Adeno-associated viruses (AAVs) have recently emerged at the forefront as gene therapy vectors; however, our understanding of host factors that influence AAV transduction in different cell types is still evolving. In the present study, we perform a genome-scale CRISPR knockout screen to identify cellular host factors that restrict AAV infection in hepatocyte cultures. We discover that Crumbs 3, which determines cellular polarity, also influences the distribution of certain carbohydrate attachment factors on the cell surface. This in turn affects the ability of virions to bind and enter the cells. This study underscores the importance of cell polarity in AAV transduction and provides a potential molecular basis for the differential infectious mechanism(s) in cell culture versus organ systems.
KEYWORDS: Adeno-associated virus, CRISPR, carbohydrate, cell polarity, parvovirus, receptor
ABSTRACT
Adeno-associated viruses (AAV) are helper-dependent parvoviruses that have been developed into promising gene therapy vectors. Many studies, including a recent unbiased genomic screen, have identified host factors essential for AAV cell entry, but no genome-wide screens that address inhibitory host factors have been reported. Here, we utilize a novel CRISPR screen to identify AAV restriction factors in a human hepatocyte cell line. The major hit from our gain-of-function screen is the apical polarity determinant Crumbs 3 (Crb3). Knockout (KO) of Crb3 enhances AAV transduction, while overexpression exerts the opposite effect. Further, Crb3 appears to restrict AAV transduction in a serotype- and cell type-specific manner. Particularly, for AAV serotype 9 and a rationally engineered AAV variant, we demonstrate that increased availability of galactosylated glycans on the surfaces of Crb3 KO cells, but not the universal AAV receptor, leads to increased capsid attachment and enhanced transduction. We postulate that Crb3 could serve as a key molecular determinant that restricts the availability of AAV glycan attachment factors on the cell surface by maintaining apical-basal polarity and tight junction integrity.
IMPORTANCE Adeno-associated viruses (AAVs) have recently emerged at the forefront as gene therapy vectors; however, our understanding of host factors that influence AAV transduction in different cell types is still evolving. In the present study, we perform a genome-scale CRISPR knockout screen to identify cellular host factors that restrict AAV infection in hepatocyte cultures. We discover that Crumbs 3, which determines cellular polarity, also influences the distribution of certain carbohydrate attachment factors on the cell surface. This in turn affects the ability of virions to bind and enter the cells. This study underscores the importance of cell polarity in AAV transduction and provides a potential molecular basis for the differential infectious mechanism(s) in cell culture versus organ systems.
INTRODUCTION
Adeno-associated viruses (AAVs) are icosahedral parvoviruses with a 4.7-kb single-stranded DNA genome (1). The AAV genome contains two alternatively spliced open reading frames: Rep, which contains genes that facilitate AAV replication and genome packaging, and Cap, which contains the structural proteins that form the viral capsid. The AAV genome is flanked by inverted terminal repeats (ITRs), which represent the only necessary packaging signal in cis, thereby allowing AAV to package exogenous DNA sequences cloned between these ITRs (1, 2).
AAV cellular entry begins with the engagement of glycan attachment factors on the cell surface (3). Distinct AAV serotypes bind different glycans, such as sialic acid, galactose, or heparan sulfate, and capsid interaction with these diverse carbohydrates plays a key role in determining cell entry and tissue tropism (4). AAV also requires additional attachment factors, including integrins, as well as the recently identified universal AAV receptor (AAVR; KIAA0319L) for cell entry (5). The latter, AAVR, was identified using a haploid genetic screen as an essential factor for viral cell entry (5). Other high-throughput screening studies using siRNA-based libraries have revealed host restriction factors involving the SUMOylation pathway, the U2 snRNP spliceosome, and DNA damage machinery (6–8). Although Genome-Scale CRISPR Knockout (GeCKO) screens have been utilized to discover host factors enabling or restricting infection by several viruses, including influenza virus and dengue virus, GeCKO screening has not yet been applied to study AAV biology (9, 10).
In the present study, we employ a CRISPR screen to elucidate novel host restriction factors in AAV transduction. Specifically, we report the discovery of the apical polarity determinant Crumbs 3 (Crb3) as a key restriction factor, and demonstrate that CRISPR knockout (KO) of Crb3 renders cultured hepatocytes more permissive to AAV. Further, we demonstrate that Crb3 enables sequestration of essential glycan attachment factors, but not AAVR from the cell surface. Ablation of Crb3 disrupts tight junction integrity and cell polarity resulting in mislocalization of glycans to the cell surface, allowing viral attachment and entry.
RESULTS
A CRISPR knockout screen identifies Crb3 as a key restriction factor for AAV infection.
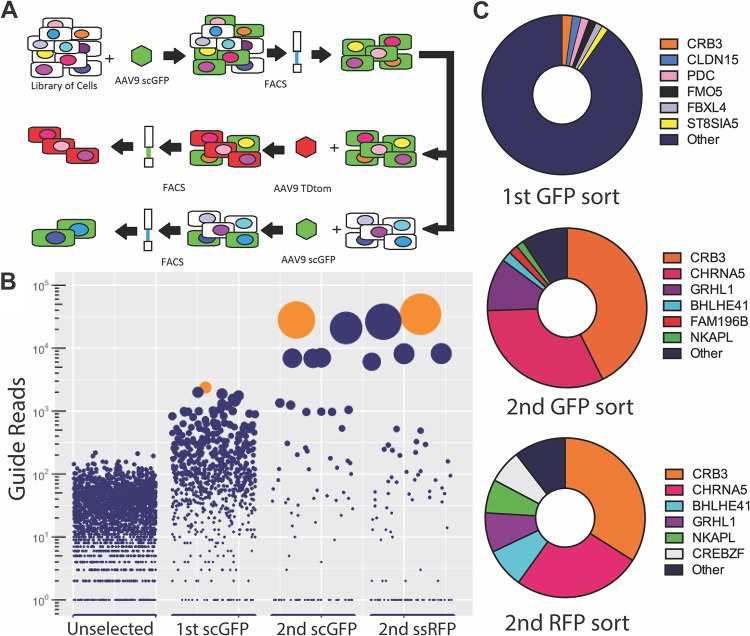
We derived a CRISPR/Cas9-based, genome-wide knockout library on the Huh7 human hepatocarcinoma cell line. These cells are poorly transduced by AAV serotypes represented by clade E or F, e.g., AAV8 or AAV9 in vitro (11, 12). Our library was derived using a human GeCKO library containing six guides for each open reading frame, with 123,411 guides (13). To elucidate host factors restricting AAV transduction, we first infected over 10 million GeCKO library cells with recombinant AAV serotype 9 packaging a self-complementary green fluorescent protein (scGFP) genome at 100 vg/cell, such that the cells should represent over 50× genome coverage of our guide library. As shown in Fig. 1A, we then subjected the transduced cells to fluorescence-activated cell sorting (FACS) in order to isolate the top 0.1% of GFP+ cells with highest expression levels. The GFP+ cells were then expanded until two populations were obtained: (i) GFP+ cells likely due to stable incorporation of AAV genomes and (ii) GFP-null cells due to loss of AAV episomes during proliferation. These two populations were further separated by FACS; then, GFP-null cells were subjected to a second round of transduction with AAV9-scGFP (100 vg/cell), and GFP+ cells were transduced using an AAV9 vector packaging a single-stranded tdTomato transgene (10,000 vg/cell) (Fig. 1A). This strategy of alternating between double-stranded (self-complementary) and conventional, single-stranded AAV vectors for transduction was adopted to screen for host factors that restrict viral entry rather than postentry events such as second-strand synthesis.
