Abstract
Activated choline metabolism is a hallmark of carcinogenesis and tumor progression, which leads to elevated levels of phosphocholine and glycerophosphocholine in all types of cancer tested so far. Magnetic resonance spectroscopy applications have played a key role in detecting these elevated choline phospholipid metabolites. To date, the majority of cancer-related studies has focused on phosphocholine and the Kennedy pathway, which constitutes the biosynthesis pathway for membrane phosphatidylcholine. Fewer and more recent studies have reported on the importance of glycerophosphocholine in cancer. In this review article, we are summarizing the recent literature on glycerophosphocholine metabolism with respect to its cancer biology and its detection by magnetic resonance spectroscopy applications.
Keywords: MR Spectroscopy (MRS) and Spectroscopic Imaging (MRSI) Methods, Methods and Engineering, Cancer, Applications, Metastases, Cancer therapy responses, Cellular and molecular cancer imaging, Cellular and molecular imaging
Graphical Abstract
Aberrant choline metabolism is a hallmark of oncogenesis and cancer progression, characterized by increased phosphocholine, glycerophosphocholine, and total-choline-containing compounds. These oncometabolites can be detected with MRS techniques in preclinical studies and clinical settings. Detection of glycerophosphocholine by MRS can be used for diagnosis, prognosis, and treatment response monitoring. Until recently, most studies have focused on the elevated phosphocholine levels in cancer, while glycerophosphocholine has remained a poorly understood oncometabolite. In this review, we summarize the recent literature on glycerophosphocholine metabolism and biology in cancer and its detection using MRS techniques.
Introduction
Cancer cells are capable of adapting to their environment1 and of inducing changes in stromal2, vascular, and immune cells3 that support their continued growth, and enable them to evade treatment.4 Different types of cancer cells often display common metabolic pathways,5 such as increased glycolytic activity,6 although their genetic make-up may be quite different from one another. Activated choline metabolism, which is a common metabolic pathway as well, has emerged as a hallmark of cancer7 and is characterized by increased levels of phosphocholine (PC), glycerophosphocholine (GPC) and total choline-containing compounds (tCho). Cancers with an activated choline metabolism have been referred to as having a ‘cholinic phenotype’.8,9 Several studies have shown that an interplay between malignant transformation10 and adaptation to the tumor microenvironment11 is causing the aberrant choline metabolism in cancer cells.
Magnetic resonance spectroscopy (MRS) has been utilized extensively for studying the alterations in choline metabolism of cancer.9,12,13 Non-invasive MRS techniques are currently being explored in the clinical setting for diagnosis, prognosis, and treatment response monitoring in cancer.9,14,15 Several water-soluble intermediate metabolites in choline phospholipid metabolism can be observed by MRS, which detects PC, GPC, and free choline (Cho) (Fig. 1), or an overlapping signal from these three choline metabolites commonly referred to as total choline (tCho), in all cancers tested so far.16,17 An increased PC level has been observed in most cancers including breast,17–20 ovarian,21 prostate,22,23 cervical,21,24 brain,25–27 and endometrial cancer.28 Some studies in cell cultures of breast and ovarian cancer cells showed a relative decrease in GPC level as compared with the PC level, proposing to use a high PC/GPC ratio as an indicator of cancer progression.10,29,30 However, a different trend of increasing GPC along with increasing PC was shown in clinical human breast cancer samples.31 In vivo MRS detection of the tCho signal was reported in some studies to be able to serve as an imaging biomarker of breast cancer in the clinical setting.13,32 MRS of the tCho signal has also been explored for monitoring chemotherapeutic response in patients with a decrease in the tCho signal being associated with response to chemotherapy.33 In addition to 1H MRS, 31P MRS is also being used for the analysis of biological samples as well as in in vivo applications. 31P MRS detects phosphomonoesters (PMEs) such as PC and phosphoethanolamine (PE), and phosphodiesters (PDEs), consisting of GPC and glycerophosphoethonolamine (GPE). Initially, it was not possible to resolve the individual phosphorus metabolites PC, PE, GPC, and GPE, but with the advancement of 31P MRS techniques, it has become feasible to identify and quantify PE, PC, GPE, and GPC in vivo.34
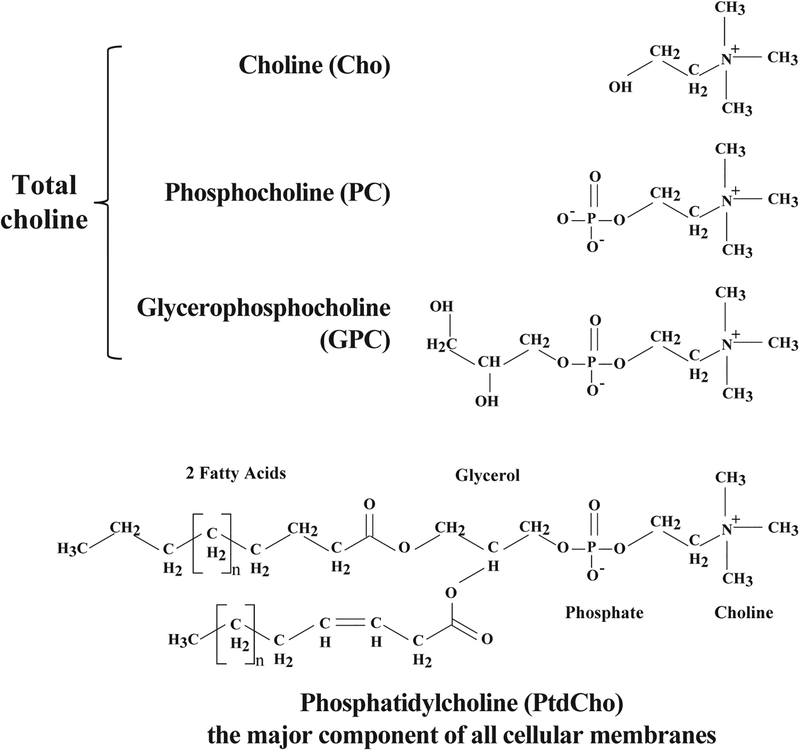
Figure 1:
Chemical structures of choline (Choline), phosphocholine (PC), glycerophosphocholine (GPC), and membrane phosphatidylcholine (PtdCho).
Until today, most studies have investigated the elevated PC levels in cancer and the related Kennedy pathway in which PC is produced. This pathway is, to a large part, driven by overexpression and activation of the enzyme choline kinase alpha (CHKα).16 CHKα has been studied extensively and several choline kinase inhibitors have been developed.35 However, although the CHKα inhibitor TCD-717 has recently completed phase I clinical trials in patients with advanced solid tumors (https://clinicaltrials.gov/ct2/show/ NCT01215864), no CHKα inhibitors are currently clinically available.35 There is also growing evidence that alternative mechanisms of CHKα function are critical for tumor growth, which are not necessarily related to CHKα’s role in producing PC, but rather CHKα’s scaffolding or regulatory roles.36–38 Several research teams are still exploring the use of CHKα inhibitors in cancer treatment.
More recent studies have demonstrated the importance of GPC in cancer, emphasizing the roles of enzymes that produce and degrade GPC. Here we are providing a summary of the recent literature on the roles of GPC in cancer, the enzymes directly involved in GPC production and degradation, and GPC detection by MRS applications.
The GPC breakdown pathway in cancer
Phosphatidylcholine (PtdCho) is the most abundant phospholipid in mammalian cell membranes.39 PtdCho is synthesized from free choline through the Kennedy pathway,40 in which free choline is phosphorylated to PC41,42 with the help of the CHK. PC is then converted to cytidine diphosphate (CDP)-choline by adding a CDP group to PC. In the next step, diacylglycerol (DAG)-cholinephosphotransferase catalyzes the final reaction using diacylglycerol (DAG)43,44 as a lipid anchor, and CDP-choline to produce PtdCho. In a catabolic pathway, PtdCho is broken down to 1-acyl-GPC and GPC, which is subsequently converted to free choline, thus completing the choline cycle39 as shown in Figs. 1 and 2.
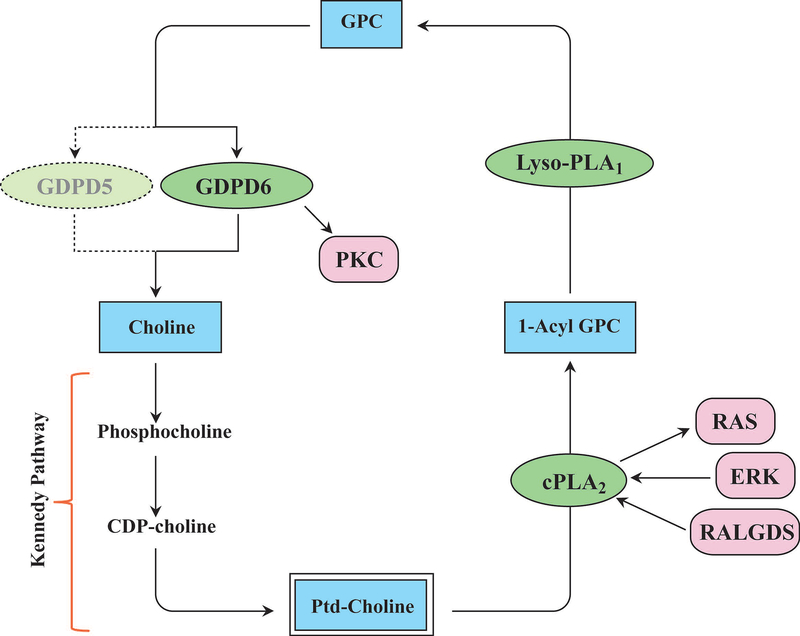
Figure 2:
Enzymes involved in the glycerophosphocholine metabolic pathway, and control of glycerophosphocholine metabolism by oncogenic signaling pathways. Enzymes are shown in green ovals, metabolites in blue boxes, and signaling pathways in red boxes. (Abbreviations: CDP, cytidine diphosphate; cPLA2, cytoplasmic phosphatidylcholine-specific phospholipase A2; ERK, extracellular signal regulated kinase; GDPD, glycerophosphodiester phosphodiesterase; GPC, glycerophosphocholine; Lyso-PLA1, lyso-phospholipase A1; PKC, Protein kinase C; RALGDS, RAL GTPase guanine nucleotide dissociation stimulator).
There are three enzymes that are contributing to the GPC pathway that breaks down PtdCho to free choline (Fig. 2). The first step in the GPC breakdown pathway is the hydrolysis of PtdCho by the enzyme cytosolic phosphoplipase A2 (cPLA2), which removes one fatty acid to produce 1-acyl-GPC.45 This is followed by a second hydrolysis step by the enzyme lysophopsholipase A1 (lyso-PLA1), which removes the second fatty acid to produce GPC.46,47 GPC is then converted to free choline and glycerol-3-phosphate by the enzyme glycerophosphocholine phosphodiesterase (GPC-PDE, EC 3.1.4.2).48–50 These three key enzymes directly regulate GPC levels in mammalian cells48–50 and can therefore contribute to the increased GPC levels that have been detected in human cancers. Here we briefly describe the basic features of these three enzymes in the GPC breakdown pathway (Fig. 2).
Phospholipases:
Phospholipases are enzymes that catalyze the hydrolysis of fatty acyl-ester bonds of phospholipids in mammalian cells51 as shown in Fig. 3A. Phospholipases can be categorized into two sets of enzymes based on their enzymatic activity: the acyl hydrolases and the phosphodiesterases.51 Phospholipases with acyl hydrolase activity are phospholipase A1 (PLA1), phospholipase A2 (PLA2), phospholipase B (PLB), lysophospholipase A1 (Lyso-PLA1), and lysophospholipase A2 (Lyso-PLA2).51 Phospholipases with phosphodiesterase activity are phospholipase C (PLC)52 and phospholipase D (PLD).51
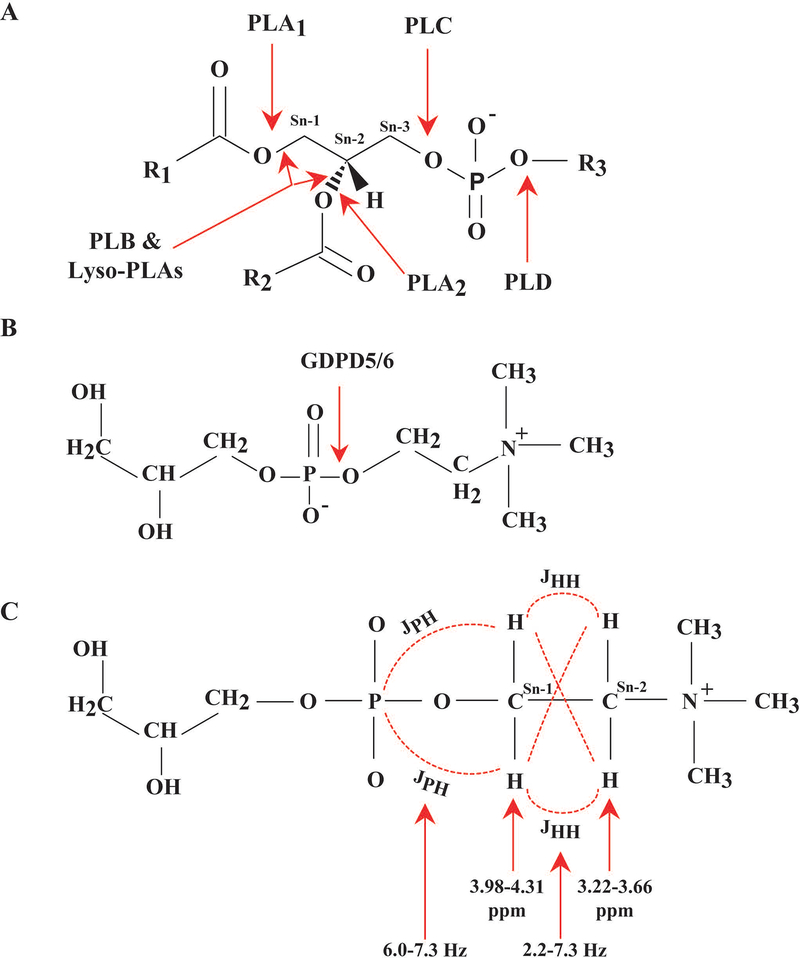
Figure 3:
(A) Phospholipid molecule along with respective phospholipase (PL) cleavage sites. PLA1 hydrolyzes the ester bond at the Sn-1 position, and PLA2 hydrolyzes at the Sn-2 position. PLC hydrolyzes the glycerol-oriented phosphodiester-bond, and PLD hydrolyzes the alcohol-oriented phosphodiester-bond. PLB and Lyso-PLAs can hydrolyze at both the Sn-1 and Sn-2 positions showing both PLA1 and PLA2 activity. (B) Enzyme activity of GDPD5 and GDPD6. (C) Partial chemical structure of PE, PC, GPE, and GPC including chemical shift (ppm) values of the 1H spins of the Sn−1CH2 and Sn−2CH2 groups with their J‐coupling constants.
Cytosolic phosphatidylcholine-specific phospholipase A2:
There are five distinctive enzyme subtypes within the PLA2 superfamily, which are cytosolic PLA2 (cPLA2), calcium-independent PLA2 (iPLA2), secreted PLA2 (sPLA2), lysosomal PLA2, and platelet-activating factor acetylhydrolase (PAF-AH).52–54 The cPLA2 family consists of six members, i.e. cPLA2α, cPLA2β, cPLA2γ, cPLA2δ, cPLA2ε, and cPLA2ζ. In mammalian systems, only cPLA2α, cPLA2β, cPLA2γ, and cPLA2δ are expressed.52,55 All members of the PLA2 superfamily catalyze the hydrolysis of fatty acids at the sn-2 position of glycerophospholipids, which produces free fatty acid and lyso-phospholipid (Fig. 3A).54 Several of the fatty acids and lipids produced by PLA2s serve as substrates for various intracellular biochemical pathways for producing lipid mediators, such as prostaglandins, leukotrienes, and thromboxanes.56 In some cases, these lipid mediators function as critical mediators of cell growth,57 cell differentiation,58–61 chronic inflammation,62 and malignant transformation.59 Cytosolic PLA2α (cPLA2α), a 85-kDa protein63 with affinity for hydrolyzing arachidonic acid (AA), is one of the important members of this superfamily.54 cPLA2α was shown to be the only phospholipase with complete specificity for AA.57,64 The AA released by cPLA2α is then metabolized by cyclooxygenase (COX) enzymes and subsequently converted to biologically active eicosanoid lipid mediators such as prostaglandin E2 (PGE2).57 COX2 has been linked to the development and progression of various cancers, including breast cancer,58,59,61 bladder cancer,65 cutaneous squamous cancer,66 and hepatocellular carcinoma (HCC).67 The enzyme cPLA2α plays an important role in various biological processes, such as inflammation,68 cell growth, and cancer development.69–71 cPLA2α is overexpressed in cancer and drives tumorigenesis by producing increased substrate for the COX2-PGE2 and related pathways.72–74 It was recently observed that cPLA2α is overexpressed in breast cancer and correlates with poor prognosis in breast cancer patients.75 In the same study, cell culture experiments showed that the overexpression of cPLA2α was associated with increased migration and invasion in breast cancer cells.75 Knocking down cPLA2α in triple-negative MDA-MB-231 breast cancer cells inhibited transforming growth factor-β (TGF-β)-induced epithelial-to-mesenchymal transition (EMT) via the Phosphatidyl-inositol-3-kinases/serine-threonine kinase B (PI3K/AKT) signaling pathway.75 Various reports have demonstrated the role of cPLA2α in cancer60,61,76,77 and inflammation,62 indicating that inhibiting cPLA2α and subsequently decreasing AA availability may serve as a therapeutic approach for treating cancer.71,76,77 The critical role of cPLA2α in carcinogenesis makes this enzyme a potential target for anti-cancer treatment. However, although several cPLA2α inhibitors have been reported as anticancer agents in preclinical studies, there are currently no chemotherapeutic drugs that are cPLA2α inhibitors available for cancer treatment. The fact that cPLA2α is expressed ubiquitously in several important human tissues including spleen, thymus, prostate, testis, ovary, small intestine, heart, brain, skeletal muscles78 may increase the propensity for side effects when developing therapeutic cPLA2α inhibitors. Research on cPLA2α in cancer and cPLA2α as therapeutic target for cancer treatment are active fields of investigation, and will hopefully provide answers as to the clinical translatability of cPLA2α -related research soon (Table 1). Targeting cPLA2α in cancer would also likely result in MRS-detectable differences in GPC levels, which would be worthwhile testing in future studies.
Table 1:
Glycerophosphocholine concentration and associated enzyme expression and activity levels in the most frequent human cancers
| Cancer Type | GPC level | Expression level | ||||
|---|---|---|---|---|---|---|
| Normal | Cancer | GDPD5 | GDPD6 | cPLA2 | Lyso-PLA1 | |
| Breast | 0.04±0.04 mmol/L330 | 0.28±0.20 mmol/L330 | ↑48 | ↑96 | ↓297 | ↓297 |
| Prostate | 0.29±0.26 mmol/kg279 | 0.57±0.87 mmol/kg279 | ND | ND | ↑331 | ND |
| Ovarian | Unaltered332 | Unaltered332 | ↑96 | ↓332 | ND | |
| Lung | Increased in tumor tissue versus normal tissue 333 | ↑334 | ND | ↑335,336 | ND | |
| Colorectal | ND | ND | ↑337 | ND | ↑338,339 | ND |
In cancer, cPLA2α expression varies depending on the genetic profile of the cancer cells.79 Distinct choline metabolic profiles were shown to be associated with differences in gene expression profiles for basal-like and luminal-like breast cancer xenograft models.79 Higher GPC as compared to PC levels were reported in basal-like xenografts whereas the opposite was observed in luminal-like xenografts.79 Moreover, cPLA2α was shown to be involved in development of resistance against endocrine therapy.61 cPLA2α expression in breast cancer is associated with epidermal growth factor receptor (EGFR) expression, mediates estrogen-dependent breast cancer cell growth, and correlates with an adverse prognosis in luminal cancers.60 cPLA2α activation also correlates with HER2 overexpression in breast cancer cells.60 Hence, monitoring cPLA2α activity can provide valuable information in breast cancer diagnosis and for evaluating treatment response. For this purpose, the Delikatny group recently synthesized and preclinically tested several phospholipase-activatable ‘smart’ fluorescent imaging probes, which are specifically activated by PLC only or cPLA2 only.80,81 Recently, the same group developed a second series of cPLA2 based imaging agents with improved activity.82 In these smart imaging agents, AA was coupled to red-shifted fluorophores, which maximized light tissue penetration, and which achieved cPLA2 specificity by employing AA, because cPLA2 is the only phospholipase with specificity for AA-containing phospholipids.64 Because of their deep tissue penetration capabilities, these novel near-infrared fluorescent smart probes can be used in vivo. One of the agents has shown the potential to be imaged in vivo in triple-negative breast cancer mouse models in which cPLA2 was overexpressed.82 Continuing the development of cPLA2-activatable imaging probes to determine in vivo cPLA2 activity will help with future translation of cPLA2 inhibitors as therapeutic targets for cancer treatment. Smart cPLA2 imaging probes are able to identify tumors in which cPLA2 activity is high, and they could be used for monitoring in vivo on-target efficacy of cPLA2 inhibitors.
Lyso-phospholipase A1:
All enzymes with phospholipase A1 activity, including lyso-PLA1, catalyze the hydrolysis of fatty acids at the sn-1 position of phospholipids producing 2-acyl-lysophospholipids, or, in the case of lyso-PLA1, glycerophospho-alcohols such as GPC, glycerophosphoethanolamine (GPE), or glycerophosphoinositol (GPI) among others and fatty acids.83 The PLA1 isozymes consist of two morphologically different families: The intracellular PLA1 family84 and the extracellular PLA1 family.85 Extracellular PLA1 enzymes belong to the lipase family with a molecular weight of 50–60 kDa.85 Lyso-PLAs are enzymes that hydrolyze lysophospholipids by removing the only remaining acyl-chain.86 Lyso-PLAs can be divided into two sub-classes based on their molecular weights, i.e. small molecular weight lyso-PLAs of <30 kDa and high molecular weight lyso-PLAs of >50 kDa. Small molecular weight lyso-PLAs can be further divided into lyso-PLA1 and lyso-PLA2, both of which are similar in size and function.86 High molecular weight lyso-PLAs display lyso-PLA activity along with other hydrolytic or lipolytic activities.86 Examples of high molecular weight lyso-PLAs include group IV calcium-dependent cPLA2, phosphatidic acid-preferring PLA1 (PA-PLA1), transacylase/lysophospholipase (TA/LysoPLA) and triacylglycerol lipase lysophospholipase.86
Mammalian cells and tissues, including rabbit heart, pig gastric mucosa,87 rat and beef liver, murine macrophage and human HL60 cells,88 have been reported to contain high lyso-PLA activity as compared to phospholipase activity, which acts as a safeguard to maintain optimally low levels of lysophospholipids in cells and tissues.86 High concentrations of lysophospholipids resulting from altered membrane phospholipid metabolism are associated with disrupted membrane conformation, and disrupted activities of membrane bound enzymes such as guanylate and adenylate cyclase.89,90 They are also associated with various diseases such as atherosclerosis, inflammation, and myocardial ischemia.86,89,90 Low, non-toxic levels of lysophospholipids act as lipid second messengers and transduce signals from membrane receptors.86 As multiple enzymes share lyso-PLA activity,86 understanding the nature of lyso-PLAs is a complex undertaking. Lyso-PLA1 and lyso-PLA2, as outlined above, have similar molecular weights and functions. These two are expressed together in mammalian cells, including heart and liver cells. Both lyso-PLA1 and lyso-PLA2 function independent of bivalent ions such as Ca2+ and Mg2+.86 Both of these proteins display 64% sequence similarity and contain a conserved catalytic triad consisting of serine, histidine and aspartate, emphasizing that both lyso-PLA1 and lyso-PLA2 share similar catalytic activity.86
Lyso-PLA1 can efficiently hydrolyze 1-palmitoyl lyso-PtdCho substrate that consists of a 9:1 equilibrium mixture of 1-palmitoyl lyso-PtdCho and 2-palmitoyl lyso-PtdCho, and hence with the fatty acid located predominately at the sn-1 position, producing free palmitic acid, suggesting that lyso-PLA is able to function both as a lyso-PLA1 and lyso-PLA2.91 A recent study showed that treating the mouse macrophage cell line RAW264.7 with parasite lyso-PLA induced the expression of interleukin 25 (IL-25), elevated BRAF and ERK1/2 mRNA levels, and phosphorylated BRAF and extracellular signal regulated kinase 1/2 (ERK1/2).91 Up-regulation of IL-25 induced expression of mesenchymal cell markers and promoted cell migration, suggesting an association of lyso-PLA with cell migration.91 A significant body of work has been published on high molecular weight lyso-PLAs, while low molecular weight lyso-PLA1 and lyso-PLA2 have yet to be explored, including their effects on cellular GPC levels (Table 1).
