Abstract
Background
This study aimed to determine independent risk loci of Graves’ disease (GD) in the thyroglobulin (TG) region.
Methods
In this two-staged association study, a total of 9,757 patients with GD and 10,626 sex-matched controls were recruited from Chinese Han population. Illumina Human660-Quad BeadChips in the discovery stage and TaqMan SNP Genotyping Assays in the replication stage were used for genotyping. Trend test and logistic regression analysis were performed in this association study.
Results
In the discovery stage, rs2294025 and rs7005834 were the most highly associated susceptibility loci with GD in TG. In the replication phase, 7 SNPs, including rs2294025 and rs7005834, were selected for fine-mapping. Finally, we confirmed that rs2294025 and rs7005834 were the independent risk loci of GD in the combined populations. At the same time, there was no significant difference between the risk allele frequencies of rs2294025 and rs7005834 in different clinical phenotypes of GD.
Conclusions
The fine mapping study of thyroglobulin identified two independent SNPs (rs2294025 and rs7005834) for GD susceptibility. However, no significant differences for rs2294025 and rs7005834 were observed, between the different clinical phenotypes of GD, including gender, Graves’ ophthalmopathy (GO), and serum levels of thyrotropin receptor antibody, thyroid peroxidase antibody, and thyroglobulin antibody. These results provide a deeper understanding of the association mechanism of thyroglobulin and GD risk.
Keywords: Thyroglobulin, Graves’ disease (GD), single nucleotide polymorphism (SNP)
Introduction
Graves’ disease (GD) is one of the most common types of organ-specific autoimmune thyroid disease (AITD). The main characteristics of GD are hyperthyroidism, various degrees of diffuse goiter, ophthalmopathy, and with a small number of pretibial myxedema (1). The etiology of GD is incompletely understood and GD affects up to 2% of a population in iodine-sufficient areas (2). GD is caused by interaction of multiple complex factors, such as genetic, environmental and endogenous factors (1,3). There is no doubt that environmental factors contribute to the development of GD in susceptible individuals, and the genetic basis plays an important role in this process. The importance of genetic factors can be seen from the aggregation of GD in the family (4). It has been shown that the consistency rate among monozygotic twins was significantly higher than that among dizygotic twins for Graves’ disease (5). These studies suggest that nearly 80% of the risk of GD is genetic (2).
TG, located on chromosome 8q24, is a 660-kDa glycoprotein that supports the production of thyroid hormones (6,7), accounting for 75–80% of total thyroid protein (8). TG can infiltrate the blood circulation, expose to the immune system, and is the main autoantigen of AITD. In addition, the best model for human autoimmune thyroiditis is to immunize mice with TG (9), since TG may be the earliest cause of the disease (10). TG may be one of the AITD susceptible genes, and its linkage studies showed that there was a significant linkage peak on chromosome 8q in the TG gene region (11). However, the number of studies on the correlation between TG and GD alone was less and the conclusion have been less consistent. In North American Caucasians, it has been reported that a polymorphism in exon 12 (rs853326G/A) resulted in a Val (G allele) to Met (A allele) substitution and the A allele frequency of GD patients was higher than control group (12). The haploid allele combination analysis of TG polymorphic loci, E10SNP24-E10SNP158-E12SNP-E33SNP, was performed between the hyperthyroidism group and the healthy group. The disease-linked haplotypes G-T-A-C, G-T-G-C and T-C-G-C was significantly different (P<0.05) (13). In case control studies, several SNPs in exons 10, 12, and 33 were significantly associated with AITD (14) or with GD relapse (15). In a Japanese population, rs2958692 in TG was associated with the evolution of GD, the refractory of GD (16). It is confirmed that in Italians, rs2069561 in TG was associated with GD (17). But in China, there was no differences between the frequencies of allele and genotype of TG between GD patients and controls (18). Case-control research showed that in Tunisia, the Tgms2 (TG microsatellite 2) or SNPs of TG were not associated with GD (19).
The previous genome-wide association study (GWAS) provided reliable evidence for the association between TG gene polymorphism and GD in Chinese Han ethnic groups (20). In this study, a fine mapping study was further carried out, based on GWAS data to determine the possible independent susceptibility SNP(s) of TG region in patients with GD. The association between the susceptibility alleles and genotypes of TG and the different phenotypes in patients with GD was also studied.
Methods
Subject
In this study, 9,757 GD patients and 10,626 controls of sex-matched were recruited (Table 1). The project was approved by the local ethics committee. All unrelated Chinese Han participants were recruited from various hospitals in China and signed informed consent forms.
Table 1. General information of patients.
| Disease status | N | Sex ratio (M/F) | Age (years) |
|---|---|---|---|
| GWAS | |||
| GD | 1,442 | 320/1,122 | 39±14 |
| Control | 1,468 | 349/1,119 | 45±9 |
| Replication | |||
| GD | 8,315 | 2,057/6,258 | 40±14 |
| Control | 9,158 | 1,841/7,317 | 46±12 |
| Combined | |||
| GD | 9,757 | 2,377/7,380 | 39±14 |
| Control | 10,626 | 2,190/8,436 | 45±12 |
The diagnosis of GD was according to the issues as followed: (I) the symptoms, signs and the laboratory evidence of hyperthyroidism, (II) diffuse goiter, while (III) at least one of the following situations: tested thyrotropin receptor antibody (TRAb) positivity, 131I intake increased or exophthalmos (20,21). Diagnosis of hyperthyroidism ophthalmopathy and goiter in accordance with the Guidelines of the American Thyroid Association. The most majority of GD patients were treated with antithyroid drugs (ATD) or radioactive iodine (131I) for more than 1 year. Other causes of hyperthyroidism will be excluded.
All the control subjects were over 35 years old and matched with the cases by area and gender. Individuals with GD or family history of GD and other autoimmune diseases had been excluded. Given that GD and other AITD is common in young women, the age over 35 years old could reduce the number of later developments of GD in the control group.
The levels of TSH, free triiodothyronine (FT3) and thyroxine (FT4) were tested by chemiluminescence immunoassay. In our laboratory, quantitative ELISA (RSR Limited, Cardiff, UK) was used to test the levels of thyroid globulin antibody (TGAb), thyroid peroxidase antibody (TPOAb) and thyrotropin receptor antibody (TRAb). The level of TGAb ≥65 u/mL, TPOAb ≥10 u/L, TRAb ≥1.5 u/L were defined positive, respectively.