FIG 1.
A high-throughput screen for AAV restriction factors reveals Crumbs 3 (Crb3) as a lead candidate. (A) Schematic of iterative FACS sorting of a Huh7 GeCKO library. The library was transduced with rAAV9-scGFP and sorted to isolate highly GFP-positive cells, which were then grown out and sorted again to separate out stably GFP-positive cells from those that became GFP negative over time. GFP-positive cells were subjected to a second sort with rAAV9-ssRFP, while GFP-negative cells were sorted again with rAAV9-scGFP. (B) Guide enrichment across iteratively sorted cells, where bubbles represent guides populations, bubble size represents relative enrichment, and bubbles highlighted in orange represent Crb3. (C) Guide representation following first- and second-round sort populations, where the proportion of the pie chart represents the proportion of reads mapping to guides for the indicated genes.
Genomic DNA was isolated from unselected, first-round sorted, and both red fluorescent protein (RFP) and GFP second-round sorted populations that were then subjected to high-throughput sequencing and bioinformatic analysis to determine guide enrichment (Fig. 1B). Briefly, regions surrounding the target guide RNA from integrated lentiviral genomes were amplified via PCR, and an additional PCR step was carried out to add MiSeq adaptors containing index sequences. The latter step allowed deconvolution of unselected, first-round scGFP, second-round scGFP, and second-round single-stranded RFP (ssRFP) sequences. Guide representation was quantified using the MaGeCK software platform, which identifies genes corresponding to guide sequences, quantifies their relative representation across samples, and performs statistical analysis (14). The first round of selection demonstrated significant enrichment of many guides, including glycan-modifying enzymes. These included ST8SIA5, implicated in capping galactosylated glycans with sialic acid, as well as GALNT5 a GalNac transferase involved in the initiation steps of mucin O-glycosylation (Fig. 1B and Data Set S1 in the supplemental material) (15, 16). This observation is noteworthy, since AAV9 utilizes terminal galactosylated glycans for cell surface attachment (11). The two top hits yielded by our first-round sort and expansion were both tight-junction components, CLDN15 and Crb3, with the latter target being highly enriched in particular (Fig. 1B and C; Data Set S1). The second-round RFP and GFP sorts independently confirmed Crb3 as our lead candidate, with nearly 50% of total guide reads representing Crb3 following these sorts (Fig. 1B and C; Data sets S2 and S3).
Crb3 and Cldn15 knockout increases hepatocyte transduction by different AAV vectors.
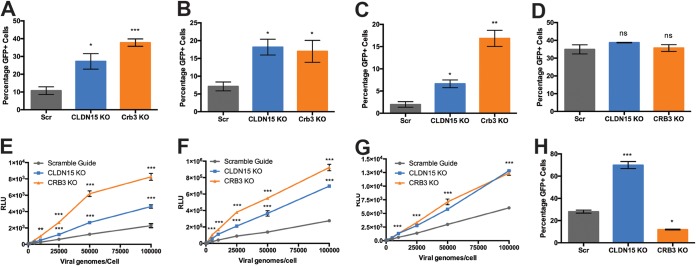
To investigate the top two hits, Crb3 and Cldn15 further, we generated clonal cell lines of Crb3 KO and Cldn15 KO using CRISPR/Cas9-mediated gene ablation in Huh7 cells. Crb3 KO and Cldn15 KO Huh7 cells were transduced alongside Scrambled guide controls (Scr) with hepatotropic AAV scGFP vectors AAV9, AAV8, and AAV3 and then subjected to flow cytometric analysis (Fig. 2A to C). AAV9 scGFP revealed that GFP-positive cells were increased by nearly 3- and 4-fold for Cldn15 and Crb3, respectively, while AAV8 demonstrated a roughly 2.5-fold increase for both cell lines. Meanwhile, AAV3 scGFP-transduced GFP-positive cells increased by roughly 3-fold for Cldn15 KO and by nearly 1 log for Crb3 KO. To rule out differential promoter or transgene expression across these lines, cells were transfected with the CBh-GFP AAV packaging plasmid cassette, and subsequent flow cytometric analysis demonstrated no significant difference in GFP expression (Fig. 2D). Further, these lines were subject to transduction with AAV1, AAV2, and AAV9 ssLuc vectors at a range of doses, revealing roughly 3- to 5-fold increases in luciferase gene expression for both lines across different vector amounts and serotypes (Fig. 2E to G).
FIG 2.
Crb3 and CLDN15 KO enhance transduction with hepatotropic AAV serotypes. (A to C) Flow cytometric analysis after transduction of Scr, CLDN15 KO, and Crb3 KO with self-complementary vectors AAV9 at 50,000 viral genomes/cell (A), AAV8 at 20,000 viral genomes/cell (B), and AAV3 at 10,000 viral genomes/cell (C). (D) Flow cytometric analysis of GFP expression after transfection of pTR-CBh-scGFP in Scr, CLDN15, and Crb3 KO cell lines. (E to G) Transduction of Scr, CLDN15 KO, and Crb3 KO Huh7 with various doses of AAV1 (E), AAV2 (F), and AAV9 (G) packaging CBA-luciferase. (H) Flow cytometric analysis of transduction of Scr, CLDN15 KO, and Crb3 KO lines with rAd5-CMV-GFP. A two-tailed unpaired t test was used (*, P < 0.05; **, P < 0.01; ***, P < 0.005).
