Recombinant vaccines based on vaccinia virus and particularly attenuated strains such as MVA are in human clinical trials, but due to the complexity of these large vectors much remains to be understood about the design parameters that alter their immunogenicity. Previous work had found that MVA vectors should be designed to express stable protein in order to induce robust immunity by CD8+ (cytotoxic) T cells. Here, we found that the primacy of stable antigen is not generalizable to all designs of MVA and may depend where a foreign antigen is inserted into the MVA genome. This unexpected finding suggests that there is an interaction between genome location and the best form of antigen for optimal T cell priming in MVA and thus possibly other vaccine vectors. It also highlights that our understanding of antigen presentation by even the best studied of vaccine vectors remains incomplete.
KEYWORDS: CD8+ T cells, CTL, cytotoxic T cells, MVA, modified vaccinia virus Ankara, antigen presentation, antigen processing, live vector vaccines, vaccinia virus
ABSTRACT
A variety of strains of vaccinia virus (VACV) have been used as recombinant vaccine vectors with the aim of inducing robust CD8+ T cell immunity. While much of the pioneering work was done with virulent strains, such as Western Reserve (WR), attenuated strains such as modified vaccinia virus Ankara (MVA) are more realistic vectors for clinical use. To unify this literature, side-by-side comparisons of virus strains are required. Here, we compare the form of antigen that supports optimal CD8+ T cell responses for VACV strains WR and MVA using equivalent constructs. We found that for multiple antigens, minimal antigenic constructs (epitope minigenes) that prime CD8+ T cells via the direct presentation pathway elicited optimal responses from both vectors, which was surprising because this finding contradicts the prevailing view in the literature for MVA. We then went on to explore the discrepancy between current and published data for MVA, finding evidence that the expression locus and in some cases the presence of the viral thymidine kinase may influence the ability of this strain to prime optimal responses from antigens that require direct presentation. This extends our knowledge of the design parameters for VACV vectored vaccines, especially those based on MVA.
IMPORTANCE Recombinant vaccines based on vaccinia virus and particularly attenuated strains such as MVA are in human clinical trials, but due to the complexity of these large vectors much remains to be understood about the design parameters that alter their immunogenicity. Previous work had found that MVA vectors should be designed to express stable protein in order to induce robust immunity by CD8+ (cytotoxic) T cells. Here, we found that the primacy of stable antigen is not generalizable to all designs of MVA and may depend where a foreign antigen is inserted into the MVA genome. This unexpected finding suggests that there is an interaction between genome location and the best form of antigen for optimal T cell priming in MVA and thus possibly other vaccine vectors. It also highlights that our understanding of antigen presentation by even the best studied of vaccine vectors remains incomplete.
INTRODUCTION
Vaccinia virus (VACV) was one of the first vectors for recombinant vaccines and candidates have now progressed to clinical trials. The generation of strong CD8+ T cell immunity to foreign antigens encoded in a VACV vector is well agreed upon, in mouse (1–3), nonhuman primate (4–6), and human (7, 8) models. However, while the bulk of historic studies aimed to understand antigen presentation from VACV were done with virulent strains such as Western Reserve (WR), these viruses have an unacceptable risk profile and are not suitable for use as human vaccines. Safer alternatives have emerged and among these is modified vaccinia virus Ankara (MVA); however, there is a lack of studies that examine MVA and WR in parallel, and such studies are required to unify a broad literature.
MVA is a hyperattenuated VACV strain that lacks immune evasion proteins and virulence factors and does not productively replicate in human tissue (9–11). Clinical trials with MVA as a smallpox vaccine have demonstrated safety, even in HIV- positive individuals (12, 13). These characteristics, along with the capacity to insert up to 25 kbp of foreign genes (10, 14), makes MVA an ideal vector for recombinant vaccines, a potential that is being pursued through the development pipeline into clinical trials (15–24). Further work is progressing to improve immunogenicity and assess the risks of broad release of MVA as a recombinant vaccine (25, 26), but these are large vectors, leaving many aspects of their biology incompletely studied.
A key design parameter for vaccines that induce CD8+ T cells is the form of antigen expressed, be that full-length protein and other stable polypeptides or, conversely, rapidly degraded antigen forms, such as ubiquitin-antigen fusion proteins, artificial polyepitopes, and minimal epitope constructs (the latter are referred to here as minigenes). This is an important consideration because of the manner in which each of these is processed and presented on major histocompatibility complex (MHC) class I to activate or prime CD8+ T cells (27). Epitope minigenes bypass the requirement for proteasomal processing from larger polypeptides and so enter the antigen presentation pathway very efficiently in infected cells and are therefore present in great abundance on MHC. However, these short peptides are rapidly degraded, being shown to have a half-life of under 10 s (28). For this reason, although minigenes, and indeed all rapidly degraded polypeptides, prime well by the direct presentation pathway, i.e., via vaccine-infected dendritic cells (DCs), they do not survive long enough to be picked up and cross presented by uninfected DCs (29–31). Epitopes from stable antigens are directly presented on virus-infected DCs at various levels, but in many cases they can also be cross primed (27). Direct presentation is linked closely with translation, and so the structures or functions of full-length proteins are not associated with the levels of presentation by this pathway, unlike factors such as the efficiency of processing and the rate of translation (27, 32–35). Cross presentation is more likely to be influenced by the features of proteins, but with the exception of stability this has not been systematically explored. Further, the relative roles of these two pathways remain difficult to dissect for stable proteins, so it is safest to assume that both pathways may be used; but if neither is efficient, the responses elicited may be modest (36, 37). Finally, we note that while epitope minigenes are themselves not realistic vaccine constructs, because they prime responses to a single epitope presented by a single MHC they are a good model for all rapidly degraded antigen forms, as noted above.
Many studies over 20 years have shown that minigenes (or rapidly degraded polypeptides) encoded by VACV WR induce a CD8+ T cell immune response that is always as strong and often significantly stronger than a corresponding full-length stable antigen (37–41). Importantly, this finding holds irrespective of the nature or function of the full-length protein and implies that for recombinant VACV strain WR (rWR), optimizing antigens for effective direct presentation is always adequate and often ideal. In contrast to this, for MVA the prevailing view in the field is that minigenes and other rapidly degraded polypeptides prime poorly. This is based on a key study that directly examined priming requisites for this vector and concluded that stable antigens are best because cross priming is dominant (42). Notably, that report also examined multiple antigens, always finding the same result, suggesting that the finding would generalize broadly. This has been more recently supported by evidence that a second wave of presentation of antigens, which for MVA occurs exclusively by cross presentation, is required for CD4+ T cell help and full development of CD8+ T cell responses (43). Further, mouse knockout models have found that molecules required for cross presentation impact the immunogenicity of MVA more than WR (44). However, many studies have found that MVA can infect DCs, and initial activation of CD8+ T cells by direct priming has been visualized (42, 43, 45–48). In addition, effective priming of a minigene by MVA has been noted as an incidental finding elsewhere (37).
The purpose of this study was to reconcile these opposing findings for WR and MVA, as well as the possible discrepancy across findings with MVA. To do this, we revisited the preferred form of antigen for CD8+ T cell priming by recombinant VACV (rVACV) based on strains WR and MVA. Our approach at the outset was to examine a broad range of antigens, which included: herpes simplex virus (HSV) glycoprotein B (gB), a highly immunodominant viral glycoprotein; influenza virus PB1F2, a very weakly immunogenic intracellular viral protein; VACV B8, a dominant native antigen of VACV; and ovalbumin, a classic model antigen for which published data already exist for WR and MVA. By choosing this broad array of antigen/epitope pairs, we sought to identify unifying patterns.
RESULTS
A minimal epitope construct of HSV gB498 is more immunogenic than the full-length protein when expressed both from VACV WR and from MVA.
The first foreign antigen/epitope we examined was gB of HSV, from which the highly immunodominant epitope gB498 is derived (49). Recombinant VACV strain WR (rWR) viruses expressing full-length gB and an endoplasmic reticulum (ER)-targeted minigene (minigene-gB498) from the J2R gene under the p7.5 promoter have been published previously (50, 51). ER-targeted epitope minigenes deliver minimal epitope sequences directly into the ER, and so their presentation in infected cells is generally independent of the transporters associated with antigen presentation, but they behave similarly to cytosolic epitope minigenes and other rapidly degraded antigen forms in terms of priming pathway preference (30, 52). J2R encodes the thymidine kinase (TK), and this function is lost in viruses made in this way due to insertional inactivation. This insertion site is referred to here as the TK locus. For this study, we generated recombinant MVA (rMVA) viruses that were matched to the rWRs above in the forms of antigen, site of insertion, and promoter. As controls, we used viruses that had insertions in the TK locus but that expressed no foreign viral protein.
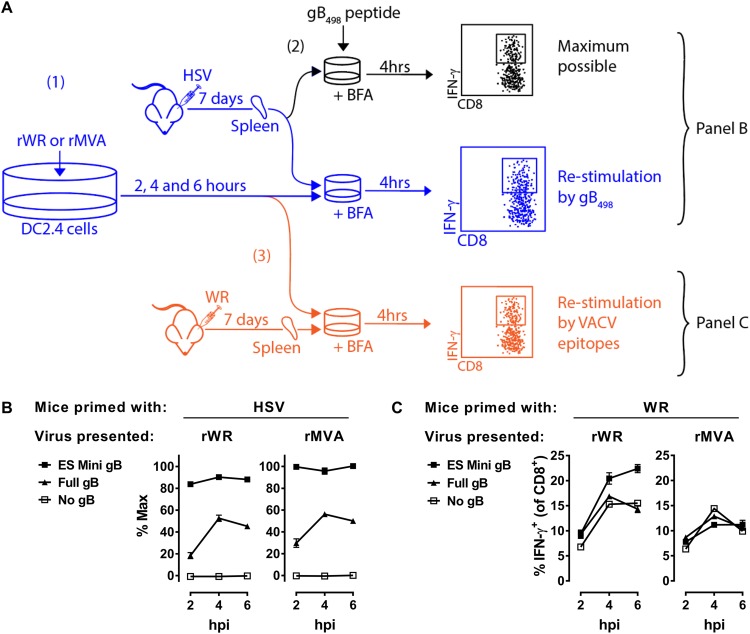
One of the limitations of using epitope minigenes is that their expression cannot be detected by conventional methods, such as Western blotting. For this reason, we needed a way to detect the epitopes presented in association with MHC class I (MHC-I) on cells infected with these viruses to ensure that all were being expressed. Further, by detecting the level of presentation, we have some indication of how well each of the viruses might perform in direct priming, assuming that the vectors can infect the relevant DCs in vivo. To do this for HSV gB498, we used an assay based on the ability of cells infected with our rVACV in vitro to restimulate CD8+ T cells from mice acutely infected with HSV (Fig. 1A). We used the C57BL/6-derived cell line DC2.4, and after 2, 4, or 6 h of infection with the rVACVs, cocultured these cells with splenocytes taken from a mouse 7 days after infection with HSV. The coculture was done in the presence of brefeldin A and restimulation of CD8+ T cells was determined by the detection of intracellular gamma interferon (IFN-γ) by flow cytometry (Fig. 1A depicted in blue). We cultured another aliquot of the same splenocytes with 1 × 10−7 M gB498 peptide and again measured the intracellular IFN-γ to establish a maximum possible response (Fig. 1A, depicted in black). This allowed the response from rVACV-infected cells to be plotted as a percentage of the maximum possible response. This was necessary for standardization across experiments. As expected, for both rWR and rMVA, cells infected with viruses expressing minigene-gB498 were the better at restimulating CD8+ T cells from HSV-infected mice than those infected with the viruses expressing full-length gB (Fig. 1B). At the same time, viruses with no form of gB failed to restimulate the HSV-immune splenocytes, showing there was no cross-reactivity between HSV- and VACV-specific CD8+ T cells. Finally, to ensure that these results were not due to differing efficiencies of the infections or other factors that might impact antigen presentation in general across the various viruses, portions of the same batches of infected cells used above were tested for their ability to stimulate CD8+ T cells from VACV WR-infected mice (Fig. 1A, depicted in orange). Restimulation of WR-immune CD8+ T cells was similar among the cultures infected with the three rWRs and also across those infected with the three rMVA (Fig. 1C). These controls suggest that the infections were all similarly efficient, at least within a VACV strain. Taken all together, we interpret the results of these experiments as showing that (i) full-length gB and minigene-gB498 were expressed and presented on MHC-I as anticipated from the rWR and rMVAs but (ii) that minigene-gB498 was a more efficient form of antigen in terms of direct presentation of the gB498 epitope on infected cells.
