Abstract
Background
Skeletal muscle atrophy, characterized by accelerated protein degradation, occurs in such conditions as unloading, immobilization, fasting, and denervation. Effective treatments for skeletal muscle atrophy are not yet available. Considering that microRNAs (miRs) may play an important role in the regulation of muscle atrophy, in the present study, we aimed to examine the effect of miR-125b-5p-based therapeutic strategies on skeletal muscle atrophy, and to explore the underlying mechanisms.
Methods
Fasting-induced atrophic mouse C2C12 myotubes and denervated rat tibialis anterior (TA) muscles were used as in vitro and in vivo models of skeletal muscle atrophy, respectively. The morphological parameters of skeletal muscle were measured by immunostaining-based quantification. The interaction between miR-125b-5p and TRAF6 3'-UTR was detected by luciferase reporter analysis. The mRNA and protein expressions were determined by real-time qPCR and Western blot analysis respectively. The miR mimics/agomir and miR inhibitor/antagomir were transfected into C2C12 myotubes and TA muscles respectively to alter the expression of miR-125b-5p.
Results
The expression of miR-125b-5p was down-regulated in both atrophic C2C12 myotubes and denervated TA muscles. The interaction between miR-125b-5p and TRAF6 3'-UTR was identified. Overexpression of miR-125b-5p protected skeletal muscle samples from atrophy in vitro and in vivo by targeting TRAF6 through inactivation of several ubiquitin-proteasome system (UPS)- and autophagy-lysosome system (ALS)-related proteins.
Conclusions
Overexpression of miR-125b-5p may provide a promising therapeutic approach to treat muscle atrophy.
Keywords: Fasting, denervation, skeletal muscle atrophy, miR-125b-5p, TRAF6
Introduction
Skeletal muscle atrophy occurs in various conditions, including unloading, limb immobilization, fasting, denervation, and cachexia (1-3). An imbalance between anabolic and catabolic processes, i.e., protein degradation exceeding protein synthesis (4,5), results in a series of pathological changes, such as reduced cross-sectional area (CSA) of muscle fibers, myofibrillar sarcomeres disruption, myofibril degradation, disturbance of myotube formation, and fibrillation (6-8). Peripheral neuro-disconnection of skeletal muscle is the cause of denervated muscle atrophy (9), where an increased rate of proteolysis is believed to be mediated through the ubiquitin-proteasome system (UPS) and autophagy-lysosome system (ALS) (10-12).
The UPS, a complex pathway activated upon denervated-dependent skeletal muscle atrophy, contributes to the loss of muscle mass. In different models of skeletal muscle atrophy, the muscle-specific E3 ubiquitin ligases, such as MAFbx/atrogin-1 and MuRF1, are up-regulated (13-15), and overexpression of either MAFbx or MuRF1 in skeletal muscle fibers may enhance proteolytic function, leading to muscle atrophy (3,16). It has been reported that mice knocked out of MAFbx or MuRF1 are resistant to various types of muscle atrophy, suggesting that these two ligases are essential to atrophy-related protein degradation (17-19).
The ALS was once thought to be activated in muscle cells during catabolic conditions (20), and autophagy has aroused significant concern for its role in metabolic homeostasis and disease progression of skeletal muscle. Beclin1 is an essential autophagy protein, and its increased expression in autophagy impairs the function of skeletal muscle (21). Mitochondrial regulation is critically important to mediate mitophagy and to maintain muscle function, whereas inactivation of the genes coding for Parkin, Pink1, or Bnip3 induces mitochondrial abnormalities (22,23). Accumulating evidence shows that the mitochondrial network is usually remodeled through the ALS during skeletal muscle atrophy (24-26).
As a member of the tumour necrosis factor (TNF) receptor-associated factor (TRAF) family, TNF receptor adaptor protein 6 (TRAF6) is involved in the activation of various signaling pathways, including NF-κB, MAPK, and phosphatidylinositide 3-kinase/Akt (PI3K/Akt) (27-29). Different from other members of the TRAF family, TRAF6 is a unique E3 ubiquitin ligase and adaptor protein. Following fasting, denervation, or cancer cachexia, TRAF6 expression is increased in atrophic skeletal muscle (9,30,31), where both the UPS and ALS pathways are coordinately up-regulated. Therefore, attenuation of skeletal muscle atrophy in TRAF6-knockout (TRAF6mko) mice may be considered to result from inhibition of both UPS and ALS (27).
MicroRNAs (miRNAs, miRs) are short, noncoding RNAs that down-regulate gene expression by binding to complementary sequences in the 3'-untranslated regions (3'-UTR) of target mRNAs. Recently, miRNAs have emerged as key regulators of many biological processes including skeletal muscle metabolic homoeostasis. Abundant evidence suggests that dynamic changes of miRNAs are involved in the regulation of skeletal muscle atrophy (32-35). For example, the decreased expression of miR-23a induces the translation of TRIM63/MuRF1 and Fbxo32/atrogin-1, whereas overexpression of miR-23a prevents diabetes-induced muscle cachexia and attenuates renal fibrosis lesions via muscle-kidney crosstalk (36); miR-29b promotes atrophy of myotubes, whereas its inhibition attenuates multiple stimuli-induced muscle atrophy (37). In addition, miR-378 acts as a vital coordinator of autophagy and apoptosis in maintaining normal muscle homeostasis (38). Our previous work (9) has also shown that miR-351 targeting TRAF6 alleviated dexamethasone-induced myotube atrophy and denervation-induced muscle atrophy.
It has been reported that miR-125b participates in neuronal differentiation of mouse P19 cells by targeting RNA binding protein LIN-28, and promotes neuronal differentiation of human cells by inhibiting multiple target genes (39,40). Moreover, the effect of miR-125b on insulin-like growth factor II (IGF-II) negatively regulates myoblast differentiation in vitro and muscle regeneration in vivo (41). However, the regulation of miR-125b on skeletal muscle atrophy has not been fully studied. To determine the specific regulation of miR-125b-5p in this aspect, in the current study, we determined that miR-125b-5p was down-regulated in fasting-induced atrophic C2C12 myotubes or denervated tibialis anterior (TA) muscles, and we also investigated the interaction between miR-125b-5p and TRAF6, showing that miR-125b-5p directly targeted the 3'-UTR of Traf6. Furthermore, we found that overexpression of miR-125b-5p alleviated fasting-induced atrophy of C2C12 myotubes and inhibited denervated-induced muscle atrophy in TA muscles, and concomitantly the expression of the UPS and ALS related-proteins was down-regulated. Inversely, inhibition of miR-125b-5p accelerated the progress of muscle atrophy via up-regulating the expression of UPS and ALS related-proteins. Collectively, our data revealed that miR-125b-5p might relieve skeletal muscle atrophy by targeting TRAF6 in vitro and in vivo.
