Abstract
Background
The papillary renal cell carcinoma (pRCC) is a rare subtype of renal cell carcinoma with limited investigation. Our study aimed to explore a robust signature to predict the prognosis of pRCC from the perspective of mutation profiles.
Methods
In this study, we downloaded the simple nucleotide variation data of 288 pRCC samples from The Cancer Genome Atlas (TCGA) database. “GenVisR” package was utilized to visualize gene mutation profiles in pRCC. The PPI network was conducted based on the STRING database and the modification was performed via Cytoscape software (Version 3.7.1). Top 50 mutant genes were selected and Cox regression method was conducted to identify the hub prognostic mutant signature in pRCC using “survival” package. Mutation Related Signature (MRS) risk score was established by multivariate Cox regression method. Receiver Operating Characteristic (ROC) curve drawn by “timeROC” was conducted to assess the predictive accuracy of overall survival (OS) and Kaplan-Meier analysis was then performed. Relationships between mutants and expression levels were compared by Wilcox rank-sum test. Function enrichment pathway analysis for mutated genes was performed by “org.Hs.eg.db”, “clusterProfiler”, “ggplot2” and “enrichplot” packages. Gene Set Enrichment Analysis was exploited using the MRS as the phenotypes, which worked based on the JAVA platform. All statistical analyses were achieved by R software (version 3.5.2). P value <0.05 was considered to be significant.
Results
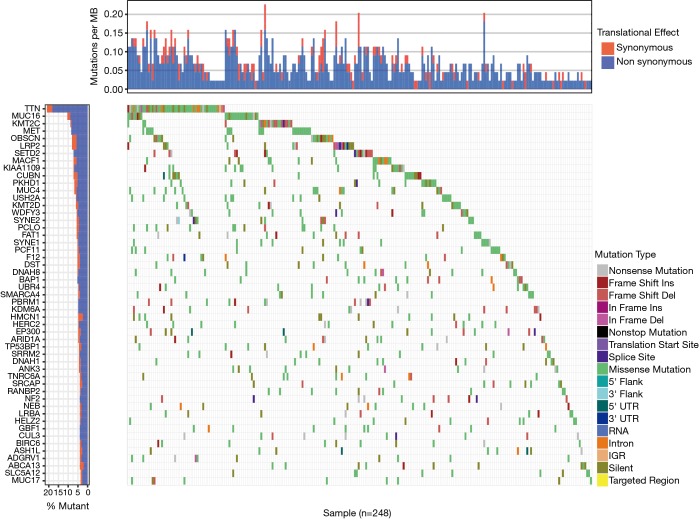
The mutation landscape in waterfall plot revealed that a list of 49 genes that were mutated in more than 10 samples, of which 6 genes (TTN, MUC16, KMT2C, MET, OBSCN, LRP2) were mutated in more than 20 samples. Besides, non-synonymous was the most frequent mutation effect, and missense mutation was one of the most common mutation types in mutated genes across 248 samples. The AUC of MRS model consisted of 17 prognostic mutant signatures was 0.907 in 3-year OS prediction. Moreover, pRCC patients with high level of MRS showed the worse survival outcomes compared with that in low-level MRS group (P=0). In addition, correlation analysis indicated that 6 mutated genes (BAP1, OBSCN, NF2, SETD2, PBRM1, DNAH1) were significantly associated with corresponding expression levels. Last, functional enriched pathway analysis showed that these mutant genes were involved in multiple cancer-related crosstalk, including PI3K-AKT signaling pathway, JAK-STAT signaling pathway, extracellular matrix (ECM)-receptor interaction or cell cycle.
Conclusions
In summary, our study was the first attempt to explore the mutation-related signature for predicting survival outcomes of pRCC based on the high-throughput data, which might provide valuable information for further uncovering the molecular pathogenesis in pRCC.
Keywords: Simple nucleotide variation, prognosis, papillary renal cell carcinoma (pRCC), high-throughput data
Introduction
Renal cell carcinoma (RCC), which is the second most common malignancy of the urinary system, comprising almost 2–3% among all human malignant neoplasms (1-3). It is noticeable that RCC is a comprehensive term for a range of different malignancies with disparate genetic drivers and histology, which lead to distinctive clinical features and treatment responses (4,5). Accounting for 15–20% of RCC, papillary renal cell carcinoma (pRCC) consists of two main sub-types: type 1 appears as papillae and tubular structures covered by small basophilic cells with small oval nuclei; type 2 is featured by large eosinophilic cells with large spherical nuclei (6,7). It was reported that the greater stage, more aggressive tumor behavior and worse prognosis were detected in type 2 pRCC (8,9).
The biological manifestations of pRCC have heterogeneity. For instance, pRCC in some patients appears as indolent, bilateral and multifocal lesions, while other patients have solitary lesions with stronger invasiveness (6,7,10). Heterogeneity of oncology might suggest that multiple abnormal gene regulations and corresponding signal pathway changes are existed during the tumorigenesis and development of pRCC. However, limited research of pRCC gene abnormalities and regulation has resulted in great challenges in effective individualized treatment options for advanced pRCC. For example, tyrosine kinase inhibitors of VEGF pathways are significantly inferior in pRCC patients compared to clear cell renal cell carcinoma patients (11). Therefore, further studies of the correlation between pRCC gene abnormalities and clinical manifestations of patients are particularly important for future precision treatment of pRCC.
