the UC was followed by fractionation, we further classify the study by the technique that was employed as 1D gel (UC_1D), 2D gel (UC_2D), OFF gel fractionation (UC_OFF), anion exchange chromatography (UC_AEX) or a combination of fractionation techniques (UC_Frac).
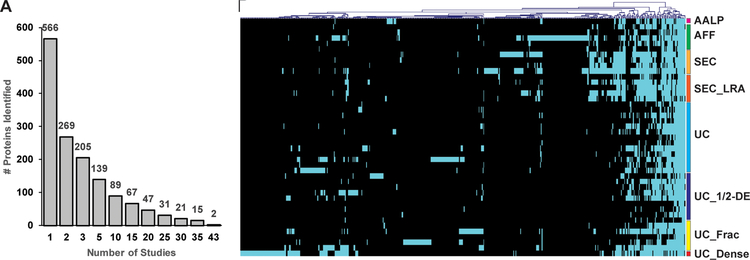
Overall, 566 proteins have been reported to be associated with HDL. Strikingly, only 2 proteins, APOA1 and apolipoprotein L1 (APOL1) were identified in all studies (Figure 1A) and only 21 proteins were found by ~75% (30) studies. Moreover, less than 25 % of proteins (139) were identified by at least 5 different studies, while more than 50 % (297) were detected by a single study (Figure 1A).
Figure 1.
Proteins associated with HDL in 37 reported studies. A) Plot showing the number of proteins associated with HDL shared by multiple studies. B) Heat map of proteins identified in each study. The studies were manually grouped based on the HDL isolation methodology. The proteins were grouped by hierarchical clustering. Teal indicates presence of the protein, black absence of the protein.
Hierarchical clustering analysis of the identified proteins and isolation methodologies shows that only a small subset of proteins is consistently found even when the same isolation technique is used. Thus, even studies employing the same isolation methodology identify divergent sets of proteins in addition to a proportionally smaller ensemble of core proteins (Figure 1B).
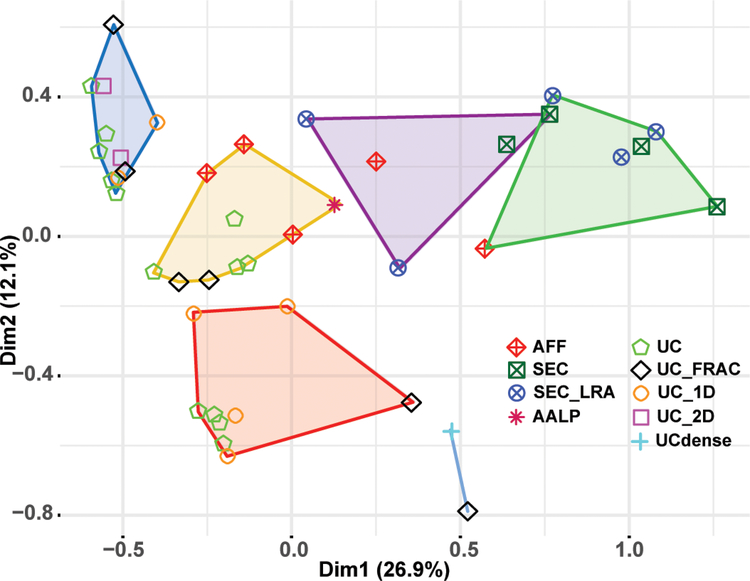
We applied an unbiased multiple correspondence analysis (MCA), a version of principal component analysis for categorical data, to detect relationships between studies with similar profiles of identified proteins, and to test whether certain groups of proteins contribute to the variance across different HDL studies. MCA was performed with a subset of proteins shared by at least 5 different studies (139 proteins) (Supplemental Table 3). We then applied hierarchical clustering based on the eigenvalues obtained from MCA to determine which studies identified similar protein ensembles. MCA (individual symbols) together with hierarchical clustering (colored polygons) clearly demonstrate the method of HDL isolation plays an important role in determining the ensemble of proteins detected in HDL (Figure 2). Studies using the same isolation technique tend to cluster together, although there are notable differences within a single isolation technique and there is overlap between different techniques. The yellow cluster includes HDL isolation by UC (green and black symbols) and some AFF (crossed red diamonds) studies, while the purple cluster comprises HDL isolation by AFF, SEC_LRA (crossed blue circles) and SEC (green crossed squares). Thus, the studies employing AFF span both UC-based as well as SEC_LRA approaches, while SEC_LRA studies bridge AFF and SEC studies. For UC-based methods, the analysis also shows a clear distinction in identified proteomes, with studies employing UC and 1D and 2D electrophoresis segregated from other UC studies (blue cluster), likely due to the smaller number of proteins identified when these techniques were applied (Figure 2).
Figure 2.
Multiple correspondence analysis (MCA) and hierarchical clustering of individual studies. MCA was performed with a subset of proteins that were identified in at least 5 different studies (n = 139). Individual studies were grouped by hierarchical clustering based on the eigenvalues for each study obtained from the MCA (e.g. based on similarity of the protein ensemble identified). The clusters of closely related studies are represented by colored polygons. Each study is further denoted by an individual symbol corresponding to the HDL isolation methodology used.
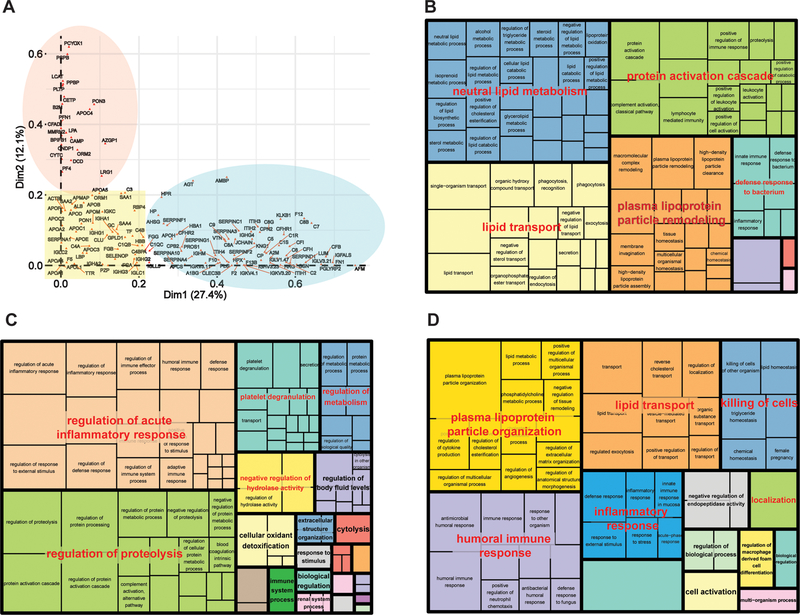
Analysis of the association of individual proteins with the first two principal components (dimensions1 and 2, Dim1 and Dim2) revealed the contribution of each protein to the distinction of the different studies (Figure 3A). Indeed, proteins with minimal correlation with either principal component (yellow square) are the ones most consistently found across the studies (Supplemental Table 4A). Proteins with the highest correlation with Dim1 (blue oval) are those mostly responsible for distinguishing the studies using AFF, SEC and SEC_LRA methods from the UC studies (Supplemental Table 4B), while those along Dim2 (red circle) contribute to the differentiation among various UC studies (Supplemental Table 4C). To gain further insight into the relationship of the isolation techniques and the identified proteins, we set a threshold of 0.2 in each dimension and performed functional annotation analysis on the 3 resulting groups of proteins. The group of proteins that do not contribute to differentiation of isolation techniques (<0.2 along both Dim1 and Dim2 in Figure 3A, yellow square) is highly enriched in functional categories related to lipoprotein biology (e.g. lipid transport, neutral lipid metabolism, lipoprotein particle remodeling, lipoprotein metabolism) as well as protein related to activation cascade and defense to bacterium (Figure 3B). In contrast, proteins mostly correlated with Dim1 and thus distinguishing AFF and SEC isolation methods from UC studies represent functional categories related to protein activation cascade, inflammatory response, immune response, exocytosis and regulation of catalytic activity (Figure 3C). The group of proteins mostly correlated with Dim2 and primarily explaining the variance among different UC-based studies represent predominantly functional categories of lipoproteins particle remodeling and lipid transport (Figure 3D).