Methods
C2C12 cell culture and transfection
Mouse C2C12 myoblasts (Cell Bank, Chinese Academy of Sciences, Shanghai, China) were cultured in growth medium consisting of Dulbecco’s modified Eagle’s medium (DMEM, Gibco-BRL, Gaithersburg, MD, USA) supplemented with 10% fetal bovine serum (FBS) (Gibco-BRL), 100 U/mL penicillin (Sigma-Aldrich, St. Louis, MO, USA), and 100 µg/mL streptomycin (Sigma-Aldrich). Myoblasts were grown to approximately 90% confluence in a 6-well plate and then transferred into another medium containing DMEM supplemented with 2% horse serum and 1% penicillin and streptomycin for 7-day differentiation into myotubes. Then, the formed C2C12 myotubes were treated with Hank’s Balanced Salt Solution (HBSS, Gibco BRL, Gaithersburg, MD, USA) for 12 and 24 h respectively, whereas C2C12 myotubes stored in the differentiation medium were used as a control.
Transfection of C2C12 myoblasts with miR-125b-5p mimics, miR-125b-5p inhibitor, or vehicle (as negative control) was performed at the fourth day of differentiation with transfection reagents (RiboBio, Guangzhou, China) according to the manufacturer’s instructions. After 3 days, the sample was switched to HBSS for 24 h as above described.
Animal surgery and transfection
Sprague-Dawley (SD) rats (weight ~200 g) were supplied by the experimental animal center of Nantong University (Nantong, Jiangsu, China). To induce denervated skeletal muscle atrophy, the left sciatic nerve of rats was cut at the mid-thigh region for 1.5 cm. The sham-operated rats underwent the same procedures but without sciatic nerve cut. Rats were killed at 0, 3, 7, 14 and 28 days after surgery respectively.
For transfection, the operated rats were randomly divided into four groups (vehicle as negative control, miR-125b-5p agomir, miR-125b-5p antagomir, miR-125b-5p agomir plus TRAF6 overexpression) to receive different injections into their TA muscles from the surgery day. In brief, miR-125b-5p agomir was injected every 3 days; antagomir was injected for 3 successive days from the surgery day. The TA muscles were taken on the 14th day post-surgery.
Immunostaining
To determine the diameter of C2C12 myotubes, the sample was fixed by 4% PFA for 30 min at room temperature, washed with phosphate buffer solution (PBS) for 15 min, and then blocked with an immunostaining blocking buffer (Beyotime, Haimen, China) for 1 h at room temperature, followed by incubation with primary antibody anti-major histocompatibility complex (MHC) (MRC OX-6, 1:200, Abcam, Cambridge, MA, USA) overnight at 4 °C. Secondary antibody goat anti-mouse IgG-FITC (1:500, Abcam) was further added to allow incubation for 2 h at room temperature. The images were captured by fluorescence microscope (Zeiss) and the diameter of C2C12 myotubes was measured by Image J software (http://rsb.info.nih.gov./ij/).
Skeletal muscle tissues were incubated with anti-laminin (ab11575, 1:200, Abcam) overnight at 4 °C, followed by incubation with secondary antibody and visualization as above mentioned. The CSA of rat TA muscle was measured by Image J software.
Luciferase reporter assay
HEK293T cells were cultured in 24-well plates in DMEM supplemented with 10% FBS and 1% penicillin and streptomycin. The cells were transfected with lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) plus TRAF6 3'-UTR wide type plasmid, TRAF6 3'-UTR mutant plasmid with miR-125b-5p, or with vehicle (negative control). After 24 h, transfected cells were collected, and luciferase activity was measured using the Dual Luciferase Assay System (Promega, Madison, WI, USA). The data were normalized to the firefly luciferase signal.
Western blot analysis
C2C12 myotubes or frozen rat TA muscles were homogenized in RIPA lysis buffer (P0013B) containing 1 mM PMSF (ST506), and protease inhibitor (P1010, all the above reagents from Beyotime). Total protein (40 µg) was separated on 10% SDS-PAGE gels (Beyotime) and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica, MA, USA) by electroblotting. The blots were blocked with 5% nonfat milk in TBS for 1 h at room temperature, and then incubated overnight at 4 °C with the respective primary antibodies. The primary antibodies included anti-myosin heavy chain (MHC, 1:2,000, R&D Systems, Minneapolis, MN, USA), anti-TRAF6 (1:1,000; Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA), anti-MuRF1 (1:1,000, Cell signaling technology, Beverly, MA), anti-MAFbx (1:1,000, Abcam), anti-ATG7 (1:1,000, Abcam), anti-BNIP3 (1:1,000, Cell signaling technology), anti-beclin-1 (1:1,000, Cell signaling technology), anti-PINK1 (1:1,000, Abcam), anti-LC3B (1:1,000, MBL, Boston, MA, USA), and anti-Tubulin (1:2,000, Proteintech, Wuhan, China). After washes with Tris-buffered saline containing 0.2–0.4% Tween-20 (TBST), the membranes were incubated with appropriate secondary antibodies (Sigma) at room temperature for 1 h. Images were obtained using a TANON imaging system (Shanghai, China).
RNA isolation and RT-qPCR
RNA was extracted from samples using TRIZOL reagent (sigma) and RNeasy mini kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. The RNA sample (1 µg) was used to synthesize first strand cDNA with a reverse transcription system using an oligo (dT) primer (Applied Biosystems, Carlsbad, CA, USA) and Omniscript reverse transcription kit (QIAGEN). Bulge-loopTM miRNA qRT-PCR Primer Sets (one RT primer and a pair of qPCR primers for each set) specific for miR-125b-5p was designed by RiboBio (Guangzhou, China). Quantitative analysis of gene expression by real-time qPCR was carried out using the CFX96 (Bio-Rad) system. Each real-time PCR (10 µL) contained 1 µL of cDNA, 5 µL of SYBR Green qPCR Master mix (QIAGEN) with forward and reverse primers at a final concentration of 10 µM. The following primers were used: Rat-Hprt1-F: TGCTGAAGATTTGGAAAAGGTGT, Rat-Hprt1-R: GCGCTTTAATGTAATCCAGCAG; Rat-Traf6-F: TGTGTGCAAAAGATGGAGCT, Rat-Traf6-R: GGCACAGCACAGTTTACACA.
