This study provides the first evidence that CRISPR/Cas9 technology can be applied to knock out the ORF57 gene from all ∼100 copies of the KSHV genome in primary effusion lymphoma (PEL) cells by coexpressing two guide RNAs (gRNAs) and Cas9 from a single expression vector in combination with single-cell cloning. The gene knockout efficiency in this system was evaluated rapidly using a direct cell PCR screening. The current CRISPR/Cas9 technology also mediated ORF57 inversion in situ in the targeted site of the KSHV genome. The successful rescue of viral lytic gene expression and infectious virion production from the ORF57 knockout (KO) genome further reiterates the essential role of ORF57 in KSHV infection and multiplication. This modified technology should be useful for knocking out any viral genes from a genome to dissect functions of individual viral genes in the context of the virus genome and to understand their contributions to viral genetics and the virus life cycle.
KEYWORDS: BCBL-1, CRISPR, Cas9, gene inversion, Kaposi's sarcoma-associated herpesvirus, knockout, ORF57, PEL cells
ABSTRACT
Kaposi’s sarcoma-associated herpesvirus (KSHV)-transformed primary effusion lymphoma cell lines contain ∼70 to 150 copies of episomal KSHV genomes per cell and have been widely used for studying the mechanisms of KSHV latency and lytic reactivation. Here, we report the first complete knockout (KO) of viral ORF57 gene from all ∼100 copies of KSHV genome per cell in BCBL-1 cells. This was achieved by a modified CRISPR/Cas9 technology to simultaneously express two guide RNAs (gRNAs) and Cas9 from a single expression vector in transfected cells in combination with multiple rounds of cell selection and single-cell cloning. CRISPR/Cas9-mediated genome engineering induces the targeted gene deletion and inversion in situ. We found the inverted ORF57 gene in the targeted site in the KSHV genome in one of two characterized single cell clones. Knockout of ORF57 from the KSHV genome led to viral genome instability, thereby reducing viral genome copies and expression of viral lytic genes in BCBL-1-derived single-cell clones. The modified CRISPR/Cas9 technology was very efficient in knocking out the ORF57 gene in iSLK/Bac16 and HEK293/Bac36 cells, where each cell contains only a few copies of the KSHV genome. The ORF57 KO genome was stable in iSLK/Bac16 cells, and, upon lytic induction, was partially rescued by ectopic ORF57 to express viral lytic gene ORF59 and produce infectious virions. Together, the technology developed in this study has paved the way to express two separate gRNAs and the Cas9 enzyme simultaneously in the same cell and could be efficiently applied to any genetic alterations from various genomes, including those in extreme high copy numbers.
IMPORTANCE This study provides the first evidence that CRISPR/Cas9 technology can be applied to knock out the ORF57 gene from all ∼100 copies of the KSHV genome in primary effusion lymphoma (PEL) cells by coexpressing two guide RNAs (gRNAs) and Cas9 from a single expression vector in combination with single-cell cloning. The gene knockout efficiency in this system was evaluated rapidly using a direct cell PCR screening. The current CRISPR/Cas9 technology also mediated ORF57 inversion in situ in the targeted site of the KSHV genome. The successful rescue of viral lytic gene expression and infectious virion production from the ORF57 knockout (KO) genome further reiterates the essential role of ORF57 in KSHV infection and multiplication. This modified technology should be useful for knocking out any viral genes from a genome to dissect functions of individual viral genes in the context of the virus genome and to understand their contributions to viral genetics and the virus life cycle.
INTRODUCTION
Kaposi’s sarcoma-associated herpesvirus (KSHV), or human herpesvirus 8 (HHV-8), is a human gammaherpesvirus closely related to Epstein-Barr virus (EBV) (1). Like other herpesviruses, KSHV establishes a life-long infection in its host, but its infection remains mostly asymptomatic in healthy individuals. However, suppression of host immunity often leads to a rise of several malignancies, including Kaposi’s sarcoma, primary effusion lymphoma (PEL), and multicentric Castleman’s disease (MCD) (2–4). The KSHV double-stranded DNA genome of ∼170 kb encodes at least ∼90 open reading frames (ORFs), several long noncoding RNAs (lncRNAs), and ∼25 mature microRNAs (miRNAs), many of which play roles in virus-induced oncogenesis (1, 5). Yet, the exact roles of many viral products in KSHV pathogenesis remain elusive due to the lack of a suitable model system.
Historically, the functions of viral genes were deduced from their homology to known viral or host counterparts or based on the presence of a previously defined functional moiety. Reporter assays were then used to confirm their predicted functional activity, frequently in the absence of viral infection. Successful cloning of the KSHV genome into a bacterial artificial chromosome (Bac) for the first time allowed rapid intended engineering of the KSHV genome, thus permitting functional analysis of numerous viral products in the context of viral infection (6, 7). Despite this advantage, there are several shortcomings of using Bac technology in KSHV research. First, it requires repeat transfer between Escherichia coli and mammalian cells after each round of mutagenesis. This laborious process often leads to undesired heterogeneity. The second disadvantage is the use of nonrelevant cells for virus propagation, including HEK293, iSLK, or Vero cells. Even though they are permissive for KSHV infection, these transformed cell lines are not suitable for studying KSHV oncogenicity or for spontaneous establishment of KSHV latency, because the viral genome in these cells is retained by selection to an antibiotic resistance gene within the inserted Bac cassette. Transfection of the KSHV Bac genome into more appropriate primary cells leads to only a short burst of lytic infection without cell transformation. As a result, the genetic studies using the KSHV Bac system are primarily limited to functional analyses of viral genes during KSHV lytic replication.
Patient-derived PEL cells represent the only naturally infected and transformed cells capable of supporting both latent and lytic infections, making them a popular KSHV research model (8). Despite their unique phenotype and wide use, PEL cells are largely omitted from KSHV genetic studies due to a high copy number of the viral genome, ranging from ∼70 copies per cell in BCBL-1 (9, 10) to ∼150 copies per cell in BCP-1 cells (ATCC CRL-2294), which has made viral genome mutagenesis extremely challenging.
The CRISPR (clustered regularly interspaced short palindromic repeats)/Cas (CRISPR-associated) system, derived from the bacterial adaptive immune system against foreign DNA, revolutionized all fields of biology (11). The widely used type II CRISPR/Cas9 of Streptococcus pyogenes consists of two major components, a single or simple guide RNA (gRNA) and a helicase/endonuclease Cas9, that together form a ribonucleoprotein complex capable of binding and cleaving target DNA at a specific location complementary to the short gRNA sequence. This finding led to the generation of a simple gene editing system adaptable to virtually any biological system. In most experiments, a single gRNA is used to create a double-strand break which can be repaired by the error prone nonhomologous end joining mechanism (NHEJ) which utilizes a random insertion or deletion of several nucleotides at the cleavage site. However, the alternative use of two or more gRNAs targeting different genomic positions could lead to deletion or replacement of DNA fragments between cleavage sites by homologous recombination (12). Numerous studies showed successful use of CRISPR/Cas9 in genetic studies of herpesviruses. However, as of today, there is no report on genetic engineering of KSHV genome in PEL cells by CRISPR/Cas9 (13, 14).
KSHV ORF57 plays profound posttranscriptional roles by promoting RNA stability, RNA splicing, and translation (15–26) and is essential for KSHV lytic replication and production of infectious virions (27–29). Although not fully understood, the observed ORF57 activities have been associated with different host cofactors for each of the known functions (21). ORF57 binds to a PAN MRE motif and stabilizes PAN RNA by interacting with PABPC-1 (17, 18, 30) and prevents hyperpolyadenylation of nuclear ORF59 RNA by interacting with RBM15 (31). ORF57 functions as a viral splicing factor in the splicing of intron-containing viral pre-mRNAs by binding to host splicing factors (32, 33). ORF57 promotes interleukin 6 (IL-6) translation by preventing IL-6 from undergoing RISC-mediated inhibition (34, 35). Recent studies also uncovered ORF57 inhibition of RNA granule formation by interacting with PKR, PACT, Ago2, and GW182 to modulate host innate responses against viral infection (36, 37). In this study, we report a successful application of CRISPR/Cas9 by using a pair of gRNAs simultaneously expressed from a modified Cas9 expression vector to completely knock out ORF57 from the KSHV genome in HEK293/Bac36, iSLK/Bac16, and BCBL-1 cells. Subsequently, by limited dilution and single cell selection, we successfully isolated several single-cell clones with ORF57 knockout (KO) from all 100 copies of the KSHV genome in BCBL-1 cells. Our study demonstrates a potential application of the novel CRISPR/Cas9 technology developed in our lab to study any pathogens by the manipulation of multiple genome copies residing in the infected cells.
RESULTS
Creation of an ORF57 KO genome from KSHV Bac36 cells using CRISPR/Cas9.
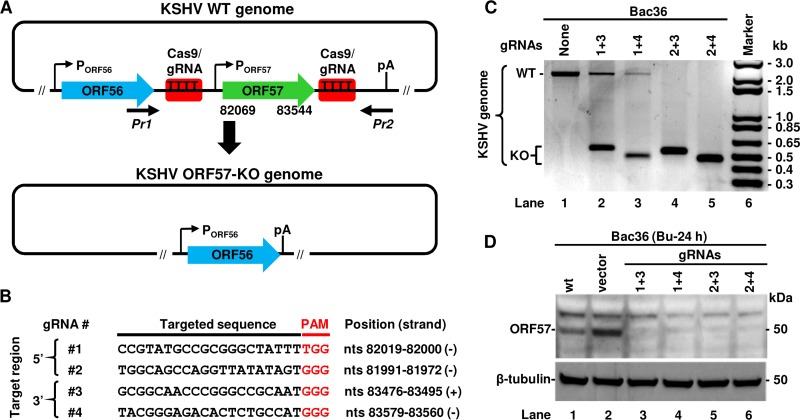
Within the KSHV genome, the ORF57 gene bears an inducible promoter responding to replication and transcription activator (RTA, or ORF50) in the early stage of KSHV lytic infection (25, 38). The ORF57 gene transcribes a monocistronic RNA from its transcriptional start site (TSS) at nucleotide (nt) 82003 (22) and uses a polyadenylation (pA) cleavage site at nucleotide 83636 for its RNA polyadenylation. The same pA site is also used for polyadenylating the bicistronic ORF56 gene transcript expressed from an ORF56 promoter (Fig. 1A) (15, 39). The primary RNA transcript of ORF57 contains two introns and can be alternatively spliced to produce an isoform RNA1 (only intron 1 spliced out) to encode a full-length 455-amino acid (aa) ORF57 protein or a noncoding isoform RNA2 (both intron 1 and intron 2 spliced out) (40).
FIG 1.
Generation of KSHV ORF57 knockout (KO) in Bac36 cells using a common CRISPR/Cas9 technology. (A) Diagram of wild-type (WT) KSHV ORF56-ORF57 locus including the promoters (arrows with P) and a common polyadenylation site (pA) used for both ORF56 and ORF57 expression. Red boxes represent the Cas9/gRNA-targeted sites in both 5′ and 3′ regions of the ORF57 gene (nt 82069 to 83544) to induce ORF57 deletion. The resulted ORF57 KO genome retains the intact ORF56 ORF and the pA site for ORF56 expression. (B) The targeted sequences of gRNAs used for ORF57 KO within the KSHV genome along with the PAM sequences shown in red and their nucleotide positions in the reference KSHV genome (GenBank accession number U75698). (C and D) The CRISPR/Cas9-mediated KO efficiency of ORF57 gene from the KSHV genome in Bac36 cells transfected with the indicated plasmids expressing various combinations of single ORF57 gRNA. The efficiency of ORF57 KO was determined by PCR (C) on total cell DNA using Pr1 plus Pr2 primers flanking the deleted region of ORF57 gene (A) or by anti-ORF57 antibody Western blotting on total cell lysates 24 h after sodium butyrate (Bu) induction of ORF57 protein expression (D). Cellular β-tubulin served as a loading control.
To create ORF57 KO without affecting ORF56 RNA polyadenylation and expression from the KSHV genome, we designed a series of paired gRNAs targeting the flanking regions of the ORF57 gene (Fig. 1A) using the online design tool developed by Zhang’s lab (41) and selected two gRNAs for each targeted region based on the predicted scores and cleavage site positions (Fig. 1B). The 5′-specific gRNAs 1 and 2 target the sites upstream of the ORF57 TSS, and the 3′-specific gRNAs 3 and 4 target the sites upstream of the ORF57 pA site (Fig. 1B). The exact target sequences of selected gRNAs together with their downstream protospacer adjacent motifs (PAMs) are shown in Fig. 1B. The use of these gRNAs would remove the ORF57 coding region without disrupting the ORF56 coding region and the downstream pA site and allow the expression of ORF56 as an essential viral primase from the ORF57 KO genome. Each selected gRNA was then cloned into a CRISPR/Cas9 expression vector to express a single gRNA. This gRNA-Cas9/Puro vector also expresses a Cas9/Puro fusion protein which is posttranslationally cleaved into a functional Cas9 to cut the gRNA-targeted genomic site and a puromycin N-acetyl-transferase to select the transfected cells.