Figure 3.
MCA correlations and functional annotation analyses of proteins reported in HDL. A) Correlation analysis of the individual proteins within the first two principal components (Dim1 and Dim2) of MCA. The yellow square indicates the group of proteins with correlation < 0.2 with either dimension. Proteins highly correlated with Dim1 (correlation > 0.2) are highlighted in the blue ellipse, while proteins with correlation > 0.2 with Dim2 are highlighted by the red ellipse. B) Functional annotation analysis of proteins that do not contribute for variance in either dimension in MCA (correlations < 0.2 with Dim1 and Dim2). C) Functional annotation analysis of proteins with correlation > 0.2 with Dim1. D) Functional annotation analysis of proteins with correlation > 0.2 with Dim2.
6. Which proteins are truly associated with HDL?
The large number of proteins reported to be associated with HDL begs the question of how so many proteins can fit on a finite number of particles given the constraint of apparent HDL size (~60 – 350 kDa). Moreover, the low consistency of the proteins identified in HDL, even within the studies using the same isolation technique, raises an urgent question of which proteins truly reside on HDL (whichever way it is defined) and which proteins are contaminants of the isolation methodology. As showed above, the protein cargo of HDL varies according to the isolation methodology. This can be, in part, a consequence of different “definitions” of HDL as well as of the capacity of some techniques to remodel HDL proteome or conversely preserve more loosely associated proteins. However, each technique also carries the potential of co-isolating proteins that do not belong to HDL, but may appear in the HDL proteome due to various reasons (imprecise density for UC, non-specific binding on the affinity resin for AFF isolation, lipid binding proteins in plasma that are not related to HDL for SEC_LRA, to name just a few). Determining which proteins belong to HDL and which proteins are contaminants is essential due to several reasons. First, it is reasonable to assume that HDL proteins may be associated with HDL biological functions. Second, HDL protein cargo maybe a marker of disease state, and therefore it is important to ensure that the biomarker protein is a stable trait of HDL and that its presence in HDL is not susceptible to variations in the isolation technique. For example, albumin (ALB), the most abundant protein in plasma (50% of plasma protein mass and ~50x higher abundance than APOA1), is generally considered a contaminant, although a previous study found ALB reduced in HDL of type I diabetes subjects [107]. ALB also co-elutes with HDL on SEC (as a dimer and tetramer) and it is known to bind free fatty acids and other lipids. Moreover, previous study suggested that albumin may associate with APOA1[110] and it has been shown that ALB may participate in the cholesterol efflux, one of key biological functions of HDL[111]. Thus, in the case of an abundant plasma protein found in HDL associated with a disease, it would be important to compare its HDL and plasma levels. Correlation between plasma and HDL levels could clarify whether or not altered HDL level is a merely consequence of its altered plasma level. For immunosorption, a number of non-specifically associated proteins may copurify together with true specifically associated proteins. It is, therefore, critically important to include negative controls to identify proteins specifically associated with the target protein, i.e. immunosorption on beads coupled with pre-immune IgG or the use of beads coupled with an indifferent protein [54]. Furthermore, strong correlation with ALB may be considered a simple test of a likely contaminant. A more direct test of protein’s association with HDL would be co-immunoprecipitation, where a protein of interest is immunoprecipitated and the precipitate is probed with specific antibody against APOA1 (to test association with APOA1 containing HDL particles) or other key HDL protein (e.g. APOE to test for presence on APOE-only particles). However, immunoprecipitation has limitations in quality of the antibody used, i.e. its specificity for the antigen and the ability to quantitatively precipitate its antigen. Besides the isolation methodology, the storage conditions may affect HDL structure and function [112,113]. However, storage is likely to impact functionality of HDL more than its composition.
7. Application of quantitative proteomics to HDL
Shotgun proteomic analysis is suitable for enumeration of HDL associated proteins, but its application to protein quantitation is limited [12]. It may be used in small studies and to create a peptide library containing information about peptides and fragments aiding further development of more quantitative methods. However, this application of shotgun proteomics may be limited as peptides appearing with high number of PSMs may not provide best signal in quantitative targeted methods [114]. In the context of translational studies, targeted methods providing robust and sensitive protein quantification are essential [115,116].
Selected reaction monitoring (SRM, also called multiple reaction monitoring (MRM)) is the standard approach in quantitative MS-based proteomics [117–119]. This method of acquisition requires selection of representative peptide candidates (at least two or three) for each protein (i.e. previous knowledge of the peptides detected well in the sample). In a typical SRM experiment, three to five fragment ions per precursor are monitored individually, using triple quadrupole mass spectrometers. With the development of instrumentation, SRM assays have become highly multiplexed allowing quantification of more than 400 proteins in single analysis [117–120]. A multiplexed SRM assay for HDL proteins (apolipoproteins A-I, C-II, C-III, E, B, and J) demonstrated similar performance of SRM to that obtained with commercially available immunoassays for each of the 6 analytes tested [12]. An SRM assay for pooled HDL samples was also developed, and 7 out of 9 proteins correlated well (Spearman rho >= 0.8) with immunoassays [121]. Moreover, 37 proteins were quantified by SRM in HDL of hemodialysis patients, and results showed elevation in a cluster of kidney disease associated-proteins [81]. A protocol to quantify by SRM 37 HDL proteins is available [37].
