Abstract
Background
To explore whether deep convolutional neural networks (DCNNs) have the potential to improve diagnostic efficiency and increase the level of interobserver agreement in the classification of thyroid nodules in histopathological slides.
Methods
A total of 11,715 fragmented images from 806 patients’ original histological images were divided into a training dataset and a test dataset. Inception-ResNet-v2 and VGG-19 were trained using the training dataset and tested using the test dataset to determine the diagnostic efficiencies of different histologic types of thyroid nodules, including normal tissue, adenoma, nodular goiter, papillary thyroid carcinoma (PTC), follicular thyroid carcinoma (FTC), medullary thyroid carcinoma (MTC) and anaplastic thyroid carcinoma (ATC). Misdiagnoses were further analyzed.
Results
The total 11,715 fragmented images were divided into a training dataset and a test dataset for each pathology type at a ratio of 5:1. Using the test set, VGG-19 yielded a better average diagnostic accuracy than did Inception-ResNet-v2 (97.34% vs. 94.42%, respectively). The VGG-19 model applied to 7 pathology types showed a fragmentation accuracy of 88.33% for normal tissue, 98.57% for ATC, 98.89% for FTC, 100% for MTC, 97.77% for PTC, 100% for nodular goiter and 92.44% for adenoma. It achieved excellent diagnostic efficiencies for all the malignant types. Normal tissue and adenoma were the most challenging histological types to classify.
Conclusions
The DCNN models, especially VGG-19, achieved satisfactory accuracies on the task of differentiating thyroid tumors by histopathology. Analysis of the misdiagnosed cases revealed that normal tissue and adenoma were the most challenging histological types for the DCNN to differentiate, while all the malignant classifications achieved excellent diagnostic efficiencies. The results indicate that DCNN models may have potential for facilitating histopathologic thyroid disease diagnosis.
Keywords: Deep convolutional neural network (DCNN), Inception-ResNet-v2, VGG-19, thyroid nodule, diagnostic efficiency, histopathology
Introduction
Thyroid nodules are common diseases presented in the clinic, and their pathological types are complex. Thyroid nodules mainly include benign tumors and malignant tumors (i.e., thyroid cancer). Benign thyroid tumors include nodular goiter and thyroid adenoma. Thyroid cancers include papillary, follicular, medullary and anaplastic carcinomas. The differential diagnosis of thyroid nodules is crucial because thyroid carcinoma requires surgery, while only follow-up is necessary in cases of benign nodules. Pathological diagnosis of resected specimens is the gold standard for tumor diagnosis. Currently, the vast majority of pathological tissue sections are acquired by pathologists, and collections of specimens accumulated over long periods are used for clinical diagnosis. Nevertheless, manual differential diagnosis of thyroid tumor histopathological images remains a challenge for three main reasons: (I) the ability to correctly diagnose samples greatly depends on the professional background and experience of the pathologist, and such experience cannot be acquired quickly; (II) the work is tedious, expensive and time-consuming; and (III) it is challenging for the human eye to distinguish subtle changes in tissues; thus, pathologists can experience fatigue, which may lead to misdiagnosis. Thus, the precise histopathologic diagnosis of thyroid nodules remains a challenging task.
Machine learning (ML) has been increasingly used in the medical imaging field and has been applied to pathological diagnoses of different diseases (1,2). Deep convolutional neural networks (DCNNs) are a type of ML, namely, a special type of artificial neural network that resembles the multilayered human cognitive system. Many researchers have investigated applications of DCNNs for assessing pathological images (3-6). Sharma et al. developed a system that used a DCNN to classify gastric carcinoma from whole-slide images (7). A DCNN was applied to the classification of breast cancer histology images and showed a satisfactory diagnostic accuracy rate (8).
In this study, we developed DCNN automated classification systems for diagnosing thyroid nodules in histopathology images using the VGG-19 and Inception-ResNet-v2 models. This study sought to reveal whether DCCNs have the potential to improve the diagnostic efficiency and increase the level of interobserver agreement in the classification of thyroid nodules.
Methods
Patients and pathological images
This study was conducted with the approval of the ethics committee of Fudan University Shanghai Cancer Center (FUSCC). The procedures were carried out in accordance with the approved guidelines. The image dataset was acquired from 806 consecutive patients who were diagnosed with thyroid tumors and underwent primary surgery at FUSCC from January 1, 2010 to July 31, 2017. Written informed consent was obtained from all patients. The pathologic slides were stained with hematoxylin and eosin (H&E) for morphological evaluation. All the images were digitized at ×100 magnification. Each image was labeled with one of 7 classes: 0: normal tissue, 1: adenoma, 2: nodular goiter, 3: PTC, 4: follicular thyroid carcinoma (FTC), 5: medullary thyroid carcinoma (MTC) and 6: anaplastic thyroid carcinoma (ATC). The labeling was performed by two senior pathologists from FUSCC who provided a diagnosis from the overall image and preliminarily specified the area of interest used for the classification. Cases of disagreement between specialists were discarded. All the images were selected so that the pathological classification could be objectively determined from the image content.
Data augmentation
Original images should be augmented to a large degree to achieve a satisfactory classification effect. In this dataset, an automatic feature extraction program was adapted to segment original images into several patches. Nuclear features are useful for differentiating between carcinoma and non-carcinoma cells and should include single nucleus information, such as color and shape, as well as nuclei organization features, such as intensity or variability. Because the nucleus has a mottled appearance with a dark color under H&E staining, we used the Laplacian of the Gaussian function (LOG) (9) to identify the nucleus and to locate cells to automatically capture the relevant region. The procedure for segmenting one image was as follows: (I) LOG was used to detect the nuclei in H&E-stained sections; (II) one nucleus was randomly selected as the center; then, the number of nuclei in the area around it was calculated (patch range: 448×448 pixels); (III) the patch was removed after the number of nuclei exceeded a threshold T (10% of the total number of nuclei in the entire original image); (IV) each original image was automatically divided into 15 patches (Figure 1).
Figure 1.
