Dengue virus (DENV) is a mosquito-borne pathogen, and infections by this virus are prevalent in over 100 tropical and subtropical countries or regions, with approximately 2.5 billion people at risk. DENV infection induces a spectrum of clinical symptoms, ranging from classical dengue fever (DF) to severe dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS). Therefore, it is important to understand the mechanisms underlying DENV pathogenesis. In this study, we reveal that the DENV membrane protein (M) interacts with the host NLRP3 protein to promote NLRP3 inflammasome activation, which leads to the activation and release of a proinflammatory cytokine, interleukin-1 beta (IL-1β). More importantly, we demonstrate that M protein can induce vascular permeability and vascular leakage and that NLRP3 is required for M-induced vascular leakage in mouse tissues. Collectively, this study reveals a distinct mechanism underlying DENV pathogeneses and provides new insights into the development of therapeutic agents for DENV-associated diseases.
KEYWORDS: DENV membrane protein, M, dengue virus, DENV, interleukin-1β, IL-1β, NLRP3 inflammasome, Nod-like receptor (NLR) family pyrin domain containing 3, NLRP3, vascular leakage, vascular permeability
ABSTRACT
Dengue virus (DENV) infection causes serious clinical symptoms, including dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS). Vascular permeability change is the main feature of the diseases, and the abnormal expression of proinflammatory cytokines is the important cause of vascular permeability change. However, the mechanism underlying vascular permeability induced by DENV has not been fully elucidated. Here, we reveal a distinct mechanism by which DENV infection promotes NLRP3 inflammasome activation and interleukin-1 beta (IL-1β) release to induce endothelial permeability and vascular leakage in mice. DENV M protein interacts with NLRP3 to facilitate NLRP3 inflammasome assembly and activation, which induce proinflammatory cytokine IL-1β activation and release. Notably, M can induce vascular leakage in mouse tissues by activating the NLRP3 inflammasome and IL-1β. More importantly, inflammatory cell infiltration and tissue injuries are induced by M in wild-type (WT) mouse tissues, but they are not affected by M in NLRP3 knockout (NLRP3−/−) mouse tissues. Evans blue intensities in WT mouse tissues are significantly higher than in NLRP3−/− mouse tissues, demonstrating an essential role of NLRP3 in M-induced vascular leakages in mice. Therefore, we propose that upon DENV infection, M interacts with NLRP3 to facilitate inflammasome activation and IL-1β secretion, which lead to the induction of endothelial permeability and vascular leakage in mouse tissues. The important role of the DENV-M-NLRP3-IL-1β axis in the induction of vascular leakage provides new insights into the mechanisms underlying DENV pathogenesis and DENV-associated DHF and DSS development.
IMPORTANCE Dengue virus (DENV) is a mosquito-borne pathogen, and infections by this virus are prevalent in over 100 tropical and subtropical countries or regions, with approximately 2.5 billion people at risk. DENV infection induces a spectrum of clinical symptoms, ranging from classical dengue fever (DF) to severe dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS). Therefore, it is important to understand the mechanisms underlying DENV pathogenesis. In this study, we reveal that the DENV membrane protein (M) interacts with the host NLRP3 protein to promote NLRP3 inflammasome activation, which leads to the activation and release of a proinflammatory cytokine, interleukin-1 beta (IL-1β). More importantly, we demonstrate that M protein can induce vascular permeability and vascular leakage and that NLRP3 is required for M-induced vascular leakage in mouse tissues. Collectively, this study reveals a distinct mechanism underlying DENV pathogeneses and provides new insights into the development of therapeutic agents for DENV-associated diseases.
INTRODUCTION
Dengue virus (DENV) is a single-strand and positive-sense RNA virus belonging to the Flavivirus genus of the Flaviviridae family. Dengue is a mosquito-borne infectious disease caused by four serotypes of DENV (DENV1 to -4) and is prevalent in over 100 tropical and subtropical countries, with approximately 2.5 billion people at risk (1–3). DENV infection induces a spectrum of clinical symptoms, ranging from classical dengue fever (DF) to severe dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS) (4, 5). The clinical manifestation of DF is fever, headache, rash, arthralgia, myalgia, and retro-orbital pain, which provides lifelong immunity to the infecting strain. Secondary infection with different DENV serotypes induces severe DHF and DSS, which is associated with life-threatening increases in hypotension, hypovolemia, vascular permeability, and shock (4, 6–8). Although a hypothesis is known as antibody-dependent enhancement (ADE) (9, 10), the pathogenesis of DHF/DSS remains largely unclear.
DENV is spherical with an envelope structure, has a single-stranded RNA genome of approximately 11 kb, and encodes a polyprotein, which is cleaved by cellular and viral enzymes into three structural proteins and seven nonstructural proteins (11). Viral structure proteins play critical roles in the DENV life cycle. The M protein production is sheared of Prm in the trans-Golgi apparatus through host Furin protease (12, 13). M protein contains 75 amino acids; the N-terminal 40 amino acids form an ectodomain containing a hydrophobic loop followed by an amphipathic helical portion, and the C-terminal 35 amino acids form a transmembrane region (14). The amphipathic helical portion interacts with envelope protein (E) to promote DENV invasion, packing mature, and release (15–18). These studies suggest that M plays an important role in DENV infection. However, the effect of M protein on host immunity is not clear.
Despite the fact that DENV pathogenesis is not completely understood, massive cytokine secretion (cytokine storm) is believed to be one of the primary contributory factors (19, 20). Clinical studies demonstrated that cytokines play a vital role in DHF pathogenesis (21, 22). Interleukin-1 beta (IL-1β), an important proinflammatory cytokine, is induced upon DENV infection (23, 24). IL-1β is not only an endogenous pyrogen inducing fever but also regulates systemic inflammation responses (25, 26). IL-1β activation requires two pathways: signal I regulates nuclear factor κB (NF-κB) signaling to induce pro-IL-1β mRNA transcription and pro-IL-1β protein synthesis (27), whereas signal II activates the inflammasome to promote IL-1β maturation and secretion (28). The best characterized inflammasome is the Nod-like receptor (NLR) family pyrin domain containing-3 (NLRP3) inflammasome (29, 30). As a crucial part of the innate immune system, the NLRP3 inflammasome plays a critical role in recognizing viral infections (31–36). Once activated, NLRP3 can recruit apoptosis-associated speck-like protein with CARD domain (ASC) and pro-caspase-1 to form the inflammasome complex. Active caspase-1 is formed by autocatalytic cleavage, which catalyzes proteolytic processing of pro-IL-1β into active IL-1β. Although the myeloid Syk-coupled C-type lectin 5A (CLEC5A) is critical for inflammasome activation upon DENV infection (23) and receptor-interacting protein (RIP) kinases and reactive oxygen species (ROS) generation in platelets are required for inflammasome activation (37), the mechanism by which DENV infection regulates the inflammasome remains largely elusive. This study reveals a distinct mechanism by which DENV infection facilitates NLRP3 inflammasome activation and IL-1β release through the viral M protein, which lead to the induction of endothelial permeability and vascular leakage in mouse tissues.
RESULTS
DENV protein synthesis is required for NLRP3 inflammasome activation.
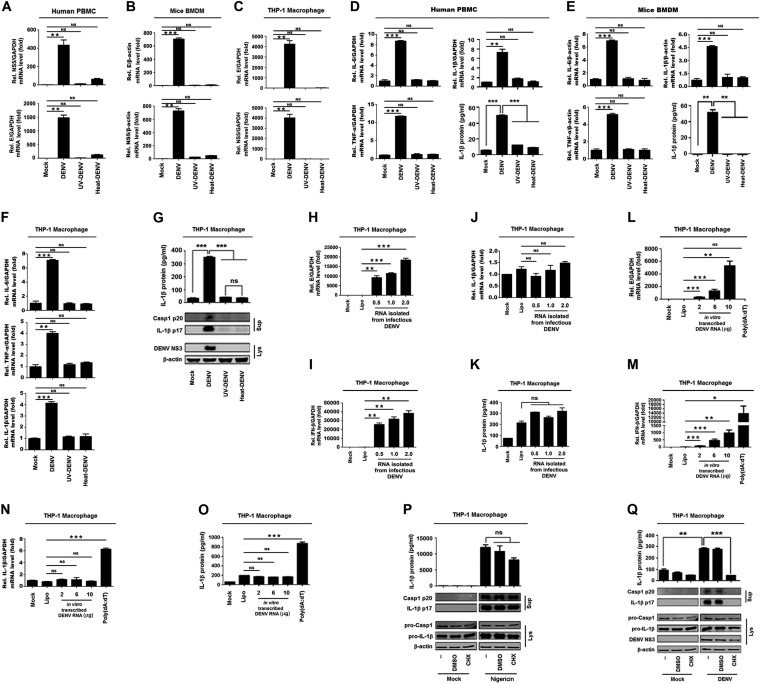
To reveal the mechanism by which DENV infection regulates the NLRP3 inflammasome, the effects of DENV2 replication, genomic RNA, and viral proteins on the inflammasome activation were determined. Human peripheral blood mononuclear cells (PBMCs), granulocyte-macrophage colony-stimulating factor (GM-CSF)-differentiated mouse bone marrow-derived macrophages (BMDMs), or phorbol-12-myristate-13-acetate (PMA)-differentiated THP-1 macrophages were treated with infectious DENV2(NGC), UV-irradiated DENV2(NGC), or heat-inactivated DENV2(NGC). Reverse transcriptase PCR (RT-PCR) analyses showed that DENV2(NGC) E and NS5 mRNAs were expressed in human PBMCs (Fig. 1A), mouse primary BMDMs (Fig. 1B), and THP-1 differentiated macrophages (Fig. 1C) infected with DENV2(NGC), whereas they were not detected in the cells treated with UV-irradiated or heat-inactivated DENV2(NGC) (Fig. 1A to C). Interestingly, quantitative RT-PCR (qRT-PCR) analyses and enzyme-linked immunosorbent assays (ELISAs) revealed that IL-1β mRNA, IL-6 mRNA, and tumor necrosis factor alpha (TNF-α) mRNA expression and IL-1β protein secretion were induced by DENV2(NGC) but not by UV-irradiated or heat-inactivated DENV2(NGC) in PBMCs, BMDMs, or THP-1 macrophages (Fig. 1D to G). These results suggest that infectious DENV can induce IL-1β, IL-6, and TNF-α expression and IL-1β secretion, whereas inactivated DENV fails to induce IL-1β, IL-6, and TNF-α expression and IL-1β secretion.
FIG 1.