Interestingly, when these different cell lines were transduced by recombinant, human adenovirus 5 (Ad5) packaging a GFP transgene, Cldn15 KO, but not Crb3 KO cells showed a significant increase in transduction (Fig. 2H). These results demonstrate that Crb3, but not Cldn15, selectively inhibits AAV transduction.
Crb3KO disrupts cell polarity and tight-junction markers.
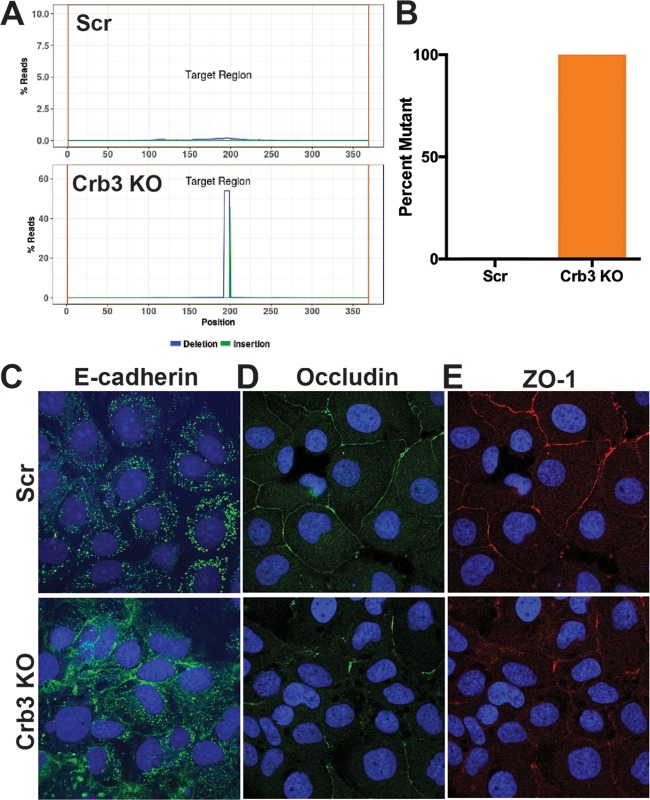
To better assess the genotype of Crb3 KO cells, Scr cells, and Crb3 KO cells were subjected to high-throughput sequencing of the Crb3 gene indel site, demonstrating that this CRISPR KO cell line had frameshift mutations across all copies of the Crb3 gene (Fig. 3A and B).
FIG 3.
Characterization of clonal Crb3 CRISPR KO cell line. (A) High-throughput sequencing of Crb3 indel site in Scr and Crb3 KO cells. (B) Quantification of the percent mutant reads for Crb3 in the Scr and clonal CRISPR Crb3 KO cell lines. (C to E) Immunofluorescent staining of Scr and Crb3 KO Huh7 cells with DAPI (blue) and E-cadherin (C), occludin (D), and ZO-1 (E).
Given the importance of Crb3 as an apical polarity determinant (17–19), as well as a component of the tight junction complex (20, 21), we next investigated the effect of Crb3 KO on these cellular components. Confocal immunofluorescence microscopy was performed to analyze the impact of Crb3 KO on E-cadherin, a marker of epithelial polarity and adherens junctions, as well as the tight-junction markers ZO-1 and occludin (18, 22). E-cadherin demonstrated marked mislocalization in Crb3 KO cells, consistent with previous in vivo studies (Fig. 3A) (18). ZO-1/occludin staining revealed disrupted tight junctions, again consistent with the previous characterization of Crb3 KO (Fig. 3B and C). Together, these data demonstrated that the absence of Crb3 in cultured hepatocytes disturbs the integrity of polarization and intercellular junctions.
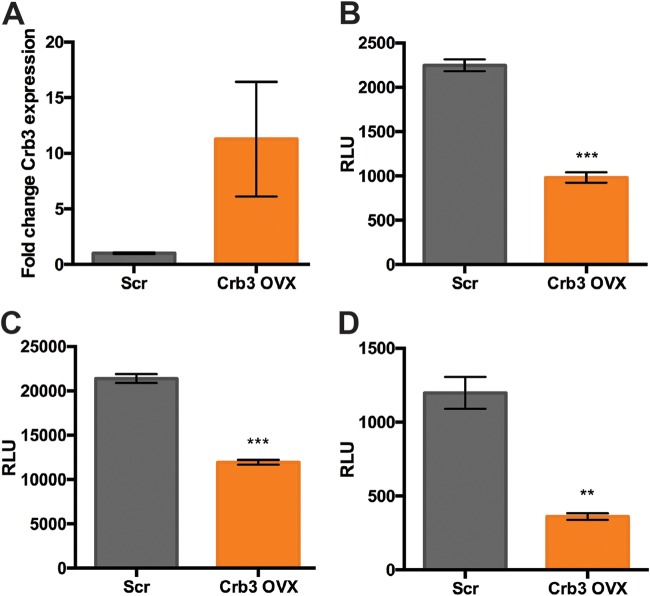
Crb3 overexpression reduces AAV transduction.
Given the putative role of Crb3 as a barrier to AAV transduction, we derived a stable, clonal Crb3 KO line and validated increased Crb3 expression via quantitative reverse transcription-PCR (qRT-PCR) (Fig. 4A). We then assessed transduction in Crb3 overexpression (OVX) and control cells with AAV1, AAV2, and AAV9 vectors packaging CBA-luciferase, finding that Crb3 OVX significantly reduced transduction with all three vectors (Fig. 4B to D). Collectively, these results support the notion that Crb3 is a universal and specific host inhibitory factor for AAV transduction in hepatocytes in vitro.
FIG 4.
Crb3 overexpression reduces AAV transduction. (A) qRT-PCR analysis of Crb3 expression in Scr and Crb3 OVX cells. (B to D) Transduction of Scr and clonal Crb3 OVX cells with AAV1 (B), AAV2 (C), and AAV9 (D) vectors packaging CBA-luciferase. A two-tailed unpaired t test was used (*, P < 0.05; **, P < 0.01; ***, P < 0.005).