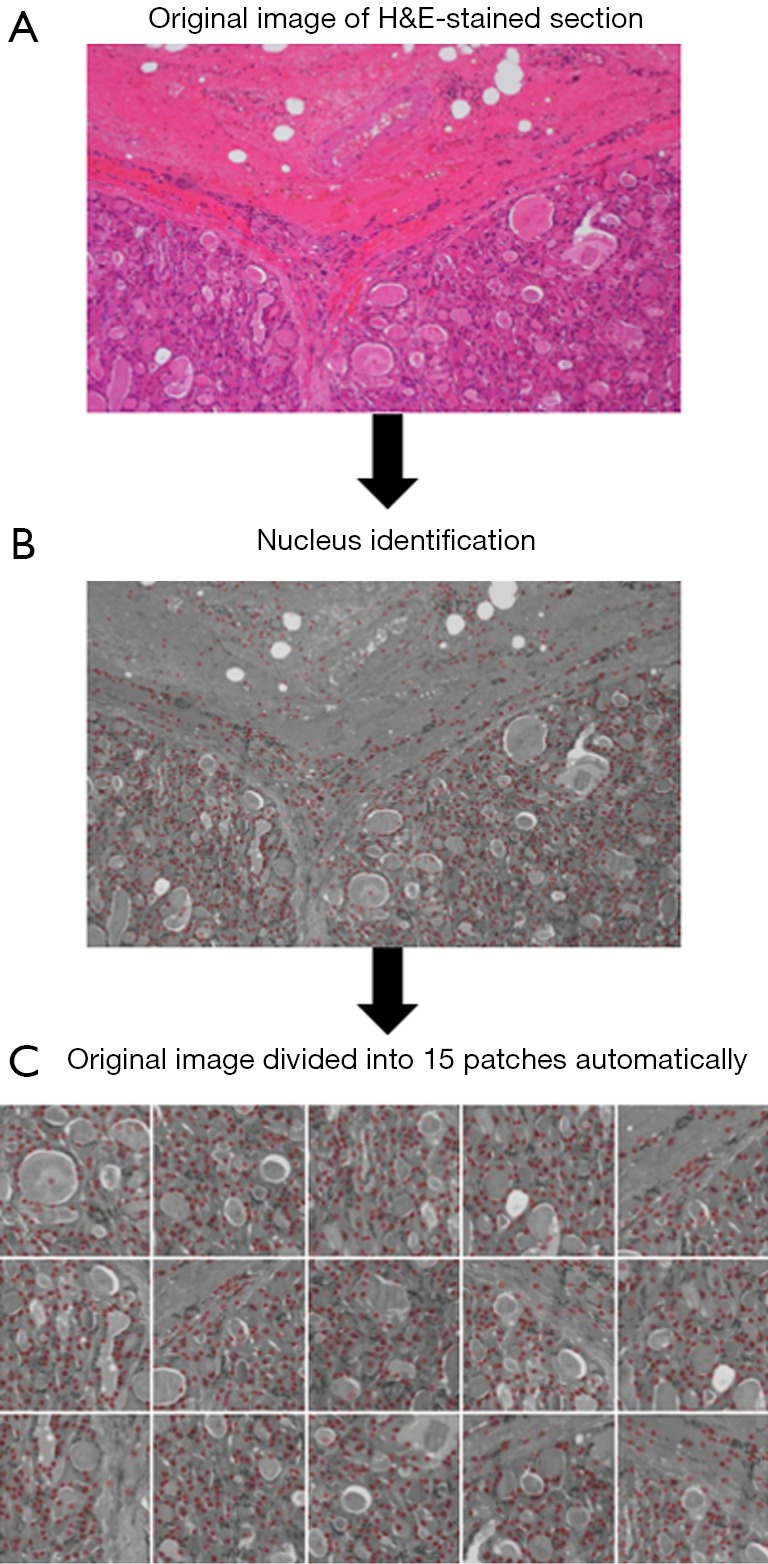
Automatic feature extraction and image segmentation program (×100). (A) An original image of the H&E stained section is input into the program. (B) LOG is used to detect nuclei which have a mottled appearance with a dark color in H&E-stained samples. (C) One nucleus is randomly selected as the center. The patch is removed after the number of nuclei exceeds a threshold T (10% of the total number of nuclei in the entire original image). Each original image was automatically divided into 15 patches.
We augmented the training data by flipping and rotating the images. Each image fragment was flipped horizontally and rotated by 0º, 90º, 180º and 270º. Through this flipping and rotating process, we increased the size of the training data eightfold. If we were instead to directly augment the training data, the required storage space would have expanded by 8 times. Thus, to save storage space, we did not augment the training data in advance but only during the training process. During each iteration of the training process, a batch of images was fetched from the training data. We randomly flipped and rotated each image in the batch. For each image, we randomly applied only one of the 8 transformations.
Network architecture
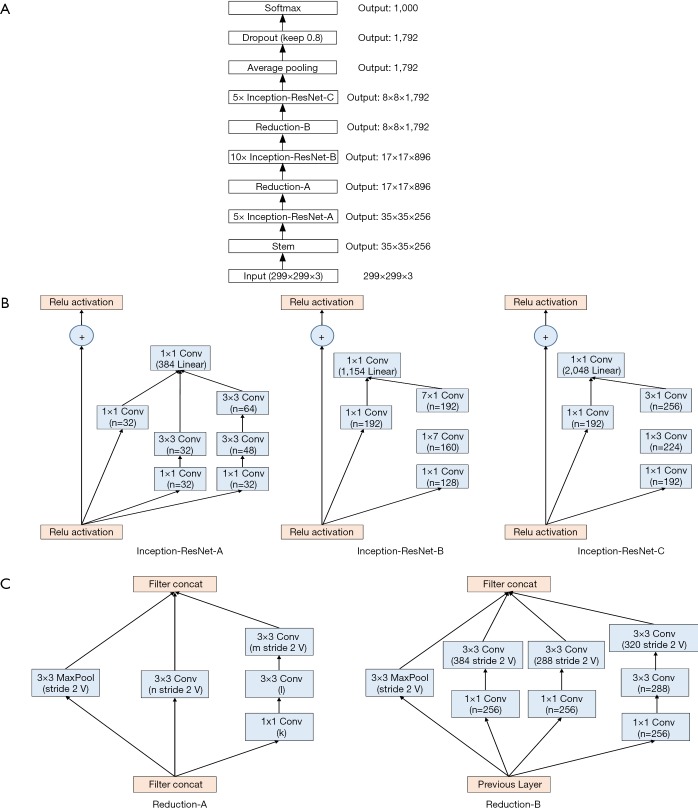
We used two DCNN models in our experiment: VGG-19 and Inception-ResNet-v2 (10). The VGG-19 architecture is described in Table 1: it comprises 16 convolutional layers and 3 fully connected layers. The convolutional layers in VGG-19 are all 3×3 convolutional layers with a stride and padding of 1; the pooling layers are all 2×2 pooling layers with a stride of 2. The default input image size in VGG-19 is 224×224. After each pooling layer, the size of the feature map is reduced by half. The last feature map before the fully connected layers is 7×7 with 512 channels, and it is expanded into a vector with 25,088 (7×7×512) channels. The VGG-19 model is shown in Figure 2.