All reactions were performed using the following thermal cycler conditions: 95 °C for 10 min, followed by 40 cycles of denaturation at 95 °C for 2 s, annealing at 60 °C for 20 s and extension at 70 °C for 10 s. The reaction was followed by a melting curve from 70 to 95 °C in 5 s increments of 0.5 °C to ensure amplification specificity. The data of TRAF6 expression normalization was accomplished using the endogenous control (Hprt1). The miR-125b-5p transcript levels were normalized to the U6 transcript levels. The relative fold-change in expression was calculated using the ΔΔCt method.
Statistical analysis
All data are expressed as means ± SD. Student’s one-way ANOVA was used to compare differences between groups. All statistical analyses were conducted with a SPSS Software Version 17.0 (SPSS Inc., Chicago, IL, USA). P<0.05 was considered as statistically significant.
Results
Decreased expression of miR-125b-5p during muscle atrophy
To determine the expression of miR-125b-5p during skeletal muscle atrophy, we chose fasting-induced atrophic C2C12 myotubes and denervated TA muscle as in vitro and in vivo models of skeletal muscle atrophy respectively.
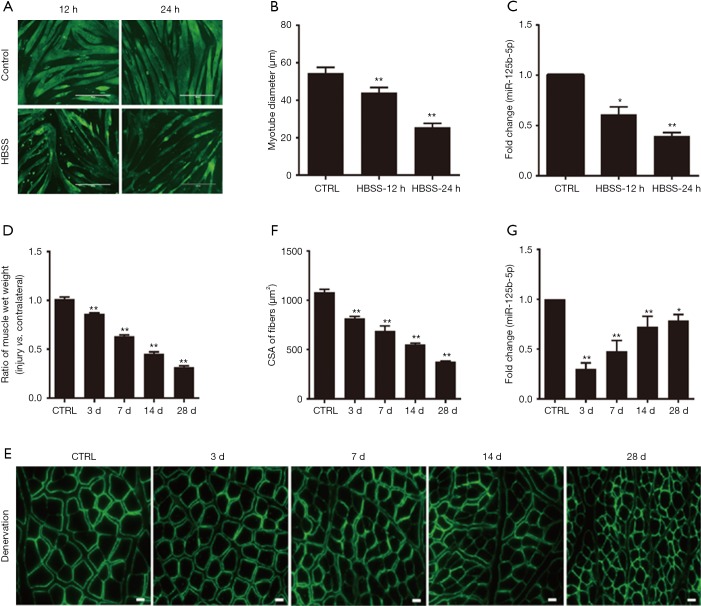
The diameter of atrophic C2C12 myotubes was significantly reduced as compared to that of normal C2C12 myotubes. The expression of miR-125b-5p in atrophic C2C12 myotubes was significantly decreased as compared to that in normal C2C12 myotubes (Figure 1A,B,C).
Figure 1.
Decreased expression of miR-125b-5p during muscle atrophy in vitro and in vivo. (A) Phenotypic changes of C2C12 myotubes following 12 or 24 h HBSS treatment (nutritional deprivation, fasting) or following no HBSS treatment (control, CTRL), as shown by MHC immunostaining (green). Scale bar, 400 µm. (B) Comparison in the average diameter of C2C12 myotubes, which had been treated as described in (A). **, P<0.01 vs. CTRL. (C) RT-qPCR data showing the expression level of miR-125b-5p in C2C12 myotubes, which had been treated as described in (A). *, P<0.05 and **, P<0.01 vs. CTRL. (D) Comparison in the wet weight ratio (the injured/contralateral side) of TA muscles of rats, which had been subjected to sciatic nerve cut before being killed at 3, 7, 14, and 28 days post-surgery. TA muscles of sham-operated rats served as CTRL. **, P<0.01 vs. CTRL. (E) Representative images of laminin immunostaining for measuring the cross-sectional area (CSA) of TA muscle fibers of rats, which had received treatments as described in (D). Scale bar, 20 µm. (F) Comparison in the CSA of rat TA muscle fibers, based on immunostaining data. **, P<0.01 vs. CTRL. (G) RT-qPCR analysis showing the decreased expression of miR-125b-5p in TA muscles of rats, which had received treatments as described in (D). *, P<0.05 and **, P<0.01 vs. CTRL. HBSS, Hank’s Balanced Salt Solution; TA, tibialis anterior; MHC, major histocompatibility complex.
The rats were subjected to sciatic nerve-cut surgery and were then killed at 3, 7, 14 and 28 days post-surgery respectively, followed by harvesting of 4 samples of atrophic TA muscles (based on different animal-killing times) respectively. The non-atrophic TA muscle of sham-operated rats (which were killed at 0 day post sham-surgery) served as control. Either the wet weight ratio or the CSA of TA muscle fibers was significantly decreased in 4 samples of atrophic TA muscles as compared to in control, non-atrophic TA muscle (Figure 1D,E,F). Likewise, the expression of miR-125b-5p in TA muscle was also significantly decreased in 4 samples of atrophic TA muscles as compared to in control, non-atrophic TA muscle (Figure 1G).
