Excessive innate immune responses can be deleterious for the host, and therefore, negative feedback is needed. Here, we describe a completely novel function for IFI44L in negatively modulating innate immune responses induced after virus infections. In addition, we show that decreasing IFI44L expression impairs virus production and that IFI44L expression negatively modulates the antiviral state induced by an analog of dsRNA or by IFN treatment. IFI44L binds to the cellular protein FKBP5, which in turn interacts with kinases essential for type I and III IFN induction and signaling, such as the kinases IKKα, IKKβ, and IKKε. IFI44L binding to FKBP5 decreased the phosphorylation of IRF-3 and IκBα mediated by IKKε and IKKβ, respectively, providing an explanation for the function of IFI44L in negatively modulating IFN responses. Therefore, IFI44L is a candidate target for reducing virus replication.
KEYWORDS: innate immunity, antiviral responses, type I and III interferon responses, signaling transduction, interferon-induced protein, IFI44L, FKBP5, IKKε kinase activity, IKKβ kinase activity, IRF-3 phosphorylation, IκBα phosphorylation, IKK kinase activity
ABSTRACT
We describe a novel function for the interferon (IFN)-induced protein 44-like (IFI44L) gene in negatively modulating innate immune responses induced after virus infections. Furthermore, we show that decreasing IFI44L expression impairs virus production and that IFI44L expression negatively modulates the antiviral state induced by an analog of double-stranded RNA (dsRNA) or by IFN treatment. The mechanism likely involves the interaction of IFI44L with cellular FK506-binding protein 5 (FKBP5), which in turn interacts with kinases essential for type I and III IFN responses, such as inhibitor of nuclear factor kappa B (IκB) kinase alpha (IKKα), IKKβ, and IKKε. Consequently, binding of IFI44L to FKBP5 decreased interferon regulatory factor 3 (IRF-3)-mediated and nuclear factor kappa-B (NF-κB) inhibitor (IκBα)-mediated phosphorylation by IKKε and IKKβ, respectively. According to these results, IFI44L is a good target for treatment of diseases associated with excessive IFN levels and/or proinflammatory responses and for reduction of viral replication.
IMPORTANCE Excessive innate immune responses can be deleterious for the host, and therefore, negative feedback is needed. Here, we describe a completely novel function for IFI44L in negatively modulating innate immune responses induced after virus infections. In addition, we show that decreasing IFI44L expression impairs virus production and that IFI44L expression negatively modulates the antiviral state induced by an analog of dsRNA or by IFN treatment. IFI44L binds to the cellular protein FKBP5, which in turn interacts with kinases essential for type I and III IFN induction and signaling, such as the kinases IKKα, IKKβ, and IKKε. IFI44L binding to FKBP5 decreased the phosphorylation of IRF-3 and IκBα mediated by IKKε and IKKβ, respectively, providing an explanation for the function of IFI44L in negatively modulating IFN responses. Therefore, IFI44L is a candidate target for reducing virus replication.
INTRODUCTION
Type I interferons alpha and beta (IFN-α and IFN-β) and type III IFN-λ and proinflammatory cytokines are central molecules in the antiviral innate immunity induced after virus infections (1). To increase the expression levels of these molecules, pathogen-associated molecular patterns (PAMPs), which include viral molecules such as glycoproteins, proteoglycans, and nucleic acid motifs, are recognized by host pattern recognition receptors (PRRs) (2–4). The PRRs include at least 3 different families such as the Toll-like receptors (TLRs), the retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs), and the nucleotide-binding oligomerization domain-containing (NOD)-like receptors (NLRs) (2, 3). Influenza A virus (IAV), which is a member of the Orthomyxoviridae family and contains an eight-segmented, negative-sense, single-stranded RNA (ssRNA) genome, is recognized by TLR-3 (double-stranded RNA [dsRNA]), TLR-7 and TLR-8 (ssRNA), RIG-I (5′ triphosphate ssRNA), and NLRP3 (5). Lymphocytic choriomeningitis virus (LCMV), the prototype member of the Arenaviridae family, which contains a negative-sense genome comprised of two ssRNA viral segments, is mainly recognized by TLR-7, RIG-I, and melanoma differentiation-associated gene 5 (MDA-5) (6). Coronaviruses (CoVs) are positive-sense ssRNA viruses recognized by MDA-5, TLR-7, and RIG-I (7–9).
The recognition of viral PAMPs by cellular PRRs leads to signaling pathways activating transcription factors, such as interferon regulatory factor 3 (IRF-3) and IRF-7 (10–12), nuclear factor kappa-light-chain enhancer of activated B cells (NF-κB) (13, 14), and ATF-2/c-Jun (15), leading to type I and III IFN and inflammatory cytokine induction. IRF-3 and IRF-7 are transcription factors phosphorylated by TANK-binding kinase 1 (TBK-1) and the inhibitor of nuclear factor kappa B (IκB) kinase IKKε (16). This posttranslational modification leads to IRF dimerization, nuclear translocation, and activation of type I and III IFNs and proinflammatory genes (17, 18). Activation of NF-κB involves the phosphorylation and subsequent degradation of IκB, a NF-κB inhibitor that binds and sequesters NF-κB in the cytoplasm of resting cells. The multisubunit IκB kinase (IKK) responsible for IκB phosphorylation contains two kinase subunits, IKKα and IKKβ, both of which are able to phosphorylate IκB, and the regulatory subunit IKKγ (19). Phosphorylation of IκB leads to its degradation, allowing NF-κB to migrate to the nucleus and activate IFN and proinflammatory cytokine expression (20). FK506-binding protein 5 (FKBP5) is a peptidyl-prolyl cis-trans isomerase that interacts with IKKα, IKKβ, and IKKγ, facilitating IKK complex assembly and leading to increased IKKα and IKKβ kinase activity, NF-κB activation, and IFN production (21). In addition, it has been shown that FKBP5 interacts with IKKε, possibly affecting its kinase activity (22).
Type I and III IFNs are secreted from infected cells and signal through different IFN receptors, leading to the activation of Janus protein tyrosine kinase 1 (JAK1) and tyrosine kinase 2 (TYK2), critical for phosphorylation and activation of signal transducer and activator of transcription 1 (STAT1) and STAT2. STAT1 is also phosphorylated by IKKε during IFN signaling, and this step is critical for the IFN-induced antiviral response (23, 24). Once phosphorylated, STAT1 and STAT2 associate with IRF-9 to form the IFN-stimulated gene factor 3 (ISGF3) complex. ISGF3 then migrates to the nucleus, binding to sequences of IFN-stimulated response elements (ISREs) present in the promoters of IFN-stimulated genes (ISGs) to increase their transcription (1, 25). Interestingly, unlike type I IFNs, type III IFNs are considered ISGs, as the expression of type III IFNs is further driven by IFN signals (26). Many ISGs control viral infections by directly targeting pathways and functions required during the virus life cycle (27). However, negative regulators of IFN production and signaling are needed for helping resolve the IFN-induced state and facilitate the return to cellular homeostasis (27–29).
IFN-induced protein 44-like (IFI44L) is a paralog gene of IFI44. IFI44L is an ISG, induced by many different viruses such as IAV and respiratory syncytial virus (30, 31). Overexpression of IFI44L has been linked to the persistence of hepatitis E virus infection (32), and it has been shown that IFI44L has weak antiviral activity against hepatitis C virus infection (33). However, a clear role of IFI44L or IFI44L single-nucleotide polymorphisms (SNPs) in the control of IFN responses and viral infections has not been defined. An IFI44L SNP leads to an alternate splice transcript form, which has been correlated with a reduction in the antibody (Ab) response to the measles vaccine (34). However, no direct correlations of IFI44L SNPs with the innate immune responses have been found.
In this study, we analyzed cellular functions for IFI44L, presenting a completely novel function for IFI44L in negatively modulating innate immune IFN responses. We found that IFI44L silencing decreases virus production and that IFI44L silencing and/or depletion and overexpression increase and decrease, respectively, the antiviral state mediated by a dsRNA analog and by IFN. Furthermore, using coimmunoprecipitation (Co-IP), we showed that IFI44L interacts with FKBP5, a protein binding to IKKβ and IKKε, and that the interaction of IFI44L with FKBP5 affects the ability of IKKε and IKKβ to phosphorylate IRF-3 and IκBα, respectively, which are crucial steps for inducing IFN and proinflammatory cytokine responses. Importantly, results from our studies demonstrate the feasibility of targeting IFI44L to modulate inflammation responses and control viral replication and, therefore, to control inflammatory and viral diseases.
RESULTS
IFI44L supports viral replication.
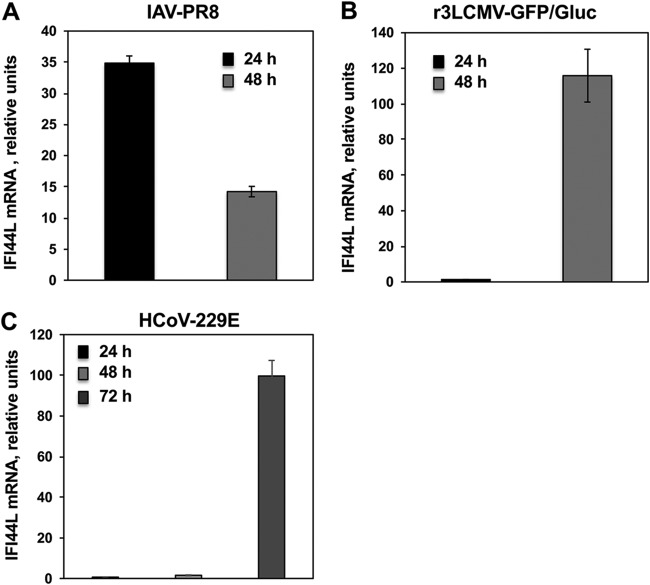
IFI44L is an ISG induced by many viruses such as IAV and respiratory syncytial virus (30, 31). To confirm that IFI44L expression is induced after virus infections, A549 cells were infected with IAV and recombinant trisegmented 3LCMV expressing green fluorescent protein (GFP) and Gaussia luciferase (r3LCMV-GFP/Gluc) (multiplicity of infection [MOI], 0.1) and IFI44L mRNA levels were analyzed at 24 and 48 hpi (Fig. 1A and B). IFI44L expression was induced in IAV-infected and r3LCMV-GFP/Gluc-A549 infected cells at 24 and 48 hpi, as expected, although whereas IFI44L induction was highest at 24 hpi in IAV-infected cells, IFI44L induction was highest at 48 hpi in r3LCMV-GFP/Gluc-infected cells (Fig. 1A and B). Alternatively, Huh-7 cells were infected with human coronavirus 229E (HCoV-229E) (MOI, 0.1) and IFI44L mRNA levels were then evaluated. After infection with HCoV-229E, the levels of IFI44L mRNA were not increased at 24 hpi. However, IFI44L mRNA levels increased by 2-fold and 100-fold at 48 and 72 hpi, respectively, in nontargeted (NT) small interfering RNA (siRNA)-transfected cells (Fig. 1C).
FIG 1.
IFI44L negatively modulates IFN responses after infection. Human A549 (A and B) or Huh-7 (C) cells were transfected with the NT control siRNA. At 36 hpt, A549 cells were infected with IAV (MOI, 0.1) (A) or with r3LCMV-GFP/Gluc (MOI, 0.1) (B) for 24 and 48 h and Huh-7 cells were infected with HCoV-229E (MOI, 0.1) (C) for 24, 48, and 72 h. Total RNAs were purified from infected cells, and expression levels of IFI44L mRNAs were evaluated by RT-qPCR and are represented as fold change values compared to the values determined with mock-infected cells.