Table 1. Architecture of the VGG-19 model.
| Layer | Patch size | Input size |
|---|---|---|
| conv×2 | 3×3/1 | 3×224×224 |
| pool | 2×2 | 64×224×224 |
| conv×2 | 3×3/1 | 64×112×112 |
| pool | 2×2 | 128×112×112 |
| conv×4 | 3×3/1 | 128×56×56 |
| pool | 2×2 | 256×56×56 |
| conv×4 | 3×3/1 | 256×28×28 |
| pool | 2×2 | 512×28×28 |
| conv×4 | 3×3/1 | 512×14×14 |
| pool | 2×2 | 512×14×14 |
| fc | 25,088×4,096 | 25,088 |
| fc | 4,096×4,096 | 4,096 |
| fc | 4,096×7 | 4,096 |
| Softmax | classifier | – |
Conv stands for the convolutional layer, pool stands for the pooling layer, and fc stands for the fully connected layer.
Figure 2.
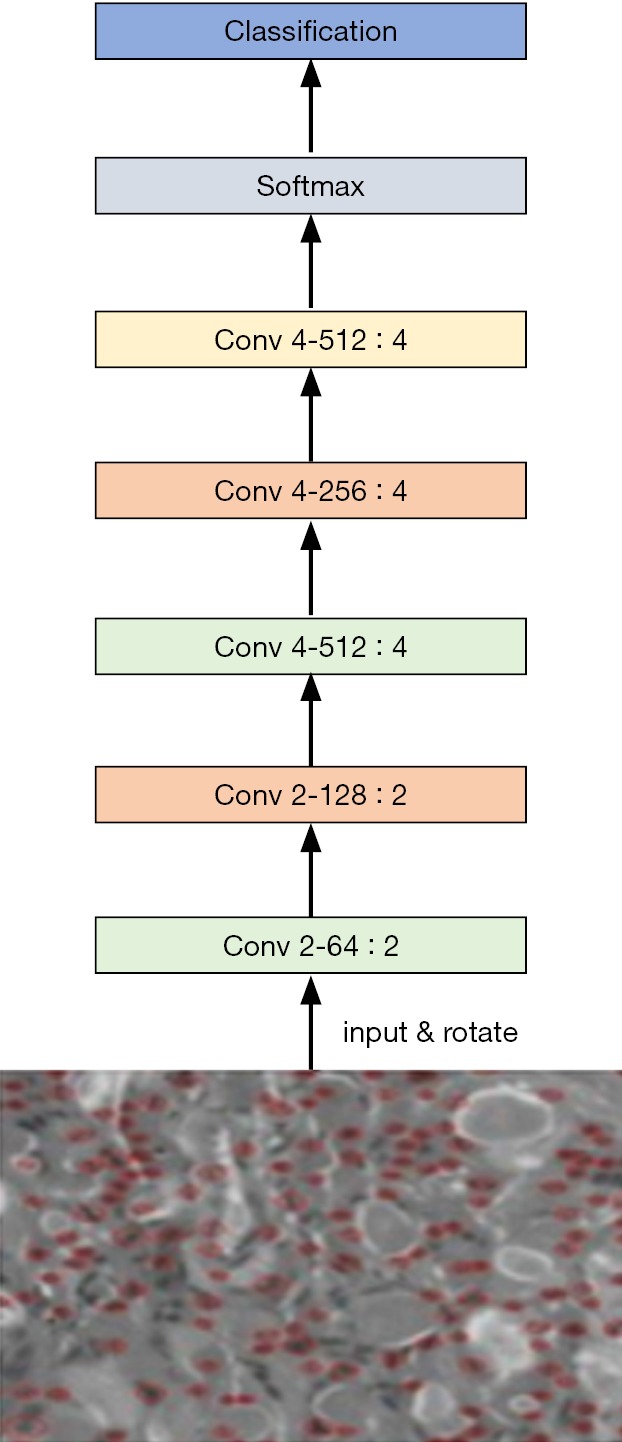
Schematic of the VGG-19 model.
The Inception-ResNet-v2 model is shown in Figure 3A, and its architecture is described in Table 2. The Inception-ResNet-v2 model included three types of inception modules: Inception-ResNet-A, Inception-ResNet-B and Inception-ResNet-C (see Figure 3B, from left to right). The inception modules are well-designed convolutional modules that both generate discriminatory features and reduce the number of parameters. Each inception module is composed of several convolutional and pooling layers in parallel. Small convolutional layers (e.g., 1×7, 7×1) are used in the inception modules to reduce the number of parameters. Two types of reduction modules also exist in Inception-ResNet-v2 that are designed to reduce the image size during training (Figure 3C). The default input size for Inception-ResNet-v2 is 299×299; thus, we resized the training data before training.
Figure 3.
Architecture of the Inception-ResNet-v2 model. (A) Schematic of the Inception-ResNet-v2 model; (B) Inception-ResNet-v2 includes of 3 types of Inception modules, labeled as A, B, C. Each inception module is composed of several convolutional layers; (C) schematic of reductions A and B, which are designed to reduce the size of the output.
