Autophagy is a homeostatic mechanism of cells to recycle components, as well as a defense mechanism to get rid of pathogens. Strategies that HSV-1 has developed to counteract autophagy have been described and involve inhibition of autophagosome formation or indirect mechanisms. Here, we present a novel mechanism that involves downregulation of two major autophagy adaptor proteins, sequestosome 1 (p62/SQSTM1) and optineurin (OPTN). These findings generate the question of why the virus targets two major autophagy adaptors if it has mechanisms to block autophagosome formation. P62/SQSTM1 and OPTN proteins have pleiotropic functions, including regulation of innate immunity, inflammation, protein sorting, and chromatin remodeling. The decrease in virus yields in the presence of exogenous p62/SQSTM1 suggests that these adaptors have an antiviral function. Thus, HSV-1 may have developed multiple strategies to incapacitate autophagy to ensure replication. Alternatively, the virus may target another antiviral function of these proteins.
KEYWORDS: HSV-1, ICP0, autophagy, mitophagy, optineurin (OPTN), p62/SQSTM1
ABSTRACT
Herpes simplex virus 1 (HSV-1) infects mucosal epithelial cells and establishes lifelong infections in sensory neurons. Following reactivation, the virus is transferred anterograde to the initial site of infection or to sites innervated by infected neurons, causing vesicular lesions. Upon immunosuppression, frequent HSV-1 reactivation can cause severe diseases, such as blindness and encephalitis. Autophagy is a process whereby cell components are recycled, but it also serves as a defense mechanism against pathogens. HSV-1 is known to combat autophagy through the functions of the γ134.5 protein, which prevents formation of the autophagophore by binding to Beclin 1, a key factor involved in the elongation of the isolation membrane, and by redirecting the protein phosphatase 1α (PP1α) to dephosphorylate the translation initiation factor 2α (eIF2α) to prevent host translational shutoff. Other viral proteins that counteract innate immunity negatively impact autophagy. Here, we present a novel strategy of HSV-1 to evade the host through the downregulation of the autophagy adaptor protein sequestosome (p62/SQSTM1) and of the mitophagy adaptor optineurin (OPTN). This down-modulation occurs during the early steps of the infection. We also found that infected cell protein 0 (ICP0) of the virus mediates the down-modulation of the two autophagy adaptors in a mechanism independent of its E3 ubiquitin ligase activity. Cells depleted of either p62 or OPTN were able to mount greater antiviral responses, whereas cells expressing exogenous p62 displayed decreased virus yields. We conclude that downregulation of p62/SQSTM1 and OPTN is a viral strategy to counteract the host.
IMPORTANCE Autophagy is a homeostatic mechanism of cells to recycle components, as well as a defense mechanism to get rid of pathogens. Strategies that HSV-1 has developed to counteract autophagy have been described and involve inhibition of autophagosome formation or indirect mechanisms. Here, we present a novel mechanism that involves downregulation of two major autophagy adaptor proteins, sequestosome 1 (p62/SQSTM1) and optineurin (OPTN). These findings generate the question of why the virus targets two major autophagy adaptors if it has mechanisms to block autophagosome formation. P62/SQSTM1 and OPTN proteins have pleiotropic functions, including regulation of innate immunity, inflammation, protein sorting, and chromatin remodeling. The decrease in virus yields in the presence of exogenous p62/SQSTM1 suggests that these adaptors have an antiviral function. Thus, HSV-1 may have developed multiple strategies to incapacitate autophagy to ensure replication. Alternatively, the virus may target another antiviral function of these proteins.
INTRODUCTION
More than 80% of the human population is already latently infected by herpes simplex virus 1 (HSV-1) (1). This virus infects primarily mucosal epithelial cells and establishes a lifelong infection in sensory neurons (1). Following reactivation, HSV-1 most commonly causes the formation of painful, vesicular lesions in the oral-facial area (1). Increased frequency of HSV-1 reactivation in elderly and immunocompromised individuals can cause severe diseases, such as herpes encephalitis and blindness due to keratitis (1).
To successfully colonize a host, HSV-1 must overcome the immune barriers of the human body, and for this, it has evolved multiple strategies, including the ability to block autophagy (2–7). Autophagy is a process for selective degradation of cellular components that serves as a homeostatic mechanism of the cells (8–11). Three major types of autophagy have been described: (i) macroautophagy, which involves the delivery of autophagic cargo inside double-membrane vesicles, called autophagosomes, to lysosomes; (ii) microautophagy, in which cytosolic materials reach the lysosomal membrane, where they are internalized; and (iii) chaperone-mediated autophagy, which involves cargo recognition by the heat shock cognate protein of 70 kDa (hsc70) and internalization into the lysosomal lumen (9, 12–14). In addition, selective macroautophagy is used by cells to maintain the integrity and number of organelles in various environments. Such specialized types of autophagy include mitophagy (removal of damaged or excess mitochondria), pexophagy (for peroxisome clearance), reticulophagy (for degradation of portions of the endoplasmic reticulum [ER]), nucleophagy (targeting portions of the nucleus), lysophagy (for lysosomes), ribophagy (for ribosomes), aggrephagy (for protein aggregates), lipophagy (for lipid droplets), proteaphagy (for inactive proteasomes), and others (9, 13, 15–18). In addition to having a role in cellular homeostasis, autophagy is a host defense mechanism that facilitates removal of pathogens. For example, autophagy removes bacteria that escape the phagosomal compartment upon phagocytosis and damaged bacterium-containing phagosomes (9, 10, 19, 20). Virions in the cytoplasm of cells or viral components are removed via a process called virophagy (4, 21–23).
The process of mammalian autophagy involves six principal steps: initiation, nucleation, elongation, closure, maturation, and degradation or extrusion (13, 24). The nascent membrane that is used for phagophore formation can originate from the Golgi complex, endosomes, ER, mitochondria, or plasma membrane. An important factor for phagophore elongation is Beclin 1, a Bcl-2 homology 3 (BH-3) domain protein that is a component of the class III phosphoinositide 3‑kinase (PI3K) complex. Beclin 1 is enzymatically inert, but it regulates the generation of phosphatidylinositol 3-phosphate [PtdIns(3)P] and recruits factors that orchestrate autophagosome formation (13, 24–28). However, autophagy can be performed independently of Beclin 1 through diverse processes. During phagophore elongation, adaptor proteins play an important role, as they deliver ubiquitinated cargos to autophagosomes for degradation (24). Among these adaptors, the best characterized is p62 (sequestosome 1 or SQSTM1). In its inactive state, p62 homodimerizes through its ubiquitin-associated (UBA) domain, and that prevents it from interacting with ubiquitin. Phosphorylation of p62 facilitates transition from dimer to monomer and binding of p62 to ubiquitin chains, so it acquires liquid-like properties. These phase-separated droplets allow the exchange of components, including LC3 and ubiquitin, with their surrounding environment (29–33). Another adaptor that has received special attention is optineurin (OPTN). OPTN regulates several cellular processes pertaining to protein trafficking and membrane cargo delivery from the trans-Golgi compartment to the plasma membrane (34). The OPTN gene originated from gene duplication of the NF-κB regulator known as NF-κB essential modulator (NEMO), and that may explain the contribution of OPTN to inflammation and innate immunity (35–40). OPTN protein carries two nearby ubiquitin binding motifs; therefore, it has preference for binding to longer poly-ubiquitin chains. As with p62, OPTN has a role in delivering ubiquitinated cargo to autophagophores, but it is also involved in clearance of damaged mitochondria (mitophagy) (35–40). Mutations of OPTN have been linked to neurodegenerative disorders (amyotrophic lateral sclerosis [ALS] and dementia) and to normal-tension glaucoma (NTG), as well as juvenile open-angle glaucoma, due to reduced survival of retinal ganglion cells (35–40).
An unequivocal mechanism of HSV-1 to counteract autophagy was discovered in the early 1990s and involved the use of γ134.5, a protein encoded by a “leaky late” gene of the virus, to prevent the host translational shutoff, mediated by activated protein kinase R (PKR), through dephosphorylation of the translation initiation factor eΙF-2α (2, 3). The γ134.5 protein has an essential role in HSV-1 replication in neurons but not in other cell types (2, 3). An additional mechanism involving the γ134.5 protein was later described and included the interaction of γ134.5 with Beclin 1, which inhibits autophagophore formation (6, 7). A virus lacking the Beclin 1 binding domain of γ134.5 failed to counteract autophagy after intracranial injection of mice and displayed impaired replication, but a minor phenotype was observed by this virus in nonneuronal cells in vitro (6, 7). Other mechanisms of HSV-1 to combat autophagy have been proposed. For example, expression of Us11, a late gene product, under an immediate early promoter in the background of the Δγ134.5 virus precluded the host translational shutoff by inhibiting PKR directly (41–44). Viral glycoprotein B suppresses the unfolded protein response (UPR) by binding to protein kinase R-like endoplasmic reticulum kinase (PERK) and preventing its activation and phosphorylation of eIF-2α (45). Finally, mechanisms by which the virus blocks innate immune responses may indirectly inhibit autophagy, as these two processes regulate one another.
Here, we discuss a novel mechanism that is used by HSV-1 to evade the functions of the adaptor proteins p62 and OPTN. This mechanism involves the immediate early gene product of the ICP0 virus that causes proteasome-dependent downregulation of both adaptor proteins. Interestingly, the ICP0 E3 ubiquitin ligase activity does not appear to be required for this process. This downregulation occurs early after infection, it requires calcium, and it depends on a cytoplasmic function of ICP0. p62 and OPTN depletion did not have an apparent effect on the infection by the wild-type virus, but it did compromise mutant viruses unable to block innate immune responses by exacerbating host responses. Wild-type virus infection was compromised by the presence of the p62 protein during the early stages of the infection, and a severe inhibition of viral gene transcription was observed. In conclusion, we have uncovered a novel HSV-1 evasion mechanism that targets major autophagy adaptor proteins. In addition, we describe a role of ICP0 for down-modulation of these adaptors that is independent of its E3 ubiquitin ligase activity but is dependent on cytoplasmic localization and calcium mobilization.
RESULTS
Down-modulation of p62/SQSTM1 and OPTN proteins during HSV-1 infection.
