Abstract
Cell-free DNA (cfDNA) and nucleosomes are two biomarkers of cell death and neutrophil extracellular trap formation that are increased in dogs with sepsis, immune-mediated haemolytic anaemia, cancer and following trauma and have diagnostic and prognostic values. cfDNA and nucleosomes are typically measured in plasma samples using DNA-specific fluorophores and ELISA assays, respectively, but their concentrations may be affected by pre-analytical variables such as sample type. The present study aimed to investigate the influence of sample type on the plasma cfDNA and nucleosome concentrations of a heterogeneous group of dogs presenting to an emergency room. Triplicate samples were collected into K2-ethylenediamine tetraacetic acid, 3.2% citrate and a specialised DNA stabilisation tube (Streck BCT), processed rapidly and frozen for batch analysis. Biomarker concentrations were compared between sample types by calculation of Spearman’s correlation coefficients, and with Deming regression, Bland-Altman plots and the Friedman test. Overall, biomarker concentrations were highly correlated between the three sample types. The most concordant results were obtained using citrate samples and the DNA stabilisation tube. Matched cfDNA concentrations between the different sample types were significantly different but there was no significant difference between the nucleosome concentrations in any of the sample types. The present study suggests that cfDNA and nucleosomes can be successfully measured in various sample types, but distinct sample types do not produce interchangeable results. This argues for use of a consistent sample type within studies and suggests standardisation may be useful for the field.
Keywords: cell-free DNA, nucleosomes, histones, neutrophil extracellular traps, sample type, dogs
Introduction
Cell-free DNA (cfDNA) is detectable in the circulation of healthy people,1–3 and normal dogs.4 In health, this cfDNA originates from various cells, but particularly from death of haematopoietic cells including those of lymphoid and myeloid origins.5 6 Some cfDNA may be also released through apoptosis, necrosis or neutrophil extracellular trap (NET) formation,7–9 through a process called NETosis.10 Plasma cfDNA has been extensively investigated as a diagnostic and prognostic biomarker in people with various disease processes.11 Increased plasma cfDNA concentrations have been identified in people with bacteraemia,12 13 sepsis,14 neoplasia15 and burns.16 In people, concentrations of cfDNA are also increased after myocardial infarction and trauma.17 18 Increased cfDNA concentrations have been identified in dogs with sepsis,19 trauma,20 neoplasia,21 gastric dilation volvulus (GDV) syndrome22 and immune-mediated haemolytic anaemia (IMHA).23 24
Concentrations of cfDNA are not specific for any one source,25 but this marker can still provide valuable insights into disease severity, prognosis and pathogenesis.26 Measurement of cfDNA provides a quantitative measure of DNA release that is applicable to multiple species and can be adapted to high-throughput screening assays.27 28 Control of pre-analytical factors is important for measurements of cfDNA concentrations.29 Plasma samples are preferable to serum because release of DNA from leukocytes following collection can falsely increase measured concentrations.29–31 Other factors that may affect cfDNA concentrations are time between collection and processing,4 32 centrifugation methods,29 and storage conditions.29 33
Nuclear DNA is coiled inside chromatin by packaging it as DNA–histone complexes consisting of DNA wound around a core of eight histone proteins. These complexes are called nucleosomes and can be released into the circulation during programmed cell death, NETosis and following cell necrosis.34 As such, cfDNA and nucleosomes share potential origins, but are distinct entities,35 with differential potential for immune cell activation through pattern recognition receptors.34 Measuring both biomarkers may provide better insights into disease processes than either alone.35
People with septic shock have significantly higher nucleosome concentrations than those with sepsis or fever,34 and plasma nucleosome concentrations correlate with organ dysfunction severity in septic critically ill people.36 Nucleosome concentrations are increased in horses with colic,37 in dogs with sepsis,19 21 and in dogs following trauma, where they correlate with injury severity and prognosis.20 Nucleosome concentrations are also increased in dogs with IMHA,24 38 a condition in which there may be a link between NETs and thrombosis.39 This is noteworthy, because nucleosomes may contribute to disease progression by promoting and propagating thrombosis.40 Histone proteins are highly conserved and hence nucleosomes are typically measured by ELISA methods that enable measurement of these complexes in multiple species. As with cfDNA, however, they are subject to pre-analytical sources of variation because they can also be released by leukocytes post-collection. It should also be recognised that assays employing the picogreen dye (such as that used in the present study) have the potential to recognise double-stranded DNA (dsDNA) that exists as free DNA fragments and also the dsDNA that is bound to histones. The picogreen assay is much more sensitive to free dsDNA fragments, compared with that bound to histones,41 but some overlap between these two biomarkers exists and may affect the interpretation of results.
Recently, sample collection tubes containing stabilising agents with the potential to reduce pre-analytical variation in cfDNA and nucleosome measurements have become available.42–45 Specifically, these tubes aim to limit post-collection release of cfDNA and nucleosomes from leukocytes through the use of preservatives that prevent cell lysis. To the author’s knowledge, none of these tubes have been evaluated in dogs to date. Given the considerable interest in measurement of cfDNA and nucleosomes in veterinary medicine and in dogs in particular, it would be valuable to know if these more expensive, specialist tubes perform better than other sample types. The present study aimed to compare the cfDNA and nucleosome concentrations measured in three plasma sample types. It was hypothesised that cfDNA and nucleosome concentrations measured in distinct sample types are different from one another.
