Abstract
The dysregulation of ubiquitin-mediated proteasomal degradation has emerged as an important mechanism of pathogenesis in several cancers. The Speckle-type POZ Protein (SPOP) functions as a substrate adaptor for the cullin3-RING ubiquitin ligase and controls the cellular persistence of a diverse array of protein substrates in hormone signaling, epigenetic control, and cell cycle regulation, to name a few. Mutations in SPOP and the resulting dysregulation of this proteostatic pathway play causative roles in the pathogenesis of prostate and endometrial cancers, whereas overexpression and mislocalization are associated with kidney cancer. Understanding the molecular mechanism of the normal function of SPOP as well as the cause of SPOP-mediated oncogenesis is thus critical for eventual therapeutic targeting of SPOP and other related pathways. Here, we will review SPOP structure, function and the molecular mechanism of how this function is achieved. We will then review how mutations and protein mislocalization contribute to cancer pathogenesis and will provide a perspective on how SPOP may be targeted therapeutically.
Keywords: SPOP, ubiquitination, ubiquitin ligase, prostate cancer, endometrial cancer, intrinsically disordered proteins, liquid-liquid phase separation
Graphical Abstract

SPOP is the substrate adaptor of the Cullin3-RING ubiquitin ligase (CRL3). SPOP recruits substrates (orange) for ubiquitination and subsequent degradation. Given its role as a tumor suppressor and proto-oncogene, SPOP inactivation, activation, amplification and mislocalization can result in oncogenesis. In this review, we discuss the mechanisms of SPOP function and how these become coopted for oncogenesis.
Introduction
The SPOP gene product Speckle-type POZ Protein (SPOP) was first identified in 1997 as a protein that exhibited a discrete speckled pattern in nuclei [1]; these punctate structures were later identified as nuclear speckles. Subsequent studies have shown that SPOP is a substrate adaptor of the cullin3-RING ubiquitin ligase (CRL3) and recruits substrates to CRL3 for ubiquitination and subsequent proteasomal degradation [2–5]. SPOP substrates include androgen receptor [6, 7], DAXX [8, 9], the BET proteins [10–12], and other important signaling cascade effectors, epigenetic modifiers, and hormone signaling effectors [13–18]; the faithful regulation of their protein levels is crucial for proper cell function. SPOP is the most frequently mutated gene in prostate cancer and is also frequently mutated in other solid tumors, such as endometrial and breast cancers; the mutation patterns and biological consequences in these cancers are different. In addition, overexpression and mislocalization of SPOP is associated with kidney cancer. Notably, SPOP was classified as an important cancer gene across 21 different types of cancers [19]. Here, we will review SPOP structure and the molecular mechanisms of its function to explain and speculate how different molecular lesions result in dysregulation and pathogenesis. We will further consider possible therapeutic interventions for these different lesions.
SPOP domain structure
Human SPOP is a 374-residue protein that is composed of three domains (Fig. 1A); the N-terminal MATH (meprin and TRAF-C homology) domain, which recognizes substrates (Fig. 1B); the central BTB (broad-complex, tramtrack and bric-a-brac) domain, which mediates SPOP dimerization as well as the interaction with the cullin3 (Cul3) complex (Fig. 1C); and the C-terminal BACK (BTB and C-terminal Kelch) domain, which acts as a second dimerization domain (Fig. 1D). The synergistic dimerization of the SPOP BTB and BACK domains promote the formation of linear, higher-order SPOP oligomers [20] (Fig. 1E,F). While SPOP is one of many Cul3 substrate adaptors, it is the only known adaptor that linearly self-associates in this manner, resulting in pronounced effects on its function as reviewed below.
Figure 1: SPOP domain and complex structure.
(A) Schematic of SPOP domain structure. (B) Crystal structures of the SPOP MATH domain (green ribbon representation) with substrate peptides reveal an extended substrate binding site. Superposition of the MATH/Puc peptide (yellow peptide, PDB code 3HQL) and the MATH/human PDX (orange peptide, PDB code 6F8F) complexes reveals similar interactions [5, 22]. (C) Crystal structure of the SPOP BTB domain dimer. The two-fold symmetric dimer (represented with one monomer in red and the other gray) is the Cul3 interaction site. (D) Crystal structure of the C-terminal SPOP BACK domain dimer (represented with one monomer in blue and the other gray, PDB code 4HS2) [28]. (E) A schematic diagram of the domain structure of SPOP monomer (left) and SPOP oligomer (far right). The linear SPOP oligomer schematic is colored based on panel A. The concentration dependent association of SPOP dimers through BACK domain interactions indicates the possibility for indefinite self-association [8]. (F) Model of SPOP oligomer created through superposition of known crystal structures: SPOP, 3HQI [5] and 4HS2 [28]. (G) Model of SPOP/Cul3 oligomer created through superposition of the SPOP/cullin-3 (PDB code 4EOZ [26]) and cullin-1/Rbx1/UbcH5 (PDB code 1LDK [59]) crystal structures with corresponding domains from the SPOP oligomer shown in panel F. The central SPOP octamer is colored as in panel A and the cullin component of the complex is colored in orange.
The MATH domain is the core substrate recognition domain of SPOP and the location of the majority of cancer associated mutations [21]. The domain consists of a sandwich of two antiparallel β-sheets that binds substrates on a long shallow groove across one face of one of the β-sheets; here, short linear motifs of substrates, termed SPOP-binding (SB) motifs, are specifically recognized (Fig. 1B). The SB motif is a five-residue motif with the consensus sequence ϕ-π-S-S/T-S/T, where ϕ is a nonpolar and π is a polar amino acid. The available crystal structures of the MATH domain bound to substrate fragments show that an individual MATH binding cleft accommodates twelve amino acids of the substrate [5]. Affinity- and specificity-providing residues line this substrate binding cleft. As a consequence, prostate cancer-related SPOP mutations occur in this region, e.g. the frequently mutated residues W131 and F133 in the binding cleft cradle the hydrophobic residue in SB motifs. These mutations ultimately result in impaired binding affinity and increased persistence of substrates in cells [21]. While the SB motif is central to substrate recognition and targeting, emerging structural data suggests that extensions of the core SB motif contribute to the favorable interactions between SPOP and substrate [22]. In addition, multivalent substrates can engage several MATH domains simultaneously, as inferred from biophysical data for substrates with higher multivalency that bind oligomeric SPOP [20, 23].
The central BTB domain of SPOP (Fig. 1C) is a common structural element found in zinc finger transcription factors as well as in Cul3 substrate adaptors [24], and the human genome contains ~205 genes that encode BTB domains [25]. BTB domains are often associated with other interaction domains, but the combination of BTB and MATH domains is rare in the human proteome and occurs only in SPOP and in its homolog SPOP-like (SPOPL) [24].