FIG 1.
Full-length gB and minigene-gB498 are expressed from rVACVs. (A) Diagram of experimental plan. DC2.4 cells were infected with rVACVs for 2, 4, or 6 h, and then the levels of HSV gB498 presentation on MHC-I were determined by coculture with HSV-immune splenocytes in the presence of brefeldin A, followed by flow cytometry to identify IFN-γ+ CD8+ T cells (blue, step 1). The HSV-immune splenocytes were separately stimulated with gB498 peptide for 4 h, and the percentages of CD8+ T cells that were IFN-γ+ were measured to provide a maximum possible gB498-specific response (black, step 2). Results from cocultures of infected cells with HSV-immune splenocytes are shown as a percentage of this maximum possible response. Finally, separate aliquots of the same infected cultures were incubated with VACV-immune splenocytes, and IFN-γ+ CD8+ T cells were again counted to determine the general levels of infection and antigen presentation (orange, step 3). (B) Data reflecting the levels of gB489 presented on triplicate cultures of rWR- or rMVA-infected cells from the experiment described above. The virus strain is shown above graphs, and the form of gB antigen expressed is in the key. (C) Data reflecting levels of VACV antigen presentation on the same triplicate cultures of rWR- and rMVA-infected cells. Means and standard errors of triplicates are shown; some errors are obscured by data points. The experiment was repeated with similar results.
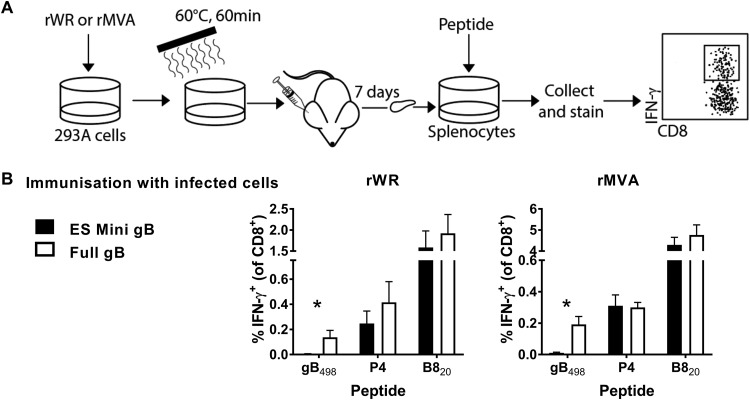
Next, we used an established vaccination method to determine whether the gB-containing antigens expressed by our viruses in infected cells might be able to cross prime CD8+ T cells in vivo (30, 37, 52, 53). As a source of antigen, human 293A cells were infected with rWR and rMVA viruses and then heat killed to destroy any infectivity, which includes residual viral inoculum as well as any replicated virus. These cells were used to vaccinate mice by intradermal (i.d.) injection of ear pinnae and, after 7 days, epitope-specific responses were determined by a short ex vivo culture with synthetic peptides and staining for CD8 and IFN-γ (Fig. 2A). We measured responses to gB498 and, to ensure similar immunization across viruses, selected VACV epitopes, including the dominant B820 and a set of less-dominant epitopes comprising A47138, A47171, L253, and A3270, for which responses were summed and are referred to as P4. All of these epitopes are shared between WR and MVA (54, 55). Both for rWR and for rMVA, only cells infected with viruses that expressed the full-length gB were able to prime a gB498-specific CD8+ T cell response above background in mice (Fig. 2B). The sizes of these responses were low, at around 0.2% of CD8+ T cells, but this is in the range published for similar types of experiments (30, 37). At the same time, rWR-infected cells primed responses to the VACV epitopes to similar levels irrespective of the form of gB expressed, and the same was observed for the pair of rMVAs. These data suggest that full-length gB but not minigene-gB498 can be cross primed in vivo. We do not interpret this result to mean that the full-length antigens are necessarily cross primed after an infection of mice with these viruses, just that to the limit of detection of this assay the minigenes are not able to be cross presented, consistent with published results for other antigens (30, 56).
FIG 2.
Cells infected with rVACVs expressing full-length gB, but not minigene-gB498, can cross prime CD8+ T cells in vivo. (A) Diagram of the experimental plan. Human 293A cells were infected with rWR or rMVA for a total of 6 h, and then counted and heat treated to eliminate any infectivity. Mice were immunized with these cells by i.d. injection and, after 7 days, epitope-specific CD8+ T cell responses were measured by in vitro culture with peptides and flow cytometry for CD8 and IFN-γ. (B) Data from the experiment described in panel A. Mice were immunized with cells infected with the VACV strain as shown above the graphs expressing the form of gB498 as shown in the key. The epitopes are shown on the x axis; P4 is the sum of responses to A47138, A47171, L253, and A3270. Means and standard errors of data from six mice combined from two independent experiments are shown (*, P < 0.05).
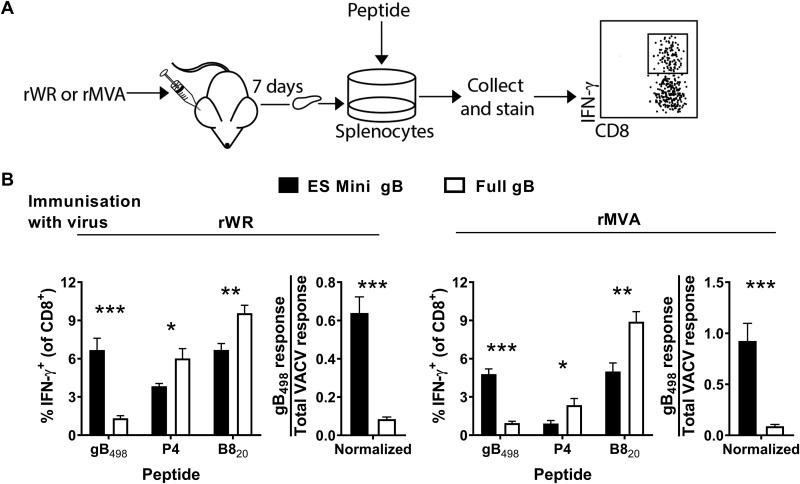
Finally, we wanted to determine for rWR and rMVA the form of HSV gB that was most immunogenic after a standard infection of mice, which is the key experiment with these viruses. To do this, mice were infected by the i.d. route with the gB498-presenting rWR and rMVA viruses, and CD8+ T cell responses to gB498, P4, and B820 in the spleen were measured 7 days later (Fig. 3A). As published previously for rWR (51), minigene-gB498 primed significantly more CD8+ T cells than full-length gB, but more surprisingly, this result was recapitulated by the rMVA viruses (Fig. 3B). Indeed, the strength of the minigene-gB498 priming from both vectors appeared to compete with native VACV epitopes, including the usually dominant B820 epitope, such that responses were reduced in mice infected with the minigene, compared with full-length gB-expressing viruses. However, we cannot be absolutely sure that reduction of response to the VACV epitopes does not reflect some disadvantage in infection or general antigen presentation for viruses expressing minigene-gB498. For this reason, we also plotted gB498-specific responses normalized against the total VACV-specific response (sum of P4 and B8), which highlighted further the difference between minigene and full-length HSV gB-expressing viruses (Fig. 3B).
FIG 3.
Minigene-gB498 is more effective than full-length gB for priming CD8+ T cells when expressed from rWR and rMVA. (A) Diagram of the experimental plan. Mice were infected with rWR and rMVA viruses expressing versions of HSV gB498 by i.d. injection and, after 7 days, epitope-specific CD8+ T cell responses were measured by in vitro culture with peptides and flow cytometry for CD8 and IFN-γ. (B) Data from the experiment described in panel A. Mice were immunized with rWRs (left) or rMVAs (right) expressing the form of gB498 as shown in the key. The peptides are shown on the x axis. The graph on the right for each VACV strain shows the gB498-specific response divided by the total VACV-specific response (i.e., the sum of P4 and B820) to account for any differences in infection across the viruses. Means and standard errors of data from six mice combined from two independent experiments are shown (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
From these data, we concluded that for the highly dominant HSV gB498 epitope, the priming requisites for rWR and rMVA are the same when similar constructs are compared, the form of gB optimized for direct presentation (Fig. 1) and not able to cross prime (Fig. 2) being the most immunogenic (Fig. 3).
Minigene-IAV PB1-F262 is more immunogenic than full-length PB1-F2 when expressed from VACV strains WR and MVA.
We were concerned that the gB498 epitope might not be representative, perhaps because of its immunodominant nature or its structure as a surface glycoprotein, so we wanted to determine whether the results above would be consistent for a very weak, cytoplasmic antigen. We chose a subdominant influenza A virus (IAV) antigen/epitope, namely, the nonstructural PB1-F2 protein and its epitope PB1-F262 (57). The viruses used were matched with those expressing HSV gB above in promoter and insertion site. The rWRs were made by others (53, 58), but the rMVAs were made for this study. The same TK– control viruses were used.
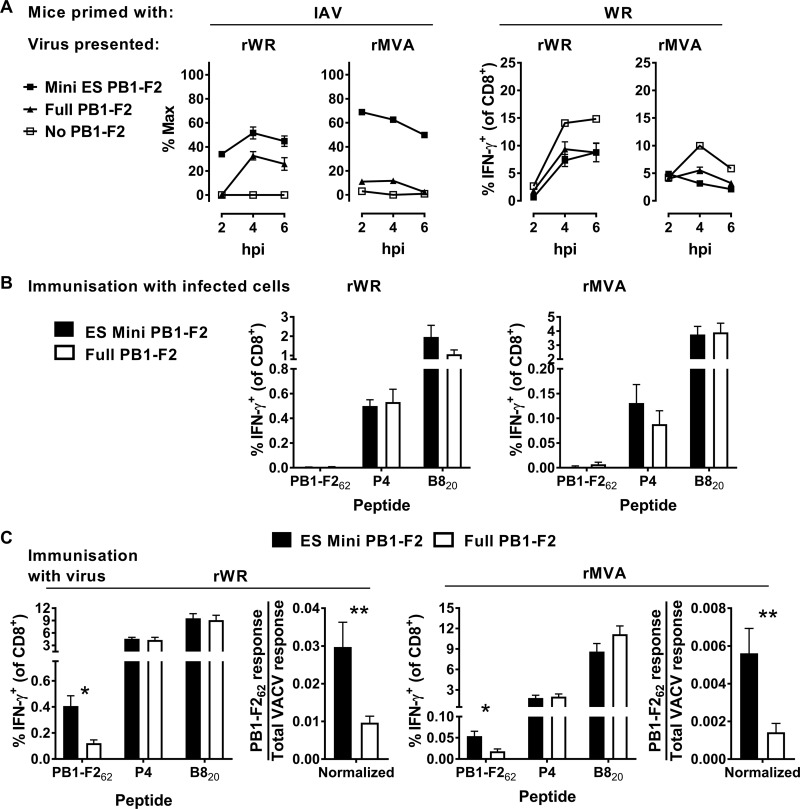
Following the experimental pattern established in the previous section, we first tested the ability of these rVACVs to express their antigens in infected cells, as reflected by the presentation of PB1-F262 on MHC-I. The experimental scheme was as shown in Fig. 1A, but this time we used splenocytes from IAV-primed mice to detect the presentation of PB1-F262. All viruses with PB1-F2, irrespective of the form, were detected by the IAV-immune CD8+ T cells, but the control WR and MVA were not detected. As expected, both for rWR and for rMVA, cells infected with viruses expressing minigene-PB1-F262 were the best at restimulating CD8+ T cells from IAV-infected mice (Fig. 4A, left). The control restimulations of WR-immune splenocytes showed that the VACV-derived epitopes were presented equally across the set of rWRs and across the set of rMVAs, so that within each strain the infection was equally efficient (Fig. 4A, right). We interpret these results, taken together, to show that these sets of viruses expressed their antigens and that there was more efficient presentation of PB1-F262 on infected cells expressing the minigene than the full-length PB1-F2.