Glycerophosphocholine phosphodiesterase:
Glycerophosphocholine phosphodiesterases (GPC-PDE), also referred to as glycerophosphodiester phosphodiesterases (GDPDs) and glycerophosphodiesterases (GDEs), are a family of enzymes that convert glycerophosphodiesters to glycerol 3-phosphate and an alcohol.92 They have been identified and characterized in various tissues including rat kidney, brain, liver, and uterus.93,94 GDPDs (GDEs) can be subdivided into two groups based on their cellular location, i.e. group one GDPDs are associated with the plasma membrane and group two GDPDs remain in the cytosol, see Fig. 4A and B.49 There are seven members in the human GDPD family, which are phylogenetically close to the E. coli GPC-PDE protein that catalyzes the hydrolysis of glycerophosphodiesters and thereby produces glycerol 3-phosphate and alcohols.49 Mammalian GDPDs show different substrate specificity, and GDE1 (no GDPD notation available), GDPD5 (GDE2) and GDPD6 (GDE5) have been reported to utilize glycerophosphodiesters as substrate and hydrolyze them to produce glycerol 3-phosphate and an alcohol, whereas GDPD2 (GDE3), GDPD1 (GDE4), GDPD4 (GDE6) and GDPD3 (GDE7) show specificity towards various other substrates as shown in Table 2. Two members of the GDE family, GDPD5 (GDE2) and GDPD6 (GDE5) have been identified to confer GPC-specific phosphodiesterase (GPC-PDE) activity to hydrolyze GPC into glycerol 3-phosphate and free choline Fig. 3B.95,96
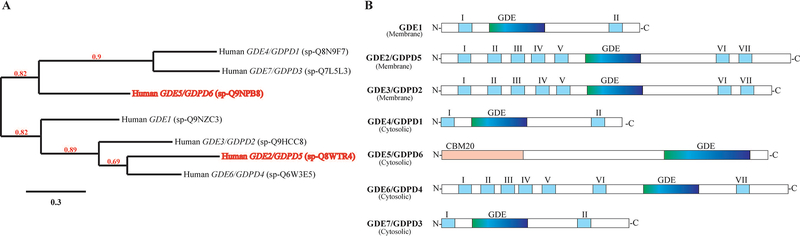
Figure 4:
Phylogenetic analysis of human GDEs/GDPDs. (A) The phylogenetic tree was built by using the Phylogeny.fr online platform using the amino acid sequences reported in the UniProtKB database. Sequences were aligned with MUSCLE (v3.8.31), and configured for highest accuracy using default parameters of MUSCLE. The phylogenetic tree was reconstructed using the maximum likelihood method implemented in PhyML program (v3.1/3.0 aLRT). Reliability for each internal branch was evaluated using the aLRT test (SH-Like). Branch support values above 50% (0.5) are shown (red numbers). The scale bar represents the amino acid percentage substitutions required for generating the corresponding tree. (B) Schematic diagram illustrating the domain structure of the seven human GDE/GDPDs. The transmembrane domains are numbered I-VII starting from the N-terminal. GDE represents the putative glycerophosphodiesterase domain containing phosphodiesterase activity. GDE1, GDE4/GDPD1 and GDE7/GDPD3 have two transmembrane domains at each terminus. GDE2/GDPD5, GDE3/GDPD2 and GDE6/GDPD4 have seven transmembrane domains. GDE5/GDPD6 contains a carbohydrate binding domain, which belong to the CBM20 family, at the N-terminus.
Table 2:
Substrate specificity of mammalian glycerophosphodiesterases
| SN | Enzyme | Enzyme activity | Substrate | End products |
|---|---|---|---|---|
| 1 | GDE1 | Glycerophosphodiesters | Glycero-phosphoinositol | Glycerol-3-phosphate and Inositol |
| Glycero-phosphoserine, Glycero-phosphoglycerol | Glycerol-3-phosphate and alcohol | |||
| Glycero-3-phospho-N-acyl ethanolamines | Glycerol-3-phosphate and N-acyl ethanolamine | |||
| Lyso-glycero-3-phospho-N-acyl ethanolamine | Glycerol-3-phosphate and N-acyl ethanolamine | |||
| Glycero-phosphocholine | Glycerol-3-phosphate and free choline | |||
| 2 | GDE2/GDPD5 | Glycerophosphodiesters | Glycero-phosphocholine | Glycerol 3 phosphate and free choline |
| Glycero-phosphoinositol | Glycerol and Inositol-1 phosphate | |||
| GPI-anchor (protein RECK, glypican 2 and 4)* | GPI anchor | |||
| 3 | GDE3/GDPD2 | Phospholipase C-like activity | Glycero-phosphoinositol | Glycerol and Inositol 1 phosphate |
| GPI-anchor (proteins RECK, glypican 2 and 4)* | GPI anchor | |||
| 4 | GDE4/GDPD1 | Lysophospholipase D-like activity | Lysophosphatidylcholine | Choline and lysophosphatidic acid |
| 5 | GDE5/GDPD6 | Glycerophosphodiesters | Glycero-phosphocholine | Glycerol 3 phosphate and free choline |
| 6 | GDE6/GDPD4 | Glycerophosphodiesters | GPI-anchor (glypican 2 and 4)* | GPI anchor |
| 7 | GDE7/GDPD3 | Lysophospholipase D-like activity | Lysophosphatidylcholine | Choline and lysophosphatidic acid |
glycosylphosphatidylinositol-anchor (reversion-inducing cysteine-rich protein with Kazal motifs’ (RECK))
The human GDPDs form two clusters in their phylogenetic tree (Fig. 4A). In the first cluster, GDPD6 (GDE5) occupies one branch on its own, making its topological structure distinctly different from the other GDPDs (Fig. 4B), as it contains an N-terminal carbohydrate binding domain (CBM20) and a C-terminal GDE domain.49 The second branch in the first cluster consists of GDPD1 (GDE4) and GDPD3 (GDE7), which both have specific activity towards glycerolysolipids instead of glycerophosphodiesters.97 Structurally, they contain two transmembrane domains, one at each terminus, along with a centrally located GDE domain.97 The second cluster consists of GDE1, which is topologically similar to GDPD1 (GDE4) and GDPD3 (GDE7), but has activity towards glycerophosphodiesters, and GDPD5 (GDE2), GDPD2 (GDE3) and GDPD4 (GDE6).49 The latter three, i.e. GDPD5, GDPD2, and GDPD4, each contain 7 transmembrane domains, which give rise to their group name “serpentine GDEs” (Fig. 4A–B).49
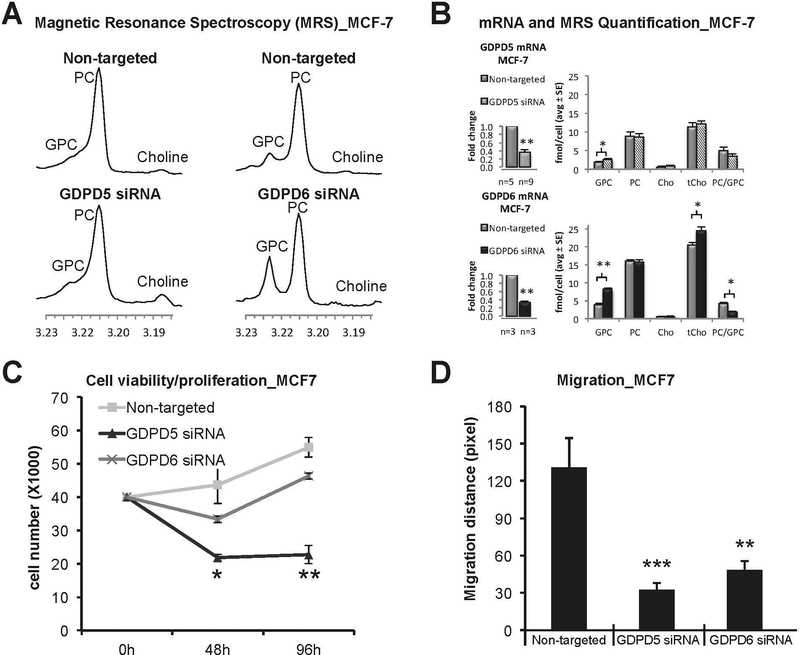
GDPD5 (GDE2) has been reported for the first time in mice as a GDPD (GDE) that regulates GPC levels osmotically in renal cells, where GPC serves as an osmoprotective organic osmolyte.95 Additionally, GDPD5 (GDE2) has a function in neuronal differentiation, as the catalytic activity of GDPD5 (GDE2) induced motor neuron differentiation98 through autonomous cleavage of a GPI anchor from the RECK protein located in the neighboring cells99 A recent report has shown that GDPD5 (GDE2) induced neuroblastoma cell differentiation by cleaving a GPI-anchored heparan sulfate proteoglycan, termed glypican-6 (GPC6), thereby releasing GPC6 and suppressing cell motility.100 GDPD5 (GDE2) has also emerged as a prognostic marker for neuroblastoma.101 Increased GPC levels were observed in a 31P MRS study of triple-negative MDA-MB-231 breast cancer xenograft models in which GDPD5 (GDE2) was constitutively silenced, and which were grown orthotopically in athymic nude mice.102 Expression levels of GDPD5 (GDE2) were reported to positively correlate with breast cancer malignancy.48 GDPD5 (GDE2), along with PLD1 and CHKα, were highly expressed in estrogen receptor negative (ER-) human breast cancer samples, which contained high PC, tCho, and decreased GPC level in contrast to estrogen receptor positive (ER+) human breast cancer samples.48 In a recent study employing high-resolution (HR) 1H MRS following siRNA silencing of GDPD5 (GDE2) or GDPD6 (GDE5) in two human breast cancer cell lines, Cao et al demonstrated that GPC levels were increased more than twofold during GDPD6 (GDE5) silencing, and marginally increased during GDPD5 (GDE2) silencing.103 Following GDPD5 (GDE2) siRNA silencing, Cao et al observed that triple-negative MDA-MB-231 breast cancer cells displayed reduced migration and invasion, while ER+ MCF-7 breast cancer cells showed reduced viability and migration103 (Fig. 5).
Figure 5:
Effects of GDPD5 and GDPD6 silencing on choline metabolite profiles. (A) Representative 1H MRS choline metabolite profiles of GDPD5 and GDPD6 siRNA treated MCF-7 cells compared to non-targeted controls. (B) GDPD5 and GDPD6 were significantly down-regulated by 65% and 67% in MCF-7 cells, respectively. Significantly increased GPC levels and decreased PC/GPC ratios were observed in GDPD6 silenced cells compared to non-targeted controls. GDPD5 silencing slightly increased GPC levels. Values are presented as mean ± standard error. *p≤0.05, **p≤0.01. The number of repeats (n) are given underneath each graph. (C) Cell proliferation. GDPD5 silencing resulted in decreased cell proliferation, while GDPD6 silencing had no significant effect on cell proliferation in MCF-7 cells compared to non-targeted controls. Values are presented as mean ± standard deviation. *p≤0.05, **p≤0.01, n=3 each. (D) Quantification of migration distance. GDPD5 and GDPD6 silencing significantly reduced cell migration in MCF-7 cells. Each assay was repeated four times. Values are presented as mean ± standard deviation. **P<0.01, ***P<0.001, n=4 each (Modified from Cao et al.103).
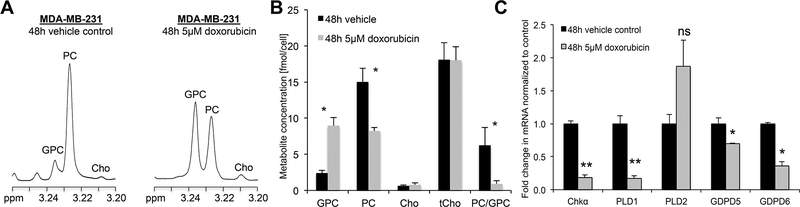
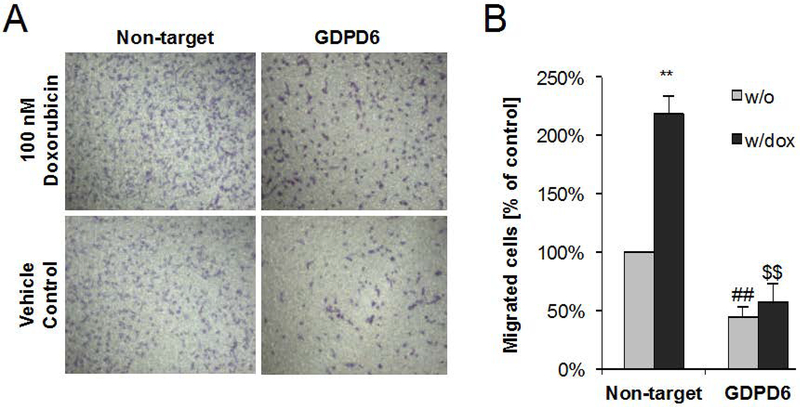
GDPD6 is a Mg2+-dependent GDE/GDPD, which shows a preference of GPC over GPE as substrate.50 GDPD6 (GDE5) consists of two highly conserved domains, a carbohydrate-binding domain, and a catalytic domain showing GDE enzyme activity.50 GDPD6 (GDE5) has been reported to act as regulatory body for skeleton muscle development.50 GDPD6 (GDE5) was demonstrated to participate in conferring migration and invasion of cancer cells via the protein kinase C-α signaling pathway.96 Increased GDPD6 (GDE5) mRNA levels have been observed in metastatic endometrial carcinomas.28,96 GDPD6 (GDE5) expression was negatively correlated with survival in endometrial and ovarian cancers.96 A recent study by the Glunde group has shown that silencing of GDPD5 (GDE2) or GDPD6 (GDE5) using siRNA can increase the efficacy of chemotherapeutic agents such as doxorubicin.104 In this study, the weakly metastatic cell line MCF-7 and highly metastatic triple negative MDA-MB-231 cells were treated with doxorubicin, which resulted in decreased PC levels and increased GPC level. Silencing of GDPD6 (GDE5) by siRNA diminished breast cancer cell migration induced by low concentrations doxorubicin. Increased GPC and decreased PC levels upon doxorubicin treatment resulted from reduced mRNA and protein expression levels of GDPD6 (GDE5), PLD1, and CHKα (Fig. 6,7).104
Figure 6:
Effects of Doxorubicin treatment in triple-negative MDA-MB-231 cells after 48 hours of treatment. (A) Representative 1H MR spectra of the choline metabolite region of the water-soluble phases of MDA-MB-231 cell extracts obtained following 48 hours of 5 μM doxorubicin treatment (treated) or vehicle (DMSO) control showing elevated GPC and decreased PC levels. (B) Quantification of choline-containing metabolite concentrations showed that the total choline containing metabolite (tCho) concentration did not change between treated and control samples while GPC increased and PC decreased, thus decreasing the PC/GPC ratio following 48 hours of 5 μM doxorubicin treatment compared to vehicle control. (C) Doxorubicin treatment affects the mRNA expression levels of genes associated with phosphatidylcholine metabolism. mRNA levels of ChKα, PLD1, GDPD5, and GDPD6 were decreases after 48 hours of treatment. There was no significant change in PLD2 mRNA level. The graphs are mean + SE. Means are calculated based on three independent experiments. *P ≤ .05, **P ≤ .01. An unpaired two-tailed t test was used for all comparisons (Modified from Cheng et al.104).
Figure 7:
GDPD6 silencing counteracts doxorubicin-promoted migration. (A) Representative microscopic images of migrated stained cells following migration through the membrane in the transwell migration assay. Treatment with 100 nM doxorubicin resulted in increased breast cancer cell migration as compared to vehicle control without drug. GDPD6 silencing decreased the number of migrated cells, and after 100 nM doxorubicin was added to the medium, the migrated cell number did not increase due to doxorubicin addition, effectively counteracting doxorubicin-promoted migration. (B) Quantification results from microscopic cell images following migration. Results are expressed as mean + SE. Means were calculated from three independent experiments. **, ##, $ $ P < 0.01. ** indicates comparison to vehicle control (DMSO) without doxorubicin; ## refers to comparison with non-target controls without doxorubicin, and $ $ refers to comparison to non-target control with doxorubicin. An unpaired two-tailed t test was used for all comparisons. (Modified from Cheng et al.104).
While important roles of GDPD5 (GDE2) and GDPD6 (GDE5) are emerging in neuroblastoma, breast cancer, and endometrial and ovarian cancers, it is important to further investigate the roles of GDPD5 (GDE2) and GDPD6 (GDE5) in cancer to evaluate their diagnostic and treatment potentials. The use of MRS to detect cellular and tissue GPC levels enables non-invasive detection of the effectiveness of GDPD5 (GDE2) or GDPD6 (GDE5) silencing or inhibition.
Oncogenic signaling pathways and transcription factors regulating the GPC pathway
The first indication that choline metabolism is regulated by oncogenic signaling pathways came from early work showing that PC levels increased following growth factor stimulation of NIH3T3 fibroblasts.105,106 The RAS/MAP kinase pathway regulates important cellular processes including cell growth, proliferation, differentiation, migration, and programmed cell death.107–110 When the RAS/MAP kinase pathway is activated by receptor tyrosine kinases (RTKs),111 it triggers a cascade of phosphorylation events leading to the phosphorylation of a variety of target proteins within the nucleus or cytoplasm or cells.107,112,113 The first part of the phosphorylation cascade relies on the activation of at least one of the four major MAP kinases, which are ERK1 and 2 (ERK1/2), ERK5, p38, and JNK.110,114 The next steps involve RAF phosphorylation of MEK1/2, which in turn phosphorylate and activate ERK1/2.110,114
Tyrosine kinases111 are crucial mediators of various signaling cascades, and have vital roles in diverse biological processes including cell proliferation, differentiation, migration, metabolism, and programmed cell death.109,111 It is well documented that tyrosine kinases are involved in the pathophysiology of cancer.109,115 Tyrosine kinases are enzymes that phosphorylate the tyrosine residues in target proteins by transferring a phosphate group from adenosine triphosphate (ATP) to the respective tyrosine residue.111,116 Tyrosine kinases can be categorized as receptor tyrosine kinases (RTKs),116 including epidermal growth factor receptor (EGFR), fibroblast growth factor receptor (FGFR) and platelet-derived growth factor receptor (PDGFR), and non-receptor tyrosine kinases (NRTKs), including SRC, Abelson tyrosine-protein kinase (ABL), focal adhesion kinase (FAK) and Janus kinase (JAK).111 The RTKs are cell surface transmembrane receptors with kinase activity, and contain a multidomain extracellular ligand-binding site with specificity to a particular ligand, a transmembrane hydrophobic helix, and a cytoplasmic tyrosine kinase domain.117 RTKs function as receptors for growth factors, cytokines, hormones, neurotrophic factors, and other extracellular signaling molecules, thereby activating the respective signaling pathways that mediate cell proliferation, differentiation, and migration, among others.111,116
Important transcription factors driving carcinogenesis as well as choline metabolism are MYC and hypoxia induced factors (HIFs).16 Several studies have shown that well-studied genes in choline phospholipid metabolism, such as CHKα, CTP:phosphocholine cytidylyltransferase CCT, and PLD are regulated by such oncogenic signaling pathways and transcription factors.16 Recent studies as reviewed below are revealing the reciprocal interactions between oncogenic signaling pathways, transcription factors, and genes in the GPC pathway, i.e. cPLA2, lyso-PLA1, GDPD5, and GDPD6.
Estrogen, progesterone, and HER2 receptor signaling:
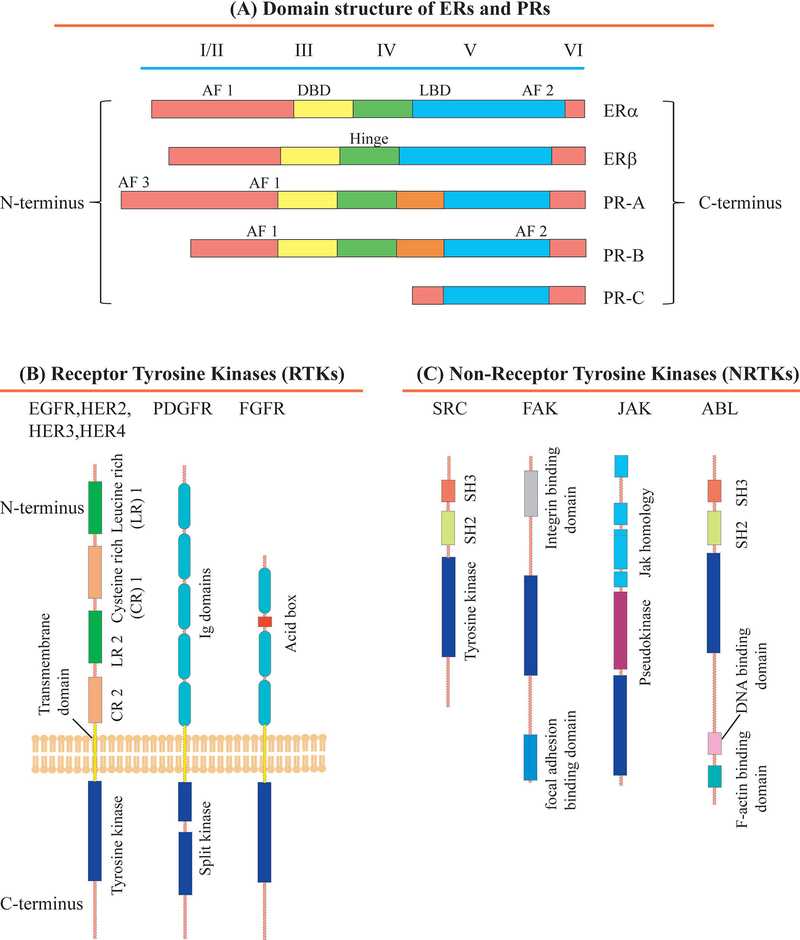
Estradiol (or estrogen), a steroid hormone, is a key player in the progression of breast cancer, as 65% of all human breast cancers are ER+, which means that their growth depends on the availability of estrogen.118 Estrogen confers its biological effects by binding to estrogen receptors, i.e. ERα and ERβ, which are structurally and functionally different from each other.118 Both of these receptors show highly homologous DNA and ligand binding domains (96% homology) along with a less homologous transcriptional activating function 1 (AF 1) domain.119 ERα also has a C-terminal AF 2 domain containing ligand-binding region as shown in Fig. 8A.119 When estrogen binds to ERα, it subsequently translocates to the nucleus, where it binds to the target gene promoter to stimulate gene transcription.120,121 ERα plays a vital role in breast cancer progression.122,123 ERα signaling has emerged as a complex signaling event as it involves various co-regulatory proteins, as well as participation of extracellular, intranuclear, and genomic molecular events.124 ER-co-regulatory proteins such as SRC-1 are differentially expressed in tumors and have altered functions that lead to tumor progression.125 A recent study reported that in mice carrying the mouse mammary tumor virus polyomavirus middle T (PyMT) transgene (Tg), SRC-1 co-regulator deficiency increased MMTV-neu-mediated tumor latency and decreased metastasis, indicating the importance of this ERα-co-regulator in breast cancer metastasis.126 ERα extra-nuclear signaling enhances the activation of PI3K, Src kinase, protein kinase C and mitogen-activated protein kinase (MAPK) pathways in the cytosol.127 Several of the kinases that are activated by ERα extra-nuclear signaling pathway are implicated in breast cancer metastasis,128 including phosphorylation of ERK and protein kinase B (AKT) for breast cancer cell migration, and Src129 and ILK1 kinases for breast cancer cell invasion and metastasis.130 Even though ERβ works as a transcription factor similar to ERα and mediates distinct physiological responses related to estrogen signaling, their physiological consequences are different.118 A recent study suggested that increased ERβ expression had a counter effect on cell proliferation,131 and that ERβ confers an anti-proliferative function in tumors.132 ERβ expression is correlated with decreased cell migration, and decreased invasive and proliferative tumors as ERβ affects integrin expression levels, thus altering adhesion and migration properties of breast cancer cells.132,133 Downregulation of ERβ expression was shown to promote EMT in prostate cancer cells.134 These studies suggest that ERβ signaling may promote anti-migratory and anti-invasive responses, however, more detailed studies of ERβ signaling in cancer are needed.
