Thrips-transmitted viruses cause devastating losses to numerous food crops worldwide. For negative-sense RNA viruses that infect plants, the arthropod serves as a host as well by supporting virus replication in specific tissues and organs of the vector. The goal of this work was to identify thrips proteins that bind directly to the viral attachment protein and thus may play a role in the infection cycle in the insect. Using the model plant bunyavirus tomato spotted wilt virus (TSWV), and the most efficient thrips vector, we identified and validated six TSWV-interacting proteins from Frankliniella occidentalis first-instar larvae. Two proteins, an endocuticle structural glycoprotein and cyclophilin, were able to interact directly with the TSWV attachment protein, GN, in insect cells. The TSWV GN-interacting proteins provide new targets for disrupting the viral disease cycle in the arthropod vector and could be putative determinants of vector competence.
KEYWORDS: Bunyavirales, insect vector, orthotospovirus, plant virology, thrips, vector biology, virus-vector interactions
ABSTRACT
The plant-pathogenic virus tomato spotted wilt virus (TSWV) encodes a structural glycoprotein (GN) that, like with other bunyavirus/vector interactions, serves a role in viral attachment and possibly in entry into arthropod vector host cells. It is well documented that Frankliniella occidentalis is one of nine competent thrips vectors of TSWV transmission to plant hosts. However, the insect molecules that interact with viral proteins, such as GN, during infection and dissemination in thrips vector tissues are unknown. The goals of this project were to identify TSWV-interacting proteins (TIPs) that interact directly with TSWV GN and to localize the expression of these proteins in relation to virus in thrips tissues of principal importance along the route of dissemination. We report here the identification of six TIPs from first-instar larvae (L1), the most acquisition-efficient developmental stage of the thrips vector. Sequence analyses of these TIPs revealed homology to proteins associated with the infection cycle of other vector-borne viruses. Immunolocalization of the TIPs in L1 revealed robust expression in the midgut and salivary glands of F. occidentalis, the tissues most important during virus infection, replication, and plant inoculation. The TIPs and GN interactions were validated using protein-protein interaction assays. Two of the thrips proteins, endocuticle structural glycoprotein and cyclophilin, were found to be consistent interactors with GN. These newly discovered thrips protein-GN interactions are important for a better understanding of the transmission mechanism of persistent propagative plant viruses by their vectors, as well as for developing new strategies of insect pest management and virus resistance in plants.
IMPORTANCE Thrips-transmitted viruses cause devastating losses to numerous food crops worldwide. For negative-sense RNA viruses that infect plants, the arthropod serves as a host as well by supporting virus replication in specific tissues and organs of the vector. The goal of this work was to identify thrips proteins that bind directly to the viral attachment protein and thus may play a role in the infection cycle in the insect. Using the model plant bunyavirus tomato spotted wilt virus (TSWV), and the most efficient thrips vector, we identified and validated six TSWV-interacting proteins from Frankliniella occidentalis first-instar larvae. Two proteins, an endocuticle structural glycoprotein and cyclophilin, were able to interact directly with the TSWV attachment protein, GN, in insect cells. The TSWV GN-interacting proteins provide new targets for disrupting the viral disease cycle in the arthropod vector and could be putative determinants of vector competence.
INTRODUCTION
Vector-borne diseases caused by animal- and plant-infecting viruses are some of the most important medical, veterinary, and agricultural problems worldwide (1, 2). The majority of viruses infecting plants are transmitted by arthropods. Understanding the molecular basis of vector competence may lead to the development of novel strategies to interrupt virus transmission by the arthropod vector. Significant progress has been made toward identification of viral determinants of transmission, but the interacting molecules in vectors remain largely elusive. For negative-sense RNA viruses, vector factors that mediate the transmission process have not been well characterized.
Bunyavirales is the largest order of negative-sense RNA viruses; 12 families have been described (https://talk.ictvonline.org/taxonomy/). The Bunyavirales contain insect-borne plant viruses that make up the family Tospoviridae (3–5). Within this family, there are 18 species and several unassigned viruses that share common features of the Orthotospovirus genus, and tomato spotted wilt orthotospovirus is the type species within this genus. Tomato spotted wilt virus (TSWV) is the best characterized orthotospovirus in terms of viral host range, genome organization, and protein functions (6, 7).
TSWV infects both monocotyledonous and dicotyledonous plants, encompassing more than 1,000 plant species worldwide (8). Due to the extremely wide host range, TSWV has caused severe economic losses to various agricultural, vegetable, and ornamental crops. The TSWV virion has a host-derived lipid bilayer into which the two glycoproteins, GN and GC, are inserted. The viral glycoproteins play an essential role in attachment to the thrips gut and fusion of the virus and host membrane (7, 9–11). Virus particles range in size from 80 to 120 nm in diameter, and the viral genome consists of three RNA molecules, L, M, and S.
Although TSWV can be maintained in the laboratory through mechanical inoculation, it is transmitted in nature by insect vectors commonly known as thrips (order Thysanoptera, family Thripidae). Seven species of Frankliniella and two species of Thrips are reported to be the vectors of TSWV (6). Among these species, the western flower thrips, Frankliniella occidentalis Pergande, is the most efficient vector of TSWV and has a worldwide distribution. TSWV is transmitted by thrips vectors in a persistent propagative manner, and the midgut cells and principal salivary glands are two major tissues for TSWV replication (12, 13). Only thrips that acquire virus during the larval stage are inoculative as adults (13–15). Because TSWV GN likely binds to a receptor at the thrips midgut brush border (9–11), we sought to identify thrips proteins that interact directly with GN, the viral attachment protein (16). Using gel overlay assays to identify first-instar larval (L1) proteins that bind to purified virions or GN, we discovered six TSWV-interacting proteins (TIPs) from F. occidentalis. Identification of these proteins using mass spectrometry was followed with secondary assays to validate the interactions and characterize protein expression in larval thrips. Two TIPs, an endocuticle structural glycoprotein and cyclophilin, interacted with GN and colocalized with GN when coexpressed in insect cells. These thrips proteins may play a role in virus entry or mediate other steps in the virus infection process in thrips. These proteins represent the first group of thrips proteins that have been described to bind to TSWV proteins, and these discoveries provide insights toward a better understanding of the molecular interplay between vector and virus.
(The data in this paper were previously presented [17]).
RESULTS
Identification of bound F. occidentalis larval proteins using overlay assays.
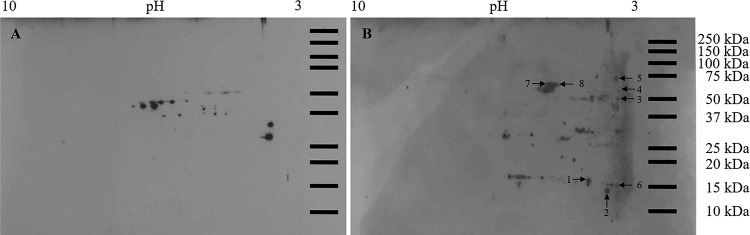
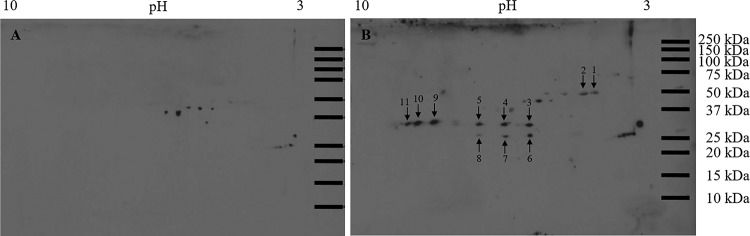
Proteins extracted from first-instar larval bodies were separated by two-dimensional (2D) electrophoresis, and overlay assays were performed with purified TSWV virions or recombinant GN glycoprotein to identify bound thrips proteins. Virion overlays identified a total of eight protein spots (Fig. 1); three occurred consistently in all four biological replicates, while five were present in three replicates. Mass spectrometry and subsequent peptide sequence analysis against a thrips whole-body transcriptome database (Fo Seq) as a reference (18) identified one to four different transcript matches per spot (Table 1). In four cases, the same putative transcript matched peptides in more than one spot. Using recombinant GN glycoprotein, 11 protein spots were detected in both biological replicates of the overlay assay (Fig. 2), and each spot was composed of a single protein (single transcript match) occurring in multiple spots. There were two different GN-interacting proteins represented by the 11 spots (Table 2). For each overlay experiment that was conducted, a control blot was included to account for nonspecific antibody binding showing that the detection levels of positively identified spots well exceeded the background level (Fig. 1 and 2). In an additional gel overlay assay using virus-free plant extract (mock purification) obtained from noninfected Datura stramonium plants, no specific antigen was detected (data not shown).
FIG 1.
Overlay assay using purified virions and F. occidentalis first-instar proteins resolved in two-dimensional gels. Total proteins (150 μg) extracted from pooled noninfected first-instar larvae (0 to 17 h old) of F. occidentalis were resolved by 2D gel electrophoresis and transferred to nitrocellulose membranes. After blocking, membranes were incubated overnight with blocking buffer (negative control) (A) or purified TSWV at 25 μg/ml (B), followed by incubation with polyclonal rabbit anti-TSWV GN antibodies. Only protein spots that consistently bound to purified TSWV in three (spots 1, 2, 4, 6, and 7) and four (spots 3, 5, and 8) biological replicates of the overlay assay were collected from three individual picking gels and subjected to ESI-mass spectrometry for protein identification. Protein spots observed in the no-overlay-control membrane represent nonspecific binding and were not collected for further analysis. Molecular mass (in kilodaltons) is shown on the y axis, and pI (as pH range) is shown on the x axis.
TABLE 1.
Identification of Frankliniella occidentalis larval proteins bound to purified virions of tomato spotted wilt virus in 2D gel overlays
| Spot no. | Fo seq contig matcha | Mascot score (P < 0.05) | % coverageb | No. of matched peptides | Peptide sequence(s) | BLASTX annotation (E < 10−20)c | Conserved motif(s) (E < 10−10) |
|---|---|---|---|---|---|---|---|
| 1 | contig01248 | 279 | 37 | 4 | R.AQQPYQQYLQNQQFQNYQQR.A, R.AAAAPILQYSNDVNPDGSFQYSYQTGDGISAQAAGFT.R, K.DAEAQVVQGSYSYTGPDGVVYTVNYIADENGYR.A, K.ALPYYNQQQATYQQQQAAYQR.P | Endocuticle structural glycoprotein | Chitin_bind_4 |
| CL4854Contig1 | 363 | 29 | 7 | R.VFFDMTVDGQPAGR.I, R.ALCTGEQGFGYK.G, R.VIPNFMCQGGDFTNHNGTGGK.S, R.KFADENFQLK.H, K.HTGPGIMSMANAGPNTNGSQFFITTVK.T, K.TSWLDNKHVVFGSVIEGMDVVK.K, K.HVVFGSVIEGMDVVK.K | Cyclophilin | Cyclophilin_ABH | |
| 2 | contig01248 | 77 | 10 | 1 | K.DAEAQVVQGSYSYTGPDGVVYTVNYIADENGYR.A | Endocuticle structural glycoprotein | Chitin_bind_4 |
| 3 | contig00018 | 196 | 25 | 4 | R.FGGALGGYNLAQTSQYHIQTDEGPER.Y, R.LEDGTVVGTYGWVDADGYLR.L, R.PYYPSSTPAVSLVSSTPR.P, R.PYYPTSTPAVVSSTPR.P | Uncharacterized, similar to cuticular protein | Chitin_bind_4 |
| contig14634 | 134 | 22 | 3 | R.GYISELPGTYDANSNSVIPEYDGIAVTHNGFR.Y, K.AGSFGYVDPFGIR.R, R.VIYYNTSPGSGFQVR.K | Uncharacterized, similar to cuticular protein | none identified | |
| CL4900Contig1 | 113 | 10 | 3 | R.GYISELPGTYDASSNSVIPEYDGIAVTHNGFR.Y, K.AGSFGYVDPFGIR.R, R.VIYYNTSPGSGFQVR.K | Uncharacterized, similar to cuticular protein | Chitin_bind_4 | |
| 4 | CL1591Contig1 | 76 | 5 | 1 | K.QESVYTAAQPAISTYK.K | Flexible cuticular protein | Chitin_bind_4 |
| 5 | CL1591Contig1 | 89 | 5 | 1 | K.QESVYTAAQPAISTYK.K | Flexible cuticular protein | Chitin_bind_4 |
| 6 | CL4854Contig1 | 615 | 30 | 9 | R.VFFDMTVDGQPAGR.I, R.ALCTGEQGFGYK.G, R.VIPNFMCQGGDFTNHNGTGGK.S, R.KFADENFQLK.H, K.FADENFQLK.H, K.FADENFQLKHTGPGIMSMANAGPNTNGSQFFITTVK.T, K.HTGPGIMSMANAGPNTNGSQFFITTVK.T, K.HVVFGSVIEGMDVVK.K, K.KVVVADCGQLS | Cyclophilin | Cyclophilin_ABH |
| 7 | CL4706Contig1 | 554 | 47 | 17 | R.GNPTVEVDLVTELGLFR.A, R.AAVPSGASTGVHEALELR.D, K.AIDNVNNIIAPELIK.S, K.EIDELMLK.L, K.LGANAILGVSLAVCK.A, K.HIADLAGNTNIILPTPAFNVINGGSHAGNK.L, K.LAMQEFMILPTGASSFK.E, K.FGLDSTAVGDEGGFAPNILNNK.E, K.EGLTLIIDAIAK.A, K.VEIGMDVAASEFYK.D, K.VEIGMDVAASEFYKDGQYDLDFK.N, K.DGQYDLDFKNPNSDK.S, K.LTDLYMEFIK.E, K.EFPMVSIEDPFDQDHWDAWTTITGK.T, K.TNIQIVGDDLTVTNPK.R, K.VNQIGSVTESIQAHLLAK.K, R.SGETEDTFIADLVVGLSTGQIK.T | Enolase | Metal_binding |
| contig12136 | 96 | 24 | 2 | R.QGDVVQGSYSLVEPDGSR.R, R.TVEYTADPVNGFNAVVHK.D | Cuticular protein | Chitin_bind_4 | |
| contig14594 | 102 | 3 | 1 | R.TVDYTADPVNGFNAVVR.K | Nuclear cap-binding protein | RRM_NCPB2d, Chitin_bind_4 |
|
| CL504Contig1 | 113 | 19 | 2 | K.AAVAVDTDYDPNPSYNYAYDIHDSLTGDAK.S, R.TVEYTADPVNGFNAVVHK.E | Cuticular protein | Chitin_bind_4 | |
| 8 | CL4706Contig1 | 407 | 43 | 15 | R.GNPTVEVDLVTELGLFR.A, R.AAVPSGASTGVHEALELR.D, K.AIDNVNNIIAPELIK.S, K.HIADLAGNTNIILPTPAFNVINGGSHAGNK.L, K.LAMQEFMILPTGASSFK.E, K.FGLDSTAVGDEGGFAPNILNNK.E, K.EGLTLIIDAIAK.A, K.VEIGMDVAASEFYK.D, K.VEIGMDVAASEFYKDGQYDLDFK.N, K.DGQYDLDFKNPNSDK.S, K.LTDLYMEFIK.E, K.EFPMVSIEDPFDQDHWDAWTTITGK.T, K.TNIQIVGDDLTVTNPK.R, K.VNQIGSVTESIQAHLLAK.K, R.SGETEDTFIADLVVGLSTGQIK.T | Enolase | Metal_binding |
| CL504Contig1 | 99 | 12 | 1 | K.AAVAVDTDYDPNPSYNYAYDIHDSLTGDAK.S | Cuticular protein | Chitin_bind_4 |
De novo-assembled contigs from F. occidentalis transcriptome derived by Roche 454/Sanger EST library hybrid (18).