The BTB domain generates a SPOP dimer that is able to engage two SB motifs within a single substrate sequence [5]. A SPOP dimer also engages two Cul3 molecules resulting in a 2:2 complex [5, 26]. The N-terminal adaptor binding domain in Cul3 binds to SPOP in a manner that highlights the structural homology among Cul1, Cul2 and Cul5 RING ligases [5]. While, all Cul3 substrate adaptors recruit substrates to the cullin single-handedly, this function is split between two proteins in other CRLs; in Cul1 RING ligases (also called SCF complexes), Skp1 connects Cul1 and the F-box protein substrate adaptor; in Cul5 RING ligases, EloC connects Cul5 to the SOCS-box protein substrate adaptor [27].
BTB and BACK domain combinations are found in over 50 human proteins [24] but only the BACK domains in SPOP and SPOPL have an atypical truncation; this allows dimerization of the SPOP BACK domain, whereas this function is lost through an insertion in the SPOPL BACK domain. BACK domain-mediated homodimerization of SPOP occurs via a less extended interface with a weaker dissociation constant than BTB dimerization [20, 28] (Fig. 1). Together, the BTB and BACK domains synergistically mediate SPOP self-association [26] into linear, higher-order oligomers (Fig. 1F), whose size distribution is directly related to SPOP concentration [20]. An increase in SPOP concentration leads to a shift of the size distribution to larger oligomers, which are always in equilibrium with small oligomers and dimers.
Importantly, each BTB domain in a SPOP oligomer recruits a Cul3 complex, generating an oligomeric CRL3 that is highly multivalent for substrates and has multiple catalytic centers [20, 26] (Fig. 1G). The BTB and BACK domain-mediated multivalency for substrates is critical for SPOP function, as we discuss below.
SPOP substrates and multivalency
The tumor suppressor activities of SPOP, as well as the tumorigenic activities of SPOP mutants, are directly tied to its ability to bind and target a growing number of substrates for proteosomal degradation (Table 1). CRL3SPOP-mediated substrate ubiquitination is critical for the proper regulation of cell apoptosis, hormone sensing, cell proliferation and tissue patterning, to name a few. SPOP substrates include hormone signaling effectors including the androgen [6, 7], estrogen [29, 30] and progesterone receptors, as well as the hormone signaling transcriptional regulator SRC3 [11, 30, 31]. SPOP targets several transcription factors including the Gli transcription factors [18, 32], which affect tissue patterning in development, the transcription factors ERG [14, 33, 34] and BRMS1 [35], the dysregulation of which play roles in the development of breast cancer, and PDX1 [22, 36], which plays roles in diabetes. Further, SPOP affects epigenetic reading and writing through its targets BRD2, BRD3, BRD4 [10–12] and SETD2 [37]. A large-scale proteomic identification of SPOP substrates revealed an interesting set of potential additional substrates [16].
Table 1:
SPOP substrates
| Substrate | SB Motifsa | Substrate Function | Reference |
|---|---|---|---|
| Androgen Receptor | 5 | Nuclear receptor related | [6, 7] |
| Estrogen Receptor | 2 | Nuclear receptor related | [29, 30] |
| Progesterone Receptor | 5 | Nuclear receptor related | [46] |
| TRIM24: Tripartite Motif Containing 24 | 4 | Nuclear receptor related | [11, 16, 53] |
| SRC3: Steroid Receptor Coactivator | 12 | Nuclear receptor related | [11, 30, 31] |
| BRD2/3/4: Bromo Domain Containing Proteins | 2/7/8 | Epigenetic/Chromatin Remodeling | [10–12] |
| BMI1: B Lymphoma Mo-MLV Insertion Region 1 Homolog | 3 | Epigenetic/Chromatin Remodeling | [54] |
| Histone Variant MacroH2A | 2 | Epigenetic/Chromatin Remodeling | [5, 54] |
| SETD2: Histone-Lysine N-Methyltransferase | 25 | Epigenetic/Chromatin Remodeling | [37] |
| GLYR1: Glyoxylate Reductase 1 Homolog | 1 | Epigenetic/Chromatin Remodeling | [16] |
| SCAF1: SR-Related CTD Associated Factor 1 | 13 | Nucleic acid transactions | [16] |
| WIZ: Widely Interspaced Zinc Finger Motifs | 2 | Nucleic acid transactions | [16] |
| DEK: DEK Proto Oncogene | 1 | Nucleic acid Transactions | [11, 16] |
| BRMS1: Breast Cancer Metastasis-Suppressor | 1 | Transcription | [35] |
| PDX1: Pancreatic And Duodenal Homeobox | 1b | Transcription | [22, 36] |
| NANOG: Homeobox Transcription Factor | 6 | Transcription | [55] |
| CAPRIN1: Cell Cycle Associated Protein 1 | 6 | Cell Cycle | [16] |
| CDC20: Cell Division Cycle Protein 20 Homolog | 2 | Cell Cycle | [56] |
| Cyclin E1 | 2 | Cell Cycle | [57] |
| cMYC: Myc Proto-Oncogene Protein | 6 | Cell Cycle | [13] |
| DAXX: Death Domain Associated Protein | 7 | Cell Cycle | [8, 9] |
| ERG: ETS Transcription Factor | 3 | Cell Cycle (Transcription) | [14, 33, 34] |
| Gli2/Gli3: Glioma-Associated Oncogene Family Zinc Finger | 8/9 | Transcription | [18, 32] |
| DUSP6/7: Dual Specificity Phosphatase | 2/2 | Phosphatase/Cell Signaling | [15] |
| PTEN: Phosphatase and Tensin Homolog | 2 | Phosphatase/Tumor Suppressor | [15] |
| INF2: Inverted Formin, FH2 and WH2 Domain Containing | 5 | Cytoskeleton | [58] |
The five residue motif is defined here as either [GAVLIMWFPC]-[STCYNQDEHR]-[ST]-[STCYNQDEHR]-[ST] or [GAVLIMWFPC]-[STCYNQDEHR]-[ST]-[ST]-[STCYNQDEHR], which includes a single site mismatch in the fourth and fifth positions
PDX1 has two mismatches from the search motif.
SB motifs are linear in nature and are thus typically localized in intrinsically disordered regions (IDRs) of SPOP substrates, which ensures that they are accessible for binding to SPOP. Indeed, many of the identified substrates contain long IDRs and multiple SB motifs (Table 1), to the extent that this seems to be a typical property of SPOP substrates. Many of these SB motifs interact only weakly with the MATH domain and have dissociation constants in the range of several micromolar to low millimolar [8, 23]. However, even such weak motifs have been shown to contribute to substrate ubiquitination [23] in the context of multivalent substrate-SPOP interactions and, therefore, shape cellular function.