Materials and methods
Study population
The present study employed a convenience sampling method to replicate typical conditions in an emergency room where samples for cfDNA and nucleosome measurements might be collected. It was expected that this would produce a range of analyte concentrations and maximise generalisability. Dogs weighing >4 kg with evidence of systemic inflammatory response syndrome (SIRS), or with a disorder expected to cause systemic inflammation (eg, metastatic hemangiosarcoma) were considered potential study subjects. The SIRS criteria used were temperature <38.1°C or >39.2°C; heart rate ≥120 beats per minute; respiratory rate ≥40 breaths per minute; white blood cell count >16×109 cells/mL or <6×109 cells/mL, or >3% band neutrophils.46
Sample collection and processing
Blood samples were collected with informed client consent from intravenous catheters or by venipuncture directly into three separate evacuated tubes containing K2-ethylenediamine tetraacetic acid (EDTA; BD Vacutainer, BD Biosciences, San Jose, CA, USA), 3.2% sodium citrate (Vacuette, Greiner Bio-One, Monroe, NC, USA) and a specific additive tube (Streck Cell-Free DNA BCT, Streck, La Vista, NE, USA) designed for cfDNA measurement. Within 10–15 min of blood collection, plasma samples were generated by centrifugation of whole blood for 10 min at 1370 g (Ultra-8V Centrifuge, LW Scientific, Lawrenceville, GA, USA). Shortly (within 5 min) after centrifugation, plasma was decanted into polypropylene freezer tubes (Polypropylene Screw-Cap Microcentrifuge Tubes, VWR, Radnor, PA, USA) by pipetting. Some plasma was deliberately left in each tube to minimise the risk of disturbing the buffy coats. The plasma samples were then frozen at −80°C pending batch analysis. At the time of batch analysis, samples were thawed at room temperature and then kept at 4°C while in use. Samples were collected within a 24-day period (7 March 2018 to 30 March 2018) with batch analysis on 4 April 2018.
cfDNA and nucleosome measurements
Concentrations of both biomarkers were measured contemporaneously. Concentrations of cfDNA were measured in triplicate using a benchtop fluorimeter (Qubit 3.0 Fluorometer, Life Technologies, Waltham, MA, USA) and relevant reagents (Quant-It HS dsDNA Kit, Life Technologies, Waltham, MA, USA) according to the manufacturers’ instructions and as previously described.19 22 Mean values of the three measurements were used for subsequent analyses. Concentrations of plasma nucleosomes were analysed using a commercial ELISA (Cell Death Detection ELISA Plus, Roche, Indianapolis, IN, USA) scaled against pooled normal canine plasma obtained from healthy canine blood donors, as previously described.20 24 38 None of the aliquots of pooled normal canine plasma had been previously thawed prior to analysis. Nucleosome concentrations were measured in duplicate and the mean values used for subsequent analyses.
Statistical methods
A priori sample size calculations were not conducted for this study. Data were assessed for normality using the D’Agostino-Pearson omnibus test. Parametric data are summarised as mean±SD while non-parametric data are summarised as median with IQR. The degree of correlation between cfDNA and nucleosome concentrations measured in different sample types was assessed by calculating Spearman’s correlation coefficient (rs). Strength of correlation was assessed as follows: <0.5 weak, 0.5–0.6 mild, 0.6–0.7 moderate, 0.7–0.8 strong, 0.8–0.9 very strong and 0.9–1.0 excellent. Since no sample type is established as the reference standard, Deming regression was used to analyse the relationship between pairs of sample types. Bland-Altman plots were constructed to identify constant and proportional biases between sample types. Median cfDNA and nucleosome concentrations were compared between sample types using the Friedman test, with Dunn’s adjustment for multiple comparisons. Alpha was set at 0.05. All analyses were conducted using commercial software (Prism 7.0e for Mac OS X, GraphPad, La Jolla, CA, USA).
Results
Patient population
In all 24 dogs were recruited, however, due to two Streck BCT tube failures (one due to breakage and one due to loss of tube vacuum), only 22 complete sets of triplicate samples were collected. The 22 dogs consisted of mixed-breed dogs (n=6), Labrador retrievers (n=4), Golden retrievers (n=3) and one each of the following English bulldog, German shepherd dog, Great Pyrenees, Hungarian vizsla, Mastiff, miniature dachshund, rottweiler, Shetland sheepdog and Staffordshire bull terrier. The mean age and bodyweight were 7.5±3.1 years (IQR 5–10.3) and 31.5±12.3 kg (IQR 23.6–39.4), respectively. On presentation to the hospital, the average temperature, heart rate and respiratory rate were 38.4°C±0.7°C (IQR 38–39), 129±22 bpm (IQR 114–140) and 28 (IQR 24–40) rpm, respectively. The median leucocyte count (n=14) was 10.6×109/mL (IQR 5.9–17.2), median neutrophil count was 8.8×109/mL (IQR 4.4–13.9), with 0% band neutrophils (IQR 0–4.5). Of the 22 dogs, 13 met at least 2 of 4 SIRS criteria (59.1%) as previously described.46 Final diagnoses in these dogs were neoplasia (n=9), gastroenteropathy (n=2), intervertebral disc disease (n=2), sepsis (n=2), trauma (n=2), acute kidney injury, fibrocartilaginous embolism, hepatopathy, peripheral neuropathy and urethral obstruction (all n=1).
cfDNA concentrations
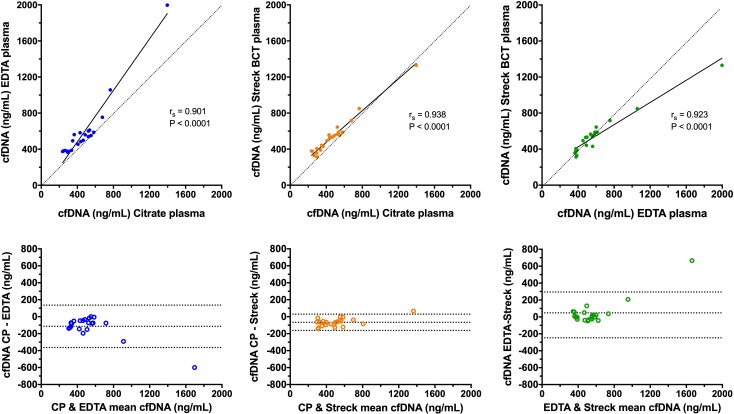
The median (IQR) cfDNA concentrations in the three sample types were as follows: citrate plasma 434 ng/mL (298–540); EDTA plasma 519 ng/mL (384–591) and additive tube plasma 532 ng/mL (388–589) (figure 1). The concentrations of cfDNA were significantly lower in the citrate plasma compared with both EDTA plasma and the additive tube plasma (both p<0.0001). There was no significant difference between cfDNA concentrations in EDTA plasma compared with additive tube plasma (p=1.0000). Calculation of Spearman’s correlation coefficient and Deming regression showed that there was excellent correlation between the cfDNA concentrations in all three of the sample types (figure 2). Bland-Altman analysis showed evidence of small constant negative biases for the citrate plasma versus other sample types but no proportional biases in any of the sample types. Consistent with the correlation analyses, the best level of agreement was found between the citrate and the additive tube plasma samples (figure 2).
