Following viral infection, the working pattern and function of the cell can be transformed through the impact on mitochondria. It still unknown how the mitochondrial response changes in cells infected with viruses in the process of virulence attenuation. EIAVDLV121 is the only effective lentiviral vaccine for large-scale use in the world. EIAVDLV34 is the parent pathogenic strain. Unlike EIAVDLV34-induced inflammation storms, EIAVDLV121 can induce high levels of apoptosis. For the first time, we found that, after the mitochondrial protein expression profile is altered, EIAVDLV34-infected cells are transformed into M1-polarized-type macrophages and cause inflammatory injury and that the intrinsic apoptosis pathway is activated in EIAVDLV121-infected cells. These studies shed light on how the mitochondrial protein expression profile changes between cells infected by pathogenic lentivirus strains and cells infected by attenuated lentivirus strains to drive different cellular responses, especially from inflammation to apoptosis.
KEYWORDS: EIAV, HIV, apoptosis, attenuation, equine, inflammation, lentivirus, mitochondrion, proteomics, vaccine
ABSTRACT
Equine infectious anemia virus (EIAV) is an equine lentivirus similar to HIV-1, targets host immune cells, and causes a life-long infection in horses. The Chinese live EIAV vaccine is attenuated from long-term passaging of a highly virulent strain in vitro. The parent pathogenic strain (EIAVDLV34) induces a host inflammatory storm to cause severe pathological injury of animals. However, the vaccine strain (EIAVDLV121) induces a high level of apoptosis to eliminate infected cells. To investigate how these processes are regulated, we performed a comparative proteomics analysis and functional study in equine monocyte-derived macrophages (eMDMs) and found that the divergent mitochondrial protein expression profiles caused by EIAV strains with different virulence led to disparate mitochondrial function, morphology, and metabolism. This in turn promoted the distinct transformation of macrophage inflammatory polarization and intrinsic apoptosis. In EIAVDLV34-infected cells, a high level of glycolysis and increased mitochondrial fragmentation were induced, resulting in the M1-polarized proinflammatory-type transformation of macrophages and the subsequent production of a strong inflammatory response. Following infection with EIAVDLV121, the infected cells were transformed into M2-polarized anti-inflammatory macrophages by inhibition of glycolysis. In this case, a decrease in the mitochondrial membrane potential and impairment of the electron transport chain led to increased levels of apoptosis and reactive oxygen species. These results correlated with viral pathogenicity loss and may help provide an understanding of the key mechanism of lentiviral attenuation.
IMPORTANCE Following viral infection, the working pattern and function of the cell can be transformed through the impact on mitochondria. It still unknown how the mitochondrial response changes in cells infected with viruses in the process of virulence attenuation. EIAVDLV121 is the only effective lentiviral vaccine for large-scale use in the world. EIAVDLV34 is the parent pathogenic strain. Unlike EIAVDLV34-induced inflammation storms, EIAVDLV121 can induce high levels of apoptosis. For the first time, we found that, after the mitochondrial protein expression profile is altered, EIAVDLV34-infected cells are transformed into M1-polarized-type macrophages and cause inflammatory injury and that the intrinsic apoptosis pathway is activated in EIAVDLV121-infected cells. These studies shed light on how the mitochondrial protein expression profile changes between cells infected by pathogenic lentivirus strains and cells infected by attenuated lentivirus strains to drive different cellular responses, especially from inflammation to apoptosis.
INTRODUCTION
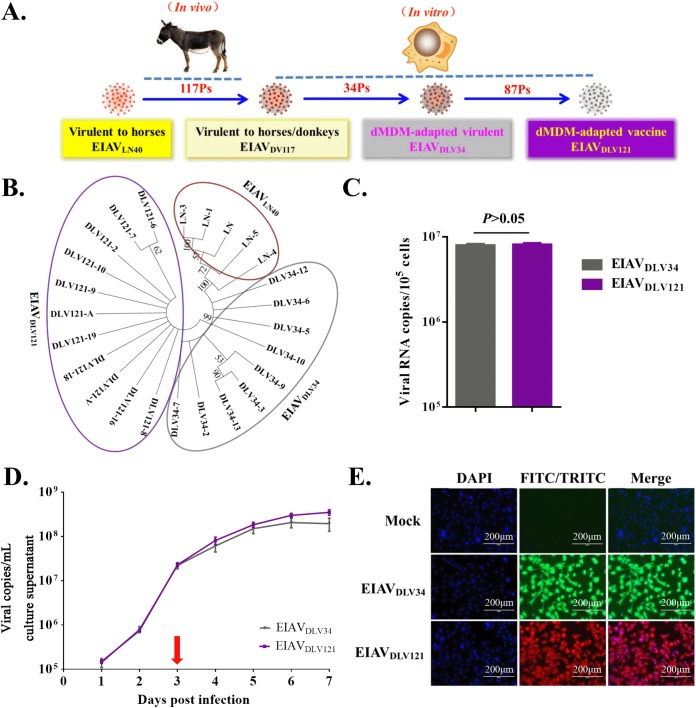
Equine infectious anemia (EIA) is a major infectious disease of equids and is characterized by periodic episodes of fever, thrombocytopenia, anemia, rapid weight loss, and generalized lymphoid proliferation. The etiologic agent of EIA is the equine infectious anemia virus (EIAV). EIAV belongs to the genus Lentivirus in the Retroviridae, a family that also includes the extensively studied human immunodeficiency virus (HIV) (1). There are many striking clinical and pathological similarities between EIA and AIDS caused by HIV infection. An effective live attenuated EIAV vaccine (EIAVDLV121) that protects horses was developed by 117 generations of passaging of an isolated virulent strain (EIAVLN40) in vivo in donkeys (EIAVDV117), followed by 121 generations of passaging in vitro in primary donkey monocyte-derived macrophages (dMDMs) (Fig. 1A). This live attenuated EIAV vaccine is the only effective live lentiviral vaccine for large-scale use in the world and has controlled the prevalence of EIAV in China (2, 3). Therefore, the EIAV attenuated vaccine system serves as a model for the development of other lentivirus vaccines. A better understanding of the host cellular response to infection by these EIAV strains should improve our knowledge regarding lentiviral pathogenicity and the protective mechanism of lentivirus vaccines. EIAVDLV34 is a dMDM-adapted virulent strain that was passaged for 34 generations in dMDMs in vitro from EIAVDV117. As with EIAVDV117, infections with EIAVDLV34 can result in acute EIA. However, the live vaccine EIAVDLV121 lost the virulence to horses. Both EIAVDLV34 and EIAVDLV121 can replicate well in equine monocyte-derived macrophages (eMDMs) in vitro, but there is a 2.3% difference between their proviral genomes. Our previous research found that EIAVDLV34 can induce a host inflammatory storm to cause severe pathological injury but that EIAVDLV121 can induce only a weak inflammatory response. In addition, the apoptosis induced in eMDMs infected with EIAVDLV121 was significantly higher than that induced in eMDMs infected with EIAVDLV34 (4, 5). This finding suggests that the host cellular inflammatory responses and apoptosis are closely related to EIAV pathogenicity and the protective mechanisms of the attenuated vaccine. However, it is still unknown how these two EIAV strains remodel the intracellular environment to induce different levels of the inflammatory response and apoptosis.
FIG 1.
Phylogenetic relationship and replication analyses of EIAVDLV34 and EIAVDLV121. (A) Process of passaging from the pathogenic virus strain to the attenuated vaccine strain. EIAVLN40, an EIAV strain isolated from Liaoning, China, has undergone 16 consecutive passages (Ps) in horses and is highly pathogenic to horses. After 117 consecutive passages in donkeys, this virus was transformed into strain EIAVDV117, which is highly virulent in both horses and donkeys. The virulence of EIAVDV117 gradually decreased after consecutive passages in dMDMs in vitro. In this study, EIAVDLV34 (the 34th generation, which is virulent for horses) and EIAVDLV121 (the 121th generation, which is avirulent and which induces protective immunity) were used for subsequent experiments. (B) Molecular phylogenetic analysis of the EIAVLN40, EIAVDLV34, and EIAVDLV121 sequences acquired through our previous studies. The evolutionary history was inferred by using a maximum likelihood method based on the general time-reversible model. Evolutionary analyses were conducted in MEGA (version 7) software (virus strains of different generations are represented by ellipses with different colors). (C) Comparison of the number of intracellular viral copies in the early phase of infection. The same infectious doses (5 × 103 TCID50/well) of the pathogenic strain EIAVDLV34 and the attenuated EIAV strain EIAVDLV121 were used to infect eMDMs in 24-well microplates. At 3 h postinfection, the number of copies of intracellular EIAV RNA was measured by qPCR. (D) Comparison of the replication kinetics of EIAVDLV34 and EIAVDLV121 in eMDMs. Stocks of these two viruses with an equal TCID50 were used to infect eMDMs, as indicated. Viruses in the culture medium were quantified as viral RNA copy numbers at various time points up to 7 dpi. (E) Detection of eMDMs infected with EIAVDLV34 or EIAVDLV121 using an IFA. eMDMs were infected with EIAVDLV34 or EIAVDLV121, as indicated. The infected cells were then washed with PBS, fixed with 4% paraformaldehyde, and incubated with EIAV-positive horse serum. Cells infected with EIAV were visualized through incubation with FITC- or TRITC-conjugated rabbit anti-horse IgG and examined using fluorescence microscopy. DAPI, 4′,6-diamidino-2-phenylindole.
The inflammatory response is correlated with macrophage polarization, which is a process by which macrophages express different functional programs, especially different mitochondrial metabolic patterns, in response to microenvironmental signals, including viral infection (6). M1-polarized macrophages are able to produce proinflammatory mediators to cause an inflammatory response, but M2-polarized macrophages are able to produce anti-inflammatory factors to dampen inflammatory and Th1 immune responses and promote tissue remodeling and repair (7). Apoptosis is a form of programmed cell death that not only eliminates pathogens but also stimulates an acquired immune response against pathogenic antigens derived from dying cells and presented by dendritic cells (DCs) (this kind of apoptosis is also known as immunogenic apoptosis) (8). Apoptosis can be triggered by two distinct, well-characterized signaling cascades, namely, the extrinsic and intrinsic pathways. In the intrinsic pathway, mitochondria play pivotal roles in driving cell death and inducing immunogenic apoptosis (9).
Here, we performed a comparative proteomics analysis of eMDMs infected either with pathogenic (EIAVDLV34) or with attenuated vaccine (EIAVDLV121) EIAV strains and found that, among all organelles, most of the resulting differentially expressed proteins were localized to mitochondria. Mitochondria are triggers of apoptosis, the transformation of cellular metabolism, and inflammatory polarization of macrophages (6, 10). The role of mitochondria in inducing different levels of the inflammatory response and the apoptosis of host cells infected by the two EIAV strains has become an interesting question. Our present study revealed that, due to alterations in mitochondrial protein (e.g., ACO2, CS, DLST, FIS1, and VDAC1) expression patterns, cells infected with the pathogenic strain EIAVDLV34 showed some characteristics in common with M1-type-polarized proinflammatory cells, such as a high level of glycolysis and increased mitochondrial fragmentation. In contrast, cells infected with the attenuated vaccine strain EIAVDLV121 showed characteristics in common with M2-type-polarized anti-inflammatory cells, including low levels of glycolysis accompanied by a normal tricarboxylic acid (TCA) cycle and increased mitochondrial fusion (6, 11). In addition, EIAVDLV121 infection promoted mitochondrial membrane permeabilization to induce additional cellular apoptosis. Thus, the findings of our study suggest a possible mechanism, based on mitochondrial function, by which the pathogenic and attenuated vaccine EIAV strains can induce different levels of the inflammatory response and the apoptosis of host cells.
RESULTS
EIAVDLV34 and EIAVDLV121 have similar replication curves and infection abilities in vitro.
EIAVLN40 is an EIAV strain that is lethal to horses. EIAVDLV34 and EIAVDLV121 are the dMDM-adapted virulent strain and the live attenuated vaccine strain, respectively (Fig. 1A) (4, 12). The results of a genetic evolution analysis showed that the three EIAV strains are located on different genetic branches on a viral phylogenetic tree (Fig. 1B) and that the differences between the proviral genomes of EIAVLN40 and EIAVDLV34, EIAVLN40 and EIAVDLV121, and EIAVDLV34 and EIAVDLV121 are 2.2%, 2.8%, and 2.3%, respectively. Because EIAVDLV34 and EIAVDLV121 can replicate well in eMDMs in vitro, these two viral strains with different virulence were used in this study. To analyze changes in the protein expression profiles of the host cells caused by the initial invasion of EIAVDLV34 or EIAVDLV121, the infectivity of these two viruses needed to be determined first. The results showed that the numbers of virus copies in the cells at 3 h after infection were 7.918 × 106/105 cells and 8.074 × 106/105 cells, respectively, and that the difference was not significant (P > 0.05) (Fig. 1C).
In cells infected with HIV, at 3 days postinfection (dpi), the production and release of virus particles are rapid, and viral proteins are concentrated in the cytoplasm. Mitochondria and other organelles are severely affected, and cell lesions occur. This period is called the cytopathic phase (13, 14). To assess the replication abilities and cytopathic phases of the pathogenic (EIAVDLV34) and the attenuated (EIAVDLV121) strains in the host cells, eMDMs, the virus copy numbers present in host cells infected with these two strains were detected using real-time quantitative PCR (qPCR) at 1, 2, 3, 4, 5, 6, and 7 days after infection. The dynamic replication curves of EIAVDLV34 and EIAVDLV121 in eMDMs were similar, and as with HIV, at 3 dpi, the release of virus particles was rapid (Fig. 1D). There was no difference in the levels of replication between EIAVDLV34 and EIAVDLV121 from 0 to 3 days after infection, and cell lesions could be observed only after the third day following infection. In addition, immunofluorescence assay (IFA) examination of EIAVDLV34- or EIAVDLV121-infected eMDMs demonstrated that in both cases up to 99% of the cells were infected at 3 days, as determined by incubation of the cells with an anti-EIAV serum and staining with a tetramethyl rhodamine isocyanate (TRITC)- or fluorescein isothiocyanate (FITC)-conjugated rabbit anti-horse IgG antibody (Fig. 1E). The period starting from 3 days postinfection with EIAV was therefore also a cytopathic phase. We then examined the differentially expressed host proteins regulated by EIAVDLV34 and EIAVDLV121 infection at the beginning of the cytotoxic phase.
Infection of eMDMs with EIAVDLV34 or EIAVDLV121 can cause differential mitochondrial protein expression profiles.