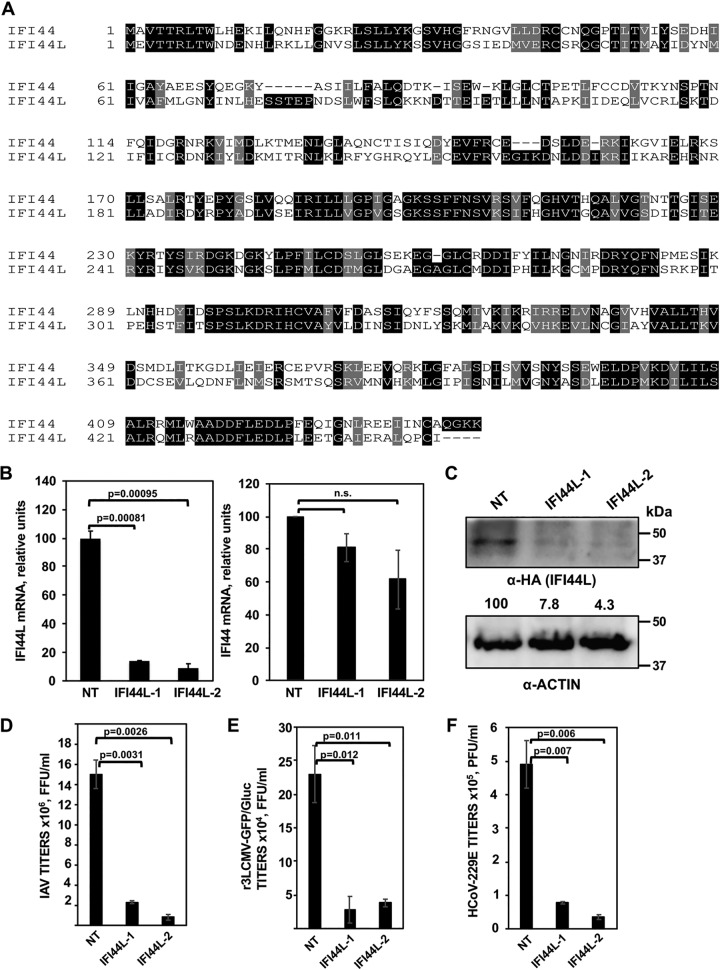
In a previous study, we found that IFI44 supports virus replication most probably due to the functions of this host cellular protein as a negative regulator of IFN responses (73). Because of these novel findings for IFI44 and because IFI44 and IFI44L are gene paralogs showing 60% homology at the amino acid level (Fig. 2A), we first assessed whether IFI44L also supports virus replication. To this end, silencing (loss-of-function) experiments were performed. Using two different siRNAs, IFI44L mRNA was silenced in A549 cells by more than 85% as determined by reverse transcription-quantitative PCR (RT-qPCR) (Fig. 2B). Importantly, IFI44 mRNA levels were not significantly decreased by the transfection of IFI44L siRNAs (Fig. 2B) since the nature of the siRNAs was specific for IFI44L. To further confirm that the IFI44L-specific siRNAs knocked down IFI44L expression at the protein level, A549 cells were silenced with the two different IFI44L siRNAs or with a nontargeted (NT) siRNA and were then transfected with a plasmid expressing IFI44L fused to a hemagglutinin (HA) epitope tag (Fig. 2C). Western blot analysis using an anti-HA-specific antibody showed a clear (more than 12-fold) reduction in the overall IFI44L expression levels in IFI44L-silenced cells by using the two IFI44L-specific siRNAs (Fig. 2C). These data indicate that the IFI44L siRNAs efficiently knocked down IFI44L expression at the mRNA (Fig. 2B) and protein (Fig. 2C) levels.
FIG 2.
IFI44L negatively affects virus production. (A) Alignment of the paralog genes IFI44 (top, NCBI reference sequence NP_006408) and IFI44L (bottom, NCBI reference sequence NP_006811). Black boxes indicate the same amino acid in both proteins, and gray boxes indicate residues with similar properties. (B) Downregulation of IFI44L (left) and IFI44 (right) in A549 cells. A549 cells were transfected with two different siRNAs specific for IFI44L or with a nontargeted (NT) siRNA control. At 36 hpt, RNA was extracted and levels of IFI44L and IFI44 mRNAs were evaluated by qPCR. P values determined by using a Student's t test to compare NT-siRNA-silenced cells and IFI44L-siRNA-silenced cells are indicated. n.s., not significant (P > 0.05). (C) At 36 h post-siRNA transfection, cells were transfected with a plasmid expressing IFI44L (pCAGGS-IFI44L-HA) during 48 h. Expression of IFI44L (using an anti-HA tag antibody, top blot) and actin (internal control, bottom) was evaluated by Western blotting. Protein levels were quantified by densitometry using ImageJ software. IFI44L protein expression levels in cells silenced with the NT siRNA were assigned a value of 100% for comparison with the level of expression in IFI44L-silenced cells (numbers below the HA blot). IFI44L expression was normalized to actin expression. Molecular weight markers (in kilodaltons) are indicated on the right. Three different experiments were performed, with similar results. (D to F) Inhibition of viral infections. At 36 h post-siRNA transfection, A549 cells (D and E) were infected with IAV (PR8 strain; MOI, 0.1) (D) or r3LCMV-GFP/Gluc (MOI, 0.1) (E) and virus titers in the cell culture supernatants were evaluated at 48 hpi. Likewise, at 36 h post-siRNA transfection, Huh-7 cells (F) were infected with HCoV-229E (MOI, 0.1) and virus titers were determined at 48 hpi. P values were determined by using a Student's t test to compare NT-siRNA-silenced cells and IFI44L-siRNA-silenced cells. Bars represent standard deviations (SDs) of results from the triplicate wells. P values determined by using a Student's t test to compare NT-siRNA-silenced cells and IFI44L-siRNA-silenced cells are indicated. Three experiments were performed with similar results.
To analyze whether IFI44L silencing decreases virus production, A549 cells were silenced with the two different siRNAs for IFI44L and the NT siRNA control and, at 36 hpt, were infected with IAV PR8 (Fig. 2D) or r3LCMV-GFP/Gluc (Fig. 2E). Alternatively, Huh-7 cells were similarly silenced and, at 36 hpt, were infected with HCoV-229E (Fig. 2F). For IAV, a significant (7-fold) reduction in virus titers was observed at 48 hpi, in cells silenced with the two different IFI44L siRNAs, compared to the NT-siRNA-silenced cells (Fig. 2D). For r3LCMV-GFP/Gluc, a significant (5-fold) reduction in virus titers was observed in IFI44L-silenced cells compared to the control cells silenced with the NT siRNA at 48 hpi (Fig. 2E). Furthermore, in HCoV-229E-infected cells, a significant (7-fold) reduction in virus titers was observed at 48 hpi, in the cells silenced with the two different IFI44L siRNAs, compared to the NT-siRNA-silenced cells (Fig. 2F). These data indicated that IFI44L supports virus replication and that this effect is broad, since similar levels of viral inhibition were observed in IFI44L siRNA-transfected cells for IAV, r3LCMV-GFP/Gluc, and HCoV-229E.
IFI44L acts as a feedback regulator of IFN responses.
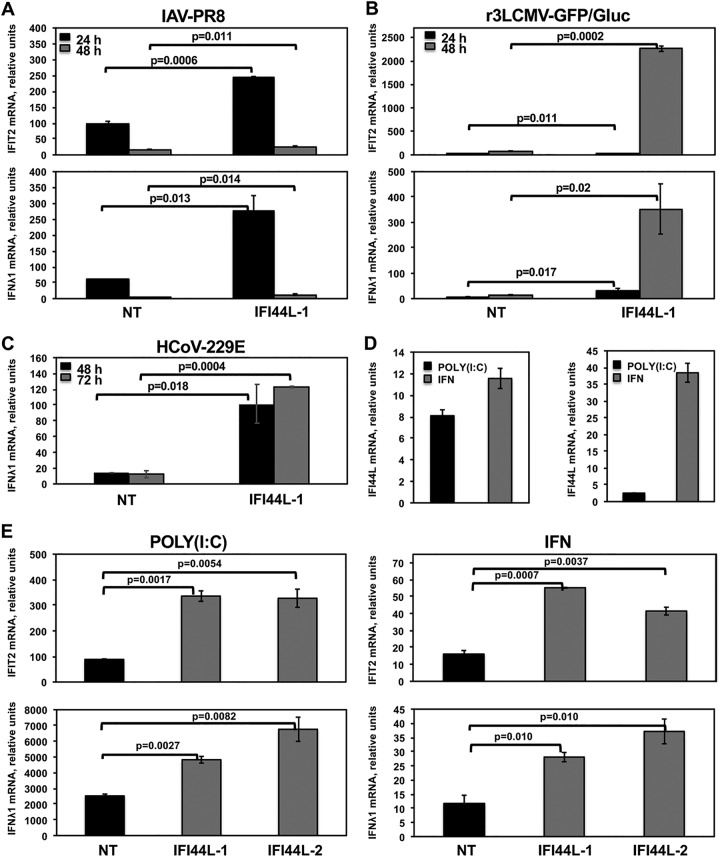
Some ISGs are known to regulate the antiviral response through a negative-feedback mechanism (28, 35). Taking into account that IAV and LCMV are sensitive to IFN responses (5, 36–38), that IFI44, a paralog ISG of IFI44L, negatively modulates innate immune responses (DeDiego et al., submitted), and that IFI44L silencing decreases virus production, we hypothesized that IFI44L could be acting as a feedback regulator of IFN responses. To analyze this hypothesis, A549 cells were silenced with an IFI44L siRNA, or with the NT siRNA control, and were then infected with IAV PR8 (Fig. 3A) or r3LCMV-GFP/Gluc (Fig. 3B) to induce host antiviral responses. Alternatively, Huh-7 cells were infected with HCoV-229E (Fig. 3C). The levels of expression of IFN-induced protein with tetratricopeptide repeats 2 (IFIT2) and IFN-λ1 (a type III IFN) ISGs were evaluated by RT-qPCR. In IAV-infected A549 cells transfected with the NT siRNA, IFIT2 mRNA levels were increased by 100-fold and 16-fold at 24 and 48 hpi, respectively, while IFN-λ1 mRNA levels were increased by 60-fold and 3-fold at 24 and 48 hpi, respectively (Fig. 3A). Interestingly, in IFI44L-silenced cells, levels of IFIT2 mRNA in IAV-infected cells were further increased by 2.5-fold and 1.5-fold at 24 and 48 hpi, respectively, and the levels of IFN-λ1 were further increased by 4-fold and 5-fold at 24 and 48 hpi, respectively, compared to NT-siRNA-transfected cells (Fig. 3A). Similarly, in cells infected with r3LCMV-GFP/Gluc, IFIT2 and IFN-λ1 were upregulated, particularly at 48 hpi (Fig. 3B). Remarkably, in cells silenced for IFI44L, the levels of IFIT2 were 1.5-fold and 25-fold higher at 24 and 48 hpi, respectively, than in control cells transfected with the NT siRNA. Likewise, the levels of IFN-λ1 were increased 7-fold and 25-fold at 24 and 48 hpi, respectively, compared to the NT-siRNA-transfected control cells (Fig. 3B). After HCoV-229E infection of Huh-7 cells, the levels of IFN-λ1 mRNA were at least 4.5-fold and 12-fold higher in cells silenced with the two different IFI44L siRNAs than in the NT-siRNA-control-transfected cells at 48, and 72 hpi, respectively (Fig. 3C). These results demonstrate that IFI44L silencing results in upregulation of IFN responses and ISGs under conditions of different viral infections, suggesting that the decrease in virus titers observed in IFI44L-silenced cells after infection was due, at least in part, to increased IFN responses in the IFI44L-silenced cells.
FIG 3.
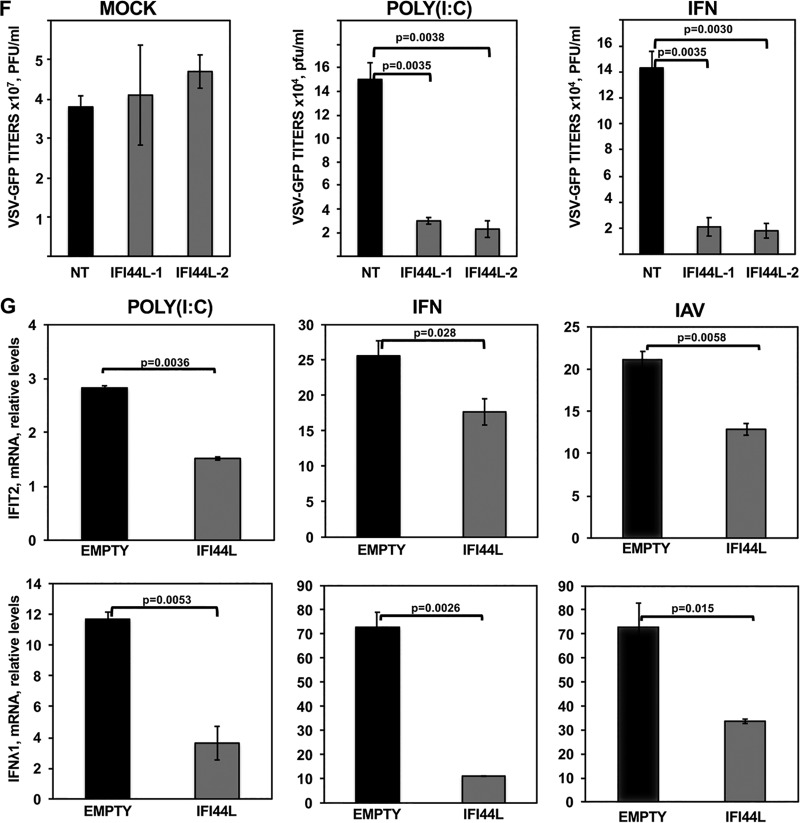
IFI44L negatively modulates IFN responses. Human A549 (A and B) or Huh-7 (C) cells were transfected with the NT control siRNA and with an siRNA specific for IFI44L. At 36 hpt, A549 cells were infected with IAV PR8 strain (MOI, 0.1) (A) or with r3LCMV-GFP/Gluc (MOI, 0.1) (B) for 24 and 48 h, and Huh-7 cells were infected with HCoV-229E (MOI, 0.1) (C) for 48 and 72 h. Total RNAs were purified from infected cells, and expression levels of IFIT2 and IFN-λ1 mRNAs were evaluated by RT-qPCR and are represented as fold change values compared to values in mock-infected cells. (D, left panel, and E) Human A549 cells were transfected with two different siRNAs specific for IFI44L (E) or with the NT siRNA control (D, left panel, and E). (D, left panel, and E) At 36 hpt with the siRNAs, cells were transfected with poly (I·C) or treated with 250 U/ml of IFN-α. Expression levels of IFI44L (D), IFIT2 (E, top panels) and IFN-λ1 (E, bottom panels) were evaluated by RT-qPCR and are represented as fold change values compared to values in mock-treated cells. (F) After poly(I·C) transfection and IFN-α treatment, cells were infected with rVSV-GFP (MOI, 0.1) and virus titers in cell culture supernatants were determined at 24 hpi. (D, right panel, and G) Human 293T cells were transfected with pCAGGS-IFI44L-HA (G, IFI44L) or with an empty pCAGGS plasmid as a control (D, right panel, and G, EMPTY). (G) At 24 hpt, cells were transfected with poly(I·C) (left panels), treated with 1,000 U/ml of IFN-α (middle panels), or infected with IAV PR8 (MOI, 1) (right panels) for 24 h. Levels of IFI44L (D), IFIT2 (G, top panels), and IFN-λ1 (G, bottom panels) were evaluated by RT-qPCR. Bars represent SDs of results determined using duplicate wells. Three different experiments were performed, with similar results. P values determined by using a Student's t test are indicated for comparisons between NT-silenced cells and IFI44L-silenced cells.