Figure 1.
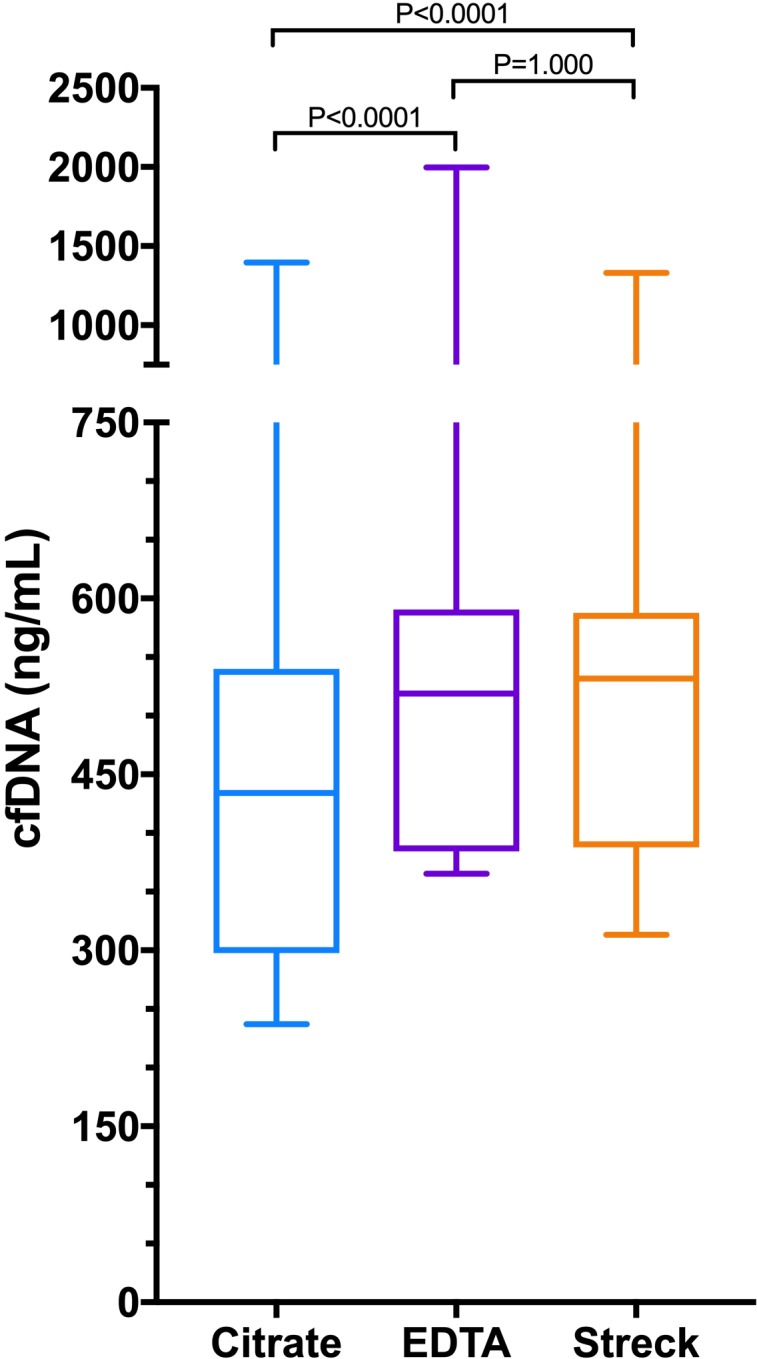
Box and whisker plots comparing cfDNA concentrations in a heterogeneous group of dogs measured in three different sample types (citrate plasma, EDTA plasma, plasma derived from Streck BCT tubes that include a preservative to limit cell lysis). The centre line represents the median, the boxes represent the IQR and the whiskers represent the minimum and maximum values. Concentrations of cfDNA were significantly different in citrate samples compared with both EDTA the Streck tube samples. There was no difference between the cfDNA concentrations in EDTA samples compared with Streck tube samples. cfDNA, cell-free DNA; EDTA, ethylenediamine tetraacetic acid.
Figure 2.
Scatterplots (upper panels) and Bland-Altman plots (lower panels) comparing the cfDNA concentrations in each of the three sample types. In the scatterplots, results from pairs of sample types have been plotted against each other combined with associated values for Spearman’s correlation coefficient (rs) and the relevant significance value. The solid black line represents the line of best fit by Deming regression. Note the rs value does not correspond to the Deming regression line. The dotted lines are lines of identity, included for reference. In the Bland-Altman plots, the mean value of the two measurements from the two sample types is plotted on the abscissa (x-axis), while the difference between the two sample types is plotted on the ordinate (y-axis). The middle of the three horizontal dotted lines represents the mean difference between the two sets of values, whereas the upper and lower dotted lines represent ±1.96 SD of the mean difference. cfDNA, cell-free DNA.