Interaction of miR-125b-5p with 3'-UTR of TRAF6
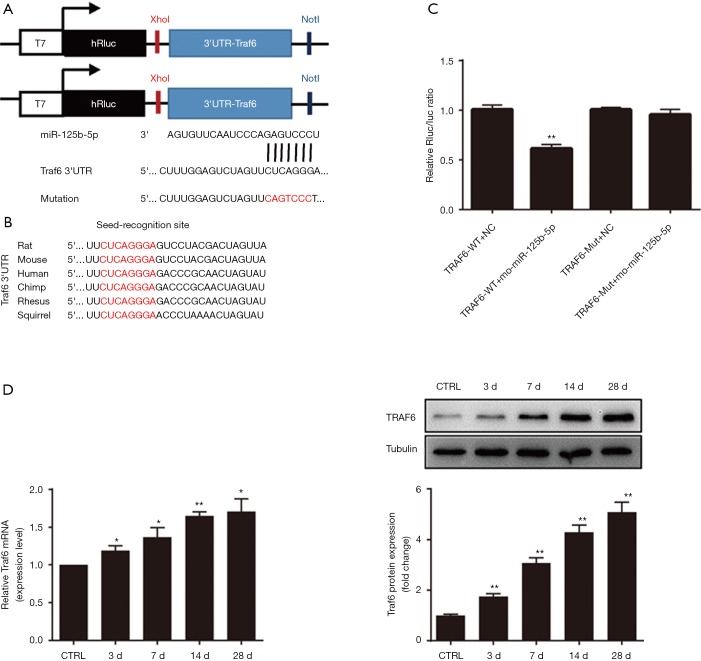
To explore the mechanism underlying the regulation of muscle atrophy by miR-125b-5p, TargetScan analysis suggested that TRAF6 might be a putative target of miR125b-5p (Figure 2A). Interestingly, all nucleotides in the seed recognition site of the Traf6 3'-UTR were conserved across several species. In other words, TRAF6 mRNA contained a highly-conserved seed recognition site which was capable of binding to the seed sequence of miR-125b-5p (Figure 2B).
Figure 2.
Traf6 acting as a direct target gene of miR-125b-5p. (A) TargetScan analysis indicating that Traf6 was the direct target of miR-125b-5p. Sketch graph indicating the mutation in the 3'-UTR sequence of Traf6 (mutant nucleotides in red). (B) Sketch illustration of the 3'-UTR of Traf6 with the seed-recognition site (labeled in red), which was capable of binding to the seed sequence of miR-125b-5p. All nucleotides in the seed recognition site were conserved across several species. (C) Comparison in relative luciferase activity after 4 different treatments: co-transfection of HEK293 cells with miR-125b-5p mimics or vehicle (negative control, NC) plus 3'-UTR of Traf6 wide type (WT)- or mutant (Mut)-containing luciferase reporter. **, P<0.01 vs. TRAF6-WT+NC. (D) RT-qPCR and Western blot analysis data comparing the expression of TRAF6 in TA muscles of rats, which had received different treatments as mentioned in Figure 1. *, P<0.05 and **, P<0.01 vs. CTRL. Also shown (inset) are representative Western blot images. TRAF6, tumour necrosis factor receptor adaptor protein 6; 3'-UTR, 3'-untranslated region; TA, tibialis anterior; CTRL, control.
The luciferase reporters were allowed to contain the wild type 3'-UTR of TRAF6 or mutant 3'-UTR of TRAF6, thus yielding 2 luciferase constructs. The miR-125b-5p mimics or vehicle (negative control), together with 2 luciferase constructs, were respectively co-transfected into HEK-293T cells, followed by measurement of the luciferase activity. For wild type 3'-UTR of TRAF6, miR-125b-5p mimics led to a significant reduction in the relative luciferase activity, but for mutant 3'-UTR of TRAF6, no decrease in the relative luciferase activity was observed (Figure 2A,C). This result suggested that miR-125b-5p might directly target TRAF6 3'-UTR. On the other hand, Western Blot analysis indicated that the expression of TRAF6 was increased in atrophic C2C12 myotubes, displaying an opposite change pattern with that of miR-125b-5p (Figure 2D vs. Figure 1G), providing further evidence for TRAF6 being a target of miR-125b-5p.
Effects of miR-125b-5p on fasting-induced atrophy of C2C12 myotubes
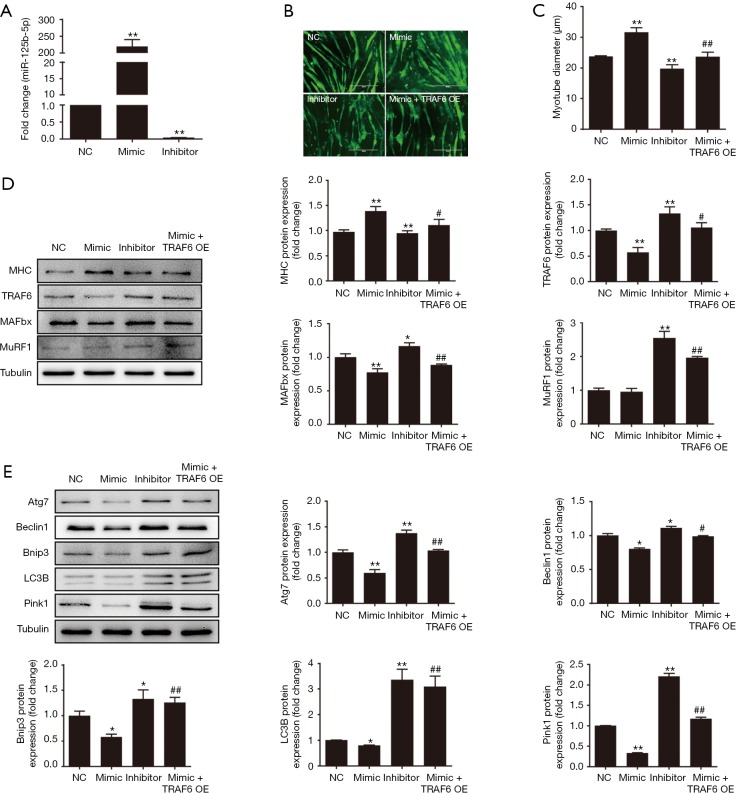
After miR-125b-5p mimics was transfected into atrophic C2C12 myotubes, the expression of miR-125b-5p was largely increased, and the extraordinary level of overexpression was nearly >200-fold over normal expression level (negative control), while transfection of atrophic C2C12 myotubes with miR-125b-5p inhibitor dramatically repressed the expression of miR-125b-5p to a near-negligible level (Figure 3A). It was understandable that the increased or decreased expression of miR-125b-5p, due to transfection with miR-125b-5p mimics or miR-125b-5p inhibitor, significantly augmented or reduced the diameter of atrophic C2C12 myotubes, as compared to unchanged expression of miR-125b-5p (Figure 3B,C). The data suggested that myotube atrophy might be relieved by high expression of miR-125b-5p.
Figure 3.