An alternative methodology for quantitative proteomics is paralleled reaction monitoring (PRM) that can be performed on quadrupole-orbitrap or quadrupole-time of flight (QTOF) mass spectrometers. Similar to SRM, PRM is a hypothesis-driven experiment, where the peptides must be selected before starting the experiment, but PRM has some potential advantages over SRM. First, it requires only selection of the precursors, as full MS2 spectrum is acquired, and a subset of fragment ions can be selected post factum to improve signal and eliminate possible noise contamination. Second, fragment ions are obtained with high resolution, further improving selectivity [122,123]. In a recent work, we compared SRM and PRM methodologies for quantification of HDL proteins, and concluded both methods are suitable for targeted quantification of HDL proteome with the needed confidence required for translational studies [124]. Moreover, both, PRM and SRM techniques were able to differentiate HDL abnormal composition in diabetic subjects with fenofibrate/rosiglitazone-induced hypoalphalipoproteinemia. This was an interesting result, given HDL proteome alterations were detected before subjects developed a striking reduction in HDL levels. Altered levels of paraoxonase/arylesterase 1 (PON1) and apolipoprotein CII (APOC2) were further confirmed by immunoblot and ELISA experiments [125]. With current instrumentation, the downside of PRM is that acquisition speed is slower than that obtained with triple quadrupole mass spectrometers and thus limits its ability to multiplex on the same scale.
Recently, another type of mass spectrometry acquisition approach called data independent acquisition (DIA) has been developed [126,127]. Like PRM experiments, DIA is currently performed in quadrupole-orbitrap or quadrupole-time of flight mass analyzers. During DIA workflow, the mass spectrometer acquires an MS1 scan, followed by a series of MS2 scans with sequential isolation of wide precursor ion windows covering the m/z range of the MS1 scan. The term DIA refers to the fact that MS2 spectra are acquired without obtaining specific precursor ion mass from a survey MS1 scan [128]. A well-known DIA acquisition method is termed sequential window acquisition of all theoretical mass spectra (SWATH-MS) [127]. Isolation windows of 10 or 25 m/z, are commonly employed in a DIA experiment, compared to PRM or SRM with isolation windows of 1 or 2 m/z. Reviews of the DIA technique, [129–131] its different data processing methodologies [132] and a tutorial on DIA/SWATH analysis [128] are available elsewhere.
DIA has some important advantages over shotgun and SRM or PRM that make it an outstanding tool for early stage studies of HDL proteome. First, it does not require a list of predefined proteins (and peptides) alleviating the need of previous knowledge or method development. Second, because full MS2 spectra are acquired across a wide range of m/z, one can re-examine the data (selecting new peptides and proteins) after acquisition [133]. A comparison between HDL proteomics of type 1 diabetes patients and controls was undertaken using DIA methodology [107]. However, this study did not presented data regarding repeatability and reproducibility of DIA methodology, and failed to validate by a different methodology the presence of one of its putative hits, factor H related protein 2, in HDL.
DIA showed reproducible results when applied to plasma proteomics [134] and was also reproducible across multiple independent laboratories [135]. Whether or not DIA will be successfully applied to HDL proteomics in a clinical setting, giving comparable results to established SRM or PRM methodologies remains to be seen.
8. The urgent need for standardization of HDL proteome quantification
The utility of targeted methods such as SRM and PRM has already been demonstrated for HDL proteome quantification [12,124]. To facilitate rapid progression from preliminary discovery studies to translational studies, a workflow employing DIA analysis either building from the list of proteins summarized here or using DIA generated chromatogram libraries (DIA combining narrow 2 m/z windows and concept of gas-phase fractionation) to rapidly select detectable proteins and corresponding best peptides to build SRM or PRM based targeted methods. Critical to high quality translational quantitative studies is inclusion of internal standards (IS) (either exogenous peptides or stable isotope labeled analogs), and quality control samples (QCs). To control for the variance introduced during the process of isolation, QC samples for the isolation should be included, i.e. the same plasma isolated multiple times in parallel with the study samples, especially when isolation has to be carried out in batches. The use of IS should ideally control for the trypsin digestion, as well as for the variation of the LCMS analyses. Labeled proteins are especially useful in this approach (e.g. 15N-APOA1) [12,124,125]. However, a stable isotope labeled peptide for each measured peptide (one per protein of interest) or a single peptide spiked in all samples to correct for mass spectrometry variability may be acceptable. In this scenario, using QC samples to control for digestion variability is necessary. A heavy labeled peptide for each protein of interest would provide the additional benefit of controlling for variabilities during the chromatographic run (i.e. ion suppression from co-eluting peptides).
In addition, all samples belonging to a given study should be digested concomitantly. Alternatively, inclusion of a single, well-characterized standard sample with each digestion and LCMS batch can be used to control for inter-batch variability [136,137]. A single standard sample could be used for absolute quantification, if the concentrations values were assigned based on independent biochemical or immunological assay or used as arbitrary reference value [137]. In summary, employment of a robust and precise quantitative methodology, together with appropriate quality controls is necessary for the successful application of HDL proteomics to translational studies. Several recent studies show that HDL proteomics is moving in that direction [136,138,139].
9. Expert Opinion
There is great need to establish new metrics for HDL protective properties. The complexity of HDL proteome parallels its functional diversity and may be related to its pleotropic functions and HDL proteins may serve as biomarkers of disease. To establish clinical relevance of HDL proteins as metric of HDL protective capacity, precise and robust quantification of HDL protein cargo in large prospective and cross-sectional cohorts is critical. So far, studies performed in different laboratories and employing a range of HDL isolation techniques indicate that consistent quantification of HDL proteins is not straightforward. Two key parameters for precise HDL proteome quantification are the reproducible isolation methodology and precise quantitative mass spectrometry method. Multiple HDL isolation methodologies are available, each one with its own advantages and limitations. The immunosorption offers gentle isolation with wide range of “non-classical” proteins (non-apolipoproteins) identified, however, it has great potential for non-specific binding and detection of contaminant proteins. Even when combined with capture of lipid binding proteins (SEC-LRA), SEC has limitation in its potential for capturing “non-HDL” and non-specific proteins, as well as limited throughput due to lengthy FPLC separation. In contrast, UC isolation can be highly reproducible, but may alter HDL proteome by stripping proteins and has limited throughput due to laborious process. The lack of consistency among different studies, even those using the same isolation technique, is currently hampering the advance of HDL field towards clinical relevance. This confounding issue, caused by co-purification/identification of non-specific proteins, can be limited in targeted methods by focus on only well-established proteins or proteins clearly associated with a disease condition. The majority of proteomic studies reported so far focused on enumerating HDL proteins and detection of differentially abundant proteins using shotgun proteomics. However, quantification of proteins by shotgun proteomics has limitations, including inherent lack of repeatability, with more than half of 550 proteins described reported by a single study, and limited quantitative response of the spectral counting and MS1-based label free quantification, and low throughput, limiting number of the samples analyzed. The later limitations of throughput and precision may be partially improved by using multiplex isobaric labeling techniques (e.g. ITRAQ, TMT) although caution in experimental design must be exercised [140]. To ensure precision, repeatability and throughput necessary for translational studies, quantitative methodologies must be employed for HDL proteome analysis. Furthermore, use of internal standards for increased precision, quality controls and standardization are further critical steps to achieving high quality, quantitative results in translational studies.