SPOP oligomerization enhances function
SPOP is the only CRL substrate adaptor that is known to self-associate indefinitely in a concentration-dependent manner and to form linear, higher-order oligomers. In fact, SPOP oligomerization is under evolutionary pressure as was shown by covariation analysis of a large number of SPOP orthologs [8]. This result indicates that the ability to form linear SPOP oligomers has been encoded in SPOP orthologs during evolution, suggesting the functional relevance of multivalency. Yet, this finding raises the question as to the underlying biological function of this linear self-association.
Given that many SPOP substrates comprise multiple SB motifs as discussed above, it is intuitive to propose that multivalent binding between SPOP and substrates impacts function. Indeed, several lines of evidence reveal the molecular basis for why SPOP oligomers have a higher activity for substrate ubiquitination than SPOP monomers or dimers. The evidence is as follows: (i) SPOP oligomerization enhances its affinity to substrates with multiple SB motifs via simultaneous engagement of the motifs by multiple MATH domains in a SPOP oligomer by avidity effects [23]. (ii) SPOP-mediated ubiquitination of substrates is enhanced by SPOP oligomerization in vitro [20, 23, 26] and in cells [8] (Fig. 2A). In fact, SPOP mutants that can only form monomers or dimers mediate nearly exclusively (multi-)monoubiquitination, i.e. the transfer of single ubiquitin moieties onto substrate lysines rather than the formation of polyubiquitin chains. SPOP oligomers mediate multiple ubiquitin transfers to create polyubiquitin chains, likely due to the longer residence time of substrates on multivalent SPOP compared to SPOP dimers or monomers [23]. (iii) Multivalency on the substrate side, i.e. the presence of multiple SB motifs in substrates, also contributes to polyubiquitination [23]. Together, this evidence shows strong support for a model in which SPOP oligomerization enhances the effectiveness of substrate ubiquitination (Fig. 2B).
Figure 2: Functional implications of SPOP oligomerization.
(A) Higher-order oligomeric SPOP ubiquitinates substrates more effectively than SPOP dimers or monomers. In vitro ubiquitination assays with CRL3SPOP and a fragment of Gli3 as a substrate (residues 1–455). Comparison of WT SPOP and self-association defective mutants mutBTB (mutations L186D, L190D, L193D, I217K), mutBACK (mutation Y353E) and a combination of both, mutBTB/BACK. All SPOP versions comprise residues 28–359. (Panel reprinted with permission from [20]). (B) Model of the role of multivalent interactions between oligomeric SPOP and multiple SB motifs in a single substrate molecule. Dimeric SPOP recruits substrates with low affinity and is shown to miss suitable steric access to lysine acceptor sites on the substrate or on ubiquitin. Oligomeric CRL3SPOP binds substrates with enhanced affinity via avidity effects and mediates effective polyubiquitination through multiple catalytic centers in the oligomeric CRL3. A SPOP tetramer is shown for clarity. SB motifs are depicted as pink bars, and the color saturation decreases for weaker motifs. (Adopted from [23] with permission.) (C) Phase separation via multivalent interactions. SPOP and DAXX undergo phase separation in vitro. Fluorescence microscopy images of a mixture of SPOP (green fluorescent, residues 28–359) and cDAXX (red fluorescent, residues 495–740). (D) The SPOP prostate cancer mutant W131G is defective for co-localization with DAXX in HeLa cells; WT SPOP colocalizes with DAXX in nuclear bodies that are distinct from nuclear speckles. SC-35 (magenta) marks nuclear speckles. (Adopted from [8] with permission.) (E) Schematic illustration of the role of phase separation in SPOP-mediated substrate turnover. SPOP phase separates with multivalent substrates and is able to target and ubiquitinate substrates localized to membrane-less organelles. SPOP cancer mutants are defective at phase separation and therefore co-localization and ubiquitination.
Importantly, the role of oligomerization for SPOP function opens the door to an interesting manner of regulation through changes in oligomer size. The close SPOP homolog SPOP-like (SPOPL) has a nearly identical BTB interface to that of SPOP but has an insertion in the BACK domain that prevents BACK-mediated dimerization [26]. SPOPL can form homodimers and SPOP-SPOPL heterodimers via its BTB domain, but its inability to dimerize via the BACK domain effectively caps SPOP oligomers. Addition of SPOPL thus decreases the size of SPOP oligomers in a SPOPL concentration-dependent manner. This size decrease is accompanied by a concomitant decrease in ubiquitination efficiency, again demonstrating the importance of SPOP oligomerization for its activity [26]. We note the possibility that SPOP oligomerization may be further influenced by other factors including post-translational modification of the SPOP self-association interfaces and the expression levels of multivalent substrates, which can stabilize SPOP oligomers in a velcro-like fashion [8].
SPOP oligomers can undergo liquid-liquid phase separation with substrates
The multivalent interactions between SPOP oligomers and substrates with multiple SB motifs have an additional potential outcome; they mediate liquid-liquid phase separation (LLPS) above a threshold, or so-called saturation concentration. Below this saturation concentration, multivalent SPOP and substrate form higher-order oligomers, i.e. their size and stoichiometry vary. Above the saturation concentration, these higher-order oligomers form a dense phase that typically appears as viscous, liquid-like droplets (Fig. 2C). The left-over light phase is characterized by a low protein concentration. Individual protein molecules in the dense phase can typically enter and leave on a second timescale [8].
LLPS is now accepted as a critical mechanism underlying the formation of membrane-less organelles in cells, or so-called biomolecular condensates [38]. The ability to undergo LLPS can also mediate the recruitment of proteins to such biomolecular condensates [39, 40]. Indeed, the same interactions that drive phase separation between SPOP and substrates in vitro mediate the colocalization of SPOP and substrates in biomolecular condensates in the nucleus, suggesting that phase separation operates as a mechanism to organize SPOP function in cells. Below the saturation concentration, SPOP is localized in nuclear speckles [1]. Recent work has shown that one SPOP substrate, DAXX, which is typically localized in PML bodies, co-localizes with SPOP in nuclear SPOP/DAXX bodies above a certain concentration (Fig. 2D) [8].
The SPOP/DAXX bodies are viscous, liquid-like and show evidence of ubiquitination activity. (i) They recruit additional components of the ubiquitination machinery such as Cul3, (ii) they contain conjugated ubiquitin, and (iii) the presence of conjugated ubiquitin depends on the ability of SPOP to recruit Cul3 to the bodies. If mutations are introduced that prevent recruitment of Cul3 to SPOP, the level of conjugated ubiquitin in the condensates decreases and cellular DAXX levels increase concomitantly [8]. Together, these observations suggest that the SPOP/DAXX bodies are active for CRL3SPOP-mediated DAXX ubiquitination (Fig. 2E).