It was found that the gene mutation played an important role in tumorigenesis and development of malignancy in recent decades (12,13). Irrespective of non-familial cancer types, cancer is often regarded as a disease of somatic mutations. Gene mutation locations and types are always the key factors determining gene function in biological behavior of malignant tumors (14-16). In pRCC, major studies were focused on the function of scattered mutant genes, whereas researches from the perspective of genome-wide mutation profiles were limited.
In recent years, a new period for cancer genomics research was formed by the flying start of the high-throughput technology. The Cancer Genome Atlas (TCGA) database, which is a pool of molecular data sets, consists cancer-causing genomic alterations among various malignant tumors (17). In our research, we downloaded the simple nucleotide variation data of pRCC samples from TCGA database. By analysis we screened for tumor mutations in pRCC, and its impact on the prognosis of patients with pRCC was investigated.
Methods
Data acquisition and preparation
We obtained the mutation data of 288 pRCC samples from TCGA database by utilizing GDC-client.exe software. Since the raw SNP data were not publicly available, we downloaded the data type of “Masked Somatic Mutation”, which was processed based on the VarScan software (18). Then, we utilized the Perl scripts in Figure S1 to extract the all mutation information in pRCC (http://fp.amegroups.cn/cms/6c072fd73aee36134dd467610b311007/atm.2019.08.113-1.xls) and “GenVisR” package was used to achieve the visualization (19). Besides, count data of the transcriptome profiles were also downloaded from the data portal, including 289 pRCC samples and matched 32 normal samples. We exploited the “edgeR” package to conduct the normalization process and the differentially expressed genes were shown by “ggplot2” in volcano plot. Last, we obtained clinical data of all pRCC samples, including age, gender, pathological stages, AJCC-TNM stages and survival outcomes with follow-up time. We integrated the mutation data with clinical information or transcriptome profiles using “merge” function in R.
Figure S1.

Exhibition of Perl scripts to extract the mutation data.
Construction of PPI network for mutation related genes
We selected the top 200 mutation genes in Figure S1 to investigate the potential molecular interactions among them. We utilized STRING database online (https://string-db.org/) to generate the PPI network of mutant genes, where the confidence score was set with 0.7 as the cutoff criteria and the disconnected nodes were hidden (20). Furthermore, we downloaded the interaction data with tsv format to modify the PPI network in Cytoscape software (Version 3.7.1) based on JAVA platform (21).
Identification of hub prognostic mutant genes
In order to identify hub mutant genes for prognosis of pRCC, we selected the top 50 mutant genes from Figure S1 with corresponding transcriptome data. We conducted the univariate Cox regression analysis to find the survival-associated mutant genes with P<0.05. Then, multivariate Cox regression model was performed to get the respective coefficients (βi) and construct the risk score with Ʃ (βi * Expi), where Expi represented the expression data of significant mutant genes from the univariate Cox analysis. Subsequently, we calculated the risk scores for each pRCC samples and classified the 288 pRCC patients into low- and high-groups (http://fp.amegroups.cn/cms/475325fb380311b80e51ecca25743a55/atm.2019.08.113-2.xls). Furthermore, we assessed the 3-year predictive value of risk score based on the hub mutant genes by Receiver Operating Characteristic (ROC) curve (22). A Kaplan-Meier plot was drawn to compare the survival difference between the two groups and P value of the log-rank test was calculated. ROC curve and survival plots were generated by “survival” packages.
Survival analysis, correlation analysis of mutants with expression levels
We selected 17 prognostic mutant genes from the Cox regression results to conduct the Kaplan-Meier analysis (Table 1). We utilized the “for cycle” R script to perform the batch survival analysis of the 17 genes via “survival” package. In addition, we merged the mutation data with the expression levels of genes. The Wilcoxon rank-sum test was used to compare the differential expression levels between wild type and mutants. A P value of log-rank test <0.05 was thought to be significant.
Table 1. 17 prognostic mutation related genes from univariate Cox regression analysis.
| Gene | HR | Z | P value |
|---|---|---|---|
| NEB | 1.535890195 | 6.119890639 | 9.36E-10 |
| HMCN1 | 1.434927891 | 3.510041483 | 0.000448 |
| NF2 | 0.628545732 | −3.033256451 | 0.0024193 |
| USH2A | 1.435435343 | 2.961594838 | 0.0030605 |
| LRP2 | 0.875104739 | −2.880651139 | 0.0039685 |
| WDFY3 | 0.531424334 | −2.669286162 | 0.0076013 |
| SETD2 | 0.463893541 | −2.650834699 | 0.0080293 |
| LRBA | 0.516620964 | −2.637850365 | 0.0083433 |
| BAP1 | 0.566327974 | −2.580426232 | 0.0098678 |
| CUBN | 0.847839001 | −2.549441457 | 0.0107896 |
| DNAH1 | 0.670617495 | −2.495101812 | 0.0125921 |
| KIAA1109 | 0.626317538 | −2.489827775 | 0.0127805 |
| HELZ2 | 1.82101297 | 2.44887956 | 0.0143301 |
| MUC16 | 1.115685673 | 2.130147912 | 0.0331594 |
| PBRM1 | 0.572329647 | −2.099277614 | 0.0357924 |
| DNAH8 | 1.237585127 | 2.098534296 | 0.035858 |
| OBSCN | 1.32128618 | 2.07721441 | 0.0377818 |
Functional analysis and Gene Set Enrichment Analysis (GSEA)
To further uncover the underlying biological pathways that these mutation genes might be associated with, we selected the top 300 mutation genes to conduct the enrichment pathway analysis. Firstly, we used the “org.Hs.eg.db” package to transfer the gene symbols to entrezIDs for subsequent analysis. Then, “clusterProfiler”, “ggplot2” and “enrichplot” packages were utilized to find the enriched Gene Ontology (GO) items (23), where FDR <0.05 was statistically significant. We described the enrichment results from three aspects, including biological process (BP), cellular component (CC), and molecular function (MF). What is more, we further identified hub Kyoto Gene and Genome Encyclopedia (KEGG) pathways with P<0.05 shown by dot plot (24).