The replication and protein synthesis of Dengue virus are essential for the activation of NLRP3 inflammasome. Human PBMCs (A, D), GM-CSF differentiated mouse BMDMs (B, E), and PMA-differentiated THP-1 macrophages (C, F, and G) were treated with infectious, UV-, or heat-inactivated DENV2(NGC) at an MOI of 5 for 24 h. The DENV E gene mRNA and NS5 gene mRNA were quantified by RT-PCR (A to C). IL-1β RNA, IL-6 mRNA, and TNF-α expression was determined by qRT-PCR, and IL-1β was measured by ELISA (D to F). IL-1β in cell supernatants was measured by ELISA and proteins in cell extracts (Lys) were analyzed by Western blotting (WB) (G). THP-1 macrophages were transfected with RNA extracted from infectious DENV2(NGC) at 0.5, 1, and 2 μg (H to K) or with in vitro-transcribed DENV2(TSV01) RNA at 2, 6, and 10 μg or treated with 10 μg/ml poly(dA·dT) (L to O). The DENV E gene mRNA (H, L), IFN-β RNA (I, M), or IL-1β RNA (J, N) was determined by RT-PCR, and IL-1β was measured by ELISA (K, O). (P, Q) PMA-differentiated THP-1 macrophages were pretreated with 100 μM CHX for 2 h and stimulated with nigericin (2 μM) for 2 h (P) or infected with DENV at an MOI of 5 for 48 h (Q). Supernatants were analyzed (top) by ELISA for IL-1β. Cell lysates were analyzed (bottom) by immunoblotting. Data are representative of three independent experiments. Mock, supernatant of C6/36 cells without DENV2 infection (A to G, P, and Q) or unprocessed cells (H to O); Lipo, transfection with Lipofectamine 2000 reagent. Values are means ± SEMs. ns, not significant; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
To determine the effect of DENV RNA on the expression of the cytokines, THP-1 differentiated macrophages were transfected with RNA extracted from infectious DENV2(NGC) or with in vitro transcribed RNA of DENV2(TSV01). Notably, in the THP-1 differentiated macrophages transfected with RNA isolated from infectious DENV2(NGC), E mRNA and beta interferon (IFN-β) mRNA were expressed (Fig. 1H and I), whereas IL-1β mRNA and protein secretion were relatively unaffected (Fig. 1J and K). Moreover, in THP-1 differentiated macrophages transfected with in vitro transcribed RNA of DENV2(TSV01), E mRNA and IFN-β mRNA were expressed (Fig. 1L and M); however, IL-1β mRNA expression and protein secretion were not influenced (Fig. 1N and O). In addition, THP-1 macrophages were treated with poly(dA·dT). We also noticed that E mRNA was not induced by poly(dA·dT) (Fig. 1L), whereas IFN-β mRNA expression and IL-1β mRNA and protein secretion were significantly induced by poly(dA·dT) (Fig. 1M to O). Collectively, these results suggest that DENV2 RNA is unable to activate IL-1β.
To determine the effects of viral proteins on inflammasome activation, THP-1 differentiated macrophages were pretreated with cycloheximide (CHX; an inhibitor of protein biosynthesis) (32) and stimulated with nigericin (an inducer of NLRP3 inflammasome) or infected with DENV2(NGC). The results indicated that IL-1β secretion, IL-1β p17 maturation, and caspase-1 p20 cleavage induced by nigericin were relatively unaffected by CHX (Fig. 1P), whereas IL-1β secretion, IL-1β p17 maturation, and caspase-1 p20 cleavage induced by DENV2(NGC) infection were inhibited by CHX (Fig. 1Q), revealing that protein synthesis is essential for DENV to induce the inflammasome. Taken together, we demonstrate that DENV replication and protein synthesis are required for the activation of the NLRP3 inflammasome.
DENV M protein promotes the activation of NLRP3 inflammasome and IL-1β.
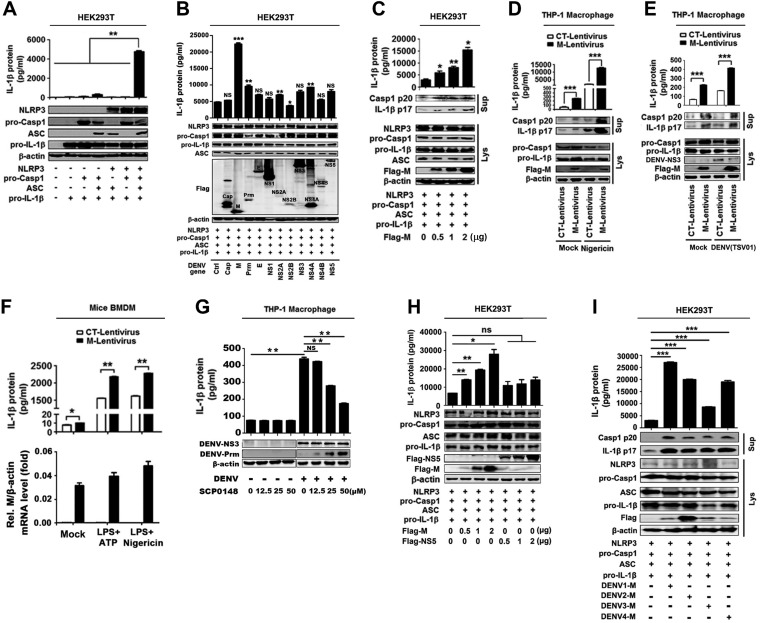
As we revealed above, DENV protein synthesis is necessary for the activation of the NLRP3 inflammasome. Next, we explored which DENV protein plays the crucial role in NLRP3 inflammasome activation by using a caspase-1 activation and IL-1β cleavage cell system as established previously (34). IL-1β secretion was detected in the presence of all components of the NLRP3 inflammasome, namely, NLRP3, ASC, pro-caspase-1, and pro-IL-1β (Fig. 2A), indicating that this system was functioning well. The cells of the system were then transfected with plasmids carrying each of the DENV2 genes, as indicated in the figure. Notably, IL-1β secretion, IL-1β p17 maturation, and caspase-1 p20 cleavage were mainly induced by M protein (Fig. 2B). We also noticed that NS2A, NS2B, and NS4B were expressed at lower levels than other proteins. We speculate that such reduction might be due to the natures of these proteins. To confirm the role of M protein in the activation of the NLRP3 inflammasome, cells of the caspase-1 activation and IL-1β cleavage system were transfected with plasmid encoding DENV2 M protein at different concentrations. The results showed that IL-1β secretion, IL-1β p17 cleavage, and caspase-1 p20 maturation were significantly induced by M protein in dose-dependent manners (Fig. 2C). In addition, to further determine the effect of M protein on the activation of the NLRP3 inflammasome, THP-1 cells were infected with M-lentivirus to generate cells stably expressing M, which were then differentiated into macrophages. IL-1β secretion, IL-1β p17 maturation, and caspase-1 p20 cleavage induced by nigericin or DENV2(TSV01) were significantly promoted by M-lentivirus (Fig. 2D and E). Moreover, IL-1β secretion induced by ATP or nigericin was further facilitated by M-lentivirus (Fig. 2F, top), and DENV2 M RNA was detected in M-lentivirus-treated mouse primary BMDMs but not in control (CT)-lentivirus-treated cells (Fig. 2F, bottom). Thus, the results suggest that DENV-M promotes NLRP3 inflammasome activation. Furthermore, we verified the role of M in such regulation by using SCP0148 (the inhibitor of Furin protease) in DENV-infected cells. Previous studies had reported that DENV Prm protein can be cleaved into M protein by the host’s Furin protease (12, 13). PMA-differentiated THP-1 macrophages were infected with DENV2(NGC) and then treated with SCP0148 at different concentrations. ELISAs revealed that the secretion of IL-1β protein in the cell supernatants was detected upon DENV infection but significantly attenuated by SCP0148 in a dose-dependent manner (Fig. 2G, top). Western blot analyses showed that the production of intracellular Prm protein was promoted by SCP0148 in a dose-dependent fashion (Fig. 2G, bottom). These results indicate that the cleavage of Prm protein into M protein is required for the promotion of IL-1β secretion and thus suggest that M is involved in NLRP3 inflammasome activation.
FIG 2.
M protein promotes the activation of the NLRP3 inflammasome. (A) HEK293T cells were transfected with the expression plasmid pro-IL-1β, pro-IL-1β, and pro-caspase-1, any three of the four plasmids, and four plasmids (pro-IL-1β, pro-caspase-1, ASC, and NLRP3) for 48 h. (Top) Supernatants were analyzed by ELISA for IL-1β. (Bottom) Cell lysates were analyzed by immunoblotting. (B) HEK293T cells were cotransfected with plasmids encoding NLRP3, ASC, pro-caspase-1 (pro-Casp1), or pro-IL-1β and with plasmids encoding Cap, M, Prm, E, NS1, NS2A, NS2B, NS3, NS4A, NS4B, or NS5 for 48 h. (Top) Supernatants were analyzed by ELISA for IL-1β. (Bottom) Cell lysates were analyzed by immunoblotting. (C) HEK293T cells were cotransfected with plasmids encoding NLRP3, ASC, pro-Casp1, and pro-IL-1β and with different concentrations of plasmid encoding DENV2 M for 48 h. (Top) Supernatants were analyzed by ELISA for IL-1β. (Bottom) Cell lysates were analyzed by immunoblotting. (D) THP-1 cells were stably infected with lentivirus-CT or lentivirus-M, differentiated into macrophages, and stimulated with 2 μM nigericin. IL-1β in cell supernatants was measured by ELISA and proteins in Lys were analyzed by WB. (E) THP-1 cells were stably infected with lentivirus-CT or lentivirus-M, differentiated into macrophages, and stimulated with DENV2(TSV01) at an MOI of 5 for 48 h. IL-1β in cell supernatants was measured by ELISA and proteins in Lys were analyzed by WB. (F) Mouse BMDMs were infected with lentivirus-CT or lentivirus-M and stimulated with LPS (1 μg/ml) plus ATP (2.5 mM) or LPS (1 μg/ml) plus nigericin (2 μM). IL-1β in supernatants was measured by ELISA and M gene mRNA was quantified by RT-PCR. (G) PMA-differentiated THP-1 macrophages were infected with DENV2(NGC) at an MOI of 5 for 12 h, and then Furin protease inhibitor (SCP0148) was added at different concentrations (12.5, 25, 50 μM) and the infection allowed to continue for 12 h. IL-1β in cell supernatants was measured by ELISA and proteins in Lys were analyzed by WB. (H) HEK293T cells were cotransfected with plasmids encoding NLRP3, ASC, pro-Casp1, and pro-IL-1β and with different concentrations of plasmid encoding DENV2 M or DENV2 NS5 (0, 0.5, 1, and 2 μg) for 48 h. (Top) Supernatants were analyzed by ELISA for IL-1β. (Bottom) Cell lysates were analyzed by immunoblotting. (I) HEK293T cells were cotransfected with plasmids encoding NLRP3, ASC, pro-Casp1, and pro-IL-1β, and transfected with plasmid encoding DENV1-M, DENV2-M, DENV3-M, and DENV4-M (2 μg) for 48 h. (Top) Supernatants were analyzed by ELISA for IL-1β. (Bottom) Cell lysates were analyzed by immunoblotting. Ctrl, empty plasmid (B); Mock, unprocessed cells (D, E, and F). Data are representative of three independent experiments. Values are means ± SEMs. ns, not significant; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
As we previously reported that Zika virus (ZIKV) NS5 pays an important role in NLRP3 inflammasome activation (38), here we determined the effect of DENV2(NGC) NS5 on NLRP3 inflammasome activation. We noticed that unlike DENV M, DENV NS5 failed to activate the inflammasome (Fig. 2H). Additionally, we explored whether different serotype DENV M proteins have broad roles in NLRP3 inflammasome activation. Notably, like DENV2-M, DENV1-M, DENV3-M, and DENV4-M also induced IL-1β secretion, IL-1β p17 maturation, and caspase-1 p20 cleavage (Fig. 2I). Taken together, our results reveal that M plays a crucial role in NLRP3 inflammasome activation.
M interacts with NLRP3 in the endoplasmic reticulum and Golgi compartment.