Figure 8:
(A) Structural and functional domains of the estrogen receptors (ERα and ERβ) and progesterone receptors (PR-A, PR-B and PR-C). Structural domains of these receptors are labeled I-VI, containing five distinct structural and functional domains: DNA-binding domain (DBD, III), hinge domain (IV), ligand-binding domain (LBD; V/VI), and three transcriptional activation function domains AF 1 and AF 3 (I/II) and AF 2 (VI). (B) Structural domains and their organization in various receptor tyrosine kinases (RTKs) such as the epidermal growth factor receptor (EGFR) family, which contains EGFR, HER2, HER3 and HER4, fibroblast growth factor receptor (FGFR) and platelet-derived growth factor receptor (PDGFR). The extracellular part of these receptors is on top and the intracellular part is on the bottom. The intracellular tyrosine kinase domains are conserved across all RTK subfamilies, while the ligand-interacting domains differ significantly due to the specificity of ligand-receptor interactions. The intra- and extracellular domains are connected by a transmembrane domain. The following abbreviations are used: CR, cysteine-rich; Ig, immunoglobulin-like; LR, leucine-rich. The LR1, CR1, LR2 and CR2 domains of the HER family are also termed domains I–IV. The PDGF receptors have a large insert in their tyrosine kinase domain and are termed split kinases. (C) Domain structure and organization in non-receptor tyrosine kinases (NRTKs) such as SRC, Abelson tyrosine-protein kinase (ABL), focal adhesion kinase (FAK), and Janus kinase (JAK). The following abbreviations are used: SH3, SRC homology 3 domain; SH2, SRC homology 2 domain; Jak, Janus homology domain.
Progesterone receptor (PR) belongs to the nuclear/steroid hormone receptor (SHR) family. The SHR family consists of ligand-dependent transcription factors that are mainly expressed in the female reproductive system and the central nervous system.135,136 PR, when bound to the steroid hormone progesterone, controls various signaling pathways associated with development, differentiation, proliferation, as well as endocrine-based cancers.137 PR is a modular protein that consists of an intrinsically disordered N-terminal domain, a central globular DNA-binding domain (DBD), and a folded C-terminal ligand-binding domain (LBD), as shown in Fig. 8A and commonly exists as three isoforms, i.e. PR-A, PR-B136 and PR-C138. The PR-A and PR-B isoforms are produced from the same gene by alternate translational start sites, and they have two transcription sites.136 PR-B is the full-length receptor with additional 164 amino acid residues at the N-terminal, which are absent in PR-A.136 These isoforms can act as either homo (A-A and/or B-B) or heterodimer (A-B), are able to regulate different groups of target genes, and show both ligand-independent and -dependent functions.136 PR-C is a highly tissue-specific, third isoform containing only N-terminal DNB and C-terminal LBD, which acts as inhibitor of PR-B in the uterus and induces labor.138 In breast cancer, the percentage of proliferating, ER/PR-positive cells increases significantly as compared to healthy women, switching from paracrine hormone signaling, where cells produce hormones that bind to receptors of other nearby cells, to autocrine mode, where cancer cells secrete hormones that bind to their own receptors.139,140
Transgenic mouse models141,142 of hormone-dependent mammary tumors as well as experiments with breast cancer cell lines showed an increase in the percentage of proliferating ER/PR positive cells due to deregulation of ERα.143 In human mammary tissues, an increase in ER/PR-positive proliferating cells has been observed in adjacent normal tissues of breast cancer patient, implying the disruption of PR-mediated paracrine signaling pathways.136,144 Currently, our knowledge about this paracrine to autocrine signaling switch or its involvement in tumorigenesis is limited.145 The signaling pathways that are affected by altered progesterone signaling during tumor growth are also not well defined,136 except for RANKL (receptor activator of nuclear factor kappa-B ligand) signaling, which was shown to be disrupted in the initial stages of progestin-sensitive mammary tumor growth in mouse models.146 In human ER/PR positive breast cancer cell lines, including T47D and MCF-7, RANKL is neither expressed nor stimulated by progesterone, however, it is upregulated in premenopausal women with breast cancer.146 Breast cancers patients with a high PR expression level display an enhanced level of RANKL expression in the tumor and adjacent normal tissue,147 and the RANKL expression level in epithelial compartments negatively correlates with tumor progression.148,149 These contradictory findings indicate that more studies are needed to understand the role of progesterone–RANKL signaling in human breast cancer.136
Overexpression of the human epidermal growth factor receptor 2 (HER2) occurs in about 15%−30% of breast cancers.150 This facilitates the activation of growth factor signaling pathways, which in turn triggers survival, proliferation, and invasion of breast cancer cells.115 Breast tumors with HER2 overexpression, referred to as HER2-positive breast cancers, are associated with aggressive tumor behavior, high recurrence rate, and reduced survival rate.150–152 HER2 is a member of the epidermal growth factor receptor (EGFR) family, which belongs to the super-family of the cell surface receptor tyrosine kinases Fig. 8B.116,117 EGFRs are activated by binding with numerous ligands, which leads to receptor dimerization and subsequent phosphorylation of the intracellular domain.153 This intracellular domain phosphorylation of receptors triggers downstream signaling pathways that result in cell proliferation and migration.153,154 Members of the EGFR family are EGFR/HER1, HER2, HER3, and HER4.155 The HER2 gene is positioned at chromosome 17, which encodes a 185-kDa transmembrane protein.156–158 The HER2 receptor’s extracellular domain remains in an active conformation, which can undergo a ligand-independent dimerization with other EGF receptors, of which the HER2/HER3 dimer combination is the most active and tumor promoting dimer. The ligand for HER2 has not yet been identified, and HER2 can be activated either by a ligand-mediated hetero-dimerization process with other receptors of the EGFR family, or by ligand-independent homo-dimerization, which occurs in the case of HER2-overexpressing tumors.159,160 Phosphorylated HER2, which arises from auto-phosphorylation of HER2 at Tyr1248, is the active form of the HER2 receptor, which is a requirement for downstream signaling.161,162 It has been observed that HER2 is present in the phosphorylated state in tumors of HER2-positive transgenic mice.163 However, there is disagreement in the literature as to the percentage of HER2-positive human breast cancer cases, which have been reported to lie within the large range of 10% - 80% of HER2 in the phosphorylated state.164
ER, PR and HER2 signaling pathways in breast cancer were shown to affect PC and GPC levels.79 ER and HER2 downregulate enzymes associated with the GPC pathway, including cPLA2 and lyso-PLA1.79,165 However, it is currently not known how ER, PR and/or HER2 signaling affect GPC levels and the GPC-regulating enzymes cPLA2, lyso-PLA1, GDPD5, GDPD6. Future studies will hopefully be able to address these unanswered questions.
Oncogenic RAS, PI3K/AKT, RALGDS signaling:
The phosphatidylinositol 3-kinase (PI3K) pathway is frequently activated in cancer cells due to mutations and epigenetic changes.166 RAL GTPase guanine nucleotide dissociation stimulator (RALGDS) is a well characterized RAS effector which induces the transformation of NIH3T3 fibroblasts.106 RALGDS along with PI3K participates in the control of cell proliferation, tumorigenesis, and survival.167 Under normal conditions, cellular functions are highly regulated by a complex network of signaling pathways, which controls proliferation, apoptosis, angiogenesis, metastasis, growth and the sensitivity of the cells toward growth signal.7,168 When this network of signaling pathways is disrupted, cells often become cancerous.7
RAS proteins are signaling proteins that, by activating critical signaling pathways, regulate proliferation.169 Many tumors express RAS proteins that are constantly activated by point mutations in the RAS genes.170,171 Activated RAS proteins drive transformation by deregulating proliferation and apoptosis, and by inducing the formation of new vasculature in the tumor.172 The RAS proteins belong to the superfamily of GTP-binding proteins, each family of which is associated with important cellular processes.173 The RAS family that regulates cell growth has three members that are activated by point mutations, i.e. HRAS, KRAS and NRAS.173 These three RAS proteins share ~85% of their amino acid sequence and function similarly.170,173 The KRAS protein is frequently expressed in most cancer cell types.174 RAS protein function requires several post-translational modifications.175 The post-translational modification of RAS starts with the addition of farnesylpyrophosphate to the cysteine residue of the carboxy-terminal tetrapeptide CAAX motif.176 In this motif, C is a cysteine, A is an aliphatic amino acid, i.e. leucine, isoleucine or valine, X is a terminal residue, i.e. methionine, serine, leucine, or glutamine.176 This is followed by addition of palmitate to one or two cysteine residues immediately upstream of the CAAX sequence.177 The AAX part of the CAAX sequence then undergoes Ras converting enzyme 1 (RCE1) mediated proteolysis, followed by methylesterification of the C-terminal prenylcysteine.177,178 As the next step, protein kinase C phosphorylates KRAS at serine-181.179 The GTP-bound active form of RAS binds and activates other effector proteins, thereby inducing cellular characteristics of transformed cells.172 The best studied effector protein of RAS is Rapidly Accelerated Fibrosarcoma (RAF), which is a member of the serine/threonine kinase family, which also includes serine/threonine-protein kinase-1 (c-RAF1), serine/threonine-protein kinase B-Raf (BRAF) and Serine/threonine-protein kinase A-Raf (ARAF).180 RAF activation by RAS starts a signaling cascade by phosphorylating and activating mitogen-activated protein kinase kinases 1 and 2 (MEK1 and MEK2), which in turn phosphorylate the mitogen-activated protein kinases (MAPKs) ERK1 and ERK2.170 The activation of ERK1 and ERK2 results in further activation of various transcription factors associated with the regulation of gene expression and cell proliferation.181
Phosphatidyl-inositol-3-kinases (PI3Ks) are members of the lipid kinase family, which phosphorylates the 3′-OH group of the inositol ring in inositol phospholipids.182 PI3K consists of a heterodimer comprising a catalytic and an adaptor/regulatory subunit.182 Binding of PI3K to phosphorylated tyrosine consensus residues of growth factor receptors results in the activation of its catalytic subunit, which leads to the formation of the second messenger phosphatidylinositol-3,4,5-triphosphate (PI3,4,5-P3) from the substrate phosphatidylinositol-4,4-bisphosphate (PI-4,5-P2).182 After that, PI3,4,5-P3 recruits a number of signaling proteins, including protein serine/threonine kinase-3′-phosphoinositide-dependent kinase 1 (PDK1) and AKT/protein kinase B (PKB), associated with cellular growth and survival.183 AKT kinase is a member of the AGC kinase, which was coined to define the subgroup of Ser/Thr protein kinases closely related to cAMP-dependent protein kinase or Protein kinase A (PKA), cGMP-dependent protein kinase (PKG), and protein kinase C (PKC), consisting of a PH domain, a catalytic kinase domain, and an extension containing a regulatory hydrophobic motif (HM).184
The PI3K/AKT pathway is crucial for cell survival under conditions of stress, including hypoxia, acidic pH, and substrate deprivation as frequently found the tumor microenvironment.185 Mammalian target of rapamycin (mTOR) is a serine/threonine kinase, which is universally expressed in mammalian cells, and which integrates the signals initiated by various stimuli to regulate protein synthesis and downstream signaling associated with cellular growth as well as metabolism.185 The activation of the PI3K/AKT/mTOR pathways due to genetic alterations has profound effects on regulating cell growth and survival resulting in increased cellular growth, metastatic capabilities and resistance to therapy.185 Thus, this complex pathway is an attractive targets for developing anticancer agents.185
Inhibition of PI3K with LY294002 is associated with a significant increase in GPC and a decrease in PC levels.186,187 Also, the inhibition of AKT causes a decrease in PC level, which is associated with a reduced CHKα expression and reduced activity emphasizing the association of PI3K/AKT in choline metabolism.188 However, it is currently not clear if and how RAS, PI3K/AKT, and mTOR signaling affect GPC levels and GPC-regulating enzymes, i.e. cPLA2, lyso-PLA1, GDPD5, GDPD6. Future studies should focus on providing these answers.
Transcription factors:
Carcinogenesis is characterized by a gradual accumulation of critical genetic and epigenetic changes, leading to initially precancerous lesions that gradually evolve into aggressive cancers.189 There are several oncogenic transcription factors, signal transducers, and transcription activators that are hyper-activated in cancer, including activator protein (AP)-1, nuclear factor (NF)-κB, and signal transducer and activator of transcription (STAT)-3 and STAT-5, respectively.189 Oncogenic transcription factors participate in the initiation and progression of cancer, as well as in the development of therapy resistance.189 There are also tumor-suppressing transcription factors that are under-activated in cancer, including p53 and retinoblastoma protein (pRb), however, not much information is available as to how to stimulate or stabilize them for the purpose of therapy.190 Transcription factors function by directly or indirectly binding to specific DNA sequences within gene regulatory regions.191,192 Extensive cross-talk among transcription factors and communication of transcription factors with target genes across different tissues and cellular contexts makes the regulatory transcription factor network highly complex.192
An important oncogenic transcription factor is AP-1, which is a dimeric protein with basic leucine zipper (bZIP), dimerization, and DNA binding domains.189 AP-1 is involved in various aspects of tumorigenesis, including enhanced proliferation, suppression of apoptosis, neoangiogenesis, and modulation of extracellular matrix (ECM) components.193 AP-1 is affected by HER2 signaling, making it a potential target for the treatment of HER2-positive breast cancer.194 Recent genomic studies suggest that different progenitor cells involved in the development of mammary glands are associated with specific transcription factor regulatory networks.195 These progenitor cells, when influenced by oncogenic events, give rise to different breast cancer subtypes, which require different treatment regimens and lead to different outcomes.195 The transcription factors GATA binding protein 3 (GATA-3) and fork-head box A1 (FOXA1) are examples of such transcription factors, which are involved in mammary morphogenesis in an ER-sensitive manner in normal development, and which are also overexpressed in luminal type tumors that are ER-positive and hence sensitive to hormonal therapy.189,196 Alterations in the ER/GATA-3/FOXA1 network enable breast cancer cells to acquire resistance to hormone therapy.197 Some transcription factors, including include Snail1, Snail2, Twist1, zinc finger E-box-binding homeobox 1 (ZEB1), ZEB2, and the nuclear factor of activated T-cells (NFAT) family,198,199 alter the EMT in carcinogenesis200 by down-regulating epithelial cadherin expression, which in turn disrupts tissue vasculature.200 STAT-3 and STAT-5 are transcription factors that modulate the tumor microenvironment and have been implicated in cancer.201,202 Another important transcription factor that is overexpressed in many cancers is HIF, which induces neoangiogenesis during tumor growth by driving up vascular endothelial growth factor (VEGF) expression, EMT, invasion, metastasis, and resistance to chemotherapy and radiation therapy.203,204 The transcriptional activity of HIF is also activated by hypoxia, which is commonly experienced by cancer cells in a growing tumor.203 The Notch transcription factor family is also involved in breast cancer by modifying the tumor microenvironment, inducing EMT, and enhancing neoangiogenesis.205
All of the above-mentioned transcription factors, which are associated with different cancer types, are vital in regulating gene expression in conjunction with other transcriptional regulators. A recent study showed that HIF1 is associated with choline metabolism in breast and prostate cancer, and that it activates CHKα by activating hypoxia response elements, thereby increasing the cellular PC and choline levels.206,207 Silencing of HIF1α and HIF2α were shown to reduce cellular GPC levels in breast cancer cells.208 It is currently not known how other transcription factors such as AP-1, GATA-3, FOXA1, Snail1/2, Twist1/2, ZEB1/2, and NFAT, STAT-3/−5, and Notch affect choline metabolism in cancer, and GPC levels in particular. Future studies should investigate the transcriptional regulation of GPC-modulating enzymes such as cPLA2, lyso-PLA1, GDPD5, GDPD6.
Interaction of the GPC metabolic pathway with other biochemical pathways
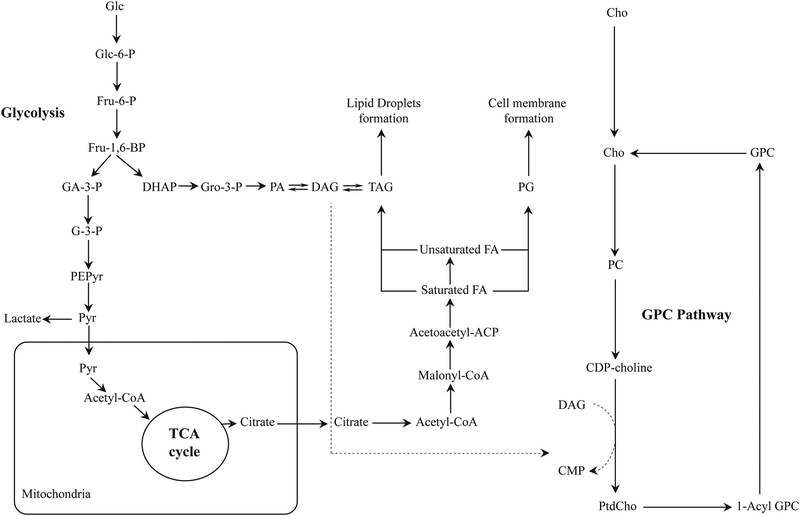
The GPC metabolic pathway interacts with multiple other biochemical pathways on several levels. In the following, we highlight two main pathways, i.e. glycolysis and triacylglycerol formation, that have a direct connection with the GPC pathway through the metabolite 1,2-diacylglycerol (DAG) as shown in Fig. 9. These three pathways are activated in cancer.
Figure 9:
Biochemical interactions between altered glycerophosphocholine (GPC), lipid and glucose metabolic pathways in cancer. Most cancer cells display increased de novo lipid synthesis, as well as activated glycolysis and GPC metabolism. Glycolysis, in which glucose is catabolized to pyruvate, provides various metabolites for lipid synthesis, including diacylglycerol (DAG). (Abbreviations: Acetoacetyl-ACP, acetoacetyl-acyl carrier protein; Cho, free choline, CDP-choline, cytidine diphosphate choline; CMP, cytidine monophosphate; DAG, diacylglycerol; DHAP, dihydroxyacetone phosphate; FA, fatty acid; Fru-6-P, fructose-6-phosphate; Fru-1,6-BP, fructose-1,6-bisphosphate; Glc, glucose; Glc-6-P, glucose-6-phosphate; GPC, glycerophosphocholine; GA-3-P, glyceraldehyde-3-phosphate; G-3-P, 3-phosphoglycerate; Gro-3-P, glycerol-3-phosphate; PEPyr, phosphoenolpyruvate; PtdCho, phosphotidylcholine; Pyr, pyruvate; PA, phosphatidate; PG, phosphoglyceride)
Glycolysis:
Glucose metabolism starts with glycolysis, which comprises a series of enzymatic degradation steps, in which glucose is catabolized to pyruvate. Glycolysis is activated in cancers by the Warburg effect, which favors glycolysis over oxidative phosphorylation under normoxic conditions. Various glycolysis intermediates can participate in the pentose phosphate pathway (PPP), which is a parallel pathway to glycolysis and produces NADPH and pentoses (5-carbon sugars) as well as ribose 5-phosphate or lead to lipid synthesis. In cancerous cell, pyruvate can either be converted to lactate, or it can be transported to the mitochondria where it participates in the tricarboxylic cycle (TCA) and is subsequently converted to fatty-acyl-coenzyme A (CoA) by fatty acid synthase. Glyceraldehyde-3-phosphate, a conversion product of fructose-1,6-bisphosphate in glycolysis, is converted to phosphatidate, which in turn is reversibly converted to DAG or TAG, as discussed in detail in the next section. DAG formed in this pathway is subsequently incorporated in the de novo synthesis of PtdCho (Fig. 9). There have not been many reports emphasizing the connection between glycolysis and the GPC metabolic pathway, and investigating this link in cancer may provide important insights.
Triacylglyceride formation in lipid droplets:
Lipid droplets have been known to influence various processes associated with tumorigenesis and in their aggressiveness. Lipid droplet formation and lipid droplet abundance have been positively correlated with degree of aggressiveness in breast cancer cell lines, from non-malignant MCF-10A to highly malignant MDA-MB-231 cells. Increased fatty acid and phospholipid synthesis, which is essential for increased proliferation of cancer cells, are the primary sources of higher lipid content in malignant cells. Additionally, breast cancer cells import free fatty acids to use them either as a substrate for energy production by β-oxidation or for storing them in the form of lipid droplets, which enables them to avoid nutrient stress due to enhanced cell proliferation.209 These lipid droplets are spherically shaped organelles with a size range of a few nanometers to hundreds of micrometers. Lipid droplets are composed of neutral lipids, including cholesterol esters, retinol esters, and triacylglycerides (TAGs) with saturated or unsaturated acyl chains, which are surrounded by a layer of phospholipids, i.e. primarily phosphatidylcholine, and different proteins. TAGs contain three fatty acid chains bound to a glycerol backbone and are synthesized by a complex pathway. This pathway requires activation of saturated or unsaturated fatty acids to fatty acyl-coenzyme A esters through acyl-CoA synthetase activity and phosphorylation of glycerol by either glycerol kinase, or cytosolic synthesis of glycerol-3-phosphate from di-hydroxy-acetone phosphate. Once fatty acyl-CoA is formed, it acylates glycerol-3-phosphate to produce 1-acylglycerol-3-phosphate, followed by another acyl-CoA-mediated acylation step to produce 1,2-diacylglycerol phosphate. 1,2-diacylglycerol phosphate is dephosphorylated to produce DAG and then is esterified to convert DAG into TAG.209 Following synthesis of TAG, de novo formation of lipid droplets takes place between two leaflets of the endoplasmic reticulum. When lipid droplets are broken down by lipolysis in non-adipose tissues, they release fatty acid from TAGs and are involved in various processes, including fatty acid oxidation, cell growth, cellular membrane synthesis, and synthesis of various lipid mediators.209 Further studies are needed to investigate the interaction of TAG and lipid droplets with the GPC pathway.
Magnetic resonance spectroscopy (MRS) techniques for detecting GPC
Significant technology advances in MRS, which include higher field strengths, new coil designs, and new pulse sequences,210,211 have enabled the detection and quantification of cellular and tumor GPC levels, among many other metabolites and amino acids.23,212,213 Recent advances in MR instrumentation, MR methodology, and development of novel contrast agents have tremendously strengthened the field of MRI and MRS.214,215 Advanced MRS techniques enable the observation of molecular, cellular, and metabolic processes in vivo, which has deepened our knowledge of cancer biology, therapeutic targets, and treatment strategies.210,215 Noninvasive MRS techniques are being developed as powerful imaging tools for preclinical studies as well as clinical applications.19,216 In the following, we discuss MR applications that are available for visualizing and quantifying catabolic and anabolic processes in cancer metabolism, with a focus on the detection of GPC.