Statistical analysis
We used “edgeR” to do the normalization and differential analysis. Univariate- and multivariate Cox regression analysis was performed based on “survival” packages. The Student’s t test was mainly used for continuous variables, while categorical variables were estimated by Chi-square test. The Wilcoxon rank-sum test was a non-parametric test mainly probable for comparing the differential distribution between two groups. Statistical analysis was achieved by R software (version 3.5.2). A P value <0.05 was thought to be significant.
Results
Data processing and landscape of mutation profiles in pRCC
We downloaded a total of 320 samples from TCGA database and there were 214 males and 77 females in the TCGAKIRP cohort, respectively. The average age was 61.49±12.07. The other clinical variables were shown in Table 2. We finally selected the 288 pRCC patients with complete simple nucleotide variation sequencing data from the total 291 pateints. We utilized the Pearl scripts to extract the mutation information in Figure S1. The results revealed a list of 49 genes that were mutated in more than 10 samples, of which 6 genes (TTN, MUC16, KMT2C, MET, OBSCN, LRP2) were mutated in more than 20 samples. We then used the “GenVisR” package to visualize the landscape of mutation profiles in pRCC (Figure 1). The waterfall plot illustrated that non-synonymous was the most frequent mutation effect, and missense mutation was one of the most common mutation types in mutated genes across 248 samples.
Table 2. Clinical baseline of patients included in study from TCGA-KIRP dataset.
| Variables | Number (%) |
|---|---|
| Vital status | |
| Alive | 40 (13.75) |
| Dead | 251 (86.25) |
| Age | 61.49±12.07 |
| Full time (days) | 977.15±888.11 |
| Gender | |
| Female | 77 (26.46) |
| Male | 214 (73.54) |
| AJCC-T | |
| T1 | 194 (66.67) |
| T2 | 33 (11.34) |
| T3 | 60 (20.61) |
| T4 | 2 (0.69) |
| Unknown | 2 (0.69) |
| AJCC-N | |
| N0 | 50 (17.18) |
| N1 | 24 (8.25) |
| N2 | 4 (1.37) |
| Unknown | 213 (73.20) |
| AJCC-M | |
| M0 | 95 (32.65) |
| M1 | 9 (3.09) |
| Unknown | 187 (64.26) |
| Stage | |
| Stage I & II | 194 (66.67) |
| Stage III & IV | 67 (23.02) |
| Unknown | 30 (10.31) |
AJCC, American Joint Committee on Cancer.
Figure 1.
Waterfall map of top 50 genes that mutated in KIRP samples. The annotation of mutation types were shown on the right with various colors.
Construction of PPI network and Mutation Related Signature (MRS) for mutant genes
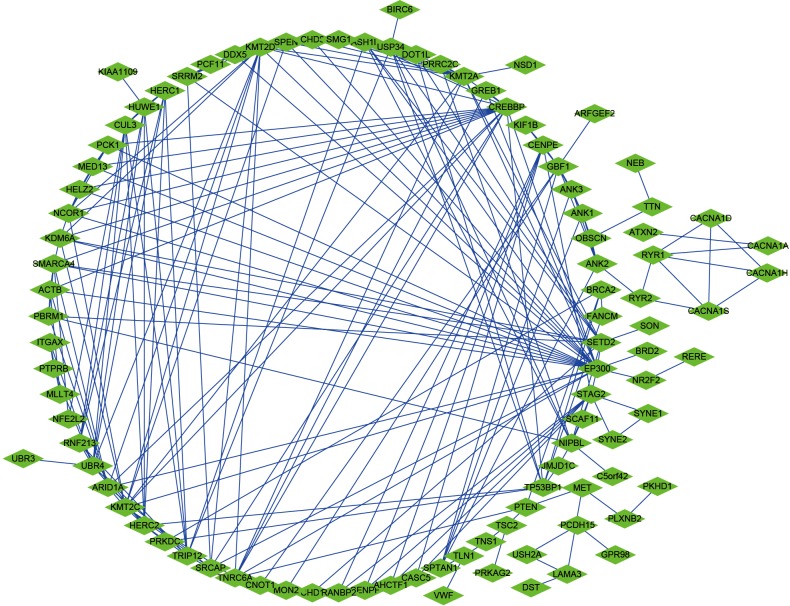
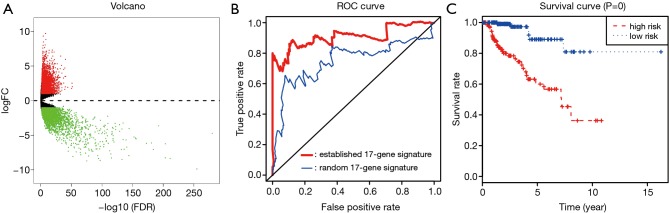
We selected a total of 600 mutated genes to construct the protein-to-protein network between mutated genes, containing 400 nodes and 1476 edges (Figure 2). We then utilized the “limma” package to conduct the normalization and differentially expressed genes (DEGs) analysis with |log(FoldChange) >1| and FDR <0.05. The significantly expressed genes between tumor and normal tissues were shown in volcano plot in Figure 3A. We then obtained the top 50 mutant genes for subsequent analysis. Univariate cox analysis was conducted with “survival” package to identify 17 hub prognostic mutant genes with