The formation of active SPOP/substrate condensates via phase separation may be advantageous by setting a maximum threshold level for substrates, above which they are degraded. Phase separation may thus be an effective mechanism for maintaining proteostasis. Another possibility is that phase separation is simply required for cellular organization. Many SPOP substrates seem to be components of biomolecular condensates. Targeting such substrates may be difficult because biomolecular condensates typically exclude many cellular constituents. Recruiting a ubiquitin ligase to biomolecular condensates via phase separation with a substrate that is already localized there may be a straightforward strategy to solve this problem.
Whether phase separation is absolutely required for SPOP function remains to be determined, but phase separation is disrupted by loss-of-function prostate cancer mutations in SPOP (Fig. 2D,E) [8]. Thus, prostate cancer mutations do not only interfere with binding of substrates to SPOP but also with their concomitant phase separation and colocalization in cells.
SPOP oligomerization as source of dominant-negative effects
SPOP is an important tumor suppressor in prostate cancer [13, 16] and across 21 different types of cancers [19]. Tumor suppressor alleles typically follow the two-hit rule, i.e. both alleles must be functionally inactivated to drive tumorigenesis. Prostate cancer patients with SPOP mutations, however, do not usually display loss of heterozygosity, i.e. the second allele is not inactivated. These findings imply that SPOP mutants have dominant-negative effects on WT SPOP molecules present in the same cell, presumably via the assembly of mixed oligomers formed by both mutant and WT SPOP [16]. These mixed oligomers must, by extension, have reduced activity to result in disease, e.g. because their valency for SB motifs in substrates is reduced.
In transgenic flies that expressed human SPOP variants in the developing fly, a SPOP mutant that was deficient in assembly via the BTB domain resulted in dominant-negative effects. In this case, the dominant-negative effects manifest as a Hedgehog gain-of-function phenotype due to reduced turnover of the Drosophila Gli ortholog, Ci [20]. No obvious dominant-negative effects were observed from SPOP prostate cancer mutants, but the reason is unclear. A possible explanation may be the inefficient assembly of mixed human and fly oligomers and therefore a subtler phenotype compared to the mutant that caps oligomer size. Nevertheless, these observations call for the systematic characterization of the molecular mechanisms of dominant-negative effects via SPOP oligomerization.
SPOP mutations in different cancers
Whether different types of cancers associated with SPOP mutations are driven by distinct mutational patterns has not been investigated thoroughly, but initial observations suggest that this is the case. Mutational inactivation of SPOP can lead to increased levels of critical SPOP substrates such as AR in prostate cancer. In contrast, endometrial cancer seems to be associated with mutations that activate SPOP activity towards some substrates. Overexpression and mislocalization of SPOP is associated with kidney cancer. We will summarize the details of the different molecular lesions and their effects below.
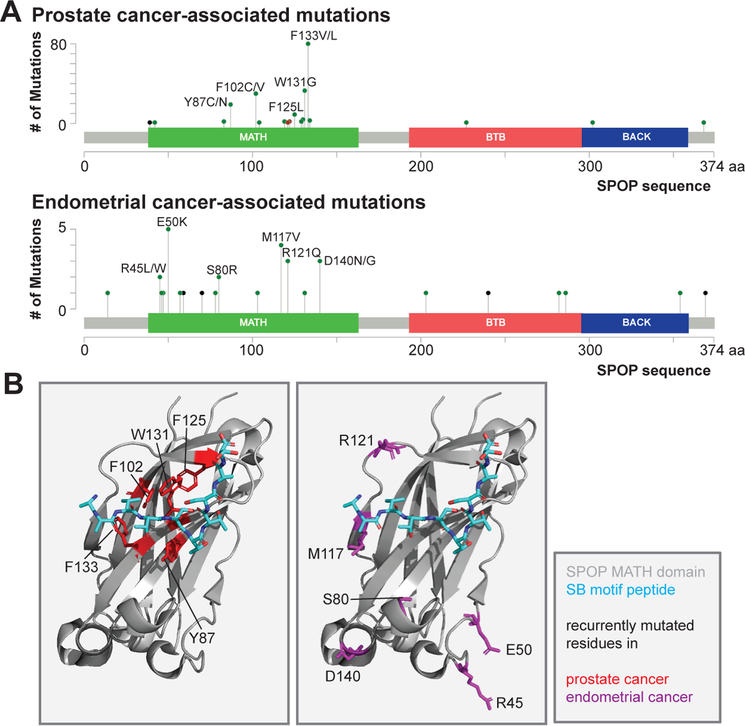
SPOP is mutated in roughly 10% of prostate cancer patients (according to data in cBio portal [41]). Several studies now directly demonstrate the role of SPOP mutations in the dysregulation of the ubiquitylome and, in turn, the progression of prostate cancer [6, 7, 16, 31]. Indeed, in the background of prostate cancer SPOP mutants a number of oncogenic substrates are stabilized due to decreased ubiquitination. These SPOP mutations are recurrent missense mutations in a specific set of amino acids (Fig. 3A) clustered in the substrate binding cleft of the SPOP MATH domain (Fig. 3B). These mutants reduce substrate binding affinity and substrate ubiquitination [6, 8, 10, 31]. Prostate cancer related SPOP mutants lead to increased cellular persistence of key oncogenic substrates, such as the androgen receptor, DEK, TRIM24, SRC3 and the BRD proteins [11, 16]. The prostate cancer SPOP mutants also increase the substrate concentration required for phase separation with SPOP [8] leading to a lack of colocalization of SPOP and substrates in cells (Figure 2D).
Figure 3: Distinct sets of SPOP missense mutations in different cancer types.
(A) Lollipop plots with mutation site and number in SPOP in prostate cancer (top) and endometrial cancer (bottom). Data from cBio portal [41]. Green, red and black lollipops indicate missense mutations, short in-frame insertions/deletions and truncations, respectively. (B) Ribbon diagram of SPOP MATH domain with recurrent missense mutations in prostate cancer (left) and endometrial cancer (right).
It was recently shown that the prostate cancer-related SPOP mutations contribute to genomic instability. Cells with SPOP mutations favor use of the relatively error-prone non-homologous end joining (NHEJ) DNA damage pathway opposed to the higher fidelity homologous recombination (HR) pathway [42]. The molecular mechanism of this change in pathway is not entirely clear but likely related to the inability of SPOP mutants to turn over a substrate in DNA damage loci. The genomic rearrangements resulting from error-prone DNA damage repair may be an important driving force in cancer pathogenesis.
SPOP has recurrent missense mutations in roughly 5% of endometrial cancer patients (cBio portal [41], and [43]). While the well-studied prostate cancer mutations are located in the substrate binding cleft of the MATH domain, the endometrial cancer mutations are mostly located outside of the cleft on the substrate-binding face of the MATH domain. These mutations are rarer but are absent in databases of normal SNPs. Recent work has reported that these mutations decrease ubiquitination of some substrates, such as TRIM24 and ER [29], while they enhance ubiquitination of others, such as the BET proteins and the AGR3 and NCOA3/SRC3 oncogenes [11]. This was unexpected because prostate cancer mutations decrease ubiquitination of all substrates, as far as is known, and result in loss of binding to individual SB motifs [21]. The recent findings on endometrial cancer mutants [11] suggest that they may instead enhance the affinity to individual SB motifs, depending on their sequence, with a concomitant change in SPOP substrate specificity, and that this may contribute to endometrial cancer pathogenesis. The underlying molecular mechanism of this change in activity towards different substrates remains unclear.