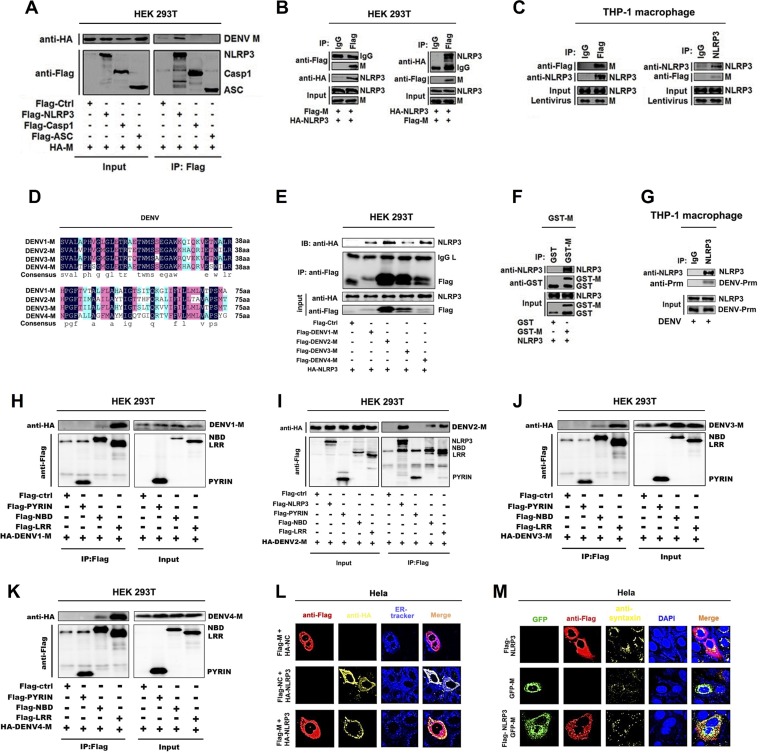
The mechanism by which M promotes inflammasome activation was evaluated. Coimmunoprecipitation assays were used for exploring the interaction of M protein with the components (NLRP3, caspase-1, and ASC) of the NLRP3 inflammation in HEK293T cells cotransfected with hemagglutinin (HA)-M and Flag-NLRP3, Flag-pro-caspase-1, or Flag-ASC. Coimmunoprecipitation (Co-IP) results indicated that, interestingly, M interacts with NLRP3 but failed to interact with ASC or caspase-1 in HEK293T cells (Fig. 3A), M and NLRP3 interacted with each other in HEK293T cells (Fig. 3B), and M and endogenous NLRP3 interacted with each other in THP-1 macrophages (Fig. 3C). Notably, sequence analyses revealed that although homologies among the 4 M proteins of DENV serotypes were not very high (Fig. 3D), all M proteins interacted with NLRP3 (Fig. 3E), which was consistent with our above-described result showing that four serotypes of DENV M activated the NLRP3 inflammasome (Fig. 2I). A glutathione transferase (GST) pulldown assay was used to prove a direct interaction of M and NLRP3. Importantly, our results showed that GST-M protein bound to NLRP3 protein (Fig. 3F). Importantly, we also proved that endogenous NLRP3 interacted with DENV2 Prm in DENV2(NGC)-infected THP-1 macrophages (Fig. 3G). It is well known that NLRP3 contains three prototypic domains: N-terminal pyrin domain (PYRIN), NACHT-associated domain (NBD), and C-terminal leucine-rich repeat (LRR) domain (39). Notably, Co-IP results indicated that the NBD domain and LRR domain interacted with all four M proteins of DENV serotypes (Fig. 3H to K). Finally, immunofluorescence assays revealed that M and NLRP3 were specifically distributed in the endoplasmic reticulum (ER) (Fig. 3L) and Golgi compartment (Fig. 3M). Together, our results demonstrate that M protein directly interacts with NLRP3 through its NBD and LRR domains.
FIG 3.
Dengue virus M only interacts with NLRP3 protein. (A) HEK293T cells were cotransfected with HA-M and Flag-NLRP3, Flag-pro-caspase-1, or Flag-ASC. Cell lysates were immunoprecipitated using anti-Flag antibody and analyzed using anti-Flag and anti-HA antibodies. Cell lysates (40 μg) were used as input. (B) HEK293T cells were cotransfected with HA-NLRP3 and Flag-M. Cell lysates were immunoprecipitated using anti-Flag antibody or anti-HA antibody and analyzed using anti-Flag and anti-HA antibodies. Cell lysates (40 μg) were used as input. (C) THP-1 macrophages were stably infected with lentivirus-M. Cell lysates were immunoprecipitated using anti-Flag antibody or anti-NLRP3 antibody and analyzed using anti-Flag and anti-NLRP3 antibodies. Cell lysates (50 μg) were used as input. (D) Sequence alignment of dengue virus M protein with different serotypes. (E) HEK293T cells were cotransfected with HA-NLRP3 and Flag-DENV1-M, Flag-DENV2-M, Flag-DENV3-M, or Flag-DENV4-M. Cell lysates were immunoprecipitated using anti-Flag antibody and analyzed using anti-Flag and anti-HA antibodies. Cell lysates (40 μg) were used as input. (F) Purified GST (10 μg) or GST-M (10 μg) was incubated with cell lysates of Flag-NLRP3-transfected HEK293T cells. (G) THP-1 macrophages were infected with DENV2(NGC) for 48 h. Cell lysates were immunoprecipitated using anti-NLRP3 antibody and analyzed using anti-DENV2-Prm antibody. Cell lysates (40 μg) were used as input. (H to K) HEK293T cells were cotransfected with HA-DENV1-M, HA-DENV2-M, HA-DENV3-M, HA-DENV4-M, and Flag-NLRP3, Flag-PYRIN, Flag-NBD, or Flag-LRR. Cell lysates were immunoprecipitated using anti-Flag antibody and analyzed using anti-Flag and anti-HA antibodies. Cell lysates (40 μg) were used as input. (L) HeLa cells were transfected with Flag-M, HA-NLRP3, or Flag-M/HA-NLRP3 for 24 h. Endoplasmic reticulum marker ER-Tracker (blue), Flag-M (red), and HA-NLRP3 (yellow) were then visualized with confocal microscopy. (M) HeLa cells were transfected with Flag-NLRP3, GFP-M, or GFP-M/Flag-NLRP3 for 24 h. Golgi apparatus marker antibodies against syntaxin 6 (yellow), Flag-tagged NLRP3 (red), and GFP-tagged M (green) and nucleus marker DAPI (blue) were then visualized with confocal microscopy. Flag-Ctrl, Flag-NC, or HA-NC indicates pcDNA3.1(+)-3×Flag or pCAggs-HA empty plasmid. Data are representative of three independent experiments.
M promotes the NLRP3 inflammasome assembly and activation.
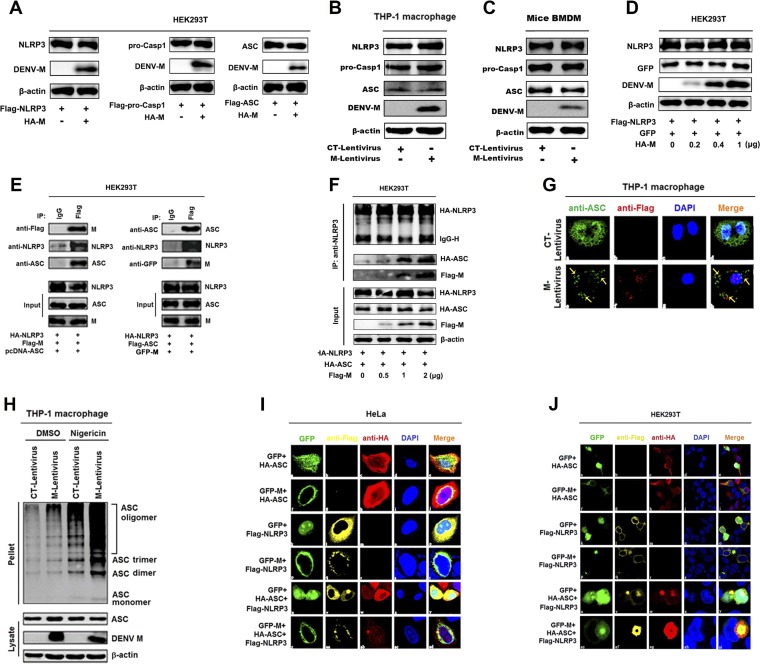
As M interacts with NLRP3, we wanted to know the role of M in the regulation of the inflammasome. Initially, we examined the effect of M protein on the expression of NLRP3, caspase-1, and ASC. The results showed that the productions of NLRP3, caspase-1, and ASC were not affected by M in HEK293T cells, THP-1 cells, and mouse BMDM cells (Fig. 4A to C); similarly, the abundance of NLRP3 was not affected by M (Fig. 4D), indicating that M is not required for production of inflammasome components. Next, we assessed the ability of M to interact with ASC in HEK293T cells cotransfected with Flag-M and HA-NLRP3 or HA-ASC. Co-IP results showed that M protein did not interact with ASC in the absence of NLRP3; however, M and ASC interacted with each other in the presence of NLRP3 (Fig. 4E), and the NLRP3-ASC interaction was enhanced by M in a dose-independent manner (Fig. 4F), suggesting that M promotes NLRP3-ASC interaction. In the process of inflammasome assembly, ASC proteins aggregate to form oligomers (40). Thus, we evaluated the role of M protein in the regulation of ASC protein aggregation in THP-1 macrophages infected with CT-lentivirus or M-lentivirus and stably expressing M protein. We revealed that in the absence of M, endogenous ASC was diffusely distributed in THP-1 macrophages, whereas in the presence of M, ASC formed small spots in the cells (Fig. 4G). Interestingly, ASC oligomerization was activated by M-lentivirus, and ASC oligomerization stimulated by nigericin was further facilitated by M-lentivirus (Fig. 4H). These results suggest that M promotes inflammasome complex assembly.
FIG 4.
Dengue virus M facilitates the assembly of NLRP3 inflammasome by interacting with NLRP3. (A) HEK293T cells were cotransfected with HA-CT or HA-M plus Flag-NLRP3, Flag-pro-Casp1, or Flag-ASC for 36 h. The indicated proteins in cell extracts were analyzed by WB. (B) THP-1 cells were stably infected with lentivirus-CT or lentivirus-M and differentiated into macrophages, and the indicated proteins in cell extracts were analyzed by WB. (C) Mouse BMDMs were infected with lentivirus-CT or lentivirus-M for 48 h, and the indicated proteins in cell extracts were analyzed by WB. (D) HEK293T cells were cotransfected with different concentrations of HA-M plus Flag-NLRP3 and GFP for 36 h. The indicated proteins in cell extracts were analyzed by WB. (E) HEK293T cells were cotransfected with Flag-M, HA-NLRP3, and pcDNA3.1(+)-ASC for 24 h. Cell lysates were immunoprecipitated using anti-Flag antibody and analyzed using anti-NLRP3, anti-ASC, and anti-Flag antibodies. HEK293T cells were cotransfected with GFP-M, HA-NLRP3, and Flag-ASC. Cell lysates were immunoprecipitated using anti-Flag antibody and analyzed using anti-NLRP3, anti-ASC, and anti-GFP antibodies. Cell lysates (40 μg) were used as inputs. (F) HEK293T cells were cotransfected with different concentrations of Flag-M plus HA-NLRP3 and HA-ASC for 24 h. Cell lysates were immunoprecipitated using anti-NLRP3 antibody and analyzed using anti-HA and anti-Flag antibodies. (G) THP-1 macrophages were stably infected with lentivirus-CT or lentivirus-M. Subcellular locations of ASC, Flag-M, and DAPI were visualized under confocal microscopy. (H) THP-1 macrophages were stably infected with lentivirus-CT or lentivirus-M and stimulated by 2 μM nigericin. ASC oligomerization was analyzed by immunoblotting. HeLa cells (I) and HEK293T cells (J) were cotransfected with GFP/HA-ASC, GFP-M/HA-ASC, GFP/Flag-NLRP3, GFP-M/Flag-NLRP3, GFP/HA-ASC/Flag-NLRP3, or GFP-M/HA-ASC/Flag-NLRP3 for 24 h. The subcellular locations of HA-tagged ASC (red), Flag-tagged NLRP3 (yellow), GFP-tagged M (green), and nucleus marker DAPI (blue) were visualized with confocal microscopy. The data are representative of three independent experiments.