To test the efficiency of ORF57 KO by the selected gRNAs, two paired vectors, one expressing a 5′-specific gRNA 1 or 2 and the other expressing a 3′-specific gRNA 3 or 4, were cotransfected initially into HEK293 cells carrying a Bac36 KSHV genome (6). HEK293-derived Bac36 cells were chosen because of their high efficiency of transfection and low viral genome copy number (estimated 1 to 2 copies/cell) (6). The cells were then selected on puromycin to enrich gRNA/Cas9-expressing cells. The efficiency of ORF57 KO from the Bac36 genome was evaluated by PCR using a primer set of Pr1 plus Pr2 flanking the targeted deletion region (Fig. 1A, also see Table S1 in the supplemental material for primer sequences). Bac36 cells without transfection of gRNA vectors, used as a negative control, showed the presence of an ∼2-kb PCR product corresponding to the wild-type (WT) KSHV genome (Fig. 1C). In contrast, the cells transfected with any combination of gRNAs showed predominantly an ∼0.5-kb PCR product corresponding to the KO genome lacking ORF57. The observed variation in the size of PCR products amplified from ORF57 KO genomes was caused by different cleavage positions of individual gRNAs (see Fig. 1B). While all combinations of gRNAs led to ORF57 KO from the KSHV genome, the ORF57 KO efficiency varied from the paired gRNAs, with gRNAs 2 and 3 or gRNAs 2 and 4 being better than gRNAs 1 and 3 or gRNAs 1 and 4 as shown by reduced detection of the ∼2-kb PCR products (Fig. 1C). To confirm the loss of ORF57 protein expression from the ORF57 KO Bac36 genome, Bac36 cells were treated with 3 mM sodium butyrate for 24 h to induce KSHV lytic replication followed by Western blotting. As shown in Fig. 1D, only the untransfected cells and the cells transfected with an empty vector exhibited detectable ORF57 protein. These data indicate that viral ORF57 gene can be easily deleted from a KSHV genome in Bac36 cells by simultaneous expression of two virus-specific gRNAs expressed from two separate vectors.
Construction of the second generation of a dual gRNA expression vector.
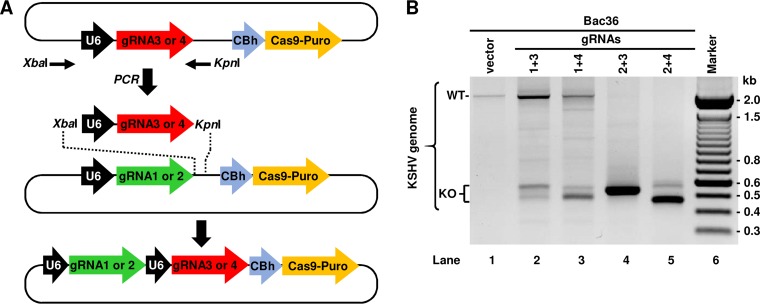
The original vectors were designed for the expression of a single gRNA often used to induce insertion/deletion mutations in typical KO experiments. However, simultaneous expression of two or more gRNAs in a cell requires successful cotransfection of two or more gRNA expression vectors into that single cell. These approaches inevitably pose problems of unequal expression due to variations in the transfection efficiency of individual vectors and cannot ensure that both 5′-specific gRNA and 3′-specific gRNA are being simultaneously delivered into the same cell. In addition, the expression of individual gRNAs could be unevenly lost during selection, as each plasmid carries its own puromycin resistance gene. Therefore, we designed a series of second-generation vectors enabling simultaneous expression of two gRNAs from a single vector. To do so, we swapped the gRNA 3 or gRNA 4 together with an upstream U6 promoter into the gRNA 1- or gRNA 2-expressing vector at XbaI and KpnI sites by PCR amplification and restriction enzyme digestion as indicated in Fig. 2A. The resulting vectors each contain two tandem gRNAs (gRNA 1 or 2 plus gRNA 3 or 4) separately under U6 promoter control and a single Cas9/Puro locus (Fig. 2A). Thus, this novel dual gRNA expression vector, when delivered to a host cell, expresses simultaneously a pair of the 5′-specific gRNA and the 3′-specific gRNA, Cas9, and a puromycin N-acetyl-transferase. To test their functionality, the second-generation plasmids were also transfected into Bac36 cells, and the efficiency of ORF57 KO was monitored by PCR. As shown in Fig. 2B, the cells transfected with the dual gRNA expression vectors exhibited ORF57 KO more efficiently for the vector expressing dual gRNAs 2 and 3 or 2 and 4 than the vector expressing gRNAs 1 and 3 or 1 and 4. Since the PAM following the gRNA 1 targeting sequence is a TGG instead of a GGG motif and was less efficient in ORF57 KO experiments in both generations of gRNA-expressing vectors, these gRNA 1-expressing vectors were excluded from the subsequent studies.
FIG 2.
Construction of the second-generation CRISPR/Cas9 vectors for expression of dual gRNAs. (A) Diagram showing construction of a CRISPR/Cas9 vector capable of simultaneous expression of two different gRNAs. First, the locus consisting of upstream U6 promoter (black arrow) and gRNA (red or green arrows) were amplified by PCR using flanking primers (black thin arrows). The resulting PCR product was inserted into a vector containing gRNA 1 or gRNA 2 using XbaI and KpnI sites introduced by PCR amplification. The final vector structure contains the two separate gRNAs each under the control of a U6 promoter followed by Cas9-Puro locus under the control of CBh (chicken β-actin hybrid) promoter. (B) Validation of functionality and potency of the second-generation gRNA vectors to facilitate ORF57 KO in Bac36 cells as determined by PCR using a Pr1 plus Pr2 primer set described in Fig. 1A. WT, wild type; KO, knockout.
Generation of ORF57 KO from all copies of the KSHV genome in BCBL-1 cells.
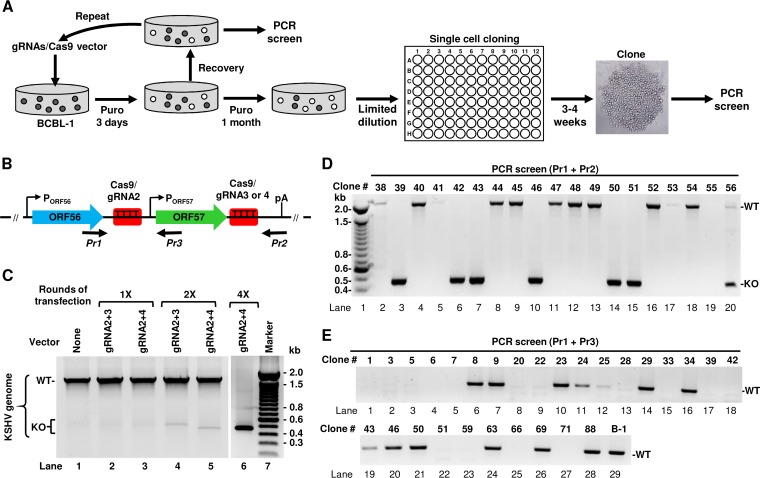
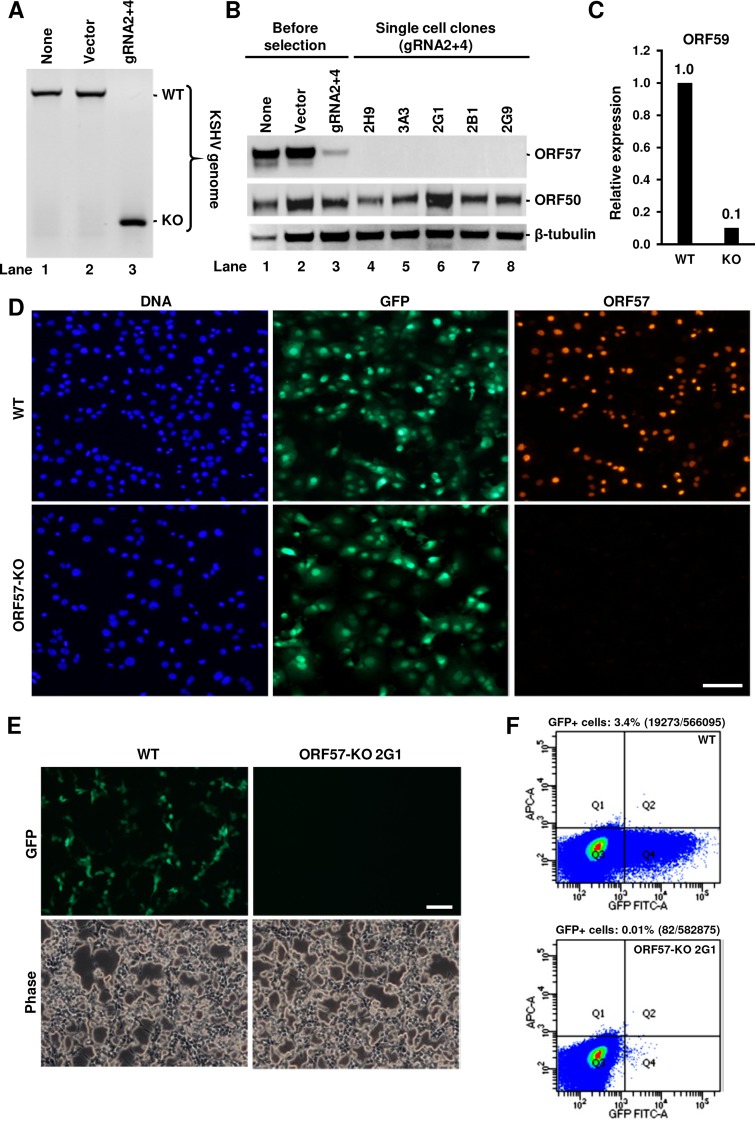
PEL cells are the only naturally KSHV-infected and -transformed human B cells capable of continuous growth in vitro. The KSHV-infected BCBL-1 cell line, which exhibits no EBV coinfection, is one of the PEL cell lines widely used in KSHV research (8). BCBL-1 cells contain a high copy number (∼70) of the KSHV genome per cell as shown in other studies (9, 10) and maintain KSHV infection at the latent stage but can be reactivated into lytic viral infection by hypoxia, oxidative stress, or chemicals (42). Because CRISPR/Cas9 has an inherent ability to attack multiple DNA strands, we decided to apply this technology to edit all copies of the KSHV genome in BCBL-1 cells and generate an ORF57 KO cell line. In our first attempt, BCBL-1 cells were transfected with the second-generation vectors expressing gRNA 2 plus 3 or gRNA 2 plus 4, followed by prolonged puromycin selection. The same work flow successfully generated the ORF57 KO from the Bac36 KSHV genome in Bac36 cells. However, such an approach led to rapid cell death, and we were unable to establish the puromycin-resistant BCBL-1 cells because the cells are refractory to vector transfection. The use of alternative DNA transfection methods, such as electroporation, led to a similar result. To increase the vector transfection efficiency, several rounds of the BCBL-1 cell transfection were carried out with second-generation gRNA-expressing vectors. After each transfection, the cells were subjected to only 3 days of puromycin selection followed by cultivation with the culture medium for 1 to 2 weeks in the absence of puromycin to allow cell viability recovery. The cells were then subjected to a new round of transfection/selection/recovery (Fig. 3A). In parallel, the efficiency of ORF57 KO was monitored by PCR screening using Pr1 plus Pr2 after each cycle (Fig. 3B). As shown in Fig. 3C, after the single round (1×) of transfection and selection, the ORF57 gene remained detectable in the cells. A substantial increase of ORF57 KO was observed in the cells after several rounds. By the fourth round (4×), the cells transfected with the gRNA 2- and 4-expressing vector showed the highest level of ORF57 KO by PCR screen (Fig. 3C). These cells sustained growth in the continuous presence of puromycin but expressed a small amount of ORF57 protein upon induction of viral lytic infection, indicating the presence of incomplete ORF57 KO cell populations.
FIG 3.
KSHV ORF57 KO in BCBL-1 cells. (A) Work flow showing multiple rounds of BCBL-1 cell transfection with the second generation of gRNA vectors followed by 3 days of puromycin selection. After achieving a detectable level of ORF57 KO, the cells were subjected to prolong puromycin selection and single-cell cloning by limited dilution in a 96-well plate. The obtained clones were expanded and used for PCR screen. (B) The KSHV ORF56-ORF57 locus, targeted sites by gRNAs (red squares), and PCR screening primers (black arrows below the diagram). (C) The efficiency of ORF57 KO in BCBL-1 cells after 1, 2, and 4 selection cycles as determined by PCR using Pr1 plus Pr2 primers. (D and E) ORF57 KO and PCR screening of isolated BCBL-1 single-cell clones derived from gRNA 2 plus 4 vector stable transfection by using Pr1 plus Pr2 (D) or Pr1 plus Pr3 (E) primers as described in panel B. WT, wild type ORF57 gene product; KO, knockout ORF57 gene product; B-1, parental BCBL-1 cell population.