FIG 4.
Presentation to, and priming of, CD8+ T cells by PB1-F262 expressed in different forms by rWR and rMVA. (A) Results obtained according to the experimental design in Fig. 1A show the extent of restimulation of CD8+ T cells from IAV-immune (left) or VACV WR-immune (right) splenocytes by cells infected by the virus strain, as shown above graphs, expressing the forms of PB1-F262 as shown in the key. For the cocultures with IAV-immune splenocytes (left), data are presented relative to the maximum possible value obtained by stimulation of the same spleen cells with PB1-F262 peptide. Means and standard errors of triplicates are shown. The experiment was repeated with similar results. (B) Results obtained according to the experimental design shown in Fig. 2A, except viruses expressed versions of PB1-F262 (as shown). Epitope-specific responses are shown as the percentage of CD8+ T cells making IFN-γ. (C) Results obtained according to the experimental design in Fig. 3A. Mice were infected with rWR and rMVA viruses expressing versions of PB1-F262 (as shown) and, 7 days later, the epitope-specific responses were measured. The graph on the right for each VACV strain shows the PB1-F262-specific response divided by the total VACV-specific response. For panels B and C, the means and standard errors of data from nine mice from three independent experiments are shown (*, P < 0.05; **, P < 0.01).
Next, we used the in vivo assay based on immunization of mice with infected and heat-treated 293A cells to determine whether full-length and minigene-PB1-F2 were able to cross prime CD8+ T cells. As above, we measured responses against PB1-F262 and as controls, VACV epitopes including B820, and the P4 set of epitopes. None of the four viruses was able to prime a PB1-F262-specific CD8+ T cell response that was clearly above background in these experiments, despite the VACV antigens being able to elicit responses at the expected levels (Fig. 4B). These data are consistent with the poor immunogenicity of PB1-F262 when mice were immunized with IAV-infected MC57G cells lacking TAP (59). Further, it is possible that expression of PB1-F2 protein is not well tolerated by cells in the context of infection with one or both rVACVs, and this is reflected in the lack of a response. Arguing against this is that the responses to the native VACV epitopes (P4 and B8) were similar for the viruses expressing full-length PB1-F2 (presumably functional) and minigene-PB1-F262 (not functional). So, for PB1-F262 we were not able to confirm whether either form of antigen was able to cross prime CD8+ T cells, probably due to the poor immunogenicity of this epitope.
Finally, mice were infected with the PB1-F262-presenting viruses to determine for each vector which form of antigen was most immunogenic in a live-virus vaccination. All responses to PB1-F262 were exceptionally weak, being just above background; however, all the average responses were above zero by more than the 95% confidence level (not shown). Further, the minigene-PB1-F262 construct primed significantly more CD8+ T cells than full-length PB1-F2, when expressed both from rWR and from rMVA (Fig. 4C). The CD8+ T cell response to the native VACV epitopes appeared to be similar across the viruses from each strain; however, to take this into account formally, we normalized PB1-F262-specific responses against the total anti-VACV response. Doing this confirmed that the responses primed by minigenes were stronger. Therefore, we conclude that minigenes are the optimum form of PB1-F262 for stimulating CD8+ T cell responses for rWR and rMVA vectors, even though responses were very low and close to the limit of detection.
With the exception of our inability to detect cross priming from infected cells, these data recapitulate what was seen for HSV gB498: there was no difference in the preferred form of antigen for priming CD8+ T cells between rWR and rMVA, and directly presented minigenes were more immunogenic than a full-length antigen.
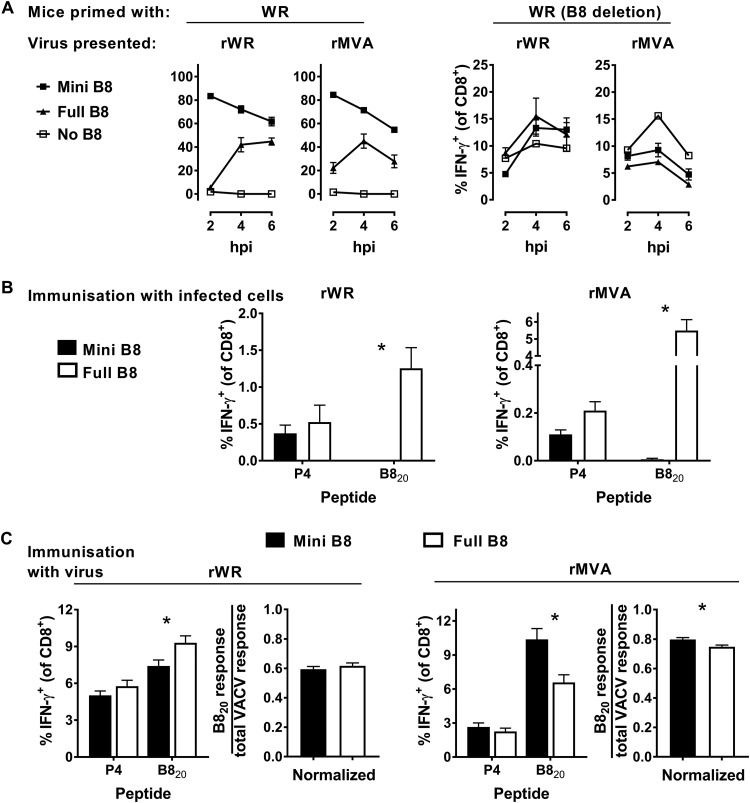
Minigene-B820 is more immunogenic than full-length B8 when expressed from MVA.
The two antigens examined thus far were expressed from the same promoter and were inserted into the TK gene, so we then extended the study to a VACV antigen expressed by its own promoter, from its native location. We chose the highly immunogenic VACV B820 epitope and compared antigen presentation and immunogenicity from wild-type viruses and from recombinants where the B8R open reading frame had been replaced with a short sequence encoding a B820 minigene. It should be noted here that B8 from MVA is truncated and predicted to be nonfunctional, so the nonrecombinant WR and MVA viruses that serve to provide full-length B8 protein are not strictly equivalent. However, (i) in well-controlled studies WR B8 fails to function in mice due to an inability to adequately bind mouse IFN-γ, so neither WR or MVA form of this protein should impact immune responses (60), and (ii) our data from Fig. 2 and 4B, in which B8 is used as a control native VACV antigen, confirm that both forms of B8 are stable enough to support very effective cross priming.
Direct presentation in vitro was used to ensure that all viruses expressed their antigens and present B820, as described above, but for these experiments the source of primed CD8+ T cells was mice infected with an IAV that expresses the B820 epitope as part of the neuraminidase stalk. Again as expected, all antigens were expressed, and cells infected with the minigene-B820-expressing viruses were better at restimulating CD8+ T cells from IAV-B820-immune splenocytes (Fig. 5A). At the same time, DC2.4 cells infected with viruses lacking B8 failed to restimulate the B820-immune splenocytes but cells infected with all viruses were able to restimulate CD8+ T cells from VACV WR-infected mice. Again, this verified that our rWR and rMVA viruses were expressing antigen and presenting B820 on infected cells as expected.
FIG 5.
Presentation to, and priming of, CD8+ T cells by B820 expressed in different forms by rWR and rMVA. (A) Results obtained according to the experimental design in Fig. 1A showing the extent of restimulation of CD8+ T cells from IAV-miniB820-immune (left) or VACV WR(delB8)-immune (right) splenocytes by cells infected by the virus strain, as shown above the graphs, expressing the forms of B820 as shown in the key. For the cocultures with IAV-miniB820-immune splenocytes (left), the data are presented relative to the maximum possible value obtained by stimulation of the same spleen cells with B820 peptide. Means and standard errors of triplicates are shown. The experiment was repeated with similar results. (B) Results obtained according to the experimental design shown in Fig. 2A, except the viruses expressed versions of B8 (as shown). Epitope-specific responses are shown as the percentage of CD8+ T cells making IFN-γ. (C) Results obtained according to the experimental design in Fig. 3A. Mice were infected with rWR and rMVA viruses expressing versions of B8 (as shown) and, 7 days later, the epitope-specific responses were measured. The graph on the right for each VACV strain shows the B8-specific response divided by the total VACV-specific response. For panels B and C, means and standard errors of data from 15 mice from five independent experiments are shown (*, P < 0.05).
Next, we used heat-killed rVACV-infected cells to test the ability of B8 from these viruses to cross prime a CD8+ T cell response. Cells infected with wild-type WR and MVA viruses, but not the recombinants expressing minigene-B820, were able to provide antigen that cross primed B820-specific CD8+ T cells (Fig. 5B). This result for the wild-type B8 versions back up the data from Fig. 2 and 4B, again confirming that not only is the truncated B8 from MVA able to be cross primed, it is better in this assay than the full-length B8 from WR. It is possible that B8 from WR is largely lost from infected cells because it is secreted and that the truncated version from MVA is either retained or perhaps remains associated with cells to a greater extent. We also noted that the mean response to the P4 set of epitopes was lower for the minigene-B820 viruses than for corresponding wild types, but this did not reach statistical significance for WR or MVA. These data together support the idea that both versions of full-length B8 are able to be cross primed but, as expected, minigene-B820 is unable to cross prime CD8+ T cells.
Having again established that the B820-presenting viruses used in this section behaved as expected in the previous experiments, we infected mice with these viruses to test them as live vaccines. When mice were infected with these viruses in the case of WR, the wild-type virus initially appeared to induce a significantly larger B820-specific CD8+ T cell response than the minigene-B820 recombinant; however, this trend was also noted for the P4 set of other VACV epitopes. To account for a possible difference in infections with these viruses, the data were normalized as a ratio of the B820-specific to the total anti-VACV response. In this analysis, there was no longer any significant difference between the two WR viruses, so it seems likely that the two forms of B820 were equally effective (Fig. 5C, left). In contrast, for MVA, the minigene-B820-expressing virus induced a very slightly larger B820-specific CD8+ T cell response than the wild-type virus (Fig. 5C, right). Furthermore, this difference between rMVAs remained significantly different when normalized against the total anti-VACV responses.
Thus, we concluded that for a VACV antigen expressed in its native condition there was no advantage from expression as a minigene by strain WR. However, this was not true for MVA, where the direct-priming minigene construct elicited a slightly, but significantly higher CD8+ T cell response. Whether this difference between strains is due to the variants of native B8 expressed by these viruses or other factors such as the extent of B8 immunodominance remains to be determined (54, 61).
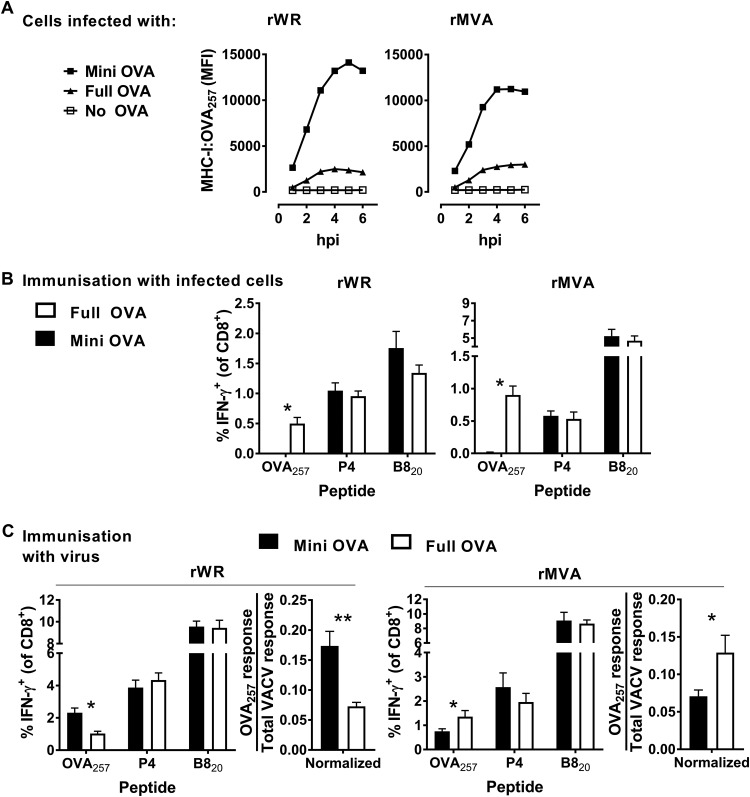
Replication of a published result for MVA expressing the OVA257 peptide from the delIII locus.