It is imperative that future studies focus on precise quantification of HDL proteins. Key questions awaiting to be answered in the field are: which proteins are truly associated with HDL; how the complex proteome correlates with HDL functions; and whether HDL protein biomarkers could predict CVD risk better than the current metrics available.
Supplementary Material
Article highlights.
HDL proteome is complex and accurate detection of HDL proteins is a challenge.
More than 550 proteins have been reported in HDL, but only a small subset of them are consistently found across multiple studies, even when the same isolation technique is used.
HDL definition and related isolation techniques are important determinants of the ensemble of proteins detected in HDL.
Translational studies are essential for establishing the relevance of HDL proteome to disease pathology.
Targeted methods providing robust and sensitive quantification are critical for translational studies.
Acknowledgments
Funding
This work was supported by grants from the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP): 2016/00696–3 and 2013/07937–8, and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq): 402683/2016–1 (to G.E.R.) and grants P30DK017047, P01HL092969, P01HL128203, and R01HL144558 from the NIH (to T.V.).
Footnotes
Declaration of interest
T. Vaisar has consulted for MedImmune LLC on studies not directly related to the subject of the review. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Reviewer disclosures
Peer reviewers on this manuscript have no relevant financial or other relationships to disclose.
References
Papers of special note have been highlighted as:
* of interest
** of considerable interest
- 1.Go AS, Mozaffarian D, Roger VL et al. Heart disease and stroke statistics--2013 update: a report from the American Heart Association. Circulation, 127(1), e6–e245 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Toth PP. High-density lipoprotein and cardiovascular risk. Circulation, 109(15), 1809–1812 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Boden WE, Probstfield JL, Anderson T et al. Niacin in patients with low HDL cholesterol levels receiving intensive statin therapy. New England Journal of Medicine, 365(24), 2255–2267 (2011s). [DOI] [PubMed] [Google Scholar]
- 4.Rader DJ, Tall AR. The not-so-simple HDL story: Is it time to revise the HDL cholesterol hypothesis? Nature Medicine, 18(9), 1344–1346 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Madsen CM, Varbo A, Nordestgaard BG. Extreme high high-density lipoprotein cholesterol is paradoxically associated with high mortality in men and women: two prospective cohort studies. European heart journal, 38(32), 2478–2486 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Voight BF, Peloso GM, Orho-Melander M et al. Plasma HDL cholesterol and risk of myocardial infarction: a mendelian randomisation study. Lancet, 380(9841), 572–580 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenson RS, Brewer HB Jr., Ansell BJ et al. Dysfunctional HDL and atherosclerotic cardiovascular disease. Nat Rev Cardiol, 13(1), 48–60 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vaisar T Proteomics investigations of HDL: challenges and promise. Curr Vasc Pharmacol, 10(4), 410–421 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- **9.Shah AS, Tan L, Long JL, Davidson WS. Proteomic diversity of high density lipoproteins: our emerging understanding of its importance in lipid transport and beyond. Journal of lipid research, 54(10), 2575–2585 (2013).Excellent review of HDL structure and speciation with discussion of the history of HDL analysis and function.
- **10.Kontush A, Lindahl M, Lhomme M, Calabresi L, Chapman MJ, Davidson WS. Structure of HDL: particle subclasses and molecular components. Handbook of experimental pharmacology, 224, 3–51 (2015).Comprehensive discussion of the HDL classifications and protein as well as lipid composition.
- 11.Riwanto M, Rohrer L, von Eckardstein A, Landmesser U. Dysfunctional HDL: from structure-function-relationships to biomarkers. Handbook of experimental pharmacology, 224, 337–366 (2015). [DOI] [PubMed] [Google Scholar]
- **12.Hoofnagle AN, Becker JO, Oda MN, Cavigiolio G, Mayer P, Vaisar T. Multiple-reaction monitoring-mass spectrometric assays can accurately measure the relative protein abundance in complex mixtures. Clinical chemistry, 58(4), 777–781 (2012).Application of targeted quantitative method to HDL. Demonstrates correlation of targeted proteomics with biochemical quantification for six HDL proteins with wide range of relativ abundance.
- 13.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Molecular & cellular proteomics : MCP, 1(11), 845–867 (2002). [DOI] [PubMed] [Google Scholar]
- 14.Asztalos BF, Niisuke K, Horvath KV. High-density lipoprotein: our elusive friend. Current opinion in lipidology, 30(4), 314–319 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Rosenson RS, Brewer HB, Chapman MJ et al. HDL measures, particle heterogeneity, proposed nomenclature, and relation to atherosclerotic cardiovascular events. Clinical chemistry, 57(3), 392–410 (2011). [DOI] [PubMed] [Google Scholar]
- *16.Karathanasis SK, Freeman LA, Gordon SM, Remaley AT. The Changing Face of HDL and the Best Way to Measure It. Clinical chemistry, 63(1), 196–210 (2017).Comprehensive review discussing HDL measurement methods in light of the most recent methodologies.
- 17.Hafiane A, Genest J. High density lipoproteins: Measurement techniques and potential biomarkers of cardiovascular risk. BBA clinical, 3, 175–188 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- *18.Shao B, Heinecke JW. Quantifying HDL proteins by mass spectrometry: how many proteins are there and what are their functions? Expert Rev Proteomics, 15(1), 31–40 (2018).Review of clinical applications of HDL proteomics.
- 19.Macheboeuf MA. Sur l’etat physiochimique de la lecithine et des esters de cholesterol dans le serum. Bull. Soc. Chim, 45, 662 (1929). [Google Scholar]
- 20.Gofman JW, Lindgren FT, Elliott H. Ultracentrifugal studies of lipoproteins of human serum. J Biol Chem, 179(2), 973–979 (1949). [PubMed] [Google Scholar]
- 21.Lewis LA, Page IH. Electrophoretic and ultracentrifugal analysis of serum lipoproteins of normal, nephrotic and hypertensive persons. Circulation, 7(5), 707–717 (1953). [DOI] [PubMed] [Google Scholar]
- **22.Havel RJ, Eder HA, Bragdon JH. The distribution and chemical composition of ultracentrifugally separated lipoproteins in human serum. The Journal of clinical investigation, 34(9), 1345–1353 (1955).First comprehensive characterization of lipoproteins isolated by ultracentrifugation.
- 23.Kostner G, Alaupovic P. Studies of the composition and structure of plasma lipoproteins. C- and N-terminal amino acids of the two nonidentical polypeptides of human plasma apolipoprotein A. FEBS Lett, 15(4), 320–324 (1971). [DOI] [PubMed] [Google Scholar]
- **24.Alaupovic P. The concept of apolipoprotein-defined lipoprotein families and its clinical significance. Current atherosclerosis reports, 5(6), 459–467 (2003).Concept of the lipoproteins classification based on protein compositions is reviewed.