To further demonstrate the key role of IFI44L in induction if IFN and ISG responses, A549 cells were silenced with the two IFI44L siRNAs and, at 36 hpt, were transfected with polyinosinic-polycytidylic acid [poly(I·C)], an analog of dsRNA mainly detected by RIG-I/MDA-5 (39), or were treated with IFN-α, and the levels of IFIT2, IFN-λ1, and IFI44L were analyzed by qPCR. In cells silenced with the NT siRNA control, levels of IFI44L mRNAs were increased by 8-fold and 12-fold after poly(I·C) and IFN treatments, respectively (Fig. 3D, left panel). Likewise, levels of IFIT2 mRNA were increased by 90-fold and 17-fold in NT-control-siRNA-transfected cells transfected with poly(I·C) and treated with IFN-α, respectively (Fig. 3E, top panels). Similarly, the levels of IFN-λ1 mRNA were increased by 2,500-fold and 12.5-fold in NT-siRNA-transfected cells transfected with poly(I·C) and treated with IFN-α, respectively (Fig. 3E, low panels). Interestingly, in cells transfected with the two different IFI44L siRNAs, IFIT2 and IFN-λ1 mRNA levels were approximately 3-fold higher after poly(I·C) and IFN treatment than in NT-siRNA-transfected cells (Fig. 3E), further demonstrating that IFI44L negatively modulates IFN responses.
To analyze the effect of IFI44L in conferring biologically relevant IFN-mediated antiviral activity to virus infection, we assessed the effect of IFI44L downregulation on viral infection. To this end, human epithelial A549 cells were transfected as indicated above with the two siRNAs specific for IFI44L and with the NT control siRNA. Then, cells were treated with poly(I·C), or with IFN-α, to induce antiviral states and, 16 h after treatment, cells were infected with rVSV-GFP, a virus highly sensitive to the antiviral state induced by IFN (40), and virus production was analyzed at 24 hpi. Recombinant vesicular stomatitis virus-GFP (rVSV-GFP) titers were approximately 300-fold lower in cells transfected with poly(I·C) or treated with IFN-α than in mock-treated cells (Fig. 3F), consistent with the induction of an antiviral state mediated by poly(I·C) and IFN-α treatments. Remarkably, and consistently with our previous results, virus titers were 5-fold lower in the cells knocked down for IFI44L than in the NT-siRNA-silenced cells (Fig. 3F). These results further confirm our previous results demonstrating that IFI44L expression decreases the induction of host antiviral responses.
To confirm these results, human 293T cells were transfected with a plasmid expressing IFI44L (pCAGGS-IFI44L-HA) or with the empty plasmid as an internal control and, at 36 hpt, cells were transfected with poly(I·C) during 16 h, treated with IFN-α during 16 h, or infected with IAV PR8 (MOI, 1) for 24 h. Then, levels of mRNAs of the ISGs IFIT2 and IFN-λ1 were analyzed by RT-qPCR (Fig. 3G, top and bottom panels, respectively). As observed in A549 cells (Fig. 3E), expression of IFIT2 and IFN-λ1 (Fig. 3G) and of IFI44L (Fig. 3D, right panel) was increased in 293T cells after poly(I·C) and IFN-α treatments. Remarkably, the levels of IFIT2 were approximately 2-fold lower in cells overexpressing IFI44L. In addition, levels of IFN-λ1 mRNA decreased 3-fold and 7-fold in poly(I·C)-treated and IFN-α-treated cells overexpressing the IFI44L protein, respectively, compared to control cells (Fig. 3G). Furthermore, after IAV infection, the levels of IFIT2 and IFN-λ1 were increased by 21-fold and 72-fold, respectively, in the empty-plasmid-transfected cells in comparison with the mock-infected cells (Fig. 3G). Interestingly, the levels of these mRNAs decreased approximately 2-fold in the IAV-infected cells overexpressing IFI44L compared to the cells transfected with empty plasmid (Fig. 3G). These results demonstrate that IFI44L overexpression decreased IFN responses mediated by poly(I·C), IFN, and IAV infections, in accordance with the silencing experiments showing that IFI44L expression negatively modulated IFN and ISG responses.
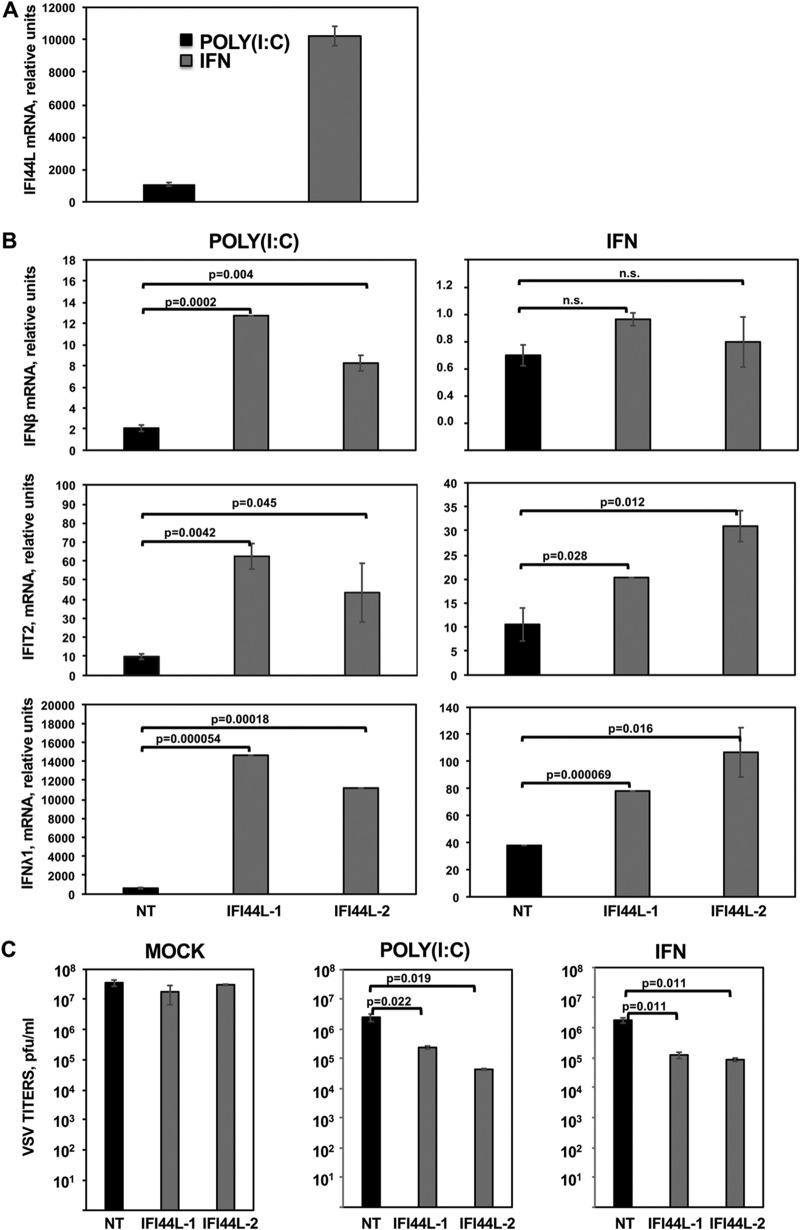
A549 cells and 293T cells are epithelial cells. To further analyze the effect of IFI44L silencing on another cell type, HAP-1 cells, which arose from a myeloid lineage, were used. First, we analyzed whether IFI44L expression is induced in this cell type by transfection with poly(I·C) or by treatment with IFN-α and by analyzing IFI44L mRNA levels by RT-qPCR (Fig. 4A). Expression of IFI44L mRNA was induced more than 1,000-fold and 10,000-fold in poly(I·C)-treated and IFN-α-treated cells, respectively (Fig. 4A), demonstrating that IFI44L is expressed and is IFN inducible in HAP-1 cells. Then, HAP-1 cells transfected with the two different siRNAs specific for IFI44L were transfected with poly(I·C) or treated with IFN-α. Both treatments induced the expression of IFIT2 and IFN-λ1, whereas only poly(I·C) transfection induced the expression of IFN-β (Fig. 4B). Notably, when IFI44L was silenced, the levels of IFN-β were increased at least 4-fold in poly(I·C)-transfected cells compared to cells transfected with the NT control siRNA (Fig. 4B). Moreover, when IFI44L expression was silenced, the levels of IFIT2 and IFN-λ1 mRNAs were increased by at least 2-fold in poly(I·C)-transfected and IFN-α-treated cells compared to cells transfected with the NT control siRNA (Fig. 4B). Correlating with the higher levels of IFN-β, IFIT2, and IFN-λ1mRNAs in IFI44L-silenced cells, rVSV-GFP titers were lower in IFI44L-silenced cells transfected with poly(I·C) or treated with IFN-α than in the control NT-transfected cells (Fig. 4C), further supporting the idea that IFI44L negatively affects antiviral responses.
FIG 4.
IFI44L expression decreases antiviral responses. (A) HAP-1 cells transfected with the NT siRNA were transfected with poly(I·C) or treated with 2,000 U/ml of IFN-α. IFI44L mRNA levels were measured by RT-qPCR. (B) HAP-1 cells transfected with NT siRNA or with two IFI44L-specific siRNAs were transfected with poly(I·C) or treated with 2,000 U/ml of IFN-α. IFN-β (top panels), IFIT2 (middle panels), and IFN-λ1 (bottom panels) mRNA levels were measured by RT-qPCR and are represented as fold change values compared to values in mock-treated cells. In panels A and B, bars represent SDs of results determined using duplicate wells. Three different experiments were performed with similar results. P values determined by using a Student's t test are indicated. (C) Cells left mock treated, transfected with poly(I·C), or treated with 2,000 U/ml of IFN-α were infected with rVSV-GFP (MOI, 0.1). Virus production was analyzed at 24 hpi. Bars represent SDs of results determined using duplicate wells. P values determined by using a Student's t test are indicated. Three different experiments were performed, with similar results.
Knocking out IFI44L negatively modulated IFN responses.
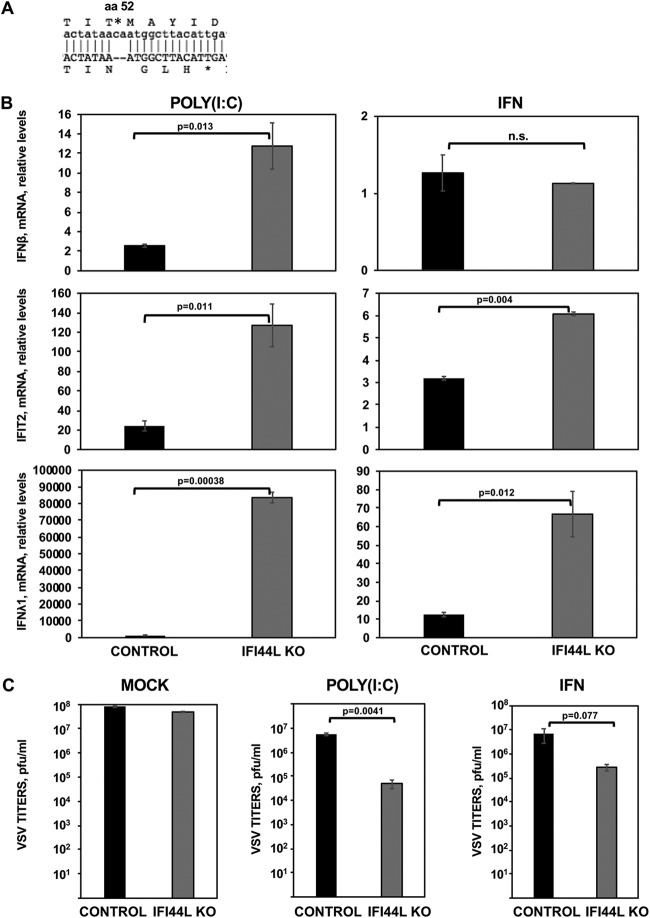
To confirm our initial downregulation results, we completely depleted IFI44L gene expression in HAP-1 cells by using the CRISPR/cas9 technology (IFI44L knockout [KO] cells). The HAP-1 cell line was chosen because this cell line is nearly haploid, making the generation of KO cells easier. Moreover, HAP-1 cells have been extensively used in similar studies (https://www.horizondiscovery.com/resources/scientific-literature). This IFI44L KO cell line encoded a 2-nucleotide frameshift deletion at the codon encoding amino acid 52 (Fig. 5A). Parental and IFI44L KO HAP-1 cells were then transfected with poly(I·C) or treated with IFN-α, and the levels of IFN-β, IFIT2, and IFN-λ1 mRNA expression were evaluated as previously described. As expected, poly(I·C) and IFN-α treatments induced IFIT2 and IFN-λ1 expression in both cell lines (Fig. 5B, middle and bottom panels). However, IFN-β was induced only after poly(I·C) treatment (Fig. 5B, top panel). Interestingly, mRNA levels of IFN-β were 5-fold higher in IFI44L KO cells treated with poly(I·C) and mRNA levels of IFIT2 were 5-fold and 2-fold higher in IFI44L KO cells treated with poly(I·C) and IFN-α, respectively, that in the parental HAP-1 control cells (Fig. 5B, top and middle panels). Furthermore, mRNA levels of IFN-λ1 were 80-fold and 6-fold higher in IFI44L KO cells treated with poly(I·C) and IFN-α, respectively, than in HAP-1 control cells (Fig. 5B, bottom panels). These data confirm that IFI44L expression decreases IFN responses.