All the data above seemed to contradict the key study on antigen presentation from MVA by suggesting that minigenes, which require direct presentation, are an optimal form of antigen to induce a CD8+ T cell response using this VACV strain. So, we next used one of the same sets of rMVAs used by Gasteiger et al. (42) to repeat a published experiment, albeit using our i.d. infection route compared to the original intraperitoneal infections. These rMVAs, expressed either ovalbumin (OVA) or an minigene-OVA257 from the delIII locus, the site of one of the major genomic deletions in MVA. This site does not exist in the WR genome, so for comparison we used rWRs that expressed the same antigens from the TK locus, maintaining consistency with the other WR viruses used here. The presentation of OVA257 can be directly measured using a monoclonal antibody (25D1.16), which detects this peptide when presented by H-2Kb, and flow cytometry (62). This is a much simpler and more strictly quantitative compared to the assays using splenocytes described for the other antigens above, so we used this method to ensure all antigens were expressed and to examine direct presentation. As has been published elsewhere, cells infected with these viruses expressing minigene-OVA257 presented substantially more OVA257 compared to those infected with rVACVs expressing full-length OVA (Fig. 6A) (42, 63).
FIG 6.
Presentation to, and priming of, CD8+ T cells by OVA257 expressed in different forms by rWR and rMVA. (A) The extent of cell surface presentation of MHC-I:OVA257 complexes was determined in DC2.4 cells after infection with OVA-, mini-OVA-, or no-OVA-expressing rWR and rMVA. MHC-I:OVA257 complexes were detected by 25D1.16 antibodies, and the MFI was determined by flow cytometry. (B) Results obtained according to the experimental design shown in Fig. 2A, except the viruses expressed versions of OVA (as shown). Epitope-specific responses are shown as the percentage of CD8+ T cells making IFN-γ. (C) Results obtained according to the experimental design in Fig. 3A. Mice were infected with rWR and rMVA viruses expressing versions of OVA (as shown) and, 7 days later, the epitope-specific responses were measured. The graph on the right for each VACV strain shows the OVA257-specific response divided by the total VACV-specific response. For panels B and C, means and standard errors of data from nine mice combined from three independent experiments are shown (*, P < 0.05; **, P < 0.01).
Next, we used heat-killed virus-infected cells to test the ability of these forms of OVA to cross prime CD8+ T cells in vivo. Irrespective of virus strain, only cells infected with viruses that express full-length OVA were able to induce a CD8+ T cell response (Fig. 6B), which is consistent with published results for rWRs (30).
Finally, mice were infected with the OVA257-expressing rWR and rMVA viruses to test the optimal form of antigen in our i.d. infection model. Consistent with all the data here for other antigens, and as published previously (41), rWR encoded minigene-OVA257 was more immunogenic than full-length OVA (Fig. 6C, left). However, unlike our data here for other antigens and consistent with the published pattern for rMVA (42), minigene-OVA257 was significantly less immunogenic than the full-length protein. Again, to formally take into account possible variations in infections across these viruses, we normalized OVA257-specific response to the total anti-VACV response (Fig. 6C). This additional analysis supported the initial comparison made with the unnormalized values. Thus, we were able to confirm previously published results for MVA, irrespective of the different route of infection, which is consistent with route not being important for determining antigen preference for MVA (42). This result confirms a contradiction between the rMVA results obtained with OVA as published previously (42) and those acquired for all three other antigens examined here.
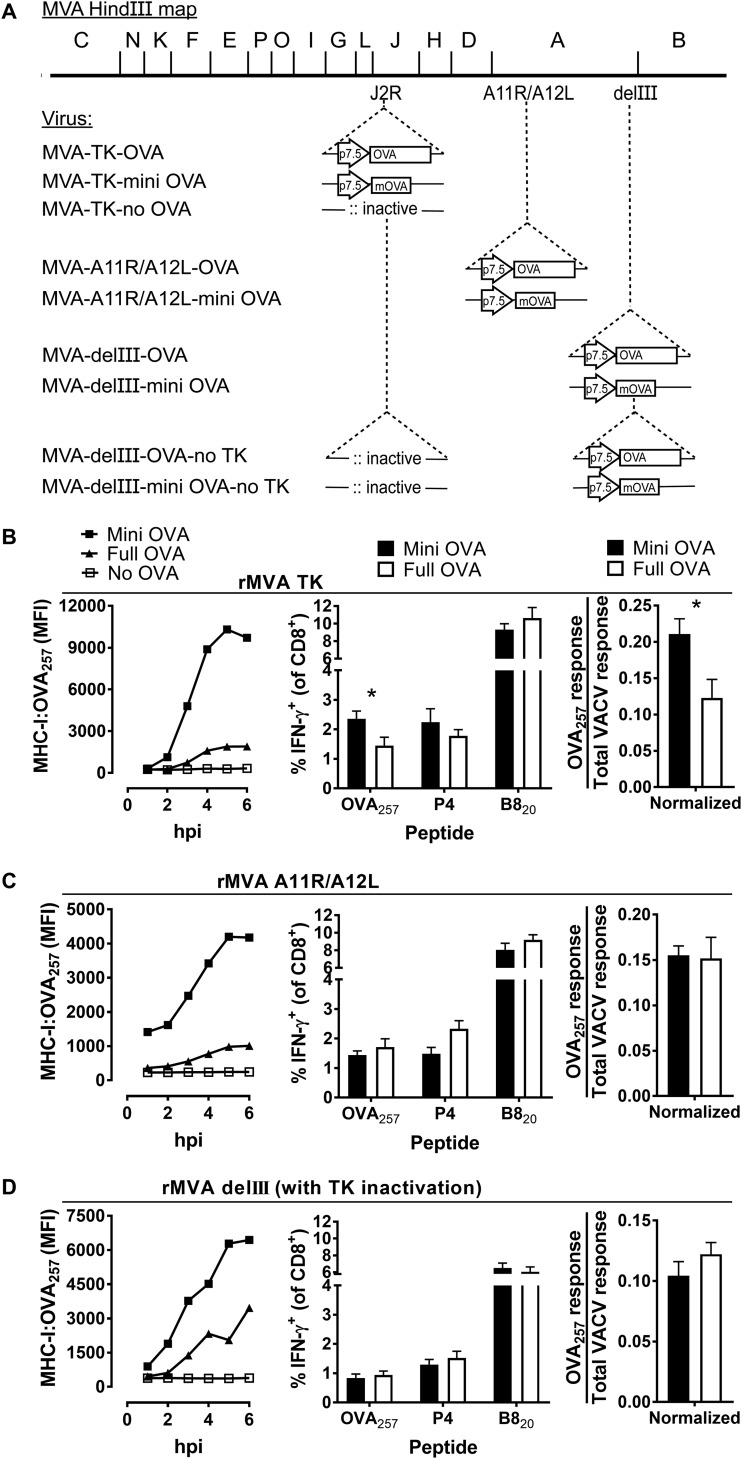
Genome location and TK gene function affect the relative immunogenicity of OVA and minigene-OVA257 expressed by rMVAs.
To address the discrepancy noted above, we considered the differences between the OVA-expressing MVAs and other foreign gene-expressing viruses used here. These were (i) the antigen, (ii) the genomic location from which antigens were expressed, and (iii) the presence of a functional TK gene, which is inactivated when this locus is used to insert foreign antigens. We examined these differences with six new viruses: an OVA and minigene-OVA257 pair of rMVAs with these antigens inserted into the TK locus; a second pair of rMVAs with these antigens expressed from the intergenic space between A11R and A12L; and finally, TK– variants of the rMVAs with OVA and mini-OVA257 expressed from the delIII region (Fig. 5A). We tested expression and direct presentation on cells infected with these pairs of rMVAs in vitro and the immunogenicity of OVA257 in mice infected with these viruses.
Expression and antigen presentation on infected DC2.4 cells was tested with the 25D1.16 monoclonal antibody and flow cytometry. We found that all of the pairs of rMVAs behaved as expected with all presenting OVA257, and cells expressing the minigene viruses presented this epitope at higher levels than those infected with their paired full-length OVA-expressing counterpart (Fig. 7B to D, left). There were some differences in the data across the various pairs, with the A11R/A12L insertion and TK– variant of MVA expressing OVA from the delIII locus apparently presenting OVA257 less efficiently than the other viruses. However, we are presenting raw mean fluorescence data and that limits direct comparisons.
FIG 7.
Presentation to, and priming of, CD8+ T cells by OVA257 expressed in different loci of rMVA and without a functional TK. (A) MVA genome maps showing HindIII fragments and the site of insertion of OVA antigenic constructs and the promoter (arrow). The “m” denotes the minigene (OVA257). (B to D, left) Extent of cell surface presentation of MHC-I:OVA257 complexes on DC2.4 cells infected with viruses. (B to D, middle) Mice were infected with rMVA viruses expressing versions of OVA257 (as shown) and, 7 days later, the responses to peptides were measured. (B to D, right) Using the data shown in the middle graphs, the OVA257-specific response was normalized by dividing by the total VACV-specific response. Means and standard errors of data from at least nine mice combined from three independent experiments are shown (*, P < 0.05).
When mice were infected with these viruses, two different patterns of immunogenicity were observed between full-length and minigene-OVA257 rMVAs. For the first pair of viruses, where antigen was expressed from the TK locus, minigene-OVA257 induced higher CD8+ T cell responses than full-length OVA (Fig. 7B, middle). This result was then tested further by normalization against the total VACV-specific responses and the difference remained in favor of minigene-OVA257 and was statistically significant (Fig. 7B, right). This was the opposite of the results obtained in the previous figure using rMVAs expressing OVA257 constructs from the delIII-based recombinants. It did, however, match the results for all the other antigen pairs that we have tested when expressed from the TK region and also for B820. This suggests that the locus of expression, rather than the antigen examined (OVA), is responsible for the apparent contradiction in previous data. For the remaining pairs of rMVAs, the full-length and minigene constructs elicited an equivalent OVA257-specific CD8+ T cell response; again, this was supported by formal normalization (Fig. 7C and D, middle and right). These data suggest that genome location and TK function can contribute to the relative immunogenicity of unstable and stable polypeptides that present OVA257 when expressed from rMVA.
DISCUSSION
This study started with the aim of carrying out a comprehensive side-by-side comparison of the antigen requisites and therefore priming pathway preferences for virulent VACV, strain WR, and for MVA. The results for rWR viruses were entirely in line with what was first shown more than 2 decades ago, specifically that short-lived constructs were the optimum antigen form to elicit CD8+ T cell responses (41). We have simply extended the data set to more antigens and expression sites in the virus and used more quantitative tools to quantify T cell responses (64). In contrast, the results for rMVAs were unexpected and so merit further discussion.
The first study of the optimum antigen forms for priming CD8+ T cells by rMVA examined multiple antigens and antigen forms, but all being expressed from delIII (42). These included full-length antigens, as well as an ubiquitylated version of tyrosinase, an epitope of tyrosinase that is derived from the leader sequence of that protein and the minigene-OVA257. There are further unpublished constructs with antigens inserted into the delIII region that all behave in the same way as shown elsewhere (I. Drexler, data not shown). Thus, there is strong evidence that whenever antigens are expressed from the delIII region of MVA, unstable antigens that can support only direct priming will induce poor CD8+ T cell responses compared to stable antigens. This was the current state of understanding in the field when we began our experiments. However, these published results contrast with the data for all of the pairs of rMVAs that we made for this study. We present data for several antigens and, in our case, three different genomic loci and come to the opposite conclusion. The differences across these two studies suggest strongly that the antigen form that primes optimal CD8+ T cell responses when expressed from rMVA differs according to the expression locus. Further support for this conclusion comes from our direct comparison of rMVAs that present the OVA257 epitope. We used a set of OVA-expressing rMVAs examined in the original study noted above and confirmed the published result with our model (Fig. 6C). This was followed by our own recombinants that differed only in that expression was from the TK locus and not delIII and found the opposite result (Fig. 7A). These results were supported by a rigorous normalization process to take into account any difference in infections (e.g., that might be caused by inaccurate virus titers used to infect the mice). In looking at these findings, it is pertinent to ask whether it was the immunogenicity of the full-length gene or the minigene (or both) that varied because this might suggest the priming pathway that is working with differing efficiency across the pairs. While a direct statistical comparison is not possible, across these two pairs of viruses (Fig. 6C to 7A) it can be seen that the immunogenicity of the minigene varies substantially, but the full-length construct elicits a similar response irrespective of expression locus. This is seen most clearly in the normalized data (Fig. 6C, far right, and Fig. 7A, right), where for full-length constructs the OVA257/total VACV-specific responses are almost identical (0.129 and 0.123), but for the minigenes they vary almost 3-fold (from 0.071 to 0.211). Taken together, the best explanation these findings, ours in the present study and those of Gasteiger et al. is that the genome location of a foreign antigen influences whether antigens designed for direct or cross priming are likely to be most immunogenic when expressed from an rMVA.