- 25.Kostner G, Alaupovic P. Studies of the composition and structure of plasma lipoproteins. Separation and quantification of the lipoprotein families occurring in the high density lipoproteins of human plasma. Biochemistry, 11(18), 3419–3428 (1972). [DOI] [PubMed] [Google Scholar]
- 26.Suenram A, McConathy WJ, Alaupovic P. Evidence for the lipoprotein heterogeneity of human plasma high density lipoproteins isolated by three different procedures. Lipids, 14(5), 505–510 (1979). [DOI] [PubMed] [Google Scholar]
- 27.Alaupovic P Apolipoprotein composition as the basis for classifying plasma lipoproteins. Characterization of ApoA- and ApoB-containing lipoprotein families. Progress in lipid research, 30(2–3), 105–138 (1991). [DOI] [PubMed] [Google Scholar]
- 28.Scanu A Studies on the conformation of human serum high-density lipoproteins HDL2 and HDL3. Proceedings of the National Academy of Sciences of the United States of America, 54(6), 1699–1705 (1965). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chapman MJ, Goldstein S, Lagrange D, Laplaud PM. A density gradient ultracentrifugal procedure for the isolation of the major lipoprotein classes from human serum. J Lipid Res, 22(2), 339–358 (1981). [PubMed] [Google Scholar]
- 30.Blanche PJ, Gong EL, Forte TM, Nichols AV. Characterization of human high-density lipoproteins by gradient gel electrophoresis. Biochimica et biophysica acta, 665(3), 408–419 (1981). [DOI] [PubMed] [Google Scholar]
- 31.Davidson WS, Silva RA, Chantepie S, Lagor WR, Chapman MJ, Kontush A. Proteomic analysis of defined HDL subpopulations reveals particle-specific protein clusters: relevance to antioxidative function. Arteriosclerosis, thrombosis, and vascular biology, 29(6), 870–876 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Z, McNamara JR, Ordovas JM, Schaefer EJ. Analysis of high density lipoproteins by a modified gradient gel electrophoresis method. Journal of lipid research, 35(9), 1698–1711 (1994). [PubMed] [Google Scholar]
- 33.Cheung MC, Albers JJ. Distribution of high density lipoprotein particles with different apoprotein composition: particles with A-I and A-II and particles with A-I but no A-II. Journal of lipid research, 23(5), 747–753 (1982). [PubMed] [Google Scholar]
- 34.Asztalos BF, Schaefer EJ. High-density lipoprotein subpopulations in pathologic conditions. The American journal of cardiology, 91(7A), 12E–17E (2003). [DOI] [PubMed] [Google Scholar]
- **35.Asztalos BF, Tani M, Schaefer EJ. Metabolic and functional relevance of HDL subspecies. Current opinion in lipidology, 22(3), 176–185 (2011).Review of HDL heteregeneity and its relation to HDL metabolism and disease.
- 36.Asztalos BF, Sloop CH, Wong L, Roheim PS. Two-dimensional electrophoresis of plasma lipoproteins: recognition of new apo A-I-containing subpopulations. Biochimica et biophysica acta, 1169(3), 291–300 (1993). [DOI] [PubMed] [Google Scholar]
- 37.Henderson CM, Vaisar T, Hoofnagle AN. Isolating and Quantifying Plasma HDL Proteins by Sequential Density Gradient Ultracentrifugation and Targeted Proteomics. Methods in molecular biology (Clifton, N.J.), 1410, 105–120 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kunitake ST, Kane JP. Factors affecting the integrity of high density lipoproteins in the ultracentrifuge. Journal of lipid research, 23(6), 936–940 (1982). [PubMed] [Google Scholar]
- 39.Munroe WH, Phillips ML, Schumaker VN. Excessive centrifugal fields damage high density lipoprotein. Journal of lipid research, 56(6), 1172–1181 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fainaru M, Glangeaud MC, Eisenberg S. Radioimmunoassay of human high density lipoprotein apo-protein A-1. Biochimica et biophysica acta, 386(2), 432–443 (1975). [DOI] [PubMed] [Google Scholar]
- 41.Murdoch SJ, Breckenridge WC. Development of a density gradient ultracentrifugation technique for the resolution of plasma lipoproteins which avoids apo E dissociation. Anal Biochem, 222(2), 427–434 (1994). [DOI] [PubMed] [Google Scholar]
- 42.Cheung MC, Wolf AC. Differential effect of ultracentrifugation on apolipoprotein A-I-containing lipoprotein subpopulations. Journal of lipid research, 29(1), 15–25 (1988). [PubMed] [Google Scholar]
- 43.Ståhlman M, Davidsson P, Kanmert I et al. Proteomics and lipids of lipoproteins isolated at low salt concentrations in D2O/sucrose or in KBr. Journal of lipid research, 49(2), 481–490 (2008). [DOI] [PubMed] [Google Scholar]
- 44.Harman NL, Griffin BA, Davies IG. Separation of the principal HDL subclasses by iodixanol ultracentrifugation. Journal of lipid research, 54(8), 2273–2281 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Albers JJ, Aladjem F. Biochemistry, 10, 3436–3442 (1971). [DOI] [PubMed] [Google Scholar]
- 46.Kostner G, Depisch A, Petek W, Holasek A. Hoppe-Seylers Z. Physiol. Chem, 352, 1440–1444 (1971). [PubMed] [Google Scholar]
- 47.Cheung MC, Albers JJ. Characterization of lipoprotein particles isolated by immunoaffinity chromatography. Particles containing A-I and A-II and particles containing A-I but no A-II. J Biol Chem, 259(19), 12201–12209 (1984). [PubMed] [Google Scholar]
- 48.McVicar JP, Kunitake ST, Hamilton RL, Kane JP. Characteristics of human lipoproteins isolated by selected-affinity immunosorption of apolipoprotein A-I. Proceedings of the National Academy of Sciences of the United States of America, 81(5), 1356–1360 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kunitake ST, La Sala KJ, Kane JP. Apolipoprotein A-I-containing lipoproteins with pre-beta electrophoretic mobility. Journal of lipid research, 26(5), 549–555 (1985). [PubMed] [Google Scholar]
- **50.Furtado JD, Yamamoto R, Melchior JT et al. Distinct Proteomic Signatures in 16 HDL (High-Density Lipoprotein) Subspecies. Arteriosclerosis, thrombosis, and vascular biology, 38(12), 2827–2842 (2018).Novel approach to determination of HDL subspecies and their composition using immunoaffinity isolation.