FIG 5.
IFI44L impairs antiviral responses. (A) Sequence alignment of HAP-1 control cells at the amino acid (aa) and nucleotide levels (top letters) and of HAP-1 IFI44L KO cells (bottom letters). The IFI44L KO cells encode a 2-nt frameshift deletion in the codon encoding amino acid 52, leading to a frameshift, and to the generation of a stop codon at codon 56. (B and C) Parental (CONTROL) and IFI44L KO HAP-1 cells were transfected with poly(I·C) or treated with 2,000 U/ml of IFN-α during 16 h. (B) Total cellular RNA was purified, and levels of IFN-β (top), IFIT2 (middle), and IFN-λ1 (bottom) mRNAs were evaluated by RT-qPCR and are represented as fold change values compared to values in mock-treated cells. Bars represent SDs of results determined using duplicate wells. Three different experiments were performed with similar results. P values determined by using a Student's t test are indicated for comparisons between WT and IFI44L KO HAP-1 cells. (C) Cells left mock treated (left), transfected with poly(I·C) (center), or treated with 2,000 U/ml of IFN-α (right) were infected with rVSV-GFP (MOI, 0.1). Virus production was analyzed at 24 hpi. Bars represent SDs of results determined using duplicate wells. Three different experiments were performed, with similar results. *, P < 0.05 (for comparisons between HAP-1 WT and IFI44L KO cells).
To evaluate the effect of IFI44L expression in the antiviral responses, parental and IFI44L KO cells were transfected with poly(I·C) or treated with IFN-α and infected with rVSV-GFP (Fig. 5C). Viral titers decreased around 10-fold in HAP-1 control cells transfected with poly(I·C) or treated with IFN-α compared to mock-treated cells (Fig. 5C), consistent with the induction of an antiviral state in these cells. Interestingly, in IFI44L KO mock-treated cells, virus titers decreased 2-fold to 3-fold compared to mock-treated control cells. Notably, rVSV-GFP titers decreased by 100-fold and 20-fold in the IFI44L KO cells transfected with poly(I·C) and treated with IFN-α, respectively, compared to parental HAP-1 cells (Fig. 5C). These results further support the idea that IFI44L negatively affects IFN responses.
IFI44L binds to FKBP5.
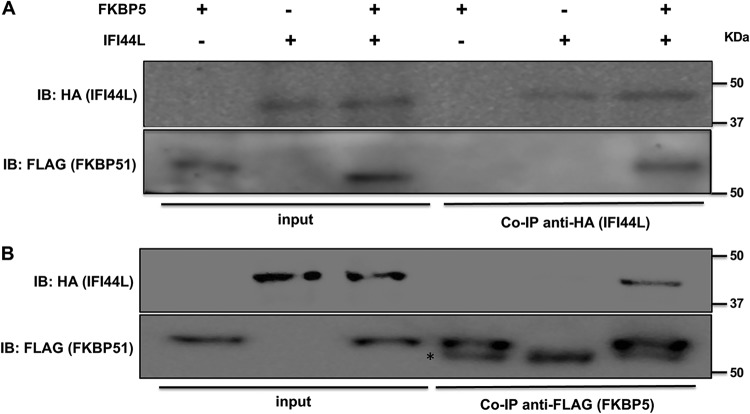
We found previously that IFI44 binds the cellular protein FKBP5 (DeDiego et al., submitted), affecting innate immune responses. Therefore, we assessed whether IFI44L also interacts with FKBP5. To this end, human 293T cells were transfected with plasmids expressing the IFI44L protein fused to a HA tag (pCAGGS-IFI44L-HA) and FKBP5 fused to a FLAG tag (pCAGGS-FKBP5-FLAG). Plasmids expressing IFI6 and IFI27 fused to the same HA tag were used as controls (data not shown) (DeDiego et al., submitted). Co-IP experiments using an anti-HA antibody-conjugated resin (to pull down IFI44L and IFI6) (Fig. 6A and data not shown) and an anti-FLAG antibody-conjugated resin (to pull down FKBP5) (Fig. 6B) were performed. As observed in Western blotting using antibodies specific for HA (to detect IFI44L) and for FLAG (to detect FKBP5) epitope tags, IFI44L specifically coimmunoprecipitated with FKBP5 by the use of affinity columns coupled to anti-HA and anti-FLAG antibodies, respectively (Fig. 6A and B). In contrast, IFI6, used as a control, did not coimmunoprecipitate with FKBP5 (data not shown). These results demonstrated an interaction between IFI44L and FKBP5 cellular proteins.
FIG 6.
IFI44L interacts with FKBP5. Human 293T cells were transiently cotransfected with plasmids encoding IFI44L (fused to an HA tag) and FKBP5 (fused to a FLAG tag). Then, Co-IP analyses using resins bound to HA-specific antibodies (to immunoprecipitate IFI44L) (A) and FLAG-specific antibodies (to immunoprecipitate FKBP5) (B) were performed and the presence of IFI44L (anti-HA antibody) and FKBP5 (anti-FLAG antibody) was analyzed by Western blotting in the initial cellular extracts (left) and after the Co-IP (right). Molecular weight markers (in kilodaltons) are indicated on the right. Three different experiments were performed, with the same results. The asterisk (*) indicates the band corresponding to the mouse IgG heavy chain. IB, immunoblot.
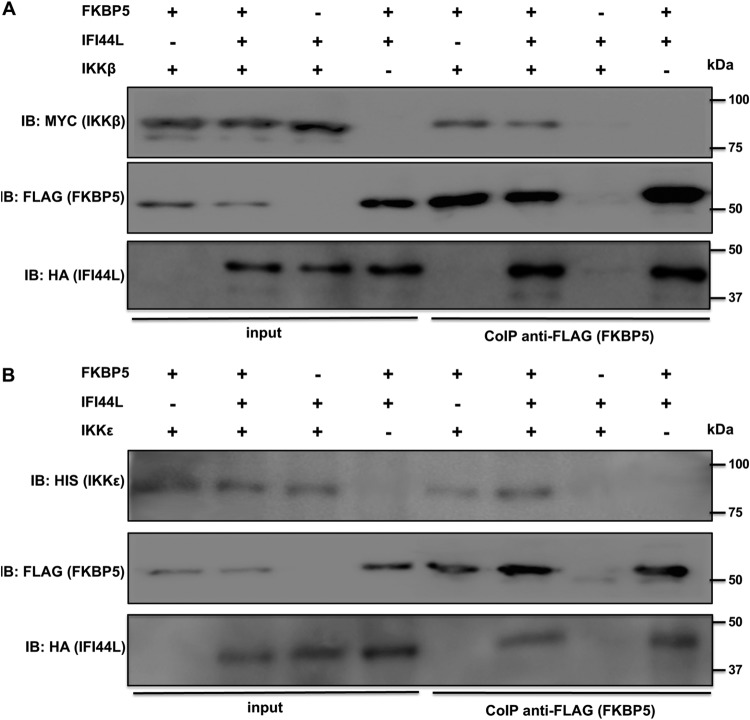
FKBP5 is an immunophilin that interacts with IKKα, IKKβ, and IKKγ and that facilitates IKK complex assembly, leading to increased IKKα and IKKβ kinase activity, NF-κB activation, and IFN production (21). In addition, FKBP5 interacts with TRAF6 and TRAF3 and induces the expression of IFN and proinflammatory cytokines (41). Furthermore, FKBP5 interacts with IKKε (DeDiego et al., submitted), likely increasing IKKε kinase activity (22) and, therefore, IFN production and signaling (16, 23). To analyze whether the binding of IFI44L to FKBP5 abolishes the binding of FKBP5 to IKKβ and IKKε, human 293T cells were transfected with pCAGGS-IFI44L, pCAGGS-FKBP5-FLAG, and pCAGGS-myc-IKKβ (Fig. 7A) or pCAGGS-His-IKKε (Fig. 7B) plasmids and were subjected to coimmunoprecipitation with an anti-FLAG resin (to pull down FKBP5). FKBP5 interacted with IKKβ and IKKε at similar levels irrespective of the presence or absence of IFI44L (Fig. 7), suggesting that the binding of IFI44L to FKBP5 does not interfere with the binding of FKBP5 to IKKβ and IKKε.
FIG 7.
Binding of IFI44L to FKBP5 does not disrupt FKBP5 interaction with IKKβ or IKKε. Human 293T cells were transiently cotransfected with plasmids expressing IFI44L-HA, FKBP5-FLAG, and either myc-IKKβ (A) or His-IKKε (B). Then, Co-IPs using a resin bound to FLAG antibody (to immunoprecipitate FKBP5) (B) were performed, and the presence of IFI44L (anti-HA tag antibody), FKBP5 (anti-FLAG tag antibody), IKKβ (anti-myc tag antibody), and IKKε (anti-His tag antibody) was analyzed by Western blotting in the initial cellular extracts (input; left) and after the Co-IP [CoIP anti-FLAG (FKBP5); right]. Molecular weight markers (in kilodaltons) are indicated on the right. Three different experiments were performed, with same results.
IFI44L decreases IKKε kinase activity.
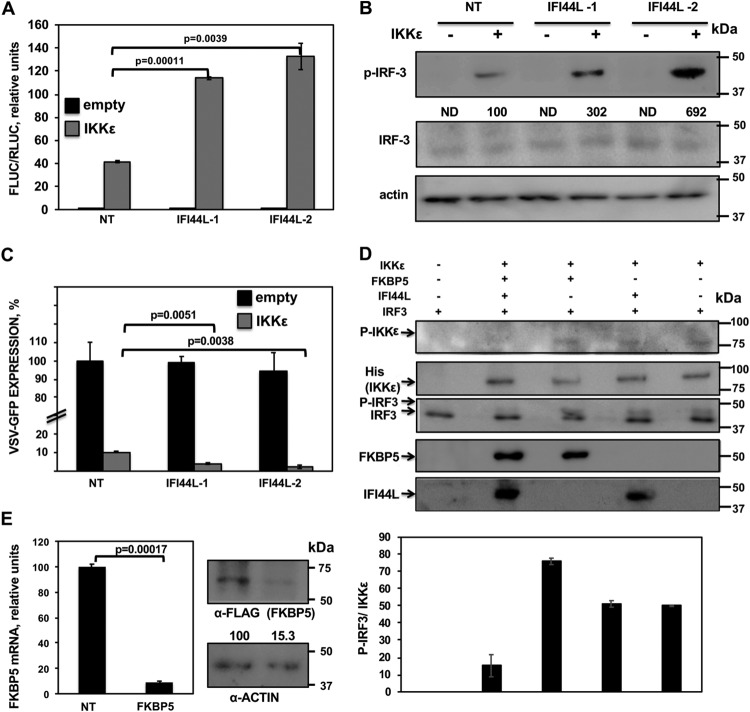
The binding of FKBP5 to IKKε might modulate the kinase activity of IKKε (22). IKKε phosphorylates the transcription factors IRF-3 and IRF-7, leading to their activation and type I and III IFN induction (16, 18). In addition, IKKε phosphorylates STAT1 during IFN signaling, a step critical for the IFN-inducible antiviral response (23, 24). To analyze whether IFI44L affects IKKε-mediated IFN production, human 293T cells were silenced with the two different siRNAs specific for IFI44L or with the NT siRNA control. Then, cells were cotransfected with plasmids expressing Firefly luciferase (Fluc) under the control of the IFN promoter, Renilla luciferase (Rluc) under the control of a constitutively expressing promoter, and IKKε. Compared to NT-siRNA-transfected cells and empty-plasmid-transfected cells, levels of Fluc expression were increased approximately 40-fold in NT-siRNA-transfected, IKKε-overexpressing cells, indicating that IKKε overexpression induced the expression of IFN. Interestingly, Fluc expression was increased by around 120-fold in cells silenced for both IFI44L siRNAs and overexpressing IKKε (Fig. 8A), demonstrating that IFI44L negatively regulates IFN production mediated by IKKε activation.