Table 2. Architecture of the Inception-ResNet-v2 model.
| Layer | Patch size/stride | Input size |
|---|---|---|
| conv | 3×3/2 | 299×299×3 |
| conv | 3×3/1 | 147×147×32 |
| Filter concat | 3×3/2 pool + 3×3/2 conv | 147×147×64 |
| Filter concat | 1×1 conv, 3×3 conv + 1×1 conv, 7×1 conv, 1×7 conv, 3×3 conv | 73×73×160 |
| Filter concat | 3×3/1 conv + max pool/2 | 71×71×128 |
| Inception-ResNet-A×5 | – | 35×35×256 |
| Reduction-A | – | 35×35×256 |
| Inception-ResNet-B×10 | – | 17×17×896 |
| Reduction-B | – | 17×17×896 |
| Inception-ResNet-C×5 | – | 8×8×1,792 |
| Average pooling | 8×8/8 | 8×8×1,792 |
| dropout | keep =0.8 | 1×1×1,792 |
| fc | 1,792×1,000 | 1,792 |
| fc | 1,000×7 | 1,000 |
Conv stands for the convolutional layer, pool stands for the pooling layer, and fc stands for the fully connected layer. All layers before fc are the base part of Inception-Resnet-v2. The patch size is the kernel size of the convolutional layer, the pooling layer or the fully connected layer. Stride is the gap between two operations. The input size is the feature map input size of the layer, and the output size of each layer is the input size of the next layer. Softmax is a function for classification. Filter concat is a module that combines different conv filters and pool filters. Inception-ResNet-A, B, C and Reduction -A, B are illustrated in Figure 2.
We applied a transfer learning approach to capitalize on the generalizability of pretrained VGG-19 and Inception-ResNet-v2 models. We used models pretrained on ImageNet to acquire the initial parameters for the customized VGG-19 and Inception-ResNet-v2 used in this study. The transfer learning approach enabled us to speed up the training convergence, reduce the training time, and increase the final classification accuracy.
The output of the original VGG-19 and Inception-ResNet-v2 networks includes 1,000 classes, but our case required only 7 classes: normal tissue, PTC, ATC, MTC, FTC, goiter and adenoma. Therefore, we changed the output channel number of the last layer from 1,000 to 7. Furthermore, differences in the contrast of the images can occur due to the staining technique used to prepare the histology slides. To prevent the varying image contrasts from influencing the training process, we normalized the contrast of the training images and used average filtering to denoise the images. Specifically, we subtracted the average of the three red, green and blue (RGB) image channels for each pixel of each image.
For training, we adopted a batch training method in which each batch contained 8 pictures. One advantage of the batch training approach is that not all the training data need to be input into the neural network simultaneously; instead, the training dataset is input into the neural network in batches. This approach reduces the storage required for training and accelerates the training process. In addition, we used stochastic gradient descent (SGD) to control the rate of gradient descent. The learning rate was set to 0.001, and the dropout rate was set to 0.4. We chose a relatively small learning rate to allow the neural network to find the best global convergence point during training. The role of the dropout layer is to prevent overfitting and increase the generalizability of the trained model.
Systematic program of histological image classification
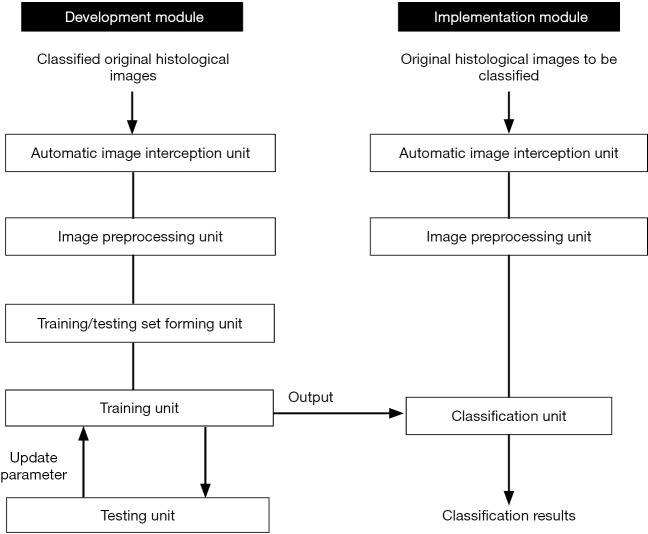
Figure 4 displays the systematic program developed for histological image classification of thyroid diseases. The model program includes a development module and an implementation module. The development module comprised an automatic image intercepting unit, an image preprocessing unit, a training/testing set forming unit, a training unit and a testing unit. The implementation module included an automatic image interception unit, a second image preprocessing unit, and a classification unit.
Figure 4.
A systematic program for histological image classification of thyroid diseases.
When constructing the DCNN model, the program detected the original classified histological images in the automatic image interception unit and then automatically generated multiple patches of 448×448 pixels, each containing a certain number of cells. Then, the patches were normalized by the image preprocessing unit. Thus, the training/testing set forming unit yielded a training set and a test set. Next, the selected and pretrained DCNN models were trained by the training unit in batches. The pretrained DCNN models were initially obtained and tested in the testing unit, and the final DCNN models were fixed when the test accuracy rate reached or exceeded a certain standard. The final models were applied in the implementation module.
When applying the final DCNN models, the original histological images were intercepted and preprocessed just as in the development module. Subsequently, these preprocessed patches were input into the final DCNN model in the classification unit for classification.
Program design and training were conducted on a desktop computer running an Ubuntu 16.04 system. The computer environment was equipped with an Ubuntu 16.04 operating system running Python, OpenCV, TensorFlow, gcc compiler, and a GTX 1080 discrete graphics GPU.
Results
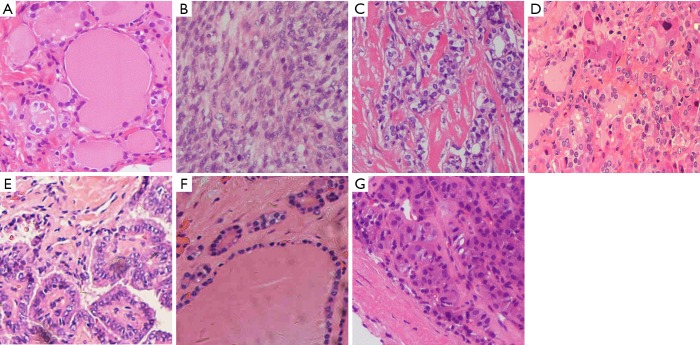
We obtained 806 H&E stained histopathology images, which included 93 images of normal tissue, 91 images of adenoma, 209 images of nodular goiter, 155 images of PTC, 76 images of FTC, 101 images of MTC and 81 images of ATC. After augmentation, 11,715 448×448 fragmented images were extracted, representing 1,125 images of normal tissue, 1,215 images of ATC, 1,110 images of FTC, 1,515 images of MTC, 2,280 images of PTC, 3,135 images of goiter and 1,335 images of adenoma. We randomly split the dataset into a training subset and a test subset for each pathology type at a ratio of 5:1. We obtained 9,796 fragmented images for training and 1,919 fragmented images for testing with no overlap from the original images, resulting in 945 normal tissue, 1,005 ATC, 930 FTC, 1,260 MTC, 1,921 PTC, 2,625 goiter and 1,110 adenoma images in the training group and 180 normal tissue, 210 ATC, 180 FTC, 255 MTC, 359 PTC, 510 goiter and 225 adenoma images in the test group (Table 3). Examples of the fragmented images from each classification are shown in Figure 5.