Genotyping, imputation and quality control in the discovery stage
In the discovery stage, genotyping was performed utilizing the Illumina Human660-Quad BeadChips (21). Also, the robust quality control was carried out for the SNPs in the discovery stages (21). Finally, the SNPs with the Hardy Weinberg equilibrium (HWE) P≥10-6, missing call rate ≤0.05, or a minor allele frequency (MAF) ≥0.01, would be kept in the association study (20,21).
As for the genotyped SNPs in the discovery stage after the robust quality control, imputation analysis was performed using the IMPUTE 2 software and the 1000 Genomes Project (June 2011) as the reference (22). After the imputation, the robust quality control was performed for the imputed SNPs, and the SNPs with the HWE P≤10−6, missing call rate ≥0.05, or a MAF ≤0.01, were excluded in the study. Finally, the association study was carried out for the imputed SNPs using ProbABEL.
Sample size and power calculations
QUANTO software (version 1.2.4) was used to calculate the sample size. According to our previous GWAS data, the significance level of genome-wide for each mutation with allele frequencies ranged from 0.01 to 0.5 and with the power at least 80%. Therefore, a minimum of 3,493 (rs7005834) and a maximum of 8,186 (rs2703004) sample pairs would be needed in this study. Therefore, a total of ~9,000 sample pairs were included in the study.
TagSNPs selection and genotyping
In discovery stage, Haploview 4.1 software and a pairwise-tagging approach were used to select the tagSNPs (23). TaqMan SNP Genotyping Assays were used to genotype the selected seven SNPs (rs2703004, rs4301434, rs2294025, rs4736437, rs10956695, rs7005834 and rs4330674) in the replication stage.
Statistical analysis
In the discovery stage and replication stage, we analyzed the SNPs by the Cochran Armitage trend test in PLINK (24). In the combined stage, Cochran Mantel Haenszel stratification analysis was utilized to check the associations. In order to detect the independent risk SNP(s), forward stepwise logistic regression analysis was carried out in the discovery stage and the combined stages using R program. Also, the conditional logistic regression analysis was performed using PLINK packages. The interactions between SNPs of TG in the different clinical phenotypes of GD, including gender, Graves’ ophthalmopathy (GO), thyroid goiter, the level of TRAb, TGAb, TPOAb was also analyzed using the χ2 test in the SPSS PASW Statistics 18 package.
Results
Association analysis with GD in the discovery stages
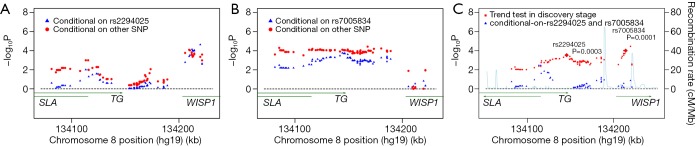
In the discovery stage, we re-examined our previous GWAS data. A ~340 kb LD block located in chromosome 8q24.22 was chosen in the Chinese population GWAS data. This block contained the strongest TG association and was separated by two recombination hotspots (recombination rate >50 cM/Mb) (Figure 1A). And then genotyping and imputation analysis in the region which encompassed TG/SLA/WISP1 was carried out. Eight hundred and eight genotyped and imputed SNPs were found in the region included TG/SLA/WISP1. After robust quality control, there were 209 SNPs with P<0.05, 98 SNPs with P<0.01, 34 SNPs with P<0.001 and one SNP with a significant P level of<0.0001 (rs10100792, P=3.58×10−5; OR, 0.74; 95% CI, 0.64–0.85) (Figure 1A). We found a cluster of SNPs highly correlated with rs2294025 between the two recombination peaks (R2>0.6) (Figure 1B). Also, 7 tagSNPs were selected for further replication (Figure 1C).
Figure 1.
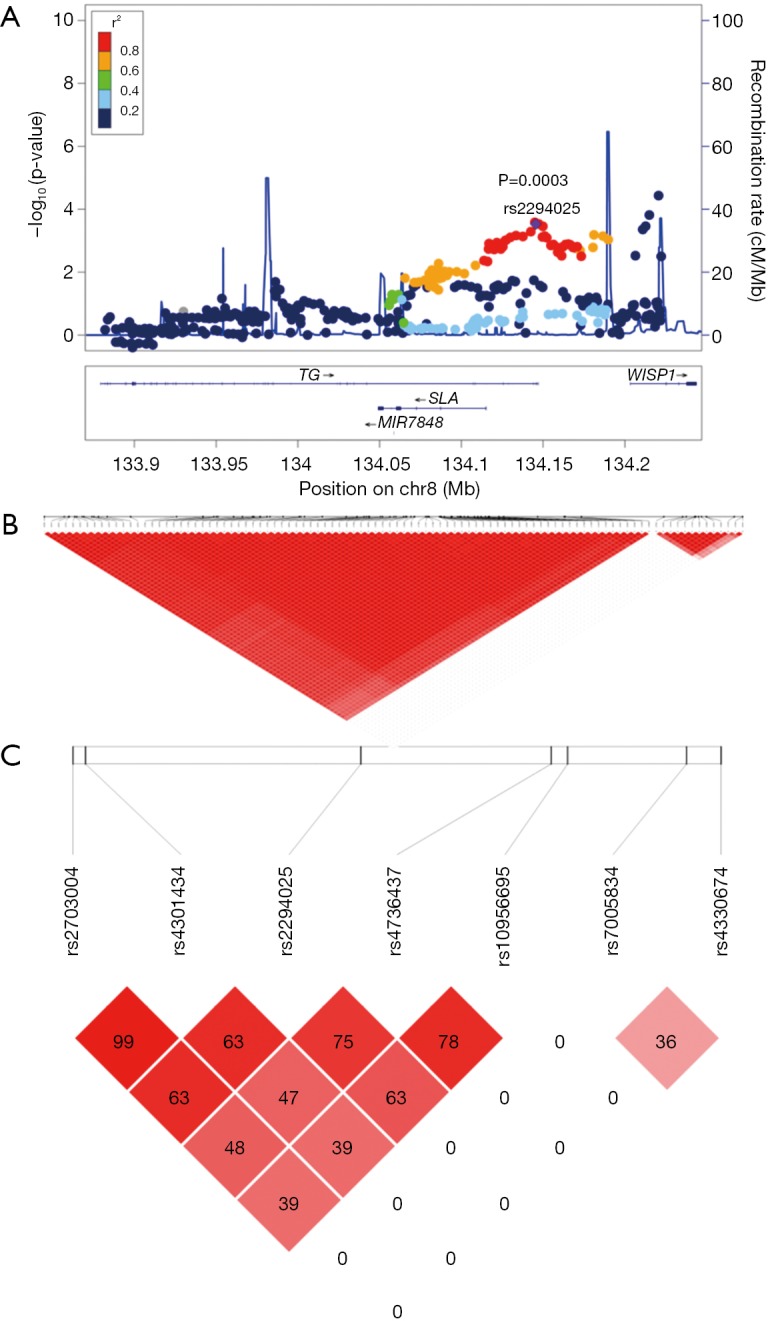
Results of association analysis for SNPs in 8q24.22 in the discovery stage. (A) In the discovery stage the regional plots of susceptibility loci associated with Graves’ disease (GD) in 8q24.22. The -log10 P values of each SNP at 8q24.22 are plotted against chromosomal position. The top SNP at 8q24.22 is rs2294025 shown as a big purple diamond. The color of each SNP spot reflects its r2 value with rs2294025 and decreasing values of r2 demonstrates with various colors from red, orange, green to sky-blue and blue. One thousand genomes from Asian samples were used to compute genetic recombination rates. Physical positions are based on GRCh37; (B) Haploview analysis results of 85 SNPs of Block 1 and 13 SNPs of Block 2, two highly linked independent susceptible blocks. The number in each small square represents the interlocking unbalance r2 value seen in two SNPS. The big red represents the complete interlocking r2=1. The higher the interlocking degree is, the closer the color is to the big red and the completely non-interlocking color is white; (C) in the replication stage, Haploview analysis of the 7 TagSNPs. The number in each small square case represents the Linkage disequilibrium r2 value between the two SNPs, and red represents the complete Linkage disequilibrium r2=1. The higher the Linkage disequilibrium is, the closer the color is to black, while the completely non-Linkage disequilibrium value is white.