FIG 8.
IFI44L modulates IKKε kinase activity. (A and B) Human 293T cells were silenced for IFI44L, and, 36 h later, cells were cotransfected with a pCAGGS-His-IKKε plasmid together with a plasmid expressing Fluc under the control of an IFN-β promoter (pIFNβ-Fluc) and a plasmid constitutively expressing Rluc under the control of a simian virus 40 (SV40) promoter. (A) At 24 hpt, levels of Fluc were determined and normalized to the levels of Rluc. Data represent means and SDs of results from triplicate wells. Experiments were repeated three times with similar results. P values determined by using a Student's t test are indicated. (B) Protein levels of p-IRF-3, total IRF-3, and actin (control) in extracts from cells treated as described for panel A were evaluated by Western blotting. Western blots were quantified by densitometry using ImageJ software (v1.46), and the amounts of p-IRF-3 were normalized to the amounts of total IRF-3 (numbers below the p-IRF-3 blot). Levels of pIRF-3 in cells silenced with the NT siRNA and transfected with the plasmid expressing IKKε were assigned a value of 100 for comparisons with pIRF-3 levels in IFI44L-silenced cells. ND, not detected. Molecular weight markers are indicated in the right (in kilodaltons). (C) Cell culture supernatants from cells treated as described for panel A were used to treat A549 cells for 24 h and infected with rVSV-GFP (MOI, 0.1). GFP expression was quantified in a microplate reader at 24 hpi. Data represent means and SDs of results from triplicate wells. Experiments were repeated three times with similar results. P values determined by using a Student's t test are indicated. (D) Human 293T cells were silenced for IFI44 or FKBP5 and were transfected with plasmids expressing His-IKKε, IFI44L-HA, and FKBP5-FLAG. At 24 hpt, IKKε complexes were purified with an anti-His antibody and assayed in a kinase assay using recombinant human IRF-3 as the substrate. Levels of total and phosphorylated forms of IRF-3 were analyzed by Western blotting using specific antibodies. Levels of IKKε were determined using an anti-His-specific antibody and levels of phospho-IKKε were evaluated using an anti-p-IKKε antibody by Western blotting. Levels of IFI44L-HA and FKBP5-FLAG proteins were detected with anti-HA-specific and anti-FLAG-specific antibodies, respectively. Protein bands were quantified by densitometry using ImageJ software (v1.46), and levels of pIRF-3 were normalized to the levels of IKKε (bottom graphic). The results show error bars and means of results from two independent experiments. Molecular weight markers (in kilodaltons) are indicated on the right. (E) Human 293T cells were transfected with NT-specific or FKBP5-specific siRNAs for 36 h. (Left) Total RNAs were extracted and expression of FKBP5 mRNA was evaluated by RT-qPCR. Error bars represent the SDs of measurements in triplicate wells. The P value for comparisons between NT-transfected cells and FKBP5 siRNA-transfected cells performed using a Student’s t test is indicated. On the right, at 36 h after siRNA transfection, cells were transfected with a plasmid expressing FKBP5-FLAG for 24 h. Western blot analyses using an anti-FLAG-specific antibody (to detect FKBP5 expression) and anti-actin antibody (internal control) were performed. Protein expression levels in cells silenced with the NT siRNA were assigned a value of 100% for comparison with the level of expression in FKBP5-silenced cells (numbers below the anti-FLAG blot). FKBP5 expression was normalized to actin expression. Molecular weight markers (in kilodaltons) are indicated on the right. Three different experiments were performed with similar results.
On the basis of these results, we next evaluated the levels of phosphorylated IRF-3 (pIRF-3) in cells silenced for IFI44L and overexpressing IKKε (Fig. 8B) (16). As expected, IKKε overexpression resulted in IRF-3 phosphorylation (Fig. 8B). Remarkably, levels of pIRF-3 were at least 3-fold higher in the cells transfected with the two IFI44L siRNAs than in the NT-control-siRNA-transfected cells (Fig. 8B), suggesting that IFI44L negatively modulates IKKε-mediated phosphorylation of IRF-3.
To further confirm that IFI44L negatively regulates the production of IFN mediated by IKKε activation, tissue culture supernatants from IFI44L-silenced cells overexpressing IKKε were collected and used to treat fresh human A549 cells to induce an antiviral state. Then, cells were infected with rVSV-GFP, and levels of GFP expression were quantified using a microplate reader (Fig. 8C). As expected, rVSV-GFP replication, as determined by GFP expression, was 20-fold lower in cells pretreated with tissue culture supernatants from IKKε-transfected cells than in cells pretreated with tissue culture supernatants from empty-plasmid-transfected cells (Fig. 8C). Interestingly, levels of GFP expression were approximately 3-fold lower in cells treated with tissue culture supernatants from IFI44L-silenced cells transfected with pCAGGS-His-IKKε than in cells treated with the supernatants from NT control siRNA cells transfected with pCAGGS-His-IKKε (Fig. 8C). These data are consistent with the Fluc results and further demonstrate that IFI44L silencing increases IFN responses after IKKε overexpression (Fig. 8A).
To analyze whether IFI44L negatively regulates IKKε kinase activity and whether such regulation would be dependent on FKBP5 expression, cells were silenced for IFI44L or FKBP5 expression and were then transfected with His-IKKε, IFI44L-HA, and FKBP5-FLAG plasmids. At 24 hpt, His-IKKε complexes were purified with an anti-His antibody and were subjected to a kinase assay using purified IRF-3 as the substrate. Expression of IKKε, FKBP5, and IFI44L (Fig. 8D) was confirmed in all the experiments. Silencing of FKBP5 was confirmed at the mRNA and protein levels (Fig. 8E). Total and phosphorylated levels of IRF-3 and IKKε were analyzed by Western blotting using specific antibodies (Fig. 8D). IKKε expression levels were similar in all the samples transfected with the IKKε-expressing plasmid (Fig. 8D, second blot). IRF-3 was phosphorylated using purified proteins from cells transfected with the plasmid encoding IKKε and not with purified proteins from cells transfected with empty plasmid, indicating that IKKε overexpression was responsible for IRF-3 phosphorylation in this assay (Fig. 8D). Interestingly, when IFI44L was expressed together with FKBP5 and IKKε, levels of p-IRF-3 normalized to the levels of IKKε were at least 5-fold lower than those seen when IFI44L was not expressed (graph panel in Fig. 8D). However, when FKBP5 was not present in the purified proteins, the presence or absence of IFI44-L had no effect on IRF-3 phosphorylation (Fig. 8D, third panel and graph). Importantly, levels of total IRF-3 were similar in all the cases (Fig. 8D, lower band in third blot). Furthermore, expression of IFI44L did not significantly affect the expression levels of IKKε (Fig. 8D, second blot) but altered the levels of phospho-IKKε (Fig. 8D, top blot), measured as a hallmark of activation (42). These results demonstrate that IFI44L negatively affects IKKε kinase activity and that this effect is dependent on the binding of IFI44L to FKBP5.
IFI44L decreases IKKβ kinase activity.
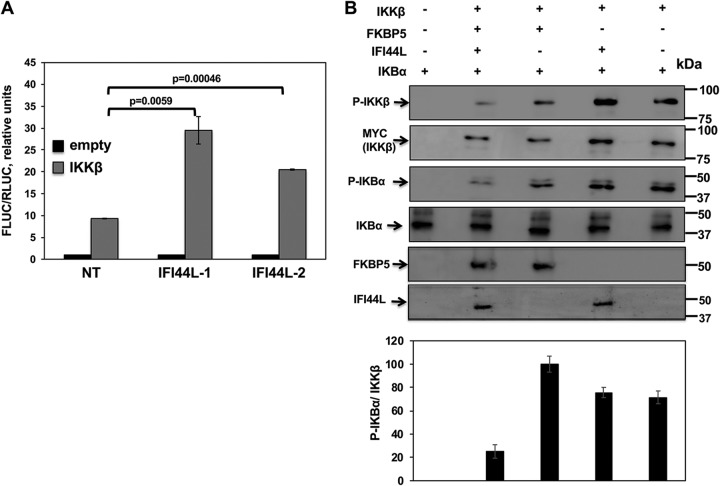
FKBP5 increases IkBα phosphorylation mediated by IKKα and IKKβ kinases (21), a crucial step for NF-κB activation (43). To assess if IFI44L would negatively modulate IKKβ activity and NF-κB activation, human 293T cells were silenced with the two different IFI44L siRNAs and cotransfected with a plasmid expressing Fluc under the control of the NF-κB promoter and with a plasmid constitutively expressing Rluc to normalize the levels of transfection, together with a plasmid expressing IKKβ or with the empty plasmid as a control. Then, 24 hpt, levels of Fluc were measured and normalized to the levels of Rluc. As expected, IKKβ overexpression induced NF-κB activation (Fig. 9A) (19). NF-κB was activated to (2-fold to 3-fold) higher levels in cells silenced for IFI44L than in NT-siRNA-transfected cells (Fig. 9A), demonstrating that IFI44L modulates IKKβ activation and, therefore, NF-κB promoter activation.
FIG 9.
IFI44L modulates IKKβ activity. Human 293T cells were transfected with two different siRNAs targeting IFI44L; 36 hpt, cells were cotransfected with pCAGGS-myc-IKKβ together with a plasmid expressing the Fluc under the control of an NF-κB-driven promoter (pNF-κB-Fluc) and a plasmid constitutively expressing Rluc under the control of the SV40 promoter. (A) At 24 hpt, levels of Fluc were determined and normalized to the levels of Rluc. Data represent means and SDs of results from triplicate wells. Experiments were repeated three times with similar results. P values determined by using a Student's t test are indicated. (B) Human 293T cells were silenced for IFI44 or FKBP5 and were transfected with plasmids expressing myc-IKKβ, IFI44L-HA, and FKBP5-FLAG. At 24 hpt, IKKβ complexes were purified with an anti-myc antibody and were used in a kinase assay with recombinant human IκBα as the substrate. Levels of phosphorylated and total IκBα were analyzed by Western blotting using specific antibodies. Levels of IKKβ were also analyzed using an anti-myc-specific antibody and phospho-IKKβ. Levels of IFI44L-HA and FKBP5-FLAG proteins were detected with anti-HA-specific and anti-FLAG-specific antibodies, respectively. Protein bands were quantified by densitometry using ImageJ software (v1.46). Levels of p-IκBα were normalized to the levels of IKKβ (bottom graphic). The results show error bars and means of results from two independent experiments. Molecular weight markers (in kilodaltons) are indicated on the right.
To analyze whether IFI44L negatively regulates IKKβ kinase activity and whether such regulation would be dependent on FKBP5 expression, human 293T cells were silenced for IFI44L or FKBP5 and were then transfected with plasmids expressing myc-IKKβ, IFI44L-HA, and FKBP5-FLAG. At 24 hpt, IKKβ proteins were purified with an anti-myc antibody, and complexes were used in a kinase assay performed with purified IκBα as a substrate. Expression of IKKβ, as well as of FKBP5 and IFI44L (Fig. 9B), was confirmed in all these experiments. IKKβ expression levels were similar in all the samples transfected with the IKKβ-expressing plasmid (Fig. 9B, second blot). As expected, IκBα was phosphorylated using purified proteins from cells transfected with IKKβ rather than using purified proteins from cells transfected with the empty plasmid, demonstrating that IKKβ overexpression was responsible for IκBα phosphorylation in our assay (Fig. 9B, third blot). Interestingly, when IFI44L was expressed together with FKBP5 and IKKβ, levels of p-IκBα normalized to the levels of IKKβ were at least 4-fold lower than those seen when IFI44L was not expressed (Fig. 9B, graph). However, when FKBP5 was not present in the purified proteins, the presence or absence of IFI44L had no effect on the levels of IκBα phosphorylation (Fig. 9B, third blot and graph). Importantly, levels of total IκBα were similar in all the cases (Fig. 9B, fourth blot) whereas the levels of phospho-IKKβ, measured as a hallmark of activation (44), were lower in cells expressing both IFI44L and FKBP5 (Fig. 9B, top blot). These results demonstrate that IFI44L negatively affected IKKβ kinase activity and that this effect was dependent on the binding of IFI44L to FKBP5. This negative effect of IFI44L on IKKβ and IKKε activity most likely contributes to the effect of IFI44L on IFN responses.
DISCUSSION
In this work, we uncovered a novel function for IFI44L in negatively regulating type I and III IFN responses induced after infection with different viruses as well as after treatments with poly(I·C) and IFN. In addition, we showed that IFI44L silencing decreases IAV, LCMV and HCoV-229E production (Fig. 2D to F), most probably due to the effect of IFI44L on IFN responses. The IFI44L mechanism of inhibition of IFN responses likely involves a novel interaction of IFI44L with cellular FKBP5 (Fig. 6), a protein which interacts with IKKα, IKKβ, and IKKε (Fig. 7) (21). This interaction decreases IRF-3 (Fig. 8D) and IkBα (Fig. 9B) phosphorylation mediated by IKKε and IKKβ, respectively, and, therefore, IFN responses (Fig. 3 and 4).