Moreover, we examined the effect of M protein on the localization of NLRP3 as speck, which is an indicator of inflammasome complex formation (41). HeLa cells and HEK293T cells were cotransfected with green fluorescent protein (GFP)/HA-ASC, GFP-M/HA-ASC, GFP/Flag-NLRP3, GFP-M/Flag-NLRP3, GFP/HA-ASC/Flag-NLRP3, or GFP-M/HA-ASC/Flag-NLRP3. In HeLa and HEK293T cells, ASC was distributed in the cytosol and nucleus (Fig. 4I and J, a to e), M and ASC were not colocalized (Fig. 4I and J, f to j), and NLRP3 was distributed in the cytosol (Fig. 4I and J, k to o). However, in the presence of M and NLRP3 or ASC and NLRP3, M or ASC colocalized with NLRP3 to form spots (Fig. 4I and J, p to y). Moreover, in the presence of M, ASC, and NLRP3, the three proteins colocalized and formed ring-like structures in HeLa cells and HEK293T cells (Fig. 4I and J, z to ad). Collectively, these results reveal that M facilitates NLRP3 inflammasome assembly by interacting with NLRP3.
M induces vascular leakage in mice by activating IL-1β.
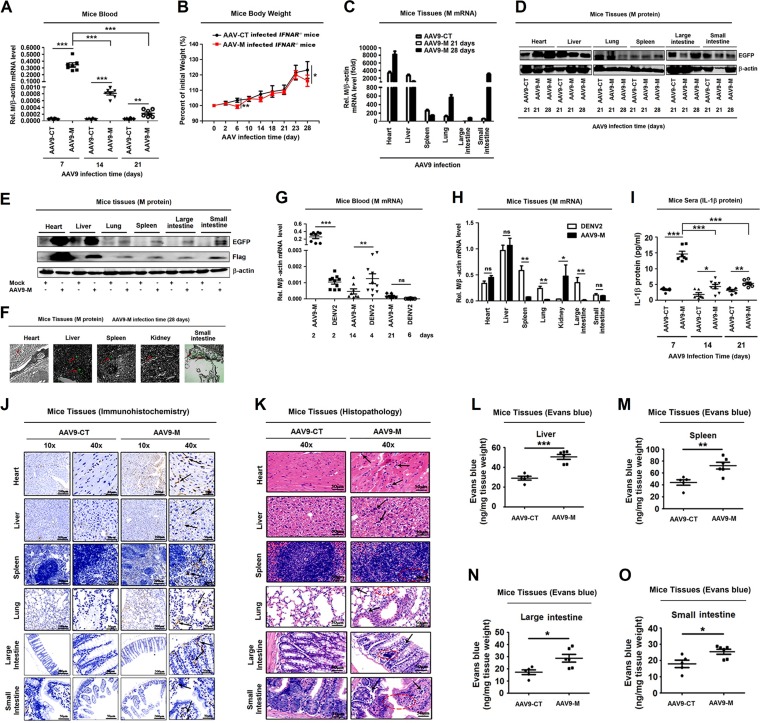
As a previous study found that DENV2 infection activates IL-1β to induce vascular permeability in platelets (37) and here we reveal that M plays an important role in NLRP3 inflammasome activation, we further determined the biologic effect of M on vascular permeability in mice. We selected adeno-associated virus (AAV9) as the vector system to express M protein in mice, as AAV9 has low immunogenicity and high expression efficiency in the host (42). In order to be consistent with the mouse model of DENV infection, IFNAR−/− C57BL/6 mice (43) were used and intravenously injected with AAV9 (42) expressing M protein (AAV9-M) or its control (AAV9-CT). M mRNA was expressed in the blood of mice infected with AAV9-M, whereas it was not detected in the control mice (Fig. 5A). We also noticed that M mRNA was highly expressed at 7 days postinfection and decreased from 14 to 21 days postinfection (Fig. 5A). We suspected that this phenomenon was due to the intravenous injection of the virus, which causes a high initial titer of the virus in the blood. With the circulation of the blood, the virus invades different tissues and is removed by lymphocytes in the blood, dropping the virus titer to lower levels. The body weights of AAV9-M mice (n = 8) were lower than those in the AAV9-CT group (n = 8) at 6 days and 28 days postinfection (Fig. 5B). M mRNA was expressed at higher levels in heart and liver, whereas it was detected at lower levels in spleen, lung, and large intestine of AAV9-M mice at 21 days and 28 days posttreatment (Fig. 5C). In small intestine, a low level of M mRNA was detected at 21 days posttreatment, but M mRNA was expressed at a higher level 28 days posttreatment (Fig. 5C). Enhanced GFP (EGFP; as an indicator of AAV9-M replication) was expressed in heart, liver, lung, spleen, large intestine, and small intestine at 28 days postinfection, indicating that M protein was successfully expressed in the mouse tissues (Fig. 5D to F). EGFP (as an indicator of M protein) and Flag-M fusion protein were expressed in heart, liver, spleen, kidney, lung, large intestine, and small intestine at 28 days postinfection, indicating that M protein was successfully expressed in the mouse tissues (Fig. 5D to F).
FIG 5.
DENV M protein induces IL-1β activation and vascular leakage in mice. IFNAR−/− C57BL/6 genetic background mice received tail vein injections of 300 μl containing 5 × 1011 vg of AAV9-EGFP or AAV9-EGFP-M. Serum was collected every 7 days for AAV9 groups from the orbits. (A) Total RNA was extracted from the plasma from mice. The RNA level of M protein was quantified by qRT-PCR. (B) Mice (n = 8 per group) were weighed every 2 days, and weight was expressed as the percentage of the initial weight. (C) The RNA levels of M protein in heart, liver, spleen, lung, large intestine, and small intestine were quantified by qRT-PCR. (D) Western blot analysis of EGFP expression in heart, liver, spleen, lung, large intestine, and small intestine after AAV9-EGFP and AAV9-EGFP-M infection. (E) Western blot analysis of EGFP and Flag-M expression in heart, liver, spleen, lung, large intestine, and small intestine in mock- and AAV9-EGFP-M-infected mice. (F) Immunofluorescence analysis of EGFP expression in heart, liver, spleen, kidney, and small intestine after AAV9-EGFP-M infection. (G) Total RNAs was extracted from blood samples from DENV2(NGC)-infected IFNAR−/− mice (n = 10) at 2, 4, and 6 days postinfection or AAV9-EGFP-M-infected IFNAR−/− mice (n = 8) at 2, 14, and 21 days postinfection. The DENV M gene mRNA was quantified by RT-PCR. (H) Total RNA was extracted from tissues, including heart, liver, spleen, lung, kidney, large intestine, and small intestine, from DENV2(NGC)-infected mice after 7 days or AAV9-EGFP-M-infected mice after 28 days. The DENV M gene mRNA was quantified by RT-PCR. (I) IL-1β in sera was measured by ELISA. Points represent the IL-1β values from each serum sample. At 28 days postinfection, mice were euthanized, and tissues were collected. (J) Immunohistochemistry analysis of IL-1β in the tissues, including heart, liver, spleen, lung, large intestine, and small intestine after AAV9-EGFP or AAV9-EGFP-M infection. Black arrows indicated the immunostaining of IL-1β. (K) Histopathology analysis of tissues, including heart, liver, spleen, lung, large intestine, and small intestine, after AAV9-EGFP or AAV9-EGFP-M infection. Black arrows indicated the infiltrated inflammatory cells; red circles indicate the aberrant cells. Evans blue dye was intravenously injected into mice 28 days after infection with AAV9-EGFP-M (n = 6) and AAV9-EGFP control (n = 5). The dye circulated for 2 h before mice were euthanatized, tissues including liver (L), spleen (M), large intestine (N), and small intestine (O) were collected, and the value of Evans blue was measured at OD610. Data are representative of two independent experiments. Values are means ± SEMs. ns, not significant; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Moreover, the levels of M mRNAs were determined and compared in AAV9-M-treated mice and DENV2(NGC)-infected mice. IFNAR−/− C57BL/6 mice were intravenously injected with AAV9-M for 2, 14, and 21 days or with DENV2(NGC) for 2, 4, and 6 days. At 2 days postinfection, M mRNA was significantly higher in AAV9-M-infected mice than in DENV-infected mice. At 14 days postinfection, M mRNA was significantly lower in AAV9-M-infected mice than in DENV-infected mice; however, at 21 days postinfection, M mRNA was very low in the two groups and had no significant differences (Fig. 5G). In addition, M mRNA expression in the tissues was determined and compared between DENV2-infected mice at 6 days postinfection and AAV9-M-infected mice at 28 days postinfection. In heart, liver, and small intestine, there were no differences in M mRNA levels between the two mouse groups. In spleen, lung, and large intestine, M mRNA was significantly higher in DENV2-infected mice than in AAV9-M-infected mice, while in kidney, M mRNA was significantly higher in AAV9-M-infected mice than in DENV2-infected mice (Fig. 5H). These results reveal that like in DENV2-infected mice, M was successfully expressed in AAV9-M-infected mice.
Notably, IL-1β was highly produced in sera of AAV9-M-infected mice at 7 days postinfection and decreased gradually from 14 to 21 days postinfection (Fig. 5I). IL-1β was detected at higher levels in heart, liver, spleen, lung, large intestine, and small intestine of AAV9-M infected mice at 28 days postinfection (Fig. 5J). More importantly, hematoxylin and eosin staining showed inflammatory cell infiltration (indicated by black arrows in Fig. 5K) and tissue injuries (indicated by the red circles in Fig. 5K) in heart, liver, spleen, lung, large intestine, and small intestine of the AAV9-M-infected group compared with tissue staining from AAV9-CT-infected mice, and Evans blue intensities in AAV9-M-treated mouse tissues were significantly higher than in the AAV9-CT-treated group (Fig. 5L to O), demonstrating that M induces vascular leakages in mouse tissues. Taken together, our data suggest that M facilitates vascular leakage in mice by activating IL-1β.
NLRP3 plays a crucial role in M-induced IL-1β activation and vascular leakage in mice.
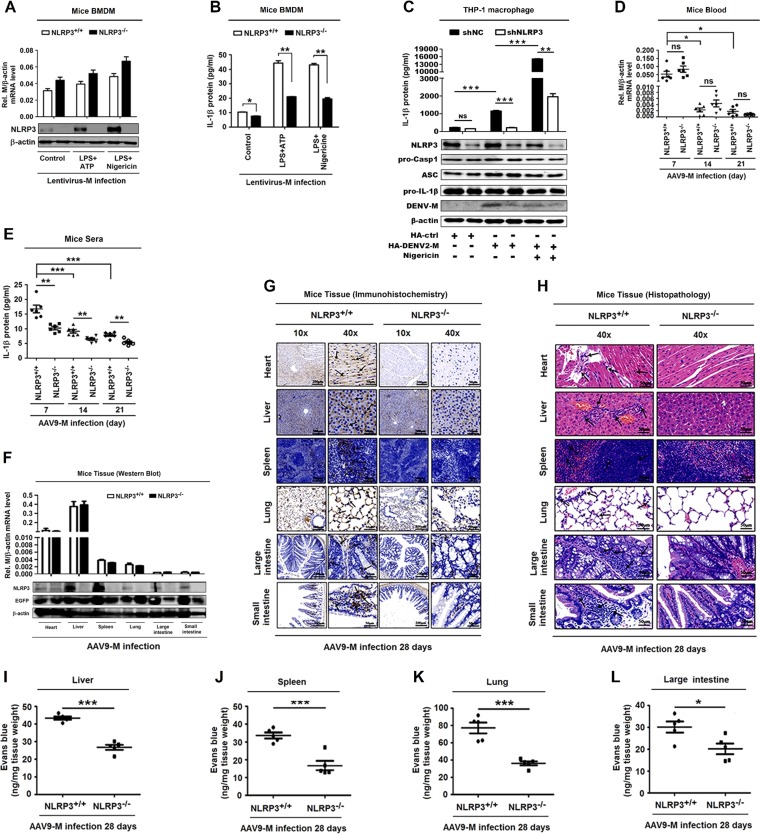
The role of NLRP3 in M-induced vascular leakage was determined in mice. Initially, the BMDMs isolated from NLRP3+/+ and NLRP3−/− mice were infected with lentivirus-M and treated with lipopolysaccharide (LPS) plus ATP or LPS plus nigericin. M mRNA was expressed in lentivirus-M-infected BMDMs of both wild-type (WT) and NLRP3−/− mice (Fig. 6A, top). NLRP3 was induced by ATP or nigericin in lentivirus-M-treated BMDMs of WT mice, whereas it was not detected in lentivirus-M-treated BMDMs of NLRP3−/− mice (Fig. 6A, bottom). These results confirm that M protein was expressed in both WT and NLRP3−/− mice and NLRP3 was knocked out in NLRP3−/− mice. IL-1β protein was significantly induced by ATP or nigericin in lentivirus-M-infected BMDMs of WT mice but slightly facilitated in lentivirus-M-infected BMDMs of NLRP3−/− mice (Fig. 6B). We also knocked down NLRP3 in THP-1 cells. Compared with that in control (shNC) cells, the level of IL-1β secretion upon M protein expression in knockdown (shNLRP3) cells was largely reduced both in mock-treated or nigericin-treated cells (Fig. 6C). These results suggest that NLRP3 is involved in M-mediated IL-1β activation.