Highest percent coverage obtained among the three picking gels used to collect protein spots for identification using ESI-mass spectrometry.
NCBI BLASTX search of the nonredundant protein database with the matching Fo nucleotide sequence contig query.
Chimeric contig (= ambiguous annotation); domains occur in opposite orientation on different ORFs.
FIG 2.
Overlay assay using recombinant GN and F. occidentalis first-instar proteins resolved in two-dimensional gels. Total proteins (150 μg) extracted from pooled healthy first-instar larvae (0 to 17 h old) of F. occidentalis were resolved by 2D gel electrophoresis and transferred to nitrocellulose membranes. The membranes were incubated overnight with blocking buffer (negative control) (A) or recombinant TSWV GN (3.5 μg/ml) (B). Using the polyclonal rabbit anti-TSWV GN, protein spots that consistently bound to the recombinant TSWV GN in two (spots 1 through 11) biological replicates of the overlay assay were collected from two individual picking gels and subjected to ESI-mass spectrometry for protein identification. Protein spots observed in the no-overlay-control membrane represent nonspecific binding and were not collected for further analysis. Molecular mass (in kilodaltons) is shown on the y axis, and pI (as pH range) is shown on the x axis.
TABLE 2.
Identification of Frankliniella occidentalis larval proteins bound to recombinant GN in 2D gel overlays
| Spot no. | Fo seq contig matcha | Mascot score (P < 0.05) | % coverageb | No. of matched peptides | Peptide sequence(s) | BLASTX annotation (E < 10−20)c | Conserved motif (E < 10−10) |
|---|---|---|---|---|---|---|---|
| 1 | CL4310Contig1 | 595 | 27 | 12 | R.AAELSSILEER.I, K.NIQADEMVEFSSGLK.G, K.GMALNLEPDNVGIVVFGNDK.L, K.GMALNLEPDNVGIVVFGNDKLIK.E, R.TGAIVDVPVGDDLLGR.V, K.TALAIDTIINQQR.F, K.YTIIVAATASDAAPLQYLAPYSGCAMGEYFR.D, K.HALIIYDDLSK.Q, R.EAYPGDVFYLHSR.L, R.EVAAFAQFGSDLDAATQQLLNR.G, K.QGQYVPMAIEEQVAVIYCGVR.G, K.IVTDFLASFNAASK | Mitochondrial ATP synthase α-subunit | AtpA |
| 2 | CL4310Contig1 | 302 | 12 | 5 | K.GMALNLEPDNVGIVVFGNDK.L, R.TGAIVDVPVGDDLLGR.V, R.VVDALGDAIDGK.G, K.HALIIYDDLSK.Q, K.IVTDFLASFNAASK | Mitochondrial ATP synthase α-subunit | AtpA |
| 3 | CL4382Contig1 | 633 | 43 | 9 | R.SSVVSQSVPVVSK.T, K.SVPQYQQQYQTVSQYQSVPQYQQQVVVK.S, K.SVPQYQQQVVVK.S, K.SAPVYSQVHHVVEQQAAPVLLR.H, R.TAFVPQYDSVSVSASAQPK.Y, K.ILSQVQEFDPAGIYR.V, R.VNFQTENGIQSAETGSVK.D, R.ASGAHLPQVPEEIQR.S, R.SLELNAAQPQKYDQDGNLVSQF | Endocuticle structural glycoprotein | Chitin_bind_4 |
| 4 | CL4382Contig1 | 137 | 15 | 3 | R.SSVVSQSVPVVSK.T, K.SVPQYQQQYQTVSQYQSVPQYQQQVVVK.S, R.VNFQTENGIQSAETGSVK.D | Endocuticle structural glycoprotein | Chitin_bind_4 |
| 5 | CL4382Contig1 | 169 | 22 | 6 | R.SSVVSQSVPVVSK.T, K.SVPQYQQQVVVK.S, R.VNFQTENGIQSAETGSVK.D, R.ASGAHLPQVPEEIQR.S, R.AAAEHGVAIVCPDTSPR.G, K.ACQAVNMPVVLQMR.E | Endocuticle structural glycoprotein | Chitin_bind_4 |
| 6 | CL4382Contig1 | 1,154 | 53 | 11 | R.SSVVSQSVPVVSK.T, K.SVPQYQQQYQTVSQYQSVPQYQQQVVVK.S, K.SVPQYQQQVVVK.S, K.SAPVYSQVHHVVEQQAAPVLLR.H, R.HVEQEIPAYQSVQHVHQPVYQSVQHVAAHHVAAPVVSR.T, R.TAFVPQYDSVSVSASAQPK.Y, K.ILSQVQEFDPAGIYR.V, R.VNFQTENGIQSAETGSVK.D, R.VNFQTENGIQSAETGSVKDIQAK.D, R.ASGAHLPQVPEEIQR.S, R.SLELNAAQPQKYDQDGNLVSQF | Endocuticle structural glycoprotein | Chitin_bind_4 |
| 7 | CL4382Contig1 | 413 | 74 | 8 | R.SSVVSQSVPVVSK.T, K.SVPQYQQQYQTVSQYQSVPQYQQQVVVK.S, K.SVPQYQQQVVVK.S, R.TAFVPQYDSVSVSASAQPK.Y, R.VNFQTENGIQSAETGSVK.D, R.VNFQTENGIQSAETGSVKDIQAK.D, R.ASGAHLPQVPEEIQR.S, R.SLELNAAQPQKYDQDGNLVSQF | Endocuticle structural glycoprotein | Chitin_bind_4 |
| 8 | CL4382Contig1 | 355 | 29 | 7 | R.SSVVSQSVPVVSK.T, K.SVPQYQQQYQTVSQYQSVPQYQQQVVVK.S, K.SVPQYQQQVVVK.S, R.VNFQTENGIQSAETGSVK.D, R.VNFQTENGIQSAETGSVKDIQAK.D, R.ASGAHLPQVPEEIQR.S, R.SLELNAAQPQKYDQDGNLVSQF | Endocuticle structural glycoprotein | Chitin_bind_4 |
| 9 | CL4382Contig1 | 155 | 5 | 2 | R.VNFQTENGIQSAETGSVK.D, R.VNFQTENGIQSAETGSVKDIQAK.D | Endocuticle structural glycoprotein | Chitin_bind_4 |
| 10 | CL4382Contig1 | 130 | 4 | 1 | R.VNFQTENGIQSAETGSVK.D | Endocuticle structural glycoprotein | Chitin_bind_4 |
| 11 | CL4382Contig1 | 150 | 13 | 7 | R.SSVVSQSVPVVSK.T, K.SVPQYQQQYQTVSQYQSVPQYQQQVVVK.S, K.SVPQYQQQVVVK.S, R.VNFQTENGIQSAETGSVK.D, R.TAFVPQYDSVSVSASAQPK.Y, R.VNFQTENGIQSAETGSVK.D, R.VNFQTENGIQSAETGSVKDIQAK.D, R.ASGAHLPQVPEEIQR.S | Endocuticle structural glycoprotein | Chitin_bind_4 |
De novo-assembled contigs from F. occidentalis transcriptome derived by Roche 454/Sanger EST library hybrid (18).
Highest percent coverage obtained among the three picking gels used to collect protein spots for identification using ESI-mass spectrometry.
NCBI BLASTX search of the nonredundant protein database with the matching Fo nucleotide sequence contig query.
Annotation of six candidate TIPs.
Our stringent sequence-filtering criteria retained four different virion-interacting proteins (for endocuticle structural glycoprotein [endoCP-V; contig01248, GenBank accession no. MH884756]; for cuticular protein [CP-V; CL4900Contig1, GenBank accession no. MH884758]; for cyclophilin [CL4854Contig1, GenBank accession no. MH884760]; and for enolase [CL4706Contig1, GenBank accession no. MH884759]) (Table 1) and two GN-interacting proteins (mitochondrial ATP synthase α [mATPase alpha; CL4310Contig1, GenBank accession no. MH884761] and endocuticle structural glycoprotein [endoCP-GN; CL4382Contig1, GenBank accession no. MH884757]) (Table 2) to move forward to validation and biological characterization. Collectively, these six protein candidates are referred to as TSWV-interacting proteins, or TIPs, and their putative identifications and sequence features are shown in Table 3. BLASTP analysis of the predicted, longest complete open reading frames (ORFs) confirmed their annotations and putative sequence homology to proteins in other insects. The three cuticle-associated TIPs (endoCP-GN, endoCP-V, and CP-V) contained predicted signal peptide sequences, indication of secreted proteins, and a chitin-binding domain (CHB4). Pairwise alignments (BLASTP) between the translated ORFs of the three cuticle TIPs and the six other gel overlay-resolved CPs or endoCPs revealed that the amino acid identities ranged from 53% to 67%, covering 30% to 49% of the queries, with E values ranging from 2.4 × 10−2 to 3.6 × 10−24. The only exception was the CP-V and contig00018 alignment, which appeared to be 100% identical along the entire length of contig00018 (E = 2.6 × 10−162) (data not shown). The other three TIPs (cyclophilin, enolase, and mATPase) contained conserved domains characteristic of their respective protein annotations (Table 3).
TABLE 3.
Final candidate list of six from larval Frankliniella occidentalis to move forward to validation and biological characterizationa
| Putative protein (abbreviation) | Fo seq contig matchb | ORF length (nucleotides/amino acids)c | Signal peptide (amino acid positions)d | Conserved domain(s)e
|
Top GenBank match accession no., protein (species) (% coverage, % identity, E value)f | ||
|---|---|---|---|---|---|---|---|
| Name | Positions (amino acids) | E value | |||||
| Cuticular protein-V (CP-V) | CL4900Contig1 | 1,302/434 | 1–18 | Chitin_bind_4 | 47–92 | 5.7 × 10−6 | XP_017786818.1, PREDICTED: cell surface glycoprotein 1 (Nicrophorus vespilloides) (94, 44, 1 × 10−85) |
| Endocuticle structural glycoprotein-GN (endoCP-GN) | CL4382Contig1 | 852/283 | 1–15 | Chitin_bind_4 | 190–246 | 7.5 × 10−11 | XP_018334183.1, endocuticle structural glycoprotein SgAbd-2-like (Agrilus planipennis) (33, 53, 4 × 10−22) |
| Endocuticle structural glycoprotein-V (endoCP-V) | contig01248 | 522/173 | 1–17 | Chitin_bind_4 | 63–119 | 1.4 × 10−18 | XP_022906571.1, endocuticle structural glycoprotein SgAbd-2-like (Onthophagus taurus) (63, 58, 2 × 10−35) |
| Cyclophilin (peptidyl-prolyl cis-trans isomerase) | CL4854Contig1 | 618/205 | None | cyclophilin_ABH_like | 44–202 | 2.3 × 10−120 | XP_019753975.1, PREDICTED: peptidyl-prolyl cis-trans isomerase (Dendroctonus ponderosae) (99, 75, 2 × 10−109) |
| PpiB | 57–196 | 2.6 × 10−63 | |||||
| Pro_isomerase | 47–201 | 1.2 × 10−62 | |||||
| Enolase | CL4706Contig1 | 1,302/433 | None | PLN00191 | 3–433 | 0 | XP_019767728.1, PREDICTED: enolase-like (Dendroctonus ponderosae) (99, 87, 0) |
| Enolase | 5–417 | 0 | |||||
| Eno | 6–431 | 0 | |||||
| Enolase_C | 143–433 | 0 | |||||
| Mitochondrial ATP synthase α-subunit | CL4310Contig1 | 1,665/554 | None | PRK09281 | 45–551 | 0 | XP_023718907.1, mitochondrial ATP synthase α-subunit, (Cryptotermes secundus) (99, 89, 0) |
| AtpA | 45–553 | 0 | |||||
| F1_ATPase_alpha | 135–416 | 0 | |||||
| ATP-synt_ab | 190–413 | 6.1 × 10−117 | |||||
Sequences were deposited into the National Center for Biotechnology Information (NCBI) GenBank database with the following accession numbers: CP-V, MH884758; endoCP-GN, MH884757; endoCP-V, MH884756; cyclophilin, MH884760; enolase, MH884759; and mitochondrial ATP synthase α, MH884761.
De novo-assembled contigs from F. occidentalis transcriptome derived by Roche 454/Sanger EST library hybrid (18).
Prediction by NCBI ORF finder (https://www.ncbi.nlm.nih.gov/orffinder/).
Prediction by SignalP (http://www.cbs.dtu.dk/services/SignalP/).
Prediction by NCBI Batch Web CD-search tool (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi, CDSEARCH/cdd v3.16); E value cutoff, 10−5. Only specific hits are shown.
BLASTP search of NCBI nr protein database against TIP ORF as of 6 August 2018; top match indicates highest alignment (maximum) score.
Classification of cuticular TIPs.