IFI44L is a genetic paralog of IFI44, most probably resulting from gene duplication. IFI44L is an ISG, induced by many different viruses such as IAV, respiratory syncytial virus, LCMV, and HCoV-229E (30, 31) (Fig. 1). Many ISGs display antiviral activity (27). However, excessive innate immune responses are deleterious to the host, and negative regulators of IFN production and signaling are needed to resolve the IFN-induced state and facilitate the return to cellular homeostasis (27–29). Here, we show a completely novel function for IFI44L as a negative regulator of IFN responses, even under conditions in which protein expression was not completely knocked down (Fig. 2B). Remarkably, we have recently shown that IFI44, a gene paralog of IFI44L, also displays that same function (DeDiego et al., submitted). In addition, other ISGs such as suppressor of cytokine signaling (SOCS) proteins SOCS1 and SOCS3 (45), IFI35 (46), ubiquitin carboxy-terminal hydrolase 18 (USP18) (47), ISG56/IFIT1 (48), optineurin (49), the ubiquitin ligases RING finger protein 5 (RNF-5) (50) and RNF-125 (51), and A20 (52) negatively modulate innate immune responses, although they operate through mechanisms and targets different from the ones we have identified for IFI44 (DeDiego et al., submitted) and IFI44L (this study).
We have shown that IFI44L silencing decreased virus production (Fig. 2D to F). As that effect applies to different viruses, the decrease was most probably due to the effect of IFI44L on negative modulation of IFN responses (Fig. 3 and 4). It is well known that IAV and LCMV are sensitive to the antiviral states induced by IFN (5, 36–38) and that many ISGs such as myxovirus resistance protein 1 (MX1), IFN-induced transmembrane (IFITM) proteins, oligoadenylate synthetase (OAS), protein kinase R (PKR), IFN-induced protein with tetratricopeptide repeats 1 (IFIT1), IFIT2, viperin, and IRF-1 restrict replication of viruses, including IAV (5). Due to the potent effect of IFN signaling in restricting virus replication, viruses encode proteins which counteract innate immune responses induced by the host, such as IAV nonstructural protein 1 (NS1) (53) and PA-X (54, 55) and LCMV nucleoprotein (NP) (56). Therefore, IAVs lacking NS1 or PA-X (54, 57) or LCMV strains possessing viral NP with altered anti-IFN function (56) induce higher IFN responses and are impaired in replication in IFN-competent systems. High doses of type I and IIII IFNs have clear effects against coronaviruses in cell cultures, in vivo, and in patients (58). In the case of coronaviruses, many proteins act as antagonists of IFN, including severe acute respiratory syndrome (SARS)-CoV nonstructural protein 1 (nsp1), papain-like protease (PLP), nsp7, nsp14, nsp15, nsp16, 3b, M, 6, and NP (58–60). As an example, the nsp1 protein of coronaviruses facilitates the efficient propagation of the virus in cells through a specific translational shutoff of host mRNAs, including IFN response genes (61, 62), and its deletion leads to virus attenuation (59, 63).
We have shown a novel interaction of IFI44L with FKBP5 (Fig. 6). FKBP5 belongs to a family of immunophilins comprising chaperones that are members of a highly conserved family of proteins, all of which are cis-trans peptidyl-prolyl isomerases (64, 65). FKBP5 binds IKKα, IKKβ, and IKKγ and facilitates IKK complex assembly, leading to increased IKKα and IKKβ kinase activity (21). In addition, it was proposed previously (22) and has been confirmed (DeDiego et al., submitted) that FKBP5 interacts with IKKε, likely modulating IKKε kinase activity. In this work, we have confirmed the interaction of FKBP5 with IKKβ and IKKε, and our data suggest that the interaction of IFI44L with FKBP5 does not abolish the binding of FKBP5 to IKKβ or IKKε (Fig. 7). Phosphorylation of IκBα by IKKβ and IKKα leads to its degradation, allowing NF-κB to migrate to the nucleus and activate IFN and proinflammatory cytokine expression (20). In addition, IKKε phosphorylates transcription factors IRF-3 and IRF-7 (16), leading to IFN induction (1), and IKKε phosphorylates STAT1, a critical step for the IFN-inducible antiviral response (23, 24). In the presence of FKBP5, IFI44L decreases IRF-3 phosphorylation mediated by IKKε (Fig. 8D), as well as IκBα phosphorylation mediated by IKKβ (Fig. 9B), identifying a likely mechanism for the role of IFI44L in negatively modulating innate immune responses.
In summary, we have described a new role for IFI44L acting as a feedback regulator of IFN responses, as well as the mechanism mediating this effect. Understanding the mechanisms of the negative regulators of innate immune responses is a very important objective, as manipulating negative regulators such as IFI44L may lead to the development of novel therapeutic approaches to control a range of pathogen infectious and also inflammatory diseases of significant clinical importance for humans.
MATERIALS AND METHODS
Cell lines.
Madin-Darby canine kidney cells (MDCK cells; ATCC CCL-34), human embryonic kidney cells (293T; ATCC CRL-11268); human lung epithelial carcinoma cells (A549; ATCC CCL-185), human hepatocellular carcinoma Huh-7 cells (kindly provided by R. Bartenschlager, University of Heidelberg, Heidelberg, Germany), and African green monkey kidney cells (Vero; ATCC CCL-81) were grown at 37°C in air enriched with 5% CO2 using Dulbecco’s modified Eagle’s medium (DMEM; Gibco) supplemented with 10% fetal bovine serum (Gibco), 100 units/ml penicillin, 0.1 mg/ml streptomycin, and 50 μg/ml gentamicin (Gibco). Human near-haploid HAP-1 parental and IFI44L knockout (KO) cells processed by using the CRISPR/cas9 technology were obtained from Horizon Discovery, Inc., and maintained at 37°C in air enriched with 5% CO2 using Iscovés modified Dulbeccós medium supplemented with 10% fetal bovine serum (Gibco), 100 units/ml penicillin, 0.1 mg/ml streptomycin, and 50 μg/ml gentamicin (Gibco). HAP-1 cells are derived from male chronic myelogenous leukemia (CML) cell line KBM-7.
Viruses.
Stocks of influenza A/Puerto Rico/8/1934 H1N1 (PR8) virus grown in MDCK cells (66); recombinant vesicular stomatitis virus (rVSV), Indiana strain, encoding green fluorescent protein (GFP) (rVSV-GFP) (40), grown in Vero cells; recombinant trisegmented LCMV (r3LCMV), Armstrong strain, expressing GFP and Gaussia luciferase (r3LCMV-GFP/Gluc) (67, 68), grown in Vero cells; and recombinant HCoV-229E grown in Huh-7 cells were previously described and used in this study.
Virus titrations.
IAV PR8 was titrated in MDCK cells (96-well format, 3 × 104 cells/well) by immunofocus assay (with results quantified as focus-forming units [FFU] per milliliter), as previously described (69). All IAV infections were performed in the presence of 1 μg/ml of tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin (Sigma). The r3LCMV-GFP/Gluc strain was titrated in Vero cells (96-well format, 3 × 104 cells/well) and GFP expression was analyzed (with results quantified as FFU per milliliter) as previously described (67, 68). HCoV-229E was titrated by plaque assay (with results quantified as PFU per milliliter) in Huh-7 cells (12-well format, 3.75 × 105 cells/well). Viral plaques were visualized and counted 3 days postinfection (dpi) using crystal violet staining. The rVSV-GFP strain was titrated by plaque assay (PFU per milliliter) in Vero cells (12-well format, 3.75 × 105 cells/well). Viral plaques were visualized and counted 1 dpi using crystal violet staining.
Plasmids.
To generate a polymerase II pCAGGS expression plasmid (70) encoding human IFI44L (GenBank accession number NM_006820.1) fused at the C-terminal to an hemagglutinin (HA) epitope tag (pCAGGS-IFI44L-HA), a plasmid encoding the wild-type human IFI44L open reading frame (ORF) was obtained from Origene and used as the template for PCR analysis (primers available upon request). pCAGGS plasmids encoding FKBP5 (GenBank accession number NM_004117.3) with a C-terminal FLAG tag epitope (pCAGGS-FKBP5-FLAG); IKKβ (GenBank accession number NM_001556) with an N-terminal myc epitope tag (pCAGGS-myc-IKKβ); and IKKε (GenBank accession number NM_014002) with an N-terminal 6×His tag (pCAGGS-His-IKKε) were previously described (DeDiego et al., submitted). Plasmid were verified by sequencing (ACGT).
Knockdown of cellular host proteins using small interfering RNAs (siRNAs).
Human A549, Huh-7, 293T, or HAP-1 cells (96-well format, 5 × 104 cells/well) were transfected independently with two different siRNAs specific for human IFI44L (Thermo Fisher Scientific s21586 and s21587) or human FKBP5 (Thermo Fisher Scientific s5215 and s5216) or with nontargeting (NT) negative-control no. 1 (Thermo Fisher Scientific AM4635). All siRNAs were transfected at a final concentration of 20 nM, using Lipofectamine RNAiMax (Thermo Fisher Scientific), according to the manufacturer’s recommendations.
Analysis of mRNAs by RT-qPCR.
IFI44L, FKBP5, IFN-λ1, and IFN-induced protein with tetratricopeptide repeats 2 (IFIT2) mRNA levels were analyzed in human A549, 293T, and HAP-1 cells. To that end, total RNA was extracted using an RNeasy minikit (Qiagen) and 100-ng volumes of total RNA were used as the templates for RT reactions with a High-Capacity cDNA transcription kit and random primers. For qPCRs, TaqMan gene expression assays (Applied Biosystems) specific for human IFI44L (Hs00915292_m1), human IFI44 (Hs00197427_m1), human FKBP5 (Hs01561006_M1), human IFN-β (Hs01077958_s1), human IFN-λ1 (Hs00601677_g1), and human IFIT2 (Hs00533665_m1) genes were used. Relative mRNA levels were quantified using the threshold cycle (2−ΔΔCT) method (71).
Protein detection by Western blotting.
Cells were collected, lysed in passive lysis buffer (Promega), and clarified by centrifugation. Then, Laemmli sample buffer (Bio-Rad) and β-mercaptoethanol were added, and samples were heated at 90°C for 5 min before 10% SDS-PAGE gels were processed. Proteins were transferred to nitrocellulose membranes (Bio-Rad) and were detected by Western blotting using primary rabbit polyclonal antibodies (pAbs) specific for the HA epitope tag (to detect overexpressed IFI44L-HA; Sigma-Aldrich H6908), FLAG epitope tag (to detect FKBP5-FLAG; Sigma-Aldrich F7425), phospho-IKKα (Ser176)/IKKβ (Ser177) (Cell Signaling 2078), phospho-IKKε (Thr501) (Rockland 600-401-267), and IRF-3 (Abcam ab25950) (diluted 1:1,000) and using mouse monoclonal antibodies (MAbs) anti-6×His (to detect His-IKKε; Thermo Scientific MA1-21315), anti-myc (to detect myc-IKKβ; Thermo Scientific 13-2500), anti-phospho-IRF-3 (Abcam ab76493), anti-phospho-IkBα (Thermo Scientific MA5-15224), anti-IkBα (Abcam ab32518), and anti-actin (Sigma-Aldrich A1978) (all diluted 1:1,000). Then, the membranes were washed 3 times with phosphate-buffered saline (PBS) containing 0.1% Tween 20 and incubated with a 1:1,000 dilution of goat anti-rabbit IgG antibodies (pAbs) or goat anti-mouse IgG antibodies (MAbs) conjugated to horseradish peroxidase (Sigma-Aldrich). Proteins in the membranes were detected by chemiluminescence using SuperSignal West Femto maximum-sensitivity substrate (Thermo Scientific), according to the manufacturer’s recommendations.
Binding of IFI44L and FKBP5 proteins.
Human 293T cells (6-well format, 106 cells/well) were transiently transfected with 2,500 ng/well of pCAGGS-FKBP5-FLAG and pCAGGS-IFI44L-HA by the use of DNA-IN for 30 h. A pCAGGS empty plasmid was used to maintain the amount of transfected DNA plasmid at a constant level. Cells were lysed in Co-IP buffer (100 mM NaCl, 0.5 mM EDTA, 20 mM Tris-HCl [pH 7.5], 1% Triton X-100, 5% glycerol) containing protease inhibitors (Thermo Scientific). Cell lysates were cleared by centrifugation and incubated overnight at 4°C with 30 μl of an anti-FLAG affinity resin (Sigma-Aldrich) to immunoprecipitate (IP) FKBP5 or with 30 μl of an anti-HA affinity resin to IP IFI44L (Pierce). After three washes in Tris-buffered saline (TBS) buffer containing 0.1% Tween 20 (for anti-HA resin) or TBS containing 0.2% SDS (for anti-FLAG resin), precipitated proteins were dissociated from the resin using disruption buffer and high temperature (95°C) and were analyzed by Western blotting as described above using anti-HA (IFI44L)-specific and anti-FLAG (FKBP5)-specific Abs.