In order to discern the independent susceptibility loci associated with GD in the TG/SLA/WISP1 region, 98 SNPs with P<0.01 were analyzed using forward stepwise regression and two-locus logistic regression analysis. The forward regression analysis showed that two SNPs (rs2294025, P=0.003; OR, 1.27; 95% CI, 1.11–1.44, and rs7005834, P=0.001; OR, 1.33; 95% CI, 1.15–1.54) were independent risk loci for GD in the GWAS cohorts. Moreover, each of the majority of SNPs, excluding 10 SNPs, were improved by rs2294025 according to the two-locus regression analysis and used the criteria of P<0.001 (Figure 2A). Interestingly, in two-locus conditional logistic regression models, rs7005834 improved all of the 10 SNPs (Figure 2B). Furthermore, the majority of other 96 SNPs were not associated with GD after conditioning on either rs2294025 or rs7005834, respectively (Figure 2A,B). However, after conditioning on both SNPs (rs2294025 and rs7005834), no other SNPs in this region were found to be independently associated with GD at a cut-off of P<0.01 (Figure 2C). Notably, rs2294025 and rs7005834 were not in the same LD block (r2=0) (Figure 1C). The above results revealed that in the Chinese Han population, rs2294025 and rs7005834 in the TG region were probably independent susceptibility loci to GD.
Figure 2.
Logistic regression results of 98 SNPs with P<0.01 at 8q24.22 in the discovery stage. (A,B) Two-locus conditional logistic regression for rs2294025 (A) or rs7005834 (B) with the other 97 SNPs with P<0.01. Conditioning on rs2294025 (A) or rs7005834 (B), the P values of each of the other 97 SNPs are shown in blue triangles. And conditioning on each of the other 97 SNPs, the P values of rs2294025 or rs7005834 are shown in red circles; (C) after conditioning on rs2294025 and rs7005834 at the same time, the P values of other SNPs in the GD susceptibility locus on 8q24.22 are shown in blue triangle. None of the other 96 SNPs on 8q24.22 improved the model with rs2294025 and rs7005834 at the level of P<0.01. The P levels of trend analysis for each of 98 SNPs are shown in red square and the top SNP rs2294025 is shown in red diamond. Estimated recombination rates (based on the eastern Asian samples from the HapMap project) plotted in cyan reflect the local linkage disequilibrium structure around the associated SNPs.
Association analysis in the replication and combined stages
In the replication stage, 8,315 Chinese Han patients with GD and 9,158 sex-matched controls were included. Seven TagSNPs were selected for genotyping. Six of seven TagSNPs could met the significant level of P<0.05 in the replication cohort, and the best association SNP was located at rs2294025 (replication stage, P=3.59×10−7, OR, 1.15; 95% CI, 1.09–1.21, Table 2).
Table 2. The association results of seven SNPs on 8q24.22.