MVAs that have a transgene inserted in the delIII region differ in two ways from the majority of our viruses in which the viral TK gene was used: the genomic site of insertion and the presence of the viral TK. In teasing these two factors apart, we found that when OVA257 antigens were inserted in the A11R/A12L intergenic space, the full-length gene and the minigene were equally immunogenic (Fig. 7C). This result was half way between those for the corresponding rMVA with delIII and TK locus insertions. This suggested that inactivation of TK was played some role in improved immunogenicity of minigenes over full-length protein expressed from the TK locus. Further support for this was provided by our results when we inactivated TK in the original delIII-inserted rMVAs (Fig. 7D). However, we also note that when B820 was expressed as a minigene from its native location in MVA, it outperformed the full protein in terms of immunogenicity (Fig. 5C). This suggests that there is a role for other factors, perhaps the antigen/epitope, e.g., if the antigen is very poorly cross primed, or the precursor frequency of T cells that recognize a particular epitope, may also be important. Taken together, however, our data establish roles both for insertion site of transgene and for TK function in determining the immunogenicity of epitope minigenes from rMVA at least for one model antigen.
The reasons why these two factors might alter priming preferences for CD8+ T cells are not clear. Differences in immunogenicity across sites and with variation due to TK function have been noted previously for virulent VACV, but in that case these differences were linked to transgene expression level (65). As noted above, in every experiment we found enhanced presentation of epitopes from minigenes over those from full-length proteins on infected DC2.4 cells. In addition, cells infected with MVAs that express full-length OVA generate enough of this protein to cross prime CD8+ T cell responses in vivo as shown in Fig. 6B for the delIII insertion and as published previously for a TK insertion (37). Having said this, we are unable to know how much of each full-length antigen is expressed and more importantly its capacity to cross prime in vivo. However, differences in immunogenicity not linked to expression have been seen previously for MVA (66). We speculate that differences in genomic insertion sites can change the interaction of rMVA with the DCs responsible for priming CD8+ T cells. In the case of rMVAs that used the delIII region, while no functional genes are disrupted, we wonder whether the transcription that is driven into the neighboring viral genes alters their expression, leading to changes in infected DCs. In support of this general concept, we have noticed that insertions that include an expressed gene into the A11R/A12L intergenic space can alter the immunogenicity of multiple native VACV epitopes, whereas a promoterless insertion did not have this effect (L. C. W. Lin and D. C. Tscharke, unpublished data). In contrast to these loci, use of the TK region leads to a loss of TK function. Indeed, this locus was originally chosen because this phenotype provided a selectable marker for recombinant viruses (67–69). VACV TK functions to phosphorylate thymidine, producing dTMP, which after a series of phosphorylation events is utilized in de novo DNA synthesis (70). In vitro, the function of this gene is not required, but the loss of TK is substantially attenuating for virulent VACVs in vivo, reducing viral loads (54, 71). However, MVA fails to replicate in vivo, and so we did not expect to find any impact of TK deletion. Further, TK deletion from WR does not substantially change the specificity of CD8+ T cell responses, despite the attenuation noted above (54). Finally, although MVA does replicate its genome in some cells, and so inhibition of this DNA synthesis might have an impact, in the case of DCs MVA aborts infection at an earlier stage, suggesting that this is not relevant for direct priming (72). It remains possible that there is some effect of TK function on DCs, perhaps prolonging their lifespan during infection. We have explored this in vitro and found no impact of TK expression from MVA on DC viability (Y. C. Wong and D. C. Tscharke, unpublished data), but our in vitro DC cultures are unlikely to model the situation in vivo very faithfully, all the more so given that a second wave of presentation has been shown to be required for full immunogenicity of VACVs (43). The role of TK and of transcription that runs into neighboring genes in MVA vaccines requires a more thorough investigation.
Our data may also have implications for our understanding of the ability of MVA to support direct priming in general. While there is one well-characterized exception, namely, minigene-IAV PA224, minigenes (either ER targeted or cytosolic) and other rapidly degraded constructs have never been found to be cross presented. This is consistent over multiple studies, and the expression of these antigens can therefore be used to establish the effectiveness of direct presentation (30, 38, 42, 51, 52, 56, 73, 74). Thus, the finding by Gasteiger et al. that multiple rapidly degraded proteins were poorly immunogenic suggested that direct priming is inefficient for MVA in general. We now find here that several rMVA minigene viruses are able to elicit CD8+ T cell responses at least as effectively as full-length proteins and often significantly better. This includes four antigens expressed from four loci in the virus, including under a natural promoter in the native location. The only exception is minigene-OVA257 expressed from delIII. The weight of evidence then suggests that MVA in general supports efficient direct priming of CD8+ T cells. There is likely to be a contribution by cross priming as well, but we are unable to determine the relative importance of these pathways for nonrecombinant MVA.
From a practical perspective, our data show that MVA can be an efficient vector for the delivery of antigens for direct presentation. Having noted this general rule, it is clear that the use of the delIII region, and perhaps others yet to be determined, to make rMVAs creates exceptions. This remains important because delIII was the original and remains a commonly used site for the introduction of genes encoding foreign antigens in rMVAs (14, 75). It is also necessary for the interpretation of any experiments where rMVA are used to dissect mechanisms of CD8+ T cell priming. Indeed, the finding that the XCR1+ DCs required for a fully functional CD8+ T cell response used exclusively cross priming was made using H-2Kbm1 mice infected with an rMVA that expressed H-2Kb and OVA from the delIII region. Finally, we have only examined systemic responses and, for the priming of resident memory populations (Trm), cross priming has been shown to be important using mouse knockout models (76).
In conclusion, we show that directly priming minigenes are optimal for CD8+ T cell priming by virulent (WR) and attenuated (MVA) rVACV, with the notable exception of rMVAs that express antigens from the DelIII region. Minigenes, while they are not likely to be used as vaccines themselves, are a model for all forms of rapidly degraded antigens. Other forms, such as ubiquitin fusions or polyepitope constructs, are more practical antigens for vaccines because they can induce responses to multiple epitopes in the context of multiple MHC allomorphs. MVAs with insertions into delIII are common, but it is not the only site used. Indeed, we are not the only group to use rMVAs with antigens expressed from the TK locus, and some of these vaccines have advanced to clinical trials, so our findings have practical implications (22, 77–83). The findings presented here in general highlight that there remain many wrinkles to iron out in our understanding of antigen presentation, even for well-studied viral vectors that have been used in human clinical trials.
MATERIALS AND METHODS
Mice.
Specific-pathogen-free, female, 7- to 14-week-old C57BL/6 mice were obtained from the Australian Phenomics Facility (Canberra, Australia) or ARC (Perth, Australia). All experiments were conducted according to relevant ethical requirements that were approved by the Australian National University Animal Ethics and Experimentation Committee (protocols F.BMB.38.08, A2011.001, A2013.037, and A2016.045).
Cells and nonrecombinant viruses.
For cross presentation assays, 293A cells (ATCC, CRL-1573) were used as antigen donor cells. The C57BL/6 mouse-derived, dendritic cell-like cell line DC2.4 was used for in vitro presentation assays (84). Unmodified Western Reserve vaccinia virus (VACV WR, ATCC VR1354) and MVA were originally a gift from B. Moss (National Institutes of Health, Bethesda, MD). Influenza A virus (IAV) strain PR8 was provided by C. Goodnow (ANU, Canberra Australia). HSV strain KOS was provided by F. Carbone (University of Melbourne, Melbourne, Australia). All strains were grown and titrated according to standard methods.
Recombinant viruses and virus construction.
The VACVs used here are listed in Table 1 with descriptions and origins, if not made for this study. With the exception of the minigene-B820 WR and MVA, which used the native B8 promoter, all antigens were expressed from the VACV p7.5 promoter. Homologous recombination between appropriately designed transfer plasmids supplied by transfection and VACV genomes provided by infection was used to make the new viruses required. Transfer plasmids were based on pSC11 and p7.5GB-ins for insertions into the TK and A11R/A12L regions, respectively (69, 85). Briefly BHK-21 or 293A cells were infected with MVA or WR, respectively, at a multiplicity of infection (MOI) of 0.05 in Dulbecco modified Eagle medium (DMEM) supplemented with 2% fetal bovine serum (FBS) and incubated at 37°C and 5% CO2 for 1 h. The inoculum was then removed, and the transfer plasmid was added in a preincubated transfection mix of plasmid, Lipofectamine 2000 (Life Technologies), and DMEM. Infected cell cultures were incubated with transfection mix for 2 days at 37°C and 5% CO2 before the virus was released by repeated freeze-thaw cycles and sonication. Recombinant viruses were isolated via serial-step-dilution plaque purification and transient dominant selection using blasticidin/green fluorescent protein (GFP) expression or direct staining for β-galactosidase (pSC11-based plasmids only) as previously described (85). PCR was used to identify recombinant viruses, and the recombinant region was verified by sequencing. The IAV strain PR8 that included B820 in the stalk of the neuraminidase protein was generated using a plasmid-based reverse genetic system as previously described for a virus that included OVA257 in the same way (86, 87).
TABLE 1.
Viruses used in this study
| Virusa | Recombinant details | Source or reference |
|---|---|---|
| MVA | Wild-type VACV strain MVA | 89 |
| WR | Wild-type VACV strain WR | 90 |
| WR-Full-gB | Full-length HSV gB in TK | 51 |
| WR-ESmini-gB | ER-targeted minigene-gB498 in TK | 50 |
| MVA-Full-gB | Full-length HSV gB in TK | This study |
| MVA-ESmini-gB | ER-targeted minigene-gB498 in TK | This study |
| WR-TK- | Insertional inactivation of TK | 69 |
| MVA-TK- | Insertional inactivation of TK | This study |
| WR-Full-PB1-F2 | Full-length IAV PB1F2 in TK | 58 |
| WR-ESmini-PB1-F2 | Minigene-IAV PB1F262 in TK | 53 |
| MVA-Full-PB1-F2 | Full-length IAV PB1F2 in TK | This study |
| MVA-ESmini-PB1-F2 | Minigene-IAV PB1F262 in TK | This study |
| IAV-miniB8 | IAV PR8 with sequence encoding B820 inserted into the neuraminidase stalk | This study |
| WR-delB8 | B8R deleted | 60 |
| WR-delB8-miniB8 | B8R replaced with minigene-B820 | 85 |
| MVA-delB8 | B8R deleted | 91 |
| MVA-delB8-miniB8 | B8R replaced with minigene-B820 | This study |
| WR-TK-OVA | Full-length OVA in TK | 41 |
| WR-TK-miniOVA | Minigene-OVA257 in TK | 92 |
| MVA-delIII-OVA | Full length OVA in delIII | 42 |
| MVA-delIII-SIINFEKL | Minigene-OVA257 in delIII | 42 |
| MVA-TK-OVA | Full-length OVA in TK | This study |
| MVA-TK-miniOVA | Minigene-OVA257 in TK | 37 |
| MVA-A11R/A12L-OVA | Full-length OVA in A11R/A12L | This study |
| MVA-A11/A12-SIINFEKL | Minigene-OVA257 in A11R/A12L | This study |
| MVA-delIII-OVA-delTK | Full-length OVA in delIII with insertional inactivation of TK | This study |
| MVA-delIII-SIINFEKL-delTK | Minigene-OVA257 in delIII with insertional inactivation of TK | This study |
The original nonrecombinant parent virus is indicated by the first letters of each name: WR, MVA, or IAV. The order of the viruses is as they appear in the main text, and they are separated into groups according to the figures and legends in which they are described.