Binding of FKBP5 to IKKβ and IKKε proteins in the presence or absence of IFI44L.
Human 293T cells (6-well format, 106 cells/well) were transiently transfected (2,000 ng each plasmid/well) with different combinations of pCAGGS-FKBP5-FLAG, pCAGGS-IFI44L-HA, and pCAGGS-His-IKKε plasmids and of pCAGGS-FKBP5-FLAG, pCAGGS-IFI44L-HA, and pCAGGS-myc-IKKβ plasmids by the use of DNA-IN for 30 h. A pCAGGS empty plasmid was used to maintain the amount of transfected DNA plasmid at a constant level. At 30 h posttransfection (hpt), cells were lysed in Co-IP buffer containing protease inhibitors (Thermo Scientific), and cleared cell lysates were incubated overnight at 4°C with 30 μl of an anti-FLAG affinity resin (Sigma-Aldrich). Resins were washed and the precipitated proteins dissociated from the resin as described above. Precipitated proteins were analyzed by Western blotting using anti-HA (IFI44L)-, anti-myc (IKKβ)-, anti-His (IKKε)-, or anti-FLAG (FKBP5)-specific Abs.
IFN response assays.
To evaluate the effect of IFI44L on the induction of IFN responses, human A549 or Huh-7 cells (96-well plate format, 5 × 104 cells/well, triplicate assays) were transfected with siRNAs specific for IFI44L, or the NT siRNA control, for 36 h. Then, cells were infected with IAV (PR8 strain) (multiplicity of infection [MOI], 0.1), r3LCMV-GFP/Gluc (MOI, 0.1), or HCoV-229E (MOI, 0.1). Alternatively, A549 and HAP-1 cells were treated with 250 U/ml and 2,000 U/ml, respectively, of universal IFN-α (Axxora) or were transfected with 250 ng/ml and 2,000 ng/ml, respectively, of polyinosinic-polycytidylic acid [poly(I·C)] (Sigma) by the use of DNA-IN for 16 h. Total RNA was extracted, and RT-qPCRs were performed as previously described. Alternatively, 293T cells (96-well plate format, 5 × 104 cells/well, triplicate assays) were transfected with 200 ng of an empty plasmid or with the pCAGGS-IFI44L-HA plasmid by the use of DNA-IN. At 24 hpt, cells were treated with universal IFN-α (Axxora) (1,000 U/ml), transfected with 1,000 ng/ml of poly(I·C) by the use of DNA-IN for 16 h, or infected with IAV for 24 h. Total RNA was extracted, and RT-qPCRs were performed as described above.
Evaluation of the effect of IFI44L on antiviral activity.
Expression of endogenous IFI44L was knocked down in human A549 cells (96-well format, 5 × 104 cells/well, triplicate assays) by transfecting IFI44L or NT siRNAs as described above. Alternatively, HAP-1 control and IFI44L KO cells were used. At 36 hpt, cells were treated with 250 U/ml (A549) or 2,000 U/ml (HAP-1) of universal IFN-α or were transfected with 250 ng/ml or 2,000 U/ml, respectively, of poly(I·C) by the use of DNA-IN. At 16 h after treatment, cells were infected with rVSV-GFP (40) and viral titers in tissue culture supernatants at 24 hpi were determined in Vero cells as described above.
Assessment of IKKε- and IKKβ-mediated activation of innate immune responses.
To measure the effect of IFI44L on the induction of IFN by overexpression of IKKε and on the activation of NF-κB mediated by IKKβ overexpression, IFI44L expression was knocked down in human 293T cells (96-well format, 5 × 104 cells/well, triplicate assays) as described above. Then, IFI44L-silenced 293T cells were cotransfected with 50 ng/well of plasmids expressing Firefly luciferase (Fluc) under the control of the IFN-β promoter (pIFNβ-Fluc) (72), 20 ng/well of a plasmid expressing Renilla luciferase (Rluc) under the control of a constitutive promoter (pRL-SV40; Promega), and 200 ng/well of pCAGGS-His-IKKε or with 50 ng/well of a plasmid expressing Fluc under the control of an NF-κB promoter (Addgene), 20 ng/well of plasmid pRL-SV40, and 200 ng/well of pCAGGS-myc-IKKβ for 24 h. Cells were lysed using Promega passive lysis buffer (20 mM Tris-HCl [pH 7.4], 5 mM EDTA, 100 mM NaCl, 1% NP-40) in the presence of a complete protease inhibitor cocktail (Thermo Scientific) for 30 min on ice. Cell lysates were clarified by centrifugation, and an equal volume of luciferase reporter buffer (Promega) was added. Fluc and Rluc protein expression levels were quantified in a LumiCount luminometer. The levels of Fluc expression were normalized to the levels of Rluc expression. In addition, tissue culture supernatants from pCAGGS-His-IKKε-transfected 293T cells were collected and used to treat A549 cells in 96-well plates (5 × 104 cells/well, triplicate assays) for 24 h. Then, cells were infected (MOI, 0.1) with rVSV-GFP. At 24 hpi, GFP intensity was measured using a fluorescent microplate reader (DTX880; Beckman Coulter) and images from GFP-expressing cells were obtained in a fluorescence microscope.
Kinase activity of IKKβ and IKKε in the presence of FKBP5 and IFI44L.
Human 293T cells (6-well format, 106 cells/well) were silenced for IFI44L or FKBP5 as described above. At 24 hpt, cells were transfected with 1,500 ng/well of pCAGGS-myc-IKKβ or pCAGGS-His-IKKε, pCAGGS-FKBP5-FLAG, and pCAGGS-IFI44L-HA plasmids. The total amounts of transfected siRNA and plasmid DNA were maintained at constant levels with NT siRNA control and empty plasmid, respectively. At 36 hpt, cells were lysed in coimmunoprecipitation (Co-IP) buffer containing protease inhibitors (Thermo Scientific) and 1 mM NaVO4. Cleared cell lysates were incubated for 4 h at 4°C with 30 μl of an anti-myc affinity resin (Sigma-Aldrich) (for myc-IKKβ complexes) or with 30 μl of nickel-Sepharose (Ni-Sepharose) Fast Flow resin (GE Healthcare) (for His-IKKε complexes). The resins were washed three times in TBS buffer containing 0.1% Tween 20 and twice in kinase buffer (Cell Signaling) and were then incubated with 0.5 μg of recombinant IkBα (Sino Biological) (for IKKβ activity) or IRF-3 (BioPharma) (for IKKε activity) for 1 h at 30°C. Proteins were dissociated from the resins using disruption buffer under high-temperature (95°C) conditions and were analyzed by Western blotting as described above.
ACKNOWLEDGMENTS
This project was funded with federal funds from the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH), Department of Health and Human Services, under CEIRS contract HHSN272201400005C to D.J.T.; through a University of Rochester Research Award to M.L.D.; and with funds from Comunidad de Madrid, Madrid, Spain (reference 2017-T1/BMD-5155), to M.L.D.
M.L.D. conceived, designed, and performed the experiments. D.J.T. and L.M.-S. provided reagents, suggestions, and resources. M.L.D. and D.J.T. provided funds. M.L.D. analyzed the data and wrote the initial manuscript draft. M.L.D., L.M.-S., and D.J.T. edited the original draft.
We declare that we have no competing interests.
REFERENCES
- 1.Levy DE, Marie IJ, Durbin JE. 2011. Induction and function of type I and III interferon in response to viral infection. Curr Opin Virol 1:476–486. doi: 10.1016/j.coviro.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jensen S, Thomsen AR. 2012. Sensing of RNA viruses: a review of innate immune receptors involved in recognizing RNA virus invasion. J Virol 86:2900–2910. doi: 10.1128/JVI.05738-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takeuchi O, Akira S. 2010. Pattern recognition receptors and inflammation. Cell 140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 4.Odendall C, Kagan JC. 2015. The unique regulation and functions of type III interferons in antiviral immunity. Curr Opin Virol 12:47–52. doi: 10.1016/j.coviro.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iwasaki A, Pillai PS. 2014. Innate immunity to influenza virus infection. Nat Rev Immunol 14:315–328. doi: 10.1038/nri3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pythoud C, Rothenberger S, Martinez-Sobrido L, de la Torre JC, Kunz S. 2015. Lymphocytic choriomeningitis virus differentially affects the virus-induced type I interferon response and mitochondrial apoptosis mediated by RIG-I/MAVS. J Virol 89:6240–6250. doi: 10.1128/JVI.00610-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li J, Liu Y, Zhang X. 2010. Murine coronavirus induces type I interferon in oligodendrocytes through recognition by RIG-I and MDA5. J Virol 84:6472–6482. doi: 10.1128/JVI.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roth-Cross JK, Bender SJ, Weiss SR. 2008. Murine coronavirus mouse hepatitis virus is recognized by MDA5 and induces type I interferon in brain macrophages/microglia. J Virol 82:9829–9838. doi: 10.1128/JVI.01199-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zust R, Cervantes-Barragan L, Habjan M, Maier R, Neuman BW, Ziebuhr J, Szretter KJ, Baker SC, Barchet W, Diamond MS, Siddell SG, Ludewig B, Thiel V. 2011. Ribose 2’-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol 12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin RT, Heylbroeck C, Pitha PM, Hiscott J. 1998. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol Cell Biol 18:2986–2996. doi: 10.1128/MCB.18.5.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoneyama M, Suhara W, Fukuhara Y, Fukuda M, Nishida E, Fujita T. 1998. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J 17:1087–1095. doi: 10.1093/emboj/17.4.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, Shimada N, Ohba Y, Takaoka A, Yoshida N, Taniguchi T. 2005. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 434:772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- 13.Lenardo MJ, Fan CM, Maniatis T, Baltimore D. 1989. The involvement of Nf-Kappab in beta-interferon gene-regulation reveals its role as widely inducible mediator of signal transduction. Cell 57:287–294. doi: 10.1016/0092-8674(89)90966-5. [DOI] [PubMed] [Google Scholar]
- 14.Visvanathan KV, Goodbourn S. 1989. Double-stranded-RNA activates binding of NF-kappa-B to an inducible element in the human beta-interferon promoter. EMBO J 8:1129–1138. doi: 10.1002/j.1460-2075.1989.tb03483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Du W, Maniatis T. 1992. An Atf/Creb binding-site protein is required for virus induction of the human interferon beta gene. Proc Natl Acad Sci U S A 89:2150–2154. doi: 10.1073/pnas.89.6.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharma S, tenOever BR, Grandvaux N, Zhou GP, Lin RT, Hiscott J. 2003. Triggering the interferon antiviral response through an IKK-related pathway. Science 300:1148–1151. doi: 10.1126/science.1081315. [DOI] [PubMed] [Google Scholar]
- 17.Servant MJ, Tenoever B, Lin R. 2002. Overlapping and distinct mechanisms regulating IRF-3 and IRF-7 function. J Interferon Cytokine Res 22:49–58. doi: 10.1089/107999002753452656. [DOI] [PubMed] [Google Scholar]
- 18.Osterlund PI, Pietila TE, Veckman V, Kotenko SV, Julkunen I. 2007. IFN regulatory factor family members differentially regulate the expression of type III IFN (IFN-lambda) genes. J Immunol 179:3434–3442. doi: 10.4049/jimmunol.179.6.3434. [DOI] [PubMed] [Google Scholar]
- 19.Karin M. 1999. How NF-kappaB is activated: the role of the IkappaB kinase (IKK) complex. Oncogene 18:6867–6874. doi: 10.1038/sj.onc.1203219. [DOI] [PubMed] [Google Scholar]
- 20.Karin M, Ben-Neriah Y. 2000. Phosphorylation meets ubiquitination: the control of NF-[kappa]B activity. Annu Rev Immunol 18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 21.Romano S, Xiao Y, Nakaya M, D'Angelillo A, Chang M, Jin J, Hausch F, Masullo M, Feng X, Romano MF, Sun S-C. 2015. FKBP51 employs both scaffold and isomerase functions to promote NF-kappa B activation in melanoma. Nucleic Acids Res 43:6983–6993. doi: 10.1093/nar/gkv615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bouwmeester T, Bauch A, Ruffner H, Angrand PO, Bergamini G, Croughton K, Cruciat C, Eberhard D, Gagneur J, Ghidelli S, Hopf C, Huhse B, Mangano R, Michon AM, Schirle M, Schlegl J, Schwab M, Stein MA, Bauer A, Casari G, Drewes G, Gavin AC, Jackson DB, Joberty G, Neubauer G, Rick J, Kuster B, Superti-Furga G. 2004. A physical and functional map of the human TNF-alpha NF-kappa B signal transduction pathway. Nat Cell Biol 6:97–105. doi: 10.1038/ncb1086. [DOI] [PubMed] [Google Scholar]
- 23.Tenoever BR, Ng SL, Chua MA, McWhirter SM, Garcia-Sastre A, Maniatis T. 2007. Multiple functions of the IKK-related kinase IKKepsilon in interferon-mediated antiviral immunity. Science 315:1274–1278. doi: 10.1126/science.1136567. [DOI] [PubMed] [Google Scholar]