| SNP | Allele/ Genotype |
Discovery (1,442 vs. 1,468) | Replication (8,315 vs. 9,158) | Combined (9,757 vs. 10,726) | Sample pairs | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr. Position Gene region | Cases, N (%) | Control, N (%) | OR (95% CI) | P | Case, N (%) | Control, N (%) | OR (95% CI) | P | Cases, N (%) | Control, N (%) | OR (95% CI) | P | ||||
| rs2703004 | A | 2,107 (73.1) | 2,230 (76.0) | 1.00 (ref) | 3,159 (72.2) | 3,161 (74.3) | 1.00 (ref) | 5,266 (72.6) | 5,391 (75.0) | 1.00 (ref) | 8,186 | |||||
| 134084800 | G | 777 (26.9) | 706 (24.0) | 1.17 (1.04–1.31) | 0.0108 | 1,215 (27.8) | 1,091 (25.7) | 1.11 (1.01–1.23) | 0.0253 | 1,992 (27.4) | 1,797 (25.0) | 1.14 (1.05–1.22) | 0.0009 | |||
| TG | AA | 764 (53.0) | 846 (57.6) | 1.00 (ref) | 1,138 (52) | 1,167 (54.9) | 1.00 (ref) | 1,902 (52.4) | 2,013 (56.0) | 1.00 (ref) | ||||||
| Intron41 | AG | 579 (40.2) | 538 (36.6) | 1.19 (1.02–1.39) | 883 (40.4) | 827 (38.9) | 1.09 (0.97–1.24) | 1,462 (40.3) | 1,365 (38.0) | 1.13 (1.25–1.07) | ||||||
| GG | 99 (6.9) | 84 (5.7) | 1.31 (0.96–1.77) | 0.0355 | 166 (7.6) | 132 (6.2) | 1.29 (1.01–1.64) | 0.0737 | 265 (7.3) | 216 (6.0) | 1.3 (1.03–1.57) | 0.0035 | ||||
| rs4301434 | A | 2,102 (72.9) | 2,227 (75.9) | 1.00 (ref) | 11,531 (72.6) | 12,565 (74.8) | 1.00 (ref) | 13,633 (72.7) | 14,792 (75.0) | 1.00 (ref) | 8,077 | |||||
| 134087450 | G | 782 (27.1) | 709 (24.1) | 1.17 (1.04–1.32) | 0.0092 | 4,345 (27.4) | 4,231 (25.2) | 1.12 (1.07–1.18) | 7.52E-06 | 5,127 (27.3) | 4,940 (25.0) | 1.13 (1.08–1.18) | 3.12E-07 | |||
| TG | AA | 760 (52.7) | 845 (57.6) | 1.00 (ref) | 4,198 (52.9) | 4,678 (55.7) | 1.00 (ref) | 4,958 (52.9) | 5,523 (56.0) | 1.00 (ref) | ||||||
| Intron41 | AG | 582 (40.4) | 537 (36.6) | 1.21 (1.03–1.4) | 3,135 (39.5) | 3,209 (38.2) | 1.09 (1.02–1.16) | 3,717 (39.6) | 3,746 (38.0) | 1.11 (1.17–1.17) | ||||||
| GG | 100 (6.9) | 86 (5.9) | 1.29 (0.95–1.75) | 0.0283 | 605 (7.6) | 511 (6.1) | 1.32 (1.16–1.49) | 1.84E-05 | 705 (7.5) | 597 (6.1) | 1.32 (1.04–1.48) | 1.20E-06 | ||||
| rs2294025 | G | 2,253 (78.1) | 2,404 (81.9) | 1.00 (ref) | 13,037 (78.4) | 14,538 (80.6) | 1.00 (ref) | 15,290 (78.4) | 16,942 (80.8) | 1.00 (ref) | 4,068 | |||||
| 134145512 | A | 631 (21.9) | 532 (18.1) | 1.27 (1.11–1.44) | 0.0003 | 3,593 (21.6) | 3,498 (19.4) | 1.15 (1.09–1.21) | 3.59E-07 | 4,224 (21.6) | 4,030 (19.2) | 1.16 (1.11–1.22) | 1.52E-09 | |||
| TG | GG | 870 (60.3) | 985 (67.1) | 1.00 (ref) | 5,124 (61.6) | 5,853 (64.9) | 1.00 (ref) | 5,994 (61.4) | 6,838 (65.2) | 1.00 (ref) | ||||||
| Intron46 | GA | 513 (35.6) | 434 (29.6) | 1.34 (1.14–1.57) | 2,789 (33.5) | 2,832 (31.4) | 1.12 (1.05–1.2) | 3,302 (33.8) | 3,266 (31.1) | 1.15 (1.12–1.2) | ||||||
| AA | 59 (4.1) | 49 (3.3) | 1.36 (0.92–2.01) | 0.0007 | 402 (4.8) | 333 (3.7) | 1.38 (1.19–1.6) | 1.55E-06 | 461 (4.7) | 382 (3.6) | 1.38 (1.09–1.58) | 9.67E-09 | ||||
| rs4736437 | T | 2,233 (77.4) | 2,379 (81.0) | 1.00 (ref) | 12,805 (77.2) | 14,548 (79.4) | 1.00 (ref) | 15,038 (77.3) | 16,927 (79.6) | 1.00 (ref) | 4,529 | |||||
| 134185668 | G | 651 (22.6) | 557 (19.0) | 1.25 (1.10–1.41) | 0.0006 | 3,777 (22.8) | 3,768 (20.6) | 1.14 (1.08–1.20) | 6.24E-07 | 4,428 (22.7) | 4,325 (20.4) | 1.15 (1.10–1.21) | 3.53E–09 | |||
| WISP1/SLA | TT | 856 (59.4) | 958 (65.3) | 1.00 (ref) | 4,959 (59.8) | 5,778 (63.1) | 1.00 (ref) | 5,815 (59.7) | 6,736 (63.4) | 1.00 (ref) | ||||||
| Intergenic | TG | 521 (36.1) | 463 (31.5) | 1.26 (1.08–1.47) | 2,887 (34.8) | 2,992 (32.7) | 1.12 (1.05–1.2) | 3,408 (35) | 3,455 (32.5) | 1.14 (1.21–1.19) | ||||||
| GG | 65 (4.5) | 47 (3.2) | 1.55 (1.05–2.28) | 0.0027 | 445 (5.4) | 388 (4.2) | 1.34 (1.16–1.54) | 3.32E-06 | 510 (5.2) | 435 (4.1) | 1.36 (1.08–1.55) | 2.84E-08 | ||||
| rs10956695 | A | 2,275 (78.9) | 2,414 (82.3) | 1.00 (ref) | 12,611 (79.2) | 13,501 (80.8) | 1.00 (ref) | 14,886 (79.2) | 15,915 (81.0) | 1.00 (ref) | 5,239 | |||||
| 134189138 | G | 609 (21.1) | 520 (17.7) | 1.24 (1.09–1.42) | 0.0009 | 3,303 (20.8) | 3,203 (19.2) | 1.10 (1.05–1.17) | 0.0004 | 3,912 (20.8) | 3,723 (19.0) | 1.12 (1.07–1.18) | 5.52E–06 | |||
| WISP1/SLA | AA | 892 (61.9) | 988 (67.3) | 1.00 (ref) | 5,015 (63.0) | 5,468 (65.5) | 1.00 (ref) | 5,907 (62.8) | 6,456 (65.8) | 1.00 (ref) | ||||||
| Intergenic | AG | 491 (34) | 438 (29.9) | 1.24 (1.06–1.45) | 2,581 (32.4) | 2,565 (30.7) | 1.1 (1.03–1.17) | 3,072 (32.7) | 3,003 (30.6) | 1.12 (1.19–1.1) | ||||||