Nucleosomes
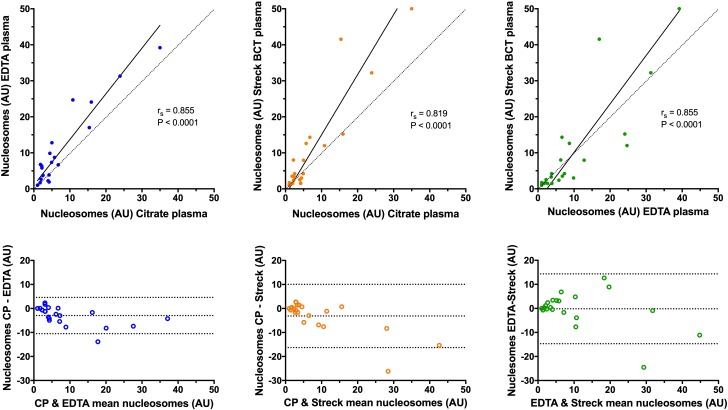
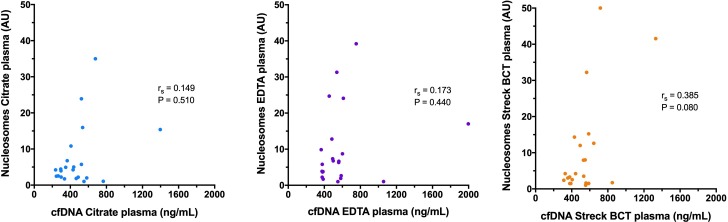
The median (IQR) nucleosome concentrations in the three sample types were as follows: citrate plasma 4.3 AU (2.1–7.8); EDTA plasma 6.5 AU (2.6–13.9) and additive tube plasma 3.9 AU (1.7–13.0) (figure 3). There were no significant differences in the concentrations of nucleosomes between any of the sample types (p≥0.150). Calculation of Spearman’s correlation coefficient and Deming regression showed that there was very strong correlation between the nucleosome concentrations in all of the sample types (figure 4). Bland-Altman analysis showed a small constant negative bias between citrate samples and the other two sample types, but no evidence of proportional biases in any of the sample types. The best level of agreement was found between the citrate and the EDTA plasma samples (figure 4). There was weak correlation between the cfDNA and nucleosome concentrations in all of the sample types, and none of the correlation coefficients were statistically significant (figure 5). The best correlation between these two biomarkers was present in the additive tube plasma samples (rs 0.385, p=0.077).
Figure 3.
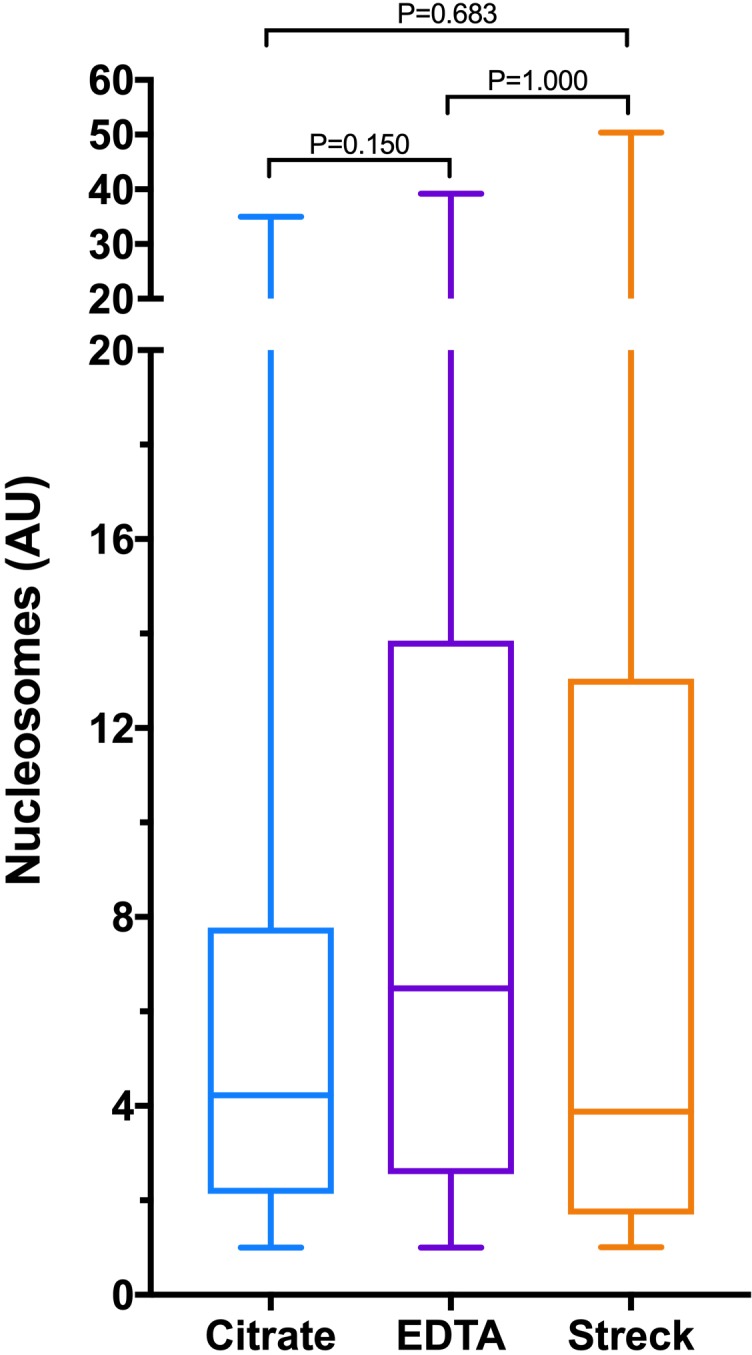
Box and whisker plots comparing nucleosome concentrations in a heterogeneous group of dogs measured in three different sample types (citrate plasma, ethylenediamine tetraacetic acid plasma, plasma derived from Streck BCT tubes that include a preservative to limit cell lysis). The centre line represents the median, the boxes represent the IQR and the whiskers represent the minimum and maximum values. There were no significant differences between the concentrations of nucleosomes in any of the sample types.
Figure 4.