Table 3. Distribution of images in the dataset.
| Pathology type | Raw image | Training data | Testing data | Total |
|---|---|---|---|---|
| Normal | 93 | 945 | 180 | 1,125 |
| ATC | 81 | 1,005 | 210 | 1,215 |
| FTC | 76 | 930 | 180 | 1,110 |
| MTC | 101 | 1,260 | 255 | 1,515 |
| PTC | 155 | 1,921 | 359 | 2,280 |
| Goiter | 209 | 2,625 | 510 | 3,135 |
| Adenoma | 91 | 1,110 | 225 | 1,335 |
| Total | 806 | 9,796 | 1,919 | 11,715 |
ATC, anaplastic thyroid carcinoma; FTC, follicular thyroid carcinoma; MTC, medullary thyroid carcinoma; PTC, papillary thyroid carcinoma.
Figure 5.
Histopathological classifications of fragmented images (HE, ×100): (A) Normal tissue; (B) ATC; (C) FTC; (D) MTC; (E) PTC; (F) goiter; (G) adenoma. ATC, anaplastic thyroid carcinoma; FTC, follicular thyroid carcinoma; MTC, medullary thyroid carcinoma; PTC, papillary thyroid carcinoma.
We trained the two models on the training data and tested them on the test data. Table 4 shows the diagnostic efficiency of the VGG-19 model on the test data. When applied to the 7 pathology types, the VGG-19 model achieved a fragmentation accuracy of 88.33% for normal tissue, 98.57% for ATC, 98.89% for FTC, 100% for MTC, 97.77% for PTC, 100% for nodular goiter and 92.44% for adenoma, exhibiting an average fragmentation accuracy of 97.34%. Using only the raw images (without data augmentation), the VGG-19 model achieved an accuracy rate of 95.70% for normal tissue, 98.77% for ATC, 98.68% for FTC, 100% for MTC, 97.42% for PTC, 100% for nodular goiter and 96.70% for adenoma. The VGG-19 model exhibited an average accuracy of 98.39% on the raw images (Table 4).
Table 4. Diagnostic efficiency of VGG-19 and Inception-ResNet-v2 on the testing data.
| Pathologic classifications | VGG-19 (%) | Inception-ResNet-v2 (%) | |||
|---|---|---|---|---|---|
| Fragment | Raw image | Fragment | Raw image | ||
| Normal tissue | 88.33 (159/180) | 95.70 (89/93) | 82.22 (148/180) | 94.62 (88/93) | |
| ATC | 98.57 (207/210) | 98.77 (80/81) | 94.76 (199/210) | 95.06 (77/81) | |
| FTC | 98.89 (178/180) | 98.68 (75/76) | 95.00 (171/180) | 92.11 (70/76) | |
| MTC | 100.00 (255/255) | 100.00 (101/101) | 98.43 (251/255) | 96.04 (97/101) | |
| PTC | 97.77 (351/359) | 97.42 (151/155) | 93.31 (335/359) | 92.90 (144/155) | |
| Goiter | 100.00 (510/510) | 100.00 (209/209) | 98.43 (502/510) | 96.65 (202/209) | |
| Adenoma | 92.44 (208/225) | 96.70 (88/91) | 91.56 (206/225) | 93.41 (85/91) | |
| Average accuracy | 97.34 (1,868/1,919) | 98.39 (793/806) | 94.42 (1,812/1,919) | 94.67 (763/806) | |
ATC, anaplastic thyroid carcinoma; FTC, follicular thyroid carcinoma; MTC, medullary thyroid carcinoma; PTC, papillary thyroid carcinoma.
The Inception-ResNet-v2 model applied to the 7 pathology types achieved a fragmentation accuracy of 82.22% for normal tissue, 94.76% for ATC, 95% for FTC, 98.43% for MTC, 93.31% for PTC, 98.43% for nodular goiter and 91.56% for adenoma. exhibiting an average fragmentation accuracy of 94.42%. On the raw images (without data augmentation), the Inception-ResNet-v2 model exhibited an accuracy rate of 94.62% for normal tissue, 95.06% for ATC, 92.11% for FTC, 96.04% for MTC, 92.90% for PTC, 96.65% for nodular goiter and 93.41% for adenoma, exhibiting an average accuracy of 94.67% on the raw images (Table 4).
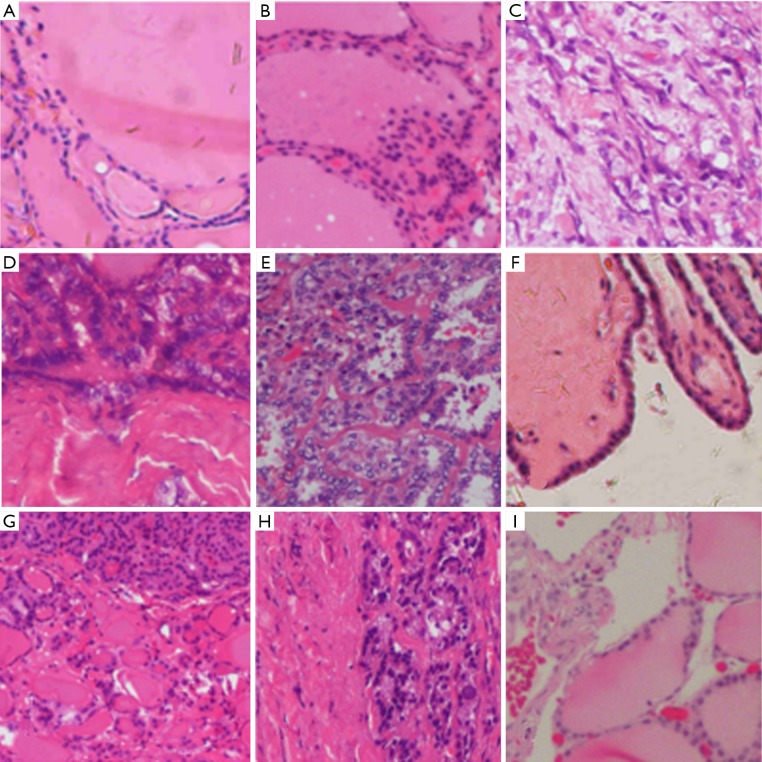
VGG-19 was clearly more accurate than was Inception-ResNet-v2 when predicting the 7 pathologic types of thyroid diseases. We further analyzed the misdiagnosed images to determine the reasons for the misdiagnosis. The specific error classifications of VGG-19 are listed in Table 5. In the thyroid normal tissue group, which showed the highest rate of misdiagnosis, 7 fragmented images were misdiagnosed as goiter and 14 as adenoma. Misclassification was frequently observed in the adenoma group, among which 3 images were misclassified as normal tissue, 5 as FTC and 9 as PTC. The ATC, FTC and PTC groups had similar accuracies for the fragmented images: 3 fragmented images of ATC were misdiagnosed as FTC; 2 fragmented images of FTC were misdiagnosed as PTC; and 7 fragmented images of PTC were misdiagnosed as FTC and 1 as goiter. No error classifications occurred in the MTC group or in the goiter group. Some typical examples of misclassified fragmented images are shown in Figure 6.