In clear cell renal cell carcinoma (ccRCC), SPOP expression is induced as a direct transcriptional target of hypoxia-inducible factors (HIFs) [15] and is overexpressed 99% of the time [44]. Furthermore, SPOP mislocalizes to the cytoplasm, where it targets proteins that contain SB motifs but are not usually SPOP substrates [15]. These targets include PTEN and other dual-specificity phosphatases (DUSPs). The dysregulated targeting of substrates through cytoplasmic accumulation of SPOP shifts the normally tumor suppressing activity of SPOP toward antiapoptotic and pro-proliferative [44], and is sufficient to induce tumorigenesis in kidney cells. Indeed, RNAi-mediated silencing of SPOP results in apoptosis of renal cell cancer cells [45].
SPOP is amplified in 5.5% of breast cancer cases and likely contributes to cancer pathogenesis by targeting nuclear hormone receptors for degradation [46]. SPOP is thus important for proteostasis and, depending on the context, can act as a tumor suppressor as well as an oncogene.
Can SPOP-related cancers be targeted therapeutically?
Given that SPOP plays important initiating or sustaining roles in several cancers, specific therapeutic interventions against the distinct molecular lesions are ultimately called for (Fig. 4). The strategies for such therapeutic interventions depend critically on whether SPOP’s tumor suppressor function is inactivated or whether it acts as a de novo oncogene in the cancer in question. Thus, we will summarize potential therapeutic strategies for different cancer types which arise from the above presented insights into the mechanisms of SPOP function.
Figure 4: Possibilities for therapeutic interventions in SPOP-related cancers.
SPOP dysfunction plays key roles in the cancer pathogenesis of subsets of patients with prostate, endometrial, breast cancer and ccRC. In prostate cancer, loss-of-function SPOP mutations lead to accumulation of substrates [6, 10, 31], which could be targeted via PROTACs. These mutants also favor the use of non-homologous end joining (NHEJ) instead of homologous recombination (HR) as the DNA damage response, potentially rendering combinations with PARP inhibitors useful [42]. In endometrial cancer, gain-of-function SPOP mutations lead to enhanced ubiquitination and turnover of BET proteins BRD2, BRD3 and BRD4, rendering cells sensitive to BET inhibitors [11]. In breast cancer, the often observed SPOP amplification could make a SPOP inhibitor useful [52]. In ccRC, hypoxia leads to HIF-mediated SPOP induction and mislocalization to the cytoplasm, where CRL3SPOP mediates ubiquitination and subsequent degradation of tumor suppressors PTEN, DUSP6 and DUSP7 [44]. A SPOP inhibitor could prevent this turnover [52]. Lastly, SPOP mutations may perturb the driving force for phase separation of SPOP with substrates [8], and small molecules could be used to readjust it.
In SPOP mutant-mediated prostate cancer, the tumor suppressor function of SPOP is mutationally inactivated and proto-oncogenic substrates including androgen receptor accumulate in the cell. Re-activation of the non-active SPOP would be a worthwhile goal but may be technically challenging. However, Proteolysis Targeting Chimeras (PROTACs [47]) and similar warheads, i.e. chimeric small molecules that can recruit ubiquitin ligases to critical oncogenic substrates for catalytic ubiquitination and subsequent turnover, may prove to be an effective therapy. Indeed, PROTACs against androgen receptor are under development [48–50]. Given that prostate cancer SPOP mutations reroute DNA damage repair to the error-prone NHEJ, PARP inhibitors may lead to synthetic lethality and could be a highly effective treatment. Classical small-molecule inhibitors of androgen signaling pathways are in clinical use and will remain being useful. These include direct androgen receptor antagonists, because of the interdependence of androgen signaling and other pathways can also include androgen synthesis inhibitors, HSP90 inhibitors and PI3K pathway inhibitors (for a review refer to [51]).
A small molecule inhibitor of SPOP that inhibits SPOP/substrate interactions has been reported [52]. Such an inhibitor could be useful in cancers with SPOP gain-of-function lesions from mutations, amplification or mislocalization, e.g. it could be used in ccRCC to prevent PTEN degradation [44]. If increased turnover of a subset of SPOP substrates by endometrial cancer SPOP mutants proves to contribute to tumorigenesis, SPOP inhibitors could also be used in endometrial cancer patients. Interestingly, it may be possible to take advantage of increased BET protein turnover by administering BET inhibitors and creating synthetic lethality, as recently suggested [11].
Lastly, while the characterization of SPOP/substrate phase separation is in its infancy, perturbation of phase separation to decrease substrate turnover could counteract the effects of SPOP amplification or activating mutations on substrate ubiquitination. Substoichiometric levels of SPOP inhibitors could achieve this goal and given the role of SPOP/SPOP and substrate/substrate interactions in mediating SPOP/substrate phase separation, additional small-molecule modulators of these protein interactions could achieve this goal. More research in this direction is required to evaluate the potential of this strategy.
Outlook
Recent progress in our understanding of the molecular mechanisms of SPOP function and dysfunction has been rapid. While SPOP was initially recognized as one of many ubiquitin ligase substrate adaptors, the recognition of its relevance soared when large-scale genome sequencing efforts revealed recurrent mutations in SPOP in cancer patients. More and more substrates have been identified, and these may contribute to disease in the context of SPOP mutations. These advances have been important for our understanding of SPOP’s role in health and disease. However, new evidence has raised new mechanistic questions. Different cancers seem to involve distinct mutations; what are their underlying molecular driving forces? Why does SPOP act as a tumor suppressor under some conditions, but as an oncogene under others? And since this context-dependence is likely related to regulation of SPOP function, how are SPOP levels regulated transcriptionally, via protein destabilization and by post-translational modification? Dysregulation of which substrates tips the balance towards oncogenesis? These questions call for the detailed biophysical and structural characterization of SPOP/substrate interactions to improve our understanding of the mechanisms of SPOP function in health and disease and to facilitate the development of targeted therapies.
Acknowledgements
We thank Nafiseh Sabri, Jill Bouchard, and Grace Usher for comments on the manuscript and insightful discussions. T.M. acknowledges funding by NIH grant R01GM112846, St. Jude Children’s Research Hospital, and the American Lebanese Syrian Associated Charities.