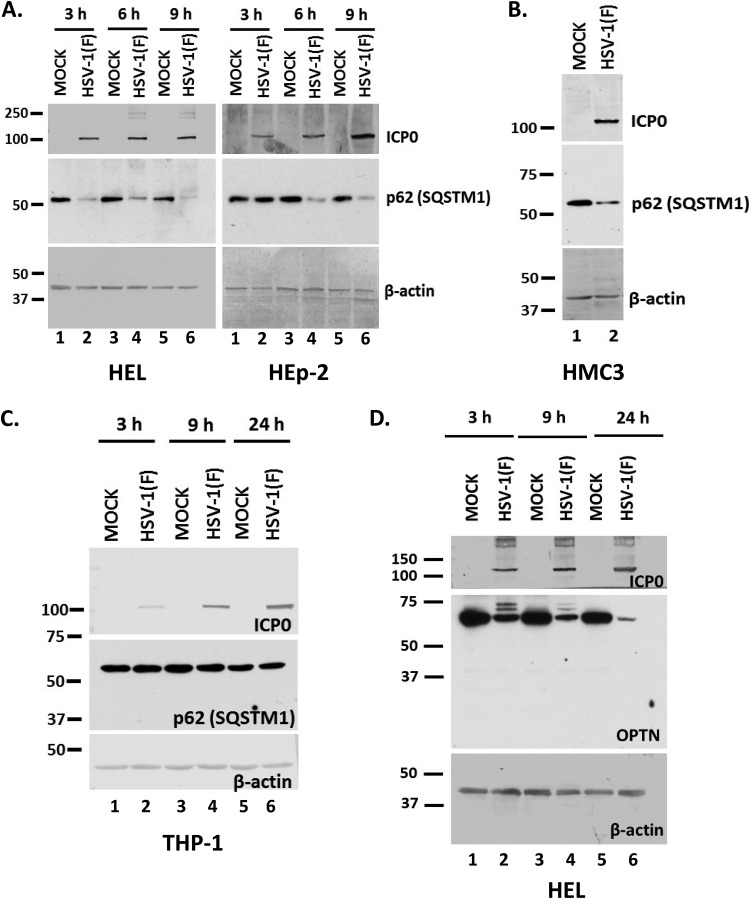
HSV-1 has developed multiple mechanisms to inhibit autophagy, and many studies have been focused on the role of the γ134.5 neurovirulence factor (3, 5–7). Our studies aimed at identifying additional mechanisms that could facilitate autophagy inhibition and immune evasion during HSV-1 infection. Using a targeted approach, we investigated the impact of HSV-1 infection on the autophagy adaptor protein sequestosome 1 (p62/SQSTM1) (32, 38). For this, we infected the human embryonic lung fibroblasts (HEL) and the human epithelial cells (HEp-2) with the HSV-1 F strain (F) (1 PFU/cell). The cells were harvested at 3, 6, and 9 h postinfection, and equal amounts of proteins were analyzed by immunoblot analysis using antibodies against the p62/SQSTM1 protein, ICP0, and β-actin. As shown in Fig. 1A, the amount of p62/SQSTM1 protein was reduced as soon as 3 h after infection of HEL cells with HSV-1(F) and by 6 h after infection of HEp-2 cells. β-Actin served as a loading control and ICP0 as a control of infection. To investigate whether the reduction of p62/SQSTM1 protein was cell type dependent, we infected human microglia cell line 3 (HMC3) and the human monocyte cell line THP-1 with HSV-1(F) (3 PFU/cell). We found that the levels of p62/SQSTM1 protein were decreased in HMC3 cells late during infection (Fig. 1B) but remained unaltered during infection of THP-1 cells (Fig. 1C). Down-modulation of p62/SQSTM1 protein was also noticed in the human glioblastoma cell line SK-N-SH, the human osteosarcoma cell line U2OS, and human embryonic kidney 293 (HEK-293) cells (data not shown). We also investigated whether other autophagy adaptors were downregulated during HSV-1 infection. Optineurin (OPTN) is an adaptor protein that has been linked to mitophagy, the autophagy of mitochondria (38). After infecting HEL cells with HSV-1(F) (3 PFU/cell), we noticed OPTN down-modulation as soon as 3 h postinfection (Fig. 1D). Slower-migrating bands of OPTN appearing after HSV-1 infection may represent posttranslational modifications.
FIG 1.
Downregulation of p62/SQSTM1 and OPTN during HSV-1 infection. (A) HEL or HEp-2 cells were infected with HSV-1(F) (1 PFU/cell), and whole-cell lysates were collected at 3, 6, and 9 h postinfection. Equal amounts of protein were analyzed by immunoblot analysis using an anti-p62 antibody. β-Actin served as a loading control, and ICP0 served as a positive control for infection. Values at the left are molecular masses (in kilodaltons). (B) HMC3 cells were infected with HSV-1(F) (3 PFU/cell) for 24 h, and p62 protein was detected in equal amounts of cell lysates as described for panel A. (C) THP-1 cells were infected with HSV-1(F) (3 PFU/cell), the cells were harvested at 3, 9, and 24 h after infection, and p62 protein was monitored in equal amounts of cell lysates as described for panel A. (D) HEL cells were infected with HSV-1(F) (3 PFU/cell), and whole-cell lysates were collected at 3, 9, and 24 h postinfection and analyzed by immunoblot analysis, using an anti-OPTN antibody and an anti-ICP0 antibody. β-Actin served as a loading control. All experiments were repeated at least two independent times, and representative Western blots are presented.
Overall, multiple autophagy adaptors appear to be downregulated during the early stages of HSV-1 infection in various cell lines.
Viral gene expression is required for downregulation of p62/SQSTM1 and OPTN.
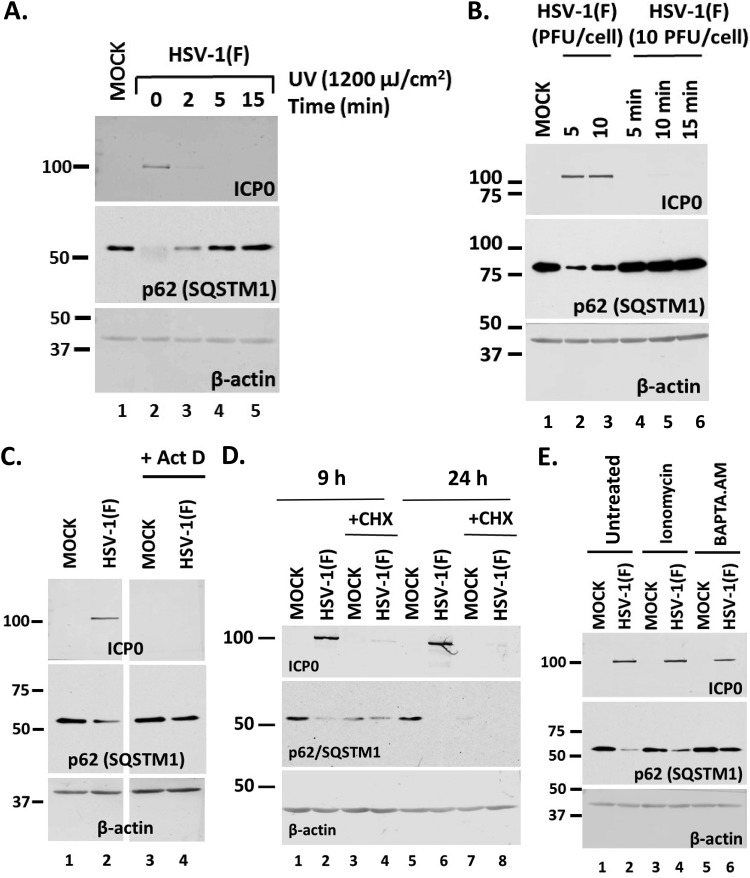
To determine the stage of the infection that could cause downregulation of the autophagy adaptors, a series of four experiments were performed. First, we inactivated the virus by UV exposure (1,200 μJ/cm2 for 2, 5, or 15 min), which damages the viral genome, and then used this virus to infect HEL cells. The cells were harvested at 24 h postinfection, and the levels of p62/SQSTM1 were monitored in equal amounts of cell lysates by immunoblot analysis. As shown in Fig. 2A, UV-inactivated virus did not cause p62/SQSTM1 downregulation. As expected, ICP0 protein was undetectable since viral gene expression could not occur from the UV-damaged genome. β-Actin was used as a loading control. Second, we heat inactivated the virus to denature virion proteins by incubation at 56°C for 5, 10, or 15 min and exposed HEL cells to this virus, as described above. As shown in Fig. 2B, heat-inactivated virus did not cause down-modulation of p62/SQSTM1, and ICP0 expression did not occur either. In the third experiment, we either blocked transcription by treatment with actinomycin D (10 μg/ml) (Fig. 2C) or blocked protein synthesis by treatment with cycloheximide (CHX) (100 μg/ml) (Fig. 2D). The cells were harvested at 9 h postinfection (10 PFU/cell), and the levels of p62/SQSTM1 protein were monitored as described above. As shown in Fig. 2D, treatment with CHX down-modulated p62/SQSTM1 both in uninfected and in infected cells, suggesting possible toxicity. In contrast, inhibition of transcription did not affect the levels of p62/SQSTM1, suggesting that viral gene transcription is essential for p62/SQSTM1 down-modulation (Fig. 2C). In the last experiment, we perturbed calcium levels by treating HEL cells with either ionomycin (10 μM), a Ca2+ ionophore that causes calcium mobilization from intracellular stores, or the calcium chelator BAPTA-AM [1,2-bis(o-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid; 10 μM]; these were added to the cultures at 1 h postinfection with HSV-1(F) (10 PFU/cell). The cells were harvested at 9 h postinfection, and the levels of p62/SQSTM1 were monitored as described above. Treatment with ionomycin did not affect the ability of the virus to cause p62/SQSTM1 downregulation, whereas calcium chelation did (Fig. 2E).
FIG 2.
Viral entry and transcription are required for down-modulation of p62 during HSV-1 infection. (A) HEL cells were infected with HSV-1(F) (10 PFU/cell) that had been exposed to UV light (1,200 μJ/cm2) for 0, 2, 5, or 15 min, and whole-cell lysates were collected at 24 h postinfection. The levels of p62 protein were analyzed in equal amounts of cell lysates using an anti-p62 antibody. ICP0 expression served as a positive control for infection. β-Actin was used as a loading control. (B) HEL cells were infected with HSV-1(F) at 5 PFU/cell (lane 2) or 10 PFU/cell (lane 3) or with viral stock (10 PFU/cell) that had been heated at 56°C for 5, 10, or 15 min (lanes 4 to 6). Whole-cell lysates were collected at 24 h postinfection, and equal amounts of protein were analyzed by immunoblot analysis, using antibodies against p62 and ICP0. β-Actin served as a loading control. (C) HEL cells were infected with HSV-1(F) (10 PFU/cell) and were left untreated or treated with actinomycin D (ActD; 10 μg/ml) at 1 h postinfection. Whole-cell lysates were collected at 9 h postinfection, and equal amounts of protein were analyzed by immunoblot analysis, using antibodies against p62 or ICP0. β-Actin served as a loading control. (D) The experiment whose results are shown was done as described for panel C except that the cells were treated with cycloheximide (CHX; 100 μg/ml), added at the moment of infection. The cells were harvested at 9 and 24 h postinfection, and equal amounts of proteins were analyzed as described above. (E) HEL cells were infected with HSV-1(F) (10 PFU/cell) and were left untreated or treated with ionomycin (10 μM) or BAPTA.AM (10 μM) at 1 h postinfection. Whole-cell lysates were collected at 9 h after infection, and equal amounts of protein were analyzed by immunoblot analysis, using antibodies against p62 or ICP0. β-Actin served as a loading control. All experiments were repeated three independent times, and representative Western blots are presented.
Our data suggest that viral gene expression and calcium are important for down-modulation of p62/SQSTM1 during HSV-1 infection.
ICP0-dependent down-modulation of p62/SQSTM1 and OPTN during HSV-1 infection.
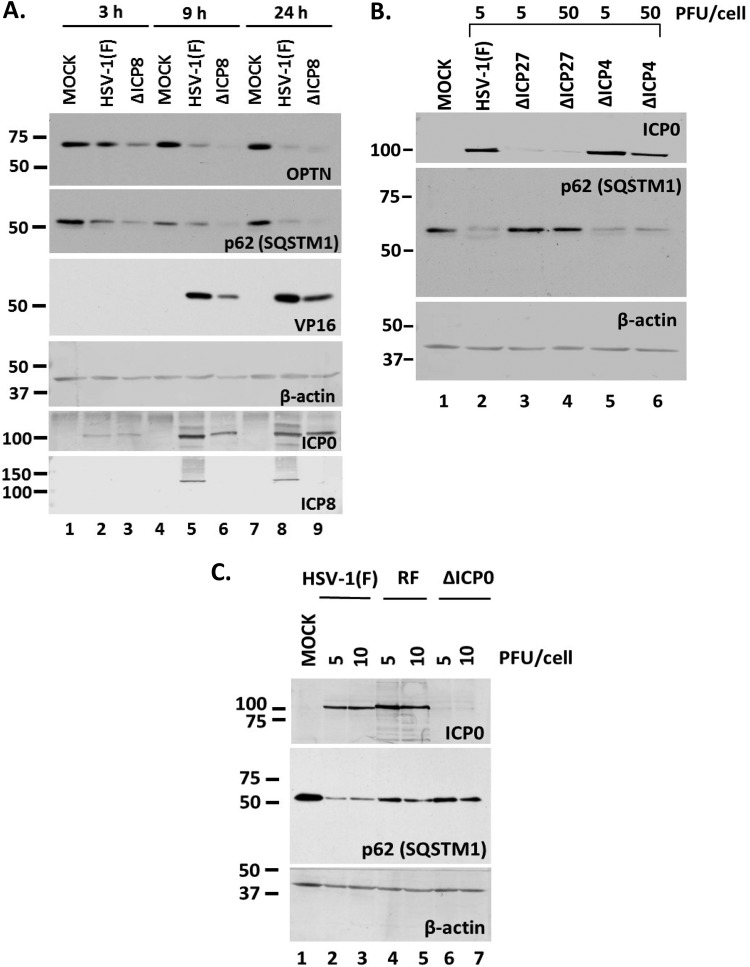
To identify viral genes involved in the downregulation of p62 and OPTN, we used a panel of mutant viruses in a series of three experiments. First, we exposed HEL cells to either the replication-deficient ΔICP8 virus or the wild-type virus (3 PFU/cell). The cells were harvested at 3, 9, and 24 h postinfection, and the levels of p62/SQSTM1 were assessed in equal amounts of cell lysates by immunoblot analysis. As shown in Fig. 3A, infection with the ΔICP8 virus caused down-modulation of p62/SQSTM1, as with the wild-type virus, as soon as 3 h postinfection. A similar down-modulation was observed for the OPTN protein. ICP0 expression was used as a control of infection, ICP8 and VP16 expression indicated the replication-defective virus, and β-actin was used as a loading control. These data suggest that virus replication is not required for downregulation of major autophagy adaptors and suggest that earlier events trigger this process.