FIG 6.
NLRP3 is crucial for M-induced IL-1β activation and vascular leakage in mice. (A and B) GM-CSF differentiated NLRP3+/+ mouse BMDM cells and NLRP3−/− mouse BMDM cells were infected with M-encoding lentivirus for 48 h and then stimulated with LPS (1 μg/ml) for 6 h and ATP (2.5 mM) for 20 min or nigericin (2 μM) for 2 h. (A) The relative M RNA level was measured by RT-PCR, and NLRP3 and β-actin proteins were determined by Western blotting. (B) IL-1β secreted in cell supernatants was analyzed by ELISA. PMA-differentiated THP-1 macrophages stably expressing short hairpin RNAs (sh-RNAs), sh-NLRP3, and transfected with HA-ctrl or HA-DENV2-M plasmid for 48 h were stimulated with nigericin (2 μM) for 2 h. (C) IL-1β in cell supernatants was measured by ELISA (top) and proteins in cell extract (Lys) were analyzed by WB (bottom). (D and E) NLRP3+/+ C57BL/6 mice (n = 6) or NLRP3−/− C57BL/6 mice (n = 6) received tail vein injections of 300 μl of M-encoding AAV9 (5 × 1011 vg) (AAV9-EGFP-M). Serum was collected every 7 days for AAV9 groups from the orbits. Total RNA was extracted from plasma samples from mice. (D) The RNA level of M protein was quantified by qRT-PCR. (E) IL-1β in the sera was measured by ELISA. Points represent the IL-1β values from each serum sample. At 28 days postinfection, mice were euthanized, and tissues were collected. (F) The levels of M RNA in heart, liver, spleen, lung, large intestine, and small intestine were quantified by qRT-PCR analyses (top), and the levels of NLRP3 protein and EGFP were determined by Western blot analyses (bottom). (G) Immunohistochemistry analysis of IL-1β in the tissues, including heart, liver, spleen, lung, large intestine, and small intestine, after AAV9-EGFP-M infection. Black arrows indicated the immunostaining of IL-1β. (H) Histopathology analysis of tissues, including heart, liver, spleen, lung, large intestine, and small intestine, after AAV9-EGFP-M infection. Black arrows indicate the infiltrated inflammatory cells. At 28 days postinfection, mice were intravenously injected with Evans blue dye. The dye was circulated for 2 h before mice were euthanized, and tissues including liver (I), spleen (J), lung (K), and large intestine (L) were collected. The value of Evans blue was measured at OD610. Data are representative of two independent experiments. Values are means ± SEMs. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
In WT and NLRP3−/− mice infected with AAV9-M, M mRNA was highly expressed in mouse sera at 7 days, reduced at 14 days, and barely detected at 21 days postinfection (Fig. 6D), and IL-1β protein was significantly higher at 7, 14, and 21 days postinfection in WT mouse sera than in sera from NLRP3−/− mice (Fig. 6E), suggesting that NLRP3 knockout suppresses IL-1β activation. We noticed that the pattern of M expression is consistent with the pattern of IL-1β production (Fig. 6D and E), supporting that M induces IL-1β production. At 28 days postinfection, M mRNA was expressed in tissues of both AAV9-M-infected WT and NLRP3−/− mice (Fig. 6F, top), NLRP3 was detected in WT mouse tissues but not in NLRP3−/− mouse tissues (Fig. 6F, bottom), and IL-1β was induced by M in WT mouse tissues but not induced in NLRP3−/− mouse tissues (Fig. 6G). These results reveal that NLRP3 is required for M-induced IL-1β activation in mouse tissues. More importantly, inflammatory cell infiltration and tissue injuries were induced by M in WT mouse tissues, but they did not occur in NLRP3−/− mouse tissues (Fig. 6H), demonstrating that NLRP3 is essential for M-mediated tissue injuries. The intensities of Evans blue in liver, spleen, lung, and large intestine of WT mice were significantly higher than in NLRP3−/− mice (Fig. 6I to L), revealing that NLRP3 is required for M-induced vascular leakages in mice. Therefore, we demonstrate that M induces vascular leakage via interacting with NLRP3 and activating the inflammasome and IL-1β (Fig. 7).
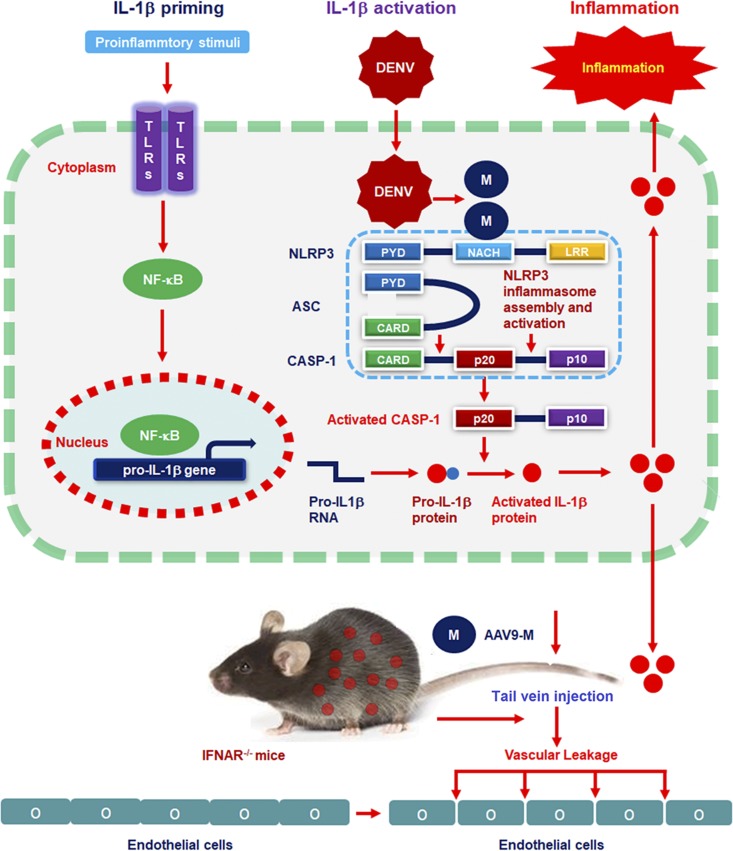
FIG 7.
A proposed model in which DENV M induces vascular leakage by activating the NLRP3 inflammasome. Upon DENV infection, the Prm protein is cleaved by Furin protease in the Golgi apparatus to form M protein. M protein then interacts with NLRP3 to facilitate NLRP3 inflammasome assembly. Once the NLRP3 inflammasome is activated, a large proportion of IL-1β is produced and released. Mature proinflammatory cytokine IL-1β then causes changes in endothelial cell permeability, including in the liver, spleen, large intestine, and small intestine.
DISCUSSION
DENV infection causes diseases ranging from self-limited DF to severe DHF and DSS (4). The change of vascular permeability is one of the main features of the diseases (6), and proinflammatory cytokines play a central role in endothelial activation and plasma leakage (19, 20). Although DENV induces IL-1β production by activating NLRP3 inflammasomes in platelets and macrophages (23, 37), the specific mechanism underlying DENV action needs to be elucidated. This study further reveals that DENV M is required for NLRP3 inflammasome activation and IL-1β release in cell lines and mouse tissues. M directly interacts with NLRP3 and facilitates inflammasome activation, thereby promoting IL-1β secretion. More importantly, we demonstrate for the first time that M induces vascular permeability and vascular leakage in mouse tissues by activating the NLRP3 inflammasome and IL-1β.
Notably, DENV replication and proteins are required for NLRP3 inflammasome activation and IL-1β release, whereas viral RNA and particles are not involved in such regulations. Previous studies showed that many viral proteins play different roles in NLRP3 inflammasome activation. Measles virus (MV) V inhibits the inflammasome (33). Influenza A virus (IAV) M2 (44) and encephalomyocarditis virus (EMCV) 2B (32) promote inflammasome activation, whereas IAV NS1 (45) and enterovirus 71 (EV71) 2A and 3C antagonize inflammasome activation (46). We have revealed that EV71 3D enhances inflammasome assembly (34) and more recently demonstrated that Zika virus (ZIKV) NS5 facilitates inflammasome activation (38). DENV contains a single open reading frame (ORF) encoding a polyprotein, which is subsequently cleaved into three structural proteins and seven nonstructural proteins (14). We noticed that M is responsible for NLRP3 inflammasome assembly and IL-1β secretion, and unlike ZIKV NS5 and DENV M, DENV NS5 failed to regulate the NLRP3 inflammasome. It is known that M interacts with E to change the pH value of the cellular surface, thus affecting virus invasion, packaging, and maturation (12, 14, 17). Here, we reveal a new function of M in the activation of the NLRP3 inflammasome and IL-1β. As a crucial component of the innate immune system, the inflammasome not only plays an important role in host defense against pathogen infections but also is involved in the progression of inflammatory diseases, such as gout and type 2 diabetes (47, 48). DENV triggers NLRP3 inflammasome assembly in platelets, and platelets may contribute to increased vascular permeability by regulating IL-1β (37). Importantly, we further demonstrate that M plays a key role in activating the NLRP3 inflammasome and IL-1β in human cell lines and in mouse primary cells and tissues.
More interestingly, M activates IL-1β secretion, thereby inducing vascular permeability and vascular leakage in mice. By studying WT mice and NLRP3−/− mice, we confirm that NLRP3 plays essential roles in M-mediated IL-1β secretion and vascular leakage in mouse tissues. It was reported that DENV NS1 induces endothelial hyperpermeability and vascular leak (43) and disrupts the endothelial glycocalyx-like layer (49). However, a recent study showed that NS1 acts on the endothelium to induce vascular leak independent of Toll-like receptor 4 (TLR4) signaling and proinflammatory cytokines (50). We reveal that M triggers vascular leakage through a distinct mechanism: M directly interacts with NLRP3 to promote ASC oligomerization and inflammasome assembly and facilitate IL-1β secretion, thereby inducing vascular leakage in mice. This study reveals a key role of M in DENV pathogenesis, provides new insights into the mechanisms underlying DENV-associated diseases, and demonstrates a direct link between NLRP3 inflammasome activation and vascular leakage in mice.
MATERIALS AND METHODS
Human blood sample analysis.