- 51.Lepedda AJ, Nieddu G, Zinellu E et al. Proteomic analysis of plasma-purified VLDL, LDL, and HDL fractions from atherosclerotic patients undergoing carotid endarterectomy: identification of serum amyloid A as a potential marker. Oxidative medicine and cellular longevity, 2013, 385214 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Diffenderfer MR, Schaefer EJ. The composition and metabolism of large and small LDL. Current opinion in lipidology, 25(3), 221–226 (2014). [DOI] [PubMed] [Google Scholar]
- 53.Santos RD, Schaefer EJ, Asztalos BF et al. Characterization of high density lipoprotein particles in familial apolipoprotein A-I deficiency. Journal of lipid research, 49(2), 349–357 (2008). [DOI] [PubMed] [Google Scholar]
- 54.Cheung MC, Vaisar T, Han X, Heinecke JW, Albers JJ. Phospholipid transfer protein in human plasma associates with proteins linked to immunity and inflammation. Biochemistry, 49(34), 7314–7322 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Collins LA, Olivier M. Quantitative comparison of lipoprotein fractions derived from human plasma and serum by liquid chromatography-tandem mass spectrometry. Proteome science, 8, 42 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Collins LA, Mirza SP, Kissebah AH, Olivier M. Integrated approach for the comprehensive characterization of lipoproteins from human plasma using FPLC and nano-HPLC-tandem mass spectrometry. Physiological genomics, 40(3), 208–215 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- **57.Gordon SM, Deng J, Lu LJ, Davidson WS. Proteomic characterization of human plasma high density lipoprotein fractionated by gel filtration chromatography. Journal of proteome research, 9(10), 5239–5249 (2010).Describes the approach to HDL proteome based on combination of SEC and lipid capture on lipid removal resin.
- 58.Heink A, Davidson WS, Swertfeger DK, Lu LJ, Shah AS. A Comparison of Methods To Enhance Protein Detection of Lipoproteins by Mass Spectrometry. Journal of proteome research, 14(7), 2943–2950 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hirowatari Y, Yoshida H, Kurosawa H, Doumitu K-i, Tada N. Measurement of cholesterol of major serum lipoprotein classes by anion-exchange HPLC with perchlorate ion-containing eluent. Journal of lipid research, (2003). [DOI] [PubMed]
- 60.Hirowatari Y, Tsunoda Y, Ogura Y, Homma Y. Analyzing of high-density lipoprotein subfractions and low-density lipoprotein subfractions in human serum with anion-exchange chromatography. Atherosclerosis, 204(2), 7 (2009). [DOI] [PubMed] [Google Scholar]
- 61.Ji X, Xu H, Zhang H, Hillery CA, Gao H-QQ, Pritchard KA. Anion exchange HPLC isolation of high-density lipoprotein (HDL) and on-line estimation of proinflammatory HDL. PloS one, 9(3) (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gordon SM, Deng J, Tomann AB, Shah AS, Lu LJ, Davidson WS. Multi-dimensional co-separation analysis reveals protein-protein interactions defining plasma lipoprotein subspecies. Molecular & cellular proteomics : MCP, 12(11), 3123–3134 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Link AJ, Eng J, Schieltz DM et al. Direct analysis of protein complexes using mass spectrometry. Nature biotechnology, 17(7), 676–682 (1999). [DOI] [PubMed] [Google Scholar]
- 64.Aebersold R, Mann M. Mass-spectrometric exploration of proteome structure and function. Nature, 537(7620), 347–355 (2016). [DOI] [PubMed] [Google Scholar]
- 65.Gillet LC, Leitner A, Aebersold R. Mass Spectrometry Applied to Bottom-Up Proteomics: Entering the High-Throughput Era for Hypothesis Testing. Annual review of analytical chemistry (Palo Alto, Calif.), 9(1), 449–472 (2016). [DOI] [PubMed] [Google Scholar]
- 66.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature biotechnology, 26(12), 1367–1372 (2008). [DOI] [PubMed] [Google Scholar]
- 67.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis, 20(18), 3551–3567 (1999). [DOI] [PubMed] [Google Scholar]
- 68.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom, 5(11), 976–989 (1994). [DOI] [PubMed] [Google Scholar]
- 69.Eng JK, Jahan TA, Hoopmann MR. Comet: an open-source MS/MS sequence database search tool. Proteomics, 13(1), 22–24 (2013). [DOI] [PubMed] [Google Scholar]
- 70.Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics, 20(9), 1466–1467 (2004). [DOI] [PubMed] [Google Scholar]
- 71.Rost HL, Sachsenberg T, Aiche S et al. OpenMS: a flexible open-source software platform for mass spectrometry data analysis. Nature methods, 13(9), 741–748 (2016). [DOI] [PubMed] [Google Scholar]
- 72.Deutsch EW, Mendoza L, Shteynberg D et al. A guided tour of the Trans-Proteomic Pipeline. Proteomics, 10(6), 1150–1159 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Keller A, Eng J, Zhang N, Li XJ, Aebersold R. A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Molecular systems biology, 1, 2005.0017 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Analytical chemistry, 75(17), 4646–4658 (2003). [DOI] [PubMed] [Google Scholar]
- 75.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Analytical chemistry, 74(20), 5383–5392 (2002). [DOI] [PubMed] [Google Scholar]
- 76.Verheggen K, Raeder H, Berven FS, Martens L, Barsnes H, Vaudel M. Anatomy and evolution of database search engines-a central component of mass spectrometry based proteomic workflows. Mass spectrometry reviews, (2017). [DOI] [PubMed]
- **77.Vaisar T, Pennathur S, Green PS et al. Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL. The Journal of clinical investigation, 117(3), 746–756 (2007).First study using HDL proteomics to demonstrate that HDL carries rich protein cargo and that may be altered in people with CVD.