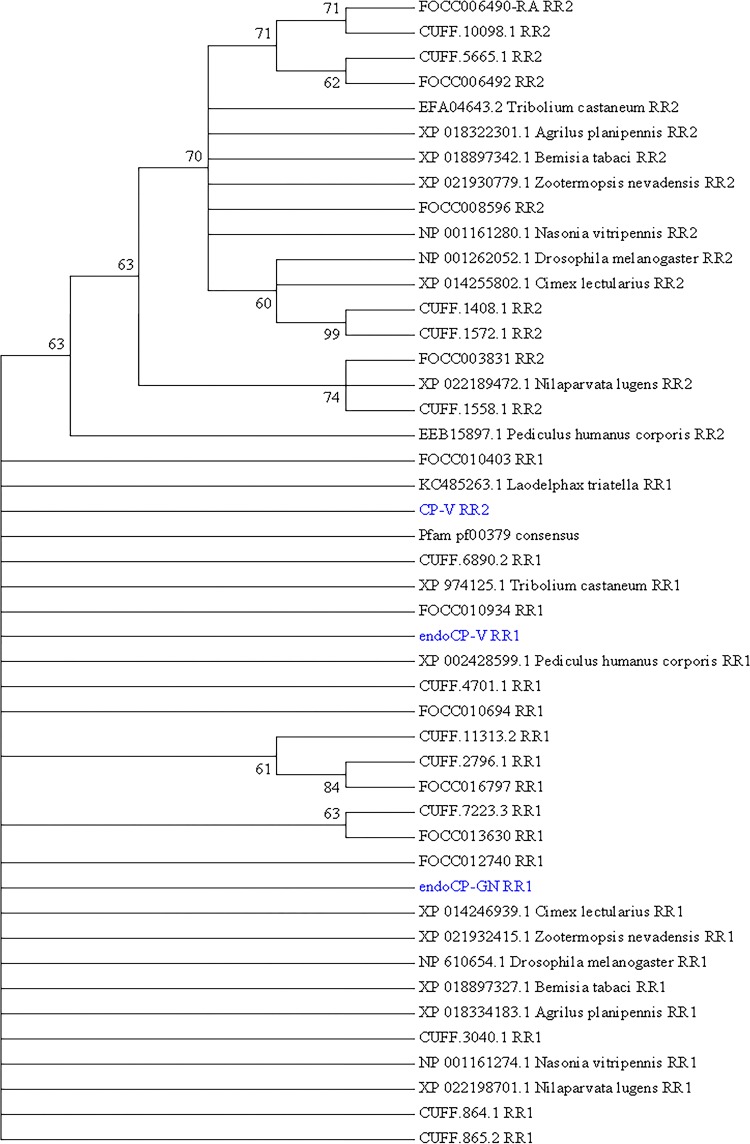
All three cuticular TIPs were classified as members of the cuticle protein-R&R (CPR) family (19) based on the occurrence of one RR extended consensus CHB4, with both endoCP-GN (E = 4 × 10−18) and endoCP-V (E = 1 × 10−26) predicted to belong to the RR1 group, and CP-V weakly supported (E = 5 × 10−6) to belong to the RR2 group of CPRs. All three sequences were phylogenetically placed into the RR1 major clade (Fig. 3) in relation to other F. occidentalis CPRs previously found to be downregulated in TSWV-infected first-instar larvae (20) and CPRs of other insect species.
FIG 3.
Phylogenetic analysis of the extended R&R (Rebers and Riddiford, CPR-RR1 and CPR-RR2) consensus, a conserved chitin-binding motif (Chitin_bind_4 [CHB4]), of the three cuticle-associated TSWV-interacting proteins (TIPs) in first-instar larvae of Frankliniella occidentalis (cuticle protein-V [CP-V], endocuticle structural glycoprotein-V [endoCP-V], and endocuticle structural glycoprotein-GN [endoCP-GN]). The analysis involved 46 sequences, as follows: the three cuticle TIPs (in blue), the “gold standard” Pfam database extended R&R consensus sequence (pf00379), 19 insect orthologous sequences obtained from NCBI GenBank (accession numbers are present), and 23 structural CPs and endoCPs (translated transcripts, designated with FOCC or CUFF identifiers) previously reported to be differentially abundant in TSWV-infected first-instar larva of F. occidentalis (20). All positions with less than 30% site coverage in the multiple alignment were eliminated, resulting in 74 amino acid positions in the final data set. Evolutionary history was inferred by the maximum likelihood (ML) method based on the Jones-Taylor-Thornton (JTT) matrix-based model, and a discrete gamma distribution was used to model the variation among sites. The bootstrap consensus tree (500 replicates) was generated by the ML algorithm, and branches corresponding to partitions reproduced in less than 60% bootstrap replicates were collapsed. The numbers shown next to branches indicate the percentage of replicate trees in which the associated taxa (sequences) clustered together in the bootstrap test. The analysis was performed with MEGA7 (73).
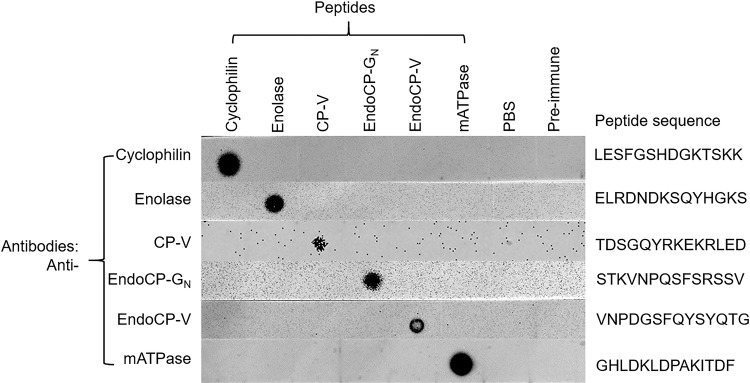
TIP antibodies bind the target peptides.
The antibodies bound to their TIP peptides in dot blot assays (Fig. 4). The affinity of each antibody to its cognate TIP peptide varied. There was high binding affinity for all antibodies tested, with the exception of CP-V, which we subsequently improved by increasing the peptide concentration and subsequent antibody incubation and chemiluminescence development times. This result demonstrates the reactivity of the TIP-peptide antibodies that were used in subsequent localization experiments with first-instar thrips larvae.
FIG 4.
Confirmation of binding specificity for antibodies produced against TSWV-interacting protein (TIP) (peptides) by dot blot analysis. Peptide antigens were diluted to 100 μg/ml (for cyclophilin, enolase, endoCP-GN, endoCP-V, and mATPase), and 2.5 mg/ml (for CP-V), and 2 μl of each peptide was used for each test. PBS buffer and preimmune serum (500,000 × dilution) were used as controls. All six diluted peptides and two controls were loaded onto six nitrocellulose membrane strips. Each strip was initially incubated with the homologous primary antibody (0.5 μg/ml, generated in mice), followed by incubation with anti-mouse-HRP (1:5,000 dilution). Each membrane strip was developed independently.
In vivo localization of TIPs in F. occidentalis in midguts and salivary glands.
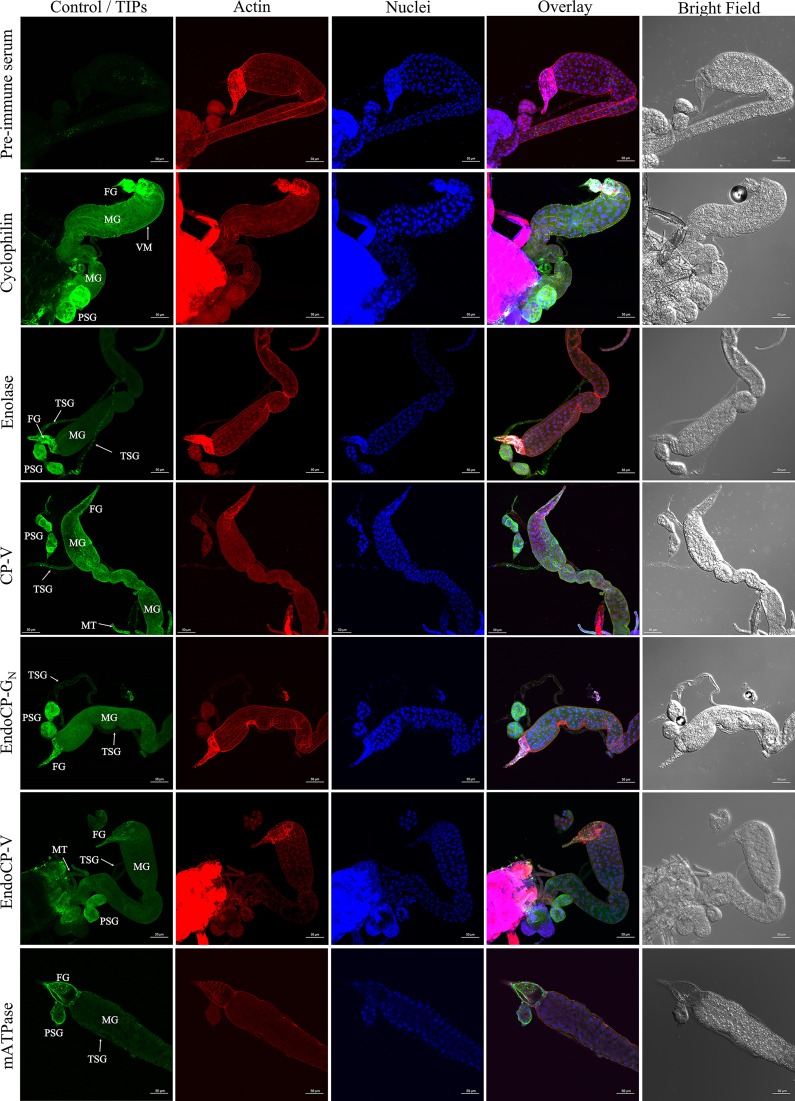
Specific antibodies raised against each confirmed TIP were used in immunolabeling experiments to localize protein expression in L1 tissues in vivo. Visualization by confocal microscopy revealed that all six TIPs were primarily localized at the foregut (esophagus), midgut (epithelial cells and visceral muscle), salivary glands (including both principal and tubular salivary glands), and Malpighian tubules (Fig. 5), and this was the case in 100% of the dissected tissues treated with TIP-specific antibodies. It was difficult to completely dissect and separate the hindgut from the carcass without damaging the tissue; therefore, the localization of TIPs in the hindgut was unclear. For each experimental replicate and unique antibody, controls of secondary antibody only and preimmune serum plus secondary antibody were included and visualized by confocal microscopy. The confocal laser settings (power and percent/gain) were adjusted to remove any background fluorescence observed with preimmune serum for each TIP, as they showed slightly higher background than did the secondary antibody control. The bright-field and merged images of these controls, depicting actin and nucleus labeling, are shown in Fig. 5 (first row).
FIG 5.
In situ detection of TSWV-interacting proteins (TIPs) in first-instar larvae of F. occidentalis. Synchronized first-instar larvae (0 to 17 h old) were kept on a 7% sucrose solution for 3 h to clean their guts from plant tissues. These larvae were then dissected and immunolabeled using specific antibodies against each TIP, as indicated. Thrips tissues incubated with preimmune mouse serum are depicted here. Confocal microscopy detection of green fluorescence (Alexa Fluor 488) represents the localization of each TIP, red represents Alexa Fluor 594-labeled actin, and blue represents DAPI-labeled nuclei. TIPs were mainly localized at the foregut (FG), midgut (MG), which includes epithelial cells and visceral muscle (VM), principal salivary glands (PSG), tubular salivary glands (TSG), and Malpighian tubules (MT). All scale bars = 50 μm.
Validation of interactions between TIPs and TSWV GN using bimolecular fluorescence complementation.
We conducted a bimolecular fluorescence complementation (BiFC) analysis of TIPs and GN in planta due to the importance of plant-expressed proteins in acquisition of virus by thrips vectors. The viral proteins that first bind and enter insect midgut cells would be those expressed and assembled into virions in the plant. The GFP-TIP fusions displayed distinct cellular localization patterns when expressed in plants (Fig. 6). Cyclophilin and mATPase appeared to be localized to the nuclei and along the cell periphery, while enolase and CP-V were present in the membranes surrounding the nuclei as well as the cell periphery. Both endoCP-GN and endoCP-V had a punctate appearance outside the nucleus. All three cuticular TIPs (CP-V, endoCP-GN, and endoCP-V) formed small bodies that appeared to be moving along the endomembranes of the cell, consistent with secretion. All TIPs were colocalized with the endoplasmic reticulum (ER) marker; however, none appeared to be colocalized with the Golgi marker (data not shown).
FIG 6.
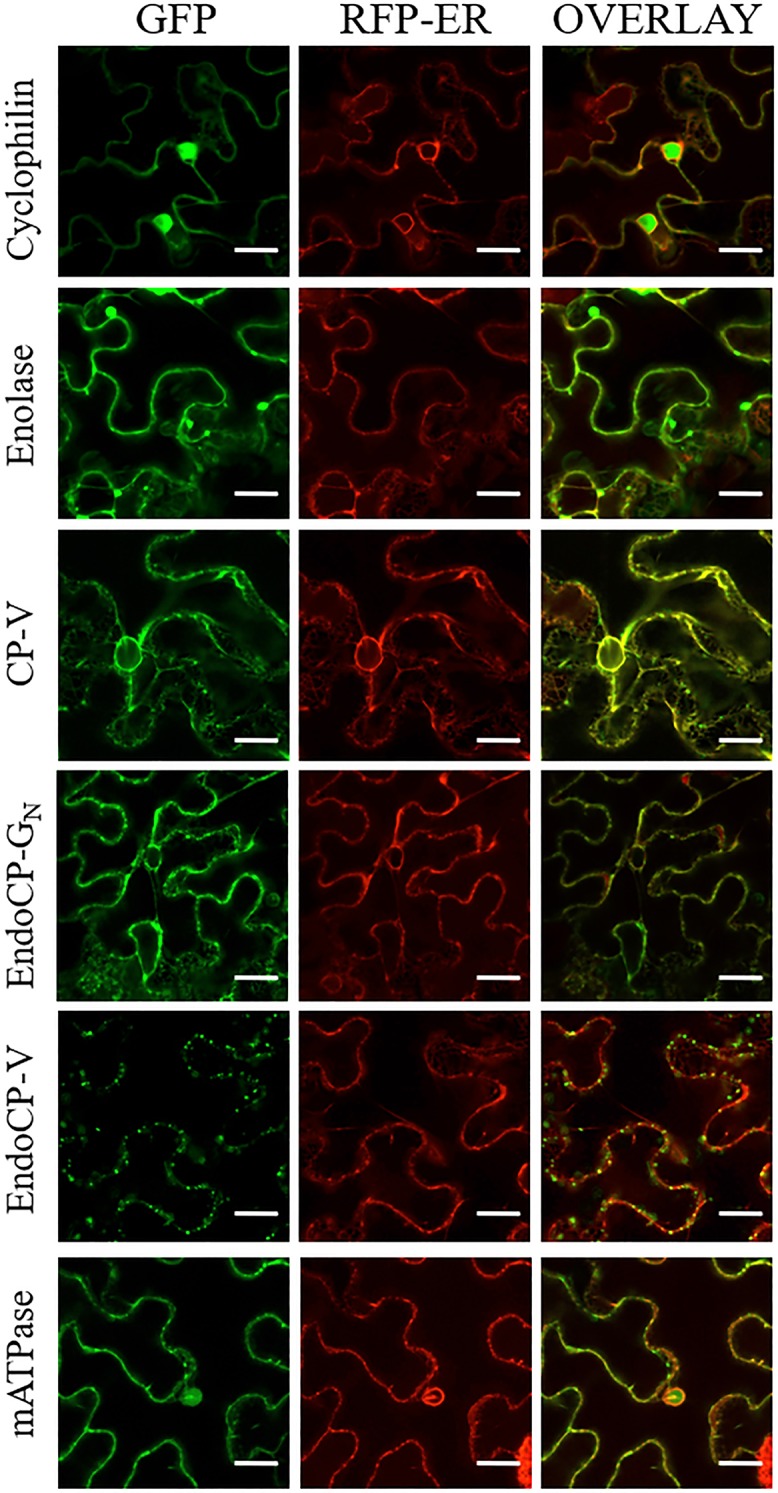
Localization of TSWV-interacting proteins (TIPs) fused to green fluorescent protein (GFP) in Nicotiana benthamiana. Plants transgenic for an RFP-ER marker were infiltrated with Agrobacterium tumefaciens strain LBA 4404 suspensions of TIP constructs. Each row indicates the specific TIP-GFP fusion in relation to the RFP-ER marker. The columns are as follows, from left the right: GFP channel, RFP channel, and the overlay between the two channels. All scale bars = 20 μm.