Subsequently, we carried out single-cell cloning and selection from these puromycin-resistant BCBL-1 cells stably transfected with the gRNA 2- and 4-expressing vector by limited dilution in 96-well plates in the presence of conditioned medium. Under these culture conditions, we observed the formation of single-cell clones within 3 to 4 weeks after plating. The obtained clones were then gradually expanded and subjected to PCR screening. To eliminate many cells required for total DNA purification, we developed a direct PCR protocol requiring only a few cells for rapid screening of many clones shortly after their appearance. With this approach, i.e., without total DNA isolation and using the Pr1 plus Pr2 primer set, we were able to screen hundreds of single-cell clones for ORF57 KO (Fig. 3D). To ensure that the selected ORF57 KO single-cell clones did not contain any copy of the WT genome, a subset of ORF57 KO single-cell clones was also screened by PCR using the Pr1 in combination with primer Pr3, an ORF57 internal primer within the ORF57 gene (Fig. 3B). With the use of such a primer set, we were able to distinguish the WT genome from the ORF57 KO genome simply by amplifying a 440-bp PCR product from the WT genome, but not from the ORF57 KO genome. As shown in Fig. 3E, several clones were negative for the 440-bp PCR amplification product with the primer set of Pr1 plus Pr3, indicating a deletion of ORF57 from all copies of the viral genome. The clones 39 (lane 17), 42 (lane 18), and 51 (lane 22) were confirmed by this primer set, but not the clones 43 (lane 19), 46 (lane 20), and 50 (lane 21) which displayed the ORF57 KO in the PCR screening by the Pr1 plus Pr2 (Fig. 3D, lanes 7, 10, and 14). After screening 107 isolated single-cell clones, we ultimately obtained 28 (26.2%) single-cell clones showing complete ORF57 KO. Together, these data demonstrated that the successful KO of the ORF57 gene from all copies of the KSHV genome in BCBL-1 cells could be achieved by using these dual gRNA-expressing vectors in combination with repeat transfection, selection, and single-cell cloning.
Verification of ORF57 expression in ORF57 KO single-cell clones.
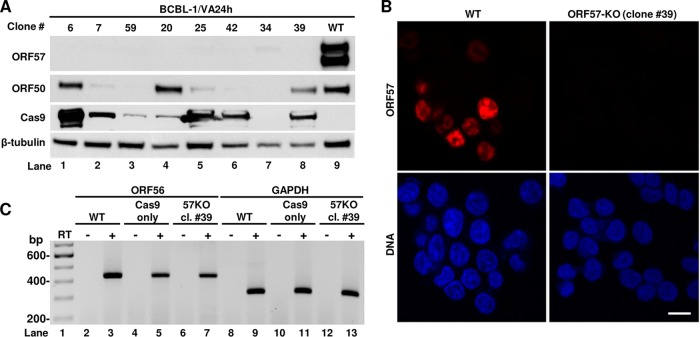
Several BCBL-1 single-cell clones containing the ORF57 KO genome in PCR screening were expanded for further verification. First, we tested for the loss of ORF57 protein expression in these single-cell clones upon induction of KSHV lytic replication by valproic acid. As shown in Fig. 4A, Western blot analysis using an anti-ORF57 antibody showed high expression of ORF57 in parental BCBL-1 cells (WT) but no ORF57 protein expression in the selected eight ORF57 KO cell clones (numbers 6, 7, 59, 20, 25, 42, 34, and 39). Given that only ∼12% of PEL cells can be reactivated for lytic KSHV infection by chemical inducers (43, 44), we next examined whether the lack of ORF57 expression in the selected ORF57 KO single-cell clones was because of their inability to be reactivated for viral lytic infection by reprobing the same blot with an antibody against KSHV ORF50, a viral lytic transactivator required for ORF57 expression. As shown in Fig. 4A, the parental BCBL-1 cells (WT) highly expressed ORF50 (lane 9). The cell clones 6, 20, and 39 (lanes 1, 4, and 8) and the cell clones 7, 25, and 42 (lanes 2, 5, and 6) lacking ORF57 expression were positive for high and low levels of ORF50 expression, respectively, but the cell clones 59 and 34 (lanes 3 and 7) did not express a detectable level of ORF50 protein. These data indicate that lack of ORF57 expression, at least in single-cell clones 6, 2, and 39, was not due to the lack of successful KSHV lytic induction but rather to the specific ORF57 KO from all copies of the virus genome. Subsequently, those cell clones lacking ORF57 expression but also showing no detectable or a low level of ORF50 expression were excluded from further studies. We also tested Cas9 expression in the selected clones. While WT or untransfected cells did not show any expression of Cas9 as expected (lane 9), almost all the cell clones, except cell clone 34 (lane 7), showed sustainable but highly variable expression of Cas9, indicating stable expression from the CRISPR/Cas9 vector (Fig. 4A). Finally, ORF57 expression in the cells was further monitored by immunofluorescence (IF) using an anti-ORF57 antibody after valproic acid reactivation. As shown in Fig. 4B, the parental BCBL-1 cells (WT) showed typical nuclear ORF57 staining in numerous cells, whereas no detectable ORF57 was in any ORF57 KO cells as represented by cell clone 39. As designed, ORF57 KO did not affect ORF56 expression, verified by reverse transcription-PCR (RT-PCR) on total RNA from induced cells. The expression of ORF56 in the single-cell clone 39 remained the same as that of viral WT genomes in the parental cells or the cells stably transfected with a Cas9-only vector (no gRNA) (Fig. 4C).
FIG 4.
Selection of BCBL-1 ORF57 KO single-cell clones expressing ORF50 (RTA, a viral transactivator), Cas9, and ORF56. (A) Western blot analysis of ORF57, ORF50, and Cas9 protein expression in selected BCBL-1 ORF57 KO clones after viral reactivation with sodium valproate (VA) for 24 h. Cellular β-tubulin served as a protein loading control. (B) Expression of ORF57 protein (red) as detected by immunofluorescence staining using anti-ORF57 antibody in BCBL-1 WT and ORF57 KO cells (clone 39) after KSHV reactivation with VA for 24 h. The cell nuclei were counterstained by DAPI. Bar, 10 μm. (C) RT-PCR analysis of ORF56 expression in ORF57 KO single-cell clone 39. Total RNA was prepared after viral reactivation with sodium valproate (VA) for 24 h, reverse transcribed in the presence (+) or absence (−) of reverse transcriptase (RT), and PCR amplified with ORF56-specific primer pair (see Table S1 in the supplemental material for details). Total RNA extracted from parental BCBL-1 cells (WT) or from a BCBL-1 cell stably transfected with a Cas9-only vector served as a control for ORF56 detection. GAPDH served as an internal RNA loading control.
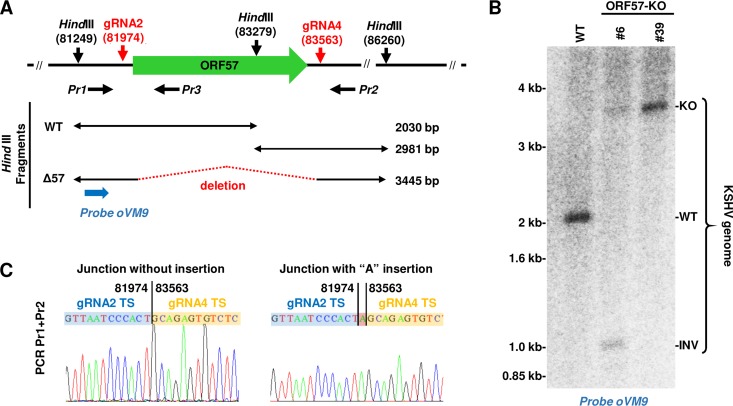
Evaluation of the KSHV ORF57 KO genome status in ORF57 KO single-cell clones led to the discovery of CRISPR/Cas9-mediated ORF57 inversion in the KSHV genome.
For further studies, two clones, clones 6 and 39, both of which showed a complete ORF57 KO phenotype by PCR screening and Western blot analysis (Fig. 3 and 4), were chosen for the evaluation of their viral genome status by restriction enzyme HindIII digestion and Southern blot analysis. As shown in Fig. 5A, there are three HindIII sites in the depicted ORF57 genome region, one of which is located within the ORF57 gene. Thus, HindIII digestion of this region would generate two cleavage products with sizes of ∼2 kb and ∼3 kb from the WT KSHV genome, but only one product (∼3.5 kb) from the ORF57 KO genome due to deletion of the ORF57 gene. By agarose gel electrophoresis, we distinguished the WT KSHV genome from the ORF57 KO genome based on the size difference of the resulting cleavage products (Fig. 5A). Accordingly, the extrachromosomal DNA enriched with viral DNA was digested by HindIII and analyzed by Southern blotting using a 32P-labeled probe, oVM9 (Pr1), upstream of the ORF57 coding region but downstream of the left HindIII site (Fig. 5A). As expected, a single ∼2-kb band was detected from the WT KSHV genome (Fig. 5B) in parental BCBL-1 cells but not from the ORF57 KO genome in both ORF57 KO cell clones 6 and 39. Instead, the ORF57 KO genome produced an expected ∼3.5-kb band (Fig. 5B). Interestingly, the KO genome in clone 6 showed a weaker ∼3.5-kb band accompanied by the appearance of an additional, unexpected ∼1-kb HindIII band, indicating the presence of two heterogenous viral genomes in clone 6 cells. To identify the sequence composition at the site of deletion in clone 39 DNA, PCR products were amplified using the primer pair of Pr1 plus Pr2 (Fig. 5A), TOPO cloned, and sequenced. As shown in Fig. 5C, we found a fusion junction of gRNA 2 and gRNA 4 target sites corresponding to a deletion junction between nucleotides 81974 and 83563 from the KSHV genome. This deletion junction is in the agreement with Cas9 cleavage sites located 3 bp upstream of the PAM sequence (45, 46). In most cases, we saw a “seamless” deletion junction. However, the insertion of 1 to 2 nontemplated mostly “A” nucleotides was also detected at the cleavage sites (Fig. 5C).
FIG 5.
Verification of the KSHV genome with ORF57 deletion in BCBL-1 ORF57 KO clones by Southern blotting. (A) Diagram depicting ORF57 locus targeted by gRNAs 2 and 4 and HindIII restriction sites. Horizontal black arrows represent the primers used in PCR screening. Below are DNA fragments predicted from the WT or ORF57 KO genome after HindIII digestion. A blue arrow represents an oligonucleotide probe used for Southern blotting. (B) Southern blot analysis of KSHV genomic DNA from original BCBL-1 cells (WT) or two separate ORF57 KO single-cell clones (#6 and #39) after HindIII digestion using a 32P-labeled oligonucleotide probe oVM9 (Pr1) shown in panel A. (C) Sequencing of the guide RNA-mediated deletion junction in the ORF57 KO genome. PCR products were obtained with the primer pair of Pr1 plus Pr2 from the single-cell clone 39 DNA and then TA cloned and sequenced. Chromatographs of two representative TA-clones show the sequence junction at the target site (TS) of guide RNA-mediated ORF57 deletion without or with additional “A” insertion. The numbers above the DNA junctions mark the nucleotide positions in the KSHV genome (GenBank accession number U75698).