Table 5. Confusion matrix of the classification results of VGG-19 (fragmented images).
| Pathologic classifications | Normal tissue | ATC | FTC | MTC | PTC | Goiter | Adenoma |
|---|---|---|---|---|---|---|---|
| Normal tissue | 159 (88.33%) | 0 | 0 | 0 | 0 | 7 (3.89%) | 14 (7.78%) |
| ATC | 0 | 207 (98.57%) | 3 (1.43%) | 0 | 0 | 0 | 0 |
| FTC | 0 | 0 | 178 (98.89%) | 0 | 2 (1.11%) | 0 | 0 |
| MTC | 0 | 0 | 0 | 255 (100%) | 0 | 0 | 0 |
| PTC | 0 | 0 | 7 (1.95%) | 0 | 351 (97.77%) | 1 (0.28%) | 0 |
| Goiter | 0 | 0 | 0 | 0 | 0 | 510 (100%) | 0 |
| Adenoma | 3 (1.33%) | 0 | 5 (2.22%) | 0 | 0 | 9 (4%) | 208 (92.44%) |
ATC, anaplastic thyroid carcinoma; FTC, follicular thyroid carcinoma; MTC, medullary thyroid carcinoma; PTC, papillary thyroid carcinoma.
Figure 6.
Typical misclassified fragmented images HE, ×100. (A,B) Misdiagnosed fragmented image of normal tissue: (A) an image misdiagnosed as goiter; (B) an image misdiagnosed as adenoma. (C) Misdiagnosed fragmented image of ATC: the image was misdiagnosed as FTC; (D,E,F) misdiagnosed fragmented images of PTC and FTC: (D) an image of PTC misdiagnosed as FTC; (E) an image of FTC misdiagnosed as PTC; (F) an image of PTC misdiagnosed as goiter; (G,H,I) misdiagnosed fragmented images of adenoma: (G) an image misdiagnosed as goiter; (H) an image misdiagnosed as FTC (containing only a portion of capsule and follicles, which are somewhat similar to FTC misdiagnosed as normal tissue). ATC, anaplastic thyroid carcinoma; FTC, follicular thyroid carcinoma; PTC, papillary thyroid carcinoma.
Discussion
Deep learning is currently the most suitable and widely used algorithm for image recognition in the artificial intelligence field. Deep learning models imitate the working mechanism of the human brain. Convolutional neural networks can be constructed to automatically extract features from input data, enabling the machine to understand the learning data, obtain information and output results. Deep learning has been applied to multiple aspects of the preoperative diagnosis of thyroid tumors. Our previous work (11) reported that the Inception-v3 network achieved promising diagnostic performance in classifying ultrasonographic images of thyroid nodules (sensitivity 93.3%, specificity 87.4%), while Ko et al. (12) revealed that there was no significant difference between experienced radiologists and a DCNN regarding the diagnostic ability to differentiate thyroid malignancies from ultrasound images (achieving area under the curve (AUC) scores of 0.805–0.860 and 0.835–0.850, respectively). A large-scale multicenter retrospective study of 2,692 patients demonstrated similar sensitivity but improved the specificity of a DCCN compared with radiologists for identifying sonographic images of thyroid nodules (13). However, DCCNs have not yet been used for histopathologic diagnosis of thyroid diseases. This article is a pioneering prospective study that utilized 2 types of DCNN algorithms and compared their performances on histological images encompassing various histological types of thyroid diseases.
From the experimental results, both the VGG-19 model and the Inception-Resnet-v2 model achieved satisfactory average accuracies for the recognition of fragmented and raw images (VGG-19: 97.3% and 98.4%; Inception-Resnet-v2: 94.4% and 94.7%). Because few reports exist of using DCNNs for the classification of thyroid tumors, for comparisons, we must refer to previous studies in which DCNNs were applied to identifying other types of tumors. For example, DCNNs have been most often used in classifying breast cancer histology images: achieving accuracies of 83.3% to 93.2% in Han’s and Araujo’s studies, respectively (8,14). Khosravi and his colleagues trained six DCNN architectures to detect H&E-stained histopathology images of different cancer tissues (lung, breast, and bladder cancers), and all the architectures performed at remarkable accuracies, varying from 96.6% to 100% (15). Thus, we can assume that DCNNs have some potential to provide reliable and accurate classifications. With the help of computer-aided diagnosis in routine H&E-stained images, pathologists may have less need for other assistive technologies, such as immune-histochemical staining, which demand more energy, time and resources.
According to the literature, the classification performances of DCCNs depend heavily on the quality and quantity of training data (16). An image of a complete pathological slice may contain numerous different tissues, including tumor tissue, normal thyroid tissue, follicular tissue, inflammatory cells, blood vessels, muscle tissue, fibrous tissue and so on, and histological images may also vary widely in color and scale batch due to different tissue preparation methods and imaging equipment. In addition, DCNN algorithms are typically trained and tested on numerous smaller images that are segmented from the original image. The DCNN images used in the current study were carefully selected by experienced pathologists, and the pathological diagnoses were definitive; therefore, the images had typical cell morphology and arrangements, which provided histological features that were well-captured by the DCNN. However, in clinical practice, this type of manual approach would be time-consuming and subjective (16,17), which could introduce bias in the algorithms. Considering the issue of data collection, we adapted an automatic feature extraction program based on the strength of nucleus recognition (which has a dark mottled appearance in slides). This procedure enabled us to obtain sufficient numbers of patches containing adequate numbers of cells that could provide adequate features for the DCNN within a reasonable amount of time. In this regard, utilization of an automatic feature extraction program could improve the efficiency of training DCNN models and avoid biases and errors during the extraction procedure.