Crb3 KO increases the cell surface presentation of galactosylated glycans but not AAVR.
Recently, AAVR has been shown to be essential for AAV cell entry and transduction (5). Therefore, we assessed the intracellular localization of AAVR in Scr and Crb3 KO cells by confocal immunofluorescence microscopy (Fig. 5A). AAVR maintained a perinuclear localization in both Scr and Crb3 KO cells, consistent with previously published immunofluorescent analysis (5). Further, Western blot analysis of whole-cell lysate from Scr and Crb3 KO cells revealed no difference in expression levels of AAVR (Fig. 5B), leading to the conclusion that Crb3 does not regulate the polarized expression or localization of AAVR in hepatocytes.
FIG 5.
Crb3 KO alters cell surface galactosylation but does not affect AAVR. (A) Immunofluorescent staining of AAVR in Scr and Crb3 KO cells. (B) Immunoblot of AAVR and actin in Scr, Crb3 OVX, and Crb3 KO cells. (C) Lectin staining for galactose expression with FITC-labeled ECL on Scr and Crb3 KO cells. (D) Quantification of ECL signal from replicates of Scr and Crb3 KO cells (n = 3). (E and F) ECL staining of Scr and Crb3 KO cells (E) and AAV9 CBA-luciferase transduction (F) of Scr and Crb3 KO cells across a range of densities, where the numbers indicate the quantity of cells seeded per well in a 24-well plate. A two-tailed unpaired t test was used (*, P < 0.05; **, P < 0.01; ***, P < 0.005).
Since cell surface glycans play a critical role in capsid binding to the cell surface, we utilized a fluorescently labeled lectin to detect levels of N-linked galactosylated glycans, the cognate AAV9 glycan attachment factor on the cell surface. As seen by epifluorescence microscopy, we found a striking increase in galactose localization on the cell surface of Crb3 KO cells relative to Scr control (Fig. 5C). Quantification of images showed a nearly 300% increase galactose-specific lectin signal on the surface of Crb3 KO cells (Fig. 5D). We further probed the surface of Scr and Crb3 KO cells using a galactose-specific lectin at a range of seeding densities to understand whether this phenotype was intrinsic to the cells or contact dependent (Fig. 5E). The previously observed increase in galactose localization in Crb3 KO cells was conserved at densities ranging from a confluent monolayer to sparse groups of cells (Fig. 5E). Notably, transduction of Scr and Crb3 KO cells with AAV9-CBA-luciferase demonstrated a significant increase in Crb3 KO regardless of cell density (Fig. 5F). These results suggest that polarity rather than tight junction integrity might play a key role in restricting the cell surface presentation of AAV attachment factors.
AAV capsids display increased cell surface attachment on Crb3KO cells.
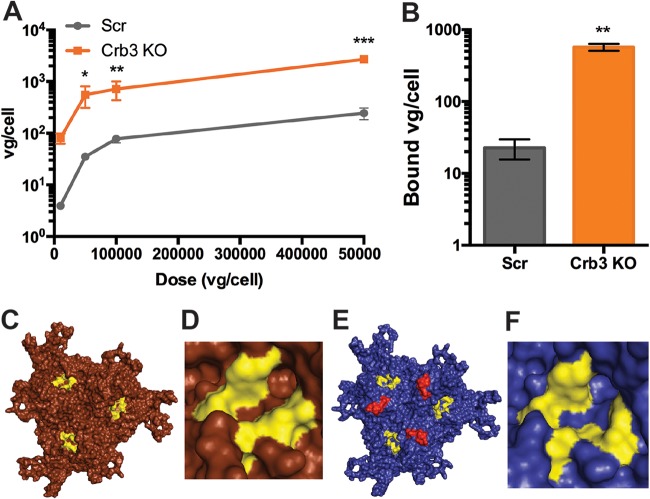
We assessed the impact of Crb3 KO mediated cell polarity disruption on viral binding. Cell surface binding of AAV9 virions in Scr and Crb3KO was assessed as previously described (23). Briefly, cells were prechilled and transduced at 4°C to allow for viral binding to the cell surface but prevent internalization. Cells were washed to remove unbound virions and then isolated DNA subject to qPCR to determine the number of vector genome copies associated per cell. Crb3 KO cells demonstrated a marked, log-fold increase in AAV9 binding regardless of the amount of incubated virions per cell (Fig. 6A).
FIG 6.
Crb3 KO augments cell surface binding of galactose-dependent AAV capsids. (A) Binding curve for AAV9 on Scr and Crb3 KO cells. (B) Binding of AAV2i8g9 on Scr and RNF121 KO cells. (C and D) Structural model of the AAV9 trimer (C) with a closeup of its galactose footprint, highlighted in yellow (D). (E and F) Structural model of the AAV2i8g9 trimer (E), along with a closeup of its engrafted galactose footprint highlighted in yellow (F). The AAV8 derived residues are highlighted in red. A two-tailed unpaired t test was used (*, P < 0.05; **, P < 0.01; ***, P < 0.005).
To establish that the increased cell surface binding of AAV9 capsids was due to increased galactose availability, we assessed the cell surface binding profile of a laboratory engineered AAV variant, AAV2i8g9 (Fig. 6B). Briefly, AAV2i8g9 is an AAV2-derived mutant with ablated heparan sulfate binding and engrafted with galactose-binding residues from the AAV9 capsid (24, 25). As seen in Fig. 4B, AAV2i8g9 capsid binding was increased by Crb3 KO by >25-fold. These results corroborate the putative role of Crb3 as an AAV restriction factor, which restricts the cell surface presentation of glycan attachment factors that would otherwise allow viral binding and entry.
DISCUSSION
Cell polarity and tight-junction integrity in epithelial cells play important roles in cellular entry for many viruses, including hepatitis C virus and adenovirus (26–29). Tight junctions and polarity restrict hepatitis C virus cellular entry in polarized hepatocytes by facilitating basolateral localization of receptors (26, 27). Meanwhile, characterization of adenoviral infection of epithelial cultures has revealed that the coxsackie B and adenovirus type 2 and 5 receptor (CAR) localizes to the basolateral membrane in human airway epithelia and that tight-junction disruption was necessary for adenoviral entry (28, 29).