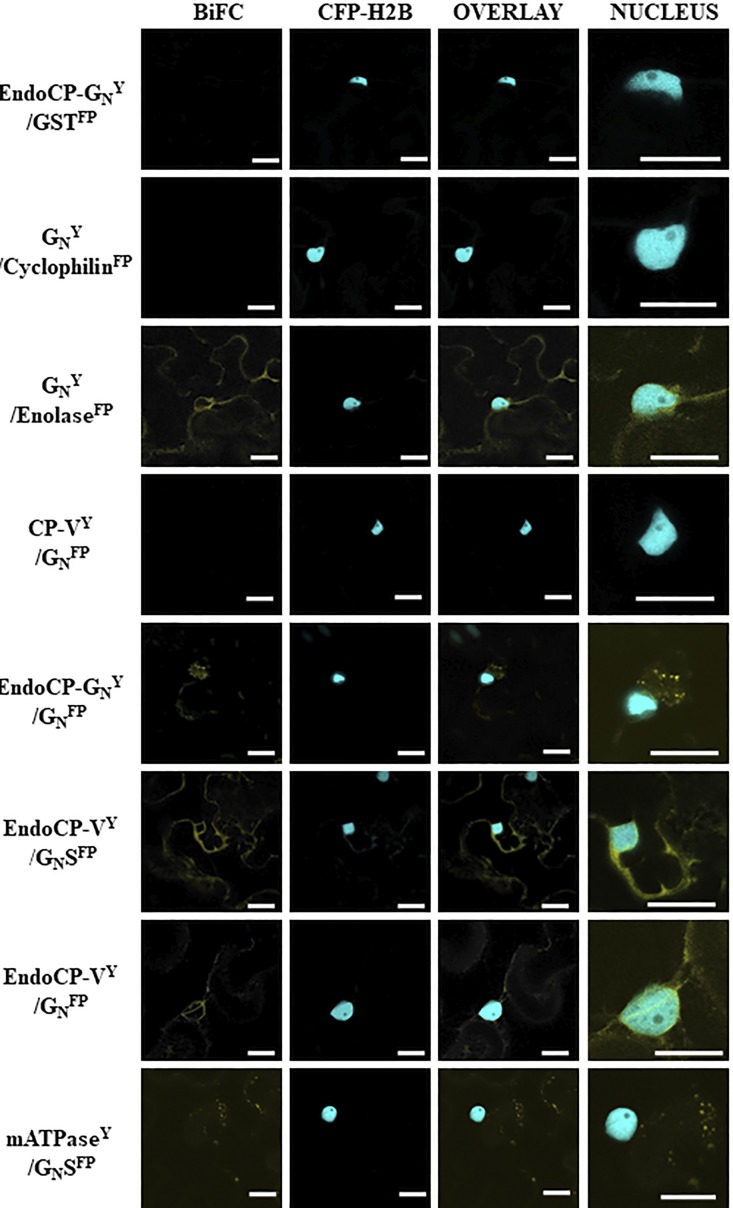
BiFC analysis validated the TSWV-TIPs interactions identified in the overlay assays between virions and GN and enolase, mATPase, endoCP-GN, and endoCP-V (Fig. 7). We used the soluble form of the viral glycoprotein (GN-S) and the insoluble form with the transmembrane domain and cytoplasmic tail in BiFC assays. The insoluble form of GN interacted with enolase, endoCP-GN, and endoCP-V. The proposed ectodomain of GN-S interacted with mATPase and endoCP-V. All of the BiFC interactions were detected in the membranes surrounding the nuclei and at the cell periphery, generally consistent with the localization patterns of the GFP-fused TIPs, as described for the localization experiment above (Fig. 6).
FIG 7.
Confirmation of interactions between TSWV proteins and TSWV-interacting proteins (TIPs) using bimolecular fluorescence complementation (BiFC) in Nicotiana benthamiana. Plants transgenic for a nuclear marker fused to cyan fluorescent protein (CFP-H2B) were infiltrated with suspensions of Agrobacterium tumefaciens transformed with plasmids encoding the GN protein (full-length or soluble form [GN-S]) and TIP proteins (endoCP-GN, cyclophilin, enolase, CP-V, endoCP-V, and mATPase) fused to either the amino or carboxy terminus of yellow fluorescent protein (YFP). The designation of Y indicates this is the N-terminal half of YFP, and FP represents the C-terminal half of YFP. The Y or FP position in the name indicates that all are carboxy-terminal fusions to the protein of interest. The positive interactors are seen by fluorescence of YFP in images shown in the BiFC column. The CFP-H2B column is indicated to give cellular reference, and the overlay between the two is also shown. The final column is the nucleus enlarged to show detail of the interacting TIPs within the cellular context. The first row is a representative negative control, including a TIP and glutathione S-transferase (all thrips and virus proteins were tested with the negative control to rule out nonspecific interactions). All scale bars = 20 μm.
Validation of interactions between TIPs and TSWV GN using split-ubiquitin membrane-based yeast two-hybrid assays.
The split-ubiquitin membrane-based yeast two-hybrid (MbY2H) system was used to validate the gel overlay interactions between the six candidate TIPs and TSWV glycoprotein GN. The presence of a transmembrane domain near the C terminus of TSWV GN makes the MbY2H system the best choice for validation of TSWV GN interactions with the candidate TIPs. The interaction between GN and endoCP-GN was consistent and strong based on the number of colonies growing on SD/–Ade/–His/–Leu/–Trp quadruple-dropout (QDO) medium, i.e., more than 1,000 colonies on all QDO plates for all three replicates (Fig. 8A), and this interaction was confirmed by a β-galactosidase assay. We detected a consistent but weak interaction (average of 15 colonies) between GN and cyclophilin, and seven of nine colonies tested by the β-galactosidase assay were positive. The remaining four TIPs showed no interaction with GN using MbY2H. Contrary to the MbY2H results, GN was determined to interact with enolase and endoCP-V in BiFC experiments. The steric constraints imposed by the position (C or N terminus) of the reporter in the MbY2H (ubiquitin half) and BiFC (yellow fluorescent protein [YFP] half) systems in yeast versus plants cells, respectively, may explain the contrasting interactions observed in these assays.
FIG 8.
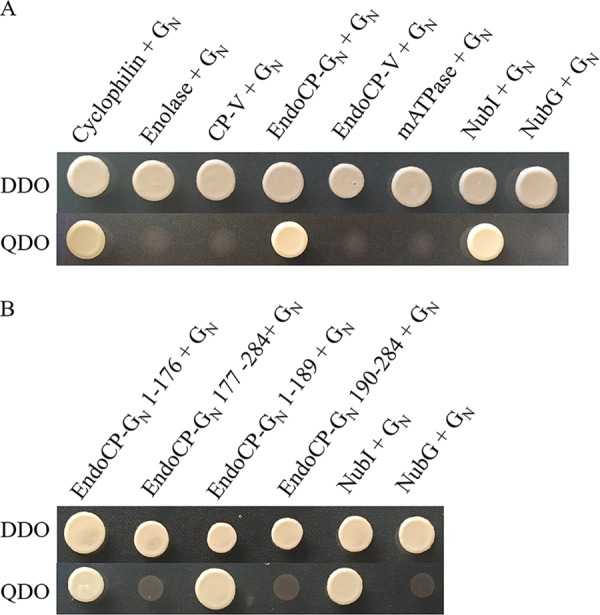
Validation of interactions between GN and TSWV-interacting proteins (TIPs) and identification of the interacting domain of endoCP-GN using a split-ubiquitin membrane-based yeast two hybrid (MbY2H) assay. (A) Interactions between GN and six TIPs. GN was expressed as GN-Cub, and TIPs were expressed as NubG-TIPs using MbY2H vectors. (B) Interactions between TSWV GN and different regions of endoCP-GN. EndoCP-GN was expressed as either the N-terminal domain (amino acids 1 to 176 and 1 to 189) that includes the nonconserved region or the C-terminal region (amino acids 177 to 284 and 190 to 284) that includes the conserved Chitin_bind_4 motif (CHB4) of endoCP-GN. Interactions between GN-Cub and NubI and between GN-Cub and NubG were used as positive and negative controls, respectively, for all MbY2H assays. Cotransformation of pTSU2-APP and pNubG-Fe65 into NYM51 was used as another positive control (data not shown). DDO, yeast double-dropout (SD/−Leu/−Trp) medium; QDO, yeast quadruple-dropout (SD/−Ade/−His/−Leu/−Trp) medium.
The nonconserved region of endoCP-GN binds TSWV GN.
Given the role of GN as the viral attachment protein in the larval thrips midgut epithelium (9–11) and the confirmed direct interaction between endoCP-GN and TSWV GN, there was interest in broadly identifying the amino acid region in the endoCP-GN sequence that binds GN. We hypothesized that the nonconserved region of endoCP-GN (N-terminal region up to amino acid 176 or 189) and not the CHB4 motif might play an important role in the interaction with TSWV GN. Using the MbY2H system, it was determined that the nonconserved region of the endoCP-GN sequence had as strong of an interaction with TSWV GN as the complete endoCP-GN sequence (Fig. 8B), with more than 500 colonies on each QDO plate for each experimental replicate, while the predicted CHB4 motif alone (amino acid positions 190 to 284) or CHB4 plus additional N-terminal amino acids (positions 177 to 284) did not show any interactions. The nonconserved endoCP-GN sequence had no matches with any reported sequences in the NCBI database.
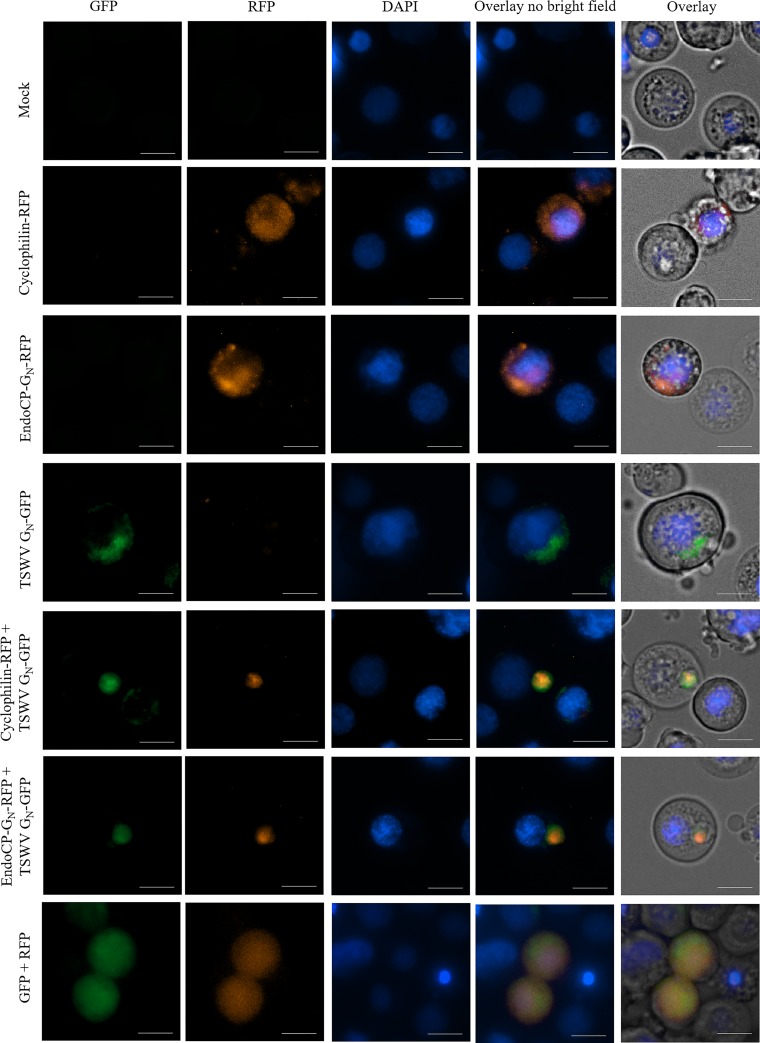
Cyclophilin and endoCP-GN colocalized with TSWV GN in insect cells.
The cellular localization of the two robust TIPs (cyclophilin or endoCP-GN fused to red fluorescent protein [RFP]) was examined for each TIP singly or together with TSWV GN-GFP in Sf9 insect cells (Fig. 9). When either RFP-fused protein was expressed alone in Sf9 cells, they were localized throughout the cytoplasm. Similarly, TSWV GN-GFP was expressed in the cytoplasm, specifically localizing to structures resembling the ER and/or Golgi, consistent with earlier studies of GN localization in animal cells (21). When cyclophilin-RFP and TSWV-GN-GFP or endoCP-GN-RFP and TSWV-GN-GFP were coexpressed in Sf9 cells, they colocalized within punctate structures, thus differing from their respective behaviors when singly expressed. The controls of coexpressed RFP and GFP (cotransfection of pHRW and pHGW) were distributed throughout the cytoplasm in single and double transfections (Fig. 9, bottom row). The control localization treatment of cyclophilin-RFP, endoCP-GN-RFP, and TSWV GN-GFP did not change when coexpressed with GFP or RFP (data not shown). Although these colocalization sites warrant further characterization, our results thus far support the validity of in vivo interactions of cyclophilin and endoCP-GN with TSWV-GN.
FIG 9.
Colocalizations of TSWV GN with endoCP-GN or cyclophilin in insect cells. Open reading frames of cyclophilin, endoCP-GN, and TSWV GN were cloned into Drosophila Gateway vectors (under the control of the Hsp70 promoter), and the resulting pHWR and pHWG expression plasmids were used for the following fusion proteins: cyclophilin-RFP, endoCP-GN-RFP, and TSWV GN-GFP. The recombinant plasmids, pHWR-cyclophilin, pHWR-endoCP-GN, and pHWG-TSWV-GN, were singly transfected or cotransfected into insect Sf9 cells. All transfection reactions were performed using the Cellfectin II reagent. Mock, no DNA treatment (top left panels) and cotransfection of pHRW and pHGW expression plasmids (bottom left panels) were used as controls. Cells were stained with DAPI 72 h posttransfection and then visualized using the Cytation 5 cell imaging multimode reader (BioTek, Winooski, VT) to detect red and green fluorescence. Exposure settings (LED intensity/integration time/camera gain) of the mock control were set as the baseline parameters for the analysis of all other treatments. Cells were visualized with a 40× objective. Scale bars = 10 μm.