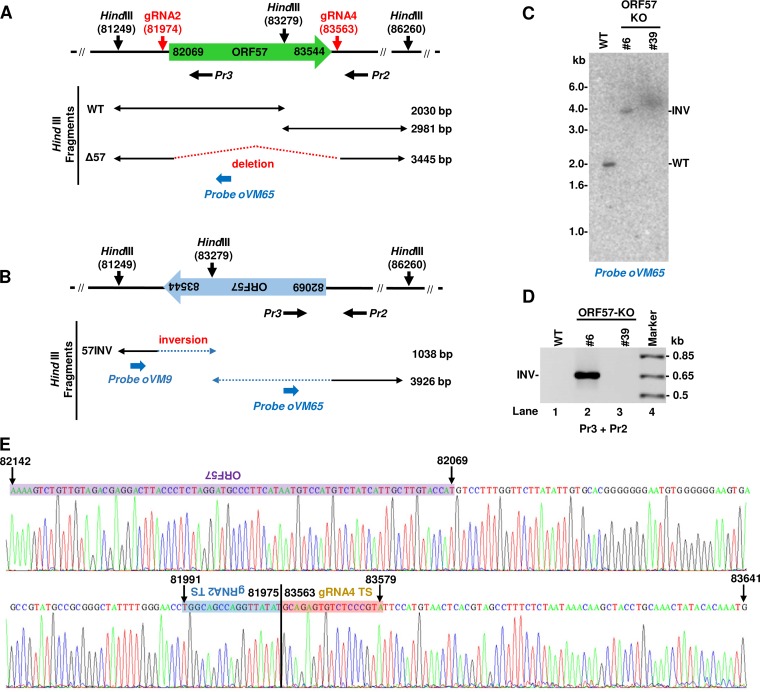
To characterize the unexpected ∼1.0-kb band from the cell clone 6 genome by Southern blotting of viral DNA after HindIII digestion, we analyzed all potential possibilities and hypothesized that this product might be derived from the ORF57 gene in an inverted orientation mediated by CRISPR/Cas9 cleavage (Fig. 6A and B). The paired gRNA-mediated Cas9 cleavage and religation in situ of the cleaved ORF57 gene in an inverted orientation to the cleaved virus genome site must have occurred immediately. Consequently, this inverted rearrangement would result in a HindIII site within the ORF57 gene being closer to the HindIII site upstream of the ORF57 gene at a distance of 1,038 bp, as diagramed in Fig. 6B and shown in a Southern blot (Fig. 5B). To confirm this hypothesis, we first applied a 32P-labeled oligonucleotide probe oVM65 to the ORF57 coding region for Southern blotting. By oVM65 reprobing the same membrane from Fig. 5B and shown in Fig. 6C, we detected a 2.0-kb band from the WT KSHV genome and a 3.9-kb band from the clone 6 virus genome, but nothing from the clone 39 virus genome, demonstrating the presence of the inverted ORF57 in the clone 6 virus genome. We further performed PCR amplification using a primer set of backward Pr3 plus backward Pr2 (Fig. 6A). The backward primer Pr3 is located within the ORF57 gene (Fig. 6A) and, in combination with the backward primer Pr2, does not amplify KSHV WT genomic DNA. However, the primer pair enables amplification only when the ORF57 gene is in the inverted orientation to make the primer Pr3 as a forward primer (Fig. 6B). By using this primer pair, we detected a PCR product at the expected size of ∼0.65 kb from cell clone 6 DNA but not from clone 39 or parental BCBL-1 (WT) cell DNA (Fig. 6D). Sanger sequencing of the 0.65-kb product by the ORF57 internal primer Pr3 confirmed the presence of the ORF57 gene and revealed that the gRNA 2 targeted sequences were indeed ligated in an inverted orientation to the gRNA 4 targeted site between nucleotides 81975 and 83563 in the clone 6 virus genome (Fig. 6E). We concluded that both ORF57 KO single-cell clones exhibit CRISPR/Cas9-mediated disruption of the ORF57 gene in the viral genome. However, cell clone 6 bears a mutant KSHV genome with the ORF57 gene in an inverted orientation derived from the CRISPR/Cas9-mediated genome cleavage and recombination.
FIG 6.
Identification of guide RNA-mediated ORF57 gene inversion (INV) in the KSHV genome in ORF57 KO clone 6 cells. Diagrams of the KSHV WT genome (A) and an inverted ORF57 gene in the clone 6 virus genome (B). Predicted DNA fragments after HindIII digestion are shown below for both virus genomes. Horizontal black arrows below the diagrams are two backward oligonucleotide primers Pr3 and Pr2 for the WT genome (A), but the primer Pr3 can be used in PCR as a forward primer with the primer Pr2 for an inverted ORF57 gene in the clone 6 virus genome (B). Oligonucleotide oVM9 described in the legend for Fig. 5 and oVM65 in blue were diagramed at relative positions in the viral genome. Oligo oVM65 was used for Southern blot analysis in this figure. (C) Southern blot analysis of the inverted ORF57 gene mediated by CRISPR/Cas9 technology. The same membrane used in the Fig. 5B Southern blot was reprobed with an ORF57 internal probe, oVM65. See the predicted size of HindIII fragments detected by this probe in panels A and B. (D) Detection and sequencing of the inverted ORF57 gene in the clone 6 cells by PCR using primers Pr3 plus Pr2 shown in panel B. The PCR products from the KSHV ORF57 single clone 6 cells were TA cloned and sequenced. (E) The primer Pr3-derived sequence chromatograph confirmed the junction sequences between the inverted target site (TS) of gRNA 2 and the TS of gRNA 4, together with the inverted ORF57 coding region (purple).
KSHV ORF57 KO led to viral genome instability and reduced expression of other KSHV genes.
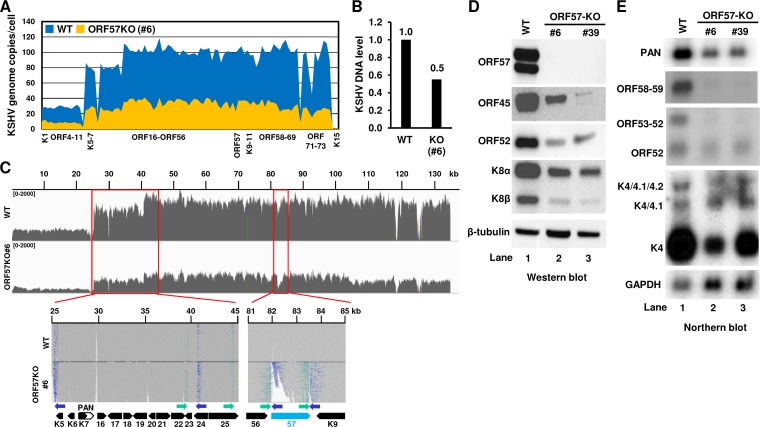
KSHV ORF57 plays an essential role in the regulation of KSHV gene expression at the posttranscriptional level (21). To test whether genomic deletion of the ORF57 gene affects viral genome instability and the expression of other viral genes, we conducted whole-genome sequencing (WGS) of the KSHV genome (WT) in parental BCBL-1 cells and the CRISPR/Cas9-mediated ORF57 KO genome in the clone 6 cells. After normalizing to the host autosomal chromosome read depth from WGS, ∼100 copies of the KSHV WT genome per cell were determined by copy number calculation from the parental BCBL-1 cells, but only ∼30 copies of the ORF57 KO genome per cell were obtained from the clone 6 cells (Fig. 7A). Quantitative PCR (qPCR) analysis of the same DNA sample used for the WGS confirmed the viral DNA reduction by 50% (Fig. 7B). Of note, we found the presence of heterogenous KSHV genomes both in the parental BCBL-1 cells and in the ORF57 KO clone 6 cells, where the K1 to ORF11 region displays only ∼25 copies/cell for the parental BCBL-1 cells and ∼10 copies/cell for the ORF57 KO clone 6 cells (Fig. 7A). Nevertheless, these data suggest that CRISPR/Cas9-mediated ORF57 KO from the KSHV genome triggers viral genome instability.
FIG 7.
ORF57 KO induces KSHV genome instability and reduces viral gene expression in BCBL-1 cells. (A) Estimated copy numbers of the KSHV WT and ORF57 KO genomes per BCBL-1 cell. Total DNA isolated from BCBL-1 cells carrying a WT or ORF57 KO (clone 6) KSHV genome was analyzed by whole-genome sequencing (WGS). The copy number of the KSHV genome in each cell clone was then calculated as described in Materials and Methods and illustrated as the mean reads depth profile of each ORF crossing the entire genome. (B) Relative KSHV genome DNA level determined by qPCR. Total DNA from WT KSHV genome and ORF57 KO genome (clone 6) cells was quantified for ORF59 DNA by qPCR. Host lncFXBO9-11 DNA served as an internal control for normalization. (C) Viral read coverage profiles from the WT genome to the ORF57 KO genome illustrated by IGV. KSHV BCBL-1 WT genome sequence (GenBank accession number MN205539) from the parental BCBL-1 cells was compared with the ORF57 KO genome sequence isolated from the clone 6 cells. Shown below are the zoomed alignment tracks with the reads colored according to pair orientations for the inverted sites identified in KSHV BCBL-1 genome when compared with the reference KSHV genome (GenBank accession number U75698). The inverted ORF57 locus in the ORF57 KO is shown on the right. (D and E) Effect of ORF57 KO and inversion on viral genome expression in ORF57 KO cells. Parental BCBL-1 WT and two stable ORF57 KO single-cell clones were reactivated for viral lytic infection by 1 mM sodium valproate for 24 h before total cell lysates and RNA were extracted separately. The expression of individual viral genes from each cell sample was examined either by Western blotting (D) for selected viral proteins or by Northern blotting (E) for indicated viral gene transcripts.
Further analyses of the two viral genomes showed a very similar nucleotide sequence composition except for the inverted ORF57 region and some minor variations (Fig. 7C). Although the KSHV BCBL-1 genome differs from the KSHV reference genome at 316 sites (see Fig. S1A, Table S2), we discovered that the ORF57 KO genome (clone 6) differs from its parental BCBL-1 genome only at 5 sites, G-to-T at nucleotide 74042 and C-to-T at nucleotide 126032, an insertion at nucleotide 126278, and two deletions from nucleotides 124843 to 124857 and nucleotides 124863 to 124871, in addition to the inverted ORF57 region (Fig. S1B). Interestingly, the G-to-T mutation at nucleotide 74042 changed the GGA (Gly) codon to a UGA stop codon for encoding a truncated ORF50 by ∼200 aa residues in most, but not all, copies of the genome. Other alterations in the ORF57 KO genome appeared to be not harmful to virus lytic induction (Fig. S1B). Despite the fact that the read orientation analyses of the KSHV BCBL-1 genome against the KSHV reference genome showed a few partially inverted sites from the ∼25- to 45-kb region, these inverted sites were common in both the WT and ORF57 KO genomes (Fig. 7C) and thus unrelated to CRISPR/Cas9 activities in the cells.
Subsequently, we compared the expression of selected viral genes both at protein and RNA levels in parental BCBL-1 cells (WT) and two ORF57 KO cell clones, clone 6 and 39, after induction of lytic KSHV infection. As shown in Fig. 7D, the expression of ORF45 (tegument protein), ORF52 (KSHV inhibitor of cGAS), and both forms (α and β) of K8 (k-bZIP) proteins was significantly reduced in the ORF57 KO cell clones compared with that of the parental BCBL-1 cells (Fig. 7D). Similarly, by examining the expression of viral genes at the RNA level in Northern blot analyses (Fig. 7E), we also observed a substantial reduction of numerous viral lytic gene transcripts, including PAN RNA and bicistronic ORF59/58, bicistronic ORF53/52, and monocistronic ORF52, and transcripts from K4.2/K4.1/K4 (39) in the ORF57 KO cell clones over the parental BCBL-1 cells (WT) (Fig. 7E). These studies further reiterate that knocking out the ORF57 gene from the KSHV genome led to profound reduction of the viral genome copies and, consequently, the expression of viral genes.
Detrimental effect of ORF57 KO on virus production in iSLK/Bac16 cells.
After demonstrating that ORF57 KO affects virus genome instability and the expression of various viral lytic genes in BCBL-1 cells, we examined the effect of ORF57 KO on KSHV virion production in iSLK/Bac16 cells carrying a KSHV-GFP genome and an inducible ORF50 transgene under the control of a Tet-On promoter; thus, an infectious KSHV-green fluorescent protein (GFP) virus could be induced from the KSHV Bac16 genome in the presence of doxycycline (7, 36, 47). Because puromycin was used to stably maintain the KSHV Bac16 genome in iSLK cells, we added a blasticidin resistance gene to our dual gRNA-expressing vector (gRNAs 2 and 4). The resulting blasticidinr-gRNA 2- and 4-expressing vector was then transfected into iSLK/Bac16 cells followed by blasticidin selection for stable transfection and single-cell cloning. As described above, the efficiency of ORF57 KO in iSLK/Bac16 cells was examined by PCR screening with the primer set of Pr1 plus Pr2 (Fig. 1A). In contrast to that in BCBL-1 cells, which contain ∼100 copies of the KSHV genome per cell, we found that a single transfection/selection cycle resulted in efficient ORF57 KO in iSLK/Bac16 cells containing only a few copies (1 to 2 copies) of the KSHV Bac16 genome per cell (48) (Fig. 8A) and a substantial reduction of ORF57 protein in the cells (Fig. 8B, lane 3). Loss of ORF57 expression from the ORF57 KO genome also led to reduced expression of ORF59, a notable ORF57 target (Fig. 8C).
FIG 8.