Effect of miR-125b-5p on muscle atrophy in vitro. (A) RT-qPCR analysis showing the increased expression of miR-125b-5p in atrophic C2C12 myotubes after transfection with miR-125b-5p mimic and the decreased expression of miR-125b-5p after transfection with miR-125b-5p inhibitor, as compared to the expression of miR-125b-5p after vehicle transfection (negative control, NC). (B) Immunofluorescent staining showing phenotypic changes of atrophic C2C12 myotubes, which were transfected with miR-125b-5p mimic, miR-125b-5p inhibitor, or co-transfected with miR-125b-5p mimic and TRAF6 overexpression (OE) lentivirus, respectively. Scale bar, 400 µm. (C) Comparison of the average diameter of C2C12 myotubes after different treatments as mentioned in (B). (D) Western blot analysis of MHC, TRAF6, MAFbx, MuRF1 protein levels in atrophic C2C12 myotubes after different treatments as mentioned in (B). Tubulin served as a loading control. (E) Western blot analysis of the ALS related-protein levels in atrophic C2C12 myotubes after different treatments as mentioned in (B). Tubulin served as a loading control. In all bar graphs, *, P<0.05 and **, P<0.01 vs. NC; #, P<0.05 and ##, P<0.01 vs. transfection with miR-125b-5p mimic. TRAF6, tumour necrosis factor receptor adaptor protein 6; MHC, major histocompatibility complex; ALS, autophagy-lysosome system.
The protein level of TRAF6 or UPS-related proteins, MuRF1 and MAFbx, in atrophic C2C12 myotubes was significantly decreased by transfection with miR-125b-5p mimic. Transfection with miR-125b-5p inhibitor, however, significantly enhanced the protein level of TRAF6 or UPS-related proteins (Figure 3D). Moreover, enhanced and reduced expression of miR-125b-5p in atrophic C2C12 myotubes, due to transfection with miR-125b-5p mimics or miR-125b-5p inhibitor, significantly decreased or increased ALS-related protein, respectively (Figure 3E). Taken together, these results indicated that miR-125b-5p relieved C2C12 myotube atrophy possibly through regulation of UPS and ALS.
Effects of miR-125b-5p on denervated atrophy of TA muscle
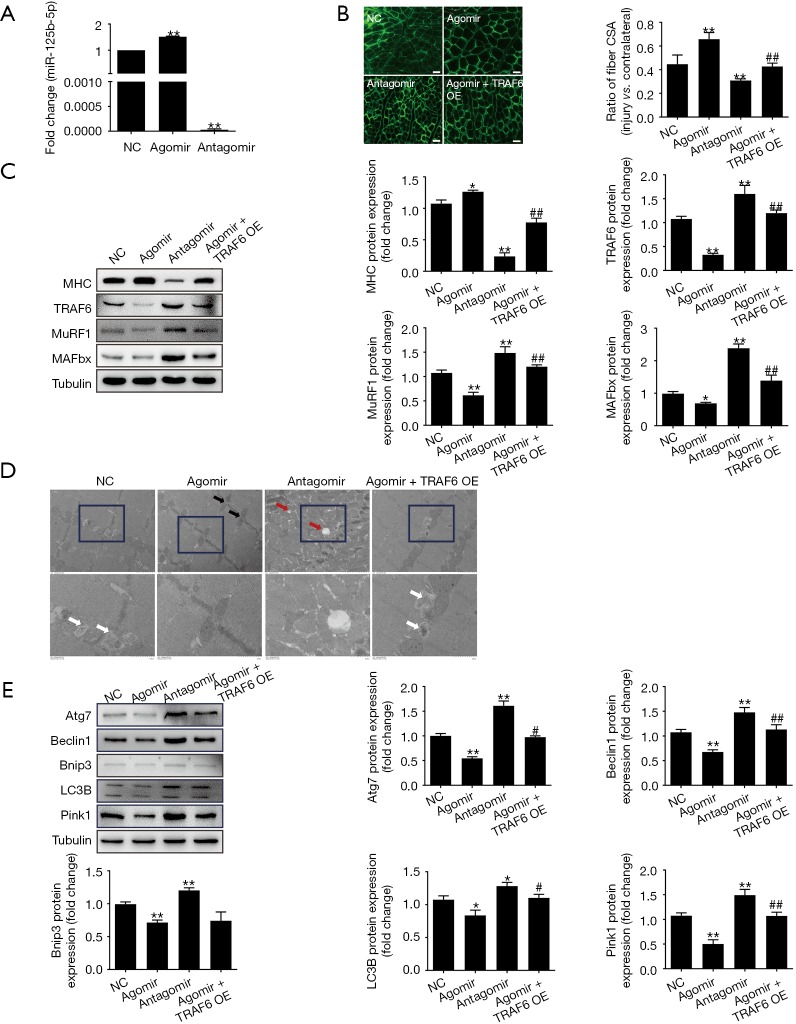
Administration of miR-125b-5p agomir significantly increased the expression of miR-125b-5p in atrophic TA muscles, and administration of miR-125b-5p antagomir significantly decreased the expression of miR-125b-5p in atrophic TA muscles, as compared to vehicle transfection (negative control) (Figure 4A). After administration of miR-125b-5p agomir, the CSA of TA muscle fibers group was significantly increased as compared to negative control, and vice versa after administration of miR-125b-5p antagomir (Figure 4B).
Figure 4.
Effect of miR-125b-5p on muscle atrophy in vivo. (A) RT-qPCR analysis showing the increased expression miR-125b-5p in denervated TA muscles after transfection with miR-125b-5p agomir, and the decreased expression in denervated TA muscles after transfection with miR-125b-5p antagomir as compared to the expression of miR-125b-5p after no transfection, (negative control, NC). (B) Comparison in the CSA ratio (the injured/contralateral side) of denervated TA muscles after transfection with miR-125b-5p agomir, miR-125b-5p antagomir, or co-transfection with miR-125b-5p agomir and TRAF6 overexpression (OE) lentivirus, respectively. Also shown are representative images of laminin immunostaining for measuring the CSA of TA muscle fibers. Scale bar, 20 µm. (C) Western blot analysis of MHC, TRAF6, MAFbx, MuRF1 protein levels in denervated TA muscles after different treatments as mentioned in (B). Tubulin was served as a loading control. (D) Electron micrographs of denervated TA skeletal muscle. Black arrows indicate mitochondrial distribution in the intermyofibrillar, white arrows indicate autophagosomes or autophagosome-engulfed mitochondria, and red arrows indicate autophagic vacuoles. (E) Western blot analysis of the ALS related-protein levels in denervated TA muscles after different treatments as mentioned in (B). Tubulin served as the loading control. In all bar graphs, *, P<0.05 and **, P<0.01 vs. NC; #, P<0.05 and ##, P<0.01 vs. transfection with miR-125b-5p agomir. TA, tibialis anterior; CSA, cross-sectional area, ALS, autophagy-lysosome system.