DISCUSSION
The availability of whole-body transcriptome data for F. occidentalis combined with improved proteomic technologies enabled us to identify for the first time thrips proteins that bind directly to the TSWV attachment protein GN. With particular relevance to viral attachment and to internalization in epithelia, two TIPs (endocuticle structural glycoprotein, endoCP-GN, and cyclophilin) were confirmed to interact directly with GN and were abundant in midgut and salivary gland tissues (22). These data may be the first indication of proteins that could act as putative cell entry receptors for tospoviruses in their insect vectors. We narrowed down the GN-binding region to the amino-terminal region of endoCP-GN excluding the conserved CHB4 domain, setting the stage for future work to decipher the essential amino acids within the nonconserved region necessary to establish the interaction. Three of the TIPs (mATPase, endoCP-V, and enolase) were validated to interact with GN in BiFC assays but not in MbY2H assays. With regard to other virus activities in host cells, the confirmed affinity of GN with diverse thrips proteins indicates that these insect proteins may be host factors involved in steps in the virus infection cycle in the invertebrate host, such as viral replication and/or virion maturation previously observed in both the animal (23, 24) and plant hosts (25, 26). Technical limitations preclude a functional analysis of the TIPs in the acquisition of virus by larval thrips. Knockdown (RNA interference) and knockout (genome editing) methods have not been developed for larval thrips, even though RNA interference methods have been developed to effectively knockdown genes in the much larger adult female thrips by the delivery of double-stranded RNA (dsRNA) directly into the hemocoel (27). Using currently available methods, larval thrips do not survive the dsRNA injection process, and even if successful, knockdown would be delayed, thus missing the narrow window of virus acquisition during early larval development.
In our initial screen, cuticular proteins were the most enriched proteins that bound to virions or GN (Tables 1 and 2; 72%). Cuticular proteins are well characterized as major components of insect hard and soft cuticles (28, 29). Soft cuticles have been documented to line the insect foregut and hindgut (30, 31), and a transmission electron microscopy study documented cuticle lining of the accessory and principal salivary gland (SG) ducts of F. occidentalis (32). In silico sequence analysis of the three cuticular TIPs (CP-V, endoCP-V, and endoCP-GN) revealed conserved CHB4 domains (R&R) suggesting their binding affinity to chitin (heteropolymer of N-acetyl-β-d-glucosamine and glucosamine), also a major component of cuticles and peritrophic membranes (PM) lining the midgut epithelium of most insects (33). Hemipteran and thysanopteran midguts lack PMs and are instead lined with perimicrovillar membranes (PMM) (34, 35); these structures have been reported to contain lipoproteins, glycoproteins, and carbohydrates (36, 37). More recently, a study reported the presence and importance of chitin in the PMM of Rhodnius prolixus (kissing bug) midguts, marking the first hemipteran midgut described to contain chitin (38). Since all three cuticular TIPs were highly expressed in the midgut and SGs of larval F. occidentalis in the present study, we hypothesize that chitin or chitin-like structures may impregnate the thrips PMM and SG linings, forming a matrix with endoCPs; however, this remains to be empirically determined. Alternatively, the thrips TIPs annotated as cuticle proteins with predicted chitin-binding domains may have yet-undescribed functions in insect biology.
Cuticular proteins are emerging as important virus interactors and responders in diverse vector-borne plant virus systems. A CP of the hemipteran vector Laodelphax striatellus was found to interact with the nucleocapsid protein (pc3) of rice stripe virus (genus Tenuivirus, family Phenuiviridae) and was hypothesized to be involved in viral transmission and to possibly protect the virus from degradation by a host immune response in the hemolymph (39). Recently, a CP of another hemipteran vector, Rhopalosiphum padi, was identified to interact with barley yellow dwarf virus-GPV (genus Luteovirus, family Luteoviridae) readthrough protein, and the transcript of this particular CP was differentially expressed in viruliferous compared to virus-free aphids (40). At the transcript level, thrips cuticular proteins of different types, including the thrips CPRs used in the present study to phylogenetically place the three cuticular TIPs, were downregulated in TSWV-infected first-instar larvae (20). Although the three cuticular TIPs identified in the present study were not reported in this earlier work to be responsive to TSWV at the transcript level, in both reports, cuticle-associated proteins were implicated in the early infection events of the thrips vector.
Cyclophilins, also known as peptidyl-prolyl cis-trans-isomerases, are ubiquitous proteins involved in multiple biological processes, including protein folding and trafficking, cell signaling, and immune responses (41). They have also been shown to promote or prevent virus infection (42, 43). For example, cyclophilin A was found to bind to viral RNA to inhibit the replication of tomato bushy stunt virus (genus Tombusvirus, family Tombusviridae) in plant leaf cells (44), while cyclophilins of the aphid vector Schizaphis graminum have been shown to play an important role in cereal yellow dwarf virus (genus Polerovirus, family Luteoviridae) transmission (45). Interactions between the thrips cyclophilin TIP as described in our work may affect similar viral processes, such as replication and maturation or transmission by the thrips vector (45, 46). The same cyclophilin was downregulated in first-instar larvae of F. occidentalis during TSWV infection (18), adding to the body of evidence that viruses modulate the expression of cyclophilins (47–52). The cyclophilin interaction with GN was consistent but weak, and this may be the reason that it was not observed in the BiFC experiments. Alternative explanations for the discrepancy in the cyclophilin-GN interaction include that the interaction does not occur in plant cells or that the weak interaction was not strong enough to fluoresce over the background to detect an in planta interaction. Others have proposed that negative-strand virus matrix proteins (structural proteins that package viral RNA) evolved from cyclophilins (53); however, bunyaviruses do not encode a matrix protein. Thus, a reason for the direct interaction between the cyclophilin with GN may be to facilitate ribonucleoprotein (RNP) packing into the virus particle, with cyclophilin serving as a surrogate matrix protein for TSWV.
Similar to cyclophilins, enolases of diverse hosts have been observed to interact with viruses. In general, enolases are essential metalloenzymes that catalyze the conversion of 2-phosphoglycerate (2-PGE) to phosphoenolpyruvate (PEP) in the glycolytic pathway for energy metabolism (54). Enolases also have defined roles as plasminogen receptors and appear to use a nonconventional strategy to localize to the cell surface because they do not have signal peptides or transmembrane domains (55, 56). The enolase TIP identified in the present study was previously reported to be upregulated in L1 thrips infected with TSWV (18), and enolase was also upregulated in response to rice stripe virus (RSV) in bodies of the planthopper vector, L. striatellus (57). In the case of flaviviruses, Aedes aegypti enolase was shown to directly interact with purified virus and recombinant envelope glycoprotein of dengue virus (58) and West Nile virus envelope protein (59). The localization of this enolase in brush border membrane vesicles of this mosquito species (60) strengthens the case for a proposed receptor role in virus entry into vector mosquito midguts. Other host-virus studies have proposed a role for enolase in antiviral defense (61), and enolases have been shown to promote extracellular matrix degradation (55). Further research is needed to determine if this TSWV-interacting enolase plays a role in virus entry to midguts and salivary glands or midgut escape.
The other TIP known to play a role in energy production is mitochondrial ATP synthase α-subunit. The multisubunit enzyme mATPase is responsible for generating the majority of cellular ATP required by eukaryotes to meet their energy needs. As with the other noncuticle TIPs, the mATPase α-subunit was previously identified to be differentially abundant (upregulated) under TSWV infection (18), as was the case for RSV-infected L. striatellus vector planthoppers (57). Mitochondria have also been previously implicated in virus-host biology. For example, African swine fever virus (genus Asfivirus, family Asfarviridae) has been shown to induce the migration of mitochondria to the periphery of viral factories (62), possibly suggesting that mitochondria supply energy for viral morphogenetic processes. The finding that two TIPs in the present study have ontologies in energy production and metabolism suggests that perturbation or direct interactions with these host proteins may be required for the successful infection of F. occidentalis by TSWV.
The discovery of six TIPs is a significant step forward for understanding thrips interactions with tospoviruses. The first evidence of TSWV protein-thrips protein interactions was presented 20 years ago (63), and the proteins described herein are the first thrips proteins documented to interact directly with the viral glycoprotein GN, involved in virus attachment to the midgut epithelial cells of the insect vector. In other eukaryotes, the six interacting proteins have biological functions that point to their putative roles in facilitating the virus infection/replication cycle by acting as a receptor or other essential step in the virus life cycle and/or host response via a defense mechanism. The virus-host systems that have defined functions for analogous TIPs include plant viruses, arboviruses, and animal/human viruses, and the findings described here provide a framework for further exploration and testing of new hypotheses regarding their roles in TSWV-thrips interactions.
MATERIALS AND METHODS
Insect rearing and plant and virus maintenance.
The F. occidentalis colony was established from insects collected on the island of Oahu, HI, and was maintained on green beans (Phaseolus vulgaris) at 22°C (±2°C) under laboratory conditions, as previously described (64). Thrips were age synchronized based on their developmental stages. For the localization and bimolecular fluorescence complementation (BiFC) experiments, wild-type and transgenic Nicotiana benthamiana plants expressing cyan fluorescent protein-histone 2B (CFP-H2B) or RFP-ER (65) were grown in a growth chamber at 25°C with a 14-h light period at 300-micromole intensity and 10-h dark cycle. TSWV (isolate TSWV-MT2) was maintained by both mechanical inoculation and thrips transmission using D. stramonium and Emilia sonchifolia, respectively (12). To avoid the generation of a virus isolate with an insect transmission deficiency, the virus was mechanically passaged only once. The single-pass mechanically inoculated symptomatic D. stramonium leaves were used for insect acquisition of TSWV. Briefly, synchronized F. occidentalis first-instar larvae (0 to 17 h old) were collected and allowed an acquisition access period (AAP) on D. stramonium for 24 h. After acquisition, D. stramonium leaves were removed, and these larvae were maintained on green beans until they developed into adults. Viruliferous adults were transferred onto clean E. sonchifolia plants for 2 days. After inoculation, thrips and inoculated E. sonchifolia plants were treated with commercial pest strips for 2 h before the plants were moved to the greenhouse for TSWV symptom development. The thrips-transmitted, TSWV-symptomatic E. sonchifolia leaves were only used for mechanical inoculation.
TSWV purification.
Mechanically inoculated D. stramonium leaves were used for TSWV purification via differential centrifugation and a sucrose gradient. Symptomatic leaves were homogenized in extraction buffer (0.033 M KH2PO4, 0.067 M K2HPO4, and 0.01 M Na2SO3) in a 1:3 ratio of leaf tissue to buffer. The homogenate was then filtered through four layers of cheesecloth, and the flowthrough was centrifuged at 7,000 rpm (7,445 × g) for 15 min using the Sorvall SLA 1500 rotor. To remove the cell debris, the pellet was resuspended in 65 ml of 0.01 M Na2SO3 and was centrifuged again at 8,500 rpm (8,643 × g) for 20 min using the Sorvall SS34 rotor. The supernatant that contained the virions was centrifuged for 33 min at 29,300 rpm (88,205 × g) using the 70 Ti rotor, and the pellet was resuspended in 15 ml of 0.01 M Na2SO3, followed by another centrifugation at 9,000 rpm (9,690 × g) for 15 min using the Sorvall SS34 rotor. The centrifugation series was repeated one additional time. The pellet was resuspended and loaded on a sucrose gradient (10% to 40% sucrose), which was centrifuged for 35 min at 21,000 rpm (79,379 × g) using the SW28 rotor. The virion band was collected and centrifuged for 1 h at 29,300 rpm (88,205 × g) using the 70 Ti rotor. The pellet was resuspended in 100 to 200 μl of 0.01 M Na2SO3. All centrifugation steps were performed at 4°C to prevent virion degradation. The purified virus was quantified using the bicinchoninic acid (BCA) protein assay kit (Thermo Fisher Scientific, Waltham, MA, USA), following the manufacturer’s instructions.
F. occidentalis L1 total protein extraction, quantification, and two-dimensional gel electrophoresis.
Total proteins from age-synchronized healthy larval thrips (0 to 17 h old) were extracted using the trichloroacetic acid-acetone (TCA-A) method (66, 67). Briefly, whole insects were ground using liquid nitrogen and were dissolved in 500 μl TCA-A extraction buffer (10% of TCA in acetone containing 2% β-mercaptoethanol). This mixture was incubated at –20°C overnight and centrifuged at 5,000 × g and 4°C for 30 min. After 3 washes with ice-cold acetone and then air drying, the pellet was resuspended in 200 μl general-purpose rehydration/sample buffer (Bio-Rad Laboratories, Hercules, CA, USA). The suspension was centrifuged at 12,000 × g for 5 min, and the protein supernatant was quantified using the BCA protein assay kit (Thermo Fisher Scientific), following the manufacturer’s instructions. For each gel, 150 μg of total protein extract was applied to an 11-cm immobilized pH gradient (IPG) strip (pH 3 to 10) for isoelectric focusing (IEF). The IEF-IPG strip equilibration and second-dimension separation of proteins were performed under the same conditions described by Badillo-Vargas et al. (18).
Overlay assays.
To identify thrips proteins that bind to TSWV virions and recombinant glycoprotein GN, we conducted gel overlay assays. For the purified virion overlays, the experiment was performed four times (biological replicates), and for the GN overlay, the experiment was performed twice. For probing the protein-protein interactions, each unstained 2D gel was electrotransferred onto Hybond-C Extra nitrocellulose membrane (Amersham Biosciences, Little Chalfont, UK) overnight at 30 V (4°C) in protein transfer buffer (48 mM Tris, 39 mM glycine, 20% methanol, and 0.037% SDS). Then, the membrane was incubated with blocking buffer (phosphate-buffered saline containing 0.05% Tween 20 [PBS-T] and 5% dry milk) for 1 h at room temperature on a rocker with a gentle rotating motion. The three following antigens were used to probe the thrips protein membranes: purified TSWV virions, recombinant glycoprotein GN (Escherichia coli expressed), and virus-free plant extract from a mock virus purification (negative control). An additional negative-control blot (no overlay) treated with antibodies alone was included in each overlay replicate. For the virus and GN treatments, 25 μg/ml and 3.5 μg/ml purified TSWV virions and recombinant GN glycoprotein, respectively, were incubated with membranes in blocking buffer at 4°C overnight with a gentle rotating motion. Membranes were washed three times using PBST and were incubated with polyclonal rabbit anti-TSWV GN antibodies at a 1:2,000 dilution in blocking buffer for 2 h at room temperature (9, 22). After washing with PBST, membranes were incubated with horseradish peroxidase (HRP)-conjugated goat anti-rabbit antibodies at a 1:5,000 dilution in blocking buffer for 1 h at room temperature. The ECL detection system (Amersham Biosciences) was used for protein visualization, following the manufacturer’s instructions. The protein spots that were consistently observed on the membranes were first compared with those protein spots that interacted with antibody-only blots (Fig. 1A and 2A) and virus-free plant extract blots, and then they were pinpointed on the corresponding Coomassie brilliant blue G-250-stained 2D gels for spot picking.
Identification of TIPs.