Magnetic resonance spectroscopy is a phenomenon that occurs owing to the interaction of nuclear spins with a strong external magnetic field ranging between 9.4 Tesla and 18.8 Tesla. Atomic nuclei have an intrinsic property referred to as spin, which arises from the number of neutrons and protons present in the nucleus. Most commonly used nuclei in MRS are nuclei with a spin of 1/2, which are 1H, 13C, and 31P. The gyromagnetic ratio of nuclei, their concentration in the area of interest, their longitudinal relaxation or spin-lattice relaxation time T1, and their transverse relaxation or spin-spin relaxation time T2, are four major factors contributing to their MRS signal intensity.217 When these nuclei are placed in the presence of an external magnetic field, they align themselves either in the same direction or opposite to it according to their energy states. When a radiofrequency pulse is applied, excited nuclei move to the higher energy state, and when the system returns to its equilibrium state, a free induction decay of radiofrequency of spins returning to equilibrium can be detected.218 Each chemical structure possesses a particular electronic environment, which causes the nuclei to resonate at slightly different frequencies. These frequencies are called chemical shifts and are denoted as the dimensionless units parts per million (ppm) in the MRS spectrum.219 The chemical shift in MRS enables the identification of metabolites in biological samples and tissues by differentiating among various chemical environments.220 It is also possible to use MRS to follow metabolites or other water-soluble molecules in biochemical reactions and biological pathways.221
1H MRS is frequently used to characterize metabolic changes in cancerous and normal tissues.222–228 MRS detection of 31P, 13C, 19F nuclei is also used for monitoring metabolic alterations, bioenergetics and metabolic fluxes in cancer.229–236 Due to the low natural abundance of 13C of 1.1 %, it has been used to isotopically label metabolic substrates which are then administered orally or by injection, and which can then be followed in the animal model or patient of interest by MRS.237 Magnetic resonance spectroscopic imaging (MRSI) spatially maps endogenous metabolites to reveal heterogeneous distributions of these metabolites in cancer tissue.215,238–241 In vivo MRS and MRSI have shown promise as diagnostic tools in clinical studies, and may improve the specificity of detecting and managing cancers in patients.14,242
In addition to MRS applications in patients or animals in vivo, MRS has been utilized for studying metabolites in cell and tissue extracts, which is possible at high spectral resolution.243 It is also possible to analyze intact tissue without any tissue extraction or processing, which can be achieved by spinning the intact sample on its axis at an angle of 54.7°, also known as the magic angle, at a high speed.244,245 Magic angle spinning averages out anisotropic interactions, which would otherwise create broad peaks due to the decreased mobility of metabolites in intact tissue.245 With magic angle spinning and the use of appropriate pulse sequences, intact tissue spectra are comparable to spectra obtained from extracted tissue.246 This technology is known as high-resolution magic angle spinning (HR MAS) MRS and is typically performed at 4° Celsius to prevent tissue degradation during the HR MAS MRS measurement247,248 Being a non-destructive technique, HR MAS MRS can be used in conjunction with other techniques, where the same sample can subsequently be used for further analysis. For example, following HR MAS MRS of an intact tissue sample, histopathology, cytology, or immunohistochemistry can be performed with the same sample.249–251
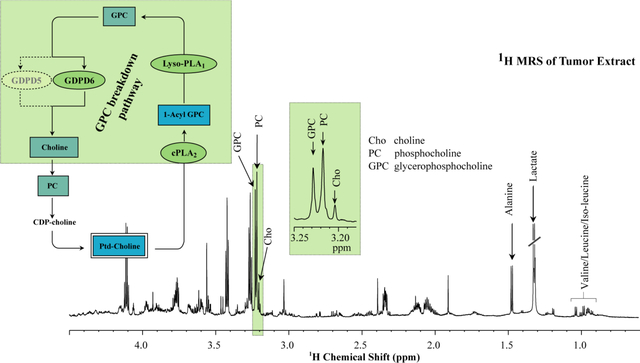
1H MRS detection of GPC:
1H MRS is the most frequently used spectroscopic imaging technique in the clinical setting because of its high sensitivity and availability. The peak emerging from water is the most intense peak in 1H MRS, but water suppression techniques have made it possible to detect some metabolite peaks at much lower abundance than the water signal.32,252 Some relatively fatty tissues, i.e. the breast, may also require fat suppression.253 1H MRS detects a signal at 3.2 ppm, which arises from the nine magnetically equivalent protons of the three methyl groups of Cho, PC, GPC, as well as betaine, and taurine in certain tissues, collectively referred to as tCho.16 In vivo 1H MRS at high magnetic field strength of 7T and higher in brain tumors has allowed for a partial spectral separation of GPC and other overlapping metabolites in the tCho signal.254 However, most cancers in other organs only allow for the detection of the unresolved tCho signal due to motion, inhomogeneity, and intense resonances from lipid and water.255
31P MRS detection of GPC:
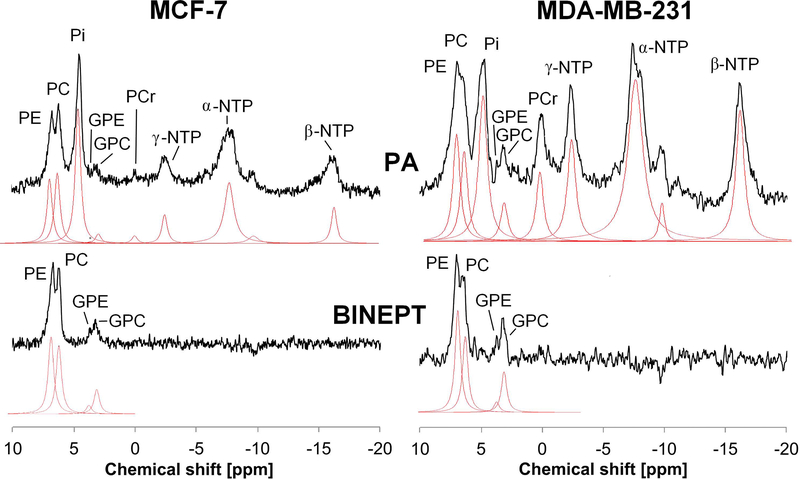
31P MRS detects the second most abundant nucleus after 1H, and has been frequently used in the study of choline phospholipid metabolism in animal models of cancer as well as cancer patients.216,256,257 31P MRS is able to detect 31P containing metabolites and has the advantage over 1H MRS that the presence of high concentrations of water and fat has no effect on 31P signal acquisition, additionally requiring a less homogeneous magnetic field. 31P MRS has an inherently low signal to noise ratio (SNR) resulting in relatively poor spatial resolution and significantly longer acquisition times, which requires dedicated coils and probe-heads.210 Advancements in hardware engineering are leading to improved SNR with reduced scan time.210 Improved pulse programming that allows the use of 1H decoupled 31P, 1H-31P cross‐polarization or 1H-31P polarization transfer experiments instead of direct detection of 31P is further increasing the spatial resolution258 (Fig. 10). Theoretically, a 2.4-fold signal enhancement (γ1H/γ31P) can be achieved by using cross polarization and polarization transfer techniques. In techniques such as refocused insensitive nuclei enhanced by polarization transfer (RINEPT),259 magnetization/polarization of excited 1H is transferred to the 31P via J-coupling (JPH) (shown in Fig 3C) during the echo-time TE1H, which increases the signal to noise ratio along with removing all signals without 1H-31P coupling. In the adiabatic version of the refocused insensitive nuclei enhanced by polarization transfer (BINEPT) sequence, TE1H is relatively long because of the small JPH coupling, which provides higher signal to noise ratio and a flat baseline, resulting in better detection of PC, GPC, PE and GPE.258 By using heteronuclear editing techniques during spectral acquisition, it is possible to further increase the sensitivity and detect PC, GPC, PE and GPE. The method is called proton observed phosphorus editing (POPE) where the optimal editing is acquired at 31P J-coupling evolution time of 1/J.260
Figure 10:
Comparison of direct detection pulse-acquire (PA, top) and polarization transfer BINEPT (bottom) 31P MR spectra of representative MCF-7 (left) and MDA-MB-231 (right) tumor xenografts. Lorentzian lines fitted using the software jMRUI (http://www.jmrui.eu/) are shown below each respective MR spectrum. The PA spectrum shows resonances from all phosphorylated metabolites, while the BINEPT spectrum shows only signals from metabolites containing 1H-31P bonds such as phosphoethanolamine (PE), phosphocholine (PC), glycerophosphoethanolamine (GPE), and glycerophosphocholine (GPC). Additional signals in the PA spectra are β-nucleoside triphosphate (NTP), nicotinamide adenine diphosphate (NAD), and diphosphodiesters (DPDE). α-NTP represents a combination of overlapping signal from α-NTP and α-nucleoside diphosphate (α-NDP). Similarly, γ-NTP is an overlapping signal from γ-NTP and β-NDP. (Adapted from Wijnen et al.258).
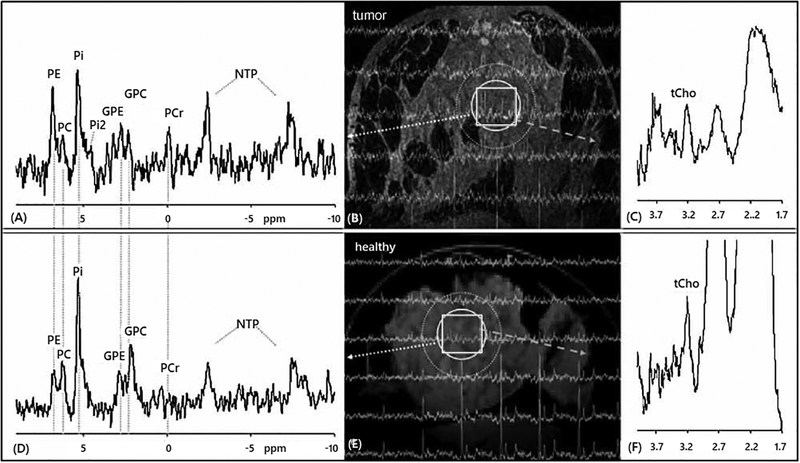
31P MRS detects the phosphomonoesters PC and phosphoethanolamine (PE) at 3.9 ppm and 4.5 ppm, and the phosphodiesters GPC and glycerophosphoethanolamine (GPE) at 0.5 ppm and −1.2 ppm, respectively.211 At a high magnetic field strength of 7T, 31P MRS of human breast tumors reliably detected GPC along with GPE, PC, and PE as shown in Fig. 11.261 A 31P MRS study at 3T demonstrated that PC, GPC, PE and GPE were detected individually when evaluating optical pathway gliomas in children.262 31P MRS has also been use to differentiate between Grade II and IV astrocytomas in patients where elevated GPC was found in Grade II astrocytoma whereas PC was increased in Grade IV Astrocytoma in an ex vivo study.263 31P MRS is also able to detect and quantify GPC, GPE, PC, and PE in various animal models of cancer, i.e. breast cancer xenograft models,79 colon cancer xenograft model,264,265 human diffuse large B-cell lymphoma xenograft model,266 human tumor kidney xenograft,267 and murine fibrosarcoma.268
Figure 11:
Localized 31P and 1H MR spectra were recorded from the breast of a breast cancer patient and a healthy control at 7 T in vivo. One slice of 31P MR spectra is mapped onto the corresponding transverse MR image obtained from the tumor-containing breast (A, B, C) and the breast of the healthy volunteer (D, E, F). Altered levels of phosphoethanolamine (PE), phosphocholine (PC) and glycerophosphocholine (GPC) are observed in the 31P MR spectrum in tumor area (A) compared with healthy control (D) (indicated by dotted circle), while 1H MR spectra of the same region (indicated by square) contained total choline (tCho) in the tumor (C) as compared with the healthy subject (F). In addition, the chemical shift of the major inorganic phosphate (Pi) resonance in the 31P MR spectra was 5.3 for both subjects, from which the pH was calculated as pH 7.5, while a second Pi resonance (Pi2) was observed in the tumor tissue corresponding to pH 6.9. Also note the absence of phosphocreatine (PCr) in the breast of the healthy subject, reflecting negligible partial volume contributions from the surrounding muscle (A). GPE, glycerophosphoethanolamine; NTP, adenosine and other nucleoside triphosphates (Adapted from Klomp et al.261).
31P MRS was employed for determining the effects of 17-Allylamino,17-demethoxygeldanamycin (17AAG), an anticancer drug inhibiting heat shock protein 90 (Hsp90), on three different colon cancer cell lines (HCT116, HT29, and SW620) and in HT29 xenografts.264 Significant increases in PC and GPC levels were observed in all cell and tumor extract following 17AAG treatment, and a significant increase in the phosphomonoester/phosphodiester ratio was also observed.264 31P MRS was also used to detect the response to combination chemotherapy in vivo in a human diffuse large B-cell lymphoma (DLCL2) xenograft model.266 The combination chemotherapy consisted of cyclophosphamide, doxorubicin, oncovin, prednisone, and bryostatin 1.266 A significant decrease in the tCho level was observed after one chemotherapy cycle and a significant decrease in the phosphomonoester to β-nucleoside triphosphate ratio was detected after the second round of therapy, demonstrating the feasibility of 31P MRS to detect chemotherapy treatment response.266 In vitro 1H and 31P HR MRS of perchloric acid extract of human prostatic tissue from benign prostatic hyperplasia and prostatic adenocarcinoma showed that the PC/total creatine (Cr), tCho/total Cr, PE/total phosphate, PC/total phosphate and GPE/total phosphate ratios were significantly increased in cancer tissue samples compared to benign prostatic hyperplasia.269
1H and 31P HR MAS MRS detection of GPC:
HR MAS MRS studies have been invaluable for the study of breast cancer tissues.31,270,271 1H HR MAS MRS is an important technique for understanding the metabolic differences between malignant and benign breast tissues. A study conducted on 76 microscopy-confirmed cancerous and 9 non-involved breast tissue samples using 1H HR MAS MRS for quantifying GPC, PC and choline revealed a high GPC/PC ratio in non-involved or benign tissues as compared to a significantly decreased GPC/PC and GPC/Cho ratio in cancerous tissues.250 In addition, 1H HR MAS MRS has also been used to predict long-term disease outcome and prognosis in breast cancer patients based on their tissue metabolic profiles. In such a study, 1H HR MAS MRS followed by multivariate principle component analysis (PCA) was performed on 29 surgically removed samples of palpable breast lesions.271 Based on the identified metabolites, a correlation was established between a given patient’s prognosis and their health status at 5 years post-surgery.271 Increased levels of GPC, taurine and creatine, combined with reduced PC and glycine levels were detected in breast tumor tissues of patients that reported back in good health at 5 years post-surgery.271
1H HR MAS MRS has also been employed for characterizing the metabolic signatures of human lung cancer.272 In this study, paired samples of tumor and adjacent tissue from 12 lung tumors were analyzed by 1H HR MAS MRS and 50 compounds were identified.272 GPC and PC were elevated in this study in lung cancer as compared to adjacent normal.272 This study shows the potential of HR MAS MRS for characterizing the metabolic phenotype, including detection of GPC, in lung cancer. In a different study, 1H HR MAS MRS obtained from 24 lung tumor and 24 control samples showed elevated levels of GPC, PC, lactate, and lipid in cancerous tissue while non-cancerous tissues contained elevated amounts of acetate, methionine and glutamate.273
1H HR MAS MRS studies on human brain tumors have been used for tumor metabolite quantification and tumor biomarker identification. Overall, 37 metabolites were identified, which accounted for the differences in astrocytoma grade II, grade III gliomas, glioblastomas, metastases, meningiomas and lymphomas.274 This study also emphasized the importance of GPC as a biomarker for tumor grade in brain tumors as the concentration of GPC decreased with increasing tumor grade while PC increased with increasing tumor grade.274 In the same study, GPE was detected in various adult brain tumors.274 In another study, 1H HR MAS MRS was used to differentiate between grade II and IV astrocytomas based on their spectral profile, which was analyzed by PCA.263 Grade II tumors contained increased levels of GPC and myo-inositol, whereas grade IV tumors had increased PC, glycine, and lipid levels.263 These studies emphasize the importance of distinguishing GPC from PC, which are difficult to resolve with in vivo MRS. In a comparative study in transgenic mice growing medulloblastoma, the authors acquired in vivo spectra at 7T and ex vivo 1H HR MAS spectra at 11.7T, and observed an increase in tCho due to increased PC only, while GPC remained stable, and free choline was reduced.275 Increased GPC levels have been used for differentiating among ependymoma, medulloblastoma, and pilocytic astrocytoma in pediatric brain tumors.276 Medulloblastoma were identified by increased levels of GPC, PC, choline, and taurine, while ependymoma were characterized by a prominent myo-inositol signal, and pilocytic astrocytomas were characterized by an increased fatty acids signal.276 In a similar study, GPC and PC levels were able to differentiate between non-enhancing grade II and grade III astrocytomas and their association with cell proliferation and angiogenesis.277 In this study, 41 patient biopsy samples (16 grade II and 25 grade III) from 24 tumors were subjected to 1H HR MAS MRS and immunohistochemistry.277 The results showed that GPC was the predominant peak in grade II tumors, while increased PC was observed in grade III, and the PC/GPC ratio was less than 1 in all cases.277 The GPC level increased with an increasing level of the cell proliferation marker Ki-67, indicating its association with tumor cell proliferation.277 Metastases to the brain have also been studied using 1H HR MAS MRS. Biopsies from human brain metastases (n=49) from different origins were investigated using 1H HR MAS MRS.213 Multivariate statistical analysis showed that metastases from malignant melanomas clustered together, while metastases from lung carcinomas overlapped with brain metastasis from other origins due to their heterogeneous nature.213 A significantly increased GPC level was observed in brain metastases originating from malignant melanomas, while brain metastases from other cancer origins showed a heterogeneous metabolic pattern.213 HR MAS MRS has also been applied in studies for assessing tumor treatment response. 1H HR MAS was used in a C6 glioma mouse model, where the effects of treatment with a glycoside and its thioglycoside analogue were evaluated.278 1H HR MAS MRS demonstrated that C6 cells treated with a higher concentration of thioglycoside resulted in significant increases in choline, PC and PC/GPC ratio.278 In the case of intact tissues, a higher concentration of thioglycoside significantly reduced tumor size and, consistent with the cell culture study, increased choline, PC and PC/GPC ratio.278 These results demonstrated that glycolipid derivatives can have significant effects on choline phospholipid metabolism, and result in cell death.278
HR MAS MRS has also been used to evaluate the occurrence and aggressiveness of prostate cancer. In a study performed with 140 tissue biopsy samples from 40 prostate cancer patients, Stenman et al observed that prostatic tissues samples with a high malignant cell fraction contained higher levels of GPC, PC/creatine, myo-inositol/scyllo-inositol, Cho/creatine, as well as higher levels of the cell proliferation marker Ki-67.212 In another study, GPC, PC, PE, and GPE were increased in cancerous human prostate tissue as compared to benign prostate tissue when quantified with a 2D 1H HR MAS total correlation spectroscopy (TOCSY) experiment.279 In the same study, the PC/GPC ratio was also significantly higher in cancerous tissue as compared to benign tissue.279 1H HR MAS MRS can also be used to study the metabolic profiles of cancer cell lines by filling the HR MAS rotor tube with cell pellets and spinning them at a lower frequency of ~2–5 kHz at 4 °C. This was applied in intact PC3 prostate cancer cells, MCF-7 breast cancer cells, and HepG2 hepatocarcinoma cells, which revealed increased levels of PC, lactate, and fatty acids in PC3 and MCF-7 cells, as compared to HepG2 cells.280 1H HR MAS MRS has been used to investigate the metabolic response of the gonadotrophin-releasing hormone blocker drug Degarelix in benign and cancerous prostate tissue samples.281 Absolute concentrations of various metabolites, including lactate, glutamine, glutamate, citrate, tCho, total creatine, taurine, myo-inositol and polyamine were measured, and PC and GPC concentrations were significantly higher in prostate cancer tissue as compare to benign tissue.281 Treatment with Degarelix resulted in a significant decrease in the levels of lactate and tCho in prostate cancer samples.281
1H HR MAS MRS has also been applied to differentiating malignant and non-malignant cervical tissue samples. Increased levels of GPC and PC have been detected in malignant versus non-malignant cervical tissue samples.282–284 Biopsy samples obtained prior to and during radiation therapy showed a positive correlation between a high tumor cell fraction as well as cell density and increased levels of PC, GPC, lactate and creatine and decreased levels of choline, glucose, and myo-inositol.285
The field of 1H and 31P HR MAS MRS is undergoing constant innovation in methodological development, with innovations occurring in acquisition, spectral processing, and metabolite quantification. A novel approach to HR MAS MRS based metabolite quantification is the use of a digital synthetic signal referred to as electronic reference to in vivo concentrations (ERETIC) signal.286 Using a digital synthetic signal as concentration reference is advantageous in cases where the addition of a reference compound may cause pH variations across samples, or where it can interact with tissue samples.287 In such a study, using a 1H, 31P, homo- and heteronuclear correlation (1H-1H and 1H-31P, respectively) HR MAS experiment, various metabolites were identified and quantified in 33 human brain tumor biopsy samples with the help of an ERETIC signal as a concentration reference.287 In this study, various metabolites including GPC, PC, choline, lactate, alanine and glutamine were identified by 1H HR MAS MRS, and tissue pH was calculated from the pH-dependent chemical shift variation in 31P HR MAS MR spectra.287 The peak assignments were confirmed with the help of 1H-1H and 1H-31P correlation spectra.287
Chemical exchange saturation transfer MRI detection of GPC:
Chemical Exchange Saturation Transfer (CEST) is an MRI contrast approach whereby rapidly exchangeable protons from hydroxyl (-OH), amine (NH2), or amide (-NH) groups of solutes are selectively saturated and detected indirectly via the attenuation of the water signal of their aqueous environment.288 Continuous transfer of protons between solute and water molecules amplifies signals emitted by these solutes; thus, CEST provides an effective means of monitoring concentrations of metabolites present at millimolar concentrations with high sensitivity.289 It is possible to detect the −OHs of choline metabolites, i.e. GPC and choline by CEST.289 Strong CEST effects from choline, for example, have been detected at 3T and 7T. At 3T in particular, these effects were identifiable from free choline without overlap from other compounds, such as glutamic and aspartic acid.290 While CEST has been used sparingly to detect specific choline metabolites, a recent report describes the implementation of CEST-MRI to detect metabolites, including GPC, in human breast cancer cell lines.289 Strong CEST contrast (MTRasym > 0.2) was generated by the−OHs of GPC at Δω = 1.0 ppm in phantom solutions of amino acids and metabolites. In cell extracts, trends in CEST-MRI contrast, which decreased in aggressive cell lines, were consistent with decreases in metabolites, including GPC, in these cell lines.289 Moreover, doxorubicin-treated cells, which contained increased GPC levels, showed a consistent increase in CEST-MRI contrast.289 These data suggest a potential future role for CEST-MRI in the detection of GPC to characterize tumor aggressiveness and monitor treatment response. Most studies on breast cancer using CEST MRI are preclinical studies, and only few clinical studies have been reported thus far. One of the biggest challenges in patient studies is the presence of strong lipid signal in breast CEST MRI, which affects the Z-spectrum. With use of an appropriate echo-time, it is possible to detect an improved CEST MRI signal.291,292 The CEST‐Dixon method has shown potential for CEST MRI in the breast at 3 T.291,292 Decomposition of water and fat resulted in homogenous fat removal from water‐only images and provided improved CEST signal.291,292
Hyperpolarized 13C MR detection of GPC:
The use of dynamic nuclear polarization (DNP) to achieve hyperpolarization in magnetic resonance imaging has enabled up to 10,000-fold improvements in sensitivity for imaging 13C-labeled substrates293 Hyperpolarized 13C MR enables the noninvasive detection and real-time monitoring of dynamic biological phenomena, including metabolism.294 This approach benefits from low background due to the low natural abundance of 13C.295 While choline metabolites are most commonly monitored via 1H MRS, 13C MRS based studies have been used to monitor choline-acetylcholine interconversion in rat brains296 and to trace the fates of choline, PC, and GPC in mammary epithelial and breast cancer cell lines.297 However, due to the low physiological concentrations of these metabolites and the relative insensitivity of 13C MRS approaches, such studies are often impractical in in vivo applications.295 Therefore, because of its ability to provide an enhanced SNR, hyperpolarized 13C MR may be of interest as a means of detecting choline and choline metabolites.42,295 This approach has not yet been used in studies of GPC; however, hyperpolarized 13C analogs of choline have been synthesized295 and used to detect the metabolism of choline to acetylcholine and to trace distributions of free choline in rat kidneys,298 suggesting that hyperpolarized 13C MR offers promise as a means of detecting GPC and monitoring its evolution. Notably, small chemical shift changes between choline and choline-derived metabolites299 and short carbon T1 relaxation times due to dipolar interactions within choline molecules represent hindrances to the use of hyperpolarized 13C in studies of choline metabolites.300 Indeed, hyperpolarized 13C probes require longer T1 relaxation times than 13C approaches at thermal equilibrium.300 To this end, deuterated choline probes have been demonstrated to show significantly increased T1.295 Deuterated choline probes are stable-isotope labeled choline analogs, i.e. [1,1,2,2-D(4),2-(13)C]choline chloride, in which with all 1H were substituted with deuterium, which increases the spin lattice relaxation time T1 to 34s at 11.4T magnetic field strength.295 This may thus, in the future, facilitate more effective visualization of labeled choline molecules as well as their chemically downstream metabolites, such as PC and GPC.