Scatterplots (upper panels) and Bland-Altman plots (lower panels) comparing the nucleosome concentrations in each of the three sample types. In the scatterplots, results from pairs of sample types have been plotted against each other combined with associated values for Spearman’s correlation coefficient (rs) and the relevant significance value. The solid black line represents the line of best fit by Deming regression. Note the rs value does not correspond to the Deming regression line. The dotted lines are lines of identity, included for reference. In the Bland-Altman plots, the mean value of the two measurements from the two sample types is plotted on the abscissa (x-axis), while the difference between the two sample types is plotted on the ordinate (y-axis). The middle of the three horizontal dotted lines represents the mean difference between the two sets of values, whereas the upper and lower dotted lines represent ±1.96 SD of the mean difference.
Figure 5.
Scatterplots comparing the cfDNA and the nucleosome concentrations in each of the three sample types. In the scatterplots, results from pairs of sample types have been plotted against each other. Also displayed are the associated values for Spearman’s correlation coefficient (rs) and the relevant significance value. Note none of the rs values were statistically significant at p<0.05. cfDNA, cell-free DNA.
Discussion
The present study aimed to compare three sample types for the measurement of cfDNA and nucleosomes in a population of dogs with disease processes predisposing to inflammation. In addition, the present study aimed to assess the utility of a specific preservative tube designed for cfDNA measurement. The data suggest that all three of the sample types can be used for measurement of cfDNA and nucleosomes. There were statistically significant differences between the concentrations measured in the three sample types for cfDNA, but not for nucleosomes. The differences in cfDNA concentrations might not be clinically relevant, however. For cfDNA measurement, the best correlation and the highest levels of agreement were found between the citrate plasma and the Streck BCT plasma samples, although all of the correlation coefficients were >0.9. For nucleosomes, the best correlation and the highest levels of agreement were found between the citrate plasma and the EDTA plasma samples. This may suggest that either EDTA or citrate plasma samples are reasonable choices for measurement of these biomarkers.
No reference standard (gold standard) method has been established for measurement of these biomarkers and hence the optimal sample type has yet to be determined. This precludes recommending a specific sample type, but data from the present study suggest that either EDTA or citrate plasma would be acceptable choices. The smallest levels of constant bias existed between the Streck and the EDTA plasma samples for both biomarkers. In contrast, the concentrations of cfDNA and nucleosomes consistently measured lower in citrate samples particularly with regard to EDTA plasma. This could be interpreted in two ways. The citrate samples may cause falsely low readings while the other sample types reflect the true concentrations in the patient, or the Streck and the EDTA samples cause falsely increased concentrations relative to the patient. Increased concentrations could be due to cell lysis that may be a source of pre-analytical error. Additional studies in dogs evaluating the quality as well as the quantity of cfDNA are warranted to determine if citrate plasma maintains cfDNA quality better than does EDTA, as has been described for people.47 Such additional studies might also evaluate whether there is an impact of diagnosis or disease severity on the sensitivity of biomarker detection in distinct sample types. For instance, future studies might investigate whether one sample type is more sensitive in sepsis, while another has greater sensitivity in trauma.
The Streck tube additive is designed to reduce the likelihood of release of cfDNA after sample collection. If those tubes work correctly in dogs, then this might suggest that the biomarker concentrations in the citrate samples are falsely low. At this time, it is not possible to determine which of these scenarios is correct. Irrespective, data from the present study argue that while different sample types might produce similar results, they are not interchangeable. Thus, any study measuring cfDNA or nucleosome concentrations in dogs should use a single sample type consistently throughout the study period.
Concentrations of cfDNA can be measured at the point of care and because they correlate with illness severity indicators such as bacteremia,19 and outcome in diseases including sepsis and IMHA,19 23 this marker may become a useful clinical tool for emergency and critical care practice. In this respect, use of a sample type that is readily available, such as EDTA or citrate, may be preferable to a more expensive, single-purpose tube such as the Streck BCT.
Although the initial descriptions of direct cfDNA measurement were performed on serum,48 plasma samples are now recommended.29 This is because serum samples can generate falsely increased concentrations,31 49 due to lysis of leukocytes during the ex vivo clotting process.50 51 Some authors have suggested that EDTA tubes may be the best choice where plasma processing must be delayed.52 In human medicine, the need for sample storage or shipping prior to processing led to the development of cfDNA stabilisation tubes such as the ones evaluated in the present study.53–55 The medical literature suggests that the Streck BCT tubes enable blood collection in a range of clinical settings,42 and extend the time available for sample processing. However, they may not offer substantial advantage over conventional sample types such as EDTA when processing occurs within 6 hours.56 Data from the present study also suggest that these stabilisation tubes perform comparably to other sample types when samples are processed and frozen rapidly after collection. Future studies in dogs might evaluate the effects of sample types in the presence of delayed processing, storage or shipping.
The lack of a significant correlation between the nucleosome concentrations and the cfDNA concentrations in these dogs is noteworthy. As discussed above, these two biomarkers are similar but distinct, in terms of their structure, origin and pathophysiological effects. The picogreen assay used in the present study has the potential to recognise both biomarkers. The lack of correlation between the two biomarker concentrations suggests that the cfDNA assay may be relatively insensitive to the nucleosomes in these plasma samples, as has been previously reported.41 In studies of other canine patient populations, these biomarkers have been determined to be positively correlated,19 20 but the strength of the association in those studies was weak. These weak associations may indicate related but distinct origins for release of these biomarkers and suggests that measuring both biomarkers may be worthwhile. Fortunately, the results of the present study confirm they can both be measured in the same sample type.
The present study is not without limitations. The sample size was relatively small, but despite this significant differences between sample types for cfDNA concentrations were detectable. The present study aimed to recruit a heterogeneous population of dogs with disease processes predisposed to inflammation to reflect a typical population of dogs in which cfDNA and nucleosomes might be measured. The final diagnoses for most dogs were not known at the time of recruitment and hence the population enrolled contained more dogs with neoplastic diseases than anticipated. The overall cfDNA and nucleosome concentrations were lower than reported for other patient populations,19 20 22 perhaps reflecting a lower disease severity in this cohort compared with others. Despite this, the cfDNA and nucleosome concentrations covered a wide range (~twofold for cfDNA and ~sevenfold for nucleosomes) and provided a solid dataset to assess for evidence of proportional biases between sample types. Inspection of the cfDNA scatterplots suggests that one sample with a high concentration of cfDNA in all three sample types may have disproportionately affected the Deming regression line (high leverage point). Reanalysis of these scatterplots without this point (not shown) did not alter the conclusions drawn, but reduced all of the correlation coefficients. Thus, data from all 22 dogs are presented here. The present study did not evaluate all of the commercially available tubes that purport to reduce pre-analytical variation. Future studies might compare the Streck BCT tube with other similar products from other manufacturers.