FIG 3.
Downregulation of p62 during HSV-1 infection is independent of viral replication but dependent on immediate early gene expression. (A) HEL cells were infected with HSV-1(F) or the ΔICP8 virus (3 PFU/cell), and whole-cell lysates were collected at 3, 9, and 24 h postinfection. Equal amounts of protein were loaded for immunoblot analysis using antibodies against OPTN, p62, ICP0, and ICP8. ICP0 served as a control of infection, whereas VP16 served as a control for viral-genome replication. β-Actin served as a loading control. (B) HEL cells were infected with HSV-1(F) (5 PFU/cell), the ΔICP27 mutant (5 or 50 PFU/cell), or the ΔICP4 mutant (5 or 50 PFU/cell), and whole-cell lysates were collected at 9 h postinfection. Equal amounts of protein were analyzed by immunoblot analysis using antibodies against p62 and ICP0. β-Actin served as a loading control. (C) HEL cells were infected with HSV-1(F) or the RF or ΔICP0 mutant at 5 or 10 PFU/cell, and whole-cell lysates were collected at 9 h postinfection. Equal amounts of protein were analyzed by immunoblot analysis, using antibodies against p62 and ICP0. β-Actin served as a loading control. All experiments were repeated three independent times, and representative Western blots are presented.
Second, we infected HEL cells with the ΔICP27 and ΔICP4 mutant viruses (5 and 50 PFU/cell), which lack the essential immediate early genes ICP27 and ICP4, respectively; therefore, viral infection is arrested at transcription initiation. Infected cells were harvested at 9 h postinfection, and the levels of p62/SQSTM1 were assessed in equal amounts of cell lysates, as described above. As shown in Fig. 3B, infection with the ΔICP4 virus caused p62/SQSTM1 protein down-modulation, whereas infection with the ΔICP27 virus did not. Among the differences known for these viruses, ICP0 expression is one. As shown in Fig. 3B, the ΔICP4 virus expressed high levels of ICP0 in order to complement the absence of ICP4, whereas in ΔICP27 virus-infected cells, ICP0 expression was reduced. These data prompted us to investigate whether ICP0 has a role in the down-modulation of p62/SQSTM1 because ICP0 is an E3 ubiquitin ligase that degrades hostile host factors (46, 47). For this, we exposed HEL cells to two different doses (5 and 10 PFU/cell) of either HSV-1(F), an ICP0-null virus (ΔICP0), or an ICP0 E3 ubiquitin ligase (RING finger [RF]) mutant (with the C116A and C156A mutations) (47). The cells were harvested at 24 h postinfection, and the levels of p62/SQSTM1 protein were assessed in equal amounts of cell lysates, as described above. As shown in Fig. 3C, the ΔICP0 virus did not cause down-modulation of p62/SQSTM1.
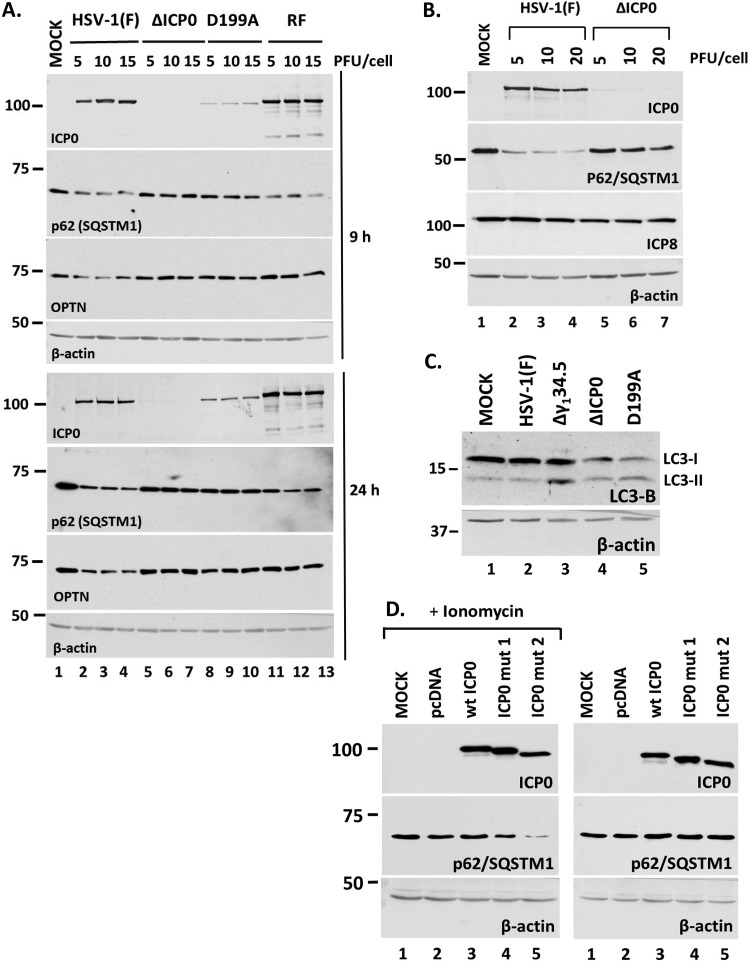
To further assess the role of ICP0, we repeated the experiment using a mutant virus in which ICP0 is retained in the nucleus due to a replacement of aspartic acid 199 with alanine (D199A mutant), in addition to the ΔICP0 virus, the ICP0 RF mutant, and the wild-type virus (48). Infections were done using three different doses of these viruses (5, 10, and 15 PFU/cell), the cells were harvested at 9 and 24 h postinfection, and the amounts of p62/SQSTM1 were analyzed by Western blotting. As shown in Fig. 4A, both the wild-type virus and the ICP0 RF mutant caused a down-modulation of p62/SQSTM1 at both time points. In contrast, the ΔΙCP0 virus did not affect p62/SQSTM1 even up to 24 h postinfection. ΔICP0 virus-infected cells accumulated amounts of ICP8 similar to those in wild-type-virus-infected cells (Fig. 4B), suggesting that lack of p62/SQSTM1 down-modulation is not due to possible replication differences. In addition, the D199A virus did not cause any reduction of the p62/SQSTM1 levels compared to those in the wild-type virus. Autophagy can result in down-modulation of p62/SQSTM1; thus, we assessed the levels of lipidated LC3-B after infection with either the wild type or the ICP0 mutant viruses as an indication of autophagy activation. Infection with the γ134.5-null virus served as a positive control since this virus cannot counteract autophagy. As shown in Fig. 4C, the ΔICP0 and D199A viruses resulted in higher ratios of LC3-II to LC3-I than in uninfected or HSV-1-infected cells, which might be indicative of autophagy activation. Thus, the small decrease of p62/SQSTM1 after infection with ICP0 mutants at late hours postinfection may be attributed to some degree of autophagy.
FIG 4.
Down-modulation of p62 during HSV-1 infection is dependent on ICP0. (A) HEL cells were infected with HSV-1(F) or the ΔICP0, D199A, or RF ICP0 mutant virus at 5, 10, or 15 PFU/cell. Whole-cell lysates were collected at 9 and 24 h after infection, and equal amounts of protein were analyzed by immunoblot analysis, using antibodies against p62 and ICP0. β-Actin served as a loading control. (B) HEL cells were infected with HSV-1(F) or the ΔICP0 virus at 5, 10, or 20 PFU/cell. The cells were harvested at 9 h postinfection, and proteins were analyzed as described for panel A. (C) HEL cells were infected with HSV-1(F) or the Δγ134.5, ΔICP0, or D199A virus at 10 PFU/cell. Whole-cell lysates were collected at 9 h after infection, and equal amounts of protein were analyzed by immunoblot analysis, using an antibody against LC3-B. β-Actin served as a loading control. (D) HEK-293 cells were either not transfected or transfected with an empty pcDNA vector, a plasmid expressing wild-type (wt) ICP0, or plasmids expressing mutated forms of ICP0 lacking either the NLS (ICP0 mut1) or the NLS and a putative NLS (ICP0 mut2). Ionomycin (5 μM) was added in one set of replicate cultures at 32 h posttransfection for 16 h. The cells were harvested at 48 h posttransfection, and the levels of p62 were determined in equal amounts of cell lysates by immunoblot analysis. All experiments were repeated at least three independent times, and representative Western blots are depicted.
Next, we asked whether ICP0 outside the context of the infection could cause p62 down-modulation. For this, in addition to using the wild-type ICP0, we developed two mutated forms that localize in the cytoplasm: one lacking the nuclear localization signal (NLS) (deletion of amino acids 447 to 511) (ICP0 mut1) and a second one lacking the NLS (deletion of amino acids 447 to 511) and a putative NLS (deletion of amino acids 244 to 277) (ICP0 mut2). ICP0 mut2 localizes quantitatively in the cytoplasm, unlike ICP0 mut1. We performed two sets of experiments. First, we transfected HEK-293 cells with either an empty vector or a plasmid expressing wild-type ICP0, ICP0 mut1, or ICP0 mut2. Second, we treated the transfected cells with ionomycin (5 μM) for 16 h to trigger calcium mobilization. The cells were harvested at 48 h posttransfection, and the levels of p62 were assessed in equal amounts of total proteins by immunoblot analysis. As shown in Fig. 4D, there was a significant down-modulation of p62 in the presence of ICP0 mut2 when cells were treated with ionomycin. A smaller effect was noticed in the cells with ICP0 mut1 in the presence of ionomycin. Interestingly, we noticed no effect by these ICP0 forms in non-ionomycin-treated cells, suggesting that calcium mobilization is important for ICP0-mediated p62 down-modulation.
Overall, downregulation of p62/SQSTM1 occurs prior to virus replication, and cytoplasmic ICP0 contributes to this process in an E3 ubiquitin ligase-independent manner.
Down-modulation of p62/SQSTM1 is proteasome dependent.
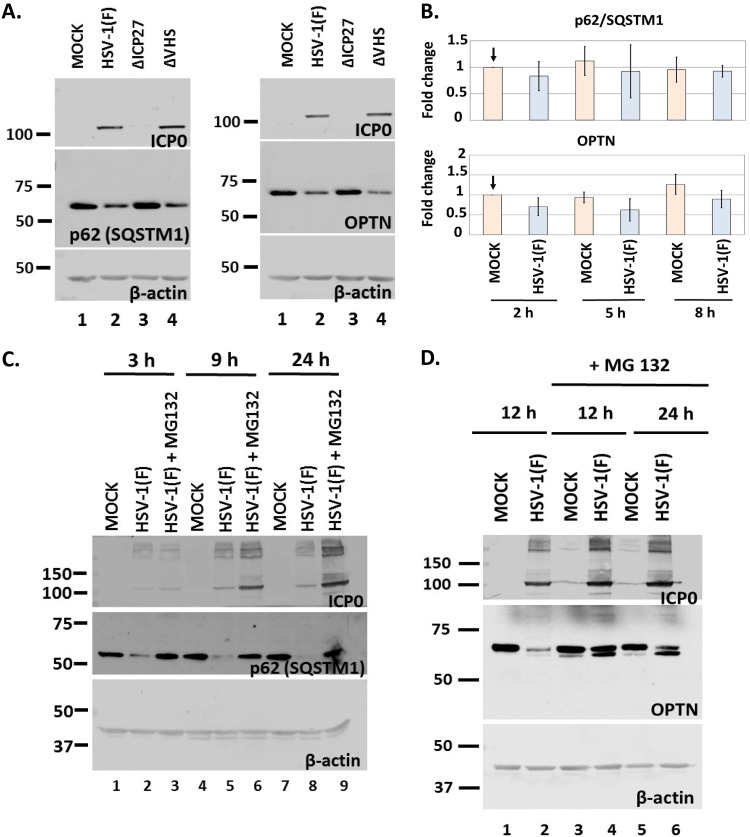
To identify the mechanism of downregulation of p62/SQSTM1 and OPTN during HSV-1 infection, we performed a series of four experiments. First, we asked if the virion host shutoff (VHS), the viral RNase, has any role in the down-modulation of p62/SQSTM1 and OPTN. For this, HEL cells were exposed to HSV-1(F), a ΔVHS virus, or a ΔICP27 virus (3 PFU/cell). The cells were harvested at 9 h postinfection, and equal amounts of cell lysates were analyzed for p62/SQSTM1 and OPTN expression. As shown in Fig. 5A, infection with the ΔVHS virus resulted in a down-modulation of p62/SQSTM1 and OPTN proteins similar to that of the wild-type virus infection. Infection with the ΔICP27 virus served as a control because this virus does not cause downregulation of the autophagy adaptors.