Detrimental effect of CRISPR/Cas9-mediated ORF57 KO on KSHV genome expression and virion production in iSLK/Bac16 cells containing a stably transfected KSHV-GFP genome. (A to C) PCR screen of ORF57 KO efficiency using flanking primers Pr1 plus Pr2 (Fig. 1A) in the untransfected iSLK/Bac16 cells (none) or the cells transfected by a Cas9-only empty vector (vector) or a gRNA 2 plus 4 expression vector (gRNA2 + 4) (A). Characterization of the ORF57 KO genome for expression of ORF50, ORF57, and ORF59 in the selected single-cell clones. The iSLK/Bac16 cells with or without transfection by a Cas9-only empty vector (vector) or a gRNA 2 plus 4 expression vector (gRNA2 + 4) as described in the legend for Fig. 3 were subjected to single-cell cloning and screening before induction of KSHV lytic replication by doxycycline and sodium butyrate. Total cell lysates and RNA before and after selection of single cell clones and viral lytic induction were prepared for Western blot analyses of KSHV ORF50 and ORF57 (B) and for RT-qPCR analysis of ORF59 (C). Cells without transfection (none) or transfected with a Cas9-only vector (vector) before selection served as controls and cellular β-tubulin served as a loading control (B). (D) WT and ORF57 KO cells with lytic induction by Dox plus Bu for 24 h were examined for GFP expression level in association with KSHV genome copies. The expression of ORF57 was examined by anti-ORF57 IF staining to determine ORF57 KO efficiency. Bar, 100 μm. Effect of ORF57 KO on infectious KSHV production. (E and F) Titration of cell-free virus production from Dox- plus Bu-induced iSLK/Bac16 cells carrying a KSHV WT or ORF57 KO (clone 2G1) genome was carried out in HEK293T cells by using the corresponding cell culture medium collected 5 days after viral lytic induction. The newly infected HEK293T cells expressing GFP as indications of GFP-KSHV virus infection were imaged by direct fluorescence (E) and then counted by FACS analysis (F) 48 h after infection. The percentages of GFP+ cells over total number of cells were calculated based on the cell counts in the Q4 region (F).
Further single-cell selection and cloning led to isolation of several single-cell clones with the complete ORF57 KO genome and showing no expression of ORF57 protein by Western blotting (Fig. 8B, lanes 4 to 8) and by anti-ORF57 IF staining (Fig. 8D). In contrast to that in BCBL-1 cells, this loss of viral gene expression was not due to the reduced viral genome copies in the examined iSLK/Bac16 single-cell clones. Because the KSHV Bac16 genome has an inserted GFP-coding gene, the similar GFP expression profiles from the WT iSLK/Bac16 and ORF57 KO cells indicate the presence of similar copy numbers of the KSHV genome in the two types of cells (Fig. 8D). Conceivably, GFP expression from the virus genome would permit easy titration of the infectious virions released from iSLK/Bac16 cells with or without ORF57 KO. Accordingly, we induced virus lytic replication and virus production from the iSLK/Bac16 cells with doxycycline (Dox) plus butyrate for 5 days. The obtained cell supernatant containing cell-free virions was then used to infect HEK293T cells. As shown in Fig. 8E, we obtained numerous KSHV-infected (GFP+) HEK293T cells 48 h after inoculation with the supernatant from iSLK/Bac16 cells carrying a WT KSHV genome, but none with the supernatant from ORF57 KO 2G1 cells. Further fluorescence-activated cell sorting (FACS) analysis of the infected GFP+ HEK293T cells showed that the cells containing the WT KSHV genome produced more infectious KSHV-GFP viruses, whereas the clone 2G1 cells bearing an ORF57 KO genome produced only a little under our experimental conditions (Fig. 8F). In general, 0.4 ml of the collected 4-ml culture supernatant (10%) from iSLK/Bac16 cells containing a WT KSHV genome, without virus concentration, infected 3.4% of HEK293T cells, whereas the same amount of culture supernatant collected from ORF57 KO 2G1 cells was not infectious for HEK293T cells, with no visible GFP expression and only 0.01% of visible GFP+ HEK293T cells detected by FACS analysis. These data provide further evidence that ORF57 is essential for KSHV replication and multiplication (27–29).
Rescuing of KSHV production from ORF57 KO iSLK/Bac16 cells.
Two rescue experiments were subsequently conducted in iSLK/Bac16-derived clone 2G1 cells containing an ORF57 KO genome, first for rescuing ORF59 expression and second for rescuing infectious KSHV production by ectopic transient expression of ORF57. As shown in Fig. 9A and B, parental iSLK/Bac16 cells (WT) and ORF57 KO cells displayed similar GFP levels, indicating no difference in KSHV-GFP genome copy numbers from the two types of cells. However, almost all WT cells were induced by doxycycline plus butyrate to express ORF59, but only a few 2G1 cells containing an ORF57 KO genome were induced to express ORF59, an ORF57 downstream target. As expected, the lack of ORF59 expression was partially rescued from the ORF57 KO genome by ectopic expression of ORF57 as determined by comparing the 2G1 cells transfected with an empty vector (FLAG only) with the cells transfected with ORF57-FLAG (Fig. 9A). Notably, we saw coexpression of ORF57-FLAG and endogenous ORF59 in the transfected 2G1 cells (Fig. 9B). The successful rescue of ORF59 expression by ectopic ORF57 in the 2G1 cells was further confirmed by the rescue of infectious KSHV production. As shown in Fig. 9C and D, we demonstrated that ectopic ORF57 expression in the 2G1 cells partially rescued the production of infectious KSHV-GFP viruses for the infection of HEK293T cells, resulting in visible GFP expression (Fig. 9C). Quantitative FACS analysis of GFP+ HEK293T cells indicated that the culture supernatant collected from the 2G1 cells containing an ORF57 KO KSHV genome was only minimally infectious but became highly infectious to HEK293T cells in the presence of ectopic ORF57, with appearance of ∼7 times more visible GFP+ HEK293T cells than in the cells without ectopic ORF57 expression (Fig. 9D). Together, these data provide a strong evidence that ORF57 is essential in KSHV lytic infection and that an ORF57 KO KSHV genome can be complemented by ectopic expression of ORF57.
FIG 9.
Rescue of infectious KSHV production from iSLK/Bac16 cells containing an ORF57 KO KSHV genome. (A and B) Rescue of ORF59 expression from an ORF57 KO genome in the clone 2G1 cells by ectopic expression of ORF57. The 2G1 single-cell clone derived from iSLK/Bac16 cells containing an ORF57 KO KSHV-GFP genome was used for transfection of an empty (FLAG only) or ORF57-FLAG expression vector. After transfection, the virus was reactivated with Dox plus Bu treatment for 24 h. The expression of ORF59 or ectopic ORF57-FLAG was monitored by IF staining by using an anti-ORF59 specific antibody alone (A) or in combination with an anti-FLAG antibody for ORF57 expression (B). The WT and ORF57 KO 2G1 cells without transfection served as controls (A). Bar, 100 μm in panel A and 20 μm in panel B. (C and D) Ectopic expression of ORF57-FLAG in ORF57 KO 2G1 cells rescues the production of infectious virions. The 2G1 cells carrying an ORF57 KO genome were transfected with an empty (FLAG only) or ORF57-FLAG-expressing vector. The virus replication was induced by Dox plus Bu treatment, and the culture supernatant containing cell-free virions was collected 5 days after induction. The production of infectious virions was determined by inoculating HEK293T cells with the collected supernatant. The number of newly infected GFP+ cells was determined 48 h after infection using live fluorescence imaging (C) or by FACS analysis (D). The 2G1 cells with transfection of an empty FLAG vector served as negative controls. Bar, 100 μm.
DISCUSSION
PEL cell lines, the only culturable KSHV-transformed cell lines, have provided enormous advantages for us to understand the molecular biology of KSHV latent and lytic infections. However, PEL cells also have disadvantages for genetic analysis of the virus genome, because PEL cells in general contain a high copy number of the KSHV genome per cell, making it difficult to modify all copies of the viral genome. In this report, we demonstrated that BCBL-1 cells, a PEL cell line, contain ∼100 copies of KSHV genome per cell by whole-genome sequencing analysis and showed the KSHV BCBL-1 genome (MN205539) differs from the reference KSHV genome from BC-1 cells (U75698) at 316 sites. By using a modified CRISPR/Cas9 system, we were able to successfully delete a viral lytic gene ORF57 from all 100 copies of the KSHV genome in BCBL-1 cells. By simultaneous expression of two gRNAs from the same vector in BCBL-1 cells and single-cell cloning, we generated multiple single-cell lines with ORF57 KO in all copies of KSHV genome, without notable off-target effects on the host genome. The new CRISPR/Cas9 system could be used easily for knocking out ORF57 or any other genes from the KSHV genome in iSLK/Bac16 cells or Bac36 cells and for constructing and characterizing new KSHV mutants as needed.
CRISPR/Cas9 technology has been applied to prevent or eliminate alpha- or betaherpesvirus infection by targeting viral essential and nonessential genes using single or combined multiple gRNAs (13, 14, 49, 50). Targeted mutations, deletions, and substitutions of herpesvirus genomes were achieved (51–53). The “curing” of latent EBV infection by CRISPR/Cas9 was also observed in Burkitt’s lymphoma cells bearing 5 to 7 copies of the EBV genome (50, 54, 55). Using EBNA-1-specific gRNA, Bigi and colleagues demonstrated that only a few copies of the EBV genome in EBV and KSHV coinfected PEL cells are required for maintaining a high copy number of the KSHV genome (56). Applications of CRISPR/Cas9 technology to study host factors contributing to KSHV infection and viral gene functions have been attempted (57–61). Like the results in the knockout of two copies of the host gene from cell lines, complete knockout of viral genes from the KSHV genome in this study and others (29, 62, 63) was easily achieved in iSLK/Bac16 or Bac36 cells, in which each cell bears only a few copies (1 to 2 copies) of the virus genome (6, 48). However, complete knock out of a viral gene from all copies of the KSHV genome in PEL cells using the conventional CRISPR/Cas9 system has been extremely challenging due to the high copy number of the viral genome per cell.
The new CRISPR/Cas9 technology developed in this report used the coexpression of two separate gRNAs and Cas9 in a cell transfected with a single expression vector. The goal of our approach is the removal of the necessity for simultaneous successful transfection of two separate gRNA-expressing vectors into a cell to achieve the Cas9-mediated gene knockout. The subsequent analysis showed that our approach resulted in 28/107 (26.2%) single-cell clones exhibiting complete ORF57 KO from all copies (∼100) of the viral genome in BCBL-1 cells. Similar efficiency of CRISPR/Cas9-mediated KO of multicopy porcine endogenous retroviruses (PERVs) was observed in a porcine kidney epithelial cell line (PK15 cells) in which each cell contains 62 copies of the PERV genome (64). By using the direct cell PCR in this report, we can rapidly screen many cell clones to achieve the targeted gene knockout in a short term.
Analysis of the viral genome in ORF57 KO cells confirmed the deletion of a targeted genomic region of 1.6 kb. Previous reports indicated that CRISPR/Cas9 is useful for mediating deletions of a specific region from a host or viral genome ranging from a few hundred to several megabases of nucleotides (12, 54). In addition to the CRISPR/Cas9-mediated deletion of the ORF57 gene, Southern blotting and WGS analyses of the clone 6 viral DNA revealed that CRISPR/Cas9 also mediated the inversion of the ORF57 gene in situ in the gRNA-cleaved sites to inactivate ORF57 expression. Since the gRNAs guide the Cas9 enzyme to cleave specific sites in a genome to generate a blunt-end double-strand break (DSB), this DSB can be repaired by the NHEJ DNA repair pathway (65–68), thus inevitably inducing an inversion of the cleaved target genes. Similar results have been observed in CRISPR/Cas9-mediated gene knockout studies, and in situ inversion of the cleaved target genes was found in zebrafish, mammalian cells, and animals (69–73).
KSHV ORF57 is an essential posttranscriptional regulator for the expression of viral lytic genes and the production of infectious virions (21, 27–29). We and others showed that ORF57 regulates the expression of vIL6 (K2), K8 (K-bZIP), K15, PAN, ORF4, ORF6, ORF8, ORF47, ORF50, ORF56, and ORF59 (15–18, 22, 26, 29, 30, 32, 33, 35, 74, 75). Unexpectedly, we found in this study that knockout of the ORF57 gene from all copies of the KSHV genome in BCBL-1 cells triggered viral genome instability and resulted in dramatic reduction of viral genome copies and expression of viral lytic genes in the cells. However, the ORF57 KO-mediated viral genome instability in BCBL-1 cells appeared not to occur in iSLK/Bac16 cells. The knockout of ORF57 from the KSHV genome in iSLK/Bac16 cells was detrimental for virus replication and the production of infectious virions. This function of ORF57 in KSHV virion production was partially rescued from the ORF57 KO genome by ectopic transient expression of ORF57. A few factors may contribute to this partial rescue. First, KSHV, like other enveloped viruses, is sensitive to lipo-based transfection reagents used for ORF57 transfection. We found a 3-fold reduction of KSHV-GFP virion production in the presence of transfection reagent (our unpublished observation). Second, inefficient and time-limited expression of ORF57 protein as a result of transient expression may not be sufficient to fully rescue the virus production. Third, although the gRNAs used in our study had no homology with any other regions of the KSHV genome, the ORF57 KO genome in BCBL-1 cells displayed an additional five sites of mutations or small insertions/deletions compared with the WT KSHV genome. One mutation changed a coding codon to a stop codon, leading to a truncated ORF50 by ∼200 aa from its transactivation domain in many copies of the ORF57 KO genome. Fourth, since the extent of off-target editing by CRISPR/Cas9 remains a matter of debate (76, 77), whether the expressed gRNAs in ORF57 KO single-cell clones could lead to any off-target editing should be further investigated.