Peptides.
Synthetic peptides made to match the sequences of the epitopes of interest (Table 2) were purchased from GenScript (Piscataway, NJ) or Mimotopes (Clayton, Victoria, Australia). Peptide master stocks were made in 100% dimethyl sulfoxide (DMSO), generally at 1 × 10−2 M, and then diluted in serum-free DMSO for use in assays at a final concentration of 1 × 10−7 M.
TABLE 2.
Synthetic peptides used in this study
| Peptidea | Originb | Sequence | MHC | Reference(s) |
|---|---|---|---|---|
| A3270 | VACV, A3270–277 | KSYNYMLL | H-2Kb | 33 |
| A47138 | VACV, A47138–146 | AAFEFINSL | H-2Kb | 61 |
| A47171 | VACV, A47171–180c | YAHINALEYI | H-2Kb | 55 |
| L253 | VACV, L253–61 | VIYIFTVRL | H-2Kb | 33 |
| B820 | VACV, B820–27 | TSYKFESV | H-2Kb | 61 |
| gB498 | HSV-1 gB498–505 | SSIEFARL | H-2Kb | 93 |
| OVA257 | Chicken, OVA257–264 | SIINFEKL | H-2Kb | 94, 95 |
| PB1F262 | IAV strain PR8, PB1F262–70 | LSLRNPILV | H-2Db | 58 |
Data from the first four peptides (A3270, A47138, A47171, and L253) were pooled and are referred to as P4.
That is, the virus/organism, protein, and position of the peptide (in subscript amino acid numbers) in protein.
Numbering from WR, the corresponding peptide from MVA is A47157–166.
In vitro assay for presentation of MHC-I–peptide complexes on infected cells.
For epitopes other than OVA257, DC2.4 cells were infected with VACVs for 2, 4, or 6 h before being placed on ice so that presentation at all time points could be tested simultaneously. Presentation was tested by coculturing the infected DC2.4 with splenocytes from an appropriately immunized mouse at a stimulator/effector ratio of 1:5 for a total of 4 h at 37°C with 5% CO2 and adding brefeldin A to 50 μg/ml after the first hour. Cells were then labeled for CD8 (anti-mouse CD8α-PE; BioLegend, clone 53-6.7) and intracellular IFN-γ (anti-mouse IFN-γ-APC; BioLegend, clone XMG1.2) staining for flow cytometric analysis (88). Events were gated on sequentially on SSC × FSC, SSC × CD8, and CD8 × IFN-γ plots to determine the percentages of CD8+ cells that were IFN-γ+. The same splenocytes were stimulated with synthetic peptide with a sequence matching the epitope of interest, and the conditions for incubation and staining were as described above. The fraction of CD8+ T cells making IFN-γ in these peptide-stimulated cultures was set at 100%, and results from cocultures with infected cells are presented relative to this maximum response. To determine presentation levels of H-2Kb-OVA257, DC2.4 cells were infected for the times noted before labeling with 25D1.16-APC antibody (BioLegend). Labeled cells were fixed in 1% paraformaldehyde before flow cytometric analysis. Events were gated on a SSC × FSC plot, and the mean fluorescence intensity (MFI) of the population was determined.
Infection of cells and immunization of mice to determine cross presentation in vivo.
Single cell suspensions of 293A cells were incubated with indicated viruses at an MOI of 5 PFU/cell in DMEM for 1 h at 37°C with shaking (200 rpm). Infected cells were resuspended in DMEM supplemented with 2% FBS, and infection continued for 5 h at 37°C with slow rotation to stop cells settling which were then counted. To inactivate the residual virus inoculum and inhibit viral replication, infected cells were resuspended in phosphate-buffered saline (PBS) and heat treated at 60°C for 60 min prior to i.d. immunization. Mice were anesthetized with isoflurane and immunized with 2 × 106 cells in 10 μl by i.d. injection of the left ear pinnae. Seven days later, the mice were euthanized, and spleens were taken to determine the extent of the responses to relevant epitopes using stimulation in vitro with synthetic peptides and staining for CD8 and IFN-γ, as described above.
Infection of mice with viruses.
Viruses were diluted in PBS, and 2 × 106 PFU was injected i.d. in the left ear pinnae. After 7 days, the mice were euthanized, and spleens were taken to determine the extent of the CD8+ T cell responses to various epitopes by stimulation with synthetic peptides and staining for CD8 and IFN-γ, as described above.
Flow cytometry and statistical analysis.
Flow cytometry data acquisition was done using an LSR-II flow cytometer (BD Biosciences). Data were analyzed as described above with FlowJo 8.8.4 software (Tree Star, Ashland, OR). Statistical analysis was performed with GraphPad Prism 7 Software. Differences between means were tested using a two-way t test and were considered significant when the P value was <0.05.
ACKNOWLEDGMENTS
We thank B. Moss, J. Yewdell, J. Bennink, F. Carbone, and C. Goodnow for viruses; G. Gasteiger for helpful discussion on the project and manuscript; and the staff of the ANU RSB Animal Services and Australian Phenomics Facility for mouse husbandry.
D.C.T. was funded by NHMRC fellowship (APP1104329) and project grants (APP1084283 and APP1023141). I.D. was supported by DFG funding GRK 1946.
REFERENCES
- 1.Qiu J, Peng S, Ma Y, Yang A, Farmer E, Cheng MA, Roden RBS, Wu TC, Chang Y-N, Hung C-F. 2018. Epithelial boost enhances antigen expression by vaccinia virus for the generation of potent CD8+ T cell-mediated antitumor immunity following DNA priming vaccination. Virology 525:205–215. doi: 10.1016/j.virol.2018.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marin-Lopez A, Calvo-Pinilla E, Barriales D, Lorenzo G, Brun A, Anguita J, Ortego J. 2018. CD8 T cell responses to an immunodominant epitope within the nonstructural protein NS1 provide wide immunoprotection against bluetongue virus in IFNAR–/– mice. J Virol 92. doi: 10.1128/JVI.00938-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altenburg AF, Magnusson SE, Bosman F, Stertman L, de Vries RD, Rimmelzwaan GF. 2017. Protein and modified vaccinia virus Ankara-based influenza virus nucleoprotein vaccines are differentially immunogenic in BALB/c mice. Clin Exp Immunol 190:19–28. doi: 10.1111/cei.13004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asbach B, Kibler KV, Köstler J, Perdiguero B, Yates NL, Stanfield-Oakley S, Tomaras GD, Kao S-F, Foulds KE, Roederer M, Seaman MS, Montefiori DC, Parks R, Ferrari G, Forthal DN, Phogat S, Tartaglia J, Barnett SW, Self SG, Gottardo R, Cristillo AD, Weiss DE, Galmin L, Ding S, Heeney JL, Esteban M, Jacobs BL, Pantaleo G, Wagner R. 2018. Priming with a potent HIV-1 DNA vaccine frames the quality of immune responses prior to a poxvirus and protein boost. J Virol 93:e01529-18. doi: 10.1128/JVI.01529-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marlin R, Nugeyre M-T, Tchitchek N, Parenti M, Hocini H, Benjelloun F, Cannou C, Dereuddre-Bosquet N, Levy Y, Barré-Sinoussi F, Scarlatti G, Le Grand R, Menu E. 2017. Modified vaccinia virus Ankara vector induces specific cellular and humoral responses in the female reproductive tract, the main HIV portal of entry. J Immunol 199:1923–1932. doi: 10.4049/jimmunol.1700320. [DOI] [PubMed] [Google Scholar]
- 6.Garcia-Arriaza J, Perdiguero B, Heeney JL, Seaman MS, Montefiori DC, Yates NL, Tomaras GD, Ferrari G, Foulds KE, Roederer M, Self SG, Borate B, Gottardo R, Phogat S, Tartaglia J, Barnett SW, Burke B, Cristillo AD, Weiss DE, Lee C, Kibler KV, Jacobs BL, Wagner R, Ding S, Pantaleo G, Esteban M. 2017. HIV/AIDS vaccine candidates based on replication-competent recombinant poxvirus NYVAC-C-KC expressing trimeric gp140 and Gag-derived virus-like particles or lacking the viral molecule B19 that inhibits type I interferon activate relevant HIV-1-specific B and T cell immune functions in nonhuman primates. J Virol 91. doi: 10.1128/JVI.02182-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Green CA, Scarselli E, Sande CJ, Thompson AJ, de Lara CM, Taylor KS, Haworth K, Del Sorbo M, Angus B, Siani L, Di Marco S, Traboni C, Folgori A, Colloca S, Capone S, Vitelli A, Cortese R, Klenerman P, Nicosia A, Pollard AJ. 2015. Chimpanzee adenovirus- and MVA-vectored respiratory syncytial virus vaccine is safe and immunogenic in adults. Sci Transl Med 7:300ra126. doi: 10.1126/scitranslmed.aac5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guardo AC, Gómez CE, Díaz-Brito V, Pich J, Arnaiz JA, Perdiguero B, García-Arriaza J, González N, Sorzano COS, Jiménez L, Jiménez JL, Muñoz-Fernández MÁ, Gatell JM, Alcamí J, Esteban M, López Bernaldo de Quirós JC, García F, Plana M, B Study RI. 2017. Safety and vaccine-induced HIV-1 immune responses in healthy volunteers following a late MVA-B boost 4 years after the last immunization. PLoS One 12:e0186602. doi: 10.1371/journal.pone.0186602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blanchard TJ, Alcamí A, Andrea P, Smith GL. 1998. Modified vaccinia virus Ankara undergoes limited replication in human cells and lacks several immunomodulatory proteins: Implications for use as a human vaccine. J Gen Virol 79:1159–1167. doi: 10.1099/0022-1317-79-5-1159. [DOI] [PubMed] [Google Scholar]
- 10.Antoine G, Scheiflinger F, Dorner F, Falkner FG. 1998. The complete genomic sequence of the modified vaccinia Ankara strain: comparison with other orthopoxviruses. Virology 244:365–396. doi: 10.1006/viro.1998.9123. [DOI] [PubMed] [Google Scholar]
- 11.Drexler I, Heller K, Wahren B, Erfle V, Sutter G. 1998. Highly attenuated modified vaccinia virus Ankara replicates in baby hamster kidney cells, a potential host for virus propagation, but not in various human transformed and primary cells. J General Virology 79:347–352. doi: 10.1099/0022-1317-79-2-347. [DOI] [PubMed] [Google Scholar]
- 12.Overton ET, Stapleton J, Frank I, Hassler S, Goepfert PA, Barker D, Wagner E, von Krempelhuber A, Virgin G, Meyer TP, Müller J, Bädeker N, Grünert R, Young P, Rösch S, Maclennan J, Arndtz-Wiedemann N, Chaplin P. 2015. Safety and immunogenicity of modified vaccinia Ankara-Bavarian Nordic smallpox vaccine in vaccinia-naive and experienced human immunodeficiency virus-infected individuals: an open-label, controlled clinical phase II trial. Open Forum Infect Dis 2:ofv040. doi: 10.1093/ofid/ofv040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greenberg RN, Overton ET, Haas DW, Frank I, Goldman M, von Krempelhuber A, Virgin G, Bädeker N, Vollmar J, Chaplin P. 2013. Safety, immunogenicity, and surrogate markers of clinical efficacy for modified vaccinia Ankara as a smallpox vaccine in HIV-infected subjects. J Infect Dis 207:749–758. doi: 10.1093/infdis/jis753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sutter G, Moss B. 1992. Nonreplicating vaccinia vector efficiently expresses recombinant genes. Proc Natl Acad Sci U S A 89:10847–10851. doi: 10.1073/pnas.89.22.10847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo ZS, Lu B, Guo Z, Giehl E, Feist M, Dai E, Liu W, Storkus WJ, He Y, Liu Z, Bartlett DL. 2019. Vaccinia virus-mediated cancer immunotherapy: cancer vaccines and oncolytics. J Immunother Cancer 7:6. doi: 10.1186/s40425-018-0495-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gross L, Lhomme E, Pasin C, Richert L, Thiebaut R. 2018. Ebola vaccine development: Systematic review of preclinical and clinical studies, and meta-analysis of determinants of antibody response variability after vaccination. Int J Infect Dis 74:83–96. doi: 10.1016/j.ijid.2018.06.022. [DOI] [PubMed] [Google Scholar]