Treatment with miR-125b-5p agomir led to an increase in MHC protein levels as well as a decrease in TRAF6 protein levels. Correspondingly, the expressions of MAFbx and MuRF-1 were down-regulated or up-regulated in TA muscles after transfection with the miR-125b-5p agomir or miR-125b-5p antagomir, respectively (Figure 4C).
It was further found (Figure 4D) that a well-organized myofibrillar structure and normal subsarcolemmal and intermyofibrillar distribution of mitochondria were visible in TA muscles due to injection of miR-125b-5p agomir. However, morphological observation of TA muscles after injection of miR-125b-5p antagomir provided the worse result even than after vehicle transfection (negative control). In addition, overexpression of miR-125b-5p (after injection of miR-125b-5p agomir) significantly decreased the expression of ALS-related protein (Figure 4E).
Discussion
The discovery of miRNAs has provided new insights into mechanisms by which skeletal muscle development, regeneration, and dysfunction are controlled (42,43). As has been reported, miR-1 expression is up-regulated in a dexamethasone-induced muscle atrophy model, and miR-1 induces the expression of MuRF1 or MAFbx via heat shock protein 70 (HSP70)/protein kinase B (Akt)/FOXO3 signaling pathway, contributing to muscle atrophy (44); miR-125b is involved in neuronal differentiation of mouse P19 cells by targeting the RNA binding protein LIN-28, and also promotes neuronal differentiation in human cells by inhibiting multiple target genes (39). Moreover, miR-125b targets DNMT3b and mediates p53 DNA methylation involving in the vascular smooth muscle cell proliferation induced by homocysteine (45). In the present study, we attempted to investigate the regulating effect of miR-125b on skeletal muscle atrophy.
miR-125b-5p was down-regulated during C2C12 myotube atrophy induced by fasting or during TA muscle atrophy induced by denervation, suggesting that miR-125b-5p may play an important role during muscle atrophy. Luciferase activity assay revealed that miR-125b-5p might directly target TRAF6 3'-UTR. The expression of TRAF6 was increased in C2C12 myotubes after fasting-induced atrophy, displaying an opposite change pattern with that of miR-125b-5p, suggesting that miR-125b-5p might negatively target TRAF6.
After transfection of atrophic C2C12 myotubes or denervated TA muscles with miR-125b-5p mimics/agomir or miR-125b-5p inhibitor/antagomir respectively, we noted that overexpression of miR-125b-5p increased the diameter of atrophic C2C12 myotubes and also increased the CSA of TA muscles, implying that miR-125b-5p might be involved in protection of muscle atrophy. By the way, the joint use of TRAF6 overexpression lentivirus might, more or less, counteract the impact of using miR-125b-5p mimics/agomir alone, providing more evidence for negative regulation of miR-125b-5p by TRAF6.
Previous studies have revealed that TRAF6 is an important adaptor protein involved in receptor-mediated activation of various signaling pathways in response to aging, denervation, and chronic diseases (9,46,47). Muscle-specific depletion of TRAF6 preserves skeletal muscle mass, fiber size, and contractile functions in response to denervation and cancer cachexia (27). TRAF6 has been found to interact with multiple components of the UPS and/or ALS in some cell types, and regulates skeletal muscle mass and activation of the UPS and/or ALS in denervated skeletal muscle (27). TRAF6 interacts with LC3B and ubiquitinylates (Lys-63-linked) beclin-1, and further regulates some autophagy formation in response to Toll-like receptor 4 signaling (48). Hence, in the present study, we detected the expression of muscle-specific E3 ubiquitin ligases: MuRF1 and MAFbx as well as ALS related-proteins: Atg7, beclin-1, Bnip3, LC3B, and PINK1. The expressions of these proteins were all inhibited by miR-125b-5p agomir. Conversely, the expressions of these UPS- and ALS-related proteins were increased by miR-125b-5p antagomir. Collectively, the data indicate that miR-125b-5p might attenuate muscle atrophy by blocking UPS- and ALS-related proteins through targeting TRAF6.
Growing evidence shows that ROS and redox disturbances may represent important signaling events in skeletal muscle atrophy (49,50), and treatment with antioxidants significantly retarded the development of skeletal muscle atrophy induced by fasting or denervation (51,52). P53, a tumor suppressor protein, acts as a regulator of cellular homeostasis, regulating oxidative stress to maintain cellular health and function in skeletal muscle (53). Of note, miR-125b blocked Bax/cytochrome C/caspase-3 apoptotic signaling pathway in rat models of cerebral ischemia-reperfusion injury by targeting p53 (54). Due to the diversity of targets for miRNAs, there may be other target genes of miR-125b-5p besides TRAF6 in the context of skeletal muscle atrophy. Hence, it would be highly interesting to identify more potential targets of miR-125b-5p in the regulation of skeletal muscle atrophy.
In conclusion, miR-125b-5p alleviates muscle atrophy via targeting of TRAF6 directly or indirectly, and the application of miR-125b-5p may represent a therapeutic approach to treat muscle atrophy.