MATERIALS AND METHODS
Design and cloning of virus-specific gRNAs.
The KSHV-specific gRNAs were designed from the 5′ and 3′ regions immediately outside the ORF57 gene (Fig. 1A and B) by an on-line CRISPR design tool developed by the Zhang lab (http://crispr.mit.edu) using KSHV genomic sequences (GenBank accession number U75698). The gRNA sequences with the top scores and limited, predicted off-target cleavage (see Table S3 in the supplemental material) were cloned into a vector pSpCas9(BB)-2A-Puro (PX459) V2.0 (catalog number 62988; Addgene) as described previously (41). The constructed plasmids in the first generation encoded a single ORF57-specific gRNA and Cas9/Puro fusion protein for stable coexpression of gRNA/Cas9 after puromycin selection of transfected cells. To construct the second generation of the vectors which simultaneously express two different gRNAs, the cloned gRNAs were amplified together with the upstream U6 promoter using a primer set of oABC1 and oABC2 containing XbaI and KpnI restriction sites. The resulting PCR products, after digestion with indicated enzymes, were then cloned into the first-generation vectors via XbaI and KpnI sites (for details, see Fig. 2A). Finally, the blasticidin-resistance gene from pCMV/Bsd (Thermo Fisher Scientific) was inserted into the second-generation vector using SbfI and NotI sites.
Cell transfection and induction of KSHV lytic infection.
KSHV-positive BCBL-1 cells (8) were cultivated in RPMI 1640 medium. HEK293 carrying a KSHV Bac36 genome or Bac36 cells (28, 78), iSLK carrying a KSHV Bac16 genome or iSLK/Bac16 cells (7), and uninfected HEK293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM). All media were supplemented with 10% defined fetal bovine serum (HyClone, GE Healthcare) and 1× penicillin-streptomycin-glutamine (Thermo Fisher Scientific). To maintain the virus genome, the Bac-carrying cells were grown under selection in the above-described medium with the addition of 150 μg/ml of hygromycin B for Bac36 cells or 150 μg/ml of hygromycin B, 1 μg/ml puromycin, and 250 μg/ml G418 for iSLK/Bac16 cells. All gRNA/Cas9 vector transfections were performed with LipoD293 In Vitro DNA transfection reagent (SignaGen Laboratories) using the standard protocol for Bac36 and iSLK/Bac16 cells or the hard-to-transfect cell protocol for BCBL-1 cells. The selection of gRNA/Cas9-expressing cells was carried out in the presence of 1 to 2 μg/ml of puromycin (InvivoGen) for Bac36 and BCBL-1 cells or 15 μg/ml blasticidin (InvivoGen) for iSLK/Bac16 cells. The KSHV lytic cycle was induced by the addition of 3 mM sodium butyrate (Bu; Sigma-Aldrich) for Bac36 cells, 1 mM sodium valproate (VA; Sigma-Aldrich) for BCBL-1 cells, or 1 μg/ml doxycycline (Dox; Clontech) plus 1 mM Bu for iSLK/Bac16 cells. The protein and RNA samples were harvested 24 to 48 h after induction.
ORF57 knockout.
Bac36 cells were transfected with two gRNA/Cas9-expressing vectors, one for a gRNA upstream of the 5′ end and the other for a gRNA downstream of the 3′ end of ORF57, followed by 2 to 3 weeks of puromycin selection. The second generation of a dual gRNA/Cas9 expression vector constructed for simultaneous expression of both 5′ and 3′ gRNAs was used to transfect BCBL-1 cells with only 2 to 3 days of puromycin selection. The selected BCBL-1 cells were recovered to full viability in the absence of puromycin before the next round of transfection and puromycin selection. The efficiency of ORF57 KO after each round was monitored by PCR. The resulting cell population with the desired KO efficiency was then subjected to final long-term (4 weeks) puromycin selection, followed by single-cell cloning. The iSLK/Bac16 cells were transfected with the second generation of a dual gRNA/Cas9 expression vector containing a blasticidin resistance gene and selected for 2 weeks in the presence of blasticidin, followed by single-cell cloning.
Single-cell cloning.
Before single-cell cloning of BCBL-1 ORF57 KO cells, the conditioned medium was prepared by collecting the culture supernatant free from parental BCBL-1 cells after 2 days of cultivation. The collected medium was then clarified with low-speed centrifugation at 100 × g for 10 min followed by filtration through 0.22-μm Millex-GS filters (Millipore). The cleared medium was used immediately or stored at 4°C for up to 1 week before use. The puromycin-resistant BCBL-1 cells were diluted to a final concentration of 5 cells/ml in culture medium consisting of two-thirds fresh complete RPMI medium and one-third conditioned medium. One hundred microliters of the diluted cell suspension was plated in each well of a 96-well plate (0.5 cell/well). After visual inspection ensuring the presence of 1 cell per well, the clones were allowed to grow for ∼3 to 4 weeks with the addition of 100 μl of fresh medium weekly to prevent the wells from drying out. After the appearance of visible clones, the cells were gradually expanded by the addition of fresh medium in 1:1 ratio every 2 to 3 days. The single-cell clones of iSLK/Bac16 were prepared in the same way without the conditioned medium.
PCR screen.
To test the ORF57 KO efficiency, total DNA from ∼1 × 106 to 5 × 106 cells was isolated using a QIAamp DNA Blood Minikit (Qiagen), treated briefly with RNase A (Thermo Fisher Scientific), and used as a template in a standard PCR using AmpliTaq polymerase (Thermo Fisher Scientific). To carry out a rapid screening of a large number of clones, a direct PCR protocol was developed by centrifuging and resuspending several hundreds of cells in 50 μl of phosphate-buffered saline (PBS) and then freezing the cells on dry ice. After thawing, 4 μl of Qiagen protease (catalog number 1017782) was added to the cells, and the reaction mixture was incubated at 56°C for 10 min. The reaction was stopped by a 5-min incubation at 95°C followed by an additional freeze/thaw cycle. The resulting product was directly used as a PCR DNA template. The sequences of primers used for the screening are shown in Table S1. To determine the sequence, the PCR products were cloned into a pCRII-TOPO vector using a TOPO TA Cloning kit (Thermo Fisher Scientific) and then sequenced.
Southern blotting.
Extrachromosomal DNA from cells was isolated using the classical Hirt method (79). Briefly, the cell pellet was lysed in lysis buffer (0.6% SDS, 10 mM EDTA, 10 mM Tris-HCl, pH 7.5) supplemented with 50 μg/ml RNase A for 2 h at 37°C. The chromosomal DNA was precipitated by the addition of 5 M NaCl to a final concentration of 1 M followed by overnight incubation at 4°C. The precipitated DNA was removed by centrifugation (11,000 × g) at 4°C for 30 min. The remaining supernatant was extracted twice using a phenol-chloroform-isoamyl alcohol solution (25:24:1). The extracted extrachromosomal DNA was precipitated overnight at −20°C by the addition of one-tenth sample volume of 3 M sodium acetate (pH 5.2) and 3 sample volumes of 100% ethanol. The precipitated DNA was spun, washed with 70% ethanol, and resuspended in water. Five micrograms of this DNA was digested with HindIII (New England Biolabs) overnight at 37°C, and the digested products were separated in a 0.8% agarose gel in 1× Tris-borate-EDTA (TBE) buffer. Separated DNA was depurinated in the gel by incubating with 0.125 M HCl for 30 min, followed by denaturation, neutralization, and capillary transfer according to a standard Southern blotting protocol. DNA transferred to a GeneScreen Plus nylon membrane (Perkin Elmer) was immobilized using UV cross-linking. The hybridization was performed in PerfectHyb Plus hybridization buffer (Sigma-Aldrich) supplemented with 100 μg/ml heat-denatured salmon sperm DNA (Thermo Fisher Scientific). The DNA oligonucleotides end-labeled with 32P by T4 PNK (New England Biolabs) were used as specific probes (Table S1). The obtained signal was captured by a Typhoon 5 laser scanner (GE Healthcare).
Whole-genome sequencing, reference genome alignment, and calculation of viral genome copy number.
Total DNA from ∼3 × 106 individual BCBL-1 cell clones was isolated using a QIAamp DNA Blood Minikit (Qiagen) as recommended by the manufacturer and subjected to whole-genome sequencing. The libraries were prepared using an Illumina TruSeq DNA sample prep kit (Illumina FC-121-1001) and sequenced on the HiSeq 4000 in paired-end mode (2 × 150 nucleotide [nt] modality). The total number of high-quality reads (Q30) ranged from 725 million to 544 million reads per sample. The obtained reads were trimmed to remove adapters and low-quality ends using Trimmomatic v0.39 (80) and mapped to a chimeric human and KSHV reference genome (hg19+U75698) using BWA-mem v0.7.15 with default parameter settings (https://arxiv.org/abs/1303.3997). Resulting BAM files were sorted using SAMtools v1.317, and PCR duplicates were marked using Picard v2.1.1 (https://broadinstitute.github.io/picard/). Realignment around indels and base recalibration were performed using the Genome Analysis Toolkit v.3.7 (GATK; Broad Institute, Cambridge, MA), followed by the GATK best practices (81, 82). Reference coverage and GC content statistics were measured using Qualimap (83). Consensus sequences were generated from BAM and VCF files using the “bcftools consensus” command from SAMtools v1.317. Mean coverage across all positions in the 22 autosomes in the human reference genome, which are largely diploid, was 20-fold. The read coverage was visualized by Integrated Genome Browser (IGV; http://software.broadinstitute.org/software/igv/home). The “color by pair orientation” was used to identify the inverted sites in the BCBL-1 viral genome compared to the reference KSHV genome (GenBank accession number U75698).
To calculate the KSHV viral genome and open reading frame (ORF) copy number, we used the GATK “DepthOfCoverage” tool to calculate the mean reads depth of each ORF annotated in the U75698 genome. We then converted the mean reads depth of each ORF to absolute copy number by dividing the mean ORF depth by one-half the mean autosomal (diploid) chromosome read depth for that sample, which approximates the expected depth for a single-copy locus.
Measurement of relative viral DNA level by qPCR.
The relative viral DNA level was also determined by qPCR on total cell DNA using a PrimeTime qPCR assay (IDT) specific for KSHV ORF59 (nucleotides 95770 to 95911) (74). The relative viral DNA level was obtained after normalizing to the level of human genomic DNA simultaneously measured by PrimeTime qPCR assay targeted to the human lnFBXO9-11 locus (see Table S1 for details).
Western blotting.
Total protein samples were prepared by direct lysis of cells in 2× Laemmli SDS sample buffer with the addition of 5% of 2-mercaptoethanol, separated on a 4% to 12% NuPAGE bis-Tris SDS-PAGE gel (Thermo Fisher Scientific), and transferred to a nitrocellulose membrane (Bio-Rad). The following antibodies were used for Western blotting: rabbit-anti RTA antibody (a generous gift from Yoshihiro Izumiya, University of California, Davis), anti-Cas9 mouse monoclonal antibody (catalog number 14697; Cell Signaling), anti-KSHV K8 (catalog no. 20015; ProMab), anti-KSHV ORF45 (a generous gift from Yan Yuan, University of Pennsylvania), anti-KSHV ORF52 (a generous gift from Fanxiu Zhu, Florida State University), rabbit anti-ORF57 antibody (28), and anti-β-tubulin mouse monoclonal antibody (catalog number T5201; Sigma-Aldrich).
RT-PCR, RT-qPCR, and Northern blotting.
Total cell RNA was isolated using TriPure isolation reagent (Roche). Before RT-PCR and RT-qPCR, the contaminant genomic DNA was removed by treatment with a TURBO DNA-free kit (Thermo Fisher Scientific) as recommended. RT-PCR was performed on 1 μg of DNase-treated RNA in a reaction mixture containing M-MLV RT (Thermo Fisher Scientific), random hexamers for cDNA synthesis, AmpliTaq (Thermo Fisher Scientific), and gene-specific primers for PCR amplification. The primers used in RT-PCR are listed in Table S1.
For RT-qPCR analysis of the ORF59 transcript, the cDNA was synthesized with a SuperScript first-strand synthesis system (Thermo Fisher Scientific) using random hexamers, followed by qPCR using a PrimeTime qPCR assay (IDT) specific for KSHV ORF59 (74). The level of endogenous GAPDH (glyceraldehyde-3-phosphate dehydrogenase) measured by a TaqMan assay (Hs02758991_g1; Thermo Fisher Scientific) was used for normalization.