The samples tested were frozen for varying amounts of time before batch analysis, but the maximum duration of storage was less than 1 month. It is possible, but unlikely that sample deterioration occurred within this time period. cfDNA concentrations are reportedly stable in serum samples repeatedly frozen and thawed. In a study by Goldshtein et al, there was a decrease in the mean cfDNA concentrations, but the difference was not statistically significant.48 A study of serum samples from human cancer patients measured nucleosomes after 6–9 months of storage at −70°C and then again ~5.5 years later. That study identified a significant decrease of 32% in measured concentrations between the first and the second analyses.57
In summary, in the present study, cfDNA and nucleosome concentrations were similar in all three sample types tested. Distinct sample types do not produce equal cfDNA concentrations, however, and should not be considered interchangeable. Thus, a consistent sample type should be used for any given study. In settings where sample processing and analysis can be conducted rapidly, there may be little advantage to using a more expensive and less widely available additive tube for cfDNA or nucleosome measurement. EDTA or citrate plasma samples appear to be reasonable choices for cfDNA or nucleosome measurement in dogs. It may be useful to consider standardisation of cfDNA and nucleosome assays in the future to maximise the comparability of results across studies and between groups, as has been recommended in human medicine.29
Footnotes
Contributors: All authors listed fulfilled the following criteria: (1) substantial contributions to conception and design, acquisition of data or analysis and interpretation of data; (2) drafting the article or revising it critically for important intellectual content. Specifically, RG designed and conducted the research, analysed the data and wrote the manuscript. All authors approved the final version of the manuscript.
Funding: This research was supported by funds from the Cornell University College of Veterinary Medicine.
Competing interests: None declared.
Ethics approval: This study was carried out in accordance with the recommendations of the Cornell University College of Veterinary Medicine (CU-CVM), Institutional Animal Care and Use Committee (Ithaca, NY, 14853). Approval for this study was granted by the local Institutional Animal Care and Use Committee (IACUC-2014–0053).
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: All data relevant to the study are included in the article.
References
- 1. Giacona MB, Ruben GC, Iczkowski KA, et al. Cell-free DNA in human blood plasma: length measurements in patients with pancreatic cancer and healthy controls. Pancreas 1998;17:89–97. 10.1097/00006676-199807000-00012 [DOI] [PubMed] [Google Scholar]
- 2. Spindler K-LG, Appelt AL, Pallisgaard N, et al. Cell-free DNA in healthy individuals, noncancerous disease and strong prognostic value in colorectal cancer. Int J Cancer 2014;135:2984–91. 10.1002/ijc.28946 [DOI] [PubMed] [Google Scholar]
- 3. Tamkovich S, Bryzgunova O, Activity P. Protease activity and cell-free DNA in blood plasma of healthy donors and breast cancer patients. J Immunoassay Immunochem 2016;37:141–53. 10.1080/15321819.2015.1069745 [DOI] [PubMed] [Google Scholar]
- 4. Burnett DL, Cave NJ, Gedye KR, et al. Investigation of cell-free DNA in canine plasma and its relation to disease. Vet Q 2016;36:122–9. 10.1080/01652176.2016.1182230 [DOI] [PubMed] [Google Scholar]
- 5. Lui YYN, Chik K-W, Chiu RWK, et al. Predominant hematopoietic origin of cell-free DNA in plasma and serum after sex-mismatched bone marrow transplantation. Clin Chem 2002;48:421–7. [PubMed] [Google Scholar]
- 6. Snyder MW, Kircher M, Hill AJ, et al. Cell-free DNA Comprises an In Vivo Nucleosome Footprint that Informs Its Tissues-Of-Origin. Cell 2016;164:57–68. 10.1016/j.cell.2015.11.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jahr S, Hentze H, Englisch S, et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res 2001;61:1659–65. [PubMed] [Google Scholar]
- 8. Yipp BG, Kubes P. NETosis: how vital is it? Blood 2013;122:2784–94. 10.1182/blood-2013-04-457671 [DOI] [PubMed] [Google Scholar]
- 9. Brinkmann V, et al. Neutrophil extracellular traps kill bacteria. Science 2004;303:1532–5. 10.1126/science.1092385 [DOI] [PubMed] [Google Scholar]
- 10. Steinberg BE, Grinstein S. Unconventional roles of the NADPH oxidase: signaling, ion homeostasis, and cell death. Sci STKE 2007;2007:pe11 10.1126/stke.3792007pe11 [DOI] [PubMed] [Google Scholar]
- 11. Butt AN, Swaminathan R. Overview of circulating nucleic acids in plasma/serum. Ann N Y Acad Sci 2008;1137:236–42. 10.1196/annals.1448.002 [DOI] [PubMed] [Google Scholar]