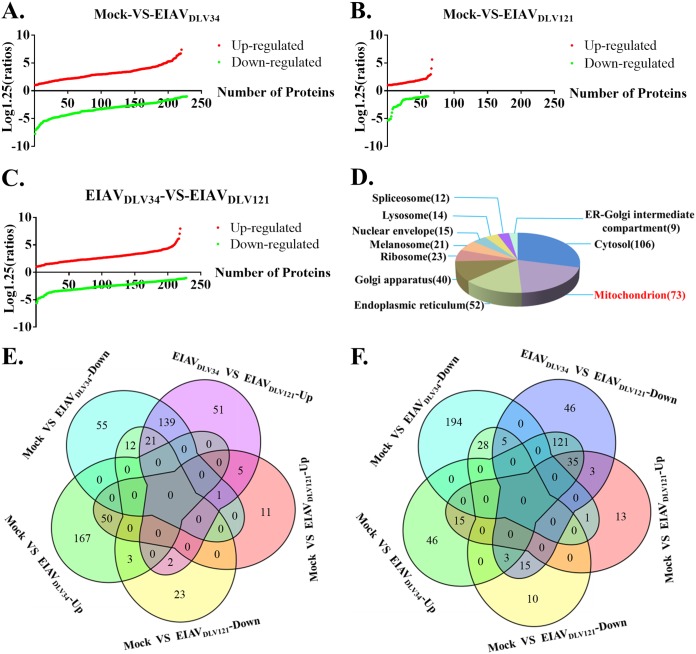
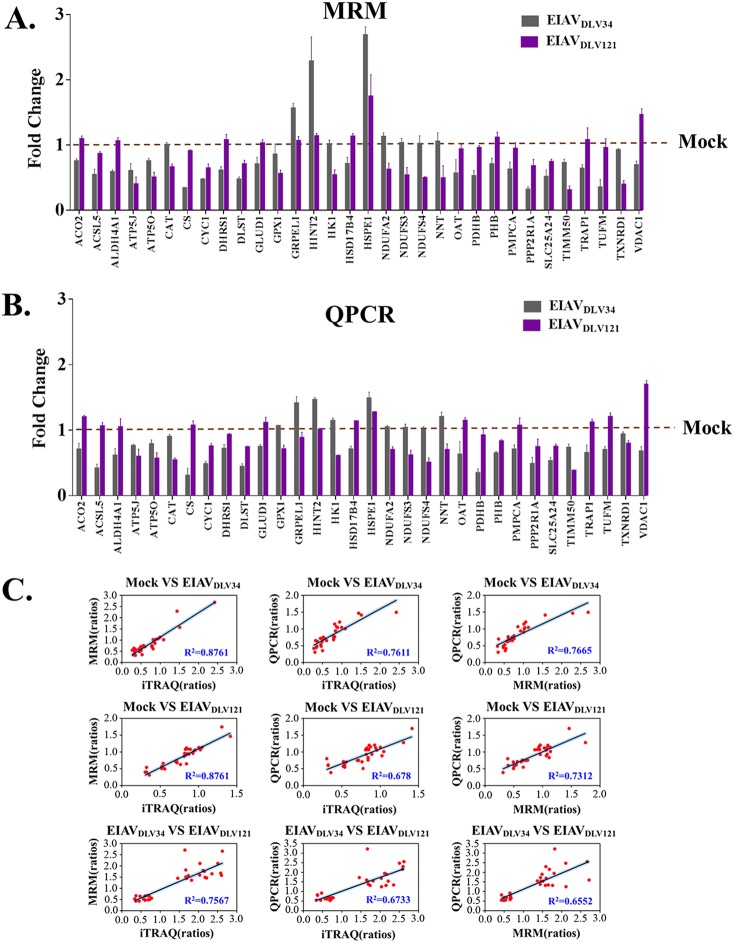
In response to virus infection, cells can alter their patterns of protein expression to adapt their cellular functions and promote or limit viral replication. The use of isobaric tags for relative and absolute quantitation (iTRAQ) is a proteomics isotope labeling quantification technique that enables the analysis of up to eight samples simultaneously in one measurement with favorable accuracy and repeatability (15). To detect changes in protein expression profiles in host cells during the cytopathic phase, three independent comparison groups of eMDMs (a mock- versus EIAVDLV34-infected group, a mock- versus EIAVDLV121-infected group, and a EIAVDLV34- versus EIAVDLV121-infected group) were prepared from horses, with each comparison group containing three flasks of cells that were infected with EIAVDLV34 or EIAVDLV121 or that were mock infected for 3 days. Equal amounts of protein from each of the three flasks of infected cells and three flasks of mock-infected cells were subjected to iTRAQ-coupled two-dimensional (2D) liquid chromatography (LC)-tandem mass spectrometry (MS/MS) analysis. A total of 2,769 proteins were detected. Of these proteins, 448 displayed significant differences in expression levels between EIAVDLV34-infected and mock-infected cells (220 were upregulated and 228 were downregulated) (Fig. 2A; see also Table S1 in the supplemental material), 128 displayed significant differences in expression levels between EIAVDLV121-infected and mock-infected cells (67 were upregulated and 61 were downregulated) (Fig. 2B; Table S2), and 447 displayed significant differences in expression levels between EIAVDLV34-infected and EIAVDLV121-infected cells (219 were upregulated and 228 were downregulated) (Fig. 2C; Table S3). To evaluate the potential roles of the 447 proteins, the proteins were analyzed by Gene Ontology (GO) cellular component (GO-CC) enrichment using the online tool Database for Annotation, Visualization, and Integrated Discovery (DAVID). The results identified differentially expressed proteins that were well distributed throughout different cellular organelles, with mitochondrial proteins being the most common (73 proteins) (Fig. 2D). The upregulated or downregulated proteins in the EIAVDLV34- versus EIAVDLV121-infected comparison group mainly overlapped the downregulated proteins (139/219, 63.5%) or upregulated proteins (121/228, 53.1%) in the mock- versus EIAVDLV34-infected comparison group. These results are illustrated in Venn diagrams (Fig. 2E and F). The iTRAQ data then needed to be validated. Multiple-reaction monitoring (MRM) is a powerful targeted mass spectrometry approach for confident quantification of the proteins/peptides in biological samples. MRM has often been used to verify iTRAQ data in the past (16, 17). Only high-confidence peptides of the target proteins exhibiting rich product ion spectra were selected for MRM (18), so in our experiment, 32 differentially expressed mitochondrial proteins obtained in the iTRAQ analysis were selected for validation by MRM and qPCR (Fig. 3A and B; Table 1; Table S4). The ratios of all these proteins or relevant mRNAs were found to be in agreement with those obtained using iTRAQ analysis (R2 > 0.64) (Fig. 3C), and indeed, the correlation between the iTRAQ and MRM data was better than that between qPCR data and either iTRAQ or MRM data (iTRAQ-MRM R2 > iTRAQ-qPCR R2 or MRM-qPCR R2). This finding may have been due to the translation process between mRNA and protein. However, all these results suggest that the iTRAQ data are reliable. These findings prompted us to further investigate changes in biological function in mitochondria following EIAVDLV34 or EIAVDLV121 infection.
FIG 2.
Comparative proteome analysis of EIAVDLV34-infected cells, EIAVDLV121-infected cells, and uninfected cells obtained through iTRAQ analysis. (A) A cumulative plot of differential protein expression ratios (log1.25 ratio) and the quantity of differentially expressed proteins between mock- and EIAVDLV34-infected cells. In total, there were 220 upregulated proteins with ratios of between 1.261 and 5.2 and 228 downregulated proteins with ratios of between 0.178 and 0.798. (B and C) The cumulative plots are similar to those in panel A, but the comparison groups are mock- versus EIAVDLV121-infected cells (B) and EIAVDLV34- versus EIAVDLV121-infected cells (C). In the mock- versus EIAVDLV121-infected comparison group, there were 67 upregulated proteins with ratios of between 1.252 and 3.508 and 61 downregulated proteins with ratios of between 0.305 and 0.799. In the EIAVDLV34- versus EIAVDLV121-infected comparison group, there were 219 upregulated proteins with ratios of between 1.256 and 5.935 and 228 downregulated proteins with ratios of between 0.284 and 0.793. (D) A pie chart of enriched GO-CC terms in the EIAVDLV34- versus EIAVDLV121-infected comparison group (P < 0.05). The areas with different colors represent different terms. ER, endoplasmic reticulum. (E) Venn diagram showing upregulated proteins from the EIAVDLV34- versus EIAVDLV121-infected comparison group with both upregulated and downregulated proteins from the mock- versus EIAVDLV34-infected and mock- versus EIAVDLV121-infected comparison groups. (F) Venn diagram showing downregulated proteins from the EIAVDLV34- versus EIAVDLV121-infected comparison group with both upregulated and downregulated proteins from the mock- versus EIAVDLV34-infected and mock- versus EIAVDLV121-infected comparison groups.
FIG 3.
MRM and qPCR verification of proteins determined by iTRAQ to be differentially expressed. (A) Alteration of the level of translation of 32 selected mitochondrial genes in eMDMs following EIAVDLV34 and EIAVDLV121 infection was examined at 3 dpi by quantitative analysis of the corresponding protein expression levels in cells infected with either EIAVDLV34 or EIAVDLV121 or in mock-infected cells. Total protein was extracted from eMDMs and reduced, alkylated, and digested with trypsin for subsequent analysis via MRM. Fold change values were calculated by using β-galactosidase for data normalization. Error bars represent the standard error from three independent experiments. (B) Alteration of the levels of transcription of 32 selected mitochondrial genes in eMDMs following EIAVDLV34 and EIAVDLV121 infection was examined by quantitative analysis of the corresponding mRNA expression levels at 3 dpi in cells infected with either EIAVDLV34 or EIAVDLV121 or mock-infected cells. Total RNA was extracted from eMDMs and reverse transcribed into cDNA for subsequent analysis via qPCR. Fold change values were calculated according to the 2−ΔΔCT method using β-actin as an internal reference gene. Error bars represent the standard error from three independent experiments. (C) Linear regression analysis of fold change ratios among iTRAQ, MRM, and qPCR in the mock- versus EIAVDLV34-infected, mock- versus EIAVDLV121-infected, and EIAVDLV34- versus EIAVDLV121-infected comparison groups. Red dots represent fold change ratios of a single gene obtained from iTRAQ and MRM analyses, iTRAQ and qPCR analyses, or MRM and qPCR analyses. R2, correlation coefficient.
TABLE 1.
Representative protein quantitative confirmation with MRM analysis
| UniProt accession no. | Gene symbol | Signature peptide(s)a | Ratiob
|
|||||
|---|---|---|---|---|---|---|---|---|
| Mock- vs EIAVDLV34-infected cells |
Mock- vs EIAVDLV121-infected cells |
|||||||
| Replicate 1 | Replicate 2 | Replicate 3 | Replicate 1 | Replicate 2 | Replicate 3 | |||
| F6Z4G8 | ACO2 | EGWPLDIR | 0.738493 | 0.792729 | 0.7350298 | 1.125217 | 1.11204 | 1.044094 |
| IVYGHLDDPANQEVER | ||||||||
| SQFTITPGSEQIR | ||||||||
| F6VV97 | ACSL5 | ASLLLENVEQGLAPGLK | 0.610584 | 0.45371 | 0.5771874 | 0.862167 | 0.838103 | 0.898917 |
| EAILEDLQK | ||||||||
| TLKPTVFPTVPR | ||||||||
| F6SD96 | ALDH4A1 | STGSVVGQQPFGGAR | 0.563822 | 0.624197 | 0.5791162 | 1.006595 | 1.087852 | 1.091489 |
| F6R3B2 | ATP5J | FEVFDKPQS | 0.60736 | 0.494156 | 0.714918 | 0.451124 | 0.279139 | 0.476522 |
| F6WLI6 | ATP5O | LVRPPVQVYGVEGR | 0.775607 | 0.712896 | 0.7796438 | 0.430852 | 0.505203 | 0.581443 |
| YATALYSAASK | ||||||||
| F7D854 | CAT | FSTVAGESGSADTVR | 1.02296 | 1.0248 | 0.96889 | 0.70658 | 0.66255 | 0.61595 |
| GAGAFGYFEVTHDITR | ||||||||
| LSQEDPDYGLR | ||||||||
| F6XMS1 | CS | DILADLIPK | 0.344118 | 0.341405 | 0.3388594 | 0.903011 | 0.928871 | 0.895671 |
| EEQVSWLSK | ||||||||
| GLVYETSVLDPDEGIR | ||||||||
| F6VRY7 | CYC1 | LSDYFPKPYPNPEAAR | 0.489579 | 0.471214 | 0.4662409 | 0.702809 | 0.660021 | 0.58153 |
| F6Q1B4 | DHRS1 | DVDGRPVDDYFSLSSVLTR | 0.65864 | 0.554938 | 0.6237483 | 1.105246 | 0.988203 | 1.143457 |
| F7D407 | DLST | ASAFALQEQPVVNAVIDDATK | 0.502914 | 0.485225 | 0.4418177 | 0.746484 | 0.645485 | 0.736709 |
| GLVVPVIR | ||||||||
| F6V4L4 | GLUD1 | HGGTIPIVPTAEFQDR | 0.755184 | 0.587322 | 0.7791074 | 1.070937 | 0.977633 | 1.042882 |
| YSTDVSVDEVK | ||||||||
| F7CTU2 | GPX1 | YVRPGGGFEPNFTLFEK | 0.893048 | 0.686364 | 0.9939122 | 0.619322 | 0.516332 | 0.552669 |
| F6Z2R9 | GRPEL1 | DLLEVADILEK | 1.648878 | 1.517436 | 1.529793 | 0.996227 | 1.080623 | 1.122609 |
| TLRPALVGVVK | ||||||||
| F7D5N4 | HINT2 | AQQAAPGGAAPTIFSR | 2.239801 | 1.942788 | 2.678114 | 1.180046 | 1.128814 | 1.109409 |
| ISQAEEGDQQLLGHLLLVAK | ||||||||
| F7BA85 | HK1 | GAALITAVGVR | 1.078111 | 1.006776 | 0.9610786 | 0.592127 | 0.452369 | 0.578759 |
| GDFIALDLGGSSFR | ||||||||
| TTVGVDGSLYK | ||||||||
| Q6RFZ6 | HSD17B4 | AYALAFGER | 0.76403 | 0.600985 | 0.7742777 | 1.107486 | 1.114815 | 1.18025 |
| IDVVVNNAGILR | ||||||||
| VVLVTGAGGGLGR | ||||||||
| F6T0A6 | HSPE1 | VLLPEYGGTK | 2.615825 | 2.624205 | 2.825523 | 1.995209 | 1.875442 | 1.372466 |
| VLQATVVAVGAGSK | ||||||||
| F6TC26 | NDUFA2 | NVSLNNFSADQVTR | 1.178114 | 1.142998 | 1.071363 | 0.708893 | 0.646 | 0.526427 |
| F6ZUJ7 | NDUFS3 | QLSAFGEYVAEILPK | 0.96614 | 1.03945 | 1.09726 | 0.424427 | 0.536103 | 0.658891 |
| SLADLTAVDIPTR | ||||||||
| F6WY50 | NDUFS4 | LDITTLTGVPEEHIK | 1.116358 | 1.060028 | 0.8917303 | 0.49342 | 0.48193 | 0.51442 |
| F6YLK4 | NNT | DGEVIFPAPTPK | 1.169349 | 0.917671 | 1.084394 | 0.331125 | 0.696631 | 0.453741 |
| FFTGQITAAGK | ||||||||
| F7C9Y5 | OAT | YGAHNYHPLPVALER | 0.708779 | 0.664383 | 0.3310578 | 0.867868 | 1.016551 | 0.9242 |
| F7A900 | PDHB | VFLLGEEVAQYDGAYK | 0.476853 | 0.619866 | 0.4847714 | 0.942635 | 0.944462 | 0.99394 |
| F6SXD7 | PHB | DLQNVNITLR | 0.620495 | 0.790164 | 0.7193452 | 1.163856 | 1.025061 | 1.160822 |
| IFTSIGEDYDER | ||||||||
| ILFRPVASQLPR | ||||||||
| F7BJX7 | PMPCA | SVAQYTGGIVK | 0.667709 | 0.500217 | 0.7151338 | 1.045826 | 0.88836 | 0.90953 |
| F7A0N8 | PPP2R1A | IGPILDNSTLQSEVKPILEK | 0.345751 | 0.276728 | 0.3418563 | 0.668381 | 0.584462 | 0.782535 |
| F6Z4N2 | SLC25A24 | QLLAGGVAGAVSR | 0.626364 | 0.493779 | 0.4313095 | 0.760115 | 0.77087 | 0.699144 |
| F6W8S4 | TIMM50 | QNLFFGSLTSR | 0.752485 | 0.76455 | 0.6664354 | 0.318215 | 0.368007 | 0.248898 |
| TVLEHYALEDDPLEAFK | ||||||||
| VLLDLSAFLK | ||||||||
| F7E2L8 | TRAP1 | AQLLQPTLEINPR | 0.696062 | 0.587367 | 0.6449818 | 1.18362 | 1.184695 | 0.867437 |
| DVGEWQQEEFYR | ||||||||
| GVVDSEDIPLNLSR | ||||||||
| F7E244 | TUFM | DLEKPFLLPVESVYSIPGR | 0.234509 | 0.469309 | 0.3555572 | 0.82113 | 0.962361 | 1.092915 |
| GITINAAHVEYSTAAR | ||||||||
| LLDAVDTYIPVPTR | ||||||||
| F6YLA3 | TXNRD1 | LELTPVAIQAGR | 0.910252 | 0.945704 | 0.9218643 | 0.433081 | 0.426849 | 0.329499 |
| VVGFHVLGPNAGEVTQGFAAALK | ||||||||
| F6PQT9 | VDAC1 | GYGFGLIK | 0.741101 | 0.711678 | 0.6336383 | 1.427767 | 1.397498 | 1.567344 |
| VTQSNFAVGYK | ||||||||
| WTEYGLTFTEK | ||||||||
Signature peptides for the target proteins were defined based on iTRAQ data, and their uniqueness was examined with a BLAST search against the proteome with Equus caballus UniProt accession number UP000002281. Only unique peptide sequences were selected for use in the MRM assays.