In this study, we carried out a CRISPR screen to identify novel host restriction factors for AAV transduction. Our results reveal that the molecular mechanism behind restriction of AAV infection in cultured hepatocytes is driven by Crb3, a determinant of apical polarity and tight-junction integrity. Although we only utilized AAV9 in our screen, we observed that Crb3 KO increases transduction with other hepatotropic AAV serotypes such as AAV3B and AAV8. We also observed that overexpression of Crb3 overexpression renders hepatocytes more resistant to AAV transduction. Interrogation of cell polarity and tight junction integrity in Crb3 KO recapitulated previous studies demonstrating the importance of Crb3 in these pathways, with polarity marker E-cadherin showing mislocalization and tight junction components ZO-1/Occludin demonstrating marked disruption (18).
While hepatocyte polarity has not been well investigated in the context of AAV transduction, polarized human airway epithelia have been shown to be less effectively transduced with AAV2 from the apical membrane due to reduced cellular entry (30–32). In contrast, AAV5 has been shown to effectively transduce from the apical membrane compared to AAV2, while AAV1 has also been demonstrated to have improved apical transduction over closely related AAV6 (33, 34). Furthermore, AAVR has recently been shown to localize to the basolateral membrane in human airway epithelia, and AAV5 has been shown to be able to transduce cells independently of AAVR (12, 35).
With regard to hepatocytes, Crb3 KO did not impact expression or localization of AAVR but demonstrated marked overexpression of galactosylated glycans on the cell surface, which are known to be cognate attachment factors for AAV9 (11). As visualized in Fig. 6C and D, the AAV9 galactose footprint (yellow) is localized at the 3-fold axis of symmetry. The AAV2i8g9 capsid was derived from an AAV2 parental capsid with AAV8 residues engrafted to ablate heparan sulfate binding (red), as well as the galactose footprint described earlier from AAV9 (yellow) (Fig. 6E and F). Both of these divergent capsids with galactose footprints demonstrated augmented binding in the context of Crb3 KO. These observations suggest that the increased binding can be specifically attributed to capsid recognition of relocalized galactosylated glycans on the surfaces of Crb3 KO cells.
The dichotomy in hepatocyte architecture in cell culture in vitro and organ systems in vivo is well documented (36, 37). We postulate that the architecture of cultured hepatocytes limits binding of some AAV capsids by restricting access to attachment factors located intracellularly or on the basolateral membrane. Disrupting the apical polarity determinant Crb3 may facilitate enhanced binding and transduction by augmenting cell surface localization of galactosylated glycans. In contrast, hepatocytes in the liver in vivo have an apical membrane that faces the bile canniculus, while the basolateral side faces blood flow (36). It is tempting to speculate that this presentation of glycan attachment factors on the luminal side of hepatocytes (in conjunction with other host factors, e.g., serum proteins) enables efficient hepatocyte binding and liver transduction commonly observed with AAV vectors.
It is also noteworthy to mention that our study might inform further studies evaluating AAV transcytosis, since AAV capsids appear to transit across cellular barriers in a serotype and cell type specific manner (32). For example, while AAV2 transduces tissue culture cells with high efficiency, AAV2 fails to transcytose across CaCo-2 (colon carcinoma) cells and MDCK (Madin-Darby kidney) cells of epithelial origin (32). Notably, AAV5 effectively crossed Caco-2 barrier epithelial monolayers; while competitive inhibition of AAV5 engagement with sialic acid reduced transduction, it did not ablate trancytosis, suggesting that AAV5 may utilize a distinct cellular entry pathway in crossing tissue layers (32). Previous work in our lab suggests that sialic acid binding affinity of AAV1 can be modulated by engrafting a minimal footprint of amino acid residues derived from AAVrh.10, which, in turn, can profoundly impact the ability to traverse the vascular endothelium/blood-brain barrier (38, 39). Together, these data suggest that glycan receptor engagement and polarized receptor localization could play integral roles in determining AAV transduction versus transcellular transport profiles in vitro and vector biodistribution in vivo. Although outside the scope of the present study, the exact mechanism by which Crumbs 3 might modulate cell surface glycan presentation remains to be determined. Specifically, additional mechanistic studies determining whether Crb3 facilitates increased expression or relocalization of glycan attachment factors on the surface of different cell types, as well as correlation with tissue architectures in vivo, are warranted. Nevertheless, it is plausible that this paradigm is relevant in the case of epithelial and endothelial barriers to AAV transport in general.
MATERIALS AND METHODS
Reagents.
Antibodies were as follows: mouse anti-actin (Abcam, ab3280), mouse anti-KIAA0319L(AAVR) (Abcam, ab105385), mouse anti-E-cadherin (BD Biosciences, 610182), mouse anti-occludin (Thermo Fisher, 33-1500), and rabbit anti-ZO-1 (Thermo Fisher, 61-7300). Propidium iodide (P4170) was purchased from Sigma-Aldrich, while Zombie Violet (423113) was obtained from BioLegend. Erythrina Cristigalli Lectin (ECL; FL-1141) was purchased from Vector Laboratories. Oligonucleotide qPCR primers and sgRNAs were purchased from Integrated DNA Technologies (Coralville, IA), with sequences listed in Table 1. The Crb3 qRT-PCR primer set has been reported earlier (40).
TABLE 1.
Oligonucleotides used in cloning and qPCR
| Target | Oligonucleotide sequence (5′–3′) |
|
|---|---|---|
| Forward | Reverse | |
| Cldn15 gRNA | caccgCGAGGTGGCGACGGGCATCA | aaacTGATGCCCGTCGCCACCTCGc |
| Crb3 gRNA | caccgCACCTACCGGCCCAGTAGCG | aaacCGCTACTGGGCCGGTAGGTGc |
| Crb3 AmpliconEZ | ATGCCATTCAGGTGGAGGTG | CCATCAGAGGCTGGTGTCTC |
| Crb3 qRT-PCR | CCTTCATCCACCAGCTCCAG | GCCAAGAGGGAGAAGACCAC |
| Luciferase qPCR | AAAAGCACTCTGATTGACAAATAC | CCTTCGCTTCAAAAAATGGAAC |
| Laminin qPCR | GTTAACAGTCAGGCGCATGGGCC | CCATCAGGGTCACCTCTGGTTCC |
| ITR qPCR | AACATGCTACGCAGAGAGGGAGTGG | CATGAGACAAGGAACCCCTAGTGATGGAG |
Cell lines.