- 12. Forsblom E, Aittoniemi J, Ruotsalainen E, et al. High cell-free DNA predicts fatal outcome among Staphylococcus aureus bacteraemia patients with intensive care unit treatment. PLoS One 2014;9:e87741 10.1371/journal.pone.0087741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huttunen R, Kuparinen T, Jylhävä J, et al. Fatal outcome in bacteremia is characterized by high plasma cell free DNA concentration and apoptotic DNA fragmentation: a prospective cohort study. PLoS One 2011;6:e21700 10.1371/journal.pone.0021700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Saukkonen K, Lakkisto P, Pettila V, et al. Cell-free plasma DNA as a predictor of outcome in severe sepsis and septic shock. Clin Chem 2008;54:1000–7. 10.1373/clinchem.2007.101030 [DOI] [PubMed] [Google Scholar]
- 15. Johnson PJ, Lo YMD. Plasma nucleic acids in the diagnosis and management of malignant disease. Clin Chem 2002;48:1186–93. [PubMed] [Google Scholar]
- 16. Chiu TW, Young R, Chan LYS, et al. Plasma cell-free DNA as an indicator of severity of injury in burn patients. Clin Chem Lab Med 2006;44:13–17. 10.1515/CCLM.2006.003 [DOI] [PubMed] [Google Scholar]
- 17. Chang CP-Y, Chia R-H, Wu T-L, et al. Elevated cell-free serum DNA detected in patients with myocardial infarction. Clinica Chimica Acta 2003;327:95–101. 10.1016/S0009-8981(02)00337-6 [DOI] [PubMed] [Google Scholar]
- 18. Lo YMD RTH, Chan LYS, Hjelm NM, et al. Plasma DNA as a prognostic marker in trauma patients. Clin Chem 2000;46:319–23. [PubMed] [Google Scholar]
- 19. Letendre J-A, Goggs R. Determining prognosis in canine sepsis by bedside measurement of cell-free DNA and nucleosomes. J Vet Emerg Crit Care 2018;28:503–11. 10.1111/vec.12773 [DOI] [PubMed] [Google Scholar]
- 20. Letendre J-A, Goggs R. Concentrations of plasma nucleosomes but not cell-free DNA are prognostic in dogs following trauma. Front Vet Sci 2018;5 10.3389/fvets.2018.00180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Letendre J-A, Goggs R. Measurement of plasma cell-free DNA concentrations in dogs with sepsis, trauma, and neoplasia. J Vet Emerg Crit Care 2017;27:307–14. 10.1111/vec.12592 [DOI] [PubMed] [Google Scholar]
- 22. Troia R, Giunti M, Calipa S, et al. Cell-Free DNA, high-mobility group box-1, and procalcitonin concentrations in dogs with gastric Dilatation-Volvulus syndrome. Front Vet Sci 2018;5 10.3389/fvets.2018.00067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jeffery U, Ruterbories L, Hanel R, et al. Cell-Free DNA and DNase activity in dogs with immune-mediated hemolytic anemia. J Vet Intern Med 2017;31:1441–50. 10.1111/jvim.14808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lawson C, Smith SA, O'Brien M, et al. Neutrophil extracellular traps in plasma from dogs with immune-mediated hemolytic anemia. J Vet Intern Med 2018;32:128–34. 10.1111/jvim.14881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Aucamp J, Bronkhorst AJ, Badenhorst CPS, et al. The diverse origins of circulating cell-free DNA in the human body: a critical re-evaluation of the literature. Biol Rev Camb Philos Soc 2018;93:1649–83. 10.1111/brv.12413 [DOI] [PubMed] [Google Scholar]
- 26. Gould TJ, Vu TT, Stafford AR, et al. Cell-Free DNA modulates clot structure and impairs fibrinolysis in sepsis. Arterioscler Thromb Vasc Biol 2015;35:2544–53. 10.1161/ATVBAHA.115.306035 [DOI] [PubMed] [Google Scholar]
- 27. Palić D, Ostojić J, Andreasen CB, et al. Fish cast nets: neutrophil extracellular traps are released from fish neutrophils. Dev Comp Immunol 2007;31:805–16. 10.1016/j.dci.2006.11.010 [DOI] [PubMed] [Google Scholar]
- 28. Chuammitri P, Ostojić J, Andreasen CB, et al. Chicken heterophil extracellular traps (HETs): novel defense mechanism of chicken heterophils. Vet Immunol Immunopathol 2009;129:126–31. 10.1016/j.vetimm.2008.12.013 [DOI] [PubMed] [Google Scholar]
- 29. El Messaoudi S, Rolet F, Mouliere F, et al. Circulating cell free DNA: preanalytical considerations. Clin Chim Acta 2013;424:222–30. 10.1016/j.cca.2013.05.022 [DOI] [PubMed] [Google Scholar]
- 30. Taback B, O'Day SJ, Hoon DSB. Quantification of circulating DNA in the plasma and serum of cancer patients. Ann N Y Acad Sci 2004;1022:17–24. 10.1196/annals.1318.004 [DOI] [PubMed] [Google Scholar]
- 31. Lee T-H, Montalvo L, Chrebtow V, et al. Quantitation of genomic DNA in plasma and serum samples: higher concentrations of genomic DNA found in serum than in plasma. Transfusion 2001;41:276–82. 10.1046/j.1537-2995.2001.41020276.x [DOI] [PubMed] [Google Scholar]
- 32. Xue X, Teare MD, Holen I, et al. Optimizing the yield and utility of circulating cell-free DNA from plasma and serum. Clin Chim Acta 2009;404:100–4. 10.1016/j.cca.2009.02.018 [DOI] [PubMed] [Google Scholar]
- 33. Wang Q, Cai Y, Brady P, et al. Real-Time PCR evaluation of cell-free DNA subjected to various storage and shipping conditions. Genet Mol Res 2015;14:12797–804. 10.4238/2015.October.19.23 [DOI] [PubMed] [Google Scholar]
- 34. Zeerleder S, Zwart B, Wuillemin WA, et al. Elevated nucleosome levels in systemic inflammation and sepsis. Crit Care Med 2003;31:1947–51. 10.1097/01.CCM.0000074719.40109.95 [DOI] [PubMed] [Google Scholar]
- 35. Marsman G, Zeerleder S, Luken BM. Extracellular histones, cell-free DNA, or nucleosomes: differences in immunostimulation. Cell Death Dis 2016;7:e2518 10.1038/cddis.2016.410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen Q, Ye L, Jin Y, et al. Circulating nucleosomes as a predictor of sepsis and organ dysfunction in critically ill patients. Int J Infect Dis 2012;16:e558–64. 10.1016/j.ijid.2012.03.007 [DOI] [PubMed] [Google Scholar]