The ratio of expression of the protein between the two samples.
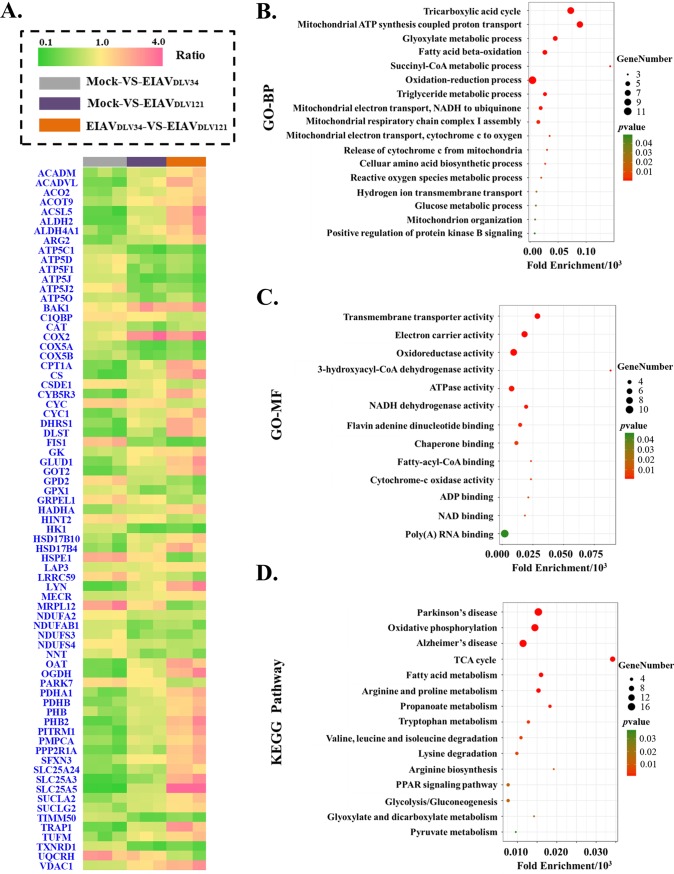
There were 45 downregulated and 12 upregulated mitochondrial proteins in the mock- versus EIAVDLV34-infected comparison group, 27 downregulated and 8 upregulated mitochondrial proteins in the mock- versus EIAVDLV121-infected comparison group, and 30 downregulated and 43 upregulated mitochondrial proteins in the EIAVDLV34- versus EIAVDLV121-infected comparison group. Each group represents three biological repeats (Fig. 4A). The 73 differentially expressed mitochondrial proteins in the EIAVDLV34- versus EIAVDLV121-infected comparison group were analyzed by GO biological process (GO-BP), GO molecular function (GO-MF), and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment to identify any enriched biological function or pathway themes. The results showed that the TCA cycle, the oxidation-reduction process, mitochondrial ATP synthesis-coupled proton transport, electron carrier activity, NADH dehydrogenase activity, and oxidative phosphorylation (OXPHOS) were the biological functions and pathways that were the most heavily affected in cells of the EIAVDLV34- versus EIAVDLV121-infected comparison group (Fig. 4B to D). Further biological function experiments were carried out to verify these bioinformatics predictions.
FIG 4.
GO and KEGG pathway enrichment results of differentially expressed mitochondrial proteins. (A) The fold change ratios of differentially expressed mitochondrial proteins are depicted as a heat map. The different comparison groups are highlighted along the bottom of the heat map and include the mock- versus EIAVDLV34-infected (gray), mock- versus EIAVDLV121-infected (purple), and EIAVDLV34- versus EIAVDLV121-infected (orange) comparison groups. Every comparison group contained three biological replicates. Each horizontal line represents a protein of one biological replicate, and the color (green, low fold change ratios; yellow, no change; pink, high fold change ratios) represents the scaled ratios. (B) Analysis of GO biological processes (GO-BP) in the bubble chart; (C) analysis of GO molecular functions (GO-MF) in the bubble chart; (D) KEGG pathway enrichment analysis in the bubble chart. P values (i.e., EASE scores) were calculated using a Benjamini-corrected modified Fisher’s exact test. EASE scores of <0.05 indicate statistical significance. Fold enrichment gives a measure of the magnitude of enrichment, and fold enrichments of 1.5 and above were considered interesting. PPAR, peroxisome proliferator-activated receptor.
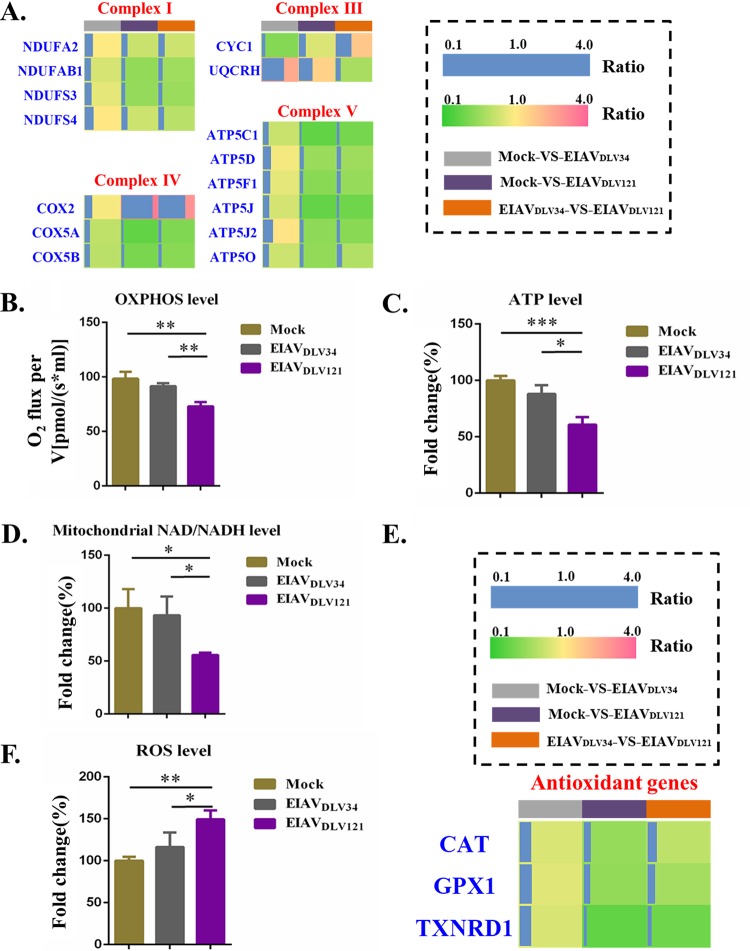
EIAVDLV34 and EIAVDLV121 infection can induce different effects on cellular OXPHOS and redox systems to produce different levels of NAD/NADH, ATP, and ROS.
Mitochondria are the primary site of reactive oxygen species (ROS) production in the cell; specifically, the electron transport chain (ETC; composed of five complexes) is a major source of ROS production by complexes I and III (19). Our data showed the differential expression of certain proteins from complexes I, III, IV, and V but not complex II in cells infected with EIAVDLV34 compared with those infected with EIAVDLV121 (Fig. 5A). Compared with mock-infected cells, cells infected with EIAVDLV34 showed no changes in the expression levels of the complex I (NADH dehydrogenase) proteins NDUFA2, NDUFAB1, NDUFS3, and NDUFS4, but the expression levels of all four of these proteins decreased in cells infected with EIAVDLV121 compared with those in mock-infected cells. Decreases in the expression levels of the subunit proteins NDUFA2, NDUFAB1, NDUFS3, and NDUFS4 from complex I would lead to decreases in the ratio of mitochondrial NAD/NADH and also in ATP synthesis and an increase in ROS production (20–24). Complex III (cytochrome bc1) transfers electrons from ubiquinol to cytochrome c (Cyto C) to form an electrochemical gradient across the mitochondrial inner membrane. UQCRH, also known as mitochondrial hinge protein, represents subunit 6 of complex III. It is essential for the formation of the complex between cytochrome c1 (CYC1) and Cyto C (25, 26). In comparison with the expression levels in mock-infected cells, we found that UQCRH expression levels increased 2.51-fold in cells infected with EIAVDLV34 and 1.57-fold in cells infected with EIAVDLV121, while CYC1 expression levels decreased 0.46-fold following EIAVDLV34 infection but showed no significant differences in cells infected with EIAVDLV121. Abnormal expression of UQCRH or CYC1 could result in the loss of OXPHOS, leading to the generation of excess ROS (27, 28). Complex IV (cytochrome c oxidase) is the terminal enzyme in the mitochondrial respiratory chain and comprises 13 subunits. It has been reported that increased expression of COX2 and abnormal expression of other subunits of complex IV disturb the activity of complex IV upon HIV-induced T-cell apoptosis (29). Our results showed a 3.50-fold increase in the expression of COX2 in cells infected with EIAVDLV121 compared to that in mock-infected cells, but there were no significant differences in COX2 expression in the mock- versus EIAVDLV34-infected comparison group. The expression of COX5A and COX5B decreased 0.72-fold and 0.70-fold, respectively, in cells infected with EIAVDLV34 and 0.31- and 0.42-fold, respectively, in cells infected with EIAVDLV121 compared with that in mock-infected cells. Complex V (ATP synthase) is a membrane-bound macromolecular complex of 625 kDa that is responsible for the synthesis of ATP, the last step in OXPHOS. Abnormal expression of protein subunits has been reported to cause ATP synthetase to work ineffectively (30–33). We found that ATP5C1 and ATP5O expression levels decreased 0.76-fold and 0.78-fold, respectively, in cells infected with EIAVDLV34, but no differential expression of other protein subunits was observed, while in cells infected with EIAVDLV121, the expression levels of the proteins ATP5C1, ATP5D, ATP5F1, ATP5J, ATP5J2, and ATP5O decreased 0.31-fold, 0.56-fold, 0.48-fold, 0.32-fold, 0.50-fold, and 0.52-fold, respectively. The expression levels of these proteins were therefore downregulated in cells infected with EIAVDLV121 compared with those in cells infected with EIAVDLV34. Overall, the abnormal expression of complexes I, III, IV, and V could inhibit OXPHOS function (Fig. 5B) and ATP synthesis (Fig. 5C), and the downregulation of the expression of complex I could also disturb NADH dehydrogenation (Fig. 5D).
FIG 5.
Assay of changes in mitochondrial OXPHOS and ROS in eMDMs after EIAVDLV34 or EIAVDLV121 infection. (A) The fold change ratios of differentially expressed proteins from mitochondrial respiratory complexes are depicted as a heat map. The different comparison groups are highlighted along the bottom of the heat map and include the mock- versus EIAVDLV34-infected (gray), mock- versus EIAVDLV121-infected (purple), and EIAVDLV34- versus EIAVDLV121-infected (orange) comparison groups. Each horizontal line represents the mean ratio for three biological replicates of one protein, and the color (green, low fold change ratios; yellow, no change; pink, high fold change ratios) represents the scaled ratios. The length of the blue line also represents the scaled ratios. (B to D) Assays of mitochondrial OXPHOS, ATP, and NAD/NADH levels in eMDMs infected with either EIAVDLV34 or EIAVDLV121 or mock-infected eMDMs. V, unit volume. (E) The fold change ratios of the differentially expressed proteins from the antioxidant system are depicted in a heat map similar to that shown in panel A. (F) Assays of mitochondrial ROS levels in eMDMs infected with either EIAVDLV34 or EIAVDLV121 or mock-infected eMDMs. Significant differences between the different groups were determined using Student's t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Error bars represent the standard error from three independent experiments.
Complexes I and III are the main generators of mitochondrial ROS, and when they work ineffectively, additional ROS are produced. Our data show that EIAVDLV121 infection but not EIAVDLV34 infection leads to decreases in the mitochondrial NAD/NADH ratio, the rate at which OXPHOS consumes oxygen, and ATP content. ROS are highly reactive molecules that can damage cellular components, such as nucleic acids, lipids, and proteins, and that can therefore alter their functions. Eukaryotic cells are equipped with an antioxidant system that serves to counterbalance the effect of ROS (34). The expression of the key antioxidant enzymes catalase (CAT), glutathione peroxidase 1 (GPX1), and thioredoxin reductase 1 (TXNRD1) was downregulated 0.54-fold, 0.52-fold, and 0.30-fold, respectively, in cells infected with EIAVDLV121 compared with that in mock-infected cells (Fig. 5E), suggesting that the antioxidant system is unable to work efficiently in these cells and is not able to balance the oxidation-reduction environment (Fig. 5F).