All blood samples from healthy donors were randomly collected from the Wuhan Blood Donation Center (Wuhan, China). Peripheral blood mononuclear cells (PBMCs) were separated by density centrifugation of fresh peripheral venous blood samples that were diluted 1:1 in pyrogen-free phosphate-buffered saline (PBS) over Histopaque (Haoyang Biotech). Cells were washed twice with PBS and resuspended in medium (RPMI 1640) supplemented with 10% fetal bovine serum (FBS), penicillin (100 U/ml), and streptomycin (100 μg/ml).
Animal study.
Wild-type C57BL/6 mice were purchased from Hubei Research Center of Laboratory Animals, Wuhan, China. NLRP3−/− C57BL/6 mice were provided by Di Wang of Zhejiang University, Hangzhou, China. IFNAR−/− C57BL/6 mice were provided by Jincun Zhao of Guangzhou Medical University, Guangzhou, China. Mice were bred and maintained under specific pathogen-free conditions at Wuhan University.
Age- and sex-matched mice were randomly assigned to animal studies. Six-week-old IFNAR−/− mice received tail vein injections of DENV2 (1 × 106 PFU/mouse) or PBS (mock). Six-week-old IFNAR−/− mice received tail vein injections of 300 μl of AAV9-EGFP-M (5 × 1011 viral genomes [vg]) or AAV9-EGFP. Six-week-old WT mice and NLRP3−/− mice received tail vein injections of 300 μl of AAV9-EGFP-M (5 × 1011 vg). Serum was collected every 2 days for DENV2 groups and every 7 days for AAV9 groups from the retro-orbital areas. One week after DENV2 injection and 4 weeks after AAV9 injection, mice were sacrificed and tissues were collected for immunohistochemistry (IHC) or histopathology analysis.
Mouse bone marrow-derived macrophages (BMDMs) were separated from 6- to 8-week-old male C57BL/6 mice and male NLRP3−/− C57BL/6 mice. Cells were cultured in RPMI 1640 medium with 10% FBS and 10% GM-CSF conditioned medium from L929 cells for 6 days.
Ethics statement.
The study was conducted according to the principles of the Declaration of Helsinki and approved by the Institutional Review Board of the College of Life Sciences, Wuhan University, in accordance with its guidelines for the protection of human subjects. Written informed consent was obtained from each participant.
All animal studies were performed in accordance with the principles described by the Animal Welfare Act and the National Institutes of Health Guidelines for the care and use of laboratory animals in biomedical research. All procedures involving mice and experimental protocols were approved by the Institutional Animal Care and Use Committee (IACUC) of the College of Life Sciences, Wuhan University.
Cell lines and cultures.
Human monocytic cell lines (THP-1), embryonic kidney cell lines (HEK293T), Aedes albopictus gut cell lines (C6/36), HeLa cells, and African green monkey kidney cells (Vero) were purchased from American Type Culture Collection (ATCC).
THP-1 cells were grown in RPMI 1640 medium and HEK293T, HeLa, and Vero cells were grown in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal calf serum, 100 U/ml penicillin, and 100 μg/ml streptomycin and maintained at 37°C in a 5% CO2 incubator. C6/36 cells were grown in minimal essential medium (MEM) supplemented with 10% fetal calf serum, 100 U/ml penicillin, and 100 μg/ml streptomycin and maintained at 30°C in a 5% CO2 incubator. THP-1 cells were stimulated with phorbol-12-myristate-13-acetate (PMA) for 12 h for differentiation into macrophages. THP-1 macrophages were infected with DENV or stimulated with nigericin, LPS, or ATP. Supernatants were collected for measurements of IL-1β (p17) and caspase-1 (p20). Cells were harvested for RT-PCR and immunoblot analyses.
Viruses.
DENV2 strain NGC (GenBank accession number KM204118.1) was provided by Xulin Chen of Wuhan Institute of Virology, Chinese Academy of Sciences. DENV2 strain TSV01 (GenBank accession number AY037116.1) was provided by Wenxin Li of College of Life Sciences, Wuhan University, China. To generate DENV stocks, C6/36 or Vero cells were incubated with DENV at a multiplicity of infection (MOI) of 0.5 for 2 h. Infected cells were cultured in fresh medium with 2% FBS for 7 days. Supernatants were harvested and centrifuged at 4,000 rpm for 10 min to remove debris and filtrated by a 0.22-μm filter membrane. DENV was aliquoted into tubes for freezing at −70°C. Virus titers were determined by plaque assay in Vero cells. For animal experiments, DENV was concentrated by ultracentrifugation at 30,000 × g for 2 h at 4°C and purified using a 20% sucrose solution at 80,000 × g overnight at 4°C.
Reagents and antibodies.
PMA, cycloheximide (CHX), nigericin, ATP, LPS, and dansylsarcosine piperidinium salt (DSS) were purchased from Sigma-Aldrich. ER-Tracker (E12353) was purchased from Thermo Fisher. TRIzol reagent was purchased from Ambion. Lipofectamine 2000 reagent was purchased from Invitrogen. The human IL-1β ELISA kit was purchased from BD Biosciences. Mouse IL-1β ELISA kit was purchased from R&D systems.
Anti-NLRP3 (D4d8T), anti-caspase-1 (D7F10), anti-IL-1β (D3U3E), anti-syntaxin 6 (C34B2), and anti-IL-1β (3A6) antibodies were purchased from Cell Signaling. The anti-GFP antibody was purchased from ABclonal. The antibody against DENV-Prm (GTX128092) was purchased from Genetex. Antibodies against Flag (F3165) and HA (H6908) were purchased from Sigma. The anti-β-actin antibody (66009) was purchased from Proteintech. The antibody against ASC (sc-271054) was purchased from Santa Cruz Biotechnology. Rabbit IgG and Mouse IgG were purchased from Invitrogen. Anti-GST (HT601) was purchased from TRANS. Anti-mouse IgG DyLight 649, anti-mouse IgG DyLight 488, anti-rabbit IgG DyLight 649, and anti-rabbit IgG fluorescein isothiocyanate (FITC) were purchased from Abbkine.
RNA extraction and qRT-PCR.
TRIzol reagent (Invitrogen, CA) was used for total RNA extraction according to the manufacturer’s instructions. RNA (1 μg) was reverse transcribed to cDNA with 0.5 μl oligo(dT) and 0.5 μl random primer at 37°C for 60 min and 72°C for 10 min. The resulting cDNA was used as the templates for RT-PCR analysis, which was performed in a LightCycler 480 thermal cycler (Roche) with heat activated polymerase at 95 for 5 min, 45 cycles of 95°C for 15 s, 58°C for 1 s, and 72°C for 30 s; the fluorescence was collected and analyzed at the 72°C step. A final melting curve step from 50°C to 95°C was used to test primer specificity. The primers used for RT-PCR are listed in Table 1.
TABLE 1.
Primers used for real-time PCR detection in this study
| Primer name | Primer sequences (5′→3′) |
|---|---|
| Human IL-1β forward | CACGATGCACCTGTACGATCA |
| Human IL-1β reverse | GTTGCTCCATATCCTGTCCCT |
| Human GAPDH forward | AAGGCTGTGGGCAAGG |
| Human GAPDH reverse | TGGAGGAGTGGGTGTCG |
| DENV2-E forward | CATTCCAAGTGAGAATCTCTTTGTCA |
| DENV2-E reverse | CAGATCTCTGATGAATAACCAACG |
| DENV2-M forward | GGAATGGGACTGGAGACACG |
| DENV2-M reverse | TGGTGTATGCCAGGATTGCT-3 |
| DENV2-NS5 forward | AGAGCCCGAATTTCCCAAGG |
| DENV2-NS5 reverse | TCCACATTTGGGCGTAGGAC |
| Mouse β-actin forward | AGAGGGAAATCGTGCGTGAC |
| Mouse β-actin reverse | CAATAGTGATGACCTGGCCGT |
| Mouse IL-1β forward | TGCCACCTTTTGACAGTGATG |
| Mouse IL-1β reverse | TGTGCTGCTGCGAGATTTGA |
| Human IL-6 forward | GTACATCCTCGACGGCATCTCA |
| Human IL-6 reverse | GCACAGCTCTGGCTTGTTCCTC |
| Human TNF-α forward | TCTCGAACCCCGAGTGACA |
| Human TNF-α reverse | GCCCGGCGGTTCA |
| Mouse IL-6 forward | AGACAAAGCCAGAGTCCTTCAGAGA |
| Mouse IL-6 reverse | GCCACTCCTTCTGTGACTCCAGC |
| Mouse TNF-α forward | ACGTGGAACTGGCAGAAGAG |
| Mouse TNF-α forward | CTCCTCCACTTGGTGGTTTG |
Plasmid construction.
Plasmids pcDNA3.1(+)-NLRP3, pcDNA3.1(+)-caspase-1, pcDNA3.1(+)-ASC, pcDNA3.1(+)-IL-1β, pcDNA3.1(+)-3×Flag-NLRP3, pcDNA3.1(+)-3×Flag-caspase-1, pcDNA3.1(+)-3×Flag-ASC, pcDNA3.1(+)-3×Flag-IL-1β, pcDNA3.1(+)-3×Flag-NBD, pcDNA3.1(+)-3×Flag-LRR, pcDNA3.1(+)-3×Flag-PYRIN, and pCAggs-HA-NLRP3/ASC were constructed previously by our laboratory. All DENV proteins were cloned into the pcDNA3.1(+)-3×Flag vector using specific primers. The cDNA templates came from the reverse transcription of RNA of the supernatants of DENV-infected C6/36 cells. To construct pCAggs-HA-M and pEGFP-C1-M, the DENV M gene was subcloned into pCAggs-HA at EcoRI and XhoI sites and into pEGFP-C1 at EcoRI and BamHI sites. The primers for plasmid construction are listed in Table 2.
TABLE 2.
Primers used for plasmid construction in this study
| Primer name | Primer sequences (5′→3′) |
|---|---|
| Capsid sense BamHI | GCGGATCCAATAACCAACGAAAAAAGGC |
| Capsid antisense EcoRI | CGGAATTCCGCCATCACTGTTGGAATCAGC |
| Envelope sense XhoI | CGCTCGAGATGCGTTGCATAGGAATATC |
| Envelope antisense XbaI | CGTCTAGAGGCCTGCACCATAACTC |
| Prm sense BamHI | CGGGATCCTTCCATTTAACCACACG |
| Prm antisense EcoRI | GCGAATTCTGTCATTGAAGGAGCGA |
| M sense BamHI | CGGGATCCTCAGTGGCACTCGTTCCAC |
| M antisense EcoRI | GCGAATTCTGTCATTGAAGGAGCGA |
| NS1 sense BamHI | CGGGATCCGATAGTGGTTGCGTTGTG |
| NS1 antisense XhoI | CGCTCGAGGGCTGTGACCAAGGAGTT |
| NS2A sense BamHI | CGGGATCCGGACATGGGCAGATTGAC |
| NS2A antisense EcoRI | GCGAATTCCCTTTTCTTGTTGGTTC |
| NS2B sense BamHI | GCGGATCCAGCTGGCCACTAAATGA |
| NS2B antisense EcoRI | GCGAATTCCCGTTGTTTCTTCACTT |
| NS3 sense BamHI | CGGGATCCGCTGGAGTATTGTGGGATG |
| NS3 antisense XbaI | GCTCTAGACTTTCTTCCAGCTGCAAAC |
| NS4A sense BamHI | GCGGATCCTCCCTGACCCTGAACCTA |
| NS4A antisense EcoRI | GCGAATTCTCTTTTCTGAGCTTCTCTGGT |
| NS4B sense BamHI | GCGGATCCGCAGCAGCGGGCATCATGA |
| NS4B antisense EcoRI | GCGAATTCCCTTCTCGTGTTGGTTGTGT |
| NS5 sense BamHI | GCGGATCCGGAACTGGCAACATAGGAGAGA |
| NS5 antisense XbaI | GCTCTAGACCACAGGACTCCTGCCTCTT |
| M sense EcoRI | GCGAATTCTCAGTGGCACTCGTTCCAC |
| M antisense XhoI | GCCTCGAGTGTCATTGAAGGAGCGACA |
| M sense EcoRI | GCGAATTCTTCAGTGGCACTCGTTCCAC |
| M antisense BamHI | GCGGATCCTGTCATTGAAGGAGCGACAG |
| M sense XbaI | GCTCTAGATCAGTGGCACTCGTTCCAC |
| M antisense BamHI | GCGGATCCTGTCATTGAAGGAGCGACAG |
| M sense EcoRI | GCGAATTCTCAGTGGCACTCGTTCCAC |
| M antisense XhoI | GCCTCGAGTGTCATTGAAGGAGCGACA |
| DENV1-M sense BamHI | GCGAATTCTCCGTAGGATGGGGCGAC |
| DENV1-M antisense EcoRI | GCGAATTCGGCCATGGATGGTGTTACCAG |
| DENV3-M sense BamHI | GCGGATCCTCAGTGGCGTTAGCTCC |
| DENV3-M antisense EcoRI | GCGAATTCTGTCATGGATGGGGTGACCAGC |
| DENV4-M sense BamHI | GCGGATCCTCAGTAGCCCTAACACCA |
| DENV4-M antisense EcoRI | GCGAATTCTCCGTAGGATGGGGCGAC |
Cell transfection.