P<0.05. Additionally, we conducted the multivariate Cox regression analysis to acquire the coefficients (βi) for hub genes, which represented the perspective weight. Based on this, the MRS could be established as: MRS = (-0.37681)*BAP1-0.20175*CUBN-0.05449*DNAH1-0.02236*DNAH8+0.65472*HELZ2+0.48385*HMCN1-0.86104*KIAA1109-0.39687*LRBA+0.01764*LRP2-0.01341*MUC16+0.52157*NEB-0.12864*NF2-0.14096*OBSCN-0.48299*PBRM1+1.13425*SETD2+0.13304*USH2A-0.44975*WDFY3. The receiver operating characteristic (ROC) curve was performed by “timeROC” package to evaluate the predictive accuracy of MRS model. The AUC of ROC was 0.907, which was higher than the AUC based on random 17-gene signature with only 0.746 indicating the high predictive accuracy for our identified MRS model (Figure 3B). What is more, pRCC patients with high level of MRS showed the worse survival outcomes compared with that in low-level MRS group (P=0), which means patients with more frequent mutant of these identified mutant genes suffered poor prognosis (Figure 3C).
Figure 2.
Construction of protein to protein interaction (PPI) network in mutated genes.
Figure 3.
Construction and assessment of TMB-related risk score for KIRP. (A) Differentially expressed genes analysis with |log(Foldchange) >1| and FDR <0.05; (B) the AUC of ROC curve was 0.907 showing the superior predictive accuracy of risk score than the random 17-gene based signature with only 0.746; (C) patients with high risk level revealed poor survival outcomes.
Kaplan-Meier survival analysis of mutant genes and correlation with gene expression
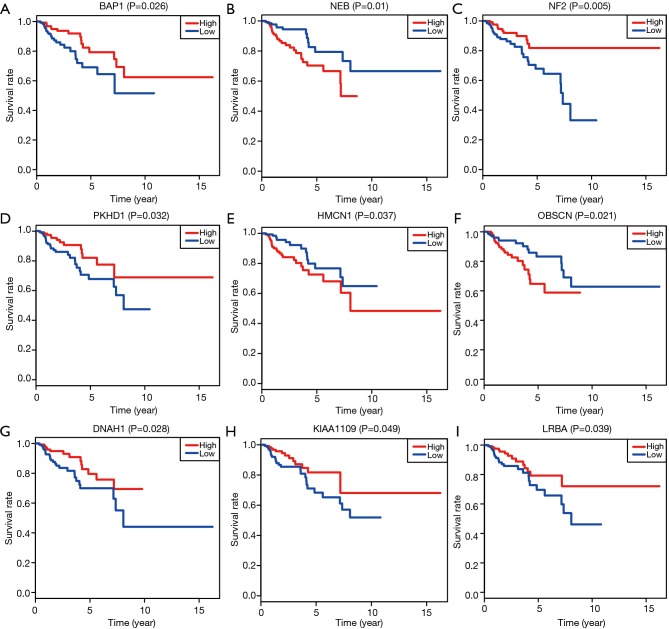
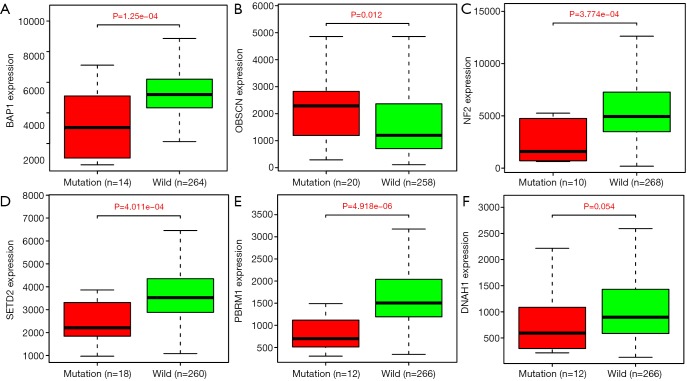
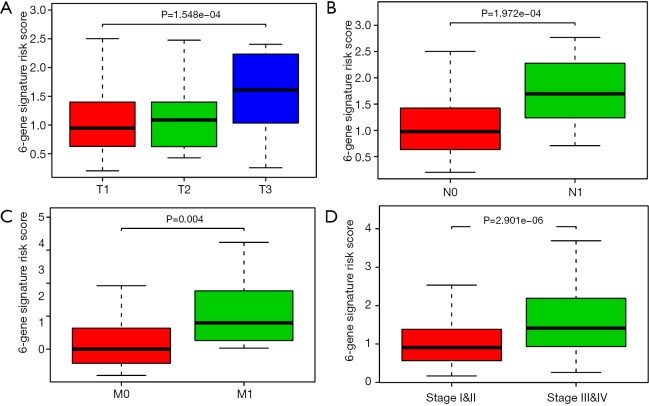
Based on the Cox regression results, we could divide the patients into two groups with low- and high-expression level according to the median value of one gene. Kaplan-Meier plot was drawn to assess the prognostic value of 17 hub mutant genes and the P value of log-rank test <0.05 was thought to be significant. We utilized the median of transcriptome data as the cutoff to divide the expression levels of one gene into high and low groups. Higher levels of NEB, HMCN1 and OBSCN were associated with poor survival outcomes, while lower levels of BAP1, NF2, PKHD1, DNAH1, KIAA1009 and LRBA correlated with a worse prognosis (Figure 4). No significant survival difference was observed in other hub mutant genes, which might result from the defined cutoff value or need large samples to validate. Moreover, the correlation analysis indicated relationships between the mutation and expression level of 6 genes (BAP1, OBSCN, NF2, SETD2, PBRM1, DNAH1), in which OBSCN, NF2 and DNAH1 mutated more frequently in pRCC than ccRCC (Figure S2). The box plots revealed that the expression levels of BAP1, NF2, SETD2, PBRM1 and DNAH1 were lower in mutant samples, while the expression level of OBSCN increased compared with normal samples (Figure 5). Additionally, we integrated the key 6 genes into one signature via multivariate Cox method and correlation analysis suggested that the 6-gene based signature correlated with higher TNM stages and advanced pathological stages (Figure S3).