Detection of GPC as a biomarker of therapy response
Currently, diagnostic radiological imaging exams using MRI, computed X-ray tomography (CT), and positron emission therapy (PET) plus biopsy-based pathological assessment of disease stage and tumor subtype are the most important clinical exams on which an appropriate treatment plan for a given cancer patient is based. Staging is based on the international tumor classification system TNM, which gives the tumor size in cm (T), number of cancer-containing, tumor-draining lymph nodes (N), and number of distant metastatic nodules (M).301–304 The treatment plan for a given cancer patient depends upon the patient’s overall health status, tumor size, tumor stage and subtype, hormone receptor status for breast305 and prostate cancers, and specific mutations. In case of localized, operable disease, surgical removal of the tumor is often the best, and in many cases curative, option. Depending upon the TNM stage, grade, and score of cancer, patients are frequently also treated with systemic chemotherapy, hormone therapy (hormone receptor positive breast and prostate cancers), and targeted therapy, or any combination thereof, along with surgical removal of the tumor and/or local radiation therapy.33,281 Chemotherapy and radiation therapy affect both healthy and cancer cells and, hence, frequently result in side effects. Chemotherapy, hormone therapy, and targeted therapy can be given as adjuvant therapy post-surgery and sometimes as neoadjuvant therapy prior to surgery to shrink tumor size pre-surgically to improve surgical outcome, which is for example often performed in locally advanced breast cancer.306 Treatment options for metastatic cancers are limited to chemotherapies, hormone therapies, and targeted therapies. Immunotherapy or biotherapy, treating cancer by improving the immune system to fight it, has an advantage over chemotherapy and radiotherapy as it affects the tumor cells specifically without causing any damage to non-cancerous tissues.307 With these different treatment options and different clinically approved agents available within each treatment category, it is important to assess treatment response throughout the entire treatment. Currently, treatment response assessment is based on Response Evaluation Criteria in Solid Tumors (RECIST)308 and the guidelines of the World Health Organization (WHO).309 These guidelines help to assess the treatment response based on tumor size changes measured by MRI, which are typically only detected at a relative late point in time, i.e. after 2–3 months and several cycles of treatment. Response assessment at 2–3 months into the selected first-line therapy option is quite late for switching to a different therapy in the case of non-responders, who will already have undergone several rounds of treatment, and may have unnecessarily suffered from toxicities and side effects, by the time of the first RECIST assessment. Several studies have shown that 1H MRS can predict the treatment response early on based on the reduction of the tCho signal in responders and increase of the tCho signal in non-responders.33,310–312 Alternatively, 31P MRS can be used to detect the PME to PDE ratio as a marker of tumor response. GPC is a part of the tCho signal and a part of the PME signal.
Response to chemotherapy:
Neoadjuvant chemotherapy for advanced breast cancer has shown an improvement in overall survival of patients,313 and is typically combined with breast‐conserving surgery along with in vivo assessment of response to chemotherapy.314 1H MRS assessment of the tCho signal in breast cancer patients, which was achieved with water and fat suppression sequences, measured prior to the start of chemotherapy and after the 3 cycle of chemotherapy, has shown potential for predicting the response to chemotherapy as shown in Fig. 12.315 A recent study showed the use of single-voxel 1H MRS to predict the response of chemotherapy treatment at 24–72 hours after administration of the first cycle of chemotherapy.316 In this study, the measured tCho levels in the tumor-containing voxels were used to differentiate patients who responded to treatment.316 This study also emphasized the importance of early assessment of response to anthracyclines in order to facilitate clinical decision‐making for further treatment.316
Figure 12:
MRS detection of tCho as an early treatment response marker to neoadjuvant chemotherapy in a responding breast cancer patient. (A) Pre-therapy T2-weighted sagittal fat suppressed image of a locally advanced breast cancer patient overlaid with the magnetic resonance spectroscopic imaging (MRSI) grid. (B) Spectrum obtained from a voxel shown in (A) with tCho signal. (C) Post-therapy MR image of the same patient after 3 cycles of neoadjuvent chemotherapy. (D) Spectrum obtained from a voxel highlighted in (C) that showed no tCho signal. (Adapted from Danishad et al315).
Preclinical evaluation of chemotherapeutic agents on cancer cells and in xenograft model obtained from patients plays critical role for further improvement of these agents. Ovarian cancer cells obtained from peritoneal effusion from 8 patients were cultured and treated with 3 anti-mitotic drugs (paclitaxel, cisplatin and carboplatin) and were subjected to 31P MR spectroscopy. A significant decrease in GPC, GPE and uridine diphospho-sugar (UDPS) was observed post-treatment317 emphasizing the metabolic differences in cancer cells after chemotherapeutic intervention. Similarly, breast cancer xenograft models representing basal-like and luminal-like breast cancer have been used for studying the treatment response to the dual PI3K/mTOR inhibitor drug called BEZ235 in tissue samples obtained from these xenografts. 31P HR MAS MRS spectra of intact tumor showed a significant increase in PC and GPC levels, and a significant decrease in PE in BEZ235 treated basal-like xenograft model when compared to the control group, while no significant metabolic changes were observed in the luminal-like xenograft model.318
Response to hormone and targeted therapy:
With our increasing knowledge of the molecular subtypes of various cancers, along with our improved understanding of the associated signaling pathways and genomic alterations in cancer, a variety of molecularly targeted therapeutic agents have been developed for the treatment of different cancers. There are three kinds of targeted therapies, which include the hormone receptor antagonists, monoclonal antibodies, and inhibitors of kinases.319 As estrogen and ERs are the determining factors for continued growth and progression of ER-positive breast cancer, targeting estrogen to inhibit the estrogen signaling pathway in ER-positive breast cancer has been used for many years.320 The use of the ER modulator tamoxifen has improved the survival rate of ER-positive breast cancer patients with early and advanced breast cancer significantly.320 Aromatase inhibitors or inactivators (AI), which are used for blocking the biosynthetic pathway of androgens, have further improved the survival rates.321 The AIs inhibit the production of estrogen from androgens via suppressing the activity of enzyme aromatase either by binding with aromatase reversibly or irreversibly.321
GPC has shown potential as a response marker for targeted cancer treatments. A recent study evaluated the effects of bevacizumab, doxorubicin or a combination of these two drugs on two different xenograft models representing luminal-like and basal-like breast cancer, showing significant differences in metabolic and transcriptomic profiles for the two models.322 The combination treatment of bevacizumab and doxorubicin inhibited tumor growth in basal-like tumors, while no effect was observed in luminal-like tumors.322 GPC levels were overall lower in the basal-like tumor model and higher in the luminal-like tumor model.322 In luminal-like tumors, GPC was significantly increased in doxorubicin and bevacizumab combination therapy as compared to bevacizumab mono-therapy.322 A similar study was performed in which PC, GPC, PE and GPE were evaluated in basal-like and luminal-like xenograft models.318 These xenograft models were treated with the dual PI3K/mTOR inhibitor BEZ235.318 The treatment response was observed using 31P HR MAS MRS, demonstrating that the basal-like xenograft model showed an increase in PC and GPC levels when treated with BEZ235.318
Resistance to hormone therapy is a major problem in treating ER+ breast cancer patients. Several studies in ER+ breast cancer cell lines, including MCF-7, MDA-MB-231, and T47D cells, as well as studies in animal models of breast cancer showed that the phosphorylation of ERs, overexpression of steroid receptor coactivator (SRC), and an enhanced activation of the HER/MAPK pathway are mechanisms responsible for hormone resistance.323
Advancements in our knowledge of activated oncogenic pathways in cancer have led to the development of small molecule inhibitors for various enzymes of oncogenic pathways. These targeted inhibitors are clinically used to treat cancer patients today, and they are improving the life span of these patients. However, tumors often recur because resistance to targeted therapy frequently develops. These resistances evolve in cancers due to additional mutations in oncogenes and by activation of critical signaling pathways. Resistance development by mutation is common in the case of tyrosine kinase inhibitors, as evolving mutations in the active site of the tyrosine kinase domain render the active site unavailable to the inhibitor. An example of this phenomenon can be found in the ABL kinase domain as it becomes resistant to the ABL inhibitor imatinib.324 As resistance to targeted therapy develops, RTK or MAPK signaling pathways are often reactivated due to alterations in upstream or downstream signaling steps.324
Response to radiation therapy:
Radiation therapy, also referred to as radiotherapy, is an effective local treatment used for treating cancers of the breast, pancreas, prostate and glioblastoma, among others. Today, radiotherapy can be used in high dose, as it is possible to specifically irradiate the tumor site, thereby sparing surrounding normal tissues. MRS has been used to study the response to radiotherapy to assess metabolic changes. For example in a study on prostate cancer, a cohort of mice with androgen-dependent CWR22 prostate tumor xenografts was treated with a single fraction of 20 Gy (Gray), and 1H MR spectra were recorded pre and 24, 48 and 96 hours post radiation.325 Significantly reduced tumor growth and a decrease in the tCho to water ratio were observed at 24 hours post radiotherapy.325 In another preclinical study, the metabolic profile of a pancreatic cancer xenograft model was observed before and after radiotherapy using 1H HR MAS MRS and principle component analysis.326 A significant decrease was observed in GPC, PC and betaine levels in the pancreatic cancer xenografts as compared to normal mouse pancreas at 2 weeks post radiotherapy.326 Intact HeLa cells, when subjected to a single dose of either high energy γ rays (40 Gy) or low energy proton beams (20 Gy) displayed increasing ratios of GPC/PC and choline/PC over time post radiotherapy.327 1H MRS has also been used for evaluating the metabolic profile of a human glioblastoma tumor xenograft model pre and post radiotherapy at multiple time points.328 In this study, a cohort of 11 mice was treated with radiotherapy using 800 cGy.328 1H MR spectra were acquired at 24 hours before radiotherapy and at 3, 7 and 14 days post-treatment.328 A significant decrease was observed in the tCho (GPC+PC) to water ratio in the treated group as compared to the untreated group at 3 and 7 day post treatment.328 These preclinical studies demonstrate the feasibility of 1H MRS for evaluating the effects of radiotherapy and show that GPC levels can change following radiotherapy. Additional studies are needed to investigate the effects of radiotherapy on GPC metabolism in greater detail.
Considerations of specificity and sensitivity of GPC detection by MRS:
MRS is not yet a routine part of clinical MR exams for diagnosis and assessing treatment response in cancer, mostly due to the lack of consistent data from large multicenter clinical trials. Technical challenges with setting up specialized clinical MRS scans reproducibly at several clinical sites have made many large multicenter clinical trials elusive thus far. Moreover, most studies have reported the use of the tCho signal in the clinical assessment of specificity and sensitivity for cancer diagnosis and treatment response evaluation because of technical challenges with resolving GPC as an individual peak. For MRS detection of tCho in a single tumor voxel in breast cancer patients at 3T, the specificity and sensitivity for detecting cancer are in the range of 95% with a positive predictive value of about 98%.329 In MRS, spectral resolution of GPC has been achieved with 1H MRS at high field (9.4T) in the brain where there are considerably fewer motion artifacts than in the body, and with specialized 31P MRS techniques outside the brain as discussed above. However, most of these studies are technical in nature and have not yet evaluated sufficiently large cohorts of cancer patients. As several novel 31P MRS techniques are now able to resolve GPC, the near future will hopefully provide answers as to the specificity and sensitivity of detecting GPC in the tumors of cancer patients for diagnosis, staging, subtyping, and assessing treatment response.
Conclusions
With our continuously growing understanding of choline phospholipid metabolism in cancer, it is becoming apparent that the GPC breakdown pathway holds promise for providing treatment targets and biomarkers that reports at an early time point on the response of a given cancer to therapy. Enzymes that are active in the GPC breakdown pathway, such as for example cPLA2, lyso-PLA1, GDPD5, and GDPD6, are frequently overexpressed in cancers and may enable cancer cell proliferation, migration, and invasion. It is likely that oncogenic signaling pathways and transcription factors affect cellular GPC levels in cancer through their effects on the GPC-modulating enzymes cPLA2, lyso-PLA1, GDPD5, GDPD6. Innovative MRS applications are continuing to help elucidate the metabolic pathways affected by cancer, including the GPC pathway in cancer. The use of GPC as a noninvasive imaging biomarker in cancer diagnosis and treatment monitoring, alone or in combination with other peaks, is still under investigation. Additional studies using innovative 31P MRS and 1H MRS techniques are necessary to further evaluate the usefulness of GPC as biomarker.
Acknowledgements
We acknowledge the support of National Cancer Institute grants R01 CA213428 and R01 CA213492.
Abbreviation
- ABL
Abelson tyrosine-protein kinase
- AF 1
activating function 1
- AI
Aromatase inhibitors
- AP-1
activator protein-1
- ATP
adenosine triphosphate
- BINEPT
adiabatic version of the refocused insensitive nuclei enhanced by polarization transfer
- bZIP
basic leucine zipper
- CCT
CTP:phosphocholine cytidylyltransferase
- CDP-Cho
cytidine diphosphate-choline
- CEST
chemical exchange saturation transfer
- CHK
choline kinase
- CHK-α
choline kinase alpha
- Choe
extracellular free choline
- Choi
intracellular free choline
- CHPT1
diacylglycerol cholinephosphotransferase 1
- CMP
cytidine monophosphate
- COX
cyclooxygenase
- cPLA2
cytosolic phospholipase A2
- CT
computed X-ray tomography
- CTP
cytidine triphosphate
- DAG
diacylglycerol
- DBD
DNA-binding domain
- DLCL2
diffuse large B-cell lymphoma
- DNP
dynamic nuclear polarization
- DPDE
diphosphodiesters
- ECM
extracellular matrix
- EGFR
epidermal growth factor receptor
- EMT
epithelial-to-mesenchymal transition
- ER
estrogen receptor
- ERETIC
electronic reference to in vivo concentrations
- ERK
extracellular signal regulated kinase
- FA
fatty acid
- FAK
focal adhesion kinase
- FGFR
fibroblast growth factor receptor
- FOXA1
fork-head box A1
- GATA-3
GATA binding protein 3
- GDE
glycerophosphodiesterases
- GDPD
glycerophosphodiester phosphodiesterases
- GPC
glycerophosphocholine
- GPC-PDE
glycerophosphocholine phosphodiesterase
- GPC6
glypican-6
- GPE
glycerophosphoethanolamine
- GPI
glycerophosphoinositol
- Gro-3-P
glycerol-3-phosphate
- HCC
hepatocellular carcinoma
- HER2
human epidermal growth factor receptor 2
- HIF
hypoxia induced factor
- HM
hydrophobic motif
- HR MAS
high-resolution magic angle spinning
- Hsp90
heat shock protein 90
- IL-25
interleukin 25
- iPLA2
calcium-independent phospholipase A2
- JNK
janus kinase
- LBD
ligand-binding domain
- Lyso-PLA1
lyso-phospholipase A1
- MAPK
mitogen-activated protein kinase
- MRS
magnetic resonance spectroscopy
- MRSI
magnetic resonance spectroscopic imaging
- mTOR
Mammalian target of rapamycin
- NAD
nicotinamide adenine diphosphate
- NFAT
nuclear factor of activated T-cells
- NF- κB
nuclear factor kappa-light-chain-enhancer of activated B cells
- NMR
nuclear magnetic resonance
- NRTK
non-receptor tyrosine kinase
- NTP
β-nucleoside triphosphate
- PAF-AH
platelet-activating factor acetylhydrolase
- PA-PLA1
phosphatidic acid-preferring phospholipase A1
- PC
phosphocholine; PCA, principle component analysis
- PC-PLC
phosphatidylcholine-specific phospholipase C
- PC-PLD
phosphatidylcholine-specific phospholipase D
- PDK1
serine/threonine kinase-3′-phosphoinositide-dependent kinase 1
- PDGFR
platelet-derived growth factor receptor
- PET
positron emission therapy
- PGE2
prostaglandin E2
- PI-4,5-P2
phosphatidylinositol-4,4-bisphosphate
- PI3,4,5-P3
phosphatidylinositol-3,4,5-triphosphate
- PI3K
Phosphatidyl-inositol-3-kinases
- PKA
cAMP-dependent protein kinase
- PKC
protein kinase C
- PKG
cGMP-dependent protein kinase
- PLA(A-D)
phospholipase (A-D)
- ppm
parts per million
- PR
Progesterone receptor
- PPP
pentose phosphate pathway
- pRb
retinoblastoma protein
- PtdCho
phosphatidylcholine
- RAF
Rapidly Accelerated Fibrosarcoma
- RANKL
receptor activator of nuclear factor kappa-B ligand
- RALGDS
RAL GTPase guanine nucleotide dissociation stimulator
- RCE1
Ras converting enzyme 1
- RECIST
response evaluation criteria in solid tumors
- RINEPT
refocused insensitive nuclei enhanced by polarization transfer
- SHR
nuclear/steroid hormone receptor
- siRNA
small interfering RNA
- SNR
signal to noise ratio
- sPLA2
secreted PLA2
- STAT-3/5
signal transducer and activator of transcription-3/5
- TA/lysoPLA
transacylase/lysophospholipase
- TE
echo time
- TGF- β
transforming growth factor-β
- TOCSY
total correlation spectroscopy
- UDPS
uridine diphospho-sugar
- VEGF
vascular endothelial growth factor
- WHO
world health organization
- ZEB1
zinc finger E-box-binding homeobox 1
References
- 1.Junttila MR, de Sauvage FJ. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature. 2013;501:346. [DOI] [PubMed] [Google Scholar]
- 2.Werb Z, Lu P. The role of stroma in tumor development. The Cancer Journal. 2015;21(4):250–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Visser KE, Eichten A, Coussens LM. Paradoxical roles of the immune system during cancer development. Nature Reviews Cancer. 2006;6:24. [DOI] [PubMed] [Google Scholar]
- 4.Son B, Lee S, Youn H, Kim E, Kim W, Youn B. The role of tumor microenvironment in therapeutic resistance. Oncotarget. 2017;8(3):3933–3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeBerardinis RJ, Chandel NS. Fundamentals of cancer metabolism. Science Advances. 2016;2(5):e1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalyanaraman B Teaching the basics of cancer metabolism: Developing antitumor strategies by exploiting the differences between normal and cancer cell metabolism. Redox Biology. 2017;12:833–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. [DOI] [PubMed] [Google Scholar]
- 8.Griffiths JR, Stevens AN, Iles RA, Gordon RE, Shaw D. 31P NMR investigation of solid tumours in the living rat. Bioscience Reports. 1981;1(4):319–325. [DOI] [PubMed] [Google Scholar]
- 9.Daly PF, Lyon RC, Faustino PJ, Cohen JS. Phospholipid metabolism in cancer cells monitored by 31P NMR spectroscopy. Journal of Biological Chemistry. 1987;262(31):14875–14878. [PubMed] [Google Scholar]
- 10.Aboagye EO, Bhujwalla ZM. Malignant transformation alters membrane choline phospholipid metabolism of human mammary epithelial cells. Cancer Res. 1999;59:80. [PubMed] [Google Scholar]
- 11.Gillies RJ, Raghunand N, Karczmar GS, Bhujwalla ZM. MRI of the tumor microenvironment. Journal of Magnetic Resonance Imaging. 2002;16(4):430–450. [DOI] [PubMed] [Google Scholar]
- 12.Baek HM, Chen JH, Nalcioglu O, Su MY. Choline as a biomarker for cell proliferation: do the results from proton MR spectroscopy show difference between HER2/neu positive and negative breast cancers? Int J Cancer. 2008;123:1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bolan P, Meisamy S, Baker E. In vivo quantification of choline compounds in the breast with 1H MR spectroscopy. Magnetic Resonance in Medicine. 2003;50:1134. [DOI] [PubMed] [Google Scholar]
- 14.He Q, Xu RZ, Shkarin P, et al. Magnetic resonance spectroscopic imaging of tumor metabolic markers for cancer diagnosis, metabolic phenotyping, and characterization of tumor microenvironment. Disease Markers. 2003;19(2–3):69–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Al-Saffar NMS, Agliano A, Marshall LV, et al. In vitro nuclear magnetic resonance spectroscopy metabolic biomarkers for the combination of temozolomide with PI3K inhibition in paediatric glioblastoma cells. PLoS ONE. 2017;12(7):e0180263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glunde K, Bhujwalla ZM, Ronen SM. Choline metabolism in malignant transformation. Nat Rev Cancer. 2011;11:835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacobs MA, Barker PB, Bottomley PA, Bhujwalla ZM, Bluemke DA. Proton magnetic resonance spectroscopic imaging of human breast cancer: A preliminary study. Journal of Magnetic Resonance Imaging. 2004;19(1):68–75. [DOI] [PubMed] [Google Scholar]
- 18.Meisamy S, Bolan PJ, Baker EH, et al. Neoadjuvant chemotherapy of locally advanced breast cancer: Predicting response with in vivo 1H MR spectroscopy—a pilot study at 4T. Radiology. 2004;233(2):424–431. [DOI] [PubMed] [Google Scholar]
- 19.Manton DJ, Chaturvedi A, Hubbard A, et al. Neoadjuvant chemotherapy in breast cancer: early response prediction with quantitative MR imaging and spectroscopy. British Journal of Cancer. 2006;94(3):427–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baek HM, Chen JH, Nie K, et al. Predicting pathologic response to neoadjuvant chemotherapy in breast cancer by using MR imaging and quantitative 1H MR spectroscopy. Radiology. 2009;251(3):653–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Booth SJ, Pickles MD, Turnbull LW. In vivo magnetic resonance spectroscopy of gynaecological tumours at 3.0 Tesla. An International Journal of Obstetrics & Gynaecology. 2009;116(2):300–303. [DOI] [PubMed] [Google Scholar]
- 22.Kurhanewicz J, Swanson MG, Nelson SJ, Vigneron DB. Combined magnetic resonance imaging and spectroscopic imaging approach to molecular imaging of prostate cancer. Journal of magnetic resonance imaging : JMRI. 2002;16(4):451–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ackerstaff E, Pflug BR, Nelson JB, Bhujwalla ZM. Detection of increased choline compounds with proton nuclear magnetic resonance spectroscopy subsequent to malignant transformation of human prostatic epithelial cells. Cancer Research. 2001;61(9):3599–3603. [PubMed] [Google Scholar]