FIG 5.
Down-modulation of p62 and OPTN protein during infection occurs at the level of protein and not of transcript. (A) HEL cells were infected with HSV-1(F) or the ΔICP27 or ΔVHS mutant (3 PFU/cell), and whole-cell lysates were collected at 9 h postinfection. Equal amounts of protein were analyzed by immunoblot analysis using antibodies against p62, OPTN, or ICP0. β-Actin served as a loading control. (B) HEL cells were not infected or infected with HSV-1(F) (5 PFU/cell), and cells were harvested at 2, 5, and 8 h postinfection. OPTN and p62 transcript levels were determined in the total amount of RNA by qPCR analysis using probes against p62 or OPTN transcripts. Primers against 18S rRNA served as a normalization control. Results from three independent assays do not demonstrate statistically significant differences. (C) HEL cells were infected with HSV-1(F) (3 PFU/cell) and were left untreated or were treated with MG132 (10 μM) at 1 h postinfection. Whole-cell lysates were collected at 3, 9, and 24 h postinfection, and equal amounts of protein were analyzed by immunoblot analysis, using antibodies against p62 and ICP0, which served as both a control for infection and a positive control for the effect of MG132 on ICP0 protein accumulation. β-Actin served as a loading control. (D) HEL cells were infected with HSV-1(F) (3 PFU/cell) and were left untreated or treated with MG132 as described for panel C. Whole-cell lysates were collected at 12 and 24 h postinfection, and protein analysis was done as described for panel C. All experiments were repeated at least three independent times, and representative Western blots are depicted.
Second, to further support the supposition that the amounts of p62/SQSTM1 and OPTN transcripts remain unaltered during infection, we exposed HEL cells to HSV-1(F) (5 PFU/cell), the cells were harvested at 2, 5, and 8 h postinfection, and the amounts of p62/SQSTM1 and OPTN transcripts in total RNA were determined by real-time PCR analysis, using TaqMan probes for the above-mentioned transcripts. As shown in Fig. 5B, both the p62/SQSTM1 and OPTN transcripts remained stable at least up to 8 h postinfection, suggesting that the decrease in the amounts of both proteins was not due to down-modulation of their transcripts.
Third, we investigated if proteasomes mediate downregulation of p62/SQSTM1 and OPTN. HEL cells were uninfected or infected with the wild-type virus, and the proteasome inhibitor MG132 (10 μM) was added in replicate cultures 1 h after the addition of virus (3 PFU/cell). The cells were harvested at 3, 9, and 24 h postinfection for analysis of p62/SQSTM1 or at 12 and 24 h postinfection for analysis of OPTN, and the amounts of both proteins were determined in equal amounts of cell lysates. As shown in Fig. 5C, MG132 prevented the down-modulation of p62/SQSTM1 protein. For OPTN, we noticed that in the presence of MG132, a faster-migrating form accumulated, which appears to originate from the slower-migrating form of the protein (Fig. 5D). Perhaps a posttranslational modification, such as proteolytic cleavage, occurs before degradation of OPTN during HSV-1 infection.
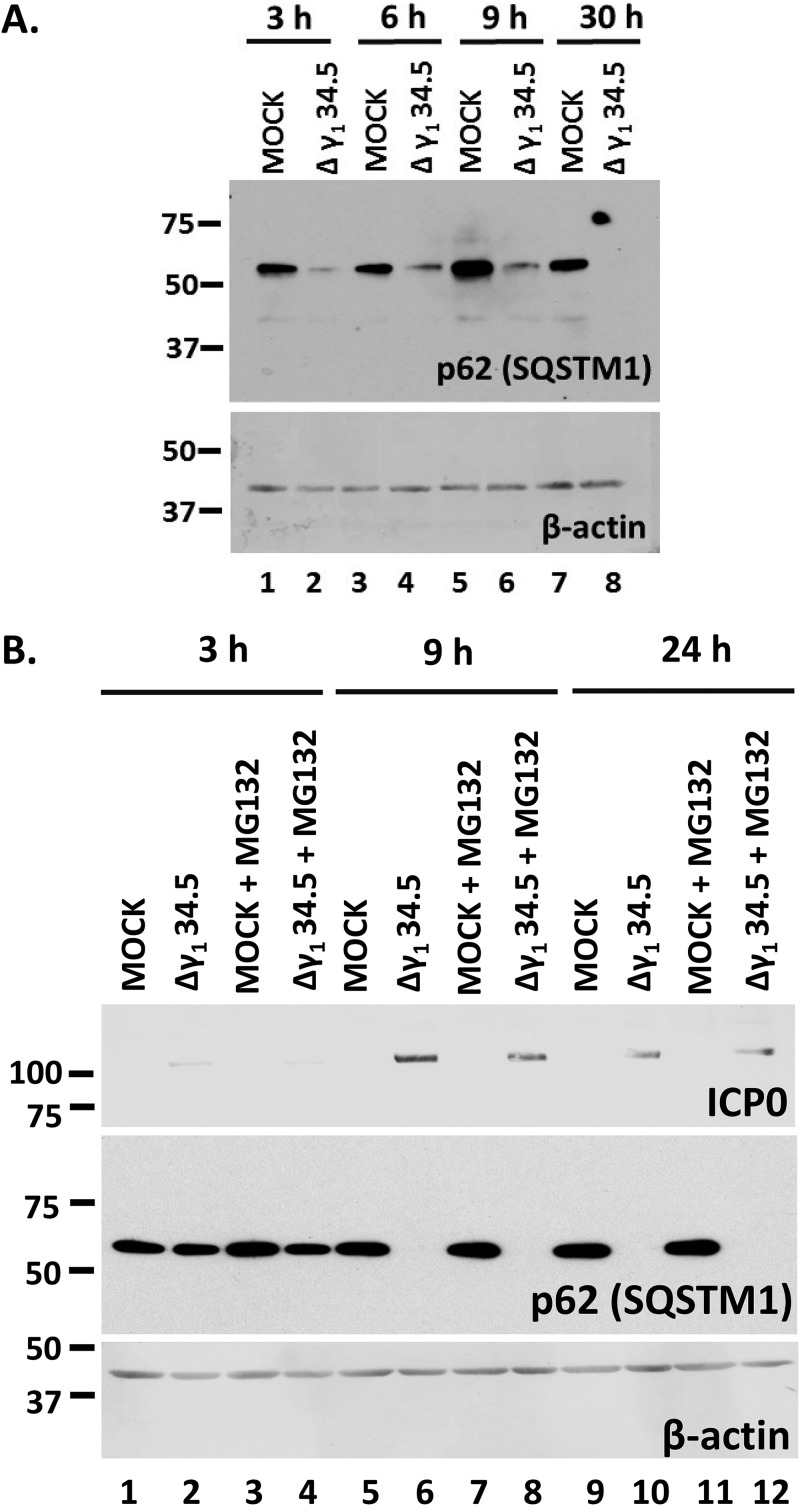
In the last series of experiments, we investigated the role of γ134.5 protein in the down-modulation of p62/SQSTM1 by infecting HEL cells with the γ134.5-null virus (3 PFU/cell). We observed that p62/SQSTM1 protein was down-modulated in Δγ134.5 virus-infected cells (Fig. 6A). However, the Δγ134.5 virus cannot block autophagy, which may be the reason for p62/SQSTM1 down-modulation, whereas HSV-1 can (Fig. 4C) (2, 6). To address whether the mechanisms of downregulation of p62/SQSTM1 are different between the two viruses, we treated Δγ134.5 virus-infected cells with MG132. We found that treatment with MG132 did not prevent the down-modulation of p62/SQSTM1 in Δγ134.5 virus-infected cells (Fig. 6B) but that it did in HSV-1-infected cells (Fig. 5C). Additional repeats of this assay demonstrated that MG132 may partially rescue p62/SQSTM1 levels in Δγ134.5 virus-infected cells but that it fully rescued the levels of this protein in wild-type-virus-infected cells.
FIG 6.
Downregulation of p62 still occurs during infection with the Δγ134.5 virus but in a proteasome-independent manner. (A) HEL cells were uninfected or infected with the Δγ134.5 virus (3 PFU/cell), and whole-cell lysates were collected at 3, 6, 9, and 30 h after infection. Equal amounts of protein were analyzed by immunoblot analysis using an anti-p62 antibody. β-Actin served as a loading control. (B) HEL cells were uninfected or infected with the Δγ134.5 virus (5 PFU/cell) and were either untreated or treated with MG132 (10 μM) at 1 h postinfection. Whole-cell lysates were collected at 3, 9, and 24 h after infection. Equal amounts of protein were analyzed by immunoblot analysis using an anti-p62 antibody and an anti-ICP0 antibody. β-Actin served as a loading control. All experiments were repeated two independent times, and representative Western blots are depicted.
Taken together, our data suggest that p62/SQSTM1 and OPTN are most likely degraded via the proteasome pathway during infection by the wild-type virus.
P62/SQSTM1 KD and OPTN KD negatively impact ΔICP0 but not wild-type virus infection.