Figure 4.
Kaplan-Meier analysis with log-rank test for identified hub TMB-related signature. TMB, tumor mutation burden.
Figure S2.
The specific mutation profiling of ccRCC cohorts. ccRCC, clear cell renal cell carcinoma.
Figure 5.
Correlation of mutation with gene expression levels.
Figure S3.
Correlation analysis of 6-gene signature risk scores with other risk clinical variables.
Functional pathway analysis of hub mutant signature and GSEA
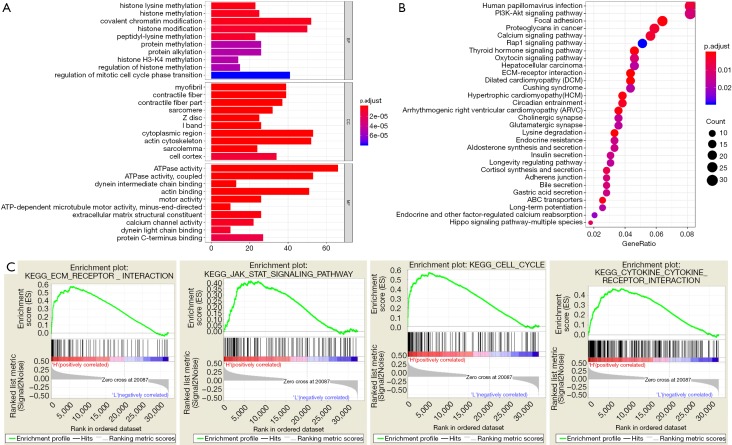
To deeply explore the potential functional pathways in pRCC that these mutant genes might be involved in, we conducted the functional pathway analysis based on the top 400 mutant genes using “org.Hs.eg.db”, “clusterProfiler” and “enrichplot” packages (Table 3). The functional enrichment results revealed that these genes were mainly associated with histone modification, covalent chromatin modification and histone methylation in BP category. In MF group, the mutant genes mainly enriched in ATPase activity, actin biding (Figure 6A). In addition, KEGG pathway analysis indicated the enrichment of mutant genes in various crosstalk in malignancy, including PI3K-AKT signaling pathway, focal adhesion, proteoglycans in cancer, calcium signaling pathway or extracellular matrix (ECM)-receptor interaction (Figure 6B). Last, the GSEA results suggested that patients in high-MRS group tend to have more relationships with ECM-receptor interaction, JAK-STAT signaling pathway, cell cycle or cytokine receptor interaction (Figure 6C).
Table 3. GO enriched items from the top 300 mutation related genes in KIRP.
| Ontology | Description | P value |
|---|---|---|
| GOTERM_BP_DIRECT | Histone lysine methylation | 2.93E-06 |
| Histone methylation | 3.79E-06 | |
| Covalent chromatin modification | 3.79E-06 | |
| Histone modification | 7.85E-06 | |
| Peptidyl-lysine methylation | 7.99E-06 | |
| Protein methylation | 4.89E-05 | |
| Protein alkylation | 4.89E-05 | |
| Histone H3-K4 methylation | 4.89E-05 | |
| Regulation of histone methylation | 5.16E-05 | |
| Regulation of mitotic cell cycle phase transition | 7.76E-05 | |
| GOTERM_MF_DIRECT | ATPase activity | 5.01E-14 |
| ATPase activity, coupled | 3.13E-11 | |
| Dynein intermediate chain binding | 4.47E-08 | |
| Actin binding | 9.95E-08 | |
| Motor activity | 1.54E-07 | |
| ATP-dependent microtubule motor activity, minus-end-directed | 2.08E-07 | |
| Extracellular matrix structural constituent | 1.67E-06 | |
| Calcium channel activity | 5.76E-06 | |
| Dynein light chain binding | 7.67E-06 | |
| Protein C-terminus binding | 1.55E-05 | |
| GOTERM_CC_DIRECT | Myofibril | 2.62E-11 |
| Contractile fiber | 6.79E-11 | |
| Contractile fiber part | 1.16E-10 | |
| Sarcomere | 1.74E-08 | |
| Z disc | 1.95E-08 | |
| I band | 1.97E-08 | |
| Cytoplasmic region | 3.93E-08 | |
| Actin cytoskeleton | 1.32E-07 | |
| Sarcolemma | 1.89E-07 | |
| Cell cortex | 1.12E-05 |
Figure 6.