| GG | 59 (4.1) | 41 (2.8) | 1.59 (1.06–2.4) | 0.0042 | 361 (4.5) | 319 (3.8) | 1.23 (1.06–1.44) | 0.0018 | 420 (4.5) | 360 (3.7) | 1.28 (1.05–1.47) | 3.35E-05 | ||||
| rs7005834 | A | 378 (13.1) | 490 (16.7) | 1.00 (ref) | 2,431 (14.6) | 2,803 (16.2) | 1.00 (ref) | 2,809 (14.4) | 3,293 (16.3) | 1.00 (ref) | 3,493 | |||||
| 134214204 | G | 2,506 (86.9) | 2,446 (83.3) | 1.33 (1.15–1.54) | 0.0001 | 14,179 (85.4) | 14,463 (83.8) | 1.13 (1.07–1.20) | 4.75E-05 | 16,685 (85.6) | 16,909 (83.7) | 1.16 (1.10–1.22) | 1.62E-07 | |||
| WISP1 | AA | 20 (1.4) | 45 (3.1) | 1.00 (ref) | 182 (2.2) | 224 (2.6) | 1.00 (ref) | 202 (2.1) | 269 (2.7) | 1.00 (ref) | ||||||
| Intron1 | AG | 338 (23.4) | 400 (27.2) | 1.9 (1.61–2.25) | 2,067 (24.9) | 2,355 (27.3) | 1.08 (0.88–1.33) | 2,405 (24.7) | 2,755 (27.3) | 1.16 (1.41–1.12) | ||||||
| GG | 1,084 (75.2) | 1,023 (69.7) | 2.38 (1.4–4.07) | 0.0003 | 6,056 (72.9) | 6,054 (70.1) | 1.23 (1.01–1.5) | 0.0002 | 7,140 (73.3) | 7,077 (70.1) | 1.34 (0.96–1.62) | 1.21E-06 | ||||
| rs4330674 | A | 344 (11.9) | 427 (14.5) | 1.00 (ref) | 609 (13.9) | 623 (14.7) | 1.00 (ref) | 953 (13.1) | 1050 (14.6) | 1.00 (ref) | 6,059 | |||||
| 134221502 | G | 2,540 (88.1) | 2,509 (85.5) | 1.26 (1.08–1.46) | 0.0031 | 3,763 (86.1) | 3,629 (85.3) | 1.06 (0.94–1.20) | 0.3406 | 6,303 (86.9) | 6,138 (85.4) | 1.13 (1.03–1.24) | 0.0098 | |||
| WISP1 | AA | 16 (1.1) | 31 (2.1) | 1.00 (ref) | 44 (2.0) | 50 (2.4) | 1.00 (ref) | 60 (1.7) | 81 (2.3) | 1.00 (ref) | ||||||
| Intron1 | AG | 312 (21.6) | 365 (24.9) | 1.66 (1.39–1.97) | 521 (23.8) | 523 (24.6) | 1.13 (0.74–1.73) | 833 (23.0) | 888 (24.7) | 1.27 (1.79–1) | ||||||
| GG | 1114 (77.3) | 1,072 (73.0) | 2.01 (1.09–3.7) | 0.0086 | 1,621 (74.2) | 1,553 (73.0) | 1.19 (0.79–1.79) | 0.6038 | 2,735 (75.4) | 2,625 (73.0) | 1.41 (0.9–1.97) | 0.0305 | ||||
In the combined population we integrated the genotyping results from the discovery and replication stage and confirmed the 7 SNPs were associated with GD. Only rs2294025 and rs4736437 reached the significance level of GWAS (combined P<5×10−8). Rs2294025 (combined P=1.52×10−9, OR, 1.16; 95% CI, 1.11–1.22, Table 2) and rs4736437 were the best association signal with GD (combined P=3.53×10−9; OR, 1.15; 95% CI, 1.10–1.21, Table 2).
Association of two independent SNPs on 8q24.22 with different phenotypes of GD
In order to further investigate the association between the two independent risk loci (rs2294025 and rs7005834) of TG and different phenotypes of GD, the genotype-phenotype association analysis was also performed in GD. Firstly, compared to the male or female control population, the two independent SNPs were associated with the patients who was male GD or female GD (rs2294025: OR, 1.15; 95% CI, 1.04–1.28; male GD vs. male control, P=7.5×10−3; OR, 1.16; 95% CI, 1.10–1.23; female GD vs. female control, P=2.77×10−7; rs7005834: OR, 1.23; 95% CI, 1.10–1.39; male GD vs. male control, P=6×10−4; OR, 1.14; 95% CI, 1.07–1.22; female GD vs. female control, P=6.38×10−5; Table 3). While the allele frequencies of two SNPs (rs2294025 and rs7005834) did not show notable differences between the male and female patients with GD (rs2294025: OR, 1.05; 95% CI, 0.96–1.14; female GD vs. male GD, P=0.2929; rs7005834: OR, 0.91; 95% CI, 0.82–1.00; female GD vs. male GD, P=0.0619, Table 3).
Table 3. Association of two independent SNPs on 8q24.22 in different phenotypes.
| SNP | rs2294025 | rs7005834 | |||||
|---|---|---|---|---|---|---|---|
| RAF (%) | P | OR (95% CI) | RAF (%) | P | OR (95% CI) | ||
| Female GD vs. female Control | 22/20 | 2.77E-07 | 1.16 (1.10–1.23) | 15/16 | 6.38E-05 | 1.14 (1.07–1.22) | |
| Male GD vs. male Control | 21/19 | 0.0075 | 1.15 (1.04–1.28) | 13/16 | 0.0006 | 1.23 (1.1–1.39) | |
| Female GD vs. male GD | 22/21 | 0.2929 | 1.05 (0.96–1.14) | 15/13 | 0.0619 | 0.91 (0.82–1.00) | |
| pTRAb+ vs. pTRAb− | 23/23 | 0.9568 | 1.00 (0.88–1.15) | 15/14 | 0.3823 | 0.93 (0.80–1.09) | |
| GO vs. nonGO | 22/21 | 0.6475 | 1.03 (0.92–1.14) | 14/15 | 0.3627 | 1.06 (0.94–1.20) | |
| TGAb+ vs. TGAb− | 26/22 | 0.1145 | 1.22 (0.95–1.55) | 16/14 | 0.2372 | 0.84 (0.63–1.12) | |
| TPOAb+ vs. TPOAb− | 23/23 | 0.6829 | 0.96 (0.80–1.16) | 14/15 | 0.4787 | 1.08 (0.87–1.34) | |
SNP, single nucleotide polymorphism; RAF, risk allele frequency; OR, odds ratio; CI, confidence interval; GD, Graves’ disease; pTRAb+, persistent TRAb positivity; GO, Graves’ ophthalmopathy.