- 37. Bauquier JR, Forbes G, Nath L, et al. Plasma HMGB-1 and nucleosome concentrations in horses with colic and healthy horses. J Vet Intern Med 2016;30:260–8. 10.1111/jvim.13811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jeffery U, Kimura K, Gray R, et al. Dogs cast NETs too: canine neutrophil extracellular traps in health and immune-mediated hemolytic anemia. Vet Immunol Immunopathol 2015;168:262–8. 10.1016/j.vetimm.2015.10.014 [DOI] [PubMed] [Google Scholar]
- 39. deLaforcade A, Bacek L, Blais M-C, et al. Consensus on the rational use of antithrombotics in veterinary critical care (curative): domain 1-Defining populations at risk. J Vet Emerg Crit Care 2019;29:37–48. 10.1111/vec.12797 [DOI] [PubMed] [Google Scholar]
- 40. Schulz C, Engelmann B, Massberg S. Crossroads of coagulation and innate immunity: the case of deep vein thrombosis. J Thromb Haemost 2013;11(Suppl 1):233–41. 10.1111/jth.12261 [DOI] [PubMed] [Google Scholar]
- 41. Chen JA, Meister S, Urbonaviciute V, et al. Sensitive detection of plasma/serum DNA in patients with systemic lupus erythematosus. Autoimmunity 2007;40:307–10. 10.1080/08916930701356317 [DOI] [PubMed] [Google Scholar]
- 42. Warton K, Yuwono NL, Cowley MJ, et al. Evaluation of Streck BCT and PAXgene Stabilised blood collection tubes for cell-free circulating DNA studies in plasma. Mol Diagn Ther 2017;21:563–70. 10.1007/s40291-017-0284-x [DOI] [PubMed] [Google Scholar]
- 43. Alidousty C, Brandes D, Heydt C, et al. Comparison of blood collection tubes from three different manufacturers for the collection of cell-free DNA for liquid biopsy mutation testing. J Mol Diagn 2017;19:801–4. 10.1016/j.jmoldx.2017.06.004 [DOI] [PubMed] [Google Scholar]
- 44. Enko D, Halwachs-Baumann G, Kriegshauser G. Plasma free DNA: evaluation of temperature-associated storage effects observed for Roche cell-free DNA collection tubes. Biochem. med. 2019;29:153–6. 10.11613/BM.2019.010904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Norton SE, Lechner JM, Williams T, et al. A stabilizing reagent prevents cell-free DNA contamination by cellular DNA in plasma during blood sample storage and shipping as determined by digital PCR. Clin Biochem 2013;46:1561–5. 10.1016/j.clinbiochem.2013.06.002 [DOI] [PubMed] [Google Scholar]
- 46. Hauptman JG, Walshaw R, Olivier NB. Evaluation of the sensitivity and specificity of diagnostic criteria for sepsis in dogs. Vet Surg 1997;26:393–7. 10.1111/j.1532-950X.1997.tb01699.x [DOI] [PubMed] [Google Scholar]
- 47. Sato A, Nakashima C, Abe T, et al. Investigation of appropriate pre-analytical procedure for circulating free DNA from liquid biopsy. Oncotarget 2018;9:31904–14. 10.18632/oncotarget.25881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Goldshtein H, Hausmann MJ, Douvdevani A. A rapid direct fluorescent assay for cell-free DNA quantification in biological fluids. Ann Clin Biochem 2009;46:488–94. 10.1258/acb.2009.009002 [DOI] [PubMed] [Google Scholar]
- 49. Thijssen MAMA, Swinkels DW, Ruers TJM, et al. Difference between free circulating plasma and serum DNA in patients with colorectal liver metastases. Anticancer Res 2002;22:421–5. [PubMed] [Google Scholar]
- 50. Jung M, Klotzek S, Lewandowski M. Changes in concentration of DNA in serum and plasma during storage of blood samples. Clin Chem 2003;49:1028–9. 10.1373/49.6.1028 [DOI] [PubMed] [Google Scholar]
- 51. Umetani N, Hiramatsu S, Hoon DSB. Higher amount of free circulating DNA in serum than in plasma is not mainly caused by contaminated extraneous DNA during separation. Ann N Y Acad Sci 2006;1075:299–307. 10.1196/annals.1368.040 [DOI] [PubMed] [Google Scholar]
- 52. Lam NYL, Rainer TH, Chiu RW. Edta is a better anticoagulant than heparin or citrate for delayed blood processing for plasma DNA analysis. Clin Chem 2004;50:256–7. 10.1373/clinchem.2003.026013 [DOI] [PubMed] [Google Scholar]
- 53. Fernando MR, Chen K, Norton S, et al. A new methodology to preserve the original proportion and integrity of cell-free fetal DNA in maternal plasma during sample processing and storage. Prenat Diagn 2010;30:418–24. 10.1002/pd.2484 [DOI] [PubMed] [Google Scholar]
- 54. Barrett AN, Zimmermann BG, Wang D, et al. Implementing prenatal diagnosis based on cell-free fetal DNA: accurate identification of factors affecting fetal DNA yield. PLoS One 2011;6:e25202 10.1371/journal.pone.0025202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hidestrand M, Stokowski R, Song K, et al. Influence of temperature during transportation on cell-free DNA analysis. Fetal Diagn Ther 2012;31:122–8. 10.1159/000335020 [DOI] [PubMed] [Google Scholar]
- 56. Kang Q, Henry NL, Paoletti C, et al. Comparative analysis of circulating tumor DNA stability in K3EDTA, Streck, and CellSave blood collection tubes. Clin Biochem 2016;49:1354–60. 10.1016/j.clinbiochem.2016.03.012 [DOI] [PubMed] [Google Scholar]
- 57. Holdenrieder S, Von Pawel J, Nagel D, et al. Long-term stability of circulating nucleosomes in serum. Anticancer Res 2010;30:1613–5. [PubMed] [Google Scholar]