Human hepatocarcinoma (Huh7) cells were obtained from the UNC Lineberger Tissue Culture Facility. HEK293 were obtained from the UNC Vector Core. Cells were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum (FBS) and 100 U/ml penicillin-streptomycin in 5% CO2 at 37°C.
Recombinant virus production.
Recombinant AAV vectors packaging a chicken β-actin (CBA) promoter-driven firefly luciferase cassette and self-complementary AAV (scAAV) vectors packaging a hybrid CBA (CBh) promoter driving GFP were generated using triple plasmid transfection in HEK293 cells as described previously (23). Viral titers were obtained as previously indicated by quantitative PCR with primers against to ITRs using a Roche LightCycler 480 (Roche Applied Sciences, Pleasanton, CA) (23).
For cloning of recombinant lentiviral cassettes, guides were annealed, phosphorylated, and ligated into BsmBI-digested lentiCRISPRv2, which was a gift from Feng Zhang (Addgene, plasmid 52961), and Crb3 cDNA was cloned into pLX304 for overexpression via Gateway Cloning, which was a gift from David Root (Addgene, plasmid 25890). Recombinant lentivirus packaging guides against Cldn15, Crb3, or Scr control guides, as well as CMV-driven overexpression of Crb3, was produced via triple plasmid transfection with psPax2 and VSVG glycoprotein for pseudotyping in HEK293 cells, as previously described (13).
Generation of CRISPR library.
The human GeCKOv2 CRISPR knockout pooled library was a gift from Feng Zhang (Addgene, 1000000048). These cassettes were used to produce recombinant lentivirus packaging puromycin resistance, Cas9, and sgRNA constructs, as described above and previously (13, 41). Huh7 cells were transduced with this recombinant lentivirus such that only one in three cells was infected to ensure that cells would only have a single knockout and then subjected to puromycin selection as described above.
FACS and flow cytometry.
For viable cell sorting, cells were washed twice with ice cold 1× phosphate-buffered saline (PBS), resuspended in 2% FBS plus PBS, and filtered to remove clumps. The cells were then stained for viability with propidium iodide (in the case of GFP-sorted populations) or Zombie Violet (in the case of tdTomato sorted populations). The cells were analyzed and sorted by using a FACSAria II (Becton Dickinson). For flow cytometric analysis, the cells were prepared as described above, fixed with paraformaldehyde, and filtered to remove clumps prior to analysis on a CyAn ADP (Beckman Coulter).
High-throughput sequencing and analysis.
Genomic DNA extracted from pre- and postselection CRISPR libraries were subjected to PCR-based amplification of guide sequences and indexing as previously described (41). Libraries were sequenced with the MiSeq platform (Illumina). Guides were identified and enrichment was quantified with MaGeCK (14), and guide enrichment data were plotted using R.
Luciferase assays.
Huh7 cells were counted and seeded overnight at equal density (3e4 cells/well) on 24-well plates. At 24 h postransduction, cells were harvested in passive lysis buffer and lysate combined with luciferin substrate from Promega (Madison, WI). Luciferase signal was then quantified by a VictorX plate reader from Perkin-Elmer (Waltham, MA).
Binding assays.
Cells were seeded overnight, prechilled at 4°C for 30 min, and incubated with rAAV-CBA-luciferase at 4°C for 1 h, followed by three washes with ice-cold 1× PBS to remove unbound virions. Then, 300 μl of ddH2O was added to each well, and the cells were subjected to three freeze-thaw cycles prior to extraction of total genomic DNA using an IBI Mini-Genomic DNA kit (IBI, Dubuque, IA). Quantification of viral genomes per cell was determined via qPCR of DNA samples with primers against the luciferase transgene and the host laminin gene.
Confocal microscopy.
Cells were seeded on slide covers in 24-well plates overnight and then fixed with 4% paraformaldehyde and permeabilized with 0.1% Triton X-100. After 30 min of blocking with 5% normal goat serum, the cells were stained with primaries, washed three times with PBS, and stained with fluorescent secondaries and DAPI (4′,6′-diamidino-2-phenylindole). After being mounted in Prolong Diamond (Invitrogen), the specimens were imaged by using a Zeiss 710 scanning confocal microscope.
Lectin staining.
Lectin staining was performed as previously described (11). Briefly, the cells were prechilled for 30 min at 4°C and then incubated with FITC-conjugated ECL for 90 min. After three washes with ice-cold 1× PBS, the cells were imaged on EVOS, and the images were quantified with ImageJ (42).
Structural modeling.
AAV9 and AAV2 viral protein (VP) structural coordinates were obtained from the RCSB Protein Data Bank (PDB codes 3UX1 and 1LP3) (43, 44). Homology modeling of the AAV2i8g9 VP3 monomer was performed using the crystal structure of AAV2 VP3 as a template via the SWISS-MODEL protein structure modeling server, and structural coordinates were mapped with WinCoot (45, 46). Three-dimensional trimer models of AAV9 and AAV2i8g9 were created using the oligomer generator function in VIPERdb-Virus Particle ExploreR2 (47). Structural models were visualized using PyMOL. The AAV9 galactose footprint (AAV9 VP1 numbering D-271, N-272, Y-446, N-470, and W-503), as well as the engrafted galactose footprint on 2i8g9, are highlighted in yellow, while the i8 motif on 2i8g9 (AAV8 VP1 numbering 588-QQNTAP-593) is shown in blue (11, 24, 48).
Statistical analysis.
Data represented as mean values plus and minus the SEM with n ≥ 3. A two-tailed unpaired Student t test was calculated with GraphPad Prism version 6. P values of <0.05 were considered significant. Asterisks are used to denote P values (*, P < 0.05; **, P < 0.01; ***, P < 0.005).
Supplementary Material
ACKNOWLEDGMENTS
We thank the UNC Chapel Hill High-throughput Sequencing Facility, Flow Cytometry Core and Microscopy Services Laboratory, as well as the Duke University Light Microscopy Core Facility.
V.J.M. and A.A. conceived the study, designed experiments and wrote the manuscript. V.J.M. carried out screening and viral infection experiments as well as bioinformatics analysis and transduction assays and analyzed the data. T.O.T., J.A.Y., and M.P. carried out clonal cell generation and assisted with transduction assays, as well as Western blots. S.M.-T. provided methodology and resources. A.A. provided conceptualization, writing, supervision, and funding acquisition.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00943-19.