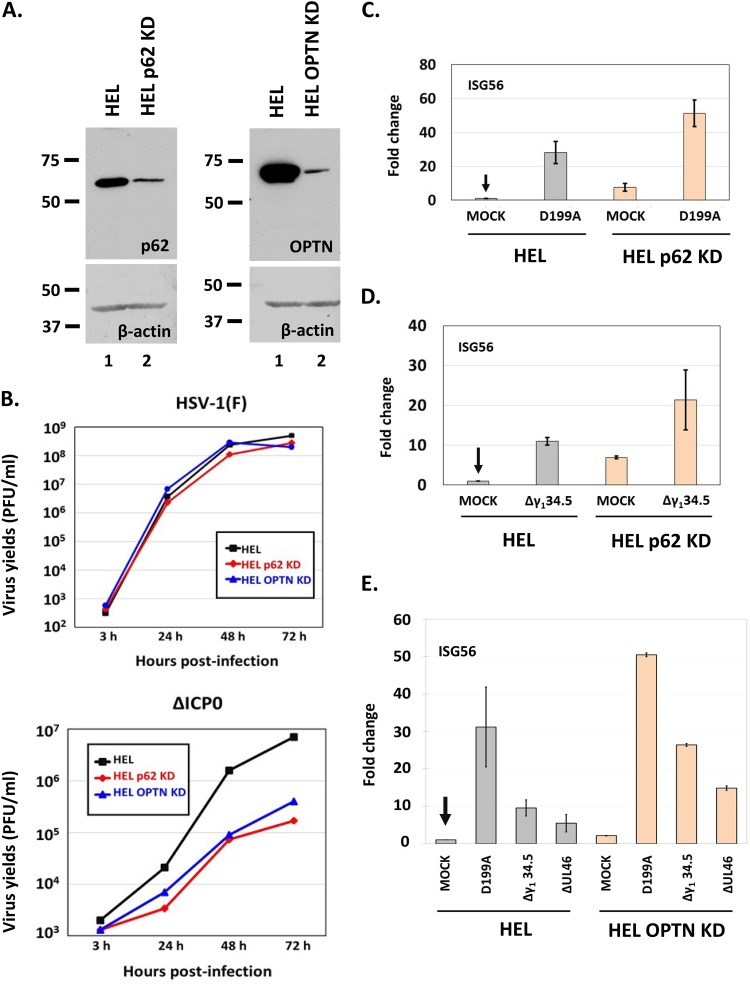
Since wild-type virus downregulates p62/SQSTM1 and OPTN early after infection, we were not expecting any benefit for the wild-type virus in cells depleted of these proteins. However, it was unclear whether mutant viruses that cannot counteract antiviral responses would be affected in cells depleted of these autophagy adaptors. To assess this, we knocked down p62/SQSTM1 and OPTN in HEL cells using lentiviral vectors carrying short hairpin RNAs (shRNAs) against their transcripts, and the efficiency of knockdown is depicted in Fig. 7A. We then exposed these cell lines to either the wild-type virus or the ΔICP0 virus (0.01 PFU/cell), harvested the cells at 3, 24, 48, and 72 h postinfection, and assessed progeny virus production by titration in Vero cells. As expected, wild-type virus infection did not display any apparent benefit in the knockdown cell lines (Fig. 7B), but the ΔICP0 virus displayed a decrease in virus yields by almost 15-fold in both cell lines (Fig. 7B). The ΔICP0 virus has defects in activating viral-gene transcription and blocking innate immune responses (46, 49–54). Thus, we asked, “how does depletion of p62/SQSTM1 and OPTN proteins impact innate immune responses to infections by defective viruses?” To assess this, we exposed HEL cells and their p62/SQSTM1 knockdown (p62/SQSTM1 KD) or OPTN KD derivatives to different mutant viruses (0.01 PFU/cell), and the cells were harvested at 12 h postinfection. Quantification of the expression of interferon-stimulated gene 56 (ISG56), a type I interferon-stimulated gene, was done by real-time PCR analysis. The mutant viruses that we chose in this study included (i) the D199A ICP0 mutant, in which ICP0 localizes to the nucleus and therefore the virus fails to block interferon regulatory factor 3 (IRF3) activation; (ii) a Δγ134.5 virus, which fails to combat innate immune responses, including autophagy; and (iii) a ΔUL46 virus, which fails to block type I interferon responses (2, 48, 55). As shown in Fig. 7C, D, and E, ISG56 expression was increased in p62/SQSTM1 KD and OPTN KD cells compared to that in their parental HEL cells after infection with the above-mentioned mutants. We also noticed increased basal levels of ISG56 expression in the uninfected knockdown cell lines compared to levels in their parental cells.
FIG 7.
Innate-immunity activation is higher upon infection of cells knocked down for p62 or OPTN. (A) Development of p62 knockdown and OPTN knockdown HEL cells with the aid of lentiviral vectors. (B) HEL cells or the p62 knockdown or OPTN knockdown derivatives were infected with either HSV-1(F) or the ΔICP0 virus (0.01 PFU/cell), and cells were collected at 3, 24, and 48 h after infection. Titration of progeny virus was then done in Vero cells for the wild-type virus or in U2OS cells for the ΔICP0 virus. (C) HEL cells or their p62 knockdown derivatives were uninfected or infected with the D199A virus (0.01 PFU/cell). Cells were harvested at 12 h postinfection, and qPCR analysis was done using primer pairs against ISG56 or the 18S rRNA, which served as a normalization control. (D) HEL cells or their p62 knockdown derivatives were uninfected or infected with the HSV-1 Δγ134.5 virus (0.01 PFU/cell). Quantification of ISG56 transcripts was done as described above. (E) HEL cells or their OPTN knockdown derivatives were uninfected or infected with the HSV-1 D199A, Δγ134.5, or ΔUL46 mutant virus (0.01 PFU/cell). Cells were harvested at 12 h postinfection, and qPCR analysis was done using primers against ISG56 transcripts or the 18S rRNA, as described above. All experiments were repeated three times, and representative results are depicted.
We conclude that p62/SQSTM1 and OPTN may negatively regulate innate immune responses following exposure of cells to mutants of HSV-1 that fail to block type I interferon responses, perhaps through regulation of autophagy.
Exogenous expression of p62/SQSTM1 negatively impacts HSV-1 virus yields.
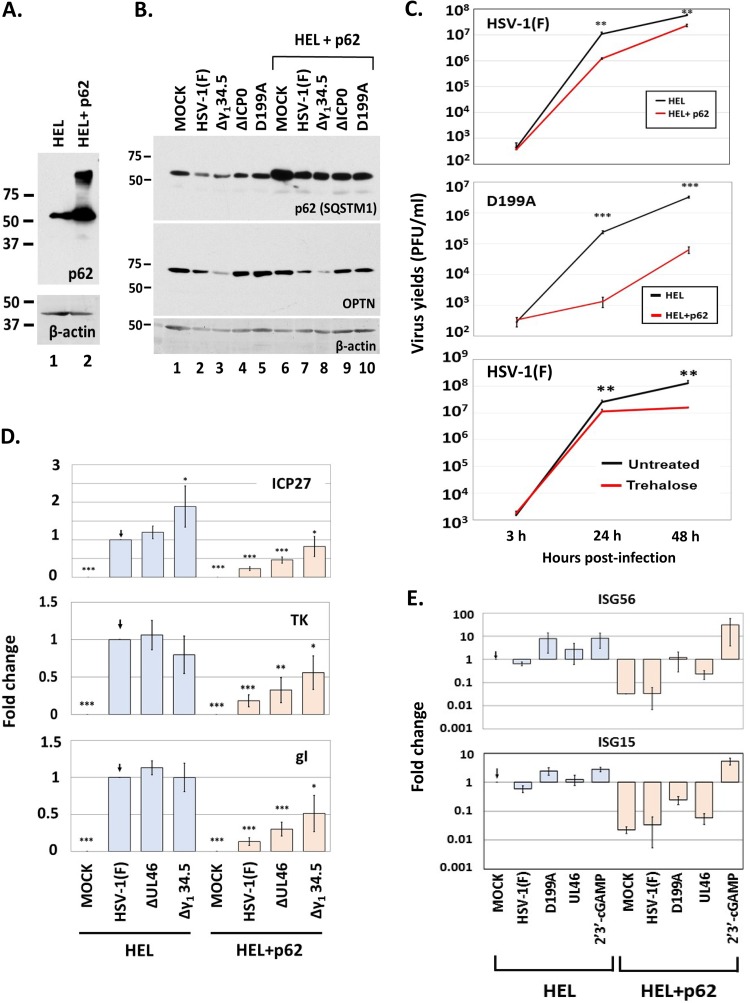
To assess the effect of p62/SQSTM1 on HSV-1 infection, we utilized a system in which HSV-1 could not effectively downregulate p62/SQSTM1. This system is an HEL cell line which expresses exogenous p62/SQSTM1 and was established with the aid of lentiviral vectors (Fig. 8A). To ensure that p62/SQSTM1 is not downregulated in this cell line, HEL and HEL with p62 were infected with either the wild-type, Δγ134.5, D199Α, or ΔICP0 virus (5 PFU/cell). The cells were harvested at 8 h postinfection, and the amounts of p62/SQSTM1 were determined in equal amounts of cell lysates. As shown in Fig. 8B, none of these viruses caused down-modulation of exogenous p62/SQSTM1, compared to the down-modulation seen in the parental cells. Subsequently, we asked whether exogenous p62/SQSTM1 affects virus yields. For this, HEL cells and their p62/SQSTM1-expressing derivatives were exposed to either HSV-1(F) or the D199A virus (0.01 PFU/cell), the cells were harvested at 3, 24, and 48 h postinfection, and progeny virus production was assessed by titration in Vero cells. As shown in Fig. 8C, exogenous expression of p62/SQSTM1 caused at least a 10-fold reduction in wild-type virus yields and at least a 15-fold reduction in D199A virus yields. Consistently, treatment with trehalose, a known autophagy activator, caused a 10-fold decrease in wild-type virus yields at 48 h postinfection. This decrease in virus yields may be attributed to a reduction in all classes of viral gene transcription that was observed in HEL cells expressing exogenous p62/SQSTM1 (Fig. 8D). Analysis of interferon-stimulated gene (ISG) transcription following infection of HEL cells plus p62 with mutants that cannot counteract innate immunity, compared to that after infections of their parental cells, indicated that these cells did not accumulate ISG transcripts as highly and that they also had a significantly lower baseline levels of ISGs (Fig. 8E). However, these cells were able to mount robust innate immune responses in the presence of cyclic G(2',5')pA(3',5')p (2′3′-cGAMP), a ligand of the stimulator of interferon genes (STING) (Fig. 8E). This is a component of innate immunity that senses foreign nucleic acids and combats pathogens, including HSV-1. Together, these data suggest that early downregulation of p62/SQSTM1 is critical for HSV-1 infection.
FIG 8.
Constitutive expression of p62 decreases viral yields. (A) Development of HEL cells constitutively expressing p62 was done using lentiviral vectors carrying the gene for p62. Whole-cell lysates from HEL cells or their derivatives overexpressing p62 were collected, and equal amounts of protein were analyzed by immunoblot analysis using an anti-p62 antibody. β-Actin served as a loading control. (B) HEL cells or their derivatives overexpressing p62 were uninfected or infected with HSV-1(F) or the Δγ134.5, ΔICP0, and D199A mutant viruses (5 PFU/cell). Whole-cell lysates were collected at 8 h postinfection, and equal amounts of protein were analyzed by immunoblot analysis using antibodies against p62 and OPTN. β-Actin served as a loading control. (C) HEL cells or their derivatives exogenously expressing p62 were infected with HSV-1(F) (0.01 PFU/cell) or the D199A virus (0.01 PFU/cell), and cells were collected at 3, 24, and 48 h after infection. Titration of progeny virus was then done in Vero cells for the wild-type virus or in U2OS cells for the D199A virus. In a parallel assay, HEL cells were infected with HSV-1(F) (0.01 PFU/cell) and treated with trehalose (300 mM), which was added to the cultures at the moment of infection. Viral yields were determined as described above. (D) HEL cells or their derivatives overexpressing p62 were uninfected or infected with HSV-1(F), the ΔUL46 mutant, or the Δγ134.5 mutant (0.5 PFU/cell). Cells were harvested at 9 h postinfection, and qPCR analysis was done using primers against ICP27, TK1, and gI or 18S rRNA. (E) HEL cells or their derivatives exogenously expressing p62 were uninfected, infected with HSV-1(F), the D199A mutant, or the ΔUL46 mutant (0.5 PFU/cell), or treated with 2′3′-cGAMP (6 μM). Cells were harvested at 9 h postinfection, and qPCR analysis was done using a primer against ISG56 or a probe against ISG15, while 18S rRNA was analyzed to serve as a normalization control. All experiments were repeated at least three times, and representative results are depicted.
DISCUSSION
Autophagy is a homeostatic mechanism for self-component recycling, but it is also used by cells to eat (phago-) an unknown (xenos) biological entity, including a pathogen, in a process known as xenophagy (56, 57). Consequently, pathogens, including HSV-1, have evolved mechanisms to counteract autophagy and frequently hijack autophagy-related factors to support their replication (4, 7, 58, 59).
A salient feature of our studies is that two major autophagy/mitophagy adaptor proteins are downregulated during the early stages of HSV-1 infection. This phenomenon was observed in multiple human cell lines, including immortalized human embryonic lung (HEL) cells, epithelial cells (HEp-2), neuroblastoma cells (SK-N-SH), microglia cells (HMC3), human osteosarcoma cells (U2OS), and others. A common characteristic of these cell lines is that HSV-1 does not display replication defects because it can counteract antiviral responses efficiently. Of these cell lines, at least the HEL cells have intact innate immunity pathways which the wild-type virus can block (55, 60). However, in the human monocytes (THP-1), in which the virus cannot counteract antiviral responses as efficiently, we noticed that p62/SQSTM1 and OPTN were not eliminated. Perhaps downregulation of the two autophagy adaptors depends on the ability of the virus to evade the host.