- 17.Sebastian S, Gilbert SC. 2016. Recombinant modified vaccinia virus Ankara-based malaria vaccines. Expert Rev Vaccines 15:91–103. doi: 10.1586/14760584.2016.1106319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gomez CE, Perdiguero B, Garcia-Arriaza J, Esteban M. 2013. Clinical applications of attenuated MVA poxvirus strain. Expert Rev Vaccines 12:1395–1416. doi: 10.1586/14760584.2013.845531. [DOI] [PubMed] [Google Scholar]
- 19.Gomez CE, Perdiguero B, Garcia-Arriaza J, Esteban M. 2012. Poxvirus vectors as HIV/AIDS vaccines in humans. Hum Vaccin Immunother 8:1192–1207. doi: 10.4161/hv.20778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boukhebza H, Bellon N, Limacher JM, Inchauspe G. 2012. Therapeutic vaccination to treat chronic infectious diseases: current clinical developments using MVA-based. Vaccines Hum Vaccin Immunother 8:1746–1757. doi: 10.4161/hv.21689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim DW, Krishnamurthy V, Bines SD, Kaufman HL. 2010. TroVax, a recombinant modified vaccinia Ankara virus encoding 5T4: lessons learned and future development. Hum Vaccin 6:784–791. doi: 10.4161/hv.6.10.13144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith CL, Mirza F, Pasquetto V, Tscharke DC, Palmowski MJ, Dunbar PR, Sette A, Harris AL, Cerundolo V. 2005. Immunodominance of poxviral-specific CTL in a human trial of recombinant-modified vaccinia Ankara. J Immunol 175:8431–8437. doi: 10.4049/jimmunol.175.12.8431. [DOI] [PubMed] [Google Scholar]
- 23.Meyer RG, Britten CM, Siepmann U, Petzold B, Sagban TA, Lehr HA, Weigle B, Schmitz M, Mateo L, Schmidt B, Bernhard H, Jakob T, Hein R, Schuler G, Schuler-Thurner B, Wagner SN, Drexler I, Sutter G, Arndtz N, Chaplin P, Metz J, Enk A, Huber C, Wolfel T. 2005. A phase I vaccination study with tyrosinase in patients with stage II melanoma using recombinant modified vaccinia virus Ankara (MVA-hTyr). Cancer Immunol Immunother 54:453–467. doi: 10.1007/s00262-004-0616-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drexler I, Staib C, Sutter G. 2004. Modified vaccinia virus Ankara as antigen delivery system: how can we best use its potential? Curr Opin Biotechnol 15:506–512. doi: 10.1016/j.copbio.2004.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garcia-Arriaza J, Esteban M. 2014. Enhancing poxvirus vectors vaccine immunogenicity. Hum Vaccin Immunother 10:2235–2244. doi: 10.4161/hv.28974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goossens M, Pauwels K, Willemarck N, Breyer D. 2014. Environmental risk assessment of clinical trials involving modified vaccinia virus Ankara (MVA)-based vectors. CGT 13:413–420. doi: 10.2174/156652321306140103221941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tscharke DC, Croft NP, Doherty PC, La Gruta NL. 2015. Sizing up the key determinants of the CD8+ T cell response. Nat Rev Immunol 15:705–716. doi: 10.1038/nri3905. [DOI] [PubMed] [Google Scholar]
- 28.Reits E, Griekspoor A, Neijssen J, Groothuis T, Jalink K, van Veelen P, Janssen H, Calafat J, Drijfhout JW, Neefjes J. 2003. Peptide diffusion, protection, and degradation in nuclear and cytoplasmic compartments before antigen presentation by MHC class I. Immunity 18:97–108. doi: 10.1016/S1074-7613(02)00511-3. [DOI] [PubMed] [Google Scholar]
- 29.Serna A, Ramirez MC, Soukhanova A, Sigal LJ. 2003. Cutting edge: efficient MHC class I cross-presentation during early vaccinia infection requires the transfer of proteasomal intermediates between antigen donor and presenting cells. J Immunol 171:5668–5672. doi: 10.4049/jimmunol.171.11.5668. [DOI] [PubMed] [Google Scholar]
- 30.Norbury CC, Basta S, Donohue KB, Tscharke DC, Princiotta MF, Berglund P, Gibbs J, Bennink JR, Yewdell JW. 2004. CD8+ T cell cross-priming via transfer of proteasome substrates. Science 304:1318–1321. doi: 10.1126/science.1096378. [DOI] [PubMed] [Google Scholar]
- 31.Shen L, Rock KL. 2004. Cellular protein is the source of cross-priming antigen in vivo. Proc Natl Acad Sci U S A 101:3035–3040. doi: 10.1073/pnas.0308345101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Croft NP, Smith SA, Wong YC, Tan CT, Dudek NL, Flesch IEA, Lin LCW, Tscharke DC, Purcell AW. 2013. Kinetics of antigen expression and epitope presentation during virus infection. PLoS Pathog 9:e1003129. doi: 10.1371/journal.ppat.1003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moutaftsi M, Peters B, Pasquetto V, Tscharke DC, Sidney J, Bui HH, Grey H, Sette A. 2006. A consensus epitope prediction approach identifies the breadth of murine T(CD8+)-cell responses to vaccinia virus. Nat Biotechnol 24:817–819. doi: 10.1038/nbt1215. [DOI] [PubMed] [Google Scholar]
- 34.Tellam J, Fogg MH, Rist M, Connolly G, Tscharke D, Webb N, Heslop L, Wang F, Khanna R. 2007. Influence of translation efficiency of homologous viral proteins on the endogenous presentation of CD8+ T cell epitopes. J Exp Med 204:525–532. doi: 10.1084/jem.20062508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu T, Guan J, Handel A, Tscharke DC, Sidney J, Sette A, Wakim LM, Sng XYX, Thomas PG, Croft NP, Purcell AW, La Gruta NL. 2019. Quantification of epitope abundance reveals the effect of direct and cross-presentation on influenza CTL responses. Nat Commun 10:2846. doi: 10.1038/s41467-019-10661-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Norbury CC. 2016. Defining cross presentation for a wider audience. Curr Opin Immunol 40:110–116. doi: 10.1016/j.coi.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 37.Wong YC, Smith SA, Tscharke DC. 2013. Systemic Toll-like receptor ligation and selective killing of dendritic cell subsets fail to dissect priming pathways for anti-vaccinia virus CD8+ T cells. J Virol 87:11978–11986. doi: 10.1128/JVI.01835-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu R-H, Remakus S, Ma X, Roscoe F, Sigal LJ. 2010. Direct presentation is sufficient for an efficient anti-viral CD8+ T cell response. PLoS Pathog 6:e1000768. doi: 10.1371/journal.ppat.1000768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Irvine KR, McCabe BJ, Rosenberg SA, Restifo NP. 1995. Synthetic oligonucleotide expressed by a recombinant vaccinia virus elicits therapeutic CTL. J Immunol 154:4651–4657. [PMC free article] [PubMed] [Google Scholar]
- 40.McCabe BJ, Irvine KR, Nishimura MI, Yang JC, Spiess PJ, Shulman EP, Rosenberg SA, Restifo NP. 1995. Minimal determinant expressed by a recombinant vaccinia virus elicits therapeutic antitumor cytolytic T lymphocyte responses. Cancer Res 55:1741–1747. [PMC free article] [PubMed] [Google Scholar]
- 41.Restifo NP, Bacik I, Irvine KR, Yewdell JW, McCabe BJ, Anderson RW, Eisenlohr LC, Rosenberg SA, Bennink JR. 1995. Antigen processing in vivo and the elicitation of primary CTL responses. J Immunol 154:4414–4422. [PMC free article] [PubMed] [Google Scholar]
- 42.Gasteiger G, Kastenmuller W, Ljapoci R, Sutter G, Drexler I. 2007. Cross-priming of cytotoxic T cells dictates antigen requisites for modified vaccinia virus Ankara vector vaccines. J Virol 81:11925–11936. doi: 10.1128/JVI.00903-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eickhoff S, Brewitz A, Gerner MY, Klauschen F, Komander K, Hemmi H, Garbi N, Kaisho T, Germain RN, Kastenmuller W. 2015. Robust anti-viral immunity requires multiple distinct T cell-dendritic cell interactions. Cell 162:1322–1337. doi: 10.1016/j.cell.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iborra S, Izquierdo HM, Martínez-López MA, Blanco-Menéndez N, Reis e Sousa C, Sancho D. 2012. The DC receptor DNGR-1 mediates cross-priming of CTLs during vaccinia virus infection in mice. J Clin Invest 122:1628–1643. doi: 10.1172/JCI60660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu L, Chavan R, Feinberg MB. 2008. Dendritic cells are preferentially targeted among hematolymphocytes by modified vaccinia virus Ankara and play a key role in the induction of virus-specific T cell responses in vivo. BMC Immunol 9:15–15. doi: 10.1186/1471-2172-9-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dai P, Wang W, Cao H, Avogadri F, Dai L, Drexler I, Joyce JA, Li X-D, Chen Z, Merghoub T, Shuman S, Deng L. 2014. Modified vaccinia virus Ankara triggers type I IFN production in murine conventional dendritic cells via a cGAS/STING-mediated cytosolic DNA-sensing pathway. PLoS Pathog 10:e1003989. doi: 10.1371/journal.ppat.1003989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kastenmüller W, Brandes M, Wang Z, Herz J, Egen JG, Germain RN. 2013. Peripheral prepositioning and local CXCL9 chemokine-mediated guidance orchestrate rapid memory CD8+ T cell responses in the lymph node. Immunity 38:502. doi: 10.1016/j.immuni.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Altenburg AF, van de Sandt CE, Li BWS, MacLoughlin RJ, Fouchier RAM, van Amerongen G, Volz A, Hendriks RW, de Swart RL, Sutter G, Rimmelzwaan GF, de Vries RD. 2017. Modified vaccinia virus Ankara preferentially targets antigen presenting cells in vitro, ex vivo, and in vivo. Sci Rep 7:8580. doi: 10.1038/s41598-017-08719-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wallace ME, Keating R, Heath WR, Carbone FR. 1999. The cytotoxic T-cell response to herpes simplex virus type 1 infection of C57BL/6 mice is almost entirely directed against a single immunodominant determinant. J Virol 73:7619–7626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blaney JE Jr, Nobusawa E, Brehm MA, Bonneau RH, Mylin LM, Fu TM, Kawaoka Y, Tevethia SS. 1998. Immunization with a single major histocompatibility complex class I-restricted cytotoxic T-lymphocyte recognition epitope of herpes simplex virus type 2 confers protective immunity. J Virol 72:9567–9574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin LC, Flesch IE, Tscharke DC. 2013. Immunodomination during peripheral vaccinia virus infection. PLoS Pathog 9:e1003329. doi: 10.1371/journal.ppat.1003329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lev A, Takeda K, Zanker D, Maynard JC, Dimberu P, Waffarn E, Gibbs J, Netzer N, Princiotta MF, Neckers L, Picard D, Nicchitta CV, Chen W, Reiter Y, Bennink JR, Yewdell JW. 2008. The exception that reinforces the rule: cross-priming by cytosolic peptides that escape degradation. Immunity 28:787–798. doi: 10.1016/j.immuni.2008.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lev A, Dimberu P, Das SR, Maynard JC, Nicchitta CV, Bennink JR, Yewdell JW. 2009. Efficient cross-priming of antiviral CD8+ T cells by antigen donor cells is GRP94 independent. J Immunol 183:4205–4210. doi: 10.4049/jimmunol.0901828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Flesch IEA, Hollett NA, Wong YC, Quinan BR, Howard D, da Fonseca FG, Tscharke DC. 2015. Extent of systemic spread determines CD8+ T cell immunodominance for laboratory strains, smallpox vaccines, and zoonotic isolates of vaccinia virus. Ji 195:2263–2272. doi: 10.4049/jimmunol.1402508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yuen TJ, Flesch IE, Hollett NA, Dobson BM, Russell TA, Fahrer AM, Tscharke DC. 2010. Analysis of A47, an immunoprevalent protein of vaccinia virus, leads to a reevaluation of the total antiviral CD8+ T cell response. J Virol 84:10220–10229. doi: 10.1128/JVI.01281-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wolkers MC, Brouwenstijn N, Bakker AH, Toebes M, Schumacher TNM. 2004. Antigen bias in T cell cross-priming. Science 304:1314–1317. doi: 10.1126/science.1096268. [DOI] [PubMed] [Google Scholar]