REFERENCES
- 1.Snyder R, Moullier P, Weitzman MD, Linden RM, JAgbandje-Mckenna M, Kleinschmidt J. 2011. Adeno-associated virusbiology adeno-associated virus: methods and protocols.
- 2.Hastie E, Samulski RJ. 2015. Adeno-associated virus at 50: a golden anniversary of discovery, research, and gene therapy success—a personal perspective. Hum Gene Ther 26:257–265. doi: 10.1089/hum.2015.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berry GE, Asokan A. 2016. Cellular transduction mechanisms of adeno-associated viral vectors. Curr Opin Virol 21:54–60. doi: 10.1016/j.coviro.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang L, Halder S, Agbandje-Mckenna M. 2014. Parvovirus glycan interactions. Curr Opin Virol 7:108–118. doi: 10.1016/j.coviro.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pillay S, Meyer NL, Puschnik AS, Davulcu O, Diep J, Ishikawa Y, Jae LT, Wosen JE, Nagamine CM, Chapman MS, Carette JE. 2016. An essential receptor for adeno-associated virus infection. Nature 530:108–112. doi: 10.1038/nature16465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hölscher C, Sonntag F, Henrich K, Chen Q, Beneke J, Matula P, Rohr K, Kaderali L, Beil N, Erfle H, Kleinschmidt JA, Muller M. 2015. The SUMOylation pathway restricts gene transduction by adeno-associated viruses. PLoS Pathog 11:e1005281-23. doi: 10.1371/journal.ppat.1005281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schreiber CA, Sakuma T, Izumiya Y, Holditch SJ, Hickey RD, Bressin RK, Basu U, Koide K, Asokan A, Ikeda Y. 2015. An siRNA screen identifies the U2 snRNP spliceosome as a host restriction factor for recombinant adeno-associated viruses. PLoS Pathog 11:e1005082-15. doi: 10.1371/journal.ppat.1005082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mano M, Ippodrino R, Zentilin L, Zacchigna S, Giacca M. 2015. Genome-wide RNAi screening identifies host restriction factors critical for in vivo AAV transduction. Proc Natl Acad Sci U S A 112:11276–11281. doi: 10.1073/pnas.1503607112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marceau CD, Puschnik AS, Majzoub K, Ooi YS, Brewer SM, Fuchs G, Swaminathan K, Mata MA, Elias JE, Sarnow P, Carette JE. 2016. Genetic dissection of Flaviviridae host factors through genome-scale CRISPR screens. Nature 535:159–163. doi: 10.1038/nature18631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heaton BE, Kennedy EM, Dumm RE, Harding AT, Sacco MT, Sachs D, Heaton NS. 2017. A CRISPR activation screen identifies a pan-avian influenza virus inhibitory host factor. Cell Rep 20:1503–1512. doi: 10.1016/j.celrep.2017.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen S, Bryant KD, Brown SM, Randell SH, Asokan A. 2011. Terminal N-linked galactose is the primary receptor for adeno-associated virus 9. J Biol Chem 286:13532–13540. doi: 10.1074/jbc.M110.210922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dudek AM, Pillay S, Puschnik AS, Nagamine CM, Cheng F, Qiu J, Carette JE, Vandenberghe LH. 2018. An alternate route for adeno-associated virus entry independent of AAVR. J Virol 92:e02213-17. doi: 10.1128/JVI.02213-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanjana NE, Shalem O, Zhang F. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li W, Xu H, Xiao T, Cong L, Love MI, Zhang F, Irizarry RA, Liu JS, Brown M, Liu XS. 2014. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol 15:1–12. doi: 10.1186/s13059-014-0554-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim Y, Kim K, Do S, Kim C, Kim S, Lee Y. 1997. Molecular cloning and expression of human α2,8-sialyltransferase (hST8Sia V). Biochem Biophys Res Commun 235:327–330. doi: 10.1006/bbrc.1997.6725. [DOI] [PubMed] [Google Scholar]
- 16.Guo H, Zhao L, Shi B, Bao J, Zheng D, Zhou B, Shi J. 2018. GALNT5 uaRNA promotes gastric cancer progression through its interaction with HSP90. Oncogene 37:4505–4517. doi: 10.1038/s41388-018-0266-4. [DOI] [PubMed] [Google Scholar]
- 17.Hayase J, Kamakura S, Iwakiri Y, Yamaguchi Y, Izaki T, Ito T, Sumimoto H. 2013. The WD40 protein Morg1 facilitates Par6–aPKC binding to Crb3 for apical identity in epithelial cells. J Cell Biol 200:635–650. doi: 10.1083/jcb.201208150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whiteman EL, Fan S, Harder JL, Walton KD, Liu C, Soofi A, Fogg VC, Hershenson MB, Dressler GR, Deutsch GH, Gumucio DL, Margolis B. 2014. Crumbs3 is essential for proper epithelial development and viability. Mol Cell Biol 34:43–56. doi: 10.1128/MCB.00999-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szymaniak AD, Mahoney JE, Cardoso WV, Varelas X. 2015. Crumbs3-mediated polarity directs airway epithelial cell fate through the Hippo pathway effector Yap. Dev Cell 34:283–296. doi: 10.1016/j.devcel.2015.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lemmers C, Michel D, Lane-Guermonprez L, Delgrossi M-H, Médina E, Arsanto J-P, Le Bivic A. 2004. CRB3 binds directly to Par6 and regulates the morphogenesis of the tight junctions in mammalian epithelial cells. Mol Biol Cell 15:1324–1333. doi: 10.1091/mbc.e03-04-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fogg VC, Liu C, Margolis B. 2005. Multiple regions of Crumbs3 are required for tight junction formation in MCF10A cells. J Cell Sci 118:2859–2869. doi: 10.1242/jcs.02412. [DOI] [PubMed] [Google Scholar]