- 24.Belkić D, Belkić K. In vivo magnetic resonance spectroscopy for ovarian cancer diagnostics: quantification by the fast Padé transform. Journal of Mathematical Chemistry. 2017;55(1):349–405. [Google Scholar]
- 25.Elkhaled A, Jalbert L, Constantin A, et al. Characterization of metabolites in infiltrating gliomas using ex vivo 1H high-resolution magic angle spinning spectroscopy. Nmr in Biomedicine. 2014;27(5):578–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Einstein DB, Wessels B, Bangert B, et al. Phase II trial of radiosurgery to Magnetic Resonance Spectroscopy–defined high-risk tumor volumes in patients with glioblastoma multiforme. International Journal of Radiation Oncology*Biology*Physics. 2012;84(3):668–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vigneron D, Bollen A, McDermott M, et al. Three-dimensional magnetic resonance spectroscopic imaging of histologically confirmed brain tumors. Magnetic Resonance Imaging. 2001;19(1):89–101. [DOI] [PubMed] [Google Scholar]
- 28.Trousil S, Lee P, Pinato DJ, et al. Alterations of choline phospholipid metabolism in endometrial cancer are caused by choline kinase alpha overexpression and a hyperactivated deacylation pathway. Cancer Research. 2014. [DOI] [PubMed] [Google Scholar]
- 29.Podo F Tumour phospholipid metabolism. NMR in Biomedicine. 1999;12(7):413–439. [DOI] [PubMed] [Google Scholar]
- 30.Iorio E, Mezzanzanica D, Alberti P, et al. Alterations of choline phospholipid metabolism in ovarian tumor progression. Cancer Research. 2005;65(20):9369–9376. [DOI] [PubMed] [Google Scholar]
- 31.Cao MD, Sitter B, Bathen TF, et al. Predicting long-term survival and treatment response in breast cancer patients receiving neoadjuvant chemotherapy by MR metabolic profiling. NMR in Biomedicine. 2012;25(2):369–378. [DOI] [PubMed] [Google Scholar]
- 32.Bolan PJ. Magnetic resonance spectroscopy of the breast: current status. Magnetic Resonance Imaging Clinics of North America. 2013;21(3):625–639. [DOI] [PubMed] [Google Scholar]
- 33.Bolan P, Kim E, Herman B, et al. MR spectroscopy of breast cancer for assessing early treatment response: Results from the ACRIN 6657 MRS trial. Journal of Magnetic Resonance Imaging : JMRI. 2017;46(1):290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van der Kemp WJ, Stehouwer BL, Boer VO, Luijten PR, Klomp DW, Wijnen JP. Proton and phosphorus magnetic resonance spectroscopy of the healthy human breast at 7 T. NMR in Biomedicine. 2017;30(2):e3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kall SL, Delikatny EJ, Lavie A. Identification of a Unique Inhibitor-Binding Site on Choline Kinase alpha. Biochemistry. 2018;57(8):1316–1325. [DOI] [PubMed] [Google Scholar]
- 36.Miyake T, Parsons SJ. Functional interactions between Choline kinase alpha, epidermal growth factor receptor and c-Src in breast cancer cell proliferation. Oncogene. 2012;31(11):1431–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Falcon SC, Hudson CS, Huang Y, et al. A non-catalytic role of choline kinase alpha is important in promoting cancer cell survival. Oncogenesis. 2013;2:e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mori N, Wildes F, Kakkad S, et al. Choline kinase-alpha protein and phosphatidylcholine but not phosphocholine are required for breast cancer cell survival. NMR Biomed. 2015;28(12):1697–1706. [DOI] [PubMed] [Google Scholar]
- 39.Mountford CE, Wright LC. Organization of lipids in the plasma membranes of malignant and stimulated cells: a new model. Trends in Biochemical Sciences. 1988;13(5):172–177. [DOI] [PubMed] [Google Scholar]
- 40.Kennedy EP, Weiss SB. The function of cytidine coenzymes in the biosynthesis of phospholipides. Journal of Biological Chemistry. 1956;222(1):193–214. [PubMed] [Google Scholar]
- 41.Cuadrado A, Carnero A, Dolfi F, Jiménez B, Lacal JC. Phosphorylcholine: a novel second messenger essential for mitogenic activity of growth factors. Oncogene. 1993;8(11):2959–2968. [PubMed] [Google Scholar]
- 42.Benilde J, Luis del P, Silvia M, Pilar E, Juan CL. Generation of phosphorylcholine as an essential event in the activation of Raf 1 and MAP kinases in growth factors‐induced mitogenic stimulation. Journal of Cellular Biochemistry. 1995;57(1):141–149. [DOI] [PubMed] [Google Scholar]
- 43.Price BD, Morris JD, Marshall CJ, Hall A. Stimulation of phosphatidylcholine hydrolysis, diacylglycerol release, and arachidonic acid production by oncogenic ras is a consequence of protein kinase C activation. Journal of Biological Chemistry. 1989;264(28):16638–16643. [PubMed] [Google Scholar]
- 44.Besterman J, Duronio V, Cuatrecasas P. Rapid formation of diacylglycerol from phosphatidylcholine: a pathway for generation of a second messenger. Proceedings of the National Academy of Sciences. 1986;83(18):6785–6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morash SC, Cook HW, Spence MW. Phosphatidylcholine metabolism in cultured cells: catabolism via glycerophosphocholine. Biochimica et Biophysica Acta (BBA) - Lipids and Lipid Metabolism. 1988;961(2):194–202. [DOI] [PubMed] [Google Scholar]
- 46.Strauss H, Leibovitz-Ben Gershon Z, Heller M. Enzymatic hydrolysis of 1-monoacyl-SN-glycerol-3-phosphorylcholine (1-lysolecithin) by phospholipases from peanut seeds. Lipids. 1976;11(6):442–448. [DOI] [PubMed] [Google Scholar]
- 47.Loo RW, Conde-Frieboes K, Reynolds LJ, Dennis EA. Activation, inhibition, and regiospecificity of the lysophospholipase activity of the 85-KDa group IV cytosolic phospholipase A2. Journal of Biological Chemistry. 1997;272(31):19214–19219. [DOI] [PubMed] [Google Scholar]
- 48.Cao MD, Döpkens M, Krishnamachary B, et al. Glycerophosphodiester phosphodiesterase domain containing 5 (GDPD5) expression correlates with malignant choline phospholipid metabolite profiles in human breast cancer. NMR in Biomedicine. 2012;25(9):1033–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Corda D, Mosca MG, Ohshima N, Grauso L, Yanaka N, Mariggiò S. The emerging physiological roles of the glycerophosphodiesterase family. The FEBS Journal. 2014;281(4):998–1016. [DOI] [PubMed] [Google Scholar]
- 50.Okazaki Y, Ohshima N, Yoshizawa I, et al. A novel glycerophosphodiester phosphodiesterase, GDE5, controls skeletal muscle development via a non-enzymatic mechanism. Journal of Biological Chemistry. 2010;285(36):27652–27663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Richmond GS, Smith TK. Phospholipases A(1). International Journal of Molecular Sciences. 2011;12(1):588–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kadamur G, Elliott MR. Mammalian phospholipase C. Annual Review of Physiology. 2013;75(1):127–154. [DOI] [PubMed] [Google Scholar]
- 53.Hu J, Tian G, Zhang N. Cytosolic phospholipase A2 and its role in cancer. Clinical Oncology and Cancer Research. 2011;8(2):71. [Google Scholar]
- 54.Burke J, Dennis E. Phospholipase A2 biochemistry. Cardiovascular Drugs and Therapy. 2009;23(1):49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kita Y, Ohto T, Uozumi N, Shimizu T. Biochemical properties and pathophysiological roles of cytosolic phospholipase A2s. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 2006;1761(11):1317–1322. [DOI] [PubMed] [Google Scholar]
- 56.Wang D, Dubois RN. Eicosanoids and cancer. Nature Reviews Cancer. 2010;10:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leslie CC. Cytosolic phospholipase A2: physiological function and role in disease. Journal of Lipid Research. 2015;56(8):1386–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bocca C, Bozzo F, Miglietta A. COX2 inhibitor NS398 reduces HT-29 cell invasiveness by modulating signaling pathways mediated by egfr and HIF1-α. Anticancer Research. 2014;34(4):1793–1800. [PubMed] [Google Scholar]
- 59.Nakanishi M, Rosenberg DW. Roles of cPLA2α and arachidonic acid in cancer. Biochimica et biophysica acta. 2006;1761(11):1335–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Caiazza F, Harvey B, Thomas W. Cytosolic phospholipase A2 activation correlates with HER2 overexpression and mediates estrogen-dependent breast cancer cell growth. Mol Endocrinol. 2010;24:953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Caiazza F, McCarthy NS, Young L, Hill AD, Harvey BJ, Thomas W. Cytosolic phospholipase A2-alpha expression in breast cancer is associated with EGFR expression and correlates with an adverse prognosis in luminal tumours. Br J Cancer. 2011;104:338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun GY, Shelat PB, Jensen MB, He Y, Sun AY, Simonyi A. Phospholipases A2 and inflammatory responses in the central nervous system. Neuromolecular medicine. 2010;12(2):133–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pickard RT, Strifler BA, Kramer RM, Sharp JD. Molecular cloning of two new human paralogs of 85-kDa cytosolic phospholipase A2. Journal of Biological Chemistry. 1999;274(13):8823–8831. [DOI] [PubMed] [Google Scholar]
- 64.Gijón MA, Leslie CC. Regulation of arachidonic acid release and cytosolic Phospholipase A2 activation. Journal of Leukocyte Biology. 1999;65(3):330–336. [DOI] [PubMed] [Google Scholar]
- 65.Czachorowski MJ, Amaral AF, Montes-Moreno S, et al. Cyclooxygenase-2 expression in bladder cancer and patient prognosis: results from a large clinical cohort and meta-analysis. PLOS ONE. 2012;7(9):e45025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jang TJ. Epithelial to mesenchymal transition in cutaneous squamous cell carcinoma is correlated with COX-2 expression but not with the presence of stromal macrophages or CD10-expressing cells. Virchows Archiv. 2012;460(5):481–487. [DOI] [PubMed] [Google Scholar]
- 67.Ogunwobi OO, Liu C. Hepatocyte growth factor upregulation promotes carcinogenesis and epithelial-mesenchymal transition in hepatocellular carcinoma via AKT and COX-2 pathways. Clinical & Experimental Metastasis. 2011;28(8):721–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Uozumi N, Kume K, Nagase T, et al. Role of cytosolic phospholipase A2 in allergic response and parturition. Nature. 1997;390:618. [DOI] [PubMed] [Google Scholar]
- 69.Sheng H, Soma V, Mu Y, Chanlu X, Paul S, Qihan D. AKT and cytosolic phospholipase A2α form a positive loop in prostate cancer cells. Current Cancer Drug Targets. 2015;15(9):781–791. [DOI] [PubMed] [Google Scholar]
- 70.Herbert SP, Odell AF, Ponnambalam S, Walker JH. Activation of cytosolic phospholipase A2α as a novel mechanism regulating endothelial cell cycle progression and angiogenesis. The Journal of Biological Chemistry. 2009;284(9):5784–5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schulte RR, Linkous AG, Hallahan DE, Yazlovitskaya EM. Cytosolic phospholipase A2 as a molecular target for the radiosensitization of ovarian cancer. Cancer letters. 2011;304(2):137–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kisslov L, Hadad N, Rosengraten M, Levy R. HT-29 human colon cancer cell proliferation is regulated by cytosolic phospholipase A2α dependent PGE2 via both PKA and PKB pathways. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 2012;1821(9):1224–1234. [DOI] [PubMed] [Google Scholar]
- 73.Gentile LB, Piva B, Capizzani BC, et al. Hypertonic environment elicits cyclooxygenase-2-driven prostaglandin E2 generation by colon cancer cells: Role of cytosolic phospholipase A2α and kinase signaling pathways. Prostaglandins, Leukotrienes and Essential Fatty Acids. 2010;82(2):131–139. [DOI] [PubMed] [Google Scholar]
- 74.Grewal S, Ponnambalam S, Walker JH. Association of cPLA2α and COX-1 with the Golgi apparatus of A549 human lung epithelial cells. Journal of Cell Science. 2003;116(11):2303–2310. [DOI] [PubMed] [Google Scholar]
- 75.Chen L, Fu H, Luo Y, et al. cPLA2α mediates TGF-β-induced epithelial–mesenchymal transition in breast cancer through PI3k/AKT signaling. Cell Death & Disease. 2017;8(4):e2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Patel MI, Singh J, Niknami M, et al. Cytosolic phospholipase A2α: A potential therapeutic target for prostate cancer. Clinical Cancer Research. 2008;14(24):8070–8079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Linkous AG, Yazlovitskaya EM, Hallahan DE. Cytosolic phospholipase A2 and lysophospholipids in tumor angiogenesis. JNCI Journal of the National Cancer Institute. 2010;102(18):1398–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Song C, Chang XJ, Bean KM, Proia MS, Knopf JL, Kriz RW. Molecular characterization of cytosolic Phospholipase A2-β. Journal of Biological Chemistry. 1999;274(24):17063–17067. [DOI] [PubMed] [Google Scholar]
- 79.Moestue SA, Borgan E, Huuse EM, et al. Distinct choline metabolic profiles are associated with differences in gene expression for basal-like and luminal-like breast cancer xenograft models. BMC Cancer. 2010;10(1):433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mawn TM, Popov AV, Beardsley NJ, et al. In vivo detection of phospholipase C by enzyme-activated near-infrared probes. Bioconjugate Chemistry. 2011;22(12):2434–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Popov AV, Mawn TM, Kim S, Zheng G, Delikatny EJ. Design and synthesis of phospholipase C and A2-activatable near infrared fluorescent smart probes. Bioconjugate chemistry. 2010;21(10):1724–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chiorazzo MG, Bloch NB, Popov AV, Delikatny EJ. Synthesis and evaluation of cytosolic phospholipase A2 activatable fluorophores for cancer imaging. Bioconjugate Chemistry. 2015;26(12):2360–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Clem B, Telang S, Trent J, Chesney J, Chand P, Tapolsky G, Inventors; Google Patents, assignee Small molecule choline kinase inhibitors, screening assays, and methods for treatment of neoplastic disorders. 2016.
- 84.van den Bosch H Intracellular phospholipases A. Biochimica et Biophysica Acta (BBA) - Biomembranes. 1980;604(2):191–246. [DOI] [PubMed] [Google Scholar]
- 85.Aoki J, Inoue A, Makide K, Saiki N, Arai H. Structure and function of extracellular phospholipase A1 belonging to the pancreatic lipase gene family. Biochimie. 2007;89(2):197–204. [DOI] [PubMed] [Google Scholar]
- 86.Wang A, Dennis EA. Mammalian lysophospholipases. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 1999;1439(1):1–16. [DOI] [PubMed] [Google Scholar]
- 87.Sunaga H, Sugimoto H, Nagamachi Y, Yamashita S. Purification and properties of lysophospholipase isoenzymes from pig gastric mucosa. Biochemical Journal. 1995;308(2):551–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Garsetti D, Holtsberg F, Steiner M, Egan R, Clark MA. Butyric acid-induced differentiation of HL-60 cells increases the expression of a single lysophospholipase. Biochemical Journal. 1992;288(3):831–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shier WT, Baldwin JH, Nilsen-Hamilton M, Hamilton RT, Thanassi NM. Regulation of guanylate and adenylate cyclase activities by lysolecithin. Proceedings of the National Academy of Sciences of the United States of America. 1976;73(5):1586–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kelly RA, O’Hara DS, Mitch WE, Smith TW. Identification of NaK-ATPase inhibitors in human plasma as nonesterified fatty acids and lysophospholipids. Journal of Biological Chemistry. 1986;261(25):11704–11711. [PubMed] [Google Scholar]
- 91.Zhou L, Shi M, Zhao L, et al. Clonorchis sinensis lysophospholipase A upregulates IL-25 expression in macrophages as a potential pathway to liver fibrosis. Parasites & vectors. 2017;10(1):295–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Larson TJ, Ehrmann M, Boos W. Periplasmic glycerophosphodiester phosphodiesterase of Escherichia coli, a new enzyme of the glp regulon. Journal of Biological Chemistry. 1983;258(9):5428–5432. [PubMed] [Google Scholar]
- 93.Dawson RM. Liver glycerylphosphorylcholine diesterase. The Biochemical journal. 1956;62(4):689–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zablocki K, Miller SP, Garcia-Perez A, Burg MB. Accumulation of glycerophosphocholine (GPC) by renal cells: osmotic regulation of GPC:choline phosphodiesterase. Proceedings of the National Academy of Sciences. 1991;88(17):7820–7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gallazzini M, Ferraris JD, Burg MB. GDPD5 is a glycerophosphocholine phosphodiesterase that osmotically regulates the osmoprotective organic osmolyte GPC. Proceedings of the National Academy of Sciences. 2008;105(31):11026–11031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Stewart JD, Marchan R, Lesjak MS, et al. Choline-releasing glycerophosphodiesterase EDI3 drives tumor cell migration and metastasis. Proceedings of the National Academy of Sciences. 2012;109(21):8155–8160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ohshima N, Kudo T, Yamashita Y, et al. New members of the mammalian glycerophosphodiester phosphodiesterase family: GDE4 and GDE7 produce lysophosphatidic acid by lysophospholipase D activity. The Journal of biological chemistry. 2015;290(7):4260–4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rao M, Sockanathan S. Transmembrane Protein GDE2 Induces Motor Neuron Differentiation in Vivo. Science. 2005;309(5744):2212–2215. [DOI] [PubMed] [Google Scholar]
- 99.Park S, Lee C, Sabharwal P, Zhang M, Meyers CLF, Sockanathan S. GDE2 promotes neurogenesis by glycosylphosphatidylinositol-anchor cleavage of RECK. Science. 2013;339(6117):324–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Matas-Rico E, van Veen M, Leyton-Puig D, et al. Glycerophosphodiesterase GDE2 promotes neuroblastoma differentiation through glypican release and is a marker of clinical outcome. Cancer Cell. 2016;30(4):548–562. [DOI] [PubMed] [Google Scholar]
- 101.Matas-Rico E, van Veen M, Moolenaar WH. GDE2/GDPD5 in neuroblastoma. Oncotarget. 2017;8(4):5672–5673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wijnen JP, Jiang L, Greenwood TR, et al. Silencing of the glycerophosphocholine phosphodiesterase GDPD5 alters the phospholipid metabolite profile in a breast cancer model in vivo as monitored by 31P MRS. NMR in Biomedicine. 2014;27(6):692–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cao MD, Cheng M, Rizwan A, et al. Targeting choline phospholipid metabolism: GDPD5 and GDPD6 silencing decrease breast cancer cell proliferation, migration, and invasion. NMR in Biomedicine. 2016;29(8):1098–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cheng M, Rizwan A, Jiang L, Bhujwalla ZM, Glunde K. Molecular effects of doxorubicin on choline metabolism in breast cancer. Neoplasia 2017;19(8):617–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Macara IG. Elevated phosphocholine concentration in ras-transformed NIH 3T3 cells arises from increased choline kinase activity, not from phosphatidylcholine breakdown. Molecular and Cellular Biology. 1989;9(1):325–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ramírez de Molina A, Penalva V, Lucas L, Lacal JC. Regulation of choline kinase activity by Ras proteins involves Ral–GDS and PI3K. Oncogene. 2002;21:937. [DOI] [PubMed] [Google Scholar]
- 107.Shapiro P RAS- MAP kinase signaling pathways and control of cell proliferation: Relevance to cancer therapy. Critical Reviews in Clinical Laboratory Sciences. 2002;39(4–5):285–330. [DOI] [PubMed] [Google Scholar]
- 108.Kurada P, White K. Ras promotes cell survival in Drosophila by downregulating hid expression. Cell. 1998;95(3):319–329. [DOI] [PubMed] [Google Scholar]
- 109.Regad T Targeting RTK signaling pathways in cancer. Cancers. 2015;7(3):1758–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Morrison DK. MAP kinase pathways. Cold Spring Harbor perspectives in biology. 2012;4(11):a011254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Paul MK, Mukhopadhyay AK. Tyrosine kinase – role and significance in cancer. International Journal of Medical Sciences. 2004;1(2):101–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Katz M, Amit I, Yarden Y. Regulation of MAPKs by growth factors and receptor tyrosine kinases. Biochimica et biophysica acta. 2007;1773(8):1161–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kyriakis JM, App H, Zhang X-f, et al. Raf-1 activates MAP kinase-kinase. Nature. 1992;358:417. [DOI] [PubMed] [Google Scholar]
- 114.Chong H, Guan K-L. Regulation of raf through phosphorylation and N terminus-C terminus interaction. Journal of Biological Chemistry. 2003;278(38):36269–36276. [DOI] [PubMed] [Google Scholar]
- 115.Yan M, Parker BA, Schwab R, Kurzrock R. HER2 aberrations in cancer: Implications for therapy. Cancer Treatment Reviews. 2014;40(6):770–780. [DOI] [PubMed] [Google Scholar]
- 116.Cadena DL, Gill GN. Receptor tyrosine kinases. The FASEB Journal. 1992;6(6):2332–2337. [DOI] [PubMed] [Google Scholar]
- 117.Vlahovic G, Crawford J. Activation of tyrosine kinases in cancer. The Oncologist. 2003;8(6):531–538. [DOI] [PubMed] [Google Scholar]
- 118.Thomas C, Gustafsson JÅ. The different roles of ER subtypes in cancer biology and therapy. Nature Reviews Cancer. 2011;11:597. [DOI] [PubMed] [Google Scholar]
- 119.Kumar V, Green S, Stack G, Berry M, Jin JR, Chambon P. Functional domains of the human estrogen receptor. Cell. 1987;51(6):941–951. [DOI] [PubMed] [Google Scholar]
- 120.McKenna NJ, Lanz RB, O’Malley BW. Nuclear receptor coregulators: cellular and molecular biology. Endocrine Reviews. 1999;20(3):321–344. [DOI] [PubMed] [Google Scholar]
- 121.McDonnell DP, Norris JD. Connections and regulation of the human estrogen receptor. Science. 2002;296(5573):1642–1644. [DOI] [PubMed] [Google Scholar]
- 122.Warner M, Nilsson S, Gustafsson JA. The estrogen receptor family. Current Opinion in Obstetrics and Gynecology. 1999;11(3):249–254. [DOI] [PubMed] [Google Scholar]
- 123.Hewitt SC, Couse JF, Korach KS. Estrogen receptor transcription and transactivation Estrogen receptor knockout mice: what their phenotypes reveal about mechanisms of estrogen action. Breast Cancer Research. 2000;2(5):345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Barnes CJ, Vadlamudi RK, Kumar R. Novel estrogen receptor coregulators and signaling molecules in human diseases. Cellular and Molecular Life Sciences CMLS. 2004;61(3):281–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.O’Malley BW. Little molecules with big goals. Science. 2006;313(5794):1749–1750. [DOI] [PubMed] [Google Scholar]
- 126.Han JS, Crowe DL. Steroid receptor coactivator 1 deficiency increases MMTV-neu mediated tumor latency and differentiation specific gene expression, decreases metastasis, and inhibits response to PPAR ligands. BMC Cancer. 2010;10(1):629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chakravarty D, Nair SS, Santhamma B, et al. Extranuclear functions of ER impact invasive migration and metastasis by breast cancer cells. Cancer Research. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Li Y, Wang JP, Santen RJ, et al. Estrogen stimulation of cell migration involves multiple signaling pathway interactions. Endocrinology. 2010;151(11):5146–5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Finn RS. Targeting Src in breast cancer. Annals of Oncology. 2008;19(8):1379–1386. [DOI] [PubMed] [Google Scholar]
- 130.Persad S, Dedhar S. The role of integrin-linked kinase (ILK) in cancer progression. Cancer and Metastasis Reviews. 2003;22(4):375–384. [DOI] [PubMed] [Google Scholar]
- 131.Ström A, Hartman J, Foster JS, Kietz S, Wimalasena J, Gustafsson JA. Estrogen receptor β inhibits 17β-estradiol-stimulated proliferation of the breast cancer cell line T47D. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(6):1566–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hartman J, Ström A, Gustafsson J-Å. Estrogen receptor beta in breast cancer—Diagnostic and therapeutic implications. Steroids. 2009;74(8):635–641. [DOI] [PubMed] [Google Scholar]