- 78.Holzer M, Birner-Gruenberger R, Stojakovic T et al. Uremia alters HDL composition and function. Journal of the American Society of Nephrology : JASN, 22(9), 1631–1641 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Holzer M, Wolf P, Curcic S et al. Psoriasis alters HDL composition and cholesterol efflux capacity. Journal of lipid research, 53(8), 1618–1624 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sreckovic I, Birner-Gruenberger R, Obrist B et al. Distinct composition of human fetal HDL attenuates its anti-oxidative capacity. Biochimica et biophysica acta, 1831(4), 737–746 (2013). [DOI] [PubMed] [Google Scholar]
- 81.Shao B, de Boer I, Tang C et al. A Cluster of Proteins Implicated in Kidney Disease Is Increased in High-Density Lipoprotein Isolated from Hemodialysis Subjects. Journal of proteome research, 14(7), 2792–2806 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Collier TS, Jin Z, Topbas C, Bystrom C. Rapid Affinity Enrichment of Human Apolipoprotein A-I Associated Lipoproteins for Proteome Analysis. Journal of proteome research, 17(3), 1183–1193 (2018). [DOI] [PubMed] [Google Scholar]
- 83.Melchior JT, Street SE, Andraski AB et al. Apolipoprotein A-II alters the proteome of human lipoproteins and enhances cholesterol efflux from ABCA1. Journal of lipid research, 58(7), 1374–1385 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Holzer M, Kern S, Birner-Grunberger R, Curcic S, Heinemann A, Marsche G. Refined purification strategy for reliable proteomic profiling of HDL2/3: Impact on proteomic complexity. Scientific reports, 6, 38533 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kopecky C, Haidinger M, Birner-Grunberger R et al. Restoration of renal function does not correct impairment of uremic HDL properties. Journal of the American Society of Nephrology : JASN, 26(3), 565–575 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mathew AV, Li L, Byun J et al. Therapeutic Lifestyle Changes Improve HDL Function by Inhibiting Myeloperoxidase-Mediated Oxidation in Patients With Metabolic Syndrome. Diabetes care, 41(11), 2431–2437 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Oberbach A, Adams V, Schlichting N et al. Proteome profiles of HDL particles of patients with chronic heart failure are associated with immune response and also include bacteria proteins. Clinica chimica acta; international journal of clinical chemistry, 453, 114–122 (2016). [DOI] [PubMed] [Google Scholar]
- 88.Trieb M, Horvath A, Birner-Gruenberger R et al. Liver disease alters high-density lipoprotein composition, metabolism and function. Biochimica et biophysica acta, 1861(7), 630–638 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vaisar T, Tang C, Babenko I et al. Inflammatory remodeling of the HDL proteome impairs cholesterol efflux capacity. Journal of lipid research, 56(8), 1519–1530 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Alwaili K, Bailey D, Awan Z et al. The HDL proteome in acute coronary syndromes shifts to an inflammatory profile. Biochimica et biophysica acta, 1821(3), 405–415 (2012). [DOI] [PubMed] [Google Scholar]
- 91.Weichhart T, Kopecky C, Kubicek M et al. Serum amyloid A in uremic HDL promotes inflammation. Journal of the American Society of Nephrology : JASN, 23(5), 934–947 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Riwanto M, Rohrer L, Roschitzki B et al. Altered activation of endothelial anti- and proapoptotic pathways by high-density lipoprotein from patients with coronary artery disease: role of high-density lipoprotein-proteome remodeling. Circulation, 127(8), 891–904 (2013). [DOI] [PubMed] [Google Scholar]
- 93.Karlsson H, Leanderson P, Tagesson C, Lindahl M. Lipoproteomics II: mapping of proteins in high-density lipoprotein using two-dimensional gel electrophoresis and mass spectrometry. Proteomics, 5(5), 1431–1445 (2005). [DOI] [PubMed] [Google Scholar]
- 94.Heller M, Stalder D, Schlappritzi E, Hayn G, Matter U, Haeberli A. Mass spectrometry-based analytical tools for the molecular protein characterization of human plasma lipoproteins. Proteomics, 5(10), 2619–2630 (2005). [DOI] [PubMed] [Google Scholar]
- 95.Mange A, Goux A, Badiou S et al. HDL proteome in hemodialysis patients: a quantitative nanoflow liquid chromatography-tandem mass spectrometry approach. PloS one, 7(3), e34107 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pedret A, Catalan U, Fernandez-Castillejo S et al. Impact of Virgin Olive Oil and Phenol-Enriched Virgin Olive Oils on the HDL Proteome in Hypercholesterolemic Subjects: A Double Blind, Randomized, Controlled, Cross-Over Clinical Trial (VOHF Study). PloS one, 10(6), e0129160 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hsieh JY, Chang CT, Huang MT et al. Biochemical and functional characterization of charge-defined subfractions of high-density lipoprotein from normal adults. Analytical chemistry, 85(23), 11440–11448 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hortin GL, Shen RF, Martin BM, Remaley AT. Diverse range of small peptides associated with high-density lipoprotein. Biochemical and biophysical research communications, 340(3), 909–915 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rezaee F, Casetta B, Levels JH, Speijer D, Meijers JC. Proteomic analysis of high-density lipoprotein. Proteomics, 6(2), 721–730 (2006). [DOI] [PubMed] [Google Scholar]
- 100.Godzien J, Ciborowski M, Armitage EG et al. A Single In-Vial Dual Extraction Strategy for the Simultaneous Lipidomics and Proteomics Analysis of HDL and LDL Fractions. Journal of proteome research, 15(6), 1762–1775 (2016). [DOI] [PubMed] [Google Scholar]
- 101.Yan LR, Wang DX, Liu H et al. A pro-atherogenic HDL profile in coronary heart disease patients: an iTRAQ labelling-based proteomic approach. PloS one, 9(5), e98368 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Watanabe J, Charles-Schoeman C, Miao Y et al. Proteomic profiling following immunoaffinity capture of high-density lipoprotein: association of acute-phase proteins and complement factors with proinflammatory high-density lipoprotein in rheumatoid arthritis. Arthritis and rheumatism, 64(6), 1828–1837 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jorge I, Burillo E, Mesa R et al. The human HDL proteome displays high inter-individual variability and is altered dynamically in response to angioplasty-induced atheroma plaque rupture. Journal of proteomics, 106, 61–73 (2014). [DOI] [PubMed] [Google Scholar]
- 104.Singh SA, Aikawa M. Unbiased and targeted mass spectrometry for the HDL proteome. Current opinion in lipidology, 28(1), 68–77 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gordon SM, Chung JH, Playford MP et al. High density lipoprotein proteome is associated with cardiovascular risk factors and atherosclerosis burden as evaluated by coronary CT angiography. Atherosclerosis, 278, 278–285 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gordon SM, McKenzie B, Kemeh G et al. Rosuvastatin Alters the Proteome of High Density Lipoproteins: Generation of alpha-1-antitrypsin Enriched Particles with Anti-inflammatory Properties. Molecular & cellular proteomics : MCP, 14(12), 3247–3257 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gourgari E, Ma J, Playford MP et al. Proteomic alterations of HDL in youth with type 1 diabetes and their associations with glycemic control: a case-control study. Cardiovascular diabetology, 18(1), 43 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rao PK, Merath K, Drigalenko E et al. Proteomic characterization of high-density lipoprotein particles in patients with non-alcoholic fatty liver disease. Clinical proteomics, 15, 10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Swertfeger DK, Li H, Rebholz S et al. Mapping Atheroprotective Functions and Related Proteins/Lipoproteins in Size Fractionated Human Plasma. Molecular & cellular proteomics : MCP, 16(4), 680–693 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gundry RL, Fu Q, Jelinek CA, Van Eyk JE, Cotter RJ. Investigation of an albumin-enriched fraction of human serum and its albuminome. Proteomics. Clinical applications, 1(1), 73–88 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sankaranarayanan S, de la Llera-Moya M, Drazul-Schrader D, Phillips MC, Kellner-Weibel G, Rothblat GH. Serum albumin acts as a shuttle to enhance cholesterol efflux from cells. Journal of lipid research, 54(3), 671–676 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Holzer M, Kern S, Trieb M, Trakaki A, Marsche G. HDL structure and function is profoundly affected when stored frozen in the absence of cryoprotectants. Journal of lipid research, 58(11), 2220–2228 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kekulawala JR, Murphy A, D’Souza W et al. Impact of freezing on high-density lipoprotein functionality. Anal Biochem, 379(2), 213–215 (2008). [DOI] [PubMed] [Google Scholar]
- 114.Bollinger JG, Stergachis AB, Johnson RS, Egertson JD, MacCoss MJ. Selecting Optimal Peptides for Targeted Proteomic Experiments in Human Plasma Using In Vitro Synthesized Proteins as Analytical Standards. Methods in molecular biology (Clifton, N.J.), 1410, 207–221 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jaffe JD, Keshishian H, Chang B, Addona TA, Gillette MA, Carr SA. Accurate inclusion mass screening: a bridge from unbiased discovery to targeted assay development for biomarker verification. Molecular Cellular Proteomics, 7(10), 1952–1962 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Smith RD. Mass spectrometry in biomarker applications: from untargeted discovery to targeted verification, and implications for platform convergence and clinical application. Clinical chemistry, 58(3), 528–530 (2012). [DOI] [PubMed] [Google Scholar]
- 117.Gillette MA, Carr SA. Quantitative analysis of peptides and proteins in biomedicine by targeted mass spectrometry. Nature methods, 10(1), 28–34 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Addona TA, Abbatiello SE, Schilling B et al. Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nature biotechnology, 27(7), 633–641 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Picotti P, Aebersold R. Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nature methods, 9(6), 555–566 (2012). [DOI] [PubMed] [Google Scholar]
- **120.Whiteaker JR, Lin C, Kennedy J et al. A targeted proteomics-based pipeline for verification of biomarkers in plasma. Nature biotechnology, 29(7), 625–634 (2011).Excellent outline of the approach to quantification of proteins using targeted proteomics approaches.