- 22.Coopman P, Djiane A. 2016. Adherens junction and E-cadherin complex regulation by epithelial polarity. Cell Mol Life Sci 73:3535–3553. doi: 10.1007/s00018-016-2260-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen S, Troupes AN, Pulicherla N, Asokan A. 2013. Multiple roles for sialylated glycans in determining the cardiopulmonary tropism of adeno-associated virus 4. J Virol 87:13206–13213. doi: 10.1128/JVI.02109-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen S, Horowitz ED, Troupes AN, Brown SM, Pulicherla N, Samulski R, JAgbandje-Mckenna M, Asokan A. 2013. Engraftment of a galactose receptor footprint onto adeno-associated viral capsids improves transduction. J Biol Chem 288:28814–28823. doi: 10.1074/jbc.M113.482380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Madigan VJ, Asokan A. 2016. Engineering AAV receptor footprints for gene therapy. Curr Opin Virol 18:89–96. doi: 10.1016/j.coviro.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mee CJ, Grove J, Harris HJ, Hu K, Balfe P, Mckeating JA. 2008. Effect of cell polarization on hepatitis C virus entry. J Virol 82:461–470. doi: 10.1128/JVI.01894-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mee CJ, Harris HJ, Farquhar MJ, Wilson G, Reynolds G, Davis C, Van Ijzendoorn SCD, Balfe P, Mckeating JA. 2009. Polarization restricts hepatitis C virus entry into HepG2 hepatoma cells. J Virol 83:6211–6221. doi: 10.1128/JVI.00246-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cohen CJ, Shieh JTC, Pickles RJ, Okegawa T, Hsieh J, Bergelson JM. 2001. The coxsackievirus and adenovirus receptor is a transmembrane component of the tight junction. Proc Natl Acad Sci 98:15191–15196. doi: 10.1073/pnas.261452898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walters RW, Grunst T, Bergelson JM, Finberg RW, Welsh MJ, Zabner J. 1999. Basolateral localization of fiber receptors limits adenovirus infection from the apical surface of airway epithelia. J Biol Chem 274:10219–10226. doi: 10.1074/jbc.274.15.10219. [DOI] [PubMed] [Google Scholar]
- 30.Duan D, Yue Y, Yan Z, McCray PB Jr, Engelhardt JF. 1998. Polarity influences the efficiency of adeno-associated recombinant virus infection in differentiated airway epithelia. Hum Gene Ther 9:2761–2776. doi: 10.1089/hum.1998.9.18-2761. [DOI] [PubMed] [Google Scholar]
- 31.Bals R, Xiao W, Sang N, Weiner DJ, Meegalla RL, Wilson JM. 1999. Transduction of well-differentiated airway epithelium by recombinant adeno-associated virus is limited by vector entry. J Virol 73:6085–6088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Di Pasquale G, Chiorini JA. 2006. AAV transcytosis through barrier epithelia and endothelium. Mol Ther 13:506–516. doi: 10.1016/j.ymthe.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 33.Zabner J, Seiler M, Walters R, Kotin RM, Fulgeras W, Davidson BL, Chiorini JA. 2000. Adeno-associated virus type 5 (AAV5) but not AAV2 binds to the apical surfaces of airway epithelia and facilitates gene transfer. J Virol 74:3852–3858. doi: 10.1128/jvi.74.8.3852-3858.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan Z, Lei-Butters DCM, Keiser NW, Engelhardt JF. 2013. Distinct transduction difference between adeno-associated virus type 1 and type 6 vectors in human polarized airway epithelia. Gene Ther 20:328–337. doi: 10.1038/gt.2012.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hamilton BA, Li X, Pezzulo AA, Abou Alaiwa MH, Zabner J. 2019. Polarized AAVR expression determines infectivity by AAV gene therapy vectors. Gene Ther 26:240–249. doi: 10.1038/s41434-019-0078-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Treyer A, Müsch A. 2013. Hepatocyte polarity. Compr Physiol 3:243–287. doi: 10.1002/cphy.c120009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gissen P, Arias IM. 2015. Structural and functional hepatocyte polarity and liver disease. J Hepatol 63:1023–1037. doi: 10.1016/j.jhep.2015.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Albright BH, Storey CM, Murlidharan G, Rivera RMC, Berry GE, Madigan VJ, Asokan A. 2018. Mapping the structural determinants required for AAVrh.10 transport across the blood-brain barrier. Mol Ther 26:510–523. doi: 10.1016/j.ymthe.2017.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Albright BH, Simon KE, Pillai M, Devlin GW, Asokan A. 2019. Modulation of sialic acid dependence influences the central nervous system transduction profile of adeno-associated viruses. J Virol 93:1–15. doi: 10.1128/JVI.00332-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alam M, Bouillez A, Tagde A, Ahmad R, Rajabi H, Maeda T, Hiraki M, Suzuki Y, Kufe D. 2016. MUC1-C represses the Crumbs complex polarity factor CRB3 and downregulates the Hippo pathway. Mol Cancer Res 14:1266–1277. doi: 10.1158/1541-7786.MCR-16-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F. 2014. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343:84–88. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dimattia MA, Nam H, Van Vliet K, Mitchell M, Bennett A, Gurda BL, McKenna R, Olson NH, Sinkovits RS, Potter M, Byrne BJ, Aslanidi G, Zolotukhin S, Muzyczka N, Baker TS, Agbandje-McKenna M. 2012. Structural insight into the unique properties of adeno-associated virus serotype 9. J Virol 86:6947–6958. doi: 10.1128/JVI.07232-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xie Q, Bu W, Bhatia S, Hare J, Somasundaram T, Azzi A, Chapman MS. 2002. The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc Natl Acad Sci U S A 99:10405–10410. doi: 10.1073/pnas.162250899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arnold K, Bordoli L, Kopp J, Schwede T. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modeling. Bioinformatics 22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 46.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carrillo-Tripp M, Shepherd CM, Borelli IA, Venkataraman S, Lander G, Natarajan P, Johnson JE, Brooks CLB III, Reddy VS. 2009. VIPERdb2: an enhanced and web API enabled relational database for structural virology. Nucleic Acids Res 37:D436–D442. doi: 10.1093/nar/gkn840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Asokan A, Conway JC, Phillips JL, Li C, Hegge J, Sinnott R, Yadav S, Diprimio N, Nam H, Agbandje-Mckenna M, Mcphee S, Wolff J, Samulski RJ. 2010. Reengineering a receptor footprint of adeno-associated virus enables selective and systemic gene transfer to muscle. Nat Biotechnol 28:79–82. doi: 10.1038/nbt.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.