VGG-19 was generally more accurate than Inception-ResNet-v2 on all the classifications. The Inception-ResNet-v2 model was mainly designed for multiscale image training, while VGG-19 is more suitable for the recognition of single-scale images. The histological images used in the training set were adjusted to a single resolution and included a similar number of cells with a dark mottled appearance; thus, the scale was relatively simple. Therefore, the average accuracy on the augmented datasets as well as the average accuracy on raw images was higher for VGG-19 than for Inception-ResNet-v2 (augmented datasets 98.39% vs. 94.67%; raw images 97.34% vs. 94.42%, respectively). Our study indicated that Inception-ResNet-v2 did not have an advantage over VGG-19 for the classification of thyroid diseases based on histological images; instead, VGG-19 performed better at this task.
After this comparison, the VGG-19 model was chosen over Inception-ResNet-v2. The pathologists at FUSCC further investigated the misdiagnosed images to analyze the reasons for failure. In this study, normal tissue was the most challenging histologic type for DCNN to differentiate, and all misdiagnosed fragments were classified as benign thyroid tumors (goiter and adenoma). Although the pathologists considered the images to be typical of normal thyroids, we reviewed the misdiagnosed images and found that several reasons could have caused the misdiagnoses. The training/test data were extracted from original images such that the fragmented images were all magnified and captured follicles of different sizes. Because goiter and adenoma both present nodular changes in the thyroid gland microscopically, when the fragmented images of normal tissue included some large follicles, the images could be misclassified (see Figure 5A vs. Figure 6A,B). As shown in Figure 6A,B, although both images intercepted large follicles, a higher intensity of the follicular epithelial cells between follicles and smaller follicles (although consistent in size) can be observed in Figure 6B than in Figure 6A, which implied that Figure 6A is more consistent with adenoma because goiter is characterized by fewer epithelial cells, given the enlarged follicles and lack of a complete capsule (18). In addition, given that the DCNN classification mechanism was mainly based on the size and staining of the nucleus, the DCNN made classification errors among normal tissue, goiter and adenoma because the follicular epithelial cells of these three pathologies share the same morphology (normal epithelial cells). Meanwhile, it was clear that the DCNN had no confusion between normal and malignant tumor tissue.
Three fragmented images of ATC were misclassified as FTC, but the fragmentation accuracy of ATC reached 98.57% in this study. Microscopically, the morphological features of ATC depend on the admixture of three main histological patterns (spindle, giant and epithelioid cells) with marked pleomorphism and numerous mitoses that show sarcomatoid, epithelioid-squamoid or other rare variant changes (19). Due to the unique cell morphology and disappearance of the follicular structure, classification errors were rare among ATC, benign tumor/normal tissue and other types of thyroid cancer. Considering the misclassifications shown in Figure 6C, we assumed that the pathological diagnosis of FTC was mainly based on capsular/vascular invasion, which presents as tumor cells infiltrating fibrovascular tissue. VGG-19 might confuse ATC with FTC due to the fibrovascular tissue of the FTC background compared with that of PTC and MTC.
Regarding the diagnostic efficiencies of PTC and FTC, VGG-19 achieved satisfactory accuracy in this study. Interestingly, the misdiagnoses of FTC and PTC overlapped at a high proportion (Table 5, Figure 6D,E). Because PTC and FTC are both included in differentiated thyroid carcinoma (DTC), cancerous cells of both types originate from thyroid follicular epithelial cells and manifest with a similar cell morphology. Additionally, a variant of PTC exists named follicular papillary thyroid carcinoma (FPTC), which has similar histopathologic features (mostly composed of follicles without papillary structure) as follicular tumors (20). Meanwhile, a papillary structure may also sometimes appear in FTC (21). In Figure 6D, the fragment included a portion of a follicular-like structure accompanied by a cancerous region derived from follicular epithelial cells; therefore, the DCNN confused the PTC patch as FTC. Similarly, the cancerous region in Figure 6E was filled with papillary structure, which accounted for the misclassification of FTC as PTC. Moreover, Figure 6F shows the misclassification of PTC as goiter. We speculate that this misclassification may have occurred because the cells in the image had low intensity along with a relatively large papilla containing colloid, which appears similar to goiter.
The diagnostic efficiency of adenoma by DCNN was comparatively lower in this experiment. More than half of the misclassifications were due to confusion between adenoma and goiter (Table 5). The epithelial cell morphology is almost the same in both adenoma and goiter; therefore, the key to the differential diagnosis relies on the following histopathologic features: (I) a complete and homogeneous capsule in adenoma; (II) smaller follicles with similar size as normal tissue in adenoma but larger follicles of various sizes in goiter; and (III) papillary hyperplasia sometimes observed in goiter (22). Therefore, when a fragmented adenoma image did not capture the abovementioned features (for example, Figure 6G presents follicles of different sizes without a complete capsule), the patch had a high misclassification risk. Another high proportion of the misclassifications corresponded to confusion between adenoma and FTC. A capsular structure exists in both adenoma and FTC, but FTC features capsular/vascular invasion, which can be better observed at low power or in whole slices. Thus, it remains difficult for the DCNN to differentiate adenoma and FTC from magnified fragments, which include only limited image information (Figure 6H). Figure 6I shows an adenoma patch misclassified as normal tissue, which might be due to the lower intensities of cells and the larger follicles compared to other adenoma patches (Figure 5G).
VGG-19 achieved a diagnostic efficiency of 100% for MTC and goiter. MTC is characterized by changes in cell morphology, including relatively uniform, short spindles or polygonal cells arranged in sheets, nests, islands, bundles, follicles or acinars (23). Thus, the distinctive cell morphology aided the DCNN in achieving its excellent diagnostic efficiency for MTC. Likewise, the goiter training data included the largest number of images (2,625 patches); thus, we assumed that the DCNN was also adequately trained and gained excellent diagnostic efficiency for these images.