HEK293T and PMA-differentiated THP-1 cells were transfected with Lipofectamine 2000 reagent (Invitrogen). To construct the NLRP3 inflammasome in HEK293T cells in vitro, the cells were seeded into 6-well plates at 5 × 105 per well for 24 h and transfected with pcDNA3.1(+)-NLRP3 (60 ng), pcDNA3.1(+)-caspase-1 (10 ng), pcDNA3.1(+)-ASC (10 ng), pcDNA3.1(+)-IL-1β (300 ng), and plasmids carrying each of the DENV2 genes (1 μg) for 48 h. Cell supernatants were collected and analyzed for mature IL-1β by ELISA, and cells were lysed and analyzed by Western blotting.
Enzyme-linked immunosorbent assay.
IL-1β in culture supernatants and mice sera was measured with an IL-1β ELISA kit (BD Biosciences, CA) and by R&D systems (Minneapolis, MN) for mice. LPS in culture supernatants was measured with an LPS ELISA kit (Expandbio).
Western blotting.
THP-1 differentiated cells were washed twice with PBS and dissolved in THP-1 lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.1% Nonidet P-40, 5 mM EDTA, and 10% glycerol). HEK293T cells were prepared in lysis buffer (50 mM Tris-HCl [pH 7.4], 300 mM NaCl, 1% Triton X-100, 5 mM EDTA, and 10% glycerol); 10% protease inhibitor (Roche) was added to the lysis buffer before using. Protein concentration was measured with a Bradford assay (Bio-Rad, Richmond, CA). Cultured cell lysates (50 μg) were electrophoresed on 8% to 12% SDS-PAGE gels and transferred to nitrocellulose membranes (Amersham, Piscataway, NJ). Nonspecific bands of control (NC) membranes were blocked with 5% skim milk for 2 h. Membranes were washed three times with phosphate-buffered saline with 0.1% Tween (PBST) and incubated with antibodies. Protein bands were visualized using a luminescent image analyzer (Fujifilm LAS-4000).
Co-immunoprecipitation assays.
HEK293T, PMA-differentiated THP-1 cells, and THP-1-M stable cells grown to 70% to 80% confluence in 10-cm dishes were cotransfected with the above-indicated plasmids for 24 to 36 h. Transfected HEK293T cells were lysed in 293T lysis buffer, and transfected THP-1 cells were dissolved in THP-1 lysis buffer. The lyses were rotated at 4°C for 30 min and centrifuged at 12,000 rpm for 15 min to remove debris. A aliquot of supernatants was used as input, and the remainder of the supernatants was incubated with antibodies overnight at 4°C and mixed with protein G Sepharose beads (GE Healthcare) for 2 h at 4°C. Immunoprecipitates were washed 4 times with respective lysis buffers, boiled in protein loading buffer, and analyzed by Western blotting.
GST pulldown assays.
To construct pGEX6p-1-M, the M gene was subcloned into pGEX6p-1 at EcoRI and XhoI sites. Plasmid pGEX6p-1-M was transfected into Escherichia coli strain BL21. After growing in ampicillin-containing LB medium at 37°C until the optical density at 600 nm (OD600) reached 0.6 to 0.8, IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a concentration of 0.2 mM, and the medium was transferred to 16°C for 12 to 16 h. Cells were harvested and sonicated in lysis buffer (PBS, 100 mM dithiothreitol [DTT], 100 mM phenylmethylsulfonyl fluoride [PMSF], 1% Triton X-100, pH 7.3). Lysates were centrifuged at 12,000 rpm for 15 min to remove debris. Supernatants were loaded into glutathione Sepharose columns (GenScript), shaken at 4°C for 2 h, slowly flowed from columns, and washed twice with PBS. Recombinant GST-M protein was eluted using elution buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 9 mg/ml reduced glutathione, and 200 μg/ml PMSF). Recombinant GST-M containing reduced glutathione was placed into PBS by using a Millipore ultrafiltration tube. Glutathione Sepharose beads (Novagen) were incubated with purified GST-M at 4°C for 2 h and then incubated with cell lysates from HEK293T cells transfected with pcDNA3.1(+)-3×Flag-NLRP3 for 4 h at 4°C. Precipitates were washed 4 times with PBS, boiled in protein loading buffer, and separated by SDS-PAGE.
Immunofluorescence.
HEK293T and HeLa cells grown on sterile coverslips were transfected with pGFP-M, pHA-NLRP3, and pFlag-ASC at 40% confluence for 48 h. Cells were fixed with 4% paraformaldehyde for 15 min, washed 3 times with ice-cold PBS containing 0.1% bovine serum albumin (BSA), and permeabilized with PBS containing 0.2% Triton X-100 for 5 min. Cells were incubated overnight with anti-HA antibody and anti-Flag antibody (1:200 in wash buffer) followed by staining with FITC-conjugated donkey anti-mouse IgG and DyLight 649-conjugated donkey anti-rabbit IgG secondary antibody (Abbkine) (1:100 in wash buffer). Nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole) for 5 min, and the cells were washed 3 times. Cells were viewed under a confocal fluorescence microscope (FluoView FV1000; Olympus, Japan).
ASC oligomerization analysis.
PMA-differentiated THP-1 cells were lysed in lysis buffer, gently shaken at 4°C for 30 min, and centrifuged at 6,000 rpm at 4°C for 15 min. Pellets were washed 3 times with PBS and resuspended in 500 μl PBS. Next, 2 mM DSS (Sigma) was added to resuspend the pellets, which were cross-linked at 37°C for 30 min. The samples were centrifuged at 6,000 rpm for 10 min. Cross-linked pellets were resuspended in 50 μl 2× SDS loading buffer, boiled for 10 min, and analyzed by Western blotting.
Quantization of vascular leakage in vivo.
Vascular leakage was quantified with the Evans blue assay. Briefly, 300 μl of 0.5% Evans blue dye was injected intravenously 4 weeks postinfection for AAV9 groups, and the dye was allowed to circulate for 2 h. Mice were euthanized and perfused with PBS. Tissues were collected, weighed, added to 1 ml formamide, and incubated at 37°C for 24 h. Evans blue concentration was quantified by measuring the OD610 and comparing the values to the standard curve. Data are expressed as nanograms Evans blue dye per milligram tissue weight.
Statistics.
All experiments were repeated at least three times with similar results. All results are expressed as the means ± the standard errors of means (SEMs). Statistical analysis was carried out using the t test for two groups and one-way analysis of variance (ANOVA) for multiple groups (GraphPad Prism 5). The data were considered statistically significant at a P value of ≤0.05.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (81730061 and 81471942), the National Health and Family Planning Commission of China (National Mega Project on Major Infectious Disease Prevention; 2017ZX10103005 and 2017ZX10202201), and the Guangdong Province Pearl River Talent Plan Innovation and Entrepreneurship Team Project (2017ZT07Y580).
We thank Di Wang of Zhejiang University, Hangzhou, China, for kindly providing NLRP3−/− C57BL/6 mice, Jincun Zhao of Guangzhou Medical University, Guangzhou, China, for kindly providing IFNAR−/− C57BL/6 mice, Xulin Chen of Wuhan Institute of Virology, Chinese Academy of Sciences, Wuhan, China, for kindly providing DENV2 strain NGC, and Wenxin Li of the College of Life Sciences, Wuhan University, China, for kindly providing DENV2 strain TSV01.
We declare no competing financial interests.