As for the three-specific antibody of thyroid, it is interesting to study whether the phenotypic heterogeneity of the two susceptibility SNPs existed between GD patients with different antibody level. In this study, after ATD treatment for more than 1 year, all GD patients were classified as TRAb-positive and TRAb-negative subtypes, TGAb-positive and TGAb-negative subtypes, TPOAb-positive and TPOAb-negative subtypes according to the level of TRAb, TGAb and TPOAb. The allele frequencies of the SNPs (rs2294025 and rs7005834) showed no significant differences between each two subtypes according to the level of three specific antibody of thyroid (heterogeneity test P>0.05; Table 3).
Meanwhile, GD patients were divided into GO (class 3–6) patients and non-GO (class 0–1) patients by the degree of GO. However, there was no heterogeneity for the allele frequencies of the SNPs (rs2294025 and rs7005834) exited between GO patients and non-GO patients.
Discussion
In 9,757 GD patients and 10,726 gender matching controls, a fine-mapping association analysis was performed for 7 SNPs in TG. In Chinese Han population, rs2294025 and rs7005834 were the two independent GD susceptibility loci. It further provides theoretical basis for the influence of ethnic and geographical variation on genetic susceptibility of GD. However, SNPs rs7005834 and rs2294025 were not associated with the different phenotypes of GD, including gender, GO, and different thyroid autoantibody level, which may provide valuable reference information for clinical prevention and treatment of GD.
During the past three decades, several immunomodulatory and thyroid specific genes as GD susceptibility genes have been identified and confirmed by linkage and association analysis, such as human leukocyte antigen D related (HLA-DR), cluster of differentiation 40 (CD40), cytotoxic T lymphocyte-associated antigen 4 (CTLA-4), thyroid-stimulating hormone receptor (TSHR) and so on (25).
The polymorphism of TG has been studied in different races, and several studies have ascertained that TG is the susceptibility gene of patients with GD, but the correlation between the susceptibility loci in the TG region and GD is still controversial. Hsiao et al. found that the TT genotype of exon 33 SNP and the GG genotype of exon 12 SNP were correlated with GD in Taiwan population (P<0.001) (26). In Japanese Ban found that in intron 41 of the TG, rs2256366 and rs2687836, were the strongest SNPs associations with GD (P = 0.002 and P = 0.0077) (27). In Chinese in TG the genotype CC of rs2069550 had been confirmed a higher risk of GD (28). At the same time, in Italian, Lombardi A confirmed that TG (rs 2069561) was associated with GD (17). However, the Tunisian family data and case-control studies have not detected the relationship between four SNPs located respectively at exon 10 (Ser715Ala), exon 12 (Met1009Val), exon 21 (Ala1483Ala) and exon 33 (Arg1980Trp) and GD (19).
In the Japanese population Ban conducted a correlation study and found that the microsatellite D8S284, D8S272 in the chromosome 8q24 and the microsatellite Tgms1and Tgms2 in intron 10 and intron 27, the exon 33 SNP of the allele frequency and genotype in GD cases and controls are no different (29). In Chinese, the allele and genotype frequencies in four common TG SNPs (T/G genotype of exon 10 SNP24 and T/C genotype of exon 10 SNP158, A/G genotype of exon 12 SNP and C/T genotype of exon 33 SNP) were found no differences between GD patients and control subjects (30).
Therefore, GD genetic susceptibility may involve different race/geographic populations genetic TG polymorphisms. Traditional research strategy is candidate gene strategy and microsatellite marker method to consider the possibility of disease susceptibility genes. Candidate gene strategy mainly aims at the area of the encoded amino acid variation, thereby the quantity of susceptible sites identified is very limited which cannot fully reflect the relationship between genes and disease (31). Linkage analysis of microsatellite markers tends to locate a very large range of susceptible segments. Thus, it is hard to confirm disease susceptibility genes in the genome (32). Therefore, a limited number of SNPs were analyzed in a small sample population. and it may be impossible for confirmation due to the differences in the pathogenic susceptibility loci or disease-causing susceptibility genes between different study populations (33).
In this study, we used a two-stage correlation study and enrolled a total of 9,757 GD patients and 10,626 control subjects in the Chinese Han population. We found that the SNPs rs2294025 and rs7005834 in TG were probably independent susceptibility loci to patients with GD for the first time. Our findings further highlight the effects of ethnic and geographic variations on genetic susceptibility to GD. The patients with GD lose immune tolerance to thyroid antigens such as TG, TPO, and TSHR genes (30). Particularly, TG is one of the most important candidate genes for GD. Serum TG level was positively correlated with the presence and severity of ophthalmopathy in GD patients. Tomoyo Mizuma found SNPs in the TG were associated with the serum levels of TG, TGAb, and TG mRNA expression (16). Haplotype analysis of these four SNPs (T/G genotype of exon 10 SNP24 and T/C genotype of exon 10 SNP158, A/G genotype of exon 12 SNP and C/T genotype of exon 33 SNP) showed that the haplotype G-C-A-C was significantly associated with the level of serum TGAb in AITD patients (P=0.028, OR=3.34) (18). Unfortunately, in this study we found that there were no significant differences for the allele frequencies of the SNPs (rs2294025 and rs7005834) between the GD patients with different phenotypes, including gender, GO, persistently positivity of TRAb, TGAb or TPOAb level. It may be that our samples were not large enough to show statistical differences. Maybe TGAb and TPOAb were the marker antibodies of AITD, but not necessarily the key pathogenic antibodies of GD, so there was no correlation between susceptibility sites and phenotypes.
We found that frequency of rs7005834 was higher in female GD patients than male GD patients. However, while the difference was not significant (P=0.0619). GD is one of the most common types of AITD with a dimorphism of gender, indicating a higher morbidity in females then males. Regulatory immune response of estrogen and androgen may play an important role (34). Testosterone prevented genesis of AITD in obese chickens (35,36). Injection of testosterone could lead the disappearance of invasive monocytes in thyroid of rats with AITD and recovery of thyroid structures (37). Krysiak et al. found that exogenous testosterone could inhibit thyroid autoimmunity (38). Chailurkit LO found that E2 was the determinant of positive TRAb, and elevated circulating E2 was related with thyroid autoimmunity in males (39). Chen et al. found that elevated E2/T ratio was related with increased AITD risk in males through an observational study (40). All these studies indicated that estrogen was a risk factor of AITD, and testosterone was a protective factor. Besides, Pyun et al. found that TG was involved in regulation of estradiol (41). In 2017, Fernández et al. found that gene polymorphism of TG was related with sexual precocity, and the puberty age with haploid GAT decreased significantly in Guzerat bulls (42). Together with our results, gene polymorphism of TG might play a role in the dimorphism of gender in GD patients.