This study demonstrated the application of DCNNs for the classification of thyroid diseases based on histology, although further potential improvements could be made by training the model with more images. Future studies should expand the training pool and integrate histomorphology with molecular biomarkers.
Conclusions
In summary, our work demonstrated that DCNN models can be applied to facilitate the differentiation of thyroid nodules using histological images in clinical settings. After training with a large dataset, the DCNN models, especially VGG-19, achieved excellent diagnostic efficiencies. The assistance of DCCN models could reduce the pathologists' workloads and improve their efficacy at determining the histopathology of thyroid tumors.
Acknowledgments
Funding: This paper was partially supported by the National Nature Science Foundation of China (No. 61772330, 61533012, and 61876109).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This work was approved by the ethical committee of FUSCC (ID: 050432-4-1212B), and all patients were required to sign informed consent forms before the related procedures.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.He L, Long LR, Antani S, et al. Histology image analysis for carcinoma detection and grading. Comput Methods Programs Biomed 2012;107:538-56. 10.1016/j.cmpb.2011.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barker J, Hoogi A, Depeursinge A, et al. Automated classification of brain tumor type in whole-slide digital pathology images using local representative tiles. Med Image Anal 2016;30:60-71. 10.1016/j.media.2015.12.002 [DOI] [PubMed] [Google Scholar]
- 3.Miki Y, Muramatsu C, Hayashi T, et al. Classification of teeth in cone-beam CT using deep convolutional neural network. Comput Biol Med 2017;80:24-9. 10.1016/j.compbiomed.2016.11.003 [DOI] [PubMed] [Google Scholar]
- 4.Cha KH, Hadjiiski L, Samala RK, et al. Urinary bladder segmentation in CT urography using deep-learning convolutional neural network and level sets. Med Phys 2016;43:1882. 10.1118/1.4944498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anthimopoulos M, Christodoulidis S, Ebner L, et al. Lung Pattern Classification for Interstitial Lung Diseases Using a Deep Convolutional Neural Network. IEEE Trans Med Imaging 2016;35:1207-16. 10.1109/TMI.2016.2535865 [DOI] [PubMed] [Google Scholar]
- 6.Teramoto A, Fujita H, Yamamuro O, et al. Automated detection of pulmonary nodules in PET/CT images: Ensemble false-positive reduction using a convolutional neural network technique. Med Phys 2016;43:2821-7. 10.1118/1.4948498 [DOI] [PubMed] [Google Scholar]
- 7.Sharma H, Zerbe N, Klempert I, et al. Deep convolutional neural networks for automatic classification of gastric carcinoma using whole slide images in digital histopathology. Comput Med Imaging Graph 2017;61:2-13. 10.1016/j.compmedimag.2017.06.001 [DOI] [PubMed] [Google Scholar]
- 8.Araújo T, Aresta G, Castro E, et al. Classification of breast cancer histology images using Convolutional Neural Networks. PLoS One 2017;12:e0177544. 10.1371/journal.pone.0177544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marr D, Hildreth E. Theory of edge detection. Proc R Soc Lond B Biol Sci 1980;207:187-217. 10.1098/rspb.1980.0020 [DOI] [PubMed] [Google Scholar]
- 10.Szegedy C, Ioffe S, Vanhoucke V. Inception-v4, Inception-ResNet and the Impact of Residual Connections on Learning. AAAI Conference on Artificial Intelligence, 2016. [Google Scholar]
- 11.Guan Q, Wang Y, Du J, et al. Deep learning based classification of ultrasound images for thyroid nodules: a large scale of pilot study. Ann Transl Med 2019;7:137. 10.21037/atm.2019.04.34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ko SY, Lee JH, Yoon JH, et al. Deep convolutional neural network for the diagnosis of thyroid nodules on ultrasound. Head Neck 2019;41:885-91. 10.1002/hed.25415 [DOI] [PubMed] [Google Scholar]
- 13.Li XC, Zhang S, Zhang Q, et al. Diagnosis of thyroid cancer using deep convolutional neural network models applied to sonographic images: a retrospective, multicohort, diagnostic study. Lancet Oncol 2019;20:193-201. 10.1016/S1470-2045(18)30762-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han Z, Wei B, Zheng Y, et al. Breast Cancer Multi-classification from Histopathological Images with Structured Deep Learning Model. Sci Rep 2017;7:4172. 10.1038/s41598-017-04075-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khosravi P, Kazemi E, Imielinski M, et al. Deep Convolutional Neural Networks Enable Discrimination of Heterogeneous Digital Pathology Images. EBioMedicine 2018;27:317-28. 10.1016/j.ebiom.2017.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kothari S, Phan JH, Stokes TH, et al. Pathology imaging informatics for quantitative analysis of whole-slide images. J Am Med Inform Assoc 2013;20:1099-108. 10.1136/amiajnl-2012-001540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu KH, Zhang C, Berry GJ, et al. Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat Commun 2016;7:12474. 10.1038/ncomms12474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cerutti JM, Delcelo R, Amadei MJ, et al. A preoperative diagnostic test that distinguishes benign from malignant thyroid carcinoma based on gene expression. J Clin Invest 2004;113:1234-42. 10.1172/JCI19617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ragazzi M, Ciarrocchi A, Sancisi V, et al. Update on anaplastic thyroid carcinoma: morphological, molecular, and genetic features of the most aggressive thyroid cancer. Int J Endocrinol 2014;2014:790834. 10.1155/2014/790834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim MJ, Won JK, Jung KC, et al. Clinical Characteristics of Subtypes of Follicular Variant Papillary Thyroid Carcinoma. Thyroid 2018;28:311-8. 10.1089/thy.2016.0671 [DOI] [PubMed] [Google Scholar]
- 21.Xu W. Comparison study of ultrasound findings between follicular papillary thyroid carcinoma and thyroid adenoma (in Chinese) Chin Mod Doctor 2016;54:110-2. [Google Scholar]
- 22.He F, Mao C, Ma M. Retrospectively Analyze of 376 Cases With Thyroid Adenoma and Nodular Goiter Pathology (in Chinese) Chin Heal Standard Management 2016;7:174-6. [Google Scholar]
- 23.Trimboli P, Giovanella L, Crescenzi A, et al. Medullary thyroid cancer diagnosis: An appraisal. Head Neck 2014;36:1216-23. 10.1002/hed.23449 [DOI] [PubMed] [Google Scholar]