REFERENCES
- 1.Wilder-Smith A, Ooi EE, Horstick O, Wills B. 2019. Dengue. Lancet 393:350–363. doi: 10.1016/S0140-6736(18)32560-1. [DOI] [PubMed] [Google Scholar]
- 2.Guzman MG, Gubler DJ, Izquierdo A, Martinez E, Halstead SB. 2016. Dengue infection. Nat Rev Dis Primers 2:16055. doi: 10.1038/nrdp.2016.55. [DOI] [PubMed] [Google Scholar]
- 3.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GR, Simmons CP, Scott TW, Farrar JJ, Hay SI. 2013. The global distribution and burden of dengue. Nature 496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martina BE, Koraka P, Osterhaus AD. 2009. Dengue virus pathogenesis: an integrated view. Clin Microbiol Rev 22:564–581. doi: 10.1128/CMR.00035-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guzman MG, Halstead SB, Artsob H, Buchy P, Farrar J, Gubler DJ, Hunsperger E, Kroeger A, Margolis HS, Martinez E, Nathan MB, Pelegrino JL, Simmons C, Yoksan S, Peeling RW. 2010. Dengue: a continuing global threat. Nat Rev Microbiol 8:S7–S16. doi: 10.1038/nrmicro2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Srikiatkhachorn A, Green S. 2010. Markers of dengue disease severity. Curr Top Microbiol Immunol 338:67–82. doi: 10.1007/978-3-642-02215-9_6. [DOI] [PubMed] [Google Scholar]
- 7.Pinheiro FP, Corber SJ. 1997. Global situation of dengue and dengue haemorrhagic fever, and its emergence in the Americas. World Health Stat Q 50:161–169. [PubMed] [Google Scholar]
- 8.Nimmannitya S. 1987. Clinical spectrum and management of dengue haemorrhagic fever. Southeast Asian J Trop Med Public Health 18:392–397. [PubMed] [Google Scholar]
- 9.Rothman AL. 2011. Immunity to dengue virus: a tale of original antigenic sin and tropical cytokine storms. Nat Rev Immunol 11:532–543. doi: 10.1038/nri3014. [DOI] [PubMed] [Google Scholar]
- 10.Halstead SB, Mahalingam S, Marovich MA, Ubol S, Mosser DM. 2010. Intrinsic antibody-dependent enhancement of microbial infection in macrophages: disease regulation by immune complexes. Lancet Infect Dis 10:712–722. doi: 10.1016/S1473-3099(10)70166-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drumond BP, Mondini A, Schmidt DJ, Bosch I, Nogueira ML. 2012. Population dynamics of DENV-1 genotype V in Brazil is characterized by co-circulation and strain/lineage replacement. Arch Virol 157:2061–2073. doi: 10.1007/s00705-012-1393-9. [DOI] [PubMed] [Google Scholar]
- 12.Keelapang P, Sriburi R, Supasa S, Panyadee N, Songjaeng A, Jairungsri A, Puttikhunt C, Kasinrerk W, Malasit P, Sittisombut N. 2004. Alterations of pr-M cleavage and virus export in pr-M junction chimeric dengue viruses. J Virol 78:2367–2381. doi: 10.1128/jvi.78.5.2367-2381.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu IM, Zhang W, Holdaway HA, Li L, Kostyuchenko VA, Chipman PR, Kuhn RJ, Rossmann MG, Chen J. 2008. Structure of the immature dengue virus at low pH primes proteolytic maturation. Science 319:1834–1837. doi: 10.1126/science.1153264. [DOI] [PubMed] [Google Scholar]
- 14.Zhang X, Ge P, Yu X, Brannan JM, Bi G, Zhang Q, Schein S, Zhou ZH. 2013. Cryo-EM structure of the mature dengue virus at 3.5-A resolution. Nat Struct Mol Biol 20:105–110. doi: 10.1038/nsmb.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsieh SC, Zou G, Tsai WY, Qing M, Chang GJ, Shi PY, Wang WK. 2011. The C-terminal helical domain of dengue virus precursor membrane protein is involved in virus assembly and entry. Virology 410:170–180. doi: 10.1016/j.virol.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin SR, Zou G, Hsieh SC, Qing M, Tsai WY, Shi PY, Wang WK. 2011. The helical domains of the stem region of dengue virus envelope protein are involved in both virus assembly and entry. J Virol 85:5159–5171. doi: 10.1128/JVI.02099-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Junjhon J, Lausumpao M, Supasa S, Noisakran S, Songjaeng A, Saraithong P, Chaichoun K, Utaipat U, Keelapang P, Kanjanahaluethai A, Puttikhunt C, Kasinrerk W, Malasit P, Sittisombut N. 2008. Differential modulation of prM cleavage, extracellular particle distribution, and virus infectivity by conserved residues at nonfurin consensus positions of the dengue virus pr-M junction. J Virol 82:10776–10791. doi: 10.1128/JVI.01180-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Panya A, Sawasdee N, Junking M, Srisawat C, Choowongkomon K, Yenchitsomanus PT. 2015. A peptide inhibitor derived from the conserved ectodomain region of DENV membrane (M) protein with activity against dengue virus infection. Chem Biol Drug Des 86:1093–1104. doi: 10.1111/cbdd.12576. [DOI] [PubMed] [Google Scholar]
- 19.de-Oliveira-Pinto LM, Gandini M, Freitas LP, Siqueira MM, Marinho CF, Setúbal S, Kubelka CF, Cruz OG, Oliveira S. A d. 2012. Profile of circulating levels of IL-1Ra, CXCL10/IP-10, CCL4/MIP-1beta and CCL2/MCP-1 in dengue fever and parvovirosis. Mem Inst Oswaldo Cruz 107:48–56. doi: 10.1590/s0074-02762012000100007. [DOI] [PubMed] [Google Scholar]
- 20.Bozza FA, Cruz OG, Zagne SM, Azeredo EL, Nogueira RM, Assis EF, Bozza PT, Kubelka CF. 2008. Multiplex cytokine profile from dengue patients: MIP-1beta and IFN-gamma as predictive factors for severity. BMC Infect Dis 8:86. doi: 10.1186/1471-2334-8-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Houghton-Trivino N, Martin K, Giaya K, Rodriguez JA, Bosch I, Castellanos JE. 2010. Comparison of the transcriptional profiles of patients with dengue fever and dengue hemorrhagic fever reveals differences in the immune response and clues in immunopathogenesis. Biomedica 30:587–597. (In Spanish). [PubMed] [Google Scholar]
- 22.Suharti C, van Gorp EC, Setiati TE, Dolmans WM, Djokomoeljanto RJ, Hack CE, Ten CH, van der Meer JW. 2002. The role of cytokines in activation of coagulation and fibrinolysis in dengue shock syndrome. Thromb Haemost 87:42–46. doi: 10.1055/s-0037-1612941. [DOI] [PubMed] [Google Scholar]
- 23.Wu MF, Chen ST, Yang AH, Lin WW, Lin YL, Chen NJ, Tsai IS, Li L, Hsieh SL. 2013. CLEC5A is critical for dengue virus-induced inflammasome activation in human macrophages. Blood 121:95–106. doi: 10.1182/blood-2012-05-430090. [DOI] [PubMed] [Google Scholar]
- 24.Chang DM, Shaio MF. 1994. Production of interleukin-1 (IL-1) and IL-1 inhibitor by human monocytes exposed to dengue virus. J Infect Dis 170:811–817. doi: 10.1093/infdis/170.4.811. [DOI] [PubMed] [Google Scholar]
- 25.Dinarello CA. 2009. Immunological and inflammatory functions of the interleukin-1 family. Annu Rev Immunol 27:519–550. doi: 10.1146/annurev.immunol.021908.132612. [DOI] [PubMed] [Google Scholar]
- 26.Sims JE, Smith DE. 2010. The IL-1 family: regulators of immunity. Nat Rev Immunol 10:89–102. doi: 10.1038/nri2691. [DOI] [PubMed] [Google Scholar]
- 27.Kostura MJ, Tocci MJ, Limjuco G, Chin J, Cameron P, Hillman AG, Chartrain NA, Schmidt JA. 1989. Identification of a monocyte specific pre-interleukin 1 beta convertase activity. Proc Natl Acad Sci U S A 86:5227–5231. doi: 10.1073/pnas.86.14.5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martinon F, Burns K, Tschopp J. 2002. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell 10:417–426. doi: 10.1016/S1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 29.Swanson KV, Deng M, Ting JP. 2019. The NLRP3 inflammasome: molecular activation and regulation to therapeutics. Nat Rev Immunol 19:477. doi: 10.1038/s41577-019-0165-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schroder K, Zhou R, Tschopp J. 2010. The NLRP3 inflammasome: a sensor for metabolic danger? Science 327:296–300. doi: 10.1126/science.1184003. [DOI] [PubMed] [Google Scholar]
- 31.Allen IC, Scull MA, Moore CB, Holl EK, McElvania-TeKippe E, Taxman DJ, Guthrie EH, Pickles RJ, Ting JP. 2009. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity 30:556–565. doi: 10.1016/j.immuni.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ito M, Yanagi Y, Ichinohe T. 2012. Encephalomyocarditis virus viroporin 2B activates NLRP3 inflammasome. PLoS Pathog 8:e1002857. doi: 10.1371/journal.ppat.1002857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Komune N, Ichinohe T, Ito M, Yanagi Y. 2011. Measles virus V protein inhibits NLRP3 inflammasome-mediated interleukin-1beta secretion. J Virol 85:13019–13026. doi: 10.1128/JVI.05942-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang W, Xiao F, Wan P, Pan P, Zhang Y, Liu F, Wu K, Liu Y, Wu J. 2017. EV71 3D protein binds with NLRP3 and enhances the assembly of inflammasome complex. PLoS Pathog 13:e1006123. doi: 10.1371/journal.ppat.1006123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen W, Xu Y, Li H, Tao W, Xiang Y, Huang B, Niu J, Zhong J, Meng G. 2014. HCV genomic RNA activates the NLRP3 inflammasome in human myeloid cells. PLoS One 9:e84953. doi: 10.1371/journal.pone.0084953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sarvestani ST, McAuley JL. 2017. The role of the NLRP3 inflammasome in regulation of antiviral responses to influenza A virus infection. Antiviral Res 148:32–42. doi: 10.1016/j.antiviral.2017.10.020. [DOI] [PubMed] [Google Scholar]
- 37.Hottz ED, Lopes JF, Freitas C, Valls-de-Souza R, Oliveira MF, Bozza MT, Da PA, Weyrich AS, Zimmerman GA, Bozza FA, Bozza PT. 2013. Platelets mediate increased endothelium permeability in dengue through NLRP3-inflammasome activation. Blood 122:3405–3414. doi: 10.1182/blood-2013-05-504449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang W, Li G, Wu D, Luo Z, Pan P, Tian M, Wang Y, Xiao F, Li A, Wu K, Liu X, Rao L, Liu F, Liu Y, Wu J. 2018. Zika virus infection induces host inflammatory responses by facilitating NLRP3 inflammasome assembly and interleukin-1beta secretion. Nat Commun 9:106. doi: 10.1038/s41467-017-02645-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strowig T, Henao-Mejia J, Elinav E, Flavell R. 2012. Inflammasomes in health and disease. Nature 481:278–286. doi: 10.1038/nature10759. [DOI] [PubMed] [Google Scholar]
- 40.Latz E, Xiao TS, Stutz A. 2013. Activation and regulation of the inflammasomes. Nat Rev Immunol 13:397–411. doi: 10.1038/nri3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martinon F, Mayor A, Tschopp J. 2009. The inflammasomes: guardians of the body. Annu Rev Immunol 27:229–265. doi: 10.1146/annurev.immunol.021908.132715. [DOI] [PubMed] [Google Scholar]
- 42.Wang D, Tai P, Gao G. 2019. Adeno-associated virus vector as a platform for gene therapy delivery. Nat Rev Drug Discov 18:358–378. doi: 10.1038/s41573-019-0012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beatty PR, Puerta-Guardo H, Killingbeck SS, Glasner DR, Hopkins K, Harris E. 2015. Dengue virus NS1 triggers endothelial permeability and vascular leak that is prevented by NS1 vaccination. Sci Transl Med 7:304ra141. doi: 10.1126/scitranslmed.aaa3787. [DOI] [PubMed] [Google Scholar]
- 44.Ichinohe T, Pang IK, Iwasaki A. 2010. Influenza virus activates inflammasomes via its intracellular M2 ion channel. Nat Immunol 11:404–410. doi: 10.1038/ni.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moriyama M, Chen IY, Kawaguchi A, Koshiba T, Nagata K, Takeyama H, Hasegawa H, Ichinohe T. 2016. The RNA- and TRIM25-binding domains of influenza virus NS1 protein are essential for suppression of NLRP3 inflammasome-mediated interleukin-1beta secretion. J Virol 90:4105–4114. doi: 10.1128/JVI.00120-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang H, Lei X, Xiao X, Yang C, Lu W, Huang Z, Leng Q, Jin Q, He B, Meng G, Wang J. 2015. Reciprocal regulation between enterovirus 71 and the NLRP3 inflammasome. Cell Rep 12:42–48. doi: 10.1016/j.celrep.2015.05.047. [DOI] [PubMed] [Google Scholar]
- 47.Szekanecz Z, Szamosi S, Kovacs GE, Kocsis E, Benko S. 2019. The NLRP3 inflammasome - interleukin 1 pathway as a therapeutic target in gout. Arch Biochem Biophys 670:82–93. doi: 10.1016/j.abb.2019.01.031. [DOI] [PubMed] [Google Scholar]
- 48.Rovira-Llopis S, Apostolova N, Banuls C, Muntane J, Rocha M, Victor VM. 2018. Mitochondria, the NLRP3 inflammasome, and sirtuins in type 2 diabetes: new therapeutic targets. Antioxid Redox Signal 29:749–791. doi: 10.1089/ars.2017.7313. [DOI] [PubMed] [Google Scholar]
- 49.Glasner DR, Ratnasiri K, Puerta-Guardo H, Espinosa DA, Beatty PR, Harris E. 2017. Dengue virus NS1 cytokine-independent vascular leak is dependent on endothelial glycocalyx components. PLoS Pathog 13:e1006673. doi: 10.1371/journal.ppat.1006673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Modhiran N, Watterson D, Muller DA, Panetta AK, Sester DP, Liu L, Hume DA, Stacey KJ, Young PR. 2015. Dengue virus NS1 protein activates cells via Toll-like receptor 4 and disrupts endothelial cell monolayer integrity. Sci Transl Med 7:304ra142. doi: 10.1126/scitranslmed.aaa3863. [DOI] [PubMed] [Google Scholar]