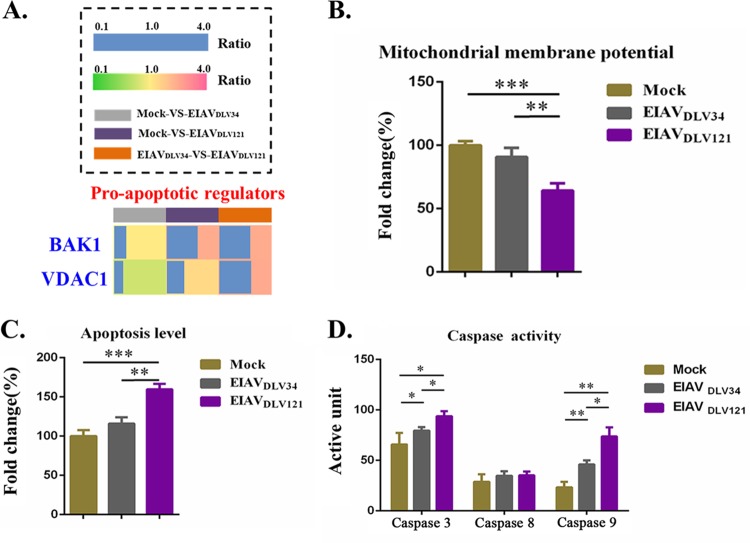
Compared with EIAVDLV34 infection, EIAVDLV121 infection can cause a decrease in Δψm and a higher rate of apoptosis.
Voltage-dependent anion channel (VDAC) proteins comprise a family of evolutionarily conserved ion channels (permeability pores) that control the movement of ATP/ADP, NADH, and apoptotic factors across the mitochondrial outer membrane (35). Overexpression of VDAC1 promoted its self-oligomerization to increase the formation of permeability pores and induce apoptosis (36–38). Moreover, if the number of BAK1 molecules (one of the proapoptotic Bcl-2 family of proteins) bound to the permeability pores increases and thus dissipates the mitochondrial membrane potential (Δψm), additional apoptosis activated by the intrinsic caspase 9-caspase 3 pathway occurs (39). Compared with mock-infected cells, those infected with EIAVDLV121 showed 2.44-fold and 1.42-fold increases in the expression levels of BAK1 and VDAC1, respectively. However, in cells infected with EIAVDLV34, the expression levels of VDAC1 decreased 0.79-fold but the levels of expression of BAK1 remained unchanged (Fig. 6A). As a result, Δψm decreased further, resulting in higher rates of apoptosis in cells infected with EIAVDLV121 than in either uninfected cells or those infected with EIAVDLV34 (Fig. 6B and C). Furthermore, infection with both EIAVDLV34 and EIAVDLV121 induced activation of the caspase 9-caspase 3 intrinsic mitochondrial apoptotic pathway, but EIAVDLV121 infection induced higher levels of activation of the caspase 9-caspase 3 pathway than EIAVDLV34 infection (Fig. 6D).
FIG 6.
Assay of eMDM apoptosis induced by the dissipation of Δψm following infection with EIAVDLV34 or EIAVDLV121. (A) The fold change ratios of BAK1 and VDAC1 expression are depicted in a heat map similar to that shown in Fig. 5A. (B to D) Assays of Δψm (B), apoptosis levels (C), and caspase activity (D) in eMDMs infected with either EIAVDLV34 or EIAVDLV121 or mock-infected eMDMs. Significant differences between the different groups were determined using Student's t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Error bars represent the standard error from three independent experiments.
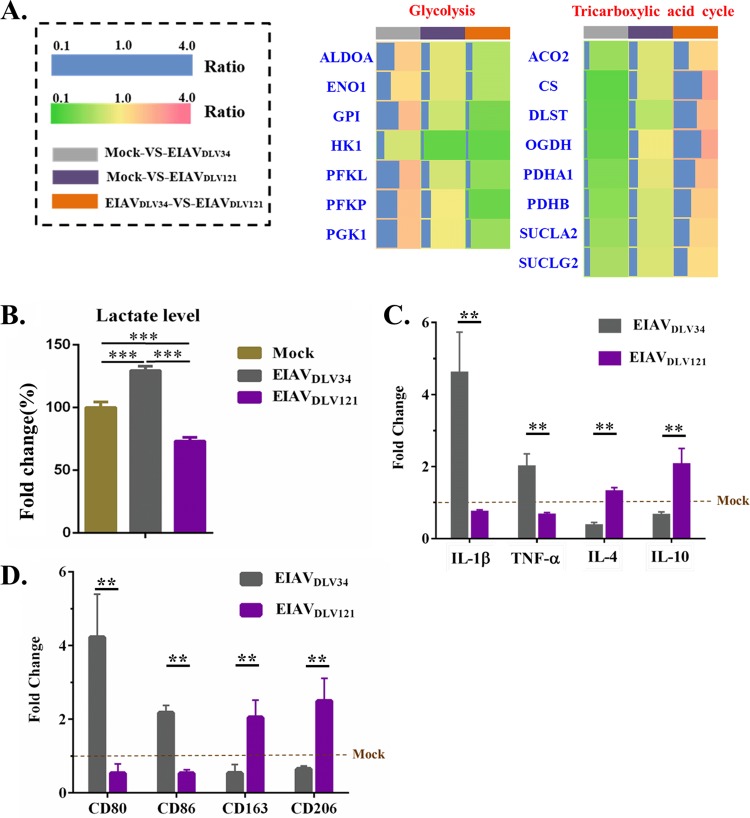
EIAVDLV34 infection can promote conversion of the cellular metabolic pattern to glycolysis, but EIAVDLV121 infection can inhibit glycolysis and maintain the function of the TCA cycle.
Reprogramming of intracellular metabolism is indispensable to the proper polarization and function of activated macrophages and can be used as a marker of macrophage polarization. M1 macrophages increase glycolysis levels and the release of lactate, whereas M2 macrophages employ mainly TCA pathways (7). In our study, at 3 days after EIAVDLV34 infection, the expression levels of the cellular glycolysis-related enzymes ALDOA, ENO1, glycosylphosphatidylinositol (GPI), PFKL, PFKP, and PGK1 increased 1.73-fold, 1.38-fold, 2.08-fold, 2.15-fold, 1.96-fold, and 1.94-fold, respectively. However, after EIAVDLV121 infection, the expression levels of these 6 enzymes remained unchanged, although another glycolysis-related enzyme, HK1, showed a 0.32-fold decrease in expression level. Interestingly, after EIAVDLV34 infection, the expression levels of enzymes related to the cellular TCA pathway, such as ACO2, CS, DLST, OGDH, PDHA1, PDHB, SUCLA2, and SUCLG2, decreased 0.56-fold, 0.30-fold, 0.33-fold, 0.35-fold, 0.45-fold, 0.50-fold, 0.58-fold, and 0.64-fold, respectively. In cells infected with EIAVDLV121, there were no significant differences in the expression levels of these TCA pathway-related enzymes, with the exception of DLST, which showed a 0.70-fold downregulation (Fig. 7A). From the expression profiles of these enzymes, we conclude that there are higher levels of glycolysis and a less effective TCA pathway in cells infected with EIAVDLV34 than in either those infected with EIAVDLV121 or uninfected cells. Furthermore, cells infected with EIAVDLV34 produced more lactate than uninfected cells, but the EIAVDLV121-infected cells produced lower levels of lactate than uninfected cells (Fig. 7B). This effect was due to the downregulation of expression of the subunits PDHA1 and PDHB of pyruvate dehydrogenase (PDH) in cells infected with EIAVDLV34. PDH promotes the overall conversion of pyruvate to acetyl coenzyme A (acetyl-CoA) but inhibits the conversion of glycolysis metabolites to lactate. Acetyl-CoA is the primary link between glycolysis and the TCA cycle. Otherwise, the first step in glycolysis metabolism is the conversion of glucose to glucose-6-phosphate (G-6-P) by hexokinases (HKs), so the downregulation of HK1 expression in EIAVDLV121-infected cells was a direct contributor to the reduction in the levels of glycolysis. In conclusion, cells infected with EIAVDLV34 exhibited metabolic characteristics of M1 macrophages, but EIAVDLV121-infected cells exhibited characteristics of M2 macrophages.
FIG 7.
Metabolic patterns in eMDMs following infection with EIAVDLV34 or EIAVDLV121. (A) The fold change ratios of differentially expressed proteins involved in the TCA cycle and glycolysis are depicted in a heat map similar to that shown in Fig. 5A. (B) Assay of lactate levels in eMDMs infected with EIAVDLV34 or EIAVDLV121 or mock-infected eMDMs. (C) Alteration of the levels of transcription of proinflammatory cytokines (IL-1β and TNF-α) and anti-inflammatory cytokines (IL-4 and IL-10) in eMDMs following EIAVDLV34 and EIAVDLV121 infection was examined by qPCR analysis of the corresponding mRNA expression levels at 3 dpi in cells infected with either EIAVDLV34 or EIAVDLV121 or mock-infected cells. The qPCR method used here is similar to that described in the legend to Fig. 3B. (D) Alteration of the levels of transcription of M1 phenotypic markers (CD80 and CD86) and M2 phenotypic markers (CD163 and CD206) in eMDMs following EIAVDLV34 and EIAVDLV121 infection was examined by qPCR analysis of the corresponding mRNA expression levels at 3 dpi in cells infected with either EIAVDLV34 or EIAVDLV121 or mock-infected cells. The qPCR method used here is similar to that described in the legend to Fig. 3B. Significant differences between the different groups were determined using Student's t test. **, P < 0.01; ***, P < 0.001. Error bars represent the standard error from three independent experiments.
To assess the polarization phenotypes of macrophages, we investigated the changes in the levels of expression of proinflammatory cytokines (intreleukin-1β [IL-1β] and tumor necrosis factor alpha [TNF-α]), anti-inflammatory cytokines (IL-4 and IL-10), M1 phenotypic markers (CD80 and CD86), and M2 phenotypic markers (CD163 and CD206) (40, 41). After EIAVDLV34 infection, the expression levels of IL-1β, TNF-α, CD80, and CD86 showed 4.58-fold, 1.98-fold, 4.22-fold, and 2.17-fold upregulation, respectively, and the expression levels of IL-4, IL-10, CD163, and CD206 showed 0.34-fold, 0.63-fold, 0.54-fold, and 0.65-fold downregulation, respectively (Fig. 7C and D). After EIAVDLV121 infection, the expression levels of IL-1β, TNF-α, CD80, and CD86 showed 0.72-fold, 0.64-fold, 0.53-fold, and 0.53-fold downregulation, respectively, and the expression levels of IL-4, IL-10, CD163, and CD206 showed 1.29-fold, 2.04-fold, 2.04-fold, and 2.49-fold upregulation, respectively (Fig. 7C and D). These results, together with the data on metabolic characteristics, indicate that macrophages transformed into the M1 type upon EIAVDLV34 infection and into the M2 type upon EIAVDLV121 infection.
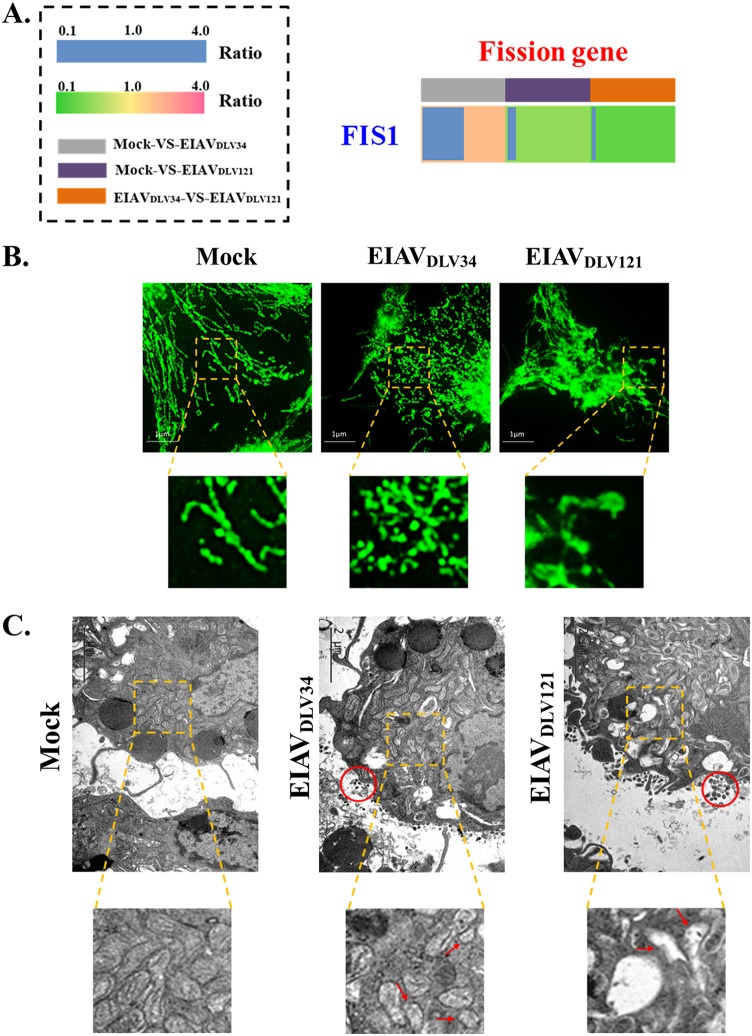
Differential expression of FIS1 in cells infected with EIAVDLV34 and EIAVDLV121 causes different mitochondrial dynamics.
The functions of mitochondria also depend both on their distribution inside the cell and on the morphology of the membrane network determined by the balance between fusion and fission events (42). Moreover, macrophages can be transformed into M1- and M2-polarized types by activators through the promotion of mitochondrial fission and fusion, respectively (11). Mitochondrial dynamics (fusion and fission) can be influenced by physiological alterations to the cellular environment during viral pathogenesis (43, 44).
Mitochondrial fission protein 1 (FIS1) is part of the mitochondrial fission machinery and plays a key role in the regulation of fission processes. Overexpression of FIS1 in various cell types leads to the protein being localized to the mitochondrial outer membrane and promotion of mitochondrial fission, and inhibition of this protein can lead to mitochondrial fusion (45). Following infection with EIAVDLV34, cellular FIS1 expression levels increased 2.08-fold (Fig. 8A), promoting mitochondrial fission and fragmentation in eMDMs (Fig. 8B and C). However, in cells infected with EIAVDLV121, FIS1 expression levels decreased 0.53-fold (Fig. 8A), leading to more mitochondrial fusion, and because Δψm was dissipated in these cells, the mitochondrial structures, including the cristae, were destroyed (Fig. 6B and 8B and C).
FIG 8.
Analysis of mitochondrial dynamics in eMDMs following EIAVDLV34 or EIAVDLV121 infection. (A) The fold change ratio of the key mitochondrial fission protein FIS1 is depicted in a heat map similar to that shown in Fig. 5A. (B) 2D-SIM immunofluorescence analysis of morphological changes in mitochondria in cells infected with EIAVDLV34 or EIAVDLV121. At 3 dpi, eMDMs infected by either EIAVDLV34 or EIAVDLV121 or mock-infected eMDMs were immunostained with antibodies specific to the translocase of the mitochondrial outer membrane 20 (TOM20; green). The zoomed images display the typical tubular mitochondrial network in uninfected cells, fragmented mitochondria in EIAVDLV34-infected cells, and expanded permeable mitochondria in EIAVDLV121-infected cells. (C) Electron micrographs of morphological changes in mitochondria in EIAVDLV34- and EIAVDLV121-infected cells. The ultrastructure of uninfected and infected (with EIAVDLV34 or EIAVDLV121) cells was examined by electron microscopy as described in Materials and Methods. In the zoomed images, normal elongated tubular mitochondria can be seen in uninfected cells, fragmented mitochondria can be seen in EIAVDLV34-infected cells, and expanded mitochondria with the loss of cristae can be seen in EIAVDLV121-infected cells. Red circles indicate the viral particles.