- 121.Yassine HN, Jackson AM, Borges CR et al. The application of multiple reaction monitoring and multi-analyte profiling to HDL proteins. Lipids in health and disease, 13(1), 13–18 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gallien S, Duriez E, Crone C, Kellmann M, Moehring T, Domon B. Targeted proteomic quantification on quadrupole-orbitrap mass spectrometer. Molecular Cellular Proteomics, 11(12), 1709–1723 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Peterson AC, Russell JD, Bailey DJ, Westphall MS, Coon JJ. Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Molecular & cellular proteomics : MCP, 11(11), 1475–1488 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- **124.Ronsein GE, Pamir N, von Haller PD et al. Parallel reaction monitoring (PRM) and selected reaction monitoring (SRM) exhibit comparable linearity, dynamic range and precision for targeted quantitative HDL proteomics. Journal of proteomics, 113, 388–399 (2015).Comparison of the targeted proteomics using SRM and PRM applied to HDL proteomics.
- 125.Ronsein GE, Reyes-Soffer G, He Y, Oda M, Ginsberg H, Heinecke JW. Targeted Proteomics Identifies Paraoxonase/Arylesterase 1 (PON1) and Apolipoprotein Cs as Potential Risk Factors for Hypoalphalipoproteinemia in Diabetic Subjects Treated with Fenofibrate and Rosiglitazone. Molecular & cellular proteomics : MCP, 15(3), 1083–1093 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Venable JD, Dong MQ, Wohlschlegel J, Dillin A, Yates JR. Automated approach for quantitative analysis of complex peptide mixtures from tandem mass spectra. Nature methods, 1(1), 39–45 (2004). [DOI] [PubMed] [Google Scholar]
- 127.Gillet LC, Navarro P, Tate S et al. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Molecular Cellular Proteomics, 11(6), O111.016717 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- **128.Ludwig C, Gillet L, Rosenberger G, Amon S, Collins BC, Aebersold R. Data-independent acquisition-based SWATH-MS for quantitative proteomics: a tutorial. Molecular systems biology, 14(8), e8126 (2018).Description of the proteomics analysis using data independent analysis.
- 129.Meyer JG, Schilling B. Clinical applications of quantitative proteomics using targeted and untargeted data-independent acquisition techniques. Expert Rev Proteomics, 14(5), 419–429 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chapman JD, Goodlett DR, Masselon CD. Multiplexed and data-independent tandem mass spectrometry for global proteome profiling. Mass spectrometry reviews, 33(6), 452–470 (2014). [DOI] [PubMed] [Google Scholar]
- 131.Sajic T, Liu Y, Aebersold R. Using data-independent, high-resolution mass spectrometry in protein biomarker research: perspectives and clinical applications. Proteomics. Clinical applications, 9(3–4), 307–321 (2015). [DOI] [PubMed] [Google Scholar]
- 132.Navarro P, Kuharev J, Gillet LC et al. A multicenter study benchmarks software tools for label-free proteome quantification. Nature biotechnology, 34(11), 1130–1136 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Aebersold R, Bensimon A, Collins BC, Ludwig C, Sabido E. Applications and Developments in Targeted Proteomics: From SRM to DIA/SWATH. Proteomics, 16(15–16), 2065–2067 (2016). [DOI] [PubMed] [Google Scholar]
- 134.Liu Y, Buil A, Collins BC et al. Quantitative variability of 342 plasma proteins in a human twin population. Molecular systems biology, 11(1), 786 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Collins BC, Hunter CL, Liu Y et al. Multi-laboratory assessment of reproducibility, qualitative and quantitative performance of SWATH-mass spectrometry. Nature Communications, 8(1), 291 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- **136.Rubinow KB, Henderson CM, Robinson-Cohen C et al. Kidney function is associated with an altered protein composition of high-density lipoprotein. Kidney Int, 92(6), 1526–1535 (2017).Largest study of HDL proteome in over 600 subjects with kidney diseas demonstrating feasibility of the targeted tandem mass spectrometry analysis of HDL in translational study setting.
- 137.Pino LK, Searle BC, Huang EL, Noble WS, Hoofnagle AN, MacCoss MJ. Calibration Using a Single-Point External Reference Material Harmonizes Quantitative Mass Spectrometry Proteomics Data between Platforms and Laboratories. Analytical chemistry, 90(21), 13112–13117 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rubinow KB, Vaisar T, Chao JH, Heinecke JW, Page ST. Sex steroids mediate discrete effects on HDL cholesterol efflux capacity and particle concentration in healthy men. J Clin Lipidol, 12(4), 1072–1082 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wang K, Zelnick LR, Hoofnagle AN et al. Alteration of HDL Protein Composition with Hemodialysis Initiation. Clin J Am Soc Nephrol, 13(8), 1225–1233 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Brenes A, Hukelmann JL, Bensaddek D, Lamond AI. Multi-batch TMT reveals false positives, batch effects and missing values. Molecular & Cellular Proteomics, mcp.RA119.001472 (2019). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.