Functional pathway analysis based on top 400 mutant genes. (A,B) GO and KEGG pathway analysis for mutated genes; (C) GSEA results showed that the TMB-related risk score correlated with ECM-receptor interaction, JAK-STAT signaling pathway, cell cycle or cytokine receptor interaction. GO, gene ontology; GSEA, Gene Set Enrichment Analysis; TMB, tumor mutation burden; ECM, extracellular matrix.
Discussion
As one of the RCC subspecies, pRCC is a heterogeneous carcinoma with no effective clinical treatment for advanced disease exist (25). It is alarming that limited cognition about the genetic basis of sporadic pRCC has hindered the exploration of new approaches to the diagnosis and treatment. In recent years, genomic landscape and pathogenesis of pRCC were comprehensively characterized profited from the rapid development of high-throughput sequencing technology (25). In our study, nucleotide variation data of 288 pRCC samples from TCGA database were collected and top 50 mutant genes were identified by utilizing “GenVisR” package and STRING database. Next, we screened hub prognostic mutant signature and established MRS risk score by multivariate Cox regression method. After the prognostic value of MRS risk score was verified, function enrichment pathway analysis of mutated genes was performed to reveal the multiple cancer-related crosstalk that genetic mutations might be involved.
Gene mutations are ubiquitous in a variety of malignancies and constitute the molecular biological basis of malignant tumor heterogeneity (26-28). In pRCC, differential expression of genes might be essential to the discrepancy in biological behavior between two different subtypes. A comprehensive molecular characterization was performed in 2016, and the results revealed that MET alterations are specifically expressed in type 1, whereas CDKN2A silencing, SETD2 mutations, TFE3 fusions and fumarate hydratase (FH) gene were discovered in type 2 (25). It was reported that MET protein which encoded by MET proto-oncogene was a transmembrane receptor tyrosine kinase (RTK) and participated in the regulation of phosphorylation of intracellular docking sites (29-31). MET was widely detected in the malignant tumors regulation by changing the phosphorylation status of mitogen-activated protein kinase (MAPK), phosphoinositide 3-kinase (PI3K)/AKT and nuclear factor-κB signaling pathways (32-36). As a methyltransferase which trimethylates histone H3 at lysine 36, SETD2 played a significant role in tumor suppression through gene mutation and silence (37,38). Particularly in metastatic renal cell carcinoma, prevalent mutations of SETD2 were 30% (39). In current research, MET were mutated in more than 20 samples. Besides, as a hub mutant gene, the expression level of SETD2 was lower in mutant samples compared with normal samples. The results are consistent with previous studies.
The giant OBSCURIN protein is encoded by OBSCN gene, which located in 1q42.13. With numerous functional domains participated in the controlling of cell adhesion, migration and cell morphology, OBSCURIN acted as a regulator in epithelial-to-mesenchymal transition (EMT) signaling pathway, which is one of the most crucial steps in neoplasm metastasis (40-45). In addition, OBSCN alterations and mutations were observed in many malignant tumors. OBSCN was always treated as a tumor suppressor because the gene inhibition might influence the cellular integration and activate cancer initiation (46). Somatic OBSCN alterations were identified in mucoepidermoid carcinoma by whole-exome sequencing and systematic genomic analyses (47). Similarly, OBSCN was mutated on at least two sites in ovarian cancer discovered by next-generation sequencing-based genomic profiling analysis (48). In our study, we found that OBSCN was mutated in more than 20 samples among 288 pRCC patients. Besides, OBSCN was identified as one of the hub prognostic mutant genes after univariate cox analysis conducted with “survival” package. However, different from the role as a tumor suppressor shown in a previous study, higher OBSCN expression level was associated with poor survival outcomes revealed by Kaplan-Meier survival analysis. Moreover, the box plots revealed that the expression level of OBSCN increased in pRCC samples compared with normal samples. Whether the different gene function is due to the tumor specificity of the PRCC or due to the bias from sample size requires a further investigation.
Remarkably, the MRS was calculated based on 17 hub prognostic mutant genes. The AUC of ROC was indicated the high predictive accuracy for the MRS model. The functional pathway analysis results revealed the associated relationships between these prognostic mutant genes and signaling pathways which closely related with cancer development. Our results revealed the frequency of mutant genes was associated with poor prognosis. This MRS model provides a reliable theoretical basis for the prognosis assessment, which might be further applied in the clinical management of pRCC.
Apart from new exploration on prognosis value of mutant genes in pRCC patients, limitations were also existed in current study. Firstly, all the mentioned results were extracted or analyzed from TCGA database through biological algorithm approaches. Selective bias might be amended if we contain more data from public other datasets and clinical patients. Secondly, the regulatory mechanism of 17 hub prognostic mutant genes in pRCC should be further studied. Finally, owing to the limited size of pRCC samples, we failed to identify the specific mutation sites that led to the abnormal gene expression which deserved further exploration.