- 133.Järvinen TAH, Pelto-Huikko M, Holli K, Isola J. Estrogen receptor β is coexpressed with ERα and PR and associated with nodal status, grade, and proliferation rate in breast cancer. The American Journal of Pathology. 2000;156(1):29–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mak P, Leav I, Pursell B, et al. ERβ impedes prostate cancer EMT by destabilizing HIF-1α and inhibiting VEGF-mediated Snail nuclear localization: Implications for gleason grading. Cancer Cell. 2010;17(4):319–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mulac-Jericevic B, Conneely OM. Reproductive tissue selective actions of progesterone receptors. 2004;128(2):139. [DOI] [PubMed] [Google Scholar]
- 136.Grimm SL, Hartig SM, Edwards DP. Progesterone receptor signaling mechanisms. Journal of Molecular Biology. 2016;428(19):3831–3849. [DOI] [PubMed] [Google Scholar]
- 137.Obr AE, Edwards DP. The biology of progesterone receptor in the normal mammary gland and in breast cancer. Molecular and Cellular Endocrinology. 2012;357(1):4–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Condon JC, Hardy DB, Kovaric K, Mendelson CR. Up-regulation of the progesterone receptor (PR)-c isoform in laboring myometrium by activation of Nuclear Factor-κB may contribute to the onset of labor through inhibition of pr function. Molecular Endocrinology. 2006;20(4):764–775. [DOI] [PubMed] [Google Scholar]
- 139.Anderson E Progesterone receptors - animal models and cell signaling in breast cancer: The role of oestrogen and progesterone receptors in human mammary development and tumorigenesis. Breast Cancer Research. 2002;4(5):197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Brisken C Hormonal control of alveolar development and its implications for breast carcinogenesis. Journal of Mammary Gland Biology and Neoplasia. 2002;7(1):39–48. [DOI] [PubMed] [Google Scholar]
- 141.Frech MS, Halama ED, Tilli MT, et al. Deregulated estrogen receptor α expression in mammary epithelial cells of transgenic mice results in the development of ductal carcinoma in situ. Cancer Research. 2005;65(3):681–685. [PMC free article] [PubMed] [Google Scholar]
- 142.Medina D, Kittrell FS, Shepard A, Contreras A, Rosen JM, Lydon J. Hormone dependence in premalignant mammary progression. Cancer Research. 2003;63(5):1067–1072. [PubMed] [Google Scholar]
- 143.Graham JD, Mote PA, Salagame U, et al. DNA replication licensing and progenitor numbers are increased by progesterone in normal human breast. Endocrinology. 2009;150(7):3318–3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Shoker BS, Jarvis C, Clarke RB, et al. Estrogen receptor-positive proliferating cells in the normal and precancerous breast. The American Journal of Pathology. 1999;155(6):1811–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Beleut M, Rajaram RD, Caikovski M, et al. Two distinct mechanisms underlie progesterone-induced proliferation in the mammary gland. Proceedings of the National Academy of Sciences. 2010;107(7):2989–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Thomas RJ, Guise TA, Yin JJ, et al. Breast cancer cells interact with osteoblasts to support osteoclast formation. Endocrinology. 1999;140(10):4451–4458. [DOI] [PubMed] [Google Scholar]
- 147.Hu H, Wang J, Gupta A, et al. RANKL expression in normal and malignant breast tissue responds to progesterone and is up-regulated during the luteal phase. Breast Cancer Research and Treatment. 2014;146(3):515–523. [DOI] [PubMed] [Google Scholar]
- 148.Azim HA, Peccatori FA, Brohée S, et al. RANK-ligand (RANKL) expression in young breast cancer patients and during pregnancy. Breast Cancer Research. 2015;17(1):24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Bhatia P, Sanders MM, Hansen MF. Expression of receptor activator of Nuclear Factor-κB ligand is inversely correlated with metastatic phenotype in breast carcinoma. Clinical Cancer Research. 2005;11(1):162–165. [PubMed] [Google Scholar]
- 150.Meric-Bernstam F, Hung MC. Advances in targeting human epidermal growth factor receptor-2 signaling for cancer therapy. Clinical Cancer Research. 2006;12(21):6326–6330. [DOI] [PubMed] [Google Scholar]
- 151.Slamon D, Godolphin W, Jones L, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244(4905):707–712. [DOI] [PubMed] [Google Scholar]
- 152.Slamon D, Clark G, Wong S, Levin W, Ullrich A, McGuire W. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235(4785):177–182. [DOI] [PubMed] [Google Scholar]
- 153.Olayioye MA. Intracellular signaling pathways of ErbB2/HER-2 and family members. Breast Cancer Research. 2001;3(6):385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nature Reviews Molecular Cell Biology. 2001;2:127. [DOI] [PubMed] [Google Scholar]
- 155.Mitri Z, Constantine T, O’Regan R. The HER2 receptor in breast cancer: Pathophysiology, clinical use, and new advances in therapy. Chemotherapy research and practice. 2012;2012:743193–743193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Fukushige S, Matsubara K, Yoshida M, et al. Localization of a novel v-erbB-related gene, c-erbB-2, on human chromosome 17 and its amplification in a gastric cancer cell line. Molecular and Cellular Biology. 1986;6(3):955–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Stern DF, Heffernan PA, Weinberg RA. p185, a product of the neu proto-oncogene, is a receptorlike protein associated with tyrosine kinase activity. Molecular and Cellular Biology. 1986;6(5):1729–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Akiyama T, Sudo C, Ogawara H, Toyoshima K, Yamamoto T. The product of the human c-erbB-2 gene: a 185-kilodalton glycoprotein with tyrosine kinase activity. Science. 1986;232(4758):1644–1646. [DOI] [PubMed] [Google Scholar]
- 159.Spears M, Taylor KJ, Munro AF, et al. In situ detection of HER2:HER2 and HER2:HER3 protein–protein interactions demonstrates prognostic significance in early breast cancer. Breast Cancer Research and Treatment. 2012;132(2):463–470. [DOI] [PubMed] [Google Scholar]
- 160.Shi Y, Huang W, Tan Y, et al. A novel proximity assay for the detection of proteins and protein complexes: quantitation of HER1 and HER2 total protein expression and homodimerization in formalin-fixed, paraffin-embedded cell lines and breast cancer tissue. Diagn Mol Pathol. 2009;18:11–21. [DOI] [PubMed] [Google Scholar]
- 161.Cicenas J, Urban P, Küng W, et al. Phosphorylation of tyrosine 1248-ERBB2 measured by chemiluminescence-linked immunoassay is an independent predictor of poor prognosis in primary breast cancer patients. European Journal of Cancer. 2006;42(5):636–645. [DOI] [PubMed] [Google Scholar]
- 162.Taniyama K, Ishida K, Toda T, et al. Tyrosine1248-phosphorylated HER2 expression and HER2 gene amplification in female invasive ductal carcinomas. Vol 152008. [DOI] [PubMed] [Google Scholar]
- 163.DiGiovanna MP, Lerman MA, Coffey RJ, Muller WJ, Cardiff RD, Stern DF. Active signaling by Neu in transgenic mice. Oncogene. 1998;17:1877. [DOI] [PubMed] [Google Scholar]
- 164.Ramić S, Asić K, Balja MP, Paić F, Benković V, Knežević F. Correlation of phosphorylated HER2 with clinicopathological characteristics and efficacy of trastuzumab treatment for breast cancer. Anticancer Research. 2013;33(6):2509–2515. [PubMed] [Google Scholar]
- 165.Giskeødegård GF, Grinde MT, Sitter B, et al. Multivariate modeling and prediction of breast cancer prognostic factors using MR metabolomics. Journal of Proteome Research. 2010;9(2):972–979. [DOI] [PubMed] [Google Scholar]
- 166.Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase–AKT pathway in human cancer. Nature Reviews Cancer. 2002;2:489. [DOI] [PubMed] [Google Scholar]
- 167.Hao Y, Wong R, Feig LA. RalGDS couples growth factor signaling to AKT activation. Molecular and cellular biology. 2008;28(9):2851–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hanahan D, Weinberg RA. The Hallmarks of Cancer. Cell. 2000;100(1):57–70. [DOI] [PubMed] [Google Scholar]
- 169.Young A, Lyons J, Miller AL, Phan V, Alarcón I, McCormick F. Ras signaling and therapies Advances in Cancer Research. Vol 102: Academic Press; 2009:1–17. [DOI] [PubMed] [Google Scholar]
- 170.Downward J Targeting RAS signalling pathways in cancer therapy. Nature Reviews Cancer. 2003;3:11. [DOI] [PubMed] [Google Scholar]
- 171.Bos JL. ras oncogenes in human cancer: A review. Cancer Research. 1989;49(17):4682–4689. [PubMed] [Google Scholar]
- 172.Shields JM, Pruitt K, McFall A, Shaub A, Der CJ. Understanding Ras: ‘it ain’t over ‘til it’s over’. Trends in Cell Biology. 2000;10(4):147–154. [DOI] [PubMed] [Google Scholar]
- 173.Lowy DR, Willumsen BM. Function and regulation of Ras. Annual Review of Biochemistry. 1993;62(1):851–891. [DOI] [PubMed] [Google Scholar]
- 174.Johnson L, Greenbaum D, Cichowski K, et al. K-ras is an essential gene in the mouse with partial functional overlap with N-ras. Genes & development. 1997;11(19):2468–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Colicelli J Human RAS superfamily proteins and related GTPases. Science’s STKE : signal transduction knowledge environment. 2004;2004(250). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Gentry LR, Nishimura A, Cox AD, et al. Divergent roles of CAAX motif-signaled posttranslational modifications in the regulation and subcellular localization of ral gtpases. Journal of Biological Chemistry. 2015;290(37):22851–22861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Oliff A Farnesyltransferase inhibitors: targeting the molecular basis of cancer. Biochimica et Biophysica Acta (BBA) - Reviews on Cancer. 1999;1423(3):C19–C30. [DOI] [PubMed] [Google Scholar]
- 178.Hancock JF, Paterson H, Marshall CJ. A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell. 1990;63(1):133–139. [DOI] [PubMed] [Google Scholar]
- 179.Barceló C, Paco N, Morell M, et al. Phosphorylation at Ser-181 of oncogenic KRAS is required for tumor growth. Cancer Research. 2014;74(4):1190–1199. [DOI] [PubMed] [Google Scholar]
- 180.Leevers SJ, Paterson HF, Marshall CJ. Requirement for ras in raf activation is overcome by targeting raf to the plasma membrane. Nature. 1994;369:411. [DOI] [PubMed] [Google Scholar]
- 181.Adjei AA. Ras signaling pathway proteins as therapeutic targets. Curr Pharm Des. 2001;7(16):1581–1594. [DOI] [PubMed] [Google Scholar]
- 182.Fruman DA, Meyers RE, Lewis CC. Phosphoinositide Kinases. Annual Review of Biochemistry. 1998;67(1):481–507. [DOI] [PubMed] [Google Scholar]
- 183.Fresno VJÁ, Casado E, de Castro J, Cejas P, Belda-Iniesta C, González-Barón M. PI3K/AKT signalling pathway and cancer. Cancer Treatment Reviews. 2004;30(2):193–204. [DOI] [PubMed] [Google Scholar]
- 184.Kumar CC, Madison V. AKT crystal structure and AKT-specific inhibitors. Oncogene. 2005;24:7493. [DOI] [PubMed] [Google Scholar]
- 185.Porta C, Paglino C, Mosca A. Targeting PI3K/AKT/mTOR signaling in cancer. Frontiers in oncology. 2014;4:64–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Wang Y, Kuramitsu Y, Baron B, et al. PI3K inhibitor LY294002, as opposed to wortmannin, enhances AKT phosphorylation in gemcitabine-resistant pancreatic cancer cells. International Journal of Oncology. 2017;50(2):606–612. [DOI] [PubMed] [Google Scholar]
- 187.Beloueche-Babari M, Jackson LE, Al-Saffar NMS, et al. Identification of magnetic resonance detectable metabolic changes associated with inhibition of phosphoinositide 3-kinase signaling in human breast cancer cells. Molecular Cancer Therapeutics. 2006;5(1):187–196. [DOI] [PubMed] [Google Scholar]
- 188.Su JS, Woods SM, Ronen SM. Metabolic consequences of treatment with AKT inhibitor perifosine in breast cancer cells. NMR in biomedicine. 2012;25(2):379–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Karamouzis MV, Papavassiliou AG. Transcription factor networks as targets for therapeutic intervention of cancer: The breast cancer paradigm. Molecular Medicine. 2011;17(11–12):1133–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Singh S, Johnson J, Chellappan S. Small molecule regulators of Rb–E2F pathway as modulators of transcription. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms. 2010;1799(10):788–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Battaglia S, Maguire O, Campbell MJ. Transcription factor co-repressors in cancer biology; roles and targeting. International journal of cancer Journal international du cancer. 2010;126(11):2511–2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Özcan S, Andrali SS, Cantrell JEL. Modulation of transcription factor function by O-GlcNAc modification. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms. 2010;1799(5):353–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Aguilera C, Nakagawa K, Sancho R, Chakraborty A, Hendrich B, Behrens A. c-Jun N-terminal phosphorylation antagonises recruitment of the Mbd3/NuRD repressor complex. Nature. 2011;469:231. [DOI] [PubMed] [Google Scholar]
- 194.Zhang X, Gamble MJ, Stadler S, et al. Genome-wide analysis reveals PADI4 cooperates with Elk-1 to activate c-Fos expression in breast cancer cells. PLoS Genetics. 2011;7(6):e1002112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Lee K, Gjorevski N, Boghaert E, Radisky DC, Nelson CM. Snail1, Snail2, and E47 promote mammary epithelial branching morphogenesis. The EMBO journal. 2011;30(13):2662–2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Hisamatsu Y, Tokunaga E, Akiyoshi S, et al. The expression of GATA-3 and FOXA1 in breast cancer: The biomarkers of hormone sensitivity in luminal-type tumors. J Clin Oncol. 2011;29 [Google Scholar]
- 197.McCune K, Bhat-Nakshatri P, Thorat MA, Nephew KP, Badve S, Nakshatri H. Prognosis of hormone-dependent breast cancers: implications of the presence of dysfunctional transcriptional networks activated by insulin via the immune transcription factor T-bet. Cancer Research. 2010;70(2):685–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Siegel PM, Muller WJ. Transcription factor regulatory networks in mammary epithelial development and tumorigenesis. Oncogene. 2010;29:2753. [DOI] [PubMed] [Google Scholar]
- 199.Casas E, Kim J, Bendesky A, Ohno-Machado L, Wolfe CJ, Yang J. Snail2 is an essential mediator of Twist1-induced epithelial mesenchymal transition and metastasis. Cancer Research. 2011;71(1):245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Giordano A, Mego M, Lee B, et al. Epithelial-mesenchymal transition in patients with HER2+ metastatic breast cancer. Journal of Clinical Oncology. 2011;29(15_suppl):623–623. [Google Scholar]
- 201.Haftchenary S, Avadisian M, Gunning PT. Inhibiting aberrant Stat3 function with molecular therapeutics: A progress report. Anticancer Drugs. 2011;22(2):115–127. [DOI] [PubMed] [Google Scholar]
- 202.Wingelhofer B, Neubauer HA, Valent P, et al. Implications of STAT3 and STAT5 signaling on gene regulation and chromatin remodeling in hematopoietic cancer. Leukemia. 2018;32(8):1713–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Bodily J, Mehta K, Laimins L. Human papillomavirus E7 enhances hypoxia-inducible factor 1-mediated transcription by inhibiting binding of histone deacetylases. Cancer research. 2011;71(3):1187–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Semenza GL. Hypoxia-inducible factors: mediators of cancer progression and targets for cancer therapy. Trends in pharmacological sciences. 2012;33(4):207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Yin L, Velazquez OC, Liu ZJ. Notch signaling: Emerging molecular targets for cancer therapy. Biochemical Pharmacology. 2010;80(5):690–701. [DOI] [PubMed] [Google Scholar]
- 206.Glunde K, Shah T, Winnard PT, et al. Hypoxia regulates choline kinase expression through Hypoxia-Inducible Factor-1α signaling in a human prostate cancer model. Cancer Research. 2008;68(1):172–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Jiang L, Greenwood TR, Artemov D, et al. Localized hypoxia results in spatially heterogeneous metabolic signatures in breast tumor models. Neoplasia. 2012;14(8):732–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Bharti SK, Mironchik Y, Wildes F, et al. Metabolic consequences of HIF silencing in a triple negative human breast cancer xenograft. Oncotarget. 2018;9(20):15326–15339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Santos CR, Schulze A. Lipid metabolism in cancer. The FEBS Journal. 2012;279(15):2610–2623. [DOI] [PubMed] [Google Scholar]
- 210.van der Velden TA, Italiaander M, van der Kemp WJ, et al. Radiofrequency configuration to facilitate bilateral breast 31P MR spectroscopic imaging and high-resolution MRI at 7 Tesla. Magnetic Resonance in Medicine. 2015;74(6):1803–1810. [DOI] [PubMed] [Google Scholar]
- 211.van der Kemp WJ, Boer VO, Luijten PR, Wijnen JP, Klomp DW. Increase in SNR for 31P MR spectroscopy by combining polarization transfer with a direct detection sequence. Magnetic Resonance in Medicine. 2012;68(2):353–357. [DOI] [PubMed] [Google Scholar]
- 212.Stenman K, Stattin P, Stenlund H, Riklund K, Gröbner G, Bergh A. 1H HRMAS NMR derived bio-markers related to tumor grade, tumor cell fraction, and cell proliferation in prostate tissue samples. Biomarker insights. 2011;6:39–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Sjøbakk T, Vettukattil R, Gulati M, et al. Metabolic profiles of brain metastases. International Journal of Molecular Sciences. 2013;14(1):2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Sinharay S, Pagel MD. Advances in magnetic resonance imaging contrast agents for biomarker detection. Annual Review of Analytical Chemistry. 2016;9(1):95–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Posse S, Otazo R, Dager SR, Alger J. MR spectroscopic imaging: Principles and recent advances. Journal of Magnetic Resonance Imaging. 2013;37(6):1301–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Semmler W, Gademann G, Bachert-Baumann P, Zabel HJ, Lorenz WJ, Kaick Gv. Monitoring human tumor response to therapy by means of P-31 MR spectroscopy. Radiology. 1988;166(2):533–539. [DOI] [PubMed] [Google Scholar]
- 217.Rabi II, Millman S, Kusch P, Zacharias JR. The molecular beam resonance method for measuring nuclear magnetic moments. The magnetic moments of 3Li6, 3Li7 and 9F19. Physical Review. 1939;55(6):526–535. [Google Scholar]
- 218.Bloembergen N, Purcell EM, Pound RV. Relaxation effects in nuclear magnetic resonance absorption. Physical Review. 1948;73(7):679–712. [Google Scholar]
- 219.Harris Robin K, Becker Edwin D, Cabral de Menezes Sonia M, Goodfellow R, Granger P. NMR nomenclature. Nuclear spin properties and conventions for chemical shifts (IUPAC Recommendations 2001). Pure Appl. Chem 2001; 73:1795–1818. [DOI] [PubMed] [Google Scholar]
- 220.Dona AC, Kyriakides M, Scott F, et al. A guide to the identification of metabolites in NMR-based metabonomics/metabolomics experiments. Computational and structural biotechnology journal. 2016;14:135–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Reo NV. NMR-based metabolomics. Drug and Chemical Toxicology. 2002;25(4):375–382. [DOI] [PubMed] [Google Scholar]
- 222.Gillies RJ, Morse DL. In vivo magnetic resonance spectroscopy in cancer. Annual Review of Biomedical Engineering. 2005;7(1):287–326. [DOI] [PubMed] [Google Scholar]
- 223.Li Y, Park I, Nelson SJ. Imaging tumor metabolism using in vivo magnetic resonance spectroscopy. The Cancer Journal. 2015;21(2):123–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Mahon MM, Williams AD, Soutter WP, et al. 1H magnetic resonance spectroscopy of invasive cervical cancer: an in vivo study with ex vivo corroboration. NMR in Biomedicine. 2004;17(1):1–9. [DOI] [PubMed] [Google Scholar]
- 225.Kim J-K, Kim D-Y, Lee Y-H, et al. In vivo differential diagnosis of prostate cancer and benign prostatic hyperplasia: localized proton magnetic resonance spectroscopy using external-body surface coil. Magnetic Resonance Imaging. 1998;16(10):1281–1288. [DOI] [PubMed] [Google Scholar]
- 226.Le HC, Lupu M, Kotedia K, Rosen N, Solit D, Koutcher JA. Proton MRS detects metabolic changes in hormone sensitive and resistant human prostate cancer models CWR22 and CWR22r. Magnetic Resonance in Medicine. 2009;62(5):1112–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Kinoshita Y, Yokota A. Absolute concentrations of metabolites in human brain tumors using in vitro proton magnetic resonance spectroscopy. NMR in Biomedicine. 1997;10(1):2–12. [DOI] [PubMed] [Google Scholar]
- 228.Dowling C, Bollen AW, Noworolski SM, et al. Preoperative proton MR spectroscopic imaging of brain tumors: correlation with histopathologic analysis of resection specimens. American Journal of Neuroradiology. 2001;22(4):604–612. [PMC free article] [PubMed] [Google Scholar]
- 229.Albers MJ, Krieger MD, Gonzalez-Gomez I, et al. Proton-decoupled 31P MRS in untreated pediatric brain tumors. Magnetic Resonance in Medicine. 2005;53(1):22–29. [DOI] [PubMed] [Google Scholar]
- 230.Glaholm J, Leach MO, Collins DJ, et al. In-vivo 31P magnetic resonance spectroscopy for monitoring treatment response in breast cancer. The Lancet. 1989;333(8650):1326–1327. [DOI] [PubMed] [Google Scholar]
- 231.Kristjansen PEG, Spang-Thomsen M, Quistorff B. Different energy metabolism in two human small cell lung cancer subpopulations examined by 31P magnetic resonance spectroscopy and biochemical analysis in vivo and in vitro. Cancer Research. 1991;51(19):5160–5164. [PubMed] [Google Scholar]
- 232.Kalra R, Wade KE, Hands L, et al. Phosphomonoester is associated with proliferation in human breast cancer: a 31P MRS study. British Journal of Cancer. 1993;67(5):1145–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Kurhanewicz J, Bok R, Nelson SJ, Vigneron DB. Current and Potential Applications of Clinical 13C MR Spectroscopy. Journal of Nuclear Medicine 2008;49(3):341–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Rivenzon-Segal D, Margalit R, Hadassa D. Glycolysis as a metabolic marker in orthotopic breast cancer, monitored by in vivo 13C MRS. American Journal of Physiology-Endocrinology and Metabolism. 2002;283(4):E623–E630. [DOI] [PubMed] [Google Scholar]
- 235.Halliday KR, Fenoglio-Preiser C, Sillerud LO. Differentiation of human tumors from nonmalignant tissue by natural-abundance 13C NMR spectroscopy. Magnetic Resonance in Medicine. 1988;7(4):384–411. [DOI] [PubMed] [Google Scholar]
- 236.Ferguson JE, Hulse P, Lorigan P, Jayson G, Scarffe JH. Continuous infusion of 5-fluorouracil with alpha 2b interferon for advanced colorectal carcinoma. British Journal of Cancer. 1995;72(1):193–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Mutlib A, Espina R, Atherton J, et al. Alternate strategies to obtain mass balance without the use of radiolabeled compounds: Application of quantitative fluorine (19F) Nuclear Magnetic Resonance (NMR) spectroscopy in metabolism studies. Chemical Research in Toxicology. 2012;25(3):572–583. [DOI] [PubMed] [Google Scholar]
- 238.Raychaudhuri B Imaging spectroscopy: Origin and future trends. Applied Spectroscopy Reviews. 2016;51(1):23–35. [Google Scholar]
- 239.Iqbal Z, Wilson NE, Thomas MA. 3D spatially encoded and accelerated TE-averaged echo planar spectroscopic imaging in healthy human brain. NMR in Biomedicine. 2016;29(3):329–339. [DOI] [PubMed] [Google Scholar]
- 240.Kurhanewicz J, Vigneron DB, Nelson SJ. Three-dimensional magnetic resonance spectroscopic imaging of brain and prostate cancer. Neoplasia (New York, NY). 2000;2(1–2):166–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Faghihi R, Zeinali-Rafsanjani B, Mosleh-Shirazi MA, et al. Magnetic resonance spectroscopy and its clinical applications: A Review. Journal of Medical Imaging and Radiation Sciences. 2017;48(3):233–253. [DOI] [PubMed] [Google Scholar]