Protein spots that were consistently identified in the 2D gel overlays were selected and manually picked for analysis. For the virion overlays, spots were picked from three replicate gels and from two replicate gels for the GN-probed membranes. The picked proteins were processed and subjected to electrospray ionization (ESI)-mass spectrometry, as previously described (18). Protein spots (peptides) that had Mascot (v2.2) scores with significant matches (P ≤ 0.05) to translated de novo-assembled contigs (all six frames) derived from mixed stages of F. occidentalis (Fo Seq 454-Sanger hybrid) (18) were identified, and NCBI BLASTX was performed on the contigs to provisionally annotate (E < 10−10) the protein and to predict conserved motifs using the contig as the query and the NCBI nonredundant protein database as the subject. The F. occidentalis sequence (Fo Seq) database used for identification was composed of a transcript collection from 454 contig sequences (26,324) and a Sanger-sequenced expressed sequence tag (EST) collection (contigs and singletons) (18, 68). The Fo Seq database is available upon request.
A second round of TIP candidate selection was conducted for stringency in moving forward to cloning and confirmation of interactions. A contig sequence was retained if it contained a complete predicted ORF (i.e., presence of both start and stop codons predicted with the ExPASy Translate Tool, https://web.expasy.org/translate/) and had at least 10% coverage by a matching peptide(s) identified for a spot as predicted by Mascot, i.e., removal of proteins identified by a single peptide with less than 10% coverage to a Fo Seq contig and/or contigs with incomplete ORFs (lacking a predicted stop codon). The translated ORFs were queried against the NCBI nonredundant protein database (BLASTP), and the CCTOP software (http://cctop.enzim.ttk.mta.hu/) (69) and SignalP 4.1 server (http://www.cbs.dtu.dk/services/SignalP/) (70) were used to predict the presence of transmembrane domains and signal peptides, respectively. Prosite (https://prosite.expasy.org/) was used to analyze putative posttranslational modifications that may have affected the electrophoretic mobility of identical proteins in the overlay assays, i.e., same peptide sequence or Fo Seq contig match identified for more than one protein spot.
Classification and phylogenetic analysis of the three confirmed cuticular TIPs.
Given the apparent enrichment of putative cuticular proteins (CP) identified in the overlay assays and the subsequent confirmation of three of those TIPs (CP-V, endoCP-GN, and endoCP-V), it was of interest to perform a second layer of protein annotations. The ORFs (amino acid sequence) of the three confirmed CP TIPs, 19 exemplar insect orthologous sequences obtained from NCBI GenBank, and a significant collection of structural CP transcripts previously reported to be differentially expressed in TSWV-infected larval thrips of F. occidentalis (20) were subjected to two complementary arthropod CP prediction tools. CutProtFam-Pred (http://aias.biol.uoa.gr/CutProtFam-Pred/home.php) (71) was used to classify each amino acid sequence by CP family; there are 12 described families for arthropods, each distinguished by conserved sequence motifs shared by members (29). CuticleDB (http://bioinformatics.biol.uoa.gr/cuticleDB/) (72) was used to distinguish what was found to be two enriched, chitin-binding CP families in our data set, CPR-RR1 and CPR-RR2, i.e., the R&R consensus motif (19). The sequences flanking the RR1 and RR2 predicted chitin-binding domains were divergent between the thrips CPs and across the entire set of CPs (thrips and other insects), making alignments using full-length ORFs ambiguous and uninformative. Therefore, the R&R consensus was used for inferring the evolutionary history of CP proteins. The flanking sequences were trimmed manually, and the 46 R&R consensus sequences (RR1 and RR2) were aligned with MEGA7 (73) using ClustalW. Phylogenetic analyses were performed in MEGA7 using the maximum likelihood (ML) method and the best substitution models determined for the data to model the variation among sites. Bootstrap consensus trees (500 replicates) were generated by the ML algorithm using the Jones-Taylor-Thornton (JTT) method for amino acid substitutions (76), with a gamma distribution. To maximize the number of informative amino acid positions in the final analysis while reducing computational time, positions with less than 30% site coverage across the 46 aligned sequences (due to 9 sequences with insertions and/or sequencing/assembly errors) were eliminated, resulting in 74 amino acid positions in the final data set.
Cloning of candidate TIPs and TSWV genes.
For the generation of full-length clones of TIPs that were used in various protein-protein assays, total RNA was extracted from L1 thrips (0 to 17 h old) using the TRIzol reagent (Thermo Fisher Scientific), according to the manufacturer’s protocol. The RNA pellet was dissolved in nuclease-free water, and 1 μg total RNA was used for cDNA synthesis using the Verso cDNA synthesis kit (Thermo Fisher Scientific). PCR was performed to amplify six identified TIP ORFs using high-fidelity polymerase in FailSafe (Epicentre, Madison, WI, USA). The designed primers used are available upon request. Amplicons were cloned into pENTR-D/TOPO (Thermo Fisher Scientific).
TSWV genes were also cloned into pENTR-D/TOPO and then recombined to different vectors using Gateway cloning techniques. Coding sequences of different glycoprotein forms (soluble [GN-S] and insoluble [GN]) were amplified from pGF7 (74). The primers used for PCR are available upon request.
Polyclonal antibodies against TIPs.
To generate antibodies to the TIPs, the protein sequence was analyzed by the antibody manufacturer (GenScript, Piscataway, NJ), using the OptimumAntigen design tool. For each TIP, a 14-amino-acid peptide was selected based on these predictions and by sequence alignments to other predicted protein sequences in GenBank. Due to the conserved CHB4 domain in endoCP-GN, endoCP-V, and CP-V, the polyclonal antibodies against these three TIPs were generated using their nonconserved region. The peptides were synthesized, and all antibodies were produced using mice (GenScript). The peptide sequences for each TIP that were used for the antibody generation were cyclophilin, LESFGSHDGKTSKK; enolase, ELRDNDKSQYHGKS; CP-V, TDSGQYRKEKRLED; endoCP-GN, STKVNPQSFSRSSV; endoCP-V, VNPDGSFQYSYQTG; and mATPase, GHLDKLDPAKITDF.
Validation of antibody binding to each TIP (peptide) using dot blot.
Peptide antibodies to the six TIPs were generated by GenScript using their standardized workflow for the generation of antibodies to peptides. Antibodies were generated by the injection of peptides in mice, and antibodies were tested for reactivity to their cognate peptide using dot blot assays. The peptides that were used to generate each antibody were diluted to 100 μg/ml using 1× PBS (pH 7.2), with the exception of the CP-V peptide that was diluted to 2.5 mg/ml, due to lower affinity of this antibody. Two microliters of each diluted peptide was spotted onto the same nitrocellulose membrane strip along with the controls of PBS and preimmune serum (500,000× dilution). A total of 6 membrane strips, one for each TIP-peptide antibody, were loaded with the same peptide samples and controls. After the membrane strips dried, each strip was incubated with blocking buffer (5% nonfat milk in Tris-buffered saline with 0.5% Tween [TBS-T]), followed by incubation with the six different primary antibodies (0.5 μg/ml; GenScript). After three washes with TBS-T (3 × 10 min), all membrane strips were incubated with secondary antibody goat anti-mouse IgG (H+L)-HRP conjugate (1:5,000 dilution; Bio-Rad Laboratories). After three washes with TBS-T, the SuperSignal West Dura extended-duration substrate (Thermo Fisher Scientific) was added onto individual membrane strips. Each membrane strip was developed separately for 5 to 10 min; however, the membrane strip that was incubated with the CP-V peptide antibody was developed for 40 min. Images were collected using the iBright imaging system (CL1000; Thermo Fisher Scientific). The reaction mixtures for the blocking, primary, and secondary antibody incubation steps were incubated for 1 h at room temperature, and the strip probed with CP-V peptide antibody was incubated for 2 h at room temperature. The entire experiment was performed three times.
Immunolabeling thrips guts, Malphigian tubules, and salivary glands.
To determine the location of TIP expression in the developmental stage of the thrips that most efficiently acquires TSWV (L1), we used the TIP antibodies in immunolocalization experiments. Treatments in the experiments included peptide antibodies to the TIPs and background controls of dissected insects incubated with (i) only secondary antibody and (ii) insects treated with preimmune serum and secondary antibody. Newly emerged larvae (0 to 17 h old) were collected from green beans and were then fed on a 7% sucrose solution for 3 h to clear plant tissue from their guts. The larvae were dissected on glass slides using cold PBS buffer and Teflon-coated razor blades. The dissected thrips were transferred into 2-cm-diameter, flat-bottomed watch glasses (BPI dishes; U.S. Bureau of Plant Industry), and the tissues were fixed for 2 h using a 4% paraformaldehyde solution in 50 mM sodium phosphate buffer (pH 7.0). The tissues were washed using PBS buffer after fixation and were incubated with PBS buffer with 1% Triton X-100 overnight. The overnight-permeabilized tissues were then washed before incubation in blocking buffer which included PBS, 0.1% Triton X-100, and 10% normal goat serum (NGS) for 1 h. After removing the blocking buffer, the dissected thrips were incubated with primary antibody of 100 μg/ml mouse-generated antibodies against each individual TIP (GenScript) that was diluted in antibody buffer (0.1% Triton X-100 and 1% NGS). After washing, 10 μg/ml goat anti-mouse secondary antibody conjugated with Alexa Fluor 488 (Thermo Fisher Scientific) was used to incubate the dissected thrips organs. Incubation was performed at room temperature for 2.5 h, 1× PBS buffer was used for washing, and every wash step included three rinses; the secondary antibody incubation was protected from light by covering the samples with aluminum foil. After removing antibodies and washing, dissected thrips were incubated for 2 h with Alexa 594-conjugated phalloidin probe (Thermo Fisher Scientific) in 1× PBS with a concentration of 4 units/ml for actin staining. After washing, the tissues were transferred onto glass slides, and SlowFade Diamond antifade mountant with 4′,6-diamidino-2-phenylindole (DAPI; Thermo Fisher Scientific) was added onto the tissues to stain the nuclei. The coverslips were slowly placed on tissues to avoid bubbles and then sealed with transparent nail polish at the edges. After blocking, the dissected thrips tissues that were only incubated with secondary antibody (without adding primary antibody) and the tissues incubated with each preimmune mouse antiserum (GenScript) were used as negative controls, respectively. All of the experiments were performed twice.
Inherent with very small tissues (<1-mm body size), there were common losses or damaged tissues during the dissection process and staining procedures; only visibly intact tissues were considered for the following data collection, and this number varied for each type of tissue (Table 4). The autofluorescent background from thrips tissues incubated with each preimmune antiserum and secondary antibodies was slightly higher than that of the thrips tissues incubated with PBS buffer and secondary antibodies (negative control) (data not shown); therefore, the confocal laser settings (power and percent gain) were adjusted to remove any background fluorescence observed for these treatments.
TABLE 4.
Number of dissected F. occidentalis first-instar larvae that were immunolabeled with specific antisera or antibodies against each TIP and were visualized by confocal microscopy from two replicates
| Antiserum or antibody | No. of dissected intacta
: |
||
|---|---|---|---|
| MG | TSG | PSG | |
| No-antibody control | 64 | 22 | 25 |
| Cyclophilin preimmune serum | 47 | 10 | 21 |
| Enolase preimmune serum | 29 | 13 | 10 |
| CP-V preimmune serum | 38 | 13 | 18 |
| EndoCP-GN preimmune serum | 11 | 2 | 7 |
| EndoCP-V preimmune serum | 28 | 12 | 10 |
| Anti-cyclophilin antibody | 26 | 15 | 10 |
| Anti-enolase antibody | 58 | 25 | 21 |
| Anti-CP-V antibody | 35 | 6 | 11 |
| Anti-endoCP-GN antibody | 91 | 32 | 35 |
| Anti-endoCP-V antibody | 50 | 20 | 20 |
| Anti-mATPase antibody | 35 | 16 | 25 |
MG, midgut; TSG, tubular salivary gland; PSG, principal salivary gland.
Split-ubiquitin membrane-based yeast two-hybrid system.
The MbY2H system was used to validate TSWV GN-TIP interactions identified in the gel overlay assays. The MbY2H system enables the validation of interactions for soluble and integral membrane proteins. TSWV GN coding sequence was cloned into the MbYTH vector pBT3-SUC, and the six TIP ORFs were cloned to vector pPR3N using the SfiI restriction site (Dualsystems Biotech, Schlieren, Switzerland). To identify the region of endoCP-GN that binds to TSWV GN using the MbY2H system, the amino acid sequence of endoCP-GN (284 aa) was used to search against the NCBI nonredundant protein database using BLASTP. The conserved CHB4 domain was located at the C terminus of endoCP-GN (amino acids 190 to 246). Therefore, the possible interacting domains, the nonconserved region of endoCP-GN (amino acids 1 to 189) and the conserved CHB4 domain (amino acids 190 to 274), were individually cloned into pPR3N using the SfiI restriction site. Based on the BLASTP results, the homologous sequences from other insect species encompassed some additional amino acids in the N-terminal direction of the CHB4 domain; therefore, we made an alternative construct that included the conserved CBH4 domain starting from amino acid 177. Hence, the coding sequence of amino acids 1 to 176 and 177 to 284 of endoCP-GN were also cloned to pPR3N using the SfiI restriction site. The primers used for cloning are available upon request.
The MbY2H assays were performed using the manufacturer’s instructions with recombinant plasmids that were confirmed by Sanger sequencing. Yeast (Saccharomyces cerevisiae strain NYM51) competent cells were freshly prepared, and the recombinant bait plasmid pBT3-SUC-GN was transformed into yeast cells. Briefly, 1.5 μg of bait plasmid was added into 100 μl of yeast competent cells with 50 μg of denatured Yeastmaker carrier DNA (TaKaRa Bio USA, Mountain View, CA) and 500 μl polyethylene glycol-lithium acetate (PEG/LiAc). The mixture was incubated at 30°C for 30 min with mixing every 10 min. Twenty microliters of dimethyl sulfoxide (DMSO) was then added to each reaction mixture, and the cells were incubated at 42°C for 20 min with repeated mixing at 5-min intervals. After centrifugation at 14,000 rpm for 15 s, the supernatant was removed, and the pellet was resuspended in 1 ml of yeast-peptone-dextrose (YPD) medium. The resuspended cells were incubated at 30°C for 90 min with shaking at 200 rpm. Then, cells were centrifuged at 14,000 rpm for 15 s and resuspended in 500 μl of sterile 0.9% (wt/vol) NaCl, which was then spread and cultured on SD/–Trp dropout medium at 30°C until the colonies were visible. Several colonies from the same SD/–Trp plate were cultured for preparing yeast competent cells. Then, each individual recombinant plasmid, pPR3N-TIP or pPR3N-partial endoCP-GN (1.5 μg/transformation reaction), was transformed into yeast competent cells expressing fused Nub-GN. The transformants were cultured on both SD/–Leu/–Trp double-dropout (DDO) and SD/–Ade/–His/–Leu/–Trp quadruple-dropout (QDO) media. The positive controls included transformation of pOst1-NubI into the yeast strain NYM51 that already expressed fused Nub-GN, as well as cotransformation of pTSU2-APP and pNubG-Fe65 into the yeast strain NYM51. The transformation of pPR3N (empty vector) into the yeast strain NYM51 that already expressed fused Nub-GN was used as the negative control. Interactions between GN-Cub and NubI and between GN-Cub and NubG were used as positive and negative controls, respectively. All transformants were spread and cultured on both DDO and QDO media and cultured at 30°C in an incubator. The entire experiment was performed three times.