Acknowledgments
Funding: This work was supported by grants from the training program of outstanding young talents in clinical medicine by Hongkou District (No. HKYQ2018-09).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Moch H, Cubilla AL, Humphrey PA, et al. The 2016 WHO Classification of Tumours of the Urinary System and Male Genital Organs-Part A: Renal, Penile, and Testicular Tumours. Eur Urol 2016;70:93-105. 10.1016/j.eururo.2016.02.029 [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. 10.3322/caac.21551 [DOI] [PubMed] [Google Scholar]
- 3.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 4.Linehan WM, Srinivasan R, Schmidt LS. The genetic basis of kidney cancer: a metabolic disease. Nat Rev Urol 2010;7:277-85. 10.1038/nrurol.2010.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Linehan WM. Genetic basis of kidney cancer: role of genomics for the development of disease-based therapeutics. Genome Res 2012;22:2089-100. 10.1101/gr.131110.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delahunt B, Eble JN. Papillary renal cell carcinoma: a clinicopathologic and immunohistochemical study of 105 tumors. Mod Pathol 1997;10:537-44. [PubMed] [Google Scholar]
- 7.Zbar B, Tory K, Merino M, et al. Hereditary papillary renal cell carcinoma. J Urol 1994;151:561-6. 10.1016/S0022-5347(17)35015-2 [DOI] [PubMed] [Google Scholar]
- 8.Pignot G, Elie C, Conquy S, et al. Survival analysis of 130 patients with papillary renal cell carcinoma: prognostic utility of type 1 and type 2 subclassification. Urology 2007;69:230-5. 10.1016/j.urology.2006.09.052 [DOI] [PubMed] [Google Scholar]
- 9.Klatte T, Pantuck AJ, Said JW, et al. Cytogenetic and molecular tumor profiling for type 1 and type 2 papillary renal cell carcinoma. Clin Cancer Res 2009;15:1162-9. 10.1158/1078-0432.CCR-08-1229 [DOI] [PubMed] [Google Scholar]
- 10.Beksac AT, Paulucci DJ, Blum KA, et al. Heterogeneity in renal cell carcinoma. Urol Oncol 2017;35:507-15. 10.1016/j.urolonc.2017.05.006 [DOI] [PubMed] [Google Scholar]
- 11.Stadler WM, Figlin RA, McDermott DF, et al. Safety and efficacy results of the advanced renal cell carcinoma sorafenib expanded access program in North America. Cancer 2010;116:1272-80. 10.1002/cncr.24864 [DOI] [PubMed] [Google Scholar]
- 12.Zhang X, Zhang Y. Bladder Cancer and Genetic Mutations. Cell Biochem Biophys 2015;73:65-9. 10.1007/s12013-015-0574-z [DOI] [PubMed] [Google Scholar]
- 13.Gumaste PV, Penn LA, Cymerman RM, et al. Skin cancer risk in BRCA1/2 mutation carriers. Br J Dermatol 2015;172:1498-506. 10.1111/bjd.13626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou C, Wu YL, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol 2011;12:735-42. 10.1016/S1470-2045(11)70184-X [DOI] [PubMed] [Google Scholar]
- 15.Lièvre A, Bachet JB, Le Corre D, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res 2006;66:3992-5. 10.1158/0008-5472.CAN-06-0191 [DOI] [PubMed] [Google Scholar]
- 16.Colussi D, Brandi G, Bazzoli F, et al. Molecular pathways involved in colorectal cancer: implications for disease behavior and prevention. Int J Mol Sci 2013;14:16365-85. 10.3390/ijms140816365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tomczak K, Czerwinska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn) 2015;19:A68-77. 10.5114/wo.2014.47136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koboldt DC, Chen K, Wylie T, et al. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 2009;25:2283-5. 10.1093/bioinformatics/btp373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skidmore ZL, Wagner AH, Lesurf R, et al. GenVisR: Genomic Visualizations in R. Bioinformatics 2016;32:3012-4. 10.1093/bioinformatics/btw325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cook HV, Doncheva NT, Szklarczyk D, et al. Viruses.STRING: A Virus-Host Protein-Protein Interaction Database. Viruses 2018. doi: . 10.3390/v10100519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoo ZH, Candlish J, Teare D. What is an ROC curve? Emerg Med J 2017;34:357-9. 10.1136/emermed-2017-206735 [DOI] [PubMed] [Google Scholar]
- 23.Gaudet P, Dessimoz C. Gene Ontology: Pitfalls, Biases, and Remedies. Methods Mol Biol 2017;1446:189-205. 10.1007/978-1-4939-3743-1_14 [DOI] [PubMed] [Google Scholar]
- 24.Kanehisa M, Furumichi M, Tanabe M, et al. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 2017;45:D353-D361. 10.1093/nar/gkw1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cancer Genome Atlas Research Network , Linehan WM, Spellman PT, et al. Comprehensive Molecular Characterization of Papillary Renal-Cell Carcinoma. N Engl J Med 2016;374:135-45. 10.1056/NEJMoa1505917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nones K, Johnson J, Newell F, et al. Whole-genome sequencing reveals clinically relevant insights into the aetiology of familial breast cancers. Ann Oncol 2019. [Epub ahead of print]. 10.1093/annonc/mdz132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Linehan WM, Schmidt LS, Crooks DR, et al. The Metabolic Basis of Kidney Cancer. Cancer Discov 2019;9:1006-21. 10.1158/2159-8290.CD-18-1354 [DOI] [PubMed] [Google Scholar]