- 57.Andreansky SS, Stambas J, Thomas PG, Xie W, Webby RJ, Doherty PC. 2005. Consequences of immunodominant epitope deletion for minor influenza virus-specific CD8+-T-cell responses. J Virol 79:4329–4339. doi: 10.1128/JVI.79.7.4329-4339.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen W, Calvo PA, Malide D, Gibbs J, Schubert U, Bacik I, Basta S, O’Neill R, Schickli J, Palese P, Henklein P, Bennink JR, Yewdell JW. 2001. A novel influenza A virus mitochondrial protein that induces cell death. Nat Med 7:1306–1312. doi: 10.1038/nm1201-1306. [DOI] [PubMed] [Google Scholar]
- 59.Chen W, Masterman KA, Basta S, Haeryfar SM, Dimopoulos N, Knowles B, Bennink JR, Yewdell JW. 2004. Cross-priming of CD8+ T cells by viral and tumor antigens is a robust phenomenon. Eur J Immunol 34:194–199. doi: 10.1002/eji.200324257. [DOI] [PubMed] [Google Scholar]
- 60.Symons JA, Tscharke DC, Price N, Smith GL. 2002. A study of the vaccinia virus interferon-gamma receptor and its contribution to virus virulence. J Gen Virol 83:1953–1964. doi: 10.1099/0022-1317-83-8-1953. [DOI] [PubMed] [Google Scholar]
- 61.Tscharke DC, Karupiah G, Zhou J, Palmore T, Irvine KR, Haeryfar SMM, Williams S, Sidney J, Sette A, Bennink JR, Yewdell JW. 2005. Identification of poxvirus CD8+ T cell determinants to enable rational design and characterization of smallpox vaccines. J Exp Med 201:95–104. doi: 10.1084/jem.20041912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Porgador A, Yewdell JW, Deng Y, Bennink JR, Germain RN. 1997. Localization, quantitation, and in situ detection of specific peptide–MHC class I complexes using a monoclonal antibody. Immunity 6:715–726. doi: 10.1016/S1074-7613(00)80447-1. [DOI] [PubMed] [Google Scholar]
- 63.Princiotta MF, Finzi D, Qian SB, Gibbs J, Schuchmann S, Buttgereit F, Bennink JR, Yewdell JW. 2003. Quantitating protein synthesis, degradation, and endogenous antigen processing. Immunity 18:343–354. doi: 10.1016/S1074-7613(03)00051-7. [DOI] [PubMed] [Google Scholar]
- 64.Flesch IEA, Hollett NA, Wong YC, Tscharke DC. 2012. Linear fidelity in quantification of anti-viral CD8+ T cells. PLoS One 7:e39533. doi: 10.1371/journal.pone.0039533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Coupar BE, Oke PG, Andrew ME. 2000. Insertion sites for recombinant vaccinia virus construction: effects on expression of a foreign protein. J Gen Virol 81:431–439. doi: 10.1099/0022-1317-81-2-431. [DOI] [PubMed] [Google Scholar]
- 66.Manuel ER, Wang ZD, Li ZQ, La Rosa C, Zhou WD, Diamond DJ. 2010. Intergenic region 3 of modified vaccinia Ankara is a functional site for insert gene expression and allows for potent antigen-specific immune responses. Virology 403:155–162. doi: 10.1016/j.virol.2010.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mackett M, Smith GL, Moss B. 1982. Vaccinia virus: a selectable eukaryotic cloning and expression vector. Proc Natl Acad Sci U S A 79:7415–7419. doi: 10.1073/pnas.79.23.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mackett M, Smith GL, Moss B. 1984. General method for production and selection of infectious vaccinia virus recombinants expressing foreign genes. J Virol 49:857–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chakrabarti S, Brechling K, Moss B. 1985. Vaccinia virus expression vector: coexpression of β-galactosidase provides visual screening of recombinant virus plaques. Mol Cell Biol 5:3403–3409. doi: 10.1128/MCB.5.12.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dubbs DR, Kit S. 1964. Isolation and properties of vaccinia mutants deficient in thymidine kinase-inducing activity. Virology 22:214–225. doi: 10.1016/0042-6822(64)90006-6. [DOI] [PubMed] [Google Scholar]
- 71.Buller RM, Smith GL, Cremer K, Notkins AL, Moss B. 1985. Decreased virulence of recombinant vaccinia virus expression vectors is associated with a thymidine kinase-negative phenotype. Nature 317:813–815. doi: 10.1038/317813a0. [DOI] [PubMed] [Google Scholar]
- 72.Chahroudi A, Garber DA, Reeves P, Liu L, Kalman D, Feinberg MB. 2006. Differences and similarities in viral life cycle progression and host cell physiology after infection of human dendritic cells with modified vaccinia virus Ankara and vaccinia virus. J Virol 80:8469–8481. doi: 10.1128/JVI.02749-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fischer MA, Tscharke DC, Donohue KB, Truckenmiller ME, Norbury CC. 2007. Reduction of vector gene expression increases foreign antigen-specific CD8+ T-cell priming. J Gen Virol 88:2378–2386. doi: 10.1099/vir.0.83107-0. [DOI] [PubMed] [Google Scholar]
- 74.Heipertz EL, Davies ML, Lin E, Norbury CC. 2014. Prolonged antigen presentation following an acute virus infection requires direct and then cross-presentation. J Immunol 193:4169–4177. doi: 10.4049/jimmunol.1302565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sutter G, Wyatt LS, Foley PL, Bennink JR, Moss B. 1994. A recombinant vector derived from the host range-restricted and highly attenuated MVA strain of vaccinia virus stimulates protective immunity in mice to influenza virus. Vaccine 12:1032–1040. doi: 10.1016/0264-410X(94)90341-7. [DOI] [PubMed] [Google Scholar]
- 76.Iborra S, Martínez-López M, Khouili SC, Enamorado M, Cueto FJ, Conde-Garrosa R, del Fresno C, Sancho D. 2016. Optimal generation of tissue-resident but not circulating memory T cells during viral infection requires cross priming by DNGR-1+ dendritic cells. Immunity 45:847–860. doi: 10.1016/j.immuni.2016.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hanke T, Blanchard TJ, Schneider J, Ogg GS, Tan R, Becker M, Gilbert SC, Hill AV, Smith GL, McMichael A. 1998. Immunogenicities of intravenous and intramuscular administrations of modified vaccinia virus Ankara-based multi-CTL epitope vaccine for human immunodeficiency virus type 1 in mice. J Gen Virol 79:83–90. doi: 10.1099/0022-1317-79-1-83. [DOI] [PubMed] [Google Scholar]
- 78.Hanke T, McMichael AJ. 2000. Design and construction of an experimental HIV-1 vaccine for a year-2000 clinical trial in Kenya. Nat Med 6:951–955. doi: 10.1038/79626. [DOI] [PubMed] [Google Scholar]
- 79.Palmowski MJ, Choi EM-L, Hermans IF, Gilbert SC, Chen JL, Gileadi U, Salio M, Van Pel A, Man S, Bonin E, Liljestrom P, Dunbar PR, Cerundolo V. 2002. Competition between CTL narrows the immune response induced by prime-boost vaccination protocols. J Immunol 168:4391–4398. doi: 10.4049/jimmunol.168.9.4391. [DOI] [PubMed] [Google Scholar]
- 80.Sanchez-Puig JM, Sanchez L, Roy G, Blasco R. 2004. Susceptibility of different leukocyte cell types to vaccinia virus infection. Virol J 1:10. doi: 10.1186/1743-422X-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schneider J, Gilbert SC, Blanchard TJ, Hanke T, Robson KJ, Hannan CM, Becker M, Sinden R, Smith GL, Hill AV. 1998. Enhanced immunogenicity for CD8+ T cell induction and complete protective efficacy of malaria DNA vaccination by boosting with modified vaccinia virus Ankara. Nat Med 4:397–402. doi: 10.1038/nm0498-397. [DOI] [PubMed] [Google Scholar]
- 82.Smith CL, Dunbar PR, Mirza F, Palmowski MJ, Shepherd D, Gilbert SC, Coulie P, Schneider J, Hoffman E, Hawkins R, Harris AL, Cerundolo V. 2005. Recombinant modified vaccinia Ankara primes functionally activated CTL specific for a melanoma tumor antigen epitope in melanoma patients with a high risk of disease recurrence. Int J Cancer 113:259–266. doi: 10.1002/ijc.20569. [DOI] [PubMed] [Google Scholar]
- 83.Cottingham MG, Andersen RF, Spencer AJ, Saurya S, Furze J, Hill AV, Gilbert SC. 2008. Recombination-mediated genetic engineering of a bacterial artificial chromosome clone of modified vaccinia virus Ankara (MVA). PLoS One 3:e1638. doi: 10.1371/journal.pone.0001638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shen Z, Reznikoff G, Dranoff G, Rock KL. 1997. Cloned dendritic cells can present exogenous antigens on both MHC class I and class II molecules. J Immunology 158:2723–2730. [PubMed] [Google Scholar]
- 85.Wong YC, Lin LC, Melo-Silva CR, Smith SA, Tscharke DC. 2011. Engineering recombinant poxviruses using a compact GFP-blasticidin resistance fusion gene for selection. J Virol Methods 171:295–298. doi: 10.1016/j.jviromet.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 86.Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. 2000. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc Natl Acad Sci U S A 97:6108–6113. doi: 10.1073/pnas.100133697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jenkins MR, Webby R, Doherty PC, Turner SJ. 2006. Addition of a prominent epitope affects influenza A virus-specific CD8+ T cell immunodominance hierarchies when antigen is limiting. J Immunol 177:2917–2925. doi: 10.4049/jimmunol.177.5.2917. [DOI] [PubMed] [Google Scholar]
- 88.Flesch IA, Wong Y, Tscharke D. 2012. Analyzing CD8 T cells in mouse models of poxvirus infection, p 199–218. In Isaacs SN. (ed), Vaccinia virus and poxvirology, vol 890 Humana Press, New York, NY. [DOI] [PubMed] [Google Scholar]
- 89.Mayr A, Hochstein-Mintzel V, Stickl H. 1975. Abstammung, Eigenschaften und Verwendung des Attenuierten Vaccinia-Stammes MVA. Infection 3:6–14. doi: 10.1007/BF01641272. [DOI] [Google Scholar]
- 90.Parker RF, Bronson LH, Green RH. 1941. Further studies of the infectious unit of vaccinia. J Exp Med 74:263–281. doi: 10.1084/jem.74.3.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kastenmuller W, Gasteiger G, Gronau JH, Baier R, Ljapoci R, Busch DH, Drexler I. 2007. Cross-competition of CD8+ T cells shapes the immunodominance hierarchy during boost vaccination. J Exp Med 204:2187–2198. doi: 10.1084/jem.20070489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bacik I, Cox JH, Anderson R, Yewdell JW, Bennink JR. 1994. TAP (transporter associated with antigen processing)-independent presentation of endogenously synthesized peptides is enhanced by endoplasmic reticulum insertion sequences located at the amino- but not carboxyl terminus of the peptide. J Immunol 152:381–387. [PubMed] [Google Scholar]
- 93.Bonneau RH, Salvucci LA, Johnson DC, Tevethia SS. 1993. Epitope specificity of H-2Kb-restricted, HSV-1-, and HSV-2-cross-reactive cytotoxic T lymphocyte clones. Virology 195:62–70. doi: 10.1006/viro.1993.1346. [DOI] [PubMed] [Google Scholar]
- 94.Falk K, Rötzschke O, Stevanovié S, Jung G, Rammensee H-G. 1991. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature 351:290–296. doi: 10.1038/351290a0. [DOI] [PubMed] [Google Scholar]
- 95.Rotzschke O, Falk K, Stevanovic S, Jung G, Walden P, Rammensee HG. 1991. Exact prediction of a natural T cell epitope. Eur J Immunol 21:2891–2894. doi: 10.1002/eji.1830211136. [DOI] [PubMed] [Google Scholar]