Northern blotting was performed as described before (16). Five micrograms of total RNA was separated in a 1% agarose gel containing 0.66 M formaldehyde in 1× 3-(N-morpholino) propanesulfonic acid (MOPS) RNA buffer (Quality Biological). After transferring and UV cross-linking, the membranes were hybridized with 32P-labeled oligonucleotide probes listed in Table S1.
Immunofluorescence staining.
VA-induced BCBL-1 cells were spun, washed with 1× PBS, and spotted onto poly-d-lysine-treated glass coverslips before fixation. The iSLK/Bac16 cells were grown directly on the glass coverslips placed in each well of a 6-well plate. The cells were fixed with 4% paraformaldehyde in 1× PBS for 20 min at room temperature, permeabilized with 0.5% Triton X-100 for 15 min, and blocked with 2% bovine serum albumin (BSA) dissolved in TPBS buffer consisting of 1× PBS and 0.02% Tween 20. Primary rabbit anti-ORF57 (28) or mouse monoclonal anti-KSHV ORF59 (catalog number 13-211-100; Advanced Biotechnologies) antibodies were used for immunostaining in combination with Alexa Fluor-labeled secondary antibody (Thermo Fisher Scientific). The cell nuclei were counterstained with Hoechst 33342 dye (Millipore Sigma) or DAPI (4′,6-diamidino-2-phenylindole; Thermo Fisher Scientific) for 5 min before mounting in Prolong Gold Antifade mounting medium (Thermo Fisher Scientific). The confocal images were collected on a Zeiss 710 microscope and analyzed by ZEN software (Zeiss).
Virus production and titration.
The production and titration of cell-free KSHV virus produced from iSLK/Bac16 cells were carried out as described previously (36). Briefly, iSLK/Bac16 cells carrying a KSHV WT or ORF57 KO genome tagged with GFP were treated with 1 μg/ml doxycycline plus 1 mM Bu to induce viral lytic infection. Five days after induction, the culture supernatants containing KSHV virions were collected and cleared of cell debris by low-speed centrifugation at 400 × g. Cleared virus inoculum (400 μl) was then applied to the monolayer of HEK293T (0.5 × 106) cells plated in a 6-well plate followed by 2-h virus adsorption at 37°C. The amount of virus in the inoculum represented by newly infected GFP+ HEK293T cells was determined 48 h after the infection by direct fluorescence microscopy and by flow cytometry (FACS) analysis of 0.5 × 106 cells.
To rescue virus production, 0.25 × 106 of ORF57 KO iSLK/Bac16 clone 2G1 cells were transfected with 2 μg of an ORF57-FLAG-expressing vector (pVM7) (84) for 2 h prior to virus reactivation with Dox plus Bu. The cells transfected with empty vector (FLAG only) were used as a negative control. Five days after virus reactivation, the culture medium was collected and cleared, and the amount of released cell-free infectious virions was determined by the infection of HEK293T cells as described above.
Data availability.
The KSHV BCBL-1 strain sequence is available in GenBank under accession number MN205539.
Supplementary Material
ACKNOWLEDGMENTS
We thank Yan Yuan (University of Pennsylvania), Yoshihiro Izumiya (University of California, Davis), and Fanxiu Zhu (Florida State University) for KSHV-specific antibodies, Jae Jung (University of Southern California) for iLSK/Bac16 cells, and Eva Gottwein (Northwestern University) for the single-cell cloning protocol.
This study was supported by Intramural Research Program of National Cancer Institute, National Institutes of Health (ZIASC010357 to Z.M.Z.). A.B. was a recipient of the Postbaccalaureate Intramural Research Training Award (POSTBAC IRTA/CRTA).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00628-19.
REFERENCES
- 1.Russo JJ, Bohenzky RA, Chien MC, Chen J, Yan M, Maddalena D, Parry JP, Peruzzi D, Edelman IS, Chang Y, Moore PS. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc Natl Acad Sci U S A 93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM. 1995. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med 332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 3.Moore PS, Gao SJ, Dominguez G, Cesarman E, Lungu O, Knowles DM, Garber R, Pellett PE, McGeoch DJ, Chang Y. 1996. Primary characterization of a herpesvirus agent associated with Kaposi’s sarcomae. J Virol 70:549–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d’Agay MF, Clauvel JP, Raphael M, Degos L, Sigaux F. 1995. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman’s disease. Blood 86:1276–1280. [PubMed] [Google Scholar]
- 5.Arias C, Weisburd B, Stern-Ginossar N, Mercier A, Madrid AS, Bellare P, Holdorf M, Weissman JS, Ganem D. 2014. KSHV 2.0: a comprehensive annotation of the Kaposi’s sarcoma-associated herpesvirus genome using next-generation sequencing reveals novel genomic and functional features. PLoS Pathog 10:e1003847. doi: 10.1371/journal.ppat.1003847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou FC, Zhang YJ, Deng JH, Wang XP, Pan HY, Hettler E, Gao SJ. 2002. Efficient infection by a recombinant Kaposi’s sarcoma-associated herpesvirus cloned in a bacterial artificial chromosome: application for genetic analysis. J Virol 76:6185–6196. doi: 10.1128/JVI.76.12.6185-6196.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brulois KF, Chang H, Lee AS, Ensser A, Wong LY, Toth Z, Lee SH, Lee HR, Myoung J, Ganem D, Oh TK, Kim JF, Gao SJ, Jung JU. 2012. Construction and manipulation of a new Kaposi’s sarcoma-associated herpesvirus bacterial artificial chromosome clone. J Virol 86:9708–9720. doi: 10.1128/JVI.01019-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Kedes D, Ganem D. 1996. Lytic growth of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat Med 2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- 9.Lallemand F, Desire N, Rozenbaum W, Nicolas JC, Marechal V. 2000. Quantitative analysis of human herpesvirus 8 viral load using a real-time PCR assay. J Clin Microbiol 38:1404–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai X, Lu S, Zhang Z, Gonzalez CM, Damania B, Cullen BR. 2005. Kaposi’s sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc Natl Acad Sci U S A 102:5570–5575. doi: 10.1073/pnas.0408192102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charpentier E, Doudna JA. 2013. Biotechnology: rewriting a genome. Nature 495:50–51. doi: 10.1038/495050a. [DOI] [PubMed] [Google Scholar]
- 12.Korablev AN, Serova IA, Serov OL. 2017. Generation of megabase-scale deletions, inversions and duplications involving the Contactin-6 gene in mice by CRISPR/Cas9 technology. BMC Genet 18:112. doi: 10.1186/s12863-017-0582-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen YC, Sheng J, Trang P, Liu F. 2018. Potential application of the CRISPR/Cas9 system against herpesvirus infections. Viruses 10:291. doi: 10.3390/v10060291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Diemen FR, Lebbink RJ. 2017. CRISPR/Cas9, a powerful tool to target human herpesviruses. Cell Microbiol 19:12694. doi: 10.1111/cmi.12694. [DOI] [PubMed] [Google Scholar]
- 15.Majerciak V, Yamanegi K, Zheng ZM. 2006. Gene structure and expression of Kaposi’s sarcoma-associated herpesvirus ORF56, ORF57, ORF58, and ORF59. J Virol 80:11968–11981. doi: 10.1128/JVI.01394-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Majerciak V, Yamanegi K, Nie SH, Zheng ZM. 2006. Structural and functional analyses of Kaposi sarcoma-associated herpesvirus ORF57 nuclear localization signals in living cells. J Biol Chem 281:28365–28378. doi: 10.1074/jbc.M603095200. [DOI] [PubMed] [Google Scholar]
- 17.Massimelli MJ, Kang JG, Majerciak V, Le SY, Liewehr DJ, Steinberg SM, Zheng ZM. 2011. Stability of a long noncoding viral RNA depends on a 9-nt core element at the RNA 5’ end to interact with viral ORF57 and cellular PABPC1. Int J Biol Sci 7:1145–1160. doi: 10.7150/ijbs.7.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sei E, Conrad NK. 2011. Delineation of a core RNA element required for Kaposi’s sarcoma-associated herpesvirus ORF57 binding and activity. Virology 419:107–116. doi: 10.1016/j.virol.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stubbs SH, Hunter OV, Hoover A, Conrad NK. 2012. Viral factors reveal a role for REF/Aly in nuclear RNA stability. Mol Cell Biol 32:1260–1270. doi: 10.1128/MCB.06420-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nekorchuk M, Han Z, Hsieh TT, Swaminathan S. 2007. Kaposi’s sarcoma-associated herpesvirus ORF57 protein enhances mRNA accumulation independently of effects on nuclear RNA export. J Virol 81:9990–9998. doi: 10.1128/JVI.00896-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Majerciak V, Zheng ZM. 2015. KSHV ORF57, a protein of many faces. Viruses 7:604–633. doi: 10.3390/v7020604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kirshner JR, Lukac DM, Chang J, Ganem D. 2000. Kaposi’s sarcoma-associated herpesvirus open reading frame 57 encodes a posttranscriptional regulator with multiple distinct activities. J Virol 74:3586–3597. doi: 10.1128/JVI.74.8.3586-3597.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bello LJ, Davison AJ, Glenn MA, Whitehouse A, Rethmeier N, Schulz TF, Barklie CJ. 1999. The human herpesvirus-8 ORF 57 gene and its properties. J Gen Virol 80:3207–3215. doi: 10.1099/0022-1317-80-12-3207. [DOI] [PubMed] [Google Scholar]
- 24.Malik P, Blackbourn DJ, Clements JB. 2004. The evolutionarily conserved Kaposi’s sarcoma-associated herpesvirus ORF57 protein interacts with REF protein and acts as an RNA export factor. J Biol Chem 279:33001–33011. doi: 10.1074/jbc.M313008200. [DOI] [PubMed] [Google Scholar]
- 25.Palmeri D, Spadavecchia S, Carroll KD, Lukac DM. 2007. Promoter- and cell-specific transcriptional transactivation by the Kaposi’s sarcoma-associated herpesvirus ORF57/Mta protein. J Virol 81:13299–13314. doi: 10.1128/JVI.00732-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pilkington GR, Majerciak V, Bear J, Uranishi H, Zheng ZM, Felber BK. 2012. Kaposi’s sarcoma-associated herpesvirus ORF57 is not a bona fide export factor. J Virol 86:13089–13094. doi: 10.1128/JVI.00606-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han Z, Swaminathan S. 2006. Kaposi’s sarcoma-associated herpesvirus lytic gene ORF57 is essential for infectious virion production. J Virol 80:5251–5260. doi: 10.1128/JVI.02570-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Majerciak V, Pripuzova N, McCoy JP, Gao SJ, Zheng ZM. 2007. Targeted disruption of Kaposi’s sarcoma-associated herpesvirus ORF57 in the viral genome is detrimental for the expression of ORF59, K8alpha, and K8.1 and the production of infectious virus. J Virol 81:1062–1071. doi: 10.1128/JVI.01558-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verma D, Li DJ, Krueger B, Renne R, Swaminathan S. 2015. Identification of the physiological gene targets of the essential lytic replicative Kaposi’s sarcoma-associated herpesvirus ORF57. J Virol 89:1688–1702. doi: 10.1128/JVI.02663-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Massimelli MJ, Majerciak V, Kruhlak M, Zheng ZM. 2013. Interplay between polyadenylate-binding protein 1 and Kaposi’s sarcoma-associated herpesvirus ORF57 in accumulation of polyadenylated nuclear RNA, a viral long noncoding RNA. J Virol 87:243–256. doi: 10.1128/JVI.01693-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Majerciak V, Uranishi H, Kruhlak M, Pilkington GR, Massimelli MJ, Bear J, Pavlakis GN, Felber BK, Zheng ZM. 2011. Kaposi’s sarcoma-associated herpesvirus ORF57 interacts with cellular RNA export cofactors RBM15 and OTT3 to promote expression of viral ORF59. J Virol 85:1528–1540. doi: 10.1128/JVI.01709-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Majerciak V, Yamanegi K, Allemand E, Kruhlak M, Krainer AR, Zheng ZM. 2008. Kaposi’s sarcoma-associated herpesvirus ORF57 functions as a viral splicing factor and promotes expression of intron-containing viral lytic genes in spliceosome-mediated RNA splicing. J Virol 82:2792–2801. doi: 10.1128/JVI.01856-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Majerciak V, Lu M, Li X, Zheng ZM. 2014. Attenuation of the suppressive activity of cellular splicing factor SRSF3 by Kaposi sarcoma-associated herpesvirus ORF57 protein is required for RNA splicing. RNA 20:1747–1758. doi: 10.1261/rna.045500.