- 28.Fang W, Ma Y, Yin JC, et al. Comprehensive Genomic Profiling Identifies Novel Genetic Predictors of Response to Anti-PD-(L)1 Therapies in Non-Small-Cell Lung Cancer. Clin Cancer Res 2019;25:5015-26. 10.1158/1078-0432.CCR-19-0585 [DOI] [PubMed] [Google Scholar]
- 29.Giordano S, Ponzetto C, Di Renzo MF, et al. Tyrosine kinase receptor indistinguishable from the c-met protein. Nature 1989;339:155-6. 10.1038/339155a0 [DOI] [PubMed] [Google Scholar]
- 30.Naldini L, Vigna E, Narsimhan RP, et al. Hepatocyte growth factor (HGF) stimulates the tyrosine kinase activity of the receptor encoded by the proto-oncogene c-MET. Oncogene 1991;6:501-4. [PubMed] [Google Scholar]
- 31.Bottaro DP, Rubin JS, Faletto DL, et al. Identification of the hepatocyte growth factor receptor as the c-met proto-oncogene product. Science 1991;251:802-4. 10.1126/science.1846706 [DOI] [PubMed] [Google Scholar]
- 32.Ponzetto C, Bardelli A, Zhen Z, et al. A multifunctional docking site mediates signaling and transformation by the hepatocyte growth factor/scatter factor receptor family. Cell 1994;77:261-71. 10.1016/0092-8674(94)90318-2 [DOI] [PubMed] [Google Scholar]
- 33.Weidner KM, Di Cesare S, Sachs M, et al. Interaction between Gab1 and the c-Met receptor tyrosine kinase is responsible for epithelial morphogenesis. Nature 1996;384:173-6. 10.1038/384173a0 [DOI] [PubMed] [Google Scholar]
- 34.Kim JY, Welsh EA, Fang B, et al. Phosphoproteomics Reveals MAPK Inhibitors Enhance MET- and EGFR-Driven AKT Signaling in KRAS-Mutant Lung Cancer. Mol Cancer Res 2016;14:1019-29. 10.1158/1541-7786.MCR-15-0506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Der Steen N, Pauwels P, Gil-Bazo I, et al. cMET in NSCLC: Can We Cut off the Head of the Hydra? From the Pathway to the Resistance. Cancers (Basel) 2015;7:556-73. 10.3390/cancers7020556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang YW, Wang LM, Jove R, et al. Requirement of Stat3 signaling for HGF/SF-Met mediated tumorigenesis. Oncogene 2002;21:217-26. 10.1038/sj.onc.1205004 [DOI] [PubMed] [Google Scholar]
- 37.Yuan W, Xie J, Long C, et al. Heterogeneous nuclear ribonucleoprotein L Is a subunit of human KMT3a/Set2 complex required for H3 Lys-36 trimethylation activity in vivo. J Biol Chem 2009;284:15701-7. 10.1074/jbc.M808431200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fahey CC, Davis IJ. SETting the Stage for Cancer Development: SETD2 and the Consequences of Lost Methylation. Cold Spring Harb Perspect Med 2017. doi: . 10.1101/cshperspect.a026468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hsieh JJ, Chen D, Wang PI, et al. Genomic Biomarkers of a Randomized Trial Comparing First-line Everolimus and Sunitinib in Patients with Metastatic Renal Cell Carcinoma. Eur Urol 2017;71:405-14. 10.1016/j.eururo.2016.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest 2009;119:1420-8. 10.1172/JCI39104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Russell MW, Raeker MO, Korytkowski KA, et al. Identification, tissue expression and chromosomal localization of human Obscurin-MLCK, a member of the titin and Dbl families of myosin light chain kinases. Gene 2002;282:237-46. 10.1016/S0378-1119(01)00795-8 [DOI] [PubMed] [Google Scholar]
- 42.Thiery JP, Acloque H, Huang RY, et al. Epithelial-mesenchymal transitions in development and disease. Cell 2009;139:871-90. 10.1016/j.cell.2009.11.007 [DOI] [PubMed] [Google Scholar]
- 43.Kontrogianni-Konstantopoulos A, Ackermann MA, Bowman AL, et al. Muscle giants: molecular scaffolds in sarcomerogenesis. Physiol Rev 2009;89:1217-67. 10.1152/physrev.00017.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fukuzawa A, Idowu S, Gautel M. Complete human gene structure of obscurin: implications for isoform generation by differential splicing. J Muscle Res Cell Motil 2005;26:427-34. 10.1007/s10974-005-9025-6 [DOI] [PubMed] [Google Scholar]
- 45.Lu W, Kang Y. Epithelial-Mesenchymal Plasticity in Cancer Progression and Metastasis. Dev Cell 2019;49:361-74. 10.1016/j.devcel.2019.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shriver M, Stroka KM, Vitolo MI, et al. Loss of giant obscurins from breast epithelium promotes epithelial-to-mesenchymal transition, tumorigenicity and metastasis. Oncogene 2015;34:4248-59. 10.1038/onc.2014.358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kang H, Tan M, Bishop JA, et al. Whole-Exome Sequencing of Salivary Gland Mucoepidermoid Carcinoma. Clin Cancer Res 2017;23:283-8. 10.1158/1078-0432.CCR-16-0720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang L, Luo M, Yang H, et al. Next-generation sequencing-based genomic profiling analysis reveals novel mutations for clinical diagnosis in Chinese primary epithelial ovarian cancer patients. J Ovarian Res 2019;12:19. 10.1186/s13048-019-0494-4 [DOI] [PMC free article] [PubMed] [Google Scholar]