We also found that the down-modulation of p62/SQSTM1 and OPTN is an early event that does not require virus replication. Among host factors, we found that calcium mobilization occurring upon entry of the virus into the cells was important, since a calcium chelator added to the cultures 1 h postinfection prevented downregulation of these autophagy adaptors (61, 62). Virus entry was not affected by this treatment, since ICP0 expression was observed. Among the viral proteins, we found that ICP0 had a role, since the levels of both the p62/SQSTM1 and OPTN proteins remained unaltered upon infection with the ICP0-null virus. Interestingly, the ICP0 E3 ubiquitin ligase activity did not contribute to downregulation of p62/SQSTM1 and OPTN, although both proteins were stabilized after treatment with the MG132 proteasome inhibitor. These data imply that ICP0 is involved in proteasome-dependent down-modulation of host proteins through a mechanism that does not involve its RING finger domain. A virus in which ICP0 is retained in the nucleus (D199A mutant) only minimally affected the levels of p62/SQSTM1 and OPTN proteins (48, 63). This may be because cytoplasmic ICP0 is required for down-modulation of p62/SQSTM1 and OPTN. The functions of ICP0 outside the nucleus remain unknown, but a few reports have linked ICP0 to surface receptor endocytosis and protein sorting (64, 65). Here, we found that a form of ICP0 that localizes in the cytoplasm may downregulate p62 outside the context of the infection when calcium mobilization is triggered by ionomycin treatment. The role of calcium on ICP0 function remains uncharacterized. Calcium may trigger signaling pathways that augment ICP0 functions, or it may activate kinases that phosphorylate ICP0. The small decrease in the amounts of p62/SQSTM1 and OPTN proteins that we noticed occasionally during D199A mutant infection might have been due to some degree of autophagy, as indicated by the ratio of LC3-II to LC3-I. Rarely, we noticed down-modulation of both adaptors in ΔICP0 virus-infected cells at late hours postinfection. This may correlate with the role of p62/SQSTM1 and OPTN in the clearance of pathogens through autophagy, as the ΔICP0 virus cannot evade the host.
Another interesting observation was that in both wild-type virus and Δγ134.5 virus-infected cells, p62/SQSTM1 and OPTN were down-modulated. In Δγ134.5 virus-infected cells, autophagy is activated, which may result in down-modulation of both autophagy adaptors, whereas in HSV-1-infected cells, it is not. These data imply that the mechanisms of downregulation of p62/SQSTM1 and OPTN after infection with these two viruses may be different. In support of this, we found that the proteasome inhibitor MG132 prevented down-modulation of both proteins in wild-type-virus-infected cells but displayed a smaller effect in Δγ134.5 virus-infected cells. These two viruses also display differences in their accumulations of the ICP0 protein. Δγ134.5 virus-infected cells accumulated significantly smaller amounts of ICP0 than the wild-type virus at all time points tested. This is consistent with the results of other studies showing reduced accumulations of ICP0 in Δγ134.5 virus-infected cells (5). These data indicate that the Δγ134.5 virus has multiple defects that result in its failure to counteract host responses, and for this reason, the mechanism of down-modulation of p62/SQSTM1 and OPTN after infection with the Δγ134.5 virus may be more complex.
The role of autophagy during HSV-1 infection is to some degree controversial, as the growth of the Δγ134.5 virus was not rescued in autophagy-deficient cells (6, 7, 56, 59, 66). This might also in part be because ICP0 does not accumulate in Δγ134.5 virus-infected cells or because the γ134.5 proteins exert their functions in neuronal cells (5–7). However, induction of autophagy negatively impacted HSV-1 replication in vitro (6, 7, 56, 59, 66). Also, autophagy appears to be important for antigen presentation and activation of adaptive immune activation during HSV-1 infection (7, 56, 67). Consistently with these studies, we found that wild-type virus growth was not affected in p62/SQSTM1 or OPTN knockdown cells, perhaps because the virus has a mechanism to down-modulate both adaptors early after infection. Interestingly, however, we found that the ICP0-null virus displayed a growth defect in p62/SQSTM1 and OPTN KD cells, and part of this defect may be attributed to the fact that these cell lines mount stronger innate immune responses. These data suggest that these autophagy adaptors may negatively regulate innate immune responses. In support of this, previous studies have shown that in p62/SQSTM1 knockdown cells, the stimulator of interferon genes (STING) was not downregulated, resulting in elevated interferon production (68). Also, p62/SQSTM1 can prevent NOD-like receptor (NLR) oligomerization via direct binding (69). In a reciprocal approach, we expressed exogenous p62/SQSTM1 in human fibroblasts so that the virus could not eliminate the protein during the early stages of the infection, and that resulted in a dramatic decrease in viral gene transcription that caused a reduction in virus yields. We also noticed that cells expressing exogenous p62/SQSTM1 displayed significantly lower basal levels of ISGs, but the extent of induction of their transcription was higher after infection with mutant viruses unable to counteract innate immune responses or after exposure to 2′3′-cGAMP, a stimulus of STING. Treatment with trehalose, a known autophagy modulator, resulted in a decrease in HSV-1 yields, suggesting that autophagy can hinder HSV-1 infection.
The mechanism by which overexpressed p62/SQSTM1 restricts HSV-1 during the early steps of the infection is unclear, and that generates another question: “why does the virus down-modulate major autophagy adaptors if it already has other mechanisms to block autophagy?” The role of γ134.5 protein has been proposed for neuronal cells. Thus, it is possible that the virus has employed multiple mechanisms to counteract autophagy in different environments. Additionally, the virus may have employed multiple strategies to target the same pathway if it inhibits the infection. Also, the γ134.5 protein inhibits mainly Beclin-dependent autophagy, but mechanisms of the virus to inhibit Beclin-independent autophagy pathways remain unknown (6). It is also possible that by degrading p62/SQSTM1 and OPTN, the virus targets a selective type of autophagy. For example, both adaptor proteins have a role in mitophagy, a process of clearance of damaged mitochondria, but the role of mitophagy during HSV-1 infection remains unknown. Finally, via their downregulation, the virus may target the pleiotropic functions of p62/SQSTM1 and OPTN. For example, p62/SQSTM1 can inhibit the NF-κB pathway, and for this reason, its overexpression in tumors promotes their survival (70). However, NF-κB is important for HSV-1 replication (71). In addition, p62 enables transcription of antioxidant proteins and detoxifying enzymes through the Keap-Nrf2 pathway (32, 33, 72). However, these antioxidant factors may negatively impact HSV-1 infection. P62/SQSTM1 also has a multifaceted role in inflammasome regulation, as it is essential for NLRP3 (NLR family pyrin domain containing 3) activation, but on the other hand, it directly binds NLRs, inhibiting their self-oligomerization (69, 73–75). Finally, p62/SQSTM1 has been found in the nucleus, where it appears to inhibit DNA damage responses (76, 77). Thus, currently, it remains unclear whether downregulation of these autophagy adaptors during HSV-1 infection is due to their role in autophagy or because of other functions that they perform (31, 32). A model summarizing our current understanding of HSV-1 infection and the interplay with p62/SQSTM1 and OPTN is summarized in Fig. 9. Perhaps by down-modulating p62/SQSTM1, the virus controls anabolic and catabolic processes to ensure sufficient resources for its replication cycle, while inhibiting unwanted cell responses. On the other hand, OPTN has a role in post-Golgi vesicle trafficking, protein sorting, and regulating innate immune components, such as the TBK1 kinase (34).
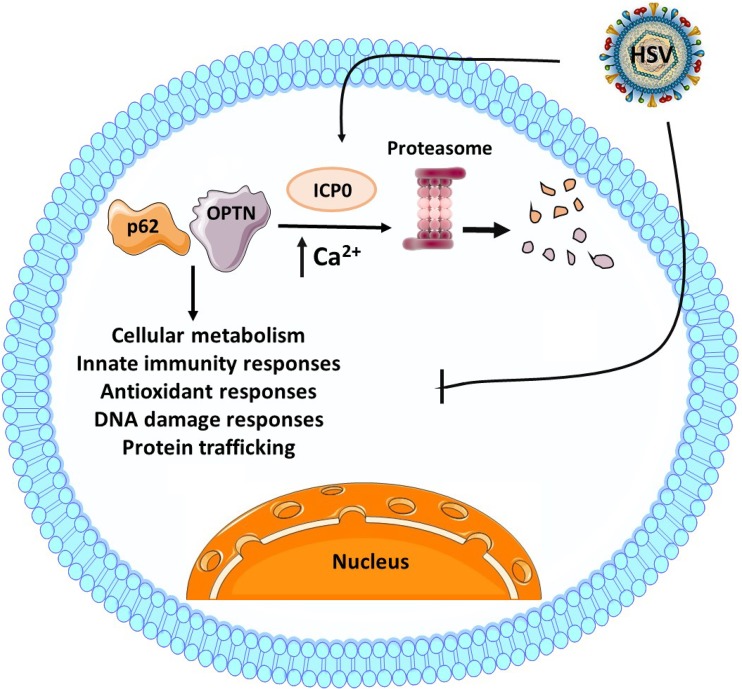
FIG 9.
Model of p62 and OPTN during HSV-1 infection. P62/SQSTM1 and OPTN proteins have been described to have various functions, including autophagy, modifying innate immunity signaling, inflammatory responses, antioxidant responses, and DNA damage responses. HSV-1 counteracts the host by evading several of these pathways. This is due, at least in part, to degradation of p62/SQSTM1 and OPTN through the proteasome, which has been described here to be dependent on ICP0 expression and Ca2+ mobilization.
In conclusion, our studies have identified two novel targets of HSV-1, the autophagy adaptor proteins p62/SQSTM1 and OPTN, whose downregulation may be a mechanism by which the virus counteracts antiviral functions. The type of antiviral functions activated by p62/SQSTM1 and OPTN during HSV-1 infection need to be investigated.
MATERIALS AND METHODS
Cell lines and virus.
The HEL cells (immortalized human embryonic lung fibroblasts transformed with human telomerase reverse transcriptase [hTERT]) were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and antibiotics. Vero, HEK-293, HEp-2, HMC3, THP-1, and SK-N-SH cells were obtained from the American Type Culture Collection (ATCC) and were cultured according to the manufacturer’s instructions. HSV-1(F) is a limited-passage isolate described before (78). The properties of the ΔICP0, R7914, ICP0 RING finger (RF), ΔICP27, d120 (ΔICP4), ΔICP8, R2621 (ΔVHS), and R3616 mutant viruses were described elsewhere (48, 79–85).
Immunoblot analysis.
The procedures for immunoblotting were described elsewhere (50, 86). Briefly, cells were solubilized in triple-detergent buffer (50 mM Tris-HCl [pH 8], 150 mM NaCl, 0.1% sodium dodecyl sulfate, 1% Nonidet P-40, 0.5% sodium deoxycholate, 100 μg of phenylmethylsulfonyl fluoride ml−1) supplemented with phosphatase inhibitors (10 mM NaF, 10 mM β-glycerophosphate, 0.1 mM sodium vanadate) and protease inhibitor cocktail (Sigma) and briefly sonicated. The protein concentration was determined with the aid of the Bio-Rad protein assay (Bio-Rad Laboratories). Ten to forty micrograms of total proteins per sample was subjected to further analysis. The mouse monoclonal antibodies to ICP0 (Santa Cruz), UL42 (Santa Cruz), ICP8 (Santa Cruz), and β-actin (Sigma) were used in a 1:1,000 dilution. The mouse monoclonal antibodies to p62/SQSTM1 (Cell Signaling) and OPTN (Santa Cruz) were used in a 1:1,000 dilution. The rabbit polyclonal antibody against LC3-B (Novus Biological) was used in a 1:2,000 dilution. Protein bands were visualized with 5-bromo-4-chloro-3-indolylphosphate (BCIP)-nitroblue tetrazolium (VWR) or with enhanced-chemiluminescence (ECL) Western blotting detection reagents (Amersham Biosciences) according to the manufacturer’s instructions. Trehalose (Acros Organics) was added to the cultures (300 mM) at the moment of infection.
Development of p62/SQSTM1 KD-, OPTN KD-, and p62/SQSTM1-expressing cell lines.