DISCUSSION
Previous research reports that infection with different viruses can drive host cells to execute distinct, virus-specific metabolic programs or mitochondrial dynamics to support or inhibit virus production (43, 46). However, to date, there has been no work on whether infection with strains of a single virus differing in virulence can cause different changes in mitochondria. We performed proteomic analysis to detect proteins expressed differentially between eMDMs infected with EIAVDLV34 and those infected with EIAVDLV121. Values obtained through the iTRAQ proteomic method have been found to be highly reproducible across biological repeats (47). In addition, the reproducibility of quantitative experiments very much depends on the standardization protocol followed during sample preparation. At least three replicates are recommended for the experiment (48). In our study, all the experiments, including iTRAQ and the verification experiments (MRM and qPCR), were carried out with three independent biological repeats (Table 1 and Fig. 3A and B; see also Tables S1 to S3 in the supplemental material). The results obtained by MRM and qPCR were all found to be well in agreement with those obtained using iTRAQ analysis (R2 > 0.64), indicating a good reproducibility of our experiments (Fig. 3C). Our results suggest that strains of EIAV with different virulence can cause different expression profiles of mitochondrial proteins in eMDMs. Furthermore, we demonstrated that these infections can cause different changes in mitochondrion-related biological functions. Mitochondrial biogenesis, fusion, and fission also have important roles in aspects of immune cell activation. More importantly, metabolic patterns are extremely relevant to immunity and inflammation in both innate and adaptive immune cells (49). We found that infection with the pathogenic virus strain EIAVDLV34 can cause a very strong cellular inflammatory reaction but that infection with the attenuated vaccine strain EIAVDLV121 does not and that these two strains of virus can induce different levels of apoptosis of eMDMs (Fig. 6C and 7C). Moreover, the proteomics and functional analysis data suggest that mitochondria play a key role in the regulation of the processes mentioned above.
It is generally accepted that a reduction in the levels of mitochondrial NAD/NADH stimulates a number of proapoptotic pathways, including the relocalization of apoptosis-inducing factor (AIF) from the outer mitochondrial membrane to the nucleus (50). In the cells infected with EIAVDLV121, the expression of four subunits of NADH dehydrogenase decreased (Fig. 5A), the activity of this complex enzyme decreased correspondingly, and NADH could not be effectively dehydrogenated into NAD (Fig. 5D). The low levels of NAD/NADH may have resulted in an increase in the rate of apoptosis in infected cells (Fig. 6C). NADH dehydrogenase can also produce ROS at the NADH-oxidizing flavin site, IF (51), which occurs when electrons leak to O2 from the fully reduced flavin. The level of steady-state reduction of NAD/NADH in situ predicts the rate of ROS production from site IF (52). Thus, the NAD/NADH ratio decreased in cells infected with EIAVDLV121, which was accompanied by a steep increase in the rate of ROS production attributed to site IF in complex I (Fig. 5F). Moreover, complex III is another main source of mitochondrial ROS generation. In complex III, ROS arise from the quinol-oxidizing site (site IIIQo), and the abnormal expression of UQCRH or CYC1 (in both EIAVDLV34- and EIAVDLV121-infected cells) could block electron transfer from cytochrome b to the quinone at center i, causing a small buildup of semiquinone at center o, which increases the conversion of O2 to superoxide (53) (Fig. 5F).
Organisms have integrated antioxidant systems that are usually effective in blocking the harmful effects of ROS. The key antioxidant enzyme CAT directly converts hydrogen peroxide (H2O2) (one of the ROS) to water and oxygen and can thereby mitigate the toxic effects of H2O2 (54). GPX1 catalyzes the reduction of H2O2, leading to the oxidation of glutathione (GSH), which can be reduced by GSH reductase using NADPH (55). TXNRD1, a member of the peroxiredoxin system, reduces thioredoxin and activates peroxiredoxin for the metabolism of H2O2 (56). However, under pathological conditions, the antioxidant systems can be overwhelmed (34). In EIAVDLV121-infected cells, the expression levels of CAT, GPX1, and TXNRD1 decreased compared with those in mock-infected cells and those in cells infected with EIAVDLV34 (Fig. 5E), meaning that additional ROS cannot be effectively removed in EIAVDLV121-infected cells.
When mitochondrial membrane permeability increases, apoptogenic proteins, including Cyto C, AIF, and Smac/DIABLO, are released from the intermembrane space into the cytosol. These proteins participate in complex processes resulting in the activation of proteases and nucleases, leading to protein and DNA degradation and, ultimately, cell death (intrinsic apoptosis) (57). The cells infected by EIAVDLV121 showed a higher mitochondrial membrane permeability and a higher intrinsic apoptosis rate than the cells infected with EIAVDLV34, so cells infected by EIAVDLV121 would be quickly eliminated by apoptosis (Fig. 6B to D). Moreover, this kind of apoptosis can stimulate an acquired immune response against viral antigens presented by DCs (8, 9).
Viruses require energy from their host cells (ATP is the main form) to drive viral replication and assembly. ATP can be produced by three major pathways in eukaryotes, namely, glycolysis, TCA cycle/OXPHOS, and beta-oxidation (58). The differential expression of enzymes involved in glycolysis and the TCA cycle in cells infected with EIAVDLV34 or EIAVDLV121 (Fig. 7A) led to different transformations of the metabolic pattern in the infected cells (Fig. 7B). In EIAVDLV34-infected cells, glycolysis was the main metabolic pattern, in which ATP can be produced quickly but in small quantities to support the rapid reproduction of viruses. Glycolysis is the main metabolic pattern of M1 macrophages. In contrast, in cells infected with EIAVDLV121, the TCA cycle was the main metabolic pattern and glycolysis was inhibited. This pattern is similar to the behavior of anti-inflammatory M2 macrophages, in which OXPHOS is the main way of generating energy as well as driving pyruvate into the TCA cycle but not to lactate (6). Moreover, our studies showed that the expression of the proinflammatory cytokines TNF-α and IL-1β was significantly upregulated in eMDMs following infection with EIAVDLV34, while following infection with EIAVDLV121, expression of the anti-inflammatory cytokines IL-4 and IL-10 was markedly upregulated (Fig. 7C). Combined with the results of analysis of macrophage polarization phenotypic markers (Fig. 7D), this finding suggests that the macrophages were transformed into the M1 type by EIAVDLV34 infection and into the M2 type by EIAVDLV121 infection. Notably, macrophages could be transformed into M1- and M2-polarized types by activators through the promotion of mitochondrial fission and fusion, respectively (11). Because of the differential expression of the VDAC1 and FIS1 proteins observed in our current research (Fig. 6A and 8A), the EIAVDLV34-infected cells showed more mitochondrial fission, but the EIAVDLV121-infected cells showed more mitochondrial fusion with permeable and indistinct mitochondrial outer membranes (Fig. 8B and C).
In general, viruses could have higher cellular adaptability and replicate more effectively in vitro with increasing numbers of passages in cells in vitro (59–62). In our study, EIAVDLV34 and EIAVDLV121 had the same replication efficiency in the first 3 days postinfection. After 3 days, EIAVDLV121 had a slightly higher replication efficiency than EIAVDLV34. We did not observe distinct differences in replication efficiency between the virulent strain EIAVDLV34 and the well dMDM-adapted strain EIAVDLV121. Excessive ROS can directly cause damage to viral components and a reduction in the release of progeny virus, while apoptosis represents a highly efficient defense mechanism against viruses, allowing the clean disposal of viral proteins and nucleic acids by the infected host (63–66). We hypothesized that the high levels of ROS and apoptosis induced by EIAVDLV121 might in turn restrict viral replication. In vivo, at the acute stage, EIAVDLV34 exhibited higher replication than EIAVDLV121, indicating that the immune system of the host controlled EIAVDLV121 infection better than EIAVDLV34 infection. The reason might be that EIAVDLV121 has a lower cellular adaptability in the in vivo environment, induces a better immune protection response, and causes greater ROS production and apoptosis in host cells than EIAVDLV34.
After infection, viral components induce changes in protein expression profiles in whole cells as well as in mitochondria (67, 68). The integrated proviral DNA of lentiviruses can affect the expression of surrounding genes in host cells by either reducing gene expression via disruptive insertion or enhancing gene expression via a promoter effect (69, 70). Promoter activity always depends on the long terminal repeat (LTR) sequence of a lentivirus (71). The HIV-1 accessory proteins Vif, Vpr, Vpu, and Nef degrade multiple cellular targets, resulting in global remodeling of the host proteome (72–75). In addition, differences in viral components can cause different changes in cellular protein expression profiles. For example, the envelopes (Envs) of the R5 and X4 strains of HIV can induce different cellular transcriptional programs (76). The diversity between the EIAVDLV34 and EIAVDLV121 nucleotide sequences is the highest in the LTR, followed by Env, and the diversity between the amino acid sequences is the highest in the S2 accessory protein, followed by the Env protein (3). In summary, we speculate that LTR, S2, and Env are the main components that induce different mitochondrial protein expression profiles in cells infected by EIAVDLV34 or EIAVDLV121. These viral components may be critical factors involved in the attenuation of equine lentivirus; however, this speculation needs further confirmation.
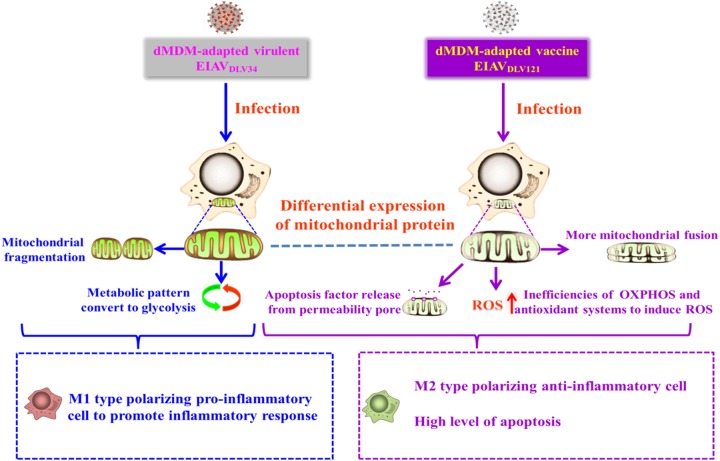
Our results showed that alteration of the expression pattern of mitochondrial proteins during infection of cells with the pathogenic virus strain EIAVDLV34 led to adaptation of the metabolic program of the cellular mitochondria, subsequent transformation of the macrophages to the M1-polarized type, and production of a strong inflammatory response. However, no significant increases in the levels of apoptosis or ROS were observed. In contrast, eMDMs infected with the attenuated vaccine strain EIAVDLV121 were transformed into M2-polarized macrophages through changes in the mitochondria, inhibiting inflammation but causing increased levels of apoptosis and ROS (Fig. 9). Our results may help to provide an understanding of cellular response to viruses with different virulence and the attenuation mechanism of the unique equine lentiviral vaccine.
FIG 9.
Schematic diagrams demonstrating the role of mitochondria in the regulation of the intracellular environment to produce an inflammatory response or apoptosis after EIAVDLV34 or EIAVDLV121 infection. (Left) After EIAVDLV34 infection, alteration of the expression pattern of mitochondrial proteins leads to the conversion of the metabolic pattern to glycolysis and mitochondrial fragmentation, which facilitate the transformation of eMDMs to M1-type-polarizing proinflammatory cells. Meanwhile, the cells keep normal apoptosis and ROS levels. Ultimately, the inflammatory response occurs. (Right) After EIAVDLV121 infection, alteration of the expression pattern of mitochondrial proteins leads to inefficiencies of OXPHOS and antioxidant systems, dissipation of the Δψm, more mitochondrial fusion, and inhibition of glycolysis, which facilitate increases of apoptosis and ROS levels and transformation of eMDMs to M2-type-polarizing anti-inflammatory cells. Ultimately, inflammation keeps balance and additional apoptosis is induced.
MATERIALS AND METHODS
Cells and viruses.
Animal experiments were approved by the Animal Ethics Committee of Harbin Veterinary Research Institute of the Chinese Academy of Agricultural Sciences (CAAS; approval number HVRI-IACUC-2017-409) and were performed in accordance with animal ethics guidelines and approved protocols. Preparations of eMDMs were obtained from equine peripheral blood mononuclear cells (PBMCs) as described previously, with a minor modification (77). Briefly, PBMCs were isolated from 200 to 300 ml of heparinized horse peripheral blood by centrifugation through a HybriMax Histopaque cushion (density = 1.077 g/cm3; Sigma, USA). Isolated PBMCs were washed with RPMI 1640 medium three times and resuspended in RPMI 1640 medium supplemented with 10% horse serum, 104 U/ml penicillin, 104 μg/ml streptomycin, 2 mM l-glutamine, 0.1 mM nonessential amino acids, 1 mM sodium pyruvate, and 0.25 mM sodium HEPES. These cells were seeded into tissue culture flasks (Corning, USA) at 5 × 106 cells/25 cm2 and incubated at 37°C in 5% CO2 for approximately 12 h. Nonadherent and loosely adherent cells were removed by mildly shaking the flasks before changing the medium, and the remaining adherent cells were further incubated for 3 days to allow differentiation into eMDMs.
Wild-type pathogenic EIAV strains isolated in China did not replicate well in cultivated cells, including primary host cells, such as dMDMs. EIAVDLV34 is a dMDM-adapted EIAV strain that was derived from the pathogenic EIAV strain EIAVDV117 by 34 passages in dMDMs. Experimental infection of horses with EIAVDLV34 resulted in the appearance of the typical symptoms of EIA in all of the inoculated animals. EIAVDLV121 is an attenuated EIAV vaccine strain that was derived from EIAVDV117 by 121 passages in dMDMs, which resulted in a loss of virulence, and that has the ability to induce protective immunity, which can effectively prevent the lethality of pathogenic strains (Fig. 1A) (4, 12). Our previous studies revealed the proviral DNA sequences of EIAVLN40, EIAVDLV34, and EIAVDLV121 (3). A molecular phylogenetic tree and genetic distances between these three strains of EIAV were constructed and analyzed using a maximum likelihood method based on the general time-reversible model with MEGA (version 7.0) software (78).
Virus titration.
The infectious titer of the EIAV strain was measured using the median (50%) tissue culture infective dose (TCID50) method as reported previously (79). At 3 days postplating, 5 × 105 eMDMs were infected with 5 × 103 TCID50 of either EIAVDLV34 or EIAVDLV121 in 24-well plates. Cell culture medium was used to mock infect eMDMs as a negative control. The culture medium was exchanged for fresh culture medium at 2 h postinfection. The cells in some wells were collected to determine the number of intracellular viral RNA copies at 3 h postinfection with EIAV. These cells were washed three times with phosphate-buffered saline (PBS) and treated with trypsin-EDTA (0.25% trypsin, 5 mM EDTA) at room temperature for 5 min to remove adherent viruses that had not entered the cells. In other wells, medium samples were collected every day from 0 to 7 dpi at 1-day intervals and stored at −80°C for further titration of infectious virus. In addition, some cells were fixed in 4% paraformaldehyde in PBS and examined for viral antigens using IFA with a 1:200 dilution of EIAV-positive horse serum, followed by an FITC- or TRITC-conjugated rabbit anti-horse IgG (Abcam, USA). Supernatant and intracellular copies of viral RNA in the eMDMs infected with EIAVDLV34 or EIAVDLV121 were quantified using real-time qPCR assays according to previously described procedures (80). Briefly, total RNA was extracted from the harvested cells or culture supernatants using the TRIzol reagent (Invitrogen, USA) or a QIAamp viral RNA minikit (Qiagen, Hilden, Germany) and was processed for cDNA synthesis using a Moloney murine leukemia virus reverse transcription kit (Invitrogen, USA) and 100 ng of RNA template. The cDNA obtained was used for qPCR analysis. The replication kinetics of the viruses were determined in three independent experiments.