Acknowledgments
Funding: This work was supported by National Natural Science Foundation of China (grant No. 81671230 and 81871554), the 973 Program (grant No. 2014CB542202 and 2014CB542203), a project funded by Jiangsu Provincial Key Medical Center and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), and 2019 Jiangsu College Students Innovation and Entrepreneurship Training Program.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The whole study protocol was approved by the Ethics Committee of Nantong University (No. 20160308-009) and all the procedures were done according to the Chinese Guidelines for the Care and Use of Laboratory Animals.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Bodine SC, Baehr LM. Skeletal muscle atrophy and the E3 ubiquitin ligases MuRF1 and MAFbx/atrogin-1. Am J Physiol Endocrinol Metab 2014;307:E469-84. 10.1152/ajpendo.00204.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kanazawa Y, Ikegami K, Sujino M, et al. Effects of aging on basement membrane of the soleus muscle during recovery following disuse atrophy in rats. Exp Gerontol 2017;98:153-61. 10.1016/j.exger.2017.08.014 [DOI] [PubMed] [Google Scholar]
- 3.Ma W, Xu T, Wang Y, et al. The role of inflammatory factors in skeletal muscle injury. Biotarget 2018;2:7 10.21037/biotarget.2018.04.01 [DOI] [Google Scholar]
- 4.Mueller TC, Bachmann J, Prokopchuk O, et al. Molecular pathways leading to loss of skeletal muscle mass in cancer cachexia--can findings from animal models be translated to humans? BMC Cancer 2016;16:75. 10.1186/s12885-016-2121-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baumann CW, Liu HM, Thompson LV. Denervation-Induced Activation of the Ubiquitin-Proteasome System Reduces Skeletal Muscle Quantity Not Quality. PLoS One 2016;11:e0160839. 10.1371/journal.pone.0160839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cea LA, Cisterna BA, Puebla C, et al. De novo expression of connexin hemichannels in denervated fast skeletal muscles leads to atrophy. Proc Natl Acad Sci U S A 2013;110:16229-34. 10.1073/pnas.1312331110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Midrio M. The denervated muscle: facts and hypotheses. A historical review. Eur J Appl Physiol 2006;98:1-21. 10.1007/s00421-006-0256-z [DOI] [PubMed] [Google Scholar]
- 8.de Castro Rodrigues A, Andreo JC, de Mattos Rodrigues SP. Myonuclei and satellite cells in denervated rat muscles: an electron microscopy study. Microsurgery 2006;26:396-8. 10.1002/micr.20258 [DOI] [PubMed] [Google Scholar]
- 9.He Q, Qiu J, Dai M, et al. MicroRNA-351 inhibits denervation-induced muscle atrophy by targeting TRAF6. Exp Ther Med 2016;12:4029-34. 10.3892/etm.2016.3856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sandri M. Protein breakdown in muscle wasting: role of autophagy-lysosome and ubiquitin-proteasome. Int J Biochem Cell Biol 2013;45:2121-9. 10.1016/j.biocel.2013.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao J, Brault JJ, Schild A, et al. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab 2007;6:472-83. 10.1016/j.cmet.2007.11.004 [DOI] [PubMed] [Google Scholar]
- 12.Tawa NE, Jr, Odessey R, Goldberg AL. Inhibitors of the proteasome reduce the accelerated proteolysis in atrophying rat skeletal muscles. J Clin Invest 1997;100:197-203. 10.1172/JCI119513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bodine SC, Latres E, Baumhueter S, et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 2001;294:1704-8. 10.1126/science.1065874 [DOI] [PubMed] [Google Scholar]
- 14.Sacheck JM, Hyatt JP, Raffaello A, et al. Rapid disuse and denervation atrophy involve transcriptional changes similar to those of muscle wasting during systemic diseases. FASEB J 2007;21:140-55. 10.1096/fj.06-6604com [DOI] [PubMed] [Google Scholar]
- 15.Huang Z, Zhu J, Ma W, et al. Strategies and potential therapeutic agents to counter skeletal muscle atrophy. Biotarget 2008;2:8 10.21037/biotarget.2018.05.02 [DOI] [Google Scholar]
- 16.Gomes MD, Lecker SH, Jagoe RT, et al. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc Natl Acad Sci U S A 2001;98:14440-5. 10.1073/pnas.251541198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clarke BA, Drujan D, Willis MS, et al. The E3 Ligase MuRF1 degrades myosin heavy chain protein in dexamethasone-treated skeletal muscle. Cell Metab 2007;6:376-85. 10.1016/j.cmet.2007.09.009 [DOI] [PubMed] [Google Scholar]
- 18.Gomes AV, Waddell DS, Siu R, et al. Upregulation of proteasome activity in muscle RING finger 1-null mice following denervation. FASEB J 2012;26:2986-99. 10.1096/fj.12-204495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Labeit S, Kohl CH, Witt CC, et al. Modulation of muscle atrophy, fatigue and MLC phosphorylation by MuRF1 as indicated by hindlimb suspension studies on MuRF1-KO mice. J Biomed Biotechnol 2010;2010:693741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bechet D, Tassa A, Taillandier D, et al. Lysosomal proteolysis in skeletal muscle. Int J Biochem Cell Biol 2005;37:2098-114. 10.1016/j.biocel.2005.02.029 [DOI] [PubMed] [Google Scholar]
- 21.McMullen CA, Ferry AL, Gamboa JL, et al. Age-related changes of cell death pathways in rat extraocular muscle. Exp Gerontol 2009;44:420-5. 10.1016/j.exger.2009.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barodia SK, Creed RB, Goldberg MS. Parkin and PINK1 functions in oxidative stress and neurodegeneration. Brain Res Bull 2017;133:51-9. 10.1016/j.brainresbull.2016.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang T, Xue L, Li L, et al. BNIP3 Protein Suppresses PINK1 Kinase Proteolytic Cleavage to Promote Mitophagy. J Biol Chem 2016;291:21616-29. 10.1074/jbc.M116.733410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Romanello V, Guadagnin E, Gomes L, et al. Mitochondrial fission and remodelling contributes to muscle atrophy. EMBO J 2010;29:1774-85. 10.1038/emboj.2010.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang Y, Chen K, Ren Q, et al. Dihydromyricetin Attenuates Dexamethasone-Induced Muscle Atrophy by Improving Mitochondrial Function via the PGC-1alpha Pathway. Cell Physiol Biochem 2018;49:758-79. 10.1159/000493040 [DOI] [PubMed] [Google Scholar]
- 26.Masiero E, Agatea L, Mammucari C, et al. Autophagy is required to maintain muscle mass. Cell Metab 2009;10:507-15. 10.1016/j.cmet.2009.10.008 [DOI] [PubMed] [Google Scholar]