The procedures for developing the p62/SQSTM1 KD, OPTN KD, and p62/SQSTM1-expressing cell lines were described elsewhere (60, 87). The PLKO.1 plasmid expressing the p62 ORF was developed by inserting the EcoRI/NotI/Klenow fragment from a hemagglutinin (HA)-p62-expressing plasmid (HA-p62 was a gift from Qing Zhong, Addgene catalog number 28027) into the BamHI/SalI/Klenow site of the PLKO.1 green fluorescent protein (GFP) cytomegalovirus (CMV) Puro (Addgene number 658-5). PLKO.1 plasmids carrying shRNAs against p62 or OPTN transcripts were purchased through Sigma-Aldrich. For the development of the respective lentiviral vectors, HEK-293 cells seeded in a 25-cm2 flask at 60% confluence were cotransfected with the PLKO.1 plasmids described above, the Gag-Pol-expressing plasmid, and the vesicular stomatitis virus G (VSV-G)-expressing plasmid at a ratio of 7:7:1 (5 μg total amount of DNA) using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instructions. At 48 h after transfection, the supernatant from the cultures was collected, filtered through a 0.45-μm-pore-size filter, and used to infect HEL cells. Puromycin selection initiated at 24 h after exposure of cells to lentiviruses and continued until only resistant clones survived. The cells expressing exogenous p62 or with the greater knockdown of p62 and OPTN were used for further experimentation.
Plasmids and transfection assays.
The pBluescript plasmid containing the full-length ICP0 was previously described (88). Standard cloning procedures were utilized to delete amino acids 447 to 511 of ICP0 (ICP0 mut1) or amino acids 447 to 511 and 244 to 277 of ICP0 (ICP0 mut2). Transfections of HEK-293 cells, seeded in 6-well plates, were performed using Lipofectamine 3000 according to the manufacturer’s instructions. Ionomycin (5 μM; Sigma) was added to transfected cells for 16 h. The cells were harvested at 48 h posttransfection, and equal amounts of proteins were analyzed by immunoblot analysis.
Real-time PCR analysis.
Procedures for real-time PCR were as described before (87). Briefly, real-time PCR analyses were performed using SYBR Green reagent (Invitrogen) or TaqMan (Applied Biosystems) according to the manufacturer’s recommendations in a 7500 fast real-time PCR system (Applied Biosystems). Predesigned probes (6-carboxyfluorescein [FAM]/MGB) for the human p62, OPTN, and ISG15 transcripts were obtained through Thermo Fisher Scientific. The 18S rRNA primers (universal primers; Ambion) were used for normalization. Primer pairs for ISG56 and for the viral ICP27, thymidine kinase (TK), and glycoprotein I (gI) genes have been described before (55, 87).
Statistical analysis.
The Prism 7 software (GraphPad) was used for statistical analysis of the NanoSight and quantitative PCR (qPCR) data. P values were calculated using a standard unpaired Student t test. P values of ≤0.05 were considered significant. Asterisks are defined as follows: *, P ≤ 0.05; **, P ≤ 0.01; and ***, P ≤ 0.001. All statistical analyses were performed using biological replicates.
ACKNOWLEDGMENTS
We thank B. Roizman (University of Chicago) for kindly providing the mutant viruses R7910, R7914, R2621, and R3616 and the ICP0 RING finger mutant. The ΔICP8 virus was a gift from D. M. Knipe (Harvard University). The ΔICP27 virus was provided by R. M. Sandri-Goldin (University of California, Irvine). A plasmid for HA-p62 was a gift from Qing Zhong (Addgene catalog number 28027).
M. Kalamvoki is funded through KUMC startup funds and the CBID COBRE grant P20 GM113117.
REFERENCES
- 1.Roizman B, Knipe DM, Whitley RJ. 2013. Herpes simplex viruses, p 1823–1897 In Knipe DM, Howley PM (ed), Fields virology, 6th ed. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Chou J, Roizman B. 1992. The gamma 1(34.5) gene of herpes simplex virus 1 precludes neuroblastoma cells from triggering total shutoff of protein synthesis characteristic of programed cell death in neuronal cells. Proc Natl Acad Sci U S A 89:3266–3270. doi: 10.1073/pnas.89.8.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chou J, Chen JJ, Gross M, Roizman B. 1995. Association of a M(r) 90,000 phosphoprotein with protein kinase PKR in cells exhibiting enhanced phosphorylation of translation initiation factor eIF-2 alpha and premature shutoff of protein synthesis after infection with gamma 134.5- mutants of herpes simplex virus 1. Proc Natl Acad Sci U S A 92:10516–10520. doi: 10.1073/pnas.92.23.10516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lussignol M, Esclatine A. 2017. Herpesvirus and autophagy: “all right, everybody be cool, this is a robbery! Viruses 9:E372. doi: 10.3390/v9120372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manivanh R, Mehrbach J, Knipe DM, Leib DA. 2017. Role of herpes simplex virus 1 gamma34.5 in the regulation of IRF3 signaling. J Virol 91:e01156-17. doi: 10.1128/JVI.01156-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orvedahl A, Alexander D, Talloczy Z, Sun Q, Wei Y, Zhang W, Burns D, Leib DA, Levine B. 2007. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe 1:23–35. doi: 10.1016/j.chom.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 7.Orvedahl A, Levine B. 2008. Autophagy and viral neurovirulence. Cell Microbiol 10:1747–1756. doi: 10.1111/j.1462-5822.2008.01175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doherty J, Baehrecke EH. 2018. Life, death and autophagy. Nat Cell Biol 20:1110–1117. doi: 10.1038/s41556-018-0201-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gatica D, Lahiri V, Klionsky DJ. 2018. Cargo recognition and degradation by selective autophagy. Nat Cell Biol 20:233–242. doi: 10.1038/s41556-018-0037-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mizushima N. 2018. A brief history of autophagy from cell biology to physiology and disease. Nat Cell Biol 20:521–527. doi: 10.1038/s41556-018-0092-5. [DOI] [PubMed] [Google Scholar]
- 11.Yang Z, Klionsky DJ. 2010. Eaten alive: a history of macroautophagy. Nat Cell Biol 12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dikic I. 2017. Proteasomal and autophagic degradation systems. Annu Rev Biochem 86:193–224. doi: 10.1146/annurev-biochem-061516-044908. [DOI] [PubMed] [Google Scholar]
- 13.Stolz A, Ernst A, Dikic I. 2014. Cargo recognition and trafficking in selective autophagy. Nat Cell Biol 16:495–501. doi: 10.1038/ncb2979. [DOI] [PubMed] [Google Scholar]
- 14.Lamb CA, Yoshimori T, Tooze SA. 2013. The autophagosome: origins unknown, biogenesis complex. Nat Rev Mol Cell Biol 14:759–774. doi: 10.1038/nrm3696. [DOI] [PubMed] [Google Scholar]
- 15.Nakatogawa H, Mochida K. 2015. Reticulophagy and nucleophagy: new findings and unsolved issues. Autophagy 11:2377–2378. doi: 10.1080/15548627.2015.1106665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klionsky DJ, Schulman BA. 2014. Dynamic regulation of macroautophagy by distinctive ubiquitin-like proteins. Nat Struct Mol Biol 21:336–345. doi: 10.1038/nsmb.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Monastyrska I, Klionsky DJ. 2006. Autophagy in organelle homeostasis: peroxisome turnover. Mol Aspects Med 27:483–494. doi: 10.1016/j.mam.2006.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang K, Klionsky DJ. 2011. Mitochondria removal by autophagy. Autophagy 7:297–300. doi: 10.4161/auto.7.3.14502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bah A, Vergne I. 2017. Macrophage autophagy and bacterial infections. Front Immunol 8:1483. doi: 10.3389/fimmu.2017.01483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitchell G, Isberg RR. 2017. Innate immunity to intracellular pathogens: balancing microbial elimination and inflammation. Cell Host Microbe 22:166–175. doi: 10.1016/j.chom.2017.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahmad L, Mostowy S, Sancho-Shimizu V. 2018. Autophagy-virus interplay: from cell biology to human disease. Front Cell Dev Biol 6:155. doi: 10.3389/fcell.2018.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi Y, Bowman JW, Jung JU. 2018. Autophagy during viral infection—a double-edged sword. Nat Rev Microbiol 16:341–354. doi: 10.1038/s41579-018-0003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tian Y, Wang ML, Zhao J. 2019. Crosstalk between autophagy and type I interferon responses in innate antiviral immunity. Viruses 11:132. doi: 10.3390/v11020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie Z, Klionsky DJ. 2007. Autophagosome formation: core machinery and adaptations. Nat Cell Biol 9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 25.Funderburk SF, Wang QJ, Yue Z. 2010. The Beclin 1-VPS34 complex—at the crossroads of autophagy and beyond. Trends Cell Biol 20:355–362. doi: 10.1016/j.tcb.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang R, Zeh HJ, Lotze MT, Tang D. 2011. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ 18:571–580. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Munz C. 2011. Beclin-1 targeting for viral immune escape. Viruses 3:1166–1178. doi: 10.3390/v3071166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vazquez CL, Colombo MI. 2010. Beclin 1 modulates the anti-apoptotic activity of Bcl-2: insights from a pathogen infection system. Autophagy 6:177–178. doi: 10.4161/auto.6.1.10743. [DOI] [PubMed] [Google Scholar]
- 29.Katsuragi Y, Ichimura Y, Komatsu M. 2015. p62/SQSTM1 functions as a signaling hub and an autophagy adaptor. FEBS J 282:4672–4678. doi: 10.1111/febs.13540. [DOI] [PubMed] [Google Scholar]
- 30.Lamark T, Svenning S, Johansen T. 2017. Regulation of selective autophagy: the p62/SQSTM1 paradigm. Essays Biochem 61:609–624. doi: 10.1042/EBC20170035. [DOI] [PubMed] [Google Scholar]
- 31.Sanchez-Martin P, Komatsu M. 2018. p62/SQSTM1—steering the cell through health and disease. J Cell Sci 131:jcs222836. doi: 10.1242/jcs.222836. [DOI] [PubMed] [Google Scholar]
- 32.Sanchez-Martin P, Saito T, Komatsu M. 2019. p62/SQSTM1: 'Jack of all trades' in health and cancer. FEBS J 286:8–23. doi: 10.1111/febs.14712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taniguchi K, Yamachika S, He F, Karin M. 2016. p62/SQSTM1—Dr. Jekyll and Mr. Hyde that prevents oxidative stress but promotes liver cancer. FEBS Lett 590:2375–2397. doi: 10.1002/1873-3468.12301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ryan TA, Tumbarello DA. 2018. Optineurin: a coordinator of membrane-associated cargo trafficking and autophagy. Front Immunol 9:1024. doi: 10.3389/fimmu.2018.01024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bansal M, Swarup G, Balasubramanian D. 2015. Functional analysis of optineurin and some of its disease-associated mutants. IUBMB Life 67:120–128. doi: 10.1002/iub.1355. [DOI] [PubMed] [Google Scholar]