Protein sample preparation and iTRAQ analysis.
After isolation and cultivation, eMDMs in 25-cm2 flasks (5 × 106 cells/flask) were infected with 5 × 104 TCID50 of either EIAVDLV34 or EIAVDLV121 for 3 days. The same number of eMDMs was mock infected with cell culture supernatant as a negative control. The culture supernatant was then removed, and the monolayers were washed twice with cold PBS, lysed with 500 μl of lysis buffer (7 M urea, 2 M thiourea, 4% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 40 mM Tris-HCl, pH 8.5), 1 mM phenylmethylsulfonyl fluoride, and 2 mM EDTA and incubated on ice for 5 min. After adding dithiothreitol (DTT) to a final concentration of 10 mM, the lysate was sonicated on ice at 200 W for 15 min and centrifuged at 25,000 × g for 20 min. The supernatant was collected and incubated with 10 mM DTT at 56°C for 1 h to reduce the disulfide bonds in the proteins and subsequently treated with 55 mM iodoacetamide in the dark for 45 min to block the alkylation of cysteine residues. The samples were then combined with 1 ml of cold acetone, incubated at −20°C overnight, and centrifuged at 25,000 × g for 20 min to collect the pellet, which was then suspended in 300 μl of 0.5 M tetraethylammonium bromide and sonicated at 200 W on ice for 15 min. The resulting protein preparations were centrifuged at 25,000 × g for 20 min, and the pellets were removed. The protein concentration of the supernatant was analyzed with a 2D Quant kit (Amersham, USA) according to the manufacturer’s instructions. Finally, the protein samples were digested with trypsin at an enzyme/protein ratio of 1:50 at 37°C overnight.
iTRAQ comparative analysis was performed on three independent comparison groups: (i) mock-infected cell samples versus EIAVDLV34-infected cell samples, (ii) mock-infected cell samples versus EIAVDLV121-infected cell samples, and (iii) EIAVDLV34-infected cell samples versus EIAVDLV121-infected cell samples. Each sample from each comparison group contained three biological repeats that were labeled with three different iTRAQ tags. In the mock- versus EIAVDLV34-infected comparison group, the three mock-infected cell samples were labeled with iTRAQ tags 113, 114, and 115 and the three EIAVDLV34-infected cell samples were labeled with iTRAQ tags 118, 119, and 121. In the mock- versus EIAVDLV121-infected comparison group, the three mock-infected cell samples were labeled with iTRAQ tags 118, 119, and 121 and the three EIAVDLV121-infected cell samples were labeled with iTRAQ tags 113, 114, and 115. In the EIAVDLV34- versus EIAVDLV121-infected comparison group, the three EIAVDLV34-infected cell samples were labeled with iTRAQ tags 113, 114, and 115 and the three EIAVDLV121-infected cell samples were labeled with iTRAQ tags 118, 119, and 121. The labeled samples of each comparison group were then mixed together prior to online 2D LC-MS/MS analysis. The labeled peptide mixtures were prepared according to a previously described method (81). Briefly, the labeled peptide mixtures were prefractionated by high-pH reverse-phase chromatography. After redissolution, the peptide mixtures were fractionated with high-pH separation using a Shimadzu ultrafast liquid chromatography system (Shimadzu, Japan) connected to a reverse-phase column (Venusil XBP C18 column; Bonna-Agela Technologies, USA). High-pH separation was performed using a four-step linear gradient, and fractions were collected and lyophilized for further analysis. The fractions of sequentially eluted peptides were redissolved in 2.0% acetonitrile with 0.1% formic acid and centrifuged at 12,000 × g for 3 min. The supernatants were harvested and analyzed using an Eksigent Nano LC 2D Plus instrument coupled with an MS, which consisted of a C18 concentration column (particle size, 5 μm; 100 μm by 20 mm; Microm BioResources Inc., Auburn, CA, USA) and a C18 separation column (particle size, 3 μm; 75 μm by 120 mm; Microm BioResources Inc., Auburn, CA, USA). MS data acquisition was performed with a triple-time-of-flight 5600 system (AB Sciex, USA). The details of the experimental conditions are the same as those given in previous studies (81).
Relative abundance quantification and identification of peptides and proteins were performed using ProteinPilot software (version 4.8; Applied Biosystems, USA). Each MS/MS spectrum was searched for against that for the Equus caballus proteome with UniProt accession number UP000002281. The protein confidence threshold cutoff was set to 1.3, with two or more peptides having values above the 95% confidence level. Analysis of the iTRAQ data was performed using ProQuant software (Applied Biosystems, Foster City, CA, USA). The cutoff value for the confidence setting was 75, and the tolerance settings for peptide identification in the ProQuant searches were 0.15 Da for MS and 0.1 Da for MS/MS. Relative quantification of proteins using the iTRAQ tags was performed on the MS/MS scans and is presented as the ratio of the area under the peaks at 113, 114, 115, 118, 119, and 121 Da, which indicate the masses of the tags corresponding to the iTRAQ reagents. The relative amount of peptide in each of the three biological repeats in each sample was calculated by dividing the mean peak area for three biological samples by the peak area for each of the three other biological samples in each comparison group (e.g., the mean peak areas for the mock-infected sample 113, 114, and 115 tags were divided by the peak areas for each of the EIAVDLV34 sample 118, 119, and 121 tags, respectively). For proteins with two or more qualified peptide matches, the average peak area ratio for a tag was calculated using the peak area ratios for peptides originating from the same tag of the same protein. The three different tags represented three biological repeats, so each protein had three ratios. To account for small differences in protein loading, these ratios were normalized using the overall ratios for all proteins in the sample, as recommended by Applied Biosystems. A two-group unequal variance t test was performed to derive lists of the proteins with differential abundance. The final list consisted of differentially expressed proteins with P values of <0.05. According to the recommendations of some previous studies on iTRAQ proteomic analysis of virus-infected target cells (82–84), differences were identified by at least two high-confidence (95%) peptides with a ratio of <0.8 or a ratio of >1.25, as calculated by ProQuant software.
GO and KEGG pathway analysis.
The differentially expressed proteins identified in this study were converted to human orthologous proteins, and the lists were submitted to DAVID at http://david.abcc.ncifcrf.gov. The differentially expressed proteins were subjected to enrichment analysis to assess the significant overrepresentation of GO-CC, GO-BP, and GO-MF terminologies and analysis of KEGG pathway categories (85). In all tests, known genes were used as the background. To measure the magnitude of enrichment, P values, indicating the significance of the overlap between various gene sets and fold enrichment, were calculated using a Benjamini-corrected modified Fisher’s exact test. Fold enrichment, together with the P value, was used to rank the enriched terms. GO-CC, GO-BP, GO-MF, or KEGG pathway categories with P values of <0.05 were considered significant, and a fold enrichment of 1.5 or above was considered interesting.
MRM-MS analysis.
To verify the expression levels of the differentially expressed mitochondrial proteins obtained by iTRAQ analysis, MRM-MS analysis was carried out. MRM is applied to the quantitative analysis of target proteins that have unique peptides based on the known or presumed amino acid sequence of the protein (86). The three groups of cell samples (mock-infected cells, cells infected with EIAVDLV34, and cells infected with EIAVDLV121), including the three biological repeats from every group, were analyzed. The samples were prepared, reduced, alkylated, and digested with trypsin according to the iTRAQ protocol and spiked with 40 fmol of β-galactosidase for data normalization. MRM analysis was performed on a Qtrap 5500 system (AB Sciex, USA) equipped with a Waters Nano Acquity ultraperformance LC system. The mobile phase consisted of solvent A (0.1% aqueous formic acid) and solvent B (98% acetonitrile with 0.1% formic acid). Peptides were separated on a BEH130 C18 column (particle size, 1.7 μm; 75 μm by 200 mm; Waters) at 300 nl/min and eluted with a gradient of 2 to 40% solvent B for 30 min and 40 to 60% solvent B for 3 min, followed by 2 min of a linear gradient to 80% solvent B and maintenance at 80% solvent B for 5 min. For the Qtrap 5500 system, a spray voltage of 2,100 V, a nebulizer gas flow rate of 20 lb/in2, and a dwell time of 10 ms were used. Multiple MRM transitions were monitored using unit resolution in both the Q1 and Q3 quadrupoles to maximize specificity.
A spectral library of the MS/MS data was searched against a database of spectra for Equus caballus sequences using the Mascot (version 2.3) program (Matrix Science, UK). The data file was imported into Skyline software (version 1.2.0.3425; Skyline), where a library was built (87). Peptides were selected for MRM method development according to the following criteria: those with unique sequences in the database, a maximum m/z of <1,250 (the limit of the quadrupole scan), a length range of 5 to 40 amino acids, a lack of methionine, the presence of carbamidomethyl on cysteine, no variable modification, and no missed cleavage of trypsin. The predicted retention time of the targeted peptides was observed with an i retention time (iRT) strategy (88). Pooled peptides were digested as described above, and preliminary MRM assays were performed to determine where these proteins were detected.
MRM assays for a given protein were successfully developed only if the protein had at least one unique peptide identified in the MS/MS spectral library (cutoff score > 0.95), had >5 fragment ions with the same elution profile and in the same ratios as in the spectral library, and had an accurate retention time (less than ±2 min of deviation against the predicted retention time). Three MRM-MS technical repeats were performed for each sample. All proteins with a P value below 0.05 and a fold change larger than 1.25 or smaller than 0.8 were considered significant.
Real-time quantitative PCR.
The relative amounts of mRNA for those proteins selected for confirmation using MRM-MS, the inflammatory cytokines, and the macrophage polarization phenotypic markers were analyzed using real-time qPCR. Monolayers of eMDMs were infected with EIAVDLV34 or EIAVDLV121 or mock infected with culture medium as described above. Total cellular RNA was extracted with the TRIzol reagent (Life Technologies, USA) according to the manufacturer’s instructions. RNA concentrations were measured spectrophotometrically at 260/280 nm (Implen, Germany). cDNA was synthesized using a PrimeScript reverse transcription reagent kit with gDNA Eraser (TaKaRa, Japan) following the manufacturer’s instructions. qPCR was performed using a Stratagene 3000 system (Agilent Technologies, Germany). The reaction volume was 20 μl and contained 2 μl of cDNA template, 10 μl of 2× SYBR green I (TaKaRa, Japan), and 0.8 μM forward and reverse primers. PCRs were performed in triplicate. For all amplifications, the cycle conditions were 95°C for 2 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. All samples were tested with three independent repeats, and the average values were taken as the quantitative result. Equine β-actin was used as an internal control. The primers for amplifying cDNAs of the 40 genes are presented in Table S4 in the supplemental material. Relative fold changes in gene expression were determined by the 2−ΔΔCT threshold cycle (CT) method (89).
NAD/NADH, OXPHOS, and ATP levels and mitochondrial ROS assay.
NAD/NADH and ATP levels in cultured cells were determined using commercial kits (EnzyChrom; BioAssay Systems, USA) according to the manufacturer’s instructions. Briefly, for the NAD/NADH assay, cell pellets were resuspended in 1.5-ml Eppendorf tubes with either 100 μl of NAD extraction buffer for NAD determination or 100 μl of NADH extraction buffer for NADH determination. The extracts were heated for 5 min at 60°C, and 20 μl of assay buffer was added, followed by the addition of the opposite extraction buffer (to neutralize the extracts). The mixtures were vortexed and then centrifuged at 13,000 × g for 5 min. Supernatants were added to a working reagent containing the assay buffer. The optical density at 565 nm was recorded at time zero and at 15 min using a microplate reader (model Elx800; Bio-Tek, USA). Differences in the absorbance from that of the standard solutions were determined and used to calculate the NADH and NAD concentrations. The ATP assay is based on bioluminescence by the reaction of ATP with d-luciferin in the presence of luciferase. Briefly, 90 μl of assay reagent (a mixture of 95 μl of assay buffer, 1 μl of substrate, and 1 μl of enzyme) was added to 100 μl of cell lysate, and the mixture was incubated at room temperature for 10 min. The luminescence of the samples was then read in a Berthold LB960 plate reader (Berthold, Germany) with an integration time of 5 s.
To determine the OXPHOS capacity of the eMDM mitochondria, uninfected eMDMs and eMDMs infected with either EIAVDLV34 or EIAVDLV121 were isolated and then subjected to high-resolution respirometry with a polarographic oxygen electrode (Oxygraph-2k; Oroboros Instruments, Innsbruck, Austria). The number of cells was kept at 2 × 106 by cell counting in each group. The respiration rates (represented by the oxygen consumption rate, O2 flux per unit volume) were assessed per the standard protocol. The measurements were performed at 37°C in RPMI 1640 medium (Invitrogen, USA).
Mitochondrial ROS were detected by staining cells with MitoSOX Red (Thermo Fisher Scientific, Waltham, USA). The details of the procedure have been described previously (90). In brief, MitoSOX Red was added to cells at a final concentration of 5 mM; after this, the cells were incubated for 30 min at 37°C to allow the loading of MitoSOX Red. The cells were then placed in a sterile flow cytometry tube at a concentration of 106 cells per 100 μl. Samples were diluted with binding buffer to a final volume of 500 μl and were run on a flow cytometer (model Cytomics FC 500; Beckman, USA) with excitation and emission maxima of approximately 510 and 580 nm, respectively.
Δψm, cellular apoptosis, and caspase 3, 8, and 9 activity analysis.
Δψm was monitored with the potentiometric dye JC-1 using a Mitoprobe assay kit (Invitrogen, USA) according to the manufacturer’s instructions. JC-1 accumulates in polarized mitochondria with a resting membrane potential and fluoresces red. However, upon the loss of Δψm, JC-1 aggregates are released from the mitochondria and fluoresce green. JC-1 at a final concentration of 2 μM was added to cells at 37°C under 5% CO2 for 15 to 30 min and then analyzed on a Cytomics FC 500 flow cytometer (Beckman, USA) with excitation at 488 nm and 633 nm using emission filters appropriate for fluorescein. Cellular apoptosis was detected by annexin V and propidium iodide (PI) double staining using an FITC-annexin V apoptosis detection kit (BD Pharmingen, USA). Briefly, cells were collected and washed with annexin V binding buffer. Cells were resuspended in 100 μl of binding buffer and stained by adding 5 μl of annexin V and 5 μl of 3 μM PI. Apoptotic cells induced with 0.5 M ethanol for 40 min were used as a positive control. After a 15-min incubation at room temperature, apoptotic cells were measured using a Cytomics FC 500 flow cytometer (Beckman, USA). Caspase 3, 8, and 9 activities were determined with caspase 3, 8, and 9 activity assay kits (Beyotime, China), respectively, which detected the production of the chromophore p-nitroanilide (pNA) after its cleavage from the peptide substrates DEVD-p-nitroanilide, IETD-p-nitroanilide, and LEHD-p-nitroanilide, respectively. Cells were lysed with lysis buffer on ice for 15 min. The supernatant was harvested by centrifugation at 16,000 × g for 10 min at 4°C. Following this step, the assays were performed on 96-well microtiter plates by incubating 40 μl of cell lysate per sample in 50 μl of reaction buffer containing 10 μl of substrate for caspase 3, 8, or 9 (2 mM acetyl [Ac]-DEVD-pNA, Ac-IETD-pNA, or Ac-LEHD-pNA, respectively). Lysates were incubated at 37°C for 2 h, and the absorbance at 405 nm was measured in an Elx800 microplate reader (Bio-Tek, USA). A negative control was performed with all the reagents except the cell lysate.