Expression of the reporter gene lacZ and the activity of expressed β-galactosidase in yeast cells derived from the MbY2H system was determined by a β-galactosidase assay kit, following the manufacturer’s protocol (Thermo Fisher Scientific).
GFP fusion protein expression and bimolecular fluorescence complementation in Nicotiana benthamiana.
To visualize protein expression and localization in plants, TSWV GN (ORFs GN and GNS) and TIP ORFs (mATPase, CP-V, endoCP-V, endoCP-GN, cyclophilin, and enolase) were expressed as fusions to autofluorescent proteins. Before launching a BiFC analysis of candidate protein interactions in planta, it is critical to determine if position of a fused fluorescent protein tag (N or C terminus of the candidate protein) affects the expression and/or localization of the fusion protein in cells. Furthermore, it was expected that the signal peptides located on the N terminus of the soluble (GN-S) and insoluble (GN) TSWV glycoprotein, and the cuticular TIPs (CP-V, endoCP-V, and endoCP-GN), would preclude the placement of tags at the N termini of these proteins. All the viral and thrips genes were moved from their entry clones into pSITE-2NB (GFP fused to the carboxy terminus of the protein of interest) or pSITE-2CA (GFP fused to the amino terminus of the protein of interest) using Gateway LR Clonase (75). After validation of plasmids by Sanger sequencing, they were transformed into Agrobacterium tumefaciens strain LBA 4404. The transformed LBA 4404 was grown for 2 days at 28°C and resuspended in 0.1 M morpholineethanesulfonic acid (MES) and 0.1 M MgCl2 to an optical density at 600 nm (OD600) between 0.6 and 1. After the addition of 0.1 M acetosyringone, the suspension was incubated at room temperature for 2 h and then infiltrated in transgenic N. benthamiana expressing an endoplasmic reticulum (ER) marker fused to the red fluorescent protein (m5RFP-HDEL) (65). Two days after infiltration, leaf tissue was mounted in water on a microscope slide for detection of GFP by confocal microscopy. Plants were infiltrated a minimum of two separate occasions with at least two leaves per plant in two different plants. A minimum of 50 cells were visualized in each plant to confirm the localization patterns of the proteins in planta. GFP fused to the N terminus of the glycoprotein (GN and GN-S), the cuticle TIPs (endoCP-GN and endoCP-V), and mATPase α produced weak signal or reduced mobility in the cell (data not shown). For the remaining proteins, there was no effect of fluorescent protein tag location on protein expression or mobility. Thus, all protein localization and BiFC validation experiments were performed with C-terminally fused TIPs for consistency in the assays.
The fusion construct design for BiFC assays was based on localization results described above and sequence analysis. Signal peptides were identified in the amino terminus of GN and three TIPs (all cuticle proteins). The signal peptide is required for proper localization and function of fusion-GFP/YFP proteins in N. benthamiana for BiFC assays. We fused half YFPs (either amino or carboxy half of YFP) to the carboxy termini of all proteins with N-terminal signal peptides using BiFC plasmids pSITE-NEN and pSITE-CEN (65). All ORFs were transferred between plasmids using Gateway LR Clonase II enzyme mix (Thermo Fisher Scientific). All clones were transformed into A. tumefaciens strain LBA 4404 and confirmed by Sanger sequencing.
Each combination of TIPs and TSWV GN and GN-S was infiltrated into N. benthamiana expressing CFP fused to a nuclear marker, histone 2B (CFP-H2B) (65). A minimum of three independent experiments with two plants and two leaves per plant for each combination of proteins were conducted. For the analysis of interactions, a minimum of 50 cells with similar localization patterns was required to confirm the interaction, and a minimum of two separate images were captured on each occasion for documentation. Glutathione-S transferase (GST) fusions to YFP halves were utilized as a nonbinding control for each of the TIPs. To be recorded as a positive interaction, levels of fluorescence of the interacting TSWV protein-TIPs had to be above those observed between each TIP and GST.
Laser scanning confocal microscopy.
Confocal microscopy was used to detect the fluorescent signal produced from TIP antibody labeling in thrips tissues and BiFC experiments in plants. All images were acquired on a Zeiss LSM 780 laser scanning confocal microscope using the C-Apochromat 40×/1.2 W Korr M27 and Plan-Apochromat 20×/0.8 M27 objectives. Image acquisition was conducted on Zen 2 Black edition v.10.0.0 at 1,024 by 1,024 pixels with a scan rate of 1.58 μs per pixel, with a pixel average of 4-bit and 16-bit depth. The laser power and percent gain settings for the detection of nuclei and actin and for the bright field were adjusted accordingly. Laser power and percent gain settings for detection of TIPs were equal to or smaller than their controls. z-stacks were taken for the localization of TIPs in thrips. Eight (TIPs localization) z-stack sections were processed using maximum intensity projection using Zen 2 Black. Zen 2 Blue edition lite 2010 v.2.0.0.0 was used for image conversion to jpeg format.
Colocalization of TIPs and TSWV GN in insect cells.
The ORFs of cyclophilin, endoCP-GN, and TSWV GN were cloned into pENTR-D/TOPO (Thermo Fisher Scientific, Grand Island, NY). The cyclophilin and endoCP-GN ENTR clones were recombined into pHWR, and the TSWV GN ENTR clone was moved into pHWG (The Drosophila Gateway collection, DGRC, Bloomington, IN). Both pHWR and pHWG have an Hsp70 promoter and gateway cloning cassette, and both RFP and GFP were expressed at the C terminus of TIPs or TSWV GN.
The recombinant expression constructs were confirmed by Sanger sequencing and then transfected into Sf9 cells. Single transfections or cotransfections were performed using Cellfectin II reagent (Thermo Fisher Scientific), following the manufacturer’s protocol. Briefly, Sf9 cells were counted and diluted to 5 × 105 cells/ml, and then 2-ml aliquots were seeded into each well of a 6-well plate. Eight microliters of Cellfectin II reagent and 3 ng of each recombinant plasmid were diluted in 100 μl Grace’s medium (Thermo Fisher Scientific). After vortexing, both diluted DNA and diluted Cellfectin II reagent were incubated at room temperature for 30 min and then were combined and incubated for an additional 30 min. Another 800 μl of Grace’s medium was added into each DNA-lipid mixture, and the entire 1-ml solution was slowly added onto Sf9 cells. The transfection mix was incubated for 5 h at 27°C, after which the solution was removed and replaced by 2 ml Sf-900 III medium. Singly transfected plasmids were pHWR-cyclophilin, pHWR-endoCP-GN, and pHWG-TSWV GN, and cotransfected plasmids were pHWR-cyclophilin and pHWG-TSWV GN; pHWR-endoCP-GN, and pHWG-TSWV GN. To rule out nonspecific interactions between the proteins of interests (TIPs or GN) and the autofluorescent protein tags, they were cotransfected with unfused RFP or GFP. In addition, a mock (no DNA) transfection was also included as a negative control.
After 72 h, Sf9 cells were resuspended and reseeded in a 24-well glass-bottom Sensoplate (Greiner Bio-One, Monroe, NC) at a 1:2 dilution. The cells were stained with DAPI and then visualized by the Cytation 5 cell imaging multimode reader with 40× PL FL and 20× PL FL objectives (BioTek, Winooski, VT). Image acquisition was performed with BioTek Gen 5 microplate reader and imager software, v3.04. Images were captured using default settings. To detect GFP and RFP, the exposure settings (light-emitting diode [LED] intensity/integration time/camera gain) of mock-transfected cells were set up as the baseline, and different treatments were set to no more than the mock settings. Other parameter settings for the detection of nuclei and bright field were adjusted accordingly. The entire experiment was performed four times.
Data availability.
The GenBank accession numbers for six TIPs are MH884760 for cyclophilin, MH884759 for enolase, MH884758 for cuticular protein (CP-V), MH884757 for endoCP-GN, MH884756 for endoCP-V, and MH884761 for mitochondrial ATP synthase α.
ACKNOWLEDGMENTS
We thank Thomas L. German and Ranjit Dasgupta for providing purified GN-S for protein overlays.
This project was supported by the following grants: USDA-NIFA 2007-35319-18326 and 2016-67013-27492, USDA-FNRI 6034-22000-039-06S, and National Science Foundation CAREER grant IOS-0953786. Ismael E. Badillo-Vargas was partially supported by the National Institute of Food and Agriculture Predoctoral Fellowship under grant KS602489.
REFERENCES
- 1.Gubler DJ. 1998. Resurgent vector-borne diseases as a global health problem. Emerg Infect Dis 4:442–450. doi: 10.3201/eid0403.980326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gubler DJ. 2002. The global emergence/resurgence of arboviral diseases as public health problems. Arch Med Res 33:330–342. doi: 10.1016/S0188-4409(02)00378-8. [DOI] [PubMed] [Google Scholar]
- 3.Beaty BJ, Bishop DH. 1988. Bunyavirus-vector interactions. Virus Res 10:289–301. doi: 10.1016/0168-1702(88)90071-8. [DOI] [PubMed] [Google Scholar]
- 4.Elliott RM. 1997. Emerging viruses: the Bunyaviridae. Mol Med 3:572–577. doi: 10.1007/BF03401814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horne KM, Vanlandingham DL. 2014. Bunyavirus-vector interactions. Viruses 6:4373–4397. doi: 10.3390/v6114373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rotenberg D, Jacobson AL, Schneweis DJ, Whitfield AE. 2015. Thrips transmission of tospoviruses. Curr Opin Virol 15:80–89. doi: 10.1016/j.coviro.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 7.Whitfield AE, Ullman DE, German TL. 2005. Tospovirus-thrips interactions. Annu Rev Phytopathol 43:459–489. doi: 10.1146/annurev.phyto.43.040204.140017. [DOI] [PubMed] [Google Scholar]
- 8.Parrella G, Gognalons P, Gebre-Selassie K, Vovlas C, Marchoux G. 2003. An update of the host range of tomato spotted wilt virus. J Plant Pathol 85:227–264. [Google Scholar]
- 9.Montero-Astúa M, Rotenberg D, Leach-Kieffaber A, Schneweis BA, Park S, Park JK, German TL, Whitfield AE. 2014. Disruption of vector transmission by a plant-expressed viral glycoprotein. Mol Plant Microbe Interact 27:296–304. doi: 10.1094/MPMI-09-13-0287-FI. [DOI] [PubMed] [Google Scholar]
- 10.Whitfield AE, Kumar NK, Rotenberg D, Ullman DE, Wyman EA, Zietlow C, Willis DK, German TL. 2008. A soluble form of the tomato spotted wilt virus (TSWV) glycoprotein GN (GN-S) inhibits transmission of TSWV by Frankliniella occidentalis. Phytopathology 98:45–50. doi: 10.1094/PHYTO-98-1-0045. [DOI] [PubMed] [Google Scholar]
- 11.Whitfield AE, Ullman DE, German TL. 2004. Expression and characterization of a soluble form of tomato spotted wilt virus glycoprotein GN. J Virol 78:13197–13206. doi: 10.1128/JVI.78.23.13197-13206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ullman DE, German TL, Sherwood JL, Westcot DM, Cantone FA. 1993. Tospovirus replication in insect vector cells: immunocytochemical evidence that the nonstructural protein encoded by the S RNA of tomato spotted wilt tospovirus is present in thrips vector cells. Phytopathology 83:456–463. doi: 10.1094/Phyto-83-456. [DOI] [Google Scholar]
- 13.Wijkamp I, van Lent J, Kormelink R, Goldbach R, Peters D. 1993. Multiplication of tomato spotted wilt virus in its insect vector, Frankliniella occidentalis. J Gen Virol 74:341–349. doi: 10.1099/0022-1317-74-3-341. [DOI] [PubMed] [Google Scholar]
- 14.Nagata T, Inoue-Nagata AK, Smid HM, Goldbach R, Peters D. 1999. Tissue tropism related to vector competence of Frankliniella occidentalis for tomato spotted wilt tospovirus. J Gen Virol 80:507–515. doi: 10.1099/0022-1317-80-2-507. [DOI] [PubMed] [Google Scholar]
- 15.Ullman DE, Cho JJ, Mau RFL, Westcot DM, Custer DM. 1992. A midgut barrier to tomato spotted wilt virus acquisition by adult western flower thrips. Phytopathology 82:1333–1342. doi: 10.1094/Phyto-82-1333. [DOI] [Google Scholar]
- 16.Sin SH, McNulty BC, Kennedy GG, Moyer JW. 2005. Viral genetic determinants for thrips transmission of tomato spotted wilt virus. Proc Natl Acad Sci U S A 102:5168–5173. doi: 10.1073/pnas.0407354102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Badillo-Vargas IE, Chen Y, Martin KM, Rotenberg D, Whitfield AE. 2019. Discovery of novel thrips vector proteins that bind to the viral attachment protein of the plant bunyavirus tomato spotted wilt virus. bioRxiv https://www.biorxiv.org/content/10.1101/416560v2. [DOI] [PMC free article] [PubMed]
- 18.Badillo-Vargas IE, Rotenberg D, Schneweis DJ, Hiromasa Y, Tomich JM, Whitfield AE. 2012. Proteomic analysis of Frankliniella occidentalis and differentially expressed proteins in response to tomato spotted wilt virus infection. J Virol 86:8793–8809. doi: 10.1128/JVI.00285-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rebers JE, Riddiford LM. 1988. Structure and expression of a Manduca sexta larval cuticle gene homologous to Drosophila cuticle genes. J Mol Biol 203:411–423. doi: 10.1016/0022-2836(88)90009-5. [DOI] [PubMed] [Google Scholar]
- 20.Schneweis DJ, Whitfield AE, Rotenberg D. 2017. Thrips developmental stage-specific transcriptome response to tomato spotted wilt virus during the virus infection cycle in Frankliniella occidentalis, the primary vector. Virology 500:226–237. doi: 10.1016/j.virol.2016.10.009. [DOI] [PubMed] [Google Scholar]
- 21.Snippe M, Smeenk L, Goldbach R, Kormelink R. 2007. The cytoplasmic domain of tomato spotted wilt virus GN glycoprotein is required for Golgi localisation and interaction with GC. Virology 363:272–279. doi: 10.1016/j.virol.2006.12.038. [DOI] [PubMed] [Google Scholar]