- 36.Boyle KB, Randow F. 2013. The role of 'eat-me' signals and autophagy cargo receptors in innate immunity. Curr Opin Microbiol 16:339–348. doi: 10.1016/j.mib.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 37.Outlioua A, Pourcelot M, Arnoult D. 2018. The role of optineurin in antiviral type I interferon production. Front Immunol 9:853. doi: 10.3389/fimmu.2018.00853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Slowicka K, Vereecke L, van LG. 2016. Cellular functions of optineurin in health and disease. Trends Immunol 37:621–633. doi: 10.1016/j.it.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 39.Toth RP, Atkin JD. 2018. Dysfunction of optineurin in amyotrophic lateral sclerosis and glaucoma. Front Immunol 9:1017. doi: 10.3389/fimmu.2018.01017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ying H, Yue BY. 2012. Cellular and molecular biology of optineurin. Int Rev Cell Mol Biol 294:223–258. doi: 10.1016/B978-0-12-394305-7.00005-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cassady KA, Gross M, Roizman B. 1998. The herpes simplex virus US11 protein effectively compensates for the gamma1(34.5) gene if present before activation of protein kinase R by precluding its phosphorylation and that of the alpha subunit of eukaryotic translation initiation factor 2. J Virol 72:8620–8626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cassady KA, Gross M, Roizman B. 1998. The second-site mutation in the herpes simplex virus recombinants lacking the gamma134.5 genes precludes shutoff of protein synthesis by blocking the phosphorylation of eIF-2alpha. J Virol 72:7005–7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lussignol M, Queval C, Bernet-Camard MF, Cotte-Laffitte J, Beau I, Codogno P, Esclatine A. 2013. The herpes simplex virus 1 Us11 protein inhibits autophagy through its interaction with the protein kinase PKR. J Virol 87:859–871. doi: 10.1128/JVI.01158-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mulvey M, Camarena V, Mohr I. 2004. Full resistance of herpes simplex virus type 1-infected primary human cells to alpha interferon requires both the Us11 and gamma(1)34.5 gene products. J Virol 78:10193–10196. doi: 10.1128/JVI.78.18.10193-10196.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mulvey M, Arias C, Mohr I. 2007. Maintenance of endoplasmic reticulum (ER) homeostasis in herpes simplex virus type 1-infected cells through the association of a viral glycoprotein with PERK, a cellular ER stress sensor. J Virol 81:3377–3390. doi: 10.1128/JVI.02191-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gu H. 2016. Infected cell protein 0 functional domains and their coordination in herpes simplex virus replication. World J Virol 5:1–13. doi: 10.5501/wjv.v5.i1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deschamps T, Waisner H, Dogrammatzis C, Roy A, Chacko S, Perera C, Prisinzano TE, Kalamvoki M. 2019. Discovery of small molecule inhibitors targeting the E3 ubiquitin ligase activity of the HSV-1 ICP0 protein using an in vitro high throughput screening assay. J Virol 93:e00619-19. doi: 10.1128/JVI.00619-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van SC, Kawaguchi Y, Roizman B. 1999. A single amino acid substitution in the cyclin D binding domain of the infected cell protein no. 0 abrogates the neuroinvasiveness of herpes simplex virus without affecting its ability to replicate. Proc Natl Acad Sci U S A 96:8184–8189. doi: 10.1073/pnas.96.14.8184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kalamvoki M, Roizman B. 2009. ICP0 enables and monitors the function of D cyclins in herpes simplex virus 1 infected cells. Proc Natl Acad Sci U S A 106:14576–14580. doi: 10.1073/pnas.0906905106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kalamvoki M, Roizman B. 2010. Interwoven roles of cyclin D3 and cdk4 recruited by ICP0 and ICP4 in the expression of herpes simplex virus genes. J Virol 84:9709–9717. doi: 10.1128/JVI.01050-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kalamvoki M, Roizman B. 2010. Role of herpes simplex virus ICP0 in the transactivation of genes introduced by infection or transfection: a reappraisal. J Virol 84:4222–4228. doi: 10.1128/JVI.02585-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hagglund R, Roizman B. 2004. Role of ICP0 in the strategy of conquest of the host cell by herpes simplex virus 1. J Virol 78:2169–2178. doi: 10.1128/jvi.78.5.2169-2178.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kalamvoki M, Roizman B. 2010. Circadian CLOCK histone acetyl transferase localizes at ND10 nuclear bodies and enables herpes simplex virus gene expression. Proc Natl Acad Sci U S A 107:17721–17726. doi: 10.1073/pnas.1012991107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Deschamps T, Kalamvoki M. 2017. Impaired STING pathway in human osteosarcoma U2OS cells contributes to the growth of ICP0-null mutant herpes simplex virus. J Virol 91:e00006-17. doi: 10.1128/JVI.00006-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Deschamps T, Kalamvoki M. 2017. Evasion of the STING DNA-sensing pathway by VP11/12 of herpes simplex virus 1. J Virol 91:e00535-17. doi: 10.1128/JVI.00535-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alexander DE, Leib DA. 2008. Xenophagy in herpes simplex virus replication and pathogenesis. Autophagy 4:101–103. doi: 10.4161/auto.5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hewitt G, Korolchuk VI. 2017. Repair, reuse, recycle: the expanding role of autophagy in genome maintenance. Trends Cell Biol 27:340–351. doi: 10.1016/j.tcb.2016.11.011. [DOI] [PubMed] [Google Scholar]
- 58.Cavignac Y, Esclatine A. 2010. Herpesviruses and autophagy: catch me if you can! Viruses 2:314–333. doi: 10.3390/v2010314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Orvedahl A, Levine B. 2009. Autophagy in mammalian antiviral immunity. Curr Top Microbiol Immunol 335:267–285. doi: 10.1007/978-3-642-00302-8_13. [DOI] [PubMed] [Google Scholar]
- 60.Kalamvoki M, Roizman B. 2014. HSV-1 degrades, stabilizes, requires, or is stung by STING depending on ICP0, the US3 protein kinase, and cell derivation. Proc Natl Acad Sci U S A 111:E611–E617. doi: 10.1073/pnas.1323414111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cheshenko N, Del RB, Woda C, Marcellino D, Satlin LM, Herold BC. 2003. Herpes simplex virus triggers activation of calcium-signaling pathways. J Cell Biol 163:283–293. doi: 10.1083/jcb.200301084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kalamvoki M, Roizman B. 2007. Bcl-2 blocks accretion or depletion of stored calcium but has no effect on the redistribution of IP3 receptor I mediated by glycoprotein E of herpes simplex virus 1. J Virol 81:6316–6325. doi: 10.1128/JVI.00311-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lopez P, Van SC, Roizman B. 2001. Requirements for the nuclear-cytoplasmic translocation of infected-cell protein 0 of herpes simplex virus 1. J Virol 75:3832–3840. doi: 10.1128/JVI.75.8.3832-3840.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Deschamps T, Dogrammatzis C, Mullick R, Kalamvoki M. 2017. Cbl E3 ligase mediates the removal of Nectin-1 from the surface of herpes simplex virus 1-infected cells. J Virol 91:e00393-17. doi: 10.1128/JVI.00393-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liang Y, Kurakin A, Roizman B. 2005. Herpes simplex virus 1 infected cell protein 0 forms a complex with CIN85 and Cbl and mediates the degradation of EGF receptor from cell surfaces. Proc Natl Acad Sci U S A 102:5838–5843. doi: 10.1073/pnas.0501253102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alexander DE, Ward SL, Mizushima N, Levine B, Leib DA. 2007. Analysis of the role of autophagy in replication of herpes simplex virus in cell culture. J Virol 81:12128–12134. doi: 10.1128/JVI.01356-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.English L, Chemali M, Duron J, Rondeau C, Laplante A, Gingras D, Alexander D, Leib D, Norbury C, Lippe R, Desjardins M. 2009. Autophagy enhances the presentation of endogenous viral antigens on MHC class I molecules during HSV-1 infection. Nat Immunol 10:480–487. doi: 10.1038/ni.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Prabakaran T, Bodda C, Krapp C, Zhang BC, Christensen MH, Sun C, Reinert L, Cai Y, Jensen SB, Skouboe MK, Nyengaard JR, Thompson CB, Lebbink RJ, Sen GC, van LG, Nielsen R, Komatsu M, Nejsum LN, Jakobsen MR, Gyrd-Hansen M, Paludan SR. 2018. Attenuation of cGAS-STING signaling is mediated by a p62/SQSTM1-dependent autophagy pathway activated by TBK1. EMBO J 37:e97858. doi: 10.15252/embj.201797858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Alegre F, Moragrega AB, Polo M, Marti-Rodrigo A, Esplugues JV, Blas-Garcia A, Apostolova N. 2018. Role of p62/SQSTM1 beyond autophagy: a lesson learned from drug-induced toxicity in vitro. Br J Pharmacol 175:440–455. doi: 10.1111/bph.14093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lu Y, Wang Q, Zhou Y, Sun L, Hu B, Xue H, Li M, Zhang K, Ren C, Duan N, Liu H, Zhang C, Li Z, Ma T. 2018. Overexpression of p62 is associated with poor prognosis and aggressive phenotypes in osteosarcoma. Oncol Lett 15:9889–9895. doi: 10.3892/ol.2018.8579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Taddeo B, Zhang W, Lakeman F, Roizman B. 2004. Cells lacking NF-kappaB or in which NF-kappaB is not activated vary with respect to ability to sustain herpes simplex virus 1 replication and are not susceptible to apoptosis induced by a replication-incompetent mutant virus. J Virol 78:11615–11621. doi: 10.1128/JVI.78.21.11615-11621.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shah SZA, Zhao D, Hussain T, Sabir N, Mangi MH, Yang L. 2018. p62-Keap1-NRF2-ARE pathway: a contentious player for selective targeting of autophagy, oxidative stress and mitochondrial dysfunction in prion diseases. Front Mol Neurosci 11:310. doi: 10.3389/fnmol.2018.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Deretic V. 2012. Autophagy as an innate immunity paradigm: expanding the scope and repertoire of pattern recognition receptors. Curr Opin Immunol 24:21–31. doi: 10.1016/j.coi.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Harijith A, Ebenezer DL, Natarajan V. 2014. Reactive oxygen species at the crossroads of inflammasome and inflammation. Front Physiol 5:352. doi: 10.3389/fphys.2014.00352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhong Z, Umemura A, Sanchez-Lopez E, Liang S, Shalapour S, Wong J, He F, Boassa D, Perkins G, Ali SR, McGeough MD, Ellisman MH, Seki E, Gustafsson AB, Hoffman HM, Diaz-Meco MT, Moscat J, Karin M. 2016. NF-kappaB restricts inflammasome activation via elimination of damaged mitochondria. Cell 164:896–910. doi: 10.1016/j.cell.2015.12.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Feng Y, Klionsky DJ. 2017. Autophagy regulates DNA repair through SQSTM1/p62. Autophagy 13:995–996. doi: 10.1080/15548627.2017.1317427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang Y, Zhang N, Zhang L, Li R, Fu W, Ma K, Li X, Wang L, Wang J, Zhang H, Gu W, Zhu WG, Zhao Y. 2016. Autophagy regulates chromatin ubiquitination in DNA damage response through elimination of SQSTM1/p62. Mol Cell 63:34–48. doi: 10.1016/j.molcel.2016.05.027. [DOI] [PubMed] [Google Scholar]
- 78.Ejercito PM, Kieff ED, Roizman B. 1968. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J Gen Virol 2:357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 79.Chou J, Kern ER, Whitley RJ, Roizman B. 1990. Mapping of herpes simplex virus-1 neurovirulence to gamma 134.5, a gene nonessential for growth in culture. Science 250:1262–1266. doi: 10.1126/science.2173860. [DOI] [PubMed] [Google Scholar]
- 80.Da Costa XJ, Jones CA, Knipe DM. 1999. Immunization against genital herpes with a vaccine virus that has defects in productive and latent infection. Proc Natl Acad Sci U S A 96:6994–6998. doi: 10.1073/pnas.96.12.6994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kawaguchi Y, Van SC, Roizman B. 1997. Herpes simplex virus 1 alpha regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J Virol 71:7328–7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Poon AP, Roizman B. 1997. Differentiation of the shutoff of protein synthesis by virion host shutoff and mutant gamma (1)34.5 genes of herpes simplex virus 1. Virology 229:98–105. doi: 10.1006/viro.1996.8425. [DOI] [PubMed] [Google Scholar]
- 83.Smith IL, Hardwicke MA, Sandri-Goldin RM. 1992. Evidence that the herpes simplex virus immediate early protein ICP27 acts post-transcriptionally during infection to regulate gene expression. Virology 186:74–86. doi: 10.1016/0042-6822(92)90062-T. [DOI] [PubMed] [Google Scholar]
- 84.Lium EK, Silverstein S. 1997. Mutational analysis of the herpes simplex virus type 1 ICP0 C3HC4 zinc ring finger reveals a requirement for ICP0 in the expression of the essential alpha27 gene. J Virol 71:8602–8614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.DeLuca NA, McCarthy AM, Schaffer PA. 1985. Isolation and characterization of deletion mutants of herpes simplex virus type 1 in the gene encoding immediate-early regulatory protein ICP4. J Virol 56:558–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dogrammatzis C, Deschamps T, Kalamvoki M. 2019. Biogenesis of extracellular vesicles during herpes simplex virus 1 infection: role of the CD63 tetraspanin. J Virol 93:e01850-18. doi: 10.1128/JVI.01850-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Deschamps T, Kalamvoki M. 2018. Extracellular vesicles released by herpes simplex virus 1-infected cells block virus replication in recipient cells in a STING-dependent manner. J Virol 92:e01102-18. doi: 10.1128/JVI.01102-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gu H, Roizman B. 2007. Herpes simplex virus-infected cell protein 0 blocks the silencing of viral DNA by dissociating histone deacetylases from the CoREST-REST complex. Proc Natl Acad Sci U S A 104:17134-9. doi: 10.1073/pnas.0707266104. [DOI] [PMC free article] [PubMed] [Google Scholar]