Cellular glycolysis analysis.
The glycolysis cell-based assay kit (Cayman Chemical, Ann Arbor, MI, USA) provides a colorimetric method for detecting l-lactate, the end product of glycolysis, which is produced and secreted by cultured cells. In this assay, lactate dehydrogenase catalyzes the reaction between NAD and lactate, yielding pyruvate and NADH. The NADH directly reduces a tetrazolium salt [2-(4-iodophenyl)-3-(4-nitrophenyl)-5-phenyl-2H-tetrazolium] to a colored formazan which absorbs at between 490 and 520 nm and which can be measured in an Elx800 microplate reader (Bio-Tek, USA). The quantity of formazan produced is proportional to the quantity of lactate in the culture medium and is thus an indirect measurement of glycolysis.
2D-structured illumination microscopy (SIM) superresolution and electron microscopy imaging.
Transmission electron microscopy (TEM) observation of mitochondria was carried out as previously described (43, 91). Uninfected eMDMs or eMDMs infected with EIAVDLV34 or EIAVDLV121 were prepared. At 3 dpi, cells were fixed and embedded. Ultrathin sections were observed under a JEM2100 transmission electron microscope (JEOL, Tokyo, Japan) at 200 kV. In the cytoplasm, the mitochondria with double membranes were clearly visible.
The cells were collected at 3 dpi, fixed, and sequentially incubated with a primary anti-translocase of the mitochondrial outer membrane 20 (TOM20; TOM20 is a marker of mitochondria) antibody (BD Biosciences, USA) at a 1:500 dilution and with a fluorescent secondary goat anti-mouse immunoglobulin-Alexa Fluor 488 antibody (Invitrogen, USA) at a 1:500 dilution. A superresolution microscope (DeltaVision OMX SR; GE Healthcare, USA) was used for mitochondrial dynamic analysis. The procedure followed that described in previous studies (92).
Statistical analysis.
Bubble map and Venn plot map analyses were performed using the OmicShare tool, a free online platform for data analysis. Pearson correlation analysis (of the data on the fold change in expression between the iTRAQ and MRM, iTRAQ and qPCR, and MRM and qPCR methods) and heat map representation (of the protein expression level) were performed using Microsoft Excel software (MS Office 2010). The quantities and ratios of differentially expressed proteins and comparisons of biological tests were analyzed and represented in GraphPad Prism (version 6.0) software (GraphPad, San Diego, CA, USA). Statistical analyses were performed using SPSS (version 17.0) software (IBM, Armonk, NY, USA). Experimental data are represented as the mean ± standard error for a minimum of three biological replicates. Student's t test was performed to examine the significance of differences between two groups. P values of less than 0.05 were considered statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by grants from the National Natural Science Foundation of China (grant 31572537), the National Natural Science Foundation of China (grant 31772720), and the National Natural Science Foundation of China (grant 81561128010), as well as the Natural Science Foundation of Heilongjiang Province of China (grant C2018071).
We declare that we have no competing interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00653-19.
REFERENCES
- 1.Leroux C, Cadore JL, Montelaro RC. 2004. Equine infectious anemia virus (EIAV): what has HIV’s country cousin got to tell us? Vet Res 35:485–512. doi: 10.1051/vetres:2004020. [DOI] [PubMed] [Google Scholar]
- 2.Liu L, Wan Y, Wu L, Sun J, Li H, Li H, Ma L, Shao Y. 2010. Broader HIV-1 neutralizing antibody responses induced by envelope glycoprotein mutants based on the EIAV attenuated vaccine. Retrovirology 7:71. doi: 10.1186/1742-4690-7-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang XF, Lin YZ, Li Q, Liu Q, Zhao WW, Du C, Chen J, Wang X, Zhou JH. 2016. Genetic evolution during the development of an attenuated EIAV vaccine. Retrovirology 13:9. doi: 10.1186/s12977-016-0240-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin YZ, Cao XZ, Li L, Li L, Jiang CG, Wang XF, Ma J, Zhou JH. 2011. The pathogenic and vaccine strains of equine infectious anemia virus differentially induce cytokine and chemokine expression and apoptosis in macrophages. Virus Res 160:274–282. doi: 10.1016/j.virusres.2011.06.028. [DOI] [PubMed] [Google Scholar]
- 5.Liu Q, Ma J, Wang XF, Xiao F, Li LJ, Zhang JE, Lin YZ, Du C, He XJ, Wang X, Zhou JH. 2016. Infection with equine infectious anemia virus vaccine strain EIAVDLV121 causes no visible histopathological lesions in target organs in association with restricted viral replication and unique cytokine response. Vet Immunol Immunopathol 170:30–40. doi: 10.1016/j.vetimm.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galvan-Pena S, O’Neill LA. 2014. Metabolic reprograming in macrophage polarization. Front Immunol 5:420. doi: 10.3389/fimmu.2014.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu L, Zhao Q, Yang T, Ding W, Zhao Y. 2015. Cellular metabolism and macrophage functional polarization. Int Rev Immunol 34:82–100. doi: 10.3109/08830185.2014.969421. [DOI] [PubMed] [Google Scholar]
- 8.Kepp O, Senovilla L, Galluzzi L, Panaretakis T, Tesniere A, Schlemmer F, Madeo F, Zitvogel L, Kroemer G. 2009. Viral subversion of immunogenic cell death. Cell Cycle 8:860–869. doi: 10.4161/cc.8.6.7939. [DOI] [PubMed] [Google Scholar]
- 9.Krysko DV, Agostinis P, Krysko O, Garg AD, Bachert C, Lambrecht BN, Vandenabeele P. 2011. Emerging role of damage-associated molecular patterns derived from mitochondria in inflammation. Trends Immunol 32:157–164. doi: 10.1016/j.it.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 10.Vakifahmetoglu-Norberg H, Ouchida AT, Norberg E. 2017. The role of mitochondria in metabolism and cell death. Biochem Biophys Res Commun 482:426–431. doi: 10.1016/j.bbrc.2016.11.088. [DOI] [PubMed] [Google Scholar]
- 11.Gao Z, Li Y, Wang F, Huang T, Fan K, Zhang Y, Zhong J, Cao Q, Chao T, Jia J, Yang S, Zhang L, Xiao Y, Zhou JY, Feng XH, Jin J. 2017. Mitochondrial dynamics controls anti-tumour innate immunity by regulating CHIP-IRF1 axis stability. Nat Commun 8:1805. doi: 10.1038/s41467-017-01919-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin YZ, Shen RX, Zhu ZY, Deng XL, Cao XZ, Wang XF, Ma J, Jiang CG, Zhao LP, Lv XL, Shao YM, Zhou JH. 2011. An attenuated EIAV vaccine strain induces significantly different immune responses from its pathogenic parental strain although with similar in vivo replication pattern. Antiviral Res 92:292–304. doi: 10.1016/j.antiviral.2011.08.016. [DOI] [PubMed] [Google Scholar]
- 13.Morrow CD, Park J, Wakefield JK. 1994. Viral gene products and replication of the human immunodeficiency type 1 virus. Am J Physiol 266:C1135–C1156. doi: 10.1152/ajpcell.1994.266.5.C1135. [DOI] [PubMed] [Google Scholar]
- 14.Ranki A, Lagerstedt A, Ovod V, Aavik E, Krohn KJ. 1994. Expression kinetics and subcellular localization of HIV-1 regulatory proteins Nef, Tat and Rev in acutely and chronically infected lymphoid cell lines. Arch Virol 139:365–378. doi: 10.1007/BF01310798. [DOI] [PubMed] [Google Scholar]
- 15.Wu WW, Wang G, Baek SJ, Shen RF. 2006. Comparative study of three proteomic quantitative methods, DIGE, cICAT, and iTRAQ, using 2D gel- or LC-MALDI TOF/TOF. J Proteome Res 5:651–658. doi: 10.1021/pr050405o. [DOI] [PubMed] [Google Scholar]
- 16.Kaur P, Rizk NM, Ibrahim S, Younes N, Uppal A, Dennis K, Karve T, Blakeslee K, Kwagyan J, Zirie M, Ressom HW, Cheema AK. 2012. iTRAQ-based quantitative protein expression profiling and MRM verification of markers in type 2 diabetes. J Proteome Res 11:5527–5539. doi: 10.1021/pr300798z. [DOI] [PubMed] [Google Scholar]
- 17.Shu Q, Cai T, Chen X, Zhu HH, Xue P, Zhu N, Xie Z, Wei S, Zhang Q, Niu L, Gao WQ, Yang F. 2015. Proteomic comparison and MRM-based comparative analysis of metabolites reveal metabolic shift in human prostate cancer cell lines. J Proteome Res 14:3390–3402. doi: 10.1021/acs.jproteome.5b00464. [DOI] [PubMed] [Google Scholar]
- 18.Gianazza E, Tremoli E, Banfi C. 2014. The selected reaction monitoring/multiple reaction monitoring-based mass spectrometry approach for the accurate quantitation of proteins: clinical applications in the cardiovascular diseases. Expert Rev Proteomics 11:771–788. doi: 10.1586/14789450.2014.947966. [DOI] [PubMed] [Google Scholar]
- 19.Drose S, Brandt U. 2012. Molecular mechanisms of superoxide production by the mitochondrial respiratory chain. Adv Exp Med Biol 748:145–169. doi: 10.1007/978-1-4614-3573-0_6. [DOI] [PubMed] [Google Scholar]
- 20.Breuer ME, Willems PH, Smeitink JA, Koopman WJ, Nooteboom M. 2013. Cellular and animal models for mitochondrial complex I deficiency: a focus on the NDUFS4 subunit. IUBMB Life 65:202–208. doi: 10.1002/iub.1127. [DOI] [PubMed] [Google Scholar]
- 21.Feng D, Witkowski A, Smith S. 2009. Down-regulation of mitochondrial acyl carrier protein in mammalian cells compromises protein lipoylation and respiratory complex I and results in cell death. J Biol Chem 284:11436–11445. doi: 10.1074/jbc.M806991200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorenkova N, Robinson E, Grieve DJ, Galkin A. 2013. Conformational change of mitochondrial complex I increases ROS sensitivity during ischemia. Antioxid Redox Signal 19:1459–1468. doi: 10.1089/ars.2012.4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoefs SJ, Dieteren CE, Distelmaier F, Janssen RJ, Epplen A, Swarts HG, Forkink M, Rodenburg RJ, Nijtmans LG, Willems PH, Smeitink JA, van den Heuvel LP. 2008. NDUFA2 complex I mutation leads to Leigh disease. Am J Hum Genet 82:1306–1315. doi: 10.1016/j.ajhg.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang P, Cheng X, Fu Z, Zhou C, Lu W, Xie X. 2013. Reduced expression of NDUFS3 and its clinical significance in serous ovarian cancer. Int J Gynecol Cancer 23:622–629. doi: 10.1097/IGC.0b013e318287a90d. [DOI] [PubMed] [Google Scholar]
- 25.Crofts AR. 2004. The cytochrome bc1 complex: function in the context of structure. Annu Rev Physiol 66:689–733. doi: 10.1146/annurev.physiol.66.032102.150251. [DOI] [PubMed] [Google Scholar]
- 26.Park ER, Kim SB, Lee JS, Kim YH, Lee DH, Cho EH, Park SH, Han CJ, Kim BY, Choi DW, Yoo YD, Yu A, Lee JW, Jang JJ, Park YN, Suh KS, Lee KH. 2017. The mitochondrial hinge protein, UQCRH, is a novel prognostic factor for hepatocellular carcinoma. Cancer Med 6:749–760. doi: 10.1002/cam4.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cape JL, Bowman MK, Kramer DM. 2007. A semiquinone intermediate generated at the Qo site of the cytochrome bc1 complex: importance for the Q-cycle and superoxide production. Proc Natl Acad Sci U S A 104:7887–7892. doi: 10.1073/pnas.0702621104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang L, Yu L, Yu CA. 1998. Generation of superoxide anion by succinate-cytochrome c reductase from bovine heart mitochondria. J Biol Chem 273:33972–33976. doi: 10.1074/jbc.273.51.33972. [DOI] [PubMed] [Google Scholar]
- 29.Tripathy MK, Mitra D. 2010. Differential modulation of mitochondrial OXPHOS system during HIV-1 induced T-cell apoptosis: up regulation of complex-IV subunit COX-II and its possible implications. Apoptosis 15:28–40. doi: 10.1007/s10495-009-0408-9. [DOI] [PubMed] [Google Scholar]
- 30.Houstĕk J, Andersson U, Tvrdík P, Nedergaard J, Cannon B. 1995. The expression of subunit c correlates with and thus may limit the biosynthesis of the mitochondrial F0F1-ATPase in brown adipose tissue. J Biol Chem 270:7689–7694. doi: 10.1074/jbc.270.13.7689. [DOI] [PubMed] [Google Scholar]
- 31.Li RJ, Zhang GS, Chen YH, Zhu JF, Lu QJ, Gong FJ, Kuang WY. 2010. Down-regulation of mitochondrial ATPase by hypermethylation mechanism in chronic myeloid leukemia is associated with multidrug resistance. Ann Oncol 21:1506–1514. doi: 10.1093/annonc/mdp569. [DOI] [PubMed] [Google Scholar]
- 32.Venkatesh S, Kumar M, Sharma A, Kriplani A, Ammini AC, Talwar P, Agarwal A, Dada R. 2010. Oxidative stress and ATPase6 mutation is associated with primary ovarian insufficiency. Arch Gynecol Obstet 282:313–318. doi: 10.1007/s00404-010-1444-y. [DOI] [PubMed] [Google Scholar]
- 33.Zhu H, Chen L, Zhou W, Huang Z, Hu J, Dai S, Wang X, Huang X, He C. 2013. Over-expression of the ATP5J gene correlates with cell migration and 5-fluorouracil sensitivity in colorectal cancer. PLoS One 8:e76846. doi: 10.1371/journal.pone.0076846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Birben E, Sahiner UM, Sackesen C, Erzurum S, Kalayci O. 2012. Oxidative stress and antioxidant defense. World Allergy Organ J 5:9–19. doi: 10.1097/WOX.0b013e3182439613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rostovtseva TK, Tan W, Colombini M. 2005. On the role of VDAC in apoptosis: fact and fiction. J Bioenerg Biomembr 37:129–142. doi: 10.1007/s10863-005-6566-8. [DOI] [PubMed] [Google Scholar]
- 36.Tomasello F, Messina A, Lartigue L, Schembri L, Medina C, Reina S, Thoraval D, Crouzet M, Ichas F, De Pinto V, De Giorgi F. 2009. Outer membrane VDAC1 controls permeability transition of the inner mitochondrial membrane in cellulo during stress-induced apoptosis. Cell Res 19:1363–1376. doi: 10.1038/cr.2009.98. [DOI] [PubMed] [Google Scholar]
- 37.Keinan N, Tyomkin D, Shoshan-Barmatz V. 2010. Oligomerization of the mitochondrial protein voltage-dependent anion channel is coupled to the induction of apoptosis. Mol Cell Biol 30:5698–5709. doi: 10.1128/MCB.00165-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharaf el Dein O, Gallerne C, Brenner C, Lemaire C. 2012. Increased expression of VDAC1 sensitizes carcinoma cells to apoptosis induced by DNA cross-linking agents. Biochem Pharmacol 83:1172–1182. doi: 10.1016/j.bcp.2012.01.017. [DOI] [PubMed] [Google Scholar]