Abbreviations
- SPOP
Speckle-type POZ Protein
- CRL
cullin RING ligase
- CRL3
cullin3 RING ligase
- DAXX
Death Domain Associated Protein
- BET
Bromodomain and Extra-Terminal Domain
- MATH
meprin and TRAF-C homology
- BTB
broad-complex, tramtrack and bric-a-brac
- BACK
BTB and C-terminal Kelch
- SRC3
Steroid Receptor Coactivator
- IDR
intrinsically disordered region
- LLPS
liquid-liquid phase separation
- LOH
loss of heterozygosity
- ccRCC
clear cell renal cell carcinoma
- PTEN
Phosphatase and Tensin Homolog
- PROTAC
Proteolysis Targeting Chimeras
- NHEJ
non-homologous end joining
- HR
homologous recombination
Footnotes
Conflict of Interest: The authors report no conflict of interest.
References
- 1.Nagai Y, Kojima T, Muro Y, Hachiya T, Nishizawa Y, Wakabayashi T & Hagiwara M (1997) Identification of a novel nuclear speckle-type protein, SPOP, FEBS Lett. 418, 23–6. [DOI] [PubMed] [Google Scholar]
- 2.Kwon JE, La M, Oh KH, Oh YM, Kim GR, Seol JH, Baek SH, Chiba T, Tanaka K, Bang OS, Joe CO & Chung CH (2006) BTB domain-containing speckle-type POZ protein (SPOP) serves as an adaptor of Daxx for ubiquitination by Cul3-based ubiquitin ligase, J Biol Chem. 281, 12664–72. [DOI] [PubMed] [Google Scholar]
- 3.Hernandez-Munoz I, Lund AH, van der Stoop P, Boutsma E, Muijrers I, Verhoeven E, Nusinow DA, Panning B, Marahrens Y & van Lohuizen M (2005) Stable X chromosome inactivation involves the PRC1 Polycomb complex and requires histone MACROH2A1 and the CULLIN3/SPOP ubiquitin E3 ligase, Proc Natl Acad Sci U S A. 102, 7635–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kent D, Bush EW & Hooper JE (2006) Roadkill attenuates Hedgehog responses through degradation of Cubitus interruptus, Development. 133, 2001–10. [DOI] [PubMed] [Google Scholar]
- 5.Zhuang M, Calabrese MF, Liu J, Waddell MB, Nourse A, Hammel M, Miller DJ, Walden H, Duda DM, Seyedin SN, Hoggard T, Harper JW, White KP & Schulman BA (2009) Structures of SPOP-substrate complexes: insights into molecular architectures of BTB-Cul3 ubiquitin ligases, Mol Cell. 36, 39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.An J, Wang C, Deng Y, Yu L & Huang H (2014) Destruction of full-length androgen receptor by wild-type SPOP, but not prostate-cancer-associated mutants, Cell reports. 6, 657–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geng C, Rajapakshe K, Shah SS, Shou J, Eedunuri VK, Foley C, Fiskus W, Rajendran M, Chew SA & Zimmermann M (2014) Androgen receptor is the key transcriptional mediator of the tumor suppressor SPOP in prostate cancer, Cancer research. 74, 5631–5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bouchard JJ, Otero JH, Scott DC, Szulc E, Martin EW, Sabri N, Granata D, Marzahn MR, Lindorff-Larsen K, Salvatella X, Schulman BA & Mittag T (2018) Cancer Mutations of the Tumor Suppressor SPOP Disrupt the Formation of Active, Phase-Separated Compartments, Mol Cell. 72, 19–36 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwon JE, La M, Oh KH, Oh YM, Kim GR, Seol JH, Baek SH, Chiba T, Tanaka K & Bang OS (2006) BTB domain-containing speckle-type POZ protein (SPOP) serves as an adaptor of Daxx for ubiquitination by Cul3-based ubiquitin ligase, Journal of Biological Chemistry. 281, 12664–12672. [DOI] [PubMed] [Google Scholar]
- 10.Dai X, Gan W, Li X, Wang S, Zhang W, Huang L, Liu S, Zhong Q, Guo J & Zhang J (2017) Prostate cancer–associated SPOP mutations confer resistance to BET inhibitors through stabilization of BRD4, Nature medicine. 23, 1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Janouskova H, El Tekle G, Bellini E, Udeshi ND, Rinaldi A, Ulbricht A, Bernasocchi T, Civenni G, Losa M, Svinkina T, Bielski CM, Kryukov GV, Cascione L, Napoli S, Enchev RI, Mutch DG, Carney ME, Berchuck A, Winterhoff BJN, Broaddus RR, Schraml P, Moch H, Bertoni F, Catapano CV, Peter M, Carr SA, Garraway LA, Wild PJ & Theurillat JP (2017) Opposing effects of cancer-type-specific SPOP mutants on BET protein degradation and sensitivity to BET inhibitors, Nat Med. 23, 1046–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang P, Wang D, Zhao Y, Ren S, Gao K, Ye Z, Wang S, Pan C-W, Zhu Y & Yan Y (2017) Intrinsic BET inhibitor resistance in SPOP-mutated prostate cancer is mediated by BET protein stabilization and AKT–mTORC1 activation, Nature medicine. 23, 1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geng C, Kaochar S, Li M, Rajapakshe K, Fiskus W, Dong J, Foley C, Dong B, Zhang L, Kwon OJ, Shah SS, Bolaki M, Xin L, Ittmann M, O’Malley BW, Coarfa C & Mitsiades N (2017) SPOP regulates prostate epithelial cell proliferation and promotes ubiquitination and turnover of c-MYC oncoprotein, Oncogene. 36, 4767–4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gan W, Dai X, Lunardi A, Li Z, Inuzuka H, Liu P, Varmeh S, Zhang J, Cheng L, Sun Y, Asara JM, Beck AH, Huang J, Pandolfi PP & Wei W (2015) SPOP Promotes Ubiquitination and Degradation of the ERG Oncoprotein to Suppress Prostate Cancer Progression, Mol Cell. 59, 917–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li G, Ci W, Karmakar S, Chen K, Dhar R, Fan Z, Guo Z, Zhang J, Ke Y, Wang L, Zhuang M, Hu S, Li X, Zhou L, Li X, Calabrese MF, Watson ER, Prasad SM, Rinker-Schaeffer C, Eggener SE, Stricker T, Tian Y, Schulman BA, Liu J & White KP (2014) SPOP promotes tumorigenesis by acting as a key regulatory hub in kidney cancer, Cancer Cell. 25, 455–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Theurillat JP, Udeshi ND, Errington WJ, Svinkina T, Baca SC, Pop M, Wild PJ, Blattner M, Groner AC, Rubin MA, Moch H, Prive GG, Carr SA & Garraway LA (2014) Prostate cancer. Ubiquitylome analysis identifies dysregulation of effector substrates in SPOP-mutant prostate cancer, Science. 346, 85–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J, Bu X, Wang H, Zhu Y, Geng Y, Nihira NT, Tan Y, Ci Y, Wu F, Dai X, Guo J, Huang YH, Fan C, Ren S, Sun Y, Freeman GJ, Sicinski P & Wei W (2018) Cyclin D-CDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance, Nature. 