There are four main limitations in this study. First, the clinical data of GD patients that we collected were not complete enough. Second, the sample size was not large enough to conclude that TG gene polymorphism was significantly correlated with clinical indicators. Third, we did not perform the follow-up study for the patients with GD recruited in our study. Fourth, we have not conducted further functional studies on these two independent risk loci.
In conclusion, our study indicated that SNPs rs2294025 and rs7005834 in TG were probably independent susceptibility loci to GD in the Chinese Han population. The influence of ethnic and geographical variation on genetic susceptibility of GD is further emphasized, which provides theoretical basis for understanding the pathogenesis of GD. However, there were no significant differences for the allele frequencies of rs2294025 and rs7005834, between the different clinical phenotypes of GD, including gender, GO, the serum level of TRAb, TGAb, or TPOAb. The frequency of rs7005834 gene in Female GD patients tends to be higher than that in male GD patients, suggesting that TG gene polymorphism may play a certain role in gender atypia of GD, and more research data are needed to demonstrate.
URLs
PLINK v1.07, http://pngu.mgh.harvard.edu/~purcell/plink/
IMPUTE2, http://mathgen.stats.ox.ac.uk/impute/impute_v2.html/
Haploview Software, https://www.broadinstitute.org/haploview/haploview
Acknowledgments
Funding: This study was funded in part by the National Natural Science Foundation of China (30971595, 30971383, 81100553,81200568, 81270863 and 31171127), National Basic Research Program of China [973] (2010CB529204 and 2012CB517604), Shanghai Science and Technology Committee (10JC1410400), Program for Graves’ Disease Innovative Research Team of Shanghai Municipal Education Commission, and Natural Science Foundation of Jiangsu Province, China (BK2009208 and SBK201221245).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the local ethics committee from Ruijin Hospital, the Central Hospital of Xuzhou, Linyi People’s Hospital, the First Affiliated Hospital of Bengbu Medical College, Medical School Hospital of Qingdao University, the Hospital Affiliated to Jiangsu University, Xin-Hua Hospital. The recruitment of patients and controls followed the ethical considerations and requirements of the Helsinki accord.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Weetman AP, McGregor AM. Autoimmune thyroid disease: further developments in our understanding. Endocr Rev 1994;15:788-830. [DOI] [PubMed] [Google Scholar]
- 2.Brix TH, Kyvik KO, Christensen K, et al. Evidence for a major role of heredity in Graves' disease: a population-based study of two Danish twin cohorts. J Clin Endocrinol Metab 2001;86:930-4. [DOI] [PubMed] [Google Scholar]
- 3.Brix TH, Kyvik KO, Hegedus L. What is the evidence of genetic factors in the etiology of Graves' disease? A brief review. Thyroid 1998;8:627-34. 10.1089/thy.1998.8.627 [DOI] [PubMed] [Google Scholar]
- 4.Stenszky V, Kozma L, Balazs C, et al. The genetics of Graves' disease: HLA and disease susceptibility. J Clin Endocrinol Metab 1985;61:735-40. 10.1210/jcem-61-4-735 [DOI] [PubMed] [Google Scholar]
- 5.Brix TH, Christensen K, Holm NV, et al. A population-based study of Graves' disease in Danish twins. Clin Endocrinol (Oxf) 1998;48:397-400. 10.1046/j.1365-2265.1998.00450.x [DOI] [PubMed] [Google Scholar]
- 6.Van Herle AJ, Vassart G, Dumont JE. Control of thyroglobulin synthesis and secretion. (First of two parts). N Engl J Med 1979;301:239-49. 10.1056/NEJM197908023010504 [DOI] [PubMed] [Google Scholar]
- 7.Van Herle AJ, Vassart G, Dumont JE. Control of thyroglobulin synthesis and secretion (second of two parts). N Engl J Med 1979;301:307-14. 10.1056/NEJM197908093010605 [DOI] [PubMed] [Google Scholar]
- 8.Shulman S. Thyroid antigens and autoimmunity. Adv Immunol 1971;14:85-185. 10.1016/S0065-2776(08)60284-9 [DOI] [PubMed] [Google Scholar]
- 9.Charreire J. Immune mechanisms in autoimmune thyroiditis. Adv Immunol 1989;46:263-334. 10.1016/S0065-2776(08)60656-2 [DOI] [PubMed] [Google Scholar]
- 10.Chen CR, Hamidi S, Braley-Mullen H, et al. Antibodies to thyroid peroxidase arise spontaneously with age in NOD.H-2h4 mice and appear after thyroglobulin antibodies. Endocrinology 2010;151:4583-93. 10.1210/en.2010-0321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tomer Y, Ban Y, Concepcion E, et al. Common and unique susceptibility loci in Graves and Hashimoto diseases: results of whole-genome screening in a data set of 102 multiplex families. Am J Hum Genet 2003;73:736-47. 10.1086/378588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ban Y, Greenberg DA, Concepcion E, et al. Amino acid substitutions in the thyroglobulin gene are associated with susceptibility to human and murine autoimmune thyroid disease. Proc Natl Acad Sci U S A 2003;100:15119-24. 10.1073/pnas.2434175100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang LQ, Wang TY, Sun QL, et al. Correlation between thyroglobulin gene polymorphisms and autoimmune thyroid disease. Mol Med Rep 2015;12:4469-75. 10.3892/mmr.2015.3978 [DOI] [PubMed] [Google Scholar]
- 14.Tomer Y, Greenberg DA, Concepcion E, et al. Thyroglobulin is a thyroid specific gene for the familial autoimmune thyroid diseases. J Clin Endocrinol Metab 2002;87:404-7. 10.1210/jcem.87.1.8291 [DOI] [PubMed] [Google Scholar]
- 15.Hsiao JY, Hsieh MC, Tien KJ, et al. Association between a C/T polymorphism in exon 33 of the thyroglobulin gene is associated with relapse of Graves' hyperthyroidism after antithyroid withdrawal in Taiwanese. J Clin Endocrinol Metab 2007;92:3197-201. 10.1210/jc.2007-0675 [DOI] [PubMed] [Google Scholar]