- 39.Uren RT, O’Hely M, Iyer S, Bartolo R, Shi MX, Brouwer JM, Alsop AE, Dewson G, Kluck RM. 2017. Disordered clusters of Bak dimers rupture mitochondria during apoptosis. Elife 6:e19944. doi: 10.7554/eLife.19944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mantovani A, Biswas SK, Galdiero MR, Sica A, Locati M. 2013. Macrophage plasticity and polarization in tissue repair and remodelling. J Pathol 229:176–185. doi: 10.1002/path.4133. [DOI] [PubMed] [Google Scholar]
- 41.Gundra UM, Girgis NM, Ruckerl D, Jenkins S, Ward LN, Kurtz ZD, Wiens KE, Tang MS, Basu-Roy U, Mansukhani A, Allen JE, Loke P. 2014. Alternatively activated macrophages derived from monocytes and tissue macrophages are phenotypically and functionally distinct. Blood 123:e110–e122. doi: 10.1182/blood-2013-08-520619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Osellame LD, Blacker TS, Duchen MR. 2012. Cellular and molecular mechanisms of mitochondrial function. Best Pract Res Clin Endocrinol Metab 26:711–723. doi: 10.1016/j.beem.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim SJ, Syed GH, Khan M, Chiu WW, Sohail MA, Gish RG, Siddiqui A. 2014. Hepatitis C virus triggers mitochondrial fission and attenuates apoptosis to promote viral persistence. Proc Natl Acad Sci U S A 111:6413–6418. doi: 10.1073/pnas.1321114111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoshizumi T, Ichinohe T, Sasaki O, Otera H, Kawabata S, Mihara K, Koshiba T. 2014. Influenza A virus protein PB1-F2 translocates into mitochondria via Tom40 channels and impairs innate immunity. Nat Commun 5:4713. doi: 10.1038/ncomms5713. [DOI] [PubMed] [Google Scholar]
- 45.James DI, Parone PA, Mattenberger Y, Martinou JC. 2003. hFis1, a novel component of the mammalian mitochondrial fission machinery. J Biol Chem 278:36373–36379. doi: 10.1074/jbc.M303758200. [DOI] [PubMed] [Google Scholar]
- 46.Claus C, Liebert UG. 2014. A renewed focus on the interplay between viruses and mitochondrial metabolism. Arch Virol 159:1267–1277. doi: 10.1007/s00705-013-1841-1. [DOI] [PubMed] [Google Scholar]
- 47.Chong PK, Gan CS, Pham TK, Wright PC. 2006. Isobaric tags for relative and absolute quantitation (iTRAQ) reproducibility: implication of multiple injections. J Proteome Res 5:1232–1240. doi: 10.1021/pr060018u. [DOI] [PubMed] [Google Scholar]
- 48.Gan CS, Chong PK, Pham TK, Wright PC. 2007. Technical, experimental, and biological variations in isobaric tags for relative and absolute quantitation (iTRAQ). J Proteome Res 6:821–827. doi: 10.1021/pr060474i. [DOI] [PubMed] [Google Scholar]
- 49.Mills EL, Kelly B, O'Neill LAJ. 2017. Mitochondria are the powerhouses of immunity. Nat Immunol 18:488–498. doi: 10.1038/ni.3704. [DOI] [PubMed] [Google Scholar]
- 50.Porcu M, Chiarugi A. 2005. The emerging therapeutic potential of sirtuin-interacting drugs: from cell death to lifespan extension. Trends Pharmacol Sci 26:94–103. doi: 10.1016/j.tips.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 51.Treberg JR, Quinlan CL, Brand MD. 2011. Evidence for two sites of superoxide production by mitochondrial NADH-ubiquinone oxidoreductase (complex I). J Biol Chem 286:27103–27110. doi: 10.1074/jbc.M111.252502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quinlan CL, Treberg JR, Perevoshchikova IV, Orr AL, Brand MD. 2012. Native rates of superoxide production from multiple sites in isolated mitochondria measured using endogenous reporters. Free Radic Biol Med 53:1807–1817. doi: 10.1016/j.freeradbiomed.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brand MD. 2010. The sites and topology of mitochondrial superoxide production. Exp Gerontol 45:466–472. doi: 10.1016/j.exger.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nicholls P. 2012. Classical catalase: ancient and modern. Arch Biochem Biophys 525:95–101. doi: 10.1016/j.abb.2012.01.015. [DOI] [PubMed] [Google Scholar]
- 55.Lee JR, Roh JL, Lee SM, Park Y, Cho KJ, Choi SH, Nam SY, Kim SY. 2017. Overexpression of glutathione peroxidase 1 predicts poor prognosis in oral squamous cell carcinoma. J Cancer Res Clin Oncol 143:2257–2265. doi: 10.1007/s00432-017-2466-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arner ES, Holmgren A. 2000. Physiological functions of thioredoxin and thioredoxin reductase. Eur J Biochem 267:6102–6109. doi: 10.1046/j.1432-1327.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- 57.Kroemer G, Galluzzi L, Brenner C. 2007. Mitochondrial membrane permeabilization in cell death. Physiol Rev 87:99–163. doi: 10.1152/physrev.00013.2006. [DOI] [PubMed] [Google Scholar]
- 58.Rich PR. 2003. The molecular machinery of Keilin’s respiratory chain. Biochem Soc Trans 31:1095–1105. doi: 10.1042/bst0311095. [DOI] [PubMed] [Google Scholar]
- 59.Karron RA, Buonagurio DA, Georgiu AF, Whitehead SS, Adamus JE, Clements-Mann ML, Harris DO, Randolph VB, Udem SA, Murphy BR, Sidhu MS. 1997. Respiratory syncytial virus (RSV) SH and G proteins are not essential for viral replication in vitro: clinical evaluation and molecular characterization of a cold-passaged, attenuated RSV subgroup B mutant. Proc Natl Acad Sci U S A 94:13961–13966. doi: 10.1073/pnas.94.25.13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fenaux M, Opriessnig T, Halbur PG, Elvinger F, Meng XJ. 2004. Two amino acid mutations in the capsid protein of type 2 porcine circovirus (PCV2) enhanced PCV2 replication in vitro and attenuated the virus in vivo. J Virol 78:13440–13446. doi: 10.1128/JVI.78.24.13440-13446.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ye C, Wu J, Tong W, Shan T, Cheng X, Xu J, Liang C, Zheng H, Li G, Tong G. 2018. Comparative genomic analyses of a virulent pseudorabies virus and a series of its in vitro passaged strains. Virol J 15:195. doi: 10.1186/s12985-018-1102-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tong C, Chen N, Liao X, Yuan X, Sun M, Li X, Fang W. 2017. Continuous passaging of a recombinant C-strain virus in PK-15 cells selects culture-adapted variants that showed enhanced replication but failed to induce fever in rabbits. J Microbiol Biotechnol 27:1701–1710. doi: 10.4014/jmb.1704.04065. [DOI] [PubMed] [Google Scholar]
- 63.Camini FC, da Silva Caetano CC, Almeida LT, de Brito Magalhães CL. 2017. Implications of oxidative stress on viral pathogenesis. Arch Virol 162:907–917. doi: 10.1007/s00705-016-3187-y. [DOI] [PubMed] [Google Scholar]
- 64.Reshi ML, Su YC, Hong JR. 2014. RNA viruses: ROS-mediated cell death. Int J Cell Biol 2014:467452. doi: 10.1155/2014/467452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Barber GN. 2001. Host defense, viruses and apoptosis. Cell Death Differ 8:113–126. doi: 10.1038/sj.cdd.4400823. [DOI] [PubMed] [Google Scholar]
- 66.Tschopp J, Thome M, Hofmann K, Meinl E. 1998. The fight of viruses against apoptosis. Curr Opin Genet Dev 8:82–87. doi: 10.1016/S0959-437X(98)80066-X. [DOI] [PubMed] [Google Scholar]
- 67.Songock WK, Kim SM, Bodily JM. 2017. The human papillomavirus E7 oncoprotein as a regulator of transcription. Virus Res 231:56–75. doi: 10.1016/j.virusres.2016.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Niller HH, Minarovits J. 2012. Similarities between the Epstein-Barr virus (EBV) nuclear protein EBNA1 and the pioneer transcription factor FoxA: is EBNA1 a “bookmarking” oncoprotein that alters the host cell epigenotype? Pathogens 1:37–51. doi: 10.3390/pathogens1010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Voigt K, Izsvak Z, Ivics Z. 2008. Targeted gene insertion for molecular medicine. J Mol Med (Berl) 86:1205–1219. doi: 10.1007/s00109-008-0381-8. [DOI] [PubMed] [Google Scholar]
- 70.Rosin-Arbesfeld R, Willbold D, Yaniv A, Gazit A. 1998. The Tat protein of equine infectious anemia virus (EIAV) activates cellular gene expression by read-through transcription. Gene 219:25–35. doi: 10.1016/s0378-1119(98)00389-8. [DOI] [PubMed] [Google Scholar]
- 71.Wang XF, Liu Q, Wang YH, Wang S, Chen J, Lin YZ, Ma J, Zhou JH, Wang X. 2018. Characterization of equine infectious anemia virus long terminal repeat quasispecies in vitro and in vivo. J Virol 92:e02150-17. doi: 10.1128/JVI.02150-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Matheson NJ, Greenwood EJ, Lehner PJ. 2016. Manipulation of immunometabolism by HIV—accessories to the crime? Curr Opin Virol 19:65–70. doi: 10.1016/j.coviro.2016.06.014. [DOI] [PubMed] [Google Scholar]
- 73.Simon V, Bloch N, Landau NR. 2015. Intrinsic host restrictions to HIV-1 and mechanisms of viral escape. Nat Immunol 16:546–553. doi: 10.1038/ni.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sugden SM, Bego MG, Pham TN, Cohen EA. 2016. Remodeling of the host cell plasma membrane by HIV-1 Nef and Vpu: a strategy to ensure viral fitness and persistence. Viruses 8:67. doi: 10.3390/v8030067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sumner RP, Thorne LG, Fink DL, Khan H, Milne RS, Towers GJ. 2017. Are evolution and the intracellular innate immune system key determinants in HIV transmission? Front Immunol 8:1246. doi: 10.3389/fimmu.2017.01246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cicala C, Arthos J, Martinelli E, Censoplano N, Cruz CC, Chung E, Selig SM, Van Ryk D, Yang J, Jagannatha S, Chun TW, Ren P, Lempicki RA, Fauci AS. 2006. R5 and X4 HIV envelopes induce distinct gene expression profiles in primary peripheral blood mononuclear cells. Proc Natl Acad Sci U S A 103:3746–3751. doi: 10.1073/pnas.0511237103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lim WS, Edwards JF, Boyd NK, Payne SL, Ball JM. 2003. Simultaneous quantitation of equine cytokine mRNAs using a multi-probe ribonuclease protection assay. Vet Immunol Immunopathol 91:45–51. doi: 10.1016/S0165-2427(02)00263-5. [DOI] [PubMed] [Google Scholar]
- 78.Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Raabe MR, Issel CJ, Montelaro RC. 1998. Equine monocyte-derived macrophage cultures and their applications for infectivity and neutralization studies of equine infectious anemia virus. J Virol Methods 71:87–104. doi: 10.1016/S0166-0934(97)00204-8. [DOI] [PubMed] [Google Scholar]
- 80.Cook RF, Cook SJ, Li FL, Montelaro RC, Issel CJ. 2002. Development of a multiplex real-time reverse transcriptase-polymerase chain reaction for equine infectious anemia virus (EIAV). J Virol Methods 105:171–179. doi: 10.1016/S0166-0934(02)00101-5. [DOI] [PubMed] [Google Scholar]
- 81.Du C, Liu HF, Lin YZ, Wang XF, Ma J, Li YJ, Wang X, Zhou JH. 2015. Proteomic alteration of equine monocyte-derived macrophages infected with equine infectious anemia virus. Proteomics 15:1843–1858. doi: 10.1002/pmic.201400279. [DOI] [PubMed] [Google Scholar]
- 82.Lu Q, Bai J, Zhang L, Liu J, Jiang Z, Michal JJ, He Q, Jiang P. 2012. Two-dimensional liquid chromatography-tandem mass spectrometry coupled with isobaric tags for relative and absolute quantification (iTRAQ) labeling approach revealed first proteome profiles of pulmonary alveolar macrophages infected with porcine reproductive and respiratory syndrome virus. J Proteome Res 11:2890–2903. doi: 10.1021/pr201266z. [DOI] [PubMed] [Google Scholar]
- 83.Liu J, Bai J, Lu Q, Zhang L, Jiang Z, Michal JJ, He Q, Jiang P. 2013. Two-dimensional liquid chromatography-tandem mass spectrometry coupled with isobaric tags for relative and absolute quantification (iTRAQ) labeling approach revealed first proteome profiles of pulmonary alveolar macrophages infected with porcine circovirus type 2. J Proteomics 79:72–86. doi: 10.1016/j.jprot.2012.11.024. [DOI] [PubMed] [Google Scholar]
- 84.Xia L, Dai L, Zhu L, Hu W, Yang Q. 2017. Proteomic analysis of IPEC-J2 cells in response to coinfection by porcine transmissible gastroenteritis virus and enterotoxigenic Escherichia coli K88. Proteomics Clin Appl 11:1600137. doi: 10.1002/prca.201600137. [DOI] [PubMed] [Google Scholar]
- 85.Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 86.Lange V, Picotti P, Domon B, Aebersold R. 2008. Selected reaction monitoring for quantitative proteomics: a tutorial. Mol Syst Biol 4:222. doi: 10.1038/msb.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ. 2010. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Escher C, Reiter L, MacLean B, Ossola R, Herzog F, Chilton J, MacCoss MJ, Rinner O. 2012. Using iRT, a normalized retention time for more targeted measurement of peptides. Proteomics 12:1111–1121. doi: 10.1002/pmic.201100463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 90.Mukhopadhyay P, Rajesh M, Hasko G, Hawkins BJ, Madesh M, Pacher P. 2007. Simultaneous detection of apoptosis and mitochondrial superoxide production in live cells by flow cytometry and confocal microscopy. Nat Protoc 2:2295–2301. doi: 10.1038/nprot.2007.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kim SJ, Syed GH, Siddiqui A. 2013. Hepatitis C virus induces the mitochondrial translocation of Parkin and subsequent mitophagy. PLoS Pathog 9:e1003285. doi: 10.1371/journal.ppat.1003285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wegel E, Gohler A, Lagerholm BC, Wainman A, Uphoff S, Kaufmann R, Dobbie IM. 2016. Imaging cellular structures in super-resolution with SIM, STED and localisation microscopy: a practical comparison. Sci Rep 6:27290. doi: 10.1038/srep27290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.