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang JG, Majerciak V, Uldrick TS, Wang X, Kruhlak M, Yarchoan R, Zheng ZM. 2011. Kaposi’s sarcoma-associated herpesviral IL-6 and human IL-6 open reading frames contain miRNA binding sites and are subject to cellular miRNA regulation. J Pathol 225:378–389. doi: 10.1002/path.2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kang JG, Pripuzova N, Majerciak V, Kruhlak M, Le SY, Zheng ZM. 2011. Kaposi’s sarcoma-associated herpesvirus ORF57 promotes escape of viral and human interleukin-6 from MicroRNA-mediated suppression. J Virol 85:2620–2630. doi: 10.1128/JVI.02144-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharma NR, Majerciak V, Kruhlak MJ, Zheng ZM. 2017. KSHV inhibits stress granule formation by viral ORF57 blocking PKR activation. PLoS Pathog 13:e1006677. doi: 10.1371/journal.ppat.1006677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharma NR, Majerciak V, Kruhlak MJ, Yu L, Kang JG, Yang A, Gu S, Fritzler MJ, Zheng ZM. 10 August 2019. KSHV RNA-binding protein ORF57 inhibits P-body formation to promote viral multiplication by interaction with Ago2 and GW182. Nucleic Acids Res doi: 10.1093/nar/gkz683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang PJ, Shedd D, Miller G. 2005. Two subclasses of Kaposi’s sarcoma-associated herpesvirus lytic cycle promoters distinguished by open reading frame 50 mutant proteins that are deficient in binding to DNA. J Virol 79:8750–8763. doi: 10.1128/JVI.79.14.8750-8763.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Majerciak V, Ni T, Yang W, Meng B, Zhu J, Zheng ZM. 2013. A viral genome landscape of RNA polyadenylation from KSHV latent to lytic infection. PLoS Pathog 9:e1003749. doi: 10.1371/journal.ppat.1003749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Majerciak V, Zheng ZM. 2016. Alternative RNA splicing of KSHV ORF57 produces two different RNA isoforms. Virology 488:81–87. doi: 10.1016/j.virol.2015.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. 2013. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Purushothaman P, Uppal T, Verma SC. 2015. Molecular biology of KSHV lytic reactivation. Viruses 7:116–153. doi: 10.3390/v7010116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davis DA, Rinderknecht AS, Zoeteweij JP, Aoki Y, Read-Connole EL, Tosato G, Blauvelt A, Yarchoan R. 2001. Hypoxia induces lytic replication of Kaposi sarcoma-associated herpesvirus. Blood 97:3244–3250. doi: 10.1182/blood.V97.10.3244. [DOI] [PubMed] [Google Scholar]
- 44.Yu F, Harada JN, Brown HJ, Deng H, Song MJ, Wu TT, Kato-Stankiewicz J, Nelson CG, Vieira J, Tamanoi F, Chanda SK, Sun R. 2007. Systematic identification of cellular signals reactivating Kaposi sarcoma-associated herpesvirus. PLoS Pathog 3:e44. doi: 10.1371/journal.ppat.0030044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang F, Doudna JA. 2017. CRISPR-Cas9 structures and mechanisms. Annu Rev Biophys 46:505–529. doi: 10.1146/annurev-biophys-062215-010822. [DOI] [PubMed] [Google Scholar]
- 46.Shou J, Li J, Liu Y, Wu Q. 2018. Precise and predictable CRISPR chromosomal rearrangements reveal principles of Cas9-mediated nucleotide insertion. Mol Cell 71:498–509. doi: 10.1016/j.molcel.2018.06.021. [DOI] [PubMed] [Google Scholar]
- 47.Yuan F, Gao ZQ, Majerciak V, Bai L, Hu ML, Lin XX, Zheng ZM, Dong YH, Lan K. 2018. The crystal structure of KSHV ORF57 reveals dimeric active sites important for protein stability and function. PLoS Pathog 14:e1007232. doi: 10.1371/journal.ppat.1007232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liang Q, Chang B, Lee P, Brulois KF, Ge J, Shi M, Rodgers MA, Feng P, Oh BH, Liang C, Jung JU. 2015. Identification of the essential role of viral Bcl-2 for Kaposi’s sarcoma-associated herpesvirus lytic replication. J Virol 89:5308–5317. doi: 10.1128/JVI.00102-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roehm PC, Shekarabi M, Wollebo HS, Bellizzi A, He L, Salkind J, Khalili K. 2016. Inhibition of HSV-1 replication by gene editing strategy. Sci Rep 6:23146. doi: 10.1038/srep23146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Diemen FR, Kruse EM, Hooykaas MJ, Bruggeling CE, Schurch AC, van Ham PM, Imhof SM, Nijhuis M, Wiertz EJ, Lebbink RJ. 2016. CRISPR/Cas9-mediated genome editing of herpesviruses limits productive and latent infections. PLoS Pathog 12:e1005701. doi: 10.1371/journal.ppat.1005701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yuen KS, Chan CP, Wong NH, Ho CH, Ho TH, Lei T, Deng W, Tsao SW, Chen H, Kok KH, Jin DY. 2015. CRISPR/Cas9-mediated genome editing of Epstein-Barr virus in human cells. J Gen Virol 96:626–636. doi: 10.1099/jgv.0.000012. [DOI] [PubMed] [Google Scholar]
- 52.Xu X, Fan S, Zhou J, Zhang Y, Che Y, Cai H, Wang L, Guo L, Liu L, Li Q. 2016. The mutated tegument protein UL7 attenuates the virulence of herpes simplex virus 1 by reducing the modulation of alpha-4 gene transcription. Virol J 13:152. doi: 10.1186/s12985-016-0600-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Y, Tang N, Sadigh Y, Baigent S, Shen Z, Nair V, Yao Y. 2018. Application of CRISPR/Cas9 gene editing system on MDV-1 genome for the study of gene function. Viruses 10:279. doi: 10.3390/v10060279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang J, Quake SR. 2014. RNA-guided endonuclease provides a therapeutic strategy to cure latent herpesviridae infection. Proc Natl Acad Sci U S A 111:13157–13162. doi: 10.1073/pnas.1410785111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Delecluse HJ, Bartnizke S, Hammerschmidt W, Bullerdiek J, Bornkamm GW. 1993. Episomal and integrated copies of Epstein-Barr virus coexist in Burkitt lymphoma cell lines. J Virol 67:1292–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bigi R, Landis JT, An H, Caro-Vegas C, Raab-Traub N, Dittmer DP. 2018. Epstein-Barr virus enhances genome maintenance of Kaposi sarcoma-associated herpesvirus. Proc Natl Acad Sci U S A 115:E11379–E11387. doi: 10.1073/pnas.1810128115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Avey D, Tepper S, Li W, Turpin Z, Zhu F. 2015. Phosphoproteomic analysis of KSHV-infected cells reveals roles of ORF45-activated RSK during lytic replication. PLoS Pathog 11:e1004993. doi: 10.1371/journal.ppat.1004993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.He M, Yuan H, Tan B, Bai R, Kim HS, Bae S, Che L, Kim JS, Gao SJ. 2016. SIRT1-mediated downregulation of p27Kip1 is essential for overcoming contact inhibition of Kaposi’s sarcoma-associated herpesvirus transformed cells. Oncotarget 7:75698–75711. doi: 10.18632/oncotarget.12359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hahn AS, Großkopf AK, Jungnickl D, Scholz B, Ensser A. 2016. Viral FGARAT homolog ORF75 of rhesus monkey rhadinovirus effects proteasomal degradation of the ND10 components SP100 and PML. J Virol 90:8013–8028. doi: 10.1128/JVI.01181-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.TerBush AA, Hafkamp F, Lee HJ, Coscoy L. 2018. A Kaposi’s sarcoma-associated herpesvirus infection mechanism is independent of integrins alpha3beta1, alphaVbeta3, and alphaVbeta5. J Virol 92:e00803-18. doi: 10.1128/JVI.00803-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Manzano M, Patil A, Waldrop A, Dave SS, Behdad A, Gottwein E. 2018. Gene essentiality landscape and druggable oncogenic dependencies in herpesviral primary effusion lymphoma. Nat Commun 9:3263. doi: 10.1038/s41467-018-05506-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ruiz JC, Hunter OV, Conrad NK. 2019. Kaposi’s sarcoma-associated herpesvirus ORF57 protein protects viral transcripts from specific nuclear RNA decay pathways by preventing hMTR4 recruitment. PLoS Pathog 15:e1007596. doi: 10.1371/journal.ppat.1007596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tso FY, West JT, Wood C. 2019. Reduction of Kaposi’s sarcoma-associated herpesvirus latency using CRISPR-Cas9 to edit the latency-associated nuclear antigen gene. J Virol 93:e02183-18. doi: 10.1128/JVI.02183-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang L, Guell M, Niu D, George H, Lesha E, Grishin D, Aach J, Shrock E, Xu W, Poci J, Cortazio R, Wilkinson RA, Fishman JA, Church G. 2015. Genome-wide inactivation of porcine endogenous retroviruses (PERVs). Science 350:1101–1104. doi: 10.1126/science.aad1191. [DOI] [PubMed] [Google Scholar]
- 65.Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. 2013. RNA-programmed genome editing in human cells. Elife 2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cho SW, Kim S, Kim JM, Kim JS. 2013. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol 31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 67.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. 2013. RNA-guided human genome engineering via Cas9. Science 339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Choi PS, Meyerson M. 2014. Targeted genomic rearrangements using CRISPR/Cas technology. Nat Commun 5:3728. doi: 10.1038/ncomms4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li J, Shou J, Guo Y, Tang Y, Wu Y, Jia Z, Zhai Y, Chen Z, Xu Q, Wu Q. 2015. Efficient inversions and duplications of mammalian regulatory DNA elements and gene clusters by CRISPR/Cas9. J Mol Cell Biol 7:284–298. doi: 10.1093/jmcb/mjv016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Canver MC, Bauer DE, Dass A, Yien YY, Chung J, Masuda T, Maeda T, Paw BH, Orkin SH. 2014. Characterization of genomic deletion efficiency mediated by clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 nuclease system in mammalian cells. J Biol Chem 289:21312–21324. doi: 10.1074/jbc.M114.564625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kraft K, Geuer S, Will AJ, Chan WL, Paliou C, Borschiwer M, Harabula I, Wittler L, Franke M, Ibrahim DM, Kragesteen BK, Spielmann M, Mundlos S, Lupianez DG, Andrey G. 2015. Deletions, inversions, duplications: engineering of structural variants using CRISPR/Cas in mice. Cell Rep 10:833–839. doi: 10.1016/j.celrep.2015.01.016. [DOI] [PubMed] [Google Scholar]
- 73.Guo Y, Xu Q, Canzio D, Shou J, Li J, Gorkin DU, Jung I, Wu H, Zhai Y, Tang Y, Lu Y, Wu Y, Jia Z, Li W, Zhang MQ, Ren B, Krainer AR, Maniatis T, Wu Q. 2015. CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell 162:900–910. doi: 10.1016/j.cell.2015.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Massimelli MJ, Majerciak V, Kang JG, Liewehr DJ, Steinberg SM, Zheng ZM. 2015. Multiple regions of Kaposi’s sarcoma-associated herpesvirus ORF59 RNA are required for its expression mediated by viral ORF57 and cellular RBM15. Viruses 7:496–510. doi: 10.3390/v7020496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vogt C, Hackmann C, Rabner A, Koste L, Santag S, Kati S, Mandel-Gutfreund Y, Schulz TF, Bohne J. 2015. ORF57 overcomes the detrimental sequence bias of Kaposi’s sarcoma-associated herpesvirus lytic genes. J Virol 89:5097–5109. doi: 10.1128/JVI.03264-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. 2013. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Smith C, Gore A, Yan W, Abalde-Atristain L, Li Z, He C, Wang Y, Brodsky RA, Zhang K, Cheng L, Ye Z. 2014. Whole-genome sequencing analysis reveals high specificity of CRISPR/Cas9 and TALEN-based genome editing in human iPSCs. Cell Stem Cell 15:12–13. doi: 10.1016/j.stem.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ye FC, Zhou FC, Yoo SM, Xie JP, Browning PJ, Gao SJ. 2004. Disruption of Kaposi’s sarcoma-associated herpesvirus latent nuclear antigen leads to abortive episome persistence. J Virol 78:11121–11129. doi: 10.1128/JVI.78.20.11121-11129.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hirt B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol 26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 80.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del AG, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, Banks E, Garimella KV, Altshuler D, Gabriel S, DePristo MA. 2013. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 43:11–33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Okonechnikov K, Conesa A, Garcia-Alcalde F. 2016. Qualimap 2: advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics 32:292–294. doi: 10.1093/bioinformatics/btv566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Majerciak V, Kruhlak M, Dagur PK, McCoy JP Jr, Zheng ZM. 2010. Caspase-7 cleavage of Kaposi sarcoma-associated herpesvirus ORF57 confers a cellular function against viral lytic gene expression. J Biol Chem 285:11297–11307. doi: 10.1074/jbc.M109.068221. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The KSHV BCBL-1 strain sequence is available in GenBank under accession number MN205539.