- 16.Mizuma T, Watanabe M, Inoue N, et al. Association of the polymorphisms in the gene encoding thyroglobulin with the development and prognosis of autoimmune thyroid disease. Autoimmunity 2017;50:386-92. 10.1080/08916934.2017.1344971 [DOI] [PubMed] [Google Scholar]
- 17.Lombardi A, Menconi F, Greenberg D, et al. Dissecting the Genetic Susceptibility to Graves' Disease in a Cohort of Patients of Italian Origin. Front Endocrinol (Lausanne) 2016;7:21. 10.3389/fendo.2016.00021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maierhaba M, Zhang JA, Yu ZY, et al. Association of the thyroglobulin gene polymorphism with autoimmune thyroid disease in Chinese population. Endocrine 2008;33:294-9. 10.1007/s12020-008-9082-x [DOI] [PubMed] [Google Scholar]
- 19.Belguith-Maalej S, Hadj Kacem H, Rebai A, et al. Thyroglobulin polymorphisms in Tunisian patients with autoimmune thyroid diseases (AITD). Immunobiology 2008;213:577-83. 10.1016/j.imbio.2008.01.004 [DOI] [PubMed] [Google Scholar]
- 20.Zhao SX, Xue LQ, Liu W, et al. Robust evidence for five new Graves' disease risk loci from a staged genome-wide association analysis. Hum Mol Genet 2013;22:3347-62. 10.1093/hmg/ddt183 [DOI] [PubMed] [Google Scholar]
- 21.Chu X, Pan CM, Zhao SX, et al. A genome-wide association study identifies two new risk loci for Graves' disease. Nat Genet 2011;43:897-901. 10.1038/ng.898 [DOI] [PubMed] [Google Scholar]
- 22.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 2009;5:e1000529. 10.1371/journal.pgen.1000529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrett JC, Fry B, Maller J, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005;21:263-5. 10.1093/bioinformatics/bth457 [DOI] [PubMed] [Google Scholar]
- 24.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559-75. 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobson EM, Huber A, Tomer Y. The HLA gene complex in thyroid autoimmunity: from epidemiology to etiology. J Autoimmun 2008;30:58-62. 10.1016/j.jaut.2007.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsiao JY, Hsieh MC, Tien KJ, et al. Exon 33 T/T genotype of the thyroglobulin gene is a susceptibility gene for Graves' disease in Taiwanese and exon 12 C/C genotype protects against it. Clin Exp Med 2008;8:17-21. 10.1007/s10238-008-0151-5 [DOI] [PubMed] [Google Scholar]
- 27.Ban Y, Tozaki T, Taniyama M, et al. Multiple SNPs in intron 41 of thyroglobulin gene are associated with autoimmune thyroid disease in the Japanese population. PLoS One 2012;7:e37501. 10.1371/journal.pone.0037501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gu LQ, Zhu W, Zhao SX, et al. Clinical associations of the genetic variants of CTLA-4, Tg, TSHR, PTPN22, PTPN12 and FCRL3 in patients with Graves' disease. Clin Endocrinol (Oxf) 2010;72:248-55. 10.1111/j.1365-2265.2009.03617.x [DOI] [PubMed] [Google Scholar]
- 29.Ban Y, Tozaki T, Taniyama M, et al. Association of a thyroglobulin gene polymorphism with Hashimoto's thyroiditis in the Japanese population. Clin Endocrinol (Oxf) 2004;61:263-8. 10.1111/j.1365-2265.2004.02096.x [DOI] [PubMed] [Google Scholar]
- 30.Tomer Y, Davies TF. Searching for the autoimmune thyroid disease susceptibility genes: from gene mapping to gene function. Endocr Rev 2003;24:694-717. 10.1210/er.2002-0030 [DOI] [PubMed] [Google Scholar]
- 31.Tesson F, Leenen FH. Still building on candidate-gene strategy in hypertension? Hypertension 2007;50:607-8. 10.1161/HYPERTENSIONAHA.107.096800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grover A, Sharma PC. Development and use of molecular markers: past and present. Crit Rev Biotechnol 2016;36:290-302. 10.3109/07388551.2014.959891 [DOI] [PubMed] [Google Scholar]
- 33.Lohmueller KE, Pearce CL, Pike M, et al. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat Genet 2003;33:177-82. 10.1038/ng1071 [DOI] [PubMed] [Google Scholar]
- 34.Lleo A, Battezzati PM, Selmi C, et al. Is autoimmunity a matter of sex? Autoimmun Rev 2008;7:626-30. 10.1016/j.autrev.2008.06.009 [DOI] [PubMed] [Google Scholar]
- 35.Fassler R, Dietrich H, Kromer G, et al. The role of testosterone in spontaneous autoimmune thyroiditis of Obese strain (OS) chickens. J Autoimmun 1988;1:97-108. 10.1016/0896-8411(88)90081-9 [DOI] [PubMed] [Google Scholar]
- 36.Gause WC, Marsh JA. Effect of testosterone treatments for varying periods on autoimmune development and on specific infiltrating leukocyte populations in the thyroid gland of obese strain chickens. Clin Immunol Immunopathol 1986;39:464-78. 10.1016/0090-1229(86)90174-1 [DOI] [PubMed] [Google Scholar]
- 37.Ansar Ahmed S, Young PR, Penhale WJ. Beneficial effect of testosterone in the treatment of chronic autoimmune thyroiditis in rats. J Immunol 1986;136:143-7. [PubMed] [Google Scholar]
- 38.Krysiak R, Kowalcze K, Okopien B. The effect of testosterone on thyroid autoimmunity in euthyroid men with Hashimoto's thyroiditis and low testosterone levels. J Clin Pharm Ther 2019. [Epub ahead of print]. 10.1111/jcpt.12987 [DOI] [PubMed] [Google Scholar]
- 39.Chailurkit LO, Aekplakorn W, Ongphiphadhanakul B. The relationship between circulating estradiol and thyroid autoimmunity in males. Eur J Endocrinol 2013;170:63-7. 10.1530/EJE-13-0455 [DOI] [PubMed] [Google Scholar]
- 40.Chen Y, Chen Y, Xia F, et al. A Higher Ratio of Estradiol to Testosterone Is Associated with Autoimmune Thyroid Disease in Males. Thyroid 2017;27:960-6. 10.1089/thy.2016.0661 [DOI] [PubMed] [Google Scholar]
- 41.Pyun JA, Kim S, Kwack K. Interaction between thyroglobulin and ADAMTS16 in premature ovarian failure. Clin Exp Reprod Med 2014;41:120-4. 10.5653/cerm.2014.41.3.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fernández ME, Loaiza Echeverri AM, Henry M, et al. Bovine thyroglobulin gene polymorphisms and their association with sexual precocity in Guzerat bulls. Reprod Domest Anim 2017;52:911-3. 10.1111/rda.12989 [DOI] [PubMed] [Google Scholar]