- 242.Montemezzi S, Cavedon C, Camera L, et al. 1H MR spectroscopy of suspicious breast mass lesions at 3T: A clinical experience. La radiologia medica. 2017;122(3):161–170. [DOI] [PubMed] [Google Scholar]
- 243.Belle JL, Harris NG, Williams SR, Bhakoo KK. A comparison of cell and tissue extraction techniques using high-resolution 1H-NMR spectroscopy. NMR in Biomedicine. 2002;15(1):37–44. [DOI] [PubMed] [Google Scholar]
- 244.Bathen TF, Sitter B, Sjøbakk TE, Tessem MB, Gribbestad I. Magnetic resonance metabolomics of intact tissue: a biotechnological tool in cancer diagnostics and treatment evaluation. Cancer Research. 2010;70(17):6692–6696. [DOI] [PubMed] [Google Scholar]
- 245.Cheng LL, Lean CL, Bogdanova A, et al. Enhanced resolution of proton NMR spectra of malignant lymph nodes using magic-angle spinning. Magnetic Resonance in Medicine. 1996;36(5):653–658. [DOI] [PubMed] [Google Scholar]
- 246.Andrew ER. The narrowing of NMR spectra of solids by high-speed specimen rotation and the resolution of chemical shift and spin multiplet structures for solids. Progress in Nuclear Magnetic Resonance Spectroscopy. 1971;8(1):1–39. [Google Scholar]
- 247.Andrew ER, Bradbury A, Eades RG. Removal of dipolar broadening of nuclear magnetic resonance spectra of solids by specimen rotation. Nature. 1959;183:1802. [Google Scholar]
- 248.Haukaas TH, Moestue SA, Vettukattil R, et al. Impact of freezing delay time on tissue samples for metabolomic studies. Frontiers in oncology. 2016;6:17–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Moestue S, Sitter B, Bathen TF, Tessem MB, Gribbestad IS. HR MAS MR spectroscopy in metabolic characterization of cancer. Current Topics in Medicinal Chemistry. 2011;11(1):2–26. [DOI] [PubMed] [Google Scholar]
- 250.Sitter B, Lundgren S, Bathen TF, Halgunset J, Fjosne HE, Gribbestad IS. Comparison of HR MAS MR spectroscopic profiles of breast cancer tissue with clinical parameters. NMR in Biomedicine. 2006;19(1):30–40. [DOI] [PubMed] [Google Scholar]
- 251.Haukaas TH, Euceda LR, Giskeødegård GF, Bathen TF. Metabolic portraits of breast cancer by HRMAS MR spectroscopy of intact tissue samples. Metabolites. 2017;7(2):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Tkáč I, Starčuk Z, Choi IY, Gruetter R. In vivo 1H NMR spectroscopy of rat brain at 1 ms Echo time. Magnetic Resonance in Medicine. 1999;41(4):649–656. [DOI] [PubMed] [Google Scholar]
- 253.Kalovidouri A, Firmenich N, Delattre BMA, et al. Fat suppression techniques for breast MRI: Dixon versus spectral fat saturation for 3D T1-weighted at 3 T. La radiologia medica. 2017;122(10):731–742. [DOI] [PubMed] [Google Scholar]
- 254.Tkáč I, Henry PG, Andersen P, Keene CD, Low WC, Gruetter R. Highly resolved in vivo 1H NMR spectroscopy of the mouse brain at 9.4 T. Magnetic Resonance in Medicine. 2004;52(3):478–484. [DOI] [PubMed] [Google Scholar]
- 255.Glunde K, Bhujwalla ZM. Metabolic tumor imaging using magnetic resonance spectroscopy. Seminars in oncology. 2011;38(1):26–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Park JM, Park JH. Human in vivo 31P MR spectroscopy of benign and malignant breast tumors. Korean Journal of Radiology. 2001;2(2):80–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Wijnen JP, van der Kemp WJ, Luttje MP, Korteweg MA, Luijten PR, Klomp DW. Quantitative 31P magnetic resonance spectroscopy of the human breast at 7 T. Magnetic Resonance in Medicine. 2012;68(2):339–348. [DOI] [PubMed] [Google Scholar]
- 258.Wijnen JP, Jiang L, Greenwood TR, van der Kemp W, Klomp DW, Glunde K. 1H/31P polarization transfer at 9.4 Tesla for improved specificity of detecting phosphomonoesters and phosphodiesters in breast tumor models. PLOS ONE. 2014;9(7):e102256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Mancini L, Payne GS, Leach MO. Comparison of polarization transfer sequences for enhancement of signals in clinical 31P MRS studies. Magnetic Resonance in Medicine. 2003;50(3):578–588. [DOI] [PubMed] [Google Scholar]
- 260.Wijnen JP, Klomp DWJ, Nabuurs CIHC, et al. Proton observed phosphorus editing (POPE) for in vivo detection of phospholipid metabolites. NMR in Biomedicine. 2016;29(9):1222–1230. [DOI] [PubMed] [Google Scholar]
- 261.Klomp DWJ, van de Bank BL, Raaijmakers A, et al. 31P MRSI and 1H MRS at 7 T: initial results in human breast cancer. NMR in Biomedicine. 2011;24(10):1337–1342. [DOI] [PubMed] [Google Scholar]
- 262.Novak J, Wilson M, MacPherson L, Arvanitis TN, Davies NP, Peet AC. Clinical protocols for 31P MRS of the brain and their use in evaluating optic pathway gliomas in children. European Journal of Radiology. 2014;83(2):e106–e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Vettukattil R, Gulati M, Sjøbakk TE, et al. Differentiating diffuse World Health Organization grade II and IV astrocytomas with ex vivo magnetic resonance spectroscopy. Neurosurgery. 2012;72(2):186–195. [DOI] [PubMed] [Google Scholar]
- 264.Chung Y-L, Troy H, Banerji U, et al. Magnetic Resonance Spectroscopic pharmacodynamic markers of the heat shock protein 90 inhibitor 17-allylamino,17-demethoxygeldanamycin (17AAG) in human colon cancer models. JNCI: Journal of the National Cancer Institute. 2003;95(21):1624–1633. [DOI] [PubMed] [Google Scholar]
- 265.Chung Y-L, Troy H, Kristeleit R, et al. Noninvasive magnetic resonance spectroscopic pharmacodynamic markers of a novel histone deacetylase inhibitor, LAQ824, in human colon carcinoma cells and xenografts. Neoplasia. 2008;10(4):303–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Huang MQ, Nelson DS, Pickup S, et al. In vivo monitoring response to chemotherapy of human diffuse large B-cell lymphoma xenografts in SCID mice by 1H and 31P MRS. Academic Radiology. 2007;14(12):1531–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Smits GA, Heerschap A, Oosterhof GO, et al. Early metabolic response to high energy shock waves in a human tumor kidney xenograft monitored by 31P magnetic resonance spectroscopy. Ultrasound in Medicine and Biology. 1991;17(8):791–801. [DOI] [PubMed] [Google Scholar]
- 268.Vaupel P, Okunieff P, Kallinowski F, Neuringer LJ. Correlations between 31P-NMR spectroscopy and tissue O2 tension measurements in a murine fibrosarcoma. Radiation Research. 1989;120(3):477–493. [PubMed] [Google Scholar]
- 269.Cornel E, Smits GA, Oosterhof G, et al. Characterization of human prostate cancer, benign prostatic hyperplasia and normal prostate by in vitro 1H and 31P Magnetic Resonance Spectroscopy. The Journal of Urology. 1993;150(6):2019–2024. [DOI] [PubMed] [Google Scholar]
- 270.Cao M, Giskeødegård G, Bathen T, et al. Prognostic value of metabolic response in breast cancer patients receiving neoadjuvant chemotherapy. BMC Cancer. 2012;12(1):39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Sitter B, Bathen TF, Singstad TE, et al. Quantification of metabolites in breast cancer patients with different clinical prognosis using HR MAS MR spectroscopy. NMR in Biomedicine. 2010;23(4):424–431. [DOI] [PubMed] [Google Scholar]
- 272.Rocha CM, Barros AS, Gil AM, et al. Metabolic profiling of human lung cancer tissue by 1H high resolution magic angle spinning (HRMAS) NMR spectroscopy. Journal of Proteome Research. 2010;9(1):319–332. [DOI] [PubMed] [Google Scholar]
- 273.Duarte IF, Rocha CM, Barros AS, et al. Can nuclear magnetic resonance (NMR) spectroscopy reveal different metabolic signatures for lung tumours? Virchows Archiv. 2010;457(6):715–725. [DOI] [PubMed] [Google Scholar]
- 274.Wright AJ, Fellows GA, Griffiths R, Wilson M, Bell BA, Howe FA. Ex-vivo HRMAS of adult brain tumours: metabolite quantification and assignment of tumour biomarkers. Molecular Cancer. 2010;9(1):66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Hekmatyar SK, Wilson M, Jerome N, et al. ¹H nuclear magnetic resonance spectroscopy characterisation of metabolic phenotypes in the medulloblastoma of the SMO transgenic mice. British journal of cancer. 2010;103(8):1297–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Cuellar-Baena S, Morales JM, Martinetto H, et al. Comparative metabolic profiling of paediatric ependymoma, medulloblastoma and pilocytic astrocytoma. International Journal of Molecular Medicine. 2010;26(6941–948). [DOI] [PubMed] [Google Scholar]
- 277.McKnight TR, Smith KJ, Chu PW, et al. Choline metabolism, proliferation, and angiogenesis in nonenhancing grades 2 and 3 astrocytoma. Journal of Magnetic Resonance Imaging. 2011;33(4):808–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.García-Álvarez I, Garrido L, Romero-Ramírez L, Nieto-Sampedro M, Fernández-Mayoralas A, Campos-Olivas R. The effect of antitumor glycosides on glioma cells and tissues as studied by proton HR-MAS NMR spectroscopy. PloS one. 2013;8(10):e78391–e78391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Swanson MG, Keshari KR, Tabatabai ZL, et al. Quantification of choline- and ethanolamine-containing metabolites in human prostate tissues using 1H HR-MAS total correlation spectroscopy. Magnetic Resonance in Medicine. 2008;60(1):33–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Bayet-Robert M, Loiseau D, Rio P, et al. Quantitative two-dimensional HRMAS 1H-NMR spectroscopy-based metabolite profiling of human cancer cell lines and response to chemotherapy. Magnetic Resonance in Medicine. 2010;63(5):1172–1183. [DOI] [PubMed] [Google Scholar]
- 281.Madhu B, Shaw GL, Warren AY, Neal DE, Griffiths JR. Response of Degarelix treatment in human prostate cancer monitored by HR-MAS 1H NMR spectroscopy. Metabolomics. 2016;12(7):120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Mahon MM, deSouza NM, Dina R, et al. Preinvasive and invasive cervical cancer: an ex vivo proton magic angle spinning magnetic resonance spectroscopy study. NMR in Biomedicine. 2004;17(3):144–153. [DOI] [PubMed] [Google Scholar]
- 283.Mahon MM, Cox IJ, Dina R, et al. 1H magnetic resonance spectroscopy of preinvasive and invasive cervical cancer: In vivo–ex vivo profiles and effect of tumor load. Journal of Magnetic Resonance Imaging. 2004;19(3):356–364. [DOI] [PubMed] [Google Scholar]
- 284.De Silva SS, Payne GS, Thomas V, Carter PG, Ind TEJ, deSouza NM. Investigation of metabolite changes in the transition from pre-invasive to invasive cervical cancer measured using 1H and 31P magic angle spinning MRS of intact tissue. NMR in Biomedicine. 2009;22(2):191–198. [DOI] [PubMed] [Google Scholar]
- 285.Lyng H, Sitter B, Bathen TF, et al. Metabolic mapping by use of high-resolution magic angle spinning 1H MR spectroscopy for assessment of apoptosis in cervical carcinomas. BMC Cancer. 2007;7(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Martínez-Bisbal MC, Monleon D, Assemat O, et al. Determination of metabolite concentrations in human brain tumour biopsy samples using HR-MAS and ERETIC measurements. NMR in Biomedicine. 2009;22(2):199–206. [DOI] [PubMed] [Google Scholar]
- 287.Esteve V, Celda B, Martínez-Bisbal MC. Use of 1H and 31P HRMAS to evaluate the relationship between quantitative alterations in metabolite concentrations and tissue features in human brain tumour biopsies. Analytical and Bioanalytical Chemistry. 2012;403(9):2611–2625. [DOI] [PubMed] [Google Scholar]
- 288.van Zijl PC, Yadav NN. Chemical exchange saturation transfer (CEST): What is in a name and what isn’t? Magnetic Resonance in Medicine. 2011;65(4):927–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Chan KW, Jiang L, Cheng M, et al. CEST‐MRI detects metabolite levels altered by breast cancer cell aggressiveness and chemotherapy response. NMR in Biomedicine. 2016;29(6):806–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Jae‐Seung L, Ding X, Alexej J, Ravinder RR. In vitro study of endogenous CEST agents at 3 T and 7 T. Contrast Media & Molecular Imaging. 2016;11(1):4–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Zhang S, Keupp J, Wang X, et al. Z-spectrum appearance and interpretation in the presence of fat: Influence of acquisition parameters. Magnetic Resonance in Medicine. 2018;79(5):2731–2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Zhang S, Seiler S, Wang X, et al. CEST-Dixon for human breast lesion characterization at 3 T: A preliminary study. Magnetic Resonance in Medicine. 2018;80(3):895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Ardenkjaer-Larsen J, Fridlund B, Gram A, et al. Increase in signal-to-noise ratio of > 10,000 times in liquid-state NMR. Proceedings of the National Academy of Sciences. 2003;100(18):10158–10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Keshari KR, Sriram R, Koelsch BL, et al. Hyperpolarized 13C pyruvate magnetic resonance reveals rapid lactate export in metastatic renal cell carcinomas. Cancer research. 2013;73(2):529–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Allouche‐Arnon H, Gamliel A, Barzilay CM, et al. A hyperpolarized choline molecular probe for monitoring acetylcholine synthesis. Contrast Media & Molecular Imaging. 2011;6(3):139–147. [DOI] [PubMed] [Google Scholar]
- 296.Katz-Brull R, Koudinov AR, Degani H. Direct detection of brain acetylcholine synthesis by magnetic resonance spectroscopy. Brain Research. 2005;1048(1):202–210. [DOI] [PubMed] [Google Scholar]
- 297.Glunde K, Jie C, Bhujwalla ZM. Molecular causes of the aberrant choline phospholipid metabolism in breast cancer. Cancer Research. 2004;64(12):4270–4276. [DOI] [PubMed] [Google Scholar]
- 298.Friesen-Waldner LJ, Wade TP, Thind K, et al. Hyperpolarized choline as an MR imaging molecular probe: Feasibility of in vivo imaging in a rat model. Journal of Magnetic Resonance Imaging. 2015;41(4):917–923. [DOI] [PubMed] [Google Scholar]
- 299.Salamanca-Cardona L, Keshari KR. 13C-labeled biochemical probes for the study of cancer metabolism with dynamic nuclear polarization-enhanced magnetic resonance imaging. Cancer & Metabolism. 2015;3(1):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Kurhanewicz J, Vigneron DB, Brindle K, et al. Analysis of cancer metabolism by imaging hyperpolarized nuclei: prospects for translation to clinical research. Neoplasia. 2011;13(2):81–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Singletary SE, Connolly JL. Breast cancer staging: working with the sixth edition of the AJCC Cancer Staging Manual. CA: A Cancer Journal for Clinicians. 2006;56(1):37–47. [DOI] [PubMed] [Google Scholar]
- 302.Mountain CF. Revisions in the international system for staging lung cancer. Chest. 1997;111(6):1710–1717. [DOI] [PubMed] [Google Scholar]
- 303.Schröder FH, Hermanek P, Denis L, Fair WR, Gospodarowicz MK, Pavone-Macaluso M. The TNM classification of prostate cancer. The Prostate. 1992;21(S4):129–138. [DOI] [PubMed] [Google Scholar]
- 304.Gunderson LL, Jessup JM, Sargent DJ, Greene FL, Stewart AK. Revised TN categorization for colon cancer based on national survival outcomes data. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010;28(2):264–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Reding KW, Bernstein JL, Langholz BM, et al. Adjuvant systemic therapy for breast cancer in BRCA1/BRCA2 mutation carriers in a population-based study of risk of contralateral breast cancer. Breast Cancer Research and Treatment. 2010;123(2):491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Schrijver WAME, Suijkerbuijk KPM van Gils CH, van der Wall E, Moelans CB, van Diest PJ. Receptor conversion in distant breast cancer metastases: A systematic review and meta-analysis. Journal of the National Cancer Institute. 2018;110(6):568–580. [DOI] [PubMed] [Google Scholar]
- 307.Borghaei H, Smith MR, Campbell KS. Immunotherapy of cancer. European Journal of Pharmacology. 2009;625(1):41–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Kim JH, Min SJ, Jang HJ, Cho JW, Kim SH, Kim HS. Comparison of RECIST 1.0 and RECIST 1.1 in patients with metastatic cancer: A pooled analysis. Journal of Cancer. 2015;6(4):387–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Palmer MK. WHO handbook for reporting results of cancer treatment. British Journal of Cancer. 1982;45(3):484–485. [Google Scholar]
- 310.Hylton NM, Blume JD, Bernreuter WK, et al. Locally advanced breast cancer: MR imaging for prediction of response to neoadjuvant chemotherapy—results from ACRIN 6657/I-SPY trial. Radiology. 2012;263(3):663–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Jacobs MA, Stearns V, Wolff AC, et al. Multiparametric magnetic resonance imaging, spectroscopy and multinuclear(23Na) imaging monitoring of preoperative chemotherapy for locally advanced breast cancer. Academic Radiology. 2010;17(12):1477–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Tozaki M, Oyama Y, Fukuma E. Preliminary study of early response to neoadjuvant chemotherapy after the first cycle in breast cancer: comparison of 1H magnetic resonance spectroscopy with diffusion magnetic resonance imaging. Japanese Journal of Radiology. 2010;28(2):101–109. [DOI] [PubMed] [Google Scholar]
- 313.Fisher B, Bryant J, Wolmark N, et al. Effect of preoperative chemotherapy on the outcome of women with operable breast cancer. Journal of Clinical Oncology. 1998;16(8):2672–2685. [DOI] [PubMed] [Google Scholar]
- 314.von Minckwitz G, Raab G, Caputo A, et al. Doxorubicin with cyclophosphamide followed by docetaxel every 21 days compared with doxorubicin and docetaxel every 14 days as preoperative treatment in operable breast cancer: the GEPARDUO study of the German Breast Group. Journal of Clinical Oncology. 2005;23(12):2676–2685. [DOI] [PubMed] [Google Scholar]
- 315.Danishad KKA, Sharma U, Sah RG, Seenu V, Parshad R, Jagannathan NR. Assessment of therapeutic response of locally advanced breast cancer (LABC) patients undergoing neoadjuvant chemotherapy (NACT) monitored using sequential magnetic resonance spectroscopic imaging (MRSI). NMR in Biomedicine. 2010;23(3):233–241. [DOI] [PubMed] [Google Scholar]
- 316.Drisis S, Flamen P, Ignatiadis M, et al. Total choline quantification measured by 1H MR spectroscopy as early predictor of response after neoadjuvant treatment for locally advanced breast cancer: The impact of immunohistochemical status. Journal of Magnetic Resonance Imaging. 2018;0(0). [DOI] [PubMed] [Google Scholar]
- 317.Abramov Y, Carmi S, Anteby SO, & Ringel I, Ex vivo 1H and 31P magnetic resonance spectroscopy as a means for tumor characterization in ovarian cancer patients. Oncology Reports. 2013;29:321–328. [DOI] [PubMed] [Google Scholar]
- 318.Esmaeili M, Bathen TF, Engebråten O, Mælandsmo GM, Gribbestad IS, Moestue SA. Quantitative 31P HR-MAS MR spectroscopy for detection of response to PI3K/mTOR inhibition in breast cancer xenografts. Magnetic Resonance in Medicine. 2014;71(6):1973–1981. [DOI] [PubMed] [Google Scholar]
- 319.Wu HC, Chang DK, Huang CT. Targeted therapy for cancer. Journal of Cancer Molecules. 2006;2(2):57–66. [Google Scholar]
- 320.den Hollander P, Savage M, Brown P. Targeted therapy for breast cancer prevention. Frontiers in Oncology. 2013;3(250). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Hong S, Didwania A, Olopade O, Ganschow P. The expanding use of third-generation aromatase inhibitors: what the general internist needs to know . Journal of general internal medicine. 2009;24 Suppl 2(Suppl 2):S383–S388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Borgan E, Lindholm E, Moestue S, et al. Subtype-specific response to bevacizumab is reflected in the metabolome and transcriptome of breast cancer xenografts. Molecular Oncology. 2013;7(1):130–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Osborne CK, Schiff R. Mechanisms of endocrine resistance in breast cancer. Annual review of medicine. 2011;62:233–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Groenendijk FH, Bernards R. Drug resistance to targeted therapies: déjà vu all over again. Molecular oncology. 2014;8(6):1067–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Dyke JP, Zakian KL, Spees WM, et al. Metabolic response of the CWR22 prostate tumor xenograft after 20 Gy of radiation studied by 1H spectroscopic imaging. Clinical Cancer Research. 2003;9(12):4529–4536. [PubMed] [Google Scholar]
- 326.He X-H, Li W-T, Gu Y-J, et al. Metabonomic studies of pancreatic cancer response to radiotherapy in a mouse xenograft model using magnetic resonance spectroscopy and principal components analysis. World journal of gastroenterology. 2013;19(26):4200–4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Grande S, Luciani AM, Rosi A, et al. Radiation effects on soluble metabolites in cultured HeLa cells examined by 1H MRS: Changes in concentration of glutathione and of lipid catabolites induced by gamma rays and proton beams. International Journal of Cancer. 2001;96(S1):27–42. [DOI] [PubMed] [Google Scholar]
- 328.Tessier AG, Yahya A, Larocque MP, Fallone BG, Syme A. Longitudinal evaluation of the metabolic response of a tumor xenograft model to single fraction radiation therapy using magnetic resonance spectroscopy. Physics in Medicine & Biology. 2014;59(17):5061. [DOI] [PubMed] [Google Scholar]
- 329.Suppiah S, Rahmat K, Mohd-Shah MN, et al. Improved diagnostic accuracy in differentiating malignant and benign lesions using single-voxel proton MRS of the breast at 3 T MRI. Clin Radiol. 2013;68(9):e502–510. [DOI] [PubMed] [Google Scholar]
- 330.Gribbestad IS, Sitter B, Lundgren S, Krane J, Axelson D. Metabolite composition in breast tumors examined by proton nuclear magnetic resonance spectroscopy. Anticancer research. 1999;19(3A):1737–1746. [PubMed] [Google Scholar]
- 331.Dong Z, Liu Y, Scott KF, et al. Secretory phospholipase A2-IIa is involved in prostate cancer progression and may potentially serve as a biomarker for prostate cancer. Carcinogenesis. 2010;31(11):1948–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Iorio E, Ricci A, Bagnoli M, et al. Activation of phosphatidylcholine cycle enzymes in human epithelial ovarian cancer cells. Cancer research. 2010;70(5):2126–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Rocha CM, Barros AS, Goodfellow BJ, et al. NMR metabolomics of human lung tumours reveals distinct metabolic signatures for adenocarcinoma and squamous cell carcinoma. Carcinogenesis. 2015;36(1):68–75. [DOI] [PubMed] [Google Scholar]
- 334.Nordstrom A, Broom O, Witek B. Metabolomics reveals the Glycerophosphodiester Phosphodiesterase GDPD5 to regulate an invasive phenotype in lung cancer cells and potentially constitute a novel drug target. The FASEB Journal. 2016;30(1_supplement):629.621–629.621. [Google Scholar]
- 335.Thotala D, Craft JM, Ferraro DJ, et al. Cytosolic phospholipaseA2 inhibition with PLA-695 radiosensitizes tumors in lung cancer animal models. PLoS One. 2013;8(7):e69688–e69688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Bennett DT, Deng X-S, Yu JA, et al. Cancer stem cell phenotype is supported by secretory phospholipase A2 in human lung cancer cells. The Annals of thoracic surgery. 2014;98(2):439–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Feng C, Zhang L, Sun Y, et al. GDPD5, a target of miR-195–5p, is associated with metastasis and chemoresistance in colorectal cancer. Biomedicine & Pharmacotherapy. 2018;101:945–952. [DOI] [PubMed] [Google Scholar]
- 338.Buhmeida A, Bendardaf R, Hilska M, et al. PLA2 (group IIA phospholipase A2) as a prognostic determinant in stage II colorectal carcinoma. Annals of Oncology. 2009;20(7):1230–1235. [DOI] [PubMed] [Google Scholar]
- 339.Wendum D, Comperat E, Boëlle PY, et al. Cytoplasmic phospholipase A2 alpha overexpression in stromal cells is correlated with angiogenesis in human colorectal cancer. Modern Pathology. 2004;18:212. [DOI] [PubMed] [Google Scholar]