553, 91–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Q, Shi Q, Chen Y, Yue T, Li S, Wang B & Jiang J (2009) Multiple Ser/Thr-rich degrons mediate the degradation of Ci/Gli by the Cul3-HIB/SPOP E3 ubiquitin ligase, Proc Natl Acad Sci U S A. 106, 21191–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, Meyerson M, Gabriel SB, Lander ES & Getz G (2014) Discovery and saturation analysis of cancer genes across 21 tumour types, Nature. 505, 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marzahn MR, Marada S, Lee J, Nourse A, Kenrick S, Zhao H, Ben-Nissan G, Kolaitis RM, Peters JL, Pounds S, Errington WJ, Prive GG, Taylor JP, Sharon M, Schuck P, Ogden SK & Mittag T (2016) Higher-order oligomerization promotes localization of SPOP to liquid nuclear speckles, EMBO J. 35, 1254–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, White TA, Stojanov P, Van Allen E, Stransky N, Nickerson E, Chae SS, Boysen G, Auclair D, Onofrio RC, Park K, Kitabayashi N, MacDonald TY, Sheikh K, Vuong T, Guiducci C, Cibulskis K, Sivachenko A, Carter SL, Saksena G, Voet D, Hussain WM, Ramos AH, Winckler W, Redman MC, Ardlie K, Tewari AK, Mosquera JM, Rupp N, Wild PJ, Moch H, Morrissey C, Nelson PS, Kantoff PW, Gabriel SB, Golub TR, Meyerson M, Lander ES, Getz G, Rubin MA & Garraway LA (2012) Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer, Nat Genet. 44, 685–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ostertag MS, Messias AC, Sattler M & Popowicz GM (2019) The Structure of the SPOP-Pdx1 Interface Reveals Insights into the Phosphorylation-Dependent Binding Regulation, Structure. 27, 327–334. e3. [DOI] [PubMed] [Google Scholar]
- 23.Pierce WK, Grace CR, Lee J, Nourse A, Marzahn MR, Watson ER, High AA, Peng J, Schulman BA & Mittag T (2016) Multiple Weak Linear Motifs Enhance Recruitment and Processivity in SPOP-Mediated Substrate Ubiquitination, J Mol Biol. 428, 1256–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stogios PJ, Downs GS, Jauhal JJ, Nandra SK & Prive GG (2005) Sequence and structural analysis of BTB domain proteins, Genome Biol. 6, R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, Sonnhammer ELL, Hirsh L, Paladin L, Piovesan D, Tosatto SCE & Finn RD (2019) The Pfam protein families database in 2019, Nucleic Acids Res. 47, D427–D432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Errington WJ, Khan MQ, Bueler SA, Rubinstein JL, Chakrabartty A & Prive GG (2012) Adaptor protein self-assembly drives the control of a cullin-RING ubiquitin ligase, Structure. 20, 1141–53. [DOI] [PubMed] [Google Scholar]
- 27.Zimmerman ES, Schulman BA & Zheng N (2010) Structural assembly of cullin-RING ubiquitin ligase complexes, Curr Opin Struct Biol. 20, 714–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Geersdaele LK, Stead MA, Harrison CM, Carr SB, Close HJ, Rosbrook GO, Connell SD & Wright SC (2013) Structural basis of high-order oligomerization of the cullin-3 adaptor SPOP, Acta Crystallogr D Biol Crystallogr. 69, 1677–84. [DOI] [PubMed] [Google Scholar]
- 29.Zhang P, Gao K, Jin X, Ma J, Peng J, Wumaier R, Tang Y, Zhang Y, An J, Yan Q, Dong Y, Huang H, Yu L & Wang C (2015) Endometrial cancer-associated mutants of SPOP are defective in regulating estrogen receptor-alpha protein turnover, Cell Death Dis. 6, e1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li C, Ao J, Fu J, Lee DF, Xu J, Lonard D & O’Malley BW (2011) Tumor-suppressor role for the SPOP ubiquitin ligase in signal-dependent proteolysis of the oncogenic co-activator SRC-3/AIB1, Oncogene. 30, 4350–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geng C, He B, Xu L, Barbieri CE, Eedunuri VK, Chew SA, Zimmermann M, Bond R, Shou J, Li C, Blattner M, Lonard DM, Demichelis F, Coarfa C, Rubin MA, Zhou P, O’Malley BW & Mitsiades N (2013) Prostate cancer-associated mutations in speckle-type POZ protein (SPOP) regulate steroid receptor coactivator 3 protein turnover, Proc Natl Acad Sci U S A. 110, 6997–7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Q, Zhang L, Wang B, Ou CY, Chien CT & Jiang J (2006) A hedgehog-induced BTB protein modulates hedgehog signaling by degrading Ci/Gli transcription factor, Dev Cell. 10, 719–29. [DOI] [PubMed] [Google Scholar]
- 33.Duan S & Pagano M (2015) SPOP Mutations or ERG Rearrangements Result in Enhanced Levels of ERG to Promote Cell Invasion in Prostate Cancer, Mol Cell. 59, 883–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.An J, Ren S, Murphy SJ, Dalangood S, Chang C, Pang X, Cui Y, Wang L, Pan Y, Zhang X, Zhu Y, Wang C, Halling GC, Cheng L, Sukov WR, Karnes RJ, Vasmatzis G, Zhang Q, Zhang J, Cheville JC, Yan J, Sun Y & Huang H (2015) Truncated ERG Oncoproteins from TMPRSS2-ERG Fusions Are Resistant to SPOP-Mediated Proteasome Degradation, Mol Cell. 59, 904–16. [DOI] [PubMed] [Google Scholar]
- 35.Kim B, Nam HJ, Pyo KE, Jang MJ, Kim IS, Kim D, Boo K, Lee SH, Yoon J-B & Baek SH (2011) Breast cancer metastasis suppressor 1 (BRMS1) is destabilized by the Cul3–SPOP E3 ubiquitin ligase complex, Biochemical and biophysical research communications. 415, 720–726. [DOI] [PubMed] [Google Scholar]
- 36.Claiborn KC, Sachdeva MM, Cannon CE, Groff DN, Singer JD & Stoffers DA (2010) Pcif1 modulates Pdx1 protein stability and pancreatic beta cell function and survival in mice, J Clin Invest. 120, 3713–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu K, Lei PJ, Ju LG, Wang X, Huang K, Yang B, Shao C, Zhu Y, Wei G, Fu XD, Li L & Wu M (2017) SPOP-containing complex regulates SETD2 stability and H3K36me3-coupled alternative splicing, Nucleic Acids Res. 45, 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shin Y & Brangwynne CP (2017) Liquid phase condensation in cell physiology and disease, Science. 357. [DOI] [PubMed] [Google Scholar]