- 24.Ng SL, Friedman BA, Schmid S, Gertz J, Myers RM, tenOever BR, Maniatis T. 2011. I kappa B kinase epsilon (IKK epsilon) regulates the balance between type I and type II interferon responses. Proc Natl Acad Sci U S A 108:21170–21175. doi: 10.1073/pnas.1119137109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoffmann HH, Schneider WM, Rice CM. 2015. Interferons and viruses: an evolutionary arms race of molecular interactions. Trends Immunol 36:124–138. doi: 10.1016/j.it.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ank N, West H, Bartholdy C, Eriksson K, Thomsen AR, Paludan SR. 2006. Lambda interferon (IFN-lambda), a type III IFN, is induced by viruses and IFNs and displays potent antiviral activity against select virus infections in vivo. J Virol 80:4501–4509. doi: 10.1128/JVI.80.9.4501-4509.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schneider WM, Chevillotte MD, Rice CM. 2014. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol 32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richards KH, Macdonald A. 2011. Putting the brakes on the anti-viral response: negative regulators of type I interferon (IFN) production. Microbes Infect 13:291–302. doi: 10.1016/j.micinf.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 29.Komuro A, Bamming D, Horvath CM. 2008. Negative regulation of cytoplasmic RNA-mediated antiviral signaling. Cytokine 43:350–358. doi: 10.1016/j.cyto.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ioannidis I, McNally B, Willette M, Peeples ME, Chaussabel D, Durbin JE, Ramilo O, Mejias A, Flano E. 2012. Plasticity and virus specificity of the airway epithelial cell immune response during respiratory virus infection. J Virol 86:5422–5436. doi: 10.1128/JVI.06757-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhai YJ, Franco LM, Atmar RL, Quarles JM, Arden N, Bucasas KL, Wells JM, Nino D, Wang XQ, Zapata GE, Shaw CA, Belmont JW, Couch RB. 2015. Host transcriptional response to influenza and other acute respiratory viral infections—a prospective cohort study. PLoS Pathog 11:e1004869. doi: 10.1371/journal.ppat.1004869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moal V, Textoris J, Ben Amara A, Mehraj V, Berland Y, Colson P, Mege JL. 2013. Chronic hepatitis E virus infection is specifically associated with an interferon-related transcriptional program. J Infect Dis 207:125–132. doi: 10.1093/infdis/jis632. [DOI] [PubMed] [Google Scholar]
- 33.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. 2011. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haralambieva IH, Ovsyannikova IG, Kennedy RB, Larrabee BR, Zimmermann MT, Grill DE, Schaid DJ, Poland GA. 2017. Genome-wide associations of CD46 and IFI44L genetic variants with neutralizing antibody response to measles vaccine. Hum Genet 136:421–435. doi: 10.1007/s00439-017-1768-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eisenacher K, Krug A. 2012. Regulation of RLR-mediated innate immune signaling—it is all about keeping the balance. Eur J Cell Biol 91:36–47. doi: 10.1016/j.ejcb.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 36.Papatriantafyllou M. 2013. Infection: the interferon paradox. Nat Rev Immunol 13:392. doi: 10.1038/nri3461. [DOI] [PubMed] [Google Scholar]
- 37.Moskophidis D, Battegay M, Bruendler MA, Laine E, Gresser I, Zinkernagel RM. 1994. Resistance of lymphocytic choriomeningitis virus to alpha/beta interferon and to gamma-interferon. J Virol 68:1951–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muller U, Steinhoff U, Reis LFL, Hemmi S, Pavlovic J, Zinkernagel RM, Aguet M. 1994. Functional role of type I and type II interferons in antiviral defense. Science 264:1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- 39.Gitlin L, Barchet W, Gilfillan S, Cella M, Beutler B, Flavell RA, Diamond MS, Colonna M. 2006. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci U S A 103:8459–8464. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stojdl DF, Lichty BD, tenOever BR, Paterson JM, Power AT, Knowles S, Marius R, Reynard J, Poliquin L, Atkins H, Brown EG, Durbin RK, Durbin JE, Hiscott J, Bell JC. 2003. VSV strains with defects in their ability to shutdown innate immunity are potent systemic anti-cancer agents. Cancer Cell 4:263–275. doi: 10.1016/S1535-6108(03)00241-1. [DOI] [PubMed] [Google Scholar]
- 41.Akiyama T, Shiraishi T, Qin JW, Konno H, Akiyama N, Shinzawa M, Miyauchi M, Takizawa N, Yanai H, Ohashi H, Miyamoto-Sato E, Yanagawa H, Yong WD, Shou WN, Inoue J. 2014. Mitochondria-nucleus shuttling FK506-binding protein 51 interacts with TRAF proteins and facilitates the RIG-I-like receptor-mediated expression of type I IFN. PLoS One 9:e95992. doi: 10.1371/journal.pone.0095992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rajsbaum R, Versteeg GA, Schmid S, Maestre AM, Belicha-Villanueva A, Martínez-Romero C, Patel JR, Morrison J, Pisanelli G, Miorin L, Laurent-Rolle M, Moulton HM, Stein DA, Fernandez-Sesma A, tenOever BR, García-Sastre A. 2014. Unanchored K48-linked polyubiquitin synthesized by the E3-ubiquitin ligase TRIM6 stimulates the interferon-IKKepsilon kinase-mediated antiviral response. Immunity 40:880–895. doi: 10.1016/j.immuni.2014.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zandi E, Rothwarf DM, Delhase M, Hayakawa M, Karin M. 1997. The IkappaB kinase complex (IKK) contains two kinase subunits, IKKalpha and IKKbeta, necessary for IkappaB phosphorylation and NF-kappaB activation. Cell 91:243–252. doi: 10.1016/s0092-8674(00)80406-7. [DOI] [PubMed] [Google Scholar]
- 44.Delhase M, Hayakawa M, Chen Y, Karin M. 1999. Positive and negative regulation of IkappaB kinase activity through IKKbeta subunit phosphorylation. Science 284:309–313. doi: 10.1126/science.284.5412.309. [DOI] [PubMed] [Google Scholar]
- 45.Song MM, Shuai K. 1998. The suppressor of cytokine signaling (SOCS) 1 and SOCS3 but not SOCS2 proteins inhibit interferon-mediated antiviral and antiproliferative activities. J Biol Chem 273:35056–35062. doi: 10.1074/jbc.273.52.35056. [DOI] [PubMed] [Google Scholar]
- 46.Das A, Dinh PX, Panda D, Pattnaik AK. 2014. Interferon-inducible protein IFI35 negatively regulates RIG-I antiviral signaling and supports vesicular stomatitis virus replication. J Virol 88:3103–3113. doi: 10.1128/JVI.03202-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sarasin-Filipowicz M, Wang XY, Yan M, Duong FHT, Poli V, Hilton DJ, Zhang DE, Heim MH. 2009. Alpha interferon induces long-lasting refractoriness of JAK-STAT signaling in the mouse liver through induction of USP18/UBP43. Mol Cell Biol 29:4841–4851. doi: 10.1128/MCB.00224-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Y, Li C, Xue P, Zhong B, Mao AP, Ran Y, Chen H, Wang YY, Yang FQ, Shu HB. 2009. ISG56 is a negative-feedback regulator of virus-triggered signaling and cellular antiviral response. Proc Natl Acad Sci U S A 106:7945–7950. doi: 10.1073/pnas.0900818106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mankouri J, Fragkoudis R, Richards KH, Wetherill LF, Harris M, Kohl A, Elliott RM, Macdonald A. 2010. Optineurin negatively regulates the induction of IFN beta in response to RNA virus infection. PLoS Pathog 6:e1000778. doi: 10.1371/journal.ppat.1000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhong B, Zhang L, Lei CQ, Li Y, Mao AP, Yang Y, Wang YY, Zhang XL, Shu HB. 2009. The ubiquitin ligase RNF5 regulates antiviral responses by mediating degradation of the adaptor protein MITA. Immunity 30:397–407. doi: 10.1016/j.immuni.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 51.Arimoto KI, Takahashi H, Hishiki T, Konishi H, Fujita T, Shimotohno K. 2007. Negative regulation of the RIG-I signaling by the ubiquitin ligase RNF125. Proc Natl Acad Sci U S A 104:7500–7505. doi: 10.1073/pnas.0611551104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wertz IE, O'Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, Wu P, Wiesmann C, Baker R, Boone DL, Ma A, Koonin EV, Dixit VM. 2004. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappa B signalling. Nature 430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 53.Hale BG, Randall RE, Ortin J, Jackson D. 2008. The multifunctional NS1 protein of influenza A viruses. J Gen Virol 89:2359–2376. doi: 10.1099/vir.0.2008/004606-0. [DOI] [PubMed] [Google Scholar]
- 54.Hayashi T, MacDonald LA, Takimoto T. 2015. Influenza A virus protein PA-X contributes to viral growth and suppression of the host antiviral and immune responses. J Virol 89:6442–6452. doi: 10.1128/JVI.00319-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jagger BW, Wise HM, Kash JC, Walters KA, Wills NM, Xiao YL, Dunfee RL, Schwartzman LM, Ozinsky A, Bell GL, Dalton RM, Lo A, Efstathiou S, Atkins JF, Firth AE, Taubenberger JK, Digard P. 2012. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science 337:199–204. doi: 10.1126/science.1222213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pythoud C, Rodrigo W, Pasqual G, Rothenberger S, Martinez-Sobrido L, de la Torre JC, Kunz S. 2012. Arenavirus nucleoprotein targets interferon regulatory factor-activating kinase IKK epsilon. J Virol 86:7728–7738. doi: 10.1128/JVI.00187-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Garcia-Sastre A, Egorov A, Matassov D, Brandt S, Levy DE, Durbin JE, Palese P, Muster T. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- 58.Kindler E, Thiel V, Weber F. 2016. Interaction of SARS and MERS coronaviruses with the antiviral interferon response. Adv Virus Res 96:219–243. doi: 10.1016/bs.aivir.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zust R, Cervantes-Barragan L, Kuri T, Blakqori G, Weber F, Ludewig B, Thiel V. 2007. Coronavirus non-structural protein 1 is a major pathogenicity factor: implications for the rational design of coronavirus vaccines. PLoS Pathog 3:e109. doi: 10.1371/journal.ppat.0030109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Totura AL, Baric RS. 2012. SARS coronavirus pathogenesis: host innate immune responses and viral antagonism of interferon. Curr Opin Virol 2:264–275. doi: 10.1016/j.coviro.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kamitani W, Narayanan K, Huang C, Lokugamage K, Ikegami T, Ito N, Kubo H, Makino S. 2006. Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. Proc Natl Acad Sci U S A 103:12885–12890. doi: 10.1073/pnas.0603144103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tanaka T, Kamitani W, DeDiego ML, Enjuanes L, Matsuura Y. 2012. Severe acute respiratory syndrome coronavirus nsp1 facilitates efficient propagation in cells through a specific translational shutoff of host mRNA. J Virol 86:11128–11137. doi: 10.1128/JVI.01700-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jimenez-Guardeno JM, Regla-Nava JA, Nieto-Torres JL, DeDiego ML, Castano-Rodriguez C, Fernandez-Delgado R, Perlman S, Enjuanes L. 2015. Identification of the mechanisms causing reversion to virulence in an attenuated SARS-CoV for the design of a genetically stable vaccine. PLoS Pathog 11:e1005215. doi: 10.1371/journal.ppat.1005215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marks AR. 1996. Cellular functions of immunophilins. Physiol Rev 76:631–649. doi: 10.1152/physrev.1996.76.3.631. [DOI] [PubMed] [Google Scholar]
- 65.Barik S. 2006. Immunophilins: for the love of proteins. Cell Mol Life Sci 63:2889–2900. doi: 10.1007/s00018-006-6215-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schickli JH, Flandorfer A, Nakaya T, Martinez-Sobrido L, Garcia-Sastre A, Palese P. 2001. Plasmid-only rescue of influenza A virus vaccine candidates. Philos Trans R Soc Lond B 356:1965–1973. doi: 10.1098/rstb.2001.0979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ortiz-Riaño E, Cheng BYH, Carlos de la Torre J, Martínez-Sobrido L. 2013. Arenavirus reverse genetics for vaccine development. J Gen Virol 94:1175–1188. doi: 10.1099/vir.0.051102-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Emonet SF, Garidou L, McGavern DB, de la Torre JC. 2009. Generation of recombinant lymphocytic choriomeningitis viruses with trisegmented genomes stably expressing two additional genes of interest. Proc Natl Acad Sci U S A 106:3473–3478. doi: 10.1073/pnas.0900088106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nogales A, Baker SF, Ortiz-Riano E, Dewhurst S, Topham DJ, Martinez-Sobrido L. 2014. Influenza A virus attenuation by codon deoptimization of the NS gene for vaccine development. J Virol 88:10525–10540. doi: 10.1128/JVI.01565-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Niwa H, Yamamura K, Miyazaki J. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 71.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 72.Kochs G, Garcia-Sastre A, Martinez-Sobrido L. 2007. Multiple anti-interferon actions of the influenza A virus NS1 protein. J Virol 81:7011–7021. doi: 10.1128/JVI.02581-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.DeDiego ML, Nogales A, Martinez-Sobrido L, Topham DJ. mBio, in press. [DOI] [PMC free article] [PubMed]