- 22.Montero-Astúa M, Ullman DE, Whitfield AE. 2016. Salivary gland morphology, tissue tropism and the progression of tospovirus infection in Frankliniella occidentalis. Virology 493:39–51. doi: 10.1016/j.virol.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 23.Elliott RM, Schmaljohn CS. 2013. Bunyaviridae In Knipe DM, Howley PM (ed), Fields virology, 6th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 24.Wang H, Alminaite A, Vaheri A, Plyusnin A. 2010. Interaction between hantaviral nucleocapsid protein and the cytoplasmic tail of surface glycoprotein GN. Virus Res 151:205–212. doi: 10.1016/j.virusres.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 25.Ribeiro D, Borst JW, Goldbach R, Kormelink R. 2009. Tomato spotted wilt virus nucleocapsid protein interacts with both viral glycoproteins GN and GC in planta. Virology 383:121–130. doi: 10.1016/j.virol.2008.09.028. [DOI] [PubMed] [Google Scholar]
- 26.Richmond KE, Chenault K, Sherwood JL, German TL. 1998. Characterization of the nucleic acid binding properties of tomato spotted wilt virus nucleocapsid protein. Virology 248:6–11. doi: 10.1006/viro.1998.9223. [DOI] [PubMed] [Google Scholar]
- 27.Badillo-Vargas IE, Rotenberg D, Schneweis BA, Whitfield AE. 2015. RNA interference tools for the western flower thrips, Frankliniella occidentalis. J Insect Physiol 76:36–46. doi: 10.1016/j.jinsphys.2015.03.009. [DOI] [PubMed] [Google Scholar]
- 28.Andersen SO, Hojrup P, Roepstorff P. 1995. Insect cuticular proteins. Insect Biochem Mol Biol 25:153–176. doi: 10.1016/0965-1748(94)00052-J. [DOI] [PubMed] [Google Scholar]
- 29.Willis JH. 2010. Structural cuticular proteins from arthropods: annotation, nomenclature, and sequence characteristics in the genomics era. Insect Biochem Mol Biol 40:189–204. doi: 10.1016/j.ibmb.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chapman RF, Simpson SJ, Douglas AE. 2013. The insects: structure and function, 5th ed Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- 31.Maddrell SHP, Gardiner BOC. 1980. The permeability of the cuticular lining of the insect alimentary canal. J Exp Biol 85:227–237. [Google Scholar]
- 32.Ullman DE, Westcot DM, Hunter WB, Mau R. 1989. Internal anatomy and morphology of Frankliniella occidentalis (Pergande) (Thysanoptera: Thripidae) with special reference to interactions between thrips and tomato spotted wilt virus. Int J Insect Morphol Embryol 18:289–310. doi: 10.1016/0020-7322(89)90011-1. [DOI] [Google Scholar]
- 33.Zhu KY, Merzendorfer H, Zhang W, Zhang J, Muthukrishnan S. 2016. Biosynthesis, turnover, and functions of chitin in insects. Annu Rev Entomol 61:177–196. doi: 10.1146/annurev-ento-010715-023933. [DOI] [PubMed] [Google Scholar]
- 34.Cristofoletti PT, Ribeiro AF, Deraison C, Rahbe Y, Terra WR. 2003. Midgut adaptation and digestive enzyme distribution in a phloem feeding insect, the pea aphid Acyrthosiphon pisum. J Insect Physiol 49:11–24. doi: 10.1016/S0022-1910(02)00222-6. [DOI] [PubMed] [Google Scholar]
- 35.Silva CP, Silva JR, Vasconcelos FF, Petretski MD, Damatta RA, Ribeiro AF, Terra WR. 2004. Occurrence of midgut perimicrovillar membranes in paraneopteran insect orders with comments on their function and evolutionary significance. Arthropod Struct Dev 33:139–148. doi: 10.1016/j.asd.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 36.Albuquerque-Cunha JM, Gonzalez MS, Garcia ES, Mello CB, Azambuja P, Almeida JC, de Souza W, Nogueira NF. 2009. Cytochemical characterization of microvillar and perimicrovillar membranes in the posterior midgut epithelium of Rhodnius prolixus. Arthropod Struct Dev 38:31–44. doi: 10.1016/j.asd.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 37.Terra WR, Costa RH, Ferreira C. 2006. Plasma membranes from insect midgut cells. An Acad Bras Cienc 78:255–269. doi: 10.1590/S0001-37652006000200007. [DOI] [PubMed] [Google Scholar]
- 38.Alvarenga ES, Mansur JF, Justi SA, Figueira-Mansur J, Dos Santos VM, Lopez SG, Masuda H, Lara FA, Melo AC, Moreira MF. 2016. Chitin is a component of the Rhodnius prolixus midgut. Insect Biochem Mol Biol 69:61–70. doi: 10.1016/j.ibmb.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 39.Liu W, Gray S, Huo Y, Li L, Wei T, Wang X. 2015. Proteomic analysis of interaction between a plant virus and its vector insect reveals new functions of hemipteran cuticular protein. Mol Cell Proteomics 14:2229–2242. doi: 10.1074/mcp.M114.046763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang H, Wu K, Liu Y, Wu Y, Wang X. 2015. Integrative proteomics to understand the transmission mechanism of Barley yellow dwarf virus-GPV by its insect vector Rhopalosiphum padi. Sci Rep 5:10971. doi: 10.1038/srep10971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumari S, Roy S, Singh P, Singla-Pareek SL, Pareek A. 2013. Cyclophilins: proteins in search of function. Plant Signal Behav 8:e22734. doi: 10.4161/psb.22734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu H, Xue Q, Cao W, Yang F, Ma L, Liu W, Zhang K, Liu X, Zhu Z, Zheng H. 2018. Foot-and-mouth disease virus nonstructural protein 2B interacts with cyclophilin A, modulating virus replication. FASEB J 32:6706–6723. doi: 10.1096/fj.201701351. [DOI] [PubMed] [Google Scholar]
- 43.von Hahn T, Ciesek S. 2015. Cyclophilin polymorphism and virus infection. Curr Opin Virol 14:47–49. doi: 10.1016/j.coviro.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kovalev N, Nagy PD. 2013. Cyclophilin A binds to the viral RNA and replication proteins, resulting in inhibition of tombusviral replicase assembly. J Virol 87:13330–13342. doi: 10.1128/JVI.02101-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tamborindeguy C, Bereman MS, DeBlasio S, Igwe D, Smith DM, White F, MacCoss MJ, Gray SM, Cilia M. 2013. Genomic and proteomic analysis of Schizaphis graminum reveals cyclophilin proteins are involved in the transmission of cereal yellow dwarf virus. PLoS One 8:e71620. doi: 10.1371/journal.pone.0071620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang X, Thannhauser TW, Burrows M, Cox-Foster D, Gildow FE, Gray SM. 2008. Coupling genetics and proteomics to identify aphid proteins associated with vector-specific transmission of polerovirus (Luteoviridae). J Virol 82:291–299. doi: 10.1128/JVI.01736-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.An P, Wang LH, Hutcheson-Dilks H, Nelson G, Donfield S, Goedert JJ, Rinaldo CR, Buchbinder S, Kirk GD, O'Brien SJ, Winkler CA. 2007. Regulatory polymorphisms in the cyclophilin A gene, PPIA, accelerate progression to AIDS. PLoS Pathog 3:e88. doi: 10.1371/journal.ppat.0030088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bleiber G, May M, Martinez R, Meylan P, Ott J, Beckmann JS, Telenti A. 2005. Use of a combined ex vivo/in vivo population approach for screening of human genes involved in the human immunodeficiency virus type 1 life cycle for variants influencing disease progression. J Virol 79:12674–12680. doi: 10.1128/JVI.79.20.12674-12680.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chatterji U, Bobardt M, Selvarajah S, Yang F, Tang H, Sakamoto N, Vuagniaux G, Parkinson T, Gallay P. 2009. The isomerase active site of cyclophilin A is critical for hepatitis C virus replication. J Biol Chem 284:16998–17005. doi: 10.1074/jbc.M109.007625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaul A, Stauffer S, Berger C, Pertel T, Schmitt J, Kallis S, Zayas M, Lopez MZ, Lohmann V, Luban J, Bartenschlager R. 2009. Essential role of cyclophilin A for hepatitis C virus replication and virus production and possible link to polyprotein cleavage kinetics. PLoS Pathog 5:e1000546. doi: 10.1371/journal.ppat.1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rits MAN, van Dort KA, Kootstra NA. 2008. Polymorphisms in the regulatory region of the cyclophilin A gene influence the susceptibility for HIV-1 infection. PLoS One 3:e3975. doi: 10.1371/journal.pone.0003975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang F, Robotham JM, Nelson HB, Irsigler A, Kenworthy R, Tang H. 2008. Cyclophilin A is an essential cofactor for hepatitis C virus infection and the principal mediator of cyclosporine resistance in vitro. J Virol 82:5269–5278. doi: 10.1128/JVI.02614-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krupovic M, Koonin EV. 2017. Multiple origins of viral capsid proteins from cellular ancestors. Proc Natl Acad Sci U S A 114:E2401–E2410. doi: 10.1073/pnas.1621061114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brown JR, Doolittle WF. 1997. Archaea and the prokaryote-to-eukaryote transition. Microbiol Mol Biol Rev 61:456–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hsiao KC, Shih NY, Fang HL, Huang TS, Kuo CC, Chu PY, Hung YM, Chou SW, Yang YY, Chang GC, Liu KJ. 2013. Surface α-enolase promotes extracellular matrix degradation and tumor metastasis and represents a new therapeutic target. PLoS One 8:e69354. doi: 10.1371/journal.pone.0069354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Plow EF, Das R. 2009. Enolase-1 as a plasminogen receptor. Blood 113:5371–5372. doi: 10.1182/blood-2009-03-208546. [DOI] [PubMed] [Google Scholar]
- 57.Zhang F, Guo H, Zheng H, Zhou T, Zhou Y, Wang S, Fang R, Qian W, Chen X. 2010. Massively parallel pyrosequencing-based transcriptome analyses of small brown planthopper (Laodelphax striatellus), a vector insect transmitting rice stripe virus (RSV). BMC Genomics 11:303. doi: 10.1186/1471-2164-11-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Munoz Mde L, Limon-Camacho G, Tovar R, Diaz-Badillo A, Mendoza-Hernandez G, Black WC. 4th, 2013. Proteomic identification of dengue virus binding proteins in Aedes aegypti mosquitoes and Aedes albopictus cells. Biomed Res Int 2013:875958. doi: 10.1155/2013/875958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Colpitts TM, Cox J, Nguyen A, Feitosa F, Krishnan MN, Fikrig E. 2011. Use of a tandem affinity purification assay to detect interactions between West Nile and dengue viral proteins and proteins of the mosquito vector. Virology 417:179–187. doi: 10.1016/j.virol.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Popova-Butler A, Dean DH. 2009. Proteomic analysis of the mosquito Aedes aegypti midgut brush border membrane vesicles. J Insect Physiol 55:264–272. doi: 10.1016/j.jinsphys.2008.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Van Lint P, Libert C. 2007. Chemokine and cytokine processing by matrix metalloproteinases and its effect on leukocyte migration and inflammation. J Leukoc Biol 82:1375–1381. doi: 10.1189/jlb.0607338. [DOI] [PubMed] [Google Scholar]
- 62.Rojo G, Chamorro M, Salas ML, Vinuela E, Cuezva JM, Salas J. 1998. Migration of mitochondria to viral assembly sites in African swine fever virus-infected cells. J Virol 72:7583–7588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bandla MD, Campbell LR, Ullman DE, Sherwood JL. 1998. Interaction of tomato spotted wilt tospovirus (TSWV) glycoproteins with a thrips midgut protein, a potential cellular receptor for TSWV. Phytopathology 88:98–104. doi: 10.1094/PHYTO.1998.88.2.98. [DOI] [PubMed] [Google Scholar]
- 64.Bautista RC, Mau RFL, Cho JJ, Custer DM. 1995. Potential of tomato spotted wilt tospovirus plant hosts in Hawaii as virus reservoirs for transmission by Frankliniella occidentalis (Thysanoptera: Thripidae). Phytopathology 85:953–958. doi: 10.1094/Phyto-85-953. [DOI] [Google Scholar]
- 65.Martin K, Kopperud K, Chakrabarty R, Banerjee R, Brooks R, Goodin MM. 2009. Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J 59:150–162. doi: 10.1111/j.1365-313X.2009.03850.x. [DOI] [PubMed] [Google Scholar]
- 66.Cilia M, Fish T, Yang X, McLaughlin M, Thannhauser TW, Gray S. 2009. A comparison of protein extraction methods suitable for gel-based proteomic studies of aphid proteins. J Biomol Tech 20:201–215. [PMC free article] [PubMed] [Google Scholar]
- 67.Cilia M, Tamborindeguy C, Fish T, Howe K, Thannhauser TW, Gray S. 2011. Genetics coupled to quantitative intact proteomics links heritable aphid and endosymbiont protein expression to circulative polerovirus transmission. J Virol 85:2148–2166. doi: 10.1128/JVI.01504-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rotenberg D, Whitfield AE. 2010. Analysis of expressed sequence tags for Frankliniella occidentalis, the western flower thrips. Insect Mol Biol 19:537–551. doi: 10.1111/j.1365-2583.2010.01012.x. [DOI] [PubMed] [Google Scholar]
- 69.Dobson L, Remenyi I, Tusnady GE. 2015. CCTOP: a consensus constrained TOPology prediction Web server. Nucleic Acids Res 43:W408–W412. doi: 10.1093/nar/gkv451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nielsen H. 2017. Predicting secretory proteins with SignalP. Methods Mol Biol 1611:59–73. doi: 10.1007/978-1-4939-7015-5_6. [DOI] [PubMed] [Google Scholar]
- 71.Ioannidou ZS, Theodoropoulou MC, Papandreou NC, Willis JH, Hamodrakas SJ. 2014. CutProtFam-Pred: detection and classification of putative structural cuticular proteins from sequence alone, based on profile hidden Markov models. Insect Biochem Mol Biol 52:51–59. doi: 10.1016/j.ibmb.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Magkrioti CK, Spyropoulos IC, Iconomidou VA, Willis JH, Hamodrakas SJ. 2004. cuticleDB: a relational database of arthropod cuticular proteins. BMC Bioinformatics 5:138. doi: 10.1186/1471-2105-5-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Adkins S, Choi TJ, Israel BA, Bandla MD, Richmond KE, Schultz KT, Sherwood JL, German TL. 1996. Baculovirus expression and processing of tomato spotted wilt tospovirus glycoproteins. Phytopathology 86:849–855. doi: 10.1094/Phyto-86-849. [DOI] [Google Scholar]
- 75.Chakrabarty R, Banerjee R, Chung SM, Farman M, Citovsky V, Hogenhout SA, Tzfira T, Goodin M. 2007. PSITE vectors for stable integration or transient expression of autofluorescent protein fusions in plants: probing Nicotiana benthamiana-virus interactions. Mol Plant Microbe Interact 20:740–750. doi: 10.1094/MPMI-20-7-0740. [DOI] [PubMed] [Google Scholar]
- 76.Jones DT, Taylor WR, Thornton JM. 1992. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The GenBank accession numbers for six TIPs are MH884760 for cyclophilin, MH884759 for enolase, MH884758 for cuticular protein (CP-V), MH884757 for endoCP-GN, MH884756 for endoCP-V, and MH884761 for mitochondrial ATP synthase α.