- 39.Dao TP, Kolaitis RM, Kim HJ, O’Donovan K, Martyniak B, Colicino E, Hehnly H, Taylor JP & Castaneda CA (2018) Ubiquitin Modulates Liquid-Liquid Phase Separation of UBQLN2 via Disruption of Multivalent Interactions, Mol Cell. 69, 965–978 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alberti S, Gladfelter A & Mittag T (2019) Considerations and Challenges in Studying Liquid-Liquid Phase Separation and Biomolecular Condensates, Cell. 176, 419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C & Schultz N (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data, Cancer Discov. 2, 401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boysen G, Barbieri CE, Prandi D, Blattner M, Chae S-S, Dahija A, Nataraj S, Huang D, Marotz C & Xu L (2015) SPOP mutation leads to genomic instability in prostate cancer, Elife. 4, e09207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Le Gallo M, O’Hara AJ, Rudd ML, Urick ME, Hansen NF, O’Neil NJ, Price JC, Zhang S, England BM, Godwin AK, Sgroi DC, Program NIHISCCS, Hieter P, Mullikin JC, Merino MJ & Bell DW (2012) Exome sequencing of serous endometrial tumors identifies recurrent somatic mutations in chromatin-remodeling and ubiquitin ligase complex genes, Nat Genet. 44, 1310–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu J, Ghanim M, Xue L, Brown CD, Iossifov I, Angeletti C, Hua S, Negre N, Ludwig M, Stricker T, Al-Ahmadie HA, Tretiakova M, Camp RL, Perera-Alberto M, Rimm DL, Xu T, Rzhetsky A & White KP (2009) Analysis of Drosophila segmentation network identifies a JNK pathway factor overexpressed in kidney cancer, Science. 323, 1218–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu X, Sun G & Sun X (2016) RNA interference-mediated silencing of speckle-type POZ protein promotes apoptosis of renal cell cancer cells, OncoTargets and therapy. 9, 2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao K, Jin X, Tang Y, Ma J, Peng J, Yu L, Zhang P & Wang C (2015) Tumor suppressor SPOP mediates the proteasomal degradation of progesterone receptors (PRs) in breast cancer cells, American journal of cancer research. 5, 3210. [PMC free article] [PubMed] [Google Scholar]
- 47.Neklesa TK, Winkler JD & Crews CM (2017) Targeted protein degradation by PROTACs, Pharmacology & therapeutics. 174, 138–144. [DOI] [PubMed] [Google Scholar]
- 48.Salami J, Alabi S, Willard RR, Vitale NJ, Wang J, Dong H, Jin M, McDonnell DP, Crew AP & Neklesa TK (2018) Androgen receptor degradation by the proteolysis-targeting chimera ARCC-4 outperforms enzalutamide in cellular models of prostate cancer drug resistance, Communications biology. 1, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Han X, Wang C, Qin C, Xiang W, Fernandez-Salas E, Yang C-Y, Wang M, Zhao L, Xu T & Chinnaswamy K (2019) Discovery of ARD-69 as a Highly Potent Proteolysis Targeting Chimera (PROTAC) Degrader of Androgen Receptor (AR) for the Treatment of Prostate Cancer, Journal of medicinal chemistry. [DOI] [PubMed] [Google Scholar]
- 50.Raina K, Lu J, Qian Y, Altieri M, Gordon D, Rossi AMK, Wang J, Chen X, Dong H & Siu K (2016) PROTAC-induced BET protein degradation as a therapy for castration-resistant prostate cancer, Proceedings of the National Academy of Sciences. 113, 7124–7129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rathkopf D & Scher HI (2013) Androgen receptor antagonists in castration-resistant prostate cancer, Cancer J. 19, 43–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guo ZQ, Zheng T, Chen B, Luo C, Ouyang S, Gong S, Li J, Mao LL, Lian F, Yang Y, Huang Y, Li L, Lu J, Zhang B, Zhou L, Ding H, Gao Z, Zhou L, Li G, Zhou R, Chen K, Liu J, Wen Y, Gong L, Ke Y, Yang SD, Qiu XB, Zhang N, Ren J, Zhong D, Yang CG, Liu J & Jiang H (2016) Small-Molecule Targeting of E3 Ligase Adaptor SPOP in Kidney Cancer, Cancer Cell. 30, 474–484. [DOI] [PubMed] [Google Scholar]
- 53.Groner AC, Cato L, de Tribolet-Hardy J, Bernasocchi T, Janouskova H, Melchers D, Houtman R, Cato AC, Tschopp P & Gu L (2016) TRIM24 is an oncogenic transcriptional activator in prostate cancer, Cancer cell. 29, 846–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hernández-Muñoz I, Lund AH, Van Der Stoop P, Boutsma E, Muijrers I, Verhoeven E, Nusinow DA, Panning B, Marahrens Y & Van Lohuizen M (2005) Stable X chromosome inactivation involves the PRC1 Polycomb complex and requires histone MACROH2A1 and the CULLIN3/SPOP ubiquitin E3 ligase, Proceedings of the National Academy of Sciences. 102, 7635–7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang J, Chen M, Zhu Y, Dai X, Dang F, Ren J, Ren S, Shulga YV, Beca F, Gan W, Wu F, Lin YM, Zhou X, DeCaprio JA, Beck AH, Lu KP, Huang J, Zhao C, Sun Y, Gao X, Pandolfi PP & Wei W (2019) SPOP Promotes Nanog Destruction to Suppress Stem Cell Traits and Prostate Cancer Progression, Dev Cell. 48, 329–344 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu F, Dai X, Gan W, Wan L, Li M, Mitsiades N, Wei W, Ding Q & Zhang J (2017) Prostate cancer-associated mutation in SPOP impairs its ability to target Cdc20 for poly-ubiquitination and degradation, Cancer Lett. 385, 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ju L-G, Zhu Y, Long Q-Y, Li X-J, Lin X, Tang S-B, Yin L, Xiao Y, Wang X-H & Li L (2018) SPOP suppresses prostate cancer through regulation of CYCLIN E1 stability, Cell Death & Differentiation, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jin X, Wang J, Gao K, Zhang P, Yao L, Tang Y, Tang L, Ma J, Xiao J, Zhang E, Zhu J, Zhang B, Zhao SM, Li Y, Ren S, Huang H, Yu L & Wang C (2017) Dysregulation of INF2-mediated mitochondrial fission in SPOP-mutated prostate cancer, PLoS Genet. 13, e1006748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ & Pagano M (2002) Structure of the Cul1–Rbx1–Skp1–F box Skp2 SCF ubiquitin ligase complex, Nature. 416, 703. [DOI] [PubMed] [Google Scholar]