- 27.Paul PK, Gupta SK, Bhatnagar S, et al. Targeted ablation of TRAF6 inhibits skeletal muscle wasting in mice. J Cell Biol 2010;191:1395-411. 10.1083/jcb.201006098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takaesu G. Two types of TRAF6-dependent TAK1 activation in the IL-1 signaling pathway. Biotarget 2018;2:2 10.21037/biotarget.2017.12.04 [DOI] [Google Scholar]
- 29.Sun H, Gong Y, Qiu J, et al. TRAF6 inhibition rescues dexamethasone-induced muscle atrophy. Int J Mol Sci 2014;15:11126-41. 10.3390/ijms150611126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paul PK, Bhatnagar S, Mishra V, et al. The E3 ubiquitin ligase TRAF6 intercedes in starvation-induced skeletal muscle atrophy through multiple mechanisms. Mol Cell Biol 2012;32:1248-59. 10.1128/MCB.06351-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun YS, Ye ZY, Qian ZY, et al. Expression of TRAF6 and ubiquitin mRNA in skeletal muscle of gastric cancer patients. J Exp Clin Cancer Res 2012;31:81. 10.1186/1756-9966-31-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soares RJ, Cagnin S, Chemello F, et al. Involvement of microRNAs in the regulation of muscle wasting during catabolic conditions. J Biol Chem 2014;289:21909-25. 10.1074/jbc.M114.561845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wada S, Kato Y, Okutsu M, et al. Translational suppression of atrophic regulators by microRNA-23a integrates resistance to skeletal muscle atrophy. J Biol Chem 2011;286:38456-65. 10.1074/jbc.M111.271270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hudson MB, Rahnert JA, Zheng B, et al. miR-182 attenuates atrophy-related gene expression by targeting FoxO3 in skeletal muscle. Am J Physiol Cell Physiol 2014;307:C314-9. 10.1152/ajpcell.00395.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohtsuka M, Tanemura M, Akamatsu H. Long noncoding RNAs regulate malignant phenotypes in colorectal cancer. Biotarget 2018;2:4 10.21037/biotarget.2018.01.01 [DOI] [Google Scholar]
- 36.Zhang A, Li M, Wang B, et al. miRNA-23a/27a attenuates muscle atrophy and renal fibrosis through muscle-kidney crosstalk. J Cachexia Sarcopenia Muscle 2018;9:755-70. 10.1002/jcsm.12296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li J, Chan MC, Yu Y, et al. miR-29b contributes to multiple types of muscle atrophy. Nat Commun 2017;8:15201. 10.1038/ncomms15201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y, Jiang J, Liu W, et al. microRNA-378 promotes autophagy and inhibits apoptosis in skeletal muscle. Proc Natl Acad Sci U S A 2018;115:E10849-58. 10.1073/pnas.1803377115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu L, Belasco JG. Micro-RNA regulation of the mammalian lin-28 gene during neuronal differentiation of embryonal carcinoma cells. Mol Cell Biol 2005;25:9198-208. 10.1128/MCB.25.21.9198-9208.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Le MT, Xie H, Zhou B, et al. MicroRNA-125b promotes neuronal differentiation in human cells by repressing multiple targets. Mol Cell Biol 2009;29:5290-305. 10.1128/MCB.01694-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ge Y, Sun Y, Chen J. IGF-II is regulated by microRNA-125b in skeletal myogenesis. J Cell Biol 2011;192:69-81. 10.1083/jcb.201007165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Diniz GP, Wang DZ. Regulation of Skeletal Muscle by microRNAs. Compr Physiol 2016;6:1279-94. 10.1002/cphy.c150041 [DOI] [PubMed] [Google Scholar]
- 43.Zhang D, Li Y, Yao X, et al. miR-182 Regulates Metabolic Homeostasis by Modulating Glucose Utilization in Muscle. Cell Rep 2016;16:757-68. 10.1016/j.celrep.2016.06.040 [DOI] [PubMed] [Google Scholar]
- 44.Kukreti H, Amuthavalli K, Harikumar A, et al. Muscle-specific microRNA1 (miR1) targets heat shock protein 70 (HSP70) during dexamethasone-mediated atrophy. J Biol Chem 2013;288:6663-78. 10.1074/jbc.M112.390369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cao C, Zhang H, Zhao L, et al. miR-125b targets DNMT3b and mediates p53 DNA methylation involving in the vascular smooth muscle cells proliferation induced by homocysteine. Exp Cell Res 2016;347:95-104. 10.1016/j.yexcr.2016.07.007 [DOI] [PubMed] [Google Scholar]
- 46.Boyce BF, Li J, Xing L, et al. Bone Remodeling and the Role of TRAF3 in Osteoclastic Bone Resorption. Front Immunol 2018;9:2263. 10.3389/fimmu.2018.02263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Z, Liu F, Wei M, et al. Chronic constriction injury-induced microRNA-146a-5p alleviates neuropathic pain through suppression of IRAK1/TRAF6 signaling pathway. J Neuroinflammation 2018;15:179. 10.1186/s12974-018-1215-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paul PK, Kumar A. TRAF6 coordinates the activation of autophagy and ubiquitin-proteasome systems in atrophying skeletal muscle. Autophagy 2011;7:555-6. 10.4161/auto.7.5.15102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vasilaki A, Richardson A, Van Remmen H, et al. Role of nerve-muscle interactions and reactive oxygen species in regulation of muscle proteostasis with ageing. J Physiol 2017;595:6409-15. 10.1113/JP274336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nuoc TN, Kim S, Ahn SH, et al. The analysis of antioxidant expression during muscle atrophy induced by hindlimb suspension in mice. J Physiol Sci 2017;67:121-9. 10.1007/s12576-016-0444-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qiu J, Fang Q, Xu T, et al. Mechanistic Role of Reactive Oxygen Species and Therapeutic Potential of Antioxidants in Denervation- or Fasting-Induced Skeletal Muscle Atrophy. Front Physiol 2018;9:215. 10.3389/fphys.2018.00215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu C, Tang L, Ni X, et al. Salidroside Attenuates Denervation-Induced Skeletal Muscle Atrophy Through Negative Regulation of Pro-inflammatory Cytokine. Front Physiol 2019;10:665. 10.3389/fphys.2019.00665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beyfuss K, Hood DA. A systematic review of p53 regulation of oxidative stress in skeletal muscle. Redox Rep 2018;23:100-17. 10.1080/13510002.2017.1416773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xie YL, Zhang B, Jing L. MiR-125b blocks Bax/Cytochrome C/Caspase-3 apoptotic signaling pathway in rat models of cerebral ischemia-reperfusion injury by targeting p53. Neurol Res 2018;40:828-37. 10.1080/01616412.2018.1488654 [DOI] [PubMed] [Google Scholar]